Abstract
Since the emergence of the pandemic COVID19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the development of vaccines has been the prime strategy to control the disease transmission. Most of the developed vaccines target the spike protein, however, the emerging variants have alterations, particularly at the same region which may pose resistance to neutralizing antibodies. In this study, we explored the variable and conserved regions of SARS-CoV-2 as a potential inclusion in a multiple-target vaccine with the exploitation of Salmonella-based vector for oral mRNA vaccine against Delta and Omicron variants. Increased IgG and IgA levels imply the induction of humoral response and the CD4+, CD8+ and IFN-γ+ sub-population level exhibits cell-mediated immune responses. The degree of CD44+ cells indicates the induction of memory cells corresponding to long-term immune responses. Furthermore, we assessed the protective efficacy of the vaccines against the Delta and Omicron variants in the hamster model. The vaccine constructs induced neutralizing antibodies and protected the viral-challenged hamsters with significant decrease in lung viral load and reduced histopathological lesions. These results reinforce the use of the conserved and variable regions as potential antigen targets of SARS-CoV-2 as well as the exploitation of bacteria-mediated delivery for oral mRNA vaccine development.
Keywords: SARS-CoV-2, Salmonella, Oral, Vaccine, mRNA, SARS-CoV-2 variants
The high mortality cases caused by the sudden emergence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019 (COVID-19) pandemic, has imposed a heavy burden on the global public health and economy. Thus, the development of effective vaccines and therapeutics are the key strategies to combat the spread of the disease. The rapid development of vaccines against COVID-19 has been remarkable and is currently being used worldwide following the emergency use approval from health authorities including World Health Organization (WHO).
Importantly, the Delta and Omicron variants have been characterized by higher viral loads and infectivity compared to the ancestral SARS-CoV-2 which increases the risk of infection and likely contributes to increased transmissibility [1,2]. The Delta variant, first detected in late 2020 in India, is found to be more transmissible than its ancestor and confers modest loss of susceptibility to neutralizing antibodies due to the alterations in the spike receptor-binding motif [[3], [4], [5]] and is less sensitive to vaccine-elicited serum neutralizing antibodies than the parental strain [3,6]. Subsequently, in late 2021, the WHO identified the emergence of the Omicron variant, currently a variant of concern (VOC), which possesses 26 to 32 mutations in the spike protein, some of which are found to be associated with its increased immune escape capability and receptor binding [[7], [8], [9]] which likely contribute to its phenotypic characteristics of increased transmissibility and risk of infection, but reduced disease severity and hospitalization compared to the previous variants [10]. A recent study has also shown markedly reduced serum antibody titres against the Omicron variant compared to ancestral virus weeks after two doses of BNT152b2 or CoronaVac [11].
The emergence of such SARS-CoV-2 variants with various mutations that may escape the protective immunity provided by the current vaccines imposes another challenge and continuously raises public health concern, especially on future vaccine efficacy. Hence, there is an urgent need in exploring alternative vaccine strategies and evaluation of different vaccines against these variants, making research on the development of next-generation vaccines for broad protective immunity highly valuable. Various types of vaccine delivery of SARS-CoV-2 antigens that induce immunity and protection efficacy against the disease are continuously being developed and evaluated. Some of the vaccine strategies include inactivated viral vaccines, live-attenuated vaccines, virus-vectored vaccines, protein subunit vaccines, virus-like particle vaccines and DNA and mRNA vaccines [12]. In this study, we specifically exploited the advantages of Salmonella-based vaccine with Semliki Forest virus (SFV) replicon which we have previously reported to efficiently amplify the target mRNA and deliver the antigens to the target tissues [13,14].
We previously studied the combination of different targets by designing a multiple-target vaccine consisting of the receptor-binding domain (RBD), heptad repeat domain (HR), membrane protein (M) and epitopes of nsp14 of SARS-CoV-2 (V-P2A) using Salmonella delivery system which induced immune response as well as protection efficacy in animal models [13,15,16]. Thus, to further explore the different SARS-CoV-2 proteins for potential inclusion in a multiple-target vaccine, we have designed and compared two Salmonella-enabled oral mRNA vaccines targeting the variable and conserved regions of SARS-CoV-2, particularly the RBD, N-terminal domain (NTD), Heptad Repeat (HR), Nucleocapsid (N) protein and selected epitopes of nsp12 (RdRp) in their ability to induce an immune response and protection in the hamster animal model against the Delta and Omicron variants.
1. Materials and methods
1.1. Experimental animals and ethics statement
Five-week-old specific pathogen-free (SPF) female BALB/c mice and female golden Syrian hamsters used in the study were provided with water and feed and maintained in the animal housing facility. Animal experiments were approved by the Jeonbuk National University Animal Ethics Committee (JBNU 2021-027). All experiments requiring the handling of live SARS-CoV-2 were performed in Biosafety Level-3 (BSL3) laboratory of the Korea Zoonosis Research Institute in South Korea.
1.2. Cells and viruses
Vero E6 and RAW 264.7 cells were cultured and maintained at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Lonza) with 10% FBS (Gibco) and 1% pen-strep (Lonza). The SARS-CoV-2 clinical isolates, hCoV-19/Korea/KDCA119861/2021 (B.1.617.2 lineage, Delta variant, Clade G) and hCoV-19/Korea/KDCA447321/2021 (B.1.1.529 lineage, Omicron variant, Clade GR) obtained from the National Culture Collection for Pathogens (NCCP) in the Korea Disease Control and Prevention Agency (KDCA) were propagated in Vero E6 cells. The titrated viral stocks were stored at −80 °C until further use.
1.3. Bacterial strains, plasmids, and primers
The strains of bacteria, plasmids, and primers used in this study are summarized in Tables S1 and S2. The bacteria were grown in Luria–Bertani broth (BD) with agitation at 37 °C with the appropriate antibiotics.
1.4. Vaccine design and construction
We designed two vaccine constructs each comprising either conserved or variable genes. The gene sequence of RBD and NTD were chosen for the variable construct and HR, N and RdRp were selected for the conserved construct. The selected sequences were amplified from the cDNA sample prepared using B.1.617.2 Delta variant. For a multicistronic expression, each gene was connected by viral self-cleaving peptide p2A by an overlap extension PCR [17]. These constructs were cloned into pJHL204, an SFV replicon-based vector [13,15]. The constructs encoding the correct DNA sequence information was verified by sequencing. A live-attenuated ST with the genotype Δlon Δcpxr ΔsifA and Δasd designated as JOL2500 was used for gene delivery. The vaccine strains were prepared by electroporation of JOL2500 with pJHL204 encoding either conserved or variable vaccine constructs. The 3D structure of the vaccine constructs was modelled using Phyre2 [18] and then subjected to Ramachandran plot analysis using PROCHECK server for measuring quality and validation [19]. The protein structures were visualised with EzMol 2.1 [20]. Additionally, B-cell and T-cell epitope prediction was done using Bepipred Linear Epitope Prediction 2.0 [21] and NetCTL 1.2 server [22] respectively.
1.5. Confirmation of the vaccine constructs expression
RAW264.7, murine macrophage cells, in 12-well plates were infected with JOL2818 (pJHL204-HR-N-RdRp; conserved), JOL2819 (pJHL204-NTD-RBD; variable) at 25 MOI for 3 h. After infection, the wells were incubated for 1 h with 100 μg/ml gentamycin-containing media to kill the external bacteria and then further incubated for 48 h. The expression of the vaccine construct was confirmed. Briefly, the cDNA was synthesized from extracted RNA and the specific construct primers were added in the PCR reaction and subjected to thermal cycling. The products were confirmed by visualization of the full-length vaccine constructs in agarose gel.
