Abstract
Site-directed mutagenesis is an invaluable technique that enables the elucidation of the contribution of specific residues to protein structure and function. The simultaneous introduction of mutations at a large number of sites (>10), singly and in multiple combinations, is often necessary to fully understand the functional contributions. We report a simple, efficient, time and cost-effective method to achieve this using commonly available molecular biology reagents and protocols, as an alternative to gene synthesis. We demonstrate this method using the Omicron Spike DNA construct as an example, and create a construct bearing 37 mutations (as compared to wild-type Spike DNA), as well as 4 other constructs bearing subsets of the full spectrum of mutations. We believe that this method can be an excellent alternative to gene synthesis, especially when three or more variants are required.
Keywords: cost-effective method, directed multiple mutagenesis, Omicron Spike, PCR based
Graphical Abstract
Graphical Abstract.

Introduction
The method of choice for the introduction of one or a few mutations located in close proximity in recombinant DNA constructs is site-directed mutagenesis. In contrast, for introducing a large number of mutations, especially those not clustered together within the region covered by a typical primer and spread over several kilobases, commercial gene synthesis is preferred [1]. However, this may be too expensive, may take too long and may not be an option in some geographical locations or for some projects. In this scenario, an alternative method combining polymerase chain reaction (PCR), overlap extension PCR, and Gibson cloning can be used to create the desired construct in a short duration and at a cost lower than that of commercial synthesis. This alternate strategy will provide a significant cost advantage especially when a series of constructs with combinations of mutations needs to be created.
The basic principle of the method lies in assembling the complete construct using a series of smaller, mutation-containing fragments. The construct is split into fragments in such a way that forward and reverse primers used to amplify a fragment will contain all of the mutations located in that fragment. The mutation-containing fragments are then seamlessly assembled using a homology-based cloning method such as Gibson assembly [2]. Mutations located within a 50 bp region are introduced using a single primer; fragment sizes typically range from 100 bp to 3 kb and an unlimited number of fragments can be combined into the final construct in one or more rounds of cloning. Due to the modular nature of construct assembly, a combinatorial library of constructs including or excluding any combination of mutations can be generated with minimal additional effort, time and cost.
We illustrate this method using the example of the Omicron Spike gene which contains 37 mutations (dispersed over nearly 3 kb) as compared to the wild-type Spike gene. We followed a similar strategy to create the Delta Spike construct previously, which was successfully used in the pseudovirus neutralization assay reported by Kumar et al. [3], validating this approach.
Overall strategy
The comprehensive list of mutations incorporated into the Omicron Spike was based on the GISAID sequence EPI_ISL_6814922 (27 November 2021) [4] and is listed below:
Spike A67V, Spike H69del, Spike V70del, Spike T95I, Spike G142D, Spike V143del, Spike Y144del, Spike Y145del, N211del, Spike L212I, Spike ins214EPE, Spike G339D, Spike S371L, Spike S373P, Spike S375F, Spike K417N, Spike N440K, Spike G446S, Spike S477N, Spike T478K, Spike E484A, Spike Q493R, Spike G496S, Spike Q498R, Spike N501Y, Spike Y505H, Spike T547K, Spike D614G, Spike H655Y, Spike N679K, Spike P681H, Spike N764K, Spike D796Y, Spike N856K, Spike Q954H, Spike N969K and Spike L981F.
The overall strategy was to split the Spike-coding sequence into fragments, each to be amplified by PCR, incorporating the desired mutations in the primer binding regions. The fragments would then be stitched together by overlap PCR, and then assembled by Gibson assembly [2] to create the complete sequence (Fig. 1). Gibson assembly uses a combination of three enzymes to ligate fragments with homologous ends and leaves no scar in the final ligated product, an essential requirement while assembling a large number of fragments. The process efficiently ligates up to five fragments, after which the efficiency drops. Since this involved a large number of fragments, at least two rounds of assembly and verification by sequencing were anticipated.
Figure 1.
Strategy for the construction of Omicron Spike incorporating 37 mutations.
