Abstract
Thermorubin (THR) is an aromatic anthracenopyranone antibiotic active against both Gram-positive and Gram-negative bacteria. It is known to bind to the 70S ribosome at the intersubunit bridge B2a and was thought to inhibit factor-dependent initiation of translation and obstruct the accommodation of tRNAs into the A site. Here, we show that thermorubin causes ribosomes to stall in vivo and in vitro at internal and termination codons, thereby allowing the ribosome to initiate protein synthesis and translate at least a few codons before stalling. Our biochemical data show that THR affects multiple steps of translation elongation with a significant impact on the binding stability of the tRNA in the A site, explaining premature cessation of translation. Our high-resolution crystal and cryo-EM structures of the 70S-THR complex show that THR can co-exist with P- and A-site tRNAs, explaining how ribosomes can elongate in the presence of the drug. Remarkable is the ability of THR to arrest ribosomes at the stop codons. Our data suggest that by causing structural re-arrangements in the decoding center, THR interferes with the accommodation of tRNAs or release factors into the ribosomal A site.
INTRODUCTION
The ever-growing antibiotic resistance among bacterial pathogens represents a major threat to public health care (1), calling for the continued search for new and improvements of existing drugs. Understanding the detailed molecular mechanisms by which various antimicrobials impede certain cellular functions is critical for their potential improvement to better suit clinical applications and circumvent development of resistance. About half of the antibiotics currently used in the clinic slow down or kill pathogenic bacteria and thereby cure infections by selectively inhibiting their ribosomes – the central components of the protein synthesis apparatus. Being the most conserved and sophisticated molecular machine of the cell, the ribosome is composed of two unequal separable subunits, small and large (30S and 50S in bacteria), which join together to form the 70S ribosome (2). Genetic information for protein synthesis is carried to the ribosome by messenger RNA (mRNA), which is read by nucleotide triplets (codons) with the help of auxiliary transfer RNA (tRNA) molecules that have two functional ends, one carrying the amino acid and the other containing the anticodon that recognizes the mRNA codons. tRNAs are indispensable adaptors that bridge the two main functional centers of the ribosome: the decoding center (DC) in the small subunit and the catalytic peptidyl transferase center (PTC) at the heart of the large subunit. While the PTC chemically links amino acids delivered by the tRNAs into polypeptides, the DC monitors the accuracy of base pairing between the codon of the mRNA and the anticodon of the tRNA, ensuring that the correct tRNAs are selected. The majority of ribosome-targeting antibiotics interfere with protein synthesis by binding at the ribosome functional centers (such as DC or PTC) and either freeze a particular conformation of the ribosome or hinder the binding of its ligands (3). In particular, DC is targeted by many chemically unrelated antibiotics, including aminoglycosides, tetracyclines, tuberactinomycins, odilorhabdins, and negamycin (3,4).
In 1964, a secondary metabolite from Thermoactinomyces antibioticus, called thermorubin (THR), was shown to exhibit a broad range of activity against both Gram-positive and Gram-negative bacteria (5,6). Despite its strong antimicrobial properties, THR is not currently used in the clinic mainly due to its poor solubility and low thermostability above 37°C (5). THR was further shown to inhibit protein synthesis in vitro (7), suggesting that the ribosome might be its primary intracellular target. Only much later, in 2012, the first structure of a vacant 70S ribosome from the Gram-negative bacterium Thermus thermophilus in complex with THR (8) revealed that, despite being structurally similar to tetracyclines, THR binds to a distinct site within DC largely overlapping with those of aminoglycosides and tuberactinomycins (9,10). The binding site of THR is located at the interface between the 30S and 50S ribosomal subunits within bridge B2a (8). The tetracyclic moiety of THR stacks between residue A1913 of Helix 69 (H69) of the 23S rRNA and the C1409-G1491 basepair at the top of helix 44 (h44) of the 16S rRNA, whereas the orthohydroxyphenyl moiety of THR stacks upon the nucleobase of U1915 in H69 (8). Importantly, THR does not affect translation in eukaryotic cells (5,6), such as yeast and fungi, which can be rationalized by the absence of the equivalent base pair in the 18S rRNA and the concomitant inability to stack with the tetracyclic moiety of the drug.
The precise mechanism of THR action during protein synthesis remains unclear. Initiation factors 1 (IF1) and 2 (IF2) interact with bridge B2a, suggesting that, in principle, ribosome-bound THR could interfere with the conformational changes of this region required for efficient delivery of initiator fMet-tRNA to the P site during translation initiation (8). Indeed, early biochemical studies demonstrated that THR stabilizes 70S ribosomes and inhibits initiation of protein synthesis under specific conditions, such as IF-dependent binding of initiator tRNA to the 70S ribosomes, but not IF-independent 70S or any type of 30S initiation complex formation (7,11). However, addition of THR does not inhibit the ability of pre-formed 70S initiation complexes (containing accommodated fMet-tRNA) to react with puromycin, suggesting that the position of the initiator fMet-tRNA in the P site is unperturbed in the presence of ribosome-bound THR (11,12). Moreover, from the previous structure, it became evident that the THR binding site in the vacant ribosome does not directly overlap with any of the tRNA binding sites (8). Nevertheless, the interaction of THR with bridge B2a induces nucleotide C1914 to flip out of H69, taking a conformation that is incompatible with the binding of aminoacyl-tRNAs into the A site (8). Thus, contrary to the concept of being a translation initiation inhibitor (7), Bulkley et al. argued that THR is likely to interfere with the accommodation of an aminoacyl-tRNA into the A site, including the very first round of translation elongation, which could be considered as inhibition of translation initiation (8). However, the progression and stalling of translating ribosomes along mRNA in the presence of THR have never been assessed. Moreover, given that many of the 16S and 23S rRNA nucleotides, especially those constituting DC and B2a bridge, change conformation upon tRNA binding, it is conceivable that the mode of THR binding to the ribosome could be different in the presence of native tRNA substrates. Finally, it is difficult to reconcile how a single nucleotide (C1914) located in a relatively unconfined pocket of the ribosome, having multiple opportunities for deflection to avoid a clash with the A-site tRNA, can prevent binding of a much larger tRNA molecule to the ribosomal A site.
In this work, we set to close these gaps and uncover the molecular mechanism underlying the mode of THR inhibition of translation. Using a combination of in vivo and in vitro techniques, we show that THR targets bacterial ribosomes, and instead of arresting the ribosome at the start codon, it does so at various codons along mRNA templates. Remarkably, on some mRNAs, THR stalls the ribosome at the stop codon. By solving the 2.7 Å-resolution X-ray crystal structure of THR in complex with the T. thermophilus 70S ribosome carrying initiator formyl-methionyl-tRNAifMet, we demonstrate that neither the binding site of the ribosome-bound drug nor the location of the P-site tRNA is affected by the presence of each other. We further determined the 2.7 Å-resolution cryo-EM structure of the Escherichia coli 70S ribosome in complex with the drug and carrying both A- and P-site tRNAs, in line with our biochemical data suggesting that THR can co-exist on the ribosome with the A-site tRNA. Additionally, we show that multiple partial reactions of the translation elongation cycle are affected by THR, with the most pronounced effect observed on the binding stability of A-site tRNA, explaining the internal ribosome stalling events. Our structural data also suggest that THR can arrest ribosomes at termination codons, likely by preventing accommodation of a release factor. Importantly, ribosome stalling at various stages of elongation and termination apparently results in the accumulation of undissociated 70S ribosomes, thereby decreasing the concentration of free ribosomal subunits available for translation initiation. Altogether, our results provide a molecular understanding of the mode of ribosome inhibition by THR.
MATERIALS AND METHODS
Reagents
All reagents and chemicals were obtained from MilliporeSigma (USA). Thermorubin was isolated from Thermoactinomyces antibioticus as reported previously (5) and provided by Dr Francis Johnson.
In vivo detection of translation inhibitors using the pDualrep2 reporter system
For the in vivo bioactivity test (Figure 1B), we used the reporter E. coli BW25113 ΔtolC-pDualrep2 strain as described previously (13,14). Briefly, 1.5 μl of the solutions of THR (10 mg/ml), erythromycin (ERY, 5 mg/ml), and levofloxacin (LEV, 25 μg/ml) in DMSO were applied onto the agar plate that already contained a lawn of the reporter strain. After overnight incubation at 37°C, the plate was scanned by ChemiDoc MP (Bio-Rad, USA) using the ‘Cy3-blot’ mode for RFP fluorescence and ‘Cy5-blot’ for Katushka2S fluorescence.
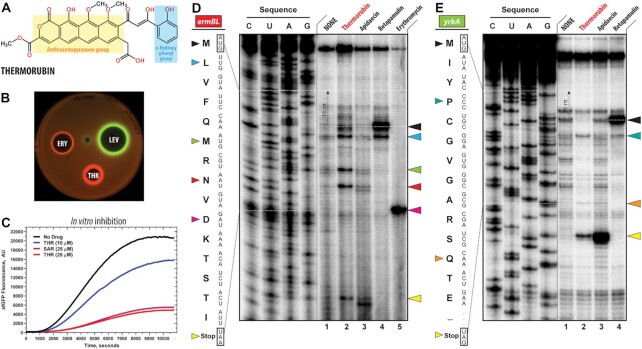
Figure 1.
