Abstract
Mesenchymal stem/stromal cells (MSCs) show tremendous potential for regenerative medicine due to their self-renewal, multi-differentiation and immunomodulatory capabilities. Largely studies had indicated conventional tissue-derived MSCs have considerable limited expandability and donor variability which hinders further application. Induced pluripotent stem cell (iPSCs)-derived MSCs (iMSCs) have created exciting source for standardized cellular therapy. However, the cellular and molecular differences between iMSCs and the cognate tissue-derived MSCs remains poorly explored. In this study, we first successfully reprogrammed human umbilical cords-derived mesenchymal stem/stromal cells (UMSCs) into iPSCs by using the cocktails of mRNA. Subsequently, iPSCs were further differentiated into iMSCs in xeno-free induction medium. Then, iMSCs were compared with the donor matched UMSCs by assessing proliferative state, differentiation capability, immunomodulatory potential through immunohistochemical analysis, flow cytometric analysis, transcriptome sequencing analysis, and combine with coculture with immune cell population. The results showed that iMSCs exhibited high expression of MSCs positive-makers CD73, CD90, CD105 and lack expression of negative-maker cocktails CD34, CD45, CD11b, CD19, HLA-DR; also successfully differentiated into osteocytes, chondrocytes and adipocytes. Further, the iMSCs were similar with their parental UMSCs in cell proliferative state detected by the CCK-8 assay, and in cell rejuvenation state assessed by β-Galactosidase staining and telomerase activity related mRNA and protein analysis. However, iMSCs exhibited similarity to resident MSCs in Homeobox (Hox) genes expression profile and presented better neural differentiation potential by activation of NESTIN related pathway. Moreover, iMSCs owned enhanced immunosuppression capacity through downregulation pools of pro-inflammatory factors, including IL6, IL1B etc. and upregulation anti-inflammatory factors NOS1, TGFB etc. signals. In summary, our study provides an attractive cell source for basic research and offers fundamental biological insight of iMSCs-based therapy.
Keywords: Mesenchymal stem/stromal cells (MSCs), Induced pluripotent stem cells (iPSCs), iPSC-derived MSCs (iMSCs), Immunomodulatory, Transcriptomics
1. Introduction
Mesenchymal stem/stromal cells (MSCs) are regarded as an ideal cell source for cell therapy due to excellently biological abilities e.g., their immunomodulatory and regenerative therapeutic potentials in vitro and in vivo. Large studies had indicated MSCs can be obtained from many different tissues, including bone marrow, cord blood, umbilical cord and placenta, adipose or other connective tissues [1]. The umbilical cord derived MSCs (UMSCs) holds great promise in regenerative medicine as non-invasive sampling and higher proliferation rate than that derived from adult tissues [2,3]. Current, over 1000 MSCs-based therapies are registered on http://www.clinicaltrials.gov/ for various diseases, such as diabetes, cancer, and liver cirrhosis etc., in which more than 150 trails are based on the UMSCs. When used in clinical trials, the conventional tissue-derived MSCs usually subjected to culture-expansion in vitro to harvest enough number of cells prior to treatment. However, studies had shown that MSCs derived from majority of the tissue sources showed great heterogeneity in proliferation, differentiation capability, and immunomodulatory potential etc., even though generally meet the criteria of International Society for Cellular Therapy (ISCT), which may explain the controversial results appeared in clinical trials treatments [4]. Thus, an alternative MSCs source for overcoming these shortcomings of connective tissue-derived MSCs is still a major challenge.
As the unlimited proliferation ability without losing their characteristics of induced pluripotent stem cells (iPSCs) perfectly addressed the need for cell number in clinical application. Large studies have shown that the iPSCs-derived MSCs (iMSCs) would be an alternative source for obtaining a functional population of MSCs for clinical application [5]. For example, Chen et al., has created one-step method without embryoid bodies (EBs) formed to direct mesengenic differentiation of human embryonic stem cells (ESCs) and iPSCs using the small molecule SB431542, of which adipogenic differentiation was limited [6]. Also, another study has presented an optimized procedure to produce MSCs from human iPSCs depends on the generation of embryoid bodies (EBs) and transforming growth factor beta 1 (TGF-β1) [7]. In addition, Wu et al., has showed anti-BMP2 Ab/BMP2 immune complex was capable of promoting osteogenic differentiation of iPSCs going through iMSCs, which may be a favorable approach for iPSC-based bone tissue engineering [8,9]. Above studies have shown that iMSCs can be successfully differentiated from iPSCs with well-defined differentiation procedures. However, to the best of our knowledge, strategies including derivation and expansion of iMSCs in serum-free, chemically defined culture conditions and use of integrating-free reprogramming methods are still crucial works towards the personalized medicine [10,11].
Several studies had indicated that iMSCs might show outstanding features in clinical application when compared that with the cognate tissue-derived parent MSCs revealed by traditional technologies [12,13]. For instance, studies have showed that iMSCs presented better cellular vitality and immunomodulatory effects when comparable to conventionally tissue-derived MSCs [14,15]. For the iMSCs self-renew and rejuvenation, Lian et al., has showed iMSCs display a higher proliferative capacity, up to 120 passages, without obvious loss of self-renewal potential and constitutively express MSCs surface antigens when compared with adult bone marrow derived MSCs [16]. Previous study has found that aging-related activities molecular such as GATA6 were greatly reduced in iMSCs compared with those in their parental cells, indicating reversal of cell aging of cellular reprogramming [17,18]. In another study, by comparing the iMSCs generated from urinary epithelial (UE) cells-derived iPSCs with the UMSCs, iMSCs showed relatively homogeneous cell populations with higher proliferative activity and greater migration ability [19]. In addition, Zhang and colleagues’ study has showed iMSCs hold lower heterogeneity and great promise in biological research and clinical applications [20]. However, another study has indicated that iMSCs showed earlier senescence-associated beta-galactosidase (SA-β-gal) activity but significantly longer telomere lengths [21]. In addition, some other studies have showed similar changes in morphology, in vitro differentiation potential, senescence-associated β-galactosidase, and DNA methylation between iMSCs and parent cells [22].
Several studies had indicated iMSCs expressed a range of immunomodulatory and anti-inflammatory factors in many disease niche [23,24]. Kim et al., has found that extracellular vesicle (EVs) from early-passage iMSCs had better immunomodulatory potency than EVs from late-passage iMSCs in TLR4-stimulated splenocytes which may provide strategies to improve regulatory function of EVs for the treatment of immune-mediated diseases [25]. Fan et al., has been demonstrated that iMSCs holding great immunomodulatory potency by activating quiescent T cells and elevate regulatory T cell response via NF-κB pathway in allergic rhinitis patients [26]. Moreover, the Cynata Therapeutics performed the world's first formal trial of an allogeneic iMSCs which met all of its clinical endpoints and produced positive safety and efficacy data for the treatment of steroid-resistant acute graft-versus-host disease (GVHD) based on the immunomodulatory effect of iMSCs [27]. However, the molecular characteristic of suppression or activation of immune system of iMSCs in immunomodulatory niche is still largely unclear.
To date, many studies have showed iMSCs presented traditional three-lineage differentiation capacity, including osteocytes, chondrocytes and adipocytes. For example, antibody-mediated osseous regeneration (AMOR) has been proved as a promising strategy for osteogenic differentiation of iMSCs [24,28]; Jungbluth et al., has indicated iMSCs combined with calcium phosphate granules (CPG) showed greater role in critical-size defects in the proximal tibias of mini-pigs in the early phase of bone healing compared to that of autologous bone marrow derived MSCs or CPG alone [29]. Holding the mesoblast lineage cell differentiation capacity of iMSCs is well described, but the ability of transdermal differentiation of iMSCs is still unexplored.
