Abstract
Background
The nonproliferating polyaneuploid cancer cell (PACC) state is associated with therapeutic resistance in cancer. A subset of cancer cells enters the PACC state by polyploidization and acts as cancer stem cells by undergoing depolyploidization and repopulating the tumor cell population after the therapeutic stress is relieved. Our aim was to systematically assess the presence and importance of this entity in men who underwent radical prostatectomy with curative intent to treat their presumed localized prostate cancer (PCa).
Materials and Methods
Men with National Comprehensive Cancer Network intermediate‐ or high‐risk PCa who underwent radical prostatectomy l from 2007 to 2015 and who did not receive neoadjuvant treatment were included. From the cohort of 2159 patients, the analysis focused on a subcohort of 209 patients and 38 cases. Prostate tissue microarrays (TMAs) were prepared from formalin‐fixed, paraffin‐embedded blocks of the radical prostatectomy specimens. A total of 2807 tissue samples of matched normal/benign and cancer were arrayed in nine TMA blocks. The presence of PACCs and the number of PACCs on each core were noted.
Results
The total number of cells in the PACC state and the total number of cores with PACCs were significantly correlated with increasing Gleason score (p = 0.0004) and increasing Cancer of the Prostate Risk Assessment Postsurgical (CAPRA‐S) (p = 0.004), but no other variables. In univariate proportional hazards models of metastasis‐free survival, year of surgery, Gleason score (9–10 vs. 7–8), pathology stage, CAPRA‐S, total PACCs, and cores positive for PACCs were all statistically significant. The multivariable models with PACCs that gave the best fit included CAPRA‐S. Adding either total PACCs or cores positive for PACCs to CAPRA‐S both significantly improved model fit compared to CAPRA‐S alone.
Conclusion
Our findings show that the number of PACCs and the number of cores positive for PACCs are statistically significant prognostic factors for metastasis‐free survival, after adjusting for CAPRA‐S, in a case–cohort of intermediate‐ or high‐risk men who underwent radical prostatectomy. In addition, despite the small number of men with complete data to evaluate time to metastatic castration‐resistant PCa (mCRPC), the total number of PACCs was a statistically significant predictor of mCRPC in univariate analysis and suggested a prognostic effect even after adjusting for CAPRA‐S.
Keywords: PACC, PGCC, polyaneuploid cancer cell, polyploid giant cancer cell, prostate cancer
1. INTRODUCTION
Therapeutic resistance in cancer is generally attributed to the existence of resistant cell clones. 1 , 2 , 3 The resistant clones are thought to be generated through either intrinsic genetic instability resulting in tumor cell heterogeneity (TCH) or a cancer stem cell (CSC) population. 4 , 5 , 6 , 7 The resistant clones allow for regrowth of the cancer cell population after treatment insult. There is a growing body of evidence that the phenomenon of therapeutic resistance may be explained by a poorly recognized but distinct cell state. This cell state is documented as the nonproliferating polyaneuploid cancer cell (PACC) state, induced by tumor microenvironmental (intrinsic) or therapeutic (extrinsic) stress. 8 , 9 , 10 A doubling of a cancer cell's aneuploid genome combined with an exit from the cell cycle enables the PACC state. The cell state can exist for an extended period of time. In response to stress, a subset of cancer cells enters the PACC state by accessing an evolutionary or developmental polyploidization program. Once that stress is relieved, cells in the PACC state can act as CSCs by undergoing depolyploidization and repopulating the tumor cell population. The role of the PACC state for therapeutic resistance adds to those of CSC and TCH. Identifying cells in the PACC state in patients may have important diagnostic, prognostic, and therapeutic implications.
Large pleomorphic cancer cells with irregular nuclei have been documented in histopathologic specimens of multiple tumor types since the 1800s. 11 These cells have been reported utilizing a variety of names: polyaneuploid cancer cells (PACCs), polyploid giant cancer cells (PGCCs), giant cancer cells (GCCs), multinucleated GCCs, blastomere‐like cancer cells, osteoclast‐like cancer cells, cells in an embryonic diapause, pleomorphic giant cells, large stem cells, and persister cells. 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 Recent data indicate that these cells do not represent a different type of cancer cell within the heterogeneous tumor cell population, but rather a specific nonproliferative, polyaneuploid cell state. 8 , 9 , 10 In response to therapeutic stress, this cell state enables survival and is responsible for driving therapeutic resistance to nearly all available treatment regimens.
