Abstract
Previous studies investigated the biochemical basis of dark-cutting conditions at elevated muscle pH (above 6), but the molecular basis at slightly above normal pH (between 5.6 and 5.8) is still unclear. The objective was to determine protein and metabolite profiles to elucidate postmortem muscle darkening at slightly elevated pH. Loins were selected based on the criteria established in our laboratory before sample collections, such as pH less than 5.8, L* values (muscle lightness) less than 38, and not discounted by the grader (high-pH beef with dark color are discounted and not sold in retail stores). Six bright red loins (longissimus lumborum) at normal-pH (average pH = 5.57) and six dark-colored strip loins at slightly elevated pH (average pH = 5.70) from A maturity carcasses were obtained within 72-h postmortem from a commercial beef purveyor. Surface color, oxygen consumption, metmyoglobin reducing activity, protein, and metabolite profiles were determined on normal-pH and dark-colored steaks at slightly elevated pH. Enzymes related to glycogen metabolism and glycolytic pathways were more differently abundant than metabolites associated with these pathways. The results indicated that oxygen consumption and metmyoglobin reducing activity were greater (P < 0.05) in darker steaks than normal-pH steaks. Enzymes involved with glycogen catabolic pathways and glycogen storage disease showed lower abundance in dark beef. The tricarboxylic acid metabolite, aconitic acid, was overabundant in darker-colored beef than normal-pH beef, but glucose derivative metabolites were less abundant. The majority of glycogenolytic proteins and metabolites reported as overabundant in the previous dark-cutting studies at high pH (>6.4) also did not show significant differences in the current study. Therefore, our data suggest enzymes involved in glycogen metabolism, in part, create a threshold for muscle darkening than metabolites.
Keywords: dark-cutting, normal-pH, mass spectrometry, meat color, metabolomics, proteomics
The objective was to determine the biochemical basis of beef muscle darkening at a slightly elevated pH. The enzymes related to glycogen metabolism and glycolytic pathways were more differently expressed in darker beef than the metabolites associated with these pathways.
Introduction
The color of meat is an important factor in consumers’ assessment of meat quality. To meat buyers, the bright cherry-red color of meat indicates freshness and wholesomeness (Sammel et al., 2002; Boykin et al., 2017; Węglarz, 2018; Salim et al., 2019; Ramanathan et al., 2020a, 2022). The color of meat is primarily determined by myoglobin, a water-soluble sarcoplasmic protein (Bendall, 1979; Faustman & Cassens, 1990; AMSA, 2012; Suman & Joseph, 2013). Myoglobin exists in three forms, namely oxy-, deoxy-, and metmyoglobin. A greater concentration of oxymyoglobin gives consumers’ desired bright cherry-red color. However, predominant deoxymyoglobin is associated with a dark meat color appearance (Ashmore et al., 1972; McKeith et al., 2016; Ramanathan et al., 2020b; Kiyimba et al., 2021). Although the prevalence of dark-cutting beef has reduced over the years, dark-cutting beef still continues to occur in the beef industry. The 2016 National Beef Quality Audit (NBQA) reported an average dark-cutter occurrence of 1.9% (Boykin et al., 2017). However, some beef packers reported an occurrence of 2% to 5% in certain months (personal communication, 2022). The dark-cutting beef is discounted during grading, and beef is not sold in retail due to its appearance and reduced shelf life. The US beef industry loses $202 million annually due to dark-cutting conditions (calculated based on 2016 NBQA and carcass discount by the American Marketing Service United States Department of Agriculture). However, the molecular and mechanistic basis for the occurrence remains unknown.
The hallmark of dark-cutting conditions is a greater than normal muscle pH. More specifically, depending on the range of ultimate pH, muscle darkening can vary from slight dark red color to a coffee-bean dark color (Mahmood et al., 2018; Lei et al., 2021; Roy et al., 2022). Thus, meat with an ultimate muscle pH greater than 5.8 results in a visually detectable dark red color (Knee et al., 2007; Holdstock et al., 2014; Mahmood et al., 2017; Steel et al., 2018). Although the trigger for dark-cutting development is not clear, the current knowledge suggests that lower glycogen levels correlate with less glycolysis and greater than normal muscle pH. More specifically, dysregulated glycogen catabolic processes postmortem cause less carbon flow, leading to less postmortem lactic acid accumulation and elevated muscle pH (Fuente-García et al., 2021; Kiyimba et al., 2021). Additionally, dark-cutting beef has increased mitochondrial respiration postmortem than normal pH (Ashmore et al., 1971; English et al., 2016; McKeith et al., 2016; Ramanathan et al., 2020b). Thus, greater than normal muscle pH in dark-cutting is conducive to enhanced mitochondrial respiration postmortem. Therefore, sustained mitochondrial respiration in dark-cutting phenotypes increases competition for available muscle oxygen and produces dark-colored muscles (Ashmore et al., 1972; Egbert & Cornforth, 1986; Ramanathan et al., 2009).
The identification of changes in protein profiles of muscle darkening at elevated muscle pH (> 5.8) noted that several proteins and metabolites involved with glycogen catabolism were less abundant, while proteins and metabolites associated with mitochondrial oxidative metabolism were overabundant in dark-cutting groups compared with normal-pH counterparts (Ramanathan et al., 2020b; Wu et al., 2020; Cônsolo et al., 2021; Fuente-García et al., 2021; Gagaoua et al., 2021; Kiyimba et al., 2021). In addition, a recent study evaluating muscle fiber characteristics showed that dark-cutting beef has greater type I fibers than normal-pH (Roy et al., 2022).
Different grade levels of dark-cutting beef are reported based on ultimate pH. Therefore, characterizing protein and metabolite profiles of muscles that produce darker color at slightly elevated pH offers valuable insights into the occurrence of dark-cutting conditions. However, limited research has characterized the protein and metabolite profiles of darker beef at slightly elevated muscle pH (<5.8). A combination of proteomics and metabolomics can provide a comprehensive and in-depth understanding of complex biological processes regulating muscle darkening at slightly elevated muscle pH. In the current study, we combined proteomics and metabolomics profiling to identify differentially abundant proteins (DAPs) and metabolites in dark-cutting beef at slightly elevated muscle pH compared with normal-pH beef. Furthermore, we utilized bioinformatics analyses to elucidate the pH-dependent effects on muscle darkening in beef.
Materials and Methods
Beef loins were purchased from a United States Department of Agriculture (USDA) Food Safety and Inspection Service inspected commercial plant. Therefore, institutional animal care and use committee approval was not requested for this study.
Sample collection and preparation
Six bright red longissimus lumborum loins at normal-pH (average pH = 5.57) and six dark-colored loins (Institutional Meat Purchasing Specification #180, NAMP, 2002; grain-finished, spray chilled) at slightly elevated pH (average pH = 5.70) from A-maturity carcasses were obtained within 72-h postmortem from a commercial beef purveyor in Amarillo, TX. Dark-colored loins were selected based on the criteria established in our laboratory before sample collections, such as pH less than 5.8, L* values (muscle lightness) less than 38, and not discounted by the grader (high-pH beef with dark color are discounted and not sold in retail stores). pH and surface color of dark-colored loins were measured in the meat plant to satisfy conditions before collection. The loins were transported on ice, and loins were also measured for pH and color after packaging. Three 2.54-cm-thick steaks from each animal were cut from the anterior end of the loin. The first steak from each loin type (normal pH and darker at slightly elevated pH) was utilized for surface color, oxygen consumption, and metmyoglobin reducing activity studies. The second steak was utilized for liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) proteomics and gas chromatography-mass spectrometry (GC-MS) non-targeted metabolomics approach. The third steak from each animal was used to determine muscle pH and proximate compositions. All analyses were conducted 96 h postmortem.
Determination of pH, proximate composition, surface color, metmyoglobin reducing activity, and oxygen consumption
The pH of each steak was recorded using a probe-type Accumet 50 pH meter (Fisher Scientific, Fairlawn, NJ). The pH probe was calibrated with buffers at pH 4 and 7. The pH probe was inserted into the meat at three locations, and the average pH was determined for each steak. The proximate compositions were determined using an Association of Official Analytical Chemist-approved (Official Method 2007.04; Anderson et al., 2007) near-infrared spectrophotometer (Foss Food Scan 78800; Dedicated Analytical Solutions, DK-3400 Hilleroed, Denmark). Protein, moisture, and fat contents were reported on a percent (%) basis.
