Abstract
Cell sheet technology has demonstrated great promise in delivering a large amount of therapeutic cells for tissue repair, including in the myocardium. However, the lack of host integration remains one of the key challenges in using cell sheets for cardiac repair. Paracrine factors secreted by mesenchymal stem cells (MSCs) have been reported to facilitate tissue repair and regeneration in a variety of ways. It has been demonstrated that paracrine factors from MSCs could enhance scaffold recellularization and vascularization. In this study, we used an in vitro cardiac matrix mimic platform to examine the effects of hMSCs preseeding on the interactions between cell sheets and cardiac matrix. The fabricated human induced pluripotent stem cells-derived cardiomyocyte sheets were attached to a decellularized porcine myocardium slice with or without preseeding of hMSCs. The hMSCs preseeding significantly enhanced the interactions between cardiomyocyte sheets and cardiac matrix in terms of cell migration distance, cell distribution, and mature vascular and cardiomyocyte marker expressions in the matrix. Growth factor and matrix metalloproteinases array analysis suggested that hMSCs- induced vascularization and MMPs regulation are the two possible mechanisms that lead to the improved CMs and cardiac matrix interactions. Further examination of these two mechanisms will enable the development of new approaches to facilitate transplanted cells for tissue repair.
Keywords: decellularized myocardium, human induced pluripotent stem cells, mesenchymal stem cells, cardiac matrix, cell sheet
Graphical Abstract
1. INTRODUCTION
The cell sheet has become an effective approach to deliver a large amount of therapeutic cells for tissue repair. For example, the human induced pluripotent stem cell (hiPSC)-engineered cardiovascular cell sheet composed by induction of cardiomyocytes and vascular cells, when applied to a rat myocardial infarction model, resulted in cell engraftment and neovascularization leading to improved heart function.1 The prevascularized cell sheet made up of human umbilical vein endothelial cells and human mesenchymal stem cells, when implanted in a rat full thickness skin wound model, has been shown to accelerate tissue repair, as evidenced by preserved skin appendages, developed epidermal tissue, reduced skin contraction, and improved cosmetic appearance.2 Similarly, a human fibroblast cell sheet, when used to demonstrate the feasibility of the islet graft model, has exhibited a three-dimensional structure of extracellular matrix and promoted the cell survival and function of human pancreatic islets.3 Compared with direct injection, cells delivered by cell sheet technology have shorter cell preparation time, higher delivery efficiency, and improved cell viability after transplantation.4–6 The cell sheet has been explored in repairing an injured heart after myocardial infarction (MI) and has shown great promise.7–9 However, the lack of host integration remains one of the key challenges in using cell sheets for cardiac repair.10,11
The ability of mesenchymal stem cells (MSCs) to produce and release paracrine factors has triggered many studies for exploiting cell therapy. Paracrine factors secreted by MSCs have been reported to facilitate tissue repair and regeneration in a variety of ways.12,13 For instance, the secretome containing secreted factors from MSCs, when injected subcutaneously in athymic mice, has been shown to enhance angiogenesis as evidenced by neovascularization.14 By injecting stromal cell-derived factor-1 in a rat MI model, Segers et al. demonstrated the recruitment of CXCR4+/c-Kit+ stem cells with increased capillary density in the damaged heart leading to improved cardiac function post-MI.15 Similarly, the secreted factors of human MSCs, when used in an ex vivo model that mimicked tissue ischemia, has significantly prevented the cell death of cardiomyocytes and endothelial cells, indicating the antiapoptotic effect of the MSC secretome.16 In our previous study, it was also found that MSC paracrine factors enhanced scaffold recellularization and vascularization.17
The objective of this proof-of-concept study is to investigate whether preseeding of MSCs would increase the integration of a human induced pluripotent stem cell (hiPSC) derived cardiomyocyte sheet into a cardiac matrix. Since the cause of MI is a significant loss of cardiomyocytes (CMs), we fabricated cell sheets using CMs derived from hiPSCs and used these cell sheets for all tests. Decellularized porcine myocardium slice (dPMS) was selected as a human cardiac matrix mimic to perform in vitro cell-matrix interaction analysis using our established method.17 The decellularized scaffolds have been extensively reported to preserve organ specific physical and chemical characteristics in extracellular matrix (ECM).18–21 Specifically, human mesenchymal stem cells (hMSCs) were first seeded into dPMS for 24 h, and then CM sheets were attached to dPMS up to 14-day culture. The interactions between CM sheets and dPMS were observed and assessed. The proliferation, phenotype changes, and growth factor release from MSCs were further examined to elucidate the potential contributing mechanisms of preseeding.
2. MATERIALS AND METHODS
2.1. Materials.
Human mesenchymal stem cells, Mesencult proliferation kit, mTeSR1 complete kit, STEMdiff cardiomyocyte differentiation kit, STEMdiff cardiomyocyte maintenance kit, STEMdiff cardiomyocyte freezing medium, and STEMdiff dissociation kit were purchased from StemCell Technologies (Vancouver, BC, Canada). Human induced pluripotent stem cells were purchased from WiCell Research Institute (Madison, WI, U.S.A.). Human growth factor antibody array C1, Human matrix metalloproteinase antibody array C1, and Protease inhibitor cocktail were obtained from RayBiotech (Norcross, GA, U.S.A.). LuminiCell tracker 540 cell labeling kit and LuminiCell tracker 670 cell labeling kit were purchased from Millipore Sigma (Burlington, MA, U.S.A.). Sodium dodecyl sulfate, Triton-X-100, and penicillin-streptomycin were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.). Matrigel GFR basement membrane matrix and Dulbecco’s phosphate-buffered saline were obtained from Corning Incorporated (Bedford, MA, U.S.A.). Glutamax, glucose solution, DMEM/F12 medium, Versene solution, live cell imaging solution, and antibiotic-antimycotic were obtained from Gibco (Carlsbad, CA, U.S.A.). Fluo-4 calcium imaging kit, Live/dead viability/cytotoxicity kit, Hoechst 33342, and Alkaline phosphatase live stain were purchased from Molecular Probes (Eugene, OR, U.S.A.). 4% Paraformaldehyde in PBS (w/v), paraffin wax (type 9), HistoPrep SH75–125D tissue embedding medium, molecular grade absolute ethanol (200 proof), anhydrous ethanol (histological grade), and xylene were purchased from ThermoFisher Scientific (Florence, KY, U.S.A.). VECTASHIELD mounting medium with DAPI was purchased from Vector Laboratories Inc. (Burlingame, CA, U.S.A.). Human/mouse SSEA-1 antibody was obtained from R&D Systems, Inc. (Minneapolis, MN, U.S.A.). GATA-4 antibody (G-4) was purchased from Santa Cruz Biotechnology (Dallas, TX, U.S.A.). All other primary antibodies, secondary antibodies, and 1X citrate buffer were purchased from Abcam (Cambridge, MA, U.S.A.). All materials were used as-received from the manufacturers.
2.2. Culture of hMSCs and hiPSCs.
Human mesenchymal stem cells (hMSCs) were expanded using a Mesencult proliferation kit supplemented with 2 mM glutamax and 1% penicillin-streptomycin (P/S). Human induced pluripotent stem cells (hiPSCs) were cultured following the feeder-independent pluripotent stem cell protocol provided by the manufacturer. Briefly, hiPSCs were seeded on the Matrigel coated 6-well plate with the complete mTeSR1 medium. Upon reaching ∼80% confluency, they were passaged using the Versene solution and split into the new 6-well plate at the ratio of 1:6. Both cell types were maintained in a humidified cell culture incubator at 37 °C and 5% CO2 with media change every 2 days for hMSCs and every day for hiPSCs. All experiments were performed using hMSCs at passages 4–6 and hiPSCs at passages 35–37.
2.3. Differentiation of hiPSCs to Cardiomyocytes.
Cardiac differentiation of hiPSCs was carried out using a STEMdiff cardiomyocyte differentiation kit following the protocol provided by the manufacturer. In brief, the hiPSCs were grown on a Matrigel-coated culture dish in the complete mTeSR1 medium for 4 days to achieve ∼95% confluency. When the cells reached the desired confluency (referred to as day 0 in the cardiac differentiation process), the differentiation of hiPSCs was initiated by incubating the cells with cardiomyocyte differentiation medium A for 2 days, switching to cardiomyocyte differentiation medium B for 2 days, and changing to differentiation medium C for another 4 days. After differentiation, the cells were cultured in STEMdiff cardiomyocyte maintenance medium for 8 days before harvesting for experiments. The cells were maintained in a humidified cell culture incubator at 37 °C and 5% CO2 with media change every day. For harvesting, the cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and dissociated using a STEMdiff dissociation kit on day 16 of differentiation to obtain the hiPSC-derived cardiomyocytes (hiPSC-derived CMs). The obtained hiPSC-derived CMs (referred as passage 0) were either used for experiments or cryopreserved in STEMdiff cardiomyocyte freezing medium until use. All experiments were performed using hiPSC-derived CMs at passages 1–2. Phase contrast and time-lapse images were taken with an inverted AxioVision A1 microscope (Carl Zeiss, Oberkochen, Germany) and AxioObserver Z1 microscope (Carl Zeiss) with incubation chamber maintained at 37 °C, respectively.
2.4. Alkaline Phosphatase in hiPSCs.
Alkaline phosphatase (AP) live stain was used to detect AP in hiPSCs in vitro according to the protocol provided by the manufacturer. Briefly, the cells were washed with prewarmed DMEM/F12 medium for 2 min and stained with 1X alkaline phosphatase live stain dye for 30 min at room temperature while protecting them from the light. After incubation, the cells were washed twice with DMEM/F12 medium for 5 min each. The fluorescence images were captured with an inverted AxioVision A1 microscope (Carl Zeiss) within 60 min after AP staining.
