Abstract
Abstract
The phylum Pseudomonadota is amongst the most represented in the environment, with a comparatively lower prevalence in the human oral cavity. The ubiquity of Pseudomonadota and the fact that the oral cavity is the most likely entry portal of bacteria from external sources underlie the need to better understand its occurrence in the interface environment-humans. Yet, the relevance oral Pseudomonadota is largely underexplored in the scientific literature, a gap that this review aims at addressing by making, for the first time, an overview of the diversity and ecology of Pseudomonadota in the oral cavity. The screening of scientific literature and human microbiome databases unveiled 1328 reports of Pseudomonadota in the oral cavity. Most of these belonged to the classes Beta- and Gammaproteobacteria, mainly to the families Neisseriaceae, Campylobacteriaceae, and Pasteurelaceae. Others also regularly reported include genera such as Enterobacter, Klebsiella, Acinetobacter, Escherichia, Burkholderia, or Citrobacter, whose members have high potential to acquire virulence and antibiotic resistance genes. This review provides evidence that clinically relevant environmental Pseudomonadota may colonize humans via oral cavity. The need for further investigation about Pseudomonadota at the environment-oral cavity interface and their role as vectors potentially involved in virulence and antibiotic resistance transmission is demonstrated.
Key points
• Neisseriaceae, Campylobacteriaceae, and Pasteurelaceae are part of the core oral microbiome
• Enterobacteriaceae, Acinetobacter, or Burkholderia are frequent in the oral microbiome
• Gut dysbiosis may be associated with colonization by ubiquitous oral Pseudomonadota
Graphical abstract
Supplementary information
The online version contains supplementary material available at 10.1007/s00253-022-12333-y.
Keywords: Human–environment nexus, Health, Saliva, Virulence factors, Antibiotic resistance, Ubiquity, One Health
Introduction
The human microbiome is a key player in the balance between health and disease. Insights into the diversity and organization of the complex microbial ecosystem that inhabits the human oral cavity are crucial to understand possible impacts on health and disease, at the oral or systemic levels (Wade 2013; Tuganbaev et al. 2022). Oral microbiomes are characterized by high richness and diversity. About 800 bacterial species have been reported in the human mouth and aerodigestive tract (i.e. pharynx, nasal passages, sinuses, and esophagus), most of which (76%) are culturable, although only 58% are officially named (The Human Microbiome Oral Database, http://www.homd.org – HOMD V3, accessed at 23 November 2022). More than half of these taxa (481 out of 789) are specifically associated with a nasal/oral or oral body site and observed to belong to 12 Bacteria and one Archaea phyla (HOMD V3).
In healthy humans, the core oral microbiome is dominated by members of six phyla, which account for more than 90% of the taxa identified (HOMD V3): Bacillota (formerly Firmicutes), Actinobacteriota (Actinobacteria), Pseudomonadota (formerly Proteobacteria), Fusobacteriota (formerly Fusobacteria), Bacteroidota (formerly Bacteroidetes), and Spirochaetota (formerly Spirochaetes) (Deo and Deshmukh 2019; Zhang et al. 2018). Nonetheless, the structure of the core microbiota, i.e. the relative proportions of each taxon, is supposed to vary. Determinant factors include the geography and diet, the anatomic and physiologic characteristics of the host, as well as the oral cavity site, and consequent biofilm formation, oxygen and nutrient availability, exposure to host immunological factors, among others (Zhigang Ren et al. 2021; Sampaio-Maia et al. 2016; Zhang et al. 2018; Wang et al. 2022b). The broad diversity of oral microbiomes was evidenced by Tierney et al. (2019) who, based on the analysis of 1473 oral metagenomes, identified 23 961 508 genes, half of which were unique in each metagenome. Such a uniqueness has led some authors to propose the oral microbiome profile as a valuable tool for biogeography investigations or forensic personal discrimination (Wang et al. 2022a, 2022b).
The interconnections between phylogenetic diversity and ecology of the microbiota inhabiting the oral cavity, and the possible relationship with host conditions (e.g. age, diet, physical condition), are supposed to have important implications in human health (Burcham et al. 2020; Sampaio-Maia et al. 2016; Tang et al. 2019; Hayes et al. 2018; Dashper et al. 2019). Indeed, the nexus between the oral and gut microbiome is a topic of interest in the exploitation of the oral microbiota (Teil Espina et al. 2019; Iwauchi et al. 2019). Considering that a milliliter of saliva may contain 8–9 log-units of microbial cells, some of which can multiply every 3–4 h, it can be estimated that 1–3 g of microbial biomass can be ingested per day (Edgar et al. 2012). Given the capacity of some of these microbial cells to survive and colonize the host’s gut, a balanced and stable oral microbiota is an essential barrier to prevent pathogen colonization and infection and, therefore, oral and/or systemic infections and/or inflammatory symptoms (Albuquerque-Souza and Sahingur 2022; Ren et al. 2021; Sampaio-Maia et al. 2016; Willis et al. 2020). The interface environment-human has received a renewed attention under the One Health perspective that considers a continuum between humans, animals, and the natural environment (Cunningham et al. 2017; Osterhaus et al. 2020). Accordingly, the human microbiome is affected by the surrounding environment, with the geography, diet, and lifestyle shaping its structure (Cunningham et al. 2017; Osterhaus et al. 2020). In particular, the oral microbiome is expected to be influenced by external conditions that include not only the range of microorganisms to which humans are exposed, for instance, via food products, but also by lifestyle and hygiene habits (Freire et al. 2020; Peters et al. 2018; Tang et al. 2019; Wang et al. 2022a, 2022b).
Pseudomonadota are reported among the predominant phyla in the natural environments, which frequently are under anthropic impacts (Chen et al. 2019; Ferro et al. 2019; Higgins et al. 2018; Nazareno Scaccia et al. 2021; Vaz-Moreira et al. 2014; 2017). In turn, Pseudomonadota include some of the most ubiquitous bacterial groups, as well as a vast array of opportunistic pathogens, for which existing antibacterial drugs may be ineffective (e.g. Acinetobacter, Pseudomonas, Enterobacteriaceae) (Ferro et al. 2019; Jordi Rello et al. 2019; Vaz-Moreira et al. 2014; Rizzatti et al. 2017; Theuretzbacher et al. 2020). Curiously, in spite of these clinically relevant features and although Pseudomonadota are the second most abundant phylum in the mouth (Wang et al. 2022b), not much attention has been given to its presence. Indeed, in discussions about the oral microbiome and its relevance, Pseudomonadota are frequently underexplored when compared with other groups (Radaic and Kapila 2021; Zhang et al. 2018). This review aims at addressing this topic by making, for the first time, an overview of the diversity and ecology of Pseudomonadota in the oral cavity.
In healthy individuals, oral Pseudomonadota are mostly represented by members of the families Neisseriaceae, Pasteulleraceae, and Campylobacteraceae, although the distribution of bacteria may be site- and subject-specific (Aas et al. 2005; Jiang et al. 2019; Zaura et al. 2009). It has been also reported that Pseudomonadota tend to increase in the oral cavity with the age and are frequently associated with inflammatory diseases (Iwauchi et al. 2019; Singh et al. 2019) and other non-infectious disorders (Costa et al. 2021; Rizzatti et al. 2017). However, further insights about Pseudomonadota diversity in the oral cavity will better elucidate the role of the oral cavity as an entry portal for environmental clinically relevant bacteria. With the increasing capacity to have holistic insights across the One Health microbiomes, new opportunities to explore the environment-oral-gut microbiome nexus emerge. This perspective not only will shed additional light into beneficial interactions and how they can promote health, but also will contribute to better understand how some antibiotic resistance or virulence determinants may have access into the human oral microbiome, or how the oral microbiome may be related with health impairment.
This review addresses the hypothesis that the human oral cavity is exposed to different groups of Pseudomonadota, some of which can also thrive in the external environment. It is also hypothesized that given their specific physiological and biochemical, intrinsic or acquired, properties some of these Pseudomonadota may be able to colonize other parts of the human body, and act as vectors of antibiotic resistance or as opportunistic infections. The approach to test the abovementioned hypotheses was to review the literature and databases available, list the most commonly reported Pseudomonadota in the oral cavity and, based on this information, infer about their ecology and distribution, presence of potential clinically relevant properties, and discuss possible implications for the human oral microbiome.
