Abstract
Background
Small extracellular vesicles (sEV) play a crucial role in immune responses to viral infection. However, the composition of sEV derived from children with viral pneumonia remains ill defined.
Methods
First, we performed mass spectrometry-based label-free proteomic analysis of urinary sEV in 7 children with viral pneumonia, 4 children with Mycoplasma pneumoniae pneumonia and 20 healthy children. Then a total of 33 proteins were selected to validate by multiple reaction monitoring analysis in an independent cohort of 20 healthy children and 29 children with pneumonia.
Results
In the discovery phase, a total of 1621 proteins were identified, while 260 proteins have differential expression in children with viral pneumonia compared to healthy children. Biological pathways primarily associated with neutrophil degranulation, carbohydrate metabolism and endocytosis were enriched in children with viral pneumonia. Finally, the abundance of eight proteins was verified to be significantly higher in children with viral pneumonia than in healthy children.
Conclusions
This pilot study with proteomic profiles of urinary sEV provided insights to the host response to viral pathogen exposure and potential diagnostic biomarkers for children with viral pneumonia, and served as the basis for understanding the fundamental biology of infection.
Impact
There were significant differences in the proteomic features of urinary sEV between children with viral pneumonia and those with Mycoplasma pneumoniae pneumonia.
Many viral infection-related proteins were identified in urinary sEV and overrepresented in children with viral pneumonia, which facilitates our understanding of the fundamental biology of viral infection.
A total of eight proteins (ANPEP, ASAH1, COL11A1, EHD4, HEXB, LGALS3BP, SERPINA1 and SERPING1) were verified as potential biomarkers for the diagnosis of viral pneumonia in children.
Introduction
Pneumonia remains a significant cause of mortality and morbidity for children worldwide.1 Viral pathogens are recognized as the leading etiology of pneumonia in children.2–4 However, the clinical presentation of viral and bacterial infection overlap significantly and usually cannot be discriminated on the basis of clinical characteristics alone.5 Therefore, how viral infection influences clinical outcomes in pediatric patients with pneumonia needs to be explored. Pneumonia is characterized by disruption of the bronchial epithelium by pathogens and the consequent host response.6 Determining these host responses will facilitate the understanding of infectious disease pathology.
Small extracellular vesicles (sEV) have been found in various body fluids and carry a variety of functional molecules including proteins, nucleic acids and lipids.7,8 There is evidence that sEV play a vital role in immune responses to viral and bacterial infection,7–9 and proteomic analysis of sEV has revealed significant changes in protein compositions under viral and bacterial infection.10–12 During influenza virus infection triggered pulmonary inflammation, sEV are released into the airways and their protein compositions changed over the course of infection, increasing expression of proteins with known anti-influenza activity.12 On the other hand, sEV have also been shown to blunt immune responses and promote viral infection. Remarkably, proteomic analyses showed that urinary sEV are significantly enriched for innate immune proteins and can contribute to host responses against pathogens.13 Thus, exploring the relationship between viral infection and sEV composition may help to decipher the mechanism of host response against viral infection and improve disease control.
In this study, we isolated urinary sEV from children with pneumonia caused by virus or Mycoplasma pneumoniae infection and healthy children (HC), performed mass spectrometry-based label-free proteomic analysis to identify virus-associated signatures and validated their expression in an independent cohort. This work reinforces that the proteomic features of urinary sEV can shed light on the host response to viral pathogen exposure and serve as the basis for understanding the fundamental biology of infection in children with pneumonia.
Methods
Patient and sample collection
The subjects included in the study were pediatric patients. Pneumonia was diagnosed based on fever, cough, breathlessness, leukocytosis, and radiological features of pneumonia. Etiologies were routinely detected as described previously.14 In brief, etiologies were determined by laboratory tests including the detection of M. pneumoniae and Chlamydia pneumoniae antibody, antigen of Streptococcus pneumoniae and influenza viruses, and throat swabs were used to test DNA or RNA of M. pneumoniae, C. pneumoniae and a wide spectrum of viruses such as influenza virus A, influenza virus B, respiratory syncytial virus V, parainfluenza virus, cytomegalovirus and adenovirus using PCR. Patients were classified mainly according to the type of pathogens. M. pneumoniae or viral-positive patients were included. In addition, a patient who was presumed to be positive for viral infection based on clinical symptoms was also included. HC were identified through laboratory test and excluded if they presented with fever or respiratory symptoms.
This study includes a discovery phase and a validation phase. As shown in Table 1, a total of 7 children with viral pneumonia (Virus), 4 children with M. pneumoniae pneumonia (MPP) and 20 HC were recruited to collect urine samples in the discovery phase. In the validation phase, an independent cohort of 12 children with viral pneumonia, 17 children with MPP and 20 HC was included, the clinical characteristics of these patients were shown in Table 2. Urine samples (≥20 ml) were collected using clinical study protocols approved by the Institutional Review Board of Shanghai Children’s Medical Center. Informed consent was obtained from the parents of all children. All samples were stored in aliquots at −80 °C until used.
Table 1.
The clinical characteristics of the participants in discovery phase.
| Characteristics | Healthy control (N = 20) | Pneumonia (N = 11) | P value* |
|---|---|---|---|
| Age (year), median (range) | 4 (3–9) | 5 (3–10) | 0.303 |
| Male sex, n (%) | 16 (80) | 4 (36) | 0.042 |
| WBC count (109/L), median (range) | 8.88 (5.84–12.07) | 8.04 (4–12.11) | 0.168 |
| Lymphocyte %, median (range) | 41.2 (20–55.1) | 29 (14–48.3) | 0.007 |
| Neutrophil %, median (range) | 50.2 (26.8–71.7) | 60.4 (37.7–83.8) | 0.027 |
| Pathogenic types | |||
| M. pneumoniae, n (%) | 4 (36) | ||
| Parainfluenza viruses, n (%) | 1 (9) | ||
| Adenovirus, n (%) | 3 (27) | ||
| Respiratory syncytial virus, n (%) | 2 (18) | ||
| Presumed viral infection based on clinical symptoms, n (%) | 1 (9) | ||
WBC white blood cells.
*P value for the sex variable was estimated by χ2 test; P value for other continuous variables was estimated by Mann–Whitney U test.
Table 2.
