Abstract
Bacterial biofilms are often defined as communities of surface attached bacteria. Biofilms are typically depicted with a classic mushroom-shaped structure that is a characteristic of Pseudomonas aeruginosa. However, it has become evident that this is not how all biofilms develop, especially in in vivo in clinical, industrial settings and in the environment where biofilms often are observed as non-surface attached aggregates. In this Review, we describe the rationale behind the 5-step model and why it fails to capture many aspects of bacterial biofilm physiology. We aim to present a simplistic developmental model for biofilm formation that is flexible enough to include all the diverse scenarios and microenvironments where biofilms are formed. With this new expanded, inclusive model, we hereby introduce a common platform for developing understanding of biofilms and antibiofilm strategies that can be tailored to the microenvironment under investigation.
Introduction
Over the past 40 years, microbiologists have categorized bacteria as displaying two life forms in nature. In one, the bacteria appear as single, independent free-floating cells (planktonic). In the other, bacteria are organized in microbial aggregates (biofilms). In addition, the word “biofilm” originally referred to biomaterial on a surface, however, more recently, non-surface attached aggregated bacteria have also been recognized as biofilms (1, 2). Likewise, non-surface associated aggregates are now recognized in clinical settings. Chronic biofilm infections are further divided into surface associated or non-surface associated infections. Surface associated infections are commonly observed in patients with implants or medical devices. Non-surface associated infections include respiratory tract infections with impaired host mucociliary clearance (in viscous airway mucus in people with cystic fibrosis (CF)), or persistent soft tissue infections that are associated with comorbidities such as diabetes and impaired vascularization of the lower limbs predisposing to non-healing wounds (3). Moreover, until recently, bacteria growing planktonically have been associated with acute infections that are generally treatable with antibiotics, though successful treatment largely depends on accurate and fast diagnosis. In cases where bacteria succeed in forming biofilms within the host, the infection is often untreatable and, sustained by low-grade inflammation, develops into a chronic state (4). However, this dogma is challenged by recent findings that suggest that the difference between bacteria in acute and chronic infections is due to metabolic activity rather than aggregation (5).
In the environment, the functional consequences of bacterial life in biofilms have been associated with enhanced protection from shear stress, desiccation, toxic compounds and protozoan grazing (6). Moreover, retention of enzymes in the biofilm matrix was proposed to improve efficiency and diversity of organic matter decomposition, and biofilm formation on plant roots and fungal cells may promote bacterial nutrient acquisition and transport, respectively (7). Pathogenic biofilms that form on plants may also have serious disease consequences(8–10). While (motile) planktonic cells are primarily found in water columns and soil pores, the predominant forms of microbial life in natural environments are linked to highly diverse biofilm communities in aquatic environments (including sediments, submerged surfaces, as free-floating flocs and on higher organisms), sediments and soil (e.g. on litter, plant roots and soil particles) (11). Likewise, biofilms dominate in industrial microbial applications, such as cleaning of wastewater and bioremediation of soil and water(12, 13).
Biofilms are associated with microbially induced corrosion in oil field pipelines, plugging pipes, fouling ship hulls creating drag and increased fuel costs, reducing heat transfer in cooling towers, and fouling manufacturing lines resulting in product contamination (13). In all these instances, the industrial system is not sterile, and so it is not necessarily an issue that bacteria are present, but more that the biofilm compromises a product or system performance. In these cases, biocide manufacturers develop clean-in-place procedures to control biofilm growth, but in reality, the biofilm is never completely eliminated, and thus, as with dental biofilms, routine cleaning and maintenance is required to keep the biofouling in check, for example in the souring of oil reservoirs by sulfate reducing bacteria (14).
A common denominator of bacterial biofilms is the distinction between surface-attached and non-surface-attached bacterial aggregates, despite new evidence showing that these share similar phenotypes (15, 16) [REF]. For both of these phenotypes the bacteria create microenvironments which in turn influence bacterial community and behavior in an interdependent and dynamic manner (17, 18). In this review we define bacterial aggregates as biofilms irrespective of attachment to a biotic or abiotic surface, and define the aggregation of bacteria as the central hallmark of bacterial biofilms (See Text Box 1).
Text box 1: Biofilm nomenclature.
BIOFILM
A microbial aggregate attached or associated with a surface and embedded in a matrix. These can include single or multiple discrete aggregates or more continuous films.
AGGREGATE
A cohesive group of microbial cells surrounded by extracellular polymeric substances, and other entrapped abiotic or biotic materials. Microbial aggregates can be surface attached, matrix-associated, or free floating in the liquid phase and display a biofilm-like phenotype.
AGGREGATION
Any biological, chemical, or physical process that allows microbial cells to form an aggregate.
Microbial Growth.
Replication of individual microbial cells that remain cohesive, whether in suspension or attached to a surface.
Autoaggregation and coaggregation.
The formation of aggregates (also known as clumps) in suspension by bacteria of the same species (autoaggregation) of by bacteria of different species (coaggregation)
Polymer depletion aggregation or depletion aggregation.
The formation of aggregates in suspension through a colloidal physics phenomenon that occurs when polymers in solution are of high enough concentration and molecular weight to initiate phase separation “forcing” microbial cells together.
Polymer bridging.
The aggregation of microbial cells in suspension caused by polymers which adhere to cell wall components forming a bridging bond between multiple cells.
ADHERENCE/ATTACHMENT
Suspended single cells or aggregates that adhere to a host cellular surface or attach to an abiotic surface, either directly to the substratum or to previously attached microbial cells or clusters.
GROWTH
Expansion of aggregates by microbial growth and concomitant EPS production, whether in suspension or attached to a surface.
ACCUMULATION
The net result of attachment, aggregation, growth, disaggregation, and detachment processes that leads to expansion or shrinkage of a biofilm or aggregate.
DISAGGREGATION
Aggregated cells, whether in suspension or associated with a surface, that shed smaller microbial aggregates or individual cells into the fluid phase.
Erosion.
Loss of single cells or very small aggregates from the aggregate or biofilm surface due to physical forces such as fluid shear or expansion of cells by growth.
Dispersal.
Specifically connotes an active and biologically-regulated release of microbial cells from an aggregate.
Cohesive fracture.
Microbial aggregates in suspension or as biofilms break apart due to internal mechanical failure, for example from forces applied by moving fluid.
Predation/Phagocytosis.
For example, by amoeba or white blood cells that physically engulf microbes from the surface of an aggregate or by fracturing or rending pieces from the aggregate.
Detachment
An overarching term encompassing all phase transfer processes in which microbial cells and extracellular polymeric substances move from the surface-attached phase to a fluid-borne phase. This term is specific to surface-attached biofilms.
Sloughing.
Release of coherent layers of surface-attached biofilm by adhesive failure (i.e., at the biofilm-substratum interface), generally by fluid shear. This mechanism is specific to surface-attached biofilms.
Removal.
Implies the response to a mechanical, chemical, or enzymatic intervention that causes attached aggregates or cells to be release from the surface.
Predation/Phagocytosis.
For example, by amoeba or white blood cells that physically engulf microbes from the surface of an aggregate or by fracturing pieces from the aggregate.
Growing biofilms in the laboratory
While bacteria have been studied in the laboratory for well over 100 years, biofilms were first studied after surface-attached bacteria were observed attached to the pacemaker lead in a patient suffering from recurrent bacteremia (19) and growing on glass slides inoculated with sea water (20). The bacteria attached to the pacemaker lead mark one of the first references to “biofilm growing bacteria” in medicine, with a subsequent explosion of interest in biofilm infections. Numerous in vitro systems have been devised to study biofilm formation (21–23) and how biofilm bacteria differ from planktonic cells, including their hallmark property of increased antibiotic tolerance, or the presence of an extracellular polymeric (EPS) matrix, a hydrogel-like substance encasing biofilm cells (24, 25). These initial findings supported the notion that microorganisms undergo significant changes in their phenotypic repertoire during the transition from planktonic to biofilm growth (Text Box 2) and revealed the potential for new ways to control or manipulate biofilms. The in vitro systems commonly used shaken, well mixed cultures, and led to most biofilm experiments being initiated by using single cell planktonic cultures with one, controlled seeding event. Likewise, the transformation of single cells into sessile biofilm communities has been thoroughly studied in closed, surface-based in vitro systems without the influx of new cells during the biofilm formation and maturation process (26–28). Such studies led to a key publication in the field describing the developmental stages of P. aeruginosa (a nosocomial pathogen), presenting the current influential “5-step biofilm model” (Figure 1) (26). The biofilm developmental stages are referred to as reversible and irreversible attachment, biofilm maturation I and II involving cluster and microcolony formation, respectively, and dispersion (26, 29). Variations of this model have also been developed for other species such as Staphylococcus aureus (30) and the soil bacterium Bacillus subtilis (31) and for algal biofilms (32).
Text box 2: Before the 5-step biofilm model.
Biofilm research in the early years primarily focused on engineering applications and observational descriptions of biofilms. However, biofilm research changed with the observation of surface-attachment specific gene regulation in vitro and the introduction of in vitro systems to study biofilm formation and phenotypes in the laboratory. This facilitated the study of specific and differential gene expression upon surface attachment in vitro (45, 46, 162) including the role of cell signaling as genetic regulation from a population (162) as well as the use of genetic tools to identify genes required for in vitro surface and subsequent biofilm formation (163–165).
The idea that biofilms are amenable to molecular genetic studies (163, 164) also opened the door to the exploration of factors beyond early surface attachment, to those contributing to biofilm architecture, metabolic interactions, phylogenetic groupings, competition and cooperation. Molecular genetic applications furthermore led to exciting progress in the development of new technologies for studying biofilm communities, advanced our understanding of the ecological significance of surface-attached bacteria, and provided new insights into the molecular genetic basis of biofilm development (41).
What followed was extensive research on genes that are required for bacteria to associate with surfaces, and investigations of differences in the transcriptional abundance of bacterial genes when growing planktonically and as biofilms. While some studies failed to detect differences in the transcriptomes of planktonic and surface associated cells (166), the majority of studies confirmed planktonic and sessile biofilm cells display distinct transcriptomic profiles, with the number of genes changing in transcript abundance upon surface-associated growth ranging from less than twenty to several hundred (167, 168). Moreover, numerous reports indicate that biofilms are heterogenous, with bacteria residing at different locations within the biofilm structure experiencing steep gradients in the concentration of nutrient resources, oxygen and waste products (such as acids produced by fermentation in oxygen-depleted zones), as well as metabolites and extracellular signaling molecules, resulting in the modulation of metabolic rates, dormancy, stress responses and mutation rates (160, 169–171). Transcriptome analyses of in vitro grown biofilms confirmed that biofilm cells experience various stresses including hypoxia or oxygen deprivation, nutrient stress, and slow growth, which increase as the biofilm grows in size (172–174). Imaging and transcriptome-imaging (parallel sequential fluorescence in situ hybridization, par-seq FISH) have provided visual evidence of the presence of subpopulations and associated gradients (i.e. chemical, signaling molecules) within the biofilm structure (119, 175–177). Additionally, changes in cell-to-cell signaling, virulence gene expression and the biosynthesis of matrix components have been reported (178–182). Notably, many of these findings have been confirmed using in vivo (animal models) biofilms, although not in human infections (183, 184).
