Abstract
Background/Aims
This study aimed to evaluate the potential of the stool microbiome and gut microbe-derived extracellular vesicles (EVs) to differentiate between patients with inflammatory bowel disease (IBD) and healthy controls, and to predict relapse in patients with IBD.
Methods
Metagenomic profiling of the microbiome and bacterial EVs in stool samples of controls (n=110) and patients with IBD (n=110) was performed using 16S rRNA sequencing and then compared. Patients with IBD were divided into two enterotypes based on their microbiome, and the cumulative risk of relapse was evaluated.
Results
There was a significant difference in the composition of the stool microbiome and gut microbe-derived EVs between patients with IBD and controls. The alpha diversity of the microbiome in patients with IBD was significantly lower than that in controls, while the beta diversity also differed significantly between the two groups. These findings were more prominent in gut microbe-derived EVs than in the stool microbiome. The survival curve tended to be different for enterotypes based on the gut microbe-derived EVs; however, this difference was not statistically significant (log-rank test, p=0.166). In the multivariable analysis, elevated fecal calprotectin (>250 mg/kg) was the only significant risk factor associated with relapse (adjusted hazard ratio, 3.147; 95% confidence interval, 1.545 to 6.408; p=0.002).
Conclusions
Analysis of gut microbe-derived EVs is better at differentiating patients with IBD from healthy controls than stool microbiome analysis.
Keywords: Inflammatory bowel diseases, Microbiota, Extracellular vesicles
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic disease that typically begins at a young age.1 Because patients with this disease generally experience relapses and remissions, it greatly affects patients’ quality of life.2 The pathogenesis of IBD is not fully understood. Genetic factors, environmental influences, gut microbes, and aberrant immune responses are thought to be involved.3 In addition, IBD is a disease that requires personalized treatment because of its wide variety of phenotypes, natural courses, and treatment responses.4
With the recent development of next-generation sequencing, analysis of the gut microbiome in relation to IBD has been actively performed. Many studies have shown that patients with IBD have a decreased diversity of gut microbiota compared to that in healthy individuals.5,6 In addition, some bacterial flora that commonly increases or decreases in patients with IBD have been reported.7 However, there are relatively few studies on microbiome biomarkers for predicting the disease activity and prognosis of IBD.
Analysis of bacterial extracellular vesicles (EVs) has recently been recognized as important in the analysis of gut microbiome related to IBD. Microbe-derived EVs are particles secreted by bacteria for communication between bacteria and host cells.8 They contain and carry various substances, such as proteins, lipids, and nucleic acids. EVs released from both pathogenic and commensal bacteria are thought to regulate immunity and corresponding signaling pathways.9 Because microbe-derived EVs contain bacterial DNA, they can be filtered from stool samples, the bacterial DNA can be extracted from them, and the microbiome can be analyzed using next-generation sequencing. In reality, it has been reported that the composition and characteristics of these microbe-derived EVs are different from the microbiome of stool.10 Since the amount of EVs varies depending on the state of the cell, it can be said that the EV quantity of a specific bacterium reflects the activity of that bacterium. Therefore, considering that the activity and metabolites of gut microbiota may be more important than simple changes in the composition of gut microbiota according to disease status, the analysis of gut microbe-derived EVs in the stool may be more useful than that of the stool microbiome itself. For example, a recent study suggested that profiling of gut microbe-derived EVs is more useful for finding novel microbial markers than profiling the stool microbiome in patients with colorectal cancer.10 In addition, it was reported that analyzing metagenomic data in connection with metabolomic data using microbe-derived EVs in the stool has good potential for the diagnosis of colorectal cancer.11 Nevertheless, studies on gut microbe-derived EVs in the stool of patients with IBD are limited.
Therefore, this study aimed to compare the stool microbiome and gut microbe-derived EVs, evaluate the efficacy of this comparison in differentiating between patients with IBD and healthy controls, and predict the occurrence of relapse in patients with IBD.
MATERIALS AND METHODS
1. Patients and definitions
Stool samples were prospectively collected from patients with IBD who had been enrolled in the Seoul National University Bundang Hospital IBD cohort. Clinical data were prospectively collected using the web-based Research Electronic Data Capture (REDCapⓇ) program (Vanderbilt University, Nashville, TN, USA). Active disease was defined as fecal calprotectin >250 mg/kg on the day that the stool was collected. For survival analysis, relapse was defined as a composite outcome of (1) new use of steroids, immunomodulators, and biologics; (2) a visit to an emergency department; (3) hospitalization; or (4) abdominal surgery. Control stool samples were obtained from healthy individuals who had abdominal symptoms but were not diagnosed with irritable bowel syndrome.12 These healthy individuals were confirmed not to have taken antibiotics or steroids within the past 3 months, or probiotics within the past 2 weeks. This study was approved by the Institutional Review Board of the Seoul National University Bundang Hospital (IRB number: B-1708/412-301), and informed consent was waived.
2. Isolation of EVs and extraction of DNA
The technique followed for isolation of EVs and DNA extraction has been described in detail previously.10 Briefly, stool samples were filtered through a cell strainer after being diluted in 10 mL of phosphate-buffered saline for 24 hours. The samples were centrifuged at 10,000 ×g for 10 minutes at 4°C to separate the EVs from the stool samples. After centrifugation, stool sample pellets contained the bacterial cells, while the supernatant contained the EVs. Bacteria and foreign particles were thoroughly eliminated from the stool sample supernatant by sterilization through a 0.22 µm filter. To extract DNA, the bacteria and EVs were boiled for 40 minutes at 100°C. To eliminate the remaining floating particles and waste, the supernatant was collected after 30 minutes of centrifugation at 13,000 rpm at 4°C. DNA was extracted using the DNeasy PowerSoil Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. DNA extracted from bacterial cells and EVs in each sample was quantified using the QIAxpert system (QIAGEN).