The presence of cells in the PACC state has been documented in virtually all cancer types, including adenocarcinomas, transitional cell tumors, squamous cell carcinomas, leukemias, lymphomas, glioblastomas, and sarcomas. 12 , 24 , 25 , 26 , 27 , 28 , 29 Most often observed in metastatic cancers or after treatment, their importance for patient prognosis has been understudied. 30 , 31 In glioma, Qu et al. 32 analyzed 76 patients and reported that the number of PACCs increased with the grade of tumors. In a study of 47 patients with anorectal melanoma, the number of PACCs was demonstrated to increase with tumor size. 33 In laryngeal cancer, Liu et al. 34 analyzed the presence of PGCCs in 102 patients and found that patients with high expression of PGCCs had a poorer prognosis. In breast cancer, Fei et al. 30 analyzed 167 histopathologic specimens, including benign tissue, primary breast tumors, and lymph node metastases, and found the highest number of PGCCs in the lymph node metastases of breast cancer patients. In a study of 30 patients, Gerashchenko et al. 35 reported that breast tumors with a higher proportion of polyploid cells were a marker of poor response to neoadjuvant chemotherapy. Lv et al. 36 investigated the presence of PGCCs with budding in 80 patients with serous ovarian tumors and found that the presence of PGCCs in the primary tumor correlated with metastasis. Zhang et al. 37 examined tissue from 159 patients with colorectal cancer and demonstrated that the presence of PGCCs with budding increased as tumors became more dedifferentiated.
The presence of cancer cells in the PACC state has been reported in prostate cancer (PCa). 12 , 38 In an autopsy study of PCa patients who had failed multiple lines of therapy, Mannan et al. 39 reported the presence of multiple cells with highly irregular polylobulated nuclei or multiple pleomorphic nuclei. Alharbi et al. 40 reported a series of 30 patients with a rare variant of PCa with focal pleomorphic giant cell features that were extremely aggressive and associated with poor outcomes. This study was undertaken to systematically assess the presence and importance of cells in the PACC state in men who underwent radical prostatectomy with curative intent to treat their presumed localized PCa.
2. MATERIALS AND METHODS
2.1. Patients
Men with National Comprehensive Cancer Network intermediate‐ or high‐risk PCa who underwent radical prostatectomy at Johns Hopkins Hospital from 2007 to 2015 and who did not receive neoadjuvant treatment were identified from the Institutional Review Board‐approved Brady Urological Institute Radical Prostatectomy database. Intermediate risk was defined as clinical stage T2b–T2c, or biopsy Gleason grade groups 2–3, or prostate‐specific antigen (PSA) 10–20 ng/ml, and high risk was defined as biopsy Gleason grade groups 4–5 or clinical stage T3, or PSA > 20 ng/ml. 41 There were 3685 men with NCCN intermediate‐ or high‐risk PCa with radical prostatectomy from 2007 to 2015 who did not receive neoadjuvant treatment. Of those, 2159 (59%) had complete follow‐up for metastasis through 2015 and represented the pool from which the case–cohort sample was drawn.
2.2. Case–cohort
Case–cohort was originally assembled to include as “cases” men with metastasis or with biochemical recurrence (BCR) with a rapid PSA doubling time (<10 months) and who were also considered to be at high risk of metastasis. 42 From the cohort of 2159 patients, a subcohort of 244 patients was selected, and 115 cases (73 with BCR and rapid PSA doubling time, and 42 with metastasis). Tissue samples from these men were analyzed for PACC; samples from 307 men were informative for PACC. To focus specifically on the risk of metastasis as the outcome of interest, we excluded 65 “men with BCR and rapid PSA doubling time but without metastasis who were included as “cases” in the original case–cohort. This resulted in a subcohort of 209 patients (including eight metastasis cases), and 30 metastasis cases not in the subcohort, all of whom were informative for PACCs.
2.3. Tissue microarrays (TMAs)
Prostate TMAs were prepared from formalin‐fixed paraffin‐embedded blocks of the radical prostatectomy specimens. A total of 2807 tissue samples of matched normal/benign and cancer were arrayed in nine TMA blocks. These TMAs were all constructed as described 42 , 43 , 44 , 45 from the index tumor (highest grade) with a 3–4‐fold sampling redundancy. On hematoxylin and eosin (H&E)‐stained tissue sections and TMA sections, identifying PACCs is difficult since they often have indistinct cell membranes, thus making it difficult to distinguish “pseudo” multinucleation from the real ones. Also, unlike many other types of adenocarcinoma and poorly differentiated carcinomas (e.g., high‐grade urothelial carcinomas, nonsmall cell lung carcinomas), easily recognizable multinucleated or bizarrely enlarged nuclei are not readily apparent in the vast majority of even very high‐grade cases. Therefore, we used immunohistochemistry against EpCAM (mouse monoclonal antibody; ab7504; Abcam) on these TMAs to visualize the epithelial cell membranes. All TMA slides were scanned on a Hamamatsu Nanozoomer and imported into Concentriq (from Proscia). The whole slide scan files were evaluated by two pathologists (L. T. and A. M. D. M.). PACCs were defined as large multinucleated or polylobated cells that are at least three times the size of a neighboring tumor cell as assessed by visual inspection. 39 The presence of PACCs and the number of PACCs on each core were noted.
