Abstract
Nicotinamide adenine dinucleotide (NAD+) precursor nicotinamide riboside (NR) has emerged as a promising compound to improve obesity-associated mitochondrial dysfunction and metabolic syndrome in mice. However, most short-term clinical trials conducted so far have not reported positive outcomes. Therefore, we aimed to determine whether long-term NR supplementation boosts mitochondrial biogenesis and metabolic health in humans. Twenty body mass index (BMI)–discordant monozygotic twin pairs were supplemented with an escalating dose of NR (250 to 1000 mg/day) for 5 months. NR improved systemic NAD+ metabolism, muscle mitochondrial number, myoblast differentiation, and gut microbiota composition in both cotwins. NR also showed a capacity to modulate epigenetic control of gene expression in muscle and adipose tissue in both cotwins. However, NR did not ameliorate adiposity or metabolic health. Overall, our results suggest that NR acts as a potent modifier of NAD+ metabolism, muscle mitochondrial biogenesis and stem cell function, gut microbiota, and DNA methylation in humans irrespective of BMI.
Nicotinamide riboside is a potent modifier of mitochondrial metabolism, satellite cell differentiation and gut microbiota.
INTRODUCTION
Activation of mitochondria is an attractive treatment option for obesity and its related metabolic complications (1). Increasing intracellular levels of nicotinamide adenine dinucleotide (NAD+), the crucial cofactor for mitochondrial energy production, has shown great promise to improve mitochondrial number and oxidative capacity and to combat obesity and its related diseases in mice (2). This observation has generated a high interest on whether mitochondrial dysfunction and metabolic health can be improved by NAD+ boosters in humans.
The intracellular NAD+ levels can be increased by supplementation with NAD+ precursors (2). Vitamin B3 forms niacin (nicotinic acid), nicotinamide (NAM), and nicotinamide riboside (NR) are naturally occurring precursors of NAD+. They are converted to NAD+ via their distinct biosynthesis routes, Preiss-Handler and salvage pathways. The clinical use of niacin and NAM is challenged by dose-dependent adverse effects such as hepatotoxicity (3). Niacin also induces cutaneous flushing through activation of G protein–coupled receptor 109A (4). However, our previous study established that niacin can improve systemic NAD+ levels and muscle mitochondrial metabolism in humans without severe side effects (5). The newest vitamin B3 family member NR (6) has been shown to increase mitochondrial biogenesis and function, protect against diet-induced obesity and insulin resistance, and improve the gut microbiota composition in mice (7–13). Therefore, NR has rapidly entered clinical trials.
To date, clinical studies have shown NR to be safe and to raise NAD+ levels in whole blood, peripheral blood mononuclear cells, urine, muscle, and brain with a dose from 300 mg/day up to 2000 mg/day (14–20). Unfortunately, most human trials have failed to show substantial improvements in adiposity and insulin sensitivity in healthy overweight or obese individuals after a 3- to 12-week NR supplementation (14–19). However, improved physical performance, muscle acetylcarnitine levels, arterial stiffness, and blood pressure or decreased circulating inflammatory cytokines have been reported in a few studies with a short-term follow-up (18, 19, 21). At present, the evidence of the efficacy of NR on tissue mitochondrial biogenesis and function still relies on data on mice as positive outcomes are lacking from clinical studies (19, 22). Until now, the number of human intervention studies with NR is still relatively low and the longest clinical trial conducted has been 12 weeks. Given that the effects of NR could be tissue specific and depend on body mass index (BMI), genetic background, and treatment duration, deep metabolic phenotyping of multiple tissues in well-controlled human interventions of longer duration is required.
We hypothesized that long-term NR supplementation could improve mitochondrial biogenesis in metabolically active tissues in humans. Therefore, we examined the effect of long-term supplementation of NR (5 months) not only on muscle and white adipose tissue (WAT) mitochondrial biogenesis (primary outcome) but also on body composition and multiple measures of metabolic health. Our design was unique: rare monozygotic (MZ) twin pairs who were discordant for BMI, allowing us to investigate whether the NR response differs in the leaner and the heavier cotwins with matched genetic background. In addition, we applied the cotwin control strategy in a small cohort of BMI-concordant MZ pairs to compare the effects of NR (one cotwin) to placebo (the other cotwin). We found that 5-month NR supplementation improves muscle mitochondrial biogenesis, muscle myoblast differentiation, and the gut microbiota composition and modulates DNA methylation, with potential effects on epigenetic control of gene expression, in both cotwins from the BMI-discordant pairs, i.e., regardless of BMI. However, these changes were not translated into improvements in adiposity or metabolic status. Together, our study underscores that long-term NR supplementation can affect various metabolic processes in humans.
RESULTS
Characteristics of study participants
Together, 20 BMI-discordant (within-pair difference in BMI, <2.5 kg/m2) and 4 BMI-concordant MZ twin pairs, aged ~40 (interquartile range, 33 to 41), participated in the study (Table 1). Four BMI-discordant twin pairs discontinued the intervention (fig. S1). Women accounted for 44 and 50% of the analyzed BMI-discordant and BMI-concordant MZ twin pairs, respectively (Table 1). All twins from the BMI-discordant pairs were supplemented with NR. Of the concordant twin pairs (Table 1), one cotwin was randomized to placebo and the other one to NR. The daily NR dose was gradually escalated by 250 mg/week to the full dose of 1000 mg/day, continuing until 5 months (Fig. 1A). The primary endpoint was the change in muscle and WAT mitochondrial biogenesis (clinicaltrials.gov entry NCT03951285). For the list of secondary endpoints, see https://clinicaltrials.gov/ct2/show/NCT03951285?term=nicotinamide+riboside&draw=4&rank=29. Figure 1A and fig. S1 present the study design and the procedures for the selection of study subjects and data analyses, respectively.
Table 1. Baseline characteristics of the twins from the BMI-discordant and BMI-concordant pairs.
Data are shown as means ± SD or median (interquartile range). P values were obtained using paired Wilcoxon signed-rank test to examine the differences on anthropometric and clinical parameters between the cotwins at baseline in the twins from the BMI-discordant (n = 16 twin pairs/32 individuals) and concordant (n = 4 twin pairs/8 individuals) pairs. au, arbitrary units; ND, not determined; HbA1c, hemoglobin A1c; HOMA, homeostasis model assessment; AUC, area under the curve; OGTT, oral glucose tolerance test; HDL, high-density lipoprotein; LDL, low-density lipoprotein; FFA, free fatty acids; BP, blood pressure; AST, aspartate transaminase; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; fP, fasting plasma; Fe, iron.
| Variable (unit) | Discordant All (n = 32 individuals) | Leaner (n = 16 individuals) | Heavier (n = 16 individuals) | P | Concordant All (n = 8 individuals) | Concordant Placebo (n = 4 individuals) | Concordant Treated (n = 4 individuals) | P |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 39.7 (33.1–41.1) | 39.7 (33.1–41.1) | 39.7 (33.1–41.1) | – | 40.5 (37.8–41.3) | 40.5 (37.8–41.3) | 40.5 (37.8–41.3) | – |
| Sex (female, %) | 44 | 44 | 44 | – | 50 | 50 | 50 | – |
| Height (cm) | 173.4 ± 10.9 | 173.5 ± 11.5 | 173.3 ± 10.7 | 0.477 | 167.4 ± 10.1 | 167.4 ± 11.1 | 167.4 ± 10.8 | 0.875 |
| BMI (kg/m2) | 30.1 ± 5.7 | 27.4 ± 4.2 | 32.8 ± 5.8 | <0.001 | 31.7 ± 6.4 | 31.5 ± 6.8 | 32.0 ± 6.9 | 0.269 |
| Weight (kg) | 90.7 ± 19.8 | 82.7 ± 17.2 | 98.6 ± 19.5 | <0.001 | 88.1 ± 13.5 | 87.2 ± 12.2 | 89.1 ± 16.6 | 0.625 |
| Waist-hip ratio | 0.96 ± 0.09 | 0.9 ± 0.1 | 1.0 ± 0.1 | <0.001 | 0.94 ± 0.04 | 0.9 ± 0.0 | 1.0 ± 0.0 | 0.250 |
| Body fat (%) | 37.1 ± 8.2 | 34.0 ± 7.8 | 40.2 ± 7.6 | <0.001 | 33.4 ± 12.8 | 34.1 ± 13.8 | 33.1 ± 14.8 | 1.000 |
| Visceral adipose tissue (cm3) | 2155 ± 878 | 1754 ± 786 | 2556 ± 799 | 0.001 | 2117 ± 693 | 2017 ± 497 | 2217 ± 921 | 0.875 |
| Subcutaneous adipose tissue (cm3) | 6461 ± 3001 | 5325 ± 2442 | 7597 ± 3155 | <0.001 | 7146 ± 4461 | 7297 ± 4799 | 6996 ± 4831 | 0.250 |
| Adipocyte cell number (per billion) | 301 (194–505) | 288 (173–383) | 357 (203–581) | 0.010 | 191 (173–267) | 233 (197–308) | 168 (147–204) | 0.125 |
| Adipocyte cell diameter (μM) | 93 ± 11 | 88 ± 10 | 97 ± 8.9 | 0.002 | 93 ± 12 | 89 ± 11 | 96 ± 12 | 0.250 |
| Adipocyte cell volume (pl) | 598 ± 201 | 508 ± 213 | 689 ± 145 | 0.007 | 559 ± 186 | 500 ± 154 | 617 ± 219 | 0.250 |
| Adipocyte cell weight (ng) | 583 (429–641) | 435 (306–572) | 627 (583–740) | 0.007 | 452 (365–674) | 407 (364–500) | 584 (427–722) | 0.250 |
|
PPAR γ , adipose tissue (au) |
0.95 ± 0.30 | 1.00 ± 0.34 | 0.91 ± 0.25 | 0.569 | ND | ND | ND | ND |
| Liver fat (%) | 1.7 (0.8–4.6) | 1.2 (0.5–2.0) | 2.3 (1.4–9.2) | 0.010 | 3.4 (1.2–8.7) | 5.2 (0.88–9.9) | 3.4 (2.0–5.4) | 0.625 |
| Lean tissue mass (kg) | 54.0 ± 10.2 | 52.0 ± 9.6 | 56.1 ± 10.5 | <0.001 | 57.5 ± 4.9 | 56.2 ± 5.5 | 58.8 ± 4.9 | 0.250 |
| Total bone mass (kg) | 2.9 ± 0.6 | 2.8 ± 0.6 | 2.9 ± 0.6 | <0.001 | 3.2 ± 0.5 | 3.2 ± 0.6 | 3.3 ± 0.5 | 0.500 |
| Basal metabolic rate (kcal/day) | 1874 ± 382 | 1781 ± 370 | 1968 ± 381 | <0.001 | 1798 ± 262 | 1782 ± 265 | 1813 ± 298 | 0.625 |
| Caloric intake (kcal/day) | 2297 ± 441 | 2244 ± 434 | 2349 ± 455 | 0.463 | 2003 ± 655 | 1872 ± 419 | 2134 ± 882 | 1.000 |
| Protein intake (g) | 96.2 (81.6–115) | 95.6 (84.5–115.3) | 96.3 (79.2–116.4) | 0.900 | 81.8 (65.3–103.5) | 81.5 (6.2–100.2) | 84.0 (65.7–110.1) | 0.625 |
| Fat intake (g) | 103.6 ± 28.5 | 97.7 ± 26.0 | 109.6 ± 30.4 | 0.562 | 78.4 ± 29.7 | 69.8 ± 17.5 | 87.1 ± 39.5 | 0.875 |
| Carbohydrate intake (g) | 211.5 ± 51.1 | 216.0 ± 50.0 | 207.0 ± 53.4 | 0.744 | 212.4 ± 68.2 | 234.9 ± 55.1 | 221.3 ± 93.2 | 0.875 |
| Niacin equivalent (mg/day) | 44.5 (36.1–49.9) | 45.4 (36.7–49.3) | 43.9 (33.5–52.9) | 0.706 | 33.0 (31.0–40.7) | 32.4 (30.6–36.5) | 35.8 (31.9–41.6) | 0.875 |
| Physical activity (total Baecke) | 7.8 ± 1.5 | 8.0 ± 1.7 | 7.7 ± 1.4 | 0.802 | 8.4 ± 1.4 | 8.1 ± 0.7 | 8.7 ± 1.9 | 0.750 |
| Alcohol consumption (doses/week) | 1.0 (0.0–2.0) | 0.9 (0–3.8) | 5.0 (1.8–9.3) | 0.009 | 1.0 (0.0–2.9) | 1.0 (0.5–1.4) | 2.0 (0.0–4.6) | 0.371 |
| Current smoker (number) | 7 | 4 | 3 | – | 3 | 2 | 1 | – |
| HbA1c (mmol/mol) | 34 (32–36) | 34 (32–35) | 35 (34–36) | 0.037 | 36 (33–37) | 35 (33–36) | 37 (35–38) | 1.000 |
| Fasting glucose (mM) | 5.6 (5.4–5.8) | 5.6 (5.3–5.7) | 5.7 (5.4–5.8) | 0.157 | 5.8 (5.1–5.8) | 5.4 (5.0–5.8) | 5.8 (5.6–5.8) | 0.375 |
| Fasting insulin (mlU/liter) | 6.0 (4.5–9.8) | 4.7 (3.8–6.7) | 7.9 (5.3–12.5) | 0.018 | 8.0 (3.9–12.9) | 7.7 (3.9–11.5) | 9.7 (4.1–14.9) | 0.375 |
| Fasting C-peptide (nM) | 0.51 (0.39–0.65) | 0.47 (0.39–0.60) | 0.56 (0.43–0.83) | 0.018 | 0.69 (0.44–1.1) | 0.69 (0.41–0.98) | 0.80 (0.44–1.2) | 0.125 |
| HOMA index | 1.5 (1.0–2.1) | 1.1 (0.94–1.6) | 2.0 (1.3–3.2) | 0.022 | 1.9 (0.96–3.4) | 1.8 (0.96–2.7) | 2.5 (1.0–3.8) | 0.250 |
| Matsuda index | 5.7 ± 2.5 | 6.8 ± 2.4 | 4.7 ± 2.1 | <0.001 | 8.2 ± 6.2 | 8.8 ± 6.9 | 7.5 ± 6.5 | 0.125 |
| Glucose AUC during OGTT | 15.7 ± 2.5 | 15.2 ± 2.5 | 16.2 ± 2.4 | 0.042 | 15.8 ± 3.6 | 14.8 ± 4.1 | 16.8 ± 3.3 | 0.125 |
| Insulin AUC during OGTT | 90 (72–135) | 85.3 (67.7–116.9) | 96.6 (78.4–175.2) | 0.006 | 95.5 (30.9–188.0) | 90.0 (30.3–161.3) | 115.2 (39.2–188.0) | 0.625 |
| Adiponectin (μg/ml) | 3.2 (2.1–4.3) | 3.2 (2.7–3.8) | 3.1 (1.6–5.2) | 0.454 | 3.4 (2.2–8.5) | 3.4 (2.8–6.8) | 5.2 (3.6–6.9) | 0.500 |
| Cholesterol (total, mM) | 4.7 ± 0.8 | 4.7 ± 0.7 | 4.7 ± 0.9 | 0.660 | 4.8 ± 0.6 | 4.7 ± 0.5 | 4.8 ± 0.8 | 0.875 |
| HDL (mM) | 1.3 (1.2–1.6) | 1.4 (1.2–1.5) | 1.3 (1.1–1.7) | 0.224 | 1.3 (1.2–1.5) | 1.3 (1.2–1.4) | 1.4 (1.2–1.6) | 0.625 |
| LDL (mM) | 3.0 ± 0.77 | 3.0 ± 0.71 | 3.1 ± 0.83 | 0.196 | 3.2 ± 0.41 | 3.1 ± 0.41 | 3.3 ± 0.45 | 0.586 |
| Triglycerides (mM) | 0.95 (0.71–1.2) | 0.81 (0.61–1.0) | 1.0 (0.73–1.3) | 0.118 | 1.0 (0.70–1.1) | 1.0 (0.88–1.0) | 0.92 (0.70–1.2) | 0.625 |
| FFA (μM) | 444 (376–551) | 415 (362–500) | 510 (428–700) | 0.542 | 398 (332–527) | 428 (325–531) | 394 (332–482) | 0.625 |
| Systolic BP (mmHg) | 133 ± 22 | 131 ± 24 | 135 ± 22 | 0.141 | 120 ± 12 | 114 ± 12 | 126 ± 7.8 | 0.098 |
| Diastolic BP (mmHg) | 83 ± 12 | 83 ± 12 | 84 ± 11 | 0.706 | 77 ± 8 | 77 ± 7.7 | 80 ± 5.9 | 0.197 |
| Pulse (per min) | 66.9 ± 10.7 | 64.5 ± 9.2 | 69.3 ± 11.8 | 0.084 | 61.9 ± 6.4 | 63.0 ± 8.0 | 60.8 ± 5.1 | 0.461 |
| Hs-CRP (mg/liter) | 1.9 (0.67–3.9) | 1.7 (0.61–3.0) | 2.3 (0.73–5.8) | 0.117 | 1.3 (0.83–5.4) | 1.6 (1.3–10.7) | 0.99 (0.55–3.3) | 0.250 |
| Total homocysteine (μM) | 11.0 (9.30–12.0) | 10.0 (9.55–12.0) | 11.0 (9.28–12.8) | 0.500 | 13.0 (11.5–13.8) | 13.0 (12.2–13.8) | 12.5 (11.5–15.0) | 1.000 |
| ALT (U/liter) | 22.0 (17.8–42.8) | 20.0 (16.5–33.5) | 33.0 (19.5–49.0) | 0.012 | 22.5 (19.8–32.0) | 23.5 (18.3–38.8) | 22.5 (21.5–28.3) | 0.581 |
| AST (U/liter) | 24.0 (21.8–31.0) | 24.0 (19.8–29.2) | 24.0 (22.8–31.0) | 0.139 | 26.5 (24.0–29.5) | 26.5 (24.8–30.0) | 26.5 (24.0–29.5) | 0.586 |
| Creatinine (μM) | 73.7 ± 12.9 | 73.2 ± 13.6 | 74.3 ± 12.7 | 0.569 | 76.6 ± 12.0 | 74.0 ± 13.2 | 79.3 ± 12.1 | 0.125 |
| Hemoglobin (g/liter) | 142 ± 9 | 142 ± 8 | 143 ± 9 | 0.659 | 136 ± 11 | 137 ± 10 | 136 ± 13 | 0.625 |
| Hematocrit (%) | 42.0 ± 2.2 | 41.9 ± 2.1 | 42.1 ± 2.4 | 0.680 | 40.6 ± 3.2 | 40.8 ± 2.9 | 40.5 ± 3.9 | 0.850 |
| Erythrocytes (×1012/liter) | 4.8 ± 0.3 | 4.8 ± 0.37 | 4.8 ± 0.31 | 0.103 | 4.5 ± 0.4 | 4.5 ± 0.4 | 4.6 ± 0.5 | 0.875 |
| MCV (fl) | 87.9 ± 3.5 | 88.4 ± 4.2 | 87.4 ± 2.6 | 0.162 | 89.0 ± 3.3 | 90.3 ± 2.1 | 87.8 ± 4.2 | 0.098 |
| MCH (pg) | 29.8 ± 1.3 | 29.9 ± 1.5 | 29.6 ± 1.2 | 0.182 | 30.1 ± 1.4 | 30.5 ± 1.0 | 29.8 ± 1.7 | 0.371 |
| MCHC (g/liter) | 338 (334–342) | 338 (334–342) | 337 (335–342) | 0.609 | 338 (335–335) | 338 (336–340) | 339 (335–343) | 0.345 |
| Leukocytes (×109/liter) | 6.4 ± 1.5 | 6.5 ± 1.8 | 6.3 ± 1.2 | 0.816 | 6.7 ± 2.3 | 7.3 ± 3.0 | 6.2 ± 1.7 | 0.423 |
| Thrombocytes (×109/liter) | 243 ± 55 | 244 ± 60 | 241 ± 52 | 0.860 | 247 ± 42 | 234 ± 29 | 260 ± 53 | 0.125 |
| fP-Fe (μM) | 19.0 ± 6.9 | 19.8 ± 7.6 | 18.1 ± 6.3 | 0.469 | 20.0 ± 7.3 | 20.0 ± 6.4 | 20.0 ± 9.1 | 0.875 |
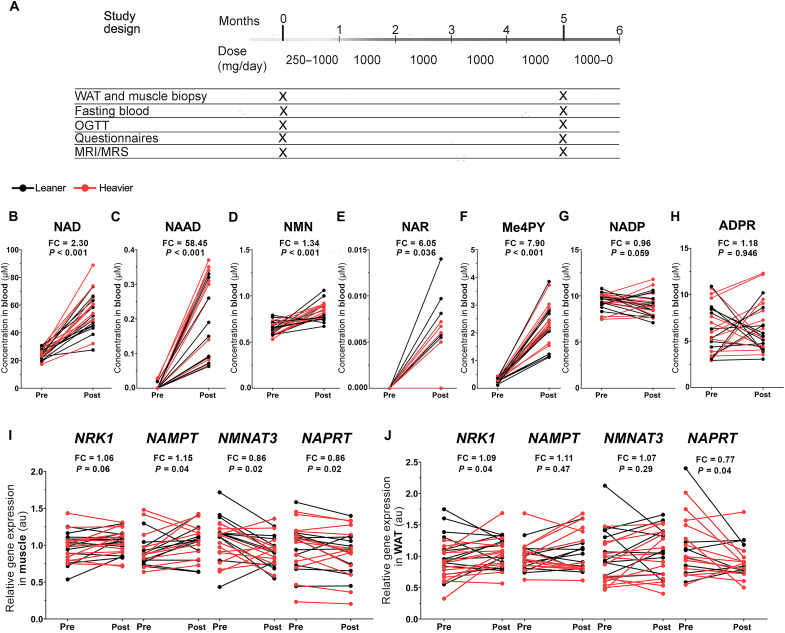
Fig. 1. NR increases whole-blood NAD+ levels and tissue NAD+ biosynthesis in the twins from the BMI-discordant pairs.
