Abstract
Despite several extraordinary improvements in cancer immunotherapy, its therapeutic effectiveness against many distinct cancer types remains mostly limited and requires further study. Different microfluidic-based cancer immunotherapy-on-a-chip (ITOC) systems have been developed to help researchers replicate the tumor microenvironment and immune system. Numerous microfluidic platforms can potentially be used to perform various on-chip activities related to early clinical cancer immunotherapy processes, such as improving immune checkpoint blockade therapy, studying immune cell dynamics, evaluating cytotoxicity, and creating vaccines or organoid models from patient samples. In this review, we summarize the most recent advancements in the development of various microfluidic-based ITOC devices for cancer treatment niches and present future perspectives on microfluidic devices for immunotherapy research.
I. INTRODUCTION
Different immunotherapies have been used successfully to treat hematological malignancies and have led to significantly improved clinical outcomes for various cancer types, such as melanoma,1 liver cancers,2 osteosarcoma,3 colorectal cancer,4 acute leukemia,5 and breast cancer.6,7 Despite increased understanding and promising clinical results, some challenges remain in current immunotherapeutic approaches, including effective cytotoxicity evaluation of immunotherapy combinations in preclinical studies, knowledge of molecular and cellular drivers of immune cell migration and immune–tumor cell interactions, and maximization of personalized medicines. Thus, preclinical strategies are being developed to model human immunity and the tumor microenvironment (TME).8 Microfluidic models provide new opportunities for investigating the intricate interactions that occur inside biological microenvironments in normal and diseased situations.9,10
Microfluidic technology originated in the 1990s and integrates concepts from different existing fields, including biology, chemistry, physics, fluid dynamics, material science, and microelectronics.11,12 The field of microfluidics has grown substantially, and the technology is becoming a powerful tool with promising application potential.13–15 Recent breakthroughs in microengineering and cell biology have enabled the creation of novel immunotherapy-on-a-chip (ITOC), cell-on-a-chip, and organ-on-a-chip techniques, which can recreate in vivo organ microenvironments for investigating complicated processes, such as tracking the interaction between human immune cells and tumor cells and assessing the effects of various drug treatments.16–18 This review summarizes recent applications of microfluidic-based devices in cancer ITOC studies. Moreover, commercial microfluidic products used for immunotherapeutic purposes are described. Finally, we discuss the current advantages, limitations, and prospects regarding future immunotherapy research utilizing microfluidic technology.
II. MICROFLUIDIC-BASED IMMUNOTHERAPY-ON-A-CHIP STRATEGIES
A. Studying cancer immunotherapeutic targets
A cascade of genetic changes causes the generation of cancer cells. Cancer cells frequently acquire the ability to suppress the immune system. Hence, placing cancer cells back on the immune system's radar has long been thought to be an essential step toward total tumor eradication and long-term anticancer protection.19 Recently, molecular profiling,20,21 measurements of gene delivery in tumor cells,22 and cancer immunotherapeutic gene targets, such as PD-L/PD-L1, CXCR2,23 KRAS+, and MET-Amp,24 human transferrin receptor (CD71),25 EpCAM,26 N-Myc,27 CCL17, and TNF-beta,28 have been used to identify biomarkers for various types of cancer via ITOC devices. In addition, the CRISPR/Cas9 technique was integrated with a microfluidic device for gene knockout or autologous dendritic cells and T-cell editing.29–31 These studies provide proof of concept that microfluidic technology is a promising approach for future gene-based therapy development.
In particular, most recent immunotherapeutic targets focus on the PD-L/PD-L1 gene to achieve immune checkpoint blockade therapy for cancer treatment. PD-L/PD-L1 is one of the crucial immunosuppressive molecules that prevents T lymphocytes from becoming activated, allowing malignancies to develop.32 In the clinic, PD-1/PD-L1 immune checkpoint blockade therapy is an effective cancer treatment method and was approved by the FDA in 2011;33 it offers significant clinical benefits and therapeutic effects in the treatment of various types of malignancies.34 In recent years, researchers have examined sensitivity to immune checkpoint therapy and identified patients' responses to specific antibody treatments using ITOC devices.
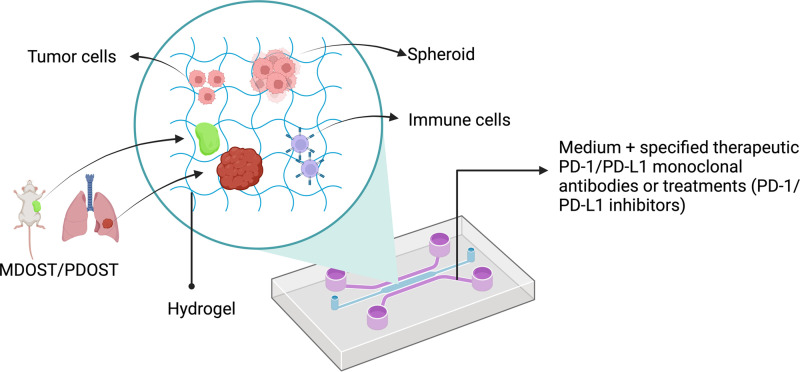
Most ITOC devices for PD-1/PD-L1 have been designed with three interconnected chambers, including a center chamber for culturing the cells/spheroids/murine- or patient-derived organotypic tumor spheroids (MDOTS/PDOTS) in a gel and two side channels for specified therapeutic monoclonal antibodies or treatments (Fig. 1). Collagen type I and Matrigel are the most common gels used as microenvironments. Many studies have successfully cocultured cancer cells with immune cells and other factors to determine the effect of PD-1/PD-L1 immune checkpoint therapy (Table I). For example, in 2020, Cui et al.39 developed “GBM-on-a-Chip,” which is a 3D microfluidics-based microphysiological system (MPS) simulating the subtype-specific in vivo glioblastoma (GBM) tumor niche, to analyze the heterogeneity of the anti-PD-1 immunotherapy response in molecularly different GBM cohorts based on DNA methylation, transcriptomic and genomic data.
FIG. 1.
Schematic of a microfluidic device for studying cancer immunotherapeutic targets (for example, PD-1/PD-L1). Created with BioRender.com.
TABLE I.
Summary of recent publications on PD-L/PD-L1 in cancer immunotherapy studies. MSC-CM, mesenchymal stem cell-conditioned medium.
| Authors, reference | Targets | Cell line | On-chip model | Time | Chip material | Hydrogel materials | Microfluidic design |
|---|---|---|---|---|---|---|---|
| Aboulkheyr Es et al. 35 | Gene expression profile analysis, including the PD1/PD-L1 gene | MCF7 | Coculturing an epithelial-like/low-expressing PD-L1 breast cancer cell line with MSC-CM | 3 days | 3D culture chip (AIM Biotech) | Collagen gel solution | The devices are made up of two parallel communication channels situated on either side of an expanded center area holding gel |
| Sehgal et al.36 | Bulk RNA-seq analysis of MC38 MDOTS after PD-1 blockade | MC38 | MC38 MDOTS treated with αPD-1, IgG2a, αCD8a, αIFN-γ, IL-6, TNF-α, and/or LCL161 in gel | 6 days | 3D culture chip (AIM Biotech) | Collagen gel solution | Three channels per microfluidic chamber: center channel used for gel loading and two side channels used for media37,38 |
| Cui et al.39 | Optimized PD-1 immunotherapy | Patient-derived GBM cells | Glioblastoma-on-a-chip | 2–3 days | SU-8, PDMS | Matrigel | The microfluidic device has a vascular-seeding channel, a hydrogel loading channel, and a medium infusion channel |
| Aboulkheyr Es et al.28 | The immune-suppressive potential of PFD therapy in breast cancer cells by treating CAF inside the TME | MCF7 | Cocultured tumor cells with CAF cells | 3 days | 3D culture chip (AIM Biotech) | Collagen gel solution | The devices are made up of two media channels that are situated on each side of a long central section known as the gel channel |
| Anastasiadou et al.40 | PD-L1 inhibition in immunotherapy and miR-34a dysregulation in cancers | U2932, SUDHL5, OMA4, and DG75 | In vitro 3D biomimetic model | 2 days | PDMS | Type 1 collagen, fibronectin | A cylindrical channel is formed in a 3D collagen matrix within a microfabricated PDMS gasket41,42 |
| Jenkins et al.37 | MDOTS/PDOTS profiling to identify biomarkers associated with a PD-1 blockade response and resistance | MC38 and B16F10 | MDOTS/PDOTS-on-a-chip | 3–6 days | PDMS and DAX-1 3D cell culture chip (AIM Biotech) | Type 1 collagen | The spheroid-collagen was inserted into the device's center gel chamber, PDOTS/MDOTS-containing collagen hydrogels were hydrated with medium with or without the specified therapeutic monoclonal antibodies |
| Lee et al.43 | Characterize the interaction of PD-1/PD-L1 and PD-L2 | COS-7 cells | Nanosensor platform integrated with a microfluidic chip | 2 days | SU-8 | N/A | The microfluidic chip includes two structural layers that are joined together by thermal diffusion. Vertical vias link the bottom layer to the top layer, which contains four channels that interact with the magneto-nanosensor chip |
In addition, according to a detailed characterization of the spheroids from animals and patients with various tumor types, many critical components of the tumor immune microenvironment (TIME) have been shown to be retained in these spheroids, including T cells that express PD-1, dendritic cells, a variety of myeloid cell types, endothelial cells (ECs), and fibroblasts. In the TIME, mesenchymal stem cells (MSCs) are involved in tumor development, migration, metastasis, and drug resistance. MSCs may change the immunosuppressive capabilities of the TIME by promoting PD-L1 expression in a wide range of malignancies. Aboulkheyr Es et al.35 used a microfluidic device to show that treating MSCs with the cytokine inhibitor pirfenidone (PFD) reduced the production and secretion of different cytokines, including CCL2 and CCL5, and therefore decreased breast cancer PD-L1 expression. They found that PFD decreases PD-L1 expression indirectly by targeting critical immune-related cytokines in tumor stromal cells, implying a new method for cancer immunotherapy in combination with another treatment. Microfluidic devices have also been used to study the inhibition of PD-L1 expression by overcoming immune suppression for oncolytic therapy.44 In 2019, by inserting stimulator U2932 EBNA2 cells transduced with either lentiviral vectors containing miR-34a or vector control into 3D biomimetic microfluidic chips, Anastasiadou et al.40 found that in EBNA2-transfected diffuse large B-cell lymphoma, miR-34a reconstitution lowered PD-L1 expression while increasing immunogenicity.
