Abstract
Objectives—
To develop and evaluate deep learning models devised for liver fat assessment based on ultrasound (US) images acquired from four different liver views: transverse plane (hepatic veins at the confluence with the inferior vena cava, right portal vein, right posterior portal vein) and sagittal plane (liver/kidney).
Methods—
US images (four separate views) were acquired from 135 participants with known or suspected nonalcoholic fatty liver disease. Proton density fat fraction (PDFF) values derived from chemical shift-encoded magnetic resonance imaging served as ground truth. Transfer learning with a deep convolutional neural network (CNN) was applied to develop models for diagnosis of fatty liver (PDFF ≥ 5%), diagnosis of advanced steatosis (PDFF ≥ 10%), and PDFF quantification for each liver view separately. In addition, an ensemble model based on all four liver view models was investigated. Diagnostic performance was assessed using the area under the receiver operating characteristics curve (AUC), and quantification was assessed using the Spearman correlation coefficient (SCC).
Results—
The most accurate single view was the right posterior portal vein, with an SCC of 0.78 for quantifying PDFF and AUC values of 0.90 (PDFF ≥ 5%) and 0.79 (PDFF ≥ 10%). The ensemble of models achieved an SCC of 0.81 and AUCs of 0.91 (PDFF ≥ 5%) and 0.86 (PDFF ≥ 10%).
Conclusion—
Deep learning-based analysis of US images from different liver views can help assess liver fat.
Keywords: attention mechanism, convolutional neural networks, deep learning, nonalcoholic fatty liver disease, proton density fat fraction, ultrasound images
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease worldwide.1,2 Currently, biopsy is considered the gold standard for liver fat grading.3 Liver biopsy, however, is costly and may lead to significant complications, including pain and bleeding. Fat grades determined using biopsy may also be unreliable due to sampling errors and subjective analysis of histology slides, which is usually performed by a single pathologist.4 Confounder-corrected chemical shift-encoded magnetic resonance imaging (CSE-MRI) can be used to determine proton density fat fraction (PDFF) and has been shown to have excellent performance for assessing liver fat.5–8 While CSE-MRI methods are accurate and noninvasive, access to this imaging modality is costly and limited. Ultrasound (US) imaging may be a good alternative to CSE-MRI because this imaging modality is noninvasive, portable, and more widely available and used, making it a potentially attractive option for NAFLD diagnosis and quantification.4
Various liver US image features, including vessel obscuration, posterior beam attenuation, and elevated liver-kidney contrast ratio, have been associated with liver fat.9,10 Accumulation of fat modifies backscattering properties of liver tissue,11 making the liver brighter in comparison to neighboring tissues, such as the kidney.12,13 In addition, blood vessels within the liver become blurred or obscured as liver fat increases due to signal attenuation. To assist radiologists in interpreting US images, various computer-aided diagnosis systems have been proposed.14,15 Currently, deep learning methods based on convolutional neural networks (CNNs) are gaining attention in medical image analysis.16 In comparison to standard approaches to image recognition, which require feature engineering, deep learning algorithms can automatically process images to extract useful features for classification. Commonly, the first convolutional blocks of a deep network extract low-level features related to image texture, while deeper layers utilize features to determine high-level concepts related to the appearance of the whole image. Moreover, several methods have been proposed to help understand how deep learning models conduct image recognition, so neural networks are no longer perceived purely as a “black box.”17 However, medical image datasets are often inadequate to develop a well-performing deep model from scratch.16,18 Consequently, transfer learning is applied with a deep model pretrained on a large set of nonmedical images to adjust the model to the medical problem of interest. Transfer learning with pretrained CNNs was applied for liver fat assessment in recent studies.19,20 For instance, deep features extracted from a pretrained CNN were used to develop regression models for PDFF quantification and fatty liver diagnosis based on a single sagittal view depicting both liver and kidney.19 Other authors fine-tuned a different CNN for fatty liver diagnosis.20 In another work, small regions of interest (ROIs) collected from homogeneous portions of the liver were used to develop a CNN from scratch for fatty liver diagnosis.21 Moreover, in a recent study, radiofrequency US data extracted from liver were utilized to train one-dimensional convolutional networks for the quantification of liver fat and NAFLD diagnosis.22
Following these promising results, we applied transfer learning with a pretrained deep CNN to assess liver fat using US images. Compared to the previous studies, we assessed the usefulness of four different US views of the liver for liver fat assessment. The rationale was that different views contain additional tissue features, such as blood vessels or kidney, which could potentially improve model performance.9 In addition, we explored whether the deep learning model logic can be visually explained by employing the class activation mapping (CAM) technique to highlight regions in liver images weighted by the models for fatty liver diagnosis.17
Materials and Methods
Study Design and Participants
This was an exploratory analysis in a prospectively conducted study at the University of California, San Diego (UCSD). An institutional review board approved this study. All procedures performed in studies involving human participants were in accordance with the Ethical Standards of the University of California, San Diego, USA and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Written informed consent was obtained. Research participants were recruited prospectively between 2016 and 2018 by a hepatologist. Inclusion criteria were age ≥18 years, known or suspected NAFLD, and willingness and ability to participate. Exclusion criteria were clinical, laboratory, or histology evidence of a liver disease other than NAFLD, excessive alcohol consumption (≥ 14 (men) or ≥7 (women) drinks/week), and steatogenic or hepatoxic medication use. Demographic and anthropometric data were recorded. Hepatic US and MRI research examinations were performed on the same day if possible or otherwise within 60 days.
US Protocol
A standardized US protocol was designed in consensus by a fellowship-trained abdominal faculty radiologist, a US medical physicist, and two experienced registered diagnostic medical sonographers. The protocol was electronically added to the scanner with a preset sequence designed to efficiently and consistently capture the required US views in the same order. US was performed (Siemens S3000, 4C1 transducer, Siemens Healthineers, Germany) on each participant by one of three experienced (>10 years) registered diagnostic medical sonographers trained in the protocol. Sonographers were free to adjust scanner parameters to capture the best-quality images for each view and participant, with participants suspending breathing on shallow expiration. US images from the following four views were included in this study’s analysis for each participant:
Three views in transverse plane: hepatic veins at the confluence with the inferior vena cava, right portal vein, and right posterior portal vein
One view in sagittal plane: liver and kidney
These views were selected based on their favorable performance in previous papers.9,10 Importantly, each view is routinely obtained during clinical US exams of the liver.
MRI Protocol and PDFF Estimation
MRI was performed at 3T (Signa HD, GE Healthcare, MN) with participants supine and an 8-channel torso phased-array coil centered over the liver. PDFF was estimated with a gradient recalled echo acquisition and magnitude-based reconstruction technique. A low flip angle was used to minimize T1 bias, and six gradient-recalled echoes were acquired at successive nominal out-of-phase and in-phase echo times to separate fat and water signals while calculating and correcting for T2* signal decay.23 Using a custom algorithm, the PDFF was computed pixel by pixel to generate PDFF maps. Blinded to US results, a trained analyst placed 1-cm radius ROIs in each of the nine Couinaud segments. The PDFF values from each ROI were averaged to yield composite per-participant PDFF values.
