Abstract
Plant-specific TCP transcription factors are key regulators of diverse plant functions. TCP transcription factors have long been annotated as basic helix–loop–helix (bHLH) transcription factors according to remote sequence homology without experimental validation, and their consensus DNA-binding sequences and protein–DNA recognition mechanisms have remained elusive. Here, we report the crystal structures of the class I TCP domain from AtTCP15 and the class II TCP domain from AtTCP10 in complex with different double-stranded DNA (dsDNA). The complex structures reveal that the TCP domain is a distinct DNA-binding motif and the homodimeric TCP domains adopt a unique three-site recognition mode, binding to dsDNA mainly through a central pair of β-strands formed by the dimer interface and two basic flexible loops from each monomer. The consensus DNA-binding sequence for class I TCPs is a perfectly palindromic 11 bp (GTGGGNCCCAC), whereas that for class II TCPs is a near-palindromic 11 bp (GTGGTCCCCAC). The unique DNA binding mode allows the TCP domains to display broad specificity for a range of DNA sequences even shorter than 11 bp, adding further complexity to the regulatory network of plant TCP transcription factors.
INTRODUCTION
Transcription factors are proteins that bind to specific DNA sequences to regulate transcription. The most basic feature of transcription factors is the possession of at least one DNA-binding domain (DBD) that can recognize sequences located in the promoter or enhancer region of regulatory genes (1). Transcription factors can be categorized into different classes according to the sequences and structural folds of their DBDs. Key questions in this area pertain to the number of classes or structural folds that exist and whether a prediction can be made regarding the interaction of a DBD with a certain DNA motif according to its sequence or structural fold.
DBDs were first categorized into five superclasses: basic domains, zinc-coordinating DNA-binding domains, helix–turn–helix (HTH) domains, β-scaffold factors with minor groove contacts and the remaining DBDs. These superclasses can be further organized into different classes and families (2); for example, the basic helix–loop–helix (bHLH) and basic region-leucine zipper (bZIP) domains belong to the basic domain class and the homeodomain belongs to the HTH domain class. With the discovery of an increasing number of structures, it is becoming difficult to categorize transcription factors into existing classes; therefore, reclassification is often required. A simple approach to defining a distinct DBD or DNA-binding motif is comparison with known structural folds in order to elucidate whether it carries new secondary structural features or DNA binding properties. One example is the ribbon–helix–helix (RHH) transcription factors that use an antiparallel β-sheet to recognize DNA, distinguishing these proteins from the well-known HTH family that insert an α-helix into the DNA major groove. Accordingly, the RHH domain has been defined as a distinct DNA-binding motif (3). Another example is BRZ-INSENSITIVE-LONGHYPOCOTYL 1(BIL1)/BRASSINAZOLE-RESISTANT 1 (BZR1) transcription factors that have a non-canonical bHLH motif, because despite using the N-terminal helix to bind to the major groove of dsDNA, the BIL1/BZR1 homodimer displays a larger tilt angle of the DNA recognition helix and a shorter second α-helix followed by a β-hairpin structure. Moreover, BIL1/BZR1 proteins recognize the NN-BRRE-core motif (NNCGTG), one of the variant G-box (CACGTG) motifs, distinguishing it from bHLH transcription factors (4). Here, we discuss the notion that plant-specific TCP transcription factors should be treated as a class possessing a distinct DNA-binding motif with new secondary structural features and DNA binding properties.
The TCP gene family was first described in the late 1990s and defined by three identified members: Teosinte branched 1 (TB1), CYCLOIDEA (CYC) and Proliferating cell factor 1/2 (PCF1/2) (5–8). These genes contain a conserved TCP domain, which was identified as the DBD of this transcription factor family (7). A study indicated that the TCP domain contains all the determinants required for DNA binding (9). Since the initial discovery, various TCP domain-containing proteins have been identified in numerous plants (10,11) and shown to participate in diverse plant growth-related processes (10), the best characterized of which are leaf development, flower shaping and shoot branching (12,13). Other studies have also demonstrated that TCP domain-containing proteins play important roles in hormone synthesis and signaling, regulation of circadian rhythm and other central biological functions (12–14).
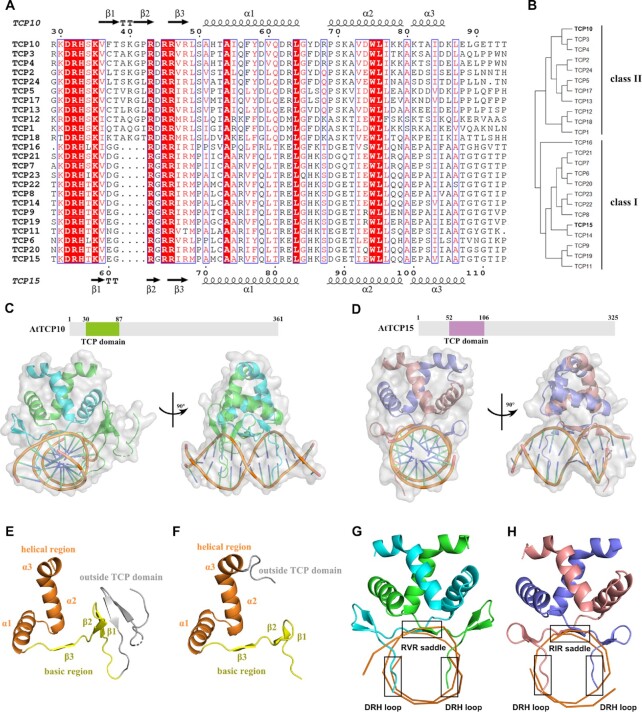
According to protein sequence conservation and other features (15), the 24 members of the TCP family in Arabidopsis thaliana can be categorized into two classes: class I (also called PCF or TCP-P) and class II (also called TCP-C). The most notable difference is that class I proteins have four amino acids fewer than class II proteins in the N-terminal basic region (Figure 1A, B). In addition, class II TCPs can be further divided into two clades: CYC/TB1-like and CIN-like.
Figure 1.
Overall complex structures of AtTCP10-DBD and AtTCP15-DBD. (A) Multiple sequence alignment of the TCP domains of 24 TCP family members from Arabidopsis. Completely conserved residues are colored with red boxes, and less conserved residues are shown as red letters. The β-strands within the TCP domain are numbered β1–β3, and the α-helices are named α1, α2 and α3. (B) Phylogenetic tree of the 24 TCP family members from Arabidopsis. The tree was constructed with the Maximum Likelihood (ML) method using the MEGAX software (33). (C) Size and domain organization of full-length AtTCP10 and overall complex structure of the AtTCP10-DBD with dsDNA in two orientations (rotated 90° along the indicated axis). Chain A (green) contains residues 3–87, while chain B (cyan) contains residues 30–87 only, covering the entire TCP domain. (D) Size and domain organization of full-length AtTCP15 and overall structure of AtTCP15-DBD with dsDNA in two orientations (rotated 90° along the indicated axis). Chain A (pink) and chain B (slate) both contain residues 52–110, covering the entire TCP domain. (E) Secondary structure of chain A of AtTCP10–DBD. (F) Secondary structure of chain A of AtTCP15-DBD. The basic region is colored yellow, and the helical region is colored orange. The region outside the TCP domain is colored gray. (G–H) Three parts of the TCP domains of AtTCP10-DBD and AtTCP15-DBD are involved in DNA binding: the RVR/RIR saddle and the two DRH loops.
TCP domain-containing proteins form dimers in solution, which is necessary for DNA binding activity, and the prevention of dimer formation can abolish DNA binding (9,12,16). Early studies revealed the consensus DNA-binding sequences as GTGGGNCC (complementary sequence: GGNCCCAC) for class I and GTGGNCCC (complementary sequence: GGGNCCAC) for class II (12,16,17), which are distinct but overlapping. These two classes share the same core sequence (GGNCC) but have different flanking nucleotides. It has been reported that one residue (Gly in class I or Asp in class II) in the N-terminal basic region determines the preference for class I or class II sequences (18); however, the manner by which this residue regulates the binding preference remains unknown.
