Plain language summary
In their recent article, Polyansky et al identify phosphatidylcholine (PC) as the most abundant lipid in the autophagosome membrane and demonstrate that eliminating de novo PC synthesis sharply impairs autophagic processing. In the absence of PC synthesis, open cup‐like structures accumulate, implicating PC as a key component in the closure of autophagosomes.
A recent report finds that specific phospholipids play a key role in closing the autophagosome.

Autophagosomes have very few transmembrane proteins. As such, lipids and peripheral membrane proteins are likely to play the most important roles in controlling the complicated membrane dynamics that underlie formation and maturation of the phagophore (the nascent autophagosome). For example, during phagophore expansion, the autophagosome adopts a cup‐like structure which ultimately closes upon itself through a membrane‐fission event. Whether lipid composition plays a role during this process, and even precisely which lipids are present as the autophagosome grows, are areas of intense investigation. In a recent study, Polyansky et al (2022) show that the yeast autophagosome membrane includes a high percentage of phosphatidylcholine (PC; Table 1). Yeast can tolerate the disruption of some lipid synthesis pathways and therefore is an ideal system to explore the impact a bulk lipid such as PC might have on autophagosome formation. Strikingly, when the authors knocked out the PC synthesizing enzymes Cho2 and Opi3, macroautophagy largely ground to a halt. Intriguingly, phagophores still formed and retain qualitatively similar structures, but their lipid compositions had shifted with the anionic phospholipid phosphatidylserine (PS) largely replacing the lost PC. The natural conclusion is that PC itself is not a major determinant of the cup‐like structure. On the contrary, in the absence of PC synthesis, open phagophores accumulate, and mature autophagosomes are lost, implying a direct role for PC in orchestrating the final closure/fission reaction.
Table 1.
Molar percent of detected lipids isolated from Atg8‐positive membranes. PC phosphatidylcholine; PE phosphatidylethanolamine; PS phosphatidylserine; PI phosphatidylinositol; PG phosphatidylglycerol; PA phosphatidic acid; SM sphingomyelin.
Polyansky et al thus contribute to a growing effort to characterize the lipidome of autophagic vesicles (Table I), which are difficult to assess due to their transient nature and numerous associations with other membranes. Phagophores are the open, cup‐shaped intermediate that expand around membranous and proteinaceous cargo until finally closing upon themselves to complete the formation of the autophagosome. Autophagosomes subsequently fuse with the vacuole or lysosome, with the entire process taking only a few minutes to complete. Successful isolation and characterization of pure autophagic membranes therefore requires strong autophagic induction, inhibition of fusion with the degradative compartment, and affinity purification against autophagosome‐associated Atg8 proteins. Polyansky et al performed lipidomics on autophagosomes isolated in this manner from S. Cerevisiae and found that in wild‐type yeast, PC contributes over 80% of the detectable glycerophospholipids. However, their approach did not allow for detection of phosphatidylethanolamine (PE). PC was also present in significant numbers in three additional studies (from 16 to 49% of detectable lipids), which is consistent with the idea that PC plays a specific role in autophagosome closure. In addition, all four studies reported a relatively high level of acyl chain unsaturation. There are some surprising differences; for example, Schutter et al (2020) reported (also in S. Cerevisiae) that PC was roughly equimolar with phosphatidylinositol (PI; 38%), while PI was only a trace lipid in this study. In Drosophila Melanogaster, PE appears to be the dominant lipid in autophagosomes, enriched over three times compared with PC (Laczkó‐Dobos et al, 2021). These discrepancies may reflect differences in the model organism used, the absolute purity of autophagosomes relative to other organelles (e.g., autolysosomes may also co‐purify when autophagosomes are isolated from human and fly cells) or the detection technique (e.g., different lipid species are analyzed in each study). Finally, lipidomics results of isolated autophagosomes could be cargo dependent.
