Abstract
Human diseases have been a critical threat from the beginning of human history. Knowing the origin, course of action and treatment of any disease state is essential. A microscopic approach to the molecular field is a more coherent and accurate way to explore the mechanism, progression, and therapy with the introduction and evolution of technology than a macroscopic approach. Non-coding RNAs (ncRNAs) play increasingly important roles in detecting, developing, and treating all abnormalities related to physiology, pathology, genetics, epigenetics, cancer, and developmental diseases. Noncoding RNAs are becoming increasingly crucial as powerful, multipurpose regulators of all biological processes. Parallel to this, a rising amount of scientific information has revealed links between abnormal noncoding RNA expression and human disorders. Numerous non-coding transcripts with unknown functions have been found in addition to advancements in RNA-sequencing methods. Non-coding linear RNAs come in a variety of forms, including circular RNAs with a continuous closed loop (circRNA), long non-coding RNAs (lncRNA), and microRNAs (miRNA). This comprises specific information on their biogenesis, mode of action, physiological function, and significance concerning disease (such as cancer or cardiovascular diseases and others). This study review focuses on non-coding RNA as specific biomarkers and novel therapeutic targets.
Keywords: Non-coding RNA, Biomarker, And Drug discovery
Introduction
History of RNA biology
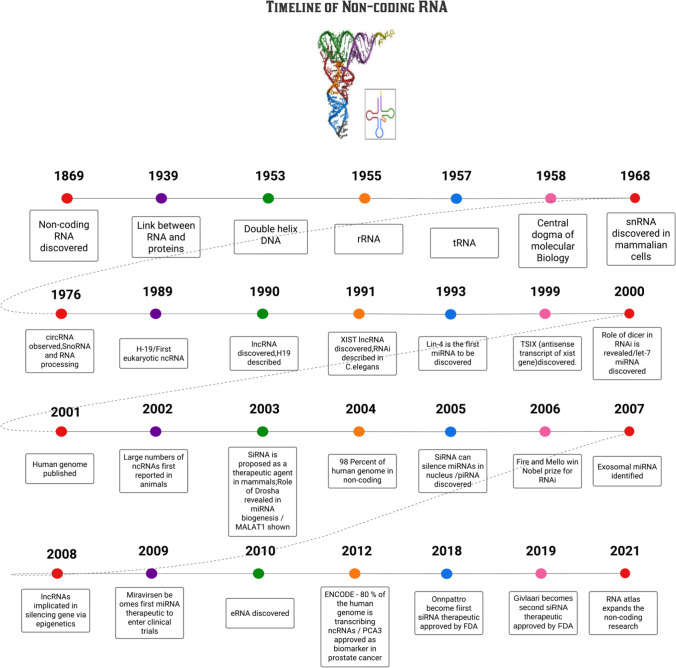
In 1958, Francis Crick established the central dogma of molecular biology by discovering the sequence of events in the passage of genetic material contained in DNA to the functioning of biological processes through proteins. However, with the development of new technologies and robust next-generation sequencing, large international consortiums such as the Functional Annotation of the Mammalian Genome (FANTOM) and the Encyclopaedia of DNA Elements (ENCODE) have described pervasive transcription (that 80% of the DNA is transcribed into RNA but only a 1.5% of that RNA translates into protein) (Carninci et al. 2005; Hangauer et al. 2013). Recent technological advances, like next-generation deep sequencing, have shown that the bulk of the genome is translated into RNAs. The universe of RNA is divided into two halves: (1) RNAs with coding potential and (2) RNAs without coding potential, sometimes known as non-coding RNAs, because of only 1 and 2% of the human genome codes for proteins (The ENCODE Project Consortium 2012). Although mRNAs have been studied in depth, most RNAs are ncRNAs. Even though ncRNAs were formerly regarded as “evolutionary junk,” new research shows that they substantially impact several molecular pathways. According to the hypothesis known as the “RNA universe,” RNA was the earliest form of life, and as DNA became more solid, RNA’s function as a messenger was left unfilled. However, it was eventually discovered that RNA is the most practical possibility in disease, epigenetics, and unknown regulatory features since it has a wide range of latent catalytic capabilities and can store genetic information (Bhatti et al. 2021). During evolution, RNA is thought to have evolved alongside proteins and DNA (Robertson and Joyce 2010). Understanding their intricate relevance in numerous biological processes, including homeostasis and development, is critical (Amaral et al. 2013). Figure 1 demonstrates the molecular events relate to non-coding RNA (Li et al. 2021a, b; Chhabra 2021).
Fig. 1.
Timeline of molecular discoveries of non-coding RNA
A relatively broad size criterion is used to classify ncRNAs into two subclasses. Small or short non-coding RNAs (ncRNAs) are ncRNAs that are less than 200 nucleotides (nt), while long non-coding RNAs are ncRNAs that are more than 200 nt (lncRNAs). These two groups are quite different from one another. LncRNAs can be as significant as several kilobases, and small ncRNAs can be as small as a few to 200 nt. The most well-known class of tiny ncRNAs, microRNAs (miRNAs), have a length of 20 nucleotides or less and have undergone substantial research (Kim et al. 2009). The other non-coding such as siRNA and piRNA. The complexity of these animals’ physiology, characteristics, and development, from lower non-chordates to humans, produces an increase in introns and intergenic sequences that are translationally modified by alternative splicing processes, leading to a further decrease in the size of this proteome (Mattick 2001).In addition, eukaryotes have more sophisticated and complex systems for RNA processing, trans induction, DNA methylation, imprinting, RNA interference (RNAi), post-transcriptional gene silencing, chromatin modification, gene editing, splicing, dosage compensation, gene regulation mechanisms, and transcriptional gene silencing (Mattick 2004). Non-coding RNA act as regulatory signal messengers for the stimuli received at sensory genetic elements (Guttman et al. 2011). The evolutionary history of prokaryotes supports their continued reliance on protein-based regulatory architecture, in contrast to eukaryotes, who have evolved new regulatory features and mechanisms to control the expression of phenotypic traits, the penetrance and expressivity of disease, and developmental programming using a variety of ncRNAs. Therefore, research on ncRNA about these linked pathways is essential to comprehend their function in health and disease (GAGEN 2005).
Distribution and types of ncRNA
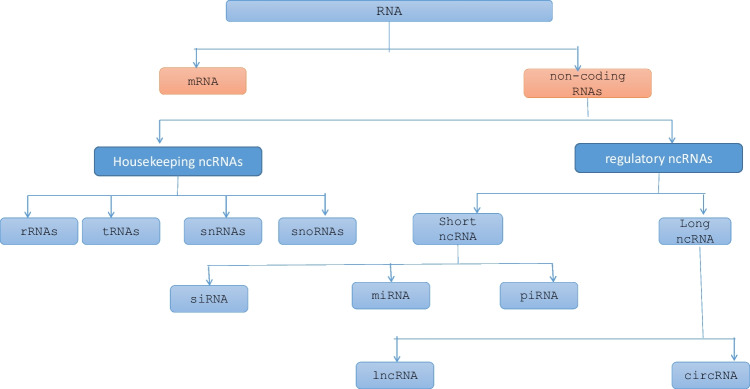
RNA comes in a variety of forms in live cells. ncRNAs are typically split into two domains based on their transcript length: short ncRNAs (under 200 nucleotides) and long ncRNAs (over 200 nucleotides). ncRNA is important in several processes, including RNA maturation, RNA processing, signaling, gene expression, and protein synthesis (Kung et al. 2013; Morris and Mattick 2014). The amount of ncRNA and the degree of species conservation are remarkably correlated. According to estimates, each cell has 107 ncRNA molecules, most of which are snRNA, snoRNA, miRNA, rRNA, and lncRNA. Although about 53,000 distinct human lncRNAs identified, only about 1000 are present in adequate quantities to legitimately support their functional significance (Djebali et al. 2012). Other types of RNA and their specificities are mentioned in this study (Bhatti et al. 2021). The overview of non-coding RNA and its functions is mentioned in Table 1. The different types of RNA are mentioned in Fig. 2.
Table 1.
Overview of non-coding RNA and its functions
| Types of RNA | Full form | Functions | References |
|---|---|---|---|
| Housekeeping ncRNA | |||
| rRNA | ribosomal RNA | Translational machinery | Fu 2014 |
| tRNA | Transfer RNA | Amino acid carriers | |
| snRNA | Small nuclear RNA | RNA processing | |
| snoRNA | Small nucleolar RNA | RNA modifications | |
| TRNA | Telomere RNA | Chromosome end synthesis | |
| Regulatory ncRNA | |||
| miRNA | MicroRNAs | RNA stability and translation control | Fu 2014 |
| lncRNA | Long non-coding RNA | Imprinting, epigenetics, nuclear structure | |
| circRNA | Circular RNA | Inhibiting miRNA activity | |
| endo-siRNA | Endogenous siRNA | RNA degradation | |
| rasiRNA | Repeat associated-derived RNA | Transcriptional control | |
| eRNA | Enhancer-derived RNA | Regulation of gene expression | |
| piRNA | PIWI-interacting RNA | Silencing transposon and mRNA decay | |
| PATs | Promoter-associated RNA | Transcription initiation and pause release | |
Fig. 2.
Different types of RNA and major non-coding RNAs
Biogenesis and functions of different types of ncRNA
RNA molecules are much more than just a blueprint for protein production. Since non-coding transcripts are expected to function similarly to proteins and can regulate the majority of cellular functions, RNA may interact with DNA, proteins, and other RNA molecules to form three-dimensional (3D) structures. The two main regulatory RNA groups—small and long ncRNAs—are partly defined by their length. Additionally, functional ncRNAs with lengths between 20 and thousands of nucleotides have grown significantly in number and classification over the past ten years. This review focuses on significant ncRNAs such as miRNA, lncRNA, and circRNA. Few other RNA will be mentioned such as piRNA, snRNA, snoRNA, and siRNA. This ncRNA will play a significant role in developmental processes and disease conditions. Numerous genes are involved in the production of ncRNAs across the whole human genome, and there may potentially be distinct transcriptional units that function independently. Transcription, nuclear maturation, export to the cytoplasm for processing, and production of functional RNA are all steps in this biogenesis process. The detailed mechanism of non-coding RNA biogenesis is mentioned in this paper (Bhatti et al. 2021). The description of specific ncRNA and the description of biogenesis are mentioned in Table 2. Non-coding RNA is an integral part of genomics and proteomics. According to the “RNA world” hypothesis, RNA may have played a role in the emergence of life, which must be able to carry and duplicate its genetic material (Joyce 1989). In contemporary organisms that have evolved to use more effective methods to copy and express their genetic material along the central axis from DNA to RNA to protein, ncRNAs seem to have retained the majority, if not all, of their original characteristics and functions. Many RNA functions are transferred to proteins while others are kept because of the exploration of selective benefits of proteins and RNA during evolution. To grasp ncRNA function and mechanism, it may be instructive to compare ncRNA function with that of proteins.
Table 2.
Description of ncRNA and its biogenesis
| Type of RNA | Full_form | Biogenesis description | Functions | References |
|---|---|---|---|---|
| miRNA | microRNA | Biogenesis of miRNAs begins with DNA sequences known as miRNA genes or clusters of genes that are only transcripted as miRNA molecules or collectively as polycistronic transcripts. MiRNAs can also be found in an intron or untranslated region (UTR) of a protein-coding gene | miRNA plays a vital role in post-transcriptional gene regulation. By inhibiting translation and destabilizing mRNA, miRNAs regulate their targets in eukaryotic cells | Annese et al. 2020 |
| lncRNA | Long non-coding RNA | lncRNAs are RNA-type molecules with a 5′ methyl-cytosine cap and a 3′ poly(A) tail transcribed by RNA polymerase II (Pol II). 31 LncRNAs are categorized into many different categories based on their various features. lncRNAs, for example, can be classified into five categories based on their genetic origins: sense, antisense, bidirectional, intronic, and intergenic. LncRNAs are categorized into three categories based on their function: rRNA, tRNA, and cRNA | A new class of epigenetic regulators called lncRNAs is crucial to regulating epigenetic processes. LncRNAs modulate histone or DNA modification, primarily methylation, and acetylation, to control epigenetic modification primarily in the nucleus, which controls gene transcription at the transcriptional level | Liu et al. 2021a, b |
| snRNA | Small nuclear RNA | snRNA can be transcribed from a promotor (similar to mRNA) and encoded within intronic sequences | Splicing of introns from primary genomic transcripts is a critical function of small nuclear RNAs | Matera et al. 2007 |
| snoRNA | Small nucleolar RNA | Except for a tiny subset of snoRNAs that RNA polymerase II transcriptions autonomously, most snoRNAs in vertebrates are encoded in the introns of protein-coding or non-coding genes. Most intronic snoRNAs are produced through co-transcription with the host gene, splicing, debranching of the intron lariat, and nucleoplasmic exonucleolytic digestion. The maturation of snoRNAs, which is co-transcriptionally induced, depends on the recruitment of ribonucleoproteins to the nascent intronic snoRNAs. Additional SnoRNPs are sent to Cajal bodies, carrying out additional maturation and processing operations. Shq1, Naf1, and NUFLP are additional auxiliary elements that contribute to snoRNP assembly and maturation. Both processing stability and nucleolar localization depend on these proteins | The function of snoRNA is to participate in rRNA processing, regulation of mRNA processing, involvement in stress response, and metabolic changes. snoRNA has a more and influential role in cancer | Liang et al. 2019 |
| siRNA | Small interfering RNA | The cascade leading to the synthesis of mature siRNA begins with transcription by RNA polymerase II (in mammals), RNA polymerase III (from an shRNA template), or RNA polymerase IV (in plants), creating double-stranded RNA (dsRNA) (dsRNA) | siRNA is frequently employed in molecular biology to silence desired genes temporarily. Upon binding to their target transcript, they trigger RNAi based on the complementarity of their sequences | Carthew and Sontheimer 2009 |
| piRNA | PIWI-interacting RNA | Lengthy RNA precursors are transcribed in the nucleus and exported into the cytoplasm. In the cytoplasm, piRNA precursors are further processed to form mature piRNAs that get loaded into Piwi proteins | piRNA has essential roles in embryonic development, the preservation of germline DNA integrity, the generation of heterochromatin, the silencing of transposon transcription, the suppression of translation, and the epigenetic regulation of sex determination | Wu et al. 2020a, b |
| circRNA | Circular RNA | CircRNAs often result from exon or intron circularization and splicing activities. Exonic circRNAs can be produced by a procedure known as back splicing, dependent on spliceosomal splicing. Exons are spliced in the opposite direction by combining an upstream and a downstream 3′ and 5′ splice site, resulting in a circular product. Exon skipping, which creates a lariat structure containing exons and introns, is another method that leads to exonic circRNAs. The intron is cut out of this precursor during self-splicing, and the lariat is circularized | CircRNAs control target gene expression by inhibiting miRNA activity as a miRNA sponge. Through several miRNA binding sites, one circRNA can control one or more miRNAs | Beermann et al. 2016 |
Comparison of miRNA, lncRNA, and circRNA in RNA biology
The mechanistic characterization of lncRNAs is far less thorough than that of miRNAs. This is partly because lncRNAs can control gene expression through intricate biochemical pathways at various levels inside the cell. Despite being present in a group of species (Guttman and Rinn 2012), such as plants (Swiezewski et al. 2009), yeast (Houseley et al. 2008), prokaryotes (Bernstein et al. 1993), and viruses (Reeves et al. 2007), lncRNAs are not as well conserved as miRNAs in terms of the nucleotide sequence. Even though lncRNAs with diverse nucleotide compositions can exhibit the same 3D structure and, consequently, the exact molecular function, this restricts the selection of cellular and animal models for researching lncRNA functions (Derrien et al. 2012). It is increasingly becoming clear that lncRNAs play a role in virtually every cellular process and that the expression of these non-coding molecules is carefully regulated in both normal conditions and several human diseases, including cancer (Tano and Akimitsu 2012).
Unlike coding genes, lncRNAs can be produced in many ways from practically any location in the human genome. Contrary to those that overlap coding genes on the antisense strand, unlike coding genes, lncRNAs can be produced in a wide range of ways from practically any location in the human genome. Contrary to those that overlap coding genes on the antisense strand, sense lncRNAs are made from segments that overlap one or more exons of another coding transcript (antisense lncRNAs); sense lncRNAs are made from segments that overlap one or more exons of another coding transcript. Other lncRNAs are produced by regulatory components like enhancers or non-coding DNA sequences like introns. Some have promoters and regulatory elements expressed from intergenic regions that do not overlap other known coding genes (Thum and Condorelli 2015). It becomes clear that just a tiny portion of the theoretically infinite number of lncRNAs that could exist have been studied thus far. However, those studied have demonstrated the capacity to control the transcriptional and post-transcriptional stages of gene expression by interacting with nucleic acids and proteins in a manner that is specific to both sequences and structures (Mercer et al. 2009; Wilusz et al. 2009). The categorization and annotation of putative lncRNAs must be carefully examined to remove protein-coding RNAs. While being categorized as non-coding molecules, some lncRNAs have recently been shown to be able to code for micro peptides (Anderson et al. 2015). Before concluding a lncRNA’s regulatory role, it is essential to prove that the skeletal muscle-specific RNA, which was previously thought to be a lncRNA, is encoded for a functional micro peptide. Evidence from recent studies revealed that conventional processes do not just regulate ncRNA expression. Circular RNAs are produced due to a back-splicing expression variation (circRNA). Since CircRNAs are made up of a covalently closed continuous loop, they lack a 5′ cap and a 3′ tail. This RNA species is more tissue-specific, moderately stable, and highly conserved (Jeck et al. 2012). The functions of each of these ncRNA were mentioned in this paper (Beermann et al. 2016). The discovery of associations between non-coding RNAs and diseases has created new therapeutic and diagnostic possibilities. Numerous miRNAs have already been effectively demonstrated to act as diagnostic or therapeutic targets for various diseases. There is specific evidence that circRNAs and lncRNAs behave similarly.
Non-coding RNA and human diseases
Functional RNA molecules known as non-coding RNA (ncRNA) cannot be translated into proteins (Djebali et al. 2012). Initially, there are only a few ncRNAs were found and studied. Later technological advancements, ncRNA types were classified into many, and each ncRNA has specific functions that lead to biomarkers and novel therapeutic approaches. Despite not all of their functions being understood, several ncRNA species play crucial roles in controlling the transcription and translation of genes and the transcription of ncRNAs. Therefore, it is no surprise that ncRNAs are crucial in normal physiologic functions, complex human traits, and human diseases (Li et al. 2018a, b). This review will mention the different types of diseases and their ncRNA as potential biomarkers and interactions in Table 3.
Table 3.
Non-coding RNA and its biomarkers
| Disease | ncRNA as biomarker | References |
|---|---|---|
| Genetic disease | ||
| Duchenne muscular dystrophy | miR-1, miR-21, miR-29, miR-30c, miR-31, miR-133, miR-181a, miR-206, miR-208a, miR-208b, miR-499;lnc-31, linc-MD1; | Salvatore et al. 2011; Hu et al. 2014; Ballarino et al. 2015; Cacchiarelli et al. 2011; Chen et al. 2006; Eisenberg et al. 2007; Giordani et al. 2014; Greco et al. 2009; Mizuno et al. 2011; Naguibneva et al. 2006; Perry and Muntoni 2016; Twayana et al. 2013; van Rooij et al. 2008; Wang et al. 2012; Yuasa et al. 2008; Zaharieva et al. 2013 |
| Myotonic dystrophy (type 1) | miR-1, miR-133a/b, miR-206;MALAT1 | Gambardella et al. 2010; Fritegotto et al. 2017; Wheeler et al. 2012 |
| Familial dysautonomia | miR-203a-3p | Hervé and Ibrahim 2016 |
| Amyotrophic lateral sclerosis | miR143-3p, miR-206, miR-208b, miR-374b-5p, miR-499;NEAT1_2 | Salvatore et al. 2011; Williams et al. 2009; Gagliardi et al. 2018 |
| Ullrich congenital muscular dystrophy | miR-30c, miR-181a | Paco et al. 2015 |
| Cystic fibrosis | miR-9, miR-93, miR-145-5p, miR-181b, miR-454, miR-509-3p;XIST, TLR8, HOTAIR, MALAT1, TLR8-AS1, BLACAT1, MEG9, BGas | Gillen et al. 2011; Hassan et al. 2012; Ramachandran et al. 2012; Balloy et al. 2017; Fabbri et al. 2014; Fabbri et al. 2017; McKiernan et al. 2014; Oglesby et al. 2013; Pierdomenico et al. 2017; Saayman et al. 2016; Sonneville et al. 2017 |
| Rett syndrome | miR-29b, miR-92, miR-122a, miR-130, miR-146a, miR-146b, miR-199a, miR-199b, miR-221, miR-296, miR-329, miR-342, miR-382, miR-409;AK081227, AK087060 | Salvatore et al. 2011; Petazzi et al. 2013; Urdinguio et al. 2010 |
| Pulmonary arterial hypertension | miR-9, miR-124, miR-130, miR-206;MEG3, LnRPT | Kim et al. 2015; Sun et al. 2017; Chen et al. 2018a, b |
| Facioscapulohumeral muscular dystrophy | miR-411; DBE-T | Harafuji et al. 2013 |
| Sézary syndrome | miR-18a, miR-21, miR-31, miR-199a2, miR-214, miR-233, miR-342, miR-486 | Salvatore et al. 2011; Qin et al. 2012; Ballabio et al. 2010; Narducci et al. 2011 |
| Lesch–Nyhan disease | miR-9, miR-181a, miR-187, miR-424 | Guibinga 2015 |
| Multiple osteochondromas | miR-21, miR-140, miR-145, miR-195, miR-214, miR-451, miR-483 | Salvatore et al. 2011; Zuntini et al. 2010 |
| Hailey–Hailey disease | miR-99a, miR-106, miR-125b, miR-181a | Manca et al. 2011 |
| Li-Fraumeni syndrome | miR-605 | Id Said & Malkin 2015 |
| Hepatoblastoma | miR-125a, miR-148a, miR-150, miR-214, miR-199a, miR-492 | Magrelli et al. 2009 |
| MELAS (mitochondrial encephalopathy syndrome) | miR-9;LINC01405, SNHG12, RP11-403P17.4, CTC-260E6.6, RP11-357D18.1 | Meseguer et al. 2015; Wang et al. 2017a, b |
| X-Chromosomal schizophrenia | let-7f-2, miR-188, miR-325, miR-509–3, and miR-510, miR-660 | Feng et al. 2009 |
| β-Thalassemia | miR-15a, miR-16–1, miR-26b, miR-96, miR-144, miR-155, miR-181a/c, miR-210, miR-320, miR-451, miR-486-3p, miR-503;DQ583499, XIST, lincRNA-TPM1, MRFS16P, lincRNA-RUNX2-2, HMI-LNCRNA, NR_001589, NR_120526, T315543 | Gasparello et al. 2017; Leecharoenkiat et al. 2017; Lulli et al. 2013; Ma et al. 2017; Morrison et al. 2018; Roy et al. 2012; Saki et al. 2016; Siwaponanan et al. 2016; Srinoun et al. 2017 |
| Cardiovascular disease | ||
| Coronary artery disease | miR-1, miR-133a/b,miR-208a/b, aHIF, ANRIL, APOA1-AS,AWPPH, BANCR,CHROME, CoroMarker, H19, HOTTIP, LIPCAR, lincRNA-p21, LINC00968, MALAT1, MIAT, NEXN-AS1, SMILR | Broadbent et al. 2007; D’Alessandra et al. 2013; Fichtlscherer et al. 2010; Hennessy et al. 2018; Hu et al. 2019; Toraih et al. 2019; Wang et al. 2016a, b, c; Xiong et al. 2019; Yang et al. 2015 |
| Cardiomyopathy | miR-1,miR-423-5p | D’Alessandra et al. 2013; Fan et al. 2015 |
| Heart failure | miR-1, miR-133a/b, miR-208a/b,miR-499, ANRIL,BACE1-AS, Chaer, Chast, CHRF, EGOT, H19, HEAT2,HRCR, HOTAIR, LIPCAR, lincRNA-ROR, LOC285194, MEG3,MHRT, MIAT, NRON,RMRP, RNY5, SOX2-OT, SRA1 | Gidlöf et al. 2013; Greco et al. 2017; Greco et al. 2016; Viereck et al. 2016; Wang et al. 2015a, b; Wang et al. 2016a, b, c |
| Aterial hypertension | AK098656, ANRIL,GAS5, Giver,Lnc-Ang362, NR_027032, NR_034083, NR_104181 | Jin et al. 2018; Bayoglu et al. 2016; Wang et al. 2016a, b, c; Leung et al. 2013a, b |
| Atrial fibrillation | miR-1, miRNA-26, miRNA-499, miRNA-328, miRNA-21, miRNA-133, miRNA-590, miRNA-206, PANCR, TCONS_00075467, KCNQ1OT1, NPPA-AS1,lncRNA-HBL1,PVT1, GAS5,LICPAR, MIAT, NRON, TCONS_00032546, TCONS_00026102 | Lu et al. 2015; Luo et al. 2013; Ling et al. 2013; Lu et al. 2010; Shan et al. 2009; Zhang et al. 2015; Holmes and Kirchhof 2016; Li, Wang, et al. 2017a, b; Shen et al. 2018; Ke et al. 2019a, b; Liu et al. 2017a, b; Zhao et al. 2020a, b; Wang et al. 2020; Yao et al. 2020; Sun et al. 2019; Wang, et al. 2015a, b |
| Atherosclerosis | LIPCAR, aHIF, ANRIL, KCNQ1OT1, MIAT, MALAT1, CoroMarker, LncPPARδ | Kumarswamy et al. 2014; Vausort et al. 2014; Cai et al. 2016; Bayes-Genis et al. 2017 |
| Acute myocardial infarction | miR-1, miR-133a/b, miR-208a/b, miR-423-5p,miR-499, miR-400, miR-320a,miR signature, aHIF, ANRIL,APF, CARL, CDR1AS, FTX, GAS5, H19, HOTAIR, KCNQ1OT1, LIPCAR, Lnc-Ang362, MALAT1, MDRL, MEG3, MHRT, Mirt1/2, n379519, NONRATT021972, NRF, PCFL, TTTY15, UCA1, UIHTC, Wisper,ZFAS1 | Cheng et al. 2010; Widera et al. 2011; Bauters et al. 2013; Liu et al. 2015a, b; Zeller et al. 2014; Vausort et al. 2014; Jakob et al. 2016; Semenza 2014; Wang et al. 2015a, b; Wang et al. 2014a, b, c; Zhang et al. 2016a, b, c; Long et al. 2018; Du et al. 2019; Zhou et al. 2018; Gao et al. 2017; Chen et al. 2020a, b; Wang et al. 2019a, b, c; Wang et al. 2014a, b, c; Wu et al. 2018; Zhang et al. 2016a, b, c; Ishii et al. 2006; Zangrando et al. 2014 l Wang et al. 2018a, b; Chen et al. 2018a, b; Wang et al. 2016a, b, c; Huang et al. 2019a, b; Chen et al. 2019; Zhang et al. 2018a, b, c; Micheletti et al. 2017 |
| Tachycardia | miR-1,miR-133a/b | SUN et al. 2015a, b |
| Takotsubo cardiomyopathy | miR-1,miR-133a/b | Jaguszewski et al. 2013 |
| Viral myocarditis | miR-208a/b,miR-499 | Corsten et al. 2010 |
| Acute coronary syndromes | miR-1, miR-133 a/b,miR-208 a/b, miR-499, miR-150, miR-132, miR-186, MACE prediction after STEMI | Cheng et al. 2010; Widera et al. 2011; Bauters et al. 2013; Liu et al. 2015a, b |
| Neurological disorders | ||
| Alzheimer’s disease | let-7b, miR-106b, miR-128, miR-34a, mIr132/212, miR-142a-5p, miR-146a-5p, miR-155-5p, miR-455-5p, miR-15/107, miR-16, miR-200b/c, miR-25, miR-29a/b-1,miR-29c, miR-33, miR-34a, miR-485-5p,miR-873-5p, miR-338-5p, BC1, BC200, LncRNA -17A, MEG3, MIAT, NDM29, NEAT1, P3Alu/SINE | Zhang et al. 2018a, b, c;Li et al. 2018a, b; Feng et al. 2018; Mus et al. 2007; Wang et al. 2019a, b, c; Massone et al. 2011; Yi et al. 2019; Ke et al. 2019a, b; Polesskaya et al. 2018 |
| Parkinson’s disease | miR-126, miR-126-5p, miR-133a/b,miR-133b, miR-153, miR-16–1, miR-183,miR-205, miR-22, miR-221, miR-27a/b, miR-342-3p, miR-34b/c, miR-494, miR-494,miR-7, miR-7/miR-153,miR-96, circSNCA, HOTAIR, NEAT1, NORAD, p21, SNHG1, U1 spliceosomal lncRNA, RP11-462G22.1, tRNA-derived fragment, UCHL1-AS, NEAT1, PINK1-AS, CDR1-AS, circDLGAP4 | Wang et al. 2017a, b;Sang et al. 2018;Kim et al. 2016;Wu et al. 2019;Lin et al. 2019;Liu & Lu 2018;Qian et al. 2019;Magee et al. 2019;Carrieri et al. 2015;de Mena et al. 2010;Fragkouli and Doxakis 2014;Zhang and Cheng 2014;Gao et al. 2018a, b;Cho et al. 2012;Espinoza et al. 2020;Kabaria et al. 2015;Xiong et al. 2014;Dong et al. 2018a, b |
| ALS (amyotrophic lateral sclerosis) | ncRNACCND1, tiRNAs (tRNA-derived RNA fragments), Lhx1as, LncMN-1, LncMN2, miR-17 ~ 92, miR-155, miR-206,miR-218, miR-375-3p, miR-375, miR-92a-3p, miR-125b-5p, miR-124-3p, miR-92a-3p, miR-20b-5p miR-223b-3p,hsa_circ_0063411, hsa_circ_0023919, hsa_circ_0088036,hsa_mir-9,ABCA12, DYRK2, POTEM, MALAT1, NEAT1, C9ORF72-AS | Ruffo et al. 2021 |
| FTD (frontotemporal disorders) | C9ORF72 (repeat expansion), MALAT1, MEG3, NEAT1, U12 snRNA, Hsrw | Ruffo et al. 2021 |
| HD (Huntington’s disease) | miR-9*, miR-10b-5p, miR-22, miR-27a,miR-34a-5p, miR-34b, miR-214, miR-125b, miR-146a, miR-150, miR-125b, miR-146a, miR-150, miR-124, miR-124a, miR-128a, miR132, miR-212/miR-132, miR-196a, miR-196a, miR-19, miR-146a, miR-432, HAR1F, HAR1R, DGCR5, MEG3, NEAT1, NEAT1-L, NEAT1-S, TUG1, TUNA, LINC00341, RPS20P22, LINC00342, HTT-AS | Chang et al. 2017;Hoss et al. 2015;Ban et al. 2017;Reynolds et al. 2018;Gaughwin et al. 2011;Prajapati et al. 2019;Ghose et al. 2011;Das et al. 2013;Das et al. 2015;Kocerha et al. 2014;Fukuoka et al. 2018;Kunkanjanawan et al. 2016;Her et al. 2017;Cheng et al. 2013;Bañez-Coronel et al. 2012;Johnson et al. 2010;Johnson et al. 2008;Johnson 2012;Chen et al. 2020a, b;Cheng et al. 2018; |
| SMA (spinal muscular atrophy) | variant of U1 snRNA (vU1), miR‐183 | Wu & Kuo 2020 |
| SCA (spinocerebellar ataxia) | ATXN8-OS,SCAANT1 | Salta & De Strooper 2017 |
| Metabolic diseases | ||
| Type 1 diabetes | LINC01370, PLUT, MALAT1, TUG1 | Lodde et al. 2020 |
| Type 2 diabetes | miR-16, CDR1,circRNA-HIPK3, hsa_circ_0054633, circANKRD36(Enhanced expression), miR-376, miR-432,miR-200, miR-184, miR-204, miR-24, miR-26, miR-148, miR-182, miR-9, miR-130a, miR-130b, miR-152, miR-187,miR-7, miR-708, miR-34a, m,iR-146a, miR-182-5p, miR-33, miR-37, miR-802, miR-122-5p,miR-106b, microRNA let-7a, let-7d, miR-29, miR-192, miR-122, miR-27a-3p, miR-27b-3p, H19, MEG3, MALAT1 | Chi et al. 2021 |
