Abstract
Intrahepatic cholangiocarcinoma (ICC) is a liver tumor featured by challenges of non-invasive early diagnosis and a higher prevalence rate in Asian countries. These characteristics necessitate the development of liquid biopsy and immunotherapy methods to improve the prognosis of patients with ICC. Herein, we conducted a pilot study on the transcriptome of tumor tissues, adjacent normal tissues, and plasma exosomes of Asian patients with ICC from northern and southern China. We identified a subgroup of immunogenic Asian ICC, which is different from Caucasian ICC and is characterized by T cell exhaustion and neutrophil extracellular traps. The levels of circ-PTPN22 (hsa_circ_0110529) and circ-ADAMTS6 (hsa_circ_0072688), potential circRNA biomarkers, were elevated in the ICC tumor tissues and plasma exosomes of this subgroup than in the other subgroups and normal controls. These circRNAs were derived from post-transcriptional backsplicing of PTPN22 and ADAMTS6 that were expressed in T cells and endothelial cells, respectively, in the ICC microenvironment. Our results revealed a subgroup of Asian ICC characterized by T cell exhaustion and neutrophil extracellular traps and marked by elevated levels of circ-PTPN22 and circ-ADAMTS6 in tumor tissues and plasma exosomes. This subgroup is potentially detectable by plasma exosomal circRNAs and treatable with immune checkpoint blockade.
Keywords: MT: Non-coding RNAs, intrahepatic cholangiocarcinoma, circular RNA, plasma exosome, neutrophil extracellular traps, T cell exhaustion, immune checkpoint blockade
Graphical abstract
Multifaceted transcriptomic analysis of Asian ICC tumor tissues, adjacent normal tissues, and plasma exosomes revealed a subgroup of immunogenic Asian ICC. This subgroup was characterized by T cell exhaustion and neutrophil extracellular traps, and was marked by elevated circ-PTPN22 and circ-ADAMTS6 in ICC tumor tissues and plasma exosomes.
Introduction
Cholangiocarcinoma has been shown to exhibit a rising prevalence, especially in Asian countries, contributing to the incidence of approximately 15% of primary liver cancer cases and 2% of cancer-related deaths worldwide.1 Intrahepatic cholangiocarcinoma (ICC) is highly malignant because of its tendency to infiltrate the nerve and lymphatic tissues, and its clinical manifestations are rarely observed in patients before the blocking of the bile duct by the tumor.2 As a result, only 20%–40% of patients with ICC present with indications for surgical resection, after which they still face a dismal postoperative prognosis, that is, a 3-year survival rate of 40%–50% and a recurrence rate of 46%–65%.3
ICC in Asian and Caucasian populations is known to arise from different etiologies. Hepatolithiasis and parasitic infections are more prevalent among Asian patients, while nonalcoholic fatty liver disease and metabolic abnormalities are associated with ICC in Western populations.4 A comparative analysis of the mutational landscape revealed a higher burden of DNA repair mutations in Asian patients with ICC.5 Furthermore, transcriptomic characteristics of Asian ICC are related to obesity, T cell infiltration, and bile acid metabolism.6
Translational therapeutic techniques, such as liquid biopsy biomarkers and immune checkpoint blockade, are expected to improve the therapy of ICC. A subset of ICC was observed with enriched immune infiltration; however, subgroup-specific biomarkers require further investigation.7 Likewise, an inflamed subgroup likely treatable with checkpoint blockade was identified in Caucasian patients with ICC.8 A subgroup of cholangiocarcinoma (intrahepatic, perihilar, and distal) presents with activated immune pathways, and elevated programmed cell death 1 (PD-1) and programmed cell death ligand 1 (PD-L1) expression.9
Circular RNAs (circRNAs) are potential biomarker candidates because of their covalently closed structure, which is stable in plasma exosomes. For example, cerebellar degenerative-related protein-1 antisense (CDR1as), which has been extensively studied in multiple cancer types,10 can serve as a biomarker for ICC.11 Similarly, circ_0005230,12 circDNM3OS,13 circ-LAMP1,14 circ0021205,15 circRTN4IP1,16 and circSETD317 can promote the proliferation, growth, or metastases of ICC. circ_0020256,18 which is associated with the tumor microenvironment, is an exosomal circRNA produced by tumor-associated M2 macrophages and promotes cancer progression. circ-CCAC1, whose expression is elevated in ICC tumor tissues and bile-derived extracellular vesicles, disrupts the endothelial barrier and induces angiogenesis.19 Similarly, circ-0000284, whose expression is elevated in ICC cancer cells and cancer cell-derived extracellular vesicles, stimulates the migration and proliferation of surrounding cells.20
In this pilot study, we conducted a multifaceted transcriptomic analysis of ICC tumor tissues, adjacent normal tissues, and plasma exosomes donated by a cohort of 14 Asian patients from northern China (n = 7) and southern China (n = 7) and compared them with Caucasian ICC, aiming to discover exosomal circRNA biomarkers of Asian ICC subgroups potentially treatable by immunotherapy.
Results
circRNA profiling in ICC tumor tissues and plasma exosomes of patients with ICC revealed potential biomarkers
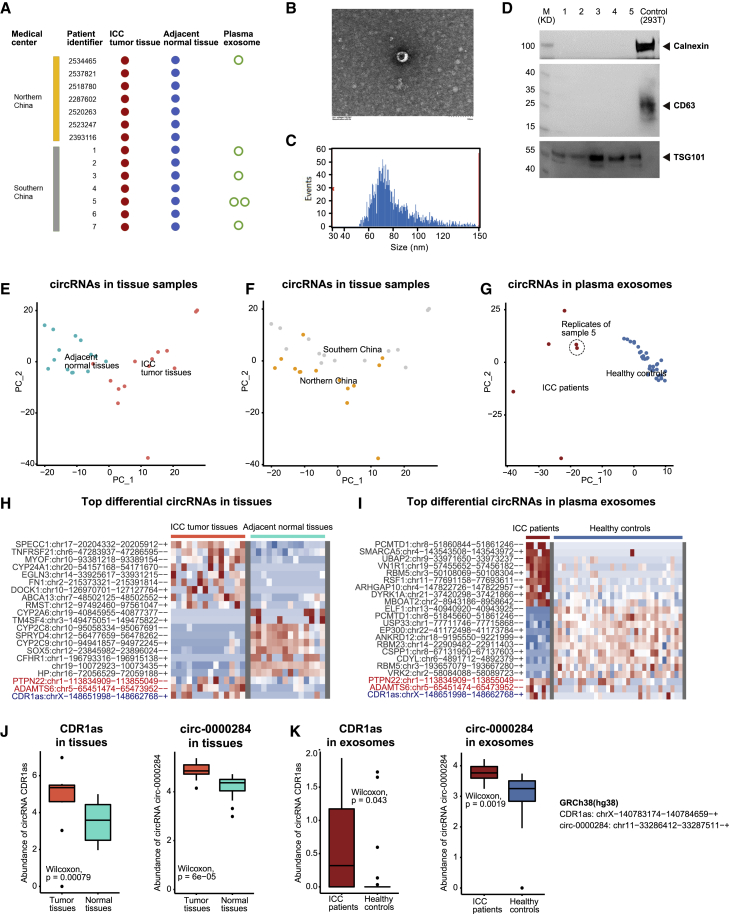
A total of 14 Asian patients with ICC donated ICC and paired adjacent normal tissues, among which six patients donated peripheral blood samples for plasma exosome extraction (Figure 1A, Table S1). The plasma exosomes of patient 5 from southern China were sampled twice to verify the reproducibility of our methods. In addition, the plasma exosomes were steadily detected using transmission electron microscopy (Figures 1B and S1A), high-sensitivity flow cytometry (Figures 1C and S1B; Tables S2 and S3), and western blotting of plasma exosome markers, including TSG101 and CD63 (positive markers) and calnexin (negative marker) proteins (Figure 1D).
