Summary
The coronavirus nucleocapsid (N) protein is known to bind to nucleic acids and facilitate viral genome encapsulation. Here we report that the N protein can mediate RNA or DNA entering neighboring cells through ACE2-independent, receptor (STEAP2)-mediated endocytosis, and achieve gene expression. The effect is more pronounced for the N protein of wild-type SARS-CoV-2 than that of the Omicron variant and other human coronaviruses. This effect is enhanced by RANTES (CCL5), a chemokine induced by N protein, and lactate, a metabolite produced in hypoxia, to cause more damage. These findings might explain the clinical observations in SARS-CoV-2-infected cases. Moreover, the N protein-mediated function can be inhibited by N protein-specific monoclonal antibodies or p38 mitogen-activated protein kinase inhibitors. Since the N-protein-mediated nucleic acid endocytosis involves a receptor commonly expressed in many types of cells, our findings suggest that N protein may have an additional role in SARS-CoV-2 pathogenesis.
Subject areas: Virology, Molecular biology
Graphical abstract
Highlights
-
•
SARS-CoV-2 N protein delivers RNA or DNA to neighboring cells through endocytosis
Virology; Molecular biology.
Introduction
The genomic RNA of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is about 30 kb long, encoding structural proteins S (Spike protein), E (Envelope protein), M (Membrane protein), and N (Nucleocapsid protein), along with more than 20 non-structural proteins (nsps) for viral RNA synthesis and packaging.1,2,3 N protein is an RNA-binding protein responsible for viral genome packaging.4 It is the most abundant protein in coronaviruses and is commonly used as a target antigen for the detection of SARS-CoV-2 infection.5 N protein contains an N-terminal RNA-binding domain (RBD), a C-terminal dimerization domain (CTD), and three intrinsically disordered regions (IDRs).6,7 In addition, N protein also interacts with membrane proteins to recruit viral genomic material essential for virion production.6
N protein has been shown to interact with various host cellular processes, including cytokine responses,8,9,10 cell-cycle arrest11 and RNAi.12 It is highly immunogenic and has been shown to induce antibodies that interfere with the viral infection process.13 Because of its physicochemical properties, the N protein phase separates into droplets containing RNA through liquid-liquid phase separation (LLPS). Such phase separation may play a role in the viral life cycle and impact host responses.14 Once N protein binds to viral RNA, the complex can recruit transforming growth factor β activated kinase 1 (TAK1) and IκB kinase (IKK) complex during LLPS, thus facilitating nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) activation. An inhibitor of LLPS, 1,6-hexanediol, can attenuate the phase separation of N protein and diminish enhanced NF-kB activation.15
In adenocarcinomic human alveolar basal epithelial (A549) cells, SARS-CoV-2 N protein has been reported to lead to excessive inflammatory responses by directly interacting with NLR family pyrin domain containing 3 (NLRP3) to form inflammasomes, as well as by promoting the maturation of pro-inflammatory cytokines.8 N protein RNA complex binds to and segregates the stress granule proteins G3BP1 and G3BP2, thereby decreasing stress granule formation.16,17
N protein plays a dual role in innate immune responses by regulating the phosphorylation and nuclear translocation of interferon regulatory transcription factor 3 (IRF3), Signal Transducer and Activator of Transcription 1 (STAT1), and Signal Transducer and Activator of Transcription 2 (STAT2).15,18 The low-dose N protein can inhibit type I interferon (IFN-I) signals and the secretion of pro-inflammatory cytokines, by binding to TRIM25 and inhibiting RIG-I ubiquitination.15 Notably, high-dose N protein has the opposite effect.15
N protein activates vasculopathy and coagulopathy in human via signal transduction mediated by Toll-like receptor 2 (TLR2), NF-kB, and P38 mitogen-activated protein kinase (MAPK), while simvastatin inhibits N-induced endothelial activation.19 Interestingly, SARS-CoV-2 N protein induces vasculopathy and coagulopathy, while N proteins from other coronaviruses such as SARS-CoV, Middle East Respiratory Syndrome coronavirus (MERS-CoV), HKU1-CoV, and influenza virus H1N1 do not.19 In addition, a recent study indicated that the phosphorylation of N protein regulates viral infectivity.17 N protein contains consensus sequences recognized by glycogen synthase kinase 3 (GSK-3), which can phosphorylate N protein. Inhibition of GSK-3 with lithium significantly abolishes SARS-CoV-2 infection in human lung epithelial cells.
COVID-19 is known to involve numerous immune dysregulations which lead to severe disease symptoms and outcomes.20,21 Among these immune dysregulations, the most life-threating is the induction of cytokine storms.22,23,24 The high expression of viral proteins and genomic material then stimulates the production of numerous pro-inflammatory cytokines and chemokines23,24,25 induced in patients with COVID-19. The burst of these pro-inflammatory cytokines and chemokines in turn activates and attracts other immune cells to participate in a storm of immune responses which cause illness in the infected patient. Although these cellular effects and clinical syndromes during infection have been well-documented,26 the details of the causal molecules and their interactions remain unknown.
Our study began with investigating the cellular effects in response to the expression of the SARS-CoV-2 N protein. We found that among three tested tissues, SARS-CoV-2 N protein specifically induces multiple cytokines and chemokines in lung cells. Unexpectedly, we further found that secreted SARS-CoV-2 N protein can deliver nucleic acids into cells through receptor-mediated endocytosis for gene expression or silencing. STEAP2 is one of the receptors. Such activity is enhanced by the chemokine regulated on activation, normal T cell expressed and secreted (RANTES) induced by N protein itself, forming a positive feedback loop. We also found that the N-protein-mediated nucleic acid delivery is enhanced by lactate, a product produced in hypoxia. Moreover, we showed that N protein expressed in a cell can disperse nucleic acid into neighboring cells, and this effect is more pronounced with the wild-type SARS-CoV-2 N protein than Omicron N protein.
Results
N protein induces cytokines and chemokines in lung cells
It is known that the infection of SARS-CoV-2 leads to numerous cellular responses.25 Among the reactions that occur during infection, the most common is the induction of cytokines and chemokines, which can cause severe life-threatening conditions. To examine the capacity of SARS-CoV-2 N protein in stimulating the production of the cytokines and chemokines, we cloned wild type N as well as its two variants (R203K G204R and Omicron BA.1) found in the GISAID database according to a previous analysis,27 and individually expressed each in A549 (lung carcinoma cells), HCT-116 (colorectal carcinoma cells) and SH-SY5Y (neuroblastoma cells). We performed secretome analysis to survey the cytokines and chemokines induced by these N proteins (Figures S1–S3).
Interestingly, N protein induced significant amounts of cytokine and chemokine secretion from A549 cells. Among these N protein-induced cytokines and chemokines, interferon-gamma-induced-protein (IP-10), interferon-λ1 (IFN-λ1), interferon-λ2/3 (IFN-λ2/3), interleukin-8 (IL-8), RANTES, and interferon-inducible T cell alpha chemoattractant (I-TAC) were also reported at heightened levels in the serum of patients with COVID-19 and have been proposed as biomarkers of disease progression.28,29,30,31 Moreover, we found that SARS-CoV-2 N protein induces the highest levels of cytokines and chemokines among the seven N proteins from human coronaviruses (Figure S4). Secretome screening of N variants (R203K G204R, which are found in N proteins of the Alpha, Lambda, Zeta, and Omicron variants) and Omicron N (P13L, R203K, G204R and deletion of E31, R32, S33) did not show prominent differences (Figure S1). However, the induction of cytokines and chemokines showed tissue specificity, as expression of the N proteins in other two cell lines (HCT-116 and SH-SY5Y) induced much less cytokines and chemokines than in A549 cells (Figures S2 and S3). The secretome screening data suggest one of the N protein’s functions is to induce cytokines and chemokines, which corresponds with the clinical observations associated with the effects of cytokine storms during SARS-CoV-2 infection.
N protein is secreted, directly binds to the cellular surface, and enters cells through endocytosis
By expressing N proteins from either the wild-type SARS-CoV-2 or Omicron variant in HPAEpiCs (a normal lung alveolar cell line isolated from human lung tissue), we found that both of N proteins can be secreted to the extracellular medium (Figure S5A). This raised the question of whether the secreted N protein from N-overexpressed cells will bind to the neighboring cells. To investigate, we treated A549 with recombinant SARS-CoV-2 N protein produced from either E. coli or mammalian cells and assessed cell surface binding. We found that recombinant N proteins produced from either E. coli or mammalian cells both bind to the A549 cell surface (Figure 1A). These data indicate that SARS-CoV-2 N protein can directly bind to cells, independent of N protein phosphorylation. Interestingly, Omicon N recombinant protein, either E.coli-derived or mammalian-derived, exhibited less cell surface binding activity (Figure 1A). Here, Table S1 shows the amino acid changes in SARS-CoV-2 N proteins among different variants of concern (VOCs) based on the information from World Health Organization (WHO) Website (https://www.who.int/activities/tracking-SARS-CoV-2-variants). Among these variants, P13L and ERS31-33 deletion are observed in all omicron variants, and R203K/G204R mutation is conserved in SARS-CoV-2 alpha, gamma, and all omicron variants. In this study, the omicron N recombinant protein used for cell surface binding is the BA.1 variant, hence, mutations of P13L, ERS31-33 deletion, and R203K/G204R of N protein are constructed to further clarify which mutation causes the reduced cell surface binding activity. Interestingly, data showed that all omicron BA.1 mutations decrease N protein binding to the cell surface suggesting that these amino acids play important role in the interaction of N protein and cell surface receptors (Figure S5B).
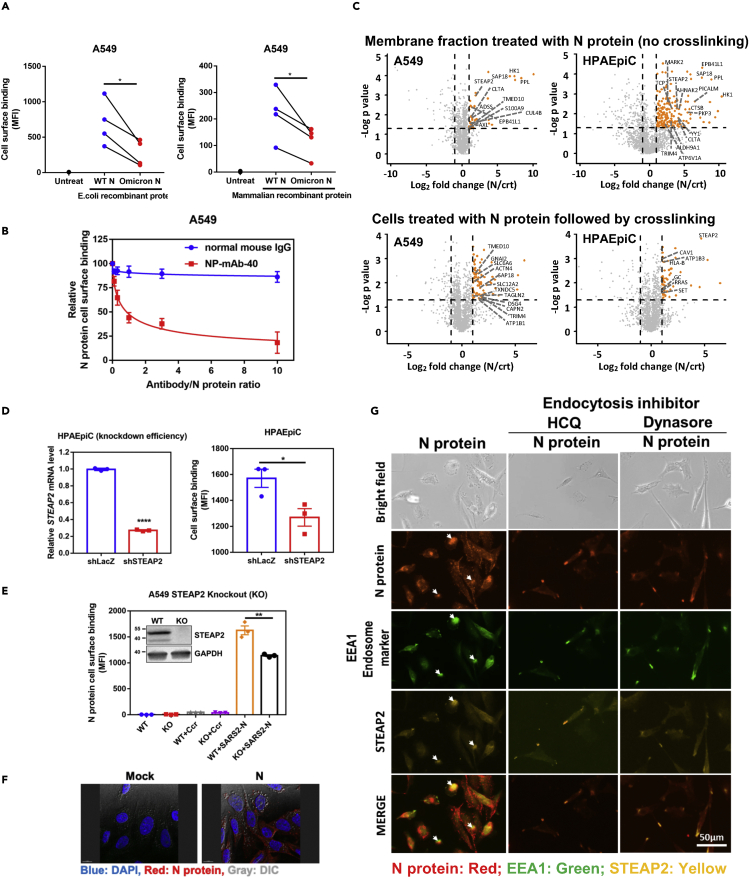
Figure 1.
N protein binds to and enters the cell through STEAP2
(A) Comparison of the cell-binding capacity of SARS-CoV-2 wild type (WT) N protein and Omicron N protein expressed in either E. coli or mammalian cells. 1 × 105 A549 cells were used to mixed with 1 μg WT N or Omicron N proteins. One hour after protein addition, allophycocyanin (APC) conjugated anti-His antibody was used to detect the cell binding capacity of WT N protein or Omicron N protein. The samples were analyzed by flow cytometry and data are shown as mean fluorescence intensity (MFI).
(B) Antibody blocking assay. Aliquots of 10 μg of SARS-CoV-2 N protein were pre-mixed with 0, 1, 3, 10, 30, and 100 μg of normal mouse IgG or anti-N monoclonal antibody (NP-mAb-40) and incubated at 4°C overnight. The antibody/N protein complex was used for the A549 cell surface binding assay. The blocking capacity of anti-N antibody was normalized to N protein only control.
(C) Membrane fractions of A549 and HPAEpiC cells were extracted and incubated with N protein conjugated beads for 3 h binding at 4°C, and pull-downed for LC-MS-MS analysis (upper panels). A549 and HPAEpiC cells were suspended and treated with N protein for 1 h on ice. After incubation, cells were crosslinked with 3 mM DTSSP for 1.5 h. Then, cells were lysed in RIPA lysis buffer, and N protein complex in the lysate was immunoprecipitated for LC-MS-MS analysis (lower panels). Y axis denotes −logP values while the X axis shows log2 fold change values. Orange dots highlight the statistically significant proteins, with p value < 0.05 (-Log p > 1.3) and fold change>2, and the enriched plasma membrane protein was labeled on the plot. Identified proteins were further sorted by HuMemProtDB.
