Abstract
Recent studies show gut microbiota-dependent metabolism of dietary phenylalanine into phenylacetic acid (PAA) is critical in phenylacetylglutamine (PAGln) production, a metabolite linked to atherosclerotic cardiovascular disease (ASCVD). Accordingly, microbial enzymes involved in this transformation are of interest. Using genetic manipulation in selected microbes and monocolonization experiments in gnotobiotic mice, we identify two distinct gut microbial pathways for PAA formation; one is catalyzed by phenylpyruvate:ferredoxin oxidoreductase (PPFOR), and the other by phenylpyruvate decarboxylase (PPDC). PPFOR and PPDC play key roles in gut bacterial PAA production via oxidative and non-oxidative phenylpyruvate decarboxylation, respectively. Metagenomic analyses revealed a significantly higher abundance of both pathways in gut microbiomes of ASCVD patients compared to controls. The present studies show a role for these two divergent microbial catalytic strategies in the meta-organismal production of PAGln. Given the numerous links between PAGln and ASCVD, these findings will assist future efforts to therapeutically target PAGln formation in vivo.
Graphical Abstract
eTOC:
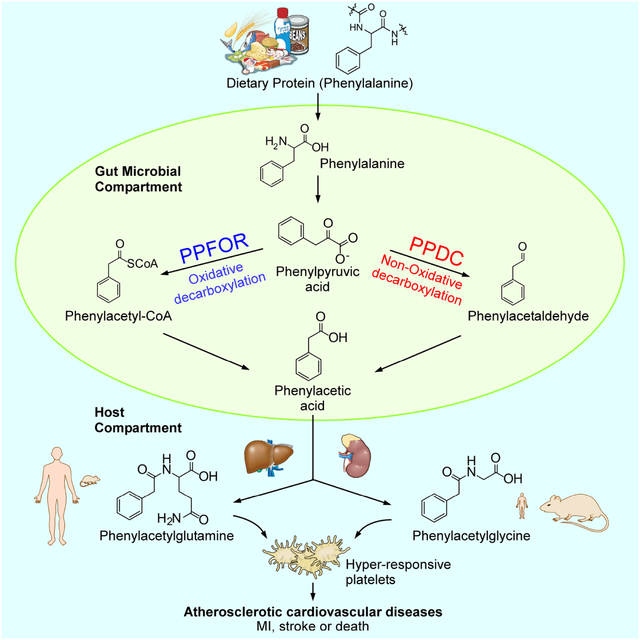
Phenylacetylglutamine (PAGln), an atherosclerotic cardiovascular disease-linked metaorganismal metabolite, is formed through the initial gut microbial transformation of dietary phenylalanine into phenylacetic acid (PAA). Zhu et al. identify two distinct gut microbial pathways for phenylacetic production: one that is catalyzed by phenylpyruvate:ferredoxin oxidoreductase (PPFOR), and the other by phenylpyruvate decarboxylase (PPDC).
Introduction
Gut microbiota derived metabolites can act alone or through pathways involving host metabolism to contribute both to health and disease processes1–3, including atherosclerotic cardiovascular disease (ASCVD)4. In fact, efforts to manipulate production of microbe-derived metabolites to improve human health is a promising area for drug discovery5. Recent large-scale clinical studies have identified phenylacetylglutamine (PAGln) as a gut microbiota dependent metabolite that is associated with both development of ASCVD and incident major adverse cardiovascular events (MACE; myocardial infarction, stroke, or death)6. Subsequent mechanistic studies have revealed PAGln directly enhances thrombosis potential in vivo via G-protein coupled receptors, including α2A, α2B, and β2-adrenergic receptors6. Furthermore, more recent studies have also shown the association between circulating levels of PAGln and stroke risk7,8. Given production of PAGln is a meta-organismal process, understanding the mechanism(s) of gut bacterial contributions to PAGln production is of interest.
The first step in the generation of PAGln is the gut bacteria-dependent transformation of dietary protein derived phenylalanine (Phe) into phynylpyruvic acid (PPY). This initial wide-spread phenylalanine deamination step can be carried out by numerous microbial enzymes including phenylalanine dehydrogenase [EC: 1.4.1.20]9, and aromatic amino acid aminotransferase [EC: 2.6.1.57]10. The gut microbes, subsequently, convert this intermediate gut microbial PPY into PAA6. Following absorption into the portal circulation, host hepatic and renal enzymes catalyze conjugation of PAA to either glutamine forming PAGln (major pathway in primates), or glycine forming phenylacetylglycine (PAGly, major pathway in rodents)6,11,12. Symbiotic gut microbes in humans13,14 and other vertebrates15 capable of generating PAA have been reported for bacteria isolates from three major phyla: Bacteroidetes16,17, Firmicutes6,18,19 and Proteobacteria20,21. However, a detailed understanding of the genetic and biochemical mechanisms of PPY conversion into PAA by human commensal bacteria has not been extensively investigated. In recent studies, we reported that PorA (ClOSPO_00147) from Clostridium sporogenes can contribute to PAA generation6,19. The results of these studies also suggested that additional microbial pathways with distinct biochemical characteristics are involved in intestinal PAA production.
Here, our studies reveal that divergent decarboxylation reactions catalyzed by two distinct catalytic strategies produce PAA from PPY: one strategy occurs via an oxidative process catalyzed by a phenylpyruvate:ferredoxin oxidoreductase (PPFOR); the second occurs via a non-oxidative catalytic strategy mediated by a phenylpyruvate decarboxylase (PPDC). Using both gain-of-function and loss-of-function bacterial mutants and microbial colonization studies in gnotobiotic mice, we show the contribution of both PPFOR and PPDC pathways in gut microbial PAA production, as well as host PAGln and PAGly generation in vivo. Furthermore, using metagenomic analyses, we show that higher abundance of both ppdc and ppfor gene homologues in the gut microbiome are each associated with the presence of ASCVD in humans.
Results
Human commensal bacteria produce phenylacetic acid (PAA) via both PorA-dependent and independent pathways.
In previous studies, we reported that the microbial porA gene (ClOSPO_00147) can produce both PAA and 4-hydroxy-phenylacetic acid (4-HPAA) from Phe and tyrosine (Tyr), respectively, in C. sporogenes. We also showed that disruption of this gene abolishes PAA and 4-HPAA formation by C. sporogenes in vitro19. To test if PorA protein is needed for PAA production by other human gut commensal bacteria, a diverse panel of bacterial strains were screened for the presence of potential PorA protein homologues in their genomes and were classified into candidate PorA-positive versus PorA-negative strains. The strains were then tested for PAA production from isotope-labelled Phe (either [13C9,15N] or [13C8] isotopologue) and non-labeled Phe as substrate (Figure S1). C. sporogenes ΔfldH mutant was used as a positive control in this screen as the wild-type C. sporogenes does not typically produce significant amounts of PAA unless the fldH gene is disrupted, leading to more PPY diverted to PorA6,19. The screen revealed that the presence of a predicted PorA (or related 2-oxoacid:ferredoxin oxidoreductase) protein homologue (Table S1) was associated with PAA production in the majority of instances. Notably, some microbes including, Klebsiella pneumoniae, Proteus mirabilis, and Acinetobacter baumannii produced PAA without harboring a recognizable PorA (or related 2-oxoacid:ferredoxin oxidoreductase) protein homologue (Figure S1).
Phenylpyruvate:ferredoxin oxidoreductase (PPFOR), but not PorA, is the major contributor for PAA generation in Bacteroides thetaiotaomicron.
A closer look at the gene neighborhood and domain structure of PorA protein in C. sporogenes suggested that it forms part of a putative α-ketoisovalerate:ferredoxin oxidoreductase (VOR) (EC 1.2.7.7). Since VOR is a member of the oxoacid:ferredoxin oxidoreductase superfamily22, it is predicted to require CoA and ferredoxin for the oxidation of the PPY and formation of phenylacetyl-CoA derivative (Figure 1A). Phenylacetyl-CoA would then be converted into PAA presumably by acyl-CoA thioesterase23 (Figure 1A). VOR was originally identified in hyperthermophilic archaea22,24,25 with α-ketoisovalerate as its natural substrate. It was also able to degrade PPY, but with only about 10–20% Kcat value of α-ketoisovalerate22. Indeed, recent studies have shown that C. sporogenes ΔporA is unable to produce branched short chain fatty acids, including isobutyrate, 2-methylbutyrate, and isovalerate from their amino acid counterparts26. We therefore looked deeper into the genomes of the PAA-superproducers – those that produced >10 μM of isotobically-labelled PAA – tested in Figure S1. In addition to potential VOR, the ‘superproducers’ also contain gene clusters coding for indolepyruvate:ferredoxin oxidoreductase (IOR) (EC:1.2.7.8). IOR is another member of the oxoacid:ferredoxin oxidoreductase superfamily, which has been reported to oxidatively decarboxylate PPY via phenylacetyl-CoA in archaea27,28. Although traditionally designated as IOR27, it is 2–3 times more active on PPY than indole-3-pyruvate (I3PY) and therefore functions also as phenylpyruvate:ferredoxin oxidoreductase (PPFOR)27,29. Compared to VOR (MTBMA_c10900-c10930), PPFOR (IOR, MTBMA_c04220–04230) from the same archaea strain Methanobacterium thermoautotrophicum was 20 times more active on PPY29. Focusing on B. thetaiotaomicron (B. theta.), a well-studied, prevalent, and genetically tractable gut microbe30,31, the BT0329–0333 cluster is annotated as VOR, while the BT0430/BT0429 is annotated as IOR (Figure 1B). In fact, BT0331 is the closest homologue in B. theta. to C. sporogenes PorA (ClOSPO_00147) with 49% identity (142/288, E-value=8e−104), while BT0430 is the closest homologue to M. marburgensis MTBMA_c04220 (IorA) with 36% identity (194/544, E-value= 7e−102). B. theta. also contains the gene cluster BT2836/BT2837, which codes for a related 2-oxoacid oxidoreductase (potentially 2-oxoglutarate:ferredoxin oxidoreductase, OGOR). Following the Gibson’s et al. phylogeny analyses of the 2-oxoacid:ferredoxin oxidoreductase superfamily32, we analyzed the phylogenetic position of B. theta BT0430/BT0429, BT0333–0329, and BT2836/BT2837 clusters together with C. sporogenes ClOSPO_00146–00149. The analyses confirmed that BT0333–0329 and ClOSPO_00146–00149 grouped with VOR, while BT0430/0429 grouped with archaeal PPFOR (IOR), and BT2836/BT2837 was OGOR (Figure 1C).
Figure 1. Phenylpyruvate:ferredoxin oxidoreductase (PPFOR) is the main contributor for phenylacetic acid (PAA) production in B. thetaiotaomicron.
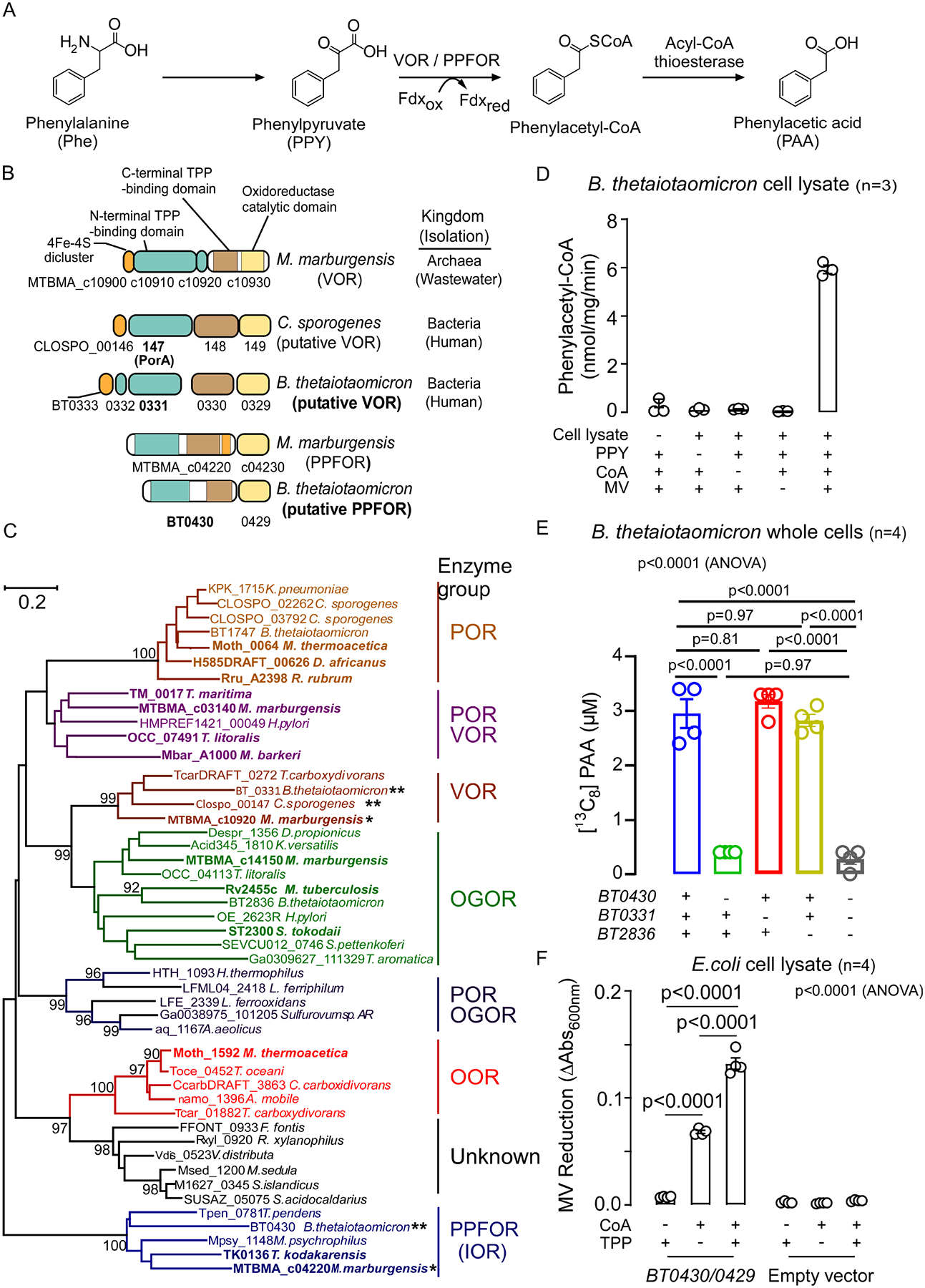
(A) Scheme of the proposed VOR/PPFOR-mediated PAA generation pathway from phenylalanine. Fdxox, Oxidized ferredoxin; Fdxred, Reduced ferredoxin.
(B) Domain structure of putative VOR of C. sporogenes, and B. thetaiotaomicron, putative PPFOR of B. thetaiotaomicron, and the identified VOR and PPFOR (IOR) of archaeon M. marburgensis.
(C) Maximum likelihood phylogenetic tree of thiamine pyrophosphate (TPP) dependent 2-oxoacids:ferredoxin oxidoreductase (OFOR) family. Bootstrap values (500 replicates) greater than 90% are shown. Groups are assigned substrate specificity based upon those enzymes (bold) within the groups that have been biochemically characterized. The preferred substrate of each OFOR enzyme group is: POR, pyruvate; VOR, α-ketoisovalerate; OGOR, α-oxoglutarate; OOR, oxalate; PPFOR (IOR), phenylpyruvate. The locus tag of the α subunit of the enzyme is shown. The scale bar denotes the number of amino acid differences per site. * identified VOR and PPFOR of M. marburgensis. ** putative VOR and PPFOR of C. sporogenes and B. thetaiotaomicron.
(D) Cell lysate of B. thetaiotaomicron oxidatively decarboxylate phenylpyruvic acid and forms phenylacetyl-CoA in the presence of coenzyme A (CoA) and the artificial electron acceptor, methyl viologen (MV). Phenylacetyl-CoA was quantified by LC-MS/MS. Data points represent the mean±SE from three independent replicates.
(E) [13C8]-PAA generation of B. thetaiotaomicron Δtdk (Control), ΔtdkΔBT0430, ΔtdkΔBT0331, ΔtdkΔBT2836 and ΔtdkΔBT0430 ΔBT0331ΔBT2836 knockout mutants in BHI media supplemented with 100 μM [13C9,15N]-Phe at 37 °C under anaerobic condition. Supernatants were sampled after 24 hours of incubation, and [13C8]-PAA was quantified using LC-MS/MS. Data points represent the mean±SE from four independent replicates. Significance was determined using One-way ANOVA followed by Tukey’s multiple comparison test.
(F) Enzymatic assay with cell lysate of E. coli BL21 DE3 overexpressing PPFOR. The PPFOR-overexpressing E.coli cell lysate was washed on 3 kDa MW cutoff filters followed by supplementation with CoA and/or TPP. MV reduction was monitored by measuring the absorption at 600 nm. Bar graphs represent the mean±SE from four independent replicates. Significance was determined using One-way ANOVA followed by Tukey’s multiple comparison test.