Why do growing phagophores incorporate PS in the absence of PC synthesis as a major phospholipid? One possibility is that PS is tolerated as a PC replacement in the architecture of the phagophore. The phagophore has two major structural domains, the highly curved rim and the sheet‐like main body of the phagophore. The diameter of the phagophore rim approaches the smallest size and the most strident curvature which are theoretically possible for pure lipid membranes (~15 nm in diameter) (Bieber et al, 2022). In these areas of high curvature, conical‐shaped lipids are likely to predominate; here, the cylindrically shaped PC may be largely excluded from the rim to concentrate in the phagophore body. In this context, it is striking that the Opi3‐deficient cells replace PC in the phagophore with PS, another largely cylindrical lipid, thus preserving the distribution of lipid shapes at the expense of an increased negative charge. Alternatively, the enrichment of PC or PS in the phagophore may be a reflection of the donor membrane from which lipids are extracted. The lipid transport protein Atg2 likely bridges contact sites between the phagophore and the endoplasmic reticulum in both yeast and mammals (Wenzel et al, 2022). As the ER is ordinarily PC‐rich, the apparent accumulation of PC in the phagophore may be a consequence of lipid transfer. Indeed, a recent study using a modified choline to allow for tracking of PC in the cell discovered that, upon starvation, virtually all newly synthesized PC in yeast cells is rapidly harvested for phagophore expansion (Orii et al, 2021), strongly implying a tight link between lipid production at the ER and autophagosome growth.
The most intriguing result from Polyansky et al is that PC is essential to close autophagosomes (Fig 1). Autophagosome closure involves the constriction of the rim, probably via the ESCRT machinery (Melia et al, 2020). In mammals, this occurs nearly simultaneously with the insertion of a singular SNARE protein, syntaxin 17, into the autophagosome membrane. PC depletion could negatively affect closure by preventing the recruitment of these key protein factors or by limiting membrane remodeling. It is challenging to distinguish a role for the lost PC versus a potentially inhibitory impact of replacement lipids. The quantitative shift from PC to PS will lead to a significant net negative charge along the phagophore membrane which may increase the bending energy required for closure or interfere with the recruitment of the closure machinery. In addition, knockout of Opi3 results in an accumulation of the PC synthesis intermediate, phosphatidylmonomethylethanolamine (PMME), which has previously been shown to conjugate with Atg8 (Sakakibara et al, 2015). This may hinder autophagy as the conjugate is not readily cleaved by Atg4 (Sakakibara et al, 2015). Importantly, Polyansky et al show that acute choline supplementation is sufficient to reverse all phenotypes by driving PC synthesis via the Kennedy pathway, thereby supporting a specific role for PC. In closing, this exciting report highlights the critical function that specific lipids can play in phagophore formation and opens the door to future studies.
Figure 1. PC is essential to close the autophagosome.
