Abstract
Strategies to minimize immune-suppressive medications after liver transplantation are limited by allograft rejection. Biopsy of liver is the current standard of care in diagnosing rejection. However, it adds to physical and economic burden to the patient and has diagnostic limitations. In this review, we aim to highlight the different biomarkers to predict and diagnose acute rejection. We also aim to explore recent advances in molecular diagnostics to improve the diagnostic yield of liver biopsies.
Keywords: Liver transplantation, immune-suppression, rejection, biomarker, molecular diagnosis, micro-RNA, donor-derived cell-free DNA, MMDX, nano-string
Abbreviations: LT, Liver transplantation; UNOS, United network for organ sharing and procurement; ILTS, International liver transplantation society; CNI, Calcineurin inhibitors; MToR, Mechanistic target of Rapamycin; MMF, Mycophenolate Mofetil; ATG, Anti-thymoglobulin; GLUT-4, glucose transport-4; ATCMR, acute T-cell mediated rejection; AMR, antibody mediated rejection; APC, antigen presenting cells; MHC, major histo–compatibility complex; FDA, Food and drug administration; AR, Acute rejection; miRNA, micro-RNA; dd cfDNA, donor-derived cell-free DNA; HNMR, high nuclear magnetic resonance; mRNA, messenger RNA; AUC, area under curve; AUROC, area under receiver operating characteristic curve; PPV, Positive predictive value; NPV, Negative predictive value; DSA, Donor specific antibodies; HLA, human leukocyte antigens; MFI, mean fluorescence intensity; MMDX, Molecular microscopic diagnostic system; RATs, rejection associated transcripts; TBB, trans-bronchial biopsies; 3BMBs, third bifurcation mucosal endo-bronchial biopsies; FFPE, formalin fixed paraffin embedded preparation; B-HOT, Banff Human Organ Transplant; MDWG, molecular diagnostic work group
Graphical abstract
LT represents the only curative option for decompensated liver disease.1 According to United network for organ sharing and procurement (UNOS), 8869 LTs were performed in the US in 2019.2 One of the challenges that affect graft survival and overall mortality in transplanted patients is graft rejection.3 Liver biopsy is considered the SOC to diagnose acute rejection and serves as the cornerstone for therapeutic interventions. However, it is invasive, expensive, and may result in complications. Also, its interpretation may be difficult in the presence of infection or recurrence of primary disease. There is a need of non-invasive biomarkers to predict the onset and severity of acute rejection and improvement in histological markers to make an accurate diagnosis.4
In this review, we aim to explore newer biomarkers and advances in molecular diagnostics and their promising use in patients with LT.
Current state of immune-suppression in liver transplantation
Transplantation of any solid organ induces a T-cell-mediated immune response, which might lead to graft rejection. Although liver has a special micro-environment that promotes tolerance, immune-suppressive (IS) medications are still necessary to avoid graft rejection and failure.5 They are used to induce and maintain graft tolerance, and also to treat rejection.6 According to the latest consensus statement by the international liver transplantation society (ILTS), many regimens are used as IS agents in LT, and include one or more of the following pharmacological classes:
-
•
Corticosteroids
-
•
Calcineurin inhibitors (CNIs): Tacrolimus and cyclosporine
-
•
Mechanistic target of Rapamycin (MTOR) inhibitors: Sirolimus and everolimus
-
•
Anti-metabolites: Mycophenolate mofetil and azathioprine
-
•
Depleting monoclonal antibodies: Anti-thymoglobulin and alemtuzumab
-
•
Non-depleting monoclonal antibodies: Basiliximab7
Each class and specific agent has unique efficacy and side–effect profile, and a tailored immune-suppression is essential for the best outcome after transplantation.6,8
Challenges with immune-suppression
Long-term follow-up of liver transplant patients shows significant morbidity and all-cause mortality related to immune-suppression. Infection is the most common cause of death in the first year post-LT. Malignancy, cardiovascular complications, and renal impairment constitute the main causes of death in later years.9 CNIs are associated with diabetes mellitus due to their effects on glucose transport-4 thus increasing insulin resistance and also affecting β-cell functions, while MTOR inhibitors are associated with hypertriglyceridemia and metabolic syndrome evolution.10 Chronic kidney disease resulting from hyaline arteriolosclerosis, glomerulo-sclerosis, and tubule-interstitial nephritis is a recognized risk in up to 70% of LT patients with chronic CNIs.11,12 Multiple reports showed that immune-suppression is linked to development of skin cancers (squamous cell carcinoma and basal cell carcinoma), lymphoproliferative disorders, colorectal carcinoma, and lung cancers in LT patients.13
The complications of immune-suppression are dependent on dose, class, and duration of the drugs. Therefore, reduction in immune-suppression is helpful in most patients starting early following transplantation and a full withdrawal of immune-suppression may be possible in select patients.7
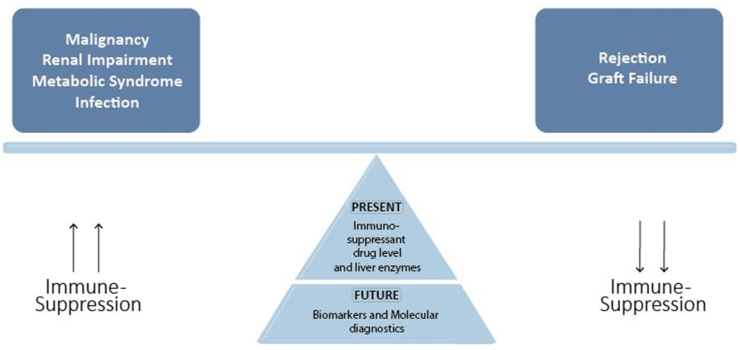
However, strategies to minimize or withdraw immune-suppression have been limited by acute graft rejection. Shaked et al. reported that 42 out of 52 patients did not tolerate reduction of immune-suppression dose to less than 50% of the baseline due to development of acute rejection.14 Figure 1 shows that balancing immune-suppression to avoid complications is necessary; however, present tools are inaccurate in evaluating the state of immune-suppression. Future direction focuses on biomarkers and molecular diagnostics to accurately identify the underlying pathways of liver rejection.
Figure 1.
Current and future strategies to adjust immune-suppression.