From each loin, a 2.5-cm-thick steak was cut, placed onto foam trays with absorbent pads, and steaks were wrapped with polyvinyl chloride film (oxygen-permeable polyvinyl chloride fresh meat film; 15,500 to 16,275 cm3 O2/m2/24 h at 23 °C, E-Z Wrap Crystal Clear Polyvinyl Chloride Wrapping Film; Koch Supplies, Kansas City, MO) and stored at 4 °C for 30 min. The surface color was measured using a HunterLab MiniScan spectrophotometer (AMSA, 2012). Following surface color measurements, each steak was cut in half. The first half was used to estimate muscle oxygen consumption, and the second half was utilized to measure metmyoglobin reducing activity. The greater postmortem muscle pH (above 5.8) seen in dark-cutting beef can influence muscle reflectance properties (AMSA, 2012). Hence, a modified method was utilized to measure oxygen consumption and metmyoglobin reducing activity (Ramanathan et al., 2019). For metmyoglobin reducing activity, samples from the interior of steak halves (approx. 3 × 3 × 1.5 cm tissue with no visible fat or connective tissue) were submerged in a 0.3% w/v solution of sodium nitrite (Sigma Aldrich, St. Louis, MO) for 20 min at 30 °C (Fisher Scientific, Model 630F, Waltham, MA) to facilitate metmyoglobin formation (Sammel et al., 2002). The sections were then removed and blotted to remove visible nitrite solution. The level of metmyoglobin content on the surface was determined by using a HunterLab Miniscan spectrophotometer. Resistance to myoglobin oxidation was a better indicator of metmyoglobin reducing activity than post-reduction values (O’Keeffe and Hood, 1982; Mancini et al., 2008). The resistance to myoglobin oxidation was reported as K/S572 ÷ K/S525. A greater number indicates greater metmyoglobin reducing activity. The steak half was bloomed at 4 °C for 1 h. Following blooming, each steak section was vacuum-packaged and incubated at 25 °C for 30 min to promote oxygen consumption. After incubation, surface color readings were taken, and the deoxymyoglobin level was measured to determine muscle oxygen consumption.
Metabolomics analysis
The metabolomics analyses were conducted at the National Institute of Health West Coast Metabolomics Center, University of California Davis, CA, USA. In brief, 10 mg of skeletal muscle tissue from normal-pH and dark-colored beef from slightly elevated pH were freeze-dried and stored at −80 °C until analysis. The samples were mixed with two 3 mm grinder glass beads (Cat.1.04015, Sigma, St. Louis, MO). Metabolites were extracted with 1,000 μL of degassed acetonitrile/isopropanol/water mixture (3:3:2, v/v/v). The mixture was homogenized for 30 s and shaken using an automatic shaker for 6 min at 4 °C. The homogenate was centrifuged at 4 °C for 2 min at 14,000 × g, and the supernatant was collected. Methyl esters (2 µL of 1 mg/mL) were added as an internal standard, and the samples were dried under a gentle stream of nitrogen gas. The dried samples were derivatized with 10 μL of methoxyamine (Thermo Fisher Scientific, Catalog number TS-45950) in pyridine and subsequently by 90 μL of N-methyl-N-(trimethylsiyl) trifluoroacetamide (Thermo Fisher Scientific, Catalog number TS-48910) for trimethylsilylation of acidic protons. The extracted metabolites were analyzed using gas chromatography-mass spectrometry (Fiehn, 2016; Ramanathan et al., 2020b).
Metabolomics data processing
GC-MS data files were preprocessed directly after data acquisition using ChromaTOF version 2.32 and stored as specific *.peg files, as generic *.txt result files, and additionally as generic ANDI MS *.cdf files (Skogerson et al., 2011). The files were exported to a data server with absolute spectra intensities and further processed by a filtering algorithm implemented in the metabolomics BinBase database. The BinBase algorithm (rtx5) settings used included: validity of chromatogram (10^7 counts s–1), unbiased retention index marker detection (MS similarity >800, validity of intensity range for high m/z marker ions), and retention index calculation by fifth-order polynomial regression. Metabolite quantification was reported as peak height using the unique ion as default. A quantification report table was produced for all KEGG compound database entries that were positively detected in more than 10% of the samples (as defined in the miniX database) for unidentified metabolites. The list of metabolite features quantified and identified in dark-cutting beef at slightly elevated pH and normal-pH beef was uploaded into MetaboAnalyst for differential metabolite expression.
Protein extraction and digestion
Skeletal muscle tissue samples of 0.5 g free of fat and connective tissue from normal-pH and dark-colored beef from slightly elevated pH (n = 6 each loin type) steaks were extracted in 5 mL of lysis buffer (6 M guanidine hydrochloride, 100 mM HEPES, 50 mM chloroacetamide (CCA), 10 mM TRIS (2-carboxyethyl) phosphine TCEP, pH 8.0) as previously described (Kiyimba et al., 2021). Protein concentration was determined using a tryptophan fluorescence assay (Wis̈niewski and Gaugaz, 2015). The extracted samples were alkylated by the addition of 10 mM iodoacetamide and incubated for 15 min at room temperature. The solutions were then diluted with three volumes of 100 mM Tris–HCl, pH 8.5, and digested at 37 °C overnight with 4 µg/mL of trypsin/LysC (Promega, Madison, WI). The samples were further digested by the second addition of trypsin/LysC (2 µg/mL) for 6 h. Further steps in sample preparation and LC-MS/MS analysis were performed as previously described (Kiyimba et al., 2021).
MS/MS database searching for identification of DAPs
The raw LC-MS/MS instrument data files were analyzed using MaxQuant software (V1.5.3.12, Max Planck Institute of Biochemistry). The MS/MS spectra from each nano-LC-MS/MS run were searched against a Uniprot Bos taurus proteome database of 23,968 protein sequences (downloaded in March 2018) using the same search parameters as previously reported (Kiyimba et al., 2021). The sequences of common contaminants were included in the searches. The obtained MaxQuant label-free quantitation (LFQ) protein intensities were imported into the Perseus v1.6.3.3 software platform (https://maxquant.net/perseus/) and analyzed for differential abundance. For this analysis, protein groups were first filtered for reverse and potential contaminants. Then LFQ intensities were analyzed within the Perseus framework, using a two-sample T-test to compare log2 transformed LFQ protein intensities. Protein expression profiles were considered significant if the p-value was less than or equal to 0.05.
Statistical and bioinformatics analyses
A completely randomized block design was employed to characterize muscle-specific differences in biochemical properties and color attributes of normal-pH and dark-colored beef at slightly elevated pH. The experiment was replicated 6 times (n = 6). Each loin from normal-pH and dark-colored beef at slightly elevated pH was considered a block. The least square means and standard error of mean were analyzed using the Proc Mixed procedure in SAS (Version 9.1, SAS Institute Inc. Cary, NC). The least squares means were separated using the pdiff option and were considered significant at P < 0.05.
Principal Component Analysis (PCA), supervised projections to latent structure-discriminant analysis, and hierarchical cluster analysis were performed to create plots and heat maps for the DAPs and metabolites using Perseus software (V.1.6.3.3,https://maxquant.net/perseus/) and MetaboAnalyst (V.5.0, http://www.metaboanalyst.ca), respectively. The metabolite data sets were normalized by a median, log-transformed, and scaled by Pareto scaling.
The online platform Metascape (https://metascape.org/) was employed to analyze the differentially abundant protein enrichment in the GO annotation and the DisGeNET database (Zhou et al., 2019). To acknowledge sampling bias, the gene list of all identified and quantified proteins in all 6 of the 6 compared samples (dark-colored beef at slightly elevated pH and normal-pH beef) was used as background uploaded into Metascape. The key pathway involved in postmortem metabolism (glycogen metabolism) was further analyzed in Cytoscape (V.3.7.1; https://cytoscape.org/) using the WikiPathway plugin (https://www.wikipathways.org; Szklarczyk et al., 2017). Protein networks were analyzed using the String database plugin in Cytoscape to explore the potential protein–protein biological interactions.
Results
Muscle surface color and biochemical characteristics
The muscle surface color and biochemical attributes of normal-pH and dark-colored beef at slightly elevated pH are summarized in Table 1. A slight elevation in muscle pH (0.13 units; 2.3% greater than normal-pH) of dark beef resulted in 11.72% decrease in lightness (L*-value difference = 5.11, P < 0.001). Dark-colored beef at slightly elevated pH had lower a*- and b*-values (P < 0.01) than normal-pH beef. The L*-value is an indicator of muscle lightness, and lower a* values indicate less red meat. Furthermore, dark-colored beef at slightly elevated pH also showed greater oxygen consumption (P = 0.03) and metmyoglobin reducing activity (MRA, P = 0.02) compared with normal-pH beef.