2.5. Intracellular Calcium Levels.
Intracellular calcium levels of hiPSC-derived CMs were measured using a Fluo-4 calcium imaging kit with an Olympus FluoView FV1000 confocal microscope (Olympus, Center Valley, PA, U.S.A.). In brief, the cells were washed with live cell imaging solution (LCIS) and incubated with Fluo-4 AM loading dye for 30 min in a humidified cell culture incubator (37 °C; 5% CO2). Next, the plate was transferred to room temperature for an additional 15 min to complete the staining procedure. Immediately after dye incubation, the cells were washed and imaged in LCIS containing 20 mM glucose solution with a confocal microscope. To take time-lapse fluorescence images, the Fluo-4 AM treated cells were excited and emitted at 488 nm and 500–545 nm respectively with an argon ion laser to acquire images in a line scanning mode (aspect ratio of 512 × 512; 254 ms per frame). The changes in fluorescence signals were analyzed using Olympus Fluoview software (Version 3.1.a) and ImageJ (NIH free image analysis software) which were then normalized to basal cell fluorescence. All imaging experiments were conducted at room temperature.
2.6. Generation of hiPSC-Derived Cardiomyocyte Sheet.
The hiPSC-derived CM sheet was generated using a 12-multiwell UpCell plate. First, the UpCell plate was coated with STEMdiff cardiomyocyte support medium for 24 h in a humidified cell culture incubator (37 °C and 5% CO2). Each well of the UpCell plate was seeded with either 0.8 × 106 or 1.6 × 106 hiPSC-derived CMs in 1 mL of STEMdiff cardiomyocyte support medium followed by incubation in a cell culture incubator for 24 h. Next, the medium was changed to 1 mL of warm STEMdiff cardiomyocyte maintenance medium to grow cells for another 24 h before harvesting the cell sheet. The cell sheet was obtained by placing the cell seeded UpCell plate to room temperature for 2 h to allow complete cell sheet detachment.
2.7. Preparation of Decellularized Porcine Myocardial Slice.
The decellularized porcine myocardial slice (dPMS) was prepared from porcine cardiac tissue following the established protocol previously described.17 In brief, fresh porcine heart was obtained from a local slaughterhouse and cut into the tissue blocks of ∼2 (length) × 2 (width) × 1 cm3 (thickness). The tissue blocks were rinsed with deionized water and treated with detergent solution containing 1% (w/v) sodium dodecyl sulfate (SDS) and 0.5% penicillin-streptomycin (P/S) at room temperature for 2.5 weeks with gentle agitation. Next, they were treated with 0.01% Triton-X-100 in 1X phosphate buffered saline (PBS) solution for 1 h and washed thoroughly with 1X PBS for 2 days to obtain decellularized myocardium tissue. The decellularized cardiac tissue was cryosectioned to obtain 600 μm thick slices. The native porcine myocardial slice (nPMS) was sliced following the same protocol and used as a control group. The removal of cells after decellularization was checked by Hematoxylin and Eosin (H&E) staining, DAPI nuclei staining, and total DNA measurement. For total DNA measurement, DNA was extracted from the native and decellularized myocardium tissues using QIAamp DNA mini kit (Qiagen, Valencia, CA, U.S.A.) and stained with Quant-iT PicoGreen dsDNA assay kit (Molecular Probes, Eugene, OR, U.S.A.). The quantitative fluorescence intensity was measured with a Synergy H1 Hybrid microplate reader (Bio-Tek Instruments, Winooski, VT, U.S.A.).
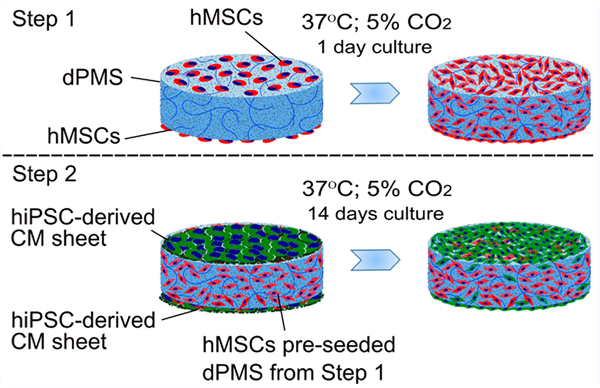
2.8. Preseeding of hMSCs into dPMS.
The dPMS was sterilized with molecular biology grade absolute ethanol for 45 min, followed by at least three extensive washes with sterilized DI water for 15 min each. Then the sterilized dPMS was immersed in respective cell growth medium, and incubated overnight at 37 °C and 5% CO2. The obtained sterile dPMS was then seeded with 2 × 105 of hMSCs in 30 μL cell culture medium on one side of the scaffold. After one side seeding, the cells were incubated in a humidified cell culture incubator (37 °C; 5% CO2) for 4 h before flipping the dPMS to receive the cell suspension on the other side at the same density. The cells were further incubated at 37 °C; 5% CO2 for 4 h before adding the growth medium to culture cells for 1 day.
2.9. Attaching hiPSC-Derived CM Sheets to dPMS.
To attach hiPSCs-derived CM sheets to dPMS, dPMS was transferred to a well of the 12-well UpCell plate containing confluent hiPSC-derived CMs. By reducing the temperature of culture from 37 °C to room temperature for 2 h, CM sheet detached from the UpCell plate and attached to dPMS. For bilateral cell sheet seeding, dPMS was first seeded with a CM sheet on one side and then flipped before transferring to a new well of the 12-well UpCell plate for cell sheet attachment on the other side. Following the same procedure, hiPSC-derived CM sheets were attached onto hMSCs preseeded dPMS. The dPMS containing the coculture of hiPSC-derived CMs and hMSCs was grown for 14 days in a 6-well transwell insert (8 μm pore size; Corning, New York, NY, U.S.A.) using the cell culture medium that consisted of the equal ratio (1:1) of growth media for hMSCs and hiPSC-derived CMs.
2.10. Tracking of Seeded Cells in dPMS.
To track the distribution of seeded cells in dPMS, the cells were prestained with either LuminiCell tracker 540 or 670 labeling kits prior to cell seeding following the directions provided by the manufacturer. In brief, the adherent cells were rinsed with warm 1X DPBS and stained with the 2 nM labeling solution in warm complete medium. After loading the dyes, the cells were incubated at 37 °C and 5% CO2 for 4 h, extensively washed with warm cell culture medium, and used for the recellularization experiments.
2.11. CM Analysis after Seeding in dPMS.
The seeding efficiency was calculated as the percentage of cells retained on dPMS 2h after seeding. The retained cell number was derived by subtracting the number of unattached cells from the initial total seeding cell number. Cell viability of seeded CMs was measured by staining the cells with calcein AM (2 μM), ethidium homodimer-1 (EthD-1; 4 μM), and hoechst 33342 (1:1000) for 30 min in cell culture incubator (37 °C and 5% CO2). Following incubation, the seeded cells were extensively rinsed with 1X DPBS to remove the unbound dyes and imaged with FluoView FV1000 confocal microscope (Olympus, Center Valley, PA, U.S.A.). Equidistant Z-stack images (7 μm apart) were taken to capture the fluorescence signal of cells from periphery toward the center of dPMS. The percentage of viable cells was calculated based on the intensities of red pixels from ethidium homodimer-1 and green pixels from calcein AM. Infiltration of CM sheet after lateral seeding was assessed by the number of images acquired in the Z-stack images. For the bilateral seeding group, the total cell distribution within the dPMS was demonstrated by staining the cell nuclei with a VECTASHIELD mounting medium containing DAPI using OCT-embedded tissue section or paraffin-embedded tissue section. Proliferation of hMSCs in dPMSs was examined by extracting DNA content from seeded cells, staining with Quant-iT PicoGreen dsDNA assay kit, and measuring the fluorescence signals using a Synergy H1 Hybrid microplate reader (Bio-Tek Instruments).
2.12. Expressions of Pluripotent and Cardiovascular Markers.
Immunofluorescence staining was used to confirm the expression of specific cell markers of the seeded cells as previously described.17 To perform immunofluorescence staining on cultured cells, the cell seeded dPMS was fixed with 4% paraformaldehyde in PBS solution for 30 min at room temperature, embedded into paraffin wax, and sliced into 6 to 8 μm portions by microtome sectioning. The paraffin-embedded sections were deparaffinized with xylene followed by dehydration with anhydrous ethanol. The heat-mediated antigen retrieval process was performed on the samples at 100 °C for 15 min using 1X citrate buffer. Next, the tissue sections were permeabilized and blocked with 0.2% Triton-X-100 in 1X PBS solution containing 5% goat serum for 1 h with gentle shaking on an orbital shaker. Following the permeabilization and blocking steps, the samples were incubated with primary antibodies at 2–8 °C for 20 h. The tested primary antibodies include Nanog (1:30), OCT4 (1:100), SOX2 (1:100), SSEA4 (1:20), TRA-1–60 (1:30), SSEA-1 (1:20), calponin (1:100), myosin light chain 2v (MLC-2v; 1:50), cardiac troponin T (cTnT; 1:100), GATA-4 (1:50), sarcomeric α-actinin (1:50), connexin 43 (CX43; 1:100), von Willebrand factor (vWF; 1:100), and α-smooth muscle actin (α-SMA; 1:100). After incubation, the samples were extensively washed with 1X PBS followed by incubation with goat-derived secondary antibodies conjugated with either Alexa Fluor 488 or 647 (1:100) for 2 h at room temperature by protecting them from light. Next, the samples were rinsed with 1X PBS and counterstained with VECTASHIELD mounting medium containing DAPI for nuclei staining. The fluorescence images were taken with an inverted AxioVision A1 microscope (Carl Zeiss). Using the same staining protocol, positive controls were prepared by staining the native cardiac tissue and negative controls were carried out on the tested samples without incubating with primary antibodies.