Recent in vitro and in vivo studies of TCPs, such as systematic evolution of ligands by exponential enrichment (SELEX), electrophoretic mobility shift assays (EMSAs), protein-binding microarrays (PBMs), yeast one-hybrid (Y1H) assays and chromatin immunoprecipitation (ChIP), have successfully summarized (12) and confirmed that TCP domain-containing proteins recognize specific GC-rich core motifs; however, the length and composition of the flanking nucleotides remain difficult to clarify. For example, ChIP has demonstrated that AtTCP15 can bind directly to the promoter regions (GGNCCC) of the CYCA2;3 and RETINOBLASTOMA-RELATED (RBR) genes, which play key roles in endoreduplication (19). In vitro studies have also shown that AtTCP15 can recognize the following DNA sequences: GTGGGNCCgN (SELEX) (20), GTGGGACC (EMSA) (15), gGGgCCCAC (PBM) (17) and TGGGCC (Y1H) (21). These non-palindromic binding sequences of different lengths are distinctly different from any known consensus DNA-binding sequences, particularly for homodimeric transcription factors. To date, no universal agreement has been reached regarding the consensus DNA-binding sequences of the two TCP classes. The structures of the TCP domain alone and in complex with dsDNA will aid our understanding of the DNA binding landscape of TCP transcription factors.
Significant effort has been made to analyze the structures and consensus DNA-binding sequences of the TCP domain, which has long been predicted to contain a basic amino acid-rich region in the N-terminus followed by a HLH domain (22). Due to the low sequence identity with any real bHLH DNA-binding domain (23), it has been suggested that TCP domain-containing proteins may contain a non-canonical bHLH domain (22,24,25). However, the basic region clearly contains helix-breaking amino acids (18), and the length of this region is much longer than that in real bHLH proteins (9). In 2010, a model of the TCP domain bound to a B-form dsDNA with the sequence GTGGTCCC was reported using a DNA-bound MyoD structure as a template (9); nevertheless, this structural model has been misleading. Recently, the apo structure of the TCP domain from OsPCF6 was solved, and the TCP domain was demonstrated to possess a low-homology bacterial RHH fold by structural comparison (26). The HLH region of the TCP domain does resemble the bacterial RHH domain, but the basic regions of the two folds are quite distinct. Both basic regions are responsible for DNA binding; however, the two domains have different DNA recognition sequences. The recognized DNA sequences are not conserved across the bacterial RHH superfamily (3), whereas the TCP domain can bind to relatively conserved DNA sequences as mentioned above.
In the present study, we determine the DNA–protein complex structures of a class I TCP domain, AtTCP15, as well as a class II TCP domain, AtTCP10, and clearly demonstrate that the TCP domain does not belong to either the bHLH or RHH family but rather defines a distinct DNA recognition and binding mechanism. The TCP domain adopts a three-site recognition mode for dsDNA, mainly through a short pair of β-strands formed in the dimer interface and two basic flexible loops from the N-terminus of each monomer. Structural analysis and comparison demonstrate that TCP proteins of both classes can bind to 11 bp DNA sequences, GTGGGNCCCAC (underlined bases are palindromic) for class I and GTGGTCCCCAC for class II. Furthermore, we determined the complex structures of the TCP domain with dsDNA of different lengths. In conjunction with EMSA and isothermal titration calorimetry (ITC) assays, we confirmed that the unique DNA binding mode allows the TCP domain to display broad specificity for a range of DNA sequences even shorter than 11 bp, which helps to explain the complex regulatory network of TCP transcription factors. The complex structures also clarify the manner by which a single residue determines the binding preference of the two TCP classes.
MATERIALS AND METHODS
Sequence alignment
The CLUSTALW website (https://www.genome.jp/tools-bin/clustalw) was used for multiple sequence alignments under default parameters. The aln file was uploaded to the ESPript 3.0 website (27) (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) for secondary structure determination of both chain A and chain B. All aligned sequences, including those of the 24 A. thaliana TCP domain-containing proteins (from AtTCP1 to AtTCP24), were downloaded from the UniProt database (https://www.uniprot.org/).
Protein expression and purification
The DNA sequence encoding truncated AtTCP10 (residues 1–87) was cloned into the pET28a vector (the protein was named AtTCP10-DBD). The leucine at position 49 was mutated to methionine (the protein was named AtTCP10-49M) for ab initio phasing due to lack of structure model for the whole TCP protein superfamily. The DNA sequence encoding truncated AtTCP15 (residues 51–113) was cloned into the pET21b vector (the protein was named AtTCP15-DBD). The Escherichia coli strain BL21 (DE3) was transformed with the AtTCP10-49M plasmid and grown in M9 Minimal Medium at 310 K. At an OD600 of 0.6–0.8, 0.5 mM isopropyl-β-d-1-thiogalactopyranoside and essential amino acids (including selenomethionine) were added, and the cells were allowed to grow at 291 K overnight. The following day, cells were collected and resuspended in buffer A (25 mM HEPES and 1 M NaCl, pH 7.0). After sonication and centrifugation, the supernatant was purified using an Ni chelation column and size-exclusion chromatography (Superdex 75, GE Healthcare) using buffer B (25 mM HEPES, 1 M NaCl and 500 mM imidazole, pH 7.0) and buffer C (25 mM HEPES and 100 mM NaCl, pH 7.0) in succession. Purified protein was concentrated and subsequently stored at 193 K after flash freezing in liquid nitrogen. The two wild-type proteins (AtTCP10-DBD and AtTCP15-DBD) were obtained in a similar manner, but Luria–Bertani (LB) broth was used as the culture medium. A final concentration of 1 mM TCEP was added to the buffers for AtTCP15-DBD.
Crystallization, data collection and structure determination
Single-stranded DNAs were purchased (Supplementary Table S2), annealed to form dsDNA and purified by size-exclusion chromatography (Superdex 75, GE Healthcare). The 12 bp class II dsDNA and AtTCP10-49M protein were mixed to a final protein concentration of 5 mg/ml. The complex crystals were obtained in 200 mM potassium sodium tartrate tetrahydrate and 20% w/v polyethylene glycol (PEG) 3350 using the sitting-drop method. The datasets were collected at the Shanghai Synchrotron Radiation Facilities (SSRF) beamline BL17U1 and processed using the XDS program package (28). The protein structure was determined by the single-wavelength anomalous diffraction (SAD) method using SHELX C/D/E in the CCP4 suite (29), and the dsDNA structure was determined by molecular replacement in Phaser-MR (30) using ideal B-form DNA autogenerated by Coot (31). The structure was further refined using the Coot (31) and phenix.refine (30) programs. The crystal refinement was completed using AtTCP10-DBD and three-site class II dsDNA. Higher resolution (1.92 Å) crystals were obtained in 10% w/v PEG 1000 and 10% w/v PEG 8000. After data collection by the beamline BL1A at the High Energy Accelerator Research Organization (KEK) and processing using HKL2000, the structure was determined by molecular replacement using AtTCP10-49M as the search model and refined as described above. The crystals of AtTCP10-DBD with one-site DNA (for crystallization) were obtained in 100 mM Bis-Tris pH 5.5, 17% w/v PEG 10000 and 100 mM ammonium acetate at a final concentration of 10 mg/ml. The crystal of AtTCP15-DBD with 12 bp class I DNA was obtained in 100 mM sodium citrate/citric acid pH 5.5, 20% w/v PEG 4000 and 10% v/v 2-propanol at a final concentration of 10 mg/ml. The two datasets were collected by the beamline BL17U1 at the SSRF. The crystals of the AtTCP15-DBD apo form were obtained in 60% v/v Tacsimate™ pH 7.0. The crystals of AtTCP10-DBD with 1 M class II DNA were obtained in 100 mM sodium HEPES pH 7.0, 10% w/v PEG 4000 and 10% v/v 2-propanol. The crystals of AtTCP10-DBD with two-site DNA were obtained in 100 mM Tris pH 8.0 and 20% w/v PEG 4000. The three datasets were collected by the beamline BL1A at the High Energy Accelerator Research Organization (KEK). The five structures were determined by molecular replacement using AtTCP10-DBD with three-site DNA as the search model. All the crystals were obtained using the sitting-drop vapor diffusion method at 291 K. Glycerol at 20% was used as a cryoprotectant.
Site-directed mutagenesis
All mutants of AtTCP10-DBD were generated using the QuikChange® mutagenesis kit (TOYOBO) with the pET28a-TCP10 plasmid as a template. The three mutant proteins (R46A + R48A, D31A + R32A + H33A and D31A + R32A + H33A + R46A + R48A) were expressed and purified in the same manner as the wild-type protein.