| Osteoporosis | DANCR, miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-205, miR-217, miR-206, miR-29b, miR-433-3p, miR-103,miR-21,miR-223, miR-146a, miR-2861, miR-214,miR-21, miR-23a, miR-24, miR-25, miR-100, miR-125b, miR-22-3p, miR-328-3p, let-7 g-5p, miR-21, miR-133a, miR-130b-3p, miR-151a-3p, miR-151b, miR-194-5p, miR-590-5p, miR-660-5p, miR-194-5p, miR-125b, miR-30, miR-5914, miR-365, miR-10b, miR-0129-3p, miR-671-5p, miR-141, miR-25, miR-21-5p, miR-93-5p, miR-100-5p, miR-125b-5p, miR-320a, miR-483-5p, miR-152-3p, miR-30e-5p, miR-140-5p, miR-324-3p, miR-19b-3p, miR-335-5p, miR-19a-3p, miR-550a-3p, miR-17-5p, miR-133a-3p | Tong et al. 2015; Foessl et al. 2019 |
| Cancer | ||
| Breast cancer | miR21, CamK-A, EPIC1, HOTAIR, LINK-A, PLK1 | Slack and Chinnaiyan 2019 |
| Lung cancer | mIR-16, miR-21, miR-34a, MALATI1 | Slack and Chinnaiyan 2019 |
| Colorectal cancer | miR-1290, CCAT1, CCAT2, HOTAIR, circCCDC66, ciRS-7 | Slack and Chinnaiyan 2019 |
| Gastric cancer | miR-506, H19, circCTNNB1, PLK1 | Slack and Chinnaiyan 2019 |
| Ovarian cancer | miR-506, FAL1, HOTAIR, PLK1 | Slack and Chinnaiyan 2019 |
| Pancreatic cancer | miR-10b, miR-50b, HOTAIR, PKN3, APN401 | Slack and Chinnaiyan 2019 |
| Prostate Cancer | miR-21, miR-221, miR-375,miR-1290, MALAT1, NEAT1, PCA3, PCAT-1, PCAT-14, SChLAP1, circAR, | Slack and Chinnaiyan 2019 |
| leukemia | miR-21,miR-155 | Slack and Chinnaiyan 2019 |
| Infectious diseases | ||
| COVID-19 | miR-1275, miR-766-3p, miR-214, miR-17 and miR-574-5p, miR-98,miR-223 | Plowman and Lagos 2021 |
| Viral hepatitis | miRNA-122 | Bhatti et al. 2021 |
| Dengue virus | miRNA-378 | Bhatti et al. 2021 |
| Japanese encephalitis virus | miR-15b | Bhatti et al. 2021 |
| Enterovirus 71 (EV71) | miR-296-5p | Bhatti et al. 2021 |
| Human immunodeficiency virus | miRNA-34a and miRNA − 217 | Bhatti et al. 2021 |
| Tuberculosis | miR-155, miR-16, miR-200, Let-7 family, miR-486, miR-223, miR-99, miR-29, miR-21, miR-193, miR-365, miR-30, miR-20, miR-146, miR-31,miR-150 | Pattnaik et al. 2022 |
| Autoimmune diseases | ||
| Rheumatoid arthritis | lncRNA AC000061, HOTAIR, GAPLINC, ZFAS1, GAS5 | Lodde et al. 2020 |
| Systemic lupus erythematous | NEAT1, Linc00513, GAS5, TUG1 | Lodde et al. 2020 |
| Multiple sclerosis | NEAT1, TUG1, RN7SK, PVT1, FAS-AS1, THRIL, GAS5, MALAT1, MEG9, NRON, ANRIL, TUG1, XIST, SOX2OT, MIAT, HULC, BACE-1AS, lncAC007278.2, IFNG-AS1-001, IFNG-AS1-003 | Lodde et al. 2020 |
| Osteoarthritis | ARFRP1, LOXL1-AS1, HOTAIR, H19, NEAT1, DANCR, etc. (up). XIST, MEG3 etc. (down). miR-455-3p, miR-411, miR-27a (up), miR-149-5p, miR-26a-5p etc.(down), Circ_0136474, CircHIPK3, ciRS-7 etc. (up), CircRNA-9119, CircSERPINE2, circANKRD36 | Ghafouri-Fard et al. 2021 |
Transposons: unexpected players in different diseases with different ncRNA
Transposable elements (TEs) are considered essential factors in the plasticity and evolution of the genome. Since TEs are so prevalent in the human genome, particularly the Alu and Long Interspersed Nuclear Element-1 (LINE-1) repeats, they are thought to be the molecular cause of several diseases. This encompasses a number of the molecular processes discussed in this article, including insertional mutation, DNA recombination, chromosomal rearrangements, changes in gene expression, and changes to epigenetic controls. Additionally, some of the more well-known and/or more recent cases of human disorders where TEs play a role are provided in this article (Chénais 2022). TEs are frequently linked to the genesis of human malignancies, whether through the insertion of LINE-1 or Alu elements that result in chromosomal rearrangements or epigenetic alterations. Numerous more clinical disorders may have their molecular roots in gene structure and/or expression changes or chromosomal recombination caused by TE. Hemoglobinopathies, metabolic, neurological, and joint disorders are among the many conditions this group of diseases represents.
Additionally, TEs may influence aging. The epigenetic derepression and mobility of TEs, which can result in disease development, appear to be significantly impacted by the pressures and environmental toxins that people are exposed to. As a result, a greater understanding of TEs may result in the development of novel possible disease diagnostic markers (Pradhan and Ramakrishna 2022).
Differences between exosomal and non-exosomal non-coding RNAs in human health and diseases
Circulating ncRNA transfer via exosomes is an intriguing method. As mediators for intercellular communication, ncRNAs can be enclosed by EVs (such as exosomes, microvesicles, and apoptotic bodies) and secreted from cells to control various diseases depending on the target cells (Li et al. 2021a). It has been demonstrated that ncRNAs exist in various bodily fluids, including serum, plasma, urine, saliva, and others, in addition to cells. The ncRNAs seen in biofluids are frequently called circulating or extracellular ncRNAs. The fact that extracellular ncRNAs are reasonably durable in plasma even though extracellular RNase activity is considerable in that environment suggests that circulating ncRNAs may be shielded from adverse circumstances. In this part, they examine how ncRNAs in exosomes and non-exosomes regulate physiological homeostasis and pathological events in health and disease (Li et al. 2021b).
Tools and methods
Investigating miRNA, lncRNA, circRNA, and other RNAs
The complete methods and investigation of ncRNA will be discussed. miRNA methods have already been thoroughly explained. Deep sequencing techniques or microarrays are the most used methods for miRNA detection. Deep sequencing is a more sensitive technique when compared to microarray-based techniques. Microarrays can lead to finding distinct RNA sequences despite using a fixed set of probes for detection (van Rooij 2011). However, the output analysis is more difficult because of the enormous volume of data and the critical requirement for bioinformatics expertise. Quantitative real-time PCR allows for the comparatively inexpensive and low-effort validation of screening results (qRT-PCR). Because the transcript is so brief, previous difficulties prompted the construction of the primer for reverse transcription. Target-specific stem-loop reverse transcription primers are currently offered on many platforms. Northern blotting and in situ hybridization are other techniques for identifying identified miRNAs. To find a miRNA’s targets, bioinformatics platforms are commonly implemented. The miRNA-related database is mentioned in Table 4. Luciferase tests are frequently used to verify expected targets of miRNAs following bioinformatics-based predictions of such targets. To completely comprehend the entire transcriptional regulatory scenario, small RNAs play a critical role in transcriptional regulation. Their abnormal expression profiles are believed to be linked to cellular dysfunction and diseases. Numerous studies are concentrating on detecting, predicting, or quantifying short RNA expression, particularly miRNAs, to better understand human health and disease.
Table 4.
miRNA based tools and databases
| Tools/databases | Description | Website | References |
|---|---|---|---|
| RNAcentral: The non-coding RNA sequence database | RNAcentral is a comprehensive database of non-coding RNA of sequences and functional annotation. It has 296 species; visualize the 2D RNA structure. It has a lot of interconnected databases | https://rnacentral.org/ | Sweeney et al. 2018 |
| RNALOSS | Designing RNA sequences with low folding energy and distribution of locally optimum secondary structures that would suggest quick and robust folding could be done using the tool RNALOSS | http://clavius.bc.edu/~clotelab/RNALOSS | Clote 2005 |
| RNAdb | The creation of a thorough mammalian non-coding RNA database (RNAdb) with over 800 different experimentally examined non-coding RNAs (ncRNAs), many of which are linked to illnesses and/or developmental processes | http://research.imb.uq.edu.au/RNAdb | Pang 2004 |
| Rfam | Rfam is a library of covariance models and numerous sequence alignments for non-coding RNA families. The user can explore numerous sequence alignments and family annotations and search a database of covariance models using a query sequence. The INFERNAL package (http://infernal.wustl.edu/) enables local searches utilizing the database as well as the flat file download | http://www.sanger.ac.uk/science/tools/rfam | Griffiths-Jones 2003 |
| EICO | They have created a specialized integrated database for researching imprinted disease genes | http://fantom2.gsc.riken.jp/EICODB/ | Nikaido 2004 |
| NONCODE | NONCODE database includes all information relates to non-coding RNAs. It has a lot of integrated databases available | http://noncode.bioinfo.org.cn | Bu et al. 2011 |
| ChIPBase v2.0 | ChIPBase also has a ChIP-Function tool and a genome browser that can predict gene functions and analyze ChIP-seq data. This research will help better understand how ncRNAs and PCGs regulate transcription | http://rna.sysu.edu.cn/chipbase/ | Zhou et al. 2017 |
| FARNA | A database about inferred functions of non-coding RNA has broad areas of human cells and tissues | http://cbrc.kaust.edu.sa/farna | Alam et al. 2017 |
| NRDTD | ncRNAs represent a novel class of drug development targets since they may influence gene expression and disease course. We created the ncRNA Therapeutic Targets Database (NRDTD), which had 165 entries of ncRNAs that were supported by clinical or experimental research as potential drug targets | http://chengroup.cumt.edu.cn/NRDTD | Chen et al. 2017 |
| BLAST | A blast is an online tool used for sequence analysis. The prediction algorithm used here is BLAST E-value | https://blast.ncbi.nlm.nih.gov/Blast.cgi | McGinnis & Madden 2004 |
| Blat | Blat is similar to the BLAST alignment tool. However, BLAT requires an exact or close match to find a hit. It finds similarities in DNA and proteins quickly. As a result, Blat is less adaptable than BLAST | https://genome.ucsc.edu/cgi-bin/hgBlat | Kent 2002 |
| Infernal | This tool will build a consensus on RNA secondary structure. The model used for infernal is the covariance model | http://infernal.janelia.org/ | Nawrocki et al. 2009 |
| FastR | This tool will detect the ncRNA | Bafna and Zhang 2004 | |
| ERPIN | ERPIN-Easy RNA profile identification. This tool searches for RNA motifs | http://rna.igmors.u-psud.fr/Software/erpin.php | Gautheret and Lambert 2001 |
| QRNA | An application extends the AMBER simulation approach with extra constraints and allows for fine-grained modification of nucleic acid structures | http://genesilico.pl/QRNAS/QRNAS_data.tar.gz | Stasiewicz et al. 2019 |
| RNAz | RNAz effectively screens for putative ncRNAs by identifying evolutionarily conserved and thermodynamically stable RNA secondary structures in numerous sequence alignments | https://www.tbi.univie.ac.at/~wash/RNAz/ | Washietl 2007 |
| Evofold | Evofold will detect the functional RNA structure using multiple sequence alignment | https://github.com/bowhan/kent/blob/master/src/hg/makeDb/trackDb/drosophila/evofold.html | Knudsen & Hein 1999 |
| MASTR | The algorithm MASTR (Multiple Alignment of STructural RNAs) iteratively enhances sequence alignment and structure prediction for a set of RNA sequences by utilizing Markov chain Monte Carlo in a simulated annealing architecture | http://mastr.binf.ku.dk/ | Lindgreen et al. 2007 |
| CSTminer | Using a possible coding score, this tool can locate statistically significant conserved blocks and determine whether they are coding or non-coding | http://www.caspur.it/CSTminer/ | Castrignano et al. 2004 |
| ESTscan | This tool will detect gene discovery and other assembly roles to find the coding regions. The algorithm used is the Hidden Markov model | https://myhits.sib.swiss/cgi-bin/estscan | Iseli et al. 1999 |
| CONC | This algorithm will predict the RNA secondary structure. It will distinguish the coding and non-coding RNA | (Zou et al. 2011),(Liu et al. 2006) | |
| CPC | The CPC webservers visualize sequence characteristics and forecast the transcript input’s coding potential | http://cpc.cbi.pku.edu.cn | Kong et al. 2007 |
| RNAfold | This tool has an extensive collection of tools like folding, designing, and analyzing RNA sequences | http://rna.tbi.univie.ac.at/ | Gruber et al. 2008 |
| Mfold | This tool will predict the secondary structure of single nucleic acids. Its easy access to RNA and DNA folding | http://www.bioinfo.rpi.edu/applications/mfold | Zuker 2003 |
| A fold | This method will be able to fold the RNA molecule that finds a conformation of energy minimization values | Zuker and Stiegler 1981 | |
| GTEx | The Genotype-Tissue Expression (GTEx) project aims to create a tissue bank and database of resources for the scientific community. The link between genetic variation and gene expression in human tissues will be investigated using GTEx. The expression datasets can be downloaded from this database for ncRNA analysis | https://gtexportal.org/home/ | Lonsdale et al. 2013 |
| DARIO | It can access numerous available ncRNA databases to quantify and annotate ncRNAs | http://dario.bioinf.uni-leipzig.de/ | Fasold et al. 2011 |
| CPSS | Quantify and annotate ncRNAs with a focus on miRNAs | http://114.214.166.79/cpss2.0/ | Wan et al. 2017 |
| RNA-CODE | Combines de novo assembly and secondary structure. Relevant to ncRNA annotation in the absence of reference genomes | http://www.cse.msu.edu/~chengy/RNA_CODE | Yuan & Sun 2013 |
| YM500v3 | A resource that emphasizes piRNAs, tRFs, snRNAs, snoRNAs, and miRNAs and comprises more than 8000 short RNA-seq datasets | http://ngs.ym.edu.tw/ym500/ | Chung et al. 2016 |
| tRF2Cancer | A web server that can find tRFs and the expression of those genes in various cancers | http://rna.sysu.edu.cn/tRFfinder/ | Zheng et al. 2016 |
| MINTbase v2.0 | MINTbase is a collection of nuclear and mitochondrial tRNA-derived fragments (or “tRFs”) discovered in various human tissues | https://cm.jefferson.edu/MINTbase/ | Pliatsika et al. 2017 |
The efficient and reasonably good next-generation sequencing approach allows the collection of large data sets with excellent accuracy. Appropriate bioinformatic procedures must be used to use the collected data and analyze for lncRNAs. Additionally, you can buy commercial arrays to look at the deregulation of a specific set of lncRNAs (e.g., Arraystar, Qiagen, Biocat). Another method to investigate the effect of lncRNAs is to use a genome-wide shRNA library to target a specific subset of lncRNAs. This library and additional studies might be used to ascertain how lncRNA inhibition influences signaling pathways or cell behavior. For instance, the lncRNA TUNA was discovered in mouse embryonic stem cells with Oct4-GFP using an shRNA library targeting 1280 lincRNA (Lin et al. 2014). The pros and cons of RNAi approaches are effectively summed up in a review written by Mohr et al. (Mohr et al. 2014).
Designing primers that only detect the ncRNA transcript is crucial for validating a screen’s results for lncRNAs. To identify coding from non-coding regions, this design is essential. A lncRNA often has modest levels of expression. In addition, lncRNA annotation is continuously evolving and may not be consistent across all databases (like Refseq, UCSC, and Ensembl). Since pseudogenes typically produce lncRNAs, the actual gene and the long non-coding transcript can be recognized using the same primers. Another difficulty arises when lncRNAs are expressed sense- or antisense-to a recognized protein-coding gene. LncRNAs are primarily found in cell nuclei. There are many challenges associated with pulling down lncRNA/protein complexes since it may provide false-positive outcomes. A highly reproducible RNA antisense purification (RAP) method was described in this paper (McHugh et al. 2015). In vitro, lncRNAs can be suppressed using a variety of compounds. It is also critical to confirm the length of annotated sequences for newly discovered lncRNAs. The rapid amplification of cDNA ends (RACE) method can amplify a lncRNA between a specific point inside the lncRNA and the sequence’s 3′ or 5′ end. The actual sequence can then be found or verified by cloning and sequencing this amplicon (Beermann et al. 2016). Detail-oriented loss- or gain-of-function studies are essential to comprehend a lncRNA’s activity in vivo (Bassett et al. 2014). Numerous lncRNA-related database was mentioned in Table 5.
Table 5.
lncRNA based tools and databases
| Tools/databases | Description | Website | References |
|---|---|---|---|
| LNCipedia | A vast and extensive class of non-coding RNA genes are LNCRNAs. 21 488 annotated human lncRNA transcripts from various sources are available on LNCipedia. The database could help start small- and large-scale lncRNA studies | https://lncipedia.org | Volders et al. 2012 |
| LNCBook | Long non-coding RNAs (lncRNAs) play essential roles in various biological functions. The integration and curation of human lncRNA and the data they are related to are the focus of LncBook. Many multi-omics data from expression, methylation, genomic variation, and lnc RNA-miRNA interaction are also integrated | http://bigd.big.ac.cn/lncbook | Ma et al. 2019 |
| Lnc2Cancer v2.0 | The new database Lnc2Cancer 2.0 presents thorough correlations between lncRNAs and human malignancies. In addition to adding new features and more data, it has recruited 4989 lncRNA-cancer correlations | http://www.bio-bigdata.net/lnc2cancer | Gao et al. 2018a, b |
| TANRIC | Long non-coding RNAs (lncRNA) have become prominent players in cancer biology. They have created TANRIC using recent large-scale RNA-seq datasets, particularly from The Cancer Genome Atlas (TCGA). It describes the lncRNA expression profiles in sizable patient cohorts with 20 cancer types | http://bioinformatics.mdanderson.org/main/TANRIC:Overview | Li et al. 2015 |
| lnCaNet | Numerous human long non-coding RNAs (lncRNAs) have been found in malignancies and implicated in various carcinogenesis processes. LnCaNet is a comprehensive database of co-expression information for cancer genes and lncRNAs | http://lncanet.bioinfo-minzhao.org/ | Liu & Zhao 2016 |
| LncRNADisease 2.0 | LncRNADisease 2.0 documents more than 200,000 lncRNA-disease correlations. The database lists the connections between lncRNAs, mRNA, and miRNA in transcriptional regulation. It incorporates connections between diseases and circular RNA that experiments have supported | http://www.rnanut.net/lncrnadisease/ | Bao et al. 2018 |
| The Cancer LncRNome Atlas | Long non-coding RNA (lncRNA) has significantly altered their understanding of cancer. Their findings imply that lncRNA expression and dysregulation are remarkably tumor-type specific. This paves the way for the creation of novel diagnostics and therapies | http://tcla.fcgportal.org/ | Yan et al. 2015 |
| SELER | Super-enhancers (SEs) are enriched in mediator binding sites, which play a vital role in the production of genes that are particular to different cell types. Through regulating SEs activity, long non-coding RNAs (SE-lncRNAs) play crucial roles in transcriptional regulation. Users can thoroughly examine the physiological and pathological activities of the data in the SELER database to fully comprehend the building blocks of living systems | http://www.seler.cn/ | Guo et al. 2019a, b |
| lncRNAdb v2.0 | lncRNAdb is a large, manually curated reference library of 287 eukaryotic lncRNAs independently published in the scientific literature. The new features include incorporating Illumina Body Atlas expression profiles, nucleotide sequence data, a BLAST search tool, and simple export | https://ngdc.cncb.ac.cn/databasecommons/database/id/23 | Quek et al. 2014 |
| LncRNAWiki | A knowledge base of human long non-coding RNAs is called LncRNAWiki 2.0. (lncRNAs). The system has substantially improved with an updated database system and curation approach. Additionally, it offers more approachable online interfaces that make data curation, retrieval, and visualization easier | https://ngdc.cncb.ac.cn/lncrnawiki1/index.php/Main_Page | Liu et al. 2021a, b |
| MONOCLdb | The antiviral response is expected to significantly influence long non-coding RNAs (lncRNAs). To detect coronavirus causing severe acute respiratory syndrome and influenza A, we used whole RNA-Seq on virally infected lungs from eight mouse strains (SARS-CoV). The interactive database MOuse NOn-Code Lung makes these data completely available (MONOCLdb) | https://www.monocldb.org/ | Josset et al. 2014 |
| CANTATAdb | Long non-coding RNAs (lncRNAs) are effective gene expression regulators in many eukaryotes. We still know very little about these compounds in plants. A database named CANTATAdb is online and offers the data for free searching, viewing, and downloading | http://cantata.amu.edu.pl/ | Szcześniak et al. 2015 |
| CPPred | The SVM classifier and several sequence features, including unique RNA features, are the foundation of the CPPred method. Most newly hypothesized novel coding RNAs (91.1%) are ncRNAs, which is consistent with earlier studies. Surprisingly, the global description of encoding properties is crucial in predicting coding capability | http://www.rnabinding.com/CPPred | Tong and Liu 2019 |
| CNIT | It remains challenging to categorize RNA transcripts into protein-coding or non-coding even as more high-throughput data has been generated by next-generation sequencing, especially for species with inadequate annotation. The CNIT (Coding-Non-Coding Identifying Tool) assesses the coding capacity of RNA transcripts more quickly and accurately. For most eukaryotic transcripts, CNIT is more accurate than CNCI and operates 200 times faster. 11 animal species’ AUC values and 27 plant species’ AUC values | http://cnit.noncode.org/CNIT | Guo et al. 2019a, b |
| LncSLdb | Long non-coding RNAs (lncRNAs) may become crucial to biological processes and cellular function. Although we still do not fully understand them, they might explain how they work. A program that handles and maintains user-gathered subcellular localization data is called lncSLdb | http://bioinformatics.xidian.edu.cn/lncSLdb | Wen et al. 2018 |
| LncATLAS | The location of long non-coding RNAs (lncRNAs) inside the cell provides essential hints about their molecular function. Based on data from RNA sequencing, LncATLAS is a comprehensive repository of lncRNA localization in human cells | https://lncatlas.crg.eu/ | Mas-Ponte et al. 2017 |
| LncLocator | Studies of long non-coding RNA (lncRNA) have drawn much interest in the discipline of RNA biology. According to recent research, their subcellular localizations include crucial information for comprehending their intricate biological roles. So far, there are no computational methods for predicting the locations of lncRNAs. The LncLocator tool will be able to locate the positions of lncRNA | http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/ | Cao et al. 2018 |
| RMDB | Since the invention of high-throughput sequencing methods, RNA structure mapping data has risen considerably. To facilitate structural, thermodynamic, and kinetic comparisons, we created an RNA mapping database (RMDB). The database now contains 53 entries outlining more than 2848 trials | https://rmdb.stanford.edu/ | Cordero et al. 2012 |
| DMfold | To predict the secondary structure with pseudoknots, “DMfold” is proposed. The Deep Learning and Improved Base Pair Maximization Principles serve as the foundation. Their code is available at the github repository | https://github.com/linyuwangPHD/RNA-Secondary-Structure-Database | Wang et al. 2019a, b, c |
| SEEKR | Most long non-coding RNAs (lncRNAs) have unknown functions, and finding one lncRNA’s function rarely reveals what the others do. A robust method for identifying connections between sequence and function in lncRNas is Kmer-based categorization | http://seekr.org/ | Kirk et al. 2018 |
| LNCediting | An RNA transcript can change a single base through the post-transcriptional process known as RNA editing. Most of these RNA editing sites are located in non-coding areas of the genome and have unclear functions. To forecast the functional impact of novel editing sites in lncRNAs, LNCediting offers specialized methods | http://bioinfo.life.hust.edu.cn/LNCediting/ | Gong et al. 2017a, b |
| Ufold | It has long been challenging to infer RNA secondary structure from nucleotide sequences. UFold suggests a unique, image-like representation of RNA sequences that Fully Convolutional Networks can parse quickly. On within-family and cross-family datasets, it dramatically outperforms earlier techniques | https://ufold.ics.uci.edu | Fu et al. 2021 |
| RNAInter | RNAInter (RNA Interactome Database) has been upgraded to version 4.0. An upgraded confidence-scoring algorithm and a more extensive data collection. Updated, user-friendly UI and > 47 million new entries. Overall, RNAInter will offer a platform that is easier to use | http://www.rnainter.org/ | Kang et al. 2022 |
| RISE | RNA regulation and function depend on RNA-RNA interactions (RRIs). There are 328,811 interactions in the RISE database, mostly involving people, mice, and yeast. mRNA and long non-coding RNAs make up more than 50% of the RRIs in RISE | http://rise.zhanglab.net/ | Gong et al. 2017a |
| IntaRNA | The prediction modes and output formats can be freely customized and upgraded with IntaRNAv2. The improved lowest energy profiles for RNA-RNA interactions are visualized using the expanded web interface. These make it possible to investigate interaction options in detail and may show many possible interaction sites | https://bio.tools/intarna | Mann et al. 2017 |
| LncRRIsearch | lncRNAs, also known as long non-coding RNAs, are essential to many biological processes. A web server called LncRRIsearch provides thorough predictions of lncRNA-lncRNA interactions. The prediction was made using RIblast, a quick and reliable technique for predicting RNA-RNA interactions | http://rtools.cbrc.jp/LncRRIsearch/ | Fukunaga et al. 2019 |
| LnChrom | The control of chromatin by long non-coding RNAs (lncRNAs) affects several biological functions and disorders. LnChrom is a database of lncRNA-chromatin interactions that have been empirically verified. There are 382 743 mouse and human interactions in it | http://biocc.hrbmu.edu.cn/LnChrom/ | Yu et al. 2018 |
| Triplexator | Triplex production offers a robust targeting mechanism for genomic locations of interest for biotechnological and gene-therapeutic applications. The first computational framework for displaying the possibilities of triplex creation is called Triplexator | https://github.com/Gurado/triplexator | Buske et al. 2012 |
| SFPEL-LPI | LncRNA-protein interactions are crucial for polyadenylation, splicing, translation, and post-transcriptional gene control. SFPEL-LPI uses a feature projection ensemble-learning frame to merge numerous features and similarities. The method predicts novel lncRNAs (or proteins) more accurately than other approaches | http://www.bioinfotech.cn/SFPEL-LPI/ | Zhang et al. 2018a, b, c |
| lncRScan | Identify the lncRNA from the complex assemblies and distinguish it from mRNA | Sun et al. 2015a, b | |
| iSeeRNA | It will detect lncRNA from large datasets | Sun et al. 2013 | |
| Annocript | It utilizes public databases and sequence analysis software to find lncRNA and confirm its high non-coding potential | Musacchia et al. 2015 | |
| LncRNA2Function | lncRNA annotation should be based on the idea that genes with comparable expression patterns can have related biological pathways and activities under different situations | Jiang et al. 2015 |
By searching current RNA-sequencing data for circular RNAs, a brand-new set of probable circRNAs can be predicted (Salzman et al. 2012). Data from long-read RNA sequencing can be utilized to look for possible circRNAs. This particular class of molecules requires a specific algorithm because their production may have involved back-splicing. Two studies demonstrate how to build a computational pipeline to identify new circRNAs (Guo et al. 2014). Using these new techniques to analyze RNA-sequencing data provides suggestions for existing circRNAs. Because the gene from which they are derived has a distinct orientation, the validation of these ncRNAs is particularly unique. Exonic circRNAs must be separated from other RNA molecules that have undergone backspacing. Divergent primers can be used in qPCRs to access the expression and access the predicted circRNAs.
