Abstract
Characterizing neuropsychiatric disorders is challenging due to heterogeneity in the population. We propose combining structural and functional neuroimaging and genomic data in a multimodal classification framework to leverage their complementary information. Our objectives are two‐fold (i) to improve the classification of disorders and (ii) to introspect the concepts learned to explore underlying neural and biological mechanisms linked to mental disorders. Previous multimodal studies have focused on naïve neural networks, mostly perceptron, to learn modality‐wise features and often assume equal contribution from each modality. Our focus is on the development of neural networks for feature learning and implementing an adaptive control unit for the fusion phase. Our mid fusion with attention model includes a multilayer feed‐forward network, an autoencoder, a bi‐directional long short‐term memory unit with attention as the features extractor, and a linear attention module for controlling modality‐specific influence. The proposed model acquired 92% (p < .0001) accuracy in schizophrenia prediction, outperforming several other state‐of‐the‐art models applied to unimodal or multimodal data. Post hoc feature analyses uncovered critical neural features and genes/biological pathways associated with schizophrenia. The proposed model effectively combines multimodal neuroimaging and genomics data for predicting mental disorders. Interpreting salient features identified by the model may advance our understanding of their underlying etiological mechanisms.
Keywords: functional network connectivity, multimodal deep learning, resting‐state functional and structural MRI, saliency, schizophrenia classification, single nucleotide polymorphism
Multimodal deep learning of imaging genetics data for characterizing mental disorders. The model processes multisource physiological data. It boosts the schizophrenia classifications accuracy by a margin compared with the existing model. The interpretation provides a handful of neurological ad genomics features that explains the underlying mechanism of the disease.
1. INTRODUCTION
Schizophrenia (SZ) is a psychotic disorder that causes impairment in people's thoughts, feelings, and behaviors. In general, people with SZ interpret reality atypically. Since the mental condition overtly manifests in adolescence and early adulthood, it is also considered a neurodevelopmental disorder (Mäki et al., 2005). People with SZ often experience hallucinations, delusions, social withdrawal, and disordered thinking. These can affect daily functioning and be highly disabling. Early identification of the disorder improves outcomes, and low‐dose antipsychotic medications may delay the onset of full‐blown psychosis (Mäki et al., 2005; Martin et al., 2017). However, SZ diagnosis is challenging for a few reasons. First, symptoms vary from person to person and change over time. Moreover, identification primarily depends on the self‐acknowledged behavioral changes in the affected individual. Another reason is that many preclinical symptoms during the prodromal period before a first episode overlap with life changes observed in healthy individuals and those who may develop other psychiatric disorders. Since diagnosis involves ruling out other disorders with similar symptoms, sole dependence on symptoms may be unreliable. Besides, the inferences are strongly modulated by the heterogeneity in the individual's psychological condition and treatment (Aboraya et al., 2006; Ward et al., 1962; Yassin et al., 2020). Therefore, research to discover the disorder's putative biological or neural substrates, which may contribute to more confident predictions or point to treatment targets, is warranted. SZ has also been linked to widespread variation at genetic loci (Jones & Cannon, 1998; Owen et al., 2004). This study aims to develop a flexible approach to link genomics and multimodal neuroimaging data to identify putative biological markers of brain disorders. We use structural and functional magnetic resonance imaging (sMRI and fMRI) and genome‐wide polymorphism collected from individuals with SZ and controls. We aim to probe the data to discover features (concepts) relevant to the disorder. Clinically, the expected insights could be neural activation patterns, structural aberrations, or genetic modifications. Strategically, we propose a model that can accurately discriminate the subjects to learn these attributes from the data. The urge for a reasonable accuracy is to provide dependable assertion about the pathophysiological mechanisms of the disorder—which result from the attributes learned by the model. Studies provide evidence for linkage between genetic risk factors for SZ and brain functional and structural changes (Passchier et al., 2020; Richards et al., 2020). Researchers suggest the shared genetic risk for SZ and SZ‐associated gray matter deficiency (Adhikari et al., 2019). Learning different modalities through a joint learning framework is promising and subject to maximizing the complementarity in the data. This shared learning approach conceivably provides crucial neurogenetic alliances for analyzing SZ. For multimodal learning, it is cardinal to incorporate a rational fusion technique to fuse multiple data modes. The proposed scheme maps the features to a common latent space to ensure a consistent fusion. We studied multiple neural networks to perform the latent space mapping and selected the best fit for each modality.
Deep neural networks (DNN) have shown considerable promise in learning features from input data in supervised and unsupervised settings (Chen et al., 2017; Tian et al., 2017; Yan et al., 2022; Zhong et al., 2016). Deep learning (DL)‐based studies have shown tremendous performance in discriminating SZ from controls (Lei et al., 2020; Oh et al., 2020; Patel et al., 2016; Qureshi et al., 2019; Yan et al., 2017; Yan et al., 2019). Using multiple (each complementary and incomplete) sources of information for classification can potentially improve the performance of the DL model. Furthermore, an optimal blending of modalities can help overall convergence and provide additional information about which aspects of each source are most relevant for the prediction. Such joint analysis can also provide a comparative significance analysis of the modalities. Multimodal DL enhances the robustness of inferences from a DL model since a learning task can explain distinct aspects of a system under investigation (Liu et al., 2018; Rashid & Calhoun, 2020). Researchers have started using multiple modalities for better prediction, which includes using structural MRI (sMRI) and resting‐state fMRI for SZ versus healthy control (HC) classification (Qureshi et al., 2017). Other studies focus on only multimodal neuroimaging fusion for SZ classification (Cetin et al., 2015; Cetin et al., 2016; Salvador et al., 2019). Our dataset includes sMRI, resting‐state fMRI, and genomic (single nucleotide polymorphism [SNP]) data in a multimodal framework to classify SZ versus HC. We first decompose the sMRI and fMRI images using independent component analysis (ICA) (Calhoun et al., 2009; Du et al., 2020). The method generates independent brain components and passes through the DL network to map the features from three modalities into a shared intermediate space. A linear attention module (Vaswani et al., 2017) is incorporated to guide the signal fusion from all modalities. The intuition behind attending to the modality‐specific signal is to scrutinize the influence of each modality on the prediction since all the modalities may not be equally critical for sample prediction. Attention controls the blending and always allows the better performer to influence the final prediction more. Results show that multimodal data fusion with attention improves classification accuracy, and the saliency model extracts highly discriminative features for characterizing SZ. The genetic information synergizes the imaging features, at both individual gene level and pathway level, to contribute to prediction. The analysis of groupwise saliency distributions reveals distinct patterns of modality‐specific influence on the decision‐making process between SZ and HC subjects. The model shows sMRI features contribute more to SZ predictions than other modalities. But the HC predictions are more influenced by fMRI and SNPs features. In general, we find that adding different modalities extends our perspective of the disorder and augments our knowledge of SZ's effects on the brain and its genetic linkages.
2. DATA PREPROCESSING
2.1. Structural MRI
sMRI scans are preprocessed by using statistical parametric mapping (SPM12, http://www.fil.ion.ucl.ac.uk/spm/). The preprocessing steps include unified segmentation and normalization of sMRI scans into gray matter, white matter, and cerebrospinal fluid (CSF). The segmentation step uses a modulated normalization algorithm to generate gray matter volume (GMV). Then a Gaussian kernel with a full width at half maximum (FWHM) = 6 mm is used to perform the smoothing on the GMV.
2.2. Functional MRI
We also use the SPM12 toolbox for preprocessing fMRI data. Before applying the preprocessing steps, we discard the first five time points of the fMRI to ensure a steady‐state magnetization. We perform rigid body motion correction using the INRI‐Align robust M‐estimation approach and apply the slice‐timing correction. fMRI imagery is then spatially normalized into the standard space Montreal Neurological Institute, using an echo‐planar imaging template, and is slightly resampled to 3 × 3 × 3 mm3 isotropic voxels. Images were then smoothed with a Gaussian kernel with FWHM = 6 mm like the sMRI data.
2.3. Genomics
The preprocessing steps for the genetic data are described in our prior work (Adhikari et al., 2019; Chen et al., 2013). There are several standard preprocessing tools for genomics data, and we used Plink (Purcell et al., 2007) for preprocessing and imputation. Linkage disequilibrium (LD) pruning was administered at r 2 < .9. The psychiatric Genomics Consortium (PGC) for SZ suggested genome‐wide association study (GWAS) (Cantor et al., 2010) score is used to select the features. The analysis selects 1280 SNPs distributed across 108 risk loci. The PGC study (He et al., 2015) reveals these SNPs express statistically significant linkage with SZ at p < 1 × 10−4.
