Abstract
The liver possesses extraordinary regenerative capacity mainly attributable to the ability of hepatocytes (HCs) and biliary epithelial cells (BECs) to self-replicate. This ability is left over from their bipotent parent cell, the hepatoblast, during development. When this innate regeneration is compromised due to the absence of proliferative parenchymal cells, such as during cirrhosis, HCs and BEC can transdifferentiate; thus, adding another layer of complexity to the process of liver repair. In addition, dysregulated lineage maintenance in these two cell populations has been shown to promote malignant growth in experimental conditions. Here, malignant transformation, driven in part by insufficient maintenance of lineage reprogramming, contributes to end-stage liver disease. Epigenetic changes are key drivers for cell fate decisions as well as transformation by finetuning overall transcription and gene expression. In this review, we address how altered DNA methylation contributes to the initiation and progression of hepatic cell fate conversion and cancer formation. We also discussed the diagnostic and therapeutic potential of targeting DNA methylation in liver cancer, its current limitations, and what future research is necessary to facilitate its contribution to clinical translation.
Keywords: Liver progenitor cell, cholangiocarcinoma, hepatocellular carcinoma, transdifferentiation, epigenetics, cellular reprogramming
Introduction
The liver is a central organ that maintains systemic tissue homeostasis by governing diverse metabolic functions including nutrient metabolism, detoxification and production/secretion of various enzymes(Monga 2015, Russell and Monga 2018). These numerous and complex tasks are facilitated by a single epithelial cell type, the “hepatocyte (HC)”, which accounts for more than 90% of the liver parenchyma. Aside from HCs, another parenchymal epithelial cell in the liver is the cholangiocyte (biliary epithelial cell, BEC), accounting for 5–7% of the total liver parenchyma. BECs accept HC-produced bile acid and transport it to the intestine through the common bile duct(Ko, Russell et al. 2019). Combined, these two epithelial cell populations account for more than 95% of hepatic parenchymal cells.
Although HCs and BECs have a dichotomy in morphology, size, function, and transcriptome in the adult liver; moreover, both are derived from a bi-potent parent cell called a “hepatoblast (HB)” during liver development. Additionally, both cell types contribute to liver regeneration primarily by self-replication as shown in the mammalian 2/3 partial hepatectomy (PHx) model(Hu and Monga 2021, Michalopoulos and Bhushan 2021). The entire mechanism is tightly orchestrated by numerous signaling pathways along with epigenetic regulators to initiate and halt parenchymal cell replication(Michalopoulos 2017, Macchi and Sadler 2020). These transient, but strictly controlled, transcriptional reprogramming events allocate limited energy to cell cycle responses and/or basal metabolic roles in the repopulating HCs(Arechederra, Berasain et al. 2020).
Unlike the 2/3 PHx model, chemical/genetic insults result in dying parenchymal cells in the regenerating tissue microenvironment. This leads to a completely different compensatory response in the liver. While intact HCs and BECs initiate self-replication (similar to the 2/3 PHx model), duct-like cells, which express BEC markers, are activated and subsequently proliferate and expand from the peri-portal region. This phenomenon results in the activation and expansion of bipotent liver progenitor cells (LPCs) generating both HCs and BECs(Ko, Russell et al. 2019, Gadd, Aleksieva et al. 2020). Unless differentiated into adult cells, active LPCs promote inflammation and stromal growth by secreting various cytokines and chemokines. Indeed, the degree of LPC activity has been demonstrated to correlate positively with liver disease severity in mouse and human(Ko, Russell et al. 2019). In transdifferentiation between HCs and BECs, epigenetic regulators determine which genes to turn on or off during regeneration and results in chromatin packing as either eu- or hetero-chromatin(Arechederra, Berasain et al. 2020, Aloia 2021, Basu and Tiwari 2021). Therefore, modulating chromatin condensation is critical for distributing transcription machinery accessibility and, as a result, transcriptional activation.
Theoretically, transdifferentiation into functional HCs and BECs are considered to only occur in end-stage cirrhotic liver when the liver has lost the ability to regenerate(Raven, Lu et al. 2017, Schaub, Huppert et al. 2018, Ko, Russell et al. 2019, Russell, Lu et al. 2019, Gadd, Aleksieva et al. 2020). Indeed, cirrhosis is counted as irreversible; 10–20% of patients progress to liver cancer without transplantation. In the cirrhotic liver, dying HCs and BECs have strong stromal expansion with irreversible fibrosis, inflammation, and continuous injury; all of which may contribute to an oncogenic microenvironment. Here, cell fate changes via chromatin structure and epigenome remodeling can potentially increase risk for malignant transformation(Sandhu, Shire et al. 2008, Nebbioso, Tambaro et al. 2018). In this regard, the Zender26 and Loude27 groups have demonstrated HC transformation into different lineage fates via distinct epigenetic reprogramming can contribute to oncogenic transformation in the liver. Particularly, experimentally proven HC-derived cholangiocarcinoma (CCA, malignant bile duct cancer), LPC-derived hepatocellular carcinoma (HCC, malignant HC cancer), and mixed ICC/HCC indicate simultaneous cellular identity reprogramming and malignant transformation(Sandhu, Shire et al. 2008, Aloia 2021, Hu, Molina et al. 2022, Ko, Kim et al. 2022). Here, identifying epigenetic regulators will be key to reduce malignant transformation of activated/primed LPCs.
In the cirrhotic liver, lineage conversion and malignant transformation are both dependent on a delicate/pathologic im/balance of transcriptional activation and repression. This balance is regulated by both transcription machinery and epigenetic modifiers. First, chromatin remodelers modify the nucleosome to close and/or expose gene loci of transcription regulation complexes. Next, a variety of repressors, enhancers, and activators are recruited to perform the necessary duty to execute lineage commitment(Greenberg and Bourc’his 2019, Macchi and Sadler 2020, Martinez-Redondo and Izpisua Belmonte 2020, Aloia 2021, Basu and Tiwari 2021). In general, epigenetic regulation for chromatin remodeling consists of 1) post-translational histone modifications such as acetylation, methylation, phosphorylation, amination, etc; 2) exchange of core histones with histone variants; 3) action of various non-coding RNAs; and 4) DNA methylation of CpG islands. These mechanisms directly impact cell fate determination. Of these, DNA methylation is among the most studied and is carried out by a single enzyme family called DNA methyltransferases (DNMTs)(Sandhu, Shire et al. 2008, Greenberg and Bourc’his 2019). DNMTs have well-known transcriptional repressor functions; these enzymes typically repress tumor suppressors and/or senescence inducers during cancer development. Additionally, DNMTs are also reported to regulate the expression of fate commitment genes. Furthermore, in addition to cell-intrinsic effects, modulation of DNMTs are also implicated in the tumor microenvironment, particularly with regards to immune responses(Segovia, San Jose-Eneriz et al. 2019, Zhang, Yang et al. 2020, Hu, Liu et al. 2021).
Overall, delineating epigenetic mechanisms for cell plasticity and subsequent malignant transformation would be fundamental since preventative modulation of key elements might reverse the irreversible pathologic progression of end-stage liver diseases into a treatable stage. In this review, we will focus on the role of DNA methylation, one of the key epigenetic transcription regulators, in these two distinct but crucial cellular events. The clinical implications of altering DNA methylation in human liver cancer will also be discussed from diverse perspectives.
Basics of DNA methylation
DNA methylation is an epigenetic modification catalyzed by DNA methyltransferases, which add a methyl group to the 5’-carbon of the pyrimidine ring of cytosine. 5-methylcytosine (5mC) is mainly found in the palindromic CpG dinucleotides (Zemach, McDaniel et al. 2010). The high-density of CpG regions are called “CpG islands” (CGIs). DNA methylations are commonly localized in intergenic regions, transposons, or gene bodies; however, CpG islands are located at the promoter regions and are enriched in over two-thirds of mammalian promoters (Larsen, Gundersen et al. 1992, Deaton and Bird 2011). Although CGIs at promoters are hot spots for DNA methylation, there are two types of unmethylated CGI promoters, which are regulated by histone modification: 1) transcriptionally activated CGI promoters are enriched in trimethylated histone H3 Lys4 (H3K4me3) (Piunti and Shilatifard 2016), and 2) transcriptionally inactive CGI promoters show enrichment of Polycomb repressive complex 2 (PRC2)-mediated trimethylated histone H3 Lys27 (H3K27me3), which are subject to reactivate gene expression response to environmental stress (Marasca, Bodega et al. 2018). Figure 1A illustrates the domain structure of DNMTs and figure 2 depicts three distinct DNA methylation processes, enzymes involved, and transcriptional consequences. DNMT family consists of DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L in the human genome (Fig. 1A). DNMT1, DNMT3A, and DNMT3B are canonical DNMT enzymes that show methyltransferase activity. However, DNMT2 and DNMT3L are non-canonical DNMT enzymes with a lack of activity because of the loss of or truncated catalytic domain.
Figure 1. Domain structure of the human DNMT and TET family proteins.
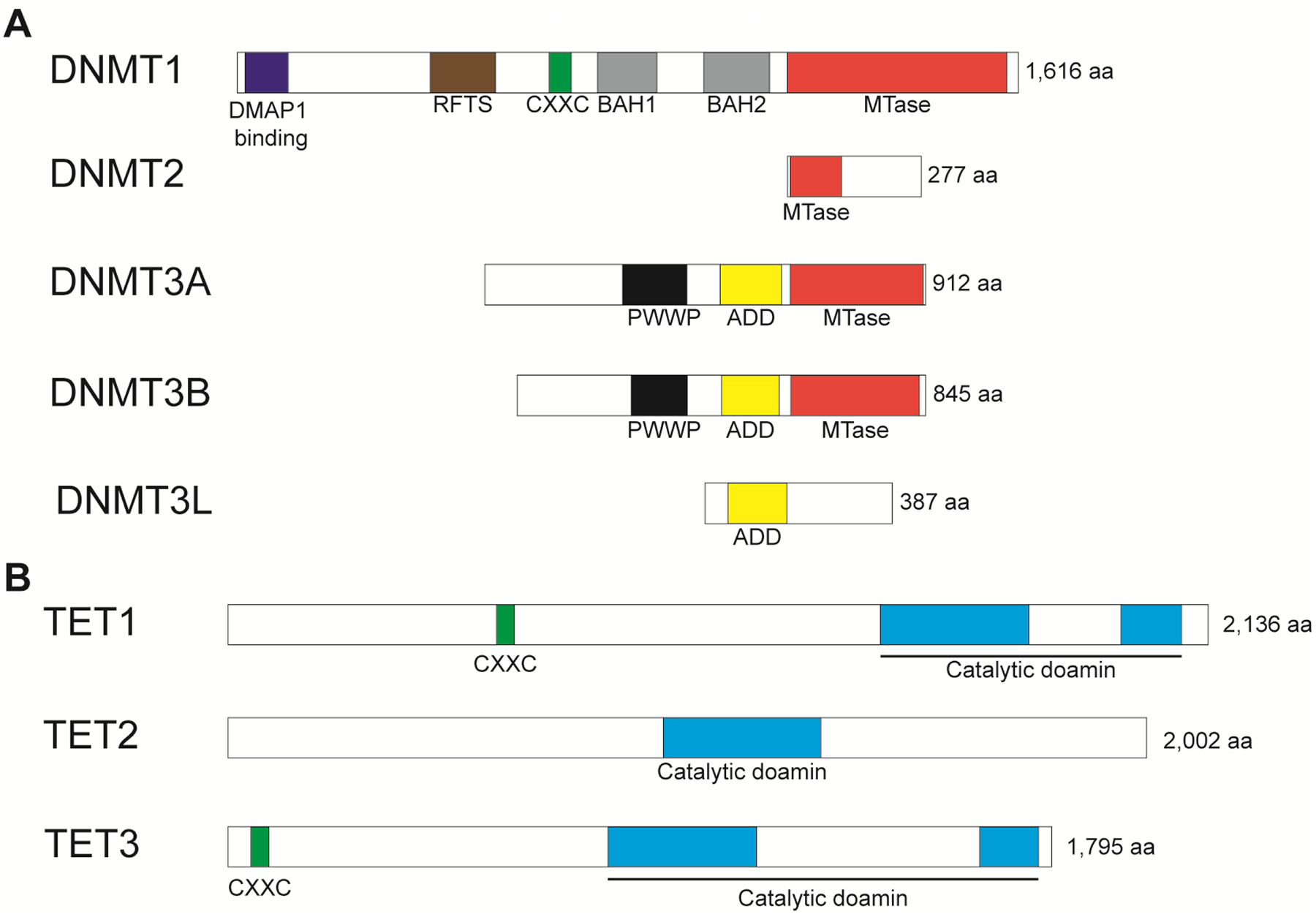
(A) Conserved domains of DNMTs showing DMAP1 binding, RFTS, CXXC, BAH, PWWP, ADD, and MTase domains. Canonical DNMTs, which are DNMT1, DNMT3A, and DNMT3B, retaining catalytic domain MTase, however, non-canonical DNMTs, which are DNMT2 and DNMT3L, showing truncated and loss of catalytic domain. (B) Conserved domains of TETs showing CXXC and catalytic domains.
Figure 2. The machinery and mechanism of DNA methylation.
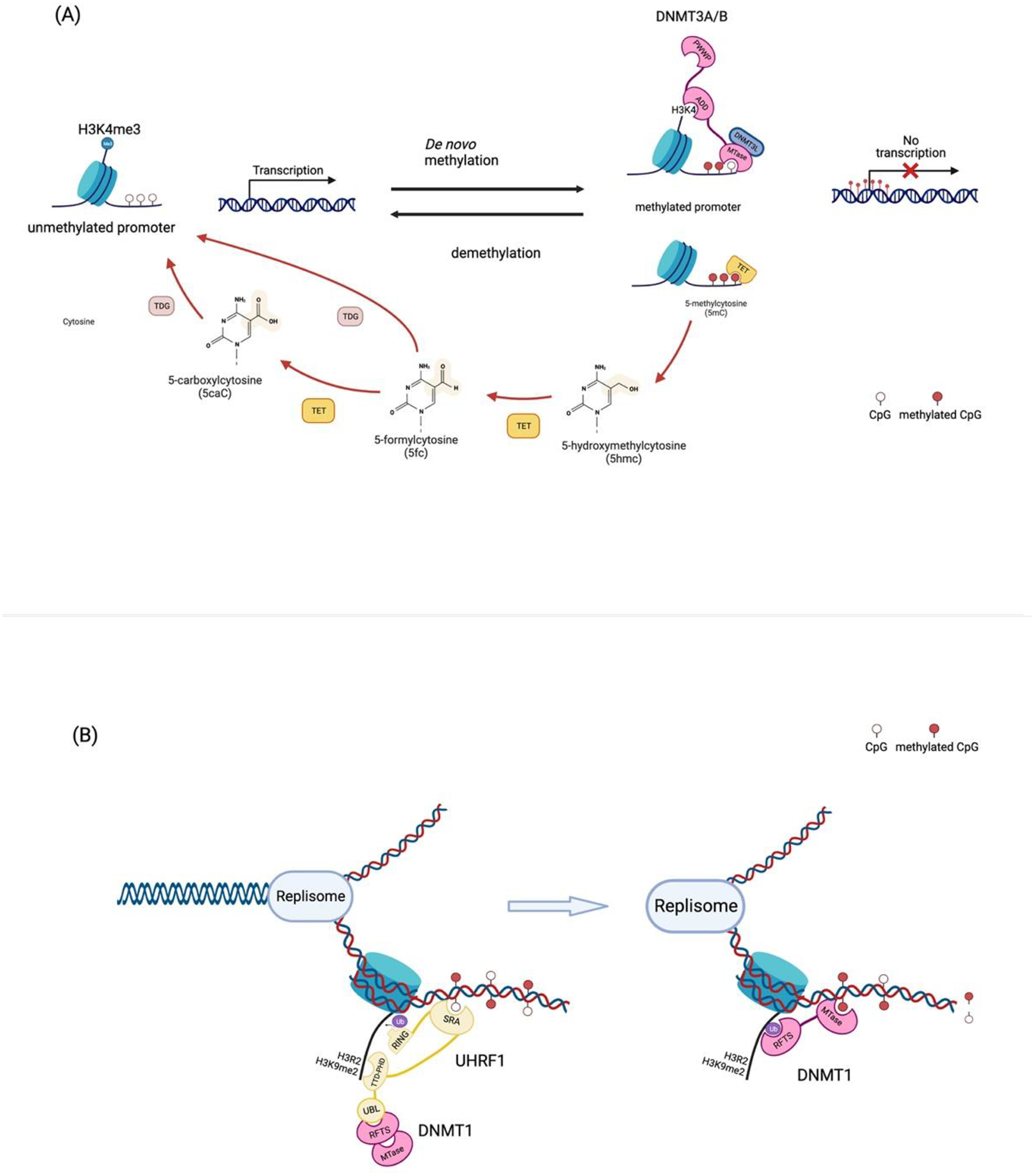
(A) de novo DNA methylation and active demethylation at the promoter region. Once the methyl groups at H3K4, which marks active/poised promoters, are removed, ADD domain of DNMT3A/B binds to H3K4. This allows the methyltransferase domain to methylate 5-cytosine of DNA. The methylated CpG island of the promoter prevents transcriptional activation. The methylated cytosine (5mC) can be catalyzed by TETs. TETs progressively oxidize 5mC to 5hmC, 5fC, and 5caC. Thymine DNA glycosylase (TDG) also carries out 5fC and 5 acC oxidization via deamination. (B) DNA methylation maintenance. TTD-PHD domain of UHRF1 can recognize and bind to H3R2 and H3K9me2. Recruitment of UHRF1 allows the SRA domain, which can recognize hemimethylated DNA, to the target site. This produces the RING domain that drives ubiquitination on the histone tail. This ubiquitination offers the docking site for RFTS of DNMT1. Binding of DNMT1 on the target site makes it possible that MTase methylates the hemimethylated DNA.
There are three types of regulatory processes in DNA methylation: 1) de novo DNA methylation, 2) maintenance of symmetrical methylation at replication foci, and 3) demethylation which are driven by different enzymes with distinct molecular structures (Fig. 2). DNMT3A/B are essential enzymes for de novo methylation that is largely guided by histone tails. Structurally, DNMT3 contains three functional domains: Pro-Trp-Trp-Pro (PWWP) domain; an ATRX-DNMT3L-DNMT3A (ADD) domain; and a C-terminal catalytic domain. The PWWP domain can bind to H3K36me3 that is enriched in the gene body of actively transcribed genes; this regulates the balance of transcription and DNA methylation in the gene bodies(Ge, Pu et al. 2004, Wagner and Carpenter 2012). The PWWP point mutation impairs the restriction of extra methylation by blocking DNMT3 anchoring to the H3K36 regions (Heyn, Logan et al. 2019, Sendzikaite, Hanna et al. 2019). The ADD domain binds to H3K4 and is repulsed by H3K4me3, a histone modification present in active promoters (Ooi, Qiu et al. 2007, Otani, Nankumo et al. 2009, Zhang, Jurkowska et al. 2010). Once the ADD domain binds to H3K4, the catalytic domain at C-terminus, the MTase domain, is released and catalyzes the DNA methylation (Guo, Wang et al. 2015). The unbound ADD domain can interact with the MTase domain and cause the autoinhibition of DNMT3 enzymes. The binding of ADD and PWWP to modified histone tails is critical for DNMT3 to identify specific target regions which is critical for toggling de novo methylation on and off. In DNMT3A/B-mediated de novo methylation, catalytically inactive DNMT3L is required for methyltransferase activity via stabilizing DNMT3A(Veland, Lu et al. 2019); specific inhibition of DNMT3L impairs overall de novo methylation activity(Jia, Jurkowska et al. 2007).
