Abstract
Current mass spectrometry methods enable high-throughput proteomics of large sample amounts, but proteomics of low sample amounts remains limited in depth and throughput. To increase the throughput of sensitive proteomics, we developed an experimental and computational framework, called plexDIA, for simultaneously multiplexing the analysis of peptides and samples. Multiplexed analysis with plexDIA increases throughput multiplicatively with the number of labels without reducing proteome coverage or quantitative accuracy. By using three-plex non-isobaric mass tags, plexDIA enables quantification of threefold more protein ratios among nanogram-level samples. Using 1-hour active gradients, plexDIA quantified ~8,000 proteins in each sample of labeled three-plex sets and increased data completeness, reducing missing data more than twofold across samples. Applied to single human cells, plexDIA quantified ~1,000 proteins per cell and achieved 98% data completeness within a plexDIA set while using ~5 minutes of active chromatography per cell. These results establish a general framework for increasing the throughput of sensitive and quantitative protein analysis.
Mass spectrometry (MS) methods can achieve deep proteome coverage1,2, low missing data3, high throughput4,5 and high sensitivity6. However, simultaneously achieving all these objectives is a considerable challenge7,8. Resolving this challenge will empower biomedical projects that are impractical with current methods8, especially those that require single-cell protein analysis9-11. Toward this goal, the throughput of sensitive protein analysis may be increased by different strategies: (1) increasing sample throughput and robustness by chemical labeling and (2) decreasing MS analysis time per sample by simultaneous (parallel) analysis of multiple peptides. These strategies are complementary, and we sought to combine them to achieve a multiplicative increase in the rate of quantifying the proteomes of limited sample amounts.
Chemical labeling is often used with data-dependent acquisition (DDA) to increase throughput via parallel sample analysis (Fig. 1a) and to control for shared artifacts, such as disturbances in peptide separation and ionization12-14. Because quantifying a mammalian proteome requires analyzing hundreds of thousands of precursor ions, and DDA methods analyze one precursor per MS2 scan, even the most optimized DDA methods require up to 1 day of liquid chromatography with tandem mass spectrometry (LC–MS/MS) for deep proteome analysis1. Non-isobaric labels, such as mTRAQ and dimethyl labeling, allow for sample multiplexing but further increase the number of precursor ions and, thus, the time needed for MS1-multiplexed DDA analysis15. In contrast, approaches using isobaric labels (such as tandem mass tags (TMTs)) do not increase the number of distinguishable precursor ions and can reduce the analysis time per sample16,17, albeit quantification with TMT is often affected by co-isolation interference13,18.
Fig. 1 ∣. Experimental design for acquisition and evaluation of plexDIA data.
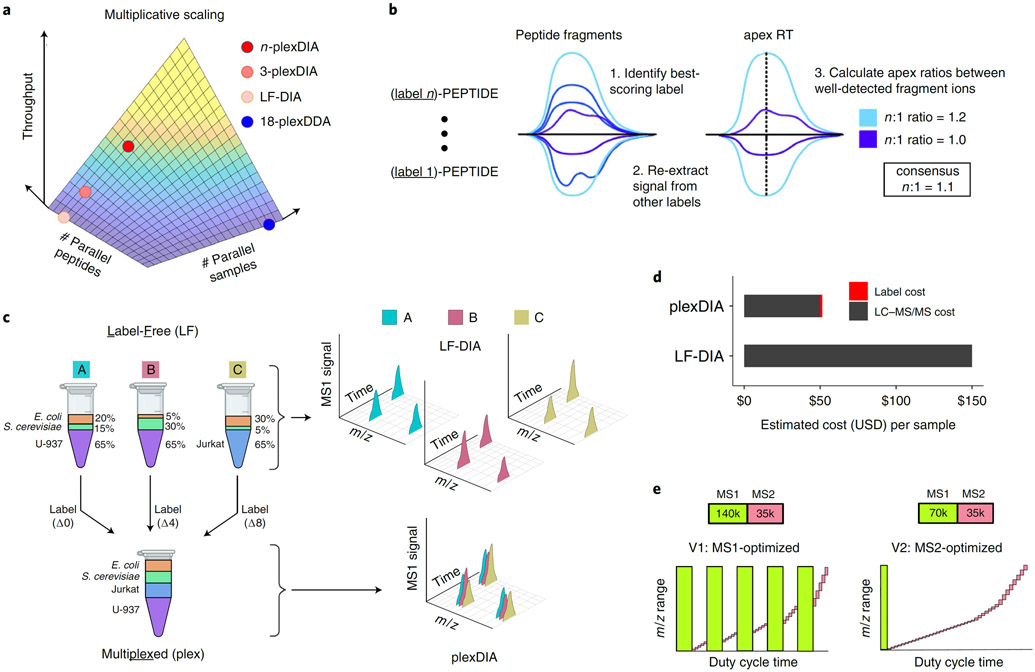
a, The throughput of MS proteomics can be increased by parallel analysis of multiple peptides or by parallel analysis of multiple samples. plexDIA aims to combine both approaches to achieve multiplicative gains. b, Precursor identifications from one label can be confidently transferred to isotopologous precursors with FDR control. The abundance of labeled precursors can be estimated by the consensus fold change relative to best quantified isotopologous precursor. c, Standards used for benchmarking LF-DIA and plexDIA quantification were prepared by mixing the proteomes of different species and cell types as shown. LF-DIA analyzed 500 ng from each of the three samples (A, B and C) separately, whereas plexDIA analyzed a mixture of these samples labeled with non-isobaric mass tags (mTRAQ). d, Analyzing samples by plexDIA is cheaper than analysis by LF-DIA because running n samples in parallel reduces the LC–MS/MS time per sample n-fold, and the cost of labeling is low. This estimate is based on a facility fee of $150 USD per hour of active gradient. e, We benchmarked the performance of plexDIA with two DIA methods, V1 and V2. V1 is an MS1-optimized method that uses frequent, high-resolution MS1 scans to facilitate accurate quantification, whereas V2 is an MS2-optimized method that takes a single MS1 scan and more MS2 scans per duty cycle.
The throughput of DDA analysis can be increased by decreasing the ion accumulation times for MS2 scans, although this results in accumulating fewer ions and, thus, limits sensitivity7. Indeed, sensitive analysis of small sample amounts requires (and is, thus, limited by) long ion accumulation times, which are typically substantially longer than the detection time required by MS detectors6,19,20. Even with short ion accumulation times for unlimited sample amounts, the requirement to serially analyze hundreds of thousands of precursor ions remains a major challenge for simultaneously achieving high throughput and deep proteome coverage by serial precursor analysis.
A fundamental solution to this challenge is the concept of isolating and analyzing multiple precursor ions simultaneously by data-independent acquisition (DIA)21. This concept has been implemented into methods for label-free DIA (LF-DIA) protein analysis22-26. Such parallel analysis of peptides decreases the time needed to analyze thousands of precursor ions and makes the throughput of optimized LF-DIA and TMT-DDA workflows similar (Fig. 1a), allowing routine quantification of about 6,000 proteins in 2 hours17. Recent DIA technologies further enabled quantification of over 8,000 proteins in 1.5 hours27, and TMTpro tags increased multiplexing to 18-plex for DDA methods4. Thus, multiplexed DDA and LF-DIA afford similar throughput (Fig. 1a).
We sought to further increase the throughput of sensitive DIA by multiplexing samples labeled with non-isobaric isotopologous mass tags, capitalizing on the fact that increasing the number of precursor ions does not increase the time needed to analyze them via tandem DIA-MS, in contrast to DDA-MS15,21. This creates a hypothetical possibility that we sought to test: the number of proteins accurately quantified by multiplexed DIA may increase multiplicatively with the number of labels used (Fig. 1a). If feasible, this possibility may enable higher throughput and more sensitive multiplexed proteomics, including single-cell proteomics, as previously suggested7,28. Although the feasibility of DIA multiplexed by SILAC29 or pulsed SILAC30,31 has been clearly demonstrated, its ability to multiplicatively increase quantitative data points remains unclear. Similarly, clever strategies have used both isobaric and isotopologous tags to multiplex DIA, but they have afforded the quantification of relatively few proteins32-34. Thus, the potential of multiplexed DIA to increase sample throughput while preserving proteome coverage and quantification accuracy has not been realized due to the increased complexity of DIA data from labeled samples33-37.
We hypothesized that an optimized experimental and analytical framework may enable n-fold multiplexed DIA to increase n-fold the number of accurate protein data points (Fig. 1a). We test this hypothesis for n = 3 using amine-reactive non-isobaric isotopologous mass tags (mTRAQ), hoping that this particular choice of mass tags can establish a framework that will, in the future, generalize to a variety of isotopologous non-isobaric mass tags with higher capacity for multiplexing. Specifically, we sought to develop a general framework and an analysis pipeline to increase the throughput of sensitive and quantitative protein analysis via plexDIA.
Results
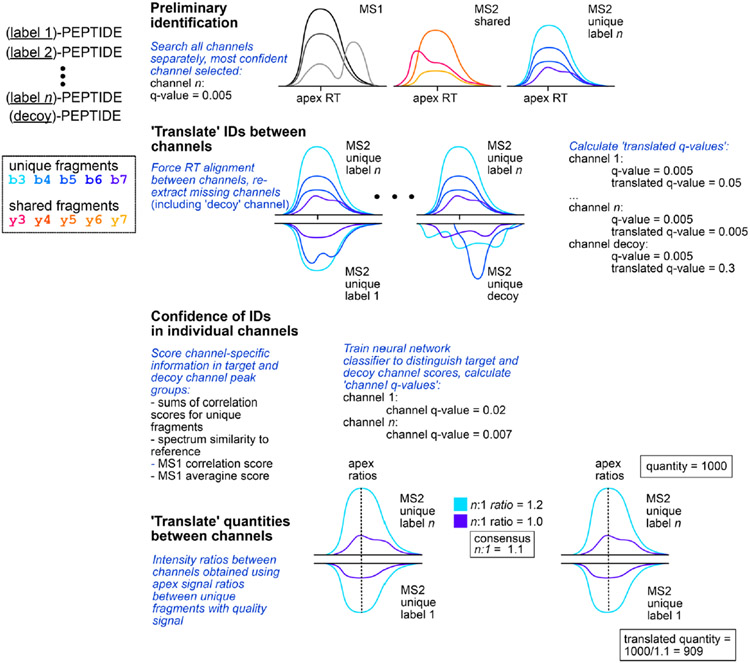
To enhance MS data interpretation, the plexDIA module of DIA-NN capitalizes on the expected regular patterns in the data, such as identical retention times and known mass shifts between the same peptide labeled with different isotopologous mass tags (Fig. 1b and Extended Data Fig. 1)7. DIA-NN uses neural networks to confidently identify labeled peptides, and these identifications are then used to re-extract data for the same peptide labeled with a different tag. Neural networks then calculate false discovery rates (FDRs) for all peptides based on a decoy channel strategy, which is empirically validated by two-species spiked experiment shown in Extended Data Fig. 2. Despite the n-fold increased spectral complexity, the plexDIA framework aims to accurately quantify peptides by calculating ratios of fragments from the most confident isotopologous precursor to the translated isotopologous precursors at the apex where the signal is greatest and the effect of interference is lowest. The mean fragment ratio is used to scale the precursor quantity of the best isotopologous precursor to the less-confident isotopologous precursors (Fig. 1b).
plexDIA benchmarks.
We sought to evaluate whether plexDIA can multiplicatively increase the number of quantitative data points relative to matched LF-DIA analysis while maintaining similar quantitative accuracy. Toward that goal, we mixed proteomes in precisely specified ratios shown in Fig. 1c, thus creating a benchmark of known protein ratios for thousands of proteins spanning a wide dynamic range of abundances, similar to previous benchmarks23. Specifically, we made three samples (A, B and C), each with an exactly specified amount of Escherichia coli, Saccharomyces cerevisiae and Homo sapiens (U-937 and Jurkat) cell lysate (Fig. 1c). A distinct aspect of this design is the incorporation of human proteomes of different cell types, which affords additional benchmarking for the reproducibility of protein identification across diverse samples and for relative protein quantification.
Each sample was either analyzed by LF-DIA or labeled with one of three amine-reactive isotopologous chemical labels (mTRAQ: Δ0, Δ4 or Δ8) (Fig. 1c). With this experimental design, plex-DIA enables threefold reduction in LC–MS/MS time per sample (Fig. 1d). The combined labeled samples were analyzed by plexDIA, and the result was used to benchmark proteomic coverage, quantitative accuracy, precision and repeatability across runs relative to LF-DIA of the same samples. LF-DIA and plexDIA were evaluated with two data acquisition methods, V1 and V2, shown in Fig. 1e. V1 included multiple high-resolution MS1 survey scans to increase the temporal resolution of precursor sampling, as previously reported3, whereas V2 included more MS2 scans to increase proteome coverage (Fig. 1e). The only difference between the duty cycles of LF-DIA and plexDIA was a 100-m/z increment in the MS1 and MS2 windows of plexDIA to account for the mass of mTRAQ added to the peptides (Methods).
plexDIA increases throughput multiplicatively.