In sum, to our knowledge, the molecular feature and cellular functional state of iMSCs is still largely undetermined [30,31]. To better compare the therapeutic efficacy between iMSC and the primary MSC, the in-depth functionality demonstration in biological research and the underlying mechanisms are urgently needed, which provides reference to optimize their phenotype and efficacy for targeting specific diseases. In the present study, we first reprogrammed human umbilical cords-derived mesenchymal stem/stromal cells (UMSCs) into iPSCs by using the mRNA of transcription factor cocktails under GMP condition. Subsequently, the iPSCs were further differentiated into iMSCs under serum-free conditions. The iMSCs were compared with the donor matched UMSCs by assessing proliferative state, differentiation capability, immunomodulatory potential through immunohistochemical analysis, flow cytometric analysis, transcriptome sequencing analysis, also combine with coculture with immune cell population assay to assess immunomodulatory effect. The present study may provide an attractive cell source for basic research and offers fundamental biological insight of iMSCs-based cell therapy.
2. Results
2.1. Generation of non-viral and non-integration human iPSCs by mRNA method
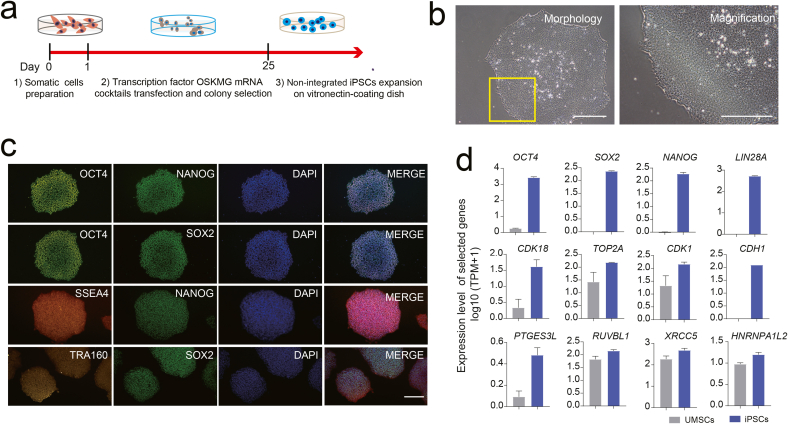
Human UMSCs at passage 3 were cultured in a 6-well plate coated with vitronectin in commercial serum-free medium. At 40–50% cell confluency, the UMSCs were transfected with pluripotent transcription factors cocktails via non-integrating, virus free, self-replicating RNA of StemRNA Reprogramming kit as shown in the schematic illustration (Fig. 1a). By day 25, the single-cell derived iPSCs colonies with edges neatly and high nucleo-cytoplasmic ratio were picked and culture-expanded in vitronectin coating and TeSR-E8 medium under Good Manufacturing Practice of Medical Products (GMP) conditions (Fig. 1b). Then, the iPSCs were confirmed by immunofluorescence staining of pluripotent proteins OCT4, SOX2, NANOG, TRA160 and SSEA4 (Fig. 1c). The mRNA expression of pluripotent genes such as OCT4, SOX2, NANOG, LIN28A and telomerase maintaining related genes such as PTGES3L, XRCC5 etc. and cell renewal related gene including CDK18, CDK1 and TOP2A etc. had a significant increase in iPSCs comparing to UMSCs revealed by bulk RNA-sequencing analyses (Fig. 1d; Supplement F. 1a-e). The above results indicated that human UMSCs could be successfully reprogrammed into iPSCs with well-defined cellular and molecular characteristic using the mRNA-based method which was in agreement with the previous study [8,9,32]. Overall, the above results strongly indicated that the iPSCs generated with the mRNA cocktails were human pluripotent stem cells with truly rejuvenation and pluripotent state and total two background iPSCs lines were established and used for further experiments in our study.
Fig. 1.
Generation and characterization of human iPSCs. a. Workflow of the generation of UMSCs-derived iPSCs through transfection with mRNA factor cocktails. b. Morphology of iPSCs during culture-expansion in vitro. Scale bar represents 100 μm. c. Immunofluorescence staining of OCT4, NANOG, SOX2, TRA160 and SSEA4. DAPI staining was done to visualize the nucleus. d. Barplot showing the relative expression level of representative pluripotent-, cell cycle- and telomere maintaining-related genes in iPSCs and UMSCs population using the RNA-sequencing data analysis.
2.2. Generation and characterization of iMSCs derived from human iPSCs
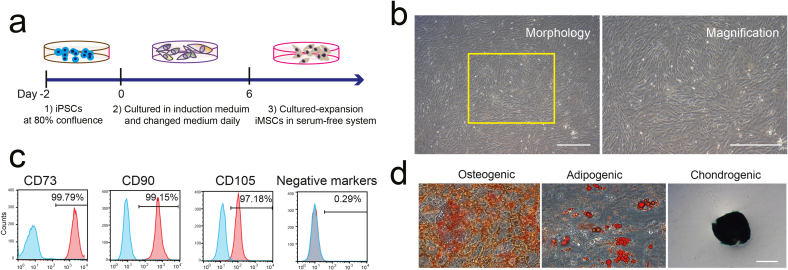
To harvest high quality of iMSCs, the human iPSCs at 80% confluency were cultured in animal component-free mesenchymal induction medium for 6 days, then the iMSCs were maintained and culture-expanded in commercial serum-free medium (Fig. 2a). Phase-contrast microscopic images of iMSCs displayed the typical fibroblastic-like MSCs morphology (Fig. 2b). The expression of pluripotent genes such as OCT4, SOX2, NANOG were significantly down regulated in iMSCs compared to iPSCs based on our RNA-sequencing data analyses (Supplement F. 1d). To further validate of iMSCs with reference to International Society of Cellular Therapy (ISCT) guidelines on MSCs criteria, we performed flow cytometric analysis and trilineage differentiation experiments. The results demonstrated that the triple positivity for the mesenchymal markers including CD73, CD90 and CD105 were significantly expressed in higher number of cells in both iMSCs and UMSCs populations both at mRNA and protein level (Fig. 2c; Supplement F. 1d; Supplement F. 2). Also, the iMSCs were successfully differentiated into osteocyte, adipocyte and chondrocyte lineages cells in specific induction medium (Fig. 2d). In total, our results demonstrated that the generated iMSCs have satisfied all the requirements to resemble human tissue source MSCs.
Fig. 2.
Differentiating the human iPSCs into iMSC. a. Schematic illustration of generation of iMSCs from human iPSCs. b. Morphology of iMSCs during culture-expansion in vitro. Scale bar represents 100 μm. c. The expression of MSC surface-positive marker (CD73, CD90, CD105) and lack expression of MSC surface-negative marker cocktails (CD34, CD45, CD11b, CD19, and HLA-DR) in culture-expanded iMSCs as assessed by FACS. d. Differentiation of adipocytes, osteocytes, and chondrocytes of iMSCs at passage 5. Scale bar 100 μm.
2.3. No significant difference in proliferative capacity between iMSCs and UMSCs
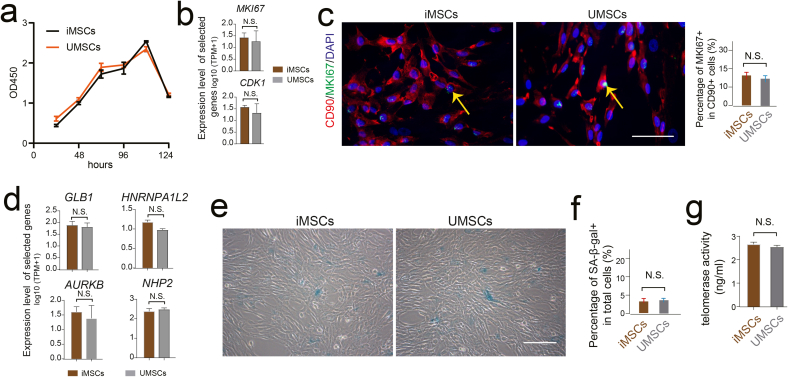
The ability of proliferation and growth is an important parameter for MSCs quality when culture-expansion in vitro. Thus, to further dissect the status of cell-renewal of iMSCs, we observed the growth curve, β-Galactosidase (β-GAL) staining and telomerase activity analysis and compared that with UMSCs. Similar pattern of growth curve was observed between iMSCs and UMSCs (Fig. 3a), also the cell cycle related gene MKI67 presented equivalent mRNA expression level (Fig. 3b). Then, immunofluorescent staining also showed there was no significant difference of MKI67 positive cell rate in CD90 positive cell populations between iMSCs and UMSCs (Fig. 3c). Further, we evaluated the mRNA expression and protein levels of β-GAL, which could indicate the status of replicative senescence. In line with the MKI67 expression pattern, there was non-significant difference of β-Galactosidase-positive cell ratio was observed between iMSCs and UMSCs population (Fig. 3d–f). Moreover, we measured the distribution of telomere maintaining related genes activities in the iMSCs and UMSC cell population. Our results showed the mRNA expression level of telomere related genes, for instance HNRNPA1L2, NHP2 etc. were similar level between iMSCs and UMSCs. And the telomerase activity of iMSCs and UMSCs measured by cell culture medium using enzyme-linked immunosorbent assay (ELISA) kit was also presented similar pattern (Fig. 3d, g). Collectively, the above results above strongly indicated that there was no significant difference of proliferative capacity between iMSCs and UMSCs in the present study.