We used the Spike-coding sequence, with the 19 amino acid deletion at the C-terminal end, as our template since this was the construct to be used in the Pseudovirus neutralization assay [5]. We used the wild-type Spike, or the Delta-plus variant Spike DNA as the template for PCR, based on the mutations required to be incorporated in a specific fragment. Three mutations were incorporated from the Delta-plus Spike DNA template and the rest were introduced using primers. We divided the Spike sequence into 16 fragments, each demarcated by the location of one or more Omicron-specific mutations, and each overlapping with the preceding and following fragments by at least 27 bp. We designed primers for the amplification of each fragment incorporating the required mutation(s) at the centre of the primer, with at least 12 bp of perfectly matched sequence to promote proper annealing and amplification during PCR (Fig. 1).
There were two regions that required a slightly different design. Fragment 8 contained nine mutations spread over ∼200 bp region, which did not align with the aforementioned strategy. Therefore, two long oligonucleotides with a 23 bp overlap were designed to generate Fragment 8 by routine overlap extension. Fragment 15 contained two mutations spread over 48 bp, which was too short to split into separate fragments, and too long for a single primer (∼74 bp). Hence, two consecutive reverse primers were designed to be used sequentially for Fragment 15, and the second PCR was done using the first PCR as a template.
A few additional modifications were employed to simplify the assembly and screening process:
Plasmids (pCMV-Spike (wt) and pCDNA-Spike (Delta-plus)) used as a template for PCRs were digested with at least two restriction enzymes in order to reduce the background from inserts during transformation.
pCMV-Spike was used as the template for PCRs, and fragments were cloned into pCDNA3.1. This allowed for the correct identification of clones by colony PCR using a combination of Spike-specific internal primers and pCDNA3.1-specific primers at each end.
The last mutation L981F was incorporated only in the final round of assembly, so as to be able to amplify one end from a pCMV-Spike template and thus allowing colony PCR-based screening as mentioned in #2 above.
The assembly was planned in such a way as to generate intermediate chimeric clones, which would contain the Omicron mutations in just the NTD, RBD or CTD (with the rest being wild-type Spike sequence).
Methods and results
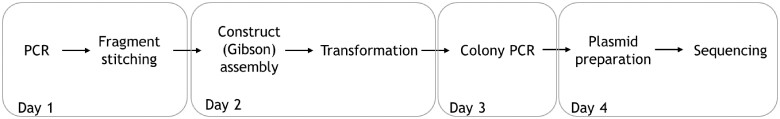
The complete Omicron Spike DNA construct was split into four parts for ease of synthesis, based on the technical limitation of the Gibson assembly process. The mutation containing fragments of each part was amplified first and assembled into a clone in the first round. A simple workflow for the generation of each clone is shown below (Fig. 2). The four parts were then combined into the complete Omicron Spike DNA construct, by following the same workflow in subsequent rounds.
Figure 2.
A simple workflow for the assembly of each part of the complete Omicron construct.
PCR amplification of fragments
Each of the fragments (1–7 and 9–16) was amplified from the appropriate template in a standard 25 µl PCR reaction (Fig. 3A, representative gel images of select PCR products are shown). Taq polymerase was used for PCR amplification of fragments for the first part (fragments less than 1000 bp). For later rounds, Q5 polymerase was used to avoid PCR errors. Additional fragments to complete the assembly of each clone were also amplified using the pCMV-Spike (wild-type) template (labelled fragments 17–23, examples shown in Fig. 3A). For example, clone 1 would include mutation containing fragments 1, 2, 3, 4 and the rest (5–16) amplified from wild-type Spike (Fragment 17). The list of primers used in this study can be found in Table 1, and details of the fragments used, along with the primers and template DNA used to amplify them, can be found in Table 2. A standard PCR reaction was assembled as follows:
Figure 3.
Representative gel images at various stages of Omicron synthesis. (A) Individual fragments were amplified by PCR using Taq Polymerase or Q5 and 8 µl was visualized on a native polyacrylamide gel (fragments <800 bp) or agarose gel. (B) Fragments stitched together by OE were visualized by native polyacrylamide gel elecrophoresis (PAGE). (C) Colony PCR was used to confirm the presence of the complete insert using two primer sets, one across the vector–insert junction at the N-terminal end of the Spike ORF (Front), and the second at the junction near the C-terminal end (End). Two representative clones are shown. (D) Plasmids amplified from positive colonies were checked for size to confirm the presence of all insert fragments, by resolving on an agarose gel. (E) For each clone assembled using Gibson assembly, the fraction of positive colonies by colony PCR, by plasmid size and finally by sequencing are indicated as a measure of efficiency of the cloning process.