2.13. Growth Factors and MMPs Arrays.
The secreted growth factors and matrix metalloproteinases (MMPs) from the cells seeded on dPMS were analyzed using human growth factor antibody array C1 and human MMP antibody array C1. Cell secretions released in the cell culture medium and retained on dPMS were collected for the array testing. The secretions retained on dPMS were obtained by homogenizing the scaffold in 1X cell lysis buffer supplemented with protease inhibitor cocktail using a VWR 200 homogenizer (VWR, Radnor, PA, U.S.A.). The homogenized tissues were incubated at 4 °C for 30 min by vigorously vortexing them every 5 min. Following incubation, the samples were centrifuged at 16 000g for 20 min at 4 °C to collect the solution that contains the secretions. The solutions obtained from cell-free dPMS was used as a negative control group. Prior to the arrays testing, the total protein concentration of samples was quantified using Bradford protein quantification assay (Bio-Rad, Hercules, CA, U.S.A.) according to the manufacturer’s recommendation. For human growth factor antibody array C1 and human MMP antibody array C1 testing, each array membrane was blocked with a blocking buffer at room temperature for 1 h and incubated with 500 μg of testing solution at 2–8 °C for 20 h. After sample incubation, the array membranes were stained with first biotinylated antibody cocktail and then horseradish peroxidase (HRP)-conjugated streptavidin at 2–8 °C for 20 h. Finally, the membranes were developed with the detection buffer and exposed with BioSpectrum 810 imaging system (Analytik Jena US LLC, Upland, CA, U.S.A.) to take the chemiluminescence images. The relative protein expression was quantitatively determined by measuring the spot signal densities from the obtained array images using a VisionWorksLS analysis software version 8.1.2 (Analytik Jena US LLC, Upland, CA, U.S.A.). The background signal density was subtracted from each densitometry data and then normalized to the positive control signals for comparing the protein expression between the tested experiment groups.
2.14. Statistical Analysis.
All experimental data are presented as mean ± standard deviation (SD) with 3–6 replicates. Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, U.S.A.). We used student’s t-test to determine statistically significant difference between two experiment groups and one-way analysis of variance (ANOVA) with Tukey’s posthoc test for comparing three or more experiment groups. A p-value less than 0.05 was considered to be statistically significant.
3. RESULTS
3.1. hiPSC-Derived CM Sheet Fabrication and Characterization.
A classical cardiac differentiation procedure was followed to derive cardiomyocytes from hiPSCs.22,23 Before inducing differentiation, the pluripotency of the cultured hiPSCs was confirmed by high expressions of pluripotency markers (Nanog, OCT4, SOX2, TRA-1–60, and SSEA-4) and AP activity (Figure 1A). The expression of SSEA-1 was not detected in our cultured hiPSCs, which has been known to be unexpressed in hiPSCs and quickly upregulates upon the initiation of differentiation.24 During differentiation, the typical cell morphology changes toward CMs as reported by others were observed and spontaneous cell beating was noticed on day 8 and the number of beating cells kept increasing afterward (Figure 1B).1,25 High expressions of cardiac-specific markers including cardiac troponin T (cTnT), GATA4, myosin light chain 2 V (MLC-2v), and calponin (a calcium binding protein for muscle contraction) were detected in the derived CMs (Figure 1C). The average percentages of GATA4+, MLC-2v+, and cTnT+ cells on day 16 of cardiac differentiation were 91.92 ± 4.31%, 84.55 ± 4.23%, and 83.81 ± 2.69%, respectively. The calcium flux in hiPSC-derived CMs was visualized and quantitated using Fluo-4 AM calcium indicator (Figure 1D). Similar spike amplitude (F/F0; 1.12 ± 0.34 vs 1.05 ± 0.27) and rate (2.31 sec per beat vs 2.42 sec per beat) were found for the beating CMs located in either center or periphery of the colony (Figure 1D). The obtained hiPSC-derived CMs were used to fabricate cell sheets using a thermal responsive culture dish. Through optimizing the cell density and detachment condition, a complete layer of CM sheet was harvested (Figure 1E). We found that the minimal number of CMs needed to generate a round cell sheet (diameter of 16.45 ± 2.77 mm) was 0.8 × 106 cells.
Figure 1.
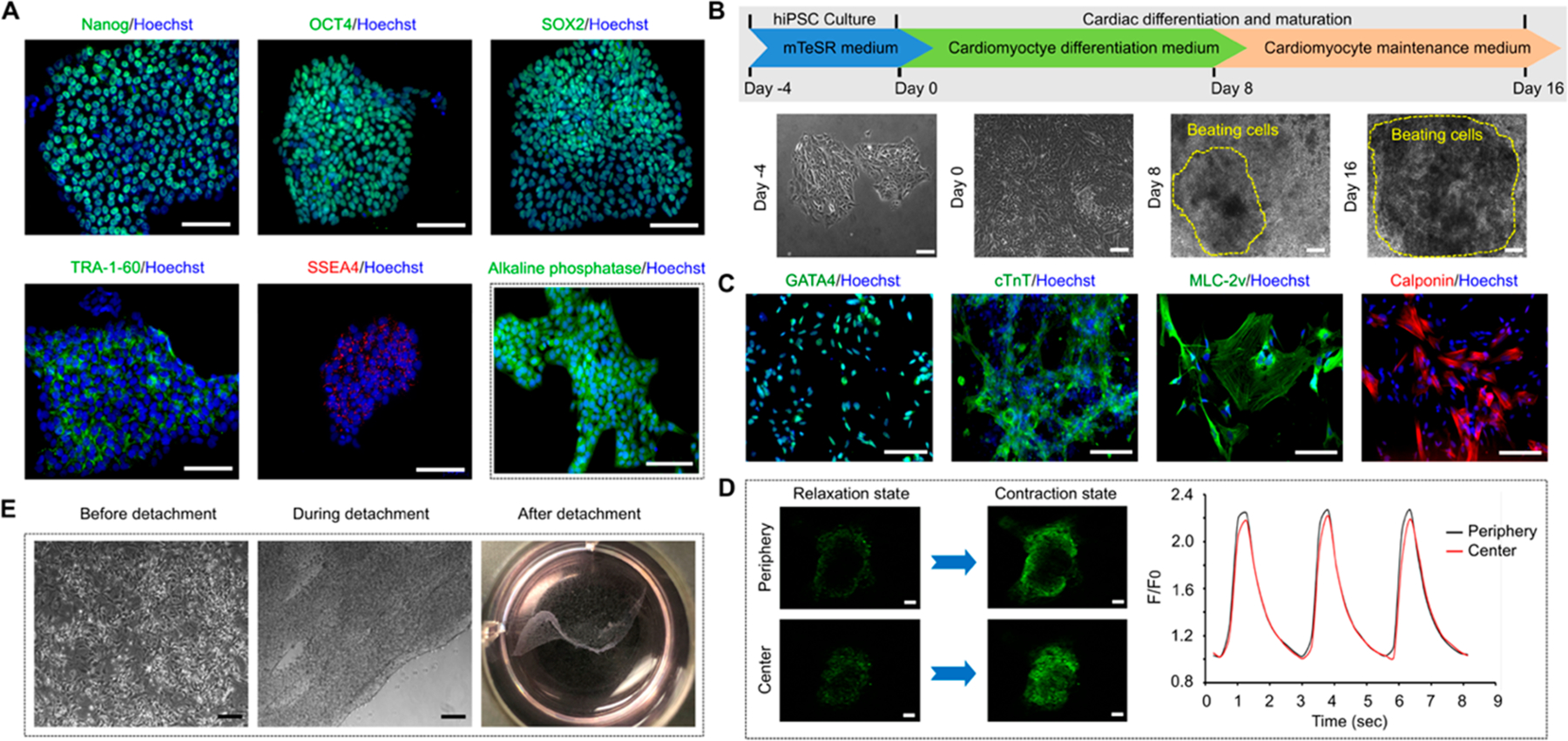
Differentiation of cardiomyocytes from hiPSCs and cell sheet generation. (A) The pluripotency markers (Nanog, OCT4, SOX2, TRA-60, SSEA4) and alkaline phosphatase activity expressions in the cultured hiPSCs. (B) Experiment timeline of the cardiomyocyte differentiation procedure and representative phase contrast images showing the morphological changes of the cells during the process. Areas defined by yellow dotted line consist of spontaneous beating cells. (C) Cardiac-specific markers (GATA4, cTnT, MLC-2v, calponin) expressions in the hiPSCs-derived CMs. (D) The fluorescence intensity changes of fluo-4 (calcium indicator) during relaxing and contracting stages of the hiPSC-derived CMs both in the center and at the periphery of the beating area. F0 is the fluo-4 fluorescence during relaxing stage. n = 3 (E) Detachment of cardiomyocyte sheet from a thermal responsive cell culture plate by lowering the culture temperature from 37 °C to room temperature for 2 h. Scale bar: 100 μm.
3.2. Interactions Between hiPSC-Derived CM Sheets and dPMS.
The dPMS was produced and characterized using our previously published method.26 Compared to the native myocardial slice at the same thickness, dPMS retained myocardial tissue structure and showed increased transparency. The sufficient removal of cells in the produced dPMS was confirmed by H&E and DAPI staining (Figure 2A). We tested to attach hiPSC-derived CM sheet to either one side or both side of dPMS (Figure 2B). Our results demonstrated that CM sheets attached firmly to both sides of dPMS (Figure 2C). Compared with individual cell seeding using cell suspension at the same total seeding cell number (0.8 × 106), the cell sheet exhibited significantly higher seeding efficiency on dPMS (93.13 ± 1.88% vs 60.63 ± 2.65%) (Figure 2D). Further increasing the total cell number contained in the cell sheet (from 0.8 × 106 cells/sheet to 1.6 × 106 cells/sheet) did not significantly affected the seeding efficiency (93.13 ± 1.88% vs 94.38 ± 0.88%) (Figure 2D). However, a noticeable reduction of cell migration distance (126.48 ± 4.09 μm vs 142.14 ± 3.75 μm on day 14) and viability (78.36 ± 1.33% vs 85.18 ± 1.78% on day 14) were detected in the dense CM sheet (1.6 × 106 cells/sheet) compared with the loose CM sheet (0.8 × 106 cells/sheet) (Figure 2E, F). We also observed that the seeded CMs ceased to further infiltrate into the dPMS shortly after seeding regardless of cell sheet density (Figure 2E).
