Abstract
As the most abundant component of the complement system, C3 and its proteolytic derivatives serve essential roles in the function of all three complement pathways. Central to this is a network of protein-protein interactions made possible by the sequential proteolysis and far-reaching structural changes that accompany C3 activation. Beginning with the crystal structures of C3, C3b, and C3c nearly twenty years ago, the physical transformations underlying C3 function that had long been suspected were finally revealed. In the years that followed, a compendium of crystallographic information on C3 derivatives bound to various enzymes, regulators, receptors, and inhibitors generated new levels of insight into the structure and function of the C3 molecule. This Review provides a concise classification, summary, and interpretation of the more than 50 unique crystal structure determinations for human C3. It also highlights other salient features of C3 structure that were made possible through solution-based methods, including Hydrogen/Deuterium Exchange and Small Angle X-ray Scattering. At this pivotal time when the first C3-targeted therapeutics begin to see use in the clinic, some perspectives are also offered on how this continually growing body of structural information might be leveraged for future development of next-generation C3 inhibitors.
Keywords: Complement, Alternative Pathway, C3, Convertase, Structural Biology, Inhibitor
1. Introduction
The complement system serves essential roles in the recognition and elimination of pathogens, clearance of physiological debris, coordination of immune responses, and numerous other aspects of homeostasis [1, 2]. However, as an early acting sentinel that can rapidly initiate the inflammatory response, inappropriate or unregulated complement activity can also be major driving force behind many human diseases that differ in severity (i.e. ‘quality of life’ illness versus life-threatening), location (i.e. localized versus systemic), and duration (i.e. acute versus chronic) [3-5]. Throughout the last three decades, a broad and increasingly nuanced understanding of the pathophysiological contributions of complement to various inflammatory, autoimmune, hematologic, and nephrologic conditions has developed alongside more recent links to etiologically-diverse diseases, including cancer and neurodegenerative disorders [3-5]. Together, this has stimulated a renewed interest the complement system among basic scientists and clinicians alike.
As the central component of the complement system, the molecule known as C3 has received considerable scientific attention since its initial isolation and characterization some 60 years ago by Müller-Eberhard and coworkers [6, 7]. Throughout this time, understanding of C3 has grown to encompass most relevant aspects of basic molecular biology including identification of its naturally-occurring genetic variants, appreciation of its relationship to other complement components, and description of the conformational changes that accompany its activation to C3b, to name but a few. Yet as causative links between the complement alternative pathway and human disease have become established with greater rigor [3-5], increasing attention has been given toward harnessing this wealth of basic information for development of therapeutic C3 inhibitors [8, 9]. With recent approvals of the C3 inhibitor pegcetacoplan (Empaveli® and Aspaveli®) by the U.S. Food and Drug Administration and European Medical Agency for treatment of Paroxsymal Noctural Hemoglobinuria (PNH), what once seemed a far off goal has now become reality.
The purpose of this review is to stimulate thought and discussion among those interested in developing the next generation therapeutic C3 inhibitors. Needless to say, it would be a formidable task to cover all of the relevant literature on C3 biochemistry, its function, and its role in human disease in one text. Fortunately, there are many authoritative and still current reviews on the complement system [1, 2] and the function of C3 specifically [10], so these issues need not be covered here. By contrast, the literature lacks a unifying review that collects and contextualizes the breadth of structural information on C3 and its fragments, and their corresponding complexes with key enzymes, regulators, effectors, and inhibitors (Table 1). Since structural studies have had a major influence on understanding the molecular details underlying C3 function, it seems both appropriate and timely to revisit this topic with an eye toward inhibitor design, and to offer some perspectives on the challenges and opportunities and that lay ahead.
Table 1.
Compilation of X-ray Crystal Structures of Human C3 or Its Fragments, Either Free or Bound to Various Ligands Involved in Activation, Regulation, or Inhibition.
| Class | C3 Protein | Ligand(s) | PDB IDa,b | Resolution (Å) |
Authors | Citation |
|---|---|---|---|---|---|---|
| C3 Proper | C3 | - | 2A73 | 3.3 | Janssen et al. | [12] |
| C3a | 4HW5 | 2.3 | Bajic et al. | [86] | ||
| C3b | - | 2I07 | 4.0 | Janssen et al. | [15] | |
| C3b | - | 5F07 | 2.8 | Forneris et al. | [43] | |
| iC3b1 | hC3Nb1 | 6YO6 | 6.0 | Jensen et al. | [87] | |
| C3c | - | 2A74 | 2.4 | Janssen et al. | [12] | |
| C3d | - | 1C3D | 1.8 | Nagar et al. | [88] | |
| C3 convertase | C3b | Factor B | 2XWJ | 4.0 | Forneris et al. | [24] |
| C3b | Factor B Factor D |
2XWB | 3.5 | Forneris et al. | [24] | |
| C3b | Factor Bb S. aureus SCIN |
2WIN | 3.9 | Rooijakkers et al. | [33] | |
| C3b | Factor Bb Factor P, monomeric formc S. aureus SCIN |
6RUR | 6.0 | Pedersen et al. | [35] | |
| C345c | Factor P, monomeric formd | 6S0B | 2.3 | van den Bos et al. | [36] | |
| Regulators | C3b | MCP, CCP(1-4) | 5FO8 | 2.4 | Forneris et al. | [43] |
| C3b | CR1, CCP(15-17) | 5FO9 | 3.3 | Forneris et al. | [43] | |
| C3b | DAF, CCP(2-4) | 5FOA | 4.2 | Forneris et al. | [43] | |
| C3b | Factor H, CCP(1-4) | 2WII | 2.7 | Wu et al. | [44] | |
| C3d | Factor H, CCP(19-20) | 2XKW 3OXU |
2.3 2.1 |
Kajander et al. Morgan et al. |
[89] [90] |
|
| C3b | mini-FHe, CCP(1-4)+(19-20) | 5O32 | 4.2 | Xue et al. | [46] | |
| C3b | mini-FHe, CCP(1-4)+(19-20) Factor I | 5O35 | 4.2 | Xue et al. | [46] | |
| Receptors | C3c | CRIg, Ig(1) | 2ICE | 3.1 | Wiesmann et al. | [23] |
| C3b | CRIg, Ig(1) | 2ICF | 4.1 | Wiesmann et al. | [23] | |
| iC3b | CR3, I-domain | 7AKK | 3.4 | Fernandez et al. | [58] | |
| C3d | CR2, CCP(1-2) | 3OED | 3.2 | van den Elsen et al. | [55] | |
| C3d | CR3, I-domain | 4M76 | 2.8 | Bajic et al. | [57] | |
| Inhibitors | C3b | Variola virus SPICE, CCP(1-4) | 5FOB | 2.6 | Forneris et al. | [43] |
| C3c | S. aureus SCIN | 3OHX | 3.5 | Garcia et al. | [22] | |
| C3c | S. aureus SCIN-B | 3T4A | 3.4 | Garcia et al. | [66] | |
| C3b | S77, Fab fragment | 3G6J | 3.1 | Katschke et al. | [68] | |
| C3b | hC3Nb1 | 6EHG | 2.7 | Jensen et al. | [67] | |
| C3c | Compstatin, 4W9A | 2QKI | 2.4 | Janssen et al. | [75] | |
| C3b | Compstatin, Cp40 | 7BAG | 2.0 | Lamers et al. | [74] | |
| C3d | S. aureus Efb-C | 2GOX | 2.2 | Hammel et al. | [18] | |
| C3d | S. aureus Ehp | 2NOJ | 2.7 | Hammel et al. | [19] | |
| C3d | S. aureus Sbi, Domain IV | 2WY7 2WY8 |
1.7 | Clark et al. | [21] | |
| C3d | Factor H, CCP(19-20) B. burgdorferi OspE |
5NBQ | 3.2 | Kolodziejczyk et al. | [91] |
As of this writing, there are 54 unique PDB depositions of crystal structures containing various forms of human C3.
In general, only the first report of a structure determination is highlighted in this Table. However, certain structures representing particularly important features or significant improvements in either resolution or model quality are also included. Structures representing alternative crystal forms, site-directed mutants, or various ligands when unbound to C3 or its fragments are omitted in the interest of space.
Further information on the nature and characterization of the monomeric form of properdin may be found in the publication of Pedersen et al. [34].
Further information on the nature and characterization of the mini-FH protein may be found in the original manuscript by Schmidt et al. [47].
2. Structural Features of Complement Component C3 and its Major Fragments
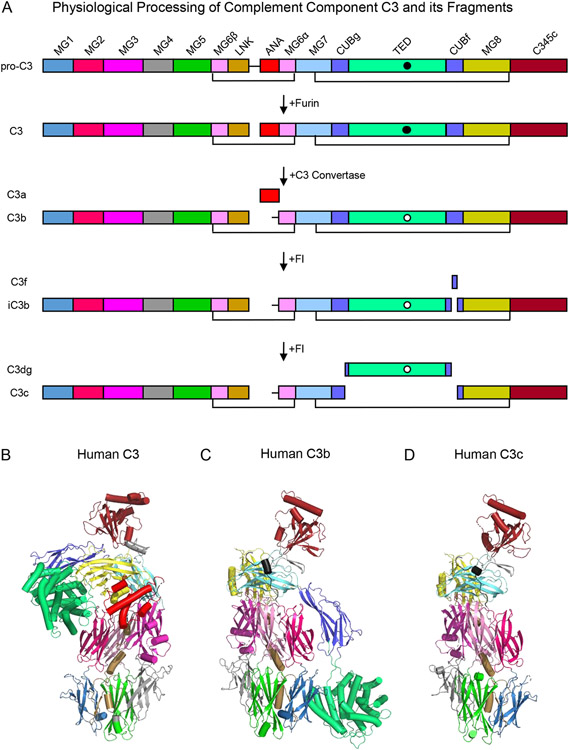
Whereas human C3 is synthesized as a 1663 amino acid pre-pro protein (UniProt Accession Number P01024), removal of its 22 residue signal peptide upon entry into the secretory pathway yields a 1641 residue pro-protein (Fig. 1A). Pro-C3 is oxidized during folding resulting in the formation of 13 pairs of disulfide bonds and is post-translationally modified with complex N-linked glycans at three distinct sites (Asn-85, Asn-939, and Asn-1617). Prior to export from the cell, cleavage of pro-C3 by furin proteases yields two distinct polypeptides conventionally known as the β-chain (residues 23-667; ~75 kDa) and α-chain (residues 672-1663; ~110 kDa) that remain covalently bound to one another through an intermolecular disulfide bond in native C3 (~185 kDa). Although the mature native C3 protein consists of 13 distinct structural domains (Fig. 1A), a series of proteolytic reactions leads to removal of either small peptide segments (e.g. C3f) or entire domains (e.g. C3a) themselves. This gives rise to various fragments of C3 that differ not only in their compositions and three-dimensional structures, but in their functional properties as well [10]. In this manner, the relatively abundant, but otherwise inert C3 molecule can be cleaved sequentially into (i) both a small chemotactic fragment (i.e. C3a; ~9kDa) and a surface-bound opsonin that serves as a platform for assembly of C3 convertases (i.e. C3b; ~176 kDa), (ii) an opsonin that no longer supports C3 convertase assembly (i.e. iC3b; ~174 kDa), and (iii) two terminal fragments (i.e. C3c; ~139 kDa and C3d; ~34 kDa), only the latter of which retains biological function as a opsonin [10]. All of these proteolytic reactions result in changes to the α-chain while the β-chain remains intact throughout the complex lifecycle of the C3 molecule (Fig. 1A).
Figure 1.
Domain Organization and Three Dimensional Structures of Complement Component C3 and its Fragments. (A) Domain structure of human C3 and its fragments. The names of each fragment are shown at the left-hand side of the panel, while the names of each individual domain are presented atop the schematic that represents pro-C3. The identities of various proteases that catalyze processing of C3 are shown by the arrows that signify each transformation. Black lines represent key disulfide bonds that covalently link the C3 polypeptide chains together throughout proteolytic processing. The intramolecular thioester found in the TED domain is represented by a filled circle in its native state and by a clear circle in its hydrolyzed state. (B) Structure of native human C3 as drawn from the PDB entry 2A73 [12]. Individual domains are colored identically to panel A, with α-helices represented as cylinders and β-strands as arrows. (C) Structure of human C3b as drawn from the PDB entry 2I07 [15]. Structure of human C3c as drawn from the PDB entry 2A74 [12]. Note that the orientation of the structures shown in panels C and D are fixed about the so-called keyring core of the molecule comprised of domains MG1-MG6.
