Background:
Nucleocapsid antigenemia in adults has demonstrated high sensitivity and specificity for acute infection, and antigen burden is associated with disease severity. Data regarding SARS-CoV-2 antigenemia in children are limited.
Methods:
We retrospectively analyzed blood plasma specimens from hospitalized children with COVID-19 or MIS-C. Nucleocapsid and spike were measured using ultrasensitive immunoassays.
Results:
We detected nucleocapsid antigenemia in 62% (50/81) and spike antigenemia in 27% (21/79) of children with acute COVID-19 but 0% (0/26) and 15% (4/26) with MIS-C from March 2020–March 2021. Higher nucleocapsid levels were associated with radiographic infiltrates and respiratory symptoms in children with COVID-19.
Conclusions:
Antigenemia lacks the sensitivity to diagnose acute infection in children but is associated with signs and symptoms of lower respiratory tract involvement. Further study into the mechanism of antigenemia, its association with specific organ involvement, and the role of antigenemia in the pathogenesis of COVID-19 is warranted.
Keywords: nucleocapsid, spike, antigenemia, COVID-19, SARS-CoV-2, pediatrics
BACKGROUND
SARS-CoV-2 proteins in the blood (antigenemia) of individuals with COVID-19 have been well described in adults but limited data are available for children, in whom clinical manifestations of SARS-CoV-2 infection are more heterogeneous. Viral nucleocapsid antigenemia has been identified as a sensitive and specific biomarker of acute infection,1–5 and both nucleocapsid and spike antigenemia have been associated with disease severity in adults.3–5 Spike protein antigenemia has also been inconsistently observed in children with SARS-CoV-2 associated multisystem inflammatory syndrome (MIS-C).6,7
Due to limited evidence, the clinical utility of detecting and quantifying SARS-CoV-2 proteins in peripheral blood from children with SARS-CoV-2 is uncertain. We, therefore, sought to determine whether antigenemia is a marker of acute SARS-CoV-2 infection or is associated with disease severity or clinical manifestations in children. We measured nucleocapsid and spike antigenemia in peripheral blood from pediatric patients with COVID-19, MIS-C, and ten healthy controls. We examined the prevalence and magnitude of antigenemia with respect to symptom onset, diagnosis and hospitalization, and association with disease severity and symptomatology.
METHODS
Patients and Specimens
Pediatric patients 0–21 years of age hospitalized at Children’s Healthcare of Atlanta (CHOA) from March 17, 2020, through March 30, 2021, were eligible for prospective blood and/or remnant plasma or sera collection (STUDY00000723) approved by the Emory University Institutional Review Board (IRB) following informed consent and assent, as applicable. A convenience sampling was collected and one specimen per patient was analyzed. Patients were classified as having COVID-19 if they had been symptomatic for no more than 14 days and had positive SARS-CoV-2 PCR testing from the nasopharynx within 14 days before to 3 days after blood sampling. Patients were not required to be hospitalized primarily for COVID-19. Patients were classified as having MIS-C if they met the CDC case definition: age less than 21 years, presenting with fever, laboratory evidence of inflammation, and evidence of severe multisystem disease, no alternative plausible diagnosis, and positive recent or current SARS-CoV-2 testing or exposure to a COVID-19 case within 4 weeks before the onset of symptoms.8 MIS-C cases with more than 14 days of symptoms at the time of blood sampling were excluded. COVID-19 severity scoring utilized the World Health Organization (WHO) ordinal scale (see Text, Supplementary Digital Content 1, http://links.lww.com/INF/E876).9 Samples collected more than 10 days after hospitalization were excluded from the analysis. Demographic, clinical, and outcome data were abstracted from the electronic medical record and entered into a Research Electronic Data Capture (REDCap) database.10,11 Controls were healthy outpatient children enrolled in a phlebotomy study (IRB00087446).