While de novo DNA methylation can take place in any CpG context across the genome, symmetrical (both strands of DNA) CpG methylation is only maintained during DNA replication by the enzyme DNMT1 in collaboration with another multidomain protein, the E3 ubiquitin-protein ligase UHRF1. Except for the conserved catalytic domain at the C-terminus, like other DNMTs, DNMT1 has unique domains which are specifically required for the maintenance of symmetrical DNA methylation(Chen and Zhang 2020): 1) DNMT1-associated protein 1 (DMAP1) binding domain, 2) a proliferating cell nuclear antigen (PCNA) binding domain, 3) a replication foci-targeting sequence (RFTS) domain, 4) a CXXC domain, and 5) two Bromo-adjacent homology (BAH) domains(Li and Zhang 2014). Each domain has a specific role and interaction partners. DMAP1 binding domain can interact with the transcriptional repressor DMAP1 and histone deacetylase 2 (HDAC2) (Rountree, Bachman et al. 2000). The PCNA binding domain and RFTS domain contribute to localizing DNMT1 to replication foci during the S phase (Chuang, Ian et al. 1997, Nishiyama, Yamaguchi et al. 2013, Qin, Wolf et al. 2015, Ishiyama, Nishiyama et al. 2017). RFTS also provides DNMT1 autoinhibitory activity by sequestering in the enzyme’s catalytic domain (Song, Rechkoblit et al. 2011, Takeshita, Suetake et al. 2011, Ishiyama, Nishiyama et al. 2017). The CXXC domain can bind to unmethylated CpGs (Pradhan, Esteve et al. 2008, Song, Rechkoblit et al. 2011). Lastly, BAH domains are involved in the localization of DNMT1 to the replication foci (Yarychkivska, Shahabuddin et al. 2018). Despite having its own DNA methyltransferase activity, UHRF1 is required for DNMT1 to retain methyltransferase function in replicating cells (Bostick, Kim et al. 2007, Sharif, Muto et al. 2007). UHRF1 is a multidomain protein that consists of a ubiquitin-like (UBL) domain, a tandem TUDOR-PHD (TTD-PHD) domain, a SET-and RING-associated (SRA) domain, and a really interesting new gene (RING) domain (Xie and Qian 2018). The TTD-PHD domains contribute to the symmetrical methylation of hemimethylated DNA through binding to H3R2 and H3K9me2 or me3 (Nady, Lemak et al. 2011, Arita, Isogai et al. 2012, Rothbart, Krajewski et al. 2012, Rothbart, Dickson et al. 2013). Importantly, the SRA domain of UHRF1 confers specific binding to DNMT1 for hemimethylated DNA at the replication foci during S-phase (Bostick, Kim et al. 2007, Sharif, Muto et al. 2007, Arita, Ariyoshi et al. 2008, Avvakumov, Walker et al. 2008, Hashimoto, Horton et al. 2008). When the SRA domain binds to hemimethylated DNA this induces allosteric changes in the RING domain, thereby activating its E3 ligase activity. This results in the accumulation of mono-ubiquitin at the H3K14, H3K18, and H3K23 (Nishiyama, Yamaguchi et al. 2013, Qin, Wolf et al. 2015); hence, generating the specific binding site for the RFTS domain of DNTM1. Consequently, the emergence of the binding site prevents the RFTS domain from masking the active site of DNMT1’s catalytic domain, together diminishing the autoinhibitory effect of DNMT1 (Ishiyama, Nishiyama et al. 2017). Furthermore, it has been discovered that UHRF1 can directly recruit DNMT1 to chromatin through the UBL domain, implying the importance of the role of UHRF1 in DNMT1-dependent DNA methylation(Li, Wang et al. 2018). Therefore, chemical/genetic inhibition of DNMT1, or UHRF, similarly impairs the maintenance of methylation status in replicating cells thereby provoking malignant transformation in various cells(Mudbhary, Hoshida et al. 2014, Kong, Chen et al. 2019). As aforementioned, once specific CpG regions are methylated, transcription of target genes is repressed. This occurs in tandem with diverse chromatin remodelers such as lymphocyte-specific helicase (LSH), H3K9 methyltransferase, histone deacetylases(Fuks, Burgers et al. 2000, Dennis, Fan et al. 2001, Fuks, Burgers et al. 2001, Deplus, Brenner et al. 2002, Esteve, Chin et al. 2006, Epsztejn-Litman, Feldman et al. 2008, Myant, Termanis et al. 2011, Tao, Xi et al. 2011), methyl-CpG-binding domain (MBD) 6 and MeCP2(Ren, Horton et al. 2018). The detailed molecular process behind methylated DNA-mediated transcriptional repression is thoroughly described in other reviews(Greenberg and Bourc’his 2019).
Given the importance of rapid and tight regulation of transcription, like other epigenetic regulations, DNA methylation is a reversible molecular event triggered by TET proteins. TET is a family of methylcytosine dioxygenases involved in the processes of oxidative demethylation of 5mC. TET enzymes catalyze the oxidation of 5mC to 5hmC, which is progressively oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Kriaucionis and Heintz 2009, Tahiliani, Koh et al. 2009, Ito, D’Alessio et al. 2010, He, Li et al. 2011, Ito, Shen et al. 2011, Wu and Zhang 2014). TET family consists of TET1, TET2, and TET3 (Fig. 1B). The catalytic domain at the C-terminus of TET consists of a double-stranded β-helix (DSBH) domain and a cysteine-rich domain(Pastor, Aravind et al. 2013). In addition, the full-length of TET1 and TET3 contain a CXXC domain, which binds to unmethylated CpG dinucleotides(Xu, Xu et al. 2012). This confers the specificity of TETs to bind CGIs(Xu, Wu et al. 2011). However, other TET proteins which lack the CXXC domain such as TET1s, TET2, TET3o, or TET3s, need additional factors to be recruited to chromatin. Therefore, DNA demethylation can occur by multiple facets such as DNA replication-dependent dilution of DNA methylation (Howell, Bestor et al. 2001), and also TET enzymatic activity.
In baseline liver, DNMT1 expression is strongly detected in all CD45+ immune cells, confirming important roles for hepatic immune response(Wang, Malnassy et al. 2021). While all HCs showed faint DNMT1 expression, a subset of BECs express DNMT1; in contrast, DMNT1 is massively overexpressed in ICC cells, implying the crucial contribution in malignant transformation(Hu, Molina et al. 2022). In murine liver injury models, hepatic expression of DNMTs and TETs are heterogeneous based on the injury types and disease stage. This indicates rapid and dynamic alterations of 5mC/5hmC in the liver are responsive to various chronic injury conditions(Pogribny, Tryndyak et al. 2009, Booth, Branco et al. 2012, Page, Paoli et al. 2016). Therefore, understanding DNA methylation in each cell type of the liver in a context-dependent manner would be necessary to understand the insight of respective enzymes in the diseased liver.
DNA methylation in liver cell plasticity
During HC-to-BEC trans-differentiation, and vice versa, origin cells revert to LPC status; transiently express HB genes; downregulate original lineage-specific genes; and acquire lineage commitment of destination cell fate. Table 1 summarizes currently available liver cell plasticity models with experimental strategies and roles for epigenetic regulators on fate conversion. This process is conducted in collaboration with diverse histone modifiers and transcription regulator complexes. Specifically, these modulators hinder the expression of genes involved in lineage fate conversion and repress HB/LPC-related genes. Since DNA methylation-mediated gene silencing takes place postnatally during liver development(Reizel, Sabag et al. 2018) for cell-lineage-specific genes, the reprogramming of DNA methylation is necessary for cell fate conversion in the adult liver. Figure 3 depicted the roles of epigenetic regulators including DNMTs and TETs during liver cell fate conversion in animal models.
Table 1.
Summary of liver cell plasticity models and effect of diverse epigenetic modulation
| Animal | Conversion | Injury | Promoter for Lineage tracing | Epigenetic Factors | Downstream Targets | Functions | |
|---|---|---|---|---|---|---|---|
| Mouse | HC→BEC | Chemical injury | DDC | AAV8-TBG-Cre (Yanger, Zong et al. 2013) | Arid1a (Li, Yang et al. 2019) | YAP1 transcription activation↑ | LPC-to-HC↑ |
| Hnf4α-DreERT2;Sox9-CreERT2(Han, Wang et al. 2019) | |||||||
| Alb-Cre ERT2(Han, Wang et al. 2019) | |||||||
| Mx1-Cre (Nagahama, Sone et al. 2014) | |||||||
| BDL | AAV8-Tbg-Cre (Yanger, Zong et al. 2013) | ||||||
| Hnf4α-DreERT2;Sox9-CreERT2(Han, Wang et al. 2019) | |||||||
| Alb-Cre ERT2(Han, Wang et al. 2019) | |||||||
| Mx1-Cre (Nagahama, Sone et al. 2014) | |||||||
| DAPM | Mx1-Cre (Nagahama, Sone et al. 2014) | ||||||
| TAA | Mx1-Cre (Nagahama, Sone et al. 2014) | ||||||
| Alb-Cre (Sekiya and Suzuki 2012) | |||||||
| CCl4 | Mx1-Cre (Nagahama, Sone et al. 2014) | ||||||
| Genetic modulation | NICD | AAV8-Tbg-Cre (Yanger, Zong et al. 2013) | |||||
|
TetO-YAPS127A TetO-YAPS127A;AAV8-Tbg-Cre-Rbpj(fl/fl) |
AAV-Tbg-Cre (Yimlamai, Christodoulou et al. 2014) | ||||||
| CAGGS-GFP-IRES-SOX9 (Yoshii, Shimata et al. 2022) | |||||||
| BEC→HC | Chemical injury | CCl4 (6–24 weeks) | OPN-Cre ERT2(manco, Clerbaux et al. 2019) | ||||
| DDC (4–24 weeks)(Deng, Zhang et al. 2018),(3 weeks)(Aloia, McKie et al. 2019) | CK19-Cre ERT2(Deng, Zhang et al. 2018) | Tet1 (Aloia, McKie et al. 2019) | ErbB-MAPK & YAP1 pathways↑ | LPC proliferation↑, LPC-to-HC↑ | |||
| TAA (24–52 weeks) | CK19-Cre ERT2(Deng, Zhang et al 2018) | ||||||
| CDE (3 weeks)(Minnis-Lyons, Ferreira-Gonzalez et al. 2021), (10 days)(Ko, Choi et al. 2016) | OPN-Cre ERT2(Minnis-Lyons, Ferreira-Gonzalez et al. 2021) | Bet (Ko, Choi et al. 2016) | BEC-to-LPC↑, LPC proliferation↑ | ||||
| Chemical injury + Genetic modulation | AhCre+Mdm2 (fl/fl) | CK19-Cre ERT2(Lu, Bird et al. 2015) | |||||
| MCD+AAV8-Tbg-p21 | CK19-Cre ERT2(Minnis-Lyons, Ferreira-Gonzalez et al. 2021) | ||||||
| DDC+AAV8-Tbg-Cre-β1-integrin(fl/fl)(Raven, Lu et al. 2017)/AAV8-Tbg-p21(Raven, Lu et al. 2017, Aloia, McKie et al. 2019) | CK19-Cre ERT2(Raven, Lu et al. 2017) | Tet1 (Aloia, McKie et al. 2019) | ErbB-MAPK & YAP1 pathways↑ | LPC proliferation↑, LPC-to-HC↑ | |||
| MCD+AAV8-Tbg-Cre-β1-integrin(fl/fl)/AAV8-Tbg-p21 | CK19-Cre ERT2(Raven, Lu et al. 2017) | ||||||
| TAA+AAV8-Tbg-Cre-β1-integrin(fl/fl)(Raven, Lu et al. 2017) | CK19-Cre ERT2(Raven, Lu et al. 2017) | ||||||
| CDE+AAV8-Tbg-Cre-Ctnnb1(fl/fl)(Ko, Russell et al. 2019, Russell, Lu et al. 2019) | CK19-Cre ERT2(Russell, Lu et al. 2019) | Hdac1 (Ko, Russell et al. 2019) | LPC-to-HC↑ | ||||
| Zebrafish | Pharmacogenetic HC ablation | NTR/Mtz-driven pan-HC ablation | fabp10a-Cre (Choi, Ninov et al. 2014) , Tp1-Cre (Choi,Ninov et al. 2014, He, Lu et al. 2014, Huang, Chang et al. 2014) | Bet (Ko, Choi et al. 2016) | myca | BEC-to-LPC↑, LPC proliferation↑ | |
| hdac1 (KO, Russell et al. 2019) | sox9b↓ | LPC-to-HC↑ | |||||
| dnmt1 (He Zhou et al. 2022) | p53↓→mTORC1↑ | BEC-to-LPC↑ | |||||
| p53↓→ BMP signaling↑ | LPC-to-HC↑ | ||||||
Figure 3. Schematic of epigenetic regulations in hepatobiliary fate conversions in animal models.
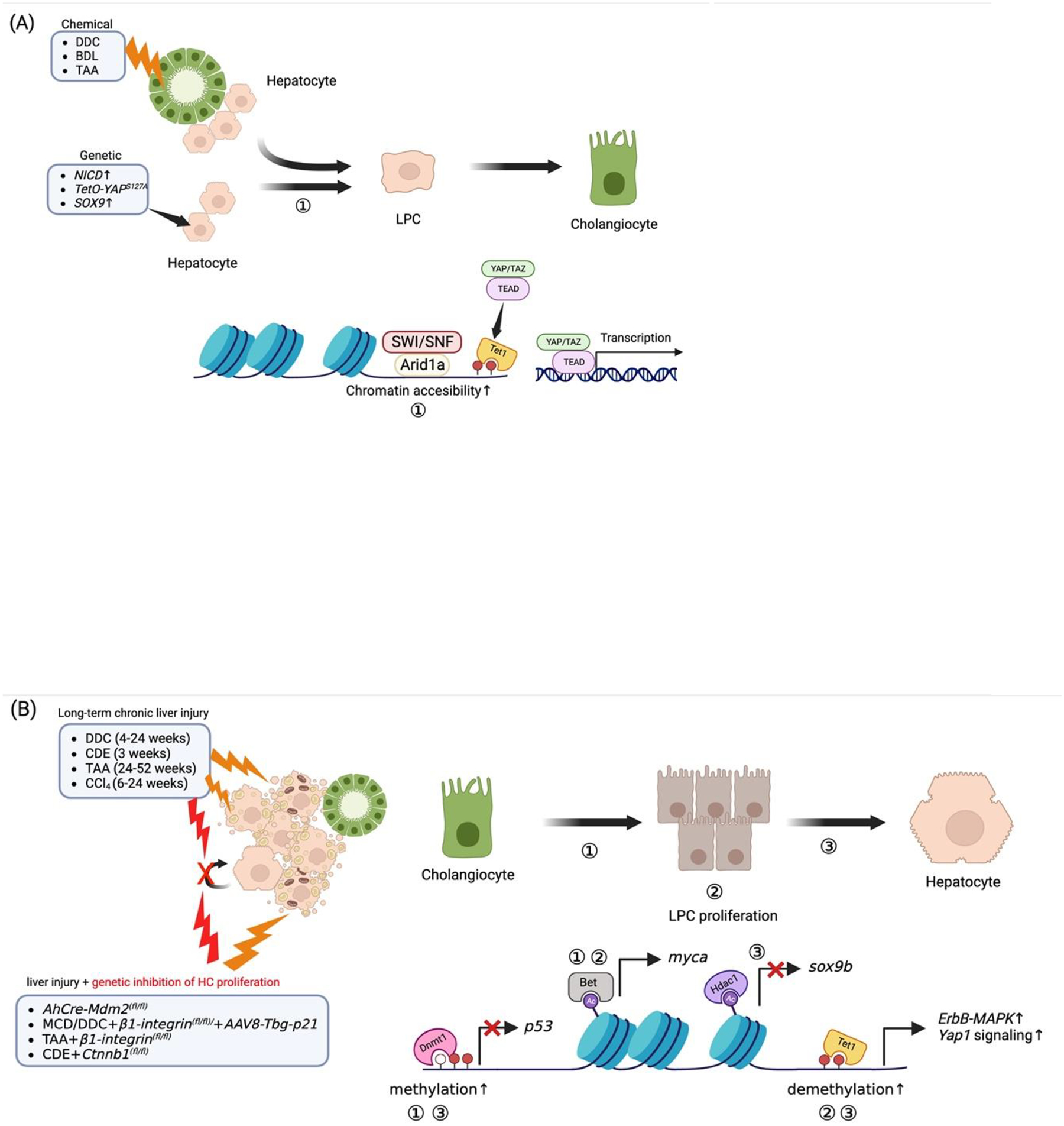
Liver insults derived from chemicals and/or genetic modulation induce dedifferentiation of HC or BECs to multipotent LPCs. (A) In HC-to-BEC, the Arid1a complex binds and opens the chromatin, thereby increasing chromatin accessibility. This event allows Yap1/Taz-Tead complex to bind to their target genes. Hence, this event promotes the activation of Yap1/Taz-Tead signaling during HC dedifferentiation into LPC. (B) In BEC-to-HC, the epigenetic reader BET and Dnmt1 positively regulate the LPC formation via the upregulation of myca and downregulation of p53 (in zebrafish), respectively. Bet-myca and Tet1-mediated ErbB-MAPK and Yap1 activation contribute to LPC proliferation and expansion. Subsequently, the epigenetic writer Hdac1 suppresses sox9b expression contributing to HC lineage differentiation; a mechanism of HC differentiation that also involves Tet1 and Dnmt1.
BEC-to-HC transdifferentiation
There are several reports on the role of DNA methylation in the setting of BEC-to-HC fate conversion. Aloia et al. recently showed that the global level of 5hmC in ductal cells in the liver following DDC diet was increased through TET1-mediated demethylation (Aloia, McKie et al. 2019). Additionally, they revealed that BEC-specific Tet1 elimination impairs BEC-derived HC repopulation in the mouse. They observed that BEC-specific Tet1 deletion significantly diminished initial ductal reaction (DR) and subsequent BEC/LPC-derived HC repopulation. By analyzing TET1-specific peaks, they identified demethylated TSS regions of mTOR, ErbB, MAPK, and Hippo-YAP1 target genes with respective change to expression. Interestingly, these targets have been reported as critical regulators in mouse and/or zebrafish BEC (LPC)-to-HC conversion models. While Fxr-mTor-Akt axis is required for the survival of LPCs(Jung, Kim et al. 2021), EGFR (receptor for ErbB)-MAPK-Sox9(So, Kim et al. 2021) and YAP1-Notch-Sox9(Yimlamai, Christodoulou et al. 2014) axis are BEC/LPC lineage-specific for ducts to become HCs. However, other repressors such as HDACs and/or DNMTs may diminish liver developmental genes and biliary lineage genes at a later stage. Along this line, the Shin group described the critical roles of Hdac1/Lsd1 in murine and zebrafish BEC-to-HEC transition through epigenetically repressing sox9 transcription, a crucial BEC/LPC lineage commitment gene(Ko, Russell et al. 2019). Recently, He et al. demonstrated Dnmt1 contributes to the BEC-to-HC differentiation by repressing p53 in the zebrafish model (table 1)(He, Zhou et al. 2022). Although p53 is a well-known DNMT1 target in a variety of cancer cells, this study demonstrated that a 20-fold higher dose of DNMT inhibitor (DNMTi, 100 mM Azacytidine) may cause chemical DNA damage leading to p53 expression in regenerating liver(Mudbhary, Hoshida et al. 2014). Another caveat of this report entails that changes to the methylome are restricted to the gene body region, but not around the p53 TSS region(Aloia, McKie et al. 2019). This is not common for DNMT1 activity and needs to be carefully repeated in the mammalian system. Therefore, the comprehensive investigation of all DNMT functions in liver cell plasticity and the identification of their direct targets will be fundamental studies.
