Abstract
A major hallmark of Alzheimer’s disease (AD) is the accumulation of extracellular aggregates of amyloid-β (Aβ). Structural polymorphism observed among Aβ fibrils in AD brains seem to correlate with the clinical subtypes suggesting a link between fibril polymorphism and pathology. Since fibrils emerge from a templated growth of low-molecular-weight oligomers, understanding the factors affecting oligomer generation is important. Membrane lipids are key factors to influence early stages of Aβ aggregation and oligomer generation, which cause membrane disruption. We have previously demonstrated that conformationally discrete Aβ oligomers can be generated by modulating the charge, composition, and chain length of lipids and surfactants. Here, we extend our studies into liposomal models by investigating Aβ oligomerization on large unilamellar vesicles (LUVs) of total brain extracts (TBE), reconstituted lipid rafts (LRs), or 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). Varying the vesicle composition by specifically increasing the amount of GM1 gangliosides as a constituent, we found that only GM1-enriched liposomes induce the formation of toxic, low-molecular-weight oligomers. Furthermore, we found that the aggregation on liposome surface and membrane disruption are highly cooperative and sensitive to membrane surface characteristics. Numerical simulations confirm such a cooperativity and reveal that GM1-enriched liposomes form twice as many pores as those formed in the absence GM1. Overall, this study uncovers mechanisms of cooperativity between oligomerization and membrane disruption under controlled lipid compositional bias, and refocuses the significance of the early stages of Aβ aggregation in polymorphism, propagation, and toxicity in AD.
Graphical Abstract
INTRODUCTION
Alzheimer’s disease (AD) is a neurodegenerative disorder associated with deposition of extracellular plaques composed of amyloid-β (Aβ) aggregates in the brain. Aβ peptide is generated by the sequential cleavage of transmembrane amyloid precursor protein (APP) by β and γ secretases and is subsequently released into the extracellular space.1-3 Monomeric Aβ is intrinsically disordered and undergoes near spontaneous aggregation toward high-molecular-weight insoluble fibrils involving sigmoidal growth kinetics.4-6 The low-molecular-weight soluble oligomers generated during aggregation are known to be the primary toxic species in early stages of AD pathology that impair hippocampal synaptic plasticity and cause blockage of long-term hippocampal potentiation (LTP).7-9 A few mechanisms by which the oligomers impart toxicity are membrane disruption via pore formation, release of reactive oxygen species (ROS), astrocytosis, and microglial activation.10-13 It has been long hypothesized that being closely associated with Aβ, membrane lipids, and surfactants are likely to interact and generate conformationally diverse low-molecular-weight oligomers.12,14-17 Lipids play an important role in the early stages of Aβ aggregation that dictates oligomer generation. 18-20 We demonstrated that micelle-forming lipids including fatty acids, lysophospholipids, and gangliosides can induce distinct conformational oligomers that have discrete cellular and pathological functions.21 Many of these oligomers are toxic to neuroblastoma cells18 and induce cerebral amyloid angiopathy (CAA) in transgenic CRND8 mice.17
Extensive investigations in the past have revealed that the kinetics and structural dynamics of Aβ aggregation are influenced by membrane components and constitution. Liposomes containing anionic phospholipids, sphingomyelins, and sterols have been reported to cause rapid amyloid formation.22-25 Furthermore, aggregation rates of Aβ are modulated differently depending on the surface charges on small unilamellar vesicles (SUVs) containing negatively charged phosphoglycerol (PG) and neutral phosphocholine (PC) or on large unilamellar vesicles (LUVs) containing a mixture of PC/PS or PC/PG lipids.26,27 In addition, other membrane components such as cholesterol and gangliosides have also been known to influence membrane Aβ interaction.28,29 Accelerated membrane disruption by Aβ has been observed in ganglioside-containing model membrane systems.13 Aβ has been observed to preferentially bind to regions containing GM1 in raft-like lipid vesicles enriched with GM1 and cholesterol and augment aggregation,24,30-35 and morphologically distinct Aβ fibril polymorphs have been known to form in the presence of GM1 containing model vesicles.36 Furthermore, cell membrane and its components also facilitate membrane disruption and pore formation by Aβ aggregation.13,37,38 However, since the formation of low-molecular weight oligomers is influenced the most by lipids, it remains unclear whether oligomerization and membrane disruption are discrete events that are temporally decoupled from one another or the two have a synergistic relationship. To address this question, here we enriched GM1 ganglioside in varying amounts on LUVs and SUVs of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), reconstituted lipid rafts (LR), and total brain extract lipid (TBE) to see the dynamics of Aβ42 (referred from here on as Aβ) oligomerization and membrane disruption. We observed that a high percentage of GM1 ganglioside doping generates distinct low-molecular-weight oligomers of Aβ that can be isolated in a lipid-complexed form. More interestingly, oligomerization and membrane disruption seem to be cooperative. Numerical simulations uncover that GM1 doping forms trimeric oligomers that form pores, which further assists aggregation of oligomers toward high-molecular-weight species. On the contrary, addition of preformed aggregates to the vesicles forms pores in a more abrupt manner. These results provide new mechanistic insights into the possible role of gangliosides in the membrane surface toward synergistic Aβ oligomerization and toxicity.
EXPERIMENTAL PROCEDURES
Materials.
Size exclusion chromatography (SEC) column (Superdex-75 HR 10/30) was purchased from GE Life Sciences (Marlborough, MA). DMPC, 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine (POPE), sphingomyelin, cholesterol, and total brain lipid extract (TBE) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL. Tris base, Tris hydrochloride, and SDS were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific, Inc. (Waltham, MA). Other chemicals, reagents, and consumables were purchased from either VWR, Inc. (Radnor, PA) or Thermo Fisher Scientific, Inc. (Waltham, MA). The monoclonal antibody Ab5 was obtained from Dr. Levites at the University of Florida (Gainesville, FL). Liposome extrusion system was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). The plasmid, pET-Sac Aβ(M1–42) was obtained from ADDGENE.
Recombinant Aβ Expression and Purification.
Recombinant Aβ (Aβ(M1–42)) was recombinantly expressed in BL21(DE3) PlysS Star Escherichia coli cells. Cells were grown in LB broth and induced for 16 h and subsequently harvested and lysed by sonication to obtain inclusion bodies. Inclusion bodies were resuspended in 6 M urea and filtered with a 0.2 μm hydrophilic PVDF filter. The filtrate was directly subjected to high-performance liquid chromatography (HPLC) using a Zorbax C8 column preheated at 80 °C. Purified Aβ was lyophilized and stored at −80 °C for further use.39 To obtain monomers, HPLC-purified Aβ (0.5–1 mg) was resuspended in 490 μL of nanopure water and allowed to stand for 30 min. NaOH was then added to the mixture to a final concentration of 10 mM and was allowed to stand for 10 min at room temperature. The mixture was then loaded onto a Superdex-75 HR 10/30 SEC column pre-equilibrated with 20 mM Tris pH 8.00 and attached either to an AKTA FPLC system (GE Healthcare, Buckinghamshire) or a BioLogic DuoFlow system (BioRad) fractionating at a flow rate of 0.5 mL/min at 25 °C. Monomers were eluted between fractions 24 and 28. The buffer pH maintained at 8.0 provides slightly better yields than that at pH 7.0. The molar concentration of each monomer fraction was determined by UV absorbance collected using a Cary 50 UV–Vis spectrometer (Agilent Technologies, Inc., Santa Clara, CA) and subsequently applying Beer–Lambert’s law ε = 1450 cm−1 M−1 at 276 nm. The purity and integrity of the peptide were confirmed using matrix-40 assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry. Purified monomers were stored at 4 °C and used for the experiment within the same day of purification.
Liposome Preparation.
LUVs were prepared as done previously.13,41-44 DMPC, POPC/POPE/ sphingomyelin/cholesterol in 33/33/10/20% by weight (for LR), and TBE liposomes were constructed from a 1:1 chloroform/methanol solution of lipids stocks. The solution was gently dried under nitrogen flow and then placed in a vacufuge with desiccant overnight to further evaporate any residual solvent. The dried lipid film was then rehydrated with either a buffer solution (10 mM phosphate or 20 mM tris buffer, pH 8.0) or a buffer solution containing doping agent 10–50% (by weight) of GM1 or GM3 for DMPC or LR and TBE, respectively, to yield a final concentration of 1 mg/mL. The hydrated lipids were vortexed for 1–1.5 h at 37 °C and subjected to 15 freeze–thaw cycles with liquid nitrogen and water bath at ~50 °C. The resulting solution was extruded 25 times through a 200 nm (for LUVs) polycarbonate nucleopore membrane filter (Whatman) with a mini extruder to obtain unilamellar vesicles. The size of the vesicles was confirmed with DLS collected using a Zetasizer Nano S instrument (Malvern, Inc., Worcestershire, U.K.) as described below.
Thioflavin-T Kinetics.
Aβ monomers (25 or 10 μM) was incubated with 0.3 mg/mL DMPC/TBE/raft-like reconstituted (LR) LUVs/SUVs in either 20 mM Tris or 10 mM sodium phosphate buffer (pH 8.00) in the presence of 50 mM NaCl and 50 μM ThT. Physiological 150 mM salt concentration was avoided due to precipitation of lipids and to avoid parallel on-pathway fibrillation reactions of Aβ42. Kinetics were read in corning black 96-well plates in a Biotek Synergy well plate reader at 37 °C monitored every 30 min with shaking for 10 s before every read. The fluorescence data were processed and normalized from 0 to 1 using Origin 8.0 as done earlier.45
Isolation of Oligomers.
Aβ oligomers were generated by incubating freshly purified Aβ monomer (25μM) with the specified LUVs/SUVs in the conditions listed below: 0.3 mg/mL DMPC LUVs; 0.3 mg/mL lipid raft LUVs; 0.3 mg/mL TBE LUVs. Additionally, 50 mM NaCl was added to all reactions prior to incubation at 37 °C under quiescent conditions for 5 h. The samples were then pelleted by centrifugation at 18 000g for 20 min, and the soluble supernatant was subjected to SEC as described above. Fractions of 500 μL were collected, and Aβ oligomers were found to be in the 16–17th fraction. The molar concentration after isolation was determined by UV–vis spectroscopy, as described above. Samples were either stored at 4 °C and used for experimentation within 72 h or lyophilized and kept at −80 °C for extended storage prior to experimentation. The size of the oligomers was confirmed with DLS.
