Abstract
Machine learning has been leveraged for image analysis applications throughout a multitude of subspecialties. This position paper provides a perspective on the evolutionary trajectory of practical deep learning tools for genitourinary pathology through evaluating the most recent iterations of such algorithmic devices. Deep learning tools for genitourinary pathology demonstrate potential to enhance prognostic and predictive capacity for tumor assessment including grading, staging, and subtype identification, yet limitations in data availability, regulation, and standardization have stymied their implementation.
Keywords: Artificial intelligence, Computational pathology, Genitourinary pathology, Digital pathology
Introduction
Multiple innovations and increasing use of digital pathology workflows has led to more research in the area of computational pathology and artificial intelligence (AI)-based tools for genitourinary pathology diagnositcs. Increased emergence of deep learning tools has followed suit with advances in technology, e.g., increased image capture and processing speeds,1 enabling the development of programs tasked to evaluate differences in cancer grading. Prostate cancer identification and grade-assessment has been the initial and primary focus for AI-based approaches in GU pathology, with comparative paucity seen in the development and interrogation of such tools purposed for other GU foci, e.g., bladder, testis or kidney.2, 3, 4, 5 This review paper from the Genitourinary Pathology Society (GUPS) seeks to distinguish the merits of digital pathology-based AI tools developed for GU practice through discussion of their diagnostic, prognostic, and therapeutic potential while offering insight on future utility and challenges currently preventing full implementation.
Digital pathology: Roadway to computational pathology
The rapidly blossoming arena of computational pathology has enabled the objective diagnosis of whole slide images (WSIs) through optimization of digital pathology (DP) workflow.6 These innovations have allowed for an integrated digital workflow in laboratories where pathologists have access to all the necessary clinical information and images related to the case. These applications and viewing solutions are web-based and allow for integration of AI applications. These applications also allow for remote workflows, clinical decision-support education, and research.7,8 AI-based machine learning (ML) approaches brought forth through computational pathology have allowed for sophisticated image analysis software to translate WSI pixels into numeric data for mathematical models, i.e., algorithms, to aid pathologist interpretations (Table 1).9
Table 1.
Limitations to diagnostic AI implementation.
| Limitations | Discussion | Current and future solutions |
|---|---|---|
| Data | ||
| Data standardization | A considerable volume of data is generated for algorithm development, often derived from multiple sources, presented via multiple file formats, and analyzed through multiple AI models. Current analytical methods are non-standardized, consequently predisposing to variations in classification and poor predictive capacity. | Fostering of a single open-source file format, similar to DICOM in radiology, to facilitate expeditious access and interrogation. |
| Data availability and cost | • Paucity of WSI datasets with pathologist annotations for ground-truth determination limits employment of supervised learning techniques. The lack of WSI datasets impeding the analytic capacity of deep learning techniques is emphasized in GUP primarily pertaining to immunohistochemical (IHC) staining. • Labeled WSI data is expensive, difficult to acquire, and time-consuming to produce. WSI data storage costs have posed barriers to digital implementation in many laboratories. |
• “Transfer learning,” e.g., pretrained networks, and data augmentation techniques may be utilized to mitigate the cumbersome nature of network training and data shortages, though are not currently capable of acting as substitutes for pathologist-annotated data • Increased utilization of unsuperised learning techniques, which do not require labeld data • Circumvention of restrictions brought forth by data privacy and proprietary techniques through open-source accessability in conjunction with capital leverage for pathologist annotations |
| Data size | Workflow (hardware and infrastructure) limitations, e.g., large network bandwith requirements to handle large WSI file sizes | Advances in WSI scanning technology and digital data transmission coupled with decreased costs of implementation on the horizon |
| Data quality | • High-resolution image reduction techniques, e.g., patch extraction, may compromise data quality. Higher-level structural information, e.g., tumor extent or shape, may only be captured through analysis of larger regions. • Clinical translation of algorithms requires generalizability throughout a wide breadth of patient populations and clinical institutions. IHC / H&E staining of tissue sections can vary significantly across laboratories and at intra-laboratory level. Analysis performed on low-quality tissue, histology slides, or staining will ultimately compromise the validity of data |
• Focused spatial correlation amongst patches, multi-level magnification patch extraction, utilization of larger patch sizes. • Normalization techniques, e.g., scale normalization for multiple image aquisition devices with varying pixels sizes, stain normalization, pixel-wise and patch-wise and semantic segmentation CNN training for enhanced region of interest detection, flexible thresholding techniques which compromise for variations in input data luminance. |
| Data utilization capacity | Deep learning systems are currently only able to classify WSI specimens with a single diagnosis. | • Removal of biological restricitons during algorithmic training • “Artificial General Intelligence (AGI)” of the future will consist of advanced algorithms employing multiple levels of classification and segmentation in conjunction with a litany of diagnostic deductive variables, mimicking the process of human conciousness. |
| Regulation / medico-legal / accountability and/or liability | • Demonstration of algorithm reproducibility on large patient populations containing outliers and non-representative individuals has caused difficulties for AI development. • “Black Box” transparency concerns surround the uninterpretable pathway of algorithmic classification deduction. Segmentation, e.g., extraction, of image objects correlated with clinical endpoints are hidden from pathologist interpretation. |
• AI Models of the future may be used to develop "universal" tumor grading systems applicable to the entire GU system through combination of prognostic, morphologic, tumor marker, and clinical course data. • Rule extraction’, through which information about histopathologic features used by an algorithm during its previously hidden segmentation process, may mitigate such concerns. |
The first documented efforts of computer-aided diagnosis (CAD) in GU pathology are over 10 years old.10 Initial studies published from 2010 to 2013 presented promising data on 58 biopsy11 and 15–20 radical prostatectomy cases,10,12 demonstrating 90% accuracy in distinguishing benign from malignant cases in 1 study12 and a respective sensitivity and specificity of 0.87 and 0.90 in another.10 Many algorithms for image analysis employ context-based gland quantification to distinguish benign prostatic tissue from prostate cancer.10,13
Recent investigations have focused on AI-based prostate cancer detection using needle biopsy slides,5,14, 15, 16, 17, 18, 19, 20, 21, 22 radical prostatectomy slides,23 tissue microarrays,16,24, 25, 26 or, rarely, a combination of radical prostatectomy and TMA slides.16,27 The current era of technological advancement has fostered development of computational tools purposed for the precise, reproduceable extraction and analysis of WSI data for pathologist-assisted diagnostics, biomarker discovery, and individualized precision medicine.28,29 Such techniques allow for the extraction of information from digitized images of patient specimens in combination with associated metadata.
Machine learning models have been developed to assist GU pathology related diagnoses through the identification of basic morphological patterns.30,31 Such models recognize histopathological patterns of interest vis-a-vis supervised training with scores of labeled, i.e., pathologist-annotated, WSI data pertaining to features associated with a known outcome (Fig. 1A). Modeled from neural networks within the human brain that enable self-iterated improvement in cognitive deliberation, deep learning has emerged as an evolutionary progression from machine learning by which programmable artificial neural networks (ANNs) are enabled to independently interpret or predict outcomes from unstructured or unlabeled WSI data inputs (Fig. 1B). These networks can self-ascertain a plethora of patterns present in a WSI input without assistance of a predefined output set. ANN architecture is supported by a scaffolding of “nodes,” (artificial neurons), including an input layer, multiple hidden layers, and an output layer (Fig. 1C).
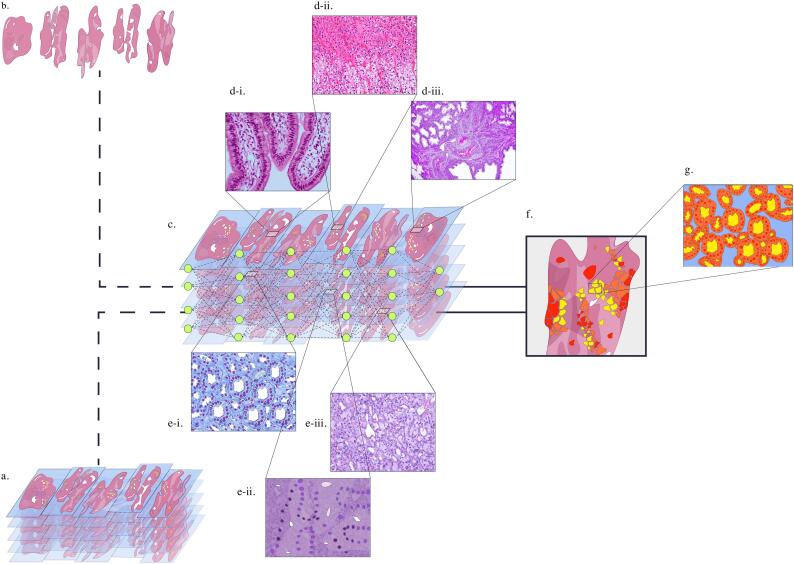
Fig. 1.
Algorithm training and feature extraction (prostate cancer).
A. Labeled WSIs used for CNN training.
B. Unmarked/non-annotated WSI data.
C. CNN self-identifying and classifying unmarked WSI input data.
D. (I–III): Non-cancerous histopathological patch classification from feature extracted normal glandular epithelium, smooth muscle, and prostatic blood vessels in stroma (in order from I to III).
E. (I–III): Cancerous histopathological patch classification from feature extracted infiltrative margins indicative of Gleason pattern (GP) 3, irregular masses of neoplastic glands indicative of GP 4, and occasional gland formation indicative of GP 5 (in order from I to III).
G. Colorized CNN output distinguishing feature-classified areas of prostate cancer corresponding to Gleason pattern score where yellow = GP3, orange = GP4, and red = GP5.
H. Automated intelligent segmentation demonstrating detection of digitized histological patterns in a region of tissue classified as GP3 where red = epithelial nuclei, orange = epithelial cell cytoplasm, and yellow = lumen.
Output results from several, often independent, steps of computation, weighting (strength of neural node connection), and assessment. Multiple layers of ANNs are used in deep learning to extract higher-level input features from WSI (Fig. 1D & E).32, 33, 34, 35 With no predefined output or outcome, the algorithm is enabled to determine what it defines as natural patterns present in the WSI input, i.e., “feature extraction.” Convolutional neural networks (CNNs), widely used for WSI analysis, have demonstrated efficacy in varying WSI analysis applications through utilization of an end-to-end learning model trainable from databases containing up to millions of WSIs.14,36 Fundamental deep neural architecture is reflected by multiple configuring layers of CNNs, with a convolutional layer functioning as a feature extractor peppered by neurons of varying weights arranged into complex feature maps.36 Histopathological classification of feature extracted tissue, e.g., cancerous vs. non-cancerous, occurs via automated object-based segmentation of similar image pixels within WSI data inputs (Fig. 1D, E).