The usefulness of measuring PD-L1 expression levels is also a research focus. When circulating tumor cell (CTC) PD-L1 levels are compared to tumor biopsy results, it may be possible to identify individuals who are likely to respond to treatment.45–47 Several microfluidic-based techniques have been developed to isolate PD-L1 and evaluate its expression on CTCs from patient biopsies, such as the Vortex HT chip containing parallel channels with rectangular trapping reservoirs,45,48 low-shear spiral microfluidic technology combined with immunocytochemistry,49 an ultra-stable nanobubble (NB) microfluidic-based reconstruction process,50 and an epitope-independent microfluidic device for enriching CTCs.51
B. Understanding cellular immune responses through dynamic interplay in the TME
The TME is a complex niche that plays a crucial role in the clinical behavior and response of many cancer types to various treatment methods.52 In particular, the TME contains not only cancer cells but also host-interacting cells, including ECs, stromal fibroblasts, and a variety of immune cells. Their interaction in the TME influences tumor development and metastasis. Many microfluidic devices (Table II and Fig. 2) have been developed to model the dynamic interplay between tumor immune cells,60 tumor ECs,61 tumor immune cell monocyte/macrophage migration,62–65 tumor vasculature,66,67 and tumor lymph nodes68 in the TME. Because they can be used to expose underlying processes of cancer growth and to estimate the success of experimental oncological medicines such as small molecules or targeted therapies, these ITOC devices are beneficial supplements to preclinical animal models.
TABLE II.
Summary of recent publications on the cellular dynamics of immune response.
| Authors, reference | Targets | Cell line | On-chip model | Time | Chip material | Hydrogel materials | Microfluidic design |
|---|---|---|---|---|---|---|---|
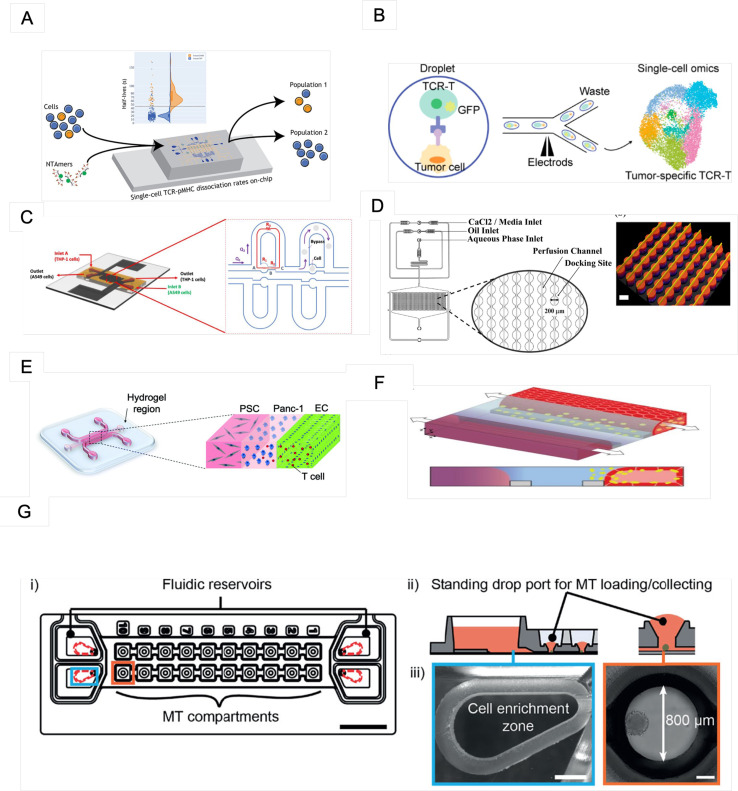
| Jemmes et al.53 | Characterizing pMHC-TCR interactions | SupT1 | Analyzed TCR-pMHC dissociation rates on live CD8+ T cells [Fig. 2(a)] | 6–10 h | SU-8, PDMS | N/A | The device has four fluid inputs (each with its own built-in filter), five fluid outputs, and eight autonomous chambers |
| Wang et al.54 | Identify TCR-pMHC interactions at a single-cell level | T2, TCR-T | Using a cellular reporter system and a fluorescence-activated droplet approach to perform function of tumor antigen-specific TCRs on single cells | SU-8, PDMS | N/A | Droplet-generating chip and droplet-sorting chip [Fig. 2(b)] | |
| Nguyen et al.55 | Investigate effects of antitumor NK cells on tumor and cardiac microtissues | HCT116 NK cells | Triple culture of 3D colorectal tumor microtissues, cardiac microtissues, and human-derived NK cells in the same microfluidic network | 3 days | Akura Flow MPS discovery platform | Hydrogel | The microfluidic chip has two separate microfluidic channels with medium reservoirs at both ends; up to ten fluidically coupled microtissues can be accommodated in each channel [Fig. 2(g)] |
| Pendharkar et al.56 | Pair single cells and carry out electrofusion efficiently | A549, THP-1 | Single-cell trapping and performing cell electrofusion | 4 days | SU-8, PDMS | N/A | Microfluidic flip-chips have a three-layered structure with PDMS channels as the top layer, a through-hole membrane as the middle layer, and titanium electrodes as the bottom layer [Fig. 2(c)] |
| Berger Fridman et al.57 | Cell morphology and EMT; oxidative stress and apoptosis; immunomodulation | MCF7, CCD-1129SK, human monocyte/macrophage cells, peripheral blood mononuclear cells | On-chip generation of Alg and Alg/Alg-S TME scaffolds | 3 days | SU-8, PDMS | Alg/Alg-S hydrogel | The chip design was composed of an array layer, featuring a flow-focusing junction for rapid droplet generation and an array with 1000 docking sites, an upper layer that spans the array and facilitates efficient perfusion of the TME scaffolds; perfusion is possible through the array channels [Fig. 2(d)] |
| De Haan et al.58 | Transendothelial T-cell migration in 3D | HMEC-1, A375, primary CD3+ T cells | Coculture with tumor cells to study T-cell invasion into tumor microenvironments [Fig. 2(f)] | 2 days | OrganoPlate (3-lane; MIMETAS) | 4 mg/ml collagen I gel | 40 chips with long gel and perfusion channels |
| Mollica et al.59 | Observe and quantify T-cell infiltration across the vasculature | HUVECs, PANC-1, PSCs | Pancreatic cancer cells, organ-specific stromal cells, and endothelial cells were cocultured inside a microfluidic device [Fig. 2(e)] | 5 days | 3D culture chip (AIM Biotech) | 3 mg ml−1 collagen solution | The device was made with a cyclic olefin polymer sealed with an oxygen-permeable membrane and consists of two lateral channels and a central region, which is divided by an array of triangular pillars placed at a distance of 100 μm |
FIG. 2.
Recent microfluidic designs for understanding the cellular dynamics of immune responses. (a) The mCPU platform was used to analyze TCR-pMHC dissociation rates. Reproduced with permission from Jammes et al., ACS Sens. 7, 159 (2022). Copyright 2022 American Chemical Society. (b) Coencapsulation of cells in droplets. Reproduced with permission from Wang et al., Anal. Chem. 94, 918 (2022). Copyright 2022 American Chemical Society. (c) Fluidic resistance model for the cell trapping process. From Pendharkar et al., Cells 10, 2855 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (d) Microfluidic design with a magnified view of the array docking sites. Reproduced with permission from Berger Fridman et al., Acta Biomater. 132, 473 (2021). Copyright 2021 Elsevier B.V. (e) The PDAC-TME model comprises cancer cells (PANC-1, blue) sown in the central gel area, bordered by two medium channels containing endothelial cells (ECs, green) and pancreatic stellate cells (PSCs, gray). From Mollica et al., Biomater. Sci. 9, 7420 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (f) 3D coculture comprising ECs and T cells (yellow). From De Haan et al., Int. J. Mol. Sci. 22, 8234 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (g) (i) The iMPSs modified based on the Akura™ Flow platform. Scale bar: 10 mm. (ii) Reservoir and adjacent MT compartments. (iii) One reservoir's cell enrichment zone (scale bar: 1 mm). From Nguyen et al., Front. Immunol. 12, 781337 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License.
Recently, there has been a trend in integrating microfluidic devices with automatic systems to handle various processes, such as measurement, imaging, image processing, and data analysis.56,57,69 This enables the device to independently conduct sophisticated fluidic processing steps. For example, a study by Jammes et al.53 demonstrated a fully autonomous microfluidic cellular processing unit (mCPU) to measure the dissociation rate between T-cell receptors (TCRs) on CD8+ T cells and peptide-major histocompatibility complexes (pMHCs). In particular, the mCPU system can assess the dissociation of single-cell surface markers and performs all essential fluid management, microscopic imaging, image processing, and data analysis, resulting in a closed-loop system capable of assessment at the single-cell level. The affinity of surface markers has been proposed as a key metric that correlates with T-cell activities, while the function of mechanical force in the dissociation rate is still being debated.
