Summary
Two engineered mycobacteriophages were administered intravenously with antibiotics to a male with treatment-refractory Mycobacterium abscessus pulmonary infection and severe cystic fibrosis lung disease. Evidence of phage-induced lysis was observed using molecular and metabolic assays within days of phage treatment initiation, followed by computerized tomography assessed improvement by day 81, and conversion of airway cultures to predominately negative by day 118. Pan-genome analysis of M. abscessus isolates pre- and post-phage treatment demonstrated genetic stability, with a general decline in diversity and no increased resistance to phage or antibiotics. Anti-phage neutralizing antibody titers to one phage increased with time, but did not prevent clinical improvement throughout the course of treatment. The subject received lung transplantation on day 379, and systematic culturing of the explanted lung did not detect M. abscessus. This study describes the course and associated markers of successful phage treatment of M. abscessus in advanced lung disease.
Keywords: Cystic Fibrosis, Mycobacterium abscessus, bacteriophages, lipoarabinomannan, immunoglobulin, lung transplant, CFTR modulator therapy, nontuberculous mycobacterial (NTM) lung disease
Introduction
Bacteriophages (phages) are viruses that selectively infect bacteria and can replicate lytically, resulting in bacterial death. Lytic phages are increasingly being evaluated for therapeutic use in difficult-to-treat infections (Hatfull et al., 2021). Mycobacterium abscessus is a nontuberculous mycobacteria (NTM) with high levels of intrinsic and acquired antibiotic resistance that limit treatment options and contribute towards poor outcomes (Daley et al., 2020). While M. abscessus has been cited as a potentially important target of phage therapy (Senhaji-Kacha et al., 2020), and is among the most common infections referred for phage therapy (Aslam et al., 2020), there are few reports of its use to date since a 2019 case study described treatment of a disseminated M. abscessus subspecies massiliense infection post-lung transplantation (Dedrick et al., 2021b; Dedrick et al., 2019).
Persons with cystic fibrosis (pwCF) represent the most vulnerable population for NTM lung disease (Nick et al., 2021). M. abscessus infections have been associated with a greater decline in lung function compared to typical CF airway pathogens (Qvist et al., 2015b), and when refractory to treatment impose a contraindication to lung transplant (Orens et al., 2006; Weill et al., 2015). In advanced CF lung disease, antibiotic treatment of M. abscessus infection may be unsuccessful due to structural changes to the lung, in particular cavities and regions of dense consolidation and parenchymal collapse, which limit therapeutic concentrations of antimicrobials. M. abscessus is also known to form biofilms within the CF airway (Qvist et al., 2015a). In microenvironments within the damaged lung, mycobacteria may survive for extended periods without oxygen or nutrients by shifting into a non-replicating “persister” state that may further thwart conventional antimicrobials and host defense (Yam et al., 2020).
Phage treatment of M. abscessus in the CF airway poses both similar and unique challenges compared to antibiotic therapy. M. abscessus clinical isolates vary greatly in their phage susceptibilities, and therefore therapeutic phages need to be personalized for each patient (Dedrick et al., 2021a; Dedrick et al., 2021c). Moreover, most phages that infect M. abscessus are temperate, with capacity to integrate into bacterial genomes, and must be engineered to have lytic but not lysogenic behavior (Dedrick et al., 2019). Co-infection by closely related lineages of M. abscessus, as well as other species of NTM, raises the potential for selection of a more phage-resistant isolate (Aslam et al., 2020; Senhaji-Kacha et al., 2020), although phage resistance occurs relatively infrequently in M. abscessus (Dedrick et al., 2021c). Development of neutralizing antibodies against the phage can also potently negate the therapy (Dedrick et al., 2021b; Roach et al., 2017). In a broader disease context, if substantial killing of M. abscessus was achieved, the resulting niche could become occupied by other opportunistic pathogens.
Pre-existing bacterial co-infections – most commonly due to Pseudomonas aeruginosa and Staphylococcus aureus – reduce the sensitivity of airway cultures to detect M. abscessus and may obscure markers of clinical response to successful treatment (Whittier et al., 1997). Given the relatively low sensitivity of NTM cultures in the CF airway, there is a need for improved culture independent markers of treatment response to assess efficacy and treatment endpoints. Experiences with phage therapy against P. aeruginosa, S. aureus, and Burkholderia dolosa in CF lung disease have demonstrated examples of improvement, but without clear evidence of eradication of the target infection (Chan et al., 2021). In some cases, phage therapy resulted in the emergence of phage-resistant and antibiotic-resistant isolates, as well as new pathogens (Chan et al., 2021).
Herein we describe the compassionate use of two engineered mycobacteriophages for a young pwCF and M. abscessus subspecies abscessus lung disease, whose infection was refractory against intensive multidrug antibiotic treatment. Extensive collection and biobanking of isolates and clinical samples throughout the duration of his infection pre- and post-phage therapy has enabled an in-depth assessment of the effect of phage treatment on M. abscessus infection in the CF lung.
Results
Clinical Presentation of A Treatment-refractory M. abscessus Infection in Advanced CF Lung Disease.
This individual is a 26-year-old man with CF (genotype H199Y/2184insA) with advanced bronchiectasis, M. abscessus lung disease, and chronic lung infection with multi-drug resistant (MDR) P. aeruginosa and methicillin-resistant S. aureus (MRSA). He had been successfully treated for pulmonary M. avium infection 5 years earlier without evidence of reoccurrence. As an adult his clinical course was notable for frequent pulmonary exacerbations, requiring hospitalization and treatment with intravenous (IV) antibiotics.
Sputum cultures first turned positive for M. abscessus subspecies abscessus with an intact erm(41) gene in May 2016 (day -1582 prior to initiation of phage treatment). Over the next year recurrent positive cultures and clinical features of increased cough, accelerated lung function decline, increased frequency of pulmonary exacerbations and worsening radiographic findings met criteria for NTM lung disease (Figure 1A, 1B) (Daley et al., 2020). Throughout this infection, the preponderance (69 of 71) of his M. abscessus isolates demonstrated only a rough colony morphology. He was initiated on 4-drug therapy on day -1236 (Figure 1D). Over the subsequent 4.7 years, he was maintained on 4–5 drugs, which were rotated based on drug availability, susceptibility testing, tolerance of side-effects, and response to treatment. During his frequent hospitalizations, this NTM therapy was modified to allow for concurrent treatment of P. aeruginosa and MRSA, although at no time was he treated with fewer than 4 drugs against M. abscessus.
Figure 1. Clinical Summary From Time of Estimated Initial Infection Through Day 500 of Phage Treatment.
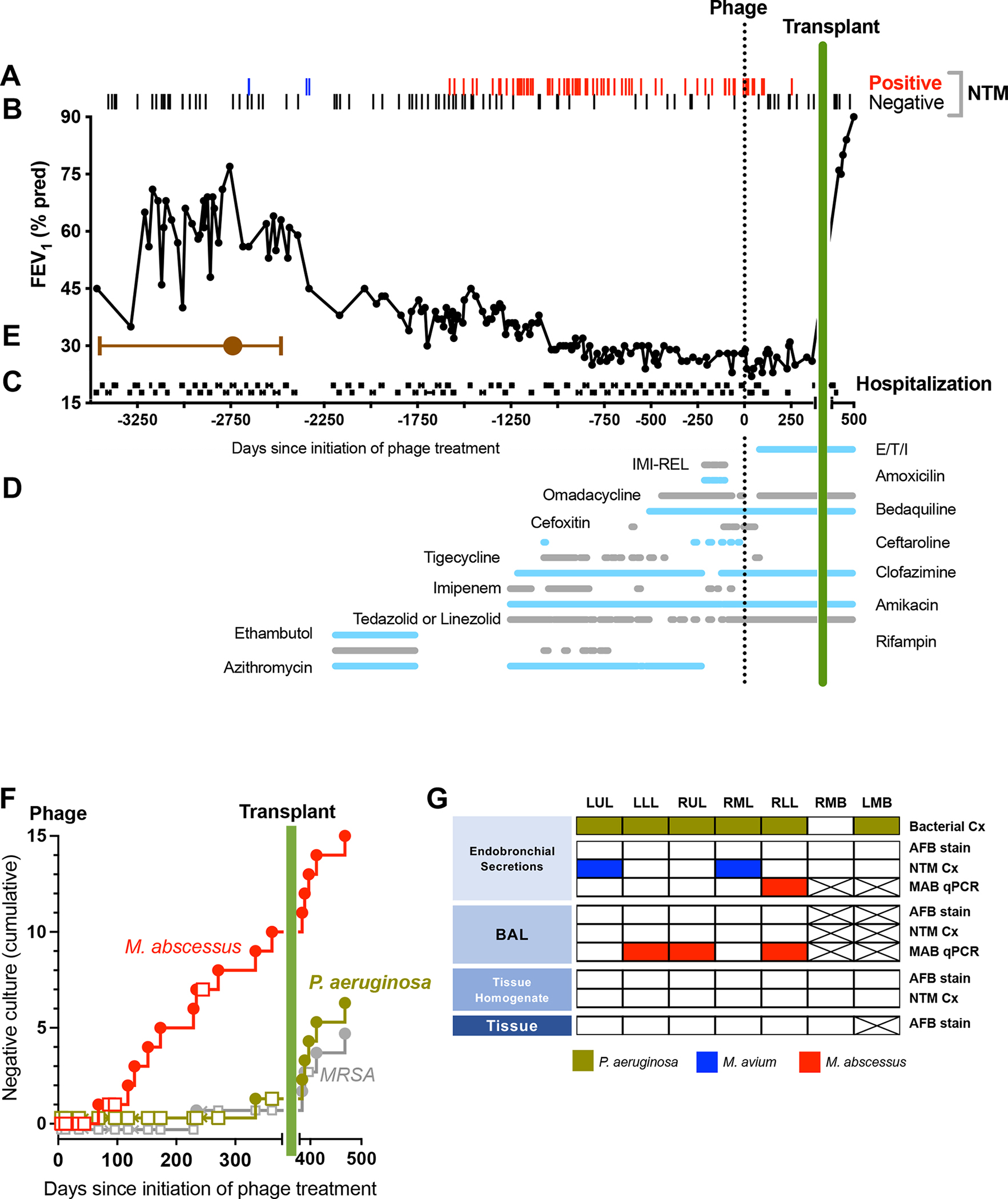
Clinical parameters are plotted versus time in days, with initial phage administration labeled as Day 0.
(A) Airway culture results (n=155) are depicted with red bars (positive M. abscessus, n=71), blue bars (positive M. avium, n=2) or gray bars (negative culture, n=82). Cultures prior to transplant (day 379) were obtained from expectorated sputum, and following transplant were obtained from BAL.
(B) Forced expiratory volume in 1 sec (FEV1, percent predicted) plotted versus days.
(C) Hospitalizations (black horizontal lines) plotted versus days.
(D) Treatment course. The type and duration of CFTR modulator and various antibiotics (horizontal lines) directed towards M. avium or M. abscessus plotted versus days. Agents used in order of most recent to earliest introduction: 1) Elexacaftor/tezacaftor/ivacaftor (E/T/I), 2) Imipenem-cilastatin-relebactam (IMI-REL), 3) Amoxicillin, 4) Omadacycline, 5) Bedaquiline, 6) Cefoxitin, 7) Ceftaroline, 8) Tigecycline, 9) Clofazimine, 10) Imipenem, 11) Amikacin, 12) Tedizolid or linezolid (oral or IV), 13) Ethambutol, 14) Rifampin, 15) Azithromycin.
(E) Timeframe of initial infection. The estimated date of the most common recent ancestor based on divergence dating analysis was pre-phage day -2494 (brown circle) with a 95% Highest Posterior Density (HPD) interval from day -1945 to day -3435 (brown bars).
(F) Cumulative negative airway cultures following initiation of phage therapy. The occurrence of negative culture over time was plotted for M. abscessus (negative cultures red circle, positive cultures red square), P. aeruginosa (negative culture green circle, positive culture green square), and MRSA (positive culture grey circle, negative culture grey square). The proportion of negative cultures was greater for M. abscessus compared to P. aeruginosa or MRSA (p=0.007 by Fisher’s Exact Test).
(G) Systematic analysis of explanted lung tissue for M. abscessus. Endobronchial secretions, BAL, homogenized tissue and tissue pathology from the left upper lobe (LUL), left lower lobe (LLL), right upper lobe (RUL), right middle lobe (RML), right lower lobe (RLL), right mainstem bronchi (RMB) and left mainstem bronchi (LMB) were analyzed by bacterial culture for usual CF pathogens (Bacterial Cx), AFB stain, NTM cultures (NTM Cx), species-specific qPCR for M. abscessus DNA (MAB qPCR). Positive results are indicated for P. aeruginosa (green box), M. avium (blue box) or M. abscessus (red box). Negative results are indicated by white boxes, and assays not performed by crossed-out box.