2.4. Statistical analyses
Descriptive statistics were used to compare cases and subcohort, including Wilcoxon's rank‐sum test for continuous variables and Fisher's exact test or χ 2 test for categorical variables. Correlations between PACCs and clinical variables were performed with linear regression or analysis of variance. The primary outcome was metastasis confirmed by imaging, and metastasis‐free survival (MFS) was measured from the date of radical prostatectomy. Multivariable Cox proportional hazards regression models, with weights for the case–cohort design and robust variance estimator defined by Barlow 46 were fit to MFS to evaluate the hazard ratio (HR) and 95% confidence interval (CI) associated with PACCs, adjusted for established prognostic factors or Cancer of the Prostate Risk Assessment Postsurgical (CAPRA‐S) score. The CAPRA‐S score combines the pathologic Gleason score, pathologic stage, surgical margin status, and preoperative PSA in an algorithm with values ranging from 0 to 12; scores of 6 or higher are considered to indicate a high risk of BCR. 47 Improvement in multivariable model fit for the addition of PACCs was assessed with the pseudo‐likelihood ratio test for the change in deviance from the full versus the reduced model. 48 In addition to the primary outcome of MFS, follow‐up for the development of metastatic castration‐resistant PCa (mCRPC) was available for 33 of the men with metastases. Time from diagnosis of metastasis to mCRPC was analyzed using standard proportional hazards regression, and improvement in model fit was assessed with the likelihood ratio test. All analyses were performed with SAS v. 9.4 (SAS Institute).
3. RESULTS
Since the great majority of prostatic adenocarcinomas do not frequently show bizarre nuclear atypia with extremely large nuclei or multinucleation that is readily apparent by H&E staining, we performed IHC against EpCAM to facilitate the recognition of cellular plasma membranes. This greatly helped in the ability to confidently identify PACCs (Figure 1 and Supporting Information: Figure 1), which in this study were limited to cells with multiple nuclei bounded by a single plasma membrane. Table 1 compares the metastasis cases to men without metastasis. Cases had the expected higher risk profile, differing significantly for all variables except age. Although many patient tumor samples had none, PACCs were significantly more frequent in cases with metastasis, 20 of 38 cases (52.6%), versus 68 of 201 (33.8%) controls, p = 0.029.
Figure 1.
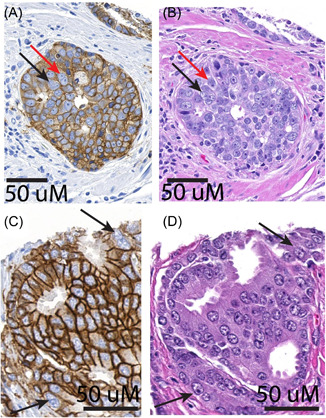
TMA spot stained with EpCAM to distinguish cells in the PACC state. TMA was stained with EpCAM (A, C) and H&E (B, D) on adjacent slices. Arrow indicates a cell in the PACC state, determined from the EpCAM stain as a cell with two nuclei or a lobulated nucleus within a single plasma membrane. The H&E in the same region does not adequately distinguish between cell–cell boundaries; thus, it is difficult to discern if there is multinucleation. H&E, hematoxylin and eosin; PACC, polyaneuploid cancer cell; TMA, tissue microarray. [Color figure can be viewed at wileyonlinelibrary.com]
Table 1.
Characteristics of patients with or without metastases (n = 239)
| Variables | Metastases (n = 38) | No metastases (n = 201) | p Value |
|---|---|---|---|
| Age, median (IQR) | 59 (56–63) | 59 (54–65) | 0.747 |
| PSA, median (IQR) | 7.0 (5.3–8.7) | 5.5 (4.3–8.3) | 0.028 |
| Race, n (%) | |||
| White | 32 (16.8) | 159 (83.3) | 0.167 |
| Black | 6 (20.7) | 23 (79.3) | |
| Other | 0 (0) | 16 (100.0) | |
| Prostatectomy year, median (IQR) | 2008 (2007–2009) | 2009 (2008–2011) | 0.014 |
| Prostatectomy Gleason score, n (%) | <0.0001 | ||
| 6 | 0 (0 | 43 (100.0 | |
| 3 + 4 | 1 (1.1) | 88 (98.9) | |
| 4 + 3 | 9 (17.3) | 43 (82.7) | |
| 8 | 6 (30.0) | 14 (70.0) | |
| 9–10 | 22 (68.8) | 10 (31.2) | |
| Pathology stage, n (%) | <0.0001 | ||
| Organ confined | 6 (4.8) | 118 (95.2) | |
| Extraprostatic extension | 10 (13.3) | 65 (86.7) | |
| Seminal vesicle involvement | 10 (45.5) | 12 (54.5) | |
| Lymph node involvement | 12 (85.7) | 2 (14.3) | |
| Surgical margins, n (%) | 0.002 | ||
| Negative | 23 (12.2) | 165 (87.8) | |
| Positive | 14 (30.4) | 32 (69.6) | |
| NCCN risk, n (%) | 0.0001 | ||
| Intermediate | 17 (9.6) | 160 (90.4) | |
| High | 15 (31.3) | 33 (68.8) | |
| Salvage treatment, n (%) | <0.0001 | ||
| None | 21 (11.0) | 170 (89.0) | |
| ADT only | 4 (66.7) | 2 (33.3) | |
| Radiation only | 8 (32.0) | 17 (68.0) | |
| ADT + radiation | 5 (35.7) | 9 (64.3) | |
| CAPRA‐S, median (IQR) | 7 (4–8) | 2 (1–4) | <0.0001 |
| Cores with PACCs, median (IQR) | 1 (0–2) | 0 (0–1) | 0.016 |
| Cores with PACCs, n (%)a | 0.029 | ||
| 0 | 18 (11.9) | 133 (88.1) | |
| ≥1 | 20 (22.7) | 68 (77.3) | |
| Total number of PACCs, median (IQR) | 1 (0–3) | 0 (0–1) | 0.011 |
| Total number of PACCs, n (%)a | 0.029 | ||
| 0 | 18 (11.9) | 133 (88.2) | |
| ≥1 | 20 (22.7) | 68 (77.3) |
Abbreviations: ADT, androgen deprivation therapy; CAPRA‐S, Cancer of the Prostate Risk Assessment Postsurgical; IQR, interquartile range; NCCN, National Comprehensive Cancer Network; PACCs, polyaneuploid cancer cells.