Figure 1.
Landscape of circRNAs in tumor tissues and plasma exosomes of Asian ICC
(A) Sampling and sequencing of the Asian ICC patient cohort. (B) Representative plasma exosomes detected by transmission electron microscopy. (C) The size of plasma exosomes analyzed by high-sensitivity flow cytometry. (D) Western blotting of plasma exosome markers, including TSG101, CD63, and calnexin proteins. (E) Principal-component analysis (PCA) of the circRNA profile in ICC tumor tissues and adjacent normal tissues, labeled by sample types. (F) PCA of the circRNA profile in ICC tumor tissues and adjacent normal tissues, labeled by medical centers. (G) PCA of the circRNA profile in plasma exosomes of patients with ICC and healthy controls (provided by ExoRbase), labeled by sample types. (H) Top differential circRNAs in ICC tumor tissues versus adjacent normal tissues. (I) Top differential circRNAs in plasma exosomes of patients with ICC versus healthy controls. (J) The abundance of CDR1as and circ-0000284 in ICC tumor tissues and adjacent normal tissues. (K) The abundance of CDR1as and circ-0000284 in plasma exosomes of patients with ICC and healthy controls.
To minimize the bias of every single algorithm,21 we retained the circRNAs predicted by ≥ 2 among the four top-cited circRNA prediction algorithms, including circRNA_finder,22 find_circ,23 Circexplorer,24 and CIRI225 (Figure S2A). The proportion of overlap between circRNAs in ICC tumor tissues and adjacent normal tissues was higher than that between the host genes; a similar phenomenon was observed with the plasma exosomal circRNAs (Figures S2B and S2C). Dynein axonemal heavy chain 14 (DNAH14) was increasingly backspliced in tumor tissues compared with that in adjacent normal tissues, which was consistent with our previous observations in pan-cancer tissues (Figure S2D).26
We observed that the ICC tumor tissues and adjacent normal tissues had distinct circRNA profiles (Figure 1E). Particularly, differences between sample types (principal component 1) were greater than those between northern and southern China populations (principal component 2) (Figure 1F). Similarly, circRNAs in plasma exosomes of patients with ICC were distinct from those of healthy controls (Figure 1G). We regarded the overlapping between upregulated circRNAs in ICC tumor tissues (Figure 1H) and plasma exosomes of patients with ICC (Figure 1I) as potential ICC biomarkers for liquid biopsy, among which the repeatedly reported CDR1as11 and circ-000028420 were also observed (Figures 1J and 1K).
A subgroup of immunogenic Asian ICC was characterized by T cell exhaustion and neutrophil extracellular traps
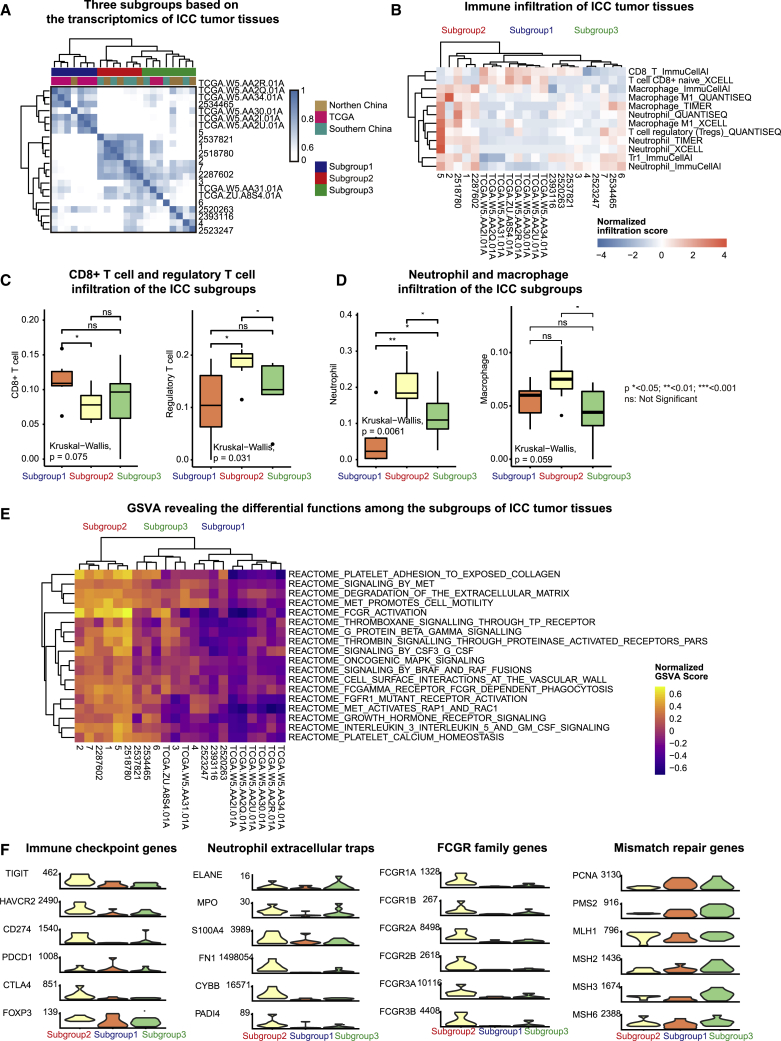
To reveal the characteristics of Asian patients with ICC, we included an additional eight pairs of ICC tumor tissues and adjacent normal tissues donated by Caucasian patients with ICC from The Cancer Genome Atlas (TCGA) (Figure 2A). The correlation between ICC tumor tissue samples revealed three subgroups among Asian and Caucasian patients with ICC. Subgroup 1 was mostly detected among Caucasian patients. Subgroup 2 was found in northern and southern China populations, showing a strong correlation among samples. Subgroup 3, which was detected in both Caucasian and Asian patients, demonstrated higher heterogeneity.
Figure 2.
Transcriptomic analysis of immune infiltration in Asian ICC tumor tissues
(A) Unsupervised hierarchical clustering of the transcriptome of Asian and Caucasian ICC tumor tissues. (B) Immune infiltration in the three ICC subgroups, predicted by ImmuCellAI, xCell, TIMER, and quanTIseq. (C) Infiltration of CD8+ T cells and regulatory T cells in ICC subgroups, predicted by ImmuCellAI. (D) Infiltration of neutrophils and macrophages in ICC subgroups, predicted by ImmuCellAI. (E) Gene set variation analysis (GSVA) of ICC tumor tissues based on the Reactome database. (F) Expression of immune checkpoint genes, neutrophil extracellular trap-associated genes, FCGR family genes, and mismatch repair genes.
