Summary
Monkeypox virus (MPXV) resides in two forms; mature and enveloped, and depending on it, distinct proteins are displayed on the viral surface. Here, we expressed two MPXV antigens from the mature, and one from the enveloped form, and tested their reactivity to sera of 11 MPXV recoverees while comparing to sera from recently and past vaccinated individuals. 8 out of 11 recoverees exhibited detectable neutralization levels against Vaccinia Lister. Sera from all recoverees bound strongly to A35R and H3L antigens. Moreover, the responses to A35R were significantly higher within the recoverees compared to both recently and past vaccinated donors. Lastly, A35R- and H3L-specific IgG+ B cells ranging from 0.03-0.46% and 0.11–0.36%, respectively, were detected in all recoverees (A35R), and in 9 out of 11 recoverees (H3L). Therefore, A35R and H3L represent MPXV immune targets and could be used in a heat-inactivated serological ELISA for the identification of recent MPXV infection.
Subject areas: Immunology, Immunological methods, Virology
Graphical abstract
Highlights
-
•
MPXV recoverees develop antibodies that target MPXV antigens A35R and H3L
-
•
Antibody response to A35R is higher in MPXV recoverees than in vaccinees
-
•
A35R and H3L-specific IgG+ B cell are more common in MPXV recoverees than vaccinees
-
•
Recent and past VACV vaccinated donors display similar antibody response to VACV
Immunology; Immunological methods; Virology
Introduction
Monkeypox virus (MPXV) is a member of the Orthopoxvirus genus and is responsible for Monkeypox disease.1,2,3 The recent MPXV outbreak is the largest recorded outbreak in non-endemic countries to date.4 As of December 28, 2022, over 82,000 people outside Africa were infected in 103 non-endemic countries, with the majority of cases detected in Europe and the Americas.5,6 The infection results in blisters, fever and discomfort, with case mortality rate currently nearing 0.1% (for the 2022 outbreak).6 Diagnosis of MPXV infection is primarily based on polymerase chain reaction (PCR) to detect the MPXV nucleic acids.7 However, as infection rates continue to increase there is a need for rapid antigen-based serological assays that can be performed at non-BSL3 point of care sites. Moreover, serological assays can promote the understanding of both T cell and B cell responses and lead to the isolation of neutralizing antibodies that can be later examined as therapeutics. Lastly, serological assays can highlight potential targets for vaccine candidates.8,9,10,11
MPXV expresses approximately 25 membrane proteins on the mature virion (MV), a form that is dominant during inter-host transmission, and additional 6 proteins on the enveloped virion (EV), a form that is dominant during intra-host transmission.12,13 Studies conducted on the related Vaccinia virus (VACV) show that entry to host cells is mediated through interactions with glycosaminoglycans, and through fusion with plasma membrane at neutral pH (mostly EV), or through low pH-expedited endocytosis (mostly MV).14,15 Furthermore, several VACV proteins have been identified as important for viral attachment and entry,14,15 some of whom were found to be targeted by antibodies elicited in immunized mice, infected macaques and both infected and vaccinated humans.16,17,18 However, the main serological and B cell markers accompanying MPXV infection in humans are still not characterized. In the current report we examined the reactivity of three MPXV antigens to MPXV convalescent sera and compared it to the responses elicited by Vaccinia virus-based vaccine.
Results
Sera from MPXV recoverees bind VACV and exhibit some VACV neutralization
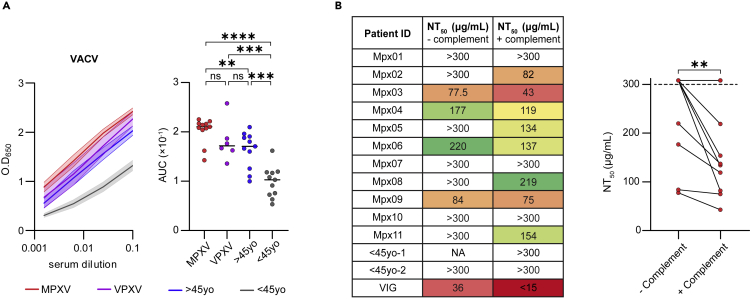
To investigate the antibodies elicited following MPXV natural infection, we recruited a cohort of 11 MPXV recoverees 33–62 days post infection. All recoverees were diagnosed in Israel between May and June 2022 (Table 1). All the donors were males between the ages of 23 and 39, who were PCR-confirmed to be infected with MPXV, and exhibited a variety of symptoms including rash, disease characteristic blisters and fever (for 8 out of 11, Table 1). MPXV is highly similar to VACV, which is the prototype of this viral family.13 Therefore, we first tested binding of heat-inactivated sera from MPXV recoverees to heat-inactivated VACV by enzyme-linked immunosorbent assay (ELISA) as previously described.19 In parallel, we included sera samples of uninfected male donors, who were recently vaccinated against MPXV (Table S1) with live non-replicating Modified Vaccinia Ankara (JYNNEOS, n = 6, named “VPXV”). In addition, we included two volunteer groups; uninfected male donors below 45 years old (samples collected before the Monkeypox outbreak, n = 11, named “uninfected <45yo”), and uninfected male donors above 45 years old (samples collected before the Monkeypox outbreak, n = 11, named “uninfected >45yo”). The latter are expected to have been vaccinated in the past with a Vaccinia-based vaccine, as smallpox vaccination was standard protocol in Israel for individuals born until 1977. Heat-inactivated sera from all MPXV recoverees, and both recent and past vaccinated (i.e., VPXV and >45yo, respectively) reacted with VACV IHDJ strain (Figure 1A). The indistinguishable serological responses between recently vaccinated VPXV group and past vaccinated >45yo donors agrees with reports about the longevity of Vaccinia immunization.20 Sera from uninfected <45yo donors had significantly lower binding to the inactivated VACV.
