Summary
The conquest of the Canary Islands by Europeans began at the beginning of the 15th century and culminated in 1496 with the surrender of the aborigines. The collapse of the aboriginal population during the conquest and the arrival of settlers caused a drastic change in the demographic composition of the archipelago. To shed light on this historical process, we analyzed 896 mitogenomes of current inhabitants from the seven main islands. Our findings confirm the continuity of aboriginal maternal contributions and the persistence of their genetic footprints in the current population, even at higher levels (>60% on average) than previously evidenced. Moreover, the age estimates for most autochthonous founder lineages support a first aboriginal arrival to the islands at the beginning of the first millennium. We also revealed for the first time that the main recognizable genetic influences from Europe are from Portuguese and Galicians.
Subject areas: Molecular biology, Genomics, Anthropology, Human Geography
Graphical abstract
Highlights
-
•
The most complete maternal genetic characterization of the extant Canarian population
-
•
The aboriginal arrival to the islands is estimated at the beginning of the millennium
-
•
The Portuguese and Galicians are the most prominent influence in the Canary Islanders
-
•
Maternal genetic connectivity was detected between the Americas and the Canaries
Molecular biology; Genomics; Anthropology; Human Geography
Introduction
Promoted by technological advances in navigation and aggressive maritime trade, the southern European kingdoms of the Iberian Peninsula and Italy rediscovered the Canary Islands for the European world at the beginning of the 14th century.1 This archipelago of seven main islands, located near the Atlantic shore of northern Africa, was already inhabited.2,3 Because they were the most exposed to pro-slavery forays, the islands of Lanzarote, Fuerteventura, and El Hierro were frequently visited by explorers.4,5 Along the 15th century, the seven islands were gradually conquered and colonized by the Europeans under the auspices of the Castile Crown of Spain. The last step in this one-century long and difficult conquest was the surrender of Tenerife, the largest of the islands, in 1496.6
Archaeological, anthropological, and linguistic studies have suggested that the North African Berbers were the most probable ancestors of the aboriginal Canarians.7,8,9,10,11,12,13,14,15,16,17,18 Regrettably, soon after the conquest, most of the aboriginal cultural expressions disappeared and the aborigines have not lived to our days as a distinct population. However, genetic studies have confirmed the close affinities between the Canarian aborigines and the North African Berbers, demonstrating that a substantial part of the aboriginal population was assimilated by the European colonizers albeit with an important sex asymmetry.19,20 In support of this, the mtDNA U6b1a and U6c1, and the Y chromosome E-M81 and E-M78 lineages with strong northern African affiliation have been found in aboriginal remnants20,21,22,23 and in the current inhabitants of the Canary Islands.19,24 Besides, it has been estimated that the contribution of aboriginal maternal lineages to the current Canarian population could be 40% on average23,24,25 with a striking maximum of up to 70% in the population of La Gomera, one of the smallest islands.26 On the contrary, the contribution of aboriginal paternal lineages has been estimated to be less than 10%.19,20 This sexual asymmetry has been explained because of military strategy employed in the conquest of several islands.19,20 Congruently, the pioneer studies based on autosomal markers27 and more recently using genome-wide autosomal data28,29,30,31 support that the footprints of the aboriginal genetic ancestry are still present in the current inhabitants at variable proportions (up to 34%).
The analyses that have been conducted so far on the current inhabitants were based, at best, on mtDNA sequences of the 403-base pair hypervariable region 1 (HVR1) along with a few coding sites in nearly a thousand unrelated donors in total,24,26,32 and on less than 30 hierarchically selected Y chromosome single-nucleotide polymorphisms.19 These low-resolution analyses introduce uncertainty and ascertainment bias, hindering the comparisons. The advent of next-generation sequencing (NGS) technologies provides an opportunity to circumvent these limitations and to expand the knowledge of the recent evolutionary history of Canary Islanders. However, a paucity of studies have incorporated the NGS to anthropological studies of this population, with the analysis of the genome30 and the mitogenome23 of ancient DNA (aDNA) samples obtained from aboriginal remains from some of the islands, and of targeted autosomal regions in the DNA of current inhabitants.31
From a genetics perspective, one aspect lacking sufficient attention is the resulting genetic admixture in the Canary Islands population after the conquest and a fine-grained analysis of their impact on the genetic background of current inhabitants. The main reason is that the European colonization of the Canary Islands after the conquest was a complex process and involved Spanish, Portuguese, Italians, and Flemish colonizers, but also sub-Saharan Africans and Moorish slaves.33 Moreover, the settlement was also different in each of the islands, depending mainly on the availability of resources suitable for sugarcane production (Gran Canaria, Tenerife, La Palma, and La Gomera) or not (El Hierro, Lanzarote, and Fuerteventura).34
Here, we obtained and analyzed the complete mitogenomes sequenced by NGS technology of almost the same sample size as the total of previous studies considered together24,26,32 of putative Canarian descendants from the seven main islands (n = 896). This allowed us, for the first time, to more precisely apportion the post-conquest genetic inputs from the main different European populations influencing the islands. In addition, by assigning previously undetermined aboriginal lineages to specific haplogroups, we were able to notably increase the estimate of founder lineages surviving in the maternal gene pool of the current-day population and assessed their coalescence age to provide potential scenarios for the settlements of the archipelago.
Results
Sequencing summary and classification of mitogenomes
The mean (±SD) number of mtDNA reads recovered per sample was 17,853 ± 14,789. The average (±SD) mtDNA depth of coverage was 47X ± 33, and an average of 89.78% ± 21.49% of the mitogenome was covered at ≥10X. As for the coverage depth of detected variants after filtering, the mean (±SD) per variant call was 52X ± 40. The haplogroup classification quality provided by Haplocheck had a mean value of 0.98 (range: 0.81–1.00) across samples. The dataset was composed of twelve macro-haplogroups (A: Figure S1, H: Figure S2 and Figure S3, I: Figure S4, J: Figure S5, K: Figure S6, L: Figure S7, M: Figure S8, T: Figure S9, U: Figure S10 and Figure S11, V: Figure S12, W: Figure S13, and X: Figure S14), reflecting the level of admixture found in the Canary Islands population. With a frequency above 10%, we found H, J, T, and U, the latter being the most predominant in the population (29.35%). U6b1a was the most frequent haplogroup (14.06%), followed by J2a2d1a (7.14%) (Table S3).
Sources and continuity of the aboriginal mtDNA gene pool in the current population
The significant increase in the number of complete sequences sampled in the current-day Canary Island population has allowed us to assign some ambiguous aboriginal HVR1 sequences described in the literature to specific mtDNA haplogroups. In this sense, the unusual variant C16290T detected in aboriginal remains from La Palma22 has been found in the mitogenome of an individual sampled in this study from the same island. This sequence was assigned to a sub-group of haplogroup H characterized by the 3531-transition, closely related to a Denmark sample (Figure S2). The HVR1 motif 16069-16126-16278-16366 found in aborigines from Tenerife and assigned to haplogroup J∗21 had a perfect match with a mitogenome of the current population from the same island, and was classified into the J1c2e2 haplogroup (Figure S5). The African L3d haplotype 16124-16223-16256-16311 found in pre-Hispanic remains from La Gomera26 has been now identified in current inhabitants of the island and classified as belonging to the L3d1b3a haplogroup (Figure S7). Finally, the rare HVR1 combination 16093–16192 found in aboriginal remains from El Hierro and classified by diagnostic variant analyses (12308 HinfI+) as U∗21 has been found here only in a mitogenome from the same island, as part of a U5a1b4 insular cluster with 9.36% frequency and having an exact match with a Danish sequence (Figure S10).
The large sample size of this study also raised the number of matches between the sequences available from the aboriginal population and from the current population of the Canary Islands (Table S1). The mean number of matches (±SE) between those was 60.43% (±2.01), with a maximum of 67.67% for El Hierro and a minimum of 50% for Lanzarote. We took this as sufficient evidence of the large continuity of aboriginal lineages in the current population. Although still supporting a large continuity of lineages, the mean value of matches was lower (51.50% ± 5.61) when the frequency of lineages is taken into account (Table S1). In addition, the populations of La Palma, Tenerife, and Gran Canaria retained a significantly lower mean aboriginal maternal substrate than the populations from El Hierro, La Gomera, Fuerteventura, and Lanzarote (37.53% vs. 61.90%, respectively; Unpaired t-test, p = 0.0099) (Table S1), which may be explained by differences in the aboriginal assimilation taking place along the European colonization. In agreement with this, samples from cemetery remains dated between the 15th–17th and 18th centuries in Gran Canaria35 and Tenerife,36 respectively, point to an early substitution of the aboriginal population. If the spatial distribution of aboriginal haplogroups over time (aboriginal remains vs. current population) is assessed, an increase in the distribution range of certain aboriginal lineages along the west–east axis associated with the post-conquest era was observed. For example, the haplogroups H1e1a, H1a0, and H4a1 were only detected in aboriginal remains from the eastern islands, while they were currently present in the western islands as well. On the contrary, H1cf and J2a2d1a were predominant in the western islands only, while they are also present in the current population at the eastern islands (Table S1).