EMSA experiments
The annealed three-site, two-site and one-site dsDNAs were used for EMSA experiments. A total of 200 ng of dsDNAs (24 pmol) was mixed with purified TCP proteins in buffer D (25 mM HEPES, 100 mM NaCl and 2.5% glycerol, pH 7.0). A final concentration of 1 mM TCEP was added to the buffer for AtTCP15-DBD. The final concentration of dsDNAs is 2.4 μM and the concentrations of protein are 2.4 μM (molar ratio 1:1), 4.8 μM (molar ratio 2:1) and 9.6 μM (molar ratio 4:1). The binding reaction mixtures (10 μl) were incubated at 4°C for 20 min and subsequently subjected to 7% non-denaturing polyacrylamide gel electrophoresis (PAGE) in 0.5× TBE buffer at 100 V for 40 min. The gels were stained with ethidium bromide for 10 min, and images were acquired using a fluorescence imaging system.
ITC assay
To determine the binding affinities of the proteins for DNA, 0.5 mM dsDNA was titrated into 0.1 mM protein using an ITC200 (GE Healthcare) at 298 K. All proteins and DNA were resuspended in buffer C (25 mM HEPES and 100 mM NaCl, pH 7.0). A concentration of 1 mM TCEP was added to the buffer for AtTCP15-DBD and the DNA. Triplicate experiments were performed independently. The thermograms were integrated in the Origin software and fitted to a one-site binding model. The dissociation constant (KD) values were calculated from triplicate thermograms [mean ± standard deviation (SD)].
Protein structure comparison
The protein structure was uploaded to the Dali server (32) (http://ekhidna2.biocenter.helsinki.fi/dali/) for a heuristic PDB search to compare it with other structures in the Protein Data Bank. The structures of the most highly ranked proteins were downloaded from the PDB and aligned in Pymol.
RESULTS
The DNA-binding sequences for TCPs
We endeavored to determine the structure of the homodimeric TCP protein in complex with dsDNA by firstly choosing the class II TCP protein AtTCP10 (residues 1–87) and a 12 bp class II dsDNA (5′-ATGTGGTCCCCC-3′ and 5′-TGGGGGACCACA-3′; italicized letters refer to the sticky ends) following the known class II consensus DNA-binding sequence GTGGNCCC. The leucine at position 49 of AtTCP10 was mutated to methionine for selenomethionyl protein preparation, which was generated for phasing the crystal structures, and the protein was named AtTCP10-49M. After solving the complex structure of AtTCP10-49M with a 12 bp class II DNA using the SAD method and analyzing the protein–DNA interface, we designed a three-site class II dsDNA (5′-ATGTGGTCCCCACT-3′ and 5′-TAGTGGGGACCACA-3′) that contained the core sequence GTGGTCCCCAC, with the consideration that all the homodimer proteins should have the ability to bind to palindromic DNA sequences. Further structural and biochemical studies confirmed the core sequence as the consensus DNA-binding sequence for class II TCPs.
Overall structures of AtTCP10-DBD and AtTCP15-DBD
Full-length AtTCP10 and AtTCP15 have 361 and 325 residues, respectively and the proteins we purified and used for structural and biochemical studies contained AtTCP10 residues 1–87 (named AtTCP10-DBD) (Figure 1C) and AtTCP15 residues 51–113 (named AtTCP15-DBD) (Figure 1D). Through trial-and-error experiments, we obtained crystals of the dsDNA–AtTCP10-DBD and dsDNA–AtTCP15-DBD complexes that diffracted to a high resolution. Here, we present the first two crystal structures of the class II TCP domain of AtTCP10 at 1.92 Å bound to a three-site class II dsDNA (5′-ATGTGGTCCCCACT-3′ and 5′-TAGTGGGGACCACA-3′) (Figure 1C, X-ray data statistics in Supplementary Table S1) and the class I TCP domain of AtTCP15 at 3 Å bound to a 12 bp class I dsDNA (5′-ATGTGGGTCCCC-3′ and 5′-TGGGGACCCACA-3′) (Figure 1D, X-ray data statistics in Supplementary Table S1).
In the complex structure of AtTCP10-DBD with a three-site class II dsDNA, each chain of dimeric AtTCP10-DBD consists of a basic region and a helical region (Figure 1E); however, the basic region contains three short β-strands (Figure 1A, E), which is a completely different structure from that of the bHLH DNA-binding motif that forms an α-helix in the basic region (34). The electron density maps of the two chains of the homodimer are not identical; the longer chain A, which is colored green, contains residues 3–87, while the shorter chain B, which is colored cyan, includes residues 30–87, and both contain the 58 amino acids of the conserved TCP domain (residues 30–87) (Supplementary Figure S1A–C, E). Within the TCP domains, both chains possess three β-strands, numbered β1–β3. Moreover, β1 and β2 in each chain form a β-hairpin, while the two β3 strands from each chain form an intermolecular paired β-sheet consisting of the three amino acids RVR (residues 46–48) (Figure 1A). Chain A of the homodimer possesses two further β-strands outside the TCP domain, named β′ and β″ (residues 15–18 and 21–24), which form an extended β-sheet with the β1 and β2 strands, while the electron density of β′ and β″ is missing in chain B (Supplementary Figure S1D). These two additional β-strands may not exist in all TCP members due to the lack of sequence conservation in this region (Supplementary Figure S2). The helical region is composed of two α-helices linked by a loop. Remarkably, the tilt angle between the two α1 helices (120°) is much larger than that of MYC2 (58°) (23) and other bHLH transcription factors (Supplementary Figure S3).
The structure of AtTCP15-DBD is similar to that of AtTCP10-DBD within the helical region, while there are some differences in the basic region (Figure 1F). Chain A (colored pink) and chain B (colored slate) of AtTCP15-DBD both include residues 52–110, which contain the entire class I TCP domain. As mentioned above, the most notable difference is that the class I TCP domain has four amino acids fewer than the class II TCP domain. Structure comparison of the two classes demonstrates that the four amino acids missing from class I TCPs reside in the β1–β2 hairpin region (Figure 1A). In class I TCP proteins, the β1 and β2 strand each consists of two amino acids, whereas in class II, the β1 and β2 strand each consists of three amino acids, which leads to shortening of the β-hairpin and a longer distance between the β-hairpin and the α1 helix (Figure 1G, H).
The three-site recognition mode
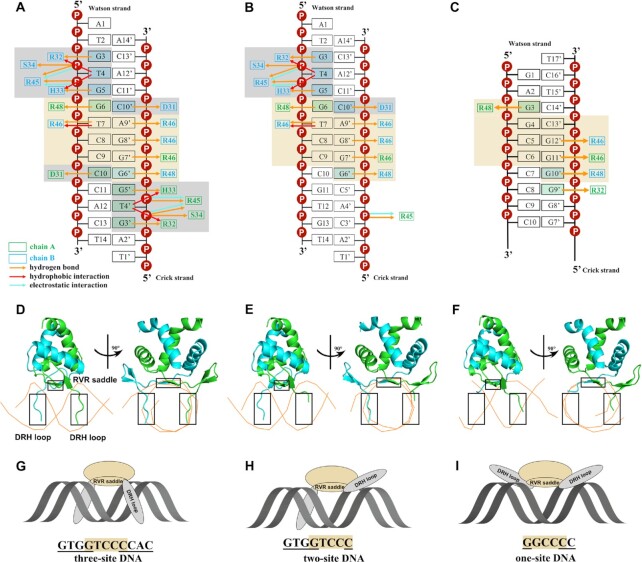
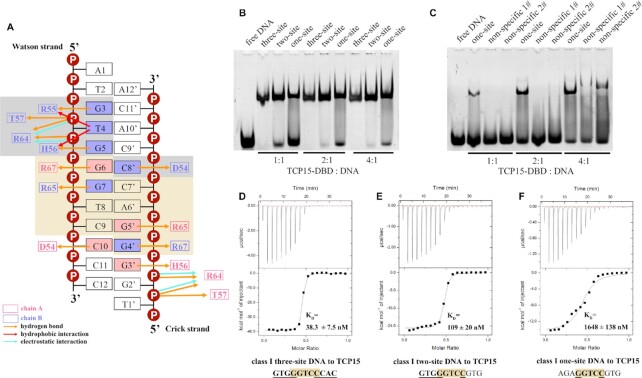
Taking the complex structure of AtTCP10-DBD with a three-site class II dsDNA (5′-ATGTGGTCCCCACT-3′ and 5′-TAGTGGGGACCACA-3′) as an example, we illustrate the DNA–protein interface. The DNA–protein interaction involves an 11 bp region (5′-GTGGTCCCCAC-3′ and 5′-GTGGGGACCAC-3′) in the three-site class II dsDNA (Figure 2A, D, G). Overall, the TCP domain adopts a three-site recognition mode for DNA, with three parts of the protein being involved in base-specific recognition: the two β3 strands (named the RVR saddle: residues 46–48) and the two N-terminal loops (named the DRH loops: residues 31–33) of both chains.
