Abstract
Disordered cellular development, abnormal neuroanatomical formations, and dysfunction of neuronal circuitry are among the pathological manifestations of cortical regions in the brain that are often implicated in complex neurodevelopmental disorders. With the advancement of stem cell methodologies such as cerebral organoid generation, it is possible to study these processes in vitro using 3D cellular platforms that mirror key developmental stages occurring throughout embryonic neurogenesis. Patterning-based stem cell models of directed neuronal development offer one approach to accomplish this, but these protocols often require protracted periods of cell culture to generate diverse cell types, and current methods are plagued by a lack of specificity, reproducibility, and temporal control over cell derivation. Although ectopic expression of transcription factors offers another avenue to rapidly generate neurons, this process of direct lineage conversion bypasses critical junctures of neurodevelopment during which disease-relevant manifestations may occur. Here, we present a directed differentiation approach for generating hPSC-derived cortical organoids with accelerated lineage specification to generate functionally mature cortical neurons in a shorter timeline than previously established protocols. This novel protocol provides precise guidance for the specification of neuronal cell type identity as well as temporal control over the pace at which cortical lineage trajectories are established. Furthermore, we present assays that can be used as tools to interrogate stage-specific developmental signaling mechanisms. By recapitulating major components of embryonic neurogenesis, this protocol allows for improved in vitro modeling of cortical development while providing a platform that can be utilized to uncover disease-specific mechanisms of disordered development at various stages across the differentiation timeline.
Basic Protocol 1:
3D human pluripotent stem cell (hPSC) neural induction
Support Protocol 1:
2D Neural rosette formation assay
Support Protocol 2:
2D Neural rosette selection and 3D Neurosphere generation
Support Protocol 3:
Enzymatic dissociation, 2D neural stem cell (NSC) expansion, and cryopreservation
Basic Protocol 2:
3D Neural spheroid expansion
Basic Protocol 3:
Accelerated cortical organoid patterning and terminal differentiation
Keywords: human pluripotent stem cell, organoids, cortical neurons, neural stem cells, neurosphere
INTRODUCTION
Pluripotent stem cells (PSCs) have greatly improved our understanding of human brain development and disease by providing an alternative to traditional model organism research through which species-specific aspects of human neurodevelopment can be studied. Beginning with embryonic stem cells, developmental neuroscience research groups were first able to generate 2-dimensional (2D) cultures of neuronal precursors in vitro (Elkabetz et al., 2008; Koch et al., 2009; Li et al., 2011; Shi et al., 2012; Zhang et al., 2001) with features comparable to animal-derived progenitors isolated from primary tissue explants (Temple et al., 1995; Svendsen et al., 1998; Carpenter et al., 1999). Although these models were initially primitive, the discovery of novel small molecules has enabled the development of more efficient directed patterning approaches, facilitating the differentiation of PSCs into diverse neuronal subtypes with high efficiency, specificity, and reproducibility (Tchieu et al., 2017) and thus permitting early exploration of therapeutic interventions for neurological diseases in vitro (Kriks et al., 2011). Additionally, recent advancements in stem cell technologies have also led to the development of complex 3-dimensional (3D) brain-like structures referred to as cerebral organoids. Through the formation of self-organized 3D tissues with distinct cytoarchitectures, electrophysiological networks and developmental kinetics, cerebral organoids recapitulate many of the fundamental aspects of human embryonic neurogenesis and offer a robust model system for investigating human neurodevelopment and disease (Kadoshima et al., 2013; Lancaster et al., 2013; Pasca et al., 2015; Qian et al., 2016; Quadrato et al., 2107; Velasco et al., 2019). As such, cerebral organoids have been widely adopted in the field and remain at the forefront of current neuroscience research with great promise for therapeutic applications (Akkerman and Defize, 2017; Wang et al., 2017).
Despite their utility, current methods for cerebral organoid generation require protracted periods of culture to generate terminally differentiated neurons and face challenges of variability and reproducibility (Lancaster et al., 2013; Quadrato et al., 2107). To address these concerns, several groups have reported approaches to enhance and accelerate the process of neuronal differentiation from pluripotent stem cells that include the modulation of key developmental signaling pathways downstream of primary neural induction, such as the activation of Wnt/β-Catenin signaling to promote the proliferation of dorsal telencephalic progenitors (Delepine et al., 2021; Lancaster et al., 2017; Nicoleau et al., 2013; Sakaguchi et al., 2015; Watanabe et al., 2005) or the use of FGF/MAPK/ERK and NOTCH signaling pathway inhibition to influence early cell cycle exit and terminal specification of neural progenitors (Chambers et al., 2012; Gantner et al., 2021; Qi et al., 2017). However, attempts to accelerate the neuronal differentiation process through extensive small molecule treatments and pathway perturbations have resulted in cumulative stressful effects that generate widespread cell death (Qi et al., 2017).
Hence, to overcome these challenges, this article presents an optimized workflow for the accelerated differentiation of functional 3D cortical organoids from human PSCs with built-in assays at multiple stages throughout the differentiation timeline, thereby enabling the expedited investigation of human cortical development with stage-specific interrogation of cellular mechanisms of corticogenesis. Basic protocol 1 provides a method for 3D spheroid generation and early neural induction from hPSCs, while support protocols 1 and 2 detail a 2D neural rosette formation assay and a 3D neurosphere generation assay, which both facilitate characterization of early features of neurodevelopment. Next, basic protocol 2 details a method for 3D neural progenitor expansion, while support protocol 3 details the process of 2D neural stem cell (NSC) isolation, maintenance, expansion, and cryopreservation to interrogate intermediate stages of corticogenesis. Finally, basic protocol 3 describes an optimized approach to 3D accelerated cortical organoid lineage patterning and terminal differentiation into functional cortical neurons.
CAUTION: The following procedures are performed in a Class II biological hazard flow hood or a laminar-flow hood under sterile conditions. Proper aseptic technique should always be maintained.
STRATEGIC PLANNING
This protocol requires a fundamental knowledge of mammalian tissue and cell culture practices, and while embryonic stem cell experience is not a necessity, it is advantageous to consult the literature for technical guidelines on stem cell methodology from reputable sources. Human pluripotent stem cells, either embryonic derived or induced by reprogramming factors, can be purchased by commercial biorepositories such as WiCell (www.wicell.org) or The Coriell Institute for Medical Research (www.coriell.org). For a detailed protocol on pluripotent stem cell maintenance, we recommend a prior Current Protocols resource (Rivera et al., 2020). It has recently been suggested that the culture conditions and growth medias for pluripotent stem cell maintenance have a profound effect on the differentiation success into cortical neurons and organoids (Cornacchia et al., 2019; Watanabe et al., 2022); however, we have observed successful cortical organoid and neuronal differentiation with all the culture media proposed herein. For a more detailed method on 3D suspension aggregate generation from human pluripotent stem cells please refer to our previous work (Whye et al, 2022). This protocol introduces a method for the in vitro generation of pure populations of cortical neurons; however, long-term functional maturation may rely on non-cell autonomous influences by co-culture with astrocytes (Odawara et al., 2014; Odawara et al., 2016; Odawara et al., 2018) or by using a physiologically enhanced medium such as BrainPhys (Autar et al, 2022; Bardy et al., 2015). Technical manuals for astrocyte co-culture systems and BrainPhys media kits can be found from industry-leading companies such as StemCell Technologies (www.stemcell.com). In addition, several technical methods for cell identity characterization and functional assessment exist, which this protocol also does not specifically address. We recommend adhering to the user’s own preferences for methods involving RNA (RT-qPCR; sn/scRNA-Seq) and protein (Immunocytochemistry; Western Blot) analyses. Suggestions from existing Current Protocols include immunofluorescence staining on dissociated neurons (Song et al., 2021) or whole organoids (Lewis-Israeli et al., 2022), and electrophysiological characterization using patch clamp (Song et al., 2021). Additional functional characterization can be achieved using calcium imaging techniques (Galiakberova et al, 2022; Jiang et al., 2018; Sharma et al., 2020; Volfely et al., 2018) or multi-electrode array (MEA) platforms from companies such as Axion Biosystems (www.axionbiosystems.com) or Maxwell Biosystems (www.mxwbio.com).
BASIC PROTOCOL 1 3D HPSC NEURAL INDUCTION
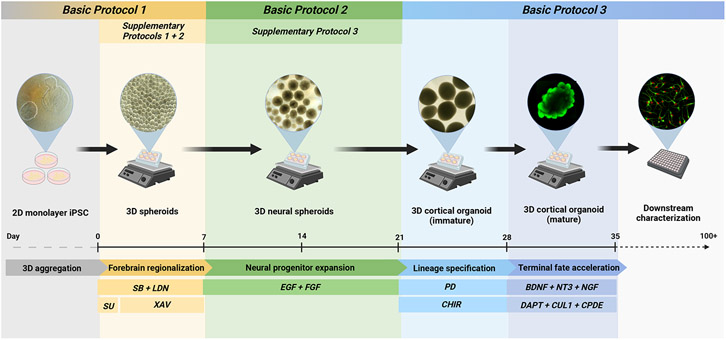
An overview of the 35-day accelerated 3D cortical organoid differentiation process is depicted by the schematic in Figure 1. The method for generating 3D aggregate suspensions from pluripotent stem cells is as previously described (Whye et al., 2022); however, the media recipes differ. The initial step for cortical organoid generation requires aggregation of hPSCs in a FGF2-rich stem cell medium for a period of 24 hours to generate robust, homogenous populations of spheroids. The spheroids are then treated with a neural induction media that employs a combination of dual Smad inhibition by SB431542 (which blocks Smads 2/3) and LDN193189 (which blocks Smads 1/5/8) along with Wnt inhibition by XAV939. Additionally, a brief pulse of the FGF receptor inhibitor SU-5402 is also utilized during the first 24 hours of the neural induction period to block downstream signaling cascades under the influence of FGF signaling that may cause cells to drift away from an anterior neural identity. Together, these patterning cues enable the acquisition of an anterior dorsal neuroectoderm identity marked by robust expression of transcription factors SOX2, PAX6, and FOXG1 (Figure 2., Table 1). After the week-long neural induction period concludes, neural spheroids can either be further manipulated in culture to generate 2D neural rosettes (Support Protocol 1), 3D neurospheres (Support Protocol 2), and 2D neural stem cells (Support Protocol 3) for downstream assaying, or they can be expanded by the administration of morphogens FGF-2 and EGF for continued differentiation (Basic Protocol 2).
Figure 1.
Schematic overview of the 35-day differentiation pipeline described in these protocols. 2D monolayer cultures of hPSCs are enzymatically dissociated and re-aggregated in suspension, and dynamic motion is applied to generate homogenous populations of spheroids. Through combinatorial signaling activity, spheroids are initially patterned towards a neural lineage that then undergo an expansion phase prior to cortical neuron cell fate commitment by accelerated and terminal differentiation. At multiple stages throughout the protocol, dissociation into 2D culture (or 3D re-aggregation) can be performed to achieve cell characterization and other complementary assays. Brightfield and fluorescent images represent the cell-stage transitions throughout this protocol.
Figure 2.
