Abstract
Marburg virus (MARV) infection results in severe viral hemorrhagic fever with mortalities up to 90%, and there is a pressing need for effective therapies. Here, we established a small interfering RNA (siRNA) conjugate platform that enabled successful subcutaneous delivery of siRNAs targeting the MARV nucleoprotein. We identified a hexavalent mannose ligand with high affinity to macrophages and dendritic cells, which are key cellular targets of MARV infection. This ligand enabled successful siRNA conjugate delivery to macrophages both in vitro and in vivo. The delivered hexa-mannose-siRNA conjugates rendered substantial target gene silencing in macrophages when supported by a mannose functionalized endosome release polymer. This hexa-mannose-siRNA conjugate was further evaluated alongside our hepatocyte-targeting GalNAc-siRNA conjugate, to expand targeting of infected liver cells. In MARV-Angola-infected guinea pigs, these platforms offered limited survival benefit when used as individual agents. However, in combination, they achieved up to 100% protection when dosed 24 h post infection. This novel approach, using two different ligands to simultaneously deliver siRNA to multiple cell types relevant to infection, provides a convenient subcutaneous route of administration for treating infection by these dangerous pathogens. The mannose conjugate platform has potential application to other diseases involving macrophages and dendritic cells.
Keywords: hemorrhagic fever, Marburg virus, mannose, GalNAc, siRNA, endosome release, conjugate
Graphical abstract
MARV infection causes high mortalities, and effective treatments are in pressing need. We established a hexavalent mannose ligand conjugate platform that successfully delivered siRNAs to macrophages and dendritic cells. Combination of this mannose-siRNA alongside our hepatocyte-targeting GalNAc-siRNA conjugate provided full protection against MARV infection in guinea pigs.
Introduction
Viral hemorrhagic fevers are a group of life-threatening infectious diseases caused by four virus families.1,2 Among them, Marburg virus disease (MVD) resulted in 24%–88% fatality rates in past outbreaks according to World Health Organization (WHO). To date, there are no vaccines or antiviral therapies approved for MVD despite it still being endemic in central Africa.3 Marburg virus (MARV) is a negatively stranded RNA virus that can be transmitted to humans from infected animals such as bats and nonhuman primates.4 The mononuclear phagocyte system, including macrophages, monocytes, Kupffer cells, and dendritic cells, are primary target cells of MARV infection, and other cells, such as hepatocytes and fibroblast-like cells, are also target cells5 The liver is the main target organ for MARV replication as the asialoglycoprotein receptor (ASGPR) expressed on hepatocytes enhances the infection of MARV.6,7,8 MARV infection cause damage in multiple organs besides the liver, such as lymph nodes, spleen, lungs, gastrointestinal tract, kidneys, heart, and central nervous system.4 Cell death in both infected cells and non-infected cells contributes to the MARV-induced tissue damage.9 The MARV genome is a 19-kb RNA encoding seven viral proteins, including nucleoprotein (NP), VP35 (polymerase cofactor), VP40 (matrix protein), glycoprotein (GP), VP30 (transcription activator), VP24 (secondary matrix protein), and an RNA-dependent RNA polymerase (L polymerase),10 which are potential targets for anti-MARV therapy development.
Short interfering RNAs (siRNAs) trigger specific and precise target gene silencing in a sequence-dependent manner through the naturally occurring RNA interference (RNAi) machinery inside.11 Inhibition of hemorrhagic fever virus (HFV) with siRNA has been reported both in vitro and in vivo.12,13,14 One of the key challenges in developing RNAi-based therapies is the development of efficient delivery platforms to promote cellular uptake of siRNAs. We previously developed a liver-targeting lipid nanoparticle formulation that enabled the first approved RNAi therapy.15 This platform was also shown to be effective for siRNA-based treatment of MARV-infected rodents and nonhuman primates.12,13 The LNP-siRNA modality has been widely investigated for various diseases.16,17 Since then, ligand-siRNA conjugates have emerged as another siRNA-based modality, also resulting in approved RNAi therapies.18 These conjugates are compatible with subcutaneous administration and generally more stable than LNP-siRNA, which is preferred in less developed countries and tropical climates. However, the well-adopted N-acetylgalactosamine (GalNAc)-siRNA conjugates deliver payloads predominantly via the asialoglycoprotein receptor (ASGPR), which is highly expressed on hepatocytes but minimally on other cell types.19 While this specificity is helpful for hepatocyte-based indications, it limits the ability to treat MARV infection, which affects both hepatocytes and the mononuclear phagocytic system (macrophages, Kupffer cells, and dendritic cells).5,20,21 A ligand-siRNA conjugate delivery platform that additionally targets these phagocytic cells could potentially address this limitation.
Mannose has been identified as a ligand for CD206 (also known as the mannose receptor) found on the surface of macrophages and immature dendritic cells.22,23 After binding mannose-rich glycoconjugates, receptor internalization mediated by endocytosis and phagocytosis of the bound ligands occurs in macrophages.24 Inspired by our GalNAc-siRNA conjugate platform, we developed a multivalent mannose conjugate platform to enable targeted delivery of siRNAs to CD206-expressing macrophages and dendritic cells. In vitro gene silencing mediated by mannose-siRNA was demonstrated in human CD14+ monocyte-derived macrophages. Importantly, combination treatment of our GalNAc-siRNA and mannose-siRNA conjugate targeting MARV NP protein conferred 100% protection against lethal MARV infection in a guinea pig model.
Results
Binding of monovalent and multivalent mannose ligands by human CD14+ monocyte-derived macrophages in vitro
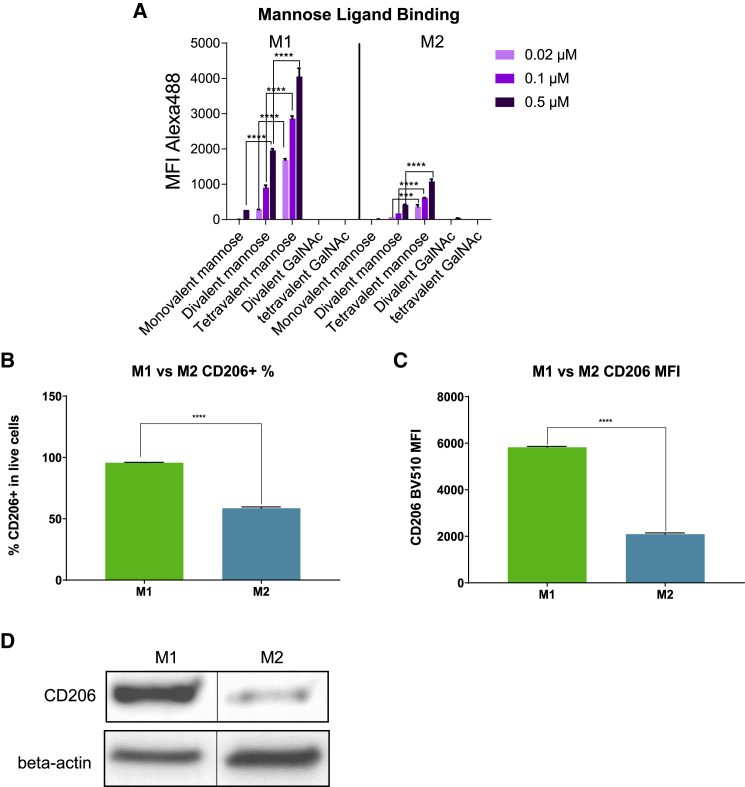
To identify a potent mannose ligand for siRNA conjugation, we designed a series of biotinylated mannose ligands with different valencies (mono-, di-, tetravalent) (Figure S1). These ligands were complexed with Alexa Fluor 488 (AF488)-conjugated streptavidin and incubated with human CD14+ monocyte-derived M1 and M2 macrophages. The binding affinity of each ligand complex was determined by quantification of AF488 fluorescence using flow cytometry. As shown in Figure 1A, dose-dependent increase of AF488 mean fluorescence intensity (MFI) was observed in both M1 and M2 macrophages, indicating ligand binding and cellular uptake. No appreciable binding of divalent or tetravalent GalNAc ligands was observed in either cell type, consistent with the absence of ASGPR expression. This indicated that the detected fluorescence resulted from mannose ligand binding and not passive non-specific fluorescent complex uptake.
Figure 1.
Binding of mono-, di-, and tetravalent mannose ligands to M1 and M2 macrophages
(A) Biotinylated mono-, di- or tetravalent mannose ligands were complexed with AF488-labeled streptavidin and incubated with human CD14+ monocyte-derived M1 and M2 macrophages. The mean fluorescence index (MFI) of AF488 was quantified using flow cytometry to determine the binding affinity of each ligand (n = 2, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, two-way ANOVA analysis). The error bars represent standard deviation.
(B) Expression of CD206 in M1 was quantified by flow cytometry (n = 3, “∗∗∗∗” = p < 0.0001, Welch’s t test). The error bars represent standard deviation.
(C) Expression of CD206 in M2 was quantified by flow cytometry (n = 3, ∗∗∗∗p < 0.0001, Welch’s t test). The error bars represent standard deviation.
(D) Expression of CD206 on M1 and M2 detected by western blot with beta-actin as a loading control. The blank lanes between the samples were removed to allow better comparison.