Table 1.
MPXV recoverees’ clinical data
| Patient ID | Age | Sex | PCR confirmation | Rash | Fever | Number of blisters | Time from infection to sample collection (days)a |
|---|---|---|---|---|---|---|---|
| Mpx01 | 30 | Male | Yes | Yes | Yes | 10–20 | 38 |
| Mpx02 | 33 | Male | Yes | Yes | Yes | <10 | 33 |
| Mpx03 | 39 | Male | Yes | Yes | Yes | >20 | 62 |
| Mpx04 | 37 | Male | Yes | Yes | Yes | 10–20 | 48 |
| Mpx05 | 39 | Male | Yes | Yes | Yes | 10–20 | 47 |
| Mpx06 | 34 | Male | Yes | Yes | No | <10 | 60 |
| Mpx07 | 26 | Male | Yes | Yes | No | 10–20 | 47 |
| Mpx08 | 36 | Male | Yes | Yes | Yes | 10–20 | NA |
| Mpx09 | 23 | Male | Yes | Yes | Yes | 10–20 | 51 |
| Mpx10 | 24 | Male | Yes | Yes | Yes | 10–20 | 51 |
| Mpx11 | 32 | Male | Yes | Yes | No | 10–20 | NA |
Date of infection was provided by the patient upon clinical examination. NA, non applicable.
Figure 1.
Sera from MPXV recoverees bind VACV and exhibit some VACV neutralization
(A) Left: binding curves as measured by ELISA at O.D.650nm demonstrating serum response against VACV IHDJ strain. MPXV recoverees are in red (n = 11), VPXV are in purple (n = 6), uninfected >45yo are in blue (n = 11) and uninfected <45yo are in gray (n = 11). The mean serum response and standard error of the mean (SEM) are depicted in bold line and shadow, respectively, for each group. Four consecutive serum dilutions, starting from 1:10, for every group of donors were tested. Right panel: Area under the curve (AUC) values depicting each donor separately. Statistical analysis was performed using One-way ANOVA. ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001.
(B) Left panel: NT50 values of purified IgG from 11 MPXV recoverees as measured in a PRNT using VACV Lister, with and without the addition of complement. Right panel: graphical representation of the NT50 values. Dashed line represents the PRNT’s measurement cutoff values of 300 μg/mL. NA – non applicable. Statistical analysis was performed using Ratio paired t-test. ∗∗p< 0.01.
We next asked whether the donors exhibit neutralizing antibodies in their sera. Owing to the strict biosafety requirements for working with authentic MPXV, as a model we used Vaccinia Lister virus (VACV Lister) and determined neutralization of protein-A-purified IgG from MPXV recoverees in a plaque reduction neutralization test (PRNT). It has been already shown that VACV vaccination elicits serum neutralization against VACV Lister,21,22 therefore, we focused on the MPXV recoverees’ samples and tested whether they exhibit neutralization in this assay as well. Briefly, VACV Lister was pre-incubated with MPXV recoverees purified IgG and used to infect Vero cells. Purified IgG from uninfected donors <45yo were used as negative controls, whereas pooled purified IgG from multiple VACV vaccinated donors (Vaccinia immune globulin, ‘VIG’23) was used as positive control. Surprisingly, only four MPXV recoverees exhibited any measurable neutralizing activity, with all of them having markedly less neutralization than VIG (Figures 1B and S1). It has been demonstrated that neutralization of both the MV and the EV forms of Orthopoxviruses requires the addition of complement.24,25 Therefore, we repeated the assay while including complement serum. This time, additional four MPXV recoverees demonstrated measurable levels of neutralization (Figure 1B). For the four donors who exhibited neutralization without complement, neutralizing activity was improved by 10–44%, further emphasizing the importance of complement components in this assay. Nevertheless, even after complement serum was added, most of the donors demonstrated relatively weak neutralizing activity, with only one donor MPX03 exhibiting NT50< 50 μg/mL. We conclude that although MPXV infection induces strong serological responses in ELISA, 33–62 days post infection, at this timepoint most recoverees do not exhibit strong neutralizing abilities against VACV Lister.