3. PROPOSED ARCHITECTURE
The proposed hybrid framework comprises two major submodules (i) Parcellation of MRI images and selection of risk genetic variables and (ii) DNN to learn multimodal features for SZ classification. Each submodule is comprised of multiple subnetworks. The subnetwork selection for distinct modalities is experimental and is driven by primary intuition based on the characteristic of input data. We initially selected subnetworks that we thought would be suitable for the modality‐specific subnetworks. Next, we ran the experiments using several state‐of‐the‐art models applicable for identical purposes, that is, carrying out the feature extraction and selecting the best performers for the final subnetworks. Figure 1 illustrates different parts of our framework. The submodules are elaborated in the Sections 3.1 and 3.2. The first submodule decomposes the MRI data using group ICA. It selects risk SNPs from genomic variables, and the subnetworks in submodule (ii) take features for three modalities and feed them through a modality‐specific deep subnetwork. Submodule (ii) generates the latent representations for the input features. Then, the representations are aggregated and used as input to the prediction network for classifying the subject. The DNN incorporates three neural networks designed for each modality with modality‐wise specifications. The prediction layer consists of multiple fully connected (FC) linear layers. The architectural details are provided in the following paragraphs. We use subject and sample interchangeably to address the input data in the description.
FIGURE 1.
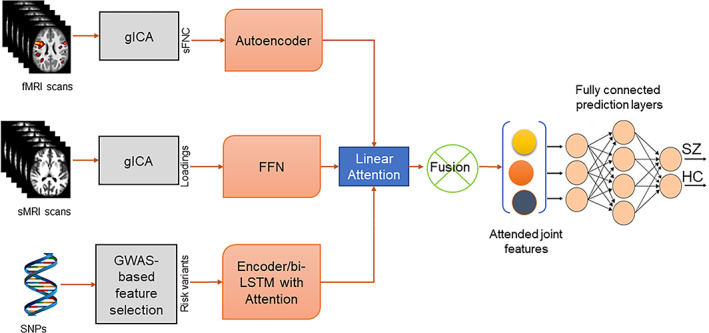
Our multimodal architecture. It has two submodules, (i) group independent component analysis (gICA) and genetic variable selection (ii) deep neural networks (DNN) for learning the modality features. The submodule (i) runs two separate gICA for structural and functional magnetic resonance imaging (sMRI and (fMRI) with distinct settings, for example, number of independent components expected. Then it computes the static functional network connectivity (sFNC) matrix for each subject from fMRI gICA and collects the ICA loading matrix from sMRI. Then, the genotyping on the genomics sequence generates single nucleotide polymorphism (SNPs). The DNN submodule consists of four subnetworks. sFNC features are extracted and learned by an AE with mean square error loss. The next subnetwork is a multilayer feed forward network for learning the ICA loading features. For genomics, we use a bi‐directional long short‐term memory (LSTM) unit with attention mechanism. After that, a fusion mechanism is applied to combine the latent features from three modalities using an attention model. The latent features are weighted by the attention score. Finally, the joint features are sent through a series of fully connected layers followed by a SoftMax prediction layer. The overall model is optimized using an Adam optimizer.
3.1. Group ICA and selection of genetic variables
We use a fully automated ICA‐based pipeline from the GIFT toolbox called Neuromark (Du et al., 2020) on sMRI and fMRI to extract functional and structural components of the brain. Neuromark is a robust analysis pipeline that can capture image features while retaining individual‐level variability. To construct spatial templates, we used two large HC datasets, collected by the human connectome project (HCP; http://www.humanconnectomeproject.org/) and the genomics superstruct project (GSP; https://www.nitrc.org/projects/gspdata). After that, we run the spatially constrained ICA from the NeuroMark pipeline on the combined subjects from three datasets, including FBIRN (Keator et al., 2016), COBRE (Aine et al., 2017), and MPRC (Adhikari et al., 2019). The selection of intrinsic connectivity networks (ICNs) in this study is based on the Neuromark template. The construction of Neuromark template is described as Group ICA with model order as 100 was performed on the GSP and HCP—HC datasets respectively, and the identified independent components (ICs) from the two datasets were then matched by comparing the corresponding group‐level spatial maps. Those pairs are considered as consistent and reproducible across GSP and HCP datasets if their spatial correlation ≥0.4. A correlation value ≥0.25 has been shown to represent a significant correspondence (p < .005, corrected) between components and here we used a higher threshold because we would like to identify more reliable and consistent ICs. The reproducible ICs pairs were further evaluated by examining their peak activations and low‐frequency fluctuations of their corresponding time courses. In total, 53 pairs of ICs were selected, arranging into 7 functional networks based on their anatomic and functional prior knowledge. The ICA with priors from NeuroMark (Du et al., 2020) resulted in 53 ICNs from the fMRI data after quality control. The quality‐controlled ICA results and various extensions have been implemented in several previous studies (Fu, Iraji, et al., 2021;Gupta et al., 2019; Saha et al., 2022). A similar approach was used for source‐based morphometry (ICA on gray matter maps) (Gupta et al., 2019; Saha et al., 2022), resulting in 30 structural components along with their loading values. The total number of connectivity networks extracted was 53 covering the whole brain. These ICNs were distributed into seven functional domains (Fu, Iraji, et al., 2021; Fu, Sui, et al., 2021): subcortical (SC), auditory (AD), sensorimotor (SM), visual (VS), cognitive‐control (CC), default‐mode (DM), and cerebellar domain (CB). After back reconstruction, we computed the static functional network connectivity (sFNC) matrix for each subject. The square matrix (53 × 53) represents the Pearson correlation between the time course of ICNs. We vectorize the sFNC matrix using the upper diagonal entries to ease the encoder's training process. Likewise, the sMRI ICs are extracted from the ICA on sMRI data. The provided priors are estimated from 6500 subjects used in this study (Abrol et al., 2017). The ICA on sMRI yields individual‐level structural networks and corresponding loadings. For the confidence of the structural networks, the network approximation scheme (infomax) was repeated 20 times in ICASSO, in which the best run was selected to ensure stability. The process extracted 30 structural components along with their loading values. For the sMRI modality, we used the loadings of the structural networks as the structural features. The third modality includes the set of SNPs selected from genomic data based on GWAS‐significant SZ risk SNPs identified by the large PGC study.
3.2. Deep multimodal neural networks
We use an autoencoder (AE) (Goodfellow et al., 2016) for the fMRI modality consisting of encoder and decoder subnetworks. The AE has been a successful neural network for a pattern learning task (Li et al., 2017; Zhuang et al., 2015). We also experimented with feed‐forward network (FFN), but AE outperforms FFN by a significant margin. The encoder subnetwork consists of five linear layers, and the decoder also has the same number of layers. The decoder reconstructs the input from the encoded features map. The encoder compresses the input sequence to reduce the dimensions and generate latent embeddings sequentially. We use a loss function to measure how well the decoder reconstructs the inputs, that is, reconstruction loss. In our case, we use the mean squared error loss for training the encoder and decoder. This network has been established as a consistent performer in representation learning (Chen et al., 2016; Kodirov et al., 2017; Kusner et al., 2017; Liou et al., 2014). The weights for the linear layers are uniformly initialized using the Xavier initialization function in Pytorch (Paszke et al., 2019). A dropout layer follows each layer to minimize the overfitting with a probability of 0.2. The subnetwork used rectified linear units (ReLU) (Agarap, 2018) activation function for capturing the nonlinearity in the data.
The sMRI subnetwork was constructed as a FFN with five FC layers. FFN is an easy‐to‐implement neural network effective for learning the representation from linear data streams (Ruck et al., 1990; Zhao et al., 2012). Also, it reduces model complexity and saves considerable running time on the model. The FC layers mostly have 30 hidden nodes followed by a dropout layer with a probability of 0.2 except for the last layer. We avoid compression in this network because the input dimension is shallow compared with the other modalities. The last layer of the subnetwork has 100 hidden nodes to match up with the latent feature size from other modalities.
We tried both compression networks as fMRI and a bi‐directional long short‐term memory (bi‐LSTM) unit for the genetic modality. The bi‐LSTM unit is incorporated with the attention mechanism (Vaswani et al., 2017). Although the SNP data are not temporally dependent, the neighboring SNPs may show LD. So, a potential neighborhood structure is present in the data, and we intend to capture it to help the prediction. We apply light pruning to the SNP data and use bi‐LSTM to capture these localized semantics which might help differentiate the disorder. The attention weights are used to help learn the representations of the neighborhood. However, we observed that the neighborhood information did not support the predictor and yielded a similar performance as MLP. Intuitively, we focused on MLP because of its less complex implementation and considerably faster runtime than other approaches.
Attention module: This subnetwork performs the fusion between embeddings generated from modality‐wise subnetworks. It consists of one linear layer followed by a SoftMax layer. It takes a concatenated two‐dimensional tensor (number of modalities‐by‐embedding sizes). The first dimension represents the number of modalities and the second one is the size of embeddings. The module attends the embeddings and learns an attention weight for each modality iteratively. Then, it multiplies the input tensor with the attention weights and expands as the input tensor's shape.
