Abstract
The electronic health record (EHR) provides valuable data for understanding physical and mental health conditions in autism. We developed an approach to identify charts of autistic young adults, retrieved from our institution’s de-identified EHR database.
Clinical notes within two cohorts were identified. Cohort 1 charts had at least one International Classification of Diseases (ICD-CM) autism code. Cohort 2 charts had only autism key terms without ICD-CM codes, and at least four notes per chart. A natural language processing tool parsed medical charts to identify key terms associated with autism diagnoses and mapped them to Unified Medical Language System Concept Unique Identifiers (CUIs). Average scores were calculated for each set of charts based on captured CUIs. Chart review determined whether patients met criteria for autism using a classification rubric.
In Cohort 1, of 418 patients, 361 were confirmed to have autism by chart review. Sensitivity was 0.99 and specificity was 0.68 with positive predictive value (PPV) of 0.97. Specificity improved to 0.81 (sensitivity was 0.95; PPV was 0.98) when the number of notes was limited to four or more per chart. In Cohort 2, 48 of 136 patients were confirmed to have autism by chart review. Sensitivity was 0.95, specificity was 0.73, and PPV was 0.70.
Our approach, which included using key terms, identified autism charts with high sensitivity, even in the absence of ICD-CM codes. Relying on ICD-CM codes alone may result in inclusion of false positive cases and exclusion of true cases with autism.
Lay Summary
Clinical notes in patient charts contain valuable data for helping to understand health problems in autistic adults. We describe a computer-based method to identify charts belonging to autistic young adults. The computer program looked for key terms (“autism,” “Asperger’s”, etc.) in clinical notes, and was able to identify notes belonging to people with autism with good accuracy.
Introduction
Autism spectrum disorder (ASD) is a complex and common condition that affects 1 in 44 children (Maenner et al., 2021). It is estimated that more than 700,000 youth on the autism spectrum will enter adulthood over the next decade (Shattuck, 2019). Autistic adults1 have increased rates of major psychiatric disorders including depression, anxiety, bipolar disorder, obsessive-compulsive disorder, schizophrenia, and suicide attempts, and nearly all medical conditions, including immune, gastrointestinal, sleep disorders, seizures, obesity, dyslipidemia, hypertension, and diabetes (Croen et al., 2015). A 25-year outcome study of autistic adults noted a median number of 11 medical conditions per person (Jones et al., 2016). Critical gaps exist in mental health services for autistic individuals throughout the lifespan (Maddox et al., 2021). Additional studies have emphasized the negative impact of mental health and sleep disturbances on quality of life in transition-age autistic youth and adults (Lawson et al., 2020), and the poor long-term overall outcomes for autistic adolescents and adults (Steinhausen et al., 2016).
The electronic health record (EHR) provides an opportunity to facilitate many aspects of clinical research aimed at characterizing health-related issues impacting autistic individuals across the lifespan. These include conducting studies of the prevalence of physical and mental health conditions (Bishop-Fitzpatrick, 2018; Davignon et al., 2018), examining how health conditions change over time and in response to interventions (Singer et al., 2022), facilitating the conduct of pragmatic clinical trials (Cowie et al., 2017), and recruitment of people for research studies. Most studies harnessing the EHR have relied on the International Classification of Diseases (ICD; World Health Organization, 1992) billing codes to identify cases of ASD (Croen et al., 2015; Bush et al., 2017; Davignon et al., 2018; Bishop-Fitzpatrick et al. 2018; Failla et al., 2021). However, there are limitations to the use of these ICD codes in identifying cases of autism. While clinicians often refer to autism diagnoses in their notes, they often do not submit billing codes for autism. Instead, clinicians submit billing codes for the diagnosis most relevant to the patient’s medical care during a given visit and their specialty, such as obesity, depression, or sleep medicine. Therefore, while diagnostic codes are useful for defining cases, they may not be uniformly used by subspecialists, and thereby miss cases of autism in the EHR.
Examining clinical notes for evidence of an autism diagnosis increases the likelihood of detecting autism. However, this process is time-intensive. An alternative option to identifying autism in clinical notes is to develop algorithms focused on the clinical notes, rather than on billing codes (Brooks et al., 2021). Natural language processing (NLP) algorithms that identify key terms have the potential to identify autistic patients and to be more sensitive than ICD codes alone. These NLP algorithms can have a higher predictive value than ICD codes for detecting autistic features (e.g., limited eye contact, repetitive behaviors) in autistic individuals [Lingren et al., 2016]. Algorithms using NLP may also provide more specificity over ICD codes, as these codes are sometimes applied to cases of suspected but not confirmed autism in order to obtain services (e.g., behavioral therapy).
The goal of our project was to present an efficient strategy for researchers to identify cases of autism in their EHR, so that they can conduct studies characterizing health conditions in autism, and the effects of interventions to improve these health conditions. To further this goal, we examined the value of incorporating an NLP tool instead of relying solely on ICD codes, to improve the accuracy of finding cases of autism in a de-identified, EHR-derived cohort of patients.
Methods
Our research was prospectively reviewed and approved by the Vanderbilt Institutional Review Board. Vanderbilt University Medical Center (VUMC) serves a large geographic area stretching from Southern Kentucky to Northern Alabama (approximately 65,000 square miles) and provides both primary and specialty care, including assessment and evaluation of autism. The diagnosis of autism is made by psychology, developmental medicine, and psychiatry. Approximately 2,900 patients with autism are served annually, 70% with private insurance, 29.98% with public insurance, and the remainder are self-pay. Approximately 77% are male and 23% are female.Charts (collection of records belonging to an individual patient) were retrieved from the VUMC Synthetic Derivative (SD). The SD is a de-identified database of our institutional EHR system that contains unique individual records [Roden et al., 2008]. The de-identification of SD records was achieved primarily through the application of a commercial electronic program, which was applied and assessed for acceptable effectiveness in scrubbing identifiers. Patient names are permanently replaced with a tag [NAMEAAA, BBB] to maintain the semantic integrity of the text and real dates have been replaced with randomly generated dates. In addition, all researchers are required to sign data use agreements indicating they will not attempt to re-identify any patients whose de-identified data are included in the research database used in this study. Charts were limited to patients ages 20-25 years old at the time of retrieval, although their records were analyzed to include younger ages, back to birth. This age group was selected given the focus of our research question—to identify autistic young adults for future studies of the natural history of health conditions and the impact of treatments on these health conditions. We currently have approximately 3.4 million patients’ records in our SD implemented with the schema from Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM). These data elements include diagnostic codes, demographics, clinic notes and clinical communications. All charts in the SD had names redacted and dates shifted by up to 1 calendar year consistently within each record but differing across all records to provide anonymity.
There were two cohorts identified. In Cohort 1, charts had at least one instance of an International Classification of Diseases, Ninth or Tenth Revision Clinical Modification (ICD-9-CM or ICD-10-CM) code for autism as listed below. In Cohort 2, charts were selected based on autism key terms and did not have ICD-CM codes.
