Abstract
In comparison to previously known severe respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, the newly emerged Omicron (B.1.1.529) variant shows higher infectivity in humans. Exceptionally high infectivity of this variant raises concern of its possible transmission via other intermediate hosts. The SARS-CoV-2 infectivity is established via the association of spike (S) protein receptor binding domain (RBD) with host angiotensin I converting enzyme 2 (hACE2) receptor. In the course of this study, we investigated the interaction between Omicron S protein RBD with the ACE2 receptor of 143 mammalian hosts including human by protein–protein interaction analysis. The goal of this study was to forecast the likelihood that the virus may infect other mammalian species that coexist with or are close to humans in the household, rural, agricultural, or zoological environments. The Omicron RBD was found to interact with higher binding affinity with the ACE2 receptor of 122 mammalian hosts via different amino acid residues from the human ACE2 (hACE2). The rat (Rattus rattus) ACE2 was found to show the strongest interaction with Omicron RBD with a binding affinity of -1393.6 kcal/mol. These distinct strong binding affinity of RBD of Omicron with host ACE2 indicates a greater potential of new host transmissibility and infection via intermediate hosts. Though expected but the phylogenetic position of the mammalian species may not dictate the Omicron RBD binding to the host ACE2 receptor suggesting an involvement of multiple factors in guiding host divergence of the variant.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10142-023-00962-z.
Keywords: ACE2 receptor; COVID-19; Host adaptation; Omicron, Receptor binding domain; SARS-CoV-2
Introduction
In December 2019, a novel coronavirus, severe respiratory syndrome coronavirus 2 (SARS-CoV-2), had emerged causing coronavirus disease 2019 (COVID-19) pandemic in Wuhan, China (Gorbalenya et al. 2020; Zhu et al. 2020). Middle East Respiratory Syndrome (MERS) CoV and SARS-CoV-2 are exceptionally dangerous viruses that have been observed to spread from bats to people via intermediate hosts such as palm civets and dromedary camels (Guan et al. 2003; Azhar et al. 2014). Owing to the first reported sequence of SARS-CoV-2 from Wuhan, the reference variant has been named Wuhan-Hu-1 (Pavan et al. 2022). The same nomenclature has been used in the present study.
Since the declaration of SARS-CoV-2 as a pandemic in February 2020, the World Health Organization (WHO) has been closely monitoring its progress till date. The WHO has categorised the emerging variants of SARS-CoV-2 into currently circulating variants of concern (VOCs), previously circulating VOCs, variants of interest (VOI), variants under monitoring (VUMs), and formerly monitored variants (FMV). According to WHO’s most recent update on July 18, 2022, there is only one variant listed under currently circulating VOCs; 4 variants under previously circulating VOCs, 8 variants under previously circulating VOI, and 15 variants are under formerly monitored variants (https://www.who.int/activities/tracking-SARS-CoV-2-variants).
The WHO categorised Omicron variant (B.1.1.529), a highly mutated novel version of SARS-CoV-2 initially found in South Africa, as one of the VOCs on November 26, 2021. Currently, Omicron is the only VOC and there are seven subvariants of it that are considered under monitoring. Reportedly, Omicron carries more than 50 mutations, 32 of which are specific to the Spike (S) protein with respect to the Wuhan-Hu-1 variant. These changes involve the substitution, deletion, and insertion of amino acids (Poudel et al. 2022). The receptor binding domain (RBD) contains some of these substitutions and mutations, posing interesting intervention in altered interactions with host receptor protein, the angiotensin I converting enzyme 2 (ACE2) (Samanta et al. 2022). These mutations at the RBD led to increased binding affinity with human ACE2 (hACE2) in a unique fashion with potential implications in high infectivity and immune/vaccine evasion (Nasrin and Ali 2021; Samanta et al. 2022). The entry of the virus can be a potential therapeutic target which can be used in combination with other approaches for better results (Lian et al. 2022).