Sera and purified IgG from MPXV recoverees bind recombinantly expressed MPXV antigens
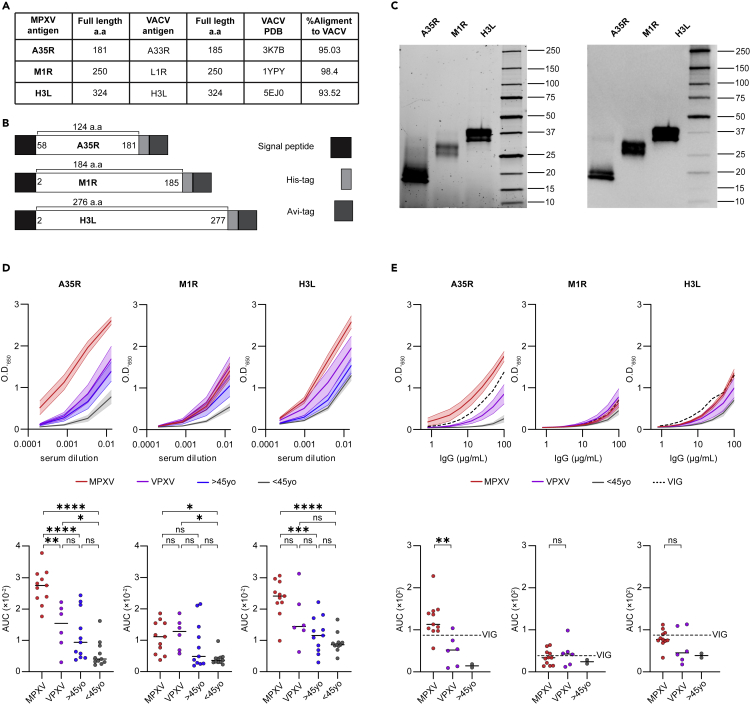
Next, we wished to map the targets for antibodies elicited following MPXV infection. We focused on three MPXV antigens that were previously described to be implicated in immune response against either MPXV or VACV.3,16,17,18,26,27 The transmembrane proteins were expressed in their soluble forms, i.e., without their corresponding transmembrane regions. Overall, we produced one antigen from the EV form, A35R (95.03% homologous to VACV antigen A33R) amino acids 58–181, and two antigens from the MV form: M1R (98.4% homologous to VACV antigen L1R) amino acids 2–185, and H3L (93.52% homologous to VACV antigen H3L) amino acids 2–277 (Figures 2A and 2B). The antigens were cloned into pcDNA3.1(−) expression vector, containing an 8xHistidine tag at their C-termini followed by Avi-tag, a 15-amino acid biotinylation sequence (Figure 2B). The antigens were expressed in mammalian Expi293F cells and purified on Nickel beads (Figure 2C).28 In silico alignment of the AlphaFold-predicted structures of the recombinantly expressed MPXV antigen truncations to the atomic structures of their corresponding VACV homologs, predicted that MPXV and VACV antigens are likely to achieve similar conformations (Figure S2A).
Figure 2.
Sera and purified IgG from MPXV recoverees bind recombinantly expressed MPXV antigens
(A) Table listing the three MPXV antigens A35R, M1R and H3L produced in this study. Published PDB IDs of VACV homologs are given.29,30,31 Amino-acid sequence alignment scores between MPXV and VACV antigens were calculated using the Clustal Omega web tool.32
(B) Construct design of the three recombinantly expressed antigens, A35R, M1R and H3L. Signal peptide, His-tag and Avi-tag are in black, light gray and dark gray, respectively. The amino acid section produced for each antigen is stated.
(C) SDS-PAGE (left) and western blot using mouse anti-Avi-tag antibody (right) of the recombinantly expressed MPXV antigens A35R, M1R and H3L after purification. The antigens are indicated on top, the protein marker is on the right of each gel/blot, and protein sizes are indicated on the right in kDa.
(D) Serum binding to MPXV antigens A35R, M1R and H3L as detected by ELISA. MPXV recoverees are in red (n = 11), VPXV are in purple (n = 6), uninfected >45yo are in blue (n = 11) and uninfected <45yo are in gray (n = 11). Upper panel: The bold lines and the shadowed areas on the graph represent the mean serum response and SEM, respectively, of 4 consecutive serum dilutions for every group of donors, starting from 1:75 dilution. Lower panel: AUC values for every individual donor. Statistical analysis was performed using One-way ANOVA. ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001.
(E) Purified IgG binding to MPXV antigens A35R, M1R and H3L as detected by ELISA. MPXV recoverees are in red (n = 11), VPXV are in purple (n = 6), uninfected <45yo are in gray (n = 2), and VIG is represented by the dashed black line. Eight consecutive IgG dilutions starting from 100 μg/mL were tested (x axis). Upper panel: The bold lines and the shadowed areas on the graph represent the mean IgG response and SEM, respectively, for every group of donors. Lower panel: AUC values for every individual donor. Statistical analysis was performed between MPXV and VPXV groups using Welch’s t test. ∗∗p< 0.01.
The three purified antigens were used to coat ELISA plates and reacted with sera from MPXV recoverees, VPXV vaccinees, >45yo, and <45yo (Figure 2D). Sera from MPXV recoverees bound A35R in a dose dependent manner, and significantly stronger than all uninfected, including both vaccinated, groups. The second antigen, M1R, was recognized stronger by MPXV recoverees compared to uninfected <45yo, yet the response was similar to that of the two vaccinated groups (although this response was lower in the >45yo group). As to the third antigen, H3L, binding was not significantly different between MPXV and VPXV groups, however, the response was significantly higher in MPXV recoverees compared to both >45yo and <45yo groups (Figure 2D). The elite MPXV sera response to A35R and H3L was further demonstrated by the fact that the median reactivity to A35R and H3L was 2.5- and 2.1-fold higher, respectively, compared to M1R, which seems to be less immunodominant following infection (Figure S2B). This difference was not observed within the VPXV vaccinated sera, that bound similarly to all three antigens. Notably, A35R reactivity was significantly stronger within >45yo group and higher by 1.35-fold compared to M1R (Figure S2B). Similar results were obtained with protein-A-purified IgG (Figure 2E). Here too, the majority of MPXV recoverees bound A35R antigen stronger, even when compared to VIG positive control, with the median response of the MPXV group being higher than VIG throughout all IgG concentrations tested. We conclude that both A35R and H3L are targets for antibodies elicited following MPXV infection, with the response to the soluble 124-amino acid truncation of A35R being significantly higher in recently infected MPXV patients than in recently or past vaccinated individuals, who received a Vaccinia virus-based vaccine.