Figure 1.
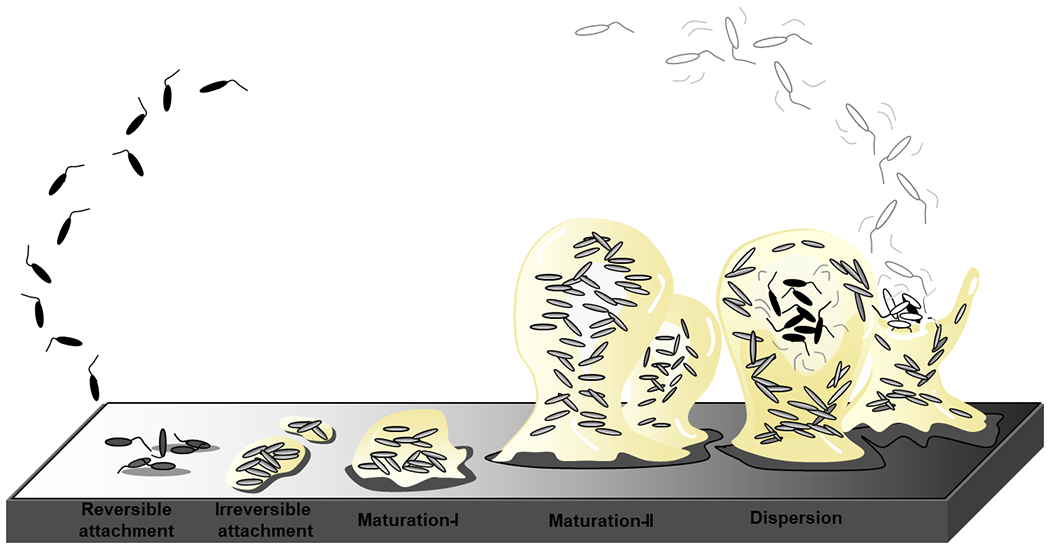
The stages of biofilm development as depicted in (26). The formation of biofilms is a cyclic process that occurs in a stage-specific and progressive manner. The process is initiated following surface contact by single planktonic cells. Several developmental steps are discernable as reversible attachment, irreversible attachment and biofilm maturation (maturation-I and -II)(26, 43). During reversible attachment, bacteria attach to the substratum via the cell pole or via the flagellum (step I), followed by longitudinal attachment. Transition to the irreversible coincides with a reduction in flagella reversal rates, reduction in flagella gene expression and the production of biofilm matrix components. This stage is also characterized by attached cells demonstrating drug tolerance(44). Biofilm maturation stages are characterized by the appearance of cell clusters that are several cells thick and are embedded in the biofilm matrix (maturation-I stage) which subsequently fully mature into microcolonies (maturation-II stage)(26, 43). Dispersion has been reported to coincide with the decrease in and degradation of matrix components, with dispersed cells being motile and demonstrating increased drug susceptibility relative to biofilm cells. The biofilm matrix is shown in beige.
While the schematic conceptual biofilm developmental model based on P. aeruginosa in vitro biofilm formation is easy to understand and has been widely generalized to describe all biofilms, this model does not necessarily describe the complexity of biofilms in real-world industrial, natural and clinical settings. Importantly, this model does not reflect the relevant microenvironments that develop within these biofilms. It is, therefore, important to consider the substantial differences between the processes occurring in a laboratory flow cell and those leading to biofilm formation in the real-world scenarios including the rhizosphere, oil pipelines, chronic wounds, the respiratory tract, at an air-water interface (i.e. a pellicle), around a prosthetic joint, in a wastewater granule, biofilms grown in microcosm ecosystems (33–35) (Liu et al., 2021; Zhao et al., 2020), and on ex vivo organoid associated biofilms (Wu et al., 2021). In such diverse systems, the processes of attachment, aggregation, interaction with biotic or abiotic materials and interfaces (e.g., roots, tissue, a gas phase, environmental polymers, corrosion deposits), growth and maturation, and detachment/dispersal are potentially quite different and do not necessarily occur sequentially. Given the variety of systems and conditions, we propose it would be useful to expand the existing model to include a wider spectrum of real-world scenarios.
In this Review, we describe the origin of the current biofilm model, its shortcomings and discuss differences in biofilm formation by diverse types of bacteria in varied experimental systems, both in vitro and in vivo, focus on new findings such as the common lack of an attachment surface, differences in matrix properties and transcriptional profiles etc. that warrant amending the current model. We suggest a revised conceptual model that encompasses the three major steps of biofilm life, aggregation, growth and disaggregation independently of surfaces and initiation from single-cell planktonic bacteria, to present a revised and simple model that we believe represents a broader range of biofilm systems.
The origin of the 5-step biofilm model
Numerous studies support the notion that biofilm formation starts with the initial surface attachment of single free floating, planktonic cells and that biofilm cells differ from their planktonic counterparts in the genes and proteins that they express (Box 2). Given the profound changes that microorganisms undergo during their transition from planktonic organisms to cells that are part of a complex, surface-attached community, it is not surprising that the transition from the planktonic to the biofilm mode of growth is a complex and highly regulated process, that is often regarded to be developmental. However, while it was widely accepted that the transition to a surface lifestyle was a highly regulated process, it remained unknown whether subsequent surface associated growth progressed simply as an accumulation of cells due to cell division or instead coincided with distinct events indicative of progressive or transitional changes over the course of biofilm formation. In an effort to better understand the progression of biofilm formation, in 2002, researchers (36) made use of a combination of direct observation by microscopy, evaluation of biofilm morphology, matrix polymer production, activation of quorum sensing-regulated genes and the quantitative analysis of protein abundance. The analysis led to the realization that over the course of biofilm formation, P. aeruginosa displays multiple phenotypes with distinct physiological characteristics (structural and metabolic changes) that can be correlated to distinct episodes or stages of biofilm development (Figure 1). These stages were referred to as reversible and irreversible attachment, maturation (maturation-I and -II stages), and dispersion, with each biofilm developmental stage corresponding to unique patterns of protein production and gene expression (37–42). The distinction between reversible and irreversible attachment was based on the time scale of the fate (whether it remains attached or detaches) of a cell over a few minutes once it contacts a surface.
In P. aeruginosa, the reversible attachment stage is characterized by cells attaching to a surface by a single pole (Fig. 1). Most surface contact is unstable, and cells are often seen returning to the bulk phase. Once rod-shaped cells commit to a more stable surface existence, cells attach via their longitudinal axis. This phenomenon is referred to as ‘irreversible attachment’ (Fig. 1). Reports, furthermore, suggest irreversible attachment initiates a cascade of changes in the bacterial cells. Apparent changes following bacterial attachment include cessation of flagella-mediated motility while at the molecular levels, changes include surface-induced gene activation of P. aeruginosa algC, a gene involved in lipopolysaccharide core biosynthesis and in the biosynthesis of the exopolysaccharide alginate (45, 46), induction of genes involved in the biosynthesis of the Psl matrix polymer (47), as well as genes linked to antibiotic resistance, including β-lactamase (48), phenazine (49), SagS and BrlR (44). The findings suggest that committing to the surface associated mode of growth not only coincides with the production of biofilm matrix components that enable cells to more firmly cement themselves to the surface, but also with biofilm antimicrobial tolerance, a hallmark characteristic of biofilms, as an early adaptive response to the sessile lifestyle. Once attached, cells will grow into a more complex multicellular mature form, which in some bacterial species including P. aeruginosa is characterized by the presence of differentiated, mushroom- or pillar-like structures or microcolonies interspersed with fluid-filled channels (50) (Fig. 1). The structuring of biofilms in microcolonies with water channels has been shown to be dependent on intercellular small messenger molecules (acylated homoserine lactones, AHLs) that are used for bacterial communication (51), rhamnolipids (52, 53), and regulatory proteins, mostly 2-component regulatory systems (54–56). However, in P. aeruginosa even cell signaling knockout mutants have been shown to form such channels suggesting their structure is determined by the interplay between intrinsic bacterial regulation and environmental conditions (57). As biofilms develop three-dimensional structure, resident bacteria near the base undergo increasingly physical separation from the bulk liquid interface and essential sources of energy or nutrients, with biofilm cells experiencing an ever-changing micro-environment. Changes are driven by cellular crowding, chemical gradients, and nutrient competition, leading to stratification within the biofilm and the creation of subpopulations (58, 59). Thus, bacteria residing at different locations within the biofilm structure experience concentration gradients of nutrient resources, oxygen and waste products (such as acids produced by fermentation in oxygen-depleted zones) as well as extracellular signaling molecules (58–60). Supporting evidence is provided by the observation that resident biofilm cells express genes linked to oxygen deprivation, general stress and stationary phase conditions, nutrient stress, and slow growth (58–62). Importantly, cells can leave the biofilm structure and return to the planktonic mode of growth by a process referred to as dispersion (63). Dispersion is an active event in which sessile, matrix-encased biofilm cells escape from the biofilm, leaving behind eroded biofilms and biofilms with central voids (36, 37, 64–66). Not surprisingly, dispersion (also referred to as seeding dispersal) is also considered to be the next stage of biofilm formation, an active event that leads to bacterial dissemination and the colonization of new locations(67). An additional key regulator of the biofilm developmental life cycle is the ubiquitous bacterial second messenger c-di-GMP, with high c-di-GMP levels favoring the biofilm mode of growth, while low levels have been associated with planktonic and dispersed cells (68–70). c-di-GMP is required to mediate surface sensing, repress motility upon surface attachment, and increase biosynthesis of biofilm matrix components, with reduced c-di-GMP levels contributing to dispersion (68–71).
The findings above suggested an expanded model of biofilm development by P. aeruginosa that detailed progression of biofilm formation and stage-specific formation of biofilms (37). While the model represented developmental stages specifically for P. aeruginosa biofilms, the model has become widely used to represent biofilm formation by diverse biofilm-forming microorganisms in various settings, as for example biofilms growing in extreme environments (72) and microalgal biofilms (73).
The developmental model of microbial biofilm formation was adopted quickly by the scientific community to serve as the major conceptual framework for biofilm research on which to base empirical research and scientific inference due to its elegant simplicity. As discussed below, the ability to extrapolate this model to biofilms outside the laboratory - in nature, engineered systems, and medicine has been limited because the diversity and complexity of biofilm structures and processes in real world, non-laboratory systems was not taken into consideration. Also importantly, the logistical difficulty in studying the initiation and development of biofilms in vivo in real time means that the bulk of our understanding is extrapolated from snapshots in time. The study of suspended aggregates in the bulk liquid is even more challenging since these do not stay in the same place for time scales relevant in developmental process observation.