Throughout the last two decades, the structures of essentially all major fragments of C3 have been solved by X-ray crystallography. Collectively, this work provides important insight into the physical changes that underlie the distinct functions associated with C3 and its fragments. Some of the most fundamental features of C3 biochemistry can be understood by examining the structures of C3, C3b, and C3c, including (i) how the α and β chains relate to one another in the fully-matured molecule, (ii) how the intramolecular thioester required for opsonization remains protected in native C3, yet becomes accessible upon activation to C3b, and finally (iii) the nature of the conformational changes that occur upon activation, thereby allowing C3b to act as a hub for protein-protein interactions within the complement system. The structure of C3 reveals that its 13 individual domains fold into a highly asymmetrical shape, wherein 6 domains are derived from the α and β chain, respectively (Fig. 1A, B). Although there are many aspects of C3 structure deserving of commentary but must be omitted in the interest of brevity, two features stand out insofar as protein architecture is concerned. The first of these is the MG6 domain, which is comprised of residues from both the α and β chain and which includes the lone interchain disulfide bond in native C3. The second of these is the CUB domain of the α-chain, which contains a large insertion that separates the CUBg and CUBf segments in the polypeptide sequence and which encodes the thioester-containing TED domain. Indeed, this entire CUB-TED region plays a crucial role in both preserving then exposing the functionality of the intramolecular thioester during activation of C3.
The intramolecular thioester is found within the eponymous TED domain and conveys the unusual ability of the protein to covalently opsonize surfaces and other biomaterial upon activation to C3b [10]. A necessarily labile moiety, the thioester arises from an isoglutamyl-cysteine bond between the sidechains of Cys-1010 and Gln-1013 [11]. The thioester is susceptible to reaction with small nucleophiles (e.g. methylamine) and is therefore prone to hydrolysis, particularly during purification and storage of C3 in laboratory settings. However, its access to bulk solvent is masked in the structure of native C3 wherein the thioester lies at a hydrophobic interface of the TED domain and MG8 (Fig. 1B) [12]. Upon activation through spontaneous tick-over to C3(H2O) or following proteolytic cleavage to C3b, the exposed thioester is converted to a free thiolate anion on Cys-1010 and an acyl-imidazole intermediate between the sidechains of Gln-1013 and His-1126 [13, 14]. This secondary step allows for preferential reaction of C3b with hydroxyl groups of nearby proteins but especially with carbohydrates found on biological surfaces (e.g. bacterial and fungal cells), thus facilitating their opsonization with C3b. In addition, C3b covalently linked to these molecules and surfaces sets the stage for amplification of complement activity by serving as a platform molecule for assembly of the C3 and C5 convertases. These latter events will be described in detail at a later point in the text.
Comparison of the native C3 structure with C3b reveals a striking example of protein conformational change (Fig. 1B, C). Whereas the keyring-like core of the molecule comprised of domains MG1-MG6 is more or less unchanged during activation, most other domains experience structural rearrangements to varying degrees [15]. These range from minor repositioning, as typified by domains MG7 and MG8, to large scale movements, such as those seen for the CUB and TED domains [15]. It is noteworthy that the TED domain undergoes the greatest changes of all, which results in an ~85 Å translation and full exposure of the intramolecular thioester away from its position in native C3 [15]. Although little is known about the speed with which these changes occur upon C3 activation, such reorientation of the TED domain has been suggested to bring the highly reactive acyl-imidazole intermediate [13, 14] into immediate proximity of hydroxyl groups borne by the target surface while minimizing its quenching by water [15]. It is also fitting that those regions of the C3 molecule that experience obvious structural changes are also those that undergo proteolytic processing during the stepwise activation and degradation of the protein (Fig. 1A). This feature can be appreciated most readily by examining the structure of C3c [12], which represents the structural scaffold of the C3 molecule absent those domains most directly involved in its key effector functions of promoting inflammation (i.e. C3a) and opsonization (i.e. TED) (Fig. 1D).
One of most enigmatic features of the C3 protein in physiological settings is its paucity of ligands prior to its activation to C3b. Aside from molecules specifically selected for their C3-binding activity, such as the peptidomimetic Compstatin [16] and various anti-C3 monoclonal antibodies [17], the panel of C3 interactors is limited to a handful S. aureus immune evasion proteins Efb-C [18], Ehp [19], and Sbi [20], the C3 convertases themselves, and little else. By contrast, C3b is a hub of interactions within the complement system and has well over two dozen endogenous ligands alone [10]. The physical basis for this property is apparent from comparing the structure of native C3 with that of C3b. In this regard, all binding sites for known interactors, including the alternative pathway pro-protease Factor B (FB), regulatory proteins like Factor H (FH), and various complement receptors, are either masked or otherwise unavailable in native C3 [15]. The exceptions to this rule not surprisingly include each of the immune evasion proteins listed above, all of which bind a site on the TED domain that is freely accessible in native C3 [18, 19, 21], as well as the scissile bond that is cleaved by C3 convertases to simultaneously release C3a and generate C3b. Even ligands like the S. aureus immune evasion protein SCIN [22] and the Complement Receptor of the Ig Superfamily (CRIg) [23], whose binding sites are retained in the C3c fragment, require conformational rearrangements from the native C3 structure to assemble or unmask their sites. Thus, the structural changes that occur upon C3 activation to C3b not only serve to expose the thioester with maximal effect, they also manifest an array of protein-protein interaction sites that drive a majority of downstream C3b functions.
3. Structural Features of C3 Convertase Formation, Stabilization, and Function
Although C3 is known to spontaneously activate through the process known colloquially as tick-over, this activation route is comparatively inefficient to the enzyme catalyzed route involving C3 convertases [1, 10]. The C3 convertases themselves are intrinsically labile, multi-subunit complexes that in the most typical situations assemble on target surfaces in a step-wise manner [1]. In the first step, a scaffold molecule covalently linked to the target surface, a pro-protease to provide the eventual catalytic functionality, and a divalent cation to facilitate the interaction between the two protein components all come together to form a structure known as the pro-C3 convertase (or pro-convertase for short). In the second step, a separate enzyme activates the pro-convertase, thereby releasing a fragment of the pro-protease and generating the corresponding C3 convertase with full catalytic activity. Despite the fact that they recognize the same native C3 substrate, C3 convertases come in two varieties that reflect at the molecular level whether their formation was initiated by either the classical or lectin pathways (i.e. C4b2a via C4 and C2) or by the alternative pathway (i.e. C3bBb via C3 and FB) [1]. Since far more is known at the structural level regarding the formation and function of the alternative pathway C3 convertase (Table 1), this topic will be examined in greater detail below.
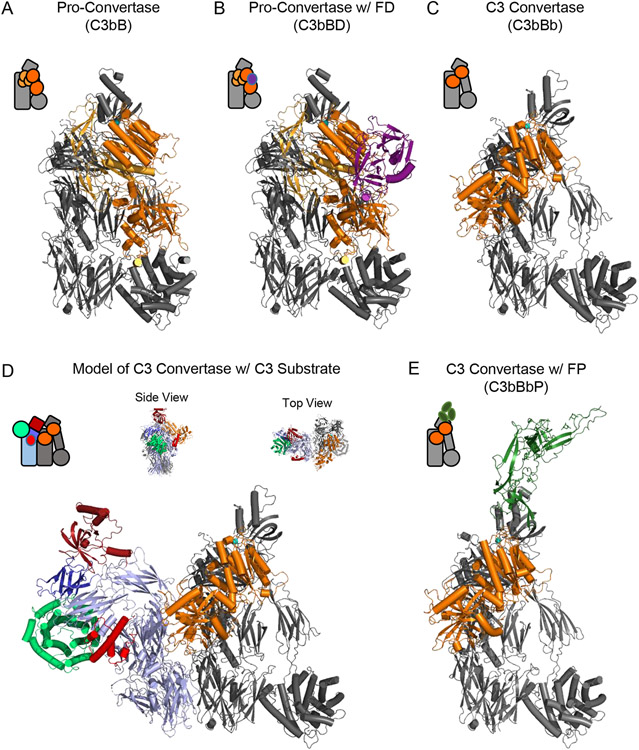
When C3b is generated and covalently attaches to a surface, it acts as a platform to recruit the pro-protease FB out of circulation and thereby form the pro-convertase, C3bB. The structure of C3bB shows that the FB binding site is derived primarily from the C345C and CUB domains of C3b, and to a lesser extent its MG2, MG3, MG7 domains and the nascent N-terminus of the α’-chain (Fig. 2A) [24]. This site is not exposed in the structure of native C3, which provides an explanation for how native C3 and FB can circulate at high concentrations in the bloodstream without leading to inadvertent generation of the pro-convertase. Conversely, the residues on FB that form contacts with C3b are derived from all five structural domains of FB [24]. Among these, the von Willebrand factor type-A (VWA) domain within the Bb region of FB stands out as it contributes the so-called metal-ion dependent adhesion site (MIDAS). The MIDAS site lies at the interface between the VWA domain of FB and the C345C domain of C3b and is responsible for the Mg2+-dependence of the alternative pathway C3 convertase in physiological systems. In this particular case, Ni2+ was substituted for Mg2+ as it leads to a marked stabilization of the complex that facilitates structural and functional studies [25].
Figure 2.
Structural Features of the Alternative Pathway C3 Convertase. (A) Structure of the alternative pathway C3 pro-convertase as drawn from the PDB entry 2XWJ [24]. C3b is colored grey while FB is colored in shades of orange, with its Ba fragment in light orange and the Bb fragment in dark orange. A divalent cation, which in physiological conditions is Mg2+, is represented as a blue sphere. (B) Structure of the alternative pathway C3 pro-convertase bound to FD as drawn from the PDB entry 2XWB [24]. C3b and FB are colored as in panel A, while FD is colored purple. (C) Structure of the alternative pathway C3 convertase as drawn from the PDB entry 2WIN [33]. Proteins are colored identically to panel A. Note the loss of the Ba domain of FB that occurs following FD cleavage. The S. aureus immune evasion protein SCIN [31, 32, 92] is omitted from this image for clarity, although it was used to facilitate crystallization of this otherwise labile complex [33]. (D) Model of the alternative pathway C3 convertase bound to its native C3 substrate. C3b and Bb are colored identically to panel C, while native C3 appears with color scheme similar to that used in Figure 1. Both side and top-down views of the model are shown above the primary image. (E) Structure of the alternative pathway C3 convertase bound to a fragment of properdin as drawn from the PDB entry 6RUR [35]. C3b and Bb are colored identically to panel C, while the properdin fragment is colored green. The S. aureus immune evasion protein SCIN has likewise been omitted from this image for clarity. Simplified representations of each structure are shown in the top left corner of each panel.
The pro-convertase C3bB has no enzymatic activity, and instead must be cleaved by the protease, Factor D (FD), to generate the fully active C3 convertase, C3bBb. FD is found in the bloodstream in a fully-matured, yet auto-inhibited state, which presumably contributes to the specificity of FD for the C3b-bound form of FB [26, 27]. The structure of the Ni2+-stabilized C3bB complex bound to a catalytic mutant of FD provides insight into how this occurs (Fig. 2B) [24]. Whereas the structure of FD on its own is characterized by binding of a self-inhibitory motif that distorts its active site residues from their expected positions [26, 27], the structure of C3bB-bound FD reveals that this inhibitory loop is displaced and that a catalytically-competent active site is present [24]. There are several other features of this structure worth noting. First, although FD binding to its C3bB substrate displaces its self-inhibitory loop, the primary interaction between FD and C3bB occurs at an exosite that is relatively far removed from the FD active site proper. This FD exosite appears to be a crucial regulatory element that limits formation of the fully-active convertase [24]. Second, the scissile loop of FB that is cleaved by FD remains disordered even following FD binding. This is consistent with the poor catalytic profile of FD and its relatively inefficient processing of idealized substrates [28]. Finally, although FD is specific for the C3b-bound form of FB, FD itself forms no direct interactions with C3b. This element of specificity arises from binding of FB in a conformation seen only in the pro-convertase, and not in the structure of unbound FB [29].