Regarding the genomic area, these primers do not amplify toward one another but are somewhat away from one another. The circle can be amplified without amplifying the genomic areas (Jeck and Sharpless 2014). The functional circRNA can be accessed through previous RNA studies, which are still evolving. Other new approaches should be implemented for the circRNA. New tools and approaches to small ncRNA and circRNA were mentioned in Tables 6 and 7.
Table 6.
Small ncRNA-based tools and databases
| Tools/databases | Description | Website | References |
|---|---|---|---|
| ncPRO-seq | Discovering new ncRNA species by impartially detecting known small ncRNAs | http://ncpro.curie.fr/ | Chen et al. 2012 |
| CoRAL | Organizing the short ncRNA into functional groups according to biologically comprehensible characteristics other than sequence; Describe ncRNA in less well-known species | http://wanglab.pcbi.upenn.edu/coral | Leung et al. 2013a, b |
| DASHR 2.0 | A database that incorporates mature products from all the main RNA classes and human small ncRNA genes | http://lisanwanglab.org/DASHR | Kuksa et al. 2019 |
Table 7.
circRNA-based tools and databases
| Tools/databases | Description | Website | References |
|---|---|---|---|
| CIRI | A first and essential step in understanding the synthesis and function of circRNAs is the thorough discovery of these molecules using high-throughput transcriptome data. For the first time, they detect and experimentally validate the prevalence of intronic/intergenic circRNAs as well as segments particular to them in the human transcriptome by applying CIRI to ENCODE RNA-seq data | https://sourceforge.net/projects/ciri/ | Gao et al. 2015 |
| CIRCexplorer | This tool is used to identify the fragments mapped to circRNA. It will identify and quantify the circ-RNAs to understand their function | https://github.com/YangLab/CIRCexplorer | Ma et al. 2021 |
| KNIFE | This tool is used to identify the circRNA and will read as a fastq file for further analysis. This tool is implemented using python and R | https://github.com/lindaszabo/KNIFE | Szabo et al. 2015 |
| find_circ | This tool identifies and finds the particular circRNAs implemented in python and R. The input sequence would read as fastq | https://github.com/rajewsky-lab/find_circ | Memczak et al. 2013 |
| MapSplice2 | This tool will map the accurate reads in the splice junction. For the alignment of RNA-Seq reads to splice junctions, the exact algorithm Mapslice is used | http://www.netlab.uky.edU/p/bioinfo/MapSplice2 | Wang et al. 2010 |
| segment | This tool will map the short sequence reads to the reference genome | https://www.bioinf.uni-leipzig.de/Software/segemehl/ | Hoffmann et al. 2009 |
| circRNA_Finder | This tool is used to identify the circular RNA in a coordinate-based expression filter. This tool was implemented in Perl | https://github.com/orzechoj/circRNA_finder | Di Liddo et al. 2019 |
| ACFS | From single- and paired-ended RNA-Seq data, Acfs enables de novo, accurate, and quick identification and abundance quantification of circRNAs. It is primarily for alignment purposes | https://github.com/arthuryxt/acfs | You & Conrad 2016 |
| NCLscan | This tool will detect the numerous non-colinear transcripts of circRNA | https://github.com/TreesLab/NCLscan | Chuang et al. 2015 |
| DCC | This tool will be able to detect and quantify the circRNA. It will be used for the mapping and alignment process. It is implemented in python packages | https://github.com/dieterich-lab/DCC | Cheng et al. 2015 |
| UROBORUS | It is a computational pipeline to detect the circRNA using RNA-Seq data | https://github.com/WGLab/UROBORUS | Song et al. 2016 |
| circBase | circBase allows users to access, download, and browse consolidated and unified data sets of circRNAs and the evidence demonstrating their expression within the genomic context. Additionally, circBase offers to find both known and novel circRNAs in sequencing data. The organism used is Human, Mouse, worm, fly, and coelacanth | http://www.circbase.org/ | Glažar et al. 2014 |
| circRNADb | The goal of circRNADb is to serve as a platform for biological information about circRNA molecules and related biological processes. The database allows the user to study a specific circular RNA of interest and continuously update the database through data search, browsing, downloading, submitting, and feedback. The dataset primarily used in exonic circRNAs and organisms is human | http://reprod.njmu.edu.cn/circrnadb | Chen et al. 2016 |
| Circ2Traits | It is a collection of databases about disease-circRNA association. The first complete database of possible links between circular RNAs and human disease | http://gyanxet-beta.com/circdb/ | Ghosal et al. 2013 |
| CircNet | According to the search, the CircNet database is the first openly accessible database to include tissue-specific circRNA expression profiles and circRNA-miRNA-gene regulation networks. It not only adds to the most recent catalog of circRNAs but also offers a complete examination of the expression of both previously known and newly discovered circRNAs | http://circnet.mbc.nctu.edu.tw/ | Liu et al. 2015a, b |
| circRNABase | The interaction database of circRNA-miRNA. The organism involved in this database is human, mouse, and worm | http://web.archive.org/web/20130922084530/starbase.sysu.edu.cn/mirCircRNA.php | Li et al. 2014 |
| MiOncoCirc | Using exome capture sequencing, 2093 clinical human cancer samples were found to contain circRNA | https://nguyenjoshvo.github.io/ | Zhao et al. 2020a, b |
| CSCD | A database that reports anticipated cellular location, RBP locations, and ORFs focuses on differentiating cancer-specific circRNAs from noncancerous circRNAs | http://gb.whu.edu.cn/CSCD | Xia et al. 2018 |
| CircRiC | This study characterizes circRNAs in cancer cell lines and investigates potential circRNA biogenesis mechanisms and their therapeutic relevance. We also offer a data portal to help with related biomedical research | https://hanlab.uth.edu/cRic | Ruan et al. 2019 |
| circMine | In order to view, search, analyze, and download data freely and to submit new data for further integration, circMine offers user-friendly web interfaces. It can be a valuable tool for finding significant circRNA in various diseases. It has 1,821,448 items of 1107 samples from 31 different human body sites, 136,871 circRNAs, 87 diseases, and 120 circRNA transcriptome datasets | http://hpcc.siat.ac.cn/circmine | Zhang et al. 2021a, b |
| CircAtlas | An integrated database of 1070 vertebrate transcriptomes and one million precise circular RNAs | http://circatlas.biols.ac.cn/ | Wu et al. 2020a, b |
| CIRCpedia v2 | A thorough circRNA annotation from more than 180 RNA-seq datasets from six distinct species is stored in a database | http://www.picb.ac.cn/rnomics/circpedia | Dong et al. 2018a, b |
| TSCD | They have conducted a thorough analysis to identify the characteristics of tissue-specific (TS) circRNAs in humans and mice. We found 302 853 TS circRNAs in the human and mouse genomes overall, with the brain having the highest density of TS circRNAs | http://gb.whu.edu.cn/TSCD | Xia et al. 2016 |
Identifying non-coding RNAs (ncRNAs), which play a significant function in the cell, is a crucial topic in biological study. The discovery of ncRNAs is now conceivably feasible, thanks to recent developments in computational prediction technology and bioinformatics. This study introduces three key computational methods for ncRNA identification: homologous search, de novo prediction, and deep sequencing data mining. There are two methods for detecting the ncRNA identification Homologous information and machine learning approaches (i.e., common features)aforementioned computational detection techniques are mostly intended for short non-coding RNAs like miRNAs, tRNAs, siRNAs, and piRNAs. However, conventional methods like PT-PCR and Northern Blot are expensive. The calculation methods can never perform well when dealing with long non-coding RNAs (lncRNA). To the current knowledge, the primary lncRNA detection method is RT-PCR or CHIP-SEQ (Wang et al. 2013). The primary software tools and ncRNA discovery method tools are mentioned in Table 8. The techniques used for ncRNA discovery are mentioned in Table 9.
Table 8.
Common techniques, databases, and tools used in ncRNA
| Tools/databases | Description | Website | References |
|---|---|---|---|
| starBase v2.0 | The interaction database of circRNA-miRNA. The organism involved in this database is human, mouse, and worm | http://starbase.sysu.edu.cn/ | Li et al. 2014 |
| miRTarBase | A source of microRNA-target interactions that have been verified through experiments | http://mirtarbase.cuhk.edu.cn/php/index.php | Chou et al. 2018 |
| miRmine | A repository of profiles of human miRNA expression | http://guanlab.ccmb.med.umich.edu/mirmine | Panwar et al. 2017 |
| EVmiRNA | A database specializing in extracellular vesicle miRNA expression patterns | http://bioinfo.life.hust.edu.cn/EVmiRNA#!/ | Liu et al. 2019 |
| miRGate | A curated library of miRNA-mRNA targets in humans, mice, and rats | http://mirgate.bioinfo.cnio.es/miRGate/ | Andrés-León et al. 2015 |
| miRBase | A database of 271 organisms’ microRNA sequences, including 48,860 mature microRNAs and 38,589 hairpin precursors | http://www.mirbase.org/ | Kozomara et al. 2018 |
| DIANA-TarBase v8 | A reference database for indexing miRNA targets that has been supported by the experiment | http://www.microrna.gr/tarbase | Karagkouni et al. 2018 |
| miRCancer | Currently, a database lists more than 9000 connections between 57,984 miRNAs and 196 types of human cancer | http://mircancer.ecu.edu/ | Xie et al. 2013 |
| Somalia 2.0 | A repository of microRNA (miRNA) target sites and cancer-related somatic alterations may change the interactions between competing endogenous RNAs and miRNAs (ceRNA) | http://compbio.uthsc.edu/SomamiR/ | Bhattacharya and Cui 2015 |
| OncomiR | A platform that explores the deregulation of miRNAs in cancer | http://www.oncomir.org/ | Wong et al. 2018 |
| miRCancerdb | An accessible resource to research target genes involved in the emergence of cancer under the regulation of microRNAs | https://mahshaaban.shinyapps.io/miRCancerdb/ | Ahmed et al. 2018 |
| miR2Disease | A database to offer a thorough resource on microRNA dysregulation in different human diseases | http://www.miR2Disease.org | Jiang et al. 2009 |
| MiRscan | An algorithm for determining the genes for microRNAs from pairs of conserved sequences that may fold back RNA | http://hollywood.mit.edu/mirscan/index.html | Lim 2003 |
| miRanda | This tool will predict the miRNA targets | http://34.236.212.39/microrna/home.do | Betel et al. 2008 |
| RNAhybrid | This tool will predict the miRNA target with unique features such as G: U base pairs in the seed region and a seed-match speed-up | https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid | Kruger & Rehmsmeier 2006 |
| TargetScan | This tool will be able to detect the miRNA targets. It will predict the miRNA, which is a functional site | http://www.targetscan.org/ | Agarwal et al. 2015 |
| PicTar | PicTar predicts targets for single microRNAs and combinations of microRNAs with high accuracy | http://pictar.mdc-berlin.de/ | Krek et al. 2005 |
| TargetFinder | TargetFinder is an interactive tool for choosing effective antisense oligonucleotides (AOs). A selection based on target mRNA secondary structures and mRNA accessible site tagging (MAST). TargetFinder is a helpful tool in the selection of AO target sites because of its graphical, user-friendly design | https://github.com/carringtonlab/TargetFinder | Bo and Wang 2004 |
| TarBase | This tool will detect the miRNA targets. Tarbes is a database of experimentally verified miRNA targets in the fruit fly, worm, zebrafish, and human/mouse | http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?%20r=tarbasev8 | SETHUPATHY 2005 |
| RNA22 | They outline a web-based tool for interactively exploring and visualizing miRNA target prediction methods in context. RNA22-GUI is now available for Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens | https://cm.jefferson.edu/rna22/ | Loher and Rigoutsos 2012 |
| GenMiR + + (Generative model for miRNA regulation) | MicroRNAs regulate a considerable fraction of mammalian genes by inhibiting protein translation. The computational prediction of miRNA genes and the target mRNAs has received much attention. Here, we offer a new Bayesian model and learning algorithm that considers gene expression patterns | http://www.psi.toronto.edu/genmir/ | Huang et al. 2007 |
| PolymiRTS | MicroRNA (miRNA) polymorphisms impair miRNA function, altering physiological and behavioral traits and causing disease. It is now possible to locate miRNA-mRNA binding sites because of polymiRTS | http://compbio.uthsc.edu/miRSNP/ | Bhattacharya et al. 2013 |
| miRDB | Numerous gene targets are regulated by small non-coding RNAs called microRNAs (miRNAs). Given their functional significance, miRNAs are the subject of extensive research. An online database system for functional annotation and target prediction for miRNAs is called miRDB | http://www.mirdb.org/ | Wang 2008 |
| miRGator | MicroRNA (miRNA)-associated gene expression, target prediction, disease association, and genomic annotation are all included in the miRGator database. It attempts to make miRNA functional research easier. The reference database miRGator v2.0 is used to study miRNA expression and function | http://mirgator.kobic.re.kr/ | Cho et al. 2011 |
| miRecords | miRecords is an integrated database for animal miRNA-target interactions. 11 well-known miRNA target prediction systems create predicted miRNA targets, which miRecords keeps. In seven animal species, the database has 1135 records of verified interactions between 301 miRNAs and 902 target genes | http://c1.accurascience.com/miRecords/ | Xiao et al. 2009 |
| mirSOM | Small non-coding RNAs called microRNAs bind to the mRNA of the target gene to control transcriptional activities. Animal miRNA target prediction is difficult because of the imperfection of this binding in animals. The prediction of miRNA targets may be more accurate due to machine learning. This tool is a miRNA target prediction tool that depends on the self-organizing map (SOM) | https://bioinformatics.uef.fi/mirsom/ | Heikkinen et al. 2011 |
| miRWalk | An open-source tool called miRWalk generates predicted and verified miRNA-binding sites for well-known genes using an easy-to-use interface. Python, MySQL, and an HTML/Javascript database are used to access the database | http://mirwalk.umm.uni-heidelberg.de/ | Sticht et al. 2018 |
| mirDIP | miDIP will be able to predict the 152 million for miRNA target prediction | http://ophid.utoronto.ca/mirDIP/ | Tokar et al. 2018 |
| psRNATarget | The high-throughput analysis of next-generation data focuses on the psRNATarget server’s architecture. Three streamlined, user-friendly interfaces are included in the server front end. Along with providing online tools for bulk downloading, keyword searching, and results sorting. It reports the number of small RNA/target site pairs | http://plantgrn.noble.org/psRNATarget/ | Dai and Zhao 2011 |
| miRTarCLIP | They developed a methodical strategy for mining miRNA-target sites from CLIP-seq and PAR-CLIP sequencing data and then linked the technique with a graphical web-based browser, which offers an intuitive user interface and thorough MTI annotations. Additionally, they demonstrated the effectiveness of miRTarCLIP as a tool for comprehending miRNAs using actual-world situations | http://mirtarclip.mbc.nctu.edu.tw/ | Chou et al. 2013 |
| MiRTDL | Genes that are linked to a variety of disorders are regulated by microRNAs. A new convolutional neural network-based miRNA target prediction algorithm is called miRTDL. It has much greater sensitivity, specificity, and accuracy, measuring 88.43, 96.44, and 89.98%, respectively | http://nclab.hit.edu.cn/CCRM/ | Shuang et al. 2016 |
| miRBShunter | According to the evidence, the classical rule about the seed matching between miRNA and target mRNAs is broken in roughly 60% of miRNA-binding activity. The in-house Ago2-dataset and an Auroglial dataset in stem cells were used to evaluate and experimentally validate miRBShunter. Overall, we offer suggestions for selecting a good peak detection algorithm and a novel technique for identifying miRNA-targets | https://github.com/TrabucchiLab/miRBShunter | Bottini et al. 2017 |
| miRTar2GO | MicroRNAs (miRNAs) control gene expression by identifying and attaching to mRNAs’ complementary regions. With more lax miRNA-target binding characteristics, miRTar2GO is developed to predict miRNA target sites. It enables the prediction of miRNA targets specific to different cell types | http://www.mirtar2go.org/ | Ahadi et al. 2016 |
| MMIA | Comprehensive human genome coverage is used by MMIA, along with categorization into different disease-associated genes, canonical pathways, and Gene Ontology | http://cancer.informatics.indiana.edu/mmia | Nam et al. 2009 |
| mirConnX | A web-based program called mirConnX can infer, show, and parse mRNA and miRNA gene regulatory networks. It builds a disease-specific, genome-wide regulatory network using analysis of gene expression data and sequencing information. It is a valuable tool for developing and exploring hypotheses because of its user-friendly design and extensive database | http://www.benoslab.pitt.edu/mirconnx | Huang et al. 2011 |
| MAGIA | A web program called MAGIA (miRNA and genes integrated analysis) is used to collectively analyze target predictions, miRNA, and gene expression data. MAGIA is accessible freely | http://gencomp.bio.unipd.it/magia | Sales et al. 2010 |
| TargetMinner | This tool will be able to detect the miRNA target prediction. They outperform ten other target prediction algorithms with their approach. Based on a pool of 90 features, we attain a much higher sensitivity and specificity of 69 and 67.8%, respectively. The issue of systematic detection of non-target mRNAs is still unresolved | http://www.isical.ac.in/%C2%A0bioinfo_miu/ | Bandyopadhyay and Mitra 2009 |
| ExprTarget | One important mechanism of gene regulation is the attachment of a class of tiny RNA molecules known as microRNAs to mRNA transcripts. Understanding gene regulation networks requires a comprehensive library of miRNA-regulated targets. ExprTarget considerably enhances both the sensitivity and specificity of miRNA target prediction | http://www.scandb.org/apps/microrna/ | Gamazon et al. 2010 |
| MirZ | MirZ web server makes statistical analysis and data mining tools available to experimental and analytical biologists that use the most recent databases of predicted miRNA target sites and sequencing-based miRNA expression profiles for species ranging from Caenorhabditis elegans to Homo sapiens | http://www.mirz.unibas.ch/ | Hausser et al. 2009 |
| mimiRNA | MicroRNAs are small non-coding RNAs that control gene expression by blocking their target mRNA genes. Their expression patterns offer substantial therapeutic and diagnostic potential and therefore are tissue- and disease-specific. To comprehend these patterns, a reliable collection of miRNA and mRNA expression data is needed. mimiRNA will be able to solve this problem | http://mimirna.centenary.org.au/ | Ritchie et al. 2009 |
| ViennaRNA | A crucial intermediate level of description of nucleic acids is their secondary structure. It captures most folding energy and is frequently well-conserved during evolution. Among the new features is a comprehensive toolbox for evaluating RNA-RNA interactions and constrained ensembles of structures. A popular collection of software tools relevant to RNA secondary structure is called the ViennaRNA Package | http://rna.tbi.univie.ac.at/ | Lorenz et al. 2011 |
| HMDD | Many miRNA-disease association entries are manually compiled from literature for the new Human MicroRNA Disease Database (HMDD v3.0). HMDD is openly available | http://210.73.221.6/hmdd | Huang et al. 2019a, b |
| mirPath | An online software package called DIANA-miRPath v3.0 is used to evaluate the regulatory functions of miRNAs and identify regulated pathways. The capabilities and database have been greatly expanded to accommodate all KEGG molecular pathway analyses and several Gene Ontology components | http://snf-515788.vm.okeanos.grnet.gr/ | Vlachos et al. 2015 |
| ExoCarta | Different types of cells release vesicles called exosomes into the extracellular surroundings. Exosomes include RNA, proteins, and lipids. Hence, it is essential to understand their molecular contents. ExoCarta includes biological pathways of exosomal proteins and dynamic protein–protein interaction networks. Based on the number of investigations, users can download the most often detected exosomal proteins | http://www.exocarta.org/ | Keerthikumar et al. 2016 |
| SeqBuster | SeqBuster is the first bioinformatic tool to comprehensively characterize miRNA variants by integrating different analysis modules into a novel platform (isomiRs). SeqBuster was used to analyze small-RNA datasets from human embryonic stem cells, and the results showed that most miRNAs contain a variety of isomiRs, some of which are linked to stem cell development. SeqBuster’s thorough description of the isomiRs may make it easier to spot miRNA variants important for both physiological and pathological processes | https://pypi.org/project/seqcluster/ | Pantano et al. 2009 |
| TransmiR | For more specific details on regulating transcription factor (TF)-miRNA, see the TransmiR v2.0 database. By manually curating 3730 TF-miRNA regulations across 19 species from 1349 papers and reviewing more than 8000 articles, more than 1.7 million tissue-specific TF-miRNA regulations based on ChIP-seq data were also added. TransmiR v2.0 would be a helpful tool for researching how miRNAs are regulated | http://www.cuilab.cn/transmir | Tong et al. 2019 |
| dbDEMC | This database is anticipated to be a valuable resource for identifying cancer-related miRNAs, which will aid in advancing human cancer categorization, diagnosis, and treatment | http://www.picb.ac.cn/dbDEMC | Yang et al. 2010 |
| miTALOS | Nearly all biological activities require microRNAs, which act as signaling pathway regulators. The expression of miRNA target and pathway genes varies across human tissues. The tool miTALOS v2 sheds light on how particular miRNAs regulate biological pathways in different tissues | http://mips.helmholtz-muenchen.de/mitalos | Preusse et al. 2016 |
| miRT | By compiling information from several experimental investigations that validate miRNA TSSs and make the whole datasets available for download, we present a novel database of validated miRNA TSSs in this study called miRT. We offer miRT as a web server that can convert TSS loci between various genome structures. For cutting-edge research on miRNA regulation, miRT may be a helpful tool | http://mirstart.mbc.nctu.edu.tw/ | Bhattacharyya et al. 2012 |
| miRandola | Small non-coding RNAs called microRNAs are crucial in the control of many biological processes. They are commonly dysregulated in cancer and have demonstrated considerable promise as diagnostic and prognostic indicators. miRandola is a comprehensive catalog of extracellular circulating miRNAs that have been extensively curated | http://mirandola.iit.cnr.it/ | Russo et al. 2012 |
| miRNEST | A comprehensive resource for microRNAs is the MiRNEST database. The user interface was enhanced, and download and upload options were added in version 2.0. In-depth miRNA predictions using deep sequencing libraries, assessments of the degradation of plants, and categorization of pre-miRNAs were also incorporated | http://rhesus.amu.edu.pl/mirnest/copy/ | Szcześniak & Makałowska 2013 |
| miR-EdiTar | The database of anticipated A-to-I edited miRNA binding sites known as miR-EdiTar is presented in this work. The database includes projected miRNA binding sites that A-to-I editing may modify and sites that A-to-I editing may cause to become miRNA-binding sites | http://www.tau.ac.il/~elieis/miR_editing | Laganà et al. 2012 |
| SM2miR | The development of miRNA therapeutics will be aided by SM2miR’s complete archive on the effects of small compounds on miRNA expression | http://210.46.85.180:8080/sm2mir/index.jsp | Liu et al. 2012 |
| YM500 | Researchers can use this interactive web interface to search this database for these four categories of analytical results. In addition to integrating data from dbSNP to help researchers distinguish between isomiRs and SNPs, YM500 enables researchers to specify the criteria for isomiRs. Integrating miRNA-related data with preexisting evidence from hundreds of sequencing datasets is made possible by a user-friendly interface. The discovered new miRNAs and isomiRs have the potential to be used in biotechnological and fundamental research | http://driverdb.tms.cmu.edu.tw/ym500v3/ | Cheng et al. 2012 |
| isomiRex | To discover isomiRs and provide an on-demand graphical display of the differentially expressed miRNAs, we provide the open-access web platform isomiRex. The platform can handle many read counts and reports the annotated microRNAs from the plant, animal, and viral NGS datasets | http://bioinfo1.uni-plovdiv.bg/isomiRex/ | Sablok et al. 2013 |
| PHDcleav | Predicting the Dicer cleavage sites in pre-miRNA using the website PHDcleav. With the help of this tool, researchers can examine the effects of genetic variants and SNPs in miRNA on the Dicer cleavage site and gene silencing. Additionally, it would be helpful in the identification of miRNAs in the human genome and the creation of pre-miRNAs specific to Dicer for effective gene silencing | http://crdd.osdd.net/raghava/phdcleav/ | Ahmed et al. 2013 |
| PASmiR | To give thorough, searchable descriptions of miRNA molecular regulation in various plant abiotic stressors, PASmiR, a literature-curated and web-accessible database, was created. Users can receive miRNA-stress regulatory entries using the plant species, abiotic stress, and miRNA identifier as keywords in the PASmiR interface. There are presently 1038 regulatory connections between 682 miRNAs and 35 abiotic stressors in 33 plant species represented by data from almost 200 published research in PASmiR | http://pcsb.ahau.edu.cn:8080/PASmiR | Zhang et al. 2013 |
| microTSS | The missing piece to incorporating the regulation of miRNA transcription into the modeling of tissue-specific regulatory networks is MicroTSS, which is easily adaptable to any cell or tissue sample | http://www.microrna.gr/microTSS/ | Georgakilas et al. 2014 |
| Chimera | A chimera is a web-based tool for small RNA-Seq data processing using microRNA (miRNA). Chimera produces data on miRNA expression based on counts for later statistical analysis. In order to map sequences to miRNA hairpin sequences, cleaning, trimming, and size selection are made automatically | http://wwwdev.ebi.ac.uk/enright-dev/chimira/ | Vitsios and Enright 2015 |
| MirGeneDB | Individual gene sequences for microRNAs are conserved throughout the animal kingdom, making them unique. Genuine miRNAs may be easily distinguished from the countless other short RNAs produced by cells using particular and mechanistically understood characteristics. Previous microRNA annotations notably omitted > 2000 bona five microRNAs and had several false positives. MirGeneDB is a robust platform for microRNA-based research, offering a more substantial and in-depth understanding of the biology and evolution of miRNAs as well as biomedical and Biomarker research | http://mirgenedb.org/ | Fromm et al. 2019 |
| DREAM | A web server for identifying mature microRNA editing events is a website for detecting RNA editing linked to microRNAs. Raw reads from microRNA sequencing can be used as input. Custom scripts analyze the statistical significance, interpret the data, and look for potential modification spots | http://www.cs.tau.ac.il/~mirnaed/ | Alon et al. 2015 |
| Islamic Bank | A web server for identifying mature microRNA editing events is a website for detecting RNA editing linked to microRNAs. Raw reads from microRNA sequencing can be used as input. Custom scripts analyze the statistical significance, interpret the data, and look for potential modification spots | http://mcg.ustc.edu.cn/bsc/isomir/ | Zhang et al. 2016a, b, c |
| TissueAtlas | Human tissue miRNA TSI levels are firmly (P = 108) linked with rat tissue miRNA TSI values. There were 1,364 miRNAs in 61 tissue biopsies from various organs. MiRNA abundance clustering showed that tissues, including various brain regions, grouped | https://ccb-web.cs.uni-saarland.de/tissueatlas/ | Ludwig et al. 2016 |
| miRNAme Converter | The central archive for miRNAs is the miRBase database. Inconsistencies in mature miRNA names result from name changes in various miRNA releases. The problems brought on by these inconsistencies are addressed by the software and online interface known as miRNAmeConverter. The primary method enables a high-throughput automatic translation of mature mi RNA names that are independent of species | http://163.172.134.150/miRNAmeConverter-shiny | Haunsberger et al. 2016 |
| mirSTP | Understanding how miRNAs are regulated in development and disease requires the identification of miRNA transcription start sites (miRNA TSSs). Researchers at Vanderbilt University created mirSTP, which offers a probabilistic method for determining active miRNA Tsss from developing transcriptomes | http://bioinfo.vanderbilt.edu/mirSTP/ | Liu et al. 2017a, b |
| ParSel | Tumor micro-RNAs (miRNA) can determine a patient’s prognosis and response to therapy. By examining their cooperative involvement in gene regulation, this methodology aims to rank the miRNAs. It uses parallel processing to examine a sizably large number of combinatorial scenarios | https://github.com/debsin/ParSel | Sinha et al. 2017 |
| isomiR2 function | Non-coding RNAs (ncRNAs) have been implicated in post-transcriptional control by changing plants’ transcriptional landscape. A standalone, quickly deployable tool called isomiR2Function enables the high-throughput and reliable discovery of both templated and untemplated isomiRs | https://github.com/347033139/isomiR2Function | Yang et al. 2017 |
| miRsig | To fully utilize the potential of miRNA in diagnostic, prognostic, and therapeutic applications, it is crucial to decode the patterns of miRNA regulation in illnesses. Computational predictions of potential miRNA-miRNA interactions have only been made in a few research. To understand how these interactions contribute to the development of the disease, further research is needed. We created miRsig, an online tool for analyzing and visualizing the core miRNA-miRNA interaction associated with disease-specific signatures | http://bnet.egr.vcu.edu/miRsig/ | Nalluri et al. 2017 |
| misalign | This computational method detects miRNA in animals based on structure and sequence alignment. This tool has greater specificity and sensitivity compared to other tools | http://bioinfo.au.tsinghua.edu.cn/miralign | Wang et al. 2005 |
| CID-miRNA | CID-miRNA uses secondary structure-based filtering algorithms and an algorithm based on stochastic context-free grammar trained on human miRNAs to identify miRNA precursors in a given DNA sequence | http://mirna.jnu.ac.in/cidmirna/ | Tyagi et al. 2008 |
| MiRank | To identify the novel miRNA using a rank-based approach | Xu et al. 2008 | |
| mirTool | To investigate miRNA through small RNA transcriptome | http://centre.bioinformatics.zj.cn/mirtools/ | Zhu et al. 2010 |
| snoSeeker | This tool has been developed to screen snoRNA genes in the human genome | Yang et al. 2006 | |
| MiRanalyzer | This tool has been developed to identify novel miRNA and known miRNA using High-throughput sequencing experiments | http://bioinfo2.ugr.es/miRanalyzer/miRanalyzer.php | Hackenberg et al. 2011 |
| UEA sRNA workbench | The UEA sRNA workbench is a collection of tools that replaces the web-based UEA sRNA Toolkit, although it is available for download and has several improved and extra features | http://srna-workbench.cmp.uea.ac.uk | Stocks et al. 2012 |
| MicroPC | Users can thoroughly compare and predict plant miRNAs and their targets using a first online resource. It provides a foundation for the ongoing investigation into miRNA’s conservation, use, and evolution across plant species and classification | http://www.biotec.or.th/isl/micropc | Mhuantong and Wichadakul 2009 |
| HHMMiR | Modeling of miRNA hairpins | Kadri et al. 2009 | |
| miReader | Discovering the novel miRNA without using genomic/reference sequences. The algorithm used is Multi-boosting | http://scbb.ihbt.res.in/2810-12/miReader.php | Jha and Shankar 2013 |
| miRPlex | To predict miRNA only through sRNA datasets without using reference genome | https://www.uea.ac.uk/computing/mirplex | Mapleson et al. 2013 |
| miRdentify | This tool will predict the miRNA in different species | http://www.ncrnalab.dk/#mirdentify/mirdentify.php | Hansen et al. 2014 |
| deepSOM | This tool will be able to detect the pre-miRNA precursor using sequence. The algorithm used is supervised machine learning | http://fich.unl.edu.ar/sinc/blog/web-demo/deepsom/ | Stegmayer et al. 2017 |
| Mirnovo | This tool will be able to detect the miRNA from small RNA seq data using a machine-learning algorithm | http://wwwdev.ebi.ac.uk/enright-dev/mirnovo/ | Vitsios et al. 2017 |
Table 9.