Table 1.
List of primers used in this study
| S.no | Primer | Sequence |
|---|---|---|
| 1. | F1.2 pCDNA | CCCAAGCTGGCTAGCGTTTAAACTTAAGCTTCACCATGTTCGTTTTCCTTGTTCT |
| 2. | R3.2 pCDNA | ATCTGCAGAATTCCACCACACTGGACTAGTGGATCCTTAGCAACATGATCCGCAAGAGCA |
| 3. | Omi F1Rev | AGTTCCAGAGATAACATGAAACCAAGT |
| 4. | Omi F2Fwd | ACTTGGTTTCATGTTATCTCTGGAACT |
| 5. | Omi F2Rev | TATGTTTGACTTTTCAATGGAAGCGAAGTA |
| 6. | Omi F3Fwd | TACTTCGCTTCCATTGAAAAGTCAAACATA |
| 7. | Omi F3Rev | GTTATTCTTATGATCCAGGAAGGGATC |
| 8. | Omi F4Fwd | GATCCCTTCCTGGATCATAAGAATAAC |
| 9. | Omi F4Rev | TCCCTGGGGCAGGTCTTCCGGTTCGCGCACAATTATTGGAGTGTG |
| 10 | Omi F5Fwd | CACACTCCAATAATTGTGCGCGAACCGGAAGACCTGCCCCAGGGA |
| 11 | Omi F5Rev | GTTGAACACCTCATCGAAGGGACACAG |
| 12 | Omi F6Fwd | CTGTGTCCCTTCGATGAGGTGTTCAAC |
| 13 | Omi F6Rev | GCACTTGAAGGTGAAGAACGGGGCCAGGTTGTAGAGGAC |
| 14 | Omi F7Fwd | GTCCTCTACAACCTGGCCCCGTTCTTCACCTTCAAGTGC |
| 15 | Omi F7Rev | ATTGTAGTTACCGCTGACCTTAGAATCGAGTTTATTAGAGTTCCA |
| 16 | Omi F8FwdOE | AAGGTCAGCGGTAACTACAATTACCTGTACCGCTTGTTTAGGAAGTCAAACCTGAAGCCTTTCGAGAGGGATATTTCAACCGAAATCTATCAAGCGGGTAATAAACCGTGTA |
| 17 | Omi F8RevOE | CACGCGATAAGGTTGATGACCCACGCCATAGGTTGGACGGAAGCTGTAAGAACGCAGGGGGAAGTAGCAGTTAAATCCTGCCACACCGTTACACGGTTTATTACCCGCTTGA |
| 18 | Omi F9Fwd | GGCGTGGGTCATCAACCTTATCGCGTG |
| 19 | Omi F9Rev | CACTCCAGTTCCTTTGAGGCCATTGAA |
| 20 | Omi F10Fwd | TTCAATGGCCTCAAAGGAACTGGAGTG |
| 21 | Omi F10Rev | GGAGTTATTCACATATTCCGCTCCGAT |
| 22 | Omi F11Fwd | ATCGGAGCGGAATATGTGAATAACTCC |
| 23 | Omi F11Rev | TCTCCTGTGTGATTTAGTTTGTGTCTG |
| 24 | Omi F12Fwd | CAGACACAAACTAAATCACACAGGAGA |
| 25 | Omi F12Rev | TGTAAGGGCGCGTTTCAATTGGGTACA |
| 26 | Omi F13Fwd | TGTACCCAATTGAAACGCGCCCTTACA |
| 27 | Omi F13Rev | GAAGCCTCCGAAATACTTAATGGGCGG |
| 28 | Omi F14Fwd | CCGCCCATTAAGTATTTCGGAGGCTTC |
| 29 | Omi F14Rev | GACTGTCAGGCCTTTAAACTTCTGGGC |
| 30 | Omi F15Fwd | GCCCAGAAGTTTAAAGGCCTGACAGTC |
| 31 | Omi F15Rev1 | GCTTCACCAGAGTATTCAAAGCCTGAGCATTATGGTTCACCACGTC |
| 32 | Omi F15Rev2 | GATAGCGCCGAATTTTGAAGAGAGTTGCTTCACCAGAGTATTCAAA |
| 33 | Omi F16Fwd | CAACTCTCTTCAAAATTCGGCGCTATC |
| 34 | Omi F16.1Fwd | TTGAACGACATCTTTAGTCGCCTTGAT |
| 35 | Omi F16.1Rev | ATCAAGGCGACTAAAGATGTCGTTCAA |
| 36 | AS507 | GGGCAAACAACAGATGGCT |
| 37 | AS575 | gttgggttttatcgccggt |
| 38 | AS645 | tccttgcttgccctcaagatcca |
| 39 | AS145 | GCGAAATTAATACGACTCACTATAGG |
Table 2.
