Abstract
Protein-carbohydrate interactions play important roles in various biological processes such as organism development, cancer metastasis, pathogen infection, and immune response, but they remain challenging to study and exploit due to their low binding affinity and noncovalent nature. Here we site-specifically engineered covalent linkages between proteins and carbohydrates under biocompatible conditions. We show that sulfonyl fluoride reacts with glycans via proximity-enabled reactivity, and to harness this a bioreactive unnatural amino acid (SFY) containing sulfonyl fluoride was genetically encoded into proteins. SFY-incorporated Siglec-7 cross-linked with its sialoglycan ligand specifically in vitro and on the surface of cancer cells. Through irreversible cloaking of sialoglycan at the cancer cell surface, SFY-incorporated Sigelc-7 enhanced the killing of cancer cells by natural killer cells. Genetically encoding the chemical cross-linking of proteins to carbohydrates (GECX-sugar) offers a solution to address the low affinity and weak strength of protein-sugar interactions.
Graphical Abstract
Editorial Summary:
Protein-carbohydrate interactions remain challenging to study due to their low binding affinity and noncovalent nature. Now, a genetically encoded bioreactive unnatural amino acid containing sulfonyl fluoride has been shown to cross-link a protein with its bound glycan, offering a solution to probe and exploit protein-carbohydrate interactions.
Introduction
Interactions between carbohydrates and proteins play important biological roles in living systems. Proteins interact with carbohydrates of glycoproteins, glycolipids, and polysaccharides presented on the cell surface to influence biological activity and recognition.1 Protein-carbohydrate interactions are involved in a broad range of biology processes, such as cell-cell communication, organism development, tumor cell metastasis, bacteria and virus invasion, and immune response.2, 3, 4 Despite a central role for molecular encounters, protein-carbohydrate interactions are challenging to study due to their dynamic nature, transient interaction, and often large number of interacting partners involved.5 Carbohydrate structure is not genetically encoded, making it not amenable to common genetic techniques and difficult to achieve monosaccharide specificity. Another salient feature adding to the difficulty is the generally low affinity of the single protein-carbohydrate interaction, with equilibrium dissociation constant Kd most often in the millimolar and some in micromolar range.1, 6 Thus it has been extremely difficult to generate high affinity protein binders for carbohydrates. In particular, many cancer cells overexpress distinct surface glycans and pathogenic microbes are covered with glycans not found in eukaryotic cells, but anti-glycan antibodies with high affinity and specificity remain lacking.4, 7, 8 Therefore, to facilitate basic research of glycobiology and to exploit glycan-targeted diagnosis and therapy,9 an effective approach is highly desired to increase the affinity of proteins for carbohydrates and to stabilize their transient interactions specifically.
The ability to covalently cross-link proteins with carbohydrates under mild cellular and in vivo settings would offer a unique solution to these challenges. Recent success in creating covalent linkages between proteins in vivo has enabled the capture of elusive protein-protein interactions, bioconjugation of amine reagents to proteins, as well as the development of covalent protein drugs for cancer immunotherapy.10, 11, 12, 13, 14, 15 However, carbohydrates contain mainly the weak nucleophilic hydroxyl group, which are difficult to react with under mild aqueous conditions. Unlike amino acid residues of proteins, many of which have distinct functional groups in their side chains to distinguish, the functional groups of monosaccharides are more or less the same. Efficient differentiation between the multiple hydroxyl groups of an unprotected carbohydrate is also difficult without enzyme catalysis.16 These chemical features of carbohydrates make them challenging not only to synthesize but also to selectively target with biocompatible chemistry.17
Sialic acid–binding immunoglobulin-like lectin 7 (Siglec-7) is an inhibitory transmembrane receptor mainly expressed on human natural killer (NK) cells.18 Siglec-7 recognizes sialic acid via its extracellular V-set immunoglobulin domain and signals through its cytosolic immunoreceptor tyrosine-based inhibitory motif (ITIM) to attenuate NK cell activation.19 The preferred carbohydrate ligand for Siglec-7 is Neu5Acα2–8Neu5Ac-containing glycans with generally low binding affinity.20, 21 Siglec-7 natively contributes to the NK cell discrimination between self and non-self, but some pathogens and cancers can up-regulate sialoglycan to evade immune surveillance and NK cell-mediated killing.22 Strategies to block such exploitation would potentially be valuable for developing glycan-based immunotherapy.
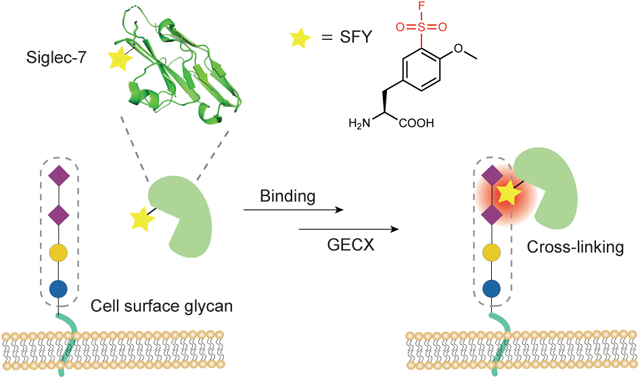
Here we developed a biocompatible method, genetically encoded chemical cross-linking of proteins with sugar (GECX-sugar), to generate covalent linkages between proteins and carbohydrates with residue specificity. We identified that sulfonyl fluoride was able to cross-link sugar via proximity-enabled reactivity, and genetically encoded into proteins a unique latent bioreactive unnatural amino acid (Uaa) SFY bearing the sulfonyl fluoride. The SFY-incorporated Siglec-7 covalently and specifically cross-linked its substrate sialoglycan in vitro and on cancer cell surface. Moreover, through covalent binding with sialoglycan on cancer cell surface, SFY-incorporated Siglec-7 enhanced the killing of cancer cells by NK cells. The site-specific covalent linkage between protein and carbohydrate enabled by GECX-sugar will facilitate the study of protein-carbohydrate interactions and open innovative avenues for engineering covalent protein-glycan complex for research and therapeutic purposes.
Results
Identification of sulfonyl fluoride to cross-link carbohydrate using plant-and-cast cross-linkers
It is challenging to covalently target carbohydrate under mild physiological conditions. The dominant functional group of carbohydrates, the hydroxyl group, is a weak nucleophile and difficult to chemically differentiate from water. Recently we have succeeded in covalently targeting amino acid side chains in vivo via proximity-enabled reactivity,23 so we expect that carbohydrate could also be covalently targeted via this mechanism.
To identify functional groups that react with carbohydrate with reactivity driven by proximity effect, we developed a strategy involving the use of plant-and-cast small molecule cross-linkers to cross-link protein-carbohydrate complex (Fig. 1a). The plant-and-cast small molecule cross-linkers contain a highly reactive succinimide ester, which reacts with Lys side chain and plants the cross-linker on proteins.24 The other end of the cross-linker contains a less reactive functional group and is cast to react with nearby functional groups of protein or carbohydrate via proximity-enabled reactivity. The covalent cross-link of protein with the bound carbohydrate can be readily determined with Western blot under denatured conditions, indicating that the test functional group on the cross-linker is able to react with carbohydrate.
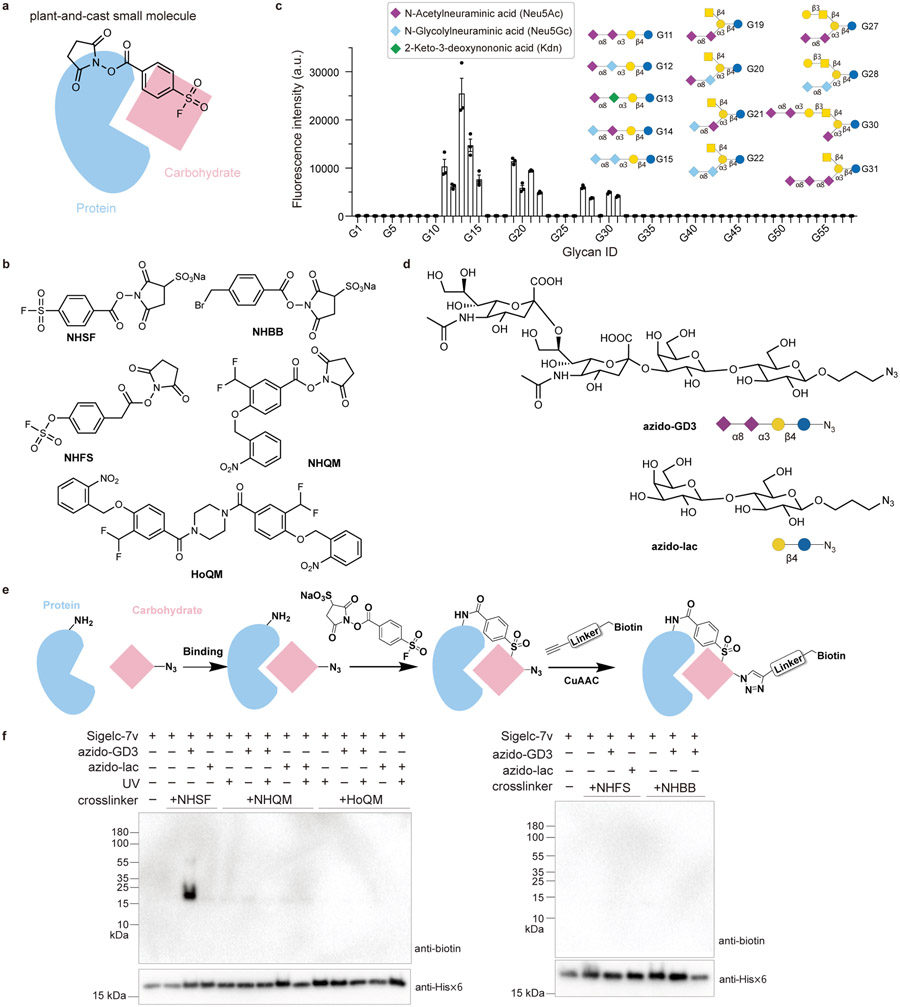
Fig. 1. Sulfonyl fluoride was identified as a suitable functional group for cross-linking carbohydrate through proximity-enabled reactivity using a strategy involving plant-and-cast small molecule cross-linkers.
