Abstract
Aminoacyl-tRNA synthetases (aaRSs) catalyze the ligation of amino acids to cognate tRNAs by consuming one molecule of ATP. Magnesium is essential for the enzymes’ activity. Certain class II aaRSs, such as lysyl-tRNA synthetase (LysRS) and seryl-tRNA synthetase (SerRS), recognize ATP together with three magnesium ions in the active site. The detailed role of how these magnesium ions facilitate the ATP recognition by the enzyme is unclear. Here, we report analyses of a crystal structure of human LysRS, in which the two enzymatic pockets of the LysRS dimer are in different states. One pocket is vacant of ATP, and the other is in an intermediate state of ATP recognition. Interestingly, only one magnesium ion instead of three is bound in both states. Compared with our previously solved LysRS structures, we proposed the order of binding for the three magnesium ions. These structures also reveal multiple intermediate ATP-bound states during the amino acid activation reaction, providing critical insights into the mechanisms of the magnesium-dependent enzyme activity of class II aaRSs.
Keywords: Aminoacyl-tRNA synthetase, aaRS, Lysyl-tRNA synthetase, LysRS, Metal activated enzyme, Crystal structure, Protein translation
Highlights
-
•
Crystal structures of human LysRS in different ATP binding states are analyzed.
-
•
The binding order of the three magnesium ions critical for LysRS activity is proposed.
-
•
These analyses provide insights into the mechanism of magnesium-dependent enzyme activity in Class II aaRSs.
1. Introduction
Aminoacyl-tRNA synthetases (aaRSs) are essential enzymes for all cellular life, playing a central role in the translation of the genetic code [1,2]. Despite the differences in the substrate amino acids, all aaRSs catalyze the shared two-step process to attach specific amino acids to their corresponding tRNAs [1]. In the first step, namely, the amino acid activation step, the enzyme binds a specific amino acid and an ATP molecule and catalyzes the formation of the aminoacyl-adenylate (aa-AMP) intermediate. In the second step, aaRS binds its cognate tRNA and further transfers the amino acid from the aa-AMP to the evolutionarily conserved 3’ adenosine terminus of the tRNA molecule [1].
Based on the two distinct architectures of active sites, the aaRSs, including 20 canonical synthetases and three later discovered synthetases, SepRS, PylRS, and ND-GluRS, are classified into two groups—named class I and class II aaRSs [3]. The catalytic domains of the class I enzymes are made up of the Rossmann fold, which is a structural motif commonly used for nucleotide binding [[4], [5], [6]]. In the active site of class I aaRSs, ATP is held in an extended conformation by the two signature motifs “HIGH” and “KMSKS”, which are located on top of the β-sheet of the Rossmann fold [[7], [8], [9]]. The amino acid is bound at a pocket formed by one side of the β-sheet, with its carboxylate group pointing to the α-phosphate of ATP. The two conserved motifs not only hold ATP rigidly before reaction but also stabilize the putative pentacoordinate transition state at the α-phosphate [10]. Class I aaRSs do not share any conserved metal binding site in the active site, which is distinct from the class II aaRSs.
Class II enzymes have a core made of a seven-stranded β-sheet with flanking α-helixes [[11], [12], [13]]. As a universal feature of class II aaRSs, the amino acid is bound at the deep pocket in the middle of the catalytic domain with its carboxylate group facing outside. ATP is bound in a bent conformation directly outside of the amino acid pocket [13]. Through this bent ATP conformation, the alpha phosphate is closely facing the carboxylate group and ready to activate the oxygen atom, under the facilitation of a nearby positively charged arginine residue and three commonly conserved magnesium ions [13]. The three magnesium binding sites are another important structure for class II aaRS to catalyze amino acid activation [14]. The magnesium ions assist the reaction cooperatively with the conserved arginine residue. In accordance with the presence of magnesium binding sites in the active site, class II aaRS are much more dependent on magnesium to catalyze aminoacylation and ATP/PPi exchange [15].
In addition to the necessity of magnesium involvement in the reaction catalysis, the binding order of the three magnesium ions and the potential role of magnesium ions in ATP recognition remain unclear. To understand these mechanisms, here we report detailed analyses of our recently and previously solved crystal structures of human LysRS as a typical example of class II aaRS. With these structures, the binding order of the magnesium ions is proposed. We also show several intermediate states of the ATP recognition process by LysRS, providing critical insights into the mechanisms of the magnesium-dependent enzyme activity of class II aaRSs.