Figure 2.
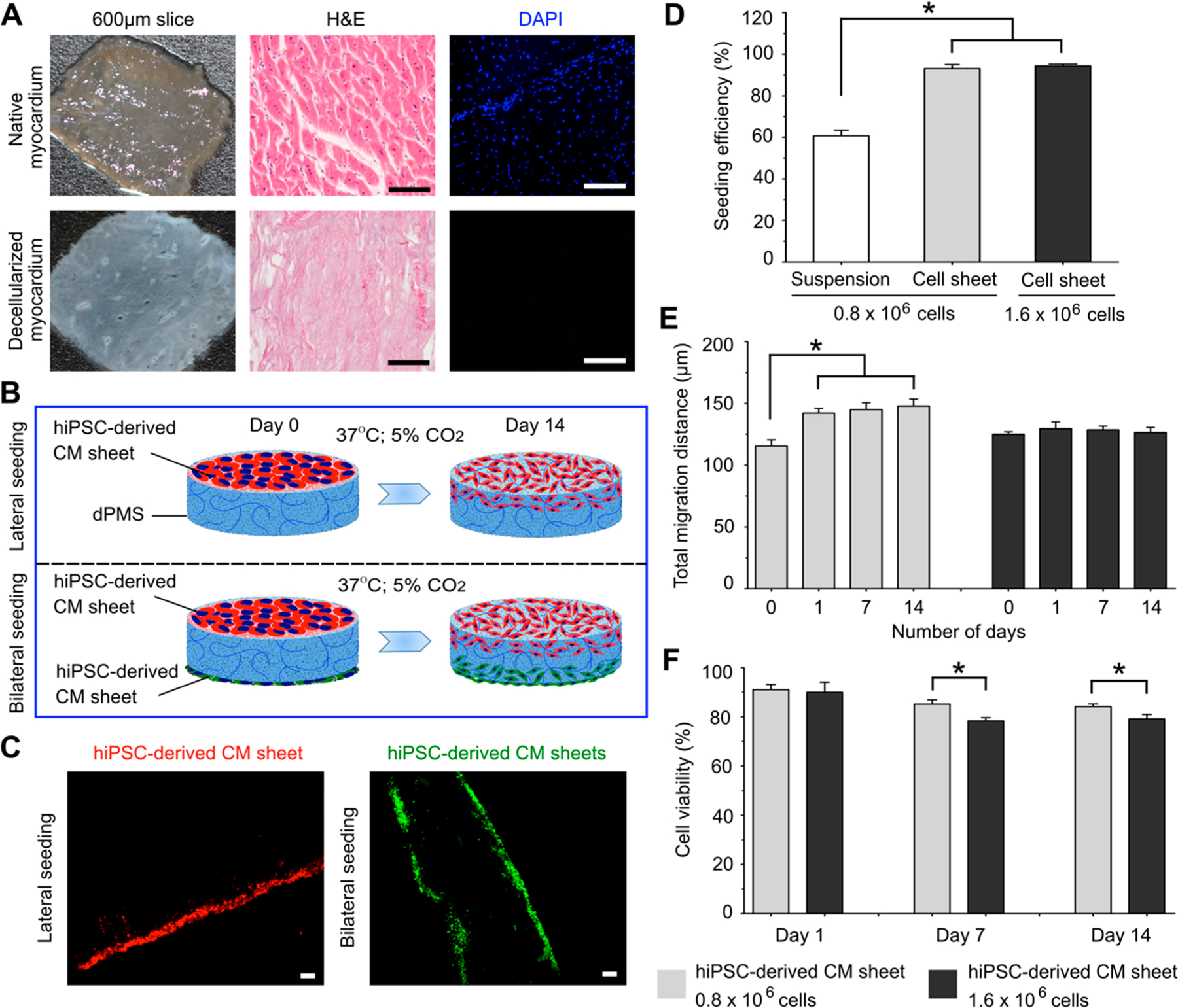
hiPSC-derived CM sheet and dPMS interactions. (A) Comparison of native porcine myocardial slice (nPMS) and decellularized porcine myocardial slice (dPMS) with the same thickness (600 μm). (B) Schematic demonstration of lateral and bilateral seeding of hiPSC-derived CM sheets onto dPMS. (C) CMs distributions 1 day after lateral and bilateral seeding of prestained hiPSC-derived CM sheets onto dPMS. (D) Cell seeding efficiency of using cell suspension and two types of CMs sheets (0.8 × 106 total cells/sheet and 1.6 × 106 total cells/sheet). (E) Migration distance of the CMs from the seeded CM sheet in dPMS. There are no statistically significant differences in terms of total migration distance between the groups of CM sheet (0.8 × 106 cells) and CM sheet (1.6 × 106 cells) at all the assessed time points (day 0, 1, 7, and 14). (F) Cell viability after seeding hiPSC-derived CM sheets to dPMS. *Represents the statistically significant difference among the experiment groups with p-value less than 0.05 (n = 6). Scale bar: 100 μm.
3.3. Improved Interactions Between CM Sheets and dPMS after hMSCs Preseeding.
We have previously developed a bilateral seeding approach to incorporate hMSCs into dPMS at high efficiency.17 Therefore, the same method was used in this study to preseed hMSCs to dPMS before CM sheet attachment (Figure 3A). One day after hMSCs preseeding, hiPSC-derived CM sheets were attached to both sides of dPMS and cultured for up to 14 days. Using cell nucleus staining, we found a significant increase of cell numbers in the dPMS (Figure 3B). Since CMs have limited capability of proliferation, hMSCs proliferation should be the main contributor of the increased total cell number in dPMS. To further analyze the seeded cells, we prestained hMSCs and CMs and tracked their distribution in the dPMS during the culture period. We found that preseeded hMSCs infiltrated throughout the dPMS 1 day after bilateral seeding while most of CMs stayed at the surfaces of dPMS with a small portion migrated to the center (Figure 3C). Close contact of both cell types was identified in the dPMS and most of them were located near to the surface of dPMS (Figure 3C). We then examined the cardiovascular markers expressions in the seeded cells (Figure 3D). For the examined endothelial cell markers (von Willebrand factor and α-smooth muscle actin) and cardiac-specific markers (cardiac troponin T, GATA4, connexin 43, and sarcomeric α-actinin), the expressions were highly regulated in cells seeded on dPMS for 14 days compared with 1 day. Some cells that positively expressed vWF and α-SMA were found to change their morphology from round to spindle shape during the culture period. The gap junction protein, CX 43, was not detected at day 1 but found to be expressed in the cell membrane between the adjacent cells at day 14 of culture. The costaining results further demonstrated that the cells expressing cardiac markers (cTnT and GATA4) were located near the surface of the dPMS while the endothelial marker (vWF) expressing cells were found throughout the dPMS.
Figure 3.
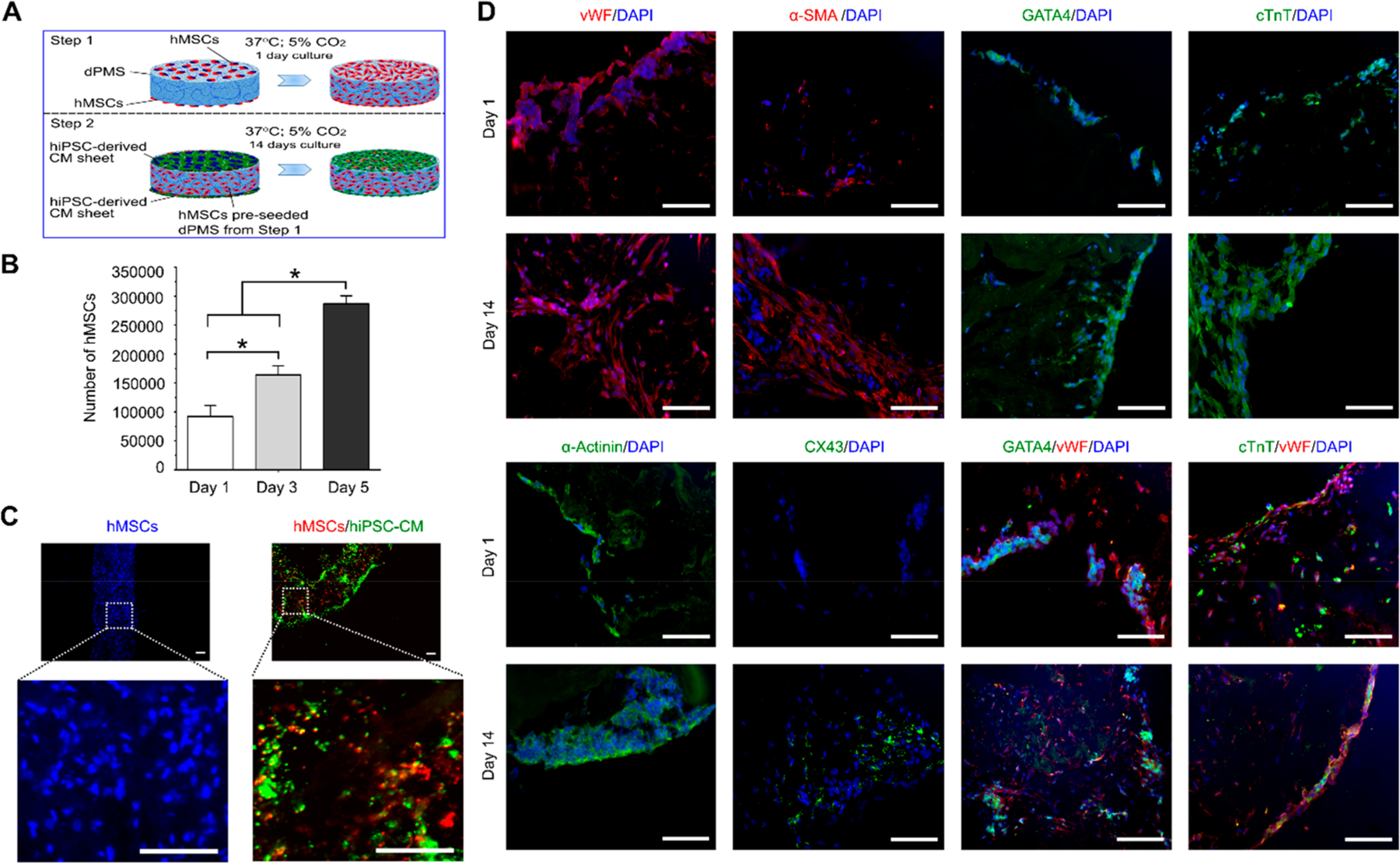
hiPSC-derived CM sheets and hMSCs preseeded dPMS interactions. (A) Schematic illustration to demonstrate the procedure of attaching hiPSC-derived CM sheets to dPMS that was preseeded with hMSCs. (B) Number of Proliferative cells (hMSCs) in dPMS after CM sheet attachment (n = 3). (C) Cell distribution in dPMS after hMSCs preseeding (left, 1 day after preseeding) and CM sheet attachment (right, at day 14 after seeding CM sheets). Blue: DAPI; Red: LuminiCell tracker 670; Green: LuminiCell Tracker 540. (D) Expressions of cardiac markers (GATA4, cTnT, sarcomeric α-actinin, and CX43) and vascular markers (vWF and α-SMA) in cells within dPMS at day 1 and 14 after attaching CM sheets to hMSCs preseeded dPMS. *Represents the statistically significant difference among the experiment groups with p-value less than 0.05. Scale bar: 100 μm.
