Abstract
Objective/background:
In vivo imaging and quantification of the microstructures of small airways in three dimensions (3D) allows a better understanding and management of airway diseases, such as asthma and chronic obstructive pulmonary disease (COPD). At present, the resolution and contrast of the currently available conventional optical coherence tomography (OCT) imaging technologies operating at 1300 nm remain challenging to directly visualize the fine microstructures of small airways in vivo.
Methods:
We developed an ultrahigh-resolution diffractive endoscopic OCT at 800 nm to afford a resolving power of 1.7 μm (in tissue) with an improved contrast and a custom deep residual learning based image segmentation framework to perform accurate and automated 3D quantification of airway anatomy.
Results:
The 800-nm diffractive OCT enabled the direct delineation of the structural components in the small airway wall in vivo. We further first demonstrated the 3D anatomic quantification of critical tissue compartments of small airways in sheep using the automated segmentation method.
Conclusion:
The deep learning assisted diffractive OCT provides a unique ability to access the small airways, directly visualize and quantify the important tissue compartments, such as airway smooth muscle, in the airway wall in vivo in 3D.
Significance:
These pilot results suggest a potential technology for calculating volumetric measurements of small airways in patients in vivo.
Keywords: Optical coherence tomography, quantitative imaging, deep learning, airway smooth muscle, small airway disease
I. Introduction
SMALL airways are the critical site of pathology and important therapeutic target for several lung diseases[1, 2], and are a major cause of global healthcare morbidity and mortality with rising prevalence rates[3, 4]. Previous studies revealed that small airway walls were thickened in fatal asthma compared to those in nonfatal asthma[5, 6]. In COPD, the disease predominantly involves small airway remodeling[1, 7]. Small airway pathology often occurs in the early course of COPD before the onset of symptoms and before discernable changes in spirometry[8, 9]. Despite their importance, evaluating small airway pathologies in vivo remains challenging[2], hindering the effective diagnostics, therapeutics, and clear understanding of the basic mechanisms of these diseases[10, 11]. Due to the relative inaccessibility to biopsy, the current methods for studying small airway pathology rely on indirect measures (such as pulmonary function testing)[12], ex vivo histopathology, or conventional radiographic techniques[13] which do not provide the resolution and contrast sufficient to exam the fine microstructures of small airways[14].
The ability to directly visualize volumetric morphology on critical tissue components in the airway wall in vivo will have a profound impact on understating the development and progression of lung disease. Given the unique capability afforded by endoscopic OCT for non-invasive and high-resolution optical biopsy, 3D imaging of airway pathology in vivo becomes possible[15, 16]. It has been demonstrated that the 1300-nm endoscopic OCT could be potentially useful for measuring small airway wall thickness in patient[17, 18]. However, the resolution (about 10 μm) and imaging contrast afforded by 1300-nm endoscopic OCT is suboptimal for accurately assessing the microstructures in the walls of the small airways.
In addition, the high-speed endoscopic OCT generates a large amount of volumetric imaging data (up to tens of GBs), making manual image quantification extremely laborious if not impossible. Neural networks are currently standard methods for automated segmentation of medical images to cope with the increasing volume of images and the shortage of available human expertise[19-22]. Nowadays, the well-established deep learning frameworks can be conveniently customized and optimized for automated OCT image quantification with considerable accuracy[23-25]. However, the adoption of such an advanced tool for the anatomic quantification of OCT airway images was limited by the insufficient capability of conventional endoscopic OCT to directly resolve airway microstructures, such as airway smooth muscle.
To address the limitations of traditional 1300-nm OCT, in this study, we developed an 800-nm diffractive endoscopic OCT with a considerably higher resolution (about 1.7 μm in tissue) and improved image contrast, enabling direct visualization of 3D airway microstructures in vivo[26-33]. Furthermore, for the first time we trained and optimized a deep residual learning based automated segmentation network and demonstrated that critical tissue compartments of small airways in sheep could be segmented, quantified and visualized in three dimensions with an accuracy similar to our experienced investigators. The capability of deep learning assisted ultrahigh-resolution diffractive endoscopic OCT to comprehensively image and anatomically quantify small airways longitudinally provides an unprecedented opportunity to image previously unexplored aspects of airway pathology in vivo and may serve as an intravital imaging tool to elucidate the etiology, progression of disease, and response to therapy in human lung diseases.
II. Materials and Methods
A. Ultrahigh-resolution Diffractive OCT System
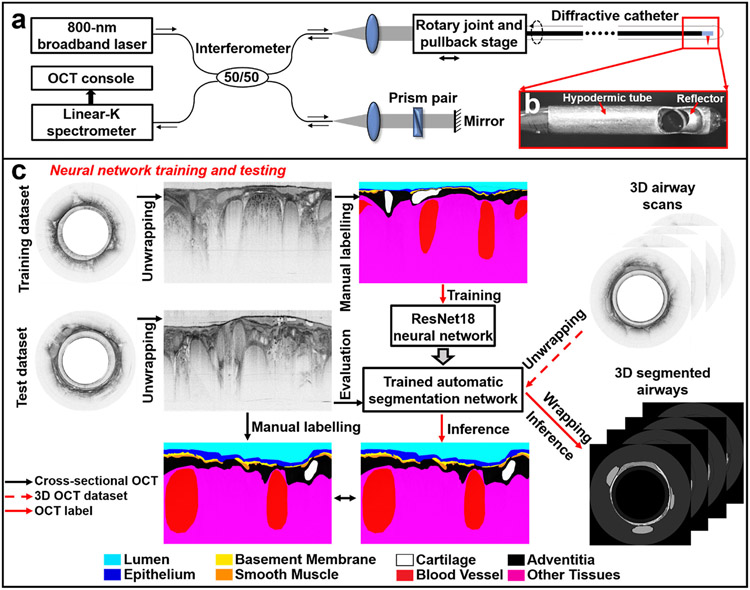
In vivo OCT images of sheep airways were acquired using a custom-built bench-top diffractive OCT system based on an 800-nm SD-OCT platform (Fig. 1a) and a diffractive imaging catheter (Fig. 1b), the details of which was reported elsewhere[27, 28, 31, 34]. The diffractive catheter had a small diameter of 1.3 mm (and 1.8 mm with the protective plastic sheath). Such an OCT system achieved excellent achromatic performance with a measured ultrahigh axial resolution of about 2.4 μm in air (or 1.7 μm in tissue), which represents a 4X improvement over conventional 1300-nm OCT. Meanwhile, our diffractive catheter provided a transversal resolution of ~7.2 μm. The image acquisition rate was 20 frames per second using a home-built broadband rotational joint[30]. 3D imaging was achieved by pulling back the circumferentially rotating catheter, with a pullback speed of 0.4 mm/s (corresponding to an image-to-image pitch of about 20 μm).
