Abstract
Artificial Intelligence (AI) is a mathematical process of computer mediating designing of algorithms to support human intelligence. AI in hepatology has shown tremendous promise to plan appropriate management and hence improve treatment outcomes. The field of AI is in a very early phase with limited clinical use. AI tools such as machine learning, deep learning, and ‘big data’ are in a continuous phase of evolution, presently being applied for clinical and basic research. In this review, we have summarized various AI applications in hepatology, the pitfalls and AI's future implications. Different AI models and algorithms are under study using clinical, laboratory, endoscopic and imaging parameters to diagnose and manage liver diseases and mass lesions. AI has helped to reduce human errors and improve treatment protocols. Further research and validation are required for future use of AI in hepatology.
Keywords: artificial intelligence, machine learning, deep learning, hepatology, NAFLD
Abbreviations: ACLF, acute on chronic liver failure; AI, artificial intelligence; ALD, alcoholic liver disease; ALT, alanine transaminase; ANN, artificial neural network; AST, aspartate aminotransferase; AUD, alcohol use disorder; CHB, chronic hepatitis B; CHC, chronic hepatitis C; CLD, chronic liver disease; CNN, convolutional neural network; DL, deep learning; FIB-4, fibrosis-4 score; GGTP, gamma glutamyl transferase; HCC, hepatocellular carcinoma; HDL, high density lipoprotein; ML, machine learning; MLR, multi-nomial logistic regressions; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; NLP, natural language processing; RF, random forest; RTE, real-time tissue elastography; SOLs, space-occupying lesions; SVM, support vector machine
In recent years, the development of Artificial Intelligence (AI) in the fields of gastroenterology and hepatology has made remarkable progress. The use of AI is studied in gastroenterology for the endoscopic evaluation of Barrett's oesophagus, oesophageal and gastric malignancies, colorectal polyp detection and characterization, evaluation of inflammatory bowel disease and capsule endoscopy for obscure gastrointestinal bleed1 (Table 1). With the increased development and usage of AI in gastroenterology, research in the field of hepatology also has accelerated. AI in hepatology can be used to detect liver fibrosis, diagnose non-alcoholic fatty liver disease (NAFLD), differentiate focal liver lesions, diagnose hepatocellular cancer, prognosticate chronic liver disease (CLD) and facilitate transplant sciences. However, multiple issues are to be sorted out to establish the AI's full functionality and clinical use. Here, we review the practical applications and ongoing research of AI in hepatology. AI has impacted health-related systems by accurately and rapidly elucidating pathology, radiology and endoscopy images, reducing medical errors and improving workflow.2 AI has aided non-experts also to reach a diagnosis by increasing the performance quality of images. The use of labelled ‘big data’ along with exceptionally enhanced computers and cloud storage, has enabled a giant leap in the development of AI sciences. This will impact at three different strata: for physicians, via quick and precise data interpretation; for healthcare, by increasing work efficiency; and for patients, by allowing them to promote healthy living using their data.
Table 1.
Glossary of Common Terms and Definitions Used in Artificial Intelligence Domains.
| Artificial intelligence (AI) | A mathematical process of computer mediating designing of algorithms and models to augment, recreate and support the natural human intelligence and decision-making. AI lets machines perform tasks that would normally require human intelligence. |
| Algorithms | Collection of specific mathematical formulae that would form the basis of a computational learning methodology. |
| Data | Data is a conglomerate of information used for processing, which can be in the form of numbers, alphabets, images or videos. |
| Machine learning (ML) | Type of learning through computer-based predictions using mathematical algorithm by analysis of provided data. It helps in predictions about unseen data using learned data. |
| Deep learning (DL) | Advanced and complex form of machine learning that uses multiple algorithms arranged in complex neural networks. Due to interweaving of multiple algorithms, infinite patterns and complex data can be generated from data sets. |
| Artificial or Convolutional neural network (ANN/CNN) | Neural networks are complex models generated by arranging together algorithms in layers. The most superficial layer draws out the most readily accessible data, analyses it, and then triggers selected deeper layers. Deeper layers in turn extract finer data and then trigger even deeper layers. Once the deepest layer is triggered, the complex neural network will make a prediction. |
| Training dataset | Data sets that are used for initial development of complex algorithms. This data is analysed again and again in a repetitive fashion until output is generated that matches the reference label. |
| Validation dataset | Data sets used to fine-tune algorithms and adjust the parameters of a model. |
| Supervised learning | Refers to use of labelled data to train algorithms. Once trained, the algorithm can be used to generate labels for new, unseen data. Most commonly used in medical research for AI tools. |
| Unsupervised learning | Refers to use of unlabelled data to train algorithms. Complex method and not used widely for medical applications. |
Abbreviations: AI, artificial intelligence; ANN, artificial neural network; CNN, convolutional neural network; DL, deep learning; ML, machine learning.
Artificial intelligence - definition, terminology and concepts
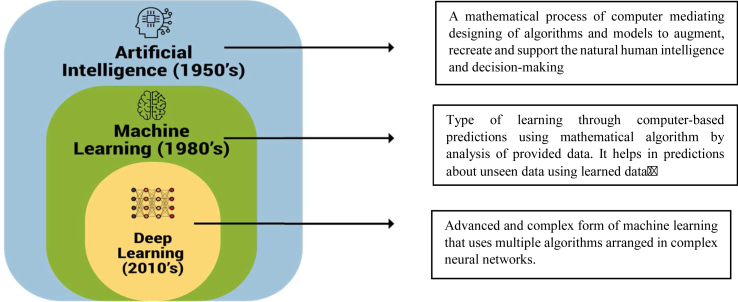
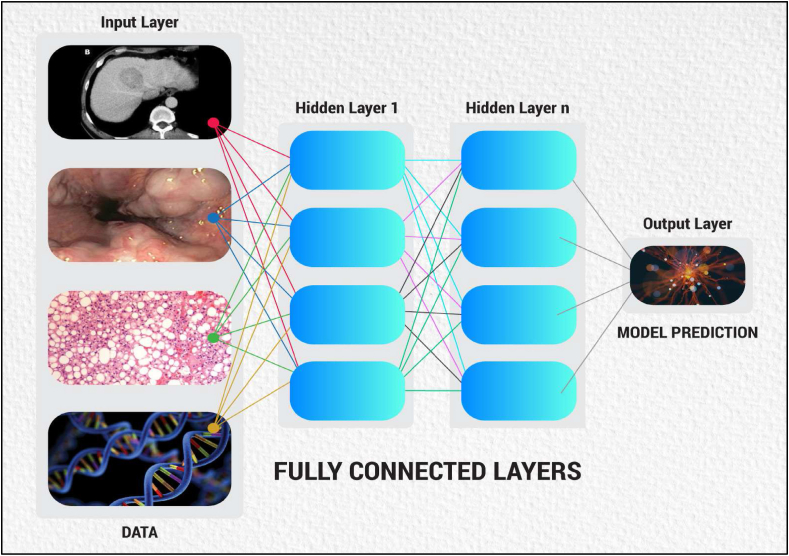
Artificial Intelligence is an umbrella term that mainly infers the use of mathematics using computers to generate software, performing functions as natural human intelligence such as problem-solving and decision-making.3 The glossary of terms1,4,5,6,7,8 used in the AI platform are listed and defined in Table 2. The hierarchy patterns of various types of AI are shown in Figure 1. The overview of the deep learning (DL) tool is demonstrated in Figure 2.