HC-to-BEC transdifferentiation
In the context of HC-to-BEC conversion, understanding regulatory mechanisms is not fully elucidated. Since DNA methylation permits murine HC-driven ICC through the repression of HC commitment genes(Hu, Molina et al. 2022), DNMTs may contribute to fate conversion of HC into the biliary lineage. An important study on HC-to-BEC reprogramming by Merrell et al. observed a link between chromatin accessibility and transcriptional regulation(Merrell, Peng et al. 2021). Here, open chromatin regions in hepatocytes showed an enrichment of binding sites for important regulators of HC identity, including HNF4α, C/EBPα/β, and FOXA. Counter to this, open chromatin regions in BECs and reprogrammed cells showed the enrichment of binding sites for important factors of biliary identity (TEAD, and HNF1β). Comprehensively, during the HC-to-BEC reprogramming, 63% of BEC-specific open chromatin peaks were newly opened, and 61% of HC-specific open chromatin peaks were closed in the reprogrammed cells. This suggests significant changes in chromatin accessibility are strongly correlated to the changes in the transcriptional network during HC-to-BEC reprogramming. Hence, this study emphasized that epigenetic regulation plays a crucial role in the massive transcriptional change during the reprogramming from HC-to-BEC. This outlines the crucial roles of diverse transcriptional repressors, such as DNMTs, in this setting to be elucidated by future studies.
DNA methylation in liver cancer
Aberrant DNA methylation has been widely recognized as a key feature of human liver cacner(Kulis and Esteller 2010, Jusakul, Cutcutache et al. 2017, Nakaoka, Saito et al. 2017, O’Rourke, Lafuente-Barquero et al. 2019, Dhayat and Yang 2020, Izykowska 2020). A growing body of evidence implicates modulating DNA methylation in altering tumor growth (Baylin 2005, Kulis and Esteller 2010, Dhayat and Yang 2020) and cellular lineage commitment in the liver (Cheedipudi, Genolet et al. 2014, Moris, Pina et al. 2016, Folguera-Blasco, Cuyas et al. 2018). Table 2 lists publications that discuss the impact of DNA methylation modulation in various murine liver cancer models.
Table 2.
Effect of DNA methylation modulation in murine liver cancer models
| Cancer type | Model | Target | Manipulation | Effect on tumor | Mechanism | Reference |
|---|---|---|---|---|---|---|
| CCA | Orthotopic xenograft | Tet1 | sh-Tet1 | CCA growth↓ | cell growth↓ and apoptosis↑ | (Bai, Zhang et al. 2021) |
| Orthotopic xenograft | Dnmt1 | CM272 | CCA growth↓ | - | (Colyn, Barcena-Varela et al. 2021) | |
| JnkΔhep + DEN + CCl4 | CCA-enriched genes↓ | Metabolic reprogramming (carbohydrate/cholesterol metabolism↓) | ||||
| myrAkt+NICD HDTVI | Dnmt1 | Azacytidine | HC-to-BEC fate conversion↓, ICC formation↓ | HC lineage-specific genes↑, downstream targets of tp53↑ | (Hu, Molina et al. 2022) | |
| sh-Dnmt1 | ||||||
| Akt+YAP1S127A HDTVI | Azacytidine | HC-to-BEC/ICC transformation↓ | ||||
| ApoE-rtTA-YAP & Nf2−/− | Tet1 | sh-Tet1 | CCA proliferation↓, CCA tumorigenesis↓ | Yap1 target genes↓ | (Wu, Mei et al. 2022) | |
| Deletion of Tet1 | ||||||
| HCC | Xenograft | Dnmt1 | CM272 | HCC growth↓, angiogenesis↓ | G9a expression↓, HK2↓, Fbp1↑, Gnmt↑ | (Barcena-Varela, Caruso et al. 2019) |
| Dnmt1 | Guadecitabine | HCC growth↓, angiogenesis↓ | Dab2ip’ Dlec1, Gstp1, Rassf1, Runx3, and Socs1 | (Jueliger, Lyons et al. 2016) | ||
| Dnmt1 | Guadecitabine | HCC growth↓ | - | (Kuang, El-Khoueiry et al. 2015) | ||
| - | Decitabine + ACRBP-specific CTL | HCC growth↓, HCC apoptosis↑ | - | (Ge, Zhang et al. 2021) | ||
| Tet1 | miR-29a↑→TET1↓ | HCC growth↑, metastasis↑ | Socs1↓→Jak/Stat3 signaling↑→Mmp9↑ | (Chen, Yin et al. 2017) |
DNA Methylation and background etiologies leading to HCC
Progressing from chronic disease to neoplasia is a complex biological process. In the liver, chronic fibrosis is preceded by cirrhosis which in turn leads to HCC. However, the onset of carcinogenesis in the liver is complicated further by underlying pathologies that lead to primary HCC. These underlying pathologies tend to result in specific mutations in genes (e.g., TERT promoter, CTNNB1, TP53, AXIN1, ARID1A, etc.) that promote cell survival, growth thus disease pathogenesis(Nault and Zucman-Rossi 2016, Harding, Zhu et al. 2019, Rebouissou and Nault 2020, Paradis and Zucman-Rossi 2022). However, whether specific chronic liver pathologies result in specific mutational signatures is still being studied.
Cirrhosis results from the liver’s attempts at repair during chronic disease, regardless of etiology, leading to hepatocytes becoming senescent(Rudolph, Chang et al. 2000). Hepatocytes that avoid this contribute to HCC pathogenesis(Farazi, Glickman et al. 2003). Here, studies have found, via methylome sequencing, several genes can become either hypermethylated or hypomethylated; thus, corresponding to disease pathogenesis which has been summarized elsewhere(Villanueva, Portela et al. 2015, Martinez-Quetglas, Pinyol et al. 2016, Cancer Genome Atlas Research Network. Electronic address and Cancer Genome Atlas Research 2017, Rebouissou and Nault 2020). DNMT enzyme expression regulating these methylation events is known to be dysregulated during HCC pathogenesis(Saito, Kanai et al. 2001, Saito, Kanai et al. 2003, Mudbhary, Hoshida et al. 2014, Barcena-Varela, Caruso et al. 2019, Bayo, Fiore et al. 2019). Here, DNMTs can play a role in silencing tumor suppressor genes and genes that maintain hepatocyte differentiation(Fernandez-Barrena, Arechederra et al. 2020, Rebouissou and Nault 2020). Therefore, the link between DNMT dysregulation and pathogenesis of HCC is strongly defined and a broader understanding of this mechanism can open the door to new therapeutic approaches.
Given the marked reduction of viral infection-related liver diseases and the explosive increase in obesity, non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) are the most prevalent etiology progressing to HCC(Byrne and Targher 2015, Lazarus, Mark et al. 2022). NAFLD/NASH is one of among multiple pathologies that are categorized as dysregulated metabolic syndromes, and is progressively increasing in incidence in western countries like the United States(Byrne and Targher 2015, Lazarus, Mark et al. 2022). Broadly, NAFLD is a precursor to oxidative damage and inflammation that results in HCC(Lazarus, Mark et al. 2022). Interestingly, several studies have identified specific patterns of methylation with regards to NASH or NAFLD related HCCs(Kuramoto, Arai et al. 2017, Kurokawa, Yoneda et al. 2022). For example, PPARγ hypermethylation was detected in NAFLD patients indicating its potential utility for HCC surveillance(Hardy, Zeybel et al. 2017). Also, studies have found that genes like PPARα/δ or TGFβ1 can demonstrate either hyper- or hypomethylation, respectively, depending on NAFLD disease severity(Zeybel, Hardy et al. 2015). PPARα/PPARδ and TGFβ1 are anti- and pro-fibrosis genes, respectively, so their differential methylation is important to disease progression. Specifically with regards to NASH driven HCC, Kuramoto et al. identified hypomethylation/overexpression of genes ACOT7, FOXN3, MAML3, PCNT, PMPCA, RASA2, SPG7 and WHSC1; and hypermethylation/reduced expression of genes RF4, DHX36, FLCN, FYTTD1, MICAL3, MRPS24, NQO2, PKP4, PLAG1, PROSER3, TRAPPC10, WDR6, ZC3H14 and ZZEF1. Of the targets identified, MAML3 was discussed as a potential regulator of the Wnt/β-catenin pathway and may be responsible for driving NASH towards oncogenesis(Alves-Guerra, Ronchini et al. 2007). Clearly, NAFLD-related HCC produces a unique epigenetic landscape that clinicians can potentially utilize regarding disease surveillance and diagnosis.
Alcohol-related liver disease (ARLD) also results in robust fibrosis and cirrhosis leading to oxidative damage. While ARLD can result in changes to the methylation landscape, these alterations are more likely associated with overall cirrhosis(Hlady, Tiedemann et al. 2014, Habash, Sehrawat et al. 2022). However, one recent study found hypomethylation in the FKBP5 gene specifically in ARLD patient tissues(Kusumanchi, Liang et al. 2021). The FKBP5 protein regulates Yap1/Tead1 expression of the cytokine CXCL1 and potentiates inflammation in the underlying fibrosis. Also, PPARα/δ and TGFβ1 are hypermethylated, while Collagen1A1 is hypomethylated in ARLD tissues compared to normal livers(Zeybel, Hardy et al. 2015). While PPARα/δ are also hypermethylated in NAFLD patients, the overall degree of methylation in ARLD patients compared to NAFLD patients is significantly lower(Zeybel, Hardy et al. 2015). Thus, the ARLD methylome may be unique enough from NAFLD to distinguish when complexed together with current diagnostic approaches.
Classically, chronic viral HBV/HCV infection has been implicated in alternating the methylation landscape that progressively results in HCC(Nagashio, Arai et al. 2011, Um, Kim et al. 2011, Nishida, Kudo et al. 2012, Hlady, Tiedemann et al. 2014, Kuramoto, Arai et al. 2017, Wijetunga, Pascual et al. 2017, Hama, Totoki et al. 2018, Kurokawa, Yoneda et al. 2022). With regards to DNA methylation, HBV infection is associated with a progressive increase in methylation events during disease pathogenesis from early dysplasia to HCC in CpG loci(Um, Kim et al. 2011). For example, APC and RASSF1A demonstrate hypermethylation from HBV associated cirrhosis at all stages to progressed HCC. While hypermethylation of the tumor suppressor RASSF1A correlates with disease progression, the authors indicate that increased methylation of APC, a regulator of the Wnt/β-catenin pathway, is counterintuitive and most likely non-specific. In contrast, hypermethylation of the tumor suppressor, P16 is only observed in neoplastic stages(Um, Kim et al. 2011). This indicates that as HBV related HCC progresses the capacity of neoplastic cells to enter the cell cycle increases as P16 is silenced. Meanwhile, SPRY2, ERK, GNMT, and PTEN do not demonstrate differential methylation indicating their importance in the natural selection, survival, and proliferation of neoplastic cells(Um, Kim et al. 2011). On the other hand, HCV infection has been observed to have a stronger role in altering methylation patterns in cirrhotic patients compared to other etiologies which enhances the progression to HCC(Hlady, Tiedemann et al. 2014). One study found that methylation in common tumor suppressor genes (e.g., HIC1, GSTP1, SOCS1, RASSF1, CDKN2A, APC, RUNX3, and PRDM2) in HCV-infected patients was associated with shorter time to development of HCC, and the number of methylated genes in HCV patients were found to be an independent risk factor for developing HCC(Nishida, Kudo et al. 2012). Other groups have gone on to find unique epigenetic patterning comparing cirrhotic and HCC patients with chronic hepatitis which has the potential for differentiating patient risk for HCC development(Kurokawa, Yoneda et al. 2022).
Together, multiple environmental factors can influence the epigenetic landscape in the liver. These events have been demonstrated to potentiate pathogenesis during chronic liver injury leading up to oncogenic transformation. Understanding how each particular etiology (e.g., viral, chemical, etc.) specifically alters the methylome in the liver can provide information for HCC surveillance. The aberrant methylation may inform what personalized therapeutics patients can be treated with for preventive measures ultimately preventing an increasing burden on both patients and the healthcare system.
Common epigenetic landscape characteristics regarding methylation and HCC
Recent studies have investigated whether the epigenetic landscape among HCC patients is consistent. What is consistent, however, is that underlying etiologies provide for unique epigenetic fingerprinting for HCC patients(Nishida, Kudo et al. 2012, Hlady, Tiedemann et al. 2014, Kuramoto, Arai et al. 2017, von Felden, Garcia-Lezana et al. 2020, Kurokawa, Yoneda et al. 2022). Taking into consideration the background etiologies discussed, the methylation status of HCC samples has been relatively thoroughly investigated(Villanueva, Portela et al. 2015, Cancer Genome Atlas Research Network. Electronic address and Cancer Genome Atlas Research 2017, Hama, Totoki et al. 2018, Kurokawa, Yoneda et al. 2022). As noted earlier, enhanced methylation is associated with reduced expression of tumor suppressor genes in HCC pathogenesis. Genes regulating cell cycle, in addition to those that regulate epithelial-to-mesenchymal transition, tend to be targets of hypermethylation(Fernandez-Barrena, Arechederra et al. 2020, Kurokawa, Yoneda et al. 2022). Recently, Zinc Finger Proteins have been identified as targets of hypermethylation, and can potentially differentiate between pre-malignant cirrhosis and early stage HCCs(Goncalves, Goncalves-Reis et al. 2022, Sun, Gan et al. 2022). With regards to disease progression, Kurokawa et al. identified HNF4A, FABP1, and SGK1 can become hypermethylated during premalignant fibrosis, but become hypomethylated once progressed to HCC. This study demonstrates the complex dynamics of methylation status and disease pathogenesis which should not be taken for granted.
Studies have identified that hypomethylation can result in enhanced gene expression and potential genetic instability(Shen, Wang et al. 2013, Villanueva, Portela et al. 2015, Hama, Totoki et al. 2018). Some hypomethylated regions may even pertain to specific transcriptional machinery such as C/ebpβ(Xiong, Wu et al. 2019), or members of the homeobox transcription factor family(Goncalves, Goncalves-Reis et al. 2022), or oncogenes like Ras(Maryam and Idrees 2018). In addition, recent studies have investigated the role of hypomethylation of Repetitive Elements (Res) in hepatocellular carcinogenesis. Specifically, investigators have consistently found that hypomethylation of the RE Long Interspersed Nuclear Elements 1 (LINE-1) is associated with genomic instability and worse overall prognosis(Baba, Yagi et al. 2018, Anwar, Hasemeier et al. 2019, Zheng, Hlady et al. 2019). LINE-1 elements compose of nearly 17% of the human genome and have the capacity to act at high frequency as retrotransposons if hypomethylated(de Koning, Gu et al. 2011, Lee, Iskow et al. 2012). Inadvertent hypomethylation of LINE-1 elements during carcinogenesis in the liver has the capacity to promote chromosomal dysregulation and promote oncogenic transformation. Studying how these, and other transposable elements, become activated in the background of chronic liver disease can lead to increased understanding of HCC oncogenesis and potential new therapeutic approaches.
Common DNMT and other enzymes potentially mutated in HCCs
Dysregulated expression of DNMT enzymes is known to result in oncogenesis. Increased expression of DNMT1 has been observed in HCC patient tissues(Saito, Kanai et al. 2003, Mudbhary, Hoshida et al. 2014, Barcena-Varela, Caruso et al. 2019). This potentially leads to enhanced methylation and silencing of tumor suppressor genes. Additionally, other studies have identified that DNMT3a and DNMT3b are also upregulated in HCC patients(Saito, Kanai et al. 2001). One study analyzed HCC patient samples from TCGA data and found many patients have at least one mutation in genes that contribute to editing of the epigenome(Bayo, Fiore et al. 2019). Nearly 50% of these patients demonstrated upregulation in genes that contribute to changes in the epigenome while 20% were downregulated when comparing normal tissue to HCC tissues(Bayo, Fiore et al. 2019). Considering how methylation status can confer disease pathogenesis it is crucial to understand how the expression of enzymes responsible for changes to the epigenome is altered during pre-malignancy to late-stage disease. This information will provide a better understanding of the role of these molecules in HCC development and progression.
DNA methylation in CCA pathogenesis (Fig. 4)
Figure 4. Schematic of the regulatory mechanism of DNA methylation in CCA tumorigenesis.
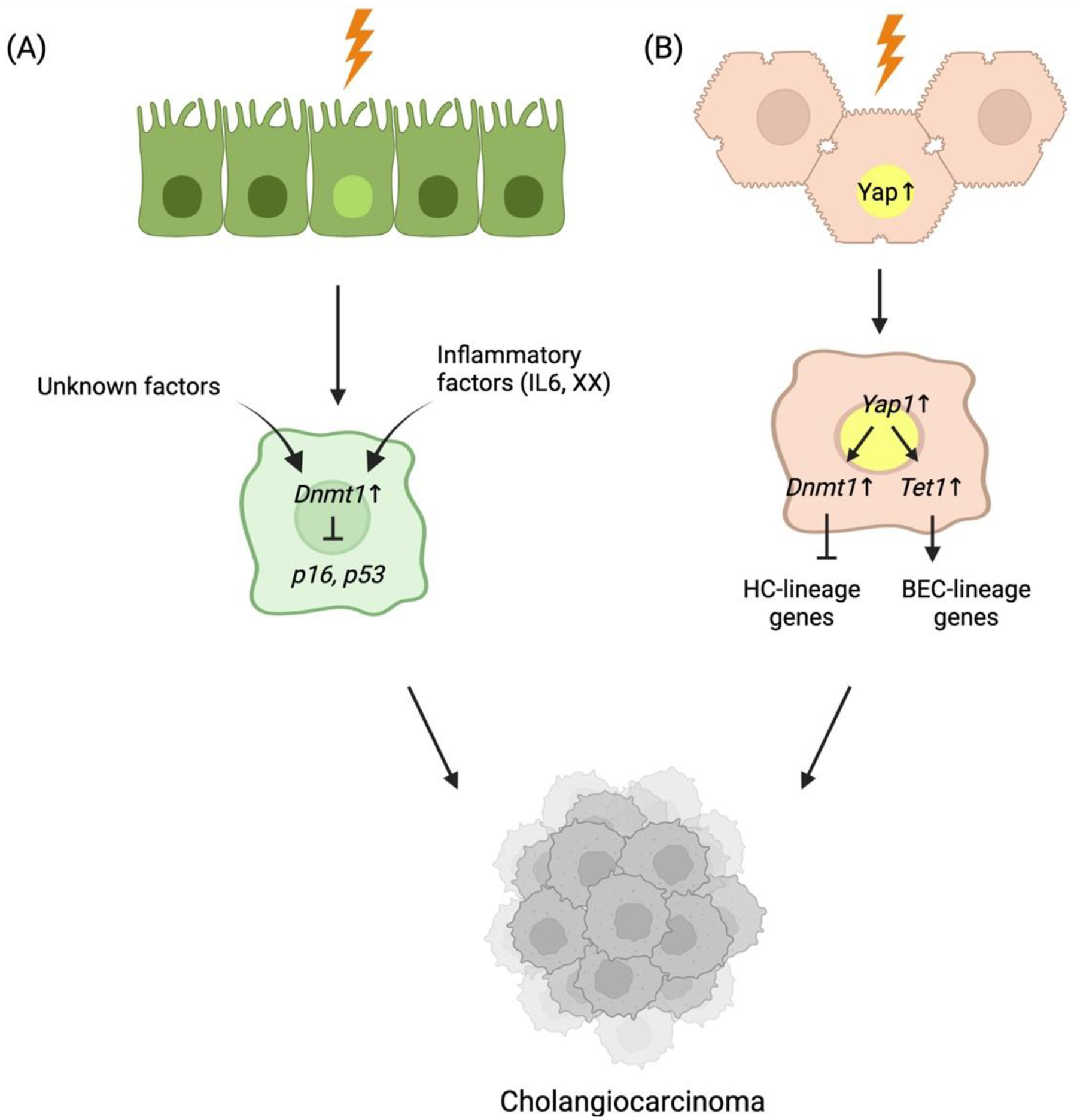
Under chronic liver injury, HCs or BECs transform into cholangiocarcinoma. (A) under biliary injuries, inflammatory factors, such as IL-6 and/or other unknown factors induce Dnmt1 expression. The increased Dnmt1 suppresses tumor suppressor genes, such as p16 and p53, thereby contributing to tumorigenesis of CCA. (B) In HC, overactivation of Yap1 induced by injuries in HC can trigger induction of Dnmt1 and Tet1 expression. Dnmt1 prevents the transcriptional activity of HC-lineage transcriptional factors, such as HNF4α and FOXA2, ultimately diminishing the HC-lineage identity. In contrast, Tet1 promotes the expression of biliary lineage-specific Yap1 targets driving CCA tumorigenesis.