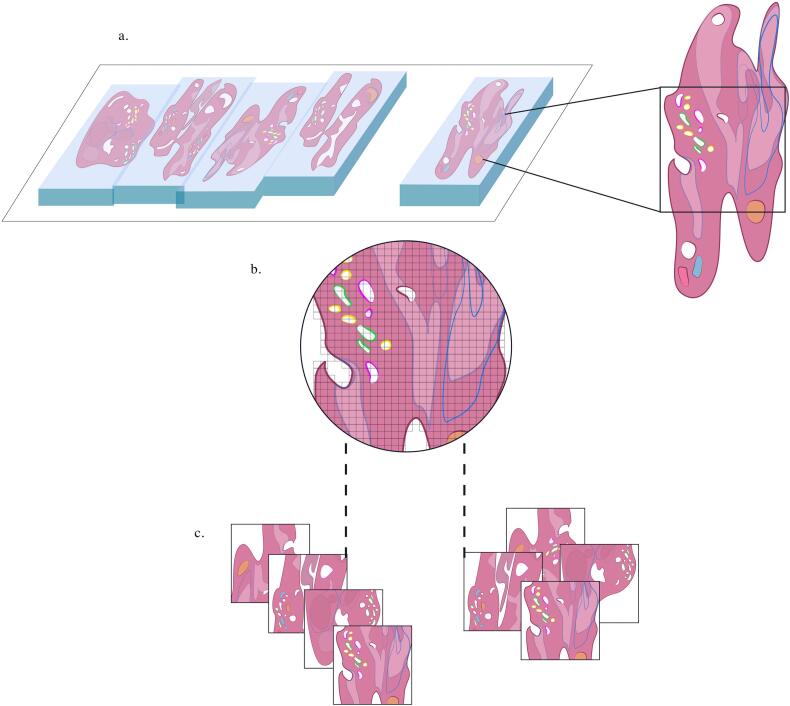
GU specialist annotation via digital segmentation tools is performed to define “ground-truth” classifications to object boundaries that may be used to evaluate neural network performance (Fig. 2A). Small-prototype image regions (patches) are extracted from WSIs to train CNNs to detect features of interest (Fig. 2B, C). During training, an algorithmic model uses labeled data from WSIs tagged with designating properties, characteristics, and classifications via ground-truth annotations by expert pathologists.
Fig. 2.
Annotation and patch extraction (prostate).
A. Genitourinary specialist annotated prostate WSIs for CNN training.
B. Series of “patches” selected from annotated WSI regions for iterative patch-based training of CNN parameters.
C. Example of data augmentation in which mirroring and cropping of extracted patched images occurs to increase training dataset variation.
Lack of substantial training data may cause an algorithmic model to “fit” too precisely to a limited dataset, thereby greatly hindering model performance in accurately classifying new or unlabeled data. There are a number of preclinical analytical factors that may contribute to variability in algorithm performance. Deep learning frameworks curated for generalized assessment of histopathology analysis address a multitude of challenges plaguing automated WSI analysis including, though not limited to large WSI dimensions, insufficiency of WSI samples for training, staining variability across laboratories, and the appropriate extraction of clinically relevant features and meaningful information from WSIs.37 Significant headway has been made towards realizing generalizability for deep learning applications in genitourinary pathology within the realm of prostate cancer diagnostics.
AI solutions for GU pathology
Prostate cancer
Histopathological diagnosis of prostatic adenocarcinoma, typically executed via light-microscopy examination of hematoxylin and eosin (H&E) stained tissue sections, requires interpreting of a constellation of features38 sometimes beguiling malignancy and navigating the challenges presented by biopsies with limited tissue presence.39 Furthermore, pre-analytic (e.g., tissue artifact) and analytic (e.g., minimal or limited adenocarcinoma of the prostate in needle core tissue) confounders40 have contributed to the resulting inter-observer variability associated with this routine diagnostic process.41 Analytical and pre-analytical factors cannot be mitigated by AI and digital workflow alone and carefull assessment of these factors is needed in order to implement AI solutions in the laboratory (Table 1, Table 2, Table 3).
Table 2.
Deep learning concepts in GUP.
| Algorithm training | During training, an algorithmic model uses labeled data tagged with designating properties, characteristics, and classifications describing an object of interest, such as WSI, to learn - thereby increasing the model’s propensity for correct predictive diagnostics, among other decision-making processes. Algorithms are classified as “supervised” (requiring human intervention), or unsupervised. Supervised machine learning techniques are utilized in numerous medical specialties, with recent application in pathology. During training, an algorithmic model uses labeled data tagged with designating properties, characteristics, and classifications describing an object of interest, such as WSI, to learn - thereby increasing the model’s propensity for correct predictive diagnostics, among other decision-making processes. |
| Artificial neural network (ANN) | Deep learning techniques are a subset of machine-learning algorithms which use artificial neural networks (ANN) to develop independent interpretative or predictive ability from unstructured or unlabeled data. With no predefined output set, the algorithm is enabled to determine what it defines as natural patterns present in the GUP input (WSIs). Such techniques use multiple layers of artificial neural networks to extract higher-level input features. |
| Convolutional neural network (CNN) | CNNs used in WSI analysis share the fundamental architecture of deep neural networks (input layer, multiple hidden layers, output layer). Each CNN layer responds to different pattern-forming features within a WSI. Features such as object boundaries can be elicited during automatic segmentation through CNN, i.e., “feature extraction”. Manual annotation through digital segmentation tools is also performed by GUP specialists for defining “ground-truth” classifications for object boundaries to benchmark neural network performance. |
| Area-based measurements | Quantify the parameters of basic elements forming the blueprint of a WSI. Pixel-based assessment is applied to area-based measurements wherein the color or intensity of staining in each pixel within a designated area is quantified through algorithm(s). |
| Cell-based measurements | Cell-based measurements identify and enumerate cells or nuclei through morphometry-based assessment, through which similar pixels (e.g., in size or shape) are grouped to predefined cell-structure profiles meeting specific criteria. |
Table 3.
GU prostate study limitations.
| Limitations | Discussion |
|---|---|
| The studies of biopsies assess individual biopsies, not the full set of biopsies (typically 12) taken during a prostate biopsy session | • Biopsy samples should be representative of prostate cancer. More specifically, they should represent tumor regions with increased risk of progression. |
| None of the studies have assessed correlation of grade by AI with clinical outcome | • Model-fitting is improved if validation is based on the patient rather than cores or patches. • Performance of Deep Learning in distinguishing low-grade from high-grade prostate cancer by “leave-out” analytical components is best through excluding patches/cores within iterative studies, using solely patient-based data. |
| The number of pathologists involved in selecting assessment sets vary from 1 to 15 | • The larger the number of pathologists, the more likely the algorithm is to be applicable to clinical samples. • Likewise, increased institutional-origin of samples consequently enhances algorithm robustness. In studies, the number of institutions from which the tissue originated, ranged from 1 to 3. |
| None of the studies report whether variants of each grade that might be challenging to grade are explicitly included | • Although the highest-grade component is conventionally regarded as the part which cancer cells might progress, there are exceptions to this presumption. • Exceptions may belie the assumption of the largest area of tumor involvement being the most likely to be progressive. |
| The methods and materials used in different studies are additional sources of variance | • Sample nature varies within differing studies. Tissue sources included tissue microarrays, needle biopsy and radical prostatectomy samples. • The unit of image (patch size) varies. • Investigators have used differing CNN programs, including MobileNet and NASNetLarge. NASNetLarge demonstrated increased accuracy in comparison to other programs including ResNet, Inveption V3, Xception, and MobileNet. |
| Additional sampling inconsistencies | Sampling varies within prostate tumor site. Tumors in the anterior/transition zone are under-sampled by transrectal biopsies. |
| Only limited data is available on radical prostatectomy and transurethral specimens |
Complications in routine pathology reporting, interpretation, and histopathological assessment of Gleason score have fomented the last decade of shifting practices in prostate cancer grading. The turn of the millennium brought forth greater understanding of morphological patterns spearheading revisionary guidelines for prostate grading by the International Society of Urological Pathologists in 2005.42 Extension of Gleason pattern (GP) 4 to incorporate poorly formed glands (those without lumens, those with rare lumens, elongated compress glands, and elongated nests) and cribriform cancers, in conjunction with discouragement for reporting Gleason scores of <5, particularly from needle biopsies (for which further changes included inclusion of minute, high grade foci), led to greater reporting of high-grade cancer as a function of often minimal findings of poorly formed or fused bands. High inter-observer variability has marred the diagnosis of poorly formed glands, even among expert urologic pathologists (κ=0.34) 43, who have expressed concern pertaining to the compromised significance of GS 7 following the 2005 guideline shift in pattern classification. In 2012, Egevad et al studied the shift in approach to Gleason grading among 337 pathologists. Findings demonstrated that, for the first time, general pathologists graded more aggressively than did uropathology experts. Authors hypothesized that if aggressive grading was perceived corollary to expertise in prostate pathology, such association may incline general pathologists to favor aggressive grading for adherence to newly cemented ideals of best practice and avoidance of criticism.42
Prostate carcinoma grade from needle biopsies bears clinical importance.44 Patients with cancer of the lowest grade (WHO grade group 1, or Gleason score 6) are candidates for active surveillance. Cancers of intermediate grade are candidates for definitive therapy—radical prostatectomy or radiation therapy. Patients with cancers of the highest grade can be enrolled in clinical trials using neoadjuvant therapy to potentially decrease the size and stage of the tumor. Though grade is an important metric for assessment and categorization of patients with prostate cancer, grade assignment by manual microscopy has limitations. Visual interpretation of histological slides, typically 3 levels for each of 12 biopsies, is time consuming. Moreover, grade reproducibility is subject to inter- and intra-pathologist subjectivity. Multiple studies revealed the kappa values of inter-pathologist variance to range between 0.27 and 0.7.45,46 Particularly challenging is distinguishing tangentially sectioned GP 3 from GP 4 (kappa = 0.27).46 Intra-pathologist variance is also notable in this instance. Reproducibility of grade ranging from 65% to 100% (mean 81.5%) has been reported, with lowest reproducibility recorded for the cribriform variant of GP 4.46 Ever more strikingly, many inter-observer studies exclusively evaluate GU expert assessment of prostate cancer while skirting inclusion of a wider breadth of practicing pathologists.47, 48, 49 Inclusion of a more diverse range of non-specialist community pathologists in such studies may demonstrate far lower kappa values.
Prognosis has also been subjected to incongruous consensus and confusion resulting from emphasis of higher-grade groupings, such as GP 5 patterns present within GP 8 tumors. Many issues remain regarding diagnosis and quantification of high-grade patterns.50
There are currently no recommendations suggesting incorporation of histologic features (apart from gland architecture) into tumor grading or surgical pathology reports, despite such features conferring worse prognosis than Gleason score alone.51
Cribriform gland with irregular border is an important feature in GP 4, as the diagnosis of cribriform GP4 (CrP4) is independently associated with adverse clinical outcomes, with a growing body of evidence strongly suggesting its presence as impactful upon clinical management of prostate adenocarcinoma, yet inter-observer consensus remains non-reassuring for expert delineation of varying gland structures, particularly with fused and ill-defined cribriform patterns.47,52, 53, 54 The inherently error-prone nature of subjective human manual assessments predisposes to inconsistency in identification.54, 55, 56 Cribriform growth patterns are associated with clinically worse biochemical recurrence-free, metastasis-free, and cancer-specific survival.56 Therein emerges a series of broken steps, parlayed from diagnostics into prognostics and patient care, where machine learning tools have emerged to deliver robust, replicable identification of cribriform glands and other histopathological features that may soon find themselves cemented within standard grading procedure. Machine learning tools have recently demonstrated promising results for the automated detection of cribriform patterns in prostate histology images.49,55,56 For the first time, researchers employed quantitative machine-based methods to investigate the prognostic utility of invasive cribriform adenocarcinoma (ICC) within specific Gleason grade groups, with results suggestive of ICC morphology fraction of tumor area (cribriform area index) having a strong prognostic role in patients with Gleason grade 2 cancer with little GP4 due to higher concordance index with biochemical recurrence than patients without evidence of ICC. Doubling of cribriform area index in those with ICC morphology was prognostic after controlling for Gleason grade, surgical margin positivity, pre-operative prostate-specific antigen level, pathological T stage, and age, yielding a hazard ratio of 1.19.56
AI tools for image analysis have allowed a more objective diagnostic recognition of prostatic adenocarcinoma, as such tools are not as subject to inter-observer variance which may stem from innumerable biases.42 As variance in inter-pathologist grading for prostate cancer is significant (eliciting a kappa value as low as 0.3), gold-standards of differing grades must be identified.46,57 Deep learning algorithms may be validated against expert-annotated, “gold-standard,” WSIs to assess accuracy, precision and reproducibility of the algorithmic model’s ability to identify variations in tissue grading.
