Abstract
Neisserial surface protein A (NspA) is currently being investigated with humans as a candidate vaccine for the prevention of meningococcal disease. Although NspA is highly conserved, the ability of anti-NspA antibodies to bind to or elicit complement-mediated bactericidal activity against diverse Neisseria meningitidis serogroup B strains is controversial. To evaluate strain differences in NspA surface accessibility and susceptibility to bactericidal activity, we prepared murine immunoglobulin G2a anti-NspA monoclonal antibodies (MAbs) and evaluated their functional activity against 10 genetically diverse N. meningitidis serogroup B strains. By colony Western blot, all 10 strains expressed NspA as detected by one or more MAbs. By flow cytometry, two MAbs were found to bind to the bacterial surface of 6 of the 10 strains. In addition, two strains showed variable NspA surface accessibility for the MAbs despite being uniformly positive for NspA expression by colony Western blotting. Only 4 of the 10 strains were susceptible to anti-NspA complement-mediated bacteriolysis. Passively administered MAb protected infant rats from developing bacteremia after challenge with N. meningitidis serogroup B strain 8047 (surface binding positive, susceptible to anti-NspA bacteriolysis), was poorly protective against strain BZ232 (surface binding variable, resistant to bacteriolysis), and did not protect against strain M986 (surface binding negative, resistant to bacteriolysis). Finally, NspA does not appear to be critical for causing bacteremia, as an NspA knockout from strain 8047 was highly virulent in infant rats. Taken together, these findings suggest that an NspA-based vaccine will need to incorporate additional antigens to elicit broad protection against N. meningitidis serogroup B.
Neisseria meningitidis serogroup B is an important cause of meningitis and sepsis. Efforts to develop an N. meningitidis serogroup B vaccine have been hampered by poor immunogenicity of the polysaccharide capsule, which is cross-reactive with host polysialic acid, and the potential danger of eliciting anticapsular autoantibodies. Alternative approaches using noncapsular antigens are being investigated (8). However, the ability of most noncapsular antigens to elicit broadly protective responses is limited by antigenic heterogeneity (15, 17, 20, 21, 24), and/or variable expression of the surface-exposed epitopes (3, 11, 22). One possible exception is Neisserial surface protein A (NspA), first described in 1997 by Martin and colleagues (13). NspA is highly conserved (4, 12, 13, 16, 19; D. Martin, C. R. Rioux, A. Villeneuve, J. Hamel, and B. R. Brodeur, Abstr. 11th Int. Pathog. Neisseria Conf., p. 198, 1998), appears to be expressed in all N. meningitidis serogroup B strains tested (13, 16), and elicits protective antibody responses in mice against strains of N. meningitidis serogroups A, B, and C (13). Murine monoclonal antibodies (MAbs) prepared against rNspA are also reported to be broadly cross-reactive and protective against N. meningitidis serogroup B strains (4, 12, 13).
Despite gene conservation and expression of NspA by all meningococcal strains tested to date, our laboratory noted differences among N. meningitidis serogroup B strains in bacterial surface binding, as determined by flow cytometry and complement-mediated bactericidal activity of mouse polyclonal anti-rNspA antisera (16). Binding and bactericidal activity were lowest for strains producing the greatest amount of capsular polysaccharide (16). While other factors may be important, these data suggest that the capsule can limit the accessibility of NspA surface epitopes and/or block complement-mediated bacteriolysis evoked by anti-NspA antibody binding. Therefore, an NspA-based vaccine might have only limited ability to elicit protective immunity against highly encapsulated strains, the very strains expected to have the greatest virulence (7, 10, 14, 23).
In contrast, Martin et al. (11th Int. Pathog. Neisseria Conf., 1998), using anti-NspA MAbs, reported that NspA was accessible on the cell surface and elicited complement-mediated bacteriolysis of nearly all meningococcal strains tested and that the MAbs could passively protect mice challenged with different strains (4).
To resolve the discrepancies among these observations, we prepared a panel of murine MAbs to rNspA. Herein, we report the ability of these MAbs to bind to the bacterial surface, to elicit complement-mediated bactericidal activity, and to passively protect infant rats challenged with N. meningitidis serogroup B strains.
MATERIALS AND METHODS
Bacterial strains.
The 10 N. meningitidis serogroup B strains chosen for this study (Table 1) were selected to be genetically diverse and representative of the 17 strains examined in an earlier study (16). The N. meningitidis serogroup B collection included five strains previously considered to be NspA surface positive using polyclonal anti-rNspA antisera and five strains considered NspA surface negative. The five surface-positive strains included three that were susceptible to anti-rNspA complement-mediated bacteriolysis and two that were resistant. All five surface-negative strains were also resistant to bacteriolysis with polyclonal anti-rNspA antisera. The 10 strains were isolated over a period of 25 years from patients residing in different countries. Based on electrophoretic typing (ET), the strains represent a broad range of genetic diversity for N. meningitidis serogroup B strains causing disease (5, 18).
TABLE 1.
N. meningitidis serogroup B strains
| Strain | Country of origin | Year isolated | Reference | Serologic classification | ET complexd | NspA sequencee | Previously reported anti-NspA reactivitya
|
|
|---|---|---|---|---|---|---|---|---|
| Surface binding | Bactericidal activity | |||||||
| 8047 | U.S. | 1978 | Zollinger, CHORIb | 2b:P1.5,2 | ND | yes | + | + |
| CU385 | Cuba | 1980 | Sacchi et al. | 4,7:P1.19,15 | 5 | yes | + | + |
| BZ198 | The Netherlands | 1986 | Caugant et al. | NT:P.NSTc | 154 | no | + | + |
| S3446 | U.S. | 1973 | Sacchi et al. | 19,14:P1.23,14 | 8 | no | + | − (static) |
| 1000 | USSR | 1989 | CHORI | NT:P1.5 | 61 | no | + | − |
| BZ232 | The Netherlands | 1964 | Caugant et al. | NT:P1.2 | 76 | yes | − | − |
| NG3/88 | Norway | 1988 | Caugant et al. | 8:P1.1 | A4 complex | yes | − | − |
| NGP165 | Norway | 1974 | CHORI | NT:P1.2 | 37 | yes | − | − |
| MC58 | United Kingdom | 1985 | McGuinness et al. | 15:P1.7,16 | 5 | yes | − | − |
| M986 | U.S. | 1963 | Sacchi et al. | 2a:P1.5,2 | 165 | yes | − | − |
Previously reported results from Moe et al. using murine polyclonal antisera (16). Surface binding was determined by flow cytometry. Static, strains that were inhibited but not killed in the assay (≥50 but <100% survival at 60 min).
CHORI, determined by our laboratory.
NT, nonserotypeable. NST, nonserosubtypeable.
Determined by electrophoretic typing (ET) (6); ND, not determined.
Previously reported (16).