Despite intensive therapy, the participant had persistently positive cultures for M. abscessus subspecies abscessus, worsening radiographic features, and declining lung function, which indicated a lack of microbiologic response (Figure 1A, 1B). In the 12 months prior to phage administration, he required 11 hospital admissions ranging from 11 to 21 days in duration (Figure 1C). He had been declined for lung transplant by three independent lung transplant programs in part due to a lack of therapeutic options for treatment–refractory M. abscessus lung disease.
Complete Genome of M. abscessus Isolated in Early Infection
An isolate collected -1555 days prior to phage administration (i.e., ‘-1555’ sample) was sequenced to provide a complete genome of an early time point isolate. Long and short reads were assembled into one circularized chromosome of 5,018,523 bp. The complete genome contained genes involved in resistance to general antibiotics (mdtH, stp, sugE) and specific antibiotics such as beta-lactams (penP), isoniazid and ethambutol (iniA), bicyclomycin (bcr), doxorubicin (drrA), fosmidomycin (fsr), and tetracycline (tetA) (Seemann, 2014) which is typical of the species. The genome was free of intact prophages and no plasmids were detected.
Identification of Phages Active Against M. abscessus
An M. abscessus isolate grown from sputum collected on day -1437 was screened for sensitivity to a panel of phages using plaque assays (Figure 2A). The M. abscessus pre-phage day -1437 isolate is sensitive to two host range mutants (HRMs) of an engineered lytic derivative of phage BPs (i.e. BPsΔ33HTH_HRM10 and BPsΔ33HTH_HRMGD03) (Dedrick et al., 2019)(Figure 2A). This engineering removed part of the repressor gene, rendering them lytic. Phage D29 is a lytic phage grouped in Cluster A and is a presumed naturally occurring derivative of a parent temperate phage (Dedrick et al., 2017; Ford et al., 1998); D29_HRMGD40 is an HRM of D29 isolated on an M. abscessus strain GD40, which unlike the D29 parent infects several M. abscessus clinical isolates (Dedrick et al., 2021c). M. abscessus isolate pre-phage day -1437 was efficiently killed by incubation with BPsΔ33HTH_HRM10 and D29_HRMGD40 alone or in combination, over a wide range of bacterial and phage concentrations (Figure 2B). Upon challenge of 108 colony forming units (CFU) of pre-phage day -1437 isolate with either phage separately or both together at a multiplicity of infection (MOI) of 10, 2–6 surviving colonies were recovered (Fig. 2B), but all were found to be fully sensitive to both phages upon retesting (Figure 2C and D). Phages BPsΔ33HTH_HRM10 and D29_HRMGD40 were thus chosen as a therapeutic phage cocktail, amplified and purified to high titer as described previously (Dedrick et al., 2019), and shown to be sterile and endotoxin-free.
Figure 2. Phage Therapy Did Not Select for Phage-resistant Isolates.
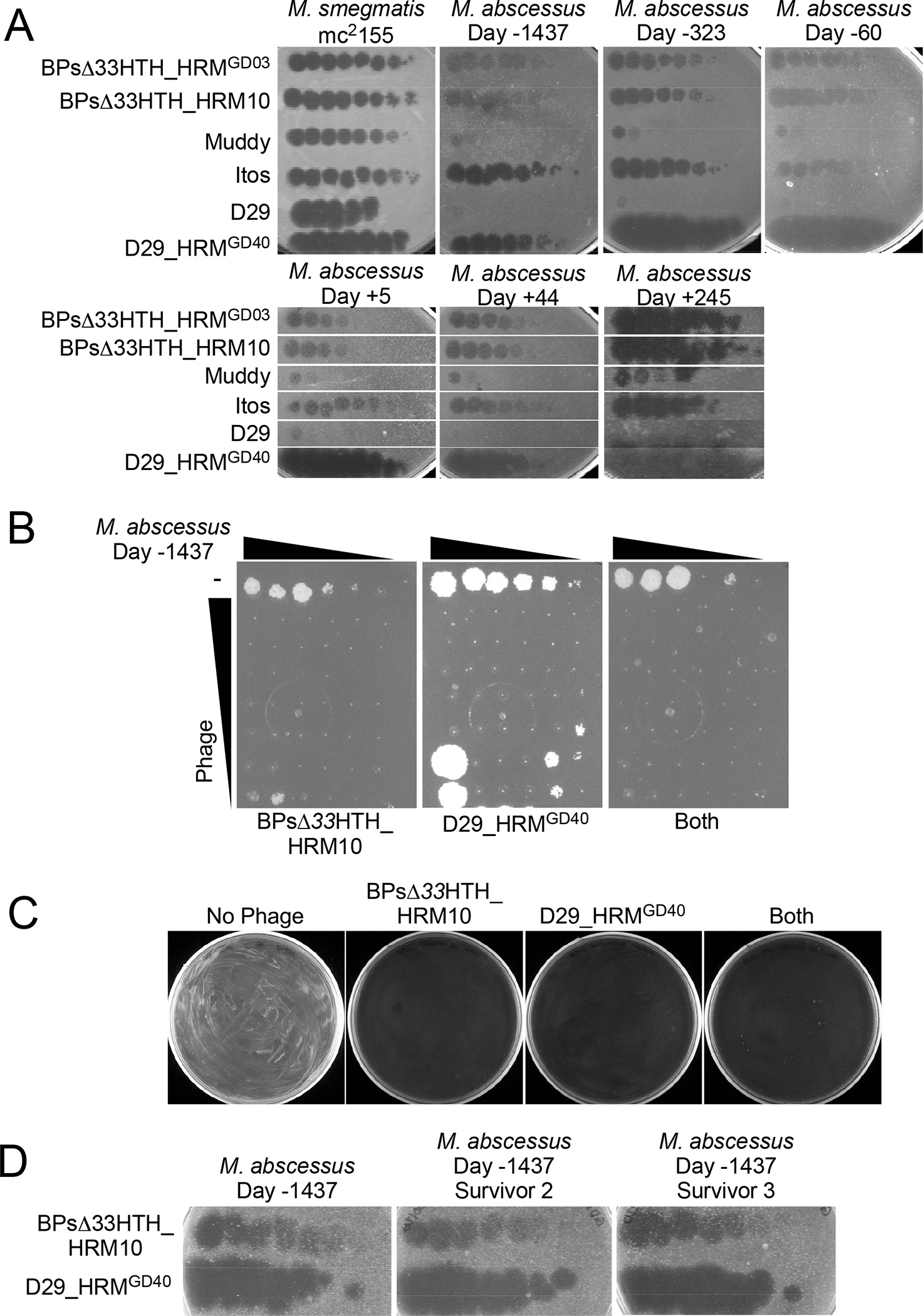
(A) Efficiencies of plating of phages on M. smegmatis mc2155 and six M. abscessus isolates from the subject -1437, -323 and -60 days prior to initiation of phage therapy, as well as 5, 44, and 245 days after therapy started. Phage lysates were 10-fold serially diluted and spotted onto top agar overlays of each strain. Phages BPΔ33HTH_HRMGD03, BPΔ33HTH_HRM10, Muddy, Itos, D29 and D29_HRMGD40 infect each M. abscessus isolate with similar efficiencies to M. smegmatis mc2155. Phage BPΔ33HTH_HRM10, which was used therapeutically previously (Dedrick et al., 2021b; Dedrick et al., 2019), and D29_HRMGD40 were chosen as therapeutic candidates; phage Itos infects efficiently but does not kill the M. abscessus strains tested well.
(B) Killing of M. abscessus strain collected -1437 days prior to initiation of phage by BPΔ33HTH_HRM10, D29_HRMGD40 or both phages. Each column has a 10-fold serial dilution of the isolate (left column, 2×105 CFU) and each row has 10-fold serial dilutions of either BPΔ33HTH_HRM10 (top row, 109 PFU), D29_HRMGD40 (top row, 107 PFU) or both phages.
(C) Survival assay showing efficient killing of M. abscessus strain collected -1437 days prior to initiation of phage with either BPΔ33HTH_HRM10 or D29_HRMGD40 or both phages together.
(D) Phage susceptibility assays of two survivors recovered from a challenge with BPΔ33HTH_HRM10 and D29_HRMGD40 of M. abscessus strain collected -1437 days prior to initiation of phage shows both are fully phage-sensitive. Phages were tested as in panel A.
Clinical Response to Administration of Mycobacteriophage
Phage dosing was initiated in September 2020 when the individual was near baseline clinical status during hospitalization for a pulmonary exacerbation. Approximately 109 plaque-forming units (PFU/ml) of BPsΔ33HTH_HRM10 and 108 PFU of D29_HRMGD40 were administered intravenously twice daily through the time of this writing (March 2022), and have been tolerated well, with no adverse events attributed to the therapy. He was discharged from the hospital on day 5 post initiation of phage therapy. He was readmitted on four occasions in the year following initiation of phage for signs and symptoms consistent with a typical pulmonary exacerbation, which on one occasion was complicated by sepsis due to MRSA infection of his implanted venous access port. NTM sputum cultures (n=17) were obtained between day 5 and 362 post-initiation of phage therapy. NTM cultures for M. abscessus subspecies abscessus remained largely positive (6 of 7) through day 96. However, 9 of the 10 most recent cultures (days 116 to 362) were negative, with a single isolate recovered on day 245 (Figure 1A). In contrast, over the same interval, only 1 of 13 (7.7%) airway cultures were negative for either P. aeruginosa or MRSA (Figure 1F). From the start of antibiotic treatment (day -1236) to the initiation of phage, 77.9% (53 of 68) of sputum cultures were positive for M. abscessus, compared to 41.1% (7 of 17) in the year following phage initiation (p=0.006 by 2-tail Fisher’s Exact Test), and 10% (1 of 10) in the most recent 280 days of phage and antibiotic treatment compared to start of antibiotics in addition to phage (p=0.0001). On day 89 the patient was also initiated on the CFTR modulator elexacaftor/tezacaftor/ivacaftor (E/T/I)(Heijerman et al., 2019) following FDA approval of the drug for the H199Y CFTR gene mutation (NIH, 2021). Over the year post-phage initiation his FEV1 ranged between 23–31 (% predicted), which was not different from prior to phage treatment. However, his hospital-free time increased (Figure 1C). Based on evidence that the M. abscessus infection was controlled, he was listed for lung transplant a year after the initiation of phage therapy, and was successfully transplanted on day 379.
Systematic Analysis of Explanted Lung Tissue and Post-transplant Course
At transplant the participant’s lungs were surveyed for evidence of M. abscessus. Bronchoalveolar lavage (BAL), swabs of unprocessed endobronchial secretions, gross pathology and homogenized tissue from each lobe and the mainstem bronchovascular margins were stained for acid fact bacilli (AFB), cultured for NTM and bacteria, and analyzed by real-time quantitative PCR (qPCR) (Rocchetti et al., 2017) (Figure 1G). All sample types were negative for AFB by stain in all lobes. Cultures were positive for P. aeruginosa from all lobes and the left mainstem bronchi, and M. avium grew from two lobes. M. abscessus DNA was detected in BAL from 3 lobes and in endobronchial secretions from one lobe by qPCR, but M. abscessus did not grow from any of the matched samples (Figure 1G). Gross pathology of the explanted lungs did not demonstrate granulomas, and AFB was not observed in any section (Supplemental Figure 1). An additional 5 airway cultures sampled via BAL of the transplanted lungs through day 500 of phage treatment (121 days post-transplant) have been negative for M. abscessus, with no clinical evidence of disseminated infection. The participant has experienced an uncomplicated recovery from transplant, with normalization of lung function (Figure 1B), and has remained on phage and antibiotic treatment.
Radiologic Changes During Phage Therapy
Serial high-resolution computerized tomography (CT) scans demonstrated severe diffuse varicose upper-lobe cystic bronchiectasis with a left apical cavity. Multifocal mucoid impaction, widespread mosaic attenuation, and scattered poorly defined nodules were present. Focal tree-in-bud pattern was most marked in the right lower lobe. Selected radiographic features associated with NTM infection were compared between a pre-treatment scan and scans from post-phage therapy day 81 and 229 (Figure 3). At day 81 clear examples of decreased size of consolidative nodules associated with phage treatment were observed, however this improvement was not uniform, and areas of worsening consolidation and mucous plugging were present as well. By post-phage day 229, additional areas of improvement were apparent.