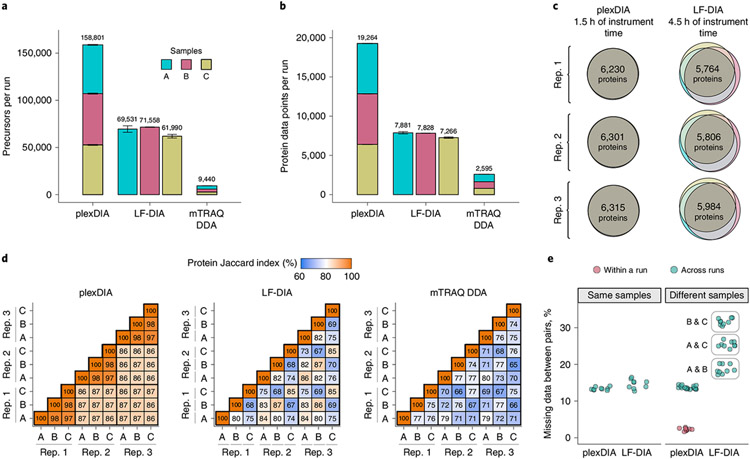
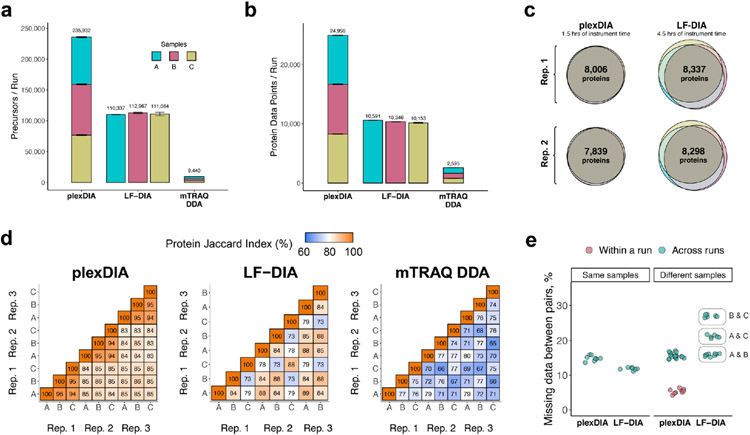
To directly benchmark the analysis of 500-ng protein samples by plexDIA relative to LF-DIA, the multiplexed and LF samples described in Fig. 1c were analyzed in triplicate by LC–MS/MS on a Q Exactive orbitrap (first generation, Thermo Fisher Scientific) with a 60-minute active nano-LC gradient. The throughput increases for duty cycles V1 (Fig. 2) and V2 (Extended Data Fig. 3) were similar, except that V2 achieved greater proteome coverage with both plexDIA and LF-DIA. The parallel data acquisition by all DIA methods resulted in a greater number of identified peptides and proteins compared to the DDA runs (Fig. 2a,b). Both V1 and V2 resulted in approximately 2.5-fold more precursors and protein data points for plexDIA than for LF-DIA per unit time (Fig. 2a,b and Extended Data Fig. 3a,b).
Fig. 2 ∣. plexDIA proteome coverage and overlap between samples and runs.
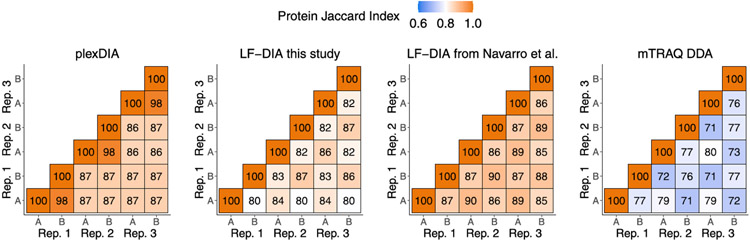
a, Number of distinct precursors identified from 60-minute active gradient runs of plexDIA, LF-DIA and mTRAQ DDA at 1% FDR. The DIA analysis employed the V1 duty cycle shown in Fig. 1c. Each sample was analyzed in triplicate (n = 3), and the results are displayed as mean; error bars correspond to standard error. b, Total number of protein data points for plexDIA, LF-DIA and mTRAQ DDA at 1% global protein FDR (n = 3). c, Venn diagrams of each replicate for plexDIA and LF-DIA display protein groups quantified across samples A, B and C. The mean number of protein groups intersected across samples A, B and C is 6,282 for plexDIA and 5,851 for LF-DIA. d, The similarity between the quantified proteins across samples is quantified by the corresponding pairwise Jaccard indices to display data completeness. e, Distributions of missing data for protein groups between pairs of runs of either the same sample (that is, replicate injections) or between different samples. All analyses used MBR. The corresponding results for the V2 duty cycle are shown in Extended Data Fig. 3.
plexDIA increases data completeness across samples.
Next, we sought to compare LF-DIA and plexDIA in terms of the consistency of protein quantification across samples. The systematic acquisition of ions by DIA is well-established as a strategy for increasing the repeatability of peptide identification relative to shotgun DDA24. In addition to consistent data acquisition, plexDIA may further reduce the variability between samples and runs and, thus, further increase the consistency (overlap) between quantified proteins relative to LF-DIA.
Indeed, both SILAC and isobaric labeling reduce missing data by enabling the quantification of peptides identified in at least one sample from a labeled set18,38. Similarly, plexDIA takes advantage of the precisely known mass shifts in the mass spectra for a peptide labeled with different tags to propagate peptide sequence identifications within a run. Specifically, confidently identified precursors in one channel (label) are matched to corresponding precursors in the other channels. This is the default analysis used with standards A, B and C. plexDIA has an additional mode for the special case when some proteins are present only in some samples of labeled sets. In such cases, plexDIA can enable sample-specific identification for each protein by using multiple MS1-based and MS2-based features to rigorously evaluate the spectral matches within a run and explicitly assign confidence for the presence of each protein in each sample. Such a special case is exemplified by a plexDIA set in which one sample has both yeast and bacterial proteins, whereas another sample has only yeast proteins (Extended Data Fig. 2). These analytical capabilities are described in the Methods.
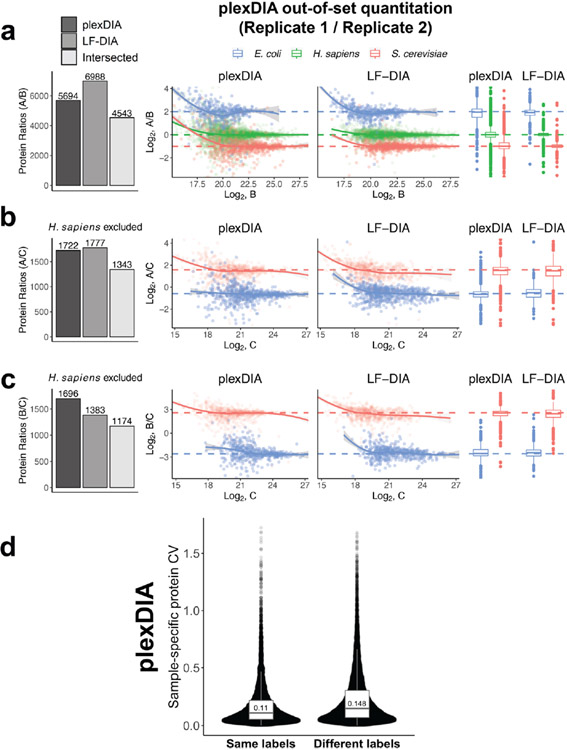
To assess whether plexDIA can improve data completeness, the protein groups quantified across samples A, B and C were plotted as Venn diagrams for each replicate of plexDIA and LF-DIA (Fig. 2c). On average, the protein groups quantified in common across samples A, B and C were 6,282 for plexDIA and 5,851 for LF-DIA. The corresponding numbers for the V2 method are 7,923 for plexDIA and 8,318 for LF-DIA (Extended Data Fig. 3c). Thus, a three-plex plexDIA increased the rate of quantifying protein ratios across all three samples by 3.22-fold for the V1 method and by 2.86-fold for the V2 method, per unit time.
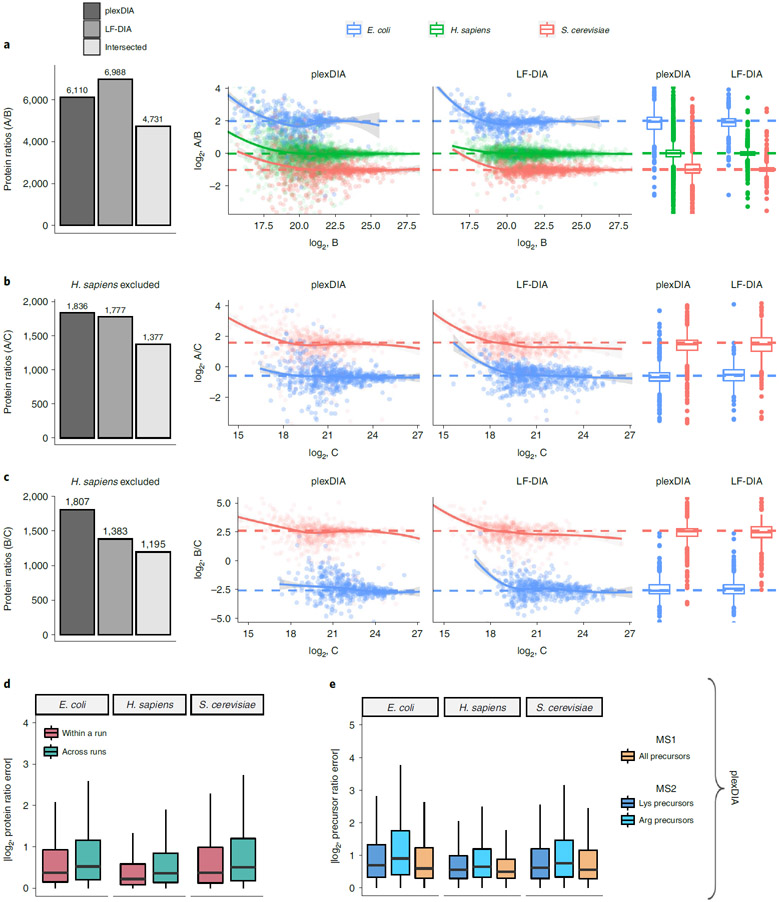
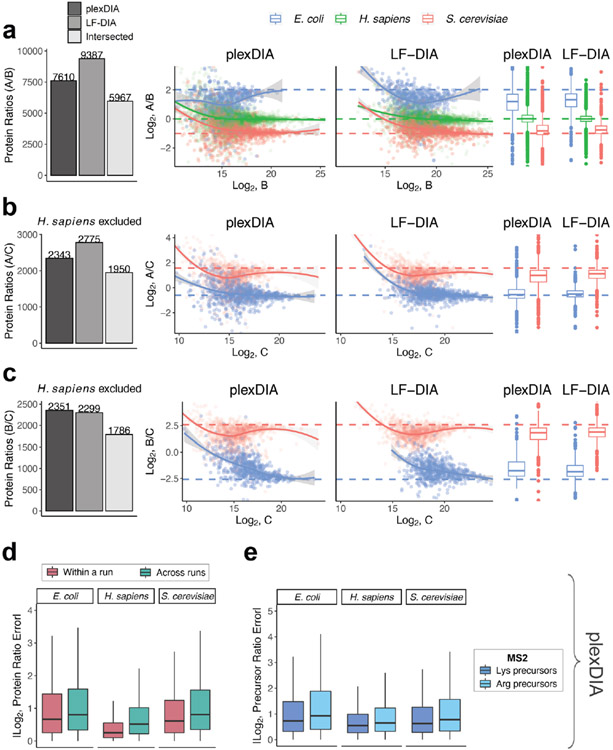
We further benchmarked the consistency of identified proteins both from the repeated analysis of the same sample (such as replicate injections of sample A) and from the analysis of different samples (such as comparing samples B and C). Consistent with previous reports for DIA data completeness, both LF-DIA and plexDIA identified largely the same proteins from replicate injections, quantified by high Jaccard indices and only about 13–15% non-overlapping proteins, as shown in Fig. 2d,e. This overlap is similar to the overlap of a high-quality LF-DIA dataset by Navarro et al.23, as shown in Extended Data Fig. 4. The overlap between the proteins identified in distinct samples remained similarly high for plexDIA, whereas it was substantially reduced for the LF-DIA analysis (Fig. 2d,e). This increased repeatability for plexDIA likely arises from the fact that samples A, B and C are analyzed in parallel as part of one set. Such a multiplexed approach also reduces missing data within a plexDIA set to 2–3% (Fig. 2d,e). The larger the difference in protein composition between two samples, the higher the fraction of missing data for LF-DIA. In contrast, the missing data for plexDIA was low across all pairs of samples (Fig. 2e). The advantages of improved data completeness by plexDIA are especially pronounced when comparing the number of protein ratios from plexDIA and LF-DIA for samples, which differ more in protein abundance—for example, B and C; sample C has sixfold more E. coli and sixfold less S. cerevisiae relative to sample B. As a result, LF-DIA allowed the quantification of only 1,383 ratios between E. coli and S. cerevisiae proteins, whereas plexDIA allowed the quantification of 1,807 protein ratios (Fig. 3a-c).