Thermorubin inhibits protein synthesis both in vivo and in vitro. (A) Chemical structure of thermorubin (THR). (B) Induction of a two-color dual reporter system sensitive to inhibitors of the ribosome progression or inhibitors of DNA replication. Spots of erythromycin (ERY), levofloxacin (LEV) and thermorubin (THR) were placed on the surface of an agar plate containing E. coli ΔtolC cells transformed with the pDualrep2 plasmid (14). Induction of expression of Katushka2S (red) is triggered by translation inhibitors, while RFP (green) is induced upon DNA damage. (C) Time-courses of inhibition of sfGFP synthesis by THR (blue and red curves) and sarecycline (SAR, magenta curve) in the in vitro cell-free translation system. AU, arbitrary units. (D, E) Ribosome stalling by THR and other protein synthesis inhibitors on ermBL(D) and yrbA(E) mRNA templates as revealed by reverse transcription inhibition (toe-printing) in a recombinant cell-free translation system. Nucleotide sequences of wild-type ermBL and yrbA genes, and the corresponding amino acid sequence, are shown on the left in each panel. Sequencing lanes (C, U, A, G) are on the left of each gel. Due to the large size of the ribosome, the reverse transcriptase used in the toe-printing assay stops 16 nucleotides downstream of the codon located in the P site. Black arrowhead marks the translation start site (retapamulin control, lane 4). Blue, green, red, teal, and orange arrowheads point to the THR-induced arrest sites within the coding sequences of ermBL or yrbA mRNAs. Magenta arrowhead points to the erythromycin-specific stalling site on ermBL only. Yellow arrowheads point to the translation stop site (apidaecin control, lane 3). Note that, similar to apidaecin, THR induces ribosome stalling at the stop codons of ermBL and yrbA mRNAs. Experiments were repeated twice independently with similar results.
In vitro translation inhibition assay
To test the ability of THR to inhibit protein synthesis in vitro (Figure 1C), we used the PURExpress translation system (New England Biolabs) reconstituted from purified components. Translation of superfolder green fluorescent protein (sfGFP) was carried out according to the manufacturer's protocol. The assembled reactions (5 μl) were supplemented with 5 ng of a plasmid encoding sfGFPRNA. Varying concentrations of THR (0, 10 or 25 μM) or sarecycline (SAR, 25 μM) were added to the reaction mixtures, which were then placed in a 384-well black-wall plate, and the progression of each reaction was monitored over 3 h at 37°C by a TECAN microplate scanner with the excitation and emission wavelengths set at 488 nm and 520 nm, respectively.
Toe-printing analysis
The synthetic DNA templates encoding the amino acid sequences for Rst1, Rst2, ErmBL and YrbA open reading frames (ORFs) were generated by polymerase chain reaction (PCR) and AccuPrime Taq DNA Polymerase (Thermo Fisher Scientific, USA). The sequences of the primers used for PCR are shown in Supplementary Table S1. The toe-printing analysis of drug-dependent ribosome stalling analysis (Figure 1D, E; Figure S1) was carried out using Rst1, Rst2, ErmBL and YrbA mRNA templates as previously described (15,16) with minor modifications. Toe-printing reactions were carried out in 5-μl aliquots containing a PURExpress transcription-translation coupled system (New England Biolabs, USA) to which the test template was added (17). The reactions were incubated at 37°C for 20–25 min. Reverse transcription on the templates was carried out using radioactively labeled primer NV1 (Supplementary Table S1). Primer extension products were resolved on 6% sequencing gels as described previously (18). The final concentrations of drugs were: 50 μM THR, 50 μM apidaecin (API), 50 μM retapamulin (RET), and 50 μM erythromycin (ERY).
Sucrose gradient ribosome/polysome profiling
Overnight culture of the E. coli JW5503 ΔtolC (KanR) strain grown at 37°C was seeded into 30 ml of fresh LB media at 1:100 dilution. The cell cultures were further grown in triplicates to log-phase at 37°C before THR was added to growth media at 10x MIC (62.5 μg/ml), and the cultures were incubated at 37°C for 1 h. DMSO was used as a negative control. The cells were pelleted at 10,000 rpm for 10 min at 4°C, and supernatants were discarded. The following cell lysis and sucrose gradient polysome profiling were accomplished as described previously with minor modifications (19). Briefly, the pelleted cells were lysed by the freeze-thaw procedure in liquid nitrogen using lysis buffer containing lysozyme. Next, to complete cell lysis, 10% sodium deoxycholate (15 μl) was added to the lysate, which was then centrifuged at 10,000 rpm for 10 min at 4°C. The recovered supernatants were transferred to clean 1.5-ml tubes and diluted 100-fold with Milli-Q ultra-pure water. To normalize the loading onto sucrose gradients, the absorbance for each sample was determined at 260 nm, and the required volumes were calculated. 13.8 A260 units of each sample were loaded onto an 11-ml 10–40% sucrose gradient and centrifuged at 35,000 rpm for 4 h at 4°C in a SW41 rotor (Beckman Coulter, USA). The resulting sucrose gradient polysome profiles were recorded at 254 nm using ÄKTA Purifier as a continuous-flow UV detector (GE Healthcare, USA). We used the UNICORN software for quantitative analysis of the observed peaks (GE Healthcare, USA). All experiments and controls were repeated in triplicates with nearly identical profiles (Supplementary Figure S2).
Inhibition of in vitro translation of canonical and leadless mRNA templates
Two DNA templates for subsequent in vitro transcription (Supplementary Table S2) were first amplified by PCR. The canonical template contains T7 promoter, 27-nt-long leader (5'-UTR), full-length fLuc gene, and 30-nt-long 3'-UTR. The leaderless template contains no 5'-UTR, and the T7 promoter is immediately followed by the full-length fLuc gene and 30-nt-long 3'-UTR. The PCR-generated DNA templates were used directly in the in vitro cell-free transcription-translation coupled PURExpress system (New England Biolabs). Inhibition of protein synthesis on leader vs. leaderless mRNAs was carried out in the absence or presence of THR at 37°C for 40 min. We used 1% DMSO as a negative control, and 0.5x MIC (3.10 μg/ml), 1× MIC (6.25 μg/ml) and 10× MIC (62.5 μg/ml) of THR. The activity of in vitro synthesized luciferase (Supplementary Figure S3) was measured by VICTOR X5 Multilabel Plate Reader (Perkin Elmer, USA).
X-ray crystallographic structure determination
Wild-type 70S ribosomes from T. thermophilus (strain HB8) were prepared as described previously (20–23). Synthetic mRNA with the sequence 5’-GGC-AAG-GAG-GUA-AAA-AUG-UAA-3’ (24MStop) containing Shine-Dalgarno sequence followed by the P-site methionine and the A-site stop codons (bold) was obtained from Integrated DNA Technologies (Coralville, IA, USA). Wild-type deacylated initiator tRNAifMet was overexpressed and purified from E. coli as described previously (24–27). The complex of the wild-type T. thermophilus 70S ribosome with mRNA, vacant A site, and P-site tRNAifMet was formed as described previously (22,23,25) in the presence of 500 μM THR.
Collection and processing of the X-ray diffraction data, model building, and structure refinement were performed as described in our previous reports (22,23,25,26,28,29). Diffraction data were collected at beamlines 24ID-C and 24ID-E at the Advanced Photon Source (Argonne National Laboratory). A complete dataset was collected using 0.979Å X-ray irradiation at 100 K from multiple regions of the same crystal, using 0.3-degree oscillations. Raw data were integrated and scaled using the XDS software package (Feb 5, 2021) (30). Molecular replacement was performed using PHASER from the CCP4 program suite (version 7.0) (31). The search model was generated from the previously published structures of the T. thermophilus 70S ribosome with bound mRNA and aminoacylated tRNAs (PDB entry 6XHW (25)). Initial molecular replacement solutions were refined by rigid-body refinement with the ribosome split into multiple domains, followed by positional and individual B-factor refinement using the PHENIX software (version 1.17) (32). Non-crystallographic symmetry restraints were applied to four parts of the 30S ribosomal subunit (head, body, spur, and helix 44) and four parts of the 50S subunit (body, L1-stalk, L10-stalk and C-terminus of the L9 protein). Structural models were built in Coot (version 0.8.2) (33). The statistics of data collection and refinement are compiled in Supplementary Table S3.
Sample preparation for cryo-EM, data acquisition and image processing
The E. coli 70S ribosomes, isolated from strain MRE600, were prepared as previously described with some modifications (34). Briefly, the cells were washed in buffer containing 20 mM Tris–HCl pH 7.4, 10.5 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA, 6 mM β-mercaptoethanol, and DNase I (16 U/g cells) and then lysed using a microfluidizer LM20-30 (Microfluidics, Westwood, MA). The lysate was cleared by centrifugation at 16,000 rpm (30,600 × g) for 1 h at 4°C and filter-sterilized through a 0.45 μm filter. To remove fine particles and residual debris from the supernatant, the lysate was spun at 30,000 rpm (104,000 × g) in a Type 45Ti rotor (Beckman Coulter, USA) for 30 min at 4°C. To isolate ribosomes, the lysate was layered onto a 1.1 M sucrose cushion buffer (20 mM Tris–HCl pH 7.4, 500 mM NH4Cl, 10.5 mM MgCl2, 0.5 mM EDTA, 6 mM β-mercaptoethanol) and spun at 43,000 rpm (214,000 × g) in a Type 45Ti rotor (Beckman) for 19 h at 4°C. Ribosome pellets were resuspended in 20 mM Tris–HCl pH 7.4, 10.5 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA and 6 mM β-mercaptoethanol. Ribosomes were then purified through 10–40% sucrose density gradients in a SW32 rotor (Beckman) at 23,000 rpm (90,000 × g) at 4°C for 13 h. The fractions containing 70S ribosomes were collected, diluted to adjust Mg2+ concentration to 10 mM, and concentrated by centrifugation at 43,000 rpm (214,000 × g) at 4°C. Pure 70S ribosomes were resuspended and brought to the final buffer containing 10 mM Tris–HCl pH 7.4, 10 mM MgCl2, 60 mM NH4Cl, and 6 mM β-mercaptoethanol, flash-frozen in liquid nitrogen and stored at –80°C.
