Summary
Tissue repair processes maintain proper organ function following mechanical or infection-related damage. In addition to antibacterial properties, mucosal associated invariant T (MAIT) cells express a tissue repair transcriptomic program and promote skin wound healing when expanded. Herein, we use a human-like mouse model of full-thickness skin excision to assess the underlying mechanisms of MAIT cell tissue repair function. Single-cell RNA sequencing analysis suggested that skin MAIT cells already express a repair program at steady state. Following skin excision, MAIT cells promoted keratinocyte proliferation, thereby accelerating healing. Using skin grafts, parabiosis, and adoptive transfer experiments, we show that MAIT cells migrated into the wound in a T cell receptor (TCR)-independent but CXCR6 chemokine receptor-dependent manner. Amphiregulin secreted by MAIT cells following excision promoted wound healing. Expression of the repair function was probably independent of sustained TCR stimulation. Overall, our study provides mechanistic insights into MAIT cell wound healing function in the skin.
Keywords: MAIT cells, tissue repair, skin excision, amphiregulin, scRNA-seq, TCR signaling, CXCR6 chemokine receptor
Graphical abstract

Highlights
-
•
MAIT cells accelerate wound closure in a human-like skin injury model
-
•
MAIT cells migrate into the inflamed skin via CXCR6/CXCL16 and independently of MR1
-
•
MAIT cell pro-repair function is independent of sustained TCR signaling
-
•
Amphiregulin production by MAIT cells accelerates wound closure
MAIT cells have tissue repair properties, but the underlying mechanisms are unclear. du Halgouet et al. demonstrate that MAIT cells accelerate wound closure by increasing epithelial proliferation. MAIT cells are recruited into the injured skin in a CXCL16/CXCR6-dependent MR1-independent manner, and their pro-repair effect is related to amphiregulin production.
Introduction
Restoring skin barrier following damage is key to maintain its function. The first step of skin healing is an inflammatory phase preventing infection and promoting debris clearance. Then, proliferation and migration of keratinocytes, endothelial cells, and fibroblasts creates new tissue. The final remodeling and reorganization phase lasts for months.1 Delayed or improper healing may result in pain and infection, up to cutaneous carcinogenesis and limb amputations. Understanding the fine tuning of skin healing is therefore crucial.
Anti-infectious and pro-inflammatory functions of T cells are well described, but several T cell subsets are also involved in skin homeostasis. Upon skin injury, type 17 commensal-specific CD8+ T cells express type 2 cytokines leading to tissue repair.2 Skin γδ T cells also promote wound healing through secretion of various molecules including keratinocyte growth factors and interleukin-17 (IL-17).3,4 Recently, mucosal associated invariant T (MAIT) cells have been shown to have tissue repair potential,5–6 but the in vivo mechanisms involved are unclear.
In humans, MAIT cells represent the most abundant T cell subset with a single specificity.7,8 MAIT cells recognize an unstable compound, 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), stabilized and presented by the major histocompatibility complex (MHC) class 1-related molecule MR1.9 5-OP-RU derives from the riboflavin (vitamin B2) synthesis pathway present in most bacteria and yeasts but not in animal cells.10 In mice, MAIT cells encompass MAIT1 (expressing the transcription factor Tbet) and MAIT17 (expressing the transcription factor RORɣt) cell subsets.11 These subsets secrete different effector molecules, from interferon (IFN)-ɣ and cytotoxic molecules for MAIT1 cells to IL-17, (G)M-CSF ([granulocyte] monocyte colony stimulating factor), and tissue repair mediators for MAIT17 cells (reviewed in Legoux et al.12).
MAIT cells are numerous in human tissues, representing 2%–10% of T cells in the gut, around 4% in the lungs, and up to 15% in the liver.13 Tissue homing is driven by the master transcription factor promyelocytic leukemia zink finger (PLZF), which downregulates Klf2 and its target CD62L.14 Parabiosis experiments, which join the vascular systems of two mice, demonstrate that MAIT cells reside in the lungs, liver, and spleen at steady state, similar to invariant natural killer T (iNKT) cells.15,16 Altogether, these innate-like T cell populations resemble mainstream tissue-resident memory (TRM) cells that remain in tissues following resolution of infection and confer protection upon reinfection.17
MAIT cells represent 0.5%–2% of T cells in the human skin,13 reaching up to 40% in the mouse skin with a high variability range (1%–40% depending on cage origin).5,15 The increase of MAIT cell numbers in Tcrd−/− mice5 suggests competition for peripheral niches, but deletion of the T cell receptor (TCR) delta locus also influences the TCR alpha chain rearrangement process.18 In vivo, after antigen-driven expansion, MAIT cells promote skin wound healing in C57BL/6 mice.5 In vitro, MAIT cell culture supernatant promotes proliferation of an intestinal epithelial cell line.6 Whether this holds true in vivo remains unknown. Moreover, the mechanisms triggering MAIT cell repair function have not been determined. In vitro analyses suggest that MAIT cells acquire tissue repair program following TCR triggering.19–6 In vivo, whether recognition of microbiota-derived 5-OP-RU or any endogenous ligand occurs during wound healing is unknown. Additional signals may be required for eliciting MAIT cell tissue repair function, such as cytokines, tissue cues, or activating signal duration.12
To address the mechanisms of MAIT cell repair function, we turned to a human-like mouse model of full-thickness skin excision. We showed that wound closure was accelerated in the presence of MAIT cells and analyzed the mechanisms leading to MAIT cell accumulation at the wound site. We tested whether TCR triggering was necessary for either MAIT cell accumulation or tissue repair function. Finally, we explored the mechanisms favoring wound healing and showed a key role of MAIT cell-derived amphiregulin (Areg). Thus, our work unraveled migration and effector mechanisms leading to MAIT cell-dependent skin repair.
Results
MAIT cells accelerate wound closure
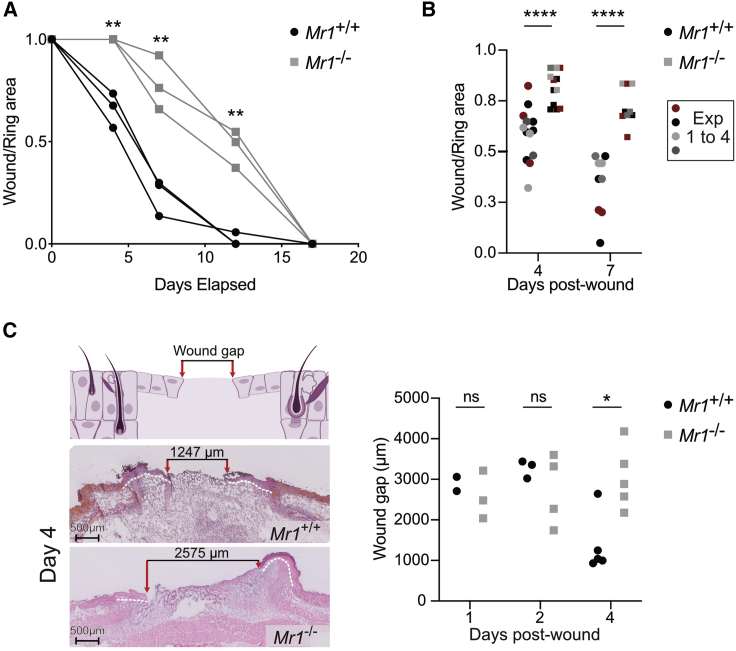
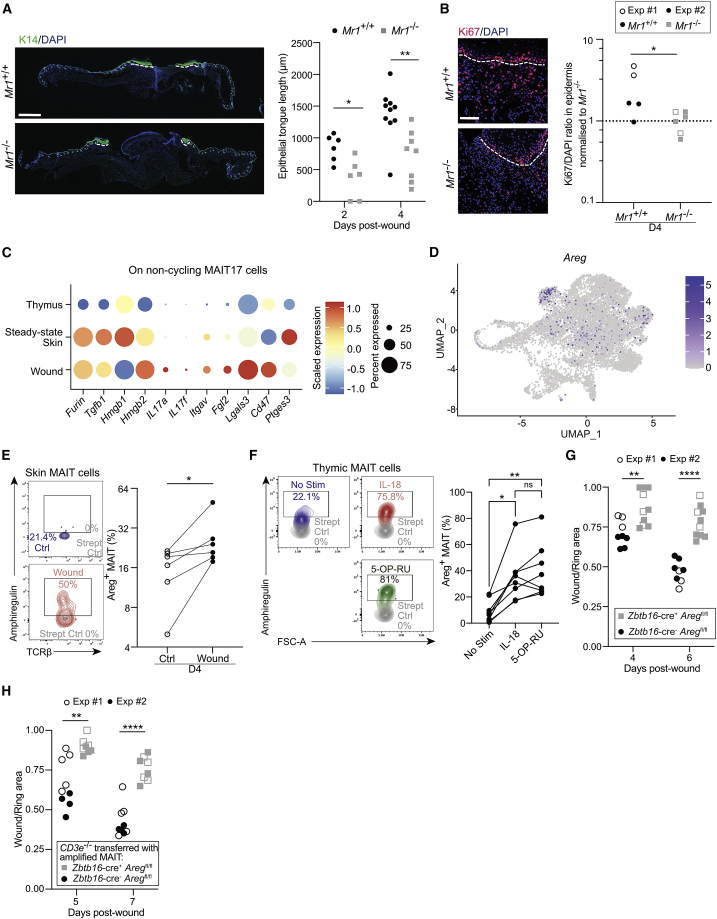
Until now, the pro-repair functions of MAIT cells in vivo have been observed after increasing their numbers in the skin, either in S. epidermidis-associated Tcrd−/− animals or after topical application of 5-OP-RU in C57BL/6 mice.5 To dissect the mechanisms by which MAIT cells improve skin wound healing in immunocompetent non-manipulated animals, we took advantage of the B6-MAITCAST mouse strain with higher MAIT cell numbers than the C57BL/6 mice.20 Of note, in the skin, MAIT cell numbers depend rather on the housing cages than on the strain used.5,15 To prevent skin contraction occurring in mice but not in humans, full-thickness excision punches were splinted using a silicon ring.21 We compared wound healing in the presence or absence of MAIT cells using Mr1+/+ and Mr1−/− mice, respectively. Wound closure (assessed by the wound on ring area ratio) was faster in Mr1+/+ as compared with Mr1−/− animals, as early as day 4 (Figures 1A and 1B). Analysis of hematoxylin and eosin-safran-colored tissue sections evidenced smaller wound gaps at day 4 in Mr1+/+ as compared with Mr1−/− animals (Figure 1C).
Figure 1.
MAIT cells accelerate wound closure
Full-thickness wounds were performed on B6-MAITCAST mice and splinted with a silicone ring to prevent epithelial contraction.
(A) Longitudinal follow-up of wound surface (ratio wound over ring areas) for Mr1+/+ (black circle) or Mr1−/− (gray square) littermates. Blind experiment. t test.
(B) Wound surface at days 4 and 7. Pooled data from four independent experiments (n4 = 13; n7 = 9). Mann-Whitney test.
(C) Hematoxylin and eosin-saffron staining of Mr1+/+ and Mr1−/− wounds 4 days after excision and longitudinal follow-up of wound gap (distance between the epithelial tongues). Pooled data from two independent experiments analyzed blindly (n1 = 2/3; n2 = 3/4; n4 = 5). Mann-Whitney test.
To test whether the MR1 molecule itself was involved in wound healing, we assessed wound closure in Mr1+/+ and Mr1−/− mice devoid of T cells (Cd3e−/− mice). As expected in the absence of T cells, wound closure was delayed (50% closure at day 14 instead of day 6 in B6-MAITCast strain), but no difference was observed between Mr1+/+ and Mr1−/− backgrounds (Figure S1A). In the T cell compartment, MR1 deficiency only affects MAIT cells.8,22 Hence, MAIT cells accelerate wound healing early on in this human-like excision model.