Validated Pipeline Approach.
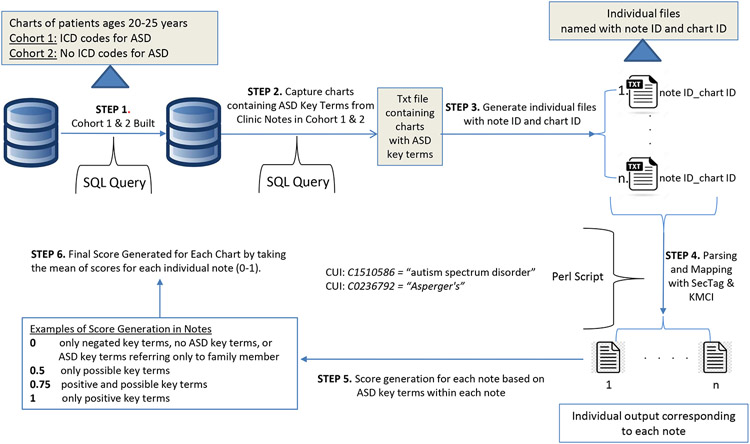
A Validated Pipeline Approach was developed under Unix/Linux working environment with data retrieved from VUMC SD. The approach used for Cohorts 1 and 2 is illustrated in Figure 1.
Figure 1. Validated Pipeline Approach to Identify ASD Charts.
Patients with ICD-CM codes for autism were retrieved from the VUMC SD using SQL (Step 1) and those with ASD key terms were placed in a text file (Step 2). Individual files were generated using a bash script (Step 3) and KMCI was applied (Step 4) to parse medical terms and map them to UMLS CUIs. A score was calculated based on defined rules for capturing ASD key terms (Step 5), and a final score was calculated for each chart by taking the mean of the scores for each individual note (Step 6).
VUMC SD = Vanderbilt University Medical Center Synthetic Derivative; KMCI = KnowledgeMap Concept Indexer; UMLS = Unified Medical Language System; CUI = Concept Unique Identifier
Cohort 1.
A total of 418 patients, ages 20-25 years, containing at least one ICD9 code for autism: 299, 299.0, 299.00, 299.01, 299.8, 299.80, 299.81, 299.9, 299.90, 299.91 and ICD10 code for autism: F84.0, F84.5, F84.8, F84.9 were retrieved from the VUMC SD using Netezza SQL (Figure 1, Step 1). Of these 418 patient charts, 19 charts did not have any autism key terms identified in their clinic notes during the 1st step of our validation pipeline. The other 399 charts contained both ICD-CM codes and autism key terms [autism, autistic, autism spectrum disorder (ASD), Asperger (which captured Asperger syndrome, Asperger’s syndrome, Asperger disorder and Asperger’s disorder], pervasive developmental disorder (which captured pervasive developmental disorder-not otherwise specified), ASD, or PDD (which captured PDD-NOS) identified from their 14,797 clinic notes. These 399 charts and their corresponding notes were placed in a txt file (Figure 1, Step 2).
Individual files from these 399 charts, containing a note ID and a chart ID, were generated using a bash script (Figure 1, Step 3). Notes containing only a questionnaire checkbox listing autism-related diagnoses were excluded from the validation pipeline as they did not provide definitive information.
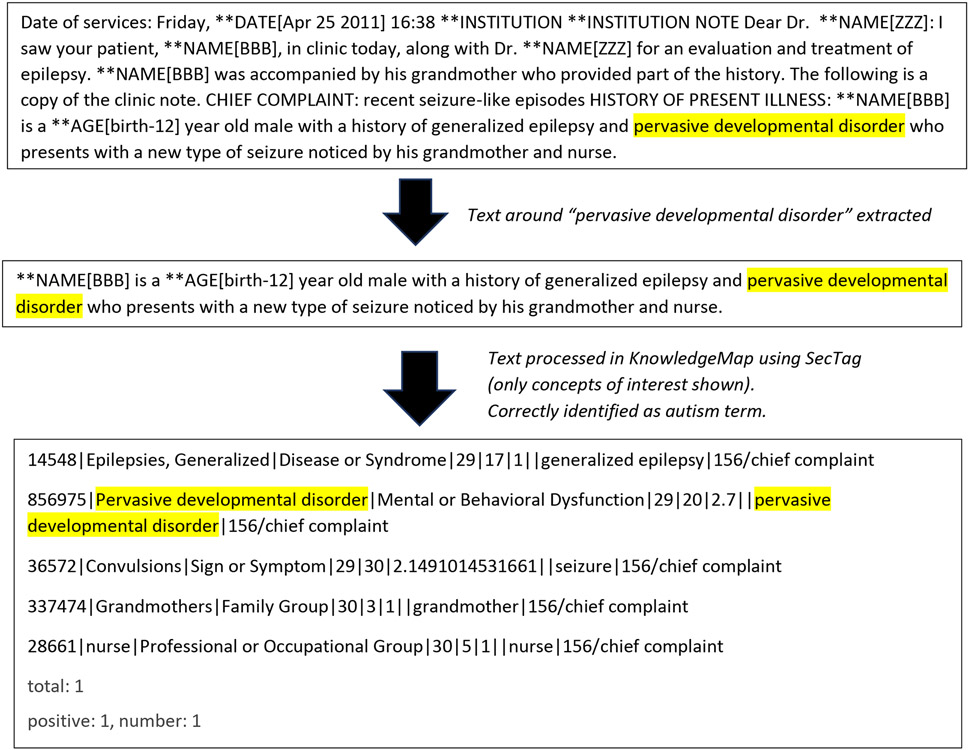
To validate and score the charts, the KnowledgeMap Concept Indexer (KMCI), an NLP tool based on named entity recognition, was applied [Denny et al., 2003; Denny et al., 2005] (Figure 1, Step 4). The KMCI parses medical charts to identify medical terms (e.g., pervasive developmental, Asperger) mentioned in text and then maps these to Unified Medical Language System Concept Unique Identifiers (UMLS CUIs). The tool utilizes term normalization and word variant generation, along with concept co-occurrence data derived from PubMed, to resolve ambiguous mappings. The KMCI approach also takes into account both explicit and implicit note sections (e.g., family history; diagnostic testing) that are recognized using SecTag. (Denny JC et al., 2009). It identifies affirmed and possible concepts and handles negation using an embedded NegEx algorithm (Chapman et al., 2001a and 2001b). The output of KMCI for each individual note was analyzed by a UNIX bash script to capture any autism CUIs matched to the list of our autism key terms. Key terms included ‘autism spectrum disorders’=CUI: C1510586, ‘asperger syndrome’=CUI: C0236792. See Supplemental Table 1 for full list of key terms and CUIs and Figures 2A, 2B, and 2C for examples of the process.
Figure 2A. SecTag Processing of a Section of Text containing an autism key term.