Fig. 3 ∣. Quantitative accuracy and precision of plexDIA and LF-DIA.
a, Bars correspond to the number of quantified protein ratios between samples A and B by plexDIA, by LF-DIA or by both methods (intersected proteins). To improve visibility, the scatter plot x and y axes were set to display data points between the 0.25% and 99.75% range. b, Same as a but for samples A and C. c, Same as a but for samples B and C. The protein ratios displayed in a–c are estimated from a single replicate, and two more replicates are shown in Extended Data Fig. 6. d, A comparison between the errors within and across plexDIA sets indicates similar accuracy. The error is defined as the difference between the mixing and the measured protein ratios for all pairs of samples, A/B, A/C and B/C. The absolute values of these errors are displayed for samples within a plexDIA set (for example, run2 A / run2 B) and for samples across sets (for example, run1 A / run2 B). The corresponding accuracy within and across plexDIA for the V2 methods is shown in Extended Data Fig. 5. e, Absolute precursor ratio errors were calculated for samples A/B, A/C and B/C and combined to compare ratio errors for MS1 and MS2 quantification. The MS2 quantification of precursors having C-terminal lysine or arginine is shown separately. Boxplots: The box defines the 25th and 75th percentiles, and the median is marked by a solid line. Outliers are marked as individual dots outside the whiskers.
Quantitative accuracy of plexDIA is similar to LF-DIA.
To benchmark the quantitative accuracy and precision of plexDIA and LF-DIA, we compared the measured protein ratios between pairs of samples to the ones expected from the study design (Fig. 1). Because each sample contains a known amount of E. coli, S. cerevisiae and H. sapiens protein lysate, and most peptides are unique to each species, the protein ratios between pairs of samples correspond to the corresponding mixing ratios23,24. The expected ratios allow for rigorous benchmarking of the accuracy and precision of plexDIA and LF-DIA. H. sapiens protein group ratios were excluded from analyses involving sample C as they would compare U-937 (A and B) to Jurkat (C) cell lines; therefore, deviations from expected ratios would be a combination of quantitative noise and cell-type-specific differences in protein abundance.
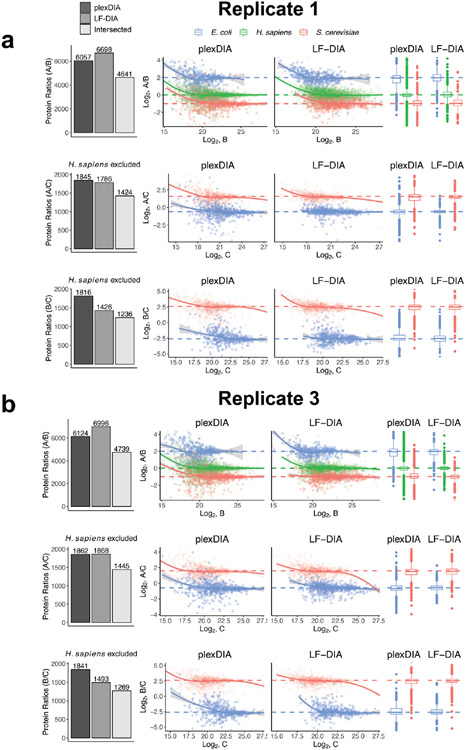
For well-controlled comparisons between the quantitative accuracy of LF-DIA and plexDIA, we used the set of protein ratios quantified by both methods. The comparison produced using the V1 method is shown in Fig. 3a-c and Extended Data Fig. 6, whereas the results from V2 are shown in Extended Data Fig. 5. These results indicate that, on average, plexDIA has similar accuracy and precision to LF-DIA. Consistent with the expectation that labeling helps to control for nuisances, the results indicate that plexDIA quantification within a set is slightly more accurate than across sets (Fig. 3d). However, the difference is small, and accuracy across different plexDIA sets is high (Extended Data Fig. 7a-c).
By design, plexDIA allows quantifying precursors based on MS2-level and MS1-level data, and we evaluated the quantitative accuracy for both levels of quantification (Fig. 3e). Because both lysine and N-terminal amine groups are labeled by the amine-reactive mTRAQ labels, both b-fragment and y-fragment ions of lysine peptides are sample specific and, thus, contribute to MS2-level quantification. In contrast, only b-ions are sample specific for arginine peptides, and, thus, only b-ions are used for their MS2-level quantification. As a result, the MS2-level quantification accuracy for arginine peptides is slightly lower (Fig. 3e). The small magnitude of this difference is likely attributable to the fact that mTRAQ stabilizes b-ions39. The accuracy of MS1 quantification by V1 is high for all peptides and slightly higher than the accuracy of MS2 quantification (Fig. 3e). The MS2-optimized duty cycle (V2) resulted in deeper proteome coverage and lower accuracy for both LF-DIA and plexDIA (Extended Data Fig. 5). However, different duty cycles implemented on different instruments will likely improve the accuracy and coverage by MS2-optimized methods.
Repeatability of plexDIA is similar to LF-DIA.
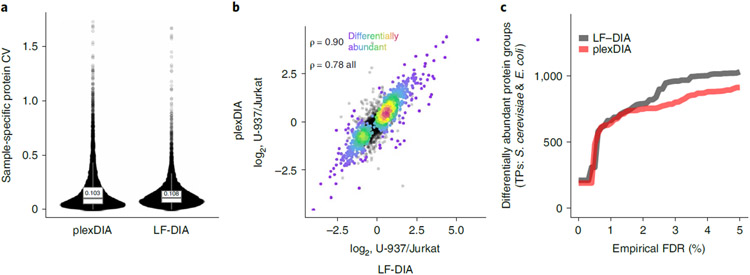
To assess the repeatability of plexDIA and LF-DIA quantitation, we computed the coefficient of variation (CV) for proteins quantified in triplicate runs for each method using MaxLFQ abundances40. We required each protein group to be quantified three times for plexDIA and LF-DIA, and then the CVs for the overlapping sample-specific protein groups (n = 12,863) were plotted in Fig. 4a. The results indicate that plexDIA and LF-DIA have relatively consistent quantitation and similar quantitative repeatability, with median CVs for repeated injections of 0.103 and 0.108, respectively. Repeatability of plexDIA was also compared for triplicates of the same labeled samples and for triplicates in which each replicate had samples with alternating labels. Median CVs for the triplicates were 0.110 and 0.148 for ‘same labels’ and ‘different labels’ experiments, respectively (Extended Data Fig. 7d).
Fig. 4 ∣. Using plexDIA to estimate differential protein abundance.
a, Quantitative repeatability was estimated by calculating CVs for MaxLFQ protein abundances (12,863 sample-specific protein data points) calculated across triplicates for plexDIA and LF-DIA. b, Proteins found to be differentially abundant between U-937 and Jurkat cells by LF-DIA were plotted as ratios of U-937/Jurkat for plexDIA and LF-DIA and colored by density. The Spearman correlation shown was calculated to quantify the agreement between the estimated relative protein levels of differentially abundant proteins at 1% FDR. Non-differentially abundant proteins are plotted in black; the Spearman correlation of all proteins (n = 2,728) and differentially abundant proteins at 1% FDR (n = 1,078) is 0.78 and 0.90, respectively. c, Number of differentially abundant proteins between samples A and B as a function of the empirical FDR. The y axis shows the number of true positives (only S. cerevisiae and E. coli proteins, which are differentially abundant), and the x axis shows the FDRs estimated from the human proteins identified to be differentially abundant. The differential abundance was estimated using three replicates from each method, and, thus, LF-DIA took three times more instrument time per sample than plexDIA. TP, true positive.
Estimating differential protein abundance by plexDIA and LF-DIA.
We investigated the agreement of differential protein abundance between U-937 and Jurkat cell lines with plexDIA and LF-DIA. Differential protein abundance was estimated from LF-DIA data, and the differentially abundant proteins at 1% FDR were used to assess the agreement between U-937 and Jurkat protein ratios estimated by plexDIA and LF-DIA (Fig. 4b). The estimates by the two methods are similar, as indicated by a Spearman correlation of 0.90 for differentially abundant proteins (n = 1,078 at 1% FDR) and a Spearman correlation of 0.78 for all intersected human proteins (n = 2,728) (Fig. 4b).
We also compared the ability of plexDIA and LF-DIA to recall true differentially abundant proteins as a function of each method’s empirical FDR. Our experimental design from Fig. 1c provides strong ground truth. It dictates that, between samples A and B, only S. cerevisiae and E. coli are differentially abundant because they were spiked in at different ratios (1:2 and 4:1, respectively), whereas human proteins are not because they are present in a 1:1 ratio and compare the same cell type (U-937 monocytes). Therefore, true positives (S. cerevisiae and E. coli proteins) and true negatives (H. sapiens proteins) are known. With this prior knowledge, we compared the number of true positives for LF-DIA and plexDIA as a function of the empirical FDR (Fig. 4c). Both methods used three replicates and performed similarly at 1% empirical FDR, with 643 proteins and 663 proteins found to be differentially abundant for plexDIA and LF-DIA, respectively. The slight increase of true positives for LF-DIA at higher empirical FDR may be due to its slightly higher precision as visible in Fig. 3. In conclusion, plexDIA achieved similar statistical power as LF-DIA while using three times less instrument time and expense.
Cell division cycle analysis with plexDIA.
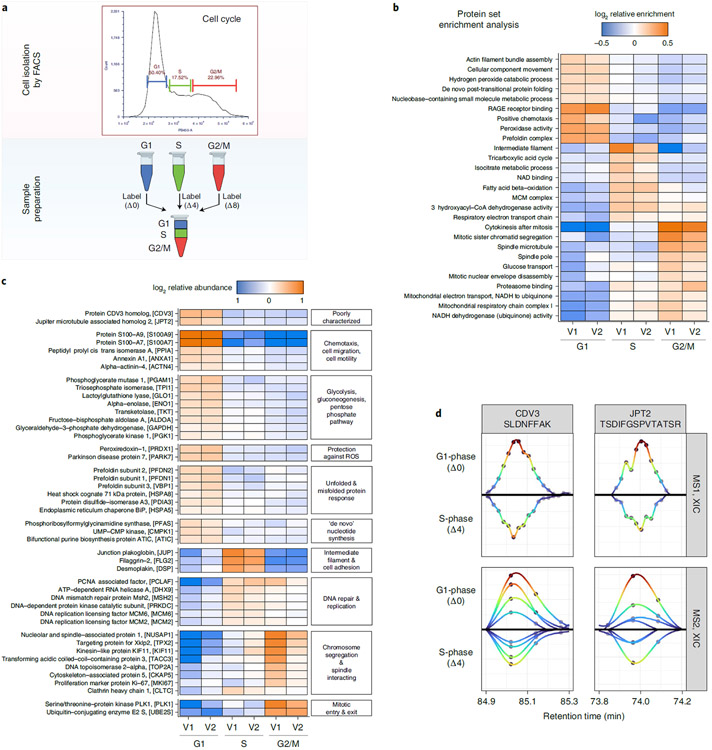
Next, we applied plexDIA to quantify protein abundance across the cell division cycle (CDC) of U-937 monocyte cells. The CDC analysis allows further validation of plexDIA based on well-established biological processes during the CDC while simultaneously offering the possibility of new discoveries. The ability of plexDIA to analyze small samples made it possible to isolate cells from different phases of the CDC based on their DNA content (Fig. 5a). The cell isolation used fluorescence-activated cell sorting (FACS), which allowed us to analyze cell populations from G1, S and G2/M phases without the artifacts associated with blocking the CDC to achieve population synchronization41.
Fig. 5 ∣. Cell cycle analysis with plexDIA.
a, U-937 monocytes were sorted by FACS based on DNA content to separate into G1, S and G2/M cell cycle phases. The samples were prepared as a plexDIA set and then analyzed with MS1-optimized and MS2-optimized data acquisition methods (referred to as V1 and V2, respectively). b, PSEA of cell cycle phases from plexDIA data. c, A subset of proteins found to be differentially abundant at 1% FDR across cell cycle phases was grouped by function and then plotted to show the relative abundances across cell phases. d, XICs at MS1 and MS2 for precursors from poorly characterized proteins, CDV3 and JPT2.
The peptides from the sorted cells were labeled with non-isobaric isotopologous labels and then combined and analyzed both by MS1-optimized (V1) and MS2-optimized (V2) plexDIA methods (Fig. 5a). By using different data acquisition methods, we aimed to (1) reduce systematic biases that may be shared by technical replicates and (2) evaluate the agreement between MS1-based and MS2-based quantification by plexDIA in the context of a biological experiment. Analyzing the V1 and V2 data with DIA-NN resulted in 4,391 unique protein groups and 4,107 gene groups at 1% global FDR. These data were filtered to include only proteotypic peptides, and then gene-level information was used for downstream protein set enrichment analysis (PSEA) and differential protein abundance analysis.
To identify biological processes regulated across the phases of the CDC, we performed PSEA using data from both V1 and V2 (Fig. 5b). The V1 and V2 data indicated very similar PSEA patterns and identified canonical CDC processes, such as the activation of the MCM complex during S phase and chromatid segregation and mitotic nuclear envelope disassembly during G2/M phase (Fig. 5b). These expected CDC dynamics and the agreement between V1 and V2 results demonstrate the utility of plexDIA for biological investigations. Furthermore, the PSEA indicated metabolic dynamics in the tricarboxylic acid (TCA) cycle and fatty acid beta-oxidation. These results provide direct evidence for the suggested coordination among metabolism and cell division42,43.