Fig. 3.
No significant difference in proliferative capacity between iMSCs and UMSCs. a. The cell viability of iMSCs and UMSCs detected by the CCK-8 assay. b. The mRNA expression level of the representative cell cycle-related genes in iMSCs and UMSCs. c. Immunofluorescence staining of CD90 and MKI67 in iMSCs and UMSCs. DAPI staining was done to visualize the nucleus. Scale bar represents 100 μm. Column chart showing the percentage of MKI67-positive cells in CD90-positive cells of iMSCs and UMSCs. d. The mRNA expression level of the representative β-Galactosidase (β-GAL) related genes and telomere maintaining-related gene in iMSCs and UMSCs. e. β-galactosidase staining in iMSCs and UMSCs. Scale bar represents 100 μm. f. Column chart showing the percentage of β-GAL positive cells in total cells of iMSCs and UMSCs. g. The protein levels of telomerase activity determined by ELISA in the culture medium of iMSCs and UMSCs.
2.4. iMSCs exhibited similarity to resident MSCs with higher neural differentiation potential
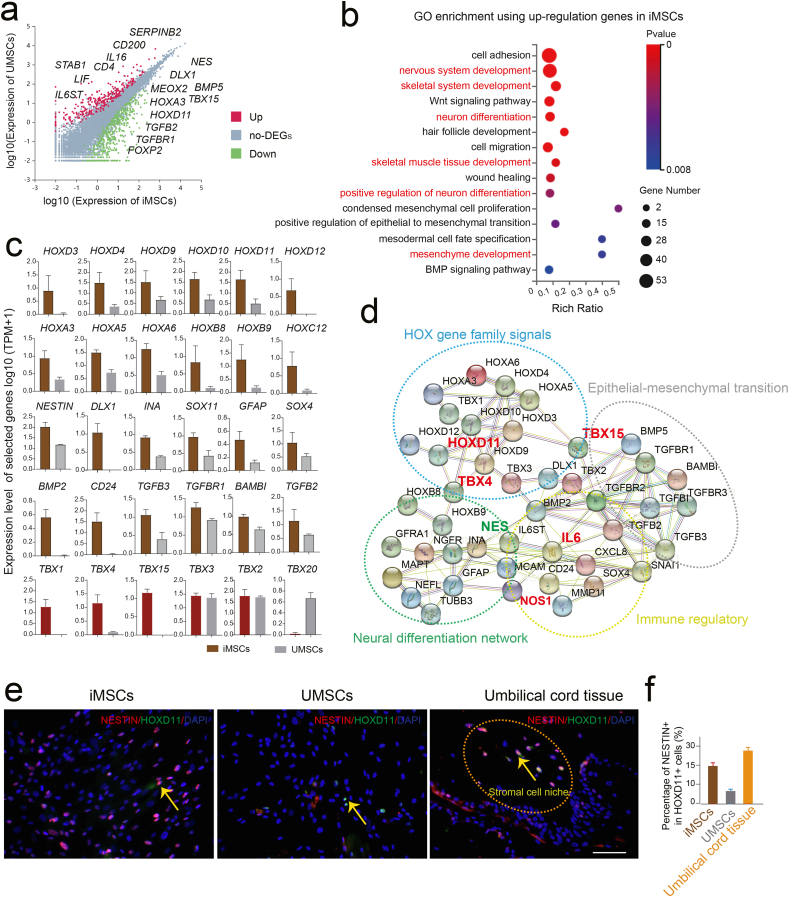
To further explore the cellular and molecular difference between iMSCs and UMSCs, we extracted the differentially expressed genes (DEGs) using the RNA-seq data analysis (Fig. 4a). Interestingly, the GO enrichment analysis of upregulation genes in iMSCs compared to UMSCs showed the embryonic tissue patterning pathway and neural differentiation related functional terms were significantly enriched in iMSCs (Fig. 4b). Previous study has showed that homeobox (Hox) genes were key regulators of the multiorgan development in embryogenesis and has a crucial role in nervous system development and function [33]. Recent studies have shown that the Hox code might be an attribute of a distinct subpopulation of tissues resident mesenchymal stromal cells (MSC), which played a crucial role in stroma renewal and tissue regeneration [34,35]. In our study, the Hox gene family members, including HOXD3, HOXD4, HOXD9, HOXD10, HOXD11, HOXD12 also HOXA3, HOXA5, HOXA6, as well as HOXB8, HOXB9 and HOXC12 were highly expressed in iMSCs compared to UMSCs (Fig. 4c). Previous study has showed transcription factor T-box family members, such as TBX16 promoting mesoderm development by activating posterior Hox genes in zebrafish gastrulae [36]. Similar results were also observed in our study, the TBX1, TBX2, TBX3, TBX4, and TBX15 were highly expressed in iMSCs and interacted with Hox genes (Fig. 4c and d). In addition, previous study has showed that naive human umbilical cord blood (hUCB)-derived multipotent mesenchymal stem cells (MSCs) expressed both mesodermal and ectodermal markers prior to neural induction [37]. In our study, we also found the neural differentiation related genes, for instance NESTIN, DLX1, SOX4 and GFAP etc. as well as the epithelial-mesenchymal transition related genes including BMP2, CD24, TGFB2, TGFB3 and TGFBR1 etc. were highly expressed in iMSCs; which in line with the functional terms by the GO enrichment analysis (Fig. 4b–d). Moreover, the immunostaining showed that the NESTIN was heterogeneously co-expressed with HOXD11, which showed relatively similar percentage of positive cells in iMSCs and in situ stromal cell area of human umbilical cord tissue; while obviously reduced in UMSCs was observed (Fig. 4e and f). Collectively, in the present study, we first revealed that iMSCs have highly expressed Hox family genes that indicated iMSCs might be more similar with the resident MSCs from the original tissue microenvironment, which indicated iMSCs might be more easily to differentiate into ectodermal neural lineage cells by activating NESTIN signals pathway in specific induction niche compared to UMSCs.
Fig. 4.
iMSCs exhibited higher neural differentiation potential. a. The selected representative upregulation genes in DEGs of iMSCs and UMSCs respectively. b. Selected GO terms of upregulation genes in iMSCs compared to UMSCs. c. Barplot showing the relative expression level of selected genes in iMSCs and UMSCs. d. The regulatory network of the selected genes using the STRING databased. e. Immunofluorescence staining of NESTIN and HOXD11 in iMSCs, UMSCs and the umbilical cord tissue, respectively. DAPI staining was done to visualize the nucleus. Scale bar represents 100 μm. f. Column chart showing the percentage of NESTIN-positive cells in HOXD11-positive cells of iMSCs, UMSCs and the umbilical cord tissue, respectively.
