Abstract
Transcranial scanning ultrasound combined with intravenously injected microbubbles (SUS+MB) has been shown to transiently open the blood–brain barrier and reduce the amyloid‐β (Aβ) pathology in the APP23 mouse model of Alzheimer's disease (AD). This has been accomplished through the activation of microglial cells; however, their response to the SUS treatment is incompletely understood. Here, wild‐type (WT) and APP23 mice were subjected to SUS+MB, using nonsonicated mice as sham controls. After 48 h, the APP23 mice were injected with methoxy‐XO4 to label Aβ aggregates, followed by microglial isolation into XO4+ and XO4− populations using flow cytometry. Both XO4+ and XO4− cells were subjected to RNA sequencing and transcriptome profiling. The analysis of the microglial cells revealed a clear segregation depending on genotype (AD model vs. WT mice) and Aβ internalization (XO4+ vs. XO4− microglia), but interestingly, no differences were found between SUS+MB and sham in WT mice. Differential gene expression analysis in APP23 mice detected 278 genes that were significantly changed by SUS+MB in the XO4+ cells (248 up/30 down) and 242 in XO− cells (225 up/17 down). Pathway analysis highlighted differential expression of genes related to the phagosome pathway and marked upregulation of cell cycle‐related transcripts in XO4+ and XO4‐ microglia isolated from SUS+MB‐treated APP23 mice. Together, this highlights the complexity of the microglial response to transcranial ultrasound, with potential applications for the treatment of AD.
Keywords: Alzheimer's disease, methoxy‐XO4, microglia, RNA sequencing, transcriptomics, ultrasound
Abbreviations
- AD
Alzheimer's disease
- Aβ
amyloid‐beta
- BBB
blood–brain barrier
- FACS
fluorescence activated cell sorting
- MBs
microbubbles
- NFκB
nuclear factor kappa light chain enhancer of activated B cells
- PBS
phosphate buffered saline
- PCA
principal components analysis
- SUS
scanning ultrasound
- WT
wild type
1. INTRODUCTION
Alzheimer disease (AD) is the most common cause of dementia worldwide. The disease is characterized by progressive and irreversible neurodegeneration. However, given the complexity of the disease combined with a lack of knowledge on how to treat AD efficiently, there is an acute requirement to develop novel treatment strategies. 1
At a histopathological level, AD is characterized by the accumulation of extracellular amyloid‐β (Aβ) plaques, intraneuronal tau deposits and increased microglial activation. 2 A broad range of studies have revealed how microglial cells assume both a protective role (through shielding, recognition, and removal of Aβ) and a detrimental role (through removal of synapses or the release of neurotoxic factors), driving the progression of AD. 3 Transcriptomic studies on microglia have advanced our understanding of the pathogenesis of AD at the level of transcriptional network dynamics, highlighting important molecular players depending on the different phases of the disease. 4 , 5 , 6 Microglia are known to phagocytose aggregated forms of Aβ, and it has been proposed that deficiencies in this process may contribute to late‐onset AD 7 and metabolic labeling in humans indicated that clearance of Aβ is impaired in AD. 8 Recently, it has been shown that Aβ‐containing microglia differ in their transcriptional signature in comparison to microglia that have not internalized the peptide. 9
An obstacle to treating AD is the blood–brain barrier (BBB), which prevents large molecules such as antibodies from entering the brain, with IgG having 0.1% transfer across the barrier. 10 Approaches to modify anti‐Aβ antibodies to increase levels in the brain are in development, 11 along with other approaches to circumvent the BBB.
Studies in animal models of AD have indicated that repeated transient BBB openings that are achieved throughout the entire brain using transcranial ultrasound in a scanning mode together with intravenously injected microbubbles (SUS+MB) significantly clear amyloid plaques. One study reported that plaque reduction can occur as fast as 48 h after BBB opening, 12 and we have shown that this process occurs through microglial phagocytosis. 13 Ultrasound‐mediated bioeffects (including microglial activation) have also been demonstrated by specifically targeting the hippocampus, 14 , 15 but the therapeutic benefit seems to be most pronounced when the brain is treated more globally. 13 Of note, this clearing process requires BBB opening 16 and is even effective at reducing Aβ pathology in 22‐month‐old senescent mice. 17 Combination treatments with ultrasound for delivery of anti‐Aβ antibodies such as Aducanumab that has been recently approved by the Food and Drug Administration (FDA) 18 or an anti‐pyroglutamylated Aβ antibody, 19 led to more effective plaque removal and behavioral improvements than in those observed in mice that were treated with either ultrasound alone or antibodies alone. 18
Ultrasound‐mediated BBB opening has also been achieved in a small safety trial that revealed tolerability in patients with mild AD when a small region of the frontal cortex was targeted. 20 A subsequent clinical study found that the BBB could be opened in parts of the hippocampus, 21 with a modest reduction in the amyloid PET signal following three treatments with ultrasound over a 6‐month period. 22 A recent clinical trial opened the BBB in the frontal lobes bilaterally and resulted in a modest reduction in the amyloid PET signal and significant improvement in neuropsychiatric symptoms. 23 In all these studies, BBB opening by ultrasound was shown to be safe and reversible in that the BBB was fully restored after 24 h.
Several mechanisms have been proposed to explain how BBB opening leads to amyloid plaque reduction, including the uptake of endogenous immunoglobulins 24 or albumin binding to amyloid, 13 followed by microglial phagocytosis of Aβ and lysosomal digestion. Here, to gain a better understanding of how the combination of SUS treatment and Aβ internalization affects microglial physiology, we analyzed the transcriptional profile of microglia isolated from APP23 mice (a model of AD) that had been subjected to SUS+MB. By using a fluorescent dye to detect Aβ internalization within the microglia, we identified differences between the microglial cells from mice treated with or without ultrasound, as well as between cells that had internalized Aβ or not.
2. RESULTS
2.1. XO4 and FACS‐based isolation of Aβ‐positive and Aβ‐negative microglia
To understand the different effects of ultrasound‐mediated BBB opening on plaque‐phagocytic and non‐phagocytic microglia in AD, we applied SUS+MB or sham (i.e., mice were anesthetized and injected with microbubbles but not exposed to ultrasound) to the brains of APP23 mice or WT littermate controls (Figure 1a,b). In addition, to be able to distinguish between microglial cells that had internalized Aβ and those that had not, we used the fluorescent Congo‐red derivative methoxy‐XO4 to stain Aβ within microglia when injected into live mice, as previously done. 9 , 25 This allowed us to use a fluorescence activated cell sorting (FACS)‐based technique to separate and isolate XO4+ (Aβ phagocytic) and XO4− (non‐phagocytic) microglia following both SUS+MB and sham treatment paradigms (Figure 1c,d).
FIGURE 1.
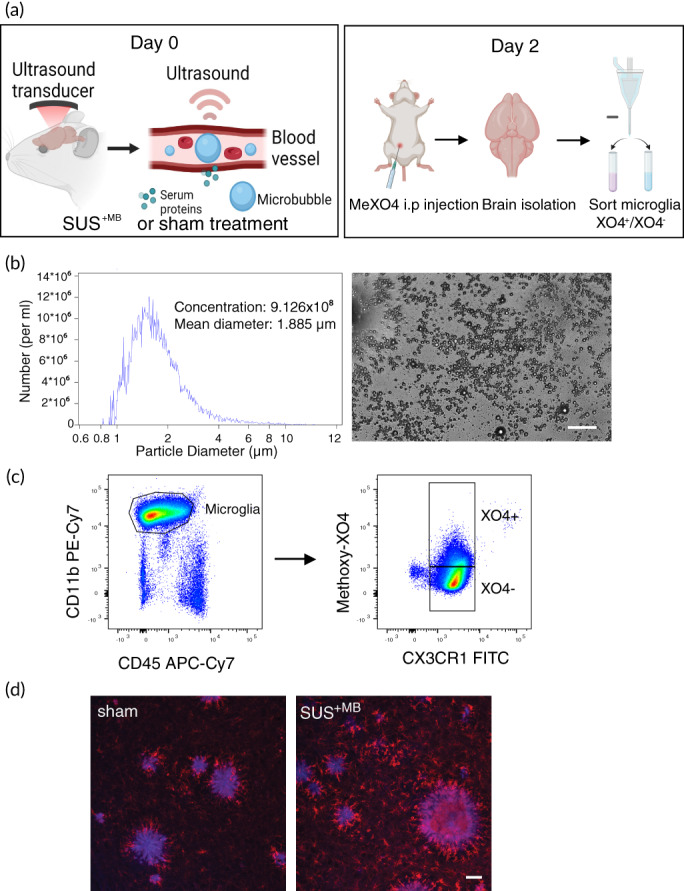
Experimental design and gating strategy to isolate XO4+ and XO4− microglia. (a) Scanning ultrasound (SUS+MB) or sham (no ultrasound) treatment was applied to APP23 transgenic and wild‐type (WT) mice. Two days post‐treatment, the mice received a single injection with methoxy‐XO4 (that binds Aβ) 2 h before euthanasia and collection of brain tissue. The brains of the mice were harvested and homogenized to form a single‐cell suspension, followed by FACS‐based isolation of XO4+ and XO4− microglial cells. (b) In‐house prepared microbubbles were used for scanning ultrasound (SUS+MB) and their size and concentration were measured using a Coulter Counter. (c) The gating strategy used to isolate microglial cells into XO4+ and XO4− populations via FACS with CD11b and CD45 antibodies to isolate a pure population of microglia, and methoxy‐XO4 fluorescence to isolate microglial cells that contain methoxy‐XO4 bound to Aβ. (d) Methoxy‐XO4 (blue) binds to Aβ plaques in the brains of APP23 mice, with Iba1‐positive microglia in red. Scale bar: 50 μm
2.2. Genotype, treatment, and Aβ internalization induce distinct phenotypes in microglia
Sorted microglial cells (both XO4+ and XO4−) from the four experimental groups (Figure 1a) were subjected to RNA isolation, followed by RNA sequencing (RNAseq) and bioinformatic analysis. Principal component analysis (PCA) revealed a clear segregation between the samples of different genotypes, being either of APP23 mutant or WT origin (Figure 2a). In addition, both PCA and hierarchical clustering analysis (Figure 2b) segregated distinct microglial populations induced by Aβ uptake (XO4+ vs. XO4− cells), as well as treatment (SUS+MB vs. sham‐treated animals), which were markedly accentuated in the APP23 samples. Of note, there was no effect of SUS+MB treatment in the microglial transcriptome of WT mice. Thus, we subsequently focused our analysis on the effects of ultrasound ± Aβ internalization in APP23‐derived microglia only.