Currently, many researchers are concentrating on adoptive cell therapy (ACT), whereby cancer patients are administered immune cells with direct anticancer activity. ACT employs genetically engineered or endogenous tumor-reactive lymphocytes to express tumor antigen receptors. CD8+ T cells are the most often employed lymphocytes in ACT.70 Two major alternatives have been established. In one strategy, which was initially described in 1993 by Zelig Eshhar, T cells are first programmed to express chimeric antigen receptors (CARs) made from tumor-reactive monoclonal antibody single-chain variable fragments. Since 2017, the Food and Drug Administration (FDA) has authorized CAR-T-cell therapy for specific malignancies. Another alternative is to create T cells using the TCR technique, which involves genetically modifying T cells that generate highly selective TCRs for malignancies. Many microfluidic devices have been developed to identify, screen, purify, quantify, and manufacture CARs71–73 and TCRs.54,70,74
Recently, T-cell dynamics have also gained much attention in the study of immunological processes. The extravasation and subsequent infiltration process of T cells have been observed and successfully quantified underflow in real-time by using the following models: the Endothelium-on-a-Chip Model, which is a high-throughput artificial membrane-free microfluidic platform;58 a three-channel microfluidic 3D pancreatic tumor model;59 an in vitro microfluidic cell extravasation assay with an incorporated microvascular network;75 and a lymph node-on-a-chip with the flow to model T-cell migration and T-cell-dendritic cell interaction.76 For instance, Mollica et al.59 created a 3D microfluidic-based pancreatic ductal adenocarcinoma (PDAC)-TME model to evaluate T-cell infiltration across the vasculature. T-cell migration into PDAC cells implanted in a collagen gel was observed, facilitating the evaluation of T-cell transmigration into the TME with respect to the upregulation and presence of PSCs, PDAC cells, and EC linings.
C. Evaluating immune cell-mediated cytotoxicity
In antitumor immunotherapy studies, natural killer (NK) cells have been explored for their potential capacity to cause cytotoxicity in cancer cells.55,77–80 Several microfluidic-based devices (Table III and Fig. 3) have been developed to evaluate NK-cell cytotoxicity in a 3D extracellular matrix microenvironment, including the following: an injection-molded plastic array culture (CACI-IMPACT) device for 3D cytotoxicity assays through spatiotemporal analysis of lymphocytes and cancer cells embedded in 3D ECM,86 a 3D in vitro tumor vasculature model for real-time monitoring of immune cell infiltration and cytotoxicity,81,87 microspheres88 and droplet microfluidic-based devices.89–92 By using droplet-based microfluidics, Antona et al.84 generated NK-92 cells, target cells (K562), antibodies, and IFN-ɣ sensing beads. The ratio of IFN-ɣ-releasing cells after 12 h was investigated, and the released IFN-ɣ and NK-92 cell cytolytic activity were also analyzed. The device successfully created droplet-contained cells, similar to an independent screening chamber, to evaluate IFN-ɣ secretion and cytotoxicity, allowing the detection of IFN-ɣ at a few femtograms per bead.
TABLE III.
Summary of recent publications on immune cell-mediated cytotoxicity studies.
| Authors, reference | Targets | Cell line | On-chip model | Time | Chip material | Hydrogel materials | Microfluidic design |
|---|---|---|---|---|---|---|---|
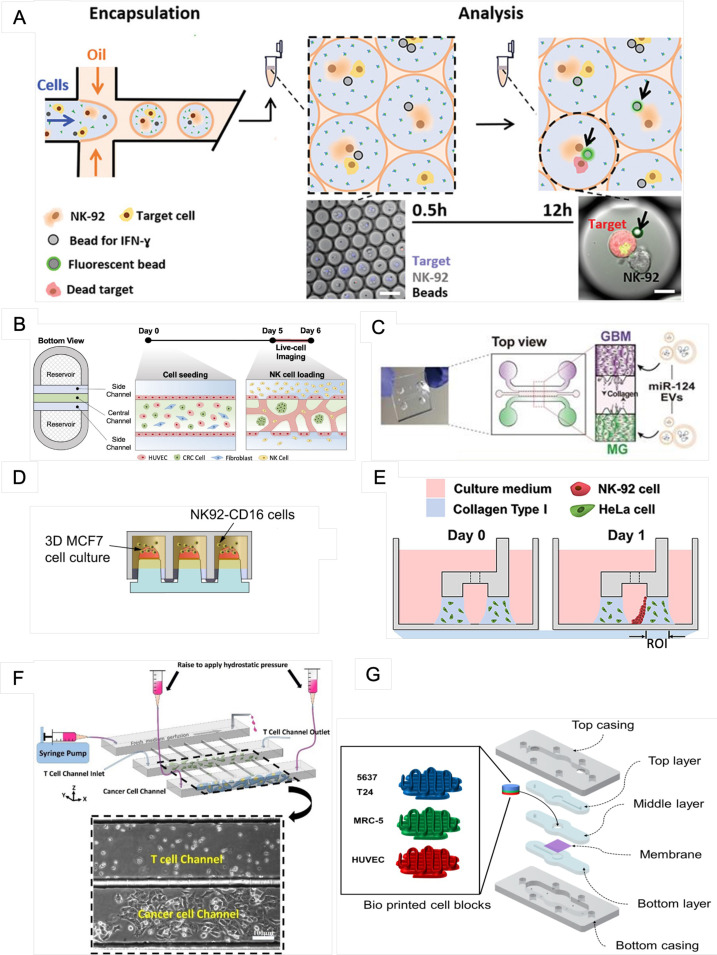
| Song et al.81 | Observe cytotoxic activity of NK cells on colorectal cancer | HUVECs, lung fibroblasts, primary NK cells, peripheral blood mononuclear cells | In vitro tumor vasculature model on the injection-molded microfluidic platform | 1 day | Polystyrene | Fibrin gel | The platform is composed of three parallel fluid guide rails, and rapid and robust fluid patterning driven by capillary forces enables high-throughput experiments [Fig. 3(b)] |
| Hong et al.82 | Determine the EV-mediated miR-124 delivery-induced synergistic antitumor effects by suppressing the growth of human GBM cells and inhibiting M2 microglial polarization | HEK293T, U373MG and U87MG, SV40, NK-92, SNU-201, SNU-466, SNU-489, SNU-626, and SNU-1105 | A tri-culture of glioblastoma cells, microglia, and NK cells in a 3D microfluidic device | 3–4 days | PDMS | 3 mg ml−1 type I collagen | The microfluidic chip consists of one gel channel at the center and two medium channels on both sides [Fig. 3(c)] |
| Gopal et al.77 | Demonstrate NK-cell-mediated cell cytotoxicity in combination with monoclonal antibodies | MiaPaCa-2, MCF-7, MDA-MB-231, NK92-CD16 | 3D coculture of NK92-CD16 cells with pancreatic and breast cancer cells | 8 days | Polystyrene 330-micropillar chips (MBD Korea Co.) | Matrigel | The chips were coated with polydopamine and hydro droplet containing cells on the top [Fig. 3(d)] |
| Kim et al.83 | Predict immunotherapeutic effects | T24 and 5637 cells, MRC-5 cells, THP-1 cells | The bladder cancer-on-a-chip was fabricated with either T24 or 5637 cells and the MRC-5, HUVEC, and THP-1-cell lines [Fig. 3(g)] | 3 days | PDMS | N/A | The microfluidic device consisted of a tissue culture platform, a nutrient supply channel, and a waste removal chamber |
| Antona et al.84 | Investigation of IFN-ɣ release and NK-cell cytotoxicity in a sensitive, quantitative, and multiplexed way | NK-92, K562 | Droplet-based microfluidics is employed to quantitatively investigate IFN-ɣ secretion from single NK cells in correlation with their cytotoxic activity against a specific target [Fig. 3(a)] | 12 h | PDMS | N/A | The water inlet of the chip was connected to a spiral channel used to separate suspended beads/cells in the aqueous phase prior to droplet generation at the T-junction |
| Chen et al.85 | Investigate the behaviors of GP33 antigen-specific CTLs against GP33±murine hepatic cancer cells (HEPA1-6) in the presence of elevated IFP and narrowed interstitial paths | BNL, HEPA1-6, 2C, and P14 CD8+ T cells | Cocultured cancer cells and immune cells in collagen gels confined in an array of microwells | 2–3 days | SU-8, PDMS | Type I collagen gel | The microfluidic platform consisted of three main channels running in parallel and an array of “slit” channels connecting adjacent main channels [Fig. 3(f)] |
| Park et al.86 | Infiltration, migration, and cytotoxic activity of NK-92 cells against HeLa cells in the collagen matrix | HeLa, NK-92 | 3D coculture of cytotoxic lymphocytes with cancer cells [Fig. 3(e)] | 1 day | Polystyrene | Type I collagen | Microfluidic rail-based microstructures are embedded in microwells with a 96-well plate format. The microstructure in a single well consists of two low rails for primary hydrogel patterning and one high rail to form a channel for secondary fluid patterning after hydrogel cross-linking |
| Ayuso et al.87 | Study natural killer cell response | MCF7, HUVECs, NK-92 | 3D coculture of NK cells with cancer cells | 3 days | SU-8, PDMS | 4.0 mg/ml collagen type I | Microdevice comprised a central microchamber to inject a 3D hydrogel and three parallel PDMS rods |
FIG. 3.
Recent microfluidic designs for evaluating immune cell-mediated cytotoxicity. (a) The droplet-based microfluidic platform for the combinatorial evaluation of IFN-ɣ release and NK-cell cytotoxicity. From Antona et al., Adv. Funct. Mater. 30, 2003479 (2020). Copyright 2020 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (b) In vitro tumor vasculature model on the injection-molded microfluidic platform. From Song et al., Front. Immunol. 12, 733317 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (c) A 3D microfluidic coculture device to recreate the interaction between GBM and microglia. From Hong et al., Theranostics 11, 9687 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (d) 3D MCF-7-cell aggregates on the surface of the 384-pillar plate and NK92-CD16 cell-mediated cytotoxicity were determined using the 384-pillar/well sandwich platform. From Gopal et al., Commun. Biol. 4, 893 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (e) The 3D cytotoxicity test procedure. Under low rails, HeLa cells buried in collagen were patterned (Day 0). NK-92 cells were injected into a microchannel created by the hydrogel after 24 h of culture. NK-92 cells were placed on a collagen block (Day 1) and cultivated for an additional 24 h by tilting the device at an angle of 90°. From Park et al., Front. Immunol. 10, 1133 (2019). Copyright 2019 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (f) The microfluidic platform for modeling physical impediments in the tumor interstitium, which consists of three high main channels joined by high slit channels. Scale bar: 100 μm. From Chen et al., Sci. Rep. 10, 13662 (2020). Copyright 2020 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (g) The whole structure of the bioprinted cell block. From Kim et al., Int. J. Mol. Sci. 22, 8887 (2021). Copyright 2021 Author(s), licensed under a Creative Commons Attribution (CC BY) License.