3. Bacterial metagenomic analysis using DNA from the EVs
Bacterial genomic DNA was amplified using 16S_V3_F (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3’) and 16S_V4_R (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’) primers specific to the V3–V4 hypervariable regions of the 16S rRNA gene. Libraries were prepared using polymerase chain reaction products according to the MiSeq system guide (Illumina, San Diego, CA, USA) and quantified using QIAxpert. Each amplicon was then quantified, and equimolar ratios were pooled and sequenced on a MiSeq system (Illumina), according to the manufacturer’s recommendations.
4. Analysis of the microbiome derived from the stool and bacterial EVs
Paired-end reads that matched the adapter sequences were trimmed using Cutadapt (version 1.1.6) with a minimum overlap of 11 bases, a maximum error rate of 15%, and a minimum length of 10 bases.13 The resulting FASTQ files containing paired-end reads were merged using CASPER (version 0.8.2) with a mismatch ratio of 0.27 and quality-filtered using the Phred (Q) score-based criteria described by Bokulich.14,15 The reads shorter than 350 bp and longer than 550 bp after merging were discarded. To identify chimeric sequences, a reference-based chimera detection step was conducted using VSEARCH (version 2.3.0) against the SILVA gold database.16,17 The sequence reads were clustered into operational taxonomic units using VSEARCH with an open clustering algorithm under a threshold of 97% sequence similarity. The representative sequences of the operational taxonomic units were classified using the EzBioCloud 16S rRNA gene sequence database with UCLUST (parallel_assign_taxonomy_uclust.py script in QIIME version 1.9.1) under default parameters.18,19
5. Statistical analyses
Group comparisons for diversity metrics were conducted and graphed using R software version 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria). Alpha diversity indices (observed operational taxonomic units, Shannon index, and phylogenetic diversity) were compared using the decimal log-transformed relative abundance of the microbiome between groups using the Wilcoxon rank-sum test (R package “microbiome version 1.9.19”). Group distances for beta diversity indices (weighted-UniFrac metric) were generated through the permutational analysis of variance using 1000 Monte Carlo permutations (R packages “phyloseq version 1.30.0” and “vegan version 2.5.6”). Principal coordinate analysis plots were generated for visualization. Discriminate taxa (abundance >0.05%, prevalence >50%) between groups were identified using the Welch’s t-test. Statistical associations between taxa and disease status were tested using the phylogenetic-tree-based microbiome association test, optimal microbiome-based association test (OMiAT), and R Bioconductor package for the differential analysis of sequence read count data (edgeR), adjusting for age, sex, smoking, body mass index, and Bristol Stool Form Scale.20,21 Adjusted p-values controlling the false discovery rate were reported where appropriate. Enterotyping was performed based on the Dirichlet multinomial mixture model using a genus-abundance matrix of stool microbiome and gut microbe-derived EVs (R package “DirichletMultinomial version 1.28.0”).22 The Laplace approximation was used to determine the optimal clusters of participants.
We compared the cumulative risk of relapse according to enterotype using the Kaplan-Meier survival analysis and the log-rank test. The Cox proportional hazards regression was performed to evaluate independent predictors of relapse in patients with IBD. Variables with a p-value <0.2 in univariable analysis were included in the multivariable Cox proportional hazards regression model. All analyses were performed using Stata version 16.0 (StataCorp LLC, College Station, TX, USA). Two-sided p-values <0.05 were considered statistically significant.
RESULTS
1. Comparison of stool microbiome and gut microbe-derived EVs between patients with IBD and healthy controls
The baseline characteristics of the study participants are shown in Supplementary Table 1. There were some differences between the patients with IBD and healthy controls. The proportion of male patients and current smokers was significantly higher in patients with IBD than that in controls. The body mass index was significantly higher in the control group than that in the IBD group. The distribution of the Bristol Stool Form Scale also differed between the two groups.
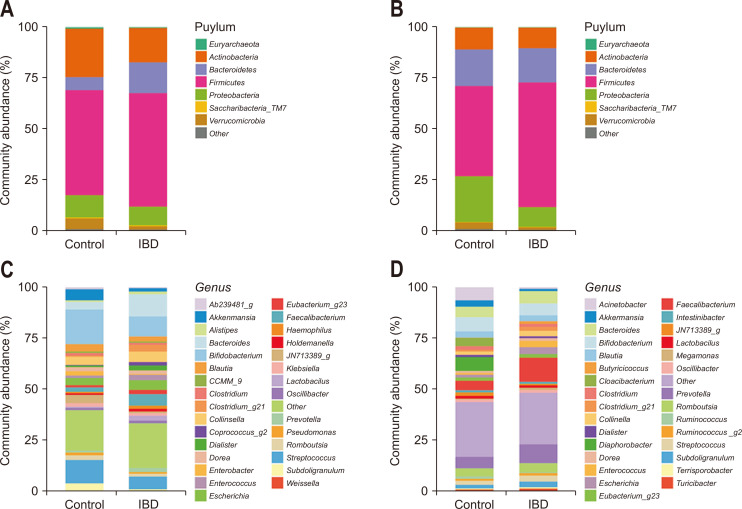
Fig. 1 shows the taxonomic composition of the stool microbiome and gut microbe-derived EVs in controls and patients with IBD at the phylum and genus levels. The taxa showing significantly different means of relative abundances by disease status in the stool microbiome and gut microbe-derived EVs analyses are shown in Tables 1 and 2, respectively. There were some differences in the significantly associated taxa between the two analyses.