The clinical characteristics of the participants in validation phase.
| Characteristics | Healthy control (n = 20) | Pneumonia (n = 29) | P value* |
|---|---|---|---|
| Age (year), median (range) | 8 (3–14) | 4 (1–10) | 0.0002 |
| Male sex, n (%) | 11 (55) | 17 (59) | 0.97 |
| Pathogenic types | |||
| M. pneumoniae, n (%) | 17 (59) | ||
| Influenza A virus, n (%) | 1 (8.3) | ||
| Influenza B virus, n (%) | 1 (8.3) | ||
| Epstein–Barr virus, n (%) | 2 (16.7) | ||
| Adenovirus, n (%) | 3 (25) | ||
| Cytomegalovirus, n (%) | 2 (16.7) | ||
| Respiratory syncytial virus, n (%) | 3 (25) | ||
*P value for the sex variable was estimated by χ2 test; P value for the age variable was estimated by Mann–Whitney U test.
sEV isolation and identification
Purification of sEV from urine was performed using the Exosome Isolation Q3 kit (Wayen Biotechnologies, China) according to the manufacturer’s protocol as described previously.15 Protein content of sEV was measured by bicinchoninic acid assay. Particle content and size distribution were measured using a qNano Tunable Resistive Pulse Sensing (TRPS) nanoparticle analyzer (iZON Science, New Zealand). The ultrastructure of sEV was analyzed by the Tecnai G2 Spirit BioTwin transmission electron microscope (FEI, OR) with a lanthanum hexaboride cathode operated at 80 keV.
Liquid chromatography tandem mass spectrometry
sEV were lysed in buffer containing 7 M urea, 2% SDS and 1× protease inhibitor cocktail, digested using the standard filter aided proteome preparation protocol and the peptides were desalted using Monospin column as described previously.15,16
Liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed with a Q Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer coupled to an EASY-nLC 1200 system (Thermo Scientific, MA). The peptides were reconstituted with 0.1% formic acid, transferred to a C18 analytical column (2 μm, 100 Å, 50 μm × 15 cm, nanoViper) and separated using the EASY-nLC 1200 system at a flow rate of 300 nL/min. Data-dependent acquisition mode was applied for the tandem mass spectrometry detection. MS/MS spectra were acquired with a scan resolution of 60,000 (FWHM) and 350–2000 m/z of mass-to-charge ratio. Peptides were fragmented using higher-energy collisional dissociation at 28% normalized collision energy.
Proteomic data analysis
Raw data were processed with MaxQuant (version 1.5.8.3) using the settings for the human protein database UniProt/SwissProt. Reverse and potential contaminant proteins were removed. Carbamidomethyl (C) was set as a fixed modification, whereas oxidation (M) and acetyl (Protein N-term) were variable modifications. The minimal peptide length was set to seven amino acids, and two missed cleavages were set as maximum. The results were filtered at 0.01 false discovery rate (FDR) for peptide and 0.05 FDR for protein. The proteomic data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD036472.
Proteins were quantified according to the value of label-free quantitation (LFQ) intensity using the LIMMA method. LIMMA stands for “linear models for microarray data”, contains functionality for fitting a broad class of statistical models such as linear regression and analysis of variance.17 LIMMA has been frequently used to analyze proteomic data.18
Multiple reaction monitoring (MRM) analysis
The 33 proteins identified by LC-MS/MS analysis were selected as candidate urinary sEV biomarker proteins (Supplementary Table S10). A peptide assay library was generated for these proteins using label-free LC-MS/MS and Skyline analysis software (version 19.1).19 Skyline was used to manually select the optimal peptide transitions. Selected peptides were analyzed by scheduled MRM analysis. In brief, the MRM data were acquired on a SCIEX QTRAP 6500 coupled to an Ekspert nanoLC 425 liquid chromatography system using MRM scanning in positive mode (Framingham, MA). Ion spray voltage was set to 5500 and source temperature was set to 150 °C. Curtain gas was set to 35. Source Gases 1 and 2 were set to 20 and 15, respectively. Raw data were analyzed in the Skyline software and peak areas were used for quantitative analysis.
Bioinformatics analysis
Principal component analysis (PCA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using R packages “pca3d” and “clusterProfiler”, respectively. Other functional enrichment analysis and protein–protein network analysis were conducted using the Metascape web tool (metascape.org).20 Only significant pathways with P < 0.05 were considered for analysis. The network was visualized using Cytoscape.21 The gene set enrichment analysis (GSEA) method (http://www.gsea-msigdb.org/) was used to identify significantly enriched or depleted groups of proteins.
Statistical analysis
Statistical significance of sEV number or size was determined with unpaired two-tailed Student’s t test. Statistical significance of clinical variables was determined with χ2 test or Mann–Whitney U test. Statistical graphics were drawn using R packages “ggplot2” and “ggpubr”.
Results
Urinary sEV abundance and size are not affected by disease status
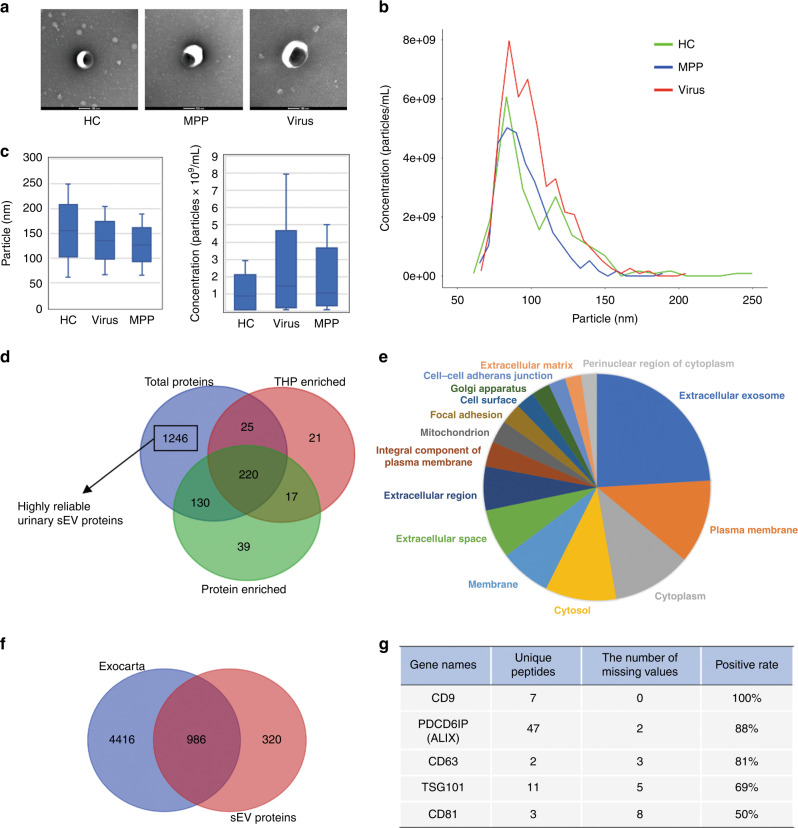
Urine samples from the 20 HC were pooled into five samples (HC1–HC5; four subjects per sample) and used for sEV isolation along with 11 individual urine samples from children with viral pneumonia and those with MPP. The morphology of the sEV preparations was determined by transmission electron microscopy, which revealed typical cup-shaped vesicles (Fig. 1a). The diameter of most particles distributed between 50 and 150 nm (Fig. 1b), coinciding with the size range of sEV defined by the International Society for Extracellular Vesicles. These data confirm that the preparations do indeed contain abundant levels of sEV. In addition, the number and size of urinary sEV detected through TRPS were comparable in HC and those with MPP or viral pneumonia (Fig. 1c).