(A) Study protocol. The daily NR dose was gradually escalated from 250 mg/day by 250 mg/week to achieve the final treatment dose of 1 g/day. At the end of the study, the dose was decreased by 250 mg/week. Clinical examinations and collection of fasting blood samples and muscle and WAT biopsies were performed at baseline and after 5 months. (B to H) Whole-blood NAD metabolite levels before versus after NR (n = 14 twin pairs/28 individuals). (I and J) Expression of genes involved in NAD+ biosynthesis in muscle (I) and WAT (J) before versus after NR (n = 10 to 13 twin pairs/21 to 26 individuals). Arbitrary unit (au) indicates the relative target gene expression normalized to the expression of reference genes. Lines connect the pre- and post-values of each individual, with black denoting the leaner and red denoting the heavier cotwins. Fold change (FC) indicates the mean of the post-NR value divided by the pre-NR value. P values were calculated using paired Wilcoxon signed-rank test. See also Table 2 and figs. S1 and S2.
On average, both the BMI-discordant and BMI-concordant twin groups crossed the cutoff for obesity (BMI mean 30.1 ± SD 5.7 and 31.7 ± 6.4, respectively). The heavier cotwins of the BMI-discordant twin pairs (BMI 32.8 ± 5.8) had expectedly higher measures of adiposity, insulin resistance, basal metabolic rate, liver fat, and enzyme alanine aminotransferase (ALT) than their leaner cotwins (BMI 27.4 ± 4.2) (Table 1). The total intake of food or macronutrients or physical activity did not differ between the leaner and the heavier cotwins of the BMI-discordant pairs, but the heavier cotwins consumed more alcohol (Table 1). Both twins from the BMI-concordant pairs randomly assigned to receive either placebo (BMI 31.5 ± 6.8) or NR (BMI 32.0 ± 6.9) appeared very similar in their baseline characteristics (Table 1). Overall, most of the clinical values were within reference values in both twin-pair groups (Table 1).
NR increases whole-blood NAD+ concentrations and tissue NAD+ biosynthesis
To elucidate the compliance and the effect of NR on NAD+ metabolism in the twins from the BMI-discordant pairs, we analyzed NAD metabolome by targeted liquid chromatography–mass spectrometry on the whole-blood samples. At baseline, blood NAD metabolites did not differ between the leaner and the heavier cotwins (fig. S2, A to F). NR boosted whole-blood NAD+ levels by 2.3-fold (Fig. 1B) in all twins from the BMI-discordant pairs. In line, nicotinic acid adenine dinucleotide (NAAD), the validated biomarker for NR supplementation and an enhanced rate of NAD+ synthesis (14), was elevated upon NR (Fig. 1C). NR also significantly increased the levels of nicotinamide mononucleotide (NMN), the phosphorylated form of NR (Fig. 1D), and nicotinic acid riboside (NAR), the deamidated form of NR (Fig. 1E). N-methyl-4-pyridone-5-carboxamide (Me4py) was elevated by eightfold (Fig. 1F), suggesting an enhanced elimination of NR’s degradation product NAM via methylation. Of the other NAD metabolites, nicotinamide adenine dinucleotide phosphate (NADP) trended to decrease (Fig. 1G), while adenosine diphosphate ribose (ADPR) did not significantly change (Fig. 1H). The heavier and the leaner cotwins were similar in their response to NR (the delta changes, i.e., the changes from baseline to 5 months), except that the increase in Me4py levels was more pronounced in the heavier cotwins compared with the leaner cotwins (Table 2). Overall, NR was effectively metabolized, and it had a great potency to increase blood NAD metabolites.
Table 2. The delta response (the change from baseline to 5 months) to NR in the twins from the BMI-discordant pairs.
Data are shown as means ± SD (normally distributed variables) or median (interquartile range, for skewed variables). P values were obtained using paired Wilcoxon signed-rank test to examine the differences on delta change (the change from baseline to 5 months) between the leaner and the heavier cotwins from the BMI-discordant pairs (n = 16 twin pairs/32 individuals). 16S, 16S rRNA; CYTB, cytochrome b; DLOOP, D-loop region; NDUFB8, NADH:ubiquinone oxidoreductase subunit B8; MT-ND5, mitochondrially encoded NADH dehydrogenase 5; SDHB, succinate dehydrogenase complex iron sulfur subunit B; UQCRC2, ubiquinol–cytochrome c reductase core protein 2; MT-CO1, mitochondrially encoded cytochrome c oxidase 1; CLPP, caseinolytic mitochondrial matrix peptidase proteolytic subunit; HSPD1, heat shock protein family D (Hsp60) member 1; CPT1β, carnitine palmitoyltransferase 1B; PAX3, paired box 3; MYF5, myogenic factor 5.
| Leaner Δ (n = 3-16 individuals) | Heavier Δ (n = 3-16 individuals) | P | |
|---|---|---|---|
| NAD metabolites | |||
| NAD+, whole blood (μM) | 30.1 ± 20.0 | 33.4 ± 12.1 | 0.519 |
| NAAD, whole blood (μM) | 0.15 (0.08–0.34) | 0.26 (0.11–0.31) | 0.533 |
| NMN, whole blood (μM) | 0.12 ± 0.15 | 0.23 ± 0.08 | 0.139 |
| NAR, whole blood (μM) | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 1.000 |
| Me4py, whole blood (μM) | 1.6 ± 0.59 | 1.9 ± 0.46 | 0.014 |
| NADP, whole blood (μM) | −0.82 (−1.7 to −0.69) | −0.15 (−0.88 to 0.60) | 0.175 |
| ADPR, whole blood (μM) | −0.85 ± 3.1 | 0.42 ± 2.8 | 0.557 |
| Expression of NAD+ biosynthesis genes | |||
| NRK1, muscle (au) | 0.08 ± 0.14 | 0.04 ± 0.13 | 0.424 |
| NAMPT, muscle (au) | 0.13 (−0.04 to 0.29) | 0.19 (0.09–0.18) | 0.742 |
| NMNAT3, muscle (au) | −0.24 ± 0.27 | −0.06 ± 0.26 | 0.129 |
| NAPRT, muscle (au) | −0.10 ± 0.20 | −0.10 ± 0.21 | 1.000 |
| NRK1, adipose tissue (au) | 0.01 ± 0.28 | 0.19 ± 0.23 | 0.068 |
| NAMPT, adipose tissue (au) | 0.06 (−0.08 to 0.23) | −0.04 (−0.13 to 0.20) | 0.922 |
| NMNAT3, adipose tissue (au) | 0.01 (−0.25 to 0.24) | 0.09 (0.04–0.23) | 0.110 |
| NAPRT, adipose tissue (au) | −0.05 (−0.24 to 0.12) | −0.15 (−0.50 to −0.05) | 0.938 |
| Anthropometric and body composition | |||
| BMI (kg/m2) | 0.89 ± 1.5 | 1.0 ± 1.2 | 0.224 |
| Weight (kg) | 2.7 ± 4.3 | 2.6 ± 3.4 | 0.339 |
| Body fat (%) | 1.5 ± 2.6 | 0.62 ± 2.2 | 0.404 |
| Visceral adipose tissue (cm3) | 250 ± 307 | 101 ± 560 | 0.610 |
| Subcutaneous adipose tissue (cm3) | 683 ± 1240 | −46 ± 1295 | 1.000 |
| Adipose tissue (cm3) | 933 ± 1476 | 55 ± 1760 | 1.000 |
| Adipocyte cell number (per billion) | −123 (−150 to −4.0) | −6.2 (−337 to 95) | 0.599 |
| Adipocyte cell diameter (μM) | 4.5 ± 16 | 1.0 ± 17 | 0.454 |
| Adipocyte cell volume (pl) | 111 ± 356 | 110 ± 17 | 0.639 |
| Adipocyte cell weight (ng) | −6.3 (−113 to 186) | 38 (−159 to 365) | 0.639 |
| PPARγ, adipose tissue (au) | 0.13 ± 0.26 | 0.15 ± 0.22 | 0.765 |
| Waist-hip ratio | 0.02 ± 0.05 | 0.00 ± 0.03 | 0.211 |
| Liver fat (%) | 0.05 (−0.25 to 0.55) | −0.10 (−0.70 to 1.10) | 0.635 |
| Lean tissue mass (g) | 158 ± 1234 | 747 ± 1322 | 0.193 |
| Total bone mass (g) | 10.5 ± 32.2 | 6.6 ± 17.4 | 0.940 |
| Basal metabolic rate (kcal/day) | 30.8 ± 54.0 | 47.0 ± 44.4 | 0.313 |
| Caloric intake (kcal/day) | 20 ± 627 | −135 ± 687 | 0.562 |
| Niacin equivalent (mg/day) | −4.0 (−12.5 to 2.1) | 0.47 (−14.7 to 11.2) | 0.528 |
| Physical activity (total Baecke) | 0.0 ± 1.5 | −0.3 ± 0.6 | 0.401 |
| Alcohol intake (doses/week) | 0.49 (0.0–0.94) | −0.19 (−0.50 to 0.13) | 0.074 |
| Glucose homeostasis | |||
| HbA1c (mmol/mol) | 1.0 (0.0–1.0) | 0.0 (−2.0 to 1.5) | 0.037 |
| Fasting glucose (mM) | 0.10 (−0.10 to 0.20) | 0.20 (−0.10 to 0.33) | 0.089 |
| Fasting insulin (mlU/liter) | 1.2 (−0.1 to 4.1) | 1.5 (−1.2 to 3.6) | 0.670 |
| Fasting C-peptide (nM) | 0.05 (0.01–0.15) | 0.11 (−0.0 to 0.18) | 0.599 |
| HOMA index | 0.35 (−0.11 to 1.0) | 0.37 (−0.22 to 0.98) | 0.804 |
| Matsuda index | −1.4 ± 2.1 | −1.0 ± 1.8 | 0.305 |
| Glucose AUC during OGTT | 0.3 ± 2.2 | 0.6 ± 1.4 | 0.635 |
| Insulin AUC during OGTT | 14.8 (−8.8 to 30.7) | 11.0 (−1.6 to 64.8) | 0.635 |
| Adiponectin (ng/ml) | 17 (−111 to 516) | −105 (−796 to 364) | 0.151 |
| Cardiovascular health | |||
| Cholesterol (total, mM) | 0.03 ± 0.33 | 0.07 ± 0.32 | 0.889 |
| HDL (mM) | −0.08 (−0.17 to 0.03) | −0.05 (−0.18 to 0.08) | 0.231 |
| LDL (mM) | 0.03 ± 0.35 | −0.08 ± 0.59 | 0.551 |
| Triglycerides (mM) | 0.13 (−0.11 to 0.26) | 0.15 (−0.08 to 0.27) | 0.231 |
| FFA (μM) | −9.48 (−89.3 to 47.7) | 36.5 (−154 to 66.5) | 0.542 |
| Systolic BP (mmHg) | −4.0 ± 17.5 | −1.7 ± 17.5 | 0.615 |
| Diastolic BP (mmHg) | −0.5 ± 9.5 | 1.1 ± 13.9 | 0.624 |
| Pulse (per min) | 2.1 ± 9.1 | 0.14 ± 12.9 | 0.575 |
| Hs-CRP (mg/liter) | 0.13 (−0.39 to 0.65) | −0.07 (−0.43 to 0.71) | 0.391 |
| Total homocysteine (μM) | 2.0 (0.15–2.5) | 0.90 (−0.93 to 1.2) | 0.024 |
| Mitochondrial parameters | |||
| Mitochondrial number, muscle (number/muscle fiber area/10 μm2) | 12.6 ± 23.0 | 12.0 ± 29.9 | 1.000 |
| Mitochondrial area, muscle (relative area %) | 0.67 (0.01–1.12) | 0.91 (−0.33 to 2.2) | 1.000 |
| Mitochondrial perimeter (μm) | −0.02 ± 0.28 | 0.04 ± 0.35 | 0.966 |
| Mitochondrial diameter (μm) | 0.01 (−0.02 to 0.02) | −0.02 (−0.02 to 0.02) | 0.413 |
| Mitochondrial form factor | −0.09 (−0.17 to 0.25) | 0.07 (−0.06 to 0.27) | 0.365 |
| Mitochondrial aspect ratio | −0.05 (−0.25 to 0.26) | 0.09 (−0.08 to 0.24) | 0.240 |
| 16S, muscle (au) | 0.27 (0.25–0.53) | 0.30 (0.01–0.68) | 0.846 |
| CYTB, muscle (au) | 0.31 (0.16–0.37) | 0.18 (0.11–0.47) | 0.622 |
| DLOOP, muscle (au) | 0.29 ± 0.16 | 0.31 ± 0.24 | 0.922 |
| 16S, adipose tissue (au) | −0.09 ± 0.42 | −0.19 ± 0.40 | 0.542 |
| CYTB, adipose tissue (au) | −0.06 (−0.52 to 0.06) | −0.04 (−0.34 to 0.08) | 0.635 |
| DLOOP, adipose tissue (au) | −0.11 ± 0.41 | −0.11 ± 0.34 | 0.934 |
| SIRT1, muscle (au) | 0.15 (−0.27 to 0.32) | 0.48 (0.27–0.73) | 0.064 |
| SIRT3, muscle (au) | −0.28 (−0.94 to 0.25) | 0.15 (−0.09 to 0.23) | 0.054 |
| ERRα, muscle (au) | 0.15 ± 0.23 | 0.15 ± 0.19 | 0.413 |
| NRF1, muscle (au) | −0.08 ± 0.25 | −0.16 ± 0.32 | 0.547 |
| PGC1α, muscle (au) | −0.07 ± 0.35 | −0.04 ± 0.38 | 0.791 |
| TFAM, muscle (au) | 0.16 ± 0.34 | 0.28 ± 0.36 | 0.244 |
| MFN2, muscle (au) | 0.14 ± 0.15 | 0.06 ± 0.21 | 0.520 |
| NDUFB8, muscle (au) | −0.09 ± 0.25 | −0.19 ± 0.27 | 0.770 |
| MT-ND5, muscle (au) | −0.10 (−0.59 to 0.20) | −0.17 (−0.44 to 0.01) | 0.893 |
| SDHB, muscle (au) | −0.04 ± 0.24 | 0.12 ± 0.55 | 0.734 |
| UQCRC2, muscle (au) | 0.01 ± 0.19 | 0.02 ± 0.23 | 0.685 |
| COX4, muscle (au) | −0.07 (−0.18 to 0.16) | 0.05 (−0.14 to 0.26) | 0.938 |
| MT-CO1, muscle (au) | −0.06 ± 0.31 | −0.02 ± 0.42 | 0.820 |
| ATP5A, muscle (au) | 0.10 ± 0.22 | 0.14 ± 0.16 | 0.910 |
| CLPP, muscle (au) | −0.01 ± 0.17 | 0.04 ± 0.19 | 0.557 |
| HSPD1, muscle (au) | −0.04 ± 0.26 | 0.00 ± 0.25 | 0.733 |
| CPT1β, muscle (au) | −0.15 ± 0.63 | −0.03 ± 0.73 | 0.765 |
| ERRα, adipose tissue (au) | −0.07 ± 0.46 | −0.02 ± 0.27 | 0.339 |
| NRF1, adipose tissue (au) | 0.08 (−0.03 to 0.16) | 0.11 (−0.09 to 0.21) | 0.787 |
| TFAM, adipose tissue (au) | −0.02 ± 0.39 | −0.05 ± 0.26 | 1.000 |
| MFN2, adipose tissue (au) | −0.07 ± 0.21 | −0.05 ± 0.25 | 0.898 |
| NDUFB8, adipose tissue (au) | −0.30 (−0.52 to −0.07) | −0.36 (−0.42 to −0.11) | 0.588 |
| MT-ND5, adipose tissue (au) | −0.19 (−0.38 to 0.02) | −0.17 (−0.43 to 0.01) | 0.622 |
| SDHB, adipose tissue (au) | −0.05 (−0.25 to 0.09) | −0.07 (−0.13 to 0.11) | 0.742 |
| UQCRC2, adipose tissue (au) | 0.30 ± 0.38 | 0.16 ± 0.26 | 0.426 |
| COX4, adipose tissue (au) | 0.04 (−0.39 to 0.30) | −0.01 (−0.15 to 0.11) | 1.000 |
| MT-CO1, adipose tissue (au) | 0.02 (−0.14 to 0.11) | −0.13 (−0.30 to −0.17) | 0.734 |
| ATP5A, adipose tissue (au) | −0.04 ± 0.32 | 0.06 ± 0.23 | 0.365 |
| CLPP, adipose tissue (au) | −0.05 (−0.28 to 0.08) | −0.02 (−0.13 to 0.08) | 0.359 |
| HSPD1, adipose tissue (au) | 0.03 (−0.05 to 0.14) | −0.01 (−0.07 to 0.17) | 0.320 |
| Satellite cells | |||
| PAX7, muscle (au) | −0.31 ± 0.30 | −0.26 ± 0.45 | 0.688 |
| PAX7+ cells (PAX7+ cells/muscle fiber area) | −6.4 ± 3.6 | −3.9 ± 2.6 | 0.578 |
| PAX7/MYOG (ratio) | −0.63 ± 0.75 | −1.1 ± 0.22 | 0.500 |
| PAX3/MYOG (ratio) | −934 (−5960 to −603) | −2420 (−5370 to −1270) | 0.750 |
| MYF5/MYOD (ratio) | −0.03 (−0.52 to 0.32) | 0.19 (0.01–1.52) | 0.500 |
| MYMK/MYOD (ratio) | 1000 (685–2340) | −319 (−508 to 1630) | 0.250 |
To investigate the influence of NR on tissue NAD+ biosynthesis, we measured mRNA expression of NAD+ biosynthetic enzymes from muscle and WAT. At baseline, in comparison to the leaner cotwins of the BMI-discordant pairs, the heavier cotwins exhibited significantly lower expression of NMN adenylyltransferase 3 (NMNAT3), one of the NAMN/NMN adenylyltransferase isoforms, in muscle and WAT (fig. S2, G and H). The expression of the NR-metabolizing enzyme NR kinase 1 (NRK1) was also lower in WAT of the heavier cotwins (fig. S2H). In the twins from the BMI-discordant pairs, NR increased the muscle expression of NAM phosphoribosyltransferase (NAMPT), the enzyme converting NAM toward NAD+, and trended to up-regulate NRK1 while decreasing NMNAT3 (Fig. 1I). In WAT, NRK1 expression was significantly elevated, while NAMPT and NMNAT3 were unchanged upon NR (Fig. 1J). NR down-regulated the Preiss-Handler pathway enzyme nicotinic acid phosphoribosyltransferase (NAPRT) in both muscle and WAT (Fig. 1, I and J), indicating decreased niacin utilization and/or increased reliance on the salvage pathway enzymes upon NR. The NR-induced alterations in the mRNA expression of NAD+ biosynthetic enzymes were similar in both cotwins, except that NR tended to increase WAT NRK1 expression more in the heavier cotwins compared with the leaner cotwins (Table 2). As a whole, NR promoted both muscle and WAT NAD+ biosynthesis via the salvage pathways.