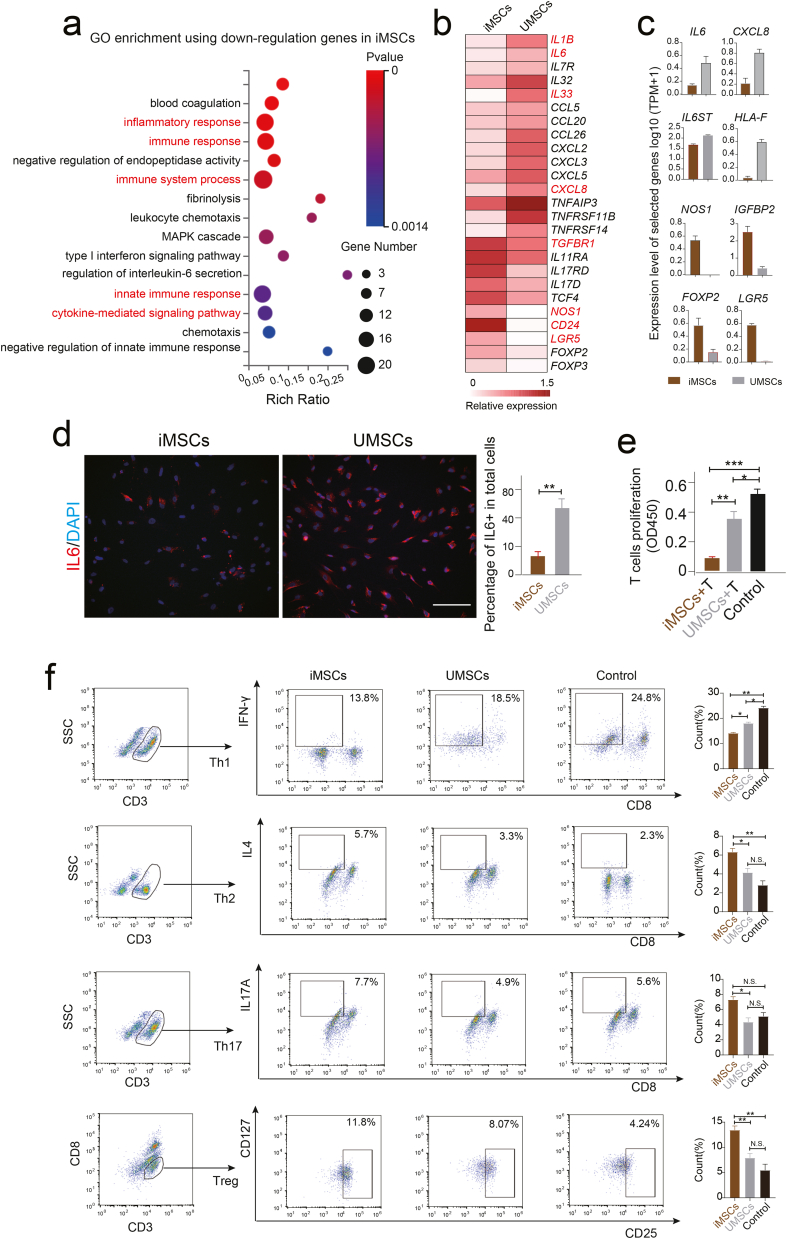
2.5. iMSCs showed higher immunosuppressive capability compared to UMSCs
To systematically dissect the molecular and function difference between iMSCs and UMSCs, we also performed the GO enrichment analysis using the downregulation genes of iMSCs revealed by bulk RNA sequencing. Interestingly, the immunoregulatory related functional terms including “inflammatory response”, “immune response”, “cytokine-mediated signaling pathway” and “regulation of interleukin-6 secretion” etc. were significantly enriched in iMSCs (Fig. 5a). Interestingly, the pro-inflammatory cytokines including IL6, IL6ST, CXCL8, IL1B etc. showed higher expression level in UMSCs while anti-inflammatory factors such as NOS1, CD24, FOXP3, FOXP2, TGFBR1 and TGFB2 etc. were highly expressed in iMSCs (Fig 4c; Fig. 5b and c). In line with mRNA level, the immunofluorescent staining showed pro-inflammatory protein IL6 was significantly lower in iMSCs populations (Fig. 5d). To further analyzed anti-inflammatory effect of iMSCs, the T cells co-culture and flow cytometric analysis were performed in vitro. We observed that iMSCs profoundly inhibited the proliferation of T cells compared to UMSCs and control group revealed by the Transwell system and CCK-8 assay (Fig. 5e; Supplement F. 3). Also, the Treg cells (CD3+CD8−CD25+CD127low/-), and Th17 (CD3+CD8−IL17A+) and TH2 (CD3+CD8−IL4+) ratio were higher in iMSCs groups while Th1(CD3+CD8−IFN-γ+) was relative higher in UMSCs revealed by flow cytometric (Fig. 5f). All the above results strongly indicated iMSCs might own supper immunosuppressive signals compared to their parent UMSCs, even though cellular and molecular mechanism of immunosuppressive of iMSCs is still need to be addressed using single-cell omics technologies in the future.
Fig. 5.
iMSCs showed higher immunosuppressive function compared to UMSCs in vitro. a. Selected representative GO terms of down-regulation genes in iMSCs compared to UMSCs. b. Heatmap showing the expression level of representative immunoregulation related genes in iMSCs and UMSCs. c. The expression levels of selected representative immunoregulation genes in iMSCs and UMSCs. d. Immunofluorescence staining of IL6 in iMSCs and UMSCs. DAPI staining was done to visualize the nucleus. Scale bar represents 100 μm. Column chart showing the percentage of IL6 positive cells in total cells of iMSCs and UMSCs. e. Column chart showing the proliferative activity of T cells detected by the CCK-8 assay. T cells cultured alone as the control group. f. T cell subpopulation analysis by flow cytometry using specific antibody including CD3, CD8, CD25, CD127, IFN-γ, IL4 and IL17A. Column chart showing the percentage of T cell subpopulations. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3. Discussion
Mesenchymal stem/stromal cells (MSCs) are increasingly used for regenerative medicine due to their multilineage differentiation and immunomodulatory capabilities. However, currently, MSCs face several limitations including fewer number of cells with low expand ability potentials, allogenic immunogenicity which hinder their further application [31,38]. Large studies had showed human iPSCs-derived MSCs owned great advantages than tissue source MSCs, and might be regard as an alternative cell source for stem cell-based therapy in clinical research [18]. However, the molecular and functional characteristics of iMSCs is still largely unknown [10]. Also, study of comparison of iMSCs and parent primary tissue MSCs on rejuvenation, differentiation and immunomodulatory capabilities still need more works.
In the current study, we first successfully reprogrammed GMP grade human UMSCs into iPSCs using the non-virus non-integrated mRNA-based method. Validation of pluripotent gene such as OCT4, SOX2 etc. at mRNA and protein level by immunofluorescence staining and RNA-sequencing, our results indicated that the iPSCs obtained in our study are true pluripotent stem cells using a safer and more regulatory friendly method, which would be a great stem cell model being capable of differentiating into any other kind of cell population in the human body for basic research and clinical application.
Subsequently, the foot-print free iPSCs were differentiated into MSCs in accordance with GMP principles, and were compared with the donor matched UMSCs in self-renewal, multi-differentiation capability, immunomodulatory potential assessed by growth curve analysis, flow cytometric, transcriptome sequencing methods etc. The results showed that the iMSCs were similar with their parental UMSCs in cell surface maker expression profile, including CD73, CD90 and CD105, cell proliferation by growth curve analysis, cellular rejuvenation state by β-Galactosidase staining and telomerase activity analysis, mesodermal differentiation potential, e.g. osteocytes, chondrocytes and adipocytes. The results obtained in our study indicated primary iMSCs showed comparable cell characteristics in terms of morphology, cell proliferation, conventional differentiation, and surface phenotype features to autologous tissue origin MSCs which were in line with previous study [39,40]. Recent study has showed the iMSCs could maintain their MSC characteristics even at later passages (P15), during which the tissue source MSCs started losing their MSC characteristics; also the iMSCs showed superior migration ability [19]. Moreover, current, the iMSCs or UMSCs culture-expanded in our study by using 2D culture systems in vitro which was largely different with MSCs survival niche in vivo. Thus, more works are still need to be done for the theme validation in the future.