To first test if B. theta. uses the catalytic strategy of CoA-dependent 2-oxoacid decarboxylation for PAA production, we measured phenylacetyl-CoA formation during incubation of PPY with B. theta. cell lysates. Since 2-oxoacid:ferredoxin oxidoreductase enzymes are sensitive to inactivation by oxygen, the assay was performed under anaerobic conditions. Indeed, we observed that B. theta. cell lysate forms phenylacetyl-CoA in the combined presence of the substrate (PPY), coenzyme A (CoA) and methyl viologen (MV), an alternative electron acceptor to ferredoxin (Figure 1D). Subsequently, to measure the contribution of VOR, PPFOR, and OGOR to PAA production in B. theta., knockout mutants were individually generated in each gene cluster. For these studies, we used a B. theta. mutant that lacks the thymidine kinase gene (tdk) as our background strain for genetic manipulation. When B. theta. mutants (the control Δtdk, ΔtdkΔBT0331, ΔtdkΔBT0430, ΔtdkΔBT2836, and ΔtdkΔBT0430ΔBT0331ΔBT2836) were grown in the presence of [13C9,15N]-Phe, [13C8]-PAA formation was not affected in either B. theta. ΔtdkΔBT0331, nor B. theta. ΔtdkΔBT2836, while it was significantly reduced (~86%, p<0.0001) in the B. theta. ΔtdkΔBT0430 mutant (Figure 1E) without an apparent effect on growth (Figure S2). Furthermore, the triple knockout mutant ΔtdkΔBT0430ΔBT0331ΔBT2836 had comparable reduction in [13C8]-PAA to the ΔtdkΔBT0430 mutant. These results suggest that in B. theta., PPFOR (BT0430/BT0429) but not VOR (BT0333–0329) nor OGOR (BT2836–2837) plays a major role in PAA production.
To further validate the ability of PPFOR (BT0430/0429) to facilitate PAA production, BT0430/0429 was heterologously overexpressed in E. coli and gain of function activity was tested in vitro using MV reduction as a readout. The PPFOR-expressing E. coli cell lysate was washed using 3 kDa MW cutoff centrifugal filters and tested for PPFOR activity. The PPFOR-expressing E. coli cell lysate reduced MV in the presence of its substrate (PPY), CoA and the cofactor thymidine pyrophosphate (TPP), while the cell lysate of E. coli transformed with the empty vector did not reduce MV (Figure 1F). This agrees well with the known dependency of 2-oxoacid oxidoreductases on TPP and coenzyme A25,27,28. The residual activity of the PPFOR-expressing lysate observed in the absence of added TPP (Figure 1F) is likely due to the tightly bound TPP that could not be removed from the enzyme through repeated washing. This is in agreement with a previous study showing some TPP-dependent microbial enzymes bind to TPP in a tight (almost irreversible) manner33. The phenylacetyl-CoA produced from oxidative decarboxylation by PPFOR was further confirmed by liquid chromatography with tandem mass spectrometry (LC-MS/MS), using an authentic phenylacetyl-CoA standard (Figure S3A). The induced expression of BT0430/0429 was confirmed by SDS-PAGE and proteomic analysis of the cell lysate and purified protein (Figure S3B, C). To characterize PPFOR substrate specificity, a group of structurally distinct α-oxo acids were individually incubated with fresh cell lysates of induced E. coli expressing the recombinant PPFOR (BT0430/0429) under anaerobic conditions and the rate of MV reduction was quantified. The recombinant PPFOR displayed the maximum activity on PPY (100%) as substrate, while the activity on each of indole-3-pyruvic acid (I3PY) and 4-hydroxy-phenylpyruvic acid (4-OH-PPY) as substrates was approximately 24%, and that of pyruvic acid was only 2% of the maximum activity (Table S2).
Inhibiting PPFOR/VOR-mediated oxidative PPY decarboxylation is not sufficient to eliminate the gut microbial production of PAA.
Nitazoxanide (NTZ) is used to treat protozoal infections in mammals34,35, and it inhibits microbial pyruvate:ferredoxin oxidoreductase, which participate in energy metabolism in microaerophilic/obligate anaerobes36,37. Since both VOR and PPFOR are related to pyruvate:ferredoxin oxidoreductase, we first tested whether NTZ inhibits PPFOR and VOR-mediated oxidative PPY decarboxylation. As shown in Figure 2A, NTZ caused strong inhibition of the MV reduction by cell lysates of C. sporogenes ΔfldH and B. theta., indicating its strong in vitro potency against PPFOR/VOR-activities. Further, we observed that NTZ also inhibited C. sporogenes ΔfldH and B. thetaiotaomicron growth at 2.5, and 10 μM final concentrations, respectively (Figure 2B). Given that P. mirabilis (and other PAA-producing Proteobacteria strains (Figure S1)) did not harbor predicted VOR or PPFOR homologues in their genomes, we tested if NTZ would affect PAA-production from this commensal. NTZ did not inhibit [13C8]-PAA production from [13C9,15N]-Phe, nor did it affect P. mirabilis growth under either aerobic or anaerobic conditions (Figure 2C). These results suggest a second possible pathway (that is not sensitive to NTZ) involved in bacterial PAA generation from Phe.
Figure 2. Inhibiting PPFOR/VOR-mediated oxidative phenylpyruvic acid decarboxylation is not sufficient to eliminate the gut microbial production of phenylacetic acid (PAA).
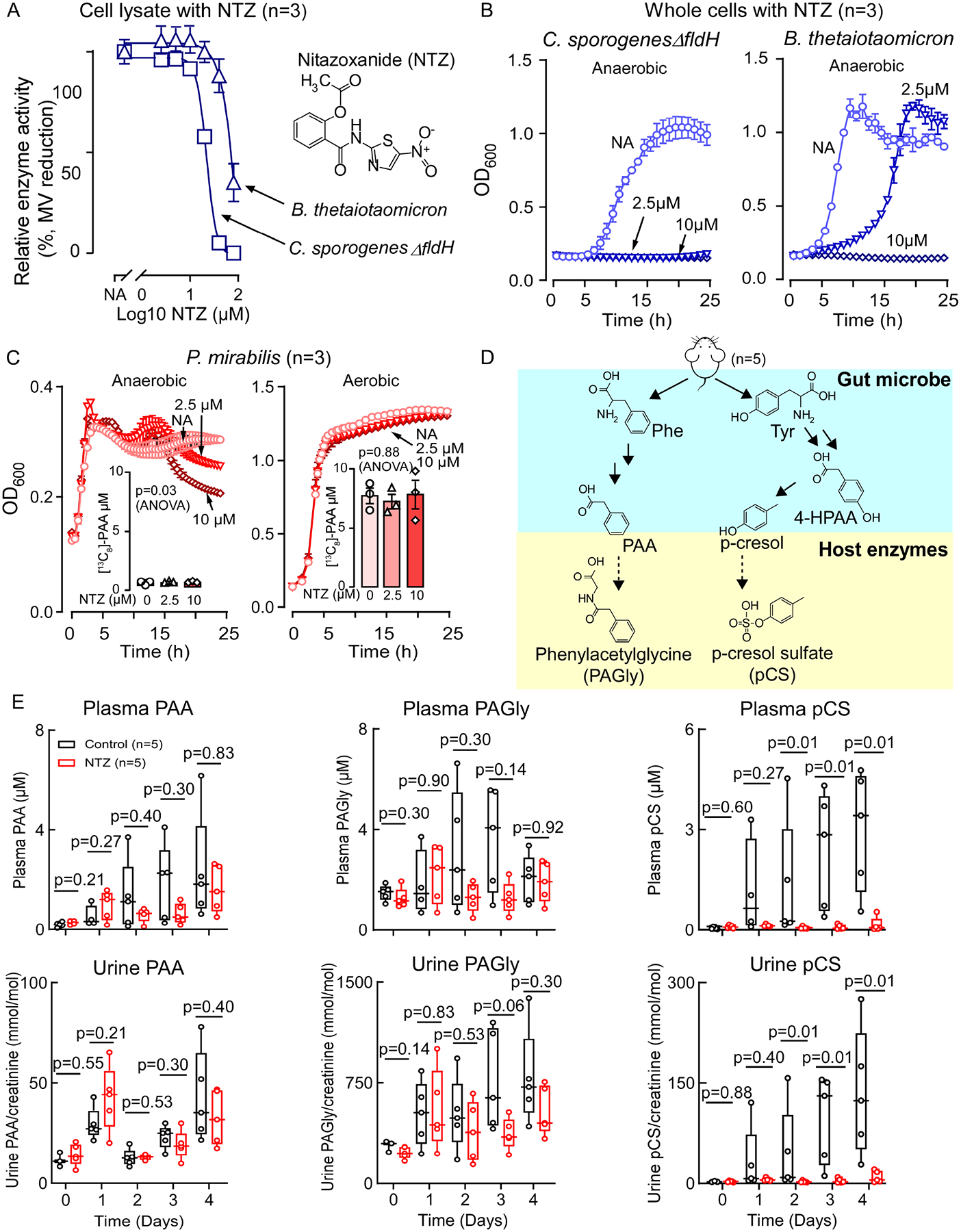
(A) Nitazoxanide (NTZ) inhibits B. thetaiotaomicron and C. sporogenes ΔfldH cell lysate mediated phenylpyruvic acid oxidative decarboxylation under anaerobic condition. Relative activity was measured by the reduction of methyl viologen (MV) monitored by measuring the absorption at 600 nm. Data points represent the mean±SE from three independent replicates.
(B) NTZ inhibits B. thetaiotaomicron and C. sporogenes ΔfldH growth under anaerobic condition.
(C) NTZ does not reduce P. mirabilis growth or PAA generation. P. mirabilis [13C8]-PAA production was measured after 24 hours of growth. Data points represent the mean±SE from three independent replicates.
(D) Scheme for phenylacetylglycine (PAGly) and p-cresol sulfate (pCS) meta-organismal production from diet-derived phenylalalnine (Phe) and tyrosine (Tyr) through the initial microbial tranformation into PAA, and 4-hydroxy-phenylacetic acid (4-HPAA), respectively. PAA is then converted by host enzymes to PAGly. For pCS production, 4-HPAA is converted by other microbial enzymes into p-cresol followed by sulfonation by host enzymes to pCS.
(E) NTZ does not significantly lower plasma and urine levels of PAA or PAGly, but does reduce pCS levels. Mice (n=5) were maintained on high protein diet (60% w/w egg white) only (Control, black) or on high protein diet supplemented with NTZ at 0.06% w/w (NTZ, red). Urine and plasma were collected at the indicated time points and analyzed by LC-MS/MS. Significance at each time point was determined using Wilcoxon rank sum test. Box and whisker plots are showing the median, lower and upper quartiles, and lower and upper extremes of data points.
Next we used NTZ as a tool to help determine how these distinct pathways for PAA generation contribute to gut commensal bacterial PAA production in vivo by testing whether NTZ inhibits generation of PAA and its downstream metabolite, phenylacetylglycine (PAGly), in a mouse model (Figure 2D, E). We reasoned that if PPFOR/VOR-mediated PPY oxidative decarboxylation is the only contributor to gut microbial PAA generation, NTZ would significantly reduce plasma and urine levels of PAGly in mice. We found that NTZ exposure did not cause a statistically significant reduction in plasma or urine levels of either PAA or PAGly (Figure 2E). We noted, however, that NTZ treatment completely suppressed plasma and urine levels of another meta-organismal metabolite, p-cresol sulfate (pCS) to nearly undetectable levels (p=0.01 for both plasma and urine, Figure 2E). pCS is generated initially by gut microbial transformation of Tyr into p-cresol via 4-HPAA intermediate38 followed by host sulfonation of the p-hydroxyl group39. Given that human commensal monoculture and in vivo mouse studies all suggest that microbiota employ catalytic strategies to generate PAA beyond the oxidative decarboxylation pathway, we therefore sought to identify alternative microbial genes involved in PAA and PAGly generation.
Phenylpyruvate decarboxylase (PPDC) plays an important role in PAA generation in Proteus mirabilis via non-oxidative decarboxylation.
We hypothesized that an alternative pathway of PAA production could involve non-oxidative decarboxylation of PPY by phenylpyruvate decarboxylase (PPDC) (EC: 4.1.1.43), as demonstrated in a facultative anaerobe found in soil, Azospirillum brasilense, and within yeast, Saccharomyces cerevisiae40,41. In these multi-step transformations, dietary Phe would first be deaminated into PPY, which is then decarboxylated by PPDC into phenylacetaldehyde. PAA might then be produced from phenylacetaldehyde by an aldehyde dehydrogenase or aldehyde:ferredoxin oxidoreductase42,43. Similar to PPFOR (IOR), A. brasilense PPDC is also active on indole-3-pyruvic acid as a substrate, and was originally annotated as indolepyruvic acid decarboxylase (IPDC)40. Using A. brasilense PPDC (Uniprot accession number P51852) as the query protein sequence for BLAST search, we identified a P. mirabilis protein, HMPREF0693_2975, which shares 24.1% amino acid identity (Figure 3A). Moreover, HMPREF0693_2975 also showed sequence homology to other members of the TPP-dependent decarboxylase family, which plays an important role in 2-oxoacid non-oxidative decarboxylation44. All enzymes analyzed of this family shared a well-conserved TPP-binding motif GD(or E)GX24NN, and an E(or D)-D-H catalytic triad with two invariant H residues (Figure 3A)45–47. To test if HMPREF0693_2975 is involved in PAA generation by P. mirabilis, we first generated a P. mirabilis knock out mutant (ΔHMPREF0693_2975) via marker-exchange mutagenesis48. While wild type P. mirabilis readily generated PAA when incubated in defined medium supplemented with Phe as the sole nitrogen source, the P. mirabilis ΔHMPREF0693_2975 mutant showed marked reduction in PAA production without apparent growth defect (Figure 3B, C), indicating that HMPREF0693_2975 plays a major role in the transformation of Phe into PAA in P. mirabilis. Likewise, when P. mirabilis ΔHMPREF0693_2975 mutant was grown in LB medium, 80% reduction in PAA production was observed when compared to the wild-type strain (Figure S4).
Figure 3. Phenylpyruvate decarboxylase (PPDC) is responsible for phenylacetic acid (PAA) production in P. mirabilis.
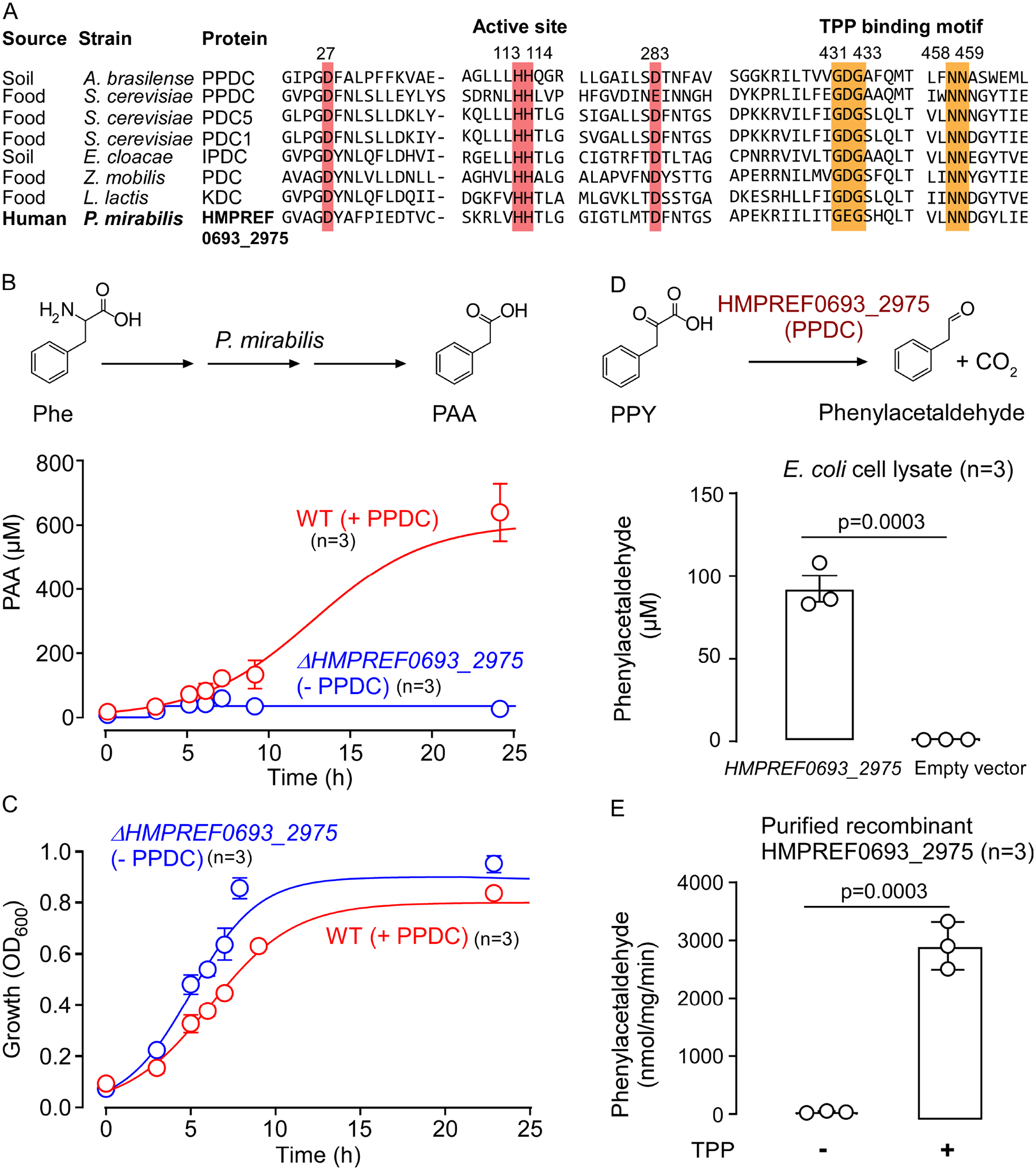
(A) The conserved active site and thiamine pyrophosphate (TPP) binding motif of HMPREF0693_2975 of P. mirabilis and characterized 2-oxoacids decarboxylases. The enzymes included in the analysis are: PPDC of Azospirillum brasilense, PPDC of Saccharomyces cerevisiae, pyruvate decarboxylase isozyme 1 (PDC1) and isozyme 2 (PDC5) of S. cerevisiae, indolepyruvate decarboxylase (IPDC) of Enterobacter cloacae, and branched-chain keto acid decarboxylase (KDC) of Lactococcus lactis.