Figure 3. Radiologic Changes During Phage Therapy.
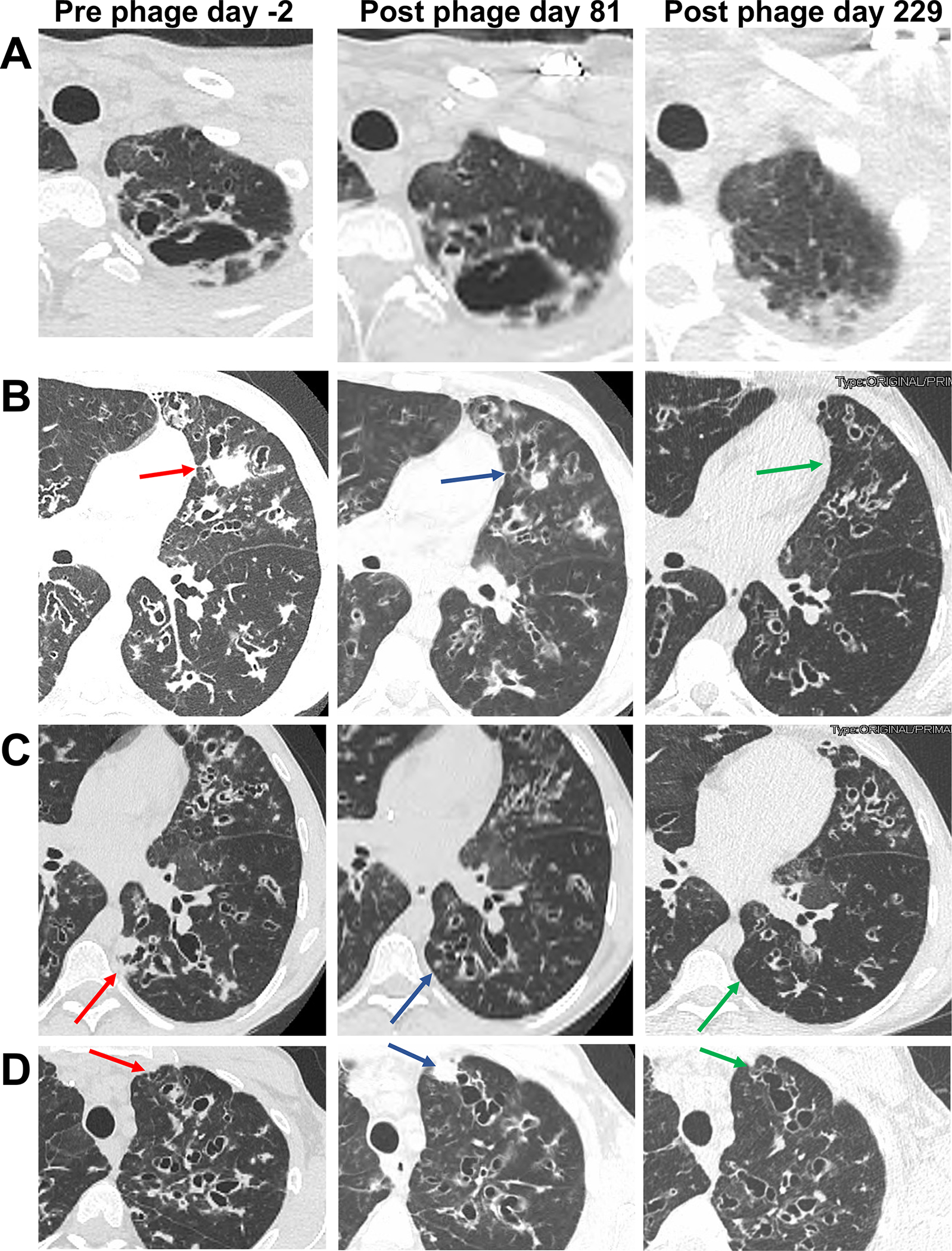
(A) Left: Axial CT pre-treatment demonstrates a left apical cavity. Middle: Little change was observed at day 81 of treatment. Right: Near resolution of the cavity was observed at day 229.
(B) Left: Axial CT pre-treatment shows widespread bronchiectasis and mucoid impaction, and a consolidative mass in the lingula (red arrow). Middle: At day 81 of treatment the bronchiectasis and mucoid impaction was largely unchanged, but the lingular mass decreased substantially (blue arrow). Right: At day 229 the consolidation was resolved (green arrow), and the mucoid impaction throughout the left mid-lung region was substantially improved.
(C) Left: Axial CT pre-treatment shows widespread bronchiectasis and mucoid impaction, and a consolidative mass in the left lower-lobe (red arrow). Middle: At day 81 of treatment the bronchiectasis and mucoid impaction was somewhat improved, and the left lower-lobe mass decreased substantially (blue arrow). Right: At day 229 the consolidation was resolved (green arrow) and bronchiectatic airways were generally reduced in size.
(D) Left: Axial CT pre-treatment shows widespread bronchiectasis and mucoid in the left upper-lobe (red arrow). Middle: At day 81 of treatment the bronchiectasis and mucoid impaction was largely unchanged, with the new appearance of a consolidative nodule in the anterior left upper lobe (blue arrow). Right: At day 229 the consolidative nodule present at day 80 was resolved (green arrow).
Culture-independent Markers Demonstrate Evidence of M. abscessus Lysis
Molecular quantification of M. abscessus DNA from total DNA of 17 sputum samples ranging from -136 days pre-phage to 361 days post-phage was analyzed by real-time qPCR (Rocchetti et al., 2017) as a complement to traditional cultures (Figure 4A). Compared to a standard curve with known concentrations of M. abscessus, sputum samples from days -136 to 82 demonstrated consistent detection of M. abscessus DNA corresponding with positive cultures during that time period (Figure 4B, Supplemental Figure 2). Sputum samples from day -136 to day 3 had M. abscessus DNA concentrations approaching the level of detection (LOD), and an increase in M. abscessus DNA was observed at days 47 and 82 just prior to culture conversion. Nine sputum samples collected between days 130 to 361 did not show detectable levels of M. abscessus DNA (below the LOD) consistent with primarily negative cultures in that time frame. The participant was unable to produce sputum following lung transplant.
Figure 4. Culture-independent Markers Demonstrate Evidence of M. abscessus Lysis and Host Response.
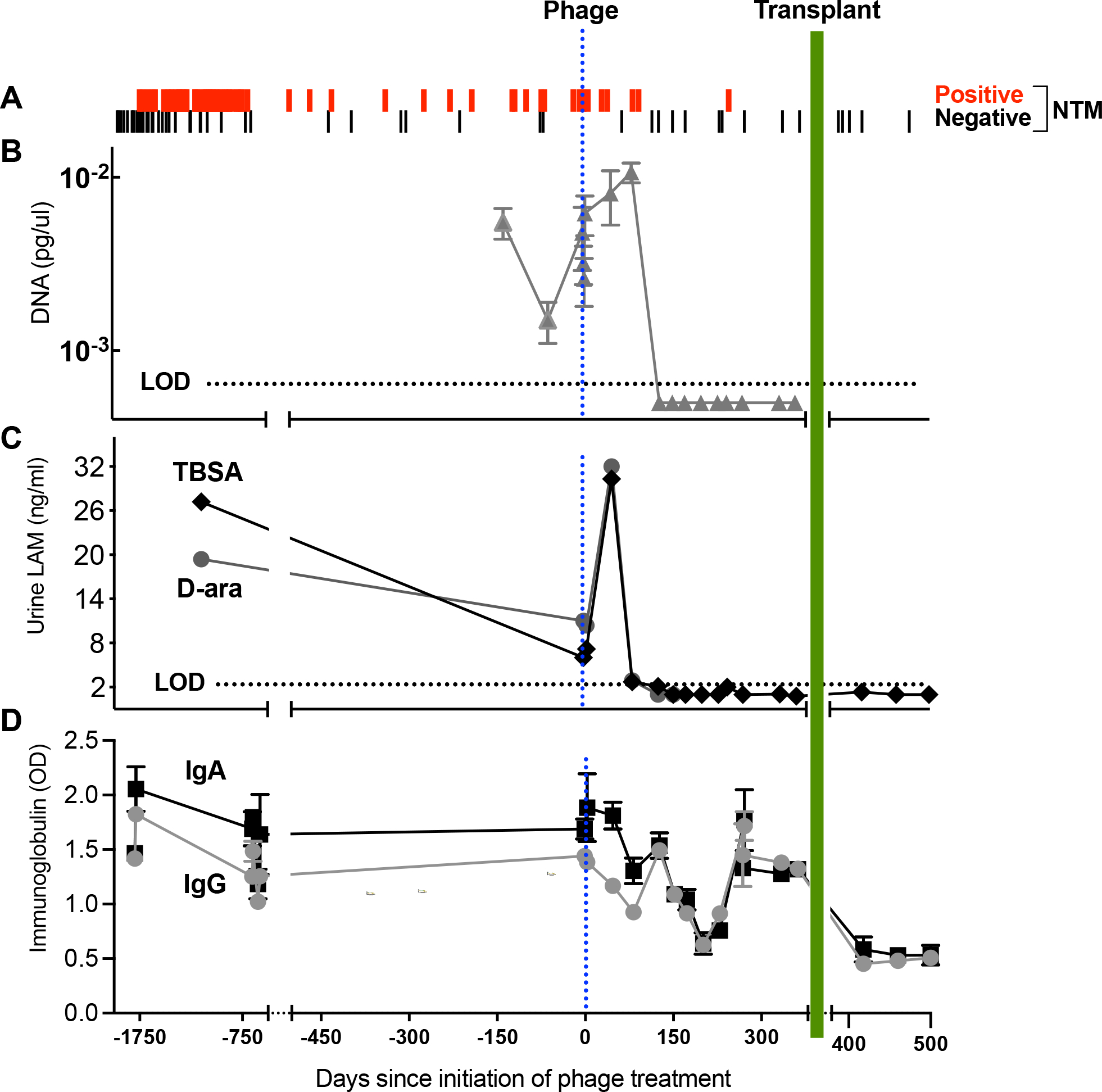
(A) Airway culture versus date in days pre- or post-phage treatment initiation. Sputum culture results were predominantly positive (red lines) prior to initiation of phage treatment. Post-phage treatment, 6 of 7 cultures were positive through day 96. From day 116 the culture turned predominantly negative (black line) with 9 of 10 negative through day 362. Over this interval a single persister isolate was recovered on day 245. Post-transplant an additional 5 cultures from BAL were negative through day 500 of phage treatment.
(B) qPCR analysis of M. abscessus DNA in sputum. The y-axis shows M. abscessus DNA concentrations (pg/ul) extrapolated from a standard curve.
(C) Urine LAM. The LAM components D-ara (grey squares) and TBSA (brown diamonds) plotted versus days. Levels of both components peaked 47 days after initiation of phage, and by day 152 were both below LOD, continuing post-transplant.
(D) Serum anti-M. abscessus immunoglobulins plotted versus days. Measured immunoglobulins were IgA (black square) and IgG (gray circle). Evidence for a decline in immunoglobulins during each phase of treatment was tested by simple linear regression models fit to IgA and IgG using a three-level categorical variable for pre-phage treatment, phage treatment, and the post-transplant interval as the only covariate. T-tests on the regression coefficients, or linear combinations, were used to compare the mean levels in each period. Compared to pre-phage titers, a significant decline in IgA was detected over the course of phage therapy prior to transplant (p=0.05) while mean IgG values trended lower (p=0.17). Both IgG and IgA declined further post-lung transplant when compared to the year prior on phage treatment (p<0.001 for both).
NTM cell wall lipoarabinomannan (LAM), a lipoglycan found in all mycobacterial species, is a sensitive marker of NTM airway infection when analyzed by GC/MS in the urine of pwCF (De et al., 2020). Urine LAM components D-Arabinose (D-ara) and tuberculostearic acid (TBSA) were measured in samples ranging from -1132 days pre-phage initiation to 500 days post-phage initiation (Figure 4C, Supplemental Figures 2 & 3). Detection of the LAM components increased in the urine following phage administration and peaked at day 47, consistent with an increase in M. abscessus cell wall lysis greater than when on antibiotic treatment alone. By day 82, urine LAM levels were below pre-phage initiation values, and by day 152 were largely undetectable, consistent with an overall reduction in NTM burden as also indicated by sputum culture results (Figure 4A).