3.4. Growth Factors and MMPs Secreted by Cell-seeded dPMS.
To examine whether the paracrine factors secreted by the preseeded hMSCs are the cause of the improved interactions between CMs sheet and dPMS, we used human growth factor array and human MMP array to measure the expression changes during the culture process. For human growth factor analysis, we chose 41 types of growth factors that have been well investigated in cardiovascular research and have shown to play key roles on affecting cell functions such as proliferation (e.g., EGF, G-CSF),27,28 migration (e.g., IGF-1 and IGFBP-1),29,30 differentiation (e.g., TGF-β, and IGFBP-4),31,32 and angiogenesis (e.g., VEGF and bFGF).33 The dPMSs seeded with both hMSCs and CMs were used as our samples. We measured the accumulative amounts of these target growth factors in both cell culture medium and scaffold during the 14 days of culture period. We found that the amount of 5 growth factors were significantly increased in either medium or scaffold including EGF, HGF, IGFBP-2, PDGF-β, and PDGF-AB (Figure 4A). Out of the rest 36 types of growth factors, the presence of 12 growth factors were increased from day 1 to day 14 in both medium and scaffold, which include Amphiregulin, GCSF, GM-CSF, IGFBP-2, IGF-1 R, M-CSF, M-CSF R, NT-4, PDGF-Rβ, VEGFR3, TGF-β1, and TGF-β2 (Figure 4B). In scaffolds, significantly higher amounts of PDGF-BB, PDGF-AB, HGF, VEGF-1, TGF-β1, and TGF-β2 were detected on day 14 compared with day 1 (Figure 4C). In the medium, we identified the significant increased amount of IGFBP-2 (Figure 4D). During the culture process, we noticed the degradation of the scaffolds seeded with hMSCs and CMs and with hMSCs alone (Figure 5A). Many studies have reported that MMPs and TIMPs are the main contributors to ECM degradation.34,35 Therefore, we performed the human MMP array analysis to examine the secretion patterns of MMPs and TIMPs from the seeded cells. We measured the amount of MMPs (MMP-1, -2, -3, -8, -9, -10, and -13) and TIMPs (TIMP-1, -2, and -4) released into the medium on day 1, 7, and 14 after seeding cells to dPMS (Figure 5B,C). When dPMSs were seeded with hMSCs, significantly higher amounts of MMP-1, TIMP-1, and TIMP-2 were detected in the medium. On day 14, the amounts of MMP-10, MMP-13, and TIMP-4 in medium were also significantly increased when compared with their amounts on day 1 (Figure 5D). When dPMSs were seeded with both hMSCs and CMs, significantly higher amounts of MMP-1, TIMP-1, and TIMP-2 were also detected in the medium. On day 14, the amounts of MMP-2 and TIMP-4 in medium were increased as compared with their amounts on day 1 (Figure 5E).
Figure 4.
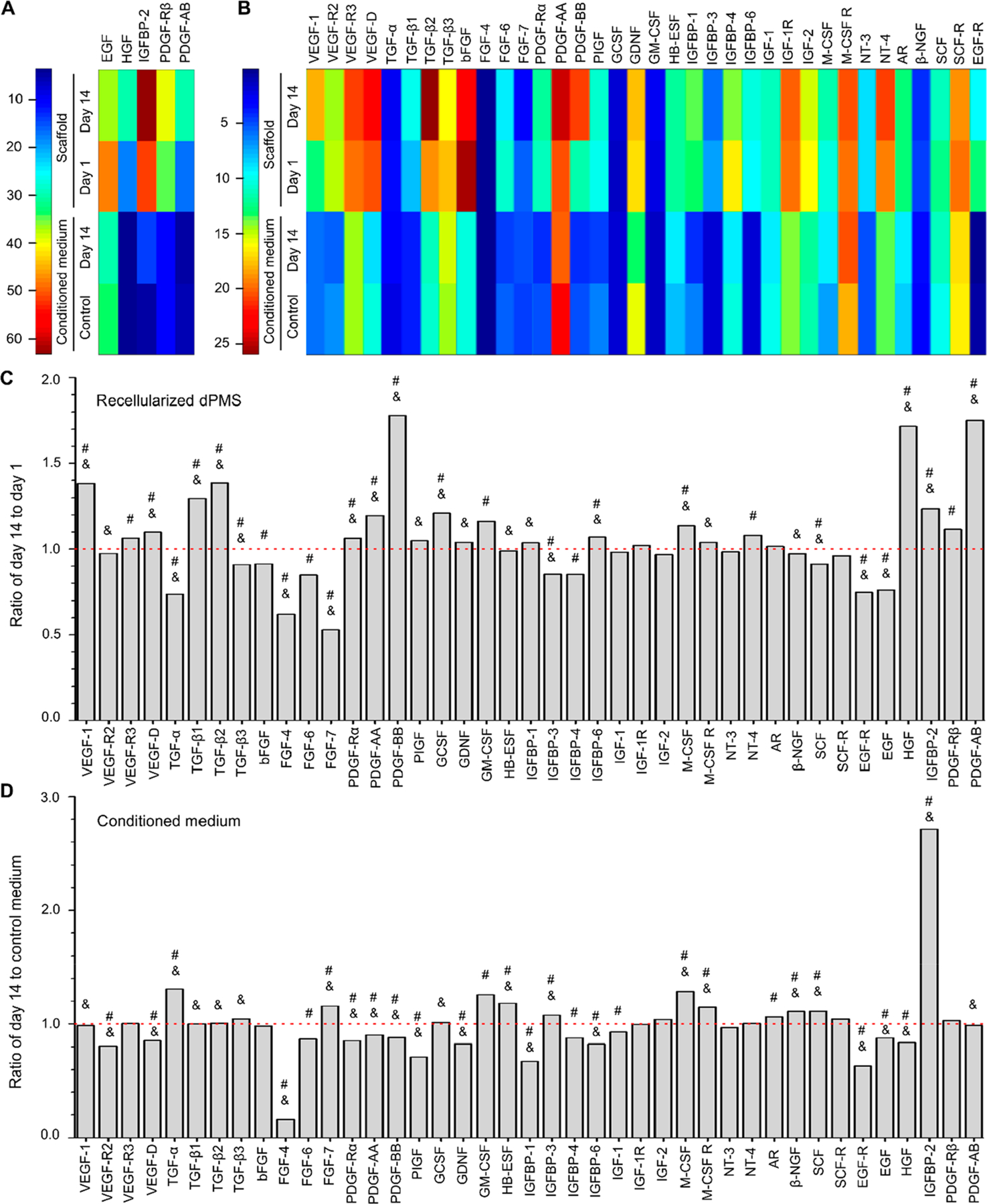
Growth factor secretion of cells in dPMS. Heat map analysis to demonstrate the expression level changes of specific human growth factors at large (A) and small scales (B). In the heat map, each column represents an individual growth factor and each row represents the tested sample. n = 4. The color bars denote the heat intensity for the expression value, with red indicating upregulated growth factors and blue indicating downregulated growth factors. Semiquantitative analysis of growth factors retained in the dPMS (C) and released into the conditioned medium (D) The y-axis represents the fold-change of growth factors from samples collected at day 14 after cell seeding to dPMS compared to day 1 (indicated by the red dotted line) (n = 4). #Represents the expression level ratio significantly different from 1 and marks the growth factor that has significantly different expression level ratios between in recelluarized dPMS and conditional medium. The full form of abbreviated growth factors is provided in the SI (Table S1).
Figure 5.
