Abstract
The family of aldehyde dehydrogenases (ALDHs) contains 19 isozymes and is involved in the oxidation of endogenous and exogenous aldehydes to carboxylic acids, which contributes to cellular and tissue homeostasis. ALDHs play essential parts in detoxification, biosynthesis, and antioxidants, which are of important value for cell proliferation, differentiation, and survival in normal body tissues. However, ALDHs are frequently dysregulated and associated with various diseases like Alzheimer's disease, Parkinson's disease, and especially solid tumors. Notably, the involvement of the ALDHs in tumor progression is responsible for the maintenance of the stem‐cell‐like phenotype, triggering rapid and aggressive clinical progressions. ALDHs have captured increasing attention as biomarkers for disease diagnosis and prognosis. Nevertheless, these require further longitudinal clinical studies in large populations for broad application. This review summarizes our current knowledge regarding ALDHs as potential biomarkers in tumors and several non‐tumor diseases, as well as recent advances in our understanding of the functions and underlying molecular mechanisms of ALDHs in disease development. Finally, we discuss the therapeutic potential of ALDHs in diseases, especially in tumor therapy with an emphasis on their clinical implications.
Keywords: aldehyde dehydrogenase, biomarker, cancer stem cell, disease, solid tumor, therapeutic resistance
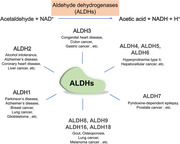
The human aldehyde dehydrogenase (ALDH) superfamily comprises 19 isozymes and can be divided into 11 distinct families. ALDHs are a group of nicotinamide adenine dinucleotide phosphate (NADP+)‐dependent enzymes that could catalyze the reversible oxidation of endogenous and exogenous aldehydes to their corresponding carboxylic acids. However, ALDHs are frequently dysregulated and associated with various diseases like Alzheimer's disease, Parkinson's disease, and especially solid tumors. Different ALDH isoenzymes are believed to be potential biomarkers or therapeutic targets in different diseases.
1. INTRODUCTION
The human aldehyde dehydrogenase (ALDH) superfamily comprises 19 isozymes and can be divided into 11 distinct families, which are respectively located in the cell cytoplasm, mitochondria, nucleus, and endoplasmic reticulum. 1 Biologically, ALDHs are a group of nicotinamide adenine dinucleotide phosphate (NADP+)‐dependent enzymes that could catalyze the reversible oxidation of endogenous and exogenous aldehydes to their corresponding carboxylic acids, 2 which protects living organisms against oxidative stress. ALDHs also take a role in scavenging reactive oxygen species (ROS) from aldehydes accumulation, thereby reducing oxidative stress in cells especially in stem cells. 3 , 4 Another vital function of ALDHs is catalyzing retinoic acid (RA) metabolism, which is crucial for normal embryonic development and epithelial differentiation. 5
Throughout the past decades, pioneering studies support that the aberrant expression of ALDHs is related to several human non‐tumor diseases, for example, Alzheimer's disease (AD) 6 and alcohol intolerance, 7 which are caused by the loss of activity or deficiency of enzyme expression and accumulation of toxic aldehydes. Another ALDH‐related disorder is caused by ALDH gene mutations, especially single‐nucleotide polymorphisms, 8 contributing to enzyme inactivation, cellular dysfunction, interruption of normal metabolic pathways, and susceptibility to human diseases, such as Sjögren–Larsson syndrome (SLS) and hyperprolinemia type II (HPII). Polymorphisms within these genes may differentially affect the risk of disease development across the ethnic groups evaluated. These increase the possibility of ALDHs as surrogate markers for identifying disease pathogenesis and clinicopathological characteristics.
It is noteworthy that aberrant ALDH activity or expression has been detected in different solid tumors, including breast cancer, 9 colorectal cancer (CRC), 10 lung cancer, 11 head‐and‐neck squamous cell carcinoma (HNSCC), 12 , 13 prostate cancer (PCa), 14 pancreatic cancer, 15 bladder cancer, 16 and glioblastoma (GBM). 17 High ALDH activity is enriched in stem‐ /progenitor‐like cells that recapitulate tumor heterogeneity. Cancer stem cells (CSCs) are rare cancer cells within a tumor with sustained self‐renewal and differentiation abilities. ALDHs overexpressed in CSCs have been confirmed to promote tumor growth, metastasis, therapeutic resistance, and immune escape. 18 , 19 , 20 Therefore, ALDHs stand out as biomarkers for CSCs in several cancers. Clinically, ALDHs are also considered indicators of poor prognosis in solid cancers. A systematic review and retrospective analysis of 1557 patients with advanced or metastatic solid cancers shows that high ALDH1 expression is distinctly associated with poorer overall survival (OS), especially in breast cancer, HNSCC, cervical cancer (CC), and ovarian cancers. 21 Therefore, clinical assessment of ALDHs during tumor progression improves our understanding of CSC evolution dynamics and paves the way for therapeutic applications. For another, targeting ALDH isoenzymes or ALDH‐related pathways hold promising therapeutic implications via suppressing cancer progression, particularly for eradicating the CSC populations.
Here we summarize the research on ALDHs‐related diseases, especially ALDHs as promising biomarkers of CSCs and powerful predictors of clinical prognosis in selected solid tumors that have been extensively studied or discussed. And we then highlight the current in our understanding of the molecular mechanisms of ALDHs in disease development, emphasizing preclinical/clinical approaches to target ALDHs in solid tumors.
2. FUNCTIONS OF ALDHS IN DISEASE
The ALDH family is involved in several biological processes essential for cell survival along with cell protection, such as the detoxification of toxic aldehydes, protection from oxidative stress, and regulation of RA metabolism. However, ALDHs are frequently dysfunctional and related to different diseases especially in solid tumors.
2.1. ALDHs in aldehydes detoxification
The detoxification role of ALDHs is critically important for homeostasis. Endogenous aldehydes are generated during amino acids, vitamins, alcohols, neurotransmitters metabolism, and lipid peroxidation (LPO), 22 whereas exogenous aldehydes are derived from the metabolism of a wide range of environmental agents. 2 , 23 Specifically, aldehyde accumulation may lead to oxidative stress and DNA damage via ROS production and LPO. However, ALDHs activate cellular antioxidant and free radical scavenging systems to attenuate oxidative stress and reduce DNA damage. 3 Singh et al. found that blood acetaldehyde levels and blood glucose levels are absolutely increased in global Aldh1b1 knockout mice, indicating that Aldh1b1 plays a pivotal role in the link between alcohol consumption and diabetes. 24 Individuals who possess the ALDH2*2 mutation (rs671) genotype lose enzyme activity and are unable to metabolize acetaldehyde, thereby contributing to oxidative stress and alcohol‐induced flushing reaction. Furthermore, alcoholics with the ALDH2*2 allele have an increased risk of suffering esophageal cancer (EC), HNSCC, CRC, and late‐onset AD. 25 , 26
In tumors, evidence points to that ALDHs such as ALDH1A1, ALDH1A3, and ALDH3A1 are critical drivers of chemotherapeutic and radiotherapeutic resistance in many solid tumors by detoxifying cytotoxic drugs and mitigating oxidative stress. 23 , 27 For example, silencing of the ALDH1A1 gene in human breast cancer cells increases their sensitivity to paclitaxel by triggering the production of ROS, and similar results are obtained with other anticancer agents such as doxorubicin, sorafenib, and staurosporine. 28 Another example is that high levels of ALDH1A1 lead to the acquisition of epithelial–mesenchymal transition (EMT) and CSC properties as well as erlotinib resistance through clearance of reactive chlorine species/ROS in lung cancers. Knockdown or pharmacological inhibition of ALDH1A1 overcomes erlotinib resistance in vitro and in vivo. 29
2.2. ALDHs in RA signaling
RA signaling plays significant roles in embryonic stem cells, 30 hematopoietic stem cells, cancer cells, and others. 31 RA and its derivatives exert critical roles in regulating gene transcription involving cellular differentiation and proliferation 32 as well as in maintaining epithelial homeostasis and immune response. 33 Initially, dietary vitamin A or retinol (ROLs) are absorbed by cells and reversibly oxidized to retinaldehydes (RALs) by retinol dehydrogenases (Figure 1). Specific ALDH isozymes (ALDH1A1, ALDH1A2, ALDH1A3, and ALDH8A1) then catalyze the NADP+‐dependent oxidation of both all‐trans‐retinal and 9‐cis‐retinal to all‐trans RA (atRA), 9‐cis‐RA, and 13‐cis‐RA. This reaction is a tightly regulated, irreversible process. 34 Newly synthesized RA can remain in the cell and bind to cellular RA–binding proteins (CRABPs). When bound to CRABPI, RA is targeted for degradation, whereas CRABPII translocates to the nucleus upon RA binding. RA could be translocated to the nucleus, where it binds and activates heterodimers of nuclear RA receptors (RARα, RARβ, and RARγ) and retinoic X receptors (RXRα, RXRβ, and RXRγ) on the RA response elements (RARE), which then induce the transcriptional activity of many genes involved in cell differentiation, cycle arrest, and apoptosis. 35 , 36 , 37 Liu et al. showed that the concentration of serum RA determined by enzyme‐linked immunosorbent assay is significantly lower in type 2 diabetes mellitus (T2DM) patients than in normal glucose tolerance subjects. 38 , 39 To date, Han et al. confirmed that dysregulated serum RA levels can be used as biomarkers along with glucose for predicting future T2DM development in Korean subjects. 40 In addition, studies have also revealed that circulating RA levels are negatively correlated with the development of coronary artery disease. 41
FIGURE 1.
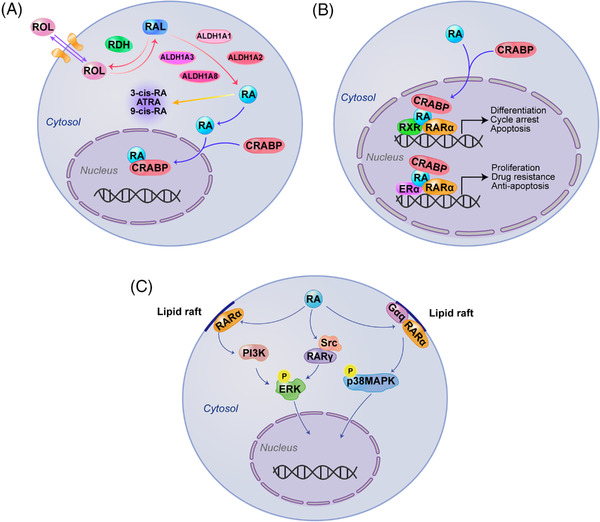
Aldehyde dehydrogenases (ALDHs) in retinoic acid (RA) signaling. (A) When retinols (ROLs) are absorbed by cells and reversibly oxidized to retinaldehydes (RALs) by retinol dehydrogenases (RDHs). Specific ALDH isozymes (ALDH1A1, ALDH1A2, ALDH1A3, and ALDH8A1) then catalyze the irreversibly nicotinamide adenine dinucleotide phosphate (NADP+)‐dependent oxidation of RALs to all‐trans RA (atRA), 9‐cis‐RA, and 13‐cis‐RA. (B) CRABPII transports RA to the nucleus where RA binds and activates heterodimers of nuclear RA receptors (RARs) or estrogen receptor α (ERα), respectively, on the RA response element (RARE), which can induce the transcriptional activity of target genes. (C) In response to RA, RARα translocates to lipid rafts in the cell membrane and interacts with Gαq proteins to activate the p38 mitogen‐activated kinase (p38MAPK) pathway. Besides, RARα and RARγ, via interaction with phosphoinositide 3‐kinase (PI3K) and non‐receptor tyrosine kinase (Src), respectively, can induce extracellular regulated–protein kinase (ERK) activation
In cancer, a well‐known example is that ALDH1A3/RA signaling activates the downstream factors homeobox transcription factor A1 (HOXA1) gene, which possesses a RARE sequence that is previously shown to be inducible by RA 35 and is hypomethylated in breast cancer cell line MDA‐MB‐468 cells to be a tumor suppressor. 42 In this respect, RAs have considerable clinical significance for the treatment of specific diseases. 43
However, there are also reports demonstrating that RA can bind with other receptors, including estrogen receptor α or peroxisome proliferator‐activated receptors (PPAR) β/δ, which then activates oncogenes such as c‐MYC and cyclin D1 and thereby promotes tumor cell proliferation, drug resistance, and inhibition of apoptosis. 44 A case in point is that RA treatment leads to parallel activation of RAR and PPAR β/δ to promote cancer cell survival in MMTV‐neu transgenic breast cancer mice. 45 , 46 Recently, a report also shows that atRA produced by ALDH1A1 transcriptionally activates functional RAREs in class III β‐tubulin (TUBB3) promoter, stimulating proliferation and sphere formation in patient‐derived bladder cancer cells. 47 Generally, retinoid receptors are present in the nucleus. In some cases, however, they can be found in the cytoplasm, where they display nongenomic functions in a ligand‐dependent or independent manner, and as monomers or in complex with various factors. 48 , 49 Indeed, studies from several laboratories demonstrate that extranuclear effects of RA and RARs appear to be involved in different mechanisms and kinase cascades, including activation of p38 mitogen‐activated kinase (MAPK), extracellular regulated–protein kinase or p42/p44MAPK, phosphoinositide 3‐kinase (PI3K), and protein kinase B. As proof, in response to RA, RARα translocates to the lipid rafts in cell membrane where it interacts with Gαq proteins and activates the MAPK pathway in breast tumor cells. 50 , 51 In addition, RA formed by ALDHs in cancer may affect cell‐autonomous pathways and trigger an immune cell fate switch. In sarcoma mouse models, Aldh1a1 or Aldh1a3 boosts atRA levels, which skews intra‐tumoral monocyte differentiation toward immunosuppressive macrophages. Pharmacological blockade of RA signaling in the tumor microenvironment increases immunostimulatory dendritic cells, enhances T‐cell‐dependent antitumor immunity, and synergizes with immune checkpoint inhibitors (ICIs). 33
3. ALDHS AS DIAGNOSIS MARKERS IN NON‐TUMOR DISEASES
A sheer number of studies support that aberrant ALDH levels are related to several metabolic diseases and neurological abnormalities, which are mainly caused by dysregulated expression or enzyme inactivation.
3.1. ALDH1
ALDH1A1: Parkinson's disease (PD) is the second most common neurodegenerative disorder, characterized by the progressive loss of dopamine (DA) neurons in the substantia nigra pars compacta (SNpc). 52 ALDH1A1 is an important molecular marker for the subpopulations of DA neurons in the SNpc that show differential susceptibility in PD‐related DA neurodegeneration, suggesting that the downregulation of ALDH1A1 expression may weaken its protective function against DA neurodegeneration in the ventral tier of SNpc. In support of this notion, a genetic deletion of Aldh1a1 exacerbates SNpc DA neuron loss in α‐synuclein transgenic mice, a murine model of PD‐related disease. However, the overexpression of ALDH1A1 is protective by preventing aldehyde risk factors, which indicates that ALDH1A1 is a potential therapeutic target in preventing PD pathogenesis. 53 Besides in SNpc of PD patients, the mRNA expression of ALDH1A1 is also reduced in the peripheral blood of PD patients, which is classified as an optimal predictor for PD risk in a panel with four other genes. 54 , 55 However, it remains to be further determined by the large sample size. In contrast to PD, the role of ALDH1A1 in AD pathogenesis remains unclear. AD is the most frequent neurodegenerative disease. 56 Although many studies have shown that ALDH1A1 is selectively expressed in the dopaminergic neurons, Fragoso et al. revealed that ALDH1A1 protein is also highly expressed in the human hippocampus, a region that is progressively degenerated in the brains of AD patients. 57 Furthermore, by analyzing clinical human AD and age‐matched control brain tissues, Nikhil et al. showed that ALDH1A1 protein expression and activity are independently regulated and may not necessarily correlate with each other. During AD pathogenesis, ALDH1A1 activity is the highest at the initial stage, which can protect from accruing oxidative stress‐induced damage, but it declines significantly at the severe stage. In contrast, the ALDH1A1 protein level is significantly higher in severe AD tissues, but vastly compromised ALDH1A1 activity, which is presumably due to increased neurotoxicity. 58 In addition, to gain insight into how ALDH1A1 neurons regulate behaviors of AD patients, by using a mutant Aldh1a1 mouse with AD, Li et al. found that Aldh1a1 neurons play an essential role in encoding a delay of gratification, and genetic deletion of Aldh1a1 causes impulsive behaviors in AD mice, which pinpoint a cellular point of entry to understand impulsive behaviors of AD patients. 59 Recently, an ongoing clinical trial (NCT04878549) is being investigated to find a new approach by analyzing a 5‐gene transcription signature, including ALDH1A1 for the diagnosis of enteric fever induced by acute undifferentiated febrile infection, which indicates that ALDH1A1 may be a new marker for disease diagnosis.
ALDH1A2: ALDH1A2 is involved in the conversion of retinol into RA, which is an essential regulator of diaphragm/lung and cardiovascular development during human embryogenesis. 60 In several animal models, downregulated RA results in cardiovascular, diaphragmatic, and associated pulmonary defects, which are consistent with the phenotype observed in patients. 61 Furthermore, variants in the ALDH1A2 can affect downstream RA‐induced gene expression and cause lethal multiple congenital anomaly syndrome that is associated with pulmonary hypoplasia and respiratory failure. 62 Lee et al. demonstrated that the synthesis of RA by ALDH1A2 marks mesoderm patterning and that specifies atrial cardiomyocytes from human pluripotent stem cells. This discovery provides new insights that can recreate aspects of cardiovascular disease in vitro and develop new therapeutic applications. 63
ALDH1B1: ALDH1B1 is involved in ethanol detoxification, and modifications in this enzyme may contribute to alcohol‐related diseases. Some human ALDH1B1 polymorphism carriers are highly susceptible to alcohol‐associated diseases in Whites. 64 , 65 In agreement with these reports, Singh et al. generated global Aldh1b1 knockout mice that have an increase in blood acetaldehyde levels and blood glucose levels, indicating that Aldh1b1 has a potentially pivotal role in the link between alcohol consumption and diabetes. 24
3.2. ALDH2
ALDH2 gene polymorphism is a potential risk marker for an array of cardiovascular anomalies and neurodegenerative diseases. The ALDH2*2 mutant (rs671) genotype, which is widely present in East Asians, 66 , 67 has significantly reduced the ALDH2 activity and induced the accumulation of aldehyde toxicity. A recent meta‐analysis of 5315 individuals by Chen et al. shows that the ALDH2∗2 polymorphism may be a potential risk factor for the development of AD, 68 which is consistent with Kamino et al. 69 and Ma et al. 70 Another study provides evidence that ALDH2‐related signaling can be activated by melatonin and restore mitophagy and cardiomyocyte homeostasis in a mice model with AD. This is supported by the observation that the inhibition of ALDH2 eliminates melatonin‐mediated cardio‐protection. 71 Notably, the group of Mochly‐Rosen also provides an important protection potential of ALDH2 against cardiac injuries, 72 which is later confirmed by others. 73 , 74 , 75 Upregulation of cellular ALDH2 activity can markedly reduce cytotoxic aldehydes and efficiently inhibit cardiac injury as well as significantly limit infarction size during cardiac ischemia in mouse models of myocardial infarction (MI) that can be reversed by the inhibition of ALDH2. Therefore, a clinical assessment of ALDH2 as a marker will be beneficial to the diagnosis of MI patients (NCT05360602). People with Asian flushing syndrome suffer alcohol intolerance and unfavorable cardiovascular conditions due to deficient activity of variant ALDH2∗2 allele. 76 A clinical trial has been completed and evaluated whether the alcohol dehydrogenase inhibitor, fomepizole can treat symptoms associated with ALDH2 deficiency (NCT00661141).