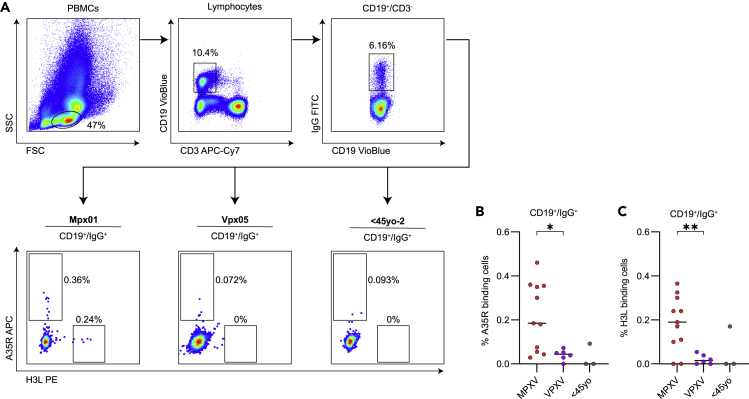
IgG+ B cells from MPXV recoverees bind A35R and H3L antigens
Antibodies are produced by B cells following antigen encounter and activation.33 We therefore evaluated the ability of B cells collected from the 11 MPXV recoverees to specifically bind A35R and H3L antigens. For this purpose, 5–6 million PBMCs from whole blood were stained for CD3, CD19, IgG as described before,28,34 as well as with biotinylated, streptavidin fluorophore conjugated A35R and H3L antigens (Figure 3A). We were able to record A35R-specific IgG+/CD19+ cells in all MPXV recoverees, frequencies ranging between 0.03% and 0.46% of the total IgG+/CD19+ cells (Figure 3B). Moreover, the levels of A35R-specific cells were significantly higher in MPXV recoverees compared to VPXV group. Similar levels of H3L-specific IgG+/CD19+ cells were detected. Although two donors did not exhibit any detectable H3L-specific IgG+/CD19+ cells, here too, the frequency of H3L-specific IgG+/CD19+ cells was significantly higher in MPXV recoverees compared to VPXV group (Figure 3C). MPXV recoverees and VPXV vaccinees exhibited similar frequencies of CD3+ cells ranging from 42%-81%, CD19+ cells ranging from 2%-12.7% and IgG+ B cells (gated from CD19+ population) ranging from 5%-30% (Figure S3). In this analysis we could not include samples from >45yo, as well as most <45yo, because for these samples PBMCs were not available.
Figure 3.
IgG+ B cells from MPXV recoverees bind A35R and H3L antigens
(A) Flow cytometry gating strategy for staining of A35R- and H3L-specific IgG+ B cells. Representative plots for MPXV recoveree (Mpx01), VPXV (Vpx05) and uninfected <45yo (<45yo-2) are shown.
(B) Frequency of A35R-specific IgG+/CD19+ cells of MPXV recoverees are in red (n = 11), VPXV are in purple (n = 6) and uninfected <45yo are in gray (n = 3). Statistical analysis was performed between MPXV and VPXV groups using Unpaired t test. ∗p< 0.05, ∗∗p< 0.01.
(C) The same as (B), but for H3L-specific IgG+/CD19+ cells.
Discussion
In the present report we demonstrate that MPXV recoverees produce both antibodies and B cells against MPXV antigens A35R and H3L. The B cell response to both these antigens was higher compared to Vaccinia-based vaccinated donors. This might be because of slight differences between the two viruses, MPXV versus VACV. Another possibility is differences in the sequences of MPXV antigens and their VACV versions (A35R 95.03% homology and H3L 93.52% homology).
A35R, is one of the 6 proteins expressed on the EV form of the virus, which is believed to be responsible mostly for cell-to-cell viral spread.35 Although the EV form of poxviruses is considered more protected and more difficult to neutralize by antibodies,36 antibodies against EV proteins were found to mediate VACV neutralization after Vaccinia vaccination and MPXV infection16,37 and removal of EV-directed antibodies abolished VIG neutralization.38 Although most of the MPXV recoverees in our cohort did not exhibit high neutralizing activity against VACV in a plaque assay 1–2 months after infection, all of them bound a 124-amino acid truncation of A35R antigen, significantly stronger than did vaccinated or uninfected donors. We found no correlation between A35R binding to MPXV sera and Vaccinia Lister neutralizing activity, which might be because of the slight differences between the two viruses, or because the plaque assay is mostly detecting MV-related viral inhibition, rather than EV-related viral inhibition. We found anti-A35R B cells in peripheral blood of all MPXV recoverees, at significantly higher levels than in vaccinated donors.
H3L antigen is expressed on the MV form, promoting binding to host cells and infectivity.39 It was identified as a target for both T cells and B cells17,27 in vaccinated mice and humans, and anti-H3L antibodies were able to elicit protection from a lethal challenge in mice.40 In agreement with that, in our study, antibodies from MPXV recoverees and vaccinees (past and recent) bound H3L. MPXV recoverees also exhibited H3L-specific IgG+ B cells, which were in higher frequency compared to recent vaccinees. Amongst the two MPXV recoverees who did not have any detectable H3L-specific IgG+ B cells was donor Mpx06 who showed neutralizing activity against the VACV Lister in plaque assay, even without the addition of complement. This might suggest that other MPXV specific antibodies and T cells were elicited by infection and contributed to viral clearance.