Electrophoresis and Immunoblotting.
Samples were run in partial-denaturing, SDS PAGE gel by diluting samples in 1× Laemmli loading buffer, without boiling, onto either 4–12% NuPAGE or 4–20% Bis-Tris BioRad TGX gels. For molecular weight determination, prestained molecular-weight markers (Novex Sharp Protein Standard, Life Technologies) were run in parallel with samples on the gel. Proteins were transferred onto a 0.2 μm immunoblot membrane (BioRad) using a thermo scientific transfer cassette for 15 min. Subsequently, the immunoblot with protein was boiled for 1 min in a microwave oven in 1× PBS, followed by blocking for 1.5 h at 25 °C in 1× PBS containing 5% nonfat dry milk with 1% Tween 20. Blots were then probed overnight at 4 °C with a 1:6000 dilution of Ab5 monoclonal antibody, which detects amino acids 1–16 of Aβ. Following primary incubation, blots were probed with a 1:6000 dilution of anti-mouse, horseradish peroxidase-conjugated secondary antibody for 1.5 h at 25 °C before being imaged using a Super Signal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific).
Dye Leak Assay.
Lipid stocks, DMPC, POPE/POPC/sphingomyelin/cholesterol (in proportions described above for LR), and TBE stored in 1:1 chloroform/methanol were dried under liquid nitrogen and vacuufuged overnight as described previously13,43,44,46 and rehydrated with 15 mM 6-carboxy-fluorscein (6 -FITC) in 10 mM sodium phosphate buffer pH 8.00. The rehydrated lipid–dye mixture was subjected to 15 freeze–thaw cycles and subsequent extrusion with 200 nm polycarbonate nucleopore membrane filter (Whatman) with on a mini extruder to obtain dye-filled LUVs. The excess dye in the solution was separated from dye-filled LUVs using 7 kDa desalting columns preequilibrated with 10 mM sodium phosphate buffer pH 8.00 centrifuged at 500g for 30 s. The size of the LUVs was confirmed using DLS as mentioned below. The leakage of dye was confirmed by comparing the fluorescence intensity (λEx: 490nm; λEm: 595nm) of intact dye-encapsulated liposomes and 2- to 3-fold increased intensity upon complete rupture of liposome upon addition of 0.2% Triton X-100.44 The percent dye leak is calculated by the difference between the dye leak intensity of LUVs with the protein and blank divided by the difference between the dye leak intensity of LUVs with Triton X-100 and blank LUVs as done previously.13
Dynamic Light Scattering (DLS) Analysis.
DLS was obtained with a Zetasizer Nano S instrument (Malvern, Inc., Worcestershire, U.K.) by running a total of 15 runs for 10 s each for every sample after equilibration for 30 s. The data were exported using manufacturer’s software and plotted using OriginLab 8.0
Fourier Transform Infrared (FTIR) Spectroscopy.
FTIR spectra were obtained with an Agilent FTIR instrument (Cary-630) with dial-path accessory. Lyophilized protein samples (Aβ isolated oligomers/monomers) (45–50 μg) were resuspended in 5 μL of D2O and samples were scanned from 1500 to 1800 cm−1 at a resolution of 4 cm−1. A total accumulation of 1024 spectral scans was obtained per sample, and data were processed by subtracting the blank D2O spectra and baseline correction using OriginLab8.
Circular Dichroism (CD) Spectroscopy.
CD spectra of the oligomers/monomers were obtained using a Jasco (Easton, MD) J-815 spectropolarimeter. An average of 6–16 spectral scans were obtained in the far-UV region (260–190 nm) at a rate of 50 nm/min (8 s response time, 1 nm bandwidth, 0.1 nm data pitch). Savitzky-Golay algorithm with a convolution width of 15 was used to smoothen the spectra in the Jasco spectrum analysis program.
Cell Viability XTT Assay.
Cell viability was measured using 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay kit (Biotium) using our previously established protocol.45 Briefly, experiments were carried out in human neuroblastoma SH-SY5Y cell lines (ATCC) grown in DMEM and Ham’s F12K (1:1) medium containing 10% FBS and 1% penicillin/streptomycin. Cells were maintained at incubator conditions set to a temperature of 37 °C and 5.5% CO2. The cells were seeded at a density of 30 000 per well in a clear bottom 96-well plate 24 h prior to oligomers incubation. Oligomers were incubated at 2.5 μM concentration for 24 h prior to performing XTT assay. All experiments were done in triplicates, and statistical analysis and data processing were carried out using Origin 8.0.
Model Simulations.
We have used a system of ordinary differential equations (ODEs) to simulate and fit the experimental data as shown previously.47-49 Parameter estimation is solved as an optimization problem in ODE systems by minimizing the objective function that calculates the deviation between simulated and experimental data. Optimization methods can be gradient-independent or gradient-based; the former method is theoretically less susceptible to stochastic noise than the latter method. Hence, in this case of stochastic optimization, we have used gradient-free metametaheuristics50,51 as our algorithm for parameter estimation. The following modeling abstraction was used in this study.
Parameter Estimation in ODE Models.
Parameter estimation in ODE models is a popular method used in the biochemical domain. It is solved as an optimization problem where an objective function that calculates the deviation between the simulated and experimental data is minimized. A plethora of prior works have used heuristic global and local search methods in stochastic systems to estimate optimal parameter values from this optimization problem. However, the embedded noise can bring in errors in the gradient estimation in such methods. On the other hand, derivative-free optimization methods avoid the computation of derivatives of the objective function, and hence, they are less prone to stochastic noise. Our Aβ competing pathways model using the ensemble kinetic simulation-based method also needs a gradient-free parameter optimization algorithm as it consists of several biochemical reactions from the competing pathways that makes the gradient-based models less effective. In fact, we reported the performance of different optimization methods for the competing pathways simulation in ref 50 and validated that derivative-free methods work best in this context.
Parameter optimization algorithms are either deterministic or stochastic. In the case of stochastic optimization, metaheuristics are generally used.52 In such cases, other heuristics are guided and modified to solve the optimization problem that can provide better solutions than local optimization algorithms. Metaheuristics are based on intensification, i.e., searching in the local space and ensuring a good solution is found, and also diversification, i.e., generation of the best results in the global space. The algorithm converges with the achievement of global optimality when a good balance between these two components is ensured. As a result, metaheuristics are more computationally expensive and might fail in some cases. Various types of metaheuristics are differential evolution, simulated annealing, genetic algorithms, harmony search, bee algorithms, and so on.50,53-55
Parameter Identifiability.
Parameter identifiability relies on the idea of agreement between the experimental data and the parameterized model-predicted observables. It is measured by an objective function, known as the weighted sum of squared errors (SSE53) and maximum likelihood estimator is used to estimate the parameters. The likelihood profile of the ith parameter pi is LP(pi), and the fitted parameters are , for I = 1, …, m by considering m number of parameters as follows
The objective function SSE(pi) with respect to all other parameters, i.e., pi≠j in the neighborhood of the original estimated parameter value , is reoptimized to calculate the likelihood profile for each fitted parameter. The confidence level is a probability that reports the confidence interval, i.e., the interval within which the true value of a parameter is located. The parameter is identifiable only when the reoptimized SSE(pi) exceeds a specific confidence level within the same range. For n data points, the likelihood contour CLC and likelihood ratio CLR are determined as
where the upper α-critical values for the F-ratio and Chi-squared distribution are regarded as and Xα2, respectively.56 In some cases, the count of parameters in the model is greater than the number of experimental points used for fitting or the mapping of the internal model states to the observed values can be insufficient. In such cases, with a dearth of data points or insufficient mapping of the observed and the experimental values, the parameters do not properly rely on the data and are called nonidentifiable.
Model Assumptions.
To prevent overfitting, we have considered a simple model with a minimal set of reactions to represent the competing pathway simulation. Ai and F are used to represent the on-pathway i-mer and fibrils, respectively. BAi, CAi, DAi, and EAi represent the off-pathway i-mers that belong to the first, second, third, and fourth pores, respectively, whereas, L represents the 0 or 50% GM1-doped pseudomicelles. The assumptions of our simple model are as follows:
A12 or on-pathway 12-mer is considered equivalent to the nucleus of an on-pathway species. In our model, we assumed that the prenucleation species vary in size from A1 to A11 while A12 is the nucleus. We assume that all postnucleated species in the on-pathway are represented as on-pathway fibrils, F.79
Similarly, BAi, CAiDAi and EAi (i = j, …, 24) are considered to be the smaller off-pathway oligomers that aggregate on the respective pores through secondary nucleation. These kinetically trapped oligomers lack the energy to aggregate further. This model was already validated in Rana et al.47
The kinetically trapped 24-mer for each of the respective pores is assumed to be highly unstable and dissociates into an on-pathway fibril and a 12-mer of that respective pore as shown in Rana et al.78
The total ThT signal is the sum of the on-pathway ThT signal and the total weighted concentration of the smaller off-pathway kinetically trapped oligomers.
The FITC signal is the weighted sum of the concentrations of all of the off-pathway oligomers.
Given the initial concentration of monomers (A1), the model estimates the best values for all of the rate constants considered in the reactions; the concentration of liposomes (L) was also estimated by the parameter fitting algorithm since the molar mass of L varies experimentally. Additionally, the number of pores and minimum on-pathway oligomer size capable of creating pores on the liposome surface are considered as model parameters; these two parameters are varied for different combinations (as discussed in the next section) to identify the best combination that fit the experimental data in terms of the reported SSE.
Control (On-Pathway) Reactions.
First, the ThT aggregation data for control Aβ (10 μM) in the absence of liposomes were mapped to the concentration of the on-pathway fibrils. The forward and backward nucleation and the forward and backward fibrillation rate constants were calculated, and SSE was recorded. For modeling simplicity, aggregates beyond 12-mers were considered to be fibrils and the rate constants were modeled for on-pathway fibril formation (Table S1), which is the basis for modeling other reactions. The following reactions were considered
| (1) |
The on-pathway reaction fluxes are as follows
To reduce the number of species considered in the on-pathway reactions, we abstracted all postnucleation species (A12 onwards) as on-pathway fibrils denoted by F. Here, the forward and backward rate constants ( and , respectively) for all prenucleation reactions were considered the same to reduce the number of parameters and based on our prior work.17,47 Similarly, the forward and backward rate constants ( and , respectively) for all postnucleation reactions were also considered the same. The intensity of the ThT data was mapped to the sum of the concentrations of the on-pathway fibrils as follows:
where kon is a scaling constant used for fitting ThT intensity (ThTon) to the fibril concentration.