There have been extensive studies linking inflammation and the development of CCA via the persistent secretion of inflammatory cytokines(Wu, Yiang et al. 2018). Among various inflammatory cytokines, interleukin 6 (IL-6) expression is correlated with regulating tumor suppressor gene expression, including Dnmt1 in vitro and in vivo (Wehbe, Henson et al. 2006, Isomoto, Mott et al. 2007, Meng, Wehbe-Janek et al. 2008, Isomoto 2009, Braconi, Huang et al. 2010). These studies revealed that sustained IL6/STAT3 signaling induces Dnmt1-mediated hypermethylation at the promoter regions of tumor suppressor genes such as p53, provoking BEC transformation into CCA, reflecting the well-described mitogenic role of IL6/STAT3 stimulation in CCA cells. Given the importance of inflammatory signatures in the transcriptome-based classification of human CCA(Sia, Hoshida et al. 2013), comprehensive investigations of response to immune checkpoint inhibitor (ICI) along with DNMTi and/or IL6 inhibitor in IL6/STAT3-Dnmt1 activated CCA cluster would be informative.
The most frequently observed hypermethylated regions in CCA are representative tumor suppressors such as p16INK4a(Braconi, Huang et al. 2010), FoxM1(Cao, Liu et al. 2020, Pogribna, Koonce et al. 2020), TP53(Zhang, Chen et al. 2018, Li, Wang et al. 2019) or PTEN(Kumar, Raeman et al. 2018, Zhang, Liu et al. 2018), which typically induce death/senescence of malignant cells. Hypermethylation of these cell cycle inhibitors results in unstoppable oncogenic proliferation in various cancer including CCA. Importantly, hypomethylation of p16INK4a has been demonstrated in more than 80% of clinical CCA. Pharmacologic inhibition of CDK4, a direct binding target of p16, suppresses CCA growth, especially when combined with either an mTOR inhibitor(Song, Liu et al. 2019) or FAK inhibitor(Zeng, Zhou et al. 2022), suggesting a potentially relevant combination therapy with precision medical approaches. Thus, a substantial investigation of targeting these genes as broad-spectrum supportive therapy in tandem with targeted therapy would be fundamental.
Intriguingly, recent studies revealed the essential roles of DNA methylation in HC transformation into iCCA cells. The Hippo-YAP1/TAZ signaling pathway plays a crucial role in the iCCA development, especially through epigenetic remodeling (Yimlamai, Christodoulou et al. 2014, Nishio, Sugimachi et al. 2016, Hyun, Al Abo et al. 2021, Song, Xu et al. 2021, Cigliano, Zhang et al. 2022, Li, Wu et al. 2022). Two recent studies reported DNA methylation is dysregulated under activated Yap1/Taz-Tead signaling in murine iCCA models (Hu, Molina et al. 2022, Wu, Mei et al. 2022). Hu et al. found that YAP1/TEAD activity is indispensable for the pathologic fate conversion of HC into biliary lineage by inducing DNMT1 expression in ICC cells. Importantly, they found that DNMT1 is preferentially localized to the promoter regions of HC lineage-specific genes regulated by HNF4α, FOXA2, CEBPɑ/β, and HNF1α; thus, trans-differentiation into the biliary lineage is promoted and demonstrates the distinct roles of DNMT in HC-to-iCCA lineage reprogramming.
Recently, Wu et al. also described indispensable roles for Tet1-dependent DNA demethylation in YAP1-mediated HC-driven ICC formation(Wu, Mei et al. 2022). They revealed that TET1 directly binds to YAP1 target genes through the interaction with TEAD, thereby resulting in demethylation and a subsequent increase in expression of target genes. This effect only occurs at the distal intergenic regions which are enriched in the enhancer mark with H3K4me1 rather than promoter regions. Taken together, not only is DNMT1-mediated DNA methylation of promoters contributes to HC identity loss, but this also results in the gain of BEC identity through Tet1-mediated DNA demethylation; all essential mechanisms in the context of YAP1-dependent HC-to-ICC transformation. Given that DNA methylation is diluted and lost during DNA replication without the maintenance of methylation by Dnmt1, the findings of Hu et al. suggest that Dnmt1-dependent methylation is essential for the transformed identity and the dysregulated cell cycle of cancer cells during their clonal expansion. However, to maintain DNA methylation on the newly replicated DNA in iCCA development, de novo methylation on the template DNA is a prerequisite. The mechanism by which de novo methylation is triggered at target regions remains unknown. Thus, Dnmt3a/b may be elaborately regulated in a context-specific manner at the very early stages of HC-to-iCCA transformation.
Overall, DNA methylation may play heterogeneous roles in ICC based on the molecular drivers, stages, underlying etiologies and/or cellular origins. Given the tremendously heterogeneous methylome in clinical ICCs, it would be important to elucidate the distinct roles of DNA methylation regarding molecular targets and stages to consider FDA-approved DNMTi to appropriately test in potential responders in the clinic.
Diagnostic and therapeutic potential of DNA methylation in liver cancer
Tables 3 and 4 summarize the literature on aberrant DNA methylation patterns in clinical HCC and CCA, respectively.
Table 3.
Summary of aberrant DNA methylation in human HCC.
| Methylation | Regulator | Target | Biologic function (regulation) | Sensitivity (%) | Specificity (%) | # of samples | Effect on tumor | Reference |
|---|---|---|---|---|---|---|---|---|
| Hypo | – | CEBPB | transcription factor, HC proliferation↑ | P < 2.2e-16 | – | 33 | Proliferation↑ | (Xiong, Wu et al., 2019) |
| – | IFNGR2 | IFN-γ pathway | – | HBV viraemia, Redox | ||||
| – | SLC45A4 | sucrose transport | – | homeostasis | ||||
| – | LITAF | transcription factor, NFκB signaling | – | |||||
| – | SRC | Regulation of telomere maintenance hippo signaling | – | – | ||||
| – | CCL20 | cell-cell signaling, inflammatory response | P = 1.70e-14 | 62 | – | (Shen et al., 2012) | ||
| Hyper | – | HOXA1 | transcription factor | 71 | 90 | 135 | – | (Chalasani et al., 2021) |
| – | EMX1 | transcription factor | ||||||
| – | TSPYL5 | cell proliferation, protein kinase B signaling, protein ubiquitination | ||||||
| – | ZNF334 | transcription factor | 88 | – | 25 | Tumor suppressor | (Sun et al., 2022) | |
| – | HIC1 | Wnt signaling↓, transcription, DNA damage response↑ | 64.8 | 93.6 | 177 | HCC-related tumor suppressor genes | (Nishida et al., 2012) | |
| – | GSTP1 | Apoptosis↓, MAPK signaling↓ | 75 | 91.9 | 177 | (Nishida et al., 2012, Song et al., 2013) | ||
| – | SOCS1 | Regulatory T cell differentiation ↑ | 57.4 | 93.6 | 177 | (Nishida et al., 2012) | ||
| – | RASSF1 | G1/S cell cycle transition, Ras signaling transduction | 68.8 | 92.5 | 177 | (Nishida et al., 2012, Um et al., 2011) | ||
| – | P16 (CDKN2A) | G2/M cell cycle transition and cell-matrix adhesion | 62.6 | 95.4 | 177 | (Nishida et al., 2012, Shen et al., 2012, Song et al., 2013) | ||
| – | APC | regulation of cell death↑, regulation of cell migration↑ | 84.1 | 93.1 | 177 | (Nishida et al., 2012) | ||
| – | RUNX3 | cell cycle↓ | 62.5 | 93.6 | 177 | |||
| – | PRDM2 | histone methylation, transcription regulation | 72.2 | 83.3 | 177 | |||
| DNMT1 | CDH1 | cell-cell adhesion↓ and cell migration↓ | 57.1 | – | 42 | EMT↑, metastasis↑ | (Lim et al., 2008) | |
| – | DAB2IP | angiogenesis↓, PI3K signaling↓ | P = 3.58e-17 | – | 62 | – | (Shen et al., 2012) | |
| – | BMP4 | BMP signaling pathway | P = 5.36e-16 | – | ||||
| – | ZFP41 | transcription factor | P = 3.25e-15 | – | ||||
| – | ZNF154 | transcription factor | P = 1.14e-12 | – | ||||
| – | ZNF540 | transcription factor | P = 2.03e-12 | – | ||||
| – | ETS2 | transcription factor | P < 2.2e-16 | – | 33 | – | (Xiong, Wu et al., 2019) | |
| – | DACH1 | DNA biosynthesis↓, fibroblast proliferation↓ | P < 2.2e-16 | – | – | |||
| – | SMPD3 | lipid metabolic process, cell cycle | P < 0.001 | – | 71 | Tumor suppressor | (Revill et al., 2013) | |
| – | NEFH | microtubule cytoskeleton organization | P < 0.001 | – |
Table 4.
Summary of aberrant DNA methylation in human CCA.
| Methylation | Regulator | Target | Biologic function (regulation) | Sensitivity (%) | Specificity (%) | # of samples | Effect on tumor | Reference |
|---|---|---|---|---|---|---|---|---|
| Hypo | – | PCLAF | cell cycles, DNA repair, DNA replication | P < 0.05 | – | 36 | hOCT1↓ | (Liu et al., 2022) |
| – | TTYH3 | chloride transport | P < 0.05 | – | TCGA | CCA migration, invasion, and proliferation↑ | (Xue et al., 2021) | |
| – | MTHFD1 | nucleotide biosynthetic process, neutrophil homeostasis | – | – | 102 | – | (Moruzzi et al., 2017) | |
| – | IMP3 | rRNA processing, ribosome biogenesis | 82 | 100 | 72 | – | (Gao et al., 2014) | |
| – | LINE-1 | RNA-mediated transposition | P < 0.001 | 172 | CCA differentiation, lymphatic invasion, and survival | (Jeong et al., 2017) | ||
| – | CDH3 | TGFβ2↓, IGFR-WNT/β-CATENIN pathway↑, cell-cell adhesion↑ | 51 | – | 59 | CCA Prognosis | (Sakamoto et al., 2015) | |
| – | EGFR | PI3K/MAPK/JNK signaling↑, DNA replication/repair↑, apoptosis↓ | 44.6 | – | 65 | (Limpaiboon et al., 2005) | ||
| Hyper | – | 14-3-3σ | cyclin-dependent protein serine/threonine kinase activity | 60 | 100 | 79 | CCA Prognosis | (Wu et al., 2020) |
| – | HOXA5 | transcription factor | P < 0.05/P < 0.01 | – | 209/34 | MXD1↓ → CCA↑ | (Xiong et al., 2022) | |
| – | PTEN | PI3K inhibition | 35 | 93 | 29 | CCA Prognosis | (Sriraksaet al., 2011) | |
| – | APOB | lipid metabolism | P < 0.05 | – | 36 | Survival | (Xu et al., 2021) | |
| – | EFHD2 | Unknown, cadherin/metal ion binding | P <0.019 | – | 118 | Survival | (Peng, Meng, Sheng, & Gao, 2021) | |
| – | PHYHIPL | Unknown, protein binding | P < 0.0001 | – | 118 | Survival | (Peng, Meng, Sheng, & Gao, 2021) | |
| – | CNRIP1 | Cannabinoid Receptor signaling receptor activity | 70 | 100 | 112 49 |
CCA migration, invasion, and proliferation↑ by inhibiting PKM2 activity | (Chen et al., 2021) (Andresen et al., 2015) | |
| – | EBF1 | transcription factor | 75 | – | 72 | CCA progression↑ | (Armartmuntree et al., 2021) | |
| – | miR-212–3p | MUC13 expression in CCA | 100 | – | 26 | MUC13↑→ CCA↑ | (Tiemin et al., 2020) | |
|
MGMT
MGMT |
p21,p27 | DNA methylation/repair/ligation/modifi cation, apoptosis i | – | – | 4 | p21↑, p27↑,CYCLINE1↑ | (Chen et al., 2020) | |
| – | 33 | 100 | 72 | (Yang, House, Guo, Herman, & Clark, 2005) | ||||
| – | 11 | 100 | 79 | (Lee, Kim, Jung, Yang, & Kang, 2002) | ||||
| – | DLEC1 | cell proliferation | 23 | 100 | 172 | CCA proliferation | (Kim et al., 2019) | |
| – | HTATIP2 | transcription factor | 14 | 54 | apoptosis↓ | (Nanok, Jearanaikoon, Proungvitaya, & Limpaiboon, 2018) | ||
| – | UCHL1 | proteolysis | 57 | – | 54 | Survival | (Nanok et al., 2018) | |
| 5-Aza | F2 | protein phosphorylation | – | – | 228 | Survival | (Chen et al., 2019) | |
| 5-Aza | AHSG | inflammatory response and phagocytosis | – | – | 228 | Survival | (Chen et al., 2019) | |
| 5-Aza | ALDH8A1 | retinoic acid metabolism | – | – | 228 | Survival | (Chen et al., 2019) | |
| 5-Aza | SERPIND1 | peptidase activity | – | – | 228 | Survival | (Chen et al., 2019) | |
| 5-Aza | AGXT | l-serine/cysteine/alanine/glyoxylate metabolic process, NOTCH signaling pathway | 228 | Survival | (Chen et al., 2019) | |||
| – | CDO1 | L-cysteine catabolic process, taurine | 76 | 92 | 81 | CCA prognosis | (Nakamoto et al., 2018) | |
| – | biosynthetic process | 77 | 100 | 39 | (Andresen et al., 2012) | |||
| – | 77 | 98 | 49 | (Andresen et al., 2015) | ||||
| 5-Aza | GATA5 | transcription factor and endodermal cell fate commitment | – | – | 152 | CCA growth↑ and metastasis↑ via WNT/β-CATENIN pathway | (Liu et al., 2018) | |
| – | HOXA1 | transcription factor | 89 | 100 | 9 | – | (Prachayakul et al., 2017) | |
| – | RASSF1A | G1/S cell cycle transition, Ras signaling transduction | 56 | 100 | 9 | – | (Prachayakul et al., 2017) | |
| – | P16 (CDKN2A) | G2/M cell cycle transition and cell-matrix adhesion | 25 | – | 36 | (Lee et al., 2002, Liu et al., 2012, Prachayakul et al., 2017, Tannapfel et al., 2000, Xiaofang, Kun, Shaoping, Zaiqiu, & Hailong, 2012) | ||
| – | NEUROG1 | transcription factor; cell cycle and differentiation | 100 | 100 | 9 | – | (Prachayakul et al., 2017) | |
| DNMT1/5-Aza | SOX17 | transcription factor, WNT/β-CATENIN pathway↑, cell differentiation↑ and cell growth↑ | P < 0.0001 | – | 48 + 37 + 6 | CCA prognosis and survival | (Merino-Azpitarte et al., 2017) | |
| – | miR-191/TP53 | tumor suppressor; cell growth↓ and DNA damage↑ | 76 | - | 152 | miR-191→ | (Li et al., 2017) | |
| 68 | - | Tet1↓→ p53↑→ CCA↑ | ||||||
| – | DCLK1 | cell differentiation and intracellular signal transduction | 87 | 100 | 93 | CCA prognosis | (Andresen et al., 2012) | |
| – | SFRP1 | apoptotic↑ and cell cycle↓ | (Andresen et al., 2012) | |||||
| – | CNRIP1 | regulation of signaling receptor activity | (Andresen et al., 2015) | |||||
| – | VIM | collagen biosynthetic process↑ | (Andresen et al., 2015) | |||||
| – | PYCARD | adaptive immune response↑, IL-8/IL-10 production↑ | 39 | – | 36 | Survival | (Xiaofang et al., 2012) | |
| – | CCND2 | cell proliferation↑ and apoptosis↓ | 74 | 100 | 45 | CCA proliferation | (Shin et al., 2012) | |
| – | CDH13 | cell-matrix adhesion↑, Rho/Rac pathway↑, EGFR pathway↑ | ||||||
| – | GRIN2B | calcium-mediated signaling and glutamate receptor signaling | ||||||
| – | TWIST1 | TNF ligand↑ PI3K signaling↑, cellular senescence↑ | ||||||
| – | RUNX3 | cell cycle↓ | 33 | 100 | 111 | CCA proliferation | (Kim et al., 2007) | |
| – | MLH1 | meiotic chromosome segregation, DNA repair | 45 | 100 | 65 | – | (Limpaiboon et al., 2005) | |
| – | DAPK1 | defense response to tumor cell, positive regulation of autophagy and apoptosis | 6 | 100 | 36 | CCA autophagy and apoptosis | (Liu et al., 2007, Xiaofang et al., 2012) | |
| – | 8 | 100 | 79 | (Lee et al., 2002) | ||||
| – | GSTP1 | negative regulation of apoptosis, negative regulation of MAP kinase/ERK1&2/NF-kB cascade | 6 | 100 | 79 | – | ||
| – | CDH1 | cell-cell adhesion↓ and cell migration↓ | 22 | 100 | 79 | CCA migration and invasion | (Lee et al., 2002) | |
| – | 30 | 95 | 111 | (Kim et al., 2007) |
Approaches to use DNA methylation as an HCC diagnostic
Epigenetics, especially regarding methylation, can provide a distinct differentiating fingerprint between patients, or more broadly etiological conditions. In fact, methylation status combined with advancements in technologies are sensitive enough to differentiate between cirrhosis and various HCC stages(Goncalves, Goncalves-Reis et al. 2022). Complexed with whole genome sequencing, identifying differentially methylated patterns can identify associations specific to early stages of HCC(von Felden, Garcia-Lezana et al. 2020, Goncalves, Goncalves-Reis et al. 2022, Kurokawa, Yoneda et al. 2022). Using differential methylation patterning can even now provide sufficient sensitivity and specificity in liquid biopsies regarding detection and even pathology site origin(Cohen, Javed et al. 2017). Studies that incorporate methylation for diagnosis or prediction can be powerful tools when combined with traditional methods for HCC surveillance. As this is the direction of several clinical trials (NCT04856046, NCT03694600, NCT03804593) it is promising that routine assessment of patient differential methylation may assist in overall surveillance and early detection for HCC.
Researchers and clinicians alike are optimistic about the potential of using methylome sequencing as a diagnostic, especially for the staging of HCC. This is demonstrated by the numerous clinical trials initiated and still ongoing which involve multiple modalities of testing for differential methylation. These studies have determined several methylated targets that can reach from 70% to 95% sensitivity and 89% to 92% specificity in detecting and staging HCC(Chalasani, Ramasubramanian et al. 2021, Goncalves, Goncalves-Reis et al. 2022). While promising, these studies acknowledge there are more gaps to be filled with regard to incorporating additional confounding factors such as sex, age, and race. Also, other studies are planning to incorporate differential alpha-fetoprotein levels together with differential methylation status to increase sensitivity and specificity.