2. Materials and methods
2.1. Structure analysis
Protein preparation, crystallization, and structure determination were reported elsewhere [[16], [17], [18], [19]]. The four human LysRS structures, including 4YCU (state 0: no magnesium), 6ILD (state 1: 1st magnesium ion, and state 2: 1st magnesium ion + flipped AMPcPP), 4DPG (state 3: 1st magnesium ion + AMPcPP), and 3BJU (complete state: 3 calcium ions + ATP), were analyzed with PyMol software (www.pymol.org). The “align” function in PyMol performs a sequence alignment followed by a structural superimposition and then carries out several cycles of refinement to reject structural outliers. Because this function does a good job on proteins with good sequence similarity (identity >30%), in this study, the structural superimpositions were performed using the “align” function with the software default parameters. After the structures were superimposed, the distances and angles were measured with the “measurement” wizard.
The direct results of the crystallographic experiments are electron density maps. We used the program “likelihood-weighted maps using a model” in the Phenix package to generate electron density maps in.ccp4 format for opening in PyMol [20]. The 2Fo-Fc map and the Fo-Fc map were generated and analyzed. The calculation of both maps involves combining the observed diffraction data (Fo) with the diffraction data calculated from the atomic model (Fc). The Fo-Fc map, also known as a difference map, is used to show which parts of a model are overfitted or missing, while the 2Fo-Fc map includes the Fo-Fc map and electron density around the model. The “isomesh” function in PyMol was used to create the electron density map figures. We contoured the 2Fo-Fc map at 1.0 sigma and the Fo-Fc map at ±3.0 sigma to show how well the electron density fits around the structural model.
3. Results
3.1. Crystal structure reveals asymmetric substrate binding in human LysRS
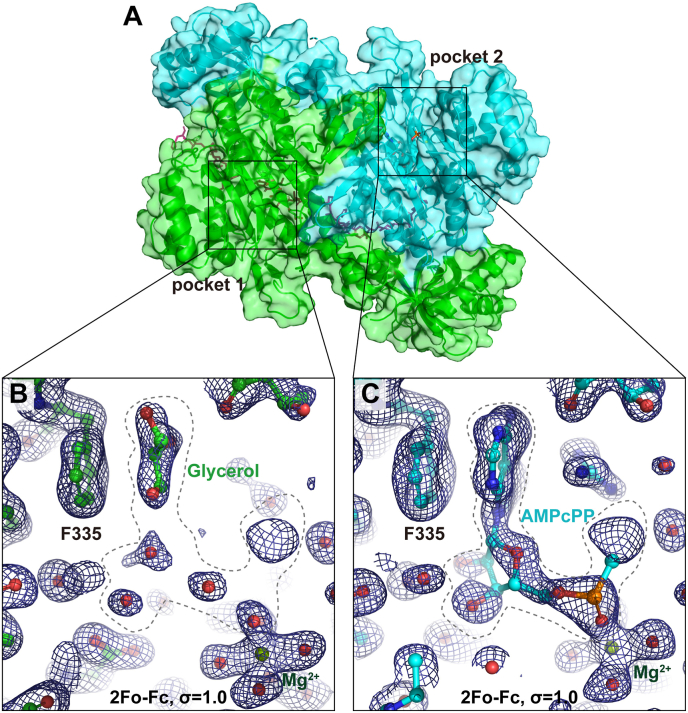
We recently solved a human LysRS crystal structure in complex with a peptide from the N terminus of the scaffold protein AIMP2 at 1.88 Å (Fig. 1A) [16]. The structure was from protein co-crystallized with 5 mM l-lysine, 5 mM ATP analogue α,β-methyleneadenosine-5′-triphosphate (AMPcPP), and 15 mM magnesium chloride. The resolved structure reveals complete electron density for l-lysine in the active center of each LysRS molecule (Fig. S1). However, the electron densities in the ATP binding pockets are apparently weaker than those for l-lysine (Fig. 1B and C). In particular, the densities in active center 1 are scattered into small pieces, which could only fit in solvent molecules (Fig. 1B). The densities in active center 2 are larger and allow fitting in a major part of AMPcPP, including the adenosine moiety and the α-phosphate group (Fig. 1C and Fig. S2A). The increased occupancy of AMPcPP in active center 2 tightens the loop of class II aaRS signature motif 2 by 0.6 Å, and stabilizes the side chain of the conserved Arg553 residue on class II aaRS signature motif 3 (Fig. S2B). These results show that the two LysRS catalytic centers in the same LysRS dimer are in different states for ATP binding (Fig. 1B–C and Fig. S2). It recalls the previous discovery of the functional asymmetry in E. coli LysRS explored by molecular dynamics [21], and is also consistent with the half-of-the-site models in several other dimeric aaRSs [[22], [23], [24], [25], [26]].