Assays
Nucleocapsid protein was measured on the Quanterix HD-X platform (Quanterix, Billerica, MA) using the SIMOA SARS-CoV-2 N Protein Antigen assay following 1:3 dilution of serum or plasma in diluent provided with the assay kit. Protein concentration was determined from a calibration curve constructed from concurrently assayed standards. Measured concentrations above 0.099 pg/mL were considered positive.12 Spike protein was assayed using the R-PLEX SARS-CoV-2 Spike assay (Meso Scale Diagnostics, MD). Samples were added to the kit undiluted except where insufficient sample volume was available in which case a 1:1.5 dilution with the provided diluent was performed. A calibration curve was constructed from concurrently assayed standards. Samples with signal more than 2.5 times the signal of the 0 pg/mL calibrator were considered positive.13
Data Analysis and Statistics
Clinical data were exported from REDCap and analyzed in MATLAB (Mathworks, Inc.). Comparisons between groups were made using the Wilcoxon rank sum test. 95% confidence intervals (CIs) for sample proportions were computed using the formula for standard error. Regression of scatter data and goodness of fit metrics were obtained using the MATLAB fit function.
RESULTS
Patient Sampling
Peripheral blood specimens from 81 COVID-19 and 26 MIS-C patients were included (Table 1). There was not a significant difference in the days of hospitalization at the time of sample collection between COVID-19 and MIS-C groups (P = 0.1, see Figure, Supplementary Digital Content 2, http://links.lww.com/INF/E876).
TABLE 1.
Clinical Characteristics of Patients with COVID-19, MIS-C and Healthy Controls
| COVID-19 (n = 81) | MIS-C (n = 26) | Controls (n = 10) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Total | Total | |||||||
| Age, years, median (IQR) | 14 | (5.8–16) | 81 | 9 | (5–13) | 26 | 10.8 | (4.3) | 10 |
| Sex, female, n (%) | 39 | (48.1) | 81 | 11 | (42.3) | 26 | 6 | (60.0) | 10 |
| Race, n (%) | |||||||||
| Asian | 2 | (2.5) | 81 | 1 | (3.8) | 26 | 0 | (0.0) | 10 |
| Black | 35 | (43.2) | 81 | 19 | (73.1) | 26 | 4 | (40.0) | 10 |
| White | 41 | (50.6) | 81 | 5 | (19.2) | 26 | 5 | (50.0) | 10 |
| Declined | 4 | (4.9) | 81 | 1 | (3.8) | 26 | 0 | (0.0) | 10 |
| Other | 0 | (0) | 81 | 0 | (0) | 26 | 1 | (10.0) | 10 |
| Ethnicity, n (%) | |||||||||
| Hispanic | 22 | (27.2) | 81 | 3 | (11.5) | 26 | 3 | (30.0) | 10 |
| Not Hispanic | 59 | (72.8) | 81 | 23 | (88.5) | 26 | 7 | (70.0) | 10 |
| Immune compromise, n (%) | |||||||||
| Diabetes | 6 | (7.4) | 81 | 0 | (0) | 26 | |||
| Sickle Cell Trait | 0 | (0) | 81 | 1 | (3.8) | 26 | |||
| Sickle Cell Disease | 3 | (3.7) | 81 | 1 | (3.8) | 26 | |||
| Lupus | 2 | (2.5) | 81 | 0 | (0) | 26 | |||
| Leukemia | 3 | (3.7) | 81 | 0 | (0) | 26 | |||
| Lymphoma | 1 | (1.2) | 81 | 0 | (0) | 26 | |||
| Other genetic disorders | 2 | (2.5) | 81 | 0 | (0) | 26 | |||
| Solid Organ Transplant | 1 | (1.2) | 81 | 0 | (0) | 26 | |||
| Timing of sampling | |||||||||
| Day of illnessa, mean (SD) | 5.7 | (3.3) | 81 | 5.7 | (3) | 26 | |||
| Day of hospitalization,mean (SD) | 2.3 | (1.9) | 81 | 1.7 | (1.7) | 26 | |||
| Symptoms, n (%) | |||||||||
| Fever | 60 | (74.1) | 81 | 25 | (96.2) | 26 | |||
| Respiratory symptoms | 51 | (63) | 81 | 10 | (38.5) | 26 | |||
| Anosmia or Ageusia | 7 | (8.6) | 81 | 1 | (3.8) | 26 | |||
| Gastrointestinal symptoms | 48 | (59.3) | 81 | 20 | (76.9) | 26 | |||
| Neurological symptoms | 26 | (32.1) | 81 | 12 | (46.2) | 26 | |||
| Ocular symptoms | 5 | (6.2) | 81 | 10 | (38.5) | 26 | |||
| Chest radiograph, n (%) | |||||||||
| Infiltrates | 33 | (51.6) | 64 | 12 | (52.2) | 23 | |||
| Pleural effusion | 10 | (15.9) | 63 | 6 | (27.3) | 22 | |||
| Outcomes | |||||||||
| WHO ordinal scale,bmean (SD) | 3.9 | (1.1) | 81 | ||||||
| Duration of hospitalization, mean (SD) | 6.1 | (5) | 81 | 6.9 | (3.8) | 26 | |||
| Days of ICU, mean (SD) | 4.9 | (4.4) | 35 | 5.1 | (2.2) | 17 | |||
| ICU admission, n (%) | 36 | (44.4) | 81 | 18 | (69.2) | 26 | |||
| Low flow O2, n (%) | 39 | (48.1) | 81 | 15 | (57.7) | 26 | |||
| Mechanical Ventilation, n (%) | 3 | (3.7) | 81 | 1 | (3.8) | 26 | |||
| Vasopressors, n (%) | 4 | (4.9) | 81 | 13 | (50) | 26 | |||
| Death, n (%) | 1 | (1.2) | 81 | 0 | (0) | 26 | |||
Day of illness describes days from the onset of COVID-19 symptoms for COVID-19 patients and days from the onset of MIS-C symptoms for MIS-C patients, many of whom did not have a distinct preceding acute COVID-19 syndrome.
