Abstract
The microbiota has received plenty of attention in recent years due to its influence on host health and productivity. The striped eggs have reduced hatching performance and resulted in economic loss. The reasons are still unknown. Microbiota is one of the potentially important factors contributing to striped egg formation. This study investigates the relationship between the microbiota and striped eggs. The litter samples, feed samples, and cloacal swab samples of female ducks that produce striped eggs and normal eggs were performed for microbial diversity and composition using 16S rRNA sequencing. The results showed that there was no significant difference between feed microbiota and cloacal swab microbiota by alpha diversity, whereas, the number of microorganisms in the litter samples of female ducks that produced striped eggs was less than those of female ducks with normal eggs. There were compositional differences in litter microbiota of female ducks between the striped egg and the normal eggs. Among them, the abundance of Staphylococcus, Corynebacterium, and Brevibacterium in the litter of female ducks that produced striped eggs was significantly higher than that produced normal eggs. And these differential bacteria maybe affect the health of female ducks and cause abnormalities in the formation process of duck eggs. Therefore, the reduction of harmful bacteria may protect the reproductive health of female ducks and decrease the proportion of striped eggs. It provides an important reference to explore why female ducks produce striped eggs.
Key words: duck, striped eggs, microbiota, litter microbiota
INTRODUCTION
Poultry eggs are rich in protein, lipids, and other nutrients, making them a relatively inexpensive source of high-quality protein (Gao et al., 2021). With the increasing population of China, the demand for duck eggs is also increasing (Hou and Liu, 2022). The importance of good quality eggs is self-evident. In recent years, it has been found that there is a phenomenon of duck striped eggs, that is, oblique striped strips of different depths can be seen on the surface of duck eggs (Sang et al., 2022). Striped eggs had lower eggshell weight, thick albumen weight, and thin albumen crude protein than normal eggs, which caused lower hatchability (Yuan et al., 2013). So, it brings great trouble to the breeding process and causes considerable economic losses. Until now, there are few studies on the specific causes of striped eggs.
Egg quality is affected by many factors such as feed, heredity, age, and environment (Nasri et al., 2020). There are many kinds of microorganisms anywhere, and the microbes in different environments have different functions (Tropini et al., 2017). It has been reported that intestinal microbes and their metabolites act as signaling molecules linking the gut, liver, brain, and reproductive tract (Nicholson et al., 2012), which in turn has a direct or indirect impact on poultry health and egg quality (Zhan et al., 2019; Feng et al., 2021). And in rearing, it is also important for the influence of microbiota in the environment on animal production. Wang et al. showed that fresh and reused litter affects the chicken GI microbiota, which may impact the host's nutritional status and intestinal health (Wang et al., 2016). When laying hens are infected with pathogenic bacteria like Salmonella, intestinal Salmonella will spread to the oviduct through the cloaca (Gantois et al., 2009). Furthermore, Salmonella alters the expression of toll-like receptors, NOD-like receptors, β-defensins, and cytokine family genes in the oviduct, resulting in the decline of egg quality (Zhang et al., 2019). Studies have shown that duck excrement contains a large number of different pathogenic bacteria (Jeong et al., 2021; Wang et al., 2021), which will affect the composition of litter microorganisms in the duck house. It has a certain impact on the health of ducks and egg quality.
Previous studies on striped eggs mainly focused on the analysis of the differences between striped and normal laying ducks, but there are few studies about the effects of environmental microbiota on striped eggs. This study explored the influence of cloacal swabs, litter, and feed microbiota on striped eggs through 16S rRNA sequencing, and provided a research basis for studying the causes of striped egg production.
MATERIALS AND METHODS
Animals and Sample Collection
A total of 260 female Pekin ducks from a purebred line were used in the present study. All the ducks were reared individually in each cage from the beginning and managed according to the Pekin duck rearing standards. The cage is 1 meters long, 0.8 meters wide, and 1 meters high. The duck house is semienclosed with 17 h of light a day. The duck house were auto ventilated by monitoring ammonia, and temperature ranged from 15°C to 25°C during experiment. The ducks were provided ad libitum commercial diets: a diet containing 21% crude protein (CP), 2,800 MJ/kg dietary metabolizable energy (ME). When the female ducks were 66 wk old, the eggs were collected for 2 or 3 consecutive days. The striped eggs could be classified into 1 to 4 categories (SANG et al., 2022). Based on the level of laying striped eggs, 15 female ducks that produced striped eggs were assigned to the striped egg group, and 15 female ducks that produced normal eggs were assigned to the normal egg group. All samples were collected when the female ducks had mostly finished feeding for the day. The litter sample of the cage where the female duck was located was taken, and the sterile cotton swab was used to sample the litter at 5 points.