(D) To knock-down (KD) STEAP2 expression, HPAEpiC cells were infected with lentivirus carrying STEAP2 shRNA followed by puromycin selection for 14 days. The STEAP2 mRNA expression levels were assessed by qRT-PCR, and the relative KD efficiency of shSTEAP2 was compared to shLacZ control (left-hand side panel). N protein binding capabilities to HPAEpiC STEAP2 KD cells and shLacZ control KD cells were assessed by flow cytometry analysis, and data were shown as mean fluorescence intensity (MFI). (right-hand side panel).
(E) Western blot analysis of STEAP2 in wild type (WT) and knock-out (KO) A549 cells were shown. N protein binding to A549 STEAP2 KO cells was assessed by flow cytometry analysis and shown as mean fluorescence intensity (MFI). Ccr (crotonyl-CoAcarboxylase/reductase, a bacterial protein) binding was used as a control.
(F) SARS-CoV-2 N protein enters alveolar cells. HPAEpiC cells were treated with 10 μg SARS-CoV-2 N protein overnight and then stained with anti-N antibody. The localization of N protein (Red) was checked by fluorescence microscope and cell morphology was observed by dimensional interference contrast (DIC). Nuclei of cells were stained by DAPI (blue).
(G) N protein entering cells by endocytosis and N protein co-localization with STEAP2. HPAEpiC alveolar cells were seeded on 8 well slides. Cells were pretreated with endocytosis inhibitors HCQ, or Dynasore. Then the cells were treated with N protein overnight. After treatment, the cells were stained by specific antibodies to detected N protein (red), endosome marker (EEA1) (green), and STEAP2 (yellow). Cells were observed under fluorescent microscopy (Invitrogen tech.). Scale bar: 50 μm. All data are shown as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001; t test. See also Figure S5.
We further assessed the cell surface binding capabilities of N proteins from other human coronaviruses (SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-NL63, HCoV-HKU1, and HCoV-229E). Notably, SARS-CoV-2 N protein exhibited the highest cell surface binding affinity as compared to other human coronavirus N proteins (Figure S5C). Among the seven coronavirus N proteins, SARS-CoV-2 N showed the highest binding affinity to A549 cells at concentrations less than 10 μg/mL (Figure S5C), followed by HCoV-NL63 and HCoV-OC43 N proteins. SARS-CoV-1 and HKU1 N proteins showed less binding affinity as compared to SARS-CoV-2, and MERS N protein showed no binding to the A549 cell surface. That SARS-CoV-2 N exhibited the highest cell surface binding affinity at low concentrations may have important pathogenic implications.
To probe the specificity of binding, we pretreated N protein with mouse anti-SARS-CoV-2 N sera or anti-N protein monoclonal antibody NP-mAb-40.32 Neutralizing data showed that either anti-N sera (Figure S5D) or anti-N monoclonal antibody (Figure 1B) treatment efficiently suppressed SARS-CoV-2 N protein binding to the A549 cell surface. To clarify if SARS-CoV-2 N protein binding is cell-type dependent, we used five human cell lines (293T kidney cells, HCT-8 colon cells, HPAEpiC and A549 lung alveolar cells, and HeLa cervical cells) and two mouse cell lines (4T1 breast cells and LL2 lung cells) in N protein binding tests. Our data show that N protein binds to all tested cell lines (Figure S5E), suggesting the cell binding is cell-type independent and SARS-CoV-2 N protein binding receptors are likely broadly expressed in most cell types.
To identify the N protein binding receptors, we performed an N protein pull-down experiment followed by proteomic analysis. Here, two pull down strategies were used. The first strategy extracts the membrane fractions of lung cells (A549 and HPAEpiC), which are then incubated with a bead-conjugated- antibody-bond N protein to pull down N-protein complex. The second is to pretreat lung cells with N protein followed by crosslinking, and the crosslinked N protein complex was pull-downed from the lysate by antibody-bond beads for LC-MS/MS analysis.33,34 A control experiment was performed by using a bacterial protein (Crt) instead of N protein to exclude non-specific binding proteins. The subcellular localization of the putative membrane proteins was further annotated according to HuMemProtDB.35 To identify the statistically significant N protein interacting partners, we quantitatively compared the protein abundance identified from the N protein-interacting proteins and control ones using > 2-fold change (N/Crt) and enrichment score (p value < 0.05). In the first experiment, the volcano plot from proteomic analysis reveals the significant enrichment of 11 and 35 N protein-interacting plasma membrane proteins in A549 and HPAEpiC membrane fraction, respectively (Figure 1C, upper panel; and Table S2). With cross-linking, 12 and 7 N protein-interacting plasma membrane proteins were identified and significantly enriched in A549 and HPAEpiC cells, respectively (Figure 1C, lower panel; and Table S3). Interestingly, among these N protein-interacting membrane proteins, STEAP2, a metalloreductase localized in the plasma membrane and Golgi complex, was the only common receptor protein significantly enriched in the pull-downed experiments.
Next, we used lentivirus-mediated shRNA knockdown and CRISPR/Cas9 knockout to confirm whether STEAP2 serves as a receptor for N surface binding. Results showed that STEAP2 knockdown in HPAEpiC cells (Figure 1D) or knockout in A549 cells (Figure 1E) significantly decreased N protein binding to the cell surface. The control protein (Ccr, also a bacterial protein) showed no binding as expected. Consistently, we also found that STEAP2 expression levels in HEK293T, HCT-8, and HPAEpiC cells are positively correlated with N protein surface binding (Figure S5F). Interestingly, STEAP2 expression in A549 cells is lower as compared to these cells but A549 cells still show strong N protein binding. In addition, STEAP2 knockout cells did not abolish N protein binding to the cell surface suggesting that other cell surface receptors might exist. Interestingly, STEAP2 overexpression in A549 cells also increased N protein binding to the cell surface (Figure S5G). These data suggest that STEAP2 is not the only surface binding receptor.
Since STEAP2 has been reported to internalize from the cell surface by endocytosis,36 we asked whether N protein binding to surface STEAP2 can trigger internalization. Using immunofluorescent staining, we found that recombinant SARS-CoV-2 N protein was indeed internalized (Figure 1F). Furthermore, we treated HPAEpiC cells with two endocytosis inhibitors, Dynasore (a cell permeable dynamin inhibitor) and HCQ (hydroxycloroquine) to block the endocytosis of the cells. Notably, the co-localization signal of N protein (red), STEAP2 (yellow), and EEA1 (endosome marker) (green) in the cytosol was reduced by Dynasore or HCQ treatment (Figure 1G). These results suggest that N protein binds to the cell surface and enters the cell through the receptor (e.g. STEAP2)-mediated endocytosis.
N protein mediates the intercellular nucleic acid transfer
N protein is a known RNA-binding protein. Recent report indicates that high-resolution cryo-electron tomography analysis of intact SARS-CoV-2 virions shows a characteristic shell-like architecture about 15 nm in diameter in individual viral ribonucleoproteins (vRNPs), comprising multi-copies of N proteins and RNA.7 Also, by negative-stain electron microscopy, the soluble RNP complexes revealed similar particles.37 Recently, an NMR study showed purified N protein formed phase-separated liquid condensates with RNA.38 We next assessed whether RNA packaging affects N protein binding to the cell surface. To this end, we synthesized short RNA (FAM-labeled 20 n.t. double-strand siRNA or 50 n.t. single-strand SARS-CoV-2 packaging sequence RNA fragments) and long single-stranded mRNA (>500 nt, e.g. spike receptor binding domain mRNA, spike full-length mRNA or luciferase-P2A-EGFP mRNA) to manipulate RNA binding to N protein. We found that the short RNA did not affect cell surface binding, while long RNA-bound N proteins markedly increased the cell binding ability (Figure 2A).
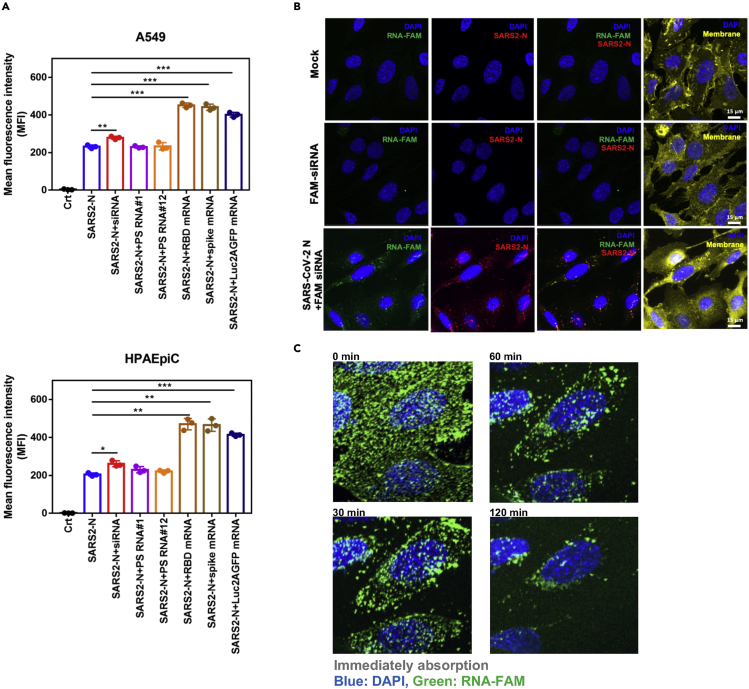
Figure 2.
N protein delivers nucleic acids into cells
(A) N protein-RNA complex binding to the cell surface. Aliquots of 10 μg SARS-CoV-2 N protein were incubated with 1 μg of indicated RNAs for 1 h at 4°C, and added to A549 or HPAEpiC cultures. SARS-CoV-2 N protein only without RNA was used as a control. The samples were analyzed by flow cytometry and data are shown as mean fluorescence intensity (MFI). Data are shown as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; t test.
(B) The observation of N protein-RNA enters into cells. HPAEpiC were seeded onto 8-well glass slides (40,000 cells/well). SARS-CoV-2 N protein 10 μg and 40 μg RNA-FAM (green) were mixed for 1 h at 4°C. cells were treated with SARS-CoV-2 N-RNA-FAM mixture for 1 h. The groups of non-treated cells and RNA-FAM only were as controls. After treatment, N protein was detected by anti-N antibody (Red). The localization of RNA-FAM was green. DAPI (blue) indicates cell nuclei. Scale bar: 15 μm.
(C) Lattice light sheet microscopy time lapse imaging of N protein-RNA complex entering into HPAEpiC cells. SARS-CoV-2 N protein 10 μg was mixed with 40 μg RNA-FAM (fluorescein) for 1 h at 4°C and then treated with ice-cooled alveolar cells. The signals of RNA-FAM and Hochest 33,342 were monitored by lattice light sheet microscopy at different time points.
We next assessed whether N protein can bind to RNA molecules and carry them into cells. We pre-mixed N protein and FAM-labeled control siRNA (RNA-FAM) and observed N-RNA complex signal indicating it can enter into cells (Figure 2B). No signal was detected in the mock and RNA-FAM only groups. We used lattice light-sheet microscopy to perform a time-lapse study visually tracing the biological process of N-protein-mediated RNA-FAM entry to the cell (Figure 2C, Video S1 in Supplementary Item entitled “N protein carries RNA entering into cells”, related to Figure 2C). Initially, the RNA-FAM signal was widely dispersed. As time progressed, the RNA-FAM signal moved from outside to the inside of the cells, clearly demonstrating that SARS-CoV-2 N protein had the ability to carry and tRNA into the cells.
The video about the observation of SARS-CoV-2 N protein carries RNA-FAM entering cells is attached in the supplementary item. Detailed materials or reagents used in this study are listed in key resources table.
N protein-mediated nucleic acid transfer can achieve gene silencing or expression
Having determined that N protein can bind and carry RNA into cells, we next asked whether RNA N-protein-mediated into cells can perform biological functions. Here, we used N protein to carry siRNAs that were designed for targeting specific genes, e.g. JUN and IFNAR1. After N protein/siRNA complex treatment, we used qRT-PCR to detect expression levels. Interestingly, both JUN and IFNAR1 expression was significantly reduced by the N-protein-mediated siRNA group as compared to solvent control, N protein only, or siRNA only control (Figure 3A). We further found that N protein was able to deliver GFP mRNA into cells and express protein, which we detected with immunofluorescent staining (Figure S6A). These results indicate that RNA carried into the cell by N protein are functionally active, suggesting N-protein-mediated RNA entry may be involved in pathogenic functions during virus infection.