Table 1.
Comparison of color and biochemical properties of normal-pH and dark-colored beef at slightly elevated pH
| Parameters | Normal-pH | Dark-colored beef | SEM | P-value | |
|---|---|---|---|---|---|
| a) Proximate composition1 | Moisture (%) | 71.7 | 71.2 | 1.2 | 0.22 |
| Protein (%) | 23.7 | 23.7 | 0.8 | 0.51 | |
| Fat (%) | 4.7 | 4.8 | 0.2 | 0.21 | |
| b) Surface color2 | L* | 43.59 | 38.48 | 0.31 | <0.001 |
| a* | 28.05 | 25.28 | 0.28 | <0.001 | |
| b* | 21.42 | 18.94 | 0.24 | <0.001 | |
| c) Biochemical properties3 | pH | 5.57 | 5.70 | 0.10 | <0.01 |
| Oxygen consumption | 0.66 | 0.86 | 0.058 | 0.03 | |
| Metmyoglobin reducing activity | 0.82 | 1.32 | 0.012 | 0.02 |
1Proximate composition was determined using a NIR-based Food Scan.
2Surface color was measured using a HunterLab MiniScan spectrophotometer after exposing the steaks to atmospheric oxygen for 1 h at 4 °C. L* represents lightness, and a lower number represents darker meat color; a* value represents redness, and a lower number indicates less red color.
3pH was measured using a probe-type pH meter and measured 96 h postmortem; deoxymyoglobin content of bloomed and vacuum-packaged steaks was used as OC. A greater number indicates more oxygen consumption; Metmyoglobin reducing activity was determined as resistance to form initial metmyoglobin formation, and a greater number indicates greater metmyoglobin reducing activity.
Proteomics analysis
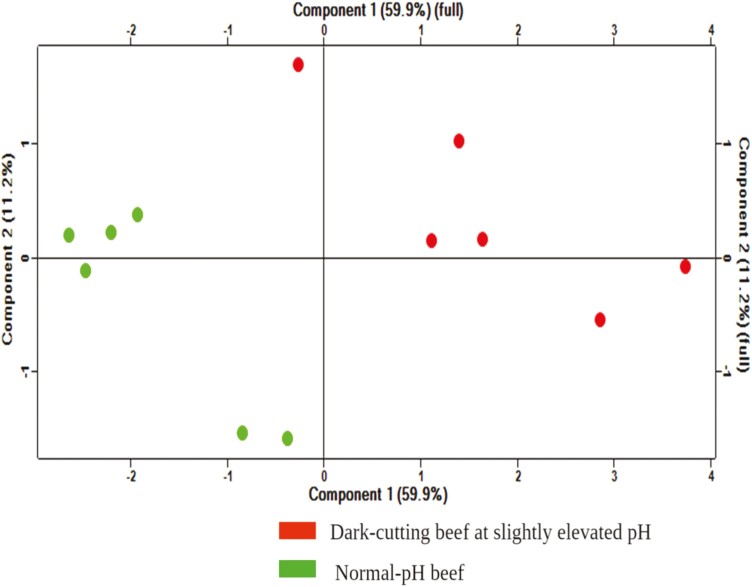
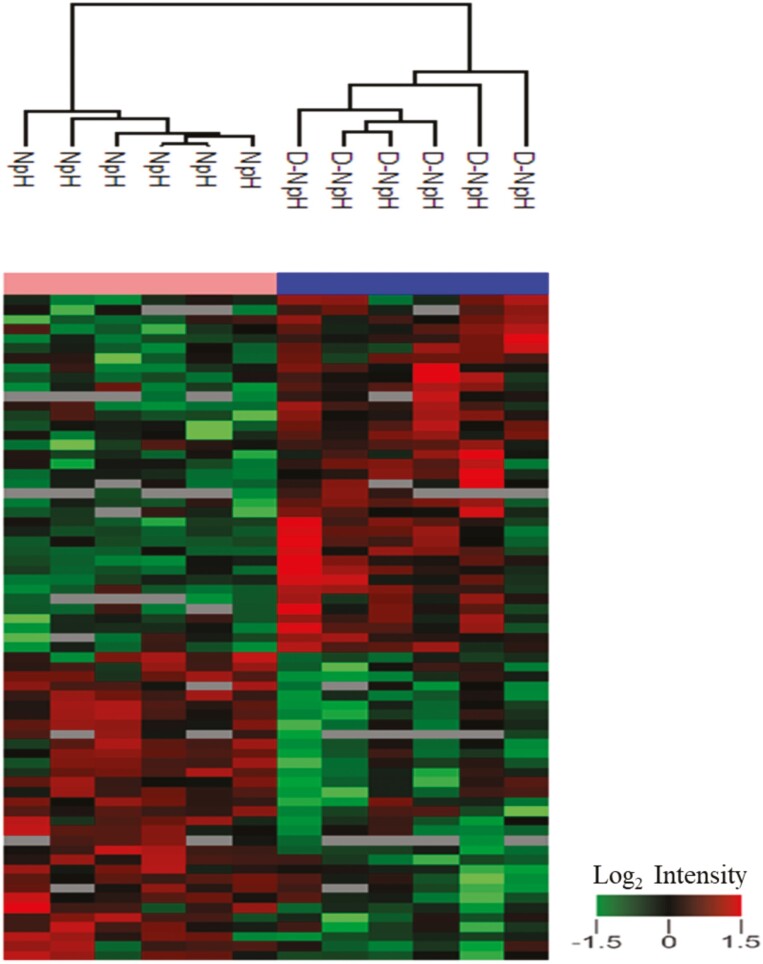
The LC-MS/MS proteomic profiling analysis identified 1,080 proteins in the proteomes of dark-colored beef at a slightly elevated pH than normal-pH beef. Among these, 23 proteins were overabundant (Table 2), and 13 proteins were less abundant (Table 3) in dark-colored beef than normal-pH beef (P < 0.05; fold change > 1.3). PCA (Figure 1) showed that 71.1% variability was explained by the first two components, with 59.9% and 11.2% total variation, respectively. The clusters of dark-colored beef at slightly elevated pH showed distinctive separation from the normal-pH beef clusters. In addition, the hierarchical clustering analysis (Figure 2) revealed distinctive clusters of protein groups co-segregating together in dark-colored beef at slightly elevated pH and normal-pH beef.
Table 2.
Proteins overabundant in dark-colored beef at slightly elevated pH compared with normal-pH beef (fold-changes: dark color/normal-pH, P < 0.05).