Recent studies have indicated HOX-positive cells may represent a distinct subpopulation of adult resident mesenchymal stromal cells (MSCs) in vivo, which is crucial for reconstructing stroma and tissue regeneration [34,35]. Moreover, large studies had indicated that the large family of Hox gene has a key role in nervous system development and function [33,41]. Interestingly, in our study, the iMSCs were highly expressed of HOX family genes compared to tissue source UMSCs. And, in line with previous study, we found the iMSCs also exhibited higher neural differentiation ability by activating NESTIN related gene network. https://pubmed.ncbi.nlm.nih.gov/?term=Fukuta+M&cauthor_id=25464501 Fukuta and colleagues’ study has showed iMSCs can be obtained from iPSCs through a neural crest lineage stage [42]. Previous study identified several subgroups both in cultured UMSCs and the cognate human umbilical cord tissue at single-cell resolution [43]. Also, previous study has indicated iMSCs showing enhanced ability to promote nerve growth by producing many of neurotrophic and neuroprotective factors [44]. In addition, previous study has showed mesenchymal stem cells derived from human iPS cells via mesoderm and neuroepithelium have different features and therapeutic potentials [45]. Further, study had showed LNGFR+THY-1+ human iPSCs-derived neural crest-like cells have the potential to develop into mesenchymal stem cells which may indicated the functional subgroups existed in different differentiation stage cell populations under specific induction system [46]. Thus, if there are functional subpopulations existed in the iMSCs population remain poorly defined. The relationship between NESTIN-positive single cells and HOX-positive single cells also need to be addressed using single cell omics technologies in future work.
Current, the immunomodulatory effect of iMSCs which play crucial role in regenerative medicine is still controversial theme [26]. For example, previous study has showed the immunomodulatory signatures of canine iMSCs are similar to the adipose tissue and bone marrow derived MSCs [47]. However, some other studies had showed iMSCs has better immunomodulatory effects than conventional MSCs, for example, Schnabel et al., has reported iMSCs had similar immunogenic properties but more potent immunomodulatory effects than tissue source MSCs [48]. Also, previous study has showed iMSCs combined with rapamycin induced islet allograft tolerance through suppressing Th1 and enhancing regulatory T-cell differentiation by downregulation of proinflammatory cytokines interleukin-2 (IL-2) and interferon-γ and upregulation of the anti-inflammatory factor TGFβ, respectively [49]. Moreover, Fang and colleagues’ study further confirmed the suppressive effects of iMSCs on the differentiation of human T helper cells by decreasing the levels of Th17 cells, IL-17A and p-STAT3 [50]. In our study, we revealed the iMSCs showed higher immunosuppressive capability compared to UMSCs using the GO enrichment analysis which were in line with previous study [49,51]. Further, the immunosuppressive effect of iMSCs were demonstrated by inhibiting the proliferation of T cells using the co-culture system (Fig. 5e; Supplement F. 3). In addition, previous study has showed iMSCs display remarkable inhibition of NK-cell proliferation and cytolytic function by down-regulating the expression of different activation markers and ERK1/2 signaling [52]. The IL10 mRNA expression was not detected but the immunosuppressive activity was significantly increased in iMSCs compared to UMSCs which might indicate more complex process of interaction of iMSCs and immune cells. Thus, the cellular and molecular mechanism of immunosuppressive of iMSCs is still need to be addressed in future work. In addition, the safety of iMSCs was assessed by subcutaneous injection in BALB/c-nu nude mice, no teratoma was observed with following up for more than 24 weeks in our study (data not shown), which was consistent with previous study [53]. Totally, our novel non-virus non-integration approach of generating iMSCs will be a good source of autologous cells for regenerative disease therapy.
4. Conclusion
In conclusion, we have developed unlimited, safe, autologous iMSCs, which were highly expressed of MSCs positive-makers CD73, CD90 CD105 and low or no expressed of negative-maker cocktails CD34, CD45, CD11b, CD19, and HLA-DR and successfully differentiated into ectoderm, endoderm and mesoderm lineage. The iMSCs were similar with their parental UMSCs in cell proliferative state, and in cell rejuvenation state also in telomerase activity feature. However, iMSCs were similar with resident MSCs which presented better neural differentiation potential than UMSCs by activation of NESTIN related pathway. Moreover, iMSCs owned enhanced immunosuppression capacity through downregulated pro-inflammatory factors IL6 etc., and upregulated anti-inflammatory factors TGFβ et al. signals. In summary, our study provides an attractive cell source for basic research and offers fundamental biological insight of clinical-grade autologous iMSCs-based therapy.
Methods
Ethics
This study was approved by the Institutional Review Boards on Ethics Committee of Animal Research Center of Shenzhen Luohu hospital (Permit No. 20220406). All the donors signed informed consent and volunteered to donate samples.
UMSCs isolation and culture-expansion
The umbilical cord tissues were obtained from two donors after written informed consent. Then, the umbilical cord tissue was undergone mechanical separation and seeded in the dish as previously reported [54]. Briefly, the umbilical cord tissue was first washed using DPBS 1–2 times, then the cord vessels were mechanically separated and discarded, and the remaining tissue were cut into 1 mm3 or smaller fragments. These fragments of tissue were seeded in a 35 mm dish and cultured at 37 °C in a 5% CO2 incubator. The culture medium was half-changed every day until cells reached 80–90% confluence and passaged for further use.
Reprogramming UMSCs into iPSCs using mRNA cocktails
The UMSCs were reprogrammed mainly based on previous report with some modifications [55]. Briefly, the UMSCs at passage 3 were seeded into a 6-well plate coated with vitronectin in commercial serum-free MSCs culture medium. At 40–50% cell confluency, the UMSCs were transfected with StemRNA Reprogramming kit (STEMCELL TECHNOLOGIES, 05930) including pluripotent transcription factors such as OCT4, SOX2, KLF4, C-MYC and GLIS1 as shown in the schematic illustration (Fig. 1a). By day25, the single-cell derived iPSCs primary colony with edges neatly and high nucleo-cytoplasmic ratio were picked and culture-expanded in vitronectin coating and TeSR-E8 medium (STEMCELL TECHNOLOGIES) under Good Manufacturing Practice of Medical Products (GMP) conditions for further use in our study.
Differentiation of human iPSCs into iMSCs
iPSCs at passage 10 were cultured in 6-well plate and maintained in TeSR-E8 medium. When the iPSCs reached 70%–80% confluency, the TeSR-E8 medium was removed and fresh animal component-free (ACF) mesenchymal induction medium (STEMCELL TECHNOLOGIES, STEMdiff™) was added to the 6-well plate for 6 days followed by serum-free MSCs culture medium and passaged with TrypLE™ Express and marked as passage 1, and the above medium was changed every day. These iMSCs were further maintained and expanded in serum-free MSCs culture medium until at passage 5 for further experiments.
RNA sequencing and data filtering
RNA sequencing and bioinformatic analysis of iMSCs, UMSCs and iPSCs were performed as previously reported [56]. Briefly total RNA was isolated from iMSCs, UMSCs and iPSCs samples using the extracted with TRIzol™ reagent (Thermo Fisher, 1596018). Then, the sequencing libraries were constructed and sequenced on BGI-SEQ2000 platform (Beijing Genomics Institute). The raw RNA-seq reads in FASTQ format were quality checked with FASTQC algorithm, and low-quality reads were trimmed using the FASTX-Toolkit. High-quality reads were aligned to the human genome using HISAT2 and assembled against human mRNA annotation using feature counts. Differentially expressed genes (DEGs) were performed by using analysis package DESeq2 in online system of BGI ( “https://biosys.bgi.com/"), in which p value < 0.05 and |log2FC| >1 were used to identify significantly differentially expressed genes.
Differentiation of adipocytes, osteocytes, and chondrocytes
The differentiation of adipocytes, osteocytes, and chondrocytes of iMSCs and UMSCs at passage 5 was performed with the commercial kit, including Differentiation was performed with the OriCell® Chondrogenesis Differentiation Kit, OriCell® Adipogenesis Differentiation Kit, and OriCell® Osteogenesis Differentiation Kit as previously described [57].
