Abstract
Biological time keeping, or the duration and tempo at which biological processes occur, is a phenomenon that drives dynamic molecular and morphological changes that manifest throughout many facets of life. In some cases, the molecular mechanisms regulating the timing of biological transitions are driven by genetic oscillations, or periodic increases and decreases in expression of genes described collectively as a “molecular clock.” In vertebrate animals, molecular clocks play a crucial role in fundamental patterning and cell differentiation processes throughout development. For example, during early vertebrate embryogenesis, the segmentation clock regulates the patterning of the embryonic mesoderm into segmented blocks of tissue called somites, which later give rise to axial skeletal muscle and vertebrae. Segmentation clock oscillations are characterized by rapid cycles of mRNA and protein expression. For segmentation clock oscillations to persist, the transcript and protein molecules of clock genes must be short‐lived. Faithful, rhythmic, genetic oscillations are sustained by precise regulation at many levels, including post‐transcriptional regulation, and such mechanisms are essential for proper vertebrate development.
This article is categorized under:
RNA Export and Localization > RNA Localization
RNA Turnover and Surveillance > Regulation of RNA Stability
Translation > Regulation
Keywords: decay, oscillation, segmentation clock, somitogenesis, translation
Genetic oscillators play crucial roles in development. The hairy/Enhancer of split gene family (Hes/her) undergo cell‐autonomous genetic oscillations to regulate the timing of vertebrate segmentation and neural cell proliferation and differentiation. mRNA stability and translation are tightly regulated to maintain oscillatory expression.
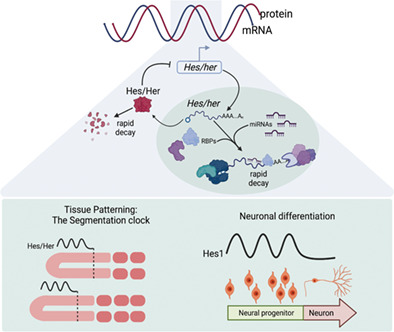
1. INTRODUCTION
Many rhythmic biological processes are controlled by a molecular “clock,” in which one or more genes are expressed in an oscillatory manner with a predictable period (Oates et al., 2012; Zhang et al., 2014). Such biological clocks have been implicated in periodic root branching in Arabidopsis (Moreno‐Risueno et al., 2010), the molting cycle in Caenorhabditis elegans (Kim et al., 2013), circadian rhythms in all diurnal and nocturnal species (Patke et al., 2020), the timing of mitosis to tightly regulate cell proliferation and tissue morphogenesis (Evans et al., 1983; Murray, 2004), and tissue patterning during somitogenesis, a fundamental vertebrate developmental process (Kageyama et al., 2012). While some oscillatory networks, like the Kai cyanobacterial circadian clock and circadian redox clock, are driven solely by dynamic post‐translational modifications (Milev et al., 2018; Snijder & Axmann, 2019), other genetic oscillators are often regulated by a negative feedback loop, in which a core oscillator gene encodes a transcriptional repressor that inhibits expression of downstream oscillators, including its own gene; this ultimately confers cell‐autonomous control of genetic oscillations (Bessho, Miyoshi, et al., 2001; Brend & Holley, 2009; Patke et al., 2020; Shimojo et al., 2008; Webb et al., 2016). At the beginning of each gene expression cycle, the core oscillating gene promoter is activated and produces oscillatory gene mRNA and protein. As oscillatory protein levels rise and reach a critical threshold, oscillatory gene transcription is inhibited, preventing further production of mRNA and protein. As oscillatory mRNA and protein are degraded over time, transcriptional repression is released, allowing for another cycle of expression to begin. Collectively, this form of autoregulation can generate a self‐sustained negative feedback loop (Bessho, Miyoshi, et al., 2001; Brend & Holley, 2009; Patke et al., 2020; Shimojo et al., 2008; Figure 1a). Whereas circadian clocks oscillate for a period of 24 h to regulate the daily rhythms of many organisms and their organ systems (Patke et al., 2020; Reinke & Asher, 2019), ultradian clocks with periods of less than 24 h, like the vertebrate segmentation clock, can cycle on the order of minutes and require rapid and robust gene regulation.
FIGURE 1.
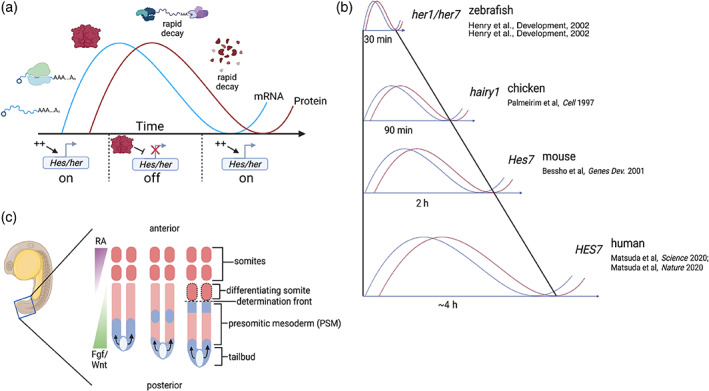
Negative feedback loops and somitogenesis. (a) Negative feedback loop autoregulation. Core oscillators, often encoding transcriptional repressors, participate in negative feedback loops to sustain autoregulatory genetic oscillations. Within each gene expression cycle, segmentation clock mRNA and protein are produced in increasing amounts, and increased protein levels correspond to decreased transcriptional activation as segmentation clock protein inhibits its own expression. As segmentation clock mRNA and protein are both degraded, repression of segmentation clock gene expression is released, allowing for another cycle of expression to begin. Collectively, this forms a self‐sustained negative feedback loop. (b) Species‐specific segmentation clock periods. Clock periodicity varies widely across vertebrates. However, the tempo of oscillations corresponds to the timing of somite formation in all species. The examples shown are classic hairy enhancer of split orthologs, but other genes also oscillate. The tempo of oscillations is determined by species‐specific biochemical rates of gene expression. (c) Oscillating PSM cells receive positional information from anterior–posterior gradients. Core segmentation clock oscillators, like zebrafish her1, are expressed in the PSM and tailbud. As cells in the tailbud proliferate, the tailbud extends and cells become displaced into the posterior PSM. Once in the posterior PSM, cells initiate robust oscillatory gene expression. These cell‐autonomous oscillations appear as traveling waves across the PSM from posterior to anterior (shown in blue), and are coordinated by Notch‐mediated cell–cell communication. At the determination front, which is established by opposing Fgf/Wnt and retinoic acid signaling gradients, cells transition from a presomitic to a somitic cell state, and a new somite boundary is formed. Although negligible over just one oscillation, PSM size changes over developmental time, gradually shrinking as the tailbud ceases to proliferate and somite formation continues. Image created using Biorender.com
Vertebrate segmentation, or somitogenesis, is a critical developmental process in which the embryonic mesoderm is sequentially divided into segments, or somites, along with the head‐to‐tail axis of all developing vertebrate embryos. Somitic cells organize to form the dermomyotome and sclerotome, which collectively form the mature trunk dermis, axial skeletal muscle, and vertebral column (Christ et al., 1978, 1986; Keynes & Stern, 1988). The sequential formation of somites is governed by a genetic oscillator called the segmentation clock, which is a network of genes that iteratively undergo waves of expression in the presomitic mesoderm (PSM) to establish boundaries between adjacent somites. Segmentation clock period is species‐specific and varies across vertebrates (zebrafish, ~30 min; chick, ~90 min; mice, ~2 h; human, ~4–5 h; Matsuda, Hayashi, et al., 2020; Matsuda, Yamanaka, et al., 2020; Palmeirim et al., 1997), and at the core of the segmentation, clock are transcriptional repressors encoded by the Hes/her gene family whose expression is oscillatory among all vertebrates examined to date (Bessho, Sakata, et al., 2001; Oates & Ho, 2002; Palmeirim et al., 1997) (Figure 1b). Hes/her genes sustain cell‐autonomous clock oscillations through an autoregulatory negative feedback loop, which is widely regarded as the evolutionarily conserved pace‐making unit of the segmentation clock (Bessho, Sakata, et al., 2001; Oates & Ho, 2002; Palmeirim et al., 1997). Additionally, other canonical Notch, Wnt, and Fgf pathway genes also oscillate in the PSM and have been implicated in initiating and synchronizing segmentation clock oscillations to propagate waves of clock gene expression across the PSM (Ay et al., 2014; Krol et al., 2011; Webb et al., 2016). An analogy to waves of clock gene expression is the “stadium wave”, where spectators, representing individual cells, generate coordinated rhythmic waves by briefly standing up (transcriptional activation), raising their arms (transcript/protein production), sitting down (transcriptional repression), and lowering their arms (transcript/protein decay) with a defined periodicity. In zebrafish embryos, Notch signaling and oscillatory Notch ligand‐encoding genes like deltaC, help coordinate the oscillation phase between neighboring cells (Delaune et al., 2012; Mara et al., 2007; Soza‐Ried et al., 2014). Similarly, the chick and mouse Notch pathway gene Lunatic Fringe (LFNG/Lfng) oscillates in the PSM and is important for synchronizing segmentation clock dynamics and patterning the anterior skeleton (Okubo et al., 2012; Shifley et al., 2008). Like Notch pathway genes, some Fgf and Wnt pathway genes are periodically expressed, though the specific genes that oscillate in each pathway vary between species (Hubaud & Pourquie, 2014; Krol et al., 2011; Mara & Holley, 2007). In a segmenting embryo, cells proliferate in the tailbud and eventually become anteriorly displaced toward the posterior PSM (Kanki & Ho, 1997; Mara et al., 2007; Mara & Holley, 2007). Once cells are in the posterior PSM, robust, cell‐autonomous oscillatory expression of segmentation clock genes initiates. As the tailbud extends and somites form, PSM cells interpret their shifting axial position relative to these anterior and posterior landmarks using opposing signaling gradients or “wavefronts”: Fgf/Wnt signaling, which originates posteriorly from the tailbud, and retinoic acid signaling, which originates anteriorly from formed somites. Together, these gradients provide dynamic positional information to PSM cells (Aulehla & Pourquie, 2010; Hubaud & Pourquie, 2014). Although not well understood, the period of molecular oscillation lengthens in the anterior PSM (Shih et al., 2015; Soroldoni et al., 2014). In a region of the anterior PSM termed the “determination front”, where levels of both Fgf/Wnt and retinoic acid signaling are low, PSM cells transition from a presomitic to somitic state, molecular oscillations cease, and a mature somite is segmented from the PSM by the formation of a somite boundary (Aulehla & Pourquie, 2010; Hubaud & Pourquie, 2014; Figure 1c).
In order for molecular oscillations to regulate the timing of somite formation, rates of each step in the oscillation, from initial transcription and translation to decay, must be precisely regulated to ensure the correct size and number of somites are produced for the respective organism (Gomez et al., 2008; Holley et al., 2000; Keynes & Stern, 1988; Lewis, 2003; Matsuda, Hayashi, et al., 2020; Palmeirim et al., 1997; Schroter & Oates, 2010). The pace of genetic oscillations required to sustain clock periodicity has been investigated computationally and experimentally, whereby the rates of different steps in the gene expression pathway, from transcription, to splicing, to translation and decay, have been mathematically modeled and/or experimentally perturbed to determine the relative contribution of each step on clock periodicity (Ay et al., 2014; Giudicelli et al., 2007; Lewis, 2003). Computational and modeling studies predict that transcriptional and translational time delays (the amount of time from transcription or translation initiation to the emergence of a mature mRNA or protein, respectively) and degradation rates of both transcript and protein are the parameters that have the largest influence on clock period (Ay et al., 2014; Giudicelli et al., 2007). Experimental evidence from in vivo studies assessing the impact of timing of mRNA production, splicing, protein synthesis, and protein degradation closely mirrors in silico predictions (Hirata et al., 2004; Hoyle & Ish‐Horowicz, 2013; Palmeirim et al., 1997; Takashima et al., 2011). Importantly, it is widely appreciated that transcriptional regulation alone is not sufficient to produce genetic oscillations. Real‐time in vivo segmentation clock reporters designed to recapitulate clock dynamics must not only contain critical transcriptional regulatory regions that drive oscillatory expression but must also contain features, typically 3′UTR sequences and protein motifs, that destabilize reporter mRNAs and proteins, respectively (Aulehla et al., 2008; Delaune et al., 2012; Masamizu et al., 2006; Yoshioka‐Kobayashi et al., 2020). These observations underscore that multiple post‐transcriptional regulatory mechanisms promote proper oscillatory expression.
Defects in segmentation clock gene expression that are sufficient to disrupt tissue‐level clock periodicity can result in defects in somite patterning, which is a phenotypic readout commonly employed in genetic and reporter‐based studies that have characterized the molecular mechanisms regulating clock period. Loss of segmentation clock gene function across multiple vertebrate species results in severe segmentation defects, characterized by irregular somite boundaries and fused vertebrae and ribs in juvenile and adult animals (Bessho, Sakata, et al., 2001; Choorapoikayil et al., 2012; Henry et al., 2002). Human congenital skeletal defects, like spondylocostal dysostosis, have been attributed to mutations in the human segmentation clock genes HES7, DLL3, and LFNG; therefore, segmentation clock oscillations are critical for muscle and axial skeletal organization (Bulman et al., 2000; Sparrow et al., 2006, 2008). Previously published reviews have provided comprehensive examinations of cellular signaling pathways and transcriptional regulation involved in coordinating segmentation clock oscillations (Hubaud & Pourquie, 2014; Kageyama et al., 2012; Oates, 2020; Oates et al., 2012). For this review, we provide perspective on the current understanding of post‐transcriptional mechanisms governing the segmentation clock and highlight those that may be shared with other developmental timing and oscillatory gene expression processes.