The alternative pathway C3 convertase is notoriously challenging to study, since it is intrinsically labile and dissociates irreversibly within minutes of formation in vitro [30]. However, inclusion of the S. aureus immune evasion protein, SCIN, which stabilizes convertases in an enzymatically inactive but kinetically stabilized form [31, 32], allows for formation of dimeric C3bBbSCIN complexes (i.e. (C3bBbSCIN)2) amenable to crystallographic analysis [33]. New understanding of the C3bBb convertase was obtained through this approach (Fig. 2C) [33]. Most significantly, loss of the Ba fragment upon its proteolysis by FD results in rearrangement of the catalytic Bb fragment relative to its position in either C3bB or C3bBD (Fig. 2A-C). This involves elimination of all C3b contacts arising from the Ba fragment (i.e. to MG2, MG3, MG7, and the α’-chain N-terminus) in addition to others from the Bb fragment (i.e. to MG2 and CUB) that are seen in the pro-convertase structures [24]. Nevertheless, the interactions between the VWA domain of the Bb fragment and the C345C domain of C3b are maintained through the MIDAS site and accessory protein-protein contacts [33]. This arrangement appears to allow the proteolytic Bb fragment to rotate in conjunction with the C345C domain, which itself exhibits inherent flexibility relative to the remainder of the C3b molecule. Presumably, this relative freedom of rotation permits the Bb protease active site to come in contact with the scissile bond of the C3 substrate [33]. Although native C3 is not a component of this structure, the location of the C3 binding site within the convertase can be extrapolated from the dimerization interface between the two copies of C3b found in each (C3bBbSCIN)2 complex. In this regard, superposition of the native C3 structure onto the second copy of C3b shows good agreement between the rigid keyring core of the proteins. This model strongly suggests that C3b domains MG4 and MG5, and very likely MG3 and MG6-8 are critical determinants of C3 substrate recognition by C3bBb (Fig. 2D) [33]. At the present time, however, confirmation of these predictions awaits further structural studies of the C3 convertase bound to its bona fide substrate, C3.
The labile nature of the C3 convertase provides an intrinsic means of regulating its enzymatic activity, yet this feature can be offset to some extent by action of the protein known as properdin. Properdin is the only known positive regulator of the alternative pathway. Although properdin has been ascribed pattern recognition receptor-like properties, its best understood function lies in stabilization of the C3bBb convertase [1]. Unfortunately, structural studies into its function are complicated by the fact that properdin is highly post-translationally modified and exists as polydisperse multimers, not to mention all of the previously stated challenges associated with studying C3 convertases. These issues can be circumvented in part through extensive recombinant protein engineering and use of stabilizing nanobodies [34-36], thereby permitting crystallographic and functional studies. Structures of monomeric properdin analogues bound to the SCIN-stabilized C3bBb convertase show that properdin interacts primarily with the C345C domain of C3b, and to a lesser extent the Bb fragment (Fig. 2E) [35, 36]. Significantly, this binding site lies in close enough proximity to contact the Bb domain and promote its retention on C3b, while not appearing to impede binding of the C3 substrate to the convertase (Fig. 2D). As properdin naturally exists as planar, rigid oligomers of extended conformations [37], the combination of these properties has been proposed to facilitate clustering alternative pathway convertases on susceptible surfaces. Given the self-amplifying nature of the alternative pathway, it is easier to understand how these particular structural features of properdin foster high-level alternative pathway activity in physiological systems.
4. Structural Considerations Underlying Regulation of C3b Function
The alternative pathway is capable of self-amplification and is responsible for a large majority of downstream complement activity in physiological settings [38, 39]. Thus, it is not surprising that unregulated alternative pathway activity can become a pathophysiological driving force in many human diseases [3-5]. Nevertheless, the fact that most of these diseases are relatively uncommon in the general population indicates that alternative pathway activity is precisely regulated in vivo. Unraveling the many layers of regulation that control alternative pathway activity has required biochemical and functional studies spanning more than 50 years [40]. It has also befitted from more recent crystallographic studies on the convertase itself and a dedicated suite of regulatory molecules. Points of interest arising from both of these approaches will be summarized in the paragraphs that follow.
There are two intrinsic regulatory mechanisms that act at the level of the convertase components themselves [1]. First, the complex sequence of events required to form the fully active alternative pathway C3 convertase represents a key point of regulation (Fig. 2). It is this requirement of not one, but two activation steps prior to convertase formation that allows the precursor proteins (i.e. C3 and FB) to exist quiescently in the same environment prior to initiation of complement activity via a specific biochemical cue. Once it is formed, though, the C3 convertase is subject to a second point of intrinsic regulation. In the absence of other factors (e.g. substitution of Ni2+ for Mg2+), the C3 convertase is intrinsically labile and decays with a half-life of approximately one minute in physiological settings [30]. This step is also irreversible, as the Bb fragment released cannot reform an active convertase. Although these latter properties are influenced to some degree by the action of properdin, together they provide important contextual and temporal restrictions on alternative pathway activity in both in vitro and in vivo settings.
There are likewise extrinsic control mechanisms that rely on the action of a dedicated suite of proteins known as the Regulators of Complement Activation (RCA; [41, 42]). RCA proteins share a common modular architecture based upon repeating complement control protein (CCP) domains. Although up to 30 CCP domains may be found in an RCA molecule, their regulatory functions localize to a discrete and contiguous stretch of repeats through which they bind directly to C3b [41, 42]. RCA proteins inhibit alternative pathway activity through two distinct mechanisms [1, 41, 42]. On one hand, decay accelerating activity not only promotes the irreversible dissociation of the Bb fragment from C3b, it also blocks pro-convertase formation by competing with FB binding to C3b. The RCA proteins Decay-Accelerating Factor (DAF) and Complement Receptor 1 (CR1) are localized at the cellular surface and protect self-cells via their decay-accelerating activity, while Complement Factor H (FH) does likewise following its capture out of circulation via interactions with self-cell associated carbohydrates and other polyanionic structures. On the other hand, cofactor activity arises when RCA proteins bind C3b and facilitate recruitment of the degradative protease known as Complement Factor I (FI). Although the RCA proteins themselves are not altered in this process, cleavage of C3b by FI generates iC3b, which no longer supports FB binding or downstream convertase formation. The RCA proteins Membrane Cofactor Protein (MCP) and CR1 exhibit cofactor activity at the self-cell surface, while surface-adsorbed FH does as well. As a group, this collection of RCA proteins provides an important self-protective mechanism against the alternative pathway by diminishing C3 convertase stability and effectively reducing the levels of C3b available for convertase formation.
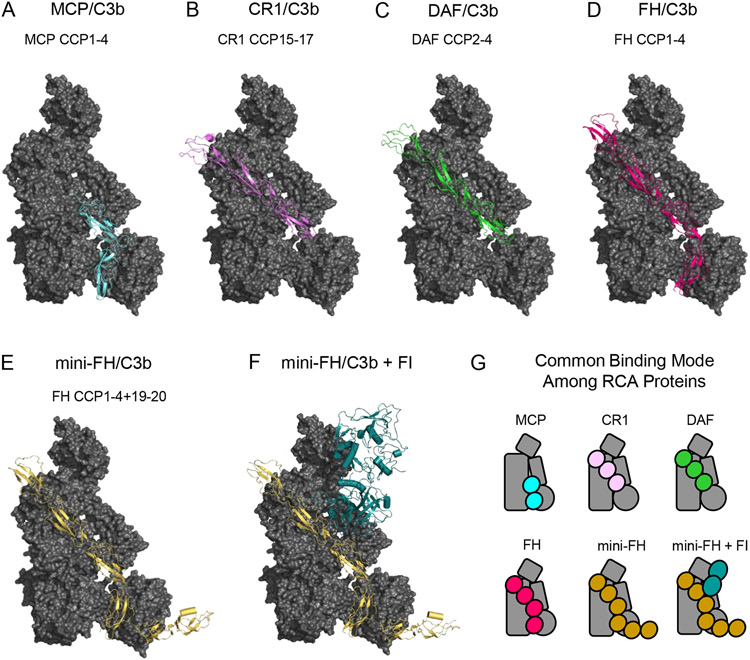
Although they differ in distribution (i.e. cell surface-retained versus soluble) and manifestation of inhibitory mechanism, all RCA proteins share important structural features beyond that of their repeating CCP domain architecture. In this regard, structural studies of C3b bound to the minimal regulatory fragments of MCP (domains CCP(1-4); [43]), CR1 (domains CCP(15-17); [43]), and DAF (domains CCP(2-4); [43]), as well that of C3b bound to FH (domains CCP(1-4); [44]) reveal that all RCA proteins employ a common C3b-binding mode (Fig. 3A-D, G). At the most fundamental level, each of these regulators binds in an elongated fashion and in an identical orientation relative to C3b [43]. A deeper interpretation of these structures reveals four apparently generic RCA-binding sites on the C3b surface, denoted sites CCPi-iv; these are derived from the nascent N-terminus of the α’-chain and MG7 (CCPi), MG6 and MG7 (CCPii), MG2 and CUB (CCPiii), and MG1 and TED (CCPiv), respectively [43]. Regardless of where the minimal regulatory fragment lies in any given RCA protein, the N-terminal most domain visible in its C3b-bound structure occupies the site closest to the generic CCPi site [43]. Indeed, the common binding mode shared among RCA proteins that exhibit decay accelerating activity only (i.e. DAF), cofactor activity only (i.e. MCP), or those with both (i.e. FH and CR1) suggests that these proteins may have diverged from a common evolutionary precursor [43].
Figure 3.
Structural Features of C3b-binding Complement Regulatory Molecules. (A) Structure of C3b bound to CCP domains 1-4 of Membrane Cofactor Protein (MCP) as drawn from PDB entry 5FO8 [43]. C3b is colored as a grey surface, while MCP is colored light blue. (B) Structure of C3b bound to CCP domains 15-17 of Complement Receptor 1 (CR1) as drawn from PDB entry 5FO9 [43]. C3b is colored as a grey surface, while CR1 is colored pink. (C) Structure of C3b bound to CCP domains 2-4 of Decay Accelerating Factor (DAF) as drawn from PDB entry 5FOA [43]. C3b is colored as a grey surface, while DAF is colored green. (D) Structure of C3b bound to CCP domains 1-4 of Complement Factor H (FH) as drawn from PDB entry 2WII [44]. C3b is colored as a grey surface, while FH is colored magenta. (E) Structure of C3b bound to CCP domains 1-4 and 19-20 of mini-FH as drawn from PDB entry 5O32 [46]. C3b is colored as a grey surface, while mini-FH is colored gold. (F) Structure of C3b bound to mini-FH and Complement FI as drawn from PDB entry 5O35 [46]. C3b and mini-FH appear identically to panel E, while FI is colored dark blue. (G) Simplified representations of the structures shown in panels A-F. Note the common binding surface recognized by each of these C3b-binding regulatory molecules.