Different techniques used for ncRNA discovery
| Techniques | Description | References |
|---|---|---|
| SHAPE (selective 20-hydroxyl analyzed by primer extension) | It is a method to decipher the lncRNAs secondary structure | Wilkinson et al. 2006 |
| PARS (parallel analysis of RNA structure) | It is a technology recently developed with the Illumina platform (nextPARS) that can study changes in lncRNA structure that may occur in carcinogenesis in order to produce results with higher throughput and sample multiplexing | Saus et al. 2018 |
| Frag-Seq (fragmentation sequencing) | It combines RNA-seq with methods that determine nuclease accessibility at the single base resolution to provide an assay for exploring RNA structure at the transcriptome-wide level | Uzilov and Underwood 2016 |
| ICE-seq (inosine chemical erasing sequencing) | It is a method that can show how A-to-I editing of lncRNAs may become dysregulated in cancer, allowing for meaningful effects on their secondary structure and, later, on the interaction with other RNA molecules | Sakurai et al. 2010 |
| BRIC-seq (50-bromo-uridine immunoprecipitation chase–deep sequencing) | It is a technique that can accurately calculate the half-life of RNA in cells under pathological and physiological settings | Imamachi et al. 2014 |
| FISSEQ (fluorescent in situ sequencings) | It is a technique that uses SOLiD sequencing to detect spatial alterations in lncRNAs that occur during cancer | Lee et al. 2015 |
| Gro-seq (global run-on assay sequencing) | It provides details about the location, orientation, and density of RNAs being actively translated by RNA pol II using an NGS-based approach | Gardini 2017 |
| Tiling arrays | A technique using probes to find transcripts from particular genomic regions | Mockler and Ecker 2005 |
| Microarrays | A technique for quickly analyzing the transcriptome’s global or parallel expression that uses many oligonucleotide probes | Yan et al. 2012 |
| RNA-seq | A method that is now the most popular sequencing technology for RNA expression detection and new RNA discovery | Wang et al. 2009 |
| RNA capture sequencing | An alternative method that combines tilling arrays and RNA-seq | Mercer et al. 2011 |
| Smart-seq | A full-length cDNA amplification technique-based method for scRNA-seq | Ramsköld et al. 2012 |
| DP-seq | A heptamer-primer-based scRNA-seq technique | Bhargava et al. 2013 |
| Quartz-seq | A technique for scRNA-seq that lowers background noise | Sasagawa et al. 2013 |
| SUPeR-seq | RNA sequencing using a single-cell global polyadenylated tail method | Fan et al. 2015 |
| RamDA-seq | a technique for studying single cells based on the full-length whole RNA sequencing | Hayashi et al. 2018 |
| Small RNA-seq | This is the type of RNASeq that distinguish small RNA from larger RNA to understand better these novel RNAs | Landgraf et al. 2007 |
| Small-seq | A technique that will detect the small RNAs in a single cell | Hagemann-Jensen et al. 2018 |
| SLAM-seq | A technique that uses s4U-to-C conversion brought on by nucleophilic substitution chemistry to separate nascent RNA from total RNA | Herzog et al. 2017 |
| TimeLapse-seq | A technique that uses the s4U-to-C conversion caused by an oxidative nucleophilic aromatic substitution process to discriminate between nascent and whole RNA | Schofield et al. 2018 |
| AMUC-seq | A process for converting s4U into a cytidine derivative using acrylonitrile to differentiate between nascent and total RNA | Chen et al. 2020a, b |
| GRID-seq | A procedure that seeks to fully identify and pinpoint the locations of all possible chromatin-interacting RNAs | Li et al. 2017a, b, c |
| iMARGI | An approach that provides in situ mapping of the RNA-genome interactome | Yan et al. 2019a, b |
| ChAR-seq | A genome-wide RNA-to-DNA interaction map produced by chromatin-associated RNA sequencing | Bell et al. 2018 |
| CLASH | An old technique for detecting direct RNA-RNA hybridization employs UV cross-linking | Kudla et al. 2011 |
| RIPPLiT | A method for examining the 3D conformations of RNAs that are stably linked with specific proteins at the transcriptome level | Metkar et al. 2018 |
| MARIO | A technique that uses a biotin-linked reagent to detect RNA-RNA interactions near every protein that binds to RNA | Nguyen et al. 2016 |
| PARIS | High throughput and resolution investigation of RNA interactions and structures using psoralen | Lu et al. 2016 |
| LIGR-seq | A technique for mapping RNA-RNA interactions in vivo at a global level | Sharma et al. 2016 |
| SPLASH | A technique that provides genome-wide paired RNA-RNA pairing data | Aw et al. 2016 |
| RIC-seq | In this method, using RNA in situ conformation sequencing to map all RNA-RNA interactions at the intra- and intermolecular level | Cai et al. 2020 |
| RNA proximity sequencing | A process using high-throughput RNA barcoding of the particles in water-in-oil emulsion droplets | Morf et al. 2019 |
| FISSEQ | A technique that will provide in situ RNA information at high throughput levels | Lee et al. 2015 |
| CeFra-seq | A process for physically isolating subcellular spaces and locating their RNAs | Taliaferro et al. 2014 |
| APEX-RIP | A technique can map organelle-associated RNAs in living cells using proximity biotinylation and protein-RNA cross-linking | Kaewsapsak et al. 2017 |
Applications of CRISPR/Cas9-mediated non-coding RNA editing in the targeted therapy of human diseases
Genome editing, also known as gene editing, refers to a range of scientific techniques that enable the modification of an organism’s DNA. These techniques enable adding, removing, or modifying genetic material at specific genomic regions. There are several genome editing methods, including ZFNs, TALENs, and CRISPR/Cas9. Comparison of these three approaches was mentioned in this article (Li et al. 2021a, b). The detailed structure and mechanism of these three different approaches were mentioned in this article (Li et al. 2021a, b).
CRISPR-Cas9, which stands for clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9, is a well-known example. The CRISPR/Cas9 system has evolved and developed quickly as a reliable, practical, user-friendly, and widely applied gene editing tool in just a few years. CRISPR/Cas9 has significantly impacted a wide range of industries, including agriculture, biotech, and healthcare. However, no industry has been affected by the technology more profoundly than cancer research, as indicated by the accumulating data in the rapidly expanding publications. The discovery and application of more specific Cas9 variants, limiting the duration of CRISPR/Cas9 activity, the use of inducible Cas9 variants, and the application of anti-CRISPR proteins (Zhang et al. 2021a, b). Further research is required to fully comprehend the governing principles of CRISPR/Cas9 specificity and to increase the sensitivity of off-target identification. Second, on-target mutagenesis typically occurs in double-strand breaks brought on by single-guided RNA/Cas9, leading to massive deletions (over several kilobases) and complex genomic rearrangements at the targeted loci, which can have pathogenic effects (Zhang et al. 2021a, b).
The research evidence accumulated to date has shown significant contributions made by genome editing systems to exploit therapeutic approaches for various types of human diseases, with the CRISPR/Cas9 system being particularly successful by directly affecting target gene loci or generating tools with multiple functions. There are other diseases these approaches were found to therapeutic drugs of clinical drugs mentioned in this article (Li et al. 2021a, b). The advancement of cell imaging, gene expression regulation, epigenetic modification, therapeutic drug development, functional gene screening, and gene diagnosis has also been aided by gene editing technologies at the same time. Innovative genome editing complexes and more focused nanostructured vesicles have improved efficiency and reduced toxicity during the delivery process, bringing genome editing technology closer to the clinic. It is reasonable to assume that genome editing technology has the potential to ultimately elucidate biological mechanisms behind disease development and progression, providing novel therapies and ultimately promoting the development of the life sciences, with further investigation into this technology (Li et al. 2020; Li et al. 2021a, b).
Non-coding RNA therapeutics
Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), as well as other types of non-coding RNAs (ncRNAs), are intriguing targets for therapeutic intervention in the treatment of cancer and a variety of other diseases. Many antisense oligonucleotides and small interfering RNAs have been used in the clinical use of RNA-based treatments over the past ten years, and several of these have acquired FDA approval. Trial findings, however, have been mixed up to this point, with some studies claiming strong effects and others showing minimal efficacy or toxicity. Clinical trials are being conducted on alternative entities like antimiRNAs, and interest is growing in lncRNA-based therapies (Winkle et al. 2021).In this review, the existing therapeutic RNA and clinical trial drugs will be mentioned in Table 10.
Table 10.
Existing ncRNA therapeutic drugs and clinical trial drugs
| Drug_name | Mode of drug admin | Target | Mechanism of Action | Disease | Company | Status of the drug | References |
|---|---|---|---|---|---|---|---|
| Fomivirsen | IVT (intravitreal administration) | CMV mRNA | Downregulates IE2 | Cytomegalovirus (CMV) retinitis with acquired AIDS | Ionis Pharmaceutical, Novartis | FDA-approved (1998) | Vitravene Study Group 2002 |
| Mipomersen | SC (subcutaneous Injection) | apo-B-100 mRNA | Downregulates ApoB | Homozygous familial hypercholesterolemia | Kastle Therapeutics, Ionis Pharmaceuticals, Genzyme |
FDA approved (2013) |
Santos et al. 2015 |
| Nusinersen | ITH | SMN2 pre-mRNA | Splicing modulation | Spinal muscular atrophy | Ionis Pharmaceuticals, Biogen |
FDA approved (2016) |
Mercuri et al. 2018 |
| Eteplirsen | IV (intravenous) | Exon 51 of DMD | Splicing modulation | Duchenne muscular dystrophy | Sarepta Therapeutics | FDA approved | Mendell et al. 2013 |
| Inotersen | SC (subcutaneous Injection) | TTR mRNA | Downregulates transthyretin mRNA | Familial amyloid polyneuropathy | Ionis Pharmaceuticals |
FDA approved (2018) |
Benson et al. 2018 |
| Golodirsen | IV(intravenous) | Exon 53 of DMD | Splicing modulation | Duchenne muscular dystrophy | Sarepta Therapeutics |
FDA approved (2019) |
Scaglioni et al. 2021 |
| Milasen | Intrathecal | CLN7 | Splicing modulation | Batten disease | Boston Children’s Hospital |
FDA approved (2018) |
Kim et al. 2019 |
| Casimersen | IV (intravenous) | Exon 45 of DMD | Splicing modulation | Duchenne muscular dystrophy | Sarepta Therapeutics |
FDA approved (2021) |
Shirley 2021 |
| 1018 ISS | IV (intravenous) | TLR9 | Enhancement of cytotoxic effector mechanisms | Non-Hodgkin’s lymphoma | Dana-Farber Cancer Institute, Brigham and Women’s Hospital, Massachusetts General Hospital, University of Rochester |
(Phase II) |
Friedberg et al. 2005 |
| Apatorsen (OGX-427) | IV (intravenous) | HSP27 | Inhibits expression of heat shock protein (Hsp27) | Urologic cancer, bladder cancer, prostate cancer, urothelial cancer, non-small-cell lung cancer, and other cancers | Achieve Life Sciences PRA Health Sciences |
(PhaseI), (Phase-II) |
Chi et al. 2016 |
| Cenersen (EL625) | IV (intravenous) | TP53 | Blocks the effects of p53 | Acute myelogenous leukemia, lymphoma | Eleos, Inc | NCT00074737(Phase II) | Cortes et al. 2011 |
| ARRx (AZD5312) | IV (intravenous) | AR | Suppression of human AR expression | Prostate Cancer | AstraZeneca |
(Phase I/II) |
De Velasco et al. 2019 |
| Custirsen (OGX-011) | IV (intravenous) | ApoJ | Inhibition of clusterin expression | Prostate cancer, breast cancer, non-small-cell lung cancer | NCIC Clinical Trials Group, Achieve Life Sciences |
(Phase I),NCT00138658(Phase-II) |
Laskin et al. 2012;Chi et al. 2005 |
| Patisiran | IV (intravenous) | TTR mRNA | Transthyretin activity is decreased | Polyneuropathy caused by hATTR amyloidosis | Alnylam |
FDA approved (2018) |
Adams et al. 2018 |
| Givosiran | SC (subcutaneous injection) | ALS1 mRNA | ALAS1 is decreased in expression | Acute hepatic porphyria | Alnylam |
FDA approved (2020) |
Balwani et al. 2020 |
| Lumasiran | SC (subcutaneous injection) | HAO1 mRNA | Reduced glycolate oxidase activity | Primary hyperoxaluria type 1 | Alnylam |
FDA approved (2020) |
Liebow et al. 2016 |
| Inclisiran | SC (subcutaneous injection) | PCSK9 | Proprotein convertase subtilsin/kexin type 9 is downregulated | Atherosclerotic cardiovascular disease | Novartis |
FDA approved (2021) |
Lamb 2021 |
| TKM-080301 | Intra-arterial/IV | PLK1 | Activation of PLK1 is reduced | Hepatocellular cancer, cancer with hepatic metastases, | National Cancer Institute, Arbutus Biopharma Corporation |
(Phase-I), NCT02191878(Phase-II) |
El Dika et al. 2018;Steegmaier et al. 2007 |
| Atu027 | IV(intravenous) | PNK3 | The expression of PNK3 is silenced | Solid tumors, pancreatic cancer | Silence Therapeutics GmbH, Granzer Regulatory Consulting & Services |
(Phase-I), NCT01808638 (Phase -II) |
Schultheis et al. 2014;Schultheis et al. 2020 |
| siG12D LODER | Locally implanted through EUS biopsy procedure | KRASG12D | Blocks the expression of KRAS | Pancreatic cancer | Silenseed Ltd |
(Phase -I), (Phase-II) |
Golan et al. 2015; Zorde Khvalevsky et al. 2013 |
| ARO-HIF2 | IV (intravenous) | HIF2A | HIF2A deregulation | Clear cell renal cell carcinoma | Arrowhead Pharmaceuticals |
(Phase-I) |
Cho & Kaelin 2016 |
| APN401 | IV (intravenous) | CBLB | Inhibition of Cbl-b enhances natural killer cell and T cell-mediated antitumor activity | Brain cancer, melanoma, pancreatic cancer, renal cell cancer | Wake Forest University Health Sciences, National Cancer Institute |
(Phase-I) |
Triozzi et al. 2015 |
| Vutrisiran | SQ (subcutaneous injection) | TTR | It lowers the expression of the TTR protein | Transthyretin-mediated amyloidosis with or without cardiomyopathy | Alnylam |
(Phase 3) |
Habtemariam et al. 2020; Sekijima et al. 2005 |
| Pegaptanib | Intravitreal | The heparin-binding domain of VEGF-165 | inhibiting VEGF-165 | Neovascular age-related macular degeneration | OSI Pharmaceuticals |
FDA-approved (2004) |
Gragoudas et al. 2004 |
| Defibrotide | IV (intravenous) | Adenosine A1/A2receptor | Adenosine A1/A2 receptor activation | Veno-occlusive disease in liver | Jazz Pharmaceuticals | FDA-approved (2020) | Richardson et al. 2017;Lee et al. 2021 |
| NOX-A12 | IV (intravenous) | CXCL12 | Interferes with CXCR4-CXCL12 interactions | Pancreatic cancer, colorectal cancer, multiple myeloma | NOXXON Pharma AG, Merck Sharp & Dohme Corp | NCT01521533(Phase- I),NCT01521533(Phase-II),NCT03168139(Phase-I/II) | Park et al. 2019 |
| NOX-E36 | IV (intravenous)/SQ (subcutaneous) | CCL2 | Suppresses the pro-inflammatory chemokine CCL2 by specifically binding to it | Diabetic nephropathy | NOXXON Pharma AG | Phase-II | Menne et al. 2016 |
| BNT162b2 | IM (intramuscular injection) | Immunogenicity and antibody response to SARS-CoV-2 S antigens | Expression of SARS-CoV-2 S antigens | COVID-19 | BioNTech and Pfizer | FDA-approved (2020) | Polack et al. 2020 |
| mRNA-1273 | IM (intramuscular injection) | Immunogenicity and antibody response to SARS-CoV-2 S antigens | Expression of SARS-CoV-2 S(Spike protein) antigens | COVID-19 | Moderna | FDA-approved (2020) | Baden et al. 2020 |
| CVnCoV | IM (intramuscular injection) | Immunogenicity and antibody response to SARS-CoV-2 S antigens | Expression of SARS-CoV-2 S antigens | COVID-19 | CureVac AG | NCT04652102 (Phase -III) | Kremsner et al. 2022 |
| AZD8601 | Epicardial | VEGF-A | VEGF-A expression is restored | Ischemic heart disease | AstraZeneca |
(Phase- II) |
Anttila et al. 2020 |
| MRT5005 | Inhalation | CFTR | CFTR expression is restored | Cystic fibrosis | Translate Bio |
(Phase -I/II) |
Yan et al. 2019a, b |
| mRNA-3704 | IV (intravenous)/SQ (subcutaneous) | MUT | MUT expression is restored | Methylmalonic aciduria | Moderna |
(Phase -I/II) |
Chandler and Venditti 2019 |
| BNT111 | IV (intravenous)/SQ(subcutaneous) | Targets four non-mutated TAAs (NY-ESO-1, MAGEA3, tyrosinase, and TPTE | Immune response induction against the four malignant melanoma-associated antigens (tyrosinase, New York-ESO-1 (NY-ESO-1), Melanoma-associated antigen A3 (MAGE-A3), and transmembrane phosphatase with tensin homology (TPTE)) | Advanced melanoma | BioNTech SE | NCT02410733 (Phase -I) | Sahin et al. 2020 |
| Miravirsen | SC (subcutaneous injection) | miR-122 | miRNA-inhibitor | HCV | Roche/Santaris | NCT01200420(Phase- II) | Janssen et al. 2013 |
| RG-012 (lademirsen) | SC (subcutaneous injection) | miR-21 | miRNA-inhibitor | Alport syndrome | Sanofi | NCT03373786(Phase- II) | Gomez et al. 2014 |
| Cobomarsen | IV/SQ | miR-155 | miRNA-inhibitor | Cutaneous T cell lymphoma/mycosis Fungoides | miRagen | NCT03713320, NCT02580552(Phase -II) | Bedewy et al. 2017 |
| MRG-110 | Intradermal | miR-92a | miRNA-inhibitor | Wound healing | miRagen | NCT03603431(Phase -I) | Bonauer et al. 2009 |
| AZD4076 | SC (subcutaneous injection) | miR-103/107 | miRNA-inhibitor | T2D with NAFLD | AstraZeneca | NCT02826525(Phase- I/IIa) | Trajkovski et al. 2011 |
| RGLS4326 | SC (subcutaneous injection) | miR-17 | miRNA-inhibitor | Autosomal dominant polycystic kidney disease | Regulus Therapeutics Inc | NCT04536688(Phase -I) | Lee et al. 2019 |
| CDR132L | IV (intravenous)/SQ (subcutaneous) | miR-132 | miRNA-inhibitor | Heart failure | Cardio Pharmaceuticals GmbH | NCT04045405(Phase -I) | Täubel et al. 2020 |
| TargomiRs | IV (intravenous)/SQ (subcutaneous) | miR-16 | miRNA-mimic | Malignant pleural mesothelioma | EnGeneIC Limited | NCT02369198(Phase- I) | van Zandwijk et al. 2017 |
| Remlarsen | Intradermal | miR-29 | miRNA-mimic | Keloids, scleroderma | miRagen | NCT03601052(Phase -II) | Cushing et al. 2011 |
| MRX34 | IV (intravenous)/SQ (subcutaneous) | miR-34a | miRNA-mimic | Melanoma, liver cancer | miRNA Therapeutics, Inc | NCT01829971(Phase -I) | Beg et al. 2017 |
Challenges in using ncRNA as biomarkers and therapeutic targets
Non-coding RNAs may be potential biomarkers and therapeutic targets because mounting data suggests they are critical regulators of the pathophysiological processes leading to many diseases. However, its clinical use has not been examined and may face numerous difficulties. First, non-coding RNAs are still being developed as biomarkers. Although RT-PCR, next-generation sequencing, and microarray analysis have been utilized in research examining the connection between non-coding RNAs and disease-specific traits, most of these investigations are still experimental. However, no research has examined the viability of choosing lncRNA/circRNA as novel biomarkers (Zhang et al. 2017a, b). The discovery of tissue- or organ-specific biomarkers would be beneficial for the early diagnosis, treatment, and intervention of organ failure, perhaps increasing the chance of disease-specific survival.