Skin MAIT cells are type 17, express a tissue repair program, and increase in numbers at the wound site
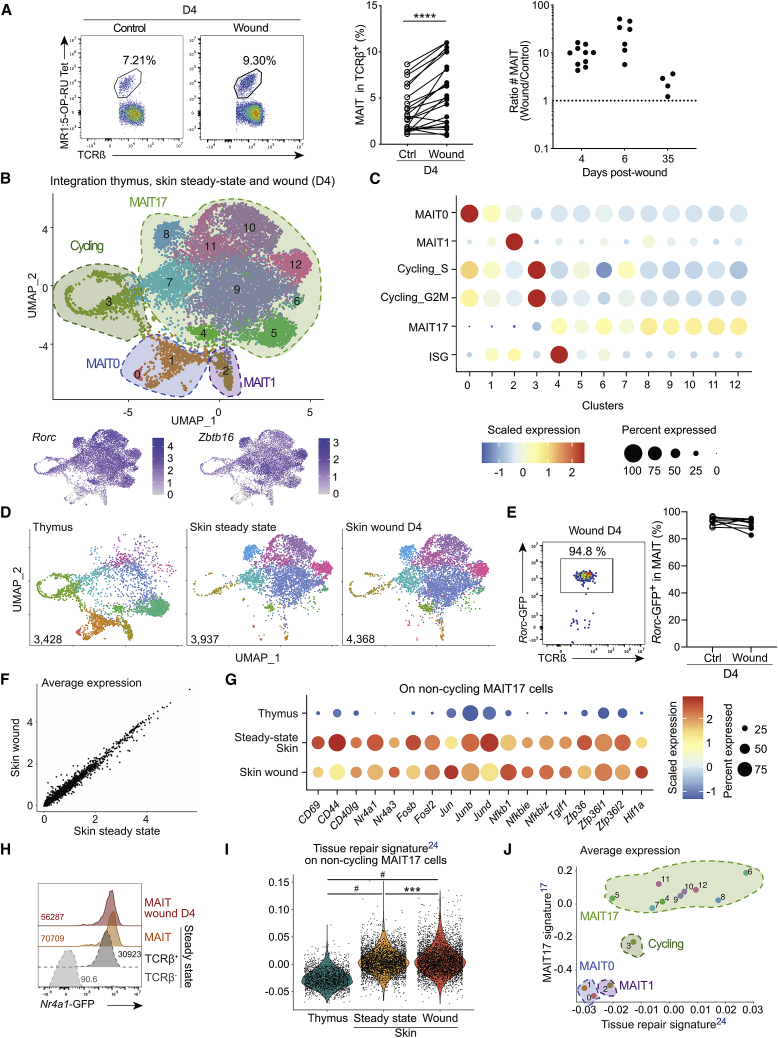
To better understand MAIT cell involvement in wound healing, we analyzed their number and phenotype in the skin. Four days after excision, the percentage of TCRβ+MR1:5-OP-RU Tet+ MAIT cells (Figure S2A) was significantly increased at the wound site as compared with contra-lateral control skin (Figure 2A, left and middle panels). To account for the variability of MAIT cell numbers between animals,5 the MAIT cell number at the wound site was normalized to the number from the same surface of the control site. MAIT cell numbers at the wound site increased up to 10-fold at days 4 and 6 and returned to basal numbers by day 35 (Figure 2A, right panel). Thus, skin repair is associated with a large and early increase of MAIT cell numbers following excision.
Figure 2.
Skin MAIT cells accumulate in the wound and constitute a homogeneous type 17 T cell population with a tissue repair program
(A) Flow cytometry staining (left), frequency (middle), and number (left, ratio wound over control numbers) of skin MAIT cells from wound and control sites at various time points. Pooled data from seven (n = 22) and four (nD4 = 10, nD6 = 7, and nD35 = 4) independent experiments for frequencies and numbers, respectively. Wilcoxon test.
(B) MAIT cells sorted from thymus, wound (D4), and steady-state skin were analyzed by scRNA-seq and integrated. UMAP (Uniform Manifold Approximation and Projection, top) and features plot for Zbtb16 and Rorc expression (bottom) are displayed.
(C) Cluster were defined by signature enrichment (Table S1; STAR Methods).
(D) UMAP from (B) split according to dataset origin.
(E) Rorc-GFP reporter expression by MAIT cells from wound and control sites. Pooled data from three independent experiments (n = 8). Please also see Figure S2B.
(F) Average gene expression from MAIT cells in wound site and steady-state skin.
(G) Differentially expressed genes in non-cycling MAIT17 cells from skin (wound and steady state) as compared to thymus. The average expression was calculated on scaled data after subsetting MAIT17 clusters from (B).
(H) Nr4a1-GFP reporter expression by skin MAIT cells from wound (red) and control (orange) skin sites, by steady-state non-MAIT TCRβ+ (dark gray) and by TCRβ− (light gray) cells. Data are representative of two independent experiments (n = 5).
(I) Tissue repair23 signature score on non-cycling MAIT17 cells. Tukey’s multiple comparison test. Please also see Figure S2E.
(J) Average expression of tissue repair23 and MAIT1715 signatures on clusters from (B). Please also see Figure S2F.
To provide an unbiased view of MAIT cell functions in the skin, we characterized their transcriptome at steady state and 4 days after excision using single-cell RNA sequencing (scRNA-seq) from B6-MAITCAST mice. The two datasets were integrated together with a thymic dataset24 to provide a reference (Figure 2B, top panel). Therefore, 12 clusters were defined. Based on expression of Zbtb16 and Rorc (Figure 2B, bottom panel), as well as gene sets from the literature (Figure 2C; Table S1), MAIT0 (clusters 0 and 1), MAIT1 (cluster 2), cycling MAIT (cluster 3, mainly thymus), and MAIT17 (clusters 4–12) cells were identified. Despite low expression of the MAIT17 signature, cluster 3 belonged to the MAIT17 cell subtype, as seen by Rorc expression (Figure 2B). Importantly, MAIT1, MAIT17, and cycling cells from the three datasets merged, which demonstrated successful integration.
Analyzing the differentially expressed genes between all clusters (Table S2, adjusted p value < 0.05) allowed detailed description of MAIT17 cell clusters. The thymus-specific cluster 5 (Figure 2D; Table S2) expressed the ribosomal Rpl and Rps genes, indicating active protein synthesis. On the contrary, the ribosomal genes were downregulated in the skin-specific cluster 6, suggesting resting cells. Many IFN-related genes (including Isg15, Cxcl10, Ifit1, Stat1, and Bst2) were expressed in cluster 4, defining IFN-stimulated gene-MAIT cells both in the thymus and skin, as described for thymic iNKT cells25 (Figure 2C). Gzmb and Gzmc had the highest fold change in cluster 7 (Table S2), suggesting cytotoxic capacities. The skin-specific cluster 8 (Figure 2D) overexpressed genes associated with tissue repair such as Il17a, Il17f, Areg, the hypoxia-induced factor 1-alpha (HIF1α) Hif1a, Itgav, and Fgl2 (Table S2). No specific function was identified in the remaining MAIT17 cell clusters (9–12) which represent variations of a common program. Thus, MAIT17 cells span various transcriptional states and effector functions, including a skin-specific subset expressing repair mediators.
In the skin, most MAIT cells were type 17 both before and after excision (Figures 2B and 2D), as confirmed at the protein level using a Rorc-GFP reporter mouse (Figure 2E) and intracellular staining for the transcription factors RORγt and Tbet (Figure S2B). Moreover, scRNA-seq datasets from wound and steady-state skins fully overlapped and were evenly distributed in the different clusters (Figures 2D and S2C). Furthermore, gene expression was highly correlated between the two datasets (Figure 2F), suggesting that the functional program responsible for accelerated wound closure was already expressed at a steady state. To better understand skin MAIT17 cell specificities, we compared skin and thymic MAIT17 cell clusters (Table S3). Skin MAIT17 cells were more activated with overexpression of Cd69, Cd44, Nr4a1, and Nr4a3, as well as Jun, Fos, and Nfkb pathways (Figure 2G; Table S3). TCR signaling in MAIT cells from the skin was confirmed by flow cytometry using the Nr4a1-GFP reporter mouse (Figure 2H). Additionally, skin MAIT17 cells overexpressed the TGF-β-induced factor homeobox Tgif1 (Figure 2G). Accordingly, skin MAIT17 cells overexpressed genes identified in skin TRM cells expressing the TGFBR2 receptor26 (Figure S2D). These results suggest that skin MAIT cells rely on TGF-β for retention and functions, similarly to mainstream TRM cells.26,27,28,29 Zfp36, Zfp36l1, and Zfp36l2 were also upregulated in skin MAIT17 cells (Table S3). The corresponding proteins regulate the stability of mRNAs encoding cytokines and other immune mediators.30
Finally, overexpression of Hif1a (Figure 2G) suggested that skin MAIT17 cells have tissue repair capacity.31 To formally assess this hypothesis, we tested the enrichment of a tissue repair signature from commensal-specific skin CD8+ T cells23 previously used to assess MAIT cell tissue repair potential.19 All MAIT17 cells from the skin overexpressed the repair signature as compared with thymic cells (Figure 2I). This was confirmed using three other tissue repair gene sets expressed by Areg-producing regulatory T cells responsible for muscle32 or lung33 repair or demonstrated to have an in vivo repair function in full-thickness wounds (identified in the TiRe database34) (Figure S2E). On average, MAIT17 cells from cluster 8 highly expressed all four tissue repair signatures, as did the skin-specific clusters 6, 9, 10, and 12 (Figures 2I and S2F). MAIT17 cells from the thymus (cluster 5) slightly overexpressed the tissue repair signatures as compared with immature MAIT0 cells (Figures 2I and S2F). Thus, the MAIT17 cell program is associated with tissue repair functions, and skin location reinforced this program.
Altogether, the number of MAIT cells increased in the skin after excision, but their transcriptome was not modified. Notably, wound closure was correlated (R2 = 0.40) with the increase in T cell numbers (Figure S2G). The correlation was slightly higher (R2 = 0.44) with the increase of MAIT cell numbers in the wound (but not with the normalized percent of MAIT cells within T cells) (Figure S2H), further suggesting the involvement of MAIT cells in wound healing.
MAIT cells are recruited to the inflamed skin
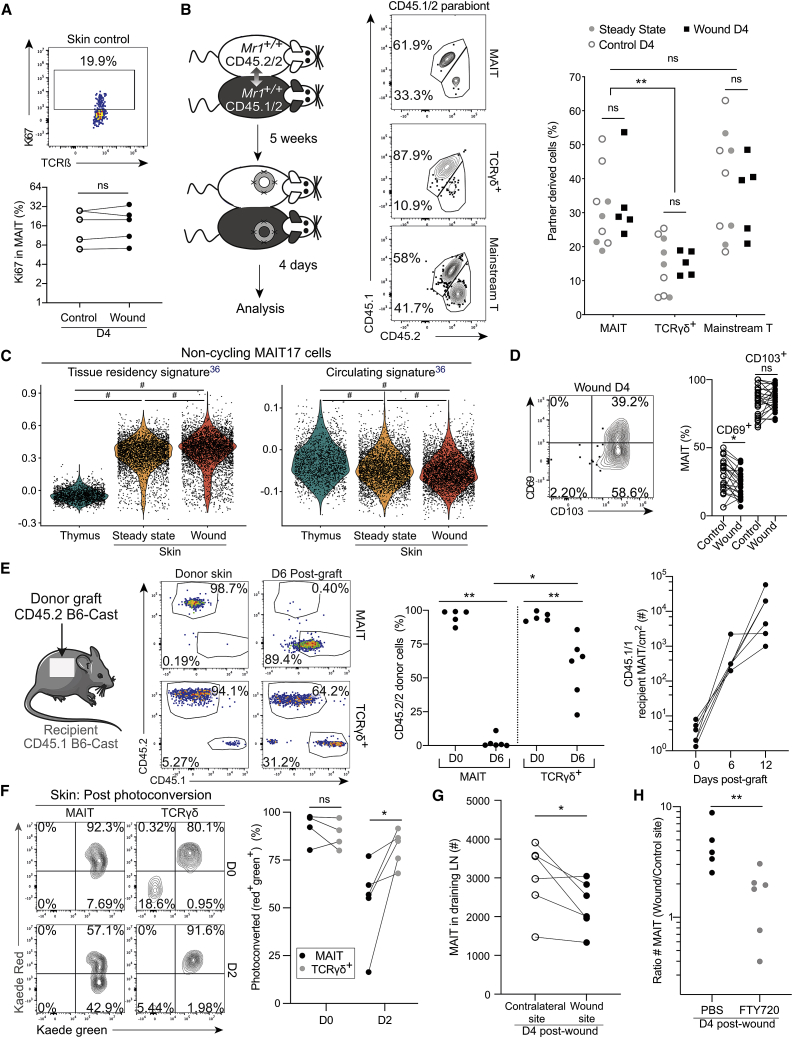
Increased MAIT cell number in the wound could result from either in situ proliferation or recruitment. Ki67 staining showed that MAIT cells proliferated similarly at wound and control sites (Figure 3A), suggesting that proliferation alone was not responsible for the increased number of MAIT cells in the wound. To determine whether MAIT cells were recruited, we performed skin excision on parabiotic pairs (Figure 3B, left panel). By contrast to lung MAIT cells which were mostly of host origin (Figure S3A), skin MAIT cells had exchanged between the two parabionts after 5 weeks of parabiosis: up to 50% of MAIT cells were partner derived in steady state, control, or wound skins (Figure 3B). This high exchange rate was not a technical artifact as skin γδ T cells remained in their original parabiont as expected.35 Still, higher expression of a residency signature (and slightly less expression of a circulating signature)36 was observed in MAIT17 cells from the skin as compared with the thymus (Figure 3C). The shorter retention time of MAIT cells in the skin as compared with other organs could be related to their variable expression pattern of CD103 and CD69: while CD69 and CD103 are both necessary for virus-specific skin TRM persistence,37 MAIT cells were mainly CD103+ but expressed low to medium amounts of CD69 (Figure 3D).