To determine the cryo-EM structure of the E. coli 70S-THR complex with tRNAs, we incubated 2 μM E. coli 70S ribosomes, 8 μM 24-MF mRNA, 8 μM fMet-tRNAifMet in 1× ribosome buffer (5 mM Tris–HCl pH 7.4, 60 mM NH4Cl, 10 mM MgCl2, 6 mM β-mercaptoethanol) at 37°C for 10 min. Then, 200 μM THR was added and the complex was incubated at room temperature for 10 min. Finally, 15 μM Phe-tRNAPhe was added and incubated for an additional 10 min at room temperature.
Quantifoil R2/1 gold 200 mesh grids (Electron Microscopy Sciences) were glow-discharged for 30 s in an (H2O2)-atmosphere using the Solarus 950 plasma cleaner (Gatan). The mixture (4 μl), containing 2 μM E. coli 70S-THR carrying A- and P-site tRNAs, was applied onto grids, blotted in 85% humidity at 22°C for 18 seconds, and plunged-frozen in liquid nitrogen-cooled ethane using a Leica EM GP2 cryo-plunger. Grids were transferred into a Titan Krios G3i electron microscope (ThermoFisher Scientific) operating at 300 keV and equipped with a Falcon3 direct electron detector camera (ThermoFisher Scientific). The image stacks (movies) were acquired with a pixel size of 0.86 Å/pixel. Data collection was done in the EPU software (ThermoFisher Scientific) setup to record movies with 39 fractions with a total accumulated dose of 40 e−/Å2/movie. A total of 10,032 image stacks were collected with a defocus ranging between –1 to –2.3 μm. The statistics of data acquisition are summarized in Supplementary Table S4.
Data processing was done in cryoSPARC 3.3.2 (35). The image stacks were imported into cryoSPARC and gain-corrected. Image frames (fractions) were motion-corrected with dose-weighting using the patch motion correction, and patch contrast transfer function (CTF) estimation was performed on the motion-corrected micrographs. Based on relative ice thickness, CTF fit, length, and curvature of motion trajectories, 9,852 micrographs were selected for further processing (Supplementary Figure S4).
982,190 particles were picked using the circular ‘blob’ picker in cryoSPARC and were filtered based on defocus adjusted power and pick scores to 972,828 particles. Then, particles were subjected to two rounds of reference-free two-dimensional (2D) classification. After discarding bad particles, 613,586 particles were selected from 2D classification and used to generate the ab-initio volume. Using ‘heterogeneous refinement’ in cryoSPARC with five groups, particles were further classified into two class averages. Both class averages represent the 70S ribosome with density for bound tRNAs. Particles were therefore combined (565,280) and classified based on focused 3D variability analysis (3DVA) with a mask around the A-site tRNA which allowed to remove 55,534 ‘bad’ particles with broken density for tRNAs. This approach yielded two main class averages containing 310,702 ribosome particles with A- and P-site tRNAs, and 199,044 ribosome particles containing P-site tRNA and no A-site tRNA. Particles from each class were re-extracted to full-size (512 × 512-pixel box), followed by non-uniform and CTF refinement in cryoSPARC. The Fourier Shell Correlation (FSC) curves were calculated using the cryo-EM validation tool in Phenix 1.19.2 for even and odd particle half-sets masked with a ‘soft mask’ excluding solvent (36). The nominal resolution of the reconstructions using the FSC-cutoff criterion of 0.143 is 2.7 Å for the 70S-THR ribosome complexed with A-site Phe-tRNAPhe and deacylated P-site tRNAifMet, and 2.8 Å for the 70S-THR ribosome complexed with P-site tRNAifMet (Supplementary Figure S5).
Model building and refinement of the E. coli 70S ribosome
As a starting model, the 30S and 50S subunits were taken from the high-resolution structure of the E. coli 70S ribosome (PDB entry 7K00 (37)) and rigid-body docked into the 2.7 Å-resolution cryo-EM map of the 70S-THR with A- and P-site tRNAs using UCSF Chimera 1.14 (38). The fMet-tRNAifMet and Phe-tRNAPhe were taken from PDB entry 6XHW (25), rigid-body fitted into the EM density, and adjusted in Coot (39). The complete model of the E. coli 70S ribosome, including modified nucleotides in rRNAs and tRNAs, ordered solvent and bound THR, A-site Phe-tRNAPhe and P-site tRNAifMet was real-space refined into the EM map for five cycles using Phenix 1.19.2 (40) with global energy minimization and group ADP refinement strategies along with base pair restraints for rRNA and tRNAs, together with Ramachandran and secondary structure restraints. The resulting model of the E. coli 70S-THR with Phe-tRNAPhe in the A site and deacylated tRNAifMet in the P site was validated using the comprehensive validation tool for cryo-EM in Phenix 1.19.2 (36). The cryo-EM data collection, refinement, and validation statistics are compiled in Supplementary Table S4.
Figure generation
All figures showing atomic models were rendered using the PyMol software (www.pymol.org), UCSF Chimera 1.14 (38) or Chimera X 1.2 (41) and assembled with Adobe Illustrator (Adobe Inc.).
Sample preparation for rapid kinetics measurements
E. coli 70S ribosomes, initiation factors (IF1, IF2 and IF3), elongation factors (EF-Tu and EF-G), aminoacylated initiator fMet-tRNAifMet, aminoacylated [14C]Phe-tRNAPhe, tRNAPhe(Prf16/17) and [14C]Val-tRNAVal were prepared as described previously (42–45). MF-mRNA containing AUG-UUU nucleotide sequence encoding for Met-Phe dipeptide and MVF-mRNA containing AUG-GUU-UUU nucleotide sequence encoding for Met-Val-Phe tripeptide were obtained by T7 transcription. The full sequences for the DNA templates used for T7 transcription of MF- and MVF-mRNAs are:
MF-mRNA: CGAATTTAATACGACTCACTATAGGGAATTCAAAAATTTAAAAGTTAACAGGTATACATACTATGTTTACGATTACTACGATCTTCTTCACTTAATGCGTCTGCAGGCATGCAAGC
MVF-mRNA: CGAATTTAATACGACTCACTATAGGGAATTCAAAAATTTAAAAGTTAACAGGTATACATACTATGGTTTTTATTACTACGATCTTCTTCACTTAATGCGTCTGCAGGCATGCAAC
The sequences of T7 promoters are italicized; sequences coding for Met-Phe or Met-Val-Phe are bold. All reactions were performed in the TAKM7 buffer (50 mM Tris–HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl and 7 mM MgCl2) unless otherwise stated.
For a typical 1-ml reaction of initiation complex formation, 1 μM 70S ribosomes were incubated with 3 μM mRNA, 2 μM fMet-tRNAifMet, 1.5 μM of IF1, IF2 and IF3 in the TAKM7 buffer supplemented with 1 mM GTP and 2 mM DTT for 1 hour at 37°C. Ternary complexes of aminoacyl-tRNA•EF-Tu•GTP were prepared by preincubation of EF-Tu with 1 mM GTP, 3 mM phosphoenolpyruvate, 2 mM DTT, 1% pyruvate kinase in the TAKM7 buffer for 15 min at 37°С, followed by addition of aminoacyl-tRNA and incubation for 1 minute. Pre-translocation ribosome complexes were formed by mixing 70S initiation and ternary complexes with subsequent incubation for 2 min at 37°C. For purification, the concentration of Mg2+ ions was increased to 21 mM, and the mixture of pre-translocation complexes was layered onto a 400-μl 1.1 M sucrose cushion (prepared in the TAKM21 buffer), followed by centrifugation in an SW55 rotor (Beckman Coulter, USA) at 55,000 rpm for 3 h at 4°C. The pellet was dissolved in the TAKM21 buffer, flash frozen in liquid nitrogen, and stored at –80°C. fMet-tRNAifMet, Phe-tRNAPhe and Val-tRNAVal were purified by HPLC and stored at –80°C. To prepare Phe-tRNAPhe(Prf16/17), 8 μM of deacylated tRNAPhe(Prf16/17) was incubated with 0.2 mM phenylalanine, 3 mM ATP, 6 μM β-mercaptoethanol, 40 nM phenylalanine-specific tRNA-synthetase, and 40 nM tRNA nucleotidyltransferase in the TAKM7 buffer for 30 min at 37°C. Phe-tRNAPhe(Prf16/17) was purified on HiTrap Q column with 0.2 to 1 M NaCl gradient in 10 mM Tris–HCl pH 7.6, 10 mM MgCl2 and stored at –80°C. Where necessary, 33 μM THR (final concentrations after mixing are given throughout) was added to the 70S ribosomes, initiation, or pre-translocation complexes and incubated on ice for 2 min.