In both M1 and M2 cells, the binding activity of mannose ligands correlated positively with valency, with tetravalent ligand showing the highest binding affinity, and monovalent ligand the least (Figure 1A). Similar advantages for multivalent ligands have been reported for GalNAc-siRNA platforms.25,26,27,28 Interestingly, all three mannose ligands showed higher binding affinity to M1 than M2, which is consistent with higher expression of target mannose receptor (CD206) in M1 than M2 differentiated with the current induction protocol (Figure 1B–1D).
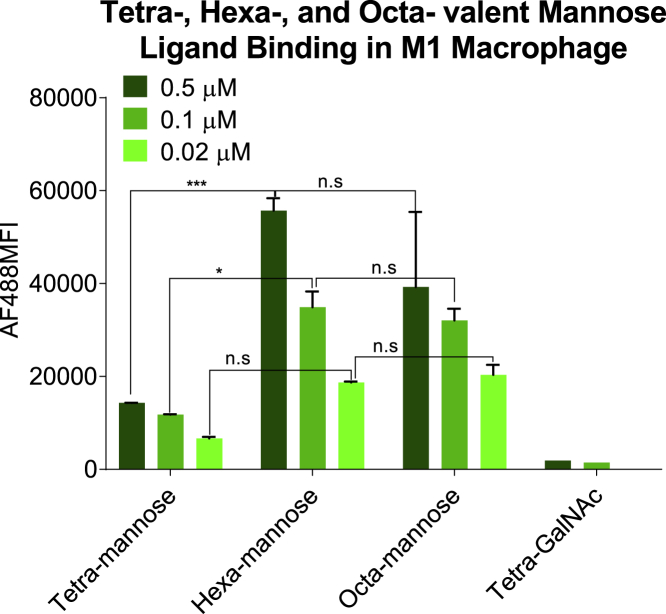
This prompted us to investigate even higher valencies, and we synthesized and tested the binding affinity of hexa- and octa-valent mannose ligand (Figures S1–S8) against the tetravalent ligand in M1 macrophage. As shown in Figure 2, hexavalent mannose ligand exhibited further increased binding, while increasing to octa-valency appeared to show a slight decrease.
Figure 2.
Binding of tetra-, hexa-, and octa-valent mannose ligands to M1 macrophages
Biotinylated tetra-, hexa-, or octa-valent mannose ligands were complexed with AF488-labeled streptavidin and incubated with M1 macrophages. The MFI of AF488 was quantified using flow cytometry to determine the binding affinity of each ligand (n = 2; ∗p < 0.05; ∗∗∗p < 0.01; n.s., p > 0.05, two-way ANOVA analysis). The error bars represent standard deviation.
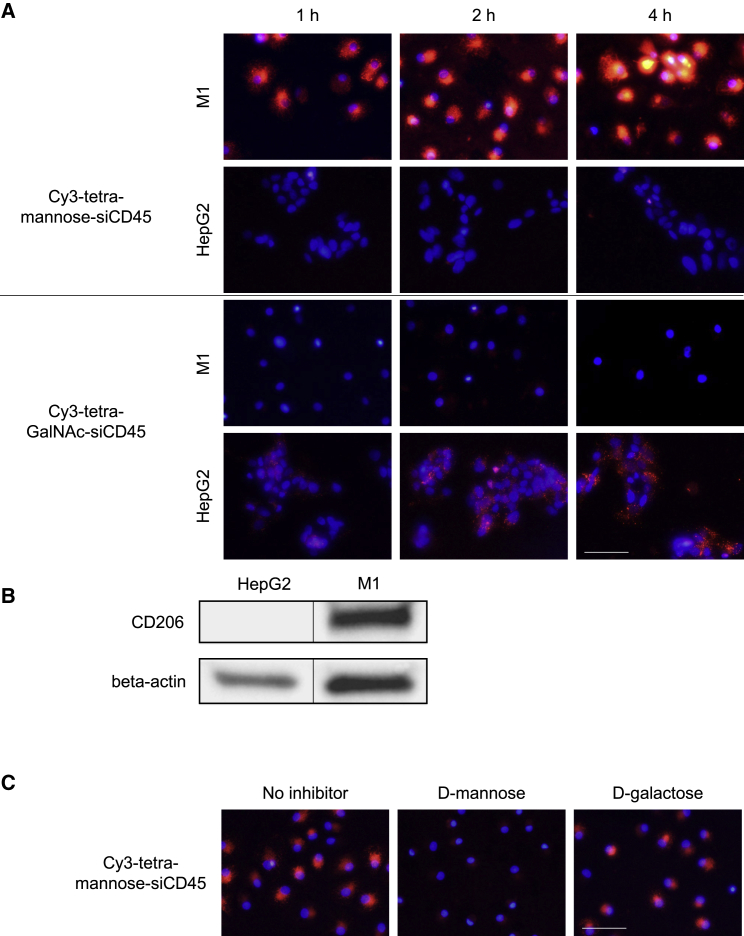
Uptake of fluorescent-labeled mannose-siRNA conjugate by macrophage and dendritic cells in vitro
To test the efficacy of mannose-enabled siRNA delivery, a chemically modified siRNA targeting mouse CD45 was conjugated with the tetravalent mannose ligand (Figure 3A). The same CD45 siRNA conjugated with a tetravalent GalNAc ligand was employed as a negative control. The conjugates were labeled with cyanine 3 (Cy3) fluorescence to allow visualization and quantification of siRNA uptake. M1 macrophages and HepG2 cells were incubated with the conjugates, and siRNA uptake was analyzed using fluorescent microscopy. Consistent with the ligand binding results, Cy3-tetra-mannose-siRNA successfully delivered to M1 macrophages but not HepG2 cells expressing lower levels of CD206 (Figure 3B). Conversely, Cy3-tetra-GalNAc-siRNA showed uptake by HepG2 cells (which express the ASGPR receptor) but was not detectable in M1 macrophages. We further compared the delivery of Cy3-tetra-mannose-siRNA to Cy3-hexa-mannose-siRNA in M1 cells. Consistent with the ligand binding data, the hexavalent mannose ligand-conjugated siRNA showed higher delivery activity than the tetravalent ligand conjugate (Figure S9).
Figure 3.
Uptake of fluorescent-labeled mannose-siRNA conjugate by M1 macrophage
(A) Cy3-labeled tetra-mannose-siCD45 or tetra-GalNAc-siCD45 were incubated with M1 macrophages or HepG2 cells. The delivery of conjugates was determined by detection of Cy3 fluorescence under a fluorescence microscope. Cell nuclei were stained with DAPI. Scale bar, 100 μm.
(B) Expression of CD206 in M1 and HepG2 cells were measured by western blot. The blank lanes between the samples were removed to allow better comparison.
(C) M1 macrophages were treated with D-mannose or D-galactose before incubating with Cy3-tetra-mannose-siCD45. The delivery of conjugates was determined by detection of Cy3 fluorescence under a fluorescence microscope. Cell nuclei were stained with DAPI. Scale bar, 100 μm.
To demonstrate the involvement of the mannose receptor in conjugate delivery, we conducted a competition binding experiment using D-mannose, a natural ligand for CD206. M1 macrophages were first incubated with D-mannose or D-galactose before treating with Cy3-tetra-mannose-siRNA. As shown in Figure 3C, competitive binding of D-mannose substantially inhibited uptake of the mannose-siRNA conjugate, illustrated by reduced fluorescence. D-galactose, which is not a ligand for the mannose receptor, showed no competitive binding effect. These results confirmed the involvement of the mannose receptor in mannose-siRNA conjugate delivery to human CD14+ monocyte delivery M1 macrophages.
In addition to macrophages, CD206 is reportedly expressed in dendritic cells (DCs), although expression is substantially reduced upon maturation of these cells.29,30,31,32 We therefore sought to test uptake in both immature DCs (iDCs) and mature DCs (mDCs), again characterizing the effect of ligand valency. M1 and M2 macrophages were included as positive controls. Cells were incubated with Cy3-tetra-mannose-siRNA, Cy3-hexa-mannose-siRNA, or Cy3-tetra-GalNAc-siRNA. The resulting fluorescence was quantified using flow cytometry and indicated effective uptake of Cy3-tetra-mannose-siRNA and Cy3-hexa-mannose-siRNA but not Cy3-tetra-GalNAc-siRNA. Consistent with the ligand binding data and reported receptor expression, both hexavalent and tetravalent mannose-siRNA conjugates showed superior delivery to M1 compared with M2 (Figure 4). Similar delivery selectivity was observed when comparing iDCs with mDCs. Of note, hexa-mannose-siCD45 was more efficient than tetra-mannose-siCD45 in delivering to M1, M2, and iDCs, particularly at the lower dose. These results further confirmed that multivalent mannose conjugates mediate effective uptake in CD206-expressing cells.
Figure 4.
Uptake of fluorescent-labeled mannose-siRNA conjugate by macrophages and dendritic cells
Cy3-labeled hexa-mannose-siCD45, tetra-mannose-siCD45, or tetra-GalNAc-siCD45 were incubated with M1, M2, iDC, or mDC derived from human CD14+ monocytes. The MFI of Cy3 was quantified using flow cytometry to determine the uptake of each conjugate. The error bars represent standard deviation.