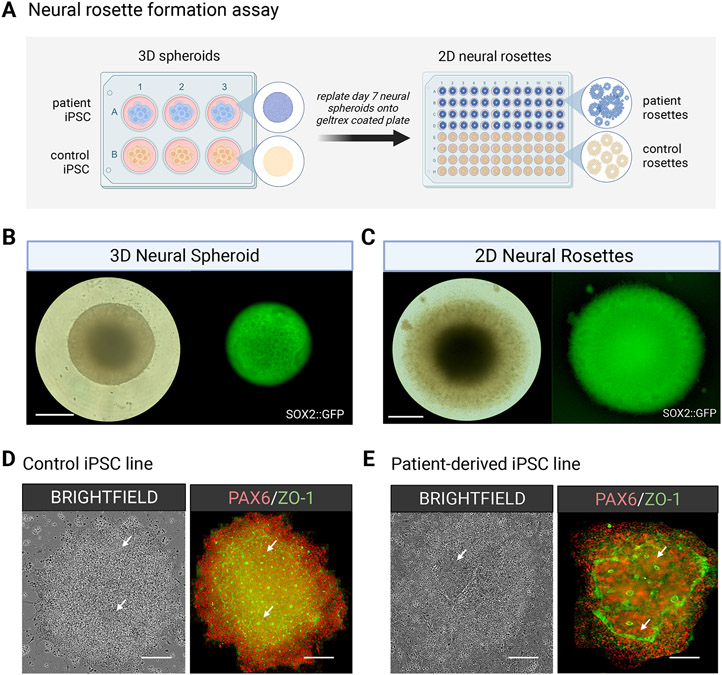
After a 7-day neural induction, 3D neural spheroids can be plated onto an ECM substrate to generate 2D neural rosette structures that can be assayed in a high throughput fashion to evaluate neurogenic potential or to identify early differences in neuronal development. (A) Schematic workflow of a neural rosette formation assay on control and disease lines. (B) Representation of a single neural spheroid in 3D suspension and (C) multiple neural rosettes within a single spheroid after attachment in 2D adherent conditions on an ECM matrix substrate. Scale bar = 330μm. (D) Brightfield and fluorescent images of immunostained neural rosettes in a control line versus (E) a patient line possessing a neurodevelopment related disease mutation. Scale bar = 200μm.
Table 1.
NSC & NPC antibodies for ICC/IF
| Component | Dilution |
|---|---|
| SOX1 (R&D systems; cat. no., AF3369) | 1:500 |
| SOX2 (Cell signaling; cat. no., 3579S) | 1:500 |
| PAX6 (Biolegend; cat. no., 901301) | 1:500 |
| NESTIN (EMD Millipore; cat. no., MAB5326) | 1:100 |
| FOXG1 (AbCam; cat. no. 18259) | 1:500 |
Materials
SOX2-GFP iPSC line (Coriell Institute, cat. no. AICS-0074-026)
Vitronectin (VTN-N) Recombinant Human Protein, Truncated (Thermo Fisher Scientific, cat. no. A14700; prepare and store 200μL aliquots at −80°C)
Essential 8™ medium kit (Thermo Fisher Scientific, cat. no. A1517001)
Essential 8™ flex medium kit (Thermo Fisher Scientific, cat. no. A28585-01)
mTeSR1 (StemCell Technologies, cat. no. 85850)
mTeSR Plus (StemCell Technologies, cat. no. 100-0276)
Stemflex™ medium kit (Thermo Fisher Scientific, cat. no. A3349401)
ROCK Inhibitor (Y-27632), 10mM solution in DMSO (Cayman Chemical 10005583)
Versene (Thermo Fisher Scientific, cat. no. 15040066)
1X Dulbecco’s Phosphate Buffered Saline (DPBS) without Ca2+, Mg2+ (Corning, cat. no. 20-031-CV)
Accutase™ Cell Detachment Solution (Innovative Cell Technologies, Inc., cat. no. AT104-500)
Corning Cell Lifter (Corning, cat. no. 3008)
Corning Small Cell Scraper (Corning, cat. no. 3010)
Bright-Line Hemacytometer Set, Hausser Scientific, Chamber and Cover Glass (2) Only, Chamber Depth:0.1 mm (Hausser Scientific, cat. no. 1492)
Trypan Blue Stain 0.4% (Thermo Fisher Scientific, cat. no. 15250061)
Neural Induction basal medium (see recipe in Reagents and Solutions)
Stage 0 Aggregation medium (see recipe in Reagents and Solutions)
Stage 1a Induction medium (see recipe in Reagents and Solutions)
Stage 1b Induction medium (see recipe in Reagents and Solutions)
Costar® 6-well Clear Flat Bottom Ultra-Low Attachment Multiple Well Plates, Individually Wrapped, Sterile (Corning Life Sciences, cat. no. 3471)
Serological Pipets
Biosafety cabinet
Inverted light microscope
37°C/5% CO2 incubator
Corning® LSE™ Orbital Shaker (Corning Life Sciences, cat. no. 6780-NP)
Flat Platform with Rubber Mat (Corning Life Sciences, cat. no. 480108)
Protocol steps with step annotations
Single cell dissociation of hPSCs & 3D spheroid aggregation in suspension (Day −1)
Enzymatic digestion of hPSCs
Aspirate growth medium (E8, E8 Flex, StemFlex, mTeSR or mTeSR plus) from the culture dish with a glass Pasteur pipet once hPSCs have reached a confluence of ~80-85%.
Rinse the dish with 1X D-PBS w/o Ca2+Mg2+ (175μL/cm2) briefly and remove.
-
Add Accutase solution (100μL/cm2) to the dish and incubate at room temperature for 5-10 min.
Be sure to maintain the proper temperature of Accutase solution (15-25°C). Do not place inside of a 37°C/5% CO2 incubator as prolonged exposure to this temperature inactivates the enzyme. Using a microscope, ensure that all cells have detached from the dish. Be careful not to leave Accutase solution on for too long as this will cause cell lysis.
Tap the side of the dish vigorously during the incubation process to dislodge cells from the surface. When cells have completely detached from the dish, dilute Accutase with a double volume of 1X D-PBS solution (200μL/cm2).
Transfer cell solution from dish into a separate conical centrifuge tube.
Centrifuge cell solution at 400 × g for 5 min.
Prepare Neural Induction basal medium and Stage 0 Aggregation medium (see recipe in Reagents and Solutions).
-
Aspirate off supernatant in conical tube and use a p1000 to resuspend cell pellet in 1mL Stage 0 Aggregation medium.
It is critical to ensure that complete dissociation of cells has occurred to achieve a single-cell solution that is necessary for optimal cell aggregation. Gently triturate the cell pellet 5-10 times so that the cell pellet is fully resuspended in the solution.
Using a serological pipet, add 9mL Stage 0 Aggregation medium to raise the volume of the cell suspension in each conical tube to 10ml.
Once the pellet is fully re-suspended in 10ml proceed to cell counting.
Take a 10μL sample of the cell suspension and make a 1:20 dilution by mixing with 190μL of 1X D-PBS in a 1.7mL microcentrifuge tube.
Perform a 1:1 dilution with Trypan Blue and conduct a manual cell count using a hemocytometer.
Seeding of hPSCs in suspension for 3D spheroid aggregation
-
13.
Adjust the cell concentration to 1 x 106 cells/ml.
While it is desired to achieve an approximate cell concentration of 1 x 106 cells/ml for optimal aggregation, acceptable ranges exist between 0.75 x 106 cells/ml to 1.25 106 cells/ml
-
14.
Pipet 5 ml of adjusted cell solution (1 x 106 cells/ml) into each well of ultra-low attachment 6-well plate to seed 5 million cells per well.
-
15.
Place ultra-low attachment multiwell dishes on an orbital shaker platform inside of 37°C/5% CO2 incubator and set the speed to 100 RPM.
-
16.
Check the cells 24hr later under a microscope to ensure that proper aggregation has occurred (refer to Figure 1 schematic for an example).
Generated spheroid populations will become visible to the naked eye ~24 hours post seeding; however, microscopic analysis should be conducted to confirm proper aggregation. A population size covering approximately ¾ of the surface area of the well (~ few hundred) should be present with an average aggregate size ≈ 200μm. If the aggregate size is well above or below 200μm then the viability of the aggregates may diminish throughout the differentiation process, and efficient organoid generation will not occur.
-
17.
After confirming that proper aggregation has occurred, place the dish inside of a biosafety cabinet and proceed to stage 1.
Stage 1: Neuroectodermal induction (Days 0-7)
Day 0: Replace Stage 0 Aggregation medium with Stage 1a Induction medium
Prepare fresh Stage 1a Induction medium (see recipe in Reagents and Solutions).
-
Swirl suspension cells in a clockwise motion to force spheroids to settle in the center of each well.
Spheroids will be large enough that they are visible; therefore, the use of a dissecting microscope (or equivalent) for handling is NOT required.
-
Using a glass Pasteur pipet, carefully aspirate along the outer edges of the well.
Be careful not to suction the centralized population of floating spheroids. It is okay to leave some residual media behind in the well.
Replace with 5mL of Stage 1a Induction medium per well.
-
Return the plate back to the orbital shaker inside of the 37 C incubator and increase the speed of the orbital shaker platform to 120RPM.
Failure to raise the speed of the orbital shaker may result in organoid fusion over the time course of differentiation
Day 2 Replace Stage 1a Induction medium with Stage 1b Induction medium
-
6.
Prepare fresh Stage 1b Induction medium (see recipe in Reagents and Solutions).
-
7.
Repeat steps 2-3.
-
8.
Replace with 5mL of Stage 1b Induction medium per well.
Days 4-7: Replenish Stage 1b Induction medium
-
9.
Prepare fresh Stage 1b Induction medium (see recipe in Reagents and Solutions).
-
10.
Repeat steps 2-3.
-
11.
Replace with 5mL of Stage 1b Induction medium per well.
Day 7: Proceed to Basic Protocol 2 NPC Expansion for continued cortical organoid differentiation or Support Protocol 1 for Neural rosette formation assay
Understanding the results
Combined SMAD inhibition targeting the TGFβ and BMP pathways has long been established as an efficient method to direct pluripotent stem cells towards a neural cell fate (Chambers et al. 2009; Chambers et al., 2012; Mica et al., 2013). Coupled with Wnt signaling antagonism through either the administration of the recombinant protein DKK-1 or small molecule inhibitors (XAV939, IWP-2, IWR-1), this neuronal cell specification can be further shifted towards an anterior telencephalic regional identity (Eiraku et al., 2008; Kadoshima et al., 2013; Nicoleau et al., 2013; Pasca et al., 2015; Rosebrock et al., 2022; Watanabe et al., 2005) that later develops into the pyramidal layers of the cortex. Human pluripotent stem cell growth medias (such as mTeSR, mTeSR plus, Essential 8, Essential 8 Flex, and StemFlex) are widely formulated to contain the morphogen basic fibroblast growth factor (bFGF or FGF2), which helps to maintain pluripotency and acts as a mitogen for cell proliferation (Dakhore et al., 2018). The media volumes and cell densities listed in Basic Protocol 1 have been optimized for the robust formation of 3D aggregates in suspension; however, inherent line-to-line variability can exist depending on the growth properties of the cell line and its corresponding genetic background. Therefore, a tailored approach to Stage 0 aggregation may require different PSC growth media depending on the growth kinetics of different cell lines. We have also found that for some challenging disease hPSC lines an additional troubleshooting step to stage 0 aggregation recipe may call for the early application of the GSK3b inhibitor, CHIR99021 (3uM final), which acts to stimulate proliferation by canonical Wnt pathway activation. However, it has previously been established that prolonged exposure to FGF2 and Wnt signaling can cause hPSCs to drift towards a caudal neuromesodermal progenitor identity (Whye et al., 2022); therefore, if this troubleshooting approach is utilized then it is suggested to follow with sustained FGF receptor inhibition (by SU-5402) for the full duration of the 7-day neural induction phase in stage 1. 3D neural spheroids can be characterized at day 7 by immunostaining for the makers SOX1, SOX2, PAX6, and NESTIN (Table 1). After the neural induction stage, 3D neural spheroids can be processed as 2D neural rosettes (Support Protocol 1), 3D neurospheres (Support Protocol 2), and 2D NSCs (Support Protocol 3) or terminally differentiated into cortical organoids (Basic Protocol 2 & 3).