To further explore the proteome remodeling during the CDC, we identified differentially abundant proteins across G1, S and G2/M phase (Fig. 5c). From the 4,107 proteins identified across V1-acquired and V2-acquired data, 400 proteins were found to be differentially abundant between cell cycle phases at 1% FDR. Some of these proteins are displayed in Fig. 5c, organized thematically based on their functions. Consistent with results from PSEA, we found good agreement between V1 and V2 and expected changes in protein abundance, such as polo-like kinase 1 and ubiquitin-conjugating enzyme E2 peaking in abundance during G2/M phase.
In addition to the differential abundance of classic CDC regulators, we found that some poorly characterized proteins were also differentially abundant, such as proteins CDV3 and JPT2. To further investigate these proteins, we examined the extracted-ion chromatogram (XIC) for representative peptides from these proteins (Fig. 5d). The XIC demonstrates consistent quantitative trends and co-elution among precursors and peptide fragments labeled with different mass tags. This consistency among the raw data bolsters the confidence in the findings by plexDIA, such as differential abundance of CDV3 and JPT2.
Single-cell analysis with plexDIA.
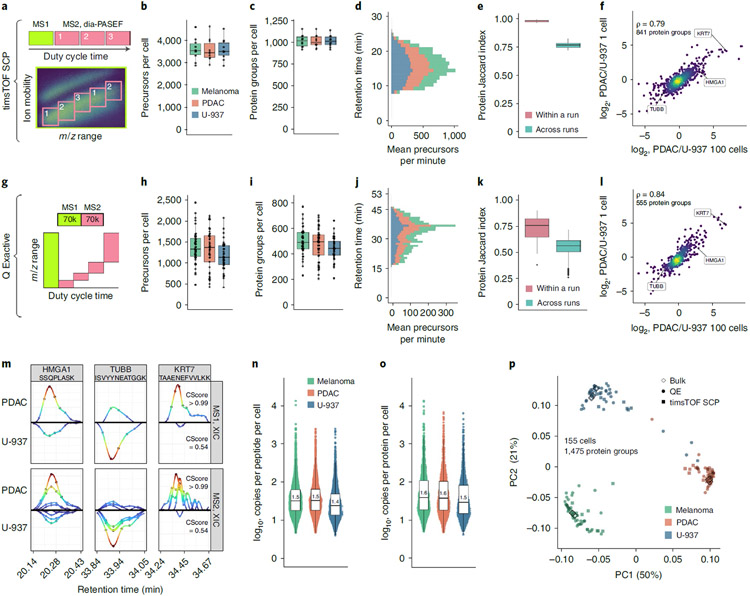
Next, we evaluated the potential of plexDIA to quantify proteins from single human cells. Single cells from melanoma (WM989-A6-G3), pancreatic ductal adenocarcinoma (PDAC) and monocyte (U-937) cell lines were prepared and combined into plexDIA sets using nano-ProteOmic sample Preparation (nPOP)44.
We aimed to test its generalizability to different types of MS detectors, an orbitrap and a time-of-flight (TOF) detector, and its ability to take advantage of ion mobility technology, such as trapped ion mobility spectrometry45. The technologies were implemented by analyzing single-cell plexDIA samples using two commercial platforms: timsTOF SCP (Fig. 6a-f) and Q Exactive classic (Fig. 6g-l). Both platforms achieved high quantitative accuracy and data completeness. To support high sample throughput, both platforms used short chromatographic gradients to separate the peptides (Fig. 6d, j), which, in the case of timsTOF SCP, allowed quantifying about 1,000 proteins per cell while using about 10 minutes of total instrument time (only 5 minutes of active chromatography) per single cell. Thus, plexDIA increases sample throughput by 3- to 12-fold over the top-performing single-cell proteomics methods that do not use isobaric mass tags46,47.
Fig. 6 ∣. Single-cell protein analysis with plexDIA.
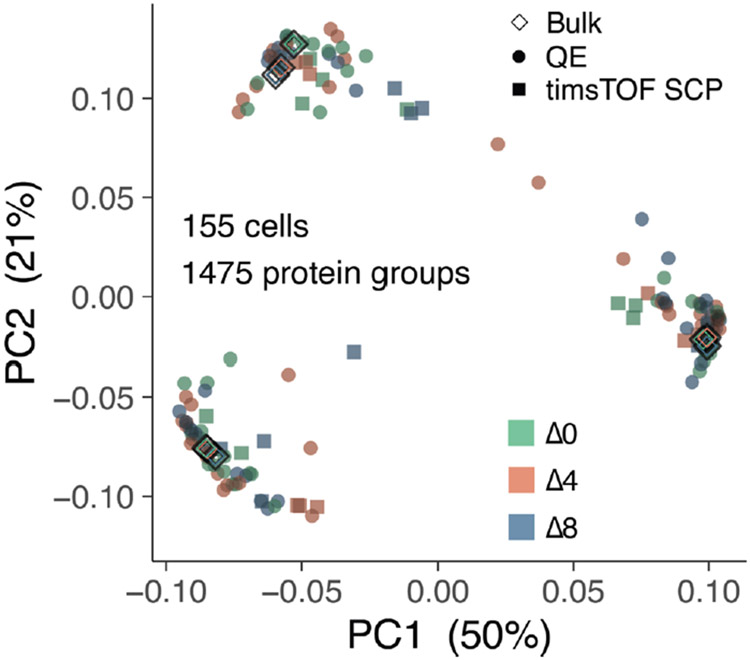
a, Cartoon visualizing the duty cycle used to analyze single-cell plexDIA sets with timsTOF SCP. Number of precursors (b) and protein groups (c) identified per single cell—data for (n = 30) single cells. d, Mean number of precursors identified from each cell type per minute of chromatographic gradient. e, Data completeness measured by Jaccard index within and between plexDIA sets—data for (n = 10) sets. f, Comparison of protein fold changes estimated from bulk samples (100 cells, x axis) or single cells (y axis). Panels g–l show analogous results to a–f but for data from a Q Exactive classic. Panels h and i show data for (n = 125) single cells, and panel k shows data for (n = 48) sets. m, XICs for precursors (MS1 level) and for peptide fragments (MS2 level) for peptides from differentially abundant proteins, HMGA1, TUBB and KRT7; data are from single cells analyzed by Q Exactive. Median number of copies for each peptide (n) and protein group (o), per single cell; data are from single cells analyzed by Q Exactive. Each distribution in n has (n > 1,800) peptides, and each distribution in o has (n > 700) proteins. p, PCA of 155 single cells, including both the cells analyzed by timsTOF SCP or by Q Exactive. The single cells are projected together with plexDIA triplicates of 100-cell bulk samples analyzed by Q Exactive. All peptides and proteins shown are at 1% FDR. Boxplots: The box defines the 25th and 75th percentiles, and the median is marked by a solid line. Outliers are marked as individual dots outside the whiskers. PC, principal component; QE, Q Exactive.
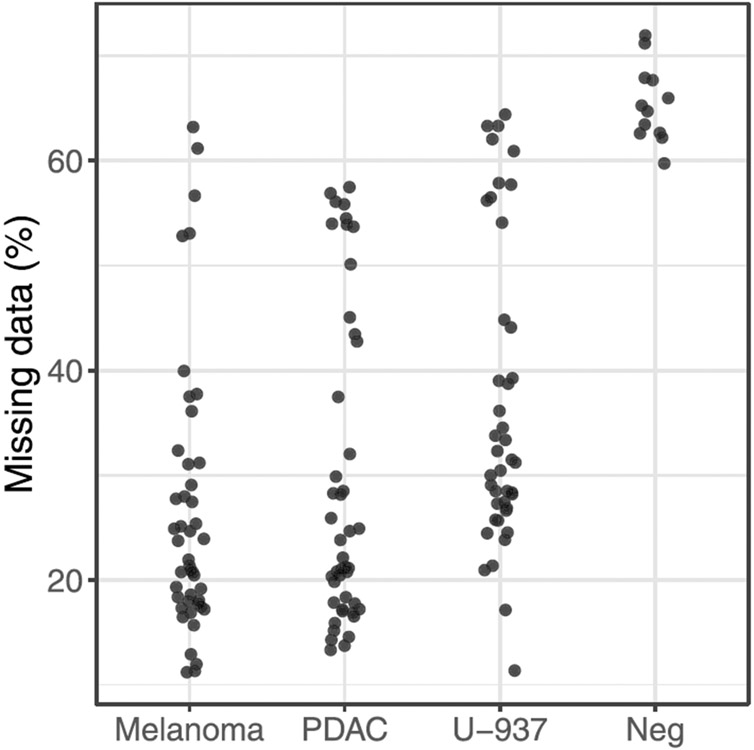
As observed with bulk samples, plexDIA resulted in high data completeness among single-cell proteomes (Fig. 6e,k). It exceeded 98% within labeled sets analyzed by timsTOF SCP (Fig. 6e) and remained over 50% even between plexDIA sets analyzed by Q Exactive (Fig. 6k). This high level of data completeness is enabled by leveraging the co-elution of isotopologues with precisely known mass offsets (Fig. 1b). Still, about 5% of the single cells had similar missing data to negative controls and were removed from downstream analysis as sample preparation likely failed (Extended Data Fig. 8).
plexDIA quantified protein fold changes spanning a 1,000-fold dynamic range and exhibited good agreement with corresponding fold changes quantified from 100-cell bulk samples (Fig. 6f,l). To explore the raw data supporting these measurements, we plotted both MS1-level and MS2-level XICs from pairs of isotopologous precursors (Fig. 6m). The data indicate that (1) the isotopologously labeled precursors co-elute and apex synchronously, and (2) the lowly abundant precursors whose identification depended on the plexDIA module have precursors, fragments and intensities in excellent agreement with the more abundant isotopologues and with the bulk measurements (Fig. 6l,m). These findings demonstrate that plexDIA may improve the sensitivity of single-cell proteomic analysis and, thus, increase data completeness, especially across cells with very different proteomes.
Sampling and detecting a sufficient number of precursor copies is key for accurate precursor quantification; otherwise, quantification accuracy is undermined by counting noise19,48. Because peptide fragmentation is usually incomplete, approaches like plexDIA that can perform MS1-level quantification are likely to count more copies per peptide than approaches relying on MS2-level or MS3-level quantification6. To evaluate this expectation, we estimated the number of peptide and protein copies that the orbitrap counted from single cells (Fig. 6n,o). The total number of peptide molecules counted by the Q Exactive instrument per single cell was about 300,000 and up to 1,000,000 for some single cells. The median number of copies sampled per peptide per single cell was about 30 (Fig. 6n,o). These estimates rely on orbitrap physics49,50 and were not extended to the single-cell measurements by the timsTOF SCP, which we expect to sample more protein copies per cell.
Single-cell plexDIA data acquired from Q Exactive and timsTOF SCP instruments were projected using a weighted principal component analysis (PCA) (Fig. 6p). To evaluate if the cell type separation is consistent with relative protein levels measured in bulk samples, we also projected 100-cell bulk plexDIA standards acquired on Q Exactive (Fig. 6p). We found strong agreement between single-cell samples and 100-cell bulk samples. Similarly, single-cell data acquired by Q Exactive and timsTOF SCP clustered by cell type, not platform type. To ensure that clustering was not an artifact of label-specific biases, we plotted the same PCA, except colored by the mTRAQ label that was used for tagging each single cell and found little to no dependence of labels on clustering (Extended Data Fig. 9).
Discussion
Although multiple methods allow increasing proteomics throughput, plexDIA is distinct in simultaneously allowing high sensitivity, depth and accuracy. plexDIA enables a multiplicative increase (threefold with three labels) in the rate of consistent protein quantification across limited sample amounts while preserving proteomic coverage, quantitative accuracy, precision and repeatability of LF-DIA. Similarly to other labeling methods, such as TMT-DDA, parallel analysis of multiple samples by plexDIA saves LC–MS/MS time. Currently, the commercially available labels for plexDIA are low-plex (mTRAQ, TMT0/TMT/shTMT or dimethyl labeling12), compared to 18-plex isobaric TMTpro labels available for DDA4. This current plex disadvantage is offset by the parallel precursor analysis enabled by plexDIA. Indeed, quantifying about 8,000 proteins per sample took 0.5 hours for three-plexDIA (Extended Data Fig. 5) and 1.1 hours for a highly optimized 16-plex TMTpro workflow51.
Furthermore, three-plexDIA affords higher sensitivity because it does not require offline fractionation and does not incur associated sample losses. We expect that the plexDIA framework will motivate the development of higher plex mass tags for plexDIA that are optimized for different applications, such as for single-cell proteomics7.
The parallel sample and peptide analysis by plexDIA becomes increasingly important for lowly abundant samples because they require long ion accumulation times that undermine the throughput of serial acquisition methods, such as TMT-DDA, even when the vast majority of MS2 scans result in confident peptide identifications7,52. Thus, plexDIA is particularly attractive for the analysis of nanogram samples as it may afford accurate and deep proteome quantification without using two-dimensional peptide separation (offline fractionation)7,19,28.