Mutants of strains 8047 and BZ198 (8047ΔNspA and BZ198ΔNspA, respectively), in which the nspA gene was inactivated, were prepared by transforming the parent strain with the plasmid pBSUDNspAERM (gift from J. Abu-Bobie, Chiron Corp., Siena, Italy). The plasmid contains a truncated nspA gene and the ermC gene (erythromycin resistance). The bacteria were transformed by selecting a few colonies grown overnight on chocolate agar plates (Remel, Lenexa, Kans.) and mixing them with 20 μl of phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin (BSA) and 1 μg of plasmid DNA. The mixture was spotted onto a chocolate agar plate, incubated for 6 h at 37°C, 5% CO2, and then diluted in PBS-BSA and spread on chocolate agar plates containing 7 μg of erythromycin per ml. The presence of the disrupted nspA gene in the genome of both strains was confirmed by PCR using the following primers: 5′-ACAGCAGGATCCTTTAACGGATTC-3′ and 5′-GTGGATGAAGCTTTGGACATTTC-3′. Lack of NspA expression was confirmed by whole-cell enzyme-linked immunosorbent assay (ELISA) as described below and Western blots of outer membrane proteins prepared from the knockout strains and resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as described previously (16).
Preparation of MAbs.
Female CD1 mice (Charles River, Hollister, Calif.) were vaccinated with microvesicles prepared from a transformed Escherichia coli strain that expressed recombinant NspA from a multicopy plasmid containing the nspA gene under the control of its own promoter (plasmid pGMS1.0) (16). The mice were given three 100-μl injections, each separated by 3 weeks, containing 10 μg of protein and 10 μg of CpG oligonucleotides (5′-TCCATGACG TTCCTGACGTT-3′). The first two doses were given subcutaneously together with aluminum phosphate (0.5%, wt/vol), and the final dose was given without adjuvant and administered intraperitoneally (i.p.). Three days later, the animals were sacrificed and their spleen cells were fused with myeloma cells (P3X63-Ag8.653) at a ratio of 1 spleen cell to 1.7 myeloma cell. After two weeks of incubation in hypoxanthine-aminopterin-thymidine selective medium, hybridoma supernatants were screened for antibody binding activity by ELISA using encapsulated N. meningitidis serogroup B strain BZ198 as the target antigen (see description below). Specificity of antibody binding to NspA was demonstrated by lack of binding in a replicate ELISA performed with BZ198ΔNspA. Hybridomas secreting NspA-specific antibody were cloned by limiting dilution and then expanded and frozen for subsequent use in tissue culture.
Antibodies from four cell lines were characterized in detail. The subclasses of the MAbs were determined using an antibody capture ELISA and alkaline phosphatase-conjugated polyclonal antibody specific for each of the mouse immunoglobulin G (IgG) subclasses, IgM, IgA, and κ and λ light chains (Southern Biotechnology Associates, Inc., Birmingham, Ala.) (9). The MAbs produced by the hybridoma clones were harvested from tissue culture media by ammonium sulfate precipitation (55%, wt/vol) and, after exhaustive dialysis in PBS buffer, purified by affinity chromatography using a Poros G/M column (Applied Biosystems, Foster City, Calif.). The concentration of the purified MAb was determined spectrophotometrically assuming a 1-mg/ml antibody solution having an absorbance of 1.35 at 280 nm.
Binding of antisera to the surface of live encapsulated meningococci.
The ability of the MAbs to bind to the surfaces of live N. meningitidis serogroup B strains was determined using a flow cytometric detection of indirect fluorescence assay, performed as described previously (16). Positive-control antibodies included meningococcal-specific serotyping or subtyping MAbs (MN14C11.6, MN16C13F4, Rijksinstituut Voor Volksgezondheid en Mileu, Bilthoven, The Netherlands) and SEAM 12, an antipolysaccharide MAb that is specific for encapsulated group B strains (9). The negative control consisted of a mouse IgG MAb (VIG10) of irrelevant specificity.
Complement-dependent bactericidal antibody activity. (i) Low inoculum assay.
After overnight growth on chocolate agar, 5 to 10 colonies were inoculated into Mueller-Hinton broth (starting optical density at 620 nm [OD620] of ∼0.1) and the test organism was grown for approximately 2.5 h to an OD620 of ∼0.6 (early log phase). One of the test strains, CU385, was grown in an identical manner except that glucose (0.25%, wt/vol) was added to the broth. In the absence of glucose in the broth, the resulting bacteria from this strain did not show an increase in CFU/ml during the bactericidal assay (see below). To prepare the bacteria for the assay, the cells were washed once in Gey's balanced salt solution (Gibco BRL, Rockville, Md.) containing 1% (wt/vol) BSA (Gey's-BSA). After an appropriate dilution, 12 μl containing approximately 300 to 400 CFU was added to a final reaction volume of 60 μl, consisting of 20% (vol/vol) complement and serial twofold dilutions of MAbs in Gey's-BSA buffer. The complement source was human serum from a healthy adult (MAS) with no detectable anticapsular antibody to group B polysaccharide as tested by ELISA and no detectable intrinsic bactericidal activity against the test strain when the serum was tested at a final concentration of 20 or 40%. After incubation at 37°C for 60 min, 20 μl of the reaction mixture was transferred to a Mueller-Hinton plate as previously described (16). Serum bactericidal titers (BC50) were defined as the antibody concentration resulting in a 50% decrease in CFU per ml after 60 min of incubation of bacteria in the reaction mixture, compared to the control CFU per ml at time zero. Typically, bacteria incubated with the negative-control antibody and complement showed a 150 to 200% increase in CFU/ml during the 60 min of incubation.
(ii) High-inoculum bactericidal assay.
This assay was adapted from that previously described by Amir et al. for measuring bactericidal activity against Haemophilus influenzae type b (2). The assay is essentially the same as the low-inoculum assay except that the final reaction mixture (120 μl to 1 ml) contained a fixed concentration (100 μg/ml) of anti-NspA MAb AL12 diluted in Gey's-BSA buffer, 20% (vol/vol) human serum as a complement source and ∼108 CFU/ml log-phase meningococcal cells diluted in Gey's-BSA. Controls included the test organism incubated in the absence of antibody (complement alone) or MAb in the presence of heat-inactivated (56°C for 30 min) complement. The reaction vials were incubated at 37°C on a Clay Nutator orbital mixer (Fisher Scientific, Pittsburgh, Pa.) for 60 min, and serial dilutions were plated onto chocolate agar plates. The results were expressed as the log decrease in meningococcal cells as compared to that of control bacteria incubated with MAb and inactivated complement.
Expression of NspA by colony blot assay.
In order to determine whether particular bacterial colonies were expressing NspA, the colonies were transferred from the surface of chocolate agar plates onto nitrocellulose filters (Bio-Rad, Richmond, Calif.) by blotting. The filters were washed once with PBS buffer containing 0.1% (wt/vol) Tween 20 (Sigma, St. Louis, Mo.) and 0.1% (wt/vol) sodium azide (wash buffer) and then blocked with wash buffer containing 1% (wt/vol) nonfat dry milk (blocking buffer). NspA was detected by adding a solution of anti-NspA MAb AL12 (∼0.1 μg/ml) in blocking buffer, incubating at ambient temperature for 2 h, and then washing the filters with wash buffer. Bound antibody was detected by adding rabbit anti-mouse IgG-, IgA-, and IgM-alkaline phosphatase conjugated polyclonal antibody (Zymed, South San Francisco, Calif.) diluted in wash buffer containing 1% (wt/vol) BSA. After incubation for 1 h at ambient temperature, the filters were washed as described above, and Sigma Fast BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) substrate was added.