(B, C) Quantification of PAA production and growth of P. mirabilis wild type and ΔHMPREF0693_2975 knockout mutant in M9 media with phenylalanine (Phe) as the sole nitrogen source under aerobic condition. OD600 and PAA in the supernatant were measured at the indicated time points. Data points represent the mean±SE from three independent replicates.
(D) Enzymatic reaction of recombinant PPDC (HMPREF0693_2975). Scheme of PPDC reaction (top) and enzymatic assay with cell lysate of E. coli BL21 DE3 overexpressing PPDC (bottom). PPDC non-oxidatively degrades phenylpyruvic acid (PPY) to form phenylacetaldehyde. Bar graphs represent the mean±SE from three independent replicates. Phenylacetaldehyde was quantified by LC-MS after derivatization with 2,4-dinitrophenylhydrazine. Significance was determined using Student’s t-test.
(E) PPDC is a thiamine-pyrophosphate (TPP) dependent enzyme. Bar graphs represent the mean±SE from three independent replicates. Significance was determined using Student’s t-test.
In complementary studies, we heterologously expressed recombinant HMPREF0693_2975 in E. coli BL21 DE3. LC-MS/MS analyses of the cell lysate from E. coli mutant overexpressing the putative PPDC confirmed the presence of phenylacetaldehyde, after derivatization with 2,4-dinitrophenylhydrazine (DNPH) (Figure 3D, S5A). Conversely, cell lysate from E. coli transformed with an empty vector showed no detectable level of phenylacetaldehyde (p<0.0001, Figure 3D). Further, when recombinant PPDC was purified (Figure S5B), the recovered apoPPDC (depleted of cofactor TPP) was inactive. However, addition of supplemental TPP restored its catalytic activity (Figure 3E). Subsequent biochemical characterization of purified recombinant PPDC of P. mirabilis demonstrated a broad substrate specificity with maximum decarboxylation activity using either PPY (100%) or pyruvic acid (91%) as substrate. The purified recombinant protein also displayed decarboxylase activity on branched-chain 2-oxoacids and aromatic 2-oxoacids such as benzoylformic acid, 4-OH-PPY and I3PY (Table S2, Figure S5C). These studies thus confirmed that HMPREF0693_2975 of P. mirabilis functions as a PPDC. We also note that protein sequence BLAST analysis suggests that P. mirabilis has a second copy of a PPDC homologue (HMPREF0693_3235) with unknown function. When HMPREF0693_3235 was overexpressed in E. coli and incubated in the presence of the 2-oxoacids listed in Table S2, no evidence of catalytic activity with any substrate was found (data not shown).
PPFOR mediated oxidative decarboxylation and PPDC mediated non-oxidative decarboxylation both contribute to intestinal PAA generation
Our cumulative studies have thus identified that at least two distinct human gut commensal enzymes potentially contribute to the generation of PAA (and thus PAGln and PAGly within the host); PPFOR (with possibly some contribution by structurally-related 2-oxoacid oxidoreductases like VOR) and PPDC, which catalyze oxidative and non-oxidative PPY decarboxylation, respectively. However, the relative contributions of these activities to PAA generation in humans is unknown. We therefore searched the Human Microbiome Project (HMP) (https://img.jgi.doe.gov/) reference genomes database (1585 genomes) to investigate the phylogenetic distribution of predicted PPFOR, VOR and PPDC homologs in human microbiota. PPFOR was found mainly in obligate anaerobes of Bacteroides and Firmicutes from the HMP reference genome database (200 out of 1585 genomes, Figure 4A, Table S3). Beyond obligate anaerobes of Bacteroides and Firmicutes, putative VOR homologs (359 out of 1585 genomes, Figure 4A, Table S3) were also found in the microaerophilic bacterium H. pylori, which belongs to Proteobacteria. PPDC homologs were commonly found in aerobes and facultative anaerobes of Proteobacteria, Firmicutes and Actinobacteria (126 of 1585 genomes, Figure 4A, Table S3). Among the aerobes/facultative anaerobes screened in Figure S1, PPDC homologues were identified in A. baumannii, K. pneumoniae, and Staphylococcus aureus.
Figure 4. Phenylacetic acid (PAA) is produced by gut microbiota via oxidative and non-oxidative phenylpyruvic acid decarboxylation.
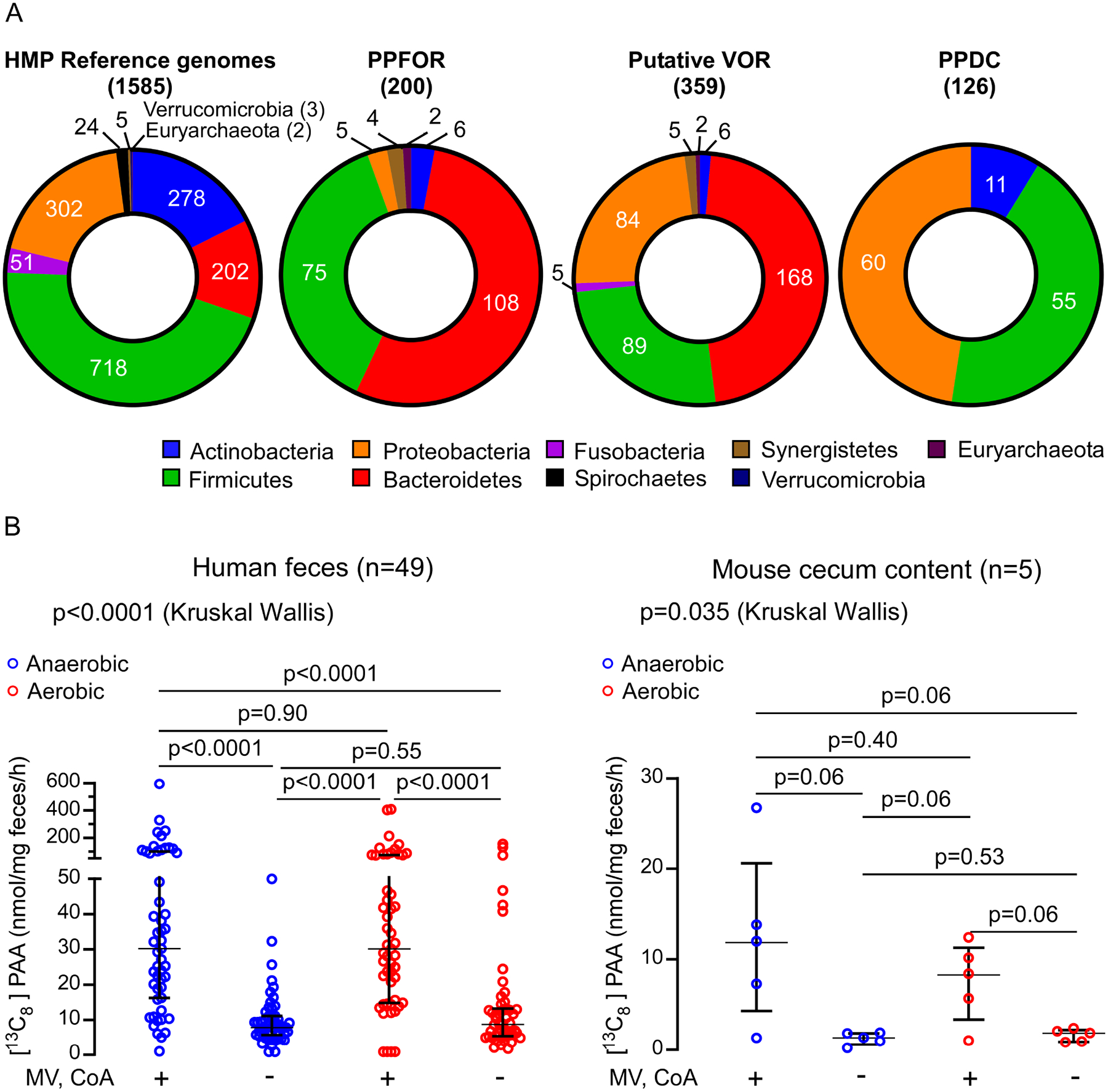
(A) Distribution of PPFOR, VOR and PPDC homologs in Human Microbiome Project (HMP) reference genomes.
The pie charts show the total number of microbial genomes harboring the corresponding subject (PPFOR, VOR, or PPDC homologues) classified according to the phyla. The leftmost chart shows the total number of microbial genomes included in the analyses for each phylum. Analyses were performed using the COG functions; COG3961 (for PPDC), COG4231 and COG1014 (for PPFOR) and COG0674, COG1013, and COG1014 (for VOR). The gene cluster architecture of PPFOR and VOR homologs are inspected manually for each hit. See Supplementary Tables S3, for detailed gene locus tag, strain information and percent identity for each hit to the corresponding subject (PPFOR of B.thetaiotaomicron, VOR of C. sporogenes and PPDC of P. mirabilis).
(B) PAA is produced both anaerobically and aerobically by human fecal microbiome (left, n=49) and mouse cecum microbiome (right, n=5). Human fecal and mouse cecal slurry supernatants were incubated with [13C9,15N]-Phe. [13C8]-PAA was quantified by LC-MS/MS. The reaction was done under anaerobic (blue) and aerobic (red) conditions. MV and CoA were included in the assay to facilitate PPFOR/VOR mediated oxidative decarboxylation. Each mouse cecum was done in three independent replicates and the averages of these replicates are plotted. Medians with interquartile ranges are shown. Significance was determined using Kruskal Wallis test.
The human intestines are a complex environment colonized by microorganisms with different oxygen requirements. Accordingly, we next sought to determine whether the oxidative PPFOR or non-oxidative PPDC decarboxylation catalytic strategies are major contributors to intestinal PAA production in human fecal polymicrobial communities. PAA production was quantified by incubating lysates of human fecal polymicrobial communities from multiple different healthy volunteers (n=49 subjects) with [13C9,15N]-Phe as substrate under both aerobic and anaerobic conditions. In a parallel set of experiments, supplemental CoA and MV were used to facilitate oxidative decarboxylation. The product [13C8]-PAA was quantified using LC-MS/MS after 1 hour of incubation at 37 °C. Notably, [13C8]-PAA was produced under both anaerobic and aerobic conditions (Figure 4B, left) in the absence of CoA and MV. However, CoA and MV supplementation significantly enhanced [13C8]-PAA generation, both in the absence and presence of air (p<0.001, p=0.004, respectively) (Figure 4B). Mass spectrometry analyses did not detect the two intermediates, [13C8]-phenylacetaldehyde or [13C8]-phenylacetyl-CoA, possibly due to their low concentration with rapid further transformation. In parallel studies, we performed similar experiments using freshly harvested mouse cecal contents (n=5). For these studies, immediately following mouse sacrifice, the animals were transferred to an anaerobic chamber, ceca recovered, lysed, bisected, and then half the harvested material was incubated with [13C9,15N]-Phe under anaerobic conditions, and the remainder incubated with [13C9,15N]-Phe in air. The results were consistent with those observed with human fecal samples (Figure 4B, right). In vitro Human fecal and mouse cecum lysate enzymatic assays thus suggest that PPY decarboxylation by both oxidative and non-oxidative catalytic strategies contribute to gut commensal bacteria PAA production (Figure 4B).
Bacterial PPDC and PFFOR activity results in PAGln and PAGly formation within the colonized host
To further confirm the role of these two distinct pathways in PAA production (and possibly PAGln and PAGly production), we opted to construct stable gain-of-function mutants for both PPFOR, and PPDC enzymes in a mouse commensal E. coli and test the activity upon colonizing germ-free mice with these engineered E. coli strains. The mouse commensal E. coli MP1 was chosen as the background strain as previous analyses have shown it is able to colonize the murine gut better than human-derived E. coli strains49. For the PPFOR pathway, the two B. theta. genes BT0429, and BT0430 were inserted in the E. coli chromosome as a single operon driven by the synthetic J23100 promotor (http://parts.igem.org/Part:BBa_J23100). Likewise for PPDC, the P. mirabilis HMPREF0693_2975 gene was also chromosomally expressed downstream of the J23100 promotor. Both E. coli MP1 mutant strains (E. coli ppfor+, and E. coli ppdc+) had similar growth rates as the wild-type when grown in LB media (Figure S6A). However, only E. coli ppfor+ was capable of producing significant amounts of PAA compared to the wild-type (Figure S6C). Consistent with the results of PPDC expression in E. coli BL21, the E. coli MP1 ppdc+ mutant produced phenylacetaldehyde in the culture media (Figure S6C), suggesting that PPDC is also functional in E. coli MP1. However, this strain might be lacking a functional aldehyde dehydrogenase or aldehyde:ferredoxin oxidoreductase capable of performing the subsequent conversion of the phenylacetaldehyde to PAA.
Subsequently, we sought to confirm that both microbial PPFOR and PPDC pathways can function in vivo to produce PAA and recapitulate the meta-organismal production of PAGln and PAGly. Thus, germ-free mice were inoculated with either wild-type E. coli MP1 or E. coli ppfor+ (gain of function mutant) under gnotobiotic housing conditions. In an independent experiment, germ-free mice were alternatively colonized with the wild-type P. mirabilis versus P. mirabilis ΔHMPREF0693_2975 (ppdc−, loss of function mutant) (Figure 5A, B). In both cases, the wild-type and the mutant strains had similar degree of colonization, as indicated by plating fecal samples on LB agar plates (Figure S6B). Notably, host circulating PAGly levels, both in serum and urine, were significantly higher in mice colonized with P. mirabilis vs P. mirabilis Δppdc, and E. coli ppfor + vs E.coli (Figure 5). Similar trends were also observed for the urine levels of PAGln. However, as the phenylacetic acid is converted mainly in the rodents into PAGly rather than PAGln6,11,12, the overall urine concentrations of PAGln were more than two-orders of magnitude below the PAGly levels, while the serum concentrations of PAGln were below the limit of detection (<10 nM).
Figure 5. Bacterial PFFOR and PPDC activity results in phenylacetylglycine (PAGly) and phenylacetylglutamine (PAGln) formation in circulation of monocolonized murine models.