Because they differ from conventional medications, such as small-molecule and protein medicines, which are also known to work primarily on protein targets, RNA-based therapies are considered the next generation of therapeutics. First, RNA aptamers can produce pharmacological effects by blocking the activity of a particular protein target. Second, for controlling a specific disease, antisense RNAs (asRNAs), miRNAs, and siRNAs can be created to specifically target mRNAs or functional ncRNAs. Thirdly, to cure a monogenic condition, gRNAs may be used to precisely alter the target sequences of a particular gene. Thus, RNA therapies can potentially increase the number of druggable targets. On-coding RNAs are promising “next-generation” biomarkers since the issues mentioned earlier and difficulties can be resolved. Non-coding RNAs may one day serve as innovative treatment targets with the help of a more profound knowledge of the mechanism underlying those specific diseases.
Conclusion and future perspectives
The attractive new field of ncRNA research demonstrates a higher level of nature’s diversity. The complexity of ncRNA research results from the more significant than specified based on ncRNAs in cellular biology. Nevertheless, even though ncRNAs have recently been discovered, there have been significant advancements in clinical applications and diagnostic methods. This research will likely expand into a new area of more potent and particular medications and personalized medicine techniques, elevating patient care to a new level. Rapid developments in bioinformatics, sequencing technologies, proteomics, and microarrays have identified a wide variety of non-coding RNAs (ncRNA), which comprise most cellular mechanism regulators principally linked to eukaryotic complexity. It seems more difficult to comprehend the unique function of these non-coding RNAs with these varied ncRNAs having integrated, complicated networks and biological pathways. The use of ncRNA therapies in formal drug development will increase.
Further information has to be obtained, possible ncRNA medicines’ pharmacokinetics and dynamics need to be examined, and thorough toxicological studies are required. To advance the field, more tools are required. There will be more phase I/II clinical studies. This study aims to investigate and advance knowledge of the mechanisms and functions of ncRNAs in human health and disease and to pave the way for novel clinical diagnostic and therapeutic approaches. When dealing with the enormous quantity of ncRNAs that need to be analyzed, ML outperforms since it can quickly address the fundamental problem. By categorizing healthy and disease samples, the current analysis of ncRNAs using ML demonstrates reasonable accuracy, indicating that the differentiation pattern is apparent in those instances. Therefore, future research should concentrate on increasing the likelihood that the ML models will recognize the distinctive pattern of each disease. However, the use of ncRNAs may significantly rise in the following years, which will contribute to the development of successful precision medicine and more individualized therapies.
Acknowledgements
The authors would like to thank the Vellore Institute of Technology (VIT), Vellore, Tamil Nadu, India, for providing the necessary research facilities and encouragement to carry out this work.
Abbreviations
- ncRNA
Non-coding RNA
- lncRNA
Long non-coding RNA
- miRNA
MicroRNA
- circRNA
Circular RNA
- snRNA
Small nuclear RNA
- snoRNA
Small nucleolar RNAs
- siRNA
Small interfering RNA
- rRNA
Ribosomal RNA
- piRNA
PIWI-interacting RNA
- FANTOM
Functional Annotation of the Mammalian Genome
- ENCODE
Encyclopedia of DNA Elements
- Nt
Nucleotide
Author contribution
TL and GPDC contributed to designing the study and data acquisition, analysis, and interpretation. GPDC supervised the entire study and edited the manuscript. The manuscript was reviewed and approved by all the authors.
Data availability
No supporting data is available in this study.
Code availability
Not applicable.
Declarations
Ethical approval and consent to participate
Not applicable.
Human and animal ethics
Not applicable.
Consent for publication
No consent.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang C-C, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin K-P, Vita G, Attarian S, Planté-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH, Kim BJ, Oh J. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med. 2018;379(1):11–21. doi: 10.1056/nejmoa1716153. [DOI] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam J-W, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. ELife 4. 10.7554/elife.05005 [DOI] [PMC free article] [PubMed]
- Ahadi A, Sablok G, Hutvagner G. miRTar2GO: a novel rule-based model learning method for cell line specific microRNA target prediction that integrates Ago2 CLIP-Seq and validated microRNA–target interaction data. Nucleic Acids Res. 2016;45(6):e42–e42. doi: 10.1093/nar/gkw1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F, Kaundal R, and Raghava GP (2013) PHDcleav: a SVM based method for predicting human Dicer cleavage sites using sequence and secondary structure of miRNA precursors. BMC Bioinforma 14(S14). 10.1186/1471-2105-14-s14-s9 [DOI] [PMC free article] [PubMed]
- Ahmed M, Nguyen H, Lai T, Kim DR. miRCancerdb: a database for correlation analysis between microRNA and gene expression in cancer. BMC Res Notes. 2018;11(1):103. doi: 10.1186/s13104-018-3160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam T, Uludag M, Essack M, Salhi A, Ashoor H, Hanks JB, Kapfer C, Mineta K, Gojobori T, Bajic VB. FARNA: knowledgebase of inferred functions of non-coding RNA transcripts. Nucleic Acids Res. 2017;45(5):2838–2848. doi: 10.1093/nar/gkw973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Erew M, Eisenberg E. DREAM: a webserver for the identification of editing sites in mature miRNAs using deep sequencing data. Bioinformatics. 2015;31(15):2568–2570. doi: 10.1093/bioinformatics/btv187. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics. 2013;12(3):254–278. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang C-L, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-León E, González Peña D, Gómez-López G, Pisano DG (2015) miRGate: a curated database of human, mouse and rat miRNA–mRNA targets. Database 2015. 10.1093/database/bav035 [DOI] [PMC free article] [PubMed]
- Annese T, Tamma R, De Giorgis M, Ribatti D (2020) microRNAs biogenesis, functions and role in tumor angiogenesis. Front Oncol 10. 10.3389/fonc.2020.581007 [DOI] [PMC free article] [PubMed]
- Anttila V, Saraste A, Knuuti J, Jaakkola P, Hedman M, Svedlund S, Lagerström-Fermér M, Kjaer M, Jeppsson A, Gan L-M. Synthetic mRNA Encoding VEGF-A in Patients Undergoing Coronary Artery Bypass Grafting: Design of a Phase 2a Clinical Trial. Mol Ther - Methods Clin Dev. 2020;18:464–472. doi: 10.1016/j.omtm.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AYL, Zhang T, Susanto TT, Fu Z, Nagarajan N, Wan Y. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol Cell. 2016;62(4):603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- Baden LR, El Sahly HM, Essink B (2020) Efficacy and safety of the mRNA-1273 SARS CoV-2 vaccine. N Engl J Med 384(5). 10.1056/nejmoa2035389 [DOI] [PMC free article] [PubMed]
- Bafna V, Zhang S (2004) FastR: fast database search tool for non-coding RNA. Proc IEEE Comput Syst Bioinform Conf 52–61. 10.1109/csb.2004.1332417 [DOI] [PubMed]
- Ballabio E, Mitchell T, van Kester MS, Taylor S, Dunlop HM, Chi J, Tosi I, Vermeer MH, Tramonti D, Saunders NJ, Boultwood J, Wainscoat JS, Pezzella F, Whittaker SJ, Tensen CP, Hatton CSR, Lawrie CH (2010) MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood 116(7):1105–1113. 10.1182/blood-2009-12-256719 [DOI] [PMC free article] [PubMed]
- Ballarino M, Cazzella V, D’Andrea D, Grassi L, Bisceglie L, Cipriano A, Santini T, Pinnarò C, Morlando M, Tramontano A, Bozzoni I. Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol Cell Biol. 2015;35(4):728–736. doi: 10.1128/MCB.01394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloy V, Koshy R, Perra L, Corvol H, Chignard M, Guillot L, Scaria V. Bronchial Epithelial Cells from Cystic Fibrosis Patients Express a Specific Long Non-coding RNA Signature upon Pseudomonas aeruginosa Infection. Front Cell Infect Microbiol. 2017;7:218. doi: 10.3389/fcimb.2017.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwani M, Sardh E, Ventura P, Peiró PA, Rees DC, Stölzel U, Bissell DM, Bonkovsky HL, Windyga J, Anderson KE, Parker C, Silver SM, Keel SB, Wang J-D, Stein PE, Harper P, Vassiliou D, Wang B, Phillips J, Ivanova A. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N Engl J Med. 2020;382(24):2289–2301. doi: 10.1056/nejmoa1913147. [DOI] [PubMed] [Google Scholar]
- Ban J-J, Chung J-Y, Lee M, Im W, Kim M. MicroRNA-27a reduces mutant hutingtin aggregation in an in vitro model of Huntington’s disease. Biochem Biophys Res Commun. 2017;488(2):316–321. doi: 10.1016/j.bbrc.2017.05.040. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25(20):2625–2631. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- Bañez-Coronel M, Porta S, Kagerbauer B, Mateu-Huertas E, Pantano L, Ferrer I, Guzmán M, Estivill X, Martí E. A Pathogenic Mechanism in Huntington’s Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity. PLoS Genet. 2012;8(2):e1002481. doi: 10.1371/journal.pgen.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2018;47(D1):D1034–D1037. doi: 10.1093/nar/gky905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, Higgs DR, Miska EA, Ponting CP (2014) Considerations when investigating lncRNA function in vivo. ELife 3. 10.7554/elife.03058 [DOI] [PMC free article] [PubMed]
- Bauters C, Kumarswamy R, Holzmann A, Bretthauer J, Anker SD, Pinet F, Thum T. Circulating miR-133a and miR-423-5p fail as biomarkers for left ventricular remodeling after myocardial infarction. Int J Cardiol. 2013;168(3):1837–1840. doi: 10.1016/j.ijcard.2012.12.074. [DOI] [PubMed] [Google Scholar]
- Bayes-Genis A, Voors AA, Zannad F, Januzzi JL, Mark Richards A, Díez J. Transitioning from usual care to biomarker-based personalized and precision medicine in heart failure: call for action. Eur Heart J. 2017;39(30):2793–2799. doi: 10.1093/eurheartj/ehx027. [DOI] [PubMed] [Google Scholar]
- Bayoglu B, Yuksel H, Cakmak HA, Dirican A, Cengiz M. Polymorphisms in the long non-coding RNA CDKN2B-AS1 may contribute to higher systolic blood pressure levels in hypertensive patients. Clin Biochem. 2016;49(10–11):821–827. doi: 10.1016/j.clinbiochem.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Bedewy AML, Elmaghraby SM, Shehata AA, Kandil NS. Prognostic Value of miRNA-155 Expression in B-Cell Non-Hodgkin Lymphoma. Turkish J Haematol. 2017;34(3):207–212. doi: 10.4274/tjh.2016.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann J, Piccoli M-T, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang Y-K, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35(2):180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JC, Jukam D, Teran NA, Risca VI, Smith OK, Johnson WL, Skotheim JM, Greenleaf WJ, Straight AF. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife. 2018;7:e27024. doi: 10.7554/eLife.27024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, Planté-Bordeneuve V, Barroso FA, Merlini G, Obici L, Scheinberg M, Brannagan TH, Litchy WJ, Whelan C, Drachman BM, Adams D, Heitner SB, Conceição I, Schmidt HH, Vita G. Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N Engl J Med. 2018;379(1):22–31. doi: 10.1056/nejmoa1716793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HD, Zopf D, Freymann DM, Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci USA. 1993;90(11):5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava V, Ko P, Willems E, Mercola M, Subramaniam S (2013) Quantitative transcriptomics using designed primer-based amplification. Sci Rep 3(1). 10.1038/srep01740 [DOI] [PMC free article] [PubMed]
- Bhattacharya A, Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA–ceRNA interactions. Nucleic Acids Res. 2015;44(D1):D1005–D1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2013;42(D1):D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, Das M, Bandyopadhyay S. miRT: A Database of Validated Transcription Start Sites of Human MicroRNAs. Genomics Proteomics Bioinforma. 2012;10(5):310–316. doi: 10.1016/j.gpb.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti GK, Khullar N, Sidhu IS, Navik US, Reddy AP, Reddy PH, Bhatti JS. Emerging role of non-coding RNA in health and disease. Metab Brain Dis. 2021;36(6):1119–1134. doi: 10.1007/s11011-021-00739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo X, Wang S. TargetFinder: a software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics. 2004;21(8):1401–1402. doi: 10.1093/bioinformatics/bti211. [DOI] [PubMed] [Google Scholar]
- Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a Controls Angiogenesis and Functional Recovery of Ischemic Tissues in Mice. Science. 2009;324(5935):1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- Bottini S, Hamouda-Tekaya N, Tanasa B, Zaragosi L-E, Grandjean V, Repetto E, Trabucchi M (2017) From benchmarking HITS-CLIP peak detection programs to a new method for identification of miRNA-binding sites from Ago2-CLIP data. Nucleic Acids Res gkx007. 10.1093/nar/gkx007 [DOI] [PMC free article] [PubMed]
- Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, Seedorf U, Rust S, Eriksson P, Hamsten A, Farrall M, Watkins H. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2007;17(6):806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, Xiao H, Liao Q, Luo H, Zhao G, Zhao H, Liu Z, Liu C, Chen R, Zhao Y. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2011;40(D1):D210–D215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: Detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22(7):1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D, Legnini I, Martone J, Cazzella V, D’Amico A, Bertini E, Bozzoni I. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med. 2011;3(5):258–265. doi: 10.1002/emmm.201100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yang Y, Chen X, He D, Zhang X, Wen X, Hu J, Fu C, Qiu D, Jose PA, Zeng C, Zhou L. Circulating “LncPPARδ” From Monocytes as a Novel Biomarker for Coronary Artery Diseases. Medicine. 2016;95(6):e2360. doi: 10.1097/md.0000000000002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Cao C, Ji L, Ye R, Wang D, Xia C, Wang S, Du Z, Hu N, Yu X, Chen J, Wang L, Yang X, He S, Xue Y. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature. 2020;582(7812):432–437. doi: 10.1038/s41586-020-2249-1. [DOI] [PubMed] [Google Scholar]
- Cao Z, Pan X, Yang Y, Huang Y, Shen H-B. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinforma (Oxford England) 2018;34(13):2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schönbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) (2005). The transcriptional landscape of the mammalian genome. Science 309(5740):1559–1563. 10.1126/science.1112014 10.1126/science.1112014
- Carrieri C, Forrest ARR, Santoro C, Persichetti F, Carninci P, Zucchelli S, and Gustincich S (2015) Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci 9. 10.3389/fncel.2015.00114 [DOI] [PMC free article] [PubMed]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrignano T, Canali A, Grillo G, Liuni S, Mignone F, Pesole G. CSTminer: a web tool for the identification of coding and noncoding conserved sequence tags through cross-species genome comparison. Nucleic Acids Res. 2004;32(Web Server):W624–W627. doi: 10.1093/nar/gkh486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RJ, Venditti CP. Gene Therapy for Methylmalonic Acidemia: Past, Present, and Future. Hum Gene Ther. 2019;30(10):1236–1244. doi: 10.1089/hum.2019.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K-H, Wu Y-R, Chen C-M (2017) Down-regulation of miR-9* in the peripheral leukocytes of Huntington’s disease patients. Orphanet J Rare Dis 12(1). 10.1186/s13023-017-0742-x [DOI] [PMC free article] [PubMed]
- Chen C-J, Servant N, Toedling J, Sarazin A, Marchais A, Duvernois-Berthet E, Cognat V, Colot V, Voinnet O, Heard E, Ciaudo C, Barillot E. ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics. 2012;28(23):3147–3149. doi: 10.1093/bioinformatics/bts587. [DOI] [PubMed] [Google Scholar]
- Chen G, Huang S, Song F, Zhou Y, He X. Lnc-Ang362 is a pro-fibrotic long non-coding RNA promoting cardiac fibrosis after myocardial infarction by suppressing Smad7. Arch Biochem Biophys. 2020;685:108354. doi: 10.1016/j.abb.2020.108354. [DOI] [PubMed] [Google Scholar]
- Chen J, Guo J, Cui X, Dai Y, Tang Z, Qu J, Raj JU, Hu Q, Gou D. The Long Noncoding RNA LnRPT Is Regulated by PDGF-BB and Modulates the Proliferation of Pulmonary Artery Smooth Muscle Cells. Am J Respir Cell Mol Biol. 2018;58(2):181–193. doi: 10.1165/rcmb.2017-0111OC. [DOI] [PubMed] [Google Scholar]
- Chen J, Hu Q, Zhang B-F, Liu X-P, Yang S, Jiang H. Long noncoding RNA UCA1 inhibits ischaemia/reperfusion injury induced cardiomyocytes apoptosis via suppression of endoplasmic reticulum stress. Genes Genomics. 2019;41(7):803–810. doi: 10.1007/s13258-019-00806-w. [DOI] [PubMed] [Google Scholar]
- Chen J-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang D-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chen R, Zhang T, Lin S, Chen Z, Zhao B, Li H, Wu S. Relationship of cardiovascular disease risk factors and noncoding RNAs with hypertension: a case-control study. BMC Cardiovasc Disord. 2018;18(1):58. doi: 10.1186/s12872-018-0795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Han P, Zhou T, Guo X, Song X, Li Y (2016) circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 6(1). 10.1038/srep34985 [DOI] [PMC free article] [PubMed]
- Chen X, Sun Y-Z, Zhang D-H, Li J-Q, Yan G-Y, An J-Y, You Z-H (2017) NRDTD: a database for clinically or experimentally supported non-coding RNAs and drug targets associations. Database 2017. 10.1093/database/bax057 [DOI] [PMC free article] [PubMed]
- Chen Y, Wu F, Chen Z, He Z, Wei Q, Zeng W, Chen K, Xiao F, Yuan Y, Weng X, Zhou Y, Zhou X. Acrylonitrile-Mediated Nascent RNA Sequencing for Transcriptome-Wide Profiling of Cellular RNA Dynamics. Advanced Science. 2020;7(8):1900997. doi: 10.1002/advs.201900997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chénais B. Transposable Elements and Human Diseases: Mechanisms and Implication in the Response to Environmental Pollutants. Int J Mol Sci. 2022;23(5):2551. doi: 10.3390/ijms23052551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Spengler RM, Keiser MS, Monteys AM, Rieders JM, Ramachandran S, Davidson BL. The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum Mol Genet. 2018;27(24):4303–4314. doi: 10.1093/hmg/ddy331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2015;32(7):1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- Cheng P-H, Li C-L, Chang Y-F, Tsai S-J, Lai Y-Y, Chan AWS, Chen C-M, Yang S-H. miR-196a Ameliorates Phenotypes of Huntington Disease in Cell, Transgenic Mouse, and Induced Pluripotent Stem Cell Models. Am J Human Genet. 2013;93(2):306–312. doi: 10.1016/j.ajhg.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-C, Chung I-F, Huang T-S, Chang S-T, Sun H-J, Tsai C-F, Liang M-L, Wong T-T, Wang H-W. YM500: a small RNA sequencing (smRNA-seq) database for microRNA research. Nucleic Acids Res. 2012;41(D1):D285–D294. doi: 10.1093/nar/gks1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, Dong X, Qin S, Zhang C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci. 2010;119(2):87–95. doi: 10.1042/cs20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R (2021) The journey of noncoding RNA from bench to clinic. Translational Biotechnology 165–201. 10.1016/b978-0-12-821972-0.00016-2
- Chi KN, Eisenhauer E, Fazli L, Jones EC, Goldenberg SL, Powers J, Tu D, Gleave ME. A phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2’-methoxyethyl antisense oligonucleotide to clusterin, in patients with localized prostate cancer. J Natl Cancer Inst. 2005;97(17):1287–1296. doi: 10.1093/jnci/dji252. [DOI] [PubMed] [Google Scholar]
- Chi KN, Yu EY, Jacobs C, Bazov J, Kollmannsberger C, Higano CS, Mukherjee SD, Gleave ME, Stewart PS, Hotte SJ. A phase I dose-escalation study of apatorsen (OGX-427), an antisense inhibitor targeting heat shock protein 27 (Hsp27), in patients with castration-resistant prostate cancer and other advanced cancers. Ann Oncol. 2016;27(6):1116–1122. doi: 10.1093/annonc/mdw068. [DOI] [PubMed] [Google Scholar]
- Chi T, Lin J, Wang M, Zhao Y, Liao Z, Wei P (2021) Non-coding RNA as biomarkers for type 2 diabetes development and clinical management. Front Endocrinol 12. 10.3389/fendo.2021.630032 [DOI] [PMC free article] [PubMed]
- Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, Sun L, Ma B, Ding J, Vancraenenbroeck R, Lobbestael E, Baekelandt V, Taymans J-M, He P, Troncoso JC, Shen Y, Cai H. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2012;22(3):608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kaelin WG. Targeting HIF2 in Clear Cell Renal Cell Carcinoma. Cold Spring Harb Symp Quant Biol. 2016;81:113–121. doi: 10.1101/sqb.2016.81.030833. [DOI] [PubMed] [Google Scholar]
- Cho S, Jun Y, Lee S, Choi H-S, Jung S, Jang Y, Park C, Kim S, Lee S, Kim W. miRGator v2.0: an integrated system for functional investigation of microRNAs. Nucleic Acids Res. 2011;39(Database issue):D158–162. doi: 10.1093/nar/gkq1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-H, Lin F-M, Chou M-T, Hsu S-D, Chang T-H, Weng S-L, Shrestha S, Hsiao C-C, Hung J-H, Huang H-D (2013) A computational approach for identifying microRNA-target interactions using high throughput CLIP and PAR-CLIP sequencing. BMC Genomics 14(S1). 10.1186/1471-2164-14-s1-s2 [DOI] [PMC free article] [PubMed]
- Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, Huang W-C, Sun T-H, Tu S-J, Lee W-H, Chiew M-Y, Tai C-S, Wei T-Y, Tsai T-R, Huang H-T, Wang C-Y, Wu H-Y, Ho S-Y, Chen P-R, Chuang C-H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T-J, Wu C-S, Chen C-Y, Hung L-Y, Chiang T-W, Yang M-Y. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2015;44(3):e29–e29. doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I-F, Chang S-J, Chen C-Y, Liu S-H, Li C-Y, Chan C-H, Shih C-C, Cheng W-C. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2016;45(D1):D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clote P. RNALOSS: a web server for RNA locally optimal secondary structures. Nucleic Acids Res. 2005;33(Web Server):W600–W604. doi: 10.1093/nar/gki382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero P, Lucks JB, Das R. An RNA Mapping DataBase for curating RNA structure mapping experiments. Bioinformatics. 2012;28(22):3006–3008. doi: 10.1093/bioinformatics/bts554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating MicroRNA-208b and MicroRNA-499 Reflect Myocardial Damage in Cardiovascular Disease. Circ: Cardiovasc Genetics. 2010;3(6):499–506. doi: 10.1161/circgenetics.110.957415. [DOI] [PubMed] [Google Scholar]
- Cortes J, Kantarjian H, Ball ED, DiPersio J, Kolitz JE, Fernandez HF, Goodman M, Borthakur G, Baer MR, Wetzler M. Phase 2 randomized study of p53 antisense oligonucleotide (cenersen) plus idarubicin with or without cytarabine in refractory and relapsed acute myeloid leukemia. Cancer. 2011;118(2):418–427. doi: 10.1002/cncr.26292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lü J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45(2):287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, Rubino M, Marenzi G, Colombo GI, Achilli F, Maggiolini S, Capogrossi MC, Pompilio G. Diagnostic Potential of Plasmatic MicroRNA Signatures in Stable and Unstable Angina. PLoS ONE. 2013;8(11):e80345. doi: 10.1371/journal.pone.0080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(suppl_2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das E, Jana NR, Bhattacharyya NP. MicroRNA-124 targets CCNA2 and regulates cell cycle in STHdh(Q111)/Hdh(Q111) cells. Biochem Biophys Res Commun. 2013;437(2):217–224. doi: 10.1016/j.bbrc.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Das E, Jana N, Bhattacharyya N. Delayed Cell Cycle Progression in STHdhQ111/HdhQ111 Cells, a Cell Model for Huntington’s Disease Mediated by microRNA-19a, microRNA-146a and microRNA-432. MicroRNA. 2015;4(2):86–100. doi: 10.2174/2211536604666150713105606. [DOI] [PubMed] [Google Scholar]
- de Mena L, Coto E, Cardo LF, Díaz M, Blázquez M, Ribacoba R, Salvador C, Pastor P, Samaranch Ll, Moris G, Menéndez M, Corao AI, Alvarez V. Analysis of theMicro-RNA-133andPITX3genes in Parkinson’s disease. Am J Med Genetics Part B: Neuropsychiatr Genetics. 2010;9999B:n/a–n/a. doi: 10.1002/ajmg.b.31086. [DOI] [PubMed] [Google Scholar]
- De Velasco MA, Kura Y, Sakai K, Hatanaka Y, Davies BR, Campbell H, Klein S, Kim Y, MacLeod AR, Sugimoto K, Yoshikawa K, Nishio K, Uemura H. Targeting castration-resistant prostate cancer with androgen receptor antisense oligonucleotide therapy. JCI Insight. 2019;4(17):122688. doi: 10.1172/jci.insight.122688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Liddo A, de OliveiraFreitasMachado C, Fischer S, Ebersberger S, Heumüller AW, Weigand JE, Müller-McNicoll M, Zarnack K. A combined computational pipeline to detect circular RNAs in human cancer cells under hypoxic stress. J Mol Cell Biol. 2019;11(10):829–844. doi: 10.1093/jmcb/mjz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma X-K, Li G-W, Yang L. CIRCpedia v2: An Updated Database for Comprehensive Circular RNA Annotation and Expression Comparison. Genomics Proteomics Bioinforma. 2018;16(4):226–233. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Han L-L, Xu Z-X. Suppressed microRNA-96 inhibits iNOS expression and dopaminergic neuron apoptosis through inactivating the MAPK signaling pathway by targeting CACNG5 in mice with Parkinson’s disease. Mol Med (Cambridge Mass) 2018;24(1):61. doi: 10.1186/s10020-018-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Yang S-T, Liu J, Zhang K-X, Leng J-Y. Silence of LncRNA GAS5 Protects Cardiomyocytes H9c2 against Hypoxic Injury via Sponging miR-142–5p. Mol Cells. 2019;42(5):397–405. doi: 10.14348/molcells.2018.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, Flanigan KM, Neely LA, Whitney D, Beggs AH, Kohane IS, Kunkel LM. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci. 2007;104(43):17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Dika I, Lim HY, Yong WP, Lin C, Yoon J, Modiano M, Freilich B, Choi HJ, Chao T, Kelley RK, Brown J, Knox J, Ryoo B, Yau T, Abou-Alfa GK. An Open-Label, Multicenter, Phase I, Dose Escalation Study with Phase II Expansion Cohort to Determine the Safety, Pharmacokinetics, and Preliminary Antitumor Activity of Intravenous TKM-080301 in Subjects with Advanced Hepatocellular Carcinoma. Oncologist. 2018;24(6):747. doi: 10.1634/theoncologist.2018-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza S, Scarpato M, Damiani D, Managò F, Mereu M, Contestabile A, Peruzzo O, Carninci P, Santoro C, Papaleo F, Mingozzi F, Ronzitti G, Zucchelli S, Gustincich S. SINEUP Non-coding RNA Targeting GDNF Rescues Motor Deficits and Neurodegeneration in a Mouse Model of Parkinson’s Disease. Mol Ther. 2020;28(2):642–652. doi: 10.1016/j.ymthe.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri E, Borgatti M, Montagner G, Bianchi N, Finotti A, Lampronti I, Bezzerri V, Dechecchi MC, Cabrini G, Gambari R. Expression of microRNA-93 and Interleukin-8 duringPseudomonas aeruginosa–Mediated Induction of Proinflammatory Responses. Am J Respir Cell Mol Biol. 2014;50(6):1144–1155. doi: 10.1165/rcmb.2013-0160oc. [DOI] [PubMed] [Google Scholar]