WHO ordinal scale was designed for patients with COVID-19 and therefore was not calculated for patients with MIS-C or healthy controls in this study.
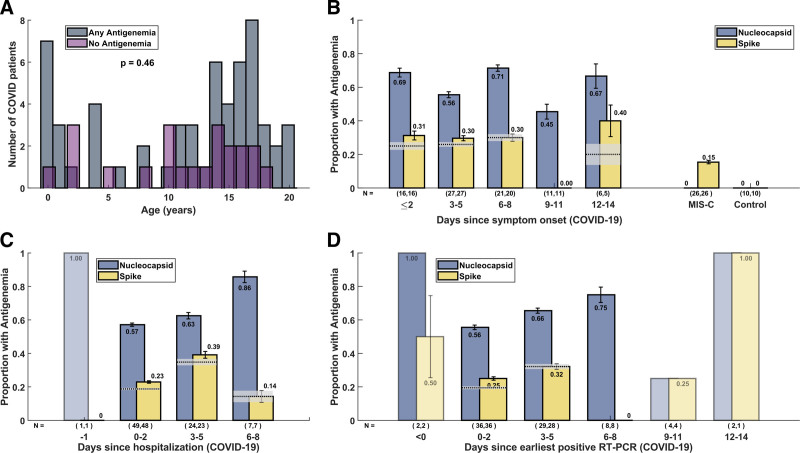
Prevalence of Antigenemia
81 COVID-19 samples were assayed for nucleocapsid protein and 79 samples were assayed for spike protein (due to insufficient volume of 2 samples). Age was not significantly different in COVID-19 patients with and without antigenemia (P = 0.46, Figure 1A), and nucleocapsid antigen levels did not differ in subgroups based on age (see Figure, Supplementary Digital Content 3, http://links.lww.com/INF/E876). Nucleocapsid antigenemia was present in 50 of 81 samples and spike antigenemia was present in 21 of 79 samples, corresponding to an overall sensitivity for acute COVID-19 of 62% for nucleocapsid and 27% for spike, respectively. Prevalence of nucleocapsid antigenemia was similar when samples were analyzed by days following hospitalization, symptom onset, and earliest positive real-time PCR (RT-PCR) (Figure 1B–D). Among 36 (44.4%) patients admitted to the ICU during their hospitalization, 21 (58%) had nucleocapsid antigenemia. Nine (26%) of 35 patients with spike measurements had spike antigenemia.
FIGURE 1.
Characteristics of COVID-19 patients and timing of blood sampling. (A) Histogram of ages of COVID-19 patients with and without antigenemia. The distribution of ages between these two groups was not significantly different by rank-sum test (P = 0.46). Panels B-D demonstrate the proportion and 95% CIs of COVID-19 patients with nucleocapsid or spike antigenemia stratified by time of blood sample collection with respect to (B) symptom onset, (C) hospital admission, and (D) earliest positive SARS-CoV-2 RT-PCR. MIS-C and control measurements are depicted in panel B for comparison. The proportion of patients with concurrent nucleocapsid and spike antigenemia is demonstrated with a dotted horizontal line (proportion) and gray box (95% CI). For groups with N < 5 CIs were not calculated and faded bars are displayed.