In vitro gene silencing by mannose-siRNA conjugates in M1 macrophage
To investigate whether the ligand binding affinity and conjugate delivery translate to gene silencing activity, we treated M1 macrophages with hexa-mannose-conjugated CD45 siRNA. As endosome release has been reported to be a significant bottleneck in siRNA conjugate delivery, we also supplemented the siRNA treatment with a mannose-conjugated, pH-responsive endosome release polymer (ERP) (Figures S10 and S11) to address this challenge.28 The same CD45 siRNA conjugates with a tetravalent GalNAc ligand and a GalNAc functionalized ERP were employed as controls. In the absence of ERP, neither GalNAc conjugates nor mannose conjugates enabled any appreciable target gene silencing in the treated cells (Figure 5). Treatment of hexa-mannose-siCD45 in combination with mannose-ERP but not GalNAc-ERP resulted in target gene inhibition in treated M1 macrophages. RNAi activity was not appreciable in the tetra-GalNAc-siCD45 treatment groups, regardless of the presence of GalNAc or mannose-ERP. These results suggest that mannose ligands support, and are indeed critical to, the delivery of both siRNA conjugates and ERP to M1 macrophages. The mannose ligand must be present on both entities to ensure their uptake and concomitant gene silencing.
Figure 5.
In vitro gene silencing by mannose-siRNA conjugates in M1 macrophages
M1 macrophages were treated with 20 μg/mL of hexa-mannose-siCD45 or tetra-GalNAc-siCD45 either alone or in combination with 100 μg/mL of mannose- or GalNAc- ERP. Cell lysates were collected at 24 h post conjugate treatment and the target gene silencing activity was assessed by quantification of mRNA with QuantiGene assay (n = 3, ∗∗∗p < 0.01, two-way ANOVA analysis). The error bars represent standard deviation.
In vivo delivery and activity of hexa-mannose-siRNA conjugate
To examine in vivo delivery and activity of hexa-mannose-siRNA conjugates, we elicited macrophages in the mouse peritoneal cavity by induction with thioglycolate.33,34 Animals were then treated with fluorescent-labeled (Alexa Fluor 647 [AF647]) hexa-mannose-siCD45 conjugate and mannose-ERP. The peritoneal cells of treated animals were collected at 24 h post treatment and analyzed with flow cytometry with AF647 for siRNA delivery and CD45 for gene silencing activity. As shown in Figure 6A, fluorescence was detected in >80% CD206+ peritoneal macrophages in AF647-hexa-mannose-siCD45-treated animals, regardless of the presence/absence of mannose-ERP. This compares with ∼50% delivery to the CD45+ cells, and the majority (∼80%) of CD45+ cells with conjugate delivery were also CD206+ (Figure S12). The delivered siCD45 enabled a trend of target gene silencing in both groups, with the mannose-ERP showing a slight benefit to RNAi activity (Figure 6B), but neither treatment achieved statistically significant gene knockdown. We hypothesized that the expression of CD206 in these thioglycolate-elicited peritoneal macrophages was not high enough to allow sufficient siRNA conjugate and/or ERP uptake. Nonetheless, these results demonstrate successful delivery of hexa-mannose-conjugated siRNA to macrophages in the thioglycolate mouse model.
Figure 6.
In vivo delivery and activity of hexa-mannose-siRNA conjugate
(A and B) C57BL/6 female mice (n = 4) were induced with 4% thioglycolate to elicit peritoneal macrophages. Animals were injected subcutaneously with a single dose of vehicle control (saline), AF647-hexa-mannose-siCD45 (10 mg/kg), AF647-hexa-mannose-siCD45 (10 mg/kg) + mannose-ERP (50 mg/kg), or mannose-ERP alone (50 mg/kg). The peritoneal cells of treated animals were collected at 24 h post siRNA and ERP treatment and analyzed with flow cytometry for the delivery of siRNA conjugate (AF647 fluorescence, A) and target gene silencing (CD45, B). (C) Female Balb/c mice (n = 4) were injected subcutaneously with a single dose of vehicle control (saline), AF647-hexa-mannose-siCD45 (3 mg/kg), or AF647-unconjugated-siCD45 (3 mg/kg). The single cells were isolated from the mouse livers at 1.5 h post siRNA treatment and analyzed with flow cytometry for the delivery of siRNA conjugate (AF647 fluorescence). Cells were also stained with ASGPR and F4/80 to identify the hepatocyte and Kupffer cell populations, respectively. The error bars stand for standard error of mean (SEM).
We further explored the delivery of hexa-mannose-siRNA conjugates to liver macrophages (Kupffer cells) that express CD206.35 Mice were then treated with a single subcutaneous dose of AF647-labeled hexa-mannose-siCD45 conjugate or unconjugated siCD45 at 3 mg/kg. At 1.5 h post conjugate treatment, liver cells were collected via perfusion and enzymatic digestions and stained with anti-ASGPR and anti-F4/80 to identify hepatocytes and Kupffer cell populations, respectively. The cells were analyzed using flow cytometry to quantify the siRNA delivery. Hexa-mannose-siCD45 and unconjugated siCD45 showed similarly low delivery to hepatocytes and total nonparenchymal cells (NPCs). In Kupffer cells, though, substantially more accumulation of hexa-mannose-siCD45 (>70%) than unconjugated siCD45 (∼30%) was observed. These results demonstrated the delivery of our hexa-mannose-siRNA conjugate to the liver Kupffer cells.
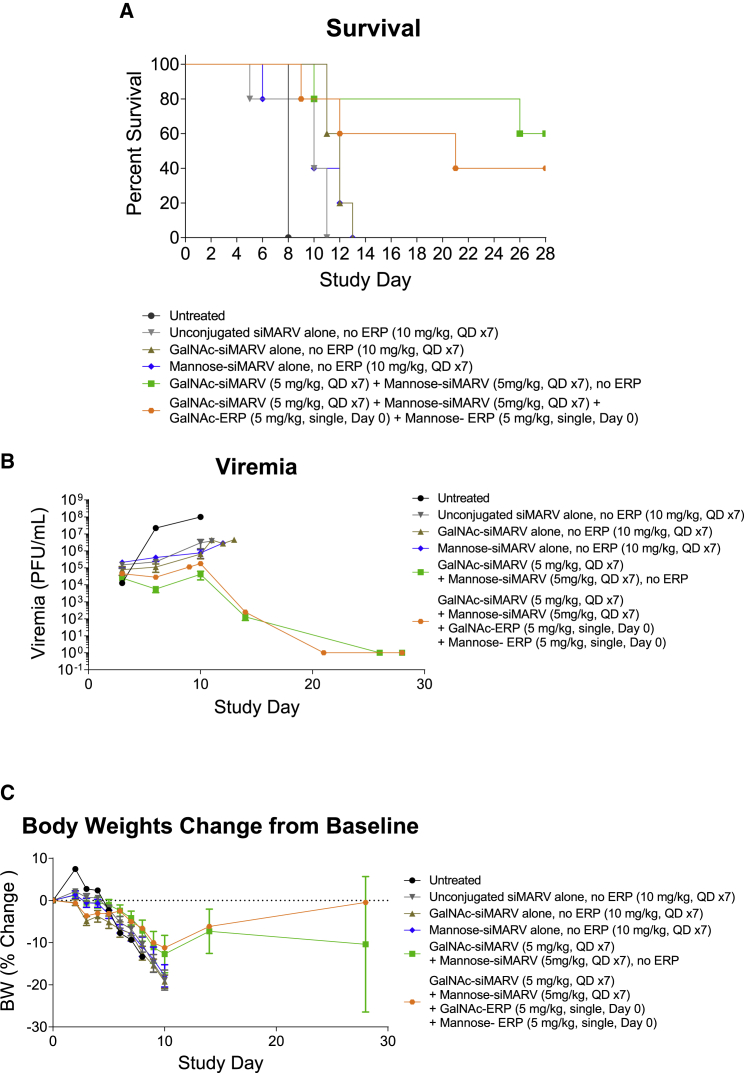
Combination treatment of hexa-mannose-siRNA and GalNAc-siRNA demonstrated antiviral potency in a guinea pig virus challenge model
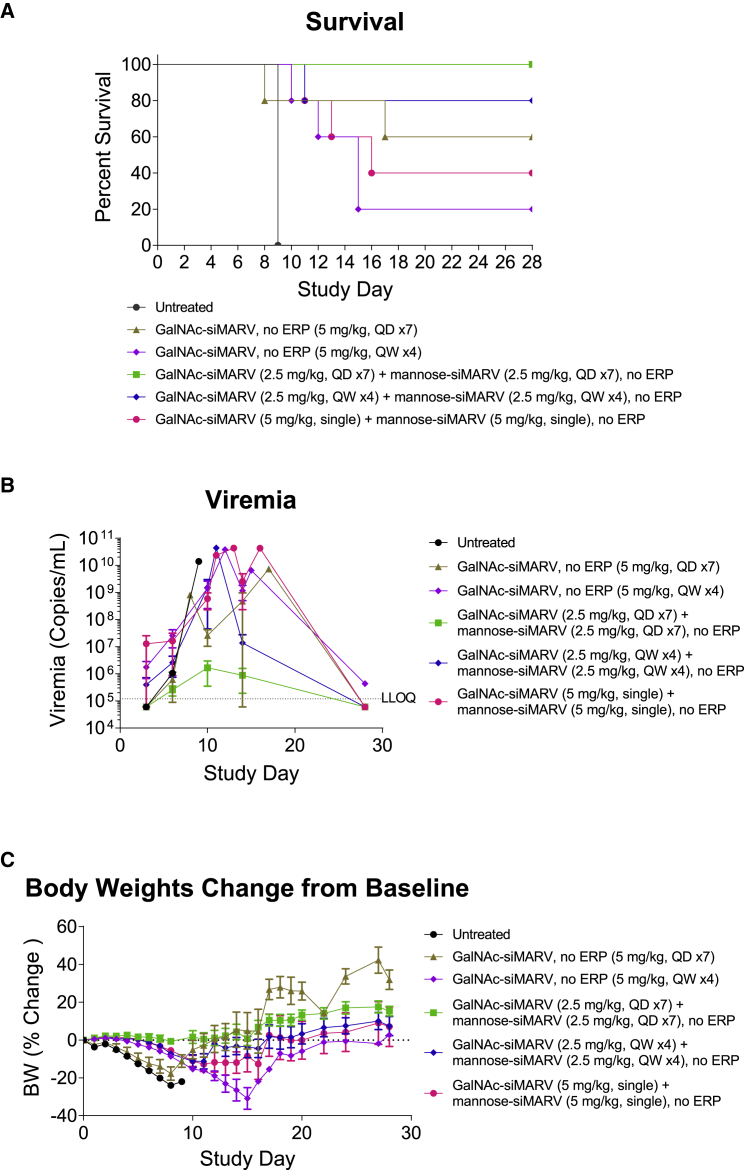
We further evaluated the mannose-siRNA delivery platform in a guinea pig model of MARV (Angola variant) infection.36 Taking the siRNA sequence targeting MARV NP previously identified for use with LNP delivery,12 we applied a heavy modification pattern comprising 2′ O-methyl (OMe) and fluorine (F) to render it suitable for conjugate delivery.37 To allow for targeting of both hepatocytes and macrophages/dendritic cells (collectively, the most relevant cell types in MARV infection), we coupled both tetra-GalNAc and hexa-mannose ligands to the siRNA. These conjugates were tested both alone and in combination in the guinea pig MARV infection model. Animals were challenged with a lethal dose of MARV and treated with conjugates at 24 h post infection. As shown in Figure 7A, unconjugated siMARV, tetra-GalNAc-siMARV, or hexa-mannose-siMARV treatment alone provided no appreciable protection against virus-induced mortality when dosed at 10 mg/kg/day for 7 days. Combination treatment of tetra-GalNAc-siMARV (5 mg/kg, once per day (QD)) and hexa-mannose-siMARV (5 mg/kg, QD) for 7 days substantially improved the survival rate to 60%, accompanied with reduced viremia and improved clinical scores (Figure S13). We also explored the effect of ERPs in this study by supplementing the treatment with GalNAc-ERP (5 mg/kg) and mannose-ERP (5 mg/kg) on the first day of conjugate treatment immediately after injection of siRNA conjugates. Combination of ERP showed no further improvement