One recent potential cancer therapeutic agent is tumor antigen-specific cytotoxic T lymphocytes (CTLs).93 Through allogeneic recognition and antigen specificity, the migratory characteristics and the anticancer response of tumor antigen-specific CTLs targeting hepatic cancer cells were studied by Chen et al.85 The microfluidic device was designed with slit channels to mimic the small capillary channels limited by fibrous encasement, and enhanced interstitial fluid pressure was achieved by applying hydrostatic pressure to the tumor. The authors observed that CTL antigen-specificity against the targeted tumor cells impacted their cytotoxic effectiveness but did not influence the success rate of CTLs seeking to penetrate the tumor core.
D. Exploring clinical utility through the patient-derived sample
Cancer vaccines have gained popularity among the many types of cancer immunotherapies due to their capacity to induce long-lasting tumor regression and antitumor immune responses. In a simplified microfluidic vaccine production platform, monocytes were isolated from patient blood and encapsulated in alginate gel droplets, resulting in much less blood product handling and accelerated vaccine production.94 Different antigen- and adjuvant-loaded monocytes may be encapsulated in separate gel droplets that suit the cells' optimal conditions to form a cocktail vaccine specific to the medical profile of a particular patient. For example, in a 4T1 breast tumor model, anti-PD-1 antibodies were embedded in gel droplets, resulting in a cocktail vaccination with increased anticancer effectiveness. Another study used glass capillary microfluidics to produce nanoparticles,95 which were shown to give a significant maturation signal to immature dendritic cells, leading to a higher presence of CD8+ T-lymphocytes in the TME and resulting in a synergistic effect with the anti-CTLA-4 therapy for melanoma cancer.
Although increasingly effective tumor nanovaccines are being created, their use is severely restricted by safety, delivery, side effects, and production volume.96,97 These issues can be solved by utilizing a microfluidic device to swiftly combine precursors within microfluidic channels and precisely control fluids, resulting in more uniform, faster, and precise particle production. Recently, by constructing a unique peptidic microarchitecture (PMA) vaccine carrier, Du et al.98 presented a technique for enhancing tumor vaccine immunogenicity. The PMA-neoantigen successfully cleared MC38 tumors in vivo when paired with immune checkpoint inhibition or a phosphatidylinositol 3 kinase (PI3K) inhibitor. The microfluidic chip was utilized to make PMA particles that were more homogeneous and accurate. In an allograft mouse tumor model, these cancer vaccination cocktails successfully and simply combined diverse treatment techniques and demonstrated good therapeutic effectiveness.
The continued integration of novel engineering tools and biological concepts, such as 3D printing, automated handling, in situ multisensors, or organoids, and patient-specific induced pluripotent stem cells into organ-on-a-chip platforms will significantly advance their biomedical applications.99 MDOTS/PDOTS cultivated on a 3D microfluidic device can be used to quickly investigate the effects of immunotherapy and targeted treatment in an ex vivo system, allowing for the discovery of therapeutic combinations. Therefore, the number of known biomarkers can be greatly increased.36,37 Preclinical research can be used to establish and steer next-generation clinical trials for lung, ovarian, kidney, multiple myeloma, and breast cancer patients38,100–103 to increase their success rate. The ex vivo model may provide a new way to select the optimum therapy for an individual patient based on the ex vivo response of cancer medication treatment alternatives and newer immunotherapeutic therapies, such as CAR-T-cell therapy, pending further research. As a vital aspect of cancer patients' long-term treatment, this precision medicine method is predicted to provide a transformational strategy for eliminating inefficient or superfluous medications, minimizing excess toxicity and costs, and limiting tumor cell cross-resistance to drugs. This method may also be utilized for preclinical testing of new treatments, investigating the formation of drug-resistant cancer subpopulations, discovering novel therapeutic targets, and establishing ex vivo models of different metastatic tumors. Organ-on-a-chip models are also used to study interactions between cells or organs and chemotherapeutic treatment,104 therapeutic antibodies,105 or the TME, including the extracellular matrix102,106–108 and blood vessels.109 The efficacy of a drug or treatment can be determined in patient-specific models, introducing the potential for single-patient clinical trials. Several studies have demonstrated designs for an integrated microfluidic chip for improving cancer treatment (Table IV and Fig. 4).
TABLE IV.
Summary of recent publications on clinical utility through patient-derived samples. HNSCC, head and neck squamous cell carcinoma.
| Authors, reference | Targets | Cell line | On-chip model | Time | Chip material | Hydrogel materials | Microfluidic design |
|---|---|---|---|---|---|---|---|
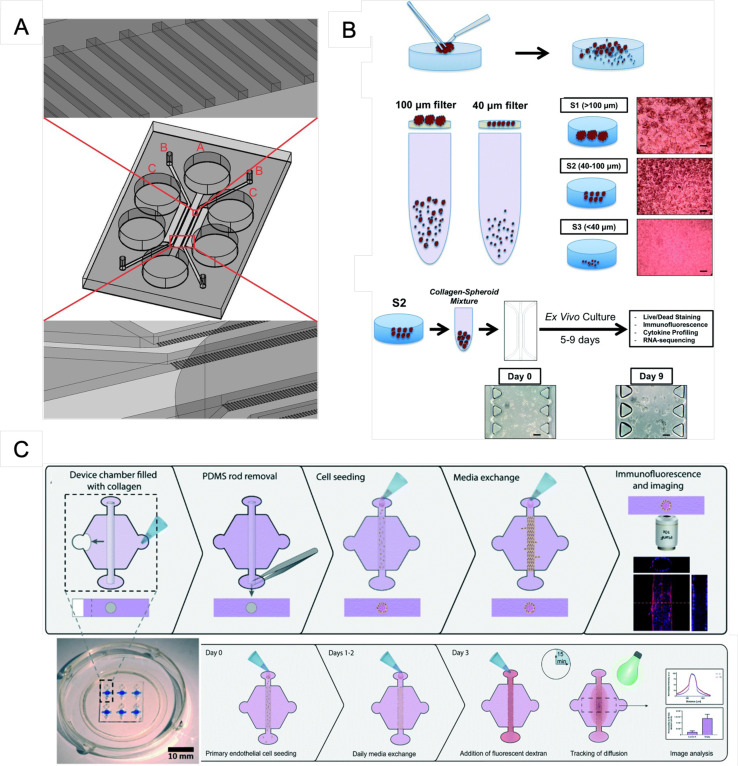
| Hafezi et al.110 | Molecularly engineer TCR-T cells that could retain their polyfunctionality seen in patients while minimizing the associated risk of organ rejection | HepG2.2.15, HepG2, peripheral blood mononuclear cells | Labeled TCR-T cells were added to the microfluidic device containing dissociated HepG2-Env target cells | 3–5 days | PDMS, SU-8 | Matrigel | The device comprising a gel region with adjoining media channels separated from the gel channel by trapezoidal posts111 |
| Virumbrales-Munoz et al.109 | Generate organotypic primary patient-specific blood vessel models using normal and tumor-associated primary CD31+ selected cells | Normal (NEnC) and tumor-associated primary CD31+ selected cells (TEnC) | Microfluidic tubular vessel models | 3–4 days | PDMS | Collagen type I | The device has a hexagonal chamber with two side ports for hydrogel injection and two differently sized ports that hold a PDMS rod and are designed to enable passive pumping for fluid flow through the lumen [Fig. 4(c)] |
| Zhou et al.112 | Streamlining the on-chip separation, capture, immunofluorescence assay and/or in situ culture of isolated cells | HeLa, Hep G2, CTCs | Integrated CTC procurement and on-chip culture | 11 days | SU-8, PDMS | N/A | The device consists of two main parts: a separation channel and a micropost-array chamber for cell trapping and culture |
| Al-Samadi, et al.113 | Isolated cancer cells, patients' serum, and immune cells; test the efficacy of immunotherapy on HNSCC patient samples | Human tongue SCC, HNSCC patient samples, human peripheral blood mononuclear cells | Tongue cancer cell line (HSC-3) embedded in a human tumor-derived matrix (Myogel/fibrin) and immune cells | 3 days | PDMS, SU-8 | Myogel/fibrin gel | Microfluidic structure contains a deep central chamber for the immune cells, which is connected to the neighboring cancer cell containing channels via microchannels. The channels incorporating the cancer cells are linked to the immune cell containing channel from both sides [Fig. 4(a)] |
| Aref et al.38 | Evaluation of murine- and patient-derived organotypic tumor spheroids | MDOTS/PDOTS | MDOTS/PDOTS-containing tumor, immune, and stromal cells were cultured in the chip with therapeutic monoclonal antibodies [Fig. 4(b)] | 5–9 days | 3D cell culture chip (AIM Biotech) | Type I and IV collagen | The device consists of three microfluidic chambers each with a central gel channel flanked by two media channels |
FIG. 4.
Recent microfluidic design for exploring clinical utility through patient-derived samples. (a) Microfluidic chip design for loading immune cells into channel A and cancer cells into the two channels labeled B. The channels labeled C are used for hydration. Reproduced with permission from Al-Samadi et al., Exp. Cell Res. 383, 111508 (2019). Copyright 2019 Elsevier Inc. (b) Workflow for MDOTS and PDOTS. A tumor specimen is obtained and dissociated physically and enzymatically; macroscopic tumors, single cells, and spheroids were all studied. The spheroids are harvested and embedded in collagen before being delivered into a microfluidic channel for culture. Scale bars: 100 μm. From Aref et al., Lab Chip 18, 3129 (2018). Copyright 2018 Author(s), licensed under a Creative Commons Attribution (CC BY) License. (c) Permeability calculations are based on the seeding technique and dextran diffusion experiments. The microchamber was filled with a collagen hydrogel polymerized around a PDMS rod, which was then removed, resulting in a tubular-shaped gap in the hydrogel. From Virumbrales-Munoz et al., Lab Chip 20, 4420 (2020). Copyright 2020 Author(s), licensed under a Creative Commons Attribution (CC BY) License.