Model Development and Evaluation
We developed two types of models. First, a regression model was designed to quantify liver fat; the ground truth was PDFF as a continuous variable. Second, we designed two classifiers to diagnose fatty liver; the PDFF values for classification were set to 5 and 10%, respectively—5% is a common threshold for diagnosing NAFLD, while 10% is a common threshold for enrolling patients in clinical trials.24,25 All models were developed by transfer learning with the ResNet-50 CNN pretrained on the ImageNet dataset.26,27 Images were cropped to remove nonrelevant data, such as dark borders and annotations. To generate more data for the training and to promote learning that would be insensitive to image perturbations expected to occur in clinical practice, small image shifts (up to 20 pixels) in the lateral direction were applied before image cropping. In addition, liver images were reflected horizontally.
To enable the CNN to extract features from the US images, the images were resized using bicubic interpolation to 224 × 224 pixels and preprocessed following the settings originally designed for the pretrained CNN.26,27 Next, the images were input to the network, and the neural features were extracted from the CNN’s global average pooling block, which averages image representations generated by the preceding convolutional blocks, producing 2048 features. Based on the extracted features, a logistic regression algorithm with L1 loss was used to develop diagnosis models, and the Lasso regression method was used to develop PDFF quantification models.19 Similarly, the Lasso method due to L1 (absolute value of magnitude) regularization promotes the selection of sparse and efficient feature sets for regression.5 In addition, we used the CAM technique to highlight regions weighted by the models for fatty liver diagnosis on liver US images for each view separately for the classification cutoff of PDFF of 5% (Appendix 1).17
We initially developed the classification and regression models for each of the four US views separately. To leverage the combined information from the four views, we applied a multiview learning approach; the outputs of models trained separately for each view were averaged to give the final estimate (hereafter referred to as “averaged multiview model”).
Participant-specific leave-one-out cross-validation was applied to assess the performance of all models (all four single-view diagnoses, all four single-view quantifications, multiview diagnosis, multiview quantification). For each training round, the training set comprised data from 134 participants, and the test results were calculated using the data from the single left-out participant. For each training round, we additionally applied a stratified 4-fold cross-validation, and the grid search method was used to select the better-performing algorithm hyperparameters. Cost functions for the classifiers were weighted inversely proportional to the corresponding class frequency to address class imbalance. For the Lasso regression, the cost function was adjusted for each participant using weights inversely proportional to the PDFF value frequencies.
Statistical Analysis
To evaluate the performance of the model-based classifiers, we performed receiver operating characteristic (ROC) analyses and calculated areas under the curve (AUC). Differences in mean AUC values calculated using the bootstrap method were compared with the Welch’s t-test with Bonferroni correction. In addition, using the point on the ROC curve closest to the upper left corner (0, 1) as a diagnostic cutoff, we calculated accuracies, sensitivities, and specificities of the classifiers.7 For each regression model, we calculated the Spearman correlation coefficient (SCC) and Pearson’s correlation coefficient (PCC) to assess the monotonicity and linearity between the models’ outputs and PDFF values, respectively. To compare the regression results produced by different models, we used the Meng test.6,28 In addition, Bland–Altman analysis was applied to compare the performance of the multiview learning model and the better-performing single-view models. MATLAB 2019a (Mathworks, MA) and Python 3.5.2 were used for calculations. The pretrained network was implemented in Keras 2.2.4 with Tensorflow backend.29
The class activation maps were reviewed retrospectively and qualitatively by two authors (MB and MA) in consensus without blinding to explore how the highlighted areas relate to features known to be used by radiologists in assessing steatosis subjectively.
Results
Participants Characteristics
A total of 135 adult participants (76 female, 59 male) recruited in chronological order met the eligibility criteria and were included in the analysis. Their mean age and body mass index values were 52 ± 13 years and 31 ± 5 kg/m2, respectively. The distribution of the per-participant PDFF values in the study cohort is presented in Figure 1. Seventeen participants had PDFF<5%, 51 had PDFF<10%, and 5 had PDFF>30%. This distribution of PDFF is representative of the local clinical population with known or suspected NAFLD. Figure 2 shows three sets of four US images corresponding to livers with PDFF values of 7, 19, and 39%, respectively. Significant vein blurring and lower liver/kidney contrast can be observed due to the increasing fat accumulation.
Figure 1.
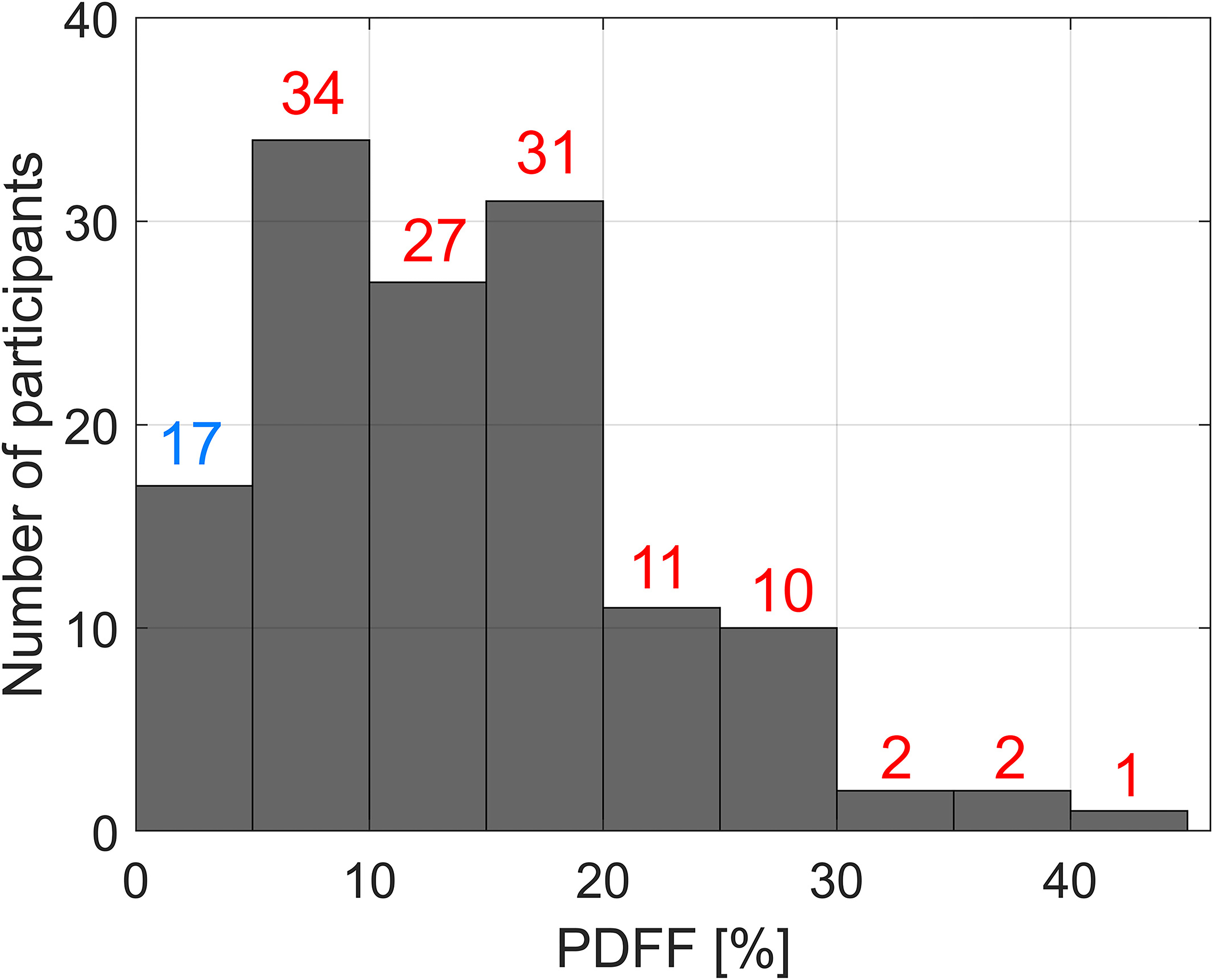
The distribution of the magnetic resonance proton density fat fraction (PDFF) values in the dataset. A PDFF cutoff value of ≥5% was used as the reference standard for the diagnosis of fatty liver disease.