Prediction model: The final subnetwork is a stack of three FC layers followed by a SoftMax layer. The FC layers have 100 hidden nodes, and the dropout is set to 0.2. It takes the attended combined data and predicts the subject's label.
We run the multimodal network for hyperparameter tuning and prediction stability over multiple repetitions. We determined these parameters by running random search (Bergstra & Bengio, 2012) hyperparameter optimization and nested cross‐validation.
3.3. Fusion
Several data fusion techniques have been introduced in the literature (Ramachandram & Taylor, 2017). The fusion methods are primarily divided into three subcategories early, mid/intermediate, or late fusion. These provide distinct pros and cons depending on their application domains. We developed several baseline models to implement those data fusion. Early fusion integrates multiple input modalities into a standard feature vector; late fusion combines the predictions from various models, and mid fusion fuses the latent features (intermediate states) of modality‐wise subnetworks to perform the downstream task. We propose mid fusion administered by an attention module for modality amalgamation.
3.3.1. Mid fusion/intermediate
We incorporate intermediate fusion that combines the learned feature representations from intermediate layers of modality‐specific subnetworks. A linear attention module governs the fusion process by learning a significance score for each modality. It weighs the contributions of each information source for generating an optimal mixture of relevant insights. In our case, this controlled aggregation is imperative to any downstream task, for example, classifying a subject's label. This study uses distinct subnetworks to extract latent compressed features from each input stream. The fusion submodule collects complementary information among different data modes to maximize the knowledge about the sample. Thus, we aggregate the latent features from sMRI, fMRI, and SNPs for the fusion and send them to the final subnetwork. The process assists the model in capturing the relation/interaction between multiple modalities. The fusion architecture primarily maps the input to a latent space by computing the modality‐wise embeddings. Then use this joint embedding for the prediction. The notable advantages of attentive fusion are (1) it provides a joint training of all available modalities, (2) the joint learning leverages the synergies between distinct sources of information, and (3) with the influence of neighboring subnetworks, the multimodal framework shows a faster convergence. The aggregation is performed following this equation:
| (1) |
where h is a modality‐specific neural network, we can describe it as h m: ℝ dm → ℝ d (m represents the modalities, m = 1, …, M and d is the dimension). The joint feature u is defined as u ϵ ℝ d. The w's are the learnable scaler for each modality. The final prediction value is p where:
| (2) |
and y is a neural network described as y: ℝ d → ℝ c, and c is the number of classes.
3.3.2. Early fusion
The input modalities are aggregated in early fusion before running it through a DNN. Our early fusion baseline includes a stack of linear layers—multilayer perceptron (MLP) as a DNN unit. We concatenate the input data from three modalities for each sample and send them through the MLP. The training and validation scheme is consistent with the mid fusion training described below.
3.3.3. Late fusion
Late fusion uses a distinct network for each modality. Each modality‐specific DNN predicts the subject's label separately, and at the end, the predictions are merged using max‐voting (Morvant et al., 2014) to determine the final label of the subject. The modality‐specific subnetworks are trained separately and validated for the best performance in terms of the evaluation metric. We capture the predictions of the best‐performing subnetwork on the test data. Then the predictions are combined through a max‐voting technique where the samples are assigned with labels predicted by majority modalities.
3.4. Training scheme for the joint fusion model
In our study, we explore all three fusion techniques. The performance analysis evident mid/intermediate fusion as the best performing model. The parameters and built‐in model selections are consistent across three fusion techniques. The framework uses a cross‐entropy loss and the Adam (Kingma & Ba, 2014) optimizer with a learning rate of 1 × 10−4 and a batch size of 32 for training in Pytorch. The Adam optimizer updates the weights in the proposed DNN by back‐propagating the cross‐entropy loss computed from the final layer's prediction and the subject's label. All the FC layers are followed by the dropout layer with p = .2 to minimize the overfitting. Our experimentations include a variety of p values for dropout layers yet; multiple experiments suggest a p = .2 value for the dropout layers provides the best performance. We also added early stopping for balancing training and validation loss, which eventually regularizes the model. The dataset is divided into training, testing, and validation set. We first partition the data into 80% for train and 20% for evaluation. The evaluation data is divided into two equal sets testing and validation. The test set is held out from the training and validation phase. The model never comes across those samples in their training phase. In the training and validation phase, the splits are randomly selected. Then, the training and validation run for 300 epochs, saving the best model based on accuracy. We loaded it back for computing the test accuracy by applying the previously saved model to test data. To avoid the fluctuation of the results, we ran the model multiple repetitions (10 times) on the same data splits and reported the average performance of the model. This joint training scheme allows modalities to interact and facilitates modality‐specific subnetworks to converge properly by leveraging the learning from other subnetworks. Moreover, we implement a multimodal regularization technique for the completeness of the experiments, which focuses on removing bias by maximizing functional entropies (Gat et al., 2020). We designed our implementation based on the existing regularizer and utilities.
4. RESULTS
4.1. Experiments
We used subjects selected from three different datasets, COBRE (Aine et al., 2017), fBIRN (Keator et al., 2016), and MRPC (Adhikari et al., 2019). Our deep model uses sMRI, fMRI, and genome‐wide polymorphism data for the classification. The combined dataset has 437 subjects with 275 HC and 162 SZ subjects. Due to heterogeneity in data preprocessing and the lack of consistent datasets used in previous studies, we generate several unimodal, bi‐modal baselines to compare the performance of our proposed multimodal classification framework. The analysis covers several data fusion techniques such as joint fusion, late fusion, additive, multiplicative, and a weighted combination of latent features.
4.2. Evaluation
The evaluation metrics we use to compare the model's performance are described below. The variables are true‐positive rate (TPR), false‐positive rate (FPR), and accuracy (ACC), which are defined as:
i. TPR = TP/(TP + FN).
ii. FPR = FP/(TP + FP), and.
iii. ACC = (TP + TN)/(TP + FP + FN + TN).
Here, TP stands for true positives: no. of SZ subjects classified as SZ; FN stands for false negatives: no. of SZ classified as HC, TN (true negatives): no. of HC subjects classified as HC, FP (false positives): no. of HC classified as SZ. The performance of different architectures we evaluated on our datasets is given in Table 1. Our proposed mid fusion with the attention model has shown the best performance with an accuracy of 92%, an average precision of 0.568, and an F1 score of 0.573 (boldface). It outperforms other baselines and other approaches to classifying SZ. The unimodal and bimodal performances are evident in the complementary information among these biological information sources. A modality like genomics shows inadequate efficacy solely. However, when the source is mixed up with other modalities, it improves the overall performance. The observation reinforces the intuition behind proposing a multimodal exploration. Thus, the weaker or less informative modalities are also useful in the presence of other active sources in the joint learning framework. For reference, we added results from all three fusion techniques early, late, and mid fusion. However, mid fusion with attention shows significant improvement compared with other fusion techniques.
TABLE 1.
Our experiments and comparisons with baselines
| No. | Fusion type | Modalities | Acc | AP | F1 score |
|---|---|---|---|---|---|
| 1 | Unimodal | fMRI | 0.81 | 0.481 | 0.451 |
| 2 | Unimodal | sMRI | 0.78 | 0.441 | 0.431 |
| 3 | Unimodal | Genetic (SNPs) | 0.68 | 0.339 | 0.310 |
| 4 | Bimodal | sMRI + fMRI | 0.83 | 0.470 | 0.438 |
| 5 | Bimodal | SNPs + sMRI | 0.78 | 0.411 | 0.402 |
| 6 | Bimodal | SNPs + fMRI | 0.81 | 0.423 | 0.387 |
| 7 | Early fusion | fMRI + sMRI + SNPs | 0.73 | 0.401 | 0.395 |
| 8 | Late fusion | fMRI + sMRI + SNPs | 0.78 | 0.426 | 0.411 |
| 9 | Functional entropies (Kingma & Ba, 2014) | fMRI + sMRI + SNPs | 0.72 | 0.369 | 0.335 |
| 10 | Mid fusion | fMRI + sMRI + SNPs | 0.88 | 0.495 | 0.432 |
| 11 | Mid fusion with attention | fMRI + sMRI + SNPs | 0.92 | 0.568 | 0.573 |
Note: Our proposed mid fusion with the attention model has shown the best performance with an accuracy of 92%, an average precision of 0.568, and an F1 score of 0.573 (boldface).
Abbreviations: fMRI, functional magnetic resonance imaging; sMRI, functional magnetic resonance imaging; SNPs, single nucleotide polymorphisms.
