Abstract
Nontypeable Haemophilus influenzae (NTHI) has four loci, lic-1 to lic-3 and lgtC, that generate phase-variable lipooligosaccharide (LOS) structures. lic-1, which is required for the expression of phosphorylcholine (ChoP), is the best characterized and is associated with an enhanced ability of H. influenzae to persist within the nasopharynges of infant rats. Recent data indicate that LOS impacts various aspects of NTHI virulence in the chinchilla model of nasopharyngeal colonization and otitis media (OM). In this study the effects of ChoP expression and the sequences of lic-1 to lic-3 and lgtC of NTHI strain 2019 were evaluated in the chinchilla OM model. Nasopharyngeal colonization data showed that a switch from the ChoP− to the ChoP+ phenotype was observed as early as day 3 after intranasal inoculation. Chinchillas colonized by strains with the ChoP+ phenotype demonstrated a significantly higher level of NTHI 2019 per milliliter of nasal lavage fluid than chinchillas colonized with predominantly the ChoP− variant (P < 0.05). The concentration of cells with the ChoP+ phenotype in the middle ear was 3 log units higher than that of cells with the ChoP− variant (P < 0.01). There was a statistically significant association between ChoP+ expression in the nasal lavage and the development of OM with culture-positive middle ear fluids in this model. These data suggest that expression of the ChoP+ phenotype promotes enhanced nasopharyngeal colonization and development of OM.
Otitis media (OM) or inflammation of the middle ear (ME), in one of its various clinical forms, is one of the most common childhood diseases. Nontypeable Haemophilus influenzae (NTHI) is a major OM pathogen and accounts for 25 to 30% of all cases of this disease. The initial event in the pathogenesis of NTHI OM is the colonization of the host mucosal surface; however, the bacterial factors that contribute to the colonization of the nasopharynx, retrograde ascension of the eustachian tube, and invasion of the ME during the natural progression of the disease are not well characterized. NTHI cells are frequently isolated from the upper respiratory tract, especially the nasopharynges of healthy children, with a reported rate of colonization of approximately 80% (8). Recent data suggested that there is a strong relationship between the frequency of colonization and the incidence of OM in children (5).
NTHI lipooligosaccharide (LOS) is a major virulence determinant and may play a role in colonization and invasion of mucosal surfaces in the respiratory tract (3, 16, 23). NTHI LOS is analogous to the lipopolysaccharide (LPS) of enteric gram-negative bacteria in that it contains lipid A linked by 3-deoxy-d-manno-octulosonic acid to a heterogeneous sugar polymer (7). NTHI LOS, however, differs from classic enterobacterial LPS in that it does not contain repeating O-antigen units and is therefore more similar to that derived from Neisseria and Bordetella species (14). Moreover, H. influenzae cells demonstrate a propensity to alter or modulate their surface-expressed antigens including LOS (10, 12). NTHI cells express, on their outer surfaces, a number of LOS core oligosaccharide epitopes, and the expression of these epitopes is subject to frequent, reversible phase variation. Four chromosomal loci, lic-1 to lic-3 and lgtC, which contain long stretches of 4-bp tandem repeats within their 5′ coding regions, have been reported to generate phase-variable LOS structures (9, 11, 19). lic-1 functions to add phosphorylcholine (ChoP) to the LOS molecule (22), lic-2 and lgtC are necessary for the expression of Galα1-4Gal (20), and the effect of variation in lic-3 is unknown. Phase variation may represent a mechanism whereby NTHI evades the host immune response or concurrently modulates its surface in order to colonize different anatomical sites, each with a unique complement of host cell receptors (15, 21). Elucidating the role of LOS phase variation is important both for understanding OM pathogenesis and for designing a candidate vaccine (6).