NTM infection status can be analyzed by measuring anti-M. avium complex (Ravnholt et al., 2019) and anti-M. abscessus antibodies in plasma (Ferroni et al., 2005) using antigens common to all mycobacteria. Anti-mycobacterial immunoglobulins bind avidly to LAM and other components of the mycobacterial cell wall, and a decline in IgA has been seen with treatment of NTM (Jhun et al., 2017). Anti-M. abscessus IgA and IgG antibodies were measured in the serum (Figure 4D) in samples ranging from pre-phage day -1795 to post-phage day 500. Compared to pre-phage titers, mean IgA titers declined and mean IgG titers trended lower over the course of phage therapy, consistent with a decreased burden of infection. A further incremental decline in both IgG and IgA was apparent post-lung transplant when compared to the year prior on phage treatment.
Serum and sputum samples were examined for the presence of phage DNA by conventional PCR, and all samples were negative. The lack of a serum signal is consistent with prior reports of rapid clearance of phages in the bloodstream (Schooley et al., 2017). Although 18 sputum samples were tested over a 12-month period, it is plausible that phage particles were present within a narrow time frame that was not captured by sputum sampling, or that phage DNA was present below the detection limit. Phage DNA was also not detected by PCR in explanted lung samples.
Reduced Genetic Diversity of M. abscessus subspecies abscessus Isolates Following Phage Therapy
A total of 40 isolates collected on 36 occasions ranging from pre-phage day -1555 to post-phage day 245 were subjected to whole genome sequencing for analysis of genetic changes and response to phage treatment (Figure 5A). M. abscessus isolates recovered prior to antibiotic treatment revealed multiple sublineages that evolved from a common ancestor within the dominant circulating clone 1 of M. abscessus subspecies abscessus seen worldwide in CF and non-CF populations (Bryant et al., 2016; Davidson et al., 2021b; Davidson et al., 2014). The estimated time of initial infection based on divergence dating analysis was November 2013 (pre-phage day -2494; 95% Highest Posterior Density (HPD) Interval: pre-phage day -3435, pre-phage day -1945), approximately 30 months prior to the first detection by culture (Figure 1E, Figure 5A, Supplemental Figure 5). With initiation of antibiotic therapy directed against M. abscessus (pre-phage day -1236), a discernable shift in the population occurred with the emergence of new sublineages A, B & C (Figure 5A). Notable mutations appeared in genes coding for potential virulence factors such as MntH (Champion et al., 2011) and transcriptional regulators, FeoA, RnE (Lau et al., 2016; Taverniti et al., 2011). With initiation of phage therapy, the M. abscessus population became genetically less diverse and isolates of only sublineage C (Figure 5A) were recovered prior to sputum culture conversion to negative, with the exception of one isolate recovered at post-phage day 245 from sublineage B that had not been observed since pre-phage day -60 (Figure 5A). Together, the isolate population demonstrated a mean substitution rate of 2.7×10−7 mutations per site/year in the core genome (Supplemental Figure 5), which is comparable to previous estimates for M. abscessus subspecies abscessus (Bryant et al., 2016). The greatest genetic distance between any two isolates was only 11 single nucleotide polymorphisms (SNPs) in the core genome.
Figure 5. Genetic Diversity and Antibiotic Sensitivity of M. abscessus Isolates Following Phage Therapy.
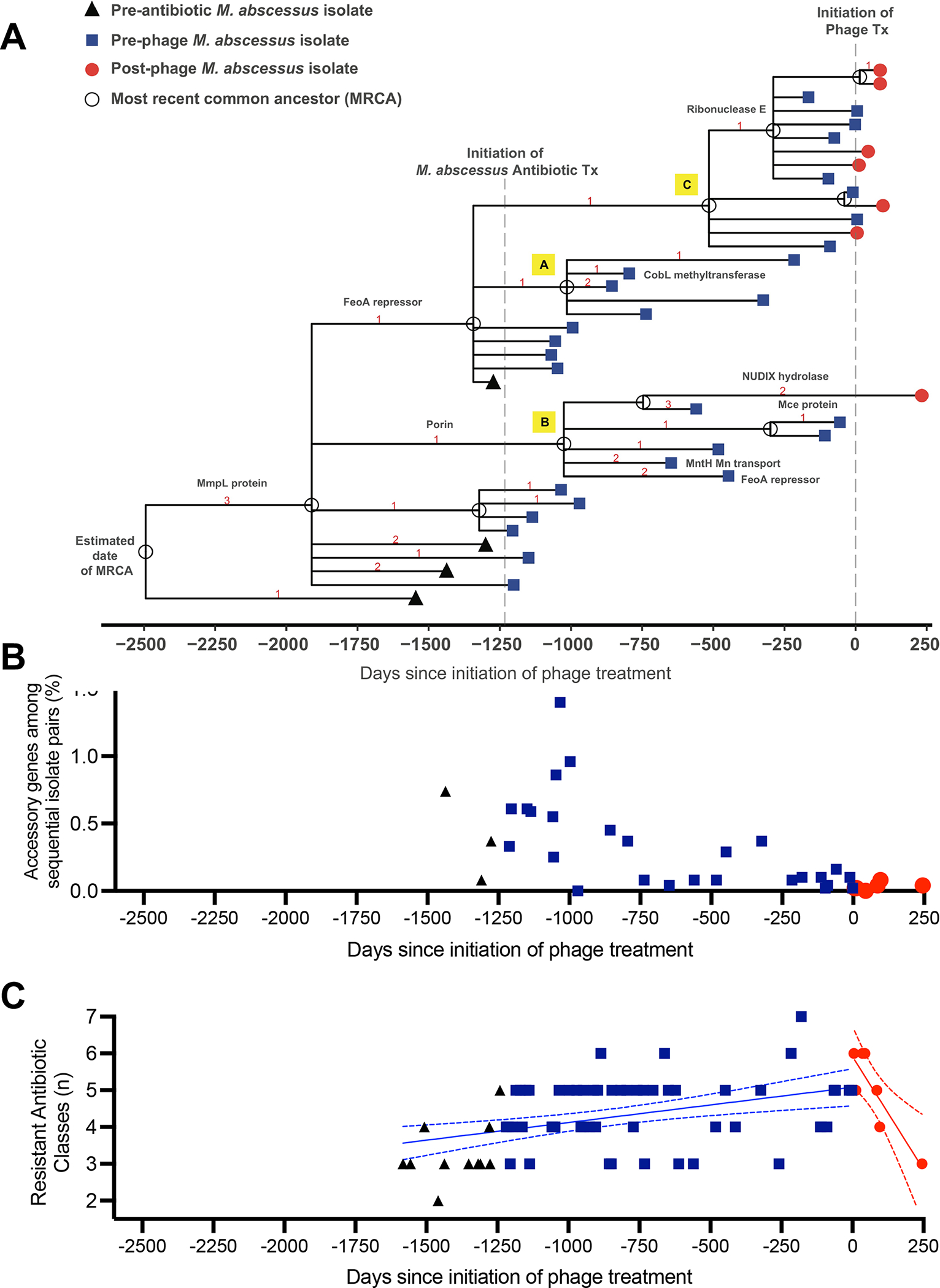
(A) Genetic lineage map of M. abscessus isolates (n=40) over the course of infection and treatment. A time scaled phylogeny was created based on genome wide SNP data using BEAST v1.10.4. The x-axis represents days since the initiation of phage treatment. Isolates are color-coded symbols on branch tips, and the most recent common ancestors (MRCA) are labeled as open circles on nodes. Red numbers indicate the number of nonsynonymous mutations that occurred on each evolutionary branch. Protein annotations for selected mutations are labeled on branches.
(B) Gene content comparisons of sequential M. abscessus isolates to assess genomic variation over time. For each pair of sequential isolates (x-axis), pan genome analysis was performed to identify accessory genes unique to only one sample. The y-axis (% accessory genes) is defined as the total # of accessory genes/ sum of all genes from both isolates in the pair. Isolate pairs post-phage were significantly less diverse than isolates pre-phage (p=0.0055 by Wilcoxon 2-sample test). Black triangles represent isolate pairs prior to initiation of M. abscessus antibiotic treatment, blue squares following start of antibiotics, and red circles following initiation of phage therapy. The decrease in accessory genes over time suggests a reduction in genetic diversity and selection for a homogeneous population.
(C) Antibiotic resistance of M. abscessus isolates does not increase following phage therapy. Number of resistant antibiotic classes were plotted versus days pre- and post-phage initiation. Black triangles represent isolates prior to initiation of M. abscessus antibiotic treatment, blue squares represent values following start of antibiotics, and red circles following initiation of phage therapy. Simple linear regression demonstrated a significant increase in slope of number of resistant classes over time prior to phage (blue line ± 95%CI, p=0.0007). Following initiation of phage, the slope was significantly negative (red line ± 95%CI, p=0.009).
Genomic variation over time was also assessed with gene content comparisons of sequential M. abscessus isolates (Figure 5B; Supplemental Figure 6). For each pair of sequential isolates, pan genome analysis was performed to identify genes unique to only one sample in the pair (i.e. accessory genes; Figure 5B, y-axis), The range of accessory genes among all sequential isolate pairs was 0–1.4%, with no evidence of plasmid acquisition or incorporation of viral DNA within the bacterial genomes. A reduction in accessory gene content was observed approximately one year after initiation of antibiotic treatment, and isolate pairs following phage initiation were significantly less diverse than those recovered prior to phage therapy (p=0.0055 by Wilcoxon 2-sample test). The decrease in accessory genes over time suggests a reduction in genomic diversity and selection for a homogeneous population as a result of both antibiotic and phage treatments.
Antibiotic Resistance of M. abscessus subspecies abscessus Isolates Did Not Increase Following Phage Therapy
All M. abscessus isolates recovered over the course of infection (n=71) were tested against a 20-drug panel of antibiotics used in the treatment of NTM infection (Supplemental Table 1). Most antibiotics tested were consistently resistant or demonstrated little variability in activity over the course of infection and treatment. Representative antibiotics of their class with available Clinical & Laboratory Standards Institute (CLSI) interpretive guidelines (Clinical and Laboratory Standards Institute, 2018) for M. abscessus were amikacin, clarithromycin, cefoxitin, doxycycline, imipenem, linezolid, moxifloxacin and trimethoprim/sulfamethoxazole (TMP/SMX). The number of resistant classes of these antibiotics increased in isolates throughout the course of the infection, with considerable variability between isolates (Figure 5C). Following the addition of phage to the antibiotic treatment, the resistance of post-phage isolates did not increase further, suggesting the combination of phage and antibiotics did not result in greater antibiotic resistance.
Phage Therapy Did Not Select for Phage-resistant Isolates
Emergence of phage resistance is a potential limiting factor that may reduce the efficacy of any phage administration. M. abscessus isolates from sputum recovered post-phage day 5, 44 and 245, as well as those collected pre-phage day -323 and -60 were tested for phage susceptibility (Figure 2A). All isolates were susceptible to both BPsΔ33HTH_HRM10 and D29_HRMGD40 and had the same phage infection profiles as did the initial strain (Figure 2D). Thus, emergence of phage resistance was not observed, even in the isolate recovered more than 8 months following initiation of treatment.
Antibody Recognition of Phages Pre- and Post-phage Treatment
Phage treatment can also fail due to anti-phage neutralizing antibodies formed rapidly by the host in response to IV phage therapy (Dedrick et al., 2021b). Serum collected pre-phage day -1 and post-phage days 44, 149 and 269 were tested for IgG binding to both phages used (Figure 6A). Surprisingly, this individual had a substantial IgG response to both phages even before phage treatment, which increased modestly in post-treatment serum, as seen in the half-max titers (Figure 6A). Serum tested at post-treatment days 3 through 500 showed a gradual increase in serum-mediated neutralization of BPsΔ33HTH_HRM10 infection, with substantial inactivation 242 days post-treatment and beyond (Figure 6B). However, only mild neutralization of D29_HRMGD40 was observed (Figure 6B). These observations suggest that this individual had naturally encountered phages that induced immunological cross-reactivity to the two phages administered here, but this cross-reactivity was not sufficiently closely related to BPs or D29 to have initiated neutralizing activity. Furthermore, he did not mount a substantial additional antibody response to BPsΔ33HTH_HRM10 until relatively late in treatment, at which point sputum cultures had become predominantly negative. These antibodies do not appear to cross-react to neutralize D29_HRM10GD40, which presumably remained active throughout the treatment course.
Figure 6. Antibody Binding to Phages Pre- and Post-Phage Treatment Demonstrates Late Development of Anti-phage Neutralizing Antibodies.