Rapid kinetics measurements
To monitor the time course of aminoacyl-tRNA interaction with the A site of the ribosome, we rapidly mixed 0.1 μM Phe-tRNAPhe(Prf16/17)•EF-Tu•GTP ternary complex with 0.4 μM initiation ribosome complexes programmed with MF-mRNA at 20°C. To study the pre-steady-state kinetics of translocation, we rapidly mixed 0.06 μM ribosome pre-translocation complexes containing deacylated tRNAifMet in the P site and fluorescently labeled fMet-Phe-tRNAPhe(Prf16/17) (46) in the A site with 2 μM EF-G in the presence of 1 mM GTP at 37°C. Fluorescence was recorded using SX-20 stopped-flow apparatus (Applied Photophysics). Proflavine fluorescence was excited at 460 nm and measured after passing through a KV495-nm cut-off filter. Light-scattering experiments were carried out at 430 nm, followed by detection at a 90° angle without the cut-off filter. Samples were rapidly mixed in equal volumes (60 μl). Time courses were obtained by averaging 5-to-7 individual transients. Data were evaluated by fitting with a two-exponential function with characteristic time constants (kapp1, kapp2), amplitudes (A1, A2) and final signal amplitude (F∞) according to equation F = F∞ + A1*exp(–kapp1*t) + A2*exp(–kapp2*t), where F is the fluorescence at time t. All calculations were performed using the GraphPad Prism 9.3.1 software (GraphPad Software, Inc).
A-site peptidyl-tRNA dissociation constant measurements
Experiments and calculations were carried out according to the previously published methodology (47) in the HAKM10 buffer (50 mM HEPES pH 7.5, 70 mM NH4Cl, 30 mM KCl and 10 mM MgCl2) at 37°C. Briefly, to induce the dissociation of fMet-[14C]Val-tRNAVal from the A site of purified pre-translocation complexes (0.25 μM), the Mg2+ concentration was adjusted from 21 to 10 mM in the sample, and the amount of peptidyl-tRNA bound to the A site was determined by nitrocellulose filtration at different incubation times. Dissociation elemental rate constant (koff) and equilibrium dissociation constant (Kd) were calculated from time courses of dissociation by numerical integration (47). The following kinetic model was used: A ⇔ B + C, and B ⇒ D, where A denotes ribosomes with peptidyl-tRNA bound to the A site; B, unbound peptidyl-tRNA; C, ribosomes with unoccupied A site; D, hydrolyzed peptidyl-tRNA. The elemental association rate constant (kon) was calculated from the ratio Kd = koff/kon.
RESULTS AND DISCUSSION
Thermorubin targets translating ribosomes in vivo
The main goal of this study was to explore the inhibitory properties of the anthracenopyranone antibiotic thermorubin (THR, Figure 1A) and uncover its mechanism of action. Although THR has long been known to inhibit protein synthesis in a crude cell lysate (7), it was also shown to exhibit potent activity against aldose reductase (48), suggesting that ribosome might not necessarily be the only cellular target for this drug. To assess whether protein synthesis is the primary target of THR in vivo, we used an E. coli-based reporter system designed to rapidly screen inhibitors targeting either protein synthesis or DNA replication (14). In this assay, sub-inhibitory concentrations of antibacterial compounds that stall translation, such as the macrolide erythromycin (ERY), induce expression of the far-red fluorescent protein reporter Katushka2S (Figure 1B, ERY, red pseudocolor ring). Compounds that trigger the SOS response, such as inhibitors of DNA gyrase (e.g. levofloxacin (LEV)), induce expression of the Red Fluorescent Protein (RFP) reporter (Figure 1B, LEV, green pseudocolor ring). Both reporters are encoded by the same pDualrep2 plasmid in the E. coli BW25113 cells. The test antibiotic is applied as a drop or a disc onto a lawn of these cells, and its intracellular target could be determined based on the reporter being induced. When E. coli cells carrying the reporters were exposed to THR, the expression of the far-red fluorescent protein Katushka2S, but not of RFP, was observed, indicating that the drug specifically arrests ribosomes and targets protein synthesis in vivo (Figure 1B, THR, red pseudocolor ring).
Thermorubin is a potent inhibitor of protein synthesis in vitro
To further confirm that THR acts as a translation inhibitor, we tested its ability to interfere with in vitro protein synthesis. The addition of THR to the cell-free transcription-translation coupled system reconstituted from purified components resulted in a dose-dependent inhibition of the synthesis of superfolder green fluorescent protein (sfGFP) (Figure 1C). In these experiments, addition of 25 μM THR decreased sfGFP synthesis by ∼10 fold, comparable to the inhibitory properties of the tetracycline-class antibiotic sarecycline (SAR), which we used as a positive control. This data suggests that THR is a potent translation inhibitor and provides a promising foundation for future ribosome-targeting drug development.
It remains unclear, however, which step of translation – initiation, elongation, or termination – is inhibited by THR. Therefore, we used primer extension inhibition assay (toe-printing) to answer this question. This method is based on the reverse-transcriptase-dependent extension of a radioactively-labeled DNA primer annealed to the 3’-end of the translated mRNA template. Primer extension reaction terminates when the reverse transcriptase encounters a stalled ribosome. This technique allows us to unambiguously identify drug-induced ribosome stalling site(s) along the mRNA with single nucleotide precisions (16,49). A large set of ribosome-targeting antibiotics has been previously tested by toe-printing using ermBL, yrbA, rst1 or rst2 mRNA templates (16) because their sequences contain codons for almost all proteinogenic amino acids. Therefore, we have chosen the same experimental system to directly compare our THR results with well-established modes of action of other antibiotics.
The addition of THR to the cell-free translation system programmed with ermBL, yrbA, rst1 or rst2 mRNAs resulted in predominant ribosome stalling at various codons within the corresponding open reading frame (ORF) but not at the start codon (Figure 1D, E; Figure S1). These data suggest that, unlike the known initiation inhibitor retapamulin (Figure 1D, lane 4, black arrowhead) (16), THR is not a bona fide initiation inhibitor, as speculated before (7). Interestingly, the ribosomes translate at least two codons before they stall in the presence of THR (Figure 1D, lane 2, blue arrowhead), excluding the possibility of THR acting during the first elongation cycle (at least on the mRNAs tested), as is the case with retapamulin. Also, depending on the mRNA used, THR can cause ribosome stalling at multiple internal codons (Figure 1D, lane 2, blue, green, and red arrowheads; Figure 1E, lane 2, teal and orange arrowheads), which is yet very distinct from the internal codon stalling caused by the macrolide antibiotic erythromycin on the ermBL mRNA (Figure 1D, lane 5, magenta arrowhead). Importantly, our experimental toe-printing data showing that THR allows for ribosome progression contradicts the previous structure-inspired idea that THR-dependent re-arrangement of nucleotide C1914 prevents binding of an A-site tRNA and, thus, indiscriminately inhibits every elongation cycle (8). Our data suggest that THR can either co-exist on the ribosome with an A-site tRNA or bind and dissociate from the ribosome during the elongation and act in a context-specific fashion, i.e. preferably bind when particular codons are present in the A site. Perhaps, the most unexpected result of the toe-printing analysis was manifested on the yrbA mRNA template (and to a lesser extent on ermBL, rst1, and rst2 mRNAs), where THR caused the most prominent ribosome stalling at the stop codon (Figure 1D, E, lane 2, yellow arrowhead; Supplementary Figure S1), suggesting that in addition to being an elongation inhibitor on some ORFs, THR acts as a termination inhibitor on the other ones. Thus, the mode of action of THR is similar to a known translation termination inhibitor, apidaecin (Figure 1D, E, lane 4, yellow arrowhead) (50). Subsequent ribosome profiling experiments are needed to provide additional insights into the context-specificity of THR action by analyzing the transcriptome-wide distribution of ribosomes along the actively translated genes.
Ribosome stalling during elongation or termination steps of protein synthesis would lead to the accumulation of undissociated 70S ribosomes, decreasing the number of free 30S and 50S ribosomal subunits, thereby indirectly inhibiting translation initiation. To address this possibility, we performed sucrose gradient ultracentrifugation profiling of crude ribosomes isolated from either untreated or THR-exposed E. coli cells (Supplementary Figure S2). As expected and in agreement with the previously published data (11), we observed a substantial (approximately 4-fold) increase in the amount of 70S particles at the expense of 30S and 50S subunits upon THR treatment in vivo (Supplementary Figure S2, blue versus red traces). However, this result does not allow us to distinguish whether the accumulated 70S-THR ribosome particles are stalled during translation, or whether they correspond to vacant non-translating ribosomes stabilized by THR. The idea that THR can indirectly inhibit translation initiation by depleting the pool of free 30S subunits available for initiation via tethering of 30S and 50S subunits to yield vacant 70S particles was discussed as an unlikely scenario in the previous studies (12). However, we decided to further explore this possibility by exploiting the observation that vacant 70S ribosomes can initiate translation exclusively on leaderless mRNAs (51,52). Therefore, if THR is an inhibitor of canonical translation initiation on 30S subunits, it is not expected to inhibit leaderless mRNA translation. To this end, we compared the extents of inhibition by THR using the in vitro synthesis of luciferase encoded by the leaderless versus canonical mRNAs (Supplementary Figure S3). We observed nearly equal inhibition of translation of both canonical (Supplementary Figure S3A) and leaderless (Supplementary Figure S3B) mRNAs, suggesting that, in the presence of THR, ribosomes stall during the elongation and/or termination but not the initiation stage of translation, which is also manifested by THR-induced accumulation of 70S particles (Supplementary Figure S2).
Thermorubin does not interfere with the P-site tRNA
The exact location of the THR binding site in the context of the vacant 70S ribosome was known from the previous 3.2 Å-resolution structure (8). While laying the foundation for understanding the mechanisms of THR action, nevertheless, this structure represents a physiologically irrelevant complex because, in the cell, antibiotics interact primarily not with vacant but with translating ribosomes associated with ligands, such as mRNAs, tRNAs, nascent protein chain and/or translation factors. Therefore, to address the possibility that the THR binding site on the ribosome is somewhat different or altered in the presence of native tRNA substrate(s), we set to determine structures of bacterial ribosome-THR complexes carrying either a single P-site tRNA or both A- and P-site tRNAs.