Fig. 1. Characterization and proteomic analysis of urinary sEV.
a Representative TEM images of sEV from healthy children (HC) and those with M. pneumoniae (MPP) or viral (Virus) pneumonia. Bars, 100 nm. b Size distribution of particles determined by TRPS. c Graph comparing the size and number of particles between healthy and pneumonia group. d Venn diagram displaying the number and overlaps of proteins identified from urinary sEV in the present study (total proteins) and those enriched in THP- and soluble protein-enriched fractions from published results. e Pie chart showing the distribution of the terms identified from GO cellular components analysis of the sEV proteome. The area of each segment is proportional to the number of proteins in the sEV proteome. Overrepresented categories with p < 0.05 are shown. f Venn diagram displaying the number and overlap of proteins identified from urinary sEV in the present study and from the Exocarta database. g List of conventional sEV markers identified by mass spectrometry. Missing values mean that the LFQ intensity is not detected.
Proteomic analysis of urinary sEV
Label-free quantitative proteomic analysis of sEV was performed. A total of 1621 proteins with ≥2 unique peptides in all groups were identified (Supplementary Table S1). Tamm-Horsfall protein (THP) and other soluble proteins are considered as the main contaminants in the separated urinary sEV preparations. To eliminate the contaminating effect of THP and other soluble proteins, we compared the 1621 proteins from this study with those in urinary THP- and soluble protein-enriched fractions identified by density gradient centrifugation from a previously published study.22 From this analysis, 1246 proteins were found to be exclusively present in the urinary sEV and were likely to be highly reliable urinary sEV proteins and were subjected to subsequent analysis (Fig. 1d and Supplementary Table S2). Enrichment analysis with gene ontology cellular component revealed that most of these 1246 proteins belong to the categories extracellular exosome, cytoplasm, plasma membrane, extracellular space and region (Fig. 1e and Supplementary Table S3). Of these 1246 proteins, 986 were present in the Exocarta database of published sEV proteomic results, including 80 of the 100 most frequently identified proteins (Fig. 1f). sEV markers were also identified by mass spectrometry including CD9, CD81, TSG101, PDCD6IP (ALIX) and CD63 in 50–100% of the samples (Fig. 1g), as expected. These data confirm that the quality of the urinary sEV proteomic data is good.
Viral pneumonia significantly affects the proteome of urinary sEV
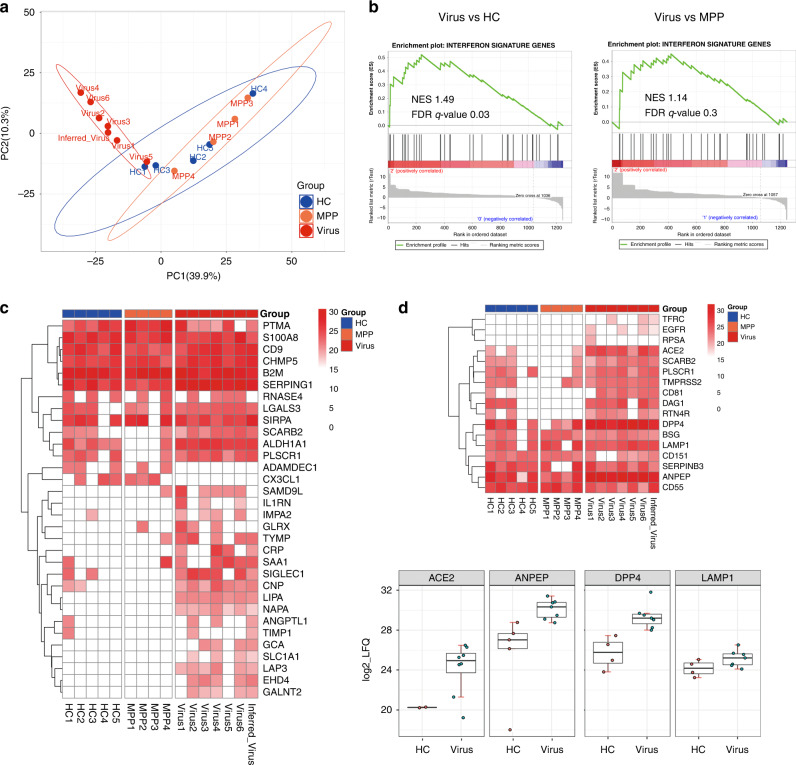
To determine the effect of pathogen infection on the urinary sEV content of children, we performed PCA of the 1246 sEV proteins, which clearly separated the samples into two groups based on virus infection, suggesting that viral pneumonia accounts for the significant differential proteomic features of sEV, rather than MPP and HC (Fig. 2a). Since viral infection increases expression level of the interferon-stimulated genes (ISGs), we analyzed whether urinary sEV derived from children with viral pneumonia were significantly enriched in ISGs as compared with that of HC and children with MPP. GSEA, with the reference gene set containing 422 ISGs obtained from a published result (Supplementary Table S3),23 demonstrated that ISGs were positively correlated with the gene signature of sEV in the viral pneumonia group, and negatively correlated with that of HC and children with MPP (Fig. 2b). A total of 32 ISGs were enriched in urinary sEV, of which 18 (SAMD9L, IL1RN, IMPA2, GLRX, TYMP, CRP, SAA1, SIGLEC1, CNP, LIPA, NAPA, ANGPTL1, TIMP1, GCA, SLC1A1, LAP3, EHD4 and GALNT2) were overrepresented in the viral pneumonia group (Fig. 2c). Interestingly, we also detected 17 receptor or co-receptor proteins for viral entry in the urinary sEV proteome, among which TFRC, EGFR and RPSA were unique to viral pneumonia and ACE2, ANPEP, DPP4 and LAMP1 were overexpressed in the viral pneumonia group (Fig. 2d), suggesting that these receptors may play an important role in the process of viral infection in children. These results indicate that the proteomic data of urinary sEV provide mechanistic insights into virus infection in children with pneumonia.