a) Scheme of the strategy: when the plant-and-cast small molecule cross-linker is added to protein-carbohydrate complex, the succinimide ester of the cross-linker reacts rapidly with Lys sidechains of the protein, placing the less reactive test functionality in close proximity to carbohydrate. If the functionality reacts with carbohydrate driven by proximity-enabled reactivity, the carbohydrate will be covalently cross-linked to the protein for detection. b) Chemical structures of five cross-linkers tested to cross-link protein with carbohydrate. c) Binding analysis of the refolded Siglec-7v with the glycosphingolipid glycan microarray, confirming Siglec-7v binding with the Neu5Acα2–8Neu5Ac-terminating glycan ligands. n=3 independent samples. The bar height and error bar represent mean ± SEM. d) Chemical structures of azido-GD3 for binding with Siglec-7v and the negative control azido-lac. e) Scheme showing the cross-linking and detection procedures. Siglec-7v was incubated with azido-GD3 for binding, after which the cross-linker was added to cross-link. Biotin was subsequently appended onto azido-GD3 via click chemistry for detection of the cross-linked GD3. f) Among the five tested cross-linkers, only NHSF cross-linked Siglec-7v with azido-GD3.
Based on our experience with targeting amino acid side chains via proximity-enabled reactivity, we designed and synthesized five plant-and-cast cross-linkers containing sulfonyl fluoride,24,25 benzyl bromide,26, 27, 28 fluorosulfate,29 and photocaged quinone methide (QM),30, 31 respectively (Fig. 1b). Functional groups of NHSF, NHBB, and NHFS are relatively inert but can react with nucleophilic side chains of Lys, His, or Tyr in close proximity. NHQM, upon photo-activation, generates QM, which is able to react with nine nucleophilic amino acid side chains including the very weak ones of Gln and Asn.30 The fifth cross-linker, HoQM, was also tested since it has photocage QM at both ends,32 which may increase the planting sites on protein and thus casting more QM to the carbohydrate for reaction.
To cross-link protein-carbohydrate interactions, we chose to work with Siglec-7, a transmembrane receptor expressed on human immune cells to regulate immune function through recognizing sialoglycans. We cloned and expressed the extracellular, sialoglycan binding V-set domain of Siglec-7 in E. coli, referred to as Siglec-7v hereafter. The Siglec-7v was purified from inclusion bodies in high concentrations of guanidine and refolded using step-wise dialysis. The intact Siglec-7v was analyzed with electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). A major peak was observed at 15937.5 Da, corresponding to intact Siglec-7v with disulfide bond formed (Supplementary Fig. 1). A glycosphingolipid glycan microarray featuring 58 glycan epitopes was used for binding analysis of the refolded Siglec-7v (Supplementary Fig. 2). Consistent with the commercially available human Siglec-7v expressed from mouse cells (Supplementary Fig. 3), the refolded Siglec-7v bound with glycan ligands containing terminal 2,8-linked sialic acid only, and not with other glycan ligands lacking terminal sialic acid (Fig. 1c, Supplementary Table 2). These results confirmed that the Siglec-7v expressed as inclusion bodies in E. coli was properly refolded and had the correct binding properties.
From the binding assay of Siglec-7v, we chose G11 as the model glycan for Siglec-7v cross-linking study. G11 was called GD3 ganglioside sugar, which is a tumor-associated carbohydrate antigen.33,34, 35 To facilitate the detection of GD3, we added an azido group at the lactose terminal, referred to as azido-GD3 (Fig. 1d), which was synthesized with a chemo-enzymatic method (Supplementary Fig. 4), purified using HPLC, and verified with ESI-MS (Supplementary Fig. 5). As a negative control for binding with Siglec-7v, an azido-lactose (azido-lac) was also synthesized, which lacked the two terminal Neu5Acα2–8Neu5Ac compared with azido-GD3 (Fig. 1d).
We then investigated if the various functional groups synthesized onto the cross-linkers could achieve protein-carbohydrate cross-linking using Siglec-7v and azido-GD3 as the model system. Siglec-7v (60 μM) was incubated with 2 mM azido-GD3 for 1 h at room temperature to allow binding, and each cross-linker was subsequently added to the solution to crosslink (Fig. 1e). For samples treated with cross-linker NHQM or HoQM, UV light was applied to activate the cross-linker. Alkyne-biotin was subsequently clicked with azido-GD3 in the samples, and the biotin signal was detected with Western blot using streptavidin-HRP. A strong band was clearly detected only in samples treated with NHSF (Fig. 1f), but not with other four cross-linkers, suggesting that NHSF crosslinked Siglec-7v with azido-GD3. We have previously demonstrated that NHSF could form cross-links between Lys residues and His, Ser, Thr, Tyr, and Lys side chains within and between proteins.24 The results here further suggest that NHSF could be applied to cross-link protein-carbohydrate interactions via the sulfonyl fluoride group.
NHSF covalently targets specific protein-carbohydrate interactions via proximity-enabled reactivity
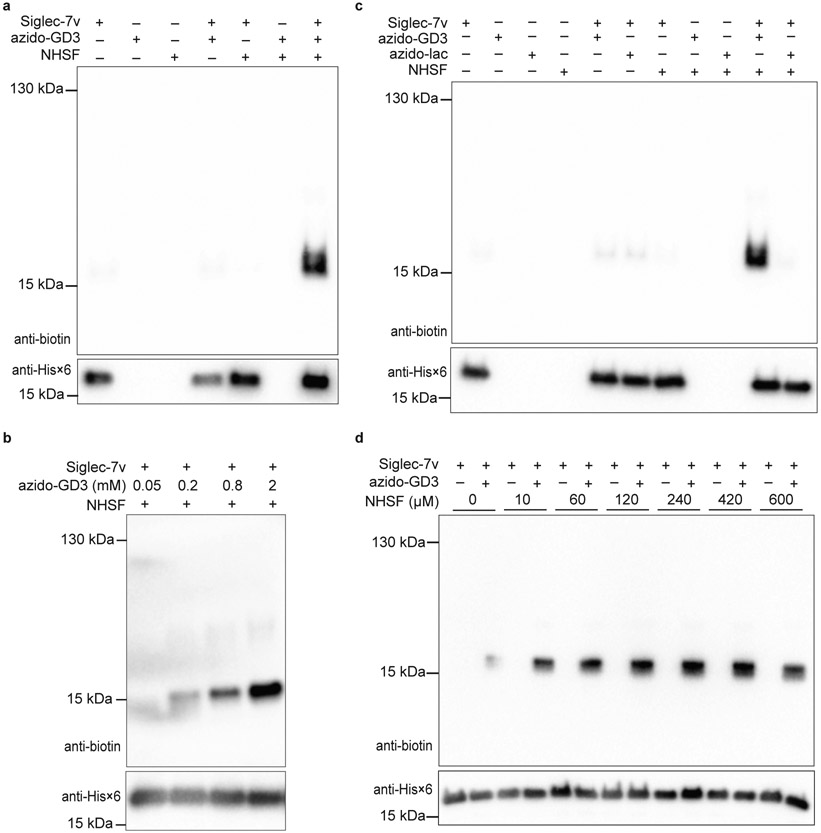
To study the specificity of NHSF-mediated carbohydrate cross-linking, we investigated the cross-linking of Siglec-7v with azido-GD3 by NHSF in more detail. The cross-linking band was observed only when Siglec-7v, azido-GD3, and NHSF were all present; No cross-linking was detected with either one or two components withdrawn from incubation (Fig. 2a). The minimum azido-GD3 concentration for cross-linking with 60 μM Siglec-7v was around 0.8 mM (Fig. 2b), consistent with the reported low binding affinity of Siglec-7 for sialoglycan. In addition, when the negative control glycan ligand, azido-lac, was used in place of azido-GD3, the cross-linking band with Siglec-7v was no longer detected (Fig. 2c). These results indicated that only when Siglec-7v bound with its glycan substrate would the cross-linking by NHSF occur. Therefore, NHSF cross-linked specific protein-carbohydrate interactions, presumably because sulfonyl fluoride is a relatively weak electrophile and protein-carbohydrate interactions, which put the sulfonyl fluoride and carbohydrate in close proximity, triggered the cross-linking reaction. Various concentrations of NHSF ranging from 10 μM to 600 μM could enable the specific cross-linking of Siglec-7v with azido-GD3 (Fig. 2d), and cross-linking efficiency was dependent on NHSF concentration applied.