2. 3′UTR‐MEDIATED REGULATION OF SEGMENTATION CLOCK GENE TRANSCRIPTS
mRNA stability, localization, and translation are commonly modulated by cis‐regulatory elements or structural motifs present within the untranslated regions (UTRs) of mRNAs (Jambor et al., 2015; Lau et al., 2010; Lecuyer et al., 2007; Sandberg et al., 2008; Vejnar et al., 2019; Wu & Bartel, 2017). Importantly, trans‐acting factors that bind 3′UTR elements can influence mRNA fate depending on the affinity of the trans factor to the mRNA and the availability of other interacting factors belonging to large, multivalent complexes that work in concert to degrade, stabilize, translocate, and/or regulate translation of an mRNA (Arvola et al., 2020; Atasoy et al., 1998; Azuma‐Mukai et al., 2008; Bulbrook et al., 2018; Chou et al., 2006; Enwerem et al., 2021; Landthaler et al., 2008; Moraes et al., 2006; Park‐Lee et al., 2003; Pullmann et al., 2007). Multiple studies investigating post‐transcriptional control of oscillating genes have demonstrated the importance of 3′UTR‐mediated regulation, uncovering specific cis‐elements and potential trans factors that modulate oscillatory gene transcript stability. Rapid mRNA clearance is crucial for the persistence of genetic oscillations, and studying the role of specific regulatory motifs within oscillatory gene mRNA 3′UTRs helps define post‐transcriptional mechanisms that promote molecular oscillations.
Reporter‐based studies of segmentation clock transcript 3′UTRs have been conducted to analyze the expression dynamics of segmentation clock mRNAs in several vertebrate genetic model systems (Davis et al., 2001; Gallagher et al., 2017; Hilgers et al., 2005; Nitanda et al., 2013; Riley et al., 2013; Tietz et al., 2020; Wahi et al., 2017). This was first demonstrated in Xenopus laevis embryos for the segmentation clock gene hairy2a, whereby the expression patterns produced from reporter constructs containing different regions of the hairy2a gene were analyzed to identify the minimal regions required to recapitulate the endogenous striped hairy2 expression pattern (Davis et al., 2001). Results from these experiments revealed that the hairy2a 3′UTR was necessary to reconstitute the endogenous expression pattern and drive the rapid decay of reporter transcripts. Similar findings have been reported in zebrafish, mouse, and chick embryos (Kawamura et al., 2016; Riley et al., 2013; Tietz et al., 2020; Wahi et al., 2017), and more recent studies have identified specific cis‐regulatory elements within segmentation clock gene mRNA 3′UTRs that influence transcript stability (Riley et al., 2013; Tietz et al., 2020; Wahi et al., 2017). These studies have motivated future experiments aimed at assessing the role of specific mRNA regulatory factors on segmentation clock mRNA decay and translation, and thus, the tempo of genetic oscillations.
2.1. miRNA‐mediated regulation of oscillatory expression
miRNAs are well‐described small noncoding RNA molecules that negatively regulate gene expression by promoting mRNA decay and/or repressing translation of their target transcript (Behm‐Ansmant et al., 2006; Naeli et al., 2022; Pillai et al., 2004, 2005). miRNA‐mediated regulation has been implicated in a broad range of developmental processes, including the maternal to zygotic transition in zebrafish and Xenopus embryos (Giraldez et al., 2006; Lund et al., 2009), muscle differentiation (Goljanek‐Whysall et al., 2011), and development of multiple organ systems (Ason et al., 2006; Bhaskaran et al., 2009; Zhao et al., 2007). Previous studies have investigated the role of miRNA‐mediated segmentation clock regulation by identifying miRNAs expressed in the PSM of developing embryos and analyzing expression of reporter genes containing segmentation clock gene 3′UTRs in the presence and absence of specific miRNAs (Riley et al., 2013; Wahi et al., 2017). miR‐125a‐5p is expressed in the PSM of chick and mouse embryos, and binding sites for the miR‐125a‐5p seed sequence are present in the 3′UTR of the oscillating gene Lunatic fringe (Lfng; Riley et al., 2013; Wahi et al., 2017). Expression of a reporter gene containing either the chick or mouse Lfng 3′UTR was strongly downregulated following exogenous overexpression of miR‐125a‐5p, and this effect was abrogated upon mutation of the putative miR‐125 binding sites in both 3′UTRs (Riley et al., 2013). In chick embryos, morpholino‐mediated interference of the miR‐125a‐5p:LFNG 3′UTR interaction (Choi et al., 2007) resulted in defects in somite patterning and oscillatory expression of endogenous segmentation clock genes (Riley et al., 2013). Surprisingly, CRISPR/Cas9 mutagenesis of miR‐125a‐5p in mouse embryos had no observed effect on endogenous segmentation clock gene oscillatory expression, and homozygous mutant embryos were morphologically wild type (Wahi et al., 2017). Collectively, these data suggest that, while miR‐125a‐5p is able to regulate Lfng mRNA stability, closely related miRNAs or other trans‐acting factors may compensate for loss of function of miR‐125a‐5p and bind the Lfng 3′UTR to regulate transcript decay. While the exact role of miR‐mediated regulation is not fully understood, computational models suggest that miR‐125a‐5p‐dependent decay is important for minimizing fluctuations and fine‐tuning Lfng oscillatory expression (Jing et al., 2015). Future experimental work assessing the impact of mutating the miRNA seed sequences in the endogenous Lfng 3′UTR will address the impact of miRNA‐mediated regulation on Lfng expression and vertebrate segmentation.
Hes/her oscillatory expression is also involved in regulating neurogenesis, whereby oscillatory expression of the mammalian Hes1 gene maintains a multipotent neural progenitor fate and facilitates the proliferation of neural stem cells (Shimojo et al., 2008). During neuronal differentiation, Hes1 oscillations are terminated through downregulation of Hes1 expression, promoting the expression of proneural genes (Hatakeyama et al., 2004; Ohtsuka et al., 1999), and 3′UTR analyses have revealed that miRNA‐mediated mRNA decay is important for regulating Hes1 expression in multiple animal models. Specifically, deletion of the seed‐complementary sequence for miR‐9 in a mouse Hes1 3′UTR‐containing luciferase reporter gene, which matches endogenous Hes1 oscillatory expression, significantly increased Luciferase protein expression and reduced the number of reporter gene oscillations, compared to reporters carrying the wild‐type Hes1 3′UTR sequence (Bonev et al., 2012). In addition, direct interference between miR‐9 and the endogenous Hes1 3′UTR miR‐9 seed sequence using oligonucleotides that bind the 3′UTR increased endogenous Hes1 mRNA levels, while miR‐9 expression was unaffected (Bonev et al., 2012). Furthermore, overexpression of miR‐9 showed a dampening effect on endogenous Hes1 oscillatory expression in murine neural progenitor cells. Damped Hes1 oscillatory expression ultimately leads to increased expression of proneural genes, causing premature neuronal differentiation (Ishibashi et al., 1995; Tan et al., 2012). miR‐9 overexpression and downregulation has been shown to disrupt oscillatory expression of zebrafish her6 and Xenopus hairy1 (hairy‐related genes) in the neural progenitor cells of each species (Bonev et al., 2011; Soto et al., 2020), indicating that miR‐9‐mediated regulation is highly conserved and important for sustained oscillatory expression of neural stem cell maintenance genes.
To ensure cell‐autonomous oscillatory expression, HES1 protein, in addition to repressing its own expression, also represses transcription of pri‐miR‐9, forming a double‐negative feedback loop (Bonev et al., 2012). As a result, peak levels of HES1 protein correspond to low transcription of pri‐miR‐9, and conversely, pri‐miR‐9 expression is high when HES1 protein levels are lowest, resulting in anti‐phase oscillatory expression between HES1 protein and pri‐miR‐9 transcripts. Thus, Hes1 and pri‐miR‐9 are observed to oscillate out of phase, which in turn sustains neural progenitor fate. In mouse and chick PSMs, mature miR‐125a‐5p is expressed uniformly throughout the PSM, suggesting this miR is not expressed in an oscillatory manner (Riley et al., 2013). However, many mature miRNAs are known to have relatively high stability, whereas primary miRNAs are considered short‐lived intermediates (Gantier et al., 2011). This is observed for mature miR‐9, which progressively accumulates over multiple cycles of pri‐mir‐9 transcriptional activation and is speculated to reach a critical threshold to help ensure precise inhibition of Hes1 expression during neuronal differentiation (Bonev et al., 2012). Thus, it would be interesting to determine whether pri‐miR‐125a is in fact transcribed in an oscillatory manner, further elucidating the mechanism of miR‐125a‐5p‐mediated oscillatory Lfng expression.
Despite the impact of miRNA overexpression and depletion on Lfng and other oscillating gene transcripts, miRNA‐mediated regulation is not universal for all segmentation clock mRNAs nor all vertebrate species. In cells expressing a luciferase reporter gene containing the mouse Hes7 3′UTR, overexpression of mir‐125a‐5p does not affect Luciferase mRNA or protein levels, compared to expression changes observed using the Lfng 3′UTR (Riley et al., 2013). This indicates that different segmentation clock gene mRNAs are subject to distinct mechanisms of post‐transcriptional regulation. Furthermore, despite the presence of predicted miRNA target sites in the zebrafish her1 3′UTR, embryos that are deficient in Dicer‐dependent miRNA processing have a normal her1 expression pattern (Gallagher et al., 2017). A broader investigation into additional post‐transcriptional regulatory factors, such as the RNA binding proteins discussed below, will further elucidate the molecular mechanisms involved in maintaining genetic oscillations during development.
2.2. Analyzing 3′UTR‐dependent segmentation clock gene mRNA decay dynamics using inducible reporter assays
Oscillating gene transcript expression is dynamic. Because transcripts are repeatedly and rapidly transcribed and degraded, discerning newly transcribed mRNAs from mRNAs that are actively being translated or have been marked for decay becomes a major challenge, particularly in whole embryo lysates. Thus, analysis of steady‐state segmentation clock mRNA levels can confound interpretations of oscillating gene transcript dynamics. The advent of vertebrate PSM cell culturing methods, in which stem cells are differentiated into PSM cells or PSM explants are cultured in vitro, can allow techniques such as nuclear or transcriptional run‐on assays in combination with transcriptional inhibitors to be used in the future to measure segmentation clock mRNA decay rates (Diaz‐Cuadros et al., 2020; Hubaud et al., 2017; Matsuda, Yamanaka, et al., 2020; Webb et al., 2014). However, analysis of segmentation clock gene transcript dynamics in whole embryos using chemical inhibitors can be complicated by (1) poor penetration or difficult delivery of transcriptional inhibitors to segmenting embryos, and (2) the rapid nature of oscillatory expression, particularly in zebrafish, that is on a time scale that is not amenable to inhibitor treatments, which can require 24 h of treatments for effective transcriptional inhibition. Using inducible reporter assays has thus been instrumental in investigating mechanisms regulating segmentation clock transcript decay, as rapid and specific induction and inhibition of reporter mRNA in whole embryos can overcome potential secondary effects introduced by global transcription inhibition (Figure 2a).
FIGURE 2.
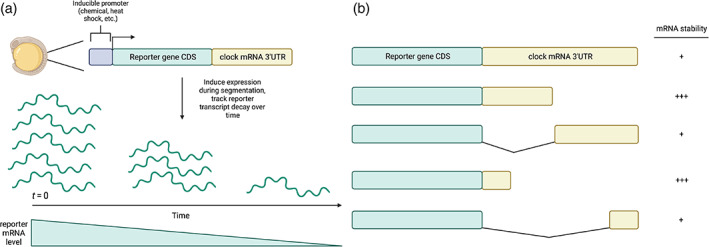
In vivo reporter assays and 3′UTR deletion analysis. (a) Inducible reporter systems and measuring mRNA decay rates. Inducible reporter assays are utilized to measure and compare reporter transcript stability in the context of varied 3′UTR sequences. Depending on the promoter sequence used, these constructs can be chemically or heat shock‐induced in segmenting embryos, when the appropriate segmentation clock transcript regulatory factors are expressed. To calculate reporter transcript decay rates, RNA is extracted from embryos collected at regular intervals post‐induction and reporter mRNA is subsequently quantified across time points using real‐time PCR. (b) 3′UTR fragmentation and reporter transcript decay analysis. Because 3′UTR sequences are rich in motifs that may or may not influence stability, the generation of a set of reporters containing varying portions of a 3′UTR can help to identify smaller regions that influence reporter stability. Upon identification of minimal regions that influence reporter stability, motif analysis followed by mutagenesis of potential regulatory elements can uncover miRNA and/or RBP binding sites that are the primary regulators of mRNA stability. Image created using Biorender.com
Inducible segmentation clock reporters were first introduced into chick embryos using the Tet‐Off system (Gossen et al., 1995; Hilgers et al., 2005). Using this technique, it was observed that the chick LFNG 3′UTR confers rapid transcript degradation, a feature observed for mouse Lfng as well (Nitanda et al., 2013). Early in vivo studies of segmentation clock reporter dynamics relied on electroporation of Tet‐Off inducible reporter constructs into segmenting embryos, followed by reporter quantification post‐induction. Transient introduction of reporter constructs is subject to variability due to mosaicism; therefore, transgenic lines carrying stably‐integrated reporter constructs have recently been developed in zebrafish to better quantify mRNA half‐lives conferred by the zebrafish her1 and dlc 3′UTRs in segmenting embryos (Tietz et al., 2020). These experiments revealed that both transcript 3′UTRs impose rapid degradation of reporter transcripts, consistent with 3′UTR analyses conducted in other vertebrates (Davis et al., 2001; Nitanda et al., 2013; Riley et al., 2013), and suggest that strong destabilizing cis‐regulatory elements reside in segmentation clock transcript 3′UTRs.