Fig. 1. 800-nm diffractive OCT system and workflow for neural network training and testing.
(a) Schematic of the diffractive spectral-domain OCT system operating at 800 nm with a customized linear-K spectrometer and an achromatic diffractive catheter of a 1.3-mm diameter. (b) Photo of the distal end of a diffractive catheter encased within a hypodermic tube. (c) For neural network training, unwrapped OCT images in the training dataset were manually labeled by an experience OCT reviewer. After that, the trained network first underwent the performance evaluation on the test dataset, the segmentation results were compared with the manually labeled ground truth. Then, the trained network was tested on 3D airway scans to automatically recognize and segment airway microstructures for volumetric visualization and quantification of small airways of sheep.
B. Study Flow and 3D Segmentation Method
In our study, we investigated the feasibility of the technology for imaging microstructures of small airways in sheep. Correlation studies were performed first to identify the correlated OCT and histology pairs[32]. Members of our research team (YW, JT) learned to accurately determine the small airway microstructures in OCT images based on correlated histologic images. These investigators then manually labelled the microstructures and tissue compartments of small airways in cross-sectional OCT images using Semantic Segmentation Editor, an open-sourced web-based interactive image annotation tool[35].
To facilitate the volumetric visualization and anatomic quantification of small airway microstructures, we customized a popular deep residual learning architecture (ResNet18) and trained an automated airway OCT segmentation framework[36]. First, we manually labeled 618 OCT cross-sectional images randomly selected from 10 sheep airways (n=5 for sheep), which were used for training a neural network for automated segmentation of small airway compartments (Fig. 1c). Then, 46 histology-correlated OCT images were used as the test dataset for evaluating the performance of the trained neural network (versus the manual segmentations of OCT investigators) through calculating the intersection over union (IoU) values between the predicted labels and manual ground truth labels (Fig. 1c).
During the training process, the randomly initialized weight parameters were updated for layer segmentation by minimizing a loss function. We implemented and compared three widely-used loss functions, including binary cross entropy, Dice loss function, and Kullback-Leibler divergence loss function[37]. We selected the Kullback-Leibler divergence loss function due to its better test performance in terms of IoU (see Results). The barebone ResNet neural network built upon PyTorch 0.4.0 and Python 3.5 was used[36], both training and inference were performed on a Nvidia GTX 1080 Graphic Card on a desktop PC running Windows 10.
C. Sheep Studies
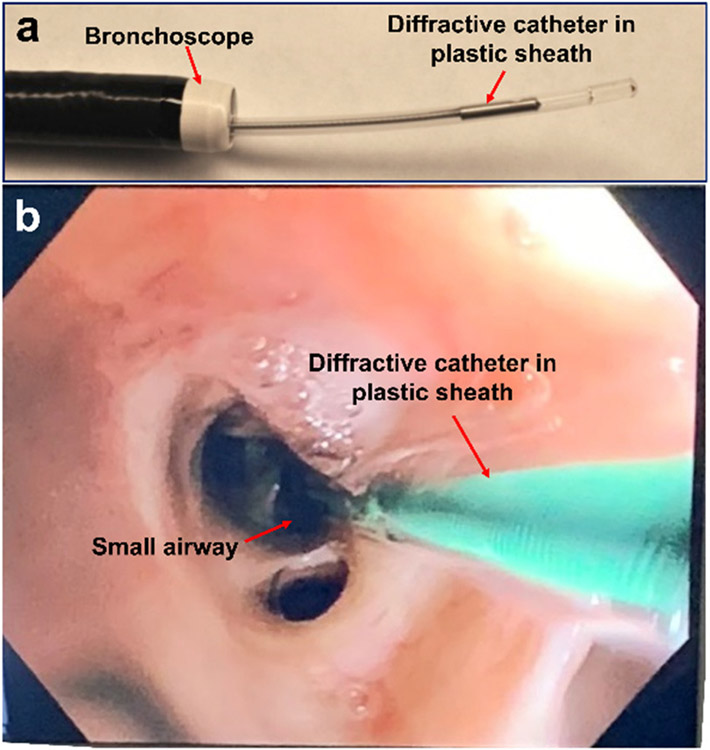
The in vivo sheep airways imaging protocol was approved by the Animal Care and Use Committee of the Johns Hopkins University. Sheep were anesthetized and OCT imaging were performed as previously reported (n=27)[32]. Briefly, the imaging catheter was deployed to the small airway through the 2.2-mm working channel of a bronchoscope (Olympus BF-P40) (Fig. 2). After OCT imaging, the sheep was sacrificed, the lungs were harvested and fixed, and the imaged airway sections were dissected for histological processing and correlation. Each histological slide contained a 10-μm thick tissue sample sectioned with close match with the microstructure architectures and orientations of cross-sectional or en face OCT images. Histological slides were stained with haemotoxylin and eosin, Masson’s trichrome, or a-smooth muscle actin (aSMA).