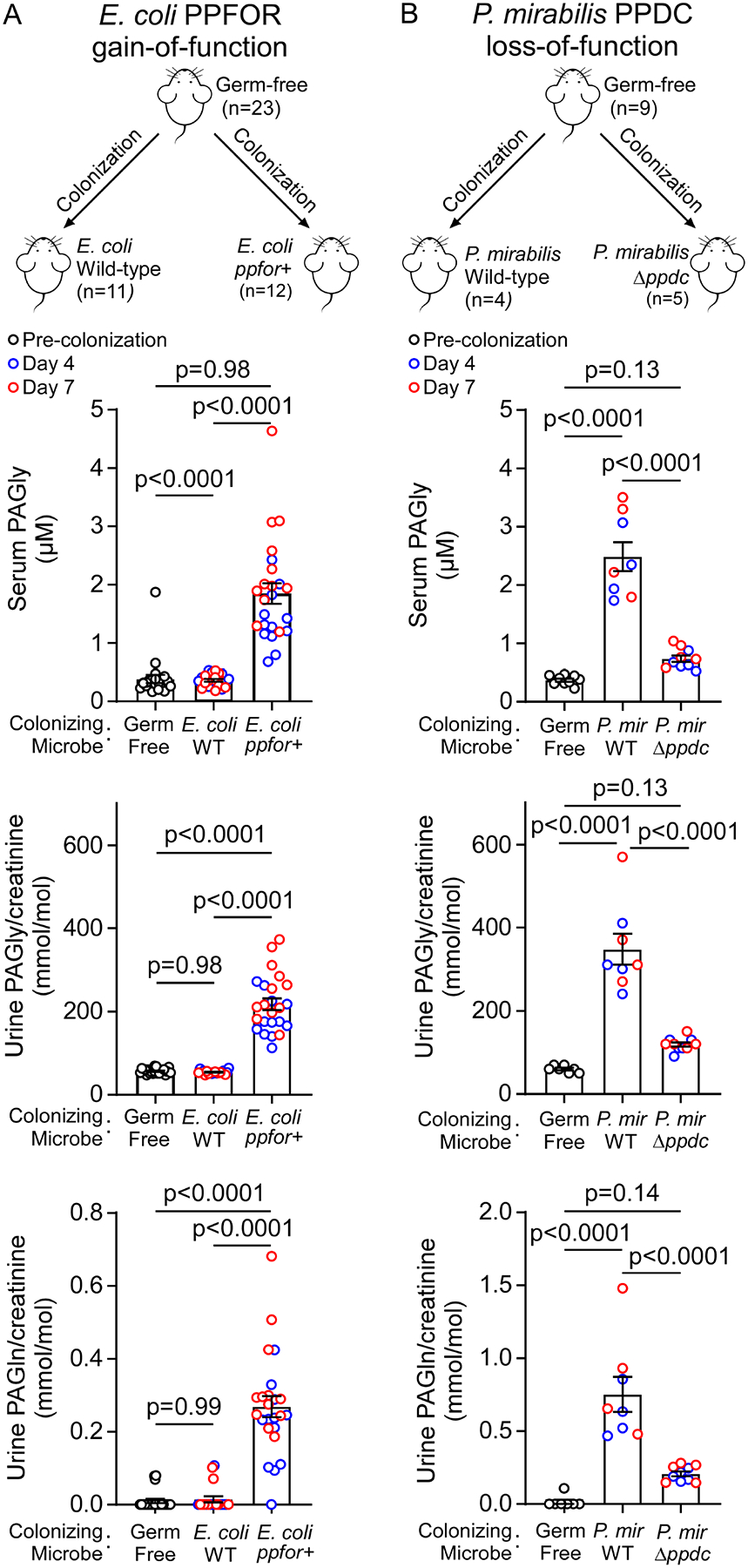
(A) Gain of PPFOR function in E. coli enabled the production of PAGly and PAGln in circulation in monocolonized germ free mice. Male (M) and female (F) germ-free C57BL/6 mice were colonized with E. coli wild-type (n=11; F=6, M=5) or E. coli expressing BT0429/BT0430 genes (ppfor+) (n=12, F=7, M=5).
(B) Loss of P. mirabilis PPDC function reduced PAGly and PAGln in circulation in monocolonized germ-free mice after colonization. Female germ-free C57BL/6 mice (n=9) were randomized, colonized with P.mirabilis wild-type (n=4) or P. mirabilis ΔHMPREF0693_2975 (Δppdc) (n=5).
Mice were maintained on sterile drinking water and chow diet. Serum and urine levels of PAGly were measured using LC-MS/MS before colonization (Germ-free; black open circles) then at day 4 (blue open circles), and day 7 (red open circles) post colonization. Data is shown as individual data points and mean±SE. No difference was seen between males and females, and therefore, the collective data is shown. Significance was determined using One-way ANOVA (p<0.0001 for seum and urine analyses in both A, and B) followed by Tukey’s multiple comparison test. For some mice, urine samples were only able to be collected at one time point (either 4 or 7 days).
Elevated levels of ppdc and ppfor gene homologues in the gut microbiome were associated with atherosclerotic cardiovascular disease (ASCVD) in humans
In a previously published study by Jie et al.50, the authors performed metagenomics analyses on stool samples from a case/control cohort of 218 patients with ASCVD versus 187 control (nonASCVD) individuals to investigate the correlation between the gut microbiome composition and the prevalence of ASCVD. To further investigate the relationship between ppfor and ppdc gene abundances and ASCVD in humans, we re-examined the deposited metagenomics data from this study50. As shown in Figure 6, individuals with higher abundances in their gut metagenomes of either the ppdc or the ppfor gene homologues were more likely to have ASCVD (Figure 6A, and C, p<0.0001 each). Compared to subjects in the lowest tertile of ppfor or ppdc abundance, patients in the highest tertile (T3) demonstrated a significantly increased ASCVD prevalence [ppfor: 3rd vs 1st tertile odds ratio, 8.92, 95% confidence interval, 5.15–15.90, p<0.0001; ppdc: 3rd vs 1st tertile odds ratio, 3.30, 95% confidence interval, 2.03–5.43, p<0.0001]. Following a multivariable logistic regression model to adjust for available ASCVD risk factors (age, sex, systolic blood pressure, HDL-C, LDL-C, triglycerides, and body mass index), elevated (3rd vs 1st tertile) ppdc and ppfor abundances each remained independently associated with ASCVD (Figure 6B, and D).
Figure 6. The association of human fecal microbial abundances of ppdc gene, and ppfor gene and atherosclerotic cardiovascular disease (ASCVD).
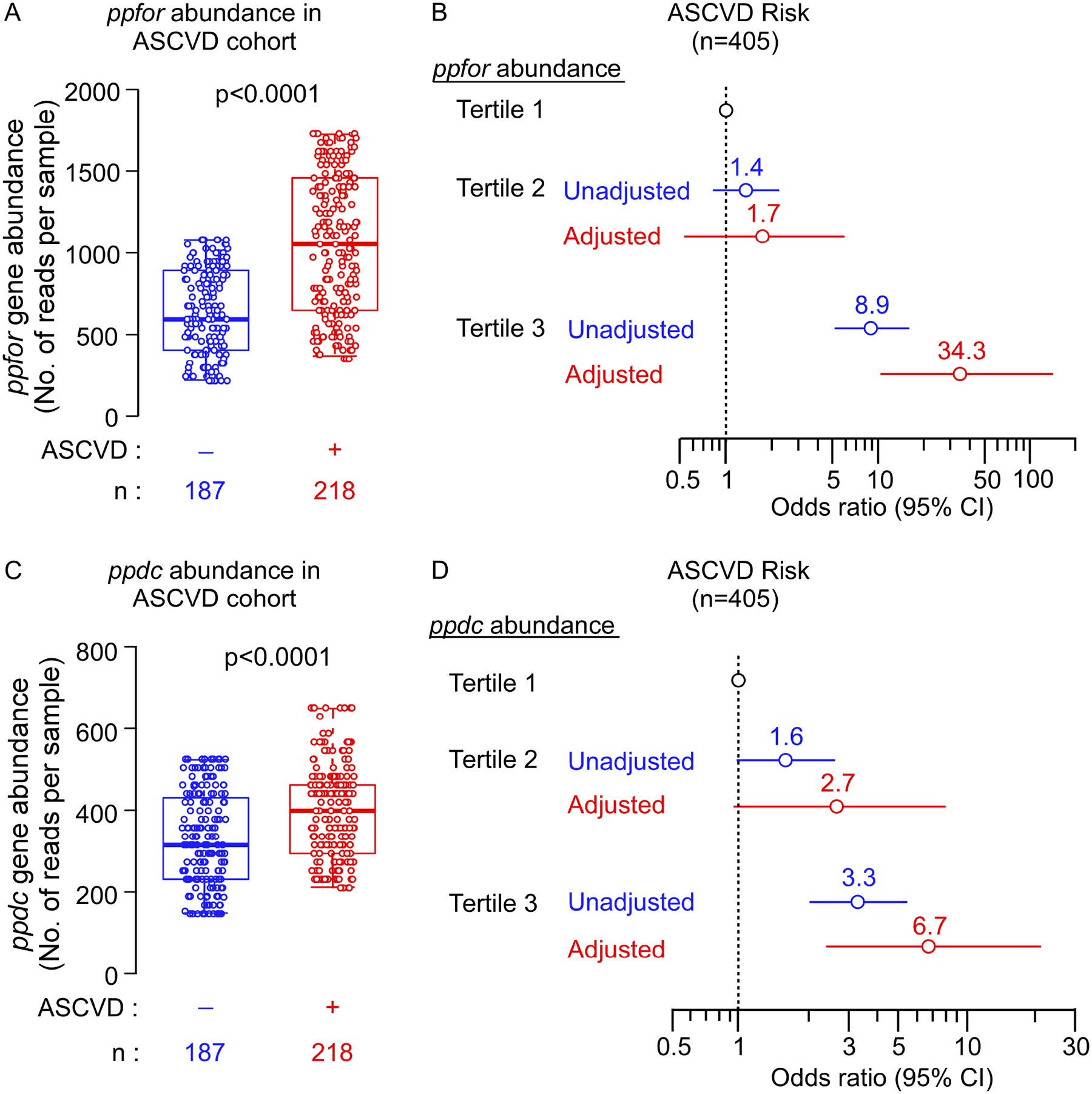
The fecal metagenomics data from Jie et al study50 (N=405, Controls = 187, ASCVD patients = 218) was used to investigate the abundance of ppfor and ppdc gene homologues in control indviduals versus ASCVD patients.
(A, C) Box–Whisker (5–95%) plots of ppfor (A) and ppdc (C) gene abundance in the gut metagenome of control indviduals versus ASCVD patients. P values were calculated using Wilcoxon-rank sum test.
(B, D) Forest plots indicating the ASCVD prevelance according to the tertiles of ppfor (B) and ppdc (D) gene abundance. The multivariable logistic regression model for odds ratio in (B), and (D) included adjustments for age, sex, systolic blood pressure (SBP), HDL cholesterol, LDL cholesterol, triglycerides, and body mass index (BMI). The 5–95% confidence interval is indicated by line length.
Discussion
The gut microbe-derived metabolite PAGln has recently been shown to contribute to ASCVD risk by enhancing platelet responsiveness and thrombosis potential through adrenergic receptor signaling6. While a role for microbiota in the initial production of PAA in the meta-organismal production of PAGln in vivo is known, the microbial enzymes underlying this process remain unclear. In this study, we have identified and characterized key human commensal enzymes that contribute to intestinal PAA production, and PAGln and PAGly generation in vivo. Our collective biochemical and genetic characterizations suggest a scheme illustrated in Figure 7, wherein gut microorganisms generate PAA via at least two distinct TPP-dependent pathways. For anaerobes, such as B. theta., oxidative decarboxylation is carried out by PPFOR, transforming PPY ultimately into a phenylacetyl-CoA intermediate, which is further transformed into PAA (presumably by acyl-CoA thioesterase)23. In contrast to the oxidative decarboxylation catalytic strategy of PPFOR, aerobes and facultative anaerobes such as P. mirabilis utilize PPDC to decarboxylate PPY in a TPP-dependent non-oxidative fashion to form phenylacetaldehyde, which may then be further oxidized (presumably by a phenylacetaldehyde dehydrogenase or phenylacetaldehyde:ferredoxin oxidoreductase) to form PAA (Figure 7)42,43. The catalytic strategy proposed in Figure 7 for both PPFOR and PPDC is based on what is known about the general reaction mechanism for the TPP-dependent pyruvate decarboxylase51 and the 2-oxoacid:ferredoxin oxidoreductase superfamily32. Following gut microbiota generation of PAA, its absorption via portal blood in the host then enables delivery to enzymes in the liver and kidneys to complete the meta-organismal transformation by conjugation with either glutamine to form PAGln (primary reaction in primates) or glycine to form PAGly (primary reaction in rodents)6,11,12,52. Our experiments with NTZ both in vitro and in vivo suggested that inhibiting the PPFOR/VOR-mediated conversion of PPY into PAA is not sufficient to abolish the gut microbial generation of PAA and subsequent PAGly production. In parallel, the meta-organismal metabolite, pCS, was almost completely abolished by NTZ administration. In our previous study, we demonstrated PorA (CLOSPO_00147) in C. sporogenes contributes to both PAA (from Phe) and 4-HPAA (from Tyr) production through the PPY and 4-OH-PPY intermediates, respectively19. However, since our current studies observed that NTZ has antibiotic-like activity, the inhibition of pCS production in vivo may be due not only to the suppression of 4-HPAA production from 4-OH-PPY through the gut microbial PPFOR/VOR pathway but also to suppression (via antibiotic-like activity) of gut anaerobes harboring p-hydroxyphenylacetate decarboxylase needed for the subsequent conversion of 4-HPAA into p-cresol38. Moreover, NTZ showed bactericidal properties against Clostridium difficile53, a model organism for p-cresol production by 4-HPAA decarboxylase54. Future studies are thus needed to investigate the mechanism(s) through which NTZ mediates its effects (e.g. inhibition of microbial enzyme pathways versus antimicrobial activities). Our subsequent recapitulation of both PPFOR and PPDC pathways in monocolonized germ-free mice through both gain-of-function and loss-of-function mutants suggests that both PPFOR and PPDC pathways can contribute to PAGln and PAGly production in hosts (Figure 7). Furthermore, our association analyses between the abundance of ppdc and ppfor genes and the prevalence of ASCVD suggests that both genes are associated with increased risk for ASCVD, even following adjustments for other comorbidities and risk factors for ASCVD. Given that the majority of gut microbes are obligate anaerobes, and that PPFOR has higher prevalence in obligate anaerobes compared to PPDC, one might speculate a larger role for PPFOR versus PPDC in the production of PAGln and PAGly under normal conditions. This will require further testing in future studies. Furthermore, it is likely that the contribution of each pathway may vary as the community structure is altered. These findings illustrate the functional redundancy commonly found in the gut microbiome55,56, which represents a theoretical difficulty in “drugging the microbiome”. If one microbial pathway is blocked, a functionally similar species may fill the niche, leaving the community functional outcome unchanged. Nevertheless, the identification of these two pathways is the first step in paving the way for future work to seek development of mitigating strategies to reduce gut microbial contribution of enteric PAA generation in PAGln-facilitated ASCVD.
Figure 7. Scheme of the two proposed gut bacterial phenylacetic acid (PAA) generation pathways.
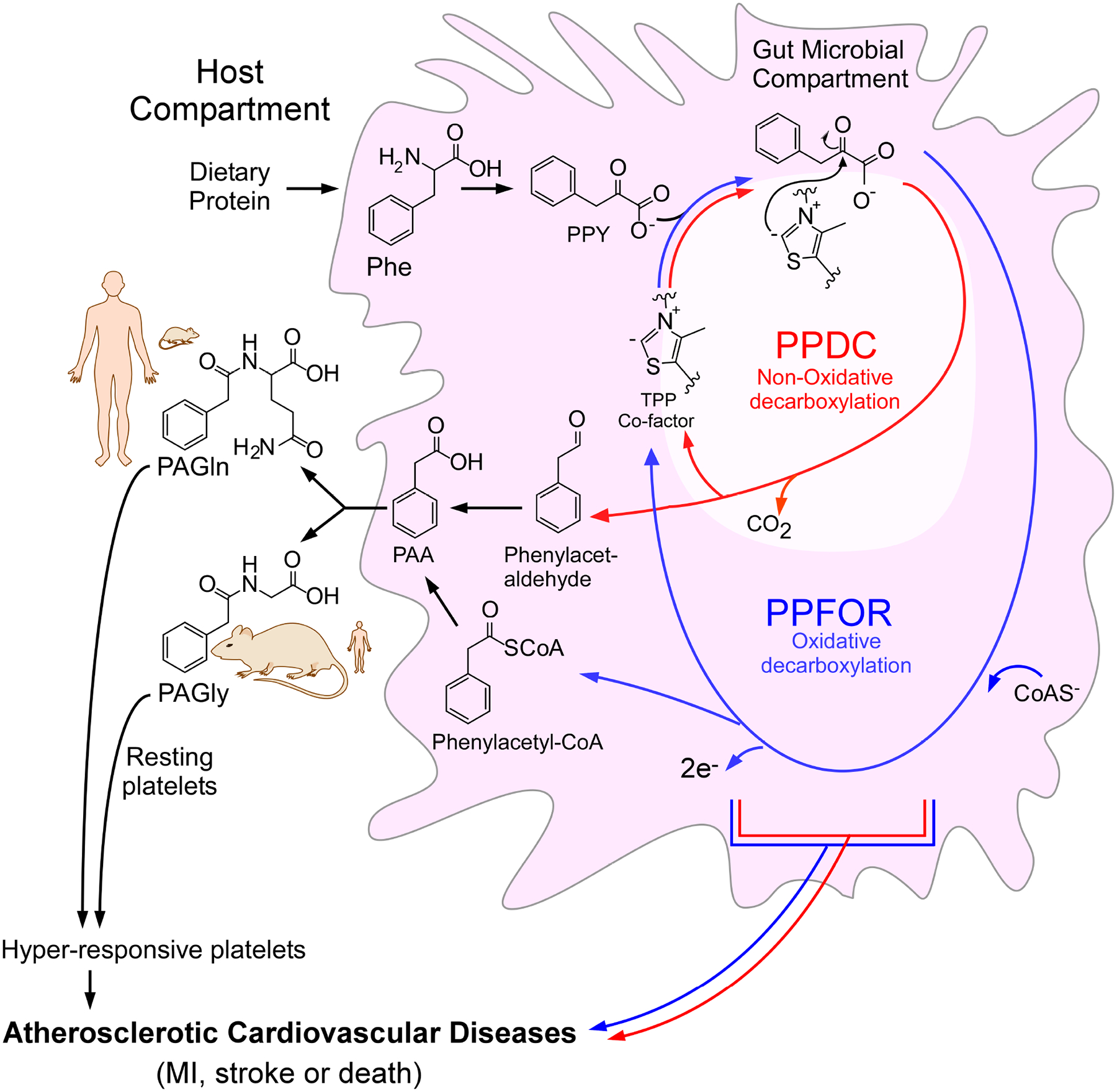
Gut bacteria convert dietary phenylalanine (Phe) into phenylpyruvic acid (PPY). This initial step is then followed by conversion of PPY into PAA through two distinct thiamine pyrophosphate (TPP)-dependent microbial pathways; an oxidative decarboxylation pathway (blue) mediated by PPFOR, and non-oxidative decarboxylation pathway (red) mediated by PPDC enzyme with phenyacetyl-CoA and phenylacetaldehyde as intermediates, respectively. PAA is converted by host liver and kidney enzymes to either phenylacetylglycine (PAGly, dominant pathway in rodents) or phenylacetylglutamine (PAGln, dominant pathway in primates). PAGly and PAGln contribute to atherosclerotic cardiovascular disease (ASCVD) risk through enhancing platelet responsiveness and thrombosis.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stanley L. Hazen (hazens@ccf.org).