Fig. 2. Cross-linking of Siglec-7v with azido-GD3 by NHSF was dependent on concentration and the specific protein-carbohydrate binding.
Top panels are western blots for GD3 via detection of biotin; bottom panels are western blots for Siglec-7v via detection of its C-terminal Hisx6 tag. a) Cross-linking was dependent on the presence of Siglec-7v, azido-GD3, and NHSF. b) Cross-linking was dependent on the concentration of azido-GD3. c) Cross-linking was dependent on the specific binding of azido-GD3 to Siglec-7v. d) Cross-linking was dependent on the concentration of NHSF. Faint background bands in the anti-biotin blots were due to low level reaction of alkyne-biotin with protein Siglec-7v nonspecifically, a common background when using azide-alkyne for click labeling as previously reported.36,37
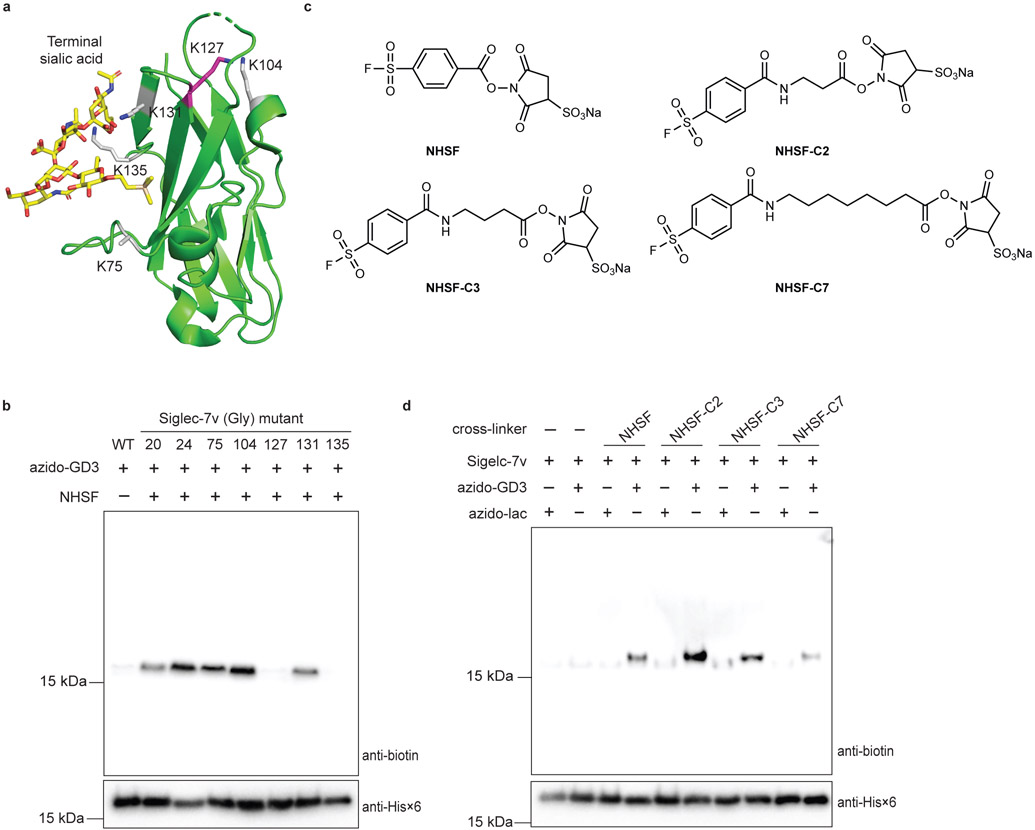
To assess if NHSF cross-linking of carbohydrate was distance dependent, we first determined which Lys residue of Siglec-7v was NHSF planted on via the succinimide ester to cross-link azido-GD3. We individually mutated all Lys residues on the Siglec-7v to Gly (Fig. 3a),38 and tested cross-linking of these Siglec-7v mutants with azido-GD3 by NHSF. Mutation of Lys to Gly at the right site would abolish the NHSF planting on Siglec-7v to reach and cross-link with azido-GD3. Mutations at sites Lys20, 24, 75, 104, or 129 did not affect cross-linking of azido-GD3 to the Siglec-7v, while mutations at Lys127 and Lys135 showed negligible cross-linking (Fig. 3b). Previously, Lys135 has been shown to directly interact with the glycan ligand via hydrogen bonding, and mutation of Lys135 to Ala abolishes glycan binding to Siglec-7.38 Our results of Siglec-7v(Lys135Gly) mutant consistently confirm the direct binding role of Lys135 and further support that NHSF cross-linking of azido-GD3 is dependent on specific binding of the glycan to the receptor. Excluding Lys135, Lys127 thus should be the site on which NHSF was planted to cross-link azido-GD3. NHSF has been shown to cross-link Cα-Cα of Lys and Ser on bovine serum albumin at a distance of 8.9 - 21.2 Å.24 Consistently, Lys127 on Siglec-7v showed reachable distance to the terminal sialic acid, which was about 14 Å (Fig. 3a). Therefore, these results indicate that NHSF cross-linked azido-GD3 in close distance via proximity-enabled reactivity.
Fig. 3. Cross-linking site on Siglec-7v and distance dependence of the cross-linker indicate that sulfonyl fluoride of NHSF reacted with carbohydrate via proximity-enabled reactivity.
a) Crystal structure of Siglec-7v binding with α(2,8)-disialygangioside GT1b (PDB: 2HRL). NHSF cross-linking site, Lys127, on Siglec-7v is shown in magenta stick. All other Lys sites are shown in grey stick. b) NHSF cross-linking of azido-GD3 with Siglec-7v Lys to Gly mutants. c) Structures of NHSF analogs with different linker lengths. d) Cross-linking of Siglec-7v with azido-GD3 by the NHSF analogs. Faint background bands in the anti-biotin blots were due to low level reaction of alkyne-biotin with protein Siglec-7v nonspecifically, a common background when using azide-alkyne for click labeling as previously reported.36,37
To further evaluate distance dependence of NHSF cross-linking and to optimize cross-linking efficiency, we next altered the length of the cross-linker. Three additional analogs of NHSF were synthesized with increasing numbers of methylene in the linker to study the effects of linker length and flexibility on the cross-linking efficiency (Fig. 3c). Siglec-7v was incubated with either azido-GD3 or azido-lac with different cross-linkers. Western blot analysis of the incubation products showed that NHSF-C2 increased the cross-linking band intensity 1.5-fold in comparison with NHSF (Fig. 3d). However, further increase of the linker length in NHSF-C3 and NHSF-C7 lowered the cross-linking efficiency. These results further corroborate the distance dependence and proximity-driven nature of NHSF cross-linking of azido-GD3.
Design and genetic incorporation of SFY into proteins in E. coli
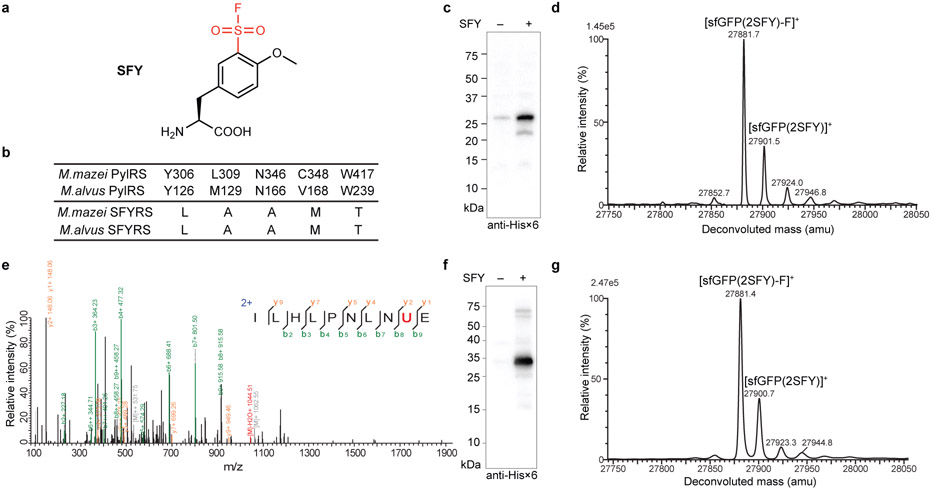
To introduce the identified sulfonyl fluoride group into proteins, we designed and synthesized the unnatural amino acid (Uaa) o-sulfonyl fluoride-O-methyltyrosine (SFY) (Fig. 4a). The sulfonyl fluoride group was placed at the meta rather than the para position of the phenyl ring, because we previously found a functional group introduced at the meta position has larger reaction area than at the para position possibly due to the rotation of the phenyl ring.25 The methoxy group was included to reduce the reactivity of sulfonyl fluoride, which would avoid potential cytotoxicity and increase reaction specificity. SFY contains sulfonyl fluoride, which is more reactive than fluorosulfate in the previously genetically encoded latent bioreactive Uaa fluorosulfate-L-tyrosine (FSY).29
Fig. 4. Genetic incorporation of SFY into proteins in E. coli.