Among the 26 patients with MIS-C and 10 healthy controls, none had nucleocapsid antigenemia (Figure 1B). Four (15%) of 26 MIS-C patients had spike antigenemia. Median duration of illness at time of blood sampling was 5 days (range 0–14 days, IQR 3–8 days) for COVID-19 and 5 days (range 0–13 days, IQR 4–8 days) for MIS-C.
Clinical Outcomes and Disease Severity
Median nucleocapsid concentration was higher in the group with chest radiograph infiltrates (median 23.7 pg/mL) compared to the group without (0.76 pg/mL) and the distributions were significantly different (P = 0.008, Figure 2A). The significant difference remained when comparing the subsets of samples obtained early (0–2 days) following admission (P = 0.002, Figure 2B) or later after symptom onset (6–11 days; P = 0.004, see Figure, Supplementary Digital Content 4, http://links.lww.com/INF/E876). No significant differences in spike protein concentration were observed in these same comparisons. No clear differences in nucleocapsid antigenemia or spike antigenemia were observed among COVID-19 patients classified as WHO ordinal scale 3, 4, or 5 and greater. Similarly, antigenemia was identified in patients with and without admission to the intensive care unit (ICU), vasopressors, and infiltrates on chest radiographs (Figure 2). Subgroups were examined early in hospital admission (Figure 2B,D) and at intervals from symptom onset (see Figure, Supplementary Digital Content 4, http://links.lww.com/INF/E876).
FIGURE 2.
Comparison of serum or plasma antigen concentrations in COVID-19 patients grouped by severity, clinical features and symptoms. (A) Nucleocapsid measurements for all included COVID-19 patients. (B) Nucleocapsid measurements for COVID-19 patients were sampled within the first 3 days of hospitalization. (C) Spike measurements for all included COVID-19 patients. (D) Spike measurements for COVID-19 patients were sampled within the first 3 days of hospitalization. Boxplots denote median (central mark), interquartile range (IQR; top and bottom of box), and range (whiskers) excluding outliers (defined as points which exceed the 75th percentile by more than 1.5 times IQR). P values >0.05 except where indicated.
Symptomatology
Sixty (74.1%) of 81 COVID-19 patients had fever, 51 (63.0%) had respiratory symptoms, 48 (59.3%) had gastrointestinal symptoms, and 26 (32.1%) had neurologic symptoms (Table 1 and see Table, Supplementary Digital Content 5, http://links.lww.com/INF/E876). When comparing patients with and without respiratory symptoms, significantly higher levels of nucleocapsid antigenemia were observed during the first 3 days of hospitalization (P = 0.016; Figure 2B). Spike antigenemia was more prevalent in those with gastrointestinal symptoms (16/46 = 34.8%) compared to those without gastrointestinal symptoms (5/33 = 15.2%) and the distribution was significantly higher for all samples (P = 0.043) as well as the subset collected within the first 3 days of hospitalization (P = 0.044, Figure 2C,D).
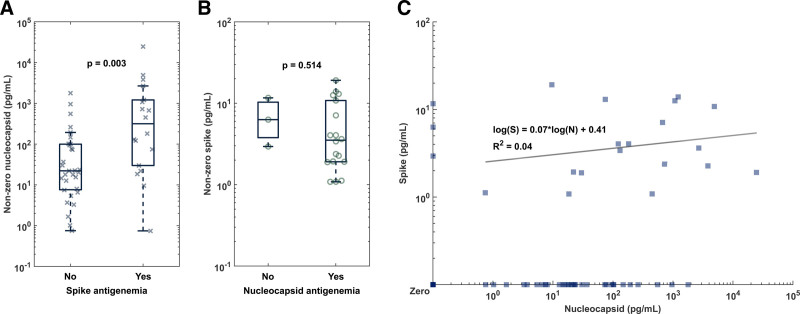
Relationship Between Nucleocapsid and Spike Antigenemia
Among patients with both spike and nucleocapsid measurements available, 18 of 49 (36.7%) patients with nucleocapsid also exhibited spike antigenemia (Figure 3A). Nucleocapsid concentration was higher in the group with concurrent spike antigenemia (median 315.9 pg/mL) as compared to the group with nucleocapsid and no spike antigenemia (median 22.1 pg/mL; P = 0.003, Figure 3A). Spike antigenemia without nucleocapsid antigenemia was rare and found in only 3 (14.3%) of 21 total patients with spike antigenemia (Figure 3B). No clear relationship was observed in comparisons of quantitative levels in samples with concurrent spike and nucleocapsid antigenemia (R2 = 0.0401 for linear fit to log-log scatter plot).