Limitations of the conceptual biofilm life cycle model
We have identified at least four limitations of the conceptual model described above:
It has not yet been determined whether biofilm formation can be described as a true developmental process when we consider biofilms formed outside of the flow cell and by species other than P. aeruginosa and Staphylococcus aureus as model biofilm species.
The model does not capture the wide variety of biofilm architectures observed in real world systems such as microbial mats which can be highly stratified along horizontal layers (74).
The model does not incorporate the diversity of aggregation (see below) and detachment mechanisms now recognized in the field by both motile and non-motile organisms such as Staphylococcus aureus, although the model has been adapted to accommodate this organism (30).
The model does not consider the succession of events in biofilms formed in open systems with a continuous influx of new colonizers. Indeed even for dental biofilms, where it is recognized that biofilm progression proceeds as an ecological succession with new species proliferating in different parts of the biofilm, the single species model is commonly depicted (75). Likewise, applying the 5-step biofilm model to industrial systems is limited. These systems are so large and complex that it is likely that all stages of attachment, growth and detachment occur simultaneously at various points in the system. The tidy description of how biofilm forms in a simple, rich media laboratory system does not necessarily capture the complexity of biofilm in most industrial or environmental systems, where surface characteristics such as scale or corrosion, the chemical properties of the bulk fluid and the fluid dynamics will all influence how the biofilm attaches, grows, and detaches. This also applies to infection sites, where it remains unknown if the site is seeded with single cells or aggregates or if the bacteria are trapped within host material in a complex environment, rather than just forming aggregates by clonal expansion. Furthermore, there are no in situ sensors that can be incorporated into these complex systems to directly monitor biofilm on surfaces, in fluid suspensions or associated with host materials. Sections of the system can be sampled during upgrades or replacement, but these only give a snapshot in time at specific locations. While sampling fluids can give clues that biofilms might be present through capturing and releasing shed cells or aggregates, all that is known is that these originated upstream in the system.
Most host-associated biofilms are subject to strong host selection. In the gut of humans and higher animals, biofilm diversity is regulated through compounds excreted from the gut epithelial cells (76). This is also the case for other host-associated biofilms, including those formed on plant roots that are shaped by plant exudates to attract growth promoting or nutrient capturing bacteria (77) and biofilms formed on algae specifically select for a stable core set of functional genes (78). Selective host-microbe interactions may lead to very complex, and less sequential, events of bacterial attachment and detachment that are not considered in the 5-step model.
The paradigmatic value of this model was first challenged by the research community as early as 2009 (79), questioning the validity of the model and the concept of biofilm formation as a developmental process. Based on definitions by several researchers (80, 81), and reviewed elsewhere (79), development coincides with changes in form and function that are part of the normal life cycle of the cell. This is regulated by a dedicated hierarchically ordered genetic pathway and stage-specific transitions in response to environmental cues. If biofilm formation is indeed a regulated developmental process, the formation of biofilms would require genetic pathways that evolved to facilitate cooperation among members of the biofilm. While it is undeniable that a community of cells form a biofilm, that biofilm formation coincides with surface structure and temporal changes and that several regulators affecting biofilm formation have been identified (82–84), no such genetic pathway regulating these morphological changes and stage-specific transitions in a hierarchically ordered manner has yet been identified (79). However, in the same year, another group (38) reported a previously uncharacterized signal transduction network regulating committed biofilm developmental steps by P. aeruginosa following attachment, in which phospho-relays and response regulators appeared to be key components of the regulatory machinery that coordinates gene expression during biofilm development in response to environmental cues. More specifically, the signaling network is composed of several two-component regulatory systems (TCS) named SagS, BfiSR, BfmRS, and MifRS (38). TCS activation occurred sequentially (SagS<BfiSR<BfmSR<MifSR) over the course of biofilm formation, while inactivation of these systems arrested biofilm formation at distinct developmental stages. ΔsagS and ΔbfiS biofilms arrested at the irreversible attachment stage, while biofilms formed by ΔbfmR and ΔmifR arrested at the maturation-1 and -2 stages of biofilm development, respectively (38, 55, 85–87).
While the discovery of the signal transduction network strongly supported the idea that formation of biofilms was a biologically regulated developmental process, at least for P. aeruginosa grown under laboratory conditions, other concerns remained, including the validity of the representation of the biofilm structure or architecture being composed of mushroom-like microcolonies. In fact, several reports demonstrated that even in P. aeruginosa, the biofilm architecture varied with growth conditions as well as the growth medium. For instance, Klausen et al. (88) demonstrated that while P. aeruginosa PAO1 biofilms grown on glucose minimal medium demonstrated the typical mushroom-shaped multicellular biofilm structures, growth in minimal medium containing citrate, casamino acids or benzoate as carbon source led to the formation of flat unstructured biofilms (Figure 2). In multispecies biofilms, different medium composition impacts not only biofilm morphology, but also species composition (89). In addition to growth medium and nutrient sources, other variations in growth conditions have been reported to influence the biofilm architecture. While P. aeruginosa forming mushroom-shaped biofilms has been associated with growth under relatively low flow rate conditions (90), static growth conditions favor the formation of pellicles that form at the air–liquid interface (91). However, at higher flow rates, structures such as streamers and ripples can form, demonstrating the remarkable ability of biofilms to adapt to the physical conditions under which they grow (57, 92).
Figure 2,
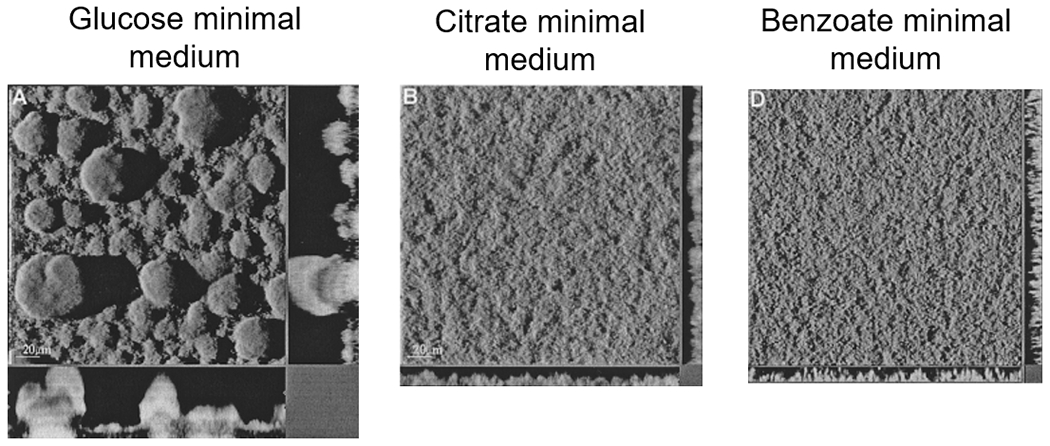
P. aeruginosa grown in flow cells under flow conditions but with different carbon sources shows remarkably different three-dimensional architectures (Sauer, K 2021)
Additionally, the organisms composing the biofilm also have a marked effect on the biofilm structure (Figure 2) (73). For example, in comparison to pure cultures of laboratory grown biofilms of either K. pneumoniae or P. aeruginosa, biofilms containing both species were thicker (93, 94). Moreover, in a mixed species biofilm composed of four bacterial soil isolates, removal of one biofilm member completely changed biofilm morphology, species structural organization and relative abundance, even when the species removed was initially low abundant and intrinsically weak in biofilm formation capability (95). Biofilms of Gram-positive bacteria S. aureus (96) and Streptococcus pneumoniae (97), while having a heterogenous appearance indicative of the presence of water channels, lack the distinct microcolonies that had become an iconic feature of the biofilm architecture. In contrast, studies with pneumococcal biofilms formed under static conditions were used to investigate chronic otitis media with effusion, since fluid/flow is severely disrupted in the middle ear during infection. Biofilm structures were also dependent on bacterial strains but were smaller (5-15μm) resembling the appearance of pneumococcal biofilms from ex vivo middle ear mucosa samples from children with chronic otitis media (98, 99). Similarly, non-typeable Haemophilus influenzae (NTHi), a Gram-negative bacterium, also formed biofilm aggregates in these otitis media samples, which are recapitulated in a chinchilla model of OM (100). NTHi biofilms also formed on differentiated airway epithelial cultures from patients with primary ciliary dyskinesia (PCD)(101). These smaller aggregated structures suggest that biofilms with highly complex 3D architectures may be less likely to form in the host microenvironments(102). Importantly, even discrete biofilm aggregates may still be able to induce inflammation and tissue destruction that leads to sustained chronic infection because they are associated with tolerance to antibiotic therapy and persistence in the host despite robust innate immune responses.
Mixed species biofilms taken from the environment are structurally very diverse. As an example, microbial mats are thick and layered, whereas bacterial aggregates on sand grains are thin and small (74). In addition, environmental biofilms often form on biodegradable material, and thus the nutrients are provided not only from ‘above’, potentially impacting growth zones and structure of the biofilm. Biofilm structure and community members can also be strongly influenced by the underlying substratum as observed in marine biofilms examined ex vivo (103).
Biofilms in the absence of an attachment surface
As reviewed by several groups (3, 104) many of the chronic bacterial infections linked to biofilms that involve aggregated bacteria and antimicrobial recalcitrance, may not involve hard surface attachment, even if a surface is present. (Figure 3). Likewise, biofilms in the environment are often free-floating, including diverse bacterial aggregates (granules) formed in wastewater treatment plants (105) or those in marine, lake and river habitats, commonly referred to as ‘marine-snow’ (11). Zetsche et al.(106) used a sophisticated experimental system to hold a sinking diatom aggregate in place while measuring aggregate’s internal oxygen profiles, as well as the flow field by particle imaging velocimetry, showing the presence of strong oxygen gradients, similar to those seen in attached biofilms. In a similar manner to attached biofilms they found that although the irregular surface of the aggregate influenced flow and mass transfer locally, there was no flow within the EPS, which in this field is referred to as “transparent exopolymeric particles” (TEP). Similarly, granular activated sludge aggregates of microorganisms create strong gradients of oxygen, pH, substrates and metabolites leading to micoenvironments enabling simultaneous aerobic and anaerobic digestion in the same aggregate, with concomitant redox gradients based physiological stratifications (107).
Figure 3.