Fig. 1.
A human LysRS crystal structure with asymmetric substrate binding states. (A) Overall view of the structure (pdb: 6ILD) in one asymmetric unit. The LysRS dimer is shown as a transparent surface and cartoons (green and cyan). The cocrystallized AIMP2 N-terminal peptide is shown as pink sticks. (B–C) Electron density maps of the enzymatic pockets 1 and 2 of the LysRS dimer are shown as blue meshes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. A primary magnesium binding site in LysRS prior to ATP binding
Previous research showed that LysRS interacts with ATP together with three metal ions, such as magnesium, calcium, or manganese (complete state of substrate binding) [19,27], which was also revealed in other class II aaRS, for example, AlaRS, AspRS, and SerRS [[28], [29], [30]]. ATP contributed to an important part of the metal coordination complex formation through the oxygen atoms of the phosphate groups (Fig. 2A). Two conserved acidic residues had been previously reported to be critical to hold metal ions together with bound ATP, but whether they could bind metal ions without ATP was not known.
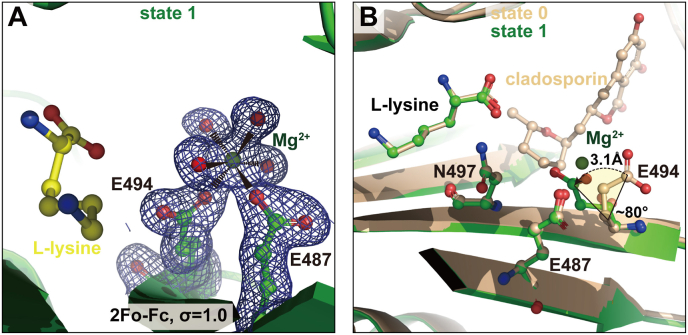
Fig. 2.
LysRS binds the first magnesium ion independent of ATP. (A) Electron density map of the first magnesium in LysRS pocket 1 (state 1). (B) Superimposition of the state 1 structure (green) with the previously solved LysRS structure (pdb: 4YCU, wheat) without magnesium (state 0), showing that the conserved residue Glu494 undergoes a conformational change to bind the first magnesium ion. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Notably, in pocket 1 of the present LysRS structure, the scattered densities share some similar patterns with pocket 2 at the ATP binding region (Fig. 1B and C). Although there is a possibility that the scattered densities might be attributed to trace amounts of ATP, the occupancy of ATP in pocket 1 must be very low. For the sake of expression, this state will be denoted as state 1. In contrast, a clear density for one magnesium ion in the active center is observed (Fig. 1, Fig. 2B). The magnesium is coordinated by two oxygen atoms from the carboxyl groups of Glu487 and Glu494 of LysRS together with four water molecules, presenting a typical octahedral arrangement (Fig. 2B).
We then compared this structure with one of our previously solved human LysRS structures, which was cocrystallized with l-lysine and the ATP competitive inhibitor cladosporin under a condition without magnesium (denoted as state 0). The conformation of residue Glu487 is almost identical in the two structures with a root-mean-square deviation (RMSD) of 0.139 Å at this residue, while the side chain of Glu494 shifts by 3.1 Å (Cδ-Cδ) and rotates by ∼80° (Fig. 2C). The conformational change of Glu494 is likely caused by the binding of the first magnesium, although we cannot rule out the possibility that it is caused by cladosporin. The binding of magnesium neutralizes the electrostatic repulsion that would have pushed the side chain of Glu494 away from Glu487 without the binding of magnesium ions.