A total of 90 samples were collected, 45 from each of the striped egg and normal egg groups, comprised of the cloacal swabs, litter, and feed. Each duck was collected the cloacal swabs, the litter, and the feed. Sterile cotton swabs were taken from inside the cloaca of the female duck and placed in 2 mL cryopreserved tubes. And the samples were stored in liquid nitrogen. Subsequently, the leftover feed of one day's feed trough of the female duck was taken and put in a sterile sealed bag. The samples were stored at −80°C. Meanwhile, the litter samples of the cage where the female ducks were housed were taken. Sterile cotton swabs were used to sample the litter at five points. The litter was stored in liquid nitrogen.
DNA Extraction and Sequencing
Total bacterial genomic DNA samples were extracted using the Fast DNA Spin Kits for soil (MP Biomedicals, Santa Ana, CA) according to the manufacturer's instructions. The completeness of the DNA sample was then assessed by 1% agarose gel electrophoresis. The V3 to V4 hypervariable region of the bacterial 16S rRNA gene was amplified with the primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’) (Munyaka et al., 2015). The PCR was carried out on a Master cycler Gradient (Eppendorf, Germany) using 25 μL reaction volumes, containing 12.5 μL 2 × Taq PCR MasterMix, 3 μL BSA(2 ng/μL), 1 μL Forward Primer (5 μM),1 μL Reverse Primer(5 μM), 2 μL template DNA, and 5.5 μL ddH2O. Cycling parameters were 95°C for 5 min, followed by 28 cycles of 95°C for 45 s, 55°C for 50 s, and 72°C for 45 s with a final extension at 72°C for 10 min. The raw paired-end (PE) reads data was obtained through the Illumina Miseq platform.
Bioinformatics Analysis
After sample splitting of PE reads obtained by MiSeq sequencing, quality control and filtering of double-ended reads were performed based on sequencing quality. In brief, use FASTP and FLASH to optimize and filter the data. The bases of the tail quality value of reads below 20, the reads below 50 bp, and the reads containing N bases were filtered. The sequence information for each sample is presented in Supplementary Material Table S1.
Qualified tags were denoized into amplicon sequence variants (ASVs) by DADA2 (Callahan et al., 2016, p. 2) or Deblur (Amir et al., 2017). The BLAST tool (Altschul et al., 1990) was used to classify all sequences into different taxonomic groups against the Silva138 database (Quast et al., 2013). The Alpha diversity analysis of microbial communities reflects the species richness and the diversity of microorganisms in the samples including the Chao index, Sobs index, Shannon index, and Simpson index. QIIME (v1.8.0) was used to calculate Alpha diversity based on the ASV information (Caporaso et al., 2010). To examine the similarity between different samples, each sample analyzed clustering and PCoA by R (v3.6.0) (Wang et al., 2012).
Statistical Analysis
The community structure data of microbial flora was presented in the form of an average. The Chao index, Sobs index, Shannon index, and Simpson index were analyzed by one-way ANOVA, and Duncan's multiple comparisons were performed for those with significant differences. Kruskal-Wallis rank sum test was used for the differences in microbial community structure. R (v3.6.0) software was used for basic statistical analysis.
RESULTS
Sequencing Output and Alpha Diversity of the Samples
The 16S rRNA gene sequencing analysis produced a total of 3,929,490 quality-filtered sequences from 90 samples with an average length of 416 bp. Denoizing was performed on the optimized data, and the statistical sequence information was shown in Table S2. Comparing ASVs numbers between groups, the number of ASVs in cloacal swabs of female ducks that produce striped eggs and normal eggs was significantly different, the number of ASVs in the litter of female ducks was similar, and the number of ASVs in the feed was little difference (Table S3). We subsequently classified the ASVs into 1,666 species, 983 genera, 395 families, 207 orders, 85 classes, and 31 phyla. The Alpha diversity indices showed a significant difference in ASVs richness between striped duck eggs and normal duck eggs (Figure 1, Table S4). Noticeably, there were no significant differences in cloacal swabs and feed, while there were significant differences in the litter between the Sobs index and the Chao index (P < 0.05).
Figure 1.
Comparison of the microbiome Alpha diversity in the cloacal swabs, the little and the feed of female ducks that produced striped eggs and normal eggs, respectively. (A) Sobs index, (B) the Chao index, (C) Shannon index (D) Simpson index of Alpha diversity at the ASV level. Boxes denote the interquartile (IQR) between the first and third quartiles (25th and 75th percentiles, respectively) and the line inside denotes the median. Whiskers denote the lowest and highest values within 1.5 times and the IQR from the first and third quartiles, respectively. Anal_Nor, Litter_Nor, and Feed_Nor are cloacal swab samples, litter samples, and feed samples of female ducks that produced normal eggs, respectively. Anal_Str, Litter_Str, and Feed_Str are cloacal swab samples, litter samples, and feed samples of female ducks that produced striped eggs.