Rejection and graft survival
Rejection in LT is a major cause of morbidity and may lead to graft failure and mortality. Graft rejection may present as acute T-cell-mediated rejection (ATCMR), antibody-mediated rejection (AMR), and chronic rejection. Each subtype has different pathophysiological characteristics and clinical presentation and requires different management modalities.3
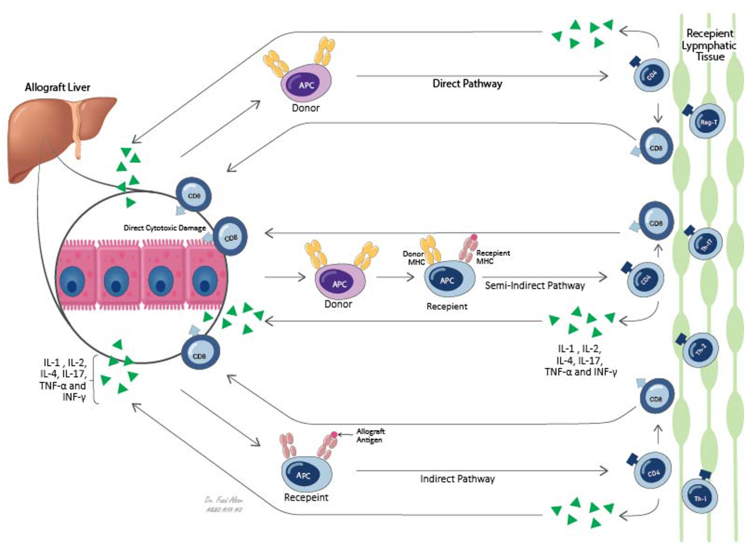
ATCMR occurs in 15–25% of LT patients in the first year following LT. It presents as elevated aminotransferases, bilirubin, and cholestatic enzymes, it may be asymptomatic in 7–11% of patients, especially after 1 year post-transplantation.15, 16, 17, 18 ATCMR has two overlapping phases: allograft recognition and effector responses. Allograft recognition has 3 pathways (Figure 2): (1) direct pathway in which the donor antigen presenting cells (APCs) migrate to the lymphoid tissues where the recipient naive CD4+ T-lymphocyte recognizes both the presented antigen and the major histo–compatibility complex (MHC) class II on donor APCs as allogeneic. Similarly, donor class I MHC molecules also are recognized by CD8+ T-cell.19 (2) Indirect pathway in which the recipient APCs present phagocytosed allogeneic material shed by donor cells to the recipient CD4+ T-helper cells. (3) Semi-indirect pathway in which a transfer of donor MHC molecules from donor APCs to recipient dendritic cell occurs by exocytosis, and then, antigen presentation takes place through the recipient APCs.20 These pathways lead to naive CD4+ T-cell to differentiate to Th1, Th2, Th17, and Reg-T-cells and migration of both CD4+ T-cells and CD8+ cells to the liver graft.21 The second phase involves cellular damaged by the recruited T-cells. CD8+ T-cell is the main mediator of the direct cytotoxic damage via perforin/granzyme pathway and FAS/FASL pathway leading to direct hepatocyte damage, aided by the various cytokines produced by CD4+ T-cell subtypes which include IL-1, IL-2, IL-4, IL-17, TNF-α, and INF-γ.20
Figure 2.
Allograft recognition pathways and immune effector pathways. 3 pathways has been identified; in direct pathway donor APCs, MHC class II molecules are recognized by recipient CD4+ T-cells as foreign and recipient CD8+ T-cells recognize MHC class I molecules on donor APCs. In indirect pathway, recipient APCs present graft antigen to recipient CD4+ T-cells, semi-indirect pathway combines both pathways as recipient APCs present exocytosis transferred donor MHC molecules to recipient CD4+ T-cells. Effector phase depends on hepatocyte cytotoxicity by activated CD8+ T-cell augmented by cytokines release from CD4+ T-cells.
AMR is poorly understood in LT, with incidence less than 1% in ABO matched grafts.22 Banff group defines AMR with the presence of four criteria: (1) endothelial hypertrophy, portal capillary dilatation, micro-vasculitis, and involvement of the central vein; (2) C4d deposition in portal microvascular endothelium; (3) high titer of donor specific antibodies; (4) no other cause to explain the presentation.23
Chronic graft rejection is a late complication that usually develops months to years after transplantation. It is uncommon complication that occurs in 3.3% of LT recipients,24,25 mostly related to non-optimized immune-suppression, characterized by ductopenia and parenchymal fibrosis and may lead to graft failure requiring re-transplantation.26 Graft rejection leads to decreased overall graft survival, increase in hospital admission and mortality, irrespective of the time and type of rejection from transplantation.27,28
Currently, there are no direct measures to evaluate the state of immune-suppression in the graft. We monitor IS drug levels and biochemical markers of liver inflammation with elevated liver enzymes and rely on liver biopsy to diagnose rejection as the SOC. However, liver enzymes cannot differentiate between rejection, infection, or biliary obstruction. Biopsies are expensive, time consuming, and carry a risk of bleeding. It led to the search for a more affordable, easier to implement, and safer non-invasive biomarkers. The goal is to predict and diagnose rejection to guide clinical interventions early in the course for a better outcome.29,30
History of biomarkers for rejection
A biomarker is a measurable quantity that can evaluate a physiological process, assess, and predict a pathological one. Ideally, it needs to be non-invasive, inexpensive, and easy to perform with a high specificity and sensitivity, and reproducible with validated studies.31
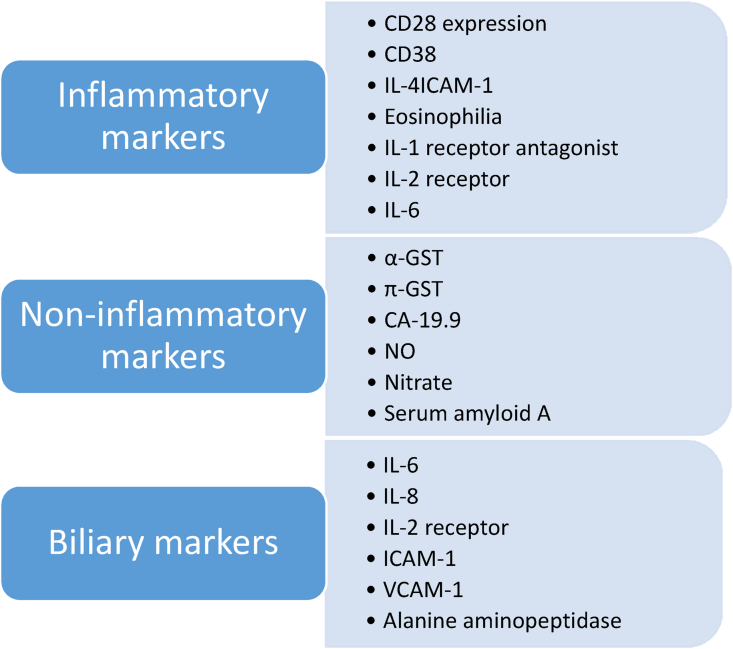
Search of biomarkers to detect liver rejection started 40 years ago with Neopterin, a catabolic product of purine metabolism and a marker of macrophage activity which was the first one to be studied.32 Further search for biomarkers focused on cytokines and inflammatory markers such as IL-2R, TNF-α, CD-28 expression, IL-15 and β2-microglobulin, biliary markers, and other markers not related to inflammation. However, all of them failed to be implemented into clinical use due to their inability to differentiate ATCMR from other conditions (infection and biliary obstruction) and the absence of validation in large multicenter studies, Figure 3 summarizes categories of the these biomarkers.33
Figure 3.