Oral Pseudomonadota in the scientific literature and in public databases
A total of eighty-seven publications surveying the microbiota in the human oral cavity, and reporting target populations, samples’ characteristics, and identification methods (Tables S1 and S2), were screened for the presence of Pseudomonadota. In addition, Pseudomonadota taxa reports in the oral cavity were searched in the databases NIH Human Microbiome Project (https://www.hmpdacc.org/) (n = 24) and expanded Human Oral Microbiome Database (http://www.homd.org/) (n = 53) (Table S1 and S2). The bacterial groups were listed and categorized according to the number of reports (number of times that a taxonomic group was identified) and frequency (quotient between the total number of identifications of a specific taxonomic group and the total number of taxa reported) (Table S3). The data was organized at the genus level, meaning that each name referred to the sum of all taxa within that genus, regardless the identification to the species level. In studies based on the 16S rRNA gene amplicon sequencing, the absence/presence of a given taxon was considered, disregarding relative abundance values.
Genera that were reported more than five times were characterized for their environmental distribution, and possible carriage of antibiotic resistance and/or virulence determinants. Common habitats were compiled based on the section of ecology and habitats of Bergey’s Manual (Garrity et al. 2005) and Nørskov-Lauritsen and Kilian (2006) for the genus Aggregatibacter. Antibiotic resistance and virulence genetic determinants were searched based on whole genome sequences available in the Pathosystems Resource Integration Center (PATRIC, https://www.patricbrc.org, accessed January 2021), through the Specialty Genes tab—The Antibiotic Resistance Database (ARDB) (Liu and Pop 2009) and the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al. 2013), and Victors virulence factors database (Sayers et al. 2019) and the Virulence Factor Database (VFDB) (Chen et al. 2016), respectively. The results were filtered for 100% sequence identity and subject/query coverage, and duplicates were deleted. The genes or gene-product designations were browsed in CARD and VFDB databases for verification of the antibiotic resistance and virulence mechanisms, respectively. The phylogenetic relationship between taxa reported in the oral cavity and human gut microbiota was assessed based on the 16S rRNA gene sequences available for the type strains of the species (https://lpsn.dsmz.de; Parte et al. 2020), used as query for mega BLAST search using the filter human gut microbiome containing 9759 16S ribosomal RNA gene sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi; (Zhang et al. 2000) (accessed at 7th May 2022).
Pseudomonadota in the oral cavity
The literature reviewed was published between 2005 and 2020 and relied on 16S rRNA gene amplicon sequencing (n = 76), mainly based on Illumina or 454 pyrosequencing, and on culture-based methods (n = 6) or targeted methods based on PCR or probe hybridization (n = 7) (Table S1). Most of the studies used samples of saliva or mouth washes, tooth or dental plaques, or soft tissues such as gingiva, cheek, tongue, or tonsils (Table S2). The number of individuals analysed per study ranged between 2 and 2338, with ages between 18 and 80 years, although the information about the studied group was not clearly provided in 12 publications. Some studies included individuals with specific conditions, such as cancer patients (6 articles) or smoker groups (6 articles), or an indigenous tribe (1 article). Based on this search, it was possible to list 1328 observations that corresponded to a total of 313 genera, of which 77 were reported three or more times (i.e. n = 27 three times, n = 10 four times, and n = 40 five times or more) (Table S2, S3). The remaining were reported only once (n = 191) or twice (n = 45) (Table S3a). The fact that distinct analytical and identification methods were used in different studies might have led to distinct identifications at the species level, a bias that we believe was considerably reduced at the genus or higher taxonomic ranks, as we used in this study (Table S3).
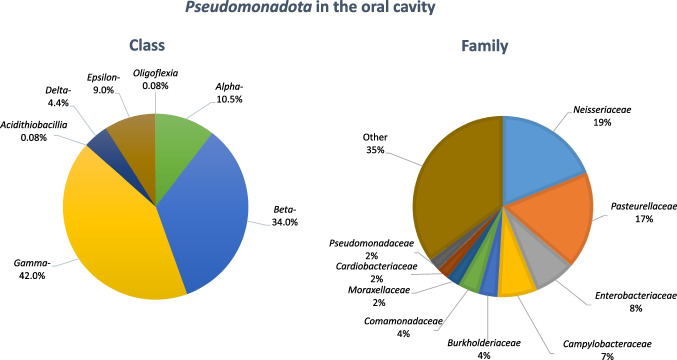
Most of the 1328 observations of Pseudomonadota in the oral cavity (Fig. 1; Table S3a) belonged to the classes Gammaproteobacteria (n = 558) and Betaproteobacteria (n = 452). Members of the classes Alphaproteobacteria (n = 139), Epsilonproteobacteria (n = 119), and Deltaproteobacteria (n = 57) were reported fewer times, and the classes Acidithiobacillia and Oligoflexia were mentioned only three times. More than half of the 1328 observations were included in four families, specifically Neisseriaceae (n = 263, class Betaproteobacteria), represented by 10 genera, Pasteurellaceae (n = 243, class Gammaproteobacteria) distributed by 13 genera, Enterobacteriaceae (n = 100, class Gammaproteobacteria) that included 12 genera, and Campylobacteraceae (n = 99, class Epsilonproteobacteria) that although represented by two genera, most (n = 98) referred to the genus Campylobacter (Table S3a). These observations agree with previous studies that refer to Neisseriaceae, Pasteurellaceae, and Campylobacteraceae as the most common Pseudomonadota of the oral microbiome (Zaura et al. 2009). However, among the genera that were reported five times or more (Table S3c) (n = 926), it noteworthy the occurrence of other Pseudomonadota, such as members of the families Enterobacteriaceae (n = 48, 5%), Burkholderiaceae (n = 43; 5%), Comamonadaceae (n = 9, 1%), Moraxellaceae (n = 31, 3%), and Pseudomonadaceae (n = 29, 3%) that include bacteria with potential clinical relevance, recurrently reported in the oral cavity (Table S3c, Table 1). For example, members of the family Enterobacteriaceae were reported in 25 distinct publications (Table S2, S3). The occurrence of taxa that do not belong to the core oral microbiome may be favoured in some conditions. For example, a recent study suggested that when compared to healthy controls, chronic kidney disease patients presented the proliferation of clinically relevant Enterobacteriaceae in the oral cavity, potentially harbouring acquired antibiotic resistance genes (Costa et al. 2021). Another example was provided by a mice model study, which showed that periodontitis was associated with the accumulation in the oral cavity of Enterobacteriaceae, specifically of the genera Klebsiella and Enterobacter (Kitamoto et al. 2020).
Fig. 1.
Pseudomonadota in the oral cavity number of reports identified at the class or family levels
Table 1.
