Abstract
Iron is an essential requirement for the survival and virulence of most bacteria. The bacterial ferrous iron transporter protein FeoB functions as a major iron transporter in prokaryotes, but its biochemical mechanism has not been fully elucidated. In the present study, we compared enzymatic properties of the cytosolic portion of pathogenic bacterial FeoBs to elucidate each bacterial strain-specific characteristic of the Feo system. We show that bacterial FeoBs are classified into two distinct groups which possess either a sole GTPase or an NTPase with a substrate promiscuity. This difference in nucleotide specificity alters cellular requirements for monovalent and divalent cations. While the hydrolytic activity of the GTP-dependent FeoBs was stimulated by potassium, the action of the NTP-dependent FeoBs was not significantly affected by the presence of monovalent cations. Mutation of Asn11, having a role in potassium-dependent GTP hydrolysis, changed nucleotide specificity of the NTP-dependent FeoB, resulting in loss of ATPase activity. Sequence analysis suggested a possible association of alanine in the G5 motif for the NTP-dependent specificity in FeoBs. This demonstration of the distinct enzymatic properties of bacterial FeoBs provides important insights into mechanistic details of Feo iron transport processes, as well as offers a promising species-specific anti-virulence target.
Keywords: Feo, ferrous iron transport, GTPase, ATPase, Potassium
INTRODUCTION
Iron is essential for most living organisms, and its acquisition presents challenges for human pathogens competing with their host for the element [1]. While iron-containing proteins are necessary for fundamental physiological processes, including respiration and DNA repair, overload of free iron is highly toxic, resulting in biomolecular damage to DNA, proteins, and lipids via Fenton reactions [2]. To control intracellular iron concentrations, a fine-scale regulatory system of iron transport such as Feo, the ferrous iron transporter, is required [3–5].
The Feo iron transport system is an evolutionarily ancient ferrous iron transporter, widely distributed among archaea and bacteria. The Feo system plays an important role in bacterial survival and virulence in mammalian hosts, as has been demonstrated by assessing the effects of deletion or mutation of the feo genes in several bacterial species [3,6–9]. In particular, the Feo system contributes to the oxygen-dependent regulation of the ferric-uptake regulator (Fur) by increasing the iron pool available for Fur binding, maintaining iron homeostasis, and altering transcription of iron-regulated genes [10], including virulence factors.
The Feo system is composed of two cytosolic proteins, FeoA and FeoC, and an iron permease, FeoB. FeoB is a multidomain transmembrane protein with an N-terminal cytoplasmic GTPase and GDP dissociation inhibitor (GDI) domain, and a C-terminal transmembrane domain. The cytosolic domain has a nucleotide hydrolytic activity which is used for regulation of iron transport by utilizing energy from the nucleotide hydrolysis. There are two important enzymic characteristics of FeoB: 1) nucleotide hydrolysis activity and 2) potassium-stimulated activity. For the nucleotide hydrolytic activity, the FeoB protein was initially considered as an ATPase, as demonstrated in a study of ATPase inhibitors in Helicobacter pylori [6]. However, in Escherichia coli, sequence and biochemical analyses showed that the FeoB protein is solely a GTPase, not an ATPase. Recently, we have shown that Vibrio cholerae and H. pylori FeoBs possess dual nucleotide specificity, suggesting Feo as a more diverse system than previously recognized. In regards to its potassium-stimulated properties, FeoB has a slow intrinsic GTPase turnover rate and weak nucleotide-binding affinity [11]. However, the GTP hydrolytic activity of Streptococcus thermophilus FeoB has been shown to be activated by potassium [12,13]. Recently, we showed that V. cholerae FeoB is not stimulated by potassium, questioning if potassium stimulation is a shared attribute of bacterial FeoBs [3].
Feo is an important virulence factor among various bacterial strains, making it a good anti-virulence target. In particular, since the Feo system is highly dependent on nucleotide binding and hydrolysis, understanding the mechanistic role of this domain is prerequisite. Therefore, it is necessary to widen the scope of the systems studied in order to learn more about the structure-reactivity relationship. In the present study, we compared enzymatic properties of pathogenic bacterial cytosolic FeoBs, originated from foods and environments. The results showed that bacterial FeoBs can be classified into two distinct groups which possess either sole GTPase or promiscuous NTPase activity. This difference in nucleotide specificity altered the requirements for monovalent and divalent cations. Mutation of Asn11, having a role in potassium-binding, abolished potassium-dependent GTPase activation in E. coli, whereas it altered nucleotide specificity in S. mutans FeoB. Sequence analysis suggested possible association of alanine in the G5 motif for the dual nucleotide-specificity of FeoBs.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are identified in Table 1. All strains were maintained at −80°C in Luria-Bertani (LB; 1% tryptone, 0.5% yeast extract and 1% NaCl, w/v) broth containing 20% (v/v) glycerol. Single colonies were grown in LB broth at 37°C. Ampicillin was used at a concentration of 100 μg/ml to select for and maintain plasmids.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning strain F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK−mK+), λ− | [40] |
| E. coli BL21(DE3) | Protein expression strain. : F− ompT hsdSB (rB−, mB−) gal dcm (DE3) | [41] |
| Plasmids | ||
| pET21a | Bacterial expression vector | Novagen |
| pEcNFeoB | pET21a carrying the cytosolic domain of E. coli FeoB | This study |
| pSmNFeoB | pET21a carrying the cytosolic domain of S. mutans FeoB | This study |
| pEcNFeoB-N11A | pEcNFeoB with a N11A mutation | This study |
| pSmNFeoB-N11A | pSmNFeoB with a N11A mutation | This study |
Plasmid construction
pET21a-based expression vectors encoding the cytosolic N-terminus of FeoB (NFeoB) from Escherichia coli (amino acids 1–274, EcNFeoB), Salmonella enterica serovar Typhimurium (amino acids 1–274, StNFeoB), Pseudomonas aeruginosa (amino acids 1–271, PaNFeoB), Streptococcus mutans (amino acids 1–273, SmNFeoB), Staphylococcus aureus (amino acids 1–272, SaNFeoB), Bacillus cereus (amino acids 1–269, BcNFeoB), or Listeria monocytogenes (amino acids 1–272, LmNFeoB) with a hexa-histidine tag were generated. All bacterial NFeoBs included a linker (KLAAALE) along with a hexa-histidine tag at the C-terminus. DNA sequences of all constructs were confirmed through Sanger DNA sequencing.
Site-directed mutagenesis
The expression plasmids pEcNFeoB and pSmNFeoB were used to generate expression constructs encoding a single amino acid substitution at position Asn11 (EcNFeoB-N11A and SmNFeoB-N11A) by site-directed mutagenesis. For mutagenesis, the QuikChange-Phusion protocol was slightly modified using Q5 DNA polymerase (M0491, New England Biolabs, Ipswich, MA, USA) instead of Phusion DNA polymerase [14]. The primers used for plasmid construction and site-directed mutagenesis are listed in Table 2.
Table 2.
Primers used for cloning and site-directed mutagenesis
| Primer | Sequence (5’–3’) |
|---|---|
| pEcNFeoB | Forward: GCAGCAGCACATATGATGAAAAAATTAACCATTGGCTTAATTG |
| Reverse: TGCTGCTGCAAGCTTTACCGCAGTGGTGAAACGGCTGGGTTC |
|
| pStNFeoB | Forward: GGAATTCCATATGATGAAAAAATTAACCATTGGTTTAattg |
| Reverse: CCCAAGCTTGACCGCCCTGGTGAAGCGGCTGG | |
| pPaNFeoB | Forward: GGGAATTCCATATGATGACCGCATTGACc |
| Reverse: CCCAAGCTTGAGCCATTGGGTCA | |
| pSmNFeoB | Forward: GCAGCACATATGATGACAGAAATTGCTTTGATTG |
| Reverse: TGCTGCAAGCTTAATTTTATCAGAAGCA | |
| pSaNFeoB | Forward: GCAGCAGCTAGCATGGAAAATTATTGTATTTTAGG |
| Reverse: TGCTGCAAGCTTTATTCTAGAGCTGAAATATTGCT | |
| pBcNFeoB | Forward: Reverse: CTAGCTAGCaTGAATAAGGTAGCTTTGCTA |
| pLmNFeoB | Forward: GGGAATTCCATATGATGAGTCAAAATACATATTGCTTAC |
| Reverse: ACGCGTCGACGAGTTTATCGGATAGTGGAATA | |
| pEcNFeoB-N11A | Forward: AATTGGTGCCCCAAATTCTGGCAAGACAACG |
| Reverse: GCCAGAATTTGGGGCACCAATTAAGCCAATG | |
| pSmNFeoB-N11A | Forward: GGGAATCCCGCCAGCGGAAAGACAAGTTTA |
| Reverse: CTTTCCGCTGGCGGGATTCCCAATCAAAG |
Protein expression and purification
For overexpression of each protein, the respective plasmids were transformed into E. coli BL21(DE3) cells (Merck, Darmstadt, Germany) under ampicillin selection. Transformed cells were grown in LB medium at 37°C in a rotary shaker at 200 rpm and were induced with 0.5 mM IPTG at OD600 0.4–0.5. After overnight induction, the cells were harvested by centrifugation at 7,500 g for 5 min at 4°C. Cell pellets were suspended in 2 ml of 25 mM Tris-HCl (pH 8.0) containing 100 mM NaCl and 1 mM phenylmethylsulfonyl fluoride (PMSF) per gram wet weight. The suspended cells were lysed by sonication, and supernatant was collected by centrifugation at 30,000 g for 30 min at 4°C. The histidine-tagged proteins were purified on a Ni2+-charged His-Bind resin (Millipore, Burlington, MA, USA) using 20 mM imidazole as the wash buffer, and gradually increasing imidazole concentration (40 mM to 250 mM), as the elution buffer, in 25 mM Tris-HCl (pH 8.0) containing 100 mM NaCl, 10% (v/v) glycerol, and 10 mM beta-mercaptoethanol.