on the survival or viremia in the challenged animals, which could be due to suboptimal dosing levels or regimen. To optimize the dosing regimen, we performed a second study to further compared a daily dosing schedule with weekly and single dose at 5 mg/kg total siRNA starting from 24 h post infection. Consistent with the first study, the GalNAc/mannose conjugate combination treatment again provided superior protection compared with the GalNAc conjugate alone, in both daily and weekly dosing regimens (Figures 8A and S14). In particular, the daily dosing group with 5 mg/kg of total GalNAc/mannose conjugate (1:1) achieved 100% survival with no clinical sign of disease. The viremia and body weight data agreed well with the animal survival results (Figures 8B and 8C). We did not test ERP in this study as it would require further optimization on conjugate dosing regimen. Together, the mannose/GalNAc combination ligand platform appears to offer superior protection in the MARV guinea pig model, a fact we currently ascribe to effective delivery of siRNA to all relevant cell types.
Figure 7.
Daily dosing of GalNAc- and mannose-conjugated anti-MARV siRNA reduced MARV-induced mortality in guinea pig
Guinea pigs (n = 5 for conjugate treatment group, n = 1 for untreated group) were challenged with a lethal dose of MARV Angola strain. Twenty-four hours post viral exposure, animals were subcutaneously injected with unconjugated siMARV, tetra-GalNAc-siMARV, hexa-mannose-siMARV alone, combination of tetra-GalNAc-siMARV + hexa-mannose-siMARV (1:1) (10 mg/kg total siRNA, mixed in one vial; daily dosing, seven doses). In one group, treatment of tetra-GalNAc-siMARV + hexa-mannose-siMARV (1:1) (10 mg/kg total siRNA, mixed in one vial; daily dosing, seven doses) was immediately followed with GalNAc-ERP + mannose-ERP (1:1) (10 mg/kg total ERP, mixed in one vial, single subcutaneous dose on the first day of conjugate treatment). The antiviral efficacy was evaluated by survival rates (A), viremia (B), and body weights (C). The error bars stand for standard error of mean (SEM).
Figure 8.
Effect of dosing regimen of anti-MARV GalNAc and mannose-conjugated siRNA in guinea pig model
Guinea pigs (n = 5 for conjugate treatment group, n = 1 for untreated group) were challenged with a lethal dose of MARV Angola strain. Twenty-four hours post viral exposure, animals were treated with tetra-GalNAc-siMARV alone or in combination with hexa-mannose-siMARV (1:1) (5 mg/kg total siRNA; daily subcutaneous dosing, seven doses, or weekly subcutaneous dose, four doses). One group with a single 10-mg/kg total siRNA dose of combination GalNAc and mannose conjugates was also tested. The antiviral efficacy was evaluated by survival rates (A), viremia (B), and body weights (C). The error bars stand for standard error of mean (SEM).
Discussion
RNAi therapeutics have demonstrated potential in addressing a variety of diseases and viral infections. They act by degrading specific targeted mRNA transcripts and silence the downstream expression of encoded proteins. Synthetic siRNAs are common RNAi triggers composed of two RNA oligonucleotides that are complementary in sequence and form a duplex. They are designed to enable post-transcriptional gene suppression through sequence-specific degradation of the selected target mRNA. The first approved RNAi product used an LNP delivery system for delivery of an siRNA targeting the transthyretin (Ttr) gene.15 This delivery platform has also achieved high efficacy in MARV infection in preclinical studies.12,13 However, the requirement of intravenous (i.v.) dosing, and, to a lesser degree, cold chain storage make the platform’s use more challenging in developing countries where MARV infection is still endemic. A ligand-siRNA conjugate format would be preferable for this indication, given its subcutaneous administration route and higher thermo-stability than LNP-siRNA. Widely used GalNAc-siRNA platforms are limited to hepatocyte targeting, while MARV infection affects multiple cell types in addition to hepatocytes, such as macrophages and dendritic cells. Development of a ligand-siRNA conjugate platform that would additionally access these cells is thus desirable.
Mannose receptor (CD206) is a membrane lectin primarily expressed in macrophages and dendritic cells.22,38,39,40 It recognizes a range of carbohydrates presented on the surfaces of pathogens and thus plays a role in immune recognition. Compared with other ligands, such as peptides and antibodies for targeting macrophages and dendritic cells,41,42 mannose is a natural ligand for CD206 and has been utilized for conjugation with small molecules and functionalizing nanoparticles for macrophage-targeting delivery.43,44,45,46,47,48,49,50 Here, we applied our experience in developing GalNAc-siRNA conjugates to establish a mannose-siRNA conjugate delivery platform.28 We synthesized a range of mannose ligands and demonstrated that binding affinity generally increased with valency, with a hexavalent ligand having the highest. This advantage in multivalency agrees with the previous finding in GalNAc ligands, but the underling mechanism could be partially different. The GalNAc-targeted receptor ASGPR is formed from two carbohydrate recognition domains with a dominant trimeric configuration.25,26,27 Thus, ligand multivalency allows increased interaction with the receptor. CD206, on the other hand, is a monomeric receptor with one major domain (C-type lectin-like domain-4, CTLD4) for mannose binding.40 It is possible that the multivalent mannose ligands would trigger some clustering effect of the receptor on the cell membrane surface and improve uptake.51 A detailed account of how multivalency affects the binding of mannose ligands has yet to be elucidated.
Receptor expression level is one of the key factors affecting delivery efficiency. We compared CD206 expression in M1 versus M2 macrophages differentiated from human CD14+ monocytes using a previously reported protocol.52 To our surprise, in contrast to results in other reports,53,54,55 M1 macrophages differentiated in this study expressed higher levels of CD206 than M2. Of note, macrophage activation resulted in a spectrum of heterogeneous cell populations, and the gene expression profiles are substantially influenced by the differentiation protocol.56,57,58 For example, the Roche CellXVivo human M1 macrophage differentiation kit could also induce CD206+ M1 macrophages from human CD14+ monocytes. Importantly, CD206 expression levels in our M1 and M2 cells were in accordance with the binding affinity of mannose ligands and uptake of mannose-siRNA conjugates. Similarly, uptake of mannose-siRNA conjugates was more effective in iDCs than mDCs, consistent with the reported CD206 expression level.29,30,31,32 These results indicated that delivery of mannose-siRNA conjugate was mediated by the CD206 receptor, which was further confirmed with our ligand competition experiment.