AI-assisted prostate cancer detection & grading
The first effort to utilize deep neural networks for prostatic adenocarcinoma detection in needle biopsy tissue, reported in 2016, involved the digitization and annotation of H&E-stained prostate biopsy tissue slides from 254 patients which were separated into training, experimental, and validation sets.14 Slide image patches were used to train CNNs for cancer detection. Mean ROC for the median analysis was 0.98–0.99 for the 90th percentile analysis. Up to 32% of cancer-negative slides could be excluded, potentially streamlining diagnostic sign-outs (Table 3).
Arvaniti et al. trained MobileNet, a deep CNN derivative, to grade prostate cancer using a discovery cohort of primary prostate carcinomas as 0.6 mm diameter cores in tissue microarrays from 641 patients. MobileNet was assessed on a test cohort of tissue microarray cores from 245 prostatectomies. The test cohort had been graded independently by 2 pathologists. Using Cohen’s quadratic kappa statistic, agreement between the AI model and each pathologist was 0.75 and 0.71, respectively. Agreement between the 2 pathologists was 0.71. Since the clinical outcome of each patient was known, the accuracy of the model in categorizing each cancer into low risk vs. intermediate risk was assessed. The model was significantly more accurate in distinguishing low-risk from intermediate-risk (P=.098) than was either pathologist (P=.79 and P=.29, respectively).25 However, misclassifications at tissue borders from preparation artifacts were noted along with incorrectly annotated stromal tissue as GP 3, potentially from occasional inclusion of stromal tissue in training regions annotated as GP 3.
Nagpal et al. used images from 752 tissue biopsies from 4 sources to train a deep learning system. In a study of biopsies from 752 patients, agreement of the model-based grade was reported, with grading by 2 GU pathologists and separately by general pathologists. Agreement occurred in 72% (68%–75%) and 58% (54%–61%), respectively. The model was less likely to over-grade WHO grade group 1 than grade group 2. The model was also less likely to over-grade WHO grade group 1 and more likely to undergrade higher grades in comparison to general pathologists. ROC curves distinguished model-based grade groups 1 and 2 from grade groups 3 through 5 (AUC = 0.97).58
A CNN used in Bulten’s study of 5759 biopsies from 1243 patients had a kappa of 0.85 compared with 3 GU pathologists, superior to the kappa of 0.82 from a pathologist panel.59
Strom reported the performance of a study based on needle biopsies from 976 patients in which the mean kappa was 0.62, similar to the kappa of 15 participating GU pathologists (kappa range 0.60–0.73).5
Bulten et al.60 demonstrated that use of AI improved overall grading of prostate biopsies. A group of 14 GU pathologists graded a set of 160 prostate biopsies. Fourteen pathologists graded 160 biopsies with and without AI algorithms. Using AI, the 14-member panel of pathologists agreed significantly. Quadratically weighted Cohen’s kappa was 0.87 vs. 0.799 when graded independently.
The performance of AI algorithms has, within the parameters of these studies, demonstrated equivalence to the capabilities of GU specialists while surpassing those of general pathologists. However, as methods and materials employed were not subject to standardization (e.g., tissue sample variations from microarray, needle biopsy, and radical prostatectomy sources), evaluation of their results as a conglomerate, rather than a collection of individual exploratory endeavors, is speculative and therefore potentially misleading. Additionally, deep learning models were trained by WSIs of individual biopsies with varying image patch sizes, rather than complete sets of 12 tissue cores typically obtained during clinical assessment for prostate cancer. Studies ostensibly eschewed clinical relevance by neglecting to assess the correlation of AI-grading with clinical outcome.
Prostate biopsy samples must properly represent the lesional tissue which could be prostate cancer or benign mimickers of prostate cancer, not just the areas conferring increased risk of progression. Convention often assumes such areas to be tissue regions of the highest-grade, however there are exceptions which belie this presumption. Information indicating the explicit inclusion of grading variants potentially posing challenges to the grading process was not provided within our selection of studies. Sample site variance must also be accounted for, as tumors in located in the anterior/transition zone of the prostate are notably under-sampled by transrectal biopsies.61,62
Given inter-pathologist variance in grading, the gold-standard or “ground truth” of grade is more robust if established by more than 1 pathologist. 26 The number of pathologists involved in selecting assessment sets in our study selection varied from 1 to 15.25,58, 59, 60 The larger the number of pathologists used for selection, the more likely the algorithm is to be applicable to clinical samples. Likewise, increase in the institutional origin of samples consequently enhances algorithm robustness. In these studies, the number of institutions from which the tissue originated ranged from 1 to 3.25,58, 59, 60
Model-fitting may be homed for clinical implementation if validation is based on the patient rather than tissue cores or image patches. Performance of deep learning in distinguishing low-grade from high-grade prostate cancer by “leave-out” analytical components is best accomplished through excluding patches/cores within iterative studies and using solely patient-based data.26
Differing deep CNN models were used by investigators, including MobileNet25 and NASNetLarge.27 In the latter study, NASNetLarge was more accurate than other programs, which included ResNet, Inveption V3, Xception, and MobileNet. Conversely, Arvaniti found that MobileNet was less likely to overfit than other evaluated programs (VGG-16, Inception-V3,3 ResNet-50, and DenseNet-121).25
Substantial progress has occurred over the last 2 years, with reports which variably included the following characteristics: large numbers of cases; use of training, validation, and test sets; interpretations by expert GU pathologists to establish “ground truth” for diagnosis; use of slides from multiple institutions; use of differing scanners, including scanners from other institutions.5,18,22,27,63 Achieving standardization of such characteristics is corollary to achieving generalizability of computer-aided diagnostic tools.
High throughput deep learning facilitating generalizability
Two large-capacity, high-speed WSI scanning systems, the Ultra-Fast Scanner, i.e., “UFS”, (Philips IntelliSite Pathology Solution, Amsterdam, Netherlands) and the Aperio AT2 System® (Leica Biosystems,™Wetzlar, Germany), were used in a 2019 Campanella et al.18 study to evaluate a deep learning system trained on WSIs of 12 132 prostate needle core biopsies from Memorial Sloan Kettering (MSK). Following training, the system was evaluated by 12 727 prostate needle core biopsies sourced from institutions around the world.
The Aperio AT2 System® scanned most biopsy samples produced at MSK. A smaller subset of 1274 biopsies were scanned by the UFS at the same location. Data from WSIs captured by these devices, in conjunction with that of global-origin WSIs, allowed for a robust assessment of generalizability. The investigators found that approximately 10 000 slides were necessary for effective training of their system.64 Authors found variations in brightness, contrast, and sharpness affected predictive performance in both WSI devices, with PIPS demonstrating a 3%-point change in the area under the curve (AUC) accordingly. Additionally, 6 false negatives were characterized by very low tumor volume.40 In 72 cases, the algorithm falsely identified foci of small glands as cancer in instances where small glands had hyperchromatic nuclei and at least a few cells with prominent nucleoli. The investigators concluded that their prostate model would allow removal of >75% of slides from pathologist workloads without sensitivity losses, and that their system could allow non-subspecialized (non-GU pathologists) to diagnose prostate cancer confidently and efficiently. As WSIs in this study were tied 1:1 with synoptic data elements, primarily benign vs. adenocarcinoma, the weakly supervised framework used provided a scalable mechanism by which datasets may be created, thereby addressing an issue plaguing the current development of AI tools for GU pathology. However, as investigators did not specifically distinguish which of the 75% of cases were removeable, the projected clinical and operational applicability of their study is still left to conjecture.
Another high-volume study from the year 2020 assessed how a deep learning system trained on 36 644 WSIs, 7514 of which had cancerous foci (Paige Prostate Alpha®, Paige AI, New York, New York.) influenced pathologists in the detection/diagnosis of prostatic adenocarcinoma. 304 expertly annotated prostate needle biopsy WSIs were used to establish ground truth for evaluation of the deep learning system.21 General pathologists had an average sensitivity of 74% and specificity of 97% without the system, and an increased sensitivity to 90% with no change in specificity with the system. The authors concluded that this system could serve as an effective second-read, e.g., quality assurance, tool. Moreover, the system could be used in settings where GU pathology subspecialists were not present, including countries with large-scale healthcare disparities. Finally, the Paige Prostate® system has been validated in 2 additional institutions,65,66 including one in a separate country.65 Earlier this year, Perincheri et al.66 employed Paige Prostate® to review 1876 prostate core whole slide images, with a sensitivity of 97.7% and specificity of 99.3%. The authors noted an area for possible improvement of the algorithm was the enhanced identification of out-of-focus scans. Regarding carcinomas missed by the algorithm, 80% were <1 mm and 2 had foamy gland features, indicative of false-negative results.40 An additional study65 highlighted Paige Prostate® to display high sensitivity (99%) and specificity (93%) at part-specimen level. Paige Prostate® also identified 4 patients whose diagnoses were upgraded from benign/suspicious to malignant. The platform software was recently authorized for use as adjunct (supplement) by the FDA for detection of areas suspicious for carcinoma in digital images of prostate needle biopsy tissue sections.67
Also in 2020, Strom et al.5 reported on employing neural networks in assessing 6682 digitized prostate needle biopsies from 976 patients. An external validation set contained 87 biopsies evaluated by 23 global GU pathologists. The system achieved ROC AUC of 0.997 for diagnosing benign vs. prostatic adenocarcinoma on the independent test set and 0.986 on the external validation set. They concluded their AI system could detect prostatic adenocarcinoma comparable to international experts in GU Pathology. Strengths of this investigation were large cohort sizes and the inclusion of deceptively benign-appearing prostatic adenocarcinomas such as pseudo-hyperplastic and atrophic pattern variants, mimickers of prostatic adenocarcinoma, and sections with thick cuts, fragmentation, and poor staining. These sections, with pre-analytic artifacts, can confound prostatic adenocarcinoma detection.40 Evaluations for all AI systems should include cases with pre-analytic artifacts, benign mimickers of prostatic adenocarcinoma,68 and deceptively benign-appearing prostatic adenocarcinomas.40,69
Pantanowitz et al.22 covered development of an AI-based algorithm trained on 549 H&E-stained slides, with an internal test set of 2501 slides and an external test set of 1627 slides digitized on a second scanner. The algorithm achieved an AUC of 0.997 for prostatic adenocarcinoma detection in the internal test set and 0.991 in the external validation set. The AI tool was further tested on 11 429 slides (from 941 cases) as a second-read system. In that test, 10.9% of slides diagnosed by the pathologist as benign were flagged as cancer, 9% of which (n=51), upon pathologist re-review, prompted orders for additional sections or stains. Re-review of those cases was estimated to consume a minimal amount of time. One case initially diagnosed as benign by the pathologist was flagged by the AI tool and was subsequently diagnosed as low-volume Gleason score 6 prostatic adenocarcinoma. This study highlights the usefulness of an AI-based algorithm as a quality assurance second-read procedure, in a routine practice setting. Such AI-based second-read quality assurance procedures can be less time-consuming than manual human second-read procedures, although comparative time data are not yet available.
Tolkach et al.27 described in 2020 the development of deep learning models for the detection of prostate cancer tissue in WSIs of 379 radical prostatectomy cases in a training cohort, with 2 TMA validation cohorts from 2 different institutions. Accuracy of prostatic adenocarcinoma detection was 97.3%–98%.
