Abstract
Nicotinic acetylcholine receptors (CHRNs) expression and their critical role in various types of cancer have been reported. However, it is still unclear which CHRNs and their associated genes play essential roles in metastasis in melanoma patients. Here, we performed bioinformatics analyses on publicly available bulk RNA sequencing (RNA-seq) data of patients with melanoma to identify the CHRNs highly expressed in metastatic melanoma. We found that CHRNA1 was highly expressed in metastatic melanoma samples compared to primary melanoma samples and was strongly associated with CHRNB1 and CHRNG. These muscle-type CHRNs (CHRNA1, CHRNB1, and CHRNG) were correlated with the ZEB1 and Rho/ROCK pathway-related genes in metastatic melanoma samples. Pairwise correlations and enrichment analyses revealed that CHRNA1 was significantly associated with myogenesis/muscle contraction and cell cycle genes. Kaplan-Meier curves illustrated the involvement of CHRNA1, four of its correlated genes (DES, FLNC, CDK1, and CDC20), and the myogenesis gene signature in the prognosis of melanoma patients. Following the bulk RNA-seq analysis, single-cell RNA-seq (scRNA-seq) analysis showed that the CHRNA1-expressing melanoma cells are primarily metastatic and had high expression levels of CHRNB1, CHRNG, and myogenesis/cell cycle-related genes. Our bioinformatics analyses of the bulk RNA-seq and scRNA-seq data of patients with melanoma revealed that CHRNA1 and its correlated myogenesis/cell-related cycle genes are critical prognosis-related markers of metastatic melanoma.
Keywords: CHRNA1, Melanoma, Metastasis, Myogenesis, Prognosis, scRNA-seq, TCGA
Highlights
-
•
CHRNA1 is highly expressed in patients with metastatic melanoma compared to primary melanomas.
-
•
CHRNA1 expression is highly correlated to CHRNB1 and CHRNG in metastatic melanoma samples.
-
•
The muscle-type CHRNs (CHRNA1, CHRNB1, and CHRNG) are associated with the ZEB1 EMT-TF and Rho/ROCK-related genes.
-
•
CHRNA1 is associated with myogenesis/cell cycle-related genes in TCGA-SKCM samples and CHRNA1-expressing melanoma cells.
-
•
CHRNA1 and its correlated genes are involved in melanoma patients' survival.
1. Introduction
Melanoma, caused by the malignant transformation of melanocytes, is a well-known severe type of skin cancer because it is highly invasive and metastatic [1,2]. Although melanoma is not an epithelial tumor, in the early stage of invasion, melanoma cells have been reported to undergo epithelial-mesenchymal transition (EMT)-like phenotype switching, which is orchestrated by EMT-inducing transcription factors (EMT-TFs), such as ZEB, TWIST, and SNAIL family proteins [3,4]. Melanoma phenotype switching by the transition to a dedifferentiated state has been thought to rarely cause various divergent differentiations, including rhabdomyosarcomatous differentiation, which expresses striated muscle genes at the metastatic sites [5]. After breaking the basement membrane, melanoma cells change their morphology and adopt three different migration modes (single cell, streaming, and collective modes) according to the surrounding environment [6,7]. The invasive front of melanoma is characterized by amoeboid movement, which is dependent on the contractility of actomyosin maintained by the Ras homolog gene family (RHO)/Rho-associated kinase (ROCK) and JAK/STAT3 signaling pathways [6,7]. Many signaling pathways, such as TGF-β, Wnt, MAPK, and PI3K-AKT, have been reported to regulate tumor invasion and metastasis, including melanoma [1,8].
Acetylcholine (ACh), the first identified neurotransmitter, is known to regulate essential cell functions, such as cell differentiation, proliferation, and migration, as a signaling molecule in non-neuronal cells [9]. ACh signaling is transmitted through two types of ACh receptors: cholinergic receptor nicotinic α, β, γ, δ, and ε subunits (CHRNs), which are ligand-gated cation channels [10], and cholinergic receptor muscarinic M (1–5) (CHRMs), which are G protein-coupled receptors [11]. Functional analysis of CHRMs in melanoma progression is limited to M3 [12], whereas the expression and detailed functional analysis of α5 and α9 CHRNs in melanoma cell lines have been reported [13]. CHRNs are classified into neuronal (α2 to α10 and β2 to β4) and muscular (α1, β1, γ, δ, and ε) receptors expressed in humans except for α8 [14,15]. Previously, CHRNs were thought to function only in the nervous system and at the neuromuscular junction; however, recent studies have reported that CHRNs and ACh are expressed in almost all types of cells, including cancer cells [9,10,15]. Since their first detection in lung cancer cells, CHRNs have been proven to be involved in tumor growth, metastasis, angiogenesis, and EMT [10,15]. Overexpression of CHRNA9 induces breast cancer growth and metastasis through EMT [16]. CHRNA7 stimulates lung cancer cell proliferation and invasion through the Ras/ERK/MAPK and JAK2/STAT/PI3K pathways [17], and the knockdown of CHRNA5 inhibits the proliferation and invasion of the lung [18] and prostate cancers [19]. In melanoma cells, overexpression of CHRNA5 promotes melanoma progression via Notch1, whereas knockdown of this receptor inhibits melanoma cell proliferation and invasion, as does inhibition of the PI3K/AKT and ERK1/2 signaling pathways [20]. CHRNA9 stimulates melanoma cell proliferation and migration through the AKT and ERK signaling pathways [21]. While previous studies using cell culture systems have shown that specific CHRNs are crucial for the process of metastasis, a comprehensive bioinformatics analysis of which types of CHRNs play essential roles in metastatic melanoma has not yet been performed.
Rapid progress in sequencing technology has made it possible to analyze gene expression at the whole cancer cell or the single-cell level by using gene expression information from individual patients [22]. The Cancer Genome Atlas (TCGA), the world's largest publicly available database, provides genomic information, RNA sequencing (RNA-seq) data, and various clinical outcomes for over 30 human cancers (https://portal.gdc.cancer.gov/). In addition, recent studies using single-cell RNA sequencing (scRNA-seq) have demonstrated tumor heterogeneity, therapy resistance, and tumor and immune ecosystems in human patients [[23], [24], [25], [26], [27]]. Therefore, data-driven analyses that integrate bulk RNA-seq and scRNA-seq data are expected to discover new mechanisms in cancer. In this study, we performed combinational bioinformatics analyses for bulk RNA-seq and scRNA-seq data of patients with melanoma. We identified CHRNA1 as the highest differentially expressed CHRNs in metastatic compared to primary melanoma patients obtained from the TCGA and GSE65904 datasets. In patients with metastatic melanoma, CHRNA1 was significantly correlated to ZEB1 EMT-TF, RHO/ROCK genes, and myogenesis/cell cycle-related genes. Survival analysis proved CHRNA1 and its correlated genes as prognosis-related markers of metastatic melanoma. Additionally, single melanoma cells expressing CHRNA1 were derived from metastatic sites and were enriched with myogenesis and cell cycle-related gene signatures.
2. Material and methods
2.1. Data collection
We used the R software (4.1.2) to perform all the bioinformatics analyses in this study. We retrieved the RNA-seq data and clinical characteristics of skin cutaneous melanoma (SKCM) patients from the TCGA database using the TCGABiolinks package (2.22.2) [28]. TCGA-SKCM samples included 103 primary melanoma and 367 metastatic melanoma samples. Another bulk RNA-seq dataset of patients with melanoma was obtained from the Gene Expression Omnibus (GEO) database (GSE65904) using the GEOquery package (2.62.2) [29]. There were 16 primary and 188 metastatic melanoma samples in the GSE65904 dataset. Furthermore, we used two melanoma scRNA-seq datasets (GSE115978 and GSE116237) to assign the characteristics of melanoma cells that expressed CHRNA1. Transcriptomic data of four samples of rhabdomyosarcomatous melanoma were collected from a previous study [5].