Materials availability
All plasmids, bacterial mutant strains, and reagents generated in this study are available from the lead contact upon completing Material Transfer Agreement.
Data and code availability
For Figure 6, data were obtained through reanalysis of Jie et al., 2017 study datasets (European Bioinformatics Institute (EBI) accession number ERP023788)50.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cultivation of wild type bacteria and mutants
Bacteroides thetaiotaomicron VPI-5482 wild type and mutants (Δtdk, ΔtdkΔBT0331, and ΔtdkΔBT0430, ΔtdkΔBT2836, and ΔtdkΔBT0430ΔBT0331ΔBT2836 genotype) were generally cultivated in either mega media57 or Brain Heart Infusion (BHI, Millipore). To characterize the growth of the B. theta mutants, strains were grown in BHI broth for 24 h at 37°C in an anaerobic chamber (Coy Laboratory Products) as the start culture. The start culture was then transferred to fresh BHI broth (inoculum size 1:100, v/v) containing 100 μM of [13C9,15N]-Phe. Growth curves were carried out in flat-bottom 96-well plates, with 200 μl per well, under anaerobic conditions at 37 °C. OD600 values were measured every 30 min using a Tecan Sunrise microplate spectrophotometer. [13C8]-PAA in the supernatant after 24 hours of incubation was quantified using LC-MS/MS. Clostridium sporogenes ATCC 15579 ΔfldH mutant was cultivated anaerobically as B. thetaiotaomicron except that Reinforced Clostridial Medium (RCM, Difco) was used instead of mega media. BHI, RCM and mega media were always supplemented with vitamin K (menadione), hematin, and l-cysteine at concentrations of 1 mg l−1, 1.2 mg l−1 and 0.5 g l−1, respectively. These additives were filter-sterilized and added to the media inside the anaerobic chamber after autoclaving. The hematin stock solution was prepared as 1.2 mg/ml in 0.2 M l-histidine. Proteus mirabilis ATCC 29906 wild type and mutant (P. mirabilis Δppdc) were either cultivated in a defined M9 medium or Luria broth (LB). The defined M9 medium contained NaCl (0.5 g l−1), KH2PO4 (3 g l−1), Na2HPO4·7H2O (12.8 g l−1), MgSO4·7H2O (0.5 g l−1), CaCl2·2H2O (0.15 g l−1), Na2MoO4·2H2O (0.5 mg l−1), FeCl3 (50 μM), and glucose (0.4%, v/v). 2 mM of Phe was used as sole nitrogen source. The LB medium contained tryptone (10 g l−1), yeast extract (5 g l−1) and NaCl (10 g l−1). To characterize the growth of P. mirabilis ATCC 29906 wild type strain and mutant, they were cultivated in 5 ml LB overnight at 37°C in a shaking incubator (250 rpm) and the pellet was collected by centrifugation at 3,000×g for 10 min. Cell pellets were then re-suspended in 1 ml of M9 media. Finally 2 ml of the M9 medium supplemented with 2 mM Phe was inoculated with 40 μl of washed culture and incubated at 37 °C. A 200 μl sample was taken from each tube at different time points. Optical density at 600 nm was recorded immediately for every sample at each time point in flat-bottom 96-well plates, with 200 μl per well. PAA in the supernatant was quantified using LC-MS/MS. Escherichia coli MP1 strain49 was a kind gift from Dr. Mark Goulian at University of Pennsylvania. This strain with its gain-of-function mutants were routinely cultured in LB media. Testing for PAA and phenylacetaldehyde production for the wild-type E. coli and mutants was done after 48 h of culturing in LB media under aerobic conditions.
Other bacterial strains which were used in Figure S1 including (Bacteroides caccae ATCC 43185, Bacteroides cellulosilyticus DSM 14838, Bacteroides ovatus ATCC 8483, Bacteroides uniformis ATCC 8492, Alistipes indistinctus YIT 12060, Lachnoclostridium (Clostridium) asparagiforme DSM 15981, Hungatella (Clostridium) hathewayi DSM 13479, Collinsella aerofaciens ATCC 25986, Akkermansia muciniphila ATCC BAA-835, Collinsella intestinalis DSM 13280, Lachnoclostridium (Clostridium) symbiosum ATCC 14940, Marvinbryantia formatexigens I-52, Blautia (Ruminococcus) gnavus ATCC 29149, Eubacterium rectale ATCC 33656, Bifidobacterium dentium ATCC 27678, Escherichia fergusonii ATCC 35469, Klebsiella pneumoniae WGLW1, Staphylococcus aureus HI022, and Acinetobacter baumannii ATCC 19606), were routinely cultured in mega media.
Mice
All animal model studies were approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic.
Murine NTZ challenge
Swiss Webster female mice (9 weeks, n=10) (Taconic, US) were maintained in microisolator cages under a strict 14h light: 10h dark cycle, ambient temperature (20–26 °C) and humidity 30–70% for the durations of the experiment. They were fed a natural ingredient rodent diet (Envigo, TD.130104). On the day of the experiment, the diet was supplemented with 60% (w/w) egg white (Judes Gluten Free NONGMO, Spray Dried Pasteurized) to enhance protein (and phenylalanine) content. Half of the mice received NTZ in their diet for 5 days at a dose of 0.6 mg per gram of the diet. Given that the mice were 9 weeks old with an average weight of 33 g, and that their daily food intake is approximately 5 g/30 g body weight58, the theoretical daily dose of NTZ was in the range of 100 mg/Kg which is similar to the typical dose of NTZ used in previous studies59–61. Whole blood was collected via saphenous vein into heparin treated capillary tubes. Urine and blood samples were collected daily through day 0 to day 4, and PAA, PAGly, pCS, and urinary creatinine were determined using LC-MS/MS.
Monoclonization experiments in germ-free mice
Germ free mice (C57BL/6) were bred at Cleveland Clinic Gnotobiotic facility and maintained in Allentown sealed positive pressure isolator cages under a strict 14h light: 10h dark cycle, ambient temperature (20–26 °C) and humidity 30–70% for the duration of the experiment. They were fed autoclaved water and autoclaved chow diet ad libitum. At the start of the experiment, blood, urine and feces were collected. Mice were gavaged with 0.2 ml of the bacterial stock. Four and seven days after colonization, blood, urine and feces were collected. Blood was processed for serum. Urine and feces were collected by spontaneous emission.
Collection of Human fecal and mouse cecal samples for PAA generation assay
Human fecal samples were collected as part of the EGGS study (NCT03039023; http://www.clinicaltrials.gov;62) from 49 healthy volunteers who had not received antibiotics or probiotics within 1 month of donation and provided written informed consent. All study protocols and informed consent for human subjects were approved by the Cleveland Clinic Institutional Review Board. Samples were diluted to make a 10% (w/v) fecal slurry by resuspension of the feces in 10% (w/v) glycerol solution, and aliquots were stored in cryogenic vials at −80 °C until use.
Swiss Webster female mice (8 weeks of age, n=5) were sacrificed by CO2 asphyxiation. Mouse ceca were harvested and immediately transferred to anaerobic chamber, followed by homogenization with 600 μl PBS buffer containing 10% glycerol.
METHOD DETAILS
Mass spectrometry quantification of the analytes
Liquid chromatography with in-line tandem mass spectrometry (LC-MS/MS) was used for the quantification of PAA, PAGly, PAGln and creatinine in mouse plasma, serum and urine. Samples (10 μl) were mixed with methanol (40 μl), spiked with internal standards, followed by vortexing and centrifugation to precipitate protein. Urine samples were diluted 1:40 with deionized water before processing. The supernatants were then transferred to glass vials with micro-inserts. For the plasma and urine samples, LC-MS/MS analyses were performed using 8050 triple quadruple mass spectrometer (Shimadzu Scientific Instruments, Columbia, MD, USA) with electrospray ionization. Samples were injected onto a reverse-phase Kinetex C18 column (50 × 2.1 mm, 2.6 μm, Cat # 00B-4462-AN, Phenomenex, Torrance, CA). Solvent A (0.1% acetic acid in water) and B (0.1% acetic acid in acetonitrile) were run using the following gradient: 0.0 min (0% B); 0.0–2.0 min (0% B); 2.0–5.0 min (0%B→20%B); 5.0–6.0 min (20%B→60%B); 6.0–7.5 min (60% B→70%B); 7.5–8.0 min (70%B→100%B); 8.0–9.5 min (100%); 9.5–10 min (100%B→0%B); 10.0–15.0 min (0% B) with a flow rate of 0.4 ml/min and a 1 μl injection volume. Electrospray ionization with multiple reaction monitoring (MRM) was used with the following transitions: m/z 193.8→ 76.1 for PAGly, m/z 265.2→130.2 for PAGln and m/z 270.1→130.2 for D5-PAGln (internal standard) in positive ion mode and m/z 135.0→91.0 for PAA, m/z 137.0→92.0 for [13C2]- PAA (internal standard), m/z 186.7→ 107.0 for pCS and 193.9 →114.1 for D7-pCS (internal standard) in negative ion mode. The following ion source parameters were applied: nebulizing gas flow, 3 l/min; heating gas flow, 10 l/min; interface temperature, 300°C; desolvation line temperature, 250°C; heat block temperature, 400°C; and drying gas flow, 10 l/min. For data analysis, Lab Solution software (Shimadzu) was used.
For urine creatinine, sample aliquots were injected (0.5 μl) onto a normal-phase Luna Silica column (2.0 × 150 mm, 5 μm, Cat # 00F-4274-B0, Phenomenex, Torrance, CA) and resolved with a linear gradient between 0.1% propionic acid in water and 0.1% acetic acid in methanol at flow rate of 0.2 ml/min. Creatinine was analyzed in positive mode and monitored using MRM transition: m/z 114→44.3 for creatinine and m/z 117→47.2 for [D3]-creatinine (internal standard).
For measuring isotopically labeled-PAA in bacterial cultures, the spent culture media were centrifuged and the supernatants were passed through 3 kDa centrifugal filters (Amicon Ultra-0.5 ml Centrifuge filters, Ultra-cel-3K, Merck Millipore). Filtrate (20 μl) was subjected to stable-isotope-dilution LC-MS/MS analysis for quantification of [13C8]- or [13C6]-PAA. Ice cold methanol containing internal standard ([13C2]- PAA; 80 μl) was added to the supernatant, followed by vortexing and centrifuging (21,000 × g; 4°C for 15 min). The clear supernatant was transferred to glass vials with microinserts. LC-MS/MS analysis was performed as described above for plasma and urine samples using the same column and solvent gradient in the negative ion mode using MRM at the following transitions: m/z 135.0→91.0 for PAA; m/z 143.0→98.0 for [13C8]- PAA; m/z 141.0→97.0 for [13C6]- PAA and m/z 137.0→92.0 for [13C2]- PAA (internal standard).
To quantify phenylacetyl-CoA, sample aliquots were injected onto the same normal-phase Luna Silica column and resolved with a linear gradient between 5 mM ammonium acetate in water (solvent A) and 5 mM ammonium acetate in 99% acetonitrile (solvent B) at a flow rate of 0.35 ml/min starting with 90% solvent A, and increasing gradually to 90% solvent B. HPLC column effluent was introduced into a 5500 triple quadruple mass spectrometer (AB SCIEX, Framingham MA, USA) using electrospray ionization in the negative-ion mode. Phenylacetyl-CoA was monitored using MRM: m/z 884.3→408.2. [13C2]-PAA acid was used as internal standard.
To quantify phenylacetaldehyde-2,4-dinitrophenylhydrazone derivative, sample aliquots were injected onto a reverse-phase Prodigy C18 HPLC column (2.0 × 150 mm, 5 μm, Cat # 00F-3300-B0, Phenomenex), eluted at a flow rate of 0.2 ml/min and resolved using a linear gradient between 5 mM ammonium acetate in water and 5 mM ammonium acetate in acetonitrile/water (95:5, v/v). HPLC column effluent was introduced into a 5500 or 4000 triple quadruple mass spectrometer (AB SCIEX, Framingham MA, USA) using electrospray ionization in the negative-ion mode. Both 2,4-dinitrophenylhydrazine derivatives of phenylacetaldehyde and phenylpropionaldehyde were monitored using MRM: m/z 299.0→268.9 for phenylacetaldehyde-2,4-dinitrophenylhydrazone; and m/z 313.0→282.9 for phenylpropionaldehyde-2,4-dinitrophenylhydrazone (internal standard).
Screening human commensals for the presence of potential PorA homologues and correlation with phenylacetic acid production
The publicly available genomes of C. sporogenes ATCC 15579 and the other 21 cultured strains shown in Figure S1 were selected on the JGI-IMG/M website. C. sporogenes CLOSPO_00147, identified in Dodd et al. 201719 as PorA, was used as the reference sequence for a protein BLAST with the selected genomes. Potential PorA or related 2-oxoacid:ferredoxin oxidoreductase protein homologues (e-value < 1e-5) were identified in 9 of the 21 genomes. All identified homologues were manually inspected for gene neighborhood. The only homologues identified in C. intestinalis (COLINT_03157) and C. aerofaciens (COLAER_01233) were found in gene neighborhood which lacked the conserved functions and therefore, these two strains were considered PorA-negative. All selected strains were cultured for 48 h at 37 °C in mega media supplemented with 100 μM of either 13C9,15N-Phe, or 13C6-Phe. They were then incubated anaerobically only (for obligate anaerobes), under both aerobic and anaerobic conditions (for facultative anaerobes) or aerobically only (for the strict aerobe A. baumannii). Samples were then taken, centrifuged and processed for LC-MS/MS analyses.
Phylogenetic analysis of OFOR family members
Phylogenetic analysis of OFOR was carried out following the procedure described by Gibson et al32. 47 OFOR sequences were retrieved from Uniprot database. An alignment was performed using the PROMALS3D server63,64, with the structures of PFOR of Desulfocurvibacter africanus and OOR of Moorella thermoacetica serving as secondary structure references. The aligned sequences were trimmed around conserved blocks of residues from the OFOR α and β subunits using Gblocks65,66, from which a maximum-likelihood tree was generated using MEGA7 package67.
Phenylacetyl-CoA assay of B. thetaiotaomicron cell lysate
B. thetaiotaomicron WT cells were cultured in 40 ml of mega media inside an anaerobic chamber using a 0.8 ml inoculum from an overnight start culture (OD600=1.2~1.5). Bacterial cells were then harvested and resuspended in 2 ml O2-free PBS buffer (pH 7.8, 1:20 v/v) that has been flushed with argon and kept in the anaerobic chamber till use. Cells were disrupted by sonication (level 6; 5 times for 10 s each) on ice with a probe sonicator (Branson Sonifier 250) in the anaerobic chamber. The cell debris was removed by centrifugation (3000 × g, 10 min) and the supernatant was used for the assay. The protein concentration was determined by Bradford method (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as a standard. The enzyme assay mixture (100 μl) was composed of PBS buffer (pH 7.8), 40~60 μg of centrifuged and clarified cell lysate, 1 mM CoA, and 1 mM MV as the artificial electron acceptor. The reaction was initiated by the addition of the substrate (PPY) at 2.5 mM final concentration and incubated at 37 °C anaerobically for 3 hours. The product, phenylacetyl-CoA, was quantified using LC-MS/MS.