FIGURE 2.
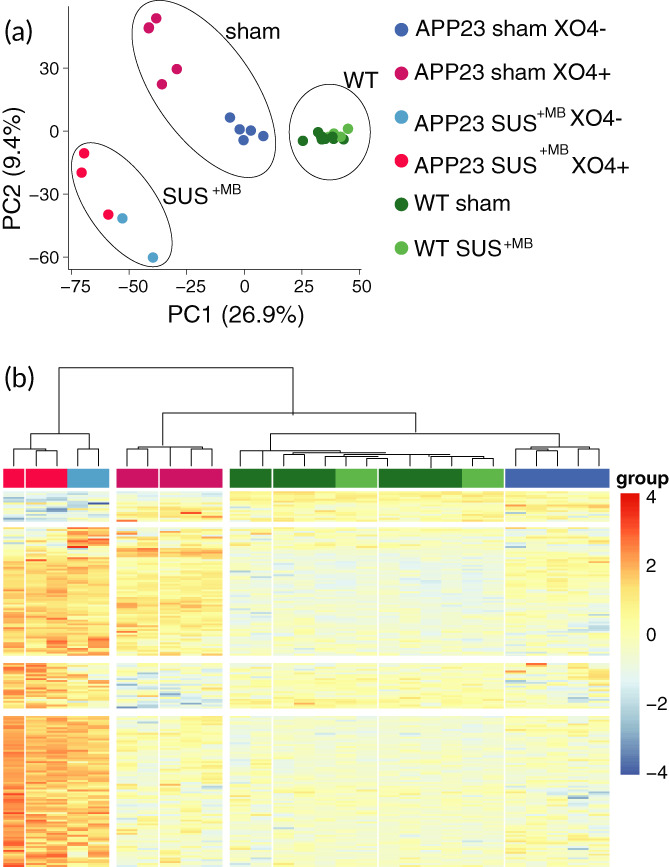
Cluster analysis distinguishes between genotype‐, treatment‐, and Aβ‐dependent microglial phenotypes. (a) Principal component analysis (PCA) reveals the presence of a WT microglia cluster independent of SUS+MB treatment that is segregated from cells of APP23 origin (further clustered dependent on both the SUS+MB treatment and their Aβ content). Ovals indicate treatment groups: SUS+MB, sham, and WT mice. (b) Hierarchical clustering of the differential genes (SUS+MB vs. sham) reveals a clear segregation between samples of APP23 or WT origin, SUS+MB treatment or sham, and whether the cells contain Aβ or not (methoxy‐XO4+ or methoxy‐XO4−).
2.3. SUS treatment induces an increased number of up‐regulated genes in microglia
To gain insight into the response of APP23 microglia to the SUS treatment regime, we further analyzed the transcripts obtained from XO4+ and XO4− cells. Our analysis identified 397 differentially enriched genes (FDR ≤ 0.05), with 155 genes being specific for XO4+ cells, 199 genes specific for XO4− microglia, and 123 genes being independent of the Aβ signature. Analyzing the treatment‐dependency patterns, we observed that most of the up‐regulated genes were induced by SUS+MB, with a total of 353 enriched genes across all the Aβ internalization levels (Figure 3a), and only 44 genes that were down‐regulated following SUS+MB treatment (Figure 3b). These enrichment patterns are highlighted in more detail by the volcano plots, with both the XO4+ cells (Figure 4a) and XO4− cells (Figure 4b) exhibiting increased numbers of differentially enriched genes induced by the SUS+MB treatment. The top 50 differentially expressed genes in the SUS+MB versus sham groups for both XO4+ and XO4− microglia are presented in Tables 1 and 2 with columns representing log2 fold change (logFC), average log2 expression (AvgExp), moderated t‐statistic, adjusted p‐value and B‐statistic for each gene. The B‐statistic is the posterior log odds of differential expression. Log fold change is defined as SUS SUS+MB to sham ratio, with a positive value indicating that the gene is upregulated by SUS+MB treatment.
FIGURE 3.
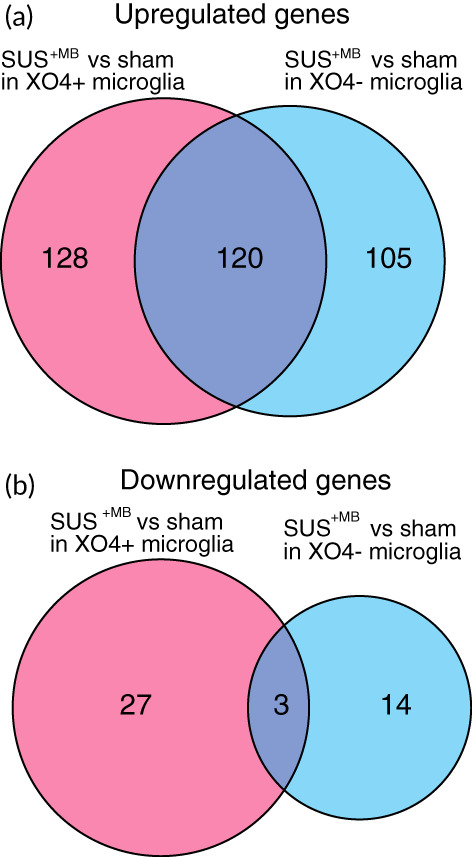
SUS+MB treatment leads to an increase in the number of differentially regulated genes in XO4+ microglia, when compared with sham‐treated APP23 mice. (a) A Venn diagram depicting the number of genes up‐regulated by SUS+MB distribute similarly between XO4+ and XO4− cells, with many genes up‐regulated in both. (b) A larger number of genes were down‐regulated in the XO4+ cells compared with XO4− cells following SUS+MB, with few genes down in both groups (adjusted p < 0.05).
FIGURE 4.
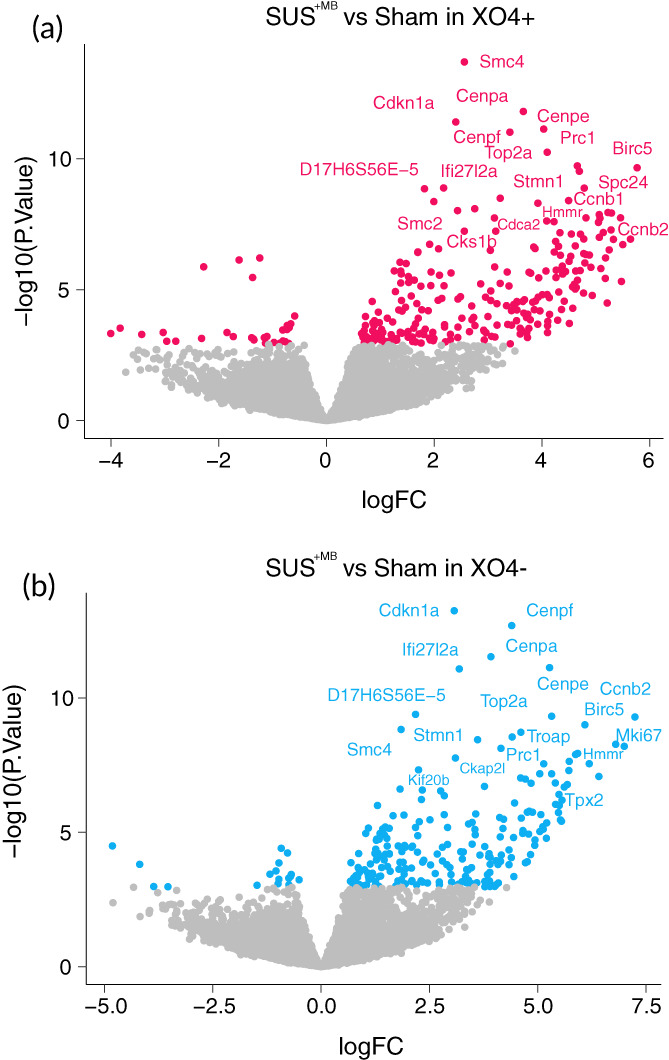
Volcano plots reveal dysregulation of genes in microglia from APP23 mice following SUS+MB or sham treatment. (a) XO4+ microglia show a large number of up‐regulated genes. The gene names of the 20 transcripts with the largest fold‐change (logFC) are shown. (b) XO4− microglia also show a large number of up‐regulated genes. The gene names of the 20 transcripts with the largest fold‐change (logFC) are shown.
TABLE 1.