Figure 2.
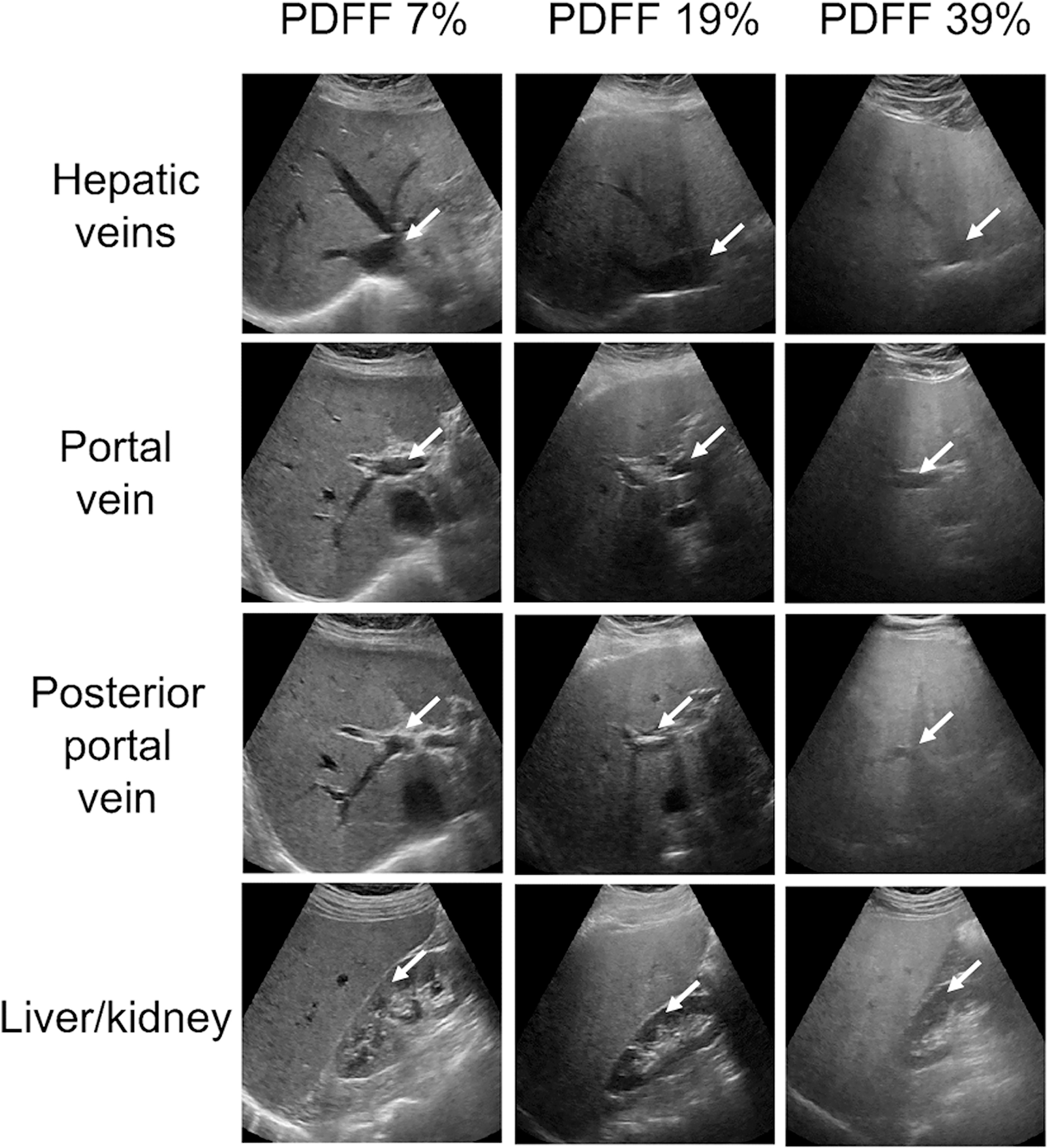
Three sets of ultrasound images (four views) corresponding to livers with different PDFF values of 7, 19, and 39%, respectively. Increasing fat accumulation causes blurring of veins and can obscure the liver/kidney interface. PDFF, proton density fat fraction. Blood vessels and kidney region were indicated with white arrows.
Performance
Table 1 lists the performance results obtained for the diagnosis models developed for each view separately and by employing the multiview learning approaches. In the case of the single liver views and the PDFF cutoff of 5%, the largest AUC value of 0.90 was achieved for the view of the right posterior portal vein in the transverse plane. The lowest AUC value of 0.76 was achieved for the view including the liver/kidney. The highest AUC value of 0.91 was achieved by the averaged multiview model but was not significantly higher than for the right posterior portal vein view (P-value >.01). Results obtained for the classifiers developed for the PDFF cutoff of 10% are presented in Table 2. The highest AUC value, 0.83, was obtained for the view of the hepatic veins, while for the remaining views, AUC values ranged from 0.77 to 0.79. The averaged multiview model achieved an AUC value of 0.86. ROC curves for the better-performing methods are presented in Figure 3.
Table 1.
Performance (Mean ± Standard Deviation) of the Models Developed using Ultrasound Images Corresponding to Different Liver Views and Both Multiview Methods
| View or model | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
|
| ||||
| Hepatic veins | 0.85 ± 0.03 | 0.80 ± 0.04 | 0.80 ± 0.05 | 0.82 ± 0.06 |
| Right portal vein | 0.83 ± 0.04 | 0.68 ± 0.09 | 0.66 ± 0.11 | 0.82 ± 0.09 |
| Right posterior portal vein | 0.90 ± 0.03 | 0.81 ± 0.03 | 0.79 ± 0.04 | 0.94 ± 0.05 |
| Liver/kidney | 0.76 ± 0.04 | 0.81 ± 0.06 | 0.83 ± 0.07 | 0.65 ± 0.08 |
| Averaged multiview model | 0.91 ± 0.03 | 0.81 ± 0.04 | 0.80 ± 0.05 | 0.88 ± 0.05 |
Model averaging corresponds to averaging all of the outputs of models developed for each view separately. Cutoff for training was set to fat fraction of 5%. AUC—area under the receiver operating characteristic curve. Accuracy, sensitivity, and specificity were calculated using a threshold defined as the point on the AUC curve closest to the left upper corner.