4.3. Model interpretation
The secondary goal of our study is to introspect the model's learning. The intuition is to explore the insights learned by the model for predicting the disease. These insights are potentially informative about the disorder and can be transmuted as crucial biomarkers for the group distinction. Furthermore, interpreting the modality‐wise knowledge gathered by the framework helps extract which structural and functional components of the brain and genes are consistently playing a significant role in characterizing a subject as SZ or HC. The analysis may create new focuses for further research by providing reliable sources of dichotomy. We implement a saliency analysis of the fully trained model to explore the feature‐wise contribution from each modality. Saliency explains individual predictions by attributing each input feature according to how much it changed the prediction (negatively or positively; Ismail et al., 2021; Simonyan et al., 2013). For the experiments, we use the Captum library (Kokhlikyan et al., 2020). Saliency is defined as , or the gradients of the prediction of the correct class with respect to the input. Captum's attribution modules are divided into three subcategories based on the applied component of a DNN, namely primary, neuron, and layer attributions. Primary attribution is the traditional feature importance algorithms that attribute output predictions to model inputs. Layers attribute output predictions to all neurons in a hidden layer, and neuron attribution methods allow us to attribute a hidden neuron to the inputs of the model. However, both neuron and layer attributions are slight modifications of the primary attribution algorithms (Kokhlikyan et al., 2020). It is a widely used method for interpreting neural networks and has been shown to be an effective measure under scrutiny (Adebayo et al., 2018; Ismail et al., 2021). Saliency maps are essentially heat maps of each input sample, indicating the relevancy of each feature to the sample's true class. The saliency experiments compute a scalar weight for each input attribute from different modalities. This weight is known as the saliency score or importance weight. We calculated the mean saliency values across the subjects and the folds for consistency.
4.3.1. Saliency analysis on fMRI features
Figure 2 illustrates the mean saliency score for each static FNC pair. We print the mean saliency score for each pair of components (connection) in a square matrix annotated with their corresponding neuro domain. We threshold the connection‐wise saliency score to extract the highly salient pairs. These 10 most salient pairs of components illustrate the most contributing functional connections in the brain. We observe at least one endpoint of salient connection is sourced from the subcortical region of the brain (Figure 2b). Therefore, the highly contributing links are between subcortical and other brain regions. Moreover, those pairs' average sFNC strength is consistently higher in the SZ subjects than in HC. The observations indicate SZ subjects have consistent interactions between subcortical to other domains, which might be related to impaired cognition, and the subject's experience of unusual information processing systems. It perhaps infers the development of necessary associations with other domains by allocating significant resources of brain circuitry to the subcortical. It also encourages domain‐specific future studies. For instance, a subcortical‐focused connectivity study is justifiable to further experiment with our observed connections—steady connectivity in SZ. Table 2 summarizes the components involved in the salient static functional connections of the brain. The connectogram in Figure 3 illustrates the connectivity strength between the salient component pairs. We observe a strong positive connection between IC 14, IC 3. A strong negative connection between IC 6 and IC 1. Figure 4 shows the mean connectivity strength in HC and SZ subgroups. We observe that patient‐control connectivity differences in six pairs were statistically significant at p ˂ .01.
FIGURE 2.
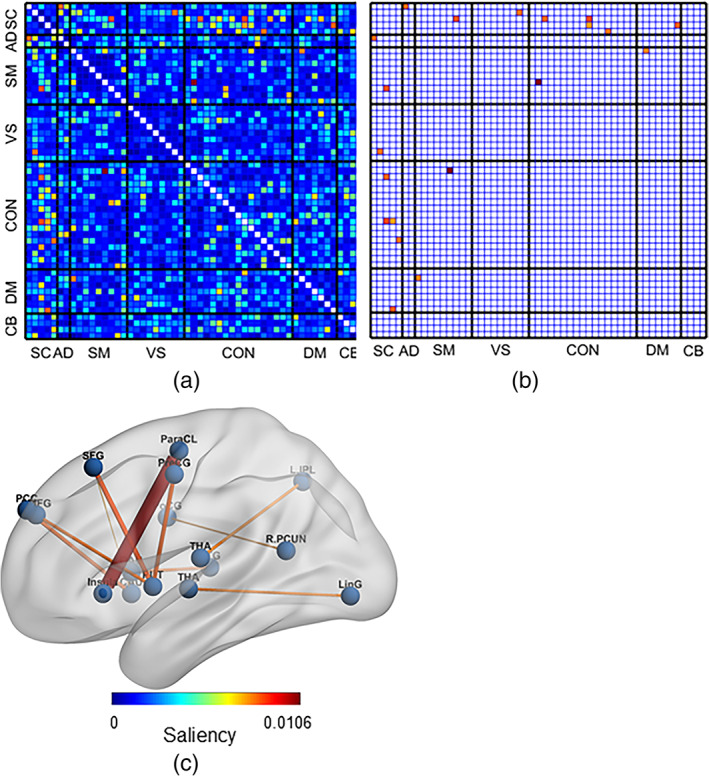
The figure visualizes the saliency analysis of functional magnetic resonance imaging features—static functional network connectivity (sFNC) pairs. (a) Each cell of the matrix represents the saliency score for a pair of static functional networks (connections). It illustrates saliencies for all the sFNC pairs (1378). (b) The figure represents the 10 best salient pairs for schizophrenia prediction. (c) Overlaid the salient connections on the AAL brain template. We observe the salient pairs mainly include components from the subcortical region; thus, the predominant connectivity's between subcortical and other domains, for example, subcortical (SC), auditory (AD), sensorimotor (SM), visual (VS), cognitive‐control (CC), default‐mode (DM), and cerebellar domain (CB), and so forth. However, we do not observe any significant connections (pairs) within the subcortical domain: The most influential connections are interdomain.
TABLE 2.
Salient pairs of components (connections) are observed from the saliency analysis of fMRI data and their neuro domains
| ID | Comp. I | Dom. I | Comp. II | Dom. II | HC‐SZ direction |
|---|---|---|---|---|---|
| 1 | Para central lobule | SMN | Insula | CON | HC > SZ |
| 2 | Putamen | SCN | Superior frontal gyrus | CON | HC < SZ |
| 3 | Putamen | SCN | Precentral gyrus | SMN | HC < SZ |
| 4 | Caudate | SCN | Posterior cingulate cortex | DMN | HC < SZ |
| 5 | Putamen | SCN | Superior medial frontal gyrus | CON | HC < SZ |
| 6 | Caudate | SCN | Superior temporal gyrus | ADN | HC < SZ |
| 7 | Thalamus | SCN | Left inferior parietal lobule | CON | HC < SZ |
Note: Comp. I and comp. II represent the pairs of components portraying functional connectivity. Dom. I and II demonstrate the domain the components belong to.
Abbreviations: fMRI, functional magnetic resonance imaging; HC, healthy control; SZ, Schizophrenia.
FIGURE 3.
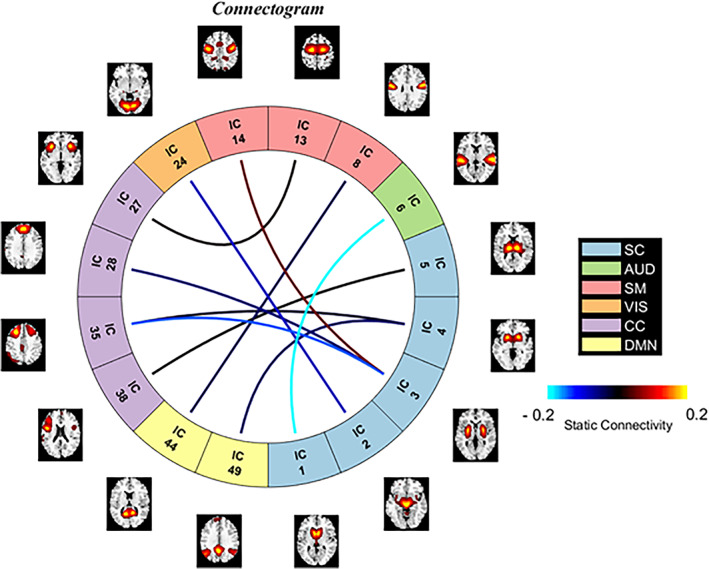
Connectogram for the 10 most salient static functional network connections. The figure illustrates the connections and participating functional networks along with their neural domains. The color of connections represents the strength of the connectivity. The corresponding domains are also presented using distinctive colors. The blue and red lines stand for negative and positive functional connectivity, respectively.
FIGURE 4.