Binary classification of total PACCs and cores with PACCs gives the same distribution
The total number of PACCs and the total number of cores with PACCs were significantly correlated with increasing Gleason score (p = 0.0004) and increasing CAPRA‐S (p = 0.004), but no other variables.
In univariate proportional hazards models of MFS, year of surgery, Gleason score (9–10 vs. 7–8), pathology stage, CAPRA‐S, total PACCs, and cores positive for PACCs (both evaluated as a continuous variable or dichotomized at ≥1 vs. 0) were all statistically significant (Table 2). Note that dichotomizing total PACCs and cores positive for PACCs gave the same distribution so that the HRs, 95% CIs, and p values are the same.
Table 2.
Univariate proportional hazards models of metastasis‐free survival in a case–cohort of intermediate‐ and high‐risk men
| Variables | HR (95% CI) | p Value |
|---|---|---|
| Year of surgery | 1.50 1.12, 2.02) | 0.007 |
| Age | 0.99 (0.93, 1.05) | 0.671 |
| PSA (per 1 ng/ml) | 1.03 (0.996, 1.07) | 0.079 |
| Gleason score | <0.0001 | |
| 7–8 | 1.0 | |
| 9–10 | 13.48 (5.40, 33.65) | |
| Pathologic stage | <0.0001 | |
| Organ‐confined | 1.0 | |
| Extraprostatic extension | 3.95 (1.27, 12.32) | |
| Seminal vesicle involvement | 17.05 (3.83, 75.83) | |
| Lymph node involvement | 61.46 (11.58, 326.33) | |
| CAPRA‐S (per 1 unit) | 2.32 (1.79, 3.01) | <0.0001 |
| Total PACCs (per 1 PACC) | 1.38 (1.09, 1.73) | 0.006 |
| Total PACCs (≥1 vs. 0)a | 2.70 (1.24, 5.86) | 0.012 |
| Cores with PACCs (per 1 core) | 1.40 (1.004, 1.94) | 0.047 |
| Cores with PACCs (≥1 vs. 0)a | 2.70 (1.24, 5.86) | 0.012 |
Abbreviations: CAPRA‐S, Cancer of the Prostate Risk Assessment Postsurgical; CI, confidence interval; HR, hazard ratio; PACCs, polyaneuploid cancer cells.
Binary classification of total PACCs and cores with PACCs gives the same distribution, so results from the proportional hazards model are also the same.
The multivariable models with PACCs that gave the best fit included CAPRA‐S. Adding either total PACCs or cores positive for PACCs (both expressed as a continuous variable) to CAPRA‐S significantly improved model fit compared to CAPRA‐S alone, based on the pseudo‐likelihood ratio test (Table 3). The best‐fitting model included total PACCs, HR = 2.00 (95% CI: 1.40, 2.87), and CAPRA‐S, HR = 2.51 (95% CI: 1.81, 3.48).
Table 3.
Multivariable proportional hazards models of metastasis‐free survival in a case–cohort of intermediate‐ and high‐risk men
| Variables | HR (95% CI) | p Value | p Value for increase in PLRTa compared to CAPRA‐S alone |
|---|---|---|---|
| Model 1 | |||
| CAPRA‐S (per 1 unit) | 2.50 (1.81, 3.46) | <0.0001 | <0.0001 |
| Total PACCs (per 1 PACC) | 2.00 (1.40, 2.85) | 0.0001 | |
| Model 2 | |||
| CAPRA‐S (per 1 unit) | 2.23 (1.73, 2.88) | <0.0001 | 0.076 |
| Total PACCs (≥1 vs. 0)b | 1.97 (0.54, 7.24) | 0.306 | |
| Model 3 | |||
| CAPRA‐S (per 1 unit) | 2.35 (1.79, 3.09) | <0.0001 | 0.0003 |
| Cores with PACCs (per 1 core) | 2.15 (1.23, 3.78) | 0.007 | |
| Model 4 | |||
| CAPRA‐S (per 1 unit) | 2.23 (1.73, 2.88) | <0.0001 | 0.076 |
| Cores with PACCs (≥1 vs. 0)b | 1.97 (0.54, 7.24) | 0.306 | |
Abbreviations: CAPRA‐S, Cancer of the Prostate Risk Assessment Postsurgical; CI, confidence interval; HR, hazard ratio; PACCs, polyaneuploid cancer cells; PLRT, pseudo‐likelihood ratio test.