To further confirm by western blotting, 150 μl of RIPA lysis buffer was added to the cells and kept on ice for 15 min. The cell lysates were subjected to sonication and the collected supernatants were mixed with equal volume of 2× SDS sample buffer and run on a 12% and 15% gel. The samples were transferred to a nitrocellulose membrane and blocked with 5% skim milk at room temperature for 2 h. 1:500 diluted primary antibodies raised in rabbits were added and incubated at 4 °C overnight. The anti-rabbit IgG-HRP secondary antibody was added at 1:6000 and further incubated for 1 h at 37 °C.
The in vitro expression of the vaccine constructs was also detected by immunofluorescence assay (IFA) as described in the supplementary material using the individual rabbit antibodies diluted at 1:200. Cells infected with JOL2865 (vector control) served as the negative control.
1.6. Mice immunization and immunogenicity study
Groups of mice (N = 8) were immunized with JOL2818 (variable), JOL2819 (conserved), mixed (JOL2818 + JOL2819) and JOL2865 (vector control) orally by administering directly into the stomach using a bulb-tipped gastric gavage needle at a dose of 1 × 108 CFU in 100 μl PBS given at two doses at a 2-week interval. Another group of mice (N = 8) did not receive any treatment and served as control. At three weeks after the second dose, single-cell suspensions of splenocytes were prepared from four mice from each group as described previously [13]. Sera, intestinal lavage and lung homogenate samples were also collected for ELISA. Salmonella-induced clinical signs such as weight loss were monitored.
1.7. FACS analysis
For fluorescence-activated cell sorting (FACS) analysis, splenocytes collected at 3 weeks after the final immunization were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin in a 96-well plate at 1 × 105 cells/well. The cells were stimulated with 400 ng of individual recombinant proteins for 48 h or with 1 μg/ml of each of the SARS-CoV-2 peptide pools (PepTivator SARS-CoV-2 Prot SDelta, SOmicron and N, Miltenyi-Biotec, Germany) for 24 h. PE-anti-CD3e antibodies, FITC-anti-CD8a, PerCPVio700-anti-CD4 and FITC-anti-CD44 were used for cell surface staining and PE-anti-IFN-γ for intracellular cytokine staining. T-cell subpopulations CD3+CD4+, CD3+CD8+ CD4+IFN-γ+, CD8+IFN-γ+ and CD44+IFN-γ+ were gated and analysed using the MACSQuant analysis system.
1.8. Cytokine measurement my qPCR
For cytokine measurement, isolation of the RNA was performed using a commercial kit (GeneAll) according to the manufacturer's instructions. The detailed methods were performed as previously described [13]. Briefly, cDNA was synthesized from 1 μg of purified RNA using RT Master Premix (Elpis Biotech) with oligo d(T)15 primer according to the manufacturer's instructions. The relative gene expression of cytokines TNF-α, IFN-γ, IL-10 and IL-4 were measured by qPCR using StepOnePlus Real-Time PCR System (Applied Biosystems) using standard conditions. SYBR Green/ROX qPCR Master Mix (ELPIS Biotech) was used together with the primers listed in Table S2 [23]. The reaction was carried out at 95 °C for 7 min pre-incubation followed by 40 cycles of 30 s each at 95 °C, 55 °C and 72 °C. The specificity of the PCR amplification and absence of contamination was confirmed by melting peak analysis with a temperature gradient of 0.1 °C s-1 from 65 °C to 97 °C. Changes in the mRNA levels in immunized mice were determined by the 2-ΔΔCT method using β-actin as the housekeeping gene.
1.9. Hamster immunization and challenge study
Female golden Syrian hamsters were divided into four groups consisting of 4 hamsters per group for SARS-CoV-2 Delta variant challenge and 3 hamsters per group for SARS-CoV-2 Omicron variant challenge and were orally administered with two doses of 2 × 108 CFU of each vaccine strains at a 2-week interval. Animals were monitored for any Salmonella-induced clinical signs including weight loss. 2 weeks after the booster dose, each animal was challenged with 1 × 104 PFU of either SARS-CoV-2 Delta or Omicron variant via the intranasal route under ketamine-xylazine anaesthesia. All hamsters were monitored for five days, and body weight changes were noted.
On day 5 post-challenge, animals were euthanized. Blood was extracted by cardiac puncture and the sera samples were collected after coagulation and centrifugation and frozen at −80 °C until use. Lungs were also collected and kept at −80° for further analysis while sections of lungs were also fixed in 10% neutral-buffered formalin for histopathological analysis.
1.10. Viral titration and RNA isolation
Lung samples collected from golden Syrian hamsters were homogenized with 1 ml of ice-cold DMEM (Lonza) in 2 ml tubes and the tissue homogenate supernatants were titrated on Vero E6 monolayers in 96-well plates. Briefly, serially diluted lung homogenate samples were inoculated on the cells and incubated at 37 °C for 72 h. The cells were observed under a microscope for cytopathic effect (CPE). The TCID50 ml−1 value was calculated with Reed-Muench method [24] and expressed in log10 TCID50/ml. The viral replication was also analysed by immunofluorescence assay.
For viral load in lung tissue homogenates, RNA was extracted using a commercial kit (GeneAll) according to the manufacturer's instructions and eluted with 50 μl nuclease-free water. 1 μg of RNA was used for real-time RT-qPCR to detect and quantify specific SARS-CoV-2 N gene.
1.11. Enzyme-linked immunosorbent assay (ELISA)
Purified recombinant proteins (RBD, NTD, HR, N, RdRp) were coated onto 96-well high-binding polystyrene plates (Greiner Bio-one) at 2.5 μg/ml in 100 mM bicarbonate-carbonate coating buffer pH 9.6 overnight at 4 °C. The wells were then blocked with 200 μl of 5% skim milk for 1 h at 37 °C and then washed three times with 0.05% PBST. Heat-inactivated serum samples from mice and hamsters, intestinal lavage and lung homogenate samples were diluted in 2% skim milk and 100 μl of the respective serum dilutions were added to the wells. The sera were incubated in the plates for at least 1 h at 37 °C. The ELISA plates were again washed thrice in PBST, followed by the addition of 100 μl of 1:3000 HRP-conjugated goat anti-mouse IgG, IgG1, Ig2a or IgA antibody (Southern Biotech) in PBST. 1:10,000 dilution of goat anti-hamster IgG (H + L) HRP (Southern Biotech) was used for hamster sera. The plates were incubated at 37 °C for 1–2 h, washed thrice in 0.05% PBST and the assay was developed using OPD substrate for 10–20 min in the dark. The optical densities (OD) were read at 492 nm in microplate reader (Tecan) after stopping the reaction with 3 M H2SO4.
1.12. Live virus neutralization assay
Neutralizing antibody titer in hamster sera was determined by microneutralization assay (MN). Briefly, 2-fold serial dilutions of sera were prepared and incubated at 37 °C for 2 h with 200TCID50 of either SARS-CoV-2 Delta variant or Omicron variant. Antibody-virus mixtures were then added onto confluent Vero E6 cells grown in 96-well plated and incubated for 72 h. Cytopathic effect (CPE) was observed under a microscope and was analyzed by immunofluorescence assay. The highest sera dilution that resulted in complete inhibition of CPE was recorded as neutralizing antibody titer and expressed as MN50.