We analyzed the infiltration of adaptive and innate immune cells in these ICC tumor tissues by using ≥2 algorithms, including ImmuCellAI,27 xCell,28 quanTIseq,29 and TIMER30 (Figure 2B). We found that subgroup 2 showed decreased infiltration by CD8+ T cells, compared with that of subgroup 1, and increased infiltration of regulatory T cells (Figures 2C and S3A), neutrophils (Figures 2D and S3B), and M1 macrophages (Figures 2D and S3C), compared with that of subgroups 1 and 2.
Gene set variation analysis (GSVA) further revealed the differential immunologic features among these ICC subgroups (Figure 2E). Compared with subgroups 1 and 3, subgroup 2 showed prominent immunogenic features, such as Fc gamma receptor (FCGR)-dependent phagocytosis, multiple processes in platelet activation and thrombosis, and signaling mediated by granulocyte colony-stimulating factor (G-CSF), interleukin 3, interleukin 5, granulocyte-macrophage colony-stimulating factor (GM-CSF), and G protein beta gamma. Specific oncogenic signaling pathways that potentially interact with these immune pathways were also activated in subgroup 2, including growth hormone receptor signaling, fibroblast growth factor receptor 1 (FGFR1) mutant receptor activation, and signaling mediated by B-Raf proto-oncogene (BRAF) and Raf-1 proto-oncogene (RAF) fusions and mitogen-activated protein kinase 1 (MAPK). Particularly, the MET proto-oncogene (MET) signaling promoted cell motility via the activation of Ras-related protein 1 (RAP1), and Rac family small GTPase 1 (RAC1), which indicated an increased tendency for metastases when combined with extracellular matrix degradation.
Further, we investigated the expression of classical marker genes associated with these immune and oncogenic features of subgroup 2 (Figure 2F). Upregulated immune checkpoint genes, including T cell immunoreceptor with Ig and ITIM domains (TIGIT), hepatitis A virus cellular receptor 2 (HAVCR2), CD274 (encoding PD-L1), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), and forkhead box P3 (FOXP3), implicated T cell exhaustion and are possible therapeutic targets for immune checkpoint blockade in subgroup 2. Increased neutrophil infiltration, platelet activation, and thrombosis indicated neutrophil extracellular traps in subgroup 2. We found the upregulation of genes associated with neutrophil extracellular traps, including myeloperoxidase (MPO), S100 calcium-binding protein A4 (S100A4), fibronectin 1 (FN1), NADPH oxidase 2 (NOX2), and peptidyl arginine deiminase 4 (PADI4) in subgroup 2. Moreover, subgroup 2 had an exclusive expression of FCGR1A, FCGR1B, FCGR2A, FCGR2B, FCGR3A, and FCGR3B, all of which belong to the FCGR family that is highly expressed in neutrophils and macrophages. In addition, mismatch repair (MMR) genes associated with the immunogenicity of tumors, such as proliferating cell nuclear antigen (PCNA), PMS1 homolog 2, mismatch repair system component (PMS2), MutS homolog 2 (MSH2), MutS homolog 3 (MSH3), and MutS homolog 6 (MSH6) were downregulated in subgroup 2.
Elevated circ-PTPN22 and circ-ADAMTS6 levels in ICC tumor tissues and plasma exosomes marked this immunogenic Asian ICC subgroup
To discover potential exosomal circRNA biomarkers, we explored the circRNAs with elevated levels in both ICC tumor tissue plasma exosomes of the immunogenic Asian ICC subgroup, especially those mechanistically associated with T cell exhaustion and neutrophil extracellular traps.
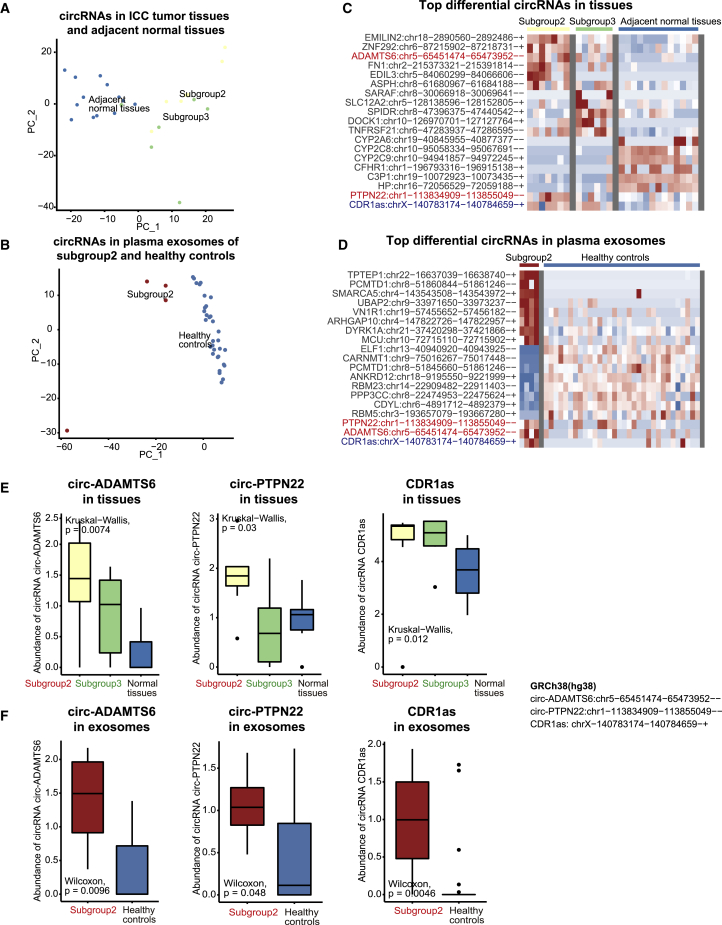
We observed that the subgroup of immunogenic Asian ICC was distinguished not only by gene expression but also by the circRNA profile (Figure 3A) and plasma exosomes (Figure 3B). Overlapping between the elevated circRNAs in the ICC tumor tissues of subgroup 2 (Figure 3C) and those in the plasma exosomes of subgroup 2 (Figure 3D) revealed potential circRNA biomarkers for the immunogenic Asian ICC. Among these potential biomarkers, circRNAs derived from protein tyrosine phosphatase non-receptor type 22 (PTPN22) and ADAM metallopeptidase with thrombospondin type 1 motif 6 (ADAMTS6) had a potential functional association with the immunogenicity of ICC tumor tissue. circ-PTPN22 (hsa_circ_0110529), circ-ADAMTS6 (hsa_circ_0072688), and CDR1as levels were upregulated in ICC tumor tissues (Figure 3E) and plasma exosomes of patients with ICC in subgroup 2 (Figure 3F). Compared with CDR1as, circ-PTPN22 and circ-ADAMTS6 performed better in distinguishing between subgroup 2 and subgroup 3.
Figure 3.