Differences in the Diversity and Composition Feature of Cloacal Microbiota
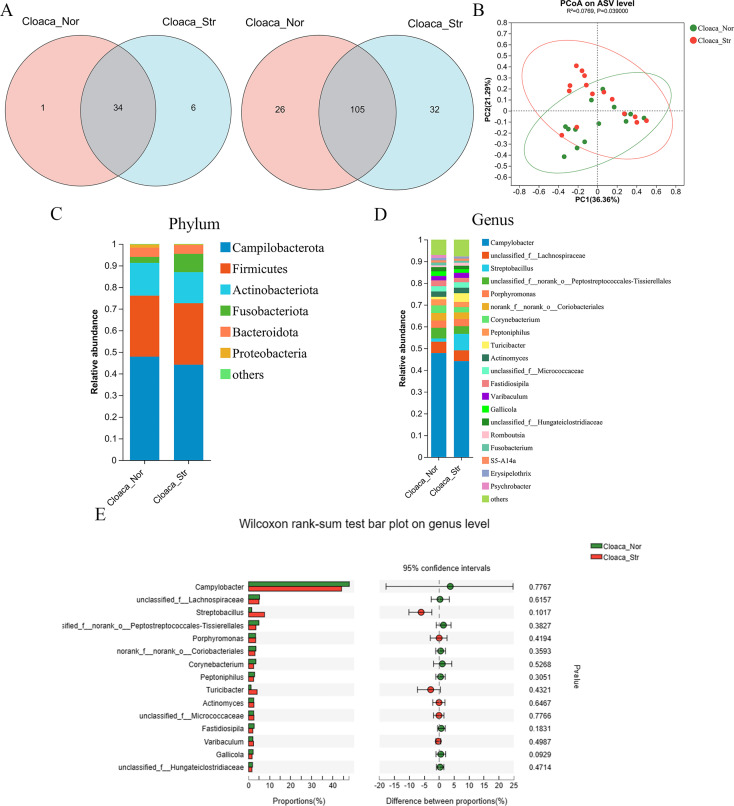
A Venn diagram was created to show the number of orders and genera shared or unique to cloacal swabs of female ducks that produced striped eggs and normal eggs (Figure 2A). At the order level, the microbiota of the Cloaca_Nor and Cloaca_Str groups shared 34 orders, whereas 1 and 6 orders were uniquely identified in the Cloaca_Nor and Cloaca_Str. At the genus level, the cloacal microbiota of the two groups shared 105 genera, with 26 and 32 genera uniquely identified in the Cloaca_Nor and Cloaca_Str group, respectively. The principal coordinates analysis (PCoA) of ASVs indicated that the microbiota is an insignificant difference between the striped egg ducks and normal egg ducks (Figure 2B).
Figure 2.
Compositions of the cloacal microbiota of female ducks that produced the striped eggs and normal eggs. (A) The number of orders and genera shared by the two groups are shown in Venn diagrams. (B) The principal coordinates analysis (PCoA) plot at the ASV level. (C) Relative abundances in the dominant phyla across the cloacal microbiota. (D) Relative abundances of dominant genera across the cloacal microbiota. Only the genera with an abundance of >1.0% in any segment were plotted. (E) The genera represented at significantly different levels in the microbiota of the two groups were shown in an extended error bar plot. The differences in the compositions were tested using Wilcoxon rank-sum test, and P < 0.05 was marked with “*”.
Campylobacter, Firmicutes, and Actinobacteriota were the dominant phyla in cloacal swabs (Figure 2C). At the genus level, Campylobacter (47.87% in the Cloaca_Nor group and 44.17% in the Cloaca_Str group), Lachnospiraceae (5.20% in the Cloaca_Nor group and 4.95% in the Cloaca_Str group), and Streptobacillus (1.51% in the Cloaca_Nor group and 7.57% in the Cloaca_Str group) were enriched in cloacal swabs (Figure 2D). Then the difference analysis found that there was no significant difference between Cloaca_Nor and Cloaca_Str among the top 15 genera (Figure 2E).