First, we crystallized T. thermophilus (Tth) 70S ribosomes in the presence of mRNA, initiator tRNAifMet and THR (Figure 1A) and determined its structure at 2.7 Å resolution (Figure 2A, 3A; Table S3). The better-quality electron density maps allowed us to visualize and unambiguously model the drug as well as the 16S and 23S nucleotides adjacent to its binding site. The inclusion of mRNA and P-site tRNA in our ribosome complexes likely provided additional stabilization of the ribosome that, in turn, contributed to the ∼0.5 Å higher resolution compared to the previous structure at 3.2 Å (8). Most importantly, the binding position of THR in our structure is identical to the one observed previously for THR bound to the vacant Tth 70S ribosome in the absence of mRNA and P-site tRNA (Figure 3B) (8). This suggests that the presence of the initiator tRNA in the P site does not affect the general mode of THR binding to the ribosome.
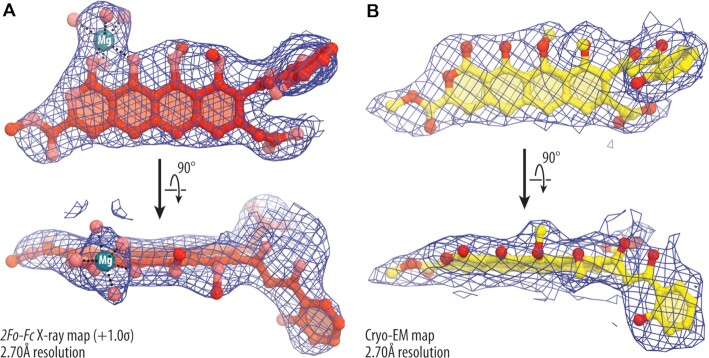
Figure 2.
X-ray and cryo-EM maps of ribosome-bound thermorubin. (A) 2Fo-Fc Fourier electron density map of THR in complex with the T. thermophilus 70S ribosome (blue mesh) carrying only the P-site tRNA viewed from two different angles. The refined model of THR is displayed within its respective electron density map contoured at 1.0σ. (B) Cryo-EM map of the ribosome-bound THR in complex with the E. coli 70S ribosome (blue mesh) carrying both A- and P-site tRNAs. Carbon and oxygen atoms are colored red and salmon in (A), and yellow and red in (B), respectively. Magnesium ion is colored in teal. Note that the locations of all chemical moieties of THR can be unambiguously determined from the electron density map.
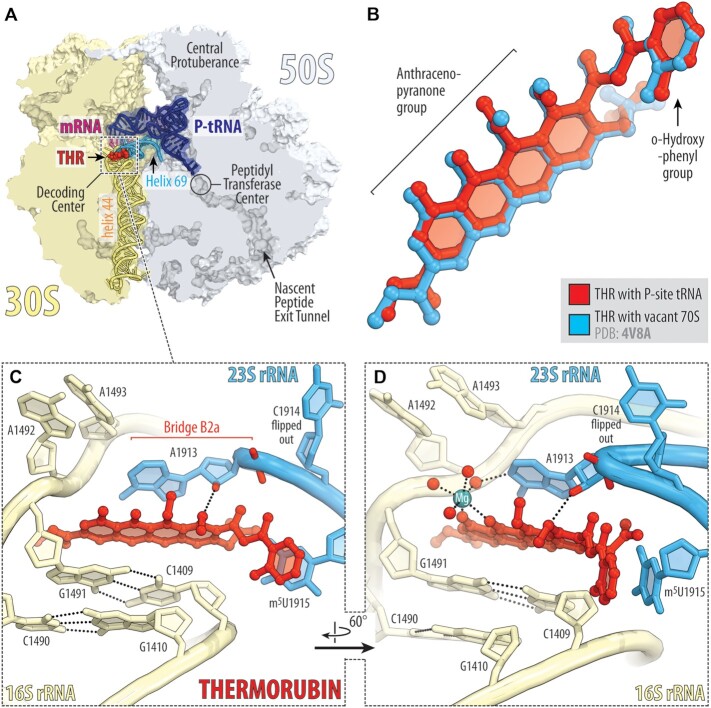
Figure 3.
X-ray crystal structure of 70S-THR complex with P-site tRNA. (A) Overview of the drug-binding site (red) in the T. thermophilus 70S ribosome carrying mRNA (magenta) and deacylated initiator tRNA in the P site (navy) viewed as a cross-cut section through the nascent peptide exit tunnel. The 30S subunit is shown in light yellow; the 50S subunit is light blue. Helices 69 and 44 of the 23S and 16S rRNAs are highlighted in blue and pale yellow, respectively. (B) Superposition of the ribosome-bound THR in the presence of P-site tRNA (red) with the previous structure of THR bound to a vacant ribosome (blue, PDB entry 4V8A (8)). The structures were aligned based on helix 44 of the 16S rRNA. (C, D) Close-up views of the THR bound in the decoding center of the 70S ribosome (E. coli numbering of the 23S and 16S rRNA nucleotides is used). Potential H-bond interactions are indicated with dashed lines. Note that binding of THR to the 16S–23S bridge B2a causes nucleotide C1914 to flip out of its usual location in the absence of A-site tRNA. Also note that THR binding, even with the empty A site, induces nucleotides A1492 and A1493 of the 16S rRNA to flip out of helix 44.
In our structure, THR binds in the loop at the top of h44, where it forms extensive π-π stacking interactions with the surrounding nucleobases of rRNAs (Figure 3C). The aromatic tetracyclic moiety of the drug is sandwiched between the C1409:G1491 base pair of the 16S rRNA on one side and nucleobase A1913 of the 23S rRNA on the other (Figure 3C). Despite the abundance of hydrogen-bond (H-bond) donors and acceptors in the THR molecule, there is only a single H-bond that directly involves the drug with the 2’-hydroxyl of residue A1913 of the 23S rRNA. Other electrostatic interactions of THR with the ribosome are mediated by a water-coordinated magnesium ion visible in the electron density map (Figures 2A and 3D). In the drug-free Tth 70S ribosome, nucleotide C1914, located at the tip of H69, stacks upon the next nucleotide m5U1915 (Supplementary Figure S6A). However, in the presence of THR, the m5U1915 nucleobase stacks with the aromatic orthohydroxyphenyl moiety of the drug, thereby taking the place of C1914 and causing it to flip out (Figure 3D; Figure S6B, C). THR binding to the DC of the ribosome also re-arranges nucleotide A1913, which becomes engaged in extensive stacking interactions with the drug and can no longer interact with the incoming A-site tRNA (Supplementary Figure S6C). Similar re-orientations of nucleotides A1913 and C1914 were observed previously in the 70S-THR structure featuring the vacant 70S ribosome (8).
Thermorubin and A-site tRNA can co-exist on the ribosome
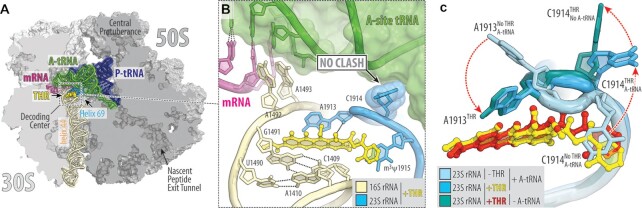
In principle, the flipped-out conformation of C1914 is incompatible with a tRNA in the ribosomal A site (Supplementary Figure S6B), as suggested before (8). However, our toe-printing data contradict this hypothesis and indicate that the ribosome is able to translate ORFs in the presence of THR (Figure 1D, E; Figure S1) – a result that could hardly be reconciled if THR is incompatible with an accommodated A-site tRNA. This observation suggests that, in the presence of THR, accommodation of an aa-tRNA into the ribosomal A site either causes re-orientation of nucleotide C1914 and pushes it out of the way, or repositions the drug into a previously unseen conformation, or both.
Therefore, in order to directly address the possibility of A-tRNA-induced C1914 re-orientation, we determined the 2.7 Å-resolution cryo-electron microscopy (cryo-EM) structure of the E. coli 70S ribosome carrying Phe-tRNAPhe and tRNAifMet in the A and P sites, respectively, and also bound with THR (Figure 4A; Table S4). Contrary to the previous hypothesis and in silico predictions (Supplementary Figure S6B), our cryo-EM structure reveals fully accommodated Phe-tRNAPhe and THR both bound to the ribosome side-by-side (Figures 2B and 4A; Figure S7). Interestingly, in the presence of THR, we observed a ∼90-degree deflection of nucleotide C1914 towards the solvent side relative to its flipped-out conformation seen in the previous structure, allowing Phe-tRNAPhe to fully accommodate into the A site (Figure 4B, C). At the same time, most of the interactions of THR with the ribosome were retained in the presence of the A-site tRNA, and its binding site remained undisturbed (Figure 4C). This structure shows that an aa-tRNA can accommodate into the ribosomal A site even in the presence of ribosome-bound THR, rationalizing our in vitro toe-printing data (Figure 1D, E).
Figure 4.