Potential circRNA biomarkers of the immunogenic Asian ICC subgroup
(A) PCA of the circRNA profile in ICC tumor tissues and adjacent normal tissues, labeled by subgroups. (B) PCA of the circRNA profile in plasma exosomes of ICC tumor tissues and healthy controls (provided by ExoRbase). (C) Top differential circRNAs in subgroup 2 versus subgroup 3 and adjacent normal tissues. (D) Top differential circRNAs in plasma exosomes of subgroup 2 versus healthy controls. (E) The abundance of circ-ADAMTS6, circ-PTPN22, and CDR1as in subgroups 2 and 3, and adjacent normal tissues. (F) Expression levels of ADAMTS6 and PTPN22 in subgroups 2 and 3, and adjacent normal tissues. (G) The abundance of circ-ADAMTS6, circ-PTPN22, and CDR1as in plasma exosomes of subgroup 2 and healthy controls.
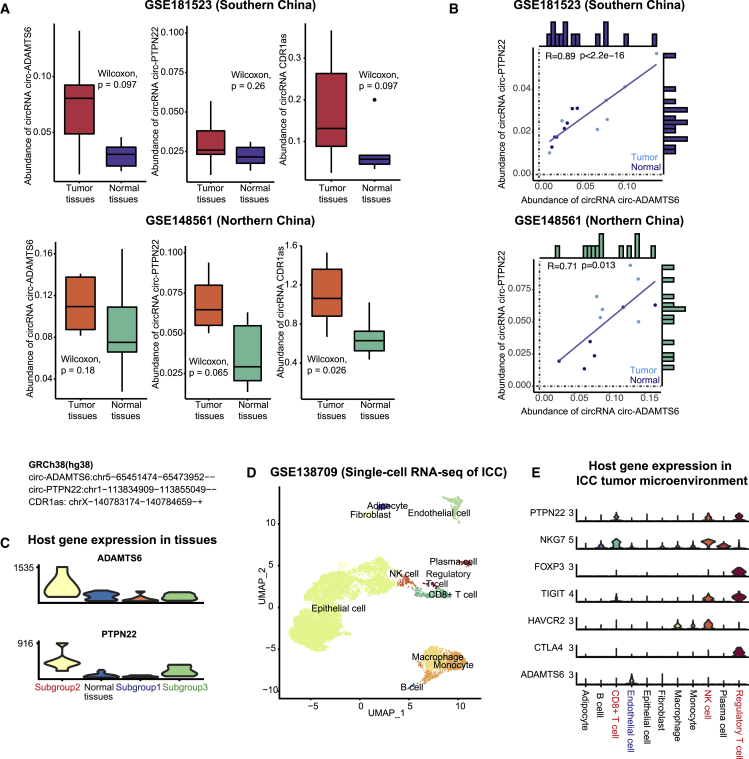
Moreover, circ-PTPN22, circ-ADAMTS6, and CDR1as were also more abundant in the ICC tumor tissues compared with the adjacent normal tissues collected by the GEO circRNA datasets: GSE181523 (Southern China, n = 7)31 and GSE148561 (Northern China, n = 6)32 (Figure 4A). This difference in the levels of circ-PTPN22 was less significant because its expression in ICC tumor tissues of subgroup 3 was as low as in the adjacent normal tissue (Figure 3E). Although the immune subgroups among these samples were unknown, circ-PTPN22 and circ-ADAMTS6 potentially determined the identical subgroups as they were significantly co-expressed (Figure 4B). Also, among the circRNAs that have been reported as ICC biomarkers, CDR1as and circ-0000284 levels were elevated in the ICC tumor samples in all three datasets, while circ-CCAC1, circ_0005230, circSETD3 levels were upregulated in one of the three datasets (Figures S4A–S4C, Table S4). This validation implied that circ-PTPN22 and circ-ADAMTS6 were potentially robust ICC biomarkers similar to CDR1as and circ-0000284, which were previously reported in several studies.
Figure 4.
Validation of circRNA biomarkers and host genes in Asian ICC tumor tissues collected by independent studies
(A) Abundance of circ-ADAMTS6, circ-PTPN22, and CDR1as in the ICC tumor tissues and adjacent tumor tissues collected by the GSE181523 (southern China) dataset and the GSE148561 (northern China) dataset. (B) Correlation between the abundance of circ-ADAMTS6 and circ-PTPN22 in ICC tumor tissues and adjacent normal tissues collected by the GSE181523 and GSE148561. (C) Expression of ADAMTS6 and PTPN22 in subgroups 1, 2, and 3, and adjacent normal tissues. (D) UMAP embedding of the single-cell transcriptome of ICC tumor tissues. (E) Expression of PTPN22, ADAMTS6, and immune checkpoint genes in different cell types.
Interestingly, PTPN22 and ADAMTS6, the host genes of circRNA biomarkers, were also upregulated in ICC tumor tissues of subgroup 2 (Figure 4C). Single-cell transcriptome of the GEO dataset, GSE138709, revealed the epithelial cells (cholangiocytes and cholangiocarcinoma cells), endothelial cells, CD8+ T cells, regulatory T cells, natural killer cells (NK cells), B cells, plasma cells, monocytes, macrophages, adipocytes, and fibroblasts in ICC tumor tissues (Figure 4D).33 PTPN22 was expressed in regulatory T cells, CD8+ T cells, and NK cells, whereas ADAMTS6 was exclusively expressed in endothelial cells. A subpopulation of these regulatory T cells highly expressed FOXP3, TIGIT, and CTLA4 (Figure 4E).
Validation of circ-PTPN22 and circ-ADAMTS6 as biomarkers in Asian ICC tumor tissues of the immunogenic subgroup
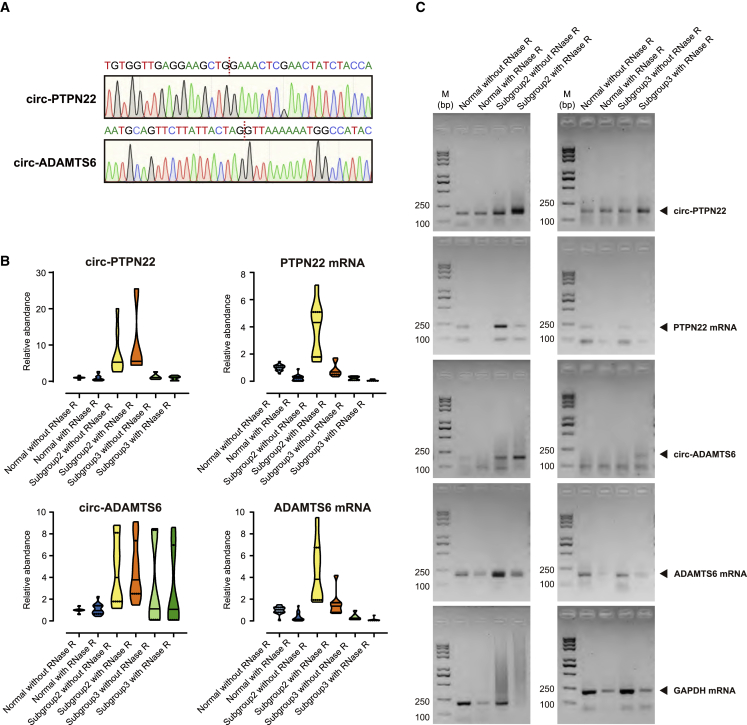
Finally, we validated the upregulation of circ-PTPN22 and circ-ADAMTS6 in ICC tumor tissues of subgroup 2 by using quantitative RT-PCR (qRT-PCR) (Table S5). The Sanger sequencing of the PCR products demonstrated the junction sites of circ-PTPN22 and circ-ADAMTS6 backsplicing (Figure 5A). As expected, these circRNAs were protected from RNase R degradation by their covalently closed structures, while the linear mRNAs of their host genes were susceptible to degradation. Consistent with the results of RNA sequencing (RNA-seq), circ-PTPN22, circ-ADAMTS6, and their host genes were expressed at a higher level in subgroup 2 than in subgroup 3 and adjacent normal tissues (Figures 5B and 3E, Table S6). This experimental validation used multiple samples in our cohort of patients with ICC from southern and northern China (Figures 5C and S5).