Differences in the Diversity and Composition Feature of Litter Microbiota
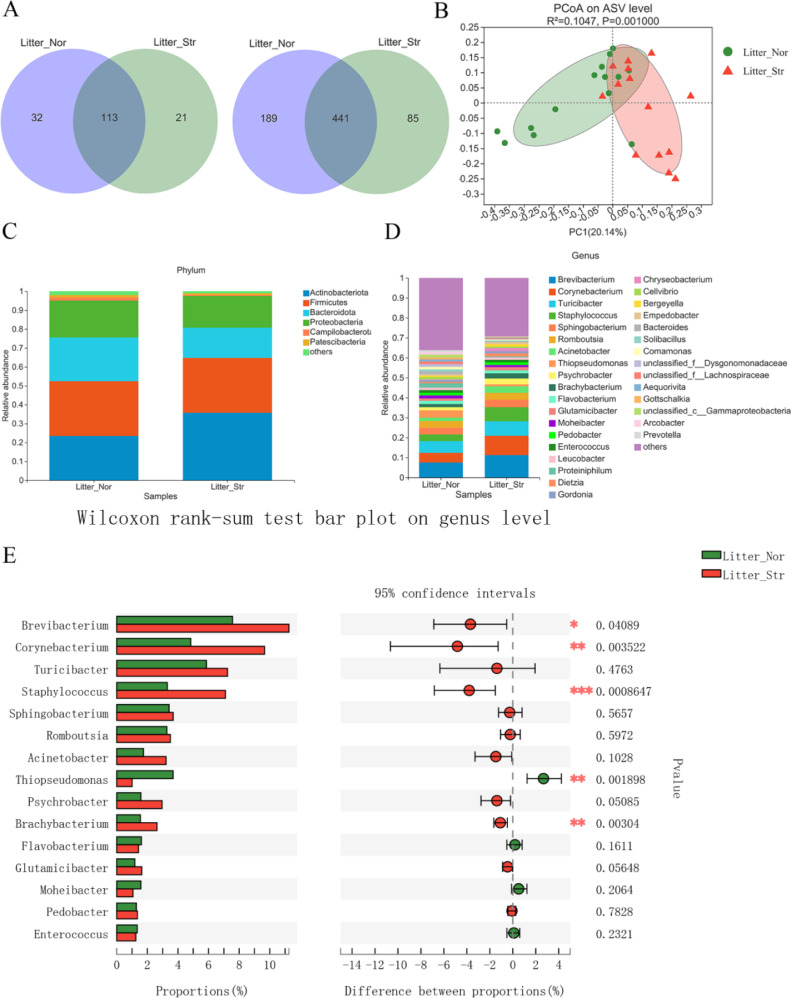
At the order level, the microbiota of the Litter_Nor and Litter_Str groups shared 113 orders, 32 orders were unique to Litter_Nor and 21 orders were unique to Litter_Str (Figure 3A). At the genus level, the microbiota of the Litter_Nor and Litter_Str groups shared 441 genera, whereas 189 and 85 genera were uniquely identified in the Litter_Nor group and the Litter_Str group. The PCoA of ASVs indicated that the litter microbiota was different in female ducks that produced striped eggs and normal eggs (Figure 3B).
Figure 3.
Compositions of the litter microbiota of female ducks that produced striped eggs and normal eggs. (A) The number of orders and genera shared by the two groups are shown in Venn diagrams. (B) The principal coordinates analysis (PCoA) plot at the ASV level. (C) Relative abundances in the dominant phyla of the litter microbiota. (D) Relative abundances of dominant genera of the litter microbiota. Only the genera with an abundance of > 1.0% in the litter were plotted. (E) The genera represented at significantly different levels in the microbiota of the two groups were shown in an extended error bar plot. The differences in the compositions were tested using Wilcoxon rank-sum test, and P < 0.05 was marked with “*”.
Firmicutes, Actinobacteria, and Bacteroidota were the main components of the microbial community, and the total proportion of the three was more than 70% (Figure 3C). At the genus level, the 4 dominant genera detected in both groups were Brevibacterium (7.56% in the Litter_Nor group and 11.25% in the Litter_Str group), Corynebacterium (4.84% in the Litter_Nor group and 9.66% in the Litter_Str group), Turicibacter (5.86% in the Litter_Nor group and 7.24% in the Litter_Str group) and Staphylococcus (3.31% in the Litter_Nor group and 7.10% in the Litter_Str group) (Figure 3D).
Then, we compared the relative abundance of microbiota between the two groups and found significantly different phyla and genera (P < 0.05). Between Litter_Nor and Litter_Str, Actinobacteria, Bacteroidota, Campylobacter, Patescibacteria, Desulfobacterota, and Fibrobacterota were significantly different (Figure S1). At the genus level, the abundance of Corynebacterium, Brevibacterium, staphylococcus, Thiopseudomonas, and Brachybacterium in the litter of female ducks that produced striped eggs was significantly higher than that produced normal eggs (Figure 3E).
Differences in the Diversity and Composition Feature of Feed Microbiota
At the order level, the microbiota of the Feed_Nor and Feed_Str groups shared 117 orders, whereas 27 orders were unique to Feed_Nor and 30 orders were unique to Feed_Str (Figure 4A). At the genus level, there were shared 431 genera in the feed microbiota, whereas 154 and 158 genera were uniquely identified in the Feed_Nor group and the Feed_Str group (Figure 4A). PCoA was used to compare the total microbial composition of the Feed_Nor and Feed_Str groups at the ASV, but the microbial communities were not separated (Figure 4B).