Figure 3.
MAIT cells are recruited into the inflamed skin
(A) Ki67 expression by MAIT cells. Data are from two (n = 5) independent experiments. Wilcoxon test.
(B) Parabiosis protocol (left) and CD45.2/2 and CD45.1/2 staining (middle). Percentage of partner-derived MAIT, ɣδ T, and mainstream T cells in the skin (right) at steady-state and in the wound and control sites 4 days after excision. Data are from three independent experiments (nsteady state+control = 9; nexcision = 5). Sídák multiple comparison test. Please also see Figure S3A.
(C) Tissue residency and circulating36 signature scores on non-cycling MAIT17 cells. Tukey’s multiple comparison test.
(D) CD69 and CD103 expression by MAIT cells. Pooled data from six independent experiments (nCD69 = 23; nCD103 = 27). Wilcoxon test.
(E) Graft protocol and CD45.2 and CD45.1 staining (left). CD45.2 donor cell frequency in MAIT and ɣδ T cells from the donor skin (D0) and after 6 days in the graft (middle). Absolute number of recipient CD45.1+ MAIT cells in grafts from D0, D6, and D12 (right, grafts from same donor are linked). Pooled data from two independent experiments (nD0 = 5; nD6 = 3/6; nD12 = 6). Mann-Whitney and Wilcoxon tests as appropriate.
(F) Example of Kaede green and red expression (left) and frequency of photoconverted cell (right) in skin MAIT and ɣδ T cells. Pooled data from two independent experiments (nD0 = 4, nD2 = 5). Paired t test.
(G) The number of MAIT cells in the inguinal and brachial LNs draining the wound or the control sites. Pooled data from two independent experiments (n = 6). Paired t test. Please also see Figures S3B and S3C.
(H) Numbers of MAIT cells (ratio wound over control sites) 4 days after excision in FTY720- or PBS-treated mice. Pooled data from two independent experiments (nPBS = 5; nFTY720 = 6). Mann-Whitney test. Please also see Figure S3D.
As skin MAIT cells exchanged at steady state, parabiotic pairs were not suitable to study their trafficking after excision. We turned to a skin graft model (Figure 3E, left panel). Six days after grafting, all MAIT cells within the graft originated from the recipient, while around 70% of ɣδ T cells remained of graft origin (Figure 3E, middle panel), demonstrating possible MAIT cell recruitment into the skin. Moreover, MAIT cell numbers increased over time in the graft (Figure 3E, right panel), confirming their influx into the healing skin.
To assess whether MAIT cells were recruited from other organs or surrounding skin similarly to cutaneous ɣδ T cells,38,39 we used the Kaede photoconvertible mouse. This mouse ubiquitously expresses the Kaede GFP, which shifts from green to red light emission after violet light illumination.40 Following whole-body illumination (D0), all T cells including γδ T and MAIT cells were photoconverted (Green+Red+) in the skin (Figure 3F). ɣδ T cells remained largely photoconverted at day 2, confirming their residency in the skin. In contrast, skin MAIT cells were replaced by non-converted ones as soon as day 2 (Figure 3F). These results indicate recruitment from outside the skin as full-thickness skin comprising dermis and epidermis was photoconverted at day 0.
About 1%–10% of photoconverted MAIT and γδ T cells were recovered from the draining inguinal and brachial lymph nodes (LNs) at day 2 (Figure S3B), suggesting migration from the skin to the draining LNs, as described for γδ T cells.41 Additionally, MAIT cell numbers were lower in the LNs draining the excision as compared with the contra-lateral sites (Figure 3G) and the LN of non-excised mice (Figure S3C). To test whether MAIT cells were recruited from the LNs to the wound, we treated excised mice with FTY720. This S1P receptor agonist blocks T cell egress from the LNs,42 resulting in strong T cell decrease in the blood (Figure S3D). FTY720 blockade inhibited the increase of MAIT cell numbers in the wound (Figure 3H), suggesting traffic through the LNs, despite the expression of a tissue residency transcriptional program in the skin.
Cognate stimulation is not required for MAIT cell recruitment and repair functions
The full-thickness excision punch puts skin microbiota in direct contact with the wound. As the MAIT cell cognate ligand is produced by most skin bacteria,5,43 we studied whether MAIT cell recruitment to the skin relied on ligand recognition. As a surrogate, we used Mr1−/− mice which do not present antigen to MAIT cells. MAIT cells infiltrated Mr1−/− skin grafted onto a Mr1+/+ mouse, reaching similar numbers to those of Mr1+/+ grafts (Figure 4A). In parabiosis experiments linking Mr1+/+ and Mr1−/− mice, MAIT cell numbers in the skin of the Mr1−/− parabiont almost reached those of the Mr1+/+ parabionts, both at control and wound sites (Figure 4B). Thus, MR1 expression on skin resident cells is not necessary for MAIT cell migration into the skin.
Figure 4.
MAIT cell recruitment and skin healing are independent of MR1
(A) Grafts were performed on Mr1+/+ animals. MAIT cell staining (left) and numbers (right) in Mr1−/− and Mr1+/+ grafts before transplant (D0) and longitudinally after grafting. Pooled data from 2 independent experiments (Mr1+/+: nD0 = 3, nD6,12 = 4; Mr1−/−: nD0 = 3, nD6 = 4, nD12 = 6). Unpaired t test.
(B) Mr1−/− and Mr1+/+ mice were parabiosed for 5 weeks. MAIT cell staining in the skin of the Mr1−/− parabiont skin (left) and numbers at wound and control sites (right) for Mr1−/− and Mr1+/+ parabionts and Mr1−/− control mice. Pooled data from two independent experiments (nMr1−control = 3; nMr1−parabiont = 9; nMr1+parabiont = 9). Tukey multiple comparison test.
(C) Percent of wound closure in Mr1−/− and Mr1+/+ parabionts and control Mr1−/− mice. Pooled data from two independent experiments (nMr1−/−alone = 3; nMr1−/−paired = 8; nMr1+/+paired = 8). Mann-Whitney and Wilcoxon tests as appropriate.
(D) Mean fluorescence intensity of GFP expression on MAIT cells at wound and control skin sites in Nr4a1-GFP animals. Pooled data from two independent experiments (n = 6). Paired t test.
(E) MAIT cell numbers at wound and control sites 4 days after transfer into Cd3e−/−Mr1+/+ and Mr1−/− mice. Pooled data from two independent experiments (nMr1+/+ = 4; nMr1−/− = 4). Mann-Whitney test. Please also see Figures S4A–S4C.
(F) Longitudinal follow-up of wound surface of transferred Cd3e−/−Mr1+/+ and Mr1−/− mice and non-transferred Cd3e−/−Mr1+/+ control mice. Pooled data from two independent experiments with one blinded (nWithTransfer = 4/4; nWithoutTransfer =8). Please also see Figure S4D.
MAIT cell infiltration in the Mr1−/− parabiont was associated with increased wound closure as compared with mice devoid of MAIT cells (Figure 4C), suggesting that MAIT cell TCR triggering is not necessary for wound healing. This result is consistent with the decrease of Nr4a1-GFP reporter expression in MAIT cells following excision (Figure 4D). Still, wound closure was delayed in Mr1−/− as compared with the Mr1+/+ parabionts (Figure 4C), which might be related to a lower number of MAIT cells (Figure 4B, right panel). Since other hematopoietic cells exchange during parabiosis, MR1 presentation may have occurred in the wound of Mr1−/− animals or in the Mr1+/+ parabiont before migration. To formally demonstrate that MAIT cell repair function was independent of cognate stimulation, we transferred in vitro expanded MAIT cells into excised Cd3e−/− mice. MR1:5-OP-RU tetramer+-enriched thymic cells were expanded using 5-OP-RU at day 0 and IL-2 for 10–15 days (Figure S4A; STAR Methods). Except for high expression of Ki67 and CD69, which are linked to in vitro activation, expanded MAIT cells retained their MAIT17 phenotype (RORɣt+Tbet−) after expansion, with high CXCR6 expression and more than 50% of the cells being CD103+ (Figure S4B). More than 96% of transferred cells were MAIT cells (Figure S4B), which was still the case in vivo after 4 days (Figure S4C). Transferring MAIT cells accelerated wound closure (Figure S4D). Expanded MAIT cells were then transferred into Cd3e−/− Mr1+/+ and Mr1−/− animals. In this system, only transferred MAIT cells expressed MR1. Again, MAIT cell recruitment into the wound site (Figure 4E) and wound closure (Figure 4F) were independent of TCR triggering, as confirmed by the absence of Nr4a1-GFP expression by transferred cells both at transfer and 4 days later (Figure S4E). Thus, MAIT cell recruitment and involvement in wound healing do not rely on sustained cognate interactions with MR1-presenting cells.
CXCR6 is necessary for MAIT cell recruitment to the wound site
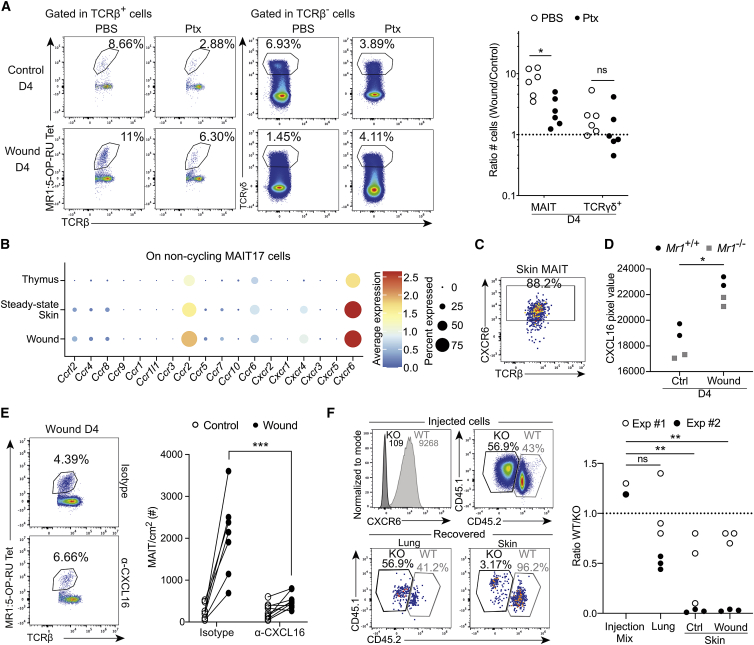
Since MAIT cell migration to the wound was independent of MR1 expression, we assessed the role of chemokines, as proposed for MAIT cell recruitment to the lungs during F. tularensis infection.44 We first blocked G protein interactions with chemokine receptors—thereby preventing chemokine signaling—by injecting pertussis toxin (Ptx) before skin excision. Like mainstream T cells (TCRβ+ cells excluding MAIT and iNKT cells; Figure S5A), MAIT cell recruitment to the wound was strongly decreased (Figure 5A; raw numbers in Figure S5B). Consistent with their residency profile, γδ T cell numbers were not modified, indicating that Ptx injection did not impair cell viability. Analysis of the scRNA-seq datasets showed that Cxcr6 and to a lesser extent Ccr2 were specifically expressed by skin MAIT17 cells (Figure 5B). At the protein level, CXCR6 was expressed by most skin MAIT cells (Figure 5C), while CCR2 was expressed by only half of them (Figure S5C). CXCL16, the ligand for CXCR6, was upregulated in the total skin lysate after excision both in Mr1+/+ and Mr1−/− animals (Figure 5D). By contrast, CCL2, the ligand for CCR2, was highly expressed, independently of the excision (Figure S5D).