Limitations of the study
Our study has several limitations; first, the MPXV samples were analyzed at a relatively early timepoint after Monkeypox infection, which could potentially result in low Vaccinia virus neutralization in PRNT. Furthermore, owing to the worldwide shortage of Monkeypox vaccines, only six samples from recently vaccinated donors were available. Moreover, these samples were collected after only one vaccine dose, and not the required two dose regimen (as well because of shortage in vaccine doses and Israeli Ministry of Health guidelines at the time the study was conducted). Another potential caveat of our work is the utility of the ELISA assay for the diagnosis of Monkeypox infections; although the response to the recombinantly expressed protein A35R was higher in all MPXV recoverees compared to both vaccinated groups, we could not rule out that this might be because of elevated antibody titers in the MPXV recoverees group, at the point of sample collection. More work is needed to determine the dynamics of the humoral response following Monkeypox infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD19 VioBlue | Miltenyi Biotec | Cat#130-113-172; RRID:AB_2725999 |
| Anti-IgG FITC | Miltenyi Biotec | Cat#130-118-340; RRID:AB_2733672 |
| Anti-CD3 APC-Cyanine7 | Biolegend | Cat#317342; RRID:AB_2563410 |
| Anti-IgG HRP | Jackson ImmunoResearch Labs | Cat#109-035-088; RRID:AB_2337584 |
| Anti-Avi-tag | Avidity | Cat#AbC |
| Vaccinia immune globulin | Omrix | Cat#Omr-IgG-am ™ 5% IV |
| Bacterial andVirusStrains | ||
| Vaccinia Lister | Israeli Ministry of Health | N/A |
| Vaccinia IHDJ | Prof. Ehud Katz | N/A |
| Biological Samples | ||
| MPXV recoverees donors whole blood | This study | Helsinki approval number 0384-22-TLV |
| Recently vaccinated donors whole blood | This study | Helsinki approval number 0384-22-TLV |
| Uninfected healthy donors whole blood | Israeli Blood bank | protocol number 0004554–2 |
| Guinea pig complement | Sigma-Aldrich | S-1639 |
| Chemicals,Peptides, andRecombinantProteins | ||
| Streptavidin APC | Miltenyi Biotec | Cat#130-106-792; RRID:AB_2661578 |
| Streptavidin PE | Miltenyi Biotec | Cat#130-106-789; RRID:AB_2661577 |
| Ficoll-Paque PLUS | Cytiva | Cat#17-1440-03 |
| Ni Sepharose beads | Cytiva | Cat#17-5318-01 |
| Protein-A beads | Cytiva | Cat#17-5199-01 |
| A35R | This study | N/A |
| M1R | This study | N/A |
| H3L | This study | N/A |
| CriticalCommercialAssays | ||
| MINI PLASMID PRESTO Kit | Geneaid | Cat#IMPDH300 |
| NucleoSpin Plasmid Kit | MACHEREY-NAGEL | Cat#MAN-740588.250 |
| Pierce™ BCA Protein Assay Kit | Thermo Scientific™ | Cat#TS-23227 |
| Experimantal Model:CellLines | ||
| EXPI 293F | Thermo Scientific™ | N/A |
| Vero cells | ATCC | ATCC-CCL-81 |
| Recombinant DNA | ||
| Truncated MPXV A35R sequence | This study/Genescript | N/A |
| Truncated MPXV M1R sequence | This study/Genescript | N/A |
| Truncated MPXV H3L sequence | This study/AZENTA | N/A |
| pcDNA3.1(−) plasmid | ThermoFisher Scientific | N/A |
| Software andAlgorithms | ||
| FlowJo v.10.8.1 | FlowJo | https://www.flowjo.com/ |
| GraphPad Prism v.9.4.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Biorender | Biorender.com | https://app.biorender.com/ |
| Adobe illustrator v.26.5 | Adobe | https://www.adobe.com/il_en/products/illustrator.html |
| PyMOL v.2.5.2 | PyMOL | https://pymol.org/2/ |
| AlphaFold2 | AlphaFold | https://alphafold.ebi.ac.uk/ |
| SnapGene v.6.0.2 | SnapGene | https://www.snapgene.com/ |
| Other | ||
| BirA 500 Kit | Avidity | Cat#BirA500; Lot: B12920 |
| Monkeypox reference genome 2018 | NCBI GenBank | Accession no. MN648051 |
| ExpiFectamine™ 293 Transfection Kit | Gibco™ | Cat#A14524 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Natalia T Freund (nfreund@tauex.tau.ac.il).
Materials availability
Monkeypox virus antigens generated in this study will be available upon request from the lead contact with a completed Materials and Transfer Agreement.
Experimental model and subject details
Ethics statement
All donors provided a written informed consent prior to participating in this study. All participants enrolled in this study were males. MPXV recoverees between the ages of 23 and 39 tested positive in a PCR assay for MPXV infection collected from saliva, blood, anus, or blisters. All 11 MPXV recoverees have not received a Vaccinia virus (VACV) based vaccine. A single sample of 170 mL of whole blood was collected 33–62 days post infection, after the donors were considered recovered. Recently vaccinated donors (VPXV) were recruited 26–39 days after receiving a single dose of the JYNNEOS vaccine. A single sample of 30 mL of whole blood was collected. Tel Aviv University Institutional Review Board (IRB) approved all studies involving patient enrollment, sample collection, and clinical follow-up (protocol number 0005243–1). Donors were followed by the Dermatology Division of Ichilov Tel Aviv Sourasky Medical Center (Helsinki approval number 0384-22-TLV). The uninfected >45yo and uninfected <45yo samples were collected from the Israeli Blood bank (protocol number 0004554–2). Both <45yo and >45yo samples were obtained between 2020-2021, before the current Monkeypox outbreak. <45yo donors are not expected to have been previously vaccinated with a VACV based vaccine.