Body weight and fat percentage increased during the study
We next determined the impact of NR on body composition in the twins from the BMI-discordant pairs. During the study, body weight (~3 kg) and the whole-body fat percentage increased significantly, and the size of all WAT depots tended to elevate in twins from the BMI-discordant pairs (Table 3). There were no significant changes in adipocyte number, diameter, volume, and weight upon NR (Table 3). However, the expression of peroxisome proliferator–activated receptor γ (PPARγ), the essential transcription factor controlling adipogenesis, was up-regulated after NR supplementation (Table 3). Lean tissue (muscle) and bone mass and liver fat content remained unaltered (Table 3). Basal metabolic rate was significantly enhanced probably due to the increase in body weight (Table 3). Food diaries and questionnaires showed no marked changes in the intake of food, macronutrients, niacin equivalent, alcohol, or physical activity (Table 3). The changes in the analyzed parameters from baseline to 5 months did not differ between the leaner and the heavier cotwins (Table 2). Together, body weight and fat percentage increased during the NR intervention, but we did not observe clear changes in the self-reported lifestyle measures.
Table 3. The effect of NR on body composition and lifestyle factors in the twins from the BMI-discordant pairs.
Data are shown as means ± SD (normally distributed variables), median (interquartile range, for skewed variables), or proportion (%, for categorical variables). P values were obtained using paired Wilcoxon signed-rank test to examine the effect of NR supplementation on anthropometric and clinical parameters in the twins from the BMI-discordant pairs (n = 16 twin pairs/32 individuals).
| Variable (unit) | All Pre (n = 32 individuals) | All Post (n = 32 individuals) | P | Leaner Pre (n = 16 individuals) | Leaner Post (n = 16 individuals) | P | Heavier Pre (n = 16 individuals) | Heavier Post (n = 16 individuals) | P |
|---|---|---|---|---|---|---|---|---|---|
| BMI (kg/m2) | 30.1 ± 5.7 | 31.1 ± 6.1 | 0.001 | 27.4 ± 4.2 | 28.3 ± 4.9 | 0.038 | 32.8 ± 5.8 | 33.8 ± 6.1 | 0.007 |
| Weight (kg) | 90.7 ± 19.8 | 93.3 ± 20.4 | <0.001 | 82.7 ± 17.2 | 85.4 ± 18.6 | 0.010 | 98.6 ± 19.5 | 101.2 ± 19.6 | 0.010 |
| Waist-hip ratio | 0.96 ± 0.09 | 0.97 ± 0.09 | 0.172 | 0.94 ± 0.08 | 0.96 ± 0.08 | 0.065 | 0.98 ± 0.09 | 0.98 ± 0.10 | 0.918 |
| Body fat (%) | 37.1 ± 8.2 | 38.2 ± 8.2 | 0.008 | 34.0 ± 7.8 | 35.5 ± 8.1 | 0.016 | 40.2 ± 7.6 | 40.9 ± 7.6 | 0.117 |
| Visceral adipose tissue (cm3) | 2155 ± 878 | 2440 ± 926 | 0.014 | 1754 ± 785 | 2097 ± 724 | 0.014 | 2555 ± 799 | 2811 ± 1005 | 0.541 |
| Subcutaneous adipose tissue (cm3) | 6461 ± 3001 | 7049 ± 3027 | 0.061 | 5325 ± 2442 | 6179 ± 2891 | 0.056 | 7597 ± 3155 | 7990 ± 3001 | 0.610 |
| Adipocyte cell number (per billion) | 301 (194–505) | 230 (140–374) | 0.092 | 288 (173–383) | 211 (128–265) | 0.169 | 357 (203–581) | 352 (202–471) | 0.454 |
| Adipocyte cell diameter (μM) | 93 ± 11 | 95.4 ± 16.4 | 0.440 | 88 ± 10 | 92.4 ± 16.5 | 0.524 | 97 ± 8.9 | 98.4 ± 16.3 | 0.804 |
| Adipocyte cell volume (pl) | 598 ± 201 | 709 ± 381 | 0.328 | 508 ± 213 | 618 ± 350 | 0.639 | 689 ± 145 | 799 ± 401 | 0.489 |
| Adipocyte cell weight (ng) | 583 (429–641) | 573 (379–833) | 0.328 | 435 (306–572) | 462 (358–659) | 0.639 | 627 (583–740) | 614 (459–954) | 0.489 |
| PPARγ, adipose tissue (au) | 0.95 ± 0.30 | 1.1 ± 0.23 | 0.025 | 1.00 ± 0.34 | 1.2 ± 0.23 | 0.206 | 0.91 ± 0.25 | 1.1 ± 0.23 | 0.092 |
| Liver fat (%) | 1.7 (0.8–4.6) | 2.1 (0.6–5.4) | 0.657 | 1.2 (0.5–2.0) | 1.9 (0.4–2.4) | 0.638 | 2.3 (1.4–9.2) | 2.2 (0.7–10.6) | 0.787 |
| Lean tissue mass (kg) | 54.0 ± 10.2 | 54.5 ± 10.5 | 0.197 | 52.0 ± 9.6 | 52.1 ± 9.6 | 0.980 | 56.1 ± 10.5 | 56.8 ± 11.1 | 0.083 |
| Total bone mass (kg) | 2.9 ± 0.6 | 2.9 ± 0.6 | 0.106 | 2.8 ± 0.6 | 2.8 ± 0.6 | 0.495 | 2.9 ± 0.6 | 2.9 ± 0.6 | 0.130 |
| Basal metabolic rate (kcal/day) | 1874 ± 382 | 1913 ± 390 | <0.001 | 1781 ± 370 | 1812 ± 377 | 0.016 | 1968 ± 381 | 2015 ± 387 | 0.003 |
| Caloric intake (kcal/day) | 2297 ± 441 | 2240 ± 469 | 0.719 | 2244 ± 434 | 2265 ± 525 | 0.900 | 2350 ± 456 | 2215 ± 421 | 0.597 |
| Protein intake (g) | 96.2 (81.6–115) | 87.5 (78.1–108) | 0.454 | 95.6 (84.5–115) | 86.6 (76.5–109) | 0.782 | 96.3 (79.2–116) | 88.4 (81.7–106) | 0.562 |
| Fat intake (g) | 103.6 ± 28.5 | 98.5 ± 22.6 | 0.405 | 97.7 ± 26.0 | 98.3 ± 27.8 | 0.816 | 109.6 ± 30.4 | 98.7 ± 16.7 | 0.211 |
| Carbohydrate intake (g) | 211.5 ± 51.1 | 210.0 ± 57.9 | 0.789 | 216.0 ± 50.0 | 216.4 ± 54.9 | 0.980 | 207.0 ± 53.4 | 203.6 ± 61.8 | 0.597 |
| Niacin equivalent (mg/day) | 44.5 (36.1–49.9) | 40.1 (35.4–49.8) | 0.295 | 45.4 (36.7–49.3) | 38.2 (30.8–50.7) | 0.144 | 43.9 (33.5–52.9) | 42.1 (37.0–49.8) | 0.782 |
| Physical activity (total Baecke) | 7.8 ± 1.5 | 7.6 ± 1.4 | 0.562 | 8.0 ± 1.7 | 8.0 ± 1.5 | 0.552 | 7.7 ± 1.4 | 7.3 ± 1.2 | 0.090 |
| Alcohol intake (doses/week) | 1.0 (0.0–2.0) | 2.1 (0.2–5.0) | 0.345 | 0.9 (0.0–3.8) | 2.1 (0.7–4.8) | 0.147 | 5.0 (1.8–9.3) | 2.0 (0.0–5.6) | 0.551 |
| Current smoker (number) | 7 | 7 | – | 4 | 4 | – | 3 | 3 | – |
Insulin sensitivity decreased during the study
To understand whether the increased body weight resulted in alterations in glucose homeostasis in the twins from the BMI-discordant pairs, we ran a comprehensive set of measures during an oral glucose tolerance test. Glucose, insulin, C-peptide, homeostasis model assessment, and Matsuda index indicated impairments in glucose homeostasis (although remaining within normal reference ranges) in all twins after NR supplementation (Table 4). No significant differences were observed regarding the changes from baseline to 5 months between the leaner and the heavier cotwins, except that hemoglobin A1c increased slightly more in the leaner cotwins than in the heavier cotwins (Table 2). Overall, our data suggest that insulin sensitivity decreased during the study.
Table 4. Glucose homeostasis in the twins from the BMI-discordant pairs upon NR supplementation.
Data are shown as means ± SD (normally distributed variables) or median (interquartile range, for skewed variables). P values were obtained using paired Wilcoxon signed-rank test to examine the effect of NR supplementation on glucose metabolism related parameters in the twins from the BMI-discordant pairs (n = 16 twin pairs/32 individuals).
| Variable (unit) | All Pre (n = 32 individuals) | All Post (n = 32 individuals) | P | Leaner Pre (n = 16 individuals) | Leaner Post (n = 16 individuals) | P | Heavier Pre (n = 16 individuals) | Heavier Post (n = 16 individuals) | P |
|---|---|---|---|---|---|---|---|---|---|
| HbA1c (mmol/mol) | 34 (32–36) | 35 (33–36) | 0.353 | 34 (32–35) | 35 (33–36) | 0.032 | 35 (34–36) | 35 (32–37) | 0.693 |
| Fasting glucose (mM) | 5.6 (5.4–5.8) | 5.7 (5.5–5.9) | 0.027 | 5.6 (5.3–5.7) | 5.7 (5.3–5.8) | 0.483 | 5.7 (5.4–5.8) | 5.8 (5.6–5.9) | 0.018 |
| Fasting insulin (mlU/liter) | 6.0 (4.5–9.8) | 7.7 (6.2–13.4) | 0.023 | 4.7 (3.8–6.7) | 7.0 (5.9–9.0) | 0.069 | 7.9 (5.3–12.5) | 12.1 (7.1–14.6) | 0.169 |
| Fasting C-peptide (nM) | 0.51 (0.39–0.65) | 0.63 (0.50–0.77) | 0.002 | 0.47 (0.39–0.60) | 0.55 (0.48–0.65) | 0.022 | 0.56 (0.43–0.83) | 0.74 (0.59–0.84) | 0.036 |
| HOMA index | 1.5 (1.0–2.1) | 2.0 (1.5–3.3) | 0.016 | 1.1 (0.9–1.6) | 1.7 (1.4–2.4) | 0.073 | 2.0 (1.3–3.2) | 3.0 (1.7–3.8) | 0.135 |
| Matsuda index | 5.7 ± 2.5 | 4.4 ± 1.9 | <0.001 | 6.8 ± 2.4 | 5.3 ± 2.0 | 0.068 | 4.7 ± 2.1 | 3.6 ± 1.4 | 0.042 |
| Glucose AUC during OGTT | 15.7 ± 2.5 | 16.1 ± 2.7 | <0.001 | 15.2 ± 2.5 | 15.4 ± 2.6 | 0.286 | 16.2 ± 2.4 | 16.9 ± 2.8 | 0.208 |
| Insulin AUC during OGTT | 90 (72–135) | 116 (88–160) | <0.001 | 85 (68–117) | 112 (86–127) | 0.217 | 97 (78–175) | 154 (91–204) | 0.173 |
| Adiponectin (μg/ml) | 3.2 (2.1–4.3) | 3.2 (2.0–4.1) | 0.946 | 3.2 (2.7–3.8) | 3.3 (2.7–3.9) | 0.495 | 3.1 (1.6–5.2) | 2.8 (1.7–4.4) | 0.277 |
Next, we examined whether cardiovascular health was affected by NR. Except for the small, clinically not meaningful decrease in high-density lipoprotein and triglycerides, no changes in blood lipid values were detected upon NR (table S1). Neither were there any changes in blood pressure, pulse rate, nor the inflammation marker high-sensitivity complement-reactive protein (CRP) (table S1). The NR’s effect on these clinical variables was similar in the leaner and the heavier cotwins (Table 2).
NR boosts mitochondrial biogenesis in muscle but not in WAT
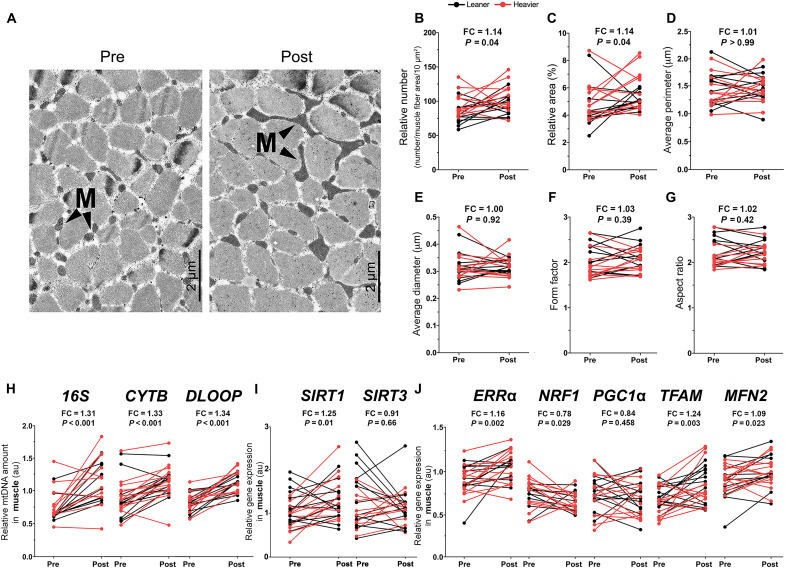
Given that NR significantly elevated whole-blood and tissue NAD+ biosynthesis in the twins from the BMI-discordant pairs, we determined the effect of NR on mitochondrial biogenesis in these twins. Transmission electron micrographs (TEMs) of muscle samples showed that mitochondria became more abundant (~14%) in the intermyofibrillar region of type I muscle fibers upon NR (Fig. 2, A and B). In addition, mitochondria covered a larger cross-sectional area of the muscle fiber area after NR supplementation (Fig. 2C). Morphological analysis of mitochondria [perimeter, diameter, form (i.e., branching) factor, or aspect (i.e., length-to-width) ratio] did not reveal any alterations upon NR (Fig. 2, D to G). In line with TEM findings, NR clearly elevated (~30%) muscle mitochondrial DNA (mtDNA) amount (Fig. 2H) and muscle expression of the following transcription factors regulating mitochondrial biogenesis: sirtuin 1 (SIRT1), estrogen-related receptor α (ERRα), transcription factor A (TFAM), and mitofusin 2 (MFN2) (Fig. 2I). In addition, NR significantly elevated the expression of muscle oxidative phosphorylation (OXPHOS) complex subunits cytochrome c oxidase subunit 4 and adenosine triphosphate synthase α (ATP5A) but down-regulated one complex I subunit (fig. S3, A and B). The leaner and the heavier cotwins were similar in their response to NR in terms of their muscle mitochondrial number, mtDNA amount, and gene expression levels except that the changes in the expression of SIRT1 and sirtuin 3 tended to differ between the heavier and the leaner cotwins (Table 2). Collectively, NR markedly increased muscle biogenesis, which was associated with the up-regulation of SIRT1/ERRα/TFAM/MFN2.