A list of fragments generated during the construction of the Omicron Spike clone, the primers and templates used for their generation
| Fragment | Primers Used | Input source |
|---|---|---|
| 1 | F1.2 pCDNA+ Omi F1Rev | Wild-type |
| 2 | Omi F2Fwd+ Omi F2Rev | Wild-type |
| 3 | Omi F3Fwd+ Omi F3Rev | Delta-plus |
| 4 | Omi F4Fwd+ Omi F4Rev | Wild-type |
| 5 | Omi F5Fwd+ Omi F5Rev | Wild-type |
| 6 | Omi F6Fwd+ Omi F6Rev | Wild-type |
| 7 | Omi F7Fwd+ Omi F7Rev | Delta-plus |
| 8 | Omi F8FwdOE+ Omi F8RevOE | – |
| 9 | Omi F9Fwd+ Omi F9Rev | Wild-type |
| 10 | Omi F10Fwd+ Omi F10Rev | Delta-plus |
| 11 | Omi F11Fwd+ Omi F11Rev | Wild-type |
| 12 | Omi F12Fwd+ Omi F12Rev | Wild-type |
| 13 | Omi F13Fwd+ Omi F13Rev | Wild-type |
| 14 | Omi F14Fwd+ Omi F14Rev | Wild-type |
| 15 | Omi F15Fwd+ Omi F15Rev | Wild-type |
| 15′ | Omi F15Fwd+ Omi F15Rev2 | Fragment 15 |
| 16 | Omi F16Fwd+ 3.2 pCDNA | Wild-type |
| 17 | Omi F5Fwd + R3.2 pCDNA | Wild-type |
| 18 | F1.2 pCDNA+ Omi F5Rev | Wild-type |
| 19 | Omi F9Fwd + R3.2 pCDNA | Wild-type |
| 20 | F1.2 pCDNA+ Omi F9Rev | Wild-type |
| 21 | Omi F13Fwd + R3.2 pCDNA | Wild-type |
| 22 | F1.2 pCDNA+ Omi F13Rev | Wild-type |
| 23 | Omi F2Fwd + Omi F5Rev | Clone 1 plasmid 3 |
| 24 | Omi F8FwdOE + R3.2 pCDNA | Wild-type |
| 25 | Omi F8FwdOE+ Omi F8RevOE | Wild-type |
| 26 | Omi F13Fwd + R3.2 pCDNA | Clone 4 plasmid 25 |
| 27 | Omi F9Fwd + OmiF16.1Rev | Clone 25 |
| 28 | OmiF16.1Fwd + R3.2 pCDNA | Wild-type |
| 29 | F1.2 pCDNA+ Omi F8RevOE | Clone 1.2, plasmid 13 |
| 30 | F1.2 pCDNA + Omi F4Rev | Clone 1.2, plasmid 13 |
| 31 | F1.2 pCDNA+ Omi F4Rev | Wild-type |
| 32 | Omi F5Fwd+ Omi F8RevOE | Clone 1.2, plasmid 13 |
| 33 | Omi10FFwd +R3.2 pCDNA | OmicronFL Clone 30 |
Challenge: The overlap PCR to generate Fragment 8 was inefficient, and prevented efficient assembly into Clone 2. This was corrected in the subsequent round of assembly by using the long oligonucleotides as just primers.