Figure 5.
CXCR6 is necessary for MAIT cell recruitment into the skin
(A) MAIT and ɣδ T cell staining (left) and numbers (wound over control sites) (right) in skin control and wound (D4) sites following Ptx injection. Pooled data from two independent experiments (n = 6). Mann-Whitney test. Please also see Figures S5A and S5B.
(B) Chemokine receptor gene expression by non-cycling MAIT17 cells from integrated single-cell datasets as in Figure 1B.
(C) CXCR6 expression by MAIT cells. Data are representative of 10 independent experiments.
(D) CXCL16 protein quantity in total skin lysate from wound (D4) or steady-state skin of Mr1+/+ and Mr1−/− mice. Pooled data from two independent experiments (n = 4). Mann-Whitney test.
(E) MAIT cell staining (left) and numbers (right) in control and wound (D4) skin sites following ɑ-CXCL16 or isotype control i.p. injection. Pooled data from two independent experiments (n = 7/10). Mann-Whitney test.
(F) Expanded MAIT cells were deleted for Cxcr6 by CRISPR-Cas9 modification. Congenic marker and CXCR6 expression on the injected pool (top left) or recovered cells (bottom left). Quantitation of recovered Cxcr6−/− cells (ratio of Cxcr6−/− over Cxcr6+/+) at different sites (right). Pooled data from two independent experiments (ninjection = 2; nother = 6). Tukey’s multiple comparison test.
We therefore focused on the CXCR6-CXCL16 interaction. In vivo blocking of CXCL16 significantly reduced recruitment of MAIT cells into the wound site (Figure 5E). To formally demonstrate the role of CXCR6 in MAIT cell recruitment into the wound, we deleted Cxcr6 using CRISPR-Cas9 technology. Thymic MAIT cells were expanded in vitro as above (Figure S4A) before delivery of a ribonucleoprotein complex containing Cas9 and a guide RNA targeting Cxcr6. Deletion was highly efficient as shown by the loss of CXCR6 expression (Figure 5F, histogram). A 50/50 mixture of Cxcr6−/− and Cxcr6+/+ MAIT cells was injected into Cd3e−/− mice 1 day after excision. Four days after transfer, most MAIT cells found in the control and wound sites were Cxcr6+/+ (Figure 5F), indicating that CXCR6 is necessary for MAIT cell recruitment into the skin.
MAIT cell-derived Areg promotes wound closure
To determine how MAIT cells promote tissue repair, we analyzed cytokines produced in vivo at wound sites of Mr1+/+ and Mr1−/− animals (Figure S6A). In Mr1+/+ animals, we observed an upregulation of several molecules involved in antibacterial responses (resistin, CXCL10), epithelial proliferation, or angiogenesis (PDGF-BB, angiopoietin-1 and angiopoietin-2, FGF-21, WISP-1/CCN4) and inflammation through recruitment or differentiation of immune cells (CXCL1, CXCL10, IL-1β, IL-17A, (G)M-CSF). These results suggest that the whole repair process was increased at day 4 in Mr1+/+ as compared with Mr1−/− animals. Accordingly, we assessed the effect of MAIT cells on epidermal proliferation using K14 (keratinocyte marker) and Ki67 immunofluorescence staining45 (Figures 6A and 6B). The length of the epithelial tongue was increased from day 2 in Mr1+/+ animals (Figure 6A), demonstrating that MAIT cells are involved early in the repair process. Ki67 staining in the epidermis (but not in the dermis, Figure S6B) was increased 4 days after excision (Figure 6B), indicating that MAIT cells stimulated early epithelial proliferation. To assess whether MAIT cells also affected angiogenesis, endothelial cells were stained using a CD31 antibody. The number of vessels was similar between Mr1+/+ and Mr1−/− wound sections (Figure S6C). Thus, MAIT cells at early time points mainly favor the proliferation of epithelial cells.
Figure 6.
MAIT cell-derived Areg exerts a tissue repair function
(A) Representative immunofluorescence images of wounds from Mr1+/+ and Mr1−/− animals (DAPI in blue, K14 in green) (left). Scale bar represents 100 μm. The epidermal tongues are underlined with white dashed lines and their length is quantified (right, D2/D4, 2 tongues per slide). Pooled data from one (D2: n = 3) and two independent experiments (D4: n = 5/4) analyzed blindly. Mann-Whitney test.
(B) Representative immunofluorescence images of wounds from Mr1+/+ and Mr1−/− animals (DAPI in blue, Ki67 in red). The white dashed line separates the epidermal tongue and the underlying dermis (left). Proliferation in the epidermis is quantified by the Ki67/DAPI ratio and normalized to the average expression in Mr1−/− animals for each experiment (right). Data are from two independent experiments (n = 5/6) analyzed blindly. Unpaired t test. Please also see Figure S6B.
(C) Dot plot showing RNA expression of repair molecules by non-cycling MAIT17 cells from integrated single-cell datasets as in Figure 1B.
(D) Feature plot of Areg expression projected on the UMAP of the integrated datasets.
(E) Ex vivo Areg staining on skin MAIT cells (blue: control skin; red: wound skin; gray: full staining except the biotinylated anti-Areg antibody). Pooled data from two independent experiments (n = 6). Wilcoxon test.
(F) Areg expression by thymic enriched MAIT cells following 36 h of in vitro activation by 5-OP-RU or IL-18. One experiment (n = 8) representative of 2. Dunn’s multiple comparison test.
(G) Wound surfaces at days 4 and 6 after excision on Zbtb16-cre−Aregfl/fl (black) and Zbtb16-cre+Aregfl/fl (gray). Pooled data from two independent experiments with one blind (full symbols) (ncre− = 8; ncre+ = 10). Mann-Whitney tests.
(H) Wound surfaces after excision (D5 and D7) of Cd3e−/− animals transferred with thymic MAIT cells expanded from Zbtb16-cre−Aregfl/fl (black) and Zbtb16-cre+Aregfl/fl (gray) littermate mice. Pooled data from two independent experiments with one blindly analyzed (full symbols) (ncre− = 9; ncre+ = 8). Mann-Whitney tests.
MAIT cell effect could be either direct, through secretion of growth factors, or indirect, by recruiting other immune subsets (as seen in Meierovics et al.46). The numbers of mainstream T cells (TCRβ+MR1:5-OP-RU-Tet−CD1d:αGalCer-Tet−), γδ T cells, iNKT cells, Langherans cells as well as Ly6Chi and Ly6Clo monocytes were similarly modified in Mr1+/+ and Mr1−/− animals throughout wound closure (Figure S6D). These results suggest that MAIT cells did not impact the recruitment of these populations and that the effect observed on keratinocyte proliferation was direct.
We therefore focused on direct MAIT cell effector functions. scRNA-seq analysis identified several pro-repair mediators described as secreted by MAIT cells19–6 and overexpressed in skin versus thymic samples, including Furin, Tgfb1, and Hmgb1 (Figure 6C). We further identified pro-repair mediators specific to cluster 8 (Figure 2A; Table S2) and upregulated in skin non-cycling MAIT17 cells (Figure 6C). Il17a and Areg, both described to favor tissue repair (reviewed in McGeachy et al.47 and Zaiss et al.48), were among the most differentially expressed genes in cluster 8 (Figures 6C and 6D; Table S2). The role of IL-17 had been already well studied during tissue repair,4,49 and we could not obtain consistent ex vivo IL-17 staining on skin MAIT cells (data not shown). Therefore, we decided to focus on Areg, as tissue repair signatures associated to Areg-producing regulatory T cells were overexpressed by skin MAIT cells (Figure S2F). Areg is an epidermal growth factor-like molecule mediating keratinocyte proliferation.48 Ex vivo intracellular staining (no restimulation) confirmed that a higher frequency of skin MAIT cells produced Areg at the wound as compared with the control site (Figure 6E). In humans, MAIT cells expressed Areg following TCR stimulation,50 but in our study, wound closure seemed independent of a sustained TCR triggering (Figure 4), suggesting that Areg could be secreted following other, TCR-independent, stimuli. We tested this hypothesis in vitro using ex vivo thymic MAIT cells as the numbers of skin MAIT cells were too low for in vitro testing and the transferred MAIT cells (Figure S4A) were of thymic origin. IL-18, which is secreted during wound healing,51 induced similar amounts of Areg expression as compared with 5-OP-RU stimulation (Figure 6F).
To determine whether Areg secretion was important for skin repair, we compared wound closure in Aregfl/fl Zbtb16-cre+ and Zbtb16-cre− animals. The wound surface was increased at days 4 and 6 in Zbtb16-cre+ animals, indicating that Areg expression by Zbtb16-expressing cells was involved in skin wound healing (Figure 6G). PLZF is expressed by MAIT cells, iNKT cells,52 a subset of γδ T cells,53 and transiently by one-third of embryonic cells during development.54 To formally investigate the involvement of MAIT cell-derived Areg in skin wound healing, we expanded thymic MAIT cells from Aregfl/fl Zbtb16-cre+ or Zbtb16-cre− mice as above (Figure S4A). The resulting >95% pure MAIT cell populations were transferred into excised Cd3e−/− Mr1+/+ animals. Wound closure was significantly delayed when Areg-deficient MAIT cells were transferred (Figure 6H). Thus, Areg production by MAIT cells is central to their tissue repair function in skin wound healing.
Discussion
In this study, we have assessed the in vivo mechanisms underlying MAIT cell repair functions in an immunocompetent host, using a human-like skin damage model. We showed that MAIT cells express a tissue repair program at steady state in the skin and improve wound closure. The repair function relied on MAIT cell recruitment into the wound, from distant sites including secondary lymphoid organs through the CXCR6-CXCL16 axis. Recruitment and wound healing function were independent of concomitant TCR stimulation. MAIT cell presence was associated with increased epithelial proliferation in the epidermis. Lastly, Areg production by MAIT cells was key to their tissue repair function.
Our previous work has shown that MAIT cells are tissue resident in the lungs, liver, and spleen.15 Here, parabiosis experiments suggest that MAIT cells reside for shorter periods in the skin as compared with the lungs, in contrast to viral-specific TRM.55 Previous results have shown no exchange of skin MAIT cells between parabionts within 13 weeks. These contradictory data may result from S. epidermidis colonization 7 weeks before parabiosis, therefore generating bona fide MAIT cell TRM in the skin.5 However, skin MAIT17 cells in our study overexpressed a residency signature as compared with their thymic counterparts, both at steady state and at the wound site. The presence of photoconverted MAIT cells in the draining LN 2 days following photoconversion suggests that some skin MAIT cells recirculate, as shown for conventional antiviral TRM.56 During wound healing, MAIT cells are recruited into the skin in a CXCR6-CXCL16-dependent manner. This mechanism is likely shared for MAIT cell recruitment to other organs as intranasal instillation of CXCL16 together with 5-OPRU drives MAIT cell accumulation into the lungs.44 However, the role of CXCR6 is difficult to study in vivo as it is necessary for full MAIT cell maturation in the thymus.57 In vitro expansion allowing CRISPR-Cas9-based genetic modification followed by adoptive transfer solved this issue. As human MAIT cells also express CXCR6,58 recruitment could happen in pathological settings in which CXCL16 is produced such as non-alcoholic fatty liver disease, renal fibrosis, or certain cancers.59
Egress from secondary lymphoid organs was necessary for MAIT cell accumulation in the wound. Although MAIT cells have been mainly described in mucosal tissues, our results suggest that LNs (or any tissue with an S1PR1-dependent egress mechanism) may act as a reservoir.60 Accordingly, MAIT cell numbers decreased in the LN draining the wound, but this drop was too low to account for the large increase of MAIT cell number at the wound site. The constant albeit low proliferation of skin MAIT cells may contribute to increasing their numbers in the wound, but additional reservoirs likely exist. One hypothesis is that tissue-resident MAIT cells (from skin or other organs) return to the circulation, similarly to skin TRM upon reactivation.56 Alternatively, another pool of non-resident MAIT cells may exist. MAIT cells being negative for CD62L and CCR7, they probably do not circulate from the blood directly to the LN. Instead, they would behave as effector memory T cells, migrating from blood to tissues, exiting tissues through the lymph in an S1PR1-dependent mechanism before going back to blood.17 In humans, the existence of a circulating pool of MAIT cells is supported by the overlap of the TCR repertoire between MAIT cells from the thoracic duct and the blood.61 In contrast with our results, FTY720 treatment of mice instilled with F. tularensis live vaccine did not hamper MAIT cell accumulation in the lungs,44 suggesting different mechanisms in this model. Additionally, the increase of skin MAIT cell number following S. epidermidis association was similar in WT and LN-deficient animals (Lta−/−),5 but the impact on skin wound healing was not studied. Altogether, pools of MAIT cells with different circulation profiles may be present in mice. Whether these pools are functionally different and exchange to some extent at steady state or during pathologies remain to be determined.