Fig. 2. NR boosts muscle mitochondrial biogenesis in the twins from the BMI-discordant pairs.
(A) TEM images of muscle intermyofibrillar mitochondria (M) in one representative study participant before and after NR. Magnification, ×2000. Scale bars, 2 μm. (B to G) Quantification of (B) the number of mitochondria per 10 μm2 relative to the muscle fiber area, (C) percentage of mitochondrial surface area per total muscle fiber area, (D) average perimeter of mitochondria, (E) average diameter of mitochondria, (F) form factor (the branching of mitochondria), and (G) aspect ratio (the length-to-width ratio of mitochondria) in muscle biopsies (n = 11 twin pairs/22 individuals). (H) Muscle relative mtDNA amount presented as a ratio of mtDNA genome per nuclear genome before versus after NR (n = 11 to 12 twin pairs/22 to 25 individuals). 16S, 16S rRNA; CYTB, cytochrome b; DLOOP, D-loop region. (I and J) Gene expression of SIRT1 and SIRT3 (I) and other key regulators of mitochondrial biogenesis (J) in muscle before versus after NR (n = 10 to 12 twin pairs/21 to 28 individuals). Arbitrary unit indicates the relative gene expression normalized to the expression of reference genes. SIRT3, sirtuin 3. Lines connect the pre- and post-values of each individual, with black denoting the leaner and red denoting the heavier cotwins. Fold change indicates the mean of the post-NR value divided by the pre-NR value. P values were calculated using paired Wilcoxon signed-rank test. See also Table 2 and fig. S3.
We next investigated the effect of NR on WAT mitochondria in the twins from the BMI-discordant pairs. NR did not significantly influence mtDNA amount and had only a minor effect on the gene expression profile in WAT (fig. S3, C to F). The changes in these parameters did not differ between the leaner and the heavier cotwins (Table 2).
NR promotes muscle satellite cell differentiation
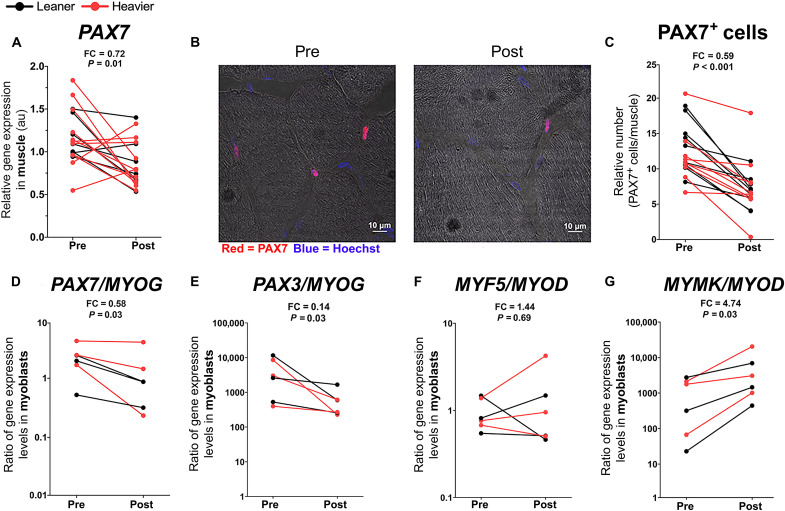
As NR has been previously shown to increase the number and the function of muscle stem cells, i.e., satellite cells in mice (10), we investigated the effect of NR on muscle satellite cells in the twins from the BMI-discordant pairs. Muscle expression of the stem cell marker paired box 7 (PAX7) and the number of PAX7+ satellite cells were significantly reduced upon NR (Fig. 3, A to C). To further characterize the muscle stem cells, we created primary myoblast cultures from the muscle biopsies. Myoblasts derived from the NR-supplemented twins showed reduced stemness and increased differentiation, as demonstrated by a significant decrease in the expression ratio of stemness markers PAX7 and paired box 3 over the late myogenic differentiation marker myogenin (MYOG) (Fig. 3, D and E). However, the expression ratio of other myogenic regulatory factors, myogenic factor 5 and myoblast determination protein 1 (MYOD), remained unaltered upon NR (Fig. 3F). The reduction in the muscle PAX7+ satellite cell number coupled with the myoblasts’ phenotypic shift toward differentiation suggested that NR may activate satellite cells and their fusion to muscle fibers (23). In human myoblasts, MYOD initiates the fusogenic program, for example, by up-regulating the expression of myomaker (MYMK), the essential myogenic fusion factor (24). Given that myoblasts derived from the NR-supplemented twins exhibited a significantly elevated expression ratio of MYMK and MYOD (Fig. 3G), our results suggest that NR likely drives the human myoblast fusion. NR’s effect on muscle PAX7 expression, satellite cell number, or myoblast PAX7/MYOG, paired box 3/MYOG, myogenic factor 5/MYOD, and MYMK/MYOD ratios was similar in both cotwins (Table 2). Overall, our data indicate that NR likely facilitated the activation of satellite cells towards differentiation and fusion to the existing myofibers, resulting in the observed decline in the satellite cell number in the twins from the BMI-discordant pairs.
Fig. 3. NR promotes muscle satellite cell differentiation in the twins from the BMI-discordant pairs.
(A) Muscle gene expression level of satellite cell marker PAX7 before versus after NR (n = 9 twin pairs/18 individuals). (B) Immunostaining of PAX7+ satellite cells in muscle cryosections before versus after NR in one representative study participant. PAX7 (red, satellite cells); Hoechst (blue, nuclei). Scale bars, 10 μm. (C) Muscle PAX7+ satellite cell quantification before versus after NR (n = 10 twin pairs/20 individuals). (D to G) Ratios of PAX7/MYOG (D), PAX3/MYOG (E), MYF5/MYOD (F), and MYMK/MYOD (G) mRNA expression in myoblasts before versus after NR (n = 3 twin pairs/6 individuals). Y axis is on a logarithmic scale. PAX3, paired box 3; MYF5, myogenic factor 5. Lines connect the pre- and post-values of each individual, with black denoting the leaner and red denoting the heavier cotwins. Fold change indicates the mean of the post-NR value divided by the pre-NR value. P values were calculated using paired Wilcoxon signed-rank test. See also Table 2.
NR may affect the epigenetic control of gene expression in muscle and WAT
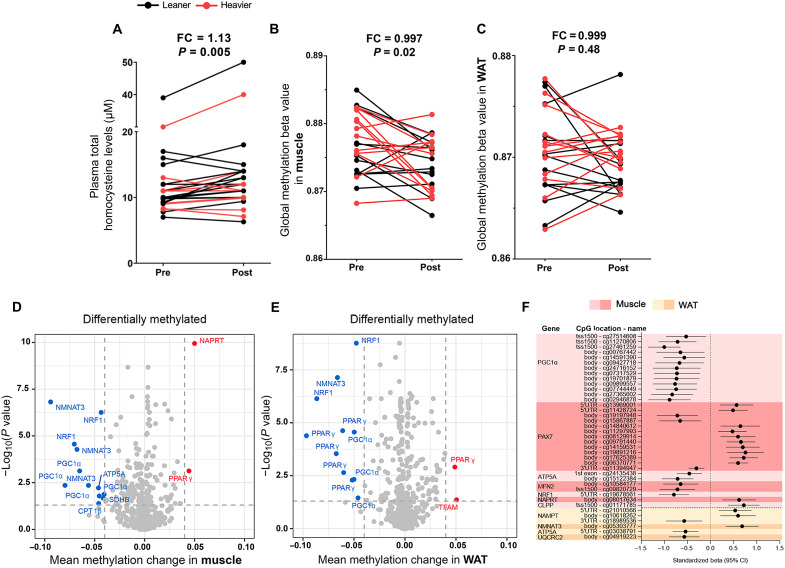
The supplementation with NAD+ precursors has been suggested to induce a decline in methyl groups due to enhanced elimination of NAM via methylation (25). Thus, we measured the levels of circulating homocysteine, increased levels of which can be considered as a marker for compromised cellular methylation status. NR slightly but significantly increased the levels of total plasma homocysteine, especially in the leaner cotwins from the BMI discordant pairs (Fig. 4A, Table 2, and table S1), but not beyond the normal range, suggesting a reduction in the cellular methylation capacity. These findings prompted us to investigate whether NR affected tissue DNA methylation. NR significantly reduced global DNA methylation level in muscle, but not in WAT (Fig. 4, B and C), with similar effects in both cotwins (median within-pair difference of 0.001, P = 0.47 in muscle, and −0.001, P = 0.52 in WAT). Overall, our results suggest that long-term NR supplementation influences global DNA methylation in a tissue-specific manner.
Fig. 4. NR modifies the epigenetic control of gene expression in the twins from the BMI-discordant pairs.
(A to C) Plasma total homocysteine (A) and global DNA methylation levels in (B) muscle and (C) WAT before versus after NR (n = 12 to 13 twin pairs/24 to 26 individuals). Lines connect the pre- and post-values of each individual, with black denoting the leaner and red denoting the heavier cotwins. Fold change indicates the mean of the post-NR value divided by the pre-NR value. (D and E) Volcano plots showing statistical significance (y axis) and the magnitude of the change in mean methylation beta value (x axis) in (D) muscle and (E) WAT upon NR (n = 12 to 14 twin pairs/25 to 28 individuals). Each dot represents a single CpG site (n = 619 and 518 in muscle and WAT, respectively). Highlighted are CpGs with FDR P < 0.05 (dashed horizontal line). CPT1β, carnitine palmitoyltransferase 1β; SDHB, succinate dehydrogenase complex iron sulfur subunit B. (F) Significant associations between CpG site methylation and gene expression in muscle (red) and WAT (yellow) upon NR (n = 7 to 9 twin pairs/14 to 18 individuals). Standardized beta values show the SD values in methylation changes associated with 1 SD of gene expression change. CI, confidence interval; CLPP, caseinolytic mitochondrial matrix peptidase proteolytic subunit; UQCRC2, ubiquinol–cytochrome c reductase core protein 2. P values for plasma total homocysteine levels were calculated using paired Wilcoxon signed-rank test. Statistical analyses of methylation data are described in Materials and Methods. See also Table 2 and tables S1 to S3.
Next, we performed differential DNA methylation analysis for selected genes involved in NAD+ biosynthesis, mitochondrial biogenesis, protein quality control, fatty acid oxidation, OXPHOS, and satellite cell identity to determine the effect of NR on muscle and WAT CpG methylation in the twins from the BMI-discordant pairs. NR altered the methylation of 173 of 619 (28%) and 210 of 518 (41%) CpG sites in muscle and WAT, respectively (Fig. 4, D and E). While NR induced both hypo- and hypermethylation, most of the analyzed CpG sites were hypomethylated (~60% in both tissues) after NR. The hypomethylated CpGs were typically located at the open sea, whereas the hypermethylated sites were mostly observed at CpG islands. Most of the hypomethylated CpGs were detected in PPARγ coactivator 1α (PGC-1α), NMNAT3, and nuclear respiratory factor 1 (NRF1) in muscle and in PPARγ and NRF1 in WAT. The leaner and the heavier cotwins did not differ in their response to NR at their tissue CpG site methylation (table S2).
As DNA methylation may influence gene expression, we assessed associations between gene expression and CpG methylation of the respective genes. In muscle, a total of 33 statistically significant associations were observed (table S3). All 13 associations observed between PGC-1α expression and methylation were negative (Fig. 4F), suggesting that hypomethylation of PGC-1α may increase its transcription in muscle. In addition, 13 significant, mostly positive associations were detected between expression and methylation of PAX7. Thus, hypomethylation of PAX7 may repress its expression in muscle. Other detected associations between gene expression and DNA methylation were represented by only up to two CpG sites at each of the genes, and the correlations were negative for ATP5A, MFN2, and NRF1 and positive for NAPRT and caseinolytic mitochondrial matrix peptidase proteolytic subunit (Fig. 4F). In WAT, there were only six associations between the gene expression and CpG site methylation at the respective genes, three of which were annotated to NAMPT and one CpG to each of the other three genes NMNAT3, ATP5A, and ubiquinol–cytochrome c reductase core protein 2 (Fig. 4F). Although it is difficult to draw any firm conclusions on the effects of DNA methylation on gene expression, these results suggest that epigenetic control could be one of the mechanisms via which NR regulates the expression of genes participating in mitochondrial biogenesis and quality control, satellite cell stemness, NAD+ biosynthesis, and OXPHOS.
NR alters plasma metabolomic profile
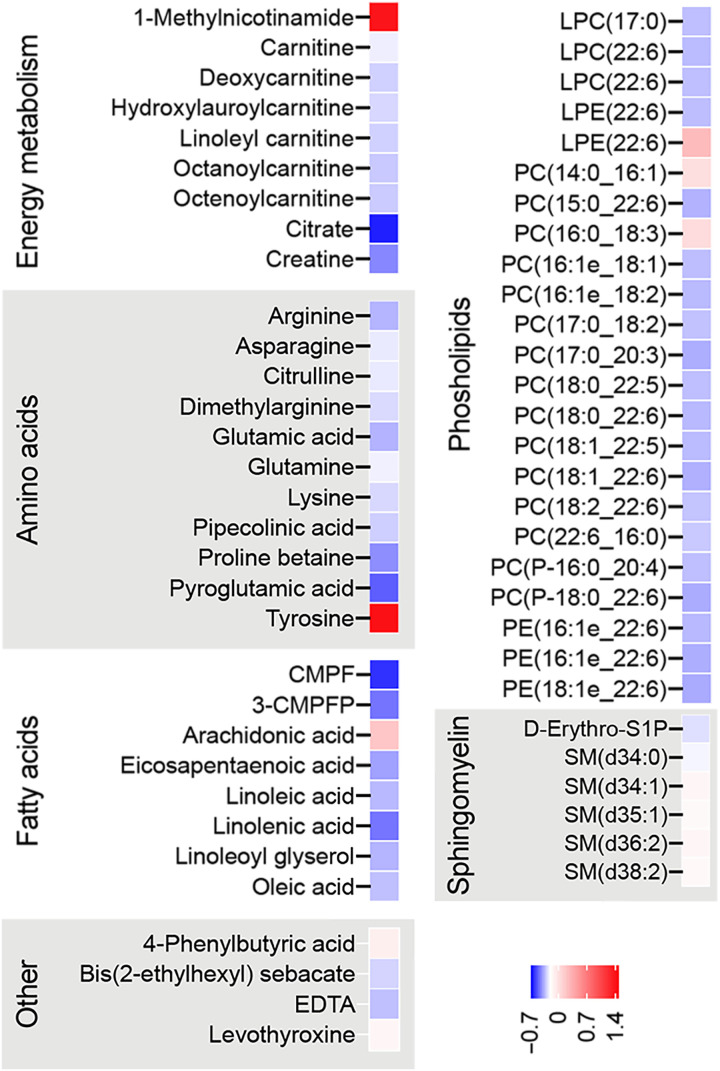
To understand whether NR affects the global plasma metabolomic profile, we performed a nontargeted metabolomic analysis of fasting plasma samples from the twins of the BMI-discordant pairs. With regression analysis, we found 460 significantly altered metabolites upon NR. Among these, 72 metabolites were identified with a level 1 or 2 in the Metabolomics Standard Initiative reporting standards (table S4). Of the 72 identified metabolites, the most significantly increased metabolite was methylNAM (Fig. 5), the NAM waste product, in line with the whole-blood NAD metabolome (Fig. 1F) and global DNA methylation results (Fig. 4, A and B). In addition, the levels of tyrosine, the amino acid precursor of brain catecholamines, were significantly elevated upon NR (Fig. 5). Most of the decreased metabolites fell into the following categories: energy metabolism, amino acids, fatty acids, phospholipids, and sphingomyelins. The content of metabolites central for the efficient mitochondrial energy production—carnitine, acylcarnitines, and polyunsaturated fatty acids—decreased significantly (Fig. 5), suggesting their lowered synthesis/excretion and/or increased uptake/utilization by the tissues. NR decreased the plasma levels of several amino acids including gluconeogenic amino acids (Fig. 5), indicating also their diminished synthesis/excretion and/or increased uptake/utilization by the tissues such as the liver, which is the main tissue responsible for amino acid metabolism in the postprandial state. Given that the plasma levels of 3-carboxy-4-methyl-5-propyl-2-furanopropionic acid, a putative biomarker of fatty fish intake (26), were significantly decreased (Fig. 5), we cannot exclude the possibility that the lowered levels of polyunsaturated fatty acids partially reflected a decline in fish intake. The decrease in phosphatidylcholines and 1,5-diaminonaphthalene was more pronounced in the heavier cotwins, but otherwise, the metabolite responses were similar in both cotwins (on the 1 or 2 metabolite identification level; table S5). Overall, our results revealed that the main plasma metabolites modulated by NR supplementation are related to mitochondrial energy metabolism, lipids, and amino acids.
Fig. 5. NR alters plasma metabolomic profile in the twins from the BMI-discordant pairs.