Fragment stitching
In order to improve the efficiency of Gibson assembly by reducing the number of inserts (to a maximum of three excluding vectors), we stitched short PCR fragments using the method of Hilgarth et al. [6]. Briefly, each PCR fragment was purified on a DNA cleanup column to get rid of primers, combined in equimolar amounts and stitched together in overlap extension 1 (OE1). Subsequently, the stitched fragment was amplified using the forward primer of the first fragment, and the reverse primer of the second fragment in OE2 (Fig. 3B).
| OE1 | |
|---|---|
| PCR fragments after column clean-up (100 ng/kb each) | 10 µl |
| 2 × Q5 mix | 12.5 µl |
| Water | 2.5 |
| Total | 25 µl |
| PCR Programme for OE1: | |
| 94°C: 30 s; [94°C: 15 s; 72°C: (−0.5°C/cycle) 30 s/kb] for 9 cycles; [94°C: 15 s; 67°C: (−0.5°C/cycle) 15 s, 72°C: 30 s/kb] for 5 cycles; 72°C: 2 min; Hold at 10°C | |
| OE2 | |
| OE1 product | 2 µl |
| 2 × Q5 mix | 12.5 µl |
| 2 µM FP | 0.35 µl |
| 2 µM RP | 0.35 µl |
| Water | 9.8 µl |
| Total | 25 µl |
| PCR Programme for OE2: | |
| 94°C: 30 s; [94°C: 15 s ; 67°C: (−0.5°C/cycle) 15 s; 72°C 30 s/kb] for 17 cycles; [94°C: 15 s; 59°C: 15 s; 72°C: 30 s/kb] for 23 cycles; 72°C: 2 min; Hold at 10°C | |
Construct assembly Part 1—Gibson assembly of Clones 1, 2, 3 and 4
pCDNA3.1 was digested with BamHI and HindIII, treated with Antarctic Phosphatase and purified using a DNA cleanup column, and checked for background by transformation before using in the assembly reaction. Each of the clones was assembled with equimolar amounts of cut vector and inserts, except in the case of fragments shorter than 400 bp, for which a 3- to 5-fold molar excess was used. The fragments that were incorporated into each of the clones are listed below and the details of the fragments can be found in Table 2. A typical Gibson assembly reaction was assembled as described below, incubated at 50°C for 1 h, and transformed into 200 µl competent cells. At least 40–50 colonies were obtained for each of the 4 reactions.
| Gibson assembly | |
|---|---|
| Clone number | Fragments used |
| Clone 1 | 1 + 2OE, 3 + 4OE, 17 and vector |
| Clone 2 | 18, 6 + 7OE, 8, 19 and vector |
| Clone 3 | 20, 10, 11 + 12OE, 21 and vector |
| Clone 4 | 22, 14 + 15’OE, 16, vector |
| Reaction: | |
| Cut vector: | 0.4 µl (50 ng) |
| Insert: | Equimolar amount of inserts (0.5–1.5 µl of each unpurified PCR product as estimated from gel images) |
| Gibson mix: | 15 µl |
| Water: | Remaining |
| Total | 20 µl |
Colony PCR screening
For each clone, five colonies were screened by colony PCR using two sets of primers: AS145 and AS645/Omi F4Rev (Front), and AS575 and AS507 (End), to confirm insertion of the entire Spike sequence (Fig. 3C, colony PCR results for clones 1 and 4 as examples). Each colony was resuspended in 20 µl of water, lysed by heating and 1.5 µl of the supernatant was used as input for a 10 µl PCR reaction. Plasmid was prepared from at least two positive clones, size-verified on a gel (Fig. 3D) and sent for sequencing with primers expected to cover the mutated region.
Challenge: Since Fragment 8 could not be generated efficiently, 10 colonies were screened for clone 2, and 4 plasmids were sequenced. However, no mutations in Fragment 8 were incorporated. In addition, for clone 3, all of the sequencing reactions failed. Therefore, we altered the strategy to include repeats of these two segments in the subsequent steps.