Reactions Involving Liposomes.
First, the forward and backward prenucleation and postnucleation rate constants computed from the controls were used for the on-pathway species for oligomerization reactions (shown below), which facilitated the reduction of the number of estimable parameters in this phase. To model the oligomerization reactions, both the ThT aggregation kinetics data and the FITC dye-leak data were considered. In our models, the initial concentrations of the liposomes were varied as their molar mass could not be precisely calculated. A sequential, multiple pore formation model was considered as opposed to one expanding pore although both are possible; since we do not have enough evidence to discount one over the other, we chose the former arbitrarily. Two possible scenarios were considered based on experimental evidence: pores formed by (i) a pre-nucleation oligomer (Aj) and/or (ii) on-pathway fibrils (i.e., a postnucleated oligomer denoted as F). These were modeled for the first pore BAi by
| (2) |
| (3) |
while for subsequent holes (CAi, DAi, EAi), this is modeled by reactions of type
| (4) |
| (5) |
Here, Aj denotes the minimum prenucleation oligomer that can form a pore and the value of the i-mer was identified through our parameter fitting mechanism. Moreover, the values of the rate constant combination (k1con and k1con′) can suggest which mechanism is more likely for the first pore formation (i.e., through prenucleation oligomer or postnucleation fibrils); similarly, the rate constant combination (k2con and k2con′) suggests which mechanism is more likely for the second pore formation and so on. The cooperativity between pore formation and aggregation was captured by considering further oligomerization reactions assisted by the edge of the pore up to 24-mers denoted by reactions of type
| (6) |
We additionally consider a bulk oligomerization in the presence of fibrils by reactions of type
| (7) |
The cooperativity among pores is captured by (k1con and k1con′), etc. For example, note that the second hole (CAi) is formed only after the first pore is formed. This is ensured by reactions of type
| (8) |
| (9) |
where the presence of BAi is necessary for the formation of Ci. Additionally, if the rate constant pair (k1con and k1con′) is less than (k2con and k2con′), this will suggest higher cooperativity in hole formation; in other words, the second hole formation (controlled by k2con and k2con′) is faster than the formation of the first hole. Finally, to map the concentration values to the ThT and FITC signals, we considered all of the species weighted by the oligomer size for FITC signal (denoted by summations ranging from 1 to 24) while only weighted values of postnucleated oligomers were considered in the ThT signal (denoted by summations ranging from 12 to 24).
The reactions considered for the oligomerization phase are as follows
| (10) |
| (11) |
| (12) |
| (13) |
where BAi denotes the first hole with an oligomer of size i-mers and L is the liposome.
The reaction fluxes involving the first pore are as follows
| (14) |
| (15) |
| (16) |
| (17) |
The reaction fluxes involving the second pore are as follows
| (18) |
Fitting of data for 0% GM1-enriched liposomes were done only with the above reactions. The number of pores was varied from one to three, and the first oligomer size was varied from two (since 1-mer cannot produce off-pathway species) to six for a total of 3 × 5 = 15 combinations in the case of 0% GM1, as shown in Table S6. The combination with two pores and first oligomer size of two recorded the least SSE, i.e., 0.0473. Here, Ci denotes the second hole with an i-mer. Similarly, for the following reactions, Di denotes the third hole, Ei denotes the fourth hole, and so on.
| (19) |
| (20) |
| (21) |
| (22) |
The reaction fluxes involving the third pore are as follows
| (23) |
| (24) |
| (25) |
| (26) |
The reaction fluxes involving the fourth pore are as follows
| (27) |
Fitting of data for 0% GM1-enriched liposomes were done only with the above reactions, i.e., four pores.
Then, ThT data were mapped to the sum of the concentrations of the on-pathway fibrils and all of the off-pathway oligomers beyond 12-mers as follows
The FITC dye leak data were mapped to the concentration of the off-pathway oligomers as follows
The model is extendable to any number of holes, and the curve fitting with the experimental data infers the optimal number of holes to be considered. For example, for 0% GM1, we first experimented with four holes (Bi, Ci, Di, Ei), which was then systematically reduced to one hole (Bi); in this case, two holes gave the best global fit with ThT and FITC dye leak data.
RESULTS
TBE and LR LUVs Enriched with GM1 Ganglioside Promote the Formation of Aβ Oligomers.
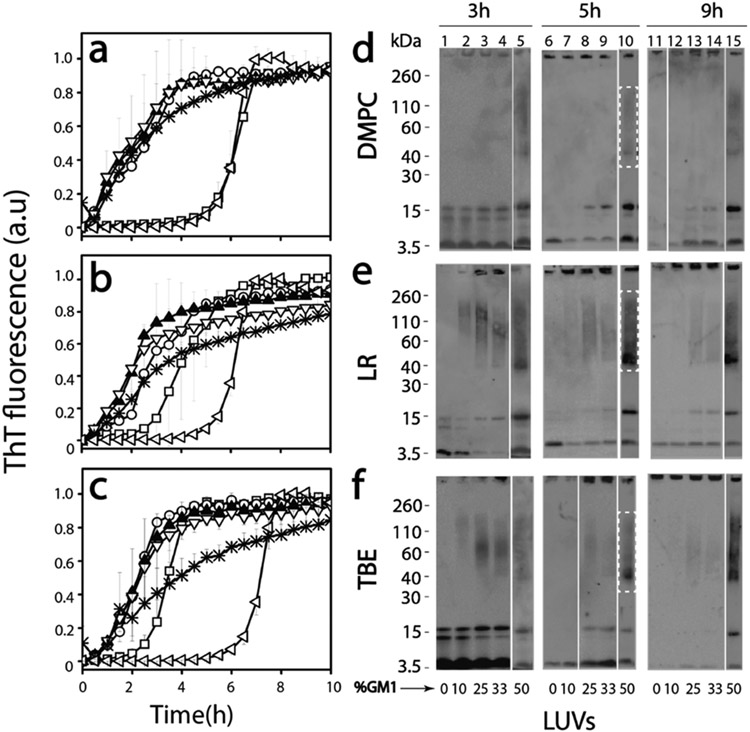
First, to obtain insights into the effect of GM1 ganglioside-enriched vesicles on the temporal dynamics of Aβ aggregation, freshly purified, seed-free Aβ monomers (25 μM) buffered in 20 mM Tris (Ph 8.0) containing 50 mM NaCl and 50 mM thioflavin-T (ThT) were incubated with 0.3 mg/mL pre-prepared LUVs of DMPC, LR, or TBE individually at 37 °C. The three liposomal systems were chosen to capture a diverse set of membrane compositions. The liposomes were made by increasing the amount of GM1 gangliosides added (% by weight) from 0 to 50%. The aggregation kinetics was monitored by ThT fluorescence on a 96-well plate reader. The control Aβ in the absence of liposome (◁ in Figure 1a-c, respectively) followed a typical sigmoidal pattern with a lag time of ~5 h. Surprisingly, incubation of Aβ with LUVs of DMPC without GM1 showed similar or slightly decreased lag time to that of Aβ in the absence of vesicles (□; Figure 1a). Incubation of Aβ with LR or TBE LUVs without GM1 gangliosides showed decreased lag times of 2–3 h (□; Figure 1b,c). However, LUVs enriched with increasing amounts of GM1 ganglioside showed a significant decrease in lag times and an increase in fluorescence intensity within 2 h of incubation (○, ▲, ▽, and * for 10, 25, 33, and 50% GM1 doping, respectively; Figure 1a-c). With micellar systems, we have previously reported the generation of discrete Aβ oligomer.17 Therefore, to investigate whether similar oligomer generation is facilitated by GM1-enriched LUVs, the incubated reactions were monitored by immunoblotting in parallel. The samples from the reactions in Figure 1a-c were electrophoresed under partial denaturing conditions after 3, 5, and 9 h of incubation and visualized via immunoblotting using the monoclonal antibody Ab5. Aβ incubated with unenriched LUVs showed monomeric, dimeric, and trimeric bands after 3 h (lane 0; Figure 1d-f). After 5 and 9 h, the dimeric and trimeric bands disappeared with a concomitant appearance of high-molecular-weight bands that failed to enter the gel that are possibly fibrils (5 and 9 h, 0%; Figure 1d-f). Similarly, incubation of Aβ with increasing amounts of GM1 also showed dimer and trimer bands along with monomers in case DMPC LUVs (Figure 1d: 10, 25, and 33%) upon 3 h of incubation. The transition from dimer and trimer to higher-molecular-weight fibrils has been observed to decrease with the increase in GM1 percentage of the LUVs. Furthermore, faint oligomeric bands ranging from 40 to 160 kDa emerged after 5 h of incubation (50%; Figure 1d), which were stable till 9 h of incubation (Figure 1d; lane 15). Immunoblots of Aβ incubated with increasing GM1-enriched LUVs in LR and TBE showed dimer and trimer bands for LUVs with lower GM1 content (Figure 1e,f). The intensity of these oligomeric bands was greater for 50% GM1 containing LR and TBE LUVs compared to 25 or 33%. Also, these oligomers were present up to 9 h of incubation (Figure 1e,f). In all samples, bands near 4.6 and >260 kDa were also observed, which indicate the presence of monomers and high-molecular-weight fibrils, respectively (Figure 1d-f). Furthermore, while the increase in GM1 percentage showed a gradual increase in the oligomer band intensity for TBE and LR LUVs, oligomer bands in DMPC LUV-incubated samples were only visible for 50% GM1-doped samples. These results suggest that both vesicles composition in conjunction with an increase in GM1 ganglioside content plays a role in the generation of Aβ oligomers.
Figure 1.
(a–c) Normalized ThT fluorescence kinetics of buffered 25 μM Aβ without (◁; control) or with DMPC (a), LR (b), and TBE (c) LUVs each of them enriched with 10 (○), 25 (▲), 33 (▽), and 50 (*) % GM1 ganglioside (by wt.) or without (□) GM1 in the presence of 50 mM NaCl in 20 mM tris buffer pH 8.00. (d–f) Partially denaturing SDS PAGE immunoblots of 25 μM Aβ in the presence of DMPC, LR, and TBE LUVs, respectively, enriched without or with 10, 25, 33, and 50% GM1. Gels were run at intervals of 3, 5, and 9 h, respectively.