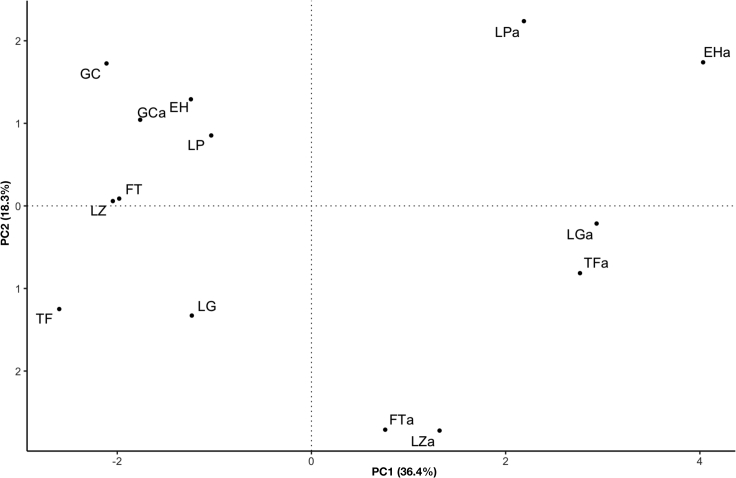
Analysis of match-based distances focusing on the aboriginal lineages allowed us to deduce relationships between the aboriginal and current-day insular populations (Table S1). This measure indicates that the aboriginal insular populations were more differentiated than the extant populations (ANOVA, p = 4.7 × 10−9). This could be explained due to the greater inter-insular migration after the conquest. The best example is the case of El Hierro, a small island with impoverished population at the time of the European contacts and with only three different aboriginal mtDNA lineages that today harbors 16 autochthonous lineages, the majority of them with genealogical links in the other islands (Table S1). In spite of the differentiation, the principal component analysis plot reflected the relationships between the aboriginal samples from the geographically close islands of El Hierro and La Palma, La Gomera and Tenerife, and between Fuerteventura and Lanzarote (Figure 1). These genetic relationships among the island populations had a resemblance of what is found in the current population. Another important peculiarity revealed by this analysis is the relationship of the aboriginal and current samples from Gran Canaria, once again supporting a continuity of mtDNA lineages over time. As for the geographic origin of the aboriginal lineages, as expected, the bulk of the sequence matches were with the populations from the Atlantic border of the Maghreb (Moroccans, Saharawi, and Mauritanians; 45.45%) or from the central area of the Maghreb (Algerians and Tunisians; 36.36%) (Table S1). Note, however, that the overlap of mtDNA lineages between both regions was considerable given that 46% of the pairwise matches with the aboriginal lineages of Canary Islanders were detected in both regions. In addition, there were exclusive matches with Europe (8.08%), involving populations mainly from the Iberian Peninsula, France, and Italy (Table S1). Considering that there are limitations in the comparisons because there is a bias toward the existence of mitogenome data from Western Europe and North American populations, the most relevant matches were those found within the H1e1a haplogroup, considered an autochthonous founder lineage,23 and others within H1a0 and H1bw, which also belong to the Eurasian macro-haplogroup H and which are absent from the northern African samples. Only 6.06% of the aboriginal lineages match exclusively with lineages that are most commonly found in sub-Saharan African populations (Table S1). Thus, the aboriginal haplotypes (16125-16213-16223 and 16187-16223-16327) previously found in Gran Canaria and assigned to the haplogroups L2 and L3e1, respectively, have exclusive matches in the Chad. On the other side, the aboriginal L2d2 and L2e2 haplotypes detected in Tenerife have their closest relatives in Cape Verde,37 and São Tomé and Principe,38 two western African archipelagos linked to the Portuguese slave trade since the 15th century. An additional aboriginal HVR1 haplotype from Tenerife (16081-16093-16175-16223-16278-16320), classified as belonging to the L3e2 haplogroup21 but that we would classify instead as L2c here according to the HVR2 region (73-93-146-150-182-195-198-263-325), has also been exclusively found in the São Tomé and Principe archipelago.38
Figure 1.
Genetic relationships between pre-Hispanic and current-day populations
PCA plot based on distance matches between pre-Hispanic (EHa = El Hierro; LPa = La Palma; LGa = La Gomera; TFa = Tenerife; GCa = Gran Canaria; FTa = Fuerteventura; LZa = Lanzarote) and current-day populations (EH = El Hierro; LP = La Palma; LG = La Gomera; TF = Tenerife; GC = Gran Canaria; FT = Fuerteventura; LZ = Lanzarote).
The lineages that are more typically assigned to sub-Saharan African populations represented 4.23% ± 1.61% of the sequences in the aboriginal population of the archipelago,23 with only 1.06% ± 0.37% surviving in the extant Canary Islands population. However, this difference was not significant (Unpaired t-test, p = 0.079). Nevertheless, for the total set of these lineages of putative sub-Saharan African assignation, the frequency in the current population (7.23% ± 1.10%) was slightly higher (Unpaired t-test, p = 0.0378) than that found in the aboriginal sequences available (4.23% ± 1.61%). In addition, we detected that the aboriginal lineages of putative sub-Saharan African assignation were significantly more abundant (Unpaired t-test, p = 0.0091) in the island populations that were involved in the sugarcane production (Gran Canaria, La Gomera, La Palma, and Tenerife) compared to those where sugar mills were not established (Lanzarote, Fuerteventura, and El Hierro). For these lineages, this difference among groups of islands is still significant in the current populations (Unpaired t-tests, p = 0.013). Note that the available evidence from samples from historical cemetery remains in Tenerife36 and Gran Canaria35 from the 15th to 17th and the 18th centuries—where sub-Saharan African mtDNA lineages accounted for 16.39%–28.57% of the retrieved sequences although they are likely not representative of the general population of the islands at that time—supports that the sub-Saharan African influences in the archipelago peaked during the boom of the Atlantic slave trade.
Finally, there were a few aboriginal lineages that only had exact matches in the Near Eastern populations. The case of the autochthonous founder lineage U6b1a (Table S1), which has been detected in one individual in Lebanon but not in North Africa, has been described.23 In addition, the subsequent complete sequencing of this Lebanese sample39 revealed that it had the variant 15697 that defines an exclusive Canarian subclade (Figure S11). Other rare coincidence is the HVR1 motif 16189–16316 that was identified in prehistoric remains from Tenerife and classified as belonging to haplogroups HV/R21 that exactly matches HV sequences from Lebanon and Iraq (Table S1). Even more surprising is the case of the N1b haplogroup control sequence 16145-16176G-16223-16297-16311-16390, documented in prehistoric remains from La Gomera,21 for which an exact match, including the infrequent transition 16297, was detected in an Armenian sequence (Table S1). In a recent study on aboriginal Canarian remains, an mtDNA genome extracted from a Tenerife mummy30 was classified as J1c3 with a retro mutation at 16126. Its closest sequence within this group has been found in Denmark (Figure S5).