ChoP, which decorates the LOS on the NTHI cell surface, has recently been implicated in the pathogenesis of NTHI diseases (22). Choline is present on host cell membrane lipids. NTHI and Streptococcus pneumoniae, major respiratory tract pathogens, uptake choline from the host and incorporate it as ChoP. LOS undergoes phase variation in expression of the ChoP epitope, and the frequency of on or off switching is about 10−2 to 10−3 per generation (22). A recent study by Weiser and coworkers suggests that decoration of the LOS with ChoP is associated with an enhanced ability of H. influenzae type b strains to persist within the nasopharynx in the infant rat model. In addition, sequence analyses of lic-1 of NTHI isolated from human respiratory tract secretions showed predominately lic-1 in frame (ChoP+) (21).
We have previously established the chinchilla model of OM with NTHI and have evaluated the ability of NTHI strain 2019 to colonize the chinchilla nasopharynx for up to 22 days after intranasal (IN) inoculation (2, 3). In this study, we evaluated the effects of the phase-variable expression of ChoP by, as well as the sequences of lic-2, lic-3, and lgtC of, NTHI 2019 on nasopharyngeal (NP) colonization and development of OM in the chinchilla model.
MATERIALS AND METHODS
Animals.
A total of 35 chinchillas (Chinchilla lanigera) weighing 250 to 400 g and free of ME disease, as determined by otoscopy and tympanometry, were used for this study.
Bacteria and preparation of inoculum.
NTHI strain 2019, originally obtained from Michael Apicella, Department of Microbiology, University of Iowa, has been previously described (3, 13). In order to prepare the inoculum, NTHI strain 2019 was grown on chocolate agar for 18 h, washed in phosphate-buffered saline, suspended in sterile pyrogen-free saline, and adjusted to a density of approximately 5 × 107 CFU/ml. The inoculum concentration was determined by standard colony plate count. Colonies grown on chocolate agar were evaluated for the phenotype and genotype of ChoP expression by colony immunoblotting and DNA sequencing. NTHI 2019 is a 98% lic-1 off variant upon initial subculture from the frozen stock culture and for up to two passages in vitro on chocolate agar. In addition, NTHI 2019 is a lic-2 on, lic-3 on, lgtC on variant, as determined by the genotypic analysis described below.
IN challenge and assessment of ChoP expression during NP colonization and OM development.
The biological consequence of expression of ChoP by NTHI for the induction of OM and persistence of NP colonization in the chinchilla model were assessed subsequent to IN challenge. Chinchillas were challenged IN with 0.3 ml of an NTHI 2019 suspension containing approximately 1.5 × 107 CFU. The inoculum was delivered by passive inhalation and was divided equally between the nares as previously described (4). Five chinchillas, preselected and randomized, were evaluated by tympanocentesis or ME lavage and NP lavage on days 1 to 4, 6, 7, and 10 after inoculation with NTHI 2019 as previously described (18). Tympanocentesis was performed on all chinchillas with ME effusions (MEEs) by aspiration with a tuberculin syringe fitted with a 25-gauge needle. If no MEE was present, the MEs were lavaged with 0.5 ml of sterile saline. Subsequent to tympanocentesis, NP lavage was performed on each chinchilla. Chinchillas were not subjected to repeat tympanocentesis or nasal lavage. Tympanocentesis and ME lavage were always performed before NP lavage to prevent contamination of the ME. For determination of the proportion of ChoP phenotypes of NTHI recovered from the samples, the sample fluids were serially diluted and plated on chocolate agar in triplicate. One plate was used for plate counts, and the other two were used for evaluation of ChoP expression by means of colony immunoblotting and DNA sequencing for genotype determination. All analyses were performed on the initial subculture without serial passage.
ME pressure (MEP) of each chinchilla was measured by means of tympanometry. Normal chinchilla MEP was considered to be between −60 and +40 daPa (17).
Colony immunoblotting.