Secondary Structure Transitions during Aggregation Reveal Potential Intermediates in GM1 Enriched Samples.
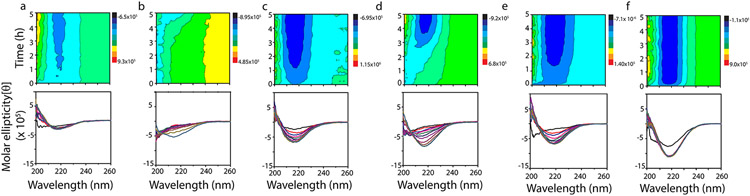
To investigate conformational changes of Aβ during aggregation, far-UV CD spectroscopy was used. Samples containing LUVs with no or enriched with 50% GM1 gangliosides from Figure 1 were analyzed. To see whether there are differences in the early oligomer formation among different LUVs due to change in their surface characteristics, we monitored the reaction for the initial 5 h. In all reactions as expected, Aβ showed conformational conversion from a random coil to β-sheet upon aggregation (Figure 2), consistent with the ThT fluorescence and immunoblot results in Figure 1. Aβ incubated with DMPC LUVs enriched with 50% GM1 showed an immediate conversion from random coil (λmin = 200 nm) to β-sheet (λmin = 218 nm; dark blue region in the contour plot) (Figure 2a), while those with no GM1 showed slow conversion from a persistent random coil structure to β-sheet (Figure 2b), also consistent with ThT aggregation kinetics. LUVs of LRs enriched with 50% GM1 cause a more rapid transition of random coil to β-sheet than the unenriched ones (Figure 2c,d). Aβ incubated with LUVs of TBE however shows a gradual transition from a random coil to α-helical within the first 1.5 h followed by the transition to a β-sheet signal (Figure 2e,f). The α-helical intermediate was more apparent in TBE LUVs enriched with 50% GM1 (Figure 2e). In addition, it is evident that the β-sheet intensity at 218 nm for DMPC-incubated samples was comparatively lower than those for the LR and TBE LUV-incubated samples (Figure 2a,c,e). This is consistent with the oligomer band intensity observed in the immunoblots (Figure 1d-f). Furthermore, among the GM1-enriched vesicles, DMPC showed the slowest transition from random coil to β-sheet and TBE was the only one in which an α-helical intermediate was observed.
Figure 2.
Far-UV CD contour and time course plots (every 30 min for up to 5 h) for buffered (20 mM tris buffer pH 8.0, 50 mM NaCl) 25 μM monomeric Aβ incubated with LUVs of 50 and 0% GM1-enriched DMPC (a and b, respectively), 50 and 0% GM1-enriched LR (c and d, respectively), and 50 and 0% GM1-enriched TBE (e and f, respectively) at 37 °C under quiescent conditions.
Aβ Oligomers Isolated from GM1-Enriched Vesicles Show Distinctive Biophysical Characteristics.
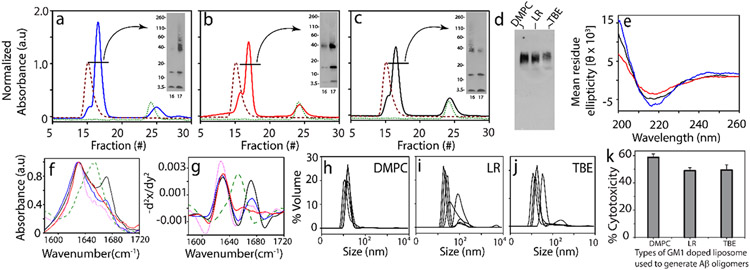
To see if oligomers generated in the presence are isolable, freshly purified Aβ monomers (25 μM) were incubated with 50% GM1 LUVs (DMPC, LR, and TBE respectively) under 37 °C quiescent conditions. To isolate the oligomeric species from the reactions containing monomeric or fibrillar species, samples after 5 h were centrifuged at 18 000g for 20 min. The supernatant was then subjected to fractionation by size exclusion chromatography (SEC) using a Superdex-75 column. Fractionation of all three LUVs showed two peaks near the void volume with a small first peak at fraction 15 and a larger one between fractions 16 and 18 (solid line; Figure 3c). In addition, a third peak at an included volume at fraction 24 was observed (solid line; Figure 3c).
Figure 3.
(a–c) SEC chromatogram for isolation of Aβ oligomers generated in the presence of 50% GM1-enriched DMPC (blue), LR (red), and TBE LUVs (black), respectively; LUV control at 0.3 mg/mL (red) and control Aβ (green) at 5 h (inset: SDS PAGE immunoblots of SEC-isolated oligomer fraction 16–17). (d) Native PAGE immunoblot for SEC-isolated Aβ oligomers generated in the presence of 50% GM1-enriched DMPC, LR, and TBE LUVs, respectively, (e) CD spectra of fraction 17 of SEC-isolated Aβ oligomers generated in the presence of 50% GM1-enriched DMPC (blue), LR (red), and TBE LUVs (black), respectively. (f) FTIR spectra of SEC-isolated Aβ oligomers generated in the presence of 50% GM1-enriched TBE (black), DMPC (blue), and LR (red) LUVs; homotypic Aβ fibril (pink) and BSA control (green), respectively. (g) Negative of double derivative of the FTIR spectra (Figure 3f-j) DLS for fraction 17 of SEC-isolated Aβ oligomers generated in the presence of 50% GM1-enriched DMPC, LR, and TBE LUVs, respectively. (k) XTT assay performed on SHY5Y neuroblastoma cells upon incubation with isolated Aβ oligomers from 50% GM1-enriched DMPC, LR, and TBE LUVs, respectively, expressed in terms of % of dead cells. n = 3 independent cell cultures on isolated oligomers, statistically significant at p < 0.05 based on one-way ANOVA.
The first fraction at 15 corresponded to free vesicles (purple dashed line; Figure 3c), while the fraction at 24 corresponded to monomeric Aβ (green dotted line; Figure 3c). After 5 h of incubation, the aliquots of fractions 16 and 17 were subjected to electrophoresis under partial denaturing conditions (with 1% SDS and without sample boiling) and visualized by immunoblotting (Figure 3a-c, insets). In all samples, monomeric bands near 4.6 kDa, multiple oligomeric bands near 15 kDa, and 38–110 kDa and some high molecular weight, nonfibrillar bands that failed to enter the gel were visible in the immunoblots. Fractions from DMPC showed more disperse oligomers between 38 and 110 kDa (Figure 3a), while those from LR and TBE showed more compact band patterns centered around 38 kDa corresponding to ~8-mer. To see whether the low-molecular-weight oligomeric bands observed were due to dissociation of the oligomers due to SDS treatment during electrophoresis, the isolated oligomeric samples were run under nondenaturing conditions (no SDS, no sample boiling) in PAGE gel followed by immunoblotting. By doing so, homogeneous oligomeric bands without the presence of any other band were observed suggesting that the monomeric and other low oligomer bands were probably due to dissociation of oligomers by SDS treatment (Figure 3d). The secondary structure of isolated oligomers was investigated by far-UV CD and FTIR spectroscopy. All oligomers were found to have β-sheet structure evident from minima at 217 nm in far-UV CD with the exception of those derived from LR, which showed a small extent of helical structure (shoulder at 222 nm) (Figure 3e). Similarly, the amide I band of the FTIR signature was investigated to gain more insights into the type of β-sheet (parallel or antiparallel) within the oligomers generated with GM1-enriched LUVs. The absorbance maxima for all three oligomer samples showed a band near 1630 cm−1 without a 1690 cm−1 band indicative of a parallel β-sheet structure57 (Figure 3f). However, only oligomers generated with TBE and DMPC LUVs enriched with 50% GM1 showed a second band near 1671 cm−1 (Figure 3f,g), which is indicative of turn conformation.58,59 It can be inferred that TBE-catalyzed oligomers have some structural differences compared to those from LR or DMPC-generated LUVs, which parallels the observation of conformational transitions with TBE LUVs (Figure 2). The size of isolated oligomers analyzed by DLS revealed that these oligomers are 18–20 nm in diameter (Figure 3h-j). However, the presence of polydispersity in these oligomers suggests the possible co-elution of some amounts of LUVs with the oligomers. Indeed, we found that only about 0.05 mg/mL (~17%) of the starting amount of lipids remains associated with the isolated oligomer (Figure S3). Furthermore, Aβ oligomers were tested for their toxicities on SHY5Y neuroblastoma cells by XTT assay.21 All three oligomers were toxic with 50% cell viability. DMPC-generated oligomers had a slightly higher cytotoxicity compared to LR and TBE (Figure 3k). Overall, these data suggest that LUVs with different surface properties and charge could lead to the generation of structurally distinctive neurotoxic oligomers as observed in micellar systems.21
GM1-Enriched Vesicles Induce Cooperative Aβ Oligomerization and Membrane Pore Formation.
