Highlights
-
•
24 differentially expressed miRNAs were identified in the poly I:C-stimulated fish.
-
•
7 and 17 miRNAs were upregulated and downregulated after poly I:C treatment.
-
•
Majority of DE miRNAs were enriched in metabolism and immune signal pathways.
Keywords: Gibel carp, Poly I:C, MicroRNA, Transcriptome, Immune response
Abstract
Polyinosinic-polycytidylic acid (poly I:C) is a synthesized analogue of viral double-strand RNA and considered as a potential immunostimulant in aquaculture. MicroRNAs (miRNAs) have been reported to play important roles in the development of the immune system and in regulation of host antiviral responses. In our earlier study, it was found that poly I:C pre-treatment could stimulate the resistance against cyprinid herpesvirus 2 (CyHV-2) infection and enhance the antiviral immune response in gibel carp. To understand the role of miRNAs in regulating the host response to poly I:C treatment, we investigated the expression profiles of miRNAs in the head kidney of poly I:C-treated gibel carp with small RNA sequencing technology. When compared with the untreated group, a total of 24 differentially expressed miRNAs were identified in the poly I:C-stimulated fish, among which, 7 and 17 miRNAs were upregulated and downregulated, respectively. Analysis of target genes of these differentially expressed miRNAs found that most targeted mRNAs were involved in catalytic activity, peptidase activity and endopeptidase activity, and were enriched in the metabolic, protein processing in endoplasmic reticulum and oxidative phosphorylation signaling pathways, suggesting that poly I:C could alter the expression of metabolism-related miRNAs in the kidney of gibel carp. Besides, it was noted that some immune-related miRNAs, including inflammation-related miRNAs (miR-192 and miR-731) and interferon-related miRNAs (miR-194a and miR-122), were downregulated after poly I:C treatment. In summary, it was found that poly I:C could regulate the cellular levels of specific miRNAs involved in metabolism and immune responses in the head kidney of gibel carp, which may increase the capacity of the immune cells to fight against pathogens infection.
1. Introduction
Gibel carp, Carassius auratus gibelio, is one of the most important popular economic fish species cultured in China. It is artificially cultivated by heterologous sperm stimulating the eggs of crucian carp (Triploid square carassius auratus ♀ × Diplont Xingguo red carp ♂) [1]. With rich nutrition and high growth performance, gibel carp has become one of the most cultured fishes in the region of Yangtze River in China, and the annual production reached 2.7 million tons [2]. The fish is characterized by fast growth, large individual size, strong resistance to stress, making it a good material for fish physiology and immunity studies. However, with the rapid development of gibel carp industry, the incidence of various diseases is increasing, and consequently results in serious economic loss. For example, Herpesviral haematopoietic necrosis disease (HVHND) caused by cyprinid herpesvirus 2 (CyHV-2) infection has posed a great threat for the gibel carp industry in China [3]. Unfortunately, there are lack of effective drugs and control measures for this disease until now.
Polyinosinic-polycytidylic acid (poly I:C), a kind of synthesized analogue of viral double-strand RNA, activates the antiviral pattern recognition receptor Toll-like receptor 3 (TLR3), induces the host antiviral response including the secretion of proinflammatory cytokine and activation of type I IFNs [4], [5]. Nowadays, poly I:C has been used in maintaining good health and improving disease resistance in fish, making it as a potential immunostimulant and vaccine adjuvant to protect fish from several aquatic virus infection. For instance, poly I:C injection could substantially reduce the cumulative mortality of sevenband grouper (Epinephelus septemfasciatus) after viral nervous necrosis (VNN) challenge, and the surviving fish were highly protected from re-challenge [6]. In our earlier study, the efficacy of poly I:C on protecting gibel carp against CyHV-2 infection has been determined, wherein poly I:C treatment decreased the cumulative mortality and induced powerful antiviral immune response [7]. Besides, increasing evidences have demonstrated that poly I:C can be used as a candidate substance for introducing trained innate immunity, which is a newly proposed concept that innate immune defense stimulation could increase nonspecific resistance to infection by homologous or heterologous pathogens [8]. As reported, feeding with poly I:C could induce long-term immune responses against Edwardsiella piscicida infection in turbot (Scophthalmus maximus) [9].
MicroRNA is a small, highly conserved, non-coding RNA that negatively regulates the target's post-transcriptional gene expression [10], [11]. Nowadays, emerging evidences have showed that the host miRNAs are involved in many biological processes including cell proliferation, differentiation, responses to viral infection, and the stress responses [12], [13]. Previously, we found that poly I:C pre-treatment could stimulate the resistance against CyHV-2 infection and enhance the antiviral immune response in gibel carp [7]. To further understand the role of poly I:C in regulating the host response, we investigated the expression profiles of miRNAs in the head kidney of poly I:C-treated gibel carp with small RNA sequencing technology. The differentially expressed miRNAs were identified, their target genes were subsequently predicted and their potential functions were annotated. As far as we know, this study describes for the first time the miRNA transcriptomic response to poly I:C stimulation in gibel carp.