SecTag first identifies text based on standard sections, such as Chief Complaint. In the example below. A concept of interest is assigned a CUI. CUIs associated with autism key terms, such as pervasive developmental disorder, can then be identified. CUI = Concept Unique Identifier
Figure 2B. SecTag Processing of a Section of Text containing an autism key term that was incorrectly negated.
In the example below. The NegEx algorithm identified text (“not even be feasible”) that incorrectly negated the key term of autism spectrum. This sentence was awkwardly constructed. Note received score of 1.
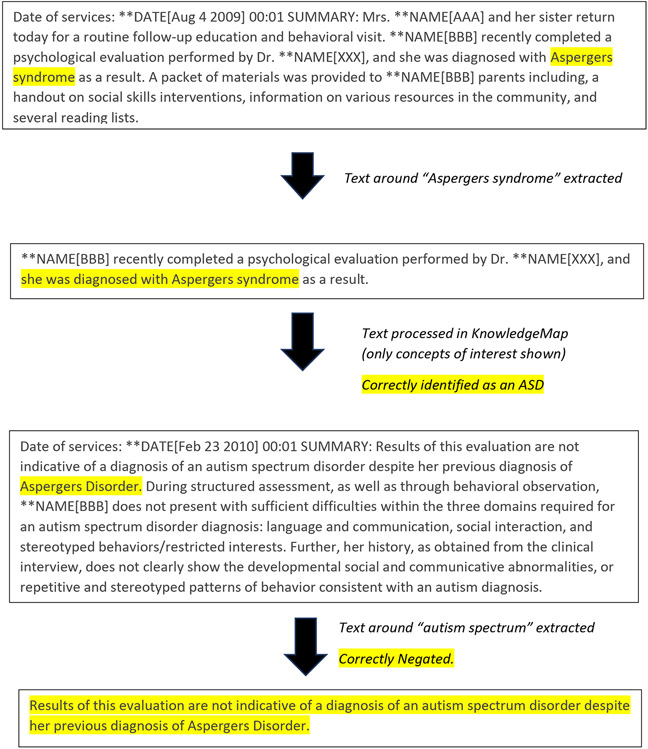
Figure 2C. Text illustrating how an autism diagnosis may evolve over time.
In this example, an individual was initially diagnosed with Asperger syndrome (Aug 4 2009) although a later diagnosis (Feb 23 2010) was no longer compatible with the diagnosis.
A score was then calculated based on the following defined rules for capturing autism key terms (Figure 1, Step 5). A note was scored ‘1’ for only positive autism key terms, ‘0’ for only negated autism key terms (“does not have autism”), ‘0.5’ for only possible (“has possible autism”), and an ‘NA’ for autism referring to a family history (“brother with autism”) or other uses of the term [e.g., cardiac atrial septal defect (ASD)]. If a combination of autism key terms were present in a note, an average score was determined for that note. For example, a note containing positive and possible key terms would receive a score of 0.75 while a note with positive and negated key terms would receive a score of 0.5. The rationale for including a scoring system was to account for the probabilistic nature of autism terms in notes. For example, over time, note in a chart might reflect a possible diagnosis of autism and then an autism case might become more definitive or eliminated with cumulative positive or negative notes after evaluation by psychology, psychiatry, or developmental medicine.
To account for potential differences in the number of available notes per patient, a final score for each patient’s chart was calculated by taking the mean of the scores for each individual note (Figure 1, Step 6). Any chart consisting exclusively of only NA autism terms (e.g., “brother with autism”) or having no key terms was assigned a score of 0 and were considered to not represent true cases of autism.
Cohort 2.
A total of 1,026 patients without autism ICD-9-CM or ICD-10-CM codes, but with clinic notes identified by a list of autism key terms above, ages 20-25 years, were identified. As with Cohort 1, those 2,349 notes with autism key terms [autism, autistic, autism spectrum disorder, Asperger, pervasive developmental disorder, ASD, and PDD- abbreviation for pervasive developmental disorder) were retrieved from the Vanderbilt Synthetic Derivative using Netezza SQL. After optimizing ROC curves using Cohort 1 data, the Cohort 2 analysis was limited to patients with at least four notes (n = 136), which included 997 total notes. Notes were further classified into 3 categories as in Cohort 1—comprehensive, simple, and questionnaire (with questionnaire notes excluded) and scores were calculated for each note, and then averaged for each chart.
Manual Chart Review
Each chart in Cohort 1 and Cohort 2 was reviewed to determine whether the patient met criteria for autism. We used a chart review rubric (Table 1) to facilitate ascertainment of true cases within the SD set. The rubric allowed for the reviewers, who were trained in the diagnosis of autism, to stratify cases into low-evidence, medium-evidence, and high-evidence subsets. Cases were defined as follows: Low-evidence cases were required to have at least one affirmative mention of an autism-specific diagnosis in a chart of any type. Medium-evidence cases were distinguished by the presence of either a psychological evaluation form with an explicit autism diagnosis OR at least two affirmative mentions of an autism-specific diagnosis by specific provider(s) (neurology, psychiatry, developmental pediatric, behavior therapy, occupational therapy, speech language pathology). High-evidence cases were distinguished by the presence of any of the following: a psychology evaluation with confirmatory Autism Diagnostic Observation Schedule (ADOS), a clinic visit by an autism specialist, a clinic visit for medication management for autism, or two mentions of the patient being treated in a dedicated autism clinic, even without the actual clinic note. Cases in which the psychological evaluation or developmental medicine evaluation did not substantiate the diagnosis of autism were considered excluded cases. For both Cohorts 1 and 2, notes were limited to those on or prior to 7/17/2016. This date was selected because our team had previously carried out a manual review of charts in Cohort 1 to establish a “gold-standard” autism-specific dataset using the rubric above, with an end-date of 7/17/2016 (Singer et al., 2022) and we wanted to be consistent in our inclusion of additional charts. We reviewed all charts that were identified by our Pipeline and that had not been previously reviewed in this “gold-standard” dataset.
Table 1.
Diagnosis of ASD in the VUMC Synthetic Derivative
|
|
|
Data Analysis.
Each cohort was first described in terms of demographics and chart classification, based on manual chart review. These classifications included: (1) high evidence of autism diagnosis by manual chart review; (2) mid evidence of autism diagnosis by manual chart review; (3) low evidence of autism diagnosis by manual chart review; (4) no data in manual chart review to support or refute an autism diagnosis- -this category included no mention of autism in the chart, limitation of autism to relative, or use of a different ASD term (cardiac atrial septal defect); (5) exclusion of an autism diagnosis, or (6) unable to confirm.
The scores derived from our autism diagnosis validating pipeline outlined in the Methods section were analyzed using R (The R Foundation for Statistical Computing, Vienna, Austria). Each chart classification was characterized in terms of the score distribution, including average and median score and quartiles. The distribution of scores in each cohort was characterized using box plots generated with the ggplot2 package in R version 3.35 (Wickham, 2006).