| Gene name | Protein name | Molecular function | Regulation | Fold change (dark color/normal-pH) | P-value |
|---|---|---|---|---|---|
| ATP1A2 | Sodium/Potassium-transporting ATPase subunit alpha-2 | ATPase, ATPase-coupled cation transmembrane transporter activity | ↑ | 1.7 | 0.02 |
| CYB5R3 | NADH-Cytochrome b5 reductase 3 | ADP, AMP, FAD, and NAD binding | ↑ | 1.4 | 0.02 |
| NDUFA7 | NADH dehydrogenase[Ubiquinone] 1 alpha subcomplex subunit 7 | NADH dehydrogenase(ubiquinone) activity, mitochondrial electron transport | ↑ | 1.5 | 0.02 |
| OXCT1 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | CoA transferase activity | ↑ | 1.4 | 0.005 |
| ACTN4 | Alpha-actin-4 | Actin binding, muscle contraction | ↑ | 1.3 | 0.02 |
| TPM2 | Tropomyosin beta chain | Actin binding, muscle contraction | ↑ | 1.3 | 0.002 |
| XIRP1 | Xin actin binding repeat-containing protein 1 | Actin filament binding, muscle contraction, structure, and associated activity | ↑ | 5.0 | 0.01 |
| SYNPO2L | Synaptopodin 2-like protein | Actin binding, sarcomere organization, muscle contraction | ↑ | 1.5 | 0.02 |
| SLMAP | Sarcolemmal membrane-associated protein | Muscle contraction | ↑ | 1.5 | 0.01 |
| JSRP1 | Junction sarcoplasmic reticulum protein 1 | Skeletal muscle contraction | ↑ | 1.3 | 0.01 |
| LDB3 | LIM domain-binding protein 3 | Actin binding and muscle structure development | ↑ | 1.3 | 0.03 |
| CSRP3 | Cysteine and glycine-rich protein 3 | Actin-binding, skeletal muscle development | ↑ | 2.1 | 0.03 |
| HSPB7 | Heat shock protein beta-7 | Protein C-terminus binding and response to unfolded protein | ↑ | 1.8 | 0.02 |
| DNAJB4 | DNAJ homolog subfamily B member 4 | ATPase activator, chaperon, and unfolded protein binding | ↑ | 1.3 | 0.008 |
| GLRX3 | Glutaredoxin-3 | Glutathione oxidoreductase activity | ↑ | 1.3 | 0.05 |
| ANXA3 | Annexin A3 | Calcium ion binding | ↑ | 1.3 | 0.03 |
| S100A2 | Protein S100-A2 | Calcium-dependant protein binding | ↑ | 2.4 | 0.05 |
| REEP5 | Receptor expression-enhancing protein 5 | Endoplasmic reticulum organization | ↑ | 1.7 | 0.016 |
| RBM10 | RNA-binding protein 10 | miRna, RNA, and metal ion binding | ↑ | 1.6 | 0.007 |
| MAPRE2 | Microtubule-associated protein RP/EB family member 2 | Microtubule, protein kinase, and identical protein binding | ↑ | 1.5 | 0.03 |
| CNBP | CCHC-type zinc finger nucleic acid-binding protein | mRNA binding and translation regulatory activity | ↑ | 1.4 | 0.02 |
| DTNA | Dystobrevin alpha | PDZ, phosphate, and Zinc ion binding | ↑ | 1.5 | 0.007 |
| IPO5 | Importin-5 | GTPase inhibitor activity, nuclear import signal receptor activity, and RNA binding | ↑ | 1.6 | 0.03 |
Table 3.
Proteins less abundant in dark-colored beef at slightly elevated pH compared with normal-pH (fold-changes: dark color/normal-pH, P < 0.05).
| Gene name | Protein name | Molecular function | Regulation | Fold change (dark color/normal-pH) | P-value |
|---|---|---|---|---|---|
| PYGM | Glycogen phosphorylase, muscle form | Glycogen phosphorylase activity, glycogen metabolic process | ↓ | −1.4 | 0.004 |
| PFKM | ATP-dependant6-phosphofructokinase, muscle type | Canonical glycolysis, AMP, ATP binding, and 6-phosphofructokinase activity | ↓ | −1.3 | 0.02 |
| PHKB | Phosphorylase b kinase regulatory subunit beta | Glycogen metabolic process, calmodulin-binding | ↓ | −1.3 | 0.004 |
| PHKA1 | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle | Glycogen metabolic processes, calmodulin binding, and phosphorylase activity | ↓ | −1.3 | 0.003 |
| PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle | Glycogen biosynthesis activity and phosphorylase kinase activity | ↓ | −1.5 | 0.001 |
| AGL | Glycogen debranching enzyme | Glycogen biosynthesis activity | ↓ | −1.3 | 0.001 |
| EPM2A | Protein-tyrosine-phosphatase | Glycogen binding and glycogen synthase activity | ↓ | −1.6 | 0.02 |
| CALM1 | Calmodulin-1 | Adenylate cyclase activator, calcium ion binding, and channel inhibitor activity | ↓ | −1.3 | 0.006 |
| GC | Vitamin D binding protein | Vitamin D binding | ↓ | −1.4 | 0.02 |
| BANF1 | BAF nuclear assembly factor 1 | DNA binding | ↓ | −1.3 | 0.04 |
| FBN1 | Fibrillin 1 | Calcium ion, heparin binding and hormone activity | ↓ | −1.3 | 0.004 |
| SDR39U1 | Epimerase family protein SDR39UI | Oxidoreductase activity | ↓ | −1.8 | 0.006 |
| CZIB | CXXC motif-containing zinc binding protein | Zinc ion binding | ↓ | −1.7 | 0.03 |
Figure 1.
Principal component analysis (PCA) of proteins differentially abundant in dark-colored beef at slightly elevated pH vs. normal-pH beef. PCA of quantified proteins at the total protein level. Red and green dots represent protein groups from dark-colored beef at slightly elevated pH vs. normal-pH beef, respectively.
Figure 2.
Hierarchical clustering analysis of protein abundance profiles in dark-colored beef at slightly elevated pH vs. normal-pH beef. The changes in protein abundance profiles of dark-colored beef at slightly elevated pH vs. normal-pH beef were quantified as described in the “materials and method” section. Red and green colors represent overabundant and less abundant proteins in dark-colored beef at slightly elevated pH compared with normal-pH beef. Heat map color legend represents the log2 transformed ratio of dark-colored beef at slightly elevated pH vs. normal-pH beef.
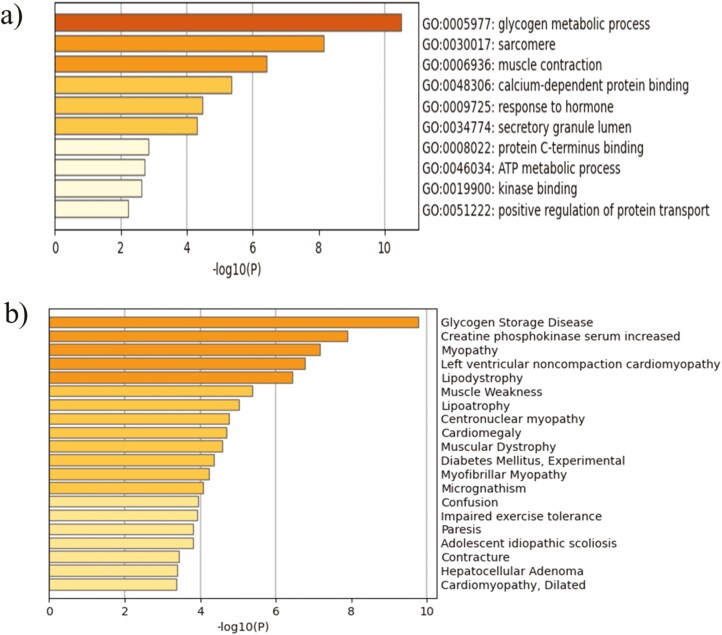
Gene ontology (GO) and pathway enrichment analyses were employed using Metascape to explore the functional annotations of the proteins within clusters. The changes in protein abundance showed enrichment in several GO processes, including glycogen metabolism, muscle contraction, sarcomere organization, calcium-dependant protein binding, and ATP metabolic processes (P < 0.001; Figure 3a). In addition, the platform of diseases-associated genes and variants analysis (DisGeNET) showed enrichment for proteins associated with the glycogen storage disease (P < 0.001; Figure 3b).
Figure 3.
Metascape functional characterization of differentially abundant proteins in dark-colored beef at slightly elevated pH vs. normal-pH beef. Proteins with significantly altered abundance in dark-colored beef at slightly elevated pH vs. normal-pH beef were analyzed for functional enrichment using Metascape. a) Statistically enriched biological processes. b) Statistically enriched terms in the DisGeNET platform of diseases-associated genes and variants. Top clusters with their representative enriched term are shown.