Fig. 2. Proteomic analysis of urinary sEV reveals viral-related signature.
a Principal component analysis (PCA) of the 16 samples based on their proteomic expression profiles. The first and second components segregated the samples and account for 39.9 and 10.3% of the variability, respectively. b Gene set enrichment analysis (GSEA) of the sEV proteomic data using a reference gene set containing 422 ISGs. The leading edge (most significant genes) are shown as vertical bars accumulated below the peak of the green enrichment score plot, indicating the upregulated genes of GSEA characterized by the highest normalized enrichment score (NES). The analysis demonstrates that ISGs are enriched in the Virus group with respect to HC or MPP group. FDR, false discovery rate. c Heatmap of all enriched genes in the GSEA. d Heatmap of identified viral receptor proteins and boxplots of the significantly upregulated viral receptor proteins in the Virus group compared with the HC group.
Identification of a viral-associated signature from the proteome of urinary sEV
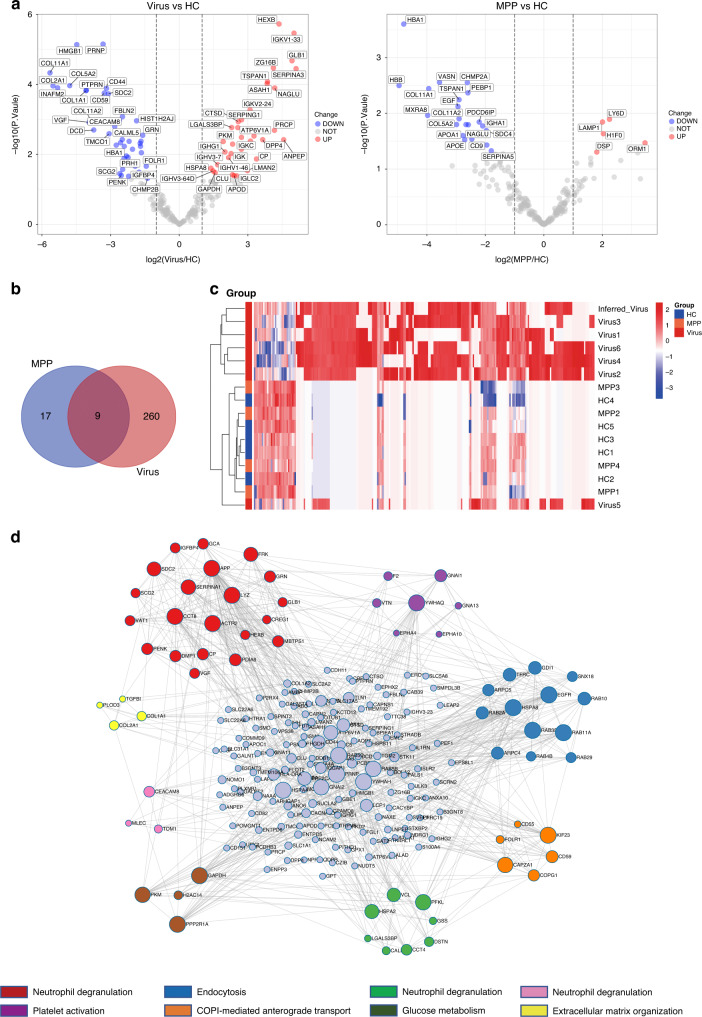
We identified differentially expressed proteins (DEPs) from the sEV of children with viral pneumonia and those with MPP compared with HC. First, a total of 221 proteins with LFQ values >0 in at least three samples in each group were subjected to quantitative comparison. Using this method, we identified 75 and 26 proteins with significant up- or downregulation in children with viral pneumonia and children with MPP, respectively (Fig. 3a and Supplementary Tables S5 and S6). Second, we identified 193 unique proteins with LFQ values >0 in at least three samples in the viral pneumonia group, but no unique proteins in the MPP group. Integrating the results of both analyses identified a total of 269 DEPs in children with viral pneumonia and 26 DEPs in those with MPP compared with HC. Of the 269 viral-related DEPs, 9 were also found in the MPP group, whereas 260 were specific to the viral pneumonia group (Fig. 3b). Unbiased hierarchical clustering analysis of the 260 viral-associated DEPs correctly clustered together the samples from children with viral infection (Fig. 3c and Supplementary Table S7). Then we performed a protein–protein interaction enrichment analysis of the 260 viral-associated DEPs using the Metascape web tool that applies the molecular complex detection (MCODE) algorithm to identify network components. The protein network analysis showed that 216 of 260 DEPs formed interactions and 8 MCODE networks were identified (Fig. 3d). Of the eight MCODE networks, three were enriched for the functional term related to neutrophil degranulation (Fig. 3d) and others were enriched for terms associated with endocytosis, protein transport, glucose metabolism, extracellular matrix organization and platelet activation respectively (Fig. 3d), all important processes or pathways involved in viral infection. Overall, these results show that a viral-associated signature of urinary sEV proteome can facilitate understanding of the fundamental biology of children with viral pneumonia.
Fig. 3. Identification of viral-associated differentially expressed proteins (DEPs).
a Volcano plots are depicted with the log2-fold change (FC) for each of the 221 proteins with LFQ value > 0 in at least three samples in each group. The left image indicates log2FC of protein abundance in Virus relative to HC. The right image indicates log2FC of protein abundance in MPP relative to HC. Red, blue and gray circles show proteins which have significant increases, decreases or no differences, respectively. b Venn diagram displaying the number and overlap of differentially expressed and unique proteins in the Virus and MPP groups compared with HC. c Heatmap of 260 viral-associated DEPs. d Protein–protein interaction network of the viral-associated DEPs constructed by Metascape and visualized using Cytoscape. A total of 216 proteins are shown in the PPI network. Sizes of dots are proportional to the score. Eight modules are indicated in different colors and shown around this network.