FIGURE 3.
Comparison of the relationship between nucleocapsid and spike antigenemia. (A) Detectable nucleocapsid levels in samples without and with concurrent spike antigenemia demonstrates overall higher levels of nucleocapsid when spike is also present. (B) Isolated spike antigenemia was rare. (C) There does not appear to be a correlation between spike and nucleocapsid levels in specimens with both proteins concurrently as indicated by a low R2 of 0.04 by linear regression of the log-transformed antigen levels.
DISCUSSION
Children exhibit overall lower severity and more heterogeneous manifestations of SARS-CoV-2 infection compared to adults.14,15 Thus, the clinical significance of SARS-CoV-2 antigenemia in adults may not apply to children. While nucleocapsid protein antigenemia is a highly sensitive and specific marker of acute SARS-CoV-2 infection in adults,1–5 our data show lower prevalence in hospitalized children even early after symptom onset. This finding is consistent with a prior report in which nucleocapsid protein was detectable in 10 of 22 children with COVID-19.6 However, our data contrast with another report by Sigal et al. describing a sensitivity of 89% in 36 COVID-19 patients.7 One reason for this difference may be the more stringent set of inclusion criteria employed by Sigal and colleagues where patients were only considered to have acute COVID-19 if they exhibited at least two symptoms (fever, chills, rigors, myalgia, headache, sore throat, new olfactory and taste disorder) in addition to either severe respiratory illness or other respiratory symptoms (cough, shortness of breath, hypoxia or difficulty breathing).7 Both our study and the study by Sigal et al. are limited by the inclusion of only hospitalized patients thus not necessarily reflecting the majority of pediatric COVID-19 cases, who do not require hospitalization. However, because our study did not utilize such a stringent case definition it may provide broader insight into antigenemia in pediatric COVID-19. Not all COVID-19 cases in our cohort were hospitalized primarily for COVID-19, so our analysis may include some cases more typical of non-hospitalized patients. Our sample set also included a high (44.4%) percentage of COVID-19 patients admitted to the ICU during their hospitalization, which may indicate a bias toward enrolling individuals with prolonged hospitalization. Nevertheless, the observed rates of nucleocapsid and spike antigenemia in ICU vs. non-ICU patients were similar.
The association between nucleocapsid antigenemia and radiographic and clinical features of lower respiratory tract disease may suggest a pathophysiologic connection between antigenemia and infection of pneumocytes and bronchial epithelium.16 A hypothesis compatible with these observations is that viral replication in respiratory epithelium is uniquely efficient at shedding viral nucleocapsid antigen into the circulation as compared to other organs, perhaps due to the inherent barrier and immunologic properties of bronchial and alveolar tissues.
While the specific mechanism resulting in SARS-CoV-2 antigenemia has not been described, it is presumptively a consequence of viral replication. SARS-CoV-2 tropism is determined by the expression of the ACE2 receptor, which is found in numerous tissues including the lung, gastrointestinal tract, kidney, and vascular endothelium. Reduced ACE2 expression as well as a more robust immune response has been postulated as an explanation for lower severity of COVID-19 in children and may similarly be a reason for lower rates of antigenemia.17 Reduced tropism, better immune response and regulation, and the absence of pre-existing endothelial dysfunction may also confer a lower propensity for endotheliitis related to SARS-CoV-2 infection which could impact antigenemia.18 Further investigation of the pathophysiology of viral proteins in blood from various tissues and the role of circulating antigen as trigger or consequence of endothelial activation may clarify why less antigenemia is observed in children.
We observed higher levels of nucleocapsid protein in samples with concurrent spike antigenemia, but also found instances of nucleocapsid only and, more rarely, spike protein only. The absence of a clear relationship between levels of spike and nucleocapsid antigenemia in these samples could suggest distinct mechanisms resulting in shedding of each of these proteins into the blood. Interestingly, while our data potentially link nucleocapsid antigenemia to lower respiratory tract disease, Yonker et al. provided evidence in one of the only other reports of SARS-CoV-2 antigenemia in children that the virus in the GI tract may be linked to the pathophysiology of MIS-C. This included RT-PCR of stool samples and plasma assays of viral antigen and zonulin, a marker of gut mucosal barrier breakdown, and led to the conclusion that gut mucosal inflammation resulting in spike antigenemia may be central to explaining MIS-C.6 Our data did not corroborate the observation of a high prevalence of spike antigenemia in MIS-C patients, although we did observe a higher frequency of spike antigenemia in COVID-19 patients with GI symptoms. The impact of SARS-CoV-2 infection in specific end organs on antigenemia remains a topic requiring further study.