III. COMMERCIAL IMMUNOTHERAPY-ON-A-CHIP PRODUCTS
Recently, versatile analytic methods have provided more quantitative knowledge of cancer progression and microenvironmental interactions that allow improvements in targeted cancer therapy. Powerful systems have been developed, such as the LabSat™ autostainer114 for TME analysis and microfluidic 3D bioprinting (RX1™ bioprinter, Aspect Biosystems),115 to print a core-shell fiber design that recapitulates the immune “cold” TME and permits monitoring of immune cell infiltration into the cancer cell core of the fiber. Furthermore, the Opto™ Cell Therapy Development Workflow for Berkeley Lights systems was designed to screen hundreds of individual T-cell targets for CAR-T-cell phenotypic and functional screening, as well as the finding of TCRs linked with certain T-cell behaviors. Subsequent live-cell export for the downstream study of T cells of interest will assist scientists in hastening the discovery of effective cancer medicines.116 AI has also been integrated with a microfluidic device (the Dexter™ microfluidic platform) to incorporate the sorting of single cells and discover functional connections between various immune cells and malignancies.117 The mechanistic change coordinated by immune activation was demonstrated by additional AI-guided matrix coupling with polyfunctional modulation properties. This method will allow for the identification of driver modulation associated with the projected therapeutic response as well as the modeling of next-generation cell-based immunotherapies.
Table V shows the list of commercial systems using microfluidics for cancer immunotherapy research.
TABLE V.
Microfluidic-based systems recently released on the market and their applications.
| Products | Applications | Company | Reference |
|---|---|---|---|
| LabSat autostainer LabSat Frozen | The automated staining instrument, which is based on a microfluidic technology, is capable of performing immunohistochemistry assays on frozen sections in approximately 12 min, while maintaining high staining quality and reproducibility | Lunaphore Technologies, Switzerland | 114 |
| Opto Cell Therapy Development Workflow | Antigen recognition mediated by CARs Differentiate CAR-T-cell subgroups TCR-mediated antigen recognition | Berkeley Lights, USA | 116 |
| RX1 bioprinter | Direct extrusion of biological fibers with varied diameters | Aspect Biosystems, Canada | 118 and 119 |
| Dexter microfluidic platform | Combining a controlled microfluid system and AI for real-time surveillance of dynamic tumor–immune interaction at the single-cell level | Shilps Sciences Inc., USA | 117 |
| CellSqueeze | Squeezing cells quickly through a microfluidic chip Temporarily exposing the cell membrane and enabling biologic substances or cargo to seep into the cell before it reseals. | SQZ Biotech, USA | 120 |
| DAX01 AIM Biotech 3D Cell Culture Chips | 3D cell culture | AIM Biotech, Singapore | 38 |
| Akura Flow MPS discovery platform | Preclinical efficacy and toxicity testing applications in a multitissue, microfluidic assay format | InSphero, Switzerland | 55 |
| OrganoPlate 3-lane | Perfused tubules, cell–cell interaction, 3D cell migration, angiogenesis, vascularization. | MIMETAS, The Netherlands | 58 |
| Polystyrene 330 micropillar chips Polystyrene 384 pillar plates | 3D cell culture | MBD Korea Co., South Korea | 77 |
IV. STRENGTHS AND WEAKNESSES OF USING MICROFLUIDIC TECHNOLOGY IN IMMUNOTHERAPY-ON-A-CHIP RESEARCH
In recent decades, the application of microfluidic technologies for addressing biological questions has emerged as a potent tool for cancer immunotherapy research. Microfluidic devices are designed to create 3D cellular models that almost exactly reproduce the microenvironment and offer various benefits over conventional in vitro systems. It is a physiologically appropriate technique for researching biological processes that underpin cancer development and therapy. The microfluidic device can be designed with multiple channels that adjust depending on users and the biological experiment. Moreover, pressure, flow velocity, oxygen concentration levels, and temperature are easy to maintain and control. This allows for more accurate replications of genuine physiological circumstances in the human body and simplifies the investigation of cellular responses under tightly regulated environments. Furthermore, microfluidic platforms can be fabricated using different materials based on experimental requirements of techniques, such as microscopy, surface modification, or biological/biochemical assays. Using a microfluidic device also reduces the number of necessary samples, materials, and reagents. Hence, the cost of consumables required for experiments dramatically decreases.
Despite the apparent advantages of microfluidic technology, several limitations remain. For example, microfluidic devices might not fully replicate realistic environments due to the artificial character of the constructed microenvironment, and these devices can exhibit drug sorption or toxicity depends on the materials used. Moreover, the microfluidic design for organ-on-a-chip models is complex because of the organ size (mass and volume) and their scale with fluid quantity and flow. Hence, to overcome this limitation, there is an unmet need for a protocol to replicate an accurate scale of the organ relative to the other components of the system, and it is crucial to have standardization to help researchers follow methods, control the resulting quality, and compare their findings. Using only a small fluid volume may also be considered a drawback for several experiments requiring many samples. In addition, human immune system studies on a microfluidic chip also challenge the researcher. With many vital cell activities, including immune and adaptive cells, a lack of one of the cellular components or an incorrect ratio may result in a difference from the natural human body. Furthermore, maintaining the correct number of different cells and their activity on a tiny device is complicated. Traditional cell seeding procedures frequently result in the loss of cells due to adhesion or sedimentation. Thus, it is reasonable to conclude that substantial advancements in microfluidic chips are necessary before they can reproduce the complexity of living species.
V. CONCLUSIONS
Microfluidic devices have tremendous potential to facilitate biologically relevant experiments and the investigation of many hitherto underexplored aspects of cell biology. In cancer immunotherapy, personalized medicine has attracted more attention due to the complexities of the cancer characteristics of different individuals and cancer types. Microfluidic devices are highly suitable for personalized targets, such as creating gene/protein profiles, performing drug screening, assessing drug combinations and drug resistance, analyzing immunotherapeutic effects, and performing cytotoxicity studies. Furthermore, there is an increasing number of device designs, and standardization in the field may be established soon.
ACKNOWLEDGMENTS
This work was funded by the National Science and Technology Council (NSTC), Taiwan, and the Young Scholar Program (MOST 110-2636-B-006-013, MOST 110-2740-B-006-002, MOST 111-2740-B-006-002, and MOST 111-2636-B-006-010 to T.-Y.T.). This work was also supported in part by the Higher Education Sprout Project, Ministry of Education to the Headquarters of University Advancement at the National Cheng Kung University. The authors are grateful for the support provided by the “Bioimaging Core Facility of the National Core Facility for Biopharmaceuticals, NSTC, Taiwan.”
Note: This paper is part of the special issue on Microfluidics and Nanofluidics for Immunotherapy
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
T.K.N.N. took the lead in writing the manuscript. C.-H.K. and T.-Y.T. provided input and critical revision. All authors provided critical feedback and helped shape the manuscript. All authors have read and approved the submitted version of the manuscript.