Functional enrichment analysis of the DEPs specific to children with viral pneumonia
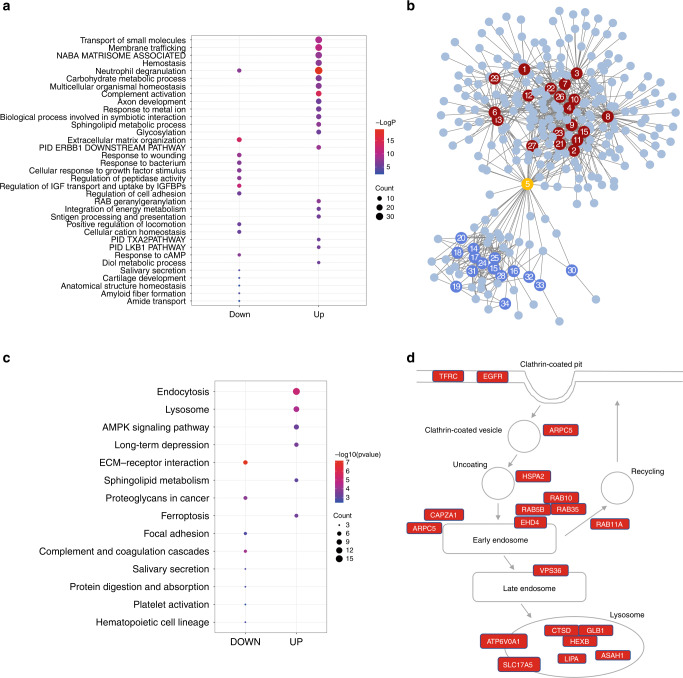
We next performed functional enrichment analysis of the upregulated and downregulated viral-associated DEPs separately using Metascape. This analysis showed that the up- and downregulated proteins were enriched for distinct biological terms (Fig. 4a, b and Supplementary Table S8). Network analysis showed that the up- and downregulated protein-enriched terms form two distinct clusters (Fig. 4b). The upregulated proteins were mapped to many terms related to metabolism including carbohydrate metabolic process, sphingolipid metabolic process, diol metabolic process, glycosylation, integration of energy metabolism, RAB geranylgeranylation and the LKB1 pathway (Fig. 4a). Downregulated proteins, on the other hand, were enriched for biological terms specific to extracellular matrix organization, regulation of IGF transport and uptake by IGFBPs, amyloid fiber formation, response to wounding, response to bacterium, cellular cation homeostasis, regulation of cell adhesion, positive regulation of locomotion, amide transport, cartilage development, response to cAMP, regulation of peptidase activity, anatomical structure homeostasis, and cellular response to growth factor stimulus (Fig. 4a). We also found that a subset of upregulated and downregulated proteins was commonly mapped to neutrophil degranulation (Fig. 4a, b), including 31 upregulated proteins (SERPINA3, ALAD, ANPEP, ASAH1, ATP6V0A1, CTSD, FRK, GLB1, HEXB, HSPA8, PFKL, SERPINA1, PKM, PRCP, RAB5B, RAB5C, VCL, CREG1, IQGAP1, MLEC, TOM1, ARPC5, ACTR2, VAT1, CCT8, RAB10, GCA, COMMD9, CAB39, RAB4B, and ANO6) and 15 downregulated proteins (CD44, CD59, CEACAM8, CD55, GRN, HMGB1, LYZ, CALML5, AMBP, APP, COL1A1, COL1A2, FOLR1, CHMP2B, PRNP) in the viral pneumonia group, coinciding with the crucial role of neutrophils in viral infections. Monitoring the changes of these proteins in urinary sEV may reveal the dynamics of neutrophil activity.
Fig. 4. Function enrichment analysis of viral-associated DEPs.
a Bubble plot of enrichment analysis of the upregulated (UP) and downregulated (DOWN) viral-associated DEPs using Metascape. The colors of the nodes are shown from red to blue in descending order for the –log10 P value. The sizes of the nodes are shown from small to large in ascending order for the number of proteins. b The network of function enrichment terms for the viral-associated DEPs. Proteins are visualized as circles without numbers. Enrichment terms are visualized as circles with numbers. The red and blue numbered circles represent terms that mapped to up- and downregulated proteins, respectively. The orange numbered circle represents the common term that mapped to all proteins assessed. The numbers represent the function enrichment terms as follow: 1, transport of small molecules; 2, membrane trafficking; 3, Naba matrisome associated; 4, hemostasis; 5, neutrophil degranulation; 6, carbohydrate metabolic process; 7, multicellular organismal homeostasis; 8, complement activation; 9, axon development; 10, response to metal ion; 11, biological process involved in symbiotic interaction; 12, sphingolipid metabolic process; 13, glycosylation; 14, extracellular matrix organization; 15, PID ERBB1 downstream pathway; 16, response to wounding; 17, response to bacterium; 18, cellular response to growth factor stimulus; 19, regulation of peptidase activity; 20, Regulation of IGF transport and uptake by IGFBPs; 21, regulation of cell adhesion; 22, RAB geranylgeranylation; 23, integration of energy metabolism; 24, antigen processing and presentation; 25, positive regulation of locomotion; 26, cellular cation homeostasis; 27, PID TXA2 pathway; 28, PID LKB1 pathway; 29, response to cAMP; 30, diol metabolic process; 31, salivary secretion; 32, cartilage development; 33, anatomical structure homeostasis; 34, amyloid fiber formation; 35, amide transport. c Bubble plot of KEGG enrichment analysis of the upregulated (UP) and downregulated (DOWN) viral-associated DEPs using clusterProfiler. The colors of the nodes are shown from red to blue in descending order of –log10 P value. The sizes of the nodes are shown from small to large in ascending order for the number of proteins. d Schematic of the endolysosome pathway. Genes significantly upregulated are marked with red.