Table 2.
Performance (Mean ± Standard Deviation) of the Models Developed Using ultrasound Images Corresponding to Different Liver Views and Both Multiview Methods
| View or model | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
|
| ||||
| Hepatic veins | 0.83 ± 0.03 | 0.79 ± 0.03 | 0.81 ± 0.05 | 0.76 ± 0.05 |
| Right portal vein | 0.77 ± 0.03 | 0.75 ± 0.03 | 0.70 ± 0.04 | 0.82 ± 0.05 |
| Right posterior portal vein | 0.79 ± 0.03 | 0.73 ± 0.03 | 0.73 ± 0.05 | 0.75 ± 0.05 |
| Liver/kidney | 0.77 ± 0.03 | 0.72 ± 0.03 | 0.69 ± 0.05 | 0.76 ± 0.05 |
| Averaged multiview model | 0.86 ± 0.03 | 0.85 ± 0.02 | 0.83 ± 0.03 | 0.88 ± 0.04 |
Model averaging corresponds to averaging all of the outputs of models developed for each view separately. Cutoff for training was set to fat fraction of 10%. AUC – area under the receiver operating characteristic curve. Accuracy, sensitivity, and specificity were calculated using a threshold defined as the point on the AUC curve closest to the left upper corner.
Figure 3.
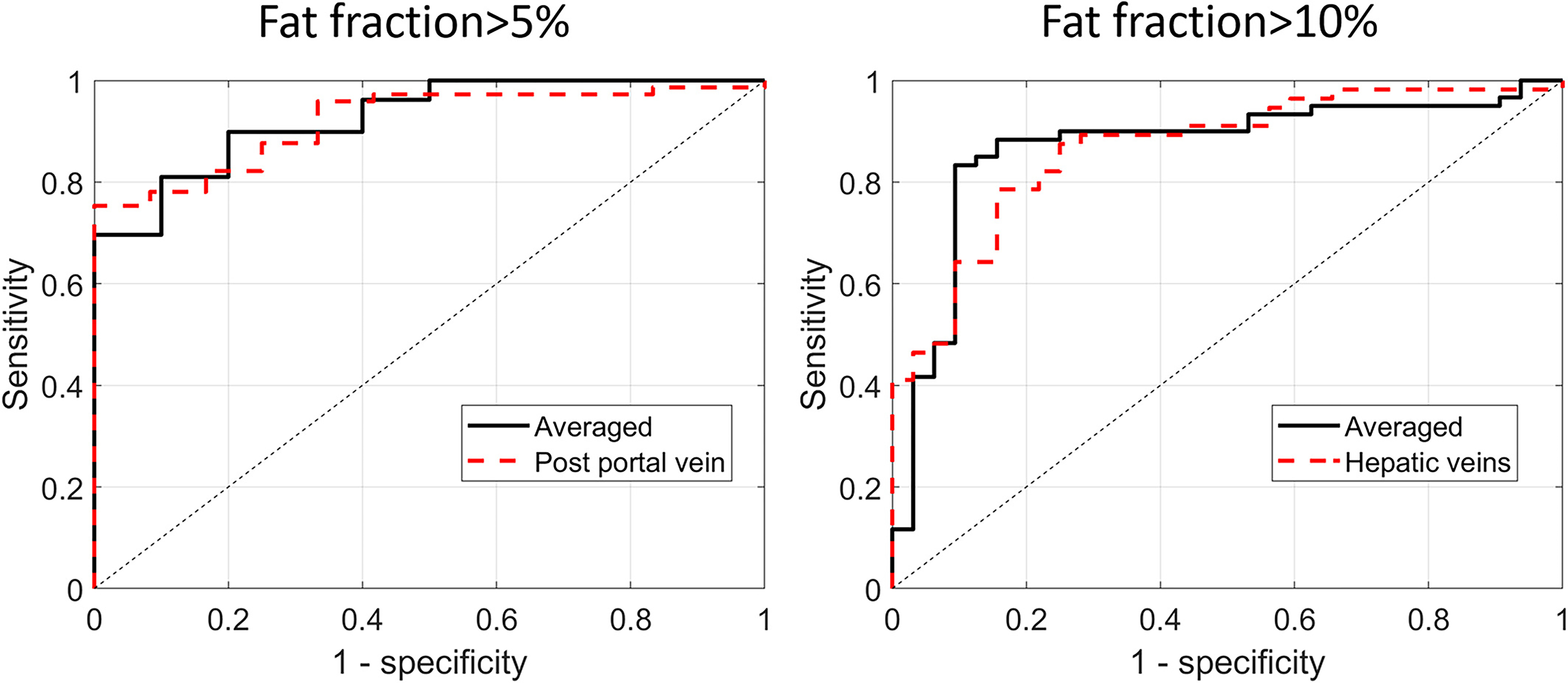
The receiver operating characteristic curves obtained using better performing single and multiview approaches to fatty liver diagnosis for the fat fraction cutoff set to (A) 5% and (B) 10%. The areas under the curves were equal to 0.90 (cutoff of 5%) and 0.83 (cutoff of 10%) for the posterior portal vein and hepatic veins views, respectively. The areas under the curves were equal to 0.91 (cutoff of 5%) and 0.86 (cutoff of 10%) for the averaged multiview model.
Table 3 lists the performance results obtained for the PDFF quantification models developed for each view separately and by employing the multiview learning approach. Correlation coefficients, SCC and PCC, calculated for all regression models were statistically significant (P-values <.01). Among the four individual views, the highest SCC of 0.78 and PCC of 0.77 were achieved by the view that included the right posterior portal vein. The averaged multiview model achieved SCC and PCC values equal to 0.81 and 0.78, respectively. However, the differences between the multiview quantification model and the model utilizing the right posterior portal vein view were not significant (P-value >.01). The relationships and Bland–Altman plots of MRI-PDFF and those using machine learning approaches are shown in Figure 4. The models underestimated PDFF values when the MRI-PDFF exceeded 30%, resulting in bias values of approximately 15% for the higher PDFF values as presented in Bland–Altman plots in Figure 4.
Table 3.
Fat Fraction Quantification Performance of the Models Developed Using Ultrasound Images Corresponding to Different Liver Views and Both Multiview Methods
| View or model | SCC | PCC |
|---|---|---|
|
| ||
| Hepatic veins | 0.75 | 0.72 |
| Right portal vein | 0.54 | 0.51 |
| Right posterior portal vein | 0.78 | 0.77 |
| Liver/kidney | 0.58 | 0.61 |
| Averaged multiview model | 0.81 | 0.78 |
Model averaging corresponds to averaging all of the outputs of models developed for each view separately. All Spearman and Pearson’s correlation coefficients (SCC and PCC, respectively) were statistically significant (P-values <.01).