Purity of the protein samples was estimated by SDS-PAGE stained with Coomassie brilliant blue R-250. The total protein concentration was determined using Biorad protein assay dye reagent (Biorad, Hercules, CA, USA).
Nucleotide hydrolysis assay
A Malachite Green-based colorimetric assay was used to measure inorganic phosphate released by NFeoB nucleotide hydrolysis activity [15]. In most cases, enzymes (0.5 or 1 μM) were mixed with various concentrations of nucleotides in 20 mM Tris, pH 8.0, 5 mM MgCl2, and 200 mM KCl and incubated at 37°C for 1 hour.
For determining the effect of monovalent cations, 200 mM of each salt (LiCl, NaCl, NH4Cl, and RbCl) was added instead of KCl, and enzymes were mixed with 650 μM of ATP or GTP. For divalent cations, 5 mM of each salt (CoCl2, MnCl2, FeSO4, and CaCl2) was added instead of MgCl2, and enzymes were mixed with 650 μM of ATP or GTP. The effect of temperature on nucleotide hydrolysis was tested at 16, 23, 30, 37, and 44°C with 650 μM of ATP or GTP in the presence of 200 mM KCl and 5 mM MgCl2. The concentrations of monovalent and divalent cations (200 mM and 5 mM, respectively) were chosen as representative physiological concentrations in bacteria.
The reaction was terminated by adding the Malachite Green solution in 1 M HCl and incubating at room temperature (23°C) for 30 min. The absorbance at 620 nm was measured using a xMark microplate reader (BioRadad, Hercules, CA, USA). For the determination of kinetic parameters, the initial rate data were fitted to the Michaelis-Menten equation by nonlinear regression using GraphPad Prism 8. All experiments were performed in triplicate.
Sequence alignment and molecular docking prediction
Sequence alignment of bacterial NFeoBs was conducted using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) [16]. A figure was prepared using ESPript web-based server (http://espript.ibcp.fr/ESPript/ESPript/) [17]. Homology modeling of SmNFeoB was performed using the fully automated structure modeling method implemented in the SWISS-MODEL workspace (http://swiss-model.expasy.org/) [18]. Interactions of proteins and ligands in 2-dimensions were conducted using PoseView (BioSolveIT, Germany) [19].
RESULTS
Bacterial FeoBs are classified into two distinct groups by nucleotide specificity
E. coli FeoB contains G-protein motifs and only possesses GTP hydrolytic activity [11]. Bacterial FeoBs have been widely recognized as GTPases, and most research has focused on the GTP hydrolysis mechanism. However, in our recent study, we showed that V. cholerae FeoB contains a dual nucleotide-specific NTPase domain essential for ferrous iron uptake [3]. This result was also the case for H. pylori FeoB.
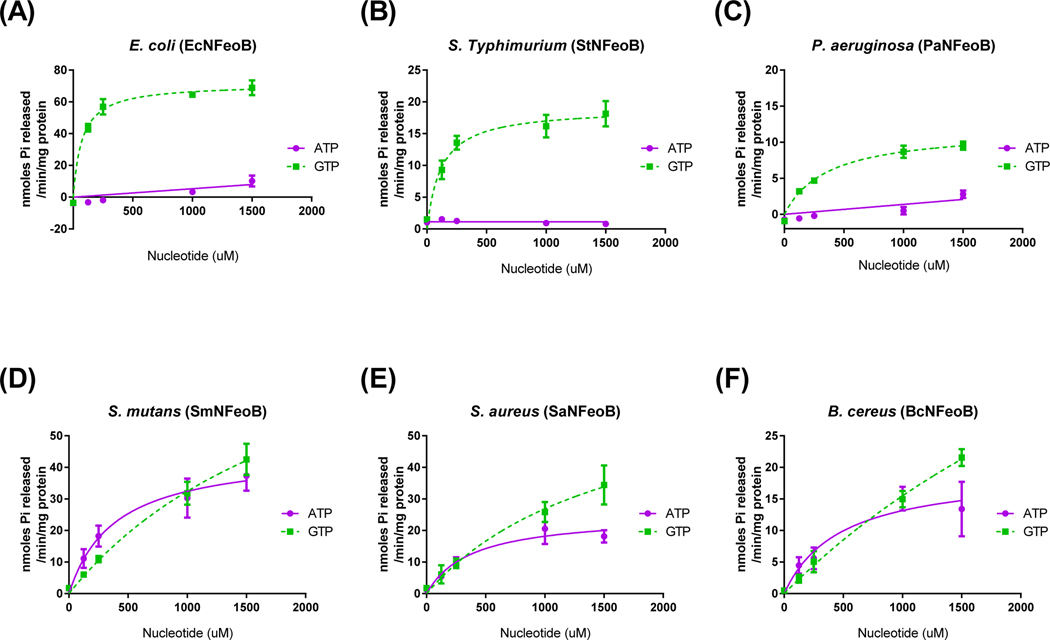
In order to broaden the enzymatic characteristics of pathogenic bacterial Feo systems especially present in foods and environments, we compared nucleotide specificity of the cytosolic N-terminus of FeoBs (NFeoBs) originated from various bacterial strains including Escherichia coli (EcNFeoB), Streptococcus mutans (SmNFeoB), Staphylococcus aureus (SaNFeoB), Bacillus cereus (BcNFeoB), Salmonella enterica serovar Typhimurium (StNFeoB) and Pseudomonas aeruginosa (PaNFeoB). Interestingly, bacterial NFeoBs were clearly divided into two groups possessing solely GTP-dependent or both ATP- and GTP-dependent hydrolases (Fig. 1). Like E. coli, S. Typhimurium and P. aeruginosa exhibited only GTP hydrolytic activity. However, the NFeoB activities of S. aureus, S. mutans, and B. cereus, which are Gram-positive strains, were specific to both ATP and GTP, and their ATP preference was even higher than that of NFeoBs from V. cholerae or H. pylori (refer to Shin et al. [3]). While catalytic efficiency for ATP could not be determined at saturating conditions due to the high absorbance of substrate in V. cholerae and H. pylori NFeoBs, the values for ATP in S. mutans, S. aureus and B. cereus NFeoBs could be determined (6.18 × 10−5, 3.63 × 10−5 and 2.13 × 10−5 s−1μM−1, respectively). These results indicated that individual bacterial FeoBs may utilize different energy-generating sources involving ATP and GTP for ferrous iron transport.
Figure 1.
Bacterial FeoBs are classified into two separate groups by nucleotide specificity. The reaction rates for E. coli (A), S. Typhimurium (B), P. aeruginosa (C), S. mutans (D), S. aureus (E), and B. cereus (F) FeoBs with ATP (●) and GTP (■) as substrate were measured as described in Materials and Methods. The error bars represent the standard deviation.