Despite several predictions, the precise route of SARS-CoV-2 transmission to humans, including the intermediate host, remains elusive. There are limited reports on SARS-CoV-2 animal reservoirs or the potential that the virus may spread to other species that coexist with or are near humans in the household, rural, agricultural, or zoological contexts (Lam et al. 2020). Coronaviruses are key multi-host viruses, and SARS-CoV-2 is one such with potentially diverse infective host range potential including a multitude of animals (Guan et al. 2003; Wang and Eaton 2007; Shaw et al. 2020). The betacoronavirus family that includes SARS-CoV-2 have been identified to infect economically significant animals like cows (Saif 2010) and pigs (Vlasova et al. 2020), as well as mice (Wang et al. 2015), rats (Lau et al. 2015), rabbits (Lau et al. 2012), and wildlife like antelope and giraffe (Hasoksuz et al. 2007). Therefore, it is envisaged that a range of species is expected to be affected by SARS-CoV-2. In vitro experiments have also revealed that many more animals might get infected with SARS-CoV-2 (Zhao et al. 2020; Lam et al. 2020). The basic premise for the wide SARS-CoV-2 host range is the possibility of using orthologues of ACE2 for cell entry, as the sequence of ACE2 is substantially conserved among vertebrates (Lam et al. 2020).
The identification of animals who are vulnerable to SARS-CoV-2 infection is crucial in order to grasp the virus’s probable host range. Studying the structural interaction between the S protein RBD and host ACE2 provides the predictive basis for the same. A recent study proposed a wide variety of probable hosts for SARS-CoV-2 based on RBD:ACE2 interaction (Lam et al. 2020). The Omicron variant with highly mutated S protein has been found to establish unique interaction between RBD and the hACE2 receptor. Therefore, it is hypothesised that this variant by forming new interactions with host ACE2 proteins might infect new hosts expanding the viral host range. The present study assesses the patterns of interactions and binding affinity of RBD of Wuhan-Hu-1 and Omicron variants of SARS-CoV-2 with a broad range of 143 mammals, including hACE2 receptors to identify the most susceptible potential hosts for the variant.
Materials and methods
Retrieval of hACE2 and S protein RBD complex structure
The RBD of the B chain of the SARS-CoV-2 S protein binds with the A chain of the ACE2 receptor in a host cell during infection. As a result, analysis was restricted to the RBD region of the S protein. The crystal structure of the SARS-CoV-2 S protein RBD coupled to the hACE2 receptor (PDB ID: 6M0J) was obtained from the Protein Data Bank (http://www.rcsb.org/) as a reference (Lan et al. 2020). Using BIOVIA discovery studio, the hACE2 structure was obtained from the reference complex (BIOVIA, Dassault Syst´emes, BIOVIA Discovery Studio, San Diego, 2021).
Retrieval of ACE2 protein sequences
This study included a total of 143 mammalian species including primates, rodents, tree shrews, rabbits and hares, pangolins, odd-toed, marsupials, insectivores, even-toed, carnivores, bats, afrotheria, and armadillos. For further in silico analysis, the ACE2 protein sequences for 142 species were extracted from the NCBI gene database (https://www.ncbi.nlm.nih.gov/) and saved in FASTA format. In Supplementary Table S1, all information regarding ACE2 sequences of the selected hosts is provided.
In silico structural homology modelling
Using BIOVIA Discovery Studio, the structure of hACE2 was retrieved from the PDB structure (PDB ID: 6M0J). The 3D structural models of ACE2 proteins of other 142 mammalian species were built by using the homology modelling service SWISS-MODEL (https://swissmodel.expasy.org/) (Waterhouse et al. 2018).
Model validations
Each 3D model of ACE2 receptors generated on the SWISS-MODEL server was checked to confirm the quality using two separate web servers. The Ramachandran plot, verify 3D score, and ERRAT score had been used by the SAVES version 6.0 server (https://saves.mbi.ucla.edu/) to analyse the models. The Protein Structure Analysis (ProSA) web server (https://prosa.services.came.sbg.ac.at/prosa.php) was used to evaluate the models using the Z-score which represents the overall model quality.
Protein–protein docking
ACE2 receptor of all species was docked with the RBD of the Wuhan-Hu-1 and Omicron variants of SARS-CoV-2 by using the ClusPro web-server (https://cluspro.org) (Kozakov et al. 2017). The RBD of Wuhan-Hu-1 and Omicron variants were docked against the modelled ACE2 receptors from all mammalian species following the previously described procedure (Samanta et al. 2022).