2.2. Determination of the differentially expressed genes (DEGs)
TCGA-SKCM RNA-seq data in HTSeq-Counts format were used for the DEG analysis. DEGs between the metastatic and the primary TCGA-SKCM samples were identified using the edgeR package (3.36.0) [30]. The trimmed mean of M-values (TMM) method was used to normalize the HTSeq-count matrix. We considered log2FC > 1.5 and false discovery rate (FDR) < 0.05, to obtain significant DEGs. The ENSEMBL gene names were converted into gene symbols using the biomaRt package (v 2.50.2) [31].
2.3. Pairwise correlations
Pearson's r correlation test was performed to evaluate the correlations between the expression levels of the different CHRNs in TCGA-SKCM and GSE65904 datasets. Correlation matrices were visualized using the psych package (2.1.9) (https://cran.r-project.org/web/packages/psych/index.html). Additionally, pairwise correlations between muscle-type CHRNs expressions and ZEB1 EMT-TF and Rho/ROCK pathway-related genes (RHOA, RHOB, RHOC, ROCK1, and ROCK2) expressions in metastatic melanoma samples were analyzed. We considered p-value <0.05 and (r ≤ −0.1 |r ≥ 0.1) for significant correlations. We identified the genes correlated with CHRNA1 in TCGA-SKCM patients under p-value <0.05 and r ≥ 0.3.
2.4. Enrichment analysis
Genes significantly correlated with CHRNA1 (p-value <0.05 and r ≥ 0.3) were used for enrichment analysis. Gene Ontology (GO) enrichment analysis (Biological process) was performed using the enrichR (3.0) interface of the Enrichr database [32]. To perform gene set enrichment analysis (GSEA) of cancer hallmarks, we used the fgsea package (1.20.0) [33]. A line enrichment plot for the HALLMARK_MYOGENESIS pathway was visualized using the plotEnrichment function.
For protein-protein interaction (PPI) analysis, CHRNA1-correlated genes were analyzed using Cytoscape software (v 3.9.1) [34]. We used the built-in StringApp [35] to construct a PPI network and reclaim the functional enrichment of the CHRNA1-correlated genes. The densely interconnected regions within our PPI network were clustered using the molecular complex detection (MCODE) algorithm [36], and four PPI subnetworks (node >3) were identified.
2.5. Survival analysis
We employed the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) to investigate the correlation of CHRNA1 to the survival of patients with metastatic melanoma using the GSE19234 dataset and scan cutoff mode. Genes included in the MCODE PPI subnetworks were explored for their relationships with survival in melanoma patients. Overall survival analyses based on the median expression cutoff were performed using the Gene Expression Profiling Interactive Analysis (GEPIA) tool [37]. Kaplan-Meier survival curves were constructed for the chosen genes, and hazard ratios (HRs) based on the Cox proportional-hazards (PH) model and log-rank p-values were calculated. A p-value <0.05 was considered significant. TCGA-SKCM samples were classified into five groups based on the stage of the melanoma (Stages 0, I, II, III, and IV). To create a survival object, we used the Surv function from the Survival package (3.2–13) (https://cran.r-project.org/web/packages/survival/index.html). Then, the survival curve was fitted separately by melanoma stages using the survfit function. The log-rank test was used to examine the survival distribution of the melanoma stage groups, and a p-value <0.05 was considered significant. Finally, the Kaplan-Meier curve was visualized using the ggsurvplot function from the Survminer package (0.4.9) (https://cran.r-project.org/web/packages/survminer/index.html).
2.6. The scRNA-seq analysis
The GSE115978 scRNA-seq dataset contained 2018 malignant cells out of 7186 cells from 31 patients with melanoma [27]. Only the count matrices of melanoma cells expressing CHRNA1 were isolated and analyzed using the Seurat package (4.0.4) [38]. We filtered cells with fewer than 200 genes and mitochondrial counts >5%. Cells of different patients were integrated using canonical correlation analysis, normalized using the LogNormalize method, and scaled. Principal component analysis (PCA) was performed using ten ncps and cells clustered with 0.4 resolution. Two clusters were recognized and represented using UMAP (dims = 1:10). We used the Wilcoxon rank-sum test to identify marker genes of the two recognized scRNA clusters. To perform the GSEA of cancer hallmarks, we used the GSVA package (1.42.0) [39], and the pheatmap package (1.0.12) (https://cran.r-project.org/web/packages/pheatmap/index.html) was used to visualize the results. The cell cycle phases of each cluster's cells were addressed based on G2/M and S markers expressions using the CellCycleScoring function [25]. For validation, an extra scRNA-seq dataset (GSE116237) was analyzed. The GSE116237 dataset comprised 674 cells from patient-derived xenograft (PDX) models [26]. We followed the default protocol of the Seurat package, and cells with fewer than 200 genes and mitochondrial counts >5% were excluded. We identified six clusters of melanoma cells at 0.3 resolution. The cell clusters were visualized using UMAP (dims = 1:10).
2.7. Profiling of the muscle-related genes in rhabdomyosarcomatous melanoma and TCGA-SKCM samples
We merged the HTSeq-FPKM expressions of the TCGA-SKCM samples and the transcripts per kilobase million (TPM) expressions of the rhabdomyosarcomatous melanoma samples into one matrix after trimming the unshared genes. High and low CHRNA1-expressing TCGA samples were identified based on the median expression cutoff value of the CHRNA1. The log2 values were calculated to unify the scale of the merged expression matrix. Then the expressions of the muscle-related genes were compared and visualized using the ComplexHeatmap package (2.10.0) [40].
3. Results
3.1. CHRNA1 showed higher expression levels in metastatic than primary melanomas
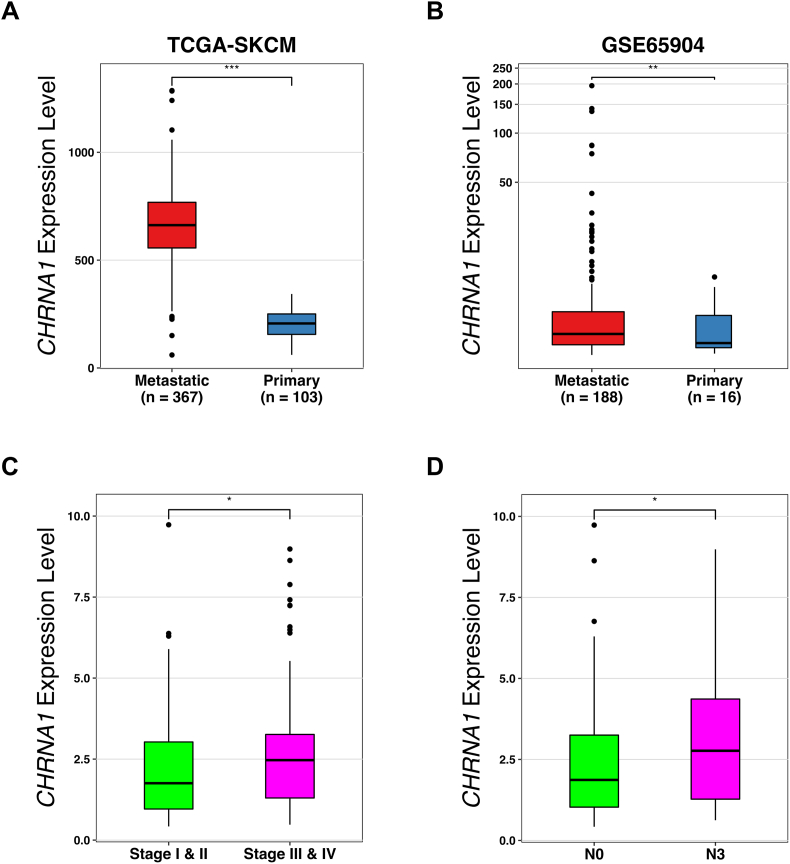
To investigate the expression of CHRNs and their roles in melanoma metastasis, we performed comprehensive computational analyses on human melanoma samples obtained from publicly available databases. Differential gene expression analysis between 367 metastatic and 103 primary melanoma samples obtained from the TCGA-SKCM database identified 1382 DEGs with log2FC > 1.5 and FDR <0.05. We detected 12 of the 16 CHRNs among the DEGs (Table 1). Interestingly, CHRNA1 showed the highest difference in expression between metastatic and primary melanomas with log2FC > 1.5 (p-value = 1.07 × 10−5) (Table 1 and Fig. 1A). We further found in another independent dataset (GSE65904) that CHRNA1 expression in metastatic melanoma was significantly higher than that in primary melanoma (p-value = 0.0065) (Fig. 1B). In addition, melanoma patients with high CHRNA1 expression were significantly correlated with late melanoma stages (stages Ⅲ and Ⅳ) (p-value = 0.024) (Fig. 1C) and with a high rate of spread to lymph nodes (N3) (p-value = 0.038) (Fig. 1D). Altogether, our results reveal that muscle-type CHRNA1 is associated with metastasis of human melanoma.