Figure 3.
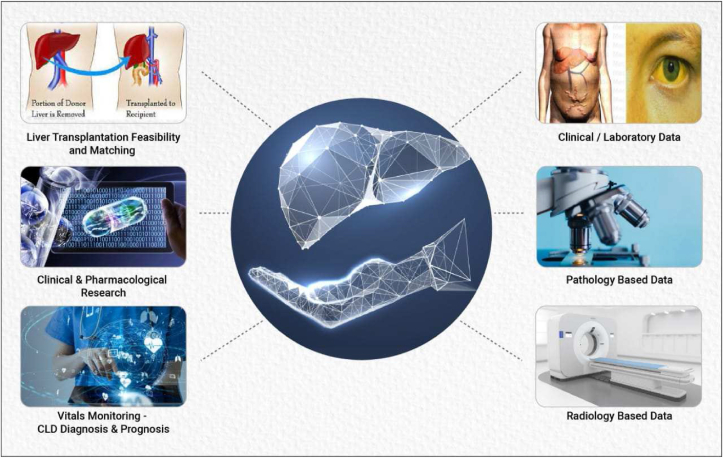
Graphical presentation showing applications of AI in Hepatology.
Table 2.
Applications of AI in Various Endoscopic Procedures.
| Procedure | Application |
|---|---|
| Upper endoscopy | 1. Early dysplasia detection in Barrett's oesophagus 2. Real-time image segmentation in volumetric laser endomicroscopy (VLE) in Barrett's oesophagus 3. Oesophageal squamous dysplasia detection 4. H pylori infection detection 5. Gastric cancer detection and depth of invasion delineation |
| Colonoscopy | 1. Real-time polyp detection 2. Polyp classification (Neoplastic vs non-neoplastic) 3. Polyp characterization and detection of depth of invasion 4. Diagnosis of inflammatory polypoidal lesions on endocytoscopic images |
| Wireless capsule endoscopy (WCE) | 1. Lesion (polyp, bleeding, ulcers) detection and classification 2. Intestinal motility assessment 3. Celiac disease and Tropical sprue assessment |
| Endoscopic Ultrasound (EUS) | 1. Differentiate chronic pancreatitis from pancreatic cancer 2. To diagnose the percentage of necrosis in peripancreatic walled-off necrotic collection 3. EUS elastography 4. Differentiate autoimmune pancreatitis from chronic pancreatitis |
Figure 1.
Arrangement of the hierarchy of artificial intelligence domains.
Figure 2.
Overview of a Deep Learning tool using data in Input layer then running Inter-neuron connections, finally showing Model predictions in the Output layer.
Radiomics and radiogenomics
The term “Radiomics” is defined as a process that uses computers to extract a large amount of information from different types of images, form various quantifiable features, and using AI algorithms build models to predict the outcomes pertaining to the diagnosis, treatment and prognosis of clinical problems especially cancer.9 Data is drawn out from medical images using high-throughput mining and applied within clinical management flowcharts to improve diagnostic, predictive and prognostic accuracy. This is especially becoming useful in cancer research. Radiomic analysis uses image-based signatures for precision diagnosis and treatment, providing a robust tool in modern medicine. Radiomics is useful not only for liver malignant lesions like HCC and non-HCC malignancy but also for benign conditions like NASH, NAFLD and portal hypertension. Presently, the branch of radiomics lacks the standard evaluation of multiple published investigations. Hence, detailed guidelines and criteria need to be set to apply radiomics in clinical medicine.10 The gene–expression profile of a tumour helps to predict the biological behaviour and plan further management after tumour resection. For all practical purposes, the only way to assess gene expression is to use tissue obtained through biopsy or tumour resection, with the disadvantages being haemorrhage and tumour metastasis, although very rare. So, preoperative evaluation of the tumour-gene expression preferably though non-invasive routes are the ideal target. With the genomic revolution in the early 1990s, medical research has been driven to study the basis of human disease on a genomic level and devise precise cancer therapies tailored to a tumour's specific genetic makeup. The novel approach of using radiomics to extract genomic data has been termed “radiogenomics”. It aims to correlate the genotype (gene expressions and mutations) and phenotype (imaging characteristics), to facilitate a deeper understanding of tumour biology and study the intrinsic tumour heterogeneity.11 Imaging characteristics can then behave as molecular surrogates that can help to predict gene-expression-associated treatment responses of various forms of cancer. These findings pave a way for the futuristic role of diagnostic and interventional radiologists in using radiographic images for the genetic assessment of cancer patients.12
Applications of ai in hepatology
Recently, there has been a great amplification of AI-based applications and softwares in the field of Gastroenterology and Endoscopy13 (Table 2). We will discuss the various applications of AI in Hepatology14 under the following subheadings-
-
A.
Liver fibrosis in chronic hepatitis B (CHB)
-
B.
Liver fibrosis in chronic hepatitis C (CHC)
-
C.
Alcoholic liver disease
-
D.
NASH and non-alcoholic fatty liver disease (NAFLD)
-
E.
Diagnosis of DILI, PSC and PBC.
-
F.
Prognosis of CLD, portal hypertension, oesophageal varices and ACLF
-
G.
Diagnosis and characterization of liver space-occupying lesions (SOLs)
-
H.
Diagnosis and management of hepatocellular carcinoma (HCC)
-
I.
Liver transplantation
A. Liver fibrosis in chronic hepatitis B (CHB)
Viral hepatitis is a significant cause of CLD. Liver fibrosis and CLD are risk factors for hepatocellular carcinoma (HCC) and hence death. It is practically impossible to perform a liver biopsy in all patients; hence AI algorithms have been developed for non-invasive evaluation of liver fibrosis. Some of the studies done using AI algorithms will be mentioned in the following sections. Wang D. et al15 proposed a bayesian learning algorithm to develop a three-layer artificial neural network (ANN) in patients with CHB. Age, platelet count, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and gamma-glutamyl transferase (GGTP) were the most critical factors in the predictive model. Similarly, using the non-invasive fibrosis-4 score (FIB-4 score), in the discovery dataset (n = 490) of CHB patients, a learning algorithm known as Gradient Boosting (GB) clearly proved the superiority of other methods as well as FIB-4 score (P < 0.001) in the prediction of advanced hepatic failure and cirrhosis.16 DL and radiomics can be used for quantitative analyses of liver fibrosis17 in CHB cirrhosis. To improve the staging of fibrosis, machine learning (ML)-based classification of real-time tissue elastography (RTE) was developed based on four classical classifiers (i.e. Support vector machine, Naïve Bayes, Random Forest and K-Nearest Neighbour).18 Wang K. et al19 by a prospective multi-centre study, have shown that radiomics of shear wave elastography (DLRE) performed better for liver fibrosis prediction in patients with CHB when compared to two-dimensional shear wave elastography (2D-SWE) or serum markers. Radiomics fibrosis index (RFI), a new DL-based model developed using gadoxetic acid-enhanced MRI, was found to be superior to AST: platelet ratio and fibrosis-4 (FIB-4) index, for staging liver fibrosis.20 These AI models predict the risk of liver fibrosis with high accuracy and hence can help to prevent unnecessary liver biopsies.