ELISA.
The whole-cell ELISA assay was performed as described by Abdillahi and Poolman (1). Briefly, bacterial cells grown overnight at 37°C in 5% CO2 on chocolate agar plates were resuspended in sterile PBS buffer and then inactivated by heating to 56°C in a water bath for 30 min. The suspension was adjusted to an OD620 of 0.1, and 100 μl of the suspension was added to wells of flat-bottom 96-well microtiter plates (Nalge Nunc International, Rochester, N.Y.). The liquid in the wells was allowed to evaporate at ambient temperature in a fume hood. Plates containing the dried bacteria were stored at ambient temperature until use. Before adding antibodies, the plates were washed once with wash buffer and blocked by adding blocking buffer and incubating at 37°C for 1 h. After removal of the blocking buffer, test antibodies diluted in blocking buffer were added to the wells and incubated at 4°C overnight. The plates were washed five times with wash buffer followed by the addition of rabbit anti-mouse IgG-, IgA-, and IgM-alkaline phosphatase conjugated polyclonal antibody (Zymed) diluted in wash buffer containing 1% (wt/vol) BSA. After 1 h of incubation at ambient temperature, the plates were washed five times with wash buffer and developed with p-nitro phenylphosphate substrate (1 mg/ml; Sigma) in 1 M diethanolamine, pH 9.8, containing 0.5 mM magnesium chloride. The OD405 was measured using a microtiter plate reader.
The specificity of the anti-NspA MAbs for NspA was determined using the following solid-phase antigens: microvesicles prepared from E. coli expressing rNspA and microvesicles prepared from E. coli transformed with the parent plasmid lacking the nspA gene, lauroyl sarcosinate-insoluble outer membrane proteins prepared from N. meningitidis serogroup B strains BZ198 and BZ198ΔNspA, and recombinant HisTag NspA purified by Ni-NTA Sepharose metal affinity chromatography. Methods used for preparing the antigens were the same as described previously (16). The antigen preparations were diluted in PBS buffer (10 to 100 μg of total protein per ml), added to the wells of microtiter plates, and allowed to bind at 4°C overnight. After washing the plates with wash buffer, the plates were blocked with PBS buffer containing 1% (wt/vol) BSA. The anti-NspA and control MAbs were diluted in wash buffer containing 1% (wt/vol) BSA or, for HisTag NspA, PBS buffer containing 1% (wt/vol) BSA and 0.3% (wt/vol) Empigen BB (Calbiochem, La Jolla, Calif.). After overnight incubation at 4°C, the plates were washed, incubated with secondary antibody, and developed as described above.
Animal protection.
The ability of the anti-rNspA antiserum to confer passive protection against N. meningitidis serogroup B bacteremia was tested in infant rats (16). In brief, 5- to 8-day old pups from litters of outbred Wistar rats (Charles River, Raleigh, N.C.) were randomly redistributed to the nursing mothers. Three strains, M986, BZ232, and 8047, were tested. Each strain had been serially passaged three times in infant rats. After the third passage, the blood cultures from the rats were grown overnight on chocolate agar. The bacteria were suspended in sterile skim milk and stored frozen at −80°C. On the day before challenge, freshly thawed bacteria were inoculated onto chocolate agar and grown overnight at 37°C in 5% CO2. On the morning of the challenge, colonies were picked, inoculated into a broth culture, and grown and prepared as described above for the bactericidal assay. In two experiments using strains 8047 and BZ232, the animals were pretreated i.p. with 100 μl of different concentrations of test or control MAbs or buffer alone. Two hours later, the animals were challenged i.p. with approximately 103 to 104 CFU of N. meningitidis serogroup B bacteria. To enhance the sensitivity of the assay, in the third experiment with strain M986, the bacteria were suspended in different concentrations of test or control MAbs immediately before the challenge and 100 μl of the mixture was administered i.p. Heparinized blood specimens were obtained by heart puncture 18 h after the bacterial challenge. In one experiment with strain M986, blood samples were also obtained 6 h after challenge. Aliquots of 1, 10, and 100 μl of blood were plated onto chocolate agar. The number of CFU per milliliter of blood was determined after overnight incubation of the plates at 37°C in 5% CO2.
RESULTS
Production of anti-NspA MAbs.
Polyclonal sera from individual mice immunized with E. coli microvesicles containing rNspA were evaluated for antibody binding by ELISA and complement-mediated bactericidal activity. As the target antigen, the ELISA included encapsulated N. meningitidis serogroup B cells from strain BZ198 or the same strain in which the NspA gene had been deleted (BZ198ΔNspA). With anti-rNspA antisera, strain BZ198 gave the best binding in the whole-cell ELISA compared to that of the other strains. The bactericidal assay included two test N. meningitidis serogroup B stains, CU385 and 1000 (Table 1). Based on the results of the ELISA and bactericidal assays, a high-responder mouse was selected for the fusion. From this fusion, we obtained five hybridoma cell lines producing antibodies that were positive by ELISA for binding to strain BZ198 but were negative for binding with the NspA knockout strain, BZ198ΔNspA. Each hybridoma cell line was subsequently cloned and grown on a larger scale in tissue culture to produce larger amounts of antibody for further characterization. The MAbs expressed by each of the five clones were designated AL4, AL5, AL8, AL11, and AL12, and were IgG2a(κ). Because of low binding and functional activity of AL8, the results reported below are limited to the remaining four MAbs.
By ELISA, the four MAbs bound to microvesicles prepared from the E. coli strain containing recombinant NspA, which had been used to immunize the mouse for preparation of the hybridomas. In contrast, only background binding was observed when the MAbs were tested at 10-fold higher concentrations against microvesicles prepared from the same E. coli strain transformed with the plasmid without the nspA gene (Fig. 1A). Similarly, the four MAbs bound to lauroyl sarcosinate-insoluble outer membrane proteins prepared from N. meningitidis serogroup B strain BZ198 but were negative when tested at 10-fold higher concentrations with outer membrane proteins prepared from strain BZ198ΔNspA, which does not express NspA (Fig. 1B). Finally, the four MAbs bound by ELISA to affinity-purified HisTag-NspA. In contrast, a negative-control MAb, VIG10, with an irrelevant specificity, showed only background binding when tested at 10-fold higher concentrations (Fig. 1C). The above results show that the four MAbs are specific for binding to NspA. Note that much higher concentrations of the MAbs were required for binding to HisTag-NspA than to NspA present in the E. coli or Neisseria membrane preparations. This result likely reflects the loss of conformational epitopes in the HisTag-NspA eluted from the affinity column in the presence of 8 M urea.
FIG. 1.