a) Structure of SFY. b) Amino acid sequences of the evolved MmSFYRS, MaSFYRS, and the corresponding WT PylRS. c) Western blot analysis of SFY incorporation into sfGFP(2TAG) by Mm-tRNAPyl/MmSFYRS pair. d) ESI-TOF MS spectrum of intact sfGFP(2SFY) protein expressed by Mm-tRNAPyl/MmSFYRS pair. e) Tandem MS spectrum of Z(24SFY) expressed by Mm-tRNAPyl/MmSFYRS pair. U represents SFY. f) Western blot analysis of SFY incorporation into sfGFP(2TAG) by Ma-tRNAPyl/MaSFYRS pair. g) ESI-TOF MS spectrum of intact sfGFP(2SFY) protein expressed by Ma-tRNAPyl/MaSFYRS pair.
We next evolved a mutant pyrrolysyl-tRNA synthetase (PylRS) specific for SFY to genetically incorporate it into proteins. A PylRS mutant library was generated by mutating residues Ala302, Leu305, Tyr306, Leu309, Ile322, Asn346, Cys348, Tyr384, Val401, and Trp417 of the Methanosarcina mazei PylRS using the small-intelligent mutagenesis approach,39 and subjected to selection as described.40, 41 A hit showing SFY-dependent phenotype was identified, which contained the following mutations (306L/309A/346A/348M/417T) and was named as MmSFYRS (Fig. 4b).
To evaluate the incorporation specificity of SFY into proteins in E. coli, we expressed the superfold green fluorescent protein (sfGFP) gene containing a TAG codon at position 2 (sfGFP-2TAG) with the tRNAPyl/MmSFYRS pair in E. coli. When 1 mM SFY was added in growth media, full-length sfGFP(2SFY) was significantly produced (Fig. 4c). The purified sfGFP(2SFY) was analyzed with ESI-TOF MS (Fig. 4d). A peak observed at 27901.5 corresponds to intact sfGFP containing SFY at site 2 (expected 27900.9 Da). Another peak measured at 27881.7 Da corresponds to sfGFP(2SFY) lacking F (expected 27881.9 Da), suggesting some F elimination during MS measurement, which has also been observed for fluorosulfate-L-tyrosine (FSY) incorporated into proteins.29 No peaks corresponding to sfGFP containing other amino acids at site 2 were observed. We also incorporated SFY at position 24 of the Z protein in E. coli, and analyzed the purified Z protein with tandem MS (Fig. 4e). A series of b and y ions unambiguously indicate that SFY was incorporated at site 24 of the Z protein. These results indicate that the tRNAPyl/MmSFYRS pair incorporated SFY into proteins with high specificity in E. coli.
We further transplanted the mutations of MmSFYRS into Methanomethylophilus alvus PylRS to generate MaSFYRS, as transplanting mutations from Methanosarcina barkeri PylRS mutant into M. alvus PylRS has been shown to increase the Uaa incorporation efficiency.31 Expression of sfGFP-2TAG gene together with the Ma-tRNAPyl/MaSFYRS pair in E. coli also showed SFY dependent production of full-length sfGFP (Fig. 4f). ESI-TOF MS analysis of the purified sfGFP(2SFY) yielded similar peaks as observed in Fig. 4d, confirming that Ma-tRNAPyl/MaSFYRS also had high specificity in incorporating SFY into proteins in E. coli (Fig. 4g). To compare incorporation efficiency, we then incorporated SFY into GFP at permissive sites 2, 40, and 182, respectively, using the Ma-tRNAPyl/MaSFYRS or the Mm-tRNAPyl/MmSFYRS pair. Through quantifying the fluorescence intensity of the expressed GFP in E. coli cells, we found that the Ma-tRNAPyl/MaSFYRS increased the SFY incorporation efficiency in E. coli over the Mm-tRNAPyl/MmSFYRS more than 10-fold, reaching a yield of over 50 mg/L GFP (Supplementary Fig. 6). The Ma-tRNAPyl/MaSFYRS pair was thus used in subsequent experiments.
Siglec-7v(SFY) enables cross-linking of sialoglycan in vitro and on mammalian cell surface
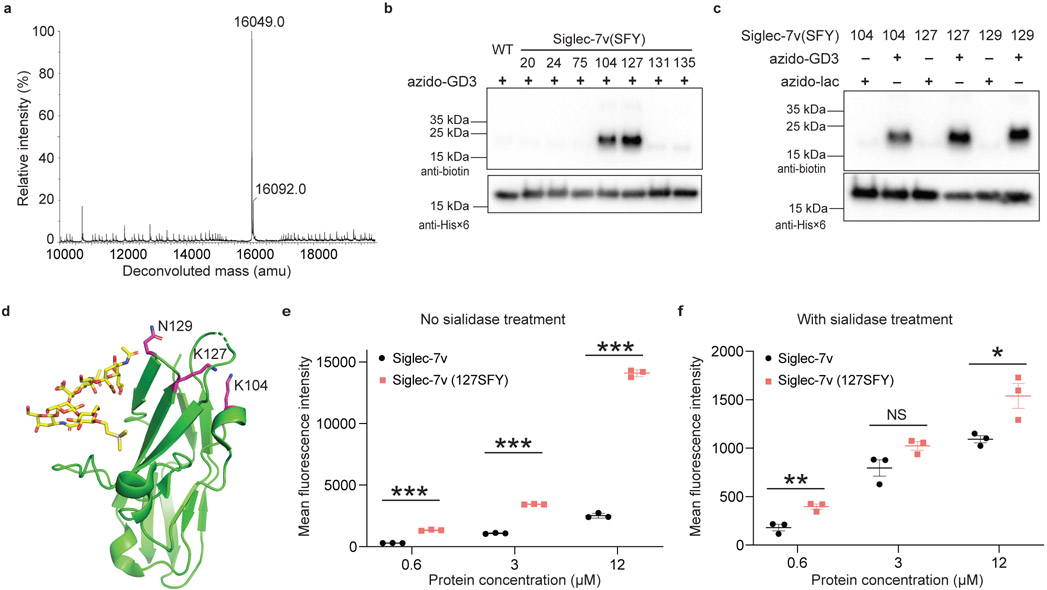
To prepare SFY incorporated Siglec-7v proteins, we expressed Siglec-7v(104TAG) gene with the Ma-tRNAPyl/MaSFYRS in E. coli in the presence of 1 mM of SFY. The expressed Siglec-7v(104SFY) protein was purified and refolded similarly as WT Siglec-7v. The Siglec-7v(104SFY) protein was produced with a yield of 5 mg/L, and the WT Siglec-7v yielded 20 mg/L. The intact mass of the purified Siglec-7v(104SFY) was analyzed with ESI-TOF MS (Fig. 5a). A major peak was measured at 16049.0 Da, corresponding to the intact Siglec-7v(104SFY) protein lacking F (expected 16049.6 Da), suggesting loss of F during MS measurement.
Fig. 5. Sigelc-7(SFY) cross-linked with azido-GD3 in vitro and with sialoglycan on cell surface.
a) ESI-MS spectrum of intact Siglec-7v(104SFY) confirmed SFY incorporation. b) Cross-linking of azido-GD3 with Siglec-7v with SFY incorporated at indicated Lys sites. c) Sigelc-7(SFY) cross-linked with azido-GD3 but not azido-lac. d) Crystal structure of Siglec-7v in complex of GT1b (PDB: 2HRL), showing Lys104, Lys127, and Gln129 in magenta stick, at which SFY incorporation led to cross-linking of azido-GD3. e-f) Flow cytometric quantification of Siglec-7v protein bound on SK-MEL-28 cell surface. After washing, more Siglec-7v(127SFY) bound with sialoglycan on SK-MEL-28 cell surface than WT Siglec-7v (e). When the cells were pretreated with sialidase to remove cell surface sialoglycan (f), the binding markedly decreased and the difference became small or nonsignificant. The line and error bar represent mean ± SEM; n = 3 independent batches of Siglec-7v proteins. * p < 0.05; ** p < 0.01, *** p < 0.001; NS, not significant; two-sided t test. P values in panel e: 0.000002 for 0.6 μM, <0.000001 for 3 μM, and <0.000001 for 12 μM. P values in panel f: 0.0069 for 0.6 μM, 0.074 for 3 μM, and 0.029 for 12 μM.