Apportioning other genetic influences in the Canary Islanders
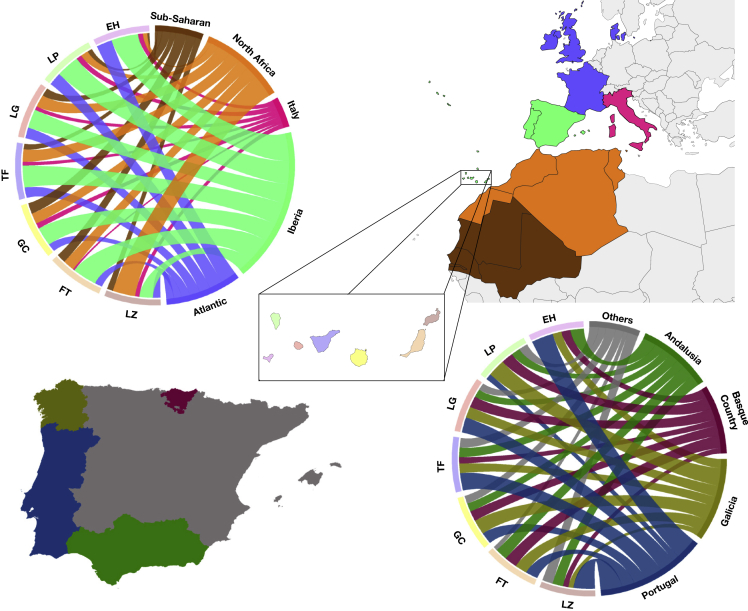
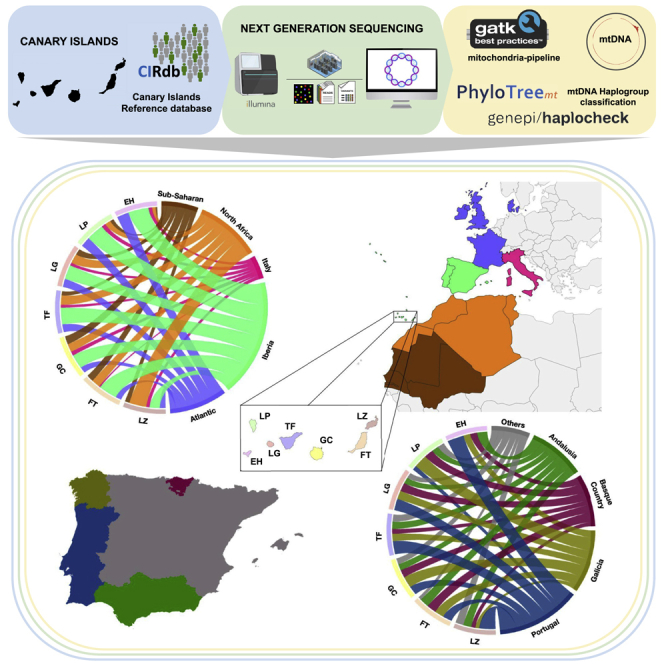
We have evidenced the continuity of a sizable proportion (around 50%) of the aboriginal maternal gene pool in the current population of the Canary Islands. In this section, we focus on estimating the maternal genetic contribution of all other population sources based on match affinities (Table S2). As expected, the main contribution (±SD) to the Canary Islands has been from the Iberian Peninsula (39.14% ± 8.12%), followed by populations from the North Africa (21.57% ± 10.53%), the Atlantic (19.29% ± 5.65%), the sub-Saharan Africans (12.29% ± 5.41%), and the Italians (7.57% ± 2.37%) (Table S4 and Figure 2). Pairwise matches for the Atlantic were mainly with Danish. We interpreted this result as Norman and Flemish origin due to the fact that Denmark is much better represented in the GenBank mtDNA sequences than France or Belgium.40,41 Regarding the pairwise matches of Canary Islanders with Africa, it should be noted that 31.79% of them did not originate directly from Africa but from America. However, the significant sampling bias that could be associated with the availability of mitogenome data in African vs. African American populations must be taken into account, which might limit the interpretations of these findings. Consistent with the relationship with the American continent, an individual from Gran Canaria belonging to an Amerindian lineage (A2) (Figure S1), which together with the individual from Fuerteventura also classified within this lineage23 add up to two individuals related to this Amerindian haplogroup found in the current population of the Canary Islands. Another interesting case already pointed out23 is related to the J2a2d1a1 lineage, in our study reclassified to J2a2d1a, composed of two individuals from the Canary Islands and Brazil, thus linking the archipelago with South America. Given the larger number of available sequences of individuals from the current population, we considerably increased the number of private Canarian sequences within this lineage, suggesting that it might be an autochthonous founder lineage. Also, as expected from past demographic studies,42 Portuguese (28.43% ± 10.71%) and Galicians (21.43% ± 7.55%) together represented about half of the pairwise matches with the Iberian populations, reaching as much as 58% in El Hierro (Table S4). The pairwise matches of the North Africa populations were highest with Lanzarote (42%). Moreover, the pairwise matches of the Atlantic populations were higher in the westernmost islands of El Hierro (29%) and La Palma (24%). This result is in agreement with the records suggesting that the Norman settlement in El Hierro43 and the Flemish in La Palma44 are particularly important. Finally, the prevalence of pairwise matching of putative sub-Saharan African sequences was highest in Gran Canaria (20%). Taken together, based on mtDNA pairwise matches, we conclude that the western islands of Tenerife, La Gomera, La Palma, and El Hierro are more related to Iberian populations and the Atlantic area (especially evident for La Palma and El Hierro). On the other hand, the eastern islands of Gran Canaria, Fuerteventura, and Lanzarote showed comparatively more mtDNA affinities with African populations.
Figure 2.
Maternal genetic contributions of continental populations
Circos plot based on the pairwise matching affinities between individuals from the Canary Islands and African and European populations (EH = El Hierro; LP = La Palma; LG = La Gomera; TF = Tenerife; GC = Gran Canaria; FT = Fuerteventura; LZ = Lanzarote).
Coalescence ages of the aboriginal lineages
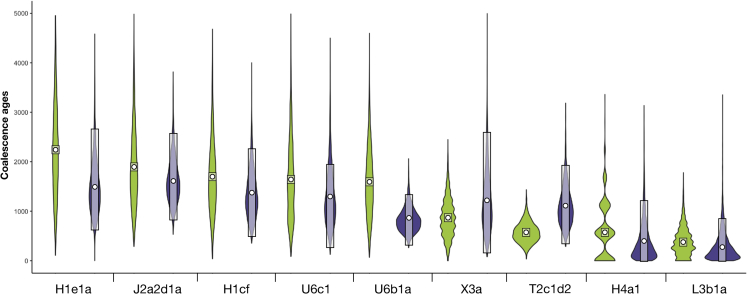
The archaeological date of the aboriginal settlement of the Canary Islands, well BCE, has been used as a calibration point to test the validity of the mtDNA substitution rate.45 However, it is difficult to sustain the idea of a permanent settlement on the islands beyond the change of era.46 Congruently, the majority of coalescent age estimates of the autochthonous founder lineages based on the mtDNA substitution rate proposed by Soares et al.45 were older when compared to archaeological dates (Table S5). On the contrary, using the mtDNA substitution rate proposed by Cabrera,47 either with the rho statistic or Beast approaches, the coalescence ages of founder lineages agree all with the same archaeological time window (Table S5), ranging between 2,268 (95% CI: 2,235–2,300) years ago (ya) for H1e1a and 380 (95% CI: 370–389) ya for L3b1a for the rho statistic, and between 1,628 (95% CI: 841–2,556) ya and 297 (95% CI: 0–906) ya for the J2a2d1a and L3b1a lineages, respectively, for Beast. In spite of the estimates derived with rho tend to be larger than those of Beast (except for T2c1d2 and X3a), the values calculated by the two approaches were highly correlated (Pearson r = 0.76; p = 0.017) and showed large overlaps in the confidence intervals (Figure 3). Thus, the relative chronological positions of lineages as estimated by both approaches had a reasonable equivalence, with the exception of those from U6b1a, T2c1d2, and X3a (Table S5). Regarding the oldest and youngest lineages (based on point estimates), except for H1e1a and J2a2d1a, both approaches roughly supported similar results. Considering as the reference the pointwise coalescence age estimates, a continuous age range was observed given the overlapping of the confidence intervals.
Figure 3.
Coalescent age estimates of the autochthonous founder lineages
Violin plots for Bayesian inference estimates using the rho statistic (green) and the Beast (purple) for each putative autochthonous Canarian mtDNA lineage. Confidence interval at 95% is shown for both approaches (the Beast highest posterior density interval, HPD, as calculated by Tracer is shown).
Demographic reconstruction in the putative autochthonous lineages
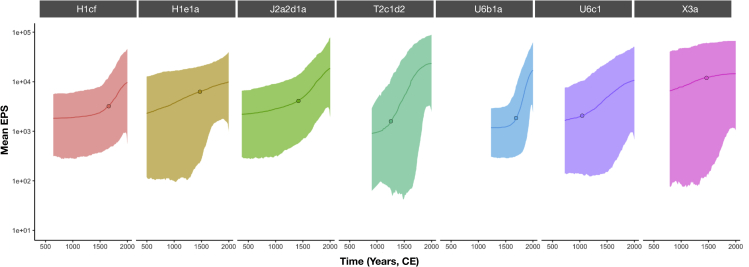
The assessment of Ne estimations through time let us identify trend changes across different lineages. Except for the young lineages L3b1a and H4a1, for which evident demographic changes over the period were not observed, all other putative autochthonous lineages exhibited a marked increase in effective population size (Ne) over time (Figure 4). Besides, H1e1a and X3a showed a smooth demographic increase over time, in contrast to the rest of the lineages that show a strong growth in Ne. H1cf and U6b1a exhibited a recent change in growth rate, dated between 200 and 300 ya. J2a2d1a, H1e1a, and X3a showed a slope change in their respective demographic trends about 500 ya. Finally, U6c1 and T2c1d2 revealed a slope change in their estimated population trend around 1,000 and 670 ya, respectively. Thus, all but two lineages showed changes in demographic trends in the 15th century CE or later.
Figure 4.
Demographic reconstruction of the autochthonous founder lineages
Bayesian skyline plots of the effective population size for each putative autochthonous Canarian mtDNA lineage (considering data from historical remains and current-day population). Average of estimates are shown as solid lines and 95% HPD intervals as shaded areas. The cutoff point of each demographic reconstruction was calculated with the KneeArrower R package.