Fig. 2. Diffractive OCT catheter in bronchoscope.
(a) The diffractive OCT catheter deployed through the working channel in bronchoscope. (b) The diffractive OCT catheter in small airway under the bronchoscopic view.
D. Image Representation
Cross-sectional OCT intensity images (wrapped and unwrapped) were converted to a logarithmic scale and displayed in an 8-bit gray scale. Volumetric images were represented by an 8-bit gray scale with varied lengths according to the size of the specimen imaged. As an alternative approach for volumetric representations of data obtained with the diffractive OCT, we also presented images in “unwrapped” 2D en face format with tissue thickness encoded using a color scale or gray scale. This provided a visual morphological representation of each tissue compartment for real-time qualitative assessment.
III. Results
A. Identification of the Microstructures of Small Airways
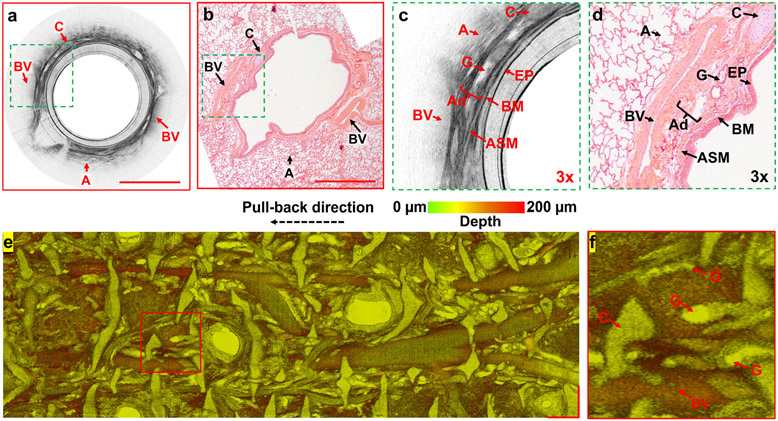
In vivo endoscopic imaging of small airways with the 800-nm diffractive OCT system provided direct visualization of the small airway wall and its major tissue compartments in vivo in the sheep model. A representative cross-sectional OCT image and its magnified view (Figs. 3a and c) were compared with the correlated histology and its magnified view (Figs. 3b and d), respectively. We were able to clearly identify the microstructures of sheep small airways, including the epithelium, basement membrane, airway smooth muscle, adventitia, submucosal glands, cartilage, blood vessels, and alveoli. Our correlation studies measuring each tissue compartment of airway in the OCT and histology revealed a good correlation, i.e., r=0.61 (p<0.001) for the epithelium, r=0.82 (p<0.001) for the basement membrane, r=0.76 (p<0.001) for the airway smooth muscle, r=0.86 (p<0.001) for the adventitia, r=0.81 (p<0.001) for the cartilage and r=0.76 (p<0.001) for the whole airway wall[32]. It is worthwhile to note that excluding submucosal glands and blood vessels in the correlation studies is because their collapsed structures in histological micrographs and the difficulties to accurate identify the walls of glands (due to the mucus) and vessels (due to the shadowing effect) in OCT images. To assess the global architecture of small airways, an 18-mm long sheep small airway was imaged and an en face view was constructed using color-coded depth projection[38], which clearly depicted the complex, interwoven network of microstructures and important tissue compartments that make up the airway wall (Figs. 3e and f).
Fig. 3. In vivo imaging of sheep small airways.
(a, b) Representative cross-sectional OCT image and correlated haemotoxylin and eosin histology, (c, d) two 3X zoomed views of the area boxed with dashed green outlines in (a, b). (e) En face image and (f) its zoomed view (marked with the red line boxed area in (e)) of an unwrapped sheep small airway of 18-mm long, by axially projecting a 200-μm thick tissue slice (starting from airway lumen surface) with color-coded depth information. A: alveoli, Ad: adventitia, ASM: airway smooth muscle, BM: basement membrane, BV: blood vessel, C: cartilage, EP: epithelium, G: submucosal glands. Scale bars: 1 mm.
B. Volumetric Visualization and Quantification of Tissue Compartments in Small Airways of Sheep
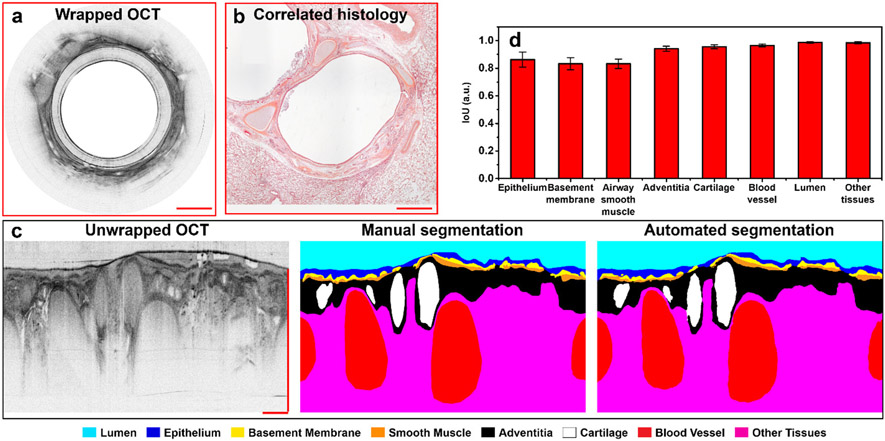
We trained a deep residual learning based neural network to automatically segment each small airway tissue compartment from the OCT images. The performance of our segmentation network was first evaluated using the test dataset. The automated segmentation results demonstrate a high similarity to the ones that were manually labelled (ground truth) by one of the experienced OCT reviewers, who segmented the OCT images by referring to corresponding histology (Figs. 4a-c). An average IoU of ~0.92 with an IoU of more than 0.8 for each tissue compartment in airway wall and airway lumen was achieved by using the Kullback-Leibler divergence loss function (Fig. 4d).