2. Materials and methods
2.1. Fish and poly I:C administration
Healthy gibel carp (average weight of 20 ± 2 g) were obtained from a commercial farm located in Yancheng city, Jiangsu province, China. The fish were maintained in the 70 cm × 50 cm × 46 cm fiberglass tanks supplied aerated fresh water under a 12 h:12 h light: dark photoperiod. The average water temperature was kept at room temperature and the oxygen saturation of the water was approximately 8.2 mg/L. The fish were fed once daily with commercial diet at the dose of their 2% weight. After acclimation for 7 days, the fish were randomly distributed into two groups (n=20), the poly I:C group (S group) was intraperitoneally injected with 100 μL of poly I:C (Sigma-Aldrich Co. USA) at a dose of 10 μg/g fish body weight [6], while the control group (D group) was intraperitoneally injected with 100 μL of PBS. At 24 h post-injection (hpi), 5 fish from each group were chosen, anaesthetized with Tricaine methanesulfonate (MS-222), and the head kidney tissues were sampled. To minimize biological variance, all the samples from each group were pooled together and divided equally into 3 replicates as independent replicates. The samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
2.2. Small RNA library construction and Illumina sequencing
To construct small RNA libraries, total RNA of each group of head kidney was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA purity was checked using the Nanodrop 2000 spectrophotometer (USA), and the concentration was measured in the Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed by the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Using the qualified total RNA as input material, sequencing libraries were generated using Small RNA Sample Pre Kit (NEB, USA). Then, the quality and output of the sequencing libraries were evaluated by the Agilent 2100 bioanalyzer and real-time PCR system. Finally, libraries were sequenced on the HiSeq 2000 platform (Illumina Inc., San Diego, CA, USA) at Novogene (TianJin, China) and single-end reads were generated.
2.3. Sequencing data analysis and identification of miRNAs
Raw reads were filtered by removing low-quality reads, reads with 5′ primer contaminants, reads with 3′ primer and reads with poly A. The remaining sequences were trimmed of their adapter sequences and sequences ranged from 18∼35 nt in length were chosen. All the high-quality reads were mapped to the Carassius auratus (goldfish) genome (Accession No. GCA_003368295.1) reference sequences by Bowtie 2. The fish is a close relative of gibel carp with high genetic similarity, and its genome was used as the reference genome to transfer the annotation. Small RNAs derived from rRNAs, tRNAs, snRNAs and snoRNAs deposited at the Rfam and NCBI GenBank databases were identified by NCBI blast. Identification was performed by comparing the data from two groups against the known maize mature miRNAs and their precursors of all fish in the miRNA database miRbase 20.0. Sequences with perfect matches were identified as known miRNAs, and novel miRNAs were predicted using miREvo and mirdeep 2 software based on the step-loops or hairpin structures.
2.4. Differentially expression of miRNAs
Fold-change and p-value was calculated from the normalized expression according the Bayesian method developed by Audic and Claverie. The differentially expressed miRNAs were identified with the screening threshold of p-value < 0.05 [14].
2.5. Validation of MicroRNA expression
The expression level of the DE miRNAs was validated by qRT-PCR. Total RNA of the head kidney from each treatment group was extracted (n = 3 pooled samples, each pooled sample was prepared from five individual samples). RNAs were reverse-transcribed using the Mir-XTM miRNA First-Strand Synthesis Kit (TaKaRa) and qRT-PCR was subsequently performed using the Mir-X miRNA qRT-PCR TB Green Kit (TaKaRa) according to the manufacturer's instructions. U6 RNA was used as an internal standard. All samples were evaluated in triplicate on the CFX96 Real-time PCR Detection System (Bio-Rad, USA), and the relative expression level was calculated with the 2−△△Ct method. All the expression data were subjected to a one-way ANOVA, and statistical significance was assumed at p < 0.05.
2.6. Function analysis of the differentially expressed miRNAs
The mRNA targets of miRNAs on gibel carp genome were forecasted using MiRanda software with parameters: S≥150 and ΔG≤−30 kcal/moL. The data predicted by both algorithms were combined and the overlaps were considered as the miRNA targets. Furthermore, the biological function was annotated with mapping targets genes to each term of the Gene Ontology database using the software DAVID, and the associated signal pathways were determined via a KEGG pathway analysis by the software KOBAS 3.0.
3. Results
3.1. Overview of the high-throughput sequencing data
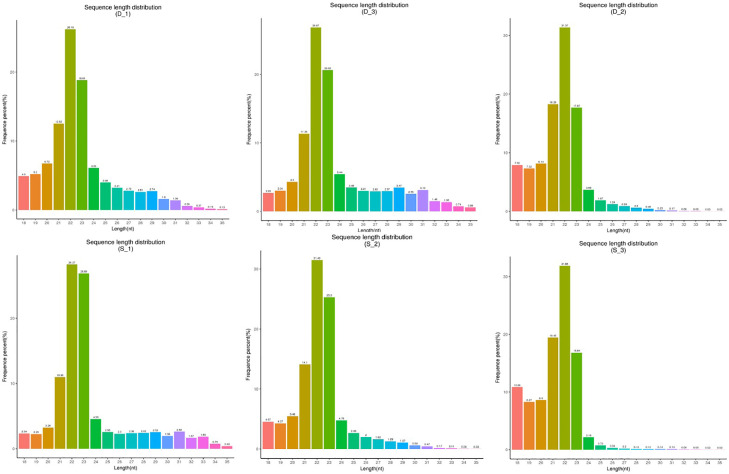
To investigate the miRNA expression profiles of gibel carp after poly I:C stimulation, small RNA libraries of the head kidney samples from two groups were constructed and sequenced. After quality filtering, about 12∼13 million clean reads were generated from each of the three samples (Table 1). The length distribution of the clean reads was analyzed, and a similar trend of the length distribution was observed in the six samples. As shown in Fig. 1, most of the small RNAs were 21-23 nt in length, and 22 nt was the dominant size.