Two groups of bacterial FeoBs exhibit different potassium-dependent activation
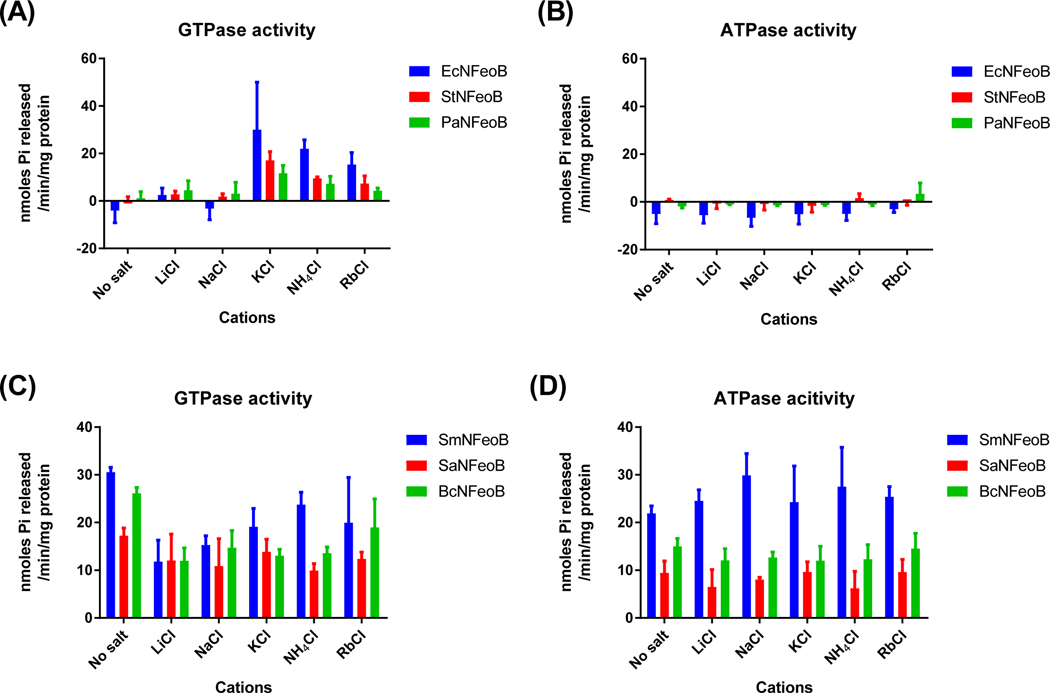
GTPase activity of FeoB in E. coli is stimulated by potassium, which may bind at a site adjacent to the GTP-binding pocket [12,13]. Unlike E. coli NFeoB, GTPase and ATPase activities of V. cholerae NFeoB are not stimulated by potassium [3]. To test whether different nucleotide specificity reflects divergent monovalent cation-dependent activation, we analyzed ATP and GTP hydrolytic activities of bacterial FeoBs with addition of 200 mM Li+, Na+, K+, NH4+, or Rb+. As expected, the GTP-specific group containing E. coli, S. Typhimurium and P. aeruginosa NFeoBs showed much higher GTPase activity with the addition of 200 mM KCl compared with sodium ions or no additional salt (Fig. 2A, B). Rb+ and NH4+, which have similar or larger ionic radii to K+, also significantly stimulated the GTPase activity of NFeoBs. No ATPase activity of EcNFeoB, StNFeoB and PaNFeoB in the presence of monovalent cations was observed, even with the addition of potassium.
Figure 2.
Two groups of bacterial FeoBs show different potassium-dependent activation. While the GTPase activity of the GTP-specific group was highly stimulated by 200 mM potassium or ammonium ions (A), the NTP-specific group was not stimulated by cations (C and D). The GTP-specific group did not exhibit ATPase activity with addition of any cations (B). The enzyme reaction buffer alone (no salt) contained 5 mM MgCl2 and 10 mM NaCl. The error bars represent the standard deviation.
Similar to V. cholerae FeoB, the NTP-specific group containing SmNFeoB, SaNFeoB, and BcNFeoB did not exhibit monovalent cation preference for its nucleotide hydrolysis activity (Fig. 2C). GTPase activity of these NFeoBs was higher without addition of any monovalent cations (P < 0.01 by one-way ANOVA for SmNFeoB and BcNFeoB) or was unaffected by monovalent cations (P = 0.24 for SaNFeoB). However, ATPase activity of the NFeoBs was similar in the presence or absence of these ions (P = 0.54, 0.45, and 0.55 by one-way ANOVA for SmNFeoB, SaNFeoB, and BcNFeoB, respectively). These findings suggested structural differences in the active sites between the GTP-specific and NTP-specific groups. Also, these differences implied that additional residues besides the G-protein motifs may play important roles in nucleotide hydrolysis followed by in vivo iron transport.
Two groups of bacterial FeoBs show different divalent metal-dependent activation
In general, divalent cations, especially magnesium ions, are required for proper NTP hydrolytic activity by coordinating β- and γ-phosphates [20]. These metal ions differ in the extent of their interactions with the nucleic bases and in their coordination within a specific enzyme conformation [21]. To investigate interactions involving metal ions with the ATP and GTP bases coordinated within FeoB, we compared the effects of divalent metal ions on NTP hydrolytic activity of each bacterial NFeoB.
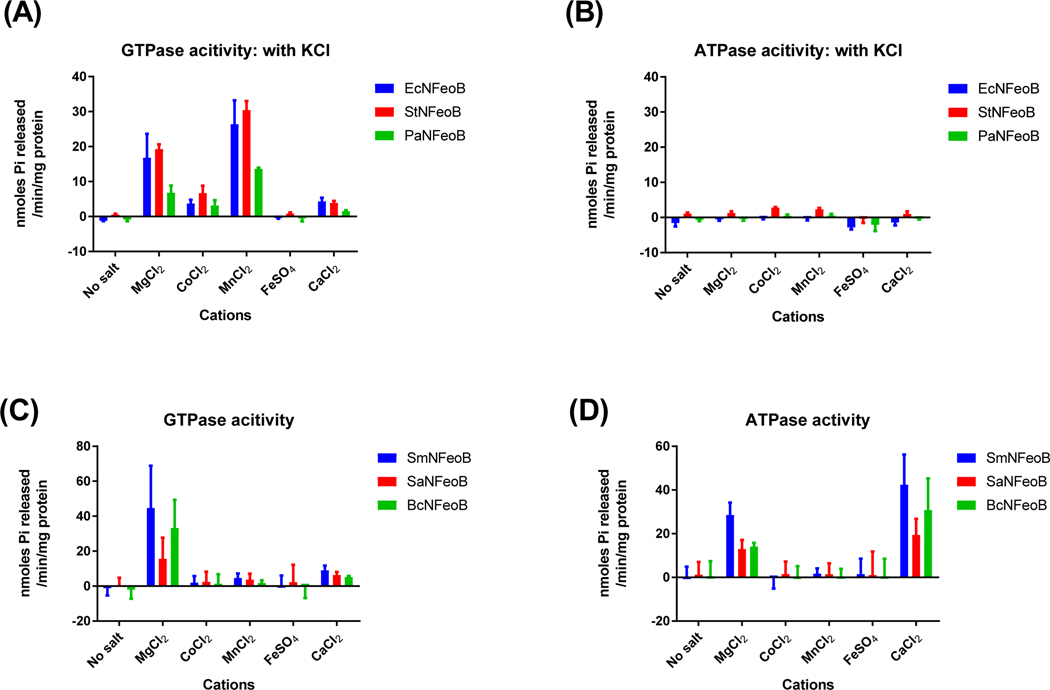
Without addition of potassium, the ATPase or GTPase activity in the group of GTP-specific NFeoBs was not stimulated by any metal ions (Fig. S1). The ATPase activity of PaNFeoB was high with Mn2+, but the difference was not statistically significant (by one-way ANOVA, P = 0.15). With the addition of potassium, the GTPase activity of the GTP-specific NFeoBs was accelerated by Mg2+ or Mn2+ (Fig. 3A, B). In particular, Mn2+ showed slightly higher activation compared to Mg2+ (P < 0.01 for StNFeoB and PaNFeoB; P = 0.16 for EcNFeoB by Student’s t-test).
Figure 3.
Two groups of bacterial FeoBs show different divalent metal-dependent activation. While the GTPase activity of the GTP-specific group was highly stimulated by 5 mM Mg2+ or Mn2+ ions (A), the NTP-specific group was stimulated only by Mg2+ for GTPase (C) and Ca2+ for ATPase (D). The GTP-specific group did not show ATPase activity with addition of any divalent ions (B). The enzyme reaction buffer contained either 10 mM NaCl and 200 mM KCl for the GTP-specific group (A and B), or only 10 mM NaCl for the NTP-specific group (C and D). The error bars represent the standard deviation.