Binding of the anti-NspA Mabs by ELISA. (A) Binding to microvesicles prepared from an E. coli strain expressing recombinant NspA (closed bars) or from the same E. coli strain transformed with the plasmid without the nspA gene (open bars). Data for the microvesicles containing rNspA were obtained at an antibody concentration of 60 ng/ml, whereas the negative control microvesicles were tested at 600 ng/ml. (B) Binding to lauroyl sarcosinate-insoluble outer membranes prepared from N. meningitidis serogroup B strains BZ198 (closed bars) or BZ198ΔNspA (open bars), in which the gene encoding NspA had been inactivated. The antibody concentrations were the same as those for panel A. (C) Binding to Ni-NTA Sepharose metal affinity-purified HisTag-NspA. The anti-NspA MAbs were tested at 5 μg/ml, the irrelevant MAb was tested at 50 μg/ml, and the polyclonal anti-HisTag NspA antisera were tested at a dilution of 1:108,000.
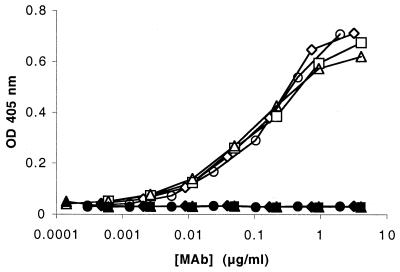
Figure 2 shows binding of the MAbs to heat-killed, whole bacterial cells from strain BZ198 as measured by ELISA. All four MAbs showed similar dose-response binding. All four MAbs showed only background binding (OD405 < 0.05) when bacterial cells from strain BZ198ΔNspA were employed as the target instead of the respective BZ198 wild-type strain.
FIG. 2.
Whole-bacterial-cell ELISA comparing binding activities of the anti-NspA MAbs AL4 (◊), AL5 (□), AL11 (▵), and AL12 (○) to N. meningitidis serogroup B strain BZ198. The respective filled symbols show the results from a comparable experiment with strain BZ198ΔNspA as the solid-phase antigen.
Anti-rNspA MAb cell surface binding as determined by indirect fluorescence flow cytometry.
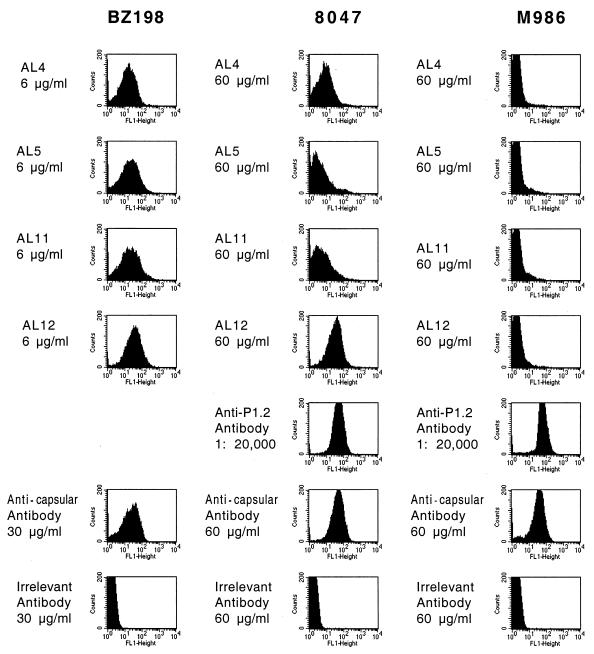
The four purified MAbs were tested by flow cytometry for their ability to bind to live cells from various N. meningitidis serogroup B strains. Representative data for three strains, BZ198, 8047, and M986, are shown in Fig. 3. The anticapsular MAb SEAM 12 bound to all three strains, whereas an irrelevant MAb, VIG10, showed only background binding. With strain BZ198, the four MAbs were all positive for binding when tested at 6 μg/ml. In contrast, with strain M986, all four MAbs were negative for binding when tested at 60 μg/ml. Note that with this strain, a positive-control anti-PorA P1.2 MAb (MN16C13F4) bound strongly when tested at a dilution of 1:20,000 (Fig. 3). With strain 8047, AL12 was positive at 10 and 60 μg/ml (data only shown for the higher dose), and AL4 was positive only when tested at the higher antibody concentration (60 μg/ml). AL5 and AL11 were negative at the highest concentrations tested (60 μg/ml). Thus, based on the flow cytometry results, the four clones appear to be different. Also, MAbs AL4 and AL12 showed the best binding of the four MAbs when tested with strain 8047.
FIG. 3.
Binding of anti-rNspA and control MAbs to live encapsulated N. meningitidis serogroup B strains BZ198, 8047, and M986 as determined by indirect-fluorescence flow cytometry. The anti-NspA MAbs include AL4, AL5, AL11, and AL12. The control MAbs include a murine MAb with an irrelevant specificity (VIG10), an anticapsular-specific murine MAb (SEAM 12), and, for strains 8047 and M986, an N. meningitidis subtype MAb anti-PorA P1.2 (Rijksinstituut Voor Volksgezondheid en Mileu, Bilthoven, The Netherlands). A subtyping anti-PorA MAb was not available for strain BZ198.
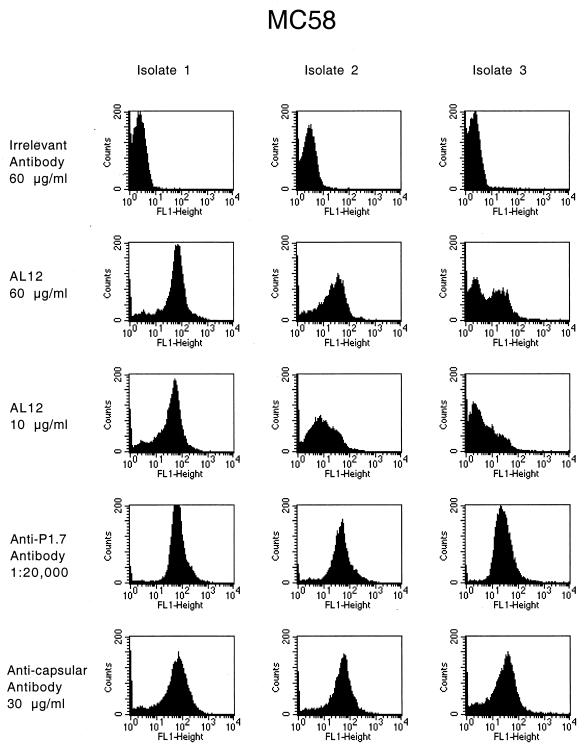
Variation in anti-NspA surface binding of AL4 and AL12 was apparent for two strains, BZ232 and MC58. Representative results with strain MC58 are shown in Fig. 4 for three separate subcultures of MC58 tested in the same experiment. Isolate 1 was strongly positive for surface binding with AL12 at 10 μg/ml, the lowest concentration tested. In contrast, isolate 3 showed much lower binding even when tested at 60 μg/ml. Also, there appeared to be a mixed population of positive and negative binding. Binding of isolate 2 appeared to be homogenous but less than isolate 1 and more than isolate 3. The binding results with AL4 were similar (data not shown). The control anti-PorA (P1.7) and anticapsular MAbs showed comparable respective results of binding to bacteria from all three subcultures. Clonal bacteria obtained from positive and negative subcultures were positive or negative irrespective of whether the parent subculture was positive or negative (data not shown).
FIG. 4.