Table 1.
Overview of the filtered data.
| Sample | total_reads | N% > 10% | low quality | 5_adapter_contamine | 3_adapter_null or insert_null | with ployA/T/G/C | clean reads |
|---|---|---|---|---|---|---|---|
| S1 | 13183812 (100.00%) | 406 (0.00%) | 151238 (1.15%) | 73269 (0.56%) | 507761 (3.85%) | 8094 (0.06%) | 12443044 (94.38%) |
| S2 | 13689259 (100.00%) | 506 (0.00%) | 183090 (1.34%) | 81769 (0.60%) | 0 (0.00%) | 8946 (0.07%) | 13414948 (98.00%) |
| S3 | 12067459 (100.00%) | 1465 (0.01%) | 468153 (3.88%) | 47953 (0.40%) | 243318 (2.02%) | 5573 (0.05%) | 11300997 (93.65%) |
| D1 | 13517921 (100.00%) | 558 (0.00%) | 192422 (1.42%) | 151313 (1.12%) | 0 (0.00%) | 15610 (0.12%) | 13158018 (97.34%) |
| D2 | 13690605 (100.00%) | 1392 (0.01%) | 389641 (2.85%) | 91032 (0.66%) | 196705 (1.44%) | 9350 (0.07%) | 13002485 (94.97%) |
| D3 | 14229229 (100.00%) | 493 (0.00%) | 205814 (1.45%) | 163621 (1.15%) | 1050704 (7.38%) | 16334 (0.11%) | 12792263 (89.90%) |
Fig. 1.
Length distribution of clean reads.
3.2. Identification of miRNA in gibel carp's head kidney
To classify and annotate small RNA sequencing results, the clean reads were aligned with Rfam, cDNA sequences, species repeat library. The tRNAs, snRNAs, rRNAs, and other RNAs were removed and the filtered sequences were aligned with miRBase database. As shown in Table S1, the reads classified into the known miRNAs accounted for 36.7%–56.64% of the total reads, while the novel RNAs accounted for only 0.06%–0.12% of the total unique clean reads by using miRDeep2 software prediction (Table S2).
3.3. Profiles of differentially expressed miRNAs
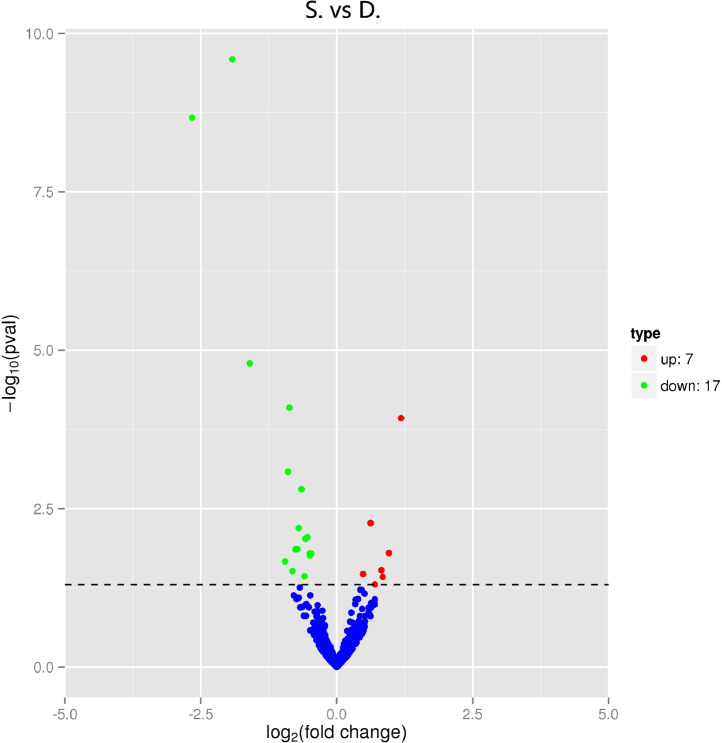
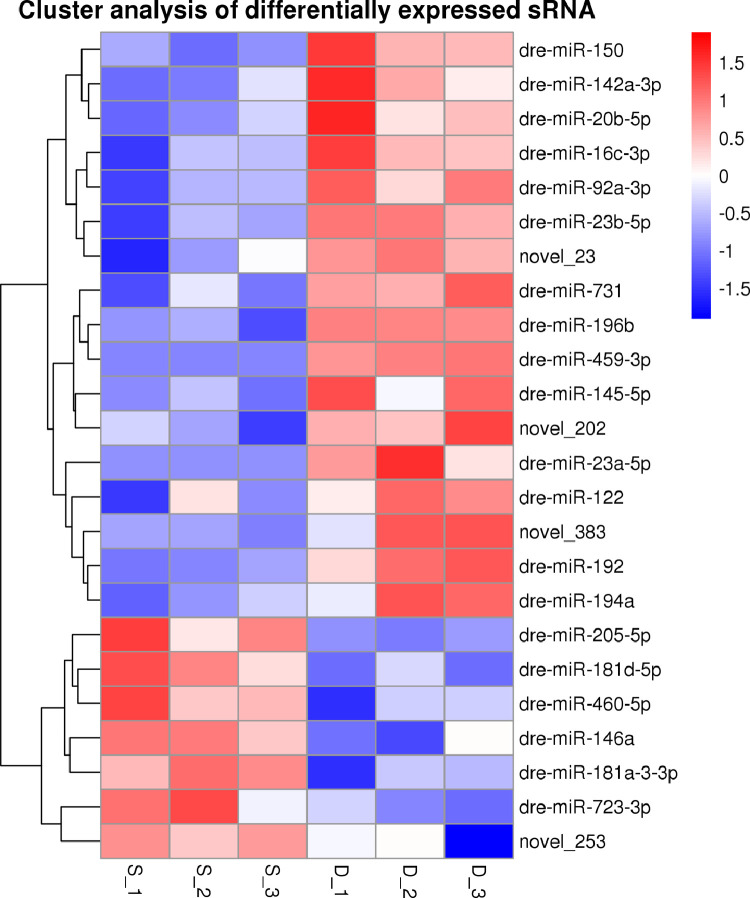
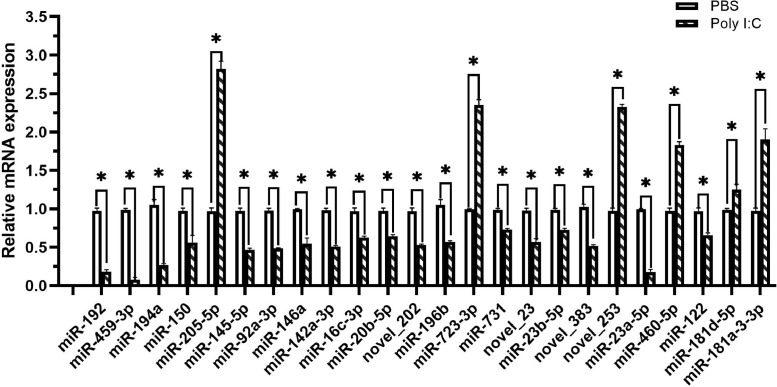
The differentially-expressed miRNAs, including and novel miRNAs between two groups, were analyzed. The expression of miRNA in two groups was shown by plotting a log2-ratio figure and scatter plot (Fig. 2). A total of 24 differentially expressed miRNAs including 20 known miRNAs and 4 novel miRNAs were significantly differentially expressed (p<0.05), including 7 and 17 miRNAs were upregulated and downregulated. As shown in Fig. 3, miR-192, miR-459-3p and miR-194a were down-regulated in the poly I:C group. Of the four novel differentially-expressed miRNAs, novel_383, novel_202 and novel_23 miRNAs were five were downregulated, whereas novel_253 were upregulated. The profiles of differentially expressed miRNAs were shown in Table 2. To confirm those differentially expressed miRNAs between two groups by the sequencing, five DE miRNAs were selected to be verified by qRT-PCR. The results showed that the expression profiles of these DE miRNAs were consistent with those obtained (Fig. 4).