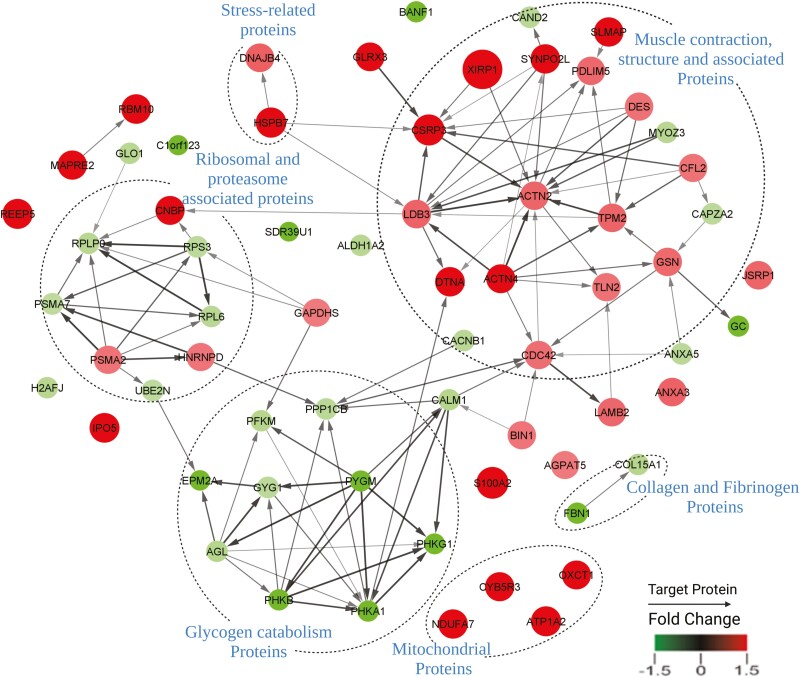
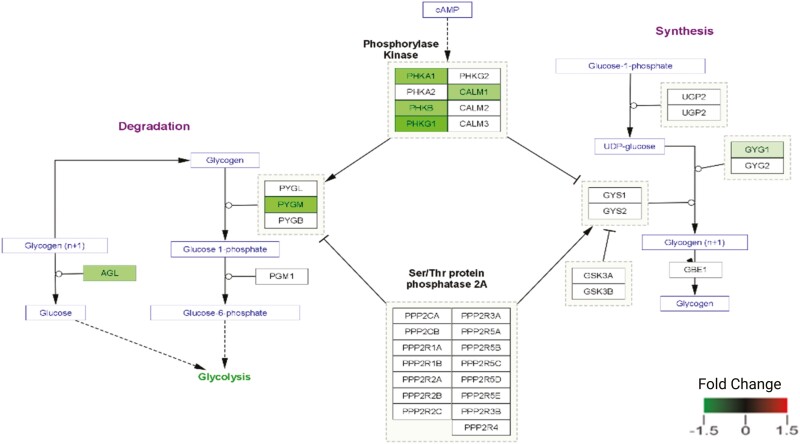
To further understand how the DAPs might participate in dark color development, we analyzed the extent to which these proteins might cooperate in specific cellular pathways. Results showed enrichment in several metabolic clusters, including seven enzymes (Figures 4 and 5) involved in the glycogen catabolic pathway were less abundant in the dark-colored beef. Annotation of the protein network (Figure 5) revealed distinctive interactive clusters of proteins associated with glycogen catabolism, muscle contraction, stress-related, and ribosomal and proteasome proteins. In the glycogen catabolic cluster, several nodes were of less abundant proteins in dark-colored beef at slightly elevated pH. While in the muscle contraction and stress-related protein–protein interaction network clusters, several nodes comprised overabundant proteins in dark-colored beef at slightly elevated pH.
Figure 4.
STRING database analysis of proteins up-and down-regulated in dark-colored beef at slightly elevated pH vs. normal-pH beef. The proteins with significant changes in protein abundance between dark-colored beef at slightly elevated pH vs. normal-pH samples were used to query potential protein–protein interactions in the string database as described in the methods. Protein interactions were confirmed with connections, while non-interacting proteins had no connections between them. The red color represents overabundant proteins and the green color represents less abundant proteins in dark-colored beef at slightly elevated pH vs. normal-pH beef. The size of the circle represents fold change in abundance, while the arrowheads indicate the target of the protein interactions.
Figure 5.
Differentially abundant proteins in the dark-colored beef at slightly elevated pH vs. normal-pH beef involved with the glycogen catabolism pathway The green color represents less abundant proteins in dark-colored beef at slightly elevated pH vs. normal-pH beef. The greater the color intensity, the greater the protein abundance.
Metabolomics analysis
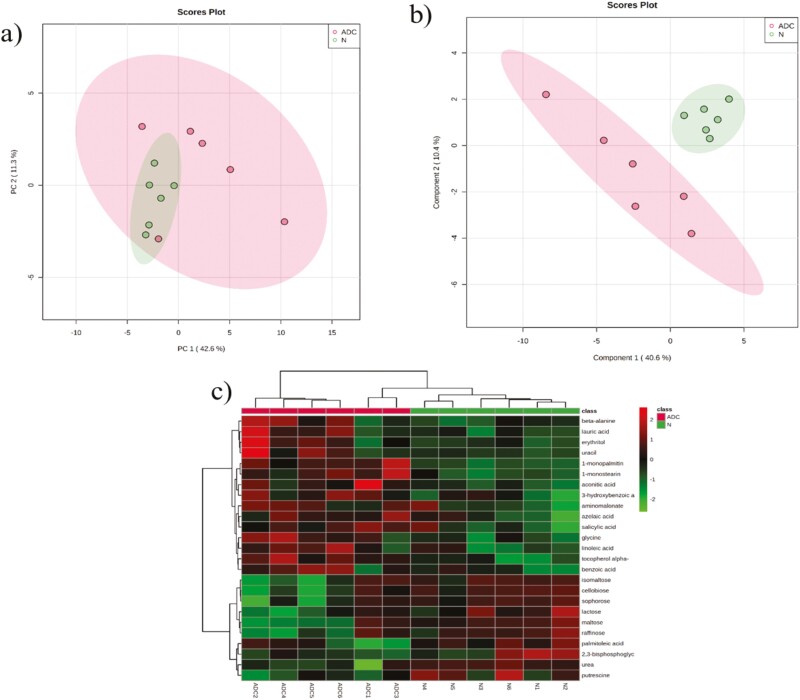
A GC-MS-based non-targeted metabolomics approach was utilized to evaluate how the changes in protein profiles impact dark-colored beef at a slightly elevated pH metabolome. A total of 174 known compounds were identified in the metabolite library. Among these, three were significantly overabundant (Table 4), while five were significantly less abundant with ≥2 fold (Table 4) in dark-colored beef. PCA (Figure 6a), partial least squares discriminant analysis (Figure 6b), and hierarchal clustering (Figure 6c) did not show distinctive clusters of metabolite groups co-segregating together in dark-colored beef at slightly elevated pH compared with normal-pH beef. Furthermore, several metabolites involved with glycolysis, tricarboxylic acid cycle, and adenine nucleotides did not show significant abundance in dark-colored beef at slightly elevated pH compared with normal-pH beef (Figures 1–3, supplementary file). However, glucose, fructose, fructose-6-phosphate, and glucose-6-phosphate were numerically lower in slightly elevated beef than in normal-pH beef.
Table 4.
Differentially abundant metabolites in normal-pH and dark-colored beef at slightly elevated pH (fold-changes: dark color/normal-pH > 2 or < 0.5, P < 0.05).
| Metabolite | Fold change (dark color/normal-pH) | Abundance (dark color/normal-pH) | P-value | FDR | Role |
|---|---|---|---|---|---|
| 1-Monopalmitin | 2.31 | over | 0.0001 | 0.02 | Fatty acid |
| 1-Monostearin | 2.27 | over | 0.002 | 0.14 | Fatty acid |
| Aconitic acid | 2.01 | over | 0.02 | 0.35 | TCA metabolite |
| Cellobiose | 0.36 | less | 0.02 | 0.35 | carbohydrate metabolism |
| Lactose | 0.30 | less | 0.05 | 0.35 | carbohydrate metabolism |
| Maltose | 0.35 | less | 0.02 | 0.35 | carbohydrate metabolism |
| Raffinose | 0.34 | less | 0.04 | 0.35 | galactose metabolism |
| Sophorose | 0.35 | less | 0.04 | 0.35 | galactose metabolism |
Abbreviation: FDR—false discovery rate P-value.
Figure 6.
a and b) PCA and partial least squares discriminant analysis of metabolite present in dark-cutting beef at slightly elevated muscle pH (ADC; pH = 5.70) and normal-pH beef (N; pH = 5.60). c) Hierarchical cluster analysis of differentially abundant metabolites in dark-colored beef at slightly elevated pH (ADC) relative to normal-pH (N). Heat map color legend represents the log2 transformed ratio of dark-colored beef at slightly elevated pH color vs. normal-pH beef.
Discussion
Meat color deviation from bright cherry-red leads to economic losses and limits consumer acceptance and marketability (Ramanathan et al., 2022). Previous research noted that a lower abundance of glycolytic enzymes and metabolites in dark-colored beef is associated with greater than normal muscle pH (>6.4; Mahmood et al., 2018; Ramanathan et al., 2020b; Cônsolo et al., 2021; Kiyimba et al., 2021; Sentandreu et al., 2021). Nevertheless, limited knowledge is currently available on protein and metabolite profiles of dark-colored beef at slightly elevated muscle pH. In the current study, we utilized an integrative approach combining proteomics and metabolomics profiling to identify DAPs and metabolites in dark-colored beef at slightly elevated muscle pH compared with normal-pH beef.