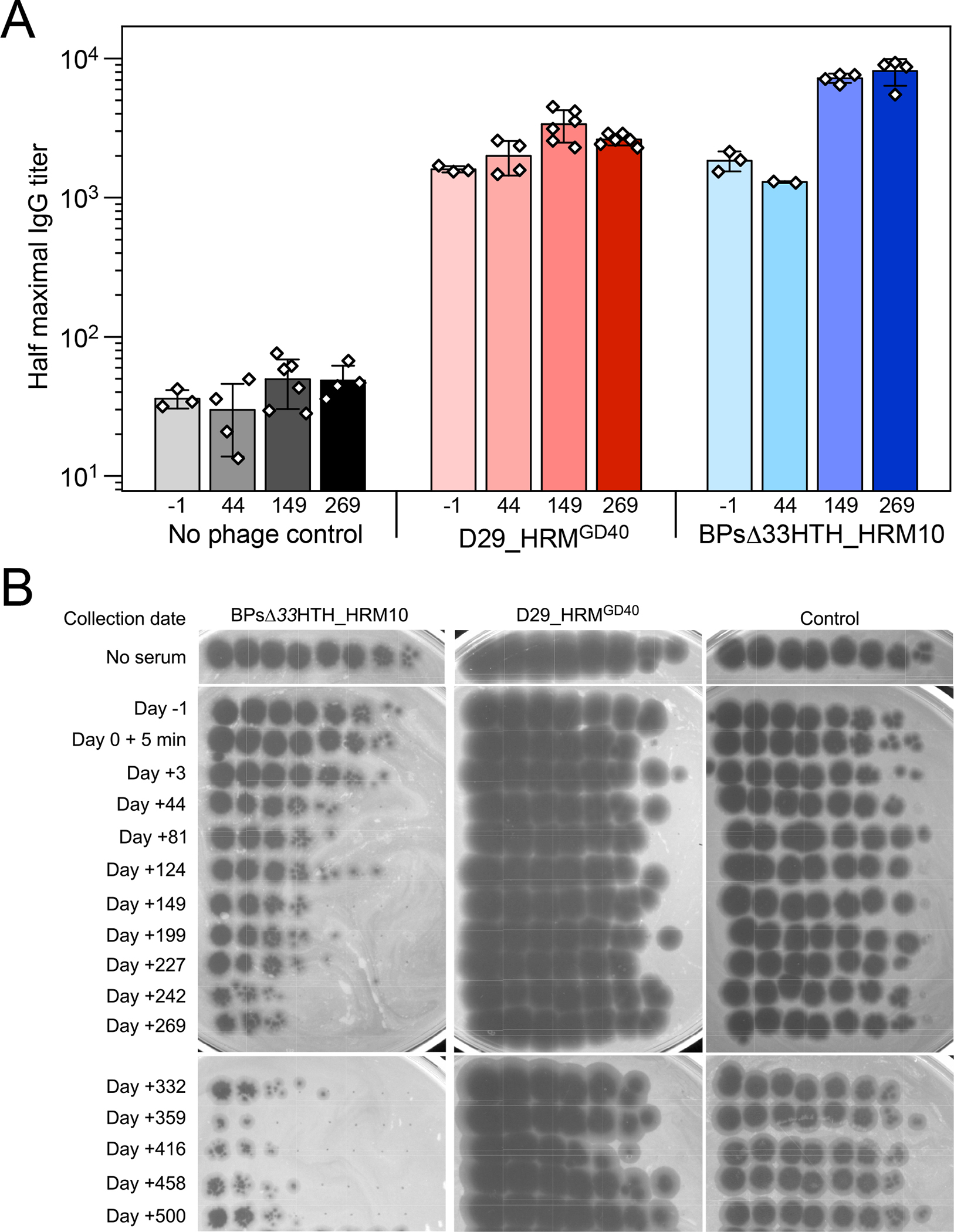
(A) Half maximal titers of IgG antibodies in pre-phage serum collected at day -1 and indicated times post-phage initiation for phages BPsΔ33HTH_HRM10 and D29_HRMGD40. Titers for two to six replicates are displayed as white data points on top of a mean bar ± s.d.
(B) Serum collected 1 day prior to phage initiation and indicated times post-phage initiation were tested for phage neutralization of BPsΔ33HTH_HRM10, D29_HRMGD40, and a control phage Fionnbharth Δ45–47. Data are representative of duplicate or triplicate assays.
Discussion
In the setting of severe infections that may possess resistance to multiple classes of antibiotics, treatment with phages is increasingly considered. Chronic pulmonary infection with M. abscessus is a particularly appealing therapeutic target, as current guidelines recommend combinations of 3 or more drugs for at least 12 months of therapy beyond initial culture conversion to negative (Daley et al., 2020). This antibiotic treatment is often unsuccessful as measured by eradication of infection (Daley et al., 2020; Martiniano et al., 2014), likely due to the capacity of M. abscessus to form biofilms or to survive within host cells, combined with physical sequestration in mucous plugs or cavities, which together may effectively shield the pathogen from therapeutic concentrations of antibiotics. Treatment is even more challenging in the context of CF lung disease, as co-infection with other bacterial pathogens reduces the sensitivity of airway cultures to monitor response to therapy. Given the capacity of M. abscessus to disseminate to skin, soft tissue and bone following immunosuppression, uncontrolled infection is cited as a contraindication to transplant (Orens et al., 2006; Weill et al., 2015).
Currently, there are no reports of successful phage treatment of M. abscessus infection of the lung. In 2019, beneficial use of mycobacteriophages was reported for the treatment of a 15-year-old girl with CF who developed disseminated M. abscessus subspecies massiliense infection following lung transplantation (Dedrick et al., 2019). In this case, the phage cocktail consisted of three mycobacteriophages, including BPsΔ33HTH_HRM10 which was also used in the current case, and is a genetically modified derivative of a temperate phage that kills its host more efficiently. The phage regimen resulted in reduction in size of skin nodules, improvement of the sternal wound infection, reduction in abdominal lymph nodes evaluated by PET-CT scan, and general clinical improvement. No resistance to phages was observed, and no neutralizing immune reactions occurred, possibly due to the use of immunosuppressive drugs to support the newly transplanted lungs (Dedrick et al., 2019). In a recent report of treatment of pulmonary infection due to M. abscessus subspecies massiliense in the context of non-CF bronchiectasis, phage treatment failed due to robust IgM- and IgG-mediated neutralizing antibody responses (Dedrick et al., 2021b). In some cases, prospective patients have died before receiving treatment (Dedrick et al., 2019).
In the current case, there is strong evidence that biologically significant lysis of M. abscessus occurred in response to mycobacteriophage administration. Currently the “gold standard” for determining response to treatment of M. abscessus lung disease is conversion of sputum cultures from positive to negative. However, in the context of CF, the sensitivity of NTM sputum cultures is particularly low, given the requirement for bacterial decontamination of P. aeruginosa and other co-pathogens, which markedly reduces the viability of M. abscessus as well (Whittier et al., 1997). In this individual, the sensitivity of individual sputum culture from the time of first positive M. abscessus to the initiation of phage treatment was only 70.3% (64 positive out of 91). However, at no time over 4.2 years of infection prior to phage initiation were there more than 2 consecutive negative cultures (Figure 1). Given the severe cavitary nature of his lung disease, it cannot be known if M. abscessus had been completely eradicated through testing of airway samples. However, in combination with antibiotics, the burden of infection was decreased below limits of detection by both sputum culture and the culture-independent assays employed. The CF Foundation and the European CF Society recommend that individuals with CF receiving NTM treatment with sequential negative cultures may be eligible for transplant listing (Floto et al., 2016). Thus, with evident response to treatment the participant was offered a lung transplant after having previously been declined. Analysis of the explanted lung tissue supports the conclusion that viable M. abscessus was sterilized from the lung, as AFB stains and cultures were negative even in samples where species-specific qPCR detected M. abscessus DNA from the same sample. Continued negative airway cultures through the first 4 months post-transplant augment this claim.
At no point during therapy was there evidence of genetic adaptation of M. abscessus to the phage. Longitudinal genetic analysis of 40 M. abscessus isolates over nearly 5 years was consistent with previous M. abscessus studies suggesting a primarily clonal infection (Bryant et al., 2016; Davidson et al., 2021b; Davidson et al., 2014) with the emergence of adapted sublineages over time (Bryant et al., 2021; Kreutzfeldt et al., 2013). Despite the limited genetic changes in the core genome among isolates, there were examples of nonsynonymous mutations observed in the same genes across multiple sublineages. For example, a gene encoding an FeoA transcriptional repressor protein acquired independent mutations in the most recent common ancestor (MRCA) of sublineages A and C and a branch of sublineage B during the course of antibiotic treatment (Figure 5A). Similar examples of within-host parallel evolution of transcriptional regulators have been observed during chronic M. abscessus infections in the CF airway that likely reflect adaptations to the lung environment (Bryant et al., 2021). Pan genome analysis revealed a substantial reduction in accessory genes after antibiotic and phage treatment consistent with selection for a persistent and homogeneous population (Figure 5B, Supplemental Figure 6). Only isolates from sublineage C were observed after the initiation of phage therapy, suggesting that the phage cocktail may have been more efficient at killing subpopulations A and B over C (Figure 5A). However, one isolate of sublineage B was recovered after nearly five months of negative cultures, but did not show increased antibiotic or phage resistance suggesting that it may have evaded phage treatment through sequestration in a lung cavity or airway biofilm.
In cases of bacteriophage treatment of P. aeruginosa and Acinetobacter baumanii, the development of phage resistance has been reported, as well as the emergence of new bacterial isolates with different antibiotic susceptibility profiles from the original infecting strain (Aslam et al., 2020; Schooley et al., 2017). It is proposed that the use of more than one phage simultaneously, as well as the combined treatment with antibiotics, are useful strategies to prevent this occurrence (Aslam et al., 2020). The relationship between use of phage therapy and conventional antibiotics is complex and not completely understood. Under various conditions, in vitro studies of combined use of phage and antibiotics have identified additive, synergistic, antagonistic, or no effects (Gu Liu et al., 2020). Enhanced susceptibility to antibiotics by phage may occur via modification of cell wall permeability and anti-biofilm mechanisms (Chakraborty and Kumar, 2019; Li et al., 2016; Senhaji-Kacha et al., 2020). In the current case, the final isolates recovered did not demonstrate increased antibiotic resistance, supporting the conclusion that phage treatment did not promote antibiotic resistance, and that the persister isolate recovered on day 245 likely originated from a cavity or occluded airway where antibiotic and phage therapy failed to penetrate.
Given the extremely long duration of antibiotic and phage treatment associated with treatment of M. abscessus, the emergence of anti-phage neutralizing antibodies is of potential concern. In previous reports, the lack of antibody development was associated with successful treatment in the setting of post-transplant immunosuppression in one case (Dedrick et al., 2019), and the rapid development of antibodies correlated with treatment failure in another (Dedrick et al., 2021b). In the current case, sputum cultures converted to negative prior to development of significant neutralizing antibodies to one of the phages. Based on in vitro assays, D29_HRMGD40 remained active throughout, even though BPsΔ33HTH_HRM10 was substantially compromised by neutralization by day 149 (Figure 6). Thus, even though a two-phage cocktail was chosen to minimize any potential phage resistance, it also presented the possibility that even if neutralization to one phage was observed, the other phage may be less immunogenic. We note that by day +245, when D29_HRMGD40 was effectively the sole active phage, no resistance to that phage was observed (Figure 2), consistent with other reports of infrequent phage resistance in M. abscessus (Dedrick et al., 2021c), a potentially substantial advantage for the therapeutic use of mycobacteriophages.