Table 1.
Differentially expressed CHRNs in metastatic vs. primary TCGA-SKCM samples.
| CHRNs | log2FC | logCPM | p-value |
|---|---|---|---|
| CHRNA1 | 1.557 | 3.299 E+00 | 1.070E-05 |
| CHRNA3 | 0.867 | −2.122E-01 | 3.380E-04 |
| CHRNA5 | 0.133 | 1.719 E+00 | 2.771E-01 |
| CHRNA6 | 1.033 | 1.865 E+00 | 1.495E-03 |
| CHRNA7 | 0.126 | −1.026 E+00 | 6.064E-01 |
| CHRNA9 | 0.422 | −3.998E-02 | 2.948E-01 |
| CHRNA10 | −0.653 | −5.548E-01 | 3.360E-08 |
| CHRNB1 | −0.229 | 3.180 E+00 | 9.781E-03 |
| CHRNB2 | −0.615 | 2.237 E+00 | 1.982E-02 |
| CHRNB4 | 0.271 | −5.207E-01 | 3.054E-01 |
| CHRND | 1.189 | −1.708 E+00 | 5.634E-04 |
| CHRNE | −0.187 | 1.199 E+00 | 3.396E-01 |
Fig. 1.
CHRNA1 expression level is higher in metastatic versus primary melanomas. (A, B) Box plots for the CHRNA1 differential expression between metastatic and primary melanomas from TCGA-SKCM (A) and GSE65904 datasets (B). (C, D) Box plots for TCGA-SKCM patients with high CHRNA1 expression levels (expression > median value) grouped by pathological stages (C) and N classification (D). * p-value <0.05, ** p-value <0.01, and *** p-value <0.001 according to Student t-Tests. N = tumor spread degree to lymph nodes.
3.2. Correlations of muscle-type CHRNs with EMT-TFs- and Rho/ROCK pathway-related genes in melanoma patients
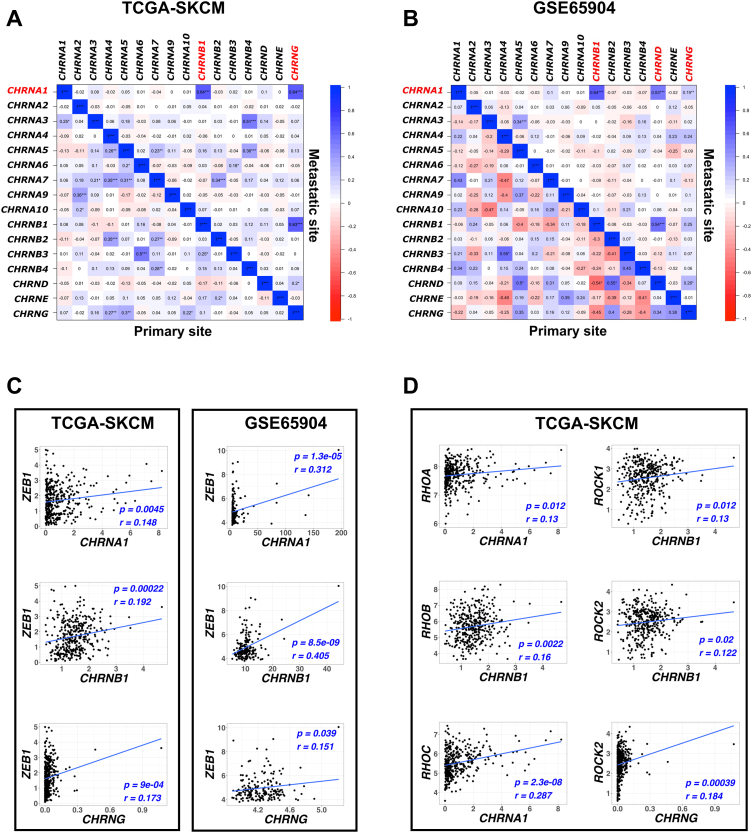
To determine which CHRNs are associated with CHRNA1 in melanoma patients, Pearson's correlation test was applied to TCGA-SKCM and GSE65904 samples (Fig. 2). We found that both CHRNB1 and CHRNG were significantly correlated with CHRNA1 (p-value <0.001) in metastatic melanoma samples (Fig. 2A and B). To further investigate the involvement of these muscle-type CHRNs (CHRNA1, CHRNB1, and CHRNG) in human melanoma metastasis, we analyzed the pairwise correlations between these CHRNs and EMT-TFs expressions in patients with metastatic melanoma. We found that the expressions of the three CHRNs were positively correlated with ZEB1 expression in both TCGA-SKCM and GSE65904 samples (Fig. 2C). Besides, we examined the correlation between the three CHRNs and the Rho GTPase (RHOA, RHOB, and RHOC)/ROCK (ROCK1 and ROCK2) genes in metastatic melanoma samples to determine their involvement in melanoma movement. Results showed that some RHO/ROCK genes were correlated with individual CHRNs (CHRNA1, CHRNB1, and CHRNG were correlated with RHOA/RHOC, RHOB/ROCK1/ROCK2, and ROCK2, respectively) (Fig. 2D). These findings explain the crucial role of CHRNA1, CHRNB1, and CHRNG in melanoma movement metastasis.
Fig. 2.