In addition, we also performed functional enrichment analysis of the viral-associated DEPs with clusterProfiler.24 Using this method, we found that the upregulated proteins were enriched for KEGG pathways specific to endocytosis, lysosome, long-term depression, ferroptosis, AMPK signaling pathway and sphingolipid metabolism, whereas the downregulated proteins were specifically mapped to pathways of ECM-receptor interaction, complement and coagulation cascades, focal adhesion, salivary secretion, protein digestion and absorption, platelet activation, proteoglycans and hematopoietic cell lineage (Fig. 4c and Supplementary Table S9). The most significant enrichment pathways were endocytosis and lysosome with enrichment in 25 upregulated proteins including HSPA8, ARPC5, RAB11A, CAPZA1, HSPA2, EGFR, RAB10, RAB35, EHD4, TFRC, RAB5B, VPS36, ARPC4, RAB5C, ACTR2, HEXB, GLB1, ASAH1, CTSD, ATP6V0A1, LIPA, CTSO, GBA, CTSF and SLC17A5 (Fig. 4d), suggesting that both pathways were activated. Collectively, the proteomic profiling of urinary sEV highlights important biological pathways associated with the host response to virus infection in children with viral pneumonia.
Validation of urinary sEV proteins by multiple reaction monitoring (MRM) analysis
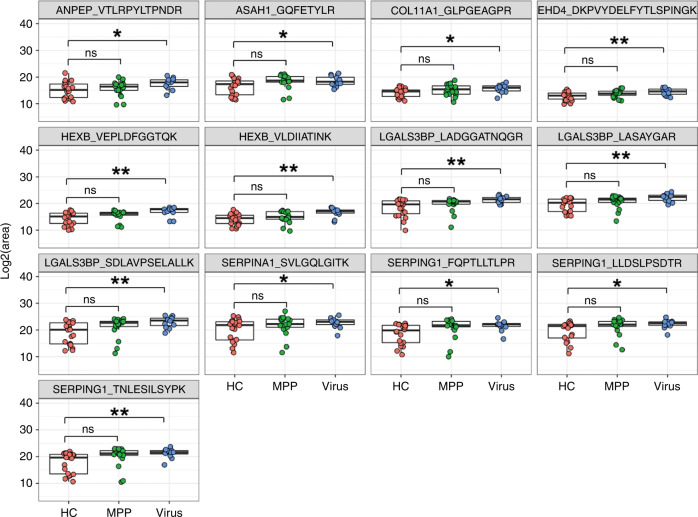
A total of 33 urinary sEV proteins functionally associated with viral pneumonia (Supplementary Table S10) for targeted detection using MRM in an independent cohort of 12 children with viral pneumonia, 17 children with MPP and 20 HC (Table 2). As a result, a total of 41 peptides representing 19 proteins were successfully detected by MRM analysis and used for quantitative analysis (Supplementary Table S11). The abundance of 13 peptides representing 8 proteins (ANPEP, ASAH1, COL11A1, EHD4, HEXB, LGALS3BP, SERPINA1, SERPING1) were significantly higher in the viral pneumonia group but not in the MPP group than in the healthy control group (Fig. 5), which is consistent with the results of label-free proteomics in discovery phase, suggesting that these urinary sEV proteins or peptides, if validated in a large cohort, may serve as potential biomarkers for the diagnosis of viral pneumonia in children.
Fig. 5. Validation of expression levels of selected urinary sEV proteins by multiple reaction monitoring.
The peak areas of thirteen peptides representing eight proteins were measured by multiple reaction monitoring. ns, not significant; *P < 0.05; **P < 0.01.
Discussion
It was previously shown that the sEV composition, including host protein and RNA, changes dramatically during infection, especially by intracellular pathogens such as viruses.25,26 Previous studies suggest that sEV from virus-infected cells play a role in promoting the innate and acquired immune response and the composition of sEV can be used to reflect the pathological condition during viral infection. In the present study, we performed proteomic analysis of urinary sEV derived from children with pneumonia, found that the proteome of urinary sEV was significantly different in children with viral pneumonia, but not those with MPP, compared with HC, and identified the molecular signature associated with viral infection and validated several proteins as potential biomarkers for the diagnosis of children with viral pneumonia.
Interferons (IFNs) are key cytokines in response to viral infection. IFN stimulation leads to the expression of several ISGs that exert a wide variety of antiviral activities.27 Remarkably, we identified more than 30 ISGs in urinary sEV, of which 18 were overrepresented in the viral pneumonia group, suggesting that the increase in expression of these ISGs in urinary sEV can be used as an indicator of the emergence of viral infections. Consistent with our results, sEV have been shown to carry ISGs and exert antiviral effects by delivering these ISGs to recipient cells.28,29 For example, sEV derived from human brain microvascular endothelial cells were found to deliver several ISGs including ISG15, ISG56, and Mx2 to macrophages, thereby help to repair the antiviral state in HIV-infected macrophages.30 Hence, we hypothesize that urinary sEV containing ISGs may have antiviral effect in children with viral pneumonia through transferring of ISGs to recipient cells.
We also identified 17 viral receptor proteins enriched in urinary sEV, illustrating that sEV may participate in the regulation of viral invasion by directly binding to viruses. In fact, previous studies have reported that some viral receptors are located on the membrane of sEV.9,31 The most compelling research is focused on ACE2 located on the sEV membrane.32–34 In particular, viral infection can promote the transfer of ACE2 from the plasma membrane to the sEV membrane.33 Our results suggest that viral infection may induce expression of ACE2 in sEV present in body fluids such as urine. However, whether the other highly expressed receptor proteins in urinary sEV from children with viral pneumonia are directly triggered by the virus infection requires further in-depth study.
Function enrichment analysis showed significant changes in the expression of many urinary sEV proteins related to neutrophil degranulation in children with viral pneumonia as compared to HC, suggesting that the proteome of sEV can reflect the signature for neutrophil activity during the progression of pneumonia. Neutrophils, the first-line effector cells to be recruited in lung inflammation, are increased in the circulation and tissues during bacterial or viral infections, and play an essential beneficial role in host defense. Therefore, neutrophil activity can be regarded as an indicator of the severity of the disease, and dysregulation of these cells has the potential to cause tissue damage.35,36 However, whether the urinary sEV loaded with these proteins are directly derived from neutrophils requires further exploration. Furthermore, it is worth considering whether the changes in their expression within sEV are consistent with those in cells and tissues. Importantly, sEV derived from neutrophils have been shown to play an essential role in the pathogenesis of lung inflammation.37,38 Therefore, we speculated that these neutrophil-related proteins identified in sEV played an important role in the pathogenesis of children with viral pneumonia.