Developing liquid biopsies are the most effective and approachable techniques that can provide availability to a large percentage of populations. A recent study demonstrated the specificity and sensitivity of distinct methylation patterns between cirrhosis and HCC patients when compared to normal samples(Goncalves, Goncalves-Reis et al. 2022). Here, the authors used a metadata approach combining the information from several datasets and trained their algorithm such that they can not only differentiate between cirrhosis and HCC, but stage as well as specific target genes that may be the “canary in the coal mine” to determine progression from chronic disease to HCC. While using cell free DNA is the most approachable method for HCC surveillance(von Felden, Garcia-Lezana et al. 2020), these technologies require fine tuning prior to implementation in the clinic. To do this, clinical trials are needed to set standards which are currently being conducted in at least one trial that aims to determine the levels of methylation to prevent recurrence or metastasis in resectable HCC (NCT04856046).
Finally, on the direction of the future and cancer detection, NCT05573217 is a new clinical trial that investigates cell-free methylation markers in liver cancer patients treated with systemic therapies to measure treatment response. Understanding how the epigenetic landscape shifts in HCC patients treated with systemic therapies is exciting as this information can potentially predict whether specific therapies are more effective than others, and when patients need to be shifted from one therapeutic intervention to another.
Diagnostic significance of DNA methylation in clinical CCA
CCA still remains difficult to diagnose early and shows a poor survival rate with poor response to therapies(Sandhu, Shire et al. 2008, Limpaiboon 2012). Like the other neoplasms, early detection is one of the most critical factors determining the prognosis of CCA patients. Often, CCA develops without underlying cirrhosis and lacks specific clinical signs, even compared to HCC. Thus, there is a dire need for the discovery of a more reliable, accurate, and effective prognostic strategy to detect the early stages of CCA. In this vein, detecting aberrant DNA methylation would be powerful since it is available in early CCA stages. These include intraductal papillary neoplasm of the liver/bile ducts (IPNL/B) and biliary intraepithelial neoplasia (BilIN), precancerous lesions of CCA and premalignant diseased liver. In addition, there are numerous reports demonstrating the efficacy of detecting hypermethylated targets using liver tissue or liquid biopsies, such as bile juice and serum, from CCA patients. According to tissue analysis, besides hypermethylated p16INK4a, several genes were suggested as suitable CCA biomarkers with significant sensitivity (>75%) and sensitivity (>90%) (e.g., CDO1, HPP1, SEMA3B, HOXA1 and SFRP1). While blood biopsy is the safest and most non-invasive source for hypermethylated biomarker detection, limited studies have been reported so far. Cell-free DNAs (cfDNAs) have been pursued as a biomarker to identify clinical CCAs since they can be released into circulating blood from tumor cells via metabolic processes (e.g., necrosis or apoptosis) and retain tumor-specific features (e.g., genetic mutations, epigenetic alterations, or chromosomal rearrangements(Warton and Samimi 2015, Lissa and Robles 2016, McAnena, Brown et al. 2017, Lu, Bi et al. 2018))(Mody, Kasi et al. 2019). Furthermore, recent reports provide evidence to support cfDNA as a prognostic biomarker(Wasenang, Chaiyarit et al. 2019, Yang, Ghoz et al. 2021). Wasenang et al. found significant differences in DNA methylation levels of OPCML and HOXD9 from CCA patients(Wasenang, Chaiyarit et al. 2019) exhibiting 62.5% sensitivity and 100% specificity. Additionally, Yang et al. analyzed plasma samples from 53 eCCA and 117 control cohorts44. In this study, they determined nine markers with a sensitivity of 63%−86% and a specificity of 88%−98%: ZNF781, CYP26C1, RYR2, HOXA1, EMX1, ST8SIA1, PTGDR, PRKCB, and BMP3. Interestingly, based on data sets from the Cancer Genome Atlas, all these markers were significantly hypermethylated and involved in cancer-related biological pathways, Implying the efficacy of cfDNAs detecting CCA-specific hypermethylation as diagnostic. Altogether, numerous DNA methylation biomarkers have been suggested to improve the sensitivity of detecting CCA while specificity needs to be more carefully examined in larger patient cohorts with appropriate controls. Carefully considering specimens, such as those from HCC and PDAC, can provide a reliable and effective surveillance method for patients in premalignant stages at high-risk for developing CCA.
Clinical trials using DNMT inhibitors in liver cancer
Fundamentally, the process of methylation leads to the silencing of genes which is counterintuitive to tumor progression. However, as discussed earlier, several groups have identified that tumor suppressor genes are the most likely genes to be methylated which in turn promotes tumor cell survival(Hlady, Tiedemann et al. 2014, Fernandez-Barrena, Arechederra et al. 2020, Goncalves, Goncalves-Reis et al. 2022, Kurokawa, Yoneda et al. 2022, Sun, Gan et al. 2022). In this direction, translational scientists have asked whether inhibiting DNMTs can reverse the methylation of tumor suppressor genes. In the liver specifically, DNMTi are rarely used for therapeutic purposes. Upon FDA approval, the DNMTi 5-azacytidine (Fig. 5A) under the trade name Vidaza has been used as a therapeutic for other malignancies for several years(Sekeres, Schuster et al. 2022). DNMT1 is inhibited by 5-azacytidine after its incorporation into DNA as a cytidine analog(Stresemann and Lyko 2008, Derissen, Beijnen et al. 2013, Kordella, Lamprianidou et al. 2021). With the incorporation of 5-azacytidine, the DNA structure is altered such that DNMT1 can no longer effectively methylate target DNA(Stresemann and Lyko 2008, McCabe, Brandes et al. 2009). 5-Azacytidine can be deactivated by cystine deaminase which is robustly expressed in the liver making the utilization of 5-azacytidine for liver pathologies, unfortunately, not very effective(Toh, Lim et al. 2019). Attempts to increase 5-azacytidine’s bioavailability are ongoing, but some have derived analogs that increase the utility in the liver. 5-aza-4-thio-2-deoxycytidine (Aza-Tdc) is an example that has been implemented in the NCT03366116 clinical trial to treat solid cancers which include the recruitment of patients with liver cancers. Aza-Tdc is a DNMT1 inhibitor by forming covalent bond with DNMT1 after Aza-Tdc is incorporated into the genome(Thottassery, Sambandam et al. 2014, Parker and Thottassery 2021) (Fig. 5B). With less off-target effects and increased specificity for DNMT1, Aza-Tdc is a promising drug that is understudied in the liver and can potentially contribute to broadening the therapeutic toolset(Thottassery, Sambandam et al. 2014, Parker and Thottassery 2021).
Figure 5. Molecular structures of DNMTi used in the clinical setting.
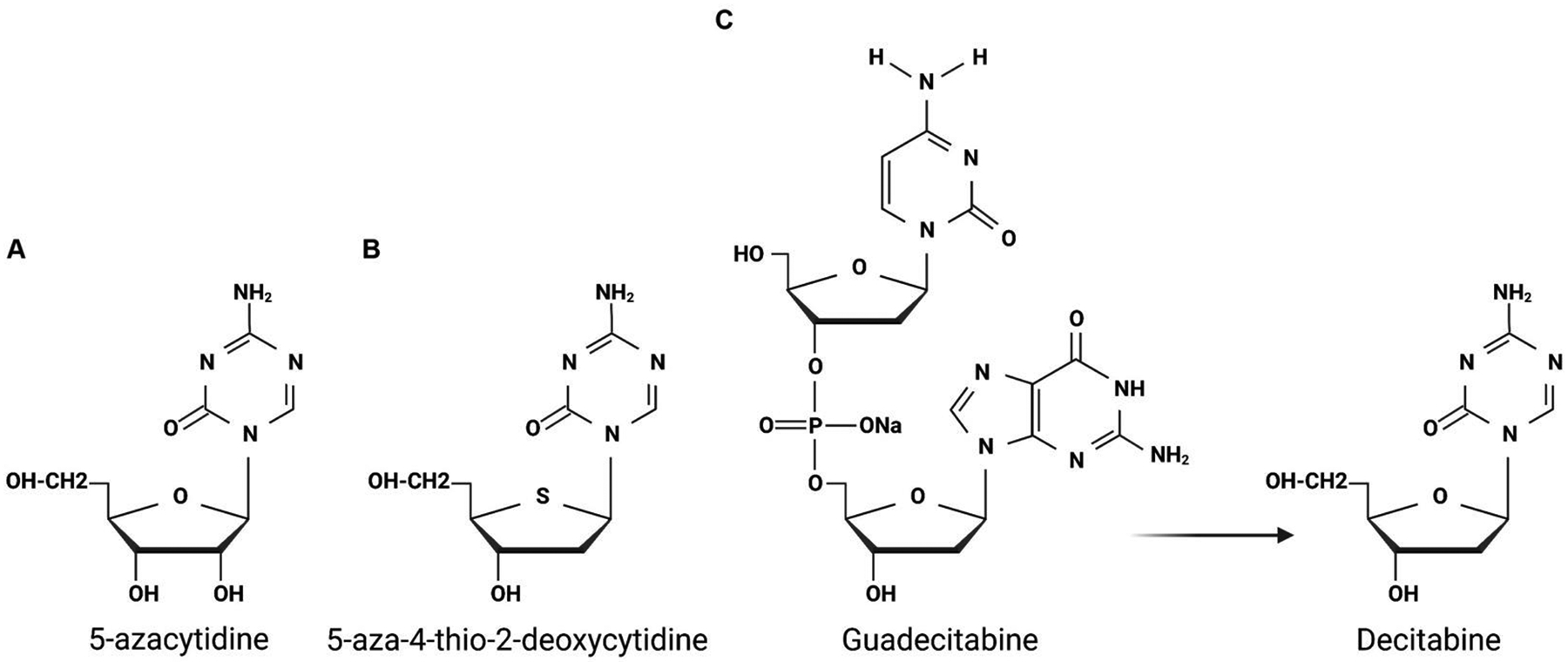
(A) Structures of 5-azacytidine and (B) 5-aza-4-thio-2-deoxycytidine. (C) Molecular structure of Guadecitabine and how it can be converted to its bioactive component, Decitabine.
Guadecitabine, another 5-azacytidine analog, was developed to circumvent deactivation. It is converted into its bioactive metabolite, Decitabine, and is more stable in the liver since it is not readily deactivated by cystine deaminase(Bennett and Licht 2018) (Fig 5C). Decitabine has been used in a completed phase 2 clinical trial that aimed to treat colorectal metastasis to the liver (NCT02316028). During cell replication, Decitabine, similar to 5-azacytidine and other analogs, is incorporated into DNA and causes genome-wide hypomethylation which in turn is expected to reverse the suppressed expression of tumor suppressor genes(Derissen, Beijnen et al. 2013, Kordella, Lamprianidou et al. 2021). Interestingly, pre-clinical studies demonstrated that Guadecitabine in cell lines can not only activate the expression of tumor suppressor genes, but also the expression of endogenous retroviruses(Liu, Zhang et al. 2018). Endogenous retroviruses are small pieces of genetic material that are normally silenced, and when reactivated can lead to a significant innate inflammatory response(Chiappinelli, Strissel et al. 2015, Roulois, Loo Yau et al. 2015, Liu, Ohtani et al. 2016, Stone, Chiappinelli et al. 2017). Thus, it reasoned that DNMTi treatment may work in tandem with immune checkpoint inhibitors (ICIs). Since the current standard of care in HCC and many other malignancies has recently implemented ICIs, clinical trials have suggested combining ICIs with Guadecitabine (NCT03257761 and NCT01799083)(Liu, Zhang et al. 2018, Moufarrij, Srivastava et al. 2020). One Phase 1b trial combines the use of Guadecitabine and the PD-1 checkpoint inhibitor Durvalumab for primary HCC and other hepatobiliary malignancies (NCT03257761). While promising, minimal in vivo pre-clinical work in the liver has been conducted using Guadecitabine, thus a full understanding of the molecular mechanisms influenced by Guadecitabine is needed. Therefore, using Guadecitabine may not only reactivate the expression of tumor suppressor genes, but also induce an inflammatory response which in combination with checkpoint inhibitors may be a viable approach for patients.
Concluding remarks
It is widely accepted that epigenetic alterations are critical in the onset and progression of various liver diseases, including liver cancer. Truly, differential DNA methylation is a delicate, two-way street in the liver that requires sensitive regulation. While there are beneficial consequences regarding specific cellular responses, such as proliferation and fate conversion that ultimately contribute to restoring normal liver function, this process can become dysregulated and result in tumorigenesis. Given the high prevalence of DNA hypermethylation in liver cancer, this field has grown significantly in the last decade, and technological advances have dramatically accelerated related research. Accordingly, these advances have led to the identification of many methylation targets that provide high sensitivity and specificity in liver cancer surveillance. While promising, since DNMTs/TETs lack specific binding motifs, other than CpG sequences, the full understanding of their function is complicated due to the lack of understanding of genetic tropism of DNA (de)methylation in the liver(Irizarry, Ladd-Acosta et al. 2009). Indeed, a recent study demonstrated the heterogeneity of DNA methylation located at the promoter and intergenic regions; thereby attributing diverse, distinct features of clinical CCAs(Jusakul, Cutcutache et al. 2017). These findings highlight the need for more comprehensive and unbiased complexed methylome and transcriptome analysis. As aforementioned, despite numerous publications reporting hypermethylation targets in liver cancer, upstream regulators triggering DNMT/TET expression or activity remain largely unknown. Furthermore, redundancy and crosstalk with other epigenetic transcription repressors such as HDACs should be comprehensively investigated. This is especially important in the context when DNMT/TET activity is inhibited as this may inform on the full therapeutic effect of DNMT inhibition in liver cancer. Since the anti-tumor effect of DNMT inhibition in liver cancer is minimal in both animal studies and clinical trials, a full understanding of epigenetic modifications should provide insight into how to improve these approaches. Therefore, a substantial understanding of context-dependent upstream regulators, as well as the identification of functional methylation targets, capable of rescuing the effect of DNMT inhibition, will be essential for considering the use of FDA-approved DNMTi in liver cancer. Finally, using hypermethylation targets for disease surveillance and prediction is a promising discovery and will be fundamental for managing premalignant liver diseases and early-stage liver cancer.
Acknowledgments:
Funding was provided by NIH grant 1R01CA258449 and PLRC Pilot & Feasibility grant PF 2019-05 to S.K and PF 2022-04 to E.D through 1P30DK120531 to the Pittsburgh Liver Research Center.
Abbreviation
- HC
hepatocyte
- BEC
biliary epithelial cell
- HB
hepatoblasts
- PHx
hepatectomy
- LPC
liver progenitor cell
- CCA
cholangiocarcinoma
- HCC
hepatocellular carcinoma
- ICC
intrahepatic cholangiocarcinoma
- DNMT
DNA methyltransferase
- 5mC
5-methylcytosine
- CGI
CpG island
- H3K4me3
trimethylated histone H3 Lys4
- PRC2
Polycomb repressive complex 2
- H3K27me3
trimethylated histone H3 Lys27
- PWWP
Pro-Trp-Trp-Pro
- ADD
ATRX-DNMT3L-DNMT3A
- H3K36me3
trimethylated histone H3 Lys36
- MTase
methyltransferase
- UHRF1
Ubiquitin Like With PHD And Ring Finger Domains 1
- DMAP1
DNMT1-associated protein 1
- PCNA
proliferating cell nuclear antigen
- RFTS
replication foci-targeting sequence
- BAH
Bromo-adjacent homology
- HDAC2
histone deacetylase 2
- UBL
ubiquitin-like
- TTD-PHD
tandem TUDOR-PHD
- SRA
SET-and RING-associated
- RING
really interesting new gene
- H3R2
histone H3 Arg2
- H3K9me2
dimethylated histone H3 Lys9
- H3K14
histone H3 Lys14
- H3K18
histone H3 Lys18
- H3K23
histone H3 Lys 23
- LSH
lymphocyte-specific helicase
- MBD
methyl-CpG-binding domain
- MeCP2
methyl-CpG binding protein 2
- TET
ten-eleven translocation
- 5hmC
5-Hydroxymethylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- DSBH
double-stranded β-helix
- DDC
3,5-Diethoxycarbonyl-1,4-Dihydrocollidine
- DR
ductal reaction
- TSS
transcription start site
- mTOR
mammalian target of rapamycin
- MAPK
mitogen-activated protein kinase
- YAP
Yes1 Associated Transcriptional Regulator
- NOTCH
Neurogenic locus notch homolog protein 1
- SOX9
SRY-Box Transcription Factor 9
- LPC
liver progenitor cell
- ERFR
Epidermal Growth Factor Receptor
- FXR
Farnesoid X receptor
- AKT
AKT Serine/Threonine Kinase 1
- TP53
Tumor protein P53
- HNF4A
Hepatocyte Nuclear Factor 4 Alpha
- C/EBP
CCAAT Enhancer Binding Protein Beta
- FOXA
Forkhead Box A1
- TEAD
TEA Domain Transcription Factor 1
- HNF1β
Hepatocyte nuclear factor-1beta
- TERT
Telomerase Reverse Transcriptase
- CTNNB1
Catenin Beta 1
- ARID1A
AT-Rich Interaction Domain 1A
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- PPAR
Peroxisome Proliferator Activated Receptor
- TGFβ1
Transforming Growth Factor Beta 1
- ARLD
Alcohol-related liver disease
- FKBP5
FKBP Prolyl Isomerase 5
- CXCL1
C-X-C Motif Chemokine Ligand 1
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- LINE-1
Long Interspersed Nuclear Elements 1
- RE
Repetitive Element
- IL6
Interleukin 6
- STAT3
Signal Transducer And Activator Of Transcription 3
- ICI
immune checkpoint inhibitor
- IPNL/B
intraductal papillary neoplasm of the liver/bile ducts
- BilIN
biliary intraepithelial neoplasia
- cfDNA
cell-free DNAs
- PDAC
Pancreatic ductal adenocarcinoma
- DNMTi
DNMT inhibitors
- Aza-TdC
5-aza-4 -thio-2 -deoxycytidine
- PD-1
Programmed Cell Death 1
- MCD
methionine- and choline-deficient
- TAA
thioacetamide
- CDE
choline-deficient ethionine-supplemented
- BDL
bile duct ligation
- DAPM
methylene dianiline
- CCL4
carbon tetrachloride
- NICD
Notch intracellular domain
- Rbpj
Recombination Signal Binding Protein For Immunoglobulin Kappa J Region
- IRES
Internal ribosome entry site
- NTR
nitroreductase
- Mtz
Metronidazole
- CK19
Keratin 19
- Alb
Albumin
- BET
bromodomain and extra-terminal domain
- HDTVI
hydrodynamic tail vein injection
- AAV8
adeno-associated virus serotype 8
- BDL
bile duct ligation
- DEN
diethylnitrosamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
None of the authors have any interests to declare related to this study.