Figure 4.
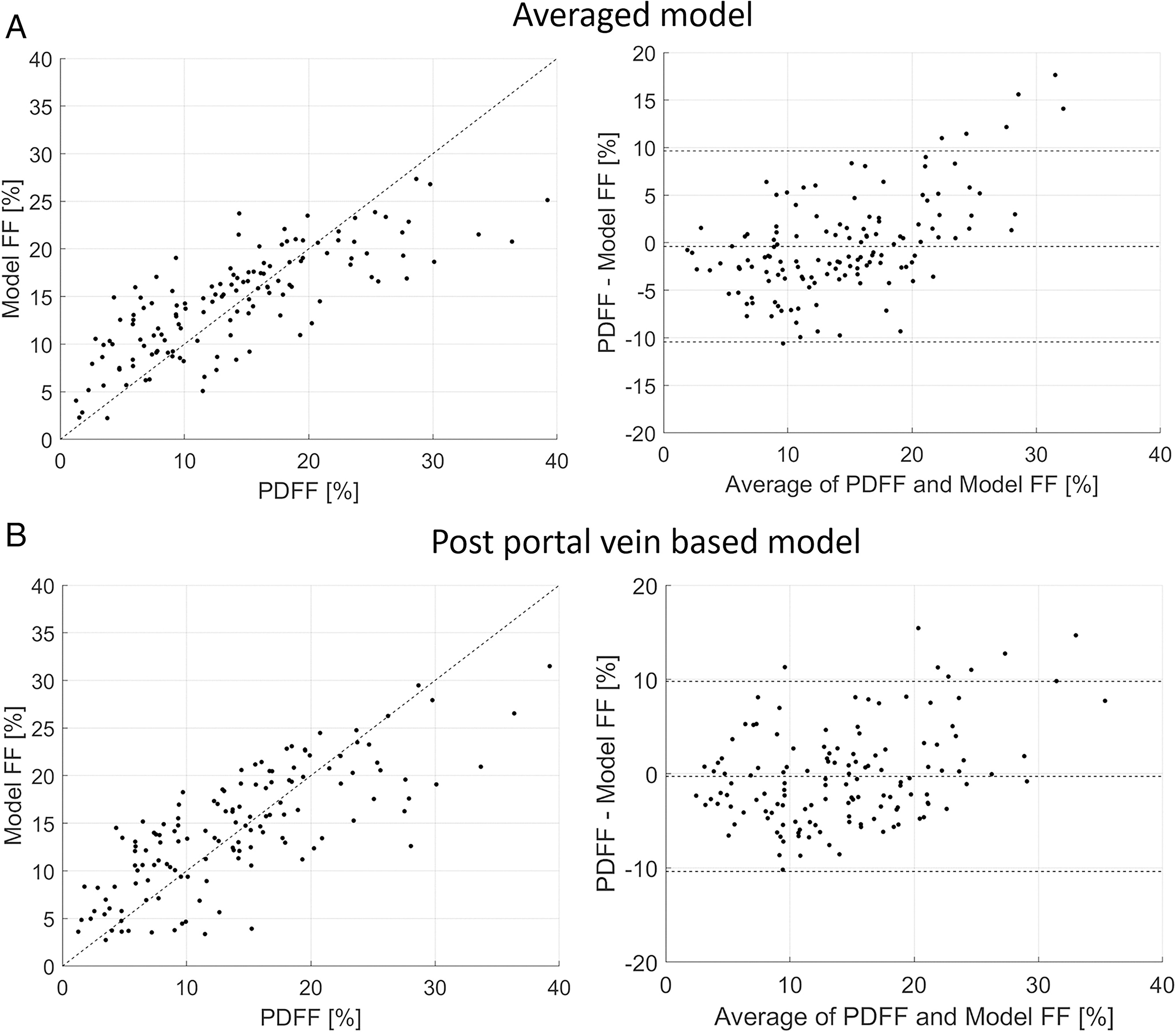
The relationships and Bland–Altman plots obtained in the case of the Lasso regression for (A) the averaged multiview model (SCC = 0.81, PCC = 0.78) and (B) the composite multiview model (SCC = 0.73, PCC = 0.70). PDFF, proton density fat fraction, SCC, Spearman correlation coefficient, PCC, Pearson’s correlation coefficient.
Figure 5 shows representative class activation maps generated using individual-view deep learning models on a patient with an average PDFF of 17%. This case was correctly assessed by all four classifiers. Regions highlighted with red and blue correspond to positive and negative weights, respectively, on the attention maps. As depicted, blood vessel-liver and liver-kidney regions are highlighted in blue, whereas superficial portions of the liver are highlighted in red. In comparison, Figure 6 shows activation maps obtained for misclassified cases, where the CAM technique performed worse at highlighting important image features.
Figure 5.
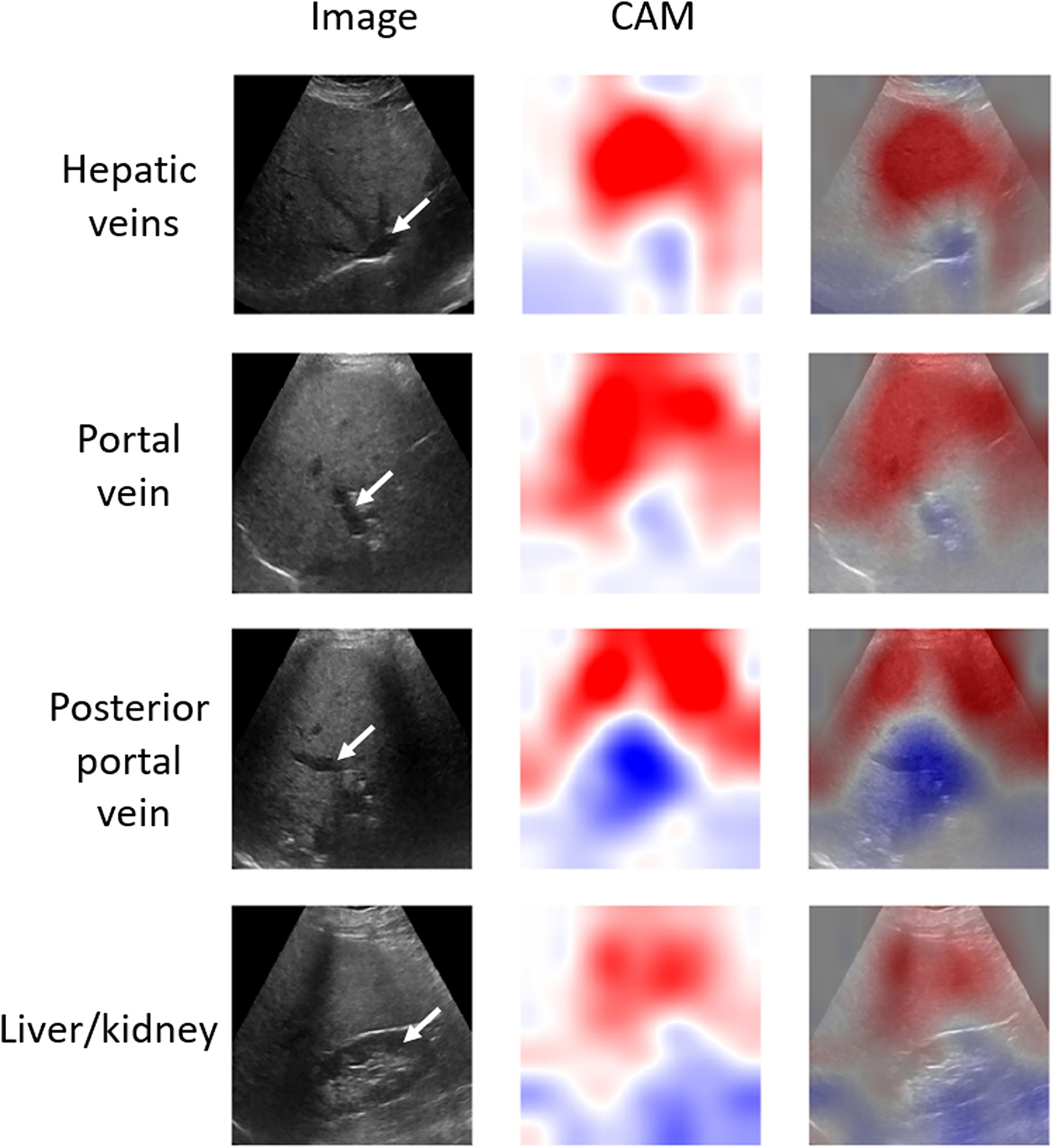
Activations maps generated for different views of a correctly classified case with proton density fat fraction of 17% using classifiers trained for each view separately. Blood vessels and kidney region were indicated with white arrows. Red color is related to positive weights in the class activation mapping method, while blue regions correspond to negative weights. The models assign strongly positive weights to upper parts of images and assign strongly negative weights to portions of the liver with characteristic image features (eg, blood vessels or liver/kidney regions). Notice rib shadows in the posterior portal vein and liver/kidney views. CAM stands for the class activation map.