Fig. 1.
Taxonomic composition of the stool microbiome and gut microbe-derived extracellular vesicles (EVs) in patients with inflammatory bowel disease (IBD) and healthy controls. (A) Phylum level (stool microbiome); (B) phylum level (gut microbe-derived EVs); (C) genus level (stool microbiome); and (D) genus level (gut microbe-derived EVs).
Table 1.
Taxa Showing a Significant Difference in Abundance in the Stool Microbiome between Patients with Inflammatory Bowel Disease and Healthy Controls
| Taxon | Log-transformation t-statistic |
FDR adjusted p-value |
|---|---|---|
| Phylum Firmicutes | ||
| Family Lachnospiraceae | 2.40 | 0.030 |
| Genus Clostridium_g21 | 3.69 | <0.001 |
| Species Ruminococcus gnavus | 3.69 | <0.001 |
| Genus Eubacterium_g5 | –7.06 | <0.001 |
| Species Eubacterium hallii | –7.06 | <0.001 |
| Genus Agathobacter | 2.50 | 0.020 |
| Species Agathobacter rectalis | 2.50 | 0.020 |
| Genus Blautia | -4.52 | <0.001 |
| Species Blautia obeum | –3.62 | 0.001 |
| Species Blautia wexlerae | –2.95 | 0.007 |
| Genus DQ057459_g | –4.47 | <0.001 |
| Genus EU457907_g | –2.53 | 0.020 |
| Genus Fusicatenibacter | –3.85 | <0.001 |
| Species Fusicatenibacter saccharivorans |
–3.85 | <0.001 |
| Genus KE159538_g | –2.40 | 0.030 |
| Genus Roseburia | 3.70 | <0.001 |
| Species Roseburia faecis | 3.52 | <0.001 |
| Species Roseburia inulinivorans | 2.37 | 0.030 |
| Genus Ruminococcus_g4 | 2.63 | 0.006 |
| Species Ruminococcus lactaris | 2.09 | 0.050 |
| Species Ruminococcus torques | 2.54 | 0.003 |
| Family Leuconostocaceae | –3.45 | <0.001 |
| Genus Weissella | –3.48 | <0.001 |
| Species Weissella cibaria | –3.19 | <0.001 |
| Species Weissella koreensis | –2.48 | 0.030 |
| Family Christensenellaceae | −3.02 | <0.001 |
| Genus AB239481_g | −3.02 | <0.001 |
| Family Mogibacterium_f | −5.65 | <0.001 |
| Family Peptostreptococcaceae | −3.14 | 0.006 |
| Genus Romboutsia | −2.80 | 0.020 |
| Family Streptococcaceae | −4.76 | <0.001 |
| Genus Streptococcus | −4.76 | <0.001 |
| Species Streptococcus oligofermentans |
−2.74 | 0.010 |
| Species Streptococcus salivarius | −2.87 | 0.010 |
| Species Streptococcus sanguinis | −2.83 | 0.008 |
| Family Veillonellaceae | 2.83 | <0.001 |
| Genus Veillonella | 2.83 | <0.001 |
| Species Veillonella dispar | 2.83 | <0.001 |
| Family Ruminococcaceae | ||
| Genus Caproiciproducens | 3.79 | <0.001 |
| Genus Faecalibacterium | 4.50 | <0.001 |
| Genus Pseudoflavonifractor | 3.70 | <0.001 |
| Species Flavonifractor plautii | 3.71 | <0.001 |
| Genus Ruminococcus_g2 | −3.38 | <0.001 |
| Species Ruminococcus bromii | −3.38 | 0.001 |
| Phylum Bacteroidetes | 6.00 | <0.001 |
| Family Bacteroidaceae | 5.37 | <0.001 |
| Genus Bacteroides | 5.37 | <0.001 |
| Species Bacteroides dorei | 1.72 | 0.040 |
| Species Bacteroides fragilis | 2.92 | 0.010 |
| Species Bacteroides ovatus | 2.55 | 0.006 |
| Species Bacteroides plebeius | 3.52 | 0.040 |
| Species Bacteroides vulgatus | 5.36 | <0.001 |
| Phylum Actinobacteria | −2.95 | 0.008 |
| Phylum Verrucomicrobia | −3.03 | 0.006 |
| Family Akkermansiaceae | −3.03 | 0.006 |
| Genus AM500802_g | −5.65 | <0.001 |
| Genus Akkermansia | −3.03 | 0.006 |
| Species Akkermansia muciniphila |
−3.03 | 0.007 |
The change of column (log transformation) shows the multiplicative change in taxa abundance between patients with inflammatory bowel disease and healthy controls. Negative numbers represent decreased abundance in patients with inflammatory bowel disease compared with healthy controls.
FDR, false discovery rate.
Table 2.