Colonies were lifted from the chocolate agar onto nitrocellulose membranes and immunoblotted to detect the ChoP+ phenotype as described previously (21). Briefly, 20 to 200 colonies from each lavage sample were probed with a 1:10,000 dilution of monoclonal antibody (MAb) HAS, which is specific for ChoP (Statens Serum Institut, Copenhagen, Denmark). Alkaline phosphatase-conjugated mouse immunoglobulin M (diluted 1:800) (Roche Molecular Biochemicals, Indianapolis, Ind.) was used as the secondary antibody. Colonies were also probed with MAb 6E4, which is a non-phase-varying 3-deoxy-d-manno-octulosonic acid-like epitope reactive with NTHI 2019, as a positive control (1). MAb 3F11, which does not react with NTHI 2019, served as a negative control (1). Michael Apicella kindly provided both MAbs.
Genotypic analysis.
Chromosomal DNA of NTHI colonies of known ChoP phenotypes was extracted from one of the three plates prepared from each sample. A single colony was harvested into 50 μl of sterile water, boiled for 10 min, and mixed with 50 μl of chloroform, and the mixture was centrifuged for 5 min at 13,000 × g. The supernatant containing genomic DNA was stored at −70°C. Isolated DNA was used as a template to amplify the 5′ regions of lic-1, lic-2, lic-3, and lgtC by PCR with primers previously described (20, 21). The PCR was performed in a 50-μl reaction mixture consisting of (final concentrations) 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCL, a 100 μM concentration of each deoxynucleoside triphosphate (dNTP), a 0.2 μM concentration of each primer, and 1 U of Taq polymerase. PCR amplification was performed using a PTC-100 thermocycler (MJ Research Inc., Watertown, Mass.) as follows. A 30-s cycle at 94°C for denaturation was followed by 30 cycles consisting of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C. The final cycle consisted of 30 s at 94°C, 30 s at 55°C, and 5 min at 72°C. The product was checked by electrophoresis on a 1% agarose gel stained with ethidium bromide. The PCR products were purified by using a Microcon-100 (Amicon, Inc., Beverly, Mass.) to remove primers and dNTPs. The numbers of CAAT repeats in lic-1, lic-2, and lic-3 and the number of GACA repeats in lgtC for the PCR fragments were determined by DNA sequencing with sequencing primers (20), a fluorescently labeled dye terminator cycle sequencing reaction kit (ABI PRISM) with AmpliTaq DNA polymerase FS (Applied Biosystems Inc., Foster City, Calif.), and an automated DNA sequencer (ABI 373 DNA sequencer; Applied Biosystems Inc.). The DNA sequencing was performed by the Neurobiotechnology Center, The Ohio State University, Columbus, Ohio.
Statistical analysis.
The Mann-Whitney rank sum test was used in the analysis of the differences in NTHI concentration in nasal lavage and in ME lavage between chinchillas with ChoP+ and ChoP− phenotypes and the differences in MEP between chinchillas with ChoP+ and ChoP− nasal isolates. Chi-square analysis was used to examine the association of ChoP expression in nasal lavage and the development of OM. Significance was accepted for all analyses when P values were <0.05.
RESULTS
The effect of ChoP expression on NP colonization.