1.13. Lung histopathology
Formalin-fixed lung samples were processed for paraffin embedding. The paraffin blocks were cut into 4-μm-thick sections and mounted on glass slides. Sections were further processed for haematoxylin and eosin staining. Stained sections were viewed under the microscope to study the histopathological changes.
1.14. Statistical analysis
All statistical analyses were performed by Student's t-test and ANOVA using GraphPad Prism 5 Software. The p-value <0.05 was considered significant.
2. Results
2.1. Design and construction of SARS-CoV-2 vaccines
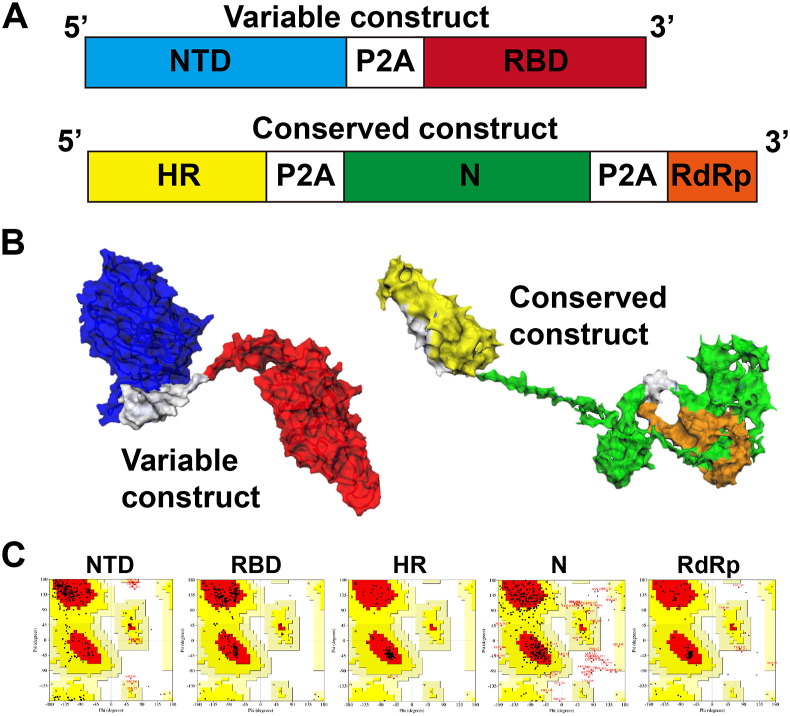
We designed two vaccine constructs such as conserved and variable from the gene sequence of B.1.617.2 Delta variant shown in Fig. 1 A and B. The constructs were cloned into pJHL204, an SFV replicon-based vector that allows the abundant expression of target gene mRNA by a self-replicating mechanism. Further, the self-cleaving peptides allow the separation of polyprotein into individual proteins post-translationally. The protein structure of the constructs was simulated (Fig. 1A and B) and Ramachandran plot analysis revealed 89.3%–100% of amino acid residues are located in the favoured and allowed regions (Fig. 1C), that implies the 2° structure formation and structural stability of selected antigen candidates. Also, B-cell and T-cell epitopes prediction from servers are listed in Tables S3 and S4 and depicted in Fig. S1. The resulting vaccine constructs were electroporated into JOL2500, a live-attenuated Salmonella Typhimurium with the genotype Δlon, Δcpxr, ΔsifA, and Δasd for the vaccine delivery. The resulting vaccine strains were designated as JOL2818 (conserved) and JOL2819 (variable), respectively. A vector control designated as JOL2865 was also prepared by electroporation of JOL2500 with pJHL204.
Fig. 1.
Vaccine design and construction. (A) Graphical representation of designed vaccine constructs. (B) 3D structure of the variable and conserved vaccine constructs showing the individual genes: NTD (blue), RBD (red), HR (yellow), N (green), RdRp (orange) and P2A peptide (white). (C) Ramachandran plot analysis showing residues located in the favoured and allowed regions for structure validation.
2.2. Expression profile of vaccine constructs
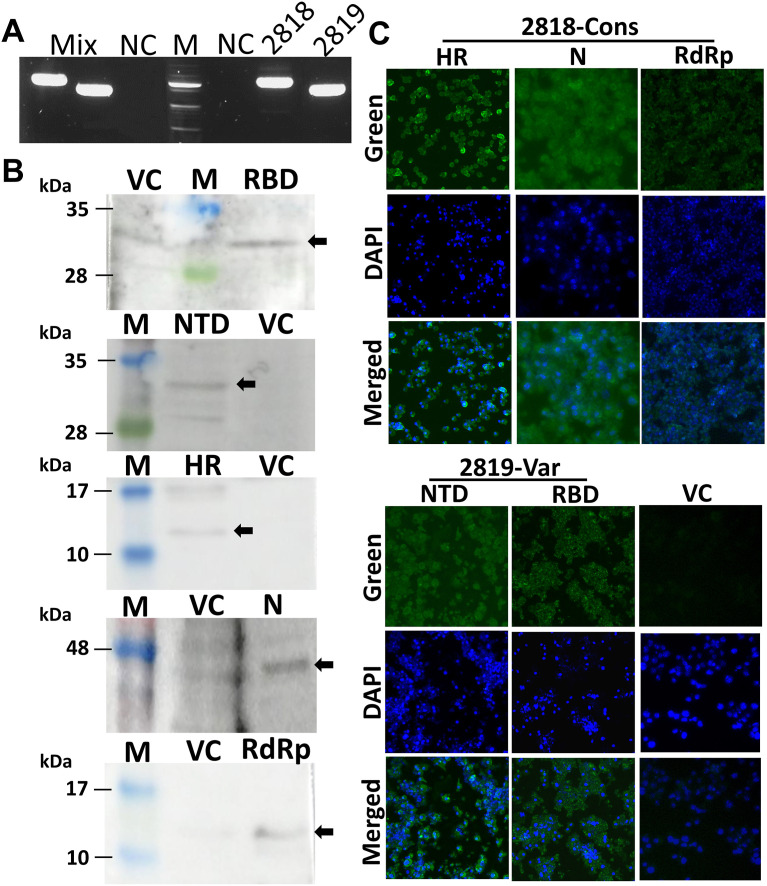
The RT-PCR analysis showed the amplification of the full-length vaccine constructs from the synthesized cDNA with a size of 2.2 kb for HR-N-RdRp (conserved vaccine; JOL2818) and a size of 1.6 kb for NTD-RBD (variable vaccine; JOL2819) (Fig. 2 A). The mixture of each vaccine strain (mix; JOL2818 + JOL2819) also revealed the amplification of both vaccine constructs while no amplification was observed in cells infected with the vector control (Fig. 2A). The protein expression of the vaccine constructs was further confirmed by western blotting and immunofluorescence assay using the hyperimmune antisera against the specific proteins. The western blot analysis of the cell lysates revealed protein bands at approximately 32, 32, 14, 44, 14 kDa for RBD, NTD, HR, N and RdRp, respectively (Fig. 2B). Immunofluorescence assay showed bright green fluorescence manifested by infected cells with conserved and variable vaccines, whereas the expression was not detected in cells infected with the vector control (Fig. 2C). These results confirmed that the expression of all five antigens were efficiently delivered by the Salmonella vector system.
Fig. 2.