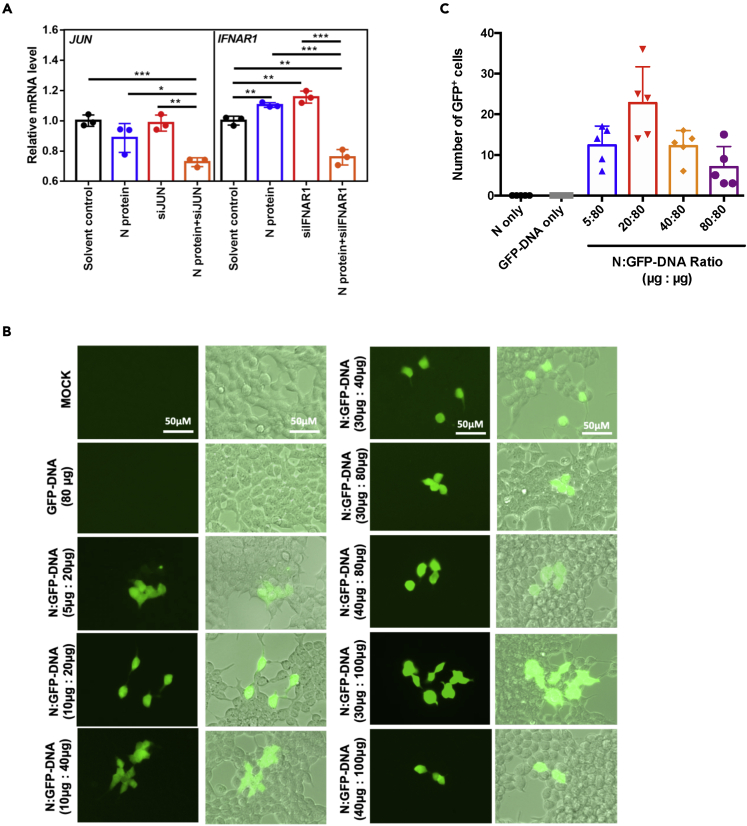
Figure 3.
Actions from N protein delivered nucleic acids
(A) 293T cells were seeded in 48-well plate for overnight attachment. Aliquots of 10 μg SARS-CoV-2 N protein were incubated with 100 μL of indicated siRNA (10 μM) or 1 h at 4°C. N protein-siRNA complex was directly added into well and expression of the siRNA targeted genes was assessed by qRT-PCR after 48 h of treatment.
(B) 293T cells were seeded onto 8-well glass slides (55,000 cells/well). SARS-CoV-2 N protein was pre-mixed with GFP-DNA (pmax-GFP) for 1 h at 4°C at indicated ratios. The N protein-GFP DNA mixtures were added to 293T cells. After 48 h, the GFP expression was observed by fluorescence microscopy. The groups of non-treated cells (MOCK) and GFP-DNA only (GFP-DNA 80 μg) were as controls.
(C) Effect of N protein:GFP-DNA ratio. N protein and GFP-DNA were mixed by indicated conditions and incubated for 1h at 4°C. These WT N protein/GFP-DNA mixtures were added into 293T cells overnight (24 h) and the formation of mixtures and the number of GFP+ cells were counted by fluoresce microscopy. The best mass ratio of N protein to DNA for GFP expression was around 1:4. All data are shown as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; t test.
Next, we tested whether N-protein-mediated DNA entry was possible. We added complexes consisting of different ratios of N protein and GFP plasmid DNA to cells (Figure 3B). In contrast to the control groups lacking N protein, we detected strong GFP expression in all test groups of N protein to GFP-DNA ratios, finding strongest expression in the mass ratio 1: 4 (N protein: GFP-DNA) group (Figure 3C). Our data show that SARS-CoV-2 N protein can deliver both types of nucleic acids into cells and achieve gene expression.
Lactate and RANTES enhance N protein-carried nucleic acid expression
Since lactate is a metabolite reported in patients infected with SARS-CoV-2,39 we tested whether lactate can elevate the efficiency of N-protein-carried DNA expression. Indeed, we found that 50 mM lactate enhanced N-carried DNA expression (Figure 4A). In contrast, pyruvate and glutamate, other related metabolites, showed no such effect (Figure S7).
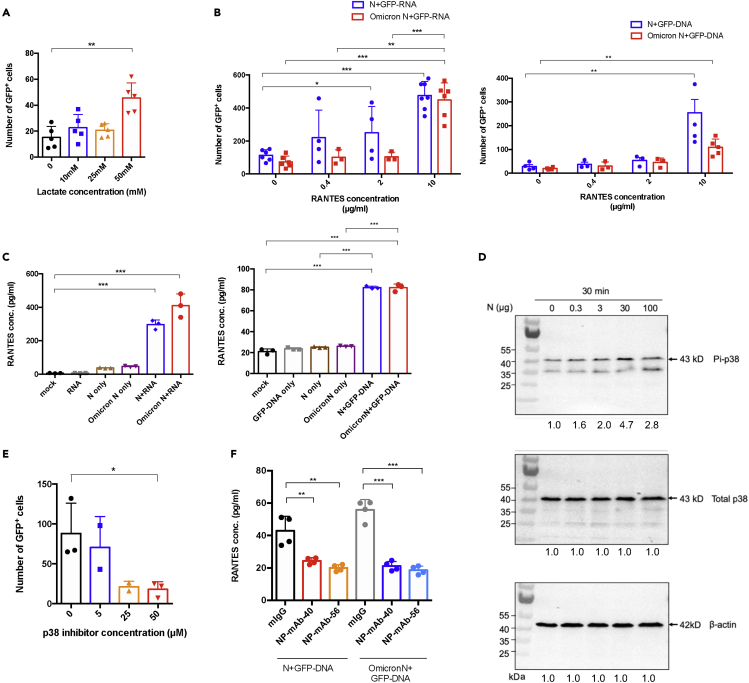
Figure 4.
Lactate and RANTES enhance N protein-carried nucleic acid expression
(A) Lactate effect. 293T cells were pretreated with lactate at indicated concentrations. After treatment, N-GFP DNA mixtures were added to 293T cells. The GFP+ cells were counted after 24 h.
(B) Dose-dependent effect of RANTES. 293T cells were treated with an indicated dose of RANTES overnight. After treatment, Pre-mixed 10 μg mammalian cells-expressed WT N or Omicron N and 40 μg GFP-RNA (Left panel), GFP-DNA (Right panel) was added to cells overnight, and the number of GFP+ cells were counted under fluorescent microscopy.
(C) N-RNA and N-DNA complexes induced RANTES. HPAEpiC cells were stimulated by mammalian expressed N protein, GFP-RNA (or DNA), and N+GFP-RNA (or DNA) mixture for 24 h. After treatment, protein secretion of RANTES was detected by flow cytometry analysis.
(D) N protein was added to HPAEpic cells at indicated dosages for 30 min. After treatment, the cells were harvested and phosphorylation of p38 was detected by Western blotting. Quantification of band intensities was normalized by non-treated cells.
(E) 293T cells were pretreated by p38 inhibitor (SB203580) at indicated dosages overnight. After treatment, the N-GFP-DNA mixtures were added to 293T cells. GFP+ cells number were counted after 24 h.
(F) Mammalian cells-expressed N protein 4 μg pretreat with anti-N monoclonal antibody (NP-mAb-40 or 56) 10 μg/mL, and then mix with 12 μg GFP-DNA for 1 h, then the N+DNA/antibody complex are added to HPAEPic cells for 48 h. RANTES concentration in the supernatant is measured and detected by anti-RANTES beads by flow cytometry analysis. All data are shown as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; t test. See also Figures S6–S9.
Given our finding that N protein expression induces several cytokines and chemokines (Figure S1), we tested if these cytokines enhance N protein delivery of DNA and expression of genes. Among the cytokines or chemokines tested (IP-10, IL-28A, IL-29, RANTES), we found that only RANTES significantly enhanced N protein-delivered GFP-RNA or GFP-DNA expression in a dose-dependent manner (Figures 4B, S6A, and S6B). Since we showed that the level of cytokines was significantly elevated in N-protein transfectants (Figure S1), we tested whether N-protein-nucleic acid complex also affects cytokine induction, potentially forming a positive feedback loop. The N protein-GFP-RNA or N protein-GFP-DNA complexes were added to HPAEpiCs for 24 h. Results indicated that such complexes significantly enhanced the production of RANTES (Figure 4C). Thus, our data suggest that a positive feedback loop exists wherein RANTES enhances N-protein-mediated nucleic acid delivery, and N-nucleic acid enhances RANTES production.
Since RANTES production was increased through the phosphorylation of p38 in influenza virus-infected human bronchial epithelial cells,40 we examined whether the level of phospho-p38 is also increased by N protein treatment. Indeed, we found that N-protein-induced p38 phosphorylation in alveolar cells in a dose-dependent manner (Figures 4D and S8). Moreover, we found that the inhibition of p38 by SB203580 significantly reduced N-protein-mediated GFP-DNA expression (Figure 4E).
Next, we tested whether N-protein-assisted nucleic acid delivery could be blocked by an anti-N protein antibody. To answer this question, several monoclonal anti-N antibodies were tested to check their efficiency in blocking N-protein-mediated GFP-DNA expression (Figure S9). We found several candidates, including NP-mAb-40 and NP-mAb-56, which significantly reduced N-protein-mediated GFP-DNA expression. We also evaluated the efficacy in blocking RANTES induction by the N-DNA complex. We find that upon the addition of NP-mAb-40 or NP-mAb-56, N-DNA complex-mediated RANTES secretion is significantly reduced (Figure 4F). In sum, we have demonstrated that through p38 signaling, N protein induces RANTES expression, which enhances nucleic acid entry and expression. Importantly, this phenomenon can be blocked by specific monoclonal anti-N antibodies.
N protein mediates the dispersion of nucleic acid to neighboring cells
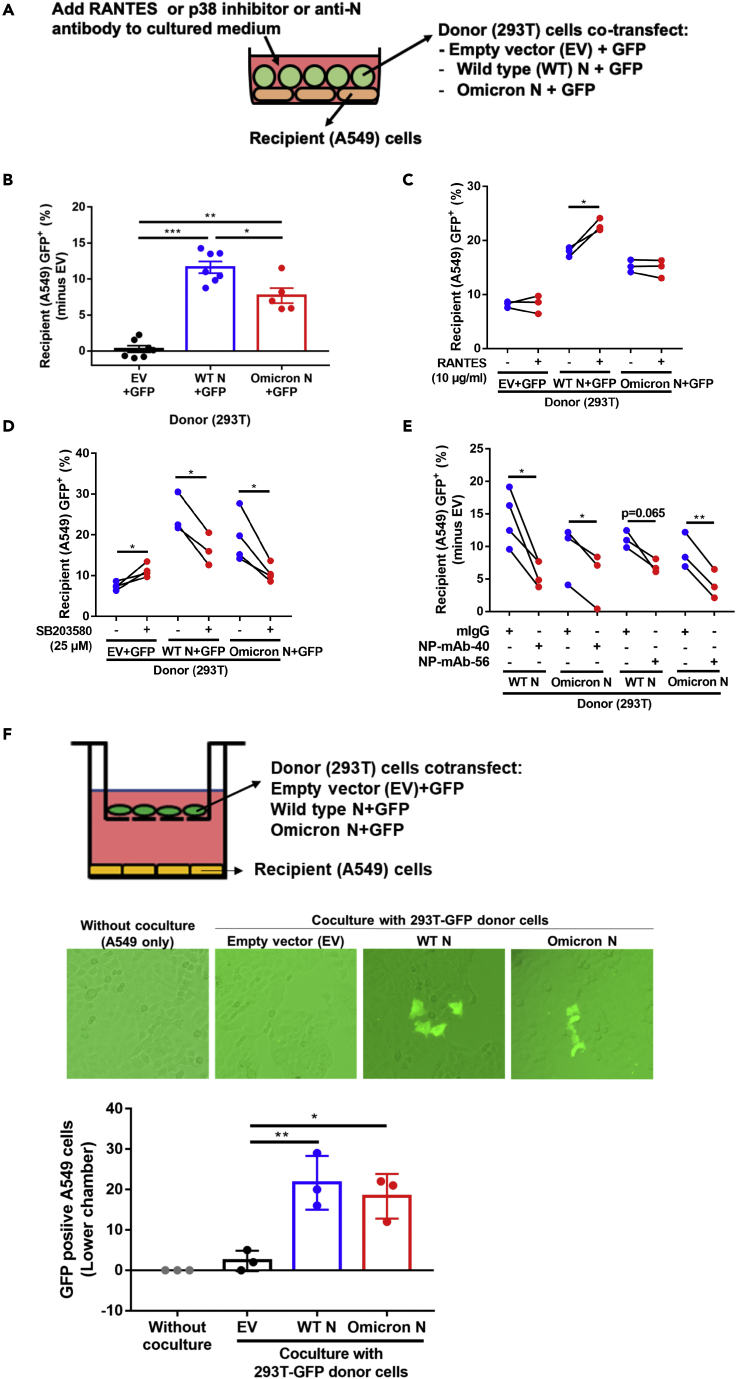
To demonstrate the physiological relevance of this N protein function, we designed a co-culture system (Figure 5A) to test whether N protein might mediate intercellular delivery and expression of nucleic acids in neighboring cells. As donor cells, we used 293T cells that ectopically expressed GFP with or without SARS-CoV-2 N protein through pre-transfected plasmids. 24 h after transfection, the donor cells were co-cultured with the recipient A549 cells. We detected GFP expression in the recipient cells using flow cytometry double gated with cytokeratin 18 as an A549 cell marker and SV40 large T antigen as a 293T cell marker (Figure S10). In addition, Q-PCR for GFP-RNA and N protein ELISA were performed to further check their existence in the medium of donor cells during co-culture. (Figure S11).