Construct assembly Part 2—Gibson assembly to generate clone 1.2 (combining clones 1 and 2) and Clone 3.4 (clones 3 and 4)
For Parts 2 and 3 of the construct assembly, protocols identical to those described for part 1 above, were followed. The fragments assembled for each are listed below.
| Gibson assembly | |
|---|---|
| Clone number | Fragments used |
| Clone 1.2 | 1, 23, 6 + 7OE, 25, 19 and vector |
| Clone 3.4 | 10, 11 + 12OE, 26 and vector |
We obtained positive clones for both in this round, as confirmed by sequencing. A six-fragment assembly (five inserts and vector) proved to be successful as seen in the generation of clone 1.2.
Construct assembly Part 3—Gibson assembly to generate full-length clone
| Gibson assembly | |
|---|---|
| Clone number | Fragments used |
| Full-length clone | 29, 27, 28 and vector |
| NTD alone | 30,17 and vector |
| RBD alone | 31, 32, 19 and vector |
| NTD+RBD | 29, 19 and vector |
| CTD alone | 20, 33 and vector |
We obtained at least one positive clone for each of the above constructs, as confirmed by sequencing.
Challenges: In some instances, we observed errors in the primer binding regions which we attribute to errors in the oligonucleotides during synthesis, especially for long oligonucleotides (such as those for Fragment 8). We overcame this by using a shorter oligonucleotide for a second round of PCR to eliminate specific errors.
The quote obtained for the commercial synthesis of this construct from a reputed vendor was INR 1 Lakh (USD 1330) and the time from order to delivery of the sequence verified construct was 45 days at our location, possibly due to import restrictions and pandemic-related delays. The in-house synthesis of the final sequence verified construct was completed in under 29 days at an estimated cost of INR 50 000 (USD 665) (Tables 3 and 4). In addition to the complete Omicron Spike gene, four additional variants were synthesized using no additional reagents. The time required for in-house synthesis was primarily limited by the time required for commercial oligo synthesis/delivery and turnaround time for sequencing, which required 50% of the total time to synthesize and sequence-validate the full-length construct. The cost saving was in part due to the use of lab-made enzymes and cloning reagents.
Table 3.
Timeline for the synthesis of the Omicron Spike construct
| Day | Activity | |
|---|---|---|
| Bench work | 1 | Strategy finalized |
| 2 | Primers ordered | |
| 3 | ||
| 4 | ||
| 5 | Primers received | |
| 6 | Fragment PCRs round 1 | |
| 7 | OE and Gibson assembly round 1 | |
| 8 | Col PCR 1 | |
| 9 | Plasmid prep, sent for sequencing | |
| 10 | ||
| 11 | ||
| 12 | ||
| 13 | Sequencing results; Fragment PCRs round 2 | |
| 14 | Round 2 cloning | |
| 15 | ||
| 16 | Col PCR 2 | |
| 17 | Plasmid prep, sent for sequencing | |
| 18 | ||
| 19 | ||
| 20 | Sequencing results | |
| 21 | Fragment PCRs round 3 | |
| 22 | Round 3 cloning | |
| 23 | Col PCR 3 | |
| 24 | ||
| 25 | ||
| 26 | Plasmid prep, sent for sequencing | |
| 27 | ||
| 28 | ||
| 29 | Sequencing results for final clone |
Table 4.