Interestingly, proteomic profiling of urinary sEV can provide information indicating the metabolic state of children with viral pneumonia. We found that 79 upregulated proteins in urinary sEV of the viral pneumonia group were primarily mapped to three metabolic processes: the carbohydrate metabolic process, the sphingolipid metabolic process and glycosylation. This result is in agreement with the conclusion that viruses can target proteins of the metabolic pathway and change the host metabolic process, thereby facilitating the growth of the viruses.39,40 In-depth analysis of these modified metabolic pathways has the potential to be the key to understanding the pathogenesis of children with viral pneumonia. Glycosylation, especially, plays an important role in virus biology.41–44 Viruses use the host glycoprotein synthesis mechanism to modify proteins presented on their surface, which can help to regulate viral infection, invasion, and recognition of host receptors.44 We found that 16 upregulated proteins in urinary sEV (ABO, ENTPD5, FUT6, GALNT1, GALNT2, GBA, PLOD3, GAL3ST1, B3GNT3, POMGNT1, B3GNT8, TMEM106A, IHH, AK1, ATP6V1A and NPR1) belong to the glycosylation pathway, suggesting that these proteins are involved in viral protein glycosylation in children with viral pneumonia, and changes in their expression may directly or indirectly reflect viral activity in the body. Several studies have shown that sEV can promote the transmission of viruses in vivo by delivering viral proteins and nucleic acids.11,45,46 However, our study suggests that the impact of sEV on virus invasion may rely on the transmission of proteins involved in glycosylation between cells to promote the process of glycosylation of viral proteins and accelerate viral spread.
In addition to the important role of glycosylation in virus invasion mentioned above, viruses can take advantage of the endolysosome mechanism to infect cells.47–49 Regulation of this pathway may directly affect the viral potential to invade. Surprisingly, the proteomic analysis of urinary sEV revealed that many proteins related to the endolysosome pathway were significantly elevated in children with pneumonia, implying that the endolysosome pathway was activated to facilitate the spread of the virus between cells in children with viral pneumonia. Similar to the above speculation, we infer that sEV may transfer these proteins associated with endolysosomes between cells to regulate the efficiency of viral endocytosis in the recipient cells.
In a subsequent validation phase, MRM analysis revealed that the abundance of eight proteins was significantly increased in children with viral pneumonia compared to HC. These proteins (ANPEP, ASAH1, COL11A1, EHD4, HEXB, LGALS3BP, SERPINA1, SERPING1) are involved in important biological processes associated with viral infection. For example, ANPEP acts as a receptor for viral spike glycoprotein, ASAH1and HEXB are lysosomal enzymes, the three proteins participate in the regulation of viral invasion and secretion. EHD4, SERPING1 and LGALS3BP are ISGs, among which EHD4 located on endosome membrane may be involved in viral entry and LGALS3BP promotes cell adhesion and stimulates host defense against viruses. SERPING1 and SERPINA1 belong to serpin family, inhibit complement activation and elastase, respectively, the two proteins may protect the tissue from damage caused by an exacerbated immune response. We hold opinion that these urinary sEV proteins, if validated in a large cohort, will be used as biomarkers for diagnosing viral pneumonia in children.
Although the current study has shed lights on the involvement of sEV in many pathways that can regulate viral infection, several aspects can be improved including increasing the sample size to improve the statistical power and allow more conclusive statements to be made on the associations between protein expression and pneumonia, and mechanistic studies to assess the impacts of these sEV proteins on viral infection.
Conclusions
To our knowledge, this is the first study of urinary sEV proteome of children with pneumonia. We showed that the proteomic profile of urinary sEV can reflect body change at the molecular level caused by pathogenic infection in children. Children with pneumonia caused by different pathogens exhibit significant overlap in clinical phenotypes, hindering precise treatment and leading to overuse of antibiotics. We successfully validated several urinary sEV proteins that were exclusively increased in children with viral pneumonia as potential biomarkers, indicating that urinary sEV may be used for further development for the purpose of distinguishing and diagnosing pediatric pneumonia caused by different pathogens.
Supplementary information
Acknowledgements
We are grateful to all the children who took part in the study.
Author contributions
All authors have met the Pediatric Research authorship requirements. L.Y., J.W. and Q.Z. conceived and designed the experiments. J.C., Y.Y., Q.P., J.W. and L.Y. collected clinical samples. J.C., D.J. and S.W. conducted sEV isolation, LC-MS/MS experiments and data analysis. D.J. and Q.Z. drafted the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by Grant from the Shanghai Municipal Health Commission (202140111).
Data availability
The datasets generated during the current study are available in the ProteomeXchange Consortium repository (www.proteomexchange.org) with the dataset identifier PXD036472.
Competing interests
D.J., S.W. and Q.Z. are employees of Wayen Biotechnologies, Inc. The other authors declare that they have no conflicts to declare.
Ethics approval and consent to participate
The study procedure was reviewed and approved by the Ethics Committee of Shanghai Children’s Medical Center. Written informed consent was provided by the patients of the children subjects.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Juan Cheng, Dongrui Ji.
Contributor Information
Jinhong Wu, Email: wujinhong630@126.com.
Lin Yang, Email: lynyang73@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41390-022-02431-y.