Analysis of direct contact residues of S protein RBD: ACE2 of all species
The direct contact residues between RBD and ACE2 receptors of all species were examined using PDBSum webserver (https://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html) (Laskowski 2001). In order to evaluate and further analyse the interactions between RBD and ACE2 of each species, the best cluster models of docking between those two proteins were chosen and uploaded into PDBSum as described in the previous study (Samanta et al. 2022).
Phylogenetic tree constructions
The sequences of ACE2 proteins from various species were aligned using MGEA X. The MGEA X Neighbor-Joining strategy was used for the phylogenetic study, which entailed many comparisons. The phylogenetic tree was generated as previously described (Laskar and Ali 2021a; Samanta et al. 2022). The tree was then visualised by an online tool, iTOL (Letunic and Bork 2021).
Results and discussion
Validation of models
The 3D structure of the RBD of the Omicron variant was generated by homology modelling using the SWISS-MODEL web server and authenticated using SAVES ver. 6.0 and the ProSA web servers. Similarly, the 3D structures of the ACE2 protein of 142 mammalian hosts were generated and validated. The sequence coverage, resolution, identity, and similarity to the query sequence derived using the SWISS-MODEL are provided in Supplementary Table S2. The QMEAN and GMQE values for each model were checked in order to examine the best-fit biological models. The QMEAN value often called “degree of nativeness,” indicates how closely the model corresponds to the experimental structures. The QMEAN value of a decent model should be at least − 4.0 and close to zero (Benkert et al. 2011). The QMEAN scores of the projected protein models were found to fall within the intended range, demonstrating the high quality of the modelled structures (Supplementary Table S2). Furthermore, the obtained values of GMQE were within the typical range of 0 to 1, further validating the structures (Biasini et al. 2014). All the host ACE2 models had appropriate parameters, as checked by SAVES version 6.0 and the ProSA web servers (Supplementary Table S3). According to the Ramachandran plot, the ACE2 models of all variants have more than 78% of their residues in the permitted zone (Supplementary Fig. S1) (Prajapat et al. 2016). These findings further confirmed the high quality of the protein model. To visualise the interaction of the S protein RBD of Wuhan-Hu-1 and Omicron variants, these 3D structures of ACE2 receptors of 142 hosts were considered.
Omicron RBD shows a stronger affinity for a large number of host ACE2
Protein–protein docking was used to predict the interactions of molecules between the ACE2 receptor and the RBD of the S protein of SARS-CoV-2 using the ClusPro web server. The binding affinities of ACE2 receptor of most of the hosts were found to be higher than that of hACE2 for the RBD of the Omicron variant. In Supplementary Table S4, the binding affinities between the ACE2 receptors and RBD of the Omicron variant are shown in terms of the lowest energy. The rat (Rattus rattus) ACE2 interaction with the Omicron RBD had the lowest energy score of − 1393.6 kcal/mol among all the 143 tested hosts, including humans. This indicates that ACE2 of rat binds to the Omicron RBD more strongly than ACE2 in other hosts, implying a higher risk of infection. Previous studies have also reported a strong binding affinity of Omicron RBD for ACE2 receptors of several other mammalian hosts including rat, mouse, palm-civet and least horseshoe bats, and a weak yet detectable binding affinity for lesser hedgehog tenrec (Li et al. 2022).
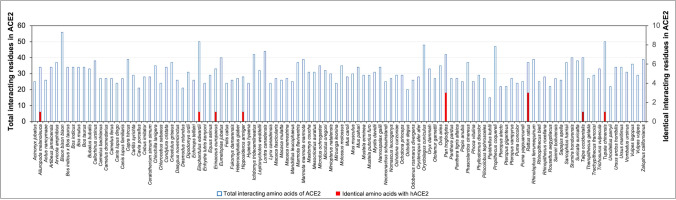
After these docking studies, an intriguing finding was made where Omicron variant RBD was found to have higher binding affinities to ACE2 receptor of some domestic animals like dogs and cats and some zoo animals like pandas, leopards, bears, tiger, bison, and lion than hACE2 (− 1216.2 kcal/mol) (Supplementary Table S4). Among the 143 selected hosts, 122 had higher binding affinities to the RBD of Omicron than humans, and only 20 had lower binding affinities than humans (Fig. 1). It indicates that Omicron may transmit to these hosts via ACE2-mediated cell entry causing a threat to a higher risk of infection. Our results are further corroborated by a study in Spain which showed the transmission of the Omicron variant from infected humans to domestic cats and dogs. It should be noted that the animals exhibited a lesser level of virulence (Sánchez-Morales et al. 2022).