Binding of anti-NspA MAb AL12 to bacteria grown from different subcultures of strain MC58 as determined by indirect-fluorescence flow cytometry. The murine control MAbs include a MAb of irrelevant specificity, an anti-PorA P1.7 MAb, and an anticapsular MAb.
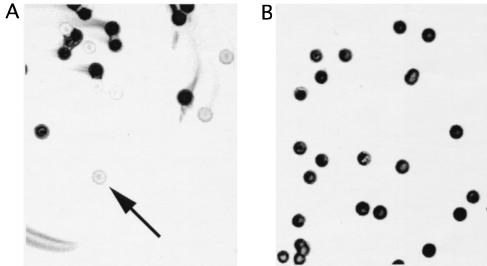
The variable binding of AL4 and AL12 to bacteria from different strain MC58 cultures did not result from failure to express NspA. As shown in Fig. 5B, all bacterial colonies tested from subculture 3 with negative or mixed surface binding with AL4 and AL12 by flow cytometry were positive for NspA expression when tested by colony Western blotting using AL12 as the detecting antibody. For comparison, representative positive and negative colonies from a mixture of strains 8047 and 8047ΔNspA are shown in Fig. 5A.
FIG. 5.
Colony Western blots using anti-NspA MAb AL12 as the detecting antibody. (A) Mixture of strains 8047 and 8047ΔNspA. The arrow indicates an example of a strain 8047ΔNspA colony. (B) Strain MC58 CFU from the flow experiment depicted in Fig. 3 in which the strain was negative for AL4 and AL12 binding (isolate 3). All colonies tested in the Western blot were positive for AL12 binding.
Table 2 summarizes the results from flow experiments measuring the ability of the different MAbs to bind to the surfaces of live bacteria from each of the 10 strains. All five strains that had been positive for binding with the polyclonal antiserum were positive for binding with MAbs AL4 and AL12. One strain, NG3/88, that had been negative for binding with polyclonal anti-NspA antibody in our previous study, was positive for surface binding with these two MAbs (compare Tables 1 and 2). Two strains that had been negative with the polyclonal antisera in our previous study (MC58 and BZ232) showed variable binding with the MAbs, and two other strains (NGP165 and M986) were negative with both the polyclonal and monoclonal anti-NspA antibodies.
TABLE 2.
Summary of anti-NspA MAb surface reactivity for genetically diverse N. meningitidis serogroup B strains
| Strain | Surface reactivity (flow) of anti-NspA MAbs and/or complement-mediated BCAa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AL4
|
AL5
|
AL11
|
AL12
|
|||||
| Flowb | BCA | Flow | BCA | Flow | BCA | Flow | BCA | |
| 8047 | + | >60 | − | 45 | − | 15 | ++ | 8 |
| CU385 | ++ | >120 | + | >120 | + | >120 | ++ | >120 |
| BZ198 | ++ | 26 | ++ | 5 | ++ | 4 | ++ | 5 |
| S3446 | ++ | >60 | ++ | >60 | ++ | 60 | ++ | 32 |
| 1000 | + | >120 | − | >120 | − | >120 | ++ | 110 |
| BZ232 | variable | >120 | − | >120 | − | >120 | variable | >120 |
| NG3/88 | ++ | >120 | + | >120 | ++ | >120 | ++ | >120 |
| NGP165 | − | >120 | − | >120 | − | >120 | − | >120 |
| MC58 | variable | >120 | variable | >120 | variable | >120 | variable | >120 |
| M986 | − | >120 | − | >120 | − | >120 | − | >120 |
| BZ198ΔNspA | − | >60 | − | >60 | − | >60 | − | >60 |
BCA, bactericidal activity (concentration of MAb that when incubated for 60 min with bacterial cells and 20% human complement yielded a 50% decrease in CFU per ml compared to that at time zero).
Surface binding was measured by flow cytometry. With one exception, all strains were tested at MAb concentrations of 10 and 60 μg/ml. The exception, strain BZ198, was tested at 6 and 60 μg/ml. Results were scored as follows: −, less than 50% of total fluorescent events have intensity above background when tested at 60 μg of MAb per ml; +, ≥50% of total fluorescent events have intensity above background at 60 μg/ml; ++, ≥50% of total fluorescent events have intensity above background at 6 or 10 μg/ml; variable, subcultures ranged from − to ++ (see text).
Bactericidal activity.
The ability of the anti-rNspA MAbs to elicit complement-mediated bacteriolysis of the 10 strains in the low inoculum is summarized in Table 2. MAb AL4, which was positive for surface binding by flow cytometry for six strains, was bactericidal only against strain BZ198. MAb AL12, which was positive for binding by the flow assay for the same six strains as AL4, was bactericidal against four strains. However, for two of these four strains, the antibody concentrations of AL12 required for eliciting bacteriolysis were high (32 and 110 μg/ml). The remaining two MAbs tested, AL5 and AL11, were bactericidal only against strains 8047 and BZ198 (AL5) and 8047, BZ198, and S3446 (AL11).
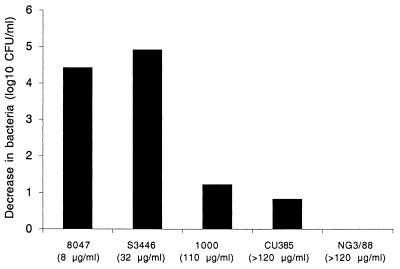
Figure 6 summarizes the bactericidal activity of AL12 when tested in the high inoculum assay in which a fixed concentration of antibody (100 μg/ml) is incubated with ∼108 CFU/ml of log-phase meningococcal cells and 20% complement. The five strains tested in this assay included one strain that was susceptible to bacteriolysis in the low-inoculum assay (8047), two strains that were moderately resistant (S3346 and 1000), and two strains that were resistant (CU385 and NG3/88). By flow cytometry, all five test strains showed strong binding of AL12 to the bacterial surface. The three strains with BC50 > 100 μg/ml in the low-inoculum assay were poorly killed when retested under the high inoculum conditions (no killing of NG3/88, and 0.8 and 1.2 log10 killing of CU385 and 1000, respectively). In contrast, the two strains with BC50 of 8 and 32 μg/ml in the low-inoculum assay were efficiently killed under the conditions of the high-inoculum assay (4.4 log10 killing of 8047 and 4.9 log killing of S3446).
FIG. 6.
High-inoculum bactericidal assay. The decrease (log10) in the number of bacteria resulting from bacteriolysis in the presence of complement and anti-NspA MAb AL12, compared to the number of control bacteria incubated with antibody alone or antibody with heat-inactivated complement, is shown. The numbers in parentheses indicate the BC50 concentrations of AL12 for each strain measured in the low-inoculum bactericidal assay.