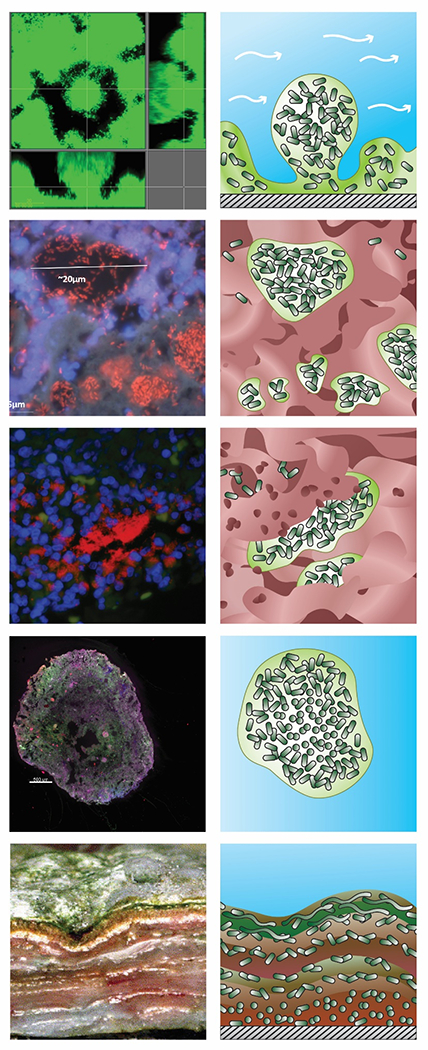
Variety of biofilm structures underscoring differences between in vitro and in vivo or environmental biofilms. Original images are shown in the left column and a schematic drawing of the structure and its organization in the right column with shading denoting water (blue), aggregated microbial cells (dark green) and their extracellular polymeric substances (light green), host cells and other material including mucus or tissue (red), and attachment surface (hatched grey). A: Mushroom structure of Pseudomonas aeruginosa biofilm in vitro in a flow cell. B: Mucus embedded aggregates of P. aeruginosa surrounded by polymorphonuclear leukocytes in a cystic fibrosis lung (108) C: Wound-embedded aggregates of P. aeruginosa surrounded by polymorphonuclear leukocytes(109). D: Aerobic granules from a full-scale AquaNereda® wastewater treatment process (image courtesy of Kylie Bodle and Cat Kirkland). E: Striated microbial mat from a Brazilian lake(110). (Jill Story assisted with figure preparation).
Two “classic” chronic infections linked to aggregated rather than surface-associated bacteria include the infection of soft tissues such as the chronic lung infection of people with cystic fibrosis (CF) (108) and chronic dermal wounds (111). Similarly, in other biofilm associated respiratory infections, such as chronic otitis media, rhinosinusitis, or biofilms on differentiated ciliated cells from people with primary ciliary dyskinesia (PCD), aggregates (~10-20 μm) may adhere to mucosal epithelia or grow as aggregates in effusion, mucus, and airway surface liquid. The bacterial aggregates seen in these infections are not necessarily modeled well by flow biofilm experimental systems, although shear is present in the bronchial airways (112).
Osteomyelitis with and without an implant also belongs to this category. In the case of osteomyelitis with implants, it is generally assumed that the bacteria are attached to the implant surface, and so can be described by the current biofilm model. This has led to much research into designing antibacterial and anti-adhesive surfaces. However, current studies show that even though the bacteria can be associated with the surface, the implant does not have to be colonized to cause a persisting infection (113, 114). Samples from implant-associated infections show that bacteria can be present both in peri-prosthetic tissue and on the implant, but not necessarily both (115). Importantly, detached aggregates recapitulated the antibiotic tolerance observed in surface attached biofilms (116).
Aggregates have also been reported for non-infectious biofilms. Consider the microbiota in the oral cavity, such as on the teeth or on the skin where the majority of bacteria are organized as small aggregates (117, 118). Bacteria clearly attach and grow on the enamel surface of teeth, however these structures tend not to be the three-dimensional mushroom structures seen in flow cells (119). Observation of polymicrobial aggregates in human saliva demonstrates a progression in structural and community complexity and in the flow cells that these aggregates attach to when forming surface adhered biofilms (119, 120). Similarly, on the skin bacteria are distributed in small heterogeneous distributed aggregates and as single cells (121).
In addition to infectious and other host associated biofilms, bacteria in the environment are present as both surface attached colonies and free floating or embedded aggregates. In biological wastewater treatment processes, dense multispecies aggregates of microorganisms self-assemble in both aerobic and anaerobic processes. The overarching observation is that the environmental microbiota are dominated by heterogeneous patterns of aggregated bacteria (11) rather than continuous films of bacteria over large (centimeter) areas, however to a certain extent this is an issue of scale. Algal biofilms on ship hulls may appear macroscopically continuous and in localized areas as a uniform flat layer but patchy under microscopic examination (122).
Aggregate formation
As outlined above, a shortcoming of the current biofilm model is that it does not account for non-surface-attached aggregates that are often observed in clinical or environmental settings. Other than their observation when the model was first published in 2002, little was known about aggregates. Since then, several publications have reported on surface independent bacterial aggregation, with bacteria in aggregates displaying similar phenotypes to bacteria present in surface-attached communities, such as increased tolerance to antibiotic and host defense as well as matrix production and slow growth (123–127).
Examples of different types of aggregates include bacteria embedded in host material such as mucus in CF lungs, slough in the chronic wound bed or external material flocs in wastewater treatment plants and soil. Host fluids including synovial fluid and human serum can induce rapid (within minutes) aggregation in both Gram positive and negative bacteria in vivo and in vitro (124, 128, 129), suggesting that host components such as fibronectin form bridging connections. Such suspended biofilm-like aggregates have also been seen ex vivo (130), and in shaken in vitro cultures (131–133). Bacteria in a shaken, liquid culture have, until recently, been assumed to be entirely planktonic, independent single cells (or short chains or clusters). However, recent publications challenge the conceptual separation between planktonic and biofilm bacteria by showing that both S. aureus and P. aeruginosa can grow as a mixture of planktonic and aggregate cells in liquid batch cultures (132–134).
Based on several laboratory studies, literature currently points to five mechanisms for the formation of free-floating aggregates (Figure 4) that are discussed here in the order they were recognized. The first is the detachment of pieces of attached biofilm due to changes in hydrodynamic shear, nutrient reduction, physical abrasion or exogenously added or endogenously produced dispersal agents (135). Loss of biofilm bacterial cells due to this process has often been referred to as sloughing. The second is through growth in the planktonic phase (134). As cells divide, the daughter cells remain with the mother cells rather than dispersing, presumably through interactions of self-recognizing surface adhesion molecules or simultaneous production of EPS. The presence of surface adhesins may also contribute to the co-aggregation of cells in the planktonic state, leading to the formation of aggregates in the absence of growth. More recently, it has been proposed that aggregation can occur in the liquid phase mediated by host polymers such as mucin and DNA (136). One potential mechanism is depletion aggregation, which occurs as entropic forces between uncharged or like-charged polymers forces particles (single bacterial cells in the case of our discussion) in the suspension to “push out” polymers between the cells as they come close together forcing the formation of aggregates (137). Another possible aggregation mechanism is that bacteria bind to molecules in host fluids through surface adhesion interactions. For example, staphylococci have been shown to aggregate in synovial fluid, which has been a proposed mechanism for initiating periprosthetic joint infection (124). This aggregation is a binding interaction between bacterial factors such as adhesin proteins and host factors such as fibrinogen, fibronectin, and hyaluronic acid (138, 139). Notably, aggregate formation in liquid or in response to host polymers includes both co-aggregation without bacterial growth as well as clonal growth of trapped bacteria, coinciding with continued increasing aggregate size. Moreover, very little is known about how cells leave aggregates, including whether aggregates disassemble by dispersion or sloughing, or simply turn into single cells.
Figure 4.
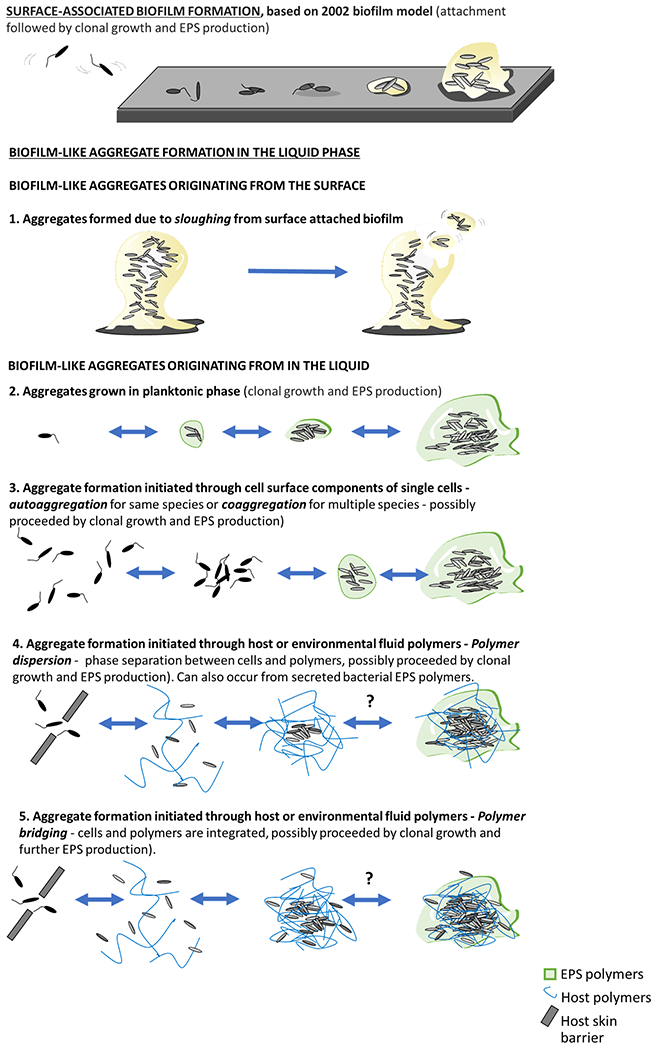
Microbial aggregate formation mechanisms. The top panel shows the “standard” model for biofilm formation starting from the attachment of single planktonic cells to a smooth surface followed by cell division and production of EPS to form 3D surface attached aggregate structures. Below are different mechanisms for generating free floating biofilm-like aggregates. The first is detachment of aggregates from attached biofilms. The second is from clonal growth (division) in the liquid, which may occur with or without facilitation by bacterially produced EPS matrices The third is aggregation of individual cells in a process called autoaggregation for a single species or coaggregation for multiple species, in which bacteria attach to each other through mutual attraction of surface molecules such as adhesins or EPS bridging interactions. The fourth is bridging aggregation which can also be mediated by host polymers, as appears to be the case in synovial fluid (140). Another mechanism of aggregation, the fifth,, is “polymer depletion aggregation” when bacteria are in the presence of non-absorbing polymers (141) and is due to entropic ordering of the colloidal system. Polymer depletion aggregation can be facilitated through bacterially produced EPS or host derived polymers (136).