In addition to identifying their conserved mode of C3b binding, structural investigation of RCA proteins also illustrates the physical basis for their function. Correlating this portfolio of structural information with pre-existing functional studies on the same RCA fragments leads to a model wherein binding to sites CCPi-iii imparts decay accelerating activity while binding to sites CCPii-iv imparts cofactor activity. For example, the RCA binding sites CCPi-ii are occupied by domains CCP(2-3) of DAF [43], which are also known to interact with Bb [45]. It remains unknown how this latter interaction promotes dissociation of Bb from C3b, however. Adjacent to this, RCA binding sites CCPii-iii share significant overlap with the binding site of FB seen in the C3bB pro-convertase structure (c.f. Fig. 2A, B and Fig. 3B-D) [24]. Presumably, this leads to direct competition between any regulators binding to this region and FB, thereby diminishing further convertase formation. Finally, the roles of RCA binding sites CCPii-iv in promoting cofactor activity can be inferred from the structure of a designer RCA protein, mini-FH, bound to both C3b and FI [46]. Mini-FH is a fusion protein consisting of the regulatory domains of FH (i.e. CCP(1-4)) linked to the FH cell surface targeting domains (i.e. CCP(19-20)), but exhibits enhanced affinity and regulatory potency when compared to native FH [47]. The structure of mini-FH bound to C3b alone (Fig. 3E) is almost indistinguishable from that of FH domains CCP(1-4) bound to C3b (Fig. 3D, G) [44, 46], illustrating that its regulatory domains occupy RCA binding sites CCPi-iv. However, the structure of the ternary complex shows extensive interactions between mini-FH CCP(2-3) and the serine protease domain of FI (Fig. 3F, G) [46]. As mini-FH CCP(2-3) occupy RCA binding sites CCPii-iii, it appears these domains help recruit FI to C3b and properly orient the FI active site toward the scissile bonds found within the C3b CUB domain [46]. The role of mini-FH CCP(4), which occupies RCA site CCPiv, therefore seems to rest in binding TED and restricting its translation away from the keyring core. It is worth noting that multiple studies indicate flexibility between the CUB-TED region of C3b and the remainder of the molecule [48-50]. Consequently, the CCPiv interaction also seems to properly orient the scissile bonds within CUB to the FI active site, culminating in the cleavage of C3b to its degradation product, iC3b.
5. Structural Basis for Recognition of C3b and its Fragments by Complement Receptors
Opsonization of biomaterial with C3b sets the stage for C3 convertase formation and amplification of the complement response via the alternative pathway [1, 10]. It likewise provides a route through which C3 fragment-opsonized material can be engulfed, destroyed, and in certain cases processed for antigen presentation by phagocytic cells [1, 10]. This essential effector function of complement is known as opsonophagocytosis and is predicated upon recognition of C3b and its degradation products by cell surface-retained complement receptors. Complement receptors cannot simply be high-affinity binders, though. They must also be selective for the activated forms of C3, since the major classes of phagocytic cells continually encounter the high concentrations of native C3 found in circulation. This all-important selectivity is achieved through two different mechanisms. First, the complement receptor binding sites are only revealed following the structural transitions that accompany C3 activation to C3b [1, 10]. Second, the step-wise degradation of C3b to iC3b and ultimately C3d provides increased access of certain binding sites to the large, surface bound receptors that recognize them [1, 10]. Not surprisingly, structural studies have contributed to understanding this process at the molecular level, as will be described below.
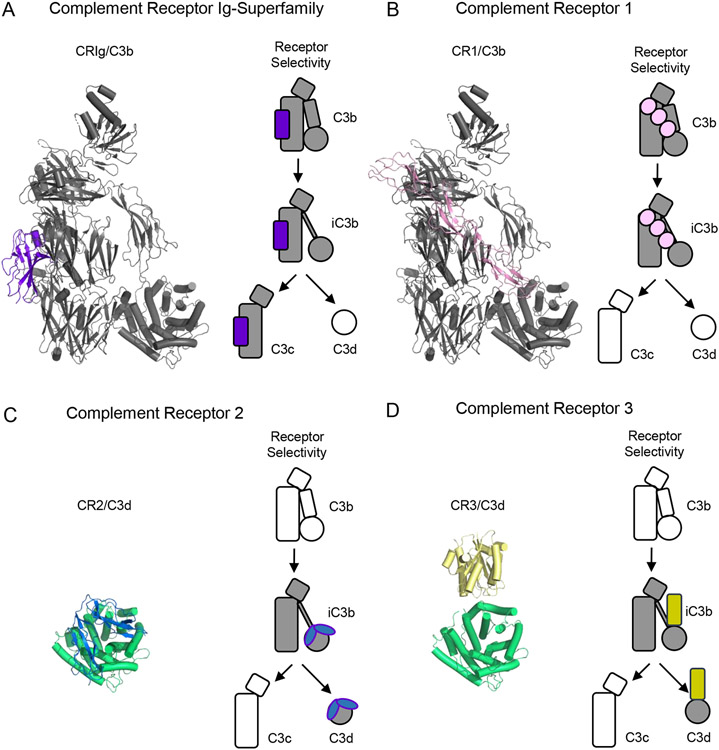
The Complement Receptor of the Ig superfamily (CRIg) is expressed on tissue resident phagocytes and acts as a high-affinity receptor for C3b and iC3b [51]. As its name suggests, the extracellular region of CRIg consists of either one or two Ig repeats depending on alternative splicing of the transcript [51]. The structure of C3b bound to an Ig-domain from CRIg shows that its binding site is comprised of MG3-MG6 as well as the LNK domains of the C3b β-chain (Fig. 4A) [23]. While these domains are accessible in the structure of native C3 [12], a slight rotation of the MG3 domain along with translation of the LNK domain are needed to complete the CRIg-binding site. This explains the inability of CRIg to bind native C3 [51]. By contrast, CRIg maintains high-affinity binding to both iC3b and C3c [51]. Although the functional relevance of its interaction with C3c is unknown, the structure of the same CRIg Ig-domain bound to C3c is almost identical to that of C3b [23]; this suggests that there are no major changes to the CRIg binding site as C3b is processed sequentially to iC3b and then C3c. A final noteworthy feature of CRIg is that its C3b-binding domain is also a potent inhibitor of the alternative pathway [23]. Since CRIg does not display any RCA-like activity, the most likely explanation for this observation is that the CRIg binding site on C3b is essential for substrate binding to the C3 convertase [23]. Indeed, this is apparent from the model of the C3-bound convertase presented in an earlier section (Fig. 2D) [33].
Figure 4.
Structural Features of Complement Receptors that Bind to C3-derived Opsonins and Fragments. (A) Structure of C3b bound to a domain from the Complement Receptor of the Ig-superfamily (CRIg) as drawn from PDB entry 2ICF [23]. C3b is colored grey, while CRIg is colored purple. (B) Structure of C3b bound to CCP domains 15-17 from Complement Receptor 1 (CR1) as drawn from PDB entry 5FO9 [43]. C3b is colored grey, while CR1 is colored pink. (C) Structure of CCP domains 1-2 from Complement Receptor 2 (CR2) bound to C3d as drawn from PDB entry 3OED [55]. C3d is colored green, while CR2 is colored blue. (D) Structure of the I-domain from Complement Receptor 3 (CR3) bound to C3d as drawn from PDB entry 4M76 [57]. C3d is colored green, while CR3 is colored yellow. Simplified representations of each receptor and the various fragments of C3 that it recognizes are shown at the right of each panel. Shaded images signify interactions known to occur, while hollow images signify activated forms of C3 that are not recognized by the receptor in question.
Complement Receptor 1 (CR1) is more widely distributed than CRIg and is found on the surface of practically all types of peripheral blood cells [52]. The opsonin-binding profile of CR1 includes not only C3b and iC3b, but C4b as well [1, 52]. CR1 is a large molecule that in its most common variant contains 30 CCP repeats in its extracellular region [52]. However, functionally redundant regions with nearly identical sequences have been identified within CR1 [52], allowing useful inferences to be gained from studies on the prototypic region of domains CCP(15-17). The structure of CR1 domains CCP(15-17) bound to C3b shows an extended binding site formed from the nascent N-terminus of the α’-chain, along with the MG2, MG6, MG7, and CUB domains (Fig. 4B) [43]. As with many other C3b-binding molecules, this binding site is masked in native C3 and requires both proteolytic release of the C3a fragment and the accompanying conformational change to assemble [12, 15]. Although the main features of this binding site must be retained upon conversion to iC3b, it is interesting to note that iC3b is lower-affinity ligand for CR1 than C3b [52]; since the CUB domain contributes to the CR1 binding site, the lower affinity of CR1 for iC3b most likely comes from changes to the CUB domain that occur following its sequential cleavages by FI [50]. Curiously, CR1 is the only FI cofactor that supports all three proteolytic cleavages of C3b needed to generate C3c and C3dg [53]. CR1 is therefore unique among complement receptors, as it exhibits RCA properties as well [1, 10, 52].
In contrast to either CRIg or CR1, the remaining receptors for C3-derived opsonins do not bind C3b [10]. Instead, their binding sites only become fully accessible upon cleavage of C3b to iC3b. The physical basis for this long-standing observation can be understood from a series of non-crystallographic structural studies. Both electron microscopy and small angle X-ray scattering analyses of iC3b are consistent with a TED domain dislodged from its position against the keyring core [46] when compared to the crystal structures of C3b alone [15, 43]. This suggests that a fundamental change in the flexibility of the CUB region must occur upon C3b cleavage to iC3b. Strong evidence regarding the nature and extent of this change comes from peptide-level resolution hydrogen-deuterium exchange studies comparing C3b to iC3b [50]. Whereas peptides within the CUB domain exhibit varying degrees of solvent protection in C3b, these same peptides are almost uniformly solvent-exposed in iC3b [50]. The simplest interpretation of these results is that site-specific cleavage by FI within the CUB domain causes it to unfold [50]. Ultimately, this leaves behind a flexible tether that connects what will become the C3c fragment to the covalently-bound opsonin fragment that will become C3d [50].
Although the complement receptors selective for iC3b and C3d share a common ligand-binding profile, they exhibit divergent structural features themselves. Complement Receptor 2 (CR2) is expressed on the surface of B lymphocytes and follicular dendritic cells [1, 10]. Its extracellular region consists of 15 CCP repeats and therefore bares some architectural similarities with CR1. The iC3b and C3d binding activity of CR2 resides in domains CCP(1-2) [54]. The structure of this CR2 fragment bound to C3d reveals that the CCP domains from the receptor fold back upon one another and recognize a binding site on the opposing face of the C3d molecule from its former thioester site (Fig. 4C) [55]. This ensures that CR2 binding to iC3b or C3d would not be impeded by their covalent linkage to a surface. Moreover, the selectivity of CR2 for iC3b and C3d versus C3b can be explained an obvious steric clash with MG1 when this CR2 binding site is inspected in C3b crystal structures [55]. This steric clash is presumably relieved by unfolding of the CUB domain upon cleavage of C3b to iC3b [50]. In contrast to CR2, Complement Receptor 3 (CR3) is expressed primarily by myeloid-lineage cells [56]. CR3 is an integrin and its iC3b and C3d binding activity resides solely within its I-domain. The structure of the CR3 I-domain bound to C3d shows that its binding site also lies opposite the former thioester (Fig. 4D) [57]. However, the CR3 binding site on C3d is slightly offset from that of CR2 such that it would sterically clash with the intact CUB domain of C3b instead of MG1 [57]. But similarly to CR2, this steric clash would be removed as the CUB domain unfolds upon cleavage of C3b to iC3b [50], thus explaining its selectivity for these late-stage opsonins [1, 10]. It also seems likely that binding of CR3 to this site would be facilitated by the conformational flexibility that a fully unfolded CUB domain provides, since it allows the C3c-like moiety to move away from the surface-bound C3d domain [46, 50]. Indeed, a very recent crystal structure of iC3b bound to the CR3 I-domain shows examples of two different orientations of the C3c-like core relative to the C3d domain [58]. This further alludes to the need for increased structural flexibility in iC3b to support its recognition by cell surface-exposed complement receptors.
6. Structural Themes Arising from Complement Inhibitors Targeting C3 or its Fragments
The interconnected events of C3 activation, degradation, and effector function are tightly controlled spatially and temporally, and require over a dozen proteins in addition to C3 itself [1, 10]. Although deciphering these intricate mechanisms at the structural and functional level required decades of experimentation, it also revealed many individual steps that could be attractive targets for inhibitor development in the process. Nevertheless, since high-level complement activity and its downstream effector events are predicated upon the activation of C3, any discussion of inhibitors acting at this level in the pathway must begin with molecules that block either the enzyme (i.e. convertase or its C3b scaffold) or the substrate (i.e. native C3) involved in this all-important reaction. In this regard, structural studies on numerous naturally-occurring and manmade inhibitors have provided clues into the various ways that targeted inhibition at the C3 level can be achieved.