Top 50 differentially expressed genes in XO4+ microglia between SUS+MB versus sham‐treated APP23 mice
| Gene symbol | logFC | AvgExp | t | Pval | adj PVal | B |
|---|---|---|---|---|---|---|
| Birc5 | 5.76502 | 1.7672889 | 9.02935 | 2.22E‐10 | 3.14E‐07 | 13.48506 |
| Cdkn3 | 5.64183 | 1.5432145 | 6.744088 | 1.18E‐07 | 3.49E‐05 | 7.515466 |
| Kif11 | 5.49926 | 0.2461131 | 6.576116 | 1.91E‐07 | 5.14E‐05 | 6.97426 |
| Ncapg | 5.47342 | −0.1786981 | 5.464827 | 4.88E‐06 | 5.93E‐04 | 3.988828 |
| Cdc20 | 5.45743 | 1.0739871 | 7.403038 | 1.82E‐08 | 8.52E‐06 | 9.133173 |
| Nuf2 | 5.32476 | −0.3289444 | 6.736156 | 1.21E‐07 | 3.49E‐05 | 7.284114 |
| Ccnb2 | 5.2893 | 1.9141487 | 7.553196 | 1.19E‐08 | 6.74E‐06 | 9.804982 |
| Aurkb | 5.28106 | 0.1698159 | 7.026936 | 5.26E‐08 | 2.05E‐05 | 8.075848 |
| Cit | 5.24623 | 0.3082589 | 6.415315 | 3.04E‐07 | 7.16E‐05 | 6.501784 |
| Hmmr | 5.22306 | 1.186993 | 7.569951 | 1.14E‐08 | 6.74E‐06 | 9.797002 |
| Esco2 | 5.20983 | 1.0709507 | 4.81764 | 3.25E‐05 | 2.80E‐03 | 2.353105 |
| Melk | 5.17911 | −0.554891 | 6.172445 | 6.16E‐07 | 1.18E‐04 | 5.824797 |
| Cdc25c | 5.17482 | −0.7504202 | 5.556821 | 3.73E‐06 | 4.68E‐04 | 4.188965 |
| Kif23 | 5.13702 | 0.6650897 | 6.945323 | 6.63E‐08 | 2.30E‐05 | 7.956364 |
| Nek2 | 5.09204 | −0.8391415 | 5.851377 | 1.57E‐06 | 2.54E‐04 | 4.972077 |
| Troap | 5.07497 | −1.0475974 | 7.38968 | 1.89E‐08 | 8.52E‐06 | 8.82398 |
| Dlgap5 | 5.0637 | −0.3794831 | 6.798489 | 1.01E‐07 | 3.16E‐05 | 7.445236 |
| Tk1 | 5.06171 | 1.4926731 | 7.504487 | 1.37E‐08 | 7.36E‐06 | 9.280781 |
| Mcm10 | 5.04637 | −1.0895909 | 7.260236 | 2.71E‐08 | 1.10E‐05 | 8.355957 |
| Saa3 | 4.93308 | −1.1465763 | 5.045849 | 1.67E‐05 | 1.68E‐03 | 2.844773 |
| Plac8 | 4.9074 | 1.316802 | 5.889764 | 1.40E‐06 | 2.33E‐04 | 5.218269 |
| Clspn | 4.87938 | 1.3914605 | 6.258953 | 4.79E‐07 | 9.81E‐05 | 6.258822 |
| Mki67 | 4.81598 | 2.6575306 | 7.401992 | 1.82E‐08 | 8.52E‐06 | 9.451222 |
| Cenpk | 4.81418 | 1.0168275 | 6.273493 | 4.59E‐07 | 9.60E‐05 | 6.240974 |
| Casc5 | 4.79524 | 1.9047316 | 6.021465 | 9.56E‐07 | 1.71E‐04 | 5.65795 |
| Ccnb1 | 4.78376 | 0.8942972 | 8.35344 | 1.33E‐09 | 1.33E‐06 | 11.79537 |
| Ndc80 | 4.77516 | 0.9359373 | 6.74738 | 1.17E‐07 | 3.49E‐05 | 7.549301 |
| Aspm | 4.77431 | 0.7784471 | 6.30262 | 4.22E‐07 | 8.98E‐05 | 6.4019 |
| Mns1 | 4.77059 | −0.9456452 | 5.116901 | 1.35E‐05 | 1.40E‐03 | 2.98948 |
| Stil | 4.71375 | −0.4286108 | 6.896241 | 7.63E‐08 | 2.46E‐05 | 7.747276 |
| Ankle1 | 4.70824 | −0.8978755 | 5.791278 | 1.87E‐06 | 2.91E‐04 | 4.711205 |
| Spc24 | 4.69279 | 1.3423116 | 8.911432 | 3.03E‐10 | 3.80E‐07 | 12.94538 |
| Gpsm2 | 4.65782 | −0.7681863 | 5.503262 | 4.36E‐06 | 5.35E‐04 | 4.048151 |
| Prc1 | 4.6556 | 2.8666675 | 9.092631 | 1.89E‐10 | 3.04E‐07 | 13.79044 |
| Cenpi | 4.63601 | 0.7782963 | 5.284361 | 8.29E‐06 | 9.00E‐04 | 3.53967 |
| Rab13 | 4.6245 | −0.436683 | 5.777597 | 1.95E‐06 | 2.91E‐04 | 4.733482 |
| Cep55 | 4.61759 | 0.1648057 | 6.537217 | 2.14E‐07 | 5.62E‐05 | 6.96793 |
| Hist1h2ae | 4.61674 | 0.6086809 | 5.236377 | 9.54E‐06 | 1.03E‐03 | 3.448414 |
| Cdc6 | 4.59923 | −0.7363785 | 5.380778 | 6.25E‐06 | 7.20E‐04 | 3.650384 |
| Foxm1 | 4.57058 | 0.2001768 | 4.66003 | 5.15E‐05 | 4.18E‐03 | 1.887387 |
| Sgol2a | 4.56722 | 0.8256839 | 5.301333 | 7.89E‐06 | 8.65E‐04 | 3.661214 |
| Tpx2 | 4.5441 | 1.3968306 | 6.899334 | 7.56E‐08 | 2.46E‐05 | 8.057218 |
| Shcbp1 | 4.5333 | −0.2503641 | 6.219989 | 5.36E‐07 | 1.06E‐04 | 6.048152 |
| Chaf1a | 4.5075 | −0.3486608 | 4.197819 | 1.95E‐04 | 1.29E‐02 | 0.699981 |
| Sgol1 | 4.50239 | 1.1036653 | 6.068962 | 8.32E‐07 | 1.54E‐04 | 5.751895 |
| Ckap2l | 4.49338 | 2.7278769 | 7.949218 | 3.99E‐09 | 3.22E‐06 | 10.8567 |
| Rad51ap1 | 4.43737 | 1.7528958 | 5.761704 | 2.04E‐06 | 3.00E‐04 | 4.897387 |
| C330027C09Rik | 4.41274 | 1.2903056 | 4.872646 | 2.77E‐05 | 2.50E‐03 | 2.500517 |
| Ccnf | 4.38312 | 0.9642677 | 5.68638 | 2.55E‐06 | 3.43E‐04 | 4.605822 |
| Ckap2 | 4.37671 | 0.4264924 | 5.925534 | 1.26E‐06 | 2.20E‐04 | 5.33635 |
Note: The logFC changes are reflected by a colour gradient ranging from red (high), to yellow (intermediate) expression.
TABLE 2.