In our study, MAIT cell recruitment and tissue repair function did not rely on antigen presentation by MR1. This result seems contradictory with the tissue repair program induced by TCR triggering both in humans and mice.19–6 In our study, skin MAIT cells expressed high amounts of Nur77 and a strong tissue repair program at steady state. Moreover, MAIT cells were expanded in vitro by adding 5-OP-RU once at the beginning of the culture. Thus, our data are consistent with TCR stimulation being necessary for program acquisition but not for actual tissue repair function. TCR stimulation at steady state is reminiscent of the tonic TCR signaling of pro-repair Vγ5Vδ1 T cells.3,62 Still, the actual triggering of MAIT cell repair function is probably not dependent on TCR signaling as MAIT cells do not express Nur77 either at the time of transfer or after migration into the wound. Similarly, regulatory CD4+ T cells in the lung following influenza infection secrete Areg even after TCR deletion.33 Also, iNKT cells deleted for their TCR secrete IFN-γ following lipopolysaccharide stimulation in vivo.63 Licensing mechanisms independent of TCR signaling have been described for commensal-specific H2-M3-restricted CD8+ T cells in the skin. At steady state, these type 17 cells express type 2 repair mediators at the RNA level only. Following tissue injury, alarmins promote secretion of the repair cytokines.2 A similar translational checkpoint likely exists for MAIT cells, explaining the identical transcriptional program of skin MAIT cells at steady-state and wound sites. This hypothesis is supported by the expression of transcripts encoding ZFP36, ZFP36L1, and ZFP36L2 which regulate the stability of mRNA for cytokines or other immune mediators.30 The repair functions may be elicited by cytokines such as IL-18, IL-4, IL-2, IL-7, and IL-21, whose receptors are expressed by skin MAIT cells (scRNA-seq, not shown). Accordingly, Areg secretion can be induced following IL-18 stimulation. Altogether, these results suggest that skin MAIT cells are in a poised functional state requiring additional signals to exert their tissue repair program.
The pro-repair effect of MAIT cells was mediated at least in part through Areg. In the skin, Areg produced in an autocrine manner promotes keratinocyte proliferation.64,65 Accordingly, MAIT cell presence increased the size of the epidermal tongue as well as keratinocyte proliferation. Areg is also expressed by lung MAIT17 cells at steady state (M.S. et al.,15 unpublished data) and following L. longbeachae infection.19 This molecule is produced by various cells of the immune system including innate cells as well as regulatory T cells, gingival γδ T cells,66 ILC2,67 and tumor-infiltrating CD8+ T cells.68 The multicellular origin of Areg suggests that multiple cell types may exert the same function. However, in our model, MAIT cell deletion alone delayed wound closure, suggesting non-redundant function in the early steps of skin healing. Skin MAIT cells also express RORγt and probably secrete IL-17, a key effector molecule for the repair function of epidermal γδ T cells.4,49 The IL-17 secreted by MAIT and γδ T cells could act similarly by inducing HIF1α in epithelial cells and a subsequent shift toward glycolysis to promote their migration.49 Although a non-redundant role of IL-17 was demonstrated using IL-17R-deficient epithelial cells, Rorc deletion in γδ T cells also affects other effector molecules regulated by RORγt. Consequently, other effector molecules and other T cell subsets such as MAIT cells are likely important for skin wound healing. Understanding the relative contributions of IL-17 and Areg derived from one or another cell type, including at steady state, would help understanding the fine tuning of epithelial repair.
In summary, our work shows that MAIT cells play a pivotal role in skin wound healing. MAIT cell implication in different types of healing delay such as diabetic wounds would therefore be of interest. Understanding whether MAIT cells have such function in other tissues will assess their full effector potential to be able to manipulate them toward pro-inflammatory or pro-repair functions.
Limitations of the study
One limitation of our study is the extensive digestion process at 37°C that may modify the transcriptional pattern of skin cells. Therefore, we favored in vivo experiments to validate the results obtained using single-cell suspensions. Moreover, the transcriptome analysis did not distinguish MAIT cells preexisting in the skin from those recruited to the wound. Whether both subsets perform in vivo repair functions is unknown. The MAIT cell transfer into Cd3e−/− mice shows that the recruited ones do elicit the repair process. An additional question which would help improve our understanding of MAIT cell function is their precise location in the skin and their relationship with other cell types.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Antibodies for Cytometry | ||
| Amphiregulin (Biot) | R&D system | Cat#BAF989; RRID: AB_2060662 |
| Aqua L/D | ThermoFisher | Cat#L34957 |
| B220 (AF700) | eBioscience | Cat#56-0452-82; RRID: AB_891458 |
| CCR2 (AF700) | R&D system | Cat#FAB5538N; RRID: AB_2725739 |
| CD103 (BV786) | BD | Cat#564322; RRID: AB_2738744 |
| CD103 (PerCP-eF710) | Invitrogen | Cat#46-1031-82; RRID: AB_2573704 |
| CD11b (APC) | eBiosciences | Cat#17-0112-83; RRID: AB_469344 |
| CD11c (PETR) | LifeTechnologies | Cat#MCD11C17; RRID: AB_1464845 |
| CD19 (AF700) | Biolegend | Cat#115528; RRID: AB_493735 |
| CD19 (FITC) | eBioscience | Cat#11-0191-85; RRID: AB_464966 |
| CD44 (BV605) | Biolegend | Cat#103047; RRID: AB_2562451 |
| CD45.1 (PE-Cy7) | Biolegend | Cat#110730; RRID: AB_1134168 |
| CD45.2 (AF700) | Biolegend | Cat#109822; RRID: AB_493731 |
| CD24 (FITC) | Invitrogen | Cat# 11-0242-82; RRID: AB_464988 |
| CD69 (PC7 & PE-Dazzle 594) | Biolegend | Cat#104512; RRID: AB_493564 |
| CXCR6 (PETR) | Biolegend | Cat#151117; RRID: AB_2721700 |
| Ki67 (PE-Cy7) | Biolegend | Cat#652426; RRID: AB_2632694 |
| Ly6C (BV785) | Biolegend | Cat#128041; RRID: AB_2565852 |
| RORgt (BV786) | BD | Cat#564723; RRID: AB_2738916 |
| Tbet (APC) | Invitrogen | Cat#17-5825-82; RRID: AB_2744712 |
| TCRb (APC-Cy7) | Biolegend | Cat#109220; RRID: AB_893626 |
| TCRgd (BV605) | Biolegend | Cat#118129; RRID: AB_2563356 |
| Tet CD1d (BV421) | NIH tetramer core facility | N/A |
| Tet MR1 (APC) | NIH tetramer core facility | N/A |
| Tet MR1 (PE) | NIH tetramer core facility | N/A |
| Antibodies for Immunohistochemistry and Immunofluorescence | ||
| DAPI | Sigma | Cat#MBD0015 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Fisher | Cat#A-21206; RRID: AB_2535792 |
| Goat anti-Chicken IgY (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 | Fisher | Cat#A32933; RRID: AB_2762845 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Cyanine3 | Fisher | Cat#A10522; RRID: AB_1500704 |
| Keratin 14 Polyclonal Chicken Antibody, Purified Antibody | Biolegend | Cat#905301; RRID: AB_2565048 |
| Ki-67 (D3B5) Rabbit mAb | Ozyme | Cat#9129S; RRID: AB_2687446 |
| Purified Chicken IgY Isotype Ctrl Antibody | BIOLEGEND | Cat#402101 |
| Purified Rat Anti-Mouse CD31 | BD | Cat#550274; RRID: AB_393571 |
| Purified Rat IgG2a κ Isotype Control | BD | Cat#559073; RRID: AB_479682 |
| Rabbit (DA1E) mAb IgG XP® Isotype Control | Ozyme | Cat#3900S; RRID: AB_1550038 |
| Chemicals, peptides, and recombinant proteins | ||
| 24G2 | Institut Curie, produced in house | N/A |
| Alt-R S.p. HiFi Cas9 Nuclease V3 | IDT | Cat#1081061 |
| Anti-mCXCL16 | R&D Systems | Cat#MAB503; RRID: AB_2276752 |
| Anti-PE microbeads | Miltenyi | Cat#130-048-801 |
| Anti-FITC microbeads | Miltenyi | Cat# 130-048-701 |
| B-mercaptoethanol | Sigma | Cat#M3148 |
| BCA Assay | ThermoFisher | Cat#23227 |
| Bovine Serum Albumin | SIGMA | Cat#A7906 |
| Brefeldin A | SIGMA | Cat#B6542 |
| CO2 independent medium | Gibco | Cat#18045088 |
| DAPI | Sigma | Cat#MBD0015 |
| Debris removal solution | Miltenyi | Cat#130-109-398 |
| Dispase | Corning | Cat#354235 |
| DNAse 1 | Roche | Cat#5401020001 |
| EDTA | Gibco | Cat#15575-038 |
| Fetal Calf Serum | Eurobio | Cat#CVFSVF00-01 |
| Fluorescent Mounting Medium | Dako | Cat#53023 |
| FTY-720 | Sigma | Cat#SML0700-25mg |
| gelatine from cold water fish skin | Sigma | Cat#67765 |
| GolgiPlug | BD | Cat#555029 |
| GolgiStop | BD | Cat#554724 |
| HEPES | Gibco | Cat#15630-056 |
| Liberase TL | Sigma | Cat#10104159001 |
| Live and Dead AQUA | Thermo Fischer Scientific | Cat#L34957 |
| LS columns | Miltenyi | Cat#130-042-401 |
| Maristamat | Sigma | Cat#M2699 |
| Monensin | Invitrogen | Cat#00-4505-51 |
| Non-essential amino acid | ThermoFisher | Cat#11140050 |
| OCT | Tissue-Tek | Cat#16-004004 |
| Paraformaldehyde | EMS | Cat#15710 |
| PBS | Eurobio scientific | Cat#CS1PB501-01 |
| Penicillin/Streptomycin | Gibco | Cat#15140-122 |
| Pertussis Toxin | Gibco | Cat#PHZ1174 |
| Protease inhibitors | Roche | Cat#11697498001 |
| Rat IgG2A Isotype Control | R&D Systems | Cat#MAB006; RRID: AB_357349 |
| RBC lysis buffer | Biolegend | Cat#420302 |
| RPMI 1640 GlutaMAX | Gibco | Cat#61870036 |
| skim milk powder | Régilait | Cat#B0110287 |
| Sodium Chloride | VWR Chemicals | Cat#27810-295 |
| Sodium Pyruvate (NaPyr) | ThermoFisher | Cat#11360070 |
| Triton X-100 | Sigma | Cat#11332481001 |
| Tween | Sigma | Cat#P9416 |
| 5-OP-RU | Curie Institute | Soudais et al., 2015 |
| Critical commercial assays | ||
| Cytofix/Cytoperm Solution Kit | BD | Cat#554714 |
| FoxP3 Transcription factor Permeabilization buffer | Thermofischer | Cat#00-5523-00 |
| mrmIL-2 | Peprotech | Cat#212-12 |
| rmIL-18 | R&D | Cat# 9139-IL |
| Proteome array | R&D | Cat#ARY028 |
| Solution P3 Primary Cell 4DNucleofector X kit S | Lonza | Cat#VAXP-3032 |
| Deposited data | ||
| Skin single cell datasets | This paper | GSE20734869 |
| Thymic single cell dataset | Legoux et al.24 | E-MTAB-7704 |
| Experimental models: Organisms/strains | ||
| B6 Aregflox/flox (Aregtm2a(EUCOMM)Hmgu) | Bred in Institut Curie - provided by D. Zaiss | Minutti et al.70 |
| Zbtb16-GFPcre | Institut Curie | Constantinides et al.54 |
| Cd3e-/-Mr1+/+ and Mr1-/- | Institut Curie | Malissen et al.71 |
| CD45.1/1 and CD45.1/2 B6-MAITCAST | Institut Curie | N/A |
| CD45.2/2 B6-MAITCASTMr1+/+ or Mr1-/- | Institut Curie | Cui et al.20 and Treiner et al.22 |
| Kaede B6 | Rachel Golub, Institut Pasteur | Tomura et al.40 |
| Nr4a1-GFP B6-MAITCAST | Institut Curie | Zikherman et al.72 |
| Rorc-GFP B6-MAITCAST | Institut Curie | Lochner et al.73 |
| Oligonucleotides | ||
| Alt-R CRISPR-Cas9 crRNA: Mm_CXCR6.1_AA: /AltR1/rCrU rGrUrA rCrGrA rUrGrG rGrCrA rCrUrA rCrGrA rGrUrU rUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A |
| Alt-R® CRISPR-Cas9 tracrRNA | IDT | Cat#1072534 |
| Software and algorithms | ||