Figure 5.
N protein-assisted nucleic acid dispersion and expression in the co-culture environment
(A)The co-culture system consisted of A549 as recipient cells, and 293T pre-transfected with two plasmids, one expressing GFP and the other expressing SARS-CoV-2 N protein or the pcDNA3.1 empty vector.
(B–E) After 24 h co-culture of the donor cells and recipient cells, cell pool was stained with cytokeratin 18 (an A549 marker) and SV40 large T antigen (a 293T marker). A549 cells in the cell pool were gated from cytokeratin 18 positive and large T antigen negative. A549 GFP positive percentage was further assessed by flow cytometry analysis. Effects of SARS-CoV-2 N variants (B), treatment with RANTES (C), the p38 inhibitor SB203580 (D), or anti-N neutralizing antibody (E) were accessed by adding these effectors to the medium. Experiments are performed in three to five biological replicates. ∗, p value <0.05 (paired two-tailed student’s t-test).
(F) SARS-CoV-2 N protein promotes gene delivery by cell-free diffusion to neighboring cells. A549 cells were plated in the lower chamber, while 293T donor cells co-transfected with plasmids expressing EGFP and indicated N proteins in the upper chamber. After 3 days of co-culture, GFP positive A549 cells were observed and counted. See also Figures S10 and S11.
As shown in Figure 5B, GFP-positive A549 recipient cells markedly increased after co-culturing with wild-type SARS-CoV-2 or Omicron N-protein-expressing donor cells as compared to the control group where donor cells lacked N protein. These results demonstrated that N protein can mediate the intercellular transfer and expression of nucleic acids. Compared to the wild-type SARS-CoV-2 N protein, the Omicron N protein showed weaker nucleic acid transfer levels in the co-culture system, which may be associated with the clinical differences between wild-type SARS-CoV-2 and Omicron infection cases. The differential activity between wild-type SARS-CoV-2 and Omicron N proteins in mediating nucleic acid transfer can be explained by the weaker cell surface binding ability of Omicron N protein (Figure 1A). In addition, RANTES (10 μg/mL) consistently enhanced SARS CoV-2 wild-type N-protein-mediated intercellular gene expression (Figure 5C), but had no effect on the Omicron N-protein-mediated intercellular gene expression. Moreover, the p38 inhibitor, SB203580 (25 μM) reduced both the wild-type and Omicron N-protein-mediated gene expression under this condition (Figure 5D), suggesting that p38 might exert other effects in addition to inducing RANTES production.
Since anti-N monoclonal antibodies can block N protein endocytosis (Figure 1F), reduce N-protein-mediated protein expression (Figure S9), and reduce RANTES secretion (Figure 4F), we next test whether anti-N antibody could diminish N-protein-mediated enhanced intercellular gene expression in a co-culture condition. Consistently, we observed that both NP-mAb-40 and NP-mAb-56 monoclonal antibodies inhibited either wild-type N protein or Omicron N-protein-mediated intercellular gene expression (Figure 5E). These results may point to an important direction for clinical studies. Extending these findings, we used a transwell co-culture system to test whether cell-cell contact is required for N-mediated intercellular nucleic acid transfer. Data showed that GFP mRNA or DNA from transfected 293T donor cells in the upper well is transferred to untransfected A549 recipient cells in the bottom well separated by a 1-μm pore size filter without donor-to-recipient cell-cell contact (Figure 5F). This result suggested that cell-cell contact is not required for N-mediated intercellular nucleic acid transfer.
Discussion
Among all the SARS-CoV-2 proteins, N protein is the most abundant during infection,41 can be detected in patient serum,42 and is commonly targeted as a marker for rapid infection testing.43 So far, its main known function is to bind to viral genomic RNA and facilitate packaging. Here we demonstrated that N protein can assist nucleic acid entry into neighboring cells, mediating biological functions including gene expression and RNAi. Since nucleic acid binding by N protein is non-specific, such a function may expand the damage inflicted by viral infection. Moreover, this function can be enhanced by RANTES, a chemokine induced by N protein itself which attracts T cells to produce more RANTES.44,45 This result is clinically relevant as RANTES production has been reported in patients undergoing a cytokine storm.46 We also found that lactic acid enhances N-protein-mediated nucleic acid delivery, which relates to the clinical observation that patients with severe COVID-19 commonly develop hypoxic conditions known to increase lactate production.47 Finally, we observed that the Omicron N protein showed weaker activity in the delivery of nucleic acids to neighboring cells through endocytosis.
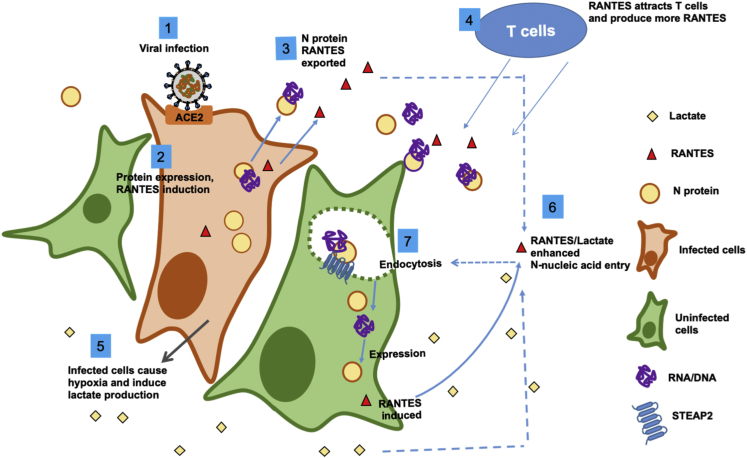
In sum, our data strongly suggest that SARS-CoV-2 N protein can facilitate intercellular nucleic acid transfer and gene expression and that such actions are enhanced by the chemokine RANTES and hypoxic metabolite lactate, forming a positive-feedback loop. We propose the following working hypothesis (Figure 6): Upon infection of SARS-CoV-2, expression of N protein promotes p38 MAPK activation and RANTES chemokine secretion. N protein complexed with nucleic acids are secreted outside the cell. Since N protein has a strong binding affinity toward RNA and DNA, it may bind with sub-genomic RNA, mRNA, or DNA fragments prior to being secreted out of the infected cell, then enter neighboring cells via STEAP2-mediated endocytosis. The cytokines induced by N protein, e.g. IP-10 and RANTES, recruit T cells,48,49 which produce more RANTES.50 In parallel, infected lung cells may cause hypoxia, promoting lactate production. Lactate and RANTES may then enhance the spreading of N-protein-RNA and N-protein-DNA complexes to neighboring cells causing further tissue damage and hypoxia. The dispersion effect, which is characteristically diminished with Omicron N protein as compared to wild-type SARS-CoV-2 N protein, may underlie infection dispersion and can be reduced by monoclonal anti-N antibodies or p38 inhibitors.
Figure 6.
Working hypothesis
A working hypothesis for SARS-CoV-2 N protein is proposed: Expression of SARS-CoV-2 N protein promotes p38 MAPK activation and RANTES chemokine secretion (1,2). N protein/nucleic acid complexes are secreted (3). The chemokines induced by N protein attract T cells, which produce more RANTES (4). The infected cells may cause local hypoxia and produce lactate (5). RANTES and lactate enhance N-protein-nucleic acid endocytosis by neighboring cells (6,7), and cause more tissue damage. The RNA/DNA carried by N protein could be originated from the virus or the host cell.
STEAP2-receptor
Our study shows that SARS-CoV-2 N protein can bind to the cell surface via STEAP2, which serves as the first non-ACE2 receptor for SARS-CoV-2. N-protein-RNA or N-protein-DNA complex can then enter cells through endocytosis. However, knocking out STEAP2 in A549 did not completely abolish N protein binding, suggesting that there are likely other receptors mediating N protein endocytosis. STEAP2 belongs to the Six-Transmembrane Epithelial Antigen of the Prostate (STEAP) protein family and may serve as a metalloreductase, but its enzymatic function is not detected at the plasma membrane,36 indicating that the protein might perform alternative functions. STEAP2 has a broad subcellular localization between the endocytic and secretory pathways, as it can internalize from the cell surface and traffic to the ER-Golgi.51,52 It may function as a receptor to deliver or sort endogenous and exogenous lipids and proteins.
Our results suggest a mechanism in which N protein hijacks the STEAP2 endocytic pathway to deliver nucleic acids into neighboring cells. One potential scenario is one in which infected cells produce N protein that may deliver RNA molecules (including sub-genomic RNA) to neighboring cells to prepare for impending infection. In another non-mutually exclusive scenario, N-protein-delivery of non-specific RNA or DNA fragments to neighboring cells spreads and amplifies infection damage. We demonstrated this strategy using a co-culture system (Figure 5), showing that pre-transfected donor (293T) cells that produce N protein can deliver the GFP-encoding nucleic acids to the co-cultured recipient (A549) cells. To further postulate, N protein may even deliver viral genomic RNA through STEAP2 or other receptor-mediated endocytosis to spread infection via this ACE2-independent pathway. Another possibility is that infected cells respond by producing cytokines and chemokines which attract T cells that, in turn, elicit more production of these molecules, causing a cytokine storm that further enhances N-protein-mediated nucleic acid delivery.
It is known that SARS-CoV-2 S protein binds to ACE2, which serves as the entry receptor to the host cell.53,54,55,56,57 Two major pathways of enveloped viral spreading have been reported: the first is through cell-free particles, and the second is through cell-to-cell contact.58,59 The latter is often dependent on tight cell-to-cell contacts or virological synapses.59,60 A recent study indicates that ACE2 is not required during SARS-CoV-2 spreading,61 and the spike protein can also mediate cell-to-cell viral transmission by cell-cell fusion.61 Our study may provide a possible pathway for N-protein-mediated RNA/DNA dispersion.
RANTES and lactate enhance N-carried DNA/RNA expression
N protein may contribute to the viral infection process in multiple different ways.62 Our data indicate that N protein significantly increased the secretion of RANTES in A549 cells (Figure 4C). We also found that N protein significantly induced the phosphorylation of p38 in alveolar cells and that inhibiting p38 phosphorylation reduces N-protein-mediated DNA expression in neighboring cells. Together, these results suggest that N protein may induce RANTES via the p38 pathway. This implication is reminiscent of a previous report that describes how platelet-activating factor (PAF) can induce the secretion of RANTES through the p38 signaling pathway in airway smooth muscle cells.63
We also found that lactate enhances N-protein-mediated nucleic acid entry and expression in cells. Lactate is used as an indicator for predicting the outcome of COVID-19 infection,39,64,65 particular in elderly patients.66 Data from 2860 patients indicated that elevated serum lactate (blood lactate concentration >2 mmol/L)67 was associated with significantly higher ICU admission and 3-month mortality.67 Within patients with COVID-19, elevated lactate concentration is associated with ICU mortality.67 Furthermore, lactate activates human macrophages and stimulates RANTES secretion through Notch signaling.68 The RANTES-CCR5 axis recruits leukocytes to inflammatory foci69,70 and may contribute to the activation of glycolysis and secretion of lactate in cancer cells.68 Here we showed that the pretreatment of lactate (50 mM) significantly enhanced the efficiency of N-mediated DNA/RNA expression.
Clinical relevance
A pertinent clinical finding in our study is that N protein mediates nucleic acid transfer between neighboring cells and that this is more pronounced with the wild-type SARS-CoV-2 N protein than the Omicron N protein. This result may correspond with and, in part, underlie the clinical report that the Omicron variant exhibits less severe symptoms and decreased viral spread within lung tissue. Furthermore, we show that the N-protein-mediated spreading of nucleic acid can be reduced by p38 inhibitors (Figure 5D) or monoclonal antibodies (Figure 5E), suggesting that such drugs may have therapeutic potential so as to prevent or mitigate the spreading of and damage caused by viral components. In our previous study,32 we found that NP-mAb-40, which was used as a neutralizing antibody in this study, cannot bind to the RNA-binding domain (RBD) and dimerization domain (DD) of N protein, suggesting that the epitopes for NP-mAb-40 recognition may be located in other domains, including the N-terminal domain (NTD), linker region or C-terminal domain. Interestingly, variations in NTD (P13L and ERS31-33 deletion) and the linker region (R203K and G204R) dramatically inhibited N protein binding to the cell surface (Figure S5B), supporting our previous result.
A recent report indicates the recurrence of PCR-positive tests in patients after recovery from COVID-19 even though there is no infectious virus detected in their body.71 This observation was explained by the finding that a fraction of the viral RNA can be reverse-transcribed and integrated into the DNA, generating viral-host chimeric transcripts.71 We posit that N protein may also mediate the spreading of these residual viral transcripts. During the review of this article, a study also reported that SARS-CoV-2 virus-infected cells can transfer N protein to bind to the surface of neighboring uninfected cells by diffusion without cell-cell contact.72 This report is consistent with our results and support our finding that N protein can further mediated nucleic acid transfer into neighboring cells, leading to the possibility that this event might occur during virus infection. However, further study is needed to prove this hypothesis.