Diversity and characteristics of the genera of bacteria belonging to the phylum Pseudomonadota, whose occurrence in the oral cavity was reported more than five times in the examined literature. In a total of 1328 genera of Pseudomonadota, 583 were reported more than five times and were affiliated to (class, number of genera, number of observations) the following: Alphaproteobacteria, 4, 27; Betaproteobacteria, 11, 327; Deltaproteobacteria, 3, 24; Epsilonproteobacteria, 2, 109; Gammaproteobacteria 20, 439. Virulence and antibiotic resistance characteristics of members of these genera were collected from the public database PATRIC (https://www.patricbrc.org/) and the following sources: ARDB, CARD, Victors, VFDB. Ecology and habitats were reviewed from the Bergey’s Manual (Garrity et al. 2005) and (Nørskov-Lauritsen and Kilian 2006) for Aggregatibacter. Additional information is provided as a supplementary file
| Reported diversity in the oral cavity | Ecology and habitats | Genome-based characterization of genus members | |||||
|---|---|---|---|---|---|---|---|
| Top | Genus Class Family Nº reports-genus/total |
Reported species (nº) | Common habitats | Virulence1 | Other taxa (%)2 | Antibiotic resistance1 | Other taxa (%)2 |
| 1 |
Neisseria Neisseriaceae Beta- 165/1328 |
N. subflava (17) N. flavescens (8) N. mucosa (7) N. elongata (7) N. sicca (5) N. bacilliformis (5) N. weaveri (4) N. oralis (4) N. lactamica (4) N. flava (4) N. polysaccharea (3) N. pharyngis (3) N. meningitidis (3) N. perflava (2) N. gonorrhoeae (2) N. cinerea (2) N. dentiae (1) N. canis (1) N. animalis (1) Neisseria spp. (82) |
Mucous membrane surface (oropharynx, nasopharynx, throat) |
Adherence Invasion Motility Transport Iron transport Protease Stress proteins Protease Host response evasion Endotoxin |
Neisseria 100% |
Target modification Efflux |
Neisseria 100% |
| 2 |
Haemophilus Pasteurellaceae Gamma- 110/1328 |
H. parainfluenzae (16) H. haemolyticus (5) H. paraphrohaemolyticus (4) H. sputorum (3) H. pittmaniae (3) H. influenzae (3) H. aegyptius (3) H. parahaemolyticus (2) H. haemoglobinophilus (1) H. ducreyi (1) Haemophilus spp. (69) |
Mucous membranes surface (upper respiratory tract, oral cavity) |
Invasion Iron transport Transport Protease Host response evasion Endotoxin Others |
Haemophilus 96.7% |
Efflux Inactivation Target modification Other |
Salmonella 62.7% Serratia 15.4% Haemophilus 7.5% Mannheimia 6.0% |
| 3 |
Campylobacter Campylobacteraceae Epsilon 98/1328 |
C. gracilis (10) C. concisus (10) C. showae (9) C. rectus (7) C. curvus (4) C. sputorum (2) C. lari (1) C. jejuni (1) C. insulaenigrae (1) C. hyointestinalis(1) C. hominis (1) C. helveticus (1) C. fetus (1) C. ureolyticus (1) Campylobacter spp. (48) |
Reproductive organs, intestinal tract and oral cavity (humans and animals) |
Adherence Invasion; Motility/Chemotaxis Host response evasion Secretion system; Others |
Campylobacter 100% |
Efflux Inactivation Target modification |
Campylobacter 98.7% |
| 4 |
Aggregatibacter Pasteurellaceae Gamma- 85/1328 |
A. actinomycetemcomitans (9) A. segnis (9) A. aphrophilus (6) A. paraphrophilus (5) Aggregatibacter spp. (56) |
Dental surfaces Pharynx, also peritoneum, pleura and bone |
None | None | None | None |
| 5 |
Kingella Neisseriaceae Beta- 53/1328 |
K. denitrificans (6) K. kingae (3) K. oralis(6) Kingella spp.(38) |
Mucous membranes, upper respiratory tract and oral cavity, also urogenital (humans and other primates) | None | None |
Efflux Inactivation Target modification |
Neisseria 22.2% Morganella 22.2% Vibrio 22.2% Salmonella 22.2% Serratia 11.1% |
| 6 |
Cardiobacterium Cardiobacteriaceae Gamma- 30/1328 |
Card. hominis (6) Card. valvarum (5) Cardiobacterium spp. (19) |
Mucous membranes (nose, mouth, and throat and also gastrointestinal tract) | None | None | None | None |
|
Lautropia Burkholderiaceae Beta- 30/1328 |
L. mirabilis (10) Lautropia spp. (20) |
Oral cavity (gingival surface) | None | None | None | None | |
| 7 |
Pseudomonas Pseudomonadaceae Gamma- 29/1328 |
P. aeruginosa (6) P. fluorescens (3) P. stutzeri (2) P. beteli(1) P. fragi (1) P. luteola (1) P. otitidis (1) P. pseudoalcaligenes (1) Pseudomonas spp. (13) |
Natural environments (water, soil, plants), and animal tissues. Include human pathogens |
Adherence Invasion Motility/Chemotaxis Transport Iron transport Protease Stress proteins Quorum sensing Protease Lipase Host response evasion Secretion system Endotoxin Toxins |
Pseudomonas 100% |
Efflux Inactivation Target modification Reduced permeability |
Pseudomonas 92.7% |
| 8 |
Eikenella Neisseriaceae Beta- 27/1328 |
E. corrodens (9) Eikenella spp. (18) |
Upper respiratory tract, oral cavity and subgingival plaque (periodontitis) |
None | None | None | None |
| 9 |
Actinobacillus Pasteurellaceae Gamma- 23/1328 |
Act. capsulatus (1) Act. equuli (1) Act. minor (1) Act. pleuropneumoniae (1) Act. porcinus (1) Act. scotiae (1) Act. succinogenes (1) Act. ureae (1) Actinobacillus spp. (15) |
Mucous membranes of different carrier animals (humans, sheep, cattle, horses, pigs, other mammals and birds) |
Motility Transport Iron transport Others |
Actinobacillus 99.9% |
Efflux Inactivation Target modification |
Vibrio 41.2% Serratia 22.1% Mannheimia 17.6% Escherichia 12.2% |
|
Enterobacter Enterobacteriaceae Gamma- 23/1328 |
Ent. cloacae (4) Ent. aerogenes (3) Ent. sakazakii (3) Ent. gergoviae (2) Ent. hormaechei (2) Ent. amnigenus (1) Ent. cancerogenus (1) Ent. mori (1) Ent. xiangfangensis (1) Enterobacter spp.(5) |
Natural environments (water, soil, plants), and animal tissues. Include human pathogens |
Adherence Invasion Transport Iron transport Motility/Chemotaxis Secretion system; Toxins Lipase Host response evasion Other |
Salmonella 49.0% Escherichia 24.8% Shigella 20.9% |
Efflux Inactivation Target modification Other |
Klebsiella 21.2% Vibrio 18.1% Enterobacter 13.8% Salmonella 13.0% Escherichia 13.1% Pseudomonas 7.0% |
|
| 10 |
Klebsiella Enterobacteriaceae Gamma- 23/1328 |
Kl. pneumoniae (10) Kl. oxytoca (4) Kl. singaporensis (1) Kl. michiganensis (1) Klebsiella spp. (7) |
Natural environments (water, soil, plants), and animal tissues Include human pathogens |
Transport Iron transport Others |
Salmonella 33.7% Escherichia 32.8% Shigella 31.7% |
Efflux Inactivation Target modification Reduced permeability Other |
Klebsiella 43.1% Escherichia 15.7% Vibrio 12.6% Enterobacter 7.9% Salmonella 6.5% |
|
Serratia Yersiniaceae Gamma- 17/1328 |
S. marcescens (6) S. ficaria (2) S. liquefaciens (1) S. rubidaea (1) S. odorifera (1) Serratia spp. (6) |
Natural environments (water, soil, plants), and animal tssues. Include human pathogens |
Transport Iron transport Others |
Yersinia 98.6% |
Efflux Inactivation Target modification Other |
Klebsiella 25.2% Escherichia 15.6% Salmonella 10.6% Serratia 23.1% Vibrio 11.7% |
|
| 11 |
Acinetobacter Moraxellaceae Gamma- 16/1328 |
Ac.baumannii (5) Ac. johnsonii (1) Ac. schindleri (1) Ac. junii(1) Acinetobacter spp. (8) |
Natural environments (water, soil, plants), and animal tissues. Include human pathogens |
Adherence Invasion Transport Iron transport Secretion systems Motility/Chemotaxis Toxins Host response evasion Others |
Pseudomonas 75.0% Staphylococcus 13.5% |
Efflux Inactivation Target modification Other |
Acinetobacter 76.3% Vibrio 5.9% |
| 12 |
Escherichia Enterobacteriaceae Gamma- 12/1328 |
Esch. coli (4) Escherichia spp. (8) |
Gastrointestinal tract of warm-blooded animals. Also in natural environments (water, soil, plants) |
Adherence Invasion Transport Iron transport Protease Lipase Secretion systems Motility/Chemotaxis Toxins Host response evasion Others |
Escherichia 69.8% Shigella 25.0% |
Efflux Inactivation Target modification Reduced permeability Other |
Escherichia 97.0% |
| 13 |
Helicobacter Helicobacteraceae Epsilon- 11/1328 |
H. pylori (7) H. trogontum (1) Helicobacter spp. (3) |
Gastrointestinal tract, oral cavity and internal organs of humans and animals |
Adherence Motility Endotoxin Toxin Secretion systems Acid resistance Others |
Helicobacter 100% |
Efflux Inactivation Other |
Helicobacter 86.1% Campylobacter 9.1% |
|
Simonsiella Neisseriaceae Beta- 11/1328 |
Sim. muelleri (6) Simonsiella spp. (5) |
Mucous membranes, oral cavity of warm-blooded animals | None | None | None | None | |
|
Sphingomonas Sphingomonadaceae Alpha- 11/1328 |
Sph. yabuuchiae (2) Sphingomonas spp. (9) |
Natural environments (water, soil, plants) |
Adherence Motility Iron transport Secretion systems Host response evasion Protease Others |
Pseudomonas 86.3% Streptococcus 13.7% |
Efflux Inactivation Target modification |
Pseudomonas 90.5% Vibrio 7.1% |
|
| 14 |
Desulfobulbus Desulfobulbaceae Delta- 10/1328 |
D. elongates (1) D. mediterraneus (1) D. rhabdoformis (1) Desulfobulbus spp. (7) |
Natural environments (anoxic black mud, sewage, fresh- or brackish water) and gastrointestinal tract of animals | None | None | None | None |