Historic biomarkers of liver rejection.
ImmuKnow, a marker approved by FDA in 2002, is a molecular assay using intra-cellular concentration of adenosine triphosphate (ATP) in CD4+ T-helper cell to assess immune system activity.34 It was proposed that CD4+ cellular activity correlates with the state of immune-suppression. It can predict rejection with a higher ATP level and infection with a lower level. Despite its initial promising results, pooled results in a meta-analysis failed to show its predictive value in acute rejection with a sensitivity of 0.43, specificity of 0.75, and positive likelihood ratio of 1.30 (95% CI: 0.74–2.28). Another meta-analysis showed that low levels of ATP may identify patients with increased risk of infection; however, no correlation was found with acute rejection.35,36
Era of new biomarkers
Advances in molecular biology have led to the evolution of modern fields of biomarkers such as metabolic, transcription factors, and genetic biomarkers.33 We systematically reviewed the literature to identify prominent studies exploring the new biomarkers, which are summarized in Table 1.
Table 1.
New Biomarkers of Rejection in Liver Transplantation.
| Author | Biomarker | Sample type | population | Outcome | |
|---|---|---|---|---|---|
| Metabolic biomarkers | Duarte37 | Phospholipid and triglyceride content | Liver biopsy | 6 adults | Identification of liver graft metabolic profile by HNMR |
| Cortes38 | Phospholipid, histidine and Bile acid biosynthesis | Liver biopsy | 124 adults | Predicting graft rejection by HNMR | |
| Transcription Factors | Muthukumar39 | miRNA-22, miR-34a, miRNA-122, miR-148a, miRNA-192, miRNA-193b, miRNA-194, miRNA-210 and miRNA-885-5p | Serum | 91 adults | Diagnosis of acute rejection, differentiating AR from other inflammatory conditions |
| Shaked40 | hsa-miRNA-483-3p and hsa-miRNA-885-5p | Serum | 69 adults | Differentiating AR from other inflammatory conditions | |
| Genetic biomarkers | Levitsky41 | 36-gene probe | serum | 186 adults | Diagnosis and prediction of AR |
| Goh42 | dd cfDNA | Serum | 20 adults | Diagnosis of AR | |
| Schütz43 | dd cfDNA | serum | 115 adults | Early detection of AR | |
| Zhao44 | dd cfDNA | serum | 49 pediatric | Diagnosis of AR |
Abbreviations: AR, Acute rejection; miR, micro-RNA; dd cfDNA, donor-derived cell-free DNA; HNMR, high nuclear magnetic resonance.
Metabolic biomarkers
Metabolic biomarkers depend on detecting multiple metabolic products or intermediary products to assess pathophysiologic processes during liver rejection.45 One of the earliest reports described the use of Hi-nuclear magnetic resonance to evaluate the metabolic profile of grafted liver before and after LT.37 Another report described the different metabolic patterns associated with graft dysfunction and development of rejection after transplantation.38 Despite their promising prospective, no metabolic profiling technique has yet been transferred to clinical practice.
The use of fecal microbiota to predict acute rejection is being explored. An ongoing clinical trial is exploring the use of fecal microbiota in patients who underwent living donor LT to predict the development of acute rejection in the first year after transplantation.46 More studies encompassing different cohorts are needed to validate the use of microbiota as a biomarker for acute rejection.
Transcription factors
Transcription factors associated with liver rejection is another field of study in evolution. Micro-RNA is a small non-coding RNA sequence regulating transcription processes47 and is associated with acute liver rejection by upregulating pro-inflammatory TGF-β pathway and downregulating FOXP3 pathway in regulatory T-cells.48 A recent study using small-RNA sequencing and miRNA microarray on 91 LT serum samples identified nine micro-RNA sequences that showed statistically significant association with acute rejection and has the ability to distinguish acute rejection from other inflammatory conditions.39 Another study performed on serum samples of 69 liver recipients and compared to biopsies showed that 2 of the studied micro-RNA sequences were statistically significant in prediction, diagnosis, and differentiating acute rejection from other inflammatory conditions.40 Transcription biomarkers are the only biomarkers being studied with the potential of differentiating acute rejection from other inflammatory causes. Further studies are needed to validate the current results and assess their ability to be transferred to clinical application.
Genetic biomarkers
Genetic biomarkers are designed to recognize genomic signaling association with acute rejection and involves both messenger RNA (mRNA) and donor-derived cell-free DNA (dd cfDNA). A recent study has identified a mRNA probe consisting of 36 genes associated with high specificity and negative predictive values (sensitivity 0.57, specificity 0.82, PPV 0.47, and NPV 0.87, AUROC 0.925) in detecting acute rejection. In addition, it showed the ability to predict the occurrence of acute rejection even before development of liver enzyme derangement.49 It needs further validation in randomized trials.