The pseudo‐likelihood ratio test is based on the change in deviance when one of the PACC variables is added to a model of CAPRA‐S alone.
Binary classification of total PACCs and cores with PACCs gives the same distribution, so results from the proportional hazards model are also the same.
Among 38 patients with metastases, 32 had complete data on PGCCs, CAPRA‐S, and time to mCRPC. Table 4 shows univariate analyses for CAPRA‐S and total PACCs (expressed as a continuous variable), and the multivariable model of total PACCs and CAPRA‐S. Cores positive for PACCs were not statistically significant in a univariate model or adjusted for CAPRA‐S (data not shown). Total PACCs were statistically significant in the univariate model, HR = 1.23 (95% CI: 1.06, 1.43), p = 0.007, but was no longer significant when adjusted for CAPRA‐S, HR = 1.17 (95% CI: 0.995, 1.38), p = 0.057. The small sample size resulted in a lack of statistical power, which may have influenced the result.
Table 4.
Univariate and multivariable proportional hazards models of time from metastasis to mCRPC in a cohort of intermediate‐ and high‐risk men with metastasis (n = 32)
| Variables | HR (95% CI) | p Value | p Value for increase in LRTa compared to CAPRA‐S alone |
|---|---|---|---|
| Univariate | |||
| CAPRA‐S (per 1 unit) | 1.14 (0.97, 1.33) | 0.106 | n/a |
| Total PACCs (per 1 PACC) | 1.22 (1.05, 1.42) | 0.011 | n/a |
| Multivariable | |||
| CAPRA‐S (per 1 unit) | 1.08 (0.93, 1.27) | 0.322 | 0.075 |
| Total PACCs (per 1 PACC) | 1.17 (0.995, 1.38) | 0.057 | |
Abbreviations: CAPRA‐S, Cancer of the Prostate Risk Assessment Postsurgical; CI, confidence interval; HR, hazard ratio; LRT, likelihood ratio test; n/a, not applicable; PACC, polyaneuploid cancer cells.
The likelihood ratio test is based on the change in deviance when total PACCs are added to a model of CAPRA‐S alone.
4. DISCUSSION
The major cause of death related to PCa is the development of therapy‐resistant metastatic disease. The possible mechanisms of therapy resistance in PCa have been broadly investigated with multiple candidates such as SOX2 activation, MYC and RAS coactivation, and ERG gene rearrangements. 49 , 50 , 51 However, the PACC state may represent an inclusive and unifying explanation for therapy resistance mechanisms, that is, underrecognized. This cell state is induced by the tumor microenvironment or therapeutic stress, can exist for an extended period of time, and can act as a CSC by undergoing depolyploidization and repopulating the tumor cell population when stress is relieved. To systematically study this phenomenon, one needs to go back to the fundamental approach to tumor pathogenesis: cell morphology.
The PACC state has two defining characteristics: polyploidy and relatively large size. Polyploidy does not necessarily mean “multinucleation” and can be pronounced as a single large nucleus; however, multinucleated cells are often polyploid. Because of the increased genomic content, polyploid cells are physically larger than the neighboring tumor cells. 12 The presence of cells in the PACC state has been shown to be associated with worse prognosis, higher tumor grade, poor differentiation, and advanced disease stage in various tumor types including PCa. 30 , 32 , 34 , 36 , 37 , 40 There is also evidence in castration‐resistant PCa that cells in the PACC state drive resistance to taxane‐based chemotherapy. 52
In this study, we investigated the presence of cells in the PACC state and their clinical importance in patients who underwent radical prostatectomy with curative intent to treat their presumed localized PCa. Our findings show that the number of PACCs and the number of cores positive for PACCs are statistically significant prognostic factors for MFS, after adjusting for CAPRA‐S, in a case–cohort of intermediate‐ or high‐risk men who underwent radical prostatectomy. In addition, despite the small number of men with complete data to evaluate time to mCRPC, the total number of cells in the PACC state was a statistically significant predictor of mCRPC in univariate analysis and suggested a prognostic effect even after adjusting for CAPRA‐S. To our knowledge, our study is the first to describe the adverse clinical implications of the presence and of cells in the PACC state in a stratified cohort of PCa patients based on the metastasis status. Assessing the prognostic value of PACCs for mCRPC by employing a larger cohort, prospective analyses of the predictive value of PACCs for adverse clinical outcome, and ultimately, whole‐genome and RNA‐sequencing of the genetic material of these cells by using microdissection methods are of interest in understanding the biology of the PACC state. It should be pointed out, however, that none of the PACC‐associated variables in Table 2 was as strong as the Gleason score, pathological stage, or CAPRA‐S.