Figure 2.
The three-site recognition mode of AtTCP10-DBD. (A–C) Schematic representations of all the interactions of AtTCP10-DBD with (A) three-site class II DNA, (B) two-site class II DNA and (C) one-site class II DNA. The residues in chain A are colored green and those in chain B are colored cyan. The orange, red and blue arrows represent hydrogen bonds, hydrophobic interactions and electrostatic interactions, respectively. The brown colored boxes show the RVR saddle and the interacting base pairs. The gray colored boxes show the DRH loop and the interacting base pairs. (D–F) Cartoon representations of AtTCP10-DBD with (D) three-site class II DNA, (E) two-site class II DNA and (F) one-site class II DNA in two orientations. DNA is represented as orange tubes passing through the phosphates of the DNA backbone. The RVR saddle and two DRH loops are in black boxes. The main difference is that two DRH loops interact with three-site DNA, one DRH loop interacts with two-site DNA, and for one-site DNA, the two DRH loops have barely any base interactions. (G–I) Model of the three-site recognition mode.
The homodimeric AtTCP10-DBD, resembling a saddle, ‘rides’ on the dsDNA. The recognized 11 bp DNA sequence (GTGGTCCCCAC) is nearly palindromic, except for the central three base pairs (Figure 2A). In the ‘seat’ part of the saddle (RVR saddle), the positively charged side chains of Arg46 and Arg48 in the β3 strands are oriented into the DNA major groove (Figure 2A, D; Supplementary Figure S4A) and recognize five base pairs (GTCCC, colored brown). Arg48A (Arg48 in chain A, green) and Arg48B (Arg48 in chain B, cyan) recognize G6 and G6′ symmetrically, while Arg46A (Arg46 in chain A, green) and Arg46B (Arg48 in chain B, cyan) recognize G7′, G8′ and A9′ in the Crick strand as well as T7 in the Watson strand, spanning the central three base pairs, which is exactly the non-palindromic part of the 11 bp DNA sequence (GTGGTCCCCAC). The four side chains of arginine tightly grasp the DNA bases (especially G6′, G7′, G8′ and G6) like four fingers, explaining why the TCP domain prefers GC-rich DNA sequences.
The residues in the two ‘stirrups’ (DRH loops) of the dimer symmetrically interact with the four palindromic base pairs (GTGG, colored gray) flanking the center TCC sequence (Figure 2A, D; Supplementary Figure S4B). D31, R32 and H33 recognize C10′/C10, G3/G3′ and G5/G5′ separately. The hydrophobic interactions of Arg32 and His33 with T4/T4′, in addition to the hydrogen bond formed between Asp31 and His33, help to stabilize the flexible loop in the DNA major groove. The contacts between the protein residues and the DNA phosphate backbone do not directly contribute to base recognition but may play a supporting role, including the electrostatic interactions of Arg45 with T4/T4′ and G5/G5′, and the hydrogen bonds formed between Arg45 and T4/T4′ as well as G5/G5′ and between Ser34 and T4 /T4′ (Figure 2A; Supplementary Figure S5). The above analysis shows that the base-specific interactions of AtTCP10-DBD involve the 11 bp core sequence 5′-GTGGTCCCCAC-3′. From structural analysis, it can be seen that three parts of the protein are involved in the base-specific recognition of three parts of the 11 bp sequence, and it appears that these interactions are relatively independent, especially for the two DRH loops. Thus, we designed two further representative DNA sequences, in which the bases interacting with one or two DRH loops were mutated.
Subsequently, we obtained the two structures of the TCP domain–DNA complexes: AtTCP10-DBD with a two-site class II dsDNA (5′-ATGTGGTCCCGTGT-3′ and 5′-TACACGGGACCACA-3′) (Figure 2B, E, H) and AtTCP10-DBD with a one-site class II dsDNA (for crystallization, 5′-GAGGCCCCCCCATAATA-3′ and 5′-ATTATGGGGGGGCCTCT-3′) (Figure 2C, F, I). The interaction between the two-site DNA and the protein involves an 8 bp region (5′-GTGGTCCC-3′ and 5′-GGGACCAC-3′) (Figure 2B). The RVR saddle still recognizes the five base pairs GTCCC; however, only one of the two DRH loops interacts with the base pairs GTGG; the other DRH loop does not recognize specific bases (Figure 2E, H). With respect to the one-site dsDNA (for crystallization) in complex with the protein, the interaction spans a 6 bp region (5′-GGCCCC-3′ and 5′-GGGGCC-3′) (Figure 2C). The RVR saddle recognizes the five base pairs GGCCC; however, the two DRH loops barely form any base interactions (Figure 2F, I). We analyzed the structures of all the bound DNA using CURVES+ (Version 3.0) (35) to check for any DNA bending that may influence the interaction with proteins, and no unusual properties were found (Supplementary Figure S6).
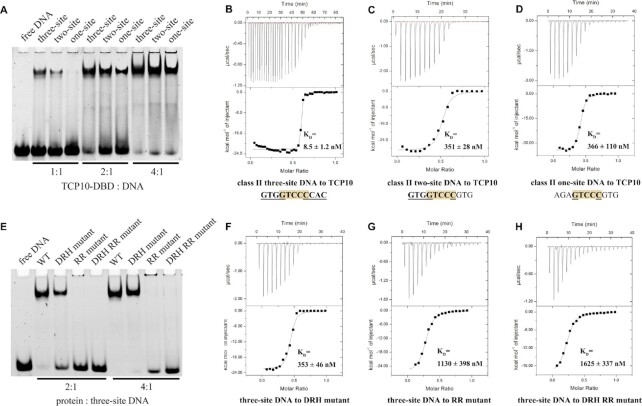
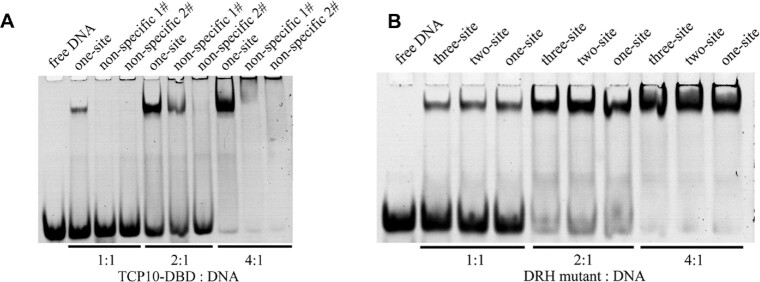
EMSA results for AtTCP10-DBD in complex with three-site, two-site and one-site class II DNA demonstrate that all the three DNA sequences can be recognized by AtTCP10-DBD, but the three-site DNA possesses the tightest binding (Figure 3A). The one-site DNA used was different from that used for crystallization (Supplementary Table S2). ITC results support the conclusion of tightest binding for three-site DNA, since the KD values for the three DNAs bound to the protein are 8.5, 351 and 366 nM, respectively (Figure 3B–D). The two-site and one-site class II DNA have markedly reduced affinities in comparison with the three-site class II DNA. We subsequently mutated all the bases in the three-site DNA and designed non-specific 1# DNA (5′-ATACTAAAAATGTT-3′ and 5′-TAACATTTTTAGTA-3′) and non-specific 2# DNA (5′-ATACAAAAAAAGTT-3′ and 5′-TAACTTTTTTTGTA-3′). Clear band shifts were observed for the one-site DNA as compared with the non-specific DNA, indicating that AtTCP10-DBD has a specific interaction with the one-site DNA (Figure 4A).
Figure 3.
EMSA and ITC results for three-site, two-site and one-site class II dsDNA with wild-type (WT) AtTCP10-DBD and the mutants. (A) EMSA results for AtTCP10-DBD in complex with three-site class II DNA, two-site class II DNA and one-site class II DNA. The molar ratio of protein to DNA is 1:1 (lanes 2–4), 2:1 (lanes 5–7) and 4:1 (lanes 8–10). (B–D) ITC results for (B) three-site class II DNA, (C) two-site class II DNA and (D) one-site class II DNA titrated into AtTCP10-DBD. (E) EMSA results for WT AtTCP10-DBD, DRH mutant, RR mutant and DRH RR mutant in complex with three-site class II DNA. The molar ratio of protein to DNA is 2:1 (lanes 2–5) and 4:1 (lanes 6–9). (F–H) ITC results for three-site class II DNA titrated into the (F) DRH mutant, (G) RR mutant and (H) DRH RR mutant.