Donor-derived cell-free DNA (dd cfDNA) is a new promising biomarker under investigation for use to detect cellular rejection in solid organ transplantation. It was hypothesized that shedding of dd cfDNA from graft may correlate with acute rejection. The utility of dd cfDNA has been explored in renal and heart transplant with promising results. In a report of pediatric renal transplantation, dd cfDNA showed sensitivity 86% and specificity 100% in the diagnosis of acute rejection (AUC 0.996)50 and is under investigation in patients with LT.51 In the setting of LT, dd cfDNA studied on 20 patients demonstrated a high sensitivity of 83.3% in detecting acute rejection with AUC 0.98.42 Another study showed that dd cfDNA has sensitivity of 90.3% (95% CI 74.2%–98.0%) in the detection of acute rejection and correlates earlier with the development of rejection compared to aminotransferases or cholestatic biomarkers. It also showed that due to a short half-life (<1.5 h), it reflected more accurately with the response to treatment (sensitivity and specificity of 97.1 and 89.3 compared to 95.7 and 82.1 for aminotransferases, respectively).43 To our knowledge, only one report analyzed the diagnostic utility of dd cfDNA in pediatric liver transplant patients. It showed that dd cfDNA yielded specificity of 81.9% and sensitivity of 81.8% in detecting acute rejection; however, its ability to differentiate acute rejection from other causes of liver injury was not explored in that study.44
Dd cfDNA has a limited ability to accurately differentiate acute rejection from other causes of graft injury such as viral infection (CMV, HCV, HBV), biliary complications, or ischemic injury.51 This limitation is more pronounced in LT than other solid organ transplants. A report of 171 heart transplant recipients showed that dd cfDNA showed a sensitivity of 95% and specificity of 92% and AUROC of 0.92 at a threshold of 0.5%.52 Another report of 93 renal transplant recipients showed that urinary dd cfDNA has the ability to distinguish acute rejection from viral infection with a sensitivity of 96%, specificity of 91%, and AUROC of 0.745 at a threshold of 0.84%.53 This disparity may be explained by the ability of hepatocytes to have unlimited proliferation compared to stable cells as in renal parenchyma and cardiac muscle. Consequently, we see higher baseline levels of cfDNA in liver transplant recipients in the absence of hepatocyte damage. In the study by Beck et al., dd cfDNA was <6.8% in stable liver transplant recipients, <2.5% in kidney transplant recipients, and <3.4% in heart transplant recipients.54 Hepatocyte proliferative ability and a higher baseline level of dd cfDNA may explain its limitation to differentiate AR from other causes of liver injury in liver transplant recipients compared to other solid organ recipients.55
None of the available biomarkers has yet reached optimum clinical applicability. Multiple obstacles hinder bench to bedside transition for these biomarkers. As this field is still in its infancy, there are no standardized laboratory methods to evaluate each biomarker, resulting in heterogeneity of study outcomes.56 Variability of expression of the biomarkers between patients and intra-patient variability during the clinical course affect their reliability.57 Furthermore, sample complexity in genetic biomarkers hinders the identification of valid biomarkers. Debey et al., showed that the presence of globin-mRNA complex leads to variability in analyzing gene expression by microarrays.58,59 Large prospective studies with well-defined study designs are needed before the acceptance of biomarkers in clinical practice.
Donor specific antibodies
Donor-specific antibodies (DSA) are immunoglobulins complimentary to polymorphic proteins that are different between the recipient and the graft donor, most DSAs are directed against donor human leukocyte antigen proteins, other DSAs such as angiotensin-II type-1 receptor antibodies, anti-glutathione S-transferase antibodies, and MHC-1-related chain A antibodies have been recognized. DSAs can be either preformed before LT or it can appear de novo after transplantation.60,61
DSAs are also associated with increased risk of graft rejection (acute and chronic). One meta-analysis showed that compared to patients who do not develop de novo DSAs, patients who develop them have a higher risk of developing acute rejection (OR 6.43, CI 3.17–13.04 P < 0.001). This association was more pronounced in pediatric population (OR 10.2, CI 4.65–22.33; P < 0.001).62
Banff work group updated criteria for diagnosing AMR include positive DSA, defined as mean fluorescence intensity (MFI) > 5000. MFI is a different metric for assessing DSA activity and was shown to have an association with clinical significance. MFI should be interpreted with caution as there is a lack of standard cut-off value and can be misleading if used as a single measure to evaluate AMR.23,63
No studies evaluating biomarkers in patients receiving multiple organ transplantation were identified.
Search for molecular diagnostics
Biopsy with histopathology can help in differentiating the cause of liver damage or formulate a differential diagnosis, stage the damage, and guide the therapeutic strategies.64 Biopsy can help differentiating the types of acute rejection (ATCMR, AMR, or chronic rejection) based on several criteria and stage each subtype.23
Despite being the SOC procedure, biopsy is far from ideal. Biopsy has two major limitations, inadequacy of liver biopsy sample, and variability of histopathological interpretation. Liver biopsy gives a representative sample of the entire liver tissue and Banff working group recommends a liver biopsy with a 16-gauge needle passing twice through the liver with more than 11 portal tracts to accurately represent the liver pathology. Reports showed that about 84% of biopsies fails to pass this threshold, and other reports showed that despite reaching this biopsy size, biopsy may miss the diagnosis suggesting that a sample of 11–16 portal tracts are necessary to adequately represent liver tissue.65 Liver biopsy interpretation is subjective and intra-observer and inter-observer bias has been shown to affect interpretation even when using Banff criteria. A report of 102 liver biopsy samples showed 27% discordance between experienced pathologists’ assessment of the biopsies for acute rejection and assessment by the initial pathologist.66 Due to these shortcomings of liver biopsy and the fact that till now there are no reliable biomarkers for diagnosis of acute rejection, many groups started to search for ways to improve diagnostic yield of liver biopsy and avoid inter and intra-observer bias.
Molecular microscopic diagnostic system
The earliest application for microarrays was tested in the early 2000's in the setting of kidney transplantation.67 The use of microarrays analysis in the setting of LT was validated in INTERLIVER study. The study aimed to analyze rejection-associated transcripts using molecular microscopic diagnostic system (MMDx) in identifying injury and rejection in liver biopsies using machine learning models.68 MMDx is a recent application of machine learning which uses microarrays to interpret human biopsies to improve the diagnostic value of liver biopsy. The validated computer models were trained to compare the histologic findings to database-stored biopsies. The models can provide data on rejection-related molecular changes. Additionally, it can provide an index which correlates with non-adherence or under immune-suppression.69 It can also predict survival of an organ with similar histological and molecular changes. MMDx shows higher reliability in kidney biopsies, and it showed diagnostic utility in both TCMR and AMR.70 MMDX can correlate histological finding to liver chemistries which is helpful in assessing the severity of rejection and monitoring response to treatment. This led to more comprehensive reporting of biopsies which will open new horizons in the early management and prediction of acute rejection.68 MMDX systems have also been studied to monitor dd cf-DNA in the serum to detect accurately diagnose acute rejection.71,72
Multiple challenges are still facing MMDX. It depends on the databases of biopsies which in themselves may be imprecise. Earlier models showed discrepancy between histological and molecular patterns of rejections. Biopsies used to train the models cannot distinguish between inflammation due to rejection and inflammation due to non-rejection etiologies that creates inconsistencies in the system.73 To overcome these challenges, multiple clinical trials are conducted in different solid organ transplantations to improve the system. Table 2 summarizes these trials.