Likewise, the risk of PD, 77 hepatic steatosis, 78 and coronary artery disease 79 seems to be increased in individuals with the ALDH2*2 polymorphism, which is also confirmed by large‐scale meta‐analysis for risk assessment to develop essential hypertension. 80
3.3. ALDH4
ALDH4A1: Interest in ALDH4A1 stems from the involvement of proline and hydroxyproline metabolism in many aspects of human health and disease. Pathogenic variants in the ALDH4A1 gene cause HPII, a metabolic disorder of the proline degradation pathway that can result in retardation and convulsion. Associated proteins of ALDH4A1 can potentially be used as pharmacological chaperones to stabilize the misfolded variants of ALDH4A1 in patients with HPII. 81 In addition, patients with ALDH4A1 gene deficiency suffer from schizoaffective disorders and schizophrenia, and the potential molecular mechanism is also associated with abnormal proline metabolism. 82 A recent study discovers that circulating ALDH4A1 is significantly elevated in the plasma of mice and humans with atherosclerosis, and the administration of ALDH4A1 antibody can protect against atherosclerosis progression. These results support that ALDH4A1 can be used as a putative disease biomarker and therapeutic target. 83
3.4. Other ALDH isozymes
Studies using cell cultures and animal models have provided evidence that corneal ALDH3A1 and lens ALDH1A1 protect the eyes against cataract formation through non‐catalytic (light filtering) and catalytic (detoxification) mechanisms. Upon UVB exposure, ALDH3A1 gene knockout mice exhibit accelerated anterior lens subcapsular opacification, confirming a protective role of ALDH3A1 against cataract formation. 84 Later, they show new evidence in vivo that ALDH3A1 is critical for maintaining corneal transparency for vision. Collectively, ALDH3A1 plays an important role in preventing cataract formation and holds great promise to be a therapeutic target. 85 Mutations in the ALDH3A2 gene cause SLS, which will result in an abnormal accumulation of toxic fatty aldehydes in the brain and skin. 86 Mutations in the ALDH5A1 gene cause succinic semialdehyde dehydrogenase (SSADH) deficiency, which is a genetic disease caused by the abnormal metabolism of γ‐aminobutyric acid (GABA). 87 Mutations in the ALDH6A1 cause methylmalonic acidemia, which is a devastating metabolic disorder with a poor prognosis. 88 Mutations in the ALDH7A1 cause pyridoxine‐responsive epilepsies, which is an inborn error of lysine catabolism that presents with refractory epilepsy in newborns. 89 It also shows that variants in ALDH7A1 may impact patients’ risk of developing osteoporosis. 90 Mutation in the ALDH16A1 gene can influence uric acid homeostasis and is associated with the pathogenesis of gout in humans and Mast syndrome. 91 , 92 Mutation in ALDH18A1 is related to cutis laxa syndromes as well as hyperammonemia due to amino acid abnormalities. 93 , 94 As discussed before and shown in Table 1, we summarize the findings on ALDHs as diagnostic markers in non‐tumor diseases.
TABLE 1.
ALDHs as potential diagnostic markers in non‐tumor diseases
| ALDH isoenzymes | Disease | Ref. |
|---|---|---|
| ALDH1A1 | Cataract formation, Parkinson's disease, Alzheimer's disease | 52 , 54 , 55 , 57 , 58 , 59 |
| ALDH1A2 | Congenital heart disease, congenital anomaly syndrome | 61 , 62 |
| ALDH1B1 | Alcohol‐induced hypersensitivity, diabetes | 24 , 64 , 65 |
| ALDH2 | Alcohol intolerance, hypertension, liver cirrhosis, Alzheimer's disease, Parkinson's disease, coronary heart disease, myocardial infarction | 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 |
| ALDH3A1 | Cataract formation | 84 |
| ALDH3A2 | Sjögren–Larsson syndrome | 86 |
| ALDH4A1 | Hyperprolinemia type II, schizophrenia, atherosclerosis | 81 , 82 |
| ALDH5A1 | Succinic semialdehyde dehydrogenase deficiency | 87 |
| ALDH6A1 | Methylmalonic academia | 88 |
| ALDH7A1 | Pyridoxine‐dependent epilepsy, osteoporosis | 89 , 90 |
| ALDH16A1 | Gout; Mast syndrome | 91 , 92 |
| ALDH18A1 | Cutis laxa (CL) syndromes, hyperammonemia | 93 , 94 |
4. ALDHS AS CSC AND PROGNOSIS MARKERS IN TUMORS
Many studies have shown that CSCs are responsible for maintaining tumor heterogeneity, fueling tumor growth, and therapeutic resistance. For two decades, ALDHs have emerged as excellent biomarkers that can be used for the isolation and characterization of the CSCs population in solid tumors. The ALDEFLUOR assay 95 , 96 and immunostaining have proven useful in the identification and isolation of CSCs. 2 , 97 In the beginning, high ALDH activity enriched in CSCs is exclusively attributed to ALDH1A1, whereas in recent years this high activity has been associated with other isoforms, 98 such as ALDH1A3 99 in breast cancer, ALDH1B1 100 in colon cancer, ALDH3A1 101 in gastric cancer (GC), and ALDH7A1 102 in PCa. In addition, ALDHs have long been regarded as potential prognostic markers in several cancer types, for example, ALDH3A1 in GBM, 17 ALDH1L2 in melanoma, 103 and ALDH18A1 in hepatocellular carcinoma (HCC). 104 Next, we summarize the current knowledge regarding ALDHs as CSC markers or prognostic markers in selected solid cancers that have been extensively studied.
4.1. ALDH1
ALDH1A1: ALDH1A1 is a rate‐limiting enzyme involved in cellular RA synthesis as well as the oxidation of acetaldehydes and LPO‐derived aldehydes. 105 Previously, ALDH1A1 is a major contributor to ALDH1 enzyme activity that could be detected by the ALDEFLOUR assay. The activity of ALDH1A1 is a reliable marker of CSCs in several types of solid tumors, including HNSCC, 12 , 13 , 106 lung cancer, 11 PCa, 14 and bladder cancer. 16 CSCs with high ALDH1A1 activity in these tumors are endowed with highly tumorigenic potential, enhanced capability of self‐renewal, and recapitulation of parental tumor heterogeneity. A well‐known example is that Ginestier et al. reported that ALDH1A1 is a general marker of both normal and cancer human mammary stem/progenitor cells. The marker can be inherited by progeny with the broadest self‐renewal capacity and lineage differentiation potential. These findings provide support for the “CSC hypothesis” and open new avenues for the study of normal breast organogenesis and breast carcinogenesis. 9 , 107 Later, Morimoto et al. investigated that ALDH1A1‐expressing breast cancers are shown to be more likely negative for ER and progesterone receptor (PR) expression, but positive for epidermal growth factor receptor 2 (HER2) and Ki‐67, correlating with more aggressive breast cancer subtypes. 108 Charafe‐Jauffret et al. reported that ALDH1A1 contributes to aggressive behaviors by promoting tumor invasion in vitro and tumor metastasis in mouse xenografts. Moreover, ALDH1A1 expression is an independent predictive factor for early metastasis and decreased survival in inflammatory breast cancer. 109 In two clinical trials to eliminate the activity of breast cancer stem cells (BCSCs) with bevacizumab (VEGF inhibitor) (NCT01190345) or reparixin (CXCR1/2 inhibitor) (NCT01861054), ALDH1A1 is one of the markers to measure the effect of drugs on BCSC activity. Consistent with breast cancer, ALDH1A1‐positive PCa and lung cancer cells display higher colony formation ability and sphere formation efficiency in vitro as well as greatly tumorigenic potential in vivo than ALDH1A1‐negative cells. 11 , 14 By analyzing clinicopathologic parameters, such as tumor stage, tumor grade, and lymph node metastasis (LNM), Kalantari et al. revealed that high ALDH1A1 expressions are significantly associated with PCa tumorigenesis and aggressive behaviors. 110 Furthermore, ALDH1A1 is a marker of both normal stem cells and CSCs in colon tissues. When tracking the colon stem‐cell populations, ALDH1A1‐positive cells are a small subpopulation of cells (≤5%) localized at the bottom of normal crypts where stem cells reside but increase during the stepwise progression to colon cancer. 10 , 111
Accumulating data have confirmed that ALDH1A1 not only stands out as a CSC biomarker but is also associated with tumor metastasis and poor prognosis in different tumor types, such as breast cancer, 9 PCa, 14 HNSCC, 13 bladder cancer, 16 GBM, 112 clear cell renal cell carcinoma (ccRCC), 113 and GC. 114 In breast tumors, consistent conclusions have shown that higher ALDH1A1 expression is associated with larger tumor size, higher histological grade, higher rates of LNM, higher expression of HER2, and lower expression of ER and PR. 115 , 116 , 117 , 118 Moreover, Kida et al. harbored the idea that ALDH1A1 expression is observably higher in triple‐negative breast cancer (TNBC) and HER2 subtypes compared to luminal subtype. 115 Recently, ALDH1A1 drives nuclear factor‐κB and MAPK signaling, which promotes the expansion of myeloid‐derived suppressor cells (MDSCs) to thwart immune surveillance. 119 Thus, shorter disease‐free survival (DFS), recurrence‐free survival (RFS), and/or OS are reported for patients with high levels of ALDH1A1 in breast tumors. 115 , 120 What's more, Zhou et al. are indicative of that ALDH1A1 is an independent prognostic factor in HNSCC patients, and the expression level of PDL‐1 may be involved in ALDH1A1‐mediated poor prognosis in patients. 121 Additionally, ALDH1A1 contributes to the invasiveness of GBM and is an independent predictor of poor clinical outcomes in patients. 112 , 122 However, the clinical relevance of ALDH1A1 in CRCs remains controversial, although an association between the increased expression of ALDH1A1 and clinicopathological parameters, such as larger tumor size, higher histological grade, greater possibility of LNM has been observed across several studies. 105 , 123 , 124 , 125 Lugli et al. showed that 987 (76.7%) of the 1287 CRC tumors have negative cytoplasmic ALDH1A1 expression which indicates that ALDH1A1 expression is not associated with patient survival. 125 However, Kahlert et al. found that the nuclear expression of ALDH1A1 is dramatically associated with shortened OS and DFS. 126 Finally, Holah et al. concluded that the epithelial expression of ALDH1 may be associated with poor prognosis, whereas its stromal expression may be associated with good prognosis in CRC patients. 123 By analyzing 4662 PCa cases and 3114 controls, Cao et al. found that genetic variants of ALDH1A1 in retinol metabolism pathway are closely related to tumor risk. The mutant (rs1330286) genotype in ALDH1A1 is associated with a low risk of PCa, whereas the mutant (rs4646653) genotype in ALDH1A3 is strongly related to a high risk of PCa, indicating that genetic variants in ALDH1A1 and ALDH1A3 may play different roles in the tumorigenesis. 127
ALDH1A2: ALDH1A2 is involved in mediating synthesis of RA. Previous studies have demonstrated that ALDH1A2 serves as a candidate tumor suppressor. Low expression of ALDH1A2 is an unfavorable prognostic biomarker for survival in PCa, 128 HNSCC, 129 breast cancer, 130 , 131 ovarian cancer, 132 and CC 133 patients. Recently, Choi et al. delineated that ALDH1A2 is the most prominently downregulated gene among ALDH family members in ovarian cancer cells, based on microarray analysis. Besides, low ALDH1A2 expression is associated with shorter DFS and OS for ovarian cancer patients. Furthermore, functional assays confirm that the low expression of ALDH1A2 increases the potential of proliferation and invasion in ovarian cancer cells. 132 , 134 An immunohistochemistry (IHC) analysis of ALDH1A2 expression displays that its low expression is correlated with a shorter RFS in human PCa specimens, presumably due to its promoter hypermethylation. 128 Notably, the ALDH1A2 gene is found to be hypermethylated via the DNMT1 or DNMT3B gene, consequently, promoting oncogenic activity. 132 However, previous studies have found that ALDH1A2 and ALDH1B1 might be major contributors to the ALDH1 activity in non‐small cell lung cancers (NSCLCs) and a high expression of ALDH1A2 mRNA is significantly associated with poorer survival in patients, 135 suggesting that ALDH1A2 may play different regulatory roles in different cancers.
ALDH1A3: ALDH1A3 is also one of important regulators in RA synthesis. It is universally acknowledged that ALDH1A3 takes an indispensable role in the generation and maintenance of CSCs in various solid cancers, that is, breast cancer, 99 melanoma, 136 ovarian cancer, 137 PCa, 138 GBM, 17 lung cancer, 139 pancreatic cancer, 140 and CRC. 141 ALDH1A3 is overexpressed in CSCs, always characterized by higher self‐renewal and tumor‐initiating capacity as well as drug resistance and worse prognosis in patients. In a report, including 58 human cancer cell lines, the ALDH1A3 mRNA expression is positively related to its ALDEFLOUR activity, implying that ALDH1A3 is one of the predominate ALDH isoenzymes to maintain the ALDHbright populations. 141 In PCa cells, ALDH1A3 is higher in primary PCa with luminal phenotype than in benign prostatic hyperplasia (BPH) tissues and normal tissues. 127 , 138 , 142 Notably, several studies further investigated that high ALDH1A3 expression is correlated with worse progression‐free survival (PFS) for patients after prostatectomy 143 and longer progression time to castration resistance for patients taking adjuvant hormonal therapy. 144 , 145 Besides, some groups have proposed that ALDH1A3 is an important breast CSC and prognosis marker. 42 , 99 , 146 Marcato et al. published that the knockdown of ALDH1A3 robustly reduces ALDEFLOUR activity in breast cancer cells and tumors. More importantly, ALDH1A3 is positively correlated with breast cancer subtypes, tumor grade, and metastasis. 42 , 99 And in TNBCs and basal‐like subtypes, cancer cells have higher levels of ALDH1A3 expression, implying the prognostic value of ALDH1A3 in breast cancer patients. 147 Interestingly, ALDH1A3 is highly expressed in mesenchymal glioma stem‐like cells (Mes‐GSCs) with a more aggressive phenotype, and the inhibition of ALDH1A3 attenuates the growth of Mes‐GSCs and sensitizes Mes‐GSCs to radiotherapy, suggesting that ALDH1A3 is a promising biomarker for Mes‐GSCs. 17 , 148 , 149 , 150 Mechanistically, forkhead box D1 protein triggers self‐renewal and tumorigenicity of Mes‐GSCs both in vitro and in vivo by regulating the transcriptional activity of ALDH1A3. 150 Further studies indicate that ALDH1A1 or ALDH1A3 as markers of GSCs may be correlated with distinct molecular subtypes of high‐grade glioma (HGG) tumors, with ALDH1A3 being a marker of the mesenchymal subtype 17 , 150 , 151 and ALDH1A1 being a marker of the classical subtype. 122 Surprisingly, as reported by Luo et al., ALDHbright melanoma‐initiating cells (MICs) highly express ALDH1A1 and ALDH1A3 isoenzymes, which have enhanced tumorigenic potential compared to ALDH‐negative cells 136 and may be associated with distinct phenotypes of MICs. ALDH1A1 is predominantly expressed in human melanoma tumor samples, whereas ALDH1A3 is predominantly expressed in human melanoma cell lines. 152
ALDH1B1: ALDH1B1 is a mitochondrial enzyme that can oxidize a broad range of substrates, including short‐ and medium‐chain aliphatic aldehydes, RALs and LPO‐derived products. 153 A growing number of studies have revealed that the aberrant expression of ALDH1B1 has been observed in several human cancers such as CRC, 154 pancreatic cancer, 15 NSCLC, 135 GC, and HCC, 155 and it is involved in tumorigenesis and metastasis as well as clinical prognosis. By analyzing ALDH1B1 mRNA and protein levels, consistent conclusions suggest that ALDH1B1 levels distinguish CRC tissues from normal tissues. 156 , 157 Specifically, ALDH1B1 protein is 5.6‐fold higher than ALDH1A1 in CRC patients. 154 In addition, Singh et al. showed that ALDH1B1 is a major contributor to ALDEFLOUR activity in highly tumorigenic colon cancer cells and can promote tumorigenesis by modulating the Wnt/β‐catenin, Notch and PI3K/Akt signaling pathways. 100 In summary, ALDH1B1 is an excellent colon cancer biomarker. Afterward, ALDH1B1 has been shown to be a progenitor or stem‐cell marker during pancreas development and carcinogenesis. 15 , 158 ALDH1B1 is abundantly expressed in human pancreatic cancer and the high expression of ALDH1B1 contributes to cancer progression and dreadful prognosis. 105 , 159 Interestingly, ALDH1B1 expression is lower in HCC when compared with normal tissues, and lower ALDH1B1 expression is associated with an unfavorable prognosis in terms of OS and RFS, 155 consistent with its roles in GC patients. 114 Polymorphisms of ALDH1B1 can cause marked reductions in acetaldehyde metabolism ability and consequently result in flushing syndrome and ethanol avoidance. Studies reported that tobacco and/or alcohol consumption in carriers of the ALDH1B1 polymorphism increases the risk of oral squamous cell carcinoma (OSCC). 160 A clinical trial (NCT04270201) to assess the relationship between ALDH1B1 polymorphism frequency and OSCC risk in the Brazilian population is underway. Further research using large prospective patient cohorts are warranted to determine the prognostic value of ALDH1B1 in certain cancers.
ALDH1L1: ALDH1L1, also known as 10‐formyltetrahydrofolate (10‐formyl‐THF) dehydrogenase, one of the folate‐metabolizing enzymes, catalyzes the oxidization of 10‐formyl‐THF to generate CO2 with concomitant NADPH production. 161 Studies have established that ALDH1L1 is often strongly and universally downregulated or silenced in a multitude of human solid cancers, including HCC, 162 , 163 , 164 lung adenocarcinoma (LUAD), 165 ccRCC, 166 neuroblastoma (NB), 167 and breast cancer. 168 Data indicate that the ALDH1L1 gene promoter is frequently hypermethylated in cancer, with low mRNA levels predicting poorer clinical outcomes. 165 , 166 , 168 , 169 , 170 For example, the ALDH1L1 gene is hypermethylated in breast tumors after acquisition of chemoresistance 171 and may be important for tumor cell survival in response to metabolic stress. 172 Overall, ALDH1L1 could be a candidate tumor suppressor for aggressive cancers. However, the prognostic role of ALDH1L1 may be cancer‐type‐specific. Li et al. reported that ALDH1A3 and ALDH1L1 are potentially major contributors to ALDH1 activities in GC, and high mRNA expressions of ALDH1A3 and ALDH1L1 predict poorer OS in GC patients. 114 Genetic variation analyses show that the ALDH1L1 variant rs2276724 genotype is of prognosis value in hepatitis B virus–related HCC patients and the carriers of rs2276724 are more probably with a favorable prognosis. 173 Further well‐designed, comprehensive, and large sample size studies are therefore needed to confirm these results.