C3 activation can be potently inhibited by molecules that interfere with convertase dynamics (i.e. its formation and/or stability) similarly to endogenous RCA proteins [1, 41, 42]. Two such examples are the Vaccinia virus complement control protein, typically referred to as VCP [59], and the Variola virus small-pox inhibitor of complement enzymes, typically referred to as SPICE in the literature [60]. Although both viral proteins are orthologs of human MCP and DAF, SPICE exhibits more potent decay-accelerating and FI-cofactor activities than does VCP [60, 61]. The structure of SPICE bound to C3b shows that it occupies generic RCA-binding CCPi-iv sites on the C3b surface (Fig. 5A) [43]. Thus, its overall C3b-binding pose most closely resembles that of FH CCP(1-4) (Fig. 3D) [44] and its engineered variant mini-FH (Fig. 3E) [46, 47]. In the specific case of mini-FH, however, the additional interactions with the TED domain contributed by FH CCP(19-20) result in enhanced affinity for C3b and inhibitory potency against the alternative pathway when compared to either FH CCP(1-4) or native FH [47]. Thus, the successful design of mini-FH using structure/function information as the primary guide suggests that a similar approach could be taken to produce novel classes of complement inhibitors.
Figure 5.
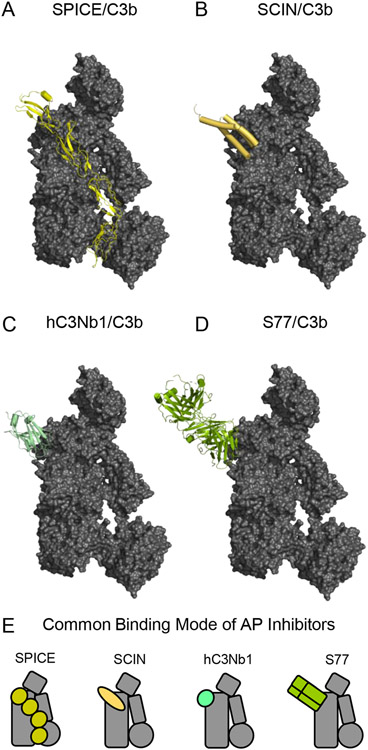
Structural Features of C3b-binding Inhibitors of the Alternative Complement Pathway. (A) Structure of C3b bound to CCP domains 1-4 of the Variola virus complement inhibitor SPICE as drawn from PDB entry 5FOB [43]. C3b is colored as a grey surface, while SPICE is colored yellow. (B) Structure of C3b bound to the S. aureus immune evasion protein SCIN as drawn from PDB entry 3OHX [22]. C3b is colored as a grey surface, while SCIN is colored pale orange. (C) Structure of C3b bound to the nanobody hC3Nb1 as drawn from PDB entry 6EHG [67]. C3b is colored as a grey surface, while hC3Nb1 is colored light green. (D) Structure of C3b bound to the F(ab) fragment of monoclonal antibody S77 as drawn from PDB entry 3G6J [68]. C3b is colored as a grey surface, while S77 is colored olive green. (E) Simplified representations of the structures shown in panels A-D. Note the common binding surface recognized by each of these C3b-binding Alternative Pathway inhibitors.
Other well characterized inhibitors of C3 activation employ mechanisms distinct from RCA proteins. SCINs are a family of small (~10 kDa) immune evasion proteins secreted by Staphylococcus aureus that bind directly to C3b and inhibit multiple processes required for C3 cleavage by the convertase [31, 32, 62-65]. Structural studies of SCIN proteins bound to C3b and C3c show that their primary binding site is formed by the nascent N-terminus of the α’-chain, along with the MG6 and MG7 domains (Fig. 5B) [22, 66]. Although this binding site largely overlaps with the generic RCA-binding CCPi-CCPii sites on C3b [43], SCINs do not exhibit canonical RCA-like activities. Instead, SCINs compete with FB and slightly diminish convertase formation [32]. They likewise compete with FH and block convertase decay [31, 32]. While convertases bound by SCINs are more stable, they are catalytically inactive [31, 32]. In addition to this, SCIN proteins also contain an N-terminal extension that mediates pseudo-dimerization of C3b via a binding site within the MG7 and MG8 domains of a second C3b molecule [22, 33, 66]. Structural interpretations of the resulting SCIN-mediated pseudodimers give rise to a currently accepted model for substrate recognition by the C3 convertase (Fig. 2D) [10, 33]. However, dimerization per se is not required for SCINs to inhibit C3 cleavage, as SCIN deletion mutants lacking the N-terminal extension still exhibit inhibitory activity [66]. Moreover, SCIN-inhibited convertases prepared under limiting C3b conditions also bind well to native C3 even though they cannot cleave it [32, 64]. Collectively, these observations are consistent with the idea that SCINs inhibit the alternative pathway C3 convertase by blocking access of its proteolytic component to the scissile bond of C3 [33].
The primary SCIN binding site overlaps with not only FB and FH [22, 44], but those of other complement regulatory components such as CR1 and DAF [4]. This suggests that other molecules targeting this so-called functional hotspot on C3b may also exhibit inhibitory activities [32, 65]. Indeed, two such inhibitors have been reported in the literature. The camelid-derived, anti-C3 nanobody known as hC3Nb1 binds with subnanomolar affinity to C3, C3b, and iC3b and inhibits the alternative pathway [67]. The structure of hC3Nb1 bound to C3b reveals that this nanobody recognition site is comprised of surface-exposed loops in MG7, and to a far lesser extent the nascent N-terminus of the α’-chain (Fig. 5C) [67]. Consistent with the properties attributed to other molecules that bind MG7, hC3Nb1 blocks assembly of the pro-convertase, degradation of C3b by FI, and even C3 binding to the pre-formed convertases to a degree [67]. Unlike hC3Nb1, the antigen-binding fragment of an antibody known as S77 does not bind to native C3, but binds with low-nanomolar affinity to C3b, iC3b, and C3c [68]. The structure of S77 bound to C3b shows that this inhibitor also recognizes MG7 (Fig. 5D), whereby it displays a similar series of functional properties to hC3Nb1 [68]. Since SPICE, SCIN, hC3Nb1, and S77 all share a similar binding site on C3b and they all inhibit C3 activation via the alternative pathway, it seems that targeting this specific area of C3b represents a generalizable strategy for developing potent C3 inhibitors (Fig. 5E).
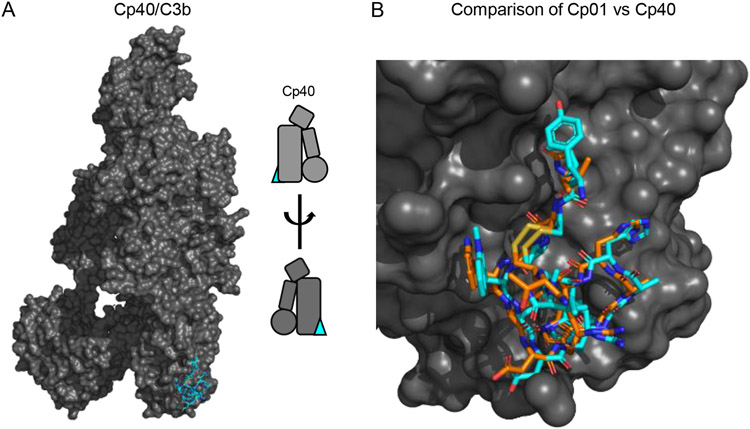
Of all the inhibitors that act at the level of C3, perhaps the best understood is a family of molecules known as Compstatins [10, 69-71]. Compstatins bind to native C3 and its C3c-containing fragments, and inhibit C3 activation via all three complement pathways [10, 69, 70]. The parental compound of Compstatin was identified from a phage display screen for novel ligands of human C3 and consists of 13 residues held together in a cyclic structure [16]. A single chemical modification and two amino acid substitutions produced an analogue known as 4W9A/Cp01 and which has ~10-fold improved affinity (KD=1.2 μM)over the parental compound [72]. Further improvements based on the Compstatin scaffold that incorporate non-natural amino acids and backbone methylation to limit conformational flexibility resulted an analogue with ~5,000-fold enhanced affinity (KD=500 pM) over the initial molecule [73]. A recently reported structure of a prototypic third generation Compstatin known as Cp40 bound to C3b shows the same interacton site as that of 4W9A/Cp01 bound to C3c (Fig. 6A) [74]. However, it also reveals important differences accounting for the striking improvement in its potency (Fig. 6B) [74]. Chief among these are improved hydrophobic contact with the binding site mediated by incorporation of the N-terminally-extended D-Tyrosine, as well as shielding effects on structural solvent molecules [74]. Separately, both of these structures show that the Compstatin binding site lies at the interface of the MG4 and MG5 domains [75]. This site is fully accessible in native C3 and explains why Compstatins are not selective for activated forms of C3. Nevertheless, this binding site appears critical for interaction of the C3 substrate with the C3 convertase (Fig. 2D) [33]. Indeed, this likely explains functional studies which show that Compstatin inhibits C3 activation by binding to native C3 and blocking its interaction with the C3 convertase, as opposed to inhibiting the C3 convertase assembly itself [74].
Figure 6.
Structural and Functional Features of Compstatin Class Complement Inhibitors. (A) Structure of C3 bound to the third-generation Compstatin Cp40 as drawn from the PDB entry 7BAG [74]. C3b is colored as a grey surface, while Cp40 is shown in stick convention and colored blue. A simplified representation of the structure is inset at the right of the panel. (B) Comparison of bound structures for the second-generation Compstatin Cp01 and Cp40 as drawn from the PDB entries 2QKI [75] and 7BAG [74], respectively. Both inhibitors are drawn in stick convention with the carbon atoms of Cp01 in orange and those of Cp40 in blue. Note that this image was constructed by superimposing the structure of Cp01 bound to C3c upon that of Cp40 bound to C3b using the keyring core of C3 as the basis for calculations.
7. Exploring New Targets: a Case for Conformation-Specific Ligands as Inhibitors?
Although there are currently greater than 50 PDB depositions describing crystallographic structure determinations of human C3, its fragments, and various complexes (Table 1), many relevant aspects of C3 function cannot be discerned through crystallography alone. Indeed, most questions related to protein dynamics, conformational flexibility, and changes therein in response to biochemical transformations or ligand binding can only be meaningfully addressed through solution based methods. Obtaining solution structural information on molecules the size of C3 and its fragments is more feasible now since improvements in instrumentation, experimental workflow, and data analysis platforms have led to a rapid maturation of Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) and Small Angle X-ray Scattering (SAXS) approaches. However, it is deployment of these solution methods in combination with crystallography that has proven most useful for revealing unexplored structure/function features of C3 and its fragments. One such example identified discrete conformational substates in C3b that have divergent ligand-binding and functional properties [76]. Since selectively targeting one conformation over another may provide opportunities for future development of complement inhibitors, the studies related to this discovery will be described in the paragraphs that follow.
The immune evasion protein Efb is secreted from Staphylococcus aureus and consists of two functional domains [77, 78]. Whereas its N-terminal domain is intrinsically disordered and binds to fibrinogen, the C-terminal domain (hereafter Efb-C) adopts a helical bundle fold, binds to C3, and is itself a potent inhibitor of the alternative pathway [18, 78-80]. Efb-C recognizes a site within the TED domain that is freely accessible in the structure of native C3 [12, 18], and forms high-affinity interactions with C3, C3b, iC3b, and C3d [18, 50]. More recent surface plasmon resonance measurements of Efb-C binding to site-specifically immobilized C3b estimate the equilibrium dissociation constant in the low picomolar range (KD=110 pM) (Fig. 7A); this extremely tight interaction is characterized by a very slow dissociation rate constant of 1.87x10−4 s−1 and an associated half-life of approximately 1 hr. Despite this high-affinity interaction, the Efb-C binding site observed in its co-crystal structure with C3d is sterically occluded by the proximity of the MG1 and TED domains in the structure of C3b alone [15, 18]. A potential explanation for this discrepancy is that the CUB-TED region of C3b rotates away from its keyring core, thus relieving the potential of steric clash with Efb-C by conformational change. This possibility is consistent with previous electron microscopy studies noting distinct conformational subsets in C3b [48], as well as with the observation that Efb-C binding leads to enhanced exposure of the epitope for the anti-C3b monoclonal antibody, C3-9 [18, 81].