REFERENCES
- Aloia L (2021). “Epigenetic Regulation of Cell-Fate Changes That Determine Adult Liver Regeneration After Injury.” Front Cell Dev Biol 9: 643055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L, McKie MA, Vernaz G, Cordero-Espinoza L, Aleksieva N, van den Ameele J, Antonica F, Font-Cunill B, Raven A, Aiese Cigliano R, Belenguer G, Mort RL, Brand AH, Zernicka-Goetz M, Forbes SJ, Miska EA and Huch M (2019). “Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration.” Nat Cell Biol 21(11): 1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Guerra MC, Ronchini C and Capobianco AJ (2007). “Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival.” Cancer Res 67(18): 8690–8698. [DOI] [PubMed] [Google Scholar]
- Andresen K, Boberg KM, Vedeld HM, Honne H, Hektoen M, Wadsworth CA, Clausen OP, Karlsen TH, Foss A, Mathisen O, Schrumpf E, Lothe RA and Lind GE (2012). “Novel target genes and a valid biomarker panel identified for cholangiocarcinoma.” Epigenetics 7(11): 1249–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen K, Boberg KM, Vedeld HM, Honne H, Jebsen P, Hektoen M, Wadsworth CA, Clausen OP, Lundin KE, Paulsen V, Foss A, Mathisen O, Aabakken L, Schrumpf E, Lothe RA and Lind GE (2015). “Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma.” Hepatology 61(5): 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar SL, Hasemeier B, Schipper E, Vogel A, Kreipe H and Lehmann U (2019). “LINE-1 hypomethylation in human hepatocellular carcinomas correlates with shorter overall survival and CIMP phenotype.” PLoS One 14(5): e0216374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arechederra M, Berasain C, Avila MA and Fernandez-Barrena MG (2020). “Chromatin dynamics during liver regeneration.” Semin Cell Dev Biol 97: 38–46. [DOI] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y and Shirakawa M (2008). “Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism.” Nature 455(7214): 818–821. [DOI] [PubMed] [Google Scholar]
- Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M and Shirakawa M (2012). “Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1.” Proc Natl Acad Sci U S A 109(32): 12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armartmuntree N, Jusakul A, Sakonsinsiri C, Loilome W, Pinlaor S, Ungarreevittaya P, Yong CH, Techasen A, Imtawil K, Kraiklang R, Suwannakul N, Kaewlert W, Chaiprasert T, Thanan R and Murata M (2021). “Promoter hypermethylation of early B cell factor 1 (EBF1) is associated with cholangiocarcinoma progression.” J Cancer 12(9): 2673–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH and Dhe-Paganon S (2008). “Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.” Nature 455(7214): 822–825. [DOI] [PubMed] [Google Scholar]
- Baba Y, Yagi T, Sawayama H, Hiyoshi Y, Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N and Baba H (2018). “Long Interspersed Element-1 Methylation Level as a Prognostic Biomarker in Gastrointestinal Cancers.” Digestion 97(1): 26–30. [DOI] [PubMed] [Google Scholar]
- Bai X, Zhang H, Zhou Y, Nagaoka K, Meng J, Ji C, Liu D, Dong X, Cao K, Mulla J, Cheng Z, Mueller W, Bay A, Hildebrand G, Lu S, Wallace J, Wands JR, Sun B and Huang CK (2021). “Ten-Eleven Translocation 1 Promotes Malignant Progression of Cholangiocarcinoma With Wild-Type Isocitrate Dehydrogenase 1.” Hepatology 73(5): 1747–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena-Varela M, Caruso S, Llerena S, Alvarez-Sola G, Uriarte I, Latasa MU, Urtasun R, Rebouissou S, Alvarez L, Jimenez M, Santamaria E, Rodriguez-Ortigosa C, Mazza G, Rombouts K, San Jose-Eneriz E, Rabal O, Agirre X, Iraburu M, Santos-Laso A, Banales JM, Zucman-Rossi J, Prosper F, Oyarzabal J, Berasain C, Avila MA and Fernandez-Barrena MG (2019). “Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma.” Hepatology 69(2): 587–603. [DOI] [PubMed] [Google Scholar]
- Basu A and Tiwari VK (2021). “Epigenetic reprogramming of cell identity: lessons from development for regenerative medicine.” Clin Epigenetics 13(1): 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB (2005). “DNA methylation and gene silencing in cancer.” Nat Clin Pract Oncol 2 Suppl 1: S4–11. [DOI] [PubMed] [Google Scholar]
- Bayo J, Fiore EJ, Dominguez LM, Real A, Malvicini M, Rizzo M, Atorrasagasti C, Garcia MG, Argemi J, Martinez ED and Mazzolini GD (2019). “A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets.” J Hepatol 71(1): 78–90. [DOI] [PubMed] [Google Scholar]
- Bennett RL and Licht JD (2018). “Targeting Epigenetics in Cancer.” Annu Rev Pharmacol Toxicol 58: 187–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W and Balasubramanian S (2012). “Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution.” Science 336(6083): 934–937. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S and Jacobsen SE (2007). “UHRF1 plays a role in maintaining DNA methylation in mammalian cells.” Science 317(5845): 1760–1764. [DOI] [PubMed] [Google Scholar]
- Braconi C, Huang N and Patel T (2010). “MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes.” Hepatology 51(3): 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne CD and Targher G (2015). “NAFLD: a multisystem disease.” J Hepatol 62(1 Suppl): S47–64. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Electronic address, w. b. e. and N. Cancer Genome Atlas Research (2017). “Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma.” Cell 169(7): 1327–1341 e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Liu L, Cao X, Cui Y, Zou C, Chen A, Qiu Y, Quan M, Ren K, Chen X and Cao J (2020). “The DNMT1/miR-34a/FOXM1 Axis Contributes to Stemness of Liver Cancer Cells.” J Oncol 2020: 8978930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani NP, Ramasubramanian TS, Bhattacharya A, Olson MC, Edwards VD, Roberts LR, Kisiel JB, Reddy KR, Lidgard GP, Johnson SC and Bruinsma JJ (2021). “A Novel Blood-Based Panel of Methylated DNA and Protein Markers for Detection of Early-Stage Hepatocellular Carcinoma.” Clin Gastroenterol Hepatol 19(12): 2597–2605 e2594. [DOI] [PubMed] [Google Scholar]
- Cheedipudi S, Genolet O and Dobreva G (2014). “Epigenetic inheritance of cell fates during embryonic development.” Front Genet 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wu H, Feng X, Chen Y, Lv Z, Kota VG, Chen J, Wu W, Lu Y, Liu H, Zhang Y, Zheng S and Wu J (2021). “DNA Methylation of Cannabinoid Receptor Interacting Protein 1 Promotes Pathogenesis of Intrahepatic Cholangiocarcinoma Through Suppressing Parkin-Dependent Pyruvate Kinase M2 Ubiquitination.” Hepatology 73(5): 1816–1835. [DOI] [PubMed] [Google Scholar]
- Chen D, Wu H, He B, Lu Y, Wu W, Liu H, Feng X, Chen J and Wu J (2019). “Five Hub Genes Can Be The Potential DNA Methylation Biomarkers For Cholangiocarcinoma Using Bioinformatics Analysis.” Onco Targets Ther 12: 8355–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Z, Chen J, Du Y, Song W, Xuan Z, Zhao L, Song G, Song P and Zheng S (2020). “Downregulation of MGMT promotes proliferation of intrahepatic cholangiocarcinoma by regulating p21.” Clin Transl Oncol 22(3): 392–400. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yin D, Zhang Y, Yu L, Li XD, Zhou ZJ, Zhou SL, Gao DM, Hu J, Jin C, Wang Z, Shi YH, Cao Y, Fan J, Dai Z and Zhou J (2017). “MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis.” Cell Death Dis 8(6): e2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z and Zhang Y (2020). “Role of Mammalian DNA Methyltransferases in Development.” Annu Rev Biochem 89: 135–158. [DOI] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Budhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Chan TA, Baylin SB and Strick R (2015). “Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses.” Cell 162(5): 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi TY, Ninov N, Stainier DY and Shin D (2014). “Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish.” Gastroenterology 146(3): 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G and Li BF (1997). “Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1.” Science 277(5334): 1996–2000. [DOI] [PubMed] [Google Scholar]
- Cigliano A, Zhang S, Ribback S, Steinmann S, Sini M, Ament CE, Utpatel K, Song X, Wang J, Pilo MG, Berger F, Wang H, Tao J, Li X, Pes GM, Mancarella S, Giannelli G, Dombrowski F, Evert M, Calvisi DF, Chen X and Evert K (2022). “The Hippo pathway effector TAZ induces intrahepatic cholangiocarcinoma in mice and is ubiquitously activated in the human disease.” J Exp Clin Cancer Res 41(1): 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, Schmidt CM, Yip-Schneider MT, Allen PJ, Schattner M, Brand RE, Singhi AD, Petersen GM, Hong SM, Kim SC, Falconi M, Doglioni C, Weiss MJ, Ahuja N, He J, Makary MA, Maitra A, Hanash SM, Dal Molin M, Wang Y, Li L, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Goggins MG, Hruban RH, Wolfgang CL, Klein AP, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B and Lennon AM (2017). “Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers.” Proc Natl Acad Sci U S A 114(38): 10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colyn L, Barcena-Varela M, Alvarez-Sola G, Latasa MU, Uriarte I, Santamaria E, Herranz JM, Santos-Laso A, Arechederra M, Ruiz de Gauna M, Aspichueta P, Canale M, Casadei-Gardini A, Francesconi M, Carotti S, Morini S, Nelson LJ, Iraburu MJ, Chen C, Sangro B, Marin JJG, Martinez-Chantar ML, Banales JM, Arnes-Benito R, Huch M, Patino JM, Dar AA, Nosrati M, Oyarzabal J, Prosper F, Urman J, Cubero FJ, Trautwein C, Berasain C, Fernandez-Barrena MG and Avila MA (2021). “Dual Targeting of G9a and DNA Methyltransferase-1 for the Treatment of Experimental Cholangiocarcinoma.” Hepatology 73(6): 2380–2396. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA and Pollock DD (2011). “Repetitive elements may comprise over two-thirds of the human genome.” PLoS Genet 7(12): e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM and Bird A (2011). “CpG islands and the regulation of transcription.” Genes Dev 25(10): 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Zhang X, Li W, Feng RX, Li L, Yi GR, Zhang XN, Yin C, Yu HY, Zhang JP, Lu B, Hui L and Xie WF (2018). “Chronic Liver Injury Induces Conversion of Biliary Epithelial Cells into Hepatocytes.” Cell Stem Cell 23(1): 114–122 e113. [DOI] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q and Muegge K (2001). “Lsh, a member of the SNF2 family, is required for genome-wide methylation.” Genes Dev 15(22): 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, de Launoit Y and Fuks F (2002). “Dnmt3L is a transcriptional repressor that recruits histone deacetylase.” Nucleic Acids Res 30(17): 3831–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derissen EJ, Beijnen JH and Schellens JH (2013). “Concise drug review: azacitidine and decitabine.” Oncologist 18(5): 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayat SA and Yang Z (2020). “Impact of circulating tumor DNA in hepatocellular and pancreatic carcinomas.” J Cancer Res Clin Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H and Bergman Y (2008). “De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes.” Nat Struct Mol Biol 15(11): 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF and Pradhan S (2006). “Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication.” Genes Dev 20(22): 3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi PA, Glickman J, Jiang S, Yu A, Rudolph KL and DePinho RA (2003). “Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma.” Cancer Res 63(16): 5021–5027. [PubMed] [Google Scholar]
- Fernandez-Barrena MG, Arechederra M, Colyn L, Berasain C and Avila MA (2020). “Epigenetics in hepatocellular carcinoma development and therapy: The tip of the iceberg.” JHEP Rep 2(6): 100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folguera-Blasco N, Cuyas E, Menendez JA and Alarcon T (2018). “Epigenetic regulation of cell fate reprogramming in aging and disease: A predictive computational model.” PLoS Comput Biol 14(3): e1006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L and Kouzarides T (2000). “DNA methyltransferase Dnmt1 associates with histone deacetylase activity.” Nat Genet 24(1): 88–91. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M and Kouzarides T (2001). “Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription.” EMBO J 20(10): 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd VL, Aleksieva N and Forbes SJ (2020). “Epithelial Plasticity during Liver Injury and Regeneration.” Cell Stem Cell 27(4): 557–573. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yang M, Jiang Z, Woda BA, Mercurio AM, Qin J, Huang X and Zhang F (2014). “IMP3 expression is associated with poor outcome and epigenetic deregulation in intrahepatic cholangiocarcinoma.” Hum Pathol 45(6): 1184–1191. [DOI] [PubMed] [Google Scholar]
- Ge YY, Zhang QM, Liu C, Zeng X, Nong WX, Chen F, Bi SQ, Guo WW, Luo B and Xie XX (2021). “Combined treatment with epigenetic agents enhances anti-tumor activity of T cells by upregulating the ACRBP expression in hepatocellular carcinoma.” Am J Transl Res 13(7): 7591–7609. [PMC free article] [PubMed] [Google Scholar]
- Ge YZ, Pu MT, Gowher H, Wu HP, Ding JP, Jeltsch A and Xu GL (2004). “Chromatin targeting of de novo DNA methyltransferases by the PWWP domain.” J Biol Chem 279(24): 25447–25454. [DOI] [PubMed] [Google Scholar]
- Goncalves E, Goncalves-Reis M, Pereira-Leal JB and Cardoso J (2022). “DNA methylation fingerprint of hepatocellular carcinoma from tissue and liquid biopsies.” Sci Rep 12(1): 11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC and Bourc’his D (2019). “The diverse roles of DNA methylation in mammalian development and disease.” Nat Rev Mol Cell Biol 20(10): 590–607. [DOI] [PubMed] [Google Scholar]
- Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y and Xu Y (2015). “Structural insight into autoinhibition and histone H3-induced activation of DNMT3A.” Nature 517(7536): 640–644. [DOI] [PubMed] [Google Scholar]
- Habash NW, Sehrawat TS, Shah VH and Cao S (2022). “Epigenetics of alcohol-related liver diseases.” JHEP Rep 4(5): 100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama N, Totoki Y, Miura F, Tatsuno K, Saito-Adachi M, Nakamura H, Arai Y, Hosoda F, Urushidate T, Ohashi S, Mukai W, Hiraoka N, Aburatani H, Ito T and Shibata T (2018). “Epigenetic landscape influences the liver cancer genome architecture.” Nat Commun 9(1): 1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang Y, Pu W, Huang X, Qiu L, Li Y, Yu W, Zhao H, Liu X, He L, Zhang L, Ji Y, Lu J, Lui KO and Zhou B (2019). “Lineage Tracing Reveals the Bipotency of SOX9(+) Hepatocytes during Liver Regeneration.” Stem Cell Reports 12(3): 624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding JJ, Zhu AX, Bauer TM, Choueiri TK, Drilon A, Voss MH, Fuchs CS, Abou-Alfa GK, Wijayawardana SR, Wang XA, Moser BA, Urunuela A, Wacheck V and Bendell JC (2019). “A Phase Ib/II Study of Ramucirumab in Combination with Emibetuzumab in Patients with Advanced Cancer.” Clin Cancer Res 25(17): 5202–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy T, Zeybel M, Day CP, Dipper C, Masson S, McPherson S, Henderson E, Tiniakos D, White S, French J, Mann DA, Anstee QM and Mann J (2017). “Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease.” Gut 66(7): 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE and Cheng X (2008). “The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix.” Nature 455(7214): 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Lu H, Zou Q and Luo L (2014). “Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish.” Gastroenterology 146(3): 789–800 e788. [DOI] [PubMed] [Google Scholar]
- He J, Zhou Y, Qian C, Wang D, Yang Z, Huang Z, Sun J, Ni R, Yang Q, Chen J and Luo L (2022). “DNA methylation maintenance at the p53 locus initiates biliary-mediated liver regeneration.” NPJ Regen Med 7(1): 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C and Xu GL (2011). “Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA.” Science 333(6047): 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Logan CV, Fluteau A, Challis RC, Auchynnikava T, Martin CA, Marsh JA, Taglini F, Kilanowski F, Parry DA, Cormier-Daire V, Fong CT, Gibson K, Hwa V, Ibanez L, Robertson SP, Sebastiani G, Rappsilber J, Allshire RC, Reijns MAM, Dauber A, Sproul D and Jackson AP (2019). “Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions.” Nat Genet 51(1): 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlady RA, Tiedemann RL, Puszyk W, Zendejas I, Roberts LR, Choi JH, Liu C and Robertson KD (2014). “Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis.” Oncotarget 5(19): 9425–9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM and Chaillet JR (2001). “Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene.” Cell 104(6): 829–838. [DOI] [PubMed] [Google Scholar]
- Hu C, Liu X, Zeng Y, Liu J and Wu F (2021). “DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application.” Clin Epigenetics 13(1): 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Molina L, Tao J, Liu S, Hassan M, Singh S, Poddar M, Bell A, Sia D, Oertel M, Raeman R, Nejak-Bowen K, Singhi A, Luo J, Monga SP and Ko S (2022). “NOTCH-YAP1/TEAD-DNMT1 Axis Drives Hepatocyte Reprogramming into Intrahepatic Cholangiocarcinoma.” Gastroenterology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Molina L, Tao J, Liu S, Hassan M, Singh S, Poddar M, Bell A, Sia D, Oertel M, Raeman R, Nejak-Bowen K, Singhi A, Luo J, Monga SP and Ko S (2022). “NOTCH-YAP1/TEAD-DNMT1 Axis Drives Hepatocyte Reprogramming Into Intrahepatic Cholangiocarcinoma.” Gastroenterology 163(2): 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S and Monga SP (2021). “Wnt/-Catenin Signaling and Liver Regeneration: Circuit, Biology, and Opportunities.” Gene Expr 20(3): 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Chang A, Choi M, Zhou D, Anania FA and Shin CH (2014). “Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model.” Hepatology 60(5): 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J, Al Abo M, Dutta RK, Oh SH, Xiang K, Zhou X, Maeso-Diaz R, Caffrey R, Sanyal AJ, Freedman JA, Patierno SR, Moylan CA, Abdelmalek MF and Diehl AM (2021). “Dysregulation of the ESRP2-NF2-YAP/TAZ axis promotes hepatobiliary carcinogenesis in non-alcoholic fatty liver disease.” J Hepatol 75(3): 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash J, Sabunciyan S and Feinberg AP (2009). “The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores.” Nat Genet 41(2): 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, Hojo H, Shimamura S, Ishikawa F, Tajima S, Tanaka K, Ariyoshi M, Shirakawa M, Ikeguchi M, Kidera A, Suetake I, Arita K and Nakanishi M (2017). “Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance.” Mol Cell 68(2): 350–360 e357. [DOI] [PubMed] [Google Scholar]
- Isomoto H (2009). “Epigenetic alterations in cholangiocarcinoma-sustained IL-6/STAT3 signaling in cholangio- carcinoma due to SOCS3 epigenetic silencing.” Digestion 79 Suppl 1: 2–8. [DOI] [PubMed] [Google Scholar]
- Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S and Gores GJ (2007). “Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing.” Gastroenterology 132(1): 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC and Zhang Y (2010). “Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification.” Nature 466(7310): 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C and Zhang Y (2011). “Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.” Science 333(6047): 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izykowska K (2020). “Methylation patterns of cutaneous T-cell lymphomas.” Exp Dermatol. [DOI] [PubMed] [Google Scholar]