ALDH1L2: ALDH1L2 is an important mitochondrial paralog of ALDH1L1 and the product of a separate gene. 174 Although both ALDH1L1 and ALDH1L2 are involved in folate metabolism, their compartmentalization results in distinct effects on overall cellular metabolism, regulating either folate pools and purine biosynthesis (cytosolic ALDH1L1) or NADPH production and oxidative stress (mitochondrial ALDH1L2). ALDH1L2 is highly expressed and presents as an independent prognostic factor for OS and RFS in melanoma, pancreatic ductal adenocarcinoma (PDAC), CRC, and GBM. 103 , 175 Several studies have shown that the depletion of ALDH1L2 markedly reduces NADPH/NADP+ and glutathione/oxidized glutathione redox couple ratios, reduces circulating tumor cells (CTCs) in the blood, and inhibits distant metastasis. 103 , 175 , 176 On the other hand, evidence suggests that ALDH1L2 combats oxidative stress by increasing total cellular NADPH, which is responsible for GSC maintenance and growth. 177
4.2. ALDH2
ALDH2 is known for its role in the metabolism of ethanol‐derived acetaldehydes. 178 , 179 Notably, ALDH2 dysfunction has been widely reported to be associated with tumorigenesis and tumor progression, which is often considered a feasible prognostic marker in different solid tumor types. Low ALDH2 expression is always responsible for poorer survival and more aggressive behaviors in liver cancer patients. ALDH2 can participate in inhibiting HCC cells metastasis via the adenosine 5′‐monophosphate‐activated protein kinase pathway, 180 promoting HCC cell's autophagy and repressing its immune escape by blocking the ROS/Nrf2 axis. 181 Remarkably, it is indicating that the polymorphisms of ALDH2 might have an important influence on liver cancer. The ALDH2*2 mutant (rs671) genotype is a shared missense mutation in the ALDH2 gene with severely reduced activity of the ALDH2 enzyme, 182 which has been found to increase protein turnover and promote hepatocarcinogenesis in vivo. 183 More recently, the polymorphism of ALDH2 gene is reported to be a risk factor with a prognostic value for alcohol‐related cancers, including HNSCC, EC, HCC, CRC, GC, and breast cancer. 184 Furthermore, a lower expression of ALDH2 in these tumors is associated with poorer OS and PFS.
However, there are also several studies emphasize that ALDH2 overexpression or high activity is associated with cancer progression and multidrug resistance. 185 Chen et al. reported that ALDH2 promotes the expression of cancer stem genes (e.g., Nanog, Oct4, and Sox2), leading to the proliferation, migration, and invasion of CD133+CD24+ Huh‐7 liver CSCs. 186 Wang et al. found that the expression of ALDH2 is also upregulated in NSCLC cells, which is associated with cancer cell stemness property and paclitaxel resistance. 187 More implementation is needed to outline a possible role for ALDH2 in cancer.
4.3. ALDH3
ALDH3A1: ALDH3A1 participates in oxidation of alcohol‐derived acetaldehydes and in the metabolism of corticosteroids, biogenic amines, neurotransmitters, and LPO. 188 In recent years, ALDH3A1 has been shown to be upregulated in several cancer types, for example, GC, 101 lung cancer, 189 , 190 HCC, 191 colon cancer, 192 PCa 193 and can be used as a biomarker to predict poor clinical outcomes. It has been demonstrated by Wu et al. that ALDH3A1 is upregulated in GC stem cells, which contributes to gastric carcinogenesis, and the high expression of ALDH3A1 protein is associated with a poorly differentiated state in GC tissues. 101 In addition, the IHC analysis of GC patient specimens demonstrates that ALDH3A1 expression is closely correlated to clinical features, including cancer dysplasia and grade, LNM, and cancer stage. 101 , 194 Collectively, ALDH3A1 is a known CSC and prognosis marker for GC. More recently, publicly available data from The Cancer Genome Atlas (TCGA) find that ALDH3A1 is highly expressed in LUAD patients with metastasis, linked to the characteristics of CSCs and EMT. Besides, high ALDH3A1 expression predicts a poor prognosis in LUAD. 195 Several groups have provided supportive evidence that ALDH1A1 and ALDH3A1 are expressed at significantly high levels in NSCLC that indicates poorer OS. 189 , 196 , 197 Correspondingly, findings indicate that ALDH3A1 boosts tumor initiation and progression in PCa. ALDH3A1 shows high expression levels under sphere‐forming conditions in vitro by DU145 prostate CSCs. 193 As the association of ALDH1A1 and ALDH7A1 with PCa progression has been definitively demonstrated, 102 , 138 it is proposed that high levels of ALDH3A1, ALDH1A1, and ALDH7A1 are involved in prostate tumorigenesis, though the detailed contribution of them to PCa requires further investigation.
ALDH3A2: ALDH3A2 catalyzes the oxidation of fatty aldehydes, and mutation in the ALDH3A2 gene results in SLS. 86 Analysis of clinicopathological features reveals an association of reduced ALDH3A2 with advanced and poorly differentiated cancers. For instance, gene set enrichment analysis and IHC results indicate that high ALDH3A2 expression is associated with superior OS of GC patients. Moreover, ALDH3A2 is negatively correlated with immune checkpoints, including cytotoxic T‐lymphocyte‐associated antigen 4, programmed cell death protein 1 in immune cells, and its ligand PD‐L2 in GC cells. 198 More recently, Antonowicz et al. supported that the loss of ALDH3A2 expression is linked to malignant transformation and progression and is associated with poorer survival in esophageal adenocarcinoma. 199 Surprisingly, ALDH3A2 represents a very intriguing isoform as its expression is downregulated but increased in response to various treatment trials in PCa. 200 Altogether, these findings potentially hint at a tumor suppressor role of ALDH3A2.
ALDH3B1: ALDH3B1 is generally thought to detoxify aldehydes from alcohol metabolism and LPO. 201 ALDH3B1 is significantly engineered to be overexpressed in lung cancer 202 and is positively correlated with patient tumor size and histological grade, indicating that ALDH3B1 is an independent prognostic biomarker of lung cancer patients. 203 ALDH3B1 and ALDH16A1 are preferentially overexpressed in HGG and can promote the proliferation and migration of glioma cells by regulating cell cycle and EMT processes. Furthermore, high expressions of ALDH3B1 and ALDH16A1 are positively associated with worse treatment response and shorter OS in GBM patients. 204
ALDH3B2: ALDH3B2 may play a major role in the detoxification of aldehydes generated by alcohol metabolism and LPO. 205 Previous data suggest that ALDH3B2 gene polymorphisms are associated with susceptibility to CRC and ESCC. 206 Furthermore, ALDH3B2 protein expression is significantly higher in the RCC tissues compared to the normal renal tissues, and ALDH3B2 independently predicts worse OS in patients. 207 More recently, Wang et al. demonstrated that high expressions of ALDH3B2 and integrin beta 1 are strong inferior prognostic biomarkers in cholangiocarcinoma (CCA) patients. 208
4.4. ALDH4
ALDH4A1: ALDH4A1, also known as delta‐1‐pyrroline‐5‐carboxylate (P5C) dehydrogenase, is involved in the metabolism of the amino acid arginine, proline and l‐valine. 34 ALDH4A1 can serve as a potential disease indicator, in which circulating ALDH4A1 is increased during atherosclerosis in mice and humans and anti‐ALDH4A1 antibodies can protect against atherosclerosis progression. 83 However, the precise role of ALDH4A1 in cancer progression and patient prognosis remains unclear. Previously, ALDH4A1 expression is transcriptionally activated by p53 to prevent the proline‐dependent ROS production, which confers a survival advantage to U373MG GBM cells and H1299 NSCLCs. 209 Additionally, polymorphisms of ALDH4A1, ALDH18A1, ALDH3B2, ALDH1L2, ALDH1A2, and ALDH2*2 are significantly associated with an elevated nasopharyngeal carcinoma risk. 210
4.5. ALDH5
ALDH5A1: ALDH5A1, which encodes for SSADH, is an enzyme involved in mitochondrial glutamate metabolism. 211 ALDH5A1 converts succinic semialdehyde into succinate, fueling the TCA cycle and thereby limiting the γ‐hydroxybutyrate (GHB) production. 212 It is reported that high ALDH5A1 expression is associated with stem‐cell‐like properties and aggressive behaviors within the context of GBM. On the contrary, the inhibition of ALDH5A1 leads to the accumulation of GHB that induces GBM stem‐like cell differentiation and reduces the aggressive phenotype. 213 RNA sequencing analysis of human breast ductal carcinoma in situ (DCIS) cells demonstrates that ALDH5A1 is overexpressed at both the mRNA and protein levels. Two independent drugs, disulfiram (DSF) 214 and valproic acid, 215 inhibit ALDH5A1 activity to reduce proliferation of DCIS. 216 More recently, the analysis results from the TCGA database show that ALDH5A1 is an excellent prognostic factor in patients with primary papillary thyroid cancer (PTC), manifesting that the high expression of ALDH5A1 predicts a worse prognosis. Mechanically, ALDH5A1 can enhance the migration and invasion ability of thyroid cancer cells through EMT transition, suggesting that ALDH5A1 may emerge as a new target for PTC therapy. 217 Conversely, mRNA and protein expressions of ALDH5A1 are lower in HCC and ovarian cancer that predicts poorer OS in patients. 218 A recent study by Kong et al. discovers that ALDH5A1 is upregulated upon the knockdown of a prostate‐specific antigen in PCa cells, accompanied by reduced tumorigenesis and metastasis in vitro and in vivo. 219 Hence, ALDH5A1 may act as an oncogene or tumor suppressor in various human cancers and is strongly associated with the clinical outcomes.
4.6. ALDH6
ALDH6A1: ALDH6A1 is a mitochondrial methylmalonate semialdehyde dehydrogenase that plays a vital role in the valine and pyrimidine catabolic pathways. ALDH6A1 catalyzes the irreversible oxidative decarboxylation of methylmalonate semialdehyde to acetyl‐CoA and propionyl‐CoA. 220 Extensive clinical analyses have shown that the decreased expression of ALDH6A1 is correlated to tumorigenesis and inferior outcomes in different kinds of cancers, which may serve as a potential diagnostic and prognostic biomarker. ALDH6A1 expression is suppressed in HCC, which is accompanied by an elevation of NO levels and a reduction of ROS levels that may support abnormal HCC cell growth. 221 Expressions of 4‐aminobutyrate aminotransferase and ALDH6A1 are significantly reduced in ccRCC patients, which are correlated with poorer survival. Overexpression of ALDH6A1 reduces cell growth and migration and impairs tumor metabolism of ccRCC cells. 222 In addition, ALDH6A1 is significantly reduced in metastatic PCa compared with normal and primary PCa, an indication of that ALDH6A1 may be a predictive biomarker for metastatic PCa. 223 Consistent with these reports, ALDH6A1 also functions as a tumor suppressor in bladder cancer. ALDH6A1 is remarkably downregulated in bladder cancer tissues and cell lines. Low expression of ALDH6A1 is positively associated with advanced cancer subtype and cisplatin resistance, hinting at a poorer outcome in bladder cancer patients. 224
4.7. ALDH7
ALDH7A1: ALDH7A1 could function in the protection of cells from oxidative stress by metabolizing a number of LPO‐derived aldehydes and function in lysine catabolism. 225 ALDH1A1 and ALDH7A1 are previously found to contribute to ALDEFLOUR activity in prostate CSCs. 102 Besides, a high expression of ALDH7A1 is suggested to predict disease progression and metastasis. van den Hoogen et al. delineated that the mRNA expression levels of several ALDH isoforms, including ALDH7A1, are evaluated in PCa clinical specimens and cell lines. Notably, strong ALDH7A1 expression triggers PCa bone metastasis. 102 , 226 Indeed, the knockdown of ALDH7A1 results in a significant reduction of ALDHbright populations and significantly inhibits clonogenicity and cell invasion of human PCa cells in vitro. 102 Subsequent studies demonstrate that ALDH1A3, ALDH7A1, and ALDH18A1 isoforms are more robustly expressed in PCa than in BPH and normal samples. 138 Similarly, low ALDH7A1 expression is associated with a lower incidence of cancer recurrence and superior RFS and OS in surgically resected NSCLC patients. 227 Recently, significantly reduced methylation levels of ALDH7A1 gene are observed in lung SCC with idiopathic pulmonary fibrosis compared with patients without fibrosis, which predicts poor outcome. 228 Taken together, ALDH7A1 has the potential to be a clinically useful biomarker in cancers.
4.8. ALDH8
ALDH8A1: ALDH8A1 is critical for catalyzing RALs to the corresponding RA. Bioinformatics analysis by Chen et al. reveals that ALDH8A1 has higher levels of promoter DNA methylation in CCA. 229 And in liver cancer, an eight‐gene signature expression, including ALDH8A1, is significantly downregulated in cancer tissues, and low expression can be regarded as an inferior prognostic biomarker. 230
4.9. ALDH9
ALDH9A1: ALDH9A1 is an enzyme that encodes γ‐trimethylaminobutyraldehyde dehydrogenase that catalyzes γ‐aminobutyraldehyde to GABA. 231 However, knowledge about the aberrant ALDH9A1 expression on cancer development remains elusive. The abundance of ALDH1A1, ALDH4A1, ALDH6A1, ALDH7A1, and ALDH9A1, which are known to be involved in xenobiotic metabolism signaling, is elevated in RCC. 232 In another comparative study, Stevenson et al. showed that ALDH1A1, ALDH2, and ALDH9A1 proteins are present in LNM specimens from breast cancer, PDAC and PCa, indicating a potential common role of these proteins in the development of LNM. 233
4.10. ALDH16
ALDH16A1: The function of ALDH16A1 reported in cancer is limited. Analysis by Wang et al. manifested that ALDH16A1 is considered a prognostic biomarker in gliomas. They show that ALDH2, ALDH5A1, ALDH6A1, ALDH1L2, ALDH1B1, ALDH18A1, ALDH1A1, ALDH8A1, and ALDH1B1 expressions are higher in isocitrate dehydrogenase (IDH) mutant gliomas and demonstrated to be favorable factors for patients with low‐risk scores, whereas ALDH16A1, ALDH3B1, ALDH3A1, ALDH7A1, ALDH1A3, ALDH1L1, and ALDH1A2 expressions are increased in IDH wildtype and are associated with a worse prognosis risk. 204
4.11. ALDH18
ALDH18A1: ALDH18A1 is a mitochondrial enzyme that catalyzes the conversion of l‐glutamate to P5C, an important intermediate step in proline metabolism. 234 ALDH18A1 overexpression locks melanoma cells into a more proliferative or tumorigenic state via proline biosynthesis. 235 ALDH18A1 knockdown decreases intracellular proline levels and impairs melanoma cell viability and tumor progression. Thus, the proline synthesis pathway can be therapeutically targeted by inhibiting ALDH18A1 in melanoma. 236 ALDH9A1 and ALDH18A1 are dramatically upregulated in CTCs of PCa compared to primary tumors. 237 Moreover, the high expression of ALDH18A1 predicts a poor clinical outcome in HCC, 104 NSCLC, 238 , 239 and breast cancer. 240 ALDH18A1 is overexpressed in highly proliferative luminal B compared to low proliferative luminal A breast cancer subtypes. 240 A recent study reveals that ALDH18A1 forms a positive feedback loop with MYCN and is involved in the regulation of the proliferation, self‐renewal and tumorigenicity of NB cells. Besides, the high expression of ALDH18A1 is linked to poorer OS and event‐free survival in the Kaplan–Meier plot. Targeting ALDH18A1 with a specific inhibitor, YG1702, dictates MYCN expression suppression and attenuates NB growth in preclinical xenograft models. 241
Based on the previous discussion, we further summarize the findings about characteristics of ALDHs in tumors in Table 2. However, the precise roles of ALDH isoenzymes in cancer cells with a stem‐cell‐like phenotype or clinical prognostic significance require further elucidation as they may greatly vary by cancer types and tissues of origin.
TABLE 2.