We then determined if SFY could enable Siglec-7v to cross-link the bound glycan ligand. Inspired by NHSF-mediated cross-linking of azido-GD3 through Lys on Siglec-7v protein, we incorporated SFY into individual Lys sites of Siglec-7v, including sites 20, 24, 75, 104, 127, 131, and 135. Each SFY incorporated Siglec-7v mutant was incubated with 2 mM azido-GD3 for 1 h at room temperature, followed with click labeling of alkyne-biotin and Western blot analysis using streptavidin-HRP (Fig. 5b). Cross-linking of azido-GD3 was detected for Siglec-7v(104SFY) and Siglec-7v(127SFY). Since site 127 showed positive cross-linking of azido-GD3 in both NHSF- and SFY-mediated cross-linking, we also tested its adjacent site 129. Indeed, Siglec-7v(129SFY) also cross-linked azido-GD3 (Fig. 5c). These three siglec-7v mutants (104SFY, 127SFY, and 129SFY) all did not crosslink the control ligand azido-lac, which lacked the two terminal Neu5Acα2–8Neu5Ac (Fig. 5c). Moreover, addition of 3’-sialyllactose, another control ligand lacking the very terminal Neu5Ac, did not reduce the crosslinking of siglec-7v(127SFY) with azido-GD3 (Supplementary Fig. 7). On the basis of the crystal structure of Siglec-7 in complex with α(2,8)-disialylated ganglioside GT1b,38 sites 104, 127, and 129 are in close proximity to the terminal sialic acid Neu5Ac of the bound GT1b, accounting for the observed SFY cross-linking of azido-GD3 (Fig. 5d). These results indicate that Siglec-7v mutants with SFY incorporated at the appropriate sites were able to cross-link the bound carbohydrate in a proximity based manner.
We further explored if SFY incorporated Siglec-7v could cross-link sialoglycan on mammalian cell surface. SK-MEL-28 is a human melanoma cell line with a high level of sialylation on cell surface.42 We incubated SK-MEL-28 cells with different concentrations of WT Siglec-7v or Siglec-7v(127SFY), followed with washing. Siglec-7v proteins bound to cell surface were stained with a fluorescently labeled antibody specific for the Hisx6 tag appended at the C-terminus of Siglec-7v, and quantified with flow cytometry. Remarkably, cells incubated with Siglec-7v(127SFY) showed higher mean fluorescence intensity (MFI) over those incubated with WT Siglec-7v in all protein concentrations tested (Fig. 5e, Supplementary Fig. 8). The cell MFI difference reached 5.6-fold when 12 μM of protein was used. This binding difference suggested that WT Siglec-7v which bound in non-covalent mode would dissociate from cell surface sialoglycan during washing, whereas Siglec-7v(127SFY) could remain bound due to the covalent cross-linking. To confirm that the strong fluorescence signal of Siglec-7v(127SFY) was mainly due to its cross-linking with sialoglycan on cell surface, we pre-treated the cells with sialidase to remove the terminal sialic acids on cell surface,43 which Siglec-7v proteins prefer binding with. The pretreated SK-MEL-28 cells were then incubated with WT Siglec-7v or Siglec-7v(127SFY), washed, and stained for quantification of bound Siglec-7v. Drastically lower MFI were measured for cells incubated with WT Siglec-7v or Siglec-7v (127SFY), and only tiny MFI difference were detected between them (Fig. 5f). Taken together, these results demonstrate that Siglec-7v(127SFY) was able to cross-link the sialoglycan on mammalian cell surface.
Siglec-7v(127SFY) enhances NK cell killing of cancer cells
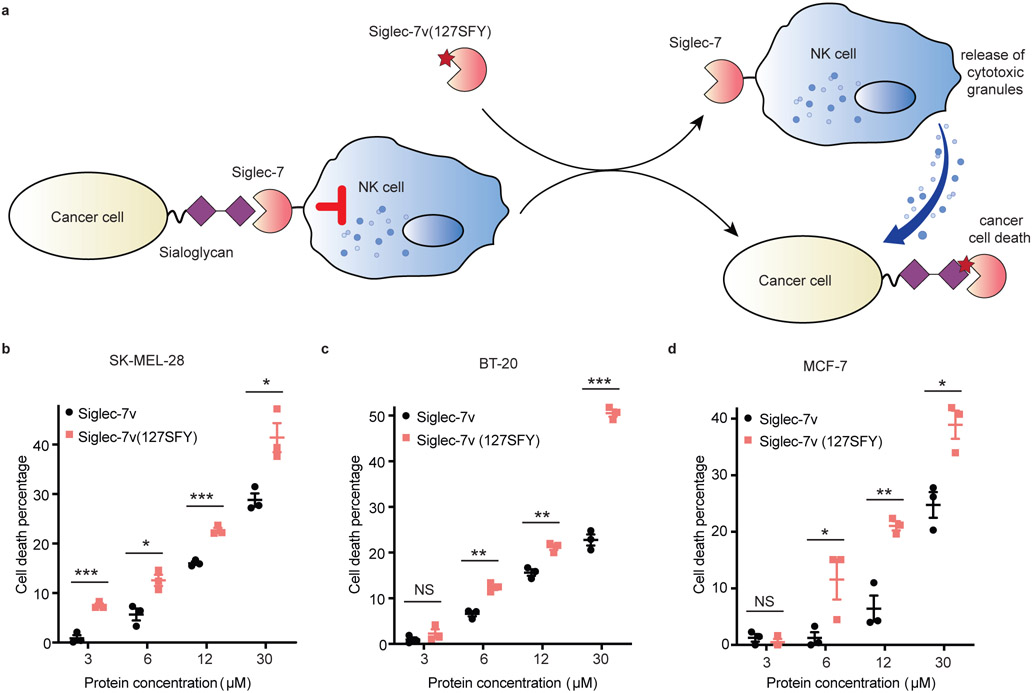
Many tumors upregulate cell surface sialic acids, which bind with Siglec-7 on human NK cells, inhibiting NK cell cytotoxicity and evading immune-surveillance. Since Siglec-7v(127SFY) could irreversibly cross-link with cell surface sialoglycan, we reasoned that it would competitively block the interaction of tumor cell surface sialoglycan with Siglec-7 of NK cells, thus enhancing NK cell killing of tumor cells (Fig. 6a).
Fig. 6. Siglec-7v(SFY) enhanced NK cell killing of cancer cells.
a) Scheme showing the use of Siglec-7v(127SFY) to block the interaction between sialoglycan on tumor cell surface and Siglec-7 of NK cells. Decreasing the inhibitory signal of Siglec-7 on NK cells would enhance NK killing of tumor cells. b-d) Cytotoxicity assay of three hypersialylated cancer cell lines showed that Siglec-7v(127SFY) enhanced NK-92 cell killing over the WT Siglec-7v. The line and error bar represent mean ± SEM; n = 3 independent batches of Siglec-7v proteins. * p < 0.05; ** p < 0.01; *** p < 0.001; NS, not significant; two-sided t test. P values in panel b: 0.00086 for 3 μM, 0.014 for 6 μM, 0.00045 for 12 μM, and 0.018 for 30 μM. P values in panel c: 0.30 for 3 μM, 0.0019 for 6 μM, 0.0040 for 12 μM, and 0.000043 for 30 μM. P values in panel d: 0.44 for 3 μM, 0.049 for 6 μM, 0.0038 for 12 μM, and 0.014 for 30 μM.
To test this hypothesis, we incubated Siglec-7v(127SFY) with three hypersialylated human cancer cell lines, SK-MEL-28 (melanoma), BT-20 (breast carcinoma), and MCF-7 (breast adenocarcinoma),44 respectively for 2 h to allow binding and cross-linking, using WT Siglec-7v as the control. The cells were washed and then subjected to incubation with human NK-92 cells. NK-92 is a cytotoxic human NK cell line that is currently in clinical trials for cancer treatment.45 Cancer cell viability was evaluated with propidium iodide staining and quantified with flow cytometry. The percent of cancer cells killed by NK-92 cells was calculated (Fig. 6b-d). For all three cancer cell lines tested, Siglec-7v(127SFY) enhanced NK-92 killing of cancer cells over WT Siglec-7v in the same concentration. The percent of dead cancer cells increased with the concentration of Siglec-7v applied. Siglec-7v(127SFY) was thus more potent than WT Siglec-7v, requiring the latter in higher concentration to reach similar level of cancer cell killing. These results indicate that covalent binding of Siglec-7v(127SFY) with sialoglycan on cancer cell surface more effectively enhanced NK killing of cancer cells than the WT Siglec-7v.