Fig. 2.
The “volcano plot” pictures of differentially expressed genes between two groups. S, poly I:C group; D, PBS group.
Fig. 3.
Cluster analysis of differentially expressed miRNAs. S, poly I:C group; D, PBS group. n = 3.
Table 2.
Profiles of differentially expressed miRNAs.
| sRNA | Sequence (5’-3’) | S_readcount | D_readcount | log2FoldChange | pval |
|---|---|---|---|---|---|
| miR-192 | AUGACCUAUGAAUUGACAGCC | 335.2857673 | 1557.371 | -1.9189 | 2.57E-10 |
| miR-459-3p | CAGGGAAUCUCUGUUACUGGGIG | 0.42516078 | 28.9946 | -2.6595 | 2.14E-09 |
| miR-194a | UGUAACAGCAACUCCAUGUGG | 116.5116478 | 484.1212 | -1.5981 | 1.61E-05 |
| miR-150 | UCUCCCAAUCCUUGUACCAGUG | 56129.97815 | 106702.5 | -0.86647 | 8.08E-05 |
| miR-205-5p | UCCUUCAUUCCACCGGAGUCUG | 173.6053103 | 67.05123 | 1.1829 | 0.000118 |
| miR-145-5p | GUCCAGUUUUCCCAGGAAUCCC | 25566.67315 | 51039.57 | -0.89689 | 0.000826 |
| miR-92a-3p | UAUUGCACUUGUCCCGGCCUGU | 76153.65866 | 122518.2 | -0.64767 | 0.001563 |
| miR-146a | UGAGAACUGAAUUCCAUAGAUGG | 226491.6234 | 142400 | 0.62413 | 0.005348 |
| miR-142a-3p | UGUAGUGUUUCCUACUUUAUGGA | 273.7438465 | 464.5156 | -0.69594 | 0.006408 |
| miR-16c-3p | UCCAAUAUUGCUCGUGCUGCUGA | 3635.883715 | 5397.797 | -0.53795 | 0.008881 |
| miR-20b-5p | CAAAGUGCUCACAGUGCAGGUAG | 1526.532019 | 2337.852 | -0.5746 | 0.009429 |
| novel_202 | CCGAUUYCUUUUGGUGUUCAGAGU | 139.7295229 | 248.6544 | -0.72343 | 0.013784 |
| miR-196b | UAGGUAGUUUCAAGUUGUUGGG | 40.79465644 | 75.73866 | -0.75703 | 0.01391 |
| miR-723-3p | AAGACAUCAAUUAAAUCUGUGCU | 37.54857976 | 15.10451 | 0.9614 | 0.01585 |
| miR-731 | AAUGACACGUUUUCUCCCGGAUCG | 9926.116722 | 14288.33 | -0.4957 | 0.016154 |
| novel_23 | GCUGGCUAUGGAUUCUGUGUCC | 5228.596212 | 7367.426 | -0.46995 | 0.016182 |
| miR-23b-5p | GGGUUCCUGGCGUGCUGAUUU | 229.1196567 | 329.343 | -0.49295 | 0.017346 |
| novel_383 | ACAGGUGUGGAACAUUAAUUCU | 0.53416085 | 7.950229 | -0.95084 | 0.021525 |
| novel_253 | CAAAUUAUGAUGUGCUGUCACC | 72.24234109 | 34.31526 | 0.82388 | 0.029598 |
| miR-23a-5p | GAAUUCCUGGCAGAGUGAUUU | 0.70416021 | 5.201512 | -0.81201 | 0.030579 |