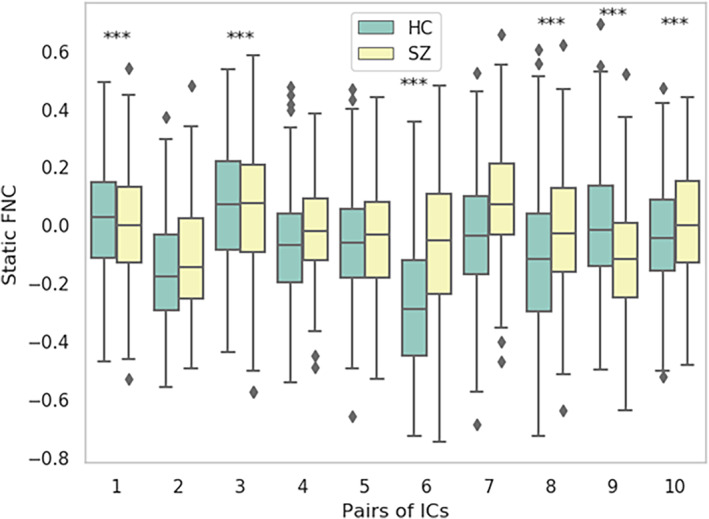
The boxplot demonstrates the max, min, and median of static functional connectivity strength in the 10 most salient static functional network connectivity (sFNC) pairs. The pairs are sorted in terms of their saliency score. We ran a two‐sample t‐test to test the statistical significance of healthy control (HC)‐schizophrenia (SZ) group differences. Pairs 1, 3, 6, 8, 9, and 10 were found to be statistically significant at p < .01 (asterisks)
4.3.2. sMRI features saliency analysis
From the average saliency scores for sMRI features, some of the components show significantly higher saliency than others. We select the most salient 5 out of 30 components for further investigation. The components are (i) caudate, (ii) anterior cingulate cortex and medial frontal cortices (mpFC), (iii) inferior and mid frontal gyrus, (iv) precuneus and posterior cingulate cortex, and (v) calcarine. Figure 5a shows the mapping of the structural components on the brain marked with their corresponding saliency score. Figure 5b is the bar plot for the component's saliency, and Figure 5c shows the mean loading value of these components across HC and SZ subgroups. The group differences are tested using a two‐sample t‐test controlled for age, sex, and site. sMRI components (i), (ii), (iii), and (v) show statistically significant HC‐SZ difference at a p < .01 and denoted by an asterisk sign. HC subjects have a stronger expression (loading) of components (ii), (iii), and (v) and a weaker of the caudate than SZ. It might result from lower gray matter in these structural components in SZ (ii, iii, and v) versus control subjects. Similarly, the caudate seems to be having weaker expression in the sMRI of HC subjects. Structural components' prominence (loading) in SZ and HC subjects are different. The group differences in distinct components are indicative of the distinct expression patterns of SZ and HC subjects on these salient structural components are probably relatable to the disorder's symptoms. In that case, they will potentially help advance our understanding of SZ pathophysiology.
FIGURE 5.
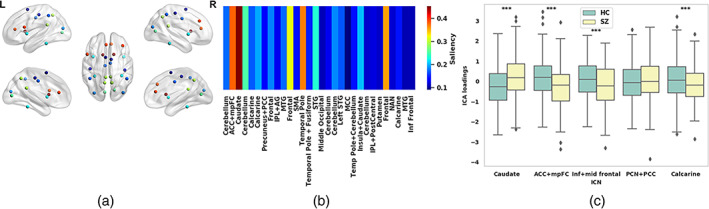
The saliency analysis of structural magnetic resonance imaging (sMRI) features. (a) Overlaying the structural components on the brain map annotated by their corresponding saliency. The subfigure includes five different views of the brain and the red colored features are the most salient for the schizophrenia (SZ) prediction. (b) We visualize the feature‐wise saliency on a bar plot, the x‐axis represents 30 sMRI features/components. We compute the mean saliency of each component across the subjects and found a subset of components ICN 3, 2, 26, 7, and 28 (sorted high to low saliency) are distinctively more salient than the others. (c) We picked the best five salient sMRI components here and the boxplot shows the max, min, and median loading values in the SZ and healthy control (HC) groups. We run a two‐sample t‐test to check the statistical significance of the HC‐SZ group differences. Four components (asterisks) show statistically significant differences at p < .01, which include the caudate, anterior cingulate (ACC) and medial prefrontal (mpFC) cortices, inferior and mid frontal gyrus, and calcarine sulcus
Saliency analysis of genetic features (SNPs)
From the saliency map computed on SNPs, we select SNPs showing higher importance scores across at least six folds of cross‐validation (total folds = 10). The higher importance score indicates a saliency value greater than the mean saliency of a particular fold. It returns 430 SNPs out of 1280, which consistently influences the prediction. These SNPs are annotated into 92 unique genes, and we conducted pathway analysis on these genes using the David functional analysis tool (Huang et al., 2007). We noted two clusters of genes involved in the dopaminergic synapse (p = .03) and postsynaptic density (p = .008), respectively shown in Table 3 . The genes included in these clusters are well documented to be associated with SZ (Föcking et al., 2015; Roberts et al., 2009). The mentionable genes involved in these pathways are the GRIN2A—glutamate receptor that contributes to the slow phase of excitatory postsynaptic current, long‐term synaptic potentiation, and learning by similarity. CACNA1C is related to calcium signaling. GRM3 is also a glutamate receptor, CHRNA3 is the cholinergic receptor; these are important in neurotransmitter signaling and involved in SZ (Fujii et al., 2003; Mössner et al., 2008; Petrovsky et al., 2010). Figure 6 visualizes the joint and marginal distributions of saliency in HC and SZ subjects. Figure 6a–c represents the joint contour plot estimated by the kernel density estimator using the data points. The marginal distribution on the top illustrates the data on the x‐axis and the distributions on the right‐hand side represent the y‐axis data. From the distributions, we observe the HC saliencies follow a light‐tailed distribution for fMRI and genetic modalities but are comparatively fat‐tailed for sMRI information. That means the saliency values are higher and low variance in fMRI than in sMRI and SNPs. In other words, HC prediction uses more fMRI information than other modalities. However, the saliencies in SZ subjects show light‐tailed (low variance) distribution for sMRI and competitively higher variance for fMRI and SNPs. It indicates sMRI source of information influences these samples' predictions more. It does not suggest the absence of other classification tasks. The attention values are normalized across the modalities, and we collected them after finishing the training process for all the specified epochs. Figure 7 demonstrates the results. The boxplot has three boxes (modalities) for each modality: instead, it explains how the predictor uses the contributions from different modalities while classifying the distinct samples. We analyze the attention weights from all three modalities while training to investigate the phenomena more precisely. We used an attention model to weigh the latent features from each modality before concatenating them in backpropagation. The results show a consistent pattern in modality‐specific information usage. We plot the attention weights from three modalities for all 437 subjects. The attention values portray the contribution of modalities in the group (SZ/HC). The bars represent the mean and SD for the attention values. We observe the mean attention for the HC group is higher in fMRI, and for SZ, it is significantly higher in sMRI. The observation could be interpreted as a distinctive distribution of information mounted in these modalities relevant to the disorder. The model consistently relies on structural modality rather than others for classifying the SZ samples, which might indicate a greater impact on structural features of SZ. In other words, structural features better differentiate the samples (SZ and HC), making them a better predictor of the disorder. Figure 8 represents the interaction between the modalities throughout the training process and the effect of modality coupling and decoupling on classification. We analyze the interaction between modalities through the connection between their latent space representations. For each subject, we measure the correlation among the latent space representation of its sMRI, fMRI, and genomic feature. Increasing similarities (correlation) between modalities could be seen as coupling those information sources—maneuvering towards similar data distributions. We observe that the correlation between fMRI and genetic information consistently increases as the model learns. With the learning from the data ~50 epochs, these two modalities become coupled and keep expanding their inter‐relationship throughout the training processes. The coupling might represent these sources are uniformly descriptive about the subject. The sMRI and genetic modality show a reasonable positive correlation across the epochs. We also observe that the sMRI and fMRI coupling has a negative correlation after a few epochs, but fMRI and genetics are in the opposite direction. The intermodality coupling and decoupling nature for characterizing the disorder suggests complementary and overlapping links between the neural and genomic systems. The uniformity or nonuniformity of data sources can be leveraged in cases where all data modalities are unavailable or may not be convenient for acquisition. The knowledge base could be exploited for handling missing modality issues common in multimodal data frameworks.
TABLE 3.
The cluster of genes or the most salient pathways associated with schizophrenia
| Cluster | Relevance | p‐value | Genes | List total | Pop hits | Pop total | Fold enrichment |
|---|---|---|---|---|---|---|---|
| KEGG_PATHWAY | Dopaminergic synapse | .028748 | GRIN2A, AKT3, PPP2R3A, and CACNA1C | 37 | 128 | 6879 | 5.809966 |
| GOTERM_CC_DIRECT | Postsynaptic density | .007676 | GRM3, CHRNA3, GRIN2A, CACNA1C, and NRGN | 78 | 184 | 18,224 | 6.348941 |
FIGURE 6.