A major advantage of genetically encoding SFY is the ability to introduce the sulfonyl fluoride into the Siglec-7v protein site specifically. Although sulfonyl fluoride could be installed on Siglec-7v through pretreating Siglec-7v with NHSF, this approach resulted in the installation of sulfonyl fluoride at multiple Lys sites nonselectively. Consequently, NHSF-pretreated Siglec-7v (Siglec-7v-SF) failed to bind with azido-GD3 in vitro (Supplementary Fig. 9a) and with sialoglycan on BT20 or SK-MEL-28 cells (Supplementary Fig. 9b). As expected, NHSF-pretreated Siglec-7v also had no effect in enhancing NK cell killing of cancer cells (Supplementary Fig. 9c). These results indicate the importance of site specificity enabled by genetic encoding.
Discussion
By applying plant-and-cast cross-linkers onto protein-carbohydrate complex, we identified sulfonyl fluoride to react with glycan via proximity-enabled reactivity under mild biocompatible conditions. A unique latent bioreative Uaa SFY bearing sulfonyl fluoride was then designed and genetically incorporated into proteins via genetic code expansion. SFY-incorporated Siglec-7v specifically cross-linked its sialoglycan ligand in vitro and on cancer cell surface. Moreover, through covalently cloaking sialoglyan on cancer cell surface, Siglec-7v(SFY) significantly enhanced NK cell killing of cancer cells over the noncovalent WT Siglec-7v.
Protein-carbohydrate interactions are noncovalent in nature. Through developing the GECX-sugar technology, here we changed this paradigm and enabled the site-specific introduction of covalent linkages into interacting protein-carbohydrate. The latent bioreactive Uaa SFY is genetically encoded into the protein to achieve residue specificity for the covalent linkage. The reaction of SFY with glycan is enabled by the close proximity of SFY side chain to the glycan hydroxyl group when protein binds to glycan. Therefore, through placing SFY into different sites of the protein, monosaccharide selectivity for the bound glycan can also be achieved for the covalent linkage. This site-specificity for both protein and glycan of GECX-sugar will enable the precise engineering of covalent linkages to cross-link protein to the interacting carbohydrate.
The irreversible cross-linking of protein with carbohydrate enables infinite affinity. GECX-sugar thus should provide innovative routes to the generation of glycan-specific binders and to the identification of the weak and transient protein-carbohydrate interactions. SFY reacts with the hydroxyl group of sialic acid due to the SuFEx reactivity of sulfonyl fluoride. As all monosaccharides contain the hydroxyl group, we expect that SFY can be incorporated into other glycan binding proteins to covalently target various glycans. Combining the specificity of glycan binding protein with SuFEx-enabled covalency, such covalent binders can be directedly evolved into artificial glycan antibodies, which have long been lacking for basic glycobiology research. Their direct binding with the glycan product will be particularly useful for monitoring the alteration of protein glycosylation and glycan antigen in cancer research and in the clinical management of cancer patients.4 In addition, carbohydrate-protein interactions are involved in a wide range of biological processes. For instance, O-linked β-N-acetylglucosamine (O-GlcNAc) is an abundant posttranslational modification of Ser and Thr on nuclear, cytoplasmic, and mitochondrial proteins, with signaling importance comparable to protein phosphorylation.46 The glyco-immune checkpoint involves the family of Siglec receptors binding with glycans to modulate various immune cell responses, including T cell, NK cell, macrophage, neutrophil, and myeloid-derived suppressor cells.47 Infectious bacteria (e.g. Mycobacterium tuberculosis, Helicobacter pylori, Schistosoma mansoni) or viruses (e.g. HIV, influenza virus) also use glycans to bind host cell receptors for cellular uptake or immune escape.48 Identification of the receptor-ligand pair underlying these carbohydrate-protein interactions in cellular and in vivo settings would be critical to understand the biological process as well as to develop therapeutics. GECX-sugar can potentially advance all these research fields.
In contrast and complementary to metabolic pathway engineering which modifies the carbohydrate,49, 50 GECX-sugar is able to covalently target endogenous carbohydrates and thus suitable for in vivo studies and therapeutic applications. Covalently targeting endogenous proteins has recently enabled the generation of covalent protein drugs with extraordinary efficacy superior to antibodies in cancer immunotherapy.13 GECX-sugar now expands the covalent targeting mechanism from endogenous proteins to endogenous carbohydrates. Cross-linking of protein to the unmodified carbohydrate converts the binding protein into an irreversible inhibitor for the native protein-carbohydrate interaction, which can be exploited for glycan-targeted diagnostic and therapeutic applications, such as enhancing NK cell killing of cancer cells demonstrated here.
In summary, GECX-sugar enables site-specific introduction of covalent linkages between proteins and carbohydrates, offering a solution to the long-standing challenge of low affinity and weak interaction. GECX-sugar will thus advance the basic study of glycobiology and inspire innovative avenues for protein diagnostics and therapeutics via effectively targeting glycan.
Methods
Synthetic procedures.
All synthetic procedures are described in the Supplementary Information.
Reagent and materials.
HRP conjugated anti-Hisx6 tag antibody was purchased from Proteintech (Cat No. HRP-66005). Streptavidin-HRP was purchased from Thermo Fisher Scientific (Cat No. PI21130). Alexa Fluor 647 conjugated Hisx6 tag antibody was purchased from Thermo Fisher Scientific (Cat No. MA1-135-A647). IL-2 was purchased form Peprotech Inc (Cat No. 200-02). Vibrio cholerae sialidase was purchased form Sigma (Cat No. 11080725001). CellTrace far red dye was purchased from Life Technologies Corporation (Cat No. C34572). Propidium iodide was purchased from Fisher Scientific (Cat No. 5018262).
Cell culture.
BT-20 (CCLZR058), MCF-7 (CCLZR278), SK-MEL-28 (CCLZR370) were purchased from UCSF Cell and Genome Engineering Core (CGEC) and authenticated by CGEC using cell line short tandem repeat (STR). NK-92 (CRL-2407) was purchased from and STR authenticated by American Type Culture Collection (ATCC). MCF-7, BT-20 and SK-MEL-28 were cultured in Eagle's Minimum Essential Medium (EMEM) containing 10 % fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). NK-92 Cells were cultured in a humidified incubator at 37 °C and 5% CO2. NK-92 cells were maintained in Minimum Essential Medium α (MEM α) modified with nucleosides and supplemented with 10% FBS, 10% horse serum, 0.1 mM β-mercaptoethanol, 0.02 mM folic acid, 0.2 mM inositol and 200 IU/mL human recombinant IL-2.
Molecular cloning.
Primers were synthesized by Integrated DNA Technologies (IDT), and all plasmids were sequenced by GENEWIZ. All reagents were obtained from New England Biolabs. Primers for cloning are listed in Supplementary Table 1.
Siglec-7v
MQKSNRKDYSLTMQSSVTVQEGMSVHVRCSFSYPVDSQTDSDPVHGYWFRAGNDISWKAPVATNNPAWAVQEETRDRFHLLGDPQTKNCTLSIRDARMSDAGRYFFRMEKGNIKWNYKYDQLSVNVTALTHHHHHHH
Positions K20, K24, K75, K104, K127, N129, I130, K131, K135 are highlighted in red.
The siglec-7v gene was synthesized by IDT. Residue 20, 24, 75,104, 127, 129, 131 or 135 of siglec-7v was mutated to an amber stop codon TAG, respectively, via site-directed mutagenesis using primers in Supplementary Table 1.
sfGFP (2SFY)
MUKGEELFTGVVPILVELDGDVNGHKFSVRGEGEGDATNGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKRHDFFKSAMPEGYVQERTISFKDDGTYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNFNSHNVYITADKQKNGIKANFKIRHNVEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSVLSKDPNEKRDHMVLLEFVTAAGITHGMDELYKGSHHHHHH
Red U: amber codon TAG at 2nd position
pEvol-MmSFYRS
pEvol-MmSFYRS plasmid was generated by introducing the MmSFYRS encoding gene into pEvol vector via homologous recombination. Briefly, the SFYRS gene was amplified with primers MmSFYRS-SpeI-F and MmSFYRS-SalI-R, purified, and ligated into pEvol vector (linearized with SpeI and SalI) with Exnase™ II.
pEvol-MaPylRS-wt
Sequence alignment indicates that the active sites of Methanosarcina mazei PylRS (MmPylRS) and Methanomethylophilus alvus PylRS (MaPylRS) are highly conserved. MaPylRS and its derivatives usually present better solubility than those synthetases derived from MmPylRS, which may lead to higher incorporation efficiency. To enhance the incorporation efficiency of SFY, we examined the incorporation of SFY using the Ma-tRNAPyl/PylRS pair. A pEvol-MaPylRS plasmid encoding an orthogonal pair of wt-MaPylRS and a mutant MaPylT was first constructed. Briefly, the wild-type MaPylRS gene 51 was chemically synthesized, amplified with MaSFYRS-SpeI-F/MaSFYRS-SalI-R primers, and introduced into the pEvol vector via homologous recombination. Then a mutant Ma-pyrrolysyl-tRNA gene, MaPylT(6), was introduced into pEvol vector via site-directed mutagenesis with MaPylT(6)-F/R primers. The resultant plasmid was named as pEvol-MaPylRS-wt and used as the template to generate pEvol-MaSFYRS.
pEvol-MaSFYRS
Mutations carried by MmSFYRS were directly transplanted into MaPylRS via PCR-amplification with primers (MaSFYRS-R1, -F2, -R2, -F3, -R3, -R4) and then ligated into the pEvol vector via multiple-fragment homologous recombination. To further improve the incorporation efficiency of SFY, the mutant MaPylT(6) was swapped with the wild-type MaPylT by using site-directed mutagenesis with MaPylT(wt)-F/R primers to afford the pEvol-MaSFYRS plasmid. The wt-MaPylT tRNA afforded much higher incorporation efficiency than the mutant MaPylT(6) tRNA (Supplementary Figure 6).