Figure 6.
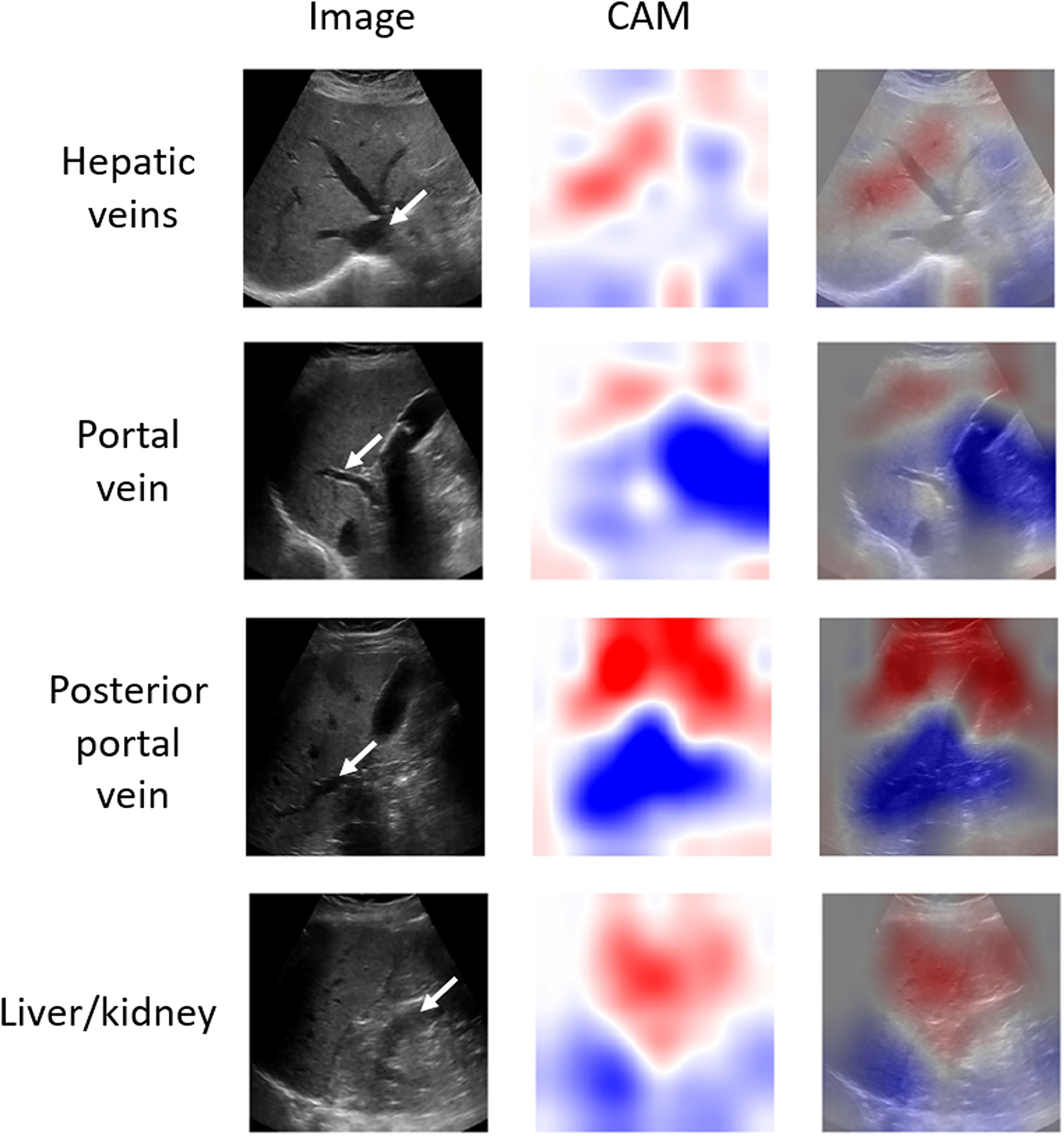
Activations maps generated for different views of misclassified cases using classifiers trained for each view separately. Blood vessels and kidney are indicated with white arrows. In comparison to the maps obtained for a correctly classified case from Figure 5, these maps show lower performance at highlighting characteristic image features (eg, blood vessels or liver/kidney regions), which could be explained by the presence of a confounder, such as the posterior shadowing of the gallbladder in portal vein and posterior portal vein views. CAM stands for the class activation map.
Discussion
Our study illustrates the feasibility of deep learning methods for assessing liver fat with four US views of the liver routinely obtained for clinical care, using contemporaneous MRI-PDFF as the reference standard. By using transfer learning with a pretrained CNN, we developed well-performing models for fatty liver diagnosis and liver fat quantification. Our study shows that all four liver views can yield good individual diagnostic performance. Thus, each view provides useful image features for liver fat assessment, and those features can be extracted by deep learning. Moreover, combining outputs from the individual-view models may improve the diagnostic performance as illustrated by the slightly better performance scores obtained by the multiview approach in comparison to the single-view methods. This may also result from the fact that different views were acquired from different parts of the liver; therefore, the overall estimates might be closer to those obtained using MRI, which reflects whole-liver PDFF by averaging PDFF values from each anatomic liver segment.