Then, receiver operator characteristic (ROC) curves were constructed to determine optimal cutpoints for identifying autism diagnoses, along with corresponding sensitivity and specificity. The cutpoint package in R version 1.1.1 (Thiele & Hirschfeld, 2021) was used to generate ROC curves and define optimal cutpoints for identifying autism cases based on selecting the average score that empirically maximized the sum of the sensitivity and specificity in each EHR-derived dataset.
For Cohorts 1 and 2, the age at first mention of autism from patients’ clinic notes was compared using a two-sample independent t-test. The proportion of visits by an autism specialist (psychology, developmental medicine, psychiatry) and the top six co-occurring conditions, along with their frequency, was compared for Cohort 1 vs. Cohort 2 using the chi-square test of proportions. If multiple visits from an autism specialist, or codes for a co-occurring condition were recorded in the same chart, each visit type or co-occurring condition was counted only once. Significance level was set at p < 0.05.
Results
Cohort 1
In Cohort 1, there were 315 men and 103 women (with an average age of 22.3 years (standard deviation of 1.66). Racial composition was as follows: White (87.6%), Black (7.4%), Asian (2.1%), Native American (0.2%), Mixed (1.2%), Unknown (0.7%) and other (0.7%). Ethnicity was as follows: non-Hispanic (94.4%), Hispanic (4.8%) and Unknown (0.71%).
Of the 418 patients, 361 (86%) patients were confirmed to have autism by chart review: high-evidence in 84 (20%), mid-evidence in 163 (39%), and low-evidence in 114 (27%). In 11 (2.6%) patients, autism was excluded by chart review. In 27 (6.5%) patients, there were no data to support or refute an autism diagnosis in the patient—examples included having a family member with autism, having a cardiac ASD (atrial septal defect) or, in the case of 19 patients, no reference to autism in the chart. In 19 (4.5%) other patients, data were too limited to confirm a diagnosis of autism. The patients excluded by chart review were comparable in age and sex (22.5 years; 82% male) to those who were confirmed to have autism by chart review (22.3 years; 77% male) and did not differ in race or ethnicity (all p values > 0.1).
Scores and number of notes for the six categories of patients defined by chart review are summarized in Table 2. Box plots for high, mid, and low evidence are depicted in Figure 3. Patients with confirmed autism (high, mid, and low evidence) had higher average scores compared to those in the categories of exclusion, no diagnosis, or limited data (t = 8.39; p < 0.0001). In the high evidence group, there were no scores below 0.83.
Table 2.
Score and Number of Notes by Category of ASD Diagnosis in Each Cohort
| Category, ASD Diagnosis (no of patients) |
Average Score (Median; Range) |
Average Number of Notes (Median; Range) |
|---|---|---|
| Cohort 1 (418) | ||
| ASD High Evidence (84) | 0.96 (0.97; 0.83-1) | 58.3 (36.5; 5-268) |
| ASD Mid Evidence (163) | 0.95 (0.98; 0.49-1) | 34.9 (18; 2-298) |
| ASD Low Evidence (114) | 0.94 (0.98; 0.50-1) | 15.1 (8; 1-123) |
| ASD Low Evidence with Four or More Charts (89) | 0.95 (0.97; 0.73-1) | 18.8 (12; 4-123) |
| ASD Excluded (11) | 0.88 (0.93; 0.50-1) | 7.27 (6; 1-15) |
| No Evidence (27) | ||
| ICD Code only (19) | No score assigned | No notes |
| Family History or Cardiac ASD only (8) | 0.31 (0; 0-1) | 1.63 (0.5; 0-9) |
| Cannot Confirm (19) | 0.71 (0.83; 0-1) | 8.58 (4; 0-46) |
| Cohort 2 (136) | ||
| ASD Mid Evidence (12) | 0.90 (1; 0-1) | 13.9 (7; 4-49) |
| ASD Low Evidence (36) | 0.93 (1; 0-1) | 8.05 (6; 4-37) |
| ASD Excluded (14) | 0.77 (0.83; 0-1) | 7.58 (6; 4-19) |
| No Evidence (59) | 0.37 (0; 0-1) | 7.49 (5; 4-33) |
| Cannot Confirm (15) | 0.77 (0.88; 0-1) | 10.1 (5; 4-64) |
Figure 3. Boxplots of Average Scores for Charts in Cohorts 1 and 2.
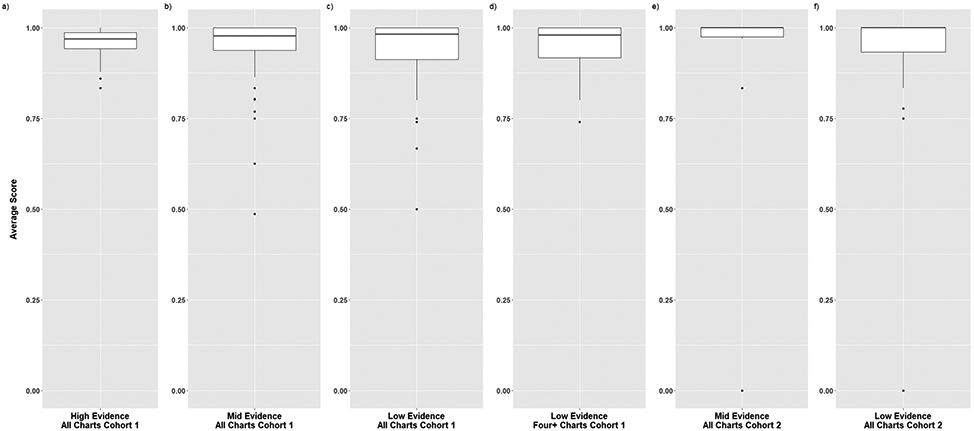
The distribution of average scores for each chart is displayed for Cohort 1 (all charts in high, mid, and low evidence categories and four or more charts in the low evidence category) and Cohort 2 (all charts in the mid and low evidence categories). The solid line depicts the median value and the rectangle depicts the interquartile range (IQR; lower and upper quartile of data). The lines extending from the boxplot show the quartiles +/− (1.5)(IQR) and the dots show potential outliers.