Endosomal escape is a major bottleneck for siRNA conjugate activity, and previous studies have shown the majority of delivered GalNAc-siRNA remains trapped in these cellular compartments, although sufficient material can reach the cytoplasm to mediate RNAi. The precise mechanics of this process have not been fully elucidated.59 This results in the requirement for high doses of GalNAc-siRNA to provide sufficient activity. We previously reported that co-administering a GalNAc functionalized ERP micelle provides markedly higher gene silencing potency in GalNAc-siRNA-treated animals28 This polymer is capable of forming small micelle structures spontaneously in aqueous media when reaching the critical micelle concentration (CMC). When entering an endosome with an acidic pH environment, the polymer micelle will be protonated and trigger endosomal membrane destabilization to promote siRNA conjugate escape. This strategy could be essential for extrahepatic cell targeting where the receptors are expressed at a lower level or ligand binding is less efficient. During the investigation of RNAi activity of our mannose-siRNA conjugates in vitro, we observed successful uptake in macrophages without appreciable gene silencing when treated with conjugate alone. The lack of activity was successfully addressed by combination treatment of mannose-ERP. It is notable that both the siRNA and the ERP had to be derivatized with mannose to observe activity, suggesting that delivery of both entities is mediated by the mannose receptor.
Encouraged by the in vitro delivery and activity results, we tested the mannose conjugate platform in vivo. In a murine model where thioglycolate is used to elicit peritoneal macrophages, successful delivery of mannose conjugates to these cells was achieved, although gene silencing activity was not appreciable. Even with the addition of the mannose-ERP, activity was not significantly improved. This is not entirely unexpected since peritoneal macrophages are known to express low levels of CD206.60 Low expression of target receptor may not support uptake of both mannose-siRNA and mannose-ERP. We also demonstrated delivery of mannose-siRNA conjugates to the liver Kupffer cells, which are liver-resident macrophages.
Results in a guinea pig model of MARV infection were considerably better. Hepatocytes, monocytes, and macrophages are all known to be infected by the virus, the latter cells becoming activated in the process.61 As expected, targeting the virus in these cell types individually with hepatocyte-targeting GalNAc-siRNA, or macrophage/DC-targeting mannose-siRNA treatment, does not provide full protection against the infection. Although some level of viremia reduction was observed in both single-treatment arms, combination of the two platforms rendered the most effective protection, consistent with the concept of targeting the virus in all relevant cell populations.5,20,21 We also observed better protection from the 5-mg/kg daily dosing regimen than the 10-mg/kg daily dosing regimen. This could be explained by the MARV virus compromising liver function during infection, which in turn is likely to be sensitive to an aggressive dosing regimen with high siRNA dose burden. This may also explain why no benefit was appreciated when supplementing the treatment with GalNAc-ERP and mannose-ERP. Indeed, weekly dosing of mannose-siRNA and GalNAc-siRNA combination at 5 mg/kg also provides reasonably good therapeutic benefit. Further optimization of dosing regimen and evaluation of impacts to liver function in the context of treated infection are the focus of ongoing work. We are also planning to combine our RNAi approach with other anti-MARV treatments to improve the therapeutic effect.62
Our mannose-siRNA delivery platform offers therapeutic potential in other indications involving macrophages and DCs. For example, macrophages play an essential pathological role in rheumatoid arthritis,63,64,65 Kupffer cells (liver macrophages) are actively involved in multiple liver diseases and liver injury,66 and macrophages are also an essential factor in cancer pathogenesis.67 Our subcutaneously administered macrophage/DC-targeting delivery platform potentially allows siRNA conjugates to be redirected to address these challenging diseases.
Materials and methods
Synthesis of mannose and GalNAc ligands
The ligands utilized in this manuscript were prepared using established organic chemistry techniques, including purification by automated flash chromatography and product confirmation with a combination of analytical high-pressure liquid chromatography (HPLC), liquid chromatography-mass spectrometry (LC-MS), mass spectrometry (MS), and proton nuclear magnetic resonance spectrometry (NMR) as appropriate. Complete synthetic protocols for all ligands contained in this manuscript can be found in the supplemental information.
Synthesis of mannose-conjugated and GalNAc-conjugated siRNAs
For sense strand synthesis, ligand succinates (Figure S8) were loaded onto 1,000 long-chain aminoalkyl controlled pore glass (CPG) using standard amide coupling chemistry. Loading was determined by Dimethoxytrityl (DMTr) assay at UV-visible (UV-vis) 504 nm. The resulting GalNAc- or mannose-loaded CPG solid support was employed in automated oligonucleotide synthesis using standard procedures. Nucleotide deprotection followed by removal from the solid support (with concurrent galactosamine acetate deprotection) afforded the GalNAc- or mannose-oligonucleotide conjugate (Figure S2). In some cases, fluorescent (Cy3 or AF647) labeling was conducted at the 5′ end of the sense strand.
For antisense strand synthesis and duplex formation, antisense strands were prepared by automated oligonucleotide synthesis using standard procedures. Nucleotide deprotection followed by removal from the solid support afforded the deprotected antisense strand. Annealing of the sense and antisense strands using standard techniques afforded siRNA conjugates.
The CD45 siRNA sequences are cited from the 5′ to 3′ end as follows:
Sense strand: mC∗mUmGmGfCmUfGfAfAmUmUmUmCmAmGmAmGmC∗mA.
Antisense strand: mU∗fG∗mCmUmCfUmGfAfAmAmUmUmCfAmGfCmCmAmG∗mU∗mU.
2′-O-methyl nucleotides are depicted as “m” + UPPER CASE; 2′-Fluoro nucleotides are depicted as “f” + UPPER CASE. Phosphorothioate linkers are depicted as “∗.”
The MARV siRNA sequences are cited from the 5′ to 3′ end as follows:
Sense strand: mG∗mA∗mUmUmCmUfCmAfGfGfAmCmUmUmCmUmUmAmUmUmA
Antisense strand: mU∗fA∗mAmUmAfAmGfAfAmGmUmCmCfUmGfAmGmAmAmUmC∗mU∗mA.
Mannose and GalNAc biotin ligand preparation
Biotinylated ligands were prepared according to procedures outlined in the supplemental data (Figures S1 and S7).
Mannose- and GalNAc-targeted ERP synthesis and micelle preparation
Polymers were synthesized by Syngene International using synthetic methodology analogous to that previously described (Figure S10).68 The polymer was dissolved in 10 mM phosphate/200 mM sucrose PBS (pH 7), up to 40 mg/mL followed by successive filtration (three or four times) through a 0.2-μm sterile filter. This produced stable polymer micelles with an approximate particle diameter of 15 nm and polydispersity of <0.16.
HepG2 cell culture
HepG2 cells (ATCC, Manassas, VA) were propagated as monolayers in 175-cm2 culture flasks at 37°C in a humidified atmosphere containing 5% CO2, using Minimum Essential Medium (MEM) (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 1× non-essential amino acids, and 0.15% sodium bicarbonate. All components were from Thermo Fisher Scientific (Waltham, MA).
Differentiation of M1, M2, iDC, and mDC from human CD14+ monocytes
Human Buffy coat was purchased from BioIVT. CD14+ monocytes were isolated from buffy coat using the StraightFrom Buffy Coat CD14 MicroBead Kit (Miltenyi Biotech, Auburn, CA) following the manufacturer’s protocol. The isolated CD14 monocytes were differentiated to M1 and M2 macrophages following a previously published protocol.52 In brief, monocytes were cultured in RPMI medium containing 10 mM HEPES, 2 mM L-glutamine, and 10% FBS. To trigger M1 macrophage differentiation, cells were stimulated with 50 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) on days 0 and 3 and further induced with 50 ng/mL interferon (IFN)-γ on day 6. For M2 macrophage differentiation, cells were treated with 50 ng/mL macrophage colony-stimulating factor (M-CSF) on day 0 and 3 and further induced with 20 ng/mL interleukin (IL)-4 in combination with 20 ng/mL IL-10 on day 6. Differentiated M1 and M2 were tested for ligand binding on day 7. iDC differentiation was stimulated with treatment of 500 U/mL GM-CSF + 1,000 U/mL IFNα2b on day 0 and day 3. mDC differentiation was triggered with treatment of 500 U/mL GM-CSF + 1,000 U/mL IFNα2b on day 0 and 500 U/mL GM-CSF + 1,000 U/mL IFNα2b + 1 μg/mL LPS in combination with 1 μg/mL LPS on day 3. iDCs and mDCs were collected on day 5 for conjugate delivery test. All components were from Thermo Fisher Scientific (Waltham, MA).
Receptor binding assay
Biotinylated mannose ligands were reconstituted in dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO) and functionalized by incubating with Alex Fluor 488-labeled streptavidin (Thermo Fisher Scientific, Waltham, MA) in Tyrode buffer (containing 10 mM HEPES, 5.6 mM glucose, 10 mM KCl, 35 mM NaCl, 0.4 mM MgCl, 1.0 mM CaCl2, and 0.1% BSA, pH 7.3) at 4°C overnight at a molar ratio of 4.5:1 of biotinylated ligands to biotin binding sites.