B. Liver fibrosis in chronic hepatitis C (CHC)
Hepatitis C virus is a significant and one of the commonest causes of CLD presently. Hepatitis C has an 85% likelihood of chronicity after an acute infection. In patients with CHC, followed for 20 years, progression to cirrhosis occurs in about 20–25% patients. Various AI modules have been framed to study cirrhosis due to CHC, some of which are being mentioned. Using 414 liver biopsies from transplant explants (training dataset) and testing on the remaining 96 biopsies (validation set), with a cut-off of >0.4, ANN given by Piscaglia et al21 provided an accurate prediction of significant fibrosis based on clinical variables, hence avoiding unnecessary liver biopsies, particularly in the setting of liver transplantation. In another study by Hasheem et al,22, using ML algorithm on 39,567 patients with CHC, four parameters – age, AST, platelet count and albumin were found to be statistically significant for advanced fibrosis. Konerman et al23 demonstrated AI models, constructed using two ML methods (Boosting and Random Forest) and logistic regression in CHC patients, which helped to target costly therapies in patients who needed it urgently. Analysis of 72,638 patients with CHC of the National Veterans Health Administration data showed that boosted survival tree-based models using longitudinal information were statistically better than cross-sectional or linear models for cirrhosis prediction in CHC.24 An ANN was created by Takayama et al25 that identified patients with CHC who responded to therapy with pegylated interferon a-2b plus ribavirin with 82% sensitivity and 88% specificity, in the era before the use of directly acting antivirals (DAA) for CHC.
C. Alcoholic liver disease
Alcohol-related liver disease is an area of hepatology, in which very few studies using AI have been done. Studies have demonstrated the prediction of hepatic fibrosis in patients with alcohol use disorder (AUD). Using a group of 31 NAFLD patients with BMI below 30 and a group of Alcoholic liver disease (ALD) patients with cirrhosis (ALDC n = 51) or without cirrhosis (ALDNC n = 51), serum transaminases, cell death markers and (adipo-) cytokines were assessed. ML techniques based on ALT/AST ratio, adipokines and cytokines helped to distinguish NAFLD and ALD.26 Tumour necrosis factor (TNF)-alpha and adiponectin were significantly lower in NAFLD patients. ALDC patients had a significantly higher serum concentrations of cell death markers, hyaluronic acid, adiponectin, and TNF-alpha as compared to ALDNC.
D. NASH and non-alcoholic fatty liver disease (NAFLD)
The incidence of NAFLD is increasing worldwide nowadays, and it has become one of the most common causes of cirrhosis. One of the primary goals of ML application development in hepatology is diagnosing fatty liver disease and staging liver fibrosis, hence substituting pathological analysis. Supervised ML classifiers were trained by Vanderbeck et al27 using a digital library of pathology images of 47 liver biopsies from patients with normal liver and with NAFLD patients. The classification algorithm performed with 89% overall accuracy and identified steatosis, bile ducts, portal veins, and sinusoids with high precision and recall. Accurate localization of microscopic liver anatomy landmarks facilitates the detection of other histological lesions.
A widely used pathologist score (Kleiner score) uses ballooning, inflammation, steatosis and fibrosis as the main histopathology features of NASH/NAFLD. Automated DL-based scores using the above findings enables pathologist-like scoring of NASH models.28 Sowa J-P et al used ML techniques to analyse specific liver serum parameters, hyaluronic acid (HA) and cell death markers of 126 patients undergoing bariatric surgery for morbid obesity. Out of these serum markers, a fibrosis scoring system could be generated using AI, even if only marginally fibrotic tissue is available.29 Based on findings that six predictors including hypertension, alanine aminotransferase (ALT), high-density lipoprotein (HDL), triglyceride, haemoglobin A1c, white blood cell count are important parameters for NAFLD diagnosis in general polulation, a ‘NAFLD ridge score’ was developed as a robust and straightforward reference comparable to existing NAFLD scores to exclude NAFLD patients in epidemiological studies.30 ML methods using history, demographic details and laboratory values of the patient can predict non-alcoholic steatohepatitis (NASH) in NAFLD patients.31 It can help to make predictive models of patients having higher chances of developing cirrhosis.
While ultrasound is the primary modality used for treating the NAFLD, due to unavailability of the skilled sonographers, especially in resource-scarce regions, the quality of diagnosis is severely affected. To address this problem, DL methods for classifying the fatty liver in ultrasound images were used. The performance analysis of the proposed framework shows that the NAFLD in ultrasound images can be detected with an accuracy of 90.6%.32 Liver biopsy is presently the gold standard modality for the diagnosis of NAFLD. To remove the intra- and inter-observer variability, a transfer-based DL algorithm, AlexNet-CNN was developed using liver biopsy images in mice and compared to conventional non-DL algorithms - ANN, multi-nomial logistic regressions (MLR), support vector machine (SVM) and random forest (RF). It was shown that AlexNet-CNN could automatically score liver fibrosis stages with a level of accuracy similar to conventional non-DL algorithms.33 A supervised DL model with a convolutional neural network (CNN) architecture using high discrimination capability of histological tissue alterations of NAFLD showed a classification accuracy of 95%. The classification capability of the new CNN model showed superior classification accuracy compared with a pre-trained AlexNet model, a visual geometry group (VGG)-16 deep architecture and a conventional multi-layer perceptron (MLP).34
E. Diagnosis of DILI, PSC and PBC
Diagnostic dilemma exists for some hepatic conditions like drug-induced liver injury (DILI), primary sclerosing cholangitis (PSC), primary biliary cirrhosis (PBC) even after thorough clinical and biochemical examinations. DILI is a condition with serious consequences, especially with the rampant use of complementary and alternative medicines (CAM). DILI can present with various clinicopathologic presentations like acute hepatitis, chronic hepatitis, granulomatous hepatitis, cholestatic hepatitis, steatohepatitis, vascular disorders or tumours leading to a lot of overlap in diagnosis. Hence AI tools35 with prediction models can help to predict the group of patients which can be predisposed to DILI and also can play an important role in diagnosis. Kristina et al36 demonstrated a ML tool to detect PSC compatible cholangiographic imaging using 3D-MRCP images with high sensitivity. Eaton et al37 gave a PSC Risk estimate tool (PREsTo) using 9 biochemical variables to predict the outcome in patients with PSC, after excluding those with advanced PSC and cholangiocarcinoma. This ML tool using ‘Gradient Boosting’ can be an excellent non-invasive method for prediction of decompensation, when compared to MELD score and Mayo Risk Score. Similarly, a risk score for PBC38 was developed using ML. This unsupervised ML tool identified novel subgroups of PBC patients and provided prognosis based on serum albumin levels. It showed that UDCA induced increase of S. Alb >1.2. Lower limit of normal is associated with improved liver transplant-free survival.