Evaluating AI-assistance to understand overreliance
Employing expert-level AI assistance was evaluated in by Steiner et al.2 Accuracy of tumor detection with and without AI assistance was evaluated in 240 prostate needle core biopsies. Prostatic adenocarcinoma diagnostic recognition was higher with AI assistance, with an absolute increase in 1.5%. Accuracy for unassisted review by 20 general pathologists was 92.7%, assisted reviews were 94.2%, while AI algorithm alone obtained 95.8%. Specificity was increased with AI assistance in 12 cases and decreased in 2 cases. Regarding the 2 false-positive AI calls, AI assistance was appropriately disregarded by the pathologists. Sensitivity was increased with AI assistance in 38 cases and decreased in 16 cases. There were 6 cases with false-negative AI calls, which upon review exhibited small tumor foci–typical of false-negative results.40 Understanding how to avoid overreliance on AI assistance is an important goal. Efforts must be made to use these tools as synergistic decision-support tools. The latter may be challenging as some of these tools are “locked systems” and it is “all or nothing.” Additionally, the investigators found a decreased amount of review time with AI assistance, with 13% less time spent per biopsy for assistance-associated reviews vs. unassisted reviews. One of the possible reasons for this decrease is faster localization of small regions-of-interest in needle core tissue. Additional issues to consider are that most of the commercially available software is focused on the applications in prostate biopsies and not on radical prostatectomy or transurethral resection specimens.
Urothelial carcinoma
Diagnostic information, including grading (low vs. high), staging (non-invasive vs. lamina propria vs. muscularis propria invasion), divergent differentiation, lymphovascular invasion and presence of carcinoma in situ, is crucial for management and prognostication of patients with urothelial carcinomas.70 However, evaluation of urothelial carcinoma invasion and grading presents challenges, with considerable inter-observer variability among pathologists.71,72 However, studies evaluating deep learning techniques in grading urothelial carcinomas are scarce.73, 74, 75 One such study involved 328 TURBT specimens with consensus reading by 3 expert GU pathologists for developing deep learning algorithms for urothelium recognition and grading.75 The AI-grading algorithm demonstrated moderate agreement with consensus reading (kappa=0.48), similar to the agreement among pathologists (kappa=0.35, 0.38, and 0.52, respectively). The AI algorithm correctly graded 76% of low-grade cancers and 71% of high-grade cancers according to consensus reading, an indication that deep learning can be used for the fully automated detection and grading of urothelial cell carcinoma.
Domain knowledge enhancing feature extraction
Domain knowledge, i.e., expert knowledge, used during training data preparation, has demonstrated to enhance the performance of feature selection in machine learning tools. Domain experts may select specific features pertinent to a classification task, as demonstrated by a study utilizing automatic pipelines for the purpose of feature extraction for 3 invasive patterns characteristic of the T1 stage of bladder cancer, e.g., desmoplastic reaction, retraction artifact, and abundant pinker cytoplasm. Six supervised machine learning models trained on pathological features from 1177 H&E-stained images of bladder cancer tissue achieved an accuracy of 91%–96% in distinguishing non-invasive (Ta) and invasive (T1) urothelial tumors.76 Interestingly, CNN models that automatically extract features from images produced an accuracy of 84%, indicating that feature extraction driven by domain knowledge outperforms CNN-based automatic feature extraction.
More studies are needed on large patient populations from multiple centers to overcome compounding factors such as variability in H&E staining and image acquisition. Rather than correlating with “consensus” grading by pathologists, future studies should correlate grading by AI algorithms with clinical outcomes. In addition, deep learning can also uncover subvisual features or molecular correlations of prognostic significance that can be unrecognized by pathologist eyes. Harmon et al. used deep learning models to assess morphological features of primary urothelial carcinomas in cystectomy specimens, merging them with microenvironment (lymphocyte infiltration) features to derive a final patient-level AI score to predict the probability of pelvic lymph node metastasis.77 The AUC of AI score (0.866) was significantly better than AUC for the clinicopathological model consisting of age, T-stage, and lymphovascular invasion (0.755, P=.021).
Future studies should also develop algorithms to assist the identification of other pathological features, such as muscularis propria invasion, the main crossroad towards a more aggressive therapy (e.g., radical cystectomy, neoadjuvant chemotherapy, or chemoradiation).78 Additional future development in computational pathology should be focused in the area of cytopathology, particularly in the area of urine cytology. Automated image analysis of biomarkers such as PD-L1 should also be further developed with deeper integration into a digital workflow
As the first step to develop an automated method to improve the staging accuracy, 3 anatomic layers, i.e., urothelium, lamina propria, and muscularis propria, must be automatically recognized. Niazi et al.79 used a modified U-Net model, a CNN-based semantic segmentation framework, to develop a deep learning algorithm for segmentation of the bladder wall. The best performing algorithm achieved an accuracy comparable to the inter-observer variability among pathologists, raising the possibility that an algorithm for mitosis detection could be used for tumors where mitosis counts play an important role in grading, such as neuroendocrine tumors, and possibly urothelial carcinomas.
Renal carcinoma
Accounting for 60%–85% of all RCC and representing 2%–3% of all cancers with an annual incidence increase of 5%, clear cell renal cell carcinoma (ccRCC) is the most common malignant tumor of epithelial origin in the kidney and the most dominant type among 40 subtypes of RCC. Low late-stage survival rates are due in part to high resistance to chemotherapy and radiotherapy. Though ccRCC is asymptomatic in its early stages, 25%–30% of patients have metastasis at the time of diagnosis. Patients with localized ccRCCs removed by nephrectomy have a high risk of metastatic relapse (20%–30%), with more than 40% of patients eventually dying from the disease.80 Early detection of ccRCC is therefore vital. Correct classification of ccRCC grade and stage is essential for guiding clinical management, molecular-based therapies, and prognosis.81 Fuhrman grade (incorporating nuclear size, nucleolar prominence, and nuclear membrane irregularities) is widely accepted as a prognostic factor in RCC despite poor inter-observer agreement,81 a major limitation to the examination of H & E images by pathologists along with the time required to diagnose.82 Diagnostic challenges have occurred with the presence of additional morphological features, e.g., sarcomatoid or spindle cell pattern, and greater eosinophilic cytoplasmic staining in higher grade ccRCC. Computational pathology approaches have shown that it is possible to overcome these limitations and to identify subtle morphological differences between clinical groups.82
Recently, computational methods have been helpful in using gene expression profiling to separate out the various stages of ccRCC. A study by Bhalla et al. analyzed gene expression of 523 samples to identify genes differentially expressed in early and late stages of ccRCC, achieving a maximum accuracy of 72.64% and 0.81 ROC using 64 genes on validation dataset.83 However, classification models still lack generalizability for the accurate and reliable prediction of ccRCC tumor stages.84 In addition, most available studies concerning renal diagnostics and prognostics focus on ccRCC, due to its dominance, which has yielded greater data availability for further ccRCC algorithm training.85 As a result, there is only limited data available on primary RCC subcategories (sucha s chromophobe, papillary RCC etc) apart from clear cell.
AI-assisted renal tumor classification and staging
Fenstermaker et al.86 developed a CNN model for the identification and presence of RCC on histopathology specimens and differentiation of RCC subtype (clear cell, chromophobe, papillary) and grade (Fuhrman grades 1 through 4). The model was trained on 3000 normal and 12 168 RCC tissue samples from 42 patients (digital H&E-stained images from the Cancer Genome Atlas). The model achieved an overall accuracy of 99.1% for distinguishing normal parenchyma from RCC, with 97.5% accuracy in distinguishing between subtypes. Accuracy for Fuhrman grade prediction was 98.4%.
In an earlier (2019) study, Tabibu et al.82 developed a CNN for the automatic classification of RCC subtypes along with the identification of features that predict survival outcome. Histopathological WSIs and clinical information from the Cancer Genome Atlas were used for training. 1027 ccRCC, 303 Papillary RCC, and 254 Chromophobe WSIs were selected, with a corresponding 379, 47, and 83 normal tissue images per each respective RCC subtype. CNNs were able to distinguish clear cell and chromophobe RCC from normal tissue with a classification accuracy of 99.39% and 87.34%, respectively. Further distinguishing between ccRCC, chromophobe, and papillary RCC by CNN achieved a classification accuracy of 94.07%. Generated risk index (based on tumor shape and nuclei) was found to have significant association with patient survival outcome.
In 2018 and 2020, Singh et al. focused on distinguishing early and late stages of papillary RCC through development of machine learning models using features extracted from single and multi-omics data from the Cancer Genome Project. Gene expression and DNA methylation data were integrated in the latter study which demonstrated slightly better performance (MCC 0.77, PR-AUC 0.79, accuracy 90.4).85,87,88
Another recent study employed machine learning algorithms to predict the probability of RCC recurrence within 5 and 10 years after nephrectomy.80 Data from 6849 patients was collected from patients listed in a Korean RCC web-database, from which analytical data from 2814 patients was used to predict recurrence.
Investigators in 1 study developed an automated computational pipeline to extract image features to delineate TFE2 Xp11.2 translocation RCC (TFE3-RCC), an aggressively progressive and challenging diagnosis often misdiagnosed with other RCC subtypes, from ccRCC. AUCs ranged from 0.84 to 0.89 when evaluating the classification models against an external validation set.89
Much work needs to be done in the area of renal cell diagnosis and classification to create meaningful and robust clinical tools for the practicing pathologists. However, there are a number of promising research studies as discussed here which will set the stage of creating clinical grade-AI tools in the future.
Testicular germ cell and sex cord tumors
Pathological features crucial for directing management of patients with testicular cancer include histological subtypes (and their quantification) and staging.90,91 Distinction between seminoma and its mimics, such as atypical seminoma vs. embryonal carcinoma, seminoma vs. solid yolk sac tumor, and quantification of seminoma and non-seminomatous components, are critical for risk stratification and post-operative management.92,93 Lymphovascular invasion would upstage an otherwise organ-confined tumor from pT1 to pT2. However, diagnosis of lymphovascular invasion is notoriously challenging. Building AI tools for rapid detection of lymphovascular invasion is not trivial but a stepwise approach combining immunohistochemistry and morphology-based algorithms may help in the creation of an AI-based clinical tool. No studies so far have attempted to develop AI algorithms to address these challenges, which would offer plentiful opportunities for future research. For instance, future tools may be developed to address quantification of non-seminomatous components and staging based on different recognition of testicular/paratesticular anatomy.
AI-assisted testicular tumor classification and staging
One recent study developed a deep learning algorithm for testicular germ cell tumors, to quantify tumor infiltrating lymphocytes (TILs) on H&E-stained tissue sections.94 The correlation coefficient between manually annotated (3 pathologists) and algorithm-counted TILs was good (overall F-score = 0.88). While on WSIs (n=89), mean Kappa value between the algorithm and pathologists (Kappa 0.35) was comparable to inter-pathologist (Kappa 0.33). Regarding seminomas, tumors with highest TIL-tertile quantified by algorithm had no relapse, while TIL quantifications performed visually by 3 pathologists on the same tumors were not significantly associated with outcome. Deep learning-based algorithms can be used for the objective detection of TILs in testicular germ cell tumors more objectively and may thereby demonstrate utility as a prognostic biomarker for disease relapse.
Limitations to diagnostic AI implementation in GU pathology
Implementation of algorithmic AI tools for GU pathology has been marred by many limitations stemming from accountability, availability, and reliability of WSI data used in their creation.