Together, these results show that the two class II aaRS conserved acidic residues are able to bind a magnesium ion prior to ATP and might be a precondition for later proper ATP interaction.
3.3. Intermediate states of ATP recognition in LysRS
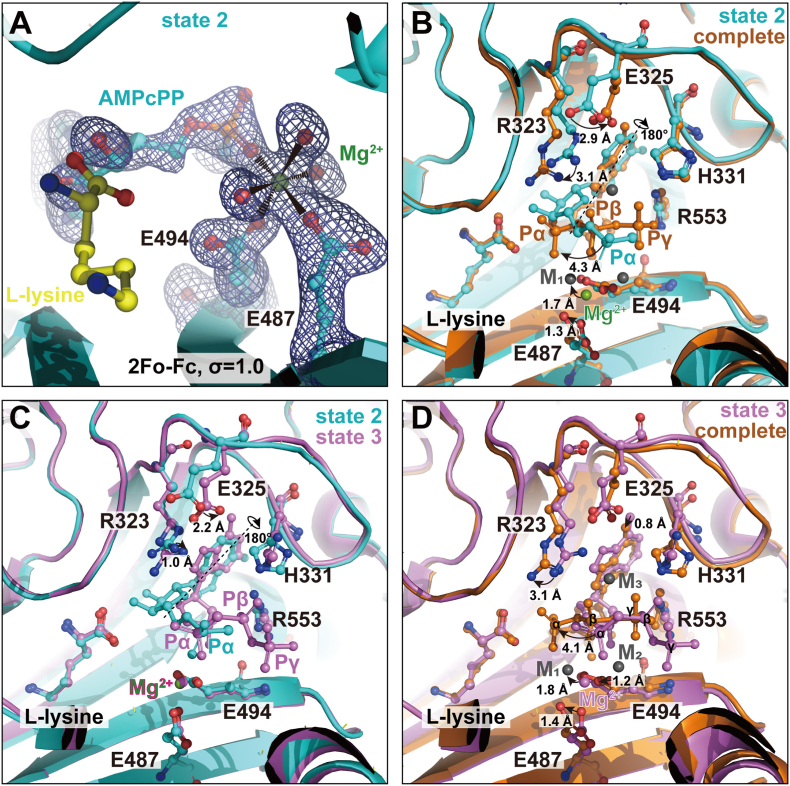
The primarily bound magnesium is also observed in active center 2 (Fig. 1, Fig. 3A). One oxygen atom of the α phosphate group replaces one water molecule and forms a coordination complex with magnesium together with residues Glu487 and Glu494 and three water molecules (Fig. 3A). The densities for the β and γ phosphate groups of AMPcPP are very weak (Fig. 1C), suggesting that the stable binding of the ligand by LysRS in this pocket is also not yet reached; thus, the crystal structure captures an intermediate state of ligand recognition (denoted as state 2).
Fig. 3.
Magnesium ions facilitate ATP recognition by LysRS. (A) Electron density map of the first magnesium and ATP analog, AMPcPP, in LysRS pocket 2 (state 2). (B) Superimposition of the state 2 structure (cyan) with the human LysRS structure (pdb: 3BJU, orange) at the complete state for ATP binding. (C) Superimposition of the state 2 structure (cyan) with the human LysRS structure (pdb: 4DPG, pink) at another intermediate state for ATP binding (state 3). (D) Superimposition of the state 3 structure (pink) with the complete state structure (orange) for ATP binding. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We therefore superimposed the intermediate state structure with the previously solved LysRS-ATP structure in the complete state (Fig. 3B). The primary magnesium in the intermediate state is 1.7 Å away from metal ion 1 in the LysRS-ATP complete structure. The α-phosphate group moves by 4.3 Å from intermediate state 2 to the complete state. Last, the adenine group is ∼180° flipped. Accordingly, the Glu325 residue moves by 2.9 Å to form an H–bond with the amide group of the flipped ATP, and the Arg323 residue extends by 3.1 Å to interact with the α phosphate in the complete state (Fig. 3B).