Our data demonstrate that plexDIA reduces the amount of missing data between diverse samples both within and across runs. This reduction stems from buffering sample-to-sample variability in protein composition. Furthermore, we introduced an approach for matching precursors within a run, which reduces missing data to a mere 2–3% in bulk samples (Fig. 2) and 2% in single-cell samples (Fig. 6). Thus, plexDIA analysis of samples with variable protein composition or abundance results in less missing data. This opens the potential for further gains. For example, small samples could be labeled and then combined with a labeled carrier sample to improve proteomic coverage of the smaller samples. Such non-isobaric carrier design will naturally extend the isobaric carrier concept20,28,53 and its benefits to DIA analysis for deep single-cell proteomics analysis. Indeed, the dynamic range, accuracy and data completeness of the single-cell protein data obtained by plexDIA (Fig. 6) can enable interpreting natural variation across the proteomes of single cells54,55. plexDIA offers a framework that may scale to n labels and, thus, increase throughput n-fold and increase the fraction of proteins quantified across all samples. Crucially, plexDIA maintains accurate quantification and good repeatability. Here we demonstrate this potential for n = 3. Increasing n beyond three offers clear benefits but also faces challenges. One challenge is the increased potential for interference, which can be resolved by increasing the resolving power of MS scans and improving data analysis. Another challenge is sampling enough ions from each peptide given the finite capacity of MS detectors, which can be relieved by sampling smaller m/z ranges—for example, quantification relying on small MS2 windows or split m/z ranges at MS1. The capacity of MS detectors is less limiting for small samples, such as single cells, and, thus, increasing the number of labels holds much potential for single-cell proteomics, as previously discussed11,28.
plexDIA can enable further gains in sensitivity for single-cell proteomics7, beyond the results demonstrated in Fig. 6. This may be achieved by including an isotopologous carrier channel, wherein a concentrated standard or pooled sample is used (1) to increase the sensitivity and, thus, identification numbers and data completeness in other channels and (2) to provide a reference signal for quantification. The quantitative aspect here has a double benefit. Quantification accuracy and robustness can be improved by (1) using MS1-level and MS2-level signals that are minimally affected by interference and by (2) calculating quantities relative to the internal standard, which is likely to also substantially reduce the batch effects associated with LC–MS performance variation. This makes the technology introduced by plexDIA highly promising not just for very deep profiling of selected samples using offline fractionation but also for large-scale experiments, wherein batch effects are a substantial challenge. Another avenue of plexDIA is increasing the throughput of applications seeking to quantify protein interactions, conformations and activities. For example, plexDIA is readily compatible with the recently reported covalent protein painting that enables analysis of protein conformations in living cells56,57. Because there are no fundamental limitations preventing the creation of non-isobaric labels that would allow a higher degree of multiplexing with DIA, we expect plexDIA to enable even higher throughput in the future. Given these considerations, we think that plexDIA will eventually become the predominant DIA workflow, preferable over LF approaches for most applications.
Methods
Cell culture.
U-937 (monocytes) and Jurkat (T cells) were cultured in RPMI 1640 Medium (Sigma-Aldrich, R8758), and HPAF-II cells (PDACs, American Type Culture Collection (ATCC), CRL-1997) were cultured in EMEM (ATCC, 30-2003). All three cell lines were supplemented with 10% FBS (Gibco, 10439016) and 1% penicillin–streptomycin (Gibco, 15140122) and grown at 37 °C. Melanoma cells (WM989-A6-G3, a kind gift from Arjun Raj, University of Pennsylvania) were grown as adherent cultures in TU2% media, which is composed of 80% MCDB 153 (Sigma-Aldrich, M7403), 10% Leibovitz L-15 (Thermo Fisher Scientific, 11415064), 2% FBS, 0.5% penicillin–streptomycin and 1.68 mM calcium chloride (Sigma-Aldrich, 499609). All cells were harvested at a density of 106 cells per ml and washed with sterile PBS. For bulk plexDIA benchmarks, U-937 and Jurkat cells were resuspended to a concentration of 5 × 106 cells per ml in LC–MS water and stored at −80 °C.
E. coli and S. cerevisiae were grown at room temperature (21 °C), shaking at 300 r.p.m., in Luria Broth (LB) and yeast-peptone-dextrose (YPD) media, respectively. Cell density was measured by OD600, and cells were harvested mid-log phase, pelleted by centrifugation and stored at −80 °C.
Preparation of bulk plexDIA samples.
The harvested U-937 and Jurkat cells were heated at 90 °C in a thermal cycler for 10 minutes to lyse by mPOP58. Triethylammonium bicarbonate (TEAB) was added to a final concentration of 100 mM (pH 8.5) for buffering, and then proteins were reduced in tris(2-carboxyethyl)phosphine (TCEP, Supelco, 646547) at 20 mM for 30 minutes at room temperature. Iodoacetamide (Thermo Fisher Scientific, A39271) was added to a final concentration of 15 mM and incubated at room temperature for 30 minutes in the dark. Next, benzonase nuclease (Millipore, E1014) was added to 0.3 U μl−1 of Trypsin Gold (Promega, V5280) to 1:25 ratio of substrate:protease and LysC (Promega, VA1170) to 1:40 ratio of substrate:protease and then incubated at 37 °C for 18 hours. E. coli and S. cerevisiae samples were prepared similarly; however, instead of lysis by mPOP, samples were lysed in 6 M urea and vortexed with acid-washed glass beads alternating between 30 seconds vortexing and 30 seconds resting on ice, repeated for a total of five times.
After digestion, all samples were desalted by Sep-Pak (Waters, WAT054945). Peptide abundance of the eluted digests was estimated by NanoDrop A280, and then the samples were dried by speed vacuum and resuspended in 100 mM TEAB (pH 8.5). U-937, Jurkat, E. coli and S. cerevisiae digests were mixed to generate three samples, which we refer to as samples A, B and C; the mixing ratios are described in Extended Data Fig. 10. Samples A, B and C were split into two groups: (1) was kept label free, and (2) was labeled with mTRAQ Δ0, Δ4 or Δ8 (SciEx, 4440015, 4427698 and 4427700), respectively. An appropriate amount of each respective mTRAQ label was added to each sample A–C, following the manufacturer’s instructions. In short, mTRAQ was resuspended in isopropanol and then added to a concentration of 1 U per 100 μg of sample and left to incubate at room temperature for 2 hours. We added an extra step of quenching the labeling reaction with 0.25% hydroxylamine for 1 hour at room temperature, as is commonly done in TMT experiments where the labeling chemistry is the same6,50. After quenching, the mTRAQ-labeled samples (A–C) were pooled to produce the final multiplexed set used for benchmarking plexDIA.
Preparation of single-cell plexDIA samples.
Single cells were thawed from liquid nitrogen storage in 10% DMSO and culture media at a concentration of 1 × 106 cells per ml. Cells were first washed twice in PBS to remove DMSO and media and then were suspended in PBS at 200 cells per μl for sorting and sample preparation by nPOP, as detailed by Leduc et al.44. In brief, single cells were isolated by cellenONE and prepared in droplets on the surface of a glass slide, including lysing, digesting and labeling individual cells. In each droplet, single cells were lysed in 100% DMSO; proteins were digested with Trypsin Gold at a concentration of 120 ng μl−1 and 5 mM HEPES (pH 8.5); peptides were chemically labeled with mTRAQ; and then, finally, single cells were pooled into a plexDIA set for subsequent analysis. Cells were prepared in clusters of three for ease of downstream pooling into plexDIA sets, and a total of 48 plexDIA sets were prepared per single glass slide.
Modifications were made to the original nPOP sample preparation protocol to enable single-cell plexDIA. We found the mTRAQ labeling reaction, unlike TMT labeling reagents, requires the presence of aqueous phase; therefore, an additional 15 nl of 200 mM TEAB buffer (pH 8.5) was added to each droplet, and the droplets were not dried down before adding mTRAQ labels. mTRAQ labels were dissolved at a concentration of 0.02 U μl−1 in ethanol, and 30 nl of this solution was added to each single cell. To dispense ethanol with the cellenONE, the cellenONE pipette aspirated 5 μl of air, followed by 20 μl of ethanol before aspirating 10 μl of mTRAQ. Lastly, when quenching the labeling reaction, only one addition of 20 nl of 5% hydroxlamine was performed. All prior and subsequent sample preparation steps were carried out as specified by Leduc et al.44.
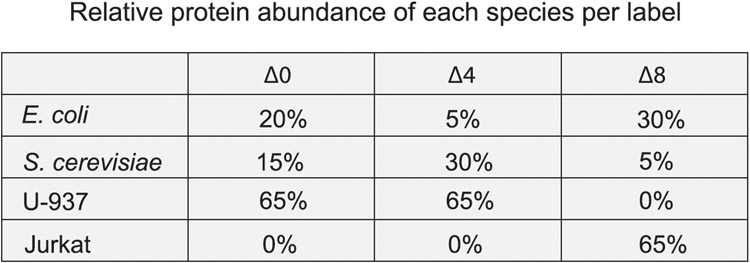
Each plexDIA set was composed of a single PDAC, melanoma and U-937 cell, except if a negative control was present in place of a cell. For samples run on the Q Exactive, every fourth set contained a negative control that received all the same reagents but did not include a single cell. This resulted in 132 single cells prepared with 12 total negative controls. Ten additional plexDIA sets were run on the timsTOF SCP for a total of 30 single cells (no negative controls). Cell types were labeled with randomized mass tags designs in the plexDIA sets to avoid any systematic biases with labeling. Specifically, each cell type was labeled with each mass tag as described in the single-cell metadata file.
CDC, FACS and sample preparation.
U-937 monocytes were grown as described above and harvested and aliquoted to a final 1-ml suspension of approximately 1 × 106 cells in RPMI 1640 Medium. Then, DNA was stained by incubating the cells with Vybrant DyeCycle Violet Stain (Invitrogen, V35003) at a final concentration of 5 μM in the dark for 30 minutes at 37 °C, as per the manufacturer’s instructions. Next, the cells were centrifuged and then resuspended in PBS to a density of 1 × 106 cells per ml. The cell suspension was stored on ice and protected from light until sorting began.
The cells were then sorted with a Beckman CytoFLEX SRT. The population of U-937s was gated to select singlets based on FSC-A and FSC-H; this population of singlets was then sub-gated based on DNA content using the PB-450 laser (ex = 405 nm / em = 450 nm). The G1 population is the most abundant population in actively dividing cells, and the G2/M populations should theoretically have double the intensity (DNA content), whereas the S-phase lies in between. Populations of G1, S and G2/M cells were collected based on these sub-gates and sorted into 2-ml Eppendorf tubes.
After sorting, the cells were centrifuged at 300g for 10 minutes; PBS was removed; and then the cells were resuspended in 20 μl of HPLC water to reach a density of approximately 4,000 cells per μl. The cell suspensions were lysed using the mPOP method, which involves freezing at −80 °C and then heating to 90 °C for 10 minutes. Next, the cell lysates were prepared exactly as described in the ‘Sample preparation’ section. In brief, the cell lysate was buffered with 100 mM TEAB (pH 8.5), and then proteins were reduced with 20 mM TCEP for 30 minutes at room temperature. Next, iodoacetamide was added to a final concentration of 15 mM and incubated at room temperature for 30 minutes in the dark, and then benzonase nuclease was added to 0.3 U μl−1. Trypsin Gold and LysC were then added to the cell lysate at 1:25 and 1:40 ratio of protease:substrate, and then the samples were incubated at 37 °C for 18 hours. After digestion, the peptides were desalted by stage tipping with C18 extraction disks (Empore, 66883-U) to remove any remaining salt that was introduced during sorting59. G1 cells were labeled with mTRAQ Δ0; S cells were labeled with mTRAQ Δ4; and G2/M cells were labeled with Δ8 and then combined to form a plexDIA set of roughly 2,000 cells per cell cycle phase (label). The combined set was analyzed with 2-hour active gradients of MS1-optimized (V1) and MS2-optimized (V2) methods as described in the ‘Acquisition of bulk data’ section.
Acquisition of bulk data.
Multiplexed and LF samples were injected at 1-μl volumes via Dionex UltiMate 3000 UHPLC to enable online nLC with a 25 cm × 75 μm IonOpticks Aurora Series UHPLC column (AUR2-25075C18A). These samples were subjected to electrospray ionization (ESI) and sprayed into a Q Exactive orbitrap for MS analysis. Buffer A is made of 0.1% formic acid (Pierce, 85178) in LC–MS-grade water. Buffer B is made of 80% acetonitrile and 0.1% formic acid mixed with LC–MS-grade water.