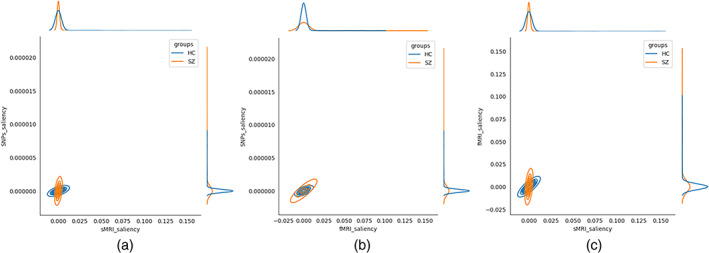
Visualization of joint and marginal distributions of subject wise saliency for functional magnetic resonance imaging (fMRI), structural MRI (sMRI), and single nucleotide polymorphisms (SNPs) modalities (a) The subfigure shows the distribution of saliency by sMRI vs SNPs. The subfigure also plots the joint kernel density of saliency scores. (b) Between fMRI versus SNPs saliency, and (c) fMRI versus sMRI
FIGURE 7.
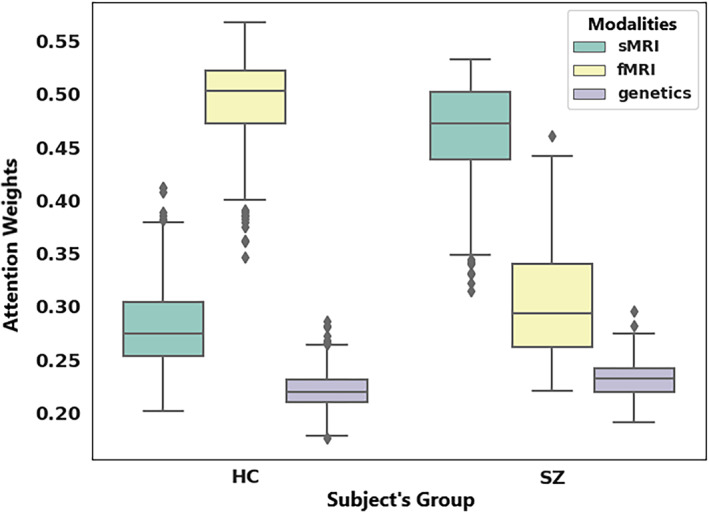
Box plots depicting modality‐wise attention weights for healthy control (HC) and schizophrenia (SZ)
FIGURE 8.
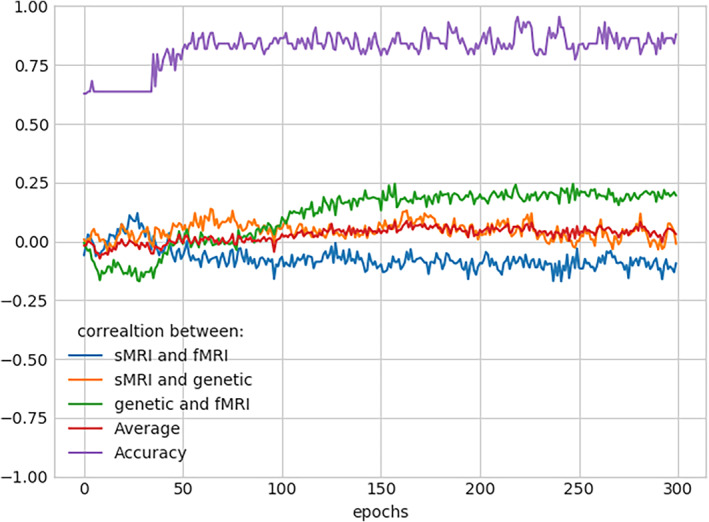
The line plots demonstrate the correlation between the subject's feature embedding from different modalities. It depicts Pearson's correlation between structural magnetic resonance imaging (sMRI), functional MRI (fMRI), and genetic data across all train epochs. Also, it includes epoch‐wise accuracies for reference
5. CONCLUSION
The physiological data sources can carry differential information about a disease. So, integrating multiple disease effect sites helps perform a more comprehensive study of the disorder. In this analysis, our goal is to develop a unified framework that can process the data from distinct biological sources and provide a generalized understanding of the disease. We aim to discover relevant patterns in the brain's structural features, functional mechanisms, and genomic pathways that lead to a coherent deciphering of the psychosis. We employ DL models to learn the attributes and design experiments to characterize the disorder. The neural network extracts unimodal/multimodal concepts that are relevant to SZ. We implement several DL techniques to achieve a legitimate model performance. That validates the neurobiological relevance of the signal the model used to characterize the health condition. Moreover, our post hoc analysis aspires to explain those concepts through saliency analysis. It provides a relevant subset of features from each modality based on their contribution score. Further analysis of those subsets of crucial features provides distinctive patterns between HC and SZ. A subgroup of sMRI and fMRI features are reported, which exhibit substantial HC/SZ group differences across distinct brain regions. Besides, the analysis of modality‐specific contributions for SZ classification helps understand how these various biological domains are impacted. It will help future studies select the SZ‐effected physiologies in limited availability. The communication pattern between these modalities is primarily explained in modality coupling and decoupling across the learning curve. The curves demonstrate how the effects in sMRI, fMRI, and genomics complement each other. Also, the reported neurogenetic features might be related to psychosis‐associated clinical and cognitive symptoms. The proposed data fusion model can discover impeccable biomarkers for the disorder, and the preceding interpretation recommends features that can potentially explain the underlying mechanism of the disease.
AUTHOR CONTRIBUTIONS
Vince D. Calhoun and Md Abdur Rahaman proposed using deep multimodal learning to process imaging genetics data. Md Abdur Rahaman and Noah Lewis developed the methodology. Md Abdur Rahaman ran the experiments and drafted the article. Noah Lewis and Jiayu Chen helped to design a few experiments and edited the article. Vince D. Calhoun supervised the whole project, thoroughly edited the article, and gave valuable feedback on the results. Armin Iraji, Zening Fu, and Theo G. M. van Erp provided the data, ran the preprocessing, and edited the article. All authors have approved the final version of the submission.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Rahaman, M. A. , Chen, J. , Fu, Z. , Lewis, N. , Iraji, A. , van Erp, T. G. M. , & Calhoun, V. D. (2023). Deep multimodal predictome for studying mental disorders. Human Brain Mapping, 44(2), 509–522. 10.1002/hbm.26077
DATA AVAILABILITY STATEMENT
Based on the IRB data privacy agreement, we are not allowed to share any subject‐specific data. However, the datasets are available online in the referred studies and can be requested from the corresponding authors suggested in the studies. The preprocessing pipelines are public, as referred to in the article. All the preprocessing tools are available online at http://trendscenter.org/software and the model's architectural details are available from the corresponding author.
REFERENCES
- Aboraya, A. , Rankin, E. , France, C. , el‐Missiry, A. , & John, C. (2006). The reliability of psychiatric diagnosis revisited: The clinician's guide to improve the reliability of psychiatric diagnosis. Psychiatry (Edgmont), 3(1), 41–50. [PMC free article] [PubMed] [Google Scholar]
- Abrol, A. , Damaraju, E. , Miller, R. L. , Stephen, J. M. , Claus, E. D. , Mayer, A. R. , & Calhoun, V. D. (2017). Replicability of time‐varying connectivity patterns in large resting state fMRI samples. NeuroImage, 163, 160–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adebayo, J. , Gilmer, J. , Muelly, M. , Goodfellow, I. , Hardt, M. , & Kim, B. (2018). Sanity checks for saliency maps. Advances in Neural Information Processing Systems, 31, pp. 9525–9536. [Google Scholar]
- Adhikari, B. M. , Hong, L. E. , Sampath, H. , Chiappelli, J. , Jahanshad, N. , Thompson, P. M. , Rowland, L. M. , Calhoun, V. D. , du, X. , Chen, S. , & Kochunov, P. (2019). Functional network connectivity impairments and core cognitive deficits in schizophrenia. Human Brain Mapping, 40(16), 4593–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarap, A. F. (2018). Deep learning using rectified linear units (ReLU). arXiv preprint arXiv:1803.08375, 9525–9536.