Figure 5.
Quantitative RT-PCR analysis of circRNA biomarkers and host genes in Asian ICC tumor tissues
(A) Sanger sequencing of the PCR product of circ-PTPN22 and circ-ADAMTS6. Red dotted lines indicate the junction site of backsplicing. (B) qRT-PCR analysis of PTPN22 mRNA, ADAMTS6 mRNA, circ-PTPN22, and circ-ADAMTS6 abundance in ICC subgroups and adjacent normal tissues. (C) Agarose gel electrophoresis of the PCR product of PTPN22 mRNA, ADAMTS6 mRNA, circ-PTPN22, and circ-ADAMTS6.
In addition, we used PCR to validate that amplicons of backsplicing junction sites of circ-PTPN22 and circ-ADAMTS6 were from the transcriptome instead of the genome, which supported the hypothesis that backsplicing is a post-transcriptional process (Table S7). The amplicons of circ-PTPN22 and circ-ADAMTS6 were detected in RNA (cDNA library) but not in genomic DNA. In contrast, those of host genes, PTPN22 and ADAMTS6 presented in both the RNA and genomic DNA. The negative control showed that the circRNAs and host gene mRNAs were absent without MMLV Reverse Transcriptase treatment (Figure S6).
Overall, these validations showed that circ-PTPN22 and circ-ADAMTS6 were expressed in higher levels in ICC tumor tissues of subgroup 2 compared with subgroup 3 and adjacent normal tissues, and had covalently closed structures generated by post-transcriptional backsplicing at the junction sites.
Discussion
This study integrated the transcriptomes of plasma exosomes, tumor tissues, and adjacent normal tissues of ICC, and revealed that PTPN22 and circ-ADAMTS6 in plasma exosomes were associated with a subgroup of immunogenic Asian ICC characterized by T cell exhaustion and neutrophil extracellular traps.
Neutrophil extracellular traps were likely to be associated with T cell exhaustion in this subgroup of immunogenic ICC (Figures 2D–2F). Triggered by G-CSF and reactive oxygen species catalyzed by NOX2, the biological processes of neutrophil extracellular traps include increased neutrophil recruitment, PADI4-mediated decondensation of chromatin, accumulation of neutrophil elastase and MPO, and the release of histones into the extracellular space. In particular, neutrophil extracellular traps have been found to promote cancer-associated thrombosis, degradation of the extracellular matrix, and cancer metastases.34,35,36
Most recently, an analysis of single-cell full-length transcriptome has provided a resource of circRNA expression in different cell types.37 This data resource demonstrated that circ-PTPN22 was expressed in regulatory and central memory T cells, whereas circ-ADAMTS6 was expressed in multiple types of stromal cells. These results add to our observation that host genes were co-expressed with circ-PTPN22 and circ-ADAMTS6 and were highly expressed in regulatory T cells, CD8+ T cells, and endothelial cells, respectively (Figures 4C and 4E).
The host gene, PTPN22, encodes a lymphoid-specific intracellular phosphatase that regulates the T cell receptor signaling pathway, whose polymorphism serves as a regulator of the threshold of T cell activation.38 More specifically, inhibition of PTPN22 enhanced T cell infiltration and anti-PD-L1 treatment39 and promoted the activation of CD8+ T cells and the response to checkpoint blockade. The expression of PTPN22 was positively correlated with regulatory T cells, M1 macrophages, and the level of immune checkpoint genes in multiple types of cancer, which was consistent with our observations of subgroup 2.40 As a backspliced product of PTPN22, multiple circRNAs have been reported to promote proliferation and inhibit T cell infiltration in pancreatic cancer,41 be associated with epithelial-mesenchymal transformation and metastases of gastric cancer,42 and serve as potential biomarkers of rheumatoid arthritis43 and systematic lupus erythematosus.44
Another host gene, ADAMTS6, encodes a secreted multidomain matrix-associated zinc metalloendopeptidase that participates in tissue remodeling. Thus, the ADAMTS family plays an important role in neutrophil extracellular traps where inflammation and thrombosis occur in microvessels,45 and its dysregulation contributes to tumor invasion and metastases.46 circRNAs derived from the backsplicing of ADAMTS6 have been revealed to regulate interleukin 1-induced apoptosis of human chondrocytes in osteoarthritis.47,48
As for the potential prognostic value, patients in subgroup 2 demonstrated prolonged overall survival compared with those in subgroup 3, although this difference was not significant due to the small sample size (Figure S7A). High expression of CDR1as in ICC tumor tissues was associated with a poor prognosis, which was consistent with the results of most previous studies (Figure S7B). In the TCGA-CHOL cohort, patients whose ICC tumor tissues highly expressed PTPN22 and ADAMTS6 showed prolonged disease-free survival (Figure S7C).
Our study has several limitations. Although the current sample size was sufficient to identify the immunogenic Asian ICC, due to its limited size, our results require future validation with a larger cohort to establish the ICC subgroups. Because the purification of exosomal RNA from the plasma was elaborate, some plasma samples matched with the tissue samples were not sequenced (Figure 1A).
In summary, this pilot study revealed a subgroup of immunogenic Asian ICC characterized by T cell exhaustion and neutrophil extracellular traps and marked by an elevated abundance of circ-PTPN22 and circ-ADAMTS6 in plasma exosomes. This subgroup of patients with ICC is potentially detectable by plasma exosomal circRNAs and treatable with immune checkpoint blockade.
Materials and methods
Patient recruitment and sample collection
This study was approved by the Ethics Committee of Peking Union Medical Hospital (northern China) and Hunan Provincial People’s Hospital (southern China). Surgically resected tumor tissues and adjacent normal tissues and plasma exosomes extracted from peripheral blood were donated by 14 patients (n = 7 from northern China, n = 7 from southern China) who provided informed consent. Pathology departments confirmed the diagnoses made in the Peking Union Medical Hospital and Hunan Provincial People’s Hospital.