The results of the high-inoculum assay provide further support for the functional activity of the anti-NspA antibody against some but not all strains. Since even among susceptible strains not all bacteria are killed under the conditions of this assay, we were also able to examine large numbers of surviving colonies for NspA expression by colony Western blotting. Bacteria surviving after complement-mediated killing with AL12 in the high-inoculum assay were tested for NspA expression by blotting ∼1,000 CFU on nitrocellulose filters and probing for NspA expression by Western blotting using AL12 as the primary antibody. All surviving colonies appeared to be strongly positive. Furthermore, repeated selection (two times) of CFU surviving in the high-inoculum assay with strain S3446 (4.9 and 5.7 log10 killed per selection) did not result in the identification of NspA-negative CFU. As a control for determining whether NspA-negative CFU could be detected in a background of NspA-positive CFU, blotting was performed on a mixture (100:1) of 8047 wild type and 8047ΔNspA. After incubation with AL12 and complement, approximately 90% of the surviving CFU examined were negative for NspA expression. Figure 5A shows an example of a Western blot of colonies from a plate containing a mixture of strains 8047 and 8047ΔNspA probed with AL12.
In the high-inoculum bactericidal assay of strains 8047 and S3446, we observed 4- to 5-log10 decreases in CFU after 60 min of incubation with the MAb and complement. Given the uniform expression of NspA among >1,000 CFU surviving after the bactericidal assay, we can estimate that the occurrence of an NspA-negative CFU to be <1 in 107 to 108 bacteria. By comparison, when similar selection experiments were performed with strain 8047 using the anti-PorA P1.2 MAb, CFUs negative for P1.2 were obtained at a frequency of approximately 1 in 108 bacteria. Finally, for strains CU385, 1000, and NG3/88, which were moderately resistant to anti-NspA-induced bacteriolysis, we also probed approximately 100 to 1,000 CFU after incubation with AL12 and complement. The CFUs tested from all of the strains showed uniform strong expression of NspA. Thus, resistance of these strains to anti-NspA bacteriolysis was not a result of failure to express NspA or the inability of the MAb to bind to the NspA epitope.
Passive protection by anti-rNspA MAb AL12.
The ability of anti-rNspA MAb AL12, the most active MAb in the flow and bactericidal assays, to confer passive protection against meningococcal bacteremia was assessed in an infant rat model. Three experiments were performed (Table 3). In experiment 1, the animals were pretreated with test or control MAbs at time zero and challenged 2 h later with 2.3 × 103 CFU of strain 8047 (positive with AL12 in the flow assay at 10 μg/ml and susceptible to bacteriolysis with a BC50 of 8 μg/ml). In experiment 2, the protocol was similar but the animals were challenged with 15 × 103 CFU of strain BZ232 (variable binding in the flow assay and resistant to bacteriolysis). In experiment 3, the animals were challenged with 2 × 103 CFU of strain M986 (negative for binding by the flow assay and resistant to bacteriolysis). Also, in order to maximize the likelihood of observing protection against strain M986 in experiment 3, the bacteria were premixed with the anti-rNspA or control MAbs immediately prior to the i.p. challenge. In all three experiments, the positive-control anticapsular MAb (SEAM 3) was protective and there was no protection observed in the animals treated with the irrelevant negative control MAb as determined by comparing the levels of bacteremia in the respective animals treated with PBS-BSA (P > 0.2 by t test of the respective geometric mean CFU/ml of blood).
TABLE 3.
Ability of anti-NspA MAb AL12 to passively protect infant rats challenged i.p. with different meningococcal B strains
| Experiment no. | Strain (challenge dose per rat in CFU) | MAb pretreatmenta | Dose (μg) per rat | Results of blood culture at 18 h
|
|
|---|---|---|---|---|---|
| No. positive/total | CFU/ml (geometric mean, 103)b | ||||
| 1 | 8047 (2.3 × 103) | Anticapsular | 10 | 2/7 | 0.05 |
| AL12 Anti-NspA | 50 | 0/7 | <0.01 | ||
| AL12 Anti-NspA | 5 | 4/7 | 1.4 | ||
| Irrelevant | 50 | 7/7 | 115 | ||
| PBS-albumin | None | 7/7 | 79 | ||
| 2 | BZ232 (15.0 × 103) | Anticapsular | 50 | 0/5 | <0.01 |
| Anticapsular | 10 | 0/5 | <0.01 | ||
| AL12 Anti-NspA | 50 | 4/5 | 0.2 | ||
| AL12 Anti-NspA | 10 | 4/5 | 0.5 | ||
| Irrelevant | 50 | 5/5 | 11.5 | ||
| PBS-albumin | None | 5/5 | 3.9 | ||
| 3 | M986 (2 × 103) | Anticapsular | 2 | 0/6 | <0.01 |
| AL12 Anti-NspA | 50 | 5/5 | >500 | ||
| AL12 Anti-NspA | 2 | 6/6 | >500 | ||
| Irrelevant | 2 | 5/5 | >500 | ||
In experiments 1 and 2, animals were pretreated with the MAb i.p. at time zero. Two hours later, the animals were challenged i.p. with bacteria. In experiment 3, to increase the sensitivity of the assay, the bacteria and MAb were mixed together immediately before the i.p. challenge. In this experiment, blood was also obtained for culture at 6 h. Results for this time are provided in the text.
For calculation of geometric mean CFU per milliliter, animals with sterile cultures were assigned a value of 10 CFU/ml. The geometric mean CFU per milliliter of animals receiving AL12 were compared to that of the combined group of negative-control animals given irrelevant MAb or PBS-BSA: experiment 1, 50 μg of AL12 versus controls, P < 0.001; 5 μg of AL12 versus controls, P < 0.005; experiment 2, 50 μg of AL12 versus controls, P < 0.01; 10 μg of AL12 versus controls, P < 0.01.
In experiment 1, none of the animals pretreated with anti-NspA MAb AL12 (50 μg per rat) and challenged by strain 8047 developed bacteremia. At a MAb dose of 5 μg per rat, four of the seven animals had bacteremia but the geometric mean CFU/ml of the treated animals was less than 1/50 of that of control animals pretreated with the irrelevant MAb or PBS-BSA (P < 0.005). In experiment 2, four of five animals pretreated with 50 μg of AL12 per rat and four of five animals pretreated with 10 μg per rat had bacteremia 18 h after challenge by strain BZ232. However, at both antibody doses, the respective geometric mean CFU/ml of blood of the treated animals was lower that that of control animals pretreated with an irrelevant MAb or PBS-BSA (P < 0.01). In experiment 3, there was no evidence of protection 18 h after challenge with strain M986, despite premixing the bacteria with the MAb immediately before the challenge. Although not shown in Table 3, in experiment 3, blood cultures were also obtained 6 h after challenge. At this time point, all animals had bacteremia but some protection was present in animals treated with the 50-μg dose of AL12 (geometric mean CFU per ml of blood of 1 × 103 versus 34.7 × 103 in the control group which was given the irrelevant MAb; P = 0.0008).
Virulence of strain 8047 in which the nspA gene has been disrupted.
Table 4 summarizes the results of challenging infant rats with strain 8047ΔNspA. All three animals given a challenge dose of approximately 8 × 103 CFU of strain 8047ΔNspA had bacteremia present 18 h later. The geometric mean CFU/ml for these pups was similar to that observed in control rats challenged with the 8047 wild-type strain. All three animals challenged with a 106-CFU dose of strain 8047ΔNspA were dead at 18 h postchallenge.
TABLE 4.