Thi Kim Ngan Ngo: Writing – original draft (lead). Cheng-Hsiang Kuo: Supervision (equal); Writing – review & editing (supporting). Ting-Yuan Tu: Supervision (equal); Writing – review & editing (lead).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.van Not O. J., de Meza M. M., van den Eertwegh A. J. M., Haanen J. B., Blank C. U., Aarts M. J. B., van den Berkmortel F. W. P. J., van Breeschoten J., de Groot J.-W. B., Hospers G. A. P., Ismail R. K., Kapiteijn E., Piersma D., van Rijn R. S., Stevense-den Boer M. A. M., van der Veldt A. A. M., Vreugdenhil G., Bonenkamp H. J., Boers-Sonderen M. J., Blokx W. A. M., Wouters M. W. J. M., and Suijkerbuijk K. P. M., Eur. J. Cancer 167, 70 (2022). 10.1016/j.ejca.2022.02.026 [DOI] [PubMed] [Google Scholar]
- 2.Deng M., Li S., Wang Q., Zhao R., Zou J., Lin W., Mei J., Wei W., and Guo R., Ann. Med. 54, 803 (2022). 10.1080/07853890.2022.2048416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogiwara Y., Nakagawa M., Nakatani F., Uemura Y., Zhang R., and Kudo-Saito C., Cancer Lett. 537, 215690 (2022). 10.1016/j.canlet.2022.215690 [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Li L., Zhang B., and Xu Z. P., J. Colloid Interface Sci. 617, 315 (2022). 10.1016/j.jcis.2022.02.132 [DOI] [PubMed] [Google Scholar]
- 5.Ogasawara M., Miyashita M., Yamagishi Y., and Ota S., Ther. Apher. Dial. 26, 537–547 (2022). 10.1111/1744-9987.13828 [DOI] [PubMed] [Google Scholar]
- 6.Liang S., Xu K., Niu L., Wang X., Liang Y., Zhang M., Chen J., and Lin M., OncoTargets Ther. 10, 4273 (2017). 10.2147/OTT.S139986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thongchot S., Jirapongwattana N., Luangwattananun P., Chiraphapphaiboon W., Chuangchot N., Sa-nguanraksa D., O-Charoenrat P., Thuwajit P., Yenchitsomanus P., and Thuwajit C., Mol. Cancer Ther. 21, 727–739 (2022). 10.1158/1535-7163.MCT-21-0823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tentler J. J., Tan A. C., Weekes C. D., Jimeno A., Leong S., Pitts T. M., Arcaroli J. J., Messersmith W. A., and Eckhardt S. G., Nat. Rev. Clin. Oncol. 9, 338 (2012). 10.1038/nrclinonc.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elani Y., Biochem. Soc. Trans. 44, 723 (2016). 10.1042/BST20160052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen C., Shay C., and Teng Y., in The New Frontier of Three-Dimensional Culture Models to Scale-Up Cancer Research, edited by Guest P. C. (Springer US, New York, 2022), pp. 3–18. [DOI] [PubMed] [Google Scholar]
- 11.Whitesides G. M., Nature 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 12.Hamdallah S. I., Zoqlam R., Erfle P., Blyth M., Alkilany A. M., Dietzel A., and Qi S., Int. J. Pharm. 584, 119408 (2020). 10.1016/j.ijpharm.2020.119408 [DOI] [PubMed] [Google Scholar]
- 13.Adzamli I. K., Seltzer S. E., Slifkin M., Blau M., and Adams D. F., Invest. Radiol. 25, 1217 (1990). 10.1097/00004424-199011000-00011 [DOI] [PubMed] [Google Scholar]
- 14.Menz W. and Guber A., Minim. Invasive Neurosurg. 37, 21 (1994). 10.1055/s-2008-1053444 [DOI] [PubMed] [Google Scholar]
- 15.Martynova L., Locascio L. E., Gaitan M., Kramer G. W., Christensen R. G., and MacCrehan W. A., Anal. Chem. 69, 4783 (1997). 10.1021/ac970558y [DOI] [PubMed] [Google Scholar]
- 16.Parlato S., De Ninno A., Molfetta R., Toschi E., Salerno D., Mencattini A., Romagnoli G., Fragale A., Roccazzello L., Buoncervello M., Canini I., Bentivegna E., Falchi M., Bertani F. R., Gerardino A., Martinelli E., Natale C., Paolini R., Businaro L., and Gabriele L., Sci. Rep. 7, 1093 (2017). 10.1038/s41598-017-01013-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh D. E. I., Dongeun B. D., Matthews A. M., Montoya-Zavala M., and Hsin H. Y., Science 328, 1662 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballerini M., Jouybar M., Mainardi A., Rasponi M., and Ugolini G. S., in Organ-on-Chips for Studying Tissue Barriers: Standard Techniques and a Novel Method for Including Porous Membranes Within Microfluidic Devices, edited by Rasponi M. (Springer, New York, NY, 2022), pp. 21–38. [DOI] [PubMed] [Google Scholar]
- 19.Ma C. and Fan R., Front. Oncol. 4, 266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer A., Gupta K., Sharma S., Hari K., Lee Y. F., Ramalingam N., Yap Y. S., West J., Bhagat A. A., Subramani B. V., Sabuwala B., Tan T. Z., Thiery J. P., Jolly M. K., Ramalingam N., and Sengupta D., J. Clin. Med. 9, 1206 (2020). 10.3390/jcm9041206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyes M., Vickers D., Billman K., Eisenhaure T., Hoover P., Browne E. P., Rao D. A., Hacohen N., and Blainey P. C., Sci. Adv. 5, eaau9223 (2019). 10.1126/sciadv.aau9223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadl A., Rastgoo A., Haghighipour N., Bolhassani A., Asgari F., and Soleymani S., PLoS One 13, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rayes R. F., Mouhanna J. G., Wang C., Milette S., Wong C., Usatii M., Giannias B., Bourdeau F., Mot R., Chandrasekaran A., Moraes C., Huang S., Quail D., Walsh L., Sangwan V., Bertos N., Fiset P.-O., Cools-Lartigue J., Ferri L. E., and Spicer J. D., Cancer Res. 79, 2799 (2019). [Google Scholar]
- 24.Lim Z., Wu X., Hafez M., Albandar H., Zhu L., Yang H., Mackay S., Chen J., Zhou J., and Ma P., J. Thorac. Oncol. 14, S718 (2019). 10.1016/j.jtho.2019.08.1531 [DOI] [Google Scholar]
- 25.Lyons V. J., Helms A., and Pappas D., Anal. Chim. Acta 1076, 154 (2019). 10.1016/j.aca.2019.05.040 [DOI] [PubMed] [Google Scholar]
- 26.Czaplicka M., Nicinski K., Nowicka A., Szymborski T., Chmielewska I., Trzcinska-Danielewicz J., Girstun A., and Kaminska A., Cancers 12, 3315 (2020). 10.3390/cancers12113315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layer J. P., Kronmuller M. T., Quast T., van den Boorn-Konijnenberg D., Effern M., Hinze D., Althoff K., Schramm A., Westermann F., Peifer M., Hartmann G., Tuting T., Kolanus W., Fischer M., Schulte J., and Holzel M., Oncoimmunology 6, e1320626 (2017). 10.1080/2162402X.2017.1320626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aboulkheyr Es H., Zhand S., Thiery J. P., and Warkiani M. E., Integr. Biol. 12, 188 (2020). 10.1093/intbio/zyaa014 [DOI] [PubMed] [Google Scholar]
- 29.Preece R., Pavesi A., Gkazi S. A., Stegmann K. A., Georgiadis C., Tan Z. M., Aw J. Y. J., Maini M. K., Bertoletti A., and Qasim W., Mol. Ther. Methods Clin. Dev. 19, 149 (2020). 10.1016/j.omtm.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henn D., Zhao D., Bonham C. A., Chen K., Greco A. H., Padmanabhan J., Sivaraj D., Trotsyuk A., Barrera J. A., Januszyk M., Stanley Qi L., and Gurtner G. C., Wound Repair Regen. 29, A31 (2021). 10.1111/wrr.12921 [DOI] [Google Scholar]
- 31.Chang B., Goff A., Sicher I., Loo J., Dunn T., Clary N., Zamarayeva A., and Calero-Garcia M., Cytotherapy 23, S152 (2021). 10.1016/S1465324921005302 [DOI] [Google Scholar]
- 32.Wang X., Teng F., Kong L., and Yu J., OncoTargets Ther. 9, 5023 (2016). 10.2147/OTT.S105862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei S. C., Duffy C. R., and Allison J. P., Cancer Discov. 8, 1069 (2018). 10.1158/2159-8290.CD-18-0367 [DOI] [PubMed] [Google Scholar]
- 34.Ju X., Zhang H., Zhou Z., and Wang Q., Am. J. Cancer Res. 10, 1 (2020). [PMC free article] [PubMed] [Google Scholar]
- 35.Aboulkheyr Es H., Bigdeli B., Zhand S., Aref A. R., Thiery J. P., and Warkiani M. E., J. Cell. Physiol. 236, 3918 (2021). 10.1002/jcp.30135 [DOI] [PubMed] [Google Scholar]