Limitations of Study
Limitations in interpreting the efficacy of phage therapy in this subject are typical of the complexity of cases of this nature. As we are aware of no previous reports of successful treatment of pulmonary M. abscessus infection with phage, it is impossible to know if the findings of this single case are representative of others who may receive this treatment. In addition, there may be unrecognized factors that modified the response in various ways. Phages were administered in addition to guideline-based treatment of CF lung disease (Mogayzel et al., 2013; Ren et al., 2018) as well as intensive antibiotic therapy for M. abscessus, MDR P. aeruginosa, and MRSA. While prior to phage therapy the choice of antibiotic therapy was varied, the combination of drugs used in conjunction with the phage was relatively constant. On day 89 of phage therapy, the CFTR modulator E/T/I was introduced as on-label treatment, as a result of approval for use with the H199Y CFTR gene mutation through a label expansion by the FDA (NIH, 2021). This label expansion was not based on results from clinical trials, but rather from in vitro studies confirming a minimum of 10% increase in chloride transport over baseline in FRT cells expressing the H199Y CFTR gene mutation (NIH, 2021). There are no previous reports of E/T/I use in a pwCF with the H199Y/2184insA CFTR genotype, and the extent of clinical benefit is not known. It is likely that E/T/I allowed for a degree of improved host response in terms of improved airway clearance and nutrition, however, this individual did not demonstrate an improvement in lung function or a reduction in sputum production with E/T/I, and at the time of transplant the degree of inflammation in his airways was severe. There are no known direct antibiotic properties of E/T/I against M. abscessus, and culture-independent markers of NTM lysis indicated a response prior to E/T/I initiation. In some pwCF, initiation of highly effective modulator therapy (HEMT) with the first CFTR modulator, ivacaftor was linked to a decrease in the relative abundance and apparent eradication of P. aeruginosa (Hisert et al., 2017)(Yi et al., 2021). However, this individual remained infected with P. aeruginosa and MRSA, with M. avium present in the explanted lung tissue, supporting the conclusion that eradication of M. abscessus was a specific response to phage therapy and not due to general improvement following E/T/I. Nevertheless, it is entirely possible that partial restoration of CFTR function by E/T/I may have resulted in beneficial alterations in the airway microenvironment that contributed towards more successful response to phage therapy.
Conclusions
Together our results derived from objective orthogonal markers provide a roadmap for successful phage treatment in the context of advanced CF lung disease. Understanding the time span associated with initial response, radiologic improvement and culture conversion following the start of treatment are essential for future therapeutic applications and clinical trial designs, as is the use of complementary culture-independent markers of M. abscessus lysis which can overcome the poor sensitivity of sputum culture and the inability to measure disease in areas of dense consolidation or parenchymal cavities. The biology of M. abscessus in the airway may serve to avoid some of the pitfalls encountered in the treatment of other pathogens common to the CF airway. In particular, most cases of M. abscessus infection are characterized by closely related lineages with relatively high genetic stability from a single common ancestor (Davidson et al., 2021b). This may limit the emergence of phage resistant isolates that have occurred in phage therapy of P. aeruginosa, where extensive genetic diversity and plasticity frequently exist due to selection for hypermutator genotypes and reinfection with new environmental clones during the course of infection (Rossi et al., 2021). It is likely that improvement in lung function would require administration of phage with antibiotics earlier in the course of infection, with the risk that anti-phage neutralizing antibodies may develop and limit the option for later treatment. However, the use of phages in very advanced lung disease with the objective to sufficiently control M. abscessus as a bridge to lung transplant may be considered based on this report.
STAR+METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jerry A. Nick (nickj@njhealth.org).
Materials Availability
Isolates of M. abscessus analyzed in this study will be made available on request following completion of a material transfer agreement.
Data Availability
Standardized datasets:
Whole genome sequence data have been deposited to the National Center for Biotechnology Information (NCBI) under BioProject PRJNA319839.
Additional datasets from this study are 1). Video files incorporating a complete set of axial, coronal and sagittal reconstructions post phage days -2, 81 and 229, 2). Gross and microscopic pathological photographs of the explanted lungs, 3). Raw data from Figures 1A, 4A, 4B, 4C, 4D, 6 and Supplemental Table 1. These files are deposited to Mendeley Data at: https://data.mendeley.com/datasets/jtdvbm2tzx/draft?a=b138c41d-bc1b-458c-8d2a-98267555428d
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subject
The individual described in this case study is a 26-year-old man with CF (genotype H199Y/2184insA), advanced bronchiectasis, and M. abscessus lung infection. He was diagnosed with CF at birth by newborn screening and developed typical complications, including chronic lung infection with multi-drug resistant (MDR) P. aeruginosa and methicillin-resistant S. aureus (MRSA), CF-related diabetes, sinus disease, exocrine pancreatic insufficiency, and malnutrition requiring supplemental feeding via percutaneous gastrostomy tube. Sputum cultures first turned positive for M. abscessus ssp. abscessus in May 2016, and he was initiated on antibiotic therapy in May 2017 with an initial combination of amikacin (nebulized), imipenem, linezolid and clofazimine. Azithromycin was not active in vitro against his M. abscessus but was administered for other indications. He was initiated on a mycobacteriophage cocktail in September, 2020, with continuation of antibiotics. He was transplanted after 379 days of treatment, and was followed for 121 days post-transplant (500 days total of phage treatment). The investigators were granted an expanded access investigational new drug (IND) for compassionate use of this mycobacteriophage cocktail by the FDA (IND 26902). The IND, treatment protocol, and informed consent were approved by the Biomedical Research Alliance of New York, LLC (BRANY) Institutional Review Board (20-07-463-528). The participant was enrolled in the CF Foundation Patient Registry (HS-1683-528), the PREDICT (NCT02073409)(HS-2798) and PATIENCE (NCT02419989)(HS-2833) trials which allowed for retrospective and prospective data and specimen collection relating to the diagnosis and treatment of his M. abscessus. Collection and banking of NTM isolates was approved by the National Jewish Health Institutional Review Board (HS-3149), as well as sampling of sputum, saliva, breath, blood and urine (BRANY IRB File HS-1234-528). Consent for analysis of explanted lung tissue was approved by the Colorado Multiple Institutional Review Board (COMIRB 11-1664) as well as written and verbal authorization for use and disclosure of protected health information (PHI). Assessments of safety and response were made weekly either at the Adult CF Clinic at National Jewish Health or at home, as well as during hospitalizations. Assessments of radiologic changes, microbiologic changes and laboratory monitoring were performed in the context of clinical care.
Phage selection and administration
A mycobacteriophage cocktail, consisting of engineered phages BPsΔ33HTH_HRM10 and D29_HRMGD40, was identified as lytic towards an M. abscessus isolate recovered early in the course of infection (October, 2016, 1532 days prior to phage initiation). Following informed consent, a dose of 109 to 108 plaque-forming units (PFU/ml) diluted in PBS was administered by IV twice daily through an existing mediport, and continues to the present.
NTM isolate and sample collection
NTM cultures were tested by the Advanced Diagnostic Laboratory (National Jewish Health, Denver CO, USA), which is a CLIA-certified lab that serves as a national NTM reference laboratory. Specimens were digested/decontaminated using a standard NaOH-NALC procedure, and the sediment used to prepare an auramine smear and inoculate various growth medium including: Lowenstein-Jensen Slant, Middlebrook 7H11 Agar/ Mitchison 7H11 Selective Agar biplate(s) and MGIT (Mycobacteria Growth Indicator Tube) incubated for up to six weeks. Positive AFB culture growth was assessed from solid growth media at 1, 3, and 6 weeks. MGIT specimens were continuously monitored. Upon growth, genomic DNA was isolated for subsequent PCR amplification. A targeted segment of the DNA-directed RNA polymerase subunit beta (rpoB) gene was performed and consensus sequence determined, constructed and blasted against a known database to determine molecular identification. All M. abscessus isolates (n=71) were tested for antibiotic susceptibility against a 20-drug panel via broth microdilution. Representative antibiotics of their class with available Clinical & Laboratory Standards Institute (CLSI) interpretive guidelines (Clinical and Laboratory Standards Institute, 2018) for M. abscessus were amikacin, clarithromycin, cefoxitin, doxycycline, imipenem, linezolid, moxifloxacin and trimethoprim/sulfamethoxazole (TMP/SMX). The number of resistant classes were tabulated for each isolate, based on the published breakpoints in MIC.
Analysis of explanted lung tissue
At the time of transplantation, the explanted lungs were surgically removed and stored at 3C in Custodial HTK Solution Transport Medium. Within 48 hours, grossly purulent endobroncial secretions were sampled from large bronchi of all lobes and processed as sputum for culture and culture-independent analysis. Distal airways from each lobe were sampled by lavage with saline and processed as sputum. Tissue from each lobe and the mainstem bronchi were dissected from regions with obvious disease and weighed, minced with scissors to 1–2 mm pieces, and combined with PBS (3C) and stainless steel beads (0.2gm, 0.9–2.0 mm) followed by homogenization by Bullet Blender (Next Advance, Inc). Tissue from each lobe was also processed for microscopy slides and stained with H&E, AFB and GMS stains.
Method Detail
Genetic analysis of M. abscessus isolates
Bacterial DNA samples were extracted using an adapted method for mycobacteria cells (Epperson and Strong, 2020), and whole genome sequencing (WGS) was performed with Illumina 2×300bp reads as described (Hasan et al., 2021). Single Nucleotide Polymorphisms (SNPs) were analyzed using a read mapping approach with bowtie2 (Langmead and Salzberg, 2012) and SAMtools mpileup (Li et al., 2009) as described previously (Davidson et al., 2021a). Pairwise SNP distances between isolates were computed with MEGA-CC 7.0.26 (Kumar et al., 2016). Estimation of mutation rate and the dates of common ancestors were calculated using Bayesian evolutionary analysis by sampling trees (BEAST v1.10.4; Suchard et al., 2018) and the following parameters: Markov chain Monte Carlo (MCMC) with a chain length of 100 million states, general time reversible (GTR) evolutionary model, strict clock, constant population size, and a minimum of 3 iterations. Statistics for clock rate and other parameters were summarized with Tracer v1.7.1 (Rambaut et al., 2018), and consensus trees were constructed with TreeAnnotator (Drummond and Rambaut, 2007). Phylogenetic trees were visualized with ggtree (Yu, 2017). Genomes were assembled with Unicycler (Wick et al., 2017), and pan genome analysis was performed with Panaroo v1.2.6 (Tonkin-Hill et al., 2020).
MinION long read sequencing and complete genome assembly
Long read sequencing and hybrid genomic assembly was performed as described (Hendrix et al., 2021) on the isolate collected pre-phage day -1555 using Oxford Nanopore Technologies MinION R9.4.1 flowcell. Nanopore reads were basecalled and demultiplexed using Guppy v3.4 (Wick et al., 2019) then assembled with Illumina reads from the same isolate using the Unicycler hybrid assembly method (Wick et al., 2017).
Urine LAM analysis by GC/MS
Urine samples were collected in sterile containers, centrifuged, frozen within one hour at −20°C, then stored in a −80°C freezer. Urine was subjected to hydrophobic interaction chromatography (HIC) over Octyl Sepharose (OS)-CL 4B. The 40% and 65% n-propanol in 0.1 M NH4OAc eluents off the HIC column were processed for GC/MS analysis as previously described (De et al., 2020). GC/MS analyses were carried out using a Thermo GC-TSQ8000 Evo Triple Quad GC mass spectrometer. Chromatograms with respective peaks were integrated manually (i.e., peak areas were defined manually and integrated areas were generated by the computer software) for the assessment of total D-ara and TBSA content.
Anti-M. abscessus IgA and IgG analysis
Heat-killed M. abscessus (ATCC 19977) was probe-sonicated and 100ng protein dried in 96-well plates. Plates were blocked with BSA and incubated overnight at 4°C with 1:10,000 dilutions of plasma. Wells were washed, and bound antibody detected with HRP-conjugated goat anti-human antibodies specific for IgG and IGA (Abcam, SouthernBiotech).
qPCR analysis for M. abscessus DNA
A real-time quantitative PCR (qPCR) assay was used to detect M. abscessus DNA in sputum and explant samples. Primer and probe sequences targeting the internal transcriber spacer (ITS) (Rocchetti et al., 2017) were ordered from IDT (Coralville, IA). TaqMan Gene Expression Master Mix (Thermofisher) was combined with forward and reverse primers (final concentration 500nM each), probe (final concentration 250nM), and water. A standard curve of M. abscessus DNA in a background of 5ng/μL of human gDNA (Takara Bio) was used for quantification including seven 10-fold dilutions ranging from 1ng/μL to 0.0001pg/μL. DNA was extracted from raw sputum, endobronchial secretions and BAL as described (Epperson and Strong, 2020). Total DNA from these samples (100ng) and standard curve samples were analyzed in triplicate with the QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, Waltham, MA) with the following cycling conditions: 50°C for 2 minutes, 95°C for 10 minutes, and 45 cycles of 95° for 15 seconds and 60°C for 1 minute.
Bacterial strains
M. smegmatis mc2155 is a laboratory strain and was grown as previously described (Jacobs-Sera et al., 2012). M. abscessus pre-phage day -1437 was grown on Middlebrook 7H9 media with OADC and 1 mM CaCl2 for 4–5 days, shaking, at 37°C. M. abscessus cultures were sonicated briefly before use as previously reported (Dedrick et al., 2019).