The gradient used for LF-DIA is as follows: 4% Buffer B (minutes 0–11.5), 4–5% Buffer B (minutes 11.5–12), 5–28% Buffer B (minutes 12–75), 28–95% Buffer B (minutes 75–77), 95% Buffer B (minutes 77–80), 95–4% Buffer B (minutes 80–80.1) and then hold at 4% Buffer B until minute 95, flowing at 200 nl min−1 throughout the gradient. Instrument control and data acquisition were performed via Xcalibur. The V1 duty cycle was comprised of 5×(1 MS1 full scan × 5 MS2 windows) as illustrated in Fig. 1b. Thus, the duty cycle has a total of 25 MS2 windows to span to full m/z scan range (380–1,370 m/z) with 0.5-Thomson (Th) overlap between adjacent windows. The length of the windows was variable for each subcycle (20 Th for subcycles 1–3, 40 Th for subcycle 4 and 100 Th for subcycle 5). Each MS1 full scan was conducted at 140,000 resolving power, 3 × 106 automatic gain control (AGC) maximum and 500-ms maximum injection time. Each MS2 scan was conducted at 35,000 resolving power, 3 × 106 AGC maximum, 110-ms maximum injection time and 27% normalized collision energy (NCE) with a default charge of 2. The RF S-lens was set to 80%. The V2 duty cycle consisted of one MS1 scan conducted at 70,000 resolving power with a 300-ms maximum injection time and 3 × 106 AGC maximum, followed by 40 MS2 scans at 35,000 resolving power with 110-ms maximum injection time and 3 × 106 AGC maximum. The window length for the first 25 MS2 scans was set to 12.5 Th; the next seven windows were 25 Th; and then the last eight windows were 62.5 Th. Adjacent windows shared a 0.5-Th overlap. All other settings were the same as the LF-DIA V1 method. All LF samples for bulk benchmarking containing S. cerevisiae, E. coli and H. sapiens were run in triplicate. However, the third run of LF-DIA sample C using the V2 method was an outlier and was omitted from analysis due to poor performance.
mTRAQ labeling increases hydrophobicity of peptides, which is why a higher % Buffer B was used during the active gradient of multiplexed samples; in addition, the scan range was shifted 100 m/z higher than LF-DIA to account for the added mass of the label. The gradient used for plexDIA is as follows: 4% Buffer B (minutes 0–11.5), 4–7% Buffer B (minutes 11.5–12), 7–32% Buffer
B (minutes 12–75), 32–95% Buffer B (minutes 75–77), 95% Buffer B (minutes 77–80), 95–4% Buffer B (minutes 80–80.1) and then hold at 4% Buffer B until minute 95, flowing at 200 nl min−1 throughout the gradient. The plexDIA V1 duty cycle was comprised of 5×(1 MS1 full scan × 5 MS2 windows), for a total of 25 MS2 windows to span to full m/z scan range (480–1,470 m/z) with 0.5-Th overlap between adjacent windows. The length of the windows was variable for each subcycle (20 Th for subcycles 1–3, 40 Th for subcycle 4 and 100 Th for subcycle 5). Each MS1 full scan was conducted at 140,000 resolving power, 3 × 106 AGC maximum and 500-ms maximum injection time. Each MS2 scan was conducted at 35,000 resolving power, 3 × 106 AGC maximum, 110-ms maximum injection time and 27% NCE with a default charge of 2. The RF S-lens was set to 30%. The plexDIA V2 duty cycle consisted of one MS1 scan conducted at 70,000 resolving power with 300-ms maximum injection time and 3 × 106 AGC maximum, followed by 40 MS2 scans at 35,000 resolving power with 110-ms maximum injection time and 3 × 106 AGC maximum. The window length for the first 25 MS2 scans was set to 12.5 Th; the next seven windows were 25 Th; and then the last eight windows were 62.5 Th. Adjacent windows shared a 0.5-Th overlap. All other settings were the same as the plexDIA V1 method. Data acquired for the CDC used 2-hour active gradients of the V1 and V2 methods.
The gradient used for mTRAQ DDA is the same used for plexDIA. However, the duty cycle was a shotgun DDA method. The MS1 full scan range was 450–1,600 m/z and was performed with 70,000 resolving power, 3 × 106 AGC maximum and 100-ms injection time. This shotgun DDA approach selected the top 15 precursors to send for MS2 analysis at 35,000 resolving power, 1 × 105 AGC maximum, 110-ms injection time, 0.3-Th isolation window offset, 0.7-Th isolation window length, 8 × 103 minimum AGC target and 30-second dynamic exclusion.
Acquisition of single-cell data.
Q Exactive.
plexDIA single-cell sets and 100-cell standards were injected at 1-μl volumes via Dionex UltiMate 3000 UHPLC to enable online nLC with a 15 cm × 75 μm IonOpticks Aurora Series UHPLC column (AUR2-15075C18A). Instrument control and data acquisition were performed via Xcalibur. These samples were subjected to ESI and sprayed into a Q Exactive orbitrap for MS analysis. Buffer A is made of 0.1% formic acid (Pierce, 85178) in LC–MS-grade water. Buffer B is made of 80% acetonitrile and 0.1% formic acid mixed with LC–MS-grade water. The gradient used is as follows: 4% Buffer B (minutes 0–2.5), 4–8% Buffer B (minutes 2.5–3), 8–32% Buffer B (minutes 3–33), 32–95% Buffer B (minutes 33–34), 95% Buffer B (minutes 34–35), 95-4% Buffer B (minutes 35–35.1) and then hold at 4% Buffer B until minute 53, flowing at 200 nl min−1 throughout the gradient. The plexDIA duty cycle was comprised of one MS1 followed by four DIA MS2 windows of variable m/z length (specifically 120 Th, 120 Th, 200 Th and 580 Th) spanning 378–1,402 m/z. Each MS1 and MS2 scan was conducted at 70,000 resolving power, 3 × 106 AGC maximum and 300-ms maximum injection time. NCE was set to 27% with a default charge of 2. The RF S-lens was set to 80%.
To generate a spectral library from 100-cell standards on the Q Exactive, the same settings were used with the exception that the duty consisted of one MS1 and 25 MS2 windows of variable m/z length (specifically, 18 windows of 20 Th, two windows of 40 Th, three windows of 80 Th and two windows of 160 Th). The MS2 scans were conducted at 35,000 resolving power, 3 × 106 AGC maximum and 110-ms maximum injection time.
timsTOF SCP.
The single-cell plexDIA sets were separated on a nanoElute liquid chromatography system (Bruker Daltonics) using a 25 cm × 75 μm, 1.6-μm C18 (AUR2-25075C18A-CSI, IonOpticks). The analytical column was kept at 50 °C. Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in acetonitrile. The column was equilibrated with 4 column volumes of mobile phase A before sample loading. The peptides were separated over 30 minutes at 250 nl min−1 using the following gradients: 2–17% B in 15 minutes, 17–25% B in 5 minutes, 25–37% B in 3 minutes and 37–85% B in 3 minutes and then maintained at 85% for 4 minutes.
Instrument control and data acquisition were performed via timsControl 3.1. The timsTOF SCP was operated in dia-PASEF mode with the following settings: mass range 100–1,700 m/z, 1/k0 Start 0.6 V s cm−2, End 1.2 V s cm−2; ramp and accumulation times were set to 166 ms; capillary voltage was 1,600 V, dry gas 3 L min−1 and dry temp 200 °C. dia-PASEF settings: Each cycle consisted of 1× MS1 full scan and 5× MS2 windows covering 297.7–797.7 m/z and 0.63–1.10 1/k0. Each window was 100 Th wide by 0.2 V s cm−2 high. There was no overlap in either m/z or 1/k0 (Fig. 6). The cycle time was 0.68 seconds. Collision-induced dissociation (CID) energy was 20–59 eV as a function of the inverse mobility of the precursor.
Spectral library generation.
The in silico predicted spectral library used in LF-DIA analysis was generated by DIA-NN’s (version 1.8.1 beta 16) deep-learning-based spectra and retention time and IMs prediction based on Swiss-Prot H. sapiens, E. coli and S. cerevisiae FASTAs (canonical and isoform) downloaded in February 2022. The spectral library used for plexDIA benchmarking was created in a similar process, with the exception of a few additional commands entered into the DIA-NN command line GUI: (1) {–fixed-mod mTRAQ 140.0949630177, nK} and (2) {–original-mods}. Two additional libraries were generated: (1) mTRAQ-labeled spectral library from FASTAs containing only E. coli and S. cerevisiae sequences; (2) mTRAQ-labeled spectral library from a FASTA containing only H. sapiens sequences. The former was used to search data shown in Extended Data Fig. 2, and the latter was used to search CDC and 100-cell standards. Triplicates of 100-cell standards of PDAC, melanoma and U-937 cells were run with the one MS1 × 25 MS2 scans method, searched using the in silico generated human-only spectral library. The results of this search generated a sample-specific library covering about 5,000 protein groups. This library was used to search single-cell plexDIA sets acquired on the Q Exactive and on the timsTOF SCP as well as 100-cell standards run on the Q Exactive with the same method used to acquire single-cell plexDIA data.
plexDIA module in DIA-NN.
A distinct feature of DIA-MS proteomics is the complexity of produced spectra, which are a mixture of fragment ions originating from multiple co-isolated precursors. This complexity has necessitated the rise of a variety of highly sophisticated algorithms for DIA data processing. Current DIA software, such as DIA-NN25, aims to find peak groups in the data that best match the theoretical information about such peptide properties as the MS/MS spectrum, the retention time and the ion mobility. Once identified correctly, the peak group—that is, the set of XICs of the precursor and its fragments in the vicinity of the elution apex—allows to integrate either the MS1-level or MS2-level signals to quantify the precursor, which is the ultimate purpose of the workflow.
Similarly to match-between-runs (MBR) algorithms, plexDIA data provide the opportunity to match corresponding ions, in this case between the same peptide labeled with different mass tags. However, the use of isotopologous mass tags, such as mTRAQ, allows to match the retention times within a run with much higher accuracy than what can be achieved across runs. Thus, the sequence propagation can be more sensitive and reliable than with MBR7. This allows to enhance sequence identifications analogously to the isobaric carrier concept introduced by TMT-based single-cell workflows53,60. With the isobaric carrier approach, a carrier channel is loaded with a relatively high amount of peptides originating from a pooled sample that facilities peptide sequence identification20,28. We implemented a similar approach in the plexDIA module integrated in DIA-NN. Once a peptide is identified in one of the channels, its exact retention time apex can be determined, which, in turn, helps identify and quantify the peptide in all of the channels by integrating the respective precursor (MS1) or fragment ion (MS2) signals.
Apart from the identification performance, plexDIA also can increase quantification accuracy. The rich complex data produced by DIA promote more accurate quantification because of algorithms that select signals from MS/MS fragment ions that are affected by interferences to the least extent25. For LF-DIA, DIA-NN selects fragments in a cross-run manner: fragments that tend to correlate well with other fragments across runs are retained, whereas those that often exhibit poor correlations due to interferences are excluded from quantification. Although this approach yields good results, a limitation remains for LF-DIA: fragment ions only affected by interferences in a modest proportion of runs are still used for quantification, thus undermining the reliability of the resulting quantities in those runs. Here, plexDIA provides a unique advantage. Theoretically, a single MS1-level or MS2-level signal with minimal interference is sufficient to calculate the quantitative ratio between the channels. In this case, if low interference quantification is possible in at least one ‘best’ channel, this quantity can be multiplied by the respective ratios across other channels to obtain accurate estimates of quantities in all channels that share at least one low interference signal with this ‘best’ channel. This idea is implemented in DIA-NN to produce ‘translated’ quantities, which have been corrected by using ratios of high-quality MS1 or MS2 signals between channels as described in Fig. 1b and Extended Data Fig. 1.
Estimating FDR.
The DIA-NN module uses a decoy-based approach, but, instead of regular decoy precursors, as used in the LF setting and likewise during the first step of the plexDIA data processing workflow, it relies on a decoy channel. Here, for each peak group match, regular or translated, DIA-NN calculates a number of scores that reflect the channel-specific signal evidence. A separate classifier based on an ensemble of neural networks, identical in architecture to the one used to obtain the regular q values25, is then trained to distinguish between sets of scores originating from target channels and the decoy channel, leading to the calculation of composite scores for the peak groups and, thus, the channel q values.
Data analysis with DIA-NN.
DIA-NN (version 1.8.1 beta 16) was used to search LF-DIA and plexDIA raw files, which is available at https://plexdia.slavovlab.net/ and https://scp.slavovlab.net/plexDIA. All LF-DIA benchmarking raw files were searched together with MBR if the same duty cycle was used; likewise, all plexDIA benchmarking raw files were searched together with MBR if the same duty cycle was used, with the exception of the CDC experiments that used V1 and V2 methods—these two runs were searched together.
DIA-NN search settings: Library Generation was set to ‘IDs, RT and IM Profiling’, Quantification Strategy was set to ‘Peak height’, scan window = 1,Mass accuracy = 10 p.p.m. and MS1 accuracy = 5 p.p.m. ‘Remove likely interferences’, ‘Use isotopologues’ and ‘MBR’ were enabled. Additional commands entered into the DIA-NN command line GUI for plexDIA: (1) {–fixed-mod mTRAQ 140.0949630177, nK}, (2) {–channels mTRAQ, 0, nK, 0:0; mTRAQ,4, nK, 4.0070994:4.0070994; mTRAQ, 8, nK, 8.0141988132:8.0141988132}, (3) {–original-mods}, (4) {–peak-translation}, (5) {–ms1-isotope-quant}, (6) {–report-lib-info} and (7) {–mass-acc-quant 5.0}. Note, (7) is only necessary for instances when MS2 quantitation is intended to be used; this command will use the pre-defined mass accuracy (for example, 10 p.p.m.) to identify precursors but restrict the mass error tolerance to the value specified for quantitation; this can help reduce the effect of interferences for MS2-level quantitation. For LF-DIA, only the following additional commands were used: (1) {–original-mods}, (2) {–peak-translation}, (3) {–ms1-isotope-quant}, (4) {–report-lib-info} and (5) {–mass-acc-quant 5.0}. The same search settings were used for single-cell Q Exactive and timsTOF SCP data; however ‘scan window’ was increased to 5.
Searching DDA data with MaxQuant.