Characteristics of aldehyde dehydrogenases (ALDHs) family in tumors
| ALDH isoenzymes | Chromosomal location | Subcellular localization | Functional activity | CSC marker | Prognostic value | Clinical trials | Ref. | |
|---|---|---|---|---|---|---|---|---|
| ALDH families | ||||||||
| ALDH1 | ALDH1A1 | 9q21.13 | Cytosol | Oxidation of RALs to RAs; oxidation of acetaldehydes and LPO‐derived aldehydes | Breast cancer, HNSCC, GBM, lung cancer, ovarian cancer, GC, PCa, bladder cancer, CRC, etc. | High expression predicts unfavorable prognosis in patients with breast cancer, HNSCC, GBM, EC, GC, lung cancer, PCa, bladder cancer, and ccRCC, etc. |
NCT01190345 (Phase II/completed) NCT01861054 (Phase II/terminated) NCT04878549 (not applicable/recruiting) |
9 , 10 , 11 , 13 , 14 , 16 , 110 , 112 , 113 , 114 |
| ALDH1A2 | 15q21.3 | Cytosol | Oxidation of RALs to RAs |
High expression predicts favorable prognosis in patients with PCa, HNSCC, breast cancer, ovarian cancer, CC, etc. High expression predicts unfavorable prognosis in patients with NSCLC, etc. |
128 , 129 , 130 , 132 , 133 , 135 | |||
| ALDH1A3 | 15q26.3 | Cytosol nucleus | Oxidation of RALs to RAs | Pancreatic cancer, PCa, melanomas, GBM, ovarian cancer, lung cancer, CRC, breast cancer, etc. | High expression predicts unfavorable prognosis in patients with pancreatic cancer, GBM, PCa, ovarian cancer, lung cancer, CRC, melanoma, breast cancer, etc. | 17 , 99 , 136 , 137 , 138 , 139 , 140 , 141 | ||
| ALDH subfamilies | ||||||||
| ALDH1 | ALDH1B1 | 9p13.1 | Mitochondria nucleus cytosol | Oxidation of acetaldehydes and LPO‐derived aldehydes | CRC, pancreatic cancer, etc. |
High expression predicts unfavorable prognosis in patients with CRC, pancreatic adenocarcinoma, NSCLC, etc. High expression predicts favorable prognosis in patients with HCC, GC, etc. |
NCT04270201 (not applicable/not yet recruiting) | 15 , 100 , 114 , 135 , 154 , 155 |
| ALDH1L1 | 3q21.3 | Cytosol | Conversion of 10‐formyl‐THF in tetrahydrofolate |
High expression predicts favorable prognosis in patients with LUAD, HCC, ccRCC, NB, breast cancer, etc. High expression predicts unfavorable prognosis in patients with GC, etc. |
114 , 162 , 165 , 166 , 167 , 168 | |||
| ALDH1L2 | 12q23.3 | Mitochondria | Conversion of 10‐formyl‐THF in tetrahydrofolate | High expression predicts unfavorable prognosis in melanoma, PDAC, CRC, GC, GBM, etc. | 103 , 175 , 176 , 177 | |||
| ALDH subfamilies | ||||||||
| ALDH2 | ALDH2 | 12q24.12 | Mitochondria | Oxidation of alcohol and LPO‐derived acetaldehydes | High expression predicts favorable prognosis in patients with liver cancer, lung cancer, breast cancer, EC, CRC, GC, HNSCC, etc. |
NCT00661141 (Phase II/Completed) (Not applicable/Not yet recruiting) |
180 , 184 | |
| ALDH3 | ALDH3A1 | 17p11.2 | Plasma membrane cytosol | Oxidation of alcohol‐derived acetaldehydes and in the metabolism of corticosteroids, biogenic amines, neurotransmitters, and LPO | GC, lung cancer, HCC, colon cancer, PCa, etc. | High expression predicts unfavorable prognosis in patients with GC, LUAD, NSCLC, HCC, PCa, colon cancer, etc. | 101 , 189 , 190 , 191 , 192 , 193 | |
| ALDH3A2 | 17p11.2 | Endoplasmic reticulum Peroxisomes | Oxidation of long‐chain aliphatic‐aldehydes into fatty acids | High expression predicts favorable prognosis in patients with GC, EAC, PCa, etc. | 198 , 199 , 200 | |||
| ALDH subfamilies | ||||||||
| ALDH3 | ALDH3B1 | 11q13.2 | Plasma membrane cytosol | Detoxification of aldehydes from alcohol metabolism and LPO | High expression predicts unfavorable prognosis in patients with lung cancer, GBM, etc. | 202 , 203 , 204 | ||
| ALDH3B2 | 11q13.2 | Lipid droplet | Detoxification of aldehydes from alcohol metabolism and LPO | High expression predicts unfavorable prognosis in patients with RCC, CCA, etc. | 207 , 208 | |||
| ALDH4 | ALDH4A1 | 1p36.13 | Mitochondria | Metabolism of the amino acid arginine, proline, and l‐valine | ||||
| ALDH subfamilies | ||||||||
| ALDH5 | ALDH5A1 | 6p22.3 | Mitochondria | Conversion of succinic semialdehyde in succinate |
High expression predicts unfavorable prognosis in patients with GBM, DCIS, PTC, etc. High expression predicts favorable prognosis in patients with HCC, ovarian cancer, etc. |
213 , 216 , 217 , 218 , 219 | ||
| ALDH6 | ALDH6A1 | 14q24.3 | Mitochondria | Oxidation of methylmalonate semialdehyde to acetyl‐CoA and propionyl‐CoA; Involvement in valine, and pyrimidine metabolism | High expression predicts favorable prognosis in patients with HCC, ccRCC, PCa, bladder cancer, etc. | 221 , 222 , 223 , 224 | ||
| ALDH7 | ALDH7A1 | 5q23.2 | Nucleus Cytosol mitochondria | PCa | High expression predicts unfavorable prognosis in patients with PCa, NSCLC, lung SCC, etc. | 102 , 226 , 227 , 228 | ||
| ALDH subfamilies | ||||||||
| ALDH8 | ALDH8A1 | 6q23.3 | Cytosol | Oxidation of RALs to RA | High expression predicts favorable prognosis in patients with liver cancer | 230 | ||
| ALDH9 | ALDH9A1 | 1q24.1 | Cytosol | Oxidation of γ‐aminobutyraldehyde to GABA | ||||
| ALDH16 | ALDH16A1 | 19q13.33 | Plasma membrane cytosol | Not fully discovered | High expression predicts unfavorable prognosis in patients with gliomas | 204 | ||
| ALDH18 | ALDH18A1 | 10q24.1 | Mitochondria | Conversion of l‐glutamate to P5C | High expression predicts unfavorable prognosis in patients with HCC, NSCLC, breast cancer, melanoma cancer, PCa, NB, etc. | 104 , 236 , 237 , 238 , 240 , 241 | ||
Abbreviations: 10‐formyl‐THF, 10‐formyltetrahydrofolate; CC, cervical cancer; CCA, cholangiocarcinoma; ccRCC, clear cell renal cell carcinoma; CRC, colorectal cancer; CSC, cancer stem cell; DCIS, breast ductal carcinoma in situ; EAC, esophageal adenocarcinoma; EC, esophageal cancer; GABA, γ‐aminobutyric acid; GBM, glioblastoma; GC, gastric cancer; HCC, hepatocellular carcinoma; HNSCC, head‐and‐neck squamous cell carcinoma; LPO, lipid peroxidation; LUAD, lung adenocarcinoma; NB, neuroblastoma; NSCLC, non‐small cell lung cancers; P5C, pyrroline‐5‐carboxylate; PCa, prostate cancer; PDAC, pancreatic ductal adenocarcinoma; PTC, papillary thyroid cancer; RA, retinoic acid; RAL, retinaldehyde.
5. THERAPEUTIC POTENTIAL OF TARGETING ALDHS
As mentioned before, ALDHs are widely accepted as functional biomarkers and regulators of stemness phenotypes in many diseases. Here, we further review the studies concerning the therapeutic potential of targeting ALDHs in diseases, especially solid tumors.
5.1. Targeting ALDHs with ALDH inhibitors
ALDH inhibitors can be classified into multi‐ALDH isoform inhibitors and isoform‐specific inhibitors which primarily inhibit one isoform. 178 , 242 The multi‐ALDH isoform inhibitors include DSF, diethylaminobenzaldehyde (DEAB), 4‐dimethylamino‐4‐methyl‐pent‐2‐ynthioic acid‐S‐methylester (DIMATE), citral, and aldehyde dehydrogenase inhibitors 1–4 and 6, dyclonine (Table 3).
TABLE 3.
Multi‐ALDH (aldehyde dehydrogenase) isoform inhibitors
| Inhibitor | Target | Cellular activity | Animal/clinical studies | Ref. |
|---|---|---|---|---|
| DSF | Irreversible ALDH inhibitor | IC50: 0.15 μM for ALDH1, and 1.45 μM for ALDH2 | DSF/Cu can inhibit ALDH‐positive NSCLC tumors. The combination of DSF and gemcitabine efficiently inhibits breast tumors; the combination of TMZ and DSF/Cu in a Phase II clinical trial (NCT03034135) rescue ALDH1A3‐mediated TMZ resistance in GBM patients | 243 , 244 , 245 , 246 , 247 |
| DEAB | A reversible substrate for ALDH1A1 and ALDH3A1, and displays competitive inhibition of ALDH1A1, ALDH1A3, ALDH1B1, and ALDH5A1 | IC50: 57 nM for ALDH1A1, 1.2 μM for ALDH1A2, 3 μM for ALDH1A3, 1.2 μM for ALDH1B1, 0.16 μM for ALDH2, and 13 μM for ALDH5A1 | Inhibition of tumor growth and pulmonary metastasis in breast cancer tumors | 95 , 248 |
| DIMATE | ALDH1 and ALDH3 subfamilies | IC50: 5 μM for ALDH1A1 and ALDH3A1 | DIMATE is effective at a dose of 14 mg/kg daily given via intraperitoneal injection in PCa tumors | 249 , 250 |
| Citral | Reversible noncompetitive ALDH inhibition | Potent inhibitory activity against ALDH1A1, ALDH1A3, and ALDH2 | Nanoparticle encapsulated citral reduces the tumor growth of ALDH1A3‐overexpressing MDA‐MB‐231 xenografts | 250 , 251 |
| Aldi‐1–4 | Covalent ALDH inhibition | IC50: range from 2.2 to 7.9 μM for ALDH1A1, 5.4 to 8.6 μM for ALDH2, and 1.7 to 12 μM for ALDH3A1 | No in vivo studies | 252 |
| Aldi‐6 | Covalent ALDH inhibition | IC50: 600 nM for ALDH1A1, 800 nM for ALDH2, and 1 μM for ALDH3A1 | The combination of Aldi‐6 and cisplatin results in a greater reduction in tumor burden in HNSCCs | 253 |
| Dyclonine | Covalent ALDH inhibition | IC50: 35 μM for ALDH2, and 76 μM for ALDH3A1 | The combination of dyclonine and sulfasalazine efficiently suppresses the growth of ALDH3A1‐expressing HNSCC or GC tumors | 252 , 254 |
Abbreviations: DEAB, diethylaminobenzaldehyde; DIMATE, 4‐dimethylamino‐4‐methyl‐pent‐2‐ynthioic acid‐S‐methylester; DSF, disulfiram; GBM, glioblastoma; GC, gastric cancer; HNSCC, head‐and‐neck squamous cell carcinoma; IC50, inhibitory concentration; NSCLC, non‐small cell lung cancers; PCa, prostate cancer; TMZ, temozolomide.
DSF is an irreversible pan‐ALDH inhibitor, and it has an inhibitory concentration (IC50) of 0.15 μM for ALDH1 and an IC50 of 1.45 μM for ALDH2. 243 , 255 DSF is a Food and Drug Administration–approved anti‐alcoholism drug in 1951. 256 In the past decades, both in vivo and in vitro experiments have shown that DSF has excellent treatment efficacy, and there are some clinical studies (Table 4) indicating that DSF is now used in patients with some diseases like alcohol‐related disorders and solid tumors. For example, DSF is used in clinical routine for the recrudescence prevention of alcohol dependency or severe alcoholism (NCT00431262) that acts as a “psychological deterrent” and causes unpleasant physical reactions after alcohol consumption. 257 Some clinical trials have shown that DSF is used in combination with other drugs, such as lorazepam for the treatment of alcohol dependency and anxiety disorders (NCT00721526). Moreover, DSF treatment can increase the expression of metalloproteinase 10 (ADAM10) in peripheral blood cells of AD mice, suggesting that DSF can be repurposed as an ADAM10 enhancer and a therapeutic approach for AD. 258 These early findings emphasize the necessity of clinical research to verify the therapeutic utility on human patients by analyzing the expression of ADAM10 in the collected blood samples (NCT03212599).
TABLE 4.
Clinical trials for disulfiram (DSF)‐based therapy
| Condition or disease | Drugs | Study phase | Status | Identifier |
|---|---|---|---|---|
| Alcoholism | Disulfiram | Not applicable | Unknown | NCT00431262 |
| Alcohol‐related disorders | Disulfiram | Not applicable | Completed | NCT00142844 |
| Cocaine‐related disorders | Naltrexone | |||
| Alcohol dependence | Disulfiram | Not applicable | Completed | NCT00721526 |
| Anxiety disorder | Lorazepam | |||
| Alcohol addiction | Disulfiram | Not applicable | Completed | NCT03212599 |
| Alzheimer's disease | ||||
| Recurrent GBM | Disulfiram/copper | Phase II | Completed | NCT03034135 |
| Temozolomide | ||||
| GBM | Disulfiram/copper | Phase II | Not yet recruiting | NCT01777919 |
| Temozolomide | ||||
| Metastatic breast cancer | Disulfiram | Phase II | Recruiting | NCT04265274 |
| Chemotherapy | ||||
| NSCLC | Disulfiram | Phase II/III | Completed | NCT00312819 |
| Chemotherapy |
Abbreviations: GBM, glioblastoma; NSCLC, non‐small cell lung cancers.
Source: clinicaltrials.gov website.
In solid tumors, DSF has been highlighted as a potential cancer therapy drug, and its cytotoxicity depends on copper (Cu). 259 , 260 , 261 Studies have shown that DSF can form a complex with Cu (DSF/Cu), which is more readily taken up by cells and exerts cytotoxic effects on a variety of cancer cells while sparing normal cells. 244 , 260 , 262 , 263 DSF/Cu can inhibit ALDH‐positive NSCLC stem cells in vitro and tumors derived from sorted ALDH‐positive CSCs in vivo. 245 A Phase I/II clinical trial has been completed and manifests surprisingly encouraging results that the combination of DSF with chemotherapy can prolong OS and PFS in patients with NSCLC (NCT00312819). 264 In addition, preclinical studies have confirmed that DSF in combination with gemcitabine or programmed death‐ligand 1 (PD‐L1) antibody efficiently inhibits 4T1 breast cancer tumorigenicity and growth by targeting ALDH1A1 enriched CSCs and MDSCs, respectively. 119 On the other hand, DSF/Cu can inhibit breast cancer metastasis by triggering apoptosis or inhibiting EMT in cultures and animal models. Based on the previous data, a Phase II clinical trial of DSF/Cu combination chemotherapy is being conducted to evaluate the therapeutic potential for patients with metastatic breast cancer (NCT04265274). A recent study highlights that ALDH1A3 is upregulated in recurrent GBM and is more resistant against temozolomide (TMZ) treatment. 151 Mechanically, ALDH1A3 participates in detoxification of LPO‐derived reactive aldehydes after TMZ treatment. TMZ in combination with DSF/Cu rescues ALDH1A3‐mediated TMZ resistance in GBM patients in a Phase II clinical trial (NCT03034135). The treatment is well tolerated, with only 1 of 23 patients (4%) experiencing dose‐limiting toxicity. 265 In addition to ALDH1A3, ALDH1A1 has also been shown to be a mediator of GBM resistance to TMZ and a reliable predictor of clinical outcomes. In an ongoing Phase II clinical trial, TMZ combined with DSF/Cu will be used as adjunctive and concurrent chemotherapy to treat newly diagnosed GBM (NCT01777919), but the results have not been published.
DEAB is a commonly used competitive and reversible ALDH inhibitor and is provided as a negative control compound in ALDEFLOUR assay. 95 DEAB is a reversible substrate for ALDH1A1 and ALDH3A1 and displays competitive inhibition of ALDH1A1, ALDH1A3, ALDH1B1, and ALDH5A1. Moreover, it has a 50% IC50 of 57 nM for ALDH1A1, 1.2 μM for ALDH1A2, 3 μM for ALDH1A3, 1.2 μM for ALDH1B1, 0.16 μM for ALDH2, and 13 μM for ALDH5A1. 95 Its role as an ALDH inhibitor has been extensively studied in breast cancer and ovarian cancer. DEAB can efficiently inhibit the growth of ovarian cancer cells and alleviate breast cancer tumor burden and pulmonary metastasis. 95 , 248 Besides, DEAB reduces the chemotherapeutic and radiotherapeutic resistance of ALDHbrightCD44high BCSCs and decreases the population of CD133+ ovarian CSCs. 266 , 267 However, its dependency on ALDH expression limits its use currently as an anticancer agent. 248
DIMATE is the most potent ALDH inhibitor targeting the ALDH1 and ALDH3 subfamilies, 249 , 268 and it has an IC50 of 5 μM for each form. 249 Specifically, it has an IC50 of 7 μM for the DU145 PCa cell line. 249 In vivo, it is effective at a dose of 14 mg/kg daily given via intraperitoneal injection. 249 Besides, DIMATE has been demonstrated to reduce tumor growth in vivo when injected intraperitoneally in melanoma models, while showing low toxicity on healthy cells. 250
Citral, which is a natural product, shows potent inhibitory activity against ALDH1A1, ALDH1A3, and ALDH2 in breast cancer cells. 251 Citral can block ALDH1A3‐mediated breast tumor growth via blocking its colony formation ability and gene expression regulation activity, 251 as well as regulating apoptosis and cell‐cycle markers expression. 269 , 270 Moreover, nanoparticle encapsulated citral is used to specifically reduce the enhanced tumor growth of MDA‐MB‐231 cells overexpressing ALDH1A3 for the beneficial effects of encapsulation in the in vivo delivery of agents. 251
Aldi‐1–4 is found in a high‐throughput screen for modulators of ALDH2 activity. 252 The crystal structure of Aldi‐1–4 shows a covalent adduct with the active site cysteine of ALDH, and thus four related compounds show similar inhibition properties and time‐dependent kinetics for ALDHs. 252 IC50 values for ALDH isozymes tested with the four compounds range from 2.2 to 7.9 μM for ALDH1A1, 5.4 to 8.6 μM for ALDH2, and 1.7 to 12 μM for ALDH3A1. 252
Aldi‐6 is subsequently developed by Kim et al. 253 It has an IC50 of 600 nM for ALDH1A1, 800 nM for ALDH2, and 1 μM for ALDH3A1. 253 Treatment with Aldi‐6 results in a markedly decrease in HNSCC cell viability in vitro, and the combination of Aldi‐6 with cisplatin leads to a greater reduction in tumor burden in vivo. 253
The oral anesthetic dyclonine is a covalent inhibitor of ALDH, and it is shown to be a weak inhibitor of ALDH2 and ALDH3A1. 254 It has an IC50 of 35 μM for ALDH2 and 76 μM for ALDH3A1. 252 The combination of dyclonine and sulfasalazine cooperatively suppresses the growth of HNSCC or GC tumors with high ALDH3A1 expression. However, monotherapy with dyclonine is not effective in vivo. 254
Recently, isoform‐specific inhibitors are under investigation in cancer research, and some inhibitors can target ALDH1A1, ALDH2, and ALDH3A1. The ALDH1A1‐specific inhibitors are Cpd3, CM026, and CM037; NCT‐501, NCT‐505, and NCT‐506; 13 g and 13 h (Table 5). Cpd3, which is the indolinedione‐based analog, has an enhanced inhibitory activity for ALDH1A1, and IC50 is 20 nM, showing modest inhibition activity for ALDH2 and ALDH3A1. 271 Cqd3 is competitive against ALDH binding, forming direct interactions with active‐site cysteine residues, particularly in ALDH1A1. 271 However, there is no in vivo study using indolinedione‐based analogs. Morgan et al. optimized the indole group of Cpd3 to theophylline or benzothienopyrimidine groups and characterized two distinct chemical classes of inhibitors, CM026 and CM037, which shows a superior selection for ALDH1A1 inhibition. 272 CM026 and CM037 have an IC50 of 0.8 and 4.6 μM for ALDH1A1 with no inhibition for other ALDH isoforms. 272 These compounds can bind within the aldehyde binding pocket of ALDH1A1 and exploit the presence of a unique glycine residue to achieve their selectivity. 272 It has been reported first that CM037 disrupts sphere formation and cell viability of ovarian cancer cells. Moreover, CM037 in combination with cisplatin moderately sensitizes ovarian cancer cells to the cytotoxic effects. 273 Furthermore, CM037 could sensitize SKOV‐3‐TR, a paclitaxel‐resistant ovarian cancer cell line, to paclitaxel treatment when they are used in combination, whereas monotherapy with either agent is ineffective. 274 Yang et al. discovered a new series of theophylline‐based analogs as potent ALDH1A1 inhibitors, and NCT‐501 is one of them. 275 NCT‐501 has an IC50 of 0.04 μM for ALDH1A1 with no inhibition of other ALDH isoforms. 275 Administration of NCT‐501 can reduce sphere formation and migratory potential of HNSCC cells, which is also cytotoxic against cisplatin‐resistant HNSCC cancer cells. 276 NCT‐501 also sensitizes SKOV‐3‐TR cells to paclitaxel treatment when they are used in combination. 274 However, NCT‐501 has limited oral bioavailability owing to metabolism by the liver before entering the systemic circulation, which limits its application in oral treatment. 275 Yang et al. also devised a series of quinoline‐based ALDH1A1 selective inhibitors, and NCT‐505 and NCT‐506 are two such compounds. 274 NCT‐505 and NCT‐506 both have an IC50 of 7 nM for ALDH1A1, showing high selectivity over other ALDH isozymes. 274 NCT‐505 and NCT‐506 also sensitize SKOV‐3‐TR cells to paclitaxel treatment when they are used in combination, 274 and subsequent studies show that NCT‐505 and NCT‐506 have reasonable systemic drug exposure when administered orally. 274 Huddle et al. explored the structural determinants of ALDH1A isoform selectivity in a series of small molecule inhibitors, and 13 g and 13 h are discovered. 277 An amount of 13 g has an IC50 of 80 nM for ALDH1A1, 250 nM for ALDH1A2, and 120 nM for ALDH1A3, whereas 13 h has an IC50 of 270 nM for ALDH1A1, 480 nM for ALDH1A2, and 130 nM for ALDH1A3. 277 A period of 13 h shows synergy with cisplatin in patient‐derived ovarian cancer spheroids, and both 13 g and 13 h deplete the CD133+ putative stem cells in a dose‐dependent manner. 277 However, the efficacy of these compounds in vivo has not yet been reported. 277
TABLE 5.