Figure 4.
EMSA results of non-specific dsDNA in complex with AtTCP10-DBD, and three-site, two-site and one-site dsDNA in complex with the DRH mutant. (A) EMSA results for one-site class II DNA, non-specific 1# DNA and non-specific 2# DNA in complex with AtTCP10-DBD. The molar ratio of protein to DNA is 1:1 (lanes 2–4), 2:1 (lanes 5–7) and 4:1 (lanes 8–10). (B) EMSA results for three-site class II DNA, two-site class II DNA and one-site class II DNA in complex with the DRH mutant. The molar ratio of protein to DNA is 1:1 (lanes 2–4), 2:1 (lanes 5–7) and 4:1 (lanes 8–10).
From the three complex structures, it can be seen that the TCP domain adopts different conformations depending on the length and composition of the binding DNA, and this conformational flexibility is mainly conferred by the two DRH loops, which makes sense because these loops are flexible in the apo structure of both classes.
The apo structure of the putative transcription factor OsPCF6, which belongs to class II TCPs, was published recently (26). The sequence identity of OsPCF6 and AtTCP10 is ∼80% within the TCP domain; thus, we compared our complex structure with the apo structure of OsPCF6 (Supplementary Figure S7A–C). The DRH loops in the apo structure of OsPCF6 possess no electron density, indicating that these loops are flexible in the apo form. For class I TCPs, we solved the apo structure of AtTCP15. AtTCP15 forms a stable dimer, and the dimeric interface has a buried surface area of ∼1317 Å2 with an interface score of 1.0 (PDBe PISA v1.52) (36). Structural comparison shows that the DRH loops and β-hairpins formed by β1 and β2 in both chains possess no electron density (Supplementary Figure S7D–F). For both TCP classes, the basic region interacts with the dsDNA major groove and forms ordered secondary structures only upon DNA binding; therefore, conformational changes in the two DRH loops probably play an important role in the process of DNA recognition and binding, especially for three-site and two-site DNA.
Both the RVR saddle and DRH loops contribute to DNA binding
To further characterize the contribution of the RVR saddle and two DRH loops to DNA recognition, we mutated the key base-interacting residues to alanine and purified three mutant proteins of AtTCP10-DBD: the DRH loop mutant (named DRH mutant, D31A + R32A + H33A), the RVR saddle mutant (named RR mutant, R46A + R48A) and the combined two-region mutant (named DRH RR mutant, D31A + R32A + H33A + R46A + R48A). EMSA experiments of wild-type (WT) AtTCP10-DBD and the three mutants with three-site DNA demonstrate that mutation of either region decreases the binding affinity for three-site DNA. In comparison with the DRH loops, mutation of the RVR saddle dramatically reduces the affinity as shown by the absence of clear band shifts for the RR or DRH RR mutant (Figure 3E). Moreover, the ITC assays illustrate that the KD of the three mutants for three-site DNA are 353, 1130 and 1625 nM, respectively (Figure 3F–H). These results indicate that both regions are crucial for DNA binding, and the RVR saddle plays a more important role than the DRH loops.
EMSA experiments of the DRH mutant in complex with the three-site, two-site and one-site class II DNA show that the three DNA sequences have similar band shifts (Figure 4B), probably because once the DRH loops are mutated, only one region (RVR saddle) can interact with the three DNA sequences, indicating that the flexibility of the two DRH loops is important for the binding of three-site and two-site DNA but not one-site DNA.
With the DNA and protein concentrations near or above the KD values, all binding events are occurring in the upper plateau of the binding curve and the EMSA experiments are in the stoichiometric binding regime, which imply that only the most rudimentary conclusions can be made about binding from these assays (37). We do realize that the EMSA and even ITC experiments are quite rough estimates of affinity parameters, while with cautious interpretation we believe they are in good agreement with our structural results.
DNA–protein interaction of class I TCP protein AtTCP15
The class I TCP protein AtTCP15 also has a three-site recognition mode, and the DNA–protein interaction patterns are similar to those of the complex of AtTCP10-DNA since the amino acids involved in DNA recognition are well conserved among the TCP members of both classes (Figure 5A). The 12 bp class I dsDNA (5′-ATGTGGGTCCCC-3′ and 5′-TGGGGACCCACA-3′) used to crystallize AtTCP15-DBD is similar to the two-site dsDNA; therefore, the interactions are also mainly in the RIR saddle (residues 65–67, corresponding to the RVR saddle in AtTCP10) and one DRH loop (residues 54–56). In the DRH loop (Figure 5A; Supplementary Figure S4D), Asp54, Arg55 and His56 form hydrogen bonds with C8′, G3 and G5. Arg55 and His56 also display hydrophobic interactions with T4. A hydrogen bond is also formed between Asp54 and His56. The electrostatic interactions and hydrogen bonds between Arg64 and T4/G5 remain. Thr57 in class I TCPs still forms a hydrogen bond with the phosphate oxygen of T4 like Ser34 in class II, while in the other DRH loop, the electron density of the side chain of Arg55 is weak. The main difference in the DNA recognition modes between the two TCP classes is in the RIR saddle (RVR saddle in class II) (Figure 5A; Supplementary Figure S4C). Arg67 symmetrically recognizes G6 and G4′, and Arg65 symmetrically recognizes G7 and G5′. There is no base-specific interaction between AtTCP15-DBD and the A6′–T8 base pair; thus, there can be any base in this position. Taken together, these data demonstrate that the class I RIR saddle binds to the perfectly palindromic sequence GGNCC. We conclude that, in comparison with the structure of AtTCP10-DBD in complex with three-site class II DNA, the class I TCP protein can bind to the palindromic DNA sequence GTGGGNCCCAC.
Figure 5.
The three-site recognition mode of AtTCP15-DBD. (A) Schematic representation of all the interactions of AtTCP15-DBD with the 12 bp class I dsDNA. The residues in chain A are colored pink and those in chain B are colored slate. The orange, red and blue arrows represent hydrogen bonds, hydrophobic interactions and electrostatic interactions, respectively. The brown colored box shows the RIR saddle and the interacting base pairs. The gray colored box shows the DRH loop and the interacting base pairs. (B) EMSA results for three-site class I DNA, two-site class I DNA and one-site class I DNA in complex with AtTCP15-DBD. The molar ratio of protein to DNA is 1:1 (lanes 2–4), 2:1 (lanes 5–7) and 4:1 (lanes 8–10). (C) EMSA results for one-site class I DNA, non-specific 1# DNA and non-specific 2# DNA in complex with AtTCP15-DBD. The molar ratio of protein to DNA is 1:1 (lanes 2–4), 2:1 (lanes 5–7) and 4:1 (lanes 8–10). (D–F) ITC results for (D) three-site class I DNA, (E) two-site class I DNA and (F) one-site class I DNA titrated into AtTCP15-DBD.
Similar to AtTCP10-DBD, EMSA results for AtTCP15-DBD in complex with three-site, two-site and one-site class I DNA also demonstrate that all three DNA sequences can be recognized by AtTCP15-DBD (Figure 5B; Supplementary Table S2), with the three-site class I DNA displaying the tightest binding. ITC assays show that the binding affinities of class I AtTCP15 with three-site, two-site and one-site class I DNA are 38.3, 109 and 1648 nM, respectively (Figure 5D–F). The EMSA results for AtTCP15-DBD in complex with one-site class I DNA and the two non-specific DNA sequences demonstrate that the one-site class I DNA has a relatively high affinity compared with the non-specific DNA, which is similar to that seen with AtTCP10-DBD (Figure 5C).