M1 and M2 macrophages were washed with Dulbecco’s PBS (DPBS) (Thermo Fisher Scientific, Waltham, MA) containing 2 mM EDTA (Thermo Fisher Scientific, Waltham, MA), resuspended in ice-cold Tyrode buffer, and seeded into sterile V-bottom 96-well plates (40,000/well). The previously prepared biotin-mannose/AF488-streptavidin complex was diluted in Tyrode buffer and mixed with the seeded macrophages to reach final concentrations of 0.5, 0.1, and 0.02 μM (based on streptavidin molar concentration). Biotin-GalNAc/AF488-streptavidin complex was included as control treatment. The cells were incubated with the mannose ligand complexes at 4°C for 1.5 h. After incubation, the cells were washed three times with ice-cold DPBS to remove any unbound ligand complex. After centrifuging at 1,200 rpm for 5 min, the cell pellet was resuspended in stain buffer prior to flow cytometry analysis.
Treatment of Cy3-labeled tetravalent mannose or GalNAc-siRNA conjugates for microscopy analysis
M1 macrophages or HepG2 cells were seeded into sterile four-well chamber slides at 80,000 cells/well and 40,000 cells/well in OptiMEM medium (Thermo Fisher Scientific, Waltham, MA), respectively. Cells were cultured at 37°C after seeding. siRNA conjugate treatment was conduct at 10 min post M1 macrophage seeding and 48 h post HepG2 cell seeding. HepG2 cells were washed with 1 mL of OptiMEM before treatment. Each was supplemented with Cy3-tetra-mannose-siCD45 or Cy3-tetra-GalNAc-siCD45 to reach the final concentration of 5 μg/mL in OptiMEM medium. The cells were incubated with the siRNA conjugate for 1, 2, or 4 h. The cells were then washed with PBS three times and fixed with 4% paraformaldehyde (PFA) (Sigma, St. Louis, MO) for 10 min at room temperature. The chambers were removed from the slides and cells mounted with one drop of mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) staining (Thermo Fisher Scientific, Waltham, MA) for the nuclei. The slides were analyzed under a fluorescence microscope.
Competitive binding assay
Human M1 macrophages were seeded into sterile four-well chamber slides at 75,000 cells/well in OptiMEM medium. OptiMEM containing D-mannose or D-galactose was added onto testing cells to reach the final concentration of 139 mM and incubated at 37°C for 5 min. Cells were then treated with Cy3-tetra-mannose-siCD45 in OptiMEM with a final concentration of 5 μg/mL and incubated for 1 h. The cells were then washed with DPBS three times and fixed with 4% PFA for 10 min at room temperature. The chambers were removed from the slides and the cells were mounted with one drop of mounting medium containing DAPI staining for the nuclei. The slides were covered, sealed, and analyzed under fluorescence microscope.
Treatment of Cy3-labeled tetravalent mannose or GalNAc-siRNA conjugates for flow cytometry analysis
M1 macrophages, M2 macrophages, iDCs, or mDCs were seeded into sterile U-bottom 96-well plates at 22,500 cells/well in OptiMEM medium. Cell were treated with Cy3-hexa-mannose-siCD45, Cy3-tetra-mannose-siCD45, or Cy3-tetra-GalNAc-siCD45 at final concentrations of 5, 1.25, or 0.312 μg/mL in OptiMEM medium. At 1 h post treatment, cells were washed with DPBS three times and resuspended in stain buffer for flow cytometry analysis.
Flow cytometry analysis
Analyses were performed on a FACSCanto II using the software FACS Diva (Becton Dickinson, San Jose, CA, United States). As a marker for viability, cells were stained with Live/Dead red or green (Thermo Fisher Scientific, Waltham, MA). The forward scatter and side scatter gate were set to include all viable cells.
When quantification of CD206 and CD45 expression was required, cells were stained with phycoerythrin (PE)-conjugated rat anti-mouse CD45 antibody (BD Biosciences, Franklin Lakes, NJ), BV510-conjugated mouse anti-human CD206 antibody (BD Biosciences, Franklin Lakes, NJ), or APC-efluor780-conjugated rat anti-mouse CD206 antibody (Thermo Fisher Scientific, Waltham, MA) before flow cytometry analysis. In brief, collected cells (from cell culture or animals) were washed and stained with Live/Dead red or green for 30 min, washed with PBS, blocked with Fc Block (CD16/21) (BD Biosciences, Franklin Lakes, NJ), and then stained with desired antibodies for 20 min before flow cytometry analysis.
Approximately 10,000–15,000 cells were counted for each sample. The expression of target receptor or genes and the binding/uptake of fluorescent-labeled ligands/conjugates were determined as increased intensity in the corresponding fluorescent channel. For biotin ligand complex binding analysis, the MFI of cells incubated with functionalized streptavidin minus the MFI of cells incubated without functionalized streptavidin (free fluorophore only) was used to determine binding/uptake.
Treatment of siRNA conjugate and ERP in M1 macrophages
M1 macrophages were seeded into sterile 96-well plates (15,000/well) and incubated with 20 μg/mL of hexa-mannose-siCD45 or tetra-GalNAc-siCD45 for 4 h at 37°C. Mannose-ERP and GalNAc-ERP were then added to the culture at the concentration of 100 ug/mL. At 24 h post siRNA treatment, cells were lysed with QuantiGene Lysis Mixture (Thermo Fisher Scientific, Waltham, MA) for gene expression quantification.
QuantiGene branched DNA assay
To evaluate gene silencing activity, cell lysates collected after the siRNA treatment were subject to QuantiGene branched DNA assay (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. In brief, cell lysates were incubated with capture probes targeting mouse Cd45 (target gene) and Gapdh (endogenous control) mRNAs at 55°C for 18–20 h. After washing, the plates were incubated with pre-amplification probes and amplification probes to amplify the signal. The excessive probes were then washed off and assay substrates were added to allow quantifying luminescence using a plate reader. The signal from Cd45 was normalized to the signal from Gapdh.
Preparation of mannose conjugates and endosomal release polymer for subcutaneous injection in mice and nonhuman primates
Mannose-siRNA conjugates were dissolved in sterile saline to a concentration of 10 mg/mL for subcutaneous injection. Polymer micelles were prepared as described above. Test articles were diluted to desired concentration and supplied in a single vial for combination treatment.
Assessment of mannose-siRNA and polymer micelle in mouse via subcutaneous administration
All animal-related procedures were conducted at Genevant Sciences Corporation, an accredited facility, according to written operating procedures, in accordance with Canadian Council on Animal Care (CCAC) Guidelines on Good Animal Practices and approved by the local Institutional Animal Care and Use Committee (IACUC). Rodent studies were performed under AUP #0618002.
C57BL/6 female mice aged 6–8 weeks (n = 4 per group) were injected intraperitoneally with 1 mL/animal of 4% thioglycolate (Sigma, St. Louis, MO) in distilled water to elicit macrophages in the peritoneum. Three days post thioglycolate stimulation, animals were injected subcutaneously in the scapular region with a single dose of vehicle control (saline), AF647-hexa-mannose-siCD45 (10 mg/kg), AF647-hexa-mannose-siCD45 (10 mg/kg) + mannose-ERP (50 mg/kg), or mannose-ERP alone (50 mg/kg), using a volume of 5 mL/kg body weight. The peritoneal cells of treated animals were collected at 24 h post siRNA and ERP treatment and analyzed with flow cytometry for siRNA delivery (AF647) and expression of CD206 and CD45.
For testing delivery of siRNAs to liver Kupffer cells. Balb/c female mice aged 6–8 weeks (n = 4 per group) were injected subcutaneously in the scapular region with a single dose of vehicle control (saline), AF647-hexa-mannose-siCD45 (3 mg/kg), or AF647-unconjugated siCD45. Single liver cells were collected using a protocol adapted from a previous publication.69 In brief, we used two-step in situ pronase/collagenase perfusion to dissociate the liver cells followed by in vitro digestion with pronase/collagenase solution. The collected single liver cells were centrifuged at 50 × g for 3 min to separate the hepatocytes and nonparenchymal NPCs. Hepatocytes were stained with anti-ASGPR antibodies (Thermo Fisher) followed by staining of PE -labeled secondary antibody (Thermo Fisher). NPCs were stained with PE-vio770-labeled anti-F4/80 antibody (Miltenyi). The siRNA delivery (AF647) and ASGPR and F4/80 expression were analyzed by flow cytometry.
Assessment of GalNAc-siRNA and mannose-siRNA in guinea pig MARV infection model
Guinea pigs (n = 5 per group) were injected intraperitoneally with 5,000 pfu/animal of guinea pig-adapted MARV (Angola variant).36 Twenty-four hours post viral exposure, animals were injected subcutaneously in the scapular region with tetra-GalNAc-siMARV or hexa-mannose-siMARV alone or in 1:1 combination (with the same total siRNA dose, mixed in the same vial for injection). Different dosing regimens, such as 10 mg/kg total siRNA daily subcutaneous dosing × 7 doses, 5 mg/kg total siRNA daily subcutaneous dosing × 7 doses, 5 mg/kg total siRNA weekly subcutaneous dose × 4 doses, or a single 10-mg/kg total siRNA dose were also tested (Figures S13 and S14). In one study (Figure S13), animals were also treated subcutaneously with GalNAc-ERP (5 mg/kg) together with mannose-ERP (5 mg/kg) (mixed in the same vial for injection) on the first day of conjugate dosing (dosing immediately after conjugate injection). The antiviral efficacy was evaluated by bodyweights, survival rates, and viremia. Guinea pigs were monitored daily and scored for disease progression with an internal MARV humane endpoint scoring sheet approved by the University of Texas Medical Branch (UTMB) IACUC. UTMB facilities used in this work are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adhere to principles specified in the eighth edition of the Guide for the Care and Use of Laboratory Animals (National Research Council).