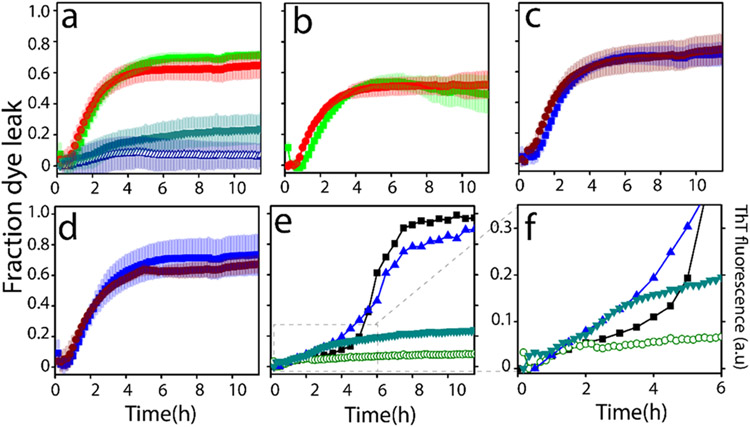
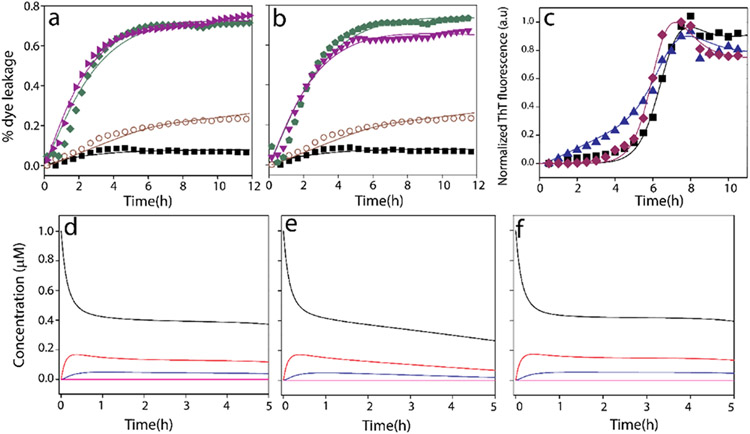
Aβ incubated with the LUVs of TBE enriched with 50% GM1 showed the presence of possible conformationally different oligomer intermediate (Figures 2 and 3). To further investigate whether these oligomers also induce membrane pore formation, dye leak assay was performed using 6-carboxy-fluorescein (6-FAM) encapsulated within TBE vesicles. Freshly purified Aβ monomers (10 μM in 20 mM Tris pH 8.0) were incubated with 6-FAM loaded TBE LUVs, and fluorescence was monitored in a 96-well plate for 12 h at 37 °C (see Experimental Procedures). Aβ monomers when incubated with TBE LUVs without GM1 showed no discernable membrane disruption (Δ; Figure 4a). However, when incubated with 50% GM1-enriched TBE LUVs, Aβ monomers showed increased FITC fluorescence at ~2 h of incubation that continued to increase up to 20% during the next 9 h (▼; Figure 4a), suggesting steady disruption of the vesicles. In contrast, preformed fibrils isolated from Aβ incubations with TBE LUVs without or with 50% GM1 showed exponential increases in pore formation (■, green; ●, red; Figure 4a). A similar pattern was also observed when the same fibrils were sonicated (■, green; ●, red; Figure 4b).
Figure 4.
Vesicle dye leak analysis monitored by 6-carboxyfluorescein (6-FAM) dye on: (a) TBE LUVs incubated with 10 μM Aβ monomers (Δ) or 2 μM isolated Aβ fibrils generated from the same liposomes (■, green); 50% GM1-enriched TBE LUVs incubated with 10μM Aβ monomers (▼, green); or 2 μM isolated Aβ fibrils generated from 50% GM1-enriched liposomes (●, red); (b) TBE LUVs incubated with 2 μM sonicated Aβ fibrils generated in the presence TBE liposomes (■, green) or 50% GM1-enriched TBE LUVs incubated with 2 μM sonicated Aβ fibrils generated in the presence of 50% GM1-enriched liposomes (●, red); (c) TBE LUVs incubated with 2 μM isolated Aβ fibrils generated in the absence of liposomes (■, blue) or 50% GM1-enriched LUVs incubated with 2 μM isolated Aβ fibrils generated in the absence of liposomes (●, brown); (d) samples in (c) but sonicated; (e) ThT fluorescence of 10μM Aβ monomers in the presence of 50% GM1-enriched TBE LUVs (◀, blue) and 50% GM3-enriched TBE LUVs (■, black); 6-FAM dye leakage of 50% GM1-enriched TBE LUVs (▼, green) and 50% GM3-enriched TBE LUVs (○, green) in the presence of 10 μM Aβ monomers; (f) zoomed-in image of Figure 4e showing the initial 6 h of the reaction.
When fibrils generated from Aβ in the absence of liposomes were incubated on TBE LUVs without or with 50% GM1 showed an exponential increase in pore formation either unsonicated (■, blue; ●, brown; Figure 4c) or sonicated (■; blue, ●, brown; Figure 4d). Together, it is evident that high-molecular-weight fibrils are able to disrupt the membranes more efficiently than low-molecular-weight oligomers, but there are several possible caveats as discussed further below. Nevertheless, the data clearly suggest that GM1 ganglioside enrichment promotes oligomers vis-à-vis membrane disruption as opposed to the unenriched liposomes. To specifically see whether glycoform distributions on the gangliosides have an effect on these properties as we had seen before with Aβ-glycopolymer interactions,60,61 TBE liposomes were also enriched with GM3 gangliosides which have significant sugar distribution differences with GM1 (Figure S2). Incubation of 50% GM3-enriched TBE LUVs showed no or minimal leakage of dye upon incubation of Aβ monomers with was observed (○; Figure 4e). This clearly indicated the specificity of interactions; while GM3-enriched liposomes showed a sigmoidal pattern of aggregation without pore formation, reactions with GM1-enriched samples showed aggregation and concomitant pore formation during the first 3 h (Figure 4e,f).
Numerical Simulations Uncover Insights into the Cooperativity in Oligomerization and Membrane Disruption.
To obtain more details on the effects of GM1 on Aβ oligomerization and membrane disruption, ordinary differential equation (ODE)-based numerical simulations were used. The basis of the models along with the reaction abstractions formulated is detailed in Experimental Procedures. We used Scatter Search optimization algorithm to fit the experimental data as it has been earlier shown that metaheuristic algorithms like scatter performs better than other algorithms to fit the Aβ aggregation.51 Sum of squared errors (SSE) was used as a metric to evaluate the models, and COmplex PAthway SImulator (COPASI)50 to solve the mathematical models. Briefly, oligomerization was considered up to the formation of 12-mers, beyond which all aggregates were considered “fibrils” for modeling simplicity. An additional reason was to identify the low-molecular-weight oligomeric species that are responsible for membrane disruption and not those that were formed late. Individual global fits of the ThT and the FITC dye-leak data of Aβ aggregation on TBE liposomal with varying gangliosides were performed. Specifically, the modeling was directed at understanding the temporal mechanisms and cooperativity by which Aβ aggregated and caused membrane disruption as a function of GM1 enrichment of liposomes.
To do so, two potential pathways of pore formation upon Aβ oligomerization on membrane surfaces were considered. Upon aggregation that generates a single pore, Aβ can then elongate/aggregate on the edge of the pore assisted by the exposed membrane components. This can either result in further enlargement of the pore or aggregates could initiate the second pore formation and so on. Since both mechanisms involve cooperativity, we arbitrarily chose the latter mechanism to model due to the lack of experimental evidence for either mechanism.
Model simulations are based on the rate constants computed (Tables S1-S5). A global fit of the ThT aggregation and FITC dye leak data showed a good fit and agreement with the experimental data (Figure 5a-c). The models showed that an increase in GM1 percentage results in more pores on the membrane surface. For example, it was found out that in the case of liposomes with 50% GM1, twice the number of pores are formed, to that of TBE liposomes with 0% GM1. Computation of various aggregate sizes formed temporally during aggregation suggested that dimeric Aβ was responsible for pore formation in the absence of GM1, while trimeric Aβ was responsible for 50% enriched GM1 liposomes. In our reaction system, the smallest and the largest oligomers were considered to be 2- and 6-mers for the control in the absence of GM1 and 2 and 8 for GM1-enriched liposomes as the lower and upper bounds. The oligomer responsible for causing the initial pore was computed by sweeping the oligomer size to fit the FITC data; this gave the least SSE (Tables S1-S5) for dimer (for 0% GM1 control) and 3-mer (for 50% GM1). However, caution needs to be exercised on the oligomer size as our models considered only limited number of species in the system, and therefore, it is possible that different oligomers within a small-molecular-weight range effect membrane disruption. The extent pore formation in the presence and absence of gangliosides is the key focus of our models and the consequent insights derived. Furthermore, cooperativity in pore formation and aggregation was also evident from the rate constants obtained. It can be observed that for 0% GM1, (k1con, k1con′) is less than (k2con, k2con′) suggesting higher cooperativity especially for 50% GM1, which showed greater cooperativity in both creation of subsequent pores and aggregation of the oligomers. This aspect of cooperativity separates the mechanism by which LUVs in the absence of GM1 form pores but do not promote robust pore-forming fibrils. However, the concentrations of the oligomers responsible for pore formation during aggregation were low in the order of ~0.5 μM at 2–3 h of incubation (Figure 5d-f) that explains the difference in the rates of pore formation between the preformed fibrils and oligomers generated in situ.
Figure 5.
Computational fits of 6-carboxyfluorescein dye leak assay of TBE LUVs (a) with 50% GM1 and 10 μM Aβ monomers (○), without GM1 and 10 μM Aβ monomers (■, black), with 50% GM1 and 2 μM Aβ fibrils (▶, purple), without GM1 and 2 μM Aβ fibril (◆, green) (b) with 50% GM1 and 10 μM Aβ monomers (○), without GM1 and 10 μM Aβ monomers (■), with 50% GM1 and 2 μM sonicated Aβ fibrils (▼, purple), without GM1 and 2 μM sonicated Aβ fibril ( , green). (c) Normalized ThT fluorescence kinetics of buffered 10 μM Aβ without (■; control) or with TBE LUVs each of them enriched with 50 (▲, blue) % GM1 ganglioside (by wt.) or without (◆, brown) GM1 in the presence of 50 mM NaCl in 10 mM sodium phosphate buffer pH 8.00; Aβ monomers (A1 (black), oligomers A2 (red) and A3 (blue), and fibrils F (pink)) distribution plots for first 5 h from the start of reactions of Aβ monomers with TBE LUVs, (d) no GM1, (e) 50% GM1 or (f) without LUVs.
, green). (c) Normalized ThT fluorescence kinetics of buffered 10 μM Aβ without (■; control) or with TBE LUVs each of them enriched with 50 (▲, blue) % GM1 ganglioside (by wt.) or without (◆, brown) GM1 in the presence of 50 mM NaCl in 10 mM sodium phosphate buffer pH 8.00; Aβ monomers (A1 (black), oligomers A2 (red) and A3 (blue), and fibrils F (pink)) distribution plots for first 5 h from the start of reactions of Aβ monomers with TBE LUVs, (d) no GM1, (e) 50% GM1 or (f) without LUVs.