A considerable volume of data is generated for algorithm development, currently derived from a litany of different sources,1 presented by a variety of WSI file formats, and is analyzable via a diversity of AI models. Lack of standardization seen throughout all stages of algorithmic construction and utilization predisposes to variations in machine learning classification with consequent poor predictive capacity. The adoption of a single open-source file format applicable to digital pathology, like that of DICOM for radiology, may facilitate expeditious access and interrogation, i.e., curation, of WSI datasets currently lacking universal image formatting.95 International standardization is needed to facilitate uniformity of image formatting for exchangeable data and designated quality control measures for WSI that may aid in leveraging accountability for AI-assisted diagnostic tools within clinical practice.
Paucity of WSI datasets with GU pathologist annotations for ground-truth determination limits employment of supervised learning techniques. The lack of WSI datasets impeding the analytic capacity of deep learning techniques is emphasized in GUP primarily pertaining to immunohistochemical (IHC) staining. Stemming from limited data sources, immunolabeled WSI datasets containing in situ molecular cell data, observable through multiplexed immunofluorescence (IF), are evermore sparce. "Transfer learning" via pretrained networks and data augmentation techniques may be utilized to mitigate the cumbersome nature of network training and data shortages, though are not currently capable of acting as substitutes for pathologist-annotated data. Increased utilization of unsupervised learning techniques, which do not require labeled data, may also mitigate shortfalls in expertly annotated WSIs that are often expensive, difficult to acquire, and time-consuming to produce. WSI data storage costs have posed barriers to digital implementation in many laboratories. Restrictions brought forth by data privacy and proprietary techniques may be circumvented with open-source accessibility. Graphical Processing Units (GPUs) are preferential to Central Processing Units (CPUs) for training and utilization of deep neural networks due to their significantly superior processing speed yet are significantly more costly. Large WSI file sizes command large network bandwidths that present hurdles in implementing infrastructure with the capacity to process swaths of data used for AI tools, yet advances in WSI scanning technology and digital data transmission coupled with decreased costs of implementation are near upon the horizon.1
High-resolution image reduction techniques, e.g., patch extraction, may compromise data quality while higher-level structural information, e.g., tumor extent or shape, may only be captured through analysis of larger tissue regions. Focused spatial correlation amongst patches, multi-level magnification patch extraction, and utilization of larger patch sizes are some of several techniques that have been developed to address these issues.
Clinical translation of algorithms requires generalizability throughout a wide breadth of patient populations and clinical institutions. IHC/H&E staining of tissue sections can vary significantly across laboratories and at intra-laboratory level. Analysis performed on low-quality tissue, histology slides, or staining will ultimately compromise the validity of data obtainable for image analysis, i.e., pre-analytical variables. High-quality tissue can be rendered useless by inadequate slide preparation from blurred vision, over- or under-staining, air-bubbles, and folded tissue. Such errors can produce inaccurate algorithms.
Normalization techniques, e.g., scale normalization for multiple image acquisition devices with varying pixels sizes, stain normalization, pixel- and patch-wise and semantic segmentation CNN training for enhanced region of interest detection, flexible thresholding techniques which compromise for variations in input data luminance.
Limitations in machine learning generalizability are not only relegated to algorithmic development for GU pathology but extend to the plethora of AI literature investigating ML used for a wide range of applications throughout the gamut of urology disciplines.96 Chen et al. reported variability and heterogeneity of documented outcomes and methodologies reported in publications within the subfields of oncology, infertility, endourology, and general urology in a recent literature review, emphasizing the crucial need for developing methods to standardize reporting to curtail limitations of generalizability in research.96
Deep learning systems for GU are currently only able to classify WSI specimens with a single diagnosis. Removal of biological restrictions during algorithmic training may serve to widen the scope of diagnostic capabilities in machine learning. "Artificial General Intelligence (AGI)" of the future may consist of advanced algorithms employing multiple levels of classification and segmentation in conjunction with a litany of diagnostic deductive variables, mimicking the process of human consciousness.
Demonstration of algorithm reproducibility on large patient populations containing outliers and non-representative individuals has caused difficulties for AI development. AI Models of the future may be used to develop "universal" tumor grading systems applicable to the entire GU system through combination of prognostic, morphologic, tumor marker, and clinical course data.
“Black Box” transparency concerns surround the uninterpretable pathway of algorithmic classification deduction. Segmentation, e.g., extraction, of image objects correlated with clinical endpoints are hidden from pathologist interpretation. Segmentation steps for deep neural network classification involve extraction of image objects correlated with clinical endpoints which are not included for review in the final output and therefore subject to distrust by pathologists who may prefer to accept more “transparent” AI algorithms. As with any proposed medical implementation that lacks information crucial to explaining its mechanics, this renders it unaccountable, with regulatory barriers inevitable to follow. “Rule extraction,” through which information about histopathological features used by an algorithm during its previously hidden segmentation process, may alleviate such concerns. Efforts towards extracting translucency from the opacity of black box algorithms are inherently efforts in preserving trust, accountability, responsibility, and patient autonomy while eliminating potential biases obscured by an epistemologically opaque methodology. Translucency may be approached through ethical and practical arguments supporting the computational realism of AI as opposed to, or in conjunction with, developing computational mechanisms that may allow self-iterative algorithms to be surveyable by humans.97 Reliability surrounding the clinical use of AI has raised concerns of potential legislative burdens incurred by developers, healthcare networks, patients, and physicians alike. The primary challenge limiting AI tools is that of an epistemological and ethical nature—one which interrogates the philosophical notion of trust in ML programs presenting outcomes which, though ultimately bearing subject to physician interpretation, remail inexplicable in instances where diagnoses are inscrutable.97
Physician liability and AI-assistive tools
Liability concerning the use of assistive AI-tools has not been formerly etched into case law due to the recent emergence of such devices in clinical settings. However, general tort law principles (those which generally favor standard of care, regardless of its outcome) may be extrapolated to garner relevant discussion on medical AI and liability, a subject that ultimately extends beyond legal ramifications for physicians who misuse such tools.98 As pathology practice is progressively ensconced by an ecosystem of healthcare systems and vendors offering machine learning tools, so too does the concept of physician liability become intertwined with these additional entities that may suffer from a proverbially shared burden of redress. An increase in liability, applied anywhere within our current ecosystem of pathology practice, may ultimately disincentivize development of AI tools along with their adoption in healthcare systems and clinical practice.99
Gerke et al. noted the high level of concern directed toward scrutinizing algorithmic performance, particularly due to the inconsistent performance of AI-tools which, despite their promise, are not immune to the introduction of human biases incurred from training.99
Under current law, physicians are subject to liability in circumstances upon which a predefined legislative and institutional standard of care is unadhered to, thereby resulting in patient injury. From the vantage point of assistive AI-tools within the scope of this framework, algorithms may choose to eschew standard protocols for a more patient-specific, i.e., personalized approach. Price et al. noted the paradoxical nature of the legislative constraints crafted to preserve patient welfare, as such limitations may inadvertently cultivate an ecosystem of practice by which adherence to legal standards are opted for in lieu of AI recommendations that may be of greater value to a patient, simply due to the latter falling outside the range of “accepted” treatment. In broad terms, physicians and healthcare systems may be liable under malpractice though not liable for the decision-making capacity of AI-tools. Algorithm designers, however, may bear the burden of faulty AI while avoiding liability from negligence or malpractice facing physicians who use such tools in an assistive capacity.99
Discussion
Recent years have welcomed a foundation of literature supporting the use of AI for urology;96,100 however, the rapid accumulation of promising clinical trials and published articles detailing urological ML applications have not been met with an equivalent proliferation of educational efforts preparing clinicians for discussions of implementation. AI literacy will be tantamount to practical competency in the emergent future, as more WSI devices obtain regulatory approval in the USA and globally,1,77,96,101, 102, 103, 104 while pathologists encroachingly find themselves in leadership roles that dictate the interdisciplinary interplay of AI across urology, pathology, radiology, metabolomics, genomics, and other disciplines.
Chen et al. reported in a comprehensive literature review that disease diagnosis was the most common application in ML for urology. 73 out of 112 articles included in the review included those which interrogated ML algorithms developed for automating and improving the detection of urological pathologies. Over 60% of publications entailed the utilization of algorithms for the detection of prostate cancer with applications in analyzing serum, urine, novel biomarker, radiomics, clinical variables, pathology slides, and electronic health records (EHR).96
As prostate cancer is the most common malignancy seen by urologists, it is foreseeable that most articles featured by Chen et al. involved new and novel algorithmic development for its detection.96 Our review followed suit, with most AI literature featuring accounts of algorithms developed for prostate cancer identification, many of which portrayed ML applications bounds beyond those focused on other GU systems. Yet, such devices, even for prostate cancer, have seen limited clinical implementation within the USA. Within the realm of GU pathology to date, only prostate recognition and grading has demonstrated capacity for clinical utilization. Most current studies evaluating ANN endeavors within other GU systems are currently relegated to academia, though still draw interest in their valuable potential. Validative efforts have been tantamount to implementation of non-AI assisted WSI for GU in clinical practice. However, limited validation studies within this realm have been circumvented by positive accounts of digital implementation shared by many departments.105, 106, 107, 108,109 Such circumvention through clinical implementation has not yet been realized for deep learning tools in GU pathology.
Human prognostic and therapeutic deliberation incorporates multidisciplinary discussions and performance status assessments.110 The incorporation of contextual clinical information into deep learning systems will allow these systems to interpret a diversity of information critical to such deliberation. Machine learning algorithms have demonstrated integrability into electronic health records (EHRs) to generate accurate predictions of short-term mortality for cancer patients, outperforming routinely used prognostic indices.111 Yet replicating the subjective nature of physician–patient interactions via AI-directed data mining of EHR has proven difficult due to the low quality and poor design of such databases. Bayesian inference has been applied to algorithmic development to suggest multiple potential diagnoses given a set of clinical findings, though there still are limitations in advising practitioners on useful next steps.112
Reproducibility of algorithms in comparison to standard histopathological assessment is an essential consideration in AI development for image analysis. “Algorithm—pathologist correlation” is utilized in this regard, with reproducibility most preferably assessed through multiple pathologists to emulate routine practice. Measures of reproducibility include pathologist–algorithm correlation and measures of inter-pathologist variability.113 It is important to maintain awareness of a “gold-standard paradox” overlying algorithmic validation. Pathologist scoring is used for the analytical validation of an image analysis tool, however such tools are used to overcome biases that are known to be exhibited during traditional pathologist assessment of tissue sections.114
Histopathological examination is widely viewed as the gold-standard procedure for achievement of a final diagnostic conclusion.115 However, histopathological examination is also highly susceptible to the introduction of artifacts which can alter otherwise normal morphologic and cytologic tissue features.115 High-quality, well-annotated, and large datasets composed from high-quality histology yield high performing deep learning models.116,117 Automated CNN-based tools, e.g., HistoQC and DeepFocus,118,119 have been developed to standardize the quality of whole slide imaging. GAN (generative adversarial network)-based approaches in eliminating noise to produce normalized H&E stained images have also been proposed.120
Although such conditions for training are ideal, large datasets are difficult to allocate for rare diagnosis or for prediction of outcomes in clinical trials using a small cohort of patients. Performance of deep learning models has also declined when data from different sources and imaging devices has been used for testing.6 AI-algorithms have still demonstrated accurate predictive capacity despite a lack of training variables or standardization of data.85
This capacity may soon extend to successful prediction of gene mutation probability in cancer from digitized H&E WSIs, as demonstrated in a recent study evaluating the performance of a deep learning model developed to determine SPOP (speckle-type POS protein) gene mutation in prostate cancer via tractable deep learning using a small dataset of 20–55 positive examples.121
Commercial software vendors have leveraged machine learning for GUP applications in immune-oncology, e.g., the OptraScan (San Jose, California) image analysis solution: OptraASSAYS® (RUO) offers prostate and kidney analysis libraries. In addition to predictive prognostics, such solutions may be utilized in future immunotherapy applications. High PD-L1 expression has demonstrated association with poor clinical outcomes in prostate cancer patients, with important roles in immunotherapy, chemotherapy, and vaccines in the treatment of prostate cancer.122,123The OptraScan platform supports interpretation of PD-L1 IHC expression on tumor and immune cells in solid tumors.