Cryo-EM structure of 70S-THR complex with both A- and P-site tRNAs. (A) Overview of the THR binding site (yellow) in the E. coli 70S ribosome containing mRNA (magenta) and full-length Phe-tRNAPhe (green) and tRNAifMet (navy) in the A and P sites, respectively, viewed as a cross-cut section through the ribosome. (B) Close-up view of THR bound in the decoding center in the presence of fully accommodated A-site tRNA. Note that nucleotide C1914 adopts a previously unseen conformation, in which it does not interfere with tRNA binding. (C) Structural re-arrangements in the decoding center (red dashed arrows) upon THR binding and tRNA accommodation. Superpositioning of the previous structure of drug-free ribosome carrying both A- and P-site tRNAs (PDB entry 6XHW (25)) with the two new 70S-THR structures with or without A-site tRNA. The structures were aligned based on helix 44 of the 16S rRNA. Note that in the presence of THR and A-site tRNA, nucleotide C1914 flips halfway out from its normal position in Helix 69 of the 23S rRNA, allowing for tRNA accommodation (light blue to blue transition). However, in the absence of A-site tRNA, nucleotide C1914 is unrestricted and fully flips out (blue to teal transition).
Thermorubin affects multiple steps of translation elongation
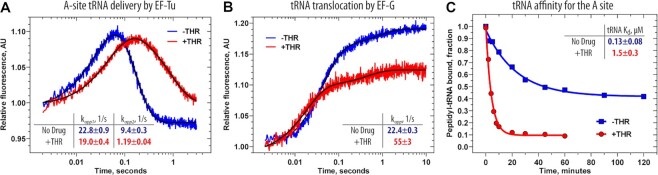
Altogether our toe-printing and structural data suggest that THR does not inhibit translation initiation and binding of tRNA to the ribosomal A site. However, the observed ribosome stalling within the ORF of mRNA templates used for toe-printing (Figure 1D, E, lane 2, blue, green, red, teal, and orange arrowheads) suggests that one or more steps of elongation could be impaired by THR. As the elongation cycle consists of (i) A-site tRNA delivery, (ii) peptide bond formation, and (iii) translocation steps, we set to assess the effects of THR on each of these individual steps using a reconstituted in vitro translation system. First, to monitor the effect of THR on tRNA delivery, we used a stopped-flow fluorescence detection assay to follow the delivery of Phe-tRNAPhe(Prf16/17) in the form of the ternary complex (EF-Tu•GTP•Phe-tRNAPhe(Prf16/17)) to the initiated ribosome complexes bearing the UUU codon in the vacant A site (Figure 5A). An increase in a characteristic biphasic fluorescence change represents the initial binding of the ternary complex to the ribosome and subsequent codon recognition, which induces GTPase activation and results in GTP hydrolysis by EF-Tu. The subsequent decay of the high fluorescence intermediate is related to the release of tRNA from the GDP-bound form of EF-Tu and accommodation of the tRNA in the A site of the ribosome (53). Two-exponential fitting of the fluorescence curves allowed us to measure the rate constants of initial ternary complex binding and subsequent tRNA accommodation. Interestingly, THR does not affect ternary complex binding (kapp1(no drug) = 22.8 ± 0.9 s−1 versus kapp1(THR) = 19.0 ± 0.4 s−1), whereas the rate of tRNA accommodation decreases by approximately eight folds (kapp2(no drug) = 9.4 ± 0.3 s−1 versus kapp2(THR) = 1.19 ± 0.04 s−1) suggesting that THR slows down aa-tRNA accommodation.
Figure 5.
Ribosome-bound THR impairs the elongation cycle. (A) Pre-steady state kinetics of A-site tRNA binding upon interaction of the ternary complex EF-Tu•GTP•Phe-tRNAPhe(Prf16/17) (0.1 μM) with the 70S initiated ribosomes (0.4 μM) containing fMet-tRNAifMet in the P site. Shown are time courses in the absence (blue) and presence (red) of THR. (B) Pre-steady state kinetics of translocation upon interaction of the 70S pre-translocation ribosome complexes (60 nM) containing deacylated-tRNAifMet in the P site and fMet-Phe-tRNAPhe(Prf16/17) in the A site with EF-G (2 μM). Color scheme is the same as in panel A. Each time course represents the average of five to seven experimental replicates. Standard deviations associated with the kinetics were calculated using GraphPad Prism software. (C) Time courses of dissociation of peptidyl-tRNA from the A site of the 70S pre-translocation ribosome complexes in the absence (blue) or presence (red) of THR.
Accommodation of the aa-tRNA into the A site of the PTC leads to peptide bond formation by fast spontaneous transfer of a peptidyl moiety of the P-site tRNA onto the A-site tRNA. Thus, the final level of dipeptide formed depends on the amount of correctly accommodated A-site tRNA. We found that THR-containing ribosomal complexes are less effective in forming fMet-[14C]Phe-dipeptidyl-tRNAPhe, with only 59% of dipeptide formed relative to the drug-free ribosomes, which is likely due to the inhibitory effect of THR on aa-tRNA accommodation.
Peptide bond formation is followed by a translocation event that comprises large-scale movements of mRNA and tRNAs through the ribosome, marking the end of each elongation cycle. During this step, the deacylated-tRNA moves from the P to the E site, which then rapidly dissociates from the ribosome. Simultaneously, peptidyl-tRNA translocates from the A to the P site maintaining codon-anticodon interactions with the mRNA that results in ribosome movement by one codon along the mRNA. To assess whether THR affects translocation, we monitored the kinetics of this process by changes in fluorescence upon addition of the translational GTPase EF-G to the pre-translocation ribosome complex containing fluorescently labeled fMet-Phe-tRNAPhe(Prf16/17) in the A site and deacylated tRNAifMet in the P site. The addition of EF-G to such complex results in a two-step change in fluorescence intensity with a predominant fast step (more than 80% of the total signal amplitude). The addition of THR increases the rate of peptidyl-tRNA translocation from the A to the P site by 2.5 fold (kNo Drug = 22.4 ± 0.3 s−1 versus kTHR = 55 ± 3 s−1), whereas the amplitude of the total fluorescence signal decreases by 30% (Figure 5B). Both of these observations indicate that A-site tRNA becomes destabilized in the presence of THR (similar to (47,54)), resulting either in dissociation of the dipeptidyl-tRNA from the ribosome before translocation or in its more rapid translocation to the P site upon addition of EF-G. These data agree with the observed slowed-down accommodation of aa-tRNA, suggesting that, although THR does not preclude tRNA binding to the ribosomal A site, THR-induced re-arrangements of rRNA nucleotides are likely responsible for the observed poor stability of the A-site tRNA. Indeed, our cryo-EM data reveals that in the presence of THR, nucleotide A1913 of the 23S rRNA is unable to form H-bond with the 2'-OH of the ribose of nucleotide 37 in the A-site tRNA rationalizing the lower stability of A-site tRNA.
Thermorubin decreases the binding affinity of tRNA to the A site
Purine nucleotide at position 37 in most tRNAs is usually heavily post-transcriptionally modified and plays a crucial role in maintaining the structure of the tRNA anticodon loop and stability of the codon-anticodon duplex (47). Removal of the modification or replacement of purine at this position with pyrimidine dramatically decreases the enthalpy of tRNA-ribosome interactions (47), suggesting that the observed loss of H-bond between this nucleotide and 23S rRNA due to THR-induced re-arrangement of A1913 (Supplementary Figure S6A, B) might influence the thermodynamic properties of A-site tRNA binding. To evaluate the effect of THR on the affinity of peptidyl-tRNA to the A site, we determined dissociation constants (47). Under appropriate conditions, A-site peptidyl-tRNA can dissociate reversibly from the purified pre-translocation ribosome complexes depending on its binding stability (55), allowing equilibrium dissociation constant (Kd) and the rate constants of tRNA dissociation from (koff) and association with the A site (kon) to be calculated. When the drug-free ribosome complexes containing A-site peptidyl-tRNAVal were incubated at 37°C and 10 mM Mg2+, ∼50% of the peptidyl-tRNA remained bound to the ribosomal A site even after 2 h of incubation (Figure 5C, blue curve). In contrast, dissociation of the same amount of peptidyl-tRNAVal from the THR-containing ribosome complexes occurred significantly faster, leaving only about 10% of the peptidyl-tRNA bound to the ribosomes after 10 min of incubation (Figure 5C, red curve). The calculated affinity of peptidyl-tRNAVal to the A site of the THR-bound ribosome is 12 times lower compared to the drug-free ribosome complexes (Kd(no drug) = 0.13 ± 0.08 μM versus Kd(THR) = 1.5 ± 0.3 μM) (Figure 5C; Table S5), yet again emphasizing that A-site tRNA binding is negatively affected by THR.