Top 50 differentially expressed genes in XO4− microglia between SUS+MB versus sham‐treated APP23 mice
| Gene symbol | logFC | AvgExp | t | Pval | adj PVal | B |
|---|---|---|---|---|---|---|
| Ccnb2 | 7.2434 | 1.914149 | 8.71212 | 5.11E‐10 | 7.22E‐07 | 12.5665 |
| Mki67 | 6.99802 | 2.657531 | 7.78265 | 6.32E‐09 | 4.76E‐06 | 10.315 |
| Cep55 | 6.79948 | 0.164806 | 7.8447 | 5.32E‐09 | 4.30E‐06 | 9.96164 |
| Nek2 | 6.41184 | −0.839141 | 6.86382 | 8.37E‐08 | 3.50E‐05 | 7.42704 |
| Hmmr | 6.18986 | 1.186993 | 7.24982 | 2.80E‐08 | 1.46E‐05 | 8.78338 |
| Birc5 | 6.08931 | 1.767289 | 8.46022 | 1.00E‐09 | 1.26E‐06 | 11.9784 |
| Troap | 5.92187 | −1.047597 | 7.56325 | 1.16E‐08 | 7.71E‐06 | 8.95771 |
| Tpx2 | 5.87434 | 1.396831 | 7.53024 | 1.27E‐08 | 7.99E‐06 | 9.61853 |
| Dlgap5 | 5.72789 | −0.379483 | 7.32031 | 2.29E‐08 | 1.29E‐05 | 8.68108 |
| Stil | 5.71934 | −0.428611 | 7.04233 | 5.03E‐08 | 2.37E‐05 | 7.82959 |
| Aurkb | 5.68337 | 0.169816 | 6.6225 | 1.67E‐07 | 5.90E‐05 | 6.88724 |
| Pbk | 5.614 | 0.578285 | 6.5491 | 2.07E‐07 | 6.86E‐05 | 6.79667 |
| Ttk | 5.56047 | −0.003396 | 6.1612 | 6.36E‐07 | 1.75E‐04 | 5.66954 |
| Fam64a | 5.54815 | 0.172748 | 5.54325 | 3.88E‐06 | 7.30E‐04 | 4.21982 |
| Bub1b | 5.52541 | 0.852802 | 5.58235 | 3.46E‐06 | 6.62E‐04 | 4.21705 |
| Aspm | 5.49894 | 0.778447 | 6.00626 | 9.99E‐07 | 2.52E‐04 | 5.51711 |
| Plk1 | 5.4907 | 0.714941 | 6.32957 | 3.90E‐07 | 1.16E‐04 | 6.2301 |
| Plac8 | 5.4786 | 1.316802 | 5.79718 | 1.84E‐06 | 4.24E‐04 | 4.89829 |
| Kif18b | 5.41274 | −0.920601 | 6.03763 | 9.12E‐07 | 2.39E‐04 | 5.27128 |
| Kif23 | 5.40663 | 0.66509 | 6.66974 | 1.46E‐07 | 5.45E‐05 | 7.16429 |
| Top2a | 5.32157 | 3.363108 | 8.73594 | 4.80E‐10 | 7.22E‐07 | 12.923 |
| Psrc1 | 5.32146 | −1.473526 | 6.94318 | 6.67E‐08 | 2.90E‐05 | 7.27514 |
| Cenpe | 5.27386 | 3.005189 | 10.3774 | 7.47E‐12 | 1.88E‐08 | 16.8919 |
| Melk | 5.20793 | −0.554891 | 5.49586 | 4.46E‐06 | 7.99E‐04 | 3.98286 |
| Brca1 | 5.19986 | −0.261344 | 5.04746 | 1.66E‐05 | 2.15E‐03 | 2.80218 |
| Ccnb1 | 5.13942 | 0.894297 | 7.24474 | 2.84E‐08 | 1.46E‐05 | 8.78185 |
| Cit | 5.12791 | 0.308259 | 5.3118 | 7.65E‐06 | 1.17E‐03 | 3.51658 |
| Ccdc158 | 5.11777 | −1.673486 | 5.38333 | 6.20E‐06 | 1.08E‐03 | 3.52871 |
| Ndc80 | 5.07311 | 0.935937 | 5.73685 | 2.20E‐06 | 4.78E‐04 | 4.73085 |
| Kif4 | 5.04925 | −0.560068 | 5.2436 | 9.34E‐06 | 1.37E‐03 | 3.39874 |
| Cenpm | 5.04778 | 0.457076 | 6.94527 | 6.63E‐08 | 2.90E‐05 | 7.85063 |
| Efcab11 | 4.93682 | −1.413903 | 4.9978 | 1.92E‐05 | 2.38E‐03 | 2.61281 |
| Sgol2a | 4.93232 | 0.825684 | 4.83884 | 3.06E‐05 | 3.63E‐03 | 2.39082 |
| Casc5 | 4.8797 | 1.904732 | 5.80704 | 1.79E‐06 | 4.21E‐04 | 5.06427 |
| Cdkn3 | 4.84763 | 1.543214 | 6.66124 | 1.50E‐07 | 5.45E‐05 | 7.39483 |
| Ncapg | 4.83529 | −0.178698 | 4.56748 | 6.73E‐05 | 6.29E‐03 | 1.65013 |
| Arfgap3 | −4.8143 | 3.227787 | −4.82253 | 3.21E‐05 | 3.68E‐03 | 1.97355 |
| Cdca3 | 4.8047 | 1.033934 | 5.83682 | 1.64E‐06 | 4.03E‐04 | 5.09039 |
| Polr3g | −4.8016 | 2.483651 | −3.08136 | 4.17E‐03 | 1.20E‐01 | ‐1.94254 |
| Nuf2 | 4.79014 | −0.328944 | 5.16747 | 1.17E‐05 | 1.63E‐03 | 3.13439 |
| Kif11 | 4.76106 | 0.246113 | 5.80768 | 1.79E‐06 | 4.21E‐04 | 4.99501 |
| Pilrb1 | 4.75962 | −0.129751 | 4.35596 | 1.24E‐04 | 1.03E‐02 | 1.05207 |
| Fxyd6 | 4.71611 | −1.097719 | 4.32485 | 1.36E‐04 | 1.09E‐02 | 0.93726 |
| Rrm2 | 4.71417 | 1.554915 | 6.78453 | 1.05E‐07 | 4.09E‐05 | 7.67497 |
| Kif14 | 4.65924 | −1.058188 | 5.27195 | 8.60E‐06 | 1.28E‐03 | 3.28961 |
| Spc25 | 4.65365 | 1.777619 | 5.18905 | 1.10E‐05 | 1.56E‐03 | 3.34028 |
| Omd | 4.62121 | 1.038544 | 5.63473 | 2.96E‐06 | 5.98E‐04 | 4.39438 |
| Cdca2 | 4.61168 | 1.731777 | 8.22232 | 1.90E‐09 | 1.95E‐06 | 11.4262 |
| Knstrn | 4.60604 | 1.515975 | 6.81994 | 9.48E‐08 | 3.83E‐05 | 7.75424 |
| Cdc20 | 4.46665 | 1.073987 | 6.07832 | 8.10E‐07 | 2.18E‐04 | 5.73238 |
Note: The logFC changes are reflected by a colour gradient ranging from red (high), to yellow (intermediate), to green (low) expression.
2.4. SUS +MB treatment induces an enrichment in microglial cell‐cycle and phagosome‐related transcriptome
We next sought to identify the functionally enriched pathways induced by SUS + MB treatment and compare them in both the XO4+ and XO4− microglia. Applying a gene ontology (GO) enrichment analysis to the SUS+MB versus sham datasets revealed the top 10 enriched pathways that included “cell cycle,” “DNA replication,” and “DNA metabolic processes” (Table 3). KEGG pathway analysis revealed that the most enriched pathways included “DNA replication” and “cell cycle,” as well as established pathways in relation to the role of microglia in AD, such as “phagosome” and the “complement and coagulation cascade” (Table 4). Inspection of the “cell cycle” and “phagosome” pathways in a treatment‐ (SUS+MB versus sham) and Aβ internalization (XO4+ versus XO4−)‐dependent manner revealed similar trends, with a stronger response found for the XO4+ microglia containing internalized Aβ (Figure 5a,b). More genes in the phagosome pathway are significantly altered by SUS+MB in XO4+ microglia (seven genes up‐regulated and three down‐regulated) than XO4− microglia (five genes up‐regulated) with two of these genes up‐regulated in both (Figure 5a). For the cell cycle pathway, there were also more genes up‐regulated in XO4+ microglia (18 genes up‐regulated) than XO4− microglia (11 genes up‐regulated with 9 of these genes up‐regulated in both; Figure 5b).
TABLE 3.
GO pathway enrichment in microglia from SUS+MB‐ versus sham‐treated APP23 mice
| Term | Ont | N | DE | p.DE | |
|---|---|---|---|---|---|
| GO:0007049 | Cell cycle | BP | 1204 | 47 | 4.25E‐15 |
| GO:0000278 | Mitotic cell cycle | BP | 657 | 32 | 1.12E‐12 |
| GO:0006260 | DNA replication | BP | 203 | 19 | 1.27E‐12 |
| GO:0022402 | Cell cycle process | BP | 838 | 36 | 1.27E‐12 |
| GO:0006259 | DNA metabolic process | BP | 633 | 31 | 2.39E‐12 |
| GO:1903047 | Mitotic cell cycle process | BP | 544 | 26 | 3.37E‐10 |
| GO:0006261 | DNA‐dependent DNA replication | BP | 113 | 13 | 4.33E‐10 |
| GO:0007059 | Chromosome segregation | BP | 251 | 18 | 4.42E‐10 |
| GO:0006974 | Cellular response to DNA damage stimulus | BP | 618 | 27 | 1.07E‐09 |
| GO:0044786 | Cell cycle DNA replication | BP | 32 | 8 | 1.92E‐09 |
| GO:0006302 | Double‐strand break repair | BP | 187 | 15 | 3.05E‐09 |
| GO:0006281 | DNA repair | BP | 401 | 21 | 4.34E‐09 |
| GO:0098813 | Nuclear chromosome segregation | BP | 196 | 15 | 5.82E‐09 |
| GO:0006310 | DNA recombination | BP | 203 | 15 | 9.39E‐09 |
| GO:0000819 | Sister chromatid segregation | BP | 153 | 13 | 1.84E‐08 |
| GO:0033260 | Nuclear DNA replication | BP | 28 | 7 | 2.07E‐08 |
| GO:0000280 | Nuclear division | BP | 288 | 17 | 2.65E‐08 |
| GO:0098687 | Chromosomal region | CC | 255 | 16 | 3.00E‐08 |
| GO:0005694 | Chromosome | CC | 983 | 32 | 3.27E‐08 |
| GO:0048285 | Organelle fission | BP | 330 | 18 | 3.36E‐08 |
Note: The p.DE values are reflected by a colour gradient ranging from white (high) to green (low) p.DE values.
TABLE 4.