MaxQuant (version 1.6.17.0) was used to search triplicate mTRAQ DDA, bulk benchmarking runs. MBR was enabled, and ‘Type’ was selected as ‘Standard’ with ‘Multiplicity’ = 3; mTRAQ-Lys0 and mTRAQ-Nter0, mTRAQ-Lys4 and mTRAQ-Nter4 and mTRAQ-Lys8 and mTRAQ-Nter8 were selected for light, medium and heavy labels, respectively. Variable modifications included Oxidation (M) and Acetyl (Protein-N-term); Carbamidomethyl (C) was selected as a fixed modification. Trypsin was selected as the protease and searched with maximum missed cleavage = 2.
Quantifying proteins for bulk plexDIA benchmarks.
MaxLFQ abundance for protein groups was calculated based on MS1 intensities (specifically the ‘MS1 Area’ column output by DIA-NN) using the DIA-NN R package25 for data acquired with the V1 method. However, for data acquired using the V2 method, MS2 quantitation (specifically the ‘Precursor Translated’ column output by DIA-NN) was used for quantitation. These protein abundances were used to calculate protein ratios across samples, which were normalized by subsetting human proteins (which are present in a 1:1 ratio, theoretically) and multiplying by a scalar such that the human protein ratios were centered on 1, and, thus, the other species (E. coli and S. cerevisiae) would be systematically shifted to account for any small loading differences across samples.
The quantitative comparisons between LF-DIA and plexDIA throughout this article are for intersected sets of proteins so that the results would not be influenced by proteins analyzed by only one method and not the other. For example, compared distributions were for the same set of proteins to avoid ‘survival biases’61.
PSEA.
PSEA was performed across the multiplexed bulk samples corresponding to cells sorted by DNA content into cell cycle phases (G1, S and G2/M). The reference human gene set database was acquired from the Gene Ontology Annotation62. The Kruskal–Wallis test was used to determine whether the hypothesis that all multiplexed samples had equivalent median protein abundances for a functionally annotated group of proteins could be rejected at a q value ≤0.05. Only protein sets with at least four proteins present were statistically tested. PSEA was run separately for the multiplexed samples analyzed by V1 and V2 methods. Protein sets were combined from both data acquisition methods if at least one method produced a q value ≤0.05.
Differential protein abundance testing.
Differential protein abundance testing was performed using precursor-level quantitation. To account for variation in sample loading amounts, precursors from each sample were normalized to their sample median. Then, each precursor was normalized by its mean across samples to convert it to relative levels. The normalized relative precursor intensities from different replicates were grouped by their corresponding protein groups and compared by a two-tailed t-test (Fig. 4b,c) or ANOVA (Fig. 5c) to estimate the significance of differential protein abundance across samples/conditions. This comparison captures both the variability between different replicates and different peptides originating from the same protein. To correct for multiple hypotheses testing, we used the Benjamini–Hochberg method to estimate q values for differential abundance of proteins and protein sets.
Relative protein fold change between U-937 cells and Jurkat cells, bulk.
Protein group abundances were calculated by MaxLFQ from triplicates of LF-DIA and plex-DIA; specifically, sample B and sample C were compared to calculate relative fold changes between H. sapiens cell lines U-937 and Jurkat. The protein groups plotted were required to be quantified in each of the triplicates of plexDIA and LF-DIA. A Spearman correlation was calculated for all protein groups and for differentially abundant protein groups.
Correcting isotopic envelope of plexDIA precursors.
mTRAQ labels, which were used in this demonstration of plexDIA, are separated by 4 Da. Because C-terminal arginine precursors are singly labeled and have a mere 4 Da separating isotopologous precursors, there is greater potential of isotopic envelope interference from lighter channels into heavier channels than there is for C-terminal lysine precursors, which would be separated by 8 Da; therefore, to improve quantitative accuracy, we correct the theoretical super-position of isotopic envelopes between channels for C-terminal arginine precursors. This can be accomplished because each precursor has a well-defined theoretical distribution of isotopes that we model with a binomial distribution; we use this theoretical distribution of isotopes to subtract and add back a precise amount of signal from heavier channels to lighter channels for MS1-level quantitation of each precursor.
Extracted ion current.
A precursor from a subset of proteins found to be differentially abundant was selected to be plotted to display the extracted ion current at MS1 and for fragments at MS2. Ion current was extracted using the DIA-NN GUI command interface by typing {–vis 25, PEPTIDE} where ‘PEPTIDE’ is the peptide sequence and ‘25’ is the number of scans to extract. MS1 and MS2 XICs were plotted to show the full elution profile. The four highest correlated fragments at MS2 were plotted; y-ions from C-terminal arginine peptide were excluded from plotting at MS2 level because these fragments are a super-position across samples as the C-terminus of arginine peptides is not labeled and, therefore, not sample specific. The lines in Figs. 5d and 6m were colored dynamically as a function of intensity.
Estimating peptide and protein copy numbers.
Precursor copy numbers at the MS1 level were estimated based on the signal-to-noise (S/N) level of individual peaks. The noise level of centroided spectra were used as reported by the Thermo firmware and extracted using a modified version of the ThermoRawFileParser63. Precursors reported by DIA-NN were matched to the S/N data based on the reported retention time with a tolerance of five scans and 12-p.p.m. mass error. The number of charges in an orbitrap is proportional to the S/N level and scales with a linear factor CN. This factor has been estimated to be CN = 3.5 for the Q Exactive orbitrap64,65 and has been confirmed by investigations with high-field orbitraps49. This proportionality constant was estimated at a resolving power of 240,000 and must be scaled by the square root ratio with the resolving power used for acquiring the spectra (R = 70,000). Precursor copy numbers are then calculated based on the number of charges z per precursor.
Analogous to the quantification, copy numbers were summed over the M and M + 1 peaks. Peptide-level copy numbers were calculated as the sum of all charge states found for a given peptide; protein-level copy numbers were calculated as the sum of all peptides not shared with other proteins (proteotypic).
Single-cell data analysis.
To increase sensitivity of single-cell analysis, Ms1. Extracted quantities output by DIA-NN were used for quantitation rather than Ms1.Area. Single cells with more than 60% missing data (no extracted MS1-level quantitation) at precursor level were considered to have failed in sample preparation and were removed from analysis. Quantitative accuracy of single-cell sets was assessed by calculating fold change between PDAC and U-937 cell types of averaged single-cell MaxLFQ protein quantities and calculating a Spearman correlation to 100-cell bulk comparisons. The 100-cell bulk comparisons consisted of triplicates in which each replicate alternated the labeling scheme. For a protein group to be included in the comparison, it was required to be quantified in at least five single cells and two-thirds of the bulk triplicates. Both the timsTOF SCP single-cell data and Q Exactive single-cell data were benchmarked to the same 100-cell Q Exactive-acquired plexDIA sets. Because missing data in DIA is related to low protein abundance, the missing MaxLFQ protein abundances in single cells and bulk were imputed with the lowest non-zero protein abundance for that protein in the same cell type and condition (bulk or single cells). The mean of each protein across the single-cell observations and bulk triplicates (respectively) was taken to represent that cell type and condition-specific protein abundance.
Single-cell sets acquired on the timsTOF SCP and Q Exactive were prepared on different days with different batches of cells. Generally, the data are quite similar as indicated by PCA (Fig. 6p), but quantitative discrepancies between bulk samples, which were acquired on the Q Exactive from one batch of cells, and single-cell sets on the timsTOF SCP from another batch of cells, may arise from real cellular differences as they were prepared from different cellular batches.
Next, 100-cell bulk plexDIA triplicates were used to identify proteins that were differentially abundant between U-937 and PDAC cells. Three proteins were chosen, and one precursor from each protein was selected to have its ion chromatogram extracted and plotted from single-cell Q Exactive-acquired data. See the ‘Extracted ion current’ subsection for more details about how this is performed.
PCA was performed on Ms1.Extracted timsTOF SCP single-cell data, Q Exactive single-cell data and Q Exactive 100-cell data. The following is a brief outline of the computational workflow. The abundance of each precursor was divided by the mean abundance of all three isotopologous precursors within the plexDIA set; then, the precursor of each labeled cell in each plexDIA was normalized to its median abundance; and then, each normalized precursor was divided by the mean of normalized precursor abundance across all labels and sets. These normalized precursor abundances were collapsed to protein group level by the median normalized abundance precursor. The protein group data were then normalized in the same way the precursors were normalized. Missing protein group data for each cell were imputed by k-nearest neighbors; the dataset was batch corrected for labels; and finally, a PCA was generated from the data. To increase the weights of proteins that are differentially abundant between the single cells in a coherent manner, we weighted each protein with the norm of its correlation vector (vector of pairwise correlations to all other proteins), as was previously described50.
Extended Data
Extended Data Fig. 1 ∣. plexDIA data processing by DIA-NN.
The plexDIA module in DIA-NN starts the data processing by splitting the input spectral library or a sequence database into multiple channels, wherein query precursor ions are generated for each of the label states. In addition, a decoy channel is generated by considering a ‘decoy’ label with a higher mass than the actual labels, for example, +12 for mTRAQ. A preliminary precursor ion identification step is then carried out, wherein a best matching peak group is found, as in label-free search, for all of the precursors, from all the channels. These peak groups are scored by the regular neural network-based classifier implemented in DIA-NN. The most confident match is then selected, across all the non-decoy channels, for each charged peptide. DIA-NN then assumes that this most confident channel pinpoints the correct retention time of the peptide. In the process we refer to as ‘translation of identifications’, DIA-NN re-extracts the signals at this retention time for the other channels, regardless of whether these have been successfully matched to some peak groups during the previous step. Scoring of these re-extracted peak groups using the previously trained neural network classifier leads to the assignment of ‘translated q-values’, which reflect the confidence level in these identifications if they were made independently from translation, and can be used for downstream data filtering. As each plexDIA acquisition measures multiple samples, DIA-NN calculates ‘channel q-values’ that reflect the confidence in the precursors being present in specific channels. This is achieved using a target-decoy method as explained in Methods. Finally, DIA-NN also takes advantage of the presence of multiple channels when quantifying precursors. Here, DIA-NN calculates the ratios between different channels using the signal ratios for selected fragment ions at the elution apex, thus minimizing the influence of any interfering signals. The ‘translated’ quantities are then calculated for all the channels except the most confident one, by dividing the quantity in the latter by the respective ratio.
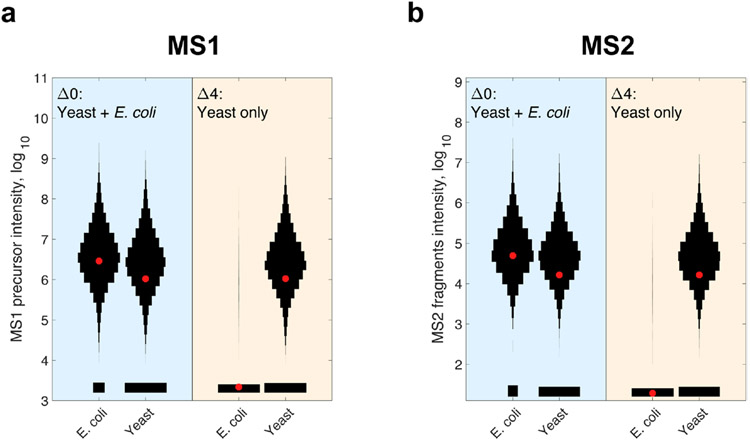
Extended Data Fig. 2 ∣. plexDIA analysis of proteins present only in one samples but missing from another.
We sought to test identification propagation by plexDIA for the case when proteins are present only in some samples and not in others. To do so, we prepared a standard in which one sample (labeled with mTRAQ, 0) had both 0.3 μg E. coli and 0.3 μg S. cerevisiae while another (labeled with mTRAQ Δ4) had only 0.3 μg S. cerevisiae. The combined set was analyzed by plexDIA using the V1 method. (a) Distributions of raw MS1 precursor intensity for E. coli and S. cerevisiae precursors at channel-q-value < 0.01. (b) Distributions of raw MS2 quantification of precursors filtered for channel-q-value < 0.01. The red asterisks correspond to the means of the distributions.
Extended Data Fig. 3 ∣. plexDIA proteomic coverage and data completeness for V2.
(a) Number of distinct precursors identified from 60 min active gradient runs for plexDIA, LF-DIA, and shotgun-DDA of mTRAQ at 1 % FDR. The DIA analysis used the V2 method, an MS2-optimized data acquisition cycle shown in Fig. 1e. Triplicates of each sample were analyzed (except sample C of LF-DIA, duplicates are analyzed), and the results displayed as mean; error bars correspond to standard error. (b) Total number of protein data points for plexDIA, LF-DIA and mTRAQ DDA at 1 % global protein FDR, (n = 3). (c) Venn-diagrams of each replicate for plexDIA and LF-DIA display protein groups quantified across samples A, B and C. The mean number of proteins groups intersected across samples A, B and C is 7,923 for plexDIA and 8,318 for LF-DIA. (d) We compute pairwise Jaccard indices to compare pairwise data completeness between plexDIA, LF-DIA and shotgun DDA for mTRAQ. All data were analyzed using match between runs. (e) Distributions of missing data between pairs of runs of either the same sample (that is, replicate injections) or between different samples.
Extended Data Fig. 4 ∣. Comparison of proteomic overlap between our runs to a high-quality DIA dataset Navarro et al.
DIA runs (including raw data from Navarro et al.) were searched with DIA-NN using match between runs. Results indicate that the data completeness from LF-DIA in this study is comparable to other high quality LF-DIA datasets.