Ribosome-bound thermorubin likely interferes with the activity of class-I RFs
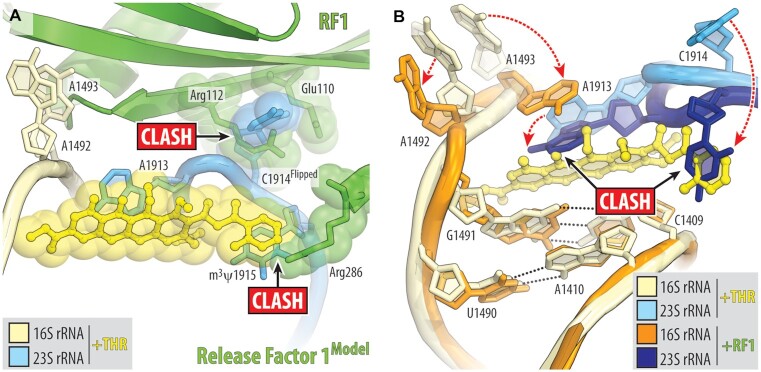
THR-dependent ribosome stalling at the stop codons of rst1, rst2, ermBL and yrbA mRNA templates used in our toe-printing assay (Figure 1D, E; Figure S1) suggests that THR can also act as an inhibitor of translation termination. In light of this finding, we explored whether THR simply prevents binding of class-I release factors (RF1 and/or RF2) to the A site or, vice versa, traps class-I RFs on the ribosome, similar to the mode of action of the peptide antibiotic apidaecin (50), thereby making translation termination impossible in either case. To distinguish between these two possibilities, we attempted to obtain structures of the 70S-RF1 complex with THR using ribosomes from T. thermophilus or E. coli and either X-ray crystallography or cryo-EM approaches. Despite numerous attempts, our structures showed that RF1 did not bind to the 70S–THR complexes, suggesting that THR sterically hinders the delivery of RF1 into the A site. Superimposition of the 70S-THR structure with the previous structures of ribosome-bound RF1 (Figure 6A) reveals a relatively small sterical hindrance between the ribosome-bound drug and Arg286 of the RF1 bound in the A site. Although even a small sterical overlap between the drug and RF1 should prevent binding of either one to the ribosome, the Arg286 residue does not interact with anything, is not restricted by the surrounding residues, and, in principle, shall be able to reorient to avoid the clash with THR. However, the THR-induced flipped-out conformation of nucleotide C1914 of the 23S rRNA produces extensive sterical clash with residues Glu110, Arg112, Lys157 and Ile293 of the A-site-bound RF1, suggesting that RF is unlikely to be able to accommodate into the A site in the presence of THR (Figure 6A) explaining why we were unable to obtain structures of such complexes. Moreover, binding of RF to the ribosome induces significant re-arrangements of nucleotides A1492, A1493 of the 16S rRNA and A1913 of the 23S rRNA, which, in turn, are incompatible with THR (Figure 6B). Altogether our structural analysis shows that although binding positions of THR and class-I RFs on the ribosome barely overlap, binding of one of these ligands to the ribosome induces mutually exclusive conformations of nucleotides around the DC, preventing binding of the other.
Figure 6.
Ribosome-bound THR likely prevents binding of release factor to the A site. (A) Superimposition of the ribosome-bound RF1 (PDB entry 3D5A (57)) with the new structure of 70S-THR complex carrying both A- and P-site tRNAs. The structures were aligned based on helix 44 of the 16S rRNA. Note that the THR-induced flipped-out conformation of C1914 would cause steric clashes with RF1 bound in the A site. (B) Structural re-arrangements in the decoding center that are required for RF1 to bind to the 70S ribosome (dashed red arrows). Note that binding of RF1/2 to the ribosome requires nucleotides A1492, A1493 of the 16S rRNA and A1913 of the 23S rRNA to adopt positions that are incompatible with ribosome-bound THR.
CONCLUSION
Altogether, our data suggest that THR is a potent inhibitor of protein synthesis that targets the elongation and/or termination steps of translation. THR could also indirectly inhibit initiation by depleting the pool of individual 30S subunits. Interestingly, the effects of THR binding on translation resemble those observed from mutations in bridge B2a (56), yet again suggesting interference with the functioning of bridge B2a to be the primary mode of action of THR. Given the current scarcity of new chemical scaffolds for new antibiotic development, we believe that the structural and biochemical data reported here will attract attention to this ‘forgotten’ antibiotic and could inform further attempts to improve THR via rational drug design. Perhaps through chemical derivatizations, the problems of THR’s poor solubility and low thermostability may be solved to make THR a more suitable candidate for clinical trials. It is intriguing, however, that a compound with such poor solubility and low thermostability is naturally produced by a bacterial host whose optimal growth temperature is between 48 and 53°C (5). This makes us wonder if THR is naturally complexed with an adjuvant, which stabilizes and solubilizes it in the cytoplasm of the producing bacterial cells. An alternate purifying strategy might help identify THR in its native form, which may be more soluble and/or potent than the parent compound.
DATA AVAILABILITY
Coordinates and structure factors were deposited in the RCSB Protein Data Bank under the accession code 8EKB for the T. thermophilus 70S ribosome crystal structure in complex with THR, mRNA, and deacylated P-site tRNAifMet. The cryo-EM map for the E. coli 70S ribosome in complex with THR, mRNA, aminoacylated A-site Phe-tRNAPhe, and deacylated P-site tRNAifMet was deposited in the Electron Microscopy Data Bank (EMDB) under the accession code EMD-28197, with the coordinates deposited in the RCSB Protein Data Bank under the accession code 8EKC.
All previously published structures that were used in this work for structural comparisons were retrieved from the RCSB Protein Data Bank: PDB entries 4V8A, 6XHW, and 3D5A.
No sequence data were generated in this study.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Mariia Rybak at the University of Texas Medical Branch for preparing the 70S-THR ribosome complex bound to Phe-tRNAPhe in the A site and tRNAifMet in the P site, and for collecting and processing the cryo-EM data. We thank Dr Francis Johnson for providing thermorubin. We thank the staff at NE-CAT beamlines 24ID-C and 24ID-E for help with X-ray diffraction data collection, especially Drs Malcolm Capel, Frank Murphy, Igor Kourinov, Anthony Lynch, Surajit Banerjee, David Neau, Jonathan Schuermann, Narayanasami Sukumar, James Withrow, Kay Perry, Ali Kaya and Cyndi Salbego. We are thankful to Dr Michael Sherman for help with cryo-EM data acquisition, the Sealy Center for Structural Biology and Molecular Biophysics of the University of Texas Medical Branch at Galveston for providing critical infrastructure and expertise, and Drs Ka-Yiu Wong and John Perkyns for computational support.
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health [P30-GM124165 to NE-CAT]. The Eiger 16M detector on 24ID-E beamline is funded by an NIH-ORIP HEI grant [S10-OD021527 to NE-CAT]. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This work used UCSF Chimera for visualization of cryo-EM volume maps and initial structure modeling [P41-GM103311 to the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco].
Author contributions: M.N.P. performed in vitro inhibition assay; V.I.M. and T.P.M. performed dual-reported assay; M.N.P. and V.I.M. performed toe-printing assays on various mRNA templates; T.P.M. performed sucrose gradient profiling; V.I.M. performed in vitro translation inhibition on canonical and leaderless mRNAs; M.N.P. and Y.S.P. performed X-ray crystallography studies; M.G.G. performed cryo-EM data analysis and structure determination; A.A.G., O.A.T. and A.P. performed pre-steady state kinetics and thermodynamics studies; Y.S.P., M.G.G., A.L.K., P.V.S. and I.A.O. supervised the experiments. All authors interpreted the results. M.N.P., A.P., A.L.K., M.G.G. and Y.S.P. wrote the manuscript.
Contributor Information
Madhura N Paranjpe, Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.
Valeria I Marina, Department of Chemistry and Institute of Functional Genomics, Lomonosov Moscow State University, Moscow 119992, Russia.
Aleksandr A Grachev, Peter the Great St.Petersburg Polytechnic University, Saint Petersburg 195251, Russia.
Tinashe P Maviza, Department of Chemistry and Institute of Functional Genomics, Lomonosov Moscow State University, Moscow 119992, Russia.
Olga A Tolicheva, Peter the Great St.Petersburg Polytechnic University, Saint Petersburg 195251, Russia.
Alena Paleskava, Peter the Great St.Petersburg Polytechnic University, Saint Petersburg 195251, Russia.
Ilya A Osterman, Department of Chemistry and Institute of Functional Genomics, Lomonosov Moscow State University, Moscow 119992, Russia.
Petr V Sergiev, Department of Chemistry and Institute of Functional Genomics, Lomonosov Moscow State University, Moscow 119992, Russia.
Andrey L Konevega, Peter the Great St.Petersburg Polytechnic University, Saint Petersburg 195251, Russia.
Yury S Polikanov, Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA; Department of Pharmaceutical Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA; Center for Biomolecular Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA.