Table 2.
Clinical Trials Evaluating Molecular Microscopic Diagnostic System.
| Study name | Status | Primary endpoint | Locality | Number of biopsies |
|---|---|---|---|---|
| INTERLIVER74 | Recruiting | Assign molecular scores of ATCMR and AMR in liver transplant biopsies | Multicenter | 800 biopsies |
| INTERHEART75 | Recruiting | Diagnosis of ATCMR and AMR in heart | Multicenter | 889 biopsies from 454 patients |
| Trifecta-Kidney cfDNA-MMDx Study72 | Recruiting | Compare DDcfDNA to MMDx, HLA, and histology in kidney | Multicenter | 700 biopsies and 2100 blood samples |
| Trifecta-Heart cfDNA-MMDx Study71 | Recruiting | Compare DD-cfDNA to MMDx to calibrate diagnosis of ATCMR and AMR in heart | Multicenter | 300 biopsies |
| INTERCOMEX76 | Recruiting | Validate the use of microarrays to reduce uncertainty in diagnosis of ATCMR and AMR in kidney | Multicenter | 1500 biopsies |
| INTERLUNGEX77 | Recruiting | Detect molecular identifiers in ATCMR and AMR in lung biopsies | Multicenter | 818 TBB and 657 3BMBs |
Abbreviations: TBB, trans-bronchial biopsies; 3BMBs, third bifurcation mucosal endo-bronchial biopsies.
Nano-string
One of the drawbacks of the MMDX systems is their dependability on a different tissue preservation technique other than conventional formalin-fixed paraffin embedded preparation (FFPE) thus limiting the practical application of MMDX.78
Nano-string® is a highly multiplexed RNA sequencing system that can analyze up to 800 target genes at the same time, it can use the standard FFPE biopsies without the need for any special preparation which gives it the advantage of being able to be studied and compared retrospectively to the archived biopsies.79 In a report evaluating liver biopsy from 133 pediatric LT, Nano-string® was used to analyze a model compromised of 194 genes to detect rejection in liver biopsies. The model was able to detect rejection (OR Log2 0.4 CI: Log2 0.2-Log2 0.6 P-value 0.05) and furthermore was able to give details on the underlying pathophysiological pathways of rejection.80
Further studies are still needed to evaluate the different RNA models to determine the most significantly associated genes with liver rejection and then to test these models prospectively to determine clinical applicability.
Banff human organ transplant gene panel
Banff molecular diagnostic work group (MDWG) did a literature review to search for genes associated with rejection in solid organ transplantation and identified 760 genes with most association. Using the Nano-string® platform, MDWG created the Banff human organ transplant (B-HOT) gene panel, to be used as research tool aiming at reaching a simplified panel to detect and classify acute rejection.81 A study evaluated B-HOT on a sample of renal transplant biopsies and found that this panel was a suitable surrogate for microarrays in the detection of genes associated with rejection. This is the first report to assess the efficacy of the new panel.82 MDWG started a multicenter cooperation to validate B-HOT panel and explore the aspects of its clinical application to improve patient care.81
Advances in molecular biology and artificial intelligence led to the development of multiple biomarkers and molecular diagnostic techniques, including micro-RNA, dd cfDNA, nano-string, and MMDX. We may need to engage expert pathologists to use both histology criteria and the molecular analyses and derive an integrated classification. Biomarkers and molecular diagnostics of liver rejection are still not ready for prime time and need more studies on multiple cohorts, including pediatric patients and simultaneous liver-kidney transplant recipients. Before any of them can be transferred from bench to bedside, we need prospective large scale multicenter studies to establish its role in the diagnosis of rejection and achieve the accuracy in predicting and preventing the occurrence of graft rejection. Only then we can achieve the optimization of immune-suppression, precision care with the prevention of long-term side-effects and graft loss, and improved survival with a better quality of life following transplantation.
Credit authorship contribution statement
All authors approved the final version of the manuscript.
Ahmed El Sabagh: Conceptualization, Writing - Original Draft, Visualization. Islam B Mohamed: Writing - Original Draft. Fuad Zain Aloor: Visualization, Designing illustrations. Ahmed Abdelwahab: Writing - Review & Editing. Manal M. Hassan: Writing - Review & Editing. Prasun K Jalal: Conceptualization, Writing - Review & Editing, Supervision, Project administration.
All authors: Critical revision of the manuscript.
Conflicts of interest
All authors have none to declare.
Acknowledgments
None.
Funding
Dora Roberts Foundation.