CHRNA1 highly correlates to CHRNB1 and CHRNG, and these muscle-type CHRNs correlate with ZEB1 EMT-TF and Rho/ROCK pathway-related genes in metastatic melanoma patients. (A, B) Correlation matrices (Pearson's correlation test r ≤ −0.1 |r ≥ 0.1) for CHRNs mRNA expressions in metastatic and primary TCGA-SKCM samples (A) and GSE65904 samples (B). * p-value <0.05, ** p-value <0.01, and *** p-value <0.001 according to Student t-Tests. (C, D) Pairwise scatter plots for the correlation between the expressions of muscle-type CHRNs and ZEB1 (C) and RHO/ROCK genes expressions (D). Significant correlation based on p-value <0.05 and Pearson's correlation test (r ≤ −0.1 |r ≥ 0.1). A blue-colored line representing the linear regression for best curve fitting was drawn. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. GO, GSEA, and PPI analyses of CHRNA1-correlated genes in melanoma
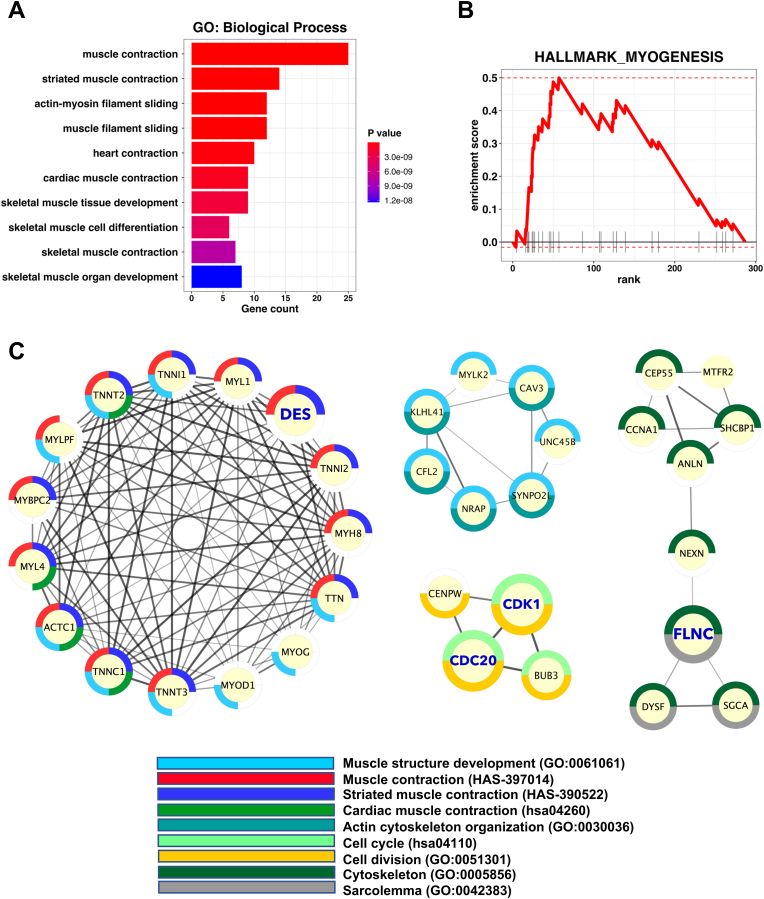
Genes positively correlated with CHRNA1 were identified under r ≥ 0.3 and p-value <0.05. The identified CHRNA1-correlated genes were presented for enrichment analyses to investigate their biological functions (Fig. 3). GO enrichment (Biological process) analysis showed that genes correlated with CHRNA1 were mainly associated with muscle and heart contraction (p-value <0.05) (Fig. 3A). Furthermore, the GSEA of cancer hallmarks confirmed the association with myogenesis (p-value <0.05) (Fig. 3B). Next, we performed PPI analysis for CHRNA1-correlated genes to explore their functional interactions. Further, we applied the MCODE algorithm to determine the highly connected PPIs (Fig. 3C). Consistent with GO enrichment and GSEA results, the top four MCODE PPI networks were muscle contraction/development, muscle development, cytoskeleton/sarcolemma, and cell cycle-related networks (Fig. 3C). These results revealed that the expression of CHRNA1 in melanoma patients is correlated with genes related to myogenesis, muscle contraction, and cell cycle.
Fig. 3.
Enrichment analysis for genes significantly correlated to CHRNA1 in metastatic TCGA-SKCM patients. (A) Bar plot showing GO enrichment (Biological Process) analyses for CHRNA1-correlated genes (r ≥ 0.3 and p-value <0.05). (B) Line enrichment plot for the GSEA of HALLMARK_MYOGENESIS. (C) MCODE densely connected PPI clusters with enrichment retrieved by StringApp.
3.4. CHRNA1, its correlated genes, and myogenesis signature were correlated to melanoma prognosis
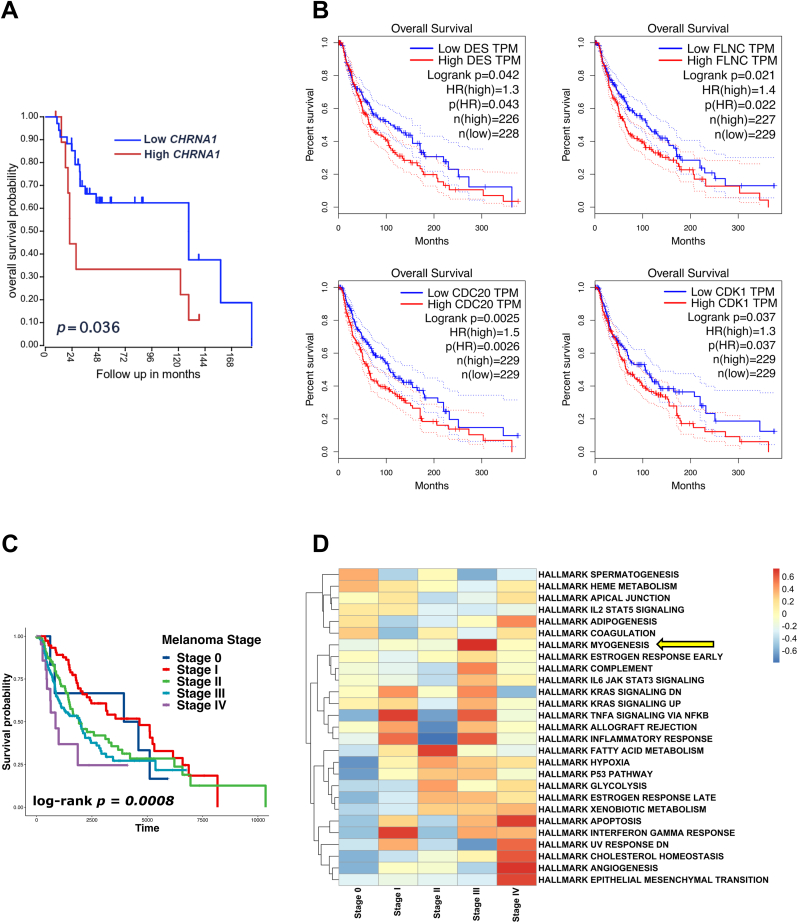
Kaplan-Meier analysis for patients with metastatic melanoma using the R2 online platform revealed that patients with high expression of CHRNA1 had a poorer prognosis than patients with low CHRNA1 expression (p-value = 0.036) (Fig. 4A). Additionally, Among the CHRNA1-correlated genes in the MCODE PPI networks, we found that DES, CDK1, CDC20, and FLNC were prognosis-related genes (log-rank p-value <0.05) (Fig. 4B). Since patients with high CHRNA1 expression were associated with late-stage melanoma (Fig. 1C), we investigated the correlation between melanoma stage and patient survival. Patients with melanoma stages III and IV had a poorer prognosis than patients with melanoma at the early stages (0, I, and II) (log-rank p-value = 0.0008) (Fig. 4C). By comparing the gene signature of every melanoma stage, we found that late-stage (III and IV) melanomas were enriched with myogenesis, angiogenesis, and EMT characteristics (Fig. 4D).
Fig. 4.