2.3. RNA binding protein motifs and segmentation clock mRNA stability
RNA binding proteins (RBPs) that bind to their cognate binding elements in 3′UTRs can recruit protein complexes that modulate mRNA stability and translation (Casolaro et al., 2008; Chou et al., 2006; Pullmann et al., 2007; Shyu et al., 1991; Vasudevan & Steitz, 2007). RNA‐binding protein motifs are prevalent in 3′UTR sequences; therefore, partitioning a full‐length 3′UTR into smaller regions for functional reporter analyses identifies critical elements that are necessary and sufficient to promote mRNA decay (Figure 2b).
Using this deletion strategy for the her1 3′UTR to generate lines carrying stably integrated inducible reporter constructs, it was discovered that the last 179 nts of the full‐length 725 nt her1 3′UTR is both necessary and sufficient to rapidly destabilize reporter transcripts (Tietz et al., 2020). Comparison of the last 179 nts of the her1 3′UTR and the full‐length dlc and her7 3′UTRs for candidate RNA‐binding protein motifs, revealed that AU‐rich elements (AREs) and Pumilio Response Elements (PREs) were shared among all three transcript 3′UTRs. Importantly, these elements are absent in regions of the her1 3′UTR that lack decay‐promoting activity in reporter assays. Both motifs are associated with well‐described negative regulators of mRNA expression, often promoting decay and repressing translation of their target transcripts (Arvola et al., 2020; Bulbrook et al., 2018; Enwerem et al., 2021). AU‐rich binding proteins (ARE‐BPs) are a large family of RBPs that can both stabilize and promote decay of mRNA targets (Chou et al., 2006; Vasudevan & Steitz, 2007), and in the case of decay, ARE‐BPs have been shown to recruit specific members of the CCR4‐Not (CNOT) complex to initiate deadenylation of their target transcripts (Lai et al., 2003). Well‐characterized destabilizing ARE‐BPs include the ARE/poly (U)‐binding/degradation factors 1 (AUF1), tristetraprolin (TTP), and KH‐type splicing regulatory protein (KSRP; Briata et al., 2005; Gratacos & Brewer, 2010; Lykke‐Andersen & Wagner, 2005; Sanduja et al., 2011). Similar to ARE‐binding proteins, Pumilio proteins are also known to regulate mRNA stability through recruitment of the CNOT complex to target transcript 3′UTRs (Arvola et al., 2020; Enwerem et al., 2021; Goldstrohm et al., 2006; Joly et al., 2013; Van Etten et al., 2012; Weidmann et al., 2014). Single mutation of the ARE or PRE in the her1 3′UTR reporter moderately stabilized reporter transcripts, whereas mutation of both elements dramatically stabilized reporter transcripts and led to >8‐fold increased half‐life relative to the unmodified full‐length her1 3′UTR reporter (Tietz et al., 2020). Results from reporter assays suggest that the ARE and PRE cooperatively promote decay and that both ARE and PRE‐mediated decay is crucial for normal her1 post‐transcriptional regulation.
RBP‐mediated post‐transcriptional regulation is implicated in several developmental processes, and misregulation of RBP function can lead to drastic developmental defects (Brinegar & Cooper, 2016; Colegrove‐Otero et al., 2005; Dash et al., 2016; Giudice & Cooper, 2014; Prashad & Gopal, 2021). Pumilio function is critical during mouse embryonic development, and loss of function of either Pum1 or Pum2 results in defects in neurogenesis (Siemen et al., 2011; Zhang et al., 2017), and Pum1/Pum2 double knockout leads to embryonic lethality during gastrulation (Lin et al., 2018), precluding an analysis of PUM requirement during segmentation. Because the RNA binding domain, or Pumilio Homology Domain is highly conserved among many species analyzed to date, from Drosophila, to fish, to mammals, and binds to a specific, well‐defined PRE sequence, candidate PUM‐regulated mRNAs can be bioinformatically predicted through the presence of PRE motifs (Goldstrohm et al., 2018). In contrast, ARE‐BPs can recognize a variety of AU‐rich sequences distinct from the defined canonical ARE and thus, are harder to predict bioinformatically. Many genes in the ARE‐BP superfamily are expressed during embryogenesis across vertebrates (Briggs et al., 2018; Collins et al., 2019; White et al., 2017), and loss of function studies in mouse embryos have demonstrated the requirement of multiple ARE‐BPs for proper development of several tissues and organ systems (Beck et al., 1998; Bell et al., 2006; Katsanou et al., 2009; Stumpo et al., 2009). Furthermore, orthologs of many ARE‐BP‐encoding genes are expressed in the PSM across multiple vertebrate species, suggesting that ARE‐BPs may play a conserved regulatory role in segmentation clock gene expression (Table 1). It is interesting to note that for the chick, mouse, and human LFNG 3′UTRs, there is at least one canonical ARE (UAUUUAU) present, with the chick LFNG 3′UTR containing two AREs (Hilgers et al., 2005). While these similarities may allude to potential shared mechanisms of segmentation clock mRNA decay, the difference in the strength and number of motifs among different species' clock transcript 3′UTRs may contribute to observed differences in mRNA decay rates. Consistent with this idea, recent massively parallel reporter assays show that ARE and PRE presence and number are frequently associated with rapid transcript decay in other contexts (Rabani et al., 2017; Siegel et al., 2022). It is interesting to consider whether the presence and strength of destabilizing 3′UTR elements in segmentation clock gene transcripts contribute to species‐specific oscillation periods.
TABLE 1.
ARE‐BP expression in vertebrate PSMs or cultured PSM cells
| Species | ARE‐BP encoding genes expressed in PSM (or cultured PSM cells) |
|---|---|
| Zebrafish (PSM) | elavl1a, elavl1b, hnrnpd, hnrnpdl, khsrp, tia1, tia1l, tial1, zfp36l1a, zfp36l1b, zfp36l2 (Rauch et al, 2003; Thisse et al, 2004; Thisse & Thisse, 2004; Wagner et al, 2018) |
| Xenopus (PSM) | elavl, hnrnpd, hnrnpdl, khsrp, tia1, zfp36l1, zfp36l2 (Gawantka et al., 1998; Treguer et al., 2013; Briggs et al., 2018) |
| Mouse (PSM) | Elavl1, Hnrnpd, Khsrp, Tia1, Tial1 Zfp36l1, Zfp36l2 (Diaz‐Cuadros et al., 2020) |
| Human (Cultured PSM cells) | ELAVL1, HNRNPD, KHSRP, TIA1, TIAL, ZFP36L1, ZFP36L2 (Diaz‐Cuadros et al., 2020) |
2.4. Functional role of segmentation clock transcript 3′UTR‐mediated post‐transcriptional regulation on somitogenesis
Reporter‐based studies have advanced our understanding of molecular mechanisms that are important for 3′UTR‐dependent mRNA regulation. However, whether disruption of 3′UTR‐mediated post‐transcriptional regulation of segmentation clock transcripts affects clock period and somitogenesis is not fully understood. This was serendipitously addressed in one study using mouse embryos, initially conducted to observe the effect of increasing transcriptional delay of Hes7 by lengthening the gene (Fujimuro et al., 2014). Knock‐in of an exogenous 10 kb DNA fragment from a human intron into a locus just downstream of the endogenous Hes7 stop codon and directly upstream of the Hes7 3′UTR disrupted oscillatory Hes7 mRNA expression and severely reduced Hes7 protein levels in mouse embryos homozygous for the insertion. Homozygous mutant neonates also exhibited segmental defects in their vertebrae and ribs, similar to phenotypes observed in Hes7 null mice (Bessho, Sakata, et al., 2001). However, upon further examination of the knock‐in allele, it was discovered that the inserted human intron was retained in mature Hes7 transcripts and led to premature poly‐adenylation within the retained human intron, ultimately producing a Hes7 transcript that lacked the endogenous 3′UTR. The replacement of the endogenous Hes7 3′UTR with an exogenous human sequence resulted in a 30% decrease in Hes7 mRNA and near undetectable levels of HES7 protein (Fujimuro et al., 2014), suggesting that loss of critical regulatory elements in the Hes7 3′UTR interfered with proper Hes7 oscillatory expression and somite patterning. More specifically, the miRNAs and/or RBPs needed to refine mRNA oscillatory expression would be unable to promote decay or regulate translation, leading to disruption of the negative feedback loop. Introducing motif‐specific mutations in endogenous 3′UTR sequences and analyzing segmentation clock mRNA and protein expression will more directly address the role of cis‐regulatory element‐dependent mRNA regulation on the tempo of clock oscillations.
3. STIMULATORS OF mRNA DECAPPING AND DEADENYLATION ARE REQUIRED FOR SUSTAINED SEGMENTATION CLOCK GENE mRNA OSCILLATIONS
In the final stages of an mRNA lifetime in eukaryotic cells, translation is terminated and transcript degradation occurs by either endonucleolytic cleavage (directed by small RNA species, like miRNAs and siRNAs; Gu et al., 2018) or 3′ and 5′ end processing, followed by exonucleolytic degradation. Deadenylation is widely regarded as the rate‐limiting and the first step to occur in deadenylation‐dependent mRNA decay, followed shortly by either 3′ to 5′ exonucleolytic decay by the exosome or, more commonly, removal of the 5′m7G cap and 5′ to 3′ Xrn1‐mediated exonucleolytic decay (Muhlrad et al., 1994; Yamashita et al., 2005; Zheng et al., 2008). Both of these processes, if left unhindered and unaided, would occur at a rate determined only by the length of the poly (A) tail and relative strength of the decapping and deadenylation complex protein interactions with the mRNA (Steiger et al., 2003). However, for transcripts targeted for rapid decay, specific activators and RNA binding proteins facilitate rapid transcript turnover by recruiting or increasing the activity of decapping and deadenylation complexes (Fenger‐Gron et al., 2005; Muhlrad et al., 1994; Nissan et al., 2010; Shyu et al., 1991). In fact, overexpression of a dominant‐negative form of Cnot7, a member of the CCR4‐NOT deadenylase complex, in zebrafish embryos disrupts segmentation clock transcript oscillatory expression and somite patterning (Fujino et al., 2018). Due to the dynamic expression of segmentation clock transcripts, it is reasonable to predict that activators of mRNA decay or factors that sequester mRNAs from the translation machinery are likely important for ensuring normal clock periodicity is maintained. A few such factors, described below, have been identified and characterized with respect to their role in segmentation clock post‐transcriptional regulation.
3.1. Highly conserved deadenylation activators promote decay of segmentation clock‐associated transcripts
The RNA binding protein Celf1 (CUGBP [CUG binding protein] Elav‐like Family Member 1), also known as Embryo Deadenylation ElemeNt Binding Protein (EDEN‐BP), is an activator of deadenylation and is known to promote rapid mRNA decay of its target transcripts (Figure 3; Cibois et al., 2010, 2013; Gautier‐Courteille et al., 2004; Moraes et al., 2006; Rattenbacher et al., 2010). It preferentially binds to GU‐rich elements but has also been shown to bind AU‐rich elements (Moraes et al., 2006; Paillard et al., 2002; Vlasova et al., 2008). Loss of Celf1 activity drastically increases the abundance of polyadenylated ARE‐containing Celf1‐target mRNAs in vitro, likely due to direct interaction between Celf1 and the deadenylase Poly (A)‐specific ribonuclease (PARN; Moraes et al., 2006). In Xenopus embryos, Celf1‐dependent deadenylation is active during early embryogenesis and celf1 expression is enriched in the paraxial mesoderm and PSM during somitogenesis (Gautier‐Courteille et al., 2004). Both in vitro and in vivo UV cross‐linking experiments demonstrated that Celf1 protein directly binds the 3′UTR of rbpj [recombination signal binding protein for immunoglobulin Kappa J region, also known as suppressor of hairless, su (H)] (Gautier‐Courteille et al., 2004). rbpj, which does not oscillate, is an important modulator of segmentation clock gene expression in Xenopus, and segmentation is impaired upon direct interference between the Celf1:rbpj mRNA interaction (Cibois et al., 2010). The rbpj 3′UTR confers rapid Celf1‐dependent deadenylation of reporter transcripts, and morpholino‐mediated knockdown of celf1 in Xenopus embryos increases the stability of endogenous rbpj mRNA (Gautier‐Courteille et al., 2004). In contrast, Xenopus oscillating genes hairy2a and esr9 are not direct targets of Celf1, suggesting that other mechanisms of segmentation clock transcript decay exist to collectively promote segmentation clock mRNA oscillations.
FIGURE 3.