In a previous study of qualitative US image features for hepatic steatosis assessed by experienced radiologists, vein blurring was among the most important individual liver features.9 Our study confirmed that that the vein-blurring liver feature is important for liver fat assessment. However, our result was obtained by using an automated algorithm rather than image interpretation by radiologists. Moreover, the model based on views presenting veins achieved higher performance than the liver/kidney view.
The results presented here confirm the usefulness of deep learning methods for fatty liver assessment presented in two previous papers19,20 in which transfer learning with a CNN was applied to classify fatty liver images. The first paper reported a high AUC value of 0.977.19 The deep learning model was developed using a set of 55 participants with histology analysis of liver biopsies as the ground truth. The second paper reported a high AUC value of 0.96 with a deep learning method using a set of 157 liver images20 for which the radiologist’s qualitative score was used as the ground truth. Due to different datasets, enrolment criteria, and reference standards, it is difficult to directly compare the results reported in previous papers with the presented study. In our study, PDFF values derived from CSE-MRI served as the ground truth. The advantage of our approach is that PDFF is quantitative, objective, noninvasive, accurate, reproducible, and representative of the whole liver.30 In comparison, histological analysis of liver biopsy specimens is subjective and prone to spatial sampling variability.3 The present study’s deep learning fat fraction estimates are limited by the accuracy of the MRI PDFF values.
To our knowledge, this is the first study assessing attention maps for CNN-based liver assessment. The CAM technique can be potentially useful for identifying the regions on liver images more important for the deep learning models. In the case of the three transverse liver views (Figure 5), the model highlighted the superficial part of the liver (red, positive weights) and hepatic-vessel interface veins (blue, negative weights). Similarly, in the case of the fourth view, the liver area and the liver-kidney area were highlighted. Our observations suggest that the models may have focused on image features known to be relevant for qualitative liver fat assessment by radiologists.9,10 On the other hand, the activation maps obtained for the misclassified cases (Figure 6) were more difficult to assess. For these maps, the important regions were not highlighted as clearly as in the case of the correctly classified images. Our qualitative results show that the CAM technique may help assess the performance of the classifier. However, an in-depth systematic study of the attention maps may be warranted to better understand their usefulness in liver image analysis. It has to be stated that, for our study, we collected only the images corresponding to the views that were reported to be useful in the previous papers.9,10 As the models need to average the attention maps to assess liver fat (see Appendix E1), the decisions taken by the models are based on the presence of particular US image features, such as veins, and the appearance of the liver. The presence of these image features is responsible for the specific activations of the deep learning models and the appearance of the generated attention maps. Therefore, in the future, it would be interesting to include images from other liver regions and investigate whether those can further improve liver fat assessment and what portions of those images are highlighted by the CAM.
The main limitation related to our study is the relatively small number of cases in the dataset, so it cannot be robustly divided into separate training, validation, and test sets. In addition, the dataset was recruited from a clinical population taken in chronological order and thus is a representative sample for diagnostic purposes but probably not for screening. Not surprisingly, there is a reduced number of participants with low fat content <5% and very high fat content >30%. However, for deep learning training methods, it is unbalanced with respect to liver fat content. We used transfer learning with a single pretrained deep CNN that served as a fixed feature extractor, and it would be interesting to assess the usefulness of different pretrained networks. Moreover, we did not assess the impact of possible confounders, such as gallbladder, rib shadows, and gallstone shadows, on the performance of the deep learning models. Such confounders can be difficult to avoid in some patients, making the models less robust in practice.
Future work may benefit from a larger study population that may even allow training a deep learning model from scratch. This might also result in more suitable and precise CAMs.31,32 For example, the so-called interpretable CNNs automatically relate convolutional filters in deep layers with image objects during training, giving more precise activation maps than the traditional CNN models (30). Furthermore, studies of repeatability and reproducibility, including comparisons of different scanners and operators, are needed to address the generalizability of the methods presented here. Variability in the acquisition of the image views and contributions of artifacts also need to be studied. Deep learning models developed using images from a particular liver view may overfit to image features that are unique to this particular view, such as the presence of veins. Given the spatial variability of liver structure and its adjacent tissues, for the model to work in a clinical setting, it would likely require a precisely controlled acquisition protocol such as that employed in this study to collect the 2D US data in comparable planes. The scanner operator would need to be trained to acquire the same views that were used to develop the computer-aided diagnosis system and to minimize artifacts. Thus, the performance of the 2D liver fat assessment deep learning method might be operator- or protocol-dependent.
Conclusion
In this study, we applied deep learning to develop efficient regression and classification models for fatty liver assessment based on US images collected from different liver views. CAMs depicted that the decisions taken by the deep models can be interpreted, and it preliminarily appears that they can be approximately related to the image features usually taken into account by radiologists.
Supplementary Material
Acknowledgments
The authors thank the research participants for making this study possible. They also gratefully acknowledge the invaluable contributions of the sonographers, Elise Housman, Susan Lynch, and Minaxi Trivedi, for their dedicated contributions and expertise, as well as the clinical coordinator Vivian Montes for her outstanding organization of this complex study. This study has received support from the National Institutes of Health (R01DK106419) and a research agreement from Siemens Healthineers, USA. Rohit Loomba receives funding support from NIEHS (5P42ES010337), NCATS (5UL1TR001442), NIDDK (R01DK106419, P30DK120515), and DOD PRCRP (CA170674P2).
Abbreviations
- AUC
area under the receiver operating characteristics curve
- CAM
class activation mapping
- CNN
convolutional neural network
- CSE-MRI
confounder-corrected chemical shift-encoded magnetic resonance imaging
- NAFLD
Nonalcoholic fatty liver disease
- PDFF
proton density fat fraction
- ROI
region of interest
- SCC
Spearman correlation coefficient
- US
ultrasound
Footnotes
Michal Byra, Aiguo Han, Andrew S. Boehringer, Yingzhen N. Zhang, William D. O’Brien, Jr, John W. Erdman, and Michael Andre declare that they have no conflicts of interest. Rohit Loomba serves as a consultant or advisory board member for Arrowhead Pharmaceuticals, Astra Zeneca, Bird Rock Bio, Boehringer Ingelheim, Bristol-Myer Squibb, Celgene, Cirius, CohBar, Conatus, Eli Lilly, Galmed, Gemphire, Gilead, Glympse bio, GNI, GRI Bio, Intercept, Ionis, Janssen Inc., Merck, Metacrine, Inc.,NGM Biopharmaceuticals, Novartis, Novo Nordisk, Pfizer, Prometheus, Sanofi, Siemens, and Viking Therapeutics. In addition, his institution has received grant support from Allergan, Boehringer-Ingelheim, Bristol-Myers Squibb, Cirius, Eli Lilly and Company, Galectin Therapeutics, Galmed Pharmaceuticals, GE, Genfit, Gilead, Intercept, Grail, Janssen, Madrigal Pharmaceuticals, Merck, NGM Biopharmaceuticals, NuSirt, Pfizer, pH Pharma, Prometheus, and Siemens. Claude Sirlin serves as consultant for Blade, Boehringer, and Epigenomics. His institution provides consultation to BMS, Exact Sciences, and IBMWatson. His institution has received grant support from Bayer, GE, Gilead, Philips, and Siemens. His institution has had lab service agreements with Enanta, Gilead, ICON, Intercept, Organovo, Nusirt, Shire, Synageva, and Takeda.