Isoform‐specific ALDH inhibitors
| Compound name | Mechanism of action | Cellular activity | Animal studies | Ref. |
|---|---|---|---|---|
| Cpd 3 | Competitive ALDH1A1 inhibition | IC50: 20 nM for ALDH1A1 | No in vivo studies | 271 |
| CM026 | Competitive ALDH1A1 inhibition | IC50: 0.8 μM for ALDH1A1 | No in vivo studies | 272 |
| CM037 | Competitive ALDH1A1 inhibition | IC50: 4.6 μM for ALDH1A1 | Ineffective in vivo | 272 , 273 , 274 |
| NCT‐501 | Competitive ALDH1A1 inhibition | IC50: 0.04 μM | Ineffective in vivo | 274 , 275 , 276 |
| NCT‐505/NCT‐506 | ALDH1A1 inhibition | IC50: 7 nM for ALDH1A1 | NCT‐505 and NCT‐506 have reasonable systemic drug exposure when administered orally | 274 |
| 13 g/13 h | Noncovalent, noncompetitive ALDH1A inhibition | IC50 (13 g): 80 nM for ALDH1A1, 250 nM for ALDH1A2, 120 nM for ALDH1A3; IC50 (13 h): 270 nM for ALDH1A1, 480 nM for ALDH1A2, 130 nM for ALDH1A3 | No in vivo studies | 277 |
| CVT‐10216 | Reversible ALDH2 inhibition | IC50: 29 nM for ALDH2 | No in vivo studies | 278 |
| ALDH423 | ALDH2 inhibition | IC50: 0.62 μM for ALDH2 | No in vivo studies | 279 |
| CB7 | Competitive ALDH3A1 inhibition | IC50: 0.2 μM for ALDH3A1 | No in vivo studies | 280 |
| CB29 | Reversible ALDH3A1 inhibition | IC50: 16 μM for ALDH3A1 | No in vivo studies | 281 |
| Acivicin | Covalent ALDH4A1 inhibition | IC50: 5.4 μM for ALDH4A1 | No in vivo studies | 282 |
Abbreviation: IC50, inhibitory concentration.
The ALDH2‐specific inhibitors are CVT‐10216 and ALDH423. CVT‐10216 is a highly selective, reversible inhibitor of ALDH‐2, and it has an IC50 of 29 nM for ALDH2. 278 A target‐specific rescoring method is applied for the search of small molecule inhibitors of the mitochondrial ALDH2, and ALDH423 has found an IC50 of 0.62 μM for ALDH2. 279 As ALDH2 is not found to be crucial during carcinogenesis, CVT‐10216 and ALDH423 have not yet been tested in cancer models. 178
The ALDH3A1‐specific inhibitors are CB7 and CB29. CB7 is identified by Parajuli et al. in a high‐throughput screen. Enzyme kinetics and crystallographic studies show that CB7 is competitive with respect to aldehyde binding and noncompetitive with respect to NADP+ binding of ALDH3A1, and it has an IC50 of 0.2 μM for ALDH3A1. 280 Treatments of the ALDH3A1‐expressing LUAD cell A549 and GBM cell SF767 with mafosfamide in combination with 10 μM CB7 enhance the antiproliferative effects over monotherapy. 280 CB29 is a reversible ALDH3A1 inhibitor, which has an IC50 of 16 μM for ALDH3A1. 281 And it also binds within the aldehyde substrate‐binding site of ALDH3A1 like CB7. 281 The therapeutic effect of CB29 is similar to that of CB7 when in combination with mafosfamide. 281 However, no in vivo study has been reported with both CB7 and CB29.
There are other ALDH isoform inhibitors like Acivicin, which is a natural product and inhibit ALDH4A1 activity by binding to the catalytic site, and it has an IC50 of 5.4 μM for ALDH4A1. 282 Cytotoxicity of Acivicin is investigated in HCC cells, which shows an IC50 for ALDH4A1 of 0.7 μM, 282 while in vivo study has not been reported nowadays.
5.2. Targeting ALDHs via inhibiting ALDH‐related molecular pathways
Targeting the ALDH‐related molecular pathways is another promising strategy to inhibit tumor progression and CSC self‐renewal. Previous studies demonstrate that multiple molecular pathways such as RA, Notch, Wnt, and TGF‐β pathways may regulate ALDHs activity (Figure 2). A good example is that Wnt/β‐catenin signaling activates ALDH1A1 transcription, and this signaling could also regulate the maintenance of CSCs and drives radio‐therapeutic resistance in PCa patients. Blocking Wnt/β‐catenin signaling by XAV939 antagonist leads to significant inhibition of the ALDH‐positive populations and re‐sensitization of the PCa cells to radiation therapy. 283 Notably, ONC201, which is a well‐tolerated compound used in a Phase I/II study for patients with advanced solid tumor (NCT02038699) (NCT02324621), 284 has been reported to downregulate CSC‐related genes, including ALDH1A1 and ALDH7A1, and suppress CSCs self‐renewal in CRC, PCa, and GBM cells. 285 Rapalink‐1, a third‐generation mTOR inhibitor, is found to block mTORC1/2 signaling, 286 accompanied by the reduction of the proportion of ALDH‐positive cells in PCa patient‐derived organoids and decreased tumor burden in xenograft mouse models. 287 CXCR1, one of the receptors for CXCL8 (IL‐8), is identified in ALDH1‐positive breast cancer CSCs. Administration inhibitor of CXCR1, reparixin, can reduce the metastatic behavior of cancer cells and eradicate the CSC populations both as a single agent and in combination with chemotherapy. 288 However, in a randomized, double‐blind, placebo‐controlled Phase II clinical trial (NCT02370238), the combination of reparixin and paclitaxel does not improve PFS of TNBC patients over paclitaxel alone, which may warrant sufficient samples in the future. Notch signaling is important for the maintenance of ALDH1A1 positive CSCs, and pharmacological inhibition of Notch pathway by using a γ‐secretase inhibitor, DAPT reduces ALDH+ tumor cells 197 , 289 in lung and breast cancer. Interestingly, ALDH and IL‐1 receptor (IL1R1) double‐positive CSCs are enriched following antiestrogen therapy and held responsible for the treatment failure in breast cancer patients, which can be attenuated with IL1R1 antagonists such as anakinra. 290 Combinatorial therapy with gossypol and phenformin, which target ALDH1L1 and oxidative phosphorylation, respectively, results in ATP depletion in GBM spheres. In addition, this dual inhibition of tumor bioenergetics markedly attenuates GBM cell stemness and invasion ability in a preclinical mouse model. 285 Noteworthy, decreased ALDH1A2 expression is associated with the acquisition of invasive capacity and stem‐cell traits due to impaired ALDH1A2‐RAR‐dependent signaling. Administration with retinoids inhibits malignancy progression in HNSCC patient‐derived xenografts. 129 Low expression of ALDH1A2 in cancers is always due to its promoter hypermethylation. 128 , 132
FIGURE 2.
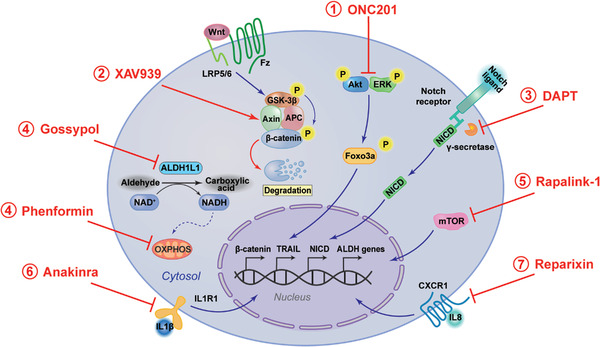
Therapies targeting aldehyde dehydrogenases (ALDHs)‐related molecular pathways. (1) ONC201 dually inhibits phosphorylation of Akt and extracellular regulated–protein kinase (ERK), leading to the dephosphorylation of transcription factor Foxo3a. Dephosphorylated Foxo3a translocates into the nucleus where it activates transcription of its target genes, including pro‐apoptotic death receptor ligand TNF‐related apoptosis‐inducing ligand (TRAIL), ALDH1A1, and ALDH7A1. (2) WNT signaling inhibitor, XAV939 stimulates β‐catenin destruction by stabilizing Axin, which finally leading to significant inhibition of the ALDH‐positive populations. (3) Pharmacological inhibition of Notch pathway by using a γ‐secretase inhibitor, DAPT reduces ALDH‐positive tumor cells. (4) Dual inhibition of ALDH1L1 and oxidative phosphorylation (OXPHOS) by gossypol and phenformin reduces tumor bioenergetics and stemness. (5) Rapalink‐1 can block mTORC1/2 signaling and reduce ALDH‐positive populations. (6) IL‐1 receptor (IL1R1) antagonist, anakinra can decrease ALDH and IL1R1 double positive cancer stem cells (CSCs). (7) Reparixin can reduce the metastatic behavior of cancer cells and eradicate the ALDH‐positive populations by inhibiting IL8‐CXCR1 signaling
5.3. Targeting ALDHs may promote immunotherapeutic efficiency
Immunotherapy has recently drawn global attention as the “new hope” for cancer treatment. 291 Several studies provided evidence that some ALDH isoforms might contribute to tumor immune surveillance. 292 Targeting of ALDH‐positive CSCs through immunological approaches is also currently underway in the treatment of cancer. In particular, ALDH3A1 enzyme‐enriched CSCs populations are positively correlated with PD‐L1 expression in melanoma and NSCLC. 190 GC mesenchymal stem cells maintain a pool of CSCs through increased the expression level of PD‐L1, which leads to the chemotherapeutic resistance of GC cells. Blocking PD‐L1 expression by neutralizing antibody in GC cells inhibits their sphere formation ability and ALDH activity. 293 , 294 More recently, Liu et al. revealed that ALDH1A1 results MDSCs infiltration in 4T1 breast tumor mouse models, and the administration of DSF robustly enhances the therapeutic efficiency of PD‐L1 monoclonal antibody in vivo. Zhang et al. found that ALDH2 mediates the immune evasion induced by alcohol in CRC by stabilizing PD‐L1 protein. 119 It is also noteworthy that the combination of ALDH2 inhibition and anti‐PD‐1 antibody enhances antitumor immunity and tumor eradication, which may serve as a novel strategy to enhance the efficacy of immune checkpoint blockade in CRC patients, especially in those who consumed alcohol. 295 The development of anti‐ALDH vaccines can be an efficient strategy to eradicate CSC populations and improve the therapy response. 296 Hassani Najafabadi et al. used a nanoparticle‐based vaccine to deliver ALDH1A1 and ALDH1A3 dual peptides to antigen‐presenting cells and generated robust ALDH‐specific T‐cell responses against ALDHbright CSCs. Combined with anti‐PD‐L1 therapy, the anti‐ALDH vaccination exerts potent antitumor efficacy in murine models of D5 melanoma and 4T1 breast cancer. 297 Similarly, ALDH dual peptides‐DC vaccination plus anti‐PD‐L1 administration result in an increased recruitment of CD3+ tumor‐infiltrating lymphocytes in the residual melanoma tumors and a further reduction of ALDHbright CSCs. Thus, targeting CSCs with these vaccines in combination with ICIs may be the new avenues for preventing tumor relapse and increasing patient survival. 298
6. OUTLOOK
Throughout the past decades, pioneering studies support that aberrant ALDH activity or expression is distinctly associated with neurological abnormalities, metabolic diseases, and especially in solid tumors. For example, ALDHs promote the oxidation of anticancer drugs, such as cyclophosphamide, into less toxic metabolites, which causes chemotherapeutic resistance. ALDHs participate in RA synthesis associated with cancer cell proliferation and immune system regulation. Besides, ALDHs play key roles in CSC‐mediated tumor recurrence and metastatic dissemination, indicating poor clinical outcomes in patients. However, the molecular mechanisms underpinning aberrant ALDH activity or expression during malignant transformation and progression are fragmented and limited, which warrants further investigation. The exploration of ALDHs more detailed functions and regulation mechanisms will also promote our understanding of other diseases like PD and AD.
Accumulating evidence indicates that ALDHs can be successfully considered diagnostic markers for certain diseases. Preclinical researches demonstrate that ALDH1A1 is an optimal predictor for PD risk. Besides, the clinical assessment of ALDH2 as a marker will be beneficial to the diagnosis of cardiac anomalies. In addition, ALDHs are considered promising biomarkers of CSCs with functional and mechanistic involvement in tumor initiation and progression as well as in modulating their response to tumor therapies. 299 Thus, progress in identifying and quantifying ALDHs as risk factors or markers is opening the way to the prevention of disease and maintenance of health.
The existence of CSCs in majority of solid tumors has important clinical implications for the development of inhibitors and design of clinical trials to assess treatment efficacy. Therefore, an ideal ALDH‐targeting inhibitor should be nontoxic and well tolerated that can be safely administered by patients and in combination with conventional therapies, such as chemotherapy, radiotherapy, and immunotherapy to improve disease control over non‐CSC, bulk tumor cells. Isoform‐specific ALDH inhibition combats the potential problem of toxicity of multi‐ALDH isoform inhibition due to the wide distribution of ALDH enzymes in normal tissues. However, this will require further in vivo testing as monotherapy due to the compensatory activity by other isoforms. Nevertheless, the multi‐ALDH inhibitors approach remains the most promising for translation into the clinic because more than one ALDH isozyme expresses in CSCs. Several strategies in ALDH inhibitor delivery systems are now employed in multiple cancer types to reduce off‐target toxicities in normal cells.
The findings of several studies indicate that CSCs with high ALDH activity harbor important alterations affecting oncogenic pathways including the Notch and Wnt pathway, mediating the resistance of cancers to therapy, tumor recurrence, and metastasis. 300 In the context of clinical oncology, several clinical trials using single‐agent and/or combination therapies have been performed to study the safety and efficacy of targeting ALDH‐related molecular pathways. 301 In addition, multiple observations highlight the interactions between ALDH‐positive CSCs and the immune system, which contributes to immunosuppression and stemness phenotype. Therefore, urgent preclinical or clinical studies may require a combination of ALDH‐specific and immune‐based therapies in the future. 302 Finally, due to the availability of large amounts of clinical information and primary clinical data, it is critical to consider repurposing “old” drugs to treat both common and rare diseases. 303 For example, the anti‐alcoholism drug DSF has attracted increasing attention for its anticancer effects. In the long term, patients will benefit from drug repurposing in translational medicine.
AUTHOR CONTRIBUTIONS
Jie Xia wrote the first draft of the manuscript. Siqin Li contributed to drawing figures. Lixing Zhang and Suling Liu contributed to manuscript revision. All authors contributed to the article and approved the submitted version.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
Not applicable.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (Stem Cell and Translational Research 2020YFA0112300); National Natural Science Foundation of China (81930075, 82073267); “Ten Thousand Plan”—National High‐Level Talents Special Support Plan (WR‐YK5202101); Program for Outstanding Leading Talents in Shanghai; Program for Outstanding Medical Academic Leader in Shanghai (2019LJ04); Program of Shanghai Academic/Technology Research Leader (20XD1400700); The innovative research team of high‐level local university in Shanghai.
Xia J, Li S, Liu S, Zhang L. Aldehyde dehydrogenase in solid tumors and other diseases: Potential biomarkers and therapeutic targets. MedComm. 2023;4:e195. 10.1002/mco2.195
Contributor Information
Suling Liu, Email: suling@fudan.edu.cn.