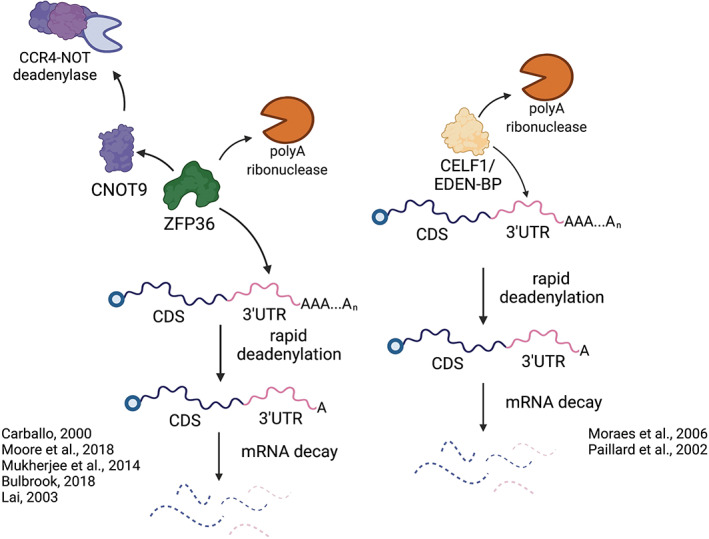
Activators of deadenylation promote rapid mRNA decay. Both CELF1/EDEN‐BP and ZFP36 proteins promote deadenylation‐dependent decay by binding 3′UTRs of target transcripts and recruiting deadenylation factors. CELF1/EDEN‐BP has been shown to directly bind transcript 3′UTRs and recruit the polyA ribonuclease (PARN) to promote rapid transcript deadenylation in Xenopus embryos. Additionally, ZFP36 proteins bind AU‐rich elements within transcript 3′UTRs and promote deadenylation through recruitment of PARN or the CCR4‐NOT complex via CNOT9. Both RNA binding proteins have been shown to regulate segmentation clock transcript stability, and further biochemical evidence will determine whether these precise interactions are also important in the context of segmentation clock transcript deadenylation and decay. Image created using Biorender.com
One class of ARE‐binding proteins, encoded by the zfp36 gene family (factors also known as TTP or Tis‐11), was found to negatively impact the expression of oscillatory genes esr5 and hairy2a and disrupted somite patterning when overexpressed in Xenopus embryos (Treguer et al., 2013). Human ZFP36 and its related proteins, ZFP36L1 and ZFP36L2, are known to directly bind mRNA and repress translation by promoting deadenylation‐dependent mRNA decay (Carballo et al., 2000; Moore et al., 2018; Mukherjee et al., 2014). ZFP36 can promote deadenylation through recruitment of the CNOT deadenylase complex, via direct interaction with CNOT9, or recruitment and activation of PARN (Figure 3; Bulbrook et al., 2018; Lai et al., 2003). Direct interactions have also been observed between human ZFP36 and the decapping factors, DCP1A and DCP2, and this interaction enhances the decapping of ARE‐containing mRNAs in vitro (Fenger‐Gron et al., 2005). It is important to note that in contrast to segmentation defects that arise from zfp36 overexpression, morpholino‐based knockdown of zfp36 expression in Xenopus embryos does not cause overt segmentation defects (Treguer et al., 2013), and this may be due to redundant functions of other RBPs and/or incomplete zfp36 knockdown. Nevertheless, it is intriguing to consider the impact of Zpf36‐mediated turnover of segmentation clock transcripts, and whether other RNA binding proteins may act in parallel to enhance deadenylation and promote mRNA decay.
3.2. Pnrc2, an enhancer of decapping, is required for segmentation clock transcript turnover
In wild‐type embryos, a striped pattern of segmentation clock mRNA expression is observed in the PSM at a fixed point in time, which arises due to coordinated oscillations of adjacent cells along with the anterior–posterior axis. In a forward genetic screen carried out in zebrafish to identify regulators of segmentation clock gene expression, the tortuga deficiency allele was recovered, which when homozygous, displays defects in expression of segmentation clock transcripts her1, her7, dlc, and other segmentation clock‐related transcripts (Dill & Amacher, 2005). Rather than typical, striped expression, tortuga mutant embryos exhibit uniform her1 and dlc mRNA expression throughout the PSM. This misexpression phenotype arises due to a defect in the clearance of segmentation clock transcripts that occurs when the function of proline‐rich nuclear receptor coactivator 2 (pnrc2), a gene deleted in the tortuga deficiency, is lost (Gallagher et al., 2017). In human cultured cells, PNRC2 has been described as a mediator between the nonsense‐mediated mRNA decay (NMD) and mRNA decapping complexes, specifically via its interactions with the NMD factor UPF1 and decapping complex protein DCP1A (Cho et al., 2009, 2013, 2015; Lai et al., 2012; Mugridge et al., 2016). Regions of the human PNRC2 protein important for PNRC2:DCP1A and PNRC2:UPF1 interactions show high sequence conservation with zebrafish Pnrc2 (Gallagher et al., 2017). In human cell culture, specific mutations within regions encoding the conserved SRC‐homology (SH3) domain and NR‐box of human PNRC2 disrupt binding to DCP1A and UPF1, respectively, which in turn leads to stabilization of reporter mRNA (Lai et al., 2012). Similarly, rescue experiments demonstrate that expression of zebrafish Pnrc2 containing the orthologous mutations within the SH3 domain and NR‐box does not rescue her1 expression defects when introduced into pnrc2 mutant embryos, in contrast to wild‐type Pnrc2 which fully restores wild‐type her1 expression in pnrc2 mutant embryos (Tietz et al., 2020). Morpholino‐mediated depletion of upf1 enhances the effects of pnrc2 depletion on her1 expression, suggesting that Upf1 facilitates Pnrc2‐mediated decay (Gallagher et al., 2017). However, additional biochemical evidence is needed to confirm whether direct interactions between Pnrc2 and other mRNA processing and decay factors are required for segmentation clock transcript decay.
Although the loss of Pnrc2‐dependent segmentation clock mRNA decay in zebrafish embryos increases transcript stability and abundance, corresponding protein levels do not appear to be increased, compared to wild‐type embryos (Gallagher et al., 2017; Tietz et al., 2020). Consistent with normal segmentation clock protein expression, pnrc2 mutant embryos form normal segments despite excess her1 mRNA, in contrast to earlier her1 overexpression studies that resulted in somite patterning defects (Giudicelli et al., 2007; Takke & Campos‐Ortega, 1999). Exogenous overexpression of her1 mRNA through microinjection or heat‐shock induction (Giudicelli et al., 2007; Takke & Campos‐Ortega, 1999) may overwhelm the mRNA decay and/or translational repression machinery, resulting in increased translation of segmentation clock protein and disruptions in clock periodicity. Additionally, it is unclear whether exogenously expressed her1 transcripts contain the complete suite of cis‐elements that are required to fully recapitulate endogenous her1 post‐transcriptional regulation. By contrast, the accumulation of endogenous her1 transcripts as a result of loss of Pnrc2‐mediated mRNA decay has alluded to the importance of translational regulation of segmentation clock transcripts. Elucidating the translation status and poly‐adenylation state of accumulated transcripts in pnrc2 mutant embryos will determine whether stabilized transcripts exist as decay intermediates or are actively translationally repressed by yet unindentified translational regulatory factors.
4. TRANSLATIONAL REGULATION OF OSCILLATORY GENE EXPRESSION
In order for segmentation clock oscillations to be sustained, the proteins encoded by core segmentation clock genes need to be degraded rapidly so that negative feedback loop‐mediated oscillatory expression is maintained (Hirata et al., 2004). The importance of HES7 protein instability in the mouse segmentation clock was demonstrated in mouse embryos that express a mutant HES7 protein that has an increased half‐life, but otherwise functions like wild‐type HES7 protein (Hirata et al., 2004). Mouse embryos expressing mutant Hes7 exhibited normal oscillations during early somitogenesis; however, after forming 3–4 normal somites, mutant embryos had fused somites, coinciding with segmentation clock gene mRNA and protein expression defects. On a molecular level, one would predict that stabilizing segmentation clock protein would prolong transcriptional repression of segmentation clock gene transcription. This would effectively dampen oscillations with each successive period and eventually perturb segmentation clock periodicity, resulting in somite patterning defects. The emergence of morphological phenotypes observed in the Hes7 mutant mouse embryos demonstrates a direct relationship between protein stability and segmentation clock periodicity. In contrast, increases in endogenous segmentation clock mRNA stability, as observed upon loss of Pnrc2‐mediated decay in zebrafish embryos, do not lead to overt segmentation defects. These observations indicate a robust post‐transcriptional mechanism of mRNA regulation exists to fine‐tune the expression of oscillatory gene transcripts.
The importance of translational regulation of gene expression, particularly at the nexus between translational repression and mRNA decay, is well appreciated (Decker & Parker, 2012). A well‐described model of translational regulation posits that RBPs and/or miRNAs that suppress the expression of target mRNAs can inhibit their translation and shuttle transcripts to cytoplasmic loci, such as processing bodies (P‐bodies), which consist of several ribonucleoprotein components, including mRNA processing and decay complex proteins (Decker & Parker, 2012; Hubstenberger et al., 2017). Transcriptomic analysis of purified P‐bodies showed that mRNAs enriched in P‐bodies collectively encode proteins that act as regulatory switches among different biological processes, including RNA processing, cell division, differentiation, and development (Hubstenberger et al., 2017). P‐body enriched mRNAs are also correlated with poor translation efficiency, compared to mRNAs that are not enriched in P‐bodies (Hubstenberger et al., 2017). Intuitively, these findings are not surprising, since the dynamics of cell division and fate specification processes, like somitogenesis and neurogenesis, are characterized by rapid and highly regulated gene expression transitions. Translational repression has been described as a method of regulation of the plant circadian clock (Juntawong & Bailey‐Serres, 2012; Missra et al., 2015), and may also be an efficient method of facilitating rapid downregulation of segmentation clock gene expression. Computational studies aimed at understanding the critical parameters needed to sustain autoinhibitory transcriptional feedback loops, such as the Hes/Her network, have shown that a translational time delay is particularly important for modulating the oscillation period (Ay et al., 2014; Murray et al., 2021). Specifically, a mathematical model that includes translational time delays to accurately model oscillation dynamics is consistent with the idea that there are translational repressive factors that help to refine the negative feedback loop so that oscillations are maintained properly (Murray et al., 2021). Importantly, experimental evidence derived from studies investigating cis‐regulatory elements that promote decay of oscillatory gene transcripts has alluded to putative translational regulatory factors (Bonev et al., 2012; Riley et al., 2013; Tietz et al., 2020).
In addition to their association with mRNA decay, RBPs, like Pumilio and ARE‐BPs, and miRNAs also have well‐described roles in translational regulation. miRNAs have been shown to regulate translation at multiple steps, including translation initiation and elongation (Fabian et al., 2010). Pumilio proteins are known to regulate translation by directly inhibiting the binding of PABP (poly (A)‐binding protein) to a target mRNA (Chritton & Wickens, 2011; Van Etten et al., 2012; Weidmann et al., 2014). Additionally, ARE‐BPs including TIA1 (Tia1 Cytotoxic Granule‐associated RNA Binding Protein) and TIAR [encoded by TIAL1 (Tia1 Cytotoxic Granule‐associated RNA Binding Protein‐like 1)], have been found to inhibit translation initiation of immune response and cancer‐associated mRNAs (Dixon et al., 2003; Gueydan et al., 1999; Piecyk et al., 2000). Expression analysis of segmentation clock gene mRNA and protein levels upon miRNA misregulation in mouse embryos (Riley et al., 2013; Wahi et al., 2017), and loss of Pnrc2‐mediated decay in zebrafish embryos (Gallagher et al., 2017; Tietz et al., 2020) suggest the translation of segmentation clock mRNAs is tightly regulated. Further exploration into translational regulatory control mechanisms of segmentation clock transcripts will fill the current knowledge gap that exists in our understanding of post‐transcriptional regulation of oscillatory gene expression.
5. CONCLUSION AND PERSPECTIVES
Rapid changes in gene expression are hallmarks of many developmental processes, some of which are observed as early as a few hours post‐fertilization, such as the maternal to zygotic transition (MZT; Vastenhouw et al., 2019). Prior to the onset of zygotic transcription, the development of metazoan embryos, particularly for animals that develop externally, relies on maternally provided gene products. As development proceeds, zygotic genome activation requires rapid clearance of maternal transcripts, marking the initiation of MZT (Vastenhouw et al., 2019). In Xenopus oocytes, Celf1/EDEN‐BP suppresses the expression of maternally deposited transcripts by promoting rapid deadenylation (Ezzeddine et al., 2002). Human CELF1/CUGBP1 protein sequence is 88% identical to Xenopus EDEN‐BP, and recombinant human CELF1/CUGBP1 can directly bind and rapidly deadenylate Xenopus maternal transcripts in Xenopus egg extracts (Ezzeddine et al., 2002; Paillard et al., 2003). Post‐transcriptional mechanisms of maternal mRNA decay have also been explored on a global scale in zebrafish embryos, in which a massively parallel reporter‐based study investigating 3′UTR elements that drive rapid degradation of maternally provided transcripts identified three predominant motifs driving decay of a subclass of transcripts: miR‐430 seed sequences, AREs, and PREs (Rabani et al., 2017). It is interesting to posit that these large‐scale mRNA decay programs have been co‐opted for use in other developmental processes which require robust and rapid modulation of mRNA expression, like in the case of the segmentation clock, and that the functions of key mRNA regulatory proteins are highly conserved. Post‐transcriptional regulation is a key mechanism to ensure proper developmental transitions, and critical regulatory factors may be re‐utilized throughout embryogenesis to quickly clear progenitor‐associated gene products and facilitate progression into more differentiated states. Future studies will further define the post‐transcriptional regulatory program that ensures proper control of developmental timing and to what extent these mechanisms are conserved.
Genetic oscillations are utilized throughout development to ensure that the timing of tissue growth and patterning is properly coordinated. In this review, we have summarized evidence of post‐transcriptional control of segmentation clock gene expression from studies conducted across vertebrates, revealing robust regulation of mRNA expression. The combination of 3′UTR‐interacting factors, deadenylation activators, and decapping enhancers facilitates precise regulation of mRNA oscillations, which in turn promotes oscillatory expression, a critical feature for the maintenance of stem cell fate. In segmenting embryos, Hes/her oscillations initiate in the posterior PSM and continue as cells are displaced anteriorly. Once cells are positioned at the determination front in the anterior PSM, oscillatory expression ceases, coinciding with pre‐somitic to somitic cell differentiation and the formation of a somite boundary (Gomez et al., 2008; Shih et al., 2015). Dynamic expression of Hes1 is also associated with progenitor fate in neural stem cells, and the termination of Hes1 oscillatory expression promotes neuronal differentiation (Shimojo et al., 2008). The question of whether the cessation of genetic oscillations is a consequence or cause of cell differentiation, and how post‐transcriptional regulators play a role in this transition, is a topic of interest among researchers studying stem cell determination processes (Hatakeyama et al., 2004; Momiji & Monk, 2009; Ohtsuka et al., 1999; Shimojo et al., 2008, 2016; Van Norman et al., 2013). Future studies aimed at uncovering the post‐transcriptional mechanisms involved in regulating genetic oscillations will provide further insight into the regulation of cell fate specification across development.