Contributor Information
Michal Byra, Department of Radiology, University of California, La Jolla, California, USA; Department of Ultrasound, Institute of Fundamental Technological Research, Polish Academy of Sciences, Warsaw, Poland.
Aiguo Han, Bioacoustics Research Laboratory, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA.
Andrew S. Boehringer, Liver Imaging Group, Department of Radiology, University of California, La Jolla, California, USA.
Yingzhen N. Zhang, Liver Imaging Group, Department of Radiology, University of California, La Jolla, California, USA.
William D. O’Brien, Jr, Bioacoustics Research Laboratory, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA.
John W. Erdman, Jr, Department of Food Science and Human Nutrition, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA.
Rohit Loomba, NAFLD Research Center, Division of Gastroenterology, Department of Medicine, University of California, La Jolla, California, USA.
Claude B. Sirlin, Liver Imaging Group, Department of Radiology, University of California, La Jolla, California, USA.
Michael Andre, Department of Radiology, University of California, La Jolla, California, USA.
References
- 1.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol 2013; 10:686. [DOI] [PubMed] [Google Scholar]
- 2.Rinella ME. Nonalcoholic fatty liver disease: a systematic review. Jama 2015; 313:2263–2273. [DOI] [PubMed] [Google Scholar]
- 3.Tapper EB, Lok AS-F. Use of liver imaging and biopsy in clinical practice. N Engl J Med 2017; 377:756–768. [DOI] [PubMed] [Google Scholar]
- 4.Schwenzer NF, Springer F, Schraml C, Stefan N, Machann J, Schick F. Non-invasive assessment and quantification of liver steatosis by ultrasound, computed tomography and magnetic resonance. J Hepatol 2009; 51:433–445. [DOI] [PubMed] [Google Scholar]
- 5.Chou Y-H, Tiu C-M, Hung G-S, Wu S-C, Chang TY, Chiang HK. Stepwise logistic regression analysis of tumor contour features for breast ultrasound diagnosis. Ultrasound Med Biol 2001; 27:1493–1498. 10.1016/S0301-5629(01)00466-5. [DOI] [PubMed] [Google Scholar]
- 6.Meng X-L, Rosenthal R, Rubin DB. Comparing correlated correlation coefficients. Psychol Bull 1992; 111:172. [Google Scholar]
- 7.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett 2006; 27:861–874. 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 8.Lapadat AM, Jianu IR, Ungureanu BS, et al. Non-invasive imaging techniques in assessing non-alcoholic fatty liver disease: a current status of available methods. J Med Life 2017; 10:19. [PMC free article] [PubMed] [Google Scholar]
- 9.Hong CW, Marsh A, Wolfson T, et al. Reader agreement and accuracy of ultrasound features for hepatic steatosis. Abdom Radiol 2019; 44:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernaez R, Lazo M, Bonekamp S, et al. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology 2011; 54:1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin SC, Heba E, Wolfson T, et al. Noninvasive diagnosis of non-alcoholic fatty liver disease and quantification of liver fat using a new quantitative ultrasound technique. Clin Gastroenterol Hepatol 2015; 13:1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall RH, Eissa M, Bluth EI, Gulotta PM, Davis NK. Hepatorenal index as an accurate, simple, and effective tool in screening for steatosis. Am J Roentgenol 2012; 199:997–1002. [DOI] [PubMed] [Google Scholar]
- 13.Webb M, Yeshua H, Zelber-Sagi S, et al. Diagnostic value of a computerized hepatorenal index for sonographic quantification of liver steatosis. Am J Roentgenol 2009; 192:909–914. [DOI] [PubMed] [Google Scholar]
- 14.Bharti P, Mittal D, Ananthasivan R. Computer-aided characterization and diagnosis of diffuse liver diseases based on ultrasound imaging: a review. Ultrason Imaging 2017; 39:33–61. [DOI] [PubMed] [Google Scholar]
- 15.Wu C-C, Yeh W-C, Hsu W-D, et al. Prediction of fatty liver disease using machine learning algorithms. Comput Methods Programs Biomed 2019; 170:23–29. [DOI] [PubMed] [Google Scholar]
- 16.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017; 42:60–88. 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016). Learning deep features for discriminative localization. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 2921–2929. [Google Scholar]
- 18.Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 2016; 35:1299–1312. [DOI] [PubMed] [Google Scholar]
- 19.Byra Michałand Styczynski G, Szmigielski C, Kalinowski P, et al. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg 2018; 13:1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy DS, Bharath R, Rajalakshmi P (2018) A novel computer-aided diagnosis framework using deep learning for classification of fatty liver disease in ultrasound imaging. 2018 IEEE 20th International Conference on e-Health Networking, Applications and Services (Healthcom). pp. 1–5 [Google Scholar]
- 21.Reddy DS, Bharath R, Rajalakshmi P (2018) Classification of non-alcoholic fatty liver texture using convolution neural networks. 2018 IEEE 20th International Conference on e-Health Networking, Applications and Services (Healthcom). pp. 1–5 [Google Scholar]
- 22.Han A, Byra M, Heba E, et al. Noninvasive diagnosis of non-alcoholic fatty liver disease and quantification of liver fat with radiofrequency ultrasound data using one-dimensional convolutional neural networks. Radiology 2020; 295:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoo T, Shiehmorteza M, Hamilton G, et al. Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology 2011; 258:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomba R MRI-PDFF treatment response criteria in nonalcoholic steatohepatitis. Hepatology 2020. 10.1002/hep.31624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castera L, Friedrich-Rust M, Loomba R. Noninvasive assessment of liver disease in patients with nonalcoholic fatty liver disease. Gastroenterology 2019; 156:1264–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009). Imagenet: a large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition, 2009. CVPR 2009. pp. 248–255 [Google Scholar]
- 27.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning.
- 28.Diedenhofen B, Musch J. Cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One 2015; 10:e0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abadi M, Barham P, Chen J, et al. TensorFlow: A System for Large-Scale Machine Learning. OSDI; 2016:265–283. [Google Scholar]
- 30.Yokoo T, Serai SD, Pirasteh A, et al. Linearity, bias, and precision of hepatic proton density fat fraction measurements by using MR imaging: a meta-analysis. Radiology 2017; 286:486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Nian Wu Y, Zhu S-C (2018). Interpretable convolutional neural networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 8827–8836. [Google Scholar]
- 32.Zhou B, Bau D, Oliva A, Torralba A. Interpreting deep visual representations via network dissection. IEEE Trans Pattern Anal Mach Intell 2018; 41:2131–2145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