F. Prognosis of CLD, portal hypertension, oesophageal varices and acute on chronic liver failure (ACLF)
Prediction of disease progression in CLD and portal hypertension is necessary for planning further management and hence prognostication. Bleeding oesophageal varices are an important cause of mortality in CLD patients. AI tools can help in the management of CLD and prediction of bleeding from varices. Two radiomics signatures (rGEV and rHRV) were developed, in a multi-centre study, using non-contrast-enhanced CT images. They act as a non-invasive complementary predictor in diagnosing gastroesophageal varices (GEV) and predicting high-risk varices (HRV) in compensated advanced CLD.39 Using laboratory parameters and liver stiffness, ML model, Extreme-gradient boosting (XGBoost) improved the endoscopic stratification to predict variceal bleeding in patients with compensated CLD with oesophageal varices.40 A DL-based model performance for CHC patients using a random forest analysis showed an excellent prediction of survival without a transplant, although less robust for predicting evolution to hepatoma at 12 and 36 months.41
An ANN model to predict the presence of oesophageal varices in patients with CHB42 was developed by Hong et al with three variables (platelet count, portal vein diameter and spleen width). An algorithmically developed formula, called the EVendo score, can predict oesophageal varices (EVs) and Varices needing treatment (VNT) based on readily available data in patients with cirrhosis. This score could avoid unnecessary procedures, especially in patients at low risk for VNT.43
An ML algorithm employing ultrasound shear wave elastography has been developed for colour analysis for CLD classification.44 A meta-analysis of AI-assisted tools using non-invasive modalities like clinical parameters, ultrasonography, elastography, computerized tomography (CT) and magnetic resonance imaging (MRI) has shown the promise in diagnosing CLD. Validations studies are warranted before their applications in clinical practice.45 Recently, a DL-based AI tool named ‘AI-Cirrhosis-ECG (ACE) score’ was proposed by Ahn et al. This score is based on cirrhosis-related ECG (Electrocardiogram) signals. This score can differentiate ECGs from patients with or without cirrhosis and can be a useful low-cost tool in the care of patients with cirrhosis.46 The VIRGIN Study47 done in China presented an analytical method to calculate a virtual Portal Pressure Gradient (vPPG) based on CT angiography and Doppler ultrasound, hence avoiding the invasive HVPG measurement for portal hypertension. Musunuri B. et al investigated the role of the ANN, which functionally mimics biological neural systems, in predicting 90-day liver disease-related mortality in ACLF patients. An accuracy of 94.12% was noticed in predicting 30-day mortality and 88.2% in predicting 90-day mortality, with an area under the curve of 0.915 and 0.921, respectively. ANN plays a very important role in predicting short-term mortality patients with high accuracy. Its application in patients with ACLF is promising as it automates and eases the method of identifying those patients at a higher risk of mortality.48
G. Diagnosis and characterization of liver space-occupying lesions (SOLs)
Various DL-based models have been developed using contrast-enhanced ultrasound, CT scan and MRI to characterize focal liver lesions49,50,51,52. But extensive validation is needed before establishing clinical use. A DL technique using Stacked Sparse Auto-encoders (SSAE) with ultrasound images suggested for diagnosing hepatic nodules (liver cysts, haemangioma, and HCC)53 has high accuracy in overall classification (97.2%) compared with three established methods: Naive Bayes, multi-support vector machine and K-Nearest Neighbor. Since MRI diagnosis is affected by subjective variations, CNNs were used to develop a DL system (DLS) to classify liver tumours based on clinical data, laboratory parameters and MR images (enhanced and unenhanced).52 Multiple prediction models were designed using CNN for 5-year metachronous liver metastasis (5YLM), applying combinations of clinical variables (age, sex, T stage, N stage) and top principal components (PCs) with logistic regression classification. The model using “1st PC (PC1) + clinical information” had a significant correlation with sex, body mass index, alcohol consumption, and fatty liver status.54
H. Diagnosis and management of hepatocellular carcinoma (HCC)
ML has various applications for the study of HCC, including diagnosis, staging, management and prognosis based on the stage of HCC. Sato et al55 developed a novel model for HCC diagnosis, showing high accuracy (87.3%) compared to a single tumour marker (alpha fetoprotein 70.7%, des-alpha-fetoprotein-L3 71.1% and gamma-carboxyprothrombin 74.9%). This model decreased the rate of liver SOLs, previously misclassified as HCC. A study by Singal et al56 showed that ML algorithms outperformed conventional regression models and markedly improved the accuracy of prediction and risk stratification for HCC development in 442 patients with compensated cirrhosis. A study by Nam JY et al57 compared a DL-based model with previous HCC prediction models, including Chinese University HCC score (CU-HCC), platelets, age, gender-hepatitis B score (PAGE-B), age, diabetes, race, aetiology of cirrhosis, sex, and severity HCC score (ADDRESS-HCC), HCC-Risk Estimating Score in CHB patients Under Entecavir (HCC-RESCUE), Toronto HCC risk index (THRI), and modified PAGE-B score (mPAGE). This model had better performance than the previous models for predicting the HCC risk in 424 patients with HBV-related cirrhosis on potent antivirals.
Multi-omics (use of multiple –“omes” such as the genome, microbiome, etc) approach was used to make a DL-based model of 360 patients with HCC, using RNA sequencing, methylation data and miRNA sequencing from The Cancer Genome Atlas.58 It was the first study to employ DL to identify multi-omics features linked to the differential survival of patients with HCC. Two DL algorithms were built, using whole-slide digitized histological data for predicting the survival of patients with HCC treated with surgery.59 This analysis included two independent cohorts. A discovery cohort (n = 194) was used to develop the algorithm and included an independent validation cohort (n = 328). This study highlights the importance of machine interactions for the appropriate construction of DL algorithms using histology slides.
Randhwa et al,60, using support vector, with MRI images as data, made an AI tool to improve radiological image classification of HCC. This can help radiologists diagnose liver tumours early. Using regularization in the vector score in the classification stage removes the overfitting problem and leads to the accurate identification of different tumour types. A DL-based assistant has been developed to help pathologists differentiate between two subtypes of primary liver cancer, HCC and cholangiocarcinoma, on haematoxylin and eosin-stained whole-slide images (WSI), and evaluated its effect on the diagnostic performance of 11 pathologists with varying levels of expertise.61 This DL-based assistant helped to increase the accuracy of pathologists.
The segmentation of HCC in CT images allows assessment of tumour load, treatment planning, prognosis and monitoring of treatment response. Since manual segmentation is a very time-consuming task and, in many cases, prone to inaccuracies, automatic tools for tumour detection and segmentation are highly desirable. One such network architecture was formed and evaluated on data provided from the radiological centre in Innsbruck, Austria. It consists of two consecutive nested fully CNN together with a joint minimization strategy. The first sub-network segments the liver, whereas the second sub-network segments the actual tumour inside the liver.62 Automatic segmentation of liver and tumours using a fully convolutional neural (FCN) network is highly advantageous as it will plan surgical management and follow-up assessment. A study by Alirr et al has shown this DL method as a promising tool for automatic analysis of the liver and its tumours.63
A systematic review by Azer et al of data analysing pathology, cellular and radiological images of HCC or liver masses using CNNs were identified and analysed.64 The review showed an optimal level of accuracy of CNNs for the segmentation and classification of HCC and other liver mass lesions. Dynamic contrast-enhanced MRI provides the most comprehensive information for differential diagnosis of liver tumours. DL tools can be used for classification and mutation prediction based on histopathology images. Chen et al65 showed that the performance level of a DL model was close to the ability of a 5-year experience pathologist, with high accuracy for differentiating benign and malignant conditions. This model has shown that four important genes, namely- FMN2, CTNNB1, TP53 and ZFX4, predicted from histopathology images, could assist in the classification and detection of gene mutation in liver cancer.