In vitro expression of vaccine strains in RAW cells. The murine macrophage cell line, RAW 264.7, was infected with the vaccine strains JOL2818, JOL2819, or vector control (VC) JOL2865. (A) Agarose gel electrophoresis analyses of PCR products for expression of full-length constructs. (B) Western blot analyses of co-expression products. Detection was performed using specific hyperimmune rabbit sera against each recombinant protein. (C) Images of immunofluorescent staining of RAW 264.7 cells showing expression of proteins. IFA was performed using antisera against the target recombinant proteins. Green fluorophore indicates protein expression and blue fluorophore indicates nuclei stained with DAPI.
2.3. Induction of vaccine-elicited antibodies and cellular immune responses in mice
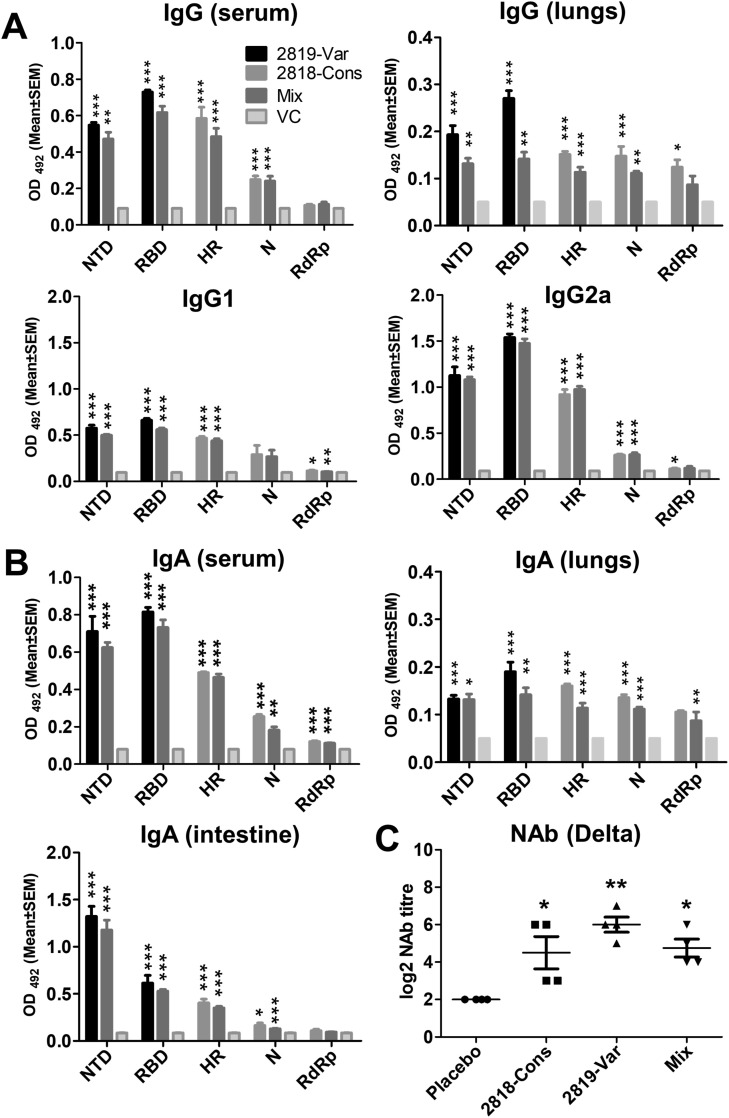
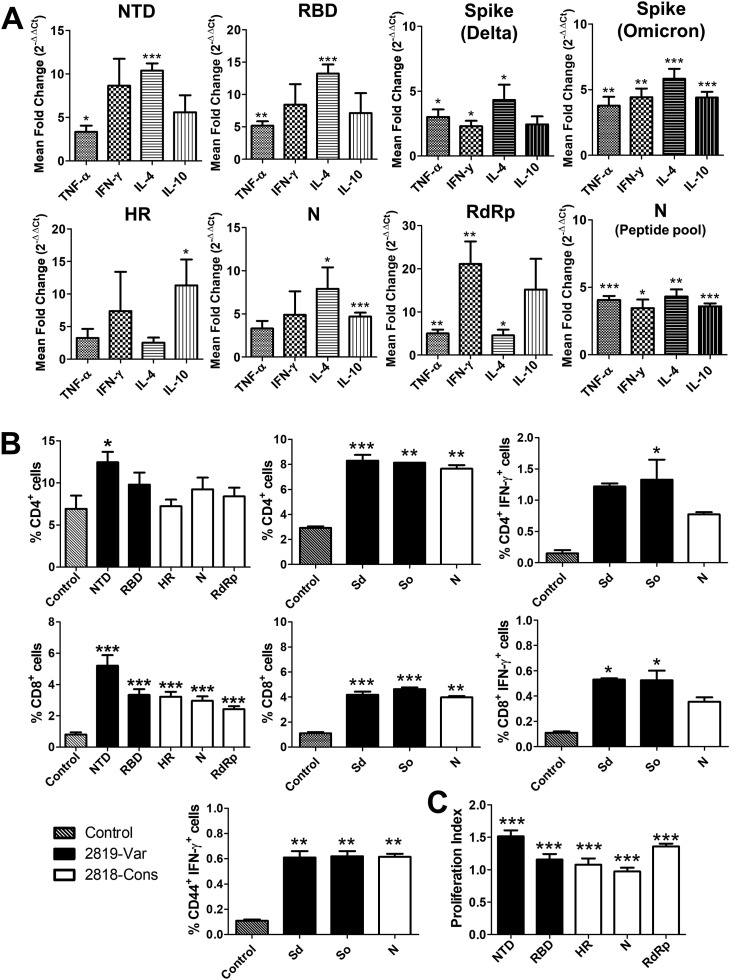
High levels of antigen-specific IgG and IgA antibodies were elicited after two doses of the oral vaccines in all the vaccinated groups (Fig. 3 A and B) with high antibodies raised against RBD and NTD and least antibodies raised against RdRp. Delta variant neutralizing antibodies from immunized mice revealed higher mean titre induced by the variable construct (Fig. 3C). Furthermore, the analysis of cell suspensions of mouse splenocytes harvested at week 3 post-immunization after stimulation with the individual recombinant proteins and peptide pools showed recall response. Changes in the expression profiles of Th1 cytokines (IFN-γ and TNF-α) and Th2 cytokines (IL-4 and IL-10) revealed upregulation and downregulation of transcripts upon stimulation. Particularly, stimulation of cell suspensions with NTD and RBD from mice immunized with the variable construct revealed upregulation of about 1.9-fold to 6.5-fold of TNF-α, 1.9-fold to 16.75-fold IFN-γ, 8.82-fold to 16.1-fold of IL-4 and 1.7-fold to 15.6-fold of IL-10 (Fig. 4 A). While cytokine expression after stimulation with HR, N, and RdRp of cell suspensions from mice immunized with conserved construct revealed upregulation of about 0.9-fold to 8.5-fold of TNF-α, 0.4-fold to 30-fold of IFN-γ, 1.0-fold to 16.4-fold of IL-4 and 5.09-fold to 22-fold of IL-10 (Fig. 4A). Stimulation with peptide pools from spike Delta and Omicron and nucleocapsid also exhibited similar pattern of upregulation of the cytokines (Fig. 4A).
Fig. 3.