- Aine, C. , Bockholt, H. J. , Bustillo, J. R. , Cañive, J. M. , Caprihan, A. , Gasparovic, C. , Hanlon, F. M. , Houck, J. M. , Jung, R. E. , Lauriello, J. , Liu, J. , Mayer, A. R. , Perrone‐Bizzozero, N. I. , Posse, S. , Stephen, J. M. , Turner, J. A. , Clark, V. P. , & Calhoun, V. D. (2017). Multimodal neuroimaging in schizophrenia: description and dissemination. Neuroinformatics, 15(4), 343–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstra, J. , & Bengio, Y. (2012). Random search for hyper‐parameter optimization. Journal of Machine Learning Research, 13(2), 281‐305. [Google Scholar]
- Calhoun, V. D. , Liu, J. , & Adalı, T. (2009). A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. NeuroImage, 45(1), S163–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor, R. M. , Lange, K. , & Sinsheimer, J. S. (2010). Prioritizing GWAS results: A review of statistical methods and recommendations for their application. The American Journal of Human Genetics, 86(1), 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin, M. S. , Houck, J. M. , Rashid, B. , Agacoglu, O. , Stephen, J. M. , Sui, J. , Canive, J. , Mayer, A. , Aine, C. , Bustillo, J. R. , & Calhoun, V. D. (2016). Multimodal classification of schizophrenia patients with MEG and fMRI data using static and dynamic connectivity measures. Frontiers in Neuroscience, 10, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin, M. S. , Houck, J. M. , Vergara, V. M. , Miller, R. L. , & Calhoun, V. (2015). Multimodal based classification of schizophrenia patients. 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE. [DOI] [PMC free article] [PubMed]
- Chen, J. , Calhoun, V. D. , Pearlson, G. D. , Perrone‐Bizzozero, N. , Sui, J. , Turner, J. A. , Bustillo, J. R. , Ehrlich, S. , Sponheim, S. R. , Cañive, J. M. , Ho, B. C. , & Liu, J. (2013). Guided exploration of genomic risk for gray matter abnormalities in schizophrenia using parallel independent component analysis with reference. NeuroImage, 83, 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , Shi, X. , Zhang, Y. , Wu, D. , & Guizani, M. (2017). Deep features learning for medical image analysis with convolutional autoencoder neural network. IEEE Transactions on Big Data, 7(4), 750–758. [Google Scholar]
- Chen, X. , Kingma, D. P. , Salimans, T. , Duan, Y. , Dhariwal, P. , Schulman, J. , Sutskever, I. & Abbeel, P. (2016). Variational lossy autoencoder. arXiv preprint arXiv:1611.02731.
- Du, Y. , Fu, Z. , Sui, J. , Gao, S. , Xing, Y. , Lin, D. , Salman, M. , Abrol, A. , Rahaman, M. A. , Chen, J. , Hong, L. E. , Kochunov, P. , Osuch, E. A. , Calhoun, V. D. , & Alzheimer's Disease Neuroimaging Initiative . (2020). NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. NeuroImage: Clinical, 28, 102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Föcking, M. , Lopez, L. M. , English, J. A. , Dicker, P. , Wolff, A. , Brindley, E. , Wynne, K. , Cagney, G. , & Cotter, D. R. (2015). Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Molecular Psychiatry, 20(4), 424–432. [DOI] [PubMed] [Google Scholar]
- Fu, Z. , Iraji, A. , Turner, J. A. , Sui, J. , Miller, R. , Pearlson, G. D. , & Calhoun, V. D. (2021). Dynamic state with covarying brain activity‐connectivity: On the pathophysiology of schizophrenia. NeuroImage, 224, 117385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z. , Sui, J. , Turner, J. A. , du, Y. , Assaf, M. , Pearlson, G. D. , & Calhoun, V. D. (2021). Dynamic functional network reconfiguration underlying the pathophysiology of schizophrenia and autism spectrum disorder. Human Brain Mapping, 42(1), 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, Y. , Shibata, H. , Kikuta, R. , Makino, C. , Tani, A. , Hirata, N. , Shibata, A. , Ninomiya, H. , Tashiro, N. , & Fukumaki, Y. (2003). Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatric Genetics, 13(2), 71–76. [DOI] [PubMed] [Google Scholar]
- Gat, I. , Schwartz, I. , Schwing, A. , & Hazan, T. (2020). Removing bias in multi‐modal classifiers: Regularization by maximizing functional entropies. Advances in Neural Information Processing System, 33, 3197–3208. [Google Scholar]
- Goodfellow, I. , Bengio, Y. , & Courville, A. (2016). Deep learning. MIT press. [Google Scholar]
- Gupta, C. N. , Turner, J. A. , & Calhoun, V. D. (2019). Source‐based morphometry: A decade of covarying structural brain patterns. Brain Structure and Function, 224(9), 3031–3044. [DOI] [PubMed] [Google Scholar]
- He, Z. H. , Hu, Y. , Li, Y. C. , Gong, L. J. , Cieszczyk, P. , Maciejewska‐Karlowska, A. , Leonska‐Duniec, A. , Muniesa, C. A. , Marín‐Peiro, M. , Santiago, C. , Garatachea, N. , Eynon, N. , & Lucia, A. (2015). PGC‐related gene variants and elite endurance athletic status in a Chinese cohort: A functional study. Scandinavian Journal of Medicine & Science in Sports, 25(2), 184–195. [DOI] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , Tan, Q. , Kir, J. , Liu, D. , Bryant, D. , Guo, Y. , Stephens, R. , Baseler, M. W. , Lane, H. C. , & Lempicki, R. A. (2007). DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research, 35(2), W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, A. A. , Corrada Bravo, H. , & Feizi, S. (2021). Improving deep learning interpretability by saliency guided training. Advances in Neural Information Processing Systems, 34, 26726–26739. [Google Scholar]
- Jones, P. , & Cannon, M. (1998). The new epidemiology of schizophrenia. Psychiatric Clinics, 21(1), 1–25. [DOI] [PubMed] [Google Scholar]
- Keator, D. B. , van Erp, T. , Turner, J. A. , Glover, G. H. , Mueller, B. A. , Liu, T. T. , Voyvodic, J. T. , Rasmussen, J. , Calhoun, V. D. , Lee, H. J. , Toga, A. W. , McEwen, S. , Ford, J. M. , Mathalon, D. H. , Diaz, M. , O'Leary, D. S. , Jeremy Bockholt, H. , Gadde, S. , Preda, A. , … FBIRN . (2016). The function biomedical informatics research network data repository. NeuroImage, 124, 1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma, D. P. and Ba, J. (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
- Kodirov, E. , Xiang, T. , & Gong, S. (2017). Semantic autoencoder for zero‐shot learning. Proceedings of the IEEE conference on computer vision and pattern recognition.
- Kokhlikyan, N. , Miglani, V. , Martin, M. , Wang, E. , Alsallakh, B. , Reynolds, J. , Melnikov, A. , Kliushkina, N. , Araya, C. , Yan, S. , & Reblitz‐Richardson, O. (2020). Captum: rA unified and generic model interpretability library for pytorch. arXiv preprint arXiv:2009.07896.
- Kusner, M. J. , Paige, B. , & Hernández‐Lobato, J. M. (2017). Grammar variational autoencoder. International conference on machine learning. PMLR.
- Lei, D. , Pinaya, W. H. L. , van Amelsvoort, T. , Marcelis, M. , Donohoe, G. , Mothersill, D. O. , Corvin, A. , Gill, M. , Vieira, S. , Huang, X. , Lui, S. , Scarpazza, C. , Young, J. , Arango, C. , Bullmore, E. , Qiyong, G. , McGuire, P. , & Mechelli, A. (2020). Detecting schizophrenia at the level of the individual: Relative diagnostic value of whole‐brain images, connectome‐wide functional connectivity and graph‐based metrics. Psychological Medicine, 50(11), 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Wang, H. , Yang, Z. , & Odagaki, M. (2017). Variation autoencoder based network representation learning for classification. Proceedings of ACL 2017, Student Research Workshop.
- Liou, C.‐Y. , Cheng, W. C. , Liou, J. W. , & Liou, D. R. (2014). Autoencoder for words. Neurocomputing, 139, 84–96. [Google Scholar]
- Liu, K. , Li, Y. , Xu, N. , & Natarajan, P. (2018). Learn to combine modalities in multimodal deep learning. arXiv preprint arXiv:1805.11730.
- Mäki, P. , Veijola, J. , Jones, P. B. , Murray, G. K. , Koponen, H. , Tienari, P. , Miettunen, J. , Tanskanen, P. , Wahlberg, K. E. , Koskinen, J. , Lauronen, E. , & Isohanni, M. (2005). Predictors of schizophrenia—A review. British Medical Bulletin, 73–74(1), 1–15. [DOI] [PubMed] [Google Scholar]
- Martin, B. , Franck, N. , Cermolacce, M. , Falco, A. , Benair, A. , Etienne, E. , Weibel, S. , Coull, J. T. , & Giersch, A. (2017). Fragile temporal prediction in patients with schizophrenia is related to minimal self disorders. Scientific Reports, 7(1), 8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvant, E. , Habrard, A. , & Ayache, S. (2014). Majority vote of diverse classifiers for late fusion. Joint IAPR international workshops on statistical techniques in pattern recognition (SPR) and structural and syntactic pattern recognition (SSPR). Springer.