| miR-460-5p | CCUGCAUUGUACACACUGUGCG | 410.392135 | 285.7737 | 0.48572 | 0.034107 |
| miR-122 | UGGAGUGUGACAAUGGUGUUUG | 526.7047585 | 837.5674 | -0.59185 | 0.036951 |
| miR-181d-5p | UACAUUCAUUGAUGUCGUUGGGUU | 7.808343033 | 7.0357 | 0.84621 | 0.037814 |
| miR-181a-3-3p | ACCAUCGAGUGUUGAGUGUACC | 43.4749099 | 23.58727 | 0.70256 | 0.049505 |
Fig. 4.
Validation of relative expression levels of of the DE miRNAs by qRT-PCR. * indicates statistically-significant differences between poly I:C and PBS samples (* p< 0.05).
3.4. Target genes prediction and function annotation
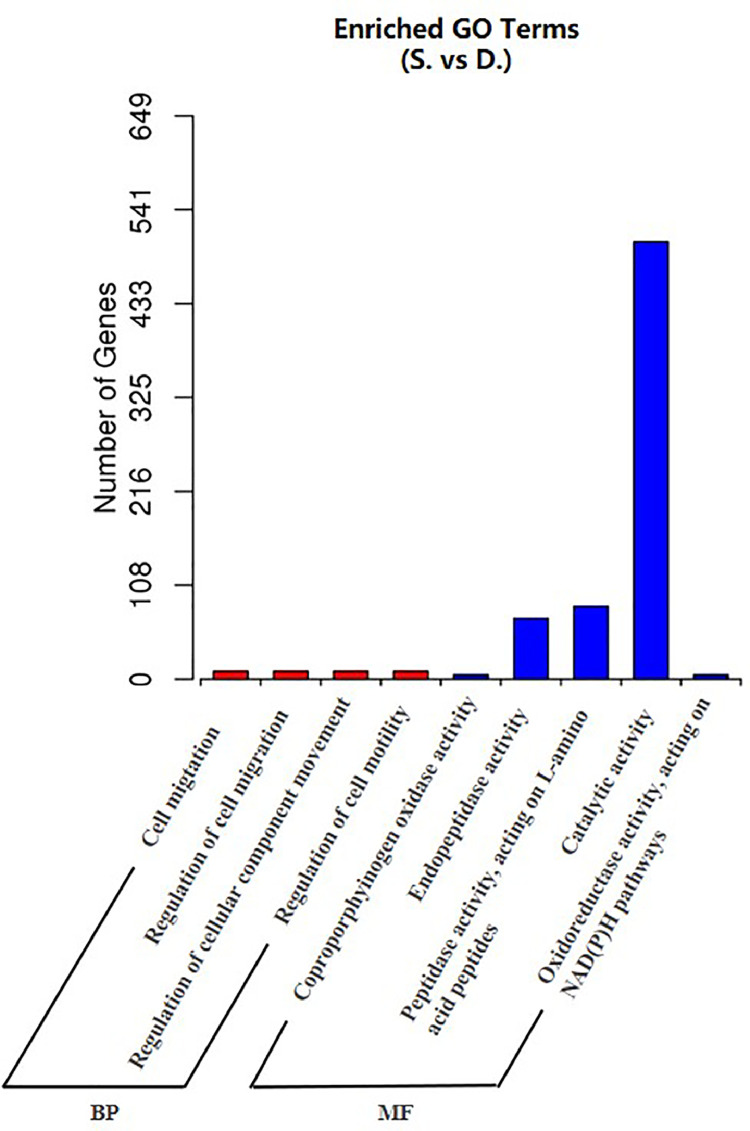
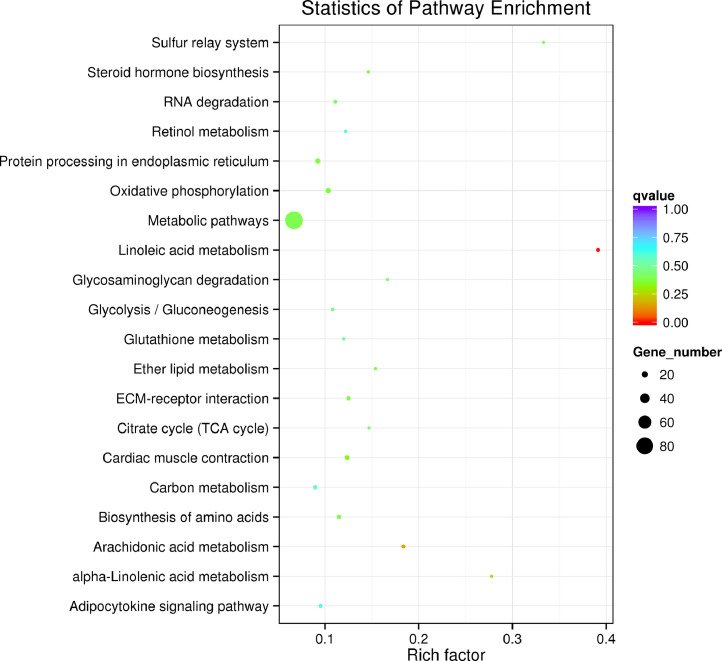
To better understand the functions of DE miRNAs of gibel carp, target gene prediction analysis was conducted using miRanda software. In total, 2074 genes were predicted as potential targets of the DE miRNAs that were identified in this study. Subsequently, GO annotation and KEGG pathway analysis were conducted. The GO analysis of identified predicted target genes revealed that most of the target genes were involved in catalytic activity, peptidase activity and endopeptidase activity classes (Fig. 5), suggesting the important role of miRNA in metabolic regulation in gibel carp. KEGG pathway analysis showed that most of the miRNA targets were enriched in metabolic, protein processing in endoplasmic reticulum and oxidative phosphorylation signal pathways (Fig. 6). Besides, it was noted that some immune-related miRNAs, including inflammation-related miRNAs (miR-192 and miR-731) and interferon-related miRNAs (miR-194a and miR-122), were downregulated after poly I:C treatment.
Fig. 5.
GO term enrichment for significantly different miRNA.
Fig. 6.
Pathway classifications of differentially expressed genes according to KEGG results.
4. Discussion
Poly I:C is a viral RNA mimic that can induce immune responses similar to that seen during viral infection [15]. For gibel carp, it was found that poly I:C could reduce the mortality of CyHV-2 infection, and improve the mRNA expression profiles of immune-related genes in our previous study [7]. Our present study focused on the miRNA-posttranscriptional regulation in the head kidney of gibel carp after poly I:C treatment. The reason why we chose the tissue is due to the fact that head kidney is the haematopoietic organ in fish, where a large number of immune cells are produced. Thus, head kidney becomes an ideal material for studying the immunogenesis