Figure 4.
Compositions of the feed microbiota of female ducks with striped eggs and normal eggs. (A) The number of orders and genera shared by the two groups were shown in Venn diagrams. (B) The principal coordinates analysis (PCoA) plot at the ASV level. (C) Relative abundances in the dominant phyla across the feed microbiota. (D) Relative abundances of dominant genera across the feed microbiota. Only the genera with an abundance of >1.0% in any segment were plotted. (E) The genera represented at significantly different levels in the microbiota of the two groups were shown in an extended error bar plot. The differences in the compositions were tested using Wilcoxon rank-sum test, and P < 0.05 was marked with “*”.
Firmicutes, Cyanobacteria, and Actinobacteria were the main components of the microbial community, and the total proportion of the three was more than 92% (Figure 4C). At the genus level, The dominant genera detected in both groups were Staphylococcus (61.65% in the Feed_Nor group and 51.53% in the Feed_Str group), Brevibacterium (2.11% in the Feed_Nor group and 1.68% in the Feed_Str group), Corynebacterium (2.53% in the Feed_Nor group and 2.92% in the Feed_Str group), and Chloroplast (13.64% in the Feed_Nor group and 21.68% in the Feed_Str group) (Figure 4D). Then, we found some significantly different phyla and genera between the 2 groups (P < 0.05). Between Feed_Nor and Feed_Str, Myxococcota was significantly different at the phylum level (P <0.05, Figure S2). At the genus level, the abundance of Dietzia in the Feed_Str was significantly higher than those in the Feed_Nor group (Figure 4E).
DISCUSSION
The hatching rate and quality of breeding eggs are important to control indexes in the current breeding process, so abnormal eggs should be decreased as much as possible. Striped eggs showed significantly lower fertility and hatchability compared with normal eggs (Yuan et al., 2013). We need to understand why female ducks produced striped eggs. The microbiota of the poultry farm environment, including pathogenic bacteria, play an important role in the microbial colonization and development of the hen's gastrointestinal tract during production (Cressman et al., 2010; Wang et al., 2016). In addition to direct effects on the egg quality and safety via vertical transmission route in the intestine-oviduct-egg, intestinal microbiota and its metabolites such as SCFAs, BA, and tryptophan derivatives are indirectly involved in regulating egg quality through the microbiota-gut-liver/brain-reproductive tract axis (Dai et al., 2022).
Alpha diversity showed that the number of microorganisms in the litter of female ducks that produced striped eggs was less than those that produced normal eggs. And there was no significant difference in the feed and cloacal swab samples. This phenomenon may be due to the maturation of some environmental microbial communities, which leads to the reduction of microbial communities through the competition of microbial communities and the filtering effect of the environment (Bearson et al., 2013; Rivera-Chávez and Bäumler, 2015; Redweik et al., 2019; Rogers et al., 2021). Lower microbial diversity indicates a reduced ability of the microbiota to maintain metabolic and cellular functionality (Lee and Hase, 2014; Polansky et al., 2015). The results of PCoA showed that there was no significant difference in the microbial community between the control group and the experimental group of the cloacal swabs and feed, while there was a significant difference between the control group and the experimental group of the litter. It may indicate that the litter microbiota is not only affected by fecal microbiota, but also by other factors such as the environment.
We identified specific microbes in the feed, litter, and cloacal swabs at the phyla and genus levels of taxa to provide novel information regarding the diversity and composition of microbiota about striped eggs. In this study, Firmicutes and Actinobacteria are common dominant phyla. Firmicutes participate in the metabolism of energy substances and play an important role in the digestion of food (Mariat et al., 2009; Ma et al., 2018). Actinobacteria has a vast pharmacological potential that remains unopposed among other microbial groups (Puttaswamygowda et al., 2019).
At the genus level, Campylobacter, Lachnospiraceae, and Streptobacillus were enriched in cloacal swabs. Brevibacterium, Corynebacterium, and Turicibacter were enriched in the litter. The feed riched in Staphylococcus, Brevibacterium, Corynebacterium, and Chloroplast. The dominant genera in the litter and feed samples were similar. It indicates that litter microbiota and feed microbiota may interact with each other (Radon et al., 2002; Weidhaas et al., 2010; O'Brien et al., 2016). No differential bacterial genera were found in the cloacal swab microbiota, suggesting that cloacal swab microbes are not the main factor responsible for striped eggs. The differences in the litter and feed microbiota between groups may be related to the striped eggs. Corynebacterium, Brevibacterium, staphylococcus, Thiopseudomonas, and Brachybacterium were significantly different between striped eggs and normal eggs.