|
Moraxella Moraxellaceae Gamma- 10/1328 |
M. oblonga (1) M. osloensis (1) M. catarrhalis (1) Moraxella spp. 7/10 |
Mucous membranes, oral cavity of warm-blooded animals |
Adherence Motility |
Helicobacter 100% |
Efflux Inactivation Target modification |
Vibrio 27.3% Escherichia 27.3% Mannheimia 9.1% Pasteurella 18.2% Psychrobacter 9.1% Others 9.1% |
|
| 15 |
Comamonas Comamonadaceae Beta- 9/1328 |
Com. testosteroni (2) Com. terrigena (1) Comamonas spp. (6) |
Natural environments (water, soil, plants) Polluted environments |
None | None |
Efflux Inactivation Target modification |
Vibrio 28.6% Escherichia 28.6% Acinetobacter 10.7% Enterobacter 10.7% Pseudomonas 10.7% Aeromonas 7.1% |
| 16 |
Aeromonas Aeromonadaceae Gamma- 8/1328 |
Aero. hydrophila (1) Aero. veronii (1) Aeromonas spp. 6/8 |
Natural environments (fresh- or brackish-water, sewage) |
Adherence Secretion systems Others |
Aeromonas 99.0% |
Efflux Inactivation Target modification Other |
Vibrio 24.7% Aeromonas 13.7% Klebsiella 13.3% Escherichia 10.1% Acinetobacter 7.1% Salmonella 7.8% |
|
Burkholderia Burkholderiaceae Beta- 7/1328 |
B. cepacia (1) Burkholderia spp. (6) |
Natural environments (water, soil, plants), and animal tissues. Include human pathogens |
Adherence Invasion Motility/Chemotaxis Secretion system Host response evasion Others |
Burkholderia 100% |
Efflux Inactivation Target modification Other |
Burkholderia 81.5% Acinetobacter 5.9% Pseudomonas 5.9% |
|
|
Citrobacter Enterobacteriaceae Gamma- 8/1328 |
Cit. koseri (2) Cit. amalonaticus (1) Cit. freundii (1) Citrobacter spp. (4) |
Gastrointestinal tract of warm-blooded animals. Also in natural environments (water, soil, plants) |
Iron transport Others |
Salmonella 54.6% Escherichia 21.7% Shigella 21.4% |
Efflux Inactivation Target modification Other |
Escherichia 19.8% Vibrio 15.2% Klebsiella 14.2% Citrobacter 11.6% Enterobacter 7.7% Salmonella 9.3% Others 5.8% |
|
|
Herbaspirillum Oxalobacteraceae Beta- 8/1328 |
Herb. hiltneri (1) Herb. frisingense (1) Herb. chlorophenolicum (1) Herb. seropedicae (1) Herb. lusitanum (1) Herb. rubrisubalbicans (1) Herb. huttiense (1) Herbaspirillum sp. (1) |
Plants (gramineous; roots, stems and leaves) | None | None | Inactivation | Escherichia 100% | |
|
Pasteurella Pasteurellaceae Gamma- 8/1328 |
Past. pneumotropica (1) Past. multocida (1) Past. mairii (1) Pasteurella spp. (5) |
Mucous membranes (upper respiratory tract) and lower genital tracts of mammals (rarely humans) and birds |
Adherence Motility Others |
Pasteurella 100% |
Efflux Inactivation Target modification |
Vibrio 21.9% Serratia 32.5% Escherichia 7.5% Manheimia 6.3% Pasteurella 10.6% Pseudomonas 5.0% Others 5.6% |
|
|
Stenotrophomonas Lysobacteraceae Gamma- 8/1328 |
Sten. maltophilia (2) Stenotrophomonas spp. (6) |
Natural environments (water, soil, plants), and animal tissues. Include human pathogens | None | None |
Efflux Inactivation Target modification Other |
Stenotrophomonas 86.3% Vibrio 13.7% |
|
| 17 |
Desulfomicrobium Desulfomicrobiaceae Delta- 7/1328 |
Des. orale (4) Desulfomicrobium spp. (3) |
Natural environments (anoxic black mud, sewage, fresh- or brackish water) and gastrointestinal tract of animals | None | None | None | None |
|
Desulfovibrio Desulfovibrionaceae Delta- 7/1328 |
Dv. desulfuricans (1) Dv. fairfieldensis (1) Dv. hydrothermalis (1) Desulfovibrio spp. (4) |
Natural environments (anoxic black mud, sewage, fresh- or brackish water) and gastrointestinal tract of animals | None | None | Inactivation |
Acinetobacter 50.0% Escherichia 50.0% |
|
|
Mannheimia Pasteurellaceae Gamma- 7/1328 |
Man. haemolytica (2) Man. varigena (1) Man. granulomatis (1) Man. ruminalis (1) Mannheimia spp. (2) |
Mucous membranes (upper respiratory tract of warm-blooded animals) | Others | Actinobacillus 100% |
Efflux Inactivation Target modification Other |
Pasteurella 33.9% Escherichia 13.3% Pseudomonas 12.5% Mannheimia 16.1% Vibrio 8.1% Others 16.1% |
|
|
Shewanella Shewanellaceae Gamma- 7/1328 |
Sh. aquimarina (1) Sh. loihica (1) Sh. japonica (1) Sh. decolorationis (1) Shewanella spp. (3) |
Nutrient rich marine environments. Occasional clinical occurrence | Others | Shigella 100% |
Efflux Inactivation Target modification |
Shewanella 23.3% Vibrio 16.5% Escherichia 14.3% Klebsiella 14.3% Salmonella 9.8% Pseudomonas 6.8% |
|
| 18 |
Achromobacter Alcaligenaceae Beta- 6/1328 |
Ach. xylosoxidans (4) Achromobacter spp. (2) |
Natural environments (water, soil, plants), and animal tissues. Occasional clinical occurrence |
Adherence Invasion Iron transport Transport Siderophore; Others |
Escherichia 54.3% Shigella 32.6% Salmonella 8.7% |
Efflux Inactivation Target modification Other |
Escherichia 50.5% Vibrio 19.0% Achromobacter 13.3% Pseudomonas 7.6% |
|
Ralstonia Burkholderiaceae Beta- 6/1328 |
Ralstonia spp. (6) | Natural environments (water, soil, plants), and animal tissues. Occasional clinical occurrence | None | None |
Inactivation Target modification |
Ralstonia 97.0% | |
|
Rhizobium Rhizobiaceae Alpha- 6/1328 |
Rh. daejeonense (1) Rh. leguminosarum (1) Rh.sullae (1) Rh.loti (1) Rhizobium sp. (2) |
Plants (symbiotic nitrogen fixation) |
Adherence Motility |
Campylobacter 100% |
Efflux Inactivation |
Agrobacterium 69.2% Salmonella 7.7% Others 7.7% |
|
| 19 |
Bordetella Alcaligenaceae Beta- 5/1328 |
Bor. pertusis (2) Bor. petri (1) Bor. parapertussis (1) Bordetella sp. (1) |
Natural environments (water, soil, plants), and animal tissues. Include pathogens |
Adherence Motility Endotoxin Host response evasion Others |
Bordetella 100% |
Efflux Inactivation Target modification Other |
Bordetella 52.7% Vibrio 39.2% |
|
Brevundimonas Caulobacteraceae Alpha- 5/1328 |
Br. Diminuta (2) Brevundimonas spp. (3) |
Natural environments, (water, soil, plants), and animal tissues. Include pathogens |
Adherence Motility |
Pseudomonas 100% | Target modification |
Acinetobacter 40.0% Vibrio 60.0% |
|
|
Methylobacterium Methylobacteriaceae Alpha- 5/1328 |
Met. fujisawaense (1) Methylobacterium spp. (4) |
Natural environments (water, poor in nutrients and/or stress conditions | None | None | None | None | |
|
Pantoea Erwiniaceae Gamma- 5/1328 |
Pan. agglomerans (1) Pantoea spp. 4/5 |
Natural environments (water, soil, plants), and animal tissues. Occasional clinical occurrence | Others |
Shigella 43.2% Salmonella 35.4% Escherichia 21.4% |
Efflux Inactivation Target modification Other |
Klebsiella 40.3% Shigella 22.4% Vibrio 14.9% Acinetobacter 7.5% Others 6.0% |
|
|
Proteus Morganellaceae Gamma- 5/1328 |
Pr. mirabilis (2) Proteus spp. (3) |
Gastrointestinal tract of warm-blooded animals. Also in natural environments (water, soil, plants) | Secretion system Others |
Escherichia 55.6% Vibrio 22.2% Shigella 11.1% Salmonella 5.6% Yersinia 5.6% |
Efflux Inactivation Target modification Reduced permeability Other |
Escherichia 36.0% Vibrio 15.9% Acinetobacter 13.7% Klebsiella 10.1% Salmonella 8.9% |
|
|
Psychrobacter Moraxellaceae Gamma- 5/1328 |
Psy. celer (1) Psychrobacter spp. (4) |
Natural environments (sea, ice), and animal tissues (skin, gills) |
Adherence Motility Iron transport Transport Others |
Neisseria 100% |
Inactivation Target modification |
Escherichia 41.2% Vibrio 23.5% Psychrobacter 17.6% Acinetobacter 5.9% Neisseria 5.9% Oligella 5.9% |
|
1Antibiotic resistance and virulence determinants were identified using the criteria of 100% of subject and query sequence coverage and 100% of sequence identity—only generic functions are indicated
2Other taxa sharing a genetic element with 100% identical amino-acid sequence (only taxa representing more than 5% are indicated)
The oral cavity as primary or transient Pseudomonadota habitat
To avoid situations of misidentification or of sporadic episodes of occurrence, the further discussion is focused at genus level identifications and on situations where consistent reports were available, i.e. genera that were reported more than five times. This procedure resulted in a list of 40 genera of the classes Gammaproteobacteria (n = 20), Betaproteobacteria (n = 11), Epsilonproteobacteria (n = 2), Deltaproteobacteria (n = 3), and Alphaproteobacteria (n = 4). For sake of simplicity, the 40 genera were ranked according to their frequency of occurrence, resulting in 19 categories designated from Top1 to Top19 (Table S3c, Table 1). In addition, the ecology and habitats, clinically relevant features (virulence and antibiotic resistance), and hypothetical horizontal gene transfer, suggested by the occurrence of identical genetic elements in other bacterial groups, are summarized in the five right-hand columns of Table 1. The association of members of almost all these genera (n = 36) to infectious disease has been demonstrated (Table 2), suggesting that specific host conditions may facilitate the opportunistic character of these bacteria.