AUTHOR CONTRIBUTIONS
Monica C. Blatnik: Conceptualization (lead); writing – original draft (lead); writing – review and editing (equal). Thomas L. Gallagher: Conceptualization (supporting); writing – original draft (supporting); writing – review and editing (equal). Sharon Amacher: Conceptualization (supporting); writing – review and editing (supporting).
FUNDING INFORMATION
Segmentation clock research in the Amacher lab is supported by NIH grant R01GM117964. Monica C. Blatnik is supported by the Pelotonia fellowship program.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLES
Mechanisms of deadenylation‐dependent decay
Animal clocks: A multitude of molecular mechanisms for circadian timekeeping
Post‐transcriptional regulation in root development
Patterning the Drosophila embryo: A paradigm for RNA‐based developmental genetic regulation
Making sense of mRNA landscapes: Translation control in neurodevelopment
ACKNOWLEDGMENT
The authors thank Dr Susan Cole for her comments and suggestions on the manuscript.
Blatnik, M. C. , Gallagher, T. L. , & Amacher, S. L. (2023). Keeping development on time: Insights into post‐transcriptional mechanisms driving oscillatory gene expression during vertebrate segmentation. WIREs RNA, 14(1), e1751. 10.1002/wrna.1751
Edited by: Carol Lutz, Associate Editor and Jeff Wilusz, Editor‐in‐Chief
Funding information National Institute of General Medical Sciences, Grant/Award Number: R01GM117964; Pelotonia
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Arvola, R. M. , Chang, C. T. , Buytendorp, J. P. , Levdansky, Y. , Valkov, E. , Freddolino, P. L. , & Goldstrohm, A. C. (2020). Unique repression domains of Pumilio utilize deadenylation and decapping factors to accelerate destruction of target mRNAs. Nucleic Acids Research, 48(4), 1843–1871. 10.1093/nar/gkz1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ason, B. , Darnell, D. K. , Wittbrodt, B. , Berezikov, E. , Kloosterman, W. P. , Wittbrodt, J. , Antin, P. B. , & Plasterk, R. H. (2006). Differences in vertebrate microRNA expression. Proceedings of the National Academy of Sciences of the United States of America, 103(39), 14385–14389. 10.1073/pnas.0603529103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy, U. , Watson, J. , Patel, D. , & Keene, J. (1998). ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. Journal of Cell Science, 111(Pt 21), 3145–3156. 10.1242/jcs.111.21.3145 [DOI] [PubMed] [Google Scholar]
- Aulehla, A. , & Pourquie, O. (2010). Signaling gradients during paraxial mesoderm development. Cold Spring Harbor Perspectives in Biology, 2(2), a000869. 10.1101/cshperspect.a000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulehla, A. , Wiegraebe, W. , Baubet, V. , Wahl, M. B. , Deng, C. , Taketo, M. , Lewandoski, M. , & Pourquie, O. (2008). A beta‐catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nature Cell Biology, 10(2), 186–193. 10.1038/ncb1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay, A. , Holland, J. , Sperlea, A. , Devakanmalai, G. S. , Knierer, S. , Sangervasi, S. , Stevenson, A. , & Ozbudak, E. M. (2014). Spatial gradients of protein‐level time delays set the pace of the traveling segmentation clock waves. Development, 141(21), 4158–4167. 10.1242/dev.111930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma‐Mukai, A. , Oguri, H. , Mituyama, T. , Qian, Z. R. , Asai, K. , Siomi, H. , & Siomi, M. C. (2008). Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proceedings of the National Academy of Sciences of the United States of America, 105, 7964–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, A. R. , Miller, I. J. , Anderson, P. , & Streuli, M. (1998). RNA‐binding protein TIAR is essential for primordial germ cell development. Proceedings of the National Academy of Sciences of the United States of America, 95(5), 2331–2336. 10.1073/pnas.95.5.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm‐Ansmant, I. , Rehwinkel, J. , Doerks, T. , Stark, A. , Bork, P. , & Izaurralde, E. (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Development, 20, 1885–1898. 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. E. , Sanchez, M. J. , Spasic‐Boskovic, O. , Santalucia, T. , Gambardella, L. , Burton, G. J. , & Turner, M. (2006). The RNA binding protein Zfp36l1 is required for normal vascularisation and post‐transcriptionally regulates VEGF expression. Developmental Dynamics, 235(11), 3144–3155. 10.1002/dvdy.20949 [DOI] [PubMed] [Google Scholar]
- Bessho, Y. , Miyoshi, G. , Sakata, R. , & Kageyama, R. (2001). Hes7: A bHLH‐type repressor gene regulated by notch and expressed in the presomitic mesoderm. Genes to Cells, 6(2), 175–185. 10.1046/j.1365-2443.2001.00409.x [DOI] [PubMed] [Google Scholar]
- Bessho, Y. , Sakata, R. , Komatsu, S. , Shiota, K. , Yamada, S. , & Kageyama, R. (2001). Dynamic expression and essential functions of Hes7 in somite segmentation. Genes & Development, 15(20), 2642–2647. 10.1101/gad.930601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran, M. , Wang, Y. , Zhang, H. , Weng, T. , Baviskar, P. , Guo, Y. , Gou, D. , & Liu, L. (2009). MicroRNA‐127 modulates fetal lung development. Physiological Genomics, 37(3), 268–278. 10.1152/physiolgenomics.90268.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev, B. , Pisco, A. , & Papalopulu, N. (2011). MicroRNA‐9 reveals regional diversity of neural progenitors along the anterior‐posterior axis. Developmental Cell, 20(1), 19–32. 10.1016/j.devcel.2010.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev, B. , Stanley, P. , & Papalopulu, N. (2012). MicroRNA‐9 modulates Hes1 ultradian oscillations by forming a double‐negative feedback loop. Cell Reports, 2(1), 10–18. 10.1016/j.celrep.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brend, T. , & Holley, S. A. (2009). Expression of the oscillating gene her1 is directly regulated by hairy/enhancer of split, T‐box, and suppressor of hairless proteins in the zebrafish segmentation clock. Developmental Dynamics: An Official Publication of the American Association of the Anatomists, 238(11), 2745–2759. 10.1002/dvdy.22100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata, P. , Forcales, S. V. , Ponassi, M. , Corte, G. , Chen, C. Y. , Karin, M. , Puri, P. L. , & Gherzi, R. (2005). p38‐dependent phosphorylation of the mRNA decay‐promoting factor KSRP controls the stability of select myogenic transcripts. Molecular Cell, 20(6), 891–903. 10.1016/j.molcel.2005.10.021 [DOI] [PubMed] [Google Scholar]
- Briggs, J. A. , Weinreb, C. , Wagner, D. E. , Megason, S. , Peshkin, L. , Kirschner, M. W. , & Klein, A. M. (2018). The dynamics of gene expression in vertebrate embryogenesis at single‐cell resolution. Science, 360(6392), eaar5780. 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinegar, A. E. , & Cooper, T. A. (2016). Roles for RNA‐binding proteins in development and disease. Brain Research, 1647, 1–8. 10.1016/j.brainres.2016.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulbrook, D. , Brazier, H. , Mahajan, P. , Kliszczak, M. , Fedorov, O. , Marchese, F. P. , & Dean, J. L. E. (2018). Tryptophan‐mediated interactions between Tristetraprolin and the CNOT9 subunit are required for CCR4‐NOT Deadenylase complex recruitment. Journal of Molecular Biology, 430(5), 722–736. 10.1016/j.jmb.2017.12.018 [DOI] [PubMed] [Google Scholar]
- Bulman, M. P. , Kusumi, K. , Frayling, T. M. , McKeown, C. , Garrett, C. , Lander, E. S. , Krumlauf, R. , Hattersley, A. T. , Ellard, S. , & Turnpenny, P. D. (2000). Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nature Genetics, 24(4), 438–441. 10.1038/74307 [DOI] [PubMed] [Google Scholar]
- Carballo, E. , Lai, W. S. , & Blackshear, P. J. (2000). Evidence that tristetraprolin is a physiological regulator of granulocyte‐macrophage colony‐stimulating factor messenger RNA deadenylation and stability. Blood, The Journal of the American Society of Hematology, 95(6), 1891–1899. 10.1182/blood.V95.6.1891 [DOI] [PubMed] [Google Scholar]
- Casolaro, V. , Fang, X. , Tancowny, B. , Fan, J. , Wu, F. , Srikantan, S. , Asaki, S. Y. , De Fanis, U. , Huang, S. K. , Gorospe, M. , Atasoy, U. X. , & Stellato, C. (2008). Posttranscriptional regulation of IL‐13 in T cells: Role of the RNA‐binding protein HuR. The Journal of Allergy and Clinical Immunology, 121(4), 853–859 e854. 10.1016/j.jaci.2007.12.1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. , Han, S. , Choe, J. , Park, S. , Choi, S. , & Kim, Y. (2013). SMG5‐PNRC2 is functionally dominant compared with SMG5‐SMG7 in mammalian nonsense‐mediated mRNA decay. Nucleic Acids Research, 41(2), 1319–1328. 10.1093/nar/gks1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. , Kim, K. , & Kim, Y. (2009). Human proline‐rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Molecular Cell, 33(1), 75–86. 10.1016/j.molcel.2008.11.022 [DOI] [PubMed] [Google Scholar]
- Cho, H. , Park, O. H. , Park, J. , Ryu, I. , Kim, J. , Ko, J. , & Kim, Y. K. (2015). Glucocorticoid receptor interacts with PNRC2 in a ligand‐dependent manner to recruit UPF1 for rapid mRNA degradation. Proceedings of the National Academy of Sciences of the United States of America, 112(13), E1540–E1549. 10.1073/pnas.1409612112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W. Y. , Giraldez, A. J. , & Schier, A. F. (2007). Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR‐430. Science, 318(5848), 271–274. 10.1126/science.1147535 [DOI] [PubMed] [Google Scholar]
- Choorapoikayil, S. , Willems, B. , Strohle, P. , & Gajewski, M. (2012). Analysis of her1 and her7 mutants reveals a spatio temporal separation of the somite clock module. PLoS One, 7(6), e39073. 10.1371/journal.pone.0039073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, C. F. , Mulky, A. , Maitra, S. , Lin, W. J. , Gherzi, R. , Kappes, J. , & Chen, C. Y. (2006). Tethering KSRP, a decay‐promoting AU‐rich element‐binding protein, to mRNAs elicits mRNA decay. Molecular and Cellular Biology, 26(10), 3695–3706. 10.1128/MCB.26.10.3695-3706.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ, B. , Jacob, H. J. , & Jacob, M. (1978). On the formation of the myotomes in avian embryos. An experimental and scanning electron microscope study. Experientia, 34(4), 514–516. [Google Scholar]
- Christ, B. , Jacob, M. , Jürgen, H. , Beate, J. , & Wachtler, B. (1986). Myogenesis: A problem of cell distribution and cell interactions. Somites in Developing Embryos, 118, 261–275. [Google Scholar]
- Chritton, J. J. , & Wickens, M. (2011). A role for the poly (A)‐binding protein Pab1p in PUF protein‐mediated repression. The Journal of Biological Chemistry, 286(38), 33268–33278. 10.1074/jbc.M111.264572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois, M. , Gautier‐Courteille, C. , Kodjabachian, L. , & Paillard, L. (2013). A gene regulation network controlled by Celf1 protein‐rbpj mRNA interaction in Xenopus somite segmentation. Biology Open, 2(10), 1078–1083. 10.1242/bio.20135629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois, M. , Gautier‐Courteille, C. , Vallee, A. , & Paillard, L. (2010). A strategy to analyze the phenotypic consequences of inhibiting the association of an RNA‐binding protein with a specific RNA. RNA, 16(1), 10–15. 10.1261/rna.1742610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove‐Otero, L. J. , Minshall, N. , & Standart, N. (2005). RNA‐binding proteins in early development. Critical Reviews in Biochemistry and Molecular Biology, 40(1), 21–73. 10.1080/10409230590918612 [DOI] [PubMed] [Google Scholar]
- Collins, J. E. , White, R. J. , Staudt, N. , Sealy, I. M. , Packham, I. , Wali, N. , Tudor, C. , Mazzeo, C. , Green, A. , Siragher, E. , Ryder, E. , White, J. K. , Papatheodoru, I. , Tang, A. , Fullgrabe, A. , Billis, K. , Geyer, S. H. , Weninger, W. J. , Galli, A. , … Busch‐Nentwich, E. M. (2019). Common and distinct transcriptional signatures of mammalian embryonic lethality. Nature Communications, 10(1), 2792. 10.1038/s41467-019-10642-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, S. , Siddam, A. D. , Barnum, C. E. , Janga, S. C. , & Lachke, S. A. (2016). RNA‐binding proteins in eye development and disease: Implication of conserved RNA granule components. WIREs RNA, 7(4), 527–557. 10.1002/wrna.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. L. , Turner, D. L. , Evans, L. M. , & Kirschner, M. W. (2001). Molecular targets of vertebrate segmentation: Two mechanisms control segmental expression of Xenopus hairy2 during somite formation. Developmental Cell, 1(4), 553–565. 10.1016/s1534-5807(01)00054-5 [DOI] [PubMed] [Google Scholar]