KEGG pathway enrichment in microglia from SUS+MB‐ versus sham‐treated APP23 mice
| Pathway | N | DE | p.DE | |
|---|---|---|---|---|
| path:mmu03030 | DNA replication | 34 | 6 | 2.13E‐06 |
| path:mmu04110 | Cell cycle | 115 | 9 | 6.69E‐06 |
| path:mmu00670 | One carbon pool by folate | 17 | 4 | 3.55E‐05 |
| path:mmu04114 | Oocyte meiosis | 90 | 5 | 3.76E‐03 |
| path:mmu05150 | Staphylococcus aureus infection | 28 | 3 | 3.95E‐03 |
| path:mmu04145 | Phagosome | 110 | 5 | 8.73E‐03 |
| path:mmu04914 | Progesterone‐mediated oocyte maturation | 72 | 4 | 9.42E‐03 |
| path:mmu00240 | Pyrimidine metabolism | 39 | 3 | 1.01E‐02 |
| path:mmu03013 | Nucleocytoplasmic transport | 96 | 4 | 2.47E‐02 |
| path:mmu01523 | Antifolate resistance | 25 | 2 | 3.32E‐02 |
| path:mmu03008 | Ribosome biogenesis in eukaryotes | 68 | 3 | 4.34E‐02 |
| path:mmu05322 | Systemic lupus erythematosus | 29 | 2 | 4.36E‐02 |
| path:mmu04610 | Complement and coagulation cascades | 30 | 2 | 4.64E‐02 |
| path:mmu05164 | Influenza A | 119 | 4 | 4.84E‐02 |
| path:mmu03440 | Homologous recombination | 34 | 2 | 5.81E‐02 |
| path:mmu03460 | Fanconi anemia pathway | 44 | 2 | 9.10E‐02 |
| path:mmu05171 | Coronavirus disease ‐ COVID‐19 | 163 | 4 | 1.19E‐01 |
| path:mmu01240 | Biosynthesis of cofactors | 106 | 3 | 1.23E‐01 |
| path:mmu05133 | Pertussis | 53 | 2 | 1.24E‐01 |
| path:mmu05140 | Leishmaniasis | 54 | 2 | 1.28E‐01 |
Note: The p.DE values are reflected by a colour gradient ranging from white (high) to green (low) p.DE values.
FIGURE 5.
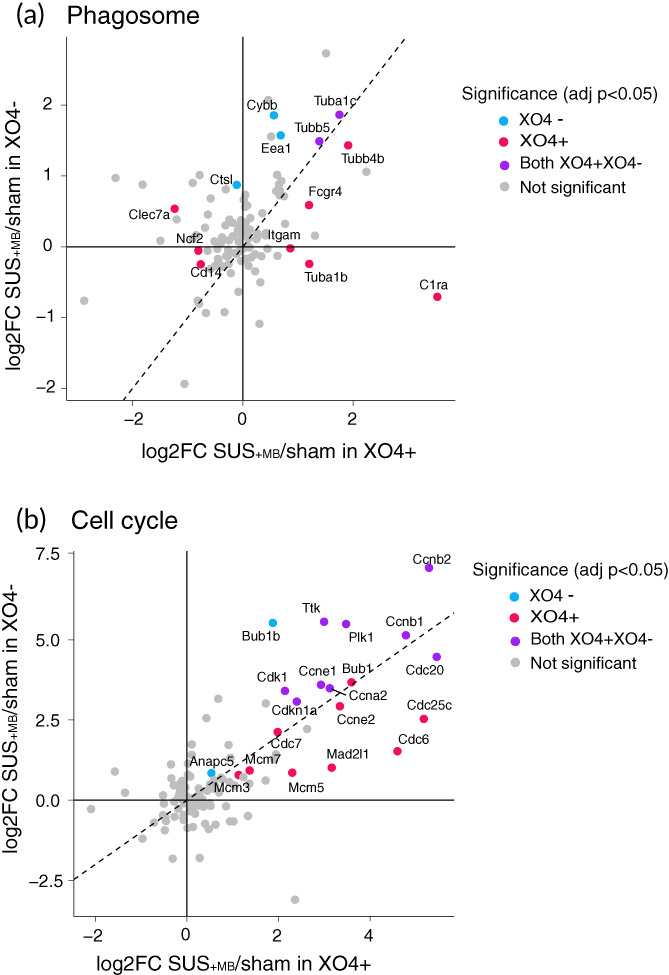
KEGG analysis reveals phagosome and cell‐cycle as top dysregulated pathways in SUS+MB treated APP23 microglia. (a) Plotting fold‐changes in gene expression (log2FC) with XO4+ (x axis) and XO4− (y axis) for phagosome genes indicates that the presence of internalized Aβ (XO4+) in microglia treated with SUS+MB leads to an additive effect on this pathway. (b) Plotting fold‐changes in gene expression (log2FC) with XO4+ (x axis) and XO4− (y axis) for cell cycle genes reveals that they are upregulated regardless of the presence of internalized Aβ (XO4+) in microglia, with cell‐cycle genes being enriched in both XO4+ and XO4− cells. The significant genes (adjusted p < 0.05) are labeled and colored according to their methoxy‐XO4 profile.
2.5. The magnitude of BBB opening after SUS treatment does not differ significantly between APP23 and WT mice
While it was not the major focus of this work, it was surprising to find that there was no effect of SUS+MB treatment on the transcriptome of WT mice at 48 h after treatment while there were many changes in the transcriptome of APP23 mice. To rule out that the differences in the transcriptomic responses between WT and APP23 mice in response to SUS+MB were due to reduced BBB opening in WT mice compared to APP23 mice, we performed an experiment whereby we injected fluorescently labeled 10 kDa dextran as a tracer to quantify the amount of BBB opening 2 h after SUS+MB treatment. The BBB appeared to open similarly in WT and APP23 mice (Figure 6a), and the fold‐change in fluorescent dextran uptake did not differ significantly by genotype (Figure 6b).
FIGURE 6.
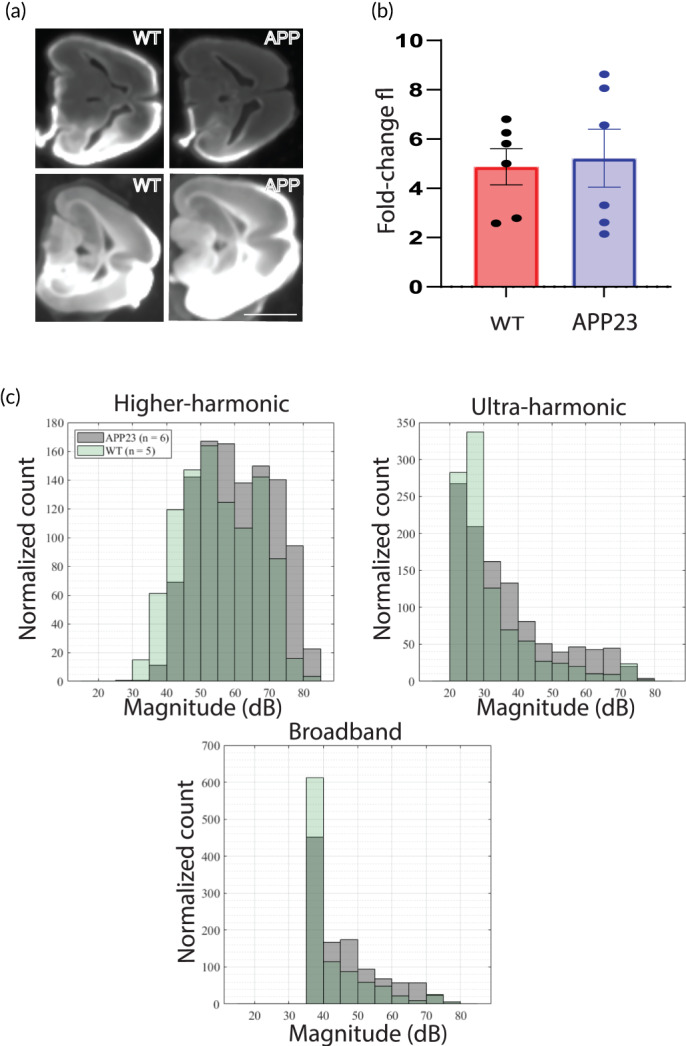
Analysis of BBB opening and acoustic emissions in wild‐type and APP23 mice. (a) Blood–brain barrier (BBB) of WT and APP23 (APP) mice in response to SUS + MB in one hemisphere was visualized by uptake of fluorescently labeled 10 kDa dextran. (b) Quantification of fluorescence in the SUS+MB treated hemisphere compared to the untreated hemisphere reveals a similar fold‐change. (c) Acoustic emissions recorded from WT and APP23 mice recorded reveals an increased higher‐harmonic emission, but similar ultra‐harmonic and broadband emission in APP23 mice compared to WT mice. Scale bar (a): 1 cm
2.6. The magnitude of cavitation signals recorded during SUS treatment is similar between APP23 and WT mice for ultraharmonic and broadband emissions
We recorded acoustic emissions with a passive cavitation detector (PCD) in order to determine whether there would be less cavitation of microbubbles in WT mice compared to APP23 mice when applying the same ultrasound settings. We extracted different frequency components corresponding to higher harmonics, ultraharmonics and broadband emissions and counted the number of ultrasound pulses of a particular magnitude. Analysis of the cavitation activity recorded during sonication of 20 spots in the right hemisphere from APP23 and WT mice revealed that the former showed an increased magnitude of higher harmonic emissions but that the ultraharmonic and broadband emissions were similar when comparing APP23 and WT mice (Figure 6c).
3. DISCUSSION
In this study, we sought to investigate the changes to the microglial transcriptomic profile induced by the application of BBB opening achieved with therapeutic ultrasound in conjunction with intravenously injected microbubbles in a mouse model of AD. This profile was obtained by analyzing the microglial transcriptome and correlating it with the presence or absence of Aβ in the microglia at 48 h after treating amyloid‐depositing APP23 mice with SUS+MB. This allowed us to identify several cellular functions that were increased by SUS+MB application in microglial populations that contained Aβ or not, such as the phagosome (reflecting uptake of Aβ for degradation), as well as the cell‐cycle pathway. Therapeutic ultrasound has a relatively larger contribution than Aβ plaque internalization on upregulating transcripts associated with the cell cycle and proliferation and this occurred independently of Aβ internalization. Our findings indicate that BBB opening by ultrasound in the context of AD modulates immunomodulatory functions for at least 48 h.