Interestingly, even though Omicron N protein also induces similar amounts of cytokines and chemokines, its cell surface binding (Figure 1A) or intercellular gene dispersion (Figure 5B) activity are weaker than that of wild-type SARS-CoV-2. While the Omicron N protein deviates from the wild-type N protein by only six amino acid residues (P13L, R203K, G204R and deletion of E31, R32, S33), the differing phenotypes elicited by Omicron N protein might be relevant in the viral evolution.
Our finding here demonstrates that N protein prominently induces cytokines and chemokines. In addition, SARS-CoV-2 N protein induces the most amounts of cytokines and chemokines compared with the six other types of known human coronaviruses (SARS-CoV, MERS, OC43, HKU1, NL-63, and 229E). Of all the types of human coronaviruses, N protein from SARS-CoV-2 exhibits the strongest cell surface binding affinity (Figure S5C). Together, these properties implicate a more important role of N protein in the pathogenesis of SARS-CoV-2 than previously recognized and justify N protein as a potential therapeutic target to be further considered and investigated. It will also be interesting to find out whether other viruses have similar functions mediated by nucleocapsid proteins.
Limitations of the study
The study is based on in vitro cell cultures with recombinant N proteins. Further confirmation requires animal study with virus and clinical patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-cytokeratin 18 antibody | Abcam | Cat#ab668 |
| anti-SV40 large T antibody | Abcam | Cat#ab234426; RRID:AB_2928058 |
| goat anti-mouse IgG-647 | Biotium | Cat#20281 |
| goat anti-rabbit IgG-555 | Biotium | Cat#20033; RRID:AB_10559671 |
| SARS/SARS-CoV-2 Nucleocapsid Monoclonal Antibody | Invitrogen | Cat#MA1-7404; RRID:AB_1018422 |
| normal mouse IgG | Santa Cruz | Cat#sc-2025; RRID:AB_737182 |
| NP-mAb-40 | Monoclonal Antibodies against Nucleocapsid Protein of SARS-CoV-2 Variants for Detection of COVID-19 | N/A |
| NP-mAb-56 | Monoclonal Antibodies against Nucleocapsid Protein of SARS-CoV-2 Variants for Detection of COVID-19 | N/A |
| p38 MAPK Antibody | Cell Signaling Technology | Cat#9212; RRID:AB_330713 |
| β-Actin Antibody | Cell Signaling Technology | Cat#4967; RRID:AB_330288 |
| Phospho-p38 MAPK (Thr180/Tyr182) Antibody | Cell Signaling Technology | Cat#9211; RRID:AB_331641 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| Anti-Na+/K+ ATPase monoclonal antibody | Abcam | Cat#ab76020; RRID:AB_1310695 |
| goat anti-rabbit-Alexa 568 antibody | Biotium | Cat#20102; RRID:AB_10854240 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 | Invitrogen | Cat#A32723; RRID:AB_2633275 |
| anti-His-APC antibody | BioLegend | Cat#362605; RRID:AB_2715818 |
| anti-His antibody | Sigma-Aldrich | Cat#H1029; RRID:AB_260015 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 (2019-nCoV) Nucleocapsid-His Recombinant Protein, Biotinylated | SinoBiological | Cat#40588-V07E |
| SARS-CoV-2 B.1.1.529 (Omicron) Nucleocapsid Protein (His Tag) | SinoBiological | Cat#40588-V07E34 |
| SARS-CoV-2 (COVID-19) Nucleocapsid protein, His Tag | Acro | Cat#NUN-C5227 |
| SARS-CoV-2 Nucleocapsid protein, His Tag (B.1.1.529/Omicron) | Acro | Cat#NUN-C52Ht |
| Lipofectamine 3000 Reagent | Thermo Fisher Scientific | Cat#L3000-015 |
| puromycin | Thermo Fisher Scientific | Cat#A1113803 |
| protein G magnetic beads | GE Healthcare | Cat#28951379 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche | Cat#4693132001 |
| PhosSTOP™ | Roche | Cat#4906837001 |
| DTSSP | Thermo Fisher Scientific | Cat#21578 |
| sequencing-grade modified trypsin | Promega | Cat#V511A |
| interleukin-28 alpha | BioLegend | Cat#754202 |
| RANTES | BioLegend | Cat#580202 |
| IP-10 | BioLegend | Cat#573502 |
| interleukin-29 | BioLegend | Cat#711204 |
| sodium DL-lactate | Sigma-Aldrich | Cat#L4263 |
| L-glutamine | Gibco | Cat#25030-081 |
| pyruvic acid | Sigma-Aldrich | Cat#107360 |
| BD Cytofix/Cytoperm™ Plus | BD Biosciences | Cat#555028 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787-100ML |
| DAPI | Invitrogen | Cat#D1306 |
| Crotonase(crt) | This paper | N/A |
| Crotonyl-CoA carboxylase/reductase(ccr) | This paper | N/A |
| 2-Mercaptoehtanol | Bio-Rad Laboratories | Cat#1610710 |
| Mini-Proten TGX | Bio-Rad Laboratories | Cat#4561086 |
| BD Cytofix/Cytoperm™ Plus | Merck Millipore | Cat#IEVH85R |
| hydroxychloroquine(HCQ) | Sigma-Aldrich | Cat#SI-H0915 |
| Dynasore | Sigma-Aldrich | Cat#D7693 |
| SB203580 | Selleckchem | Cat#S1076 |
| Critical commercial assays | ||
| EZ-PCR kit | Biological Industries | Cat#20-700-20 |
| LEGENDplex™ Human Anti-Virus Response Panel (13-plex) with V-bottom Plate | BioLegend | Cat#740390 |
| LEGENDplex™ HU Proinflam. Chemokine Panel 1 (13-plex) w/FP | BioLegend | Cat#740984 |
| Human RANTES Flex set | BD Biosciences | Cat#558324 |
| Human Soluble Protein Master Buffer Kit | BD Biosciences | Cat#558264 |
| QIAamp® DNA mini kit | QIAGEN | Cat#ID: 56304 |
| RNeasy Mini Kit | QIAGEN | Cat#74106 |
| PowerUp™ SYBR™ Green Master Mix | Applied Biosystems™ | Cat#A25741 |
| ibson Assembly® Master Mix – Assembly | NEB | Cat#E2611L |
| Experimental models: Cell lines | ||
| Human embryonic kidney 293T | American Type Culture Collection | Cat#CRL-3216 |
| human cervical cancer HeLa | American Type Culture Collection | Cat#CCL-2 |
| mouse lung cancer LL2 | American Type Culture Collection | Cat#CRL-1642 |
| human lung adenocarcinoma A549 | American Type Culture Collection | Cat#CCL-185 |
| human colon adenocarcinoma HCT-8 | American Type Culture Collection | Cat#CCL-244 |
| mouse mammary gland epithelium 4T1 | American Type Culture Collection | Cat#CRL-2539 |
| Human pulmonary alveolar epithelial cells(HPAEpiC) | ScieCellTM | Cat#3200 |
| Oligonucleotides | ||
| siRNA-FAM; 5'-CCAACTGAAACGATTTGTGCACCA CTCACTGTCTTTTTTGATGGTAGAGT-3′ |
MDBio | Cat#NO.5860029552 |
| JUN siRNA; 5′-GAGCGGACCUUAUGGCUAC-3′; 5′-GAACAGGUGGCACAGCUUA-3′; 5′-GAAACGAC CUUCUAUGACG-3′ and 5′- UGAAAGCUCAGAACU CGGA-3′ |
Dharmacon | Cat#L-003268-00-0005 |
| siIFNAR1; 5′- GCGAAAGUCUUCUUGAGAU-3′; 5′- UGAAACCACUGACUGUAUA-3′; 5′- GAAAAUU GGUGUCUAUAGU-3′ and 5′- GAAGAUAAGGCAA UAGUGA-3′ |
Dharmacon | Cat#L-020209-00-0005 |
| STEAP2 forward primers for qRT-PCR; 5'- CCTCTG CTTACCGATGAGAAGG-3′ |
This paper | N/A |
| STEAP2 reverse primers for qRT-PCR; 5'- CAGGAGGGAAAGTAAGCCAAGG -3′ |
This paper | N/A |
| PS RNA#1: Covid-19 viral RNA sequence; 5′-CCAA CTGAAACGATTTGTGCACCACTCACTGTCTTTTT TGATGGTAGAGT-3′ |
MDBio | N/A |
| PS RNA#12: Covid-19 viral RNA sequence; 5′-ATTT GTGCACCACTCACTGTCTTTTTTGATGGTAGAG TTGATGGTCAAGT-3′ |
MDBio | N/A |
| pLKO.1-LacZ shRNA | National C6 RNAi Core Facility | Cat#TRCN0000072224 |
| pLKO.1-STEAP2 shRNA | National C6 RNAi Core Facility | Cat#TRCN0000297801 |
| Recombinant DNA | ||
| pmax-GFP | Lonza | Cat#VDC-1040 |
| pcDNA3.1-SARS-CoV2 N-6xHis tag | this paper | N/A |
| pcDNA3.1-SARS-CoV2 N(R203K, G204R)-6xHis tag | this paper | N/A |
| pcDNA3.1-SARS-CoV2 N(omicron)-6xHis tag | this paper | N/A |
| pcDNA3.1-SARS N-6xHis tag | this paper | N/A |
| pcDNA3.1-MERS N-6xHis tag | this paper | N/A |
| pcDNA3.1-229E N-6xHis tag | this paper | N/A |
| pcDNA3.1-NL63 N-6xHis tag | this paper | N/A |
| pcDNA3.1-HKU1 N-6xHis tag | this paper | N/A |
| pcDNA3.1-OC43 N-6xHis tag | this paper | N/A |
| pET15b-SARS-CoV2 N(P13L)-6xHis tag | this paper | N/A |
| pET15b-SARS-CoV2 N(31-33 deletion)-6xHis tag | this paper | N/A |
| pET15b-SARS-CoV2 N(R203K, G204R)-6xHis tag | this paper | N/A |
Resource availability
Lead contact
All the information and the requests of materials in this paper should be directed to the lead contact: liaoj@gate.sinica.edu.tw.
Materials availability
All materials are described in the STAR Methods key resources table, and there is no new unique reagent in this study.
Experimental model and subject details
Cell culture and treatment
Human embryonic kidney 293T cells (American Type Culture Collection, CRL-3216), human cervical cancer HeLa cells (American Type Culture Collection, CCL-2) and mouse lung cancer LL2 cells (American Type Culture Collection, CRL-1642) were cultured in DMEM (Gibco, 11965-065) and human lung adenocarcinoma A549 cells (American Type Culture Collection, CCL-185), human colon adenocarcinoma HCT-8 cells (American Type Culture Collection, CCL-244) and mouse mammary gland epithelium 4T1 cells (American Type Culture Collection, CRL-2539) were cultured in RPMI 1640 Medium (Gibco, 22400-071), respectively. Culture medium was supplemented with 10% fetal bovine serum (Avantor, VW-89510-186) and 1% penicillin/streptomycin (Gibco, 15140-122). Human pulmonary alveolar epithelial cells (HPAEpiC, ScieCellTM, #3200) were cultured in alveolar epithelial cell medium (AEpiCM, #3201, ScieCellTM). All cells were maintained at 37°C in humid air with 5% CO2 condition and checked for mycoplasma contamination using EZ-PCR kit (Biological Industries, 20-700-20). For Endocytosis inhibition observation, cells were blocked by pretreatment of hydroxychloroquine (HCQ, Sigma-Aldrich, SI-H0915) or Dynasore (Sigma-Aldrich, D7693). E-coli expressed His-tagged wild-type SARS-CoV-2 or His-tagged Omicron N protein proteins were purchased from Sino Biological (wild-type: Cat No: 40588-V07E, Omicron: Cat No: 40588-V07E34). Mammalian cells expressed His-tagged SARS-CoV-2 wild-type or His-tagged Omicron N protein proteins were purchased from Acro Biosystem (WT Cat No: NUN-C5227, Omicron: Cat No: NUN-C52Ht).
Method details
Plasmid DNA and RNA
Wild-type (Wu-han strain) SARS-CoV-2 N proteins (Genbank: NC_045512.2), R203K/G204R, and Omicron N protein were synthesized by MISSION BIOTECH CO., LTD, Taiwan, and ligated to the pcDNA3.1 backbone using Gibson Assembly (NEB, E2611L). The sequence of other coronavirus nucleocapsid proteins including SARS-N (Genbank: AYV99827.1), MERS-N (Genbank: AHE78101.1), NL63-N (Genbank: AFV53152.1), 229E-N (Genbank: AGW80953.1), HKU1-N (Genbank: ARU07581.1), OC43-N (Genbank: YP_009555245.1) were constructed to the pcDNA3.1 backbone by Gibson Assembly. GFP-DNA used in this study, pmax-GFP plasmid was supplied by Lonza; in vitro transcribed (IVT) single strand mRNAs: SARS-CoV-2 RBD, SARS-CoV-2 Spike and Luciferase-P2A-GFP used in this study were provided by National Biotechnology Research Park (NBRP), Taipei, Taiwan. FAM-labeled siRNA, RNA#1 and RNA#12 RNA used in this study is purchased from MDBio, Inc. Taiwan and the sequence is 5′-UUCUCCGAACGUGUCACGUTT-3′.