SUPPORT PROTOCOL 1 NEURAL ROSETTE FORMATION ASSAY
Neural rosettes are in vitro neural tube-like features that form within 3D neural spheroids during the neural induction process (Elkabetz et al., 2008; Koch et al., 2009; Li et al., 2011; Shi et al., 2012; Zhang et al., 2001) described in stage 1, and rosettes can be isolated and purified in culture by plating whole differentiating spheroids onto an ECM-rich matrix such as Geltrex or Matrigel. This substrate allows for increased cell motility and re-organization into distinctive rosette structures, which can then be isolated by mechanical or enzymatic selection. Neural rosettes can be characterized by immunostaining to confirm the presence of neural stem cell markers (Table 1). Furthermore, neural rosette formation assays can be used for various downstream applications including the screening of multiple pluripotent stem cell lines to assess neurogenic potential and for early comparative phenotypic analyses between normal development and disease states (Figure 2). Additionally, neural rosettes can also be used as a selection tool to generate purified neurospheres, as described in support protocol 2 (Figure 3).
Figure 3.
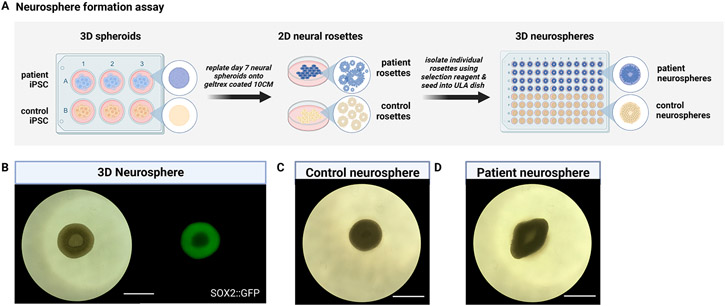
Neurosphere formation assays provide a complementary tool to neural rosette formation assays. (A) Schematic workflow of a neurosphere formation assay on control and disease lines. (B) Representation of a single neurosphere displaying patterned expression of NSC marker SOX2. Neurosphere formation assay applied to (C) a control hiPSC line versus (D) a patient line, which reveals early morphological differences. scale bar = 930μm.
Materials
3D Neural spheroids in 6-well ultra-low attachment suspension plate (generated in Basic Protocol 1)
Neural Induction basal medium (see recipes in Reagents and Solutions)
Stage 2 NSC expansion medium (see recipes in Reagents and Solutions)
Dulbecco’s Modified Eagle’s Medium, DMEM (Corning Life Sciences, cat. no. 10-013-CV)
Geltrex LDEV-free, hESC qualified, 1ml, reduced growth factor basement membrane matrix (Thermo fisher Scientific, cat. no. A1413301)
Pipet-Lite XLS+ manual 12-channel pipette, 20-300μl (Rainin, cat. no. 17013811)
TerraRack tips, 300μl, LTS (Rainin, cat. no. 17014965)
Wide orifice tips, 200μl (Genessee Scientific, cat. no. 22-425)
Falcon 96-well clear flat bottom TC microplate (Corning, cat. no. 353072)
Serological pipets
Inverted microscope
Protocol steps with step annotations
Preparation of 96-well Geltrex coated plate
-
Thaw a vial of 1ml Geltrex overnight on ice at 4° C according to manufacturer’s recommendations.
It is important to thaw the Geltrex slowly at cold temperatures to avoid polymerization of the basement membrane matrix.
-
Prepare a 1:100 dilution of thawed Geltrex on ice with cold sterile DMEM basal medium under a laminar flow hood.
It is critical to maintain cold temperatures throughout this process to prevent premature polymerization.
-
Add enough Geltrex solution (315μl/cm2) to cover the entire surface area of each well of a 96-well flat bottom plate and incubate for at least 1 hr. at 37°C/5% CO2.
Geltrex coated plates can be stored at 37°C for several days or at 4°C for a couple weeks. Do not aspirate Geltrex coated plates until ready to plate 3D neural spheroids.
Transfer 3D Neural spheroids into Geltrex coated 96-well plate
-
4.
Aspirate off Geltrex solution from the 96-well plate using a glass Pasteur pipette.
-
5.
Using a 12 insert multi-channel pipet, dispense 200μl of neural induction basal medium to each well.
-
6.
Retrieve 6-well suspension plates containing 3D neural spheroids (Basic Protocol 1) from the orbital shaker and bring under the laminar flow hood.
-
7.
Using a p200 wide orifice pipet tip, collect individual 3D neural spheroids from wells of a 6-well suspension plate and transfer directly into single wells of a 96-well Geltrex coated plate.
-
8.
Return plate to 37° C/5% CO2 incubator and allow spheroids to attach for a period of 24 hours.
Once attached to the Geltrex substrate, plated spheroids will flatten out and some cells will begin to migrate from their outer edges.
-
9.
At 48 hours, replenish neural induction medium to ensure that adequate neural rosette formation occurs.
Flower-like rosette structures will become visible throughout the cores of the attached neural spheroids between 24-96 hours. If rosettes do not appear after 96 hours, then typical neural induction has not occurred.
SUPPORT PROTOCOL 2 NEUROSPHERE GENERATION
The term “neurosphere” has traditionally been used to describe 3D spheroids derived from dissociated primary neural tissue explants which are maintained in culture through the administration of morphogens FGF-2 and EGF (Temple et al., 1995; Svendsen et al., 1998; Carpenter et al., 1999). With the development of stem cell differentiation methods, purified 3D neurospheres can now be generated directly from hPSCs (Begum et al., 2015; Hofrichter et al., 2017; Reubinoff et al., 2001). Since hPSC-derived 3D neural spheroids contain multiple heterogeneous rosette structures, some research groups have desired to generate purified 3D neurospheres that contain a singular neural rosette (Knight et al., 2018). Through enzymatic or mechanical selection, individual neural rosettes can be isolated from 2D cultures and reaggregated in 3D to generate purified neurospheres amenable to downstream assays or terminal differentiation (Wang et al., 2022). This support protocol utilizes enzymatic selection to generate neurospheres from the 2D rosettes generated in support protocol 1. Briefly, the 3D neural spheroids generated from basic protocol 1 are transferred into larger format culture conditions (e.g., 10 or 15 cm) for adherent 2D neural rosette formation. After enzymatic treatment with STEMDIFF selection reagent, individual rosettes are isolated and transferred into 96-well ultra-low attachment plates for reaggregation into 3D purified neurospheres (Figure 3A). In this high-throughput format, the neurospheres generated in this protocol can undergo comparative phenotypic analyses across normal and disease states as well as screening assays involving small molecule and signaling morphogen treatments (Figure 3B-D).
Materials
2D Neural Rosettes in 10cm Geltrex coated dishes (Support Protocol 1)
Neural Induction basal medium (see recipes in Reagents and Solutions)
Stage 2 NSC expansion medium (see recipes in Reagents and Solutions)
Dulbecco’s Modified Eagle’s Medium, DMEM (Corning Life Sciences, cat. no. 10-013-CV)
1X Dulbecco’s Phosphate Buffered Saline (D-PBS) without Ca2+, Mg2+ (Corning, cat. no. 20-031-CV)
STEMdiff Neural rosette selection reagent (StemCell Technologies, cat. no. 05832)
Nunc EasYDish 10 cm tissue culture dish (Thermo fisher Scientific, cat. no. 150466)
Olympus 1000μl wide orifice pipet tip (Genesee Scientific, cat. no. 22-428)
Costar 6-well flat bottom ultra-low attachment plates (Corning, cat. no. 3471)
Costar assay plate, 96 well, round bottom, non-treated (Corning, cat. no. 3795)
Costar universal lid corner notch (Corning, cat. no. 3099)
Olympus 200μl wide orifice pipet tip (Genesee Scientific, cat. no. 22-425)
Serological pipets
Inverted microscope
Protocol steps with step annotations
Incubation with neural rosette selection reagent
Aspirate off neural induction basal medium from a 10cm dish containing neural rosettes.
Rinse plate with enough 1x D-PBS (175μl/cm2) to cover surface area of the entire 10 cm dish.
Aspirate off 1x D-PBS and add pre-warmed neural rosette selection reagent (175μl/cm2) to the dish.
-
Incubate at 37° C for 30-60 min.
At this stage rosette morphology will become partially dissociated as cells become loosely detached from the dish. Incubation times may vary so be attentive. Over-incubation will cause peripheral neural crest cells to also detach from the dish.
-
Slowly and gently remove the rosette selection reagent from the 10cm dish with a 10ml serological pipet.
Be careful not to dislodge or aspirate loosely bound neural rosettes.
-
Using a p1000 wide orifice tip, dispense 5 ml of Stage 2 NSC expansion medium directly onto attached colonies to dislodge neural rosettes.
Gaps will appear throughout the spheroid colonies where rosettes have detached, which will be visible under a microscope. Repeat attempts to detach all rosettes may be necessary; however, too many attempts to manually dislodge rosettes should be avoided as this may remove undesired non-neuronal cells.
3D Neurosphere formation and proliferation
-
7.
Once most rosettes have been detached, transfer 5ml cell solution into a single well of a 6-well suspension plate.
-
8.
Using a p200 wide orifice tip, isolate individual floating rosettes in 50μl volume and transfer into single wells of a 96-well ULA plate.
-
9.
Raise total volume to 200μl per well with Stage 2 NSC expansion medium (see recipes in Reagents and Solutions).
-
10.
Return the 96-well ULA plate with detached neural rosettes to a 37° C/5% CO2 incubator.
Over the next 24 hours, single neural rosettes will form into round, purified neurospheres.
-
11.
Replenish Stage 2 NSC expansion with half medium exchange (100μl) every-other-day until neurospheres are ready to assay.
3D neurospheres can be maintained in a multipotent state for a period of several days to weeks but will require serial passaging over time using an enzymatic reagent such as Accutase. Try to avoid neurosphere sizes growing above 200 μm as this will cause intrinsic differentiation to occur. Optimization of 3D neurosphere maintenance should be established by the end user.
Understanding the results
Under standard dual Smad inhibition by SB431542 & LDN193189, a subpopulation of neural crest cells is generated (Elkabetz et al., 2008; Chambers et al., 2009; Kim et al., 2021) that migrates from the periphery of rosette structures when the neural spheroids are plated onto an ECM-rich substrate such as Geltrex (Figure 2 D, E). These neural crest cells have been shown to be specified by the combined activities BMP and Wnt signaling (Chambers et al., 2012; Mica et al., 2013), which can be reduced when adding the Wnt inhibitor XAV939 (Rosebrock et al., 2022) and increasing the concentration of BMP inhibitor LDN193189. As an additional measure, 2D neural rosette populations can be positively selected from 3D neural spheroids using the enzymatic STEMDIFF neural rosette selection reagent and reaggregated in 3D to generate purified neurospheres (Figure 3). 3D neurospheres generated from different hPSC lines can be assayed for their proliferative and differentiation capacities to investigate disease-specific differences as has been recently reported (Wang et al., 2022).