Heatmaps showing significant plasma metabolite log2-fold changes after NR (n = 14 twin pairs/28 individuals; nominal P < 0.05). Blue color indicates lower and red color indicates higher metabolite level upon NR. CMPF, 3-carboxy-4-methyl-5-propyl-2-furanopropionic acid; 3-CMPFP, 3-carboxy-4-methyl-5-pentyl-2-furanpropanoic acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidyl-ethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; D-Erythro-S1P, d-erythro-sphingosine-1-phosphate; SM, sphingomyelin. Related statistical analyses are described in Materials and Methods. See also tables S4 and S5.
NR modulated the gut microbiota composition
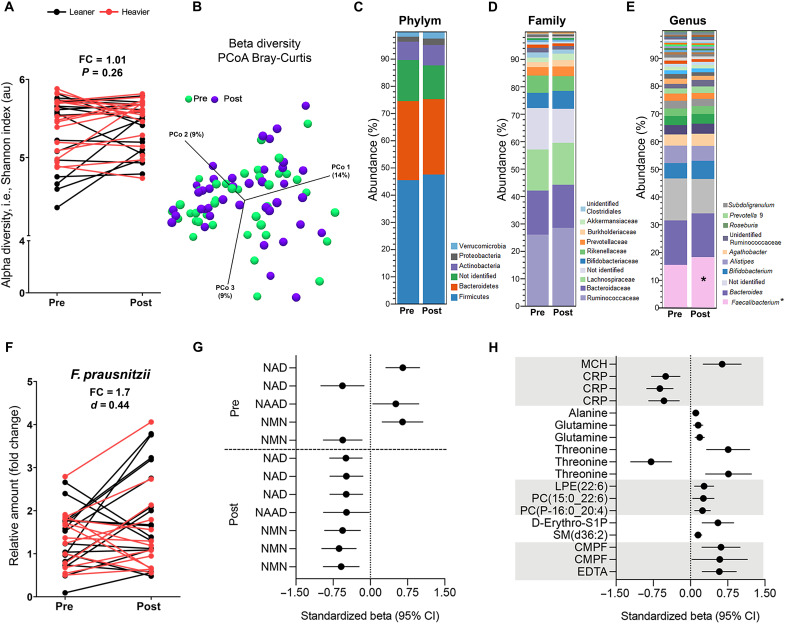
To evaluate whether NR modified the gut microbiota, 16S ribosomal RNA (rRNA) gene sequencing of fecal samples of the twins from the BMI-discordant pairs was performed. The alpha- or beta-diversity of the gut microbiota did not change during the study (Fig. 6, A and B). In contrast, NR increased the abundance of the genus Faecalibacterium by ~2%, but this increase did not pass the multiple testing correction via false discovery rate (FDR) (Fig. 6, C to E). As F. prausnitzii is the only validated species of Faecalibacterium genus, we quantified the NR-induced changes in F. prausnitzii using quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR). There was an increasing ~1.7-fold effect on F. prausnitzii upon NR (Fig. 6F). The changes for any of the analyzed gut microbiota operational taxonomic units (OTUs) did not significantly differ between the leaner and the heavier cotwins (FDR P > 0.05) (table S6). Overall, our data suggest that NR increased the abundance of F. prausnitzii, which is one of the most important commensal bacteria of the human gut microbiota (27, 28).
Fig. 6. NR improves the gut microbiota composition in the twins from the BMI-discordant pairs.
(A) Alpha-diversity of the gut microbiota before versus after NR (n = 11 twin pairs/22 individuals). Lines connect the pre- and post-values of each individual, with black denoting the leaner and red denoting the heavier cotwins. Fold change indicates the mean of the post-NR value divided by the pre-NR value. (B) Principal components analysis (PCoA) of the gut microbiota beta-diversity before versus after NR (n = 11 twin pairs/22 individuals). (C to E) Average gut microbiota abundance at the phylum (C), family (D), and genus (E) level before versus after NR (n = 11 twin pairs/22 individuals). In the legends of the bars, the phylum level shows all detected phyla in the samples and at the family and genus level. Asterisk indicates nominal P < 0.05. (F) Relative amount of F. prausnitzii presented as a fold change to 16S rRNA amount in fecal DNA before versus after NR (n = 11 twin pairs/22 individuals). Cohen’s d value of 0.44 suggests a medium effect size. (G and H) Standardized beta values showing SD values in (G) NAD metabolites associated with 1 SD in baseline abundance of Faecalibacterium or Faecalibacterium abundance change upon NR and in (H) clinical variables and plasma metabolites associated with NR-induced changes in Faecalibacterium (n = 11 twin pairs/22 individuals). Rows represent associations with Faecalibacterium OTUs. MCH, mean corpuscular hemoglobin. Statistical analyses are described in Materials and Methods. See also tables S6 and S7.
The abundance of Faecalibacterium associates with changes in blood metabolite concentrations
As the gut microbiota can influence the response of the host to vitamin B3 supplementation (29), we determined associations between OTUs and blood metabolites (table S7) in the twins from the BMI-discordant pairs. Three of five Faecalibacterium OTU baseline levels associated positively with the change in whole-blood NAD+, NAAD, and NMN upon NR, suggesting that the baseline abundance of Faecalibacterium may support the NR-induced elevation in blood NAD metabolome (Fig. 6G). In addition, the NR supplementation–induced increase in Faecalibacterium abundance was negatively associated with the change in whole-blood levels of NAD+, NAAD, and NMN (Fig. 6G), possibly reflecting competition for NR between Faecalibacterium and the host. The association analysis with blood clinical variables revealed that the Faecalibacterium increase was negatively associated with the change in high-sensitivity CRP but positively with the change in mean corpuscular hemoglobin (Fig. 6H). Last, the Faecalibacterium increase was associated positively with the change in plasma gluconeogenic amino acids (alanine, glutamine, and threonine), phospholipids, and sphingomyelins (Fig. 6H). Together, our results imply that F. prausnitzii may contribute to the regulation of NAD+, inflammation, and amino acid and lipid metabolism in humans.
NR and placebo affected adiposity and metabolic health similarly in BMI-concordant twin pairs
We lastly studied the impact of NR on whole-blood NAD metabolites, body composition, and clinical variables in comparison to placebo in four BMI-concordant twin pairs. We observed that NR treatment appeared to induce whole-blood NAD+, NAAD, and Me4py in comparison to placebo (fig. S2, I to N), confirming compliance to the NR treatment. Of the body composition parameters, body weight (~3 kg), fat percentage, liver fat percentage, and adipocyte weight and volume increased (however, statistically nonsignificantly probably due to small sample size) in the placebo-treated cotwins, while these variables remained stable in the NR-treated cotwins (table S8). We did not observe any changes in the lifestyle factors, glucose and lipid metabolism, and cardiovascular health–related variables in the placebo- or the NR-treated twins from the BMI-concordant pairs (table S8). In conclusion, on the basis of these findings, NR’s effect on body composition and metabolic health did not markedly differ from the effects of placebo.
NR supplementation was well tolerated
NR supplementation was well tolerated by the study participants. Reported side effects included muscle pain, gastrointestinal irritation, sweating, nausea, and headache but not cutaneous flushing typically caused by niacin (4). The placebo-treated twins did not report any side effects. Safety parameters including, for example, the kidney function marker creatinine, liver enzymes, and complete blood count remained unaltered in the blood samples of all study participants (tables S8 and S9). The only exception was that there was a statistically but not clinically significant reduction in hemoglobin in the twins from the BMI-discordant pairs upon NR (table S9). However, this normocytic, normochromic condition (table S9) was likely related to blood loss due to blood sampling. As a whole, these results showed that long-term NR supplementation did not markedly alter the measured safety parameters.
DISCUSSION
Boosting of NAD+ levels is currently under the focus of intensive research as a potential treatment option for metabolic diseases (2). In our study, NR effectively increased the levels of whole-blood NAD+, NAAD, NMN, and Me4py. This is in line with the previous short-term NR clinical studies (14–19). Compared to niacin, NR with the dose of 1 g/day was less potent in elevating whole-blood NAD+ levels as NR induced a twofold increase after 5 months, while niacin with the same dose induced a sixfold increase after 4 months (5). However, the levels of NR and niacin were not molar equivalents in these studies, but the molar concentration was higher for niacin. So far, short-term clinical studies have not reported the positive effects of NR on tissue NAD+ biosynthesis (14–19). We here provide the first evidence that long-term NR supplementation increases muscle and WAT NAD+ biosynthesis in humans regardless of BMI. This NAD+ biosynthesis–boosting effect occurred even in the heavier cotwins from the BMI-discordant pairs, which showed lower expression of NAD+ biosynthetic genes, NMNAT3 and NRK1, especially in WAT, compared with the leaner cotwins, at baseline. Given that the up-regulation of both salvage pathway enzymes NRK1 and NAMPT was observed upon NR, especially in muscle, NAD+ generated in tissues was likely originating both from NR and NAM, the degradation product of NR. This agrees with mouse studies showing that most of the orally consumed NR is metabolized to NAM, and a small proportion of NR is directly used for NAD+ biosynthesis via NRKs (30).
Currently, there is plenty of evidence for the efficacy of NR on muscle mitochondrial biogenesis in mice (7–11) but not in humans (19, 22). In our study, the NR-induced improvement in NAD+ metabolism led to an increase in muscle mitochondrial number and mtDNA amount after 5 months. In line with the previous mouse studies (9, 31), the mechanism was linked to the up-regulation of SIRT1/ERRα/TFAM/MFN2. The effect of NR on mitochondrial biogenesis was tissue specific as significant changes in mitochondria-related parameters were not detected in WAT. Collectively, our evidence emphasizes that a supplementation time longer than 3 months is required to detect the impact of NR on muscle mitochondrial biogenesis in healthy individuals with overweight and obesity. This notion is supported by our previous finding showing that niacin with the dose of 1 g/day significantly increased muscle mitochondrial mass in healthy normal-weight controls after 4 months (5). Overall, our current clinical trial is a proof of principle of NR’s effects on muscle mitochondrial biogenesis in humans.
NR is known to exert beneficial effects on muscle satellite cells in mice (10). Here, we show that NR reduced the number of muscle PAX7+ satellite cells likely by increasing differentiation and fusion of satellite cells to the existing myofibers. Given that we did not observe the increase in muscle mass, our data reveal that satellite cell fusion can be uncoupled from muscle hypertrophy under specific conditions in humans. It has been shown previously that during homeostasis, reduced Notch activity in satellite cells results in premature differentiation and fusion to myofibers bypassing self-renewal (23). It will be interesting to probe whether NR modulates Notch signaling in satellite cells. Our myoblast results align with a recently published study demonstrating an increased differentiation capacity of NR-supplemented human muscle precursor cells (32). One molecular mechanism mediating NR’s effect on myogenic differentiation could be the NR’s degradation product, NAM, which is shown to increase myogenesis in mouse stem cells (33). Whether the increased differentiation of satellite cells provides a functional benefit or disadvantage upon exercise and muscle damage needs to be addressed in future studies.
In mice, NR with the dose of 400 to 500 mg/kg per day has been shown to counteract the negative metabolic consequences of a high-fat diet (9), but a high dose of NR (~1000 mg/kg per day) has been shown to cause WAT dysfunction and impaired glucose homeostasis (34). So far, positive outcomes on body composition from the clinical trials are largely lacking (14–19). During our study, adiposity increased in the twins from the BMI-discordant pairs, although their self-reported lifestyle factors remained the same. The underlying mechanism may be linked to morphological and molecular changes in WAT. However, we did not detect changes in adipocyte number or size or WAT PPARγ dysregulation that have been reported in mice upon high doses of NR (34). As the body weight and fat mass tended to increase also in the placebo-treated twins, the weight gain may be related to a normally occurring increase in adiposity over time rather than to NR per se (35). In our previous study, the long-term niacin supplementation resulted in an opposite outcome related to body composition compared with the effects of NR in the current study. Niacin with the dose of 1 g/day reduced whole-body fat percentage in normal-weight study participants after 4 months (5). Similarly, as for adiposity, no significant effect on insulin sensitivity has been found in the published clinical trials with NR (16, 17, 19). Here, we report elevated fasting glucose and insulin levels and impaired insulin sensitivity in the oral glucose tolerance test in twins from the BMI-discordant pairs after a 5-month NR supplementation. As niacin is well known to raise fasting glucose levels and impair insulin sensitivity by increasing hepatic gluconeogenesis and by decreasing insulin signaling (36), our findings raise the question whether niacin and NR exhibit a similar mechanism of action on glucose metabolism in humans. Overall, on the basis of the current clinical evidence, larger and longer placebo-controlled studies are required to clarify the role of NR as a modifier of adiposity and glucose metabolism in humans. Given that most clinical studies with NR, like ours, have been performed in metabolically healthy individuals (14–16, 18, 19), human studies in patient populations with metabolic diseases are warranted in the future.
Epigenetic mechanisms such as DNA methylation have been shown to regulate gene expression upon NR in mice (37). Typically, DNA methylation in gene promoter regions is associated with transcriptional repression. However, the relationship between DNA methylation and gene expression at a given location is not straightforward. Here, we demonstrate that NR supplementation led to a muscle-specific decline in global DNA methylation likely via reduced methyl pool size due to increased elimination of NAM via methylation. Nevertheless, both hyper- and hypomethylation of CpG sites were detected in muscle and WAT, although most of the analyzed sites were hypomethylated upon NR. Notably, methylation changes at the CpG sites were associated with the altered tissue expression for several analyzed genes such as PGC-1α, PAX7, and NAMPT. Given that NAD+ precursors have been typically linked to the regulation of gene transcription via NAD+-dependent histone deacetylases (38), our study underscores that DNA methylation may also be involved in the epigenetic control of tissue metabolism and muscle stem cell identity upon NR in humans. The mechanism via which NR diminishes DNA methylation could be related to the inhibition of DNA methyltransferases through direct or indirect mechanisms, as suggested previously (37).
The disruption of the gut microbiota homeostasis, i.e., dysbiosis, is a typical feature of metabolic and muscle diseases (39). In line with the previous mouse studies (11, 40), NR ingestion slightly improved the gut microbiota composition in our study participants. Similarly, niacin has been reported to beneficially affect the gut microbiota composition in humans (41). We here show that NR supplementation increased the proportion of the only identified species of the genus Faecalibacterium, namely, F. prausnitzii, which is a gut bacterium that promotes metabolic health and anti-inflammatory responses (27, 28). In agreement with the latter notion, Faecalibacterium abundance was negatively associated with high-sensitivity CRP. Intriguingly, our correlation analyses between Faecalibacterium abundance and whole-blood NAD metabolites imply that F. prausnitzii may compete for the NAD+ precursors with the host for its own NAD+ biosynthesis and promote the in vivo efficiency of NR in humans. Supporting these conclusions, it is known that F. prausnitzii can metabolize NR and NMN (42) and that the gut microbiota are crucial for the NAD+ boosting effect of NR in mice (29). Together, our findings suggest that NR changes the gut microbiota composition in a manner that may improve health and that the NAD+ metabolism could be partly regulated through the interaction between the host and Faecalibacterium in humans.
In conclusion, NR supplementation is a potential treatment option to be tested in individuals with decreased muscle mitochondrial biogenesis and dysbiosis. As possible adverse effects, we report a declining muscle satellite cell number and a possibility for impaired glucose metabolism. As patients with chronic muscle disease typically exhibit satellite cell dysfunction, muscle performance and regeneration are important endpoints to be monitored in future NR clinical trials. Overall, our data underscore the role of NR as a potent modifier of systemic NAD+ levels, muscle mitochondrial biogenesis, satellite cell function, DNA methylation, and the gut microbiota in humans. Notably, NR modulated these metabolic processes similarly in both leaner and heavier cotwins, i.e., regardless of BMI.
Limitations of the study
The study participants volunteered to two muscle and WAT biopsy collections, yielding material sufficient for histology, electron microscopy, and RNA and DNA isolation, but not for mitochondrial respirometry and protein level analyses. The electron microscopic evaluation of tissues was limited to muscle because of improper fixation of WAT samples. The open study setting and the lack of a statistically powered placebo group may compromise our results related to body composition and metabolic health, and these endpoints need to be followed up in larger placebo-controlled trials. Although NR supplementation significantly increased blood NAD+ levels, it is not clear whether the observed effects of NR administration are specific to the boosting of NAD+ or other NAD metabolites such as methylNAM. From a mechanistic perspective, we demonstrate that NR supplementation up-regulated SIRT1/ERRα/TFAM/MFN2 and modulated epigenetic control of gene transcription, but this provides only a correlative insight into the mechanism via which NR regulates mitochondrial biogenesis in humans. Therefore, further work must be done to determine the mechanisms underlying the reported mitochondrial phenotype. In addition, we recognize that it is possible that long-term NR supplementation increases mitochondrial biogenesis as a compensatory mechanism for NR’s toxicity to mitochondria. Consequently, further studies are needed to understand whether long-term NR supplementation is truly beneficial for muscle mitochondrial function, as well as for muscle physiology such as exercise tolerance and regeneration, in humans.
MATERIALS AND METHODS
Study design
BMI-discordant and BMI-concordant MZ twin pairs were screened from two large population-based cohorts of Finnish twins (FinnTwin12 and Finntwin16, n = 5000, 1200 of which are MZ), established by J.Ka. (43) and recruited by K.H.P. This study was approved by the Ethics Committee of the Helsinki University Central Hospital (protocol number 270/13/01/2008), and the study was conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all subjects.
The primary objective of this study was to investigate whether long-term NR supplementation increases muscle and WAT mitochondrial biogenesis in humans. In addition, we aimed to understand whether this intervention could also affect adiposity and metabolic health. Our primary and secondary endpoints were prospectively selected (clinicaltrials.gov entry NCT03951285). A sample size of 10 twin pairs was determined sufficient to detect an effect size of 0.7 with 0.05 significance level and 0.80 power in mitochondrial-related parameters (44).
The study participant selection is described in fig. S1. Twenty-two MZ twin pairs with cotwins having a BMI difference of at least 2.5 kg/m2 were considered as BMI discordant. Two twin pairs did not meet the inclusion criteria presented in the next paragraph. Another four twin pairs were considered as BMI concordant (within-pair difference in BMI, <2.5 kg/m2). Four BMI-discordant twin pairs discontinued the intervention because of pregnancy, muscle pain, or unwillingness to follow the study protocol (fig. S1). Sixteen BMI-discordant and four BMI-concordant twin pairs were included in the data analyses, as shown in fig. S1.