- 36.Sehgal K., Portell A., Ivanova E. V., Lizotte P. H., Mahadevan N. R., Greene J. R., Vajdi A., Gurjao C., Teceno T., Taus L. J., Thai T. C., Kitajima S., Liu D., Tani T., Noureddine M., Lau C. J., Kirschmeier P. T., Liu D., Giannakis M., Jenkins R. W., Gokhale P. C., Goldoni S., Pinzon-Ortiz M., Hastings W. D., Hammerman P. S., Miret J. J., Paweletz C. P., and Barbie D. A., J. Clin. Invest. 131, e135038 (2021). 10.1172/JCI135038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins R. W., Aref A. R., Lizotte P. H., Ivanova E., Stinson S., Zhou C. W., Bowden M., Deng J. H., Liu H. Y., Miao D. N., He M. X., Walker W., Zhang G., Tian T., Cheng C. R., Wei Z., Palakurthi S., Bittinger M., Vitzthum H., Kim J. W., Merlino A., Quinn M., Venkataramani C., Kaplan J. A., Portell A., Gokhale P. C., Phillips B., Smart A., Rotem A., Jones R. E., Keogh L., Anguiano M., Stapleton L., Jia Z. H., Barzily-Rokni M., Canadas I., Thai T. C., Hammond M. R., Vlahos R., Wang E. S., Zhang H., Li S., Hanna G. J., Huang W., Hoang M. P., Piris A., Eliane J. P., Stemmer-Rachamimov A. O., Cameron L., Su M. J., Shah P., Izar B., Thakuria M., LeBoeuf N. R., Rabinowits G., Gunda V., Parangi S., Cleary J. M., Miller B. C., Kitajima S., Thummalapalli R., Miao B. C., Barbie T. U., Sivathanu V., Wong J. H., Richards W. G., Bueno R., Yoon C. H., Miret J., Herlyn M., Garraway L. A., Van Allen E. M., Freeman G. J., Kirschmeier P. T., Lorch J. H., Ott P. A., Hodi F. S., Flaherty K. T., Kamm R. D., Boland G. M., Wong K. K., Dornan D., Paweletz C. P., and Barbie D. A., Cancer Discov. 8, 196 (2018). 10.1158/2159-8290.CD-17-0833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aref A. R., Campisi M., Ivanova E., Portell A., Larios D., Piel B. P., Mathur N., Zhou C. S., Coakley R. V., Bartels A., Bowden M., Herbert Z., Hill S., Gilhooley S., Carter J., Canadas I., Thai T. C., Kitajima S., Chiono V., Paweletz C. P., Barbie D. A., Kamm R. D., and Jenkins R. W., Lab Chip 18, 3129 (2018). 10.1039/C8LC00322J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui X., Ma C., Vasudevaraja V., Serrano J., Tong J., Peng Y., Delorenzo M., Shen G., Frenster J., Morales R.-T. T., Qian W., Tsirigos A., Chi A. S., Jain R., Kurz S. C., Sulman E. P., Placantonakis D. G., Snuderl M., and Chen W., Elife 9, e52253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anastasiadou E., Stroopinsky D., Alimperti S., Jiao A. L., Pyzer A. R., Cippitelli C., Pepe G., Severa M., Rosenblatt J., Etna M. P., Rieger S., Kempkes B., Coccia E. M., Sui S. J. H., Chen C. S., Uccini S., Avigan D., Faggioni A., Trivedi P., and Slack F. J., Leukemia 33, 132 (2019). 10.1038/s41375-018-0178-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alimperti S., Mirabella T., Bajaj V., Polacheck W., Pirone D. M., Duffield J., Eyckmans J., Assoian R. K., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 114, 8758 (2017). 10.1073/pnas.1618333114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen D. H.-T., Stapleton S. C., Yang M. T., Cha S. S., Choi C. K., Galie P. A., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 110, 6712 (2013). 10.1073/pnas.1221526110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J. R., Bechstein D. J. B., Ooi C. C., Patel A., Gaster R. S., Ng E., Gonzalez L. C., and Wang S. X., Nat. Commun. 7, 12220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiao H., Chen X., Wang Q., Zhang J., Huang D., Chen E., Qian H., Zhong Y., Tang Q., and Chen W., Biomater. Sci. 8, 2472 (2020). 10.1039/D0BM00172D [DOI] [PubMed] [Google Scholar]
- 45.Dhar M., Che J., Wong J. M., Pao E., Yu V. S. H., Matsumoto M., Goldman J., Garon E., Sollier E., Kulkarni R., and Di Carlo D., Cancer Res. 75, 1582 (2015). 10.1158/1538-7445.AM2015-1582 [DOI] [Google Scholar]
- 46.Lin S. Y., Chang S.-C., Lam S., Irene Ramos R., Tran K., Ohe S., Salomon M. P., Bhagat A. A. S., Teck Lim C., Fischer T. D., Foshag L. J., Boley C. L., O’Day S. J., and Hoon D. S. B., Clin. Chem. 66, 169 (2020). 10.1373/clinchem.2019.307140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strati A., Zavridou M., Economopoulou P., Gkolfinopoulos S., Psyrri A., and Lianidou E., Clin. Chem. 67, 642 (2021). 10.1093/clinchem/hvaa321 [DOI] [PubMed] [Google Scholar]
- 48.Goldman J. W., Dhar M., Che J., Garon E. B., Hu-Lieskovan S., Matsumoto M., Wolf B. R., Carroll J. M., Crabtree M. J., Tucker D. A., Strunck J., Sollier E., Kulkarni R., and Di Carlo D., Mol. Cancer Ther. 14, B98 (2015). 10.1158/1535-7163.TARG-15-B98 [DOI] [Google Scholar]
- 49.Kulasinghe A., Perry C., Kenny L., Warkiani M. E., Nelson C., and Punyadeera C., BMC Cancer 17, 333 (2017). 10.1186/s12885-017-3316-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Natarajan A., Mol. Imaging Biol. 23, 1753 (2021). 10.1007/s11307-021-01694-x [DOI] [Google Scholar]
- 51.Zavridou M., Strati A., Koutsodontis G., Psyrri A., and Lianidou E., Cancer Res. 77, 1718 (2017). 10.1158/1538-7445.AM2017-1718 [DOI] [Google Scholar]
- 52.Giraldo N. A., Sanchez-Salas R., Peske J. D., Vano Y., Becht E., Petitprez F., Validire P., Ingels A., Cathelineau X., Fridman W. H., and Sautès-Fridman C., Br. J. Cancer 120, 45 (2019). 10.1038/s41416-018-0327-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jammes F., Schmidt J., Coukos G., and Maerkl S. J., ACS Sens. 7, 159 (2022). 10.1021/acssensors.1c01935 [DOI] [PubMed] [Google Scholar]
- 54.Wang S., Liu Y., Li Y., Lv M., Gao K., He Y., Wei W., Zhu Y., Dong X., Xu X., Li Z., Liu L., and Liu Y., Anal. Chem. 94, 918 (2022). 10.1021/acs.analchem.1c03678 [DOI] [PubMed] [Google Scholar]
- 55.Nguyen O. T. P., Misun P. M., Lohasz C., Lee J., Wang W., Schroeder T., and Hierlemann A., Front. Immunol. 12, 781337 (2021). 10.3389/fimmu.2021.781337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pendharkar G., Lu Y.-T., Chang C.-M., Lu M.-P., Lu C.-H., Chen C.-C., and Liu C.-H., Cells 10, 2855 (2021). 10.3390/cells10112855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berger Fridman I., Kostas J., Gregus M., Ray S., Sullivan M. R., Ivanov A. R., Cohen S., and Konry T., Acta Biomater. 132, 473 (2021). 10.1016/j.actbio.2021.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Haan L., Suijker J., van Roey R., Berges N., Petrova E., Queiroz K., Strijker W., Olivier T., Poeschke O., Garg S., and van den Broek L. J., Int. J. Mol. Sci. 22, 8234 (2021). 10.3390/ijms22158234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mollica H., Teo Y. J., Tan A. S. M., Tan D. Z. M., Decuzzi P., Pavesi A., and Adriani G., Biomater. Sci. 9, 7420 (2021). 10.1039/D1BM00210D [DOI] [PubMed] [Google Scholar]
- 60.Tu H., Wu Z., Xia Y., Chen H., Hu H., Ding Z., Zhou F., and Guo S., Analyst 145, 4138 (2020). 10.1039/D0AN00110D [DOI] [PubMed] [Google Scholar]
- 61.Qian Z., Fei J., Zong S., Yang K., Li L., Liu R., Wang Z., and Cui Y., ACS Sens. 5, 208 (2020). 10.1021/acssensors.9b02085 [DOI] [PubMed] [Google Scholar]
- 62.Lee S. W. L., Seager R. J., Litvak F., Spill F., Sieow J. L., Leong P. H., Kumar D., Tan A. S. M., Wong S. C., Adriani G., Zaman M. H., and Kamm A. R. D., Integr. Biol. 12, 90 (2020). 10.1093/intbio/zyaa007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harizaj A., De Smedt S. C., Lentacker I., and Braeckmans K., Expert Opin. Drug Deliv. 18, 229 (2021). 10.1080/17425247.2021.1828340 [DOI] [PubMed] [Google Scholar]
- 64.Pavesi A., Wong S. C., Kamm R., Lee S. W. L., and Adirani G., Cancer Immunol. Res. 7, A049 (2019). 10.1158/2326-6074.CRICIMTEATIAACR18-A049 [DOI] [Google Scholar]
- 65.Boussommier-Calleja A., Atiyas Y., Haase K., Headley M., Lewis C., and Kamm R. D., Biomaterials 198, 180 (2019). 10.1016/j.biomaterials.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campisi M., Sundararaman S. K., Kitajima S., Chiono V., Kamm R. D., and Barbie D. A., Cancer Res. 79, 958 (2019). 10.1158/1538-7445.AM2019-958 [DOI] [Google Scholar]
- 67.Campisi M., Sundararaman S. K., Shelton S. E., Knelson E. H., Mahadevan N. R., Yoshida R., Tani T., Ivanova E., Canadas I., Osaki T., Lee S. W. L., Thai T., Han S., Piel B. P., Gilhooley S., Paweletz C. P., Chiono V., Kamm R. D., Kitajima S., and Barbie D. A., Front. Immunol. 11, 2090 (2020). 10.3389/fimmu.2020.02090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shim S., Belanger M. C., Harris A. R., Munson J. M., and Pompano R. R., Lab Chip 19, 1013 (2019). 10.1039/C8LC00957K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore N., Doty D., Zielstorff M., Kariv I., Moy L. Y., Gimbel A., Chevillet J. R., Lowry N., Santos J., Mott V., Kratchman L., Lau T., Addona G., Chen H., and Borenstein J. T., Lab Chip 18, 1844 (2018). 10.1039/C8LC00256H [DOI] [PubMed] [Google Scholar]
- 70.Adriani G., Pavesi A., and Kamm R. D., Methods Cell Biol. 146, 199 (2018). 10.1016/bs.mcb.2018.05.009 [DOI] [PubMed] [Google Scholar]
- 71.Ringwelski B., Jayasooriya V., and Nawarathna D., J. Phys. D: Appl. Phys. 54, 065402 (2021). 10.1088/1361-6463/abc2f3 [DOI] [Google Scholar]
- 72.Agarwalla P., Ogunnaike E. A., Ahn S., Ligler F. S., Dotti G., and Brudno Y., Adv. Healthc. Mater. 9, 2000275 (2020). 10.1002/adhm.202000275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elsemary M. T., Maritz M. F., Smith L. E., and Thierry B., Cytotherapy 21, e4 (2019). 10.1016/j.jcyt.2019.04.013 [DOI] [PubMed] [Google Scholar]
- 74.Segaliny A. I., Li G., Kong L., Ren C., Chen X., Wang J. K., Baltimore D., Wu G., and Zhao W., Lab Chip 18, 3733 (2018). 10.1039/C8LC00818C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pavesi A., Tan A. T., Chen M. B., Adriani G., Bertoletti A., and Kamm R. D., in 2015 Annual International Conference of the IEEE Engineering in Medical and Biology Society (IEEE, 2015), p. 1853. [DOI] [PubMed] [Google Scholar]
- 76.Moura Rosa P., Gopalakrishnan N., Ibrahim H., Haug M., and Halaas O., Lab Chip 16, 3728 (2016). 10.1039/C6LC00702C [DOI] [PubMed] [Google Scholar]