SUPPORT PROTOCOL 3 ENZYMATIC DISSOCIATION, NSC EXPANSION, AND CRYOPRESERVATION
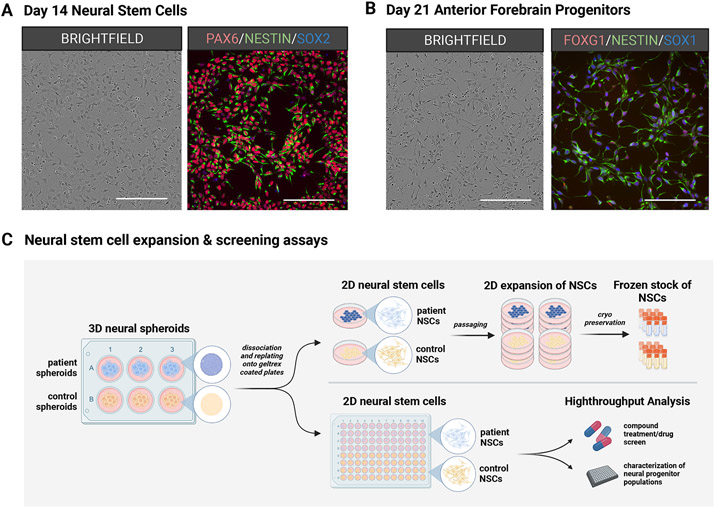
Neural stem cells (NSCs) are generated from the dissociation of rosette-containing 3D neural spheroids or purified 3D neurospheres, and they can be maintained for prolonged periods in 2D culture formats while possessing asymmetric potential for continued proliferation or terminal differentiation (Figure 4A, B). NSC populations are extremely pliable and can be passaged several times or cryopreserved to generate frozen stocks for later experimental use. NSCs can be seeded in a variety of formats that are amenable to phenotypic analysis or large-scale compound screens (Figure 4C). For example, NSC proliferation assays can be used to investigate the kinetics of cell cycle regulation across pluripotent stem cell lines and among disease states, and NSC differentiation assays can be used to evaluate the dynamics of cell lineage specification throughout neuronal development. The support protocol here details methods to expand and cryopreserve 2D NSC cultures generated from the 3D neural spheroid populations created in basic protocol 1, but the same approach can be used on purified 3D neurospheres created from support protocol 2.
Figure 4.
Neural stem and progenitor cells generated at several stages of cortical organoid development. Brightfield and fluorescent images of (A) 2D NSC populations harvested from early neural spheroids and (B) 2D neural progenitor cell (NPC) populations from differentiating cortical organoids. (C) Schematic workflow highlighting the utility of NSCs and NPCs that can be propagated and maintained in culture to generate frozen stocks or assayed for cell characterization & compound testing. Scale bar = 200μm.
Materials
3D Neural spheroids (Basic Protocol 1)
Neural Induction basal medium (see recipes in Reagents and Solutions)
Stage 2 NSC expansion medium (see recipes in Reagents and Solutions)
Geltrex LDEV-free, hESC qualified, 1ml, reduced growth factor basement membrane matrix (Thermo fisher Scientific, cat. no. A1413301)
Dulbecco’s Modified Eagle’s Medium (Corning Life Sciences, cat. no. 10-013-CV)
Accutase™ Cell Detachment Solution (Innovative Cell Technologies, Inc., cat. no. AT104-500)
DNase I Vial D2 (Worthington Biochemical, cat. no. LK003170)
Y-27632 (ROCK inhibitor; Cayman Chemical Company, cat. no. 10005583)
CryoStor CS10 freeze medium (Biolife Solutions, cat. no. 210102)
Corning 2ml internal threaded polypropylene cryogenic vial (Corning Life Sciences, cat. no. 430488)
Corning CoolCell LX freezing container (Corning Life Sciences, cat. no. 432001)
Protocol steps with step annotations
3D Neural spheroid Enzymatic Dissociation
-
Retrieve the 6-well suspension plate from the orbital shaker and collect 3D neural spheroids from each well into a single 50ml conical tube using a 5 ml pipet.
Allow 3D neural spheroids to settle to the bottom of the tube by gravity. If neural spheroids do not settle by gravity, then centrifuge at 200 × g for 5 min.
Aspirate off supernatant using a glass Pasteur pipet.
-
Using a serological pipet, rinse 3D neural spheroids with a 50ml volume of 1x D-PBS and allow spheroids to settle by gravity. Then repeat aspiration of the supernatant.
If neural spheroids do not settle by gravity, then centrifuge at 200 × g for 5 min.
Add 15 ml of Accutase (+ 50μg/ml DNase I) enzyme reagent and incubate for 15-20 min. at room temperature.
Using a 50 ml serological pipet, dilute Accutase with a double volume (30 ml) of Neural Induction basal medium (see recipes in Reagents and Solutions).
Pipet up and down several times, then centrifuge 50 ml conical tube at 400 × g for 5 min.
Aspirate off supernatant and resuspend cell pellet in 10 ml of Stage 2 NSC expansion medium + 10μM final ROCK inhibitor (Y-27632).
Perform a manual cell count using a hemocytometer.
2D NSC Expansion
-
9.
Prepare Geltrex coated 10cm dishes prior to starting this step (see Support Protocol 1).
-
10.
Seed dissociated 3D neurospheres into 10 cm dishes at concentration ranges between 50,000 – 200,000 cells/cm2 in a total volume of 15ml Stage 2 NSC expansion medium.
Desired confluence for NSC populations should be at least 50% of the total surface area. Appropriate seeding densities for NSCs expansion may vary and should be optimized for each individual cell line.
-
11.
Replace Stage 2 NSC expansion media to cultures on an every-other-day basis until they have reached between ≥ 90% confluence.
2D NSCs can be subsequently passaged for several generations using the Accutase dissociation protocol below or cryopreserved for later use.
2D NSC Enzymatic Dissociation
-
12.
Once 2D NSCs have reached ≥ 90% confluence they can be harvested for cryopreservation.
-
13.
Briefly rinse 10 cm dish with 1x D-PBS (175μl/cm2) and then aspirate off solution.
-
14.
Add Accutase reagent to the dish (100μl/cm2) and incubate for 5-10 minutes at room temperature.
-
15.
Dilute Accutase with Neural Induction basal medium (150μl/cm2).
-
16.
Centrifuge at 400 × g for 5 min.
-
17.
Resuspend the cell pellet with Neural Induction basal medium.
-
18.
Perform a manual cell count using a hemocytometer.
2D NSC Cryopreservation
-
19.
Resuspend the cell pellet in ice cold CryoStor CS10 freezing solution at a concentration of 5×106 cells/ml.
-
20.
Pipet 1 ml volume of cell solution (5x106 total) into each cryovial.
-
21.
Place cryovials into a Corning Coolcell XL freezing container, and place into −80°C ultra-low temperature freezer.
-
22.
After a period of 24 hours, transfer cryovials into liquid nitrogen storage.
Understanding the results
3D Neural spheroids (generated in Basic Protocol 1) or purified 3D neurospheres (generated in Support protocol 2) can be enzymatically dissociated with Accutase and plated as single cells on an ECM substrate like Geltrex or Laminin to generate populations of individual 2D neural stem cells (NSCs). These 2D NSCs can be maintained in culture over multiple passages or cryopreserved for later use (Figure 4). It is essential that 2D NSCs be maintained in adherent conditions with treatment by morphogens EGF & FGF2 to maintain their multipotency, otherwise they will terminally differentiate into mature neurons.
BASIC PROTOCOL 2 3D NEURAL PROGENITOR EXPANSION
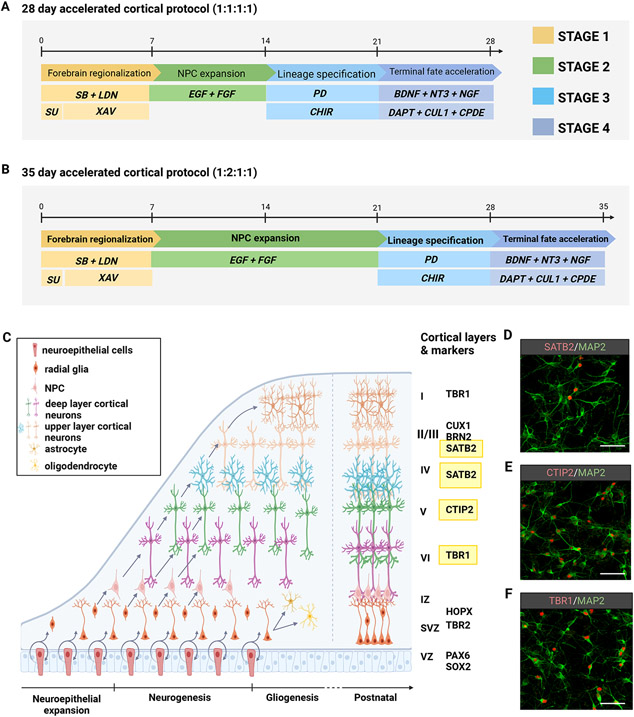
After the hPSC-derived spheroids have undergone Stage 1 neural induction (Basic Protocol 1) a Stage 2 expansion period follows through treatment with morphogens FGF2 and EGF to allow for the proliferation of progenitor cells within the developing neural spheroids. This stage of the protocol provides 2 options: A) a week-long expansion with FGF2 & EGF (Days 7-14), or B) 2 weeks of expansion with FGF2 & EGF (Days 7-21). Depending on the desired outcome, the two options will provide a 28-day protocol (Option A) that generates TBR1+ and CTIP2+ lower pyramidal layer cortical organoids (Figure 5A), or a 35-day protocol (Option B) that generates a heterogenous mixture of TBR1+ CTIP2+ SATB2+ upper and lower pyramidal layer cortical organoids (Figure 5B).
Figure 5.
Split-stream 3D protocols for the accelerated development of cortical lineages from hPSCs. (A) Schematic of a 28-day accelerated differentiation protocol with week-long periods across 4 stages. This option generates robust populations of TBR1+ cortical neurons as shown in panel F. (B) Schematic of a 35-day accelerated differentiation protocol with extension of stage 2 by an additional week. This option generates robust populations of TBR1+ CTIP2+ and SATB2+ pyramidal neurons as shown in panels D, E, and F. (C) Illustration of the cellular scaffolding and distribution of subtypes in the developing neocortex. (D-F) Immunostaining representation of upper and lower pyramidal layers of the neocortex generated by the representative protocols. Scale bar = 75μm.
Materials
3D Neural spheroids in 6-well ultra-low attachment suspension plate (Generated in Basic Protocol 1 above)
Neural Induction basal medium (see recipes in Reagents and Solutions)
Stage 2 NSC expansion medium (see recipes in Reagents and Solutions)
Protocol steps with step annotations
Stage 2: Neural progenitor expansion (Days 7-14 or Days 7-21)
Day 7: Replace Stage 1b Induction medium with Stage 2 NSC expansion medium
Prepare Stage 2 NSC expansion medium (see recipes in Reagents and Solutions).
-
Tilt 6-well plate at a 45° angle to allow aggregates to settle to the bottom of each well by gravity.
During this stage the organoids begin to grow considerably larger in size. Thus, it is no longer optimal to swirl organoids in the center of each well for aspiration.