Taxa Showing a Significant Difference in the Abundance of Gut Microbe-Derived Extracellular Vesicles between Patients with Inflammatory Bowel Disease and Healthy Controls
| Taxon | Log-transformation t-statistic |
FDR adjusted p-value |
|---|---|---|
| Phylum Firmicutes | 6.61 | <0.001 |
| Family Lachnospiraceae | ||
| Genus DQ057459_g | −2.75 | 0.020 |
| Genus Fusicatenibacter | −3.63 | 0.002 |
| Species Fusicatenibacter saccharivorans |
−3.63 | 0.002 |
| Family Veillonellaceae | ||
| Genus Dialister | −2.83 | 0.009 |
| Species Dialister invisus | −2.83 | 0.009 |
| Family Ruminococcaceae | 4.59 | <0.001 |
| Genus Butyricicoccus | 2.95 | 0.006 |
| Genus Faecalibacterium | 4.66 | <0.001 |
| Genus JN713389_g | −3.23 | 0.005 |
| Genus Sporobacter | 2.51 | 0.030 |
| Family Acidaminococcaceae | −2.98 | 0.002 |
| Genus Phascolarctobacterium | −2.98 | 0.003 |
| Species Phascolarctobacterium faecium |
−2.98 | 0.003 |
| Family Enterococcaceae | 2.25 | <0.001 |
| Genus Enterococcus | 2.25 | <0.001 |
| Species Enterococcus rivorum | 2.25 | <0.001 |
| Family Lactobacillaceae | 1.83 | 0.030 |
| Genus Lactobacillus | 1.83 | 0.030 |
| Species Lactobacillus ruminis | 1.83 | 0.030 |
| Family Erysipelotrichaceae | ||
| Genus Catenibacterium | 2.32 | 0.005 |
| Genus CCMM_g | 3.10 | 0.008 |
| Genus Holdemanella | 3.18 | 0.005 |
| Phylum Actinobacteria | ||
| Family Coriobacteriaceae | 2.47 | 0.020 |
| Genus Collinsella | 2.63 | 0.009 |
| Species Collinsella aerofaciens | 2.63 | 0.009 |
| Genus Eggerthella | −2.49 | 0.020 |
| Phylum Verrucomicrobia | −6.09 | <0.001 |
| Family Akkermansiaceae | −6.09 | <0.001 |
| Genus Akkermansia | −6.09 | <0.001 |
| Species Akkermansia muciniphila |
−6.09 | <0.001 |
| Phylum Proteobacteria | −6.76 | <0.001 |
| Family Moraxellaceae | −14.98 | <0.001 |
| Genus Acinetobacter | −14.46 | 0.007 |
| Species Acinetobacter bereziniae | −15.79 | <0.001 |
| Species Acinetobacter modestus | −12.36 | <0.001 |
| Species Acinetobacter parvus | −10.97 | <0.001 |
| Species Acinetobacter johnsonii | 3.10 | <0.001 |
| Genus Klebsiella | 2.43 | <0.001 |
| Species Klebsiella granulomatis | 3.15 | <0.001 |
| Species Klebsiella variicola | 2.89 | 0.004 |
| Genus Moraxella | −4.69 | <0.001 |
| Species Moraxella osloensis | −4.69 | <0.001 |
| Family Comamonadaceae | −14.81 | <0.001 |
| Genus Diaphorobacter | −14.80 | <0.001 |
| Species Diaphorobacter nitroreducens |
−14.80 | <0.001 |
| Family Sphingomonadaceae | 4.48 | <0.001 |
| Genus Sphingomonas | 4.48 | <0.001 |
| Family Enterobacteriaceae | ||
| Genus Enterobacter | −15.95 | <0.001 |
| Species Enterobacter kobei | −15.95 | <0.001 |
The change of column (log transformation) shows the multiplicative change in taxa abundance from between patients with inflammatory bowel disease and healthy controls. Negative numbers represent decreased abundance in patients with inflammatory bowel disease compared with healthy controls.
FDR, false discovery rate.
In the stool microbiota analysis, a significant enrichment of Bacteroidetes with a decrease in Actinobacteria and Verrucomicrobia was observed in patients with IBD at the phylum level (Table 1, Supplementary Fig. 1). The relative abundance of Lachnospiraceae, Veillonellaceae, and Bacteroidaceae was significantly higher in patients with IBD, and those of Leuconostocaceae, Christensenellaceae, Peptostreptococcaceae, Streptococcaceae, and Akkermansiaceae was higher in the controls at the family level.
Analysis of gut microbe-derived EVs revealed that Firmicutes were significantly enriched, while Verrucomicrobia and Proteobacteria were significantly decreased in patients with IBD at the phylum level (Table 2, Supplementary Fig. 2). At the family level, the relative abundance of Ruminococcaceae, Enterococcaceae, Lactobacillaceae, Coriobacteriaceae, and Sphingomonadaceae was significantly higher in patients with IBD, whereas that of Acidaminococcaceae, Akkermansiaceae, Moraxellaceae, and Comamonadaceae was significantly higher in the controls.
The association between the microbiome abundance of each taxon and disease status was further tested using TMAT, OMiAT, and edgeR. Supplementary Tables 2 and 3 show the analysis results for taxa that were significant according to Welch’s t-test, TMAT, OMiAT, and edgeR.