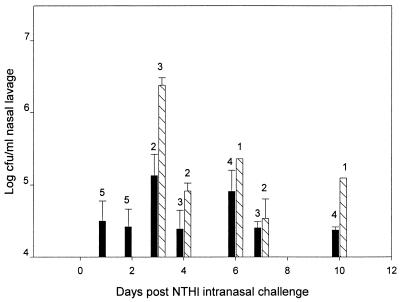
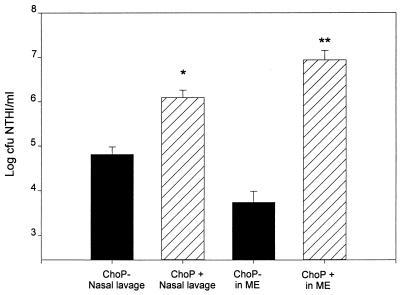
Expression of the ChoP phenotype during NP colonization for up to 10 days after IN challenge is shown in Fig. 1. The inoculum was 98% ChoP−. On days 1 and 2 after IN inoculation, although there was some evidence of expression of the ChoP+ phenotype, 90 to 98% of colonies isolated from the nasal lavage from the five chinchillas examined at each of these sample times still were ChoP−. By day 3, however, ChoP+ was the predominant phenotype (>90%) found in the NP lavage samples from three out of the five chinchillas examined. From day 3 through day 10, NP isolates from 9 out of the total of 25 chinchillas switched from ChoP− to ChoP+ (Fig. 1). The switching of >90% of the isolates to the ChoP+ phenotype was observed in seven chinchillas on days 3, 4, 6, and 7, and the switching of 50% of the isolates to ChoP+ in two chinchillas on days 7 and 10 after IN challenge was recorded. It is noteworthy that chinchillas colonized with the predominantly ChoP+ NTHI 2019 had 20 times more NTHI cells per milliliter of nasal lavage than the chinchillas colonized predominantly (90 to 99%) with ChoP− variants (P < 0.05) (Fig. 2). Switching to expression of the ChoP+ phenotype appears to result in a higher level of colonization of the nasopharynx. All the NTHI 2019 colonies isolated from the inocula and the nasal lavage fluids showed strong positive reactivity with MAb 6E4 and were negative when probed with MAb 3F11, the positive and negative controls, respectively.
FIG. 1.
Comparison of the concentrations of NTHI ChoP variants in the nasal lavage fluid after IN challenge. Each data point represents the geometric mean number of CFU of either ChoP− (▪) or ChoP+ (▧) NTHI ± the standard error of the mean per milliliter of nasal lavage fluid from one to five chinchillas. The number of each chinchilla expressing the phenotype is indicated above each bar.
FIG. 2.
Comparison of the concentrations of NTHI ChoP variants in nasal lavage and ME fluid or lavage. Data shown represent the geometric mean numbers of CFU of either ChoP− (▪) or ChoP+ (▧) NTHI ± the standard errors of the means per milliliter of nasal lavage fluid (26 versus 9 chinchillas, respectively) or MEEs or ME lavage fluid (7 chinchillas for both). ∗ and ∗∗, P < 0.05 and P < 0.01, respectively, compared to the ChoP− group.
The effect of ChoP expression in the nasopharynx on the development of OM.
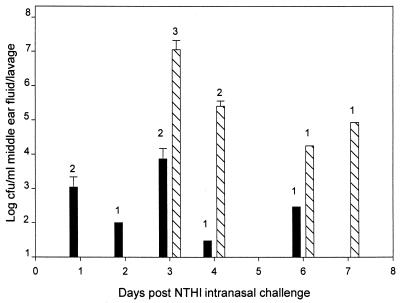
The ability of NTHI 2019 to invade the ME subsequent to IN inoculation was evaluated in relation to ChoP phenotype expression. On days 1 and 2 after IN challenge, a total of 3 out of 10 chinchillas developed culture-positive ME lavage fluid (Fig. 3) and 95% of the NTHI 2019 cells recovered from the ME lavage had the ChoP− phenotype. NTHI colonized the ME but did not induce any signs or symptoms of disease. From day 3 to day 7, however, at a time when a switch to the ChoP+ phenotype was occurring in the nasopharynx, seven chinchillas developed severe OM with culture-positive MEEs. Greater than 95% of the NTHI cells isolated from the MEEs were ChoP+. Three of the four remaining chinchillas examined during this time period did not develop OM with culture-positive ME lavage but did demonstrate colonization of the ME by NTHI. ME lavage fluids from these animals exhibited less than 104 CFU of NTHI/ml of lavage fluid, with greater than 90% incidence of the ChoP− phenotype (Fig. 3). The concentration of ChoP+ NTHI in the ME was 3 log units higher than that in MEs colonized primarily with the ChoP− variant (P < 0.01; Fig. 2). All the NTHI cells recovered from MEEs or ME lavage showed the same reactivity with MAb 6E4 and MAb 3F11 that was described previously for the nasal lavage isolates.