Using a glass Pasteur pipet, slowly aspirate from the top down from each well until only a small volume of residual media remains.
Replace with 5ml of Stage 2 NSC expansion medium per well.
Days 9-14 (or 9-21): Replenish Stage 2 NSC expansion medium
-
5.
Replenish Stage 2 NSC medium every other day as indicated above.
BASIC PROTOCOL 3 3D ACCELERATED CORTICAL LINEAGE PATTERNING AND TERMINAL DIFFERENTIATION
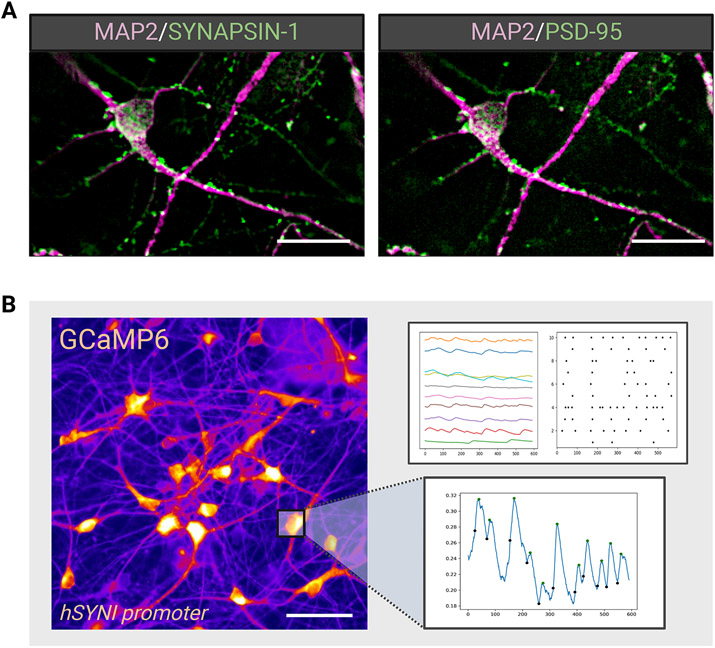
The final stages of this protocol involve the accelerated differentiation (Stage 3) and terminal specification (Stage 4) of neural patterned 3D spheroids into mature cortical organoids (see schematic in Figure 1 and Figure 5). Combinatorial pathway analysis revealed an optimal approach to accelerated cortical organoid lineage specification that capitalizes on simultaneous Wnt pathway activation (CHIR99021) with MAPK/ERK pathway inhibition (PD0325901) followed by robust NOTCH pathway inhibition (DAPT, Compound E, Culture One supplement), which results in the generation of pure populations of cortical neurons with diverse pyramidal layer identities (Figure 5D-F, Table 2). Additionally, we have identified several temporal periods for the generation of cortical neuron populations with corresponding pyramidal layer-specific identities. By utilizing a (1:1:1:1) staged approach, we generate populations of lower layer VI TBR1+ neurons and layer V CTIP2+ neurons in only 28 days (Figure 5A). By extending Stage 2 of the protocol (1:2:1:1 approach), in addition to TBR1+ neurons we also generate larger proportions of deep layer CTIP2+ neurons and upper pyramidal layer SATB2+ neurons in only 35 days (Figure 5B). To facilitate the survival and maturation of the cortical organoids during the stage 4 terminal cell fate acceleration by robust NOTCH inhibition, the added use of neurotrophic family growth factors BDNF, NT-3, and NGF is required. For the long-term functional maturation of cortical neurons, the use of astrocyte co-cultures is highly recommended (refer to strategic planning section), while Stage 5 maturation medium is employed (see recipe in Reagents and Solutions). Neurons generated by this accelerated approach will display markers of functional maturity and exhibit electrophysiological activity (Figure 6, Table 3). Additional resources for functional analysis and characterization are beyond the scope of this paper, and references cited in the Strategic planning section should be consulted.
Table 2.
Cortical layer marker antibodies for ICC/IF
| Component | Dilution |
|---|---|
| TBR1 (AbCam; cat. no. 31940) | 1:500 |
| CTIP2 (AbCam; cat. no. 18465) | 1:100 |
| SATB2 (AbCam; cat. no. 51502) | 1:100 |
| CUX1 (Proteintech; cat. no. 11733-1-AP) | 1:500 |
| TUJ1 (Biolegend; cat. no., 801202) | 1:500 |
Figure 6.
Functional maturation of accelerated cortical neurons. (A) Immunofluorescent staining reveals the presence of synaptic markers Synapsin-1 and PSD-95 and (B) Calcium reporter signaling measures the activity across populations of differentiated neurons. Scale bar = 25μm.
Table 3.
Maturation and functional neuronal antibodies for ICC/IF
| MAP2 (AbCam; cat. no. 5392) | 1:2000 |
| Synapsin 1 (Millipore Sigma; cat. no., AB1543P) | 1:250 |
| PSD95 (NeuroMabs; cat. no., 75-028) | 1:500 |
Materials
3D NPCs (Generated in Basic Protocol 2)
Neural Differentiation basal medium (see recipes in Reagents and Solutions)
Stage 3 cortical differentiation acceleration medium (see recipes in Reagents and Solutions)
Stage 4 cortical differentiation termination medium (see recipes in Reagents and Solutions)
Maturation basal medium (see recipes in Reagents and Solutions)
Stage 5 cortical maturation medium (see recipes in Reagents and Solutions)
Papain tissue dissociation kit (Worthington Biochemical, cat. no. LK003150)
Protocol steps with step annotations
Stage 3: Accelerated patterning into cortical lineages (Days 14-21 for 28-day cortical differentiation or days 21-28 for 35-day cortical differentiation)
Day 14 (or 21): Accelerated cortical differentiation
Prepare Stage 3 cortical differentiation acceleration medium (see recipes in Reagents and Solutions).
Tilt 6-well plate at a 45° angle to allow aggregates to settle to the bottom of each well by gravity.
Using a glass Pasteur pipet, slowly aspirate from the top down from each well until only a small volume of residual media remains.
Replace with 5ml of Stage 3 cortical differentiation acceleration medium per well.
Days 16-21 (or 23-28): Replenish Stage 3 accelerated cortical differentiation medium
-
5.
Replenish Stage 3 cortical differentiation acceleration medium every other day as indicated above.
Stage 4: Terminal cortical organoid differentiation (Day 21-28 for 28-day cortical organoid differentiation or Day 28-35 for 35-day cortical organoid differentiation)
Day 21 (or 28): Replace Stage 3 cortical differentiation acceleration medium with Stage 4 cortical differentiation termination medium
-
6.
Prepare Stage 4 cortical differentiation termination medium (see recipes in Reagents and Solutions).
-
7.
Tilt 6-well plate at a 45° angle to allow aggregates to settle to the bottom of each well by gravity.
-
8.
Using a glass Pasteur pipet, slowly aspirate from the top down from each well until only a small volume of residual media remains.
-
9.
Replace with 5ml of Stage 4 cortical differentiation termination medium per well.
Day 23-28 (or 30-35): Replenish Stage 4 cortical differentiation termination medium
-
10.
Replenish Stage 4 cortical differentiation termination medium every other day as indicated above until end point (Day 28 or Day 35) for dissociation, characterization, or long-term culture for functional analysis.
-
11.
Dissociate end-stage organoids with Papain Tissue Dissociation Kit according to manufacturer’s recommendations (Worthington Biochemical) or an equivalent enzyme.
-
12.
Characterize end-stage identities by immunostaining dissociated cells for cortical layer markers (Table 2).
Stage 5: Long-term cortical neuron maturation (Day 28-35+)
Day 28+ or 35+: Maintain dissociated cortical neurons in Stage 5 cortical neuron maturation medium
-
13.
Prepare Stage 5 cortical neuron maturation medium (see recipes in Reagents and Solutions).
-
14.
Plate dissociated organoids as single cells in Stage 5 cortical neuron maturation medium on substrates such as Poly-L-Ornithine and Laminin.
-
15.
Co-culture neurons with astrocytes or a physiologically enhanced medium for an extended duration of time (~ 3-12 weeks) for periodic functional analysis by electrophysiology or calcium imaging.
Consult the strategic planning section for suggestions on assay design & methods.
Understanding the results
After following the neural induction and progenitor expansion stages (represented in basic protocols 1 & 2), the resulting spheroids can be directed into several diverse cortical lineages; however, the protocol presented here specifies the generation of glutamatergic pyramidal neurons of the neocortex (Table 2). Depending upon the duration of Stage 2 in basic protocol 2, either one week (Figure 5A) or two (Figure 5B), one can generate deep layer CTIP2 (Figure 5E) and TBR1 (Figure 5F) or upper layer SATB2+ (Figure D) pyramidal neurons in a total of 4 to 5 weeks, a much shorter time frame than previous reports (Kadoshima et al., 2013; Pasca et al., 2015). This cortical neuron differentiation is further enhanced by exposing neural spheroids to the pro-dorsal Wnt activating compound CHIR99021 (a GSK3β inhibitor) and accelerated by treatment with the MAPK/ERK pathway inhibitor PD03251. Recent reports using neurodevelopmental disorder related hiPSC lines have discovered inherent cell line divergence towards a ventral forebrain cell identity despite applying similar dorsal patterning methods as described here (Eichmuller et al., 2022; Paulsen et al., 2022). This intrinsic drift may be overcome by the additional application of a small molecule antagonist of the sonic hedgehog pathway, Cyclopamine, during Stage 3 of basic protocol 3 to prevent SHH-mediated subpallial specification (Bagley et al., 2017). Notch signaling has been shown to be indispensable for the cell cycle regulation and maintenance of neural stem and progenitor cells (Elkabetz et al., 2008), and several methods using the gamma secretase inhibitor (DAPT) have been able to drive NSCs and NPCs out of the cell cycle into terminally differentiated cortical neuron fates (Qi et al., 2017; Gantner et al., 2021). Stage 4 of basic protocol 3 calls for the robust inhibition of notch signaling through combined treatment with ƴ-secretase inhibitors DAPT, Compound E (Cpd E), and Culture One supplement. This strategy drives the cortical progenitors out of the cell cycle and prevents astrogliogenesis (Zhou et al., 2010), which is known to occur following neurogenesis in the context of brain development and patterning based neural differentiation protocols (Pasca et al., 2015; Sloan et al., 2017; Marton et al., 2019). Therefore, the resulting cellular identities generated from both 28-day and 35-day protocols are exclusively glutamatergic pyramidal cortical neurons (Figure 5, D-F). Lastly, following the dissociation of the pure cortical organoids, neurons can be cultured for long-term maintenance in 2D co-culture with astrocytes to promote synaptic maturity and functional activity (Hedegaard et al., 2020; Kuijlaars et al., 2016). After several weeks, neurons will express the synaptic markers Synapsin-I and PSD-95 (Figure 6A; Table 3) and can be assayed for calcium signaling (Figure 6B) or electrophysiological activity.