Discussion
Previous studies have addressed the recent population history of the Canary Islands from a genetic perspective. However, most of them have focused on unraveling the relationships of the native Canarian aborigines with the current African populations and most lacked a fine-grained assessment of the European genetic contributions that have been occurring after the Spanish conquest of the archipelago. With an unprecedented number of complete mitogenomes sequenced for the extant Canarian population, this study provides the largest and most comprehensive maternal genetic characterization of the current Canary Islands populations. By comparing these with data from extant African and European populations, we inferred that the Portuguese are the most prominent maternal influence in the archipelago, except for Lanzarote, where the North African resemblance was the most evident. Given that the genetic diversity of the North African populations remains largely unexplored at the necessary resolution for the analyses, this has hindered to delve into a fine-grained level of analysis of the Berber vs. non-Berber influences in the Canaries. Besides, our results have shown that the gene pool of the Canary autochthonous settlers (of Berber origin) has a large continuity in the extant population. Most importantly, we also evidenced that such maternal contribution is considerably larger than previously evidenced. Moreover, age estimates for the putative autochthonous lineages supported a first arrival to the islands by first settlers at the beginning of the first millennium, although with wide confidence intervals covering several centuries before CE. Surprisingly, some of these putative aboriginal lineages had coalescence ages after European colonization.
The post-conquest settlements on the archipelago
Our findings reveal that nearly half of the maternal gene pool of the current Canary Islanders has been contributed by post-conquest influences. In addition, it has been possible to quantify, at the island scale, the contributions by different continental regions in the genetic pool of the current Canary Islands population, supporting the differential impact of population settlements throughout the archipelago in the post-conquest period.33 At the time, Portuguese competed with Normans and Spanish in the conquest of the archipelago. Thus, the Portuguese influences in the Canary Islands were important even before the conquest was completed. Specifically, the Portuguese settled in La Gomera before the Normans and the Spanish.48 In fact, the Portuguese settled in the Canary Islands at a similar rate as they did in other Macaronesian islands, such as Madeira and the Azores. This activity diminished only after 1640, when the Kingdom of Portugal became independent from the Kingdom of Spain. After the conquest, sugarcane production was the most important economic activity of the archipelago,49,50 especially for the islands with more water resources and wooded surfaces, such as Gran Canaria, Tenerife, La Palma, and La Gomera. This motivated the influx of experienced personnel to operate the sugarcane industries, especially Portuguese because of their experience in their management.51 Other European contingents arrived afterward, mainly to invest in its promotion. This was the case of the Italians and Flemings. In parallel, the arrival of slave labor, mainly from sub-Saharan Africa,52,53 occurred due to the high demand for labor and investors' interest in increasing the profitability of the industry. An example of this demographic event is supported by the high prevalence of sequences of sub-Saharan African origin in Gran Canaria, which played a fundamental role in the sugarcane production industry.54 On the other hand, Lanzarote, Fuerteventura, and El Hierro islands based their economies on conventional agriculture and livestock exploitation at the time.33 A peculiarity of the settlement of the eastern islands of Lanzarote and Fuerteventura was the impact of Moors and Moorish in their demographic development, which would fit with previous estimates55 suggesting that the population of Moorish origin was estimated at almost a third of the Canary Islands population in the 16th century. Therefore, our findings could reconcile with the important role of forced immigration of sub-Saharan Africans and Moorish slaves in the repopulation of the islands.53 It is worth highlighting that, despite the recent homogenization of the Canarian population due to frequent inter-island migrations, our results broadly fit into the historical chronicles of these settlements during and after the conquest. At the western end of the archipelago, the populations from El Hierro and La Palma have the largest proportion of Atlantic influences, mainly of Norman and of Fleming origin, respectively. The largest islands of Tenerife and Gran Canaria, which are expected to have received the largest affluence of post-Hispanic migrants, showed very similar frequencies in the demic components. The most evident differences were a major impact of the Iberian component in Tenerife (41% vs 35% in Gran Canaria), and a major impact of the sub-Saharan African component in Gran Canaria (20% vs 10% in Tenerife).
The Canary Islands have played a fundamental role as a strategic hub in the commercial connectivity between the American and European continents,56 in which the slave trade played a central place.36 The evidence supporting this slave trade is reflected both in the significant number of matches of Canarian and African American sequences and the large number of connections with sequences from the Dominican Republic in the L∗ lineage. Another finding related to this was previously noted23 in relation to the autochthonous founder lineage J2a2d1a, for which the presence of a single individual from Brazil within a lineage composed exclusively of Canarian individuals could be related to the trafficking of labor of aboriginal source during the Atlantic slave trade or alternatively to the recurrent migrations of Canarians during the last centuries.57,58,59 In line with this, other studies have shown the maternal contribution of the Canarian population to the Americans.60,61,62 On the other hand, the presence of two Amerindian individuals in the Canary Islands also points to a migratory process in the opposite direction (i.e. from America to Europe),23 explained by Rando et al.24 as the possible result of the slave trade that brought some native American slaves during the 16th century,63 or due to the return of Canarian emigrants during the 18th and 19th centuries.64 Therefore, these findings support both the active participation of Canary Islands inhabitants in the colonization of America, and the genetic connections between current Canary Islanders and the American continent.
Affinities between the aboriginal and extant Canarian populations
The genetic imprint of the aboriginal population persists in the current population of the archipelago. Here, we have evidenced that this maternal contribution is slightly higher than that observed by previous studies,23,24,25 reaching levels in the range of 51.50%–60.43% depending on the approximation used for the estimation. In addition, our results also showed a marked demographic growth of lineages that are of putative aboriginal origin since the European conquest. This finding is in line with the evidenced sexual asymmetry resulting from a strong assimilation of matrilineal lineages that would favor mating between colonizing males and aboriginal females, added to the presumably increased aboriginal male mortality during the conquest.20,65 However, we warn that this result could have been affected by an imbalance during sampling, since the number of sequences sampled belonging to the current population is much larger than the number of sequences available from aboriginal remains. The important mitogenomic differentiation observed between island populations in pre-colonial times23 could be indicative with the classic—but otherwise unproven—hypothesis that the Canarian aborigines lacked the skills that would facilitate the inter-insular maritime connections. Whatever the case, this situation changed drastically since the beginning of the Spanish conquest because the enslaved aborigines from some islands were transported to other islands as forced labor. It is also known that the conquerors allied with aboriginal factions from some islands to fight against those who resisted, some of whom were even transferred to other islands in the process. An example of the latter is well documented in the chronicles of the conquest of Tenerife and La Palma, the two last islands of the Canary archipelago to be conquered by the Castile Crown, which required the aboriginal help from Gran Canaria.66 This inter-island transference of aborigines could be the explanation for the particular distribution of certain haplogroups, such as H1e1a, H1a0, and H4a1, which have been only detected in the aboriginal remains of the eastern islands while they have been sampled only in the current population of the western islands. These within-archipelago migrations could also explain the smoothing of the genetic differences observed in the extant island populations.
The aboriginal settlement of the islands
The application of a new evolutionary rate to the autochthonous mtDNA lineages47 has substantially decreased their coalescence ages. In most cases, there was a correspondence with the archaeological data recovered in the different Canary Islands populations.46 Considering only the point coalescence age estimates, most of the autochthonous founder lineages support a first aboriginal arrival to the islands at the beginning of the first millennium, fitting with the findings of previous studies.23,46,67 However, if the wide confidence intervals are considered, the age range associated with the first aboriginal arrival could span from centuries before the CE to a time just after the onset of European colonization. The coalescence age estimates obtained for each of the lineages with the rho statistic and with Beast showed slight differences, which could be explained by the bias introduced by the analysis of in short coalescence times. However, a clear overlap is observed between the confidence intervals recovered by each approach for each lineage. Finally, with the results achieved, we were unable to provide well-founded inferences about the colonization process of the Canary Islands for three main reasons: the bias associated with age inference because of using mtDNA for short coalescence times, the large overlap between the confidence intervals associated with the point estimates for the different lineages, and the slight inconsistencies in the estimates between both approaches. Thus, future studies leveraging the information provided by other molecular markers, such as from the Y chromosome or from autosomes, together with the archaeological contextualization of the genetic data could help to shed more light on the dynamics of the colonization of the Canary Islands by the aboriginal population.