Viruses
VACV-Lister (Elstree; provided by the Israeli Ministry of Health) was propagated on the chorioallantois membranes of embryonated eggs and titrated on Vero cells (ATCC-CCL-81). Vaccinia IHDJ strain was kindly provided by Prof. Ehud Katz.
Method details
Sample collection and processing
Blood samples of 170 mL were collected at least two weeks after MPXV recovery, and 30 mL at least 26 days post vaccination. One VPXV donor (VPXV04) received a VACV based vaccine prior to the current MPXV outbreak. All samples were processed for PBMCs isolation. Briefly, each whole blood sample was diluted 1:3 in RPMI 1640 medium and separated using Ficoll gradient according to the manufacturer’s protocol. Buffy-coat layer cells were washed 3 times with RPMI, resuspended in FBS containing 10% DMSO and frozen in liquid nitrogen until later use.
Expression of MPXV antigens
Viral isolate sequenced in Israel from the 2018 MPXV outbreak, NCBI GenBank (accession no. "GeneBank: MN648051"),2 was used as a template for MPXV antigen production. The DNA sequences of the soluble domains of antigens A35R, M1R and H3L were optimized for expression in mammalian cells and added to an 8×Histidine sequence (His-tag) and biotinylation encoding sequence (Avi-tag), before being sent to an outsider vendor for production. Generated DNA sequences were cloned into the pcDNA3.1(−) vector and transfected into EXPI293F cells. Expressed antigens were purified from the cells medium using Nickel beads.28,41 Antigens concentration for subsequent tests was measured using NanoDrop Spectrometer at O.D.280nm and BCA protein assay.
SDS-PAGE and western blot
To validate proper expression of the MPXV antigens, SDS-PAGE was performed using 1 μg of each antigen containing 5% β-mercaptoethanol. Western Blotting was performed by transferring the proteins to a Nitrocellulose membrane followed by blocking the membrane for 2 hours at room temperature (RT) in blocking buffer containing PBS×1, 3% BSA, 0.05% Tween20 and 20mM EDTA. The membrane was incubated overnight at 4° with a mouse anti-Avi-tag antibody diluted 1:5000 in blocking buffer. After 5 washes with washing buffer containing PBS×1 and 0.05% Tween20, the membrane was incubated with a secondary HRP-conjugated antibody diluted 1:5000 in blocking buffer for 45 minutes at RT. Following 5 additional washes, ECL was added, and Western blot images were acquired.
Plaque reduction neutralization test
MPXV recoverees neutralization activity was assessed in a PRNT. IgG from all MPXV recoverees and from 2 uninfected <45yo donors was purified using protein-A agarose beads. For PRNT without complement, 50 plaque forming units (PFU) per well of VACV Lister were pre-incubated for 1 hour with MPXV or <45yo purified IgG at a starting concentration of 200 μg/mL followed by 6 consecutive 2-fold dilutions. Vaccinia immune globulin, ‘VIG’, was also used as positive control at a starting concentration of 500 μg/mL followed by 6 consecutive 2-fold dilutions. Pre-incubated Vaccinia and purified IgG were used to infect 5×105 Vero cells. The experiment was carried out in duplicates, and 72 hours post infection plaques were counted and NT50 was determined. For PRNT with complement, Guinea pig complement (S-1639) was added to the Vaccinia Lister, reaching a final concentration of 1% after pre-incubation with purified IgG. Subsequent steps of the PRNT with complement are similar.
Enzyme-linked immunosorbent assay
VACV ELISA: VACV IHDJ strain was diluted to a concentration of 107 PFU per mL in a carbonate bicarbonate solution and used to coat high binding ELISA plates overnight at 4°. ELISA plates were then blocked for 2 hours at RT in a TSTA solution containing 2% BSA, 50mM Tris, 142mM NaCl, 0.05% Azid and 0.05% Tween20 followed by washing the plates once in washing buffer containing PBS×1 and 0.05% Tween20. Heat inactivated sera samples were diluted 4-fold in TSTA, starting from 1:10 and added to the ELISA plates for 1 hour at RT. ELISA plates were washed 3 times with washing buffer followed by adding anti-IgG HRP-conjugated antibody diluted 1:5000 in TSTA for 45 minutes at RT. Following 5 additional washes, TMB/E was added, and the optical density (OD) was measured after 10 minutes at 650 nm.
MPXV antigen ELISA: 5 μg/mL of each of the purified antigens, A35R, M1R and H3L, were used to coat high binding ELISA plates overnight at 4°. ELISA plates were then blocked for 2 hours at RT in blocking buffer containing PBS×1, 3% BSA, 0.05% Tween20 and 20mM EDTA followed by washing the plates once in washing buffer containing PBS×1 and 0.05% Tween20. Heat inactivated sera samples or purified IgG were diluted 4- and 2-fold in blocking buffer starting from 1:75 and 100 μg/mL, respectively, and added to the ELISA plates for 1 hour at RT. ELISA plates were washed 3 times with washing buffer followed by adding anti-IgG HRP-conjugated antibody diluted 1:5000 in blocking buffer for 45 minutes at RT. Following 5 additional washes, TMB/E was added, and the OD was measured after 10 minutes at 650 nm.
Flow cytometry
5–6 million PBMC were thawed at 37°, washed in RPMI and resuspended in FACS buffer containing PBS×1, 1% FBS and 2mM EDTA. For antigen-specific B cell staining, MPXV antigens A35R and H3L were biotinylated on their Avi-tag sequence using the BIR-A biotinylation reaction, followed by their conjugation to streptavidin coupled fluorophores. The PBMCs were stained for CD3, CD19, IgG as well as for with biotinylated, streptavidin fluorophore conjugated A35R and H3L and analyzed in a Flow Cytometer.