Allele-exchange mutagenesis of BT0430, BT0331 and BT2836 in B. thetaiotaomicron Δtdk
DNA fragments (~1 kb) corresponding to the upstream and downstream regions of the target gene were amplified and a subsequent overlap PCR was used to fuse the two fragments which were then ligated into the suicide plasmid pExchange-tdk68. Escherichia coli S17–1λpir competent cells were transformed with the ligated plasmid via electroporation. E. coli S17–1 λpir harboring the plasmid was then mixed with an overnight culture of B. thetaiotaomicron Δtdk. The suspension was plated on BHI-blood agar without antibiotics and incubated aerobically at 37 °C for 1 day. The bacterial biomass was scraped up and re-suspended in TYG (Tryptone-Yeast extract-Glucose) medium. The suspension was then plated on BHI-blood agar supplemented with gentamicin at 200 ng/μl (to select against E. coli S17 donor cells) and erythromycin at 25 ng/μl. Single-crossover integrants were then selected, cultured in TYG medium overnight and plated on BHI-blood agar medium supplemented with 5-Fluoro-2’-deoxyuridine to select for the correct double cross-over mutants. The deletion mutants were screened by PCR amplification and further verified by Sanger sequencing.
Nitazoxanide (NTZ) effect on growth and PAA production
B. thetaiotaomicron was grown in 200 μl of mega media (inoculum size 1/25, v/v) containing NTZ with varying concentrations. The whole cell inhibition assay was carried out in a 96-well plate (sterile, flat-bottom) in the anaerobic chamber and OD600 values were measured every 30 min. The growth curve of C. sporogenes was examined in the same way as B. thetaiotaomicron except that reinforced clostridium media was used instead of mega media. P. mirabilis was grown in 200 μl of LB media (inoculum size 1/50, v/v) containing 100 μM [13C9,15N]-Phe and NTZ at varying concentrations, and the growth was monitored overtime in 96-well plate. After 24 h, [13C8]-PAA was determined by LC–MS/MS.
Sequence alignment of PPDC homologous proteins
Multiple sequence alignment of PPDC homologous proteins was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/)69. Members of the thiamine pyrophosphate (TPP)-dependent decarboxylase family included in the analyses were pyruvate decarboxylase isozyme 1 (PDC1, Accession: CAA97573.1) and isozyme 2 (PDC5, Accession: CAA97705.1) of S. cerevisiae41, pyruvate decarboxylase (PDC, Accession: AHJ73198.1) from Zymomonas mobilis70, branched-chain keto acid decarboxylase from Lactococcus lactis (KDC, Accession: AY548760.1)71 and indolepyruvate decarboxylase (IPDC, Accession: WP_013098183) from Enterobacter cloacae72.
Marker-exchange mutagenesis of ppdc in Proteus mirabilis ATCC 29906
ppdc (HMPREF0693_2975) gene deletion from P. mirabilis was performed as described previously48. Briefly, the SacB gene cassette was amplified from plasmid pK18mobsacB73. Similarly, ~ 2 kb fragment containing the ampicillin resistance gene cassette, R6K replication origin and RP4-oriT was PCR amplified from plasmid pExchange-tdk68. These two fragments were then fused together to form a single 3.6 kb segment. Two PCR reactions were then used to amplify ~ 1.1kb DNA fragment upstream of the HMPREF0693_2975 gene (including the first 30 codon of the gene) and 1.2 kb fragment downstream (including the last 28 codons of the gene). These up- and downstream arms were ligated to the ends of a Kanamycin resistance gene cassette through fusion PCR to form a single 3.4 kb segment. The final 3.4 kb and 3.6 kb segments were then ligated using the In-Fusion® HD Cloning kit (Clontech) to make the suicide plasmid pExSacB-2975::Kan. The ligated plasmid was transformed into E. coli S17 λpir. pExSacB-2975::Kan was then transferred to P. mirabilis through conjugation and the resulted conjugants were screened on LB agar plates containing tetracycline at 10 ng/μl (to select against E. coli S17 donor cells), kanamycin at 50 ng/μl and ampicillin at 200 ng/μl. One single-crossover integrant was then selected and re-streaked on LSW-Sucrose agar plate (tryptone 10 g/l, yeast extract 5 g/l, glycerol 5 ml/l, NaCl 0.4 g/l, sucrose 100 g/l and agar 20 g/l) supplemented with kanamycin at 50 ng/μl to select for the correct double cross-over mutants. One mutant was then selected, re-streaked, and confirmed for the loss of the conjugated plasmid through Sanger sequencing and its ability to grow in presence of kanamycin but not ampicillin.
Cloning and heterologous overexpression of PPDC, and PPFOR in E. coli BL21 DE3
The ppdc gene (HMPREF0693_2975) of P. mirabilis and ppfor genes (BT0430/BT0429) of B. thetaiotaomicron were codon optimized, chemically synthesized by GenScript and inserted into the expression vector pET16b in-between the NdeI and BamHI restriction sites. The resulting plasmids were then transformed into the expression host E. coli BL21 DE3.
For PPDC overexpression, 250 ml of LB medium were inoculated with 5 ml of an overnight culture of the recombinant E. coli, and grown at 37 °C to an OD600 of 0.6. Protein expression was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were grown overnight at room temperature. Cells were harvested by centrifugation at 6,000 × g for 10 min, and the pellets were stored at −80 °C. For cell disruption, the pellets were resuspended in 50 mM Tris-HCl, pH 7.8, with 50 mM NaCl and cells were sonicated (level 6; 3 times for 10 s each) with a probe sonicator (Branson Sonifier 250). The cell lysate was cleared by centrifugation (10,000 × g; 25 min; 4°C), and the supernatant was applied to a Ni-NTA column (GE healthcare). The recombinant protein was purified and eluted at an imidazole concentration of 250 mM. The enzyme purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining, and the protein concentration was determined by Bradford method using bovine serum albumin as a standard.
PPFOR expression was induced in the same way as PPDC, except the induction was carried out anaerobically. E. coli culture was transferred to anaerobic chamber after addition of IPTG at 0.5 mM final concentration. Cells were harvested by centrifugation at 6,000 × g for 10 min. The cell pellets were resuspended in 50 mM Tris-HCl, pH 7.8, 50 mM NaCl and immediately transferred to Hungate tubes flushed with O2-free argon. PPFOR α-subunit (BT0430) was 10xHis-tagged at the N-terminus, and the induction of which was examined by SDS-PAGE after Ni-NTA purification as described in PPDC purification. PPFOR β subunit (BT0429) was cloned without an affinity tag, and therefore, its expression was examined by LC-MS/MS of E. coli cell lysate after trypsin in-solution digestion.
For in-gel protein digestion, the band corresponding to BT0430 was cut from the gel, washed and de‐stained in 50% ethanol and 5% acetic acid in water. The gel pieces were then dehydrated in acetonitrile and dried in a Speed-vac. Proteins in the dried pieces were reduced with 0.1 ml of 65 mM Dithiothreitol (DTT) at room temperature for 30 min, followed by alkylation for another 30 min at room temperature with 0.1 ml of 162 mM iodoacetamide. Gel pieces were dehydrated two times with 0.2 ml acetonitrile and rehydrated with 0.2 mL of 100 mM ammonium bicarbonate. Proteins were digested by adding 5 μl of 10 ng/μl of trypsin in 50 mM ammonium bicarbonate, followed by incubation overnight at 37 °C. The peptides were extracted into two portions of 30 μl each containing 50% acetonitrile and 5% formic acid in water. The combined extracts were evaporated to < 10 μl in a Speed‐vac and then re‐suspended in 1% acetic acid in water to make up a final volume of ~30 μl for LC‐MS analysis.
For in solution protein digestion, proteins were reconstituted in 8 M urea in Tris-HCl 100 mM, pH 8.0, reduced with DTT and alkylated with iodoacetamide as described above followed by overnight precipitation with cold acetone (−20 °C). Samples were centrifuged at 12,000 × g for 8 minutes at 0 °C, and the supernatants were removed. Protein pellets were air dried for 30 min and dissolved in 40 μl of 100 mM tri-ethyl ammonium bicarbonate (TEAB) with 0.5 μg trypsin per sample. After overnight incubation, digested samples were centrifuged at 21,000 × g for 15 min, and 5 μg of the digest from each sample was transferred to a new tube, dried down in a SpeedVac and reconstituted in 25 μl of 0.1% formic acid in water.
Samples were analyzed by LC-MS/MS using an Orbitrap Velos (ThermoScientific) equipped with an Easy-nLC HPLC system and a Dionex (25 cm × 75 μm id) Acclaim Pepmap C18, 2-μm, 100-Å reversed-phase capillary chromatography column. Peptide digests were injected onto the reverse phase column and eluted at a flow rate of 0.5 μl/min using mobile phase A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The gradient was held at 5% B for 5 minutes, %B was increased linearly to 35% in 40 minutes, increased linearly to 70% B in 10 minutes, and maintained at 70% B for 5 minutes. A data-dependent acquisition method was used that involved full scan MS1 (300–2,000 Da) acquisition in the Orbitrap MS at a resolution of 60,000. This was followed by collision induced dissociation (CID) at 35% CE and ion trap detection on the ten most abundant ions. Dynamic exclusion was enabled for ions fragmented twice in 30 s, and these ions were placed on an exclusion list for 30 s. Dynamic exclusion was enabled where ions fragmented two times in 10 seconds and these ions were excluded for 30 seconds. Peak lists were generated with Proteome Discoverer 2.3 (Thermo Fischer Scientific).
Measuring recombinant Phenylpyruvate:ferredoxin oxidoreductase (PPFOR) activity
Recombinant PPFOR activity was routinely determined anaerobically by the PPY-dependent reduction of MV. Since PPFOR is oxygen sensitive, we used fresh cell lysate to minimize the enzyme activity loss during storage. To confirm that PPFOR enzymatic product is phenylacetyl-CoA, 10 μl of the reaction mixture were mixed with 40 μl of ice-cold methanol followed by vortexing and centrifuging (21,000 × g; 4°C for 15 min). The de-proteinated supernatant was then transferred to a glass vial with a micro-insert for LC-MS/MS. Authentic synthetic phenylacetyl-CoA was examined in the same way to compare the spectrum. The optimum pH and reaction time for monitoring products with recombinant enzyme were determined to be Glycine-NaOH buffer 100 mM, pH 10.5 (Figure S7A). The time course experiment showed that the enzyme activity slowed down after 20 min of incubation as assessed by MV reduction (Figure S7B).
A typical reaction mixture contained ~20–30 μg of cell lysate of induced E. coli BL21 DE3/pET16b-PPFOR cells, 100 mM Gly-NaOH buffer (pH 10.5), 0.1 mM MgSO4, 0.1 mM TPP, 1 mM CoA, and 1 mM MV in a final volume of 200 μl. The reaction was initiated by the addition of PPY at 2.5 mM final concentration and incubated at 37 °C for 15 min. The reaction was done in a 96-well plate and the absorbance changes at 600 nm were measured within a Tecan Sunrise microplate reader for 20 min with 30 s interval in the Coy anaerobic chamber.
To test the PPFOR dependency on CoA, and TPP, lysate of induced E. coli BL21 DE3/pET16b-PPFOR cells or the control E. coli harboring the empty pET16b, was washed on a 3 kDa cutoff centrifugal filters (Merck Millipore) 5 times to remove TPP and CoA. The washing buffer was Tris-HCl 50 mM supplemented with NaCl at 50 mM, pH 8.5. The washed lysate was then added to Tris-HCl buffer adjusted to pH 7.8. MgSO4, MV, and phenylpyruvic acid were then added at final concentrations of 0.1, 1, and 2.5 mM, respectively. The reaction was done in a 96-well plate and the absorbance changes at 600 nm (corresponding to MV reduction) were measured within a Tecan Sunrise microplate reader over a 20 min time period in the Coy anaerobic chamber. TPP and CoA were added to the test wells at final concentration of 1 mM for each.
Measuring recombinant phenylpyruvate decarboxylase (PPDC) activity
To confirm that PPDC enzymatic product is phenylacetaldehyde, 5 μl of enzyme reaction mixture was mixed with 20 μl of saturated 2,4-dinitrophenylhydrazine (DNPH) solution in acetonitrile, 0.1 mM HCl at a final volume of 50 μl and incubated for 30 min at room temperature. Samples were speed-vacuum dried and resuspended in 40 μl of acetonitrile, followed by vortexing and centrifuging (21,000 × g; 4°C for 15 min). The supernatant was transferred to a glass vial with a micro-insert for LC-MS/MS. The authentic phenylacetaldehyde was derivatized and examined in the same way to compare the collision induced dissociation (CID) spectrum. PPDC activity was monitored by quantifying phenylacetaldehyde production by LC-MS/MS analysis. For this, 25 μM of phenylpropionaldehyde was included in the derivatization as internal standard.
Several buffers were first compared in order to determine a suitable buffering system and the optimum pH for the purified PPDC. The data presented in Figure S7C demonstrated that PPDC had highest activity at pH 6.8 in 100 mM MES (2-(N-morpholino) ethanesulfonic acid) buffer, which was then chosen for the following experiments. The time course experiment showed that enzyme activity slowed down after 20 min of incubation as assessed by phenylacetaldehyde released (Figure S7D). A typical reaction mixture contained 2 μg of purified recombinant PPDC, 100 mM MES (pH 6.8), 0.1 mM MgSO4, 0.1 mM TPP in a final volume of 100 μl. The reaction was initiated by the addition of PPY at final concentration of 2.5 mM and incubated at 37 °C for 20 min unless stated otherwise.
To test for TPP dependency, purified PPDC was washed on a 10 kDa cutoff centrifugal filter columns 6~8 times to remove TPP. The purified PPDC was then incubated with vehicle versus 0.1 mM TPP in 100 mM MES buffer (pH 6.5) on ice for 20 min to saturate the enzyme with cofactor. Catalytic activity was determined as described above.
The activity of recombinant PPDC on various substrates was determined indirectly by coupled reaction monitoring the reduction of NAD+ by change in absorbance at 340 nm in the presence of aldehyde dehydrogenase under aerobic conditions. For these studies, 0.35 mM NAD+, and 0.125 unit of alcohol dehydrogenase from Baker’s yeast were included in the assay. The absorbance at 340 nm was monitored within a 96-well plate using a UV-Vis spectrophotometer. Relative activities were expressed as a percentage of the highest activity observed (in the presence of substrate phenylpyruvate).
Analyses of PPDC, PPFOR, and VOR protein homologue abundance in Human Microbiome project (HMP) references genomes
HMP references genomes were selected and analyzed through the IMG program on the Joint Genome Institute website (http://www.hmpdacc-resources.org/cgi-bin/imgm_hmp/main.cgi). The analyses were done using the COG functions; COG3961 (for PPDC), COG4231 and COG1014 (for PPFOR) and COG0674, COG1013, and COG1014 (for VOR). All the identified hits were then manually inspected for the gene neighborhood. For PPFOR homologues, only those hits which contained COG4231 and COG1014 in the same cluster were retained. Similarly, for the VOR homologues, the candidate hits were only retained if COG1013, COG1014, and COG0674 were found in the same gene cluster. We then used Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) to perform multiple sequence alignment (with default settings) for each group and the percent identity for each hit to the corresponding subject (P. mirabilis HMPREF0693_2975 for PPDC, B. thetaiotaomicron BT0430/0429 for PPFOR α and β subunits and C. sporogenes ClOSPO_00147/00148/00149 for VOR α, β, and γ subunits) was reported.
Human fecal and mouse cecal lysate PAA generation rate
To reduce enzyme inactivation by air, human fecal and mouse ceca samples were processed in the anaerobic chamber and anaerobic assays were performed first. Human fecal slurry and mouse ceca slurry were sonicated (level 7; 5 times for 10 s each) on ice with a probe sonicator (Branson Sonifier 250) inside the anaerobic chamber. The cell debris were removed by centrifugation (3000 × g, 10 min) and the supernatants were used for the assay. To test the oxidative PPY mediated PAA production, enzymatic assay was carried out in 100 mM Glycine-NaOH buffer (pH 10.5) containing 20 μl of cell lysate supernatant, 0.1 mM MgSO4, 0.1 mM TPP, 1 mM CoA, and 1 mM MV as the artificial electron acceptor in a final volume of 100 μl. To test for the non-oxidative PPY mediated PAA production, enzymatic assay was carried out in 100 mM MES buffer (pH 6.8) containing 20 μl of cell lysate supernatant, 0.1 mM MgSO4, and 0.1 mM TPP at a final volume of 100 μl. The reaction was initiated by the addition of [13C9,15N]-Phe at 100 μM final concentration and incubated at 37 °C anaerobically for 1 hour. A parallel experiment was done under aerobic condition. The product, [13C8]-PAA was quantified using LC-MS/MS.
Chromosomal insertion of PPDC and PPFOR genes in E. coli MP1
The codon-optimized ppdc and ppfor genes (corresponding to P. mirabilis HMPREF0693_2975 and B. thetaiotaomicron BT0430/Bt0429) were cloned from the synthesized pET16b expression vectors, and ligated to a suicide plasmid with upstream and downstream homology arms designed to place the inserted genes between K427_10530 and K427_10525 in E. coli MP1 chromosome. Both genetic constructs were constitutively expressed from J23100 promotor (http://parts.igem.org/Part:BBa_J23100). Likewise, T7 terminator was placed downstream of both constructs. The suicide plasmids were then transformed into E. coli S17 λpir. The recipient E. coli MP1 were transformed in advance with ampicillin resistant plasmid then conjugated with the donor E. coli S17 λpir and proceeded as described earlier in “Marker-exchange mutagenesis for Proteus mirabilis”. The final E. coli ppdc+ and E. coli ppfor+ mutants were then purified, sequence verified, and cured from the original ampicillin resistant plasmid.