Induction of humoral and mucosal responses by JOL2818 (conserved) and JOL2819 (variable). Balb/c mice were immunized orally with two doses of 1 × 107. Specific antibody responses in mice (N = 4) at week 3 post-immunization were assessed for NTD, RBD, HR, N and RdRp antigens. (A) IgG, IgG1 and IgG2a response in serum and lung of mice was determined by ELISA. The bars indicate the means of the titres from four biologically independent mice per group. (B) IgA response in sera of immunized mice at sera dilution of 1:50, sIgA response in the intestinal lavage at a sample dilution of 1:100 and IgA response in lung homogenates at 1:10 sample dilution. (C) Serum neutralizing antibody titre (NAb) from mice quantified by CPE and IFA analysis against SARS-CoV-2 Delta variant. Error bars denote SEM. Data were analysed by independent-samples T-test and ANOVA. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Fig. 4.
Cellular immune response in mice. Immune recall response from mice (N = 4) was analysed at 3 weeks post-immunization. Splenocyte suspensions were stimulated separately with individual recombinant proteins, spike and nucleocapsid peptide pools. (A) Changes in the cytokine expression profile of IFN-γ, TNF-α, IL-4 and IL-10 determined by qPCR at 48 h after stimulation with recombinant proteins and at 24 h after stimulation with peptide pools. (B) Percentages of CD4+ and CD8+ T cells quantified and presented in a bar diagram. Further percentage of CD4+IFN-γ+, CD8+IFN-γ+ and CD44+IFN-γ+ T cell expansion were presented upon stimulation with the peptide pools. (C) Bar diagram showing splenocyte proliferation index. The bars represent the mean values from four biologically independent mice per group. Error bars denote SEM. Data were analysed by independent-samples T-test and ANOVA. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Flow cytometric analysis of changes in T cells after stimulation with the recombinant proteins as well as peptide pools also revealed an increase in both CD4+ and CD8+ T cell subpopulations compared with the placebo controls (Fig. 4B). Particularly, a significant increase (p < 0.05) in CD4+ was observed after stimulation with NTD recombinant protein and the peptide pools. Additionally, changes in the CD8+ T cell sub-population were significantly increased upon stimulation of the recombinant proteins as well as peptide pools. Intracellular cytokine staining of IFN-γ positive CD4+ and CD8+ T cells and percentage of CD44+IFN-γ+ (Fig. 4B) further revealed T cell expansion and anti-viral response in vaccine-immunized groups upon stimulation with the respective peptide pools.
2.4. Induction of humoral immune response and neutralizing antibodies in hamster model
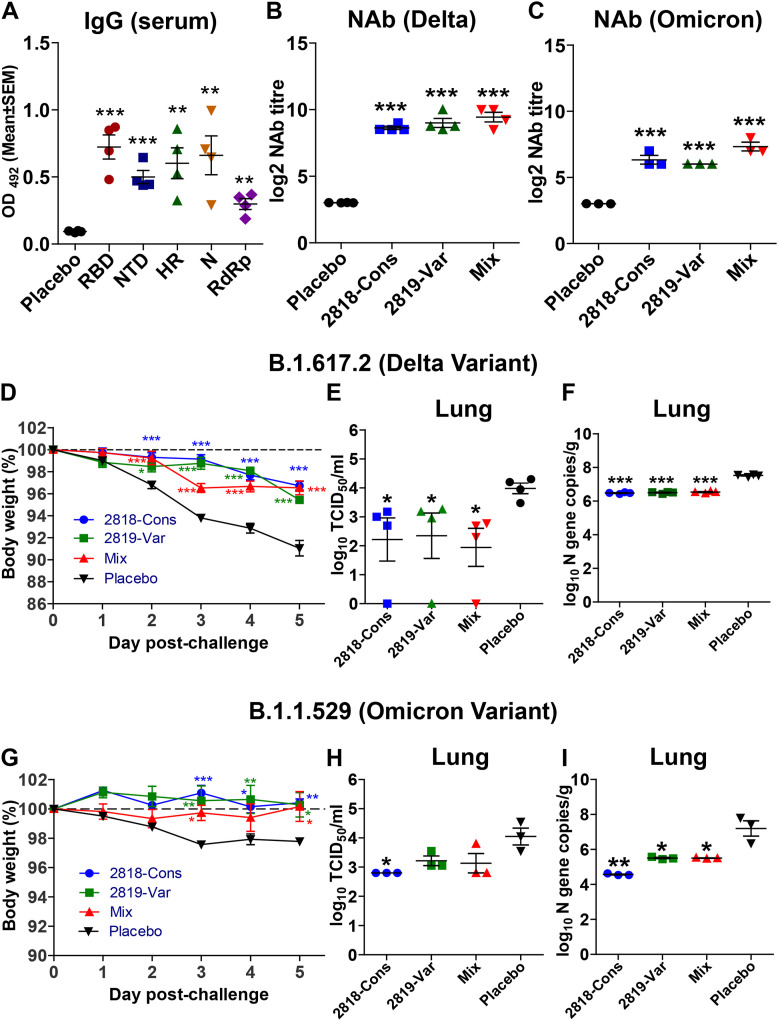
Humoral IgG immune response was elicited against the five target antigens with the highest mean of antigen-specific antibodies observed against RBD followed by N, HR, NTD, and least observed against RdRp detected by ELISA (Fig. 5 A). Furthermore, to test the ability of hamster sera to neutralize two SARS-CoV-2 variants such as Delta and Omicron, the levels of NAbs were determined by microneutralization expressed as MN50. Sera from separately immunized hamsters with the conserved and variable constructs, as well as hamsters immunized with the mixture of the two, exhibited log2-transformed MN50 titre of 9–10 against Delta variant (Fig. 5B) and showed cross-neutralization with 6–8 titre against Omicron variant (Fig. 5C).
Fig. 5.
Humoral immune response and protection efficacy evaluation by the vaccines in hamster model. Syrian hamsters were immunized with 2 doses of 2 × 108 CFU and IgG and neutralizing antibody titre were assessed 3 weeks post-immunization. (A) IgG response in sera of hamsters on individual antigens. (B) Neutralizing antibody titre (NAb) quantified by CPE and IFA analysis against SARS-CoV-2 Delta variant. (C) NAb quantified by CPE and IFA analysis against SARS-CoV-2 Omicron variant. To evaluate the protection conferred by the vaccine in a hamster model, Syrian hamsters were immunized orally with 2 doses of 2 × 108 CFU at a 2-week interval. 3 weeks post-immunization, hamsters were intranasally inoculated with 1 × 104 PFU of either the delta variant or omicron variant. (D) Percentage body weight change of hamsters after viral challenge with Delta variant. (E) Lung viral burden in Delta-challenged hamsters quantified by qPCR detection of SARS-CoV2 N gene. (F) Lung viral burden in Delta-challenged hamsters measured by TCID50. (G) Percentage body weight change of hamsters after viral challenge with Omicron variant. (H) Lung viral burden in Omicron-challenged hamsters quantified by qPCR detection of SARS-CoV2 N gene. (I) Lung viral burden in Omicron-challenged hamsters measured by TCID50. Data were presented as log10 TCID50 and log10-transformed viral N gene copies/g. Data were analysed by independent-samples T-test and ANOVA. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
2.5. Protection of immunized hamster from Delta and Omicron SARS-CoV-2 challenge
Overall, the weight loss of all immunized hamsters challenged with Delta or Omicron was less observed at 2–3 days post-infection compared to infected placebo groups (Salmonella without the plasmid) (Fig. 5D and G). Lung samples collected at 5-dpi were subjected to viral titration to determine the viral burden and revealed a significant decrease in the viral load of the Delta variant in all immunized groups with one hamster from each group showing no detectable viral titre (Fig. 5E). On the other hand, decreased infectious viral titre was also observed in all immunized hamsters with a significant decrease (p < 0.05) of the viral burden of Omicron variant in the lungs of hamsters immunized with the conserved vaccine construct (Fig. 5H). In addition, the viral N gene copies were also reduced and recorded as 6.5 copies/g and 5.8 copies/g for all vaccinated groups infected with Delta and Omicron variants, respectively (Fig. 5F and I).