- Fabbri E, Tamanini A, Jakova T, Gasparello J, Manicardi A, Corradini R, Sabbioni G, Finotti A, Borgatti M, Lampronti I, Munari S, Dechecchi M, Cabrini G, Gambari R. A Peptide Nucleic Acid against MicroRNA miR-145-5p Enhances the Expression of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) in Calu-3 Cells. Molecules. 2017;23(1):71. doi: 10.3390/molecules23010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y (2015) Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 16(1). 10.1186/s13059-015-0706-1 [DOI] [PMC free article] [PubMed]
- Fasold M, Langenberger D, Binder H, Stadler PF, Hoffmann S. DARIO: a ncRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2011;39(suppl_2):W112–W117. doi: 10.1093/nar/gkr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS. Evidence for X-chromosomal schizophrenia associated with microRNA alterations. PLoS ONE. 2009;4(7):e6121. doi: 10.1371/journal.pone.0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Liao Y-T, He J-C, Xie C-L, Chen S-Y, Fan H-H, Su Z-P, Wang Z (2018) Plasma long noncoding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol 18(1). 10.1186/s12883-017-1008-x [DOI] [PMC free article] [PubMed]
- Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T, Müller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating MicroRNAs in Patients With Coronary Artery Disease. Circ Res. 2010;107(5):677–684. doi: 10.1161/circresaha.109.215566. [DOI] [PubMed] [Google Scholar]
- Foessl I, Kotzbeck P, Obermayer-Pietsch B (2019) miRNAs as novel biomarkers for bone related diseases. J Lab Precis Med 4:2. 10.21037/jlpm.2018.12.06
- Fragkouli A, Doxakis E (2014) miR-7 and miR-153 protect neurons against MPP+-induced cell death via upregulation of mTOR pathway. Front Cell Neurosci 8. 10.3389/fncel.2014.00182 [DOI] [PMC free article] [PubMed]
- Friedberg JW, Kim H, McCauley M, Hessel EM, Sims P, Fisher DC, Nadler LM, Coffman RL, Freedman AS. Combination immunotherapy with a CpG oligonucleotide (1018 ISS) and rituximab in patients with non-Hodgkin lymphoma: increased interferon-α/β–inducible gene expression, without significant toxicity. Blood. 2005;105(2):489–495. doi: 10.1182/blood-2004-06-2156. [DOI] [PubMed] [Google Scholar]
- Fritegotto C, Ferrati C, Pegoraro V, Angelini C. Micro-RNA expression in muscle and fiber morphometry in myotonic dystrophy type 1. Neurol Sci. 2017;38(4):619–625. doi: 10.1007/s10072-017-2811-2. [DOI] [PubMed] [Google Scholar]
- Fromm B, Domanska D, Høye E, Ovchinnikov V, Kang W, Aparicio-Puerta E, Johansen M, Flatmark K, Mathelier A, Hovig E, Hackenberg M, Friedländer MR, Peterson KJ. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 2019;48(D1):D132–D141. doi: 10.1093/nar/gkz885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Cao Y, Wu J, Peng Q, Nie Q, Xie X. UFold: fast and accurate RNA secondary structure prediction with deep learning. Nucleic Acids Res. 2021;50(3):e14–e14. doi: 10.1093/nar/gkab1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1(2):190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga T, Iwakiri J, Ono Y, Hamada M (2019) LncRRI search: a web server for lncRNA-RNA interaction prediction integrated with tissue-specific expression and subcellular localization data. Front Genet 10. 10.3389/fgene.2019.00462 [DOI] [PMC free article] [PubMed]
- Fukuoka M, Takahashi M, Fujita H, Chiyo T, Popiel HA, Watanabe S, Furuya H, Murata M, Wada K, Okada T, Nagai Y, Hohjoh H. Supplemental Treatment for Huntington’s Disease with miR-132 that Is Deficient in Huntington’s Disease Brain. Mol Ther - Nucleic Acids. 2018;11:79–90. doi: 10.1016/j.omtn.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagen M. Inherent size constraints on prokaryote gene networks due to ?accelerating? growth. Theory Biosci. 2005;123(4):381–411. doi: 10.1016/j.thbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Gagliardi S, Zucca S, Pandini C, Diamanti L, Bordoni M, Sproviero D, Arigoni M, Olivero M, Pansarasa O, Ceroni M, Calogero R, Cereda C (2018) Long non-coding and coding RNAs characterization in peripheral blood mononuclear cells and spinal cord from amyotrophic lateral sclerosis patients. Sci Rep 8(1). 10.1038/s41598-018-20679-5 [DOI] [PMC free article] [PubMed]
- Gamazon ER, Im H-K, Duan S, Lussier YA, Cox NJ, Dolan ME, Zhang W. ExprTarget: An Integrative Approach to Predicting Human MicroRNA Targets. PLoS ONE. 2010;5(10):e13534. doi: 10.1371/journal.pone.0013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambardella S, Rinaldi F, Lepore SM, Viola A, Loro E, Angelini C, Vergani L, Novelli G, Botta A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48. doi: 10.1186/1479-5876-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J-X, Li Y, Wang S-N, Chen X-C, Lin L-L, Zhang H. Overexpression of microRNA-183 promotes apoptosis of substantia nigra neurons via the inhibition of OSMR in a mouse model of Parkinson’s disease. Int J Mol Med. 2018 doi: 10.3892/ijmm.2018.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Liu Y, Guo S, Yao R, Wu L, Xiao L, Wang Z, Liu Y, Zhang Y. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell Physiol Biochem. 2017;44(4):1497–1508. doi: 10.1159/000485588. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zhao F (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16(1). 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed]
- Gao Y, Wang P, Wang Y, Ma X, Zhi H, Zhou D, Li X, Fang Y, Shen W, Xu Y, Shang S, Wang L, Wang L, Ning S, Li X. Lnc2Cancer v20: updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2018;47(D1):D1028–D1033. doi: 10.1093/nar/gky1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardini A (2017) Global Run-On sequencing (GRO-seq). Methods Mol Biol (Clifton N.J.) 1468:111–120. 10.1007/978-1-4939-4035-6_9 [DOI] [PMC free article] [PubMed]
- Gasparello J, Fabbri E, Bianchi N, Breveglieri G, Zuccato C, Borgatti M, Gambari R, Finotti A. BCL11A mRNA Targeting by miR-210: A Possible Network Regulating γ-Globin Gene Expression. Int J Mol Sci. 2017;18(12):2530. doi: 10.3390/ijms18122530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Björkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum Mol Genet. 2011;20(11):2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles 1 1Edited by J. Doudna. J Mol Biol. 2001;313(5):1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- Georgakilas G, Vlachos IS, Paraskevopoulou MD, Yang P, Zhang Y, Economides AN, Hatzigeorgiou AG (2014) microTSS: accurate microRNA transcription start site identification reveals a significant number of divergent pri-miRNAs. Nature. Communications 5(1). 10.1038/ncomms6700 [DOI] [PubMed]
- Ghafouri-Fard S, Poulet C, Malaise M, Abak A, Mahmud Hussen B, Taheriazam A, … Hallajnejad M (2021) The Emerging Role of Non-Coding RNAs in Osteoarthritis. Front Immunol 12. 10.3389/fimmu.2021.773171 [DOI] [PMC free article] [PubMed]
- Ghosal S, Das S, Sen R, Basak P, Chakrabarti J (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4. 10.3389/fgene.2013.00283 [DOI] [PMC free article] [PubMed]
- Ghose J, Sinha M, Das E, Jana NR, Bhattacharyya NP. Regulation of miR-146a by RelA/NFkB and p53 in STHdh(Q111)/Hdh(Q111) cells, a cell model of Huntington’s disease. PLoS ONE. 2011;6(8):e23837. doi: 10.1371/journal.pone.0023837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidlöf O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, Erlinge D (2013) Circulating cardioenriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord 13(1). 10.1186/1471-2261-13-12 [DOI] [PMC free article] [PubMed]
- Gillen AE, Gosalia N, Leir S-H, Harris A. microRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem J. 2011;438(1):25–32. doi: 10.1042/bj20110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani L, Sandoná M, Rotini A, Puri P, Consalvi S, Saccone V. Muscle-specific microRNAs as biomarkers of Duchenne Muscular Dystrophy progression and response to therapies. Rare Dis. 2014;2(1):e974969. doi: 10.4161/21675511.2014.974969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan T, Khvalevsky EZ, Hubert A, Gabai RM, Hen N, Segal A, Domb A, Harari G, David EB, Raskin S, Goldes Y, Goldin E, Eliakim R, Lahav M, Kopleman Y, Dancour A, Shemi A, Galun E. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6(27):24560–24570. doi: 10.18632/oncotarget.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, Nakagawa N, Xin C, Newitt R, Pandya S, Xia T-H, Liu X, Borza D-B, Grafals M, Shankland SJ, Himmelfarb J, Portilla D, Liu S, Chau BN, Duffield JS. Anti–microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Investig. 2014;125(1):141–156. doi: 10.1172/jci75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Liu C, Liu W, Xiang Y, Diao L, Guo A-Y, Han L. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017;45(D1):D79–D84. doi: 10.1093/nar/gkw835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Shao D, Xu K, Lu Z, JohnLu Z, Yang YT, Zhang QC. RISE: a database of RNA interactome from sequencing experiments. Nucleic Acids Res. 2017;46(D1):D194–D201. doi: 10.1093/nar/gkx864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragoudas ES, Adamis AP, Cunningham ET, Feinsod M, Guyer DR. Pegaptanib for Neovascular Age-Related Macular Degeneration. N Engl J Med. 2004;351(27):2805–2816. doi: 10.1056/nejmoa042760. [DOI] [PubMed] [Google Scholar]
- Greco S, De Simone M, Colussi C, Zaccagnini G, Fasanaro P, Pescatori M, Cardani R, Perbellini R, Isaia E, Sale P, Meola G, Capogrossi MC, Gaetano C, Martelli F. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 2009;23(10):3335–3346. doi: 10.1096/fj.08-128579. [DOI] [PubMed] [Google Scholar]
- Greco S, Zaccagnini G, Fuschi P, Voellenkle C, Carrara M, Sadeghi I, Bearzi C, Maimone B, Castelvecchio S, Stellos K, Gaetano C, Menicanti L, Martelli F. Increased BACE1-AS long noncoding RNA and β-amyloid levels in heart failure. Cardiovasc Res. 2017;113(5):453–463. doi: 10.1093/cvr/cvx013. [DOI] [PubMed] [Google Scholar]
- Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, Castelvecchio S, Gaetano C, Finato N, Beltrami AP, Menicanti L, Martelli F. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14(1):183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. Rfam: an RNA family database. Nucleic Acids Res. 2003;31(1):439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res. 2008;36(Web Server):W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibinga G-H. MicroRNAs: tools of mechanistic insights and biological therapeutics discovery for the rare neurogenetic syndrome Lesch-Nyhan disease (LND) Adv Genet. 2015;90:103–131. doi: 10.1016/bs.adgen.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15(7). 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed]
- Guo J-C, Fang S-S, Wu Y, Zhang J-H, Chen Y, Liu J, Wu B, Wu J-R, Li E-M, Xu L-Y, Sun L, Zhao Y. CNIT: a fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019;47(W1):W516–W522. doi: 10.1093/nar/gkz400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z-W, Xie C, Li K, Zhai X-M, Cai G-X, Yang X-X, and Wu Y-S (2019b) SELER: a database of super-enhancer-associated lncRNA- directed transcriptional regulation in human cancers. Database 2019b. 10.1093/database/baz027 [DOI] [PMC free article] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtemariam BA, Karsten V, Attarwala H, Goel V, Melch M, Clausen VA, Garg P, Vaishnaw AK, Sweetser MT, Robbie GJ, Vest J. Single-Dose Pharmacokinetics and Pharmacodynamics of Transthyretin Targeting N-acetylgalactosamine–Small Interfering Ribonucleic Acid Conjugate, Vutrisiran Healthy Subjects. Clin Pharmacol Ther. 2020;109(2):372–382. doi: 10.1002/cpt.1974. [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;39(suppl):W132–W138. doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann-Jensen M, Abdullayev I, Sandberg R, Faridani OR. Small-seq for single-cell small-RNA sequencing. Nat Protoc. 2018;13(10):2407–2424. doi: 10.1038/s41596-018-0049-y. [DOI] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013;9(6):e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Venø MT, Kjems J, Damgaard CK. miRdentify: high stringency miRNA predictor identifies several novel animal miRNAs. Nucleic Acids Res. 2014;42(16):e124. doi: 10.1093/nar/gku598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harafuji N, Schneiderat P, Walter MC, Chen Y-W. miR-411 is up-regulated in FSHD myoblasts and suppresses myogenic factors. Orphanet J Rare Dis. 2013;8(1):55. doi: 10.1186/1750-1172-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan F, Nuovo GJ, Crawford M, Boyaka PN, Kirkby S, Nana-Sinkam SP, Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE. 2012;7(11):e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haunsberger SJ, Connolly NMC, Prehn JHM (2016) miRNAmeConverter: an R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics btw660. 10.1093/bioinformatics/btw660 [DOI] [PubMed]
- Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M. MirZ: an integrated microRNA expression atlas and target prediction resource. Nucleic Acids Res. 2009;37(Web Server):W266–W272. doi: 10.1093/nar/gkp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, Nikaido I (2018) Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun 9. 10.1038/s41467-018-02866-0 [DOI] [PMC free article] [PubMed]
- Heikkinen L, Kolehmainen M, Wong G. Prediction of microRNA targets in Caenorhabditis elegans using a self-organizing map. Bioinformatics. 2011;27(9):1247–1254. doi: 10.1093/bioinformatics/btr144. [DOI] [PubMed] [Google Scholar]
- Hennessy EJ, van Solingen C, Scacalossi KR, Ouimet M, Afonso MS, Prins J, Koelwyn GJ, Sharma M, Ramkhelawon B, Carpenter S, Busch A, Chernogubova E, Matic LP, Hedin U, Maegdefessel L, Caffrey BE, Hussein MA, Ricci EP, Temel RE, Garabedian MJ. The long noncoding RNA CHROME regulates cholesterol homeostasis in primates. Nat Metab. 2018;1(1):98–110. doi: 10.1038/s42255-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her L-S, Mao S-H, Chang C-Y, Cheng P-H, Chang Y-F, Yang H-I, Chen C-M, Yang S-H. miR-196a Enhances Neuronal Morphology through Suppressing RANBP10 to Provide Neuroprotection in Huntington’s Disease. Theranostics. 2017;7(9):2452–2462. doi: 10.7150/thno.18813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé M, Ibrahim EC. MicroRNA screening identifies a link between NOVA1 expression and a low level of IKAP in familial dysautonomia. Dis Model Mech. 2016;9(8):899–909. doi: 10.1242/dmm.025841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL. Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods. 2017;14(12):1198–1204. doi: 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermüller J. Fast Mapping of Short Sequences with Mismatches, Insertions and Deletions Using Index Structures. PLoS Comput Biol. 2009;5(9):e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AP, Kirchhof P (2016) Pitx2 adjacent noncoding RNA. Circ Arrhythm Electrophysiol 9(1). 10.1161/circep.115.003808 [DOI] [PubMed]
- Hoss AG, Labadorf A, Latourelle JC, Kartha VK, Hadzi TC, Gusella JF, MacDonald ME, Chen J-F, Akbarian S, Weng Z, Vonsattel JP, Myers RH (2015) miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med Genet 8(1). 10.1186/s12920-015-0083-3 [DOI] [PMC free article] [PubMed]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA Modulates Histone Modification and mRNA Induction in the Yeast GAL Gene Cluster. Mol Cell. 2008;32(5):685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Kong M, Ye Y, Hong S, Cheng L, Jiang L. Serum miR-206 and other muscle-specific microRNAs as non-invasive biomarkers for Duchenne muscular dystrophy. J Neurochem. 2014;129(5):877–883. doi: 10.1111/jnc.12662. [DOI] [PubMed] [Google Scholar]
- Hu Y-W, Guo F-X, Xu Y-J, Li P, Lu Z-F, McVey DG, Zheng L, Wang Q, Ye JH, Kang C-M, Wu S-G, Zhao J-J, Ma X, Yang Z, Fang F-C, Qiu Y-R, Xu B-M, Xiao L, Wu Q, Wu L-M. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J Clin Investig. 2019;129(3):1115–1128. doi: 10.1172/JCI98230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Athanassiou C, Benos PV. mirConnX: condition-specific mRNA-microRNA network integrator. Nucleic Acids Res. 2011;39(suppl):W416–W423. doi: 10.1093/nar/gkr276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Morris QD, Frey BJ. Bayesian Inference of MicroRNA Targets from Sequence and Expression Data. J Comput Biol. 2007;14(5):550–563. doi: 10.1089/cmb.2007.r002. [DOI] [PubMed] [Google Scholar]
- Huang S, Tao W, Guo Z, Cao J, Huang X. Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-455-5p. Gene. 2019;701:1–8. doi: 10.1016/j.gene.2019.02.098. [DOI] [PubMed] [Google Scholar]
- Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, Zhou Y, Cui Q. HMDD v3.0: a database for experimentally supported human microRNA–disease associations. Nucleic Acids Res. 2019;47(D1):D1013–D1017. doi: 10.1093/nar/gky1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Id Said B, Malkin D. A functional variant in miR-605 modifies the age of onset in Li-Fraumeni syndrome. Cancer Genet. 2015;208(1–2):47–51. doi: 10.1016/j.cancergen.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Imamachi N, Tani H, Mizutani R, Imamura K, Irie T, Suzuki Y, Akimitsu N. BRIC-seq: A genome-wide approach for determining RNA stability in mammalian cells. Methods. 2014;67(1):55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol, 138–148. https://pubmed.ncbi.nlm.nih.gov/10786296/ [PubMed]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Jaguszewski M, Osipova J, Ghadri J-R, Napp LC, Widera C, Franke J, Fijalkowski M, Nowak R, Fijalkowska M, Volkmann I, Katus HA, Wollert KC, Bauersachs J, Erne P, Luscher TF, Thum T, Templin C. A signature of circulating microRNAs differentiates takotsubo cardiomyopathy from acute myocardial infarction. Eur Heart J. 2013;35(15):999–1006. doi: 10.1093/eurheartj/eht392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob P, Kacprowski T, Briand-Schumacher S, Heg D, Klingenberg R, Stähli BE, Jaguszewski M, Rodondi N, Nanchen D, Räber L, Vogt P, Mach F, Windecker S, Völker U, Matter CM, Lüscher TF, Landmesser U (2016) Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction. Eur Heart J ehw563. 10.1093/eurheartj/ehw563 [DOI] [PubMed]
- Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV Infection by Targeting MicroRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/nejmoa1209026. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2012;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A, Shankar R. miReader: Discovering Novel miRNAs in Species without Sequenced Genome. PLoS ONE. 2013;8(6):e66857. doi: 10.1371/journal.pone.0066857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Ma R, Wang J, Wu X, Jin S, Peng J, Tan R, Zhang T, Li Y, Wang Y (2015) LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics 16(S3). 10.1186/1471-2164-16-s3-s2 [DOI] [PMC free article] [PubMed]
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37(Database):D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Lin X, Yang L, Fan X, Wang W, Li S, Li J, Liu X, Bao M, Cui X, Yang J, Cui Q, Geng B, Cai J. AK098656, a Novel Vascular Smooth Muscle Cell-Dominant Long Noncoding RNA Promotes Hypertension. Hypertension. 2018;71(2):262–272. doi: 10.1161/hypertensionaha.117.09651. [DOI] [PubMed] [Google Scholar]
- Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, Stanton LW. Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genomics. 2010;41(3):269–274. doi: 10.1152/physiolgenomics.00019.2010. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH-L, Jia H, Vanisri RR, Pandey T, Lu Z-H, Buckley NJ, Stanton LW, Lipovich L. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA. 2008;15(1):85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Tchitchek N, Gralinski LE, Ferris MT, Eisfeld AJ, Green RR, Thomas MJ, Tisoncik-Go J, Schroth GP, Kawaoka Y, Pardo-Manuel de Villena F, Baric RS, Heise MT, Peng X, Katze MG. Annotation of long non-coding RNAs expressed in Collaborative Cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014;11(7):875–890. doi: 10.4161/rna.29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce GF. RNA evolution and the origins of life. Nature. 1989;338(6212):217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of miR-34b and miR-34c enhances α-synuclein expression in Parkinson’s disease. FEBS Lett. 2015;589(3):319–325. doi: 10.1016/j.febslet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadri S, Hinman V, Benos PV (2009) HHMMiR: efficient de novo prediction of microRNAs using hierarchical hidden Markov models. BMC Bioinform 10(S1). 10.1186/1471-2105-10-s1-s35 [DOI] [PMC free article] [PubMed]
- Kaewsapsak P, Shechner DM, Mallard W, Rinn JL, Ting AY. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Elife. 2017;6:e29224. doi: 10.7554/eLife.29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Tang Q, He J, Li L, Yang N, Yu S, Wang M, Zhang Y, Lin J, Cui T, Hu Y, Tan P, Cheng J, Zheng H, Wang D, Su X, Chen W, Huang Y. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 2022;50(D1):D326–D332. doi: 10.1093/nar/gkab997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018;46(D1):D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Yang Z, Yang F, Wang X, Tan J, Liao B. Long Noncoding RNA NEAT1 Aggravates Aβ-Induced Neuronal Damage by Targeting miR-107 in Alzheimer’s Disease. Yonsei Med J. 2019;60(7):640–650. doi: 10.3349/ymj.2019.60.7.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z-P, Xu Y-J, Wang Z-S, Sun J. RNA sequencing profiling reveals key mRNAs and long noncoding RNAs in atrial fibrillation. J Cell Biochem. 2019 doi: 10.1002/jcb.29504. [DOI] [PubMed] [Google Scholar]
- Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, Gangoda L, Mathivanan S. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J Mol Biol. 2016;428(4):688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–-The BLAST-Like Alignment Tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Fiesel FC, Belmonte KC, Hudec R, Wang W-X, Kim C, Nelson PT, Springer W, Kim J (2016) miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol Neurodegener 11(1). 10.1186/s13024-016-0121-4 [DOI] [PMC free article] [PubMed]
- Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, Pendergast MK, Goldkind SF, Lee EA, Kuniholm A, Soucy A, Vaze J, Belur NR, Fredriksen K, Stojkovska I, Tsytsykova A, Armant M, DiDonato RL, Choi J, Cornelissen L. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med. 2019;381(17):1644–1652. doi: 10.1056/nejmoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-D, Lee A, Choi J, Park Y, Kang H, Chang W, Lee M-S, Kim J. Epigenetic modulation as a therapeutic approach for pulmonary arterial hypertension. Exp Mol Med. 2015;47:e175. doi: 10.1038/emm.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kirk JM, Kim SO, Inoue K, Smola MJ, Lee DM, Schertzer MD, Wooten JS, Baker AR, Sprague D, Collins DW, Horning CR, Wang S, Chen Q, Weeks KM, Mucha PJ, Calabrese JM. Functional classification of long non-coding RNAs by k-mer content. Nat Genet. 2018;50(10):1474–1482. doi: 10.1038/s41588-018-0207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen B, Hein J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15(6):446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- Kocerha J, Xu Y, Prucha MS, Zhao D, Chan AW (2014) microRNA-128a dysregulation in transgenic Huntington’s disease monkeys. Molecular. Brain 7(1). 10.1186/1756-6606-7-46 [DOI] [PMC free article] [PubMed]
- Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2018;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kremsner PG, Ahuad Guerrero RA, Arana-Arri E, Aroca Martinez GJ, Bonten M, Chandler R, Corral G, De Block EJL, Ecker L, Gabor JJ, Garcia Lopez CA, Gonzales L, Granados González MA, Gorini N, Grobusch MP, Hrabar AD, Junker H, Kimura A, Lanata CF, Lehmann C. Efficacy and safety of the CVnCoV SARS-CoV-2 mRNA vaccine candidate in ten countries in Europe and Latin America (HERALD): a randomised, observer-blinded, placebo-controlled, phase 2b/3 trial. Lancet Infect Dis. 2022;22(3):329–340. doi: 10.1016/s1473-3099(21)00677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci USA. 2011;108(24):10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuksa PP, Amlie-Wolf A, Katanić Ž, Valladares O, Wang L-S, Leung YY. DASHR 20: integrated database of human small non-coding RNA genes and mature products. Bioinforma (Oxford England) 2019;35(6):1033–1039. doi: 10.1093/bioinformatics/bty709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients With Heart Failure. Circ Res. 2014;114(10):1569–1575. doi: 10.1161/circresaha.114.303915. [DOI] [PubMed] [Google Scholar]
- Kung JTY, Colognori D, Lee JT. Long Noncoding RNAs: Past, Present, and Future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkanjanawan T, Carter RL, Prucha MS, Yang J, Parnpai R, Chan AWS. miR-196a Ameliorates Cytotoxicity and Cellular Phenotype in Transgenic Huntington’s Disease Monkey Neural Cells. PLoS ONE. 2016;11(9):e0162788. doi: 10.1371/journal.pone.0162788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganà A, Paone A, Veneziano D, Cascione L, Gasparini P, Carasi S, Russo F, Nigita G, Macca V, Giugno R, Pulvirenti A, Shasha D, Ferro A, Croce CM. miR-EdiTar: a database of predicted A-to-I edited miRNA target sites. Bioinformatics. 2012;28(23):3166–3168. doi: 10.1093/bioinformatics/bts589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb YN. Inclisiran: First Approval. Drugs. 2021;81(3):389–395. doi: 10.1007/s40265-021-01473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller R-U. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin JJ, Nicholas G, Lee C, Gitlitz B, Vincent M, Cormier Y, Stephenson J, Ung Y, Sanborn R, Pressnail B, Nugent F, Nemunaitis J, Gleave ME, Murray N, Hao D. Phase I/II trial of custirsen (OGX-011), an inhibitor of clusterin, in combination with a gemcitabine and platinum regimen in patients with previously untreated advanced non-small cell lung cancer. J Thorac Oncol. 2012;7(3):579–586. doi: 10.1097/JTO.0b013e31823f459c. [DOI] [PubMed] [Google Scholar]
- Lee EC, Valencia T, Allerson C, Schairer A, Flaten A, Yheskel M, Kersjes K, Li J, Gatto S, Takhar M, Lockton S, Pavlicek A, Kim M, Chu T, Soriano R, Davis S, Androsavich JR, Sarwary S, Owen T, Kaplan J. Discovery and preclinical evaluation of anti-miR-17 oligonucleotide RGLS4326 for the treatment of polycystic kidney disease. Nat Commun. 2019;10(1):4148. doi: 10.1038/s41467-019-11918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, Zhang K, Church GM. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10(3):442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TB, Yang K, Ko HJ, Shim JR, Choi BH, Lee JH, Ryu JH. Successful defibrotide treatment of a patient with veno-occlusive disease after living-donor liver transplantation: A case report. Medicine. 2021;100(25):e26463. doi: 10.1097/MD.0000000000026463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leecharoenkiat K, Tanaka Y, Harada Y, Chaichompoo P, Sarakul O, Abe Y, Smith DR, Fucharoen S, Svasti S, Umemura T. Plasma microRNA-451 as a novel hemolytic marker for β0-thalassemia/HbE disease. Mol Med Rep. 2017;15(5):2495–2502. doi: 10.3892/mmr.2017.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. 2013;113(3):266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung YY, Ryvkin P, Ungar LH, Gregory BD, Wang L-S. CoRAL: predicting non-coding RNAs from small RNA-sequencing data. Nucleic Acids Res. 2013;41(14):e137–e137. doi: 10.1093/nar/gkt426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Brant E, Budak H, Zhang B. CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J Zhejiang Univ Sci B. 2021;22(4):253–284. doi: 10.1631/jzus.B2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ni Y-Q, Xu H, Xiang Q-Y, Zhao Y, Zhan J-K, He J-Y, Li S, Liu Y-S (2021) Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal transduction and targeted. Therapy 6(1). 10.1038/s41392-021-00779-x [DOI] [PMC free article] [PubMed]