Secretome and RANTES concentration measurement
48 hours after transfection of different SARS-CoV-2 viral proteins or different coronavirus N proteins, supernatant from overexpressed cells were collected. Cytokine and chemokine levels were detected using LEGENDplex™ Human Anti-Virus Response Panel (13-plex) with V-bottom Plate (BioLegend, Cat number, 740390) and LEGENDplex™ HU Proinflam. Chemokine Panel 1 (13-plex) w/FP (BioLegend, Cat: 740984) according to the manufacturer’s instructions. Briefly, 25 μl of supernatant was mixed with 25 μl Assay buffer and 25 μl mixed beads, then incubated with continuous shaking at room temperature for 2 hours. After incubation, the solutions were centrifuged at 300g for 5minutes at room temperature. The bound mixtures were then mixed with Streptavidin-conjugated with PE beads and incubated at room temperature for another 1 hour with continuous shaking. The final bound mixtures of cytokines or chemokines were washed by 200 μl wash buffer, resuspended in 150 μl wash buffer, and subjected to flow cytometry for secretome analysis. To measure RANTES concentration, supernatant from treated cells were collected after treatment. The Human RANTES Flex set (BD, Cat: 558324), which contains beads conjugated with anti-RANTES antibody and secondary detection antibody, was used in combination with Human Soluble Protein Master Buffer Kit (BD, Cat: 558264) to perform Cytometric Bead Array to measure RANTES concentration.
Cell staining for flow cytometry
Wash the cells pellet with PBS once and incubate with antibody in ice-cold FACS buffer (1x PBS, with 2% FBS) on ice for 30 minutes. Wash the cell pellet with ice-cold PBS once. The labeled cells were then resuspended in ice-cold FACS buffer and subjected to flow cytometry analysis (Attune, Thermo Fisher Scientific).
Generation of STEAP2 knockdown cells
The pLKO.1 plasmids encoding shRNA with sequences targeting human STEAP2 (TRCN0000297801: 5′-GCGCGACAACAGGTTATTGAA-3′) were purchased from the National C6 RNAi Core Facility (Taipei, Taiwan). The shRNA targeting E.coli LacZ beta-galactosidase (TRCN0000072224: 5′-CGCGATCGTAATCACCCGAGT-3′) was used as a control. For knockdown experiments, lentivirus expressing shRNA was first collected from the medium after 72 hours transfection in 293T cells. Next, HPAEpiC cells were infected with the collected lentiviruses for 24 hours and selected in the cultured medium containing puromycin (2 μg/ml, Thermo Fisher Scientific, A1113803) for 14 days. Knockdown efficiency of the STEAP2 was analyzed by qRT-PCR using the designed STEAP2 primers (forward: 5'- CCTCTGCTTACCGATGAGAAGG-3′ and reverse primers: 5'- CAGGAGGGAAAGTAAGCCAAGG -3′) with PowerUp™ SYBR™ Green Master Mix (Applied Biosystems™, A25741) and QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific).
Cell surface binding and blocking assay
Cells were detached by cell dissociation buffer (Gibco, 13151-014) and suspended in ice-cold cultured medium. Cells were transferred into 96-well plate (1×105 cells in 100 μl cultured medium/well). Cells were washed with ice-cold phosphate-buffered saline (PBS, Lonza, 17-516F) and resuspended in 100 μl of ice-cold FACS buffer (PBS supplemented with 2% FBS) containing different concentrations of His-tagged control proteins (crotonyl-CoA carboxylase/reductase (ccr) or crotonase (crt)) or coronavirus N proteins. Following a 1 hour on-ice incubation, cells were washed with ice-cold PBS and stained with anti-His-APC antibody (1:300 dilution; #362605, BioLegend) for 30 minutes on ice. After staining, cells were washed with ice-cold PBS, and resuspended in 200 μl of ice-cold FACS buffer for flow cytometry analysis by Attune NxT (Invitrogen). For evaluating RNA package effect on N protein surface binding, 10 μg N protein was pre-incubated with or without 1 μg RNA for 1 hour at 4°C followed by 1 hour on-ice cell binding.
For serum neutralizing assay, 30 μg of N protein was pre-incubated with 1 μl of mouse anti-N serum at 4°C overnight. For monoclonal antibody neutralizing assay, 10 μg of N protein was pre-incubated with 1, 3, 10, 30 or 100 μg of NP-mAb-40 (Lu et al., 2021) at 4°C overnight. Next, cells were incubated with the N protein/serum or N protein/antibody mixture for 1 hour cell surface binding. For screening functional anti-N antibodies that could block N assisted nucleic acid expression, 3 μg of mammalian cells-produced wild-type N or Omicron N recombinant protein were used to pre-bind with 9 μg different antibody at 4°C for overnight. After binding, 12 μg pmax-GFP DNA were added to antibody/N protein mixture at 4°C for 1 hour. These complexes were then added to 5x104 293T cells and GFP+ cells were counted after 48-hour incubation.
Isolation and digestion of N protein interacting membrane proteins
For isolation of cell membrane proteins, 1x107 cells were detached by Cell Dissociation Buffer, then washed with cultured medium and PBS once. Cells were resuspended in hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, and 1x protease inhibitor (Roche, 04693132001) mixture, pH 7.4) and incubated on ice for 15 minutes. Cells were homogenized with a Dounce homogenizer (Wheaton). The membrane fraction was purified by two-step centrifugation. First nuclei were pelleted by centrifugation at 1000g for 10 minutes at 4°C. The supernatant was mixed with 1.8 M sucrose to yield a final concentration of 0.25 M sucrose, and was then centrifuged for 1 hour at 13,000 rpm at 4°C. The pellet of remaining membrane proteins was dissolved in 90% (v/v) formic acid (FA). Bradford assay (Bio-Rad) was used to determine the membrane protein concentration. Membrane protein samples were further vacuum-dried for −80°C storage. For Crt (a control protein of bacterial origin) or N protein-beads preparation, 10 μg of His-tag Crt or N protein was mixed with 1 μl of anti-His antibody (Sigma-Aldrich, H1029) and 50 μl of protein G magnetic beads (GE Healthcare, #28951379) in 500 μl RIPA Lysis buffer (Millipore, #20-188), and rotated at 4°C overnight. Crt or N protein-beads were washed with RIPA Lysis buffer three times. In the meantime, freeze-dried membrane proteins were resuspended in 500 μl of 1% NP40 buffer (150 mM NaCl, 50 mM Tris-HCl, 1% NP40) containing protease inhibitor, and sonicated to clear. Resolved membrane proteins were mixed with prepared Crt or N protein-beads, and rotated for 3 hours at 4°C. Pull-downed beads were washed with 500 μl of RIPA Lysis buffer three times and stored at −80°C.
For DTSSP (3,3′-dithiobis (sulfosuccinimidyl propionate), #21578, Thermo Fisher Scientific) crosslinking, cells were detached by cell dissociation buffer and resuspended in ice-cold PBS containing 2% FBS at 1x107 cells/ml density. 100 μg of Crt or N protein was mixed with cell suspension and rotated at 4°C for 1 hour surface binding. Afterwards, cells were washed with ice-cold PBS twice and resuspended in 1.2 ml of fresh prepared DTSSP linker (3 mM in PBS), and rotated at 4°C for 1.5 hours. 133.3 μl of 10X TBS buffer (0.5 M Tris, 1.5 M NaCl, pH 7.6) was added to the cell solution and incubated for 15 minutes at 4°C while rotating to stop crosslinking. Cells were washed with ice-cold PBS and lysed by protease inhibitor containing RIPA Lysis buffer (Cell Signaling Technology, #9806). His-tagged Crt or N protein bound surface proteins were immunoprecipitated using anti-His antibody (H1029, Sigma-Aldrich) as described above. To elute pull-downed membrane proteins from the beads, magnetic beads were incubated with elution buffer (1% SDS, 20 mM Tris-HCl and 150 mM NaCl) for 20 minutes at 4°C. The eluent was collected and dried by SpeedVac. (Thermo Fisher Scientific).
For protein digestion, the protein pellet was resuspended in 0.1 M triethylammonium bicarbonate (TEABC) buffer containing 6 M urea, 5 mM EDTA, and 2% (w/v) SDS. Proteins were chemically reduced by adding 5 mM tris(2-carboxyethyl) phosphine hydrochloride (TCEP) at 37°C for 30 minutes, and alkylated by 2 mM methyl methanethiosulfonate (MMTS) at room temperature for 30 minutes. To incorporate proteins into a gel directly in the Eppendorf vial, acrylamide/bisacrylamide solution (40%, v/v, 29:1), 10% (w/v) ammonium persulfate (APS), and N,N,N′,N′-tetramethylethylenediamine (TEMED) was applied to the protein solution. The gel was cut into small pieces and washed with 25 mM TEABC and 25 mM TEABC containing 50% (v/v) ACN. The gel samples were further dehydrated with 100% acetonitrile (ACN) and then completely dried by SpeedVac. Proteolytic digestion was then performed using sequencing-grade modified trypsin (Promega) (protein:trypsin 10:1, g/g) in 25 mM TEABC at 37°C overnight. Peptides were extracted from the gel using sequential extraction with 25 mM TEABC, 0.1% (v/v) Trifluoroacetic acid (TFA) in water, 0.1% (v/v) TFA in 50% ACN, and 100% ACN. The solutions were dried by SpeedVac and resuspended in 0.1% (v/v) TFA for further desalting using ZipTipsTM (Millipore).
LC-MS-MS analysis
The LC-MS/MS analysis was performed following the method previously described 71. The mass spectrometry (MS) analysis was performed on a Thermo Scientific™ Orbitrap Fusion™ Tribrid™ mass spectrometer connected to an UltiMate™ 3000 RSLCnano System (Thermo Fisher Scientific, Bremen, Germany) equipped with a nanospray interface (Proxeon, Odense, Denmark). The tryptic peptides were loaded into the PepMap C18 50 cm × 75 μm ID column (Thermo Fisher Scientific) and were separated with a 90-minutes gradient at a flow rate of 250 nL/minutes. The mass spectra of survey MS1 scan (m/z 375–1500) were acquired in the Orbitrap mass analyzer at 120,000 resolution for a maximum injection time of 50 ms with an AGC target value of 4e5. MS2 scan was performed with a precursor isolation window of 1.4 m/z and dynamic exclusion time 20 s and fragmentation in HCD mode.
The protein identification was performed by processing of MS raw files in Proteome Discoverer (ver. 2.1) by SequestHT engine 72 against the SwissProt human protein database (version 2020.10). Furthermore, protein sequence FASTA from nucleoprotein (SwissProt: P0DTC9) and short-chain-enoyl-CoA hydratase (SwissProt: P52046) were added to the protein database. The spectra search criteria include 10 ppm mass tolerance for precursor, 0.02 Da for product ions, and trypsin enzyme specificity with up to 2 missed cleavages. All search engines set static methylthiolation (+45.988 Da) on Cys residues, as well as dynamic oxidation (+15.995 Da) on Met residues and acetylation on protein N terminus (+42.016 Da). The minimal peptide length was set as 6 residues. The false discovery rate (FDR) of identification was set at 1% at peptide and protein levels. The normalization and t-test for quantitative comparison were performed by Perseus (ver. 1.6.14.0).
GFP DNA expression and siRNA delivery by N protein
293T were seeded onto Millicell® EZ slides (Merck Millipore, C86024) (50,000 cells/well) in 200 μl medium. For GFP-DNA delivery, 1 μg N protein was mixed with 4 μg GFP-DNA (pmax-GFP, Lonza) for 1 hour at 4°C at indicated ratios. For examination of the effect of cytokines and chemokines to N protein mediated GFP-DNA expression, we pretreat cells with several cytokines or chemokines including 10μg/ml interleukin-28 alpha (IL-28A) (BioLegend, Cat:754202), 10μg/ml RANTES (BioLegend, Cat:580202), 10μg/ml IP-10 (BioLegend, Cat:573502) or 10 μg/ml interleukin-29 (IL-29) (BioLegend, Cat:711204) overnight. To test whether nutrients affect the efficiency of N protein mediated GFP-DNA expression, we pretreat cells with 10, 25 or 50 mM sodium DL-lactate (Sigma-Aldrich, Cat: L4263), 25 mM L-glutamine (Gibco, Cat: 25030-081) and 25 mM pyruvic acid (Sigma-Aldrich, Cat: 107360). The N protein and GFP-DNA mixture was gently pipetted to cell condition medium. After 24 hours, the expression of GFP was assessed using Invitrogen™ EVOS™ FL Auto Imaging System (Thermo Fisher Scientific).
For siRNA delivery, 10 μg N protein was mixed with 100 μl of 10 μM JUN siRNA (Dharmacon, Cat: L-003268-00-0005), which contains a mixture of four siRNAs, (sequences: 5′-GAGCGGACCUUAUGGCUAC-3′; 5′-GAACAGGUGGCACAGCUUA-3′; 5′-GAAACGACCUUCUAUGACG-3′ and 5′- UGAAAGCUCAGAACUCGGA-3′) or 10 μM siIFNAR1 (Dharmacon, Cat: L-020209-00-0005), which contain a mixture of four different siRNAs (sequences: 5′- GCGAAAGUCUUCUUGAGAU-3′; 5′- UGAAACCACUGACUGUAUA-3′; 5′- GAAAAUUGGUG-UCUAUAGU-3′ and 5′- GAAGAUAAGGCAAUAGUGA-3′). The mixture was incubated for 1 hour at 4°C, then directly added into cultured medium and gently pipetted up and down. 48 hours after treatment, the RNA was extracted (Qiagen, Cat: 74106) and the gene expression was detected by qRT-PCR.