REAGENTS AND SOLUTIONS
Maturation basal medium
500 ml Neurobasal™-A Medium (Gibco, cat. no. 10888022)
10 ml 50X B27 supplement (Thermo Fisher Scientific, cat. no. 17504044)
5 ml 100X GlutaMAX supplement (Thermo Fisher Scientific, cat. no. 350500-061)
5 ml 100X Pen Strep (Thermo Fisher Scientific, cat. no. 15140-122)
5 ml 100X MEM Non-Essential Amino Acids (Thermo Fisher Scientific, cat. no. 11140-050)
910 μl 55mM 2-Mercaptoethanol (Thermo Fisher Scientific, cat. no. 21985-023)
500 μl 200mM L-Ascorbic Acid (Sigma, cat. no. A4403)
Store at 4°C for up to 3 weeks
Neural induction basal medium
500 ml DMEM/F12 Medium (Thermo Fisher Scientific, cat. no. 11320033)
500 ml Neurobasal™-A Medium (Gibco, cat. no. 10888022)
10 ml 100X N-2 supplement (Thermo Fisher Scientific, cat. no. 17502-048)
20 ml 50X B27 supplement minus Vitamin A (Thermo Fisher Scientific, cat. no. 12587010)
10 ml 100X GlutaMAX supplement (Thermo Fisher Scientific, cat. no. 35050-061)
10 ml 100X Pen Strep (Thermo Fisher Scientific, cat. no. 15140-122)
10 ml 100X MEM Non-Essential Amino Acids (Thermo Fisher Scientific, cat. no. 11140-050)
1.82 ml 55mM 2-Mercaptoethanol (Thermo Fisher Scientific, cat. no. 21985-023)
1 ml 200mM L-Ascorbic Acid (Sigma, cat. no. A4403)
Store at 4°C for up to 3 weeks
Neural differentiation basal medium
500 ml DMEM/F12 Medium (Thermo Fisher Scientific, cat. no. 11320033)
500 ml Neurobasal™-A Medium (Gibco, cat. no. 10888022)
10 ml 100X N-2 supplement (Thermo Fisher Scientific, cat. no. 17502-048)
20 ml 50X B27 supplement (Thermo Fisher Scientific, cat. no. 17504044)
10 ml 100X GlutaMAX supplement (Thermo Fisher Scientific, cat. no. 35050-061)
10 ml 100X Pen Strep (Thermo Fisher Scientific, cat. no. 15140-122)
10 ml 100X MEM Non-Essential Amino Acids (Thermo Fisher Scientific, cat. no. 11140-050)
1.82 ml 55mM 2-Mercaptoethanol (Thermo Fisher Scientific, cat. no. 21985-023)
1 ml 200mM L-Ascorbic Acid (Sigma, cat. no. A4403)
Store at 4°C for up to 3 weeks
Small molecule and protein stock solutions
BDNF (PeproTech, cat. no. 450-02), 20 μg/ml in PBS plus 0.1% BSA; store in 100-μl aliquots at −80°C for 12 months.
CHIR99021 (Tocris, cat. no. 4423), 10 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
Compound E (Tocris, cat. no. 6476), 1 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
CultureOneTM Supplement, 100× (Thermo Fisher Scientific, cat. no. A33202-01), store at 4°C for 12 months.
DAPT (Tocris, cat. no. 2634), 10 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
EGF (PeproTech, cat. no. AF-100-15), 20 μg/ml in PBS plus 0.1% BSA; store in 100-μl aliquots at −80°C for 12 months.
FGF2 (PeproTech, cat. no. AF-100-18B), 20 μg /ml in PBS plus 0.1% BSA; store in 100-μl aliquots at −80°C for 12 months.
GDNF (PeproTech, cat. no. 450-10), 20 μg /ml in PBS plus 0.1% BSA; store in 100-μl aliquots at −80°C for 12 months.
LDN193189 (Tocris, cat. no. 6053), 2.5 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
NGF (R&D systems, cat. no. 256-GF-100/CF), 20 μg /ml in PBS plus 0.1% BSA; store in 100-μl aliquots at −80°C for 12 months.
NT-3 (StemCell Technologies, cat. no. 78074.1), 20 μg /ml in PBS plus 0.1% BSA; store in 100-μl aliquots at −80°C for 12 months.
SB431542 (Selleck Chemicals, cat. no. S1067), 10 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
SU5402 (Selleck Chemicals, cat. no. S7667), 50 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
XAV939 (Selleck Chemicals, cat. no. S1180), 10 mM in DMSO; store in 100-μl aliquots at −80°C for 12 months.
Y-27632 (ROCK inhibitor; Cayman Chemical Company, cat. no. 10005583), 10 mM in DMSO; store in 100-μl aliquots at −20°C for 12 months.
Stage 0 aggregation medium
PSC culture medium (i.e., E8 Flex, Stemflex, mTeSR, or mTeSR plus) (see Basic Protocol 1 for catalog information)
10 μM ROCK inhibitor, Y-27632 (See Small Molecule and Protein Stock Solutions)
Prepare fresh each time point
Stage 1a induction medium
Neural induction basal medium (see Reagents and Solutions), store at 4°C for up to 3 weeks
10 μM SB431542 (see recipe for stock solution and small molecule and protein stock solutions)
0.25 μM LDN193189 (see recipe for stock solution and small molecule and protein stock solutions)
5 μM XAV939 (see recipe for stock solution and small molecule and protein stock solutions)
5 μM SU-5402 (see recipe for stock solution and small molecule and protein stock solutions)
Prepare fresh each time point
Stage 1b induction medium
Neural induction basal medium (see Reagents and Solutions), store at 4°C for up to 3 weeks
10 μM SB431542 (see recipe for stock solution and small molecule and protein stock solutions)
0.25 μM LDN193189 (see recipe for stock solution and small molecule and protein stock solutions)
5 μM XAV939 (see recipe for stock solution and small molecule and protein stock solutions)
Prepare fresh each time point
Stage 2 NSC expansion medium
Neural induction basal medium (see Reagents and Solutions), store at 4°C for up to 3 weeks
20 ng/ml EGF (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml FGF2 (see recipe for stock solution and small molecule and protein stock solutions)
Prepare fresh each time point
Stage 3 Cortical differentiation acceleration medium
Neural differentiation basal medium (see Reagents and Solutions), store at 4°C for up to 3 weeks
2.5μM PD0325901 (see recipe for stock solution and small molecule and protein stock solutions)
3μM CHIR99021 (see recipe for stock solution and small molecule and protein stock solutions)
Prepare fresh each time point
Stage 4 Cortical differentiation termination medium
Neural differentiation basal medium (see Reagents and Solutions), store at 4°C for up to 3 weeks
10μM DAPT (see recipe for stock solution and small molecule and protein stock solutions)
0.1μM Compound E (see recipe for stock solution and small molecule and protein stock solutions)
1x Culture One supplement (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml NGF (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml NT-3 (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml BDNF (see recipe for stock solution and small molecule and protein stock solutions)
Prepare fresh each time point
Stage 5 Cortical neuron maturation medium
Maturation basal medium (see Reagents and Solutions), store at 4°C for up to 3 weeks
20 ng/ml NGF (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml NT-3 (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml BDNF (see recipe for stock solution and small molecule and protein stock solutions)
20 ng/ml GDNF (see recipe for stock solution and small molecule and protein stock solutions)
Prepare fresh each time point
COMMENTARY
Background Information
Over the past two decades there has been a fundamental shift in stem cell methodology driven by an evolution in technique from simple in vitro cellular models (Chambers et al., 2009) to more sophisticated 3D organoids that reflect the complexity of the physical dynamics and cell network interactions in the developing brain (Kadosihma et al., 2013; Lancaster et al., 2013; Lancaster et al., 2014; Pasca et al., 2105; Lancaster et al., 2017; Quadrato et al., 2017). However, these methods have been largely plagued by internal variability and a lack of reproducibility across cell lines. The consensus in the field has been to improve the standardization of the differentiation process to allow for more efficient modeling of healthy and disordered development (Andrews & Nowakowski, 2019; Anderson et al., 2021), and recent reports have shown marked improvements in cerebral organoid generation (Velasco et al., 2019) that have been used to investigate neurodevelopmental disorders (Paulsen et al., 2022). However, it remains a challenge to mimic aspects of brain development in vitro in a timely manner and protocols that have attempted to accelerate this process have resulted in off-target trajectories with reduced cell viabilities (Chambers et al., 2012; Qi et al., 2017). Taking advantage of this prior knowledge, we have identified a protocol that results in the accelerated development of 3D hPSC-derived cortical organoids with high precision and increased specificity while providing greater temporal control and improved adaptability across cell lines. By staggering developmental signaling pathway modulation across 4 distinct stages of varying temporal durations (1:1:1:1 or 1:2:1:1), we generate uniquely directed cortical lineage trajectories. This protocol is also amenable to a multitude of suspension formats from individual multi-well culture dishes to large volume bioreactor platforms. Furthermore, we also introduce several assays to both improve the differentiation workflow and serve as tools for stage-specific investigation into the mechanisms of brain development and disease in vitro. However, because the cortical organoids generated by this protocol possess populations of purely excitatory neurons, they alone are not sufficient to recreate the excitatory-inhibitory balance of neuronal networks that are often disrupted by mutations in genes associated with intellectual disability and neurodevelopmental disorders (Anderson et al., 2021). Therefore, if investigation of these interactions is desired, we recommend an integrated “assembloid” approach that has been previously described to better capture these network dynamics (Bagley et al., 2017; Birey et al., 2017). Like the protocol presented here, it might be possible to achieve high specificity of ventral forebrain patterning by exchanging the pro-dorsal compound (CHIR99021) in Stage 3 of this protocol with a pro-ventral compound such as Smoothened agonist (SAG) or Purmorphamine, as has been previously reported (Maroof et al., 2013; Nicholas et al., 2013). Together, this would allow for the interrogation of cell autonomous and non-cell autonomous influences in development and disease. Nevertheless, this novel protocol allows for more adequate in vitro modeling of cortical development in a controlled and timely manner and can be utilized to reveal disease-specific phenotypes across the developmental timeline, thus enhancing our understanding of complex neurodevelopmental disorders and allowing for earlier identification of potential targets for therapeutic intervention.
Critical Parameters
hPSC culture maintenance & quality control
It is necessary to use optimal cell culture techniques and culture media systems for the derivation and maintenance of pluripotent stem cells, of which several resources exist and are available by commercial vendors such as StemCell Technologies (www.stemcell.com) or Thermofisher Scientific (www.thermofisher.com). Prior to all PSC tissue culture work should be an initial testing of cell lines for mycoplasma contamination, and lines should be routinely monitored for mycoplasma as this can have a profound effect on cell behavior. Mycoplasma testing reagents and additional resources can be found from industry-leading companies such as Invivogen (www.invivogen.com). Additional QC should involve pluripotency panels, karyotyping, STR profiling, and whole genome sequencing may be desired for cell lines that have been genetically engineered or known to have disease-relevant mutations. Signs of spontaneous differentiation in the culture should be removed by commercially available reagents (such as ReLeSR from StemCell Technologies), and karyotypically abnormal cell stocks should be discarded. Prior to the start of differentiation PSC cultures should exhibit standard growth curves, calculated by the individual, and cultures should not be too overgrown or spontaneously differentiated.
Enzymatic digestion and ROCK inhibitor treatment
To ensure adequate generation of homogenous populations of spheroids, it is critical to dissociate hPSC colonies into single cells using enzymatic reagents like Accutase according to the manufacturer’s recommendations. Despite common practice in the stem cell field, Accutase has been formulated to perform optimally at room temperature and the user is expected to adhere to this guideline from the manufacturer. While undergoing enzymatic digestion, hPSCs should be monitored, and agitated, to improve single cell dissociation while also remaining cognizant of the duration of time that the cells are exposed. Too little enzymatic incubation will result in the presence of large clumps that will contribute to heterogenous sizes and improperly formed spheroids, while too long of an incubation will result in cell lysis and failure of aggregation. In addition, the use of ROCK inhibitor is required whenever performing single cell dissociations (Watanabe et al., 2007).