Table 2.
Examples of infectious diseases associated with Pseudomonadota reported in the oral cavity.
Source: https://www.patricbrc.org (accessed at May 7th 2022) and examples of additional references: (1) Pathak et al. 2021; (2) Cross et al. 2018; (3) Shrestha et al. 2021; (4) Calheiros Cruz et al. 2022; (5) Chi et al. 2004; (6) Nseir et al. 2011; (7) Hagiya et al. 2018; (8) Danger et al. 2022; (9) Zhou et al. 2021; (10) Chen et al. 2011
| Infectious disease category | Pseudomonadota genera |
|---|---|
| Oral |
Aggregatibacter Campylobacter (1) Desulfovibrio Desulfobulbus (2) Eikenella |
| Respiratory tract (bronchitis, pneumonia, meningitis, etc.) |
Achromobacter Actinobacillus Bordetella Burkholderia Haemophilus Klebsiella Mannheimia Neisseria Pantoea (3) Pseudomonas Psychrobacter Serratia Stenotrophomonas |
| Gastrointestinal tract |
Aeromonas Campylobacter Escherichia Helicobacter Lautropia (4) Proteus |
| Blood (sepsis, bacteremia) |
Acinetobacter Aeromonas Brevundimonas (5) Citrobacter Comamonas (6) Desulfovibrio (7) Escherichia Herbaspirillum Kingella Methylobacterium Moraxella (8) Neisseria Pantoea Pasteurella Proteus Ralstonia (9) Serratia Stenotrophomonas |
| Other (arthritis, osteomyelitis, endocarditis, cellulitis, encephalitis, skin, etc.) |
Cardiobacterium (10) Citrobacter Enterobacter Haemophilus Kingella Neisseria Pasteurella Proteus Shewanella |
The assessment of the information compiled in Table 1 highlights three profiles—bacteria mainly associated with humans and other animals, environmental bacteria, and ubiquitous bacteria. Bacteria mainly associated with humans and other animals were represented mainly in the Top1–6 groups, composed of genera whose major habitat includes mucosa of oral cavity and sometimes also of the gastrointestinal and/or genital tract. Environmental bacteria were found in the Top7–19 groups, which include also genera associated with animals and humans (e.g. Top8, 13, some Top16, 17; Table 1) and others, whose natural environment (water, soils, plants) is the primary habitat. Genera such as Rhizobium, Herbaspirillum, Shewanella, or Methylobacterium are good examples of bacteria that typically thrive in the natural environment, and which presence in the oral cavity may be explained based on diet, lifestyle, or familiar context, with unknown potential clinical relevance (Table 1) (Hisham Altayb et al. 2022; Nasidze et al. 2011). Other groups, typically of environmental nature, like the strict anaerobic sulphate-reducing Deltaproteobacteria, such as members of the genera Desulfovibrio, Desulfomicrobium, and Desulfobulbus, have been reported in the oral microbiota of healthy people (Deo and Deshmukh 2019), although may be also associated with pathologies such as periodontal disease, dental plaques, or gastrointestinal inflammation (Colombo et al. 2009; Khor et al. 2021). The group of ubiquitous bacteria, with a wide distribution that span from pristine to heavily contaminated environments, and also the human and animal body, are those of major concern in the environment-human oral cavity interface. These bacterial genera, such as Pseudomonas, Escherichia, Citrobacter, Klebsiella, or Burkholderia, were in the groups Top7, 9, 11, 12, 16 (Table 1), include opportunistic pathogens, and frequently harbour acquired antibiotic resistance. The presence of these groups in the oral cavity may be associated with poor hygiene conditions, oral dysbiosis, deficient host defences, systemic diseases, or other factors; for example, chronic nail-biting habit and chronic kidney disease promote the oral carriage of Enterobacteriaceae (Baydaş et al. 2007; Costa et al. 2021). The ecology and genome plasticity of these ubiquitous bacteria may also explain the diverse array of antibiotic resistance and virulence mechanisms that characterize these groups (Table 1). The facts that members of these genera can thrive in soil, water, and plants, where human and animal excreta can be also present, and have typically highly dynamic genomes, suggest that these bacteria can serve as vectors of clinically relevant features (e.g. antibiotic resistance, virulence) from the environment to humans (Sanz-García et al. 2021; Wang et al. 2022a, 2022b). Indeed, the summary provided in Table 1 shows that bacteria of some of these genera hold a broad set of antibiotic resistance genes, generically included in the categories efflux, inactivation, target modification, reduced permeability, among others, that are shared by other Pseudomonadota. Also, virulence genes, related with factors such as adherence, invasion, transport, iron transport, secretion systems, motility/chemotaxis, toxins, host response evasion, among others are reported in bacteria of these genera. Virulence and antibiotic resistance determinants may contribute to enhance the capacity for colonizing, which can also be favoured by charity processes among the microbial community members, e.g. through extracellular antibiotic degradation. In some cases, identical gene sequences (100% sequence identity and coverage) were found in other taxa, suggesting the potential for dynamic horizontal gene transfer. Remarkably, bacteria of genera, such as Achromobacter, Acinetobacter, Aeromonas, Bordetella, Burkholderia, Campylobacter, Citrobacter, Comamonas, Enterobacter, Escherichia, Haemophilus, Klebsiella, Pantoea, Pasteurella, Proteus, Serratia, and Shewanella, harbour antibiotic resistance and/or virulence genes that are 100% identical to others reported in other taxa, whose habitats include the transition between animals, humans, soil, water, and plants. Although horizontal gene transfer is probably rare in the oral cavity (Tierney et al. 2019), the colonization by bacteria with a rich accessory genome comprised by antibiotic resistance or virulence genes, acquired somewhere else, is of concern, mainly in elderly, immune-depressed, and individuals with oral dysbiosis (Radaic and Kapila 2021). It is suggested that some Pseudomonadota with opportunistic pathogenic character and able to harbour acquired genes can reach the oral cavity and, eventually, can colonize it as well as the gut or other body habitats and ultimately cause disease. Moreover, it is noteworthy that some of these bacteria are probably able to cross the distinct One Health compartments and may represent important vectors of transmission of antibiotic resistance between the natural environment and the humans (Osterhaus et al. 2020). For some genera, specifically Aggregatibacter, Cardiobacterium, Desulfobulbus, Desulfomicrobium, Desulfovibrio, Eikenella, Herbaspirillum, Lautropia, Methylobacterium, Ralstonia, and Simonsiella, the data available on virulence or antibiotic resistance was scant. Although most of these genera have been associated with the oral cavity and human body and reported to thrive in natural environments with low anthropogenic impacts, further information about their genome plasticity and physiology may be relevant to better understand their role in the oral cavity.