mechanism of fish [16]. Here, we investigated the miRNA transcriptome profiles of head kidney of gibel carp at 24 h post poly I:C stimulation. By high-throughput sequencing, there were 24 differentially expressed miRNAs in head kidney treated with poly I:C, including 7 upregulated miRNAs and 17 downregulated miRNAs. Compared with other miRNA sequencings, the identified differentially expressed miRNAs were less abundant. There are two reasons accountable for this situation: One possible reason is that poly I:C is a small molecule with a simple structure, which has limited biological functions. Another contributing cause is that the total microRNAs mapped on the reference genome was fewer than other studies. Besides, it is noted that the majority of DE miRNAs showed down-regulated expression in head kidney. The down-regulation of these miRNAs could increase the expression of their target genes, which may have positive effects on the host's respond to the poly I:C stimulation.
Emerging evidences have showed that fish can obtain trained innate immunity using ligands to pattern recognition receptors [17]. It offers an interesting and attractive approach to increase disease resistance for fish culture. Poly I:C can activate TLR3 and trigger innate responses, thus making it a promising material for trained innate immunity. Actually, the potential of poly I:C pre-treatment to induce trained immunity has been confirmed. For example, poly I:C could confer protection against a bacterial infection with a Gram-negative pathogen in mice [8]. As reported, intracellular metabolism plays important roles during induction of trained immunity [18]. Metabolism-related signaling cascades including glycolysis, oxidative phosphorylation, the tricarboxylic acid cycle, amino acid, and lipid metabolism has been demonstrated to be interplaying pathways that are crucial for the establishment of innate immune memory [19]. Here, KEGG pathway analysis showed that most of the miRNA targets were enriched in metabolic signal pathways. This is in accordance with a previous report, in which several metabolites were accumulated in poly I:C-treated fish [20]. After poly I:C stimulation, these metabolism-related miRNAs in head kidney were induced and might regulate the intracellular metabolism via their target, thereby contributing to increasing the capacity of the innate immune cells and establishment of innate immune memory.