Acknowledgments
The authors would like to thank the Genevant animal facility and UTMB Animal Resource Center for husbandry support of laboratory animals. The authors would like to thank Ms. Brittany Fransaw for her support with the guinea pig studies. This study was supported by the National Institutes of Health (NIH) grant U19AI142785. Operations support of the Galveston National Laboratory was supported by NIH grant UC7AI094660.
Author contributions
X.Y., R.H., and J.H. designed the experiments and wrote the manuscript. R.H. and M.W. designed and synthesized the ligands. E.S. conducted the ligand binding studies. L.P. conducted the conjugate delivery and in vitro activity studies. X.Y., C.P., and K.M. conducted the rodent conjugate delivery studies. R.W.C. and V.B. performed the guinea pig activity studies. J.H. and T.W.G. provided experimental and strategic guidance.
Declaration of interests
Some authors are employees or consultants of Genevant Sciences Corporation as noted in the author affiliations and own shares or stock options in their respective companies.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.09.009.
Supplemental information
References
- 1.Iannetta M., Di Caro A., Nicastri E., Vairo F., Masanja H., Kobinger G., Mirazimi A., Ntoumi F., Zumla A., Ippolito G. Viral hemorrhagic fevers other than ebola and lassa. Infect. Dis. Clin. North Am. 2019;33:977–1002. doi: 10.1016/j.idc.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Geisbert T.W., Jahrling P.B. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10:S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 3.Peterson A.T., Lash R.R., Carroll D.S., Johnson K.M. Geographic potential for outbreaks of Marburg hemorrhagic fever. Am. J. Trop. Med. Hyg. 2006;75:9–15. doi: 10.4269/ajtmh.2006.75.1.0750009. [DOI] [PubMed] [Google Scholar]
- 4.Abir M.H., Rahman T., Das A., Etu S.N., Nafiz I.H., Rakib A., Mitra S., Emran T.B., Dhama K., Islam A., et al. Pathogenicity and virulence of Marburg virus. Virulence. 2022;13:609–633. doi: 10.1080/21505594.2022.2054760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehedi M., Groseth A., Feldmann H., Ebihara H. Clinical aspects of Marburg hemorrhagic fever. Future Virol. 2011;6:1091–1106. doi: 10.2217/fvl.11.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker S., Spiess M., Klenk H.-D. The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J. Gen. Virol. 1995;76(Pt 2):393–399. doi: 10.1099/0022-1317-76-2-393. [DOI] [PubMed] [Google Scholar]
- 7.Bechtelsheimer H., Korb G., Gedigk P. 1971. pp. 62–67. (Marburg Virus Disease). [Google Scholar]
- 8.Geisbert T.W., Daddario-DiCaprio K.M., Geisbert J.B., Young H.A., Formenty P., Fritz E.A., Larsen T., Hensley L.E. Marburg virus Angola infection of rhesus macaques: pathogenesis and treatment with recombinant nematode anticoagulant protein c2. J. Infect. Dis. 2007;196:S372–S381. doi: 10.1086/520608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olejnik J., Ryabchikova E., Corley R.B., Mühlberger E. Intracellular events and cell fate in filovirus infection. Viruses. 2011;3:1501–1531. doi: 10.3390/v3081501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towner J.S., Khristova M.L., Sealy T.K., Vincent M.J., Erickson B.R., Bawiec D.A., Hartman A.L., Comer J.A., Zaki S.R., Ströher U., et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol. 2006;80:6497–6516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.-J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thi E.P., Mire C.E., Lee A.C., Geisbert J.B., Ursic-Bedoya R., Agans K.N., Robbins M., Deer D.J., Cross R.W., Kondratowicz A.S., et al. siRNA rescues nonhuman primates from advanced Marburg and Ravn virus disease. J. Clin. Invest. 2017;127:4437–4448. doi: 10.1172/JCI96185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ursic-Bedoya R., Mire C.E., Robbins M., Geisbert J.B., Judge A., MacLachlan I., Geisbert T.W. Protection against lethal Marburg virus infection mediated by lipid encapsulated small interfering RNA. J. Infect. Dis. 2014;209:562–570. doi: 10.1093/infdis/jit465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thi E.P., Mire C.E., Lee A.C.H., Geisbert J.B., Zhou J.Z., Agans K.N., Snead N.M., Deer D.J., Barnard T.R., Fenton K.A., et al. Lipid nanoparticle siRNA treatment of Ebola virus Makona infected nonhuman primates. Nature. 2015;521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim S.A., Cox A., Tung M., Chung E.J. Clinical progress of nanomedicine-based RNA therapies. Bioact. Mater. 2022;12:203–213. doi: 10.1016/j.bioactmat.2021.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo S., Li K., Hu B., Li C., Zhang M., Hussain A., Wang X., Cheng Q., Yang F., Ge K., et al. Membrane-destabilizing ionizable lipid empowered imaging-guided siRNA delivery and cancer treatment. Exploration (Beijing). 2021;1:35–49. doi: 10.1002/EXP.20210008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonezawa S., Koide H., Asai T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020;154-155:64–78. doi: 10.1016/j.addr.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Sullivan J., Muñoz-Muñoz J., Turnbull G., Sim N., Penny S., Moschos S. Beyond GalNAc! Drug delivery systems comprising complex oligosaccharides for targeted use of nucleic acid therapeutics. RSC Adv. 2022;12:20432–20446. doi: 10.1039/d2ra01999j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debacker A.J., Voutila J., Catley M., Blakey D., Habib N. Delivery of Oligonucleotides to the liver with GalNAc – from research to registered therapeutic drug. Mol. Ther. 2020;28:1759–1771. doi: 10.1016/j.ymthe.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldmann H., Volchkov V.E., Volchkova V.A., Klenk H.D. The glycoproteins of Marburg and Ebola virus and their potential roles in pathogenesis. Arch. Virol. Suppl. 1999;15:159–169. doi: 10.1007/978-3-7091-6425-9_11. [DOI] [PubMed] [Google Scholar]
- 21.Alves D.A., Glynn A.R., Steele K.E., Lackemeyer M.G., Garza N.L., Buck J.G., Mech C., Reed D.S. Aerosol exposure to the Angola strain of Marburg virus causes lethal viral hemorrhagic fever in cynomolgus macaques. Vet. Pathol. 2010;47:831–851. doi: 10.1177/0300985810378597. [DOI] [PubMed] [Google Scholar]
- 22.Azad A.K., Rajaram M.V.S., Schlesinger L.S. Exploitation of the macrophage mannose receptor (CD206) in infectious disease diagnostics and therapeutics. J. Cytol. Mol. Biol. 2014;1:1000003. doi: 10.13188/2325-4653.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg H., Jégouzo S.A.F., Lasanajak Y., Smith D.F., Drickamer K., Weis W.I., Taylor M.E. Structural analysis of carbohydrate binding by the macrophage mannose receptor CD206. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boldescu V., Crudu V., Sucman N., Pogrebnoi S., Zviaghinţeva M., Stîngaci E., Pogrebnoi V., Macaev F. Molecular concepts of macrophage targeting. ChemJMold. 2013;8:21–31. [Google Scholar]
- 25.Westerlind U., Westman J., Törnquist E., Smith C.I.E., Oscarson S., Lahmann M., Norberg T. Ligands of the asialoglycoprotein receptor for targeted gene delivery, part 1: synthesis of and binding studies with biotinylated cluster glycosides containing N-acetylgalactosamine. Glycoconj. J. 2004;21:227–241. doi: 10.1023/B:GLYC.0000045095.86867.c0. [DOI] [PubMed] [Google Scholar]
- 26.Lee R.T., Lin P., Lee Y.C. New synthetic cluster ligands for galactose/N-acetylgalactosamine-specific lectin of mammalian liver. Biochemistry. 1984;23:4255–4261. doi: 10.1021/bi00313a037. [DOI] [PubMed] [Google Scholar]
- 27.Connolly D.T., Townsend R.R., Kawaguchi K., Bell W.R., Lee Y.C. Binding and endocytosis of cluster glycosides by rabbit hepatocytes. Evidence for a short-circuit pathway that does not lead to degradation. J. Biol. Chem. 1982;257:939–945. [PubMed] [Google Scholar]
- 28.Holland R.J., Lam K., Ye X., Martin A.D., Wood M.C., Palmer L., Fraser D., McClintock K., Majeski S., Jarosz A., et al. Ligand conjugate SAR and enhanced delivery in NHP. Mol. Ther. 2021;29:2910–2919. doi: 10.1016/j.ymthe.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wollenberg A., Mommaas M., Oppel T., Schottdorf E.M., Günther S., Moderer M. Expression and function of the mannose receptor CD206 on epidermal dendritic cells in inflammatory skin diseases. J. Invest. Dermatol. 2002;118:327–334. doi: 10.1046/j.0022-202x.2001.01665.x. [DOI] [PubMed] [Google Scholar]
- 30.Cochand L., Isler P., Songeon F., Nicod L.P. Human lung dendritic cells have an immature phenotype with efficient mannose receptors. Am. J. Respir. Cell Mol. Biol. 1999;21:547–554. doi: 10.1165/ajrcmb.21.5.3785. [DOI] [PubMed] [Google Scholar]