The TCP domain is a distinct DNA-binding motif
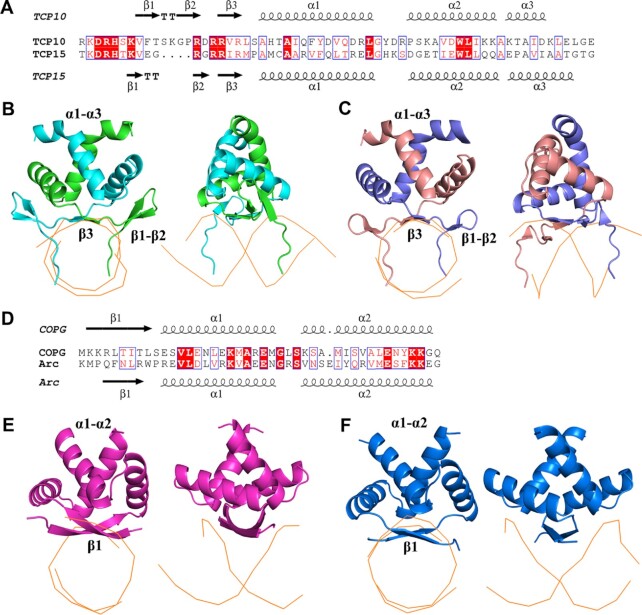
The sequence identity between the TCP domain and bHLH domain is ∼15%, below the sequence identity (25–30%) that can guarantee structural fold similarity. Also, the secondary structural features and DNA binding properties of the TCP domain are completely different from those of well-known DNA-binding domains such as bHLH, zinc finger and HTH. To search for any possible similar structural folds which are not detectable by comparing sequences, we uploaded the AtTCP10-DBD and AtTCP15-DBD coordinates to the Dali server, which can compare one query structure against those in the Protein Data Bank (http://ekhidna2.biocenter.helsinki.fi/dali/) (32). Besides the apo structure of OsPCF6, the most highly ranked structural folds identified by Dali belong to the bacterial RHH transcription factor family (3). The root mean square deviation (RMSD) between the TCP and RHH domains is ∼3 Å according to the Dali calculation. The helical regions of the two types of DNA-binding motifs appear relatively similar. Both α1 helices have ∼14 residues that are followed by a short loop and the α2 helix (Figure 6A, D). Furthermore, the orientation of the two α-helices is similar, with the tilt angle between the two α1 helices being ∼130° (Supplementary Figure S8A, B).
Figure 6.
Comparison of the TCP domains with RHH domains. (A) Sequence alignment of the TCP domains of AtTCP10 and AtTCP15. (B and C) Comparison of the DNA–protein complex structures of the TCP domains of AtTCP10 and AtTCP15. (D) Sequence alignment of the RHH domains of CopG and Arc. (E and F) Comparison of the DNA–protein complex structures of the RHH domains of CopG (purple) and Arc (blue).
However, the N-terminus of the bacterial RHH domain presents a longer antiparallel β-sheet with alternating polar and non-polar side chains. Taking the RHH domain of the CopG protein as an example, the denoted 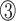 ,
, 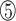 and
and 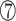 positions in the β-sheet contain residues with hydrophobic side chains that are oriented toward the hydrophobic core formed by the α-helices. In contrast, residues at the
positions in the β-sheet contain residues with hydrophobic side chains that are oriented toward the hydrophobic core formed by the α-helices. In contrast, residues at the 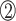 ,
, 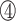 and
and 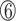 positions are oriented away from the core, and their side chains contact the DNA nucleotides in the major groove (Figure 6D–F; Supplementary Figure S8C). In comparison with the TCP domain, the β3 strand has only three residues, and the properties of the RVR/RIR residues are similar to those in the bacterial RHH ribbon region. The side chain of valine or isoleucine is oriented toward the hydrophobic core, and the arginine (positions 46 and 48) residues interact with the DNA (Figure 6A–C; Supplementary Figure S8D).
positions are oriented away from the core, and their side chains contact the DNA nucleotides in the major groove (Figure 6D–F; Supplementary Figure S8C). In comparison with the TCP domain, the β3 strand has only three residues, and the properties of the RVR/RIR residues are similar to those in the bacterial RHH ribbon region. The side chain of valine or isoleucine is oriented toward the hydrophobic core, and the arginine (positions 46 and 48) residues interact with the DNA (Figure 6A–C; Supplementary Figure S8D).
In addition to the above interactions occurring in the DNA major groove, the α2 helix of the RHH domain non-specifically contacts the DNA phosphate backbone, which is not observed with the TCP domain. On the other hand, the N-terminus of the TCP domain displays a β-hairpin formed by β1 and β2, in addition to the β3 strand. The two N-terminal DRH loops of the dimer insert into the DNA major groove and contact the DNA bases and phosphate backbone; thus, the overall interactions of the DBDs with dsDNA are very different. The TCP domain ‘rides’ on the dsDNA major groove using the RVR/RIR saddle and the two DRH loops (Figure 6B, C), whereas the bacterial RHH domain simply ‘sits’ on the DNA (Figure 6E, F) using the extended β-strands across approximately six base pairs. Moreover, the DNA-binding sequences are not conserved in the RHH domain family, while the TCP domain can bind to conserved 11 bp DNA sites as mentioned above; therefore, we believe that the TCP domain is a distinct class of DNA-binding motif.
Investigation into DNA binding specificity
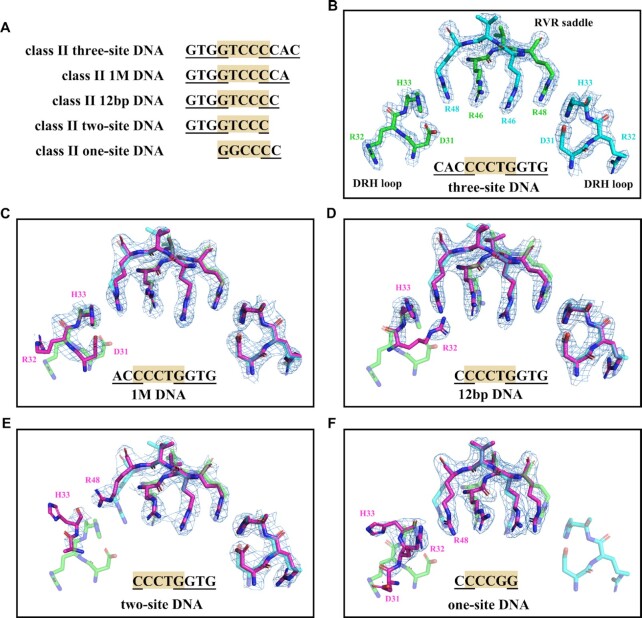
The two-site and one-site DNA are representative DNA sequences in which all the bases interacting with one or two DRH loops are mutated as compared with the three-site DNA (Figure 7A), but what if only one or two bases are mutated? The structure of AtTCP10-49M in complex with 12 bp class II DNA (5′-ATGTGGTCCCCC-3′ and 5′-TGGGGGACCACA-3′) has two bases mutated. We then obtained the complex structures of AtTCP10-DBD with 1 M class II DNA (5′-ATGTGGTCCCCAGT-3′ and 5′-TACTGGGGACCACA-3′), which has one base mutated. From these data, it is possible to understand how the mutation of bases that interact with one DRH loop influences the DNA–protein interaction.
Figure 7.
Flexibility of the two DRH loops. (A) Sequence alignment of the five class II DNA sequences recognized by AtTCP10-DBD. Only the interacting bases are shown. (B) Electron density map of the RVR saddle (residues 46–48) and DRH loops (residues 31–33) in the complex structure of AtTCP10-DBD with three-site DNA. (C) Structure comparison of AtTCP10-DBD in complex with three-site DNA and AtTCP10-DBD in complex with 1 M DNA (purple). Arg32A has weak electron density because the G–C pair is mutated. (D) Structure comparison of AtTCP10-DBD in complex with three-site DNA and AtTCP10-DBD in complex with 12 bp DNA (purple). Asp31A and Arg32A have weak electron density, and only the His33A recognizes G5′. There is a slight shift of His33A and Arg48B. (E) Structure comparison of AtTCP10-DBD in complex with three-site DNA and AtTCP10-DBD in complex with two-site DNA (purple). The interaction network of the DRH loop is destroyed, and the three amino acids DRH do not recognize specific bases. (F) Structure comparison of AtTCP10-DBD in complex with three-site DNA and AtTCP10-DBD in complex with one-site DNA (purple). The interaction network of the two DRH loops is destroyed, and only the RVR region recognizes DNA. The 2FO–FC electron density is contoured at the 1.5σ level.