Interestingly, in another our previously solved LysRS structure, the α phosphate is located similarly to the structure of state 2, while the orientation of the adenosine moiety is similar to the complete state of ATP binding (Fig. 3C and D), presenting another intermediate state (denoted as state 3). Therefore, flipping over of the adenosine moiety into the complete state occurs earlier than the movement of the α phosphate to the final state. From state 2 to state 3, the Arg323 and Glu325 residues move closer to the flipping over of the adenosine moiety by 1.0 Å and 2.2 Å to form H–bonds (Fig. 3C). From state 3 to the complete state, the side chain of Glu487 shifts by 1.4 Å, and Glu494 shifts by 1.2 Å, the magnesium moves by 1.8 Å, and the α phosphate group moves by 4.1 Å deeper toward the carboxylate group of l-lysine in the active center. The relative position of the magnesium ion becomes between the α and β phosphate groups of ATP (Fig. 3D). Notably, even in state 3, the second and third magnesium ions are still not resolved, implying that the last two magnesium ions perch between state 3 and the complete state and that they may play a role in the transition of the states.
4. Discussion
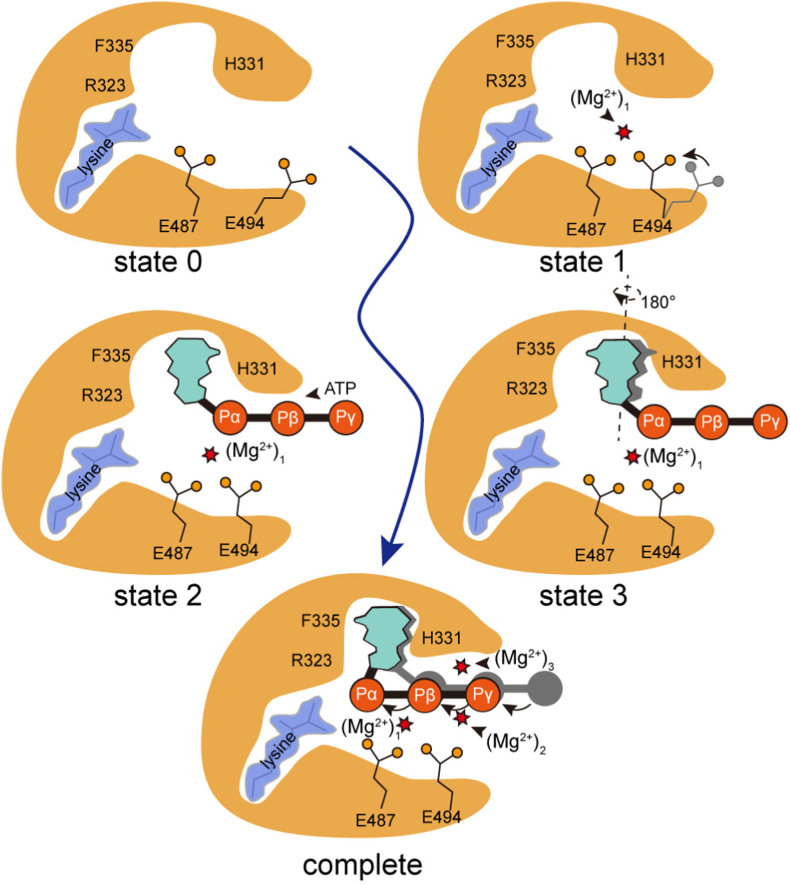
In summary, our recent human LysRS crystal structure captured two different states in ATP recognition (Fig. 1), in support of the alternating site manner, which was suggested to be a mechanism of class II aaRS to improve the catalytic efficiency [21]. In the process of ATP recognition, the conserved Glu494 rotates towards another conserved acidic residue Glu487, and they together coordinate the first magnesium (Fig. 2) prior to ATP binding. In the early state of ATP binding, all three phosphate groups are located outside of the first magnesium (Fig. 3). Then, the adenosine moiety flips by ∼180°, and the two conserved acidic residues Glu487 and Glu494 slightly shift towards the center of the active pocket. At the same time, the last two magnesium ions perch into the structure, and the phosphate groups move deeper in the pocket, which is the complete state for substrate recognition and ready for the amino acid activation reaction (Fig. 4).
Fig. 4.
Schematic diagram showing different states during the recognition of substrate ATP by LysRS.