Fig. 1.
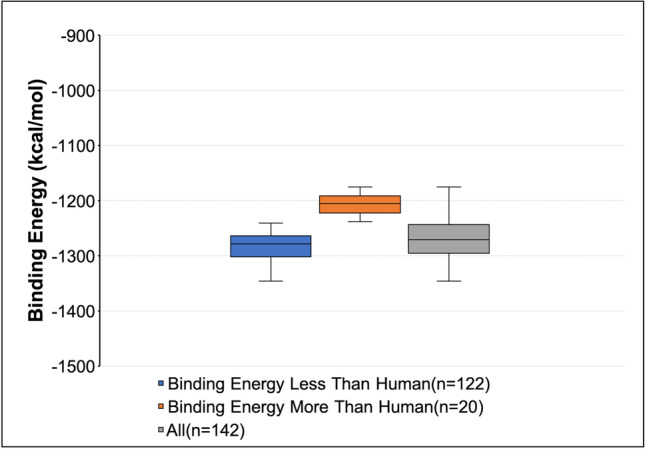
Binding energy of RBD of Omicron variant against ACE2 receptors of 142 mammalian hosts. The binding affinities of Omicron RBD against the ACE2 receptors of potential mammalian hosts were predicted by performing protein–protein docking using the ClusPro web server
Though we have summarised the ACE2:RBD interaction in terms of binding energy so far, it is important noting that this is one of the many facets of viral entry. This is just the first step of the membrane fusion process and the accruing mutations in the Omicron variant have the potential to alter the subsequent steps leading to viral infection as has been reported (Yang et al. 2022). Presently, we further dwell upon the interaction between the Omicron RBD and host ACE2.
Omicron RBD interacts with the host ACE2 in a unique way
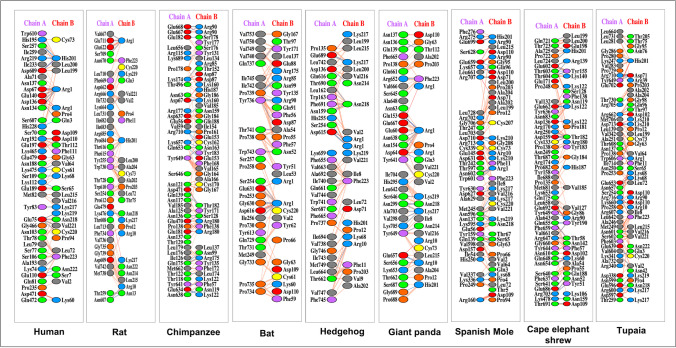
The current research also aimed to look at the pattern of contacts between the RBD of Omicron and the ACE2 of all selected hosts, since this interaction is critical for viral infectivity. The PDBSum web server was used to examine the amino acid residues implicated in direct contact between ACE2 and RBD. In 2D format, the most likely interacting residues of both interaction partners were retrieved and analysed. Supplementary Fig. S2 depicts all of these interactions. The interacting amino acids of ACE2 of all hosts to the RBD of Wuhan-Hu-1 were isolated and studied in detail to determine how extensive and diverse these interactions are. The interacting amino acid residues of ACE2 of all selected hosts with the RBD of Omicron and Wuhan-Hu-1 variants were compared. The RBD of the Omicron variant showed unique interaction using 30 amino acid residues of RBD with 41 residues on hACE2 as reported in our previous study (Samanta et al. 2022).