Extraction and characterization of plasma exosomes
Plasma exosomes were isolated using a Microfuge 20R (Beckman, Brea, CA, USA) and an Ultracentrifuge CP100MX (Hitachi, Tokyo, Japan) at 2000 × g for 30 min, 10,000 × g for 45 min, 100,000 × g for 70 min, and then 100,000 × g for 70 min at 4°C. Transmission electron microscopy HT-7700 (Hitachi, Tokyo, Japan) was used to observe the plasma exosomes identified by negative staining with phosphotungstic acid. Size distribution analysis of plasma exosomes was performed using a high-sensitivity flow cytometry N30E (NanoFCM, Xiamen, China) The side scattering (SSC) distribution histogram of the mixture was converted into its corresponding vesicle size. For western blotting, we used a 10-μg sample for each lane and the 293T cells as the control. The following antibodies were used: anti-calnexin (12186, 1:1,000) (Santa Cruz Biotechnology, Dallas, TX, USA), anti-CD63 (ab134045, 1:1,000) (Abcam, Cambridge, UK), and anti-TSG101 (ab125011, 1:1,000) (Abcam, Cambridge, UK). Peroxidase conjugated goat anti-rabbit IgG (Merck Millipore, Burlington, NJ, USA) was used as the secondary antibody and the western blot membrane was imaged using a ChemiScope 3000 Mini system (CLINX, Shanghai, China).
High-throughput RNA-seq
Total RNA from the tissue and plasma exosome samples was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and those with an RIN value > 7 were used for library preparation. Next-generation sequencing libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA). rRNA was depleted from total RNA using the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA). The rRNA-depleted RNA was fragmented and reverse transcribed. First-strand cDNA was synthesized using ProtoScript II Reverse Transcriptase with random primers and actinomycin D (New England Biolabs, Ipswich, MA, USA). Second-strand cDNA was synthesized using the Second Strand Synthesis Enzyme Mix, including dACG-TP/dUTP (New England Biolabs, Ipswich, MA, USA). The double-stranded cDNA was purified by the AxyPrep Mag PCR Clean-up kit (Axygen, Union City, CA, USA) and then treated with End Prep Enzyme Mix (Illumina, San Diego, CA, USA). Size selection of adaptor-ligated DNA was then performed by the AxyPrep Mag PCR Clean-up kit (Axygen, Union City, CA, USA), and fragments of ∼360 base pairs (bp) (with an approximate insert size of 300 bp) were recovered. The dUTP-marked second strand was digested with the Uracil-Specific Excision Reagent (USER) enzyme (New England Biolabs). Each sample was amplified by PCR for 14 cycles using P5 and P7 primers and then cleaned up by the AxyPrep Mag PCR Clean-up kit (Axygen, Union City, CA, USA). Libraries with different indices were multiplexed and loaded on the Illumina NovaSeq 6000 system (Illumina).
RNA extraction, qRT-PCR, and Sanger sequencing
Total RNA was isolated using TRIzol (Invitrogen) with DNase I treatment, and the extracted RNA was dissolved in 80 μL RNase-free distilled water and stored at −80°C. DNase I-treated RNA (∼5 μg) was treated for 30 min at 37°C with or without 3 U/μg of RNase R (Epicentre, Charlotte, NC, USA), and 1 μL RNase R-treated RNA was directly reverse transcribed using an MMLV Reverse Transcriptase 1st-Strand cDNA Synthesis Kit with a random hexamer primer (Invitrogen) according to the manufacturer’s protocol. The cDNA was diluted 10-fold before qRT-PCR analysis.
qRT-PCR experiments were performed in duplicate (technical replicates) using 2 × RealStar Green Fast Mixture (GenStar, Beijing, China) and a LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland). PCR program settings were 95°C for 10 min (initial denaturation); 95°C for 15 s, and 60°C for 1 min, for 45 cycles. Ct values were obtained by the second derivative maximum method and analyzed by the 2−ΔΔCt method.49 PCR products were visualized after electrophoresis in 2% ExRed-stained (Zomanbio, Beijing, China) agarose gel, and purified through the V-ELUTE Gel Mini Purification Kit (Zomanbio, Beijing, China). The pBLUE-T plasmid was constructed by the pBLUE-T fast cloning kit (Zomanbio, Beijing, China), and Sanger sequencing was performed.
DNA and RNA extraction, PCR, and Sanger sequencing
Total genomic DNA was extracted from tissues by DNA digestion buffer incubation (50 mM Tris-HCl pH 8.0, 100 mM EDTA pH 8.0, 100 mM NaCl, 1% SDS, 0.5 mg/mL proteinase K) overnight at 65°C with gentle shaking, and then treated with 10 μg/mL RNase A (Zomanbio, Beijing, China) at 37°C. Total RNA was isolated using TRIzol (Invitrogen) with DNase I treatment. The cDNA library was total RNA directly reverse transcribed using an MMLV Reverse Transcriptase first-Strand cDNA Synthesis Kit with a random hexamer primer (Invitrogen). The negative control was total RNA not treated with MMLV Reverse Transcriptase.
PCR experiments were performed in duplicates using the TransStart FastPfu Fly polymerase (TransGen Biotech, Beijing, China). PCR program settings were 95°C for 5 min (initial denaturation); 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, for 45 cycles. PCR products were visualized after electrophoresis in 2% ExRed-stained (Zomanbio, Beijing, China) agarose gel, and purified through the V-ELUTE Gel Mini Purification Kit (Zomanbio, Beijing, China). The pBLUE-T plasmid was constructed using the pBLUE-T fast cloning kit (Zomanbio, Beijing, China), and Sanger sequencing was performed.
TCGA-CHOL RNA-seq datasets
We downloaded the RNA-seq data of Caucasian ICC samples from TCGA. We selected Caucasian patients with ICC, whose primary cancer sites were the liver and intrahepatic bile ducts, and from whom both ICC tumor tissues and adjacent normal tissues could be obtained (TCGA.W5.AA34, TCGA.W5.AA2Q, TCGA.W5.AA30, TCGA.W5.AA2U, TCGA.W5.AA2R, TCGA.W5.A8S4, TCGA.W5.AA31, and TCGA.WU.AA2I). Moreover, we used the GEPIA2 web server (http://gepia2.cancer-pku.cn/#) for survival analysis of the TCGA-CHOL cohort.50
ExoRbase and GEO circRNA datasets
We downloaded the raw fastq data of the RNA-seq of plasma exosomes collected by exoRbase51 (http://www.exorbase.org/) from the Sequence Read Archive database. We included all 32 healthy controls from the research in China. These data were processed using the same pipeline processing for ICC sample data. We also collected the circRNA microarray data of ICC tumor tissues from two public datasets (GEO accession number: GSE181523 and GSE148561).31,32
Gene expression analysis
We adopted the identical mRNA analysis pipeline used by TCGA (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/Expression_mRNA_Pipeline/). We used fastp52 for quality control and trimming of raw RNA-seq data. STAR53 was used to map the reads to the human genome (GRCh38 reference genome and Gencode v22 annotation), and HTSeq-count54 was used to quantify the mapped reads. DESeq255 package was used to normalize gene expression and estimate the significance of differentially expressed genes in tumor tissues. The thresholds of significance were a log 2-fold change ≥2 or ≤ −2, and an adjusted p value ≤0.05 (Wald test).
circRNA expression analysis
Our pipeline combined four top-cited circRNA prediction algorithms (circRNA_finder,22 find_circ,23 Circexplorer,24 and CIRI225) using the default parameters. We retained the circRNAs predicted by ≥2 among these four circRNA prediction algorithms. Subsequently, we averaged the backsplicing junction reads generated by the different algorithms. Any circRNA with an average raw count >1,000 was removed. The circRNA nomenclature provides information on the chromosome, the two backsplicing sites, and strandness; for example, “chr1_113834909_113855049_-” (hg38, aka GRCh38 reference genome).