I. Liver transplantation
An ANN model, based on clinical and biochemical data of cirrhotic patients, having a high probability of mortality in 1 year; was developed by Banerjee et al66 for the identification of the best candidates for liver transplantation. Three-dimensional (3D) simulation software, using 3D visualization and 3D reconstruction of CT images, is a valuable tool for pre-hepatectomy assessment, virtual hepatectomy and measuring hepatic volumes. These 3D models combined with hydrodynamic analysis have been used to diagnose and manage portal hypertension.67 An ANN was used to predict survival times of 1168 patients planned for liver transplantation, by Khosravi et al.68 It estimated a survival probability of 1–5 years with an AUROC curve of 86.4% vs. 80.7% for Cox proportional hazard regression models. ANN has helped to make liver donor-recipient matching models by researchers, providing powerful technology that would ease decision-making.69 AI tools using liver segmentation will hence help to plan hepatic resection, prevent donor-recipient mismatch, improve survival of graft and patient overall and also help in the approval of new immunosuppressant drugs by playing an important role in research and development in drug trials. The salient studies of AI and ML are listed in Table 4.
Key points
Various AI-based applications and models, developed using clinical, laboratory and radiology data, play an important role in diagnosis, prediction of severity and prognostication of liver diseases (Figure 3).
Table 4.
Salient Studies of AI and ML in Liver Diseases.
| Study | Disease | Patients | Modality | AI techniques used | Salient features |
|---|---|---|---|---|---|
| Wei et al.16 | HBV/HCV | Train: 343 HBV Test: 147 HBV; 484 HCV |
Clinical and Laboratory data | Decision Tree, Random Forest, Gradient Booster |
Age, AST, ALT and platelet count were used to construct ML algorithms to predict fibrosis in HBV patients. The model was superior to FIB-4 score. |
| Wang K et al.19 | HBV | Training: 266 HBV Validation: 132 HBV |
Ultrasound | Convolutional Neural Network | Deep learning Radiomics of Elastography (DLRE) was superior to 2D-Sheer Wave Elastography and biomarkers for assessing liver fibrosis stages. |
| Konerman MA et al.23 | HCV | Train: 533 HCV Test: 183 HCV |
Clinical and Laboratory data | Logistic Regression, Random Forest |
ML-based longitudinal fibrosis prediction model shows AUROC = 0.78–0.79 for fibrosis progression and AUROC = 0.79–0.86 for clinical progression |
| Vanderbeck et al.27 | NASH/NAFLD | NAFLD-27 Healthy liver-20 |
Pathology data | Support Vector Machine | Automatic classification algorithm of steatosis had an 89% overall accuracy and identified macrosteatosis with ≥95%precision and recall |
| Yip TF et al.30 | NASH/NAFLD | Train: 146 NAFLD;354 Healthy volunteers (HV) Test: 118 NAFLD, 394 HV |
Clinical and Laboratory data | Logistic Regression, Ridge Regression, AdaBoost, Decision Tree |
ML algorithms based on ALT, HDL-C, triglycerides, HbA1C, WBC and Hypertension were used to develop NAFLD prediction scores. Overall accuracy of NAFLD ridge score- 87%, AUROC- 0.87, 92% sensitivity and 90% specificity. |
| Agarwal S et al.40 | CLD/Oesophageal varices | 828 patients having compensate advanced CLD with oesophageal varices (EV) | Laboratory data Endoscopy images Liver stiffness measurement |
Extreme Gradient Boosting (XGBoost) | The accuracy of machine learning (ML)-based model to predict future VB was 98.7 (97.4–99.5)%, 93.7 (88.8–97.2)%, and 85.7 (82.1–90.5)% in derivation (n = 497), internal validation (n = 149), and external validation (n = 182) cohorts, respectively, which was better than endoscopic classification [58.9 (55.5–62.3)%] alone. Patients stratified high risk on both endoscopy and model had 1-year and 3-year bleeding rates of 31–43% and 64–85%, respectively |
| Dong TS et al.43 | Oesophageal varices | Train: 238 Liver cirrhosis Test: 109 Liver cirrhosis |
Clinical and Laboratory data | Random Forest | EVendo score was developed to identify oesophageal varices with AUROC = 0.82, and could spare 30–40% low-risk patients from unnecessary procedures. |
| Minerali et al.74 | DILI | 1036 FDA-approved individual compounds | Biopharmaceutics Drug Disposition Classification System dataset | Bayesian machine learning models | A ML tool named MegaTox™ can predict DILI in early-stage clinical compounds and recently approved FDA drugs. |
| Eaton et al.37 | PSC outcomes | Train: 509 PSC Test: 278 PSC |
Clinical data | Gradient Boosting | A score PREsTo was created using nine variables. This model can predict decompensation and performs better than Mayo Risk score and MELD score. |
| Ahn JC et al.46 | Cirrhosis | Liver cirrhosis: 5212 Age and sex matched controls: 20,728 |
Electrocardiogram (ECG), Clinical data, Laboratory data |
Convolutional Neural Network | AI-Cirrhosis-ECG (ACE) score was created. It has an excellent performance (AUROC = 0.908) for classifying ECGs from patients with cirrhotics and controls. It is positively associated with markers of liver disease specially MELD-Na and trends show improvements after liver transplant. |
| Singal AG et al.56 | HCC risk | Train: 442 Liver cirrhosis Test: 1050 LC |
Clinical and Laboratory data | Decision Tree, Random Forest |
ML algorithm compared to conventional regression models had significantly better accuracy with >80% sensitivity for predicting HCC development |
| Briceño J et al.69 | Post liver transplantation survival | 1003 Liver transplantations (90% train, 10% test) | Clinical and laboratory data | Artificial neural network, Logistic Regression, Decision Tree, Support Vector Machine | 64 donor and recipient variables were used to train ANN to predict LT graft survival and optimize donor-recipient matching. Graft survival prediction was 90.79% with AUROC = 0.82 |
Abbreviations: AI, artificial intelligence; ALT, alanine transaminase; ANN, artificial neural network; AST, aspartate aminotransferase; CLD, chronic liver disease; HCC, hepatocellular carcinoma; HDL, high density lipoprotein; ML, machine learning; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
The limitations of AI implementation
There are certain obstacles and pitfalls, despite all the advantages of AI technology. Medicine is a field of science with multiple research gaps, hence designing perfect AI models has been difficult to date, especially in gastroenterology and hepatology (Table 3). A flawed algorithm can cause harm to a large group of patients. Instead of a single doctor's mistake harming a patient, the potential for machine algorithms causing iatrogenic risk is vast. As a result, systematic amendment, extensive simulation, validation and audit along with prospective trials and scrutiny are required when an AI algorithm is launched in clinical practice. Another critical aspect of the future of AI rests with the preservation and protection of data and personal information, maintaining privacy. Given the risks of hacking and data robbery, there will be less interest in the use of algorithms that risk revealing a patient's medical history.
Table 3.
Future Research Points of Investigations for Artificial Intelligence in Gastroenterology and Hepatology.
|
|
|
|
|
|
|
Abbreviation: AUROC, area under the receiver operating characteristic.
Now, as AI moves towards face recognition and the use of genomic imprints, it further stresses the fact that data security will be a chief concern. Developing software with full-proof security systems, especially for data transfer and storage, will help increase the confidence of patients for AI development.
High dependency on AI tools can result in the reduction of the skills of clinicians. One should remember that AI tools also have diagnostic errors. Hence, clinicians should be vigilant in the initial phase of AI implementation, disapproving the diagnosis; they believe AI has made an error. The initial use of AI tools will prolong diagnostic time, mainly due to the clinicians learning the new process and technology adapting itself to new unprocessed data. Hence there should be a dedicated team of people involved in the development of AI applications.