Among the 142 hosts, 134 showed completely unique binding residues with Omicron. For instance, rat (Rattus rattus) ACE2 interacts with RBD of Omicron with 8.1% identical residues with that of the hACE2 (Fig. 2). Other hosts like chimpanzee (Pan troglodytes), bat (Hipposideros armiger), hedgehog (Erinaceus europaeus), giant panda (Ailuropoda melanoleuca), spanish mole (Talpa occidentalis), cape elephant shrew (Elephantulus edwardii), and tupaia (Tupaia chinensis) used 7.14%, 3.6%, 3.03%, 2.94%, 2.5%, 2%, and 2% identical residues to that of hACE2 to interact with RBD of Omicron variant respectively (Fig. 3). The unique and stronger interaction of hosts ACE2 and Omicron RBD indicates the possibility of higher viral transmission.
Fig. 2.
Interactions of Omicron RBD with host ACE2 receptors obtained by protein–protein docking. The interactions between the RBD of Omicron S protein with the ACE2 receptor of hosts were analysed using PDBSum web server as described in the text. The figure represents the interaction of Omicron RBD and the potential hosts with higher affinity. [Chain A: ACE2 of hosts; chain B: RBD of Omicron; red line: salt bridges; blue line: hydrogen bond; yellow dash: non-bonded interactions]
Fig. 3.
Comparative interacting residues of host ACE2 with the Omicron RBD with respect to hACE2. By using the ClusPro server for molecular docking, the interactions between hACE2 and other mammalian host ACE2 against Omicron RBD were evaluated and analysed in PDBSum as described in the text. The number of interacting residues of host ACE2 receptors and the identical residues to that of the hACE2 were calculated
Comparative binding energy analysis of Wuhan-Hu-1 and Omicron variants
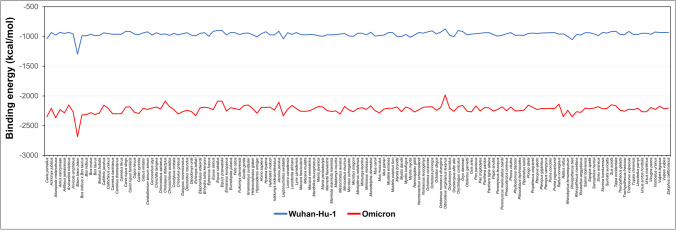
Several studies had reported that the Wuhan-Hu-1 variant is capable to infect some domestic animals like dogs (Shi et al. 2020), cats (Shi et al. 2020), hamsters (Liu et al. 2021), and rabbits (Liu et al. 2021), zoological animals (Lam et al. 2020), and wildlife (Hasoksuz et al. 2007). In this study, we compared the binding energy of each host with Wuhan-Hu-1 and Omicron variants. From the ClusPro web server results, it was evident that the binding efficiency of each host ACE2 to Omicron RBD was much greater than their binding efficiency to Wuhan-Hu-1 RBD (Fig. 4). The available data shows that the novel variant, Omicron, interacts more strongly to each selected host. These findings suggest that the Omicron variant is more potent to infect these mammalian hosts than the Wuhan-Hu-1 variant.
Fig. 4.