In fish, interferon (IFN) system could respond to viral infection and induce an "antiviral state", providing an important first line of defense against various viruses [21]. Poly I:C is known to be an immunostimulant that acts as one of the most potent IFN inducers [22]. Here, we found that wo IFN -related miRNAs (miR-194a and miR-122) were differentially expressed in poly I:C group. As reported, miRNA-194a could interact specifically with the 3′UTR of IRF7 in a negative manner, resulting in inhibition of IRF7 expression and blocking the promoter activity of type Ⅰ interferon [23]. As for miR-122, it could modulate type-I interferon expression by blocking the suppressor of cytokine signaling 1[24]. It was found that the expression of miR-194a and miR-122 were down-regulated in the head kidney of the poly I:C-stimulated gibel carp, suggesting that miR-194a and miR-122 may be promising candidate miRNAs involved in the regulation of type-I interferon system induced by poly I:C.
Previous report revealed that poly I:C could induce inflammation reaction, while appropriate inflammatory response could play a protective role to defend viral infection [25], [26]. In this study, we found that two inflammation-related miRNAs (miR-192 and miR-731) were differentially expressed at 24h h after poly I:C treatment. MiR-192 could negatively regulate the expression of its targets gene IL-1RI, resulting in the activation of the signaling cascade to arouse the immune response in reaction against pathogen in Japanese flounder [27]. MiR-731 is known to be involved in the inflammation and apoptosis processes via targeting TLR and apoptosis pathways in grass carp [28]. It was noted that both miR-192 and miR-731 were down-regulated expressed in the head kidney of gibel carp after poly I:C stimulation, which suggested that the two miRNAs may play important roles in mediating inflammation reactions induced by poly I:C stimulation in gibel carp.
5. Conclusion and future perspective
Overall, a total of 24 differentially expressed miRNAs in the head kidney of poly I:C-stimulated gibel carp compared to the control fish, including 17 downregulated miRNAs and 7 upregulated miRNAs. Functional analysis revealed that the majority of DE miRNAs were enriched in metabolism and immune signal pathways, suggesting that poly I:C could regulate the cellular levels of specific miRNAs involved in metabolism and immune responses in the head kidney of gibel carp, which may increase the capacity of the immune cells to fight against pathogens infection. As we know, this is the first report of the miRNA transcriptome of gibel carp in response to poly I:C stimulation. Further work is needed to elucidate the precise role of identified miRNA on immune regulation and the potential for the biomarkers of trained innate immunity.
Ethics statement
All experiments were performed according to the regulation of local and central government of China.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Jiangsu Provincial Key Research and Development Program (BE2021326) and the Natural Science Foundation of Jiangsu Province of China (BK20181053).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fsirep.2023.100083.
Appendix. Supplementary materials
Data availability
No data was used for the research described in the article.
References
- 1.Fan Y., Zhou Y., Zeng L., et al. Identification, structural characterization, and expression analysis of toll-like receptors 2 and 3 from gibel carp (Carassius auratus gibelio) Fish Shellfish Immunol. 2018;72:629–638. doi: 10.1016/j.fsi.2017.11.044. [DOI] [PubMed] [Google Scholar]
- 2.Gui J.F., Zhou L. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci. China Life Sci. 2010;53(4):409–415. doi: 10.1007/s11427-010-0092-6. [DOI] [PubMed] [Google Scholar]
- 3.Thangaraj R.S., Nithianantham S.R., Dharmaratnam A., Kumar R., Pradhan P.K., Thangalazhy Gopakumar S., Sood N. Cyprinid herpesvirus 2 (CyHV-2): a comprehensive review. Rev. Aquacult. 2020;13(2):796–821. [Google Scholar]
- 4.Liao Z., Su J. Progresses on three pattern recognition receptor families (TLRs, RLRs and NLRs) in teleost. Dev. Comp. Immunol. 2021;122 doi: 10.1016/j.dci.2021.104131. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q.N., Tang Y.Y., Zhou M.J., Luo S., Li Y.T., Wang G., Zhang D.Z., Yang H., Tang B.P., He W.F. Differentially expressed genes involved in immune pathways from yellowhead catfish (Tachysurus fulvidraco) after poly (I:C) challenge. Int. J. Biol. Macromol. 2021;183:340–345. doi: 10.1016/j.ijbiomac.2021.04.167. [DOI] [PubMed] [Google Scholar]
- 6.Kittipong T., Nichika S., Hirofumi Y., et al. Influence of temperature on Mx gene expression profiles and the protection of sevenband grouper, Epinephelus septemfasciatus, against red-spotted grouper nervous necrosis virus (RGNNV) infection after poly (I: C) injection. Fish Shellfish Immunol. 2014;40:441–445. doi: 10.1016/j.fsi.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J.L., Cui Z.Y., Hu G.Y., Jiang X.Y., Wang J., Qiao G., Li Q. Transcriptome analysis provides insights into the antiviral response in the spleen of gibel carp (Carassius auratus gibelio) after poly I: C treatment. Fish Shellfish Immunol. 2020;102:13–19. doi: 10.1016/j.fsi.2020.03.065. [DOI] [PubMed] [Google Scholar]