Flow cytometry analysis
The cultured-expanded iMSCs at passage 5 were harvested and dissociated into single cells by 0.125% Trypsin-EDTA. To determine cell surface antigen expression, the cells were incubated with the Human MSC Analysis Kit (BD Stemflow™), contain antibodies panel: positive markers including APC Mouse Anti-Human CD73, FITC Mouse Anti-Human CD90, PerCP-Cy™5.5 Mouse Anti-Human CD105; and PE hMSC Negative Cocktail CD34, CD45, CD11b, CD19 and HLA-DR. Upon completion of the incubation, the cells were analyzed using a flow cytometer (BD Biosciences) and gated by forward scatter and side scatter. The suspension cells of whole blood derived PBMC were selected and precultured with monoclonal anti-CD3 antibody (Biolegend, 317,301). Then, they were co-cultured with the mitomycin treatmented iMSCs and UMSCs in well plates by adding the monoclonal anti-CD28 antibody (Biolegend, 302,901) in the culture medium. Then, the coculture were stimulated with PMA/Ionomycin to obtained more activated T cells and further cultured for total 3 days, then the T cells were collected for T cell subpopulation analysis by flow cytometry using specific antibody including FITC Mouse Anti-Human CD3, BV510 Mouse Anti-Human CD8, PE Mouse Anti-Human CD8, PE-Cy™7 Mouse Anti-Human CD25, Alexa Fluor®647 Mouse Anti-Human CD127, BV421 Mouse Anti-Human IFN-γ, PE Rat Anti-Human IL4 and PE Mouse Anti-Human IL17A.
Transwell experiment
The mitomycin treatmented iMSCs and UMSC (10,000 cells/well) were plated in the upper chamber of a Transwell plate (Costar, Corning, Inc.) 4 h prior to the addition of T cells (100,000 cells/well) to the lower chamber and further cultured for total 3 days. Cultures were stimulated by the addition of 2.5 μg/ml PHA-L (eBiosciences, 00–4977) and CCK-8 reagent (Beyotime, C0039) was added for the last 2 h of coculture.
Immunofluorescence staining
iMSCs and UMSCs at passage 5, iPSCs at passage 15 were fixed in 4% paraformaldehyde solution for 20 min and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. After 120 min blocking with 3% BSA (SIGMA), cells were incubated with primary antibody overnight at 4 °C. On the next day, cells were washed, and stained with secondary antibodies (1:300, goat anti-rabbit IgG-Cy3; or 1:300, goat anti-mouse IgG-FITC) for 60 min at room temperature and then washed three times with phosphate-buffered saline (PBS). For human umbilical cord tissue, paraffin sections were cleared of paraffin, hydrated, and were blocked in 1% BSA for 2 h at room temperature and then incubated with primary antibodies at 4 °C in a humidifying box overnight, followed by incubation with secondary antibodies for 1 h in the dark at room temperature. The primary antibodies for respective cells include MKI67 (1:300, Abcam), OCT4 (1:200, Abcam), SOX2 (1:200, Abcam), NAONG (1:200, Abcam), SSEA4 (1:200, Abcam), TRA160 (1: 200, Abcam), THY1 (1:200, Abcam), IL6 (1:100, Abcam), HOXD11 (1:100, Abcam), NESTIN (1:100, Abcam). DAPI (4′,6-diamidino-2-phenylindole) (1:500) was used as counter-staining for nuclei. The images were captured and analyzed with the Olympus IX73 and Image J software.
β-Galactosidase (β-Gal) staining
β-Gal staining was performed according to the manufacturer's protocol (Solarbio, G1580). Briefly, iMSCs and UMSCs at passage 5 were seeded in 6-well plate. When cells reached 90% confluence, the medium was discarded, and the cells were washed with PBS 1 time and fixed using the fixative solution for 15 min at room temperature, and then the cells were washed with PBS 2 times and incubated at 37 °C overnight with the β-Gal staining solution. The images were captured with the Olympus IX73. The percentage was calculated from ten different view fields of each sample in three independent experiments.
ELISA detection
The culture supernatant of iMSCs and UMSCs at passage 5 was collected for the test of telomerase using the enzyme-linked immunosorbent assay (ELISA) kit (RuiFan, RF2385) according to the instructions.
Regulatory network construction
Significantly differentially expressed TFs (adjusted p value < 0.05, |log2FC| >1) between each population were selected and submitted to the STRING database to construct the potential regulatory networks [58]. TFs without any edge were removed from the network.
Gene Oncology (GO) term analysis
GO functional analysis of DEGs was performed using R package in online system of BGI. The Benjamini-Hochberg (BH) method was used for multiple test correction. GO terms with p value less than 0.05 were considered as significantly enriched.
Statistical analysis
Quantitative data were expressed as mean ± SEM. GraphPad 8.0 software was used for statistical analysis. The difference between two groups was determined by unpair t-test. We considered p value < 0.05 as statistically significant.
Data availability statement
The sequencing data with fastq format was stored in NCBI, accession number PRJNA860220 https://dataview.ncbi.nlm.nih.gov/object/PRJNA860220.
Author contribution statement
Quanlei Wang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Chongfei Chang; Yuwei Wang: Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Feilong Ma; Qiuting Deng; Yanru An: Performed the experiments; Contributed reagents, materials, analysis tools and data. Dongxiu Peng; Shun Yang; Xufeng Qi; Huiru Tang: Conceived and designed the experiments; Wrote the paper. Fei Wang; Qixiao Wang; Fei Gao: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools and data. Xiaoming Jiang; Dongqing Cai; Guangqian Zhou: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools and data. All authors read and approved the manuscript for submission.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
We would like to thank members of our laboratories for useful discussions, and sincerely thank the support provided by Cell Center of Cheerland Group Shenzhen. This study was supported by Science, Technology and Innovation Commission of Shenzhen Municipality, China, the grant number (JCYJ20180507183628543 & [2022]14th).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2022.e12683.
Contributor Information
Xiaoming Jiang, Email: jiangxiaoming@cheerlandgroup.com.
Dongqing Cai, Email: tdongbme@jnu.edu.cn.
Guangqian Zhou, Email: gqzhou@szu.edu.cn.
Appendix. ASupplementary data
The following are the supplementary data related to this article:
References
- 1.Fazeli Z., Abedindo A., Omrani M.D., Ghaderian S.M.H. Mesenchymal stem cells (MSCs) therapy for recovery of fertility: a systematic Review. Stem Cell Rev. Reports. 2018 doi: 10.1007/s12015-017-9765-x. [DOI] [PubMed] [Google Scholar]
- 2.Montesinos J.J., Flores-Figueroa E., Castillo-Medina S., Flores-Guzmán P., Hernández-Estévez E., Fajardo-Orduña G., Orozco S., Mayani H. Human mesenchymal stromal cells from adult and neonatal sources: comparative analysis of their morphology, immunophenotype, differentiation patterns and neural protein expression. Cytotherapy. 2009;11:163–176. doi: 10.1080/14653240802582075. [DOI] [PubMed] [Google Scholar]
- 3.Romanov Y.A. Searching for alternative sources of postnatal human mesenchymal stem cells: candidate MSC-like cells from umbilical cord. Stem Cell. 2003;21:105–110. doi: 10.1634/stemcells.21-1-105. [DOI] [PubMed] [Google Scholar]
- 4.Can A., Celikkan F.T., Cinar O. Umbilical cord mesenchymal stromal cell transplantations: a systemic analysis of clinical trials. Cytotherapy. 2017;19:1351–1382. doi: 10.1016/j.jcyt.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Dupuis V., Oltra E. Methods to produce induced pluripotent stem cell-derived mesenchymal stem cells: mesenchymal stem cells from induced pluripotent stem cells. World J. Stem Cell. 2021;13:1094–1111. doi: 10.4252/wjsc.v13.i8.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y.S., Pelekanos R.A., Ellis R.L., Horne R., Wolvetang E.J., Fisk N.M. Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. Stem Cells Transl. Med. 2012;1:83–95. doi: 10.5966/sctm.2011-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGarvey S.S., Ferreyros M., Kogut I., Bilousova G. Differentiating induced pluripotent stem cells toward mesenchymal stem/stromal cells. Methods Mol. Biol. 2021 doi: 10.1007/7651_2021_383. [DOI] [PubMed] [Google Scholar]