Limitations of the study
This study has certain limitations due to the heterogeneous representation in the databases of human mitogenomes from worldwide populations, reflecting a measurable bias toward representing more genetic data from European populations than from African populations. The limited data from the North African region are especially important. Because of this, the North African influences should be interpreted with caution as the genetics of the populations from this region continue to be largely uncharacterized at genomic scale.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Blood samples | CDC of the Canary Islands | Cabrera León et al.68 |
| Critical commercial assays | ||
| Blood genomicPrep Mini Spin Kit | Cytiva | 28-9042-65 |
| Qubit™ dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32851 |
| Illumina DNA Prep with Enrichment | Illumina | 20025524 |
| Nextera DNA Exome | Illumina | 20020617 |
| High Sensitivity D1000 ScreenTape Assays | Agilent | 5067-5584 | 5067-5585 | 5067-5587 |
| Genomic DNA ScreenTape Assay | Agilent | 5067-5365 | 5067-5366 |
| Deposited data | ||
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ |
| Human reference mitogenome, revised Cambridge Reference Sequence (rCRS) | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/nuccore/251831106 |
| Sequenced mitogenomes (FASTA) | This manuscript | Genbank: OP681790 - OP682685 |
| Worldwide matching mitogenomes sequences to the aboriginal and current-day Canary Island sequences | NCBI GenBank; Mitomap; Ian Logan 2020; AmtDB | www.ncbi.nlm.nih.gov/genbank; www.mitomap.org/MITOMAP; www.ianlogan.co.uk/sequences_by_group/haplogroup_select.htm; http://amtdb.org |
| Software and algorithms | ||
| GATK v.4 | McKenna et al.69 | https://github.com/broadinstitute/gatk |
| Haplocheck v.1.3.3 | Weissensteiner et al.70 | https://github.com/genepi/haplocheck |
| PhyloTree v.17 | van Oven & Kayser71 | http://www.phylotree.org |
| vt tool v.0.57721 | Tan et al.72 | https://genome.sph.umich.edu/wiki/Vt |
| ggplot2 | Wickham73 | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Circos plot | Krzywinski et al.74 | http://mkweb.bcgsc.ca/tableviewer/ |
| Beast v.2.6.4 | Bouckaert et al.75 | https://www.beast2.org/ |
| jModelTest | Darriba et al.76 | https://github.com/ddarriba/jmodeltest2 |
| Tracer v.1.7.2 | Rambaut et al.77 | https://github.com/beast-dev/tracer/releases/tag/v1.7.2 |
| Seqboot | N/A | https://evolution.genetics.washington.edu/phylip/doc/seqboot.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Carlos Flores (cflores@ull.edu.es).
Materials availability
This study did not generate new unique reagents.
Method details
Samples, library preparation, and sequencing
The study was approved by the Research Ethics Committee of the Hospital Universitario Nuestra Señora de Candelaria (CHUNSC_2020_95) and performed according to The Code of Ethics of the World Medical Association (Declaration of Helsinki).
The samples for this study were obtained after informed consent from the cohort study ‘CDC of the Canary Islands’.68 This dataset constitutes the most extensive population cohort for medical studies of the Canary Islands population, which involves nearly 7,000 randomly selected donors, aged between 18 and 75 years from the seven main islands and without gender bias. Eight hundred ninety-six DNA samples from unrelated donors were selected for the study. The samples selected for this study self-declared that the four grandparents were born in the same island and that they had no personal history of cardiovascular, metabolic, immunologic, or cancer diseases.79 The total number of individuals sequenced per island ranged from 52 to 215. By island, from west to east, the number of sequenced individuals was as follows: El Hierro, 106; La Palma, 101; La Gomera, 136; Tenerife, 175; Gran Canaria, 215; Fuerteventura, 52; and Lanzarote, 111. DNA was extracted from peripheral blood using a commercial column-based solution (Blood genomicPrep Mini Spin Kit, Marlborough, MA, USA). DNA quantifications were performed in a Qubit 3.0 Fluorometer by the QubitTM dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The samples were subjected to different whole-exome sequencing (WES) capture solutions as described elsewhere.80 The library quality controls were carried out in a TapeStation 4200 (Agilent Technologies, Santa Clara, CA, USA) and sequencing was conducted on NextSeq 550, HiSeq 4000, or NovaSeq 6000 (Illumina, Inc., San Diego, CA, USA).
Bioinformatic analysis
Processing of WES data to extract and assign the mtDNA haplogroups was carried out using an in-house pipeline based on GATK v.4.69 Briefly, the reads were aligned to the GRCh37/hg19 reference genome while the mtDNA reads realigned to the revised Cambridge Reference Sequence (rCRS), GenBank NC_012920,81,82 following GATK best practices for this circular genome as described elsewhere.78 As an additional step, and in order to avoid potential artifacts during the variant calling, variants supported by less than 70% of the reads were discarded. Based on our benchmarking results for human mtDNA classification tools and given the superior performance for WES data,78 haplogroups were classified from BAM files using Haplocheck v.1.3.3.70 mtDNA sequences were classified according to the PhyloTree v.17 (http://www.phylotree.org).71 In order to harmonize the genomic information retrieved in the BAM and VCF files (given that the former may contain variants not recovered during the variant calling step or that may have been filtered out during the quality controls) a refinement of the genotype of each sample was performed. Briefly, missing diagnostic variants or rare point mutations and indels were manually inspected across the intermediate files generated by the bioinformatic processing. For the novel mutations identified during haplogroup classification, only the variants with a depth of coverage greater than 10X were retained, with the exception of mutations that were found shared among different individuals. Multiallelic variants were split using the vt tool v.0.5772172 keeping those with higher read support (>70%). Finally, for phylogenetic reconstruction, the mutation in the 16519 hotspot and the indels around nucleotide positions 309, 522, 573, and 16182–16193 were excluded from the analyses.
Partial mtDNA sequences for population comparisons
To increase the sample size for population-based comparisons of aboriginal and extant Canarian populations, we included previously published partial mtDNA haplotypes (based on HVR1 sequences and diagnostic variants) from extant and aboriginal Canary Islanders (Table S1), provided that the recovered haplotypes could be unequivocally assigned to specific sub-haplogroups. For this reason, partial sequences as those identical to the rCRS reference sequence,82 including those assigned by diagnostic variants to haplogroups H∗ or U∗, have been omitted from all analyses as they could be assigned to different subgroups within each haplogroup. However, it should be noted that, while this approximation was only used to identify matches between sequences (see below) and not for frequency estimations, the results of this analysis have certain limitations and, therefore, care should be taken when interpreting them.
Extracting the closest continental mtDNA sequences
The closest worldwide matching complete mitogenomes to the aboriginal and current-day Canary Island sequences were obtained by searching in the following databases: NCBI GenBank (www.ncbi.nlm.nih.gov/genbank/), Mitomap (www.mitomap.org/MITOMAP), Ian Logan 2020 (www.ianlogan.co.uk/sequences_by_group/haplogroup_select.htm), and AmtDB (http://amtdb.org) (Table S2).
Population-based analysis
To compensate for the influence of the most common haplotypes on the calculation of frequency-based distance, a measure based on pairwise matches was used. For this measure, matches in pairs of sequences that differ by a maximum of two mutations were considered. The algorithm defines Ixy, the identity between populations x and y, as the double number of matches between them (2Mxy) divided by the sum of the number of different lineages in x and y (Nx + Ny). Thus, the pairwise matches could vary from zero (no matches found between two populations) to one (all the different lineages are shared between the two populations). The distance between populations (Dxy) was calculated from 1 - Ixy. Distance matches were visualized through principal components analysis (PCA) using R v.4.0.4 environment83 and the ggplot2 package.73 To assess the temporal or spatial differences between populations based on the pairwise matches or the haplogroup frequency differences, a two-tailed Fisher exact test was used. Mean haplogroup frequency comparisons were tested using unpaired t-tests. To explore the maternal genetic contributions to the Canary Islands gene pool a Circos plot74 was constructed based on the pairwise matching affinities between individuals from the Canarian archipelago and African and European populations (Table S2). To simplify the analysis, five population groups were defined: Atlantic, Iberian, Italian, North African, and Sub-Saharan African. Within North Africa, we did not distinguish between Berber and non-Berbers because the data lacks ethic or language information to be used in this analysis. Previous studies based on different markers suggest a lack of significant genetic differences between Berbers and non-Berbers in North Africa.84,85,86,87,88 However, given that the North African genetic diversity remains a largely unexplored, this should be considered as a limitation of the study. Within Iberia, five regions were established in subsequent analyses: Galicia, Portugal, Basque Country, Andalusia, and the rest of the Iberian Peninsula. With respect to Galicia and Portugal, although previous studies support that both populations are genetically close,89 we preferred to keep them separate to obtain a higher resolution in the region.
Age estimates of the aboriginal founder lineages
Coalescence ages for the putative autochthonous Canarian lineages were calculated using the rho statistic90 and two alternative mtDNA substitution rates: i) an overall interspecific rate of one mutation every 3,624 years (95% Confidence Interval [CI]: 2,973–4,275)45; and ii) using a new rate proposed by Cabrera,47 based on the most recent period of the human demographic history that assumes a time-dependency effect on the evolutionary rate, increasing the mutation rate to one mutation every 1,400 years (95% CI: 1,261–1,539). Seqboot was used to generate 3,600 bootstrapped mtDNA alignments to calculate the rho statistic for each autochthonous founder lineages by using an in-house script (https://github.com/genomicsITER/mitogenomes/tree/main/CanarymtDNA).