- 8.Ribes S., Arcilla C., Ott M., Schutze S., Hanisch U.K., Nessler S., Nau R. Pre-treatment with the viral Toll-like receptor 3 agonist poly(I:C) modulates innate immunity and protects neutropenic mice infected intracerebrally with Escherichia coli. J. Neuroinflamm. 2020;17(1):24. doi: 10.1186/s12974-020-1700-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J., Wang Z., Zhao Y., Yang J., Zhang Y., Liu Q., Yang D. Feeding with poly(I:C) induced long-term immune responses against bacterial infection in turbot (Scophthalmus maximus) Fish Shellfish Immunol. Rep. 2021;2 doi: 10.1016/j.fsirep.2021.100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asirvatham A.J., Magner W.J., Tomasi T.B. miRNA regulation of cytokine genes. Cytokine. 2009;45(2):58–69. doi: 10.1016/j.cyto.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreassen R., Hoyheim B. miRNAs associated with immune response in teleost fish. Dev. Comp. Immunol. 2017;75:77–85. doi: 10.1016/j.dci.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Blahna M.T., Hata A. Regulation of miRNA biogenesis as an integrated component of growth factor signaling. Curr. Opin. Cell Biol. 2013;25(2):233–240. doi: 10.1016/j.ceb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z., Lin Z., Pang X., Shan P., Wang J. MicroRNA regulation of Toll-like receptor signaling pathways in teleost fish. Fish Shellfish Immunol. 2018;75:32–40. doi: 10.1016/j.fsi.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhang X., Liang Z., Zhu M., Zhang M., Zhang T., Zhang Z., Cao M., Gu Y., Pan J., Yan B., Zhu H., Xue R., Cao G., Gao Y., Hu X., Gong C. Expression pattern and regulatory network of gibel carp (Carassius gibelio) miRNAs and their putative target genes in response to CyHV-2 infection. Aquaculture. 2020;523 [Google Scholar]
- 15.Vats A., Gautam D., Maharana J., Singh Chera J., Kumar S., Rout P.K., Werling D., De S. Poly I:C stimulation in-vitro as a marker for an antiviral response in different cell types generated from Buffalo (Bubalus bubalis) Mol. Immunol. 2020;121:136–143. doi: 10.1016/j.molimm.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Zhang X., Zhang Y., Qin Q., Huang X., Huang Y. Establishment of a cell line from the head kidney of giant grouper (Epinephelus lanceolatus) and its susceptibility to fish viruses. Aquacult. Rep. 2021;21 [Google Scholar]
- 17.Zhang Z., Chi H., Dalmo R.A. Trained innate immunity of fish is a viable approach in larval aquaculture. Front. Immunol. 2019;10:42. doi: 10.3389/fimmu.2019.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riksen N.P., Netea M.G. Immunometabolic control of trained immunity. Mol. Aspects Med. 2021;77 doi: 10.1016/j.mam.2020.100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domínguez-Andrés J., Joosten L.A.B., Netea M.G. Induction of innate immune memory: the role of cellular metabolism. Curr. Opin. Immunol. 2019;56:10–16. doi: 10.1016/j.coi.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Guo C., Ye J., Song M., Peng X., Li H. Poly I:C promotes malate to enhance innate immune response against bacterial infection. Fish Shellfish Immunol. 2022;131:172–180. doi: 10.1016/j.fsi.2022.09.064. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y.B., Gui J.F. Molecular regulation of interferon antiviral response in fish. Dev. Comp. Immunol. 2012;38(2):193–202. doi: 10.1016/j.dci.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Jung M.H., Jung S.J. Protective immunity against rock bream iridovirus (RBIV) infection and TLR3-mediated type I interferon signaling pathway in rock bream (Oplegnathus fasciatus) following poly (I:C) administration. Fish Shellfish Immunol. 2017;67:293–301. doi: 10.1016/j.fsi.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Guan X.L., Zhang B.C., Sun L. pol-miR-194a of Japanese flounder (Paralichthys olivaceus) suppresses type I interferon response and facilitates Edwardsiella tarda infection. Fish Shellfish Immunol. 2019;87:220–225. doi: 10.1016/j.fsi.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Li A., Song W., Qian J., Li Y., et al. MiR-122 modulates type I interferon expression through blocking suppressor of cytokine signaling 1. Int. J. Biochem. Cell Biol. 2013;45(4):858–865. doi: 10.1016/j.biocel.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Foster C.G., Landowski L.M., Sutherland B.A., Howells D.W. Differences in fatigue-like behavior in the lipopolysaccharide and poly I:C inflammatory animal models. Physiol. Behav. 2021;232 doi: 10.1016/j.physbeh.2021.113347. [DOI] [PubMed] [Google Scholar]
- 26.Farag N.S., Breitinger U., Breitinger H.G., El Azizi M.A. Viroporins and inflammasomes: A key to understand virus-induced inflammation. Int. J. Biochem. Cell Biol. 2020;122 doi: 10.1016/j.biocel.2020.105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu Q., Xu T. miR-192 targeting IL-1RI regulates the immune response in miiuy croaker after pathogen infection in vitro and in vivo. Fish Shellfish Immunol. 2016;54:537–543. doi: 10.1016/j.fsi.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Fang Y., Xu X.Y., Tao L., Shen Y., Li J. Effects of microRNA-731 on inflammation and apoptosis by targeting CiGadd45aa in grass carp. Fish Shellfish Immunol. 2020;97:493–499. doi: 10.1016/j.fsi.2019.12.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.