The circRNA profile was of high dimensionality and sparsity, a feature selection was necessary before normalization and differential analysis. Any circRNA expressed in less than two samples was removed. The circRNA profile was log normalized, and subjected to variable selection, principal-component analysis, and differential expression analysis to find the upregulated circRNAs in each ICC subgroup, adjacent normal tissues, and plasma exosomes from patients with ICC or healthy controls.
Immune infiltration prediction
We used multiple algorithms to infer the immune infiltration of tumor tissues, which included ImmuCellAI,27 xCell,28 quanTIseq,29 and TIMER.30 We adopted the results supported by ≥2 among these four algorithms.
Single-cell transcriptomic analysis
We collected the single-cell RNA-seq data of ICC tumor tissues from a public dataset (GEO accession number: GSE138709).33 Harmony56 was used to integrate the single-cell transcriptome of ICC tumor tissues from five patients. Cell types were determined based on marker gene expression and SingleR,57 a machine learning-based pipeline.
Statistical analysis
The R software (version 4.0.3) was used to conduct statistical analysis and visualization. All reported p values were two-sided with 0.05 as the threshold of significance.
Availability of data and materials
The circRNA and mRNA datasets generated in this study will be available in the GEO repository (GEO accession number: GSE188330) and SRA (BioProject accession number: PRJNA777408) upon publication. The circRNA prediction algorithms (https://github.com/Selecton98/CircRNA_4DGlab2) and the Jupyter notebooks (https://github.com/Selecton98/CircRNA_ICC) are available in the GitHub repository.
Acknowledgments
This study was supported by grants from the National High Level Hospital Clinical Research Funding (2022-PUMCH-A-236), CAMS Innovation Fund for Medical Sciences (CIFMS) (2020-I2M-C&T-B-019), Beijing Municipal Natural Science Foundation Project (Grant No. 7222130), Chen Xiao Ping Foundation for the Development of Science and Technology of Hubei Province (CXPJJH1200008-10), and Beijing Undergraduate Training Programs for Innovation and Entrepreneurship (202010023046).
Author contributions
X.W. managed the team, analyzed the data, and drafted the manuscript; G.W. designed and conducted the experiments; Y.D. improved the data analysis and revised the manuscript; Z.W. and Y.S. collected and sequenced the clinical samples; F.Y., X.C., and S.D. contributed to sample collection; Y.Z., H.X., and J.W. conceptualized the project and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.12.012.
Contributor Information
Haifeng Xu, Email: xuhf781120@sina.com.
Yongchang Zheng, Email: yong-chang_zheng@outlook.com.
Supplemental information
References
- 1.Shaib Y., El-Serag H.B. The epidemiology of cholangiocarcinoma. Semin. Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 2.Moeini A., Sia D., Bardeesy N., Mazzaferro V., Llovet J.M. Molecular pathogenesis and targeted therapies for intrahepatic cholangiocarcinoma. Clin. Cancer Res. 2016;22:291–300. doi: 10.1158/1078-0432.CCR-14-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodson R.M., Weiss M.J., Cosgrove D., Herman J.M., Kamel I., Anders R., Geschwind J.F.H., Pawlik T.M. Intrahepatic cholangiocarcinoma: management options and emerging therapies. J. Am. Coll. Surg. 2013;217:736–750.e4. doi: 10.1016/j.jamcollsurg.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H., Yang T., Wu M., Shen F. Intrahepatic cholangiocarcinoma: epidemiology, risk factors, diagnosis and surgical management. Cancer Lett. 2016;379:198–205. doi: 10.1016/j.canlet.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Cao J., Hu J., Liu S., Meric-Bernstam F., Abdel-Wahab R., Xu J., Li Q., Yan M., Feng Y., Lin J., et al. Intrahepatic cholangiocarcinoma: genomic heterogeneity between eastern and western patients. JCO Precis. Oncol. 2020;4 doi: 10.1200/PO.18.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaisaingmongkol J., Budhu A., Dang H., Rabibhadana S., Pupacdi B., Kwon S.M., Forgues M., Pomyen Y., Bhudhisawasdi V., Lertprasertsuke N., et al. Common molecular subtypes among asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell. 2017;32:57–70.e3. doi: 10.1016/j.ccell.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeuillard E., Conboy C.B., Gores G.J., Rizvi S. Immunobiology of cholangiocarcinoma. JHEP Rep. 2019;1:297–311. doi: 10.1016/j.jhepr.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Job S., Rapoud D., Dos Santos A., Gonzalez P., Desterke C., Pascal G., Elarouci N., Ayadi M., Adam R., Azoulay D., et al. Identification of four immune subtypes characterized by distinct composition and functions of tumor microenvironment in intrahepatic cholangiocarcinoma. Hepatology. 2020;72:965–981. doi: 10.1002/hep.31092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jusakul A., Cutcutache I., Yong C.H., Lim J.Q., Huang M.N., Padmanabhan N., Nellore V., Kongpetch S., Ng A.W.T., Ng L.M., et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Z., Cao Q., Zhao Z., Song C. Biogenesis, features, functions, and disease relationships of a specific circular RNA: CDR1as. Aging Dis. 2020;11:1009–1020. doi: 10.14336/AD.2019.0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X.M., Li Z.L., Li J.L., Xu Y., Leng K.M., Cui Y.F., Sun D.J. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. Eur. Rev. Med. Pharmacol. Sci. 2018;22:365–371. doi: 10.26355/eurrev_201801_14182. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y., Yao Y., Liu Y., Wang Z., Hu Z., Su Z., Li C., Wang H., Jiang X., Kang P., et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging. 2019;11:1907–1917. doi: 10.18632/aging.101872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Y., Yu T., Wang Y., Huang X., Wei X. Circular RNA circDNM3OS functions as a miR-145-5p sponge to accelerate cholangiocarcinoma growth and glutamine metabolism by upregulating MORC2. OncoTargets Ther. 2021;14:1117–1129. doi: 10.2147/OTT.S289241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y., Gao P., Wang Z., Su Z., Liao G., Han Y., Cui Y., Yao Y., Zhong X. Circ-LAMP1 contributes to the growth and metastasis of cholangiocarcinoma via miR-556-5p and miR-567 mediated YY1 activation. J. Cell Mol. Med. 2021;25:3226–3238. doi: 10.1111/jcmm.16392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu J., Chen W., Zheng L., Fang S., Zhang D., Kong C., Yang Y., Qiu R., Zhao Z., Lu C., et al. Circular RNA Circ0021205 promotes cholangiocarcinoma progression through MiR-204-5p/RAB22A Axis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.653207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J., Wang R., Tang R., Gu P., Han J., Huang W. CircRTN4IP1 regulates the malignant progression of intrahepatic cholangiocarcinoma by sponging miR-541-5p to induce HIF1A production. Pathol. Res. Pract. 2022;230 doi: 10.1016/j.prp.2021.153732. [DOI] [PubMed] [Google Scholar]