Coalescence ages were also estimated following a Bayesian approach using Beast v.2.6.4.75 We used the GTR nucleotide substitution model, as the best fitting substitution model indicated by jModelTest,76 and applied the corresponding substitution rate proposed by Cabrera.47 The clock model was set as a strict clock assuming that every branch in a phylogenetic tree evolves according to the same evolutionary rate, and a Yule model prior was applied. An UPGMA tree was used as the seed tree and each analysis was run for four billion iterations sampled every million iterations obtaining 4,000 trees. The resulting log and tree files were inspected using Tracer v.1.7.2,77 after discarding the first 10% of trees as burn-in, and confirming effective sample size (ESS) values exceeding 200 as indicated by the best practices guidelines.91 The time to the most recent common ancestor (TMRCA) was obtained with Tracer. Additionally, demographic analyses were performed to reconstruct the population dynamics of each of the autochthonous founder lineages using the Bayesian Skyline coalescent model. For each run, the same setup as described above was applied. Tracer was used to extract the Bayesian skyline results from the sampled trees of each analysis and the skyline plot was constructed by using the ggplot2 R package.73
Acknowledgments
This research was funded by Ministerio de Ciencia e Innovación (RTC-2017-6471-1; AEI/FEDER, UE), co-financed by the European Regional Development Funds ‘A way of making Europe’ from the European Union; Fundación CajaCanarias and Fundación Bancaria “La Caixa” (2018PATRI20); Cabildo Insular de Tenerife (CGIEU0000219140); Convenio Marco de Cooperación Consejería de Educación-Cabildo Insular de Tenerife 2021–2025 (CGIAC0000014697); and by the agreement OA17/008 with Instituto Tecnológico y de Energías Renovables (ITER) to strengthen scientific and technological education, training, research, development, and innovation in Genomics, Personalized Medicine, and Biotechnology. A.D.U. was supported by a fellowship from the Spanish Ministry of Education and Vocational Training (grant number FPU16/01435). We would like to thank the support from our colleagues from the Teide-HPC Supercomputing facility (http://teidehpc.iter.es/en), which was funded by INP-2011-0063-PCT-430000-ACT (INNPLANTA program) from the Spanish Ministry of Economy and Competitiveness. The authors deeply acknowledge the University Hospital Nuestra Señora de Candelaria for the support and extend our gratitude to all members of the CDC group for their work, and to all the participants for their collaboration. Finally, the authors would like to deeply thank the two anonymous reviewers for their technical advice and discussions that have improved the manuscript.
Author contributions
Conceptualization, V.G.O., L.A.R.R., V.M.C., C.F.; Data curation, V.G.O., L.A.R.R., V.M.C., A.M.B., J.M.L.S., D.J.T., R.G.M.; Methodology, V.G.O., L.A.R.R., A.M.B., J.M.L.S., A.D.U., D.J.T., A.I.C., M.C.R., A.C.L., R.G.M.; Supervision, C.F.; Writing—original draft preparation, V.M.C., V.G.O., L.A.R.R., C.F.; Writing—review and editing, V.G.O., V.M.C., L.A.R.R., C.F. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Inclusion and diversity
We support inclusive, diverse and equitable conduct of research.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105907.
Supplemental information
Data and code availability
-
•
The data generated as part of this study has been deposited in the National Center of Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). Mitogenomes can be found under the accession numbers GenBank: OP681790 to GenBank: OP682685.
-
•
All original code used for generating bootstrapped mtDNA alignments is available at: https://github.com/genomicsITER/mitogenomes/tree/main/CanarymtDNA.
-
•
Processing of WES data was carried out using an in-house pipeline based on GATK v.4 described elsewhere.78
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Aznar-Vallejo E. HID; 2008. Exploración y colonización en la configuración de la Europa Atlántica. [DOI] [Google Scholar]
- 2.Hooton E.A. Corinthian Press; 1925. The Ancient Inhabitants of the Canary Islands. [Google Scholar]
- 3.Farrujia de la Rosa A.J. Springer Science & Business Media; 2013. An Archaeology of the Margins: Colonialism, Amazighity and Heritage Management in the Canary Islands. [Google Scholar]
- 4.Bonnet Reverón B. Las expediciones a las Canarias en el siglo XIV. Rev. Indias. 1944;5 [Google Scholar]
- 5.Morales Padrón F. Los descubrimientos en los siglos XIV y XV, y los archipiélagos atlánticos. Anu. Estud. Atl. 1971;17:429–465. [Google Scholar]
- 6.Abreu-Galindo J. 1955. Historia de la conquista de las siete islas de Canaria (Goya ediciones) [Google Scholar]
- 7.Billy G. El Museo Canario; 1980. Le Peuplement Prehistorique de l’Archipel Canarien; pp. 59–74. [Google Scholar]
- 8.Bermúdez De Castro J.M. The Carabelli trait in human prehistoric populations of the Canary Islands. Hum. Biol. 1989;61:117–131. [PubMed] [Google Scholar]
- 9.Bermúdez de Castro J.M. Third molar agenesis in human prehistoric populations of the Canary Islands. Am. J. Phys. Anthropol. 1989;79:207–215. doi: 10.1002/ajpa.1330790208. [DOI] [PubMed] [Google Scholar]
- 10.Navarro-Mederos J.F.N. “El viaje de las loceras” La transmisión de tradiciones cerámicas prehistóricas e históricas de África a Canarias y su reproducción en las islas. Anu. Estud. Atl. 1999;1:61–118. [Google Scholar]
- 11.Guatelli-Steinberg D., Irish J.D., Lukacs J.R. Canary islands-north African population affinities: measures of divergence based on dental morphology. Homo. 2001;52:173–188. doi: 10.1078/0018-442x-00027. [DOI] [PubMed] [Google Scholar]
- 12.Farrujia de la Rosa A.J., Pichler W., Rodrigue A., García Marín S. The Libyco–Berber and Latino–Canarian scripts and the colonization of the canary islands. Afr. Archaeol. Rev. 2010;27:13–41. doi: 10.1007/s10437-010-9070-4. [DOI] [Google Scholar]
- 13.Tejera Gaspar A., Perera Betancor M.A. Las supuestas inscripciones púnicas y neopúnicas de las Islas Canarias. Spal. Revista de Prehistoria y Arqueología. 2011;20:175–184. doi: 10.12795/spal.2011.i20.11. [DOI] [Google Scholar]
- 14.Santana-Cabrera J., Velasco-Vazquez J., Rodríguez Rodríguez A. Vol. 19. 2012. Patrón cotidiano de actividad física y organización social del trabajo en la Gran Canaria prehispánica (siglos XI-XV): la aportación de los marcadores óseos de actividad física; pp. 125–163. (Tabona.). [Google Scholar]
- 15.Morales J., Rodríguez-Rodríguez A., González-Marrero M.d.C., Martín-Rodríguez E., Henríquez-Valido P., del-Pino-Curbelo M., del-Pino-Curbelo M. The archaeobotany of long-term crop storage in northwest African communal granaries: a case study from pre-Hispanic Gran Canaria (cal. ad 1000–1500) Veget. Hist. Archaeobot. 2014;23:789–804. doi: 10.1007/s00334-014-0444-4. [DOI] [Google Scholar]
- 16.Hagenblad J., Morales J., Leino M.W., Rodríguez-Rodríguez A.C. Farmer fidelity in the Canary Islands revealed by ancient DNA from prehistoric seeds. J. Archaeol. Sci. 2017;78:78–87. doi: 10.1016/j.jas.2016.12.001. [DOI] [Google Scholar]
- 17.Ferrando A., Manunza A., Jordana J., Capote J., Pons A., Pais J., Delgado T., Atoche P., Cabrera B., Martínez A., et al. A mitochondrial analysis reveals distinct founder effect signatures in Canarian and Balearic goats. Anim. Genet. 2015;46:452–456. doi: 10.1111/age.12302. [DOI] [PubMed] [Google Scholar]
- 18.Olalde I., Capote J., Del-Arco M.C., Atoche P., Delgado T., González-Anton R., Pais J., Amills M., Lalueza-Fox C., Ramírez O. Ancient DNA sheds light on the ancestry of pre-hispanic Canarian pigs. Genet. Sel. Evol. 2015;47:40. doi: 10.1186/s12711-015-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores C., Maca-Meyer N., Pérez J.A., González A.M., Larruga J.M., Cabrera V.M. A predominant European ancestry of paternal lineages from Canary Islanders. Ann. Hum. Genet. 2003;67:138–152. doi: 10.1046/j.1469-1809.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 20.Fregel R., Gomes V., Gusmão L., González A.M., Cabrera V.M., Amorim A., Larruga J.M. Demographic history of Canary Islands male gene-pool: replacement of native lineages by European. BMC Evol. Biol. 2009;9:181. doi: 10.1186/1471-2148-9-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maca-Meyer N., Arnay M., Rando J.C., Flores C., González A.M., Cabrera V.M., Larruga J.M. Ancient mtDNA analysis and the origin of the Guanches. Eur. J. Hum. Genet. 2004;12:155–162. doi: 10.1038/sj.ejhg.5201075. [DOI] [PubMed] [Google Scholar]