Pseudomonadota: from the oral cavity to the gut microbiome
An interconnection between the oral and gut microbiomes has been demonstrated in the literature (Khor et al. 2021; Kitamoto et al. 2020; Kitamoto and Kamada 2022). According to Kitamoto et al. (2020), bacteria can be transmitted from the oral cavity to the gut through hematogenous or enteral routes. In the hematogenous route, the bacteria have access to a systemic circulation through oral mechanical injuries (induced by, e.g. hard mastication, brushing, orthodontics, extractions, or unhealthy periodontium) with the subsequent gut colonization. In the enteral route, bacteria migrate through the gastrointestinal tract till colonizing the intestine. It is generally assumed that only part of the swallowed oral bacteria reaches and colonizes the healthy gut due to the gastric acidity. Indeed, the gut-resident- and oral microbiota are represented by distinct bacterial groups identified based on 16S rRNA gene sequence amplicon analysis (Wang et al. 2022b). Comparatively, Pseudomonadota are present at much lower proportions in the gut than in the oral cavity (< 5% vs. > 30%) (Wang et al. 2022b). However, when members of the oral bacterial community reach the gut, they may induce a considerably change in the gut microbiome composition, with systemic repercussions. For example, Nakajima et al. (2015) showed that the oral administration to mice of Porphyromonas gingivalis (phylum Bacteroidota) led to a decrease of Bacillota in the gut, while increased serum endotoxin levels were observed, suggesting the impairment of the barrier leading to the dissemination of enteric bacteria into the liver.
The search for closely related Pseudomonadota taxa in the oral cavity and of human gut suggested evidence for this nexus. The sequence identity between the 16S rRNA gene of the type strain of species reported in the oral cavity and human gut microbiome revealed values ranging from 78.1 to 98.6% (Table S4). Identity values above 97%, a threshold below which it is assumed that two organisms belong to different species (Stackebrandt and Goebel 1994), were observed for the species Enterobacter cloacae, Enterobacter sakazakii (valid name Kosakonia sacchari), Enterobacter hormaechei, Klebsilela pneumoniae, Citrobacter koseri, Citrobacter amalonaticus, and Desulfovibrio desulfuricans. In addition, although with lower sequence identity values (91.2–90.4%), significant alignments (e-values ranging from e−87 to e−179) were observed for species of the genera Neisseria, Haemophilus, Campylobacter, Kingella, Pseudomonas, Eikenella, Actinobacillus, Acinetobacter, Moraxella, and Comamonas (Table S4). The finding of significant sequence identities may be limited by multiple factors, specifically, the use of type strains 16S rRNA gene sequences, the incapacity to detect minor populations, as is the case of Pseudomonadota, due to low DNA sequencing depth of the human microbiomes, and the fact that the DNA sequences being compared do not belong to the same human microbiome (gut and oral). It must be noted that that nexus oral-gut is probably established based on minor populations. Indeed, Tierney et al. (2019) observed that although gut and oral microbiota genes are shared, that is observed to < 2.5% of the microbiome genes; i.e. considering 95% of sequence identity, only 549 610 genes were common to the oral (23 411 898 genes) and gut (21 704 828 genes) microbiomes. While the low depth of sequencing is a major limitation to reliably compare metagenomes, Tierney et al. (2019) argued that each individual holds a unique microbiome, which can be fingerprinted based on rare microbial strains. Clearly, some Pseudomonadota genera are particularly suitable candidates for such fingerprinting approach, for epidemiological and health condition evaluations. As suggested, even if in low numbers, some oral Pseudomonadota genera are likely to reach and colonize the human gut. Eventually, this situation may be favored or triggered by dysbiosis conditions, frequently associated with the increased relative abundance of Pseudomonadota in the oral or gut microbiome (Khor et al. 2021; Weiss and Hennet 2017). Indeed, the study of Atarashi et al. (2017) showed based on gnotobiotic models that Klebsiella spp. isolated from the salivary microbiota tend to colonize the gut when the intestinal microbiota is under dysbiosis, eliciting a severe gut inflammation by strongly inducing T helper 1 cells. These findings have been confirmed in subsequent studies (Kitamoto et al. 2020), who showed that the periodontitis-driven accumulation of Klebsiella spp. and Enterobacter spp. in the oral cavity might result in a consequent increase in the gut, possibly inducing colitis. Curiously, such effects were only observed in susceptible hosts, as that oral bacteria did not colonize the gastrointestinal tract of healthy animals (Kitamoto et al. 2020). These studies suggest that species of the phylum Pseudomonadota, despite constituting a small fraction of the oral microbiota, can colonize the gut, and contribute to maintain gut dysbiosis and chronic inflammation (Khor et al. 2021; Kitamoto and Kamada 2022).
Final considerations
Pseudomonadota may include oral pathobionts, some of which constitute a reservoir of virulence and antibiotic resistance genes (Table 1, Table 2). In addition to the regularly found groups, others comprising the accessory, variable, or “non-core” microbiome (Deo and Deshmukh 2019) may be of interest, as indicators of specific health conditions or of an unbalanced microbiota. By default, the non-core microbiome is highly diverse and vast, probably influenced by the individual health conditions, dietary and lifestyle choices, hygiene practices, geography, and even ethnicity (Hisham Altayb et al. 2022; Wang et al. 2022b). In our review, we reported more than 250 bacterial genera that have not been listed as part of the core microbiota of the oral cavity, and which occurrence is probably minor in abundance and shaped by external variables. However, by highlighting a short list of 40 genera that were recurrently reported in the literature, this review suggests that non-core Pseudomonadota may be more diverse and frequent in the oral cavity than formerly believed, stressing the need of further investigation.
The increasing potential of anthropogenic bacteria to colonize, invade, and persist in environment-human interfaces, mainly when subjected to strong disinfection or antimicrobial actions, deserves attention (Becerra-Castro et al. 2016; Blaustein et al. 2021; Alexander Mahnert et al. 2019; Osterhaus et al. 2020). This review supports the hypothesis that humans exposed to food products and environments where increasingly anthropogenic microbiomes pullulate may have increased probability of acquiring antibiotic-resistant and virulent bacteria. These colonization events may only manifest under a host debilitation situation. The relationship between oral dysbiosis and oral (e.g. periodontal disease or dental caries) or systemic diseases (e.g. diabetes, cancer, endocarditis, systemic infections), where the unbalanced microbiome may be the trigger for the pathology, has been demonstrated (Albuquerque-Souza and Sahingur 2022; Al-Qadami et al. 2022; Khor et al. 2021). The interplay between the external factors and the colonization or proliferation in the oral cavity of some bacterial groups may be the key to prevent and control some pathologies. In addition, this is a crucial interface to better understand the environment-humans continuum that is implicitly assumed by the One Health concept.
Supplementary information
Below is the link to the electronic supplementary material.
Author contribution
CM conceived and designed the research. IL, TBC, and VH performed the literature search and analysed taxa frequency data. All authors assessed the properties of the most cited taxa. CM analysed oral-gut taxa phylogenetic relatedness and collected examples taxa associated diseases. All authors contributed for writing, and all read and approved the manuscript.
Funding
Open access funding provided by FCT|FCCN (b-on). This study was funded by (1) FEDER through project RISK.AR “Assessing the risks associated with environmental antibiotic resistant bacteria: propagation and transmission to humans” (PTDC/CTA-AMB/28196/2017) – Programa Operacional Competitividade e Internacionalização and by (2) the European Regional Development Fund through COMPETE 2020 – Programa Operacional Competitividade e Internacionalização (POCI) and by Portuguese public funds through FCT (Fundação para a Ciência e a Tecnologia) (PTDC/MEC-MCI/29777/2017) with the institutional support of FCT UID/Multi/50016/2020.
Data Availability
Information about the sources of data supporting the results are reported throughout the text, with indication of the respective websites and access date.