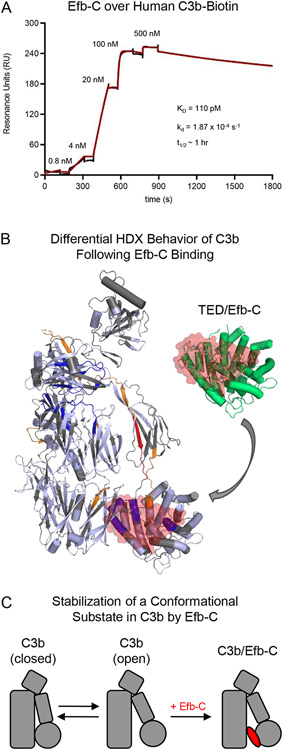
Figure 7.
Selective Stabilization of an Open Conformational State in C3b by the S. aureus Immune Evasion Protein Efb-C. (A) Characterization of Efb-C binding to site-specifically immobilized C3b as assessed by surface plasmon resonance. Human C3b that had been site-specifically biotinylated at its thioester cysteine [32, 66] was captured on a streptavidin-coated biosensor surface prior to injecting increasing concentrations of Efb-C in a single-cycle kinetic study. The reference-corrected sensorgram is shown with a black line while the outcome of fitting to a Langmuir model is shown with a red line. Efb-C binding to C3b exhibits low-picomolar affinity (KD ~110 pM) and is kinetically stable as indicated by its dissociation rate constant (10−4 s−1) (inset). (B) Differential Hydrogen/Deuterium exchange mapping of C3b as a function of Efb-C. The Hydrogen/Deuterium Exchange (HDX) profile was characterized by Mass Spectrometry for samples of C3b in the presence of either Efb-C or a non-binding mutant [76]. Peptides from C3b that exhibited increased HDX are colored either orange (moderate increase) or red (highest increase), while those that exhibited decreased HDX are colored dark blue. Peptides from C3b displayed no change are colored in pale blue, while peptides that were absent from the coverage map are colored grey. A full-scale representation of the structure of Efb-C bound to the TED domain from C3b [18] is inset at the top right of the panel, with Efb-C colored as a transparent red surface and TED colored green. Superposition of this structure onto the HDX map of C3b reveals that residues comprising the Efb-C binding site within the TED domain are highly exchange-protected, thereby demonstrating consistency between the crystallographic and in solution studies. (C) Simplified representation showing stabilization of an open conformational state in C3b upon Efb-C binding. As the interaction between Efb-C and C3b is ultra-high affinity and kinetically stable, Efb-C binding is depicted as an essentially irreversible step in this mechanism.
Differential HDX-MS studies provide better understanding of the changes in C3b arising following Efb-C binding (Fig. 7B) [18, 76]. In agreement with the Efb-C/C3d co-crystal structure, the exchange profiles for TED domain peptides corresponding to the Efb-C binding site show significant protection in the presence of Efb-C [76]. Oppositely, peptides representing the linker that connects the CUB and TED domains and within the CUB domain itself show significant enhancements in their exchange profiles [76]. Intriguingly, this property extends to two peptides within the MG8 domain that are far removed from the Efb-C binding site [15, 76]. This suggests that Efb-C binding results in increased exposure of a region of C3b extending from the TED domain up through MG8. SAXS analyses performed on samples of either C3b alone or when bound to Efb-C illustrate how this may occur at the structural level [76]. Whereas C3b on its own appears to be distributed across two conformers distinguished by a rotation of the CUB-TED region (hereafter called “closed” and “open”), the Efb-C bound form is restricted to the open conformer whose CUB-TED region is displaced from the keyring core. Since Efb-C does not readily dissociate from C3b, these results indicate that Efb-C binding preferentially stabilizes an open conformer of C3b that normally exists within a population of C3b molecules (Fig. 7C).
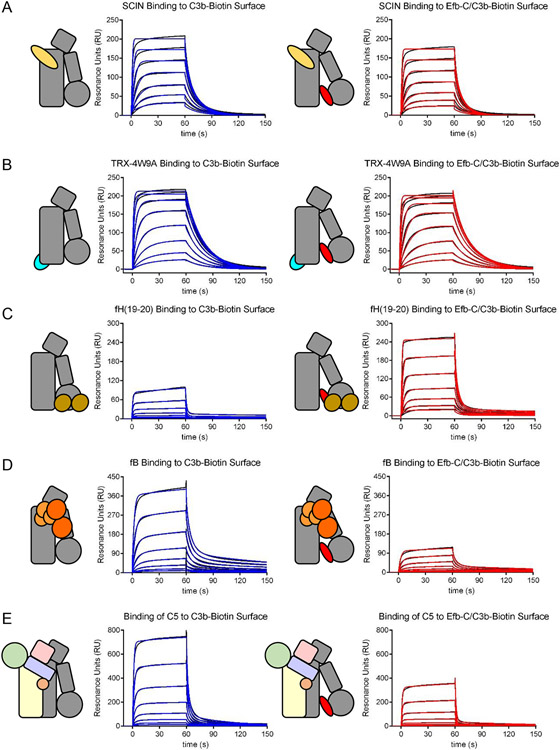
Many peptides that display significant exchange differences in the Efb-C/C3b complex versus C3b alone map to known ligand binding sites [76]. This suggests that the open conformer of C3b might have altered ligand binding properties when compared to its closed counterpart. Although no experimental tools exist at present to selectively stabilize the closed conformation of C3b, the tightly binding Efb-C protein that does not readily dissociate provides a means to assess the functional attributes of the open C3b conformer alongside a normally distributed population of C3b molecules. Thus, surface plasmon resonance can be used to investigate binding of several ligands to C3b alone or C3b that had been presaturated with Efb-C [76]. Neither the S. aureus immune evasion protein SCIN (Fig. 8A), nor a fusion protein displaying the first-generation Compstatin sequence 4W9A show any changes in binding as a function of Efb-C (Fig. 8B) [82]. However, CCP domains 19-20 from FH show a marked increase in binding in the presence of Efb-C (Fig. 8C); since this region of FH recognizes a site within the TED domain of C3b, this observation suggests that the FH(19-20) binding site is more accessible in the open C3b conformer. By contrast, binding of FB is reduced by over 75% in the presence of Efb-C (Fig. 8D) [76]. This demonstrates that Efb-C inhibits the alternative pathway by blocking formation of the pro-convertase and therefore formation of the active convertase responsible for complement amplification [76].
Figure 8.
Certain Ligands Exhibit Conformational Dependence in Their Affinity for C3b. The binding of various ligands to either C3b alone (left panels) or C3b that had been presaturated with Efb-C and therefore trapped in the open state (right panels) was assessed by surface plasmon resonance. Reference-corrected sensorgram series are shown with black lines, while the outcome of fitting to kinetic models is shown with either blue (C3b alone) or red (Efb-C/C3b) lines. (A) Comparison of binding of the S. aureus immune evasion protein SCIN. (B). Comparison of binding of a fusion protein consisting of E. coli thioredoxin and a minimal Compstatin sequence [82]. (C) Comparison of binding of CCP domains 19-20 of human FH. (D) Comparison of binding of human FB. (E) Comparison of binding of human C5. Whereas neither SCIN nor TRX-4W9A displayed sensitivity to the presence of Efb-C, FH(19-20) exhibited increased affinity for C3b in the presence of Efb-C, presumably due to increased exposure of its binding site within the TED domain. By contrast, the affinity of both FB and C5 were significantly diminished in the presence of Efb-C. This suggests that molecules which trap C3b in an open conformational state may be potent inhibitors of those aspects of complement function dependent upon FB/C3b and C5/C3b interactions.
Interestingly, a greater than 50% reduction in the interaction between C5 and C3b is also seen in the presence of Efb-C (Fig. 8E) [82]. Interpreting this observation is not as straightforward, since the structure of neither the C5/C3b complex nor the C5 convertase has been reported. However, information on the nature of this interaction can be derived from the structure of the C3b analogue, Cobra Venom Factor, bound to C5 [83]. The C5/CVF structure bares many similarities to the model of the C3 convertase bound to C3 (Fig. 2D), including binding of the C3/C5 substrate on the side of the scaffolding molecule (i.e. C3b or CVF) opposite its CUB-TED region [33, 83]. This side of C3b also harbors binding sites for CRIg and Compstatin, both of which inhibit substrate binding to C3b-containing convertases [23, 74, 75]. Curiously, there are minimal differences in the exchange profiles on this side of C3b in response to Efb-C binding (Fig. 7B) [76]. Although the peptide coverage map is not complete in this area and limits the information available, a single peptide within the MG3 domain does exhibit significant exchange enhancement and is oriented toward the putative C3/C5 substrate-binding interface [76]. While it is nearly certain that C3/C5 binding involves multiple points of contact with the C3b scaffold of the convertase [83], it is tempting to speculate that this peptide from MG3 and the region surrounding it might play an important role. Further investigation is clearly needed to fully understand the physical basis for C3/C5 substrate binding to the C3b-containing convertases. Nevertheless, these data suggest that substrate binding to the convertase is influenced by the conformational state of C3b. When considered together with the fact that the open conformer of C3b is inefficient at supporting C3 convertase assembly [65, 76], selectively targeting of conformational substates in C3b seems to be an attractive option for future inhibitor discovery.
8. Concluding Remarks
The last decades have witnessed an increased appreciation for the role of complement as a pathophysiological driving force across the human inflammatory disease landscape [3-5]. Yet throughout most of this time, the only complement specific therapeutic approved by regulatory agencies for clinical use has been the anti-C5 monoclonal antibody eculizumab (Soliris®). While debate continues as to which point of inhibition is best for a given disease or indication, there is ample evidence to suggest that therapeutic inhibition of C3 may not only be desirable but preferable to C5 blockade in various situations [8, 9, 84, 85]. Although the developmental timeline of the first C3-targeted inhibitors spanned 25 years from the initial discovery till the recent approval, the availability of these first-in-class drugs marks a new era in complement therapeutics. Indeed, with proof of efficacy established and approvals thus obtained, the future of C3 inhibitors may be as promising now as it has ever been. Despite this prevailing sense of optimism, there must also be understanding that no single type of inhibitor will be best for every indication where C3 inhibition appears warranted. Paradoxically, the very complexity of the C3 molecule from its biogenesis, to its activation, to its degradation and effector functions provides numerous opportunities for intervention (Fig. 1). With the encompassing collection of structural information on C3, its various fragments, and their complexes described here (Table 1), the field seems well-positioned to leverage these insights not only for basic research purposes, but for investigating and developing new therapeutic concepts as well.
As always, charting the course forward will require careful considerations of the benefits and limitations of various approaches. Recurring structural themes, such as multiple types of inhibitors binding to the MG7 domain of C3b (Fig. 5), and the shared C3b-binding modes of naturally-occurring regulatory molecules (Fig. 3) [43], should definitely be taken into account. Future work should likewise consider information that can help identify those strategies likely to be well-tolerated from those that will not. In this regard, harnessing the endogenous regulators already found within the body seems prudent. Capitalizing on unexpected activities within these regulators, such as CRIg inhibiting the alternative pathway (Fig. 4) [23], or the molecular engineering of gain-of-function variants, such as mini-FH (Fig. 3) [47], are merely two such examples. Finally, the biological proof of concept provided by immune evasion proteins that have arisen through millennia of host-pathogen co-evolution is something that should not be dismissed. That fact that SCIN, Efb, and related proteins from S. aureus all target C3b-containing convertases is certainly not coincidental [65]. Any molecules that employ similar mechanisms and mimic their activities ought to be powerful complement inhibitors in their own right. Future work in this area seems particularly warranted, given the finding that Efb-C selects for a conformation of C3b that is deficient in both convertase formation and substrate recognition (Figs. 7 & 8). Although there are certain to be challenges ahead and there is no guarantee than any single approach will lead to success, the many opportunities highlighted above suggest that the next generation of C3-targeted therapeutics should not take nearly as long to develop the first.