Fig. 4. Performance evaluation of the trained segmentation network with test dataset.
(a, b) Representative cross-sectional OCT image from the test dataset and its correlated histology. (c) The manual segmentation (center, ground truth) is compared with the automated segmentation results (right) from the unwrapped OCT (left). (d) The similarity (characterized with the intersection over union (IoU)) calculated by comparing 14 automated segmentation results with their corresponding manual ground truth labels for each compartment in the airway wall, the resulting IoU are presented as mean values with standard deviations. Scale bars: 500 μm.
The trained neural network was then applied to the series of OCT images of the small airways, and the segmented microstructures were then reconstructed along the longitudinal lumen direction in a 3D fashion (Fig. 5). This permitted quantitative evaluation of the architecture and volume of each tissue compartment. We have parameterized the unwrapped en face views of each tissue compartment (Figs. 6a-c) by encoding tissue thickness in color for an 18-mm long sheep small airway segment. One can clearly appreciate the longitudinally organized collagenous structures of basement membrane anchoring the similarly oriented epithelium (Figs. 6a and b). Running perpendicular to the longitudinally oriented basement membrane, is the circumferentially oriented airway smooth muscle (Fig. 6c).
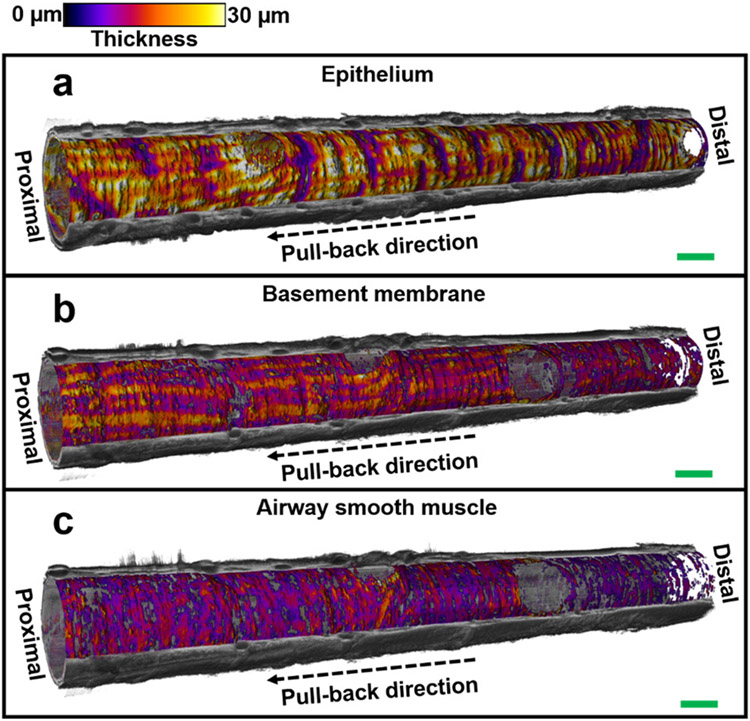
Fig. 5. 3D visualization of small airway tissue compartments.
Visualization of the volumetric architectures of tissue compartments (embedded in the cut-away view of the reconstructed 3D OCT image) in (n=1) 18-mm long sheep small airway, including epithelium (a), basement membrane (b), and airway smooth muscle (c). Thickness of each tissue compartment is represented with color and the cut-away view of 3D OCT image is represented with gray scale. Scale bars: 1 mm.
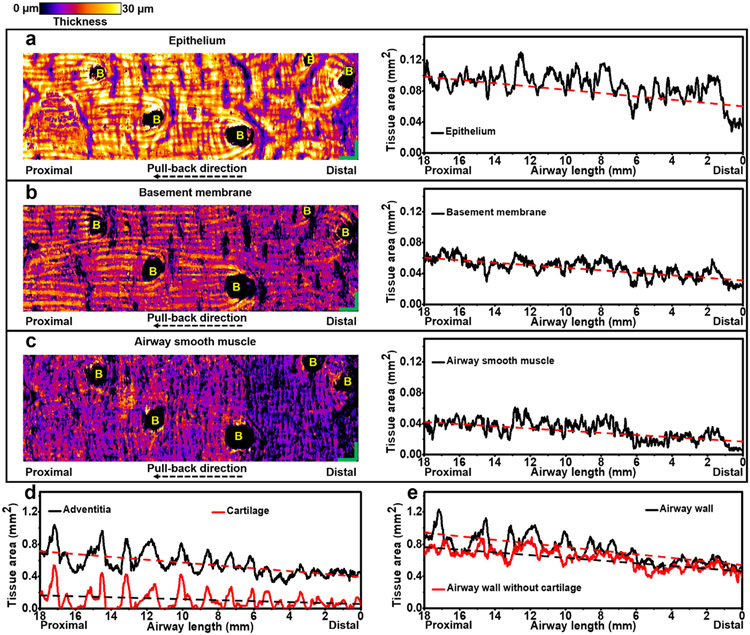
Fig. 6. Visualization and quantification of small airway tissue compartments.
(a-c) Unwrapped en face views of three tissue compartments in (n=1) representative 18-mm long sheep small airway with tissue thickness coded by color, including the epithelium (a), basement membrane (b), and airway smooth muscle (c). Quantifications of cross-sectional area of small airway tissue compartments, such as epithelium (a), basement membrane (b), airway smooth muscle (c), adventitia and cartilage (d), and airway wall with and without cartilage (e), are shown along the longitudinal airway lumen direction. The linear fitting lines (red and black dashed lines) in (a-e) indicate the increasing cross-sectional areas of each tissue compartment along the catheter pull-back direction (from distal to proximal end). 0 mm in the horizontal axes in (a-e) indicates the starting distal point of the pull-back OCT imaging. B: branch. Scale bars: 1 mm.