FIG. 3.
Comparison of NTHI concentrations of ChoP variants in ME fluid after IN challenge. Each data point represents the geometric mean number of CFU of either ChoP− (▪) or ChoP+ (▧) NTHI ± the standard error of the mean per milliliter of MEEs or ME lavage from one to five chinchillas. The number of each chinchilla expressing the phenotype is indicated above each bar.
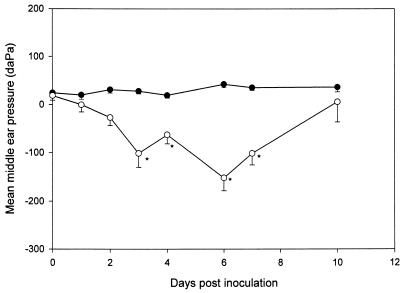
Chinchillas with >90% ChoP− NTHI cells colonizing the nasopharynx had normal MEPs between −60 and +40 daPa during the entire duration of the experiment. Chinchillas with NTHI colonies that switched to 50 to 90% ChoP+, however, demonstrated statistically significantly more-negative MEPs (−101, −63, −152, and −102 daPa) on days 3, 4, 6, and 7, respectively, than the chinchillas colonized with ChoP− NP isolates (P < 0.01) (Fig. 4).
FIG. 4.
Comparison of mean MEPs (± standard errors of the means) between ChoP− (●) and ChoP+ (○) variants after IN challenge as determined by tympanometry over a 10-day observation period. Values below −60 daPa are considered abnormal for the chinchilla. ∗, P < 0.05 compared to values for the ChoP− group.
Sequence analysis of lic-1, lic-2, lic-3, and lgtC genes.
NTHI isolates in nasal lavage samples from 35 chinchillas, MEEs from 8 chinchillas, and ME lavage samples from 6 chinchillas were analyzed for their sequences of lic-1 to lic-3 and lgtC. DNA sequencing of the 5′ region of the lic-1 gene indicated that the ChoP− variant had 26 CAAT repeats. Organisms which switched from the ChoP− phenotype in the inoculum to ChoP+ in the lavage samples either gained one (n = 27) or lost one (n = 25) CAAT repeat. All of the ChoP+ isolates from the MEEs contained 27 CAAT repeats. Isolates with either the ChoP+ or ChoP− phenotype were all lic-2 on, lic-3 on, lgtC on variants. The numbers of CAAT repeats were 17 for lic-2 and 15 for lic-3, and the number of GACA repeats was 12 for lgtC. There was no on or off switching for lic-2, lic-3, and lgtC observed during the study period (Table 1).
TABLE 1.
Genotypic analysis of lic-1 to lic-3 and lgtC of NTHI from MEE and lavage samples during NP colonization and development of OM after IN challenge with NTHI 2019
| Daya | Sample source (no. of chinchillas) | No. of CAAT repeats in lic-1 | lic-1 ChoP phenotypeb (>90%) | No. of CAAT repeats in:
|
No. of GACA repeats in lgtC | |
|---|---|---|---|---|---|---|
| lic-2 | lic-3 | |||||
| 0 | Inocula | 26 | − | 17 | 15 | 12 |
| 1 | Nasal lavage (5) | 26 | − | 17 | 15 | 12 |
| ME lavage (2) | 26 | − | 17 | 15 | 12 | |
| 2 | Nasal lavage (5) | 26 | − | 17 | 15 | 12 |
| ME lavage (1) | 26 | − | 17 | 15 | 12 | |
| 3 | Nasal lavage and MEE (3) | 27 | + | 17 | 15 | 12 |
| Nasal lavage and MEE (1) | 26 | − | 17 | 15 | 12 | |
| Nasal lavage (2) | 26 | − | 17 | 15 | 12 | |
| ME lavage (1) | 26 | − | 17 | 15 | 12 | |
| 4 | Nasal lavage and MEE (2) | 27 | + | 17 | 15 | 12 |
| Nasal lavage (3) | 26 | − | 17 | 15 | 12 | |
| ME lavage (1) | 26 | − | 17 | 15 | 12 | |
| 6 | Nasal lavage and MEE (1) | 27 | + | 17 | 15 | 12 |
| Nasal lavage (4) | 26 | − | 17 | 15 | 12 | |
| ME lavage (1) | 26 | − | 17 | 15 | 12 | |
| 7 | Nasal lavage (1) and MEE (1) | 27 | + | 17 | 15 | 12 |
| Nasal lavage (3) | 26 | − | 17 | 15 | 12 | |
| Nasal lavage (1) | 26/25c | −/+ | ||||
| 10 | Nasal lavage (4) | 26 | − | 17 | 15 | 12 |
| Nasal lavage (1) | 26/25c | −/+ | 17 | 15 | 12 | |
Day after IN inoculation.