Expanding the existing 5-step biofilm model
Visualization of biofilms and bacterial aggregates in other in vitro experimental systems, in the environment and in infections, reveals major disparities with the original model (figure 1). As evaluation of biofilms from diverse environments has become more sophisticated, major differences in the microenvironment of the individual biofilms or aggregate to substrates, oxygen and exposure to secreted products has been observed This varies depending on whether bacteria are directly adjacent to the growth medium or entrapped in some sort of biological (e.g mucus, tissue, infection wound bed) or non-biological (e.g. within corrosion or hard water deposits) material, as opposed to a self-produced biofilm matrix. The microenvironments that develop due to an interplay between microbial physiology, substrates and physicochemical conditions (redox and pH) and mass transfer, within biofilms and aggregates, plays a dominant role in determining the metabolism and behavior of the bacteria, affecting characteristics as antibiotic tolerance, growth rate, and expression of virulence factors (18, 142–144).
For these reasons, we have created and propose an updated, more encompassing model describing the three major events in biofilm formation, the aggregation, growth and disaggregation (Figure 5).
Figure 5,
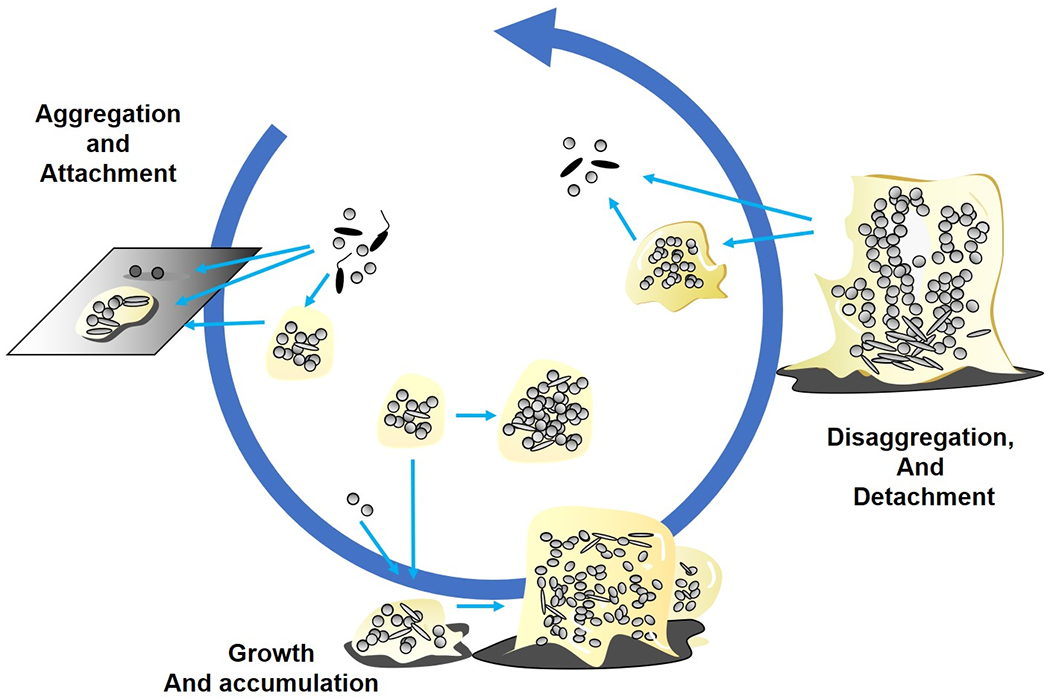
Inclusive model showing the three main events in biofilm formation encompassing in vitro, in situ and in vivo systems. Aggregation and attachment: In this event bacteria aggregate to each other or attach to surfaces, being both biotic and abiotic. Growth and accumulation: In this event, aggregated and attached bacterial colonies expand by growth and recruitment of surrounding cells. Disaggregation and detachment: In this event bacteria can leave the biofilm as aggregates and as single cells depending on the mechanism. These three events characterize and represent most if not all biofilm scenarios independently of time and maturity.
This new model represents three basic events that we believe can be used to depict most of the different scenarios for biofilm formation independent of whether the condition is in vitro, in situ or in vivo. It bridges and combines the different possibilities and pathways of biofilm aggregate development in an inclusive model. We acknowledge that this is a work in progress, based on what we know to date. Thus, we expect that the model is not final, but will undergo future revision.
In contrast to the five-step model, the present model considers open systems that may be encountered in the environment or the human airways or gut, where a continuous influx of new biofilm members is likely. Most importantly, there are no known correlations suggesting a particular biofilm structure is either “better” or “worse” in any given situation. Ex vivo and ex situ observations suggest mushroom structures and surface-attached three-dimensional structures are just as likely to occur as the aggregates observed in chronic infections and the natural environment with and without surface association. A key concept is that biofilm aggregates are heterogeneous diverse microbial communities, shaped and influenced by different environmental cues, that represent multiple discrete microenvironments.
The original model was largely derived based on data from in vitro flow cells experiments, however snapshots of biofilms from environmental systems and from in vivo and ex vivo studies suggest this type of development is not always supported in vivo or in open systems. This led to questioning the general validity of the original model. From in vitro investigations we know that flow and nutrients are important in the experimental systems shaping the three-dimensional architecture of the surface attached biofilms (26, 145). The question then becomes, how much do we really know about the microenvironment and biofilm development in environmental, in vivo and ex vivo examples? Photosynthetic mats are well described relatively flat biofilms where the penetration of sunlight and metabolic activity of the organisms leads to stratified species distribution and microenvironments (146). Suspended biofilm aggregates used for wastewater treatment such as aerobic granules are another example of a stratified biofilm. In this case the aggregates are generally spherical. While direct measurements of the microenvironment are difficult because they are free floating, stratification showing aerobes on the outside and anaerobes on the inside provides evidence of oxic and anoxic zones (147). These microenvironments allow simultaneous aerobic digestion and anaerobic denitrification of wastewater and, patchy aggregates of bacteria in industrial systems and corrosion products in tubercules (148). In iron and steel industrial pipes biofilms can cause microbially induced corrosion due to the development of microenvironments (149). These biofilms tend to be present as mound shaped aggregates on metal surfaces and consist of bacteria and corrosion products. Stratification of organisms such as iron-oxidizing and sulfur-reducing bacteria create anoxic zones within the tubercule, which become anodic relative to the surrounding metal causing pitting corrosion below the tubercule and rust deposition at the surface. These examples illustrate how the interplay between the original external environmental conditions and physiology of biofilm microorganisms leads to the creation of different biofilm structures and microenvironments in situ. Mechanical forces can also shape biofilm architecture, microbial community, and microenvironment development. Samples from river biofilms growing under higher turbulence were thinner, more compact and formed more homogenous layers than those growing under lower hydrodynamic shear (150). In a medical context in CF lung, bacteria can be present and form aggregates independently of the epithelial surface (127). Thus, the new proposed model includes a variety of conditions and biofilm developmental pathways to embrace multiple diverse habitats and microenvironments from the environment, industry and in medicine. What we do know is that the microenvironment depends on the immediate milieu surrounding a single cell, the aggregate itself and the close proximity of the aggregate (151).
As for the hallmark mushroom-shaped structures of the original developmental model, these appear to be dependent on the flow conditions, surface attachment, and carbon source of principally P. aeruginosa, where the mushroom structure forms primarily during flow conditions on a surface with glucose as the carbon source (27). For most other species, even under flow conditions and in the presence of glucose, mushroom structures do not form. In the environment outside of stromatolites and some hot spring structures, mushroom structures appear to be uncommon (152).
Conclusion
The most cited and used model (Figure 1) for biofilm development is extremely intuitive, which explains in part why it has become the preferred model to describe all kinds of biofilm formation. However, as discussed in this Review, the five-step developmental model of P. aeruginosa biofilms grown in flow cells is limited in its scope. The inclusive model considers the possibility of aggregation, and that one pathway is not mutually exclusive of another. Biofilms do not necessarily form a mushroom shaped structure as the final culminating structure, nor is there an absolute dependence on a surface. Currently, no developmental model accurately depicts biofilm formation of all microorganisms, habitats, and all microenvironments. With the inclusive model, we depict three major steps of biofilm growth irrespective of the presence of a surface: aggregation, growth, and disaggregation.
Growing evidence indicates that biofilms do not necessarily require surface attachment to form. Aggregates formed in fluids, due to clonal growth, co-aggregation, or aggregates induced by bacterial EPS or host fluids, demonstrate many of the characteristics previously attributed only to surface-associated biofilms. These aggregates are not limited to laboratory conditions but may be found as part of the human microbiota, in several chronic infection sites and in the environment (102, 153–156). Two decades of biofilm research indicate that the model depicted in Figure 1 was incomplete because it did not capture the multiple biofilm structures and phenotypes that can form with different bacteria and in different microenvironments. This has important implications for how we study biofilms specifically and bacteria in general, as different biofilm experimental systems in vitro or experimental animals in vivo cannot encompass all the factors important for different microenvironments (21). We further propose that the research question should drive the study and interpretation of the results, not the experimental system used to do the research. This is also important for how we extrapolate from the experimental situation to the native scenario. We need to understand biofilms in the context of the relevant microenvironment.
Given that free floating and surface associated aggregates are now accepted as sharing similarities to surface-associated biofilms, several questions remain to be addressed. For example, it is not known what drives aggregate formation in the absence of a surface — that is, does bacteria-bacteria adhesion involve the same mechanisms as attachment of single cells to surfaces? Also do aggregates interact with surfaces and can aggregates attach to surface biofilms, and if so how? Are the same surface properties commonly associated with initial cell-surface adhesion (stiffness and surface energy, which in turn is a function of electrostatic charge, wettability, surface tension and roughness) as important for attachment as macroscale topographical features such as edges, screw holes, expansions and contractions, threads, etc., that may physically entrap aggregates? It is well established that in vitro biofilms actively disperse, but do aggregates actively disassemble and/or disperse cells to the surroundings? These questions could be investigated by analyzing gene-expression profiles during the different stages of biofilm development in the absence and presence of a surface. How do the transcriptional profiles of bacteria in aggregates that have developed through chemical/physical interaction or growth differ from each other and from biofilms formed on surfaces? Furthermore, are successional dynamics and community assembly processes similar for aggregates and surface-associated biofilms? A fundamental open question is whether aggregation protects bacteria from antimicrobials. Recent work suggests that it is not the aggregation alone that promotes tolerance towards antimicrobial agents and host defenses, but gradients of oxygen and nutrients that may become pronounced in aggregates as they increase in size (157). The aggregate size may also determine how easily phagocytes engulf the aggregates (158). In flow cells and as depicted in the original 5 step model, this results in stratified growth with a fast-growing exterior and a dormant inner subpopulation (159). In infections host material can surround microbial aggregates causing gradients. Thus, the original 5 step model does not accurately represent the microenvironment around these aggregates, and likely also fails to capture the reality of biofilms in complex environmental and industrial systems. Concentration gradients influence and regulate bacterial physiology and metabolism, and are reciprocally controlled by the microenvironment as well as by matrix components(59, 160). However, an important question that arises is what is a threshold aggregate size for tolerance manifestation and how do the microenvironment and access to nutrients and electron acceptors influence aggregate size? The sizes of biofilms have been shown to vary much between in vitro and in vivo biofilms(102). The questions of tolerance and matrix production and physiology in general might be addressed by controlling the microenvironment, possibly in three dimensional experimental models, to move beyond the attachment surface as the main constraint controlling immediate access to nutrients and electron acceptors.