DISCUSSION
Aggregate polymorphism is increasingly becoming known as a distinguishable feature among many AD patients.62-68 It is speculated that such polymorphic fibrils are in part responsible for the observed phenotypes. Since fibrils are the end products of templated aggregate growth, we hypothesize that the conformational differences and selection among low-molecular weight oligomers are key in determining the dominant fibril polymorph. In this regard, we have previously shown that membrane lipids and surfactants modulate Aβ aggregation pathways to generate conformationally distinct oligomers capable of propagating their structure toward fibrils.17,48 Specifically, we have observed that Aβ oligomers generated in the presence of lipid micelles are structurally distinct and cause distinct phenotype in APP transgenic mice.17 Similarly, a plethora of studies point toward the effect of other membrane model systems like liposomes on the aggregation of Aβ and membrane disruption.69-74 For example, properties of membranes such as surface charges 19,75 curvature,76,77 composition,78 etc. have been shown to have profound effects on aggregation. Similarly, Aβ aggregates are known to form pores and channels in the membrane that are attributable to their biophysical characteristics.13,37,79,80 However, it remains unclear whether and how low-molecular-weight Aβ oligomers are generated upon its interaction with liposomal surfaces and whether such a generation is dependent on the membrane components. Furthermore, the coupling between oligomerization and membrane pore formation remains unclear.
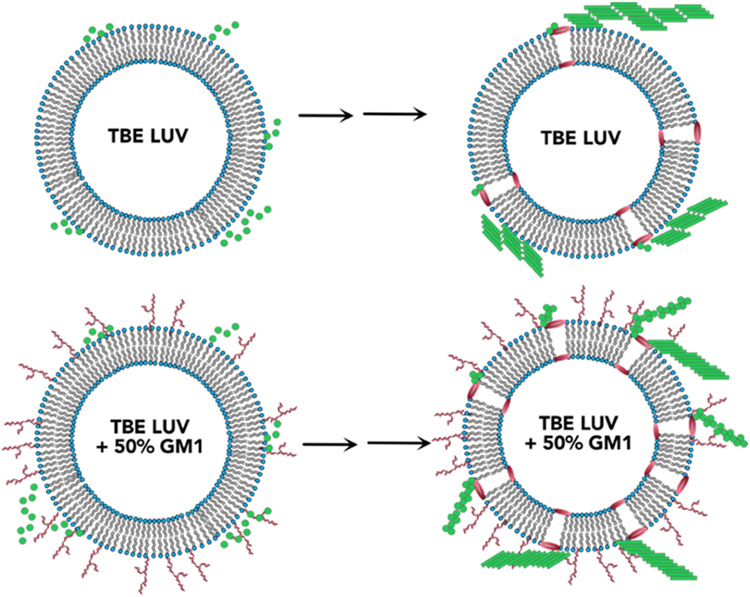
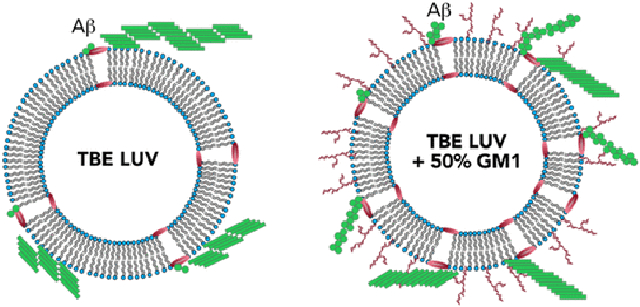
The study presented here shows that alteration of surface characteristics, especially to the degree of charge density by dilution with neutral GM1 gangliosides decisively affect the oligomerization of Aβ (Figure 6). Our findings support the previous studies by Williams et al. that membrane damage is induced by soluble monomeric and oligomeric Aβ in membrane vesicles and that presence of GM1 influences the binding of Aβ on the membrane and membrane permeation.81-83 However, we also observed significant dye-leak or membrane disruption upon addition of preformed fibrils and sonicated fibrils regardless of GM1 content. This indicates that the membrane disruption mechanisms can differ depending on the size and structure of Aβ aggregates. While LUVs without or very low amount of GM1 accelerates the aggregation of Aβ to form higher-molecular-weight fibrils in the first 5 h of incubation, LUVs enriched with high concentration of GM1 causes oligomerization of Aβ on the LUV surface, kinetically trapping the Aβ oligomers. Three different types of LUVs used, i.e., DMPC, LR, and TBE, that have different compositions were found to augment aggregation of Aβ but also showed oligomerization when enriched with 50% GM1. This implicates the significance of gangliosides in Aβ aggregation as previous studies have established in many reports.34,41,84-86 Furthermore, the GM1-enriched TBE LUVs showed somewhat modified ThT aggregation kinetics that correlated with a partially helical conformational state at an early aggregation stage. More importantly, these temporal changes also coincided with membrane disruption brought upon only by high GM1-enriched samples. It must be borne in mind that although pore formation is one of the mechanisms that explains dye leak, but other mechanisms such as membrane reorganization and deformation cannot be discounted. However, we consider the more probable mechanism of pore formation and explain it as a function of GM1 enrichment. This phenomenon may be due to altered lipid packaging or dilution of anionic charge density or both due to GM1 enrichment. It is noteworthy that the pore formation was not abrupt but rather slow and progressive in nature but only showed ~20% at the end of 11 h of incubation with Aβ monomers (Figure 4). By contrast, fibrils and sonicated fibrils of Aβ generated in the presence and absence of liposome showed rapid pore formation. Furthermore, the addition of fibrils generated from GM1-enriched liposomes too showed rapid pore formation. Two possible explanations can be rendered for these observations; (a) the oligomers formed during aggregation on the liposome surface are present in low concentrations (as computed to ~0.5 μM; Figure 5e) to effect rapid change in pore formation kinetics, or (b) not oligomers but high-molecular-weight fibrils effect membrane disruption more effectively. A third explanation could be that the mechanisms of pore formation could be either numerous small pores or a few large pores for monomers aggregating on the surface or when preformed aggregates are added, respectively. Yet another key observation is that the oligomerization and membrane disruption is also selective to the nature of sugar distributions on gangliosides. While GM1 ganglioside promotes membrane pore formation, GM3 does seem to have such an effect, nor does it promote oligomers. Collectively, the data indicate that in early stages of oligomer formation, membrane selectivity is important, to generate conformationally distinct and toxic species; however, in later stages, when the higher-molecular-weight species are already formed, the membrane is ruptured in a different mechanism than while Aβ oligomerization. Furthermore, oligomerization and pore formation seem to be cooperative and coupled to one another. As mentioned earlier, our lab and others have reported the formation of structurally distinct Aβ aggregates with equally distinct biophysical properties in the presence of GM1 gangliosides. Recently, Matsuzaki and his group reported the formation of amyloid tape fibrils with mixed parallel and antiparallel β-sheet structure in the presence of GM1 in membrane model systems.84 Therefore, it can be concluded that membrane lipid composition along with GM1 content play a role in generating oligomers within a distinct molecular weight range. This inference is further supported by our CD time course data, which show that the secondary structure of the intermediates and pathway of oligomerization are different for liposomes enriched with GM1 gangliosides. In this report, we further these findings to uncover that GM1 ganglioside enrichment in the membrane vesicles not only promotes oligomerization but also induces membrane disruption in a cooperative manner. This suggests that aggregation and modulation of membrane dynamics are coupled to one another, and such a coupling displays strong membrane compositional bias. Such cooperative mechanisms may lead to the generation of conformationally distinctoligomers and other aggregates, which are templates for the formation of polymorphic fibrils observed in patient brains.
Figure 6.
Schematic of conclusions drawn from this study showing the effect of GM1 enrichment in liposomes.
CONCLUSIONS
The findings presented in this study bring forth the significance of membrane components, specifically GM1 ganglioside, in the oligomerization of Aβ and concomitant membrane disruption. Although oligomerization and membrane disruption have been independently studied in numerous investigations prior, the coupling and synergy between the two events have seldom been investigated in greater detail. Results from this report suggest that Aβ oligomerization and membrane disruption are highly cooperative processes and are facilitated by the concentration-dependent presence of GM1 gangliosides. The data also indicate that the mechanism and extent of membrane disruption vary depending on the size and structure of Aβ aggregates. These data may reflect potential mechanisms by which Aβ and lipid components synergistically trigger cellular dysfunction via membrane disruption well before the emergence of high-molecular-weight fibrils, further portending the significance of lipid-associated low-molecular-weight oligomers in pathology.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the following agencies for their financial support: National Institute of Aging (1R56AG062292-01), National Institute of General Medical Sciences (R01GM120634) and the National Science Foundation (NSF CBET 1802793) to V.R and (NSF CBET 1802588) to P.G. They also thank the National Center for Research Resources (5P20RR01647-11) and the National Institute of General Medical Sciences (8 P20 GM103476-11) from the National Institutes of Health for funding through INBRE for the use of their core facilities.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00495.
DLS data showing size of LUVs for DMPC, LR, and TBE; structures of GM1 and GM3 gangliosides; quantification of lipid-associated within isolated oligomers; and tables showing parametric values for computing rate constants for model simulation of Aβ oligomerization (PDF)
The authors declare no competing financial interest.
Contributor Information
Jhinuk Saha, Department of Chemistry and Biochemistry, School of Mathematics and Natural Sciences, University of Southern Mississippi, Hattiesburg, Mississippi 39406, United States.
Priyankar Bose, Department of Computer Science, Virginia Commonwealth University, Richmond, Virginia 23220, United States.
Shailendra Dhakal, Center for Molecular and Cellular Biosciences, University of Southern Mississippi, Hattiesburg, Mississippi 39406, United States.
Preetam Ghosh, Department of Computer Science, Virginia Commonwealth University, Richmond, Virginia 23220, United States.
Vijayaraghavan Rangachari, Department of Chemistry and Biochemistry, School of Mathematics and Natural Sciences, University of Southern Mississippi, Hattiesburg, Mississippi 39406, United States; Center for Molecular and Cellular Biosciences, University of Southern Mississippi, Hattiesburg, Mississippi 39406, United States.