Our study is limited by retrospective analysis of clinical specimens from a cross-sectional cohort of hospitalized patients. Symptom onset was documented for every patient but may have been subject to recall bias. Samples were collected during a range of days following symptom onset, molecular diagnosis, or hospital admission, which we address by presenting analyses stratified by time from each of these events. While this provides insight into the duration of antigenemia, it comes at the expense of standardization of sample collection and does not provide the resolution longitudinal sampling would offer. Our sample collection for this study concluded before the emergence of some SARS-CoV-2 variants of concern (VOCs), including B.1.617.2 (Delta) and B.1.1.529 (Omicron) and preceded emergency use authorization of SARS-CoV-2 vaccinations for children <16 years of age. Meanwhile, most antigenemia studies among adults include a majority of unvaccinated individuals. We did not gather data on prior infection, but samples were collected during a period when re-infection was uncommon. There may be differences in antigenemia associated with VOCs as well as prior immunity from natural infection or vaccination that are not currently defined in the literature.
In addition to potentially providing clues into the pathogenesis of COVID-19, blood-based biomarkers for SARS-CoV-2 infection could have diagnostic or prognostic applications in clinical practice as we have previously discussed in adult patients.5 In contrast with studies of adult patients, our data suggest that SARS-CoV-2 antigenemia is not a sensitive marker of acute COVID-19 in hospitalized children and therefore is unlikely to have diagnostic utility in this population. Meanwhile, we did not find an association between disease severity as defined by the WHO ordinal scale but the timing and frequency of sample collection in this study is likely inadequate to fully assess prognostic value, which has been demonstrated in adults.3–5 Viral antigens in the blood are attractive candidates as biomarkers to guide the tailoring of anti-SARS-CoV-2 therapy in certain patients such as antiviral agents, monoclonal antibodies, convalescent plasma, and immune-modulating therapy. These potential clinical applications of antigen detection warrant further study.
In conclusion, our data represent a large and diverse pediatric cohort with COVID-19 or MIS-C where there is currently a paucity of data. Our data show the insensitivity of nucleocapsid antigenemia in the diagnosis of acute COVID-19 and the absence of nucleocapsid antigenemia and rare spike antigenemia in children with MIS-C. We observed an association of nucleocapsid antigenemia with clinical manifestations of respiratory tract disease. Further understanding may come from determination of the mechanisms of antigen shedding from various tissue sources and clarification of the role of antigenemia in COVID-19 pathogenesis, especially within the respiratory tract.
ACKNOWLEDGMENTS
We thank the Emory Children’s Center Vaccine Research Center (ECC-VRC) laboratory and the Children’s Healthcare of Atlanta Research Laboratory for their assistance in collecting and processing samples. We thank the participants and their families for providing samples to further our understanding of pediatric COVID-19 and MIS-C. This study was approved by the Emory University Institutional Review Board (STUDY00000723) for the collection of prospective blood samples and/or remnant plasma or sera leftover from clinical laboratory tests following informed consent and assent, as applicable. Controls were healthy outpatient children who participated in a phlebotomy study (STUDY00087446).
AUTHOR CONTRIBUTIONS
GLD performed the data analysis. GLD and CAR wrote the manuscript. HPV, GLD, and KM performed the assays. KRVH assisted with the data analysis. AL, MP, LH, EJA, and CAR collected the samples. All authors reviewed the manuscript and figures.
Supplementary Material
Footnotes
This work was supported by the National Institutes of Health [grant numbers U54 EB027690 03 and UL1TR002378] and the National Institute of Allergy and Infectious Diseases [grant number F30AI152342 to K.R.V.H.].