We further assessed the cross-sectional area of each tissue compartment longitudinally. We observed that the areas of the epithelium, basement membrane, airway smooth muscle, adventitia and cartilage increase gradually along the catheter pull-back direction (distal to the proximal direction) (Figs. 6a-d), leading to an increased cross-sectional airway wall area (Fig. 6e). Linear fitting was performed in one representative airway to indicate the trend of changes of cross-sectional areas for each tissue compartment and the entire airway wall over the length of the single airway (Fig. 6). It is worth pointing out that the oscillations on tissue areas of microcompartments in longitudinal lumen direction were not caused by breathing (since a breath hold was used during imaging), or heartbeat (since the frequency of sheep heartbeat is around 1-1.5 Hz and no such characteristic temporal frequency features were identified in the frequency analyses of the fluctuations of tissue areas along the pull-back direction). Those oscillations on measurements may be contributed by the limitations of resolutions (~1.7 μm in axial direction and ~7.2 μm in transversal direction) and quantification accuracy (an average IoU of ~0.92) of the diffractive OCT system.
C. Measuring Band Widths of Basement Membrane and Airway Smooth Muscle in the OCT and Histology
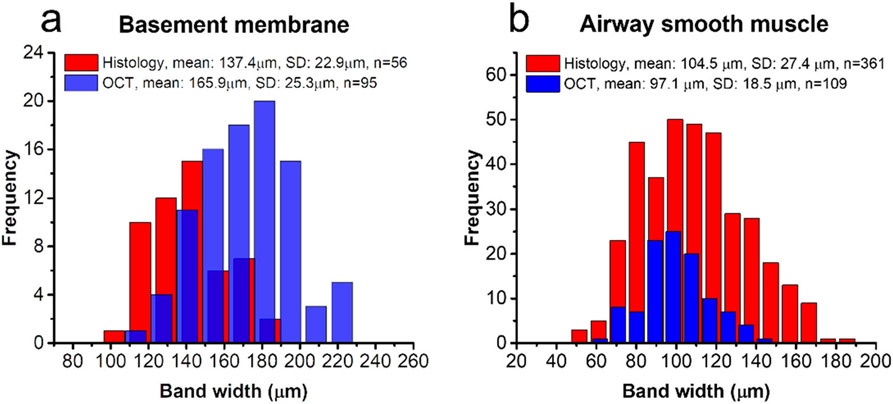
Both the volumetric architectures of basement membrane and airway smooth muscle in OCT images demonstrate good visual correlation with histology (Figs. 6a-c and Fig. 7). We further measured bandwidths of basement membrane and airway smooth muscle in the OCT and histology. The band widths were first measured directly in the histology micrographs, as shown with double-headed arrows in Figs. 7c-d. As for the OCT, the basement membrane and airway smooth muscle layers were first segmented from the volumetric datasets (n=6) with the trained neural network. Unwrapped en face images of basement membrane and airway smooth muscle were reconstructed by summing the pixels perpendicular to the lumen surface and encoding tissue thickness in gray level (Fig. 8). Thickness profiles from the 1-mm cross-sections were randomly selected on en face images. Then the band peaks and band widths were calculated using the find-peaks algorithm in Python. Band widths were obtained at the full width at half maximum (FWHM) of the peak. A minimum peak prominence of 3.5 microns was enforced in the algorithm and was chosen in consideration of the resolution limitations of the system (Fig. 8). All calculations of band width were measured from airway segments (1 mm in length and averaged 5 adjacent pixels) free from branching or excessive mucus obstruction. As shown in Fig. 9, the basement membrane and airway smooth muscle measured in the OCT and histology show closely related band widths, i.e., 165.9 ± 25.3 μm in OCT versus 137.4 ± 22.9 μm in histology for the basement membrane and 97.1 ± 18.5 μm in OCT versus 104.5 ± 27.4 μm in histology for the airway smooth muscle.
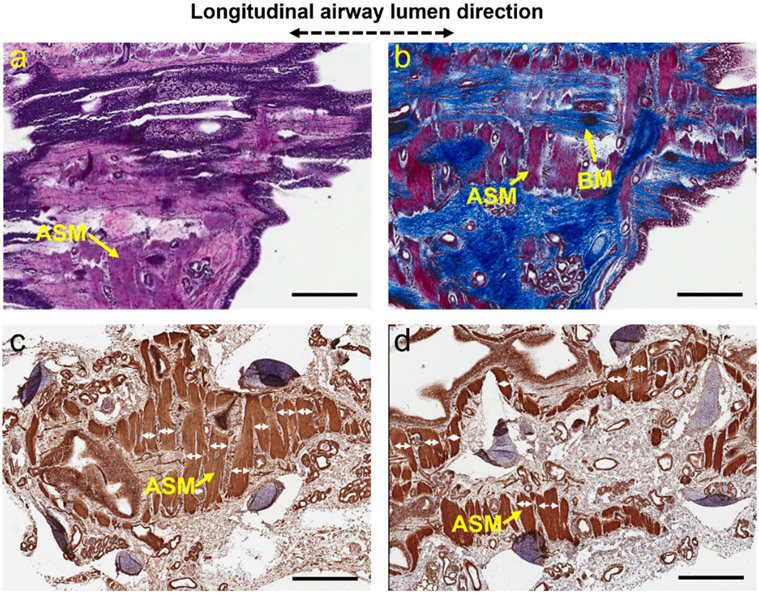
Fig. 7. 3D architectures of basement membrane and airway smooth muscle in sheep small airways.