- Mössner, R. , Schuhmacher, A. , Schulze‐Rauschenbach, S. , Kühn, K. U. , Rujescu, D. , Rietschel, M. , Zobel, A. , Franke, P. , Wölwer, W. , Gaebel, W. , Häfner, H. , Wagner, M. , & Maier, W. (2008). Further evidence for a functional role of the glutamate receptor gene GRM3 in schizophrenia. European Neuropsychopharmacology, 18(10), 768–772. [DOI] [PubMed] [Google Scholar]
- Oh, J. , et al. (2020). Identifying schizophrenia using structural MRI with a deep learning algorithm. Frontiers in Psychiatry, 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, M. J. , Williams, N. M. , & O'Donovan, M. C. (2004). The molecular genetics of schizophrenia: New findings promise new insights. Molecular Psychiatry, 9(1), 14–27. [DOI] [PubMed] [Google Scholar]
- Passchier, R. V. , Stein, D. J. , Uhlmann, A. , van der Merwe, C. , & Dalvie, S. (2020). Schizophrenia polygenic risk and brain structural changes in methamphetamine‐associated psychosis in a South African population. Frontiers in Genetics, 11, 1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszke, A. , Gross, S. , Massa, F. , Lerer, A. , Bradbury, J. , Chanan, G. , Killeen, T. , Lin, Z. , Gimelshein, N. , Antiga, L. , Desmaison, A. , Kopf, A. , Yang, E. , DeVito, Z. , Raison, M. , Tejani, A. , Chilamkurthy, S. , Steiner, B. , Fang, L. , …, Chintala, S. Pytorch: An imperative style, high‐performance deep learning library. Advances in Neural Information Processing Systems, 2019. 32, 8024–8035. [Google Scholar]
- Patel, P. , Aggarwal, P. , & Gupta, A. . (2016). Classification of schizophrenia versus normal subjects using deep learning. Proceedings of the Tenth Indian Conference on Computer Vision, Graphics and Image Processing.
- Petrovsky, N. , Quednow, B. B. , Ettinger, U. , Schmechtig, A. , Mössner, R. , Collier, D. A. , Kühn, K. U. , Maier, W. , Wagner, M. , & Kumari, V. (2010). Sensorimotor gating is associated with CHRNA3 polymorphisms in schizophrenia and healthy volunteers. Neuropsychopharmacology, 35(7), 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. R. , Bender, D. , Maller, J. , Sklar, P. , de Bakker, P. I. W. , Daly, M. J. , & Sham, P. C. (2007). PLINK: A tool set for whole‐genome association and population‐based linkage analyses. The American Journal of Human Genetics, 81(3), 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi, M. N. I. , Oh, J. , Cho, D. , Jo, H. J. , & Lee, B. (2017). Multimodal discrimination of schizophrenia using hybrid weighted feature concatenation of brain functional connectivity and anatomical features with an extreme learning machine. Frontiers in Neuroinformatics, 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi, M. N. I. , Oh, J. , & Lee, B. (2019). 3D‐CNN based discrimination of schizophrenia using resting‐state fMRI. Artificial Intelligence in Medicine, 98, 10–17. [DOI] [PubMed] [Google Scholar]
- Ramachandram, D. , & Taylor, G. W. (2017). Deep multimodal learning: A survey on recent advances and trends. IEEE Signal Processing Magazine, 34(6), 96–108. [Google Scholar]
- Rashid, B. , & Calhoun, V. (2020). Towards a brain‐based predictome of mental illness. Human Brain Mapping, 41(12), 3468–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, A. L. , Pardiñas A.F., Frizzati A., Tansey K.E., Lynham A.J., Holmans P., Legge S.E., Savage J.E., Agartz I., Andreassen O.A., Blokland G.A.M., Corvin A., Cosgrove D., Degenhardt F., Djurovic S., Espeseth T., Ferraro L., Gayer‐Anderson C., Giegling I., …, Walters J.T.R., The relationship between polygenic risk scores and cognition in schizophrenia. Schizophrenia Bulletin, 2020. 46(2): p. 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R. C. , Roche, J. K. , Conley, R. R. , & Lahti, A. C. (2009). Dopaminergic synapses in the caudate of subjects with schizophrenia: Relationship to treatment response. Synapse (New York, N.Y.), 63(6), 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck, D. W. , Rogers, S. K. , & Kabrisky, M. (1990). Feature selection using a multilayer perceptron. Journal of Neural Network Computing, 2(2), 40–48. [Google Scholar]
- Saha, D. K. , Silva, R. F. , Baker, B. T. , & Calhoun, V. D. (2022). Decentralized spatially constrained source‐based morphometry. 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI). IEEE.
- Salvador, R. , Canales‐Rodríguez, E. , Guerrero‐Pedraza, A. , Sarró, S. , Tordesillas‐Gutiérrez, D. , Maristany, T. , Crespo‐Facorro, B. , McKenna, P. , & Pomarol‐Clotet, E. (2019). Multimodal integration of brain images for MRI‐based diagnosis in schizophrenia. Frontiers in Neuroscience, 13, 1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan, K. , Vedaldi, A. , & Zisserman, A. (2013). Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034.
- Tian, K. , Zhou, S. , & Guan, J. (2017). Deepcluster: A general clustering framework based on deep learning. Joint European conference on machine learning and knowledge discovery in databases. Springer.
- Vaswani, A. , Shazeer, N. , Parmar, N. , Uszkoreit, J. , Jones, L. , Gomez, A. N. , Kaiser, Ł. , & Polosukhin, I. (2017). Attention is all you need. Advances in Neural Information Processing Systems, 30, 5998‐6008. [Google Scholar]
- Ward, C. H. , Beck, A. T. , Mendelson, M. , Mock, J. E. , & Erbaugh, J. K. (1962). The psychiatric nomenclature: Reasons for diagnostic disagreement. Archives of General Psychiatry, 7(3), 198–205. [DOI] [PubMed] [Google Scholar]
- Yan, W. , Calhoun, V. , Song, M. , Cui, Y. , Yan, H. , Liu, S. , Fan, L. , Zuo, N. , Yang, Z. , Xu, K. , Yan, J. , Lv, L. , Chen, J. , Chen, Y. , Guo, H. , Li, P. , Lu, L. , Wan, P. , Wang, H. , … Sui, J. (2019). Discriminating schizophrenia using recurrent neural network applied on time courses of multi‐site FMRI data. eBioMedicine, 47, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W. , Plis, S. , Calhoun, V. D. , Liu, S. , Jiang, R. , Jiang, T. Z. , & Sui, J. (2017) Discriminating schizophrenia from normal controls using resting state functional network connectivity: A deep neural network and layer‐wise relevance propagation method. 2017 IEEE 27th international workshop on machine learning for signal processing (MLSP). IEEE.
- Yan, W. , Qu, G. , Hu, W. , Abrol, A. , Cai, B. , Qiao, C. , Plis, S. M. , Wang, Y. P. , Sui, J. , & Calhoun, V. D. (2022). Deep learning in neuroimaging: Promises and challenges. IEEE Signal Processing Magazine, 39(2), 87–98. [Google Scholar]
- Yassin, W. , Nakatani, H. , Zhu, Y. , Kojima, M. , Owada, K. , Kuwabara, H. , Gonoi, W. , Aoki, Y. , Takao, H. , Natsubori, T. , Iwashiro, N. , Kasai, K. , Kano, Y. , Abe, O. , Yamasue, H. , & Koike, S. (2020). Machine‐learning classification using neuroimaging data in schizophrenia, autism, ultra‐high risk and first‐episode psychosis. Translational Psychiatry, 10(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Hicks, F. , & Fayek, A. R. (2012). Applicability of multilayer feed‐forward neural networks to model the onset of river breakup. Cold Regions Science and Technology, 70, 32–42. [Google Scholar]
- Zhong, G. , Wang, L. N. , Ling, X. , & Dong, J. (2016). An overview on data representation learning: From traditional feature learning to recent deep learning. The Journal of Finance and Data Science, 2(4), 265–278. [Google Scholar]
- Zhuang, F. , Cheng, X. , Luo, P. , Pan, S. J. , & He, Q. (2015). Supervised representation learning: Transfer learning with deep autoencoders. Twenty‐Fourth International Joint Conference on Artificial Intelligence.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Based on the IRB data privacy agreement, we are not allowed to share any subject‐specific data. However, the datasets are available online in the referred studies and can be requested from the corresponding authors suggested in the studies. The preprocessing pipelines are public, as referred to in the article. All the preprocessing tools are available online at http://trendscenter.org/software and the model's architectural details are available from the corresponding author.