Interestingly, the group of NTP-specific NFeoBs indicated different divalent metal-dependent activation to the GTP-specific NFeoBs, even for the GTPase activity (Fig. 3C, D). It is noted that the NTPase assay of NFeoBs in this group was conducted without addition of KCl because their activities were not dependent on monovalent cations. Unlike the acceleration of GTP hydrolysis by Mg2+ or Mn2+ as shown in the GTP-specific group, the activity of the dual nucleotide-specific group was affected by addition of only Mg2+. Its ATPase activity was stimulated by Mg2+ or Ca2+. From these results, we inferred that the NTP-specific group differs from the GTP-specific group not only in its possession of ATPase activity, but also in the catalytically productive conformation of the active sites.
Mutation of Asn11 alters nucleotide-specificity of bacterial NFeoBs
Asn11 is a conserved asparagine residue in bacterial FeoBs, as well as in potassium-dependent G-proteins [12]. This residue is critical for the coordination of potassium at the nucleotide-binding site and subsequent potassium-dependent activation [13]. However, in this study, the NTP-specific NFeoBs were not stimulated by any specific monovalent cations. Therefore, we hypothesized that mutation of Asn11 may result in loss of GTP hydrolysis activity but not ATP hydrolysis.
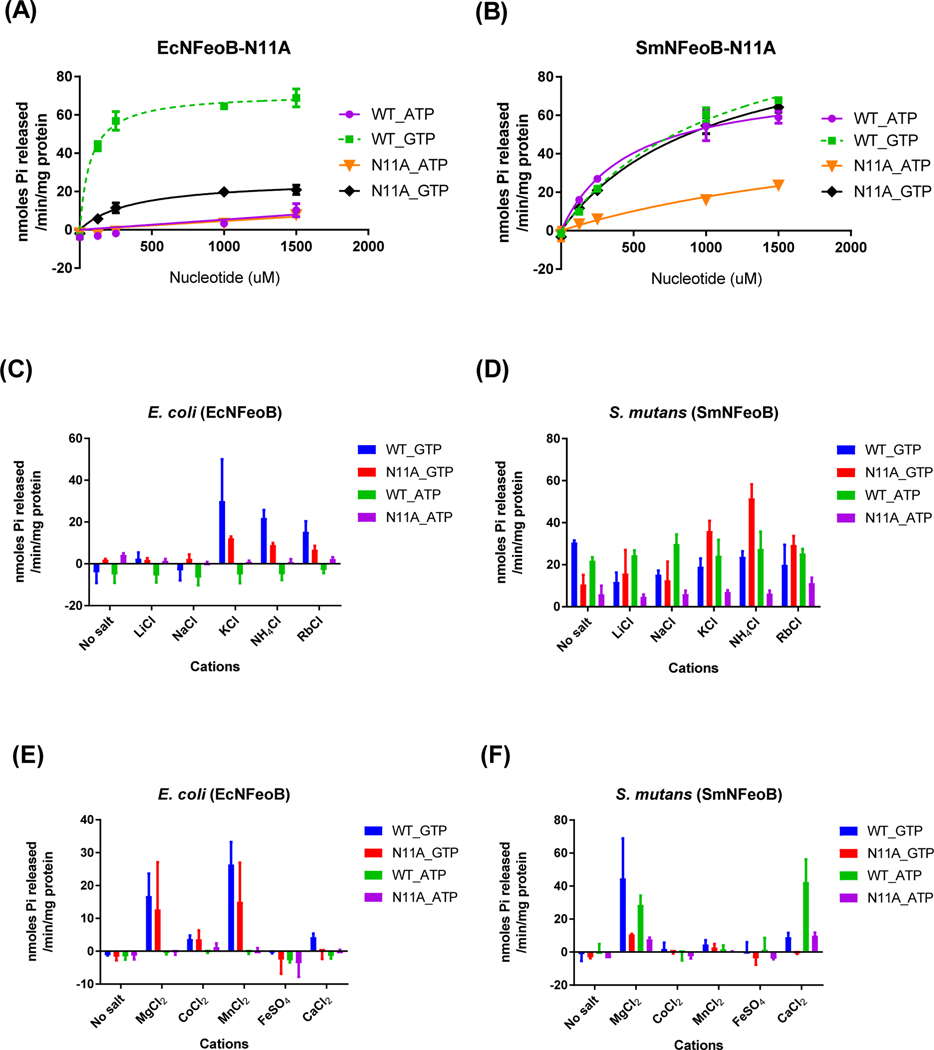
To test whether Asn11 mediates potassium binding in the groups of bacterial FeoBs, we mutated Asn11 to alanine in the GTP-specific E. coli NFeoB (EcNFeoB-N11A) and the NTP-specific S. mutans NFeoB (SmNFeoB-N11A) and analyzed their nucleotide hydrolysis specificity and cation dependence (Fig. 4). As expected, EcNFeoB-N11A mutant exhibited a drop in GTPase activity (decreased by 13.4-fold for catalytic efficiency compared to the wild type). Its ATPase activity was same as the activity of the wild type. However, in contrast to EcNFeoB-N11A, SmNFeoB-N11A mutant was shown to display similar GTPase activity to that of the wild type, whereas its ATPase activity was significantly diminished (decreased by 6.8-fold for catalytic efficiency compared to the wild type).
Figure 4.
Mutation of Asn11 alters nucleotide-specificity of bacterial FeoBs. The reaction rates for E. coli-N11A mutant (A) and S. mutans-N11A mutant (B) with ATP (● and ▼) and GTP (■ and ◆) as substrates were measured as described in Materials and Methods. Cation dependence profiles were not changed by the mutation in EcNFeoB (C), whereas the N11A mutant of SmNFeoB became more dependent on potassium (D). There was no difference in the profile of divalent cation dependence between the wild type and mutant in both EcNFeoB and SmNFeoB (E and F). (C and D) The enzyme reaction buffer alone (no salt) contained 5 mM MgCl2 and 10 mM NaCl. (E and F) The enzyme reaction buffer contained either 10 mM NaCl and 200 mM KCl (E) or only 10 mM NaCl (F). The error bars represent the standard deviation.
In order to discern if the altered nucleotide specificity also affects mono- and di-valent cation dependence, we compared the effects of cations on NTPase activity of wild type and N11A mutants of EcNFeoB and SmNFeoB. As shown in Fig. 4, GTP hydrolysis of the EcNFeoB mutant was significantly diminished compared to wild type, but the cation dependence profile, including monovalent and divalent cations, was unchanged. The NTPase activity of SmNFeoB, as one of the NTP-specific NFeoBs, was not stimulated by any monovalent cations but, interestingly, the N11A mutant became more dependent on potassium. There was no difference in the profile of divalent cation dependence between the wild type and mutant of SmNFeoB. Taken together, these results suggested that potassium dependence is a key determinant for the nucleotide specificity in FeoB, and changing a single amino acid associated with potassium binding was able to alter the nucleotide specificity.