Previous studies have shown that Corynebacterium can cause severe respiratory disease (Trost et al., 2012; Sangal and Hoskisson, 2016; David and Daum, 2017; Timms et al., 2018). Meanwhile, high levels of Corynebacterium in the air of duck houses can significantly affect duck performance, leading to increased immune suppression and disease susceptibility (Martin et al., 2010; Wu et al., 2019; Zhu et al., 2022). Staphylococci were widely distributed and were found in many parts of healthy birds, including skin, mucous membranes, and intestines (Szafraniec et al., 2020). Previous studies have found that Staphylococcus aureus can not only cause skin and soft tissue infection but also cause genital tract abscess and microbiota disorder, thereby causing the occurrence of endometritis (David and Daum, 2017). The uterus and reproductive tract of female ducks producing striped eggs also showed lesions and inflammation, and Staphylococcus may be one of the factors. In addition, previous studies have found the presence of Brevibacterium in poultry feces (Weidhaas et al., 2010; Ryu et al., 2014). Brevibacterium in the litter may come from duck feces. At present, there are many diseases caused by Brevibacterium, such as osteomyelitis and sepsis (Funke et al., 1997; Riegel, 1998; Peel et al., 2022). We hypothesize that different microbiota in the feed and litter may contribute to the production of striped eggs by influencing host health.
CONCLUSION
The 16S rRNA gene sequencing results showed that the microbial community structure was different in the litter and feed of female ducks that produced striped eggs and normal eggs. The abundance of Staphylococcus, Corynebacterium, and Brevibacterium in the litter of female ducks that produced striped eggs was significantly higher than those of normal eggs. Therefore, the increase of these different genera may affect the production function of female ducks and lead to striped egg production. These observations provide insights into the causes of the production of striped eggs, and more studies should be conducted in the future.
ACKNOWLEDGMENTS
Authors’ contributions: LZ wrote the paper and performed data analysis; SQQ performed research and analyzed data; YX and LY performed research; ZCH designed this study and revised the manuscript. All authors edited the manuscript and agreed with its final form. All authors read and approved the final manuscript
Funding: This project was supported by the National Modern Agricultural Industry Technology System (CARS-42-9), National Nature Science Foundation of China (31972525, 31572388), Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding (G20220628007).
Ethics approval and consent to participate: The animal care and experimental procedures were approved by the Animal Care and Use Committee of China Agricultural University and adhered to the university's guidelines for animal research.
DISCLOSURES
The authors declare that they have no competing interests, and the manuscript is approved by all authors for publication. This manuscript is original and has not been published in whole or in part previously.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2022.102436.
Appendix. Supplementary materials
Supplementary materials are available at Poultry Science online. Table S1-3. Sequence information statistics table after optimization. Table S4. Comparison of the microbiome Alpha diversity in the cloacal swabs, the little and the feed of female ducks that produced striped eggs and normal eggs. Figure S1-2 The phyla represented at significantly different levels in the litter and feed microbiota of the two groups were shown in an extended error bar plot.
REFERENCES
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., Knight R. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2 doi: 10.1128/mSystems.00191-16. e00191–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson S.M.D., Allen H.K., Bearson B.L., Looft T., Brunelle B.W., Kich J.D., Tuggle C.K., Bayles D.O., Alt D., Levine U.Y., Stanton T.B. Profiling the gastrointestinal microbiota in response to Salmonella: low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 2013;16:330–340. doi: 10.1016/j.meegid.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman M.D., Zhongtang Y., Nelson Michael C., Moeller Steven J., Lilburn Michael S., Zerby Henry N. Interrelations between the microbiotas in the litter and in the intestines of commercial broiler chickens. Appl. Environ. Microbiol. 2010;76:6572–6582. doi: 10.1128/AEM.00180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D., Qi G.-H., Wang J., Zhang H.-J., Qiu K., Wu S.-G. Intestinal microbiota of layer hens and its association with egg quality and safety. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M.Z., Daum R.S. Treatment of Staphylococcus aureus infections. Curr. Top Microbiol. Immunol. 2017;409:325–383. doi: 10.1007/82_2017_42. [DOI] [PubMed] [Google Scholar]