| Astrios software (Summit) | BECKMAN COULTER (Summit v62) | https://www.beckman.fr/flow-cytometry/cell-sorters/moflo-astrios-eq |
| Cytoflex software (CytExpert) | BECKMAN COULTER (V2.4) | https://www.beckman.fr/flow-cytometry/research-flow-cytometers/cytoflex/software |
| FlowJo | BD (V10.8.0) | https://www.flowjo.com/ |
| Fortessa software (BD FACSDiva software) | BD (V6) | https://www.bdbiosciences.com/en-eu/products/software/instrument-software/bd-facsdiva-software |
| Image J Software | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Prism | GraphPad (V8) | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Astrio | BECKMAN COULTER | Cat#B25982 |
| Biopsy punch (4Ø) | Stiefel | Cat#600210 |
| ChemiDoc Imaging System | BIORAD | Cat#17001402 |
| Chromium 3’ Chip | 10X Genomics | N/A |
| Cryostat | LEICA | Cat#CM1950 |
| Cytoflex LX | BECKMAN COULTER | Cat#C00445 |
| Fortessa LSR | BD | Cat#23-11617-01 |
| Hair removal cream | Veet | Cat#EA_3108955 |
| Microscope pour immunostaining | ThermoFischer | Cat#EVOS_M500 |
| Silicone sheet (5mm thick) | Grace Bio-Labs | Cat#GBL664581-5EA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marion Salou (marion.salou@curie.fr).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Unless specified otherwise, congenic B6-MAITCAST strain, on an Mr1+ or Mr1-/- background were used in this study.20 Reporter genes under the control of Rorc (RORγt),73 Nr4a1 (Nur77)72 or Zbtb16 (PLZF)54 promoters were introgressed into the B6-MAITCAST background. CD45.1/1, CD45.1/2, CD45.2/2 animals were generated by crossing CD45.1/1 B6 animals onto CD45.2/2 B6-MAITCAST mice. Photoconvertible Kaede mice were generously provided by R. Golub (Institut Pasteur, Paris).40 Mr1+/+ and Mr1-/- Cd3e-/- mice were generated in house by crossing MR1 on a B6 background with Cd3e-/- mice.71 B6 Aregflox/flox (Aregtm2a(EUCOMM)Hmgu)70 provided by D. Zaiss were crossed onto B6-MAITCast Zbtb16GFPcre mice.5 In all experiments, we accounted for the cage effect on immune cell population (e.g., MAIT cell frequency impacted by microbiota composition)5 by spreading animals and/or litters from the same breeding cages into the different experimental groups. All experiments were conducted in an accredited animal facility by the French Veterinarian Department following ethical guidelines approved by the relevant ethical committee (APAF1S no. 24245–2020021921558370-v1).
Method details
Skin excision was performed as previously described.21 Briefly, after shaving and depilation, a 4 mm full-thickness wound was performed on the back of the mouse using a biopsy punch. Silicone rings were then sutured to prevent epithelial skin contraction. A clean dressing was applied and regularly changed to avoid infection of the wounds. Macroscopic measurements were performed on pictures by calculating wound and ring areas using ImageJ. Wound size was then estimated as a wound to ring area ratio, with 1 being an open wound and 0 a fully closed wound.
Parabiotic surgery
Aged-matched congenically distinct B6-MAITCAST mice either Mr1+/+ or Mr1-/- were co-housed for a minimum of 2 weeks before being surgically joined as parabiotic pairs as previously described.15,74 Skin and lungs were collected 5 weeks later.
Skin graft
Graft surgery was performed as described.75 Briefly, either Mr1+/+ or Mr1-/- B6-MAITCAST donor skin was collected. A 2 cm2 sample was grafted onto congenically distinct Mr1+/+ B6-MAITCAST mice. Recipients with grafts from identical donors were sacrificed at different time points to follow cellular infiltration kinetics.
Kaedes photoconversion
Mice were anesthetized, shaved on the dorsal side, and exposed to violet light (395nm U.V. light 95 Watts for 60s)40 to photoconvert skin cells.
Tissue processing
Back skin was shaved, depilated (2 min of hair removal cream, Veet) and full-thickness samples (ie dermis and epidermis) were collected in C02 independent medium (Gibco). The following area were sampled: steady state skin, wound site (1 cm2 encompassing the wound and surrounding rims, meaning that the wound represents 12.6% of the sampled skin, back top left of the mouse) or contralateral control skin (1 cm2 of skin at the contralateral side as compared to the wound, i.e. back top right). The contralateral site called control is used to normalize the number of MAIT cells present in the wound to account for inter-individual variations.5
Skin single cell suspensions were obtained by putting the samples (flattened, epidermis side up) at 37°C for 45 min in 1 mL of 500 CU Dispase (Corning). The skin sample was then chopped in RPMI 1640 GlutaMAX media supplemented with 1 mM sodium pyruvate, 1 mM non-essential amino acids, 50 μM β-mercaptoethanol, 20 mM HEPES, 100 U/mL penicillin, 100 mg/mL streptomycin, 0.5 mg/mL DNase I (all products from Sigma-Aldrich), and 0.25 mg/ml Liberase TL (Roche) and incubated for 1h45 min at 37°C in a 5% CO2 incubator. After filtering on a 40 μm filter and 2 washing in PBS, BSA 0.5%, 2 mM EDTA, the cell suspension was removed of skin debris using the cell debris removal solution (Miltenyi) following manufacturer’s instructions.
Lung single cell suspensions were obtained as described.15 Blood cells were recovered by centrifugation after red blood cell lysis (Biolegend). LNs were scratched onto 40μm filter and cells were washed in PBS 1x, BSA 0.5%, 2mM EDTA before use.
Flow cytometry
Extracellular staining was performed with the relevant titrated antibodies in staining buffer (PBS 0.5% BSA, 2 mM EDTA and anti–FcR 2.4G2 produced in house) for 20 min at 4°C. Staining for transcription factors or cytokines was performed on fixed and permeabilized cells using the appropriate kits (Foxp3 Fixation kit (Thermofisher) and BD Fix/Perm kit, respectively) as per manufacturer instructions, followed by 20 min incubation at 4°C with the relevant titrated antibodies. If needed, tetramer staining was performed before the extracellular staining step, for 30 min at room temperature in staining buffer containing MR1 tetramers loaded with 5-OP-RU or 6-FP with or without CD1d tetramers loaded with PBS-57 (both tetramers from the NIH Tetramer Core Facility; Emory University, GA) and anti-TCRβ. Flow cytometry acquisition was performed on a Cytoflex (Beckman) or Fortessa cytometer (BD). FACS was performed on an Astrios cell sorter (Beckman Coulter). Data were analyzed using FlowJo software.
Ex vivo amphiregulin production
Brefeldin A (Sigma) was injected i.v.70 6 hours before sacrifice. Skin was processed as described above, except for addition of Monensin (10 μM, Invitrogen) in every buffer. To prevent extracellular secretion the resulting single cell suspension was maintained for 3h at 37°C with maristamat (10 mM, Sigma), GolgiPlug (1/1000; BD) and GolgiStop (1/1500; BD)33 before staining.
Single cell RNA sequencing
Single cell suspensions of 18 wounds (1 cm2) or steady state back skin (same location as the wound) were pooled together and live TCRβ+MR1:5-OP-RU+ were isolated by Aria cell sorter (BD) in 10% FCS CO2-independent medium. 7,000 cells for each sample were loaded onto a chromium 3’ chip following the Single Cell 3’ Kit V3 (10X Genomics). Generation and acquisition of the sequencing reads were performed according to the manufacturer recommendations (10X Genomics) by the ICGex NGS plateform of the Institut Curie.
Single cell RNA-seq preprocessing
The reads were aligned and feature-barcode matrices were generated using the Cell ranger pipeline version 3.1.0. The reference genome used is the mm10-3.0.0.
Single cell RNA analysis
All analyses were performed using R version 4.1.0 and the following packages: Seurat_4.1.0, clustree_0.4.4, ggplot2_3.3.5 and dplyr_1.0.8. Based on the distribution of the numbers of genes and molecules detected per cell, the following filters were applied to remove outliers: nFeature_RNA > 1,200 & nCount_RNA > 3,500 & nCount_RNA < 25,000 for the control site, and nFeature_RNA > 1,200 & nCount_RNA > 3,000 & nCount_RNA < 33,000 for the wound site, respectively. Cells containing more than 15% of mitochondrial genes were considered as dying cells and filtered out. Following preliminary analyses, some contaminating cells representing less than 0.15% of the total cells were removed, based on the expression of either F4/80, CD11b, CD11c, CD64, CD20 or CD206. The thymic dataset24 was filtered based on the following arguments: nFeature_RNA > 800 & nCount_RNA < 22,000 and less than 10% of mitochondrial genes. The cells with C1qc expression (more than 0.01%) were also removed. In summary, we obtained 3 datasets with comparable cell numbers (3,937 for skin distal, 4,368 for skin wound and 3,428 for thymus) and median number of features (2,458 for skin distal, 2,439.5 for skin wound and 1,812 for thymus).
All three datasets were integrated together using the corresponding Seurat vignette.76 The variable features number was set to 2,000 for the skin datasets and encompassed Tbx21 and Mki67. Given that most MAIT cells in the skin are MAIT17 cells, neither Rorc nor Zbtb16 were present in the VariableFeatures lists. This number was raised to 2500 for the thymus, to encompass Rorc and Zbtb16. Following normalization of each dataset and linear transformation (ScaleData), the anchors were identified using the default parameters, except the number of Integration Features which was raised to 4000 to encompass Zbtb16 and Rorc. The data were then integrated. A principal component analysis was run, and the number of principal components to use for downstream clustering (n=25) was determined as proposed in the Seurat Vignette (https://satijalab.org/seurat/articles/pbmc3k_tutorial.html). Graph-based clustering (Louvain) was performed using the default parameters, and a UMAP (dims=25) was constructed with a resolution of 0.4 based on the stability observed with the package clustree. The differentially expressed genes were determined using the FindAllMarkers() function (using a logistic regression, test.use = “LR”, and testing the effect of the dataset, latent.vars = “orig.ident” to correct for the batch effect). The same analysis was performed to determine the genes specific for the skin datasets, after sub-setting the non-cycling MAIT17 cells (FindMarkers()). The signatures used throughout the analysis were gene lists from the litterature15,23,25,32–36,77 and are presented in Table S1.
Migration inhibition protocols (FTY20, Pertussis toxin and ɑ-CXCL16 treatments)
FTY720 (Sigma) 0.5 mg/kg or PBS alone was injected daily from the day prior to skin excision, until organ collection.42 Pertussis toxin78 treatment (1 μg in 100 μl i.p., Gibco) was performed one day prior to skin excision and daily until organ collection. In vivo CXCL16 blocking was done by injecting i.p. 100 μg of anti-CXCL16 antibody or 100 μg of IgG isotype (R&D Systems) one day prior to skin excision and daily until organ collection.