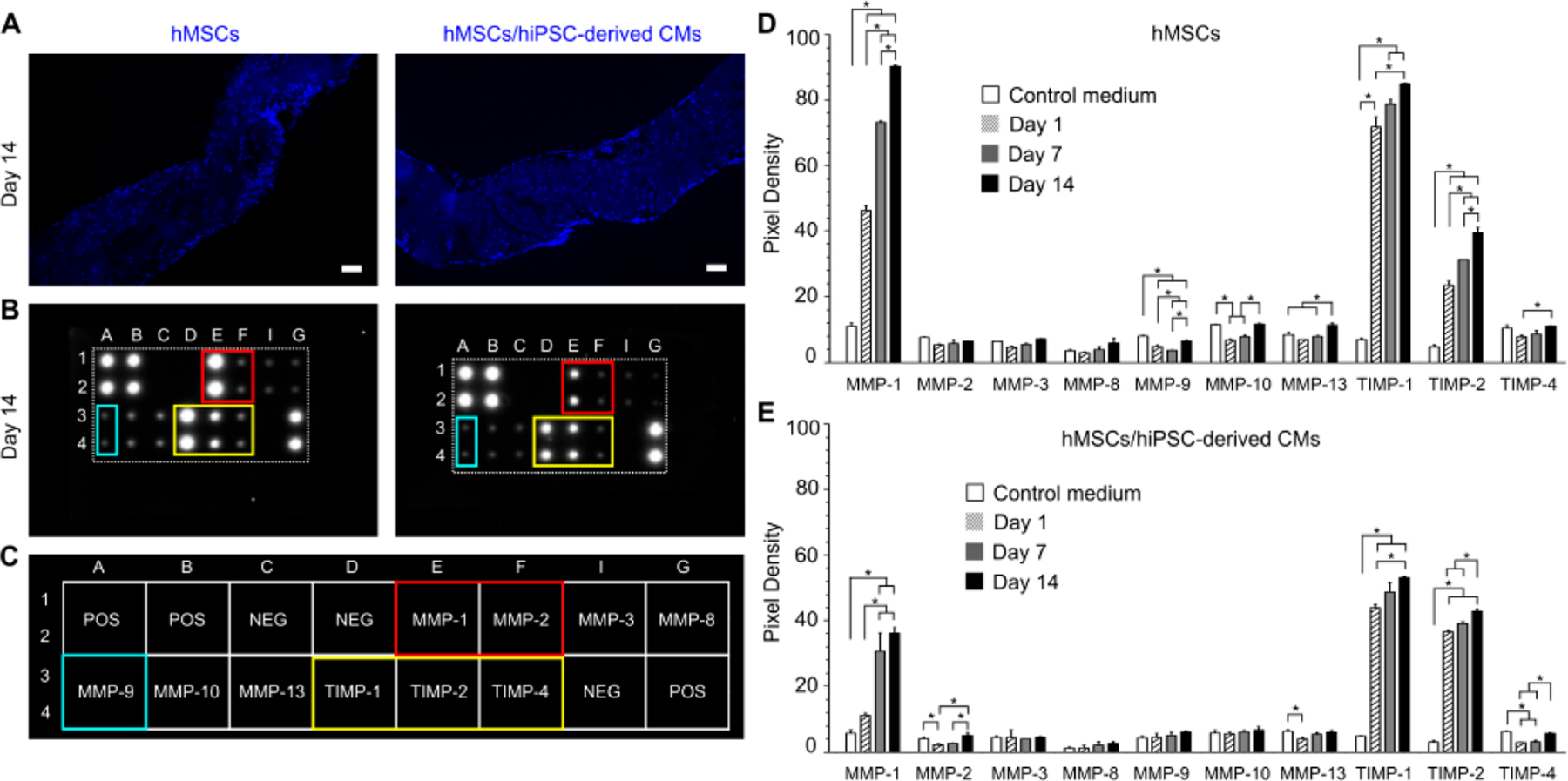
MMPs secretion from cells after seeding in dPMS. (A) Cell distribution at day 14 after bilaterally seeding hMSCs to dPMS (left) and CMs sheets to dPMS containing preseeded hMSCs (right). Blue: DAPI Nuclei staining. Scale bar: 100 μm. (B) Human MMP Array images showing the expression levels of 10 specific human MMPs in the sample conditioned medium 14 days after culture. (C) Map used to locate specific MMP antibody in the array. (D) Densitometry analysis to demonstrate the expression levels of 10 specific MMPs in medium conditioned by hMSCs seeded dPMS (n = 3). (E) Densitometry analysis to demonstrate the expression levels of 10 specific MMPs in medium conditioned by dPMS seeded with CMs sheets and hMSCs (n = 3) *Represents the statistically significant difference among the experiment groups with p-values less than 0.05.
4. DISCUSSIONS
In this study, we explored dPMS as an in vitro platform to investigate interactions between cells and cardiac matrix. The dPMS was fabricated from decellularized porcine myocardium that has been reported to retain cardiac-specific microstructure, ECM composition, and support cardiac cell attachment, growth, and migration.20,36,37 The thickness of the decellularized myocardium significantly affected the cell responses.38 Therefore, we sliced the decellularized myocardium into a thickness (600 μm) that has been proven to preserve matrix mechanical properties and support cell growth to generate the dPMS and serve as a cardiac matrix mimic. Delivering iPSCs-CMs to infarcted hearts using cell sheet technology has become a very promising approach for treating MI.39,40 MI results in the permanent loss of up to 1 billion cardiomyocytes (CMs) that have limited regenerative capacity.41 Cell sheet technology enables the delivery of a large amount of cells to the injured region of the heart at high efficiency.42 However, poor host integration of the deliver cells has been reported.10,11 Using the in vitro cell/matrix analysis platform, we tested our hypothesis that preseeding of MSCs will increase the integration of the cell sheet into the cardiac matrix.
The results obtained from this study proved our hypothesis. Similar to what has been observed in vivo, CMs delivered by cell sheet technology have a higher attachment rate to cardiac matrix than through cell suspension.43 CMs tend to remain on the surface of the cardiac matrix with very limited infiltration. Reducing the cell density of the sheet will not further enhance the interactions between the cell sheet and the cardiac matrix. MSC preseeding significantly increases the migration distances of CMs in cardiac matrix. CMs are found in the center of dPMS containing preseeded MSCs, which has not been seen in the control groups. In addition, during the 14 day culture period, upregulated expressions of mature CM markers (cTnT, CX43, and sarcomeric α-actinin) of the cells in the dPMS have been identified. It has been reported that when using the classical protocol to derive CMs from hiPSC, the obtained CMs remain largely immature when compared to human adult cardiomyocytes.44–46 The organ-specific cues of decellularized cardiac matrix have shown to facilitate the maturation of hiPSC-derived CMs both in vitro and in vivo.47,48 Taken all together, the enhanced infiltration of hiPSC-derived CMs in the dPMS caused by the preseeded MSCs increases their exposure to the cardiac differentiation cues and therefore improve their maturation.
The contributing mechanisms of the preseeded hMSCs have also been explored in this study. hMSCs may affect hiPSC-derived CMs directly or indirectly through paracrine factors and cell–cell contact mediated interactions. When seeded to dPMS bilaterally, hMSCs have shown to quickly (within 24 h) migrate throughout the matrix and kept proliferating. Close contact of hMSCs and CMs has been found in dPMS. Cell–cell contact could facilitate information sharing about internal and external conditions and coordinate their functions such as movement in response to specific situations. The observed hMSCs-CMs contact could be another contributor to the improved CMs integration to the cardiac matrix. After 14 days of culture, increasing number of cells that highly express mature vascular markers (vWF and α-SMA) have been observed in dPMS. Sincethe hiPSC-derived CMs have been verified for their cardiomyocyte phenotype prior to seeding, the cells that stained positive for vWF and α-SMA are most likely differentiated from the preseeded hMSCs. In our previous study, vascular differentiation of hMSCs in dPMS has also been detected.17,26 Recent studies have elucidated that vascular smooth muscle cells and endothelial cells can directly interact with CMs and regulate the maturation of hiPSC-derived CMs.49 Our growth factor array analysis has revealed a significantly increased angiogenic factors (e.g., VEGF and PDGF) and vascular differentiation inducers (e.g., TGF-β and IGFBP-4) in dPMS containing preseeded hMSCs. Likely, these secreted factors combined with the cardiac-specific microenvironment cues provided by dPMS cause hMSCs to differentiate into vascular cells and guide them to regulate CMs. Recently, it has been suggested that MMPs regulate many cell responses such as cell migration, proliferation and matrix invasion.50–52 Both hMSCs and CMs can secrete MMPs and TIMPs but usually with different secretion profiles.53,54 The pattern of secreted MMPs changes with different culture condition or physiological status.55 From the MMPs array analysis, we have found upregulated secretions of multiple MMPs and their inhibitors from hMSCs induced by their interactions with dPMS. The presence of MMP-1, TIMP-1, TIMP-2, and TIMP-4 in the conditioned medium has dramatically increased during the 14 day culture period. Further adding CMs sheet to the system has altered the expression patterns of MMP-2, MMP-9, MMP-10, and MMP-13. It is worth noting that the amount of MMPs secretion decreased when dPMS contained both hMSCs and CMs compared to hMSCs alone. The reasons caused the alteration of MMPs secretion and the roles of these MMPs on affecting the interactions of CMs, hMSCs and dPMS remain unclear. MSCs have been known for their capability of secreting various types of bioactive molecules such as cytokines, chemokines, immunomodulatory molecules, and growth factors in response to the local environment. Our results further suggest that the presence and communication of different type of cells could also affect the production of MSCs secretions. Likely, MSCs would change their secretion composition based on the need of neighboring cells.
Some limitations are noted in this study. The cell densities of recellularized dPMSs vary with different seeding approaches. MSCs preseeding increased the total cell number in the matrix, which may cause interaction changes among cells and between the cell–matrix. The preseeding of hMSCs could enhance the interactions between CM sheets and cardiac matrix but would not trigger CM functional contraction in the dPMS. Spontaneous beating was observed from the CMs we derived from iPSCs (Supporting Information, SI, Movie S1) and could not be detected once the CM sheet was attached to the matrix. The weight and mechanics of dPMS might be too much for the seeded CMs to overcome to contract. However, CM seeding number used in this study might be not sufficient for this thick and large scaffold. In the adult human myocardium, the cell density is about 1.0 × 108/cm3 tissue volume.56,57 Cardiomyocytes (CMs) constitute about 30–40% of the total cells in the heart,58 which are significantly higher in number than the CMs to tissue volume ratio used in this study (7.0 × 106/cm3). In addition, largely due to perfusion limitation, most of the infiltrated CMs were found within the range near to the surface of the dPMS. Perfusion culture will be exploited in our future study to improve CM distribution.