Previous studies have revealed that BBB opening by ultrasound is a reversible process and that it is fully restored after 24 h as measured by contrast‐enhanced MRI. 23 Our previous studies have observed a reduction in Aβ plaque load following an ultrasound treatment paradigm consisting of five to seven treatments repeated on a weekly basis, 13 whereas other studies have observed a response in bulk tissue (including microglia) ranging from 1 week after 6 weekly treatments, 26 and 6 27 , 28 and 24 h 27 , 29 , 30 after a single treatment. As microglia are a cell population presenting with a fast and dynamic response depending on both the intensity and time‐point after stimulation (acute vs. chronic response), we sought to observe the transcriptomic changes in these cells outside of the acute response of microglia to BBB opening. We chose a 48 h time point because the BBB is closed by then 13 , 23 ; however, the response of microglia to Aβ plaques was pronounced at this time point as shown by immunohistology, 13 , 17 , 24 and plaque clearance has been shown to peak 2 days after BBB opening as demonstrated by in vivo two‐photon microscopy. 12
Previous studies have so far investigated the cellular response to ultrasound‐mediated BBB opening in bulk tissue from WT mice and rats and found increased inflammatory transcript levels that were high at 6 h post‐treatment and mostly returned to baseline after 24 h. 27 , 28 , 29 , 31 Although these studies did not examine changes at 48 h, it would be expected that the inflammatory response would continue to resolve. Along these lines, at 48 h post‐treatment, we have not detected transcriptomic changes in microglia between the experimental groups in WT mice, in stark contrast to the changes observed 48 h post‐treatment in APP23 mice. The long‐lasting effects that we have identified in the APP23 mice could provide an important framework for future AD‐targeted therapies.
Several previous studies have investigated the effect of ultrasound application to the brain by applying omics techniques to cell populations. One study investigating ultrasound‐mediated delivery of plasmids to the brain of WT mice performed single‐cell RNA sequencing and found an upregulation of lysosomal genes in microglia 48 h after ultrasound treatment. 31 In support of this, in a SWATH quantitative proteomics screen following a series of 6 weekly sessions of SUS+MB treatments in aged C57Bl/6 (WT) mice, 32 we identified an increase in two microglial proteins (LRBA and CAGP) that are involved in phagocytosis. 26
In our analysis performed a priori to our bioinformatic data mining, we evaluated whether Aβ load (XO4+/XO4−) and treatment (SUS+MB/sham) are independent effects or interacting. Assessing the response of key pathways (phagocytosis and cell cycle), we conclude that the effects are independent and therefore can be analyzed in isolation. Our initial bioinformatics screen aimed to identify differences between microglia from WT and APP23 mice subjected to SUS+MB (with or without Aβ internalization) has revealed several interesting aspects related to the cellular response to the treatment. Thus, the WT microglia revealed the presence of a similar effect on the transcriptome in both sham‐ and SUS+MB‐treated experimental groups, as revealed by both the PCA and heatmap analysis. This could be attributed to the fast resolution of microglial response to acute stimulation. 32 The WT transcriptome was found to cluster in the proximity of the transcriptome specific to XO4− APP23 sham‐treated microglia, reflecting a particular nonphagocytic cellular state, that is, most likely a nondisease associated microglial phenotype. We found microglia from APP23 mice treated with SUS+MB to have more dysregulated transcripts than those from SUS+MB treated WT mice at the 48h time point. One potential explanation is that we were looking at a time point too late to capture transcriptomic changes in WT microglia in response to SUS+MB, whereas microglia from APP23 mice remain activated after SUS+MB due to an increased level of microglial responsiveness brought about by amyloid plaques. Another potential explanation for the difference in magnitude of the transcriptomic response between these genotypes could be that SUS+MB induced more BBB opening in the APP23 mice. While we found that there was no systematic difference in the uptake of a fluorescently labeled dextran tracer upon SUS+MB treatment in the two genotypes, there was substantial variability between mice. Future studies should measure the magnitude of BBB opening per mouse by using in vivo MRI imaging at various time points and correlate transcriptomic responses to the amount of BBB opening in each mouse as has been recently done in WT mice. 29 In addition, the response of microbubbles to ultrasound may differ between APP23 and WT mice because of differences in their cerebrovasculature, 33 or the fact that APP23 mice weigh less than their WT littermates. We performed recordings of acoustic emissions and found that APP23 mice had higher harmonics emissions than WT mice, but that ultraharmonic and broadband emissions were similar. Broadband emissions are associated with the largest magnitude and most violent cavitation activities, and these were mostly similar between WT and APP23 mice. The cause and significance of this difference in cavitation activity between WT and APP23 mice are unclear; however, it is conceivable that the increased cavitation recorded in APP23 mice might lead to an increased magnitude of transcriptomic changes at 48 h, which warrants further systematic studies, for instance, by using a cavitation controller. 27
If applied at an early stage of AD, boosting the Aβ phagocytic activity of microglia may present a promising therapeutic strategy by increasing the clearance of protein deposits. 34 A previous attempt to investigate the microglial response following ultrasound treatment focused on investigating transcripts related to the downstream effects of the NFκB pathway and damage‐associated molecules (DAMs) in bulk lysates from WT rodent brains, with most transcript levels returning to baseline after 24 h. 30 A subsequent study, however, reported no significant changes in the expression of any of the NFκB‐related genes when using a lower, more clinically relevant dose of microbubbles. 35 These opposing effects could be attributed to the specific ultrasound parameters that elicit a cavitation‐modulated inflammatory response through the microbubbles present in the blood circulation. 27 In addition, the transcriptomic response to ultrasound‐induced BBB opening was found to be dependent on the type of anesthesia used during the procedure. 31 Of note, we used ultrasound settings that we have previously demonstrated to increase microglial phagocytosis, 13 with no damage to neurons, 36 and which likely leads to lower levels of cavitation and BBB opening. Taken together, these results indicate that the magnitude of the effects of ultrasound‐mediated BBB opening are dependent on the applied ultrasound parameters, time points and methodology (such as whether RNA or protein levels are investigated, or whether heterogeneous tissue or isolated cell‐types are being analyzed).
Using a protocol that we have recently applied to reveal the transcriptional signature of microglia associated with Aβ phagocytosis, 9 in the current study, we aimed to investigate changes after SUS+MB treatment in the APP23 model of AD. Our results revealed enriched pathways in the Aβ‐containing microglia, such as cell cycle, phagosome, complement activity, and metabolism. Some of the pathways identified by our analysis have previously been associated with microglial activation in AD, supporting our results. Indeed, microglia have been observed to remove synapses in AD through a mechanism involving members of the complement system. 37 , 38 In addition, it has been proposed that the metabolism of microglia is impaired in AD, an effect that can be ameliorated by enhancing the cellular energetic and biosynthetic metabolism. 39 Increased microglial numbers in the proximity of plaques are associated with more compact plaques and reduced axonal dystrophy, 40 and we have previously reported increased microglial numbers around plaques following SUS+MB treatment. 17 Higher numbers of microglia around plaques may result from an increased proliferation or metabolic activity, as hinted at in the present study. Reactivation of the cell‐cycle machinery in microglia following ultrasound treatment is of particular interest, as it has been recently reported that repopulating microglial cells following ablation are neuroprotective in AD. 41 , 42
4. CONCLUSIONS
We have examined the effect of SUS+MB on the microglial transcriptome in the presence or absence of amyloid pathology. SUS+MB leads to temporary opening of the BBB and alters microglial gene expression in the AD brain to modulate several cellular pathways, including the cell cycle, various metabolic pathways and the phagosome. Harnessing the protective effects of microglia in the context of AD could potentially be achieved by a combination of SUS+MB‐mediated BBB opening and targeted drug delivery.
5. MATERIALS AND METHODS
5.1. Animals
In this study, we have used APP23 mice (harboring the AD Swedish K670M, N671L double mutation in the APP gene 43 ) and WT mice. The animals were maintained on a 12 h light/dark cycle and housed in a PC2 facility with ad libitum access to food and water. All experimental procedures in this study (Figure 1a) were approved by the University of Queensland Animal Ethics Committee (AEC) (QBI/412/14/NHMRC and QBI/554/17/NHMRC), and Monash University AEC (17241) and were conducted in compliance with the ARRIVE guidelines (Animal Research: Reporting in Vivo Experiments).
5.2. SUS treatment
Microbubbles comprising a phospholipid shell and octafluoropropane gas core were prepared in‐house. 1,2‐Distearoyl‐sn‐glycero‐3‐phosphocholine (DSPC) and 1,2‐distearoyl‐sn‐glycero‐3‐phosphoethanolamine‐N‐[amino(polyethylene glycol)‐2000] (DSPE‐PEG2000) (Avanti Polar Lipids) were mixed in a 9:1 molar ratio and dissolved in chloroform (Sigma), after which the chloroform solvent was evaporated under vacuum. The dried phospholipid cake was then dissolved in PBS with 10% glycerol to a concentration of 1 mg lipid/ml and heated to 55°C in a sonicating water bath. The solution was placed in a 1.5 ml glass high‐performance liquid chromatography (HPLC) vial with the air in the vial replaced with octafluoropropane gas (Arcadophta). The microbubbles were activated on the day of the experiment by agitation of the vial in a dental amalgamator at 4000 rpm for 45 s. Activated microbubbles were measured with a Multisizer 4e coulter counter which reported a mean diameter of 1.885 μm and a concentration of 9.12 × 108 microbubbles/ml. These microbubbles were also observed to be polydisperse under a microscope (Figure 1b).
For treatment delivery, an integrated focused ultrasound system (Therapy Imaging Probe System, TIPS, Philips Research) was used. This system consisted of an annular array transducer with a natural focus of 80 mm, a radius of curvature of 80 mm, a spherical shell of 80 mm with a central opening of 31 mm diameter, a 3D positioning system, and a programmable motorized system to move the ultrasound focus in the x and y planes to cover the entire brain area. A coupler mounted to the transducer was filled with degassed water and placed on the head of the mouse with ultrasound gel for coupling, to ensure unobstructed propagation of the ultrasound to the brain.
