Extended Data Fig. 7 |. Enrichment of eQTLs in disease-associated variants.
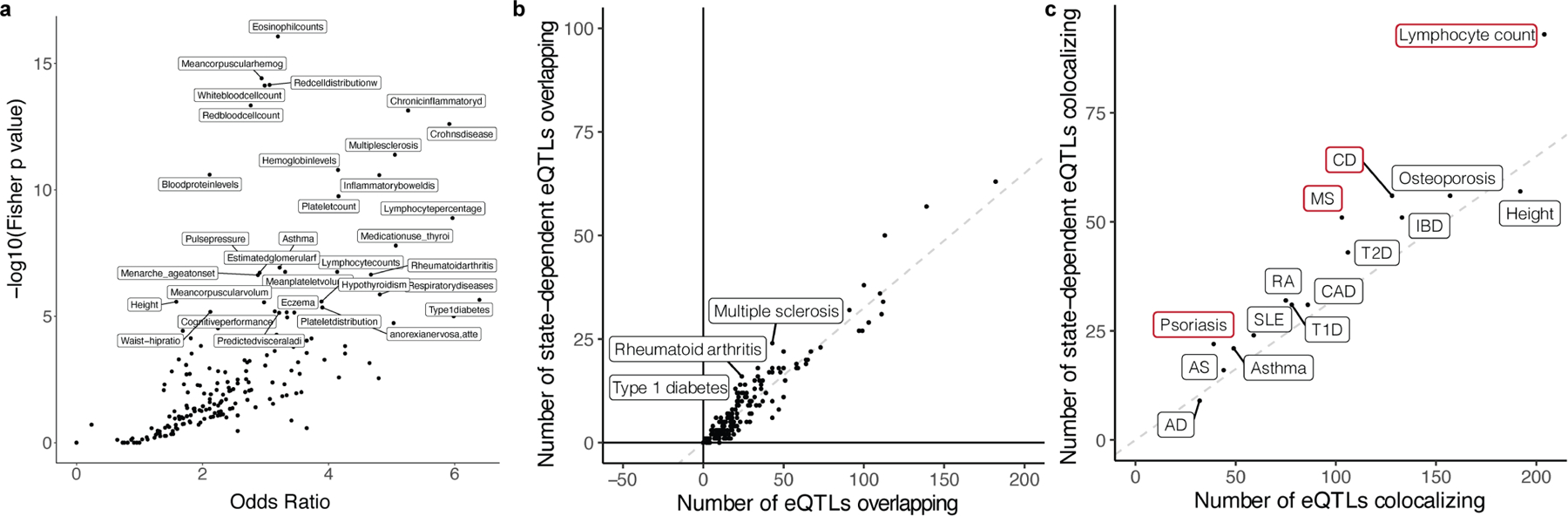
a, Dot plot of traits from the GWAS catalogue plotted based on the −log(Fisher p value) and odds ratio of the enrichment test comparing the proportion of GWAS variants colocalizing with memory-T cell eQTLs for one trait compared to all other traits. Labelled traits have p < 10−5. b, Dot plot of traits from the GWAS catalogue plotted based on the number of GWAS variants overlapping state-dependent eQTLs compared to the total number of GWAS variants overlapping eQTLs. Overlap was defined by r2 > 0.5. The dashed line represents the overall proportion of state-dependent eQTLs (2,117/6,511 = 0.33) and labelled traits have Fisher p < 0.005. c, Dot plot of traits plotted based on the number of GWAS variants colocalizing with state-dependent eQTLs compared to the total number of GWAS variants colocalizing with eQTLs under the Bayesian coloc model. The dashed line represents the overall proportion of state-dependent eQTLs (2,117/6,511 = 0.33) and traits outlined in red are significantly enriched at Fisher p < 0.01. AD=Atopic dermatitis, AS=Ankylosing spondylitis, SLE=Systemic lupus erythematosus, T(1/2)D=Type 1/2 diabetes, RA=Rheumatoid arthritis, CAD=Coronary artery disease, MS=Multiple sclerosis, IBD=Inflammatory bowel disease, CD=Crohn’s disease.