Proteomic profiling revealed seven enzymes involved in glycogen catabolic pathways were less abundant in dark-colored beef at slightly elevated pH (Table 3). The low abundant enzymes in dark-colored beef (Figures 4 and 5) are associated with glycogen degradation pathways in the muscle (Komoda and Matsunaga, 2015). More specifically, glycogen phosphorylase, muscle isoform (PYGM) catalyzes the rate-limiting step in glycogen catabolism via the phosphorolytic cleavage of glycogen to produce glucose-1-phosphate was down-regulated. Furthermore, enrichment analysis in the platform of disease-associated genes and variants (DisGeNET) also showed enrichment for proteins involved in the glycogen storage disease (Figure 3b). Thus, lower levels of these proteins can reduce dark-colored muscle’s capacity to mobilize glycogen and the ability to accumulate lactate postmortem. Therefore, our data agree with previous findings relating incidences of muscle darkening in beef with defective glycogen metabolism (Fuente-Garcia et al., 2020; Kiyimba et al., 2021).
The central dogma of molecular biology is genes regulate proteins, and proteins regulate metabolites (Crick, 1970; Morange, 2009; Shapiro, 2009). Interestingly, the changes in protein abundance profiles related to glycolytic and tricarboxylic pathways observed in the present study did not coincide with significantly abundant metabolite profiles (Figures 1–3, supplementary file). In addition, the majority of glycogenolytic proteins and metabolites less abundant in the previous dark-cutting studies (Ramanathan et al., 2020b; Kiyimba et al., 2021) at high pH (> 6.4) also did not show significant differences in the current study. Various glycolytic metabolites like glucose, glucose-6-phosphate, fructose, and fructose-6-phosphate were numerically lower in slightly elevated beef than normal-pH beef. Therefore, these data suggest enzyme activities threshold for muscle darkening. Consistent with this observation, several proteins and metabolites implicated in regulating postmortem pH decline, for example, lactate- and pyruvate-dehydrogenases, lactate, and pyruvate, respectively (Elkhalifa et al., 1984; Robergs et al., 2004; Apaoblaza et al., 2020; Gagaoua et al., 2021), were not differentially abundant in dark-colored beef at slightly elevated pH compared with normal-pH bright red steaks. Thus, the lack of significant changes in proteins and metabolites involved in muscle acidification explains a slightly elevated muscle pH observed in dark-colored beef in the present study.
When muscle glycogen content is low, there is a lack of a linear relationship between glycogen content and muscle pH decline, especially (England et al., 2016). Consistent with this disconnect between glycogen content, glycolysis, and muscle pH, we observed that glycolytic metabolite profiles were not significantly different in dark-colored beef at slightly elevated pH and normal-pH bright red steaks (Table 4 and Figure 1, supplementary file). In support, a recent study characterizing muscle properties of dark-colored beef at slightly elevated pH (approximately 0.21 pH difference) relative to normal-pH beef also revealed no differences between muscle darkening (based on L* values) and glycogen content (Ijaz et al., 2022). Furthermore, in another study, a similar glucosidic potential was observed in Canadian AB4 (dark beef at pH < 5.9) compared with AA (normal bright red color at pH 5.6; Holdstock et al., 2014). Therefore, we speculate that other pathways, besides muscle glycolysis and glycogen content, might contribute to postmortem muscle pH decline and darkening in dark-colored beef at slightly elevated pH.
Muscle contractile proteins such as alpha-actin-4, xin actin, synaptopodin 2-like protein, and sarcolemma membrane-associated protein were overabundant in slightly elevated beef than normal-pH beef (Figure 5 and Table 2). Interestingly, in previous research, muscle contractile proteins were not different in dark-cutting beef compared to normal-pH beef (Kiyimba et al., 2021). Even though the mechanistic basis for overabundance is not clear, we speculate that contractile muscle proteins might have helped to increase glycolysis as an energy-adaptive mechanism.
Although the extent of stress in dark-colored beef is unknown in this research, several stress-related proteins, such as heat shock proteins and chaperones (Table 2), were overabundant in dark-colored beef at slightly elevated pH compared with normal-pH beef. Heat shock proteins are important in protein quality control by mediating folding and refolding of misfolded proteins. Therefore, an overabundance of stress-related proteins in dark-colored beef at slightly elevated pH suggests increased oxidative stress. Previous studies also noted greater mitochondrial content in dark-cutting beef (pH > 6.4) than normal-pH beef (McKeith et al., 2016; Ramanathan et al., 2020b; Kiyimba et al., 2021). In the current research, proteins involved in mitochondrial substrate-level and oxidative phosphorylation (ATP1A2, CYB5R3, NDUFA7, OXCT1; Table 2) were overabundant in slightly elevated pH than normal-pH beef. Therefore, our data suggest that dark-colored beef at slightly elevated pH has greater mitochondrial respiratory capacity compared with normal-pH beef. Consistent with this observation, muscle oxygen consumption was greater in dark-colored beef at slightly elevated pH than normal-pH beef (Table 1). Thus, greater mitochondrial oxygen consumption can result in more deoxymyoglobin and darker meat color (Ashmore et al., 1971; English et al., 2016; McKeith et al., 2016; Ramanathan and Mancini, 2018; Ramanathan et al., 2020b, 2020c). Hence, a slightly elevated pH also can decrease the shrinkage of muscle bundles, which decreases reflectance of light and results in dark muscle.
Conclusions
The current research demonstrates the reduced abundance of proteins involved with glycogen catabolic processes and the overabundance of mitochondrial oxidative proteins in dark-colored beef at slightly elevated pH than normal-pH beef. Interestingly, the number of metabolites associated with glycogen, glycolytic, and tricarboxylic pathways was not differentially abundant compared with protein profiles. Previous dark-cutting (pH > 6.4) vs. normal-pH beef study revealed dysregulation of glycogen and glycolytic proteins and metabolite abundance. Thus, the aberrant regulation of molecular signals driving muscle darkening is highly dependent on the changes in protein expression profiles rather than metabolite profiles.
Supplementary Data
Supplementary data are available at Journal of Animal Science online.
Figure 1. Box and whisker plots of non-significantly abundant (P > 0.05) glycolytic metabolite present in dark-colored beef at slightly elevated pH (ADC) compared with normal-pH (N).
Figure 2. Box and whisker plots of non-significantly abundant (P > 0.05) tricarboxylic acid cycle (TCA) metabolites present in dark-colored beef at slightly elevated pH (ADC) and normal-pH (N).
Figure 3. Box and whisker plots of non-significantly abundant (P > 0.05) adenine nucleotide metabolites present in dark-colored beef at slightly elevated pH (ADC) and normal-pH (N).
Acknowledgment
This work was supported by the Agriculture and Food Research Institute grant 2016–09054 and the United States National Science Foundation Major Research Instrumentation (NSF MRI), EPSCoR programs (award DBI/0722494). In addition, we thank the Genomics and Proteomics Center at Oklahoma State University for providing facilities and technical support throughout the proteome analysis study. We also thank the University of California Davis, the National Institute of Health, West Coast Metabolomics Center for providing facilities and technical support throughout the metabolomics analysis.
Glossary
Abbreviations
- ATP
adenosine tri-phosphate
- ADP
adenosine di-phosphate
- GC-MS
gas chromatography-mass spectrometry
- LC-MS/MS
liquid chromatography mass spectrometry/mass spectrometry
- LFQ
label-free quantitation
Contributor Information
Frank Kiyimba, Department of Animal and Food Sciences, Oklahoma State University, Stillwater, Ok 74078, USA.
Drew Cassens, Department of Animal Science, Tarleton State University, Stephenville, TX 76402, USA.
Steven D Hartson, Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK 74078, USA.
Janet Rogers, Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK 74078, USA.
Joshua Habiger, Department of Statistics, Oklahoma State University, Stillwater, OK 74078, USA.
Gretchen G Mafi, Department of Animal and Food Sciences, Oklahoma State University, Stillwater, Ok 74078, USA.