N. meningitidis serogroup B strain 8047 in which the NspA gene has been deleted (8047ΔNspA) causes bacteremia and death in infant rats challenged i.p.
| Strain | Bacterial dose (CFU)/rat | Results of blood culture 18 h after challenge
|
|
|---|---|---|---|
| No. positive/total | CFU/ml (geometric mean, 103) | ||
| 8047 (wild type) | 7.8 × 103 | 3/3 | 123 |
| 8047ΔNspA | 8.3 × 103 | 3/3 | 109 |
| 8047ΔNspA | 8.3 × 106 | 3/3a | NDb |
Three of three were dead at 18 h.
ND, not done.
DISCUSSION
The results presented here extend our previous observations made with polyclonal antisera (16). Using a panel of anti-NspA MAbs, we have shown that large differences exist among N. meningitidis serogroup B strains in the ability of anti-NspA antibody to bind to the surface of live encapsulated bacteria, to elicit complemented-mediated bacteriolysis, and to confer passive protection in vivo. With the exception of two strains, the results were highly reproducible in replicate experiments. The two exceptional strains, BZ232 and MC58, showed variations in anti-NspA-surface binding among different subcultures (Fig. 4). Another important new finding is the ability of an NspA-knockout strain to cause bacteremia and death of infant rats after i.p. challenge (Table 3), a result indicating that NspA expression is not essential for causing bacteremia.
The underlying mechanism(s) responsible for the observed strain differences in anti-NspA surface binding remains unknown. Although NspA is highly conserved among N. meningitidis serogroup B strains (4, 13, 16), changes in one or more amino acids in the protein, particularly in surface-accessible loops, could result in decreased anti-NspA binding. However, with one notable exception (4), all strains in which the nspA gene has been sequenced show high conservation of gene segments encoding putative surface-exposed loops (100% identity for N. meningitidis serogroup B strains) (4, 16). Also, strains NGP165 and CU385 have nearly identical protein sequences (one amino acid difference, Gly versus Ala at position 150, which is not located in a putative loop segment), yet NGP165 is negative for surface binding with AL12 while CU385 is strongly positive (16). Finally, based on colony Western blot analysis, the NspA epitope defined by MAb AL12 was present in all 10 strains examined, despite negative or variable surface binding in four of the strains (Table 2). Therefore, amino acid sequence variation is unlikely to result in the differences in functional activity of the anti-NspA MAbs described in this study.
Alternative mechanisms that might explain decreased surface accessibility of NspA on live bacterial cells and/or decreased susceptibility to complement-mediated anti-NspA bacteriolysis include strain differences in the amount of NspA produced and steric interference by other variable bacterial surface structures or a combination of the two. In our previous study with polyclonal anti-NspA antisera, we found a statistically significant trend for poorer antibody surface binding among the N. meningitidis serogroup B strains that were the highest producers of capsular polysaccharide (16). However, some strains were inconsistent with this trend. For example, strain 8047 is a high producer of capsular polysaccharide but is positive for anti-NspA surface binding and susceptible to bactericidal activity, while strain NGP165, which is a low capsular polysaccharide producer, is negative for anti-NspA surface binding and resistant to bactericidal activity. Some exceptions can be explained by quantitative differences in NspA expression (i.e., high for 8047 and low for NGP165, as determined by SDS-PAGE of lauroyl sarcosinate-extracted outer membrane preparations [16]). For other strains such as CU385 that were positive for surface binding but negative for bactericidal activity and other strains with variable anti-NspA reactivity, such as MC58 and BZ232, there were no apparent differences in NspA expression. Therefore, there are likely to be other mechanisms for interfering with binding and bactericidal activity.
The data indicating variation in anti-NspA-surface binding in respective subcultures of two strains are of particular interest (Fig. 4), given that NspA expression was uniform in all CFU tested by Western blotting. Furthermore, colonies derived and expanded from each of the three subcultures could test positive or negative for NspA surface binding by the flow assay. In contrast, there was no variability of anti-PorA MAb binding to the bacterial cells from the same strains in control experiments (Fig. 4). These results imply that some other variable surface structure can inhibit antibody binding to NspA but not to PorA. Future studies will need to examine the possible effect of lipooligosaccharide phenotypes, expression of other outer membrane proteins, or sialyation of surface molecules on antibody binding to NspA epitopes.
Whatever the underlying mechanism(s), the observed strain and intrastrain variability in NspA surface accessibility appears to have important consequences for the ability of anti-NspA antibodies to elicit complement-mediated bacteriolysis. None of the strains that were negative or variable for surface binding in the flow cytometry assay were susceptible to anti-NspA complement-mediated bacteriolysis (Table 2). There were also two strains (NG3/88 and CU385), each of which was positive for surface binding with the four MAbs but resistant to complement-mediated bacteriolysis, even when tested at >10-fold-higher antibody concentrations than those used in the surface binding assay (Table 2). In the presence of complement, both strains were readily killed by the positive-control IgG anticapsular antibody. The reasons for poor bactericidal activity of the anti-NspA MAbs that bind well to these bacteria remain unknown.
The passive protection data obtained with MAb AL12 in infant rats challenged by different N. meningitidis serogroup B strains were consistent with the in vitro bactericidal results (compare Tables 2 and 3). Thus, MAb AL12 was highly protective against challenge by N. meningitidis serogroup B strain 8047 (surface binding positive and susceptible to bacteriolysis), was partially protective against strain BZ232 (surface binding variable and resistant to bacteriolysis), and failed to protect against strain M986 (negative for surface binding and resistant to bacteriolysis).
The bactericidal and passive protection results described above differ from those described by Cadieux et al., who reported that an anti-NspA MAb, Me-7, was broadly bactericidal against a panel of 14 serogroup B, C, and A strains (all but 1 serogroup A strain were killed) (4). Since we were unable to obtain MAb Me-7 from Cadieux et al. for our studies, we cannot exclude the possibility that this MAb is more broadly reactive and bactericidal than the MAbs evaluated in our panel. However, an alternative explanation for these conflicting results may lie in the differences in the methods used to assess bactericidal and passive protective activities. For example, Cadieux et al. defined protection in their mouse model as a significant decrease in bacteremia 5 h after challenge in the treated animal. Typically, the Me-7-treated mice that were considered protected had 103 to 104 bacteria per ml of blood (<25% of control animals). In contrast, the primary endpoint of protection in our experiments was bacteremia measured at 18 h. In a passive protection experiment, antibody amount is limited. Therefore, it is possible to observe protection at 6 h but not at 18 h after challenge, as observed with strain M986 (see Table 3 and Results).
In conclusion, considerable evidence indicates that NspA is highly conserved in pathogenic Neisseria meningitidis strains and that NspA is expressed in all N. meningitidis serogroup B strains tested to date (4, 13, 16). The present data with a panel of anti-NspA MAbs confirm earlier observations with polyclonal antisera that despite nspA gene conservation and expression, many N. meningitidis serogroup B strains show poor antibody binding to the surfaces of live bacteria and resist anti-NspA complement-mediated bacteriolysis. Further studies are needed to define the underlying mechanisms modulating surface accessibility of NspA. The ability of the NspA gene knockout strain to cause bacteremia and lethality in infant rats (Table 4) provides evidence that this molecule is not essential for serum resistance or for causing bacteremia once the organism has entered the blood stream. Taken together, the data underscore the likelihood that an NspA-based vaccine will need to be supplemented by additional antigens to elicit broad-based protection.