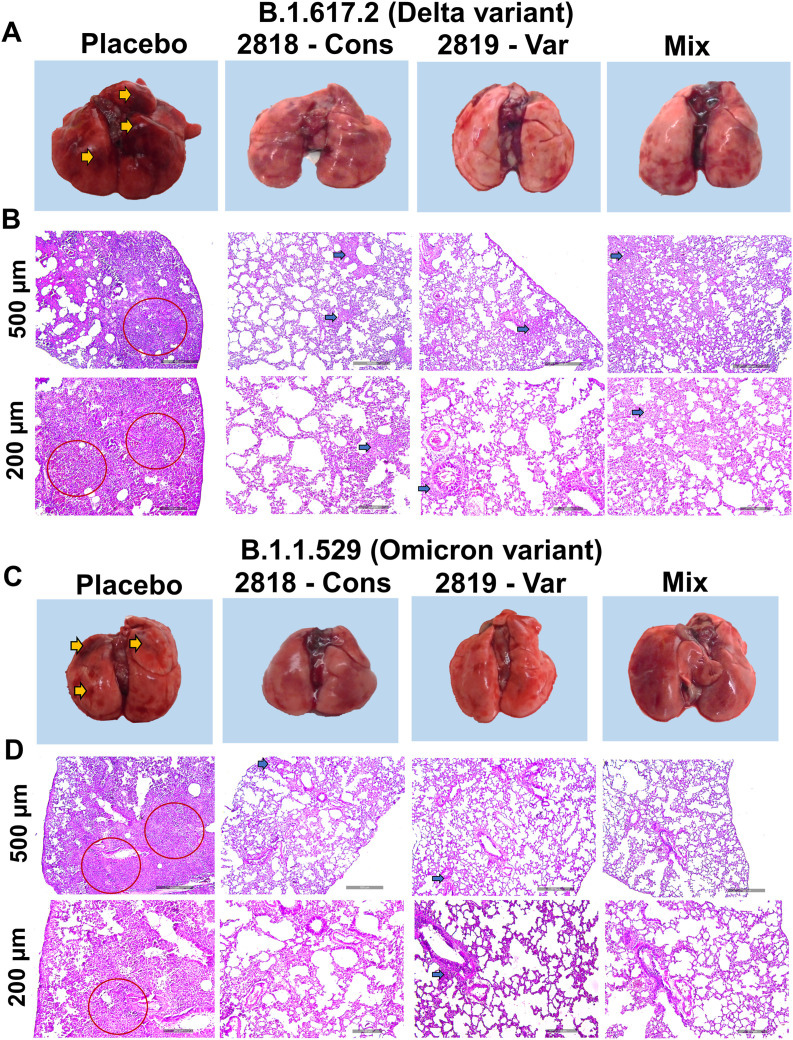
Gross analysis of the lungs showed evident signs of inflammation and pneumonitis in the placebo group of hamsters infected with either Delta variant or Omicron variant while no such severe inflammation and congestion was observed in the vaccinated hamsters (Fig. 6 A and C). Further histopathological analysis of lung sections also revealed severe infiltration of inflammatory cells in the lung parenchyma, bronchial and bronchiolar mucosa, and surrounding airways in the placebo group of hamsters infected with either SARS-CoV-2 Delta or Omicron variant. For all immunized groups, stained lung sections showed few focal lesions of congestion and mild inflammatory infiltrates, showing reduced inflammation in the lungs (Fig. 6B and D).
Fig. 6.
Gross and histopathological analysis of the lungs from immunized Syrian hamsters challenged by SARS-CoV-2. Assessment of freshly collected lungs at 5dpi from challenged hamsters. (A) Gross pathology images from the placebo group and immunized hamsters challenged with SARS-CoV-2 delta variant. (B) Haematoxylin and Eosin (H&E) stained lung sections from the placebo group and immunized hamsters challenged with SARS-CoV-2 delta variant. (C) Gross lung images from the placebo group and immunized hamsters challenged with SARS-CoV-2 omicron variant. (D) Haematoxylin and Eosin (H&E) stained lung sections from the placebo group and immunized hamsters challenged with SARS-CoV-2 omicron variant. Gross pathology images of hamster lungs show multifocal extensive areas (yellow arrow). Infected lung tissue sections showing severe lesions of interstitial pneumonia (red circle) in the placebo group. Histopathological lesions were observed in bronchioles and adjacent alveolar spaces showing mild inflammatory changes (blue arrow).
3. Discussion
The emergence of the pandemic COVID-19 has caused a great impact on global public health and the best-known strategy to combat the spread of this disease is the rapid development of effective vaccines. Most of the currently approved vaccines were based on the spike protein due to its vital role in the binding of the virus and fusion with the host cell membrane receptor thus, antibodies produced against this protein could inhibit further viral replication [25,26]. With the known role of the spike protein in SARS-CoV-2 infection and adaptive immunity, it has been identified as a target site for neutralizing antibodies, making it a key target for vaccine design [[27], [28], [29]]. However, there are concerns about the future efficacy of the current vaccines because of the evolving capability of the SARS-CoV-2 shown by the emergence of various variants carrying mutations on the spike protein, particularly in the receptor-binding domain (RBD) of the variants that could impair the efficiency of COVID-19 vaccines [30]. Thus, the development of efficient vaccines targeting multiple viral proteins such as those variable and conserved regions [31] will bring further understanding of other potential targets and exploration of other alternative vaccine strategies and delivery systems for universal variant vaccines. Thus, in this study, we specifically constructed two vaccine strains with the SARS-CoV-2 variable proteins NTD and RBD and the conserved proteins HR, N, and epitopes of RdRp, separately, and evaluated the elicitation of immune response and protection efficacy in an animal model as well as in vitro.