Our results have also indicated that hMSCs-induced vascularization and MMPs regulation are the two possible mechanisms that lead to the improved CMs and cardiac matrix interactions. Further examination of the individual and synergistic effects of these two mechanisms will enable the development of new approaches to facilitate transplanted cells for tissue repair. In the future we will further verify this conclusion in vivo by injecting hMSCs to myocardium prior to patching CMs sheets and examine the cell responses in a complex system. If our hypothesis is proven in vivo, then this strategy can be exploited into many cell-based cardiac therapies to help solve the host integration challenges. Hydrogel-based cell delivery systems can be developed to control release hMSCs and CMs sheet in a sequential manner to the infarcted area of heart for cardiac repair.
5. CONCLUSIONS
An in vitro cardiac matrix mimic using decellularized porcine myocardium slice was developed to test the effects of hMSC preseeding on CM sheets infiltration. Our results demonstrated MSC-preseeding significantly enhanced the cell migration distance, cell distribution, and mature vascular and cardiomyocyte marker expressions in the cardiac matrix. The growth factor and matrix metalloproteinases array analysis data suggested that hMSCs induced vascularization and MMP regulation are the two possible mechanisms that lead to the improved CMs and cardiac matrix interactions. This methodology might be exploited in cell-based cardiac therapies to solve the host integration challenges and might enable new approaches to facilitating transplanted cells for tissue repair.
Supplementary Material
ACKNOWLEDGMENTS
We greatly thank the financial support from the National Institute of Health (1R15HL122949, R15HL140503) and American Heart Association (19AIREA34400087).
Footnotes
The authors declare no competing financial interest.
Contributor Information
KC Pawan, Department of Biomedical Engineering, The University of Akron, Akron, Ohio 44325, United States.
Shah Mickey, Department of Biomedical Engineering and Department of Integrated Bioscience, The University of Akron, Akron, Ohio 44325, United States.
Shaik Rubia, Department of Biomedical Engineering, The University of Akron, Akron, Ohio 44325, United States.
Hong Yi, Department of Bioengineering, University of Texas at Arlington, Arlington, Texas 76019, United States.
Zhang Ge, Department of Biomedical Engineering, The University of Akron, Akron, Ohio 44325, United States.
REFERENCES
- 1.Masumoto H; Ikuno T; Takeda M; Fukushima H; Marui A; Katayama S; Shimizu T; Ikeda T; Okano T; Sakata R; Yamashita JK Human iPS cell-engineered cardiac tissue sheets with cardiomyocytes and vascular cells for cardiac regeneration. Sci. Rep 2015, 4 (1), 6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L; Xing Q; Zhai Q; Tahtinen M; Zhou F; Chen L; Xu Y; Qi S; Zhao F Pre-vascularization Enhances Therapeutic Effects of Human Mesenchymal Stem Cell Sheets in Full Thickness Skin Wound Repair. Theranostics 2017, 7 (1), 117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsushima H; Kuroki T; Adachi T; Kitasato A; Ono S; Tanaka T; Hirabaru M; Kuroshima N; Hirayama T; Sakai Y; Soyama A; Hidaka M; Takatsuki M; Kin T; Shapiro J; Eguchi S Human Fibroblast Sheet Promotes Human Pancreatic Islet Survival and Function in Vitro. Cell Transplant 2016, 25 (8), 1525–1537. [DOI] [PubMed] [Google Scholar]
- 4.Sekine W; Haraguchi YJ Thickness limitation and cell viability of multi-layered cell sheets and overcoming the diffusion limit by a porous-membrane culture insert. J. Biochips Tissue Chips 2011, 01 (01), 007 DOI: 10.4172/2153-0777.S1-007. [DOI] [Google Scholar]
- 5.Yang J; Yamato M; Nishida K; Hayashida Y; Shimizu T; Kikuchi A; Tano Y; Okano TJ Corneal epithelial stem cell delivery using cell sheet engineering: Not lost in transplantation. Drug Target 2006, 14 (7), 471–482. [DOI] [PubMed] [Google Scholar]
- 6.Alghuwainem A; Alshareeda AT; Alsowayan B Scaffold-Free 3-D Cell Sheet Technique Bridges the Gap between 2-D Cell Culture and Animal Models. Int. J. Mol. Sci 2019, 20 (19), 4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi H; Shimizu T; Yamato M; Tono K; Masuda H; Asahara T; Kasanuki H; Okano TJ Fibroblast sheets co-cultured with endothelial progenitor cells improve cardiac function of infarcted hearts. J. Artif. Organs 2008, 11 (3), 141–147. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi T; Miyagawa S; Pearson JT; Fukushima S; Saito A; Tsuchimochi H; Sonobe T; Fujii Y; Yagi N; Astolfo A; Shirai M; Sawa Y Functional and Electrical Integration of Induced Phiripotent Stem Cell-Derived Cardiomyocytes in a Myocardial Infarction Rat Heart. Cell Transplant 2015, 24 (12), 2479–2489. [DOI] [PubMed] [Google Scholar]
- 9.Sekine H; Shimizu T; Hobo K; Sekiya S; Yang J; Yamato M; Kurosawa H; Kobayashi E; Okano T Endothelial Cell Coculture Within Tissue-Engineered Cardiomyocyte Sheets Enhances Neovascularization and Improves Cardiac Function of Ischemic Hearts. Circulation 2008, 118 (14), S145–S152. [DOI] [PubMed] [Google Scholar]
- 10.Vunjak-Novakovic G; Tandon N; Godier A; Maidhof R; Marsano A; Martens TP; Radisic M Challenges in Cardiac Tissue Engineering. Tissue Eng., Part B 2010, 16 (2), 169–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu T; Sekine H; Yang J; Isoi Y; Yamato M; Kikuchi A; Kobayashi E; Okano T Polysurgery of cell sheet grafts overcomes diffusion limits to produce thick, vascularized myocardial tissues. FASEB J 2006, 20 (6), 708–710. [DOI] [PubMed] [Google Scholar]
- 12.Ranganath SH; Levy O; Inamdar MS; Karp JM Harnessing the Mesenchymal Stem Cell Secretome for the Treatment of Cardiovascular Disease. Cell Stem Cell 2012, 10 (3), 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vizoso F; Eiro N; Cid S; Schneider J; Perez-Fernandez R Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int. J. Mol. Sci 2017, 18 (9), 1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrada R; Li N; Sarojini H; An J; Lee M-J; Wang EJ Secretome from mesenchymal stem cells induces angiogenesis via Cyr61. J. Cell. Physiol 2009, 219 (3), 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segers VFM; Tokunou T; Higgins LJ; MacGillivray C; Gannon J; Lee RT Local Delivery of Protease-Resistant Stromal Cell Derived Factor-1 for Stem Cell Recruitment After Myocardial Infarction. Circulation 2007, 116 (15), 1683–1692. [DOI] [PubMed] [Google Scholar]
- 16.Iso Y; Spees JL; Serrano C; Bakondi B; Pochampally R; Song Y-H; Sobel BE; Delafontaine P; Prockop DJ Multipotent human stromal cells improve cardiac function after myocardial infarction in mice without long-term engraftment. Biochem. Biophys. Res. Commun 2007, 354 (3), 700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kc P; Shah M; Liao J; Zhang G Prevascularization of Decellularized Porcine Myocardial Slice for Cardiac Tissue Engineering. ACS Appl. Mater. Interfaces 2017, 9 (3), 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Meglio F; Nurzynska D; Romano V; Miraglia R; Belviso I; Sacco AM; Barbato V; Di Gennaro M; Granato G; Maiello C; Montagnani S; Castaldo C Optimization of Human Myocardium Decellularization Method for the Construction of Implantable Patches. Tissue Eng., Part C 2017, 23 (9), 525–539. [DOI] [PubMed] [Google Scholar]
- 19.Lu TY; Lin B; Kim J; Sullivan M; Tobita K; Salama G; Yang L Repopulation of decellularized mouse heart with human induced pluripotent stem cell-derived cardiovascular progenitor cells. Nat. Commun 2013, 4 (1), 2307. [DOI] [PubMed] [Google Scholar]
- 20.Ott HC; Matthiesen TS; Goh S-K; Black LD; Kren SM; Netoff TI; Taylor DA Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat. Med 2008, 14 (2), 213–221. [DOI] [PubMed] [Google Scholar]
- 21.Kc P; Hong Y; Zhang G Cardiac tissue-derived extracellular matrix scaffolds for myocardial repair: advantages and challenges. Regen. Biomater 2019, 6 (4), 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross S; Holliday M; Lim S; Semsarian C Characterization of the first induced pluripotent stem cell line generated from a patient with autosomal dominant catecholaminergic polymorphic ventricular tachycardia due to a heterozygous mutation in cardiac calsequestrin-2. Stem Cell Res 2019, 37, 101450. [DOI] [PubMed] [Google Scholar]
- 23.Poulin H; Martineau L; Racine V; Puymirat J; Chahine M Differentiation of lymphoblastoid-derived iPSCs into functional cardiomyocytes, neurons and myoblasts. Biochem. Biophys. Res. Commun 2019, 516 (1), 222–228. [DOI] [PubMed] [Google Scholar]