The original 5-step model has provided a unifying, yet possibly unintentionally biased understanding of biofilm morphology. This may have caused an unfortunate division of the research area, as some researchers studying biofilms that deviated from this model (flocs, granules, particle, aggregates, mats etc.), may have been excluded from interacting with and interpreting work from researchers studying in vitro model biofilms. We hope that simpler more inclusive model can help to unite the biofilm research community and allow for more cross disciplinary collaboration and knowledge sharing. Specifically, the 5-step model may become challenging when used to describe clinical manifestations and devise new in vitro test methods to evaluate medical implants, drugs and treatments, as these may fail due to lack of extrapolation. Crucially, differences in the microenvironment between in vitro and in vivo may underpin why direct extrapolation is not possible. Additionally, relying on the original biofilm model (Figure 1), healthcare professionals may have a conceptual framework that markedly differs from clinical findings and observations, leading to the erroneous conclusion that a biofilm is not present in a given clinical sample and lead to treatment regimens that will not effectively treat infections (161). It is our hope also to broaden this framework, ultimately leading to improved infection diagnostics and selection of efficient, targeted treatment regimes.
In summary, we suggest a new overall model for biofilm formation that considers the most inclusive recent insights. It demonstrates the three major events: aggregation, growth, and disaggregation. Our intent is that this simpler model will alleviate some of the misconceptions of how biofilms form in diverse environments ranging from industrial systems, to environmental habitats and medical settings. We hope that as a scientific community, we can expand on this model to facilitate an inclusive, less controversial interdisciplinary discussion on biofilms and biofilm formation.
References
- 1.Costerton JW, Geesey GG, Cheng KJ. How bacteria stick. Sci Am. 1978;238(1):86–95. [DOI] [PubMed] [Google Scholar]
- 2.McCoy WF, Bryers JD, Robbins J, Costerton JW. Observations of fouling biofilm formation. Can J Microbiol. 1981;27(9):910–7. [DOI] [PubMed] [Google Scholar]
- 3.Lebeaux D, Chauhan A, Rendueles O, Beloin C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens. 2013(2):288–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21 Suppl 1:S1–25. [DOI] [PubMed] [Google Scholar]
- 5.Kolpen M, Kragh KN, Enciso JB, Faurholt-Jepsen D, Lindegaard B, Egelund GB, et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghupathi PK, Liu W, Sabbe K, Houf K, Burmølle M, Sørensen SJ. Synergistic Interactions within a Multispecies Biofilm Enhance Individual Species Protection against Grazing by a Pelagic Protozoan. Frontiers in Microbiology. 2018;8(2649). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jass J, Roberts SK, Lappin-Scott HM. Microbes and enzymes in biofilms. Enzymes in the Environment Activity, Ecology and Applications Marcel Dekker Inc, New York, USA. 2002:307–26. [Google Scholar]
- 8.Tkacz A, Poole P. The plant microbiome: The dark and dirty secrets of plant growth. PLANTS, PEOPLE, PLANET. 2021;3(2):124–9. [Google Scholar]
- 9.ANNOUS BA SOLOMON EB, COOKE PH, BURKE A. BIOFILM FORMATION BY SALMONELLA SPP. ON CANTALOUPE MELONS**. Journal of Food Safety. 2005;25(4):276–87. [Google Scholar]
- 10.Rudrappa T, Biedrzycki ML, Bais HP. Causes and consequences of plant-associated biofilms. FEMS Microbiol Ecol. 2008;64(2):153–66. [DOI] [PubMed] [Google Scholar]
- 11.Flemming HC, Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17(4):247–60. [DOI] [PubMed] [Google Scholar]; Excellent review that quantitatively explores and proves the long-standing notion that biofilm is the predominant form of prokaryotic life.
- 12.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–64. [DOI] [PubMed] [Google Scholar]
- 13.Vishwakarma V. Impact of environmental biofilms: Industrial components and its remediation. Journal of Basic Microbiology. 2020;60(3):198–206. [DOI] [PubMed] [Google Scholar]
- 14.Jurelevicius D, Ramos L, Abreu F, Lins U, de Sousa MP, dos Santos VVCM, et al. Long-term souring treatment using nitrate and biocides in high-temperature oil reservoirs. Fuel. 2021;288:119731. [Google Scholar]
- 15.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One. 2011;6(11):e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitzilaiou E, Kuria AM, Siegumfeldt H, Rasmussen MA, Knøchel S. The impact of bacterial cell aggregation on UV inactivation kinetics. Water Res. 2021;204:117593. [DOI] [PubMed] [Google Scholar]
- 17.Bjarnsholt T, Whiteley M, Rumbaugh K, Stewart PS, Jensen PO, Frimodt-Moller N. The importance of understanding the infectious microenvironment. lancet Infect Dis. 2021;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornforth DM, Diggle FL, Melvin JA, Bomberger JM, Whiteley M. Quantitative Framework for Model Evaluation in Microbiology Research Using Pseudomonas aeruginosa and Cystic Fibrosis Infection as a Test Case. mBio. 2020;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrie TJ, Nelligan J, Costerton JW. A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation. 1982;66(6):1339–41. [DOI] [PubMed] [Google Scholar]
- 20.Zobell CE. The Effect of Solid Surfaces upon Bacterial Activity. J Bacteriol. 1943;46(1):39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simple observation results in paradigm changing idea, bacteria like to live in communities.
- 21.Thaarup IC, Bjarnsholt T. Current In Vitro Biofilm-Infected Chronic Wound Models for Developing New Treatment Possibilities. Adv Wound Care (New Rochelle). 2021;10(2):91–102. [DOI] [PubMed] [Google Scholar]
- 22.Sternberg C, Bjarnsholt T, Shirtliff M. Methods for dynamic investigations of surface-attached in vitro bacterial and fungal biofilms. Methods Mol Biol. 2014;1147:3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azeredo J, Azevedo NF, Briandet R, Cerca N, Coenye T, Costa AR, et al. Critical review on biofilm methods. Crit Rev Microbiol. 2017;43(3):313–51. [DOI] [PubMed] [Google Scholar]
- 24.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. [DOI] [PubMed] [Google Scholar]
- 25.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, et al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2012;109(50):20632–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184(4):1140–54. [DOI] [PMC free article] [PubMed] [Google Scholar]; The original key paper that started the concept of a biofilm life cycle, and is revisited in this review
- 27.Klausen M, Heydorn A, Ragas P, Lambertsen L, aes-Jorgensen A, Molin S, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48(6):1511–24. [DOI] [PubMed] [Google Scholar]
- 28.Pamp SJ, Sternberg C, Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry A. 2009;75(2):90–103. [DOI] [PubMed] [Google Scholar]
- 29.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. [DOI] [PubMed] [Google Scholar]
- 30.Moormeier DE, Bayles KW. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol. 2017;104(3):365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nature Reviews Microbiology. 2013;11(3):157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Xiao Y, Liao K, Leng Y, Lu Q. Development of microalgal biofilm for wastewater remediation: from mechanism to practical application. Journal of Chemical Technology & Biotechnology. 2021;96(11):2993–3008. [Google Scholar]
- 33.Liu J, Lu H, Wu L, Kerr PG, Wu Y. Interactions between periphytic biofilms and dissolved organic matter at soil-water interface and the consequent effects on soil phosphorus fraction changes. Sci Total Environ. 2021;801:149708. [DOI] [PubMed] [Google Scholar]
- 34.Wu BC, Haney EF, Akhoundsadegh N, Pletzer D, Trimble MJ, Adriaans AE, et al. Human organoid biofilm model for assessing antibiofilm activity of novel agents. NPJ Biofilms Microbiomes. 2021;7(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Liu H, Wang R, Wu C. Interactions between dicyandiamide and periphytic biofilms in paddy soils and subsequent effects on nitrogen cycling. Sci Total Environ. 2020;718:137417. [DOI] [PubMed] [Google Scholar]
- 36.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. Journal of Bacteriology. 2002;184(4):1140–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annual Reviews in Microbiology. 2002;56(1):187–209. [DOI] [PubMed] [Google Scholar]
- 38.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathogens. 2009;5(11):e1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]; Findings reported here demonstrated for the first time that the formation of biofilms is coordinated by a genetic pathways that regulates morphological changes of biofilms and stage-specific transitions in a hierarchical ordered manner. Components of the genetic pathways only appeared to play a role under biofilm growth conditions.
- 39.Petrova OE, Gupta K, Liao J, Goodwine JS, Sauer K. Divide and conquer: the Pseudomonas aeruginosa two-component hybrid SagS enables biofilm formation and recalcitrance of biofilm cells to antimicrobial agents via distinct regulatory circuits. Environmental Microbiology. 2017;19(5):2005–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Molecular Microbiology. 1998;28(3):449–61. [DOI] [PubMed] [Google Scholar]
- 41.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Characklis WG. Attached microbial growths-II. Frictional resistance due to microbial slimes. Water Research. 1973;7(9):1249–58. [Google Scholar]
- 43.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5(11):e1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta K, Marques CNH, Petrova OE, Sauer K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. Journal of Bacteriology. 2013;195(21):4975–87 [DOI] [PMC free article] [PubMed] [Google Scholar]; Findings reported here indicated that biofilm cells gain heightened tolerance to antimicrobial agents in a manner independent of biofilm biomass accumulation (demonstrating mature biofilm architecture)
- 45.Davies DG, Charabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181 – 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies DG, Geesey GG. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61(3):860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environmental Microbiology. 2012;14(8):1913–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagge N, Hentzer M, Andersen JB, Ciofu O, Givskov M, Hoiby N. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2004;48(4):1168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood DW, Gong F, Daykin MM, Williams P, Pierson LS. N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Journal of Bacteriology. 1997;179(24):7663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood SR, Kirkham J, Marsh PD, Shore RC, Nattress B, Robinson C. Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J Dent Res. 2000;79(1):21–7. [DOI] [PubMed] [Google Scholar]
- 51.Davies DG P MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–8. [DOI] [PubMed] [Google Scholar]
- 52.Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Espinosa-Urgel M. Resident parking only: rhamnolipids maintain fluid channels in biofilms. J Bacteriol. 2003;185(3):699–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuchma SL, Connolly JP, O’Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. Journal of Bacteriology. 2005;187(4):1441–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Molecular Microbiology. 2012;86:819–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sriramulu DD, Lünsdorf H, Lam JS, Römling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol. 2005;54(7):667–76. [DOI] [PubMed] [Google Scholar]
- 57.Purevdorj B, Costerton JW, Stoodley P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2002;68(9):4457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]; Understanding the relationship between biofilm and fluid dynamics is crucial and completely overlooked in traditional microbiological systems. It impacts not only mass transfer rates, but also the biofilm architecture, spatial organization, and detachment.