Comparative binding affinities of Wuhan-Hu-1 and Omicron RBDs against 143 host ACE2 receptors. The binding affinities of the S protein RBD of the Omicron variant against ACE2 receptors of all the mammalian hosts were calculated using the ClusPro server and compared with that of the Wuhan-Hu-1 variant
Omicron host adaptation is independent of the phylogenetic position of hosts
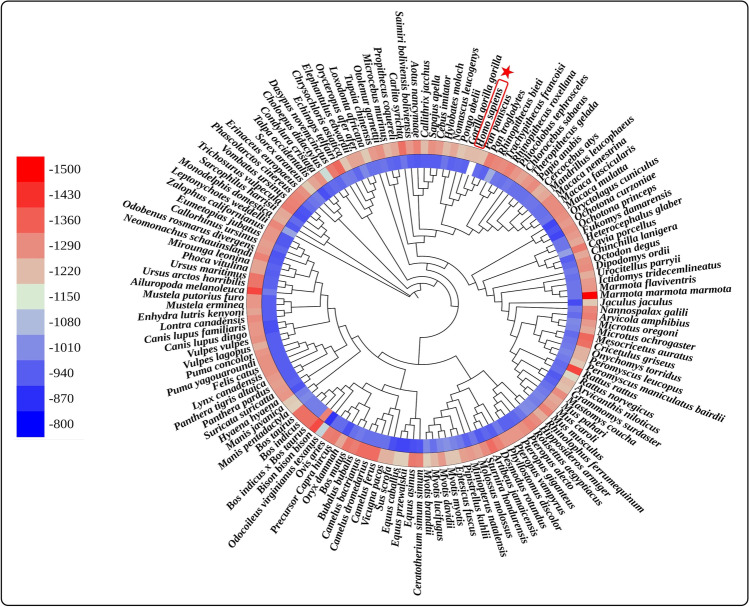
The overall sequence variation across the ACE2 of all the selected mammalian species has been studied in order to better understand the molecular basis of the varied ACE2 receptor functions. A phylogenetic tree based on the sequences of ACE2 protein was created for this purpose using MEGA X software (Fig. 5). We observed that the ability of ACE2 receptors to assist SARS-CoV-2 entrance is independent of their evolutionary clustering. For example, if we see the binding energy between the RBD of S protein and ACE2 receptor, the binding energy is not correlated with evolutionary clustering. This indicates that, despite their close evolutionary relationship to humans, those hosts do not always carry a higher risk of infection. The phylogenetic relationship has no or very little impact on the binding efficiency or infection risk suggesting alternate driving forces for evolution therein. This is expected because the SARS-CoV-2 pandemic might be altering the course of viral evolution but in terms of host evolution; so far, it remains inconsequential (Laskar et al. 2021; Laskar and Ali 2021b).
Fig. 5.
Phylogenetic tree of ACE2 protein sequences of 143 mammalian species. The circular heatmap represents the binding energy of the Omicron variant (outer circle) and Wuhan-Hu-1 variant (inner circle). The position of the human in the phylogenetic tree is indicated using a red border and star
Conclusion
The SARS-CoV-2 Omicron variant interacts with the ACE2 receptor of several mammalian hosts with stronger affinity than hACE2 involving unique amino acid residues indicating the possibility of the viral transmission in new hosts. Rat ACE2 showed the strong binding with the highest affinity towards Omicron RBD. Several other mammals including domestic and zoo animals showed binding affinity greater than hACE2. These mammals inhabit in close contact with humans, and therefore, the possible transmission of Omicron into these hosts poses a greater threat of viral spread and may aggravate the ongoing pandemic.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the Department of Biological Sciences, Aliah University, Kolkata, India, for providing the necessary facilities for this work. SSMA acknowledges the Council of Scientific & Industrial Research (CSIR) Govt. of India for the financial assistance in the form of a Junior Research Fellowship (JRF).
Author contribution
Conceptualization: Safdar Ali, Mehboob Hoque; methodology: Arijit Samanta, Syed Sahajada Mahafujul Alam, Mehboob Hoque; formal analysis and investigation: Arijit Samanta, Syed Sahajada Mahafujul Alam, Safdar Ali, Mehboob Hoque; writing—original draft preparation: Arijit Samanta, Syed Sahajada Mahafujul Alam; writing— review and editing: Arijit Samanta, Syed Sahajada Mahafujul Alam, Safdar Ali, Mehboob Hoque; resources: Arijit Samanta, Syed Sahajada Mahafujul Alam, Safdar Ali, Mehboob Hoque; Supervision: Mehboob Hoque. All authors have read and agreed to the published version of the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Arijit Samanta and Syed Sahajada Mahafujul Alam contributed equally to this work.