Machine or deep learning empowered automated diagnostic algorithms have demonstrated the capacity to assist pathologists’ decision-making process in difficult cases. Yet, outside of the realm of prostate pathology, orchestration of machine learning tools for integrability within GU reporting workflows has been overlooked. Apart from those for prostate pathology, most AI assistive tools feature in our review have been developed as standalone tools. For instance, with bladder, one tool may grade, but still another is required to assess depth of invasion. For the fully streamlined facilitation of GU reporting, a machine learning tool must: 1) recognize there is tumor, 2) grade the tumor, and 3) judge depth of invasion. These tasks be seamless and running in parallel or appropriate sequence, rather than having a pathologist select from a dropdown separately for each step in reporting the diagnosis. Constant manual selection of individual tools without general intelligence adds friction to reporting in digital sign-out workflows.
Conclusions
Machine learning tools developed for GU pathology have demonstrated promising applications in diagnosis and prognosis, identification of histologic subtypes and identification of tumor grades via analysis of histopathology specimens.
AI-algorithms have demonstrated the capacity to help aid clinicians in prognostic management and the development of precision treatment strategies. Though efforts still are necessary to improve prognostic capacity, AI applications in GU pathology will ultimately aid pathologists in tumor assessment via enhanced accuracy and efficiency of histopathological diagnostic execution.
Competing Interests statement
No support for this work has been received in the form of grants and/or equipment, drugs, or any form of compensation, financial or otherwise.
Contributor Information
Anil V. Parwani, Email: anil.parwani@osumc.edu.
Ankush Patel, Email: ankushpatel@rcsi.ie.
Ming Zhou, Email: mzhou3@tuftsmedicalcenter.org.
John C. Cheville, Email: cheville.john@mayo.edu.
Hamid Tizhoosh, Email: hamid.tizhoosh@uwaterloo.ca.
Peter Humphrey, Email: peter.humphrey@yale.edu.
Victor E. Reuter, Email: reuterv@mskcc.org.
Lawrence D. True, Email: ltrue@uw.edu.
References
- 1.Patel A., Balis U., Cheng J., Li Z., Lujan G., McClintock D., et al. Contemporary whole slide imaging devices and their applications within the modern pathology department: a selected hardware review. J Pathol Inform. 2021;12(1):27. doi: 10.4103/jpi.jpi_66_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steiner D.F., Nagpal K., Sayres R., Foote D.J., Wedin B.D., Pearce A., et al. Evaluation of the use of combined artificial intelligence and pathologist assessment to review and grade prostate biopsies. JAMA Netw Open. 2020;3(11) doi: 10.1001/jamanetworkopen.2020.23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marginean F., Arvidsson I., Simoulis A., Christian Overgaard N., Astrom K., Heyden A., et al. An artificial intelligence-based support tool for automation and standardisation of Gleason grading in prostate biopsies. Eur Urol Focus. 2021;7(5):995–1001. doi: 10.1016/j.euf.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Kartasalo K., Bulten W., Delahunt B., Chen P.C., Pinckaers H., Olsson H., et al. Artificial intelligence for diagnosis and gleason grading of prostate cancer in biopsies-current status and next steps. Eur Urol Focus. 2021;7(4):687–691. doi: 10.1016/j.euf.2021.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Strom P., Kartasalo K., Olsson H., Solorzano L., Delahunt B., Berney D.M., et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 2020;21(2):222–232. doi: 10.1016/S1470-2045(19)30738-7. [DOI] [PubMed] [Google Scholar]
- 6.Lu M.Y., Williamson D.F.K., Chen T.Y., Chen R.J., Barbieri M., Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng. 2021;5(6):555–570. doi: 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lujan G.M., Savage J., Shana'ah A., Yearsley M., Thomas D., Allenby P., et al. Digital pathology initiatives and experience of a large academic institution during the Coronavirus disease 2019 (COVID-19) pandemic. Arch Pathol Lab Med. 2021;145(9):1051–1061. doi: 10.5858/arpa.2020-0715-SA. [DOI] [PubMed] [Google Scholar]
- 8.Scarl R.T., Parwani A., Yearsley M. From glass-time to screen-time: a pathology resident's experience with digital sign-out during the coronavirus 2019 pandemic. Arch Pathol Lab Med. 2021;145(6):644–645. doi: 10.5858/arpa.2020-0511-ED. [DOI] [PubMed] [Google Scholar]
- 9.Gupta L., Klinkhammer B.M., Boor P., Merhof D., Gadermayr M. International Conference on Medical Imaging with Deep Learning–Full Paper Track. 2018. Iterative learning to make the most of unlabeled and quickly obtained labeled data in histology. [Google Scholar]
- 10.Monaco J.P., Tomaszewski J.E., Feldman M.D., Hagemann I., Moradi M., Mousavi P., et al. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise Markov models. Med Image Anal. 2010;14(4):617–629. doi: 10.1016/j.media.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle S., Feldman M., Tomaszewski J., Madabhushi A. A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Biomed Eng. 2012;59(5):1205–1218. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 12.Gorelick L., Veksler O., Gaed M., Gomez J.A., Moussa M., Bauman G., et al. Prostate histopathology: learning tissue component histograms for cancer detection and classification. IEEE Trans Med Imaging. 2013;32(10):1804–1818. doi: 10.1109/TMI.2013.2265334. [DOI] [PubMed] [Google Scholar]
- 13.Kothari S., Phan J.H., Stokes T.H., Wang M.D. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc. 2013;20(6):1099–1108. doi: 10.1136/amiajnl-2012-001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litjens G., Sanchez C.I., Timofeeva N., Hermsen M., Nagtegaal I., Kovacs I., et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somanchi S., Neill D.B., Parwani A.V. Discovering anomalous patterns in large digital pathology images. Stat Med. 2018;37(25):3599–3615. doi: 10.1002/sim.7828. [DOI] [PubMed] [Google Scholar]
- 16.Nir G., Hor S., Karimi D., Fazli L., Skinnider B.F., Tavassoli P., et al. Automatic grading of prostate cancer in digitized histopathology images: learning from multiple experts. Med Image Anal. 2018;50:167–180. doi: 10.1016/j.media.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Lucas M., Jansen I., Savci-Heijink C.D., Meijer S.L., de Boer O.J., van Leeuwen T.G., et al. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019;475(1):77–83. doi: 10.1007/s00428-019-02577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Werneck Krauss Silva V., Busam K.J., et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteban A.E., Lopez-Perez M., Colomer A., Sales M.A., Molina R., Naranjo V. A new optical density granulometry-based descriptor for the classification of prostate histological images using shallow and deep Gaussian processes. Comput Methods Programs Biomed. 2019;178:303–317. doi: 10.1016/j.cmpb.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Kott O., Linsley D., Amin A., Karagounis A., Jeffers C., Golijanin D., et al. Development of a deep learning algorithm for the histopathologic diagnosis and gleason grading of prostate cancer biopsies: a pilot study. Eur Urol Focus. 2021;7(2):347–351. doi: 10.1016/j.euf.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raciti P., Sue J., Ceballos R., Godrich R., Kunz J.D., Kapur S., et al. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod Pathol. 2020;33(10):2058–2066. doi: 10.1038/s41379-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pantanowitz L., Quiroga-Garza G.M., Bien L., Heled R., Laifenfeld D., Linhart C., et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: a blinded clinical validation and deployment study. Lancet Digit Health. 2020;2(8):e407–e416. doi: 10.1016/S2589-7500(20)30159-X. [DOI] [PubMed] [Google Scholar]
- 23.Han W., Johnson C., Gaed M., Gomez J.A., Moussa M., Chin J.L., et al. Histologic tissue components provide major cues for machine learning-based prostate cancer detection and grading on prostatectomy specimens. Sci Rep. 2020;10(1):9911. doi: 10.1038/s41598-020-66849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T.H., Sridharan S., Macias V., Kajdacsy-Balla A., Melamed J., Do M.N., et al. Automatic Gleason grading of prostate cancer using quantitative phase imaging and machine learning. J Biomed Opt. 2017;22(3):36015. doi: 10.1117/1.JBO.22.3.036015. [DOI] [PubMed] [Google Scholar]
- 25.Arvaniti E., Fricker K.S., Moret M., Rupp N., Hermanns T., Fankhauser C., et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8(1):12054. doi: 10.1038/s41598-018-30535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nir G., Karimi D., Goldenberg S.L., Fazli L., Skinnider B.F., Tavassoli P., et al. Comparison of artificial intelligence techniques to evaluate performance of a classifier for automatic grading of prostate cancer from digitized histopathologic images. JAMA Netw Open. 2019;2(3) doi: 10.1001/jamanetworkopen.2019.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolkach Y., Dohmgörgen T., Toma M., Kristiansen G. High-accuracy prostate cancer pathology using deep learning. Nat Mach Intell. 2020;2(7):411–418. [Google Scholar]
- 28.Aeffner F., Zarella M.D., Buchbinder N., Bui M.M., Goodman M.R., Hartman D.J., et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association. J Pathol Inform. 2019;10:9. doi: 10.4103/jpi.jpi_82_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosun A.B., Pullara F., Becich M.J., Taylor D.L., Fine J.L., Chennubhotla S.C. Explainable AI (xAI) for anatomic pathology. Adv Anat Pathol. 2020;27(4):241–250. doi: 10.1097/PAP.0000000000000264. [DOI] [PubMed] [Google Scholar]
- 30.Cui M., Zhang D.Y. Artificial intelligence and computational pathology. Lab Invest. 2021;101(4):412–422. doi: 10.1038/s41374-020-00514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidey-Gibbons J.A.M., Sidey-Gibbons C.J. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19(1):64. doi: 10.1186/s12874-019-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi Y. In: Artificial Intelligence and Machine Learning for Digital Pathology. Holzinger A., Goebel R., Mengel M., Müller H., editors. Springer; Cham: 2020. Black box nature of deep learning for digital pathology: beyond quantitative to qualitative algorithmic performances; pp. 95–101. (Lecture Notes in Computer Science). [Google Scholar]
- 33.Topol E.J. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 34.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 35.LeCun Y., Boser B., Denker J., Henderson D., Howard R., Hubbard W., et al. Handwritten digit recognition with a back-propagation network. Adv Neural Inform Process Syst. 1989;2 [Google Scholar]
- 36.Li C., Li X., Rahaman M., Li X., Sun H., Zhang H., et al. 2022. A comprehensive review of computer-aided whole-slide image analysis: from datasets to feature extraction, segmentation, classification, and detection approaches.https://arxiv.org/abs/2102.10553 arXiv. Preprint posted online February 21, 2021; Available from. [Google Scholar]