Inclusion criteria were as follows: (i) age > 18 years, (ii) BMI > 18.5 kg/m2 in both members of the twin pair, (iii) agreement to maintain current level of physical activity throughout the study, and (iv) agreement to avoid vitamin supplementation or nutritional products with vitamin B3 14 days before the enrolment and during the study. The exclusion criteria were as follows: (i) unstable medical conditions as determined by the investigator, (ii) clinically significant abnormal laboratory results at screening (e.g., aspartate aminotransferase and/or ALT > 2 × upper limit of normal, and/or bilirubin > 2 × upper limit of normal), (iii) subjects who would have had a planned surgery during the course of the trial, (iv) history of or a current diagnosis of any cancer (except for successfully treated basal cell carcinoma diagnosed less than 5 years before screening), (v) history of blood/bleeding disorders, (vi) immunocompromised individuals such as subjects that had undergone organ transplantation or subjects diagnosed with human immunodeficiency virus, (vii) hepatitis, and (viii) blood donation in the previous 2 months.
Data were analyzed from 16 BMI-discordant (44% females) and 4 BMI-concordant twin pairs (50% males) with a mean age of 40 (Table 1). The study participants had the following medications: hypertension (n = 2; angiotensin II receptor blocker, calcium channel blocker, and diuretic), asthma or allergy (n = 7; an inhaled corticosteroid, beta-2 agonist, and antihistamine), type 2 diabetes (n = 1; metformin and insulin), rheumatoid arthritis (n = 2; hydroxychloroquine and methotrexate), psychiatric (n = 3; selective serotonin intake inhibitor, valproate, a tricyclic antidepressant, and a neuroleptic), and oral contraceptives (n = 2). The number of smokers is shown in Table 1.
The examination protocol of this nonrandomized, open-label study is described in Fig. 1A. All twins from the BMI-discordant pairs received NR supplementation. For BMI-concordant twin pairs, one cotwin received NR and the other cotwin received placebo. The cotwins were allocated to the two groups randomly and in a double-blinded fashion. The NR dose (1 g/day) was selected on the basis of a previous study showing that a niacin supplementation of 1 g/day had a favorable effect on systemic NAD+ levels, body composition, and mitochondrial biogenesis (5). The weekly dose was initially 250 mg/week. The daily NR dose was gradually escalated by 250 mg/week so that the full dose of 1 g/day was reached in 1 month. The supplementation was continued for 4 months with the full dose. At the end of the study, the dose was slowly decreased to the initial level at a rate of 250 mg/week. Study participants were advised to take half of the NR dose in the morning and the other half in the evening. The collection of samples such as blood, WAT and muscle biopsies, saliva, and feces was performed after overnight fasting at the same time in the morning both at baseline and after 5 months. Safety laboratory tests included blood count, iron levels, and liver and kidney function tests. One-to-one phone conversations were conducted at least once a month to follow up compliance to the study and potential symptoms or side effects. All participants were instructed to continue their normal routine with no changes to their physical activity and diet. The analysis of body composition, clinical variables, and whole-blood NAD metabolites was performed for all twin pairs. Gene expression, imaging, and omic analyses were conducted only for BMI-discordant twin pairs.
Body composition, energy intake, and expenditure
Weight, height, waist circumference, whole-body fat (assessed by dual-energy x-ray absorptiometric scans), abdominal subcutaneous and visceral fat (by magnetic resonance imaging), and liver fat (by magnetic resonance spectroscopy) were measured in all study participants as described previously (45). In short, imaging for amounts of subcutaneous adipose tissue, visceral adipose tissue, and liver fat was performed on a clinical 1.5-T imager (Avanto, Siemens, Erlangen, Germany). To allow measurement of abdominal fat distribution, a stack of abdominal T1-weighted magnetic resonance images (16 slices, slice thickness of 10 mm, repetition time of 91 ms, time to echo of 5.2 ms, and flip angle of 80°) were obtained from 8 cm above to 8 cm below the L4/5 lumbar intervertebral disks using frequency selective fat excitation. The amount of visceral and subcutaneous adipose tissue was determined using the segmentation software SliceOmatic 5.0 (TomoVision, Quebec, Canada).
A point-resolved spectroscopy sequence was used for volume selection in hepatic magnetic resonance spectroscopy. A 25 mm × 25 mm × 25 mm voxel was placed in the middle of the right liver lobe, and liver spectra with time to echo of 30 ms and four averages were collected. Signal acquisition was triggered to end exhalation using a navigator belt to eliminate artifacts due to respiratory motion so that repetition time was kept at >4000 ms. Liver spectra were analyzed with jMRUI 6.0 software (46), and intensities of methylene and water resonances were determined using the AMARES algorithm (47). Signal intensities were corrected for relaxation effects, and liver fat was determined as an intensity ratio of methylene/(methylene + water). Ratios were further converted to mass fractions as described previously (48).
Muscle mass and energy expenditure were measured using the Tanita MC-980 bioelectrical impedance device. Physical activity was estimated with the Baecke questionnaire, and dietary intake from 3-day food records was analyzed with the Diet32 program (Aivo Finland Oy) based on a national Finnish database for food composition (Fineli; www.fineli.fi).
Blood laboratory examinations
Routine blood tests were performed for all study participants. Blood samples were collected after overnight fasting, and whole-blood, separated plasma, and serum samples were frozen at −80°C. Blood count, ALT, aspartate transaminase, hemoglobin A1c, glucose, insulin, C-peptide, lactate, total cholesterol, high-density lipoprotein, low-density lipoprotein, triglycerides, homocysteine, and high-sensitivity CRP were analyzed using standardized methods at the HUSLAB laboratories.
A 75-g oral glucose tolerance test with four time points (0, 30, 60, and 120 min) was performed after a 12-hour overnight fast, followed by measurements of plasma glucose with spectrophotometric hexokinase and glucose-6-phosphate dehydrogenase assay (Roche Diagnostics) and of serum insulin with time-resolved immunofluorometric assay (PerkinElmer). Free fatty acids were determined with a NEFA kit (Wako Chemicals #999-75406), apolipoprotein B levels with an Apolipoprotein B Konelab kit (Thermo Fisher Scientific, #981663), and plasma adiponectin levels with an enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, #DHWAD0).
Adipose tissue sampling
Adipose tissue biopsies were collected from all study participants at baseline and at the end of the study (Fig. 1A). The subcutaneous WAT biopsies were taken from superficial abdominal adipose tissue using a needle biopsy under local anesthesia (lidocaine). WAT samples for analyses were snap-frozen and stored in liquid nitrogen or at −80°C.
Samples used for determination of adipocyte size and number were immediately processed by collagen digestion and separation of adipocytes by centrifugation as described previously (49). In brief, the subcutaneous adipose tissue was minced and incubated for 1 hour at 37°C with continuous shaking in 10 ml of adipocyte medium [Dulbecco’s modified Eagle’s medium/F-12 (1:1; Invitrogen) with 16 μM biotin, 18 μM panthotenate, 100 μM ascorbate, and antibiotic-antimycotic (Invitrogen)], supplemented with 2% bovine serum albumin (Sigma-Aldrich) and collagenase A (2 mg/ml; Roche Diagnostics). Digestion was terminated by adding adipocyte medium supplemented with 10% newborn calf serum (Sigma-Aldrich) and centrifuging for 10 min at 600g. After washing the adipocytes with adipocyte medium, images of the adipocytes were taken with a light microscope (Zeiss, Axioplan2) using ×50 magnification. Adipocyte diameters were automatically measured with a custom algorithm for ImageJ (50), which preprocessed the image to enhance the adipocyte borders and used a circle-detection algorithm to identify the cells. The algorithm identified the adipocytes with standardized microscope settings and was validated against 2000 manually measured diameters from 20 pictures (r = 0.85, P < 0.001). Mean adipocyte volume was calculated for each individual using the following formula
V = cell volume (μm3), d = cell diameter (μm), w = cell weight (μg), and n = total adipocyte number, where m = total body fat mass (kg) and V (m3) ∗ rho (density, fat) = mean weight of a single adipocyte. Density of fat cell triglycerides = 0.915 g/ml. Adipocytes were assumed to be spheres.
Muscle sampling, TEM, and satellite cell histology and quantification
Muscle tissue biopsies were collected from all study participants at baseline and at the end of the study (Fig. 1A) via Bergström needle biopsy from the vastus lateralis muscle in sterile conditions under local anesthesia. The muscle samples for molecular analyses were snap-frozen and stored in liquid nitrogen or at −80°C.
Samples for TEM analysis were fixed in 2.5% glutaraldehyde. For plastic embedding, they were then treated with 1% osmium tetroxide dehydrated in ethanol and embedded in epoxy resin. One-millimeter section was stained with methylene blue (0.5%, w/v) and boric acid (1%, w/v) and examined with a light microscope to mark the interesting areas for TEM analyses. Thereafter, 60- to 90-nm sections were cut on grids and stained with uranyl acetate and lead citrate by the Viikki Electron Microscopy Unit of the Institute of Biotechnology (EMBI, Biocenter Finland) and viewed with a JEM-1400 transmission electron microscope (Jeol). From each study subject’s pre- and post-NR time points, 15 TEM images were taken from the intermyofibrillar area of type I muscle fibers, determined by mitochondria content, using ×2000 magnification. Total mitochondria number per 10 μm2 relative to the muscle fiber area, mitochondrial surface area per muscle fiber area (%), dimension values [average diameter (μm) and perimeter (μm)], and total muscle fiber area (μm2) were determined by painting each mitochondrion in Microscopy Image Browser (51). These values were used to calculate the total mitochondria number per 10 μm2 of muscle fiber area, mitochondrial surface area per muscle fiber area (%, mitochondria density), and major shape descriptors, branching (form factor) and length-to-width ratio (aspect ratio).
Muscle samples for histological staining were snap-frozen in liquid nitrogen–cooled isopentane. Cryosections were prepared to measure PAX7 immunostaining in muscle tissues: Nine-micrometer-thick frozen sections were postfixed in 4% paraformaldehyde in Dulbecco’s phosphate-buffered saline (DPBS) for 10 min at room temperature and washed three times for 5 min in DPBS. Slides were then incubated overnight at 4°C in mouse monoclonal Pax7 primary antibody (Developmental Studies Hybridoma Bank, #AB_528428, deposited by Kawakami A) diluted 1:100 in blocking solution (15% normal goat serum, 1.5% bovine serum albumin, and 0.5% Triton X-100 in DPBS). Slides were washed three times for 10 min in DPBS and incubated in Cy3 goat anti-mouse immunoglobulin G1 secondary antibody (1:500 dilution; Jackson ImmunoResearch, #115-165-003) and Hoechst (1 μg/ml) for 1 hour at room temperature to visualize the nuclei. Slides were washed as described above and mounted. Images were generated using Pannoramic 250 FLASH II digital slide scanner (3DHistec, Thermo Fisher Scientific) and Leica SP8 STED confocal microscope (Leica). For PAX7+ cell quantification, muscle sections were scored from a minimum of two to three sections per sample per condition and normalized to cross-sectional area of the muscle section. All quantifications were performed using Case Viewer 2.0 software (3DHistech, Thermo Fisher Scientific), and images were blinded for quantification.
Myoblast cell cultures
Muscle samples for primary muscle cell cultures, i.e., myoblast cultures, were processed immediately after excision. Samples were washed with 5 ml of isolation medium [0.05% trypsin-EDTA (SAFC Biosciences), glucose (1.8 g/liter; Sigma-Aldrich), N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid buffer (7.15 g/liter; Sigma-Aldrich), NaCl (7.6 g/liter; Sigma-Aldrich), KCl (0.224 g/liter; Sigma-Aldrich), and phenol red (1.2 mg/liter; Sigma-Aldrich)]. Thereafter, 5 ml of isolation medium was added and tissue was minced. Another 10 ml of isolation medium was added, and the sample was transferred to a 20-ml beaker with a magnetic stirrer and placed in a 37°C incubator for 20 min. The supernatant containing muscle cells was transferred into a 50-ml centrifuge tube containing 15 ml of myoblast growth medium [Ham’s F10 with GlutaMAX (Gibco) supplemented with 15% fetal bovine serum (Thermo Fisher Scientific), fetuin (500 mg/liter; Sigma-Aldrich), insulin (200 mg/liter; Sigma-Aldrich), 0.05% bovine serum albumin (Sigma-Aldrich), dexamethasone (0.39 mg/liter; Sigma-Aldrich), gentamicin (50 mg/liter; Sigma-Aldrich), and human epidermal growth factor (2 mg/liter; Thermo Fisher Scientific)]. This process was repeated twice to break down any remaining tissue. Last, the supernatant containing the isolated muscle cells was centrifuged for 10 min at 100g to collect the cells, and cell pellet was resuspended in 5 ml of myoblast growth medium. Myoblasts were seeded on 25-cm2 flasks (Corning). After reaching 70% confluence, they were subcultured in 150-cm2 flasks (Corning) and finally on six-well plates (Corning) at a density of 100,000 cells per well. When confluent, the cells were collected and stored at −80°C.
Targeted quantitative NAD metabolome analysis
The measurement of whole-blood NAD metabolome was carried out from all study participants after dual extractions as recently described (5). For analysis of Me4py (group A analyte), samples were spiked with 400 pmol of [d3]-Me4py (internal standard A). For analysis of NAD+, NADP, NMN, NAR, NAAD, and ADPR, samples were dosed with 13C-yeast extract (internal standard B) as described (18, 52).
In short, to quantify Me4py (group A analyte), 75 μl of whole blood was added to 20 μl of group A internal standard and 500 μl of 3:1 4% trichloroacetic acid:acetonitrile was added. The mixture was allowed to sit on ice for 20 min, after which samples were sonicated twice for 20 s and centrifuged at 4°C for 13 min at 16,100g. Next, the supernatant was removed and dried under vacuum overnight at room temperature. The samples were reconstituted in 2% acetonitrile/water immediately before the analytical run. To quantify NAD+, NADP, NMN, NAR, NAAD, and ADPR (group B analytes), 75 μl of whole blood was added to 20 μl of group B internal standard prepared in water and mixed with 500 μl of 3:1 4% trichloroacetic acid:acetonitrile with vortexing. After resting on ice, the samples were centrifuged as described above. Next, supernatant was removed and dried under vacuum overnight and reconstituted in 2% acetonitrile/water. After reconstitution, samples were transferred to Waters polypropylene plastic total recovery vials and stored in a Waters Acquity H class autosampler maintained at 8°C until injection. In all cases, 8 μl of extract was loaded onto the column. For one set of samples, group B analytes plus Me4py were extracted as for the group B analytes. One hundred microliters of blood was used, and 200 pmol of d3-Me4py was added to the internal standard mix. All analytes in this group except NAAD had a corresponding stable labeled internal standard. Labeled NAD+ was used as the internal standard for NAAD.
Separation and quantitation of analytes were performed with a Waters Acquity LC interfaced with a Waters TQD mass spectrometer operated in positive ion multiple reaction monitoring mode as described (52) with minor modifications. Multiple reaction monitoring transitions monitored were as follows: NAD+ 664.1>136.1; 13C10-NAD+ 674.1>136.1; NADP 744.1>136.1; 13C10-NADP 754.1>136.1; NAAD 665.1>136.1; NMN 335>123: 13C5-NMN 340>123; NAR 256>124; 13C5-NAR 261>124; ADPR 560>136.1; 13C10-ADPR 570>136.1; Me-4-py 153>136; d3-Me-4-py 156>139. Separate Hypercarb columns (Thermo Fisher Scientific) were used for the separations. The conditions for group A were as follows: solvent A: 10 mM NH4OAc with 0.1% formic acid, solvent B: acetonitrile with 0.1% formic acid, and solvent D: methanol, flow of 0.30 ml/min; gradient initial of 98% A and 2% B; 2.25 min, 98% A and 2% B; 11 min, 74.8% A, 17.9% B, and 7.3% D; 11.1 min,10% A and 90% B; 14.3 min, 10% A and 90% B; 14.4 min, 98.2% A and 2% B; end, 18.5 min. Conditions for group B were as follows: solvent A: 7.5 mM NH4OAc with 0.05% NH4OH and solvent B: acetonitrile with 0.05% NH4OH; flow of 0.353 ml/min; gradient initial of 97% A and 3% B; 1.8 min, 97% A and 3% B; 10 min, 65.5% A and 34.5% B; 11 min, 10% A and 90% B; 13.2 min, 10% A and 90% B; 13.3 min, 97% A and 3% B; end, 19 min.
MtDNA analyses and quantification
Total DNA, including mtDNA, was extracted from the snap-frozen muscle (15 mg) and WAT (150 mg) using standard phenol-chloroform extraction and ethanol precipitation and stored at 4°C until use. MtDNA content was determined by the ratio of mtDNA to genomic DNA using RT-qPCR. To detect genomic DNA, primers for nuclear β-amyloid precursor protein, beta-2-microglobulin, and hemoglobin subunit beta were used. To measure mtDNA, mitochondrial 16S rRNA, cytochrome B, and D-loop region primers were chosen. The PCR primer sequences are provided in table S10.
RT-qPCR was performed using SYBR Green MasterMix (Thermo Fisher Scientific), 10 μM primers, and 2 ng of DNA template in a 384-well plate format using CFX384 Touch Real-Time PCR Detection Systems (Bio-Rad Laboratories). Thermal cycling included initial denaturation of 3 min at 95°C, 39 cycles of 10 s at 95°C and 30 s at 62°C, final extension of 10 s at 95°C, and melting curve analysis from 65° to 95°C with 0.5°C increments. MtDNA content was presented as a relative level of mtDNA genome per nuclear genome. Data analysis was performed in qBASE+ 3.2 software (Biogazelle) using the ΔΔCt method.
Gene expression
Total RNA from myoblasts was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA from muscle tissue and WAT was isolated using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to the manufacturer’s instructions with minor modifications. In short, muscle tissue (30 mg in 600 μl of Buffer RLT Plus) was lysed using TissueLyser II (Qiagen) at 25 Hz for 2 min, and the lysate was transferred into the AllPrep DNA Mini spin column. WAT (250 mg in 1 ml of Buffer RLT Plus) was similarly lysed except for the time being 2 × 2 min. As the sample volume exceeded the recommended amount, the lysate was divided into two AllPrep DNA Mini spin columns. Thereafter, muscle and WAT samples were processed according to the manufacturer’s instructions. RNA was stored at −80°C.