- 22.Fregel R., Pestano J., Arnay M., Cabrera V.M., Larruga J.M., González A.M. The maternal aborigine colonization of La Palma (Canary Islands) Eur. J. Hum. Genet. 2009;17:1314–1324. doi: 10.1038/ejhg.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fregel R., Ordóñez A.C., Santana-Cabrera J., Cabrera V.M., Velasco-Vázquez J., Alberto V., Moreno-Benítez M.A., Delgado-Darias T., Rodríguez-Rodríguez A., Hernández J.C., et al. Mitogenomes illuminate the origin and migration patterns of the indigenous people of the Canary Islands. PLoS One. 2019;14:e0209125. doi: 10.1371/journal.pone.0209125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rando J.C., Cabrera V.M., Larruga J.M., Hernández M., González A.M., Pinto F., Bandelt H.J. Phylogeographic patterns of mtDNA reflecting the colonization of the Canary Islands. Ann. Hum. Genet. 1999;63:413–428. doi: 10.1046/j.1469-1809.1999.6350413.x. [DOI] [PubMed] [Google Scholar]
- 25.Pinto F., González A.M., Hernández M., Larruga J.M., Cabrera V.M. Genetic relationship between the Canary Islanders and their African and Spanish ancestors inferred from mitochondrial DNA sequences. Ann. Hum. Genet. 1996;60:321–330. doi: 10.1111/j.1469-1809.1996.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 26.Fregel R., Cabrera V.M., Larruga J.M., Hernández J.C., Gámez A., Pestano J.J., Arnay M., González A.M. Isolation and prominent aboriginal maternal legacy in the present-day population of La Gomera (Canary Islands) Eur. J. Hum. Genet. 2015;23:1236–1243. doi: 10.1038/ejhg.2014.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto F.M., González A.M., Hernández M., Larruga J.M., Cabrera V.M. Sub-Saharan influence on the Canary Islands population deduced from G6PD gene sequence analysis. Hum. Biol. 1996;68:517–522. [PubMed] [Google Scholar]
- 28.Pino-Yanes M., Corrales A., Basaldúa S., Hernández A., Guerra L., Villar J., Flores C. North African influences and potential bias in case-control association studies in the Spanish population. PLoS One. 2011;6:e18389. doi: 10.1371/journal.pone.0018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botigué L.R., Henn B.M., Gravel S., Maples B.K., Gignoux C.R., Corona E., Atzmon G., Burns E., Ostrer H., Flores C., et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl. Acad. Sci. USA. 2013;110:11791–11796. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Varela R., Günther T., Krzewińska M., Storå J., Gillingwater T.H., MacCallum M., Arsuaga J.L., Dobney K., Valdiosera C., Jakobsson M., et al. Genomic analyses of pre-European conquest human remains from the canary islands reveal close affinity to modern north Africans. Curr. Biol. 2017;27:3396–3402.e5. doi: 10.1016/j.cub.2017.09.059. [DOI] [PubMed] [Google Scholar]
- 31.Guillen-Guio B., Lorenzo-Salazar J.M., González-Montelongo R., Díaz-de Usera A., Marcelino-Rodríguez I., Corrales A., Cabrera de León A., Alonso S., Flores C. Genomic analyses of human European diversity at the Southwestern edge: isolation, African influence and disease Associations in the canary islands. Mol. Biol. Evol. 2018;35:3010–3026. doi: 10.1093/molbev/msy190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos C., Fregel R., Cabrera V.M., González A.M., Larruga J.M., Lima M. Mitochondrial DNA patterns in the Macaronesia islands: variation within and among archipelagos. Am. J. Phys. Anthropol. 2010;141:610–619. doi: 10.1002/ajpa.21180. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Armesto F. Clarendon Press; 1982. The Canary Islands after the Conquest: The Making of a Colonial Society in the Early Sixteenth Century. [Google Scholar]
- 34.Viña Brito A., Ronquillo Rubio M. El primer ciclo del azúcar en Canarias. Balance historiográfico. 2006 doi: 10.12795/hid.2013.i40.12. [DOI] [Google Scholar]
- 35.Santana J., Fregel R., Lightfoot E., Morales J., Alamón M., Guillén J., Moreno M., Rodríguez A. The early colonial atlantic world: new insights on the African Diaspora from isotopic and ancient DNA analyses of a multiethnic 15th-17th century burial population from the Canary Islands, Spain. Am. J. Phys. Anthropol. 2016;159:300–312. doi: 10.1002/ajpa.22879. [DOI] [PubMed] [Google Scholar]
- 36.Maca-Meyer N., Cabrera V.M., Arnay M., Flores C., Fregel R., González A.M., Larruga J.M. Mitochondrial DNA diversity in 17th-18th century remains from Tenerife (canary islands) Am. J. Phys. Anthropol. 2005;127:418–426. doi: 10.1002/ajpa.20148. [DOI] [PubMed] [Google Scholar]
- 37.Brehm A., Pereira L., Bandelt H.-J., Prata M.J., Amorim A. Mitochondrial portrait of the Cabo Verde archipelago: the Senegambian outpost of Atlantic slave trade. Ann. Hum. Genet. 2002;66:49–60. doi: 10.1017/S0003480001001002. [DOI] [PubMed] [Google Scholar]
- 38.Trovoada M.J., Pereira L., Gusmão L., Abade A., Amorim A., Prata M.J. Pattern of mtDNA variation in three populations from São Tomé e Príncipe. Ann. Hum. Genet. 2004;68:40–54. doi: 10.1046/j.1529-8817.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- 39.Matisoo-Smith E.A., Gosling A.L., Boocock J., Kardailsky O., Kurumilian Y., Roudesli-Chebbi S., Badre L., Morel J.-P., Sebaï L.L., Zalloua P.A. A European mitochondrial haplotype identified in ancient Phoenician remains from Carthage, North Africa. PLoS One. 2016;11:e0155046. doi: 10.1371/journal.pone.0155046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S., Besenbacher S., Li Y., Kristiansen K., Grarup N., Albrechtsen A., Sparsø T., Korneliussen T., Hansen T., Wang J., et al. Variation and association to diabetes in 2000 full mtDNA sequences mined from an exome study in a Danish population. Eur. J. Hum. Genet. 2014;22:1040–1045. doi: 10.1038/ejhg.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raule N., Sevini F., Li S., Barbieri A., Tallaro F., Lomartire L., Vianello D., Montesanto A., Moilanen J.S., Bezrukov V., et al. The co-occurrence of mtDNA mutations on different oxidative phosphorylation subunits, not detected by haplogroup analysis, affects human longevity and is population specific. Aging Cell. 2014;13:401–407. doi: 10.1111/acel.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spínola F.F. XV Coloquio de Historia Canario-Americana; 2004. Portugueses en Canarias en el siglo XVII. Una relación de 1626; pp. 310–320. [Google Scholar]
- 43.Navarro-Mederos J.F. Arqueología de las islas Canarias. Espac. Tiempo Forma. Ser. Prehist. Arqueol. 1997 doi: 10.5944/etfi.10.1997.4662. [DOI] [Google Scholar]
- 44.Viña Brito A. Vol. 194. 2012. pp. 161–191. (Los flamencos en Canarias en el siglo XVI : ¿Una comunidad extranjera? Especificidades en la isla de La Palma. Revista de Historia Canaria.). [Google Scholar]
- 45.Soares P., Ermini L., Thomson N., Mormina M., Rito T., Röhl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velasco-Vazquez J., Alberto-Barroso V., Delgado T., Moreno M., Lecuyer C., Richardin P. Settlement, colonization and early history of the Canary Islands: the C14 as a paradigm. Anu. Estud. Atl. 2020:1–24. doi: 10.36980/10530.9904. [DOI] [Google Scholar]
- 47.Cabrera V.M. Human molecular evolutionary rate, time dependency and transient polymorphism effects viewed through ancient and modern mitochondrial DNA genomes. Sci. Rep. 2021;11:5036. doi: 10.1038/s41598-021-84583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Álvarez J. Primera conquista y cristianización de La Gomera. Anu. Estud. Atl. 1960;6:445–492. [Google Scholar]
- 49.Ronquillo Rubio M. Organismo Autónomo de Museos y Centros Excmo; Cabildo Insular de Tenerife: 2008. Ingenios azucareros en la colonización canaria: 1487-1526. Localización espacial y organización del espacio en Gran Canaria. [Google Scholar]
- 50.Viña Brito A. Estudios Canarios: Anuario del Instituto de Estudios Canarios; 2015. El cultivo de la caña de azúcar en Canarias en los inicios de la colonización; pp. 239–264. [Google Scholar]