One of the main challenges in this study was to accurately identify cells in the PACC state. We found the EpCAM stain is helpful in visualizing the larger and atypical tumoral cells and increasing our ability to detect PACCs of various morphologies. It is clear that using a specific biomarker to highlight PACCs would be the ideal approach in studying these cells. However, there are currently no biomarkers for the PACC state, either for monitoring in vivo or for isolation, and we believe this should be an area for future research.
Another challenge that might have affected the results of this study is tumor heterogeneity, which has long been known to be present in PCa, 53 given the fact that we employed TMAs to detect PACCs in our cohort. Although we applied a 3–4‐fold sampling redundancy to reduce the margin of error, it is clear that the results of this study might have been affected by underrepresentation of PACCs because of the heterogeneous nature of PCa.
We did not determine the incremental improvement in the concordance index associated with adding PACCs to the model containing CAPRA‐S because we are not aware of a validated approach to doing so for a case‐cohort study design. However, it has been shown that if a variable is a statistically significant addition to a multivariable model, it is mathematically equivalent to demonstrating a significant improvement in model performance and that the test for significance of adding the variable has greater statistical power than a test of increase in concordance index. Since PACCs were statistically significant when added to a model with CAPRA‐S, it implies that adding the biomarker significantly improved model performance. 54 , 55
Because of its important role in disease resistance, we believe it is important to eliminate the PACC state during the treatment of aggressive PCa patients. However, there are no agents to specifically target these cells to date. Their unique biology and phenotype may create therapeutic opportunities as they may have unexpected vulnerabilities. This is a critical area of research in combination with molecular analysis of their genome.
Although there is scarce knowledge about the biology of cells the PACC state, we and others have shown their likely role in mediating disease resistance. We present additional evidence that they are significant prognostic factors for metastasis in patients with PCa who underwent radical prostatectomy with curative intent to treat their presumed localized PCa.
CONFLICTS OF INTEREST
Kenneth J. Pienta is a consultant for CUE Biopharma, Inc. and holds an equity interest in Keystone Biopharma, Inc. Bruce J. Trock has research grants through Johns Hopkins from MDxHealth, Inc., Myriad Genetics, Inc., Opko Health, Inc., and Exact Sciences, Inc. Angelo M. De Marzo is a consultant for Merck, Inc. and Cepheid Inc. He has sponsored research support from Janssen and Myriad Genetics Inc. Sarah R. Amend holds an equity interest in Keystone Biopharma, Inc. The remaining author declares no conflicts of interest.
Supporting information
Supplementary Figure 1.
ACKNOWLEDGMENTS
This work was supported by NCI Grants U54CA143803, CA163124, CA093900, and CA143055, and the Prostate Cancer Foundation to Kenneth J. Pienta; NCI Grant P50CA058236, US Department of Defense CDMRP/PCRP (W81XWH‐21‐0‐373), the Patrick C. Walsh Prostate Cancer Research Fund to Bruce J. Trock; US Department of Defense Prostate Cancer Biospecimen Network Site, Grant/Award Number: W81XWH‐18‐2‐0015; National Cancer Institute, Grant/Award Numbers: MCL U01 CA1936390 P30 CA006973; SPORE in Prostate Cancer, Grant/Award Number: P50CA58236; The Prostate Cancer Foundation to Angelo M. De Marzo; US Department of Defense CDMRP/PCRP (W81XWH‐20‐10353), the Patrick C. Walsh Prostate Cancer Research Fund, and the Prostate Cancer Foundation to Sarah R. Amend.
Trabzonlu L, Pienta KJ, Trock BJ, De Marzo AM, Amend SR. Presence of cells in the polyaneuploid cancer cell (PACC) state predicts the risk of recurrence in prostate cancer. The Prostate. 2023;83:277‐285. 10.1002/pros.24459
DATA AVAILABILITY STATEMENT
All data are available upon request.
REFERENCES
- 1. Cree IA, Charlton P. Molecular chess? Hallmarks of anti‐cancer drug resistance. BMC Cancer. 2017;17(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E. The challenge of drug resistance in cancer treatment: a current overview. Clin Exp Metastasis. 2018;35(4):309‐318. [DOI] [PubMed] [Google Scholar]
- 3. Aleksakhina SN, Kashyap A, Imyanitov EN. Mechanisms of acquired tumor drug resistance. Biochim Biophys Acta. 2019;1872(2):188310. [DOI] [PubMed] [Google Scholar]
- 4. Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8(6):1095‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dagogo‐Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81‐94. [DOI] [PubMed] [Google Scholar]
- 7. Luo M, Brooks M, Wicha M. Epithelial–mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr Pharm Des. 2015;21(10):1301‐1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pienta KJ, Hammarlund EU, Axelrod R, Brown JS, Amend SR. Poly‐aneuploid cancer cells promote evolvability, generating lethal cancer. Evol Appl. 2020;13(7):1626‐1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pienta KJ, Hammarlund EU, Austin RH, Axelrod R, Brown JS, Amend SR. Cancer cells employ an evolutionarily conserved polyploidization program to resist therapy. Semin Cancer Biol. 2022;81:145‐159. 10.1016/j.semcancer.2020.11.016 [DOI] [PubMed] [Google Scholar]
- 10. Pienta KJ, Hammarlund EU, Brown JS, Amend SR, Axelrod RM. Cancer recurrence and lethality are enabled by enhanced survival and reversible cell cycle arrest of polyaneuploid cells. Proc Natl Acad Sci USA. 2021;118(7):e2020838118. 10.1073/pnas.2020838118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Virchow R. Cellular pathology, as based upon physiological and pathological histology Classics of Medicine Library Special Edition. In: Witt RM De ed. Twenty lectures delivered in the Pathological Institute of Berlin during the months of February, March and April, Leslie B. Adams, Jr .; 1858. 1860:xxvi, 27–554.