Complementary DNA (cDNA) was synthesized from 0.5 μg of muscle tissue and myoblast RNA using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. The SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) was used to synthesize cDNA from 0.5 μg of WAT RNA according to the manufacturer’s instructions.
Expression levels of genes associated with NAD+ biosynthesis, mitochondrial metabolism, mitochondrial OXPHOS, and additional potential NR target genes were determined from tissue samples with RT-qPCR. Gene expression levels were measured from myoblasts to assess their self-renewal, differentiation, and fusion potential. The RT-qPCR primer sequences are provided in table S10. RT-qPCR was performed using SYBR Green MasterMix (Thermo Fisher Scientific), 10 μM primers, and 5 ng of DNA template in 384-well plate format with conditions identical to those used for mtDNA quantification PCR. Three reference genes were used for normalization: actin, beta-2-microglobulin, and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta. Data analysis was performed with the ΔΔCt method using the qBASE+ 3.2 software (Biogazelle).
DNA methylation analysis
High–molecular weight WAT and muscle DNA was extracted using a QIAmp DNA Mini kit (Qiagen) and bisulfite-converted by an EZ DNA Methylation kit (ZYMO Research) following the manufacturers’ protocols. DNA methylation was quantified using Infinium MethylationEPIC BeadChip arrays (Illumina) according to the manufacturer’s instructions. The data were preprocessed using R-package minfi (53). Sample quality control was performed in R-package MethylAid with default settings (54). Bad-quality probes were removed with the following criteria: (i) detection P > 0.01, (ii) bead counts of <3, (iii) zero methylation probe intensity, and (iv) ambiguously mapping probes (55). The data were normalized using functional normalization (56) followed by Beta Mixture Quantile normalization (57). CpG sites annotated to genes with gene expression data available were selected for the analysis, resulting in a final dataset of 518 CpGs in WAT and 619 in muscle.
The average long interspersed nuclear element-1 methylation levels were computed using R-package REMP (58) for each individual as a surrogate measure for global DNA methylation level. Differences in global methylation levels upon NR (baseline versus 5 months) in the twins from the BMI-discordant pairs and differences in the changes of global methylation upon NR between the leaner and the heavier cotwins were compared using Wilcoxon signed-rank test. The level of significance (two-tailed) was set at P < 0.05.
Differential methylation analysis was performed for the selected CpGs in WAT (n = 14 twin pairs/28 individuals) and muscle (n = 12 twin pairs/25 individuals) using a linear model (package limma in R-Bioconductor) to identify CpG sites with altered methylation (Benjamini-Hochberg FDR P < 0.05) upon NR supplementation in the twins from the BMI-discordant pairs. The regression models were adjusted for, sex, age, smoking, and twinship. Regression analysis was also performed to identify within-pair differences in the changes in CpG site methylation levels (changes from baseline to 5 months, FDR P < 0.05).
Plasma metabolite isolation and nontargeted metabolite profiling
We performed nontargeted metabolite profiling using two different chromatographic–mass spectrometric techniques coupled online to high-resolution mass spectrometry and further acquired data with positive and negative electrospray ionization (ESI) mode. Further information about the instruments’ setup and data acquisition parameters, in concert with molecular feature finding and peek picking, can be obtained from the previous publications (59, 60).
Briefly, plasma samples were thawed on ice, and a 100-μl aliquot of plasma was dispensed into a 96-well Captiva ND filter plate 0.2 μm PP (Captiva ND, 0.2 μm PP, Agilent Technologies) containing 400 μl of ice-cold acetonitrile. Samples were mixed with a pipette to thoroughly precipitate plasma proteins. Samples were then centrifuged at 700g for 5 min at 4°C, and the supernatants were collected to a 96-well storage plate and stored at −10°C.
A pooled sample from all biological samples per experiment was injected at the beginning of the sequence to equilibrate the analytical platform and then after every 12 samples throughout the analysis for quality control. In addition, a solvent blank was prepared and injected at the beginning of the sequence. Samples were randomized before the analysis.
The analysis of amphiphilic metabolites was carried out using an ultrahigh-performance liquid chromatography (Vanquish Flex UHPLC system, Thermo Fisher Scientific, Bremen, Germany) coupled online to high-resolution mass spectrometry (Q Exactive Classic, Thermo Fisher Scientific, Bremen, Germany) (59). The sample solution (2 μl) was injected onto a reversed-phase column (Zorbax Eclipse XDBC18, 2.1 × 100 mm, 1.8 μm; Agilent Technologies, Palo Alto, CA, USA) that was kept at 40°C. The mobile phase, delivered at 400 μl/min, consisted of water (eluent A) and methanol (eluent B), both containing 0.1% (v/v) of formic acid. The following gradient profile was used: 0 to 10 min: 2 to 100% B, 10 to 14.50 min: 100% B, 14.50 to 14.51 min: 100 to 2% B, and 14.51 to 20 min: 2% B. The sample tray was at 10°C during these analyses. Mass spectrometry was equipped with heated ESI. The positive and negative ionization modes were used to acquire the data. At the beginning and end of the sample analysis, data-dependent product ion scans (MS2) were acquired for each mode. The detector was calibrated before the sample sequence and subsequently operated at high mass accuracy [<2 parts per million (ppm)]. Continuous mass axis calibration was performed by monitoring reference ions m/z (mass/charge ratio) 214.08963 in the positive ionization mode.
For the analysis of hydrophilic compounds, an ultrahigh-performance liquid chromatography (1290 LC system, Agilent Technologies, Waldbronn, Karlsruhe, Germany) coupled online to high-resolution mass spectrometry (6540 UHD accurate-mass quadrupole-time-of-flight mass spectrometry, Agilent Technologies, Waldbronn, Karlsruhe, Germany) was used (60). The sample solution (2 μl) was injected onto a column (HILIC, Acquity UPLC BEH Amide 1.7 μm, 2.1 × 100 mm; Waters Corporation, Milford, MA, USA) that was kept at 45°C. Mobile phases, delivered at 600 μl/min, consisted of 50% (v/v) (eluent A) and 90% (v/v) (eluent B) acetonitrile, respectively, both containing 20 mM ammonium formate (pH 3). The following gradient profile was used: 0 to 2.5 min: 100% B, 2.5 to 10 min: 100% B → 0% B, 10 to 10.1 min: 0% B → 100% B, and 10.1 to 12.5 min: 100% B. The sample tray was at 10°C during these analyses. Mass spectrometry was equipped with a heated ESI source, operated in both positive and negative ionization modes. For data acquisition, a 2-GHz extended dynamic range mode was used in both positive and negative ion modes from 50 to 1600 (m/z). At the beginning and end of the sample analysis, MS2 values were acquired for each mode. The detector was calibrated before the sample sequence and subsequently operated at high accuracy (<2 ppm). Continuous mass axis calibration was performed by monitoring two reference ions from an infusion solution throughout the runs. The reference ions were m/z 121.050873 and m/z 922.009798 in the positive mode and m/z 112.985587 and m/z 966.000725 in the negative mode.
Differential plasma metabolite profile analysis and metabolite identification
Differential metabolite analysis was performed on the plasma metabolite levels of the twins from the BMI-discordant pairs (n = 14 twin pairs/28 individuals) using a linear model (package limma in R-Bioconductor) to identify metabolite features that had significantly changed levels (nominal P < 0.05) upon NR supplementation in the twins from the BMI-discordant pairs (regardless of heavy/lean status). Regression models were adjusted for leaner/heavier status, sex, age, smoking status, and twinship (shared genetic effects). Regression analysis was also carried out to identify changes in metabolite levels (changes from baseline to 5 months, nominal P < 0.05) that were different in the leaner and the heavier cotwins. The regression models were adjusted for sex, age, smoking status, and twinship.
The final dataset contained 580 and 2398 molecular features from hydrophilic interaction chromatography and reversed-phase analysis, respectively. Annotation of these significantly different metabolites found between the study groups was generated on the basis of accurate mass and isotope information, i.e., ratios, abundances, and spacing, as well as product ion spectra (MS2) against existing libraries, either in-house for level I or online spectral databases (i.e., Human Metabolome Database, MzCloud, METLIN, and ChemSpider) for levels II to IV, according to the guidelines from the Metabolomics Standard Initiative (61).
Gut microbiota sequencing and composition analyses
For stool collection at home, study subjects were provided with the supplies and instructions for the collection. Study subjects were asked to keep the samples in a cold environment and to bring the sample to the hospital. The stool was then immediately stored at −80°C. The interval between collecting and freezing the stool was within 12 to 24 hours. The sample collection at the hospital was conducted in the same fashion. For the analyses of the gut microbiota, the exclusion criteria were antibiotic course 1 month before sampling.
For the microbial community analysis, the total microbial DNA was extracted from ~80 mg of feces using Stool Extraction Kit v2 (Hain Lifescience GmbH, Nehren, Germany) and semiautomated GenoXtract (Hain Lifescience Gmbh), accompanied with preceding bead beating. Thereafter, the rRNA gene was amplified using universal primers M13_S-D-Bact-0341-b-S-17 (5′-TGTAAAACGACGGCCAGT-3′) and P1_S-D-Bact-0785-a-A-21 (5′-CCTCTCTATGGGCAGTCGGTGAT-3′) targeting the V3-V4 regions of the small subunit (SSU) rRNA gene. PCR and adding of the linker and fusion primers [0.05 μM M13_S-D-Bact-0341-b-S-17, 0.5 μM IonA_IonXpressBarcode_M13 (Thermo Fisher Scientific, #4474517), and 0.5 μM P1_S-D-Bact-0785-a-A-21] were performed as described previously (62). After purifying the PCR products with AMPure XP (Beckman Coulter) and pooling them in equimolar quantities, sequencing was performed as described before (62). The 16S rRNA gene sequences were quality-filtered and clustered to OTUs at 97% similarity using the CLC Microbial Genomics Package (Qiagen).
The rRNA gene sequences were classified using the SILVA SSU Ref database (99%, arb-silva.de/projects/ssu-ref-nr/). Statistical analyses for the gut microbiota were performed with the CLC Microbial Genomics Package and SPSS Statistics (IBM). The alpha-diversity of the gut microbiota was quantified with the Shannon index, and beta-diversity analysis was based on Bray-Curtis distance and permutational multivariate analysis of variance (PERMANOVA) for significance testing. The taxonomic differences between baseline and 5 months were analyzed with paired Wilcoxon signed-rank test in SPSS, followed by Benjamini-Hochberg FDR correction for multiple testing. The level of significance (two-tailed) was set at P < 0.05 after the multiple testing correction.
Microbiota data were further preprocessed and normalized using packages phyloseq and edgeR in R-Bioconductor. Differential analysis was performed for the gut microbiota levels of the BMI-discordant twins (n = 11 twin pairs/22 individuals) using a linear model (package limma in R-Bioconductor) (63) to identify microbial taxa that significantly changed (FDR P < 0.05) upon NR (baseline versus 5 months) in the twins from the BMI-discordant pairs (regardless of heavy/lean status). The regression models were adjusted for leaner/heavier status, sex, age, smoking status, and twinship. Regression analysis was also carried out to identify changes in microbiota (changes from baseline to 5 months, FDR P < 0.05) that were different in the leaner and the heavier cotwins as a result of NR intervention. The regression models were adjusted for sex, age, smoking status, and twinship.
F. prausnitzii RT-qPCR analysis was performed using iQ SYBR Supermix and CFX96 Real-time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) as described previously (62). The RT-qPCR program for the 16S rRNA genomic region was preincubation at 95°C for 3 min, 40 cycles at 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s. The results of F. prausnitzii were normalized to the results of 16S rRNA, and the fold change at baseline and 5 months was calculated with the ΔΔCt method. Because of great interindividual variation, the changes in the abundance of F. prausnitzii upon NR (baseline versus 5 months) were analyzed with Cohen’s d as follows
where M1 and M2 are the means for the first and second samples, and SDpooled is the pooled SD for the samples.
Association analyses
Associations between CpG methylation (outcome variable) and gene expression (explanatory variables) were identified. First, using linear models with adjustments for age, sex, smoking status, and twinship, gene expression changes between baseline and 5 months were regressed against DNA methylation changes between baseline and 5 months. Benjamini-Hochberg FDR–corrected P < 0.05 was considered significant.
Next, the associations between NAD/plasma metabolites (outcome variable) and the gut microbiota (explanatory variables) were examined. First, changes between baseline and 5 months in NAD metabolites corresponding to changes between baseline and 5 months in the gut microbiota were identified. Then, we identified NAD metabolite changes between baseline and 5 months associated to the baseline gut microbiota. Last, changes between baseline and 5 months in plasma metabolites and in blood clinical parameters, in relation to changes between baseline and 5 months in the gut microbiota, were identified. All the regression models mentioned above were adjusted for leaner/heavier status, sex, age, antibiotics usage, smoking status, and twinship.
Statistical analysis
Clinical variables, blood NAD metabolites, mitochondrial measures, and gene expression data are expressed as means ± SD for normally distributed variables and as median (interquartile range) for non-normal variables. The normality of the distribution was determined using the Shapiro-Wilk test. Variables with a P value of less than 0.05 were considered non-normally distributed. Outlier removal for gene expression and mtDNA amount data was performed in GraphPad Prism by using outlier analysis ROUT (Q = 1). Baseline differences between the cotwins (the leaner versus the heavier cotwins or the NR- versus placebo-treated cotwins), changes upon NR or placebo in all individuals (baseline versus 5 months), and differences in the cotwin responses to NR (BMI-discordant pairs) were assessed using paired Wilcoxon signed-rank test using R software, and the level of significance (two-tailed) was set at P < 0.05.
Acknowledgments
We thank the study participants for their valuable contribution to this research. We acknowledge M. Urjansson, K. Sohlo, T. Hallaranta, A. Muranen, I. Paetau, H. Perttunen-Nio, E. Silvennoinen, A. Sandelin, S. Saarinen, and M. Tuominen for their technical expertise. We also thank M. Lindman and I. Belevich (the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki) for the TEM sample preparation and help in preprocessing and analyzing the TEM images, respectively. We wish to acknowledge the core facility services Genome Biology Unit, HiLIFE and Faculty of Medicine, University of Helsinki, and Biocenter Finland and Light Microscopy Unit, Institute of Biotechnology, University of Helsinki. We also thank R. Dellinger and the Chromadex CERP Science Team.
Funding: This work was supported by Centre of Excellence funding from the Academy of Finland (#272376 to K.H.P. and #336823 to J.Ka.), Academy of Finland grants (286359, 314455, 335445, and Profi6 336449 to E.P.; 335443, 314383, 272376, and 266286 to K.H.P.; 308042 to S.P.; 297908 and 328685 to M.O.), Sigrid Jusélius Foundation (to K.H.P.), Finnish Medical Foundation (to H.A.K.L. and N.P.), Medical Society of Finland (to H.A.K.L.), Biomedicum Helsinki Foundation (to N.P.), Finnish Diabetes Research Foundation (to E.P.), Novo Nordisk Foundation (NNF20OC0060547, NNF17OC0027232, and NNF10OC1013354 to K.H.P.), Gyllenberg Foundation (to K.H.P.), Government Research Funds to Helsinki University Hospital (to K.H.P.), National Institutes of Health (R01HL147545 to C.B.), and Alfred E. Mann Family Foundation (to C.B.).
Author contributions: Conceptualization: E.P. and K.H.P. Investigation: H.A.K.L., M.K., A.He., S.G., M.S., M.S.S., S.H., S.S., N.P., T.S., J.Ku., A.Ha., J.T., and K.H.P. Formal analysis: H.A.K.L., M.K., A.He., M.M., B.W.v.d.K., and S.P. Visualization: H.A.K.L., M.K., A.He., B.W.v.d.K., and S.P. Resources: J.Ka. Supervision: E.P., K.H.P., M.-R.T., N.L., P.-H.G., M.T., M.L., C.B., P.K., S.P., and M.O. Writing—original draft: E.P., H.A.K.L., M.K., and A.He. Writing—review and editing: All authors.