Protein expression and purification.
Proteins were expressed in E. coli DH10B and purified using procedures described below.
Siglec-7v and siglec-7v (SFY)
For wildtype siglec-7v expression, the plasmid pBAD-siglec-7v was transformed into E. coli. For the incorporation of SFY into siglec-7v, the plasmid pBAD-siglec-7v(TAG) and was co-transformed with pEVOL-SFYRS into E. coli, and plated on LB agar plate supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Several colonies were picked and inoculated in 50 mL 2x YT (5 g/L NaCI, 16 g/L Tryptone, 10 g/L Yeast extract). The cells were grown at 37 °C, 220 rpm to an OD600 of 0.5, the medium was then added with either 0.2 % L-arabinose only or 0.2 % L-arabinose plus 1 mM SFY, and the expression were carried out at 25 °C, 220 rpm for 18-22 h. Cells were harvested at 3,000 g, 4 °C for 10 min. For protein purification, cells were resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM imidazole) supplemented with EDTA free protease inhibitor cocktail and 1 μg/mL Dnase. The cells were opened by sonification, after which the cell lysis solution was centrifuged at 10,000 g at 4 °C for 15 min. The pellet was suspended in guanidine buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 6 M guanidine) and centrifuged at 10,000 g at 4 °C for 15 min. The supernatant was collected and incubated with 500 μL Ni-NTA affinity resin. The resin was washed with guanidine wash buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM imidazole, 6 M guanidine) for 3 times, and then the protein was eluted twice with elution buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 300 mM imidazole, 6 M guanidine). The eluted protein was diluted into dialysis buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl) with 4 M guanidine to a final concentration of 0.1 mg/mL and dialyzed against dialysis buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl) with 2 M guanidine, and then 0 M guanidine for 8 h each at 4 °C. The refolded protein was concentrated to 1 mg/mL for further use.
sfGFP(2SFY)
For the incorporation of SFY into sfGFP, the plasmid pBAD-sfGFP(2TAG) and was co-transformed with pEVOL-SFYRS into E. coli, and plated on LB agar plate supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Several colonies were picked and inoculated in 50 mL 2x YT (5 g/L NaCI, 16 g/L Tryptone, 10 g/L Yeast extract). The cells were grown at 37 °C, 220 rpm to an OD600 of 0.5, the medium was then added with either 0.2% L-arabinose only or 0.2% L-arabinose plus 1 mM SFY, and the expression were carried out at 18 °C, 220 rpm for 18-22 h. Cells were harvested at 3,000 g, 4 °C for 10 min. For protein purification, cells were resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM imidazole, EDTA free protease inhibitor cocktail, 1 μg/mL Dnase). The cells were opened by sonification, after which the cell lysis solution was centrifuged at 10,000 g at 4 °C for 15 min. The supernatant was collected and incubated with 500 μL Ni-NTA affinity resin. The resin was washed with wash buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM imidazole) for 3 times, and the protein was eluted twice with elution buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 300 mM imidazole).
Z(24SFY)
For the incorporation of SFY into Z protein, the plasmid pBAD-Z(24TAG) and was co-transformed with pEVOL-SFYRS into E. coli, and plated on LB agar plate supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. The Z protein expression and purification was the same as described above.
Glycan microarray analysis.
Siglec-7v (20 μg/mL) was incubated with the glycan microarray at room temperature for 3 h with gentle shaking, after which the microarray was washed with TSMT buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2 and 0.05 % Tween-20) at room temperature for 3 times. Alexa Fluor 647 conjugated Hisx6 tag-specific antibody was diluted at 1:100 and incubated with the microarray at room temperature for 2 h with gentle shaking. After 3 times wash with TSMT buffer, the microarray was scanned at 635 nm with GenePix 4000B. The fluorescence intensity was measured, and data was interpreted into a two-dimensional bar chart. The y-axis is the fluorescence intensity to reveal relative protein binding signals for each glycan.
Small molecule mediated siglec-7v cross-linking in vitro.
To test if small molecule cross-linker could cross-link siglec-7v with azido-GD3 and azido-lac, 60 μM siglec-7v was incubated with 2 mM azido-GD3 or azido-lac in PBS (pH 7.4) at room temperature for 1 h. Then the solution was treated with or without 0.3 mM NHSF, NHBr, NHFS, NHQM, or HoQM at room temperature for 1 h, respectively. The NHQM or HoQM was then illuminated with or without UV for 15 min at wavelength 365 nm. After that, 200 μM alkyne-biotin, 0.5 mM CuSO4, 1 mM THPTA and 1 mM sodium ascorbate were added and the reaction mixture was incubated at room temperature in dark environment for 0.5 h. Samples were then boiled at 95 °C for 5 min and analyzed with Western blot against Hisx6 tag antibody (1:5000 dilution) or streptavidin-horseradish peroxidase (HRP).
To label the azdio-glycan/siglec-7v complex with alkyne-biotin through Cu (I)-catalyzed azide-alkyne cycloaddition click chemistry, we initially used 2 mM alkyne-biotin and an 1 h incubation time, which showed a relatively high background due to nonspecific reaction of alkyne with proteins at low level as previously reported.36,37 To optimize the labeling conditions, we reduced the alkyne-biotin from 2 mM to 200 μM and decreased the labeling time from 1 h to 0.5 h, which achieved much cleaner background.
Selection of SFY-specific synthetase (SFYRS).
DH10B cells (100 μL) harboring the pREP positive selection reporter was transformed with 122 ng of pBK-TK3 library via electroporation. The electroporated cells were subjected to selections by following procedures previously described.29,39,40 The pBK plasmids encoding the selected SFYRS gene were extracted by miniprep (QIAGEN, Cat. No. 27104) and separated from the reporter plasmids by DNA electrophoresis. The resulted pBK plasmids were analyzed by Sanger-sequencing.
Siglec-7v(SFY) cross-linking GD3 in vitro.
Purified siglec-7v(SFY) (60 μM) was incubated with 2 mM azido-GD3 or azido-lac in PBS (pH 7.4) at room temperature for 1 h. After that, click reaction was performed and samples were subjected to Western blot for biotin signal detection via streptavidin-HRP.
Sigelc-7v(127SFY) cross-linking sialoglycan on mammalian cell surface.
SK-MEL-28 cells were plated into 6-well plate and incubated for 24 h. 100 μL Vibrio cholerae sialidase (Sigma) or 100 μL PBS was added with 400 μL medium without FBS for 24 h. To test if siglec-7v(127SFY) could cross-link sialoglycan on mammalian cell surface, different concentrations of siglec-7v or siglec-7v(127SFY) was incubated with SK-MEL-28 cells pre-treated with or without sialidase in PBS (pH 7.4) at 37°C in 5 % CO2 incubator for 2 h. Cells were washed 4 times with PBS buffer and labeled with Alexa Fluor 647 conjugated Hisx6 tag monoclonal antibody (1:100 dilution) at room temperature for 1 h. Cells were harvested for flow cytometric analysis with BD LSRFortessa cell analyzer using software FlowJo.
Siglec-7v(127SFY) enhancing NK cell killing of cancer cells.
Target cells were pre-labeled with CellTrace far-red dye at room temperature for 10 min. Siglec-7v or siglec-7v(127SFY) of different concentrations was incubated with 5 x 104 target cells in PBS (pH 7.4) at 37°C in 5 % CO2 incubator for 2 h. Cells were washed 3 times by PBS and subsequentially incubated with 5 x 105 NK cells for 4 h. Propidium iodide (10 μg/mL) was added to each sample, and NK cell cytotoxicity was evaluated by flow cytometric analysis with BD LSRFortessa cell analyzer using software FlowJo. Cells were acquired after electronic gating on CellTrace far-red dye, and percentage of propidium iodide–positive cells was determined. Cell death percentage was calculated as experimental % death – control % death. Control % death was determined using the group without protein incubation.
Mass spectrometry.