CHRNA1 and its correlated genes are linked to prognosis in melanoma. (A) Kaplan-Meier survival curve for CHRNA1 in metastatic melanoma patients based on the R2 platform (http://r2.amc.nl) analysis. (B) Kaplan-Meier survival curves for genes derived from the highly interconnected (CHRNA1-correlated genes) MCODE PPIs. (C) Kaplan-Meier survival curve for TCGA-SKCM patients within different melanoma stage groups. (D) Heatmap showing the GSEA of cancer hallmarks among the different melanoma stage groups. Yellow arrow pointing to the hallmark myogenesis enrichment. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
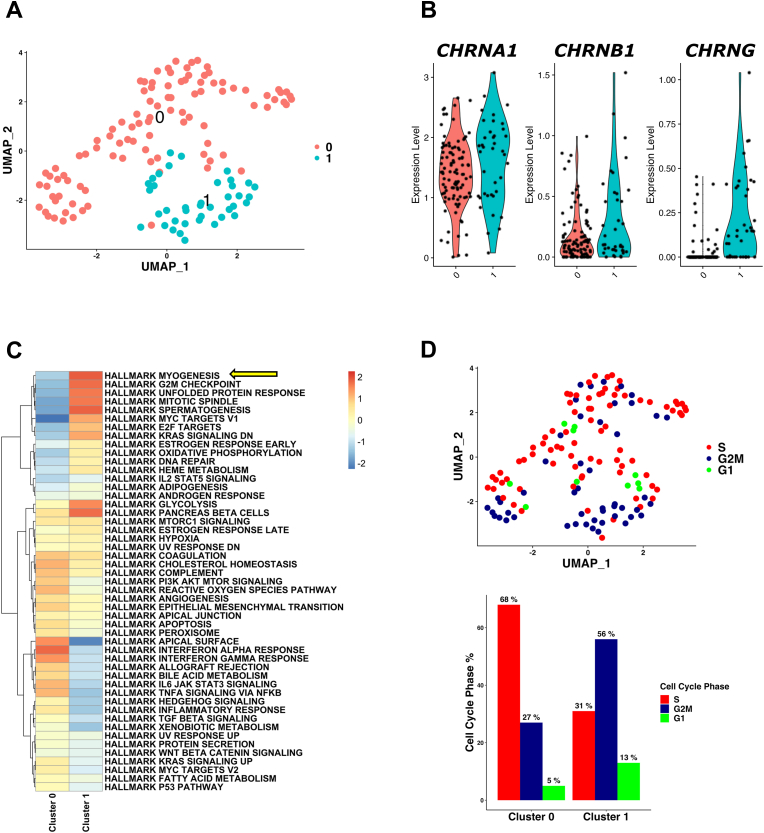
3.5. Analysis of CHRNA1-expressing melanoma cells using scRNA-seq
Using bulk RNA-seq data, CHRNA1 expression was shown to be correlated with the expressions of myogenesis- and cell cycle-related genes. We next investigated the characteristics of melanoma cells expressing CHRNA1 at the single-cell level. We re-analyzed the publicly available scRNA-seq dataset (GSE115978) of patients with melanoma (Fig. 5 and Fig. S1). First, we isolated 138 melanoma cells that expressed CHRNA1. The CHRNA1-expressing melanoma cells were represented in two clusters (clusters 0 and 1) (Fig. 5A). These cells were derived from 12 melanoma patients (Fig. S1A) and were mainly metastatic cells (Fig. S1B). The expressions of the top 10 marker genes of each scRNA cluster confirmed that these two clusters (clusters 0 and 1) had different gene expression profiles (Fig. S1C). The muscle-type CHRNs (CHRNA1, CHRNB1, and CHRNG) showed higher expressions in cluster 1 than in cluster 0 (Fig. 5B). Additionally, GSEA of cancer hallmarks revealed that cluster 1 cells were highly enriched with myogenesis, G2M checkpoint, and mitotic spindle (Fig. 5C). In line with these results, 56% of cluster 1 cells were in the G2M cell cycle phase (Fig. 5D). These results suggest that CHRNA1 expression correlates with the expression of myogenesis- and cell cycle-related genes. Similar results were obtained by analyzing another scRNA-seq dataset (GSE116237) (Fig. S2). One cluster of melanoma cells (cluster 5) showed the highest expression level of CHRNA1 (Fig. S2B), and this cluster was enriched with myogenesis, EMT, and angiogenesis signatures (Fig. S2C).
Fig. 5.
Melanoma cells expressing CHRNA1 at the scRNA-seq level had a myogenesis gene signature. (A) UMAP plot representing clusters (0 and 1) of single cells from the GSE115978 scRNA-seq dataset. (B) Violin plots for CHRNA1, CHRNB1, and CHRNG expressions in the scRNA clusters. (C) Heatmap demonstrates the characteristic GSEA of cancer hallmarks for each scRNA cluster. Yellow arrow pointing to the hallmark myogenesis enrichment. (D) UMAP plot (upper) of the cell cycle phases (S, G2M, and G1) and their percentages (lower bar plot) across the scRNA clusters. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Comparison of muscle-related gene expression between CHRNA1-expressing melanoma and rhabdomyosarcomatous melanoma
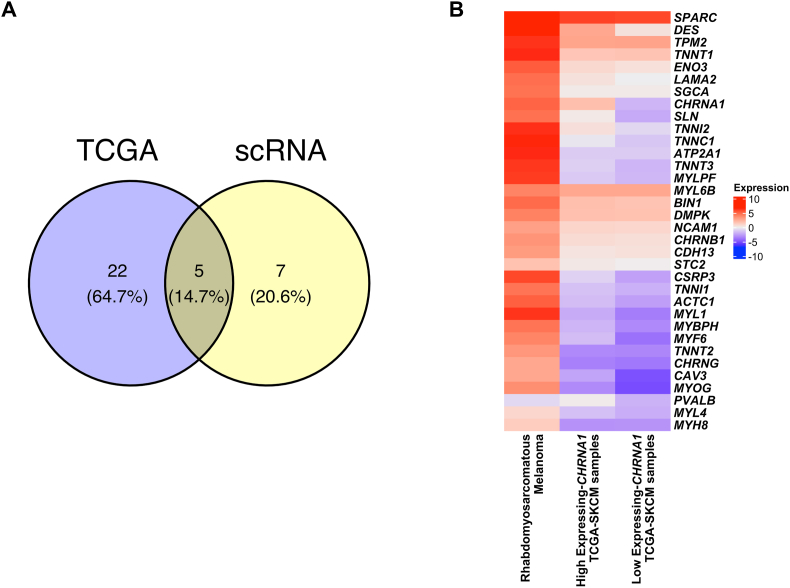
Previous studies have revealed that melanoma can change into various differentiation states, owing to its high cellular plasticity. One of these differentiated states has been reported to be rhabdomyosarcomatous melanoma, which has been shown to express many muscle-related genes. To determine whether melanomas with high expression of CHRNA1 have similar gene expression profiles to rhabdomyosarcomatous melanoma, we compared the expressions of the identified muscle-related genes that correlate with CHRNA1 in both TCGA-SKCM samples and rhabdomyosarcomatous melanoma samples (Fig. 6). 27 and 12 muscle-related genes were identified from TCGA-SKCM and scRNA-seq data as CHRNA1-correlated genes, respectively, of which only five genes were overlapped (Fig. 6A). We compared the expression of the 34 muscle-related genes between rhabdomyosarcomatous melanoma and high and low CHRNA1-expressing TCGA-SKCM samples (Fig. 6B). We found that the expression levels of almost all the muscle CHRNA1-correlated genes were lower in melanoma than that in rhabdomyosarcomatous melanoma samples (Fig. 6B). Moreover, some muscle CHRNA1-correlated genes (DES, SLN, TNNI2, and TNNC1) showed higher expression in the high CHRNA1-expressing TCGA-SKCM samples than in the samples with low expression of CHRNA1 (Fig. 6B). These results indicate that CHRNA1, together with the following genes (DES, SLN, TNNI2, and TNNC1), may function in the melanoma differentiation process to the rhabdomyosarcomatous melanoma state.