Lixing Zhang, Email: zhang_lx@fudan.edu.cn.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Jackson B, Brocker C, Thompson DC, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5(4):283‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non‐P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singh S, Brocker C, Koppaka V, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aoyama K, Saha A, Tolar J, et al. Inhibiting retinoic acid signaling ameliorates graft‐versus‐host disease by modifying T‐cell differentiation and intestinal migration. Blood. 2013;122(12):2125‐2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Labrecque J, Dumas F, Lacroix A, Bhat PV. A novel isoenzyme of aldehyde dehydrogenase specifically involved in the biosynthesis of 9‐cis and all‐trans retinoic acid. Biochem J. 1995;305(pt 2):681‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2(2):138‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vasiliou V, Malamas M, Marselos M. The mechanism of alcohol intolerance produced by various therapeutic agents. Acta Pharmacol Toxicol (Copenh). 1986;58(5):305‐310. [DOI] [PubMed] [Google Scholar]
- 8. Teixeira TM, da Silva HD, Goveia RM, et al. First description and evaluation of SNPs in the ADH and ALDH genes in a population of alcoholics in Central‐West Brazil. Alcohol. 2017;65:37‐43. [DOI] [PubMed] [Google Scholar]
- 9. Ginestier C, Hur MH, Charafe‐Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382‐3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell‐associated marker in lung cancer. Mol Cancer Res. 2009;7(3):330‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Visus C, Ito D, Amoscato A, et al. Identification of human aldehyde dehydrogenase 1 family member A1 as a novel CD8+ T‐cell‐defined tumor antigen in squamous cell carcinoma of the head and neck. Cancer Res. 2007;67(21):10538‐10545. [DOI] [PubMed] [Google Scholar]
- 13. Chen YC, Chen YW, Hsu HS, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385(3):307‐313. [DOI] [PubMed] [Google Scholar]
- 14. Li T, Su Y, Mei Y, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2009;90(2):234‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ioannou M, Serafimidis I, Arnes L, et al. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev Biol. 2013;374(1):153‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su Y, Qiu Q, Zhang X, et al. Aldehyde dehydrogenase 1 A1‐positive cell population is enriched in tumor‐initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(2):327‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644‐8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong S, Feng Y, Cheng L. Cellular reprogramming as a therapeutic target in cancer. Trends Cell Biol. 2019;29(8):623‐634. [DOI] [PubMed] [Google Scholar]
- 19. Phi LTH, Sari IN, Yang YG, et al. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu S, Cong Y, Wang D, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2(1):78‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han S, Huang T, Wu X, et al. Prognostic value of ALDH1 and Nestin in advanced cancer: a systematic meta‐analysis with trial sequential analysis. Ther Adv Med Oncol. 2019;11:1758835919830831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ursini F, Maiorino M. Lipid peroxidation and ferroptosis: the role of GSH and GPx4. Free Radic Biol Med. 2020;152:175‐185. [DOI] [PubMed] [Google Scholar]
- 23. Rodriguez‐Zavala JS, Calleja LF, Moreno‐Sanchez R, Yoval‐Sanchez B. Role of aldehyde dehydrogenases in physiopathological processes. Chem Res Toxicol. 2019;32(3):405‐420. [DOI] [PubMed] [Google Scholar]
- 24. Singh S, Chen Y, Matsumoto A, et al. ALDH1B1 links alcohol consumption and diabetes. Biochem Biophys Res Commun. 2015;463(4):768‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Priyadharshini Christy J, George Priya Doss C. Single amino acid polymorphism in aldehyde dehydrogenase gene superfamily. Front Biosci (Landmark Ed). 2015;20(2):335‐376. [DOI] [PubMed] [Google Scholar]
- 26. Zhu ZY, Liu YD, Gong Y, et al. Mitochondrial aldehyde dehydrogenase (ALDH2) rescues cardiac contractile dysfunction in an APP/PS1 murine model of Alzheimer's disease via inhibition of ACSL4‐dependent ferroptosis. Acta Pharmacol Sin. 2022;43(1):39‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oh T, Kwon M, Yu JS, et al. Ent‐peniciherqueinone suppresses acetaldehyde‐induced cytotoxicity and oxidative stress by inducing ALDH and suppressing MAPK signaling. Pharmaceutics. 2020;12(12):1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allison SE, Chen Y, Petrovic N, et al. Activation of ALDH1A1 in MDA‐MB‐468 breast cancer cells that over‐express CYP2J2 protects against paclitaxel‐dependent cell death mediated by reactive oxygen species. Biochem Pharmacol. 2017;143:79‐89. [DOI] [PubMed] [Google Scholar]
- 29. Lei HM, Zhang KR, Wang CH, et al. Aldehyde dehydrogenase 1A1 confers erlotinib resistance via facilitating the reactive oxygen species‐reactive carbonyl species metabolic pathway in lung adenocarcinomas. Theranostics. 2019;9(24):7122‐7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155(1):215‐227. [DOI] [PubMed] [Google Scholar]
- 31. Biswas AK, Han S, Tai Y, et al. Targeting S100A9‐ALDH1A1‐retinoic acid signaling to suppress brain relapse in EGFR‐mutant lung cancer. Cancer Discov. 2022;12(4):1002‐1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muzio G, Maggiora M, Paiuzzi E, Oraldi M, Canuto RA. Aldehyde dehydrogenases and cell proliferation. Free Radic Biol Med. 2012;52(4):735‐746. [DOI] [PubMed] [Google Scholar]
- 33. Devalaraja S, To TKJ, Folkert IW, et al. Tumor‐derived retinoic acid regulates intratumoral monocyte differentiation to promote immune suppression. Cell. 2020;180(6):1098‐1114.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36(2):279‐299. [DOI] [PubMed] [Google Scholar]
- 35. Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226(2):322‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Donato LJ, Suh JH, Noy N. Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res. 2007;67(2):609‐615. [DOI] [PubMed] [Google Scholar]
- 37. McGrane MM. Vitamin A regulation of gene expression: molecular mechanism of a prototype gene. J Nutr Biochem. 2007;18(8):497‐508. [DOI] [PubMed] [Google Scholar]
- 38. Liu Y, Chen H, Wang J, Zhou W, Sun R, Xia M. Association of serum retinoic acid with hepatic steatosis and liver injury in nonalcoholic fatty liver disease. Am J Clin Nutr. 2015;102(1):130‐137. [DOI] [PubMed] [Google Scholar]
- 39. Liu Y, Chen H, Mu D, et al. Circulating retinoic acid levels and the development of metabolic syndrome. J Clin Endocrinol Metab. 2016;101(4):1686‐1692. [DOI] [PubMed] [Google Scholar]
- 40. Han Y, Yang Y, Kim M, Jee SH, Yoo HJ, Lee JH. Serum retinal and retinoic acid predict the development of type 2 diabetes mellitus in Korean subjects with impaired fasting glucose from the KCPS‐II cohort. Metabolites. 2021;11(8):510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Chen H, Mu D, et al. Association of serum retinoic acid with risk of mortality in patients with coronary artery disease. Circ Res. 2016;119(4):557‐563. [DOI] [PubMed] [Google Scholar]
- 42. Marcato P, Dean CA, Liu RZ, et al. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol. 2015;9(1):17‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei Q, Dong Z. The yin and yang of retinoic acid signaling in kidney diseases. J Clin Invest. 2020;130(10):5124‐5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shaw N, Elholm M, Noy N. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator‐activated receptor beta/delta. J Biol Chem. 2003;278(43):41589‐41592. [DOI] [PubMed] [Google Scholar]
- 45. Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid‐resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci USA. 2008;105(21):7546‐7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vassalli G. Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells. Stem Cells Int. 2019;2019:3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Namekawa T, Ikeda K, Horie‐Inoue K, et al. ALDH1A1 in patient‐derived bladder cancer spheroids activates retinoic acid signaling leading to TUBB3 overexpression and tumor progression. Int J Cancer. 2020;146(4):1099‐1113. [DOI] [PubMed] [Google Scholar]
- 48.Al Tanoury Z, Piskunov A, Rochette‐Egly C. Vitamin A and retinoid signaling: genomic and nongenomic effects. J Lipid Res. 2013;54(7):1761‐1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Larange A, Cheroutre H. Retinoic acid and retinoic acid receptors as pleiotropic modulators of the immune system. Annu Rev Immunol. 2016;34:369‐394. [DOI] [PubMed] [Google Scholar]
- 50. Alsayed Y, Uddin S, Mahmud N, et al. Activation of Rac1 and the p38 mitogen‐activated protein kinase pathway in response to all‐trans‐retinoic acid. J Biol Chem. 2001;276(6):4012‐4019. [DOI] [PubMed] [Google Scholar]
- 51. Piskunov A, Rochette‐Egly C. A retinoic acid receptor RARalpha pool present in membrane lipid rafts forms complexes with G protein alphaQ to activate p38MAPK. Oncogene. 2012;31(28):3333‐3345. [DOI] [PubMed] [Google Scholar]
- 52. Kalia LV, Lang AE. Parkinson's disease. Lancet. 2015;386(9996):896‐912. [DOI] [PubMed] [Google Scholar]
- 53. Liu G, Yu J, Ding J, et al. Aldehyde dehydrogenase 1 defines and protects a nigrostriatal dopaminergic neuron subpopulation. J Clin Invest. 2014;124(7):3032‐3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fan HH, Guo Q, Zheng J, et al. ALDH1A1 genetic variations may modulate risk of Parkinson's disease in Han Chinese population. Front Neurosci. 2021;15:620929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Molochnikov L, Rabey JM, Dobronevsky E, et al. A molecular signature in blood identifies early Parkinson's disease. Mol Neurodegener. 2012;7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keren‐Shaul H, Spinrad A, Weiner A, et al. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017;169(7):1276‐1290.e17. [DOI] [PubMed] [Google Scholar]
- 57. Fragoso YD, Shearer KD, Sementilli A, de Carvalho LV, McCaffery PJ. High expression of retinoic acid receptors and synthetic enzymes in the human hippocampus. Brain Struct Funct. 2012;217(2):473‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nikhil K, Viccaro K, Shah K. Multifaceted regulation of ALDH1A1 by Cdk5 in Alzheimer's disease pathogenesis. Mol Neurobiol. 2019;56(2):1366‐1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li X, Chen W, Huang X, et al. Synaptic dysfunction of Aldh1a1 neurons in the ventral tegmental area causes impulsive behaviors. Mol Neurodegener. 2021;16(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16(2):110‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dolle P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128(7):1019‐1031. [DOI] [PubMed] [Google Scholar]
- 62. Beecroft SJ, Ayala M, McGillivray G, et al. Biallelic hypomorphic variants in ALDH1A2 cause a novel lethal human multiple congenital anomaly syndrome encompassing diaphragmatic, pulmonary, and cardiovascular defects. Hum Mutat. 2021;42(5):506‐519. [DOI] [PubMed] [Google Scholar]
- 63. Lee JH, Protze SI, Laksman Z, Backx PH, Keller GM. Human pluripotent stem cell‐derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21(2):179‐194.e4. [DOI] [PubMed] [Google Scholar]
- 64. Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, Beenfeldt Fredriksen S, Linneberg A. The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol Clin Exp Res. 2008;32(11):1984‐1991. [DOI] [PubMed] [Google Scholar]
- 65. Linneberg A, Gonzalez‐Quintela A, Vidal C, et al. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy. 2010;40(1):123‐130. [DOI] [PubMed] [Google Scholar]
- 66. Gross ER, Zambelli VO, Small BA, Ferreira JC, Chen CH, Mochly‐Rosen D. A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu Rev Pharmacol Toxicol. 2015;55:107‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang W, Wang C, Xu H, Gao Y. Aldehyde dehydrogenase, liver disease and cancer. Int J Biol Sci. 2020;16(6):921‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen J, Huang W, Cheng CH, Zhou L, Jiang GB, Hu YY. Association between aldehyde dehydrogenase‐2 polymorphisms and risk of Alzheimer's disease and Parkinson's disease: a meta‐analysis based on 5,315 individuals. Front Neurol. 2019;10:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kamino K, Nagasaka K, Imagawa M, et al. Deficiency in mitochondrial aldehyde dehydrogenase increases the risk for late‐onset Alzheimer's disease in the Japanese population. Biochem Biophys Res Commun. 2000;273(1):192‐196. [DOI] [PubMed] [Google Scholar]
- 70. Ma L, Lu ZN. Role of ADH1B rs1229984 and ALDH2 rs671 gene polymorphisms in the development of Alzheimer's disease. Genet Mol Res. 2016;15(4):grm.15048740. [DOI] [PubMed] [Google Scholar]
- 71. Wang S, Wang L, Qin X, et al. ALDH2 contributes to melatonin‐induced protection against APP/PS1 mutation‐prompted cardiac anomalies through cGAS‐STING‐TBK1‐mediated regulation of mitophagy. Signal Transduct Target Ther. 2020;5(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly‐Rosen D. Activation of aldehyde dehydrogenase‐2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun A, Zou Y, Wang P, et al. Mitochondrial aldehyde dehydrogenase 2 plays protective roles in heart failure after myocardial infarction via suppression of the cytosolic JNK/p53 pathway in mice. J Am Heart Assoc. 2014;3(5):e000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ma H, Guo R, Yu L, Zhang Y, Ren J. Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur Heart J. 2011;32(8):1025‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yasue H, Mizuno Y, Harada E. Association of East Asian variant aldehyde dehydrogenase 2 genotype (ALDH2*2*) with coronary spasm and acute myocardial infarction. Adv Exp Med Biol. 2019;1193:121‐134. [DOI] [PubMed] [Google Scholar]
- 76. Tardif R, Liu L, Raizenne M. Exhaled ethanol and acetaldehyde in human subjects exposed to low levels of ethanol. Inhal Toxicol. 2004;16(4):203‐207. [DOI] [PubMed] [Google Scholar]
- 77. Zhang X, Ye YL, Wang YN, et al. Aldehyde dehydrogenase 2 genetic variations may increase susceptibility to Parkinson's disease in Han Chinese population. Neurobiol Aging. 2015;36(9):2660.e9–13. [DOI] [PubMed] [Google Scholar]
- 78. Zhong W, Zhang W, Li Q, et al. Pharmacological activation of aldehyde dehydrogenase 2 by Alda‐1 reverses alcohol‐induced hepatic steatosis and cell death in mice. J Hepatol. 2015;62(6):1375‐1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang LL, Wang YQ, Fu B, Zhao SL, Kui Y. Aldehyde dehydrogenase 2 (ALDH2) polymorphism gene and coronary artery disease risk: a meta‐analysis. Genet Mol Res. 2015;14(4):18503‐18514. [DOI] [PubMed] [Google Scholar]
- 80. Kato N, Takeuchi F, Tabara Y, et al. Meta‐analysis of genome‐wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43(6):531‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Convertino M, Das J, Dokholyan NV. Pharmacological chaperones: design and development of new therapeutic strategies for the treatment of conformational diseases. ACS Chem Biol. 2016;11(6):1471‐1489. [DOI] [PubMed] [Google Scholar]
- 82. Nagaoka A, Kunii Y, Hino M, et al. ALDH4A1 expression levels are elevated in postmortem brains of patients with schizophrenia and are associated with genetic variants in enzymes related to proline metabolism. J Psychiatr Res. 2020;123:119‐127. [DOI] [PubMed] [Google Scholar]
- 83. Lorenzo C, Delgado P, Busse CE, et al. ALDH4A1 is an atherosclerosis auto‐antigen targeted by protective antibodies. Nature. 2021;589(7841):287‐292. [DOI] [PubMed] [Google Scholar]
- 84. Lassen N, Bateman JB, Estey T, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(‐/‐)/Aldh1a1(‐/‐) knock‐out mice. J Biol Chem. 2007;282(35):25668‐25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen Y, Jester JV, Anderson DM, et al. Corneal haze phenotype in Aldh3a1‐null mice: in vivo confocal microscopy and tissue imaging mass spectrometry. Chem Biol Interact. 2017;276:9‐14. [DOI] [PubMed] [Google Scholar]
- 86. Rizzo WB, Carney G. Sjögren‐Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2). Hum Mutat. 2005;26(1):1‐10. [DOI] [PubMed] [Google Scholar]
- 87. Didiasova M, Banning A, Brennenstuhl H, et al. Succinic semialdehyde dehydrogenase deficiency: an update. Cells. 2020;9(2):477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marcadier JL, Smith AM, Pohl D, et al. Mutations in ALDH6A1 encoding methylmalonate semialdehyde dehydrogenase are associated with dysmyelination and transient methylmalonic aciduria. Orphanet J Rare Dis. 2013;8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Engelke UF, van Outersterp RE, Merx J, et al. Untargeted metabolomics and infrared ion spectroscopy identify biomarkers for pyridoxine‐dependent epilepsy. J Clin Invest. 2021;131(15):e148272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Guo Y, Tan LJ, Lei SF, et al. Genome‐wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6(1):e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pantouris G, Dioletis E, Chen Y, Thompson DC, Vasiliou V, Lolis EJ. Expression, purification and crystallization of the novel Xenopus tropicalis ALDH16B1, a homologue of human ALDH16A1. Chem Biol Interact. 2019;304:168‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hanna MC, Blackstone C. Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1. Neurogenetics. 2009;10(3):217‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nozaki F, Kusunoki T, Okamoto N, et al. ALDH18A1‐related cutis laxa syndrome with cyclic vomiting. Brain Dev. 2016;38(7):678‐684. [DOI] [PubMed] [Google Scholar]
- 94. Marco‐Marin C, Escamilla‐Honrubia JM, Llacer JL, Seri M, Panza E, Rubio V. Delta(1)‐pyrroline‐5‐carboxylate synthetase deficiency: an emergent multifaceted urea cycle‐related disorder. J Inherit Metab Dis. 2020;43(4):657‐670. [DOI] [PubMed] [Google Scholar]
- 95. Morgan CA, Parajuli B, Buchman CD, Dria K, Hurley TD. N,N‐diethylaminobenzaldehyde (DEAB) as a substrate and mechanism‐based inhibitor for human ALDH isoenzymes. Chem Biol Interact. 2015;234:18‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Storms RW, Trujillo AP, Springer JB, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA. 1999;96(16):9118‐9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Xu J, Yang X, Deng Q, et al. TEM8 marks neovasculogenic tumor‐initiating cells in triple‐negative breast cancer. Nat Commun. 2021;12(1):4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhou L, Sheng D, Wang D, et al. Identification of cancer‐type specific expression patterns for active aldehyde dehydrogenase (ALDH) isoforms in ALDEFLUOR assay. Cell Biol Toxicol. 2019;35(2):161‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Marcato P, Dean CA, Pan D, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29(1):32‐45. [DOI] [PubMed] [Google Scholar]
- 100. Singh S, Arcaroli J, Chen Y, et al. ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/beta‐catenin, notch and PI3K/Akt signaling pathways. PLoS One. 2015;10(5):e0121648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wu D, Mou YP, Chen K, et al. Aldehyde dehydrogenase 3A1 is robustly upregulated in gastric cancer stem‐like cells and associated with tumorigenesis. Int J Oncol. 2016;49(2):611‐622. [DOI] [PubMed] [Google Scholar]
- 102. van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. The aldehyde dehydrogenase enzyme 7A1 is functionally involved in prostate cancer bone metastasis. Clin Exp Metastasis. 2011;28(7):615‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Piskounova E, Agathocleous M, Murphy MM, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ding Z, Ericksen RE, Escande‐Beillard N, et al. Metabolic pathway analyses identify proline biosynthesis pathway as a promoter of liver tumorigenesis. J Hepatol. 2020;72(4):725‐735. [DOI] [PubMed] [Google Scholar]
- 105. Singh S, Arcaroli J, Thompson DC, Messersmith W, Vasiliou V. Acetaldehyde and retinaldehyde‐metabolizing enzymes in colon and pancreatic cancers. Adv Exp Med Biol. 2015;815:281‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Clay MR, Tabor M, Owen JH, et al. Single‐marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32(9):1195‐1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Carvalho MJ, Laranjo M, Abrantes AM, Torgal I, Botelho MF, Oliveira CF. Clinical translation for endometrial cancer stem cells hypothesis. Cancer Metastasis Rev. 2015;34(3):401‐416. [DOI] [PubMed] [Google Scholar]
- 108. Morimoto K, Kim SJ, Tanei T, et al. Stem cell marker aldehyde dehydrogenase 1‐positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009;100(6):1062‐1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Charafe‐Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1‐positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16(1):45‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kalantari E, Saadi FH, Asgari M, Shariftabrizi A, Roudi R, Madjd Z. Increased expression of ALDH1A1 in prostate cancer is correlated with tumor aggressiveness: a tissue microarray study of Iranian patients. Appl Immunohistochem Mol Morphol. 2017;25(8):592‐598. [DOI] [PubMed] [Google Scholar]