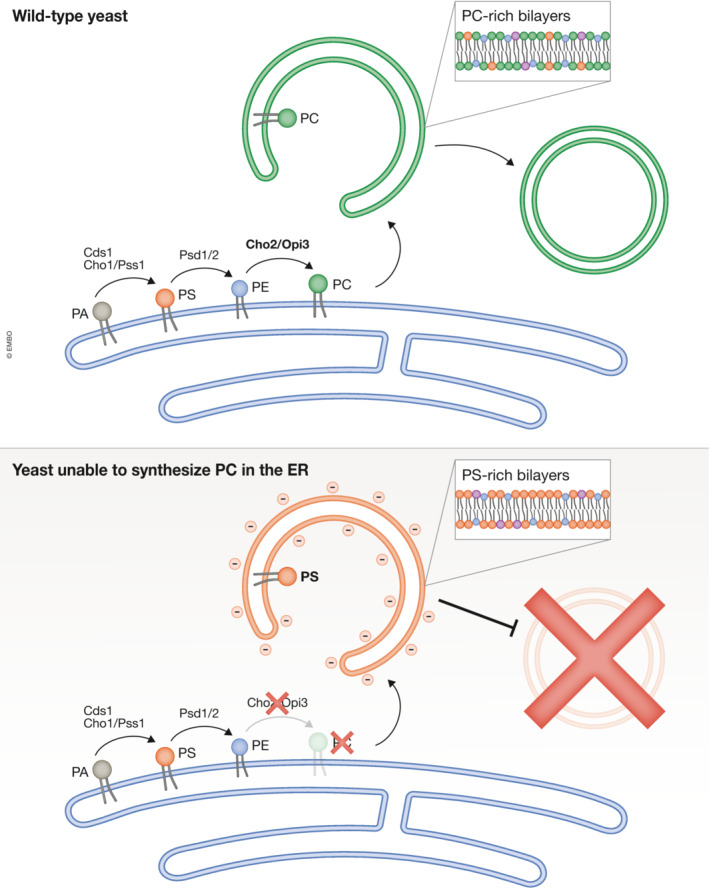
In wild‐type cells, PC is produced in the ER and eventually traffics to the phagophore, where it makes up a large percentage of the autophagosome lipid composition. However, when genes producing key PC synthesis enzymes Cho2 or Opi1 are deleted, PC levels in the yeast and in the purified autophagosomes become undetectable. Under these conditions, phagophore‐like structures still form, but they now harbor a very high percentage of PS lipids. These PS‐rich phagophores accumulate in the cell, but do not close, thereby blocking autophagy.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 GM100930 to T.J. Melia; F31 AG079606 to D.M. Fuller).
The EMBO Journal (2023) 42: e113046
See also A Polyansky et al (December 2022)
References
- Bieber A, Capitanio C, Erdmann PS, Fiedler F, Beck F, Lee C‐W, Li D, Hummer G, Schulman BA, Baumeister W et al (2022) In situ structural analysis reveals membrane shape transitions during autophagosome formation. Proc Natl Acad Sci USA 119: e2209823119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczkó‐Dobos H, Maddali AK, Jipa A, Bhattacharjee A, Végh AG, Juhász G (2021) Lipid profiles of autophagic structures isolated from wild type and Atg2 mutant Drosophila . Biochim Biophys Acta Mol Cell Biol Lipids 1866: 158868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia TJ, Lystad AH, Simonsen A (2020) Autophagosome biogenesis: from membrane growth to closure. J Cell Biol 219: e202002085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orii M, Tsuji T, Ogasawara Y, Fujimoto T (2021) Transmembrane phospholipid translocation mediated by Atg9 is involved in autophagosome formation. J Cell Biol 220: e202009194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyansky A, Shatz O, Fraiberg M, Shimoni E, Dadosh T, Mari M, Reggiori FM, Qin C, Han X, Elazar Z (2022) Phospholipid imbalance impairs autophagosome completion. EMBO J 41: e110771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara K, Eiyama A, Suzuki SW, Sakoh‐Nakatogawa M, Okumura N, Tani M, Hashimoto A, Nagumo S, Kondo‐Okamoto N, Kondo‐Kakuta C et al (2015) Phospholipid methylation controls Atg32‐mediated mitophagy and Atg8 recycling. EMBO J 34: 2703–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt D, Bozkurt S, Henning‐Domres P, Huesmann H, Eimer S, Bindila L, Behrends C, Boyle E, Wilfling F, Tascher G et al (2022) Lipid and protein content profiling of isolated native autophagic vesicles. EMBO Rep 23: e53065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutter M, Giavalisco P, Brodesser S, Graef M (2020) Local fatty acid channeling into phospholipid synthesis drives Phagophore expansion during autophagy. Cell 180: 135–149.e14 [DOI] [PubMed] [Google Scholar]
- Wenzel EM, Elfmark LA, Stenmark H, Raiborg C (2022) ER as master regulator of membrane trafficking and organelle function. J Cell Biol 221: e202205135 [DOI] [PMC free article] [PubMed] [Google Scholar]