REFERENCES
- (1).Shoji M; Golde TE; Ghiso J; Cheung TT; Estus S; Shaffer LM; Cai X-D; McKay DM; Tintner R; Frangione B; Younkin SG (1992) Production of the Alzheimer Amyloid β Protein by Normal Proteolytic Processing. Science 1992, 258, 126–129. [DOI] [PubMed] [Google Scholar]
- (2).Selkoe DJ; Podlisny MB; Joachim CL; Vickers EA; Lee G; Fritz LC; Oltersdorf T Beta-amyloid precursor protein of Alzheimer disease occurs as 110-to 135-kilodalton membrane-associated proteins in neural and nonneural tissues. Proc. Natl. Acad. Sci. U.S.A 1988, 85, 7341–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chow VW; Mattson MP; Wong PC; Gleichmann M An overview of APP processing enzymes and products. Neuromol. Med 2010, 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cohen SIA; Linse S; Luheshi LM; Hellstrand E; White DA; Rajah L; Otzen DE; Vendruscolo M; Dobson CM; Knowles TPJ Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 9758–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Arosio P; Knowles TPJ; Linse S On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys 2015, 17, 7606–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Garai K; Frieden C Quantitative analysis of the time course of Aβ oligomerization and subsequent growth steps using tetramethylrhodamine-labeled Aβ. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 3321–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Li S; Selkoe DJ A mechanistic hypothesis for the impairment of synaptic plasticity by soluble Aβ oligomers from Alzheimer’s brain. J. Neurochem 2020, 154, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Walsh DM; Klyubin I; Fadeeva JV; Cullen WK; Anwyl R; Wolfe MS; Rowan MJ; Selkoe DJ Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 2002, 416, 535–539. [DOI] [PubMed] [Google Scholar]
- (9).Hong W; Wang Z; Liu W; O’Malley TT; Jin M; Willem M; Haass C; Frosch MP; Walsh DM Diffusible, highly bioactive oligomers represent a critical minority of soluble Aβ in Alzheimer’s disease brain. Acta Neuropathol. 2018, 136, 19–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Stroud JC; Liu C; Teng PK; Eisenberg D Toxic fibrillar oligomers of amyloid- have cross- structure. Proc. Natl. Acad. Sci. U.S.A 2012, 109, 7717–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).De Felice FG; Velasco PT; Lambert MP; Viola K; Fernandez SJ; Ferreira ST; Klein WL Aβ oligomers induce neuronal oxidative stress through an N-methyl-D-aspartate receptor-dependent mechanism that is blocked by the Alzheimer drug memantine. J. Biol. Chem 2007, 282, 11590–11601. [DOI] [PubMed] [Google Scholar]
- (12).Soto C Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci 2003, 4, 49. [DOI] [PubMed] [Google Scholar]
- (13).Sciacca MFM; Kotler SA; Brender JR; Chen J; Lee D; Ramamoorthy A Two-Step Mechanism of Membrane Disruption by Aβ through Membrane Fragmentation and Pore Formation. Biophys. J 2012, 103, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Rangachari V; Dean DN; Rana P; Vaidya A; Ghosh P Cause and consequence of Aβ – Lipid interactions in Alzheimer disease pathogenesis. Biochim. Biophys. Acta, Biomembr 2018, 1860, 1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rangachari V; Moore BD; Reed DK; Sonoda LK; Bridges AW; Conboy E; Hartigan D; Rosenberry TL Amyloid-β(1–42) Rapidly Forms Protofibrils and Oligomers by Distinct Pathways in Low Concentrations of Sodium Dodecylsulfate. Biochemistry 2007, 46, 12451–12462. [DOI] [PubMed] [Google Scholar]
- (16).Kumar A; Bullard RL; Patel P; Paslay LC; Singh D; Bienkiewicz EA; Morgan SE; Rangachari V Non-Esterified Fatty Acids Generate Distinct Low-Molecular Weight Amyloid-β (Aβ42) Oligomers along Pathway Different from Fibril Formation. PLoS One 2011, 6, No. e18759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Dean DN; Das PK; Rana P; Burg F; Levites Y; Morgan SE; Ghosh P; Rangachari V Strain-specific fibril propagation by an Aβ dodecamer. Sci. Rep 2017, 7, No. 40787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kumar A; Paslay LC; Lyons D; Morgan SE; Correia JJ; Rangachari V Specific soluble oligomers of amyloid-β peptide undergo replication, and form non-fibrillar aggregates In interfacial environments. J. Biol. Chem 2012, 287, 21253–21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Khondker A; Alsop RJ; Rheinstädter MC Membrane-accelerated amyloid-β aggregation and formation of cross-β sheets. Membranes 2017, 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Korshavn KJ; Satriano C; Lin Y; Zhang R; Dulchavsky M; Bhunia A; Ivanova MI; Lee YH; La Rosa C; Lim MH; Ramamoorthy A Reduced lipid bilayer thickness regulates the aggregation and cytotoxicity of amyloid-β. J. Biol. Chem 2017, 292, 4638–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Saha J; Dean DN; Dhakal S; Stockmal KA; Morgan SE; Dillon KD; Adamo MF; Levites Y; Rangachari V Biophysical characteristics of lipid-induced Aβ oligomers correlate to distinctive phenotypes in transgenic mice. FASEB J. 2021, 35, No. e21318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Terzi E; Hölzemann G; Seelig J Interaction of Alzheimer β-amyloid peptide (1– 40) with lipid membranes. Biochemistry 1997, 36, 14845–14852. [DOI] [PubMed] [Google Scholar]
- (23).Zhao H; Tuominen EKJ; Kinnunen PKJ Formation of Amyloid Fibers Triggered by Phosphatidylserine-Containing Membranes. Biochemistry 2004, 43, 10302–10307. [DOI] [PubMed] [Google Scholar]
- (24).Ege C; Lee KYC Insertion of Alzheimer’s Aβ40 peptide into lipid monolayers. Biophys. J 2004, 87, 1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).McLaurin J; Chakrabartty A Characterization of the interactions of Alzheimer β-amyloid peptides with phospholipid membranes. Eur. J. Biochem 1997, 245, 355–363. [DOI] [PubMed] [Google Scholar]
- (26).Accardo A; Shalabaeva V; Cotte M; Burghammer M; Krahne R; Riekel C; Dante S Amyloid β peptide conformational changes in the presence of a lipid membrane system. Langmuir 2014, 30, 3191–3198. [DOI] [PubMed] [Google Scholar]
- (27).Ke PC; Zhou R; Serpell LC; Riek R; Knowles TPJ; Lashuel HA; Gazit E; Hamley IW; Davis TP; Fändrich M; et al. Half a century of amyloids: past, present and future. Chem. Soc. Rev 2020, 49, 5473–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nicastro MC; Spigolon D; Librizzi F; Moran O; Ortore MG; Bulone D; Biagio PLS; Carrotta R Amyloid β-peptide insertion in liposomes containing GM1-cholesterol domains. Biophys. Chem 2016, 208, 9–16. [DOI] [PubMed] [Google Scholar]
- (29).Yu X; Zheng J Cholesterol Promotes the Interaction of Alzheimer β-Amyloid Monomer with Lipid Bilayer. J. Mol. Biol 2012, 421, 561–571. [DOI] [PubMed] [Google Scholar]
- (30).Hayashi H; Kimura N; Yamaguchi H; Hasegawa K; Yokoseki T; Shibata M; Yamamoto N; Michikawa M; Yoshikawa Y; Terao K; Matsuzaki K; Lemere CA; Selkoe DJ; Naiki H; Yanagisawa K A Seed for Alzheimer Amyloid in the Brain. J. Neurosci 2004, 24, 4894–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yanagisawa K; Odaka A; Suzuki N; Ihara Y GM1 ganglioside–bound amyloid β–protein (Aβ): A possible form of preamyloid in Alzheimer’s disease. Nat. Med 1995, 1, 1062–1066. [DOI] [PubMed] [Google Scholar]
- (32).Matsuzaki K; Horikiri C Interactions of amyloid β-peptide (1– 40) with ganglioside-containing membranes. Biochemistry 1999, 38, 4137–4142. [DOI] [PubMed] [Google Scholar]
- (33).Mori K; Mahmood MI; Neya S; Matsuzaki K; Hoshino T Formation of GM1 ganglioside clusters on the lipid membrane containing sphingomyeline and cholesterol. J. Phys. Chem. B 2012, 116, 5111–5121. [DOI] [PubMed] [Google Scholar]
- (34).Matsuzaki K; Kato K; Yanagisawa K Aβ polymerization through interaction with membrane gangliosides. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2010, 1801, 868–877. [DOI] [PubMed] [Google Scholar]
- (35).Flagmeier P; De S; Michaels TCT; Yang X; Dear AJ; Emanuelsson C; Vendruscolo M; Linse S; Klenerman D; Knowles TPJ; Dobson CM Direct measurement of lipid membrane disruption connects kinetics and toxicity of Aβ42 aggregation. Nat. Struct. Mol. Biol 2020, 27, 886–891. [DOI] [PubMed] [Google Scholar]
- (36).Matsuzaki K Aβ–ganglioside interactions in the pathogenesis of Alzheimer’s disease. Biochim. Biophys. Acta, Biomembr 2020, 1862, No. 183233. [DOI] [PubMed] [Google Scholar]
- (37).Lin H; Zhu YJ; Lal R Amyloid β Protein (1–40) Forms Calcium-Permeable, Zn2+-Sensitive Channel in Reconstituted Lipid Vesicles. Biochemistry 1999, 38, 11189–11196. [DOI] [PubMed] [Google Scholar]
- (38).Korshavn KJ; Bhunia A; Lim MH; Ramamoorthy A Amyloid-β adopts a conserved, partially folded structure upon binding to zwitterionic lipid bilayers prior to amyloid formation. Chem. Commun 2016, 52, 882–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yoo S; Zhang S; Kreutzer AG; Nowick JS An Efficient Method for the Expression and Purification of Aβ(M1–42). Biochemistry 2018, 57, 3861–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Vivekanandan S; Brender JR; Lee SY; Ramamoorthy A A partially folded structure of amyloid-beta(1-40) in an aqueous environment. Biochem. Biophys. Res. Commun 2011, 411, 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Chi EY; Frey SL; Lee KYC Ganglioside GM1-Mediated Amyloid-beta Fibrillogenesis and Membrane Disruption. Biochemistry 2007, 46, 1913–1924. [DOI] [PubMed] [Google Scholar]
- (42).MacDonald RC; MacDonald RI; Menco BPM; Takeshita K; Subbarao NK; Hu L Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim. Biophys. Acta, Biomembr 1991, 1061, 297–303. [DOI] [PubMed] [Google Scholar]
- (43).Kremer JJ; Murphy RM Kinetics of adsorption of β-amyloid peptide Aβ(1–40) to lipid bilayers. J. Biochem. Biophys. Methods 2003, 57, 159–169. [DOI] [PubMed] [Google Scholar]