For SUS+MB applications, mice were anesthetized with ketamine (90 mg/kg) and xylazine (6 mg/kg) and the hair on their head was shaved and depilated. They were then injected retro‐orbitally with 1 μl/g body weight of microbubble solution and placed under the ultrasound transducer with the head immobilized. A heating pad was used to maintain body temperature. Parameters for the ultrasound delivery were 1 MHz center frequency, 0.65 MPa peak negative pressure, 10 Hz pulse repetition frequency, 10% duty cycle, and a 6 s sonication time per spot. The focus of the transducer was 1.5 mm × 12 mm in the transverse and axial planes, respectively. The motorized positioning system moved the focus of the transducer array in a grid with 1.5 mm spacing between individual sites of sonication so that ultrasound was delivered sequentially to the entire brain as described previously. 13 , 18 Mice typically received a total of 24 spots of sonication in a 6 × 4 raster grid pattern. For the sham treatment, mice received all injections and were placed under the ultrasound transducer, but no ultrasound was emitted. The time between injecting microbubbles and commencing ultrasound delivery was 60 ± 10 s and the duration of sonication was approximately 3 min (total time from microbubble injection approximately 4 min).
5.3. Acute isolation of microglia and FACS
Two hours prior to brain harvest, mice were injected intraperitoneally with methoxy–X04 (2 mg/ml in 1:1 ratio of DMSO to 0.9% [w/v] NaCl, pH 12) at 5 mg/kg. Mice were euthanized by CO2 and transcardially perfused with ice‐cold PBS prior to brain extraction. Whole brains, excluding the brain stem, olfactory bulbs and cerebellum, were dissected for microglial isolation. Single cell suspensions were prepared by mechanical dissociation using meshes of decreasing sizes from 250 to 70 μm and suspensions were enriched for microglia by density gradient separation. Briefly, the cell pellet was resuspended in 70% (v/v) isotonic Percoll (1× PBS + 90% [v/v] Percoll), overlayed with 37% (v/v) isotonic Percoll and centrifuged with slow acceleration and no brake at 2000 g for 20 min at 4°C. The microglia‐enriched cell population isolated from 37% to 70% interphase was diluted 1:5 in ice‐cold PBS and recovered by cold centrifugation at maximum speed for 1 min in microcentrifuge tubes. The cell pellet was then stained with antibodies to microglial cell surface markers (CD11b‐PE Cy7, 1:200 Biolegend, #101216; CD45‐APC Cy7, 1:200, BD Biosciences # 103116;) for isolation using the FACSAria™ III cell sorter (Figure 1c). CX3CR1 was stained using CX3CR1‐FITC (1:100, Biolegend, #149019). Microglia were defined as live/propidium iodide (PI)− (Sigma‐Aldrich, St. Louis, MO, #P4864), CD11b+, CD45low, CX3CR1+ cells. The XO4+ population gate was set using methoxy‐XO4‐injected WT animals. X04+ and X04− microglial populations were sorted separately for further analysis.
5.4. Immunohistology
APP23 mice were treated with SUS+MB and were perfused with PBS and drop fixed in 4% paraformaldehyde in PBS. They were then cryoprotected in 30% sucrose in PBS and sectioned at 40 μm with a freezing sliding microtome. Microglia were immunostained with anti‐Iba1 antibody (Wako, JP 1:1000) followed by incubation with an anti‐rabbit secondary antibody AlexaFluor 568 conjugate (Invitrogen). Sections were then co‐stained with 10 μM methoxy‐XO4 (Tocris Bioscience) in PBS with 20% ethanol before cover‐slipping. Images were obtained with a spinning disk confocal microscope (Nikon Diskovery) with a 20× objective, acquiring z‐stacks through the entire depth of the section (Figure 1d).
5.5. Bulk RNA‐seq and bioinformatics analysis
RNA extraction from FACS‐sorted microglia was performed using the RNeasy Micro Kit (Qiagen, #74004) and RNA quality was assessed using the Bioanalyser (Agilent RNA 6000 Pico kit; #5067–1513). The libraries were prepared using microglia RNA samples with RIN value ≥7.9 as previously described. 9 Sequencing reads were mapped to the mouse transcriptome reference genome (GRCm38) using STAR (v020201). We then established read counts for each gene using featureCounts (v1.5.2). The bulk RNA‐seq read counts were further analyzed using R (v4.0.2), limma (v3.42.2), and edgeR (v3.28.1). Data handling and plotting were performed using tidyverse (v1.3.0). In detail, we first removed lowly or nonexpressed genes with the filterByExpr function, and we calculated TMM (trimmed mean of m‐values) normalization factors to remove composition bias using calcNormFactors. To visualize dimensionality reduction of the sequencing data, we first removed unwanted variation in the data with removeBatchEffect. The PCA plot (Figure 2a) is using prcomp and ggplot to visualize the remaining variance in the data. The heatmap (Figure 2b) is plotting the DEGs from either contrast using the wardD2 method and the pheatmap function. To determine differentially expressed genes (DEGs), we first determined the gene‐wise variance trends using voom. Then, we built a linear model using the lmFit function and all combinations of genotype and treatment, batch, and sex as covariates. The Venn diagrams (Figure 3a,b) are generated with the VennDiagram package (v1.7.0). Tables 1 and 2 show the top 50 DEGs for the two contrasts of interest (ranked by absolute log fold change). Pathway and gene ontology enrichments are calculated from the respective DEG lists using the kegga and goana functions (Tables 3 and 4).
5.6. BBB opening with concurrent cavitation monitoring and dextran uptake
For concurrent cavitation monitoring, a ¼" diameter, 3.5 MHz, unfocused single‐element transducer (V3840N‐SU; Olympus NDT Inc., MA, USA) was employed as a passive cavitation detector (PCD) and placed through the central opening of the transmitting transducer. The detected microbubble emission was then amplified (Model 5662; Olympus NDT Inc.) and sampled using a 16‐bit digitizer card (M4i.4421; Spectrum Instrumentation GmbH, Grosshansdorf, Germany) at 31.25 MHz, in synchronization with the ultrasound pulse excitation of the TIPS system.
The recorded PCD signals were processed off‐line using a custom developed MATLAB (MathWorks Inc., MA, USA) script. Three PCD components, higher‐harmonics (HH), ultra‐harmonics (UH) and broadband (BB) emissions were extracted by selecting the frequency bandwidth within the frequency domain as described (with = 1 MHz):
HH: and
UH: and
BB: and
A 30 kHz interval was selected by inspecting the frequency spectrum to differentiate the PCD component. Finally, the intensity of selected components was calculated using Parseval's theorem as following:
where and denote the frequency spectrum of the PCD signal obtained by the Fourier transform and the number of frequency bins in the spectrum, respectively. Mice received 15 sonication spots directed at the right hemisphere. Immediately after the sonications, the mice were injected with 20 mg/kg 10 kDa lysine‐fixable dextran‐TMR (Invitrogen) in saline retroorbitally and after 2 h they were perfused with a saline flush followed by 20 ml of 4% paraformaldehyde. Sections were cut with a vibratome at 100 μm thickness and imaged using a LiCor infrared scanner. Two sections per mouse were quantified, comparing the fluorescence signal in the sonicated to the untreated control hemisphere.
AUTHOR CONTRIBUTIONS
Gerhard Leinenga: Conceptualization (equal); formal analysis (equal); investigation (equal); writing – original draft (equal). Liviu‐Gabriel Bodea: Conceptualization (equal); formal analysis (equal); writing – original draft (equal). Jan Schröder: Formal analysis (equal); writing – original draft (equal). Giuzhi Sun: Investigation (equal). Yichen Zhou: Formal analysis (equal). Jae Song: Data curation (equal); formal analysis (equal); writing – review and editing (equal). Jürgen Götz: Conceptualization (equal); formal analysis (equal); funding acquisition (lead); writing – original draft (equal). Alexandra Grubman: Conceptualization (equal); formal analysis (equal); funding acquisition; writing – original draft (equal). Jose M. Polo: Supervision, Formal analysis (equal); writing – review and editing (equal).
CONFLICT OF INTEREST
The authors declare that no competing interest exists.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/btm2.10329.
Supporting information
Appendix S1 DEGs_XO4neg_SUS_vs_sham.
Appendix S2 DEGs_XO4plus.
Appendix S3 DEGs_XO4plus_SUS_vs_sham.
ACKNOWLEDGMENTS
The authors would like to acknowledge Flowcore, Monash Health Translation Precinct Medical Genomics Facility, and Australian Research Laboratories/Monash Animal Research Platform, Monash University, for the provision of instrumentation, training and technical support. The authors thank Tishila Palliyaguru and Linda Cumner for assistance with animal colonies, and Rowan Tweedale for critical reading of the manuscript.
Leinenga G, Bodea L‐G, Schröder J, et al. Transcriptional signature in microglia isolated from an Alzheimer's disease mouse model treated with scanning ultrasound. Bioeng Transl Med. 2023;8(1):e10329. doi: 10.1002/btm2.10329
Gerhard Leinenga, Liviu‐Gabriel Bodea, and Jan Schröder authors contributed equally to this work.
Funding information The authors acknowledge support by the Estate of Dr Clem Jones AO, the National Health and Medical Research Council of Australia (GNT1145580, GNT1176326), and the State Government of Queensland (DSITI, Department of Science, Information Technology and Innovation) to Jürgen Götz. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government. Alexandra Grubman was funded by a NHMRC‐ARC Dementia Fellowship and Alexandra Grubman and Giuzhi Sun received funding from Yulgilbar Foundation and Dementia Australia.