- Jeong S, Lee K, Wen X, Kim Y, Cho NY, Jang JJ and Kang GH (2017). “Tumoral LINE-1 hypomethylation is associated with poor survival of patients with intrahepatic cholangiocarcinoma.” BMC Cancer 17(1): 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A and Cheng X (2007). “Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.” Nature 449(7159): 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jueliger S, Lyons J, Cannito S, Pata I, Pata P, Shkolnaya M, Lo Re O, Peyrou M, Villarroya F, Pazienza V, Rappa F, Cappello F, Azab M, Taverna P and Vinciguerra M (2016). “Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma.” Epigenetics 11(10): 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Kim M, So J, Lee SH, Ko S and Shin D (2021). “Farnesoid X Receptor Activation Impairs Liver Progenitor Cell-Mediated Liver Regeneration via the PTEN-PI3K-AKT-mTOR Axis in Zebrafish.” Hepatology 74(1): 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, Choo SP, Myint SS, Thanan R, Nagarajan S, Lim WK, Ng CCY, Boot A, Liu M, Ong CK, Rajasegaran V, Lie S, Lim AST, Lim TH, Tan J, Loh JL, McPherson JR, Khuntikeo N, Bhudhisawasdi V, Yongvanit P, Wongkham S, Totoki Y, Nakamura H, Arai Y, Yamasaki S, Chow PK, Chung AYF, Ooi L, Lim KH, Dima S, Duda DG, Popescu I, Broet P, Hsieh SY, Yu MC, Scarpa A, Lai J, Luo DX, Carvalho AL, Vettore AL, Rhee H, Park YN, Alexandrov LB, Gordan R, Rozen SG, Shibata T, Pairojkul C, Teh BT and Tan P (2017). “Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma.” Cancer Discov 7(10): 1116–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Cho NY, Choi M, Lee S, Jang JJ and Kang GH (2007). “Methylation profiles of multiple CpG island loci in extrahepatic cholangiocarcinoma versus those of intrahepatic cholangiocarcinomas.” Arch Pathol Lab Med 131(6): 923–930. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lee K, Jeong S, Wen X, Cho NY and Kang GH (2019). “DLEC1 methylation is associated with a better clinical outcome in patients with intrahepatic cholangiocarcinoma of the small duct subtype.” Virchows Arch 475(1): 49–58. [DOI] [PubMed] [Google Scholar]
- Ko S, Choi TY, Russell JO, So J, Monga SPS and Shin D (2016). “Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration.” J Hepatol 64(2): 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S, Kim M, Molina L, Sirica AE and Monga SP (2022). “YAP1 activation and Hippo pathway signaling in the pathogenesis and treatment of intrahepatic cholangiocarcinoma.” Adv Cancer Res 156: 283–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S, Russell JO, Molina LM and Monga SP (2019). “Liver Progenitors and Adult Cell Plasticity in Hepatic Injury and Repair: Knowns and Unknowns.” Annu Rev Pathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S, Russell JO, Tian J, Gao C, Kobayashi M, Feng R, Yuan X, Shao C, Ding H, Poddar M, Singh S, Locker J, Weng HL, Monga SP and Shin D (2019). “Hdac1 Regulates Differentiation of Bipotent Liver Progenitor Cells During Regeneration via Sox9b and Cdk8.” Gastroenterology 156(1): 187–202 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Chen J, Xie W, Brown SM, Cai Y, Wu K, Fan D, Nie Y, Yegnasubramanian S, Tiedemann RL, Tao Y, Chiu Yen RW, Topper MJ, Zahnow CA, Easwaran H, Rothbart SB, Xia L and Baylin SB (2019). “Defining UHRF1 Domains that Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties.” Cancer Cell 35(4): 633–648 e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordella C, Lamprianidou E and Kotsianidis I (2021). “Mechanisms of Action of Hypomethylating Agents: Endogenous Retroelements at the Epicenter.” Front Oncol 11: 650473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S and Heintz N (2009). “The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain.” Science 324(5929): 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y, El-Khoueiry A, Taverna P, Ljungman M and Neamati N (2015). “Guadecitabine (SGI-110) priming sensitizes hepatocellular carcinoma cells to oxaliplatin.” Mol Oncol 9(9): 1799–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M and Esteller M (2010). “DNA methylation and cancer.” Adv Genet 70: 27–56. [DOI] [PubMed] [Google Scholar]
- Kumar P, Raeman R, Chopyk DM, Smith T, Verma K, Liu Y and Anania FA (2018). “Adiponectin inhibits hepatic stellate cell activation by targeting the PTEN/AKT pathway.” Biochim Biophys Acta Mol Basis Dis 1864(10): 3537–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto J, Arai E, Tian Y, Funahashi N, Hiramoto M, Nammo T, Nozaki Y, Takahashi Y, Ito N, Shibuya A, Ojima H, Sukeda A, Seki Y, Kasama K, Yasuda K and Kanai Y (2017). “Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis.” Carcinogenesis 38(3): 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa S, Yoneda M, Ogawa Y, Honda Y, Kessoku T, Imajo K, Saito S, Nakajima A and Hotta K (2022). “Two differentially methylated region networks in nonalcoholic fatty liver disease, viral hepatitis, and hepatocellular carcinoma.” BMC Gastroenterol 22(1): 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumanchi P, Liang T, Zhang T, Ross RA, Han S, Chandler K, Oshodi A, Jiang Y, Dent AL, Skill NJ, Huda N, Ma J, Yang Z and Liangpunsakul S (2021). “Stress-Responsive Gene FK506-Binding Protein 51 Mediates Alcohol-Induced Liver Injury Through the Hippo Pathway and Chemokine (C-X-C Motif) Ligand 1 Signaling.” Hepatology 74(3): 1234–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R and Prydz H (1992). “CpG islands as gene markers in the human genome.” Genomics 13(4): 1095–1107. [DOI] [PubMed] [Google Scholar]
- Lazarus JV, Mark HE, Anstee QM, Arab JP, Batterham RL, Castera L, Cortez-Pinto H, Crespo J, Cusi K, Dirac MA, Francque S, George J, Hagstrom H, Huang TT, Ismail MH, Kautz A, Sarin SK, Loomba R, Miller V, Newsome PN, Ninburg M, Ocama P, Ratziu V, Rinella M, Romero D, Romero-Gomez M, Schattenberg JM, Tsochatzis EA, Valenti L, Wong VW, Yilmaz Y, Younossi ZM, Zelber-Sagi S and Consortium NC (2022). “Advancing the global public health agenda for NAFLD: a consensus statement.” Nat Rev Gastroenterol Hepatol 19(1): 60–78. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ and N. Cancer Genome Atlas Research (2012). “Landscape of somatic retrotransposition in human cancers.” Science 337(6097): 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim WH, Jung HY, Yang MH and Kang GH (2002). “Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma.” Am J Pathol 161(3): 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E and Zhang Y (2014). “DNA methylation in mammals.” Cold Spring Harb Perspect Biol 6(5): a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wu BK, Kanchwala M, Cai J, Wang L, Xing C, Zheng Y and Pan D (2022). “YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis.” Nat Cell Biol 24(3): 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J, Shi BJ, Nie J, Ding XT, Li B, Zhou GW and Zhang ZY (2017). “MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma.” Hepatology 66(1): 136–151. [DOI] [PubMed] [Google Scholar]
- Li SJ, Wang L, Sun ZX, Sun SJ, Gao J and Ma RL (2019). “LncRNA SNHG1 promotes liver cancer development through inhibiting p53 expression via binding to DNMT1.” Eur Rev Med Pharmacol Sci 23(7): 2768–2776. [DOI] [PubMed] [Google Scholar]
- Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z and Qian C (2018). “Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation.” Nucleic Acids Res 46(6): 3218–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang L, He Q, Hu C, Zhu L, Ma X, Ma X, Bao S, Li L, Chen Y, Deng X, Zhang X, Cen J, Zhang L, Wang Z, Xie WF, Li H, Li Y and Hui L (2019). “A Homeostatic Arid1a-Dependent Permissive Chromatin State Licenses Hepatocyte Responsiveness to Liver-Injury-Associated YAP Signaling.” Cell Stem Cell 25(1): 54–68 e55. [DOI] [PubMed] [Google Scholar]
- Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM and Jung G (2008). “Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter.” Gastroenterology 135(6): 2128–2140, 2140 e2121–2128. [DOI] [PubMed] [Google Scholar]
- Limpaiboon T (2012). “Epigenetic aberrations in cholangiocarcinoma: potential biomarkers and promising target for novel therapeutic strategies.” Asian Pac J Cancer Prev 13 Suppl: 41–45. [PubMed] [Google Scholar]
- Limpaiboon T, Khaenam P, Chinnasri P, Soonklang M, Jearanaikoon P, Sripa B, Pairojkul C and Bhudhisawasdi V (2005). “Promoter hypermethylation is a major event of hMLH1 gene inactivation in liver fluke related cholangiocarcinoma.” Cancer Lett 217(2): 213–219. [DOI] [PubMed] [Google Scholar]
- Lissa D and Robles AI (2016). “Methylation analyses in liquid biopsy.” Transl Lung Cancer Res 5(5): 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Ohtani H, Zhou W, Orskov AD, Charlet J, Zhang YW, Shen H, Baylin SB, Liang G, Gronbaek K and Jones PA (2016). “Vitamin C increases viral mimicry induced by 5-aza-2’-deoxycytidine.” Proc Natl Acad Sci U S A 113(37): 10238–10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang L, Li H, Hinoue T, Zhou W, Ohtani H, El-Khoueiry A, Daniels J, O’Connell C, Dorff TB, Lu Q, Weisenberger DJ and Liang G (2018). “Integrative Epigenetic Analysis Reveals Therapeutic Targets to the DNA Methyltransferase Inhibitor Guadecitabine (SGI-110) in Hepatocellular Carcinoma.” Hepatology 68(4): 1412–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zhou TF, Qiu BA, Yang YX, Zhu YJ, An Y, Zhao WC, Wu YT, Ma PF, Li JB and Xia NX (2018). “Methylation-Mediated Silencing of GATA5 Gene Suppresses Cholangiocarcinoma Cell Proliferation and Metastasis.” Transl Oncol 11(3): 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng C, Cai Y, Gu Y, Wu Y, Chen K and Wu Z (2022). “Pancancer analyses reveal the regulation and clinical outcome association of PCLAF in human tumors.” Int J Oncol 60(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Jiang H, Zhang CS, Yu SP, Wang ZQ and Su HL (2012). “Targeted drug regulation on methylation of p53-BAX mitochondrial apoptosis pathway affects the growth of cholangiocarcinoma cells.” J Int Med Res 40(1): 67–75. [DOI] [PubMed] [Google Scholar]
- Liu XF, Kong FM, Xu Z, Yu SP, Sun FB, Zhang CS, Huang QX, Zhou XT and Song ZW (2007). “Promoter hypermethylation of death-associated protein kinase gene in cholangiocarcinoma.” Hepatobiliary Pancreat Dis Int 6(4): 407–411. [PubMed] [Google Scholar]
- Lu L, Bi J and Bao L (2018). “Genetic profiling of cancer with circulating tumor DNA analysis.” J Genet Genomics 45(2): 79–85. [DOI] [PubMed] [Google Scholar]
- Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, Ridgway RA, Kendall T, Williams MJ, Jamieson T, Raven A, Hay DC, Iredale JP, Clarke AR, Sansom OJ and Forbes SJ (2015). “Hepatic progenitor cells of biliary origin with liver repopulation capacity.” Nat Cell Biol 17(8): 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi F and Sadler KC (2020). “Unraveling the Epigenetic Basis of Liver Development, Regeneration and Disease.” Trends Genet 36(8): 587–597. [DOI] [PubMed] [Google Scholar]
- Manco R, Clerbaux LA, Verhulst S, Bou Nader M, Sempoux C, Ambroise J, Bearzatto B, Gala JL, Horsmans Y, van Grunsven L, Desdouets C and Leclercq I (2019). “Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury.” J Hepatol 70(6): 1180–1191. [DOI] [PubMed] [Google Scholar]
- Marasca F, Bodega B and Orlando V (2018). “How Polycomb-Mediated Cell Memory Deals With a Changing Environment: Variations in PcG complexes and proteins assortment convey plasticity to epigenetic regulation as a response to environment.” Bioessays 40(4): e1700137. [DOI] [PubMed] [Google Scholar]
- Martinez-Quetglas I, Pinyol R, Dauch D, Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A, Rodriguez-Carunchio L, Sole M, Lujambio A, Villanueva A, Thung S, Esteller M, Zender L and Llovet JM (2016). “IGF2 Is Up-regulated by Epigenetic Mechanisms in Hepatocellular Carcinomas and Is an Actionable Oncogene Product in Experimental Models.” Gastroenterology 151(6): 1192–1205. [DOI] [PubMed] [Google Scholar]
- Martinez-Redondo P and Izpisua Belmonte JC (2020). “Tailored chromatin modulation to promote tissue regeneration.” Semin Cell Dev Biol 97: 3–15. [DOI] [PubMed] [Google Scholar]
- Maryam M and Idrees M (2018). “Study of promoter hypomethylation profiles of RAS oncogenes in hepatocellular carcinoma derived from hepatitis C virus genotype 3a in Pakistani population.” J Med Virol 90(9): 1516–1523. [DOI] [PubMed] [Google Scholar]
- McAnena P, Brown JA and Kerin MJ (2017). “Circulating Nucleosomes and Nucleosome Modifications as Biomarkers in Cancer.” Cancers (Basel) 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Brandes JC and Vertino PM (2009). “Cancer DNA methylation: molecular mechanisms and clinical implications.” Clin Cancer Res 15(12): 3927–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Wehbe-Janek H, Henson R, Smith H and Patel T (2008). “Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes.” Oncogene 27(3): 378–386. [DOI] [PubMed] [Google Scholar]
- Merino-Azpitarte M, Lozano E, Perugorria MJ, Esparza-Baquer A, Erice O, Santos-Laso A, O’Rourke CJ, Andersen JB, Jimenez-Aguero R, Lacasta A, D’Amato M, Briz O, Jalan-Sakrikar N, Huebert RC, Thelen KM, Gradilone SA, Aransay AM, Lavin JL, Fernandez-Barrena MG, Matheu A, Marzioni M, Gores GJ, Bujanda L, Marin JJG and Banales JM (2017). “SOX17 regulates cholangiocyte differentiation and acts as a tumor suppressor in cholangiocarcinoma.” J Hepatol 67(1): 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell AJ, Peng T, Li J, Sun K, Li B, Katsuda T, Grompe M, Tan K and Stanger BZ (2021). “Dynamic Transcriptional and Epigenetic Changes Drive Cellular Plasticity in the Liver.” Hepatology 74(1): 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK (2017). “Hepatostat: Liver regeneration and normal liver tissue maintenance.” Hepatology 65(4): 1384–1392. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK and Bhushan B (2021). “Liver regeneration: biological and pathological mechanisms and implications.” Nat Rev Gastroenterol Hepatol 18(1): 40–55. [DOI] [PubMed] [Google Scholar]
- Minnis-Lyons SE, Ferreira-Gonzalez S, Aleksieva N, Man TY, Gadd VL, Williams MJ, Guest RV, Lu WY, Dwyer BJ, Jamieson T, Nixon C, Van Hul N, Lemaigre FP, McCafferty J, Leclercq IA, Sansom OJ, Boulter L and Forbes SJ (2021). “Notch-IGF1 signaling during liver regeneration drives biliary epithelial cell expansion and inhibits hepatocyte differentiation.” Sci Signal 14(688). [DOI] [PubMed] [Google Scholar]
- Mody K, Kasi PM, Yang J, Surapaneni PK, Bekaii-Saab T, Ahn DH, Mahipal A, Sonbol MB, Starr JS, Roberts A, Nagy R, Lanman R and Borad MJ (2019). “Circulating Tumor DNA Profiling of Advanced Biliary Tract Cancers.” JCO Precis Oncol 3: 1–9. [DOI] [PubMed] [Google Scholar]
- Monga SP (2015). “beta-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis.” Gastroenterology 148(7): 1294–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris N, Pina C and Arias AM (2016). “Transition states and cell fate decisions in epigenetic landscapes.” Nat Rev Genet 17(11): 693–703. [DOI] [PubMed] [Google Scholar]
- Moruzzi S, Guarini P, Udali S, Ruzzenente A, Guglielmi A, Conci S, Pattini P, Martinelli N, Olivieri O, Tammen SA, Choi SW and Friso S (2017). “One-carbon genetic variants and the role of MTHFD1 1958G>A in liver and colon cancer risk according to global DNA methylation.” PLoS One 12(10): e0185792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moufarrij S, Srivastava A, Gomez S, Hadley M, Palmer E, Austin PT, Chisholm S, Diab N, Roche K, Yu A, Li J, Zhu W, Lopez-Acevedo M, Villagra A and Chiappinelli KB (2020). “Combining DNMT and HDAC6 inhibitors increases anti-tumor immune signaling and decreases tumor burden in ovarian cancer.” Sci Rep 10(1): 3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, Lachenmayer A, Revill K, Alsinet C, Sachidanandam R, Desai A, SenBanerjee S, Ukomadu C, Llovet JM and Sadler KC (2014). “UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma.” Cancer Cell 25(2): 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K, Termanis A, Sundaram AY, Boe T, Li C, Merusi C, Burrage J, de Las Heras JI and Stancheva I (2011). “LSH and G9a/GLP complex are required for developmentally programmed DNA methylation.” Genome Res 21(1): 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, Wang YX, Bronner C, Chedin F, Arrowsmith CH and Dhe-Paganon S (2011). “Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.” J Biol Chem 286(27): 24300–24311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama Y, Sone M, Chen X, Okada Y, Yamamoto M, Xin B, Matsuo Y, Komatsu M, Suzuki A, Enomoto K and Nishikawa Y (2014). “Contributions of hepatocytes and bile ductular cells in ductular reactions and remodeling of the biliary system after chronic liver injury.” Am J Pathol 184(11): 3001–3012. [DOI] [PubMed] [Google Scholar]
- Nagashio R, Arai E, Ojima H, Kosuge T, Kondo Y and Kanai Y (2011). “Carcinogenetic risk estimation based on quantification of DNA methylation levels in liver tissue at the precancerous stage.” Int J Cancer 129(5): 1170–1179. [DOI] [PubMed] [Google Scholar]
- Nakamoto S, Kumamoto Y, Igarashi K, Fujiyama Y, Nishizawa N, Ei S, Tajima H, Kaizu T, Watanabe M and Yamashita K (2018). “Methylated promoter DNA of CDO1 gene and preoperative serum CA19–9 are prognostic biomarkers in primary extrahepatic cholangiocarcinoma.” PLoS One 13(10): e0205864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka T, Saito Y and Saito H (2017). “Aberrant DNA Methylation as a Biomarker and a Therapeutic Target of Cholangiocarcinoma.” Int J Mol Sci 18(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanok C, Jearanaikoon P, Proungvitaya S and Limpaiboon T (2018). “Aberrant methylation of HTATIP2 and UCHL1 as a predictive biomarker for cholangiocarcinoma.” Mol Med Rep 17(3): 4145–4153. [DOI] [PubMed] [Google Scholar]
- Nault JC and Zucman-Rossi J (2016). “TERT promoter mutations in primary liver tumors.” Clin Res Hepatol Gastroenterol 40(1): 9–14. [DOI] [PubMed] [Google Scholar]
- Nebbioso A, Tambaro FP, Dell’Aversana C and Altucci L (2018). “Cancer epigenetics: Moving forward.” PLoS Genet 14(6): e1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N, Kudo M, Nagasaka T, Ikai I and Goel A (2012). “Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma.” Hepatology 56(3): 994–1003. [DOI] [PubMed] [Google Scholar]
- Nishio M, Sugimachi K, Goto H, Wang J, Morikawa T, Miyachi Y, Takano Y, Hikasa H, Itoh T, Suzuki SO, Kurihara H, Aishima S, Leask A, Sasaki T, Nakano T, Nishina H, Nishikawa Y, Sekido Y, Nakao K, Shin-Ya K, Mimori K and Suzuki A (2016). “Dysregulated YAP1/TAZ and TGF-beta signaling mediate hepatocarcinogenesis in Mob1a/1b-deficient mice.” Proc Natl Acad Sci U S A 113(1): E71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, Koseki H and Nakanishi M (2013). “Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication.” Nature 502(7470): 249–253. [DOI] [PubMed] [Google Scholar]
- O’Rourke CJ, Lafuente-Barquero J and Andersen JB (2019). “Epigenome Remodeling in Cholangiocarcinoma.” Trends Cancer 5(6): 335–350. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X and Bestor TH (2007). “DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.” Nature 448(7154): 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M and Shirakawa M (2009). “Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain.” EMBO Rep 10(11): 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A, Paoli P, Moran Salvador E, White S, French J and Mann J (2016). “Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape.” J Hepatol 64(3): 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis V and Zucman-Rossi J (2022). “Pathogenesis of primary liver carcinomas.” J Hepatol. [DOI] [PubMed] [Google Scholar]
- Parker WB and Thottassery JV (2021). “5-Aza-4’-thio-2’-deoxycytidine, a New Orally Bioavailable Nontoxic “Best-in-Class”: DNA Methyltransferase 1-Depleting Agent in Clinical Development.” J Pharmacol Exp Ther 379(3): 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L and Rao A (2013). “TETonic shift: biological roles of TET proteins in DNA demethylation and transcription.” Nat Rev Mol Cell Biol 14(6): 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Meng G, Sheng X and Gao H (2021). “Transcriptome and DNA methylation analysis reveals molecular mechanisms underlying intrahepatic cholangiocarcinoma progression.” J Cell Mol Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A and Shilatifard A (2016). “Epigenetic balance of gene expression by Polycomb and COMPASS families.” Science 352(6290): aad9780. [DOI] [PubMed] [Google Scholar]