Fig. 6.
Melanoma samples with high CHRNA1 expression have a profile quite similar to patients with rhabdomyosarcomatous melanoma. (A) Venn diagram represents the muscle-related genes enriched in samples with CHRNA1 high expression from TCGA-SKCM and the GSE115978 scRNA-seq datasets. (B) Heatmap showing the expression of the muscle-related genes in high & low CHRNA1-expressing TCGA-SKCM samples and the rhabdomyosarcomatous melanoma samples.
4. Discussion
Nicotine, the primary component in tobacco, causes health problems and is well known for its malignant effects on various cancers [10]. Nicotine has been reported to act on cancer cells mainly through CHRNs. Previous studies using cancer cell lines have shown that nicotine treatment promotes the malignant transformation of tumors while blocking the expression of CHRNs or antagonist treatment results in the inhibition of tumor progression [10,[17], [18], [19], [20], [21]]. Recently, CHRNA5 [20] and CHRNA9 [21] were implicated in melanoma progression and migration in melanoma cell lines. In this study, to analyze the type and role of CHRNs in melanoma metastasis, we used in silico analysis of human melanoma samples obtained from TCGA-SKCM and GSE65904 datasets. By analyzing these samples, we found that among the 16 CHRNs expressed in humans, CHRNA1, but not CHRNA5 and CHRNA9, showed the highest differential expression in metastatic melanoma samples. Although overexpression of CHRNA1 has been reported in early-stage lung adenocarcinoma [41,42], head and neck squamous cell carcinoma (SCC) [43], and glioblastoma multiforme (GBM) tumors [44], our study is the first to confirm its high expression in metastatic melanoma.
Phenotype switching, an EMT-like process, has been described in melanoma, driven by EMT-TFs [3,4]. In the current study, CHRNA1 was significantly correlated with CHRNB1 and CHRNG in metastatic melanoma samples. Furthermore, we found that the expressions of CHRNA1, CHRNB1, and CHRNG were significantly correlated with the expression of ZEB1 EMT-TF. ZEB1 has been reported to be a critical regulator for melanoma phenotype switching [3,45]. ZEB1 showed increased expression during melanoma progression, and its overexpression was correlated with poor prognosis and therapy resistance in melanoma patients [[45], [46], [47], [48]]. On the other hand, the knockdown of ZEB1 decreased the invasiveness and inhibited the growth of melanoma cells [45,46]. Another interesting observation in our research was the significant correlation between the expressions of these muscle-type CHRNs and Rho/ROCK pathway-related genes, which play a crucial role in tumor migration [6]. Previous studies have revealed that CHRNA9 and mouse chrna7 regulate TWIST1 in breast cancer cell lines and RhoA activation in neural cell lines, respectively [16,49]. Our results suggest that muscle-type CHRNs may be directly involved in melanoma metastasis via regulation of ZEB1 EMT-TF expression and Rho/ROCK pathway activation.
Enrichment and pathway analyses of genes correlated with CHRNA1 showed that these genes are involved in myogenesis, muscle contraction, and cell proliferation. CHRNA1 and four correlated genes (DES, CDK1, CDC20, and FLNC) were associated with poor prognosis in metastatic melanoma patients. Recurrence-free and overall survival analyses revealed that high expression of CHRNA1 was correlated with reduced survival in patients with lung adenocarcinoma [41] and glioblastoma [44]. DES gene encodes Desmin, an intermediate filament muscle-specific protein functioning in several types of cancer [5,[50], [51], [52], [53]]. High Desmin expression has been reported in rhabdomyosarcomatous melanoma [5] and colorectal tumors [51,53]. In contrast, patients with gallbladder cancer have lower expression levels of Desmin in cancerous tissues compared to non-tumor tissues owing to epigenetic dysregulation [52]. Consistent with our findings, overexpression of Desmin was associated with poor prognosis [51] and advanced stages [53] in patients with colorectal cancer. Upregulation of CDK1 [54] and CDC20 [55] genes has been reported in various cancers and linked to poor prognosis [56]. FLNC gene encodes Filamin-C, an actin-binding protein involved in the metastasis cancer cells [57]. Overexpression of FLNC was associated with progression, metastasis, and poor prognosis in hepatocellular carcinoma [58], esophageal squamous cell carcinoma [59], and glioblastoma [57]. Consistent with the bulk RNA-seq outputs, using scRNA-seq analysis, we found that myogenesis and cell proliferation gene signatures were enriched in cells expressing CHRNA1.
Accumulating evidence from recent experiments has shown increased expression of sarcomeric proteins, such as Myosin, Titin, MyBP-C, Obscurin, ACTN, Nebulin, Synemin, Desmin, Plectin, Nesprin, and Vimentin, in various cancers, and their overexpression has been reported to be associated with the malignant transformation of cancer [50]. In normal cells of the body, these proteins function in skeletal and cardiac muscles to contract using sarcomere structures, whereas, in cancer cells, they are thought to be involved in invasion and migration for metastasis. In our study, the CHRNA1-correlated myogenesis/sarcomere genes, including MYOG, MYH8, and DES, were highly expressed in metastatic melanoma, similar to rhabdomyosarcomatous melanoma. These results suggested that their expression may be involved in melanoma malignancy. In the future, the significance of CHRNA1 and sarcomeric gene expression in the malignant transformation of melanoma should be further investigated. Our analysis demonstrated the involvement of the CHRNA1 and its correlated myogenesis/cell cycle-related genes in melanoma metastasis and prognosis. However, further experimental confirmation of the role of CHRNA1 expression in melanoma progression should be carried out. For instance, CHRNA1 knockout or knockdown experiments in vivo or in vitro could be conducted. Immunostaining of clinical tissue samples and real-time PCR (RT-qPCR) studies would be advantageous to confirm both the CHRNA1 expression as well as the association between CHRNA1 and identified correlated genes in patients with melanoma.
5. Conclusion
In conclusion, our bioinformatics analysis of melanoma patients revealed that CHRNA1 was highly expressed in metastatic melanoma and was correlated with CHRNB1 and CHRNG. Moreover, the expression of muscle-type CHRNs (CHRNA1, CHRNB1, and CHRNG) was significantly correlated with ZEB1 EMT-TF and Rho/ROCK pathway-related genes. The gene profile of CHRNA1-expressing metastatic melanoma patients and single melanoma cells showed a high enrichment of myogenesis/cell cycle-related genes. CHRNA1, its correlated genes, and myogenesis signature correlated with prognosis in melanoma patients. Our results reveal that CHRNA1 may function in melanoma metastasis and, together with its correlated myogenesis/cell cycle genes, are critical prognosis-related markers of metastatic melanoma.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration competing of interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Mohamed Nabil Bakr reports financial support was provided by Ministry of Higher Education, Egypt.
Acknowledgments
We appreciate the valuable conversation and critical feedback from our laboratory members. M.N.B is supported by a full Ph.D. fellowship (KH447) from the Arab Republic of Egypt's Ministry of Higher Education.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2023.101425.
Contributor Information
Haruko Takahashi, Email: harukot@hiroshima-u.ac.jp.