(a, b) Two adjacent histological slides, which were stained with haemotoxylin and eosin (a) and Masson’s trichrome (b), respectively, show the volumetric architectural evolvement of basement membrane and airway smooth muscle at different depth. (c, d) Two adjacent histological slides stained with a smooth muscle actin stain (i.e., aSMA) to specifically visualize airway smooth muscle cells. Starting from the airway luminal surface, histological slides were sectioned in sequence along the longitudinal direction of the airway lumen with as close as possible orientation to en face OCT images. The histological slides shown in (a, c) were sectioned before those shown in (b, d). All these histological specimens were 10-μm thick. The band widths of airway smooth muscle (white double-headed arrows) can be measured directly on the histology micrographs. ASM: airway smooth muscle, BM: basement membrane. Scale bars: 1 mm.
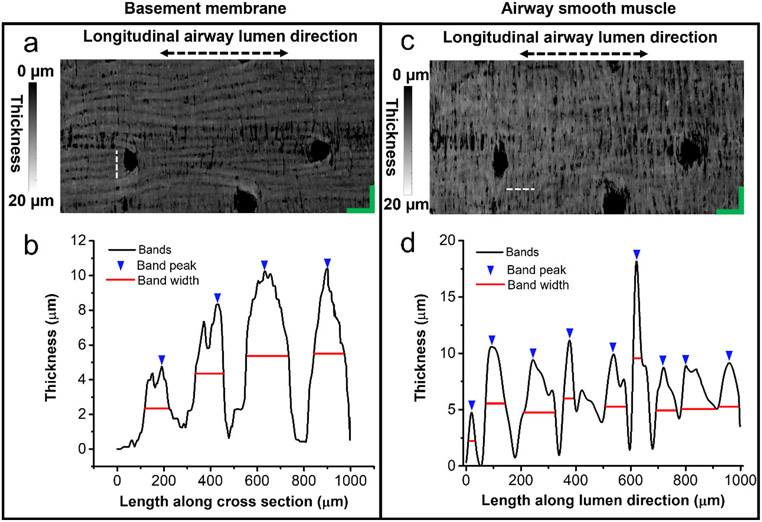
Fig. 8. Measuring band widths of basement membrane and airway smooth muscle in unwrapped en face OCT images.
(a, c) Representative en face images of basement membrane and airway smooth muscle with the tissue thickness encoded in gray level. Dashed lines: 1mm. (b, d) Thickness profiles from the 1-mm horizontal (for basement membrane) and longitudinal (for airway smooth muscle) cross-sections indicated with dashed lines in (a) and (c), respectively.
Fig. 9. Band widths of basement membrane and airway smooth muscle in the OCT versus in histology.
(a) As for the band widths of basement membrane, the average measurement from 6 histological micrographs (n=56) is compared with that from 6 en face OCT images (n=95). (b) The average airway smooth muscle band width is measured in 21 histological micrographs (n=361), versus that measured in 6 en face OCT images (n=109).
IV. Discussions
Previously, it was elegantly demonstrated that polarization-sensitive OCT operating at 1300 nm could be used for indirectly estimating smooth muscle content in medium airways (with a diameter larger than 4 mm) by assessing bulk tissue birefringence[39]. To our knowledge, this report is the first to demonstrate the capability of OCT for directly visualizing and quantifying airway smooth muscle and other critical tissue compartments in small airways (with a diameter less than 2 mm) in vivo, which may be critical sites related to asthma and COPD[1, 2, 11, 13, 40].
Recent studies have suggested that smooth muscle remodeling plays a key role in the pathogenesis of asthma and results in a significant increase in smooth muscle thickness through hyperplasia and hypertrophy[41, 42]. Direct visualization and quantification of airway smooth muscle in vivo with diffractive OCT may offer a unique opportunity for longitudinally studying the remodeling process, determining the severity, phenotyping, guiding and monitoring the response to treatment in asthma[2, 40, 41]. Furthermore, it has been reported that COPD progression closely correlates with the thickening of small airway wall tissue, owing to the increased volume in each tissue compartment by a repair or remodeling process[1, 11]. As one of the best predictors of the rapid decline in forced expiratory volume (FEV1) in COPD patients[43], nonspecific airway responsiveness was found to be strongly associated with the increased thickness in the epithelium and basement membrane[44]. The capability of diffractive OCT to visualize and assess volumetric morphological changes in each of these critical tissue compartment in small airways in vivo will likely provide critical information to elucidate the development and progression of COPD.
The current study simply demonstrated the feasibility of diffractive endoscopic OCT. It was limited to image small airways of sheep and the smallest airways imaged are approximately 1.8 mm in diameter. Nevertheless, we found that the circumferentially oriented bundle architecture of smooth muscle in the sheep small airways was similar to that of medium airways measured with birefringence OCT[39]. A systematic study is imperative for a better understanding of the 3D microstructure of small airways and validate the clinical potential of diffractive OCT for assessing airway pathology in patients. For future clinical use, 800-nm diffractive OCT technology needs further improvements. First, although the imaging speed of 20 frames per second is sufficient for demonstrating its operational feasibility, a higher speed is more desirable for clinic use to image over a longer airway segment and minimize motion artifacts. In principle, the speed can be improved by using a fast fiber-optic rotary joint and a fast imaging spectrometer. Second, training a high-performance deep learning neural network currently requires a large amount of high-quality manually labeled images, and manually labelling is onerous. Overcoming this barrier would further facilitate the clinical adoption of diffractive OCT technology, and unsupervised training or other training schemes with limited manually labeled images should be explored in the future. Additionally, we have already begun to test a new achromatic OCT microprobe that is only 1 mm in diameter, allowing in vivo imaging the smallest airways where most chronic lung diseases begin[29]. We envision the diffractive OCT technology can be applied for imaging medium and large airways which will likely require further imaging catheter modifications. Potential modifications include using a balloon to centralize the imaging probe in airways and adding an optofluidic channel for temporarily changing the local optical properties of lung tissues to improve the imaging performance[45]. Using a lung phantom will be able to help test these new catheter modifications[46].