- 37.Khened M., Kori A., Rajkumar H., Krishnamurthi G., Srinivasan B. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci Rep. 2021;11(1):11579. doi: 10.1038/s41598-021-90444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey P.A. Histopathology of prostate cancer. Cold Spring Harb Perspect Med. 2017;7(10) doi: 10.1101/cshperspect.a030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltran L., Ahmad A.S., Sandu H., Kudahetti S., Soosay G., Moller H., et al. Histopathologic false-positive diagnoses of prostate cancer in the age of immunohistochemistry. Am J Surg Pathol. 2019;43(3):361–368. doi: 10.1097/PAS.0000000000001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C., Humphrey P.A. False-negative histopathologic diagnosis of prostatic adenocarcinoma. Arch Pathol Lab Med. 2020;144(3):326–334. doi: 10.5858/arpa.2019-0456-RA. [DOI] [PubMed] [Google Scholar]
- 41.Giunchi F., Jordahl K., Bollito E., Colecchia M., Patriarca C., D'Errico A., et al. Interpathologist concordance in the histological diagnosis of focal prostatic atrophy lesions, acute and chronic prostatitis, PIN, and prostate cancer. Virchows Arch. 2017;470(6):711–715. doi: 10.1007/s00428-017-2123-1. [DOI] [PubMed] [Google Scholar]
- 42.Egevad L., Ahmad A.S., Algaba F., Berney D.M., Boccon-Gibod L., Comperat E., et al. Standardization of Gleason grading among 337 european pathologists. Histopathology. 2013;62(2):247–256. doi: 10.1111/his.12008. [DOI] [PubMed] [Google Scholar]
- 43.Zhou M., Li J., Cheng L., Egevad L., Deng F.M., Kunju L.P., et al. Diagnosis of "poorly formed glands" gleason pattern 4 prostatic adenocarcinoma on needle biopsy: an interobserver reproducibility study among urologic pathologists with recommendations. Am J Surg Pathol. 2015;39(10):1331–1339. doi: 10.1097/PAS.0000000000000457. [DOI] [PubMed] [Google Scholar]
- 44.Epstein J.I., Egevad L., Amin M.B., Delahunt B., Srigley J.R., Humphrey P.A., et al. The 2014 international society of urological pathology (ISUP) consensus conference on gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40(2):244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 45.Allsbrook W.C., Jr., Mangold K.A., Johnson M.H., Lane R.B., Lane C.G., Amin M.B., et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: urologic pathologists. Hum Pathol. 2001;32(1):74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 46.McKenney J.K., Simko J., Bonham M., True L.D., Troyer D., Hawley S., et al. The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. J Urol. 2011;186(2):465–469. doi: 10.1016/j.juro.2011.03.115. [DOI] [PubMed] [Google Scholar]
- 47.Kweldam C.F., Nieboer D., Algaba F., Amin M.B., Berney D.M., Billis A., et al. Gleason grade 4 prostate adenocarcinoma patterns: an interobserver agreement study among genitourinary pathologists. Histopathology. 2016;69(3):441–449. doi: 10.1111/his.12976. [DOI] [PubMed] [Google Scholar]
- 48.van der Kwast T.H., van Leenders G.J., Berney D.M., Delahunt B., Evans A.J., Iczkowski K.A., et al. ISUP consensus definition of cribriform pattern prostate cancer. Am J Surg Pathol. 2021;45(8):1118–1126. doi: 10.1097/PAS.0000000000001728. [DOI] [PubMed] [Google Scholar]
- 49.Zelic R., Giunchi F., Lianas L., Mascia C., Zanetti G., Andren O., et al. Interchangeability of light and virtual microscopy for histopathological evaluation of prostate cancer. Sci Rep. 2021;11(1):3257. doi: 10.1038/s41598-021-82911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sehn J.K. Prostate cancer pathology: recent updates and controversies. Mo Med. 2018;115(2):151–155. [PMC free article] [PubMed] [Google Scholar]
- 51.McKenney J.K., Wei W., Hawley S., Auman H., Newcomb L.F., Boyer H.D., et al. Histologic grading of prostatic adenocarcinoma can be further optimized: analysis of the relative prognostic strength of individual architectural patterns in 1275 patients from the canary retrospective cohort. Am J Surg Pathol. 2016;40(11):1439–1456. doi: 10.1097/PAS.0000000000000736. [DOI] [PubMed] [Google Scholar]
- 52.Ambrosini P., Hollemans E., Kweldam C.F., Leenders G., Stallinga S., Vos F. Automated detection of cribriform growth patterns in prostate histology images. Sci Rep. 2020;10(1):14904. doi: 10.1038/s41598-020-71942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Slot M.A., Hollemans E., den Bakker M.A., Hoedemaeker R., Kliffen M., Budel L.M., et al. Inter-observer variability of cribriform architecture and percent Gleason pattern 4 in prostate cancer: relation to clinical outcome. Virchows Arch. 2021;478(2):249–256. doi: 10.1007/s00428-020-02902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shah R.B., Cai Q., Aron M., Berney D.M., Cheville J.C., Deng F.M., et al. Diagnosis of "cribriform" prostatic adenocarcinoma: an interobserver reproducibility study among urologic pathologists with recommendations. Am J Cancer Res. 2021;11(8):3990–4001. [PMC free article] [PubMed] [Google Scholar]
- 55.Singh M., Kalaw E.M., Jie W., Al-Shabi M., Wong C.F., Giron D.M., et al. 2019. Cribriform pattern detection in prostate histopathological images using deep learning models. arXiv:1910.04030. [Google Scholar]
- 56.Leo P., Chandramouli S., Farre X., Elliott R., Janowczyk A., Bera K., et al. Computationally derived cribriform area index from prostate cancer hematoxylin and eosin images is associated with biochemical recurrence following radical prostatectomy and is most prognostic in gleason grade group 2. Eur Urol Focus. 2021;7(4):722–732. doi: 10.1016/j.euf.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berney D.M., Algaba F., Camparo P., Comperat E., Griffiths D., Kristiansen G., et al. The reasons behind variation in Gleason grading of prostatic biopsies: areas of agreement and misconception among 266 European pathologists. Histopathology. 2014;64(3):405–411. doi: 10.1111/his.12284. [DOI] [PubMed] [Google Scholar]
- 58.Nagpal K., Foote D., Tan F., Liu Y., Chen P.C., Steiner D.F., et al. Development and validation of a deep learning algorithm for gleason grading of prostate cancer from biopsy specimens. JAMA Oncol. 2020;6(9):1372–1380. doi: 10.1001/jamaoncol.2020.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bulten W., Pinckaers H., van Boven H., Vink R., de Bel T., van Ginneken B., et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 2020;21(2):233–241. doi: 10.1016/S1470-2045(19)30739-9. [DOI] [PubMed] [Google Scholar]
- 60.Bulten W., Balkenhol M., Belinga J.A., Brilhante A., Cakir A., Egevad L., et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod Pathol. 2021;34(3):660–671. doi: 10.1038/s41379-020-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serefoglu E.C., Altinova S., Ugras N.S., Akincioglu E., Asil E., Balbay M.D. How reliable is 12-core prostate biopsy procedure in the detection of prostate cancer? Can Urol Assoc J. 2013;7(5-6):E293–E298. doi: 10.5489/cuaj.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salami S.S., Ben-Levi E., Yaskiv O., Ryniker L., Turkbey B., Kavoussi L.R., et al. In patients with a previous negative prostate biopsy and a suspicious lesion on magnetic resonance imaging, is a 12-core biopsy still necessary in addition to a targeted biopsy? BJU Int. 2015;115(4):562–570. doi: 10.1111/bju.12938. [DOI] [PubMed] [Google Scholar]
- 63.Ishida J., Masuda T. Surgical case of lung cancer with anomalous right pulmonary vein;Report of a case. Kyobu Geka. 2020;73(3):230–232. [PubMed] [Google Scholar]
- 64.Iizuka O., Kanavati F., Kato K., Rambeau M., Arihiro K., Tsuneki M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci Rep. 2020;10(1):1504. doi: 10.1038/s41598-020-58467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anaba E.L., Cole-Adeife M.O., Oaku R.I. Prevalence, pattern, source of drug information, and reasons for self-medication among dermatology patients. Dermatol Ther. 2021;34(2) doi: 10.1111/dth.14756. [DOI] [PubMed] [Google Scholar]
- 66.Perincheri S., Levi A.W., Celli R., Gershkovich P., Rimm D., Morrow J.S., et al. An independent assessment of an artificial intelligence system for prostate cancer detection shows strong diagnostic accuracy. Mod Pathol. 2021;34(8):1588–1595. doi: 10.1038/s41379-021-00794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.U.S. Food and Drug Administration . 2021. FDA Authorizes Software that Can Help Identify Prostate Cancer [Press Release] 09/21/2021. [Google Scholar]
- 68.Trpkov K. Benign mimics of prostatic adenocarcinoma. Mod Pathol. 2018;31(S1):S22–S46. doi: 10.1038/modpathol.2017.136. [DOI] [PubMed] [Google Scholar]
- 69.Humphrey P.A. Variants of acinar adenocarcinoma of the prostate mimicking benign conditions. Mod Pathol. 2018;31(S1):S64–S70. doi: 10.1038/modpathol.2017.137. [DOI] [PubMed] [Google Scholar]
- 70.Comperat E., Varinot J., Moroch J., Eymerit-Morin C., Brimo F. A practical guide to bladder cancer pathology. Nat Rev Urol. 2018;15(3):143–154. doi: 10.1038/nrurol.2018.2. [DOI] [PubMed] [Google Scholar]
- 71.Lawless M., Gulati R., Tretiakova M. Stalk versus base invasion in pT1 papillary cancers of the bladder: improved substaging system predicting the risk of progression. Histopathology. 2017;71(3):406–414. doi: 10.1111/his.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kvikstad V., Mangrud O.M., Gudlaugsson E., Dalen I., Espeland H., Baak J.P.A., et al. Prognostic value and reproducibility of different microscopic characteristics in the WHO grading systems for pTa and pT1 urinary bladder urothelial carcinomas. Diagn Pathol. 2019;14(1):90. doi: 10.1186/s13000-019-0868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi H.K., Jarkrans T., Bengtsson E., Vasko J., Wester K., Malmstrom P.U., et al. Image analysis based grading of bladder carcinoma. Comparison of object, texture and graph based methods and their reproducibility. Anal Cell Pathol. 1997;15(1):1–18. doi: 10.1155/1997/147187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spyridonos P., Cavouras D., Ravazoula P., Nikiforidis G. Neural network-based segmentation and classification system for automated grading of histologic sections of bladder carcinoma. Anal Quant Cytol Histol. 2002;24(6):317–324. [PubMed] [Google Scholar]