Yutaka Kikuchi, Email: yutaka@hiroshima-u.ac.jp.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Schadendorf D., Fisher D.E., Garbe C., Gershenwald J.E., Grob J.-J., Halpern A., Herlyn M., Marchetti M.A., McArthur G., Ribas A., Roesch A., Hauschild A. Melanoma., Nat Rev Dis Primers. 2015;1 doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 2.Miller A.J., Mihm M.C. Melanoma. N. Engl. J. Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y., Durand S., Dalle S., Caramel J. EMT-inducing transcription factors, drivers of melanoma phenotype switching, and resistance to treatment. Cancers (Basel) 2020;12 doi: 10.3390/cancers12082154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedri D., Karras P., Landeloos E., Marine J.C., Rambow F. Epithelial-to-mesenchymal-like transition events in melanoma. FEBS J. 2022;289 doi: 10.1111/febs.16021. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira I., Droop A., Edwards O., Wong K., Harle V., Habeeb O., Gharpuray-Pandit D., Houghton J., Wiedemeyer K., Mentzel T., Billings S.D., Ko J.S., Füzesi L., Mulholland K., Prusac I.K., Liegl-Atzwanger B., de Saint Aubain N., Caldwell H., Riva L., van der Weyden L., Arends M.J., Brenn T., Adams D.J. The clinicopathologic spectrum and genomic landscape of de-/trans-differentiated melanoma. Mod. Pathol. 2021;34 doi: 10.1038/s41379-021-00857-z. [DOI] [PubMed] [Google Scholar]
- 6.Pandya P., Orgaz J.L., Sanz-Moreno V. Modes of invasion during tumour dissemination. Mol Oncol. 2017;11 doi: 10.1002/1878-0261.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark A.G., Vignjevic D.M. Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 2015;36 doi: 10.1016/j.ceb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.He H., Shao X., Li Y., Gihu R., Xie H., Zhou J., Yan H. Targeting signaling pathway networks in several malignant tumors: progresses and challenges. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.675675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wessler I., Kirkpatrick C.J. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br. J. Pharmacol. 2008;154 doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grando S.A. Connections of nicotine to cancer. Nat. Rev. Cancer. 2014;14 doi: 10.1038/nrc3725. [DOI] [PubMed] [Google Scholar]
- 11.Wess J., Eglen R.M., Gautam D. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat. Rev. Drug Discov. 2007;6 doi: 10.1038/nrd2379. [DOI] [PubMed] [Google Scholar]
- 12.Boss A., Oppitz M., Drews U. Muscarinic cholinergic receptors in the human melanoma cell line SK-Mel 28: modulation of chemotaxis. Clin. Exp. Dermatol. 2005;30 doi: 10.1111/j.1365-2230.2005.01865.x. [DOI] [PubMed] [Google Scholar]
- 13.Lucianò A.M., Tata A.M. Functional characterization of cholinergic receptors in melanoma cells. Cancers (Basel) 2020;12 doi: 10.3390/cancers12113141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalamida D., Poulas K., Avramopoulou V., Fostieri E., Lagoumintzis G., Lazaridis K., Sideri A., Zouridakis M., Tzartos S.J. Muscle and neuronal nicotinic acetylcholine receptors: structure, function and pathogenicity. FEBS J. 2007;274 doi: 10.1111/j.1742-4658.2007.05935.x. [DOI] [PubMed] [Google Scholar]
- 15.Schuller H.M. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat. Rev. Cancer. 2009;9 doi: 10.1038/nrc2590. [DOI] [PubMed] [Google Scholar]
- 16.Huang L.C., Lin C.L., Qiu J.Z., Lin C.Y., Hsu K.W., Tam K.W., Lee J.Y., Yang J.M., Lee C.H. Nicotinic acetylcholine receptor subtype alpha-9 mediates triple-negative breast cancers based on a spontaneous pulmonary metastasis mouse model. Front. Cell. Neurosci. 2017;11 doi: 10.3389/fncel.2017.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaal C., Chellappan S.P. Nicotine-mediated cell proliferation and tumor progression in smoking-related cancers. Mol. Cancer Res. 2014;12 doi: 10.1158/1541-7786.MCR-13-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun H., Ma X. α5-nAChR modulates nicotine-induced cell migration and invasion in A549 lung cancer cells. Exp. Toxicol. Pathol. 2015;67 doi: 10.1016/j.etp.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Qi J.C., Xue W.Y., Zhang Y.P., Qu C.B., Lu B.S., Yin Y.W., Liu K.L., bin Wang D., Li W., Zhao Z.M. Cholinergic α5 nicotinic receptor is involved in the proliferation and invasion of human prostate cancer cells. Oncol. Rep. 2020;43 doi: 10.3892/or.2019.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang N.N., Meng X., Qin G., An Y., Zhang Q.Q., Cheng X., Huang S. α5-nAChR modulates melanoma growth through the Notch1 signaling pathway. J. Cell. Physiol. 2020;235 doi: 10.1002/jcp.29435. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen H.D., Liao Y.C., Ho Y.S., Chen L.C., Chang H.W., Cheng T.C., Liu D., Lee W.R., Shen S.C., Wu C.H., Tu S.H. The α9 nicotinic acetylcholine receptor mediates nicotine-induced pd-l1 expression and regulates melanoma cell proliferation and migration. Cancers (Basel) 2019;11 doi: 10.3390/cancers11121991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong M., Tao S., Zhang L., Diao L.T., Huang X., Huang S., Xie S.J., Xiao Z.D., Zhang H. RNA sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 2020;13 doi: 10.1186/s13045-020-01005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J., Slowikowski K., Zhang F. Single-cell transcriptomics in cancer: computational challenges and opportunities. Exp. Mol. Med. 2020;52 doi: 10.1038/s12276-020-0422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Wang D., Peng M., Tang L., Ouyang J., Xiong F., Guo C., Tang Y., Zhou Y., Liao Q., Wu X., Wang H., Yu J., Li Y., Li X., Li G., Zeng Z., Tan Y., Xiong W. Single‐cell RNA sequencing in cancer research. J. Exp. Clin. Cancer Res. 2021;40 doi: 10.1186/s13046-021-01874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J.R., Cohen O., Shah P., Lu D., Genshaft A.S., Hughes T.K., Ziegler C.G.K., Kazer S.W., Gaillard A., Kolb K.E., Villani A.C., Johannessen C.M., Andreev A.Y., van Allen E.M., Bertagnolli M., Sorger P.K., Sullivan R.J., Flaherty K.T., Frederick D.T., Jané-Valbuena J., Yoon C.H., Rozenblatt-Rosen O., Shalek A.K., Regev A., Garraway L.A. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016:352. doi: 10.1126/science.aad0501. 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambow F., Rogiers A., Marin-Bejar O., Aibar S., Femel J., Dewaele M., Karras P., Brown D., Chang Y.H., Debiec-Rychter M., Adriaens C., Radaelli E., Wolter P., Bechter O., Dummer R., Levesque M., Piris A., Frederick D.T., Boland G., Flaherty K.T., van den Oord J., Voet T., Aerts S., Lund A.W., Marine J.C. Toward minimal residual disease-directed therapy in melanoma. Cell. 2018;174 doi: 10.1016/j.cell.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 27.Jerby-Arnon L., Shah P., Cuoco M.S., Rodman C., Su M.J., Melms J.C., Leeson R., Kanodia A., Mei S., Lin J.R., Wang S., Rabasha B., Liu D., Zhang G., Margolais C., Ashenberg O., Ott P.A., Buchbinder E.I., Haq R., Hodi F.S., Boland G.M., Sullivan R.J., Frederick D.T., Miao B., Moll T., Flaherty K.T., Herlyn M., Jenkins R.W., Thummalapalli R., Kowalczyk M.S., Cañadas I., Schilling B., Cartwright A.N.R., Luoma A.M., Malu S., Hwu P., Bernatchez C., Forget M.A., Barbie D.A., Shalek A.K., Tirosh I., Sorger P.K., Wucherpfennig K., van Allen E.M., Schadendorf D., Johnson B.E., Rotem A., Rozenblatt-Rosen O., Garraway L.A., Yoon C.H., Izar B., Regev A. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175 doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., Ceccarelli M., Bontempi G., Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sean D., Meltzer P.S. GEOquery: a bridge between the gene expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23 doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 30.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26 doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4 doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuleshov M.v., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Maayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. Korotkevich, V. Sukhov, N. Budin, B. Shpak, M.N. Artyomov, A. Sergushichev, Fast gene set enrichment analysis, (n.d.). 10.1101/060012. [DOI]