- 31.Sallusto F., Cella M., Danieli C., Lanzavecchia A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J. Exp. Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellman I., Turley S.J., Steinman R.M. Antigen processing for amateurs and professionals. Trends Cell Biol. 1998;8:231–237. doi: 10.1016/s0962-8924(98)01276-8. [DOI] [PubMed] [Google Scholar]
- 33.Gonçalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2015;111:14.1.1–14.1.16. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008;83:14–14.1.14. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elchaninov A.V., Fatkhudinov T.K., Vishnyakova P.A., Lokhonina A.V., Sukhikh G.T. Phenotypical and functional polymorphism of liver resident macrophages. Cells. 2019;8:1032. doi: 10.3390/cells8091032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross R.W., Fenton K.A., Geisbert J.B., Ebihara H., Mire C.E., Geisbert T.W. Comparison of the pathogenesis of the Angola and ravn strains of Marburg virus in the outbred Guinea pig model. J. Infect. Dis. 2015;212:S258–S270. doi: 10.1093/infdis/jiv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M., et al. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol. Ther. 2018;26:708–717. doi: 10.1016/j.ymthe.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki Y., Shirai M., Asada K., Yasui H., Karayama M., Hozumi H., Furuhashi K., Enomoto N., Fujisawa T., Nakamura Y., et al. Macrophage mannose receptor, CD206, predict prognosis in patients with pulmonary tuberculosis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-31565-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geijtenbeek T.B.H., Gringhuis S.I. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Pomares L. The mannose receptor. J. Leukoc. Biol. 2012;92:1177–1186. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 41.Qiao N., Du G., Zhong X., Sun X. Recombinant lactic acid bacteria as promising vectors for mucosal vaccination. Exploration (Beijing). 2021;1 doi: 10.1002/EXP.20210026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C., Jeong H., Bae Y., Shin K., Kang S., Kim H., Oh J., Bae H. Targeting of M2-like tumor-associated macrophages with a melittin-based pro-apoptotic peptide. J. Immunother. Cancer. 2019;7:147. doi: 10.1186/s40425-019-0610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan X., Jia L., Niu Y., Qi H., Chen X., Zhang Q., Zhang J., Wang Y., Dong L., Wang C. Targeted depletion of tumour-associated macrophages by an alendronate–glucomannan conjugate for cancer immunotherapy. Biomaterials. 2014;35:10046–10057. doi: 10.1016/j.biomaterials.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Wu H., Ji B., Qian W., Xia S., Wang L., Xu Y., Chen J., Yang L., Mao H. Targeted imaging of CD206 expressing tumor-associated M2-like macrophages using mannose-conjugated antibiofouling magnetic iron oxide nanoparticles. ACS Appl. Bio Mater. 2020;3:4335–4347. doi: 10.1021/acsabm.0c00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diebold S.S., Plank C., Cotten M., Wagner E., Zenke M. Mannose receptor-mediated gene delivery into antigen presenting dendritic cells. Somat. Cell Mol. Genet. 2002;27:65–74. doi: 10.1023/a:1022975705406. [DOI] [PubMed] [Google Scholar]
- 46.Glass E.B., Masjedi S., Dudzinski S.O., Wilson A.J., Duvall C.L., Yull F.E., Giorgio T.D. Optimizing mannose “click” conjugation to polymeric nanoparticles for targeted siRNA delivery to human and murine macrophages. Acs Omega. 2019;4:16756–16767. doi: 10.1021/acsomega.9b01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaps L., Leber N., Klefenz A., Choteschovsky N., Zentel R., Nuhn L., Schuppan D. In vivo siRNA delivery to immunosuppressive liver macrophages by α-mannosyl-functionalized cationic nanohydrogel particles. Cells. 2020;9:1905. doi: 10.3390/cells9081905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalle Vedove E., Costabile G., Merkel O.M. Mannose and mannose-6-phosphate receptor–targeted drug delivery systems and their application in cancer therapy. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201701398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irache J.M., Salman H.H., Gamazo C., Espuelas S. Mannose-targeted systems for the delivery of therapeutics. Expert Opin. Drug Deliv. 2008;5:703–724. doi: 10.1517/17425247.5.6.703. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y., Zhang Z., Pan Z., Liu Y. Advanced bioactive nanomaterials for biomedical applications. Exploration (Beijing). 2021;1 doi: 10.1002/EXP.20210089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drickamer K. Multiplicity of lectin-carbohydrate interactions. Nat. Struct. Biol. 1995;2:437–439. doi: 10.1038/nsb0695-437. [DOI] [PubMed] [Google Scholar]
- 52.Kelly A., Grabiec A.M., Travis M.A. Macrophages Methods and Protocols. Vol. 1784. Humana New York; 2018. Culture of human monocyte-derived macrophages; pp. 1–11. [DOI] [PubMed] [Google Scholar]
- 53.Raggi F., Pelassa S., Pierobon D., Penco F., Gattorno M., Novelli F., Eva A., Varesio L., Giovarelli M., Bosco M.C. Regulation of human macrophage M1–M2 polarization balance by hypoxia and the triggering receptor expressed on myeloid cells-1. Front. Immunol. 2017;8:1097. doi: 10.3389/fimmu.2017.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertani F.R., Mozetic P., Fioramonti M., Iuliani M., Ribelli G., Pantano F., Santini D., Tonini G., Trombetta M., Businaro L., et al. Classification of M1/M2-polarized human macrophages by label-free hyperspectral reflectance confocal microscopy and multivariate analysis. Sci. Rep. 2017;7:8965. doi: 10.1038/s41598-017-08121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lescoat A., Ballerie A., Jouneau S., Fardel O., Vernhet L., Jego P., Lecureur V. M1/M2 polarisation state of M-CSF blood-derived macrophages in systemic sclerosis. Ann. Rheum. Dis. 2019;78:e127. doi: 10.1136/annrheumdis-2018-214333. [DOI] [PubMed] [Google Scholar]
- 56.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. Macrophage activation and polarization: nomenclature and experimental Guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian C., Yun Z., Yao Y., Cao M., Liu Q., Hu S., Zhang S., Luo D. Heterogeneous macrophages: supersensors of exogenous inducing factors. Scand. J. Immunol. 2019;90 doi: 10.1111/sji.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown C.R., Gupta S., Qin J., Racie T., He G., Lentini S., Malone R., Yu M., Matsuda S., Shulga-Morskaya S., et al. Investigating the pharmacodynamic durability of GalNAc–siRNA conjugates. Nucleic Acids Res. 2020;48:11827–11844. doi: 10.1093/nar/gkaa670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 61.Ströher U., West E., Bugany H., Klenk H.-D., Schnittler H.-J., Feldmann H. Infection and activation of monocytes by Marburg and ebola viruses. J. Virol. 2001;75:11025–11033. doi: 10.1128/JVI.75.22.11025-11033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cross R.W., Bornholdt Z.A., Prasad A.N., Borisevich V., Agans K.N., Deer D.J., Abelson D.M., Kim D.H., Shestowsky W.S., Campbell L.A., et al. Combination therapy protects macaques against advanced Marburg virus disease. Nat. Commun. 2021;12:1891. doi: 10.1038/s41467-021-22132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szekanecz Z., Koch A.E. Macrophages and their products in rheumatoid arthritis. Curr. Opin. Rheumatol. 2007;19:289–295. doi: 10.1097/BOR.0b013e32805e87ae. [DOI] [PubMed] [Google Scholar]
- 64.Maruotti N., Annese T., Cantatore F.P., Ribatti D. Macrophages and angiogenesis in rheumatic diseases. Vasc. Cell. 2013;5:11. doi: 10.1186/2045-824X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cuda C.M., Pope R.M., Perlman H. The inflammatory role of phagocyte apoptotic pathways in rheumatic diseases. Nat. Rev. Rheumatol. 2016;12:543–558. doi: 10.1038/nrrheum.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Heide D., Weiskirchen R., Bansal R. Therapeutic targeting of hepatic macrophages for the treatment of liver diseases. Front. Immunol. 2019;10:2852. doi: 10.3389/fimmu.2019.02852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poh A.R., Ernst M. Targeting macrophages in cancer: from bench to bedside. Front. Oncol. 2018;8:49. doi: 10.3389/fonc.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prieve M.G., Harvie P., Monahan S.D., Roy D., Li A.G., Blevins T.L., Paschal A.E., Waldheim M., Bell E.C., Galperin A., et al. Targeted mRNA therapy for ornithine transcarbamylase deficiency. Mol. Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aoyama T., Kuwahara-Arai K., Uchiyama A., Kon K., Okubo H., Yamashina S., Ikejima K., Kokubu S., Miyazaki A., Watanabe S. Spleen-derived lipocalin-2 in the portal vein regulates Kupffer cells activation and attenuates the development of liver fibrosis in mice. Lab. Invest. 2017;97:890–902. doi: 10.1038/labinvest.2017.44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.