- 24.Lu HE; Tsai MS; Yang YC; Yuan CC; Wang TH; Lin XZ; Tseng CP; Hwang SM Selection of alkaline phosphatase-positive induced pluripotent stem cells from human amniotic fluid-derived cells by feeder-free system. Exp. Cell Res 2011, 317 (13), 1895–1903. [DOI] [PubMed] [Google Scholar]
- 25.Lundy SD; Zhu W-Z; Regnier M; Laflamme MA Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cells Dev 2013, 22 (14), 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah M; KC P; Copeland KM; Liao J; Zhang G A Thin Layer of Decellularized Porcine Myocardium for Cell Delivery. Sci. Rep 2018, 8 (1), 16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aghila Rani KG; Kartha CC Effects of epidermal growth factor on proliferation and migration of cardiosphere-derived cells expanded from adult human heart. Growth Factors. Growth Factors 2010, 28 (3), 157–165. [DOI] [PubMed] [Google Scholar]
- 28.Shimoji K; Yuasa S; Onizuka T; Hattori F; Tanaka T; Hara M; Ohno Y; Chen H; Egasgira T; Seki T; Yae K; Koshimizu U; Ogawa S; Fukuda K G-CSF Promotes the Proliferation of Developing Cardiomyocytes In Vivo and in Derivation from ESCs and iPSCs. Cell Stem Cell 2010, 6 (3), 227–237. [DOI] [PubMed] [Google Scholar]
- 29.Jones JI; Gockerman A; Busby WH; Wright G; Clemmons DR Insulin-like growth factor binding protein 1 stimulates cell migration and binds to the alpha 5 beta 1 integrin by means of its Arg-Gly-Asp sequence. Proc. Natl. Acad. Sci. U. S. A 1993, 90 (22), 10553–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y; Han X; Wang X; Zhu B; Li B; Chen Z; Ma G; Wan M Sustained Release of IGF-1 by 3D Mesoporous Scaffolds Promoting Cardiac Stem Cell Migration and Proliferation. Cell. Physiol. Biochem 2018, 49 (6), 2358–2370. [DOI] [PubMed] [Google Scholar]
- 31.Xue Y; Yan Y; Gong H; Fang B; Zhou Y; Ding Z; Yin P; Zhang G; Ye Y; Yang C; Ge J; Zou YJ Insulin-Like Growth Factor Binding Protein 4 Enhances Cardiomyocytes Induction in Murine-Induced Pluripotent Stem Cells. J. Cell. Biochem 2014, 115 (9), 1495–1504. [DOI] [PubMed] [Google Scholar]
- 32.Goumans M-J; de Boer TP; Smits AM; van Laake LW; van Vliet P; Metz CHG; Korfage TH; Kats KP; Hochstenbach R; Pasterkamp G; Verhaar MC; van der Heyden MAG; de Kleijn D; Mummery CL; van Veen TAB; Sluijter JPG; Doevendans PA TGF-β1 induces efficient differentiation of human cardiomyocyte progenitor cells into functional cardiomyocytes in vitro. Stem Cell Res 2008, 1 (2), 138–149. [DOI] [PubMed] [Google Scholar]
- 33.Rebouca̧ J; de S; Santos-Magalhaẽs NS Formiga FR. Cardiac Regeneration using Growth Factors: Advances and Challenges. Arq. Bras. Cardiol 2016, 107 (3), 271–275, DOI: 10.5935/abc.20160097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jabłonśka-Trypuć A; Matejczyk M; Rosochacki S Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J. Enzyme Inhib. Med. Chem 2016, 31 (sup1), 177–183. [DOI] [PubMed] [Google Scholar]
- 35.Nagase H; Visse R; Murphy G Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res 2006, 69 (3), 562–573. [DOI] [PubMed] [Google Scholar]
- 36.Wang B; Tedder ME; Perez CE; Wang G; de Jongh Curry AL; To F; Elder SH; Williams LN; Simionescu DT; Liao J Structural and biomechanical characterizations of porcine myocardial extracellular matrix. J. Mater. Sci.: Mater. Med. 2012, 23 (8), 1835–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q; Yang H; Bai A; Jiang W; Li X; Wang X; Mao Y; Lu C; Qian R; Guo F; Ding T; Chen H; Chen S; Zhang J; Liu C; Sun N Functional engineered human cardiac patches prepared from nature’s platform improve heart function after acute myocardial infarction. Biomaterials 2016, 105, 52–65. [DOI] [PubMed] [Google Scholar]
- 38.Shah M; Kc P; Zhang G In Vivo Assessment of Decellularized Porcine Myocardial Slice as an Acellular Cardiac Patch. ACS Appl. Mater. Interfaces 2019, 11 (27), 23893–23900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishigami M; Masumoto H; Ikuno T; Aoki T; Kawatou M; Minakata K; Ikeda T; Sakata R; Yamashita JK; Minatoya K Human iPS cell-derived cardiac tissue sheets for functional restoration of infarcted porcine hearts. PLoS One 2018, 13 (8), No. e0201650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L; Guo J; Zhang P; Xiong Q; Wu SC; Xia L; Roy SS; Tolar J; O’Connell TD; Kyba M; Liao K; Zhang J Derivation and High Engraftment of Patient-Specific Cardiomyocyte Sheet Using Induced Pluripotent Stem Cells Generated From Adult Cardiac Fibroblast. Circ.: Heart Failure 2015, 8 (1), 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundy SD; Gantz JA; Pagan CM; Filice D; Laflamme MA Pluripotent Stem Cell Derived Cardiomyocytes for Cardiac Repair. Curr. Treat. Options Cardiovasc. Med 2014, 16 (7), 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haraguchi Y; Shimizu T; Yamato M; Okano T Regenerative Therapies Using Cell Sheet-Based Tissue Engineering for Cardiac Disease. Cardiol. Res. Pract 2011, 2011, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekine H; Shimizu T; Dobashi I; Matsuura K; Hagiwara N; Takahashi M; Kobayashi E; Yamato M; Okano T Cardiac Cell Sheet Transplantation Improves Damaged Heart Function via Superior Cell Survival in Comparison with Dissociated Cell Injection. Tissue Eng., Part A 2011, 17 (23–24), 2973–2980. [DOI] [PubMed] [Google Scholar]
- 44.Ahmed RE; Anzai T; Chanthra N; Uosaki H A Brief Review of Current Maturation Methods for Human Induced Pluripotent Stem Cells-Derived Cardiomyocytes. Front. Cell Dev. Biol 2020, 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machiraju P; Greenway SC Current methods for the maturation of induced pluripotent stem cell-derived cardiomyocytes. World J. Stem Cells 2019, 11 (1), 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koivumak̈i JT; Naumenko N; Tuomainen T; Takalo J; Oksanen M; Puttonen KA; Lehtonen Š; Kuusisto J; Laakso M; Koistinaho J; Tavi P Structural Immaturity of Human iPSC-Derived Cardiomyocytes: In Silico Investigation of Effects on Function and Disease Modeling. Front. Physiol 2018, 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fong AH; Romero-Loṕez M; Heylman CM; Keating M; Tran D; Sobrino A; Tran AQ; Pham HH; Fimbres C; Gershon PD; Botvinick EL; George SC; Hughes CCW Three-Dimensional Adult Cardiac Extracellular Matrix Promotes Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Tissue Eng., Part A 2016, 22 (15–16), 1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadota S; Pabon L; Reinecke H; Murry CE In Vivo Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Neonatal and Adult Rat Hearts. Stem Cell Rep 2017, 8 (2), 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scuderi GJ; Butcher J Naturally Engineered Maturation of Cardiomyocytes. Front. Cell Dev. Biol 2017, 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson JL; Dwivedi A; Somerville M; George SJ; Newby AC Matrix Metalloproteinase (MMP)-3 Activates MMP-9 Mediated Vascular Smooth Muscle Cell Migration and Neointima Formation in Mice. Arterioscler. Thromb. Vasc. Biol 2011, 31 (9), e35–e44, DOI: 10.1161/atvbaha.111.225623. [DOI] [PubMed] [Google Scholar]
- 51.Webb AH; Gao BT; Goldsmith ZK; Irvine AS; Saleh N; Lee RP; Lendermon JB; Bheemreddy R; Zhang Q; Brennan RC; et al. Inhibition of MMP-2 and MMP-9 decreases cellular migration, and angiogenesis in in vitro models of retinoblastoma. BMC Cancer 2017, 17 (1), 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newby A Matrix metalloproteinases regulate migration, proliferation, and death of vascular smooth muscle cells by degrading matrix and non-matrix substrates. Cardiovasc. Res 2006, 69 (3), 614–624. [DOI] [PubMed] [Google Scholar]
- 53.Lozito TP; Tuan RS Mesenchymal stem cells inhibit both endogenous and exogenous MMPs via secreted TIMPs. J. Cell. Physiol 2011, 226 (2), 385–396. [DOI] [PubMed] [Google Scholar]
- 54.DeLeon-Pennell KY; Meschiari CA; Jung M; Lindsey ML Matrix Metalloproteinases in Myocardial Infarction and Heart Failure. Prog. Mol. Biol. Transl 2017, 147, 75–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sternlicht MD; Werb Z How Matrix Metalloproteinases Regulate Cell Behavior. Annu. Rev. Cell Dev. Biol 2001, 17 (1), 463–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y; Zhang X; Wei Y; Guo C; Li R; Zeng Q; Zhang Y Culturing of Ventricle Cells at High Density and Construction of Engineered Cardiac Cell Sheets Without Scaffold. Int. Heart J 2009, 50 (5), 653–662. [DOI] [PubMed] [Google Scholar]
- 57.Zia S; Mozafari M; Natasha G; Tan A; Cui Z; Seifalian AM Hearts beating through decellularized scaffolds: whole-organ engineering for cardiac regeneration and transplantation. Crit. Rev. Biotechnol 2016, 36 (4), 705–11. [DOI] [PubMed] [Google Scholar]
- 58.Zhou P; Pu WT Recounting Cardiac Cellular Composition. Circ. Res 2016, 118 (3), 368–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