Metagenomic screening and association analyses of ppdc and ppfor gene abundance and ASCVD prevalence.
We used the publicly available metagnomics data from a previously published ASCVD case/control cohort by Jie et al50. In this cohort, all ASCVD subjects were ethnically Han Chinese, 40–80 years old, and had either stable angina, unstable angina, or acute myocardial infarction, with ASCVD diagnosis confirmed by ≥50% stenosis in one or more major coronary arteries on coronary angiography. Controls were comprised of subjects free of clinically evident ASCVD symptoms at the time of the medical examination.
Metagenomics data was downloaded using the esearch function, implemented in the E-utilities package. Individual metagenomics reads were quality filtered using nesoni package (https://github.com/Victorian-Bioinformatics-Consortium/nesoni). Quality filtered metgenome reads were digitally normalized using khmer package74. Normalized reads were assembled into contigs using megahit assembler75. Protein coding nucleotide sequences for ppdc (annotated as phenylpyruvate decarboxylase or indolepyruvate decarboxylase) and ppfor (annotated as phenylpyruvate:ferredoxin oxidoreductase or indolepyruvate:ferredoxin oxidoreductase) were downloaded from NCBI’s Nucleotide database using the esearch function with the following parameters: –db Nucleotide -query AND “bacteria(porgon:__txid2)” and efetch. Local blast databases were constructed for ppdc and ppfor genes using the makeblastdb function in blast+ package. High speed BLASTN was used to identify the ppfor and ppdc genes in the metagenome assemblies (contigs > 300bp)76. BLASTN output (i.e. xml) was parsed for best blast hit analysis using biopython with coverage and minimum % identity of 90% and 50% for ppfor and ppdc genes, respectively. For each sample, the quality filtered individual metagenome reads were mapped to the ppfor and ppdc homologues (constructed from metagenome assemblies) using vsearch77.
QUANTIFICATION AND STATISTICAL ANALYSIS
The Wilcoxon–rank sum test or Student’s t test for continuous variables were used to determine significant difference between two groups. Kruskal-Wallis rank sum test or ANOVA were used to compare three or more groups. Statistical details for each experiment are stated in the figure legends with number of samples (or animals) “n” shown within the figures. All key in vitro experiments were repeated multiple times to ensure data reproducibility. To test for the relationship between ppfor and ppdc gene abundance and ASCVD, odds ratio for binary ASCVD and corresponding 95% confidence interval were calculated using both univariable (unadjusted) and multivariable (adjusted) logistic regression models. Adjustments were made for age, sex, systolic blood pressure (SBP), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglycerides, and body mass index (BMI). Statistical analyses were performed using either RStudio-R version 4.1.3. (2022-03-10) (Vienna, Austria), or GraphPad Prism version 9.4.0. A p-value of <0.05 was considered statistically significant.
Supplementary Material
Table S1 (Excel file): Potenital porA (or related 2-oxoacid:ferredoxin oxidoreductase) homologues in the genomes of a panel of human commensal bacteria, related to Figure 1, and Figure S1.
The publicly available genomes of C. sporogenes ATCC 15579 and the other 21 cultured strains shown in Figure S1 were selected on the JGI-IMG/M website. C. sporogenes CLOSPO_00147 (PorA), was used as the reference sequence for a protein BLAST with the selected genomes. Potential PorA or related 2-oxoacid:ferredoxin oxidoreductase protein homologues (e-value < 1e-5) were identified in 9 of the 21 genomes. All identified homologues were manually inspected for gene neighborhood. The only homologues identified in C. intestinalis (COLINT_03157) and C. aerofaciens (COLAER_01233) were found in gene neighborhood which lacked the conserved functions and therefore, these two strains were considered PorA-negative.
Table S3 (Excel file): Gene cluster homologs for PPFOR, VOR, and PPDC in Human Microbiome Project (HMP) reference genomes, related to Figure 4.
A total 1585 microbial genomes were included in the analyses. For PPFOR, analyses were performed using COG4231 and COG1014, the COG functions assigned for B. thetaiotaomicron BT0430 and BT0429 (corresponding to PPFOR α and β subunits, respectively). For VOR, analyses were performed using COG0674, COG1013, and COG1014, the COG functions assigned for C. sporogenes Clospo_00147, Clospo_00148, and Clospo_00149, respectively (Corresponding to VOR α, β, and ɣ subunits). For PPDC, analyses were performed using COG3961, the COG function assigned for P. mirabilis HMPREF0693_2975.The gene cluster architecture was inspected manually for each hit.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Clostridium sporogenes | ATCC | ATCC 15579 |
| Bacteroides caccae | ATCC | ATCC 43185 |
| Bacteroides cellulosilyticus | DSMZ | DSM 14838 |
| Bacteroides ovatus | ATCC | ATCC 8483 |
| Bacteroides thetaiotaomicron | ATCC | VPI-5482 |
| Bacteroides uniformis | ATCC | ATCC 8492 |
| Alistipes indistinctus | DSMZ | YIT 12060 |
| Lachnoclostridium (Clostridium) asparagiforme | DSMZ | DSM 15981 |
| Hungatella (Clostridium) hathewayi | DSMZ | DSM 13479 |
| Collinsella aerofaciens | ATCC | ATCC 25986 |
| Akkermansia muciniphila | ATCC | ATCC BAA-835 |
| Collinsella intestinalis | DSMZ | DSM 13280 |
| Lachnoclostridium (Clostridium) symbiosum | ATCC | ATCC 14940 |
| Eubacterium rectale | ATCC | ATCC 33656 |
| Bifidobacterium dentium | ATCC | ATCC 27678 |
| Escherichia fergusonii | ATCC | ATCC 35469 |
| Klebsiella pneumoniae | BEI Resources | WGLW1 |
| Staphylococcus aureus | BEI Resources | HI022 |
| Proteus mirabilis | ATCC | ATCC 29906 |
| Acinetobacter baumannii | ATCC | ATCC 19606 |
| Escherichia coli S17-1 λ pir | Yale CGSC | S17–1 λ pir |
| Escherichia coli BL21 (DE3) | Yale CGSC | BL21 (DE3) |
| Escherichia coli MP1 | Dr. Mark Goulian at University of Pennsylvania | MP1 |
| Proteus mirabilisΔ HMPREF0693_2975 | This study | N/A |
| E. coli ppdc + | This study | N/A |
| E. coli ppfor + | This study | N/A |
| B. thetaiotaomicron Δtdk | Koropatkin et al., 200868 | N/A |
| B. thetaiotaomicron ΔtdkΔBT0331 | This study | N/A |
| B. thetaiotaomicron ΔtdkΔBT2826 | This study | N/A |
| B. thetaiotaomicron ΔtdkΔBT0430 | This study | N/A |
| B. thetaiotaomicron ΔtdkΔBT0430ΔBT0331ΔBT0430 | This study | N/A |
| Clostridium sporogenes ΔfldH | Dodd et al., 201719 | |
| Biological samples | ||
| Human fecal samples | Wilcox et al., 202162 | (NCT03039023; http://www.clinicaltrials.gov) |
| Chemicals | ||
| Natural ingredient rodent diet | Envigo | Cat# TD.130104 |
| Gluten-free Egg white (For murine NTZ challenge) | Judes | N/A |
| Methyl viologen (MV) | Sigma-Aldrich | Cat# 856177 |
| Phenylalanine (Phe) | Sigma-Aldrich | Cat# P5030 |
| Phenylpyruvic acid (PPY) | Sigma-Aldrich | Cat# 286958 |
| Thiamine pyrophosphate (TPP) | Sigma-Aldrich | Cat# C8754 |
| 5-Fluoro-2′-deoxyuridine | Sigma-Aldrich | Cat# SIAL-F0503 |
| Coenzyme A (CoA) sodium salt | Sigma-Aldrich | Cat# SIAL-C3144 |
| 2,4-dinitrophenylhydrazine (DNPH) | Sigma-Aldrich | Cat#D199303 |
| 2-Phenylpropionaldehyde | Sigma-Aldrich | Cat#241369 |
| Phenylacetaldehyde | Sigma-Aldrich | Cat#107395 |
| Phenylacetyl coenzyme A lithium salt | Sigma-Aldrich | Cat# P2153 |
| isopropyl-β-d-thiogalactopyranoside (IPTG) | Bio Basic | Cat# C9H18O5S |
| 13C2-Phenylacetic acid (13C2-PAA) | Chem Cruz | Cat# SC-236371 |
| D3-Creatinine | CDN Isotopes | Cat# D-3689 |
| p-Cresol sulfate potassium salt | Toronto Research Chemicals (TRC) | Cat# T536805 |
| D7-p-cresol sulfate potassium salt | Cambridge Isotopes Laboratories | Cat# DLM-9786–0.01 |
| D5-Phenylacetylglutamine (D5-PAgln) | CDN Isotopes | Cat# D-6900 |
| L-13C9 15N-Phenylalaine | Sigma-Aldrich | Cat# 608017 |
| L-13C6-Phenylalaine | Cambridge isotope | Cat# CLM-1055-PK |
| Phenylacetylglycine (PAGly) | Bachem | Cat# 4016439.0001 |
| Phenylacetylglutamine (PAGln) | Santa Cruz | Cat# SC-212551A |
| Nicotinamide adenine dinucleotide sodium salt | Sigma-Aldrich | Cat# N0632 |
| Alcohol dehydrogenase from Saccharomyces cerevisiae | Sigma-Aldrich | Cat# A7011 |
| Nitazoxanide (NTZ) | Sigma-Aldrich | Cat# N0290 |
| Pyruvic acid | Sigma-Aldrich | Cat# 107360 |
| Indole-3-pyruvic acid (I3PY) | Sigma-Aldrich | Cat# I7017 |
| 4-hydroxy-phenylpyruvic Acid (4-OH-PPY) | Sigma-Aldrich | Cat# 114286 |
| 3-methyl-2-oxovaleric acid | Santa Cruz | Cat# sc-214140 |
| Benzoylformic acid | Sigma-Aldrich | Cat# B13055 |
| Ethyl 2-oxo-4-phenylbutanoate | Sigma-Aldrich | Cat# 375322 |
| 2-oxobutanoic acid | Sigma-Aldrich | Cat# K401 |
| Methyl 2-oxopropanoate | Sigma-Aldrich | Cat# 371173 |
| Ethyl thiophene-2-glyoxylate | Sigma-Aldrich | Cat# CDS001215 |
| 4-methylthio-2-oxobutanoic acid | Sigma-Aldrich | Cat# K6000 |
| Ethyl pyruvate | Sigma-Aldrich | Cat# W245712 |
| Brain Heart Infusion (BHI) powder | Millipore | Cat# VM929593 024 |
| Reinforced Clostridial Media (RCM) powder | Difco | Cat# 218081 |
| Luria Bertani broth (LB) powder | Difco | Cat# 244620 |
| Agar powder | Difco | Cat# 281230 |
| Hematin | ChemCruz | Cat# SSC-207729 |
| L-Histidine | TCI | Cat# H0149 |
| L-Cysteine | J.T.Baker | Cat# 2071-05 |
| Tryptone | MP | Cat# 1010817 |
| Yeast extract | Alfa Aesar | Cat# J60287 |
| Glycerol | Fisher Bio reagents | Cat# BP229-1 |
| Sodium chloride | Fisher Chemical | Cat# S271-500 |
| Sucrose | Sigma-Aldrich | Cat# S0389 |
| D-glucose | Acros Organics | Cat# 41095-0010 |
| Potassium Phosphate | MP | Cat# 191431 |
| Magnesium sulfate | Macron Fine Chemicals | Cat# 6070-12 |
| Sodium bicarbonate | J.T.Baker | Cat# 3509-01 |
| Calcium chloride | J.T.Baker | Cat# 1313-01 |
| Ferrous sulfate heptahydrate | MP | Cat# 194663 |
| Tween 80 | Fisher Chemical | Cat# T164-500 |
| Sodium Acetate trihydrate | Fisher Scientific | Cat# S220-1 |
| Meat Extract | Sigma-Aldrich | Cat# 70164 |
| ATCC Vitamin Mix | ATCC | Cat# MD-VS |
| ATCC Trace Mineral Mix | ATCC | Cat# MD-TMS |
| chow diet | LabDiet | Cat# 5010 |
| Experimental models: Organisms/strains | ||
| Mouse: Swiss Webster female | Taconic | SW-F |
| Mouse: Germ Free mice (C57BL/6) | Raised in in the gnotobiotic facility at Cleveland Clinic foundation | N/A |
| Recombinant DNA | ||
| pExchange-tdk | Koropatkin et al., 200868 | N/A |
| pK18mobsacB | Schafer et al., 199473 | N/A |
| pExSacB-2975::Kan | This study | N/A |
| pKEx-MP-PPDC | This study | N/A |
| pKEx-MP-PPFOR | This study | N/A |
| pET16-PPDC | chemically synthesized by GenScript | N/A |
| pET16-PPFOR | chemically synthesized by GenScript | N/A |
| Critical commercial assays | ||
| Bradford Protein Assay | Bio-Rad | Cat# 5000201 |
| Software and Algorithms | ||
| MultiQuant | Sciex | https://sciex.com/products/software/multiquant-software |
| GraphPad Prism (version 9.4) | GraphPad | https://www.graphpad.com/ |
| Lab Solutions (version 5.89) | Shimadzu | https://www.shimadzu.com/an/products/software-informatics/labsolutions-series/index.html |
| Megahit (version 1.2.9) | Li et al., 201575 | https://github.com/voutcn/megahit |
| Vsearch (Version 2.21.1) | Rognes et al., 201677 | https://github.com/torognes/vsearch |
| Nesoni (Version 0.134) | Paul Harrison | https://github.com/Victorian-Bioinformatics-Consortium/nesoni |
| Khmer (Version 2.1.2) | Crusoe et al., 201574 | https://github.com/dib-lab/khmer |
| E_utils (Version 0.6.0) | https://github.com/biocommons/eutils | |
| R (version 4.1.3) | RStudio | https://www.rstudio.com/ |
| Human Microbiome Project (HMP) genomes in The Integrated Microbial Genomes and Metagenomes (IMG/M-HMP) | The Joint Genome Institute | http://www.hmpdacc-resources.org/imgm_hmp/. |
| Clustal Omega for Multiple sequence alignment | Sievers et al., 201169 | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Deposited data | ||
| Metagenomics data (For data in Figure 6) | Jie et al., 201750 | European Bioinformatics Institute (EBI) accession code ERP023788 |
| Pyruvate decarboxylase isozyme 1 (PDC1) from S. cerevisiae | Vuralhan et al., 200341 | NCBI Accession: CAA97573.1 |
| Pyruvate decarboxylase isozyme 2 (PDC5) from S. cerevisiae | Vuralhan et al., 200341 | NCBI Accession: CAA97705.1 |
| pyruvate decarboxylase (PDC) from Zymomonas mobilis | Zverlov et al., 198970 | NCBI Accession: AHJ73198.1 |
| Branched-chain keto acid decarboxylase from Lactococcus lactis (KDC) | Smit et al., 200571 | NCBI Accession: AY548760.1 |
| Indolepyruvate decarboxylase (IPDC) from Enterobacter cloacae | Schutz et al., 200372 | NCBI Accession: WP_013098183 |
| Others | ||
| In-Fusion® HD Cloning kit | Takara | Cat# 638910 |
Highlights:
PPFOR is the main contributor for PAA production in Bacteroides thetaiotaomicron.
PPDC plays an important role in PAA generation in Proteus mirabilis.
PPFOR and PPDC contribute to PAA production and PAGln formation in colonized hosts
Microbial ppdc and ppfor homologue abundances are each associated with ASCVD
Acknowledgments
This work is supported by grants from the NIH and Office of Dietary Supplements (P01HL147823, R01HL103866) and the Foundation Leducq. M.D. reports partial support from NIH grant R21CA267711. I.N. reports partial support from NIH grant R01HL160747. Mass spectrometry studies were performed on instrumentation housed in a facility supported in part through a Shimadzu Center of Excellence award and NIH shared instrumentation grant 1S10OD016346. The authors are also thankful for the Proteomics and Metabolomics core in the Lerner Research Institute for their assistance with the proteomics analysis.