- 51.Greenfield S.M. Madeira and the beginnings of new world sugar cane cultivation and plantation slavery: a study in institution building. Ann. NY. Acad. Sci. 1977;292:536–552. doi: 10.1111/j.1749-6632.1977.tb47771.x. [DOI] [Google Scholar]
- 52.Lobo Cabrera M. Ediciones del Excmo. Cabildo Insular de Gran Canaria; 1982. La esclavitud en las Canarias orientales en el siglo XVI (Negros, moros y moriscos) [Google Scholar]
- 53.Viña Brito A., Hernández González M. 2006. Esclavos (Archivo Histórico Provincial de Santa Cruz de Tenerife) [Google Scholar]
- 54.Florido Castro A. 2000. El patrimonio industrial azucarero de Gran Canaria. Recuperación de la azucarera de San Juan de Telde como Museo Etnoagrario. XIV Coloquio de Historia Canario Americana. [Google Scholar]
- 55.Lobo Cabrera M. 2015. Los moriscos en Canarias: de esclavos a naturales (Mercurio Editorial) [Google Scholar]
- 56.Armenteros Martínez I. 2018. The Canary Islands as an Area of Interconnectivity between the Mediterranean and the Atlantic (Fourteenth-Sixteenth Centuries). Nikolas Jaspert & Sebastian Kolditz, eds. [Google Scholar]
- 57.Macías Hernández A.M. Ediciones Tabapress; 1991. Emigración Española a Ultramar, 1492-1914 A. Eiras Roel. [Google Scholar]
- 58.Hernández-González M. Vol. 126. Cuadernos Americanos: Nueva Epoca; 2008. pp. 132–172. (La emigración canaria a América a través de la historia). [Google Scholar]
- 59.Mendizabal I., Sandoval K., Berniell-Lee G., Calafell F., Salas A., Martínez-Fuentes A., Comas D. Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evol. Biol. 2008;8:213. doi: 10.1186/1471-2148-8-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sans M., Figueiro G., Ackermann E., Barreto I., Egaña A., Bertoni B., Poittevin-Gilmet E., Maytia D., Hidalgo P.C. Mitochondrial DNA in Basque descendants from the city of Trinidad, Uruguay: Uruguayan- or Basque-like population? Hum. Biol. 2011;83:55–70. doi: 10.3378/027.083.0104. [DOI] [PubMed] [Google Scholar]
- 61.Secher B., Fregel R., Larruga J.M., Cabrera V.M., Endicott P., Pestano J.J., González A.M. The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evol. Biol. 2014;14:109. doi: 10.1186/1471-2148-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baeta M., Núñez C., Cardoso S., Palencia-Madrid L., Piñeiro-Hermida S., Arriba-Barredo M., Villanueva-Millán M.J., M de Pancorbo M. Different evolutionary history for Basque diaspora populations in USA and Argentina unveiled by mitochondrial DNA analysis. PLoS One. 2015;10:e0144919. doi: 10.1371/journal.pone.0144919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lobo Cabrera M. Grupos humanos en la sociedad canaria del siglo XVI. Colección Guagua. 1979 [Google Scholar]
- 64.Castellano J.M., Macías F.J. Centro de la Cultura Popular Canaria; 1997. Historia de Canarias. [Google Scholar]
- 65.Flores C., Larruga J., González A., Hernández M., Pinto F., Cabrera V. The origin of the canary island aborigines and their contribution to the modern population: a molecular genetics perspective. Curr. Anthropol. 2001;42:749–755. doi: 10.1086/323819. [DOI] [Google Scholar]
- 66.Morales Padrón F. 1978. Canarias: crónicas de su conquista (Ayuntamiento de Las Palmas) [Google Scholar]
- 67.Morales J., Rodríguez A., Henríquez-Valido P. In: la contribución de los estudios carpológicos. Fernández J., Mujika J.A., García A.A., editors. Universidad del País Vasco; 2017. Agricultura y recolección vegetal en la arqueología prehispánica de las Islas Canarias (siglos III-XV d.C.) [Google Scholar]
- 68.Cabrera León A., Rodríguez Pérez M.C., González D.A., Domínguez Coello S., Aguirre Jaime A., Brito Díaz B., González Hernández A., Pérez Méndez L.I., y el grupo CDC Presentación de la cohorte “CDC de Canarias”: objetivos, diseño y resultados preliminares. Rev. Esp. Orientac. Psicopedag. 2008;82:519–534. doi: 10.1590/s1135-57272008000500007. [DOI] [PubMed] [Google Scholar]
- 69.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weissensteiner H., Forer L., Fendt L., Kheirkhah A., Salas A., Kronenberg F., Schoenherr S. Contamination detection in sequencing studies using the mitochondrial phylogeny. Genome Res. 2021;31:309–316. doi: 10.1101/gr.256545.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 72.Tan A., Abecasis G.R., Kang H.M. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 74.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.García-Olivares V., Muñoz-Barrera A., Lorenzo-Salazar J.M., Zaragoza-Trello C., Rubio-Rodríguez L.A., Díaz-de Usera A., Jáspez D., Iñigo-Campos A., González-Montelongo R., Flores C. A benchmarking of human mitochondrial DNA haplogroup classifiers from whole-genome and whole-exome sequence data. Sci. Rep. 2021;11:20510. doi: 10.1038/s41598-021-99895-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Díaz-de Usera A., Rubio-Rodríguez L.A., Muñoz-Barrera A., Lorenzo-Salazar J.M., Guillen-Guio B., Jáspez D., Corrales A., Íñigo-Campos A., García-Olivares V., Rodríguez Pérez M.D.C., et al. Developing CIRdb as a catalog of natural genetic variation in the Canary Islanders. Sci. Rep. 2022;12:16132. doi: 10.1038/s41598-022-20442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Díaz-de Usera A., Lorenzo-Salazar J.M., Rubio-Rodríguez L.A., Muñoz-Barrera A., Guillen-Guio B., Marcelino-Rodríguez I., García-Olivares V., Mendoza-Alvarez A., Corrales A., Íñigo-Campos A., et al. Evaluation of whole-exome enrichment solutions: lessons from the high-end of the short-read sequencing scale. J. Clin. Med. 2020;9:3656. doi: 10.3390/jcm9113656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 82.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 83.R Core Team . 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 84.Bosch E., Calafell F., Pérez-Lezaun A., Clarimón J., Comas D., Mateu E., Martínez-Arias R., Morera B., Brakez Z., Akhayat O., et al. Genetic structure of north-west Africa revealed by STR analysis. Eur. J. Hum. Genet. 2000;8:360–366. doi: 10.1038/sj.ejhg.5200464. [DOI] [PubMed] [Google Scholar]
- 85.Bosch E., Calafell F., Comas D., Oefner P.J., Underhill P.A., Bertranpetit J. High-resolution analysis of human Y-chromosome variation shows a sharpdiscontinuity and limited gene flow between Northwestern Africa and the iberian Peninsula. Am. J. Hum. Genet. 2001;68:1019–1029. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harich N., Esteban E., Chafik A., López-Alomar A., Vona G., Moral P. Classical polymorphisms in Berbers from Moyen Atlas (Morocco): genetics, geography, and historical evidence in the Mediterranean peoples. Ann. Hum. Biol. 2002;29:473–487. doi: 10.1080/03014460110104393. [DOI] [PubMed] [Google Scholar]
- 87.Bentayebi K., Abada F., Ihzmad H., Amzazi S. Genetic ancestry of a Moroccan population as inferred from autosomal STRs. Meta Gene. 2014;2:427–438. doi: 10.1016/j.mgene.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arauna L.R., Comas D. Genetic heterogeneity between Berbers and Arabs. eLS. 2017:1–7. doi: 10.1002/9780470015902.a0027485. [DOI] [Google Scholar]
- 89.Barral-Arca R., Pischedda S., Gómez-Carballa A., Pastoriza A., Mosquera-Miguel A., López-Soto M., Martinón-Torres F., Álvarez-Iglesias V., Salas A. Meta-analysis of mitochondrial DNA variation in the iberian Peninsula. PLoS One. 2016;11:e0159735. doi: 10.1371/journal.pone.0159735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Forster P., Harding R., Torroni A., Bandelt H.J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 91.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data generated as part of this study has been deposited in the National Center of Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). Mitogenomes can be found under the accession numbers GenBank: OP681790 to GenBank: OP682685.
-
•
All original code used for generating bootstrapped mtDNA alignments is available at: https://github.com/genomicsITER/mitogenomes/tree/main/CanarymtDNA.
-
•
Processing of WES data was carried out using an in-house pipeline based on GATK v.4 described elsewhere.78
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.