It is improbable that the AI systems will render clinicians input completely obsolete, as the AI systems are designed to keep human intelligence as a centre-space. They lack the technical complexity required to achieve autonomy.70 Rather than hinder the already skilled clinicians, they will aid and further support for patient management. Clinicians should embrace AI systems as adjuncts to increase the quality of care. Machines cannot replace the component of ‘human touch’; hence clinicians ethically will play a major role in decision-making for their patients based on patient's preferences and comfort. Another limitation of AI technology at present is the lack of superior quality datasets for algorithm development. Currently, with the lack of utility of ML algorithms in clinical practice, most of the evidence is from preclinical trials. Hence collections of data and storage from involvements of multiple centres with high volume load will help in collecting data. Proper validation studies then need to be done after the AI tools have been made.
Specific DL methods are considered ‘black-box’ models, so it is not easy to understand the processing of data. This has prohibited physicians from finding potential confounding variables. There has been much published about ‘black-box’ warning of algorithms.71 AI use can be ethically challenging, as patient preferences cannot be machine learnt. Also, in case of misdiagnosis, who shall be legally responsible - whether the endoscopist, the clinician or the programmer is still undecided? Moreover, the racial distinction might be necessary for certain areas (as fibrosis in viral hepatitis), and it may be an inherent bias in AI. So, different population groups must be considered in developing and validating AI tools, to increase their sensitivity, specificity and accuracy.
Key points
Use of AI technology is advantageous to the fields of gastroenterology, hepatology and medicine in general, but have some roadblocks at present. Mainly being data security, lack of human touch, concerns regarding overpowering human intelligence and lack of proper databases. These pitfalls shall be overcome in the future with proper clinical trials and studies.
The future perspectives of AI in hepatology
The field of AI at present is full of promises but relatively less in proofs and validation. Before it is used in clinical practice, AI requires rigorous studies, clinical validation in a real-world scenario and publication in peer-reviewed journals. There are concerns that machines in the future will replace doctors, but the fact is that the designers of AI modules are cautious for that to happen. Human life is too precious and cannot be experimented with as a ‘self-driven car’. Hence the usage of AI in clinical practice will be slow and cautious, first involving processes with minimal risk and complications. Gradually this will lay the foundation for high-performance medicine, which will absorb more and more human data, decreasing the reliance on human resources and eventually form a symbiotic relationship between human and machine intelligence for the betterment of humankind. The expansion of AI in gastroenterology and hepatology is crucial for the progress of these fields. Also, the use of AI is advantageous compared to traditional regression analysis methods used in research by incorporating large samples and by reducing interobserver bias.
An upcoming impact of AI is that it forms an important part of the evolution of precision and personalized medicine. It describes the revolution in health care triggered by knowledge gained from sequencing the human genome. The field has evolved to recognize how the intersection of multi-omic data combined with medical history, social/behavioural determinants, and environmental knowledge precisely characterizes health states, disease states, and therapeutic options for affected individuals. The synergy of AI and precision medicine has augmented the yield of highly personalized medical diagnostic and therapeutic information. The ultimate goal of this combination is the prevention and early detection of diseases affecting the individual, which could ultimately decrease the disease burden for the public at large, and, therefore, the cost of preventable health care for all. Genotype-guided treatment of HCC is an example of precision medicine. The genetic characterization of HCC is not well established like other malignancies due to a lack of genomic studies. However, ‘big genomic data’ and AI analysis will help identify patients with CLD who are at high risk for fibrosis or HCC. Patients with advanced HCC are presently treated with newer agents such as molecular targeted therapy or immunotherapy. However, the group of patients who will respond to the drugs is not known. If AI can predict this treatment response, precision medicine can become a reality soon.
Healthcare systems in developing countries like India have a lot of challenges, especially in the rural areas. AI helps in addressing these issues by assisting the doctors in better and quick diagnosis, delivering personalized healthcare, providing high-quality healthcare to rural areas, and helping doctors and nurses in training to handle complex medical conditions. AI can help monitor a patient's condition having chronic ailments with the help of a smartphone.72 Using clinical, genetic, molecular information from large datasets, AI can be helpful to find new therapeutic targets. Apart from the extensive number of AI applications being made, a lot of unmet needs are work on alcohol related liver injury, metabolic and autoimmune liver diseases. Hence there is a lot of scope for technical growth in the AI sub-speciality, paving the way to improve the accuracy of the AI tools. AI systems for liver segmentation and diagnosis should be widely available within the next 5 years, which will help in liver lesion characterization and aid in liver transplantation. Working in isolation from AI and data scientists will be a hindrance to the growth of clinical medicine. Hence, the adoption of coordinated research opportunities will facilitate the development of many clinically useful tools.
Key points
The future of AI is promising with the introduction of precision and personalized medicine, mainly involving data from the human genome. Overall healthcare services will improve especially in rural areas of developing and underdeveloped countries.
Summary
AI is an upcoming promising technology that is rapidly becoming an essential part of patient management. Applications of AI have expanded in all branches of medicines, especially endoscopy and hepatology. The conglomeration of data which can be clinical/laboratory, multi-omics, natural language processing (NLP) and Image recognition (both radiology-based and pathology-based) has contributed to the prediction of fibrosis, classification of liver masses and prediction of treatment response and transplant outcomes.73 In this review the majority of studies mentioned focussed on diagnosis part. There are very few studies that help to predict treatment response, post-liver transplant response, and prediction of hepatotoxicity in newer drug development and more studies are needed. AI also helps for real-time biomonitoring, by identification of patients at high risk of clinical decompensation and hospital admission, so that timely intervention can be done for high-risk patients. With the increasing advancement of image capture and storage, AI will bring striking changes to the diagnosis of various liver diseases with the ‘big data’ being available. However, there are many hurdles to overcome, which researchers will do in the near future using validation studies and molecular research. It is expected that gastroenterology and hepatology will be one of the first areas in medicine to introduce AI tools on a wide-scale basis, due to its inherent reliance on endoscopic and radiological imaging. Hence, GI and liver specialists should be proud that our field sets the ground for AI development in medicine.
Credit authorship contribution statement
Both the authors have drafted the manuscript and reviewed the relevant databases for literature relevant to this review. HR and RK conceptualized and prepared the manuscript. HR designed the images. DNR critically reviewed and edited the article.
Conflicts of interest
All authors have none to declare.
Funding
None.
Ethical requirement
This review article does not have any studies with human or animal subjects.