Declaration of Interests
S.L.H., Z.W. and J.T.A. report being named as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and/or therapeutics, and being eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland HeartLab, a wholly owned subsidiary of Quest Diagnostics, P&G, and Zehna Therapeutics. S.L.H. and J.T.A. report being paid consultants for Zehna Therapeutics. S.L.H also reports having received research funds from P&G, Roche Diagnostics, and Zehna Therapeutics. The other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information
Supplementary file 1: Figures S1–S7, Table S2, and Table S4
References
- 1.Zmora N, Soffer E, and Elinav E (2019). Transforming medicine with the microbiome. Sci Transl Med 11. 10.1126/scitranslmed.aaw1815. [DOI] [PubMed] [Google Scholar]
- 2.Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, Vanamala J, Dorrestein PC, Turnbaugh PJ, and Knight R (2014). The intestinal metabolome: an intersection between microbiota and host. Gastroenterology 146, 1470–1476. 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donia MS, and Fischbach MA (2015). HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 349, 1254766. 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witkowski M, Weeks TL, and Hazen SL (2020). Gut Microbiota and Cardiovascular Disease. Circ Res 127, 553–570. 10.1161/CIRCRESAHA.120.316242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JM, and Hazen SL (2017). Targeting of microbe-derived metabolites to improve human health: The next frontier for drug discovery. J Biol Chem 292, 8560–8568. 10.1074/jbc.R116.765388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, Cajka T, Mohan ML, Li L, Wu Y, et al. (2020). A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 180, 862–877.e822. 10.1016/j.cell.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu F, Feng X, Li X, Luo Y, Wei M, Zhao T, and Xia J (2021). Gut-Derived Metabolite Phenylacetylglutamine and White Matter Hyperintensities in Patients With Acute Ischemic Stroke. Front Aging Neurosci 13, 675158. 10.3389/fnagi.2021.675158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu F, Li X, Feng X, Wei M, Luo Y, Zhao T, Xiao B, and Xia J (2021). Phenylacetylglutamine, a Novel Biomarker in Acute Ischemic Stroke. Front Cardiovasc Med 8, 798765. 10.3389/fcvm.2021.798765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asano Y, Nakazawa A, Endo K, Hibino Y, Ohmori M, Numao N, and Kondo K (1987). Phenylalanine dehydrogenase of Bacillus badius. Purification, characterization and gene cloning. Eur J Biochem 168, 153–159. 10.1111/j.1432-1033.1987.tb13399.x. [DOI] [PubMed] [Google Scholar]
- 10.Mavrides C, and Orr W (1975). Multispecific aspartate and aromatic amino acid aminotransferases in Escherichia coli. J Biol Chem 250, 4128–4133. [PubMed] [Google Scholar]
- 11.Delaney J, Neville WA, Swain A, Miles A, Leonard MS, and Waterfield CJ (2004). Phenylacetylglycine, a putative biomarker of phospholipidosis: its origins and relevance to phospholipid accumulation using amiodarone treated rats as a model. Biomarkers 9, 271–290. 10.1080/13547500400018570. [DOI] [PubMed] [Google Scholar]
- 12.Karoum F, Chuang LW, Mosnaim AD, Staub RA, and Wyatt RJ (1983). Plasma and cerebrospinal fluid concentration of phenylacetic acid in humans and monkeys. J Chromatogr Sci 21, 546–550. 10.1093/chromsci/21.12.546. [DOI] [PubMed] [Google Scholar]
- 13.Smith EA, and Macfarlane GT (1997). Formation of Phenolic and Indolic Compounds by Anaerobic Bacteria in the Human Large Intestine. Microb Ecol 33, 180–188. 10.1007/s002489900020. [DOI] [PubMed] [Google Scholar]
- 14.Smith EA, and Macfarlane GT (1996). Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol 81, 288–302. 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- 15.Scott TW, Ward PF, and Dawson RM (1964). The formation and metabolism of phenyl-substituted fatty acids in the ruminant. Biochem J 90, 12–24. 10.1042/bj0900012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaczmarek FS, and Coykendall AL (1980). Production of phenylacetic acid by strains of Bacteroides asaccharolyticus and Bacteroides gingivalis (sp. nov.). J Clin Microbiol 12, 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayrand D (1979). Identification of clinical isolates of selected species of Bacteroides: production of phenylacetic acid. Can J Microbiol 25, 927–928. 10.1139/m79-138. [DOI] [PubMed] [Google Scholar]
- 18.Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, and Flint HJ (2013). Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res 57, 523–535. 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- 19.Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, and Sonnenburg JL (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhuiyan MS, Ellett F, Murray GL, Kostoulias X, Cerqueira GM, Schulze KE, Mahamad Maifiah MH, Li J, Creek DJ, Lieschke GJ, and Peleg AY (2016). Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci U S A 113, 9599–9604. 10.1073/pnas.1523116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erdmann GR, and Khalil SKW (1985). Quantitative analysis of phenylacetic acid in proteus mirabilis cultures by high performance liquid chromatography. Journal of Liquid Chromatography 8, 1507–1517. 10.1080/01483918508067161. [DOI] [Google Scholar]
- 22.Heider J, Mai X, and Adams MW (1996). Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J Bacteriol 178, 780–787. 10.1128/jb.178.3.780-787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, and Lima CD (2006). Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem 281, 11028–11038. 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 24.Mai X, and Adams MW (1996). Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol 178, 5890–5896. 10.1128/jb.178.20.5890-5896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kletzin A, and Adams MW (1996). Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol 178, 248–257. 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo CJ, Allen BM, Hiam KJ, Dodd D, Van Treuren W, Higginbottom S, Nagashima K, Fischer CR, Sonnenburg JL, Spitzer MH, and Fischbach MA (2019). Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science 366. 10.1126/science.aav1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai X, and Adams MW (1994). Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J Biol Chem 269, 16726–16732. [PubMed] [Google Scholar]
- 28.Ozawa Y, Siddiqui MA, Takahashi Y, Urushiyama A, Ohmori D, Yamakura F, Arisaka F, and Imai T (2012). Indolepyruvate ferredoxin oxidoreductase: An oxygen-sensitive iron-sulfur enzyme from the hyperthermophilic archaeon Thermococcus profundus. J Biosci Bioeng 114, 23–27. 10.1016/j.jbiosc.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Tersteegen A, Linder D, Thauer RK, and Hedderich R (1997). Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur J Biochem 244, 862–868. 10.1111/j.1432-1033.1997.00862.x. [DOI] [PubMed] [Google Scholar]
- 30.Mimee M, Tucker AC, Voigt CA, and Lu TK (2015). Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst 1, 62–71. 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim B, Zimmermann M, Barry NA, and Goodman AL (2017). Engineered Regulatory Systems Modulate Gene Expression of Human Commensals in the Gut. Cell 169, 547–558 e515. 10.1016/j.cell.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson MI, Chen PY, and Drennan CL (2016). A structural phylogeny for understanding 2-oxoacid oxidoreductase function. Curr Opin Struct Biol 41, 54–61. 10.1016/j.sbi.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morey AV, and Juni E (1968). Studies on the nature of the binding of thiamine pyrophosphate to enzymes. J Biol Chem 243, 3009–3019. [PubMed] [Google Scholar]
- 34.Theodos CM, Griffiths JK, D’Onfro J, Fairfield A, and Tzipori S (1998). Efficacy of nitazoxanide against Cryptosporidium parvum in cell culture and in animal models. Antimicrob Agents Chemother 42, 1959–1965. 10.1128/AAC.42.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moron-Soto M, Gutierrez L, Sumano H, Tapia G, and Alcala-Canto Y (2017). Efficacy of nitazoxanide to treat natural Giardia infections in dogs. Parasit Vectors 10, 52. 10.1186/s13071-017-1998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman PS, Sisson G, Croxen MA, Welch K, Harman WD, Cremades N, and Morash MG (2007). Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob Agents Chemother 51, 868–876. 10.1128/AAC.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren CA, van Opstal E, Ballard TE, Kennedy A, Wang X, Riggins M, Olekhnovich I, Warthan M, Kolling GL, Guerrant RL, et al. (2012). Amixicile, a novel inhibitor of pyruvate: ferredoxin oxidoreductase, shows efficacy against Clostridium difficile in a mouse infection model. Antimicrob Agents Chemother 56, 4103–4111. 10.1128/AAC.00360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito Y, Sato T, Nomoto K, and Tsuji H (2018). Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol 94. 10.1093/femsec/fiy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rong Y, and Kiang TKL (2021). Characterization of human sulfotransferases catalyzing the formation of p-cresol sulfate and identification of mefenamic acid as a potent metabolism inhibitor and potential therapeutic agent for detoxification. Toxicol Appl Pharmacol 425, 115553. 10.1016/j.taap.2021.115553. [DOI] [PubMed] [Google Scholar]
- 40.Somers E, Ptacek D, Gysegom P, Srinivasan M, and Vanderleyden J (2005). Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indole-3-acetic acid biosynthesis. Appl Environ Microbiol 71, 1803–1810. 10.1128/AEM.71.4.1803-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuralhan Z, Morais MA, Tai SL, Piper MD, and Pronk JT (2003). Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl Environ Microbiol 69, 4534–4541. 10.1128/aem.69.8.4534-4541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debnar-Daumler C, Seubert A, Schmitt G, and Heider J (2014). Simultaneous involvement of a tungsten-containing aldehyde:ferredoxin oxidoreductase and a phenylacetaldehyde dehydrogenase in anaerobic phenylalanine metabolism. J Bacteriol 196, 483–492. 10.1128/JB.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrandez A, Prieto MA, Garcia JL, and Diaz E (1997). Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett 406, 23–27. 10.1016/s0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 44.Costelloe SJ, Ward JM, and Dalby PA (2008). Evolutionary analysis of the TPP-dependent enzyme family. J Mol Evol 66, 36–49. 10.1007/s00239-007-9056-2. [DOI] [PubMed] [Google Scholar]
- 45.Schenk G, Leeper FJ, England R, Nixon PF, and Duggleby RG (1997). The role of His113 and His114 in pyruvate decarboxylase from Zymomonas mobilis. Eur J Biochem 248, 63–71. 10.1111/j.1432-1033.1997.t01-1-00063.x. [DOI] [PubMed] [Google Scholar]
- 46.Robinson BH, and Chun K (1993). The relationships between transketolase, yeast pyruvate decarboxylase and pyruvate dehydrogenase of the pyruvate dehydrogenase complex. FEBS Lett 328, 99–102. 10.1016/0014-5793(93)80973-x. [DOI] [PubMed] [Google Scholar]
- 47.Kneen MM, Stan R, Yep A, Tyler RP, Saehuan C, and McLeish MJ (2011). Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS J 278, 1842–1853. 10.1111/j.1742-4658.2011.08103.x. [DOI] [PubMed] [Google Scholar]
- 48.Howery KE, and Rather PN (2019). Allelic Exchange Mutagenesis in Proteus mirabilis. Methods Mol Biol 2021, 77–84. 10.1007/978-1-4939-9601-8_8. [DOI] [PubMed] [Google Scholar]
- 49.Lasaro M, Liu Z, Bishar R, Kelly K, Chattopadhyay S, Paul S, Sokurenko E, Zhu J, and Goulian M (2014). Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J Bacteriol 196, 1723–1732. 10.1128/JB.01296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, et al. (2017). The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8, 845. 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo DJ, and Jordan F (1983). Active site directed irreversible inactivation of brewers’ yeast pyruvate decarboxylase by the conjugated substrate analogue (E)-4-(4-chlorophenyl)-2-oxo-3-butenoic acid: development of a suicide substrate. Biochemistry 22, 3735–3740. 10.1021/bi00285a003. [DOI] [PubMed] [Google Scholar]
- 52.Moldave K, and Meister A (1957). Synthesis of phenylacetylglutamine by human tissue. J Biol Chem 229, 463–476. [PubMed] [Google Scholar]
- 53.McVay CS, and Rolfe RD (2000). In vitro and in vivo activities of nitazoxanide against Clostridium difficile. Antimicrob Agents Chemother 44, 2254–2258. 10.1128/AAC.44.9.2254-2258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selmer T, and Andrei PI (2001). p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur J Biochem 268, 1363–1372. 10.1046/j.1432-1327.2001.02001.x. [DOI] [PubMed] [Google Scholar]
- 55.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, et al. (2018). Function and functional redundancy in microbial systems. Nat Ecol Evol 2, 936–943. 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 56.Moya A, and Ferrer M (2016). Functional Redundancy-Induced Stability of Gut Microbiota Subjected to Disturbance. Trends Microbiol 24, 402–413. 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Romano KA, Vivas EI, Amador-Noguez D, and Rey FE (2015). Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 6, e02481. 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachmanov AA, Reed DR, Beauchamp GK, and Tordoff MG (2002). Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet 32, 435–443. 10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li CH, Lu ZR, Zhao ZD, Wang XY, Leng HJ, Niu Y, and Wang MP (2021). Nitazoxanide, an Antiprotozoal Drug, Reduces Bone Loss in Ovariectomized Mice by Inhibition of RANKL-Induced Osteoclastogenesis. Front Pharmacol 12, 781640. 10.3389/fphar.2021.781640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Kowrany SI, El Ghaffar AEA, Shoheib ZS, Mady RF, and Gamea GAM (2019). Evaluation of nitazoxanide as a novel drug for the treatment of acute and chronic toxoplasmosis. Acta Trop 195, 145–154. 10.1016/j.actatropica.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 61.Blagburn BL, Drain KL, Land TM, Kinard RG, Moore PH, Lindsay DS, Patrick DA, Boykin DW, and Tidwell RR (1998). Comparative efficacy evaluation of dicationic carbazole compounds, nitazoxanide, and paromomycin against Cryptosporidium parvum infections in a neonatal mouse model. Antimicrob Agents Chemother 42, 2877–2882. 10.1128/AAC.42.11.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilcox J, Skye SM, Graham B, Zabell A, Li XS, Li L, Shelkay S, Fu X, Neale S, O’Laughlin C, et al. (2021). Dietary Choline Supplements, but Not Eggs, Raise Fasting TMAO Levels in Participants with Normal Renal Function: A Randomized Clinical Trial. Am J Med 134, 1160–1169 e1163. 10.1016/j.amjmed.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pei J, Kim BH, and Grishin NV (2008). PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36, 2295–2300. 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pei J, and Grishin NV (2014). PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol Biol 1079, 263–271. 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talavera G, and Castresana J (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56, 564–577. 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 66.Castresana J (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17, 540–552. 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, and Tamura K (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koropatkin NM, Martens EC, Gordon JI, and Smith TJ (2008). Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115. 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zverlov VV, Bankovskii VK, Churikova OV, Mogutov MA, and Iur’ev MZ (1989). [Cloning the pyruvate decarboxylase gene of Zymomonas mobilis and its expression in Escherichia coli]. Mol Gen Mikrobiol Virusol, 11–13. [PubMed] [Google Scholar]
- 71.Smit BA, van Hylckama Vlieg JE, Engels WJ, Meijer L, Wouters JT, and Smit G (2005). Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation. Appl Environ Microbiol 71, 303–311. 10.1128/AEM.71.1.303-311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schutz A, Sandalova T, Ricagno S, Hubner G, Konig S, and Schneider G (2003). Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid. Eur J Biochem 270, 2312–2321. 10.1046/j.1432-1033.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 73.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, and Puhler A (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 74.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S, et al. (2015). The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res 4, 900. 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li D, Liu CM, Luo R, Sadakane K, and Lam TW (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, Ye W, Zhang Y, and Xu Y (2015). High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res 43, 7762–7768. 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rognes T, Flouri T, Nichols B, Quince C, and Mahé F (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Excel file): Potenital porA (or related 2-oxoacid:ferredoxin oxidoreductase) homologues in the genomes of a panel of human commensal bacteria, related to Figure 1, and Figure S1.
The publicly available genomes of C. sporogenes ATCC 15579 and the other 21 cultured strains shown in Figure S1 were selected on the JGI-IMG/M website. C. sporogenes CLOSPO_00147 (PorA), was used as the reference sequence for a protein BLAST with the selected genomes. Potential PorA or related 2-oxoacid:ferredoxin oxidoreductase protein homologues (e-value < 1e-5) were identified in 9 of the 21 genomes. All identified homologues were manually inspected for gene neighborhood. The only homologues identified in C. intestinalis (COLINT_03157) and C. aerofaciens (COLAER_01233) were found in gene neighborhood which lacked the conserved functions and therefore, these two strains were considered PorA-negative.
Table S3 (Excel file): Gene cluster homologs for PPFOR, VOR, and PPDC in Human Microbiome Project (HMP) reference genomes, related to Figure 4.
A total 1585 microbial genomes were included in the analyses. For PPFOR, analyses were performed using COG4231 and COG1014, the COG functions assigned for B. thetaiotaomicron BT0430 and BT0429 (corresponding to PPFOR α and β subunits, respectively). For VOR, analyses were performed using COG0674, COG1013, and COG1014, the COG functions assigned for C. sporogenes Clospo_00147, Clospo_00148, and Clospo_00149, respectively (Corresponding to VOR α, β, and ɣ subunits). For PPDC, analyses were performed using COG3961, the COG function assigned for P. mirabilis HMPREF0693_2975.The gene cluster architecture was inspected manually for each hit.
Data Availability Statement
For Figure 6, data were obtained through reanalysis of Jie et al., 2017 study datasets (European Bioinformatics Institute (EBI) accession number ERP023788)50.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