Estimated cost of consumables in the study
| Item | Approximate cost |
|---|---|
| Oligonucleotides | INR 33 000 (INR 18–22/base based on scale of synthesis) |
| Sequencing | INR 6000 (INR 500/reaction) |
| PCR consumables and kits | INR 6000 |
| Cloning consumables | INR 5000 (INR 300/reaction Gibson mix, media and miscellaneous) |
| PCR with Taq polymerase | |
|---|---|
| Input DNA | 10 ng (0.5 µl) |
| FP (10 µM) | 0.75 µl |
| RP (10 µM) | 0.75 µl |
| 10X Taq Buffer | 2.5 µl |
| 2.5 mM dNTPs | 2.0 µl |
| Taq DNA Pol | 0.5 µl |
| Water | 18 µl |
| Total | 25 µl |
| PCR Programme for Taq | |
| 95°C: 4 min; [95°C: 20 s; 48°C: 20 s; 72°C: 20 s] 5 cycles; [95°C: 20 s, 58°C: 20 s, 72°C: 20 s] 25 cycles; 72°C: 10 min; Hold at 10°C | |
| PCR with Q5 | |
|---|---|
| Input DNA | 10ng (0.5 µl) |
| FP (10 µM) | 0.75 µl |
| RP (10 µM) | 0.75 µl |
| 2 × Q5 mix | 12.5 µl |
| Water | 10.5 |
| Total | 25 µl |
| PCR Programme for Q5: | |
| 98°C: 5 min; [98°C: 10 s, 72°C: 1.30 min] 28 cycles; 72°C: 10 min, Hold at 10°C | |
Discussion
In this study, we report a strategy for simultaneous site-directed mutagenesis at multiple sites spread over several kilobases, with the added option of being able to generate a combinatorial library of mutant constructs. The method uses simple laboratory reagents and protocols and the strategy and methods are widely applicable. In addition to being an alternative for gene synthesis for complex constructs bearing multiple mutations, this technique may be used to synthesize a large collection of single as well as modular combinations of mutations. Constructs generated in this manner could be useful in a wide array of applications including but not limited to (alanine) scanning mutagenesis for identification of sites of post-translation modifications [7], fitness, function, activity or structure contributions [8–10], as well as for functional analysis of a spectrum of patient mutations in genes implicated in monogenic disorders [11].
With the specific example of the Omicron Spike gene, we were able to generate the construct containing 37 mutations spread across a ∼3.7 kb DNA fragment using simple methods and reagents, accessible to most basic molecular biology labs in the world. Despite delays in sequencing turnaround and strategic/experimental failures, we were able to complete this synthesis in half the time that it took to receive a commercially synthesized construct at our location. As suggested by the numbers in Fig. 3E, the efficiency at each step was quite high and this complex construct with the desired sequence was assembled by screening only 5–10 colonies at each intermediate stage. Along the way, we were also able to generate constructs containing mutations only in smaller domains in the full-length gene.
A previous report described the LFEAP mutagenesis method which also relied on PCR and ligation-mediated assembly to incorporate multiple mutations [12]. However, this method involved the amplification of the entire plasmid, bringing with it the increased possibility of errors, and T4 DNA ligase-mediated assembly which is usually less efficient than overlap-mediated assembly. Traditional scanning mutagenesis methods involve elegant but technically more complex strategies using specially modified templates [13]. IVA cloning is also a comparable simple method for cloning that relies on recA-independent assembly in Escherichia coli but reports lower efficiencies for multi-fragment assemblies [14]. Both IVA cloning and POE cloning [15] involve amplification of the vector in addition to the insert fragments, which may necessitate whole-plasmid sequencing for verification. A T5 exonuclease-dependent cloning method could reduce costs further; however, efficiency with multi-fragment assemblies remains to be tested in this protocol [16].
Gibson assembly was the single most important experimental method that made the current strategy possible. This method allows for seamless assembly without the requirement for or introduction of any extra or restricted nucleotide sequences. Efficient incorporation of up to five inserts in one assembly, significantly reduces the amount of time required to assemble complex constructs. Combined with the very efficient oligo stitching method reported by Hilgarth et al., technically up to ten fragments may be assembled in one shot.
In this study, we have demonstrated single or multiple amino acid changes at consecutive or spaced locations, insertions and deletions (single or up to three residues). We expect that large deletions (>10 bp) will also be technically feasible. However, introduction of repetitive sequences, or large insertions may be slightly more challenging and may require modification of the described strategy. Despite these caveats, we believe that this strategy will be of great use in resource-constrained settings.
Author contributions
Rita Rani (Conceptualization [equal], Data curation [lead], Formal analysis [lead], Investigation [lead], Methodology [lead], Resources [supporting], Supervision [equal], Validation [supporting], Writing—original draft [supporting], Writing—review and editing [equal]), Kishore V.L. Parsa (Conceptualization [supporting], Funding acquisition [supporting], Methodology [supporting], Project administration [supporting], Resources [equal], Supervision [supporting], Writing—original draft [supporting]), Kiranam Chatti (Conceptualization [supporting], Investigation [supporting], Validation [supporting], Writing—original draft [supporting], Writing—review and editing [supporting]), and Aarti Sevilimedu (Conceptualization [equal], Data curation [supporting], Formal analysis [supporting], Funding acquisition [lead], Investigation [supporting], Methodology [supporting], Project administration [lead], Resources [equal], Supervision [equal], Validation [supporting], Visualization [lead], Writing—original draft [lead], Writing—review and editing [equal]). Rita Rani and Aarti Sevilimedu wrote the manuscript with inputs from Kishore V.L. Parsa and Kiranam Chatti.