- Decker, C. J. , & Parker, R. (2012). P‐bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harbor Perspectives in Biology, 4(9), a012286. 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune, E. A. , Francois, P. , Shih, N. P. , & Amacher, S. L. (2012). Single‐cell‐resolution imaging of the impact of notch signaling and mitosis on segmentation clock dynamics. Developmental Cell, 23(5), 995–1005. 10.1016/j.devcel.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Cuadros, M. , Wagner, D. E. , Budjan, C. , Hubaud, A. , Tarazona, O. A. , Donelly, S. , Michaut, A. , Al Tanoury, Z. , Yoshioka‐Kobayashi, K. , Niino, Y. , Kageyama, R. , Miyawaki, A. , Touboul, J. , & Pourquie, O. (2020). In vitro characterization of the human segmentation clock. Nature, 580(7801), 113–118. 10.1038/s41586-019-1885-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, K. K. , & Amacher, S. L. (2005). Tortuga refines notch pathway gene expression in the zebrafish presomitic mesoderm at the post‐transcriptional level. Developmental Biology, 287(2), 225–236. 10.1016/j.ydbio.2005.07.032 [DOI] [PubMed] [Google Scholar]
- Dixon, D. A. , Balch, G. C. , Kedersha, N. , Anderson, P. , Zimmerman, G. A. , Beauchamp, R. D. , & Prescott, S. M. (2003). Regulation of cyclooxygenase‐2 expression by the translational silencer TIA‐1. The Journal of Experimental Medicine, 198(3), 475–481. 10.1084/jem.20030616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwerem, I. I. I. , Elrod, N. D. , Chang, C. T. , Lin, A. , Ji, P. , Bohn, J. A. , Levdansky, Y. , Wagner, E. J. , Valkov, E. , & Goldstrohm, A. C. (2021). Human Pumilio proteins directly bind the CCR4‐NOT deadenylase complex to regulate the transcriptome. RNA, 27(4), 445–464. 10.1261/rna.078436.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. , Rosenthal, E. T. , Youngblom, J. , Distel, D. , & Hunt, T. (1983). Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell, 33(2), 389–396. 10.1016/0092-8674(83)90420-8 [DOI] [PubMed] [Google Scholar]
- Ezzeddine, N. , Paillard, L. , Capri, M. , Maniey, D. , Bassez, T. , Aït‐Ahmed, O. , & Osborne, H. B. (2002). EDEN dependent translational repression of maternal mRNAs is conserved between Xenopus and drosophila. Proceedings of the National Academy of Sciences of the United States of America, 99(1), 257–262. 10.1073/pnas.012555499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, M. R. , Sonenberg, N. , & Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry, 79, 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Fenger‐Gron, M. , Fillman, C. , Norrild, B. , & Lykke‐Andersen, J. (2005). Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Molecular Cell, 20(6), 905–915. 10.1016/j.molcel.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Fujimuro, T. , Matsui, T. , Nitanda, Y. , Matta, T. , Sakumura, Y. , Saito, M. , Kohno, K. , Nakahata, Y. , & Bessho, Y. (2014). Hes7 3′ UTR is required for somite segmentation function. Scientific Reports, 4(1), 1–9. 10.1038/srep06462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino, Y. , Yamada, K. , Sugaya, C. , Ooka, Y. , Ovara, H. , Ban, H. , Akama, K. , Otosaka, S. , Kinoshita, H. , Yamasu, K. , Mishima, Y. , & Kawamura, A. (2018). Deadenylation by the CCR4‐NOT complex contributes to the turnover of hairy‐related mRNAs in the zebrafish segmentation clock. FEBS Letters, 592(20), 3388–3398. 10.1002/1873-3468.13261 [DOI] [PubMed] [Google Scholar]
- Gallagher, T. L. , Tietz, K. T. , Morrow, Z. T. , McCammon, J. M. , Goldrich, M. L. , Derr, N. L. , & Amacher, S. L. (2017). Pnrc2 regulates 3′UTR‐mediated decay of segmentation clock‐associated transcripts during zebrafish segmentation. Developmental Biology, 429(1), 225–239. 10.1016/j.ydbio.2017.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantier, M. P. , McCoy, C. E. , Rusinova, I. , Saulep, D. , Wang, D. , Xu, D. , Irving, A. T. , Behlke, M. A. , Hertzog, P. J. , Mackay, F. , & Williams, B. R. (2011). Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Research, 39(13), 5692–5703. 10.1093/nar/gkr148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier‐Courteille, C. , Le Clainche, C. , Barreau, C. , Audic, Y. , Graindorge, A. , Maniey, D. , Osborne, H. B. , & Paillard, L. (2004). EDEN‐BP‐dependent post‐transcriptional regulation of gene expression in Xenopus somitic segmentation. Development, 131(24), 6107–6117. 10.1242/dev.01528 [DOI] [PubMed] [Google Scholar]
- Gawantka, V., Pollet, N., Delius, H., Vingron, M., Pfister, R., Nitsch, R., Blumenstock, C., & Niehrs, C. (1998). Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mechanisms of Development, 77(2), 95–141. 10.1016/s0925-4773(98)00115-4 [DOI] [PubMed] [Google Scholar]
- Giraldez, A. J. , Mishima, Y. , Rihel, J. , Grocock, R. J. , Van Dongen, S. , Inoue, K. , Enright, A. J. , & Schier, A. F. (2006). Zebrafish MiR‐430 promotes deadenylation and clearance of maternal mRNAs. Science, 312(5770), 75–79. 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- Giudice, J. , & Cooper, T. A. (2014). RNA‐binding proteins in heart development. Advances in Experimental Medicine and Biology, 825, 389–429. 10.1007/978-1-4939-1221-6_11 [DOI] [PubMed] [Google Scholar]
- Giudicelli, F. , Ozbudak, E. M. , Wright, G. J. , & Lewis, J. (2007). Setting the tempo in development: An investigation of the zebrafish somite clock mechanism. PLoS Biology, 5(6), e150. 10.1371/journal.pbio.0050150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm, A. C. , Hall, T. M. T. , & McKenney, K. M. (2018). Post‐transcriptional regulatory functions of mammalian Pumilio proteins. Trends in Genetics, 34(12), 972–990. 10.1016/j.tig.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm, A. C. , Hook, B. A. , Seay, D. J. , & Wickens, M. (2006). PUF proteins bind Pop2p to regulate messenger RNAs. Nature Structural & Molecular Biology, 13(6), 533–539. 10.1038/nsmb1100 [DOI] [PubMed] [Google Scholar]
- Goljanek‐Whysall, K. , Sweetman, D. , Abu‐Elmagd, M. , Chapnik, E. , Dalmay, T. , Hornstein, E. , & Munsterberg, A. (2011). MicroRNA regulation of the paired‐box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proceedings of the National Academy of Sciences of the United States of America, 108(29), 11936–11941. 10.1073/pnas.1105362108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, C. , Ozbudak, E. M. , Wunderlich, J. , Baumann, D. , Lewis, J. , & Pourquie, O. (2008). Control of segment number in vertebrate embryos. Nature, 454(7202), 335–339. 10.1038/nature07020 [DOI] [PubMed] [Google Scholar]
- Gossen, M. , Freundlieb, S. , Bender, G. , Muller, G. , Hillen, W. , & Bujard, H. (1995). Transcriptional activation by tetracyclines in mammalian cells. Science, 268(5218), 1766–1769. 10.1126/science.7792603 [DOI] [PubMed] [Google Scholar]
- Gratacos, F. M. , & Brewer, G. (2010). The role of AUF1 in regulated mRNA decay. WIREs RNA, 1(3), 457–473. 10.1002/wrna.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, K. , Mok, L. , & Chong, M. M. (2018). Regulating gene expression in animals through RNA endonucleolytic cleavage. Heliyon, 4(11), e00908. 10.1016/j.heliyon.2018.e00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan, C. , Droogmans, L. , Chalon, P. , Huez, G. , Caput, D. , & Kruys, V. (1999). Identification of TIAR as a protein binding to the translational regulatory AU‐rich element of tumor necrosis factor α mRNA. Journal of Biological Chemistry, 274(4), 2322–2326. 10.1074/jbc.274.4.2322 [DOI] [PubMed] [Google Scholar]
- Hatakeyama, J. , Bessho, Y. , Katoh, K. , Ookawara, S. , Fujioka, M. , Guillemot, F. , & Kageyama, R. (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development, 131(22), 5539–5550. 10.1242/dev.01436 [DOI] [PubMed] [Google Scholar]
- Henry, C. A. , Urban, M. K. , Dill, K. K. , Merlie, J. P. , Page, M. F. , Kimmel, C. B. , & Amacher, S. L. (2002). Two linked hairy/enhancer of split‐related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development, 129(15), 3693–3704. [DOI] [PubMed] [Google Scholar]
- Hilgers, V. , Pourquié, O. , & Dubrulle, J. (2005). In vivo analysis of mRNA stability using the Tet‐off system in the chicken embryo. Developmental Biology, 284(2), 292–300. 10.1016/j.ydbio.2005.05.021 [DOI] [PubMed] [Google Scholar]
- Hirata, H. , Bessho, Y. , Kokubu, H. , Masamizu, Y. , Yamada, S. , Lewis, J. , & Kageyama, R. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nature Genetics, 36(7), 750–754. 10.1038/ng1372 [DOI] [PubMed] [Google Scholar]
- Holley, S. A. , Geisler, R. , & Nusslein‐Volhard, C. (2000). Control of her1 expression during zebrafish somitogenesis by a delta‐dependent oscillator and an independent wave‐front activity. Genes & Development, 14(13), 1678–1690. [PMC free article] [PubMed] [Google Scholar]
- Hoyle, N. P. , & Ish‐Horowicz, D. (2013). Transcript processing and export kinetics are rate‐limiting steps in expressing vertebrate segmentation clock genes. Proceedings of the National Academy of Sciences of the United States of America, 110(46), E4316–E4324. 10.1073/pnas.1308811110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaud, A. , & Pourquie, O. (2014). Signalling dynamics in vertebrate segmentation. Nature Reviews. Molecular Cell Biology, 15(11), 709–721. 10.1038/nrm3891 [DOI] [PubMed] [Google Scholar]
- Hubaud, A. , Regev, I. , Mahadevan, L. , & Pourquie, O. (2017). Excitable dynamics and yap‐dependent mechanical cues drive the segmentation clock. Cell, 171(3), 668–682.e611. 10.1016/j.cell.2017.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger, A. , Courel, M. , Benard, M. , Souquere, S. , Ernoult‐Lange, M. , Chouaib, R. , Yi, Z. , Morlot, J. B. , Munier, A. , Fradet, M. , Daunesse, M. , Bertrand, E. , Pierron, G. , Mozziconacci, J. , Kress, M. , & Weil, D. (2017). P‐body purification reveals the condensation of repressed mRNA regulons. Molecular Cell, 68(1), 144–157.e145. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Ishibashi, M. , Ang, S. L. , Shiota, K. , Nakanishi, S. , Kageyama, R. , & Guillemot, F. (1995). Targeted disruption of mammalian hairy and enhancer of split homolog‐1 (HES‐1) leads to up‐regulation of neural helix‐loop‐helix factors, premature neurogenesis, and severe neural tube defects. Genes & Development, 9(24), 3136–3148. 10.1101/gad.9.24.3136 [DOI] [PubMed] [Google Scholar]
- Jambor, H. , Surendranath, V. , Kalinka, A. T. , Mejstrik, P. , Saalfeld, S. , & Tomancak, P. (2015). Systematic imaging reveals features and changing localization of mRNAs in drosophila development. eLife, 4, e05003. 10.7554/eLife.05003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, B. , Yuan, J. , Yin, Z. , Lv, C. , Lu, S. , Xiong, H. , Tang, H. , Ye, G. , & Shi, F. (2015). Dynamic properties of the segmentation clock mediated by microRNA. International Journal of Clinical and Experimental Pathology, 8(1), 196–206. [PMC free article] [PubMed] [Google Scholar]
- Joly, W. , Chartier, A. , Rojas‐Rios, P. , Busseau, I. , & Simonelig, M. (2013). The CCR4 deadenylase acts with Nanos and Pumilio in the fine‐tuning of Mei‐P26 expression to promote germline stem cell self‐renewal. Stem Cell Reports, 1(5), 411–424. 10.1016/j.stemcr.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong, P. , & Bailey‐Serres, J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana . Frontiers in Plant Science, 3, 66. 10.3389/fpls.2012.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, R. , Niwa, Y. , Isomura, A. , Gonzalez, A. , & Harima, Y. (2012). Oscillatory gene expression and somitogenesis. Wiley Interdisciplinary Reviews: Developmental Biology, 1(5), 629–641. 10.1002/wdev.46 [DOI] [PubMed] [Google Scholar]
- Kanki, J. P. , & Ho, R. K. (1997). The development of the posterior body in zebrafish. Development, 124(4), 881–893. 10.1242/dev.124.4.881 [DOI] [PubMed] [Google Scholar]
- Katsanou, V. , Milatos, S. , Yiakouvaki, A. , Sgantzis, N. , Kotsoni, A. , Alexiou, M. , & Kontoyiannis, D. L. (2009). The RNA‐binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Molecular and Cellular Biology, 29(10), 2762–2776. 10.1128/MCB.01393-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, A. , Ovara, H. , Ooka, Y. , Kinoshita, H. , Hoshikawa, M. , Nakajo, K. , Yokota, D. , Fujino, Y. , Higashijima, S. , Takada, S. , & Yamasu, K. (2016). Posterior‐anterior gradient of zebrafish hes6 expression in the presomitic mesoderm is established by the combinatorial functions of the downstream enhancer and 3′UTR. Developmental Biology, 409(2), 543–454. 10.1016/j.ydbio.2015.11.010 [DOI] [PubMed] [Google Scholar]