Immunofluorescent staining
Cells were seeded onto Millicell® EZ slides. After treatment, cells were washed by Phosphate-Buffered Saline (PBS) gently two times, then fixed in fixation buffer (BD Biosciences, Cat.555028) for 10 minutes, and then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, T8787-100ML) in PBS for 5 minutes. Nonspecific binding sites on the cells were blocked with blocking buffer (1% BSA in PBS) for 1 hour. The expression of N protein was detected by anti-N antibody (Invitrogen, Ma1-7404). After incubation with primary antibody, the cells were then gently washed with PBS twice. Slides were incubated with goat anti-Mouse antibody conjugated with Alexa Fluor 488 (1:250; Invitrogen, A32723) for 1 hour. The nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI) (1:1000) (Invitrogen, D1306). Cells were observed under confocal microscopy (TCS SP5; Leica,). To detect Na+/K+ ATPase (as a membrane marker), cells were stained with anti-Na+/K+ ATPase monoclonal antibody (Abcam, EP1845Y) (1:200) for 90 minutes. After incubation, cells were washed by PBS and treated with secondary antibodies – goat anti-rabbit-alexa 568 nm (1:400; Biotium, 20102) -- for 1 hour. The localization of SARS2-N protein, and cell membrane were observed by confocal microscopy. For detection of co-localization of N protein, endosome, and STEAP2, cells were seeded onto Millicell® EZ slides. After treatment, cells were washed by Phosphate-Buffered Saline (PBS) gently two times, then fixed in fixation buffer (BD Biosciences, Cat.555028) for 10 min, and then permeabilized with 0.1% Triton X-100(Sigma-Aldrich, T8787-100ML) in PBS for 5 min. Nonspecific binding sites on the cells were blocked with blocking buffer (1% BSA in PBS) for 1 hour. The expression of N protein was detected by anti-N antibody (Invitrogen, Ma1-7404). The expression of endosome was detected by anti-EEA1 antibody (Genetex GTX82138). The expression of STEAP2 was detected by anti-STEAP2 antibody (Novus NBP-76823). After incubation with a primary antibody, the cells were then gently washed with PBS twice. Slides were incubated with goat anti-chicken IgY (FITC) (GTX27114), goat anti-mouse CF®568 (Biotium,20100), and goat anti-rabbit CF®640 (Biotium,20202), for 1 hour. Cells were observed under fluorescent microscopy (EVOS imaging systems).
Western blot analysis
Cells were harvested and then lysed on ice using RIPA Lysis buffer (Cell Signaling Technology, #9806) containing a protease inhibitor (Roche, No.4693132001) and a phosphates inhibitor (Roche, No.4906837001). For SDS-PAGE, cell lysates containing 20 μg of total cellular protein for each sample were denatured by treating with 1X Laemmli sample buffer (Bio-Rad Laboratories, Inc) containing 2-Mercaptoehtanol (Bio-Rad Laboratories, Inc). The reduced protein samples were run on a Mini-Proten TGX (Bio-Rad Laboratories, 4561086) at 200 V for 25 minutes in 1X TGS buffer (Omics Bio, Cat. No IB3372). The proteins were transferred to a Immobilon®-E PVDF Membrane (Millipore, IEVH85R), at 120 V for 80 minutes in 1X Western Transfer buffer (Omics Bio, Cat. No IB3352) with 20% Methanol. The blot was then blocked by incubation in 5% BSA TBST for 1 hour. The blot was then incubated with anti-p38 antibody (1:1000, Cell Signaling Technology, #9212) overnight. After incubation with primary antibody, the blot was washed by TBST and incubated with secondary antibody (1:3000, goat anti-rabbit conjugated HRP, Cell Signaling Technology, #7074). The blot was stripped in Restore western blot stripping buffer (Thermo Fisher Scientific, Cat. No 21059), according to the manufacturer’s instructions, and re-probed by using anti-total p38 antibody (Cell Signaling Technology, #9212) and β-actin antibody (Cell Signaling Technology, #4967).
Lattice light-sheet microscopy
Lattice light sheet microscope configuration: The laser output is controlled by the acousto optical turntable filter under the HHMI’s SPIM system management. The selected laser beam was transformed to multiple coherent Bessel beams (Lattice light sheet) via spatial light modulator (SLM) and polarization beam splitter. The customized mask can filter other diffraction from SLM to decrease the background of the Lattice light sheet in the Z direction. The motive of X and Z-direction of Lattice light sheet can be manipulated by the X and Z-galvo mirrors set after the customized mask. The controlled Lattice light sheet illuminated the fluorescent stained sample through the 32.8 degree. The plane of emission fluorescent signals was collected by the orthogonal detection objective. The collected signals were imaged onto the chip of the sCOMS camera.
Image acquisition: For the entry processing of N protein and RNA observation, RNA labeled with FAM fluorophore (Ex = 488 nm, Em = 500∼550 nm) and nucleus of target cells, 5x105 HPAEpiC cells which were pre-seeded on 6 wells the day before analysis were labeled with Hoechst 33342 (Ex = 405 nm, Em = 415∼485 nm). 10 μg SARS-CoV-2 N protein was pre-incubated with 40 μg RNA-FAM at 4 degrees for 1 hour. At the same time, the diluted factor 1:30000 Hoechst 33342 was also incubated with HPAEpiC cells at 37°C for 30 minutes. After the formation of the N-protein/RNA-FAM mixture, the complex was added to targeting cells and washed one time by 1x PBS to remove unbound signal and then the sample is subjected to lattice light-sheet microscopy for analysis. The setting of the Lattice light sheet adjusted to the length of 26 μm which is set in the Y-axis direction to cover the entire cell. And 405 nm and 488 nm Lattice patterns were generated from the HHMI Matlab QXGA code. The size of the illuminated plane is a 26 μm width and 145 μm long rectangle. And the region of interesting needs to cover by 0.5 μm interval step 161 layers. According to the intensity and photobleaching level of FAM, the imaging strategy used the 50 ms exposure time with each channel that 2 colors and three-dimensional (3D) images spend 16.1 seconds. In order to record the details of the entry process, the 3D localization of each signal is captured every minute until 2 hours.
Image processing: Raw 3D tiff image is 256 multiply 1408 pixels per plane and sample scanning directional interval 0.5 μm 161 layers. Total 121-time points 3D tiff files processed through the DeconvCuda software (HHMI) to deskew and deconvolute the Raw to transform the normal bottom to top-view pictures. The region of 3D image captured Width = 102.164 μm, High = 145.024 μm, and Depth = 14.283 μm. And then transformed images demonstrated by Amira software to present the relationship between time and signal locations.
Co-culture
5x105 293T cells were seeded overnight in a 6-well plate, and 2 × 105 A549 cells were seeded in a 12-well plate. 293T cells used as donor cells were co-transfected with 5 μg of GFP and 5 μg of pcDNA3.1-SARS-CoV-2 N or pcDNA3.1 empty vector. Simultaneously, A549 cells were pretreated with or without 10 μg/ml RANTES or 50 mM lactate or 25 μM SB203580 (#S1076, Selleckchem) for 24 hours. 24 hours after transfection and pretreatment, transfected 293T cells were resuspended and washed with 10 ml of 1X PBS once. Cultured medium in the plate containing A549 cells was removed, and 1x106 293T transfected cells resuspended in 1 ml fresh cultured medium containing with or without 10 μg/ml RANTES or 50 mM lactate or 25 μM SB203580 or 10 μg/ml of normal mouse IgG (#sc-2025, Santa Cruz) or 10 μg/ml of NP-mAb-4032 or NP-mAb-5632 were seeded into A549 plate. After 24 hours coculture, cells were trypsinized and washed with 1X PBS. 1x106 collected cell pool was further fixed with ice-cold 70% ethanol for 15 minutes at 4°C. Fixed cells were washed with 1X PBS once. Cells were stained with mouse anti-cytokeratin 18 antibody (1:1000 dilution, ab668, Abcam) and rabbit anti-SV40 large T antibody (1:500 dilution, ab234426, abcam) in 200 μl of FACS buffer (1X PBS containing 2% FBS) for 30 minutes at room temperature. Cells were washed with 1X PBS once. Cells were stained with goat anti-mouse IgG-647 (1:1000 dilution, #20281, Biotium) and goat anti-rabbit IgG-555 (1:1000 dilution, #20033, Biotium) in 200 μl FACS buffer for 30 minutes at room temperature. Cells were washed with 1X PBS and resuspended in 200 μl FACS buffer for FACS analysis. A549 cells were gated from the population with cytokeratin 18 positive and SV40 large T antigen negative.
For transwell coculture, 5x105 293T cells were seeded overnight in a 6-well plate. 293T cells used as donor cells were co-transfected with 5 μg of GFP and 5 μg of pcDNA3.1-SARS-CoV-2 N or pcDNA3.1 empty vector. 2 × 105 A549 cells were seeded in a 12-well plate (lower chamber). After 24 hours transfection, transfected 293T cells were washed with fresh cultured medium once. Then, 1x105 transfected 293T cells were plated inside 12-well transwell inserts of 1-μm pore size (upper chamber, #MCRP12H48, Millipore). After 3 days coculture, GFP positive cells in the bottom wells were counted.
SARS-CoV-2 N protein ELISA
For detecting the existence of SARS-CoV-2 N protein during co-culture, supernatant from ectopically expressed 293T cells were collected after transfection 48 hours and level of N protein is measured by SARS-CoV-2 Nucleocapsid Protein ELISA kit (LeadGene, Cat: 71300) following the instruction. In brief, 100 μl of supernatant were added to the 96 well ELISA plate which was pre-coated with anti-SARS-CoV-2 NP monoclonal antibodies. Then, add 100 μl 1x working HRP conjugated anti-SARS-CoV-2 N protein monoclonal antibody solution for 1 hour at room temperature. After incubation, wash six times with 300 μl wash buffer, and add 100 μl TMB (tetramethylbenzidine) substrate until the wells show obvious color. Finally, Stop the reaction by 2M H2SO4 and measure the signal by SYNERGY/LX multi-mode reader (BioTek).
Cell viability assay
The viability of cells was examined by colorimetric Cell Counting Kit-8 (CCK8) CCK8 assay kits (BIOTOOLS, Cat: TEN-cck8) according to the manual instruction. In brief, 50,000 cells per well were seeded on 96-well plate overnight. Lactate was added into the cell medium at the indicated concentration, and then the cells were treated with N protein + GFP DNA. After treatment, the GFP positive cells were counted. After cell counting, 10 μl/well (96 well plate) of CCK-8 solution were added into the cells and incubate for 1 hour at 37°C. Then the absorbance was measured at 450 nm.
Quantification and statistical analysis
All statistics were performed using two-tailed t-tests. P<0.05 was considered significant. The data shown in this study uses mean ± SEM from at least three biological replicates.
Additional resources
There is no additional resources in this paper.
Acknowledgments
We thank the National C6 RNAi Core Facility at Academia Sinica for establishing the STEAP2 knockout cell clones. We thank Mass Spectrometry Core Facility of the Biomedical Translation Research Center at Academia Sinica for providing LC-MS/MS service to identify the membrane receptor interacting with N protein. We thank the Biomedical Translation Research Center for providing the in vitro transcribed single-strand mRNA of SARS-CoV-2 RBD, SARS-CoV-2 Spike, and Luciferase-P2A-GFP. We thank our former group member, Mr. Hao-Nien Chen, for assistance in plasmid construction. We thank Hsiu-Ting Lin and Wan-Yu Chen for their assistance in monoclonal antibodies production. We thank Li-Yu Chen for the assistance in the analysis of pull-downed protein candidates.
Author contributions
JLW, IIK, and JYG designed and performed the experiments, analyzed and interpreted the data, and participated in writing the article. WCH. constructed the plasmids, performed the experiments, analyzed and interpreted the data, and participated in writing the article. BCC designed and WCT performed the experiments for lattice light sheet microscope. YJC designed and HJC performed the experiments for LC-MS/MS. HCW supervised monoclonal antibody generation and provided suggestions in writing the article. JCL designed the overall research strategy and experiments, and supervised research, and wrote and edited the article.
Declaration of interests
The authors declare no competing interests.
Published: February 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.105995.
Supplemental information
Data and code availability
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/z6bgky278p.1.