V. Conclusion
We report for the first time, noninvasive, and automated quantification and visualization of 3D subsurface microstructures in small airways of animals in vivo with the deep learning assisted diffractive OCT. With an 800-nm broadband laser source and correction of chromatic aberration by diffractive optics, the newly developed diffractive OCT technology was able to achieve superior imaging resolution and contrast (versus 1300-nm OCT) for accurate delineation of small airway microstructures in vivo, such as the epithelium, basement membrane, airway smooth muscle, in sheep. This ultrahigh-resolution endoscopic OCT further enabled us to visualize and quantify the tissue compartments in the airway wall in 3D using a custom deep residual learning based segmentation method. Given the high resolution and high segmentation accuracy, deep learning assisted diffractive OCT enables the in vivo objective assessment of airway pathology and treatment outcome in ways there were previously not possible, allowing a better understanding and management of human lung diseases, such as asthma and COPD.
Acknowledgment
The authors thank the support from Mr. Roberto Zambito for labeling histology, Mr. Sai Mu Dalike Abaxi for help measure band widths of basement membrane and airway smooth muscle, and Dr. Dawei Li for technological assistance on the diffractive OCT system. The authors declare that they have no competing interests.
This work was supported in part by the National Institutes of Health (grants R01CA153023 (X. D. Li), R01HL121788 (R. H. Brown and X. D. Li), T32HL007534 and F32HL144121 (J. Thiboutot), and in part by The Wallace H. Coulter Foundation (X. D. Li). W. Yuan and J. Thiboutot contributed equally to this work.
Contributor Information
Wu Yuan, Johns Hopkins University, Baltimore, MD 21205, USA; Department of Biomedical Engineering and Shun Hing Institute of Advanced Engineering, The Chinese University of Hong Kong, Shatin, Hong Kong SAR, China..
Jeffrey Thiboutot, Division of Pulmonary and Critical Care Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Hyeon-cheol Park, Department of Biomedical Engineering, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Ang Li, Department of Biomedical Engineering, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Jeffrey Loube, Division of Pulmonary and Critical Care Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA..
Wayne Mitzner, Division of Pulmonary and Critical Care Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA..
Lonny Yarmus, Division of Pulmonary and Critical Care Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Robert H. Brown, Department of Anesthesiology and Critical Care Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Xingde Li, Department of Biomedical Engineering, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
References
- [1].Hogg JC, et al. , "The nature of small-airway obstruction in chronic obstructive pulmonary disease," New England Journal of Medicine vol. 350, pp. 2645–2653, 2004. [DOI] [PubMed] [Google Scholar]
- [2].Van den Berge M, et al. , "Small airway disease in asthma and COPD: clinical implications," Chest vol. 139, pp. 412–423, 2011. [DOI] [PubMed] [Google Scholar]
- [3].Gibson GJ, et al. , "Respiratory health and disease in Europe: the new European Lung White Book," European Respiratory Journal vol. 42, pp. 559–563, 2013. [DOI] [PubMed] [Google Scholar]
- [4].Chung KF, et al. , "International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma," European Respiratory Journal vol. 43, pp. 343–73, Feb 2014. [DOI] [PubMed] [Google Scholar]
- [5].Carroll N, et al. , "The structure of large and small airways in nonfatal and fatal asthma," American Review of Respiratory Disease vol. 147, pp. 405–10, Feb 1993. [DOI] [PubMed] [Google Scholar]
- [6].James AL, et al. , "Airway smooth muscle thickness in asthma is related to severity but not duration of asthma," European Respiratory Journal vol. 34, pp. 1040–5, Nov 2009. [DOI] [PubMed] [Google Scholar]
- [7].Hogg JC, "Pathophysiology of airflow limitation in chronic obstructive pulmonary disease," The Lancet vol. 364, pp. 709–721, 2004. [DOI] [PubMed] [Google Scholar]
- [8].Postma DS, et al. , "Risk factors and early origins of chronic obstructive pulmonary disease," The Lancet vol. 385, pp. 899–909, 2015. [DOI] [PubMed] [Google Scholar]
- [9].Martinez FJ, et al. , "At the root: defining and halting progression of early chronic obstructive pulmonary disease," Am J Respir Crit Care Med vol. 197, pp. 1540–1551, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Elias JA, et al. , "Airway remodeling in asthma," Journal of clinical investigation vol. 104, pp. 1001–6, Oct 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McDonough JE, et al. , "Small-airway obstruction and emphysema in chronic obstructive pulmonary disease," New England Journal of Medicine vol. 365, pp. 1567–75, Oct 27 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lange P, et al. , "Lung-function trajectories leading to chronic obstructive pulmonary disease," New England Journal of Medicine vol. 373, pp. 111–22, Jul 9 2015. [DOI] [PubMed] [Google Scholar]
- [13].Hogg JC, et al. , "Small airway obstruction in COPD: new insights based on micro-CT imaging and MRI imaging," Chest vol. 143, pp. 1436–1443, May 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pare PD, et al. , "Airway imaging in disease: gimmick or useful tool?," Journal of Applied Physiology vol. 113, pp. 636–46, Aug 15 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fujimoto JG, et al. , "Optical biopsy and imaging using optical coherence tomography," Nature medicine vol. 1, pp. 970–972, 1995. [DOI] [PubMed] [Google Scholar]
- [16].Yun SH, et al. , "Comprehensive volumetric optical microscopy in vivo," Nature medicine vol. 12, p. 1429, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Coxson HO, et al. , "Airway wall thickness assessed using computed tomography and optical coherence tomography," Am J Respir Crit Care Med vol. 177, pp. 1201–1206, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen Y, et al. , "Validation of human small airway measurements using endobronchial optical coherence tomography" Respiratory medicine vol. 109, pp. 1446–1453, 2015. [DOI] [PubMed] [Google Scholar]