Funding
This research was supported by a grant R35GM140852 from the U.S. National Institutes of Health to B.V.G., by the R. Weaver and S. Weaver endowment to J.D.L, and by the NWO Spinoza prize from the Netherlands Organisation for Scientific Research (NWO) and funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 787241) to P.G.
REFERENCES
- [1].Ricklin D, Hajishengallis G, Yang K, Lambris JD, Complement: a Key System for Immune Surveillance and Homeostasis, Nat. Immunol 11 (2010) 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hajishengallis G, Reis ES, Mastellos DC, Ricklin D, Lambris JD, Novel Mechanisms and Functions of Complement, Nat. Immunol 18 (2017) 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ricklin D, Lambris JD, Compement in Immune and Inflammatory Disorders: Pathophysiological Mechanisms, J. Immunol 190 (2013) 3831–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ricklin D, Reis ES, Lambris JD, Complement in Disease: a Defence System Turning Offensive, Nat. Rev. Nephrol 2016(12) (2016) 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reis ES, Mastellos DC, Ricklin D, Mantovani A, Lambris JD, Complement in Cancer: Untangling an Intricate Relationship, Nat. Rev. Immunol 18 (2018) 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Muller-Eberhard HJ, Nilsson U, Aronsson T, Isolation and Characterization of Two beta1-Glycoproteins of Human Serum, J. Exp. Med 111 (1960) 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Muller-Eberhard HJ, Nilsson U, Relation of a beta(1)-Glycoprotein of Human Serum to the Complement System, J. Exp. Med 111 (1960) 217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ricklin D, Lambris JD, Complement in Immune and Inflammatory Disorders: Therapeutic Interventions, J. Immunol 190 (2013) 3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mastellos DC, Ricklin D, Lambris JD, Clinical Promise of Next-Generation Complement Therapeutics, Nat. Rev. Drug Disc 18 (2019) 707–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ricklin D, Reis ES, Mastellos DC, Gros P, Lambris JD, Complement Component C3 - The "Swiss Army Knife" of Innate Immunity and Host Defense, Immunol. Rev 274 (2016) 33–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thomas ML, Janatova J, Gray WR, Tack BF, Third Component of Human Complement: Localization of the Internal Thiolester Bond, Proc. Natl. Acad. Sci. USA 79 (1982) 1054–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Ekdahl-Nilsson K, Nilsson B, Gros P, Structures of Complement Component C3 Provide Insights Into the Function and Evolution of Immunity, Nature 437 (2005) 505–511. [DOI] [PubMed] [Google Scholar]
- [13].Law SKA, Dodds AW, The Internal Thioester and the Covalent Binding Properties of the Complement Proteins C3 and C4, Protein Sci. 6 (1997) 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gadjeva M, Dodds AW, Taniguchi-Sidle A, Willis AC, Isenman DE, Law SK, The Covalent Binding Reaction of Complement Component C3, J. Immunol 161 (1998) 985–990. [PubMed] [Google Scholar]
- [15].Janssen BJC, Christodoulidou A, McCarthy A, Lambris JD, Gros P, Structure of C3b Reveals Conformational Changes Underlying Complement Activity, Nature 444 (2006) 213–216. [DOI] [PubMed] [Google Scholar]
- [16].Sahu A, Kay BK, Lambris JD, Inhibition of Human Complement by a C3-Binding Peptide Isolated from a Phage-Displayed Random Peptide Library, J. Immunol 157 (1996) 884–891. [PubMed] [Google Scholar]
- [17].Alsenz J, Becherer JD, Nilsson B, and Lambris JD, Structural and Functional Analysis of C3 Using Monoclonal Antibodies., Curr. Top. Microbiol. Immunol 153 (1989) 235–248. [DOI] [PubMed] [Google Scholar]
- [18].Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV, A Structural Basis for Complement Inhibition by Staphylococcus aureus, Nat. Immunol 8 (2007) 430–437. [DOI] [PubMed] [Google Scholar]
- [19].Hammel M, Sfyroera G, Pyrpassopoulos S, Ricklin D, Ramyar KX, Pop M, Jin Z, Lambris JD, Geisbrecht BV, Characterization of Ehp: a Secreted Complement Inhibitory Protein from Staphylococcus aureus, J. Biol. Chem 202 (2007) 30051–30061. [DOI] [PubMed] [Google Scholar]
- [20].Burman JD, Leung E, Atkins KL, O'Seaghdha MN, Lango L, Bernado P, Bagby S, Svergun DI, Foster TJ, Isenman DE, van den Elsen JM, Interaction of Human Complement with Sbi, a Staphylococcal Immunoglobulin-binding Protein: Indications of a Novel Mechanism of Complement Evasion by Staphylococcus aureus, J. Biol. Chem 283 (2008) 17579–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Clark EA, Crennell S, Upadhyay A, Zozulya AV, Mackay JD, Svergun DI, Bagby S, van den Elsen JM, A Structural Basis for Staphylococcal Complement Subversion: X-ray Structure of the Complement-binding Domain of Staphylococcus aureus Protein Sbi in Complex with Ligand C3d, Mol. Immunol 48 (2011) 452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Garcia BL, Ramyar KX, Tzekou A, Ricklin D, McWhorter WJ, Lambris JD, Geisbrecht BV, Molecular Basis for Complement Recognition and Inhibition Determined by Crystallographic Studies of the Staphylococcal Complement Inhibitor (SCIN) Bound to C3c and C3b J. Mol. Biol 402 (2010) 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren Campagne M, Structure of C3b in complex with CRIg gives insights into regulation of complement activation., Nature 444 (2006) 217–220. [DOI] [PubMed] [Google Scholar]
- [24].Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P, Structures of C3b in Complex with Factors B and D Give Insight into Complement Convertase Formation, Science 330 (2010) 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fishelson Z, Muller-Eberhard HJ, C3 Convertase of Human Complement: Enhanced Formation and Stability of the Enzyme Generated with Nickel Instead of Magnesium, J. Immunol 129 (1982) 2603–2607. [PubMed] [Google Scholar]
- [26].Narayana SV, Carson M, el-Kabbani O, Kilpatrick JM, Moore D, Chen X, Bugg CE, Volanakis JE, DeLucase LJ, Structure of Human Factor D. A Complement System Protein at 2.0 A Resolution, J. Mol. Biol 235 (1994) 695–708. [DOI] [PubMed] [Google Scholar]
- [27].Volanakis JE, Narayana SV, Complement Factor D, a Novel Serine Protease, Protein Sci. 5 (1996) 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim S, Narayana SV, Volanakis JE, Crystal Structure of a Complement Factor D Mutant Expressing Enhanced Catalytic Activity, J. Biol. Chem 270 (1995) 24399–24405. [DOI] [PubMed] [Google Scholar]
- [29].Milder FJ, Gomes L, Schouten A, Janssen BJ, Huizinga EG, Romijn RA, Hemrika W, Roos A, Daha MR, Gros P, Factor B Structure Provides Insights into Activation of the Central Protease of the Complement System, Nat. Struct. Mol. Biol 14 (2007) 224–228. [DOI] [PubMed] [Google Scholar]
- [30].Pangburn MK, Muller-Eberhard HJ, The C3 Convertase of the Alternative Pathway of Human Complement. Enzymic Properties of the Biomolecular Proteinase., Biochem. J 235 (1986) 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rooijakkers SH, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJ, van Kessel KP, van Strijp JA, Immune Evasion by a Staphylococcal Complement Inhibitor that Acts on C3 Convertases, Nat. Immunol 6 (2005) 920–927. [DOI] [PubMed] [Google Scholar]
- [32].Ricklin D, Tzekou A, Garcia BL, Hammel M, McWhorter WJ, Sfyroera G, Wu Y-Q, Holers VM, Herbert AP, Barlow PN, Geisbrecht BV, Lambris JD, A Molecular Insight into Complement Evasion by the Staphylococcal Complement Inhibitor Protein Family., J. Immunol 183 (2009) 2565–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P, Structural and Functional Implications of the Alternative Complement Pathway C3 Convertase Stabilized by a Staphylococcal Inhibitor., Nat. Immunol 10 (2009) 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pedersen DV, Roumenina L, Jensen RK, Gadeberg TAF, Marinozzi C, Picard C, Rybkine T, Thiel S, Sorensen UBS, Stover D, Fremeaux-Bacchi V, Andersen GR, Functional and Structural Insight into Properdin Control of Complement Alternative Pathway Amplification, EMBO J. 36 (2017) 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pedersen DV, Gadeberg TAF, THomas C, Wang Y, Joram N, Jensen RK, Mazarakis SMM, Revel M, El Sissy C, Petersen SV, Lindorff-Larsen K, Thiel S, Laursen NS, Fremeaux-Bacchi V, Andersen GR, Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System, Front. Immunol 10 (2019) 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].van den Bos RM, Pearce NM, Granneman J, Brondijk THC, Gros P, Insights into Enhanced Complement Activation by Structures of Properdin and Its Complex with the C-terminal Domain of C3b, Front. Immunol 10 (2019) 2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pedersen DV, Pedersen MN, Mazarakis SMM, Wang Y, Lindorff-Larsen K, Arleth L, Andersen GR, Properdin Oligomers Adopt Rigid Extended Conformations Supporting Function, Elife 10 (2021) e63356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Harboe M, Ulvund G, Vien L, Fung M, and Mollnes TE, The Quantitative Role of Alternative Pathway Amplification in Classical Pathway Induced Terminal Complement Activation, Clin. Exp. Immunol 138 (2004) 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fromell K, Adler A, Aman A, Manivel VA, Huang S, Duhrkop C, Sandholm K, Ekdahl KN, Nilsson B, Assessment of the Role of C3(H2O) in the Alternative Pathway, Front. Immunol 11 (2020) 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Muller-Eberhard HJ, Schreiber RD, Molecular Biology and Chemistry of the Alternative Pathway of Complement, Adv. Immunol 29 (1980) 1–53. [DOI] [PubMed] [Google Scholar]
- [41].Kirkitadze MD, Barlow PN, Structure and Flexibility of the Multiple Domain Proteins that Regulate Complement Activation, Immunol. Rev 180 (2001) 146–161. [DOI] [PubMed] [Google Scholar]
- [42].Zipfel PF, Skerka C, Complement Regulators and Inhibitory Proteins, Nat. Rev. Immunol 9 (2009) 729–740. [DOI] [PubMed] [Google Scholar]
- [43].Forneris F, Wu J, Xue X, Ricklin D, Lin Z, Sfyroera G, Tzekou A, Volokhina E, Granneman JC, Hauhart R, Bertram P, Liszewski MK, Atkinson JP, Lambris JD, Gros P, Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3b-binding Mode, EMBO J. (2016) pii: e201593673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wu J, Wu YQ, Ricklin D, Janssen BJ, Lambris JD, Gros P, Structure of Complement Fragment C3b-factor H and Implications for Host Protection by Complement Regulators., Nat. Immunol 10 (2009) 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Harris CL, Pettigrew DM, Lea SM, Morgan BP, Decay-Accelerating Factor Must Bind Both Components of the Complement Alternative Pathway C3 Convertase to Mediate Efficient Decay J. Immunol 178 (2007) 352–359. [DOI] [PubMed] [Google Scholar]