- Li H, Liu X, Zhang L, Li X. LncRNA BANCR facilitates vascular smooth muscle cell proliferation and migration through JNK pathway. Oncotarget. 2017;8(70):114568–114575. doi: 10.18632/oncotarget.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5(1):1–23. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zheng L, Jiang A, Mo Y, Gong Q. Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. NeuroReport. 2018;29(13):1061–1067. doi: 10.1097/WNR.0000000000001057. [DOI] [PubMed] [Google Scholar]
- Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Can Res. 2015;75(18):3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang L, Chen L-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Li X, Zhou B, Chen L, Gou L-T, Li H, Fu X-D. GRID-seq reveals the global RNA–chromatin interactome. Nat Biotechnol. 2017;35(10):940–950. doi: 10.1038/nbt.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang X, Wang W, Du J, Wei J, Zhang Y, Wang J, Hou Y. Altered long non-coding RNA expression profile in rabbit atria with atrial fibrillation: TCONS_00075467 modulates atrial electrical remodeling by sponging miR-328 to regulate CACNA1C. J Mol Cell Cardiol. 2017;108:73–85. doi: 10.1016/j.yjmcc.2017.05.009. [DOI] [PubMed] [Google Scholar]
- Liang J, Wen J, Huang Z, Chen X, Zhang B, Chu L (2019) Small nucleolar RNAs: insight into their function in Cancer. Front Oncol 9. 10.3389/fonc.2019.00587 [DOI] [PMC free article] [PubMed]
- Liebow A, Li X, Racie T, Hettinger J, Bettencourt BR, Najafian N, Haslett P, Fitzgerald K, Holmes RP, Erbe D, Querbes W, Knight J. An Investigational RNAi Therapeutic Targeting Glycolate Oxidase Reduces Oxalate Production in Models of Primary Hyperoxaluria. J Am Soc Nephrol. 2016;28(2):494–503. doi: 10.1681/asn.2016030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Chang K-Y, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang C-S, Cunningham TJ, Head SR, Duester G, Dong PDS, Rana TM. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53(6):1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Hou S, Dai Y, Jiang N, Lin Y. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol Chem. 2019;400(9):1217–1228. doi: 10.1515/hsz-2018-0431. [DOI] [PubMed] [Google Scholar]
- Lindgreen S, Gardner PP, Krogh A. MASTR: multiple alignment and structure prediction of non-coding RNAs using simulated annealing. Bioinforma (Oxford England) 2007;23(24):3304–3311. doi: 10.1093/bioinformatics/btm525. [DOI] [PubMed] [Google Scholar]
- Ling T-Y, Wang X-L, Chai Q, Lau T-W, Koestler CM, Park SJ, Daly RC, Greason KL, Jen J, Wu L-Q, Shen W-F, Shen W-K, Cha Y-M, Lee H-C. Regulation of the SK3 channel by microRNA-499–potential role in atrial fibrillation. Heart Rhythm. 2013;10(7):1001–1009. doi: 10.1016/j.hrthm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gough J, Rost B. Distinguishing Protein-Coding from Non-Coding RNAs through Support Vector Machines. PLoS Genet. 2006;2(4):e29. doi: 10.1371/journal.pgen.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li Y, Lin B, Sheng Y, Yang L. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev Cell. 2017;42(4):333–348.e5. doi: 10.1016/j.devcel.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Z, Liu C, Zou D, Li Q, Feng C, Jing W, Luo S, Zhang Z, Ma L. LncRNAWiki 2.0: a knowledgebase of human long non-coding RNAs with enhanced curation model and database system. Nucleic Acids Res. 2021;50(D1):D190–D195. doi: 10.1093/nar/gkab998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Zhao Y, Li C-I, Stengel KR, Acharya P, Johnston G, Hiebert SW, Shyr Y. Identification of active miRNA promoters from nuclear run-on RNA sequencing. Nucleic Acids Res. 2017;45(13):e121. doi: 10.1093/nar/gkx318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang Q, Zhang J, Li C, Miao Y-R, Lei Q, Li Q, Guo A-Y. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D89–D93. doi: 10.1093/nar/gky985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fan Z, Zhao T, Cao W, Zhang L, Li H, Xie Q, Tian Y, Wang B. Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: An independent study of Han population. Exp Gerontol. 2015;72:230–238. doi: 10.1016/j.exger.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang S, Meng F, Wang J, Zhang Y, Dai E, Yu X, Li X, Jiang W. SM2miR: a database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics. 2012;29(3):409–411. doi: 10.1093/bioinformatics/bts698. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lu Z. Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clin Exp Pharmacol Physiol. 2018;45(8):841–848. doi: 10.1111/1440-1681.12932. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao M. lnCaNet: pan-cancer co-expression network for human lncRNA and cancer genes. Bioinformatics. 2016;32(10):1595–1597. doi: 10.1093/bioinformatics/btw017. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ding W, Yu W, Zhang Y, Ao X, Wang J. Long non-coding RNAs: Biogenesis, functions, and clinical significance in gastric cancer. Mol Ther - Oncolytics. 2021;23:458–476. doi: 10.1016/j.omto.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-C, Li J-R, Sun C-H, Andrews E, Chao R-F, Lin F-M, Weng S-L, Hsu S-D, Huang C-C, Cheng C, Liu C-C, Huang H-D. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2015;44(D1):D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodde V, Murgia G, Simula ER, Steri M, Floris M, Idda ML. Long Noncoding RNAs and Circular RNAs in Autoimmune Diseases. Biomolecules. 2020;10(7):1044. doi: 10.3390/biom10071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28(24):3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- Long B, Li N, Xu X-X, Li X-X, Xu X-J, Guo D, Zhang D, Wu Z-H, Zhang S-Y. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem Biophys Res Commun. 2018;495(1):312–318. doi: 10.1016/j.bbrc.2017.11.030. [DOI] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA package 2.0. Algorithms Mol Biol 6(1). 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed]
- Lu Y, Hou S, Huang D, Luo X, Zhang J, Chen J, and Xu W (2015) Expression profile analysis of circulating microRNAs and their effects on ion channels in Chinese atrial fibrillation patients. Int J Clin Exp Med 8(1):845–853. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4358520/ [PMC free article] [PubMed]
- Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, Zhang Y, Shan H, Luo X, Bai Y, Sun L, Song W, Xu C, Wang Z, Yang B. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122(23):2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- Lu Z, Zhang Q, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR, Chang HY. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165(5):1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulli V, Romania P, Morsilli O, Cianciulli P, Gabbianelli M, Testa U, Giuliani A, Marziali G. MicroRNA-486-3p Regulates γ-Globin Expression in Human Erythroid Cells by Directly Modulating BCL11A. PLoS ONE. 2013;8(4):e60436. doi: 10.1371/journal.pone.0060436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang N, Lin H, Xiao L, Maguy A, Qi X-Y, Li Y, Gao X, Dong D, Zhang Y, Bai Y, Ai J, Sun L, Lu H, Luo X-Y, Wang Z. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Investig. 2013;123(5):1939–1951. doi: 10.1172/jci62185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Liu F, Du X, Ma D, Xiong L. Changes in lncRNAs and related genes in β-thalassemia minor and β-thalassemia major. Frontiers of Medicine. 2017;11(1):74–86. doi: 10.1007/s11684-017-0503-1. [DOI] [PubMed] [Google Scholar]
- Ma L, Cao J, Liu L, Du Q, Li Z, Zou D, Bajic VB, Zhang Z. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019;47(D1):D128–D134. doi: 10.1093/nar/gky960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X-K, Xue W, Chen L-L, Yang L. CIRCexplorer pipelines for circRNA annotation and quantification from non-polyadenylated RNA-seq datasets. Methods. 2021;196:3–10. doi: 10.1016/j.ymeth.2021.02.008. [DOI] [PubMed] [Google Scholar]
- Magee R, Londin E, Rigoutsos I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat Disord. 2019;65:203–209. doi: 10.1016/j.parkreldis.2019.05.035. [DOI] [PubMed] [Google Scholar]
- Magrelli A, Azzalin G, Salvatore M, Viganotti M, Tosto F, Colombo T, Devito R, Di Masi A, Antoccia A, Lorenzetti S, Maranghi F, Mantovani A, Tanzarella C, Macino G, Taruscio D. Altered microRNA Expression Patterns in Hepatoblastoma Patients. Transl Oncol. 2009;2(3):157–163. doi: 10.1593/tlo.09124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca S, Magrelli A, Cialfi S, Lefort K, Ambra R, Alimandi M, Biolcati G, Uccelletti D, Palleschi C, Screpanti I, Candi E, Melino G, Salvatore M, Taruscio D, Talora C. Oxidative stress activation of miR-125b is part of the molecular switch for Hailey-Hailey disease manifestation. Exp Dermatol. 2011;20(11):932–937. doi: 10.1111/j.1600-0625.2011.01359.x. [DOI] [PubMed] [Google Scholar]
- Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017;45(W1):W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D, Moxon S, Dalmay T, Moulton V. MirPlex: a tool for identifying miRNAs in high-throughput sRNA datasets without a genome. J Exp Zool Part B Mol Dev Evol. 2013;320(1):47–56. doi: 10.1002/jez.b.22483. [DOI] [PubMed] [Google Scholar]
- Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23(7):1080–1087. doi: 10.1261/rna.060814.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E, Russo C, Florio T, Dieci G, Cancedda R, Pagano A. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41(2):308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8(3):209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5(4):316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(Web Server):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen C-K, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan PJ, Molloy K, Cryan SA, McElvaney NG, Greene CM. Long noncoding RNA are aberrantly expressed in vivo in the cystic fibrosis bronchial epithelium. Int J Biochem Cell Biol. 2014;52:184–191. doi: 10.1016/j.biocel.2014.02.022. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, Alfano L, Gomez AM, Lewis S, Kota J, Malik V, Shontz K, Walker CM, Flanigan KM, Corridore M, Kean JR, Allen HD, Shilling C, Melia KR, Sazani P. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74(5):637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- Menne J, Eulberg D, Beyer D, Baumann M, Saudek F, Valkusz Z, Więcek A, Haller H (2016) C-C motif-ligand 2 inhibition with emapticap pegol (NOX-E36) in type 2 diabetic patients with albuminuria. Nephrol Dial Transplant gfv459. 10.1093/ndt/gfv459 [DOI] [PMC free article] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, Shieh PB, Tulinius M, Mazzone ES, Montes J, Bishop KM, Yang Q, Foster R, Gheuens S, Bennett CF, Farwell W. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med. 2018;378(7):625–635. doi: 10.1056/nejmoa1710504. [DOI] [PubMed] [Google Scholar]
- Meseguer S, Martínez-Zamora A, García-Arumí E, Andreu AL, Armengod M-E. The ROS-sensitive microRNA-9/9* controls the expression of mitochondrial tRNA-modifying enzymes and is involved in the molecular mechanism of MELAS syndrome. Hum Mol Genet. 2015;24(1):167–184. doi: 10.1093/hmg/ddu427. [DOI] [PubMed] [Google Scholar]
- Metkar M, Ozadam H, Lajoie BR, Imakaev M, Mirny LA, Dekker J, Moore MJ. Higher-Order Organization Principles of Pre-translational mRNPs. Mol Cell. 2018;72(4):715–726.e3. doi: 10.1016/j.molcel.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhuantong W, Wichadakul D. MicroPC (μPC): A comprehensive resource for predicting and comparing plant microRNAs. BMC Genomics. 2009;10(1):366. doi: 10.1186/1471-2164-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting C-C, Alexanian M, Maric D, Maison D, Nemir M, Young RA, Schroen B, González A, Ounzain S, Pedrazzini T. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. 2017;9(395):eaai9118. doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Nakamura A, Aoki Y, Ito N, Kishi S, Yamamoto K, Sekiguchi M, Takeda S, Hashido K. Identification of Muscle-Specific MicroRNAs in Serum of Muscular Dystrophy Animal Models: Promising Novel Blood-Based Markers for Muscular Dystrophy. PLoS ONE. 2011;6(3):e18388. doi: 10.1371/journal.pone.0018388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Ecker JR. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85(1):1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Smith JA, Shamu CE, Neumüller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol. 2014;15(9):591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J, Wingett SW, Farabella I, Cairns J, Furlan-Magaril M, Jiménez-García LF, Liu X, Craig FF, Walker S, Segonds-Pichon A, Andrews S, Marti-Renom MA, Fraser P. RNA proximity sequencing reveals the spatial organization of the transcriptome in the nucleus. Nat Biotechnol. 2019;37(7):793–802. doi: 10.1038/s41587-019-0166-3. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TA, Wilcox I, Luo H-Y, Farrell JJ, Kurita R, Nakamura Y, Murphy GJ, Cui S, Steinberg MH, Chui DHK. A long noncoding RNA from the HBS1L-MYB intergenic region on chr6q23 regulates human fetal hemoglobin expression. Blood Cells Mol Dis. 2018;69:1–9. doi: 10.1016/j.bcmd.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci. 2007;104(25):10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchia F, Basu S, Petrosino G, Salvemini M, Sanges R. Annocript: a flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics. 2015;31(13):2199–2201. doi: 10.1093/bioinformatics/btv106. [DOI] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8(3):278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Nalluri JJ, Barh D, Azevedo V, Ghosh P. miRsig: a consensus-based network inference methodology to identify pan-cancer miRNA-miRNA interaction signatures. Sci Rep. 2017;7(1):39684. doi: 10.1038/srep39684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37(Web Server issue):W356–362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, Scala E, Fadda P, Cristofoletti C, Facchiano A, Frontani M, Monopoli A, Ferracin M, Negrini M, Lombardo GA, Caprini E, Russo G. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sézary syndrome. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25(10):1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TC, Cao X, Yu P, Xiao S, Lu J, Biase FH, Sridhar B, Huang N, Zhang K, Zhong S. Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nat Commun. 2016;7:12023. doi: 10.1038/ncomms12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido I. EICO (Expression-based Imprint Candidate Organizer): finding disease-related imprinted genes. Nucleic Acids Res. 2004;32(90001):548D–551. doi: 10.1093/nar/gkh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby IK, Chotirmall SH, McElvaney NG, Greene CM. Regulation of Cystic Fibrosis Transmembrane Conductance Regulator by MicroRNA-145, -223, and -494 Is Altered in ΔF508 Cystic Fibrosis Airway Epithelium. J Immunol. 2013;190(7):3354–3362. doi: 10.4049/jimmunol.1202960. [DOI] [PubMed] [Google Scholar]
- Paco S, Casserras T, Rodríguez MA, Jou C, Puigdelloses M, Ortez CI, Diaz-Manera J, Gallardo E, Colomer J, Nascimento A, Kalko SG, Jimenez-Mallebrera C. Transcriptome Analysis of Ullrich Congenital Muscular Dystrophy Fibroblasts Reveals a Disease Extracellular Matrix Signature and Key Molecular Regulators. PLoS ONE. 2015;10(12):e0145107. doi: 10.1371/journal.pone.0145107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC. RNAdb–a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2004;33(Database issue):D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano L, Estivill X, Martí E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2009;38(5):e34–e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar B, Omenn GS, Guan Y (2017) miRmine: a database of human miRNA expression profiles. Bioinformatics btx019. 10.1093/bioinformatics/btx019 [DOI] [PMC free article] [PubMed]
- Park EJ, Choi J, Lee KC, Na DH. Emerging PEGylated non-biologic drugs. Expert Opin Emerg Drugs. 2019;24(2):107–119. doi: 10.1080/14728214.2019.1604684. [DOI] [PubMed] [Google Scholar]
- Pattnaik B, Patnaik N, Mittal S, Mohan A, Agrawal A, Guleria R, Madan K. Micro RNAs as potential biomarkers in tuberculosis: A systematic review. Non-Coding RNA Res. 2022;7(1):16–26. doi: 10.1016/j.ncrna.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MM, Muntoni F. Noncoding RNAs and Duchenne muscular dystrophy. Epigenomics. 2016;8(11):1527–1537. doi: 10.2217/epi-2016-0088. [DOI] [PubMed] [Google Scholar]
- Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, Gomez A, Huertas D, Esteller M. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol. 2013;10(7):1197–1203. doi: 10.4161/rna.24286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierdomenico AM, Patruno S, Codagnone M, Simiele F, Mari VC, Plebani R, Recchiuti A, Romano M. microRNA-181b is increased in cystic fibrosis cells and impairs lipoxin A4 receptor-dependent mechanisms of inflammation resolution and antimicrobial defense. Sci Rep. 2017;7(1):13519. doi: 10.1038/s41598-017-14055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliatsika V, Loher P, Magee R, Telonis AG, Londin E, Shigematsu M, Kirino Y, Rigoutsos I. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2017;46(D1):D152–D159. doi: 10.1093/nar/gkx1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman T, Lagos D. Non-Coding RNAs in COVID-19: Emerging Insights and Current Questions. Non-Coding RNA. 2021;7(3):54. doi: 10.3390/ncrna7030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/nejmoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya O, Kananykhina E, Roy-Engel AM, Nazarenko O, Kulemzina I, Baranova A, Vassetsky Y, Myakishev-Rempel M. The role of Alu-derived RNAs in Alzheimer’s and other neurodegenerative conditions. Med Hypotheses. 2018;115:29–34. doi: 10.1016/j.mehy.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Pradhan RK, Ramakrishna W. Transposons: Unexpected players in cancer. Gene. 2022;808:145975. doi: 10.1016/j.gene.2021.145975. [DOI] [PubMed] [Google Scholar]
- Prajapati B, Fatma M, Maddhesiya P, Sodhi MK, Fatima M, Dargar T, Bhagat R, Seth P, Sinha S. Identification and epigenetic analysis of divergent long non-coding RNAs in multilineage differentiation of human Neural Progenitor Cells. RNA Biol. 2019;16(1):13–24. doi: 10.1080/15476286.2018.1553482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preusse M, Theis FJ, Mueller NS. miTALOS v2: Analyzing Tissue Specific microRNA Function. PLoS ONE. 2016;11(3):e0151771. doi: 10.1371/journal.pone.0151771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Ye Y, Mao H, Yao L, Sun X, Wang B, Zhang H, Xie L, Zhang H, Zhang Y, Zhang S, He X. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222 /p27/mTOR pathway in Parkinson’s disease. Exp Cell Res. 2019;384(1):111614. doi: 10.1016/j.yexcr.2019.111614. [DOI] [PubMed] [Google Scholar]
- Qin Y, Buermans HPJ, van Kester MS, van der Fits L, Out-Luiting JJ, Osanto S, Willemze R, Vermeer MH, Tensen CP. Deep-sequencing analysis reveals that the miR-199a2/214 cluster within DNM3os represents the vast majority of aberrantly expressed microRNAs in Sézary syndrome. J Invest Dermatol. 2012;132(5):1520–1522. doi: 10.1038/jid.2011.481. [DOI] [PubMed] [Google Scholar]
- Quek XC, Thomson DW, Maag JLV, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME. lncRNAdb v20: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2014;43(D1):D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Karp PH, Jiang P, Ostedgaard LS, Walz AE, Fisher JT, Keshavjee S, Lennox KA, Jacobi AM, Rose SD, Behlke MA, Welsh MJ, Xing Y, McCray PB. A microRNA network regulates expression and biosynthesis of wild-type and ΔF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci USA. 2012;109(33):13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science (New York N.Y.) 2007;316(5829):1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- Reynolds RH, Petersen MH, Willert CW, Heinrich M, Nymann N, Dall M, Treebak JT, Björkqvist M, Silahtaroglu A, Hasholt L, Nørremølle A. Perturbations in the p53/miR-34a/SIRT1 pathway in the R6/2 Huntington’s disease model. Mol Cell Neurosci. 2018;88:118–129. doi: 10.1016/j.mcn.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Richardson PG, Smith AR, Triplett BM, Kernan NA, Grupp SA, Antin JH, Lehmann L, Shore T, Iacobelli M, Miloslavsky M, Hume R, Hannah AL, Nejadnik B, Soiffer RJ. Defibrotide for Patients with Hepatic veno-occlusive disease/sinusoidal obstruction syndrome: interim results from a treatment IND study. Biol Blood Marrow Transplant. 2017;23(6):997–1004. doi: 10.1016/j.bbmt.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Ritchie W, Flamant S, Rasko JEJ. mimiRNA: a microRNA expression profiler and classification resource designed to identify functional correlations between microRNAs and their targets. Bioinformatics. 2009;26(2):223–227. doi: 10.1093/bioinformatics/btp649. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF. The Origins of the RNA World. Cold Spring Harb Perspect Biol. 2010;4(5):a003608–a003608. doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, Bhattacharya G, Lahiri A, Dasgupta UB, Banerjee D, Chandra S, Das M. hsa-miR-503 Is Downregulated in β Thalassemia Major. Acta Haematol. 2012;128(3):187–189. doi: 10.1159/000339492. [DOI] [PubMed] [Google Scholar]
- Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, Ye Y, Zhang Z, Mills T, Feng J, Liu C-J, Jing J, Cao J, Zhou B, Wang L, Zhou Y, Lin C, Guo A-Y, Chen X, and Diao L (2019) Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med 11(1). 10.1186/s13073-019-0663-5 [DOI] [PMC free article] [PubMed]
- Ruffo P, Strafella C, Cascella R, Caputo V, Conforti FL, Andò S, Giardina E (2021) Deregulation of ncRNA in neurodegenerative disease: focus on circRNA, lncRNA and miRNA in amyotrophic lateral sclerosis. Front Genet 12. 10.3389/fgene.2021.784996 [DOI] [PMC free article] [PubMed]
- Russo F, Di Bella S, Nigita G, Macca V, Laganà A, Giugno R, Pulvirenti A, Ferro A. miRandola: extracellular circulating microRNAs database. PLoS ONE. 2012;7(10):e47786. doi: 10.1371/journal.pone.0047786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman SM, Ackley A, Burdach J, Clemson M, Gruenert DC, Tachikawa K, Chivukula P, Weinberg MS, Morris KV. Long non-coding RNA bgas regulates the cystic fibrosis transmembrane conductance regulator. Mol Ther. 2016;24(8):1351–1357. doi: 10.1038/mt.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablok G, Milev I, Minkov G, Minkov I, Varotto C, Yahubyan G, Baev V. isomiRex: Web-based identification of microRNAs, isomiR variations and differential expression using next-generation sequencing datasets. FEBS Lett. 2013;587(16):2629–2634. doi: 10.1016/j.febslet.2013.06.047. [DOI] [PubMed] [Google Scholar]
- Sahin U, Oehm P, Derhovanessian E, Jabulowsky RA, Vormehr M, Gold M, Maurus D, Schwarck-Kokarakis D, Kuhn AN, Omokoko T, Kranz LM, Diken M, Kreiter S, Haas H, Attig S, Rae R, Cuk K, Kemmer-Brück A, Breitkreuz A, Tolliver C. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585(7823):107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- Saki N, Abroun S, Soleimani M, Kavianpour M, Shahjahani M, Mohammadi-Asl J, Hajizamani S. MicroRNA expression in β-thalassemia and sickle cell disease: a role in the induction of fetal hemoglobin. Cell J. 2016;17(4):583–592. doi: 10.22074/cellj.2016.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat Chem Biol. 2010;6(10):733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, Romualdi C. MAGIA, a web-based tool for miRNA and genes integrated analysis. Nucleic Acids Res. 2010;38(Web Server):W352–W359. doi: 10.1093/nar/gkq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta E, De Strooper B. Noncoding RNAs in neurodegeneration. Nat Rev Neurosci. 2017;18(10):627–640. doi: 10.1038/nrn.2017.90. [DOI] [PubMed] [Google Scholar]
- Salvatore M, Magrelli A, Taruscio D. The role of microRNAs in the biology of rare diseases. Int J Mol Sci. 2011;12(10):6733–6742. doi: 10.3390/ijms12106733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Q, Liu X, Wang L, Qi L, Sun W, Wang W, Sun Y, Zhang H. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging. 2018;10(6):1281–1293. doi: 10.18632/aging.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RD, Raal FJ, Donovan JM, Cromwell WC. Mipomersen preferentially reduces small low-density lipoprotein particle number in patients with hypercholesterolemia. J Clin Lipidol. 2015;9(2):201–209. doi: 10.1016/j.jacl.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR (2013) Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non genetic gene-expression heterogeneity. Genome Biol 14(4). 10.1186/gb-2013-14-4-r31 [DOI] [PMC free article] [PubMed]
- Saus E, Willis JR, Pryszcz LP, Hafez A, Llorens C, Himmelbauer H, Gabaldón T. nextPARS: parallel probing of RNA structures in Illumina. RNA. 2018;24(4):609–619. doi: 10.1261/rna.063073.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglioni D, Catapano F, Ellis M, Torelli S, Chambers D, Feng L, Beck M, Sewry C, Monforte M, Harriman S, Koenig E, Malhotra J, Popplewell L, Guglieri M, Straub V, Mercuri E, Servais L, Phadke R, Morgan J, Muntoni F (2021) The administration of antisense oligonucleotide golodirsen reduces pathological regeneration in patients with Duchenne muscular dystrophy. Acta Neuropathol Commun 9(1). 10.1186/s40478-020-01106-1 [DOI] [PMC free article] [PubMed]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, Simon MD. timelapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat Methods. 2018;15(3):221–225. doi: 10.1038/nmeth.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis B, Strumberg D, Kuhlmann J, Wolf M, Link K, Seufferlein T, Kaufmann J, Feist M, Gebhardt F, Khan M, Stintzing S, Pelzer U. Safety, efficacy and pharcacokinetics of targeted therapy with the liposomal rna interference therapeutic Atu027 Combined with gemcitabine in patients with pancreatic adenocarcinoma. a randomized phase Ib/IIa study. Cancers. 2020;12(11):3130. doi: 10.3390/cancers12113130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, Lange C, Giese K, Kaufmann J, Khan M, Drevs J. First-in-human phase i study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol. 2014;32(36):4141–4148. doi: 10.1200/jco.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarström P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121(1):73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu Rev Physiol. 2014;76:39–56. doi: 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2005;12(2):192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y, Yang B. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83(3):465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ. Global mapping of human RNA-RNA interactions. Mol Cell. 2016;62(4):618–626. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- Shen C, Kong B, Liu Y, Xiong L, Shuai W, Wang G, Quan D, Huang H. YY1-induced upregulation of lncRNA KCNQ1OT1 regulates angiotensin II-induced atrial fibrillation by modulating miR-384b/CACNA1C axis. Biochem Biophys Res Commun. 2018;505(1):134–140. doi: 10.1016/j.bbrc.2018.09.064. [DOI] [PubMed] [Google Scholar]