Matthieu G Gagnon, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, TX77555, USA; Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555, USA; Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, TX 77555, USA; Institute for Human Infections and Immunity, University of Texas Medical Branch, Galveston, TX 77555, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Illinois State startup funds [to Y.S.P.]; National Institutes of Health [R01-GM136936 to M.G.G.]; Welch Foundation [H-2032–20200401 to M.G.G.]; Russian Ministry of Science [075-15-2021-1396 to V.I.M., reporter analysis, in vitro translation and sucrose gradient profiling]; Russian Science Foundation [22-14-00278 to A.L.K., pre-steady state kinetics and thermodynamics of elongation reactions]. Funding for open access charge: Illinois State startup funds [to Y.S.P.]; National Institutes of Health [R01-GM136936 to M.G.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Collaborators A.R. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodnina M.V. Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 2018; 10:a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polikanov Y.S., Aleksashin N.A., Beckert B., Wilson D.N.. The mechanisms of action of ribosome-targeting peptide antibiotics. Front. Mol. Biosci. 2018; 5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin J., Zhou D., Steitz T.A., Polikanov Y.S., Gagnon M.G.. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 2018; 87:451–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Craveri R., Coronelli C., Pagani H., Sensi P.. Thermorubin, a new antibiotic from a thermoactinomycete. Clin. Med. (Northfield). 1964; 71:511–521. [PubMed] [Google Scholar]
- 6. Cavalleri B., Turconi M., Pallanza R.. Synthesis and antibacterial activity of some derivatives of the antibiotic thermorubin. J. Antibiot. (Tokyo). 1985; 38:1752–1760. [DOI] [PubMed] [Google Scholar]
- 7. Pirali G., Somma S., Lancini G.C., Sala F.. Inhibition of peptide chain initiation in Escherichiacoli by thermorubin. Biochim. Biophys. Acta. 1974; 366:310–318. [DOI] [PubMed] [Google Scholar]
- 8. Bulkley D., Johnson F., Steitz T.A.. The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge B2a of the ribosome. J. Mol. Biol. 2012; 416:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arenz S., Wilson D.N.. Blast from the past: reassessing forgotten translation inhibitors, antibiotic selectivity, and resistance mechanisms to aid drug development. Mol. Cell. 2016; 61:3–14. [DOI] [PubMed] [Google Scholar]
- 10. Blaha G.M., Polikanov Y.S., Steitz T.A.. Elements of ribosomal drug resistance and specificity. Curr. Opin. Struct. Biol. 2012; 22:750–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin F., Wishnia A.. The protein synthesis inhibitor thermorubin. 1. Nature of the thermorubin-ribosome complex. Biochemistry. 1982; 21:477–483. [DOI] [PubMed] [Google Scholar]
- 12. Lin F., Wishnia A.. The protein synthesis inhibitor thermorubin. 2. Mechanism of inhibition of initiation on Escherichiacoli ribosomes. Biochemistry. 1982; 21:484–491. [DOI] [PubMed] [Google Scholar]
- 13. Zakalyukina Y.V., Birykov M.V., Lukianov D.A., Shiriaev D.I., Komarova E.S., Skvortsov D.A., Kostyukevich Y., Tashlitsky V.N., Polshakov V.I., Nikolaev E.et al.. Nybomycin-producing streptomyces isolated from carpenter ant Camponotusvagus. Biochimie. 2019; 160:93–99. [DOI] [PubMed] [Google Scholar]
- 14. Osterman I.A., Komarova E.S., Shiryaev D.I., Korniltsev I.A., Khven I.M., Lukyanov D.A., Tashlitsky V.N., Serebryakova M.V., Efremenkova O.V., Ivanenkov Y.A.et al.. Sorting out antibiotics' mechanisms of action: a double fluorescent protein reporter for high-throughput screening of ribosome and DNA biosynthesis inhibitors. Antimicrob. Agents Chemother. 2016; 60:7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vazquez-Laslop N., Thum C., Mankin A.S.. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008; 30:190–202. [DOI] [PubMed] [Google Scholar]
- 16. Orelle C., Carlson S., Kaushal B., Almutairi M.M., Liu H., Ochabowicz A., Quan S., Pham V.C., Squires C.L., Murphy B.T.et al.. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 2013; 57:5994–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pantel L., Florin T., Dobosz-Bartoszek M., Racine E., Sarciaux M., Serri M., Houard J., Campagne J.M., de Figueiredo R.M., Midrier C.et al.. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell. 2018; 70:83–94. [DOI] [PubMed] [Google Scholar]
- 18. Meydan S., Marks J., Klepacki D., Sharma V., Baranov P.V., Firth A.E., Margus T., Kefi A., Vázquez-Laslop N., Mankin A.S.. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell. 2019; 74:481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin D., Fredrick K.. Analysis of polysomes from bacteria. Methods Enzymol. 2013; 530:159–172. [DOI] [PubMed] [Google Scholar]
- 20. Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 21. Polikanov Y.S., Blaha G.M., Steitz T.A.. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012; 336:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Polikanov Y.S., Steitz T.A., Innis C.A.. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014; 21:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A.. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015; 22:342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitt E., Blanquet S., Mechulam Y.. Crystallization and preliminary X-ray analysis of Escherichiacoli methionyl-tRNAMet(f) formyltransferase complexed with formyl-methionyl-tRNAMet(f). Acta Crystallogr. D Biol. Crystallogr. 1999; 55:332–334. [DOI] [PubMed] [Google Scholar]
- 25. Svetlov M.S., Syroegin E.A., Aleksandrova E.V., Atkinson G.C., Gregory S.T., Mankin A.S., Polikanov Y.S.. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. 2021; 17:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Syroegin E.A., Aleksandrova E.V., Polikanov Y.S.. Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes. Nat. Chem. 2022; 10.1038/s41557-022-01073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Syroegin E.A., Aleksandrova E.V., Polikanov Y.S.. Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine. Nucleic Acids Res. 2022; 50:7669–7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Syroegin E.A., Flemmich L., Klepacki D., Vazquez-Laslop N., Micura R., Polikanov Y.S.. Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol. Nat. Struct. Mol. Biol. 2022; 29:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitcheltree M.J., Pisipati A., Syroegin E.A., Silvestre K.J., Klepacki D., Mason J.D., Terwilliger D.W., Testolin G., Pote A.R., Wu K.J.Y.et al.. A synthetic antibiotic class overcoming bacterial multidrug resistance. Nature. 2021; 599:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabsch W xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 34. Basu R.S., Sherman M.B., Gagnon M.G.. Compact IF2 allows initiator tRNA accommodation into the p site and gates the ribosome to elongation. Nat. Commun. 2022; 13:3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 36. Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D., Urzhumtsev A.. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 2018; 74:814–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watson Z.L., Ward F.R., Meheust R., Ad O., Schepartz A., Banfield J.F., Cate J.H.. Structure of the bacterial ribosome at 2 a resolution. Elife. 2020; 9:e60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 39. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J.et al.. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. D Struct. Biol. 2019; 75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E.. UCSF chimerax: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maksimova E.M., Vinogradova D.S., Osterman I.A., Kasatsky P.S., Nikonov O.S., Milon P., Dontsova O.A., Sergiev P.V., Paleskava A., Konevega A.L.. Multifaceted mechanism of amicoumacin a inhibition of bacterial translation. Front. Microbiol. 2021; 12:618857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paleskava A., Maksimova E.M., Vinogradova D.S., Kasatsky P.S., Kirillov S.V., Konevega A.L.. Differential contribution of protein factors and 70S ribosome to elongation. Int. J. Mol. Sci. 2021; 22:9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pletnev P.I., Shulenina O., Evfratov S., Treshin V., Subach M.F., Serebryakova M.V., Osterman I.A., Paleskava A., Bogdanov A.A., Dontsova O.A.et al.. Ribosomal protein S18 acetyltransferase RimI is responsible for the acetylation of elongation factor tu. J. Biol. Chem. 2022; 298:101914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vinogradova D.S., Zegarra V., Maksimova E., Nakamoto J.A., Kasatsky P., Paleskava A., Konevega A.L., Milon P.. How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol. 2020; 18:e3000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Savelsbergh A., Matassova N.B., Rodnina M.V., Wintermeyer W.. Role of domains 4 and 5 in elongation factor g functions on the ribosome. J. Mol. Biol. 2000; 300:951–961. [DOI] [PubMed] [Google Scholar]
- 47. Konevega A.L., Soboleva N.G., Makhno V.I., Semenkov Y.P., Wintermeyer W., Rodnina M.V., Katunin V.I.. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal a site by stacking and Mg2+-dependent interactions. RNA. 2004; 10:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayashi K., Dombou M., Sekiya M., Nakajima H., Fujita T., Nakayama M.. Thermorubin and 2-hydroxyphenyl acetic acid, aldose reductase inhibitors. J. Antibiot. (Tokyo). 1995; 48:1345–1346. [DOI] [PubMed] [Google Scholar]
- 49. Hartz D., McPheeters D.S., Traut R., Gold L.. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988; 164:419–425. [DOI] [PubMed] [Google Scholar]
- 50. Florin T., Maracci C., Graf M., Karki P., Klepacki D., Berninghausen O., Beckmann R., Vazquez-Laslop N., Wilson D.N., Rodnina M.V.et al.. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Mol. Biol. 2017; 24:752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Balakin A.G., Skripkin E.A., Shatsky I.N., Bogdanov A.A.. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic Acids Res. 1992; 20:563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moll I., Hirokawa G., Kiel M.C., Kaji A., Blasi U.. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004; 32:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pape T., Wintermeyer W., Rodnina M.V.. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the a site of the e. coli ribosome. EMBO J. 1998; 17:7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peske F., Savelsbergh A., Katunin V.I., Rodnina M.V., Wintermeyer W.. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004; 343:1183–1194. [DOI] [PubMed] [Google Scholar]
- 55. Semenkov Y.P., Rodnina M.V., Wintermeyer W.. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 2000; 7:1027–1031. [DOI] [PubMed] [Google Scholar]
- 56. Kipper K., Hetenyi C., Sild S., Remme J., Liiv A.. Ribosomal intersubunit bridge B2a is involved in factor-dependent translation initiation and translational processivity. J. Mol. Biol. 2009; 385:405–422. [DOI] [PubMed] [Google Scholar]
- 57. Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller H.F.. Structural basis for translation termination on the 70S ribosome. Nature. 2008; 454:852–857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors were deposited in the RCSB Protein Data Bank under the accession code 8EKB for the T. thermophilus 70S ribosome crystal structure in complex with THR, mRNA, and deacylated P-site tRNAifMet. The cryo-EM map for the E. coli 70S ribosome in complex with THR, mRNA, aminoacylated A-site Phe-tRNAPhe, and deacylated P-site tRNAifMet was deposited in the Electron Microscopy Data Bank (EMDB) under the accession code EMD-28197, with the coordinates deposited in the RCSB Protein Data Bank under the accession code 8EKC.
All previously published structures that were used in this work for structural comparisons were retrieved from the RCSB Protein Data Bank: PDB entries 4V8A, 6XHW, and 3D5A.
No sequence data were generated in this study.