Contributor Information
Alexandra Grubman, Email: alexandra.grubman@monash.edu.
Jose M. Polo, Email: jose.polo@monash.edu.
Jürgen Götz, Email: j.goetz@uq.edu.au, Email: j.goetz.brisbane@gmail.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. Differential gene expression data are provided as supplementary information.
REFERENCES
- 1. Polanco JC, Li C, Bodea LG, Martinez‐Marmol R, Meunier FA, Götz J. Amyloid‐beta and tau complexity ‐ towards improved biomarkers and targeted therapies. Nat Rev Neurol. 2018;14(1):22‐39. [DOI] [PubMed] [Google Scholar]
- 2. De Strooper B, Karran E. The cellular phase of Alzheimer's disease. Cell. 2016;164(4):603‐615. [DOI] [PubMed] [Google Scholar]
- 3. Scheiblich H, Trombly M, Ramirez A, Heneka MT. Neuroimmune connections in aging and neurodegenerative diseases. Trends Immunol. 2020;41(4):300‐312. [DOI] [PubMed] [Google Scholar]
- 4. Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late‐onset Alzheimer's disease. Cell. 2013;153(3):707‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keren‐Shaul H, Spinrad A, Weiner A, et al. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017;169(7):1276‐1290 e1217. [DOI] [PubMed] [Google Scholar]
- 6. Grubman A, Chew G, Ouyang JF, et al. A single‐cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell‐type‐specific gene expression regulation. Nat Neurosci. 2019;22(12):2087‐2097. [DOI] [PubMed] [Google Scholar]
- 7. Ewers M, Biechele G, Suarez‐Calvet M, et al. Higher CSF sTREM2 and microglia activation are associated with slower rates of beta‐amyloid accumulation. EMBO Mol Med. 2020;12(9):e12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mawuenyega KG, Sigurdson W, Ovod V, et al. Decreased clearance of CNS beta‐amyloid in Alzheimer's disease. Science. 2010;330(6012):1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grubman A, Choo XY, Chew G, et al. Transcriptional signature in microglia associated with Abeta plaque phagocytosis. Nat Commun. 2021;12(1):3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leinenga G, Langton C, Nisbet R, Götz J. Ultrasound treatment of neurological diseases ‐ current and emerging applications. Nat Rev Neurol. 2016;12(3):161‐174. [DOI] [PubMed] [Google Scholar]
- 11. Weber F, Bohrmann B, Niewoehner J, et al. Brain shuttle antibody for Alzheimer's disease with attenuated peripheral effector function due to an inverted binding mode. Cell Rep. 2018;22(1):149‐162. [DOI] [PubMed] [Google Scholar]
- 12. Poon CT, Shah K, Lin C, et al. Time course of focused ultrasound effects on beta‐amyloid plaque pathology in the TgCRND8 mouse model of Alzheimer's disease. Sci Rep. 2018;8(1):14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leinenga G, Götz J. Scanning ultrasound removes amyloid‐beta and restores memory in an Alzheimer's disease mouse model. Sci Transl Med. 2015;7(278):278ra233. [DOI] [PubMed] [Google Scholar]
- 14. Burgess A, Dubey S, Yeung S, et al. Alzheimer disease in a mouse model: MR imaging‐guided focused ultrasound targeted to the hippocampus opens the blood‐brain barrier and improves pathologic abnormalities and behavior. Radiology. 2014;273(3):736‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen Y, Hua L, Yeh CK, et al. Ultrasound with microbubbles improves memory, ameliorates pathology and modulates hippocampal proteomic changes in a triple transgenic mouse model of Alzheimer's disease. Theranostics. 2020;10(25):11794‐11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leinenga G, Koh WK, Götz J. Scanning ultrasound in the absence of blood‐brain barrier opening is not sufficient to clear beta‐amyloid plaques in the APP23 mouse model of Alzheimer's disease. Brain Res Bull. 2019;153:8‐14. [DOI] [PubMed] [Google Scholar]
- 17. Leinenga G, Götz J. Safety and efficacy of scanning ultrasound treatment of aged APP23 mice. Front Neurosci. 2018;12:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leinenga G, Koh WK, Götz J. A comparative study of the effects of Aducanumab and scanning ultrasound on amyloid plaques and behavior in the APP23 mouse model of Alzheimer disease. Alzheimers Res Ther. 2021;13(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun T, Shi Q, Zhang Y, et al. Focused ultrasound with anti‐pGlu3 Abeta enhances efficacy in Alzheimer's disease‐like mice via recruitment of peripheral immune cells. J Control Release. 2021;336:443‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lipsman N, Meng Y, Bethune AJ, et al. Blood‐brain barrier opening in Alzheimer's disease using MR‐guided focused ultrasound. Nat Commun. 2018;9(1):2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rezai AR, Ranjan M, D'Haese PF, et al. Noninvasive hippocampal blood‐brain barrier opening in Alzheimer's disease with focused ultrasound. Proc Natl Acad Sci U S A. 2020;117(17):9180‐9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Haese PF, Ranjan M, Song A, et al. Beta‐amyloid plaque reduction in the hippocampus after focused ultrasound‐induced blood‐brain barrier opening in Alzheimer's disease. Front Hum Neurosci. 2020;14:593672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park SH, Baik K, Jeon S, Chang WS, Ye BS, Chang JW. Extensive frontal focused ultrasound mediated blood‐brain barrier opening for the treatment of Alzheimer's disease: a proof‐of‐concept study. Transl Neurodegener. 2021;10(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jordao JF, Thevenot E, Markham‐Coultes K, et al. Amyloid‐beta plaque reduction, endogenous antibody delivery and glial activation by brain‐targeted, transcranial focused ultrasound. Exp Neurol. 2013;248:16‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bolmont T, Haiss F, Eicke D, et al. Dynamics of the microglial/amyloid interaction indicate a role in plaque maintenance. J Neurosci. 2008;28(16):4283‐4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blackmore DG, Turpin F, Palliyaguru T, et al. Low‐intensity ultrasound restores long‐term potentiation and memory in senescent mice through pleiotropic mechanisms including NMDAR signaling. Mol Psychiatry. 2021;26:6975‐6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ji R, Karakatsani ME, Burgess M, Smith M, Murillo MF, Konofagou EE. Cavitation‐modulated inflammatory response following focused ultrasound blood‐brain barrier opening. J Control Release. 2021;337:458‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mathew AS, Gorick CM, Thim EA, et al. Transcriptomic response of brain tissue to focused ultrasound‐mediated blood‐brain barrier disruption depends strongly on anesthesia. Bioeng Transl Med. 2021;6(2):e10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mathew AS, Gorick CM, Price RJ. Multiple regression analysis of a comprehensive transcriptomic data assembly elucidates mechanically‐ and biochemically‐driven responses to focused ultrasound blood‐brain barrier disruption. Theranostics. 2021;11(20):9847‐9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kovacs ZI, Kim S, Jikaria N, et al. Disrupting the blood‐brain barrier by focused ultrasound induces sterile inflammation. Proc Natl Acad Sci U S A. 2017;114(1):E75‐E84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathew AS, Gorick CM, Price RJ. Single‐cell mapping of focused ultrasound‐transfected brain. Gene Ther. 2021. [online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bodea LG, Wang Y, Linnartz‐Gerlach B, et al. Neurodegeneration by activation of the microglial complement‐phagosome pathway. J Neurosci. 2014;34(25):8546‐8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reuter B, Venus A, Heiler P, et al. Development of cerebral microbleeds in the APP23‐transgenic mouse model of cerebral amyloid Angiopathy‐a 9.4 tesla MRI study. Front Aging Neurosci. 2016;8:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deczkowska A, Keren‐Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease‐associated microglia: a universal immune sensor of neurodegeneration. Cell. 2018;173(5):1073‐1081. [DOI] [PubMed] [Google Scholar]
- 35. McMahon D, Hynynen K. Acute inflammatory response following increased blood‐brain barrier permeability induced by focused ultrasound is dependent on microbubble dose. Theranostics. 2017;7(16):3989‐4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hatch RJ, Leinenga G, Götz J. Scanning ultrasound (SUS) causes no changes to neuronal excitability and prevents age‐related reductions in hippocampal CA1 dendritic structure in wild‐type mice. PloS One. 2016;11(10):e0164278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hong S, Beja‐Glasser VF, Nfonoyim BM, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352(6286):712‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benetatos J, Bennett RE, Evans HT, et al. PTEN activation contributes to neuronal and synaptic engulfment by microglia in tauopathy. Acta Neuropathol. 2020;140(1):7‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ulland TK, Song WM, Huang SC, et al. TREM2 maintains microglial metabolic fitness in Alzheimer's disease. Cell. 2017;170(4):649‐663 e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuan P, Condello C, Keene CD, et al. TREM2 Haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron. 2016;90(4):724‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sosna J, Philipp S, Albay R 3rd, et al. Early long‐term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre‐fibrillar oligomers in 5XFAD mouse model of Alzheimer's disease. Mol Neurodegener. 2018;13(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olmos‐Alonso A, Schetters ST, Sri S, et al. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer's‐like pathology. Brain. 2016;139(Pt 3):891‐907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Götz J, Bodea LG, Goedert M. Rodent models for Alzheimer disease. Nat Rev Neurosci. 2018;19(10):583‐598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 DEGs_XO4neg_SUS_vs_sham.
Appendix S2 DEGs_XO4plus.
Appendix S3 DEGs_XO4plus_SUS_vs_sham.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Differential gene expression data are provided as supplementary information.