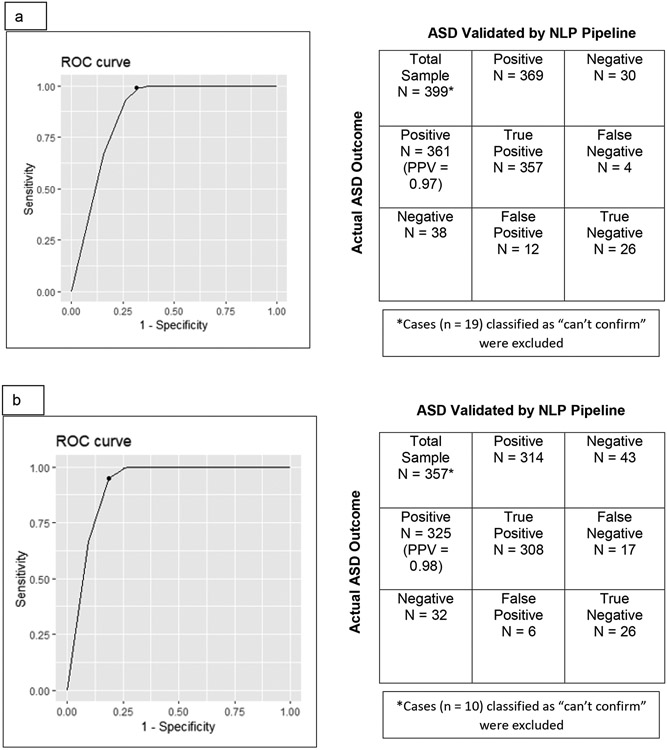
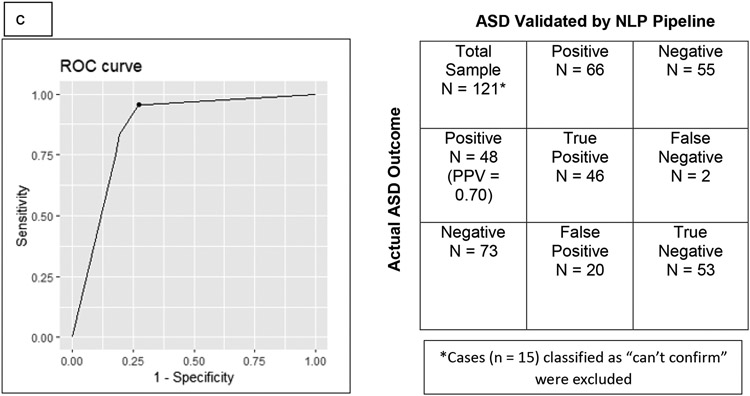
An initial ROC curve (along with a confusion matrix, which indicates positive and negative cases) was constructed for Cohort 1, shown in Figure 4A. Categories 1-5 (confirmed autism diagnosis—high, mid, and low evidence; exclusion, and no diagnosis) were included in the ROC curve analysis –those patients with limited data (Category 6) were not included. The sensitivity was 0.99, the specificity was 0.68, and the positive predictive value (PPV) was 0.97 with an optimal cutpoint score of 0.7 for detecting an autism diagnosis by our NLP-based validating pipeline.
Figure 4.
Receiver-Operator (ROC) curves, accompanied by confusion matrices (indicate positive and negative cases), were constructed for each Cohort. Figure 4a shows the ROC curve and confusion matrix for all charts in Cohort 1. Figure 4b shows the ROC curve and confusion matrix for all charts in Cohort 1 containing four or more notes. Figure 4c shows the ROC curve and confusion matrix for all charts in Cohort 2. PPV = Positive Predictive Value.
We examined the number of notes in relation to high, mid, and low evidence charts. For high evidence, there was only one chart with fewer than four notes (3 notes) and for mid evidence, there were only five charts with fewer than four notes (1 or 2 notes). For low evidence, there were 25 charts that had fewer than four notes (1-3 notes); therefore, for low evidence, Figure 3 box plots included separate plots for all charts and those with four or more notes. The median score for the 89 charts with four or more notes was 0.97 with a range of 0.73-1 (Table 2).
Given the finding that limiting low evidence charts to those with four or more notes increased the range of scores, a second ROC curve and confusion matrix were constructed for Cohort 1, shown in Figure 4B. In this ROC curve, only patients with at least four notes were included (325 had a confirmed autism diagnosis by chart review). This resulted in a slightly lower sensitivity (0.95) and similar PPV (0.98) to the initial ROC curve, but a higher specificity (0.81) and higher optimal cutpoint (0.9) and provided the rationale for limiting data in Cohort 2 to those with at least four notes.
As the use of only one instance of an ICD code may have affected our specificity, we examined the relationship between the number of ICD codes recorded per patient and the confirmation of autism. Of the 418 identified patients, 82 patients had only one ICD code and the remainder (336 patients) had 2 or more ICD codes. Of the 82 patients with only one ICD code, 49 (60%) were confirmed cases of autism as compared to 86% of confirmed cases of autism for the entire sample in Cohort 1.
Cohort 2
In Cohort 2, there were 78 men and 58 women with an average age of 22.2 years (standard deviation of 1.61). Racial composition was as follows: White (80.1), Black (12.5), Asian (1.5), Native American (0.74), Mixed (2.9), Unknown (2.2) and other (0). Ethnicity was as follows: non-Hispanic (97%), Hispanic (0.7%) and Unknown (2.2%).
Of the 136 patients, 48 (35.3%) patients were confirmed to have autism by chart review: high-evidence in none, mid-evidence in 12 (25%), and low-evidence in 36 (75%). In 14 (10.3%) patients, autism was excluded. In 59 (43.4%) patients, there were no data to support or refute an autism diagnosis. In 15 (11%) patients, data were too limited to confirm an autism diagnosis. The patients excluded by chart review were comparable in age and sex (21.8 years; 64% male) to those who were confirmed to have autism by chart review (22.5 years; 71% male), and did not differ in race or ethnicity (all p values > 0.1).
Scores and number of notes for the categories of charts are summarized in Table 2, with box plots in Figure 3. The mid and low evidence patients confirmed to have autism had higher average and median scores than those in the categories of exclusion, no diagnosis, or limited data (t = 7.29; p < 0.0001).
As with Cohort 1, those patients with limited data (Category 6) were not included. Charts that had 75% or more of the notes coded as NA (no diagnosis) were scored as 0 and were considered to not represent true cases of autism. An ROC curve and confusion matrix for Cohort 2 had a sensitivity of 0.95 and PPV of 0.70, a specificity of 0.73, and optimal cutpoint of 0.8.
Comparison of Cohorts 1 and 2 with Respect to Age at First Mention of Autism, Autism Specialists Seen, and Co-occurring Conditions (Tables 3-4).
Table 3.
Comparison of Cohorts 1 and 2: Age at First Mention of ASD, Autism Specialists Seen, and Co-occurring Conditions
| Cohort 1 | Cohort 2 | t (df) | p-value | |
|---|---|---|---|---|
| Age at First Mention of ASD (Mean, SD) | 9.3 (4.2) years | 11.4 (3.7) years | −3.5 (60.9) | <0.0001 |
| Cohort 1 | Cohort 2 | chi-square | p-value | |
| Autism Specialists Seen (Percentage) | ||||
| Developmental Medicine | 26% | 10% | 4.97 | 0.026 |
| Psychiatry | 48% | 19% | 13.44 | 0.0002 |
| Psychology | 34% | 23% | 2.01 | 0.156 |
SD = standard deviation; df = degrees of freedom
Table 4.