+, ChoP+; −, ChoP−; −/+, only 50% of colonies shifted to the ChoP+ phenotype.
26/25, n = 26 for the ChoP− colonies and n = 25 for the ChoP+ colonies.
The association of ChoP expression in nasal lavage and the development of OM.
Nine of 35 (26%) chinchillas showed a switch from the ChoP− to the ChoP+ phenotype in the NP lavage during day 3 to day 10. Seven of these nine (78%) chinchillas, in which >90% of the NTHI cells switched to ChoP+, developed severe OM with culture-positive MEEs between day 3 and day 7 postinoculation (Table 2). Only two chinchillas, one in which 50% of the NTHI cells had switched to ChoP+ by day 7 and the other by day 10, failed to undergo colonization of the ME and to develop OM.
TABLE 2.
The association of expression of NTHI 2019 ChoP in the nasal lavage with ME infection
| ME status | No. of chinchillas (%) with indicated phenotype in nasal lavagea
|
|
|---|---|---|
| ChoP+ | ChoP− | |
| OM with culture-positive MEE | 7∗ (78) | 1 (3.8) |
| Culture-positive ME lavage without OM | 0 | 6 (23) |
| Culture-negative ME lavage without OM | 2 | 19 |
| Total | 9 (26) | 26 (74) |
∗, P < 0.001 (chi-square test).
On the other hand, only 1 of 26 chinchillas (3.8%) with NP colonization with >90% ChoP− NTHI cells developed OM with culture-positive MEEs on day 3. Six demonstrated colonization of the ME but without evidence or signs of OM. No MEEs were present in these chinchillas, and the organisms were recovered by lavage of the ME. There was a statistically significant association between ChoP+ expression in nasal lavage and the development of OM. Chinchillas with ChoP+ expression in nasal lavage were more inclined to the development of OM with culture-positive MEEs in this model (Table 2).