- Feng J., Lu M., Wang J., Zhang H., Qiu K., Qi G., Wu S. Dietary oregano essential oil supplementation improves intestinal functions and alters gut microbiota in late-phase laying hens. J. Anim. Sci. Biotechnol. 2021;12:72. doi: 10.1186/s40104-021-00600-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke G., von Graevenitz A., Clarridge J.E., 3rd, Bernard K.A. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Gast R., Humphrey T.J., Van Immerseel F. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol. Rev. 2009;33:718–738. doi: 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- Gao Z., Zhang J., Li F., Zheng J., Xu G. Effect of oils in feed on the production performance and egg quality of laying hens. Animals (Basel) 2021;11:3482. doi: 10.3390/ani11123482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S., Liu L. Status, future development trend and suggestions of waterfowl industry in 2021. Chin. J. Anim. Sci. 2022;58 227-231+238. [Google Scholar]
- Jeong J., Lee J.-Y., Kang M.-S., Lee H.-J., Kang S.-I., Lee O.-M., Kwon Y.-K., Kim J.-H. Comparative characteristics and zoonotic potential of Avian Pathogenic Escherichia coli (APEC) isolates from chicken and duck in South Korea. Microorganisms. 2021;9:946. doi: 10.3390/microorganisms9050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.-J., Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- Ma C., Sun Z., Zeng B., Huang S., Zhao J., Zhang Y., Su X., Xu J., Wei H., Zhang H. Cow-to-mouse fecal transplantations suggest intestinal microbiome as one cause of mastitis. Microbiome. 2018;6:200. doi: 10.1186/s40168-018-0578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariat D., Firmesse O., Levenez F., Guimarăes V., Sokol H., Doré J., Corthier G., Furet J.-P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Kämpfer P., Jäckel U. Quantification and identification of culturable airborne bacteria from duck houses. Ann. Occup. Hyg. 2010;54:217–227. doi: 10.1093/annhyg/mep088. [DOI] [PubMed] [Google Scholar]
- Munyaka P.M., Eissa N., Bernstein C.N., Khafipour E., Ghia J.-E. Antepartum antibiotic treatment increases offspring susceptibility to experimental colitis: a role of the gut microbiota. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasri H., van den Brand H., Najjar T., Bouzouaia M. Egg storage and breeder age impact on egg quality and embryo development. J. Anim. Physiol. Anim. Nutr. (Berl) 2020;104:257–268. doi: 10.1111/jpn.13240. [DOI] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- O'Brien K.M., Chimenti M.S., Farnell M., Tabler T., Bair T., Bray J.L., Nonnenmann M.W. High throughput genomic sequencing of bioaerosols in broiler chicken production facilities. Microb. Biotechnol. 2016;9:782–791. doi: 10.1111/1751-7915.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel M.J., Torres R.S.G., Ivančić M., Hernández B.E.A. Management of intertarsal septic arthritis in an ostrich (Struthio camelus) Vet. Med. Sci. 2022;8:125–129. doi: 10.1002/vms3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky O., Sekelova Z., Faldynova M., Sebkova A., Sisak F., Rychlik I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl. Environ. Microbiol. 2015;82:1569–1576. doi: 10.1128/AEM.03473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaswamygowda G.H., Olakkaran S., Antony A., Purayil A.K. In: Recent Developments in Applied Microbiology and Biochemistry. Buddolla V., editor. Academic Press; 2019. Chapter 22 - present status and future perspectives of marine actinobacterial metabolites; pp. 307–319. Pages. [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radon K., Danuser B., Iversen M., Monsó E., Weber C., Hartung J., Donham K., Palmgren U., Nowak D. Air contaminants in different European farming environments. Ann. Agric. Environ. Med. 2002;9:41–48. [PubMed] [Google Scholar]
- Redweik G.A.J., Daniels K., Severin A.J., Lyte M., Mellata M. Oral treatments with probiotics and live salmonella vaccine induce unique changes in gut neurochemicals and microbiome in chickens. Front. Microbiol. 2019;10:3064. doi: 10.3389/fmicb.2019.03064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegel P. Bacteriological and clinical aspects of corynebacterium. Ann. Biol. Clin. (Paris) 1998;56:285–296. [PubMed] [Google Scholar]
- Rivera-Chávez F., Bäumler A.J. The pyromaniac inside you: Salmonella Metabolism in the host gut. Annu. Rev. Microbiol. 2015;69:31–48. doi: 10.1146/annurev-micro-091014-104108. [DOI] [PubMed] [Google Scholar]
- Rogers A.W.L., Tsolis R.M., Bäumler A.J. Salmonella versus the microbiome. Microbiol. Mol. Biol. Rev. 2021;85 doi: 10.1128/MMBR.00027-19. e00027–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Elk M., Khan I.U.H., Harwood V.J., Molina M., Edge T.A., Domingo J.S. Comparison of two poultry litter qPCR assays targeting the 16S rRNA gene of Brevibacterium sp. Water Res. 2014;48:613–621. doi: 10.1016/j.watres.2013.10.015. [DOI] [PubMed] [Google Scholar]