References
- Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS et al (2020) The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed]
- Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hasoksuz M, Alekseev K, Vlasova A, et al. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J Virol. 2007;81:4981–4990. doi: 10.1128/JVI.02361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SD, Bordin N, Waman VP, et al. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci Rep. 2020;10:16471. doi: 10.1038/s41598-020-71936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Laskar R, Ali S. Differential mutation profile of SARS-CoV-2 proteins across deceased and asymptomatic patients. Chem Biol Interact. 2021;347:109598. doi: 10.1016/j.cbi.2021.109598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskar R, Jilani MG, Ali S. Implications of genome simple sequence repeats signature in 98 Polyomaviridae species. 3 Biotech. 2021;11:35. doi: 10.1007/s13205-020-02583-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskar R, Ali S (2021a) Phylo-geo-network and haplogroup analysis of 611 novel coronavirus (SARS-CoV-2) genomes from India. Life Sci Alliance 4 202000925. 10.26508/lsa.202000925 [DOI] [PMC free article] [PubMed]
- Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Woo PCY, Yip CCY, et al. Isolation and characterization of a novel betacoronavirus subgroup a coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol. 2012;86:5481–5496. doi: 10.1128/JVI.06927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Woo PCY, Li KSM, et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of betacoronavirus 1 and has implications for the ancestor of betacoronavirus lineage A. J Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Han P, Huang B, et al. Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s. Cell Discov. 2022;8:65. doi: 10.1038/s41421-022-00431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q, Zhang K, Zhang Z, et al. Differential effects of macrophage subtypes on SARS-CoV-2 infection in a human pluripotent stem cell-derived model. Nat Commun. 2022;13:2028. doi: 10.1038/s41467-022-29731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu G, Wang Y, et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc Natl Acad Sci. 2021;118:e2025373118. doi: 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin T, Ali S. Genetic path of the emergence of SARS-CoV-2. Gene Cell Tissue in Press. 2021 doi: 10.5812/gct.118302. [DOI] [Google Scholar]
- Pavan M, Bassani D, Sturlese M, Moro S. From the Wuhan-Hu-1 strain to the XD and XE variants: is targeting the SARS-CoV-2 spike protein still a pharmaceutically relevant option against COVID-19? J Enzyme Inhib Med Chem. 2022;37:1704–1714. doi: 10.1080/14756366.2022.2081847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S, Ishak A, Perez-Fernandez J, et al. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts – what is known so far? Travel Med Infect Dis. 2022;45:102234. doi: 10.1016/j.tmaid.2021.102234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat R, Bhattachar I, Kumar A. Homology modeling and structural validation of Type 2 diabetes associated transcription factor 7-like 2 (TCF7L2) Trends Bioinforma. 2016;9:23–29. doi: 10.3923/tb.2016.23.29. [DOI] [Google Scholar]
- Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract. 2010;26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A, Alam SSM, Ali S, Hoque M (2022) Analyzing the interaction of human ACE2 and RBD of spike protein of SARS-CoV-2 in perspective of Omicron variant. EXCLI J 21Doc610 ISSN 1611–2156. 10.17179/EXCLI2022-4721 [DOI] [PMC free article] [PubMed]
- Sánchez-Morales L, Sánchez-Vizcaíno JM, Pérez-Sancho M, et al. The Omicron (B.1.1.529) SARS-CoV-2 variant of concern also affects companion animals. Front Vet Sci. 2022;9:940710. doi: 10.3389/fvets.2022.940710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LP, Wang AD, Dylus D, et al. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol Ecol. 2020;29:3361–3379. doi: 10.1111/mec.15463. [DOI] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova AN, Wang Q, Jung K, et al (2020) Porcine coronaviruses. In: Malik YS, Singh RK, Yadav MP (eds) Emerging and Transboundary Animal Viruses. Springer Singapore, Singapore, pp 79–110
- Wang L-F, Eaton BT. Bats, civets and the emergence of SARS. In: Childs JE, Mackenzie JS, Richt JA, editors. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. Berlin Heidelberg, Berlin, Heidelberg: Springer; 2007. pp. 325–344. [Google Scholar]
- Wang W, Lin X-D, Guo W-P, et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Wang C, White KI, et al. Structural conservation among variants of the SARS-CoV-2 spike postfusion bundle. Proc Natl Acad Sci. 2022;119:e2119467119. doi: 10.1073/pnas.2119467119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chen D, Szabla R, et al. Broad and differential animal angiotensin-converting enzyme 2 receptor usage by SARS-CoV-2. J Virol. 2020;94:e00940–e1020. doi: 10.1128/JVI.00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.