- [46].Xue X, Wu J, Ricklin D, Forneris F, Di Crescenzio P, Schmidt CQ, Granneman J, Sharp TH, Lambris JD, Gros P, Regulator-Dependent Mechanisms of C3b Processing by Factor I Allow Differentiation of Immune Responses, Nat. Struct. Mol. Biol 24 (2017) 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schmidt CQ, Bai H, Lin Z, Risitano AM, Barlow PN, Ricklin D, Lambris JD, Rational engineering of a minimized immune inhibitor with unique triple-targeting properties, J. Immunol 190 (2013) 5712–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nishida N, Walz T, Springer TA, Structural Transitions of Complement Component C3 and its Activation Products, Proc. Nat. Acad. Sci. U.S.A 103 (2006) 19737–19742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen H, Schuster MC, Sfyroera G, Geisbrecht BV, Lambris JD, Solution Insights into the Structure of the Efb/C3 Complement Inhibitory Complex as Revealed by Lysine Acetylation nand Mass Spectrometry, J. Am. Soc. Mass. Spectrom 19 (2008) 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Papanastasiou M, Koutsogiannaki S, Sarigiannis Y, Geisbrecht BV, Ricklin D, Lambris JD, Structural Implications for the Formation and Function of the Complement Effector Protein iC3b, J. Immunol 198 (2017) 3326–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Helmy KY, Katschke KJ, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, Scales SJ, Ghilardi N, van Lookeren Campagne M, CRIg: a Macrophage Complement Receptor Required for Phagocytosis of Circulating Pathogens, Cell 124 (2006) 915–927. [DOI] [PubMed] [Google Scholar]
- [52].Krych-Goldberg M, Atkinson JP, Structure-Function Relationships of Complement Receptor Type 1, Immunol. Rev 180 (2001) 112–122. [DOI] [PubMed] [Google Scholar]
- [53].Medof ME, Prince GM, Mold C, Release of Soluble Immune Complexes from Immune Adherence Receptors on Human Erythrocytes is Mediated by C3b Inactivator Independently of Beta 1H and is Accompanied by Generation of C3c, Proc. Natl. Acad. Sci. USA 79 (1982) 5047–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kalli KR, Ahearn JM, Fearon DT, Interaction of iC3b with Recombinant Isotypic and Chimeric Forms of CR2, J. Immunol 147 (1991) 590–594. [PubMed] [Google Scholar]
- [55].van den Elsen JMH, Isenman DE, A Crystal Structure of the Complex Between Human Complement Receptor 2 and Its Ligand C3d, Science 332 (2011) 608–611. [DOI] [PubMed] [Google Scholar]
- [56].Ross GD, Regulation of the Adhesion Versus Cytotoxic Functions of the Mac-1/CR3/alphaMbeta2-Integrin Glycoprotein, Crit. Rev. Immunol 20 (2000) 197–222. [PubMed] [Google Scholar]
- [57].Bajic G, Yatime L, Sim RB, Vorup-Jensen T, Andersen GR, Structural Insight on the Recognition of Surface-bound Opsonins by the Integrin I Domain of Complement Receptor 3, Proc. Natl. Acad. Sci. USA 110 (2013) 16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Fernandez FJ, Santos-Lopez J, Martinez-Barricarte R, Querol-Garcia J, Martin-Merinero H, Navas-Yuste S, Savko M, Shepard WE, Rodriguez de Cordoba S, Vega MC, The Crystal Structure of iC3b-CR3 aI Reveals a Modular Recognition of the Main Opsonin iC3b by the CR3 Integrin Receptor, Nat. Commun 13 (2022) 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kotwal GJ, Moss B, Vaccinia Virus Encodes a Secretory Polypeptide Structurally Related to Complement Control Proteins, Nature 335 (1988) 176–178. [DOI] [PubMed] [Google Scholar]
- [60].Rosengard AM, Liu Y, Nie Z, Jimenez R, Variola Virus Immune Evasion Design: Expression of a Highly Efficient Inhibitor of Human Complement, Proc. Natl. Acad. Sci. USA 99 (2002) 8808–8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sfyroera G, Katragadda M, Morikis D, Isaacs SN, Lambris JD, Electrostatis Modeling Predicts the Activities of Orthopoxvirus Complement Control Proteins, J. Immunol 174 (2005) 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jongerius I, Köhl J, Pandey MK, Ruyken M, van Kessel KP, van Strijp JA, Rooijakkers SH, Staphylococcal Complement Evasion by Various Convertase-blocking Molecules J. Exp. Med 204 (2007) 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jongerius I, Puister M, Wu J, Ruyken M, van Strijp JA, Rooijakkers SH, Staphylococcal Complement Inhibitor Modulates Phagocyte Responses by Dimerization of Convertases., J. Immunol 184 (2010) 420–425. [DOI] [PubMed] [Google Scholar]
- [64].Garcia BL, Summers BJ, Ramyar KX, Tzekou A, Lin Z, Ricklin D, Lambris JD, Laity JH, Geisbrecht BV, A Structurally Dynamic N-terminal Helix is a Key Functional Determinant in Staphylococcal Complement Inhibitor (SCIN) Proteins, J. Biol. Chem 288 (2013) 2870–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Garcia BL, Ramyar KX, Ricklin D, Lambris JD, Geisbrecht BV, Advances in Understanding the Structure, Function, and Mechanism of the SCIN and Efb Families of Staphylococcal Immune Evasion Proteins, Adv. Exp. Med. Biol 946 (2012) 113–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Garcia BL, Summers BJ, Zhuoer L, Ramyar KX, Ricklin D, Fu Z-Q, Lambris JD, Geisbrecht BV, Diversity in the C3b Contact Residues and Tertiary Structures of the Staphylococcal Complement Inhibitor (SCIN) Protein Family, J. Biol. Chem 287 (2012) 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jensen RK, Pihl R, Gadeberg TAF, Jensen JK, Andersen KR, Thiel S, Laursen NS, Andersen GR, A Potent Complement Factor C3-specific Nanobody Inhibiting Multiple Functions in the Alternative Pathway of Human and Murine Complement, J. Biol. Chem 293 (2018) 6269–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Katschke KJ Jr, Stawicki S, Yin J, Steffek M, Xi H, Sturgeon L, Hass PE, Loyet KM, Deforge KM, Wu Y, van Lookeren Campagne M, Wiesmann C, Structural and Functional Analysis of a C3b-specific Antibody that Selectively Inhibits the Alternative Pathway of Complement, J. Biol. Chem 284 (2009) 10473–10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ricklin D, Lambris JD, Compstatin: a Complement Inhibitor on its Way to Clinical Application, Adv. Exp. Med. Biol 632(273–292) (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mastellos DC, Yancopoulou D, Kokkinos P, Huber-Lang M, Hajishengallis G, Biglarnia AR, Lupu F, Nilsson B, Risitano AM, Ricklin D, Lambris JD, Compstatin: a C3-Targeted Complement Inhibitor Reaching its Prime for Bedside Intervention, Eur. J. Clin. Invest 45 (2015) 423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mastellos DC, Ricklin D, Sfyroera G, Sahu A, From Discovery to Approval: a Brief History of the Compstatin Family of Complement C3 Inhibitors, Clin. Immunol 235 (2022) 108785. [DOI] [PubMed] [Google Scholar]
- [72].Mallik B, Katragadda M, Spruce LA, Carafides C, Tsokos CG, Morikis D, Lambris JD, Design and NMR Characterization of Active Analogues of Compstatin Containing Non-Natural Amino Acids, J. Med. Chem 48 (2005) 274–286. [DOI] [PubMed] [Google Scholar]
- [73].Qu H, Ricklin D, Bai H, Chen H, Reis ES, Maciejewski M, Tzekou A, DeAngelis RA, Resuello RR, Lupu F, Barlow PN, Lambris JD, New Analogs of the Clinical Complement Inhibitor Compstatin with Subnanomolar Affinity and Enhanced Pharmacokinetic Properties, Immunobiology 218 (2013) 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lamers C, Xue X, Smiesko M, van Son H, Wagner B, Sfyroera G, Gros P, Lambris JD, Ricklin D, Novel Insights into the Picomolar Complement Inhibitor Compstatin (Cp40): Structural Determinants of Key Interactions with C3b and Elucidation of Molecule Mode-of-Action, Nat. Commun (2022) IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Janssen BJ, Halff EF, Lambris JD, Gros P, Structure of Compstatin in Complex with Complement Component C3c Reveals a New Mechanism of Complement Inhibition., J. Biol. Chem 282 (2007) 29241–29247. [DOI] [PubMed] [Google Scholar]
- [76].Chen H, Ricklin D, Hammel M, Garcia BL, McWhorter WJ, Sfyroera G, Wu YQ, Tzekou A, Li S, Geisbrecht BV, Woods VL Jr., Lambris JD, Allosteric Inhibition of Complement Function by a Staphylococcal Immune Evasion Protein, Proc. Natl. Acad. Sci. U.S.A 107 (2010) 17621–17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Boden MK, Flock J-I, Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman, Microb. Pathog 12 (1992) 289–298. [DOI] [PubMed] [Google Scholar]
- [78].Ko YP, Liang X, Smith CW, Degen JL, Hook M, Binding of Efb from Staphylococcus aureus to fibrinogen blocks neutrophil adherence, J. Biol. Chem 286 (2011) 9865–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lee LYL, Liang X, Hook M, Brown EL, Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb), J Biol Chem 279 (2004) 50710–50716. [DOI] [PubMed] [Google Scholar]
- [80].Lee LYL, Hook M, Haviland D, Wetsel RA, Yonter EO, Syribeys P, Vernachio J, Brown EL, Inhibition of Complement Activation by a Secreted Staphylococcus aureus Protein, J. Infect. Dis 190 (2004) 571–579. [DOI] [PubMed] [Google Scholar]
- [81].Hack CE, Paardekooper J, Smeenk RJT, Abbink J, Eerenber AJM, Nuijens JH, Disruption of the Internal Thioester Bond in the Third Component of Complement, (C3) which Results in the Exposure of Neodeterminants Also Present on Activation Products of C3. An Analysis with Monoclonal Antibodies, J. Immunol 141 (1988) 1602–1609. [PubMed] [Google Scholar]
- [82].Garcia BL, Skaff DA, Chatterjee A, Hanning A, Walker JK, Wyckoff GJ, Geisbrecht BV, Identification of C3b-Binding Small-Molecule Complement Inhibitors Using Cheminformatics, J. Immunol 198 (2017) 3705–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Laursen NS, Andersen KR, Braren I, Spillner E, Sottrup-Jensen L, Andersen GR, Substrate Recognition by Complement Convertases Revealed in the C5-Cobra Venom Factor Complex, EMBO J. 30 (2011) 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ricklin D, Lambris JD, Therapeutic Control of Complement Activation at the Level of the Central Component C3, Immunobiology 221 (2016) 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mastellos DC, Reis ES, Yancopoulou D, Hajishengallis G, Ricklin D, Lambris JD, From Orphan Drugs to Adopted Therapies: Advancing C3-Targeted Intervention to the Clinical Stage, Immunobiology 221 (2016) 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bajic G, Yatime L, Klos A, Andersen GR, Human C3a and C3a desArg Anaphylatoxins Have Conserved Structure, in Contrast to C5a and C5a desArg, Protein Sci. 22 (2013) 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jensen RK, Bajic G, Sen M, Springer TA, Vorup-Jensen T, Andersen GR, Complement Receptor 3 Forms a Compact High-Affinity Complex with iC3b, J. Immunol 206 (2021) 3032–3042. [DOI] [PubMed] [Google Scholar]
- [88].Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM, X-ray Crystal Structure of C3d: A C3 Fragment and Ligand for Complement Receptor 2, Science 280 (1998) 1277–1281. [DOI] [PubMed] [Google Scholar]
- [89].Kajander T, Lehtinen MJ, Hyvarinen S, Bhattacharjee A, Leung E, Isenman DE, Meri S, Goldman A, Jokiranta TS, Dual Interaction of Factor H with C3d and Glycosaminoglycans in Host-Nonhost Discrimination by Complement, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 2897–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Morgan HP, Schmidt CQ, Guariento M, Gillespie D, Herbert AP, Mertens H, Blaum BS, Svergun D, Johansson CM, Uhrin D, Barlow PN, Hannan JP, Structural Basis for Engagement by Complement Factor H of C3b on a Self Surface, Nat. Struct. Mol. Biol 18 (2011) 463–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kolodziejczyk R, Mikula KM, Kotila T, Postis VLG, Jokiranta TS, Goldman A, Meri T, Crystal Structure of a Tripartite Complex Between C3dg, C-terminal Domains of Factor H and OspE of Borrelia burgdorferi, PLOS ONE 12 (2017) e0188127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rooijakkers SHM, Milder FJ, Bardoel BW, Ruyken M, van Strijp JAG, Gros P, Staphylococcal Complement Inhibitor: Structure and Active Sites, J. Immunol 179 (2007) 2989–2998. [DOI] [PubMed] [Google Scholar]