- 12. Amend SR, Torga G, Lin KC, et al. Polyploid giant cancer cells: unrecognized actuators of tumorigenesis, metastasis, and resistance. Prostate. 2019;79(13):1489‐1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mirzayans R, Andrais B, Murray D. Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers. 2018;10(4):118. 10.3390/cancers10040118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J, Niu N, Zhang J, et al. Polyploid giant cancer cells (PGCCs): the evil roots of cancer. Curr Cancer Drug Targets. 2019;19(5):360‐367. [DOI] [PubMed] [Google Scholar]
- 15. Illidge T. Polyploid giant cells provide a survival mechanism for p53 mutant cells after DNA damage. Cell Biol Int. 2000;24(9):621‐633. [DOI] [PubMed] [Google Scholar]
- 16. Niu N, Mercado‐Uribe I, Liu J. Dedifferentiation into blastomere‐like cancer stem cells via formation of polyploid giant cancer cells. Oncogene. 2017;36(34):4887‐4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brooks PJ, Glogauer M, McCulloch CA. An overview of the derivation and function of multinucleated giant cells and their role in pathologic processes. Am J Pathol. 2019;189(6):1145‐1158. [DOI] [PubMed] [Google Scholar]
- 18. Moein S, Adibi R, da Silva Meirelles L, Nardi NB, Gheisari Y. Cancer regeneration: polyploid cells are the key drivers of tumor progression. Biochim Biophys Acta. 2020;1874(2):188408. [DOI] [PubMed] [Google Scholar]
- 19. Weihua Z, Lin Q, Ramoth AJ, Fan D, Fidler IJ. Formation of solid tumors by a single multinucleated cancer cell. Cancer. 2011;117(17):4092‐4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajaraman R, Guernsey DL, Rajaraman MM, Rajaraman SR. Stem cells, senescence, neosis and self‐renewal in cancer. Cancer Cell Int. 2006;6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dhimolea E, de Matos Simoes R, Kansara D, et al. An embryonic diapause‐like adaptation with suppressed myc activity enables tumor treatment persistence. Cancer Cell. 2021;39(2):240‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rehman SK, Haynes J, Collignon E, et al. Colorectal cancer cells enter a diapause‐like DTP state to survive chemotherapy. Cell. 2021;184(1):226‐242.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu J, Niu N, Li X, Zhang X, Sood AK. The life cycle of polyploid giant cancer cells and dormancy in cancer: opportunities for novel therapeutic interventions. Semin Cancer Biol. 2022;81:132‐144. 10.1016/j.semcancer.2021.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saini G, Joshi S, Garlapati C, et al. Polyploid giant cancer cell characterization: new frontiers in predicting response to chemotherapy in breast cancer. Semin Cancer Biol. 2022;81:220‐231. 10.1016/j.semcancer.2021.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imai Y, Morishita S, Ikeda Y, et al. Immunohistochemical and molecular analysis of giant cell carcinoma of the pancreas: a report of three cases. Pancreas. 1999;18(3):308‐315. [DOI] [PubMed] [Google Scholar]
- 26. Mosnier JF, Balique JG. Pleomorphic giant cell carcinoma of the esophagus with coexpression of cytokeratin and vimentin and neuroendocrine differentiation. Arch Pathol Lab Med. 2000;124(1):135‐138. [DOI] [PubMed] [Google Scholar]
- 27. O'Connor RC, Hollowell CMP, Laven BA, Yang XJ, Steinberg GD, Zagaja GP. Recurrent giant cell carcinoma of the bladder. J Urol. 2002;167(4):1784. [PubMed] [Google Scholar]
- 28. Shen R, Wen P. Clear cell renal cell carcinoma with syncytial giant cells: a case report and review of the literature. Arch Pathol Lab Med. 2004;128(12):1435‐1438. [DOI] [PubMed] [Google Scholar]
- 29. Schwartz HS, Eskew JD, Butler MG. Clonality studies in giant cell tumor of bone. J Orthop Res. 2002;20(2):387‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fei F, Zhang D, Yang Z, et al. The number of polyploid giant cancer cells and epithelial–mesenchymal transition‐related proteins are associated with invasion and metastasis in human breast cancer. J Exp Clin Cancer Res. 2015;34:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang S, Mercado‐Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem‐like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33(1):116‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qu Y, Zhang L, Rong Z, He T, Zhang S. Number of glioma polyploid giant cancer cells (PGCCs) associated with vasculogenic mimicry formation and tumor grade in human glioma. J Exp Clin Cancer Res. 2013;32:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu G, Wang Y, Fei F, et al. Clinical characteristics and preliminary morphological observation of 47 cases of primary anorectal malignant melanomas. Melanoma Res. 2018;28(6):592‐599. [DOI] [PubMed] [Google Scholar]