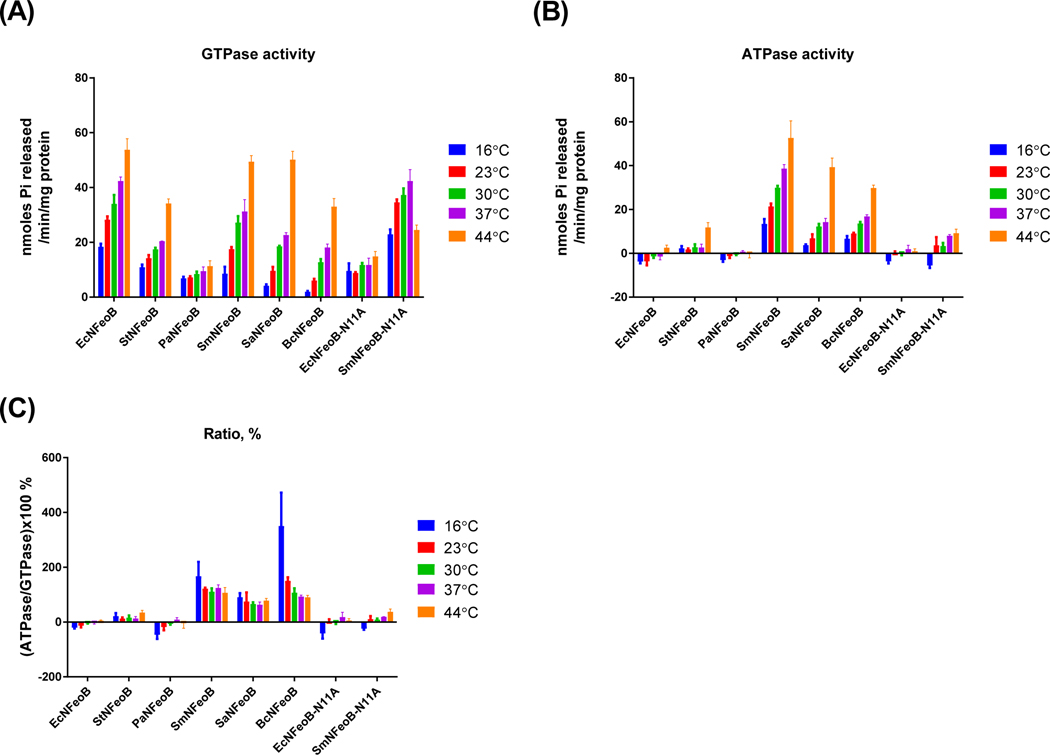
Nucleotide preference of bacterial NFeoBs is not affected by temperature
Bacteria colonize different environments in terms of nutrient sources, energy, and temperature throughout their lifecycle [22,23]. Some bacterial enzymes, such as a multidrug ABC transporter (PatA/PatB) from Streptococcus pneumoniae and an ABC transporter (CvaB) from E. coli, show adjustment of nucleotide preference for their activity at different temperatures [23,24]. Therefore, we decided to investigate the effect of temperature on the nucleotide specificity of bacterial NFeoBs (Fig. 5). All of the NFeoBs, except for SmNFeoB-N11A, showed enhanced GTP as well as ATP hydrolytic activity with increasing temperature. Accordingly, the percent ratio of ATPase versus GTPase activities was not significantly changed. Exceptionally, B. cereus NFeoB revealed increased preference for ATP than GTP with decreasing temperature. B. cereus has the ability to grow at temperatures between 4 to 5 and 10°C [25]. It is speculated that B. cereus FeoB may change its nucleotide specificity during adaptation to cold environments.
Figure 5.
Nucleotide-specificity of FeoBs are not dependent on temperature. GTPase activity (A) and ATPase activity (B) of bacterial FeoBs was measured with 650 μM of each nucleotide at different temperatures. Nucleotide preference was calculated as a percentage ratio of ATPase activity divided by GTPase activity of each FeoB (C). The error bars represent the standard deviation.
Phylogenetic analysis and sequence alignment of bacterial FeoBs
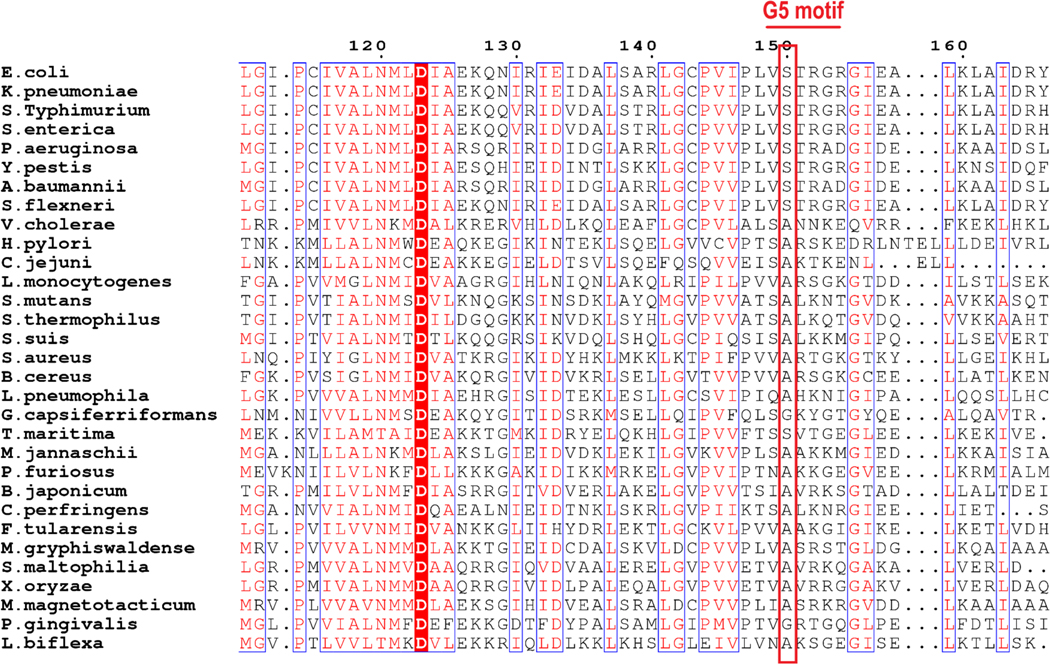
Sequence alignment of the cytosolic domain of prokaryotic FeoB proteins showed that the residue at position 148 in V. cholerae (Ser148) can be separated into either polar (serine, asparagine, and glutamine) or nonpolar (valine and isoleucine) amino acids in the aligned sequences [3]. E. coli FeoB, a member of the GTP-specific group, has valine in this position, whereas V. cholerae and H. pylori, members in the dual nucleotide-specific group, have serine instead. Based on the findings in the current study, we conducted phylogenetic analysis of NFeoBs as well as whole FeoB proteins and compared amino acid sequences.
As shown in Fig. S2, both the cytosolic domain and whole FeoB sequences of GTP-specific bacteria were grouped as gamma-proteobacteria, while NTP-specific bacteria were present in various bacterial phyla. It is noteworthy that the protein sequence of V. cholerae FeoB, a gamma-proteobacterium, is situated between the two groups in accordance with its lower ATP hydrolytic activity compared to the activity of SmNFeoB, SaNFeoB, and BcNFeoB. In order to investigate conservation of the serine residue in the G5 motif, we aligned protein sequences of FeoBs with respect to the G-protein motifs (Fig. 6). In contrast to our speculation surrounding serine conservation in the NTP-specific group, SaNFeoB and BcNFeoB have valine, as does E. coli FeoB, ruling out the possible conservation of serine for ATP hydrolysis. Alternatively, we found conservation of alanine in the NTP-specific group (Ala149 in VcNFeoB) and serine as the corresponding residue in the GTP-specific group (Ser150 in EcNFeoB).
Fig. 6.
Serine/alanine in the G5 motif is conserved in accordance with the nucleotide specificity of FeoB. Sequence alignment of the G5 motif from bacterial FeoB proteins illustrated a weak sequence conservation in the region. The second residue in the G5 motif (red box) is alanine in the NTP-specific group, or serine in the GTP-specific group. Bacterial strains in the analysis were selected based on publications where their FeoB was studied. A figure was prepared using ESPript web-based server (http://espript.ibcp.fr/ESPript/ESPript/) [17].
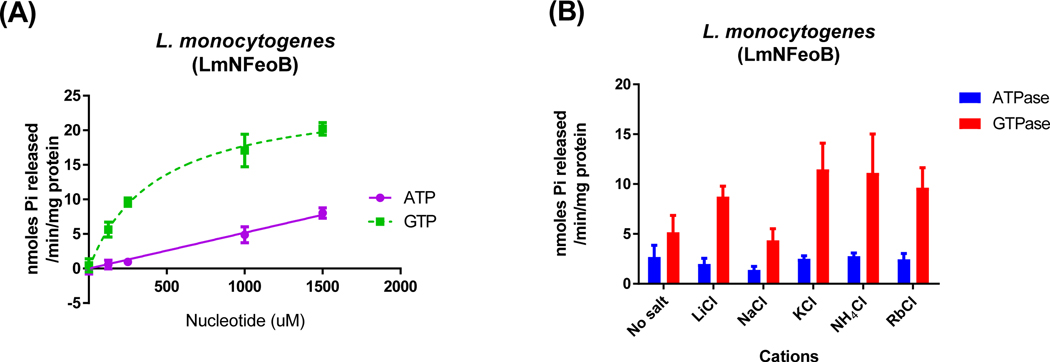
To confirm the sequence similarity and alanine conservation in the NTP-specific group, we purified the cytosolic domain of L. monocytogenes FeoB (LmNFeoB) as a possible candidate of the dual nucleotide-specific group and analyzed its enzymatic properties (Fig. 7). LmNFeoB exhibited ATP hydrolysis activity similar to the activity of VcNFeoB, but not as much as other gram-positive bacterial FeoBs. Its GTPase activity was higher than other monovalent cations but did not increase greatly with potassium compared to the activity of the GTP-specific FeoBs. This result implied that the sequence similarity and conserved alanine in the G5 motif may be determinants of nucleotide specificity. However, these attributes are not necessarily required for the ATP-dependent activity and associated potassium dependence and hence warrant extensive structural investigations.