MAIT expansion and adoptive transfer
Thymic single-cell suspensions were obtained by mechanical dissociation trough a 40 μm cell strainer. Cells were first incubated with MR1:6FP tetramer to avoid unspecific staining, stained using MR1:5-OP-RU-PE tetramer. Enrichment by positive selection on LS columns (Miltenyi) was performed after staining the cells with anti-PE microbeads (Miltenyi).
2 x 106 cells/mL were plated in complete RPMI 1640 media (10% FCS, 100U Penicillin/Streptomycin, 10 mM Hepes, 1 mM Sodium Pyruvate, 1X Non-essential Amino Acid, 50 μM β-mercaptoethanol) and stimulated with 150 nM 5-OP-RU and 10 ng/mL of rmIL-2 (Peprotech). MAIT cells were expanded for 10 to 15 days by addition of rmIL-2 (every 2 days) before adoptive transfer or genetic manipulation.
CXCR6 CRISPR-Cas9 genetic targeting
To create a CXCR6-specific RNP complex, oligos crRNA_CXCR6_AA (100 pmole) and tracrRNA (100 pmole) were first annealed using a slow ramp reaching 23°C and incubated at room temperature 10 min with 10 μg S.p Hifi Cas9 Nuclease V3. 2.106 (all reagents from IDT). Expanded MAIT cells were transfected according to the manufacturer instruction (Lonza). Briefly, 2 x 106 cells were resuspended in nucleofection solution (Lonza) with 3 μl RNP complex, transferred to nucleofection cuvette strips, electroporated using the DN110 program (4D-Nucleofector Core Unit: Lonza, AAF-1002B) and incubated in complete RPMI 1640 media at 32 °C for 24 hours to force non homologous repair recombination. Transfected cells were further cultured for 2-3 days before transfer. The efficiency of Cxcr6 deletion was evaluated for each experiment on the day of injection by flow-cytometry.
Proteome array
Skin samples (1 cm2) were chopped in PBS containing 1% Triton and protease inhibitors, flash-frozen, and then thawed before scratching on a 40 μm filter and centrifugation at 14,000 g for 5 min. 200 μg (as determined by BCA assay from ThermoFisher), of each sample was then added to each blot following manufacturer’s instruction (R&D). Blots were imaged using the ChemiDoc imaging system (BIORAD). The resulting pixel densities of the protein immune-blot dots were quantified with image J (NIH) following manufacturer’s instruction (R&D). Two measurements from each sample were obtained and the pair of duplicate spots was averaged, each Mr1+/+ average values was then divided by the average of Mr1-/- duplicates.
Immunostaining
Wounds were embedded in OCT (Tissue-Tek) and stored at -80°. Five μm OCT sections were cut using a Cryostat (LEICA). For each skin sample, Hematoxylin and eosin staining was used to determine the middle of the wound, given by the distance between first visible hair follicles on each side of the open wound. All immunostainings were done no more than 5 sections away (25 μm) from that center. Sections were air-dried for 10 min and fixed with 4% paraformaldehyde (EMS) for 5 min. Sections were washed 3 times in PBS 0.05% Tween and then blocked for one hour in permeabilization and blocking (PB) buffer (0.5% skim milk powder, 0.25% gelatine from cold water fish skin, 0.5% Triton X-100, 20 mM HEPES, 0.9% NaCl, pH 7.2 (all reagents from Sigma-Aldrich)). Primary antibodies were diluted in PB buffer and incubated overnight at 4°C, washed 3 times in PBS tween (0.05%). Slides were then stained with secondary antibodies for 1h at room temperature, washed 3 times in PBS tween (0.05%), stained with DAPI (Sigma), washed twice in PBS tween (0.05%), mounted with fluoromount (DAKO) and imaged using an EVOS-M500 microscope (ThermoFisher). Primary antibodies were used as follows: rabbit anti-K14 (1:1000; Biolegend), rabbit anti-mouse Ki67 (1:500; Cell signalling 9129S), rat anti-mouse CD31 (1:100; BD Biosciences) or isotypes. Alexa Fluor-conjugated antibodies (ThermoFisher Scientific) were used at 1:1000 as secondary antibodies.
Thymic MAIT cell in vitro activation
Thymic single-cell suspensions were obtained by mechanical dissociation trough a 40 μm cell strainer. Enrichment in mature cells (CD24-) was achieved by negative selection using LS columns (Miltenyi) after staining the cells with anti-CD24-FITC antibody (Invitrogen) and anti-FITC microbeads (Miltenyi). 1 x 106 cells/mL were plated for 36h in complete RPMI 1640 media (10% FCS, 100 U Penicillin/Streptomycin, 10 mM Hepes, 1 mM Sodium Pyruvate, 1X Non-essential Amino Acid, 50 μM β-mercaptoethanol) alone or with addition of 5-OP-RU (1.5 μM, 5-OP-RU synthesised in house) or of rmIL18 (10 ng/mL, R&D).
Quantification and statistical analysis
For each experiment, number of independent experiments, replicates and the statistical tests used are indicated in the figure legends. The following statistical test were used and calculated by GraphPad Prism v8 (GraphPad): Mann-Whitney, Wilcoxon, unpaired t-test, paired t test, sídák’s multiple comparison test, Tukey’s multiple comparison test (with p values: ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001 and ∗∗∗∗p<0.0001 and “ns” if the comparison was non-significant).
Acknowledgments
We thank the animal facility platform of the Institut Curie (V. Dangles-Marie, C. Alberti, C. Daviaud, M. Garcia, the mouse facility zootechnicians, and the genotyping platform), as well as the flow cytometry platform. We also thank the ICGex NGS platform of the Institut Curie for the help with the single-cell experiments. The ICGex NGS platform is supported by the grants ANR-10-EQPX-03 (Equipex) and ANR-10-INBS-09-08 (France Génomique Consortium) from the Agence Nationale de la Recherche (“Investissements d’Avenir” program), by ITMO-Cancer Aviesan (Plan Cancer III), and by the SiRIC-Curie program (SiRIC Grant INCa-DGOS-465 and INCa-DGOS-Inserm_12554). Data management, quality control, and primary analysis were performed by the bioinformatics platform of the Institut Curie. We thank the Pathex and anatomo-pathology platforms of the Institut Curie. We are grateful to L. Gapin, R.A. Paiva, and J. Waterfall for critical reading of the manuscript and C. Hivroz for discussion. We thank the NIH Tetramer Core Facility (Emory University) for providing CD1d and MR1 tetramers. The MR1:5-OP-RU tetramer technology was developed jointly by J. McCluskey, J. Rossjohn, and D. Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. This study received funding from the following institutions: Institut National de la Santé et de la Recherche Médicale (O.L.), Institut Curie (O.L.), Agence Nationale de la Recherche Grant MAITANR-16-CE15-0020-01 (O.L.), Agence Nationale de la Recherche Grant MAIT-repair ANR-20-CE15-0028-01 (O.L.), Agence Nationale de la Recherche ANR-10-IDEX-0001-02 PSL (O.L.), European Research Council ERC-2019-ADG-885435 (O.L.), and Fondation pour la Recherche Médicale FDT202106013036 (A.d.H.).
Author contributions
Conceptualization, A.d.H., O.L., and M.S.; formal analysis, A.d.H., M.A., M.M., and M.S.; funding acquisition, O.L.; investigation, A.d.H., A.D., A.A., V.P., T.Y., L.C., Y.E.M., H.B., F.L., L.P., R.G., and M.S.; methodology, A.d.H., M.A., O.L., and M.S.; project administration, A.d.H.; resources, D.Z., R.G., S.A., T.Y., L.C., and R.R.; supervision, O.L. and M.S.; visualization, A.d.H. and M.S.; writing – original draft, A.d.H. and M.S.; writing – review & editing, A.d.H., O.L., and M.S.
Declaration of interests
The authors declare no competing interests.
Published: January 10, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.12.004.
Contributor Information
Olivier Lantz, Email: olivier.lantz@curie.fr.
Marion Salou, Email: marion.salou@curie.fr.
Supplemental information
Data and code availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE207348 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207348).
References
- 1.Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 2.Harrison O.J., Linehan J.L., Shih H.-Y., Bouladoux N., Han S.-J., Smelkinson M., Sen S.K., Byrd A.L., Enamorado M., Yao C., et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363:eaat6280. doi: 10.1126/science.aat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jameson J., Ugarte K., Chen N., Yachi P., Fuchs E., Boismenu R., Havran W.L. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 4.MacLeod A.S., Hemmers S., Garijo O., Chabod M., Mowen K., Witherden D.A., Havran W.L. Dendritic epidermal T cells regulate skin antimicrobial barrier function. J. Clin. Invest. 2013;123:4364–4374. doi: 10.1172/JCI70064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Constantinides M.G., Link V.M., Tamoutounour S., Wong A.C., Perez-Chaparro P.J., Han S.-J., Chen Y.E., Li K., Farhat S., Weckel A., et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366:eaax6624. doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng T., Akther H.D., Hackstein C.-P., Powell K., King T., Friedrich M., Christoforidou Z., McCuaig S., Neyazi M., Arancibia-Cárcamo C.V., et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 2019;28:3077–3091.e5. doi: 10.1016/j.celrep.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin E., Treiner E., Duban L., Guerri L., Laude H., Toly C., Premel V., Devys A., Moura I.C., Tilloy F., et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franciszkiewicz K., Salou M., Legoux F., Zhou Q., Cui Y., Bessoles S., Lantz O. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunol. Rev. 2016;272:120–138. doi: 10.1111/imr.12423. [DOI] [PubMed] [Google Scholar]
- 9.Corbett A.J., Eckle S.B.G., Birkinshaw R.W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 10.Kjer-Nielsen L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 11.Rahimpour A., Koay H.F., Enders A., Clanchy R., Eckle S.B.G., Meehan B., Chen Z., Whittle B., Liu L., Fairlie D.P., et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J. Exp. Med. 2015;212:1095–1108. doi: 10.1084/jem.20142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legoux F., Salou M., Lantz O. MAIT cell development and functions: the microbial connection. Immunity. 2020;53:710–723. doi: 10.1016/j.immuni.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Provine N.M., Klenerman P. MAIT cells in health and disease. Annu. Rev. Immunol. 2020;38:203–228. doi: 10.1146/annurev-immunol-080719-015428. [DOI] [PubMed] [Google Scholar]
- 14.Mao A.-P., Constantinides M.G., Mathew R., Zuo Z., Chen X., Weirauch M.T., Bendelac A. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. USA. 2016;113:7602–7607. doi: 10.1073/pnas.1601504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salou M., Legoux F., Gilet J., Darbois A., du Halgouet A., Alonso R., Richer W., Goubet A.-G., Daviaud C., Menger L., et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med. 2019;216:133–151. doi: 10.1084/jem.20181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas S.Y., Scanlon S.T., Griewank K.G., Constantinides M.G., Savage A.K., Barr K.A., Meng F., Luster A.D., Bendelac A. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. J. Exp. Med. 2011;208:1179–1188. doi: 10.1084/jem.20102630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masopust D., Soerens A.G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dauphars D.J., Mihai A., Wang L., Zhuang Y., Krangel M.S. Trav15-dv6 family Tcrd rearrangements diversify the Tcra repertoire. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211581. e20211581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinks T.S.C., Marchi E., Jabeen M., Olshansky M., Kurioka A., Pediongco T.J., Meehan B.S., Kostenko L., Turner S.J., Corbett A.J., et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 2019;28:3249–3262.e5. doi: 10.1016/j.celrep.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y., Franciszkiewicz K., Mburu Y.K., Mondot S., Le Bourhis L., Premel V., Martin E., Kachaner A., Duban L., Ingersoll M.A., et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J. Clin. Invest. 2015;125:4171–4185. doi: 10.1172/JCI82424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn L., Prosser H.C.G., Tan J.T.M., Vanags L.Z., Ng M.K.C., Bursill C.A. Murine model of wound healing. J. Vis. Exp. 2013:e50265. doi: 10.3791/50265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treiner E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 23.Linehan J.L., Harrison O.J., Han S.-J., Byrd A.L., Vujkovic-Cvijin I., Villarino A.V., Sen S.K., Shaik J., Smelkinson M., Tamoutounour S., et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. 2018;172:784–796.e18. doi: 10.1016/j.cell.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legoux F., Gilet J., Procopio E., Echasserieau K., Bernardeau K., Lantz O. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat. Immunol. 2019;20:1244–1255. doi: 10.1038/s41590-019-0465-3. [DOI] [PubMed] [Google Scholar]