Statistical and Flow Cytometry analysis
Statistical analysis was carried out using the GraphPad Prism software version 9.4.1. Analysis of Flow Cytometry data was carried out using FlowJo software version 10.8.
Quantification and statistical analysis
Statistical analysis was carried out using the GraphPad Prism software version 9.4.1. Analysis of Flow Cytometry data was carried out using FlowJo software version 10.8. Statistical details of each experiment can be found in the figure legend associated with each figure.
Acknowledgments
We thank all the donors. We thank the members of the Freund Lab for fruitful discussions. We thank Dr Ohad Mazor from the Israel Institute for Biological Research for his support. K.P.’s research is supported in part by a fellowship from the Edmond J. Safra Center for Bioinformatics at Tel Aviv University. We thank Vice President of Research and Development of Tel Aviv University for funding. We thank Mr. Eli Gelman and Prof. Ariel Porat for their support. Images for graphical abstract were created using BioRender.
Author contributions
R.Y. planned and performed the experiments, analyzed data, prepared the figures, and wrote the manuscript together with N.T.F. N.F. conducted the patient follow up and recruited the donors and collected the samples with the help of E.T. H.T. and T.I. carried out all the neutralization studies, analyzed viral inhibition data with the help of LCM, as well as critically reviewed the data and the manuscript. M.M. helped with protein production and cloning. K.P. performed the AlphaFold predictions. D.H. and E.S. wrote and submitted the ethics protocols, helped design the study and oversaw donor enrolment into the study. N.T.F. planned and supervised the experiments, wrote the ethical protocols, analyzed the data and wrote the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Published: February 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.105957.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sklenovská N., Van Ranst M. Emergence of monkeypox as the most important orthopoxvirus infection in humans. Front. Public Health. 2018;6:241. doi: 10.3389/fpubh.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erez N., Achdout H., Milrot E., Schwartz Y., Wiener-Well Y., Paran N., Politi B., Tamir H., Israely T., Weiss S., et al. Diagnosis of imported monkeypox, Israel, 2018. Emerg. Infect. Dis. 2019;25:980–983. doi: 10.3201/eid2505.190076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum F.M., Torres-Ruesta A., Tay M.Z., Lin R.T.P., Lye D.C., Rénia L., Ng L.F.P. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat. Rev. Immunol. 2022;22:597–613. doi: 10.1038/s41577-022-00775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornhill J.P., Barkati S., Walmsley S., Rockstroh J., Antinori A., Harrison L.B., Palich R., Nori A., Reeves I., Habibi M.S., et al. Monkeypox virus infection in humans across 16 countries - april-june 2022. N. Engl. J. Med. 2022;387:679–691. doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- 5.OurWorldInData . 2022. Monkeypox Data Explorer.https://ourworldindata.org/explorers/monkeypox?tab=map&facet=none&Metric=Confirmed+cases&Frequency=Cumulative&Relative+to+population=false&country=∼OWID_WRL [Google Scholar]
- 6.CDC (2022) 2022. Monkeypox Outbreak Global Map.https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html [Google Scholar]
- 7.Nörz D., Tang H.T., Emmerich P., Giersch K., Fischer N., Schmiedel S., Addo M.M., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Rapid adaptation of established high-throughput molecular testing infrastructure for monkeypox virus detection. Emerg. Infect. Dis. 2022;28:1765–1769. doi: 10.3201/eid2809.220917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munitz A., Edry-Botzer L., Itan M., Tur-Kaspa R., Dicker D., Marcoviciu D., Goren M.G., Mor M., Lev S., Gottesman T., et al. Rapid seroconversion and persistent functional IgG antibodies in severe COVID-19 patients correlates with an IL-12p70 and IL-33 signature. Sci. Rep. 2021;11:3461. doi: 10.1038/s41598-021-83019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowe J.E., Jr. Principles of broad and potent antiviral human antibodies: insights for vaccine design. Cell Host Microbe. 2017;22:193–206. doi: 10.1016/j.chom.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaeck L.M., Lamers M.M., Verstrepen B.E., Bestebroer T.M., van Royen M.E., Götz H., Shamier M.C., van Leeuwen L.P.M., Schmitz K.S., Alblas K., et al. Low levels of monkeypox virus neutralizing antibodies after MVA-BN vaccination in healthy individuals. Nat. Med. 2022 doi: 10.1038/s41591-022-02090-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengali Z., Satheshkumar P.S., Moss B. Orthopoxvirus species and strain differences in cell entry. Virology. 2012;433:506–512. doi: 10.1016/j.virol.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moss B. Poxvirus cell entry: how many proteins does it take? Viruses. 2012;4:688–707. doi: 10.3390/v4050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moss B. Poxvirus membrane biogenesis. Virology. 2015;479–480:619–626. doi: 10.1016/j.virol.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B. Membrane fusion during poxvirus entry. Semin. Cell Dev. Biol. 2016;60:89–96. doi: 10.1016/j.semcdb.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilchuk I., Gilchuk P., Sapparapu G., Lampley R., Singh V., Kose N., Blum D.L., Hughes L.J., Satheshkumar P.S., Townsend M.B., et al. Cross-neutralizing and protective human antibody specificities to poxvirus infections. Cell. 2016;167:684–694.e9. doi: 10.1016/j.cell.2016.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies D.H., Liang X., Hernandez J.E., Randall A., Hirst S., Mu Y., Romero K.M., Nguyen T.T., Kalantari-Dehaghi M., Crotty S., et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keasey S., Pugh C., Tikhonov A., Chen G., Schweitzer B., Nalca A., Ulrich R.G. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Israely T., Paran N., Lustig S., Erez N., Politi B., Shafferman A., Melamed S. A single cidofovir treatment rescues animals at progressive stages of lethal orthopoxvirus disease. Virol. J. 2012;9:119. doi: 10.1186/1743-422X-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiura H., Schwehm M., Eichner M. Still protected against smallpox? Estimation of the duration of vaccine-induced immunity against smallpox. Epidemiology. 2006;17:576–581. doi: 10.1097/01.ede.0000229196.41862.c2. [DOI] [PubMed] [Google Scholar]