- 8.Wu K.H., Wang S.Y., Xiao Q.R., Yang Y., Huang N.P., Mo X.M., Sun J. Efficient generation of functional cardiomyocytes from human umbilical cord-derived virus-free induced pluripotent stem cells. Cell Tissue Res. 2018;374:275–283. doi: 10.1007/s00441-018-2875-1. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q., Yang B., Cao C., Hu K., Wang P., Man Y. Therapeutic antibody directed osteogenic differentiation of induced pluripotent stem cell derived MSCs. Acta Biomater. 2018;74:222–235. doi: 10.1016/j.actbio.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 10.de Matos B.M., Robert A.W., Stimamiglio M.A., Correa A. Pluripotent-derived mesenchymal stem/stromal cells: an overview of the derivation protocol efficacies and the differences among the derived cells. Stem cell Rev. reports. 2022;18:94–125. doi: 10.1007/s12015-021-10258-z. [DOI] [PubMed] [Google Scholar]
- 11.de Peppo G.M. GMP-compatible, xeno-free culture of human induced mesenchymal stem cells. Methods Mol. Biol. 2021;2286:121–129. doi: 10.1007/7651_2020_285. [DOI] [PubMed] [Google Scholar]
- 12.Gao W.-X., Sun Y.-Q., Shi J., Li C.-L., Fang S.-B., Wang D., Deng X.-Q., Wen W., Fu Q.-L. Effects of mesenchymal stem cells from human induced pluripotent stem cells on differentiation, maturation, and function of dendritic cells. Stem Cell Res. Ther. 2017;8:48. doi: 10.1186/s13287-017-0499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Q., Liu F. Derivation and characterization of mesenchymal stem cells from iPS cells. Methods Mol. Biol. 2020 doi: 10.1007/7651_2020_327. [DOI] [PubMed] [Google Scholar]
- 14.Sabapathy V., Kumar S. hiPSC-derived iMSCs: NextGen MSCs as an advanced therapeutically active cell resource for regenerative medicine. J. Cell Mol. Med. 2016;20:1571–1588. doi: 10.1111/jcmm.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitt J., Vallabhaneni K.C., Penfornis P., Pochampally R. Induced pluripotent stem cell-derived mesenchymal stem cells: a leap toward personalized therapies. Curr. Stem Cell Res. Ther. 2016;11:141–148. doi: 10.2174/1574888x10666151001114321. [DOI] [PubMed] [Google Scholar]
- 16.Lian Q., Zhang Y., Liang X., Gao F., Tse H.-F. Directed differentiation of human-induced pluripotent stem cells to mesenchymal stem cells. Methods Mol. Biol. 2016;1416:289–298. doi: 10.1007/978-1-4939-3584-0_17. [DOI] [PubMed] [Google Scholar]
- 17.Jiao H., Walczak B.E., Lee M.-S., Lemieux M.E., Li W.-J. GATA6 regulates aging of human mesenchymal stem/stromal cells. Stem Cell. 2021;39:62–77. doi: 10.1002/stem.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitzhorn L.-S., Megges M., Wruck W., Rahman M.S., Otte J., Degistirici O., Meisel R., Sorg R.V., Oreffo R.O.C., Adjaye J. Human iPSC-derived MSCs (iMSCs) from aged individuals acquire a rejuvenation signature. Stem Cell Res. Ther. 2019;10:100. doi: 10.1186/s13287-019-1209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajasingh S., Sigamani V., Selvam V., Gurusamy N., Kirankumar S., Vasanthan J., Rajasingh J. Comparative analysis of human induced pluripotent stem cell-derived mesenchymal stem cells and umbilical cord mesenchymal stem cells. J. Cell Mol. Med. 2021;25:8904–8919. doi: 10.1111/jcmm.16851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Chen M., Liao J., Chang C., Liu Y., Padhiar A.A., Zhou Y., Zhou G. Induced pluripotent stem cell-derived mesenchymal stem cells hold lower heterogeneity and great promise in biological research and clinical applications. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.716907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umrath F., Weber M., Reinert S., Wendel H.-P., Avci-Adali M., Alexander D. iPSC-derived MSCs versus originating jaw periosteal cells: comparison of resulting phenotype and stem cell potential. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21020587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Rebollo E., Franzen J., Goetzke R., Hollmann J., Ostrowska A., Oliverio M., Sieben T., Rath B., Kornfeld J.-W., Wagner W. Senescence-associated metabolomic phenotype in primary and iPSC-derived mesenchymal stromal cells. Stem Cell Rep. 2020;14:201–209. doi: 10.1016/j.stemcr.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeratunga P., Shahsavari A., Fennis E., Wolvetang E.J., Ovchinnikov D.A., Whitworth D.J. Induced pluripotent stem cell-derived mesenchymal stem cells from the tasmanian devil (Sarcophilus harrisii) express immunomodulatory factors and a tropism toward devil facial tumor cells. Stem Cell. Dev. 2020;29:25–37. doi: 10.1089/scd.2019.0203. [DOI] [PubMed] [Google Scholar]
- 24.Wu Q., Yang Y., Xie D., Li S., Liu Y., Shu L., Fu G., Xu Y., Ji P. The sialylation profile of IgG determines the efficiency of antibody directed osteogenic differentiation of iMSCs by modulating local immune responses and osteoclastogenesis. Acta Biomater. 2020;114:221–232. doi: 10.1016/j.actbio.2020.07.055. [DOI] [PubMed] [Google Scholar]
- 25.Kim H., Zhao Q., Barreda H., Kaur G., Hai B., Choi J.M., Jung S.Y., Liu F., Lee R.H. Identification of molecules responsible for therapeutic effects of extracellular vesicles produced from iPSC-derived MSCs on Sjögren's syndrome. Aging Dis. 2021;12:1409–1422. doi: 10.14336/AD.2021.0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan X.-L., Zeng Q.-X., Li X., Li C.-L., Xu Z.-B., Deng X.-Q., Shi J., Chen D., Zheng S.G., Fu Q.-L. Induced pluripotent stem cell-derived mesenchymal stem cells activate quiescent T cells and elevate regulatory T cell response via NF-κB in allergic rhinitis patients. Stem Cell Res. Ther. 2018;9:170. doi: 10.1186/s13287-018-0896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloor A.J.C., Patel A., Griffin J.E., Gilleece M.H., Radia R., Yeung D.T., Drier D., Larson L.S., Uenishi G.I., Hei D., Kelly K., Slukvin I., Rasko J.E.J. Production, safety and efficacy of iPSC-derived mesenchymal stromal cells in acute steroid-resistant graft versus host disease: a phase I, multicenter, open-label, dose-escalation study. Nat. Med. 2020;26:1720–1725. doi: 10.1038/s41591-020-1050-x. [DOI] [PubMed] [Google Scholar]
- 28.Wu Z., Liu W., Wang Z., Zeng B., Peng G., Niu H., Chen L., Liu C., Hu Q., Zhang Y., Pan M., Wu L., Liu M., Liu X., Liang D. Mesenchymal stem cells derived from iPSCs expressing interleukin-24 inhibit the growth of melanoma in the tumor-bearing mouse model. Cancer Cell Int. 2020;20:33. doi: 10.1186/s12935-020-1112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungbluth P., Spitzhorn L.-S., Grassmann J., Tanner S., Latz D., Rahman M.S., Bohndorf M., Wruck W., Sager M., Grotheer V., Kröpil P., Hakimi M., Windolf J., Schneppendahl J., Adjaye J. Human iPSC-derived iMSCs improve bone regeneration in mini-pigs. Bone Res. 2019;7:32. doi: 10.1038/s41413-019-0069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diederichs S., Tuan R.S. Functional comparison of human-induced pluripotent stem cell-derived mesenchymal cells and bone marrow-derived mesenchymal stromal cells from the same donor. Stem Cell. Dev. 2014;23:1594–1610. doi: 10.1089/scd.2013.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M., Shaw G., Murphy M., Barry F. Induced pluripotent stem cell-derived mesenchymal stromal cells are functionally and genetically different from bone marrow-derived mesenchymal stromal cells. Stem Cell. 2019;37:754–765. doi: 10.1002/stem.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schondorf D.C., Elschami M., Schieck M., Ercan-Herbst E., Weber C., Riesinger Y., Kalman S., Steinemann D., Ehrnhoefer D.E. Generation of an induced pluripotent stem cell cohort suitable to investigate sporadic Alzheimer's Disease. Stem Cell Res. 2019;34 doi: 10.1016/j.scr.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 33.Philippidou P., Dasen J.S. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulebyakina M., Makarevich P. Hox-positive adult mesenchymal stromal cells: beyond positional identity. Front. Cell Dev. Biol. 2020;8:624. doi: 10.3389/fcell.2020.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picchi J., Trombi L., Spugnesi L., Barachini S., Maroni G., Brodano G.B., Boriani S., Valtieri M., Petrini M., Magli M.C. HOX and TALE signatures specify human stromal stem cell populations from different sources. J. Cell. Physiol. 2013;228:879–889. doi: 10.1002/jcp.24239. [DOI] [PubMed] [Google Scholar]