Competing interests: C.B. is the inventor of intellectual property on the nutritional and therapeutic uses of NR. He serves as chief scientific advisor of ChromaDex, which licensed, developed, and commercialized NR technologies, and holds stock in ChromaDex. E.P. has received a speaking fee from ChromaDex. The other authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The gut microbiota 16S RNA sequencing data are available in the NCBI databank (the BioProject accession: PRJNA819007). Plasma metabolomic data have been deposited to the Metabolomics Workbench (Project ID: PR001356). The study whole blood samples were sent for measurement of NAD metabolites to C.B.’s laboratory under a material transfer agreement.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Tables S1, S8 to S10
Other Supplementary Material for this : manuscript includes the following:
Tables S2 to S7
REFERENCES AND NOTES
- 1.R. Jokinen, S. Pirnes-Karhu, K. H. Pietiläinen, E. Pirinen, Adipose tissue NAD+-homeostasis, sirtuins and poly(ADP-ribose) polymerases—Important players in mitochondrial metabolism and metabolic health. Redox Biol. 12, 246–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. Cantó, K. J. Menzies, J. Auwerx, NAD+ metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L. Rajman, K. Chwalek, D. A. Sinclair, Therapeutic potential of NAD-boosting molecules: The in vivo evidence. Cell Metab. 27, 529–547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. H. Ganji, G. D. Kukes, N. Lambrecht, M. L. Kashyap, V. S. Kamanna, Therapeutic role of niacin in the prevention and regression of hepatic steatosis in rat model of nonalcoholic fatty liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G320–G327 (2014). [DOI] [PubMed] [Google Scholar]
- 5.E. Pirinen, M. Auranen, N. A. Khan, V. Brilhante, N. Urho, A. Pessia, A. Hakkarainen, J. K. Ulla Heinonen, M. S. Schmidt, K. Haimilahti, P. Piirilä, N. Lundbom, M. R. Taskinen, C. Brenner, V. Velagapudi, K. H. Pietiläinen, A. Suomalainen, Niacin cures systemic NAD+ deficiency and improves muscle performance in adult-onset mitochondrial myopathy. Cell Metab. 31, 1078–1090. (2020). [DOI] [PubMed] [Google Scholar]
- 6.P. Bieganowski, C. Brenner, Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117, 495–502 (2004). [DOI] [PubMed] [Google Scholar]
- 7.D. W. Frederick, E. Loro, L. Liu, A. Davila Jr., K. Chellappa, I. M. Silverman, W. J. Quinn III, S. J. Gosai, E. D. Tichy, J. G. Davis, F. Mourkioti, B. D. Gregory, R. W. Dellinger, P. Redpath, M. E. Migaud, E. Nakamaru-Ogiso, J. D. Rabinowitz, T. S. Khurana, J. A. Baur, Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 24, 269–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R. Cerutti, E. Pirinen, C. Lamperti, S. Marchet, A. A. Sauve, W. Li, V. Leoni, E. A. Schon, F. Dantzer, J. Auwerx, C. Viscomi, M. Zeviani, NAD+-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 19, 1042–1049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. Canto, R. H. Houtkooper, E. Pirinen, D. Y. Youn, M. H. Oosterveer, Y. Cen, P. J. Fernandez-Marcos, H. Yamamoto, P. A. Andreux, P. Cettour-Rose, K. Gademann, C. Rinsch, K. Schoonjans, A. A. Sauve, J. Auwerx, The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.H. Zhang, D. Ryu, Y. Wu, K. Gariani, X. Wang, P. Luan, D. D’Amico, E. R. Ropelle, M. P. Lutolf, R. Aebersold, K. Schoonjans, K. J. Menzies, J. Auwerx, NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016). [DOI] [PubMed] [Google Scholar]
- 11.X. Yu, M. Xue, Y. Liu, Z. Zhou, Y. Jiang, T. Sun, H. Liang, Effect of nicotinamide riboside on lipid metabolism and gut microflora-bile acid axis in alcohol-exposed mice. Food Sci. Nutr. 9, 429–440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. A. Trammell, B. J. Weidemann, A. Chadda, M. S. Yorek, A. Holmes, L. J. Coppey, A. Obrosov, R. H. Kardon, M. A. Yorek, C. Brenner, Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci. Rep. 6, 26933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.V. V. Lozada-Fernandez, O. deLeon, S. L. Kellogg, F. L. Saravia, M. A. Hadiono, S. N. Atkinson, J. L. Grobe, J. R. Kirby, Nicotinamide riboside-conditioned microbiota deflects high-fat diet-induced weight gain in mice. mSystems 7, e0023021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. A. Trammell, M. S. Schmidt, B. J. Weidemann, P. Redpath, F. Jaksch, R. W. Dellinger, Z. Li, E. D. Abel, M. E. Migaud, C. Brenner, Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 7, 12948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. E. Airhart, L. M. Shireman, L. J. Risler, G. D. Anderson, G. A. Nagana Gowda, D. Raftery, R. Tian, D. D. Shen, K. D. O’Brien, An open-label, non-randomized study of the pharmacokinetics of the nutritional supplement nicotinamide riboside (NR) and its effects on blood NAD+ levels in healthy volunteers. PLOS ONE 12, e0186459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.C. R. Martens, B. A. Denman, M. R. Mazzo, M. L. Armstrong, N. Reisdorph, M. B. McQueen, M. Chonchol, D. R. Seals, Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD+ in healthy middle-aged and older adults. Nat. Commun. 9, 1286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O. L. Dollerup, B. Christensen, M. Svart, M. S. Schmidt, K. Sulek, S. Ringgaard, H. Stodkilde-Jorgensen, N. Moller, C. Brenner, J. T. Treebak, N. Jessen, A randomized placebo-controlled clinical trial of nicotinamide riboside in obese men: Safety, insulin-sensitivity, and lipid-mobilizing effects. Am. J. Clin. Nutr. 108, 343–353 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Y. S. Elhassan, K. Kluckova, R. S. Fletcher, M. S. Schmidt, A. Garten, C. L. Doig, D. M. Cartwright, L. Oakey, C. V. Burley, N. Jenkinson, M. Wilson, S. J. E. Lucas, I. Akerman, A. Seabright, Y. C. Lai, D. A. Tennant, P. Nightingale, G. A. Wallis, K. N. Manolopoulos, C. Brenner, A. Philp, G. G. Lavery, Nicotinamide riboside augments the aged human skeletal muscle NAD+ metabolome and induces transcriptomic and anti-inflammatory signatures. Cell Rep. 28, 1717–1728. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C. M. E. Remie, K. H. M. Roumans, M. P. B. Moonen, N. J. Connell, B. Havekes, J. Mevenkamp, L. Lindeboom, V. H. W. de Wit, T. van de Weijer, S. Aarts, E. Lutgens, B. V. Schomakers, H. L. Elfrink, R. Zapata-Perez, R. H. Houtkooper, J. Auwerx, J. Hoeks, V. B. Schrauwen-Hinderling, E. Phielix, P. Schrauwen, Nicotinamide riboside supplementation alters body composition and skeletal muscle acetylcarnitine concentrations in healthy obese humans. Am. J. Clin. Nutr. 112, 413–426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. Brakedal, C. Dolle, F. Riemer, Y. Ma, G. S. Nido, G. O. Skeie, A. R. Craven, T. Schwarzlmuller, N. Brekke, J. Diab, L. Sverkeli, V. Skjeie, K. Varhaug, O. B. Tysnes, S. Peng, K. Haugarvoll, M. Ziegler, R. Gruner, D. Eidelberg, C. Tzoulis, The NADPARK study: A randomized phase I trial of nicotinamide riboside supplementation in Parkinson’s disease. Cell Metab. 34, 396–407. (2022). [DOI] [PubMed] [Google Scholar]
- 21.C. F. Dolopikou, I. A. Kourtzidis, N. V. Margaritelis, I. S. Vrabas, I. Koidou, A. Kyparos, A. A. Theodorou, V. Paschalis, M. G. Nikolaidis, Acute nicotinamide riboside supplementation improves redox homeostasis and exercise performance in old individuals: A double-blind cross-over study. Eur. J. Nutr. 59, 505–515 (2020). [DOI] [PubMed] [Google Scholar]
- 22.O. L. Dollerup, S. Chubanava, M. Agerholm, S. D. Sondergard, A. Altintas, A. B. Moller, K. F. Hoyer, S. Ringgaard, H. Stodkilde-Jorgensen, G. G. Lavery, R. Barres, S. Larsen, C. Prats, N. Jessen, J. T. Treebak, Nicotinamide riboside does not alter mitochondrial respiration, content or morphology in skeletal muscle from obese and insulin-resistant men. J. Physiol. 598, 731–754 (2020). [DOI] [PubMed] [Google Scholar]
- 23.P. Mourikis, R. Sambasivan, D. Castel, P. Rocheteau, V. Bizzarro, S. Tajbakhsh, A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252 (2012). [DOI] [PubMed] [Google Scholar]
- 24.H. Zhang, J. Wen, A. Bigot, J. Chen, R. Shang, V. Mouly, P. Bi, Human myotube formation is determined by MyoD-Myomixer/Myomaker axis. Sci. Adv. 6, eabc4062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.W. P. Sun, M. Z. Zhai, D. Li, Y. Zhou, N. N. Chen, M. Guo, S. S. Zhou, Comparison of the effects of nicotinic acid and nicotinamide degradation on plasma betaine and choline levels. Clin. Nutr. 36, 1136–1142 (2017). [DOI] [PubMed] [Google Scholar]
- 26.K. Hanhineva, M. A. Lankinen, A. Pedret, U. Schwab, M. Kolehmainen, J. Paananen, V. de Mello, R. Sola, M. Lehtonen, K. Poutanen, M. Uusitupa, H. Mykkänen, Nontargeted metabolite profiling discriminates diet-specific biomarkers for consumption of whole grains, fatty fish, and bilberries in a randomized controlled trial. J. Nutr. 145, 7–17 (2015). [DOI] [PubMed] [Google Scholar]
- 27.E. Munukka, A. Rintala, R. Toivonen, M. Nylund, B. Yang, A. Takanen, A. Hänninen, J. Vuopio, P. Huovinen, S. Jalkanen, S. Pekkala, Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 11, 1667–1679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.H. Sokol, B. Pigneur, L. Watterlot, O. Lakhdari, L. G. Bermudez-Humaran, J. J. Gratadoux, S. Blugeon, C. Bridonneau, J. P. Furet, G. Corthier, C. Grangette, N. Vasquez, P. Pochart, G. Trugnan, G. Thomas, H. M. Blottiere, J. Dore, P. Marteau, P. Seksik, P. Langella, Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.I. Shats, J. G. Williams, J. Liu, M. V. Makarov, X. Wu, F. B. Lih, L. J. Deterding, C. Lim, X. Xu, T. A. Randall, E. Lee, W. Li, W. Fan, J. L. Li, M. Sokolsky, A. V. Kabanov, L. Li, M. E. Migaud, J. W. Locasale, X. Li, Bacteria boost mammalian host NAD metabolism by engaging the deamidated biosynthesis pathway. Cell Metab. 31, 564–579. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. Liu, X. Su, W. J. Quinn3rd, S. Hui, K. Krukenberg, D. W. Frederick, P. Redpath, L. Zhan, K. Chellappa, E. White, M. Migaud, T. J. Mitchison, J. A. Baur, J. D. Rabinowitz, Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S. Wang, T. Wan, M. Ye, Y. Qiu, L. Pei, R. Jiang, N. Pang, Y. Huang, B. Liang, W. Ling, X. Lin, Z. Zhang, L. Yang, Nicotinamide riboside attenuates alcohol induced liver injuries via activation of SirT1/PGC-1α/mitochondrial biosynthesis pathway. Redox Biol. 17, 89–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K. L. Seldeen, A. Shahini, R. Thiyagarajan, Y. Redae, M. Leiker, N. Rajabian, A. Dynka, S. T. Andreadis, B. R. Troen, Short-term nicotinamide riboside treatment improves muscle quality and function in mice and increases cellular energetics and differentiating capacity of myogenic progenitors. Nutrition 87-88, 111189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Y. Zhou, Z. Zhou, W. Zhang, X. Hu, H. Wei, J. Peng, S. Jiang, SIRT1 inhibits adipogenesis and promotes myogenic differentiation in C3H10T1/2 pluripotent cells by regulating Wnt signaling. Cell Biosci. 5, 61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.W. Shi, M. A. Hegeman, A. Doncheva, M. Bekkenkamp-Grovenstein, V. C. J. de Boer, J. Keijer, High dose of dietary nicotinamide riboside induces glucose intolerance and white adipose tissue dysfunction in mice fed a mildly obesogenic diet. Nutrients 11, 2439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Chen, Y. Ye, Y. Zhang, X. F. Pan, A. Pan, Weight change across adulthood in relation to all cause and cause specific mortality: Prospective cohort study. BMJ 367, l5584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. R. Guyton, H. E. Bays, Safety considerations with niacin therapy. Am. J. Cardiol. 99, 22C–31C (2007). [DOI] [PubMed] [Google Scholar]
- 37.A. Serrano, M. Asnani-Kishnani, C. Couturier, J. Astier, A. Palou, J. F. Landrier, J. Ribot, M. L. Bonet, DNA methylation changes are associated with the programming of white adipose tissue browning features by resveratrol and nicotinamide riboside neonatal supplementations in mice. Nutrients 12, 461 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R. H. Houtkooper, E. Pirinen, J. Auwerx, Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 13, 225–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Y. Fan, O. Pedersen, Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Y. Jiang, Y. Liu, M. Gao, M. Xue, Z. Wang, H. Liang, Nicotinamide riboside alleviates alcohol-induced depression-like behaviours in C57BL/6J mice by altering the intestinal microbiota associated with microglial activation and BDNF expression. Food Funct. 11, 378–391 (2020). [DOI] [PubMed] [Google Scholar]
- 41.D. Fangmann, E. M. Theismann, K. Turk, D. M. Schulte, I. Relling, K. Hartmann, J. K. Keppler, J. R. Knipp, A. Rehman, F. A. Heinsen, A. Franke, L. Lenk, S. Freitag-Wolf, E. Appel, S. Gorb, C. Brenner, D. Seegert, G. H. Waetzig, P. Rosenstiel, S. Schreiber, K. Schwarz, M. Laudes, Targeted microbiome intervention by microencapsulated delayed-release niacin beneficially affects insulin sensitivity in humans. Diabetes Care 41, 398–405 (2018). [DOI] [PubMed] [Google Scholar]
- 42.A. Heinken, M. T. Khan, G. Paglia, D. A. Rodionov, H. J. Harmsen, I. Thiele, Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J. Bacteriol. 196, 3289–3302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. Kaprio, The finnish twin cohort study: An update. Twin Res. Hum. Genet. 16, 157–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Heinonen, J. Buzkova, M. Muniandy, R. Kaksonen, M. Ollikainen, K. Ismail, A. Hakkarainen, J. Lundbom, N. Lundbom, K. Vuolteenaho, E. Moilanen, J. Kaprio, A. Rissanen, A. Suomalainen, K. H. Pietiläinen, Impaired mitochondrial biogenesis in adipose tissue in acquired obesity. Diabetes 64, 3135–3145 (2015). [DOI] [PubMed] [Google Scholar]
- 45.M. Graner, A. Seppalä-Lindroos, A. Rissanen, A. Hakkarainen, N. Lundbom, J. Kaprio, M. S. Nieminen, K. H. Pietiläinen, Epicardial fat, cardiac dimensions, and low-grade inflammation in young adult monozygotic twins discordant for obesity. Am. J. Cardiol. 109, 1295–1302 (2012). [DOI] [PubMed] [Google Scholar]
- 46.D. Stefan, A. Andrasecu, E. Popa, A. Lazariev, O. Strbak, Z. Starcuk, M. Cabañas, D. Ormondt, D. Graveron-Demilly, Quantitation of magnetic resonance spectroscopy signals: The jMRUI software package. Meas. Sci. Technol. 20, 104035 (2009). [Google Scholar]
- 47.L. Vanhamme, A. van den Boogaart, S. Van Huffel, Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J. Magn. Reson. 129, 35–43 (1997). [DOI] [PubMed] [Google Scholar]
- 48.A. Kotronen, V. R. Velagapudi, L. Yetukuri, J. Westerbacka, R. Bergholm, K. Ekroos, J. Makkonen, M. R. Taskinen, M. Oresic, H. Yki-Järvinen, Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia 52, 684–690 (2009). [DOI] [PubMed] [Google Scholar]
- 49.S. Heinonen, L. Saarinen, J. Naukkarinen, A. Rodriguez, G. Fruhbeck, A. Hakkarainen, J. Lundbom, N. Lundbom, K. Vuolteenaho, E. Moilanen, P. Arner, S. Hautaniemi, A. Suomalainen, J. Kaprio, A. Rissanen, K. H. Pietiläinen, Adipocyte morphology and implications for metabolic derangements in acquired obesity. Int. J. Obes. 38, 1423–1431 (2014). [DOI] [PubMed] [Google Scholar]
- 50.B. W. van der Kolk, S. Saari, A. Lovric, M. Arif, M. Alvarez, A. Ko, Z. Miao, N. Sahebekhtiari, M. Muniandy, S. Heinonen, A. Oghabian, R. Jokinen, S. Jukarainen, A. Hakkarainen, J. Lundbom, J. Kuula, P. H. Groop, T. Tukiainen, N. Lundbom, A. Rissanen, J. Kaprio, E. G. Williams, N. Zamboni, A. Mardinoglu, P. Pajukanta, K. H. Pietiläinen, Molecular pathways behind acquired obesity: Adipose tissue and skeletal muscle multiomics in monozygotic twin pairs discordant for BMI. Cell Rep. Med. 2, 100226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.I. Belevich, M. Joensuu, D. Kumar, H. Vihinen, E. Jokitalo, Microscopy image browser: A platform for segmentation and analysis of multidimensional datasets. PLOS Biol. 14, e1002340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. A. Trammell, C. Brenner, Targeted, LCMS-based metabolomics for quantitative measurement of NAD+ metabolites. Comput. Struct. Biotechnol. J. 4, e201301012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. J. Aryee, A. E. Jaffe, H. Corrada-Bravo, C. Ladd-Acosta, A. P. Feinberg, K. D. Hansen, R. A. Irizarry, Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. van Iterson, E. W. Tobi, R. C. Slieker, W. den Hollander, R. Luijk, P. E. Slagboom, B. T. Heijmans, MethylAid: Visual and interactive quality control of large Illumina 450k datasets. Bioinformatics 30, 3435–3437 (2014). [DOI] [PubMed] [Google Scholar]
- 55.W. Zhou, P. W. Laird, H. Shen, Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 45, e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. P. Fortin, A. Labbe, M. Lemire, B. W. Zanke, T. J. Hudson, E. J. Fertig, C. M. Greenwood, K. D. Hansen, Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 15, 503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. E. Teschendorff, F. Marabita, M. Lechner, T. Bartlett, J. Tegner, D. Gomez-Cabrero, S. Beck, A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y. Zheng, B. T. Joyce, L. Liu, Z. Zhang, W. A. Kibbe, W. Zhang, L. Hou, Prediction of genome-wide DNA methylation in repetitive elements. Nucleic Acids Res. 45, 8697–8711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. Noerman, M. Kokla, V. M. Koistinen, M. Lehtonen, T. P. Tuomainen, C. Brunius, J. K. Virtanen, K. Hanhineva, Associations of the serum metabolite profile with a healthy Nordic diet and risk of coronary artery disease. Clin. Nutr. 40, 3250–3262 (2021). [DOI] [PubMed] [Google Scholar]
- 60.J. Pekkinen, K. Olli, A. Huotari, K. Tiihonen, P. Keski-Rahkonen, M. Lehtonen, S. Auriola, M. Kolehmainen, H. Mykkänen, K. Poutanen, K. Hanhineva, Betaine supplementation causes increase in carnitine metabolites in the muscle and liver of mice fed a high-fat diet as studied by nontargeted LC-MS metabolomics approach. Mol. Nutr. Food Res. 57, 1959–1968 (2013). [DOI] [PubMed] [Google Scholar]
- 61.L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, T. Hankemeier, N. Hardy, J. Harnly, R. Higashi, J. Kopka, A. N. Lane, J. C. Lindon, P. Marriott, A. W. Nicholls, M. D. Reily, J. J. Thaden, M. R. Viant, Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. Lensu, R. Pariyani, E. Mäkinen, B. Yang, W. Saleem, E. Munukka, M. Lehti, A. Driuchina, J. Linden, M. Tiirola, L. Lahti, S. Pekkala, Prebiotic xylo-oligosaccharides ameliorate high-fat-diet-induced hepatic steatosis in rats. Nutrients 12, 3225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. E. Ritchie, B. Phipson, D. Wu, Y. Hu, C. W. Law, W. Shi, G. K. Smyth, limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Tables S1, S8 to S10
Tables S2 to S7