DISCUSSION
The mechanism of ChoP expression and the impact on the virulence of NTHI during disease are not clear. Genes within lic-1 chromosomal loci are directly involved in the expression of the phase-variable ChoP epitope. The reported frequency of on-off switching is 10−2 to 10−3 per generation but varies from strain to strain depending on the length of the repetitive sequence (22). Previous data indicate that during NP carriage in infant rats, there is gradual selection of H. influenzae expressing ChoP, so that by day 10 47% of the colonies from nasal lavage revert to the ChoP+ phenotype. These data indicate that ChoP expression may play a role in the persistence of the organisms on mucosal surfaces (20). The ChoP+ phenotype also renders the organism susceptible to killing by serum-derived C-reactive protein (CRP) and complement (20, 21) and places NTHI at risk in the event of inflammation and serum transudation at the site of colonization on the mucosal surface. Data from the present study were similar to previous reports from Weiser et al. regarding the importance of phase-variable expression of ChoP of H. influenzae during NP colonization in infant rats. Our results indicate that, subsequent to IN inoculation of the chinchilla nasopharynx with ChoP− NTHI, there is a selection of ChoP+ variants. This selection appears to occur much more rapidly in the chinchillas than in infant rats challenged IN with an H. influenzae type b strain and is associated with an increased incidence of OM with culture-positive MEEs displaying the ChoP+ phenotype as well as significantly negative MEPs. Moreover, the increased level of NTHI per milliliter of lavage fluid suggested that cells with the ChoP+ phenotype colonized at a higher level in the nasopharynx than ChoP− cells. It is noteworthy that, while the ChoP− variants are also capable of colonizing the ME space, they induce overt disease less frequently and severely than the ChoP+ variants. In a preliminary study, we have examined eight pairs of matched NTHI NP and ME isolates from children with chronic OM; all of them showed greater than 98% ChoP+ phenotype and a lic-1 on genotype (unpublished data). These data suggest that the microenvironment in the nasopharynx and ME cavity may facilitate the selection of the ChoP+ phenotype, which is optimally adapted to colonizing or inducing disease in these particular anatomical niches.
It is important to keep in mind that other genes within lic-2, lic-3, and lgtC also contribute to the phase-variable expression of epitopes of LOS and play an important role in the host-bacterium interactions (20). Galα1-4Gal, one of the LOS epitopes which is added by lic-2 together with lgtC, has been associated with resistance to antibody-mediated serum bactericidal activity (20). A genotypic analysis of the effect of LOS phase variation on human respiratory tract secretions by Weiser et al. indicated that 90% of NTHI cells have lic-1 in frame (ChoP+) and that there is a significant association between the expression of lgtC and that of lic-2 during NTHI pneumonias (20, 22). The terminal galactose conferring serum resistance may be especially important for the NTHI at sites of inflammation, such as the ME fluid, where antibody and complement, as well as CRP derived from serum transudation, may be present. The lic-2, lic-3, and lgtC genes were all in frame (on) in the NTHI 2019 inoculum as well as all the NP and ME isolates examined during these experiments. Only lic-1 varied from off to on. Further studies with lic-2 off and lgtC off variants are needed to explore the role of lic-2 and lgtC in NP colonization and development of OM in the chinchilla model.
In conclusion, our data indicated that phase-variable expression of ChoP and a switch to the ChoP+ phenotype contributed to increased NP colonization and the development of OM in the chinchilla model. The mechanisms responsible for the increased virulence of the ChoP+ NTHI phenotype warrant further investigation.
ACKNOWLEDGMENTS
This study was supported, in part, by grant 5-R01-DC00090-28 from the NIDCD/NIH.
We thank Lisa Routt and Kathy Holloway for manuscript preparation.
REFERENCES
- 1.Campagnari A A, Spinola S M, Lesse A J, Kwaik Y A, Mandrell R E, Apicella M A. Lipooligosaccharide epitopes shared among gram-negative non-enteric mucosal pathogens. Microb Pathog. 1990;8:353–362. doi: 10.1016/0882-4010(90)90094-7. [DOI] [PubMed] [Google Scholar]
- 2.DeMaria T F. Animal models for nontypeable Haemophilus influenzae otitis media. Pediatr Infect Dis. 1989;8:S40–S42. doi: 10.1097/00006454-198901001-00015. [DOI] [PubMed] [Google Scholar]