The S1 subunit of SARS-CoV-2, consisting of the NTD and RBD, has been extensively studied and known to play a key role in receptor binding with the host membrane which makes it the main target of vaccine development [32]. Furthermore, the S1 subunit of SARS-CoV-2 has reported mutations and deletions as observed in the emerging variants and there is evidence that this can change the antigenicity of the protein which can affect antibody neutralization and vaccine escape [8,33]. Interestingly, aside from S1-RBD, the S1-NTD also contains epitopes for CoV NAbs found in infected patients which also makes it a potential vaccine target [12] (Tables S3 and S4). Thus, in addition to RBD, we further explored the inclusion of NTD in the variable vaccine construct. While most of the current vaccines target the “extra-viral” spike protein, several studies have also shown the potentiality of other viral targets against COVID-19 such as the conserved regions of the S2 subunit, the HR1 and HR2 [31] and the “intra-viral” conserved proteins such as the nucleocapsid (N-protein) [34,35] and non-structural proteins [36] (Fig. 1). The heptad repeat (HR1 and HR2), situated in the S2 subunit of the spike protein that is responsible for viral fusion and entry, is suggested to be a potential target in universal coronavirus vaccines [12]. The N-protein is relatively conserved among mutant strains and is the most abundant viral protein [37]. It has been found to be highly immunogenic during infection [35,38] containing T cell epitopes (Table S4) and a major target for antibody responses (Fig. S1 and Table S3) [12,39]. Thus, it has been found to be an excellent immunogen to stimulate the host immune system [37] particularly the T cell responses which are regarded as critical for controlling infections and further viral dissemination [34]. The non-structural proteins (NSPs), on the other hand, have important roles in viral pathogenicity. In particular, the NSP12, also known as the RdRp, allows the virus to carry out RNA synthesis and has structural similarities among RdRp of SARS-CoV and SARS-CoV-2 [36,40,41]. Aside from the evidence of RdRp as a target for therapeutics [36,41], an epitope prediction study (Tables S3 and S4) has revealed its antigenic properties [42] suggesting it to be a potential candidate for the development of a vaccine against SARS-CoV-2 as well as other non-structural proteins [43]. We have analysed the stereochemical property of our vaccine constructs by Ramachandran plot, which interprets the structural stability of the selected antigens (Fig. 1).
In this study, we used the P2A, a viral self-cleaving peptide, to generate a multi-antigen expression in which the expression of all proteins in the vaccine constructs were confirmed by western blot and IFA (Fig. 2). We then compared the humoral and cellular immune response elicited by the vaccination of the two vaccine strains expressing the conserved genes and variable genes, orally administered using Salmonella-based delivery system. The highest titre was observed against RBD followed by NTD which is part of the SARS-CoV-2 S1 domain (Fig. 3). This suggests that the combination of RBD and NTD is more immunogenic in terms of B cell response compared to other antigens. This result and hypothesis correlate with another study which also demonstrated that the SARS-CoV-2 S1 domain is more immunogenic than RBD domain alone when immunized in mice [44]. The conserved HR in the S2 spike protein elicited high IgG titre (Fig. 3A) which demonstrates and supports its potential target in universal coronavirus vaccines [12,31]. The oral administration of the vaccine strains resulted in significantly increased mucosal and sIgA response both in the intestine and lung (Fig. 3B). The mucosal antibody response implies that the present bacterial vaccination can hinder the virus at the prime route of entry. Strong cellular immune responses were elicited by both the vaccine constructs as observed in the increased CD4+ and CD8+ T cells (Fig. 4B). Among the proteins, the NTD-specific CD4+ T cells were recorded to be the highest, suggesting its possible significant contribution in promoting B cell antibody production and helping promote the induction of tissue-resident memory CD8+ T cells. We also observed considerably higher number of IFN-γ secreting CD4 and CD8 cells were detected in immunized mice (Fig. 4B), exemplifying the anti-viral immunogenicity [45]. In furtherance, a significant increase in the CD44 sub-population with INF-γ production was detected. The CD44 exist on the surface of memory T cells that attribute to protection against forthcoming infection after immunization [46]. The results decipher the presence of memory T cells to confer long term immune response (Fig. 4B). Our study on the cytokine expression revealed induction of Th1 response with upregulation of the pro-inflammatory cytokines such as TNF-α and INF-γ (Fig. 4A). Interestingly, the epitopes of the non-structural protein RdRp, despite of its overall low immunogenicity possibly due to low protein expression, have shown the highest increase in the cytokine expression of IFN-γ (Fig. 4A) suggesting its potential in inducing cellular immune response which might have contributed to the decreased viral burden in the lungs of the hamsters immunized with the conserved vaccine.
Evaluation of the immune response and protection efficacy of the conserved vaccine and variable vaccine was further assessed in the hamster animal model. The serum IgG titre was assessed which revealed significant induction of antibodies against all the antigens after immunization in hamsters (Fig. 5A). Furthermore, the induction of neutralizing antibodies in mice (Fig. 3C) and hamsters was observed both in conserved- and variable-vaccinated hamsters (Fig. 5A and C). Although the serum from immunized hamsters with the two vaccines constructed based on the delta genomic sequence showed a cross-neutralizing titre (Fig. 5B and C) against the Omicron variant, the slight decrease in neutralization could be associated with the antigenic difference between the spike glycoproteins of the two variants [47]. The associated protection brought by immunization of the vaccine strains is supported by the significant decrease in lung viral load (Fig. 5E and H) after virus infection with Delta and Omicron variants of the immunized hamsters as well as reduced inflammation in the lungs (Fig. 6). We observed better cross-protection against the Omicron variant by the conserved construct that may be attributed to the elicitation of superior T cell response by the nucleocapsid and RdRp which may have critical control on the infection especially if the virus escapes the antibody response against HR, a part of the spike protein. A similar observation was recorded in a study of nucleocapsid-specific immunity by Dangi et al. [34] which suggests the advantage of including other viral antigens besides the spike protein for T cell responses which may have critical role in subsequent viral dissemination.
In this study we have specifically employed our previously developed eukaryotic vector, pJHL204, comprising the Semliki Forest virus (SFV) promoter and SFV RdRp for its efficient transcription through a self-replication and self-transcribing mechanism [48]. The plasmid expressing antigens was delivered by Salmonella-based vaccine strain for efficient delivery of antigens to target tissues [13,14]. With the innate ability of Salmonella to invade and proliferate in professional antigen-presenting cells (APCs), it has the advantage to directly deliver the DNA cargo to these cells and subsequently result in induction of cellular and humoral response [49]. Also, as safety concern is one of the important considerations in vaccine development especially with live-attenuated bacteria, of the vaccine delivery strain JOL2500 used in this study with genotype of Δlon, ΔcpxR, ΔsifA, and Δasd has been proven to have optimal survival in the host without any virulence [14,50]. Furthermore, vaccine production for this unique strategy can be easily scaled up and prepared rapidly at lower cost.
In summary, we have demonstrated the feasibility of targeting multiple antigens of variable and conserved regions of SARS-CoV-2 by recording induction of strong cellular and humoral responses which culminated in significant protection in the live virus-challenged hamsters. In addition, the combination of efficient eukaryotic expression and Salmonella-mediated vaccine delivery offers a unique vaccine platform and alternative strategy in combating infectious diseases such as SARS-CoV-2. Further dose optimization of mix vaccine will be useful to acquire a potential broad-spectrum protection. The findings reinforce the exploitation of bacteria-mediated delivery for the development of safe, effective and convenient oral mRNA vaccine against SARS-CoV-2 and also other infectious diseases [49].