References
- 1.Shimamura T., Goto R., Watanabe M., Kawamura N., Takada Y. Liver transplantation for hepatocellular carcinoma: how should we improve the thresholds? Cancers. 2022;14:419. doi: 10.3390/cancers14020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.OTPN/SRTR OPTN/SRTR 2019 annual data report: Liver. https://srtr.transplant.hrsa.gov/annual_reports/2019/Liver.aspx
- 3.Levitsky J., Goldberg D., Smith A.R., et al. Acute rejection increases risk of graft failure and death in recent liver transplant recipients. Clin Gastroenterol Hepatol. 2016;15:584–593.e2. doi: 10.1016/j.cgh.2016.07.035. Clin Gastroenterol Hepatol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krenzien F., Keshi E., Splith K., et al. Diagnostic biomarkers to diagnose acute allograft rejection after liver transplantation: systematic review and meta-analysis of diagnostic accuracy studies. Front Immunol. 2019;10:758. doi: 10.3389/fimmu.2019.00758. Front Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez–Fueyo A., Strom T.B. Immunologic basis of graft rejection and tolerance following transplantation of liver or other solid organs. Gastroenterology. 2011;140:51–64. doi: 10.1053/j.gastro.2010.10.059. Gastroenterology. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moini M., Schilsky M.L., Tichy E.M. Review on immunosuppression in liver transplantation. World J Hepatol. 2015;7:1355–1368. doi: 10.4254/wjh.v7.i10.1355. World Journal of Hepatology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlton M., Levitsky J., Aqel B., et al. International liver transplantation society consensus statement on immunosuppression in liver transplant recipients. Transplantation. 2018;102:727–743. doi: 10.1097/TP.0000000000002147. Transplantation. [DOI] [PubMed] [Google Scholar]
- 8.Di Maira T., Little E.C., Berenguer M. Immunosuppression in liver transplant. Best Pract Res Clin Gastroenterol. 2020;46–47 doi: 10.1016/j.bpg.2020.101681. Baillière's best practice & researchClinical gastroenterology. [DOI] [PubMed] [Google Scholar]
- 9.Watt K.D.S., Pedersen R.A., Kremers W.K., Heimbach J.K., Charlton M.R. Evolution of causes and risk factors for mortality post-liver transplant: results of the NIDDK long-term follow-up study. Am J Transplant. 2010;10:1420–1427. doi: 10.1111/j.1600-6143.2010.03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azhie A., Sheth P., Hammad A., Woo M., Bhat M. Metabolic complications in liver transplantation recipients: how we can optimize long-term survival. Liver Transplant. 2021;27:1468–1478. doi: 10.1002/lt.26219. [DOI] [PubMed] [Google Scholar]
- 11.Beckebaum S., Cicinnati V.R., Radtke A., Kabar I. Calcineurin inhibitors in liver transplantation – still champions or threatened by serious competitors? Liver Int. 2013;33:656–665. doi: 10.1111/liv.12133. [DOI] [PubMed] [Google Scholar]
- 12.Ziolkowski J., Paczek L., Senatorski G., et al. Renal function after liver transplantation: calcineurin inhibitor nephrotoxicity. Transplant Proc. 2003/09/01;35:2307–2309. doi: 10.1016/S0041-1345(03)00786-3. 2003. [DOI] [PubMed] [Google Scholar]
- 13.Chak E., Saab S. Risk factors and incidence of de novo malignancy in liver transplant recipients: a systematic review: risk factors and incidence of de novo malignancy in liver transplant recipients. Liver Int. 2010;30:1247–1258. doi: 10.1111/j.1478-3231.2010.02303.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaked A., DesMarais M.R., Kopetskie H., et al. Outcomes of immunosuppression minimization and withdrawal early after liver transplantation. Am J Transplant : Off J Am Soc Transplant Am Soc Transplant Surg. 2019;19:1397–1409. doi: 10.1111/ajt.15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nacif L.S., Pinheiro R.S., Pécora RAdA., et al. Late acute rejection in liver transplant: a systematic review. Arq Bras Cir Dig. Jul-Sep 2015;28:212–215. doi: 10.1590/S0102-67202015000300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurairajah P.H., Carbone M., Bridgestock H., et al. Late acute liver allograft rejection; a study of its natural history and graft survival in the current era. Transplantation. Apr 15 2013;95:955–959. doi: 10.1097/TP.0b013e3182845f6c. [DOI] [PubMed] [Google Scholar]
- 17.Akamatsu N., Sugawara Y., Tamura S., et al. Late-onset acute rejection after living donor liver transplantation. World J Gastroenterol. 2006;12:6674–6677. doi: 10.3748/wjg.v12.i41.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson J.A., Schlitt H.J. Subclinical T cell-mediated liver transplant rejection: the jury is still out. J Hepatol. 2018;69:570–571. doi: 10.1016/j.jhep.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Demetris A.J., Bellamy C.O., Gandhi C.R., Prost S., Nakanuma Y., Stolz D.B. Functional immune anatomy of the liver-as an allograft. Am J Transplant. Jun 2016;16:1653–1680. doi: 10.1111/ajt.13749. [DOI] [PubMed] [Google Scholar]
- 20.Ronca V., Wootton G., Milani C., Cain O. The immunological basis of liver allograft rejection. Front Immunol. 2020;11:2155. doi: 10.3389/fimmu.2020.02155. 2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth L.A., Lechler R.I., Lombardi G. Continuous acquisition of MHC:peptide complexes by recipient cells contributes to the generation of anti-graft CD8(+) T cell immunity. Am J Transplant : Off J Am Soc Transplant Am Soc Transplant Surg. 2017;17:60–68. doi: 10.1111/ajt.13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demetris A.J., Zeevi A., O'Leary J.G. ABO-compatible liver allograft antibody-mediated rejection: an update. Curr Opin Organ Transplant. 2015;20:314–324. doi: 10.1097/MOT.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demetris A.J., Bellamy C., Hübscher S.G., et al. 2016 comprehensive update of the Banff working group on liver allograft pathology: introduction of antibody-mediated rejection. Am J Transplant. 2016;16:2816–2835. doi: 10.1111/ajt.13909. Am J Transplant. [DOI] [PubMed] [Google Scholar]