Comparison of Cohorts 1 and 2: Co-occurring Conditions
| Top Six Co-occurring Conditions (ICD-9 Codes) | |
|---|---|
| Cohort 1 | Cohort 2 |
| Attention deficit disorder with hyperactivity (314.01) | Constipation, unspecified (564.00) |
| Other Convulsions (780.39) | Abdominal pain, unspecified site (789.00) |
| Constipation, unspecified (564.00) | Other Convulsions (780.39) |
| Anxiety state, unspecified (300.00) | Allergic rhinitis, cause unspecified (477.9) |
| Abdominal pain, unspecified site (789.00) | Attention deficit disorder with hyperactivity (314.01) |
| Unspecified episodic mood disorder (296.90) | Anxiety state, unspecified (300.00) |
| Percentage of Co-occurring Conditions | ||||
|---|---|---|---|---|
| Condition | Cohort 1 | Cohort 2 | chi-square | p-value |
| Attention deficit disorder with hyperactivity | 47% | 21% | 10.59 | 0.001 |
| Other Convulsions | 45% | 23% | 7.68 | 0.006 |
| Constipation, unspecified | 39% | 35% | 0.11 | 0.74 |
| Anxiety state, unspecified | 35% | 21% | 3.05 | 0.08 |
| Abdominal pain, unspecified site | 33% | 29% | 0.13 | 0.71 |
| Unspecified episodic mood disorder | 32% | 15% | 5.40 | 0.02 |
| Allergic rhinitis, cause unspecified | 15% | 20% | 0.62 | 0.43 |
The mean age at which autism was first mentioned was younger for Cohort 1 compared to Cohort 2 and visits to developmental medicine and psychiatry were also proportionally higher in Cohort 1. The top six conditions in both Cohorts 1 and 2 are shown in Table 4. Five of the Cohort 1 conditions were also among the top six co-occurring conditions identified in Cohort 2. Allergic rhinitis was the only condition included in the top six for Cohort 2 (fourth most frequent condition) that was not among the top six conditions observed in Cohort 1. Cohorts 1 and 2 had similar proportions of the majority of co-occurring conditions (Table 4), with the exceptions being that the proportion of patients with ADHD, convulsions, and mood disorder were significantly higher in Cohort 1.
Discussion
In this study, we demonstrated a practical strategy for identifying charts of autistic adults in a large de-identified EHR. Our goal was to develop an efficient EHR-based strategy for researchers to identify cases of autism. By incorporating an NLP tool to identify autism key terms, we were able to identify autism charts with high sensitivity, even in the absence of ICD codes. Specificity was improved when the number of notes was limited to four or more per chart.
Out of a total of 409 adults confirmed as having an autism diagnosis, 88% had an ICD code (i.e., Cohort 1). The remaining 12% of confirmed cases of autism were included in Cohort 2 and were identified using only key terms from notes. The age that autism was first mentioned was younger in those with ICD codes. For patients with ICD codes, there were also a higher proportion seen by specialist clinicians more likely to make autism diagnoses (e.g., developmental medicine, psychiatry). These findings reflect that children with autism receiving an ICD code are being seen at a younger age and are seen by clinicians who specialize in diagnosing autism and are more likely to bill ICD codes for autism in their practices. Confirmed cases of autism with ICD codes were comparable to those identified solely by key terms in relation to many of the top six co-occurring conditions. Five of the top conditions were shared by both cohorts (mood disorder was included in the top six conditions in Cohort 1 and allergic rhinitis was included in the top six conditions in Cohort 2). Several of these conditions were recorded with a significantly higher percentage of patients in Cohort 1 in comparison to Cohort 2—ADHD, convulsions, and mood disorder. This difference may reflect that patients in Cohort 1, who were diagnosed earlier, may have had more time to accumulate diagnoses, or reflect differing referral patterns (e.g., common referrals to psychiatry) in those diagnosed by autism specialists.
Our study is unique in that we not only focused on charts with autism-related ICD codes but also included charts without autism-related ICD codes, whose notes contained autism key terms. All charts were reviewed manually to determine whether the patients met criteria for autism. Previous studies have focused on ICD codes to identify autistic patients, which may result in the inclusion of controls without autism diagnoses, or the exclusion of cases with autism diagnoses. Our validated pipeline approach identified autism charts with high sensitivity, even in the absence of ICD codes for charts from patients with autism retrieved from the EHR. Here we discuss features of this validated pipeline approach, including the decisions made in its development, and future refinements that would optimize its performance.
Cohort Selection, and Proportion of Confirmed Cases in each Cohort
We chose to analyze two different cohorts of patient charts, with Cohort 1 containing ICD codes for autism and Cohort 2 without ICD codes for autism, as we anticipated that those containing ICD codes would have a higher pre-test probability of an autism diagnosis. We found this to be correct-- the proportion of charts in Cohort 1 having a confirmed ASD diagnosis was 86%, compared to 35% in Cohort 2.
We also found, somewhat unexpectedly, that physicians evaluating patients excluded autism in 11 patients’ charts in Cohort 1, with no evidence for autism in an additional 27 charts in Cohort 1, even in the presence of an ICD-CM diagnosis code for autism. This likely reflected that the patient had been given a provisional diagnosis of autism by someone in the healthcare field to facilitate referrals for evaluations, even though this diagnosis was not reflected in the actual clinic note and reflects one of the limitations of ICD-CM codes. It is also possible that an ICD-CM code may have been entered in error by a provider, or that the notes were not complete (did not adequately document autistic features). The timing of visits may also have played a role, where an ICD code was assigned and then a clinical evaluation subsequently excluded the diagnosis of autism.
Despite the absence of ICD-CM codes in Cohort 2, 35% of patient charts in that cohort did have key terms substantiating an autism diagnosis. This finding emphasizes that relying on ICD-CM codes alone may miss the diagnosis of autism, especially if the treating health care provider is seeing them primarily for a different condition (e.g., sleep, GI, epilepsy, depression), and supports the use of key terms derived from clinical notes (Brooks et al., 2021).
Scoring Approach
We chose to score individual notes in each chart and created an average and median score for each chart. While analyses of the cases with an autism diagnosis documented a significantly higher average score compared to those without autism, average and median scores were comparable for the high, mid, and low evidence charts in Cohort 1, and mid and low evidence charts in Cohort 2 (there were no high evidence charts in Cohort 2). This finding shows that our current methodology does not distinguish autism cases based on level of evidence.
We explored the alternative approach of programming to identify a “high evidence” chart (on the basis of a psychology note) but found that this approach had challenges. For example, a diagnosis might evolve over time, as illustrated in Figure 2C, where a child was initially given an autism diagnosis and this was subsequently revised. By using an approach that combined scores across multiple charts, we were able to ascertain a chart based on all of the clinical notes that were available in the EHR for that particular patient. While we accounted for potential differences in the number of available notes per patient across charts by creating an average, we did recognize the limitation that charts with large numbers of notes might be more accurate. This limitation was worked into the adjustment to optimize ROC curves for Cohort 1 (see below), and also in our restricting Cohort 2 charts to those containing four or more notes.
Adjustments to Optimize ROC Curves
To optimize ROC curves in both cohorts, patient charts without any notes with positive autism diagnoses were considered to not represent cases of autism and rescored manually post-hoc as zero. In other words, if key terms were absent, or only reflected a family history of autism or use of the cardiac ASD abbreviation (atrial septal defect), the patient chart was assigned a zero instead of an NA. As autism is heritable (Colvert et al., 2015; Sandin et al., 2014) and many patients with autism have a positive family history (e.g., sibling with autism), we elected not to assign zeros to individual elements within a chart that otherwise was positive for an autism diagnosis.