- 25.Baranek T., Lebrigand K., de Amat Herbozo C., Gonzalez L., Bogard G., Dietrich C., Magnone V., Boisseau C., Jouan Y., Trottein F., et al. High dimensional single-cell analysis reveals iNKT cell developmental trajectories and effector fate decision. Cell Rep. 2020;32:108116. doi: 10.1016/j.celrep.2020.108116. [DOI] [PubMed] [Google Scholar]
- 26.Christo S.N., Evrard M., Park S.L., Gandolfo L.C., Burn T.N., Fonseca R., Newman D.M., Alexandre Y.O., Collins N., Zamudio N.M., et al. Discrete tissue microenvironments instruct diversity in resident memory T cell function and plasticity. Nat. Immunol. 2021;22:1140–1151. doi: 10.1038/s41590-021-01004-1. [DOI] [PubMed] [Google Scholar]
- 27.Casey K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 2012;188:4866–4875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirai T., Yang Y., Zenke Y., Li H., Chaudhri V.K., De La Cruz Diaz J.S., Zhou P.Y., Nguyen B.A.-T., Bartholin L., Workman C.J., et al. Competition for active TGFβ cytokine allows for selective retention of antigen-specific tissue- resident memory T cells in the epidermal niche. Immunity. 2021;54:84–98.e5. doi: 10.1016/j.immuni.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay L.K., Wynne-Jones E., Freestone D., Pellicci D.G., Mielke L.A., Newman D.M., Braun A., Masson F., Kallies A., Belz G.T., et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity. 2015;43:1101–1111. doi: 10.1016/j.immuni.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Makita S., Takatori H., Nakajima H. Post-transcriptional regulation of immune responses and inflammatory diseases by RNA-binding ZFP36 family proteins. Front. Immunol. 2021;12:711633. doi: 10.3389/fimmu.2021.711633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezvani H.R., Ali N., Nissen L.J., Harfouche G., de Verneuil H., Taïeb A., Mazurier F. HIF-1α in epidermis: oxygen sensing, cutaneous angiogenesis, Cancer, and non-cancer disorders. J. Invest. Dermatol. 2011;131:1793–1805. doi: 10.1038/jid.2011.141. [DOI] [PubMed] [Google Scholar]
- 32.Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arpaia N., Green J.A., Moltedo B., Arvey A., Hemmers S., Yuan S., Treuting P.M., Rudensky A.Y. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanai H., Budovsky A., Tacutu R., Barzilay T., Abramovich A., Ziesche R., Fraifeld V.E. Tissue repair genes: the TiRe database and its implication for skin wound healing. Oncotarget. 2016;7:21145–21155. doi: 10.18632/oncotarget.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L., Sandrock I., Odak I., Aizenbud Y., Wilharm A., Barros-Martins J., Tabib Y., Borchers A., Amado T., Gangoda L., et al. Single-cell transcriptomics identifies the adaptation of Scart1+ Vγ6+ T cells to skin residency as activated effector cells. Cell Rep. 2019;27:3657–3671.e4. doi: 10.1016/j.celrep.2019.05.064. [DOI] [PubMed] [Google Scholar]
- 36.Milner J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 2017;552:253–257. doi: 10.1038/nature24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- 38.Sumaria N., Roediger B., Ng L.G., Qin J., Pinto R., Cavanagh L.L., Shklovskaya E., Fazekas de St Groth B., Triccas J.A., Weninger W. Cutaneous immunosurveillance by self-renewing dermal γδ T cells. J. Exp. Med. 2011;208:505–518. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien R.L., Born W.K. Dermal γδ T cells--What have we learned? Cell. Immunol. 2015;296:62–69. doi: 10.1016/j.cellimm.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomura M., Yoshida N., Tanaka J., Karasawa S., Miwa Y., Miyawaki A., Kanagawa O. Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proc. Natl. Acad. Sci. USA. 2008;105:10871–10876. doi: 10.1073/pnas.0802278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamizo S., Egawa G., Tomura M., Sakai S., Tsuchiya S., Kitoh A., Honda T., Otsuka A., Nakajima S., Dainichi T., et al. Dermal Vγ4(+) γδ T cells possess a migratory potency to the draining lymph nodes and modulate CD8(+) T-cell activity through TNF-α production. J. Invest. Dermatol. 2015;135:1007–1015. doi: 10.1038/jid.2014.516. [DOI] [PubMed] [Google Scholar]
- 42.Brinkmann V., Davis M.D., Heise C.E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 2002;277:21453–21457. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 43.Mondot S., Boudinot P., Lantz O. MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics. 2016;68:537–548. doi: 10.1007/s00251-016-0927-9. [DOI] [PubMed] [Google Scholar]
- 44.Yu H., Yang A., Liu L., Mak J.Y.W., Fairlie D.P., Cowley S. CXCL16 stimulates antigen-induced MAIT cell accumulation but trafficking during lung infection is CXCR6-independent. Front. Immunol. 2020;11:1773. doi: 10.3389/fimmu.2020.01773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castela M., Nassar D., Sbeih M., Jachiet M., Wang Z., Aractingi S. Ccl2/Ccr2 signalling recruits a distinct fetal microchimeric population that rescues delayed maternal wound healing. Nat. Commun. 2017;8:15463. doi: 10.1038/ncomms15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meierovics A., Yankelevich W.J.C., Cowley S.C. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA. 2013;110:E3119–E3128. doi: 10.1073/pnas.1302799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGeachy M.J., Cua D.J., Gaffen S.L. The IL-17 family of cytokines in health and disease. Immunity. 2019;50:892–906. doi: 10.1016/j.immuni.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaiss D.M.W., Gause W.C., Osborne L.C., Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42:216–226. doi: 10.1016/j.immuni.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konieczny P., Xing Y., Sidhu I., Subudhi I., Mansfield K.P., Hsieh B., Biancur D.E., Larsen S.B., Cammer M., Li D., et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science. 2022;377:eabg9302. doi: 10.1126/science.abg9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamichhane R., Schneider M., de la Harpe S.M., Harrop T.W.R., Hannaway R.F., Dearden P.K., Kirman J.R., Tyndall J.D.A., Vernall A.J., Ussher J.E. TCR- or cytokine-activated CD8+ mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. 2019;28:3061–3076.e5. doi: 10.1016/j.celrep.2019.08.054. [DOI] [PubMed] [Google Scholar]
- 51.Kämpfer H., Kalina U., Mühl H., Pfeilschifter J., Frank S. Counterregulation of interleukin-18 mRNA and protein expression during cutaneous wound repair in mice. J. Invest. Dermatol. 1999;113:369–374. doi: 10.1046/j.1523-1747.1999.00704.x. [DOI] [PubMed] [Google Scholar]
- 52.Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kreslavsky T., Savage A.K., Hobbs R., Gounari F., Bronson R., Pereira P., Pandolfi P.P., Bendelac A., von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Constantinides M.G., McDonald B.D., Verhoef P.A., Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slütter B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., Harty J.T. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2017;2:eaag2031. doi: 10.1126/sciimmunol.aag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonseca R., Beura L.K., Quarnstrom C.F., Ghoneim H.E., Fan Y., Zebley C.C., Scott M.C., Fares-Frederickson N.J., Wijeyesinghe S., Thompson E.A., et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 2020;21:412–421. doi: 10.1038/s41590-020-0607-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koay H.-F., Su S., Amann-Zalcenstein D., Daley S.R., Comerford I., Miosge L., Whyte C.E., Konstantinov I.E., d’Udekem Y., Baldwin T., et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol. 2019;4:eaay6039. doi: 10.1126/sciimmunol.aay6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 59.Korbecki J., Bajdak-Rusinek K., Kupnicka P., Kapczuk P., Simińska D., Chlubek D., Baranowska-Bosiacka I. The role of CXCL16 in the pathogenesis of cancer and other diseases. Int. J. Mol. Sci. 2021;22:3490. doi: 10.3390/ijms22073490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brinkmann V., Billich A., Baumruker T., Heining P., Schmouder R., Francis G., Aradhye S., Burtin P. Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 2010;9:883–897. doi: 10.1038/nrd3248. [DOI] [PubMed] [Google Scholar]
- 61.Voillet V., Buggert M., Slichter C.K., Berkson J.D., Mair F., Addison M.M., Dori Y., Nadolski G., Itkin M.G., Gottardo R., et al. Human MAIT cells exit peripheral tissues and recirculate via lymph in steady state conditions. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98487. e98487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chodaczek G., Papanna V., Zal M.A., Zal T. Body-barrier surveillance by epidermal γδ TCRs. Nat. Immunol. 2012;13:272–282. doi: 10.1038/ni.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vahl J.C., Heger K., Knies N., Hein M.Y., Boon L., Yagita H., Polic B., Schmidt-Supprian M. NKT cell-TCR expression activates conventional T cells in vivo, but is largely dispensable for mature NKT cell biology. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001589. e1001589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cook P.W., Mattox P.A., Keeble W.W., Pittelkow M.R., Plowman G.D., Shoyab M., Adelman J.P., Shipley G.D. A heparin sulfate-regulated human keratinocyte autocrine factor is similar or identical to amphiregulin. Mol. Cell. Biol. 1991;11:2547–2557. doi: 10.1128/mcb.11.5.2547-2557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kennedy-Crispin M., Billick E., Mitsui H., Gulati N., Fujita H., Gilleaudeau P., Sullivan-Whalen M., Johnson-Huang L.M., Suárez-Fariñas M., Krueger J.G. Human keratinocytes’ response to injury upregulates CCL20 and other genes linking innate and adaptive immunity. J. Invest. Dermatol. 2012;132:105–113. doi: 10.1038/jid.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishnan S., Prise I.E., Wemyss K., Schenck L.P., Bridgeman H.M., McClure F.A., Zangerle-Murray T., O’Boyle C., Barbera T.A., Mahmood F., et al. Amphiregulin-producing γδ T cells are vital for safeguarding oral barrier immune homeostasis. Proc. Natl. Acad. Sci. USA. 2018;115:10738–10743. doi: 10.1073/pnas.1802320115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monticelli L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G.K., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwong B.Y., Roberts S.J., Silberzahn T., Filler R.B., Neustadter J.H., Galan A., Reddy S., Lin W.M., Ellis P.D., Langford C.F., et al. Molecular analysis of tumor-promoting CD8+ T cells in two-stage cutaneous chemical carcinogenesis. J. Invest. Dermatol. 2010;130:1726–1736. doi: 10.1038/jid.2009.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Minutti C.M., Modak R.V., Macdonald F., Li F., Smyth D.J., Dorward D.A., Blair N., Husovsky C., Muir A., Giampazolias E., et al. A macrophage-pericyte axis directs tissue restoration via amphiregulin-induced transforming growth factor beta activation. Immunity. 2019;50:645–654.e6. doi: 10.1016/j.immuni.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malissen M., Gillet A., Ardouin L., Bouvier G., Trucy J., Ferrier P., Vivier E., Malissen B. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 1995;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zikherman J., Parameswaran R., Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lochner M., Peduto L., Cherrier M., Sawa S., Langa F., Varona R., Riethmacher D., Si-Tahar M., Di Santo J.P., Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORγt+ T cells. J. Exp. Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamran P., Sereti K.-I., Zhao P., Ali S.R., Weissman I.L., Ardehali R. Parabiosis in mice: a detailed protocol. J. Vis. Exp. 2013:50556. doi: 10.3791/50556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng C.-H., Lee C.-F., Fryer M., Furtmüller G.J., Oh B., Powell J.D., Brandacher G. Murine full-thickness skin transplantation. J. Vis. Exp. 2017:55105. doi: 10.3791/55105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Legoux F., Bellet D., Daviaud C., El Morr Y., Darbois A., Niort K., Procopio E., Salou M., Gilet J., Ryffel B., et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366:494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- 78.Burns D.L. Subunit structure and enzymic activity of pertussis toxin. Microbiol. Sci. 1988;5:285–287. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE207348 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207348).