Declarations
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Inês Leão, Teresa Bento de Carvalho, and Valentina Henriques contributed equally to this study.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque-Souza E, Sahingur SE. Periodontitis, chronic liver diseases, and the emerging oral-gut-liver axis. Periodontol 2000. 2022;89(1):125–141. doi: 10.1111/prd.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander Mahnert A, Moissl-Eichinger C, Zojer M, Bogumil D, Mizrahi I, Rattei T, Martinez JL, Berg G. Man-made microbial resistances in built environments. Nat Commun. 2019;10(1):968. doi: 10.1038/s41467-019-08864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, Thaiss CA, Sato M, Toyooka K, Said HS, Yamagami H, Rice SA, Gevers D, Johnson RC, Segre JA, Chen K, Kolls JK, Elinav E, Morita H, Xavier RJ, Hattori M, Honda K (2017) Ectopic colonization of oral bacteria in the intestine drives T H 1 cell induction and inflammation. Science (80) 358:359–365. 10.1126/science.aan4526 [DOI] [PMC free article] [PubMed]
- Baydaş B, Uslu H, Yavuz İ, Ceylan İ, Dağsuyu İM. Effect of a chronic nail-biting habit on the oral carriage of Enterobacteriaceae. Oral Microbiol Immunol. 2007;22:1–4. doi: 10.1111/j.1399-302X.2007.00291.x. [DOI] [PubMed] [Google Scholar]
- Becerra-Castro C, Macedo G, Silva AMT, Manaia CM, Nunes OC. Proteobacteria become predominant during regrowth after water disinfection. Sci Total Environ. 2016;573:313–323. doi: 10.1016/j.scitotenv.2016.08.054. [DOI] [PubMed] [Google Scholar]
- Blaustein R, Michelitsch LM, Glawe AJ, Lee H, Huttelmaier S, Hellgeth N, Ben Maamar S, Hartmann EM. Toothbrush microbiomes feature a meeting ground for human oral and environmental microbiota. Microbiome. 2021;9(1):32. doi: 10.1186/s40168-020-00983-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham ZM, Garneau NL, Comstock SS, Tucker RM, Knight R, Metcalf JL. Patterns of oral microbiota diversity in adults and children: a crowdsourced population study. Sci Rep. 2020;10:2133. doi: 10.1038/s41598-020-59016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calheiros Cruz G, Sousa M, Vilela S, Costa FTE, Silva FJ. Lautropia mirabilis: an exceedingly rare cause of peritoneal dialysis-associated peritonitis. Case Rep Nephrol Dial. 2022;12(2):81–84. doi: 10.1159/000524494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Kemp M, Bruun NE, Bangsborg JM, Højlyng N, Hesselbjerg A, Dargis R, Christensen JJ. Cardiobacterium valvarum infective endocarditis and phenotypic/molecular characterization of 11 Cardiobacterium species strains. J Med Microbiol. 2011;60:522–528. doi: 10.1099/jmm.0.025353-0. [DOI] [PubMed] [Google Scholar]
- Chen Q-L, Cui H-L, Su J-Q, Penuelas J, Zhu Y-G. Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 2019;24:530–541. doi: 10.1016/j.tplants.2019.02.010. [DOI] [PubMed] [Google Scholar]
- Chen ZD, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Fung C, Wong W, Liu C. Brevundimonas bacteremia: two case reports and literature review. Scand J Infect Dis. 2004;36:59–61. doi: 10.1080/00365540310018879. [DOI] [PubMed] [Google Scholar]
- Colombo APV, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa CFFA, Merino-Ribas A, Ferreira C, Campos C, Silva N, Pereira L, Garcia A, Azevedo Á, Mesquita RBR, Rangel AOSS, Manaia CM, Sampaio-Maia B (2021) Characterization of oral Enterobacteriaceae prevalence and resistance profile in chronic kidney disease patients undergoing peritoneal dialysis. Front Microbiol 12:736685. 10.3389/fmicb.2021.736685 [DOI] [PMC free article] [PubMed]
- Cross KL, Chirania P, Xiong W, Beall CJ, Elkins JG, Giannone RJ, Griffen AL, Guss AM, Hettich RL, Joshi SS, Mokrzan EM, Martin RK, Zhulin IB, Leys EJ, Podar M (2018) Insights into the evolution of host association through the isolation and characterization of a novel human periodontal pathobiont, Desulfobulbusoralis. MBio 9(2):e02061-17. 10.1128/mBio.02061-17 [DOI] [PMC free article] [PubMed]
- Cunningham AA, Daszak P, Wood JLN. One Health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc B Biol Sci. 2017;372:20160167. doi: 10.1098/rstb.2016.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger B, Ripplinger C, Blondeau J, Blondeau L, Peermohamed S. Bacteremia and polyarticular septic arthritis secondary to Moraxella bovis in a pregnant patient with HIV who injects drugs. J Assoc Med Microbiol Infect Dis Can. 2022;7(2):146–149. doi: 10.3138/jammi-2021-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper SG, Mitchell HL, Lê Cao K-A, Carpenter L, Gussy MG, Calache H, Gladman SL, Bulach DM, Hoffmann B, Catmull DV, Pruilh S, Johnson S, Gibbs L, Amezdroz E, Bhatnagar U, Seemann T, Mnatzaganian G, Manton DJ, Reynolds EC. Temporal development of the oral microbiome and prediction of early childhood caries. Sci Rep. 2019;9:19732. doi: 10.1038/s41598-019-56233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122–128. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Teil EM, Gabarrini G, Harmsen HJM, Westra J, van Winkelhoff AJ, van Dijl JM. Talk to your gut: the oral-gut microbiome axis and its immunomodulatory role in the etiology of rheumatoid arthritis. FEMS Microbiol Rev. 2019;43:1–18. doi: 10.1093/femsre/fuy035. [DOI] [PubMed] [Google Scholar]
- Edgar M, Dawes C, O´Mullane D (2012) Anatomy and physiology of salivary glands, 4th ed. Stephen Hancocks Limited
- Ferro P, Vaz-Moreira I, Manaia CM. Betaproteobacteria are predominant in drinking water: are there reasons for concern? Crit Rev Microbiol. 2019;45:649–667. doi: 10.1080/1040841X.2019.1680602. [DOI] [PubMed] [Google Scholar]
- Freire M, Moustafa A, Harkins DM, Torralba MG, Zhang Y, Leong P, Saffery R, Bockmann M, Kuelbs C, Hughes T, Craig JM, Nelson KE. Longitudinal study of oral microbiome variation in twins. Sci Rep. 2020;10:7954. doi: 10.1038/s41598-020-64747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity G, Brenner DJ, Krieg NR, Staley JR (2005) Bergey’s Manual of Systematic Bacteriology: Volume 2 : The Proteobacteria. Springer US
- Ghanyah Al-Qadami G, Van Sebille Y, Bowen J, Wardill H. Oral-gut microbiome axis in the pathogenesis of cancer treatment-induced oral mucositis. Front Oral Health. 2022;28(3):881949. doi: 10.3389/froh.2022.881949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Firestone ND, Gross EL, DiFranco JM, Hardman JH, Vriesendorp B, Faust RA, Janies DA, Leys EJ. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS ONE. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiya H, Kimura K, Nishi I, Yamamoto N, Yoshida H, Akeda Y, Tomono K. Desulfovibrio desulfuricans bacteremia: a case report and literature review. Anaerobe. 2018;49:112–115. doi: 10.1016/j.anaerobe.2017.12.013. [DOI] [PubMed] [Google Scholar]
- Hayes RB, Ahn J, Fan X, Peters BA, Ma Y, Yang L, Agalliu I, Burk RD, Ganly I, Purdue MP, Freedman ND, Gapstur SM, Pei Z. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018;4:358. doi: 10.1001/jamaoncol.2017.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Pal C, Sulaiman IM, Jia C, Zerwekh T, Dowd SE, Banerjee P. Application of high-throughput pyrosequencing in the analysis of microbiota of food commodities procured from small and large retail outlets in a U.S. metropolitan area – a pilot study. Food Res Int. 2018;105:29–40. doi: 10.1016/j.foodres.2017.10.057. [DOI] [PubMed] [Google Scholar]
- Hisham Altayb H, Chaieb K, Baothman O, Alzahrani FA, Zamzami MA, Almugadam BS. Study of oral microbiota diversity among groups of families originally from different countries. Saudi J Biol Sci. 2022;29(7):103317. doi: 10.1016/j.sjbs.2022.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwauchi M, Horigome A, Ishikawa K, Mikuni A, Nakano M, Xiao J, Odamaki T, Hironaka S. Relationship between oral and gut microbiota in elderly people. Immun Inflamm Dis. 2019;7:229–236. doi: 10.1002/iid3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak JL, Yan Y, Zhang Q, Wang L, Ge L. The role of oral microbiome in respiratory health and diseases. Respir Med. 2021;185:106475. doi: 10.1016/j.rmed.2021.106475. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Liu J, Chen L, Gan N, Yang D (2019) The oral microbiome in the elderly with dental caries and health. Front Cell Infect Microbiol 8:442. 10.3389/fcimb.2018.00442 [DOI] [PMC free article] [PubMed]