CAR’s institution has received funds to conduct clinical research unrelated to this manuscript from BioFire Inc, GSK, MedImmune, Micron, Janssen, Merck, Moderna, Novavax, PaxVax, Pfizer, Regeneron, Sanofi-Pasteur. She is co-inventor of patented RSV vaccine technology unrelated to this manuscript, which has been licensed to Meissa Vaccines, Inc. EJA has consulted for Pfizer, Sanofi Pasteur, Janssen, and Medscape, and his institution receives funds to conduct clinical research unrelated to this manuscript from MedImmune, Regeneron, PaxVax, Pfizer, GSK, Merck, Sanofi-Pasteur, Janssen, and Micron. He also serves on a safety monitoring board for Kentucky BioProcessing, Inc. and Sanofi Pasteur. His institution has also received funding from NIH to conduct clinical trials of Moderna and Janssen COVID-19 vaccines. GLD, HPV, KRVH, KM, AL, MAP, LH, SRS, JDR, and WAL report no conflicts of interest.
Wilbur A. Lam and Christina A. Rostad contributed equally to this study.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.pidj.com).
Contributor Information
Gregory L. Damhorst, Email: gregory.damhorst@emory.edu.
Hans P. Verkerke, Email: hverker@emory.edu.
Kristin R.V. Harrington, Email: kristin.harrington@emory.edu.
Kaleb McLendon, Email: kaleb.benjamin.mclendon@emory.edu.
Austin Lu, Email: austintigerlu@gmail.com.
Maria A. Perez, Email: maria.de.los.angeles.perez.lizasuain@emory.edu.
Laila Hussaini, Email: lhussai@emory.edu.
Evan J. Anderson, Email: evanderson@emory.edu.
Sean R. Stowell, Email: srstowell@bwh.harvard.edu.
John D. Roback, Email: jroback@emory.edu.
Wilbur A. Lam, Email: wilbur.lam@emory.edu.
REFERENCES
- 1.Hingrat QL, Visseaux B, Laouenan C, et al. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect. 2020;27:789.e1–789.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Ong CM, Yun C, et al. Diagnostic value of nucleocapsid protein in blood for SARS-CoV-2 infection. Clin Chem. 2021;68:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogata AF, Maley AM, Wu C, et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 Patients with severe disease. Clin Chem. 2020;66:1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Hogan CA, Verghese M, et al. SARS-CoV-2 nucleocapsid plasma antigen for diagnosis and monitoring of COVID-19. Clin Chem. 2021;68:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verkerke HP, Damhorst GL, Graciaa DS, et al. Nucleocapsid antigenemia is a marker of acute SARS-CoV-2 infection. J Infect Dis. 2022;226:1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonker LM, Gilboa T, Ogata AF, et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J Clin Investig. 2021;131:e149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigal GB, Novak T, Mathew A, et al. Measurement of SARS-CoV-2 antigens in plasma of pediatric patients with acute COVID-19 or MIS-C using an ultrasensitive and quantitative immunoassay. Clin Infect Dis. 2022;75:1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC. Multisystem Inflammatory Syndrome in Children (MIS-C) Associated with Coronavirus Disease 2019 (COVID-19). 2021. Available at: https://emergency.cdc.gov/han/2020/han00432.asp. Accessed December 14, 2021.
- 9.World Health Organization. Novel coronavirus COVID-19 therapeutic trial synopsis. WHO R&D Blueprint. 2020:12. [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quanterix. Simoa® SARS CoV‐2 N Protein Advantage Kit HD‐X Data Sheet. Available at: https://www.quanterix.com/wp-content/uploads/2020/12/SARS-CoV-2-N-Protein-Advantage-Data-Sheet-for-HD-X.pdf. Accessed June 2, 2022.
- 13.R-PLEX SARS-CoV-2 Spike. Available at: https://www.mesoscale.com/~/media/files/data%20sheets/r-plex%20sars-cov-2%20spike.pdf. Accessed July 18, 2022.
- 14.Shane AL, Sato AI, Kao C, et al. A pediatric infectious diseases perspective of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and novel coronavirus disease 2019 (COVID-19) in children. J Pediatric Infect Dis Soc. 2020;9:596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogunovic D, Merad M. Children and SARS-CoV-2. Cell Host Microbe. 2021;29:1040–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.V’Kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou J, Thomas PG, Randolph AG. Immunology of SARS-CoV-2 infection in children. Nat Immunol. 2022;23:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams PCM, Howard-Jones AR, Hsu P, et al. SARS-CoV-2 in children: spectrum of disease, transmission and immunopathological underpinnings. Pathology. 2020;52:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.