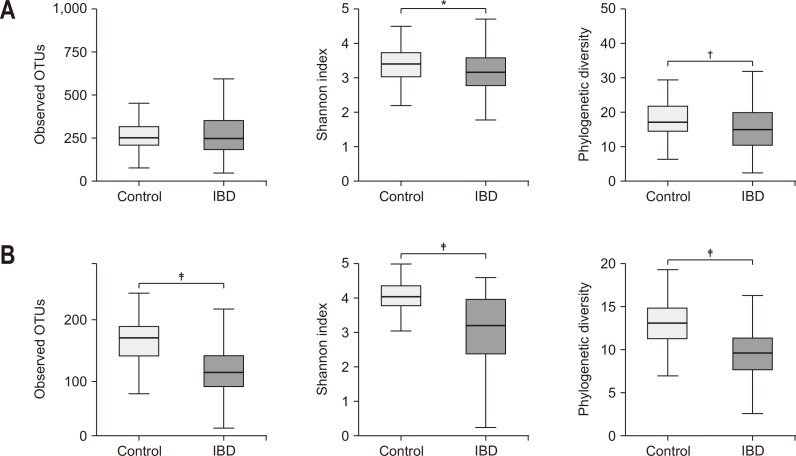
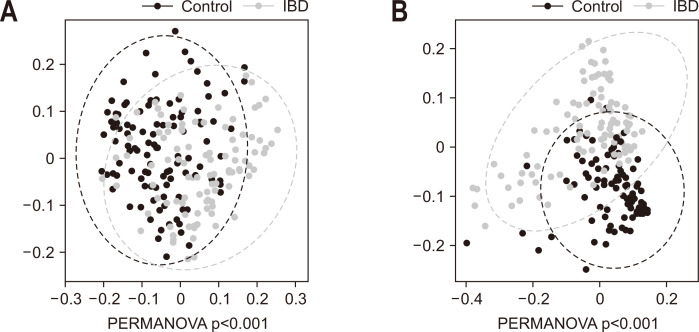
The alpha diversity of the microbiome in patients with IBD was significantly lower than that of the controls, with a more prominent difference in the gut microbe-derived EVs than in the stool microbiome (Fig. 2). The beta diversity of the microbiome was significantly different between groups (Fig. 3) and the difference was also more prominent in the gut microbe-derived EVs than in the stool microbiome.
Fig. 2.
Alpha diversity of the stool microbiome (A) and gut microbe-derived EVs (B) in patients with IBD and healthy controls.
EV, extracellular vesicle; IBD, inflammatory bowel disease; OTUs, operational taxonomic units. *p<0.05, †p<0.01, ‡p<0.001.
Fig. 3.
Principal coordinate analysis of the stool microbiome (A) and gut microbe-derived extracellular vesicles (B) in patients with inflammatory bowel disease (IBD) and healthy controls.
PERMANOVA, permutational analysis of variance.
2. Comparison of stool microbiome and gut microbe-derived EVs according to disease activity of IBD
There was no difference in the alpha or beta diversity of stool microbiome and gut microbe-derived EVs according to the disease activity of IBD (Supplementary Figs 3 and 4). Additional analyses in the ulcerative colitis and Crohn’s disease subgroups, revealed that there was no difference in the alpha or beta diversity of stool microbiome and gut microbe-derived EVs according to the disease activity of each disease (data not shown).
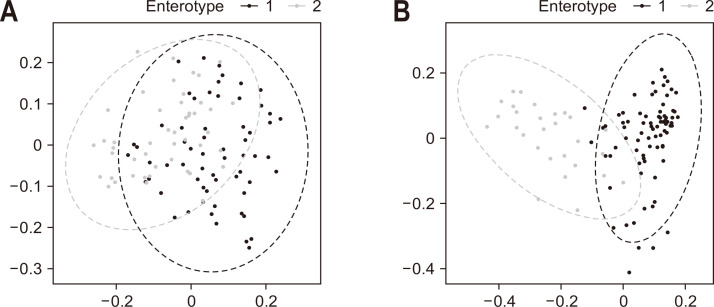
3. Enterotypes based on the stool microbiome and gut microbe-derived EVs in patients with IBD
Using cluster analysis, patients with IBD were divided into two enterotypes based on the bacterial composition of the stool (Fig. 4A) and the gut microbe-derived EVs (Fig. 4B). Enterotypes 1 and 2 were more clearly separated in the gut microbe-derived EVs than in the stool microbiome. Table 3 shows the taxa with a significant difference in abundance between enterotype 1 and enterotype 2 in the gut microbe-derived EVs of patients with IBD. Prevotella was uniquely significant for all the statistical methods considered in this study (Supplementary Table 4).
Fig. 4.
Cluster analysis based on the bacterial composition of the stools of patients with inflammatory bowel disease. (A) Enterotype based on the stool microbiome. (B) Enterotype based on gut microbe-derived extracellular vesicles.
Table 3.
Taxa Showing a Significant Difference in the Abundance between Enterotype 1 and Enterotype 2 in Gut Microbe-Derived Extracellular Vesicles of Patients with Inflammatory Bowel Disease
| Taxon | Log-transformation t-statistic | FDR adjusted p-value |
|---|---|---|
| Phylum Firmicutes | ||
| Family Ruminococcaceae | 2.36 | 0.030 |
| Family Enterococcaceae | −2.93 | 0.030 |
| Family Lachnospiraceae | −4.45 | 0.020 |
| Genus Clostridium_g21 | −3.84 | 0.008 |
| Species Ruminococcus gnavus | −3.84 | 0.010 |
| Genus Anaerostipes | −3.14 | 0.020 |
| Species Anaerostipes hadrus | −3.14 | 0.030 |
| Phylum Bacteroidetes | 9.48 | <0.001 |
| Family Prevotellaceae | 9.71 | 0.002 |
| Genus Prevotella | 9.71 | 0.002 |
| Phylum Actinobacteria | −4.49 | <0.001 |
| Family Bifidobacteriaceae | −3.98 | 0.002 |
| Genus Bifidobacterium | −3.98 | 0.007 |
| Family Coriobacteriaceae | −3.10 | 0.020 |
| Genus Collinsella | −3.09 | 0.030 |
| Phylum Proteobacteria | −3.61 | 0.001 |
The change of column (log transformation) represents the multiplicative change in taxa abundance from enterotype 1 to enterotype 2. Negative numbers represent decreased abundance of enterotype 2 compared with enterotype 1.
FDR, false discovery rate.