- 24.Angelico R., Sensi B., Manzia T.M., et al. Chronic rejection after liver transplantation: opening the Pandora's box. World J Gastroenterol. 2021;27:7771–7783. doi: 10.3748/wjg.v27.i45.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blakolmer K., Jain A., Ruppert K., et al. Chronic liver allograft rejection in a population treated primarily with tacrolimus as baseline immunosuppression: long-term follow-up and evaluation of features for histopathological staging. Transplantation. 2000;69:2330–2336. doi: 10.1097/00007890-200006150-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhary N.S., Saigal S., Bansal R.K., Saraf N., Gautam D., Soin A.S. Acute and chronic rejection after liver transplantation: what A clinician needs to know. J Clin Exp Hepatol. 2017;7:358–366. doi: 10.1016/j.jceh.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitsky J., Goldberg D., Smith A.R., et al. Acute rejection increases risk of graft failure and death in recent liver transplant recipients. Clin Gastroenterol Hepatol. 2016;15:584–593. doi: 10.1016/j.cgh.2016.07.035. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dogan N., Hüsing-Kabar A., Schmidt H.H., Cicinnati V.R., Beckebaum S., Kabar I. Acute allograft rejection in liver transplant recipients: incidence, risk factors, treatment success, and impact on graft failure. J Int Med Res. 2018;46:3979–3990. doi: 10.1177/0300060518785543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuberger J., Cain O. The need for alternatives to liver biopsies: non-invasive analytics and diagnostics. Hepat Med. 2021;13:59–69. doi: 10.2147/HMER.S278076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamali K., Schmelzle M., Kamali C., et al. Sensing acute cellular rejection in liver transplant patients using liver-derived extracellular particles: a prospective, observational study. Original research. Front Immunol. 2021-May-05:12doi. doi: 10.3389/fimmu.2021.647900. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Califf R.M. Biomarker definitions and their applications. Exp Biol Med (Maywood) 2018;243:213–221. doi: 10.1177/1535370217750088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margreiter R., Fuchs D., Hausen A., et al. Neopterin as a new biochemical marker for diagnosis of allograft rejection: experience based upon evaluation of 100 consecutive cases. Transplantation. 1983;36:650–653. doi: 10.1097/00007890-198336060-00013. Transplantation. [DOI] [PubMed] [Google Scholar]
- 33.Verhelst X.P.D., Troisi R.I., Colle I., Geerts A., van Vlierberghe H. Biomarkers for the diagnosis of acute cellular rejection in liver transplant recipients: a review: biomarkers for ACR in liver transplantation. Hepatol Res. 2013;43:165–178. doi: 10.1111/hepr.12012. [DOI] [PubMed] [Google Scholar]
- 34.Israeli M., Klein T., Sredni B., et al. ImmuKnow: a new parameter in immune monitoring of pediatric liver transplantation recipients. Liver Transplant. 2008/06/01;14:893–898. doi: 10.1002/lt.21426. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigo E., López-Hoyos M., Corral M., et al. ImmuKnow as a diagnostic tool for predicting infection and acute rejection in adult liver transplant recipients: a systematic review and meta-analysis. Liver Transplant. 2012;18:1244–1252. doi: 10.1002/lt.23497. [DOI] [PubMed] [Google Scholar]
- 36.Xiaoting L., Jun X., Wenhua L., et al. Can immune cell function assay identify patients at risk of infection or rejection? A meta-analysis. Transplantation. 2012;93:737–743. doi: 10.1097/TP.0b013e3182466248. [DOI] [PubMed] [Google Scholar]
- 37.Duarte I.F., Stanley E.G., Holmes E., et al. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Anal Chem. Sep 1 2005;77:5570–5578. doi: 10.1021/ac050455c. [DOI] [PubMed] [Google Scholar]
- 38.Cortes M., Pareja E., García-Cañaveras J.C., et al. Metabolomics discloses donor liver biomarkers associated with early allograft dysfunction. J Hepatol. Sep 2014;61:564–574. doi: 10.1016/j.jhep.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Muthukumar T., Akat K.M., Yang H., et al. Serum MicroRNA transcriptomics and acute rejection or recurrent hepatitis C virus in human liver allograft recipients: a pilot study. Transplantation. 2021 doi: 10.1097/TP.0000000000003815. Publish Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaked A., Chang B.L., Barnes M.R., et al. An ectopically expressed serum miRNA signature is prognostic, diagnostic, and biologically related to liver allograft rejection. Hepatology. 2017;65:269–280. doi: 10.1002/hep.28786. Hepatology. [DOI] [PubMed] [Google Scholar]
- 41.Levitsky J., Kandpal M., Guo K., Kleiboeker S., Sinha R., Abecassis M. Donor-derived cell-free DNA levels predict graft injury in liver transplant recipients. Am J Transplant. 2022;22:532–540. doi: 10.1111/ajt.16835. [DOI] [PubMed] [Google Scholar]
- 42.Goh S.K., Do H., Testro A., et al. The measurement of donor-specific cell-free DNA identifies recipients with biopsy-proven acute rejection requiring treatment after liver transplantation. Transplant Direct. 2019;5:e462. doi: 10.1097/TXD.0000000000000902. e462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schütz E., Fischer A., Beck J., et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: a prospective, observational, multicenter cohort study. PLoS Med. 2017;14 doi: 10.1371/journal.pmed.1002286. PLoS Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao D., Zhou T., Luo Y., et al. Preliminary clinical experience applying donor-derived cell-free DNA to discern rejection in pediatric liver transplant recipients. Sci Rep. 2021;11:1138. doi: 10.1038/s41598-020-80845-6. Sci Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortes M., García-Cañaveras J.C., Pareja E., Lahoz A. In: Biomarkers in Liver Disease. Patel V.B., Preedy V.R., editors. Springer Netherlands; 2017. Liver transplantation biomarkers in the metabolomics era; pp. 99–128. [Google Scholar]
- 46.Institute of Liver and Biliary Sciences I Role of Fecal Microbiota in Predicting Graft Rejection and Sepsis Among Recipients of Living Donor Liver Transplant in First Year. https://clinicaltrials.gov/ct2/show/NCT04621812?cond=Rejection%3B+Transplant%2C+Liver&draw=3&rank=18 Updated October 5, 2021.
- 47.Mohr A., Mott J. Overview of MicroRNA biology. Semin Liver Dis. 2015;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamdorf M., Kawakita S., Everly M. The potential of MicroRNAs as novel biomarkers for transplant rejection. J Immunol Res. 2017;2017:4072364. doi: 10.1155/2017/4072364. 4072364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levitsky J., Asrani S.K., Schiano T., et al. Discovery and validation of a novel blood-based molecular biomarker of rejection following liver transplantation. Am J Transplant. 2020;20:2173–2183. doi: 10.1111/ajt.15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puliyanda D.P., Swinford R., Pizzo H., Garrison J., De Golovine A.M., Jordan S.C. Donor-derived cell-free DNA (dd-cfDNA) for detection of allograft rejection in pediatric kidney transplants. Pediatr Transplant. Mar 2021;25 doi: 10.1111/petr.13850. [DOI] [PubMed] [Google Scholar]
- 51.Knight S.R., Thorne A., Lo Faro M.L. Donor-specific cell-free DNA as a biomarker in solid organ transplantation. A systematic review. Transplantation. Feb 2019;103:273–283. doi: 10.1097/tp.0000000000002482. [DOI] [PubMed] [Google Scholar]