Phage susceptibility screening
Standard plaque assays were used for phage susceptibility testing by spotting 10-fold serial dilutions on both M. smegmatis mc2155 and M. abscessus pre-phage day -1437 top agar overlays. Phages used for screening were obtained from the collection at the University of Pittsburgh and were propagated on M. smegmatis mc2155. Aliquots of 107 CFU M. abscessus were incubated with or without phage at a multiplicity of infection of 10 for 48 hr in liquid media and plated on solid media.
Mycobacteriophage cocktail preparation
Phages were grown on M. smegmatis mc2155 using solid media and recovered by collection of top agar (Dedrick et al., 2021c). After dialysis, the EndoZyme II (Hyglos GmbH) assay was performed according to manufacturer’s instructions and endotoxin levels were undetectable. Sterility of BPsΔ33HTH_HRM10 and D29_HRMGD40 was confirmed by Accugen, Inc. BPsΔ33HTH_HRM10 at 1×1011 PFU ml−1 and D29_HRMGD40 at 1×1010 PFU ml−1 were combined into a cocktail monthly.
Phage neutralization assays
Each serum sample was diluted (1:10) in phage buffer (10 mM Tris-HCl (pH 7.5), 10 mM MgSO4, and 68 mM NaCl) and incubated with either BPsΔ33HTH_HRM10 or D29_HRMGD40 or a control phage Fionnbharth Δ45-47 for 24 hours at room temperature. Next, 10-fold serial dilutions were made and 3 μl of each dilution were spotted onto top agar overlays of M. smegmatis mc2155 and then incubated at 37°C for 24 hours. Serum samples were not heat inactivated.
Anti-phage IgG analysis by ELISA
Enzyme-linked immunosorbent assays (ELISAs) were completed as described (Dedrick et al., 2021b). Briefly, EIA microplates were coated with either pure coating buffer (carbonate-bicarbonate, pH = 9.6) or 50 ng of total protein from highly purified phage (BPsΔ33HTH_HRM10 or D29_HRMGD40), then incubated with at least eight serum dilutions and probed with Goat Anti-Human IgG Fc (HRP) (Abcam ab98624). Sera were not heat inactivated. The plates were developed with TMB substrate and stopped with H2SO4 before quantification of OD450 (signal) and OD570 (background). After background subtraction, the OD450 values were plotted against dilution and fit with a fixed-baseline logistic curve. Half-maximal serum IgG titers were determined from the fit.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data and statistical analysis for clinical features, markers of host and pathogen response, and antibiotic resistance were performed using GraphPad Prism 9.0.2 and JMP 13 (SAS Institute, INC). Replicates and statistical details can also be found in the methods and figure legends. For virus neutralization assays, the serum and plasma samples were diluted and tested in duplicate. For ELISAs, the sera were diluted and tested in triplicate. ELISA data and statistical analyses were performed in OriginLab 2020 version 9.7.0.188.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti-Human IgG H&L (HRP) | Abcam | ab97175 |
| Goat Anti-Human IgA (HRP) | Southern Biotech | 2050-05 |
| KPL SureBlue Reserve TMB Microwell Peroxidase Substrate | seracare | 5120-0082 |
| Goat Anti-Human IgG Fc (HRP) preadsorbed | Abcam | ab98624 |
| Bacterial and virus strains | ||
| Mycobacterium smegmatis mc2155 | Snapper et al., 1990 | NC_008596 |
| Mycobacterium abscessus patient samples | This paper | National Jewish Health |
| Mycobacteriophage BPsΔ33HTH_HRM10 | Dedrick et al., 2019 | http://phagesdb.org |
| Mycobacteriophage D29_HRMGD40 | Dedrick et al., 2021 | http://phagesdb.org |
| Mycobacteriophage Muddy | Dedrick et al., 2019 | http://phagesdb.org |
| Mycobacteriophage Itos | Dedrick et al., 2021 | http://phagesdb.org |
| Mycobacteriophage BPsΔ33HTH_HRMGD03 | Dedrick et al., 2021 | http://phagesdb.org |
| Mycobacterium abscessus | ATCC | 19977 |
| Biological samples | ||
| Patient serum | This paper | National Jewish Health |
| Patient sputum | This paper | National Jewish Health |
| Patient urine | This paper | National Jewish Health |
| Airway secretions from patient explanted lungs | This paper | National Jewish Health |
| Bronchioalveolar lavage from patient explanted lungs | This paper | National Jewish Health |
| Tissue from patient explanted lungs | This paper | National Jewish Health |
| Chemicals, peptides, and recombinant proteins | ||
| Ringers Solution | Oxoid | Cat# BR0052G |
| 3,3′,5,5′-Tetramethylbenzidine (TMB) Liquid Substrate System for ELISA | Sigma Aldrich | Cat# T0440-1 |
| Ammonium Acetate | SIGMA-ALDRICH | CAS:631-61-8 |
| Octyl Sepharose CL-4B | GE Healthcare | 17-0790-01; CAS: 68652-09-5 |
| n-Propanol | Macron Fine Chemicals | CAS: 71-23-8 |
| n-Hexane | Fisher Chemical | CAS: 110-54-3 |
| Chloroform | EMD Millipore | CAS: 67-66-3 |
| Acetonitrile | SIGMA-ALDRICH | CAS:75-05-8 |
| Trifluoroacetic acid | SIGMA-ALDRICH | CAS: 76-05-1 |
| Trifluoroacetic anhydride | SIGMA-ALDRICH | CAS:407-25-0 |
| D-Arabinose (U-13C5, 99%) CLM-8477-0 | Cambridge Isotope Lab | CAS: NA; PSO 8I-382 |
| (R)-(-)-2-Octanol | SIGMA-ALDRICH | CAS:5978-70-1 |
| Sodium Hydroxide | Fisher Chemical | CAS:1310-73-2 |
| Palmitic-2,2-d2 acid | Cambridge Isotope Lab | CAS: 57-10-3 |
| N-Ethyldiisopropylamine | SIGMA-ALDRICH | CAS: 7087-68-5 |
| 2,3,4,5,6-Pentafluorobenzyl Bromide | SIGMA-ALDRICH | CAS: 1765-40-8 |
| Sulfuric acid | EMD Millipore | CAS: 7664-93-9 |
| Custodial HTK Solution Transport Medium | Essential Pharmaceuticals | |
| Poly-Prep Chromatography Columns | BIO-RAD | Cat. #731-1550 |
| HR GC Column VF-5ms | Agilent Technologies | Part No: CP8944 |
| Carbonate-bicarbonate buffer capsules | Sigma Aldrich | Cat. # C3041 |
| Critical commercial assays | ||
| EndoZyme II | Hyglos GmbH | Cat# 890030 |
| TaqMan™ Gene Expression Master Mix | Thermofisher | Cat # 4369016 |
| LA Taq DNA Polymerase with GC buffers | Takara | Cat # RR02AG |
| Q5 High-Fidelity DNA Polymerase | NEB | Cat # M0491L |
| Nextera XT Index Kit v2 Set A | Illumina | Cat # FC-131-2001 |
| MiSeq Reagent Kit v2 | Illumina | Cat # MS-102-2003 |
| Deposited data | ||
| Bacterial isolate whole genome sequences | This paper | PRJNA319839 |
| Video files incorporating a complete set of axial, coronal and sagittal reconstructions of high-resolution computerized tomography (CT) scans from each timepoint illustrated in Figure 3 (2 days prior to phage, post-phage therapy day 81, and post-phage day 229) | This paper | DOI: 10.17632/jtdvbm2tzx.1 |
| Gross and microscopic pathological photographs of the explanted lungs at time of lung transplant (day 379 of phage therapy). | This paper | DOI: 10.17632/jtdvbm2tzx.1 |
| Raw data from Figures 1A, 1F, 4A, 4B, 4C, 4D, 6 and Supplemental Table 1. https://data.mendeley.com/datasets/jtdvbm2tzx/draft?a=b138c41d-bc1b-458c-8d2a-98267555428d |
This paper | DOI: 10.17632/jtdvbm2tzx.1 |
| Oligonucleotides | ||
| MAG forward: 5’- CAC GGG GTG GAC AGG ATT TA -3’ MAG reverse: 5’- TAA GGA GCA CCA TTT CCC AG -3’ Probe: 5’- /56-FAM/TCA CCA AGT AGA TAY CCA CTA CAG A/36-TAMSp/ -3’ |
Rocchetti et al., 2017 Integrated DNA Technologies (IDT) |
N/A |
| ACTB-Fwd: 5’- CACCATTGGCAATGAGCGGTTC -3’ ACTB-Rev: 5’- AGGTCTTTGCGGATGTCCACGT -3’ oTM179: 5’- GATTCTCACTCTACCGGACTAGTC -3’ oTM180: 5’- CTTGCTGCGATGACACAAGTAAAC -3’ ClusterG-Fwd: 5’- GGCCGTGGGCAGAGGAAACC -3’ ClusterG-Rev: 5’- AGAACCTCAACACCGGCGCG -3’ |
Integrated DNA Technologies (IDT) | N/A |
| Software and algorithms | ||
| OriginLab 2021b version 9.8.5.212 | https://www.originlab.com/ | |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| MEGA-CC v7.0.26 | Kumar et al., 2016 | https://www.megasoftware.net/web_help_10/MEGA-CC.htm |
| Biorender | Biorender.com | N/A |
| BEAST v1.10.4 | Suchard et al., 2018 | https://beast.community/ |
| Tracer v1.7.1 | Rambaut et al., 2018 | https://github.com/beast-dev/tracer |
| ggtree (R package) | Yu et al., 2017 | https://bioconductor.org/packages/release/bioc/html/ggtree.html |
| Unicycler | Wick et al., 2017 | https://github.com/rrwick/Unicycler |
| Panaroo v1.2.6 | Tonkin-Hill et al., 2020 | https://github.com/gtonkinhill/panaroo |
| Other | ||
| Gen5 Microplate Reader and Imager Software | BioTek | TS Installation Version 2.06.10 |
Acknowledgments
The Authors wish to thank the individual described in this report for his willingness and enthusiasm to participate in these studies. Jackie Homra, FNP-C, Sarah Ellington, RN, and the entire faculty and staff of the Colorado Adult CF Program (National Jewish Health, Denver) the Respiratory Institute (Saint Joseph Hospital, Denver) and the UCHealth Transplant Center (University of Colorado Hospital, Aurora, CO) for their excellent care and for accommodating this study. Daniel T. Merrik, MD (University of Colorado, Denver) and Steve D. Groshong, MD, PhD (National Jewish Health) for pathological evaluation of the explant lung tissue. Idana C. Espinoza, PharmD, MPH, and the Saint Joseph Hospital Department of Pharmacy for pharmacy support throughout the study, Jon L. Koff, MD, for critical reading of the manuscript, Keira A. Cohen, MD for generous assistance with treatment protocol design, Becky Garlena, Deborah Jacobs-Sera, Haley Aull, Crystal Petrone, Marion Jones and Ibrahim Adoellail for technical support. Original Graphic Abstract figure was created with BioRender.com.
Funding provided by: R01HL146228, NICK20Y2-SVC, NICK20Y2-OUT, NICK17K0, NICK18P0, MALCO19I0 and to GFH by National Institute of Health grant R35 GM131729, Cystic Fibrosis Foundation grant HATFUL19GO, Howard Hughes Medical Institute grant GT12053, and a kind donation from the Fowler Fund for Phage Research. RMDn was supported by K01-AI125726. MS acknowledges funding from the Colorado Advanced Industries Accelerator Grant Program and a donation from Stephen and Betty Thorp.
Footnotes
Declaration of Interests
GFH is a compensated consultant for Janssen Inc.