- 36.Payumo A.Y., McQuade L.E., Walker W.J., Yamazoe S., Chen J.K. Tbx16 regulates hox gene activation in mesodermal progenitor cells. Nat. Chem. Biol. 2016;12:694–701. doi: 10.1038/nchembio.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwart I., Hill A.J., Girdlestone J., Manca M.F., Navarrete R., Navarrete C., Jen L.-S. Analysis of neural potential of human umbilical cord blood-derived multipotent mesenchymal stem cells in response to a range of neurogenic stimuli. J. Neurosci. Res. 2008;86:1902–1915. doi: 10.1002/jnr.21649. [DOI] [PubMed] [Google Scholar]
- 38.Zhao C., Ikeya M. Generation and applications of induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cell. Int. 2018;2018 doi: 10.1155/2018/9601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdal Dayem A., Lee S. Bin, Kim K., Lim K.M., Jeon T.-I., Seok J., Cho A.S.-G. Production of mesenchymal stem cells through stem cell reprogramming. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakob M., Hambrecht M., Spiegel J.L., Kitz J., Canis M., Dressel R., Streckfuss-Bömeke K. Pluripotent stem cell-derived mesenchymal stem cells show comparable functionality to their autologous origin. 2020. Cells 10. [DOI] [PMC free article] [PubMed]
- 41.Hobert O. Homeobox genes and the specification of neuronal identity. Nat. Rev. Neurosci. 2021;22:627–636. doi: 10.1038/s41583-021-00497-x. [DOI] [PubMed] [Google Scholar]
- 42.Fukuta M., Nakai Y., Kirino K., Nakagawa M., Sekiguchi K., Nagata S., Matsumoto Y., Yamamoto T., Umeda K., Heike T., Okumura N., Koizumi N., Sato T., Nakahata T., Saito M., Otsuka T., Kinoshita S., Ueno M., Ikeya M., Toguchida J. Derivation of mesenchymal stromal cells from pluripotent stem cells through a neural crest lineage using small molecule compounds with defined media. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q., Li J., Wang S., Deng Q., Wang K., Dai X., An Y., Dong G., Ke W., Chen F., Liu L., Yang H., Du Y., Zhao W., Shang Z. Single‐cell transcriptome profiling reveals molecular heterogeneity in human umbilical cord tissue and culture‐expanded mesenchymal stem cells. FEBS J. 2021;288:3069–3082. doi: 10.1111/febs.15834. [DOI] [PubMed] [Google Scholar]
- 44.Brick R.M., Sun A.X., Tuan R.S. Neurotrophically induced mesenchymal progenitor cells derived from induced pluripotent stem cells enhance neuritogenesis via neurotrophin and cytokine production. Stem Cells Transl. Med. 2018;7:45–58. doi: 10.1002/sctm.17-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eto S., Goto M., Soga M., Kaneko Y., Uehara Y., Mizuta H., Era T. Mesenchymal stem cells derived from human iPS cells via mesoderm and neuroepithelium have different features and therapeutic potentials. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouchi T., Morikawa S., Shibata S., Fukuda K., Okuno H., Fujimura T., Kuroda T., Ohyama M., Akamatsu W., Nakagawa T., Okano H. LNGFR(+)THY-1(+) human pluripotent stem cell-derived neural crest-like cells have the potential to develop into mesenchymal stem cells. Differentiation. 2016;92:270–280. doi: 10.1016/j.diff.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Shahsavari A., Weeratunga P., Ovchinnikov D.A., Whitworth D.J. Pluripotency and immunomodulatory signatures of canine induced pluripotent stem cell-derived mesenchymal stromal cells are similar to harvested mesenchymal stromal cells. Sci. Rep. 2021;11:3486. doi: 10.1038/s41598-021-82856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnabel L.V., Abratte C.M., Schimenti J.C., Felippe M.J.B., Cassano J.M., Southard T.L., Cross J.A., Fortier L.A. Induced pluripotent stem cells have similar immunogenic and more potent immunomodulatory properties compared with bone marrow-derived stromal cells in vitro. Regen. Med. 2014;9:621–635. doi: 10.2217/rme.14.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng P.-P., Liu X.-C., Ma P.-F., Gao C., Li J.-L., Lin Y.-Y., Shao W., Han S., Zhao B., Wang L.-M., Fu J.-Z., Meng L.-X., Li Q., Lian Q.-Z., Xia J.-J., Qi Z.-Q. iPSC-MSCs combined with low-dose rapamycin induced islet allograft tolerance through suppressing Th1 and enhancing regulatory T-cell differentiation. Stem Cell. Dev. 2015;24:1793–1804. doi: 10.1089/scd.2014.0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang S.-B., Zhang H.-Y., Jiang A.-Y., Fan X.-L., Lin Y.-D., Li C.-L., Wang C., Meng X.-C., Fu Q.-L. Human iPSC-MSCs prevent steroid-resistant neutrophilic airway inflammation via modulating Th17 phenotypes. Stem Cell Res. Ther. 2018;9:147. doi: 10.1186/s13287-018-0897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wruck W., Graffmann N., Spitzhorn L.-S., Adjaye J. Human induced pluripotent stem cell-derived mesenchymal stem cells acquire rejuvenation and reduced heterogeneity. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.717772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giuliani M., Oudrhiri N., Noman Z.M., Vernochet A., Chouaib S., Azzarone B., Durrbach A., Bennaceur-Griscelli A. Human mesenchymal stem cells derived from induced pluripotent stem cells down-regulate NK-cell cytolytic machinery. Blood. 2011;118:3254–3262. doi: 10.1182/blood-2010-12-325324. [DOI] [PubMed] [Google Scholar]
- 53.Chow L., Johnson V., Regan D., Wheat W., Webb S., Koch P., Dow S. Safety and immune regulatory properties of canine induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cell Res. 2017;25:221–232. doi: 10.1016/j.scr.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fong C.Y., Subramanian A., Biswas A., Gauthaman K., Srikanth P., Hande M.P., Bongso A. Derivation efficiency, cell proliferation, freeze-thaw survival, stem-cell properties and differentiation of human Wharton's jelly stem cells. Reprod. Biomed. Online. 2010;21:391–401. doi: 10.1016/j.rbmo.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Sfougataki I., Grafakos I., Varela I., Mitrakos A., Karagiannidou A., Tzannoudaki M., Poulou M., Mertzanian A., Roubelakis G M., Stefanaki K., Traeger-Synodinos J., Kanavakis E., Kitra V., Tzetis M., Goussetis E. Reprogramming of bone marrow derived mesenchymal stromal cells to human induced pluripotent stem cells from pediatric patients with hematological diseases using a commercial mRNA kit. Blood Cells Mol. Dis. 2019;76:32–39. doi: 10.1016/j.bcmd.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Ding H., Chen S., Pan X., Dai X., Pan G., Li Z., Mai X., Tian Y., Zhang S., Liu B., Cao G., Yao Z., Yao X., Gao L., Yang L., Chen X., Sun J., Chen H., Han M., Yin Y., Xu G., Li H., Wu W., Chen Z., Lin J., Xiang L., Hu J., Lu Y., Zhu X., Xie L. Transferrin receptor 1 ablation in satellite cells impedes skeletal muscle regeneration through activation of ferroptosis. J. Cachexia. Sarcopenia Muscle. 2021;12:746–768. doi: 10.1002/jcsm.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang W., Lian W., Chen J., Li W., Huang J., Lai B., Li L., Huang Z., Xu J. Rapid identification of genome-edited mesenchymal stem cell colonies via Cas9. Biotechniques. 2019;66:231–234. doi: 10.2144/btn-2018-0183. [DOI] [PubMed] [Google Scholar]
- 58.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., Von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data with fastq format was stored in NCBI, accession number PRJNA860220 https://dataview.ncbi.nlm.nih.gov/object/PRJNA860220.