Fig. 7.
L. monocytogenes NFeoB possesses NTP-specific NTPase activity. The reaction rates for L. monocytogenes NFeoB (A) with ATP (● and ▼) and GTP (■ and ◆) as substrates were measured as described in Materials and Methods. LmNFeoB did not show dependence on any monovalent cations (B), similar to the dual nucleotide specific bacterial FeoBs. The enzyme reaction buffer alone (no salt) contained 5 mM MgCl2 and 10 mM NaCl. The error bars represent the standard deviation.
DISCUSSION
The Feo ferrous iron transporter has major roles in bacterial survival and virulence in mammalian hosts. Despite its importance, mechanistic details of the iron transport process, including the source of energy involved, remain unanswered. In this study, we showed that bacterial FeoBs are present as two distinct groups which possess either sole GTPase or NTPase activity (Fig. 1). This difference in nucleotide specificity altered the requirements for monovalent and divalent cations (Figs. 2 and 3). Mutation of Asn11, having a role in potassium-binding and potassium-dependent GTP hydrolysis, abolished potassium-dependent GTPase activation in E. coli. However, the same mutation in S. mutans did not act on GTPase activity. Instead, S. mutans FeoB largely lost its ATPase activity resulting from the mutation (Fig. 4). Sequence analysis suggested a possible association of alanine in the G5 motif for the NTP-specificity in FeoBs (Fig. 6).
Two groups of bacterial FeoBs classified by nucleotide specificity
The Feo system was first identified as an ATPase based on sequence alignments with those of other ATP-utilizing enzymes, further supported by ATP hydrolysis inhibitor studies [6,26,27]. However, in 2002, Marlovits et al. showed that E. coli FeoB contains a guanine-nucleotide-specific binding site which is required for efficient ferrous iron uptake through the FeoB-dependent system [11]. Since then, FeoB has been widely considered to be a GTPase. In our previous study, we demonstrated for the first time that V. cholerae FeoB possesses ATP as well as GTP hydrolysis activities. This ATPase activity was sufficient for functional ferrous iron transport in V. cholerae. Here, we expanded the coverage of the bacterial FeoBs, especially present in foods and environments, to elucidate the biochemical characteristics of NFeoBs. On the contrary that most studies of bacterial FeoBs have focused on their GTPase activity, our results pointed out that the sole GTP-specific FeoBs are limited to gamma-proteobacteria while bacterial species in broader phyla have NTP-specific FeoBs (Fig. 1 and S2). In addition, we also found that V. cholerae FeoB, one of the gamma-proteobacteria studied in our previous research, is situated between the two groups in accordance with its lower ATP hydrolytic activity compared to the activity of other NTP-specific FeoBs.
Requirement of monovalent cations for FeoB NTPase activity
P-loop nucleoside triphosphatases (NTPases) represent the most common protein fold in cellular organisms [20]. The P-loop domains are characterized by two conserved motifs, known as the Walker A and B motifs, where the N-terminal β-strand and α-helix form an elongated flexible loop for binding the NTP’s phosphate chain [28]. These domains are typically activated by an arginine “finger” or sometimes by monovalent cations, especially potassium. The arginine “finger” promotes GTP hydrolysis by rotating the α-phosphate with respect to β- and γ-phosphates towards an eclipsed conformation, which may lower the activation energy barrier produced by the repulsion between the oxygen atoms of all three phosphate groups [29,30].
The potassium-dependent P-loop NTPases including TEES (TrmE-Era-EngA-YihA-Septin) GTPases, YchF and FeoB have been suggested to possess an ancestral trait subsequently replaced by reliance on arginine fingers [20,31,32]. Two conserved asparagines in the P-loop, another following it (GxxNxGKSxLxN), and an insertion of a specific potassium-binding ‘K-loop’ in Switch I are suggested as attributes of GTPases employing a potassium-mediated mechanism [29]. FeoB conserves these attributes, implying that the potassium-mediated mechanism may be operative. In the present study, we showed that FeoBs are not necessarily potassium-dependent, although FeoBs have highly conserved attributes (Fig. 2). Rafay et al. reported that some enzymes having the attributes of potassium-mediated GTP hydrolysis do not use potassium to accelerate GTP hydrolysis [29]. It still remains unclear whether a universal catalytic mechanism can be applied to all P-loop NTPases since there is continuum of transition states in the multitude of GTP-binding proteins [20,33]. From further detailed structural and mechanistic investigations of bacterial FeoBs, we expect to provide more comprehensive understanding of the catalytic mechanisms of the different P-loop NTPases.
Effect of Asn11 mutation on FeoB NTPase activity
A conserved asparagine residue (Asn11 in EcNFeoB and SmNFeoB) is critical for coordination of the potassium ion and induction of a structural bend in the Switch I loop so that it caps the nucleotide binding site [12,34]. Biochemical analyses of NFeoB from S. thermophilus demonstrate that mutation of Asn11 abolishes potassium-binding and potassium-dependent activation [12]. Similar to results in S. thermophilus NFeoB, we found that EcNFeoB showed a decrease in its GTP hydrolysis activity by the mutation of Asn11 to alanine (Fig. 4). However, in contrast to the N11A mutant of S. thermophilus FeoB, the cation-dependent activation was lost, and the EcNFeoB mutant retained its potassium-accelerated activity. In addition to the EcNFeoB mutation, we found that the corresponding mutation of SmNFeoB altered its nucleotide specificity by diminishing ATP hydrolysis activity. Interestingly, this alteration also modified cation-dependent GTP hydrolysis. It is noteworthy that the sequence identity of S. mutans and S. thermophilus FeoB is 77.4 % and 77.3 % for NFeoB and whole FeoB, respectively. It is still unclear why the enzymatic properties of S. mutans and S. thermophilus FeoBs are different despite the high sequence similarity. Different reaction conditions or other differences among bacterial strains might affect enzymatic characteristics. L. monocytogenes FeoB showed unexpectedly lower ATP preference than other Gram-positive bacteria, although it has a highly similar protein sequence to those strains (Fig. 7). Further structural analysis and prediction of possible determinants will be required.
Requirement of divalent cations for FeoB NTPase activity
Our data suggested that two groups of bacterial FeoBs exhibit different divalent metal-dependent activation (Fig. 3). While the GTP-specific FeoBs are either Mg2+- or Mn2+-dependent, GTPase activity of the dual nucleotide-specific FeoBs is only Mg2+-dependent. In P-loop NTPases, Mg2+ is coordinated by a threonine from the G1 motif, two trans-waters, and one oxygen atom each from the β- and γ-phosphates of the nucleotide [35]. Mg2+ and Mn2+ ions can replace each other in active centers of many enzymes, including DNA and RNA polymerases, phosphatases and kinases [36]. Structural comparison of Mg2+- and Mn2+-dependent enzymes indicates that both metal cations are bound in similar structural motifs [36]. However, Mg2+ is a harder Lewis acid than Mn2+, so Mg2+ is usually bound to oxygen, while Mn2+ is bound to nitrogen or sulfur atoms [36]. These differences may slightly change the environments of metal-binding sites, depending on the different nucleoside-specific groups. Hence, stimulation of ATP hydrolysis by addition of Ca2+ to the NTP-specific group members should be possible.
A role of G5 motif in FeoB nucleotide specificity
In our previous study on V. cholerae FeoB, we suggested that Ser148 and Asn150 in VcNFeoB may be candidates for residues that determine nucleotide specificity [3]. However, from the current results, it seems that these residues are not distinguishable between the two different nucleotide-specific groups. Instead, we found that the second amino acid residue in the G5 motif, which is serine (in the GTP-specific group; S150 in EcNFeoB) or alanine (in the NTP-specific group, A149 and A143 in VcNFeoB and SmNFeoB, respectively), is greatly conserved, at least among the bacterial strains tested in this study (Fig. 6).