References
- 1.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29:1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Q., Lu S., Corbett K.D. Structural basis for SARS-CoV-2 nucleocapsid protein recognition by single-domain antibodies. Front. Immunol. 2021;12:719037. doi: 10.3389/fimmu.2021.719037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan P., Shen M., Yu Z., Ge W., Chen K., Tian M., Xiao F., Wang Z., Wang J., Jia Y., et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021;12:4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su C.M., Wang L., Yoo D. Activation of NF-kappaB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 2021;11:13464. doi: 10.1038/s41598-021-92941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Sui L., Wu P., Wang W., Wang Z., Yu Y., Hou Z., Tan G., Liu Q., Wang G. A dual-role of SARS-CoV-2 nucleocapsid protein in regulating innate immune response. Signal Transduct. Target. Ther. 2021;6:331. doi: 10.1038/s41392-021-00742-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W., Chen J., Hu D., Pan P., Liang L., Wu W., Tang Y., Huang X.R., Yu X., Wu J., Lan H.Y. SARS-CoV-2 N protein induces acute kidney injury via Smad3-dependent G1 cell cycle arrest mechanism. Adv. Sci. 2022;9:e2103248. doi: 10.1002/advs.202103248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015;89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y.H., Li J., Liu X.E., Wang L., Li T., Zhou Y.H., Zhuang H. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J. Virol. Methods. 2005;130:45–50. doi: 10.1016/j.jviromet.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y., Ma L., Cai S., Zhuang Z., Zhao Z., Jin S., Xie W., Zhou L., Zhang L., Zhao J., Cui J. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-kappaB hyper-activation and inflammation. Signal Transduct. Target. Ther. 2021;6:167. doi: 10.1038/s41392-021-00575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., 3rd, Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo L., Li Z., Zhao T., Ju X., Ma P., Jin B., Zhou Y., He S., Huang J., Xu X., et al. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci. Bull. 2021;66:1194–1204. doi: 10.1016/j.scib.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., Jin L., Deng F., Qiu Y., Zhou X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian Y., Lei T., Patel P.S., Lee C.H., Monaghan-Nichols P., Xin H.B., Qiu J., Fu M. Direct activation of endothelial cells by SARS-CoV-2 nucleocapsid protein is blocked by simvastatin. J. Virol. 2021;95:e0139621. doi: 10.1128/JVI.01396-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond M.S., Kanneganti T.D. Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol. 2022;23:165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Chen S., Jin Y., Ji W., Zhang W., Duan G. An update on innate immune responses during SARS-CoV-2 infection. Viruses. 2021;13:2060. doi: 10.3390/v13102060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karwaciak I., Salkowska A., Karas K., Dastych J., Ratajewski M. Nucleocapsid and spike proteins of the coronavirus SARS-CoV-2 induce IL6 in monocytes and macrophages-potential implications for cytokine storm syndrome. Vaccines. 2021;9:54. doi: 10.3390/vaccines9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu R.J., Yu W.C., Peng G.R., Ye C.H., Hu S., Chong P.C.T., Yap K.Y., Lee J.Y.C., Lin W.C., Yu S.H. The role of cytokines and chemokines in severe acute respiratory syndrome coronavirus 2 infections. Front. Immunol. 2022;13:832394. doi: 10.3389/fimmu.2022.832394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 27.Yang H.C., Chen C.H., Wang J.H., Liao H.C., Yang C.T., Chen C.W., Lin Y.C., Kao C.H., Lu M.Y.J., Liao J.C. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. Proc. Natl. Acad. Sci. USA. 2020;117:30679–30686. doi: 10.1073/pnas.2007840117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Wang J., Liu C., Su L., Zhang D., Fan J., Yang Y., Xiao M., Xie J., Xu Y., et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020;26:97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuda Y., Homma T., Inoue H., Goto Y., Sato Y., Ikeda H., Onitsuka C., Sato H., Akimoto K., Ebato T., et al. Serum IL-28A/IFN-lambda2 is linked to disease severity of COVID-19. Sci. Rep. 2022;12:5458. doi: 10.1038/s41598-022-09544-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson B.K., Seethamraju H., Dhody K., Corley M.J., Kazempour K., Lalezari J., Pang A.P.S., Sugai C., Mahyari E., Francisco E.B., et al. CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14. Int. J. Infect. Dis. 2021;103:25–32. doi: 10.1016/j.ijid.2020.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Xu C., Agudelo Higuita N.I., Bhattacharya R., Chakrabarty J.H., Mukherjee P. Evaluation of I-TAC as a potential early plasma marker to differentiate between critical and non-critical COVID-19. Cell Stress. 2022;6:6–16. doi: 10.15698/cst2022.01.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu R.M., Ko S.H., Chen W.Y., Chang Y.L., Lin H.T., Wu H.C. Monoclonal antibodies against nucleocapsid protein of SARS-CoV-2 variants for detection of COVID-19. Int. J. Mol. Sci. 2021;22:12412. doi: 10.3390/ijms222212412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y.J., Roumeliotis T.I., Chang Y.H., Chen C.T., Han C.L., Lin M.H., Chen H.W., Chang G.C., Chang Y.L., Wu C.T., et al. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–244.e17. doi: 10.1016/j.cell.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 35.Dimayacyac-Esleta B.R.T., Tsai C.F., Kitata R.B., Lin P.Y., Choong W.K., Lin T.D., Wang Y.T., Weng S.H., Yang P.C., Arco S.D., et al. Rapid high-pH reverse phase StageTip for sensitive small-scale membrane proteomic profiling. Anal. Chem. 2015;87:12016–12023. doi: 10.1021/acs.analchem.5b03639. [DOI] [PubMed] [Google Scholar]
- 36.Hasegawa H., Li C., Alba B.M., Penny D.M., Xia Z., Dayao M.R., Li P., Zhang J., Zhou J., Lim D., et al. Membrane cholesterol modulates STEAP2 conformation during dynamic intracellular trafficking processes leading to broad subcellular distribution. Exp. Cell Res. 2018;370:208–226. doi: 10.1016/j.yexcr.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 37.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a Biophysical Basis for its dual functions. Mol. Cell. 2020;80:1092–1103.e4. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savastano A., Ibáñez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henry B.M., Aggarwal G., Wong J., Benoit S., Vikse J., Plebani M., Lippi G. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: a pooled analysis. Am. J. Emerg. Med. 2020;38:1722–1726. doi: 10.1016/j.ajem.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kujime K., Hashimoto S., Gon Y., Shimizu K., Horie T. p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 2000;164:3222–3228. doi: 10.4049/jimmunol.164.6.3222. [DOI] [PubMed] [Google Scholar]
- 41.Bai Z., Cao Y., Liu W., Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13:1115. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T., Wang L., Wang H., Li X., Zhang S., Xu Y., Wei W. Serum SARS-COV-2 nucleocapsid protein: a sensitivity and specificity early diagnostic marker for SARS-COV-2 infection. Front. Cell. Infect. Microbiol. 2020;10:470. doi: 10.3389/fcimb.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Ong C.M., Yun C., Mo W., Whitman J.D., Lynch K.L., Wu A.H.B. Diagnostic value of nucleocapsid protein in blood for SARS-CoV-2 infection. Clin. Chem. 2021;68:240–248. doi: 10.1093/clinchem/hvab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crawford A., Angelosanto J.M., Nadwodny K.L., Blackburn S.D., Wherry E.J. A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog. 2011;7:e1002098. doi: 10.1371/journal.ppat.1002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikolajczyk T.P., Nosalski R., Szczepaniak P., Budzyn K., Osmenda G., Skiba D., Sagan A., Wu J., Vinh A., Marvar P.J., et al. Role of chemokine RANTES in the regulation of perivascular inflammation, T-cell accumulation, and vascular dysfunction in hypertension. FASEB J. 2016;30:1987–1999. doi: 10.1096/fj.201500088R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patterson B.K., Seethamraju H., Dhody K., Corley M.J., Kazempour K., Lalezari J.P., Pang A.P., Sugai C., Francisco E.B., Pise A., et al. Disruption of the CCL5/RANTES-CCR5 pathway restores immune homeostasis and reduces plasma viral load in critical COVID-19. medRxiv. 2020 doi: 10.1101/2020.05.02.20084673. Preprint at. [DOI] [Google Scholar]
- 47.Nechipurenko Y.D., Semyonov D.A., Lavrinenko I.A., Lagutkin D.A., Generalov E.A., Zaitceva A.Y., Matveeva O.V., Yegorov Y.E. The role of acidosis in the pathogenesis of severe forms of COVID-19. Biology. 2021;10:852. doi: 10.3390/biology10090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schall T.J., Bacon K., Toy K.J., Goeddel D.V. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347:669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- 49.Loetscher M., Gerber B., Loetscher P., Jones S.A., Piali L., Clark-Lewis I., Baggiolini M., Moser B. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J. Exp. Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catalfamo M., Karpova T., McNally J., Costes S.V., Lockett S.J., Bos E., Peters P.J., Henkart P.A. Human CD8+ T cells store RANTES in a unique secretory compartment and release it rapidly after TcR stimulation. Immunity. 2004;20:219–230. doi: 10.1016/s1074-7613(04)00027-5. [DOI] [PubMed] [Google Scholar]
- 51.Korkmaz K.S., Elbi C., Korkmaz C.G., Loda M., Hager G.L., Saatcioglu F. Molecular cloning and characterization of STAMP1, a highly prostate-specific six transmembrane protein that is overexpressed in prostate cancer. J. Biol. Chem. 2002;277:36689–36696. doi: 10.1074/jbc.M202414200. [DOI] [PubMed] [Google Scholar]
- 52.Gomes I.M., Maia C.J., Santos C.R. STEAP proteins: from structure to applications in cancer therapy. Mol. Cancer Res. 2012;10:573–587. doi: 10.1158/1541-7786.MCR-11-0281. [DOI] [PubMed] [Google Scholar]
- 53.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 57.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 59.Mothes W., Sherer N.M., Jin J., Zhong P. Virus cell-to-cell transmission. J. Virol. 2010;84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong P., Agosto L.M., Munro J.B., Mothes W. Cell-to-cell transmission of viruses. Curr. Opin. Virol. 2013;3:44–50. doi: 10.1016/j.coviro.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng C., Evans J.P., King T., Zheng Y.M., Oltz E.M., Whelan S.P.J., Saif L.J., Peeples M.E., Liu S.L. SARS-CoV-2 spreads through cell-to-cell transmission. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2111400119. e2111400119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lutomski C.A., El-Baba T.J., Bolla J.R., Robinson C.V. Multiple roles of SARS-CoV-2 N protein facilitated by Proteoform-specific interactions with RNA, host proteins, and Convalescent antibodies. JACS Au. 2021;1:1147–1157. doi: 10.1021/jacsau.1c00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maruoka S., Hashimoto S., Gon Y., Takeshita I., Horie T. PAF-induced RANTES production by human airway smooth muscle cells requires both p38 MAP kinase and Erk. Am. J. Respir. Crit. Care Med. 2000;161:922–929. doi: 10.1164/ajrccm.161.3.9906059. [DOI] [PubMed] [Google Scholar]
- 64.Gupta G.S. The lactate and the lactate dehydrogenase in inflammatory diseases and major risk factors in COVID-19 patients. Inflammation. 2022;45:2091–2123. doi: 10.1007/s10753-022-01680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y., Zhang H., Mu S., Wei W., Jin C., Tong C., Song Z., Zha Y., Xue Y., Gu G. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: a retrospective and observational study. Aging (Albany NY) 2020;12:11245–11258. doi: 10.18632/aging.103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carpenè G., Onorato D., Nocini R., Fortunato G., Rizk J.G., Henry B.M., Lippi G. Blood lactate concentration in COVID-19: a systematic literature review. Clin. Chem. Lab. Med. 2022;60:332–337. doi: 10.1515/cclm-2021-1115. [DOI] [PubMed] [Google Scholar]
- 67.Bruno R.R., Wernly B., Flaatten H., Fjølner J., Artigas A., Bollen Pinto B., Schefold J.C., Binnebössel S., Baldia P.H., Kelm M., et al. Lactate is associated with mortality in very old intensive care patients suffering from COVID-19: results from an international observational study of 2860 patients. Ann. Intensive Care. 2021;11:128. doi: 10.1186/s13613-021-00911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin S., Sun L., Lyu X., Ai X., Du D., Su N., Li H., Zhang L., Yu J., Yuan S. Lactate-activated macrophages induced aerobic glycolysis and epithelial-mesenchymal transition in breast cancer by regulation of CCL5-CCR5 axis: a positive metabolic feedback loop. Oncotarget. 2017;8:110426–110443. doi: 10.18632/oncotarget.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaburagi Y., Shimada Y., Nagaoka T., Hasegawa M., Takehara K., Sato S. Enhanced production of CC-chemokines (RANTES, MCP-1, MIP-1alpha, MIP-1beta, and eotaxin) in patients with atopic dermatitis. Arch. Dermatol. Res. 2001;293:350–355. doi: 10.1007/s004030100230. [DOI] [PubMed] [Google Scholar]
- 70.Luster A.D. Chemokines--chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L., Richards A., Barrasa M.I., Hughes S.H., Young R.A., Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2105968118. e2105968118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.López-Muñoz A.D., Kosik I., Holly J., Yewdell J.W. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Sci. Adv. 2022;8:eabp9770. doi: 10.1126/sciadv.abp9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video about the observation of SARS-CoV-2 N protein carries RNA-FAM entering cells is attached in the supplementary item. Detailed materials or reagents used in this study are listed in key resources table.
Data Availability Statement
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/z6bgky278p.1.