The cobinding of metal ions and ATP to aaRS was also revealed in other class II aaRSs, for example, AlaRS, AspRS, and SerRS [[28], [29], [30]]. Similar to LysRS, the catalytic core of AspRS and AsnRS consists of a characteristic seven-strand antiparallel β-sheet surrounded by a number of α-helices (Fig. S3A). ATP and three magnesium ions are localized in similar places in their ATP-binding pockets, which are formed by conserved residues, including Arg323, Glu325, His331, Arg335 and Glu494 (Figs. S3B–D. Residues are numbered based on human LysRS). Although these residues have a systematic shift of 1–2 Å, their conformations are similar. Therefore, the mechanism shown in this study might be general to other class II aaRS, particularly aaRS that require three magnesium ions in the step of the amino acid activation reaction.
5. Conclusion
Taking LysRS as the representative class II aaRS, this paper reveals a variety of transition states of ATP binding in the amino acid activation reaction and determines the binding sequence of three magnesium ions cobound with ATP, providing important insights into the catalytic mechanism of magnesium-dependent aaRS.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China grants 22277132 and 21977107 and the State Key Laboratory of Bioorganic and Natural Products Chemistry.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2023.101426.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Dickman S.R., Boll D.J. Differential purification of methionine-tRNA synthetase and lysine-tRNA synthetase from rabbit liver. Biochem. Biophys. Res. Commun. 1977;78:1191–1197. doi: 10.1016/0006-291x(77)91419-x. [DOI] [PubMed] [Google Scholar]
- 2.Berg B.H., III. The RNA component of aminoacyl-tRNA synthetase complexes isolated from mouse liver. Absence of amino acid accepting activity. Biochim. Biophys. Acta. 1975;414:93–98. doi: 10.1016/0005-2787(75)90212-9. [DOI] [PubMed] [Google Scholar]
- 3.Eriani G., Delarue M., Poch O., et al. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 4.Argos P., Rossmann M.G., Johnson J.E. A four-helical super-secondary structure. Biochem. Biophys. Res. Commun. 1977;75:83–86. doi: 10.1016/0006-291x(77)91292-x. [DOI] [PubMed] [Google Scholar]
- 5.Walker E.J., Jeffrey P.D. Sequence comparisons in the aminoacyl-tRNA synthetases with emphasis on regions of likely homology with sequences in the Rossmann fold in the methionyl and tyrosyl enzymes. Protein Seq. Data Anal. 1988;1:187–193. [PubMed] [Google Scholar]
- 6.Nureki O., Vassylyev D.G., Katayanagi K., et al. Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science. 1995;267:1958–1965. doi: 10.1126/science.7701318. [DOI] [PubMed] [Google Scholar]
- 7.Kellermann O., Brevet A., Tonetti H., et al. Methionyl-tRNA synthetase from sheep liver. Purification of a fully active monomer derived from high-molecular-weight complexes by trypsin treatment. Evidence for immunological cross-reaction with the corresponding enzyme from sheep mammary gland. Eur. J. Biochem. 1978;88:205–210. doi: 10.1111/j.1432-1033.1978.tb12439.x. [DOI] [PubMed] [Google Scholar]
- 8.Brevet A., Kellermann O., Tonetti H., et al. Macromolecular complexes of aminoacyl-tRNA synthetases from eukaryotes. 2. Agarose gel-filtration behaviour of the extensively purified high-molecular-weight complex(es) of seven aminoacyl-tRNA synthetases from sheep liver. Eur. J. Biochem. 1979;99:551–558. doi: 10.1111/j.1432-1033.1979.tb13287.x. [DOI] [PubMed] [Google Scholar]
- 9.Kellermann O., Brevet A., Tonetti H., et al. Macromolecular complexes of aminoacyl-tRNA synthetases from eukaryotes. 1. Extensive purification and characterization of the high-molecular-weight complex(es) of seven aminoacyl-tRNA synthetases from sheep liver. Eur. J. Biochem. 1979;99:541–550. doi: 10.1111/j.1432-1033.1979.tb13286.x. [DOI] [PubMed] [Google Scholar]
- 10.Harris C.L., Marin K., Stewart D. tRNA sulfurtransferase: a member of the aminoacyl-tRNA synthetase complex in rat liver. Biochem. Biophys. Res. Commun. 1977;79:657–662. doi: 10.1016/0006-291x(77)91162-7. [DOI] [PubMed] [Google Scholar]