Cell seeding density & orbital shaker platform speed
Another major influence for spheroid generation relies on cell seeding density and shaker platform speed, both of which have been optimized as detailed in this protocol. Failure to adhere to these guidelines will result in spheroids of inadequate size, insufficient quantity, and/or irregular shapes.
Freshness of reagents & titration of small molecules
The small molecule concentrations set forth in this protocol have been validated across several hPSC lines; however, it is possible that, on occasion, a certain line may not behave according to the recommendation. In this event, the titration of small molecules and growth factors may be necessary to achieve desired outcomes. In any circumstance, the quality and freshness of reagents should be always maintained throughout the differentiation process as this has a major impact on the success of directed patterning.
Medium changes
Scheduled media changes have also been optimized and validated across several hPSC lines; however, there may be instances where more frequent media changes are required as the nutritional demands of the organoid culture increase. In this instance, it is fine to increase the frequency of media changes at the discretion of the user.
Troubleshooting
See Table 4 for troubleshooting guidelines.
Table 4.
Troubleshooting guidelines
| Protocol | Problem | Possible causes | Solution(s) |
|---|---|---|---|
| Basic Protocol 1 | Aggregates do not form from single cell suspension after enzymatic digestion |
Cells died from prolonged Accutase treatment | Frequent monitoring of time & cell dissociation during the enzymatic incubation stage |
| Basic Protocol 1 | Aggregated spheroids are heterogenous in size and are large (> 200μm). | Insufficient single cell dissociation resulting in remaining clumps Too low or too high of a seeding density plated per well of 6-well plate Orbital shaker platform speed set too low at the initial aggregation step (< 100rpm) |
Incubate Accutase at appropriate temperatures and duration of time. Use mechanical force to dissociate hPSC colonies Seed cells at recommended densities outlined in this protocol and set shaker platform instrument at indicated speeds |
| Basic Protocol 1 | Aggregated spheroids are too small (< 200μm). | Orbital shaker platform speed set too high at the initial aggregation step (> 100rpm) |
Set shaker platform instrument at indicated speeds and only increase as directed in the protocol |
| Basic Protocol 1 | Neural spheroids deteriorate in culture | Incorrect media recipe with wrong concentrations of small molecules |
Make sure to follow media recipes exactly and calculate the precise concentration of small molecules |
| Support Protocol 1 | Rosette structures fail to form | Inefficient neural patterning Improper ECM-matrix coating |
Ensure that correct media and concentrations of small molecules are used Make sure ECM substrate is properly diluted and coating conditions are followed |
| Support Protocol 2 and 3 and Basic Protocol 2 | Neurospheres fail to form, NSCs do not expand, and neural spheroids fail to grow | Absence of ROCK inhibitor Lack of FGF & EGF morphogens |
Make sure to include ROCK inhibitor compound when detaching & isolating rosette structures or NSCs Ensure that FGF2 and EGF have been properly added to the expansion medium |
| Basic Protocol 3 | Deterioration of organoids | Acidic media caused by overconsumption of nutrients Increased oxidative stress |
Monitor organoid metabolism and look for color changes indicated by phenol red indicator. Switch to an everyday feeding schedule if necessary to maintain organoid health Add additional nutrient support like Sodium Pyruvate and alternative buffers like HEPES |
| Basic Protocol 1, 2 & 3 | Contamination of cell culture | Improper sterile technique Failure to use antibiotics in the culture media |
Use superior aseptic technique when performing cell culture Use penicillin/streptomycin for all medias as indicated in the protocol |
| Basic Protocol 3 | Organoids fail to dissociate | Inadequate dissociation system | Use industry leading reagents and dissociation kits Optimize protocol beyond manufacturer’s recommendations |
| Basic Protocol 3 | Inefficient differentiation marked by lack of diverse cell types | Non-adherence to the established protocol SOPs Suboptimal hPSC line |
Make sure to follow protocol steps without deviations Test a widely utilized and commercially available hPSC line |
Time Considerations:
Basic Protocol 1: 7 days
Basic Protocol 2: 7 or 14 days
Basic Protocol 3: 14 days *Dissociated organoids can be kept for over 100 days in 2D astrocyte monolayer co-cultures
Acknowledgements
This research was conducted with support from an anonymous donor as well as support from the Rosamund Stone Zander Translational Neuroscience Center and the Intellectual and Developmental Disorders Research Center (IDDRC) Clinical Translation (CT) Core National Institutes of Health (NIH) P50 HD105351. The SOX2-GFP human induced pluripotent stem cell (hiPSC) line was procured from the Allen Institute for Cell Science through a Material Transfer Agreement (MTA) with the Coriell Institute for Medical Research. Some figure illustrations were created using BioRender (www.biorender.com) with appropriate licensing.
Footnotes
Conflict of Interest
Mustafa Sahin reports grant support from Novartis, Biogen, Astellas, Aeovian, Bridgebio, and Aucta. He has served on Scientific Advisory Boards for Novartis, Roche, Regenxbio, SpringWorks Therapeutics, Jaguar Therapeutics and Alkermes. The other authors declare no conflict of interest.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Literature Cited
- 1.Akkerman N, Defize LH. Dawn of the organoid era: 3D tissue and organ cultures revolutionize the study of development, disease, and regeneration. Bioessays. 2017. Apr;39(4). doi: 10.1002/bies.201600244. Epub 2017 Feb 21. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NC, Chen PF, Meganathan K, Afshar Saber W, Petersen AJ, Bhattacharyya A, Kroll KL, Sahin M; Cross-IDDRC Human Stem Cell Working Group. Balancing serendipity and reproducibility: Pluripotent stem cells as experimental systems for intellectual and developmental disorders. Stem Cell Reports. 2021. Jun 8;16(6):1446–1457. doi: 10.1016/j.stemcr.2021.03.025. Epub 2021 Apr 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews MG, Nowakowski TJ. Human brain development through the lens of cerebral organoid models. Brain Res. 2019. Dec 15;1725:146470. doi: 10.1016/j.brainres.2019.146470. Epub 2019 Sep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Autar K, Guo X, Rumsey JW, Long CJ, Akanda N, Jackson M, Narasimhan NS, Caneus J, Morgan D, Hickman JJ. A functional hiPSC-cortical neuron differentiation and maturation model and its application to neurological disorders. Stem Cell Reports. 2022. Jan 11;17(1):96–109. doi: 10.1016/j.stemcr.2021.11.009. Epub 2021 Dec 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017. Jul;14(7):743–751. doi: 10.1038/nmeth.4304. Epub 2017 May 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, Gorris M, Galet B, Palomares V, Brown J, Bang AG, Mertens J, Böhnke L, Boyer L, Simon S, Gage FH. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A. 2015. May 19;112(20):E2725–34. doi: 10.1073/pnas.1504393112. Epub 2015 Apr 13. Erratum in: Proc Natl Acad Sci U S A. 2015 Jun 23;112(25):E3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begum AN, Guoynes C, Cho J, Hao J, Lutfy K, Hong Y. Rapid generation of sub-type, region-specific neurons and neural networks from human pluripotent stem cell-derived neurospheres. Stem Cell Res. 2015. Nov;15(3):731–741. doi: 10.1016/j.scr.2015.10.014. Epub 2015 Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benito-Kwiecinski S, Giandomenico SL, Sutcliffe M, Riis ES, Freire-Pritchett P, Kelava I, Wunderlich S, Martin U, Wray GA, McDole K, Lancaster MA. An early cell shape transition drives evolutionary expansion of the human forebrain. Cell. 2021. Apr 15;184(8):2084–2102.e19. doi: 10.1016/j.cell.2021.02.050. Epub 2021 Mar 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O'Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Paşca SP. Assembly of functionally integrated human forebrain spheroids. Nature. 2017. May 4;545(7652):54–59. doi: 10.1038/nature22330. Epub 2017 Apr 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter MK, Cui X, Hu ZY, Jackson J, Sherman S, Seiger A, Wahlberg LU. In vitro expansion of a multipotent population of human neural progenitor cells. Exp Neurol. 1999. Aug;158(2):265–78. doi: 10.1006/exnr.1999.7098. [DOI] [PubMed] [Google Scholar]
- 11.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009. Mar;27(3):275–80. doi: 10.1038/nbt.1529. Epub 2009 Mar 1. Erratum in: Nat Biotechnol. 2009 May;27(5):485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi SH, Studer L. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012. Jul 1;30(7):715–20. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornacchia D, Zhang C, Zimmer B, Chung SY, Fan Y, Soliman MA, Tchieu J, Chambers SM, Shah H, Paull D, Konrad C, Vincendeau M, Noggle SA, Manfredi G, Finley LWS, Cross JR, Betel D, Studer L. Lipid Deprivation Induces a Stable, Naive-to-Primed Intermediate State of Pluripotency in Human PSCs. Cell Stem Cell. 2019. Jul 3;25(1):120–136.e10. doi: 10.1016/j.stem.2019.05.001. Epub 2019 May 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dakhore S, Nayer B, Hasegawa K. Human Pluripotent Stem Cell Culture: Current Status, Challenges, and Advancement. Stem Cells Int. 2018. Nov 22;2018:7396905. doi: 10.1155/2018/7396905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichmüller OL, Corsini NS, Vértesy Á, Morassut I, Scholl T, Gruber VE, Peer AM, Chu J, Novatchkova M, Hainfellner JA, Paredes MF, Feucht M, Knoblich JA. Amplification of human interneuron progenitors promotes brain tumors and neurological defects. Science. 2022. Jan 28;375(6579):eabf5546. doi: 10.1126/science.abf5546. Epub 2022 Jan 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008. Nov 6;3(5):519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008. Jan 15;22(2):152–65. doi: 10.1101/gad.1616208. Erratum in: Genes Dev. 2008 May 1;22(9):1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delepine C, Pham VA, Tsang HWS, Sur M. GSK3ß inhibitor CHIR 99021 modulates cerebral organoid development through dose-dependent regulation of apoptosis, proliferation, differentiation and migration. PLoS One. 2021. May 5;16(5):e0251173. doi: 10.1371/journal.pone.0251173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galiakberova AA, Surin AM, Bakaeva ZV, Sharipov RR, Zhang D, Dorovskoy DA, Shakirova KM, Fisenko AP, Dashinimaev EB. IPSC-Derived Human Neurons with GCaMP6s Expression Allow In Vitro Study of Neurophysiological Responses to Neurochemicals. Neurochem Res. 2022. Apr;47(4):952–966. doi: 10.1007/s11064-021-03497-6. Epub 2021 Dec 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gantner CW, Hunt CPJ, Niclis JC, Penna V, McDougall SJ, Thompson LH, Parish CL. FGF-MAPK signaling regulates human deep-layer corticogenesis. Stem Cell Reports. 2021. May 11;16(5):1262–1275. doi: 10.1016/j.stemcr.2021.03.014. Epub 2021 Apr 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedegaard A, Monzón-Sandoval J, Newey SE, Whiteley ES, Webber C, Akerman CJ. Pro-maturational Effects of Human iPSC-Derived Cortical Astrocytes upon iPSC-Derived Cortical Neurons. Stem Cell Reports. 2020. Jul 14;15(1):38–51. doi: 10.1016/j.stemcr.2020.05.003. Epub 2020 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofrichter M, Nimtz L, Tigges J, Kabiri Y, Schröter F, Royer-Pokora B, Hildebrandt B, Schmuck M, Epanchintsev A, Theiss S, Adjaye J, Egly JM, Krutmann J, Fritsche E. Comparative performance analysis of human iPSC-derived and primary neural progenitor cells (NPC) grown as neurospheres in vitro. Stem Cell Res. 2017. Dec;25:72–82. doi: 10.1016/j.scr.2017.10.013. Epub 2017 Oct 26. [DOI] [PubMed] [Google Scholar]