References
- 1.Chahal D., Byrne M.F. A primer on artificial intelligence and its application to endoscopy. Gastrointest Endosc. 2020;92:813–820. doi: 10.1016/j.gie.2020.04.074. e4. [DOI] [PubMed] [Google Scholar]
- 2.Topol E.J. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 3.Colom R., Karama S., Jung R.E., Haier R.J. Human intelligence and brain networks. Dialogues Clin Neurosci. 2010;12:489–501. doi: 10.31887/DCNS.2010.12.4/rcolom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michie D., Spiegelhalter D.J., Taylor C.C. 1994. Machine Learning, Neural and Statistical Classification. [Google Scholar]
- 5.Sathya R., Abraham A. Comparison of supervised and unsupervised learning algorithms for pattern classification. Int J Adv Res Artif Intell. 2013;2:34–38. [Google Scholar]
- 6.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L.-Q., Wang J.-Y., Yu S.-Y., et al. Artificial intelligence in medical imaging of the liver. World J Gastroenterol. 2019;25:672. doi: 10.3748/wjg.v25.i6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Berre C., Sandborn W.J., Aridhi S., et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. 2020;158:76–94. doi: 10.1053/j.gastro.2019.08.058. e2. [DOI] [PubMed] [Google Scholar]
- 9.Hu W., Yang H., Xu H., Mao Y. Radiomics based on artificial intelligence in liver diseases: where are we? Gastroenterol report. 2020;8:90–97. doi: 10.1093/gastro/goaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambin P., Leijenaar R.T., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 11.Pinker K., Shitano F., Sala E., et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imag. 2018;47:604–620. doi: 10.1002/jmri.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutman A.M., Kuo M.D. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 13.Pannala R., Krishnan K., Melson J., et al. Artificial intelligence in gastrointestinal endoscopy. Video. 2020;5:598–613. doi: 10.1016/j.vgie.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H.W., Sung J.J.Y., Ahn S.H. Artificial intelligence in liver disease. J Gastroenterol Hepatol. 2021;36:539–542. doi: 10.1111/jgh.15409. [DOI] [PubMed] [Google Scholar]
- 15.Wang D., Wang Q., Shan F., Liu B., Lu C. Identification of the risk for liver fibrosis on CHB patients using an artificial neural network based on routine and serum markers. BMC Infect Dis. 2010;10:251. doi: 10.1186/1471-2334-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei R., Wang J., Wang X., et al. Clinical prediction of HBV and HCV related hepatic fibrosis using machine learning. EBioMedicine. 2018;35:124–132. doi: 10.1016/j.ebiom.2018.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Luo Y., Huang W., et al. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med. 2017;89:18–23. doi: 10.1016/j.compbiomed.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Wang K., Lu X., Zhou H., et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729–741. doi: 10.1136/gutjnl-2018-316204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park H.J., Lee S.S., Park B., et al. Radiomics analysis of gadoxetic acid–enhanced MRI for staging liver fibrosis. Radiology. 2019;290:380–387. doi: 10.1148/radiol.2018181197. [DOI] [PubMed] [Google Scholar]
- 21.Piscaglia F., Cucchetti A., Benlloch S., et al. Prediction of significant fibrosis in hepatitis C virus infected liver transplant recipients by artificial neural network analysis of clinical factors. Eur J Gastroenterol Hepatol. 2006;18:1255–1261. doi: 10.1097/01.meg.0000243885.55562.7e. [DOI] [PubMed] [Google Scholar]
- 22.Hashem S., Esmat G., Elakel W., et al. Comparison of machine learning approaches for prediction of advanced liver fibrosis in chronic hepatitis C patients. IEEE ACM Trans Comput Biol Bioinf. 2017;15:861–868. doi: 10.1109/TCBB.2017.2690848. [DOI] [PubMed] [Google Scholar]
- 23.Konerman M.A., Zhang Y., Zhu J., Higgins P.D., Lok A.S., Waljee A.K. Improvement of predictive models of risk of disease progression in chronic hepatitis C by incorporating longitudinal data. Hepatology. 2015;61:1832–1841. doi: 10.1002/hep.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konerman M.A., Beste L.A., Van T., et al. Machine learning models to predict disease progression among veterans with hepatitis C virus. PLoS One. 2019;14 doi: 10.1371/journal.pone.0208141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takayama T., Ebinuma H., Tada S., et al. Prediction of effect of pegylated interferon alpha-2b plus ribavirin combination therapy in patients with chronic hepatitis C infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowa J.-P., Atmaca Ö., Kahraman A., et al. Non-invasive separation of alcoholic and non-alcoholic liver disease with predictive modeling. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderbeck S., Bockhorst J., Komorowski R., Kleiner D.E., Gawrieh S. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum Pathol. 2014;45:785–792. doi: 10.1016/j.humpath.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinemann F., Birk G., Stierstorfer B. Deep learning enables pathologist-like scoring of NASH models. Sci Rep. 2019;9 doi: 10.1038/s41598-019-54904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sowa J.-P., Heider D., Bechmann L.P., Gerken G., Hoffmann D., Canbay A. Novel algorithm for non-invasive assessment of fibrosis in NAFLD. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yip T.F., Ma A., Wong V.S., et al. Laboratory parameter-based machine learning model for excluding non-alcoholic fatty liver disease (NAFLD) in the general population. Aliment Pharmacol Ther. 2017;46:447–456. doi: 10.1111/apt.14172. [DOI] [PubMed] [Google Scholar]
- 31.Fialoke S., Malarstig A., Miller M.R., Dumitriu A., editors. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2018. Application of machine learning methods to predict non-alcoholic steatohepatitis (NASH) in non-alcoholic fatty liver (NAFL) patients. [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy D.S., Bharath R., Rajalakshmi P., editors. 2018 IEEE 20th International Conference on E-Health Networking, Applications and Services (Healthcom) IEEE; 2018. A novel computer-aided diagnosis framework using deep learning for classification of fatty liver disease in ultrasound imaging. [Google Scholar]
- 33.Yu Y., Wang J., Ng C.W., et al. Deep learning enables automated scoring of liver fibrosis stages. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-34300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arjmand A., Angelis C.T., Christou V., et al. Training of deep convolutional neural networks to identify critical liver alterations in histopathology image samples. Appl Sci. 2020;10:42. [Google Scholar]
- 35.Vall A., Sabnis Y., Shi J., Class R., Hochreiter S., Klambauer G. The promise of AI for DILI prediction. Front Artif Intell. 2021;4:15. doi: 10.3389/frai.2021.638410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringe K.I., Vo Chieu V.D., Wacker F., et al. Fully automated detection of primary sclerosing cholangitis (PSC)-compatible bile duct changes based on 3D magnetic resonance cholangiopancreatography using machine learning. Eur Radiol. 2021;31:2482–2489. doi: 10.1007/s00330-020-07323-5. [DOI] [PubMed] [Google Scholar]
- 37.Eaton J.E., Vesterhus M., McCauley B.M., et al. Primary sclerosing cholangitis risk estimate tool (PREsTo) predicts outcomes of the disease: a derivation and validation study using machine learning. Hepatology. 2020;71:214–224. doi: 10.1002/hep.30085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerussi A., Verda D., Bernasconi D.P., et al. Machine learning in primary biliary cholangitis: a novel approach for risk stratification. Liver Int. 2022;42:615–627. doi: 10.1111/liv.15141. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y., Huang F., Yang L., et al. Development and validation of a radiomics signature as a non-invasive complementary predictor of gastroesophageal varices and high-risk varices in compensated advanced chronic liver disease: a multicenter study. J Gastroenterol Hepatol. 2021;36:1562–1570. doi: 10.1111/jgh.15306. [DOI] [PubMed] [Google Scholar]