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A1A03033084) and by BK21 FOUR Program by Jeonbuk National University Research Grant.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2023.105101.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Riediker M., Briceno-Ayala L., Ichihara G., Albani D., Poffet D., Tsai D.H., et al. Higher viral load and infectivity increase risk of aerosol transmission for Delta and Omicron variants of SARS-CoV-2. Swiss Med Wkly. 2022;152 doi: 10.4414/smw.2022.w30133. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J Med Virol. 2022;94 doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 3.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599 doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motozono C., Toyoda M., Zahradnik J., Ikeda T., Saito A., Tan T., et al. An emerging SARS-CoV-2 mutant evading cellular immunity and increasing viral infectivity. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv. 2021 [Google Scholar]
- 6.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596 doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 7.Tian D., Sun Y., Xu H., Ye Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J Med Virol. 2022;94 doi: 10.1002/jmv.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkhout B., Herrera-Carrillo E. SARS-CoV-2 evolution: on the sudden appearance of the Omicron variant. J Virol. 2022;96 doi: 10.1128/jvi.00090-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khajotia R. Omicron: the highly mutational COVID-19 variant with immune escape. Pan Afr Med J. 2022;41 doi: 10.11604/pamj.2022.41.84.33373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohandas S., Yadav P.D., Sapkal G., Shete A.M., Deshpande G., Nyayanit D.A., et al. Pathogenicity of SARS-CoV-2 Omicron (R346K) variant in Syrian hamsters and its cross-neutralization with different variants of concern. EBioMedicine. 2022;79 doi: 10.1016/j.ebiom.2022.103997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng S.M.S., Mok C.K.P., Leung Y.W.Y., Ng S.S., Chan K.C.K., Ko F.W., et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022;28 doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21 doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jawalagatti V., Kirthika P., Park J.Y., Hewawaduge C., Lee J.H. Highly feasible immunoprotective multicistronic SARS-CoV-2 vaccine candidate blending novel eukaryotic expression and Salmonella bactofection. J Adv Res. 2022;36 doi: 10.1016/j.jare.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senevirathne A., Park J.Y., Hewawaduge C., Perumalraja K., Lee J.H. Eukaryotic expression system complemented with expressivity of Semliki Forest Virus's RdRp and invasiveness of engineered Salmonella demonstrate promising potential for bacteria mediated gene therapy. Biomaterials. 2021;279 doi: 10.1016/j.biomaterials.2021.121226. [DOI] [PubMed] [Google Scholar]
- 15.Jawalagatti V., Kirthika P., Hewawaduge C., Yang M sik, Park J.Y., Oh B., et al. Bacteria-enabled oral delivery of a replicon-based mRNA vaccine candidate protects against ancestral and delta variant SARS-CoV-2. Mol Ther. 2022;30 doi: 10.1016/j.ymthe.2022.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jawalagatti V., Kirthika P., Hewawaduge C., Park J.Y., Yang M.S., Oh B., et al. A simplified SARS-CoV-2 mouse model demonstrates protection by an oral replicon-based mRNA vaccine. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.811802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajan P., Mishra P.K.K., Joshi P. Defining the complement C3 binding site and the antigenic region of Haemonchus contortus GAPDH. Parasite Immunol. 2019;41 doi: 10.1111/pim.12611. [DOI] [PubMed] [Google Scholar]
- 18.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10 doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26 doi: 10.1107/s0021889892009944. [DOI] [Google Scholar]
- 20.Reynolds C.R., Islam S.A., Sternberg M.J.E. EzMol: a web server wizard for the rapid visualization and image production of protein and nucleic acid structures. J Mol Biol. 2018;430 doi: 10.1016/j.jmb.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giulietti A., Overbergh L., Valckx D., Decallonne B., Bouillon R., Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25 doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 24.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27 doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 25.Martínez-Flores D., Zepeda-Cervantes J., Cruz-Reséndiz A., Aguirre-Sampieri S., Sampieri A., Vaca L. SARS-CoV-2 vaccines based on the spike glycoprotein and implications of new viral variants. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7 doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvatori G., Luberto L., Maffei M., Aurisicchio L., Aurisicchio L., Roscilli G., et al. SARS-CoV-2 spike protein: an optimal immunological target for vaccines. J Transl Med. 2020;18 doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Z., Gong W. Will mutations in the spike protein of SARS-CoV-2 lead to the failure of COVID-19 vaccines? J Korean Med Sci. 2021;26 doi: 10.3346/JKMS.2021.36.E124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fathizadeh H., Afshar S., Masoudi M.R., Gholizadeh P., Asgharzadeh M., Ganbarov K., et al. SARS-CoV-2 (Covid-19) vaccines structure, mechanisms and effectiveness: a review. Int J Biol Macromol. 2021:188. doi: 10.1016/j.ijbiomac.2021.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K.H., Bhatnagar N., Jeeva S., Oh J., Park B.R., Shin C.H., et al. Immunogenicity and neutralizing activity comparison of SARS-CoV-2 spike full-length and subunit domain proteins in young adult and old-aged mice. Vaccines. 2021;9 doi: 10.3390/vaccines9040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagesha S.N., Ramesh B.N., Pradeep C., Shashidhara K.S., Ramakrishnappa T., Krishnaprasad B.T., et al. SARS-CoV 2 spike protein S1 subunit as an ideal target for stable vaccines: a bioinformatic study. Mater Today Proc. 2021;49 doi: 10.1016/j.matpr.2021.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19 doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dangi T., Class J., Palacio N., Richner J.M., Penaloza MacMaster P. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yadav R., Chaudhary J.K., Jain N., Chaudhary P.K., Khanra S., Dhamija P., et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10 doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj R. Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing. Biochem Biophys Rep. 2021;25 doi: 10.1016/j.bbrep.2020.100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thura M., En Sng JX., Ang K.H., Li J., Gupta A., Hong J.M., et al. Targeting intra-viral conserved nucleocapsid (N) proteins as novel vaccines against SARS-CoVs. Biosci Rep. 2021;41 doi: 10.1042/BSR20211491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 39.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53 doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., et al. Structural genomics of SARS-COV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12 doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muhammed Y., Yusuf Nadabo A., Pius M., Sani B., Usman J., Anka Garba N., et al. SARS-CoV-2 spike protein and RNA dependent RNA polymerase as targets for drug and vaccine development: a review. Biosaf Heal. 2021;3 doi: 10.1016/j.bsheal.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yashvardhini N., Kumar A., Jha D.K. Immunoinformatics identification of B-and T-cell epitopes in the RNA-dependent RNA polymerase of SARS-CoV-2. Can J Infect Dis Med Microbiol. 2021;2021 doi: 10.1155/2021/6627141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammed M.E.A. SARS-CoV-2 proteins: are they useful as targets for COVID-19 drugs and vaccines? Curr Mol Med. 2021;22 doi: 10.2174/1566524021666210223143243. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Wang L., Cao H., Liu C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J Med Virol. 2021;93 doi: 10.1002/jmv.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmire J.K., Tan J.T., Whitton J.L. Interferon-γ acts directly on CD8+ T cells to increase their abundance during virus infection. J Exp Med. 2005;201:1053–1059. doi: 10.1084/jem.20041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baaten B.J.G., Li C.-R., Bradley L.M. Multifaceted regulation of T cells by CD44. Commun Integr Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lechmere T., Snell L.B., Graham C., Seow J., Shalim Z.A., Charalampous T., et al. Broad neutralization of SARS-CoV-2 variants, including Omicron, following breakthrough infection with Delta in COVID- 19-vaccinated individuals. mBio. 2022;13 doi: 10.1128/mbio.03798-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komdeur F.L., Singh A., van de Wall S., Meulenberg J.J.M., Boerma A., Hoogeboom B.N., et al. First-in-human phase I clinical trial of an SFV-based RNA replicon cancer vaccine against HPV-induced cancers. Mol Ther. 2021;29 doi: 10.1016/j.ymthe.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jawalagatti V., Kirthika P., Lee J.H. Oral mRNA vaccines against infectious diseases- a bacterial perspective [Invited] Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.884862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aganja R.P., Sivasankar C., Hewawaduge C., Lee J.H. Safety assessment of compliant, highly invasive, lipid A-altered, O-antigen-defected Salmonella strains as prospective vaccine delivery systems. Vet Res. 2022;53:76. doi: 10.1186/s13567-022-01096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.