- (44).Jimah JR; Schlesinger PH; Tolia NH Liposome Disruption Assay to Examine Lytic Properties of Biomolecules. Bio-Protoc. 2017, 7, No. e2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Dhakal S; Saha J; Wyant CE; Rangachari V αS Oligomers Generated from Interactions with a Polyunsaturated Fatty Acid and a Dopamine Metabolite Differentially Interact with Aβ to Enhance Neurotoxicity. ACS Chem. Neurosci 2021, 12, 4153–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Saha A; Mondal G; Biswas A; Chakraborty I; Jana B; Ghosh S In vitro reconstitution of a cell-like environment using liposomes for amyloid beta peptide aggregation and its propagation. Chem. Commun 2013, 49, 6119–6121. [DOI] [PubMed] [Google Scholar]
- (47).Rana P; Dean DN; Steen ED; Vaidya A; Rangachari V; Ghosh P Fatty Acid Concentration and Phase Transitions Modulate Aβ Aggregation Pathways. Sci. Rep 2017, 7, No. 10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Dean DN; Rana P; Campbell RP; Ghosh P; Rangachari V Propagation of an Aβ Dodecamer Strain Involves a Three-Step Mechanism and a Key Intermediate. Biophys. J 2018, 114, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ghosh P; Vaidya A; Kumar A; Rangachari V Determination of critical nucleation number for a single nucleation amyloid-β aggregation model. Math. Biosci 2016, 273, 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Rana P; Bose P; Vaidya A; Rangachari V; Ghosh P In Global Fitting and Parameter Identifiability for Amyloid-β Aggregation with Competing Pathways, IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), 2020; pp 73–78. [Google Scholar]
- (51).Glover F Future paths for integer programming and links to artificial intelligence. Comput. Oper. Res 1986, 13, 533–549. [Google Scholar]
- (52).Brasington A; Sacco C; Halbritter J; Wehbe R; Harik R Automated fiber placement: A review of history, current technologies, and future paths forward. Compos., Part C: Open Access 2021, 6, No. 100182. [Google Scholar]
- (53).Ghosh S; Ghosh P; Basu K; Das SK In GaMa: An Evolutionary Algorithmic Approach for the Design of Mesh-Based Radio Access Networks, IEEE Conference on Local Computer Networks 30th Anniversary (LCN’05), IEEE, 2005; p 8. [Google Scholar]
- (54).Raue A; Kreutz C; Maiwald T; Bachmann J; Schilling M; Klingmüller U; Timmer J Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 2009, 25, 1923–1929. [DOI] [PubMed] [Google Scholar]
- (55).Schaber J Easy parameter identifiability analysis with COPASI. Biosystems 2012, 110, 183–185. [DOI] [PubMed] [Google Scholar]
- (56).Özdemir D The Chi-Squared, F-Distribution and the Analysis of Variance. In Applied Statistics for Economics and Business, Springer, 2016; pp 169–190. [Google Scholar]
- (57).Zou Y; Li Y; Hao W; Hu X; Ma G Parallel β-sheet fibril and antiparallel β-sheet oligomer: New insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from FTIR spectroscopy. J. Phys. Chem. B 2013, 117, 4003–4013. [DOI] [PubMed] [Google Scholar]
- (58).Byler DM; Susi H Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers 1986, 25, 469–487. [DOI] [PubMed] [Google Scholar]
- (59).Susi H; Byler DM Resolution-Enhanced Fourier Transform Infrared Spectroscopy of Enzymes. In Methods in Enzymology, Elsevier, 1986; pp 290–311. [DOI] [PubMed] [Google Scholar]
- (60).Das PK; Dean DN; Fogel AL; Liu F; Abel BA; McCormick CL; Kharlampieva E; Rangachari V; Morgan SE Aqueous RAFT synthesis of glycopolymers for determination of saccharide structure and concentration effects on amyloid β aggregation. Biomacromolecules 2017, 18, 3359–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Bristol AN; Saha J; George HE; Das PK; Kemp LK; Jarrett WL; Rangachari V; Morgan SE Effects of Stereochemistry and Hydrogen Bonding on Glycopolymer-Amyloid-β Interactions. Biomacromolecules 2020, 21, 4280–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Condello C; Lemmin T; Stöhr J; Nick M; Wu Y; Maxwell AM; Watts JC; Caro CD; Oehler A; Keene CD; Bird TD; Duinen SG; van Lannfelt L; Ingelsson M; Graff C; Giles K; DeGrado WF; Prusiner SB Structural heterogeneity and intersubject variability of Aβ in familial and sporadic Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A 2018, 115, E782–E791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Qiang W; Yau W-M; Lu J-X; Collinge J; Tycko R Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 2017, 541, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Petkova AT; Leapman RD; Guo Z; Yau W-M; Mattson MP; Tycko R Self-propagating, molecular-level polymorphism in Alzheimer’s ß-amyloid fibrils. Science 2005, 307, 262–265. [DOI] [PubMed] [Google Scholar]
- (65).Huang D; Zimmerman MI; Martin PK; Nix AJ; Rosenberry TL; Paravastu AK Antiparallel β-sheet structure within the C-terminal region of 42-residue Alzheimer’s amyloid-β peptides when they form 150-kDa oligomers. J. Mol. Biol 2015, 427, 2319–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Lu J-X; Qiang W; Yau W-M; Schwieters CD; Meredith SC; Tycko R Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 2013, 154, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Paravastu AK; Leapman RD; Yau W-M; Tycko R Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A 2008, 105, 18349–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Paravastu AK; Qahwash I; Leapman RD; Meredith SC; Tycko R Seeded growth of β-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Yip CM; McLaurin J Amyloid-β Peptide Assembly: A Critical Step in Fibrillogenesis and Membrane Disruption. Biophys. J 2001, 80, 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Lau T-L; Ambroggio EE; Tew DJ; Cappai R; Masters CL; Fidelio GD; Barnham KJ; Separovic F Amyloid-β Peptide Disruption of Lipid Membranes and the Effect of Metal Ions. J. Mol. Biol 2006, 356, 759–770. [DOI] [PubMed] [Google Scholar]
- (71).Kotler SA; Walsh P; Brender JR; Ramamoorthy A Differences between amyloid-β aggregation in solution and on the membrane: insights into elucidation of the mechanistic details of Alzheimer’s disease. Chem. Soc. Rev 2014, 43, 6692–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Vander Zanden CM; Wampler L; Bowers I; Watkins EB; Majewski J; Chi EY Fibrillar and nonfibrillar amyloid beta structures drive two modes of membrane-mediated toxicity. Langmuir 2019, 35, 16024–16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Delgado DA; Doherty K; Cheng Q; Kim H; Xu D; Dong H; Grewer C; Qiang W Distinct membrane disruption pathways are induced by 40-residueβ -Amyloid peptides. J. Biol. Chem 2016, 291, 12233–12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Qiang W; Yau W-M; Schulte J Fibrillation of β amyloid peptides in the presence of phospholipid bilayers and the consequent membrane disruption. Biochim. Biophys. Acta, Biomembr 2015, 1848, 266–276. [DOI] [PubMed] [Google Scholar]
- (75).Sabaté R; Espargaró A; Barbosa-Barros L; Ventura S; Estelrich J Effect of the surface charge of artificial model membranes on the aggregation of amyloid β-peptide. Biochimie 2012, 94, 1730–1738. [DOI] [PubMed] [Google Scholar]
- (76).Sugiura Y; Ikeda K; Nakano M High Membrane Curvature Enhances Binding, Conformational Changes, and Fibrillation of Amyloid-β on Lipid Bilayer Surfaces. Langmuir 2015, 31, 11549–11557. [DOI] [PubMed] [Google Scholar]
- (77).Terakawa MS; Lin Y; Kinoshita M; Kanemura S; Itoh D; Sugiki T; Okumura M; Ramamoorthy A; Lee Y-H Impact of membrane curvature on amyloid aggregation. Biochim. Biophys. Acta, Biomembr 2018, 1860, 1741–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Lee J; Kim YH; T Arce F; Gillman AL; Jang H; Kagan BL; Nussinov R; Yang J; Lal R Amyloid β Ion Channels in a Membrane Comprising Brain Total Lipid Extracts. ACS Chem. Neurosci 2017, 8, 1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Jang H; Arce FT; Ramachandran S; Capone R; Lal R; Nussinov R β-Barrel Topology of Alzheimer’s β-Amyloid Ion Channels. J. Mol Biol 2010, 404, 917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Qi J-S; Qiao J-T Amyloid β-protein fragment 31–35 forms ion channels in membrane patches excised from rat hippocampal neurons. Neuroscience 2001, 105, 845–852. [DOI] [PubMed] [Google Scholar]
- (81).Williams TL; Day IJ; Serpell LC The effect of Alzheimer’s Aβ aggregation state on the permeation of biomimetic lipid vesicles. Langmuir 2010, 26, 17260–17268. [DOI] [PubMed] [Google Scholar]
- (82).Williams TL; Johnson BRG; Urbanc B; Jenkins ATA; Connell SDA; Serpell LC Aβ42 oligomers, but not fibrils, simultaneously bind to and cause damage to ganglioside-containing lipid membranes. Biochem. J 2011, 439, 67–77. [DOI] [PubMed] [Google Scholar]
- (83).Williams TL; Serpell LC; Urbanc B Stabilization of native amyloid β-protein oligomers by Copper and Hydrogen peroxide Induced Cross-linking of Unmodified Proteins (CHICUP). Biochim. Biophys. Acta, Proteins Proteomics 2016, 1864, 249–259. [DOI] [PubMed] [Google Scholar]
- (84).Okada Y; Okubo K; Ikeda K; Yano Y; Hoshino M; Hayashi Y; Kiso Y; Itoh-Watanabe H; Naito A; Matsuzaki K Toxic Amyloid Tape: A Novel Mixed Antiparallel/Parallel β-Sheet Structure Formed by Amyloid β-Protein on GM1 Clusters. ACS Chem Neurosci 2019, 10, 563–572. [DOI] [PubMed] [Google Scholar]
- (85).Wakabayashi M; Okada T; Kozutsumi Y; Matsuzaki K GM1 ganglioside-mediated accumulation of amyloid β-protein on cell membranes. Biochem. Biophys. Res. Commun 2005, 328, 1019–1023. [DOI] [PubMed] [Google Scholar]
- (86).Kakio A; Nishimoto S; Yanagisawa K; Kozutsumi Y; Matsuzaki K Cholesterol-dependent formation of GM1 ganglioside-bound amyloid β-protein, an endogenous seed for Alzheimer amyloid. J. Biol. Chem 2001, 276, 24985–24990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.