- Keynes, R. J. , & Stern, C. D. (1988). Mechanisms of vertebrate segmentation. Development, 103(3), 413–429. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Grun, D. , & van Oudenaarden, A. (2013). Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nature Genetics, 45(11), 1337–1344. 10.1038/ng.2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol, A. J. , Roellig, D. , Dequeant, M. L. , Tassy, O. , Glynn, E. , Hattem, G. , Mushegian, A. , Oates, A. C. , & Pourquie, O. (2011). Evolutionary plasticity of segmentation clock networks. Development, 138(13), 2783–2792. 10.1242/dev.063834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, T. , Cho, H. , Liu, Z. , Bowler, M. , Piao, S. , Parker, R. , Kim, Y. , & Song, H. (2012). Structural basis of the PNRC2‐mediated link between mrna surveillance and decapping. Structure, 20(12), 2025–2037. 10.1016/j.str.2012.09.009 [DOI] [PubMed] [Google Scholar]
- Lai, W. S. , Kennington, E. A. , & Blackshear, P. J. (2003). Tristetraprolin and its family members can promote the cell‐free deadenylation of AU‐rich element‐containing mRNAs by poly (A) ribonuclease. Molecular and Cellular Biology, 23(11), 3798–3812. 10.1128/MCB.23.11.3798-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler, M. , Gaidatzis, D. , Rothballer, A. , Chen, P. , & Soll, S. (2008). Molecular characterization of human Argonaute‐containing ribonucleoprotein complexes and their bound target mRNAs. RNA, 14, 2580–2596. 10.1261/rna.1351608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A. G. , Irier, H. A. , Gu, J. , Tian, D. , Ku, L. , Liu, G. , Xia, M. , Fritsch, B. , Zheng, J. Q. , Dingledine, R. , Xu, B. , Lu, B. , & Feng, Y. (2010). Distinct 3′ UTRs differentially regulate activity‐dependent translation of brain‐derived neurotrophic factor (BDNF). Proceedings of the National Academy of Sciences of the United States of America, 107(36), 15945–15950. 10.1073/pnas.1002929107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer, E. , Yoshida, H. , Parthasarathy, N. , Alm, C. , Babak, T. , Cerovina, T. , Hughes, T. R. , Tomancak, P. , & Krause, H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell, 131(1), 174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lewis, J. (2003). Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Current Biology, 13(16), 1398–1408. 10.1016/s0960-9822(03)00534-7 [DOI] [PubMed] [Google Scholar]
- Lin, K. , Zhang, S. , Shi, Q. , Zhu, M. , Gao, L. , Xia, W. , Geng, B. , Zheng, Z. , & Xu, E. Y. (2018). Essential requirement of mammalian Pumilio family in embryonic development. Molecular Biology of the Cell, 29(24), 2922–2932. 10.1091/mbc.E18-06-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E. , Liu, M. , Hartley, R. S. , Sheets, M. D. , & Dahlberg, J. E. (2009). Deadenylation of maternal mRNAs mediated by miR‐427 in Xenopus laevis embryos. RNA, 15(12), 2351–2363. 10.1261/rna.1882009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke‐Andersen, J. , & Wagner, E. (2005). Recruitment and activation of mRNA decay enzymes by two ARE‐mediated decay activation domains in the proteins TTP and BRF‐1. Genes & Development, 19(3), 351–361. 10.1101/gad.1282305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara, A. , & Holley, S. A. (2007). Oscillators and the emergence of tissue organization during zebrafish somitogenesis. Trends in Cell Biology, 17(12), 593–599. 10.1016/j.tcb.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Mara, A. , Schroeder, J. , Chalouni, C. , & Holley, S. A. (2007). Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nature Cell Biology, 9(5), 523–530. 10.1038/ncb1578 [DOI] [PubMed] [Google Scholar]
- Masamizu, Y. , Ohtsuka, T. , Takashima, Y. , Nagahara, H. , Takenaka, Y. , Yoshikawa, K. , Okamura, H. , & Kageyama, R. (2006). Real‐time imaging of the somite segmentation clock: Revelation of unstable oscillators in the individual presomitic mesoderm cells. Proceedings of the National Academy of Sciences of the United States of America, 103(5), 1313–1318. 10.1073/pnas.0508658103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M. , Hayashi, H. , Garcia‐Ojalvo, J. , Yoshioka‐Kobayashi, K. , Kageyama, R. , Yamanaka, Y. , Ikeya, M. , Toguchida, J. , Alev, C. , & Ebisuya, M. (2020). Species‐specific segmentation clock periods are due to differential biochemical reaction speeds. Science, 369(6510), 1450–1455. 10.1126/science.aba7668 [DOI] [PubMed] [Google Scholar]
- Matsuda, M. , Yamanaka, Y. , Uemura, M. , Osawa, M. , Saito, M. K. , Nagahashi, A. , Nishio, M. , Guo, L. , Ikegawa, S. , Sakurai, S. , Kihara, S. , Maurissen, T. L. , Nakamura, M. , Matsumoto, T. , Yoshitomi, H. , Ikeya, M. , Kawakami, N. , Yamamoto, T. , Woltjen, K. , … Alev, C. (2020). Recapitulating the human segmentation clock with pluripotent stem cells. Nature, 580(7801), 124–129. 10.1038/s41586-020-2144-9 [DOI] [PubMed] [Google Scholar]
- Milev, N. B. , Rhee, S. G. , & Reddy, A. B. (2018). Cellular timekeeping: It's redox o'Clock. Cold Spring Harbor Perspectives in Biology, 10(5), e2027698. 10.1101/cshperspect.a027698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra, A. , Ernest, B. , Lohoff, T. , Jia, Q. , Satterlee, J. , Ke, K. , & von Arnim, A. G. (2015). A precedent for translational control of mRNAs in a core oscillator is found in the Arabidopsis circadian clock. The Plant Cell, 27(9), 2582–2599. 10.1105/tpc.15.00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momiji, H. , & Monk, N. A. (2009). Oscillatory notch‐pathway activity in a delay model of neuronal differentiation. Physical Review. E, Statistical, Nonlinear, and Soft Matter Physics, 80(2 Pt 1), 021930. 10.1103/PhysRevE.80.021930 [DOI] [PubMed] [Google Scholar]
- Moore, M. J. , Blachere, N. E. , Fak, J. J. , Park, C. Y. , Sawicka, K. , Parveen, S. , Zucker‐Scharff, I. , Moltedo, B. , Rudensky, A. Y. , & Darnell, R. B. (2018). ZFP36 RNA‐binding proteins restrain T cell activation and anti‐viral immunity. eLife, 7, e33057. 10.7554/eLife.33057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes, K. C. , Wilusz, C. J. , & Wilusz, J. (2006). CUG‐BP binds to RNA substrates and recruits PARN deadenylase. RNA, 12(6), 1084–1091. 10.1261/rna.59606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Risueno, M. A. , Van Norman, J. M. , Moreno, A. , Zhang, J. , Ahnert, S. E. , & Benfey, P. N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science, 329(5997), 1306–1311. 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugridge, J. S. , Ziemniak, M. , Jemielity, J. , & Gross, J. D. (2016). Structural basis of mRNA‐cap recognition by Dcp1‐Dcp2. Nature Structural & Molecular Biology, 23(11), 987–994. 10.1038/nsmb.3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D. , Decker, C. J. , & Parker, R. (1994). Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′‐>3′ digestion of the transcript. Genes & Development, 8(7), 855–866. 10.1101/gad.8.7.855 [DOI] [PubMed] [Google Scholar]
- Mukherjee, N. , Jacobs, N. C. , Hafner, M. , Kennington, E. A. , Nusbaum, J. D. , Tuschl, T. , Blackshear, P. J. , & Ohler, U. (2014). Global target mRNA specification and regulation by the RNA‐binding protein ZFP36. Genome Biology, 15(1), R12. 10.1186/gb-2014-15-1-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A. W. (2004). Recycling the cell cycle: Cyclins revisited. Cell, 116(2), 221–234. 10.1016/s0092-8674(03)01080-8 [DOI] [PubMed] [Google Scholar]
- Murray, P. J. , Ocana, E. , Meijer, H. A. , & Dale, J. K. (2021). Auto‐regulation of transcription and translation: Oscillations, excitability and intermittency. Biomolecules, 11(11), 1566. 10.3390/biom11111566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeli, P. , Winter, T. , Hackett, A. P. , Alboushi, L. , & Jafarnejad, S. M. (2022). The intricate balance between microRNA‐induced mRNA decay and translational repression. The FEBS Journal. 10.1111/febs.16422 [DOI] [PubMed] [Google Scholar]
- Nissan, T. , Rajyaguru, P. , She, M. , Song, H. , & Parker, R. (2010). Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Molecular Cell, 39(5), 773–783. 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitanda, Y. , Matsui, T. , Matta, T. , Higami, A. , Kohno, K. , Nakahata, Y. , & Bessho, Y. (2013). 3′‐UTR‐dependent regulation of mRNA turnover is critical for differential distribution patterns of cyclic gene mRNAs. The FEBS Journal, 281(1), 146–156. 10.1111/febs.12582 [DOI] [PubMed] [Google Scholar]
- Oates, A. C. (2020). Waiting on the fringe: Cell autonomy and signaling delays in segmentation clocks. Current Opinion in Genetics & Development, 63, 61–70. 10.1016/j.gde.2020.04.008 [DOI] [PubMed] [Google Scholar]
- Oates, A. C. , & Ho, R. K. (2002). Hairy/E (spl)‐related (her) genes are central components of the segmentation oscillator and display redundancy with the Delta/notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development, 129(12), 2929–2946. [DOI] [PubMed] [Google Scholar]
- Oates, A. C. , Morelli, L. G. , & Ares, S. (2012). Patterning embryos with oscillations: Structure, function and dynamics of the vertebrate segmentation clock. Development, 139(4), 625–639. 10.1242/dev.063735 [DOI] [PubMed] [Google Scholar]
- Ohtsuka, T. , Ishibashi, M. , Gradwohl, G. , Nakanishi, S. , Guillemot, F. , & Kageyama, R. (1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. The EMBO Journal, 18(8), 2196–2207. 10.1093/emboj/18.8.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo, Y. , Sugawara, T. , Abe‐Koduka, N. , Kanno, J. , Kimura, A. , & Saga, Y. (2012). Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans‐repression of notch signalling. Nature Communications, 3, 1141. 10.1038/ncomms2133 [DOI] [PubMed] [Google Scholar]
- Paillard, L. , Legagneux, V. , Maniey, D. , & Osborne, H. B. (2002). C‐Jun ARE targets mRNA deadenylation by an EDEN‐BP (embryo deadenylation element‐binding protein)‐dependent pathway. The Journal of Biological Chemistry, 277(5), 3232–3235. 10.1074/jbc.M109362200 [DOI] [PubMed] [Google Scholar]
- Paillard, L. , Legagneux, V. , & Osborne, H. B. (2003). A functional deadenylation assay identifies human CUG‐BP as a deadenylation factor. Biology of the Cell, 95(2), 107–113. 10.1016/s0248-4900(03)00010-8 [DOI] [PubMed] [Google Scholar]
- Palmeirim, I. , Henrique, D. , Ish‐Horowicz, D. , & Pourquie, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell, 91(5), 639–648. 10.1016/s0092-8674(00)80451-1 [DOI] [PubMed] [Google Scholar]
- Park‐Lee, S. , Kim, S. , & Laird‐Offringa, I. (2003). Characterization of the interaction between neuronal RNA‐binding protein HuD and AU‐rich RNA. The Journal of Biological Chemistry, 278(41), 39801–39808. 10.1074/jbc.M307105200 [DOI] [PubMed] [Google Scholar]
- Patke, A. , Young, M. W. , & Axelrod, S. (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nature Reviews. Molecular Cell Biology, 21(2), 67–84. 10.1038/s41580-019-0179-2 [DOI] [PubMed] [Google Scholar]
- Piecyk, M. , Wax, S. , Beck, A. , Kedersha, N. , Gupta, M. , Maritim, B. , Chen, S. , Gueydan, C. , Kruys, V. , Streuli, M. , & Anderson, P. (2000). TIA‐1 is a translational silencer that selectively regulates the expression of TNF‐α. The EMBO Journal, 19(15), 4154–4163. 10.1093/emboj/19.15.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R. S. , Artus, C. G. , & Filipowicz, W. (2004). Tethering of human ago proteins to mRNA mimics the miRNA‐mediated repression of protein synthesis. RNA, 10(10), 1518–1525. 10.1261/rna.7131604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R. S. , Bhattacharyya, S. N. , Artus, C. G. , Zoller, T. , Cougot, N. , Basyuk, E. , Bertrand, E. , & Filipowicz, W. (2005). Inhibition of translational initiation by Let‐7 MicroRNA in human cells. Science, 309(5740), 1573–1576. 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Prashad, S. , & Gopal, P. P. (2021). RNA‐binding proteins in neurological development and disease. RNA Biology, 18(7), 972–987. 10.1080/15476286.2020.1809186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullmann, R. J. , Kim, H. , Abdelmohsen, K. , Lal, A. , Martindale, J. , Yang, X. , & Gorospe, M. (2007). Analysis of turnover and translation regulatory RNA‐binding protein expression through binding to cognate mRNAs. Molecular and Cellular Biology, 27(18), 6265–6278. 10.1128/MCB.00500-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani, M. , Pieper, L. , Chew, G. L. , & Schier, A. F. (2017). A massively parallel reporter assay of 3′ UTR sequences identifies in vivo rules for mRNA degradation. Molecular Cell, 68(6), 1083–1094.e1085. 10.1016/j.molcel.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenbacher, B. , Beisang, D. , Wiesner, D. L. , Jeschke, J. C. , Von Hohenberg, M. , St. Louis‐Vlasova, I. A. , & Bohjanen, P. R. (2010). Analysis of CUGBP1 targets identifies GU‐repeat sequences that mediate rapid mRNA decay. Molecular and Cellular Biology, 30(6), 3970–3980. 10.1128/MCB.00624-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, G. J., Lyons, D. A., Middendorf, I., Friedlander, B., Arana, N., Reyes, T., & Talbot, W. S. (2003). Submission and Curation of Gene Expression Data. ZFIN Direct Data Submission. http://zfin.org