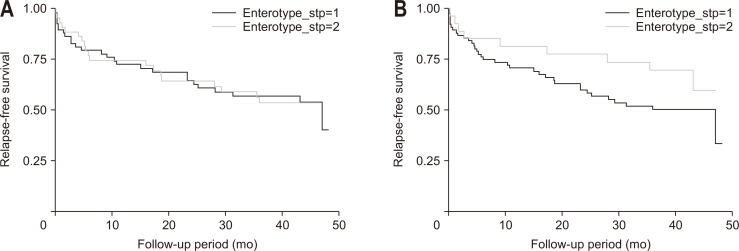
There was no significant difference in the risk of relapse between enterotype 1 and enterotype 2 based on the stool microbiome (log-rank test, p=0.926) (Fig. 5A). Enterotype 1 tended to have a higher risk of relapse than that of enterotype 2, based on the gut microbe-derived EVs. However, the difference was not statistically significant (log-rank test, p=0.166) (Fig. 5B). In multivariable analysis, elevated fecal calprotectin (>250 mg/kg) was the only risk factor for relapse identified (hazard ratio, 3.147; 95% confidence interval, 1.545 to 6.408; p=0.002) (Table 4).
Fig. 5.
Comparison of Kaplan-Meier survival curves according to enterotype. (A) Based on stool microbiome. (B) Based on gut microbe-derived extracellular vesicles.
Table 4.
Univariable and Multivariable Analyses of Predictors Associated with Relapse in Patients with Inflammatory Bowel Disease
| Variable | Univariable analysis | Multivariable analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | ||
| Age (>40 yr) | 0.617 (0.327–1.163) | 0.136 | 0.987 (0.965–1.009) | 0.247 | |
| Sex (female) | 1.065 (0.539–2.102) | 0.857 | |||
| Current smoker | 0.982 (0.456–2.112) | 0.962 | |||
| Disease duration (>2 yr) | 1.434 (0.768–2.678) | 0.257 | |||
| History of abdominal surgery | 1.140 (0.406–3.201) | 0.804 | |||
| Enterotype based on EVs | |||||
| Type 1 | Reference | ||||
| Type 2 | 0.600 (0.289–1.247) | 0.171 | 0.772 (0.335–1.777) | 0.542 | |
| C-reactive protein (>0.5 mg/dL) | 2.801 (1.485–5.283) | 0.001* | 1.379 (0.653–2.910) | 0.400 | |
| Fecal calprotectin (>250 mg/kg) | 3.418 (1.828–6.389) | <0.001* | 3.147 (1.545–6.408) | 0.002* | |
| Medication use | |||||
| Steroids | 3.033 (1.500–6.135) | 0.002* | 1.827 (0.784–4.260) | 0.163 | |
| Immune modulators | 2.227 (1.208–4.106) | 0.010* | 0.892 (0.413–1.927) | 0.772 | |
| TNF-α inhibitors | 2.226 (1.123–4.414) | 0.022* | 1.777 (0.786–4.015) | 0.167 | |
HR, hazard ratio; CI, confidence interval; EV, extracellular vesicle; TNF, tumor necrosis factor.
p-values were calculated using Cox regression. Relapse was defined as a composite outcome of (1) new use of steroids, immunomodulators, and biologics; (2) a visit to an emergency department; (3) hospitalization; or (4) abdominal surgery.*p<0.05.
DISCUSSION
In this study, there was a significant difference in the composition of the stool microbiome and gut microbe-derived EVs between patients with IBD and controls. The alpha and beta diversity of the microbiome in patients with IBD was significantly lower than that of controls. These results are consistent with previous studies.6,23 Importantly, according to the disease status, the differences were more prominent in the gut microbe-derived EVs than in the stool microbiome. This supports our hypothesis that analysis of gut microbe-derived EVs has greater potential than analysis of the stool microbiome as a tool for investigating the pathogenesis of IBD. EVs and their parental cells exhibit differences in their material profiles; therefore, the composition of the stool microbiome and gut microbe-derived EVs differs. The functional significance of nucleic acids carried in bacterial EVs is not clear.24 However, different material profiles between EVs and the bacterial microbiome suggest that the cargo loaded into EVs is carefully selected through a specific mechanism, and EVs are more likely a selective representation of the parental cells and disease state.
In this study, the microbiota that elicited significant differences between IBD and control groups were also different between the stool microbiome and gut microbe-derived EVs. In the analysis of the stool microbiome, Caproiciproducens, and Pseudoflavonifractor were high, while Blautia, Eubacterium_g5, Fusicatenibacter, Streptococcus, and Weissella were low in with IBD. In contrast, Klebsiella and Sphingomonas were more abundant, and Akkermansia, Diaphorobacter, Enterobacter, and Moraxella were less abundant in the gut microbe-derived EVs of patients with IBD. We searched the literature on the relationship between these bacteria and IBD. Among the genera that were different in stool microbiome, there were some reports of Eubacterium and Fusicatenibacter being decreased in patients with IBD.25,26 There are very few studies on other genera. Among the genera that were different in the microbiome derived from EVs, one study reported that Sphingomonas was increased in colitis-associated cancer compared to sporadic cancer.27 Another study found a decrease in Enterobacter count in pediatric patients with newly diagnosed Crohn’s disease.28 Additionally, Akkermansia abundance is known to decrease in patients with IBD.29,30 It is difficult to quantitatively evaluate whether analysis of the stool microbiome or analysis of gut microbe-derived EVs is the better model for microbiome analysis in IBD. However, considering the results of this study, we suggest that gut microbe-derived EVs may be more reliable and provide more information regarding the gut microbiome in patients with IBD.