- 111. Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5(4):e10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Schafer A, Teufel J, Ringel F, et al. Aldehyde dehydrogenase 1A1—a new mediator of resistance to temozolomide in glioblastoma. Neuro Oncol. 2012;14(12):1452‐1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang K, Chen X, Zhan Y, et al. Increased expression of ALDH1A1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med Oncol. 2013;30(2):574. [DOI] [PubMed] [Google Scholar]
- 114. Li K, Guo X, Wang Z, et al. The prognostic roles of ALDH1 isoenzymes in gastric cancer. Onco Targets Ther. 2016;9:3405‐3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kida K, Ishikawa T, Yamada A, et al. Effect of ALDH1 on prognosis and chemoresistance by breast cancer subtype. Breast Cancer Res Treat. 2016;156(2):261‐269. [DOI] [PubMed] [Google Scholar]
- 116. Demir H, Dulgar O, Gulle BT, Turna H, Ilvan S. Prognostic value of aldehyde dehydrogenase 1 (ALDH1) in invasive breast carcinomas. Bosn J Basic Med Sci. 2018;18(4):313‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liu Y, Lv DL, Duan JJ, et al. ALDH1A1 expression correlates with clinicopathologic features and poor prognosis of breast cancer patients: a systematic review and meta‐analysis. BMC Cancer. 2014;14:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu JF, Xia P, Hu WQ, Wang D, Xu XY. Aldehyde dehydrogenase 1 expression correlates with clinicopathologic features of patients with breast cancer: a meta‐analysis. Int J Clin Exp Med. 2015;8(6):8425‐8432. [PMC free article] [PubMed] [Google Scholar]
- 119. Liu C, Qiang J, Deng Q, et al. ALDH1A1 activity in tumor‐initiating cells remodels myeloid‐derived suppressor cells to promote breast cancer progression. Cancer Res. 2021;81(23):5919‐5934. [DOI] [PubMed] [Google Scholar]
- 120. Ma F, Li H, Li Y, et al. Aldehyde dehydrogenase 1 (ALDH1) expression is an independent prognostic factor in triple negative breast cancer (TNBC). Medicine (Baltimore). 2017;96(14):e6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhou AL, Wang X, Yu W, et al. Expression level of PD‐L1 is involved in ALDH1A1‐mediated poor prognosis in patients with head and neck squamous cell carcinoma. Pathol Res Pract. 2020;216(9):153093. [DOI] [PubMed] [Google Scholar]
- 122. Xu SL, Liu S, Cui W, et al. Aldehyde dehydrogenase 1A1 circumscribes high invasive glioma cells and predicts poor prognosis. Am J Cancer Res. 2015;5(4):1471‐1483. [PMC free article] [PubMed] [Google Scholar]
- 123. Holah NS, Aiad HA, Asaad NY, Elkhouly EA, Lasheen AG. Evaluation of the role of ALDH1 as cancer stem cell marker in colorectal carcinoma: an immunohistochemical study. J Clin Diagn Res. 2017;11(1):EC17‐EC23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Langan RC, Mullinax JE, Ray S, et al. A pilot study assessing the potential role of non‐CD133 colorectal cancer stem cells as biomarkers. J Cancer. 2012;3:231‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lugli A, Iezzi G, Hostettler I, et al. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer. 2010;103(3):382‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kahlert C, Gaitzsch E, Steinert G, et al. Expression analysis of aldehyde dehydrogenase 1A1 (ALDH1A1) in colon and rectal cancer in association with prognosis and response to chemotherapy. Ann Surg Oncol. 2012;19(13):4193‐4201. [DOI] [PubMed] [Google Scholar]
- 127. Cao D, Meng Y, Li S, et al. Association study between genetic variants in retinol metabolism pathway genes and prostate cancer risk. Cancer Med. 2020;9(24):9462‐9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kim H, Lapointe J, Kaygusuz G, et al. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005;65(18):8118‐8124. [DOI] [PubMed] [Google Scholar]
- 129. Seidensaal K, Nollert A, Feige AH, et al. Impaired aldehyde dehydrogenase 1 subfamily member 2A‐dependent retinoic acid signaling is related with a mesenchymal‐like phenotype and an unfavorable prognosis of head and neck squamous cell carcinoma. Mol Cancer. 2015;14:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhang W, Wang H, Qi Y, Li S, Geng C. Epigenetic study of early breast cancer (EBC) based on DNA methylation and gene integration analysis. Sci Rep. 2022;12(1):1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wu S, Xue W, Huang X, et al. Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumour Biol. 2015;36(4):2421‐2426. [DOI] [PubMed] [Google Scholar]
- 132. Choi JA, Kwon H, Cho H, Chung JY, Hewitt SM, Kim JH. ALDH1A2 is a candidate tumor suppressor gene in ovarian cancer. Cancers (Basel). 2019;11(10):1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhou R, Chen Z, Xiao ZR, Wang SL, Rong C. HPV‐related promoter methylation‐based gene signature predicts clinical prognosis of patients with cervical cancer. Front Oncol. 2021;11:753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wang Y, Shao F, Chen L. ALDH1A2 suppresses epithelial ovarian cancer cell proliferation and migration by downregulating STAT3. Onco Targets Ther. 2018;11:599‐608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non‐small‐cell lung cancer. Drug Des Devel Ther. 2015;9:5087‐5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Luo Y, Dallaglio K, Chen Y, et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30(10):2100‐2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Saw YT, Yang J, Ng SK, et al. Characterization of aldehyde dehydrogenase isozymes in ovarian cancer tissues and sphere cultures. BMC Cancer. 2012;12:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Le Magnen C, Bubendorf L, Rentsch CA, et al. Characterization and clinical relevance of ALDHbright populations in prostate cancer. Clin Cancer Res. 2013;19(19):5361‐5371. [DOI] [PubMed] [Google Scholar]
- 139. Shao C, Sullivan JP, Girard L, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non‐small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20(15):4154‐4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kong B, Wu W, Cheng T, et al. A subset of metastatic pancreatic ductal adenocarcinomas depends quantitatively on oncogenic Kras/Mek/Erk‐induced hyperactive mTOR signalling. Gut. 2016;65(4):647‐657. [DOI] [PubMed] [Google Scholar]
- 141. Feng H, Liu Y, Bian X, Zhou F, Liu Y. ALDH1A3 affects colon cancer in vitro proliferation and invasion depending on CXCR4 status. Br J Cancer. 2018;118(2):224‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Quattrini L, Sadiq M, Petrarolo G, et al. Aldehyde dehydrogenases and prostate cancer: shedding light on isoform distribution to reveal druggable target. Biomedicines. 2020;8(12):569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang S, Liang C, Bao M, et al. ALDH1A3 correlates with luminal phenotype in prostate cancer. Tumour Biol. 2017;39(4):1010428317703652. [DOI] [PubMed] [Google Scholar]
- 144. Wang S, Zhou X, Liang C, et al. ALDH1A3 serves as a predictor for castration resistance in prostate cancer patients. BMC Cancer. 2020;20(1):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Federer‐Gsponer JR, Muller DC, Zellweger T, et al. Patterns of stemness‐associated markers in the development of castration‐resistant prostate cancer. Prostate. 2020;80(13):1108‐1117. [DOI] [PubMed] [Google Scholar]
- 146. Croker AK, Rodriguez‐Torres M, Xia Y, et al. Differential functional roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and therapy resistance of human breast cancer cells. Int J Mol Sci. 2017;18(10):2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Motomura H, Nozaki Y, Onaga C, et al. High expression of c‐Met, PKClambda and ALDH1A3 predicts a poor prognosis in late‐stage breast cancer. Anticancer Res. 2020;40(1):35‐52. [DOI] [PubMed] [Google Scholar]
- 148. Fauss J, Sprang B, Leukel P, et al. ALDH1A3 segregated expression and nucleus‐associated proteasomal degradation are common traits of glioblastoma stem cells. Biomedicines. 2021;10(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Li G, Li Y, Liu X, et al. ALDH1A3 induces mesenchymal differentiation and serves as a predictor for survival in glioblastoma. Cell Death Dis. 2018;9(12):1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Cheng P, Wang J, Waghmare I, et al. FOXD1‐ALDH1A3 signaling is a determinant for the self‐renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76(24):7219‐7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wu W, Wu Y, Mayer K, et al. Lipid peroxidation plays an important role in chemotherapeutic effects of temozolomide and the development of therapy resistance in human glioblastoma. Transl Oncol. 2020;13(3):100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhang S, Yang Z, Qi F. Aldehyde dehydrogenase‐positive melanoma stem cells in tumorigenesis, drug resistance and anti‐neoplastic immunotherapy. Mol Biol Rep. 2020;47(2):1435‐1443. [DOI] [PubMed] [Google Scholar]
- 153. Stagos D, Chen Y, Brocker C, et al. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde‐metabolizing enzyme. Drug Metab Dispos. 2010;38(10):1679‐1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405(2):173‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yang CK, Wang XK, Liao XW, et al. Aldehyde dehydrogenase 1 (ALDH1) isoform expression and potential clinical implications in hepatocellular carcinoma. PLoS One. 2017;12(8):e0182208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Wang H, Li Y, Zhou D, et al. Aldehyde dehydrogenase 1B1 is a potential marker of colorectal tumors. Histol Histopathol. 2021;36(2):183‐194. [DOI] [PubMed] [Google Scholar]
- 157. Matsumoto A, Arcaroli J, Chen Y, et al. Aldehyde dehydrogenase 1B1: a novel immunohistological marker for colorectal cancer. Br J Cancer. 2017;117(10):1537‐1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Mameishvili E, Serafimidis I, Iwaszkiewicz S, et al. Aldh1b1 expression defines progenitor cells in the adult pancreas and is required for Kras‐induced pancreatic cancer. Proc Natl Acad Sci USA. 2019;116(41):20679‐20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Singh S, Arcaroli JJ, Orlicky DJ, et al. Aldehyde dehydrogenase 1B1 as a modulator of pancreatic adenocarcinoma. Pancreas. 2016;45(1):117‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lindemann A, Takahashi H, Patel AA, Osman AA, Myers JN. Targeting the DNA damage response in OSCC with TP53 mutations. J Dent Res. 2018;97(6):635‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Krupenko SA. FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism. Chem Biol Interact. 2009;178(1‐3):84‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chen XQ, He JR, Wang HY. Decreased expression of ALDH1L1 is associated with a poor prognosis in hepatocellular carcinoma. Med Oncol. 2012;29(3):1843‐1849. [DOI] [PubMed] [Google Scholar]
- 163. Krupenko SA, Oleinik NV. 10‐formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down‐regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13(5):227‐236. [PubMed] [Google Scholar]
- 164. Rodriguez FJ, Giannini C, Asmann YW, et al. Gene expression profiling of NF‐1‐associated and sporadic pilocytic astrocytoma identifies aldehyde dehydrogenase 1 family member L1 (ALDH1L1) as an underexpressed candidate biomarker in aggressive subtypes. J Neuropathol Exp Neurol. 2008;67(12):1194‐1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Oleinik NV, Krupenko NI, Krupenko SA. Epigenetic silencing of ALDH1L1, a metabolic regulator of cellular proliferation, in cancers. Genes Cancer. 2011;2(2):130‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Dmitriev AA, Rudenko EE, Kudryavtseva AV, et al. Epigenetic alterations of chromosome 3 revealed by NotI‐microarrays in clear cell renal cell carcinoma. Biomed Res Int. 2014;2014:735292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Hartomo TB, Van Huyen Pham T, Yamamoto N, et al. Involvement of aldehyde dehydrogenase 1A2 in the regulation of cancer stem cell properties in neuroblastoma. Int J Oncol. 2015;46(3):1089‐1098. [DOI] [PubMed] [Google Scholar]
- 168. Beniaminov AD, Puzanov GA, Krasnov GS, et al. Deep sequencing revealed a CpG methylation pattern associated with ALDH1L1 suppression in breast cancer. Front Genet. 2018;9:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Krupenko SA, Horita DA. The role of single‐nucleotide polymorphisms in the function of candidate tumor suppressor ALDH1L1. Front Genet. 2019;10:1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Senchenko VN, Kisseljova NP, Ivanova TA, et al. Novel tumor suppressor candidates on chromosome 3 revealed by NotI‐microarrays in cervical cancer. Epigenetics. 2013;8(4):409‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Luo Y, Huang J, Tang Y, et al. Regional methylome profiling reveals dynamic epigenetic heterogeneity and convergent hypomethylation of stem cell quiescence‐associated genes in breast cancer following neoadjuvant chemotherapy. Cell Biosci. 2019;9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Lee SH, Jeon Y, Kang JH, Jang H, Lee H, Kim SY. The combination of loss of ALDH1L1 function and phenformin treatment decreases tumor growth in KRAS‐driven lung cancer. Cancers (Basel). 2020;12(6):1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zhu G, Liao X, Han C, et al. ALDH1L1 variant rs2276724 and mRNA expression predict post‐operative clinical outcomes and are associated with TP53 expression in HBV‐related hepatocellular carcinoma. Oncol Rep. 2017;38(3):1451‐1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA. ALDH1L2 is the mitochondrial homolog of 10‐formyltetrahydrofolate dehydrogenase. J Biol Chem. 2010;285(30):23056‐23063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Krupenko SA, Krupenko NI. ALDH1L1 and ALDH1L2 folate regulatory enzymes in cancer. Adv Exp Med Biol. 2018;1032:127‐143. [DOI] [PubMed] [Google Scholar]
- 176. Miyo M, Konno M, Colvin H, et al. The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol Rep. 2017;37(1):417‐425. [DOI] [PubMed] [Google Scholar]
- 177. Quere M, Alberto JM, Broly F, et al. ALDH1L2 knockout in U251 glioblastoma cells reduces tumor sphere formation by increasing oxidative stress and suppressing methionine dependency. Nutrients. 2022;14(9):1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Koppaka V, Thompson DC, Chen Y, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64(3):520‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Li D, Zhao H, Gelernter J. Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol‐induced medical diseases in Asians. Hum Genet. 2012;131(5):725‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Hou G, Chen L, Liu G, et al. Aldehyde dehydrogenase‐2 (ALDH2) opposes hepatocellular carcinoma progression by regulating AMP‐activated protein kinase signaling in mice. Hepatology. 2017;65(5):1628‐1644. [DOI] [PubMed] [Google Scholar]
- 181. Hu J, Yang L, Peng X, et al. ALDH2 hampers immune escape in liver hepatocellular carcinoma through ROS/Nrf2‐mediated autophagy. Inflammation. 2022;45(6):2309‐2324. [DOI] [PubMed] [Google Scholar]
- 182. Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137(5):1768‐1775. [DOI] [PubMed] [Google Scholar]
- 183. Jin S, Chen J, Chen L, et al. ALDH2(E487K) mutation increases protein turnover and promotes murine hepatocarcinogenesis. Proc Natl Acad Sci USA. 2015;112(29):9088‐9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Zhang H, Fu L. The role of ALDH2 in tumorigenesis and tumor progression: targeting ALDH2 as a potential cancer treatment. Acta Pharm Sin B. 2021;11(6):1400‐1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Kallifatidis G, Smith DK, Morera DS, et al. beta‐Arrestins regulate stem cell‐like phenotype and response to chemotherapy in bladder cancer. Mol Cancer Ther. 2019;18(4):801‐811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Chen L, Wu M, Ji C, Yuan M, Liu C, Yin Q. Silencing transcription factor FOXM1 represses proliferation, migration, and invasion while inducing apoptosis of liver cancer stem cells by regulating the expression of ALDH2. IUBMB Life. 2020;72(2):285‐295. [DOI] [PubMed] [Google Scholar]
- 187. Wang W, Wang J, Liu S, et al. An EHMT2/NFYA‐ALDH2 signaling axis modulates the RAF pathway to regulate paclitaxel resistance in lung cancer. Mol Cancer. 2022;21(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376(pt 3):615‐623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59(3):340‐349. [DOI] [PubMed] [Google Scholar]
- 190. Terzuoli E, Bellan C, Aversa S, et al. ALDH3A1 overexpression in melanoma and lung tumors drives cancer stem cell expansion, impairing immune surveillance through enhanced PD‐L1 output. Cancers (Basel). 2019;11(12):1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Calderaro J, Nault JC, Bioulac‐Sage P, et al. ALDH3A1 is overexpressed in a subset of hepatocellular carcinoma characterised by activation of the Wnt/ss‐catenin pathway. Virchows Arch. 2014;464(1):53‐60. [DOI] [PubMed] [Google Scholar]
- 192. Alix‐Panabieres C, Cayrefourcq L, Mazard T, Maudelonde T, Assenat E, Assou S. Molecular portrait of metastasis‐competent circulating tumor cells in colon cancer reveals the crucial role of genes regulating energy metabolism and DNA repair. Clin Chem. 2017;63(3):700‐713. [DOI] [PubMed] [Google Scholar]
- 193. Yan J, De Melo J, Cutz JC, Aziz T, Tang D. Aldehyde dehydrogenase 3A1 associates with prostate tumorigenesis. Br J Cancer. 2014;110(10):2593‐2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zhao L, Lei H, Shen L, et al. Prognosis genes in gastric adenocarcinoma identified by cross talk genes in diseaserelated pathways. Mol Med Rep. 2017;16(2):1232‐1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Fan F, Yin R, Wang L, et al. ALDH3A1 driving tumor metastasis is mediated by p53/BAG1 in lung adenocarcinoma. J Cancer. 2021;12(16):4780‐4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Rebollido‐Rios R, Venton G, Sanchez‐Redondo S, et al. Dual disruption of aldehyde dehydrogenases 1 and 3 promotes functional changes in the glutathione redox system and enhances chemosensitivity in nonsmall cell lung cancer. Oncogene. 2020;39(13):2756‐2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70(23):9937‐9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Yin Z, Wu D, Shi J, et al. Identification of ALDH3A2 as a novel prognostic biomarker in gastric adenocarcinoma using integrated bioinformatics analysis. BMC Cancer. 2020;20(1):1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Antonowicz S, Bodai Z, Wiggins T, et al. Endogenous aldehyde accumulation generates genotoxicity and exhaled biomarkers in esophageal adenocarcinoma. Nat Commun. 2021;12(1):1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Ferrari N, Granata I, Capaia M, et al. Adaptive phenotype drives resistance to androgen deprivation therapy in prostate cancer. Cell Commun Signal. 2017;15(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Marchitti SA, Orlicky DJ, Vasiliou V. Expression and initial characterization of human ALDH3B1. Biochem Biophys Res Commun. 2007;356(3):792‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Marchitti SA, Brocker C, Orlicky DJ, Vasiliou V. Molecular characterization, expression analysis, and role of ALDH3B1 in the cellular protection against oxidative stress. Free Radic Biol Med. 2010;49(9):1432‐1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Sun H, Zhang M, Li L, Huang Z. ALDH3B1 is an independent prognostic biomarker of lung adenocarcinoma. Technol Cancer Res Treat. 2020;19:1533033820946018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Wang Z, Mo Y, Tan Y, et al. The ALDH family contributes to immunocyte infiltration, proliferation and epithelial‐mesenchymal transformation in glioma. Front Immunol. 2021;12:756606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Gao ZG, Yang Y, Han XF, Wang YL, Wang ZJ. ALDH3B2 polymorphism is associated with colorectal cancer susceptibility. J Oncol. 2020;2020:5179635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Yin J, Tang W, Long T, et al. Association of ALDH3B2 gene polymorphism and risk factors with susceptibility of esophageal squamous cell carcinoma in a Chinese population: a case‐control study involving 2,358 subjects. Oncotarget. 2017;8(66):110153‐110165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Wu Y, Wei X, Feng H, et al. An eleven metabolic gene signature‐based prognostic model for clear cell renal cell carcinoma. Aging (Albany NY). 2020;12(22):23165‐23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Wang Y, Li K, Zhao W, et al. Aldehyde dehydrogenase 3B2 promotes the proliferation and invasion of cholangiocarcinoma by increasing integrin beta 1 expression. Cell Death Dis. 2021;12(12):1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53‐inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49(3):134‐140. [DOI] [PubMed] [Google Scholar]