- 21.Orr N., Forman M., Marcus H., Lustig S., Paran N., Grotto I., Klement E., Yehezkelli Y., Robin G., Reuveny S., et al. Clinical and immune responses after revaccination of israeli adults with the Lister strain of vaccinia virus. J. Infect. Dis. 2004;190:1295–1302. doi: 10.1086/423851. [DOI] [PubMed] [Google Scholar]
- 22.Priyamvada L., Carson W.C., Ortega E., Navarra T., Tran S., Smith T.G., Pukuta E., Muyamuna E., Kabamba J., Nguete B.U., et al. Serological responses to the MVA-based JYNNEOS monkeypox vaccine in a cohort of participants from the Democratic Republic of Congo. Vaccine. 2022;40:7321–7327. doi: 10.1016/j.vaccine.2022.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbero G.J., Gray A., Scott T.F., Kempe C.H. Vaccinia gangrenosa treated with hyperimmune vaccinal gamma globulin. Pediatrics. 1955;16:609–618. [PubMed] [Google Scholar]
- 24.Benhnia M.R.E.I., McCausland M.M., Moyron J., Laudenslager J., Granger S., Rickert S., Koriazova L., Kubo R., Kato S., Crotty S. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J. Virol. 2009;83:1201–1215. doi: 10.1128/JVI.01797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaever T., Meng X., Matho M.H., Schlossman A., Li S., Sela-Culang I., Ofran Y., Buller M., Crump R.W., Parker S., et al. Potent neutralization of vaccinia virus by divergent murine antibodies targeting a common site of vulnerability in L1 protein. J. Virol. 2014;88:11339–11355. doi: 10.1128/JVI.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X., Zhang Y., Jiang W., Wang D., Lu J., Gu G., Qin C., Fang M. Protective human anti-poxvirus monoclonal antibodies are generated from rare memory B cells isolated by multicolor antigen tetramers. Vaccines (Basel) 2022;10 doi: 10.3390/vaccines10071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khlusevich Y., Matveev A., Emelyanova L., Goncharova E., Golosova N., Pereverzev I., Tikunova N. New p35 (H3L) epitope involved in vaccinia virus neutralization and its deimmunization. Viruses. 2022;14 doi: 10.3390/v14061224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mor M., Werbner M., Alter J., Safra M., Chomsky E., Lee J.C., Hada-Neeman S., Polonsky K., Nowell C.J., Clark A.E., et al. Multi-clonal SARS-CoV-2 neutralization by antibodies isolated from severe COVID-19 convalescent donors. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H.P., Garman S.C., Allison T.J., Fogg C., Moss B., Garboczi D.N. The 1.51-Angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies. Proc. Natl. Acad. Sci. USA. 2005;102:4240–4245. doi: 10.1073/pnas.0501103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su H.P., Singh K., Gittis A.G., Garboczi D.N. The structure of the poxvirus A33 protein reveals a dimer of unique C-type lectin-like domains. J. Virol. 2010;84:2502–2510. doi: 10.1128/JVI.02247-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh K., Gittis A.G., Gitti R.K., Ostazeski S.A., Su H.P., Garboczi D.N. The vaccinia virus H3 envelope protein, a major target of neutralizing antibodies, exhibits a glycosyltransferase fold and binds UDP-glucose. J. Virol. 2016;90:5020–5030. doi: 10.1128/JVI.02933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisen H.N., Siskind G.W. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 34.Watson A., Li H., Ma B., Weiss R., Bendayan D., Abramovitz L., Ben-Shalom N., Mor M., Pinko E., Bar Oz M., et al. Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis. Nat. Commun. 2021;12:602. doi: 10.1038/s41467-021-20930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolffe E.J., Weisberg A.S., Moss B. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 2001;75:303–310. doi: 10.1128/JVI.75.1.303-310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt F.I., Bleck C.K.E., Mercer J. Poxvirus host cell entry. Curr. Opin. Virol. 2012;2:20–27. doi: 10.1016/j.coviro.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Law M., Smith G.L. Antibody neutralization of the extracellular enveloped form of vaccinia virus. Virology. 2001;280:132–142. doi: 10.1006/viro.2000.0750. [DOI] [PubMed] [Google Scholar]
- 38.Bell E., Shamim M., Whitbeck J.C., Sfyroera G., Lambris J.D., Isaacs S.N. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology. 2004;325:425–431. doi: 10.1016/j.virol.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Lin C.L., Chung C.S., Heine H.G., Chang W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 2000;74:3353–3365. doi: 10.1128/jvi.74.7.3353-3365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies D.H., McCausland M.M., Valdez C., Huynh D., Hernandez J.E., Mu Y., Hirst S., Villarreal L., Felgner P.L., Crotty S. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 2005;79:11724–11733. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R., Mor M., Ma B., Clark A.E., Alter J., Werbner M., Lee J.C., Leibel S.L., Carlin A.F., Dessau M., et al. Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies. Commun. Biol. 2022;5:789. doi: 10.1038/s42003-022-03739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.