Ranjith Ramanathan, Department of Animal and Food Sciences, Oklahoma State University, Stillwater, Ok 74078, USA.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- AMSA. 2012. AMSA meat color measurement guidelines. 2nd ed. Champaign, (IL):Am. Meat Sci. Assoc. . :1–135. [Google Scholar]
- Anderson, S., Collaborators:, S.Aldana, M.Beggs, J.Birkey, A.Conquest, R.Conway, T.Hemminger, J.Herrick, C. Hurley, et al. . 2007. Determination of Fat, moisture, and protein in meat and meat products by using the FOSS FoodScan near-infrared spectrophotometer with FOSS artificial neural network calibration model and associated database: collaborative study. J. AOAC Int. 90:1073–1083. doi: 10.1093/JAOAC/90.4.1073https://academic.oup.com/jaoac/article/90/4/1073/5657890. [DOI] [PubMed] [Google Scholar]
- Apaoblaza, A., Gerrard S. D., Matarneh S. K., Wicks J. C., Kirkpatrick L., England E. M., Scheffler T. L., Duckett S. K., Shi H., Silva S. L., . et al. 2020. Muscle from grass- and grain-fed cattle differs energetically. Meat Sci. 161:309–320. doi: 10.1016/j.meatsci.2019.107996 [DOI] [PubMed] [Google Scholar]
- Ashmore, C. R., Doerr L., Foster G., and Carroll F.. . 1971. Respiration of mitochondria isolated from dark-cutting beef. J. Anim. Sci. 33:574–577. doi: 10.2527/jas1971.333574x [DOI] [PubMed] [Google Scholar]
- Ashmore, C. R., Parker W., and Doerr L.. . 1972. Respiration of mitochondria isolated from dark-cutting beef: postmortem changes. J. Anim. Sci. 34:46–48. doi: 10.2527/jas1972.34146x [DOI] [PubMed] [Google Scholar]
- Bendall, J. R. 1979. Relations between muscle pH and important biochemical parameters during the postmortem changes in mammalian muscles. Meat Sci. 3:143–157. doi: 10.1016/0309-1740(79)90016-0 [DOI] [PubMed] [Google Scholar]
- Boykin, C. A., Eastwood L. C., Harris M. K., Hale D. S., Kerth C. R., Griffin D. B., Arnold A. N., Hasty J. D., Belk K. E., Woerner D. R., . et al. 2017. National beef quality audit–2016: In-plant survey of carcass characteristics related to quality, quantity, and value of fed steers and heifers. J. Anim. Sci. 2993.– . doi: 10.2527/jas2017.1543. [DOI] [PubMed] [Google Scholar]
- Cônsolo, N. R. B., Rosa A. F., Barbosa L. C. G. S., Maclean P. H., Higuera-Padilla A., Colnago L. A., and Titto E. A. L.. . 2021. Preliminary study on the characterization of Longissimus lumborum dark cutting meat in Angus × Nellore crossbreed cattle using NMR-based metabolomics. Meat Sci. 172:108350. doi: 10.1016/j.meatsci.2020.108350 [DOI] [PubMed] [Google Scholar]
- Crick, F. 1970. Central dogma of molecular biology. Nature.. 227:561.– . doi: 10.1038/227561a0. Available from: https://www.nature.com/articles/227561a0 [DOI] [Google Scholar]
- Egbert, W. R., and Cornforth D. P.. . 1986. Factors influencing color of dark cutting beef muscle. J. Food Sci. 51:57–59. doi: 10.1111/j.1365-2621.1986.tb10835.x [DOI] [Google Scholar]
- Elkhalifa, E. A., Anglemier A. F., Kennick W. H., and Elgasim E. A.. . 1984. Effect of prerigor pressurization on postmortem bovine muscle lactate dehydrogenase activity and glycogen degradation. J. Food Sci. 49:593–594. doi: 10.1111/j.1365-2621.1984.tb12476.x [DOI] [Google Scholar]
- England, E. M., Matarneh S. K., Oliver E. M., Apaoblaza A., Scheffler T. L., Shi H., and Gerrard D. E.. . 2016. Excess glycogen does not resolve high ultimate pH of oxidative muscle. Meat Sci. 114:95–102. doi: 10.1016/j.meatsci.2015.10.010 [DOI] [PubMed] [Google Scholar]
- English, A. R., Wills K. M., Harsh B. N., Mafi G. G., VanOverbeke D. L., and Ramanathan R.. . 2016. Effects of aging on the fundamental color chemistry of dark-cutting beef. J. Anim. Sci. 94:4040–4048. doi: 10.2527/jas.2016-0561 [DOI] [PubMed] [Google Scholar]
- Faustman, C., and Cassens R. G.. . 1990. The biochemical basis for discoloration in fresh meat: a review. J. Muscle Foods. 1:217–243. doi: 10.1111/j.1745-4573.1990.tb00366.x [DOI] [Google Scholar]
- Fiehn, O. 2016. Metabolomics by Gas Chromatography-Mass Spectrometry: the combination of targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 114:4–.1. doi: 10.1002/0471142727.MB3004S114. . https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4829120/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente-García, C., Sentandreu M. A., Aldai N., Oliván M., and Sentandreu E.. . 2021. Proteomic pipeline for biomarker hunting of defective bovine meat assisted by liquid chromatography-mass spectrometry analysis and chemometrics. J. Proteomics 238:104153. doi: 10.1016/j.jprot.2021.104153. Available from: https://linkinghub.elsevier.com/retrieve/pii/S187439192100052X. [DOI] [PubMed] [Google Scholar]
- Fuente-Garcia, C., Sentandreu E., Aldai N., Oliván M., and Sentandreu M. Á.. . 2020. Characterization of the myofibrillar proteome as a way to better understand differences in bovine meats having different ultimate pH values. Proteomics. 20:2000012. doi: 10.1002/pmic.202000012. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/pmic.202000012 [DOI] [PubMed] [Google Scholar]
- Gagaoua, M., Warner R. D., Purslow P., Ramanathan R., Mullen A. M., López-Pedrouso M., Franco D., Lorenzo J. M., Tomasevic I., Picard B., . et al. 2021. Dark-cutting beef: A brief review and an integromics meta-analysis at the proteome level to decipher the underlying pathways. Meat Sci. 181:108611. doi: 10.1016/j.meatsci.2021.108611 [DOI] [PubMed] [Google Scholar]
- Holdstock, J., Aalhus J. L., Uttaro B. A., López-Campos O., Larsen I. L., and Bruce H. L.. . 2014. The impact of ultimate pH on muscle characteristics and sensory attributes of the longissimus thoracis within the dark cutting (Canada B4) beef carcass grade. Meat Sci. 98:842–849. doi: 10.1016/j.meatsci.2014.07.029 [DOI] [PubMed] [Google Scholar]
- Ijaz, M., Zhang D., Hou C., Mahmood M., Hussain Z., Zheng X., and Li X.. . 2022. Changes in postmortem metabolites profile of atypical and typical DFD beef. Meat Sci. 193:108922. doi: 10.1016/j.meatsci.2022.108922 [DOI] [PubMed] [Google Scholar]
- Kiyimba, F., Hartson S. D., Rogers J., VanOverbeke D. L., Mafi G. G., and Ramanathan R.. . 2021. Changes in glycolytic and mitochondrial protein profiles regulates postmortem muscle acidification and oxygen consumption in dark-cutting beef. J. Proteomics 232:104016. doi: 10.1016/j.jprot.2020.104016 [DOI] [PubMed] [Google Scholar]
- Knee, B. W., Cummins L. J., Walker P. J., Kearney G. A., Warner R. D., Knee B. W., Cummins L. J., Walker P. J., Kearney G. A., and Warner R. D.. . 2007. Reducing dark-cutting in pasture-fed beef steers by high-energy supplementation. Aust. J. Exp. Agric. 47:1277–1283. doi: 10.1071/EA05362. https://www.publish.csiro.au/ea/EA05362. [DOI] [Google Scholar]
- Komoda, T., and Matsunaga T.. . 2015. Metabolic pathways in the human body. In: Biochemistry for medical professionals. Elsevier. p. 25–63. doi: 10.1016/B978-0-12-801918-4.00009-8 [DOI] [Google Scholar]
- Lei, H., Yang T., Mahmood S., Abo-Ismail M., Roy B. C., Li C., Plastow G. S., and Bruce H. L.. . 2021. A genome-wide case-control association study of dark cutting in beef cattle. Can. J. Anim. Sci. 101:158–167. doi:10.1139/cjas-2019-0039. [Google Scholar]