In contrast to the survival curves according to stool enterotypes, the survival curves tended to differ according to gut microbe-derived EV enterotypes in patients with IBD; however, the difference was not statistically significant. Several studies have investigated whether the gut microbiome can predict clinical relapse in IBD, but the findings have varied.31,32 Many factors affect the clinical outcome and relapse of IBD. An elevated fecal calprotectin level was the only significant risk factor for relapse identified in the multivariable analysis, suggesting that clinical factors are more useful than complicated microbiome markers for predicting clinical relapse. Interestingly, Prevotella was a uniquely significant genus for all the statistical methods used in this study. Based on the analysis of the gut microbe-derived EVs, the relapse rate seemed to be higher in the group with less Prevotella (enterotype 1). When the human enterotype was first suggested, Prevotella, a representative short-chain fatty acid-producing bacterium, was classified as the main characteristic of a good enterotype.33 Enterotypes are strongly related to long-term dietary patterns. Long-term consumption of carbohydrates and a diet rich in protein and animal fats, are related to the abundance of Prevotella and Bacteroides, respectively.34 Nevertheless, there are few reports on whether a decrease in Prevotella in IBD is associated with a poor prognosis. Given the reports that the Mediterranean diet, with a high fiber intake, has some effect on IBD,35 Prevotella may have a protective role against flare-ups in IBD. However, a previous study suggested the potential of the Prevotella species as an intestinal pathobiont.36 Therefore, caution should be exercised when interpreting these results. To validate our hypothesis, further studies focusing specifically on Prevotella are required to assess the potential of using gut microbe-derived EVs to predict IBD flare-ups.
There was no difference in either the alpha or beta diversity of stool microbiome and gut microbe-derived EVs, according to the disease activity of IBD. Using a definition of active disease as a C-reactive protein level >0.5 mg/dL at the time of stool sampling, we did not find any significant results (data not shown). While some previous studies have shown positive results regarding whether the gut microbiome differs according to disease activity in IBD,37,38 other studies have not shown a difference.39,40 Further studies using multiomics are required to draw a conclusion regarding whether the gut microbiome differs according to disease activity in IBD.
This study has several strengths. Firstly, this study was unique. To date, most studies on EVs in IBD have focused on those originating from various host cells. In addition, most studies on bacterial EVs have focused on proteins as EV cargo. Few studies have compared gut microbe-derived EVs with the stool microbiome. Moreover, to our knowledge, no study has evaluated the relationship between gut microbe-derived EVs and clinical relapse of IBD. Despite performing meticulous survival analysis, we did not find significant biomarkers that could predict clinical relapse. All the clinical factors that affect the course of IBD were prospectively collected in a well-established cohort, and individuals were followed up for a sufficient length of time (median, 43 months). Secondly, microbial analysis was performed using robust methods. When we compared the microbiome between patients with IBD and healthy controls, we adjusted for many factors known to influence the composition of the microbiome, including age, sex, smoking, body mass index, and stool form. In addition, we applied statistical methods specifically developed for microbiome analysis.
However, our study also has some limitations. Firstly, we used 16S rRNA sequencing for microbiome analysis. Because this method provides limited sequencing depth, we have not discussed the results at the species level. Further studies using whole-genome sequencing are warranted. Secondly, although sequencing for microbiome analysis was performed using the same method, samples from patients with IBD and controls were not sequenced simultaneously. Therefore, there is a possible bias stemming from the batch effect. Third, although we adjusted for many factors, we did not collect data on history of antibiotic and probiotic use, or alcohol consumption in the patients with IBD. Therefore, we cannot rule out the possibility that these factors may have influenced the composition of the microbiome. Last, other samples such as blood and urine in these individuals were not available. Some EVs generated by gut microbes circulate throughout the body via the colonic mucosa and vascular system of the host,41 and are finally excreted in the urine. Therefore, validating the results using blood or urine samples, may provide concrete evidence in functional analysis.
In conclusion, gut microbe-derived EVs are better than the stool microbiome for differentiating patients with IBD from healthy controls.
SUPPLEMENTARY MATERIALS
Supplementary materials can be accessed at https://doi.org/10.5009/gnl220081.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation (NRF) of Korea grant funded by the Korean government (MSIT; No. 2021R1C1C1004170), and the Hyundai Motor Chung Mong-Koo Foundation.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Study concept and design: Y.S.P., H.Y. Data acquisition: Y.S.P., H.Y. Data analysis and interpretation: M.H., H.Y., N.E.K., K.K. Drafting of the manuscript: M.H., H.Y. Critical revision of the manuscript for important intellectual content: C.M.S., N.K., D.H.L. Statistical analysis: M.H., H.Y. Obtained funding: H.Y. Administrative, technical, or material support; study supervision: C.M.S., N.K., D.H.L. Approval of final manuscript: all authors.