- 58.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nature Reviews Microbiology. 2008;6(3):199. [DOI] [PubMed] [Google Scholar]
- 59.Serra DO, Hengge R. Stress responses go three dimensional–the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environmental Microbiology. 2014;16(6):1455–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. Journal of Bacteriology. 2012;194(8):2062–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heacock-Kang Y, Sun Z, Zarzycki-Siek J, McMillan IA, Norris MH, Bluhm AP, et al. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Molecular Microbiology. 2017;106(6):976–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haussler S, Fuqua C. Biofilms 2012: new discoveries and significant wrinkles in a dynamic field. Journal of Bacteriology. 2013;195(13):2947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18(10):571–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrova OE, Sauer K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Current Opinion in Microbiology. 2016;30:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davies DG. Biofilm Dispersion. In Biofilm Highlights; Springer: Berlin. 2011:1–28. [Google Scholar]
- 66.Steinberg N, Keren-Paz A, Hou Q, Doron S, Yanuka-Golub K, Olender T, et al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Science Signaling. 2020;13(632):eaaw8905. [DOI] [PubMed] [Google Scholar]
- 67.Purevdorj-Gage B, Costerton WJ, Stoodley P. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology. 2005;151(5):1569–76. [DOI] [PubMed] [Google Scholar]
- 68.Valentini M, Filloux A. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria. J Biol Chem. 2016;291(24):12547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nature Reviews Microbiology. 2017;15(5):271–84. [DOI] [PubMed] [Google Scholar]
- 71.Purcell EB, Tamayo R. Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiology Reviews. 2016;40(5):753–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin W, Wang Y, Liu L, He J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. International Journal of Molecular Sciences. 2019;20(14):3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mantzorou A, Ververidis F. Microalgal biofilms: A further step over current microalgal cultivation techniques. Science of The Total Environment. 2019;651:3187–201. [DOI] [PubMed] [Google Scholar]
- 74.Prieto-Barajas CM, Valencia-Cantero E, Santoyo G. Microbial mat ecosystems: Structure types, functional diversity, and biotechnological application. Electronic Journal of Biotechnology. 2018;31:48–56. [Google Scholar]
- 75.Hao Y, Huang X, Zhou X, Li M, Ren B, Peng X, et al. Influence of Dental Prosthesis and Restorative Materials Interface on Oral Biofilms. International Journal of Molecular Sciences. 2018;19(10):3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang C-S, Kao C-Y. Current understanding of the gut microbiota shaping mechanisms. Journal of Biomedical Science. 2019;26(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biology and Fertility of Soils. 2015;51. [Google Scholar]
- 78.Roth-Schulze AJ, Pintado J, Zozaya-Valdés E, Cremades J, Ruiz P, Kjelleberg S, et al. Functional biogeography and host specificity of bacterial communities associated with the Marine Green Alga Ulva spp. Molecular Ecology. 2018;27(8):1952–65. [DOI] [PubMed] [Google Scholar]
- 79.Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17(2):73–87. [DOI] [PubMed] [Google Scholar]
- 80.Dworkin M. Developmental biology of the bacteria: Benjamin/Cummings Pub. Co; 1985. [Google Scholar]
- 81.Brun YV, Shimkets LJ. Prokaryotic development: Asm Press; Washington, DC; 2000. [Google Scholar]
- 82.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A Signaling Network Reciprocally Regulates Genes Associated with Acute Infection and Chronic Persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7(5):745–54. [DOI] [PubMed] [Google Scholar]
- 83.Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007;189(22):8154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013;21(1):39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petrova OE, Schurr JR, Schurr MJ, Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Molecular Microbiology. 2011;81(3):767–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. Journal of Bacteriology. 2010;192:5275–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation Journal of Bacteriology. 2011;193:6614–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Molecular Microbiology. 2003;48(6):1511–24. [DOI] [PubMed] [Google Scholar]
- 89.Lee KWK, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8(4):894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10(9):2331–43. [DOI] [PubMed] [Google Scholar]
- 91.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Molecular Microbiology. 2004;51(3):675–90. [DOI] [PubMed] [Google Scholar]
- 92.Fabbri S, Li J, Howlin RP, Rmaile A, Gottenbos B, De Jager M, et al. Fluid-driven interfacial instabilities and turbulence in bacterial biofilms. Environ Microbiol. 2017;19(11):4417–31. [DOI] [PubMed] [Google Scholar]
- 93.James GA, Beaudette L, Costerton JW. Interspecies bacterial interactions in biofilms. Journal of industrial microbiology. 1995;15(4):257–62. [Google Scholar]
- 94.Murga R, Stewart PS, Daly D. Quantitative analysis of biofilm thickness variability. Biotechnol Bioeng. 1995;45(6):503–10. [DOI] [PubMed] [Google Scholar]
- 95.Liu W, Russel J, Røder HL, Madsen JS, Burmølle M, Sørensen SJ. Low-abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Environ Microbiol. 2017;19(7):2893–905. [DOI] [PubMed] [Google Scholar]
- 96.Sauer K, Steczko J, Ash SR. Effect of a solution containing citrate/methylene blue/parabens on Staphylococcus aureus bacteria and biofilm, and comparison with various heparin solutions. J Antimicrob Chemother. 2009;63(5):937–45. [DOI] [PubMed] [Google Scholar]
- 97.Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, et al. Phenotypic characterization of Streptococcus pneumoniae biofilm development. Journal of Bacteriology. 2006;188(7):2325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296(2):202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper to directly link biofilms with OM using molecular probes specific for 3 pathogens, which cause otitis media, on human middle ear mucosal epithelial biopsies from children with chronic otitis media and not on uninfected biopsies
- 99.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bakaletz LO. Bacterial biofilms in the upper airway - evidence for role in pathology and implications for treatment of otitis media. Paediatr Respir Rev. 2012;13(3):154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker WT, Jackson CL, Allan RN, Collins SA, Kelso MJ, Rineh A, et al. Primary ciliary dyskinesia ciliated airway cells show increased susceptibility to Haemophilus influenzae biofilm formation. Eur Respir J. 2017;50(3). [DOI] [PubMed] [Google Scholar]
- 102.Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sorensen SR, Moser C, Kuhl M, et al. The in vivo biofilm. Trends Microbiol. 2013;21(9):466–74. [DOI] [PubMed] [Google Scholar]
- 103.Tuck B, Watkin E, Somers A, Machuca LL. A critical review of marine biofilms on metallic materials. npj Materials Degradation. 2022;6(1):25. [Google Scholar]
- 104.Hall-Stoodley L, Stoodley P, Kathju S, Hoiby N, Moser C, Costerton JW, et al. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol. 2012;65(2):127–45. [DOI] [PubMed] [Google Scholar]
- 105.Trego AC, Mills S, Collins G. Granular biofilms: Function, application, and new trends as model microbial communities. Critical Reviews in Environmental Science and Technology. 2021;51(15):1702–25. [Google Scholar]; Focused and informative review on types, functions and applications of microbial granules
- 106.Zetsche E-M, Larsson AI, Iversen MH, Ploug H. Flow and diffusion around and within diatom aggregates: Effects of aggregate composition and shape. Limnology and Oceanography. 2020;65(8):1818–33. [Google Scholar]
- 107.Wilén B-M, Liébana R, Persson F, Modin O, Hermansson M. The mechanisms of granulation of activated sludge in wastewater treatment, its optimization, and impact on effluent quality. Applied Microbiology and Biotechnology. 2018;102(12):5005–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44(6):547–58. [DOI] [PubMed] [Google Scholar]; First paper to show, using specific 16S molecular FISH probes for P. auruginosa in biofilm aggregates in situ in the respiratory tract in individuals with CF.
- 109.Kirketerp-Moller K, Jensen PO, Fazli M, Madsen KG, Pedersen J, Moser C, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46(8):2717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vasconcelos C, Warthmann R, McKenzie JA, Visscher PT, Bittermann AG, van Lith Y. Lithifying microbial mats in Lagoa Vermelha, Brazil: modern Precambrian relics? Sedimentary Geology. 2006;185(3-4):175–83. [Google Scholar]
- 111.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, Madsen KG, Phipps R, Krogfelt K, et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 2008;16(1):2–10. [DOI] [PubMed] [Google Scholar]
- 112.Gloag ES, Wozniak DJ, Stoodley P, Hall-Stoodley L. Mycobacterium abscessus biofilms have viscoelastic properties which may contribute to their recalcitrance in chronic pulmonary infections. Sci Rep. 2021;11(1):5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jensen LK, Koch J, Dich-Jorgensen K, Aalbaek B, Petersen A, Fuursted K, et al. Novel porcine model of implant-associated osteomyelitis: A comprehensive analysis of local, regional, and systemic response. J Orthop Res. 2017;35(10):2211–21. [DOI] [PubMed] [Google Scholar]
- 114.Li C, Renz N, Trampuz A. Management of Periprosthetic Joint Infection. Hip Pelvis. 2018;30(3):138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dudareva M, Barrett L, Figtree M, Scarborough M, Watanabe M, Newnham R, et al. Sonication versus Tissue Sampling for Diagnosis of Prosthetic Joint and Other Orthopedic Device-Related Infections. J Clin Microbiol. 2018;56(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol. 2004;186(14):4486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper demonstrating aggregates are continually shed from biofilms and demonstrate biofilm tolerance to antibiotics.