The intact protein mass was obtained using electrospray ionization mass spectrometry (ESI-MS) with a QTOF Ultima (Waters) mass spectrometer, operating under positive electrospray ionization mode, connected to an LC-20AD (Shimadzu) liquid chromatography unit. For tandem mass spectrometry, peptides were separated by nano-LC Ultimate 3000 high-performance liquid chromatography system (Thermo Fisher). The cross-linking mass spectra were analyzed with pLink 2.3.52, 14
Statistics and reproducibility.
All biochemical data (Fig 1f, Fig 2, Fig 3b, Fig 3d, Fig 4c, Fig 4f, Fig 5b, Fig 5c, Supplementary Fig 7b and 9a) were obtained in triplicate with similar results. All cell staining or labelling experiments (Fig 5e-f, Fig 6b-d, Supplementary Fig 8 and 9b-c) were performed in triplicate.
Supplementary Material
Acknowledgements
L.W. acknowledges the support of the NIH (R01GM118384 and R01CA258300).
Footnotes
Competing Interests
L.W., S.L. and N.W. are inventors on a patent application filed as International Application No. PCT/US2022/031925 by The Regents of the University of California. The patent application covers unnatural amino acid SFY and its ability to crosslink carbohydrates as described in the manuscript. The other authors (B.Y. and W.S.) declare no competing interests.
Data Availability
All data supporting the results and conclusions are available within the Article and its Supplementary Information and Source Data. Source data are provided with this paper. Synthetic experimental procedures, compound characterization, NMR and HPLC are available in the Supplementary Information. Requests for materials should be addressed to L.W.
References
- 1.Sears P & Wong C Carbohydrate Mimetics: A New Strategy for Tackling the Problem of Carbohydrate-Mediated Biological Recognition. Angew. Chem. Int. Ed. Engl 38, 2300–2324 (1999). [PubMed] [Google Scholar]
- 2.Spiro RG Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R (2002). [DOI] [PubMed] [Google Scholar]
- 3.Fuster MM & Esko JD The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 5, 526–542 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Stowell SR, Ju T & Cummings RD Protein glycosylation in cancer. Annu. Rev. Pathol 10, 473–510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imberty A & Prestegard JH Structural Biology of Glycan Recognition. (2015). In “Essentials of Glycobiology”, 3rd Ed. doi: 10.1101/glycobiology.3e.030 [DOI] [Google Scholar]
- 6.Nelson RM, Venot A, Bevilacqua MP, Linhardt RJ & Stamenkovic I Carbohydrate-protein interactions in vascular biology. Annu. Rev. Cell Dev. Biol 11, 601–631 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Sterner E, Flanagan N & Gildersleeve JC Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol 11, 1773–1783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polonskaya Z, Savage PB, Finn MG & Teyton L High-affinity anti-glycan antibodies: challenges and strategies. Curr. Opin. Immunol 59, 65–71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudak JE & Bertozzi CR Glycotherapy: new advances inspire a reemergence of glycans in medicine. Chem. Biol 21, 16–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang Z et al. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat. Methods 10, 885–888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L Genetically encoding new bioreactivity. N. Biotechnol 38, 16–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B et al. Spontaneous and specific chemical cross-linking in live cells to capture and identify protein interactions. Nat. Commun 8, 2240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q et al. Developing Covalent Protein Drugs via Proximity-Enabled Reactive Therapeutics. Cell 182, 85–97.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Liu C et al. Identification of Protein Direct Interactome with Genetic Code Expansion and Search Engine OpenUaa. Adv. Biol. (Weinh) 5, e2000308 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Xuan W, Li J, Luo X & Schultz PG Genetic Incorporation of a Reactive Isothiocyanate Group into Proteins. Angew. Chem. Int. Ed. Engl 128, 10219–10222 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Jäger M & Minnaard AJ Regioselective modification of unprotected glycosides. Chem. Commun 52, 656–664 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Wang L-X & Davis BG Realizing the Promise of Chemical Glycobiology. Chem. Sci 4, 3381–3394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falco M et al. Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J. Exp. Med 190, 793–802 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crocker PR, Paulson JC & Varki A Siglecs and their roles in the immune system. Nat. Rev. Immunol 7, 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Yamaji T, Teranishi T, Alphey MS, Crocker PR & Hashimoto Y A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to alpha 2,8-disialyl and branched alpha 2,6-sialyl residues. A comparison with Siglec-9. J. Biol. Chem 277, 6324–6332 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Attrill H et al. The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design. Biochem. J 397, 271–278 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macauley MS, Crocker PR & Paulson JC Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol 14, 653–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao L & Wang L New covalent bonding ability for proteins. Protein Sci. 31, 312–322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B et al. Proximity-enhanced SuFEx chemical cross-linker for specific and multitargeting cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 115, 11162–11167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoppmann C & Wang L Proximity-enabled bioreactivity to generate covalent peptide inhibitors of p53-Mdm4. Chem. Commun 52, 5140–5143 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Xiang Z et al. Proximity-enabled protein crosslinking through genetically encoding haloalkane unnatural amino acids. Angew. Chem. Int. Ed. Engl 53, 2190–2193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen XH et al. Genetically encoding an electrophilic amino Acid for protein stapling and covalent binding to native receptors. ACS Chem. Biol 9, 1956–1961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoppmann C, Maslennikov I, Choe S & Wang L In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J. Am. Chem. Soc 137, 11218–11221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N et al. Genetically Encoding Fluorosulfate-l-tyrosine To React with Lysine, Histidine, and Tyrosine via SuFEx in Proteins in Vivo. J. Am. Chem. Soc 140, 4995–4999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J et al. Genetically Encoding Photocaged Quinone Methide to Multitarget Protein Residues Covalently in Vivo. J. Am. Chem. Soc 141, 9458–9462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J et al. Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents. J. Am. Chem. Soc 142, 17057–17068 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J et al. Photocaged Quinone Methide Crosslinkers for Light-Controlled Chemical Crosslinking of Protein-Protein and Protein-DNA Complexes. Angew. Chem. Int. Ed. Engl 58, 18839–18843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H et al. Chemoenzymatic synthesis of GD3 oligosaccharides and other disialyl glycans containing natural and non-natural sialic acids. J. Am. Chem. Soc 131, 18467–18477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu RK, Tsai Y-T, Ariga T & Yanagisawa M Structures, biosynthesis, and functions of gangliosides--an overview. J. Oleo. Sci 60, 537–544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krengel U & Bousquet PA Molecular recognition of gangliosides and their potential for cancer immunotherapies. Front. Immunol 5, 325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speers AE & Cravatt BF Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol 11, 535–546 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Yang X & Verhelst SHL Comparative analysis of click chemistry mediated activity-based protein profiling in cell lysates. Molecules 18, 12599–12608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Attrill H et al. Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J. Biol. Chem 281, 32774–32783 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Lacey VK, Louie GV, Noel JP & Wang L Expanding the library and substrate diversity of the pyrrolysyl-tRNA synthetase to incorporate unnatural amino acids containing conjugated rings. ChemBioChem 14, 2100–2105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takimoto JK, Dellas N, Noel JP & Wang L Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol 6, 733–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi T, Hoppmann C, Yang B & Wang L Using Protein-Confined Proximity To Determine Chemical Reactivity. J. Am. Chem. Soc 138, 14832–14835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Portoukalian J, Zwingelstein G & Doré JF Lipid composition of human malignant melanoma tumors at various levels of malignant growth. Eur. J. Biochem 94, 19–23 (1979). [DOI] [PubMed] [Google Scholar]
- 43.Razi N & Varki A Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl. Acad. Sci. U. S. A 95, 7469–7474 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dippold WG et al. Cell surface antigens of human malignant melanoma: definition of six antigenic systems with mouse monoclonal antibodies. Proc. Natl. Acad. Sci. U. S. A 77, 6114–6118 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suck G et al. NK-92: an ‘off-the-shelf therapeutic’ for adoptive natural killer cell-based cancer immunotherapy. Cancer Immunol. Immunother 65, 485–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joiner CM, Li H, Jiang J & Walker S Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Curr. Opin. Struct. Biol 56, 97–106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Wall S, Santegoets KCM, van Houtum EJH, Büll C & Adema GJ Sialoglycans and Siglecs Can Shape the Tumor Immune Microenvironment. Trends Immunol. 41, 274–285 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Li RE, van Vliet SJ & van Kooyk Y Using the glycan toolbox for pathogenic interventions and glycan immunotherapy. Curr. Opin. Biotechnol 51, 24–31 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Hu C-W et al. Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase. Nat. Chem. Biol 13, 1267–1273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang PV et al. Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew. Chem. Int. Ed. Engl 48, 4030–4033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borrel G et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15, 679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z-L et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun 10, 3404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results and conclusions are available within the Article and its Supplementary Information and Source Data. Source data are provided with this paper. Synthetic experimental procedures, compound characterization, NMR and HPLC are available in the Supplementary Information. Requests for materials should be addressed to L.W.