REFERENCES
- 1.Park SH. Update on the epidemiology of inflammatory bowel disease in Asia: where are we now? Intest Res. 2022;20:159–164. doi: 10.5217/ir.2021.00115.b758430e5d2f48c9825f8dcc9ae32504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller R, Mazurak N, Fantasia L, et al. Quality of life in inflammatory bowel diseases: it is not all about the bowel. Intest Res. 2021;19:45–52. doi: 10.5217/ir.2019.00135.cee0bffdd97b4e2d95e1c62e0142fec9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizoguchi E, Low D, Ezaki Y, et al. Recent updates on the basic mechanisms and pathogenesis of inflammatory bowel diseases in experimental animal models. Intest Res. 2020;18:151–167. doi: 10.5217/ir.2019.09154.ef11dc7b229745468f1359d9ce213bd1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai K, Kunisaki R, Kakuta F, et al. Phenotypic characteristics of pediatric inflammatory bowel disease in Japan: results from a multicenter registry. Intest Res. 2020;18:412–420. doi: 10.5217/ir.2019.00130.4a9bc1a0cbf44f81935d8037a247f5ab [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin SY, Kim Y, Kim WS, et al. Compositional changes in fecal microbiota associated with clinical phenotypes and prognosis in Korean patients with inflammatory bowel disease. Intest Res. 2022 Jun 14; doi: 10.5217/ir.2021.00168. https://doi.org/10.5217/ir.2021.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry D, Reinisch W. Intestinal microbiota: a source of novel biomarkers in inflammatory bowel diseases? Best Pract Res Clin Gastroenterol. 2013;27:47–58. doi: 10.1016/j.bpg.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015;34:474–490. doi: 10.1002/mas.21420. [DOI] [PubMed] [Google Scholar]
- 9.Chang X, Wang SL, Zhao SB, et al. Extracellular vesicles with possible roles in gut intestinal tract homeostasis and IBD. Mediators Inflamm. 2020;2020:1945832. doi: 10.1155/2020/1945832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J, Kim NE, Yoon H, et al. Fecal microbiota and gut microbe-derived extracellular vesicles in colorectal cancer. Front Oncol. 2021;11:650026. doi: 10.3389/fonc.2021.650026.d14d1dfcec404fc98fec3a9ebd5b3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DJ, Yang J, Seo H, et al. Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci Rep. 2020;10:2860. doi: 10.1038/s41598-020-59529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin CM, Choi YJ, Lee DH, et al. Validity and safety of ID-JPL934 in lower gastrointestinal symptom improvement. Sci Rep. 2021;11:13046. doi: 10.1038/s41598-021-92007-3.a60bc0717083457d810206a5bd4207e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 14.Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon S, Lee B, Yoon S. CASPER: context-aware scheme for paired-end reads from high-throughput amplicon sequencing. BMC Bioinformatics. 2014;15(Suppl 9):S10. doi: 10.1186/1471-2105-15-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KJ, Park J, Park SC, Won S. Phylogenetic tree-based microbiome association test. Bioinformatics. 2020;36:1000–1006. doi: 10.1093/bioinformatics/btz686. [DOI] [PubMed] [Google Scholar]
- 21.Koh H, Blaser MJ, Li H. A powerful microbiome-based association test and a microbial taxa discovery framework for comprehensive association mapping. Microbiome. 2017;5:45. doi: 10.1186/s40168-017-0262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo Presti A, Zorzi F, Del Chierico F, et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front Microbiol. 2019;10:1655. doi: 10.3389/fmicb.2019.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valter M, Verstockt S, Finalet Ferreiro JA, Cleynen I. Extracellular vesicles in inflammatory bowel disease: small particles, big players. J Crohns Colitis. 2021;15:499–510. doi: 10.1093/ecco-jcc/jjaa179. [DOI] [PubMed] [Google Scholar]
- 25.Hiippala K, Jouhten H, Ronkainen A, et al. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients. 2018;10:988. doi: 10.3390/nu10080988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeshita K, Mizuno S, Mikami Y, et al. A single species of clostridium subcluster XIVa decreased in ulcerative colitis patients. Inflamm Bowel Dis. 2016;22:2802–2810. doi: 10.1097/MIB.0000000000000972. [DOI] [PubMed] [Google Scholar]
- 27.Richard ML, Liguori G, Lamas B, et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes. 2018;9:131–142. doi: 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sila S, Jelić M, Trivić I, Tambić Andrašević A, Hojsak I, Kolaček S. Altered gut microbiota is present in newly diagnosed pediatric patients with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2020;70:497–502. doi: 10.1097/MPG.0000000000002611. [DOI] [PubMed] [Google Scholar]
- 29.Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 30.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X, Huang C, Xu J, et al. Gut microbiota is a potential biomarker in inflammatory bowel disease. Front Nutr. 2022;8:818902. doi: 10.3389/fnut.2021.818902.ae94e12d620c47a5b64e795cc957b090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun T, Di Segni A, BenShoshan M, et al. Individualized dynamics in the gut microbiota precede Crohn's disease flares. Am J Gastroenterol. 2019;114:1142–1151. doi: 10.14309/ajg.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 33.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis JD, Sandler RS, Brotherton C, et al. A randomized trial comparing the specific carbohydrate diet to a Mediterranean diet in adults with Crohn's disease. Gastroenterology. 2021;161:837–852. doi: 10.1053/j.gastro.2021.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Precup G, Vodnar DC. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: a comprehensive literature review. Br J Nutr. 2019;122:131–140. doi: 10.1017/S0007114519000680. [DOI] [PubMed] [Google Scholar]
- 37.Prosberg M, Bendtsen F, Vind I, Petersen AM, Gluud LL. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol. 2016;51:1407–1415. doi: 10.1080/00365521.2016.1216587. [DOI] [PubMed] [Google Scholar]
- 38.Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 39.Galazzo G, Tedjo DI, Wintjens D, et al. Faecal microbiota dynamics and their relation to disease course in Crohn's disease. J Crohns Colitis. 2019;13:1273–1282. doi: 10.1093/ecco-jcc/jjz049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Öhman L, Lasson A, Strömbeck A, et al. Fecal microbiota dynamics during disease activity and remission in newly diagnosed and established ulcerative colitis. Sci Rep. 2021;11:8641. doi: 10.1038/s41598-021-87973-7.888e6863e5fa45338bf1f85f53a2768b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang SC, Kim SR, Yoon YJ, et al. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small. 2015;11:456–461. doi: 10.1002/smll.201401803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.