- Pogribna M, Koonce NA, Mathew A, Word B, Patri AK, Lyn-Cook B and Hammons G (2020). “Effect of titanium dioxide nanoparticles on DNA methylation in multiple human cell lines.” Nanotoxicology 14(4): 534–553. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA, Latendresse JR, Rusyn I and Beland FA (2009). “Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet.” J Hepatol 51(1): 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachayakul V, Kanchanapermpoon J, Thuwajit C, Boonyaarunnate T, Pongpaibul A, Chobson P and Thuwajit P (2017). “DNA Methylation Markers Improve the Sensitivity of Endoscopic Retrograde Cholangiopancreatography-Based Brushing Cytology in Extrahepatic Cholangiocarcinoma.” Technol Cancer Res Treat 16(6): 1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan M, Esteve PO, Chin HG, Samaranayke M, Kim GD and Pradhan S (2008). “CXXC domain of human DNMT1 is essential for enzymatic activity.” Biochemistry 47(38): 10000–10009. [DOI] [PubMed] [Google Scholar]
- Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forne I, Pichler G, Horl D, Fellinger K, Spada F, Bonapace IM, Imhof A, Harz H and Leonhardt H (2015). “DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination.” Cell Res 25(8): 911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O’Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, Kotelevtsev Y, Ffrench-Constant C, Boulter L and Forbes SJ (2017). “Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration.” Nature 547(7663): 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouissou S and Nault JC (2020). “Advances in molecular classification and precision oncology in hepatocellular carcinoma.” J Hepatol 72(2): 215–229. [DOI] [PubMed] [Google Scholar]
- Reizel Y, Sabag O, Skversky Y, Spiro A, Steinberg B, Bernstein D, Wang A, Kieckhaefer J, Li C, Pikarsky E, Levin-Klein R, Goren A, Rajewsky K, Kaestner KH and Cedar H (2018). “Postnatal DNA demethylation and its role in tissue maturation.” Nat Commun 9(1): 2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Horton JR, Zhang X, Blumenthal RM and Cheng X (2018). “Detecting and interpreting DNA methylation marks.” Curr Opin Struct Biol 53: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM and Powers S (2013). “Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma.” Gastroenterology 145(6): 1424–1435 e1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH and Strahl BD (2013). “Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation.” Genes Dev 27(11): 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH and Strahl BD (2012). “Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation.” Nat Struct Mol Biol 19(11): 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, O’Brien C and De Carvalho DD (2015). “DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts.” Cell 162(5): 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE and Baylin SB (2000). “DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci.” Nat Genet 25(3): 269–277. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Millard M, Schreiber-Agus N and DePinho RA (2000). “Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery.” Science 287(5456): 1253–1258. [DOI] [PubMed] [Google Scholar]
- Russell JO, Lu WY, Okabe H, Abrams M, Oertel M, Poddar M, Singh S, Forbes SJ and Monga SP (2019). “Hepatocyte-Specific beta-Catenin Deletion During Severe Liver Injury Provokes Cholangiocytes to Differentiate Into Hepatocytes.” Hepatology 69(2): 742–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JO and Monga SP (2018). “Wnt/beta-Catenin Signaling in Liver Development, Homeostasis, and Pathobiology.” Annu Rev Pathol 13: 351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H and Hirohashi S (2003). “Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas.” Int J Cancer 105(4): 527–532. [DOI] [PubMed] [Google Scholar]
- Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H and Hirohashi S (2001). “Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis.” Hepatology 33(3): 561–568. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Imai K, Higashi T, Taki K, Nakagawa S, Okabe H, Nitta H, Hayashi H, Chikamoto A, Ishiko T, Beppu T and Baba H (2015). “Significance of P-cadherin overexpression and possible mechanism of its regulation in intrahepatic cholangiocarcinoma and pancreatic cancer.” Cancer Sci 106(9): 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu DS, Shire AM and Roberts LR (2008). “Epigenetic DNA hypermethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets.” Liver Int 28(1): 12–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, Karns RA, Chen F, Rezvani M, Luu HY, Mattis AN, Rougemont AL, Rosenthal P, Huppert SS and Willenbring H (2018). “De novo formation of the biliary system by TGFbeta-mediated hepatocyte transdifferentiation.” Nature 557(7704): 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia C, San Jose-Eneriz E, Munera-Maravilla E, Martinez-Fernandez M, Garate L, Miranda E, Vilas-Zornoza A, Lodewijk I, Rubio C, Segrelles C, Valcarcel LV, Rabal O, Casares N, Bernardini A, Suarez-Cabrera C, Lopez-Calderon FF, Fortes P, Casado JA, Duenas M, Villacampa F, Lasarte JJ, Guerrero-Ramos F, de Velasco G, Oyarzabal J, Castellano D, Agirre X, Prosper F and Paramio JM (2019). “Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression.” Nat Med 25(7): 1073–1081. [DOI] [PubMed] [Google Scholar]
- Sekeres MA, Schuster M, Joris M, Krauter J, Maertens J, Breems D, Gyan E, Kovacsovics T, Verma A, Vyas P, Wang ES, Ching K, O’Brien T, Gallo Stampino C, Ma WW, Kudla A, Chan G and Zeidan AM (2022). “A phase 1b study of glasdegib + azacitidine in patients with untreated acute myeloid leukemia and higher-risk myelodysplastic syndromes.” Ann Hematol 101(8): 1689–1701. [DOI] [PubMed] [Google Scholar]
- Sekiya S and Suzuki A (2012). “Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes.” J Clin Invest 122(11): 3914–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendzikaite G, Hanna CW, Stewart-Morgan KR, Ivanova E and Kelsey G (2019). “A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice.” Nat Commun 10(1): 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M and Koseki H (2007). “The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA.” Nature 450(7171): 908–912. [DOI] [PubMed] [Google Scholar]
- Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH, Yu MW, Chen CJ and Santella RM (2012). “Genome-wide DNA methylation profiles in hepatocellular carcinoma.” Hepatology 55(6): 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Wang S, Zhang YJ, Wu HC, Kibriya MG, Jasmine F, Ahsan H, Wu DP, Siegel AB, Remotti H and Santella RM (2013). “Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips.” Epigenetics 8(1): 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SH, Lee K, Kim BH, Cho NY, Jang JY, Kim YT, Kim D, Jang JJ and Kang GH (2012). “Bile-based detection of extrahepatic cholangiocarcinoma with quantitative DNA methylation markers and its high sensitivity.” J Mol Diagn 14(3): 256–263. [DOI] [PubMed] [Google Scholar]
- Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V and Llovet JM (2013). “Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes.” Gastroenterology 144(4): 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J, Kim M, Lee SH, Ko S, Lee DA, Park H, Azuma M, Parsons MJ, Prober D and Shin D (2021). “Attenuating the Epidermal Growth Factor Receptor-Extracellular Signal-Regulated Kinase-Sex-Determining Region Y-Box 9 Axis Promotes Liver Progenitor Cell-Mediated Liver Regeneration in Zebrafish.” Hepatology 73(4): 1494–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH and Patel DJ (2011). “Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.” Science 331(6020): 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H and Wong LL (2013). “Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma.” PLoS One 8(2): e55761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Liu X, Wang H, Wang J, Qiao Y, Cigliano A, Utpatel K, Ribback S, Pilo MG, Serra M, Gordan JD, Che L, Zhang S, Cossu A, Porcu A, Pascale RM, Dombrowski F, Hu H, Calvisi DF, Evert M and Chen X (2019). “Combined CDK4/6 and Pan-mTOR Inhibition Is Synergistic Against Intrahepatic Cholangiocarcinoma.” Clin Cancer Res 25(1): 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xu H, Wang P, Wang J, Affo S, Wang H, Xu M, Liang B, Che L, Qiu W, Schwabe RF, Chang TT, Vogl M, Pes GM, Ribback S, Evert M, Chen X and Calvisi DF (2021). “Focal adhesion kinase (FAK) promotes cholangiocarcinoma development and progression via YAP activation.” J Hepatol 75(4): 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraksa R, Zeller C, El-Bahrawy MA, Dai W, Daduang J, Jearanaikoon P, Chau-In S, Brown R and Limpaiboon T (2011). “CpG-island methylation study of liver fluke-related cholangiocarcinoma.” Br J Cancer 104(8): 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone ML, Chiappinelli KB, Li H, Murphy LM, Travers ME, Topper MJ, Mathios D, Lim M, Shih IM, Wang TL, Hung CF, Bhargava V, Wiehagen KR, Cowley GS, Bachman KE, Strick R, Strissel PL, Baylin SB and Zahnow CA (2017). “Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden.” Proc Natl Acad Sci U S A 114(51): E10981–E10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresemann C and Lyko F (2008). “Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine.” Int J Cancer 123(1): 8–13. [DOI] [PubMed] [Google Scholar]
- Sun D, Gan X, Liu L, Yang Y, Ding D, Li W, Jiang J, Ding W, Zhao L, Hou G, Yu J, Wang J, Yang F, Yuan S and Zhou W (2022). “DNA hypermethylation modification promotes the development of hepatocellular carcinoma by depressing the tumor suppressor gene ZNF334.” Cell Death Dis 13(5): 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L and Rao A (2009). “Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1.” Science 324(5929): 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita K, Suetake I, Yamashita E, Suga M, Narita H, Nakagawa A and Tajima S (2011). “Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).” Proc Natl Acad Sci U S A 108(22): 9055–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannapfel A, Benicke M, Katalinic A, Uhlmann D, Kockerling F, Hauss J and Wittekind C (2000). “Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver.” Gut 47(5): 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xi S, Shan J, Maunakea A, Che A, Briones V, Lee EY, Geiman T, Huang J, Stephens R, Leighty RM, Zhao K and Muegge K (2011). “Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences.” Proc Natl Acad Sci U S A 108(14): 5626–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thottassery JV, Sambandam V, Allan PW, Maddry JA, Maxuitenko YY, Tiwari K, Hollingshead M and Parker WB (2014). “Novel DNA methyltransferase-1 (DNMT1) depleting anticancer nucleosides, 4’-thio- 2’-deoxycytidine and 5-aza-4’-thio-2’-deoxycytidine.” Cancer Chemother Pharmacol 74(2): 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemin P, Fanzheng M, Peng X, Jihua H, Ruipeng S, Yaliang L, Yan W, Junlin X, Qingfu L, Zhefeng H, Jian L, Zihao G, Guoxing L, Boshi S, Ming Z, Qinghui M, Desen L and Lianxin L (2020). “MUC13 promotes intrahepatic cholangiocarcinoma progression via EGFR/PI3K/AKT pathways.” J Hepatol 72(4): 761–773. [DOI] [PubMed] [Google Scholar]
- Toh TB, Lim JJ and Chow EK (2019). “Epigenetics of hepatocellular carcinoma.” Clin Transl Med 8(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um TH, Kim H, Oh BK, Kim MS, Kim KS, Jung G and Park YN (2011). “Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis.” J Hepatol 54(5): 939–947. [DOI] [PubMed] [Google Scholar]
- Veland N, Lu Y, Hardikar S, Gaddis S, Zeng Y, Liu B, Estecio MR, Takata Y, Lin K, Tomida MW, Shen J, Saha D, Gowher H, Zhao H and Chen T (2019). “DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells.” Nucleic Acids Res 47(1): 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Mendez-Gonzalez J, Imbeaud S, Letouze E, Hernandez-Gea V, Cornella H, Pinyol R, Sole M, Fuster J, Zucman-Rossi J, Mazzaferro V, Esteller M, Llovet JM and Consortium H (2015). “DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma.” Hepatology 61(6): 1945–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Felden J, Garcia-Lezana T, Schulze K, Losic B and Villanueva A (2020). “Liquid biopsy in the clinical management of hepatocellular carcinoma.” Gut 69(11): 2025–2034. [DOI] [PubMed] [Google Scholar]
- Wagner EJ and Carpenter PB (2012). “Understanding the language of Lys36 methylation at histone H3.” Nat Rev Mol Cell Biol 13(2): 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Malnassy G and Qiu W (2021). “The Epigenetic Regulation of Microenvironment in Hepatocellular Carcinoma.” Front Oncol 11: 653037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton K and Samimi G (2015). “Methylation of cell-free circulating DNA in the diagnosis of cancer.” Front Mol Biosci 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasenang W, Chaiyarit P, Proungvitaya S and Limpaiboon T (2019). “Serum cell-free DNA methylation of OPCML and HOXD9 as a biomarker that may aid in differential diagnosis between cholangiocarcinoma and other biliary diseases.” Clin Epigenetics 11(1): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehbe H, Henson R, Meng F, Mize-Berge J and Patel T (2006). “Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression.” Cancer Res 66(21): 10517–10524. [DOI] [PubMed] [Google Scholar]
- Wijetunga NA, Pascual M, Tozour J, Delahaye F, Alani M, Adeyeye M, Wolkoff AW, Verma A and Greally JM (2017). “A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets.” Oncogene 36(14): 2030–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BK, Mei SC, Chen EH, Zheng Y and Pan D (2022). “YAP induces an oncogenic transcriptional program through TET1-mediated epigenetic remodeling in liver growth and tumorigenesis.” Nat Genet 54(8): 1202–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BK, Mei SC, Chen EH, Zheng Y and Pan D (2022). “YAP induces an oncogenic transcriptional program through TET1-mediated epigenetic remodeling in liver growth and tumorigenesis.” Nat Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H and Zhang Y (2014). “Reversing DNA methylation: mechanisms, genomics, and biological functions.” Cell 156(1–2): 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Yiang GT, Cheng PW, Chu PY and Li CJ (2018). “Molecular Targets in Hepatocarcinogenesis and Implications for Therapy.” J Clin Med 7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Fan H, Lang R, Li X, Zhang X, Lv S and He Q (2020). “Overexpression of 14-3-3delta Predicts Poor Prognosis in Extrahepatic Cholangiocarcinoma Patients.” Biomed Res Int 2020: 8435420. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xiaofang L, Kun T, Shaoping Y, Zaiqiu W and Hailong S (2012). “Correlation between promoter methylation of p14(ARF), TMS1/ASC, and DAPK, and p53 mutation with prognosis in cholangiocarcinoma.” World J Surg Oncol 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S and Qian C (2018). “The Growing Complexity of UHRF1-Mediated Maintenance DNA Methylation.” Genes (Basel) 9(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Liu W, Wang X, Wu G, Wang Q, Guo T, Huang W, Wang B and Chen Y (2022). “HOXA5 inhibits the proliferation of extrahepatic cholangiocarcinoma cells by enhancing MXD1 expression and activating the p53 pathway.” Cell Death Dis 13(9): 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Wu F, Wu Q, Xu L, Cheung OK, Kang W, Mok MT, Szeto LLM, Lun CY, Lung RW, Zhang J, Yu KH, Lee SD, Huang G, Wang CM, Liu J, Yu Z, Yu DY, Chou JL, Huang WH, Feng B, Cheung YS, Lai PB, Tan P, Wong N, Chan MW, Huang TH, Yip KY, Cheng AS and To KF (2019). “Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming.” Nat Commun 10(1): 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen D, Feng X, Hu J, Ge J, Yan C, Zhang D, Ling Z, Chen J and Wu J (2021). “Apolipoprotein B Is Associated With the Microenvironment of Cholangiocarcinoma.” Front Oncol 11: 654689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K and Shi YG (2011). “Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells.” Mol Cell 42(4): 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, Liu XS, Sugrue SP, He X, Min J, Kato Y and Shi YG (2012). “Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development.” Cell 151(6): 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Dong B, Zhao Y, Wang Y, Yang C, Xie Y, Niu Z and Zhu C (2021). “Upregulation of TTYH3 promotes epithelial-to-mesenchymal transition through Wnt/beta-catenin signaling and inhibits apoptosis in cholangiocarcinoma.” Cell Oncol (Dordr) 44(6): 1351–1361. [DOI] [PubMed] [Google Scholar]
- Yang B, House MG, Guo M, Herman JG and Clark DP (2005). “Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma.” Mod Pathol 18(3): 412–420. [DOI] [PubMed] [Google Scholar]
- Yang JD, Ghoz H, Aboelsoud MM, Taylor WR, Yab TC, Berger CK, Cao X, Foote PH, Giama NH, Barr Fritcher EG, Mahoney DW, Moser CD, Smyrk TC, Kipp BR, Gores GJ, Roberts LR and Kisiel JB (2021). “DNA Methylation Markers for Detection of Cholangiocarcinoma: Discovery, Validation, and Clinical Testing in Biliary Brushings and Plasma.” Hepatol Commun 5(8): 1448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE and Stanger BZ (2013). “Robust cellular reprogramming occurs spontaneously during liver regeneration.” Genes Dev 27(7): 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarychkivska O, Shahabuddin Z, Comfort N, Boulard M and Bestor TH (2018). “BAH domains and a histone-like motif in DNA methyltransferase 1 (DNMT1) regulate de novo and maintenance methylation in vivo.” J Biol Chem 293(50): 19466–19475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ and Camargo FD (2014). “Hippo pathway activity influences liver cell fate.” Cell 157(6): 1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii D, Shimata K, Yokouchi Y, Komohara Y, Suda H, Honda M, Yamamura K, Hibi T and Inomata Y (2022). “SOX9 contributes to the progression of ductular reaction for the protection from chronic liver injury.” Hum Cell 35(2): 721–734. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P and Zilberman D (2010). “Genome-wide evolutionary analysis of eukaryotic DNA methylation.” Science 328(5980): 916–919. [DOI] [PubMed] [Google Scholar]
- Zeng F, Zhou Y, Khowtanapanich T and Saengboonmee C (2022). “Cyclin-Dependent Kinase 4/6 Inhibitors: A Potential Breakthrough Therapy for Malignancies of Gastrointestinal Tract.” In Vivo 36(4): 1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeybel M, Hardy T, Robinson SM, Fox C, Anstee QM, Ness T, Masson S, Mathers JC, French J, White S and Mann J (2015). “Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease.” Clin Epigenetics 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu J, Li G, Wei J, Chen H, Zhang C, Zhao J, Wang Y, Dang S, Li X, Fang X, Liu L and Liu M (2018). “Fresh red raspberry phytochemicals suppress the growth of hepatocellular carcinoma cells by PTEN/AKT pathway.” Int J Biochem Cell Biol 104: 55–65. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang C, Wu C, Cui W and Wang L (2020). “DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy.” Cancers (Basel) 12(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Chen H, Zhou B, Zhang QY, Liao Y, Wang JS and Wang ZH (2018). “lncRNA HOXB-AS3 promotes hepatoma by inhibiting p53 expression.” Eur Rev Med Pharmacol Sci 22(20): 6784–6792. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W and Jeltsch A (2010). “Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail.” Nucleic Acids Res 38(13): 4246–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Hlady RA, Joyce BT, Robertson KD, He C, Nannini DR, Kibbe WA, Achenbach CJ, Murphy RL, Roberts LR and Hou L (2019). “DNA methylation of individual repetitive elements in hepatitis C virus infection-induced hepatocellular carcinoma.” Clin Epigenetics 11(1): 145. [DOI] [PMC free article] [PubMed] [Google Scholar]