- 23.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010. Mar 2;107(9):4335–40. doi: 10.1073/pnas.0910012107. Epub 2010 Feb 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Zhou Y, Bao X, Chen C, Randolph LN, Du J, Lian XL. An Ultrasensitive Calcium Reporter System via CRISPR-Cas9-Mediated Genome Editing in Human Pluripotent Stem Cells. iScience. 2018. Nov 30;9:27–35. doi: 10.1016/j.isci.2018.10.007. Epub 2018 Oct 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013. Dec 10;110(50):20284–9. doi: 10.1073/pnas.1315710110. Epub 2013 Nov 25. Erratum in: Proc Natl Acad Sci U S A. 2014 May 20;111(20):7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HM, Noh HB, Lee SH, Lee KG, Chang B, Cheong E, Lee CJ, Hwang DY. Fine-tuning of dual-SMAD inhibition to differentiate human pluripotent stem cells into neural crest stem cells. Cell Prolif. 2021. Sep;54(9):e13103. doi: 10.1111/cpr.13103. Epub 2021 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight GT, Lundin BF, Iyer N, Ashton LM, Sethares WA, Willett RM, Ashton RS. Engineering induction of singular neural rosette emergence within hPSC-derived tissues. Elife. 2018. Oct 29;7:e37549. doi: 10.7554/eLife.37549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009. Mar 3;106(9):3225–30. doi: 10.1073/pnas.0808387106. Epub 2009 Feb 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011. Nov 6;480(7378):547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuijlaars J, Oyelami T, Diels A, Rohrbacher J, Versweyveld S, Meneghello G, Tuefferd M, Verstraelen P, Detrez JR, Verschuuren M, De Vos WH, Meert T, Peeters PJ, Cik M, Nuydens R, Brône B, Verheyen A. Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci Rep. 2016. Nov 7;6:36529. doi: 10.1038/srep36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013. Sep 19;501(7467):373–9. doi: 10.1038/nature12517. Epub 2013 Aug 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014. Oct;9(10):2329–40. doi: 10.1038/nprot.2014.158. Epub 2014 Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017. Jul;35(7):659–666. doi: 10.1038/nbt.3906. Epub 2017 May 31. Erratum in: Nat Biotechnol. 2018 Oct 11;36(10):1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, Kim WR, Lipton SA, Zhang K, Ding S. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011. May 17;108(20):8299–304. doi: 10.1073/pnas.1014041108. Epub 2011 Apr 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, Widmer HR, Eggan K, Goldstein PA, Anderson SA, Studer L. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013. May 2;12(5):559–72. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, Sasai Y, Alvarez-Buylla A, Rubenstein JL, Kriegstein AR. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013. May 2;12(5):573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicoleau C, Varela C, Bonnefond C, Maury Y, Bugi A, Aubry L, Viegas P, Bourgois-Rocha F, Peschanski M, Perrier AL. Embryonic stem cells neural differentiation qualifies the role of Wnt/β-Catenin signals in human telencephalic specification and regionalization. Stem Cells. 2013. Sep;31(9):1763–74. doi: 10.1002/stem.1462. [DOI] [PubMed] [Google Scholar]
- 38.Odawara A, Katoh H, Matsuda N, Suzuki I. Physiological maturation and drug responses of human induced pluripotent stem cell-derived cortical neuronal networks in long-term culture. Sci Rep. 2016. May 18;6:26181. doi: 10.1038/srep26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odawara A, Matsuda N, Ishibashi Y, Yokoi R, Suzuki I. Toxicological evaluation of convulsant and anticonvulsant drugs in human induced pluripotent stem cell-derived cortical neuronal networks using an MEA system. Sci Rep. 2018. Jul 10;8(1):10416. doi: 10.1038/s41598-018-28835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odawara A, Saitoh Y, Alhebshi AH, Gotoh M, Suzuki I. Long-term electrophysiological activity and pharmacological response of a human induced pluripotent stem cell-derived neuron and astrocyte co-culture. Biochem Biophys Res Commun. 2014. Jan 24;443(4):1176–81. doi: 10.1016/j.bbrc.2013.12.142. Epub 2014 Jan 7. [DOI] [PubMed] [Google Scholar]
- 41.Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ. 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell. 2016. Apr 7;18(4):467–80. doi: 10.1016/j.stem.2016.03.003. Epub 2016 Mar 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O'Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Paşca SP. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015. Jul;12(7):671–8. doi: 10.1038/nmeth.3415. Epub 2015 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen B, Velasco S, Kedaigle AJ, Pigoni M, Quadrato G, Deo AJ, Adiconis X, Uzquiano A, Sartore R, Yang SM, Simmons SK, Symvoulidis P, Kim K, Tsafou K, Podury A, Abbate C, Tucewicz A, Smith SN, Albanese A, Barrett L, Sanjana NE, Shi X, Chung K, Lage K, Boyden ES, Regev A, Levin JZ, Arlotta P. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022. Feb;602(7896):268–273. doi: 10.1038/s41586-021-04358-6. Epub 2022 Feb 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci. 2004. Mar 1;117(Pt 7):1269–80. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- 45.Qi Y, Zhang XJ, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, Ganat Y, Azevedo R, Zeltner N, Brivanlou AH, Karayiorgou M, Gogos J, Tomishima M, Tessier-Lavigne M, Shi SH, Studer L. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol. 2017. Feb;35(2):154–163. doi: 10.1038/nbt.3777. Epub 2017 Jan 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017. May 4;545(7652):48–53. doi: 10.1038/nature22047. Epub 2017 Apr 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, Ben-Hur T. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001. Dec;19(12):1134–40. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 48.Rosebrock D, Arora S, Mutukula N, Volkman R, Gralinska E, Balaskas A, Aragonés Hernández A, Buschow R, Brändl B, Müller FJ, Arndt PF, Vingron M, Elkabetz Y. Enhanced cortical neural stem cell identity through short SMAD and WNT inhibition in human cerebral organoids facilitates emergence of outer radial glial cells. Nat Cell Biol. 2022. Jun;24(6):981–995. doi: 10.1038/s41556-022-00929-5. Epub 2022 Jun 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015. Nov 17;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma Y, Saha S, Joseph A, Krishnan H, Raghu P. In vitro human stem cell derived cultures to monitor calcium signaling in neuronal development and function. Wellcome Open Res. 2020. Feb 3;5:16. doi: 10.12688/wellcomeopenres.15626.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012. Oct;7(10):1836–46. doi: 10.1038/nprot.2012.116. Epub 2012 Sep 13. [DOI] [PubMed] [Google Scholar]
- 52.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012. Feb 5;15(3):477–86, S1. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strano A, Tuck E, Stubbs VE, Livesey FJ. Variable Outcomes in Neural Differentiation of Human PSCs Arise from Intrinsic Differences in Developmental Signaling Pathways. Cell Rep. 2020. Jun 9;31(10):107732. doi: 10.1016/j.celrep.2020.107732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, Caldwell MA. A new method for the rapid and long-term growth of human neural precursor cells. J Neurosci Methods. 1998. Dec 1;85(2):141–52. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- 55.Tchieu J, Zimmer B, Fattahi F, Amin S, Zeltner N, Chen S, Studer L. A Modular Platform for Differentiation of Human PSCs into All Major Ectodermal Lineages. Cell Stem Cell. 2017. Sep 7;21(3):399–410.e7. doi: 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temple S, Qian X. bFGF, neurotrophins, and the control or cortical neurogenesis. Neuron. 1995. Aug;15(2):249–52. doi: 10.1016/0896-6273(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 57.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019. Jun;570(7762):523–527. doi: 10.1038/s41586-019-1289-x. Epub 2019 Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vőfély G, Berecz T, Szabó E, Szebényi K, Hathy E, Orbán TI, Sarkadi B, Homolya L, Marchetto MC, Réthelyi JM, Apáti Á. Characterization of calcium signals in human induced pluripotent stem cell-derived dentate gyrus neuronal progenitors and mature neurons, stably expressing an advanced calcium indicator protein. Mol Cell Neurosci. 2018. Apr;88:222–230. doi: 10.1016/j.mcn.2018.02.003. Epub 2018 Feb 6. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Chiola S, Yang G, Russell C, Armstrong CJ, Wu Y, Spampanato J, Tarboton P, Ullah HMA, Edgar NU, Chang AN, Harmin DA, Bocchi VD, Vezzoli E, Besusso D, Cui J, Cattaneo E, Kubanek J, Shcheglovitov A. Modeling human telencephalic development and autism-associated SHANK3 deficiency using organoids generated from single neural rosettes. Nat Commun. 2022. Oct 6;13(1):5688. doi: 10.1038/s41467-022-33364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Wang SN, Xu TY, Miao ZW, Su DF, Miao CY. Organoid technology for brain and therapeutics research. CNS Neurosci Ther. 2017. Oct;23(10):771–778. doi: 10.1111/cns.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe M, Buth JE, Haney JR, Vishlaghi N, Turcios F, Elahi LS, Gu W, Pearson CA, Kurdian A, Baliaouri NV, Collier AJ, Miranda OA, Dunn N, Chen D, Sabri S, Torre-Ubieta L, Clark AT, Plath K, Christofk HR, Kornblum HI, Gandal MJ, Novitch BG. TGFβ superfamily signaling regulates the state of human stem cell pluripotency and capacity to create well-structured telencephalic organoids. Stem Cell Reports. 2022. Oct 11;17(10):2220–2238. doi: 10.1016/j.stemcr.2022.08.013. Epub 2022 Sep 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005. Mar;8(3):288–96. doi: 10.1038/nn1402. Epub 2005 Feb 6. [DOI] [PubMed] [Google Scholar]
- 63.Wen Y, Jin S. Production of neural stem cells from human pluripotent stem cells. J Biotechnol. 2014. Oct 20;188:122–9. doi: 10.1016/j.jbiotec.2014.07.453. Epub 2014 Aug 20. [DOI] [PubMed] [Google Scholar]
- 64.Whye D, Wood D, Kim KH, Chen C, Makhortova N, Sahin M, Buttermore ED. Dynamic 3D Combinatorial Generation of hPSC-Derived Neuromesodermal Organoids With Diverse Regional and Cellular Identities. Curr Protoc. 2022. Oct;2(10):e568. doi: 10.1002/cpz1.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, Mei A, McHenry L, Lisuk D, Grasmick JM, Silberman P, Silberman G, Jappelli R, Gage FH. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports. 2014. Feb 27;2(3):295–310. doi: 10.1016/j.stemcr.2014.01.009. Erratum in: Stem Cell Reports. 2014 Jul 8;3(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001. Dec;19(12):1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 67.Zhou ZD, Kumari U, Xiao ZC, Tan EK. Notch as a molecular switch in neural stem cells. IUBMB Life. 2010. Aug;62(8):spcone. doi: 10.1002/iub.372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.