- 52.Agbor-Enoh S., Shah P., Tunc I., et al. Cell-free DNA to detect heart allograft acute rejection. Circulation. 2021;143:1184–1197. doi: 10.1161/circulationaha.120.049098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X.-T., Chen W.-F., Li J., et al. Urine donor–derived cell-free DNA helps discriminate BK polyomavirus-associated nephropathy in kidney transplant recipients with BK polyomavirus infection. Original research. Front Immunol. 2020-August-19;11 doi: 10.3389/fimmu.2020.01763. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beck J., Bierau S., Balzer S., et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013;59:1732–1741. doi: 10.1373/clinchem.2013.210328. [DOI] [PubMed] [Google Scholar]
- 55.Grskovic M., Hiller D.J., Eubank L.A., et al. Validation of a clinical-grade Assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016/11/01;18:890–902. doi: 10.1016/j.jmoldx.2016.07.003. 2016. [DOI] [PubMed] [Google Scholar]
- 56.Terasaki P., Lachmann N., Cai J. Summary of the effect of de novo HLA antibodies on chronic kidney graft failure. Clin Transpl. 2006:455–462. [PubMed] [Google Scholar]
- 57.Min J.L., Barrett A., Watts T., et al. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genom. 2010;11:96. doi: 10.1186/1471-2164-11-96. 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Debey S., Zander T., Brors B., Popov A., Eils R., Schultze J.L. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics. May 2006;87:653–664. doi: 10.1016/j.ygeno.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 59.Roedder S., Vitalone M., Khatri P., Sarwal M.M. Biomarkers in solid organ transplantation: establishing personalized transplantation medicine. Genome Med. 2011;3:37. doi: 10.1186/gm253. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCaughan J.A., Tinckam K.J. Donor specific HLA antibodies & allograft injury: mechanisms, methods of detection, manifestations and management. Transpl Int. 2018;31:1059–1070. doi: 10.1111/tri.13324. [DOI] [PubMed] [Google Scholar]
- 61.Del Bello A., Congy-Jolivet N., Danjoux M., Muscari F., Kamar N. Donor-specific antibodies and liver transplantation. Hum Immunol. 2016/11/01;77:1063–1070. doi: 10.1016/j.humimm.2016.02.006. 2016. [DOI] [PubMed] [Google Scholar]
- 62.Beyzaei Z., Geramizadeh B., Bagheri Z., Karimzadeh S., Shojazadeh A. De novo donor specific antibody and long-term outcome after liver transplantation: a systematic review and meta-analysis. Front Immunol. 2020;11:613128. doi: 10.3389/fimmu.2020.613128. 613128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tambur A.R., Herrera N.D., Haarberg K.M., et al. Assessing antibody strength: comparison of MFI, C1q, and titer information. Am J Transplant. Sep 2015;15:2421–2430. doi: 10.1111/ajt.13295. [DOI] [PubMed] [Google Scholar]
- 64.Naini B.V.M.D., Lassman C.R.M.D.P. Liver transplant pathology. Surg Pathol Clin. 2013;6:277–293. doi: 10.1016/j.path.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Sanai F.M., Keeffe E.B. Liver biopsy for histological assessment: the case against. Saudi J Gastroenterol : Off J Saudi Gastroenterol Assoc. Apr-Jun 2010;16:124–132. doi: 10.4103/1319-3767.61244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coffin C.S., Burak K.W., Hart J., Gao Z-h. The impact of pathologist experience on liver transplant biopsy interpretation. Mod Pathol. 2006/06/01;19:832–838. doi: 10.1038/modpathol.3800605. 2006. [DOI] [PubMed] [Google Scholar]
- 67.Halloran P.F., Einecke G. Microarrays and transcriptome analysis in renal transplantation. Nat Clin Pract Nephrol. 2006/01/01;2:2–3. doi: 10.1038/ncpneph0066. 2006. [DOI] [PubMed] [Google Scholar]
- 68.Madill-Thomsen K., Abouljoud M., Bhati C., et al. The molecular diagnosis of rejection in liver transplant biopsies: first results of the INTERLIVER study. Am J Transplant. Aug 2020;20:2156–2172. doi: 10.1111/ajt.15828. [DOI] [PubMed] [Google Scholar]
- 69.Halloran P.F., Madill-Thomsen K.S. The Molecular Microscope(®) Diagnostic System meets eminence-based medicine: a clinician's perspective. Am J Transplant. Oct 2020;20:2964–2965. doi: 10.1111/ajt.15940. [DOI] [PubMed] [Google Scholar]
- 70.Madill-Thomsen K.S., Wiggins R.C., Eskandary F., Böhmig G.A., Halloran P.F. The effect of cortex/medulla proportions on molecular diagnoses in kidney transplant biopsies: rejection and injury can Be assessed in medulla. Am J Transplant. Aug 2017;17:2117–2128. doi: 10.1111/ajt.14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uo Alberta. Trifecta-Heart cfDNA-MMDx Study. https://clinicaltrials.gov/ct2/show/NCT04707872
- 72.Uo Alberta. Trifecta-Kidney cfDNA-MMDx Study. https://clinicaltrials.gov/ct2/show/NCT04239703
- 73.Randhawa P.S. The molecular microscope diagnostic system (MMDx) in transplantation: a pathologist's perspective. Am J Transplant. Aug 2020;20:1965–1966. doi: 10.1111/ajt.15887. [DOI] [PubMed] [Google Scholar]
- 74.Halloran P. Diagnostic and Therapeutic Applications of Microarrays in Liver Transplantation (INTERLIVER) https://clinicaltrials.gov/ct2/show/NCT03193151
- 75.Halloran P. Diagnostic and Therapeutic Applications of Microarrays in Heart Transplantation. https://clinicaltrials.gov/ct2/show/NCT02670408
- 76.Halloran P. Diagnostic and Therapeutic Applications in Microarrays in Organ Transplantation. https://clinicaltrials.gov/ct2/show/NCT01299168
- 77.Halloran P. Diagnostic and Therapeutic Applications of Microarrays in Lung Transplantation. https://clinicaltrials.gov/ct2/show/NCT02812290
- 78.Madill-Thomsen K., Perkowska-Ptasińska A., Böhmig G.A., et al. Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant. 2020;20:1341–1350. doi: 10.1111/ajt.15752. [DOI] [PubMed] [Google Scholar]
- 79.Adam B., Afzali B., Dominy K.M., et al. Multiplexed color-coded probe-based gene expression assessment for clinical molecular diagnostics in formalin-fixed paraffin-embedded human renal allograft tissue. Clin Transplant. 2016;30:295–305. doi: 10.1111/ctr.12689. [DOI] [PubMed] [Google Scholar]
- 80.Feng S., Bucuvalas J.C., Demetris A.J., et al. Evidence of chronic allograft injury in liver biopsies from long-term pediatric recipients of liver transplants. Gastroenterology. 2018;155:1838–1851. doi: 10.1053/j.gastro.2018.08.023. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mengel M., Loupy A., Haas M., et al. Banff 2019 Meeting Report: molecular diagnostics in solid organ transplantation–Consensus for the Banff Human Organ Transplant (B-HOT) gene panel and open source multicenter validation. Am J Transplant. 2020;20:2305–2317. doi: 10.1111/ajt.16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith R.N. In-silico performance, validation, and modeling of the Nanostring Banff Human Organ transplant gene panel using archival data from human kidney transplants. BMC Med Genom. 2021;14:86. doi: 10.1186/s12920-021-00891-5. 86. [DOI] [PMC free article] [PubMed] [Google Scholar]