- Sang Q., Sun Y., Liu Y., Hou Z. Estimation of heritability and genetic parameters of related traits in striped duck eggs. China Anim. Husbandry Vet. Med. 2022;03:830–836. [Google Scholar]
- Sangal V., Hoskisson P.A. Evolution, epidemiology and diversity of Corynebacterium diphtheriae: new perspectives on an old foe. Infect. Genet. Evol. 2016;43:364–370. doi: 10.1016/j.meegid.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Szafraniec G.M., Szeleszczuk P., Dolka B. A review of current knowledge on staphylococcus agnetis in poultry. Animals (Basel) 2020;10:1421. doi: 10.3390/ani10081421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms V.J., Nguyen T., Crighton T., Yuen M., Sintchenko V. Genome-wide comparison of Corynebacterium diphtheriae isolates from Australia identifies differences in the Pan-genomes between respiratory and cutaneous strains. BMC Genomics. 2018;19:869. doi: 10.1186/s12864-018-5147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C., Earle K.A., Huang K.C., Sonnenburg J.L. The gut microbiome: connecting spatial organization to function. Cell Host Microbe. 2017;21:433–442. doi: 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost E., Blom J., de C. Soares S., Huang I.-H., Al-Dilaimi A., Schröder J., Jaenicke S., Dorella F.A., Rocha F.S., Miyoshi A., Azevedo V., Schneider M.P., Silva A., Camello T.C., Sabbadini P.S., Santos C.S., Santos L.S., Hirata R.J., Mattos-Guaraldi A.L., Efstratiou A., Schmitt M.P., Ton-That H., Tauch A. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J. Bacteriol. 2012;194:3199–3215. doi: 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Lilburn M., Yu Z. Intestinal microbiota of broiler chickens as affected by litter management regimens. Front. Microbiol. 2016;7:593. doi: 10.3389/fmicb.2016.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-R., Lian X.-L., Su T.-T., Long T.-F., Li M.-Y., Feng X.-Y., Sun R.-Y., Cui Z.-H., Tang T., Xia J., Huang T., Liu Y.-H., Liao X.-P., Fang L.-X., Sun J. Duck wastes as a potential reservoir of novel antibiotic resistance genes. Sci. Total Environ. 2021;771 doi: 10.1016/j.scitotenv.2020.144828. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hua-Fang S., Yan H., Jin-Ya W., Yun-Xia J., Fung-Yee T.N., Hong-Wei Z. Comparison of the levels of bacterial diversity in freshwater, intertidal Wetland, and Marine Sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012;78:8264–8271. doi: 10.1128/AEM.01821-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J.L., Macbeth T.W., Olsen R.L., Sadowsky M.J., Norat D., Harwood V.J. Identification of a Brevibacterium marker gene specific to poultry litter and development of a quantitative PCR assay. J. Appl. Microbiol. 2010;109:334–347. doi: 10.1111/j.1365-2672.2010.04666.x. [DOI] [PubMed] [Google Scholar]
- Wu B., Qin L., Wang M., Zhou T., Dong Y., Chai T. The composition of microbial aerosols, PM2.5, and PM10 in a duck house in Shandong province, China. Poult. Sci. 2019;98:5913–5924. doi: 10.3382/ps/pez365. [DOI] [PubMed] [Google Scholar]
- Yuan J., Wang B., Huang Z., Fan Y., Huang C., Hou Z. Comparisons of egg quality traits, egg weight loss and hatchability between striped and normal duck eggs. Br. Poult. Sci. 2013;54:265–269. doi: 10.1080/00071668.2013.770449. [DOI] [PubMed] [Google Scholar]
- Zhan H.Q., Dong X.Y., Li L.L., Zheng Y.X., Gong Y.J., Zou X.T. Effects of dietary supplementation with Clostridium butyricum on laying performance, egg quality, serum parameters, and cecal microflora of laying hens in the late phase of production. Poult. Sci. 2019;98:896–903. doi: 10.3382/ps/pey436. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Gu T., Xu Q., Zhu G., Chen G. Effects of Salmonella enterica serovar Enteritidis infection on egg production and the immune response of the laying duck Anas platyrhynchos. PeerJ. 2019;7:e6359. doi: 10.7717/peerj.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Liao L., Su J., Liu Z., Pan S., Huang Y., Wu Y. Interactions of Muscovy duck reovirus, gut microbiota, and host innate immunity: transcriptome and gut microbiota analysis. Vet. Microbiol. 2022;264 doi: 10.1016/j.vetmic.2021.109286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials are available at Poultry Science online. Table S1-3. Sequence information statistics table after optimization. Table S4. Comparison of the microbiome Alpha diversity in the cloacal swabs, the little and the feed of female ducks that produced striped eggs and normal eggs. Figure S1-2 The phyla represented at significantly different levels in the litter and feed microbiota of the two groups were shown in an extended error bar plot.