The residue in the G5 loop plays a key role in nucleotide affinity and GDP release rate [35,37]. Serine in the position causes a rapid intrinsic GDP release rate, while the corresponding alanine decelerates GDP release rate. This change is thought to result from altering the nucleotide-protein hydrogen-bonding pattern, displacing the nucleotide base, and imparting reduced GDP affinity. More specifically, when the position has alanine, an Asn residue interacts with the GDP O6 and N7 groups by hydrogen-bonding. Meanwhile, when the position has serine, the Asn residue is stabilized by electrostatic interactions with the hydroxyl group of the serine residue, leading to a weaker nucleotide-protein affinity for the GDP to be released [37]. ATP and GTP have different functional groups in their base, which are 6-amino purine in ATP and 2-amino 6-oxo purine in GTP. Based on structural information, it is speculated that alanine in the G5 motif compared to serine would provide a higher propensity for ATP to interact with amino acid residues in FeoB.
Structural implications of ATP and GTP binding with FeoBs
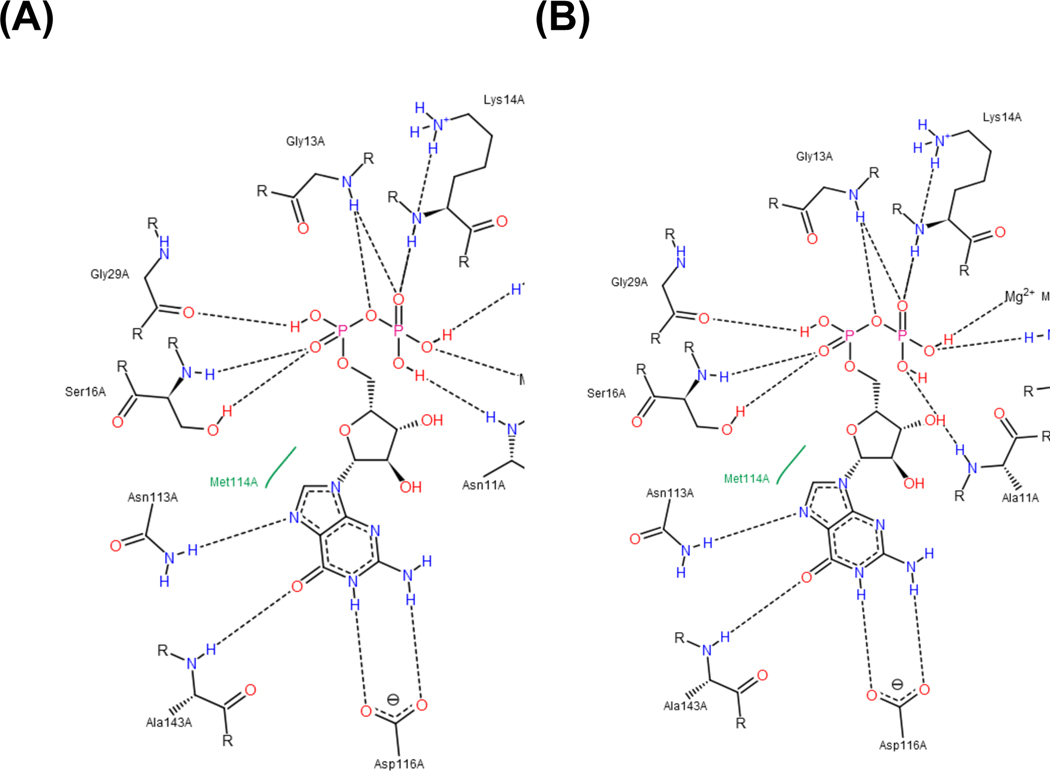
P-loop NTPases work as molecular switches between the GDP-bound “OFF” and the GTP-bound “ON” state [34]. When GDP is bound, the Switch I loop moves into its active conformation, then subsequently potassium ion promotes linking of alpha- and gamma-phosphates of the nucleotide triphosphate chain [20]. This leads to rotation of the gamma-phosphate group yielding an almost eclipsed, catalytically productive conformation of the nucleotide.
In this study, we propose: 1) adenine and guanine nucleotide bases may have different interactions with the G5 motif, and 2) the triphosphate groups of ATP and GTP may have different positional contacts with Switch I. Fig. 8 shows a positional view of GDP-binding in homology models of the SmNFeoB wild type and N11A mutant predicted from a crystal structure of S. thermophilus NFeoB (PDB ID: 3SS8). The guanine base contacts with Asn113, Ala143, and Asp116. In particular, Ala143 and Asp116 provide GTP-specificity of the nucleotide by hydrogen-bonding with 2-amino and 6-hydroxy groups, whereas the adenine base (6-aminopurine) does not interact with these residues.
Fig. 8.
Binding pocket of SmNFeoB wild type (A) and N11A mutant (B) with GDP. In the PoseView diagram, black dashed lines indicate hydrogen bonds, salt bridges, and metal interactions. Green solid lines show hydrophobic interactions. Interactions were determined by geometric criteria, as previously described in [19].
Changes in the purine-binding site can induce conformational modification in the FeoB structure. This leads to the different shapes of the triphosphate chain, affecting the catalytic activity of nucleotide hydrolysis, including interaction with switch regions, and coordination with attacking water molecules. In the present study, mutation of Asn11 caused alteration of the nucleotide specificity for SmNFeoB so as to be a sole GTPase. It is notable that the modification of a single amino acid residue beyond the purine base-binding site was able to change the entire nucleotide specificity of the protein.
Physiological implications of the two groups of FeoBs
Why might prokaryotic FeoBs possess these two distinct nucleotide specificities? Generally, Gram-positive bacteria are postulated as the earliest prokaryotes from which both archaebacteria and Gram-negative bacteria evolved [38]. In this study, we identified that Gram-positive and some Gram-negative bacterial FeoBs have the NTP-specificity, while a subset of gamma-proteobacteria appear to have lost the ability to use ATP. In addition, the cytosolic domain of FeoB is homologous to human Ras G-protein which is GTP-specific and thus has been referred to as a “living fossil” of this family of eukaryotic GTPases [39]. Based on these findings, we speculate that NTP-specific (or promiscuous nucleotide-specific) FeoB might have emerged before the divergence of bacteria and archaea, then its substrate specificity might have been directed to GTP during the process of evolution.
Why, then, has the evolutionary process been directed to GTP-specific and subsequently potassium-stimulated hydrolysis activity? Iron transport into the cell is required to be tightly regulated. Meanwhile, the utilization, or at least binding, of purine nucleotides is highly energy-consuming for the iron transport system. In terms of the consideration of environmental habitats of the Gram-negative bacteria studied here, iron availability might be limited, and potassium availability might increase in the host environment.
The Feo system plays an important role in ferrous iron acquisition in numerous bacterial species. Our results clearly indicated that this family of transporters is more diverse and more specific, according to certain bacterial species. This demonstration of distinct enzymatic properties of prokaryotic FeoBs provides important insights into mechanistic details of the Feo iron transport process, as well as offering a promising species-specific anti-virulence target.
Supplementary Material
Funding
This research was supported by the C1 Gas Refinery Program (2016M3D3A1A01913268) and the Basic Science Research Program (NRF-2019R1I1A1A01058125), both through the National Research Foundation of Korea.
Abbreviations
- Feo
ferrous iron transporter
- Fur
ferric-uptake regulator
- GDI
GDP dissociation inhibitor
- NFeoB
cytosolic N-terminus of FeoB
- EcNFeoB
Escherichia coli NFeoB
- VcNFeoB
Vibrio cholerae NFeoB
- HpNFeoB
Helicobacter pylori NFeoB
- SmNFeoB
Streptococcus mutans NFeoB
- SaNFeoB
Staphylococcus aureus NFeoB
- BcNFeoB
Bacillus cereus NFeoB
- StNFeoB
Salmonella enterica serovar Typhimurium NFeoB
- PaNFeoB
Pseudomonas aeruginosa NFeoB
- LmNFeoB
Listeria monocytogenes NFeoB
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest regarding the contents of this article.