ACKNOWLEDGMENTS
This work was supported by grants RO1 AI45642, AI46464, AI25008, and RR01271 from NIAID, NIH.
We thank Jeanette Abu-Bobie, Chiron Corp., Siena, Italy, for the gift of plasmid pBSUDNspAERM. We also thank Apurva Dave and Katherine Alter for performing antibody purification and flow cytometry assays, respectively.
REFERENCES
- 1.Abdillahi H, Poolman J T. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J Med Microbiol. 1988;26:177–180. [PubMed] [Google Scholar]
- 2.Amir J, Liang X, Granoff D M. Variability in the functional activity of vaccine-induced antibody to Haemophilus influenzae type b. Pediatr Res. 1990;27:358–364. doi: 10.1203/00006450-199004000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Arhin F F, Moreau F, Coulton J W, Mills E L. Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can J Microbiol. 1998;44:56–63. [PubMed] [Google Scholar]
- 4.Cadieux N, Plante M, Rioux C R, Hamel J, Brodeur B R, Martin D. Bactericidal and cross-protective activities of a monoclonal antibody directed against Neisseria meningitidis NspA outer membrane protein. Infect Immun. 1999;67:4955–4959. doi: 10.1128/iai.67.9.4955-4959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caugant D A, Bol P, Hoiby E A, Zanen H C, Froholm L O. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958–1986. J Infect Dis. 1990;162:867–874. doi: 10.1093/infdis/162.4.867. [DOI] [PubMed] [Google Scholar]
- 6.Caugant D A, Bovre K, Gaustad P, Bryn K, Holten E, Hoiby E A, Froholm L O. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J Gen Microbiol. 1986;132:641–652. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 7.DeVoe I W. The meningococcus and mechanisms of pathogenicity. Microbiol Rev. 1982;46:162–190. doi: 10.1128/mr.46.2.162-190.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frasch C E. Meningococcal vaccines: past, present and future. In: Cartwright K, editor. Meningococcal disease. New York, N.Y: John Wiley & Sons; 1995. pp. 245–283. [Google Scholar]
- 9.Granoff D M, Bartoloni A, Ricci S, Gallo E, Rosa D, Ravenscroft N, Guarnieri V, Seid R C, Shan A, Usinger W R, Tan S, McHugh Y E, Moe G R. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J Immunol. 1998;160:5028–5036. [PubMed] [Google Scholar]
- 10.Hammerschmidt S, Birkholz C, Zahringer U, Robertson B D, van Putten J, Ebeling O, Frosch M. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 11.Jelfs J, Munro R, Wedege E, Caugant D A. Sequence variation in the porA gene of a clone of Neisseria meningitidis during epidemic spread. Clin Diagn Lab Immunol. 2000;7:390–395. doi: 10.1128/cdli.7.3.390-395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin D, Brodeur B R, Hamel J, Couture F, de Alwis U, Lian Z, Martin S, Andrews D, Ellis R W. Candidate Neisseria meningitidis NspA vaccine. J Biotechnol. 2000;83:27–31. doi: 10.1016/s0168-1656(00)00294-7. [DOI] [PubMed] [Google Scholar]
- 13.Martin D, Cadieux N, Hamel J, Brodeur B R. Highly conserved Neisseria meningitidis surface protein confers protection against experimental infection. J Exp Med. 1997;185:1173–1183. doi: 10.1084/jem.185.7.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masson L, Holbein B E. Influence of nutrient limitation and low pH on serogroup B Neisseria meningitidis capsular polysaccharide levels: correlation with virulence for mice. Infect Immun. 1985;47:465–471. doi: 10.1128/iai.47.2.465-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milagres L G, Gorla M C, Sacchi C T, Rodrigues M M. Specificity of bactericidal antibody response to serogroup B meningococcal strains in Brazilian children after immunization with an outer membrane vaccine. Infect Immun. 1998;66:4755–4761. doi: 10.1128/iai.66.10.4755-4761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moe G R, Tan S, Granoff D M. Differences in surface expression of NspA among Neisseria meningitidis group B strains. Infect Immun. 1999;67:5664–5675. doi: 10.1128/iai.67.11.5664-5675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naess L M, Oftung F, Aase A, Wetzler L M, Sandin R, Michaelsen T E. Human T-cell responses after vaccination with the Norwegian group B meningococcal outer membrane vesicle vaccine. Infect Immun. 1998;66:959–965. doi: 10.1128/iai.66.3.959-965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizza M, Scarlato V, Masignani V, Giuliani M M, Arico B, Comanducci M, Jennings G T, Baldi L, Bartolini E, Capecchi B, Galeotti C L, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood D W, Jeffries A C, Saunders N J, Granoff D M, Venter J C, Moxon E R, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 19.Plante M, Cadieux N, Rioux C R, Hamel J, Brodeur B R, Martin D. Antigenic and molecular conservation of the gonococcal NspA protein. Infect Immun. 1999;67:2855–2861. doi: 10.1128/iai.67.6.2855-2861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenqvist E, Hoiby E A, Wedege E, Caugant D A, Froholm L O, McGuinness B T, Brooks J, Lambden P R, Heckels J E. A new variant of serosubtype P1.16 in Neisseria meningitidis from Norway, associated with increased resistance to bactericidal antibodies induced by a serogroup B outer membrane protein vaccine. Microb Pathog. 1993;15:197–205. doi: 10.1006/mpat.1993.1070. [DOI] [PubMed] [Google Scholar]
- 21.Tappero J W, Lagos R, Ballesteros A M, Plikaytis B, Williams D, Dykes J, Gheesling L L, Carlone G M, Hoiby E A, Holst J, Nokleby H, Rosenqvist E, Sierra G, Campa C, Sotolongo F, Vega J, Garcia J, Herrera P, Poolman J T, Perkins B A. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA. 1999;281:1520–1527. doi: 10.1001/jama.281.16.1520. [DOI] [PubMed] [Google Scholar]
- 22.van der Ende A, Hopman C T, Zaat S, Essink B B, Berkhout B, Dankert J. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the −10 and −35 regions of the promoter. J Bacteriol. 1995;177:2475–2480. doi: 10.1128/jb.177.9.2475-2480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel U, Hammerschmidt S, Frosch M. Sialic acids of both the capsule and the sialylated lipooligosaccharide of Neisseria meningitidis serogroup B are prerequisites for virulence of meningococci in the infant rat. Med Microbiol Immunol. 1996;185:81–87. doi: 10.1007/s004300050018. [DOI] [PubMed] [Google Scholar]
- 24.Wedege E, Hoiby E A, Rosenqvist E, Bjune G. Immune responses against major outer membrane antigens of Neisseria meningitidis in vaccinees and controls who contracted meningococcal disease during the Norwegian serogroup B protection trial. Infect Immun. 1998;66:3223–3231. doi: 10.1128/iai.66.7.3223-3231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]