The interactions between the base pairs GTGGTCCC and AtTCP10-DBD in the three complexes (1 M DNA, 12 bp DNA and two-site DNA) are the same as those seen with the three-site class II DNA (Figure 7B–E). In other words, the structures of the RVR saddle and one DRH loop are the same in the four complexes, and they interact with DNA in the same manner. In the structure of AtTCP10-DBD in complex with 1 M class II DNA, the DNA (GTGGTCCCCA) has one base pair mutated as compared with the three-site class II DNA (GTGGTCCCCAC). The only difference is that Arg32A has weak electron density because the G–C pair is mutated (Figure 7C). The 12 bp class II DNA (GTGGTCCCC) has two base pairs mutated as compared with the three-site class II DNA (GTGGTCCCCAC). In one DRH loop, Asp31A and Arg32A have weak electron density and only His33A recognizes G5′. Structural comparison shows a slight shift of His33A and Arg48B, possibly due to disruption of the stable interaction network of the DRH loop (Figure 7D). For the complex structure of AtTCP10-DBD with two-site class II dsDNA (GTGGTCCC), three base pairs are mutated, the interaction network of one DRH loop is destroyed and the three amino acids DRH do not recognize specific bases (Figure 7E). Moreover, for one-site class II DNA, the interaction network of the two DRH loops is destroyed and only the RVR region recognizes DNA (Figure 7F).
Taken together, it can be seen that mutation of the bases on one side of the three-site DNA recognized by one DRH loop fine-tunes the protein–DNA interaction and DNA binding specificity. Along with the three-site recognition mode, both sides of the three-site DNA may have mutations. This enables TCP domains to recognize multiple DNA sequences of different lengths in addition to three-site, two-site and one-site DNA. Furthermore, any possible in vivo protein–protein interactions in the two DRH loops may influence DNA recognition, which explains why previous studies have had difficulty clarifying the DNA length and sequence of the binding consensus for TCP proteins. The three-site recognition mode and flexibility of the DRH loops add further complexity to the regulatory network of TCP transcription factors.
DISCUSSION
Here, we determined several DNA–protein complex structures of the class I TCP domain AtTCP15-DBD and class II TCP domain AtTCP10-DBD and showed that the TCP domains form distinct DNA-binding families. The homodimeric TCP domains adopt a novel three-site recognition mode of dsDNA mainly through the RVR/RIR saddle in the central dimeric interfaces and two DRH loops from each monomer on the sides, which were previously proposed to be long helices of bHLH family transcription factors. The class II TCP transcription factors recognize the 11 bp near-palindromic sequence (GTGGTCCCCAC), which has been confirmed to be the DNA binding consensus for the class II TCP proteins. The palindromic 11 bp DNA sequence (GTGGGNCCCAC) is known to be the DNA binding consensus for class I TCP transcription factors.
According to previous in vitro and in vivo studies, the reported lengths of the consensus binding sequences for TCP proteins have always been shorter than 11 bp. The three-site recognition mechanism and flexibility of the DRH loops allow the TCP domains to recognize DNA sequences shorter than 11 bp. The RVR/RIR saddle, along with one of the DRH loops, should be sufficient for binding to an 8 bp DNA (two-site) but with lower affinity than for 11 bp DNA. In addition, the RVR/RIR saddle alone may recognize a 5 bp DNA (one-site).
This is consistent with the binding sites for TCP proteins previously found in the Arabidopsis genome by DAP-seq and ampDAP-seq (which uses a DNA library from which DNA modifications have been removed by PCR) (Supplementary Figure S9) (38). DAP-seq results for class I TCP proteins (AtTCP7, AtTCP9, AtTCP14, AtTCP15, AtTCP20, AtTCP21 and AtTCP22) show that the binding motif for class I TCPs is GTGGGNCCCAC (Supplementary Figure S9A). DAP-seq results for AtTCP13 and AtTCP17, and ampDAP-seq results for AtTCP24, demonstrate that the binding motif for class II TCPs is GTGGTCCCCAC (Supplementary Figure S9B). DAP-seq results for AtTCP3 and AtTCP24, and ampDAP-seq results for AtTCP13, indicate that the binding motif for class II TCPs is two-site class II GTGGTCCC (Supplementary Figure S9C). Analysis of the released DAP-seq data for class I TCP proteins suggests that ∼12% of the peaks in the promoter region have full occupancy of the three-site binding sequence GTGGGNCCCAC.
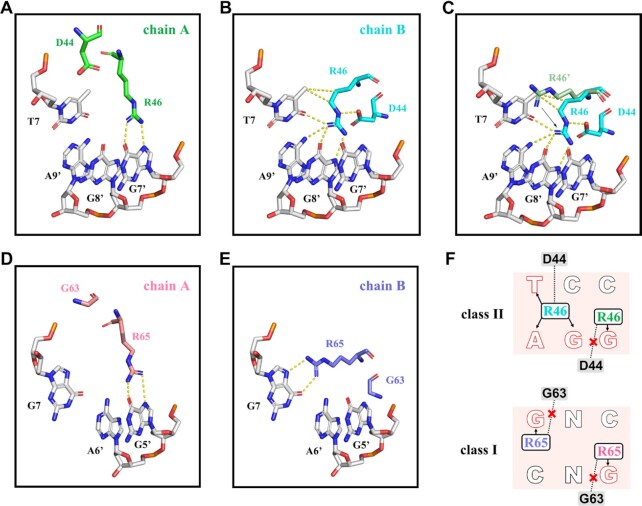
As mentioned above, the two classes of TCP domains have the ability to bind to 11 bp DNA with distinct differences in the central three base pairs, leading to varied DNA binding specificity. For the class II TCP protein, Arg46 residues from two chains recognize the non-palindromic center TCC bases (Figure 8A, B). The side chain of Arg46B is fixed by hydrophobic interactions with T7 and hydrogen bonds with bases (G8′, A9′ and T7) and Asp44 (Figure 8A), but Arg46A only interacts with G7′ (Figure 8B). It appears that the side chain orientation of Arg46B is opposite to that of Arg46A′ (symmetrical display of Arg46A) due to the influence of Asp44B (Figure 8C). In comparison with Arg46A, which only interacts with the single base G7′, Asp44B facilitates orientation of the side chain of Arg46B toward G8′ from the position of T7 (Figure 8A–C). For the class I TCP protein, Arg65 recognizes the palindromic center GNC bases symmetrically. Arg65A and Arg65B recognize G5′ and G7 (Figure 8D, E). The side chain orientations of the two Arg65 residues in class I are similar to those of Arg46A in class II. Previous studies have shown that Asp44 in class II (or Gly in class I) determines the DNA binding specificity of TCP proteins (18). Our structure proves that neither of the two Gly63 residues in class I TCP15 contact Arg65; thus, the two Arg65 residues recognize the bases G5′ and G7.
Figure 8.
Recognition mechanisms of the center three base pairs by AtTCP10-DBD and AtTCP15-DBD. (A) Arg46A of AtTCP10-DBD interacts with G7’. Asp44A has no interaction with Arg46A. (B) Arg46B of AtTCP10-DBD interacts with T7, G9′ and G8′, and also forms a hydrogen bond with Asp44B. (C) Arg46′ represents the symmetrical display of Arg46A. Arg46B is oriented toward G8′ from the position of T7 as compared with Arg46′. (D) Arg65A of AtTCP15-DBD interacts with G5′. Gly63A cannot interact with Arg65A. (E) Arg65B of AtTCP15-DBD interacts with G7. Gly63B cannot interact with Arg65B. (F) DNA recognition mechanism of class I and class II TCPs. Position 44 is an aspartic acid residue in class II TCPs, and the side chain of Arg46B is fixed by hydrophobic interactions with T and hydrogen bonds with bases (G, A and T) and Asp44. This position is a glycine in class I TCPs, and it does not interact with Arg65. Arg65 would recognize G symmetrically.
The bases T7, G8′ and A9, and the amino acids Arg46B and Asp44B, together form a stable interaction network that allows the homodimer AtTCP10 to recognize the near-palindromic sequence TCC. Asp44A does not interact with Arg46A because the DNA sequence is not suitable. We believe that even if the center three bases were TNA, both Asp44 residues could not interact with Arg46 due to steric hindrance (Figure 8F). One example showing the importance of the Asp/Gly is AtTCP16. Although TCP16 belongs to the class I TCP proteins, it prefers the class II DNA-binding sequence (18). The class I TCP proteins possess the conserved Gly, while TCP16 has a conserved Asp. The four amino acids missing from the basic region of class I TCP proteins have little influence on DNA binding; thus, Asp determines that TCP16 prefers the class II DNA sequence with the bases TCC.