Contributor Information
Rita Rani, Center for Innovation in Molecular and Pharmaceutical Sciences, Dr. Reddy’s Institute of Life Sciences, University of Hyderabad Campus, Hyderabad, 500046, India.
Kishore V L Parsa, Center for Innovation in Molecular and Pharmaceutical Sciences, Dr. Reddy’s Institute of Life Sciences, University of Hyderabad Campus, Hyderabad, 500046, India.
Kiranam Chatti, Center for Innovation in Molecular and Pharmaceutical Sciences, Dr. Reddy’s Institute of Life Sciences, University of Hyderabad Campus, Hyderabad, 500046, India.
Aarti Sevilimedu, Center for Innovation in Molecular and Pharmaceutical Sciences, Dr. Reddy’s Institute of Life Sciences, University of Hyderabad Campus, Hyderabad, 500046, India.
Funding
This work was supported by Dr. Reddy’s Institute of Life Sciences, Hyderabad.
Conflict of interest statement. The authors declare that there is no conflict of interest.
Supplementary data
Supplementary data is available at Biology Methods and Protocols online.
References
- 1. Hughes RA, Ellington AD.. Synthetic DNA synthesis and assembly: putting the synthetic in synthetic biology. Cold Spring Harb Perspect Biol 2017;9:a023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gibson DG, Young L, Chuang RY. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009;6:343–5. [DOI] [PubMed] [Google Scholar]
- 3. Kumar JV, Banu S, Sasikala M. et al. Effectiveness of REGEN-COV antibody cocktail against the B.1.617.2 (delta) variant of SARS-CoV-2: a cohort study. J Intern Med 2021;291:380–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GISAID - hCov19 variants. https://www.gisaid.org/hcov19-variants/ (17 January 2022, date last accessed).
- 5. Yu J, Li Z, He X, Gebre MS. et al. Deletion of the SARS-CoV-2 spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. 2021;95(11):e00044–21. 10.1128/JVI.00044-21. [DOI] [PMC free article] [PubMed]
- 6. Hilgarth RS, Lanigan TM.. Optimization of overlap extension PCR for efficient transgene construction. MethodsX 2020;7:100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakanishi S, Sanderson BW, Delventhal KM. et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 2008;15:881–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roscoe BP, Thayer KM, Zeldovich KB. et al. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol 2013;425:1363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weiss GA, Watanabe CK, Zhong A. et al. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci USA 2000;97:8950–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hietpas RT, Jensen JD, Bolon DNA.. Experimental illumination of a fitness landscape. Proc Natl Acad Sci USA 2011;108:7896–901. doi: 10.1073/PNAS.1016024108/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmiesing J, Lohmöller B, Schweizer M. et al. Disease-causing mutations affecting surface residues of mitochondrial glutaryl-CoA dehydrogenase impair stability, heteromeric complex formation and mitochondria architecture. Hum Mol Genet 2017;26:538–51. [DOI] [PubMed] [Google Scholar]
- 12. Zeng F, Zhang S, Hao Z. et al. Efficient strategy for introducing large and multiple changes in plasmid DNA. Sci Rep 2018;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kunkel TA, Roberts JD, Zakour RA.. [19] Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 1987;154:367–82. [DOI] [PubMed] [Google Scholar]
- 14. García-Nafría J, Watson JF, Greger IH.. IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci Rep 2016;6:27459–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hejlesen R, Füchtbauer EM.. Multiple site-directed mutagenesis via simple cloning by prolonged overlap extension. Biotechniques 2020;68:345–8. [DOI] [PubMed] [Google Scholar]
- 16. Xia Y, Li K, Li J. et al. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res 2019;47:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]