- 75.Jansen I., Lucas M., Bosschieter J., de Boer O.J., Meijer S.L., van Leeuwen T.G., et al. Automated detection and grading of non-muscle-invasive urothelial cell carcinoma of the bladder. Am J Pathol. 2020;190(7):1483–1490. doi: 10.1016/j.ajpath.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 76.Yin P.N., Kc K., Wei S., Yu Q., Li R., Haake A.R., et al. Histopathological distinction of non-invasive and invasive bladder cancers using machine learning approaches. BMC Med Inform Decis Mak. 2020;20(1):162. doi: 10.1186/s12911-020-01185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harmon S.A., Sanford T.H., Brown G.T., Yang C., Mehralivand S., Jacob J.M., et al. Multiresolution application of artificial intelligence in digital pathology for prediction of positive lymph nodes from primary tumors in bladder cancer. JCO Clin Cancer Inform. 2020;4:367–382. doi: 10.1200/CCI.19.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassan O., Murati Amador B., Lombardo K.A., Salles D., Cuello F., Marwaha A.S., et al. Clinical significance of urothelial carcinoma ambiguous for muscularis propria invasion on initial transurethral resection of bladder tumor. World J Urol. 2020;38(2):389–395. doi: 10.1007/s00345-019-02782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niazi M.K.K., Yazgan E., Tavolara T.E., Li W., Lee C.T., Parwani A., et al. Semantic segmentation to identify bladder layers from H&E images. Diagn Pathol. 2020;15(1):87. doi: 10.1186/s13000-020-01002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim H., Lee S.J., Park S.J., Choi I.Y., Hong S.H. Machine learning approach to predict the probability of recurrence of renal cell carcinoma after surgery: Prediction model development study. JMIR Med Inform. 2021;9(3) doi: 10.2196/25635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tian K., Rubadue C.A., Lin D.I., Veta M., Pyle M.E., Irshad H., et al. Automated clear cell renal carcinoma grade classification with prognostic significance. PLoS One. 2019;14(10) doi: 10.1371/journal.pone.0222641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tabibu S., Vinod P.K., Jawahar C.V. Pan-renal cell carcinoma classification and survival prediction from histopathology images using deep learning. Sci Rep. 2019;9(1):10509. doi: 10.1038/s41598-019-46718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhalla S., Chaudhary K., Kumar R., Sehgal M., Kaur H., Sharma S., et al. Gene expression-based biomarkers for discriminating early and late stage of clear cell renal cancer. Sci Rep. 2017;7(1):44997. doi: 10.1038/srep44997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li F., Yang M., Li Y., Zhang M., Wang W., Yuan D., et al. An improved clear cell renal cell carcinoma stage prediction model based on gene sets. BMC Bioinformatics. 2020;21(1):232. doi: 10.1186/s12859-020-03543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giulietti M., Cecati M., Sabanovic B., Scire A., Cimadamore A., Santoni M., et al. The role of artificial intelligence in the diagnosis and prognosis of renal cell tumors. Diagnostics (Basel) 2021;11(2):206. doi: 10.3390/diagnostics11020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fenstermaker M., Tomlins S.A., Singh K., Wiens J., Morgan T.M. Development and validation of a deep-learning model to assist with renal cell carcinoma histopathologic interpretation. Urology. 2020;144:152–157. doi: 10.1016/j.urology.2020.05.094. [DOI] [PubMed] [Google Scholar]
- 87.Singh N.P., Bapi R.S., Vinod P.K. Machine learning models to predict the progression from early to late stages of papillary renal cell carcinoma. Comput Biol Med. 2018;100:92–99. doi: 10.1016/j.compbiomed.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 88.Singh N.P., Vinod P.K. Integrative analysis of DNA methylation and gene expression in papillary renal cell carcinoma. Mol Genet Genomics. 2020;295(3):807–824. doi: 10.1007/s00438-020-01664-y. [DOI] [PubMed] [Google Scholar]
- 89.Cheng J., Han Z., Mehra R., Shao W., Cheng M., Feng Q., et al. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma. Nat Commun. 2020;11(1):1778. doi: 10.1038/s41467-020-15671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verrill C., Yilmaz A., Srigley J.R., Amin M.B., Comperat E., Egevad L., et al. Reporting and staging of testicular germ cell tumors: The international society of urological pathology (ISUP) testicular cancer consultation conference recommendations. Am J Surg Pathol. 2017;41(6):e22–e32. doi: 10.1097/PAS.0000000000000844. [DOI] [PubMed] [Google Scholar]
- 91.Cheng L., Albers P., Berney D.M., Feldman D.R., Daugaard G., Gilligan T., et al. Testicular cancer. Nat Rev Dis Primers. 2018;4(1):29. doi: 10.1038/s41572-018-0029-0. [DOI] [PubMed] [Google Scholar]
- 92.Williamson S.R., Delahunt B., Magi-Galluzzi C., Algaba F., Egevad L., Ulbright T.M., et al. The World Health Organization 2016 classification of testicular germ cell tumours: a review and update from the International Society of Urological Pathology Testis Consultation Panel. Histopathology. 2017;70(3):335–346. doi: 10.1111/his.13102. [DOI] [PubMed] [Google Scholar]
- 93.Adra N., Einhorn L.H. Testicular cancer update. Clin Adv Hematol Oncol. 2017;15(5):386–396. [PubMed] [Google Scholar]
- 94.Linder N., Taylor J.C., Colling R., Pell R., Alveyn E., Joseph J., et al. Deep learning for detecting tumour-infiltrating lymphocytes in testicular germ cell tumours. J Clin Pathol. 2019;72(2):157–164. doi: 10.1136/jclinpath-2018-205328. [DOI] [PubMed] [Google Scholar]
- 95.Dimitriou N., Arandjelovic O., Caie P.D. Deep learning for whole slide image analysis: An overview. Front Med (Lausanne) 2019;6:264. doi: 10.3389/fmed.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen A.B., Haque T., Roberts S., Rambhatla S., Cacciamani G., Dasgupta P., et al. Artificial intelligence applications in urology: reporting standards to achieve fluency for urologists. Urol Clin North Am. 2022;49(1):65–117. doi: 10.1016/j.ucl.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Durán J.M., Jongsma K.R. Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J Med Ethics. 2021;47(5):329–335. doi: 10.1136/medethics-2020-106820. [DOI] [PubMed] [Google Scholar]
- 98.Price W.N., Gerke S., Cohen I.G. Potential liability for physicians using artificial intelligence. JAMA. 2019;322(18):1765–1766. doi: 10.1001/jama.2019.15064. [DOI] [PubMed] [Google Scholar]
- 99.Maliha G., Gerke S., Cohen I.G., Parikh R.B. Artificial intelligence and liability in medicine: balancing safety and innovation. Milbank Q. 2021;99(3):629–647. doi: 10.1111/1468-0009.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hameed B.M.Z., AVL S.D., Raza S.Z., Karimi H., Khanuja H.S., Shetty D.K., et al. Artificial intelligence and its impact on urological diseases and management: a comprehensive review of the literature. J Clin Med. 2021;10(9) doi: 10.3390/jcm10091864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mata L.A., Retamero J.A., Gupta R.T., Garcia Figueras R., Luna A. Artificial intelligence-assisted prostate cancer diagnosis: Radiologic-pathologic correlation. Radiographics. 2021;41(6):1676–1697. doi: 10.1148/rg.2021210020. [DOI] [PubMed] [Google Scholar]
- 102.Khosravi P., Lysandrou M., Eljalby M., Li Q., Kazemi E., Zisimopoulos P., et al. A deep learning approach to diagnostic classification of prostate cancer using pathology-radiology fusion. J Magn Reson Imaging. 2021;54(2):462–471. doi: 10.1002/jmri.27599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goldenberg S.L., Nir G., Salcudean S.E. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol. 2019;16(7):391–403. doi: 10.1038/s41585-019-0193-3. [DOI] [PubMed] [Google Scholar]
- 104.Eun S.J., Kim J., Kim K.H. Applications of artificial intelligence in urological setting: a hopeful path to improved care. J Exerc Rehabil. 2021;17(5):308–312. doi: 10.12965/jer.2142596.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li X., Liu J., Xu H., Gong E., McNutt M.A., Li F., et al. A feasibility study of virtual slides in surgical pathology in China. Hum Pathol. 2007;38(12):1842–1848. doi: 10.1016/j.humpath.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 106.Stathonikos N., Veta M., Huisman A., van Diest P.J. Going fully digital: perspective of a Dutch academic pathology lab. J Pathol Inform. 2013;4:15. doi: 10.4103/2153-3539.114206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Al-Janabi S., Huisman A., Nap M., Clarijs R., van Diest P.J. Whole slide images as a platform for initial diagnostics in histopathology in a medium-sized routine laboratory. J Clin Pathol. 2012;65(12):1107–1111. doi: 10.1136/jclinpath-2012-200878. [DOI] [PubMed] [Google Scholar]
- 108.Pantanowitz L., Wiley C.A., Demetris A., Lesniak A., Ahmed I., Cable W., et al. Experience with multimodality telepathology at the University of Pittsburgh Medical Center. J Pathol Inform. 2012;3:45. doi: 10.4103/2153-3539.104907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saco A., Ramirez J., Rakislova N., Mira A., Ordi J. Validation of whole-slide imaging for histolopathogical diagnosis: current state. Pathobiology. 2016;83(2-3):89–98. doi: 10.1159/000442823. [DOI] [PubMed] [Google Scholar]
- 110.Kang J., Morin O., Hong J.C. Closing the gap between machine learning and clinical cancer care-first steps into a larger world. JAMA Oncol. 2020;6(11):1731–1732. doi: 10.1001/jamaoncol.2020.4314. [DOI] [PubMed] [Google Scholar]
- 111.Manz C.R., Chen J., Liu M., Chivers C., Regli S.H., Braun J., et al. Validation of a machine learning algorithm to predict 180-day mortality for outpatients with cancer. JAMA Oncol. 2020;6(11):1723–1730. doi: 10.1001/jamaoncol.2020.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gaskill O. Differential (AI) decisions: a new AI model uses electronic health records to make differential diagnoses Pathologist; 2021 Available from: https://thepathologist.com/diagnostics/differential-ai-decisions?fbclid=IwAR1XiK0AmvnMBgswXxf5IktDEFQcxec6bqePXTWhnTOWem8DAcj7BdDSJTs.
- 113.Pell R., Oien K., Robinson M., Pitman H., Rajpoot N., Rittscher J., et al. The use of digital pathology and image analysis in clinical trials. J Pathol Clin Res. 2019;5(2):81–90. doi: 10.1002/cjp2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aeffner F., Wilson K., Martin N.T., Black J.C., Hendriks C.L.L., Bolon B., et al. The gold standard paradox in digital image analysis: manual versus automated scoring as ground truth. Arch Pathol Lab Med. 2017;141(9):1267–1275. doi: 10.5858/arpa.2016-0386-RA. [DOI] [PubMed] [Google Scholar]
- 115.Taqi S.A., Sami S.A., Sami L.B., Zaki S.A. A review of artifacts in histopathology. J Oral Maxillofac Pathol. 2018;22(2):279. doi: 10.4103/jomfp.JOMFP_125_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O'Hurley G., Sjostedt E., Rahman A., Li B., Kampf C., Ponten F., et al. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol. 2014;8(4):783–798. doi: 10.1016/j.molonc.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Compton C. Garbage in, garbage out: The hidden reason laboratory test results may not be as reliable as they seem Pathologist; 2018 Available from: https://thepathologist.com/diagnostics/garbage-in-garbage-out.
- 118.Janowczyk A., Zuo R., Gilmore H., Feldman M., Madabhushi A. HistoQC: an open-source quality control tool for digital pathology slides. JCO Clin Cancer Inform. 2019;3:1–7. doi: 10.1200/CCI.18.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Senaras C., Niazi M.K.K., Lozanski G., Gurcan M.N. DeepFocus: detection of out-of-focus regions in whole slide digital images using deep learning. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0205387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shimizu H., Nakayama K.I. Artificial intelligence in oncology. Cancer Sci. 2020;111(5):1452–1460. doi: 10.1111/cas.14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schaumberg A.J., Rubin M.A., Fuchs T.J. H&E-stained whole slide image deep learning predicts SPOP mutation state in prostate cancer. BioRxiv. 2016 [Google Scholar]
- 122.Xu Y., Song G., Xie S., Jiang W., Chen X., Chu M., et al. The roles of PD-1/PD-L1 in the prognosis and immunotherapy of prostate cancer. Mol Ther. 2021;29(6):1958–1969. doi: 10.1016/j.ymthe.2021.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Serag A., Ion-Margineanu A., Qureshi H., McMillan R., Saint Martin M.J., Diamond J., et al. Translational AI and deep learning in diagnostic pathology. Front Med (Lausanne) 2019;6(185):185. doi: 10.3389/fmed.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]