- 117.Bay L, Barnes CJ, Fritz BG, Thorsen J, Restrup MEM, Rasmussen L, et al. Universal Dermal Microbiome in Human Skin. mBio. 2020;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Burmolle M, Thomsen TR, Fazli M, Dige I, Christensen L, Homoe P, et al. Biofilms in chronic infections - a matter of opportunity - monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol. 2010;59(3):324–36. [DOI] [PubMed] [Google Scholar]
- 119.Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci U S A. 2020;117(22):12375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bowen WH, Burne RA, Wu H, Koo H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018;26(3):229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ashrafi M, Novak-Frazer L, Bates M, Baguneid M, Alonso-Rasgado T, Xia G, et al. Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Scientific Reports. 2018;8(1):9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salta M, Wharton JA, Blache Y, Stokes KR, Briand J-F. Marine biofilms on artificial surfaces: structure and dynamics. Environmental Microbiology. 2013;15(11):2879–93. [DOI] [PubMed] [Google Scholar]
- 123.Alhede M, Kragh KN, Qvortrup K, lesen-Holm M, van GM, Christensen LD, et al. Phenotypes of Non-Attached Pseudomonas aeruginosa Aggregates Resemble Surface Attached Biofilm. PLoS One. 2011;6(11):e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dastgheyb SS, Hammoud S, Ketonis C, Liu AY, Fitzgerald K, Parvizi J, et al. Staphylococcal persistence due to biofilm formation in synovial fluid containing prophylactic cefazolin. Antimicrob Agents Chemother. 2015;59(4):2122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Landry RM, An D, Hupp JT, Singh PK, Parsek MR. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol Microbiol. 2006;59(1):142–51. [DOI] [PubMed] [Google Scholar]
- 126.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407(6805):762–4. [DOI] [PubMed] [Google Scholar]
- 127.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109(3):317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pestrak MJ, Gupta TT, Dusane DH, Guzior DV, Staats A, Harro J, et al. Investigation of synovial fluid induced Staphylococcus aureus aggregate development and its impact on surface attachment and biofilm formation. PLoS One. 2020;15(4):e0231791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Macias-Valcayo A, Staats A, Aguilera-Correa JJ, Brooks J, Gupta T, Dusane D, et al. Synovial Fluid Mediated Aggregation of Clinical Strains of Four Enterobacterial Species. Adv Exp Med Biol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bidossi A, Bottagisio M, Savadori P, De Vecchi E. Identification and Characterization of Planktonic Biofilm-Like Aggregates in Infected Synovial Fluids From Joint Infections. Front Microbiol. 2020;11:1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kragh KN, Alhede M, Rybtke M, Stavnsberg C, Jensen PO, Tolker-Nielsen T, et al. The Inoculation Method Could Impact the Outcome of Microbiological Experiments. Appl Environ Microbiol. 2018;84(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, et al. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS One. 2009;4(5):e5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS One. 2012;7(7):e41075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kragh KN, Alhede M, Rybtke M, Stavnsberg C, Jensen PO, Tolker-Nielsen T, et al. Inoculation method could impact the outcome of microbiological experiments. Appl Environ Microbiol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13(1):7–10. [DOI] [PubMed] [Google Scholar]
- 136.Secor PR, Michaels LA, Ratjen A, Jennings LK, Singh PK. Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in <em>Pseudomonas aeruginosa</em>. Proceedings of the National Academy of Sciences. 2018;115(42):10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Secor PR, Michaels LA, Ratjen A, Jennings LK, Singh PK. Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2018;115(42):10780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dastgheyb S, Parvizi J, Shapiro IM, Hickok NJ, Otto M. Effect of biofilms on recalcitrance of staphylococcal joint infection to antibiotic treatment. J Infect Dis. 2015;211(4):641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper demonstrating importance of host factors in rapid aggregation of S aureus into biofilm-like aggregates
- 139.Knott S, Curry D, Zhao N, Metgud P, Dastgheyb SS, Purtill C, et al. Staphylococcus aureus Floating Biofilm Formation and Phenotype in Synovial Fluid Depends on Albumin, Fibrinogen, and Hyaluronic Acid. Front Microbiol. 2021;12:655873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gilbertie JM, Schnabel LV, Hickok NJ, Jacob ME, Conlon BP, Shapiro IM, et al. Equine or porcine synovial fluid as a novel ex vivo model for the study of bacterial free-floating biofilms that form in human joint infections. PLOS ONE. 2019;14(8):e0221012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dorken G, Ferguson GP, French CE, Poon WCK. Aggregation by depletion attraction in cultures of bacteria producing exopolysaccharide. Journal of The Royal Society Interface. 2012;9(77):3490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper linking polymer depletion with bacterial aggregates.
- 142.Cornforth DM, Dees JL, Ibberson CB, Huse HK, Mathiesen IH, Kirketerp-Moller K, et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A. 2018;115(22):E5125–E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rossi E, Falcone M, Molin S, Johansen HK. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat Commun. 2018;9(1):3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15(12):740–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kragh KN, Hutchison JB, Melaugh G, Rodesney C, Roberts AE, Irie Y, et al. Role of Multicellular Aggregates in Biofilm Formation. MBio. 2016;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hawes I, Sumner D, Jungblut AD. Complex Structure but Simple Function in Microbial Mats from Antarctic Lakes. In: Hurst CJ, editor. The Structure and Function of Aquatic Microbial Communities. Cham: Springer International Publishing; 2019. p. 91–120. [Google Scholar]
- 147.Franca RDG, Pinheiro HM, van Loosdrecht MCM, Lourenço ND. Stability of aerobic granules during long-term bioreactor operation. Biotechnology Advances. 2018;36(1):228–46. [DOI] [PubMed] [Google Scholar]
- 148.Li Y, Xu D, Chen C, Li X, Jia R, Zhang D, et al. Anaerobic microbiologically influenced corrosion mechanisms interpreted using bioenergetics and bioelectrochemistry: A review. Journal of Materials Science & Technology. 2018;34(10):1713–8. [Google Scholar]
- 149.Bahrami A, Khouzani MK, Harchegani BB. Establishing the root cause of a failure in a firewater pipeline. Engineering Failure Analysis. 2021;127:105474. [Google Scholar]
- 150.Risse-Buhl U, Anlanger C, Kalla K, Neu TR, Noss C, Lorke A, et al. The role of hydrodynamics in shaping the composition and architecture of epilithic biofilms in fluvial ecosystems. Water Research. 2017;127:211–22. [DOI] [PubMed] [Google Scholar]
- 151.Kirketerp-Moller K, Stewart PS, Bjarnsholt T. The zone model: A conceptual model for understanding the microenvironment of chronic wound infection. Wound Repair Regen. 2020;28(5):593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. The Architecture of Iron Microbial Mats Reflects the Adaptation of Chemolithotrophic Iron Oxidation in Freshwater and Marine Environments. Frontiers in Microbiology. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bay L, Kragh KN, Eickhardt SR, Poulsen SS, Gjerdrum LMR, Ghathian K, et al. Bacterial Aggregates Establish at the Edges of Acute Epidermal Wounds. Adv Wound Care (New Rochelle). 2018;7(4):105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ring HC, Bay L, Nilsson M, Kallenbach K, Miller IM, Saunte DM, et al. Bacterial biofilm in chronic lesions of hidradenitis suppurativa. Br J Dermatol. 2017;176(4):993–1000. [DOI] [PubMed] [Google Scholar]
- 155.Ring HC, Thorsen J, Saunte DM, Lilje B, Bay L, Riis PT, et al. The Follicular Skin Microbiome in Patients With Hidradenitis Suppurativa and Healthy Controls. JAMA Dermatol. 2017;153(9):897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qvist T, Eickhardt S, Kragh KN, Andersen CB, Iversen M, Hoiby N, et al. Chronic pulmonary disease with Mycobacterium abscessus complex is a biofilm infection. Eur Respir J. 2015;46(6):1823–6. [DOI] [PubMed] [Google Scholar]
- 157.Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, Parker A, et al. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 2010;10:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alhede M, Lorenz M, Fritz BG, Jensen PO, Ring HC, Bay L, et al. Bacterial aggregate size determines phagocytosis efficiency of polymorphonuclear leukocytes. Med Microbiol Immunol. 2020;209(6):669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol. 2008;68(1):223–40. [DOI] [PubMed] [Google Scholar]
- 160.Díaz-Pascual F, Lempp M, Nosho K, Jeckel H, Jo JK, Neuhaus K, et al. Spatial alanine metabolism determines local growth dynamics of Escherichia coli colonies. eLife. 2021;10:e70794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bjarnsholt T, Mastroianni E, Kirketerp-Moller K, Stewart PS, Mahr AM, Dominguez Cabanes A, et al. The impact of mental models on the treatment and research of chronic infections due to biofilms. APMIS. 2021. [DOI] [PubMed] [Google Scholar]
- 162.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–8. [DOI] [PubMed] [Google Scholar]
- 163.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. [DOI] [PubMed] [Google Scholar]
- 164.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28(3):449–61. [DOI] [PubMed] [Google Scholar]
- 165.Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48(1):253–67. [DOI] [PubMed] [Google Scholar]
- 166.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413(6858):860–4. [DOI] [PubMed] [Google Scholar]
- 167.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185(7):2080–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, et al. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob Agents Chemother. 2004;48(4):1175–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ren Y, Wang C, Chen Z, Allan E, van der Mei HC, Busscher HJ. Emergent heterogeneous microenvironments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing. FEMS Microbiology Reviews. 2018;42(3):259–72. [DOI] [PubMed] [Google Scholar]
- 170.Serra DO, Hengge R. Stress responses go three dimensional – the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environmental Microbiology. 2014;16(6):1455–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Povolotsky TL, Keren-Paz A, Kolodkin-Gal I. Metabolic Microenvironments Drive Microbial Differentiation and Antibiotic Resistance. Trends Genet. 2021;37(1):4–8. [DOI] [PubMed] [Google Scholar]
- 172.Heacock-Kang Y, Sun Z, Zarzycki-Siek J, McMillan IA, Norris MH, Bluhm AP, et al. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol Microbiol. 2017;106(6):976–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liao J, Schurr MJ, Sauer K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. Journal of Bacteriology. 2013;195:3352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kowalski CH, Morelli KA, Stajich JE, Nadell CD, Cramer RA. A Heterogeneously Expressed Gene Family Modulates the Biofilm Architecture and Hypoxic Growth of Aspergillus fumigatus. mBio. 2021;12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dar D, Dar N, Cai L, Newman DK. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science. 2021;373(6556). [DOI] [PMC free article] [PubMed] [Google Scholar]; Using an innovative transcriptome-imaging approach, this article provides visual evidence of differential gene expression and heterogeneity within biofilms at the single cell level.
- 176.Nair HA, Periasamy S, Yang L, Kjelleberg S, Rice SA. Real Time, Spatial, and Temporal Mapping of the Distribution of c-di-GMP during Biofilm Development. J Biol Chem. 2017;292(2):477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Klauck G, Serra DO, Possling A, Hengge R. Spatial organization of different sigma factor activities and c-di-GMP signalling within the three-dimensional landscape of a bacterial biofilm. Open Biology. 2018;8(8):180066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lenz AP, Williamson KS, Pitts B, Stewart PS, Franklin MJ. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2008;74(14):4463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62(5):1264–77. [DOI] [PubMed] [Google Scholar]
- 180.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327(5967):866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rumbo-Feal S, Gómez MJ, Gayoso C, Álvarez-Fraga L, Cabral MP, Aransay AM, et al. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One. 2013;8(8):e72968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bielecki P, Puchalka J, Wos-Oxley ML, Loessner H, Glik J, Kawecki M, et al. In-vivo expression profiling of Pseudomonas aeruginosa infections reveals niche-specific and strain-independent transcriptional programs. PLoS One. 2011;6(9):e24235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, et al. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol. 2003;5(12):1294–308. [DOI] [PubMed] [Google Scholar]