- 77.Gopal S., Kwon S. J., Ku B., Lee D. W., Kim J., and Dordick J. S., Commun. Biol. 4, 893 (2021). 10.1038/s42003-021-02417-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chava S., Bugide S., Gupta R., and Wajapeyee N., J. Vis. Exp. 156, e60714 (2020). 10.3791/60714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kandarian F., Sunga G. M., Arango-Saenz D., and Rossetti M., J. Vis. Exp. 126, 56191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giannattasio A., Weil S., Kloess S., Ansari N., Stelzer E. H. K., Cerwenka A., Steinle A., Koehl U., and Koch J., BMC Cancer 15, 351 (2015). 10.1186/s12885-015-1321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song J., Choi H., Koh S. K., Park D., Yu J., Kang H., Kim Y., Cho D., and Jeon N. L., Front. Immunol. 12, 733317 (2021). 10.3389/fimmu.2021.733317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hong S., You J. Y., Paek K., Park J., Kang S. J., Han E. H., Choi N., Chung S., Rhee W. J., and Kim J. A., Theranostics 11, 9687 (2021). 10.7150/thno.60851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J. H., Lee S., Kang S. J., Choi Y. W., Choi S. Y., Park J. Y., and Chang I. H., Int. J. Mol. Sci. 22, 8887 (2021). 10.3390/ijms22168887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Antona S., Abele T., Jahnke K., Dreher Y., Gopfrich K., Platzman I., and Spatz J. P., Adv. Funct. Mater. 30, 2003479 (2020). 10.1002/adfm.202003479 [DOI] [Google Scholar]
- 85.Chen S.-C., Wu P.-C., Wang C.-Y., and Kuo P.-L., Sci. Rep. 10, 13662 (2020). 10.1038/s41598-020-70694-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park D., Son K., Hwang Y., Ko J., Lee Y., Doh J., and Jeon N. L., Front. Immunol. 10, 1133 (2019). 10.3389/fimmu.2019.01133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ayuso J. M., Truttschel R., Gong M. M., Humayun M., Virumbrales-Munoz M., Vitek R., Felder M., Gillies S. D., Sondel P., Wisinski K. B., Patankar M., Beebe D. J., and Skala M. C., Oncoimmunology 8, 1553477 (2019). 10.1080/2162402X.2018.1553477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu D., Yu Y., Zhao C., Shou X., Piao Y., Zhao X., Zhao Y., and Wang S., ACS Appl. Mater. Interfaces 11, 33716 (2019). 10.1021/acsami.9b12816 [DOI] [PubMed] [Google Scholar]
- 89.Dashnamoorthy R., Beheshti A., Sarkar S., Sabhachandani P., Passero F. C., Purvey S., Boissel L., Konry T., and Evens A. M., Blood 128, 4174 (2016). 10.1182/blood.V128.22.4174.4174 [DOI] [Google Scholar]
- 90.Sarkar S., Kang W., Jiang S., Li K., Ray S., Luther E., Ivanov A. R., Fu Y., and Konry T., Lab Chip 20, 2317 (2020). 10.1039/D0LC00158A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarkar S., Sabhachandani P., Ravi D., Potdar S., Purvey S., Beheshti A., Evens A. M., and Konry T., Front. Immunol. 8, 1736 (2017). 10.3389/fimmu.2017.01736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sarkar S., McKenney S., Sabhachandani P., Adler J., Hu X., Stroopinksy D., Rosenblatt J., Avigan D., and Konry T., Sens. Actuators B 282, 580 (2019). 10.1016/j.snb.2018.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Principe N., Kidman J., Goh S., Tilsed C. M., Fisher S. A., Fear V. S., Forbes C. A., Zemek R. M., Chopra A., Watson M., Dick I. M., Boon L., Holt R. A., Lake R. A., Nowak A. K., Lesterhuis W. J., McDonnell A. M., and Chee J., Front. Immunol. 11, 584423 (2020). 10.3389/fimmu.2020.584423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian Y., Xu C., Feng J., Huangfu Y., Wang K., and Zhang Z.-L., Lab Chip 21, 4414 (2021). 10.1039/D1LC00646K [DOI] [PubMed] [Google Scholar]
- 95.Fontana F., Fusciello M., Groeneveldt C., Capasso C., Chiaro J., Feola S., Liu Z., Makila E. M., Salonen J. J., Hirvonen J. T., Cerullo V., and Santos H. A., ACS Nano 13, 6477 (2019). 10.1021/acsnano.8b09613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mi Y., Hagan C. T., Vincent B. G., and Wang A. Z., Adv. Sci. 6, 1801847 (2019). 10.1002/advs.201801847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Scheetz L., Park K. S., Li Q., Lowenstein P. R., Castro M. G., Schwendeman A., and Moon J. J., Nat. Biomed. Eng. 3, 768 (2019). 10.1038/s41551-019-0436-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Du Y., Liu Y., Wang D., Bai H., Wang Z. J., He X. R., Zhang P., Tian J., and Wang J., J. Immunother. Cancer 10, e003564 (2022). 10.1136/jitc-2021-003564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma C., Peng Y., Li H., and Chen W., Trends Pharmacol. Sci. 42, 119 (2021). 10.1016/j.tips.2020.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ivanova E., Aref A., Jenkins R., Lizotte P., Huang W., Palakurthi S., Kirschmeier P., Barbie D., Paweletz C., Janne P., and Wong K.-K., Cancer Res. 77, LB-218 (2017). 10.1158/1538-7445.AM2017-LB-218 [DOI] [Google Scholar]
- 101.Larios D., Ivanova E., Aref A., Portell A., De Rienzo A., Barbie D., Paweletz C., and Bueno R., J. Thorac. Oncol. 14, S297 (2019). 10.1016/j.jtho.2019.08.596 [DOI] [Google Scholar]
- 102.Ding S., Wang Z., Hsu C., Hsu D., and Shen X., Ann. Oncol. 32, S361 (2021). 10.1016/j.annonc.2021.08.281 [DOI] [Google Scholar]
- 103.Zilberberg J., Zhang W., Siegel D. S., and Lee W. Y., Cancer Res. 77, B08 (2017). 10.1158/1538-7445.EPSO16-B08 [DOI] [Google Scholar]
- 104.Geyer M. and Queiroz K., Front. Cell Dev. Biol. 9, 761807 (2021). 10.3389/fcell.2021.761807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kerns S. J., Belgur C., Petropolis D., Kanellias M., Barrile R., Sam J., Weinzierl T., Fauti T., Freimoser-Grundschober A., Eckmann J., Hage C., Geiger M., Ng P. R., Tien-Street W., Manatakis D. V., Micallef V., Gerard R., Bscheider M., Breous-Nystrom E., Schneider A., Giusti A. M., Bertinetti-Lapatki C., Grant H. S., Roth A. B., Hamilton G. A., Singer T., Karalis K., Moisan A., Bruenker P., Klein C., Bacac M., Gjorevski N., and Cabon L., Elife 10, e67106 (2021). 10.7554/eLife.67106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu G. H., Wu J. G., Li Z. H., Shi S. Y., Wu D., Wang X. B., Xu H., Liu H., Huang Y. X., Wang R. D., Shen J., Dong Z. H., and Wang S. Q., Bio-Des. Manuf. 5, 437–450 (n.d.). 10.1007/s42242-022-00188-1 [DOI] [Google Scholar]
- 107.Dashnamoorthy R., Sarkar S., Sabachandani P., Purvey S., Beheshti A., Konry T., and Evens A. M., Blood 130, 1473 (2017). [Google Scholar]
- 108.Kerr S. C., Morgan M. M., Gillette A. A., Livingston M. K., Lugo-Cintron K. M., Favreau P. F., Florek L., Johnson B. P., Lang J. M., Skala M. C., and Beebe D. J., Integr. Biol. 12, 250 (2020). 10.1093/intbio/zyaa020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Virumbrales-Munoz M., Chen J., Ayuso J., Lee M., Abel E. J., and Beebe D. J., Lab Chip 20, 4420 (2020). 10.1039/D0LC00252F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hafezi M., Lin M., Chia A., Chua A., Ho Z. Z., Fam R., Tan D., Aw J., Pavesi A., Krishnamoorthy T. L., Chow W. C., Chen W., Zhang Q., Wai L.-E., Koh S., Tan A. T., and Bertoletti A., Hepatology 74, 200 (2021). 10.1002/hep.31662 [DOI] [PubMed] [Google Scholar]
- 111.Pavesi A., Tan A. T., Koh S., Chia A., Colombo M., Antonecchia E., Miccolis C., Ceccarello E., Adriani G., Raimondi M. T., Kamm R. D., and Bertoletti A., JCI Insight 2, e89762 (2017). 10.1172/jci.insight.89762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou J., Tu C., Liang Y., Huang B., Fang Y., Liang X., and Ye X., Analyst 145, 1706 (2020). 10.1039/C9AN02092F [DOI] [PubMed] [Google Scholar]
- 113.Al-Samadi A., Poor B., Tuomainen K., Liu V., Hyytiainen A., Suleymanova I., Mesimaki K., Wilkman T., Makitie A., Saavalainen P., and Salo T., Exp. Cell Res. 383, 111508 (2019). 10.1016/j.yexcr.2019.111508 [DOI] [PubMed] [Google Scholar]
- 114.Kehren A., Procopio M. G., Pelz B., Siddiqui Z., Roman K., Adnane S., Brajkovic S., Hoyt C., Dupouy D. G., and Soltermann A., J. Immunother. Cancer 8, A25 (2020). 10.1136/jitc-2020-ITOC7.47 [DOI] [Google Scholar]
- 115.Fortier A.-M., Bedford E., Torres S. F., Getsios S., Kinsella J. M., Wadsworth S., and Park M., Cancer Res. 81, PO006 (2021). [Google Scholar]
- 116.Stadler G. and Lovgren J., Cytotherapy 23, S95 (2021). 10.1016/S1465324921004199 [DOI] [Google Scholar]
- 117.Majumder P., Majumder B., Singh R., Sane A., Manglik R., Keshari R., Mohanasundaram M., and Lal A., J. Immunother. Cancer 9, A13 (2021). 10.1136/jitc-2021-SITC2021.013 [DOI] [Google Scholar]
- 118.Choudhury D., Anand S., and Naing M. W., Int. J. Bioprinting 4, 139 (2018). 10.18063/ijb.v4i2.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee C., Abelseth E., de la Vega L., and Willerth S. M., Mater. Today Chem. 12, 78 (2019). 10.1016/j.mtchem.2018.12.005 [DOI] [Google Scholar]
- 120.Talarico L. A., Vicente-Suarez I., Blagovic K., Chong-Ng E., Jones L., Pomerance L., and Bernstein H., Cancer Immunol. Res. 6, A61 (2018). 10.1158/2326-6074.TUMIMM17-A61 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.