References Cited:
- Aslam S, Lampley E, Wooten D, Karris M, Benson C, Strathdee S, and Schooley RT (2020). Lessons Learned From the First 10 Consecutive Cases of Intravenous Bacteriophage Therapy to Treat Multidrug-Resistant Bacterial Infections at a Single Center in the United States. Open forum infectious diseases 7, ofaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JM, Brown KP, Burbaud S, Everall I, Belardinelli JM, Rodriguez-Rincon D, Grogono DM, Peterson CM, Verma D, Evans IE, et al. (2021). Stepwise pathogenic evolution of Mycobacterium abscessus. Science 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, et al. (2016). Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, and Kumar A (2019). The extracellular matrix of mycobacterial biofilms: could we shorten the treatment of mycobacterial infections? Microb Cell 6, 105–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion OL, Karlyshev A, Cooper IAM, Ford DC, Wren BW, Duffield M, Oyston PCF, and Titball RW (2011). Yersinia pseudotuberculosis mntH functions in intracellular manganese accumulation, which is essential for virulence and survival in cells expressing functional Nramp1. Microbiology (Reading) 157, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Chan BK, Stanley G, Modak M, Koff JL, and Turner PE (2021). Bacteriophage therapy for infections in CF. Pediatr Pulmonol 56 Suppl 1, S4–S9. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2018). Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes, 3rd ed. CLSI standard document M24 (Wayne, PA, USA: ). [PubMed] [Google Scholar]
- Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Bottger EC, Brozek J, Griffith DE, Guglielmetti L, et al. (2020). Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin Infect Dis 71, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Benoit JB, Kammlade SM, Hasan NA, Epperson LE, Smith T, Vasireddy S, Brown-Elliott BA, Nick JA, Olivier KN, et al. (2021a). Genomic characterization of sporadic isolates of the dominant clone of Mycobacterium abscessus subspecies massiliense. Scientific reports 11, 15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Hasan NA, Epperson LE, Benoit JB, Kammlade SM, Levin AR, Calado de Moura V, Hunkins J, Weakly N, Beagle S, et al. (2021b). Population Genomics of Mycobacterium abscessus from United States Cystic Fibrosis Care Centers. Annals of the American Thoracic Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Hasan NA, Reynolds PR, Totten S, Garcia B, Levin A, Ramamoorthy P, Heifets L, Daley CL, and Strong M (2014). Genome sequencing of Mycobacterium abscessus isolates from patients in the united states and comparisons to globally diverse clinical strains. J Clin Microbiol 52, 3573–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P, Amin AG, Graham B, Martiniano SL, Caceres SM, Poch KR, Jones MC, Saavedra MT, Malcolm KC, Nick JA, et al. (2020). Urine lipoarabinomannan as a marker for low-risk of NTM infection in the CF airway. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Aull HG, Jacobs-Sera D, Garlena RA, Russell DA, Smith BE, Mahalingam V, Abad L, Gauthier CH, and Hatfull GF (2021a). The Prophage and Plasmid Mobilome as a Likely Driver of Mycobacterium abscessus Diversity. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Freeman KG, Nguyen JA, Bahadirli-Talbott A, Smith BE, Wu AE, Ong AS, Lin CT, Ruppel LC, Parrish NM, et al. (2021b). Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nature medicine 27, 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature medicine 25, 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Mavrich TN, Ng WL, and Hatfull GF (2017). Expression and evolutionary patterns of mycobacteriophage D29 and its temperate close relatives. BMC Microbiol 17, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Smith BE, Garlena RA, Russell DA, Aull HG, Mahalingam V, Divens AM, Guerrero-Bustamante CA, Zack KM, Abad L, et al. (2021c). Mycobacterium abscessus Strain Morphotype Determines Phage Susceptibility, the Repertoire of Therapeutically Useful Phages, and Phage Resistance. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, and Rambaut A (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson LE, and Strong M (2020). A scalable, efficient, and safe method to prepare high quality DNA from mycobacteria and other challenging cells. J Clin Tuberc Other Mycobact Dis 19, 100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferroni A, Sermet-Gaudelus I, Le Bourgeois M, Pierre-Audigier C, Offredo C, Rottman M, Guillemot D, Bernede C, Vincent V, Berche P, et al. (2005). Measurement of immunoglobulin G against Mycobacterial antigen A60 in patients with cystic fibrosis and lung infection due to Mycobacterium abscessus. Clinical Infectious Diseases 40, 58–66. [DOI] [PubMed] [Google Scholar]
- Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann JL, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, et al. (2016). US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis. Thorax 71 Suppl 1, i1–i22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford ME, Sarkis GJ, Belanger AE, Hendrix RW, and Hatfull GF (1998). Genome structure of mycobacteriophage D29: implications for phage evolution. J Mol Biol 279, 143–164. [DOI] [PubMed] [Google Scholar]
- Gu Liu C, Green SI, Min L, Clark JR, Salazar KC, Terwilliger AL, Kaplan HB, Trautner BW, Ramig RF, and Maresso AW (2020). Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan NA, Davidson RM, Epperson LE, Kammlade SM, Beagle S, Levin AR, de Moura VC, Hunkins JJ, Weakly N, Sagel SD, et al. (2021). Population Genomics and Inference of Mycobacterium avium Complex Clusters in Cystic Fibrosis Care Centers, United States. Emerg Infect Dis 27, 2836–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Dedrick RM, and Schooley RT (2021). Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu Rev Med. [DOI] [PubMed] [Google Scholar]
- Heijerman HGM, McKone EF, Downey DG, Van Braeckel E, Rowe SM, Tullis E, Mall MA, Welter JJ, Ramsey BW, McKee CM, et al. (2019). Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet 394, 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix J, Epperson LE, Durbin D, Honda JR, and Strong M (2021). Intraspecies plasmid and genomic variation of Mycobacterium kubicae revealed by the complete genome sequences of two clinical isolates. Microb Genom 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisert KB, Heltshe SL, Pope C, Jorth P, Wu X, Edwards RM, Radey M, Accurso FJ, Wolter DJ, Cooke G, et al. (2017). Restoring Cystic Fibrosis Transmembrane Conductance Regulator Function Reduces Airway Bacteria and Inflammation in People with Cystic Fibrosis and Chronic Lung Infections. Am J Respir Crit Care Med 195, 1617–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing G, et al. (2012). On the nature of mycobacteriophage diversity and host preference. Virology 434, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhun BW, Kim SY, Park HY, Jeon K, Shin SJ, and Koh WJ (2017). Changes in Serum IgA Antibody Levels against the Glycopeptidolipid Core Antigen during Antibiotic Treatment of Mycobacterium avium Complex Lung Disease. Jpn J Infect Dis 70, 582–585. [DOI] [PubMed] [Google Scholar]
- Kreutzfeldt KM, McAdam PR, Claxton P, Holmes A, Seagar AL, Laurenson IF, and Fitzgerald JR (2013). Molecular longitudinal tracking of Mycobacterium abscessus spp. during chronic infection of the human lung. PLoS One 8, e63237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular biology and evolution, msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Krewulak KD, and Vogel HJ (2016). Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev 40, 273–298. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing S (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou M, Fan X, Yan J, Li W, and Xie J (2016). Mycobacteriophage SWU1 gp39 can potentiate multiple antibiotics against Mycobacterium via altering the cell wall permeability. Scientific reports 6, 28701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniano SL, Sontag MK, Daley CL, Nick JA, and Sagel SD (2014). Clinical significance of a first positive nontuberculous mycobacteria culture in cystic fibrosis. Annals of the American Thoracic Society 11, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogayzel PJ Jr., Naureckas ET, Robinson KA, Mueller G, Hadjiliadis D, Hoag JB, Lubsch L, Hazle L, Sabadosa K, Marshall B, et al. (2013). Cystic fibrosis pulmonary guidelines. Chronic medications for maintenance of lung health. Am J Respir Crit Care Med 187, 680–689. [DOI] [PubMed] [Google Scholar]
- Nick JA, Daley CL, Lenhart-Pendergrass PM, and Davidson RM (2021). Nontuberculous mycobacteria in cystic fibrosis. Curr Opin Pulm Med 27, 586–592. [DOI] [PubMed] [Google Scholar]
- NIH U.S.N.L.o.M. (2021). LABEL: TRIKAFTA- elexacaftor, tezacaftor, and ivacaftor kit. In DAILYMED (Bethesda, MD, USA: National Library of Medicine; ). [Google Scholar]
- Orens JB, Estenne M, Arcasoy S, Conte JV, Corris P, Egan JJ, Egan T, Keshavjee S, Knoop C, Kotloff R, et al. (2006). International guidelines for the selection of lung transplant candidates: 2006 update--a consensus report from the Pulmonary Scientific Council of the International Society for Heart and Lung Transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation 25, 745–755. [DOI] [PubMed] [Google Scholar]
- Qvist T, Eickhardt S, Kragh KN, Andersen CB, Iversen M, Hoiby N, and Bjarnsholt T (2015a). Chronic pulmonary disease with Mycobacterium abscessus complex is a biofilm infection. Eur Respir J 46, 1823–1826. [DOI] [PubMed] [Google Scholar]
- Qvist T, Gilljam M, Jonsson B, Taylor-Robinson D, Jensen-Fangel S, Wang M, Svahn A, Kotz K, Hansson L, Hollsing A, et al. (2015b). Epidemiology of nontuberculous mycobacteria among patients with cystic fibrosis in Scandinavia. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society 14, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, and Suchard MA (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol 67, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnholt C, Qvist T, Kolpen M, Pressler T, Skov M, and Hoiby N (2019). Antibody response against Mycobacterium avium complex in cystic fibrosis patients measured by a novel IgG ELISA test. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society 18, 516–521. [DOI] [PubMed] [Google Scholar]
- Ren CL, Morgan RL, Oermann C, Resnick HE, Brady C, Campbell A, DeNagel R, Guill M, Hoag J, Lipton A, et al. (2018). Cystic Fibrosis Foundation Pulmonary Guidelines. Use of Cystic Fibrosis Transmembrane Conductance Regulator Modulator Therapy in Patients with Cystic Fibrosis. Annals of the American Thoracic Society 15, 271–280. [DOI] [PubMed] [Google Scholar]
- Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, Weitz JS, and Debarbieux L (2017). Synergy between the Host Immune System and Bacteriophage Is Essential for Successful Phage Therapy against an Acute Respiratory Pathogen. Cell Host Microbe 22, 38–47 e34. [DOI] [PubMed] [Google Scholar]
- Rocchetti TT, Silbert S, Gostnell A, Kubasek C, Campos Pignatari AC, and Widen R (2017). Detection of Mycobacterium chelonae, Mycobacterium abscessus Group, and Mycobacterium fortuitum Complex by a Multiplex Real-Time PCR Directly from Clinical Samples Using the BD MAX System. The Journal of molecular diagnostics : JMD 19, 295–302. [DOI] [PubMed] [Google Scholar]
- Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, Molin S, and Johansen HK (2021). Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol 19, 331–342. [DOI] [PubMed] [Google Scholar]
- Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, et al. (2017). Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. [DOI] [PubMed] [Google Scholar]
- Senhaji-Kacha A, Esteban J, and Garcia-Quintanilla M (2020). Considerations for Phage Therapy Against Mycobacterium abscessus. Frontiers in microbiology 11, 609017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, and Rambaut A (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4, vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverniti V, Forti F, Ghisotti D, and Putzer H (2011). Mycobacterium smegmatis RNase J is a 5’-3’ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol Microbiol 82, 1260–1276. [DOI] [PubMed] [Google Scholar]
- Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, et al. (2020). Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome biology 21, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill D, Benden C, Corris PA, Dark JH, Davis RD, Keshavjee S, Lederer DJ, Mulligan MJ, Patterson GA, Singer LG, et al. (2015). A consensus document for the selection of lung transplant candidates: 2014--an update from the Pulmonary Transplantation Council of the International Society for Heart and Lung Transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation 34, 1–15. [DOI] [PubMed] [Google Scholar]
- Whittier S, Olivier K, Gilligan P, Knowles M, and Della-Latta P (1997). Proficiency testing of clinical microbiology laboratories using modified decontamination procedures for detection of nontuberculous mycobacteria in sputum samples from cystic fibrosis patients. The Nontuberculous Mycobacteria in Cystic Fibrosis Study Group. Journal of Clinical Microbiology 35, 2706–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, and Holt KE (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS computational biology 13, e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, and Holt KE (2019). Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome biology 20, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam YK, Alvarez N, Go ML, and Dick T (2020). Extreme Drug Tolerance of Mycobacterium abscessus “Persisters”. Frontiers in microbiology 11, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi B, Dalpke AH, and Boutin S (2021). Changes in the Cystic Fibrosis Airway Microbiome in Response to CFTR Modulator Therapy. Frontiers in cellular and infection microbiology 11, 548613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y and Lam TTY (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8, 28–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Standardized datasets:
Whole genome sequence data have been deposited to the National Center for Biotechnology Information (NCBI) under BioProject PRJNA319839.
Additional datasets from this study are 1). Video files incorporating a complete set of axial, coronal and sagittal reconstructions post phage days -2, 81 and 229, 2). Gross and microscopic pathological photographs of the explanted lungs, 3). Raw data from Figures 1A, 4A, 4B, 4C, 4D, 6 and Supplemental Table 1. These files are deposited to Mendeley Data at: https://data.mendeley.com/datasets/jtdvbm2tzx/draft?a=b138c41d-bc1b-458c-8d2a-98267555428d