- [19].Zhang J, et al. , "Automatic and robust segmentation of endoscopic OCT images and optical staining," Biomedical optics express vol. 8, pp. 2697–2708, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].De Fauw J, et al. , "Clinically applicable deep learning for diagnosis and referral in retinal disease," Nature medicine vol. 24, p. 1342–2018. [DOI] [PubMed] [Google Scholar]
- [21].Moen E, et al. , "Deep learning for cellular image analysis," Nature methods p. 1, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Soltanian-Zadeh S, et al. , "Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning," Proceedings of the National Academy of Sciences vol. 116, pp. 8554–8563, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li D, et al. , "Parallel deep neural networks for endoscopic OCT image segmentation" Biomedical optics express vol. 10, pp. 1126–1135, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma Y, et al. , "ROSE: a retinal OCT-angiography vessel segmentation dataset and new model," IEEE transactions on medical imaging vol. 40, pp. 928–939, 2020. [DOI] [PubMed] [Google Scholar]
- [25].Yang Z, et al. , "Connectivity-based deep learning approach for segmentation of the epithelium in in vivo human esophageal OCT images," Biomedical optics express vol. 12, pp. 6326–6340, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tumlinson AR, et al. , "In vivo ultrahigh-resolution optical coherence tomography of mouse colon with an achromatized endoscope," Journal of biomedical optics vol. 11, p. 064003, 2006. [DOI] [PubMed] [Google Scholar]
- [27].Xi J, et al. , "Diffractive catheter for ultrahigh-resolution spectral-domain volumetric OCT imaging," Optics letters vol. 39, pp. 2016–2019, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yuan W, et al. , "Optimal operational conditions for supercontinuum-based ultrahigh-resolution endoscopic OCT imaging," Optics letters vol. 41, pp. 250–253, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yuan W, et al. , "Super-achromatic monolithic microprobe for ultrahigh-resolution endoscopic optical coherence tomography at 800 nm," Nature communications vol. 8, p. 1531, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Park H-C, et al. , "Broadband rotary joint for high-speed ultrahigh-resolution endoscopic OCT imaging at 800 nm," Optics letters vol. 42, pp. 4978–4981, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li K, et al. , "Super-achromatic optical coherence tomography capsule for ultrahigh-resolution imaging of esophagus," Journal of biophotonics vol. 12, p. e201800205, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Thiboutot J, et al. , "Visualization and validation of the structural components in the airway wall in vivo with diffractive optical coherence tomography," Academic Radiology vol. In Press, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yuan W, et al. , "In vivo assessment of inflammatory bowel disease in rats with ultrahigh-resolution colonoscopic OCT," Biomedical optics express vol. 13, pp. 2091–2102, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yuan W, et al. , "Optimal operational conditions for supercontinuum-based ultrahigh-resolution endoscopic OCT imaging," Optics letters vol. 41, pp. 250–253, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].(2019). Semantic segmentation editor. Available: https://github.com/Hitachi-Automotive-And-Industry-Lab/semantic-segmentation-editor [Google Scholar]
- [36].He K, et al. , "Deep residual learning for image recognition," in Proceedings of the IEEE conference on computer vision and pattern recognition 2016, pp. 770–778. [Google Scholar]
- [37].Dos Santos VA, et al. , "CorneaNet: fast segmentation of cornea OCT scans of healthy and keratoconic eyes using deep learning," Biomedical optics express, vol. 10, pp. 622–641, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nam AS, et al. , "Complex differential variance algorithm for optical coherence tomography angiography," Biomedical optics express vol. 5, pp. 3822–3832, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Adams DC, et al. , "Birefringence microscopy platform for assessing airway smooth muscle structure and function in vivo," Science translational medicine vol. 8, pp. 359ra131–359ra131, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gross NJ and Barnes PJ, "New therapies for asthma and chronic obstructive pulmonary disease," Am J Respir Crit Care Med vol. 195, pp. 159–166, Jan 15 2017. [DOI] [PubMed] [Google Scholar]
- [41].James AL, et al. , "Airway smooth muscle hypertrophy and hyperplasia in asthma," American Journal of Respiratory and Critical Care Medicine vol. 185, pp. 1058–64, May 15 2012. [DOI] [PubMed] [Google Scholar]
- [42].Girodet P-O, et al. , "Bronchial smooth muscle remodeling in nonsevere asthma," Am J Respir Crit Care Med vol. 193, pp. 627–633, 2016. [DOI] [PubMed] [Google Scholar]
- [43].Tashkin DP, et al. , "Methacholine reactivity predicts changes in lung function over time in smokers with early chronic obstructive pulmonary disease. The Lung Health Study Research Group," Am J Respir Crit Care Med vol. 153, pp. 1802–1811, 1996. [DOI] [PubMed] [Google Scholar]
- [44].Wiggs B, et al. , "A model of the mechanics of airway narrowing," Journal of Applied Physiology vol. 69, pp. 849–860, 1990. [DOI] [PubMed] [Google Scholar]
- [45].Quirk BC, et al. , "Optofluidic needle probe integrating targeted delivery of fluid with optical coherence tomography imaging," Optics letters vol. 39, pp. 2888–2891, 2014. [DOI] [PubMed] [Google Scholar]
- [46].Listewnik P, et al. , "Porous phantoms mimicking tissues—investigation of optical parameters stability over time," Materials vol. 14, p. 423, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]