- Mahmood, S., Roy B. C., Larsen I. L., Aalhus J. L., Dixon W. T., and Bruce H. L.. . 2017. Understanding the quality of typical and atypical dark cutting beef from heifers and steers. Meat Sci. 133:75–85. Oxford, UK. doi: 10.1016/j.meatsci.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Mahmood, S., Turchinsky N., Paradis F., Dixon W. T., and Bruce H. L.. . 2018. Proteomics of dark cutting longissimus thoracis muscle from heifer and steer carcasses. Meat Sci. 137:47–57. doi: 10.1016/j.meatsci.2017.11.014 [DOI] [PubMed] [Google Scholar]
- Mancini, R. A., Seyfert M., and Hunt M. C.. . 2008. Effects of data expression, sample location, and oxygen partial pressure on initial nitric oxide metmyoglobin formation and metmyoglobin-reducing-activity measurement in beef muscle. Meat Sci. 79:244–251. doi: 10.1016/j.meatsci.2007.09.008 [DOI] [PubMed] [Google Scholar]
- McKeith, R. O., King D. A., Grayson A. L., Shackelford S. D., Gehring K. B., Savell J. W., and Wheeler T. L.. . 2016. Mitochondrial abundance and efficiency contribute to lean color of dark cutting beef. Meat Sci. 116:165–173. doi: 10.1016/j.meatsci.2016.01.016 [DOI] [PubMed] [Google Scholar]
- Morange, M. 2009. The Central Dogma of molecular biology. Reson. 14:236–247. doi: 10.1007/s12045-009-0024-6. Available from: https://link.springer.com/article/10.1007/s12045-009-0024-6 [DOI] [Google Scholar]
- O’Keeffe, M., and Hood D. E.. . 1982. Biochemical factors influencing metmyoglobin formation on beef from muscles of differing colour stability. Meat Sci. 7:209–228. doi: 10.1016/0309-1740(82)90087-0 [DOI] [PubMed] [Google Scholar]
- Ramanathan, R., and Mancini R. A.. . 2018. Role of mitochondria in beef color: a review. Meat Muscle Biol. 2:309–320. doi: 10.22175/mmb2018.05.0013 Available from: https://www.iastatedigitalpress.com/mmb/article/id/9068/. [DOI] [Google Scholar]
- Ramanathan, R., Mancini R. A., and Konda M. R.. . 2009. Effects of lactate on beef heart mitochondrial oxygen consumption and muscle darkening. J. Agric. Food Chem. 57:1550–1555. doi: 10.1021/jf802933p. [DOI] [PubMed] [Google Scholar]
- Ramanathan, R., Nair M. N., Hunt M. C., and Suman S. P.. . 2019. Mitochondrial functionality and beef colour: A review of recent research. S. Afr. J. Anim. Sci. 49:9–19. doi: 10.4314/sajas.v49i1.2 [DOI] [Google Scholar]
- Ramanathan, R., Hunt M. C., Mancini R. A., Nair M. N., Denzer M. L., Suman S. P., Mafi G. G., Ramanathan R., Hunt M. C., Mancini R. A., . et al. 2020a. Recent updates in meat color research: integrating traditional and high-throughput approaches. Meat Muscle Biol. 4:1–24. doi: 10.22175/MMB.9598. Available from: https://www.iastatedigitalpress.com/mmb/article/id/9598/. [DOI] [Google Scholar]
- Ramanathan, R., Kiyimba F., Gonzalez J. M., Mafi G. G., and DeSilva U.. . 2020b. Impact of up- and down-regulation of metabolites and mitochondrial properties on pH and color of longissimus muscle from normal-pH and dark-cutting beef. J. Agric. Food Chem. 68:7194–7203. doi: 10.1021/acs.jafc.0c01884 [DOI] [PubMed] [Google Scholar]
- Ramanathan, R., Suman S. P., and Faustman C.. . 2020c. Biomolecular interactions governing fresh meat color in post-mortem skeletal muscle: a review. J. Agric. Food Chem. 68:12779–12787. doi: 10.1021/acs.jafc.9b08098 [DOI] [PubMed] [Google Scholar]
- Ramanathan, R., Lambert L. H., Nair M. N., Morgan B., Feuz R., Mafi G., and Pfeiffer M.. . 2022. Economic loss, amount of beef discarded, natural resources wastage, and environmental impact due to beef discoloration. Meat Muscle Biol. 6:13218–13219. doi: 10.22175/MMB.13218. Available from: https://www.iastatedigitalpress.com/mmb/article/id/13218/. [DOI] [Google Scholar]
- Robergs, R. A., Ghiasvand F., and Parker D.. . 2004. Biochemistry of exercise-induced metabolic acidosis. Am. J. Physiol. Integr. Comp. Physiol. 287:R502–R516. doi: 10.1152/ajpregu.00114.2004. Available from: http://www.physiology.org/doi/10.1152/ajpregu.00114.2004. [DOI] [PubMed] [Google Scholar]
- Roy, D. B., Mahmood M. S., and Bruce D. H. L.. . 2022. Are muscle fiber types different between normal and dark-cutting beef? 10.1139/CJAS-2021-0085. doi: 10.1139/CJAS-2021-0085. Available from: https://cdnsciencepub.com/doi/abs/10.1139/CJAS-2021-0085 [DOI]
- Salim, A. P. A. A., Suman S. P., Canto A. C. V. C. S., Costa-Lima B. R. C., Viana F. M., Monteiro M. L. G., Silva, T. J. P. and Conte-Junior C. A.. . 2019. Muscle-specific color stability in fresh beef from grain-finished Bos indicus cattle. Asian-Australasian J. Anim. Sci. 32:1036–1043. doi: 10.5713/ajas.18.0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammel, L. M., Hunt M. C., Kropf D. H., Hachmeister K. A., Kastner C. L., and Johnson D. E.. . 2002. Influence of chemical characteristics of beef inside and outside semimembranosus on color traits. J. Food Sci. 67:1323–1330. doi: 10.1111/j.1365-2621.2002.tb10282.x [DOI] [Google Scholar]
- Sentandreu, E., Fuente-García C., Pardo O., Oliván M., León N., Aldai N., Yusà V., and Sentandreu M. A.. . 2021. Protein biomarkers of bovine defective meats at a glance: gel-free hybrid quadrupole-orbitrap analysis for rapid screening. J. Agric. Food Chem. 69:7478–7487. doi: 10.1021/acs.jafc.1c02016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A. 2009. Revisiting the central dogma in the 21st century. Ann. N. Y. Acad. Sci. 1178:6–28. doi: 10.1111/j.1749-6632.2009.04990.x [DOI] [PubMed] [Google Scholar]
- Skogerson, K., Wohlgemuth G., Barupal D. K., and Fiehn O.. . 2011. The volatile compound BinBase mass spectral database. BMC Bioinf. 12:1–15. doi: 10.1186/1471-2105-12-321. https://link.springer.com/articles/10.1186/1471-2105-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel, C., Mcgilchrist P., Rivas P. G., Warner R., and Tarr G.. . 2018. Effect of weather conditions ante-mortem on the incidence of dark cutting in feedlot finished cattle-A retrospective analysis. New South Wales: Final report, Meat and Livestock Australia Limited, Locked Bag. pp 1–58.
- Suman, S. P., and Joseph P.. . 2013. Myoglobin Chemistry and Meat Color. Annu. Rev. Food Sci. Technol. 4:79–99. doi: 10.1146/annurev-food-030212-182623 [DOI] [PubMed] [Google Scholar]
- Szklarczyk, D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N. T., Roth A., Bork P., . et al. 2017. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45:D362–D368. doi: 10.1093/nar/gkw937. Available from: https://academic.oup.com/nar/article/45/D1/D362/2290901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Węglarz, A. 2018. Meat quality defined based on pH and colour depending on cattle category and slaughter season. Czech J. Anim. Sci. 55:548–556. doi: 10.17221/2520-cjas. [DOI] [Google Scholar]
- Wis̈niewski, J. R., and Gaugaz F. Z.. . 2015. Fast and sensitive total protein and peptide assays for proteomic analysis. Anal. Chem. 87:4110–4116. doi: 10.1021/ac504689z [DOI] [PubMed] [Google Scholar]
- Wu, S., Luo X., Yang X., Hopkins D. L., Mao Y., and Zhang Y.. . 2020. Understanding the development of color and color stability of dark cutting beef based on mitochondrial proteomics. Meat Sci. 163:108046. doi: 10.1016/j.meatsci.2020.108046 [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., Benner C., and Chanda S. K.. . 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10. doi: 10.1038/S41467-019-09234-6. Available from: https://pubmed.ncbi.nlm.nih.gov/30944313/. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.