- Jordi Rello J, Kalwaje Eshwara V, Lagunes L, Alves J, Wunderink RG, Conway-Morris A, Rojas JN, Alp E, Zhang Z. A global priority list of the TOp TEn resistant Microorganisms (TOTEM) study at intensive care: a prioritization exercise based on multi-criteria decision analysis. Eur J Clin Microbiol Infect Dis. 2019;38(2):319–323. doi: 10.1007/s10096-018-3428-y. [DOI] [PubMed] [Google Scholar]
- Khor B, Snow M, Herrman E, Ray N, Mansukhani K, Patel KA, Said-Al-Naief N, Maier T, Machida CA. Interconnections between the oral and gut microbiomes: reversal of microbial dysbiosis and the balance between systemic health and disease. Microorganisms. 2021;9:496. doi: 10.3390/microorganisms9030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG, Hayashi A, Imai J, Sugihara K, Miyoshi M, Brazil JC, Kuffa P, Hill BD, Rizvi SM, Wen F, Bishu S, Inohara N, Eaton KA, Nusrat A, Lei YL, Giannobile WV, Kamada N. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182:447–462.e14. doi: 10.1016/j.cell.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto S, Kamada N. Periodontal connection with intestinal inflammation: microbiological and immunological mechanisms. Periodontol 2000. 2022;89(1):142–153. doi: 10.1111/prd.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Pop M. ARDB-antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Arimatsu K, Kato T, Matsuda Y, Minagawa T, Takahashi N, Ohno H, Yamazaki K (2015) Oral administration of P. gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of enterobacteria to the liver. PLoS One 10:e0134234. 10.1371/journal.pone.0134234 [DOI] [PMC free article] [PubMed]
- Nasidze I, Li J, Schroeder R, Creasey JL, Li M, Stoneking M. High diversity of the saliva microbiome in Batwa Pygmies. PLoS One. 2011;6:e23352. doi: 10.1371/journal.pone.0023352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazareno Scaccia N, Vaz-Moreira I, Manaia CM. The risk of transmitting antibiotic resistance through endophytic bacteria. Trends Plant Sci. 2021;26(12):1213–1226. doi: 10.1016/j.tplants.2021.09.001. [DOI] [PubMed] [Google Scholar]
- Nørskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., <i>Aggregatibacter aphr. Int J Syst Evol Microbiol. 2006;56:2135–2146. doi: 10.1099/ijs.0.64207-0. [DOI] [PubMed] [Google Scholar]
- Nseir W, Khateeb J, Awawdeh M, Ghali M. Catheter-related bacteremia caused by Comamonas testosteroni in a hemodialysis patient. Hemodial Int. 2011;15:293–296. doi: 10.1111/j.1542-4758.2010.00524.x. [DOI] [PubMed] [Google Scholar]
- Osterhaus ADME, Vanlangendonck C, Barbeschi M, Bruschke CJM, Christensen R, Daszak P, de Groot F, Doherty P, Drury P, Gmacz S, Hamilton K, Hart J, Katz R, Longuet C, McLeay J, Morelli G, Schlundt J, Smith T, Suri S, Umali K, van Aken J, Wagenaar JA. Make science evolve into a One Health approach to improve health and security: a white paper. One Health Outlook. 2020;2(1):6. doi: 10.1186/s42522-019-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70:5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BA, McCullough ML, Purdue MP, Freedman ND, Um CY, Gapstur SM, Hayes RB, Ahn J. Association of coffee and tea intake with the oral microbiome: results from a large cross-sectional study. Cancer Epidemiol Biomarkers Prev. 2018;27:814–821. doi: 10.1158/1055-9965.EPI-18-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaic A, Kapila YL. The oralome and its dysbiosis: new insights into oral microbiome-host interactions. Comput Struct Biotechnol J. 2021;27(19):1335–1360. doi: 10.1016/j.csbj.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017:1–7. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio-Maia B, Caldas IM, Pereira ML, Pérez-Mongiovi D, Araujo R (2016) The oral microbiome in health and its implication in oral and systemic diseases. In: Advances in Applied Microbiology. Elsevier Ltd, pp 171–210 [DOI] [PubMed]
- Sanz-García F, Gil-Gil T, Laborda P, Ochoa-Sánchez LE, Martínez JL, Hernando-Amado S. Coming from the wild: multidrug resistant opportunistic pathogens presenting a primary, not human-Linked, environmental habitat. Int J Mol Sci. 2021;22:8080. doi: 10.3390/ijms22158080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers S, Li L, Ong E, Deng S, Fu G, Lin Y, Yang B, Zhang S, Fa Z, Zhao B, Xiang Z, Li Y, Zhao X-M, Olszewski MA, Chen L, He Y. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 2019;47:D693–D700. doi: 10.1093/nar/gky999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Nabin KC, Bastola C, Jahir T, Risal R, Thapa S, Enriquez D, Schmidt F. Pantoea agglomerans: an elusive contributor to chronic obstructive pulmonary disease exacerbation. Cureus. 2021;13:e18562–e18562. doi: 10.7759/cureus.18562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Torralba MG, Moncera KJ, DiLello L, Petrini J, Nelson KE, Pieper R. Gastro-intestinal and oral microbiome signatures associated with healthy aging. GeroScience. 2019;41:907–921. doi: 10.1007/s11357-019-00098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence snalysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- Sulyanto RM, Thompson ZA, Beall CJ, Leys EJ, Griffen AL. The predominant oral microbiota is acquired early in an organized pattern. Sci Rep. 2019;9:10550. doi: 10.1038/s41598-019-46923-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z-Z, Chen G, Hong Q, Huang S, Smith HM, Shah RD, Scholz M, Ferguson JF (2019) Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front Genet 10:454. 10.3389/fgene.2019.00454 [DOI] [PMC free article] [PubMed]
- Theuretzbacher U, Outterson K, Engel A, Karlén A. The global preclinical antibacterial pipeline. Nat Rev Microbiol. 2020;18(5):275–285. doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney BT, Yang Z, Luber JM, Beaudin M, Wibowo MC, Baek C, Mehlenbacher E, Patel CJ, Kostic AD. The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe. 2019;26:283–295.e8. doi: 10.1016/j.chom.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuganbaev T, Yoshida K, Honda K (2022) The effects of oral microbiota on health. Science 27;376(6596):934–936. 10.1126/science.abn1890 [DOI] [PubMed]
- Vaz-Moreira I, Nunes OC, Manaia CM. Ubiquitous and persistent Proteobacteria and other Gram-negative bacteria in drinking water. Sci Total Environ. 2017;586:1141–1149. doi: 10.1016/j.scitotenv.2017.02.104. [DOI] [PubMed] [Google Scholar]
- Vaz-Moreira I, Nunes OC, Manaia CM. Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol Rev. 2014;38(4):761–778. doi: 10.1111/1574-6976.12062. [DOI] [PubMed] [Google Scholar]
- Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Wang J, Feng J, Zhu Y, Li D, Wang J, Chi W. Diversity and biogeography of human oral saliva microbial communities revealed by the earth microbiome project. Front Microbiol. 2022;2022(13):931065. doi: 10.3389/fmicb.2022.931065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Song F, Gu H, Wei X, Zhang K, Zhou Y, Luo H. Comparative evaluation of the salivary and buccal mucosal microbiota by 16S rRNA sequencing for forensic investigations. Front Microbiol. 2022;13:777882. doi: 10.3389/fmicb.2022.777882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74(16):2959–2977. doi: 10.1007/s00018-017-2509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JR, Gabaldón T. The human oral microbiome in health and disease: from sequences to ecosystems. Microorganisms. 2020;8:308. doi: 10.3390/microorganisms8020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yangheng Zhang Y, Wang X, Li H, Ni C, Du Z, Yan F. Human oral microbiota and its modulation for oral health. Biomed Pharmacother. 2018;99:883–893. doi: 10.1016/j.biopha.2018.01.146. [DOI] [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7(1–2):203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zhigang Ren Z, Wang H, Cui G, Lu H, Wang L, Luo H, Chen X, Ren H, Sun R, Liu W, Liu X, Liu C, Li A, Wang X, Rao B, Yuan C, Zhang H, Sun J, Chen X, Li B, Hu C, Wu Z, Yu Z, Kan Q, Li L. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut. 2021;70(7):1253–1265. doi: 10.1136/gutjnl-2020-323826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Tang D, Wei S, Hu Z, Wang X, Luo D. Ralstonia mannitolilytica sepsis after elective cesarean delivery: a case report. BMC Pregnancy Childbirth. 2021;21:737. doi: 10.1186/s12884-021-04214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Information about the sources of data supporting the results are reported throughout the text, with indication of the respective websites and access date.