References
- [1].Payne SM, Mey AR, Wyckoff EE, Vibrio Iron Transport: Evolutionary adaptation to life in multiple environments, Microbiol. Mol. Biol. Rev 80 (2016) 69–90. 10.1128/MMBR.00046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Frawley ER, Fang FC, The ins and outs of bacterial iron metabolism, Mol Microbiol. 93 (2014) 609–616. 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shin M, Mey AR, Payne SM, Vibrio cholerae FeoB contains a dual nucleotide-specific NTPase domain essential for ferrous iron uptake, Proc. Natl. Acad. Sci. U.S.A (2019). 10.1073/pnas.1817964116. [DOI] [PMC free article] [PubMed]
- [4].Peng ED, Wyckoff EE, Mey AR, Fisher CR, Payne SM, Nonredundant roles of iron acquisition systems in Vibrio cholerae, Infect Immun. 84 (2016) 511–523. 10.1128/IAI.01301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weaver EA, Wyckoff EE, Mey AR, Morrison R, Payne SM, FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB, J. Bacteriol 195 (2013) 4826–4835. 10.1128/JB.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Velayudhan J, Hughes NJ, McColm AA, Bagshaw J, Clayton CL, Andrews SC, Kelly DJ, Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter, Molecular Microbiology. 37 (2000) 274–286. 10.1046/j.13652958.2000.01987.x. [DOI] [PubMed] [Google Scholar]
- [7].Pérez NM, Ramakrishnan G, The reduced genome of the Francisella tularensis live vaccine strain (LVS) encodes two iron acquisition systems essential for optimal growth and virulence, PLoS ONE. 9 (2014) e93558. 10.1371/journal.pone.0093558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Aranda J, Cortés P, Garrido ME, Fittipaldi N, Llagostera M, Gottschalk M, Barbé J, Contribution of the FeoB transporter to Streptococcus suis virulence, Int. Microbiol 12 (2009) 137–143. [PubMed] [Google Scholar]
- [9].Lau CKY, Krewulak KD, Vogel HJ, Bacterial ferrous iron transport: the Feo system, FEMS Microbiol. Rev 40 (2016) 273–298. 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- [10].Beauchene NA, Mettert EL, Moore LJ, Keleş S, Willey ER, Kiley PJ, O2 availability impacts iron homeostasis in Escherichia coli, Proc. Natl. Acad. Sci. U.S.A 114 (2017) 12261–12266. 10.1073/pnas.1707189114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM, The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria, Proc. Natl. Acad. Sci. U.S.A 99 (2002) 16243–16248. 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ash M-R, Guilfoyle A, Clarke RJ, Guss JM, Maher MJ, Jormakka M, Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B, J. Biol. Chem 285 (2010) 14594–14602. 10.1074/jbc.M110.111914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ash M-R, Maher MJ, Guss JM, Jormakka M, The structure of an N11A mutant of the G-protein domain of FeoB, Acta Cryst F. 67 (2011) 1511–1515. 10.1107/S1744309111042965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xia Y, Chu W, Qi Q, Xun L, New insights into the QuikChangeTM process guide the use of Phusion DNA polymerase for site-directed mutagenesis, Nucleic Acids Res. 43 (2015) e12–e12. 10.1093/nar/gku1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Quan A, Robinson PJ, Rapid purification of native dynamin I and colorimetric GTPase assay, in: Methods in Enzymology, Academic Press, 2005: pp. 556–569. 10.1016/S00766879(05)04049-8. [DOI] [PubMed] [Google Scholar]
- [16].Sievers F, Higgins DG, Clustal Omega, accurate alignment of very large numbers of sequences, Methods Mol. Biol 1079 (2014) 105–116. 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- [17].Gouet P, Courcelle E, Stuart DI, Métoz F, ESPript: analysis of multiple sequence alignments in PostScript, Bioinformatics. 15 (1999) 305–308. 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- [18].Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res. 46 (2018) W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stierand K, Rarey M, Drawing the PDB: Protein-ligand complexes in two dimensions, ACS Med Chem Lett. 1 (2010) 540–545. 10.1021/ml100164p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shalaeva DN, Cherepanov DA, Galperin MY, Golovin AV, Mulkidjanian AY, Evolution of cation binding in the active sites of P-loop nucleoside triphosphatases in relation to the basic catalytic mechanism, ELife. 7 (2018) e37373. 10.7554/eLife.37373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Amsler PE, Sigel H, Comparison of the metal-ion-promoted dephosphorylation of the 5’-triphosphates of adenosine, inosine, guanosine and cytidine by Mn2+, Ni2+ and Zn2+ in binary and ternary complexes, Eur J Biochem. 63 (1976) 569–581. 10.1111/j.1432-1033.1976.tb10261.x. [DOI] [PubMed] [Google Scholar]
- [22].Smoot LM, Smoot JC, Graham MR, Somerville GA, Sturdevant DE, Migliaccio CAL, Sylva GL, Musser JM, Global differential gene expression in response to growth temperature alteration in group A Streptococcus, PNAS. 98 (2001) 10416–10421. 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Orelle C, Durmort C, Mathieu K, Duchêne B, Aros S, Fenaille F, André F, Junot C, Vernet T, Jault J-M, A multidrug ABC transporter with a taste for GTP, Scientific Reports. 8 (2018) 2309. 10.1038/s41598-018-20558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhong X, Tai PC, When an ATPase is not an ATPase: at low temperatures the C-terminal domain of the ABC Transporter CvaB is a GTPase, J Bacteriol. 180 (1998) 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].de Sarrau B, Clavel T, Clerté C, Carlin F, Giniès C, Nguyen-The C, Influence of anaerobiosis and low temperature on Bacillus cereus growth, metabolism, and membrane properties, Appl Environ Microbiol. 78 (2012) 1715–1723. 10.1128/AEM.06410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kammler M, Schön C, Hantke K, Characterization of the ferrous iron uptake system of Escherichia coli., Journal of Bacteriology. 175 (1993) 6212–6219. 10.1128/jb.175.19.62126219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sestok AE, Linkous RO, Smith AT, Toward a mechanistic understanding of Feo-mediated ferrous iron uptake, Metallomics. 10 (2018) 887–898. 10.1039/C8MT00097B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Walker J. e., Saraste M, Runswick M. j., Gay N. j., Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold., The EMBO Journal. 1 (1982) 945–951. 10.1002/j.14602075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rafay A, Majumdar S, Prakash B, Exploring potassium-dependent GTP hydrolysis in TEES family GTPases, FEBS Open Bio. 2 (2012) 173–177. 10.1016/j.fob.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mittal R, Ahmadian MR, Goody RS, Wittinghofer A, Formation of a transition-state analog of the Ras GTPase reaction by Ras·GDP, tetrafluoroaluminate, and GTPase-activating proteins, Science. 273 (1996) 115–117. 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- [31].Mulkidjanian AY, Bychkov AY, Dibrova DV, Galperin MY, Koonin EV, Origin of first cells at terrestrial, anoxic geothermal fields, PNAS. 109 (2012) E821–E830. 10.1073/pnas.1117774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dibrova DV, Galperin MY, Koonin EV, Mulkidjanian AY, Ancient systems of sodium/potassium homeostasis as predecessors of membrane bioenergetics, Biochemistry Mosc. 80 (2015) 495–516. 10.1134/S0006297915050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wittinghofer A, Phosphoryl transfer in Ras proteins, conclusive or elusive?, Trends in Biochemical Sciences. 31 (2006) 20–23. 10.1016/j.tibs.2005.11.012. [DOI] [PubMed] [Google Scholar]
- [34].Wittinghofer A, Vetter IR, Structure-function relationships of the G Domain, a canonical switch motif, Annual Review of Biochemistry. 80 (2011) 943–971. 10.1146/annurevbiochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- [35].Guilfoyle A, Maher MJ, Rapp M, Clarke R, Harrop S, Jormakka M, Structural basis of GDP release and gating in G protein coupled Fe2+ transport, The EMBO Journal. 28 (2009) 2677–2685. 10.1038/emboj.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Khrustalev VV, Barkovsky EV, Khrustaleva TA, Magnesium and manganese binding sites on proteins have the same predominant motif of secondary structure, Journal of Theoretical Biology. 395 610 (2016) 174–185. 10.1016/j.jtbi.2016.02.006. [DOI] [PubMed] [Google Scholar]
- [37].Guilfoyle AP, Deshpande CN, Vincent K, Pedroso MM, Schenk G, Maher MJ, Jormakka M, Structural and functional analysis of a FeoB A143S G5 loop mutant explains the accelerated GDP release rate, The FEBS Journal. 281 (2014) 2254–2265. 10.1111/febs.12779. [DOI] [PubMed] [Google Scholar]
- [38].Gupta RS, The natural evolutionary relationships among prokaryotes, Crit. Rev. Microbiol 26 (2000) 111–131. 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- [39].Hantke K, Is the bacterial ferrous iron transporter FeoB a living fossil?, Trends in Microbiology. 11 (2003) 192–195. 10.1016/S0966-842X(03)00100-8. [DOI] [PubMed] [Google Scholar]
- [40].Grant SG, Jessee J, Bloom FR, Hanahan D, Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants, Proc. Natl. Acad. Sci. U.S.A 87 (1990) 4645–4649. 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Studier FW, Moffatt BA, Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes, J. Mol. Biol 189 (1986) 113–130. 10.1016/00222836(86)90385-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.