- 34.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13 doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doncheva N.T., Morris J.H., Gorodkin J., Jensen L.J. Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 2019;18 doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader G.D., Hogue C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4 doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fleming L.M., Yeung B., Rogers A.J., McElrath J.M., Blish C.A., Gottardo R., Smibert P., Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184 doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14 doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32 doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 41.Chang P.M.H., Yeh Y.C., Chen T.C., Wu Y.C., Lu P.J., Cheng H.C., Lu H.J., Chen M.H., Chou T.Y., Huang C.Y.F. High expression of CHRNA1 is associated with reduced survival in early stage lung adenocarcinoma after complete resection. Ann. Surg Oncol. 2013;20 doi: 10.1245/s10434-013-3034-2. [DOI] [PubMed] [Google Scholar]
- 42.Carlisle D.L., Liu X., Hopkins T.M., Swick M.C., Dhir R., Siegfried J.M. Nicotine activates cell-signaling pathways through muscle-type and neuronal nicotinic acetylcholine receptors in non-small cell lung cancer cells. Pulm. Pharmacol. Ther. 2007;20 doi: 10.1016/j.pupt.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Scherl C., Schäfer R., Schlabrakowski A., Tziridis K., Iro H., Wendler O. Nicotinic acetylcholine receptors in head and neck cancer and their correlation to tumor site and progression. ORL (Oto-Rhino-Laryngol.) (Basel) 2016;78 doi: 10.1159/000445781. [DOI] [PubMed] [Google Scholar]
- 44.Spina R., Voss D.M., Asnaghi L., Sloan A., Bar E.E. Atracurium Besylate and other neuromuscular blocking agents promote astroglial differentiation and deplete glioblastoma stem cells. Oncotarget. 2016;7 doi: 10.18632/ONCOTARGET.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richard G., Dalle S., Monet M., Ligier M., Boespflug A., Pommier R.M., Fouchardière A., Perier‐Muzet M., Depaepe L., Barnault R., Tondeur G., Ansieau S., Thomas E., Bertolotto C., Ballotti R., Mourah S., Battistella M., Lebbé C., Thomas L., Puisieux A., Caramel J. ZEB 1‐mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol. Med. 2016;8 doi: 10.15252/emmm.201505971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caramel J., Papadogeorgakis E., Hill L., Browne G.J., Richard G., Wierinckx A., Saldanha G., sborne J., Hutchinson P., Tse G., Lachuer J., Puisieux A., Pringle J.H., Ansieau S., Tulchinsky E. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24 doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Vandamme N., Denecker G., Bruneel K., Blancke G., Akay O., Taminau J., de Coninck J., de Smedt E., Skrypek N., van Loocke W., Wouters J., Nittner D., Kohler C., Darling D.S., Cheng P.F., Raaijmakers M.I.G., Levesque M.P., Mallya U.G., Rafferty M., Balint B., Gallagher W.M., Brochez L., Huylebroeck D., Haigh J.J., Andries V., Rambow F., van Vlierberghe P., Goossens S., van den Oord J.J., Marine J.C., Berx G. The EMT transcription factor ZEB2 promotes proliferation of primary and metastatic melanoma while suppressing an invasive, mesenchymal-like phenotype. Cancer Res. 2020;80 doi: 10.1158/0008-5472.CAN-19-2373. [DOI] [PubMed] [Google Scholar]
- 48.Denecker G., Vandamme N., Akay Ö., Koludrovic D., Taminau J., Lemeire K., Gheldof A., de Craene B., van Gele M., Brochez L., Udupi G.M., Rafferty M., Balint B., Gallagher W.M., Ghanem G., Huylebroeck D., Haigh J., van den Oord J., Larue L., Davidson I., Marine J.C., Berx G. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014;21 doi: 10.1038/cdd.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King J.R., Kabbani N. Alpha 7 nicotinic receptor coupling to heterotrimeric G proteins modulates RhoA activation, cytoskeletal motility, and structural growth. J. Neurochem. 2016 doi: 10.1111/jnc.13660. [DOI] [PubMed] [Google Scholar]
- 50.Yang X., Ren H., Guo X., Hu C., Fu J. The expressions and mechanisms of sarcomeric proteins in cancers. Dis. Markers. 2020;2020 doi: 10.1155/2020/8885286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Y., Peng J., Liu W., Zhang P., Huang L., Gao B., Shen T., Zhou Y., Chen H., Chu Z., Zhang M., Qin H. Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer. Mol. Cell. Proteomics. 2009;8 doi: 10.1074/mcp.M800541-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhunia S., Barbhuiya M.A., Gupta S., Shrivastava B.R., Tiwari P.K. Epigenetic downregulation of desmin in gall bladder cancer reveals its potential role in disease progression. Indian J. Med. Res. 2020;151(Supplement) doi: 10.4103/ijmr.IJMR_501_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arentz G., Chataway T., Price T.J., Izwan Z., Hardi G., Cummins A.G., Hardingham J.E. Desmin expression in colorectal cancer stroma correlates with advanced stage disease and marks angiogenic microvessels. Clin. Proteonomics. 2011;8 doi: 10.1186/1559-0275-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stauffer S., Zeng Y., Zhou J., Chen X., Chen Y., Dong J. CDK1-mediated mitotic phosphorylation of PBK is involved in cytokinesis and inhibits its oncogenic activity. Cell. Signal. 2017;39 doi: 10.1016/j.cellsig.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gayyed M.F., El-Maqsoud N.M.R.A., Tawfiek E.R., el Gelany S.A.A., Rahman M.F.A. A comprehensive analysis of CDC20 overexpression in common malignant tumors from multiple organs: its correlation with tumor grade and stage. Tumor Biol. 2016;37 doi: 10.1007/s13277-015-3808-1. [DOI] [PubMed] [Google Scholar]
- 56.Li J., Wang Y., Wang X., Yang Q. CDK1 and CDC20 overexpression in patients with colorectal cancer are associated with poor prognosis: evidence from integrated bioinformatics analysis. World J. Surg. Oncol. 2020;18 doi: 10.1186/s12957-020-01817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamil M., Shinsato Y., Higa N., Hirano T., Idogawa M., Takajo T., Minami K., Shimokawa M., Yamamoto M., Kawahara K., Yonezawa H., Hirano H., Furukawa T., Yoshimoto K., Arita K. High filamin-C expression predicts enhanced invasiveness and poor outcome in glioblastoma multiforme. Br. J. Cancer. 2019;120 doi: 10.1038/s41416-019-0413-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang B., Liu Y., Zhao J., Hei K., Zhuang H., Li Q., Wei W., Chen R., Zhang N., Li Y. Ectopic overexpression of filamin C scaffolds MEK1/2 and ERK1/2 to promote the progression of human hepatocellular carcinoma. Cancer Lett. 2017;388 doi: 10.1016/j.canlet.2016.11.037. [DOI] [PubMed] [Google Scholar]
- 59.Tanabe K., Shinsato Y., Furukawa T., Kita Y., Hatanaka K., Minami K., Kawahara K., Yamamoto M., Baba K., Mori S., Uchikado Y., Maemura K., Tanimoto A., Natsugoe S. Filamin C promotes lymphatic invasion and lymphatic metastasis and increases cell motility by regulating Rho GTPase in esophageal squamous cell carcinoma. Oncotarget. 2017;8 doi: 10.18632/oncotarget.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.