- 3.DeMaria T F, Apicella M A, Nichols W A, Leake E R. Evaluation of the virulence of nontypeable Haemophilus influenzae lipooligosaccharide htrB and rfaD mutants in the chinchilla model of otitis media. Infect Immun. 1997;65:4431–4435. doi: 10.1128/iai.65.11.4431-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeMaria T F, Murwin D M, Leake E R. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect Immun. 1996;64:5187–5192. doi: 10.1128/iai.64.12.5187-5192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y. Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics J Infect Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 6.Foxwell A R, Kyd J M, Cripps A W. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson B W, Melaugh W, Phillips N J, Apicella M A, Campagnari A A, Griffiss J M. Investigation of the structural heterogeneity of lipooligosaccharides from pathogenic Haemophilus and Neisseria species and of R-type lipopolysaccharides from Salmonella typhimurium by electrospray mass spectrometry. J Bacteriol. 1993;175:2702–2712. doi: 10.1128/jb.175.9.2702-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harabuchi Y, Faden H, Yamanaka N, Duffy L, Wolf J, Krystofik D. Nasopharyngeal colonization with nontypeable Haemophilus influenzae and recurrent otitis media. Tonawanda/Williamsville Pediatrics J Infect Dis. 1994;170:862–866. doi: 10.1093/infdis/170.4.862. [DOI] [PubMed] [Google Scholar]
- 9.High N J, Deadman M E, Moxon E R. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(1-4)beta Gal. Mol Microbiol. 1993;9:1275–1282. doi: 10.1111/j.1365-2958.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 10.Hood D W, Deadman M E, Allen T, Masoud H, Martin A, Brisson J R, Fleischmann R, Venter J C, Richards J C, Moxon E R. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]
- 11.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarosik G P, Hansen E J. Identification of a new locus involved in expression of Haemophilus influenzae type b lipooligosaccharide. Infect Immun. 1994;62:4861–4867. doi: 10.1128/iai.62.11.4861-4867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee N G, Sunshine M G, Apicella M A. Molecular cloning and characterization of the nontypeable Haemophilus influenzae 2019 rfaE gene required for lipopolysaccharide biosynthesis. Infect Immun. 1995;63:818–824. doi: 10.1128/iai.63.3.818-824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandrell R E, McLaughlin R, Aba K Y, Lesse A, Yamasaki R, Gibson B, Spinola S M, Apicella M A. Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect Immun. 1992;60:1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran A P, Prendergast M M, Appelmelk B J. Molecular mimicry of host structures by bacterial lipopolysaccharides and its contribution to disease. FEMS Immunol Med Microbiol. 1996;16:105–115. doi: 10.1111/j.1574-695X.1996.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 16.Rao V K, Krasan G P, Hendrixson D R, Dawid S, St. Geme J W., III Molecular determinants of the pathogenesis of disease due to nontypeable Haemophilus influenzae. FEMS Microbiol Rev. 1999;23:99–129. doi: 10.1111/j.1574-6976.1999.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenfeld R M, Doyle W J, Swarts J D, Seroky J, Pinero B P. Third-generation cephalosporins in the treatment of acute pneumococcal otitis media. An animal study. Arch Otolaryngol Head Neck Surg. 1992;118:49–52. doi: 10.1001/archotol.1992.01880010053015. [DOI] [PubMed] [Google Scholar]
- 18.Tong H H, Blue L E, James M A, DeMaria T F. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:921–924. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiser J N, Maskell D J, Butler P D, Lindberg A A, Moxon E R. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. J Bacteriol. 1990;172:3304–3309. doi: 10.1128/jb.172.6.3304-3309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiser J N, Pan N. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol Microbiol. 1998;30:767–775. doi: 10.1046/j.1365-2958.1998.01108.x. [DOI] [PubMed] [Google Scholar]
- 21.Weiser J N, Pan N, McGowan K L, Musher D, Martin A, Richards J. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J Exp Med. 1998;187:631–640. doi: 10.1084/jem.187.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiser J N, Shchepetov M, Chong S T. Decoration of lipopolysaccaride with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect Immun. 1997;65:943–950. doi: 10.1128/iai.65.3.943-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwahlen A, Rubin L G, Moxon E R. Contribution of lipopolysaccharide to pathogenicity of Haemophilus influenzae: comparative virulence of genetically-related strains in rats. Microb Pathog. 1986;1:465–473. doi: 10.1016/0882-4010(86)90008-2. [DOI] [PubMed] [Google Scholar]