Additional optimizations were made in Cohort 1 and Cohort 2. In Cohort 1, to optimize ROC curves, analyses were rerun limiting charts to those with four or more notes, as a higher number of notes was assumed to reduce uncertainty in diagnosis. This resulted in a slightly lower sensitivity but a higher specificity to the initial ROC curve. The vast majority of charts that were classified as high and mid-evidence already had four or more notes; it was the low evidence charts where the largest differences in notes were observed. For the low evidence charts, the lower value in the range of scores was higher, although the average and median were unchanged.
In Cohort 2, as charts were already limited to those with four or more notes, charts with 75% of notes coded as NA (no diagnosis) were scored as 0.
Given our results, we recommend that a combination of ICD codes and key terms be used when using the EHR to identify cases of ASD.
In clinical research, our goal is often to identify as many cases of autism as possible, and then confirm the diagnosis of autism in the process of conducting the study. For example, for recruitment of autistic participants for a clinical trial, once identified using the EHR, we would then confirm their diagnosis in the screening process. For an observational study, we would review their charts in more depth as part of the data collection process, confirming their diagnosis during this chart review. Therefore, to meet our goals, we would aim for a higher sensitivity. The use of a scoring algorithm is expected to improve trial efficiency. For this study, each chart required about 10-20 minutes to manually review, depending on the composition of notes. Using an average of 15 minutes per chart, for a large dataset of 5,000 charts, this would equate to 1,250 hours of effort. If an algorithm such as ours, which uses a combination of ICD codes and key terms derived from the EHR to identify cases of autism, is applied a priori much of this manual review time could be saved. Confirmation of diagnosis could then be performed during the screening process for a clinical study or more in-depth chart review.
Although not incorporated into our algorithm, we compared findings for one vs. two or more ICD codes in Cohort 1 in relation to confirmed cases of ASD. We found that 60% of patients with only one ICD code were confirmed to have ASD by chart review compared to 86% in the entire sample of Cohort 1 charts. Therefore, requiring two or more ICD codes to identify an individual as having ASD would be expected to increase specificity, although sensitivity would be lost.
We recognize that other researchers’ goals may differ. If the goal is to have better accuracy in detecting cases, a higher specificity may be appropriate. In future work, these optimizations can be incorporated into the programming of our validating pipeline to improve performance
Study Limitations
Because this was a pilot study, charts were limited to patients who were 20-25 years old at the time of data analysis. We could have planned to analyze a larger cohort with a wider age range (e.g., 18–35-year-olds), or include only children, but chose to start with a small cohort of adults to gain experience with the methodology, given that our goals were to identify autistic young adults for future studies. In future samples, charts from children, adolescents, and a wider age range of adults should be analyzed to replicate these findings. Our findings will also require confirmation in larger samples and replication in other EHR systems.
Another limitation is that our algorithm may preferentially detect autistic adults who are more severely affected, given that they may have more medical visits than those who are less severely affected. This is a general limitation of any study that focuses on the use of the EHR to identify cases. People with private insurance may also have better access to services, translating into more medical visits and documentation of their conditions in the EHR—notably, the type of insurance is not recorded in the de-identified database used in this study and effects of insurance should be evaluated in future work. People who received their autism diagnosis in childhood in a different medical system and then moved into our area/medical system would also be less likely to be picked up by our algorithms. However, some of these individuals would have been missed if we were relying solely on ICD diagnoses, and not incorporating key terms into our algorithm. Finally, because of diagnostic changes and cohort effects, results may not be applicable to middle aged and older autistic adults. Other limitations were that we were unable to comment on intellectual disability status, as it is not consistently documented in our EHR.
One of the limitations related to ICD codes is that certain codes, such as 299.8 and 299.9 may refer to other conditions besides Asperger’s syndrome and pervasive developmental disorder, including childhood psychosis and schizophrenia. However, if the goal of these algorithms is increased sensitivity, then including 299.8 and 299.9 allows for inclusion of more cases that can then be reviewed during the process of conducting a study (e.g., screening for a clinical trial or data collection). There were also limitations in the KMCI approach, with some text falsely negated. For example, as shown in Figure 2B, awkwardly constructed sentences might be incorrectly negated. This limitation is challenging, especially in large health systems with numerous health care providers who do not use a similar sentence structure for their charting. If only a few health care providers are involved in charting, programming can take their charting styles into account, but this may not be feasible when multiple health care providers are involved. The score approach was implemented, in part to overcome this limitation.
In summary, our findings support that the application of NLP methodology is feasible in the identification of autistic adults in an EHR system. Our work may also serve as a model for phenotyping other complex conditions, especially conditions lacking laboratory tests or other diagnostic measures to classify cases. A major advantage of EHR data, linked with biobanked samples, is sufficient sample size to study genetic associations across cohorts. In this case, the quality of the cohort is key for increasing accuracy and precision in the diagnosis and treatment of genetic disorders. The two autism cohorts created in our study demonstrate that we should not include cases with only ICD codes, as some cases of autism may lack ICD codes. Conversely, we should not define controls solely on the basis of their lacking ICD codes. Our work also emphasizes the importance of having an interdisciplinary team – in addition to researchers, including clinicians who are familiar with the study population, and informatics experts familiar with programming and the application of NLP in the field of medical informatics, open-source products such as KMCI.
Supplementary Material
Acknowledgements
Dr. Josh Smith from Vanderbilt University Medical Center provided input and review into portions of the manuscript related to KnowledgeMap Concept Indexer. Debbie Wofford helped with organization of the data and manual chart review. This work was directly supported by a NIH grant to Dr. Malow (K18 MH122755). We would like to acknowledge the resources graciously provided by the VUMC SD, whose projects are supported by numerous sources: institutional funding, private agencies, and federal grants including the National Center for Advancing Translational Sciences (NCATS) Clinical Translational Science Award (CTSA) Program, Award Number 5UL1TR002243.
Footnotes
Conflict of Interest
No authors have conflicts of interest
To honor the preferences, autonomy, and rights of the autistic community, we have chosen to use identify-first language (e.g., autistic person) in this article when referring to autistic adults (Bottema-Beutel, 2021). However, when referring to our algorithms for identifying autism in medical records, for accuracy and completeness, we refer to terms such as autism, autism spectrum disorder, and Asperger syndrome.