- 11.Cusack S., Berthet-Colominas C., Hartlein M., et al. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 12.Ruff M., Krishnaswamy S., Boeglin M., et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp) Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 13.Cusack S., Hartlein M., Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991;19:3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxholm H.J., Pitot H.C. Characterization of a proteolipid complex of aminoacyl-tRNA synthetases and transfer RNA from rat liver. Biochim. Biophys. Acta. 1979;562:386–399. doi: 10.1016/0005-2787(79)90103-5. [DOI] [PubMed] [Google Scholar]
- 15.Vadeboncoeur C., Lapointe J. Slow diffusion of glutamate and ATP-Mg into high-molecular-weight complexes containing the glutamyl-tRNA synthetase from bovine brain. Eur. J. Biochem. 1980;109:581–587. doi: 10.1111/j.1432-1033.1980.tb04831.x. [DOI] [PubMed] [Google Scholar]
- 16.Hei Z., Wu S., Liu Z., et al. Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex. J. Biol. Chem. 2019;294:4775–4783. doi: 10.1074/jbc.RA118.006356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang P., Han H., Wang J., et al. Structural basis for specific inhibition of tRNA synthetase by an ATP competitive inhibitor. Chem. Biol. 2015;22:734–744. doi: 10.1016/j.chembiol.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ofir-Birin Y., Fang P., Bennett S.P., et al. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell. 2013;49:30–42. doi: 10.1016/j.molcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo M., Ignatov M., Musier-Forsyth K., et al. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: implications for multisynthetase complex formation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2331–2336. doi: 10.1073/pnas.0712072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams P.D., Afonine P.V., Bunkoczi G., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang C.V., Yang D.C. High molecular weight complexes of eukaryotic aminoacyl-tRNA synthetases. Int. J. Biochem. 1982;14:539–543. doi: 10.1016/0020-711x(82)90030-1. [DOI] [PubMed] [Google Scholar]
- 22.Hilderman R.H. Characterization of a homogeneous complex of arginyl- and lysyl-tRNA synthetase: zinc and adenosine 5'-phosphate dependent synthesis of diadenosine 5',5'''-P1,P4-tetraphosphate. Biochemistry. 1983;22:4353–4357. doi: 10.1021/bi00288a001. [DOI] [PubMed] [Google Scholar]
- 23.Mirande M., Cirakoglu B., Waller J.P. Seven mammalian aminoacyl-tRNA synthetases associated within the same complex are functionally independent. Eur. J. Biochem. 1983;131:163–170. doi: 10.1111/j.1432-1033.1983.tb07244.x. [DOI] [PubMed] [Google Scholar]
- 24.Walker E.J., Treacy G.B., Jeffrey P.D. Molecular weights of mitochondrial and cytoplasmic aminoacyl-tRNA synthetases of beef liver and their complexes. Biochemistry. 1983;22:1934–1941. doi: 10.1021/bi00277a030. [DOI] [PubMed] [Google Scholar]
- 25.Brevet A., Geffrotin C., Kellermann O. Macromolecular complex of aminoacyl-tRNA synthetases from sheep liver. Identification of the methionyl-tRNA synthetase component by affinity labeling. Eur. J. Biochem. 1982;124:483–488. doi: 10.1111/j.1432-1033.1982.tb06619.x. [DOI] [PubMed] [Google Scholar]
- 26.Mirande M., Cirakoglu B., Waller J.P. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. III. Assignment of aminoacyl-tRNA synthetase activities to the polypeptide components of the complexes. J. Biol. Chem. 1982;257:11056–11063. [PubMed] [Google Scholar]
- 27.Desogus G., Todone F., Brick P., et al. Active site of lysyl-tRNA synthetase: structural studies of the adenylation reaction. Biochemistry. 2000;39:8418–8425. doi: 10.1021/bi0006722. [DOI] [PubMed] [Google Scholar]
- 28.Guo M., Chong Y.E., Shapiro R., et al. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature. 2009;462:808–812. doi: 10.1038/nature08612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt E., Moulinier L., Fujiwara S., et al. Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. EMBO J. 1998;17:5227–5237. doi: 10.1093/emboj/17.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belrhali H., Yaremchuk A., Tukalo M., et al. The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure. 1995;3:341–352. doi: 10.1016/s0969-2126(01)00166-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.