- 34. Liu HT, Xia T, You YW, et al. Characteristics and clinical significance of polyploid giant cancer cells in laryngeal carcinoma. Laryngoscope Investig Otolaryngol. 2021;6(5):1228‐1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerashchenko BI, Salmina K, Eglitis J, Huna A, Grjunberga V, Erenpreisa J. Disentangling the aneuploidy and senescence paradoxes: a study of triploid breast cancers non‐responsive to neoadjuvant therapy. Histochem Cell Biol. 2016;145(4):497‐508. [DOI] [PubMed] [Google Scholar]
- 36. Lv H, Shi Y, Zhang L, et al. Polyploid giant cancer cells with budding and the expression of cyclin E, S‐phase kinase‐associated protein 2, stathmin associated with the grading and metastasis in serous ovarian tumor. BMC Cancer. 2014;14:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang D, Yang X, Yang Z, et al. Daughter cells and erythroid cells budding from PGCCs and their clinicopathological significances in colorectal cancer. J Cancer. 2017;8(3):469‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopez‐Beltran A, Eble JN, Bostwick DG. Pleomorphic giant cell carcinoma of the prostate. Arch Pathol Lab Med. 2005;129(5):683‐685. [DOI] [PubMed] [Google Scholar]
- 39. Mannan R, Wang X, Bawa PS, et al. Polypoidal giant cancer cells in metastatic castration‐resistant prostate cancer: observations from the Michigan Legacy Tissue Program. Med Oncol. 2020;37(3):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alharbi AM, De Marzo AM, Hicks JL, Lotan TL, Epstein JI. Prostatic adenocarcinoma with focal pleomorphic giant cell features: a series of 30 cases. Am J Surg Pathol. 2018;42(10):1286‐1296. [DOI] [PubMed] [Google Scholar]
- 41. Mohler J, Bahnson RR, Boston B, et al. The NCCN Prostate Cancer Clinical Practice Guidelines in Oncology. J Nat Comp Cancer Netw. 2010;8(2):200. [DOI] [PubMed] [Google Scholar]
- 42. Vidal I, Zheng Q, Hicks JL, et al. GSTP1 positive prostatic adenocarcinomas are more common in Black than White men in the United States. PLoS One. 2021;16(6):e0241934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chaux A, Peskoe SB, Gonzalez‐Roibon N, et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25(11):1543‐1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gurel B, Iwata T, M Koh C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Faith DA, Isaacs WB, Morgan JD, et al. Trefoil factor 3 overexpression in prostatic carcinoma: prognostic importance using tissue microarrays. Prostate. 2004;61(3):215‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case‐cohort designs. J Clin Epidemiol. 1999;52(12):1165‐1172. [DOI] [PubMed] [Google Scholar]
- 47. Cooperberg MR, Hilton JF, Carroll PR. The CAPRA‐S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117(22):5039‐5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Langholz B, Jiao J. Computational methods for case‐cohort studies. Comput Stat & Data Anal. 2007;51:3737‐3748. [Google Scholar]
- 49. de Wet L, Williams A, Gillard M, et al. SOX2 mediates metabolic reprogramming of prostate cancer cells. Oncogene. 2022;41(8):1190‐1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arriaga JM, Panja S, Alshalalfa M, et al. A MYC and RAS co‐activation signature in localized prostate cancer drives bone metastasis and castration resistance. Nature Cancer. 2020;1(11):1082‐1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Udager AM, Shi Y, Tomlins SA, et al. Frequent discordance between ERG gene rearrangement and ERG protein expression in a rapid autopsy cohort of patients with lethal, metastatic, castration‐resistant prostate cancer. Prostate. 2014;74(12):1199‐1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mittal K, Donthamsetty S, Kaur R, et al. Multinucleated polyploidy drives resistance to docetaxel chemotherapy in prostate cancer. Br J Cancer. 2017;116(9):1186‐1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haffner MC, Zwart W, Roudier MP, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol. 2021;18(2):79‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pepe MS, Kerr KF, Longton G, Wang Z. Testing for improvement in prediction model performance. Stat Med. 2013;32(9):1467‐1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vickers AJ, Cronin AM, Begg CB. One statistical test is sufficient for assessing new predictive markers. BMC Med Res Methodol. 2011;11(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1.
Data Availability Statement
All data are available upon request.