- 40.Agarwal S., Sharma S., Kumar M., et al. Development of a machine learning model to predict bleed in esophageal varices in compensated advanced chronic liver disease: a proof of concept. J Gastroenterol Hepatol. 2021;36:2935–2942. doi: 10.1111/jgh.15560. [DOI] [PubMed] [Google Scholar]
- 41.Konerman M.A., Lu D., Zhang Y., et al. Assessing risk of fibrosis progression and liver-related clinical outcomes among patients with both early stage and advanced chronic hepatitis C. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong W-d, Ji Y-f, Wang D., Chen T-z, Zhu Q-h. Use of artificial neural network to predict esophageal varices in patients with HBV related cirrhosis. Hepat Mon. 2011;11:544. [PMC free article] [PubMed] [Google Scholar]
- 43.Dong T.S., Kalani A., Aby E.S., et al. Machine learning-based development and validation of a scoring system for screening high-risk esophageal varices. Clin Gastroenterol Hepatol. 2019;17:1894–1901. doi: 10.1016/j.cgh.2019.01.025. e1. [DOI] [PubMed] [Google Scholar]
- 44.Gatos I., Tsantis S., Spiliopoulos S., et al. A machine-learning algorithm toward color analysis for chronic liver disease classification, employing ultrasound shear wave elastography. Ultrasound Med Biol. 2017;43:1797–1810. doi: 10.1016/j.ultrasmedbio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Decharatanachart P., Chaiteerakij R., Tiyarattanachai T., Treeprasertsuk S. Application of artificial intelligence in chronic liver diseases: a systematic review and meta-analysis. BMC Gastroenterol. 2021;21:1–16. doi: 10.1186/s12876-020-01585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn J.C., Attia Z.I., Rattan P., et al. Development of the AI-Cirrhosis-ECG (ACE) Score: an electrocardiogram-based deep learning model in cirrhosis. Am J Gastroenterol. 2022;117:424–432. doi: 10.14309/ajg.0000000000001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi X., Li Z., Huang J., et al. Virtual portal pressure gradient from anatomic CT angiography. Gut. 2015;64:1004–1005. doi: 10.1136/gutjnl-2014-308543. [DOI] [PubMed] [Google Scholar]
- 48.Musunuri B., Shetty S., Shetty D.K., et al. Acute-on-chronic liver failure mortality prediction using an artificial neural network. Engineered Science. 2021;15:187–196. [Google Scholar]
- 49.Wu K., Chen X., Ding M. Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik. 2014;125:4057–4063. [Google Scholar]
- 50.Masuzaki R., Kanda T., Sasaki R., et al. Application of artificial intelligence in hepatology: Minireview. Artif Intell Gastroenterol. 2020;1:5–11. [Google Scholar]
- 51.Yasaka K., Akai H., Abe O., Kiryu S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology. 2018;286:887–896. doi: 10.1148/radiol.2017170706. [DOI] [PubMed] [Google Scholar]
- 52.Zhen S-h, Cheng M., Tao Y-b, et al. Deep learning for accurate diagnosis of liver tumor based on magnetic resonance imaging and clinical data. Front Oncol. 2020;10:680. doi: 10.3389/fonc.2020.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hassan T.M., Elmogy M., Sallam E.-S. Diagnosis of focal liver diseases based on deep learning technique for ultrasound images. Arabian J Sci Eng. 2017;42:3127–3140. [Google Scholar]
- 54.Lee S., Choe E.K., Kim S.Y., Kim H.S., Park K.J., Kim D. Liver imaging features by convolutional neural network to predict the metachronous liver metastasis in stage I-III colorectal cancer patients based on preoperative abdominal CT scan. BMC Bioinf. 2020;21:1–14. doi: 10.1186/s12859-020-03686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato M., Morimoto K., Kajihara S., et al. Machine-learning approach for the development of a novel predictive model for the diagnosis of hepatocellular carcinoma. Sci Rep. 2019;9:1–7. doi: 10.1038/s41598-019-44022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singal A.G., Mukherjee A., Elmunzer B.J., et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013;108:1723. doi: 10.1038/ajg.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nam J.Y., Sinn D.H., Bae J., Jang E.S., Kim J.-W., Jeong S.-H. Deep learning model for prediction of hepatocellular carcinoma in patients with HBV-related cirrhosis on antiviral therapy. JHEP Reports. 2020;2 doi: 10.1016/j.jhepr.2020.100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24:1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saillard C., Schmauch B., Laifa O., et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology. 2020;72:2000–2013. doi: 10.1002/hep.31207. [DOI] [PubMed] [Google Scholar]
- 60.Randhawa S., Alsadoon A., Prasad P., Al-Dala’in T., Dawoud A., Alrubaie A. Deep learning for liver tumour classification: enhanced loss function. Multimed Tool Appl. 2021;80:4729–4750. [Google Scholar]
- 61.Kiani A., Uyumazturk B., Rajpurkar P., et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ digital medicine. 2020;3:1–8. doi: 10.1038/s41746-020-0232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gruber N., Antholzer S., Jaschke W., Kremser C., Haltmeier M., editors. 2019 13th International Conference on Sampling Theory and Applications (SampTA) IEEE; 2019. A joint deep learning approach for automated liver and tumor segmentation. [Google Scholar]
- 63.Alirr O.I. Deep learning and level set approach for liver and tumor segmentation from CT scans. J Appl Clin Med Phys. 2020;21:200–209. doi: 10.1002/acm2.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azer S.A. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: a systematic review. World J Gastrointest Oncol. 2019;11:1218. doi: 10.4251/wjgo.v11.i12.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen M., Zhang B., Topatana W., et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ precision oncology. 2020;4:1–7. doi: 10.1038/s41698-020-0120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banerjee R., Das A., Ghoshal U.C., Sinha M. Predicting mortality in patients with cirrhosis of liver with application of neural network technology. J Gastroenterol Hepatol. 2003;18:1054–1060. doi: 10.1046/j.1440-1746.2003.03123.x. [DOI] [PubMed] [Google Scholar]
- 67.Fang C., An J., Bruno A., et al. Consensus recommendations of three-dimensional visualization for diagnosis and management of liver diseases. Hepatology International. 2020;14:437–453. doi: 10.1007/s12072-020-10052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khosravi B., Pourahmad S., Bahreini A., Nikeghbalian S., Mehrdad G. Five years survival of patients after liver transplantation and its effective factors by neural network and cox poroportional hazard regression models. Hepat Mon. 2015;15 doi: 10.5812/hepatmon.25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Briceño J., Cruz-Ramírez M., Prieto M., et al. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: results from a multicenter Spanish study. J Hepatol. 2014;61:1020–1028. doi: 10.1016/j.jhep.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 70.Tang A., Tam R., Cadrin-Chênevert A., et al. Canadian Association of Radiologists white paper on artificial intelligence in radiology. Can Assoc Radiol J. 2018;69:120–135. doi: 10.1016/j.carj.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Castelvecchi D. Can we open the black box of AI? Nature. 2016;538:20–23. doi: 10.1038/538020a. [DOI] [PubMed] [Google Scholar]
- 72.Heida A., Knol M., Kobold A.M., Bootsman J., Dijkstra G., van Rheenen P.F. Agreement between home-based measurement of stool calprotectin and ELISA results for monitoring inflammatory bowel disease activity. Clin Gastroenterol Hepatol. 2017;15:1742–1749. doi: 10.1016/j.cgh.2017.06.007. e2. [DOI] [PubMed] [Google Scholar]
- 73.Ahn J.C., Connell A., Simonetto D.A., Hughes C., Shah V.H. Application of artificial intelligence for the diagnosis and treatment of liver diseases. Hepatology. 2021;73:2546–2563. doi: 10.1002/hep.31603. [DOI] [PubMed] [Google Scholar]
- 74.Minerali E., Foil D.H., Zorn K.M., Lane T.R., Ekins S. Comparing machine learning algorithms for predicting drug-induced liver injury (DILI) Mol Pharm. 2020;17:2628–2637. doi: 10.1021/acs.molpharmaceut.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]