- Reinke, H. , & Asher, G. (2019). Crosstalk between metabolism and circadian clocks. Nature Reviews. Molecular Cell Biology, 20(4), 227–241. 10.1038/s41580-018-0096-9 [DOI] [PubMed] [Google Scholar]
- Riley, M. F. , Bochter, M. S. , Wahi, K. , Nuovo, G. J. , & Cole, S. E. (2013). Mir‐125a‐5p‐mediated regulation of Lfng is essential for the avian segmentation clock. Developmental Cell, 24(5), 554–561. 10.1016/j.devcel.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg, R. , Neilson, J. R. , Sarma, A. , Sharp, P. A. , & Burge, C. B. (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science, 320(5883), 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanduja, S. , Blanco, F. F. , & Dixon, D. A. (2011). The roles of TTP and BRF proteins in regulated mRNA decay. WIREs RNA, 2(1), 42–57. 10.1002/wrna.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroter, C. , & Oates, A. C. (2010). Segment number and axial identity in a segmentation clock period mutant. Current Biology, 20(14), 1254–1258. 10.1016/j.cub.2010.05.071 [DOI] [PubMed] [Google Scholar]
- Shifley, E. T. , VanHorn, K. M. , Perez‐Balaguer, A. , Franklin, J. D. , Weinstein, M. , & Cole, S. E. (2008). Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development, 135, 899–908. [DOI] [PubMed] [Google Scholar]
- Shih, N. P. , Francois, P. , Delaune, E. A. , & Amacher, S. L. (2015). Dynamics of the slowing segmentation clock reveal alternating two‐segment periodicity. Development, 142(10), 1785–1793. 10.1242/dev.119057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo, H. , Isomura, A. , Ohtsuka, T. , Kori, H. , Miyachi, H. , & Kageyama, R. (2016). Oscillatory control of Delta‐like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes & Development, 30(1), 102–116. 10.1101/gad.270785.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo, H. , Ohtsuka, T. , & Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron, 58(1), 52–64. 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Shyu, A. , Belasco, J. , & Greenberg, M. (1991). Two distinct destabilizing elements in the c‐fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & Development, 5(2), 221–331. 10.1101/gad.5.2.221 [DOI] [PubMed] [Google Scholar]
- Siegel, D. A. , Le Tonqueze, O. , Biton, A. , Zaitlen, N. , & Erle, D. J. (2022). Massively parallel analysis of human 3′ UTRs reveals that AU‐rich element length and registration predict mRNA destabilization. G3, 12(1), jkab404. 10.1093/g3journal/jkab404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemen, H. , Colas, D. , Heller, H. C. , Brustle, O. , & Pera, R. A. (2011). Pumilio‐2 function in the mouse nervous system. PLoS One, 6(10), e25932. 10.1371/journal.pone.0025932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, J. , & Axmann, I. M. (2019). The Kai‐protein clock‐keeping track of cyanobacteria's daily life. Sub‐Cellular Biochemistry, 93, 359–391. 10.1007/978-3-030-28151-9_12 [DOI] [PubMed] [Google Scholar]
- Soroldoni, D. , Jorg, D. J. , Morelli, L. G. , Richmond, D. L. , Schindelin, J. , Julicher, F. , & Oates, A. C. (2014). Genetic oscillations. A Doppler effect in embryonic pattern formation. Science, 345(6193), 222–225. 10.1126/science.1253089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto, X. , Biga, V. , Kursawe, J. , Lea, R. , Doostdar, P. , Thomas, R. , & Papalopulu, N. (2020). Dynamic properties of noise and Her6 levels are optimized by miR‐9, allowing the decoding of the Her6 oscillator. The EMBO Journal, 39(12), e103558. 10.15252/embj.2019103558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soza‐Ried, C. , Ozturk, E. , Ish‐Horowicz, D. , & Lewis, J. (2014). Pulses of notch activation synchronise oscillating somite cells and entrain the zebrafish segmentation clock. Development, 141(8), 1780–1788. 10.1242/dev.102111 [DOI] [PubMed] [Google Scholar]
- Sparrow, D. B. , Chapman, G. , Wouters, M. A. , Whittock, N. V. , Ellard, S. , Fatkin, D. , Turnpenny, P. D. , Kusumi, K. , Sillence, D. , & Dunwoodie, S. L. (2006). Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. American Journal of Human Genetics, 78(1), 28–37. 10.1086/498879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow, D. B. , Guillen‐Navarro, E. , Fatkin, D. , & Dunwoodie, S. L. (2008). Mutation of hairy‐and‐enhancer‐of‐Split‐7 in humans causes spondylocostal dysostosis. Human Molecular Genetics, 17(23), 3761–3766. 10.1093/hmg/ddn272 [DOI] [PubMed] [Google Scholar]
- Steiger, M. , Carr‐Schmid, A. , Schwartz, D. C. , Kiledjian, M. , & Parker, R. (2003). Analysis of recombinant yeast decapping enzyme. RNA, 9(2), 231–238. 10.1261/rna.2151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo, D. J. , Broxmeyer, H. E. , Ward, T. , Cooper, S. , Hangoc, G. , Chung, Y. J. , Shelley, W. C. , Richfield, E. K. , Ray, M. K. , Yoder, M. C. , Aplan, P. D. , & Blackshear, P. J. (2009). Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA‐binding protein, results in defective hematopoiesis. Blood, 114(12), 2401–2410. 10.1182/blood-2009-04-214619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima, Y. , Ohtsuka, T. , González, A. , Miyachi, H. , & Kageyama, R. (2011). Intronic delay is essential for oscillatory expression in the segmentation clock. Proceedings of the National Academy of Sciences of the United States of America, 108(8), 3300–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke, C. , & Campos‐Ortega, J. A. (1999). her1, a zebrafish pair‐rule like gene, acts downstream of notch signalling to control somite development. Development, 126(13), 3005–3014. [DOI] [PubMed] [Google Scholar]
- Tan, S. L. , Ohtsuka, T. , Gonzalez, A. , & Kageyama, R. (2012). MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes to Cells, 17(12), 952–961. 10.1111/gtc.12009 [DOI] [PubMed] [Google Scholar]
- Tietz, K. T. , Gallagher, T. L. , Mannings, M. C. , Morrow, Z. T. , Derr, N. L. , & Amacher, S. L. (2020). Pumilio response and AU‐rich elements drive rapid decay of Pnrc2‐regulated cyclic gene transcripts. Developmental Biology, 462(2), 129–140. 10.1016/j.ydbio.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse, B., Heyer, V., Lux, A., Alunni, V., Degrave, A., Seiliez, I., Kirchner, J., Parkhill, J. ‐P., & Thisse, C. (2004). Spatial and Temporal Expression of the Zebrafish Genome by Large‐Scale In Situ Hybridization Screening. The Zebrafish: Genetics, Genomics, and Informatics. Methods in Cell Biology, 77, 505–519. 10.1016/s0091-679x(04)77027-2 [DOI] [PubMed] [Google Scholar]
- Thisse, B., & Thisse, C. (2004). Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. http://zfin.org
- Treguer, K. , Faucheux, C. , Veschambre, P. , Fedou, S. , Theze, N. , & Thiebaud, P. (2013). Comparative functional analysis of ZFP36 genes during Xenopus development. PLoS One, 8(1), e54550. 10.1371/journal.pone.0054550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten, J. , Schagat, T. L. , Hrit, J. , Weidmann, C. A. , Brumbaugh, J. , Coon, J. J. , & Goldstrohm, A. C. (2012). Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. The Journal of Biological Chemistry, 287(43), 36370–36383. 10.1074/jbc.M112.373522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman, J. M. , Xuan, W. , Beeckman, T. , & Benfey, P. N. (2013). To branch or not to branch: The role of pre‐patterning in lateral root formation. Development, 140(21), 4301–4310. 10.1242/dev.090548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw, N. L. , Cao, W. X. , & Lipshitz, H. D. (2019). The maternal‐to‐zygotic transition revisited. Development, 146(11), dev161471. 10.1242/dev.161471 [DOI] [PubMed] [Google Scholar]
- Vasudevan, S. , & Steitz, J. A. (2007). AU‐rich‐element‐mediated upregulation of translation by FXR1 and Argonaute 2. Cell, 128(6), 1105–1118. 10.1016/j.cell.2007.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejnar, C. E. , Abdel Messih, M. , Takacs, C. M. , Yartseva, V. , Oikonomou, P. , Christiano, R. , Stoeckius, M. , Lau, S. , Lee, M. T. , Beaudoin, J. D. , Musaev, D. , Darwich‐Codore, H. , Walther, T. C. , Tavazoie, S. , Cifuentes, D. , & Giraldez, A. J. (2019). Genome wide analysis of 3′ UTR sequence elements and proteins regulating mRNA stability during maternal‐to‐zygotic transition in zebrafish. Genome Research, 29(7), 1100–1114. 10.1101/gr.245159.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova, I. A. , Tahoe, N. M. , Fan, D. , Larsson, O. , Rattenbacher, B. , Sternjohn, J. R. , Vasdewani, J. , Karypis, G. , Reilly, C. S. , Bitterman, P. B. , & Bohjanen, P. R. (2008). Conserved GU‐rich elements mediate mRNA decay by binding to CUG‐binding protein 1. Molecular Cell, 29(2), 263–270. 10.1016/j.molcel.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi, K. , Friesen, S. , Coppola, V. , & Cole, S. E. (2017). Putative binding sites for mir‐125 family miRNAs in the mouse Lfng 3′ UTR affect transcript expression in the segmentation clock, but mir‐125a‐5p is dispensable for normal somitogenesis. Developmental Dynamics, 246(10), 740–748. 10.1002/dvdy.24552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G., & Klein, A. M. (2018). Single‐cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science, 360(6392), 981–987. 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, A. B. , Lengyel, I. M. , Jorg, D. J. , Valentin, G. , Julicher, F. , Morelli, L. G. , & Oates, A. C. (2016). Persistence, period and precision of autonomous cellular oscillators from the zebrafish segmentation clock. eLife, 5, e08438. 10.7554/eLife.08438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, A. B. , Soroldoni, D. , Oswald, A. , Schindelin, J. , & Oates, A. C. (2014). Generation of dispersed presomitic mesoderm cell cultures for imaging of the zebrafish segmentation clock in single cells. Journal of Visualized Experiments, (89), 50307. 10.3791/50307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann, C. A. , Raynard, N. A. , Blewett, N. H. , Van Etten, J. , & Goldstrohm, A. C. (2014). The RNA binding domain of Pumilio antagonizes poly‐adenosine binding protein and accelerates deadenylation. RNA, 20(8), 1298–1319. 10.1261/rna.046029.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R. J. , Collins, J. E. , Sealy, I. M. , Wali, N. , Dooley, C. M. , Digby, Z. , Stemple, D. L. , Murphy, D. N. , Billis, K. , Hourlier, T. , Fullgrabe, A. , Davis, M. P. , Enright, A. J. , & Busch‐Nentwich, E. M. (2017). A high‐resolution mRNA expression time course of embryonic development in zebrafish. eLife, 6, e30860. 10.7554/eLife.30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , & Bartel, D. P. (2017). Widespread influence of 3′‐end structures on mammalian mRNA processing and stability. Cell, 169(5), 905–917.e911. 10.1016/j.cell.2017.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, A. , Chang, T. C. , Yamashita, Y. , Zhu, W. , Zhong, Z. , Chen, C. Y. , & Shyu, A. B. (2005). Concerted action of poly (A) nucleases and decapping enzyme in mammalian mRNA turnover. Nature Structural & Molecular Biology, 12(12), 1054–1063. 10.1038/nsmb1016 [DOI] [PubMed] [Google Scholar]
- Yoshioka‐Kobayashi, K. , Matsumiya, M. , Niino, Y. , Isomura, A. , Kori, H. , Miyawaki, A. , & Kageyama, R. (2020). Coupling delay controls synchronized oscillation in the segmentation clock. Nature, 580(7801), 119–123. 10.1038/s41586-019-1882-z [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Chen, D. , Xia, J. , Han, W. , Cui, X. , Neuenkirchen, N. , Hermes, G. , Sestan, N. , & Lin, H. (2017). Post‐transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes & Development, 31(13), 1354–1369. 10.1101/gad.298752.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Lahens, N. F. , Ballance, H. I. , Hughes, M. E. , & Hogenesch, J. B. (2014). A circadian gene expression atlas in mammals: Implications for biology and medicine. Proceedings of the National Academy of Sciences of the United States of America, 111(45), 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Ransom, J. F. , Li, A. , Vedantham, V. , von Drehle, M. , Muth, A. N. , Tsuchihashi, T. , McManus, M. T. , Schwartz, R. J. , & Srivastava, D. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA‐1‐2. Cell, 129(2), 303–317. 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
- Zheng, D. , Ezzeddine, N. , Chen, C. Y. A. , Zhu, W. , He, X. , & Shyu, A. B. (2008). Deadenylation is prerequisite for P‐body formation and mRNA decay in mammalian cells. The Journal of Cell Biology, 182(1), 89–101. 10.1083/jcb.200801196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