- 17.Xiong W., Zhang A., Xiao X., Liu W. CircSETD3 (hsa_circ_0000567) inhibits proliferation and induces apoptosis in cholangiocarcinoma cells via downregulation of microRNA-421 expression. Bioengineered. 2022;13:10191–10201. doi: 10.1080/21655979.2022.2061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S., Chen Z., Li Z., Li S., Wen Z., Cao L., Chen Y., Xue P., Li H., Zhang D. Tumor-associated macrophages promote cholangiocarcinoma progression via exosomal Circ_0020256. Cell Death Dis. 2022;13:94. doi: 10.1038/s41419-022-04534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y., Leng K., Yao Y., Kang P., Liao G., Han Y., Shi G., Ji D., Huang P., Zheng W., et al. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73:1419–1435. doi: 10.1002/hep.31493. [DOI] [PubMed] [Google Scholar]
- 20.Wang S., Hu Y., Lv X., Li B., Gu D., Li Y., Sun Y., Su Y. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin. Sci. 2019;133:1935–1953. doi: 10.1042/CS20190589. [DOI] [PubMed] [Google Scholar]
- 21.Hansen T.B. Improved circRNA identification by combining prediction algorithms. Front. Cell Dev. Biol. 2018;6:20. doi: 10.3389/fcell.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y., Zhang J., Zhao F. Circular RNA identification based on multiple seed matching. Briefings Bioinf. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Dong Y., Wu Z., Wang G., Shi Y., Zheng Y. Machine learning-based comparative analysis of pan-cancer and pan-normal tissues identifies pan-cancer tissue-enriched circRNAs related to cancer mutations as potential exosomal biomarkers. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.703461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao Y.R., Zhang Q., Lei Q., Luo M., Xie G.Y., Wang H., Guo A.Y. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv. Sci. 2020;7 doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., Krogsdam A., Loncova Z., Posch W., Wilflingseder D., et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q., Wang H., Li Z., Li F., Liang L., Zou Y., Shen H., Li J., Xia Y., Cheng Z., et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatol. 2022;76:135–147. doi: 10.1016/j.jhep.2021.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Zhao X., Zhang X., Zhang Z., Liu Z., Zhu J., Lyu S., Li L., Lang R., He Q. Comprehensive circular RNA expression profiling constructs a ceRNA network and identifies hsa_circ_0000673 as a novel oncogene in distal cholangiocarcinoma. Aging. 2020;12:23251–23274. doi: 10.18632/aging.104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M., Yang H., Wan L., Wang Z., Wang H., Ge C., Liu Y., Hao Y., Zhang D., Shi G., et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020;73:1118–1130. doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Masucci M.T., Minopoli M., Del Vecchio S., Carriero M.V. The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front. Immunol. 2020;11:1749. doi: 10.3389/fimmu.2020.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang D., Liu J. Neutrophil extracellular traps: a new player in cancer metastasis and therapeutic target. J. Exp. Clin. Cancer Res. 2021;40:233. doi: 10.1186/s13046-021-02013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios-Acedo A.L., Mège D., Crescence L., Dignat-George F., Dubois C., Panicot-Dubois L. Platelets, thrombo-inflammation, and cancer: collaborating with the enemy. Front. Immunol. 2019;10:1805. doi: 10.3389/fimmu.2019.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu W., Zhang J., Cao X., Cai Z., Zhao F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 2022;13:3242. doi: 10.1038/s41467-022-30963-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregersen P.K., Lee H.S., Batliwalla F., Begovich A.B. PTPN22: setting thresholds for autoimmunity. Semin. Immunol. 2006;18:214–223. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Cubas R., Khan Z., Gong Q., Moskalenko M., Xiong H., Ou Q., Pai C., Rodriguez R., Cheung J., Chan A.C. Autoimmunity linked protein phosphatase PTPN22 as a target for cancer immunotherapy. J. Immunother. Cancer. 2020;8:e001439. doi: 10.1136/jitc-2020-001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho W.J., Croessmann S., Lin J., Phyo Z.H., Charmsaz S., Danilova L., Mohan A.A., Gross N.E., Chen F., Dong J., et al. Systemic inhibition of PTPN22 augments anticancer immunity. J. Clin. Invest. 2021;131:e146950. doi: 10.1172/JCI146950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., Han P., Chen C., Xie S., Zhang H., Song Y., Hu H., Zhao Q., Lian C. circPTPN22 attenuates immune microenvironment of pancreatic cancer via STAT3 acetylation. Cancer Gene Ther. 2021 doi: 10.1038/s41417-021-00382-w. [DOI] [PubMed] [Google Scholar]
- 42.Ma S., Kong S., Gu X., Xu Y., Tao M., Shen L., Shen X., Ju S. As a biomarker for gastric cancer, circPTPN22 regulates the progression of gastric cancer through the EMT pathway. Cancer Cell Int. 2021;21:44. doi: 10.1186/s12935-020-01701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Z., Zhong Z., Miao Q., Zhang Y., Ni B., Zhang M., Tang J. circPTPN22 as a novel biomarker and ceRNA in peripheral blood mononuclear cells of rheumatoid arthritis. Mol. Med. Rep. 2021;24:617. doi: 10.3892/mmr.2021.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao Q., Zhong Z., Jiang Z., Lin Y., Ni B., Yang W., Tang J. RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus. 2019;28:520–528. doi: 10.1177/0961203319830493. [DOI] [PubMed] [Google Scholar]
- 45.Matevosyan K., Sarode R. Thrombosis, microangiopathies, and inflammation. Semin. Thromb. Hemost. 2015;41:556–562. doi: 10.1055/s-0035-1556587. [DOI] [PubMed] [Google Scholar]
- 46.Binder M.J., McCoombe S., Williams E.D., McCulloch D.R., Ward A.C. The extracellular matrix in cancer progression: role of hyalectan proteoglycans and ADAMTS enzymes. Cancer Lett. 2017;385:55–64. doi: 10.1016/j.canlet.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Shen L., Ji C., Lin J., Yang H. Regulation of circADAMTS6-miR-324-5p-PIK3R3 ceRNA pathway may be a novel mechanism of IL-1β-induced osteoarthritic chondrocytes. J. Bone Miner. Metabol. 2022;40:389–401. doi: 10.1007/s00774-021-01308-0. [DOI] [PubMed] [Google Scholar]
- 48.Fu Q., Li L., Wang B., Wu J., Li H., Han Y., Xiang D., Chen Y., Zhu J. CircADAMTS6/miR-431-5p axis regulate interleukin-1β induced chondrocyte apoptosis. J. Gene Med. 2021;23 doi: 10.1002/jgm.3304. [DOI] [PubMed] [Google Scholar]
- 49.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz430. W556–w560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S., Li Y., Chen B., Zhao J., Yu S., Tang Y., Zheng Q., Li Y., Wang P., He X., Huang S. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx891. D106–d112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The circRNA and mRNA datasets generated in this study will be available in the GEO repository (GEO accession number: GSE188330) and SRA (BioProject accession number: PRJNA777408) upon publication. The circRNA prediction algorithms (https://github.com/Selecton98/CircRNA_4DGlab2) and the Jupyter notebooks (https://github.com/Selecton98/CircRNA_ICC) are available in the GitHub repository.