Heterodimer formation of TCP proteins has often been discussed in tune with heterodimeric bHLH transcription factors, where the heterodimer is often essential for protein function, such as circadian master regulator CLOCK-BMAL1 (39). Within the TCP domain, the crucial amino acids in the dimerization region (RVR or RIR saddle and the helical region) of the two classes are conserved, which do not prohibit heterodimer formation. However, the members that can form heterodimers and their resulting stability, in addition to the requirement of heterodimer formation for certain functions, should be clarified. Another outstanding question concerns the DNA recognition mode of heterodimers; heterodimers formed between the same class should maintain the original DNA binding specificity, while heterodimers of the two classes may prefer the class II DNA sequence since there will be only one Asp interacting with the Arg. The monomer of the class I protein in the heterodimer resembles the monomer that has fewer DNA interactions in the class II homodimer.
Finally, our data propose that the TCP transcription factors should be regarded as a unique and novel class of DNA-binding proteins instead of RHH or bHLH transcription factors. The distinct DNA binding mode of both classes in vitro will aid our understanding of the DNA recognition motif gained from in vivo experiments. A whole-genome search for the DNA-binding sequences of TCP transcription factors will help to define the TCP regulatory network and understand the role of the two classes of TCP proteins in plants.
DATA AVAILABILITY
The accession numbers for the structures of AtTCP10-49M&12bp class II DNA, AtTCP10-DBD&three-site class II DNA, AtTCP15-DBD&12bp class I DNA, AtTCP10-DBD&1M class II DNA, AtTCP10-DBD&two-site class II DNA, AtTCP15-DBD and AtTCP10-DBD&one-site class II DNA (for crystallization) are RCSB PDB: 7VP1, 7VP2, 7VP3, 7VP4, 7VP5, 7VP6 and 7VP7, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the National Facility for Protein Science, Shanghai for providing us with the opportunity to test crystals and collect datasets on the SSRF BL17U1 and BL19U1 beamlines. We are grateful to the KEK Photon Factory and its staff members for their assistance in data collection. We thank the National Center for Protein Sciences at Peking University, Beijing, for assistance with crystal screening and ITC experiments. We would also like to thank Professor Jiawei Wang for his help with data processing and structure determination.
Author contributions: X.-D.S. conceived and supervised the entire project. J.-K.N. performed the preliminary work. Y.Z. carried out the gene construction, protein expression and purification, and crystal screening and optimization. Y.Z. and B.W. collected the X-ray diffraction data. Y.Z. and Y.-P.X. performed the structure determination and structure analysis. X.-D.S., Y.Z. and Y.-P.X. wrote the manuscript. G.Q. and H.C. participated in the project.
Contributor Information
Yi Zhang, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
Yong-ping Xu, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
Ju-kui Nie, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
Hong Chen, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
Genji Qin, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
Bo Wang, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
Xiao-Dong Su, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, 100871, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The present work was supported by the National Science Foundation of China (NSFC) [31670740 and 31270803].
Conflict of interest statement. None declared.
REFERENCES
- 1. Mitchell P.J., Tjian R.. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989; 245:371–378. [DOI] [PubMed] [Google Scholar]
- 2. Wingender E. Classification scheme of eukaryotic transcription factors. Mol. Biol. 1997; 31:483–497. [PubMed] [Google Scholar]
- 3. Schreiter E.R., Drennan C.L.. Ribbon–helix–helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 2007; 5:710. [DOI] [PubMed] [Google Scholar]
- 4. Nosaki S., Miyakawa T., Xu Y., Nakamura A., Hirabayashi K., Asami T., Nakano T., Tanokura M.. Structural basis for brassinosteroid response by BIL1/BZR1. Nat Plants. 2018; 4:771. [DOI] [PubMed] [Google Scholar]
- 5. Doebley J., Stec A., Hubbard L.. The evolution of apical dominance in maize. Nature. 1997; 386:485. [DOI] [PubMed] [Google Scholar]
- 6. Luo D., Carpenter R., Vincent C., Copsey L., Coen E.. Origin of floral asymmetry in Antirrhinum. Nature. 1996; 383:794. [DOI] [PubMed] [Google Scholar]
- 7. Cubas P., Lauter N., Doebley J., Coen E.. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999; 18:215–222. [DOI] [PubMed] [Google Scholar]
- 8. Doebley J., Stec A., Gustus C.. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995; 141:333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aggarwal P., Das Gupta M., Joseph A.P., Chatterjee N., Srinivasan N., Nath U.. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell. 2010; 22:1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin-Trillo M., Cubas P.. TCP genes: a family snapshot ten years later. Trends Plant Sci. 2010; 15:31–39. [DOI] [PubMed] [Google Scholar]
- 11. Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C.. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007; 65:23–33. [DOI] [PubMed] [Google Scholar]
- 12. González-Grandío E., Cubas P.. Gonzalez D.H. TCP transcription factors: evolution, structure, and biochemical function. Plant transcription factors: evolutionary, structural and functional aspects. 2015; Academic Press; 139–151. [Google Scholar]
- 13. Lopez J.A., Sun Y., Blair P.B., Mukhtar M.S.. TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 2015; 20:238–245. [DOI] [PubMed] [Google Scholar]
- 14. Nicolas M., Cubas P.. TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 2016; 33:33–41. [DOI] [PubMed] [Google Scholar]
- 15. Manassero N.G.U., Viola I.L., Welchen E., Gonzalez D.H.. TCP transcription factors: architectures of plant form. Biomol Concepts. 2013; 4:111–127. [DOI] [PubMed] [Google Scholar]
- 16. Kosugi S., Ohashi Y.. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002; 30:337–348. [DOI] [PubMed] [Google Scholar]
- 17. Franco-Zorrilla J.M., Lopez-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R.. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl Acad. Sci. USA. 2014; 111:2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Viola I.L., Reinheimer R., Ripoll R., Manassero N.G., Gonzalez D.H.. Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 2012; 287:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z.-Y., Li B., Dong A.-W.. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant. 2012; 5:270–280. [DOI] [PubMed] [Google Scholar]
- 20. Viola I.L., Uberti Manassero N.G., Ripoll R., Gonzalez D.H.. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 2011; 435:143–155. [DOI] [PubMed] [Google Scholar]
- 21. Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C.P., Van Aken O., Millar A.H., Murcha M., Whelan J.. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010; 22:3921–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kosugi S., Ohashi Y.. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997; 9:1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lian T.F., Xu Y.P., Li L.F., Su X.D.. Crystal structure of tetrameric arabidopsis MYC2 reveals the mechanism of enhanced interaction with DNA. Cell Rep. 2017; 19:1334–1342. [DOI] [PubMed] [Google Scholar]
- 24. Toledo-Ortiz G. The Arabidopsis basic/helix–loop–helix transcription factor family. Plant Cell. 2003; 15:1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J.. Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and arabidopsis. Plant Physiol. 2006; 141:1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun L., Zou X., Jiang M., Wu X., Chen Y., Wang Q., Wang Q., Chen L., Wu Y.. The crystal structure of the TCP domain of PCF6 in Oryza sativa L. reveals an RHH-like fold. FEBS Lett. 2020; 594:1296–1306. [DOI] [PubMed] [Google Scholar]
- 27. Robert X., Gouet P.. Deciphering key features in protein structures with the new endscript server. Nucleic Acids Res. 2014; 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabsch W Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 32. Holm L., Laakso L.M.. Dali server update. Nucleic Acids Res. 2016; 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones S. An overview of the basic helix–loop–helix proteins. Genome Biol. 2004; 5:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blanchet C., Pasi M., Zakrzewska K., Lavery R.. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011; 39:W68–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 37. Weeramange C.J., Fairlamb M.S., Singh D., Fenton A.W., Swint-Kruse L.J.P.S.. The strengths and limitations of using biolayer interferometry to monitor equilibrium titrations of biomolecules. Protein Sci. 2020; 29:1004–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O’Malley R.C., Huang S.-s.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016; 165:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Z., Wu Y., Li L., Su X.-D.. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix–loop– helix domains with E-box DNA. Cell Res. 2013; 23:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the structures of AtTCP10-49M&12bp class II DNA, AtTCP10-DBD&three-site class II DNA, AtTCP15-DBD&12bp class I DNA, AtTCP10-DBD&1M class II DNA, AtTCP10-DBD&two-site class II DNA, AtTCP15-DBD and AtTCP10-DBD&one-site class II DNA (for crystallization) are RCSB PDB: 7VP1, 7VP2, 7VP3, 7VP4, 7VP5, 7VP6 and 7VP7, respectively.