References
- 1.Marangu D, Zar HJ. Childhood pneumonia in low-and-middle-income countries: an update. Paediatr. Respir. Rev. 2019;32:3–9. doi: 10.1016/j.prrv.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nascimento-Carvalho AC, Ruuskanen O, Nascimento-Carvalho CM. Comparison of the frequency of bacterial and viral infections among children with community-acquired pneumonia hospitalized across distinct severity categories: a prospective cross-sectional study. BMC Pediatr. 2016;16:105. doi: 10.1186/s12887-016-0645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhuiyan MU, et al. Role of viral and bacterial pathogens in causing pneumonia among western Australian children: a case-control study protocol. BMJ Open. 2018;8:e020646. doi: 10.1136/bmjopen-2017-020646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues C, Groves H. Community-acquired pneumonia in children: the challenges of microbiological diagnosis. J. Clin. Microbiol. 2018;56:e01318–01317. doi: 10.1128/JCM.01318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klompas M. Does this patient have ventilator-associated pneumonia? JAMA. 2007;297:1583–1593. doi: 10.1001/jama.297.14.1583. [DOI] [PubMed] [Google Scholar]
- 6.Dela Cruz CS, et al. Future research directions in pneumonia. NHLBI Working Group Report. Am. J. Respir. Crit. Care Med. 2018;198:256–263. doi: 10.1164/rccm.201801-0139WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raab-Traub N, Dittmer DP. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017;15:559–572. doi: 10.1038/nrmicro.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schorey JS, Harding CV. Extracellular vesicles and infectious diseases: new complexity to an old story. J. Clin. Invest. 2016;126:1181–1189. doi: 10.1172/JCI81132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chahar HS, Corsello T, Kudlicki AS, Komaravelli N, Casola A. Respiratory syncytial virus infection changes cargo composition of exosome released from airway epithelial cells. Sci. Rep. 2018;8:387. doi: 10.1038/s41598-017-18672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedford JG, et al. Airway exosomes released during influenza virus infection serve as a key component of the antiviral innate immune response. Front. Immunol. 2020;11:887. doi: 10.3389/fimmu.2020.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiemstra TF, et al. Human urinary exosomes as innate immune effectors. J. Am. Soc. Nephrol. 2014;25:2017–2027. doi: 10.1681/ASN.2013101066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, et al. Lectin microarray combined with mass spectrometry identifies haptoglobin-related protein (HPR) as a potential serologic biomarker for separating nonbacterial pneumonia from bacterial pneumonia in childhood. Proteom. Clin. Appl. 2018;12:e1800030. doi: 10.1002/prca.201800030. [DOI] [PubMed] [Google Scholar]
- 15.Song Z, et al. Comprehensive proteomic profiling of urinary exosomes and identification of potential non-invasive early biomarkers of Alzheimer’s disease in 5xfad mouse model. Front. Genet. 2020;11:565479. doi: 10.3389/fgene.2020.565479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng J, et al. Proteomic profiling of serum small extracellular vesicles reveals immune signatures of children with pneumonia. Transl. Pediatr. 2022;11:891–908. doi: 10.21037/tp-22-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Kojima K, Mobley JA, West AB. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine. 2019;45:351–361. doi: 10.1016/j.ebiom.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhondt B, et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles. 2020;9:1736935. doi: 10.1080/20013078.2020.1736935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane M, et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe. 2016;20:392–405. doi: 10.1016/j.chom.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T, et al. Clusterprofiler 4.0: a universal enrichment tool for interpreting omics. Data. Innov. (N. Y) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshikawa FSY, Teixeira FME, Sato MN, Oliveira L. Delivery of micrornas by extracellular vesicles in viral infections: could the news be packaged? Cells. 2019;8:611. doi: 10.3390/cells8060611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolte-‘t Hoen E, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: are they close relatives? Proc. Natl Acad. Sci. USA. 2016;113:9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 29.Yao Z, et al. Label-free proteomic analysis of exosomes secreted from THP-1-derived macrophages treated with IFN-alpha identifies antiviral proteins enriched in exosomes. J. Proteome Res. 2019;18:855–864. doi: 10.1021/acs.jproteome.8b00514. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, et al. Exosomes contribute to the transmission of anti-HIV activity from TLR3-activated brain microvascular endothelial cells to macrophages. Antivir. Res. 2016;134:167–171. doi: 10.1016/j.antiviral.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crenshaw BJ, Gu L, Sims B, Matthews QL. Exosome biogenesis and biological function in response to viral infections. Open Virol. J. 2018;12:134–148. doi: 10.2174/1874357901812010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cocozza F, et al. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 spike protein-containing virus. J. Extracell. Vesicles. 2020;10:e12050. doi: 10.1002/jev2.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie, F. et al. Engineering extracellular vesicles enriched with palmitoylated ACE2 as COVID-19 therapy. Adv. Mater.33, e2103471 (2021). [DOI] [PMC free article] [PubMed]
- 34.Zhang J, et al. The interferon-stimulated exosomal HACE2 potently inhibits SARS-CoV-2 replication through competitively blocking the virus entry. Signal Transduct. Target Ther. 2021;6:189. doi: 10.1038/s41392-021-00604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galani IE, Andreakos E. Neutrophils in viral infections: current concepts and caveats. J. Leukoc. Biol. 2015;98:557–564. doi: 10.1189/jlb.4VMR1114-555R. [DOI] [PubMed] [Google Scholar]
- 36.Grudzinska FS, et al. Neutrophils in community-acquired pneumonia: parallels in dysfunction at the extremes of age. Thorax. 2020;75:164–171. doi: 10.1136/thoraxjnl-2018-212826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vargas A, Roux-Dalvai F, Droit A, Lavoie JP. Neutrophil-derived exosomes: a new mechanism contributing to airway smooth muscle remodeling. Am. J. Respir. Cell Mol. Biol. 2016;55:450–461. doi: 10.1165/rcmb.2016-0033OC. [DOI] [PubMed] [Google Scholar]
- 38.Genschmer KR, et al. Activated PMN exosomes: pathogenic entities causing matrix destruction and disease in the lung. Cell. 2019;176:113–126.e115. doi: 10.1016/j.cell.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Clippinger AJ, Alwine JC. Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends Microbiol. 2011;19:360–367. doi: 10.1016/j.tim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girdhar K, et al. Viruses and metabolism: the effects of viral infections and viral insulins on host metabolism. Annu. Rev. Virol. 2021;8:373–391. doi: 10.1146/annurev-virology-091919-102416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagdonaite I, Wandall HH. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y, Bowden TA, Wilson IA, Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. The importance of glycans of viral and host proteins in enveloped virus infection. Front. Immunol. 2021;12:638573. doi: 10.3389/fimmu.2021.638573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, Li S, Wu S, Chen L. Exosomes modulate the viral replication and host immune responses in HBV infection. Biomed. Res. Int. 2019;2019:2103943. doi: 10.1155/2019/2103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q, et al. Exosomes mediate horizontal transmission of viral pathogens from insect vectors to plant phloem. Elife. 2021;10:e64603. doi: 10.7554/eLife.64603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 2014;6:a016972. doi: 10.1101/cshperspect.a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh S, et al. Beta-coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e1514. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the ProteomeXchange Consortium repository (www.proteomexchange.org) with the dataset identifier PXD036472.