Extended Data Fig. 5 ∣. plexDIA quantitative accuracy for MS2-optimized data acquisition (V2).
As demonstrated with the MS1-optimized method in Fig. 3 of the main text, here we show quantitative accuracy of plexDIA using MS2-optimized data acquisition—specifically, we only show data from the second run of a triplicate set. (a) The number of protein groups quantified in both samples A and B is shown with barplots. plexDIA quantified 7,610 PGs, LF-DIA 9,387 PGs, and intersected between plexDIA and LF-DIA was 5,967 PGs. These 5,967 PGs were plotted to compare quantitative accuracy between plexDIA and LF-DIA for in-common protein groups. To improve visibility, the scatter plot x and y axes were set to display data points between 0.25% and 99.75% range. (b) Same as (a), but for samples A and C; human proteins were excluded because they compare two different human cell types. (c) Same as (b), but for samples B and C. (d) Absolute protein ratio errors were calculated for samples A/B, A/C and B/C and combined to compare ratio errors for samples within a plexDIA run (for example, run2 A / run2 B) to samples across runs (for example, run1 A /run2 B) with plexDIA. (e) Absolute precursor ratio errors were calculated for samples A/B, A/C and B/C and combined to compare MS2-quantified ratio errors for C-terminal lysine precursors and C-terminal arginine precursors. Boxplots: The box defines the 25th and 75th percentiles and the median is marked by a solid line. Outliers are marked as individual dots outside the whiskers. All data shown are from (n = 1) representative replicate.
Extended Data Fig. 6 ∣. Quantitative accuracy for DIA replicates using V1.
Similar to main Fig. 3, we display the results from the other replicates for a total of (n = 3) replicates. (a) Figures are the same as shown in Fig. 3 of the main text with the exception that this shows the first replicate of plexDIA and the first replicate of samples A, B and C for LF-DIA. (b) Same as (a), but for the third replicate of plexDIA and LF-DIA, samples A, B and C. Boxplots: The box defines the 25th and 75th percentiles and the median is marked by a solid line. Outliers are marked as individual dots outside the whiskers. Panel a shows data from (n = 1) replicate and panel b shows data from another (n = 1) replicate for a total of (n = 2) replicates shown in this figure.
Extended Data Fig. 7 ∣. Quantitative accuracy and repeatability across different plexDIA sets and labels.
(a) Relative protein levels between samples A,B and C estimated from samples analyzed in different plexDIA sets, that is, out-of-set quantification. The quantitative accuracy between sets (and thus runs) is comparable to the set accuracy shown in Fig. 3. The display is the same as shown in main Fig. 3, but the protein ratios are estimated across runs (for example, run 1 A / run 2 B); LF-DIA is showing protein ratios for the 2nd replicate of samples A, B and C. (b) Same as (a), but for samples A and C; H. sapiens proteins were not analyzed because they are from distinct cell types. (c) Same as (b) but for samples B and C. All data shown in panels a–c are from (n = 1) representative replicate. Boxplots: The box defines the 25th and 75th percentiles and the median is marked by a solid line. Outliers are marked as individual dots outside the whiskers. (d) Quantitative repeatability of plexDIA across across different labels. Protein CVs were estimated for the same samples labeled with the same label (as in main Fig. 4) or for the same sample labeled with different labels in different runs for example, run 1, Δ0, sample A & run 2, Δ4, sample A & run 3, Δ8, sample A. Both distributions contains CV for the same set of (n = 15,158) sample-specific protein data points per condition (Same Labels or Different Labels). The median CV when using the same label was 0.110 while the label swap had a median CV of 0.148.
Extended Data Fig. 8 ∣. plexDIA missing data in single cells and negative controls.
Percent of precursors with no MS1-level quantitation per single cell or negative control. Single cells were required to have <60% missing data to be included in downstream analysis.
Extended Data Fig. 9 ∣. Single-cell PCA colored by mTRAQ label.
Rather than colors corresponding to a cell type as performed in Fig. 6p, here colors correspond to which mTRAQ label was used to tag the single cells. This is performed to check whether labeling-induced biases affect clustering of single-cells; here there appears to be little to no effect.
Extended Data Fig. 10 ∣. Relative protein abundances for each species per label.
Distribution of relative protein abundance of each species across labels. The Δ0, Δ4 and Δ8 samples were pooled and used for quantitative benchmarking of plexDIA.
Supplementary Material
Acknowledgements
We thank A. Makarov, E. Gordon and D. Perlman for discussions and constructive comments. This work was funded by a New Innovator Award from the National Institute of General Medical Sciences from the National Institutes of Health to N.S. under award DP2GM123497, an Allen Distinguished Investigator award through the Paul G. Allen Frontiers Group to N.S. and a Seed Networks Award from CZI CZF2019-002424 to N.S. This work received further support from The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001134); the UK Medical Research Council (FC001134); and the Wellcome Trust (FC001134 and IA 200829/Z/16/Z), as well as the European Research Council (SyG 951475 to M.R.). The work was further supported by the German Federal Ministry of Education and Research (BMBF), as part of the National Research Node ‘Mass Spectrometry in Systems Medicine’ (MSCoresys), under grant agreements 031L0220A to M.R. and 161L0221 to V.D.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-022-01389-w.
Reporting summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Data, code and protocols are available at https://plexdia.slavovlab.net/ and https://github.com/SlavovLab/plexDIA. Supporting information for the single-cell plexDIA is available at https://scp.slavovlab.net/plexDIA.
Competing interests
M.W. is an employee of Bruker Corporation, which manufactures timsTOF SCP. The authors declare that they have no other competing financial interests.
Extended data is available for this paper at https://doi.org/10.1038/s41587-022-01389-w.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-022-01389-w.
Data availability
The raw data and search results are available at MassIVE: MSV000089093 Processed data and metadata are available at https://scp.slavovlab.net/Derks_et_al_2022.
References
- 1.Bekker-Jensen DB et al. An optimized shotgun strategy for the rapid generation of com- prehensive human proteomes. Cell Syst. 4, 587–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedrich C et al. Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nat. Commun 12, 3576 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xuan Y et al. Standardization and harmonization of distributed multi-center proteotype analysis supporting precision medicine studies. Nat. Commun 11, 5248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J et al. TMTpro-18plex: the expanded and complete set of TMTpro reagents for sample multiplexing. J. Proteome Res. 20, 2964–2972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messner CB et al. Ultra-fast proteomics with Scanning SWATH. Nat. Biotechnol 39, 846–854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petelski AA et al. Multiplexed single-cell proteomics using SCoPE2. Nat. Protoc 16, 5398–5425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slavov N Driving single cell proteomics forward with innovation. J. Proteome Res 20, 4915–4918 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slavov N Increasing proteomics throughput. Nat. Biotechnol 39, 809–810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slavov N Unpicking the proteome in single cells. Science 367, 512–513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A Towards resolving proteomes in single cells. en. Nat. Methods 18, 856 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Slavov N Scaling up single-cell proteomics. Mol. Cell. Proteomics 21, 100179 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S & Heck AJ Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc 4, 484–494 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Fonslow BR, Shan B, Baek M-C & Yates JR III Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 113, 2343–2394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petelski AA & Slavov N Analyzing ribosome remodeling in health and disease. Proteomics 20, e2000039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mertins P et al. iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell. Proteomics 11, M111.014423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell JD, Paulo JA, O’Brien JJ & Gygi SP Proteome-wide evaluation of two common protein quantification methods. J. Proteome Res 17, 1934–1942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muntel J et al. Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res 18, 1340–1351 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Rauniyar N & Yates JR III Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res 13, 5293–5309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Specht H & Slavov N Transformative opportunities for single-cell proteomics. J. Proteome Res 17, 2563–2916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Specht H & Slavov N Optimizing accuracy and depth of protein quantification in experiments using isobaric carriers. J. Proteome Res 20, 880–887 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venable JD, Dong M-Q, Wohlschlegel J, Dillin A & Yates JR Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 (2004) . [DOI] [PubMed] [Google Scholar]
- 22.Dong M-Q et al. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Navarro P et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat. Biotechnol 34, 1130–1136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Costa C et al. Impact of the identification strategy on the reproducibility of DDA and DIA results. J. Proteome Res 19, 3153–3161 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demichev V, Messner CB, Vernardis SI, Lilley KS & Ralser M DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinitcyn P et al. MaxDIA enables library-based and library-free data-independent acquisition proteomics. Nat. Biotechnol 39, 1563–1573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demichev V et al. High sensitivity dia-PASEF proteomics with DIA-NN and FragPipe. Preprint at https://www.biorxiv.org/content/10.1101/2021.03.08.434385v1.full (2021). [Google Scholar]
- 28.Slavov N Single-cell protein analysis by mass spectrometry. Curr. Opin. Chem. Biol 60, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minogue CE et al. Multiplexed quantification for data-independent acquisition. Anal. Chem 87, 2570–2575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y et al. Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat. Commun 8, 1212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pino LK, Baeza J, Lauman R, Schilling B & Garcia BA Improved SILAC quantification with data-independent acquisition to investigate bortezomib-induced protein degradation. J. Proteome Res 20, 1918–1927 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong X et al. Mass defect-based DiLeu tagging for multiplexed data-independent acquisition. Anal. Chem 92, 11119–11126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian X, de Vries MP, Permentier HP & Bischoff R A versatile isobaric tag enables proteome quantification in data-dependent and data-independent acquisition modes. Anal. Chem 92, 16149–16157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian X, de Vries MP, Permentier HP & Bischoff R The isotopic Ac-IP tag enables multiplexed proteome quantification in data-independent acquisition mode. Anal. Chem 93, 8196–8202 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salovska B et al. Isoform-resolved correlation analysis between mRNA abundance regulation and protein level degradation. Mol. Syst. Biol 16, e9170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haynes SE, Majmudar JD & Martin BR DIA-SIFT: a precursor and product ion filter for accurate stable isotope data-independent acquisition proteomics. Anal. Chem 90, 8722–8726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salovska B, Li W, Di Y & Liu Y BoxCarmax: a high-selectivity data-independent acquisition mass spectrometry method for the analysis of protein turnover and complex samples. Anal. Chem 93, 3103–3111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Kang U-B, Yeom J, Kim H & Lee C Quantitative analysis of mTRAQ-labeled proteome using full MS scans. J. Proteome Res 9, 3750–3758 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Cox J et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper S The synchronization manifesto: a critique of whole-culture synchronization. FEBS J. 286, 4650–4656 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Aguilar V & Fajas L Cycling through metabolism. EMBO Mol. Med 2, 338–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slavov N & Botstein D Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol. Bio. Cell 22, 1997–2009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leduc A, Huffman RG & Slavov N Droplet sample preparation for single-cell proteomics applied to the cell cycle. Preprint at https://www.biorxiv.org/content/10.1101/2021.04.24.441211v1 (2021). [Google Scholar]
- 45.Fernandez-Lima F, Kaplan DA, Suetering J & Park MA Gas-phase separation using a trapped ion mobility spectrometer. Int. J. Ion Mobil. Spectrom 14, 10.1007/s12127-011-0067-8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunner A-D et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol 18, e10798 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cong Y et al. Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell. Chem. Sci 12, 1001–1006 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slavov N Counting protein molecules for single-cell proteomics. Cell 185, 232–234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denisov E, Damoc E & Makarov A Exploring frontiers of orbitrap performance for long transients. Int. J. Mass Spectrom 466, 116607 (2021). [Google Scholar]
- 50.Specht H et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 22, 50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 17, 399–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huffman RG et al. Prioritized single-cell proteomics reveals molecular and functional polarization across primary macrophages. Preprint at https://www.biorxiv.org/content/10.1101/2022.03.16.484655v1 (2022). [Google Scholar]
- 53.Budnik B, Levy E, Harmange G & Slavov N SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 19, 161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slavov N Learning from natural variation across the proteomes of single cells. PLOS Biol. 20, e3001512 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franks A, Airoldi E & Slavov N Post-transcriptional regulation across human tissues. PLoS Comput. Biol 13, e1005535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bamberger C et al. Protein footprinting via covalent protein painting reveals structural changes of the proteome in Alzheimer’s disease. J. Proteome Res 20, 2762–2771 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slavov N Measuring protein shapes in living cells. J. Proteome Res 20, 3017–3017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 58.Specht H et al. Automated sample preparation for high-throughput single-cell proteomics. Preprint at https://www.biorxiv.org/content/10.1101/399774v1 (2018). [Google Scholar]
- 59.Keshishian H et al. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc 12, 1683–1701 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budnik B, Levy E, Harmange G & Slavov N Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Preprint at https://www.biorxiv.org/content/10.1101/102681v1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huffman G, Chen AT, Specht H & Slavov N DO-MS: data-driven optimization of mass spectrometry methods. J. Proteome Res 18, 2493–2500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huntley R et al. The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res. 43, D1057–D1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hulstaert N et al. ThermoRawFileParser: modular, scalable, and cross-platform RAW file conversion. J. Proteome Res 19, 537–542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eiler J et al. Analysis of molecular isotopic structures at high precision and accuracy by Orbitrap mass spectrometry. Int. J. Mass Spectrom 422, 126–142 (2017). [Google Scholar]
- 65.Makarov A & Denisov E Dynamics of ions of intact proteins in the Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom 20, 1486–1495 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and search results are available at MassIVE: MSV000089093 Processed data and metadata are available at https://scp.slavovlab.net/Derks_et_al_2022.