- Shirley M. Casimersen: First Approval. Drugs. 2021 doi: 10.1007/s40265-021-01512-2. [DOI] [PubMed] [Google Scholar]
- Shuang C, Guo M, Wang C, Liu X, Liu Y, Wu X. MiRTDL: A deep learning approach for miRNA target prediction. IEEE/ACM Trans Comput Biol Bioinforma. 2016;13(6):1161–1169. doi: 10.1109/TCBB.2015.2510002. [DOI] [PubMed] [Google Scholar]
- Sinha D, Sengupta D, Bandyopadhyay S (2017) ParSel: parallel selection of Micro-RNAs for survival classification in cancers. Molecular Informatics 36(7). 10.1002/minf.201600141 [DOI] [PubMed]
- Siwaponanan P, Fucharoen S, Sirankapracha P, Winichagoon P, Umemura T, Svasti S. Elevated levels of miR-210 correlate with anemia in β-thalassemia/HbE patients. Int J Hematol. 2016;104(3):338–343. doi: 10.1007/s12185-016-2032-0. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179(5):1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhang N, Han P, Moon B-S, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87–e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville F, Ruffin M, Coraux C, Rousselet N, Le Rouzic P, Blouquit-Laye S, Corvol H, Tabary O (2017) MicroRNA-9 downregulates the ANO1 chloride channel and contributes to cystic fibrosis lung pathology. Nature. Communications 8(1). 10.1038/s41467-017-00813-z [DOI] [PMC free article] [PubMed]
- Srinoun K, Nopparatana C, Wongchanchailert M, Fucharoen S. MiR-155 enhances phagocytic activity of β-thalassemia/HbE monocytes via targeting of BACH1. Int J Hematol. 2017;106(5):638–647. doi: 10.1007/s12185-017-2291-4. [DOI] [PubMed] [Google Scholar]
- Stasiewicz J, Mukherjee S, Nithin C, Bujnicki JM (2019) QRNAS: software tool for refinement of nucleic acid structures. BMC Struct Biol 19(1). 10.1186/s12900-019-0103-1 [DOI] [PMC free article] [PubMed]
- Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krššák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, Grauert M, Adolf GR, Kraut N, Peters J-M, Rettig WJ. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17(4):316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Stegmayer G, Yones C, Kamenetzky L, Milone DH. High class-imbalance in pre-miRNA prediction: a novel approach based on deepSOM. IEEE/ACM Trans Comput Biol Bioinf. 2017;14(6):1316–1326. doi: 10.1109/tcbb.2016.2576459. [DOI] [PubMed] [Google Scholar]
- Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13(10):e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics. 2012;28(15):2059–2061. doi: 10.1093/bioinformatics/bts311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Guo Z, Zhang C, Che H, Gong W, Shen Z, Shi Y, and Ge S (2019) LncRNA NRON alleviates atrial fibrosis through suppression of M1 macrophages activated by atrial myocytes. Biosci Reports 39(11):BSR20192215. 10.1042/BSR20192215 [DOI] [PMC free article] [PubMed]
- Sun K, Chen X, Jiang P, Song X, Wang H, Sun H (2013) iSeeRNA: identification of long intergenic noncoding RNA transcripts from transcriptome sequencing data. BMC Genomics 14(S2). 10.1186/1471-2164-14-s2-s7 [DOI] [PMC free article] [PubMed]
- Sun L, Liu H, Zhang L, Meng J. lncRScan-SVM: a tool for predicting long non-coding RNAs using support vector machine. PLoS ONE. 2015;10(10):e0139654. doi: 10.1371/journal.pone.0139654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Sun S, Zeng S, Li Y, Pan W, Zhang Z. Expression of circulating microRNA-1 and microRNA-133 in pediatric patients with tachycardia. Mol Med Rep. 2015;11(6):4039–4046. doi: 10.3892/mmr.2015.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Nie X, Sun S, Dong S, Yuan C, Li Y, Xiao B, Jie D, Liu Y. Long non-coding RNA MEG3 downregulation triggers human pulmonary artery smooth muscle cell proliferation and migration via the p53 signaling pathway. Cellul Physiol Biochem. 2017;42(6):2569–2581. doi: 10.1159/000480218. [DOI] [PubMed] [Google Scholar]
- Sweeney BA, Petrov AI, Burkov B, Finn RD, Bateman A, Szymanski M, Karlowski WM, Gorodkin J, Seemann SE, Cannone JJ, Gutell RR, Fey P, Basu S, Kay S, Cochrane G, Billis K, Emmert D, Marygold SJ, Huntley RP, Lovering RC. RNAcentral: a hub of information for non-coding RNA sequences. Nucleic Acids Res. 2018;47(D1):D221–D229. doi: 10.1093/nar/gky1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16(1). 10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed]
- Szcześniak MW, Makałowska I. miRNEST 2.0: a database of plant and animal microRNAs. Nucleic Acids Res. 2013;42(D1):D74–D77. doi: 10.1093/nar/gkt1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szcześniak MW, Rosikiewicz W, Makałowska I. CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol. 2015;57(1):e8–e8. doi: 10.1093/pcp/pcv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Wang ET, Burge CB. Genomic analysis of RNA localization. RNA Biol. 2014;11(8):1040–1050. doi: 10.4161/rna.32146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K, Akimitsu N (2012) Long non-coding RNAs in cancer progression. Front Genet 3. 10.3389/fgene.2012.00219 [DOI] [PMC free article] [PubMed]
- Täubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, Rode L, Weigt H, Genschel C, Lorch U, Theek C, Levin AA, Bauersachs J, Solomon SD, Thum T. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J. 2020;42(2):178–188. doi: 10.1093/eurheartj/ehaa898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116(4):751–762. doi: 10.1161/circresaha.116.303549. [DOI] [PubMed] [Google Scholar]
- Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild A-C, Tsay M, Lu R, Jurisica I. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(Database issue):D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Liu S. CPPred: coding potential prediction based on the global description of RNA sequence. Nucleic Acids Res. 2019;47(8):e43–e43. doi: 10.1093/nar/gkz087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Gu P, Xu S, Lin X. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem. 2015;79(5):732–737. doi: 10.1080/09168451.2014.998617. [DOI] [PubMed] [Google Scholar]
- Tong Z, Cui Q, Wang J, Zhou Y. TransmiR v2.0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019;47(D1):D253–D258. doi: 10.1093/nar/gky1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toraih EA, El-Wazir A, Alghamdi SA, Alhazmi AS, El-Wazir M, Abdel-Daim MM, Fawzy MS. Association of long non-coding RNA MIAT and MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events. Genet Mol Biol. 2019;42(3):509–518. doi: 10.1590/1678-4685-gmb-2018-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- Triozzi P, Kooshki M, Alistar A, Bitting R, Neal A, Lametschwandtner G, and Loibner H (2015) Phase I clinical trial of adoptive cellular immunotherapy with APN401 in patients with solid tumors. J ImmunoTherapy Cancer, 3(S2). 10.1186/2051-1426-3-s2-p175
- Twayana S, Legnini I, Cesana M, Cacchiarelli D, Morlando M, Bozzoni I. Biogenesis and function of non-coding RNAs in muscle differentiation and in Duchenne muscular dystrophy. Biochem Soc Trans. 2013;41(4):844–849. doi: 10.1042/BST20120353. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Vaz C, Gupta V, Bhatia R, Maheshwari S, Srinivasan A, Bhattacharya A. CID-miRNA: a web server for prediction of novel miRNA precursors in human genome. Biochem Biophys Res Commun. 2008;372(4):831–834. doi: 10.1016/j.bbrc.2008.05.134. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Fernández AF, Lopez-Nieva P, Rossi S, Huertas D, Kulis M, Liu C-G, Croce CM, Calin GA, Esteller M. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of Rett syndrome. Epigenetics. 2010;5(7):656–663. doi: 10.4161/epi.5.7.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilov AV, Underwood JG. High-throughput nuclease probing of RNA structures using FragSeq. Methods Mol Biol (Clifton N.J.) 2016;1490:105–134. doi: 10.1007/978-1-4939-6433-8_8. [DOI] [PubMed] [Google Scholar]
- van Rooij E. The Art of MicroRNA Research. Circ Res. 2011;108(2):219–234. doi: 10.1161/circresaha.110.227496. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci. 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey DL, Cooper WA, Kritharides L, Ridley L, Pattison ST, MacDiarmid J, Brahmbhatt H, Reid G. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18(10):1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115(7):668–677. doi: 10.1161/circresaha.115.303836. [DOI] [PubMed] [Google Scholar]
- Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, Just A, Fendrich J, Scherf K, Bolesani E, Schambach A, Weidemann F, Zweigerdt R, de Windt LJ, Engelhardt S, Dandekar T (2016) Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 8(326). 10.1126/scitranslmed.aaf1475 [DOI] [PubMed]
- Vitravene Study Group A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with AIDS. Am J Ophthalmol. 2002;133(4):467–474. doi: 10.1016/s0002-9394(02)01327-2. [DOI] [PubMed] [Google Scholar]
- Vitsios DM, Enright AJ. Chimira: analysis of small RNA sequencing data and microRNA modifications: Fig. 1. Bioinformatics. 2015;31(20):3365–3367. doi: 10.1093/bioinformatics/btv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitsios DM, Kentepozidou E, Quintais L, Benito-Gutiérrez E, van Dongen S, Davis MP, Enright AJ. Mirnovo: genome-free prediction of microRNAs from small RNA sequencing data and single-cells using decision forests. Nucleic Acids Res. 2017;45(21):e177. doi: 10.1093/nar/gkx836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Research. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders P-J, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2012;41(D1):D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Gao J, Zhang H, Jiang X, Zang Q, Ban R, Zhang Y, Shi Q. CPSS 2.0: a computational platform update for the analysis of small RNA sequencing data. Bioinformatics. 2017;33(20):3289–3291. doi: 10.1093/bioinformatics/btx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wei L, Guo M, Zou Q. Computational approaches in detecting non- coding RNA. Curr Genomics. 2013;14(6):371–377. doi: 10.2174/13892029113149990005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song T, Zhao Y, Zhao J, Wang X, Fu X. Long non-coding RNA LICPAR regulates atrial fibrosis via TGF-β/Smad pathway in atrial fibrillation. Tissue Cell. 2020;67:101440. doi: 10.1016/j.tice.2020.101440. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu C-Y, Zhou L-Y, Wang J-X, Wang M, Zhao B, Zhao W-K, Xu S-J, Fan L-H, Zhang X-J, Feng C, Wang C-Q, Zhao Y-F, and Li P-F (2015) APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188–3p. Nat Commun 6(1). 10.1038/ncomms7779 [DOI] [PubMed]
- Wang K, Liu F, Liu C-Y, An T, Zhang J, Zhou L-Y, Wang M, Dong Y-H, Li N, Gao J-N, Zhao Y-F, Li P-F. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23(8):1394–1405. doi: 10.1038/cdd.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liu F, Zhou L-Y, Long B, Yuan S-M, Wang Y, Liu C-Y, Sun T, Zhang X-J, Li P-F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114(9):1377–1388. doi: 10.1161/circresaha.114.302476. [DOI] [PubMed] [Google Scholar]
- Wang K, Long B, Zhou L-Y, Liu F, Zhou Q-Y, Liu C-Y, Fan Y-Y, and Li P-F (2014b) CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 5(1). 10.1038/ncomms4596 [DOI] [PubMed]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, MacLeod JN, Chiang DY, Prins JF, Liu J. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18):e178–e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Sun T, Li N, Wang Y, Wang J-X, Zhou L-Y, Long B, Liu C-Y, Liu F, Li P-F (2014) MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PLoS Genet 10(7):e1004467. 10.1371/journal.pgen.1004467 [DOI] [PMC free article] [PubMed]
- Wang L, Liu Y, Zhong X, Liu H, Lu C, Li C, Zhang H. DMfold: a novel method to predict RNA secondary structure with pseudoknots based on deep learning and improved base pair maximization principle. Front Genet. 2019;10:143. doi: 10.3389/fgene.2019.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhou L, Jiang P, Lu L, Chen X, Lan H, Guttridge DC, Sun H, Wang H. Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol Ther. 2012;20(6):1222–1233. doi: 10.1038/mt.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang X, Guo Y, Rong H, Liu T. The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget. 2017;8(15):24449–24456. doi: 10.18632/oncotarget.15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang X, Zhang Y, Li Z, Xie X, Wang J, Gao M, Zhang S, Hou Y. Transcriptome analysis of canine cardiac fat pads: involvement of two novel long non-coding RNAs in atrial fibrillation neural remodeling. J Cell Biochem. 2015;116(5):809–821. doi: 10.1002/jcb.25037. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhuang Q, Ji K, Wen B, Lin P, Zhao Y, Li W, Yan C. Identification of miRNA, lncRNA and mRNA-associated ceRNA networks and potential biomarker for MELAS with mitochondrial DNA A3243G mutation. Sci Rep. 2017;7:41639. doi: 10.1038/srep41639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yong C, Yu K, Yu R, Zhang R, Yu L, Li S, Cai S. Long noncoding RNA (lncRNA) n379519 promotes cardiac fibrosis in post-infarct myocardium by targeting miR-30. Med Sci Monitor. 2018;24:3958–3965. doi: 10.12659/msm.910000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21(18):3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang M, Liu H. LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer’s disease. Biosci Biotechnol Biochem. 2019;83(4):609–621. doi: 10.1080/09168451.2018.1562874. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Z, Zhang W, Wang Y. Long noncoding RNA LINC00968 promotes endothelial cell proliferation and migration via regulating miR-9-3p expression. J Cell Biochem. 2018 doi: 10.1002/jcb.28103. [DOI] [PubMed] [Google Scholar]
- Wang X-M, Li X-M, Song N, Zhai H, Gao X-M, Yang Y-N. Long non-coding RNAs H19, MALAT1 and MIAT as potential novel biomarkers for diagnosis of acute myocardial infarction. Biomed Pharmacother=Biomedecine & Pharmacotherapie. 2019;118:109208. doi: 10.1016/j.biopha.2019.109208. [DOI] [PubMed] [Google Scholar]
- Wang Y-N-Z, Shan K, Yao M-D, Yao J, Wang J-J, Li X, Liu B, Zhang Y-Y, Ji Y, Jiang Q, Yan B. Long Noncoding RNA-GAS5. Hypertension. 2016;68(3):736–748. doi: 10.1161/hypertensionaha.116.07259. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang X-J, Ji Y-X, Zhang P, Deng K-Q, Gong J, Ren S, Wang X, Chen I, Wang H, Gao C, Yokota T, Ang YS, Li S, Cass A, Vondriska TM, Li G, Deb A, Srivastava D, Yang H-T. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22(10):1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S. Prediction of structural noncoding RNAs with RNAz. Methods Mol Biol. 2007;395:503–526. doi: 10.1007/978-1-59745-514-5_32. [DOI] [PubMed] [Google Scholar]
- Wen X, Gao L, Guo X, Li X, Huang X, Wang Y, Xu H, He R, Jia C, Liang F (2018) lncSLdb: a resource for long non-coding RNA subcellular localization. Database 2018. 10.1093/database/bay085 [DOI] [PMC free article] [PubMed]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, Kempf T, Wollert KC, Thum T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51(5):872–875. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1(3):1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 Delays ALS Progression and Promotes Regeneration of Neuromuscular Synapses in Mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics — challenges and potential solutions. Nat Rev Drug Discovery. 2021 doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong NW, Chen Y, Chen S, Wang X. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinforma (Oxford England) 2018;34(4):713–715. doi: 10.1093/bioinformatics/btx627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhao Z-A, Liu J, Hao K, Yu Y, Han X, Li J, Wang Y, Lei W, Dong N, Shen Z, Hu S. Long noncoding RNA Meg3 regulates cardiomyocyte apoptosis in myocardial infarction. Gene Ther. 2018;25(8):511–523. doi: 10.1038/s41434-018-0045-4. [DOI] [PubMed] [Google Scholar]
- Wu W, Ji P, and Zhao F (2020) CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol 21(1). 10.1186/s13059-020-02018-y [DOI] [PMC free article] [PubMed]
- Wu X, Pan Y, Fang Y, Zhang J, Xie M, Yang F, Yu T, Ma P, Li W, Shu Y. The biogenesis and functions of piRNAs in human diseases. Mol Ther - Nucleic Acids. 2020;21:108–120. doi: 10.1016/j.omtn.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-Y, and Kuo H-C (2020) Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J Biomed Sci 27(1). 10.1186/s12929-020-00636-z [DOI] [PMC free article] [PubMed]
- Wu Y-Y, Chiu F-L, Yeh C-S, Kuo H-C (2019) Opportunities and challenges for the use of induced pluripotent stem cells in modelling neurodegenerative disease. Open Biol 9(1). 10.1098/rsob.180177 [DOI] [PMC free article] [PubMed]
- Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, Wei L, Han L, He C. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C (2016) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform bbw081. 10.1093/bib/bbw081 [DOI] [PubMed]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, and Li T (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37(Database):D105–D110. 10.1093/nar/gkn851 [DOI] [PMC free article] [PubMed]
- Xie B, Ding Q, Han H, Wu D. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013;29(5):638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- Xiong G, Jiang X, Song T. The overexpression of lncRNA H19 as a diagnostic marker for coronary artery disease. Revista Da Associacao Medica Brasileira (1992) 2019;65(2):110–117. doi: 10.1590/1806-9282.65.2.110. [DOI] [PubMed] [Google Scholar]
- Xiong R, Wang Z, Zhao Z, Li H, Chen W, Zhang B, Wang L, Wu L, Li W, Ding J, Chen S. MicroRNA-494 reduces DJ-1 expression and exacerbates neurodegeneration. Neurobiol Aging. 2014;35(3):705–714. doi: 10.1016/j.neurobiolaging.2013.09.027. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhou X, Zhang W. MicroRNA prediction with a novel ranking algorithm based on random walks. Bioinforma (Oxford England) 2008;24(13):i50–58. doi: 10.1093/bioinformatics/btn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Wang Z-H, Guo J-T. The research strategies for probing the function of long noncoding RNAs. Genomics. 2012;99(2):76–80. doi: 10.1016/j.ygeno.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan C-X, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Huang N, Wu W, Chen W, Jiang Y, Chen J, Huang X, Wen X, Xu J, Jin Q, Zhang K, Chen Z, Chien S, Zhong S. Genome-wide colocalization of RNA-DNA interactions and fusion RNA pairs. Proc Natl Acad Sci USA. 2019;116(8):3328–3337. doi: 10.1073/pnas.1819788116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, McCray PB, Jr, Engelhardt JF. Advances in gene therapy for cystic fibrosis lung disease. Hum Mol Genet. 2019;28(R1):R88–R94. doi: 10.1093/hmg/ddz139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-H, Zhang X-C, Huang Z-P, Zhou H, Huang M-B, Zhang S, Chen Y-Q, Qu L-H. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34(18):5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Sablok G, Qiao G, Nie Q, Wen X (2017) isomiR2Function: An integrated workflow for identifying MicroRNA variants in plants. Front Plant Sci 08. 10.3389/fpls.2017.00322 [DOI] [PMC free article] [PubMed]
- Yang Y, Cai Y, Wu G, Chen X, Liu Y, Wang X, Yu J, Li C, Chen X, Jose PA, Zhou L, Zeng C. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci. 2015;129(8):675–685. doi: 10.1042/cs20150121. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ren F, Liu C, He S, Sun G, Gao Q, Yao L, Zhang Y, Miao R, Cao Y, Zhao Y, Zhong Y, Zhao H (2010) dbDEMC: a database of differentially expressed miRNAs in human cancers. BMC Genomics 11(S4). 10.1186/1471-2164-11-s4-s5 [DOI] [PMC free article] [PubMed]
- Yao L, Zhou B, You L, Hu H, Xie R. LncRNA MIAT/miR-133a-3p axis regulates atrial fibrillation and atrial fibrillation-induced myocardial fibrosis. Mol Biol Rep. 2020 doi: 10.1007/s11033-020-05347-0. [DOI] [PubMed] [Google Scholar]
- Yi J, Chen B, Yao X, Lei Y, Ou F, Huang F. Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer’s disease through inactivating the PI3K/Akt signaling pathway. J Cell Biochem. 2019 doi: 10.1002/jcb.29108. [DOI] [PubMed] [Google Scholar]
- You X, and Conrad TO (2016) Acfs: accurate circRNA identification and quantification from RNA-Seq data. Sci Reports 6(1). 10.1038/srep38820 [DOI] [PMC free article] [PubMed]
- Yu F, Zhang G, Shi A, Hu J, Li F, Zhang X, Zhang Y, Huang J, Xiao Y, Li X, Cheng S (2018) LnChrom: a resource of experimentally validated lncRNA–chromatin interactions in human and mouse. Database 2018. 10.1093/database/bay039 [DOI] [PMC free article] [PubMed]
- Yuan C, Sun Y. RNA-CODE: a noncoding RNA classification tool for short reads in NGS data lacking reference genomes. PLoS ONE. 2013;8(10):e77596. doi: 10.1371/journal.pone.0077596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K, Hagiwara Y, Ando M, Nakamura A, Takeda S, Hijikata T. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct. 2008;33(2):163–169. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- Zaharieva IT, Calissano M, Scoto M, Preston M, Cirak S, Feng L, Collins J, Kole R, Guglieri M, Straub V, Bushby K, Ferlini A, Morgan JE, Muntoni F. Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular Dystrophy. PLoS ONE. 2013;8(11):e80263. doi: 10.1371/journal.pone.0080263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangrando J, Zhang L, Vausort M, Maskali F, Marie P-Y, Wagner DR, Devaux Y (2014) Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics 15(1). 10.1186/1471-2164-15-460 [DOI] [PMC free article] [PubMed]
- Zeller T, Keller T, Ojeda F, Reichlin T, Twerenbold R, Tzikas S, Wild PS, Reiter M, Czyz E, Lackner KJ, Munzel T, Mueller C, Blankenberg S. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J. 2014;35(31):2106–2114. doi: 10.1093/eurheartj/ehu151. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qin C, An C, Zheng X, Wen S, Chen W, … Wu Y (2021a) Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer 20(1):126. 10.1186/s12943-021-01431-6 [DOI] [PMC free article] [PubMed]
- Zhang J, Gao C, Meng M, Tang H. Long noncoding RNA MHRT protects cardiomyocytes against H2O2-induced apoptosis. Biomol Ther. 2016;24(1):19–24. doi: 10.4062/biomolther.2015.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yu L, Xu Y, Liu Y, Li Z, Xue X, Wan S, Wang H. Long noncoding RNA upregulated in hypothermia treated cardiomyocytes protects against myocardial infarction through improving mitochondrial function. Int J Cardiol. 2018;266:213–217. doi: 10.1016/j.ijcard.2017.12.097. [DOI] [PubMed] [Google Scholar]
- Zhang S, Yue Y, Sheng L, Wu Y, Fan G, Li A, Hu X, ShangGuan M, Wei C (2013) PASmiR: a literature-curated database for miRNA molecular regulation in plant response to abiotic stress. BMC Plant Biol 13(1). 10.1186/1471-2229-13-33 [DOI] [PMC free article] [PubMed]
- Zhang T, Pang P, Fang Z, Guo Y, Li H, Li X, Tian T, Yang X, Chen W, Shu S, Tang N, Wu J, Zhu H, Pei L, Liu D, Tian Q, Wang J, Wang L, Zhu L-Q, Lu Y. Expression of BC1 impairs spatial learning and memory in Alzheimer’s disease via APP translation. Mol Neurobiol. 2018;55(7):6007–6020. doi: 10.1007/s12035-017-0820-z. [DOI] [PubMed] [Google Scholar]
- Zhang T-N, Li D, Xia J, Wu Q-J, Wen R, Yang N, Liu C-F. Non-coding RNA: a potential biomarker and therapeutic target for sepsis. Oncotarget. 2017;8(53):91765–91778. doi: 10.18632/oncotarget.21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu Y, Min Z, Liang G, Mo J, Ju Z, Zeng B, Guan W, Zhang Y, Chen J, Zhang Q, Li H, Zeng C, Wei Y, Chan G-F. circMine: a comprehensive database to integrate, analyze and visualize human disease–related circRNA transcriptome. Nucleic Acids Res. 2021;50(D1):D83–D92. doi: 10.1093/nar/gkab809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yue X, Tang G, Wu W, Huang F, Zhang X. SFPEL-LPI: Sequence-based feature projection ensemble learning for predicting LncRNA-protein interactions. PLoS Comput Biol. 2018;14(12):e1006616. doi: 10.1371/journal.pcbi.1006616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, Huang Y, Zhao X, Huang L, Wang Z, Hou Y, Li J, Tian Y, Yu J, Han H, Liu Y, Gao F, Zhang Y, Wang S, Du Z. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zang Q, Xu B, Zheng W, Ban R, Zhang H, Yang Y, Hao Q, Iqbal F, Li A, Shi Q. IsomiR Bank: a research resource for tracking IsomiRs. Bioinformatics. 2016;32(13):2069–2071. doi: 10.1093/bioinformatics/btw070. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zheng S, Geng Y, Xue J, Wang Z, Xie X, Wang J, Zhang S, Hou Y. MicroRNA profiling of atrial fibrillation in canines: miR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PLoS ONE. 2015;10(3):e0122674. doi: 10.1371/journal.pone.0122674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cheng Y. miR-16-1 promotes the aberrantα-synuclein accumulation in parkinson disease via targeting heat shock protein 70. Sci World J. 2014;2014:1–8. doi: 10.1155/2014/938348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gao W, Long Q-Q, Zhang J, Li Y-F, Liu D-C, Yan J-J, Yang Z-J, Wang L-S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7(1):7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ma Z, Guo Z, Zheng M, Li K, Yang X. Analysis of long non-coding RNA and mRNA profiles in epicardial adipose tissue of patients with atrial fibrillation. Biomed Pharmacother = Biomedecine & Pharmacotherapie. 2020;121:109634. doi: 10.1016/j.biopha.2019.109634. [DOI] [PubMed] [Google Scholar]
- Zhao S, Li S, Liu W, Wang Y, Li X, Zhu S, Lei X, Xu S. Circular RNA signature in lung adenocarcinoma: a mioncocirc database-based study and literature review. Front Oncol. 2020;10:523342. doi: 10.3389/fonc.2020.523342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L-L, Xu W-L, Liu S, Sun W-J, Li J-H, Wu J, Yang J-H, Qu L-H. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016;44(W1):W185–193. doi: 10.1093/nar/gkw414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K-R, Liu S, Sun W-J, Zheng L-L, Zhou H, Yang J-H, Qu L-H. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45(D1):D43–D50. doi: 10.1093/nar/gkw965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zou Y-G, Xue Y-Z, Wang X-H, Gao H, Dong H-W, Zhang Q. Long non-coding RNA H19 protects acute myocardial infarction through activating autophagy in mice. European Rev Med Pharmacol Sci. 2018;22(17):5647–5651. doi: 10.26355/eurrev_201809_15831. [DOI] [PubMed] [Google Scholar]
- Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, Wu J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38(suppl_2):W392–W397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorde Khvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E, Giladi H, Rivkin L, Simerzin A, Eliakim R, Khalaileh A, Hubert A, Lahav M, Kopelman Y, Goldin E, Dancour A. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci. 2013;110(51):20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Lin C, Liu X-Y, Han Y-P, Li W-B, Guo M-Z. Novel representation of RNA secondary structure used to improve prediction algorithms. Genet Mol Res: GMR. 2011;10(3):1986–1998. doi: 10.4238/vol10-3gmr1181. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuntini M, Salvatore M, Pedrini E, Parra A, Sgariglia F, Magrelli A, Taruscio D, Sangiorgi L. MicroRNA profiling of multiple osteochondromas: identification of disease-specific and normal cartilage signatures. Clin Genet. 2010;78(6):507–516. doi: 10.1111/j.1399-0004.2010.01490.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No supporting data is available in this study.
Not applicable.