References
- Bishop-Fitzpatrick L, Movaghar A, Greenberg JS, Page D, DaWalt LS, Brilliant MH, & Mailick MR (2018). Using machine learning to identify patterns of lifetime health problems in decedents with autism spectrum disorder. Autism Research, 11(8), 1120–1128. 10.1002/aur.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottema-Beutel K,. Kapp SK, Lester JN, Sasson NJ, and Hand BN. (2021). Avoiding ableist language: Suggestions for autism researchers. Autism in Adulthood, 3(1): 18–29. 10.1089/aut.2020.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JD, Bronskill SE, Fu L, Saxena FE, Arneja J, Pinzaru VB, Anagnostou E, Nylen K, McLaughlin J, & Tu K (2021). Identifying children and youth with autism spectrum disorder in electronic medical records: Examining health system utilization and comorbidities. Autism Research, 14(2), 400–410. 10.1002/aur.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush RA, Connelly CD, Pérez A, Barlow H, Chiang GJ (2017). Extracting autism spectrum disorder data from the electronic health record. Appl Clin Inform. 19;8(3):731–741. doi: 10.4338/ACI-2017-02-RA-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG (2001a). Evaluation of negation phrases in narrative clinical reports. Proc AMIA Symp. 105–9. [PMC free article] [PubMed] [Google Scholar]
- Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG (2001b). A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 34(5):301–10. [DOI] [PubMed] [Google Scholar]
- Colvert E, Tick B, McEwen F, Stewart C, Curran SR, Woodhouse E, Gillan N, Hallett V, Lietz S, Garnett T, Ronald A, Plomin R, Rijsdijk F, Happé F, & Bolton P (2015). Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry, 72(5), 415–423. 10.1001/jamapsychiatry.2014.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie MR, Blomster JI, Curtis LH, Duclaux S, Ford I, Fritz F, Goldman S, Janmohamed S, Kreuzer J, Leenay M, Michel A, Ong S, Pell JP, Southworth MR, Stough WG, Thoenes M, Zannad F, & Zalewski A (2017). Electronic health records to facilitate clinical research. Clinical Research in Cardiology, 106(1), 1–9. 10.1007/s00392-016-1025-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croen LA, Zerbo O, Qian Y, Massolo ML, Rich S, Sidney S, & Kripke C (2015). The health status of adults on the autism spectrum. Autism,19(7), 814–823. 10.1177/1362361315577517 [DOI] [PubMed] [Google Scholar]
- Davignon MN, Qian Y, Massolo M, & Croen LA (2018). Psychiatric and Medical Conditions in Transition-Aged Individuals With ASD. Pediatrics, 141(Suppl 4), S335–S345. 10.1542/peds.2016-4300K [DOI] [PubMed] [Google Scholar]
- Denny JC, Irani PR, Wehbe FH, Smithers JD, & Spickard A 3rd (2003). The KnowledgeMap project: development of a concept-based medical school curriculum database. AMIA … Annual Symposium proceedings. AMIA Symposium, 195–199. [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Spickard A 3rd, Miller RA, Schildcrout J, Darbar D, Rosenbloom ST, & Peterson JF (2005). Identifying UMLS concepts from ECG Impressions using KnowledgeMap. AMIA … Annual Symposium proceedings. AMIA Symposium, 196–200. [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Spickard A 3rd, Johnson KB, Peterson NB, Peterson JF, & Miller RA (2009). Evaluation of a method to identify and categorize section headers in clinical documents. Journal of the American Medical Informatics Association: JAMIA, 16(6), 806–815. 10.1197/jamia.M3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla MD, Schwartz KL, Chaganti S, Cutting LE, Landman BA, & Cascio CJ (2021). Using phecode analysis to characterize co-occurring medical conditions in autism spectrum disorder. Autism, 25(3), 800–811. 10.1177/1362361320934561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KB, Cottle K, Bakian A, Farley M, Bilder D, Coon H, & McMahon WM (2016). A description of medical conditions in adults with autism spectrum disorder: A follow-up of the 1980s Utah/UCLA Autism Epidemiologic Study. Autism, 20(5), 551–561. 10.1177/1362361315594798 [DOI] [PubMed] [Google Scholar]
- Lawson LP, Richdale AL, Haschek A, Flower RL, Vartuli J, Arnold SR, & Trollor JN (2020). Cross-sectional and longitudinal predictors of quality of life in autistic individuals from adolescence to adulthood: The role of mental health and sleep quality. Autism, 24(4), 954–967. 10.1177/1362361320908107. [DOI] [PubMed] [Google Scholar]
- Lingren T, Chen P, Bochenek J, Doshi-Velez F, Manning-Courtney P, Bickel J, Wildenger Welchons L, Reinhold J, Bing N, Ni Y, Barbaresi W, Mentch F, Basford M, Denny J, Vazquez L, Perry C, Namjou B, Qiu H, Connolly J, Abrams D, … Savova G (2016). Electronic Health Record based algorithm to identify patients with autism spectrum disorder. PloS One, 11(7), e0159621. 10.1371/journal.pone.0159621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner MJ, Shaw KA, Bakian AV, Bilder DA, Durkin MS, Esler A, Furnier SM, Hallas L, Hall-Lande J, Hudson A, Hughes MM, Patrick M, Pierce K, Poynter JN, Salinas A, Shenouda J, Vehorn A, Warren Z, Constantino JN, DiRienzo M, … Cogswell ME (2021). Prevalence and characteristics of autism spectrum disorder among children aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2018. Morbidity and mortality weekly report. Surveillance summaries (Washington, D.C. 2002), 70(11), 1–16. 10.15585/mmwr.ss7011a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox BB, Dickson KS, Stadnick NA, Mandell DS, & Brookman-Frazee L (2021). Mental Health Services for Autistic Individuals Across the Lifespan: Recent Advances and Current Gaps. Current psychiatry reports, 23(10), 66. 10.1007/s11920-021-01278-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, & Masys DR (2008). Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clinical Pharmacology and Therapeutics, 84(3), 362–369. 10.1038/clpt.2008.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, & Reichenberg A (2014). The familial risk of autism. JAMA, 311(17), 1770–1777. 10.1001/jama.2014.4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck P Drexel University, (2019). Growing numbers of young adults on the autism spectrum. https://drexel.edu/autismoutcomes/blog/overview/2019/June/Growing-numbers-of-young-adults-on-the-autism-spectrum/. Accessed May 20, 2022. [Google Scholar]
- Singer EV, Niarchou M, Maxwell-Horn A, Hucks D, Johnston R, Sutcliffe JS, Davis LK, & Malow BA (2022). Characterizing sleep disorders in an autism-specific collection of electronic health records. Sleep Medicine, 92, 88–95. 10.1016/j.sleep.2022.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhausen HC, Mohr Jensen C, & Lauritsen MB (2016). A systematic review and meta-analysis of the long-term overall outcome of autism spectrum disorders in adolescence and adulthood. Acta Psychiatrica Scandinavica, 133(6), 445–452. 10.1111/acps.12559 [DOI] [PubMed] [Google Scholar]
- Thiele C, & Hirschfeld G (2021). cutpointr: Improved Estimation and Validation of Optimal Cutpoints in R. Journal of Statistical Software, 98(11), 1–27. 10.18637/jss.v098.i11 [DOI] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York. Retrieved from https://ggplot2.tidyverse.org [Google Scholar]
- World Health Organization (1992). International classification of diseases and related health problems (10th rev., ICD-10). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.