- 210. Liao WL, Chan FC, Chang KP, et al. Associations between ALDH genetic variants, alcohol consumption, and the risk of nasopharyngeal carcinoma in an east Asian population. Genes (Basel). 2021;12(10):1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Lee HHC, Pearl PL, Rotenberg A. Enzyme replacement therapy for succinic semialdehyde dehydrogenase deficiency: relevance in gamma‐aminobutyric acid plasticity. J Child Neurol. 2021;36(13‐14):1200‐1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Chambliss KL, Gibson KM. Succinic semialdehyde dehydrogenase from mammalian brain: subunit analysis using polyclonal antiserum. Int J Biochem. 1992;24(9):1493‐1499. [DOI] [PubMed] [Google Scholar]
- 213. El‐Habr EA, Dubois LG, Burel‐Vandenbos F, et al. A driver role for GABA metabolism in controlling stem and proliferative cell state through GHB production in glioma. Acta Neuropathol. 2017;133(4):645‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Suh JJ, Pettinati HM, Kampman KM, O'Brien CP. The status of disulfiram: a half of a century later. J Clin Psychopharmacol. 2006;26(3):290‐302. [DOI] [PubMed] [Google Scholar]
- 215. van der Laan JW, de Boer T, Bruinvels J. Di‐n‐propylacetate and GABA degradation. Preferential inhibition of succinic semialdehyde dehydrogenase and indirect inhibition of GABA‐transaminase. J Neurochem. 1979;32(6):1769‐1780. [DOI] [PubMed] [Google Scholar]
- 216. Kaur H, Mao S, Li Q, et al. RNA‐Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS One. 2012;7(12):e50249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Deng XY, Gan XX, Feng JH, et al. ALDH5A1 acts as a tumour promoter and has a prognostic impact in papillary thyroid carcinoma. Cell Biochem Funct. 2021;39(2):317‐325. [DOI] [PubMed] [Google Scholar]
- 218. Tian X, Han Y, Yu L, et al. Decreased expression of ALDH5A1 predicts prognosis in patients with ovarian cancer. Cancer Biol Ther. 2017;18(4):245‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Kong P, Zhang L, Zhang Z, et al. Emerging proteins in CRPC: functional roles and clinical implications. Front Oncol. 2022;12:873876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Sass JO, Walter M, Shield JP, et al. 3‐Hydroxyisobutyrate aciduria and mutations in the ALDH6A1 gene coding for methylmalonate semialdehyde dehydrogenase. J Inherit Metab Dis. 2012;35(3):437‐442. [DOI] [PubMed] [Google Scholar]
- 221. Shin H, Cha HJ, Lee MJ, et al. Identification of ALDH6A1 as a potential molecular signature in hepatocellular carcinoma via quantitative profiling of the mitochondrial proteome. J Proteome Res. 2020;19(4):1684‐1695. [DOI] [PubMed] [Google Scholar]
- 222. Lu J, Chen Z, Zhao H, et al. ABAT and ALDH6A1, regulated by transcription factor HNF4A, suppress tumorigenic capability in clear cell renal cell carcinoma. J Transl Med. 2020;18(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Cho SY, Kang S, Kim DS, et al. HSP27, ALDH6A1 and prohibitin act as a trio‐biomarker to predict survival in late metastatic prostate cancer. Anticancer Res. 2018;38(11):6551‐6560. [DOI] [PubMed] [Google Scholar]
- 224. Guo Q, Zhang T, Gong Y, et al. Aldehyde dehydrogenase 6 family member A1 negatively regulates cell growth and to cisplatin sensitivity in bladder cancer. Mol Carcinog. 2022;61(7):690–701. [DOI] [PubMed] [Google Scholar]
- 225. Alison MR, Guppy NJ, Lim SM, Nicholson LJ. Finding cancer stem cells: are aldehyde dehydrogenases fit for purpose? J Pathol. 2010;222(4):335‐344. [DOI] [PubMed] [Google Scholar]
- 226. van den Hoogen C, van der Horst G, Cheung H, et al. High aldehyde dehydrogenase activity identifies tumor‐initiating and metastasis‐initiating cells in human prostate cancer. Cancer Res. 2010;70(12):5163‐5173. [DOI] [PubMed] [Google Scholar]
- 227. Giacalone NJ, Den RB, Eisenberg R, et al. ALDH7A1 expression is associated with recurrence in patients with surgically resected non‐small‐cell lung carcinoma. Future Oncol. 2013;9(5):737‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Hata A, Nakajima T, Matsusaka K, et al. A low DNA methylation epigenotype in lung squamous cell carcinoma and its association with idiopathic pulmonary fibrosis and poorer prognosis. Int J Cancer. 2020;146(2):388‐399. [DOI] [PubMed] [Google Scholar]
- 229. Chen D, Wu H, He B, et al. Five hub genes can be the potential DNA methylation biomarkers for cholangiocarcinoma using bioinformatics analysis. Onco Targets Ther. 2019;12:8355‐8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. He J, Zhao H, Deng D, et al. Screening of significant biomarkers related with prognosis of liver cancer by lncRNA‐associated ceRNAs analysis. J Cell Physiol. 2020;235(3):2464‐2477. [DOI] [PubMed] [Google Scholar]
- 231. Ambroziak W, Pietruszko R. Human aldehyde dehydrogenase: metabolism of putrescine and histamine. Alcohol Clin Exp Res. 1987;11(6):528‐532. [DOI] [PubMed] [Google Scholar]
- 232. Perroud B, Ishimaru T, Borowsky AD, Weiss RH. Grade‐dependent proteomics characterization of kidney cancer. Mol Cell Proteomics. 2009;8(5):971‐985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Stevenson J, Barrow‐McGee R, Yu L, et al. Proteomics of REPLICANT perfusate detects changes in the metastatic lymph node microenvironment. NPJ Breast Cancer. 2021;7(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Hu CA, Lin WW, Obie C, Valle D. Molecular enzymology of mammalian Delta1‐pyrroline‐5‐carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J Biol Chem. 1999;274(10):6754‐6762. [DOI] [PubMed] [Google Scholar]
- 235. De Ingeniis J, Ratnikov B, Richardson AD, et al. Functional specialization in proline biosynthesis of melanoma. PLoS One. 2012;7(9):e45190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Kardos GR, Wastyk HC, Robertson GP. Disruption of proline synthesis in melanoma inhibits protein production mediated by the GCN2 pathway. Mol Cancer Res. 2015;13(10):1408‐1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Zheng Y, Miyamoto DT, Wittner BS, et al. Expression of beta‐globin by cancer cells promotes cell survival during blood‐borne dissemination. Nat Commun. 2017;8:14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Ding Z, Ericksen RE, Lee QY, Han W. Reprogramming of mitochondrial proline metabolism promotes liver tumorigenesis. Amino Acids. 2021;53(12):1807‐1815. [DOI] [PubMed] [Google Scholar]
- 239. Kocher F, Tymoszuk P, Amann A, et al. Deregulated glutamate to pro‐collagen conversion is associated with adverse outcome in lung cancer and may be targeted by renin‐angiotensin‐aldosterone system (RAS) inhibition. Lung Cancer. 2021;159:84‐95. [DOI] [PubMed] [Google Scholar]
- 240. Craze ML, Cheung H, Jewa N, et al. MYC regulation of glutamine‐proline regulatory axis is key in luminal B breast cancer. Br J Cancer. 2018;118(2):258‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Guo YF, Duan JJ, Wang J, et al. Inhibition of the ALDH18A1‐MYCN positive feedback loop attenuates MYCN‐amplified neuroblastoma growth. Sci Transl Med. 2020;12(531):eaax8694. [DOI] [PubMed] [Google Scholar]
- 242. Dinavahi SS, Gowda R, Gowda K, et al. Development of a novel multi‐isoform ALDH inhibitor effective as an antimelanoma agent. Mol Cancer Ther. 2020;19(2):447‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Eneanya DI, Bianchine JR, Duran DO, Andresen BD. The actions of metabolic fate of disulfiram. Annu Rev Pharmacol Toxicol. 1981;21:575‐596. [DOI] [PubMed] [Google Scholar]
- 244. Barceloux DG. Copper. J Toxicol Clin Toxicol. 1999;37(2):217‐230. [DOI] [PubMed] [Google Scholar]
- 245. Liu X, Wang L, Cui W, et al. Targeting ALDH1A1 by disulfiram/copper complex inhibits non‐small cell lung cancer recurrence driven by ALDH‐positive cancer stem cells. Oncotarget. 2016;7(36):58516‐58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Jin N, Zhu X, Cheng F, Zhang L. Disulfiram/copper targets stem cell‐like ALDH(+) population of multiple myeloma by inhibition of ALDH1A1 and Hedgehog pathway. J Cell Biochem. 2018;119(8):6882‐6893. [DOI] [PubMed] [Google Scholar]
- 247. Duan X, Xiao J, Yin Q, et al. Multi‐targeted inhibition of tumor growth and lung metastasis by redox‐sensitive shell crosslinked micelles loading disulfiram. Nanotechnology. 2014;25(12):125102. [DOI] [PubMed] [Google Scholar]
- 248. Matsunaga N, Ogino T, Hara Y, Tanaka T, Koyanagi S, Ohdo S. Optimized dosing schedule based on circadian dynamics of mouse breast cancer stem cells improves the antitumor effects of aldehyde dehydrogenase inhibitor. Cancer Res. 2018;78(13):3698‐3708. [DOI] [PubMed] [Google Scholar]
- 249. Fournet G, Martin G, Quash G. alpha,beta‐Acetylenic amino thiolester inhibitors of aldehyde dehydrogenases 1&3: suppressors of apoptogenic aldehyde oxidation and activators of apoptosis. Curr Med Chem. 2013;20(4):527‐533. [DOI] [PubMed] [Google Scholar]
- 250. Perez‐Alea M, McGrail K, Sanchez‐Redondo S, et al. ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene. 2017;36(41):5695‐5708. [DOI] [PubMed] [Google Scholar]
- 251. Thomas ML, de Antueno R, Coyle KM, et al. Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Mol Oncol. 2016;10(9):1485‐1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Khanna M, Chen CH, Kimble‐Hill A, et al. Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases. J Biol Chem. 2011;286(50):43486‐43494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Kim J, Shin JH, Chen CH, et al. Targeting aldehyde dehydrogenase activity in head and neck squamous cell carcinoma with a novel small molecule inhibitor. Oncotarget. 2017;8(32):52345‐52356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Okazaki S, Shintani S, Hirata Y, et al. Synthetic lethality of the ALDH3A1 inhibitor dyclonine and xCT inhibitors in glutathione deficiency‐resistant cancer cells. Oncotarget. 2018;9(73):33832‐33843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Lam JP, Mays DC, Lipsky JJ. Inhibition of recombinant human mitochondrial and cytosolic aldehyde dehydrogenases by two candidates for the active metabolites of disulfiram. Biochemistry. 1997;36(44):13748‐13754. [DOI] [PubMed] [Google Scholar]
- 256. Johansson B. A review of the pharmacokinetics and pharmacodynamics of disulfiram and its metabolites. Acta Psychiatr Scand Suppl. 1992;369:15‐26. [DOI] [PubMed] [Google Scholar]
- 257. Kleczkowska P, Sulejczak D, Zaremba M. Advantages and disadvantages of disulfiram coadministered with popular addictive substances. Eur J Pharmacol. 2021;904:174143. [DOI] [PubMed] [Google Scholar]
- 258. Reinhardt S, Stoye N, Luderer M, et al. Identification of disulfiram as a secretase‐modulating compound with beneficial effects on Alzheimer's disease hallmarks. Sci Rep. 2018;8(1):1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Skrott Z, Mistrik M, Andersen KK, et al. Alcohol‐abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552(7684):194‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Wu X, Xue X, Wang L, et al. Suppressing autophagy enhances disulfiram/copper‐induced apoptosis in non‐small cell lung cancer. Eur J Pharmacol. 2018;827:1‐12. [DOI] [PubMed] [Google Scholar]
- 261. Rae C, Tesson M, Babich JW, Boyd M, Sorensen A, Mairs RJ. The role of copper in disulfiram‐induced toxicity and radiosensitization of cancer cells. J Nucl Med. 2013;54(6):953‐960. [DOI] [PubMed] [Google Scholar]
- 262. Chen D, Cui QC, Yang H, Dou QP. Disulfiram, a clinically used anti‐alcoholism drug and copper‐binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006;66(21):10425‐10433. [DOI] [PubMed] [Google Scholar]
- 263. Denoyer D, Masaldan S, La Fontaine S, Cater MA. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics. 2015;7(11):1459‐1476. [DOI] [PubMed] [Google Scholar]
- 264. Nechushtan H, Hamamreh Y, Nidal S, et al. A phase IIb trial assessing the addition of disulfiram to chemotherapy for the treatment of metastatic non‐small cell lung cancer. Oncologist. 2015;20(4):366‐367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Huang J, Chaudhary R, Cohen AL, et al. A multicenter phase II study of temozolomide plus disulfiram and copper for recurrent temozolomide‐resistant glioblastoma. J Neurooncol. 2019;142(3):537‐544. [DOI] [PubMed] [Google Scholar]
- 266. Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem‐like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res Treat. 2012;133(1):75‐87. [DOI] [PubMed] [Google Scholar]
- 267. Chefetz I, Grimley E, Yang K, et al. A Pan‐ALDH1A inhibitor induces necroptosis in ovarian cancer stem‐like cells. Cell Rep. 2019;26(11):3061‐3075.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Quash G, Fournet G, Courvoisier C, et al. Aldehyde dehydrogenase inhibitors: alpha,beta‐acetylenic N‐substituted aminothiolesters are reversible growth inhibitors of normal epithelial but irreversible apoptogens for cancer epithelial cells from human prostate in culture. Eur J Med Chem. 2008;43(5):906‐916. [DOI] [PubMed] [Google Scholar]
- 269. Nigjeh SE, Yeap SK, Nordin N, Rahman H, Rosli R. In vivo anti‐tumor effects of citral on 4T1 breast cancer cells via induction of apoptosis and downregulation of aldehyde dehydrogenase activity. Molecules. 2019;24(18):3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Nordin N, Yeap SK, Rahman HS, et al. In vitro cytotoxicity and anticancer effects of citral nanostructured lipid carrier on MDA MBA‐231 human breast cancer cells. Sci Rep. 2019;9(1):1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Kimble‐Hill AC, Parajuli B, Chen CH, Mochly‐Rosen D, Hurley TD. Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole‐2,3‐diones. J Med Chem. 2014;57(3):714‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Morgan CA, Hurley TD. Characterization of two distinct structural classes of selective aldehyde dehydrogenase 1A1 inhibitors. J Med Chem. 2015;58(4):1964‐1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Condello S, Morgan CA, Nagdas S, et al. beta‐Catenin‐regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34(18):2297‐2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Yang SM, Martinez NJ, Yasgar A, Danchik C, Johansson C, Wang Y, et al. Discovery of orally bioavailable, quinoline‐based aldehyde dehydrogenase 1A1 (ALDH1A1) inhibitors with potent cellular activity. J Med Chem. 2018;61(11):4883‐4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Yang SM, Yasgar A, Miller B, Lal‐Nag M, Brimacombe K, Hu X, et al. Discovery of NCT‐501, a potent and selective theophylline‐based inhibitor of aldehyde dehydrogenase 1A1 (ALDH1A1). J Med Chem. 2015;58(15):5967‐5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Kulsum S, Sudheendra HV, Pandian R, Ravindra DR, Siddappa G, Nisheena R, et al. Cancer stem cell mediated acquired chemoresistance in head and neck cancer can be abrogated by aldehyde dehydrogenase 1 A1 inhibition. Mol Carcinog. 2017;56(2):694‐711. [DOI] [PubMed] [Google Scholar]
- 277. Huddle BC, Grimley E, Buchman CD, Chtcherbinine M, Debnath B, Mehta P, et al. Structure‐based optimization of a novel class of aldehyde dehydrogenase 1A (ALDH1A) subfamily‐selective inhibitors as potential adjuncts to ovarian cancer chemotherapy. J Med Chem. 2018;61(19):8754‐8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Overstreet DH, Knapp DJ, Breese GR, Diamond I. A selective ALDH‐2 inhibitor reduces anxiety in rats. Pharmacol Biochem Behav. 2009;94(2):255‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Wang B, Buchman CD, Li L, Hurley TD, Meroueh SO. Enrichment of chemical libraries docked to protein conformational ensembles and application to aldehyde dehydrogenase 2. J Chem Inf Model. 2014;54(7):2105‐2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Parajuli B, Fishel ML, Hurley TD. Selective ALDH3A1 inhibition by benzimidazole analogues increase mafosfamide sensitivity in cancer cells. J Med Chem. 2014;57(2):449‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Parajuli B, Georgiadis TM, Fishel ML, Hurley TD. Development of selective inhibitors for human aldehyde dehydrogenase 3A1 (ALDH3A1) for the enhancement of cyclophosphamide cytotoxicity. Chembiochem. 2014;15(5):701‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Kreuzer J, Bach NC, Forler D, Sieber SA. Target discovery of acivicin in cancer cells elucidates its mechanism of growth inhibition. Chem Sci. 2014;6(1):237‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Cojoc M, Peitzsch C, Kurth I, Trautmann F, Kunz‐Schughart LA, Telegeev GD, et al. Aldehyde dehydrogenase is regulated by beta‐catenin/TCF and promotes radioresistance in prostate cancer progenitor cells. Cancer Res. 2015;75(7):1482‐1494. [DOI] [PubMed] [Google Scholar]
- 284. Stein MN, Malhotra J, Tarapore RS, Malhotra U, Silk AW, Chan N, et al. Safety and enhanced immunostimulatory activity of the DRD2 antagonist ONC201 in advanced solid tumor patients with weekly oral administration. J Immunother Cancer. 2019;7(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Prabhu VV, Lulla AR, Madhukar NS, Ralff MD, Zhao D, Kline CLB, et al. Cancer stem cell‐related gene expression as a potential biomarker of response for first‐in‐class imipridone ONC201 in solid tumors. PLoS One. 2017;12(8):e0180541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. La Manna F, De Menna M, Patel N, Karkampouna S, De Filippo MR, Klima I, et al. Dual‐mTOR inhibitor rapalink‐1 reduces prostate cancer patient‐derived xenograft growth and alters tumor heterogeneity. Front Oncol. 2020;10:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Goldstein LJ, Perez RP, Yardley D, Han LK, Reuben JM, Gao H, et al. A window‐of‐opportunity trial of the CXCR1/2 inhibitor reparixin in operable HER‐2‐negative breast cancer. Breast Cancer Res. 2020;22(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Mamaeva V, Niemi R, Beck M, Ozliseli E, Desai D, Landor S, et al. Inhibiting notch activity in breast cancer stem cells by glucose functionalized nanoparticles carrying gamma‐secretase inhibitors. Mol Ther. 2016;24(5):926‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Sarmiento‐Castro A, Caamano‐Gutierrez E, Sims AH, Hull NJ, James MI, Santiago‐Gomez A, et al. Increased expression of interleukin‐1 receptor characterizes anti‐estrogen‐resistant ALDH(+) breast cancer stem cells. Stem Cell Rep. 2020;15(2):307‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18(3):175‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Gao W, Wen H, Liang L, Dong X, Du R, Zhou W, et al. IL20RA signaling enhances stemness and promotes the formation of an immunosuppressive microenvironment in breast cancer. Theranostics. 2021;11(6):2564‐2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Sun L, Huang C, Zhu M, Guo S, Gao Q, Wang Q, et al. Gastric cancer mesenchymal stem cells regulate PD‐L1‐CTCF enhancing cancer stem cell‐like properties and tumorigenesis. Theranostics. 2020;10(26):11950‐11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Shen DD, Pang JR, Bi YP, Zhao LF, Li YR, Zhao LJ, et al. LSD1 deletion decreases exosomal PD‐L1 and restores T‐cell response in gastric cancer. Mol Cancer. 2022;21(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Zhang H, Xia Y, Wang F, Luo M, Yang K, Liang S, et al. Aldehyde dehydrogenase 2 mediates alcohol‐induced colorectal cancer immune escape through stabilizing PD‐L1 expression. Adv Sci (Weinh). 2021;8(10):2003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Hu Y, Lu L, Xia Y, Chen X, Chang AE, Hollingsworth RE, et al. Therapeutic efficacy of cancer stem cell vaccines in the adjuvant setting. Cancer Res. 2016;76(16):4661‐4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Hassani Najafabadi A, Zhang J, Aikins ME, Najaf Abadi ZI, Liao F, Qin Y, et al. Cancer immunotherapy via targeting cancer stem cells using vaccine nanodiscs. Nano Lett. 2020;20(10):7783‐7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Liao F, Zhang J, Hu Y, Najafabadi AH, Moon JJ, Wicha MS, et al. Efficacy of an ALDH peptide‐based dendritic cell vaccine targeting cancer stem cells. Cancer Immunol Immunother. 2022;71(8):1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauss A, et al. Cancer stem cells‐origins and biomarkers: perspectives for targeted personalized therapies. Front Immunol. 2020;11:1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell. 2019;24(1):25‐40. [DOI] [PubMed] [Google Scholar]
- 301. Dimri M, Satyanarayana A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers (Basel). 2020;12(2):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells – a clinical update. Nat Rev Clin Oncol. 2020;17(4):204‐232. [DOI] [PubMed] [Google Scholar]
- 303. Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41‐58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


